Abstract
Retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs), which are pivotal sensors of RNA virus invasions, mediate the transcriptional induction of genes encoding type I interferons (IFNs) and proinflammatory cytokines, successfully establishing host antiviral immune response. A few excellent reviews have elaborated on the structural biology of RLRs and the antiviral mechanisms of RLR activation. In this review, we give a basic understanding of RLR biology and summarize recent findings of how RLR signaling cascade is strictly controlled by host regulatory mechanisms, which include RLR-interacting proteins, post-translational modifications and microRNAs (miRNAs). Furthermore, we pay particular attention to the relationship between RLRs and diseases, especially how RLRs participate in SARS-CoV-2, malaria or bacterial infections, how single-nucleotide polymorphisms (SNPs) or mutations in RLRs and antibodies against RLRs lead to autoinflammatory diseases and autoimmune diseases, and how RLRs are involved in anti-tumor immunity. These findings will provide insights and guidance for antiviral and immunomodulatory therapies targeting RLRs.
Keywords: RLRs, RIG-I, MDA5, Autoinflammatory diseases, Autoimmune diseases
Abbreviations: RIG-Ⅰ, retinoic acid-inducible gene I; RLR, RIG-Ⅰ-like receptor; MDA5, melanoma differentiation-associated protein 5; LGP2, laboratory of genetics and physiology 2; IFNs, interferons; SNPs, single-nucleotide polymorphisms; CADM, clinically amyopathic dermatomyositis
1. Introduction
RNA viruses compose most of the viruses related to human infectious diseases which brings forward significant public health concerns. The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is an RNA virus which can cause coronavirus disease 2019 (COVID-19) and has become a primary healthcare threat in recent years. The characteristic feature of SARS-CoV-2 infection is the inadequate production or low response of type-I IFNs, a class of cytokines essential for antiviral defense. Retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs), a typical family of cellular receptors, serve as critical cytosolic sensors of virally derived ribonucleic acids to halt viral invasion. Upon activation, these sensors participate in the composition of signaling cascades that lead to the transcription of genes encoding type I IFNs, therefore building the first line of protection against viral infections. Accordingly, viruses have evolved strategies to withstand innate defenses and promote replication, which results in virus-host conflict and causes pattern recognition receptor (PRR)-induced systemic inflammatory disease [1].
Based on decades of overexpression or knockout studies of RLRs, it is well known that RLRs, especially RIG-I and melanoma differentiation-associated protein 5 (MDA5), are essential for antiviral defense and type I IFN production during virus invasion. Apart from this classical role, it is also found that RLRs are related to autoinflammatory and autoimmune diseases and cancers. Consequently, this made us question whether RIG-I like receptor acts as a friend or foe in diseases.
2. Basic biology of RLRs
2.1. Characterization and structure of RLRs
RLRs are extensively expressed in tissues and an abundance of cell types, including immune and nonimmune cells such as cancer cells and central nervous system cells. In contrast, they pivotally function as innate immune initiators in epithelial and myeloid cells [2,3]. As one of the PRRs which detect pathogens and allow eukaryotic cells to respond to infections rapidly, RLRs detect replicating viruses in the cytoplasm, particularly at the early phase of viral infection. RLRs encompass three homologous members, including RIG-I (or DEAD box polypeptide 58, DDX58), Melanoma differentiation-associated protein 5 (MDA5, also known as interferon induced with helicase C domain 1, IFIH1), and laboratory of genetics and physiology 2 (LGP2, or DExH box polypeptide 58, DHX58) [1].
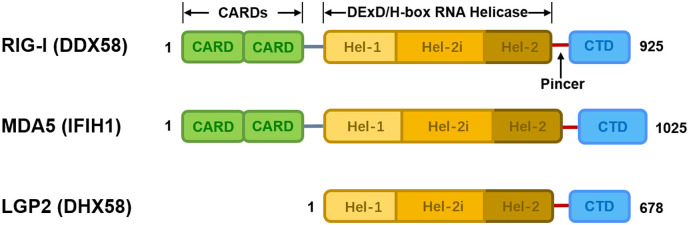
Every RLR has a central conserved DExD/H-box RNA helicase domain for ATP catalysis and activation, and a C-terminal domain (CTD) for RNA binding and unwinding [[4], [5], [6]]. RIG-I and MDA5 have two extra tandem N-terminal caspase activation and recruitment domains (CARDs), mediating the downstream signal transduction and the transcriptional activation of type I IFNs and other immediate antiviral response genes that encode proinflammatory cytokines [7,8]. LGP2, the most mysterious RLR protein, lacks CARDs and therefore cannot convey downstream signaling, but can functionally regulate RIG-I and MDA5 signaling [9,10]. The helicase domain of LGP2 shared 30% homology with RIG-I and 40% with MDA5 [11]. In resting cells, RLRs maintain a low expression level and an auto-repressed conformation via the sequestering of the RNA helicase domain by CARD. Once the virus invades or IFN exposes, the helicase domain is pulled away from CARD, and RLRs then transform into an activated conformation. RLRs levels then rapidly increase to trigger the signal transduction pathways and elicit innate immune defense, which leads to the production of proinflammatory cytokines and an extensively effective antiviral state in the host [12] (Fig. 1).
Fig. 1.
Schematic representation of RLRs.
Each RLR has a conserved central DExD/H-box RNA helicase domain and a pincer domain that connects the helicase domain to the CTD domain. The helicase domain consists of Hel-1, Hel-2i and Hel-2. RIG-I and MDA5 have two CARD domains essential for the induction of downstream signaling. The amino acid sequence length of each protein is shown.
2.2. Interactions among RLRs
Currently, LGP2 is considered to regulate the function of RIG-I and MDA5 signaling [1,13]. For effective MDA5-mediated antiviral responses against many viruses, functionally activated LGP2 is indispensable [14]. Several studies reported that LGP2-/- mice and LGP2-/- cells showed higher susceptibility and severely impaired IFN responses to certain RNA viruses (e.g., encephalomyocarditis virus) recognized by MDA5 [15,16]. Coordinated action between LGP2 and MDA5 in MDA5-dependent response was found by Annie et al., and they revealed that LGP2 stabilizes the filament assembly of MDA5 on short dsRNA and activates MDA5, amplifying the MDA5-mediated IFN response [17]. Miyamoto et al. also found that the overexpression of dsRNA-binding protein PACT stimulates the activity of the encephalomyocarditis virus (EMCV)-induced IFN-β promoter only when LGP2 and MDA5 are expressed together, rather than when MDA5 is expressed alone, suggesting that LGP2 is pivotal for triggering MDA5-dependent immune response by EMCV [18]. However, Bruns et al. reported that LGP2 is an inhibitor of MDA5-mediated IFN- β production at high concentrations but is an activator at low concentrations [19]. Furthermore, Sanchez et al. performed In vitro interaction analyses and discovered that the LGP2 has opposite regulatory effects on MDA5 and RIG-I. Through the different interactions between LGP2 CTD and PACT, LGP2 specifically promotes MDA5 signaling while suppressing RIG-I signaling [20]. In SARS-CoV-2-infected cells, the separate knockdowns of only LGP2 and MDA5 can both significantly lower IFN-β mRNA level, further suggesting that MDA5 and LGP2 collaboratively detect viral RNA and trigger innate immune responses [21]. However, further analysis is required to fully elucidate the concrete interplay among these three members of RLRs.
2.3. RNA Ligands sensed by RLRs
Although RIG-I and MDA5 have many structural similarities, they detect distinct spectrums of viruses. Generally, RIG-I mainly detects RNA species from plenty of viruses belonging to Flaviviridae, Paramyxoviridae, Orthomyxoviridae, and Rhabdoviridae, whereas MDA5 detects RNA mainly from Picornaviridae [22,23]. RIG-I recognizes most of the currently studied single-stranded (ss) RNA viruses, including all negative or minus (-)-strand RNA viruses and very few positive or plus(+)-strand RNA viruses [16]. In addition, some dsDNA viruses could also be recognized by RLRs because both the positive and negative strands of dsDNA viruses are able to produce dsRNA during transcription [24]. These viral types are summarized in table 1 (Table 1 ).
Table 1.
Representative viruses sensed by RLRs following infection.
| Viruses sensed only by RIG-I | Viruses sensed only by MDA5 | Viruses sensed by both RIG-I and MDA5 | |
|---|---|---|---|
| ssRNA (+) |
Caliciviridae Murine norovirus-1 [36] |
Picornaviridae Theiler’s murine encephalomyocarditis virus [37] Mengo virus [37] Poliovirus [38] Saffold virus [39] Caliciviridae Murine norovirus-1 36] |
Picornaviridae Encephalomyocarditis virus [40] Coxsackie B3 virus [40] Rhinovirus [41] [42] Flaviviridae West Nile virus [43] Dengue virus [44] Japanese encephalitis virus [37] Hepatitis C virus [45] [46] [47] Coronaviridae mouse hepatitis virus [48] [49] SARS-CoV-2 [50] Flaviviridae Zika virus [51] Togaviridae Sindbis virus [52] |
| ssRNA (-) |
Orthomyxoviridae Influenza A virus [44] Influenza B virus [37] Paramyxoviridae Newcastle disease virus [53] Respiratory syncytial virus [44] Nipah [54] human parainfluenza 5 mRNA [55] Arenaviridae Lassa [54] Lymphocytic choriomeningitis virus [56] Bunyaviridae Rift Valley fever virus [54] |
None |
Filoviridae Ebola [54] [57] Rhabdoviridae Rabies virus [58] Vesicular stomatitis virus [59] [60] Paramyxoviridae Measles virus [61] [62] human Metapneumovirus [63] Paramyxoviridae Sendai Virus [53] |
| dsRNA | none | None |
Reoviridae Reovirus [64] Rotavirus [65] |
| dsDNA |
Herpesviridae Epstein-Barr virus [66] Poxviridae Myxoma virus [67] |
Poxviridae Vaccina virus [68] |
Herpesviridae Herpes simplex virus 1 [69] [70] Hepadnaviridae Hepatitis B virus [71] [72] [73] |
Abbreviations: ssRNA(+): plus-strand RNA; ssRNA(-): minus-strand RNA; dsRNA: double-stranded RNA; dsDNA: double-stranded DNA.
Cellular RNA is abundant in host cells, but only virus-derived RNA can initiate the innate immune response, so there must be a molecular identification mechanism for RLRs to distinguish the molecular characteristics of self and non-self RNA. Indeed, there exists sequence-independent RNA-sensing mechanisms of RIG-I and MDA5. Whether the sensor is RIG-I or MDA5 is determined by the length and secondary structure of viral RNA [9]. RIG-I mainly binds ssRNA and 5′ mono-, di-, or tri-phosphorylated low molecular weight dsRNA. At the same time, MDA5 recognizes high molecular weight dsRNA and higher-order RNA structures with a weaker affinity than RIG-I and LGP2 [25,[26], [27], [28], [29], [30]]. More studies have shown that RIG-I recognizes 3’ poly-U/UC tract RNA of hepatitis C virus and 3’ U/A-rich RNAs of influenza virus NS1 gene [31,32]. Polyriboinosinic acid–polyribocytidylic acid is usually denoted as poly(I:C) and is a synthetic mimic of viral dsRNA polymer that simulates viral infection [33]. RIG-I and MDA5 preferentially bind to poly(I:C) smaller than 1 bp and larger than 2 kbp in vitro, respectively[10,34]. Additionally, mitochondrial dsRNA (mtdsRNA) is a kind of highly immunogenic RNA which is misprocessed and released into the cytoplasm and accumulates under pathological conditions, such as viral infections. MDA5 has been recently identified as the primary sensor for recognizing cytosolic mtdsRNA through MAVS-mediated downstream type I IFN signaling pathways, which may prime an antiviral immune response [35]. This finding further elucidates the importance of proper processing of self-RNA and the wide variety of ligands sensed by the RLRs family.
2.4. RLR signaling cascade
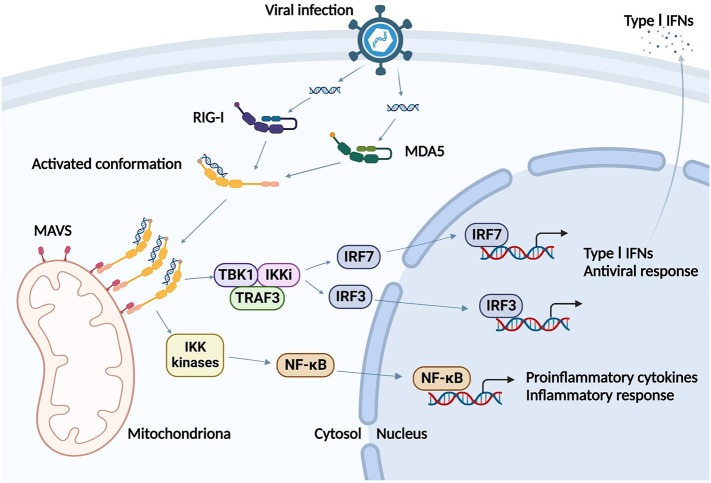
Functional studies have suggested that RNA-dependent ATP hydrolysis activity plays a critical role in distinguishing non-self RNA from self-RNA using RIG-I and MDA5 [[74], [75], [76]]. Upon sensing non-self RNA molecules, monomers of RIG-I or MDA5 soon form the tetrameric structures through the ATP hydrolysis-dependent interactions of N-terminal CARDs. Then the mitochondrial antiviral-signaling protein(MAVS), an adaptor molecule located on mitochondrial outer membrane or peroxisome or mitochondrial-associated membrane, is recruited via the interplay between RIG-I/MDA5 CARD and MAVS CARD [6,[77], [78], [79]]. MAVS then forms prion-like filaments and builds a higher-order signalosome platform for activating signalling proteins, tumor necrosis factor receptor-associated factor (TRAF), TANK-binding kinase 1 (TBK1) and I-kappa-B-kinase (IKK) kinases [80]. This results in the subsequent activation and nuclear translocation of primary transcription factors interferon regulatory factor 3/7 (IRF3/7) and transcriptional factors nuclear factor kappa-B (NF-κB), culminating in the secretion of type-I IFNs and proinflammatory cytokines including IL-1β, IL-6, TNF-α, etc., respectively [2,81,82]. Thus, a potent type-I IFN antiviral response gets potently elicited (Fig. 2).
Fig. 2.
The signaling pathways of the RLR. Upon viral infection, RIG-I and MDA5 recognize dsRNA and soon convert to activated conformations. Then RIG-I and MDA5 form homopolymers and recruit MAVS, which then activate TBK1/IKK kinases to generate type I IFNs for antiviral responses and activate NF-κB to transcribe the proinflammatory cytokines for inflammatory responses.
Abbreviations: RIG-I: retinoic acid-inducible gene I; MDA5: melanoma differentiation-associated protein 5; MAVS: mitochondrial antiviral-signaling protein; TBK1: TANK-binding kinase 1; IKK: I-kappa-B-kinase; IRF3/7: interferon regulatory factor 3/7; NF-κB: nuclear factor kappa-B; type I IFNs: type I interferons. [Figure was created with BioRender (https://BioRender.com).]
After secreted in an autocrine and paracrine manner, type I IFNs specifically bind to the IFN- α/β receptor (IFNAR), which activates the intracellular Janus kinase-signal transducer and activator of transcription (JAK–STAT) pathway, especially STAT1 and STAT2, causing downstream signal transduction [83,84]. Once phosphorylated, STAT1 and STAT2 rapidly bind to IRF9 and constitute the interferon-stimulated gene factor 3 (ISGF3) transcriptional complex 11. Next, ISGF3 binds to type I IFN-dependent gene promoters and drives the transcription and translation of IFN-stimulated genes (ISGs), which elicit both innate and adaptive immune programs within cells, therefore blocking the viral life cycle and triggering programmed cell death, especially apoptosis, in infected cells [85,86].
3. Regulation of RLR signaling cascade
Control on RLR antiviral cascade signaling is essential not only to prevent harmful and unwanted type I IFN responses in the absence of infection but also to initiate timely and sensitive pathogen recognition in the presence of pathogen invasion [87]. Moreover, the host has developed a lot of mechanisms to regulate the activity of RLRs, such as miRNAs, interacting proteins and post-translational modifications (PTMs). Through these delicate modulations, the feedback or feedforward regulatory loops of antiviral response can be successfully established.
3.1. Interacting proteins
Many dsRNA-binding proteins have been found to interact with RLRs and modulate the RLR signaling cascade, including RLR-dsRNA binding, RLR assembly, and RLR intracellular localization.
PACT, the first dsRNA-binding protein found to regulate RLR signaling cascade, directly interacts with RIG-I CTD and enhances its ATPase and helicase activities, resulting in potent initiation and maintenance of RIG-I-induced type I IFN production [88]. The overexpression of PACT stimulates about twice the production of IFN-β in SeV-infected cells [89]. PACT also facilitates RNA-induced MDA5 oligomerization and directly interacts with LGP2, thereby increasing both RIG-I- and MDA5-induced IFN production [90,91]. Interestingly, TAR-RNA-binding protein2(TRBP), the homologous protein of PACT, acts oppositely to PACT and restrains RIG-I-mediated IFN-β production induced by poly(I:C) and 5’ppp dsRNA viral mimics via specifically sequestrating RIG-I from dsRNA [92]. A coreceptor of RIG-I and MDA5 named ZCCH3 simultaneously binds with RLR-sensed RNAs and RLRs, strongly enhancing the binding activity of RLRs [93]. Nucleotide-binding oligomerization domain 1 (NOD1) [94], ribonucleoprotein PTB-binding 1 (RAVER1) [95], and DXH29 [96] can significantly and specifically amplify MDA5-dsRNA binding affinity, whereas ZFYVE1 [97] and Arf-like protein 5B (Arl5B) [98] competitively bind to virus-derived RNA and reduce MDA5-RNA binding and MDA5 oligomerization. ZFYVE1 deficiency protects the mice from death induced by MDA5-sensed EMCV, but not RIG-I-sensed VSV. Other than the combined function of ubiquitin (Ub), Ring finger protein 135 (RNF135) can additionally induce RIG-I assemblies to enhance RIG-I signaling in an RNA-length dependent manner [99]. Chaperone protein 14-3-3η promotes the antiviral immune response by facilitating the oligomerization of MDA5 and its translocation to mitochondria [100]. These findings indicate that the interacting network involved in RLR signaling is fine-tuned, complicated and comprehensive.
3.2. Post-translational modifications
Post-translational modifications (PTMs) are crucial for the activation and deactivation of RIG-I and MDA5. By regulating the activity, stability, cellular compartment, conformation and interactions with additional factors, PTMs precisely alter the functionality of RLRs. PTMs mainly include ubiquitination, phosphorylation, SUMOylation and acetylation (Table 2 ).
Table 2.
Post-translational modifications of RLRs.
| Substrate | Modification | Binding site | Key Refs | |
|---|---|---|---|---|
| TRIM25 | RIG-I | K63-linked polyubiquitination | CARD (K190, K193) | [101] |
| RNF135 | CARD, CTD (K96, K115, K788, K849, K851, K888, K907, K909) | [127] [128] | ||
| TRIM4 | CARD (K164) | [102] | ||
| MEX3C | CARD (K45, K99, K169) | [103] | ||
| TRIM65 | MDA5 | Helicase (K743) | [108] | |
| RNF125 | RIG-I, MDA5 | K48- linked polyubiquitination | CARD | [105] [106] |
| RNF122 | RIG-I | CARD (K115, K146) | [104] | |
| c-Cbl | CTD (K813) | [129] | ||
| CHIP | CARD | [107] | ||
| TRIM40 | RIG-I, MDA5 | K27/K48-linked polyubiquitination | K23, K43 and K68 in the first CARD of MDA5 | [106] |
| TRIM13 | MDA5 | Unknown | upstream of IRF3 | [130] |
| A20 | RIG-I | remove K63-linked polyubiquitin | K63-linked polyubiquitin | [109] |
| CYLD | [110] | |||
| USP3 | [131] | |||
| USP21 | [132] | |||
| USP25 | [133] | |||
| USP4 | RIG-I | remove K48-linked polyubiquitin | K48-linked polyubiquitin | [134] |
| USP15 | TRIM25 | [135] | ||
| USP17 | RIG-I, MDA5 | deubiquitination | unknown | [136] |
| PKC-α | RIG-I | phosphorylation | CARD (Ser8) | [137] [138] |
| PKC-β | CARD (Thr170) | [139] | ||
| CKII | CTD (Thr770, Ser854-855) | [140] | ||
| PP1α, PP1γ | RIG-I, MDA5 | dephosphorylation | CARD (RIG-I: S8, T170; MDA5: S88) | [141] |
| RIOK3 | MDA5 | dephosphorylation | CTD (S828) | [120] |
| TRIM38 | RIG-I, MDA5 | SUMOylation | K96 and K888 of RIG-I, K43 and K865 of MDA5 | [123] |
| PIAS2 β | MDA5 | SUMOylation | CARD | [142] |
| SENP2 | RIG-I, MDA5 | deSUMOylation | [123] | |
| hdac6 | RIG-I | deacetylation | CTD (K909) | [85,86] |
| FAT10 | RIG-I | deamidation | CARD (T55) | [143] |
Red represents positive regulation, and blue represents negative regulation.
Ubiquitination of RLRs controls their signals both positively and negatively. Several E3 ubiquitin ligases, such as tripartite motif-containing protein 25(TRIM25) [101], RNF135 [99], TRIM4 [102] and MEX3C [103], augment the activation of RIG-I through Lysine (Lys, K) 63-linked polyubiquitination. On the other hand, several E3 ubiquitin ligases, such as RNF122 [104], RNF125 [105], TRIM40 [106] and carboxyl terminus of HSC70-interacting protein (CHIP) [107], negatively regulate RIG-I activity via K48-linked polyubiquitination. The activation and oligomerization of MDA5 are induced by TRIM65-mediated and K63-linked polyubiquitination [108]. Deubiquitinases (DUBs) operate as negative regulators of RLR signaling by removing the K63-linked polyubiquitin modifications. For example, A20 [109] and cylindromatosis (CYLD) [110] both target RIG-I and efficiently inhibit RIG-I-mediated IFN-β production. Besides, ubiquitin-like (UBL) proteins play a dual-functioned regulatory role on RLRs depending on their target proteins. For instance, the UBL protein HLA-F adjacent transcription 10 (FAT10) suppresses the RIG-I-TRIM25 complex through binding and stabilizing the RIG-I protein, thereby dampening the RIG-I-mediated antiviral responses[111]. The other UBL protein, IFN-induced 15-kDa protein (ISG15), performs ISGylation (a ubiquitin-like modification) and activates MDA5 but reduces RIG-I intracellular levels [[112], [113], [114]]. Intriguingly, it has been recently found that SARS-CoV-2 utilizes autogenous papain-like protease to antagonize MDA5 ISGylation and evade host innate immunity, which may account for the SARS-CoV-2 immune evasion and the suppression of host IFN-I production[115]. The balance of polyubiquitination and deubiquitination regulate protein stability, oligomerization and RLR activity, also participating in innate antiviral responses.
Phosphorylation, another PTM, is regulated by the antagonism of kinase and phosphatase. In resting cells, RIG-I maintains a conformation of self-inhibition by phosphorylation of CARD and CTD to prevent prematurity of antiviral signals. After the invasion of the RNA virus, RIG-I is rapidly dephosphorylated to elicit innate antiviral response. Conventional protein kinase C-α (PKC-α) and PKC-β are mainly responsible for the phosphorylation of RIG-I CARD, which suppresses CARD ubiquitination by TRIM25 [[116], [117], [118]]. Similarly, the CTD domain of RIG-I and MDA5 can be phosphorylated by casein kinase II (CK II) [119] or RIO kinase 3 (RIOK3), respectively, thus disrupting their filament formation and attenuating downstream signaling [120]. In contrast, during viral infection or viral RNA binding, RIG-I and MDA5 CARD are dephosphorylated by phosphatase I α and phosphatase I γ (PP 1 α and PP 1 γ), which is crucial for their activation [121]. Based on the pivotal role of phosphorylation regulation in RLRs activation, the agonist or antagonist of kinase and phosphatase might be potential therapeutics for RLRs-related diseases.
SUMOylation is a ubiquitination-like post-translational modification that covalently attaches a small ubiquitin-like modifier (SUMO) to the Lys residues of the target protein. It also plays a vital role in protein function regulation and host antiviral defense [122]. By performing dynamic SUMOylation of RIG-I and MDA5 at K865 and K888, TRIM38 inhibits their K48-linked polyubiquitination and enhances their stability [123]. On the contrary, deSUMOylation mediated by sentrin/sumo-specific protease 2 (SENP2) prevents excessive and deleterious immunopathological damage to the host by promoting the K48-linked polyubiquitination of RIG-I and MDA5 [123]. Like TRIM38, PIAS2β is also a SUMOylation ligase, but specifically interacts with and enhances the SUMOylation of MDA5 but not RIG-I [124].
Reversible acetylation also regulates RIG-I activation by controlling its viral RNA-binding activity. The acetylation of RIG-I CTD inhibits the recognition and binding of CTD to viral RNA, leading to the inability of RIG-I to assemble into signal-active homo-oligomers. In contrast, deacetylation of CTD by histone deacetylase 6 (HDAC6) restores RIG-I’s ability to bind RNA and oligomerize [125] [126].
3.3. miRNAs
Although previous studies focusing on the role of microRNAs (miRNAs) in immune regulation are rather limited, a growing number of studies have demonstrated that miRNAs are involved in regulating the RLR signaling pathway in mammals.
miR-146a was the first miRNA reported in 2009 to regulate the RIG-I signaling pathway. It impairs RIG-I signaling, blocks RIG-I-dependent type I IFN secretion and allows viral replication in macrophages by targeting TNFR-associated factor 6 (TRAF6) and interleukin 1 receptor-associated kinase 1/2 (IRAK1/2) during vesicular stomatitis virus (VSV) infections [144]. miR-92a also inhibits macrophage-mediated anti-VSV immune responses by directly targeting RIG-I and reducing its expression [145]. Similarly, miR-218 targets and decreases RIG-I in human and mouse macrophages while facilitating the immune evasion of VSV. Targeted inhibition of miR-218 rescues the signaling activity of RIG-I and helps macrophages defend invaded viruses [146]. miR-485 is produced by the host after viral invasion and specifically targets RIG-I mRNA for degradation and results in impaired type I IFN response and uncontrolled virus replication [147].
Unlike the previously mentioned miRNAs that negatively regulate RIG-I, Zhao et al. found that miR-136 is an agonist of RIG-I and can promote IFN-β and IL-6 secretion and accumulation, leading to the strong antagonism against H5N1 influenza A virus (IAV) replication in human lung epithelial cells (A549) [148]. Moreover, microRNA let-7b specifically augments RIG-I signaling to promote the activation and nuclear translocation of IRF3 and induces the enhanced type I IFN response during early hepatitis C virus (HCV) infection [149]. Additionally, miR-145 overexpression by liposome-mediated transient transfection led to an immune response in vitro via RIG-I recognition [150].
miR-125a-3p is found to reduce MDA5 expression in CD4+ T lymphocytes of patients with systemic lupus erythematosus (SLE) depending on the dose [151]. In miiuy croaker, a non-mammal, miR-203 has a negative regulatory effect on MDA5 through directly targeting the MDA5 gene [152], and miR-145-5p also inhibits MDA5 transcription in a dose-and-time-dependent manner via degrading the MDA5 mRNAs [153]. In chicken spleens infected with Avian leukosis virus subgroup J (ALV-J), miR-34b-5p targets MDA5 and downregulates the genes in the MDA5-mediated signaling pathway to increase the replication of ALV-J and proliferation of ALV-J-infected cells [154].
Apart from RIG-I and MDA5, MAVS also gets regulated by miRNAs. Liu et al. found that overexpression of miR-33/33* leads to blunted MAVS activation, enhanced viral lethality, and reduced type I IFN secretion both in vitro and in vivo. The specific mechanism in which miR-33/33* prevents MAVS from forming functional homo-oligomers and activation is that miR-33/33* targets adenosine monophosphate activated protein kinase (AMPK) for posttranslational silence and disturbs mitochondrial homeostasis by suppressing mitophagy. Intriguingly, miR-33/33* transcription in macrophages is impeded by VSV stimulation in an IFNAR-dependent manner [155]. Little is known about the miRNAs that regulate LGP2.
Taken together, these studies discover a brand-new role of miRNAs in modulating host antiviral immune response and shed new insights for clinically treating not only viral infection but also autoinflammation in the years to come.
4. RLRs and infections
As cellular non-self RNA sensors, RIG-I and MDA5 are essential for interferon production and antiviral immune response in many infective diseases. RIG-I-/- and Mda5 -/- mice showed high susceptibility to RNA virus infections, suggesting that RIG-I and MDA5-mediated antiviral responses are vital for eliminating RNA viruses. In addition to this, RLRs are also involved in the innate immune response against parasite and bacterium infections.
4.1. MDA5 senses and responds to SARS-CoV-2
The world is now experiencing a significant public health crisis, the COVID-19 pandemic, caused by SARS-CoV-2. Once the SARS-CoV-2 virus infects pneumocytes, cytosolic MDA5 and LGP2 predominantly mediate the delayed interferon induction and then establish an antiviral milieu through triggering ISGs. Further data demonstrated that SARS-CoV-2 intermediates also specifically trigger the interferon production via the MDA5 signaling pathway [21]. Moreover, ISGylation of MDA5 CARD domains conjugated with interferon-stimulated protein ISG15 facilitates the oligomerization of MDA5, triggering the positive feedback of innate antiviral response. However, the SARS-CoV-2 papain-like protease de-ISGylates and blocks ISG15-induced activation of MDA5, also contributing to viral evasions of host immune surveillance [115]. In a prospective study of 227 ICU patients with COVID-19, rs199076 TT variation on IFIH1, the gene coding MDA5, was found to be associated with decreased inflammation and better prognosis [156]. The single-cell sequencing analysis of upper airway tissues from normal children samples indicated that the mRNA level of MDA5 in immune cells were presented higher than that in adults, and triggered prominent antiviral immunity during SARS-CoV-2 invasion. This finding provides an explanation for why low morbidity of COVID-19 happened in children rather than in adults [157]. All of these findings contribute to our understanding of the molecular basis of SARS-CoV-2, from its innate immune recognition to the signaling response it induces.
4.2. RLRs participate in anti-malaria immune response
In addition to virus infection, MDA5 is identified as a cytosolic sensory receptor recognizing Plasmodium RNA from malaria parasites and initiated a type I IFN response controlling Plasmodium parasite replication in hepatocytes [158]. Increased phosphatidylserine on the surface of Plasmodium-infected red blood cells lead to the engulfment of Plasmodium by conventional dendritic cells (cDCs), and release Plasmodium ligand bound to MDA5, triggering an early protective IFN-I response in malaria infections [159]. Moreover, Plasmodium RNA-harboring extracellular vesicles (EVs) from Plasmodium-infected RBCs fuse with responding natural killer (NK) cells and activate the MDA5 signaling pathway, contributing to the immune response to malaria infections [160]. Recently, the internalization of Plasmodium-derived microvesicles through monocytes has been found to stimulate the RIG-I cascade and result in the binding of HUR1 to chemokine CXCL10 mRNA 3′UTR, consequently restricting the synthesis and secretion of CXCL10. The decreased CXCL10 level is not suitable for malaria growth. However, fatal cerebral malaria grew with heightened serum CXCL10 levels, which accelerated the development of Plasmodium [161]. These data suggest that parasites have developed sophisticated mechanisms to utilize host molecules such as MDA5 and RIG-I to reprogram the immune response and control their own fate.
4.3. RLRs sense bacterial RNA
Aside from viruses and parasites, RLRs are also able to sense bacterial nucleic acids, especially those released into the cytoplasm of infected cells by intracellular pathogenic bacteria, leading to the expression of type I IFNs. When live Listeria monocytogenes enter macrophage cytoplasm, bacterial RNA/DNA is secreted from Listeria and detected by RIG-I, MDA5 and STING, stimulating the release of interferon β. Meanwhile, RIG-I recognizes bacterial nucleic acids, and activated inflammasome and IL-1β release (147). A recent study showed that RIG-I recognized a complex composed of Listeria monocytogenes RNAs and a small bacterial RNA-binding protein Zea upon Listeria monocytogene infection, thereby enhancing the release of IFN-β [162]. It has been well accepted that RIG-I and MDA5, but mainly RIG-I, allow Listeria-infected cells to effectively sense bacterial activity, virulence, and localization in the cytosol. The intracellular Salmonella mRNA gets detected by RIG-I rather than MDA5 to launch the production of interferon β in nonphagocytic cells, but not in immune cells [163]. Mycobacterium tuberculosis RNA/DNA was released into the macrophage cytosol, inducing IFN-β production through the crosstalk between host RIG-I and STING sensing pathway [164].
These data support a broad function of RLRs in antipathogenic immunity. Furthermore, there may be redundancy or crosstalk between different DNA/RNA sensing signaling pathways mediating the antipathogenic immune response. As a result, many studies have focused on developing synthetic agonists of RLRs, especially RIG-I, to serve as potent pan-antivirals and vaccine adjuvants via the activation of IFN signaling and immune response [[165], [166], [167]].
5. IFIH1 is identified as a susceptibility gene for autoimmune diseases
IFIH1 has been identified as a susceptible gene for a series of autoimmune diseases. Genome-wide association studies (GWASs) revealed that plenty of single nucleotide polymorphisms (SNPs) or single nucleotide variants (SNVs) in the IFIH1 gene is associated with the risk for type 1 diabetes (T1D) [168], autoimmune thyroid disease [169], psoriasis [170,171], systemic lupus erythematosus (SLE) [24], and inflammatory bowel disease [172] [173], rheumatoid arthritis [174], vitiligo [175] and multiple sclerosis [176] (Table 3).
Table 3.
Pathogenic SNPs and SNVs of DDX58 (RIG-I) and IFIH1 (MDA5).
| Gene | Mutation | Type | Effect | Disorder |
|---|---|---|---|---|
| IFIH1 | A946T | Gain-of function (GOF) | Increased expression of IFIH1 | SLE |
| IFIH1 | R779H | GOF | Increased expression of IFIH1 | SLE, AGS |
| IFIH1 | rs35337543 | Loss of function (LOF) | Disruption of the normal splicing of IFIH1 transcript and impaired function of MDA5 | T1D |
| rs35667974 | ||||
| ss107794688 | ||||
| ss107794687 | ||||
| IFIH1 | c. 1694G>T | LOF | Full or partial MDA5 defect | Early-onset inflammatory bowel disease |
| c. C2105T | ||||
| c.C373A | ||||
| c. A1909G | ||||
| IFIH1 | G821S | GOF | Spontaneous typeIIFN induction | Lupus-like nephritis |
| IFIH1 | c. 2159G>A | GOF | Increased MDA5-dsRNA binding affinity and enhanced MDA5 filament stability | AGS |
| c. 2336G>A | ||||
| c. 1009A>G | ||||
| c. 2335C>T | ||||
| c. 1178A>T | ||||
| c. 1483G>A | ||||
| IFIH1 | c. 1354G>A | GOF | Increased activation of IFNB1, which encodes IFN-β | AGS |
| c. 1114C>T | ||||
| c.2336G>A | ||||
| MDA5 | G495R | GOF | Lost tolerance to imperfect self Alu-dsRNA duplex | AGS |
| IFIH1 | K365E | LOF | Impaired IFN-β signalling | IMD95 |
| K889X | ||||
| c.2016delA Ile872Ter | ||||
| DDX58 | E373A | GOF | Constitutive activation of typeIIFNs and increased ISGs expression | SMS |
| C268F | ||||
| E383A | ||||
| IFIH1 | R822Q | GOF | Increased production of typeIIFNs | SMS |
Abbreviations: AGS, Aicardi-Goutieres syndrome; TID, type 1 diabetes; SLE, systemic lupus erythematosus; IMD95, immunodeficiency 95.
5.1. SNPs or SNVs in IFIH1 gene linked to systemic lupus erythematosus (SLE)
The rs1990760 (A946T) SNP in the IFIH1 gene is the most common coding-sequence-variant linked to autoimmune diseases. This mutation is presumed to change the expression, structure and function of MDA5. The T allele of rs1990760 is associated with anti-double-stranded DNA antibodies and low IFN-α level in serum among SLE subjects [177]. Bioinformatic analysis predicted that this variant will not change the protein structure. Indeed, it is possible that the A946T risk allele is a gain-of-function SNP and will cause augmented MDA5 expression [178]. Intriguingly, the genetic defect in a patient with rare early-onset and severe SLE is identified as de novo R779H mutation in the IFIH1 gene, which has been reported to cause Aicardi-Goutieres syndrome, a rare genetically neuroimmunological disorder [179] [180]. These data further support the role of MDA5 in the pathogenesis of SLE.
5.2. SNVs in IFIH1 gene associated with type 1 diabetes
Through association analysis, Nejentsev et al. found that four rare loss-of-function mutations in the IFIH1 gene, including two mutations in the exon and two mutations in the intron, have more potent protective effects against TID risks than other non-synonymous SNPs (nsSNPs) [181]. The mechanism may be that these protective variants might disrupt the normal splicing of IFIH1 transcript, dampen the function of MDA5 and attenuate innate immune responses against viruses. In contrast, the incidence of T1D correlates with enterovirus (especially coxsackievirus B4) infection and the overall antiviral effect of MDA5 [[182], [183], [184]]. For the development of T1D, the interaction among individual heterogeneity, constant enteroviral challenges and antiviral immune responses may play an important role [185,186]. Viral infection and type I IFN production can induce ER stress in β-cells, which leads to the generation of neoantigen epitopes. This, in turn, results in the activation of β-cell-specific autoreactive T lymphocytes and damages β-cells [184].
5.3. SNVs in IFIH1 gene involved in early-onset inflammatory bowel disease
Recently, four rare heterozygous variations in the IFIH1 gene were identified in 7 unrelated children with very early-onset inflammatory bowel disease. The variants are inherited from their parents, who have no gastrointestinal or immunological issues. These variants are believed to be loss-of-function mutations, and may lead to complete or partial MDA5 deficiency, thus impairing intestinal pathogen sensation [187]. This finding suggests the role of MDA5 in innate intestinal immunity.
5.4. IFIH1 gain-of-function mutation cause lupus-like nephritis in mice
Direct evidence linking MDA5 gain-of-function or loss-of-function variants to autoimmune phenotypes is currently limited. In mice models harboring IFIH1 G821S mutation induced by N-ethyl-N-nitrosourea (ENU), lupus-like nephritis developed with spontaneous type I IFN induction in multiple organs, which were not seen in MAVS-/- or IFNAR-/- backgrounds. And this provides the first evidence of a direct correlation between the MDA5 gain-of-function mutation and an autoimmune phenotype [172,188]. Moreover, merely overexpression of the Ifih1 gene could not cause autoimmune symptoms, indicating that mutation-induced MDA5 dysregulation is pivotal to eliciting the autoimmune response [189]. However, it remains elusive how SNPs or SNVs in IFIH1 cause these MDA5 structural and functional transformations, which may lead to exacerbated antiviral responses or constitutive activation of the type I IFN pathway that contributes to increased susceptibility to these diseases. These findings are expected to deepen our understanding of the relationship between RLRs-mediated innate immune response and autoimmune disorders. This may assist in developing the relevant therapeutics against these disorders.
6. Gain-of-function (GOF) mutations in RLRs cause auto-inflammatory diseases
Gain-of-function mutations of RLRs often make the body mistake self-derived nucleic acids for viral products and lead to aberrant type I IFN signaling production (termed IFN-I signature), causing extensive organ damage and many rare disorders characterized by IFN-I signature [190,191]. Some of these disorders are monogenic diseases, including Aicardi-Goutieres syndrome, Singleton-Merten Syndrome and Immunodeficiency 95, and are therefore good models for molecular studies.
6.1. IFIH1 mutations result in Aicardi-Goutieres syndrome
Aicardi-Goutieres syndrome (AGS) is a rare genetical neuroimmunological disease and type I interferonopathy characterized by impaired psychomotor development, dystonic posturing, spasticity, increased IFN-α in cerebrospinal fluid and up-regulated ISG gene expression in infancy. In 8 unrelated patients from AGS families, six heterozygous variants have been identified in the conserved helicase domains of the IFIH1 gene. In vitro cellular experiments demonstrated that the heterozygous IFIH1 variants led to apparent induction of interferon and ISGs genes in the absence or presence of exogenous MDA5 ligand. The biochemical assays revealed that these substitute residues increase the binding activity of MDA5 to RNA and the stability of the activated MDA5 filament [180]. However, the other three mutant MDA5 proteins encoded by novel heterozygous mutations from Japanese AGS patients lacked the response to specific agonists, although IFN-β was induced in hepatoma cells transfected with MDA5 mutants without agonist stimulation [192]. These data suggest that gain-of-function mutations in IFIH1 lead to constitutive MDA5 activation, interferon signaling, and eventually interferonopathy. One possibility for constitutive MDA5 activation in AGS patients is that the IFIH1variants alter their conformation via releasing CARD domain autorepression, thereby eliciting interferon signaling without ligand stimulation [188]. Another possibility is that MDA5 mutants misrecognized self-RNA, inducing an antiviral response [180]. However, endogenous ligands for MDA5 mutants are in need to be defined. Recent evidence further revealed that MDA5 mutants from AGS patients detected and formed filaments on the self Alu-dsRNA duplex, and then turned into a constitutive activation of MDA5 signaling because these mutants lost tolerance to imperfect duplex construct [193].
6.2. IFIH1 mutations lead to Immunodeficiency 95
Immunodeficiency-95 (IMD95) is a hereditary disease characterized by severe early-onsets of viral lung infection during childhood, caused by a loss of response of the innate immune system to viral invasions. This results in a failure of interferon production and inhibition of viral replication. Four kinds of homozygous mutations in the IFIH1 gene, including missense, nonsense, and frameshift variant, have been linked to Immunodeficiency-95 patients [187,[194], [195], [196]]. These MDA5 mutants impaired their activity to assemble and respond to the virus mimics, and failed to activate IFN-β pathway signaling, suggesting that they belong to loss-of-function variations.
6.3. RLRs identified as genetic defects of Singleton-Merten Syndrome
Singleton-Merten syndrome (SMS) is an unusual and hereditary disorder manifested with abnormal calcifications of the aorta and valves, dental inflammation, and osseous abnormality during childhood. The variants in DDX58 or IFIH1 have been proved to link to SMS [197]. Three heterozygous mutations in the DDX58 gene, E373A, E383A and C268F, were first identified in two SMS families characterized by glaucoma, aortic calcification, and skeletal anomalies. They result in the constitutive IFN-I production and ISGs gene expression [198]. So far, only one kind of IFIH1 variant (R822Q) has been identified in four SMS patients from distinct families, and this mutant strengthens MDA5-mediated IFN-β production and interferon signature genes, leading to an overt inflammatory disease [180,192,199]. It will be intriguing to investigate how the distinct amino acid substitutions in MDA5 alter the conformation and function of MDA5 and lead to SMS and AGS.
Why changes in the function of one single gene can lead to so many different phenotypes of autoinflammation or autoimmune diseases remains to be further explored. One possible hypothesis is that these conformational changes induced by amino acid substitutions enable MDA5 and RIG-I to easily acquire activity to self dsRNA [197]. Some also consider that genetically relevant factors may play a driving role in the onset and development of diseases.
7. RLRs expression pattern in autoimmune disease
In addition to SNPs and SNVs of RLRs, a recent study showed that RIG-I expression of peripheral T lymphocyte increases in patients with DM as well as T cell lymphopenia, presenting a negative correlation between RIG-I and T cell count. Upregulated RIG-I induced apoptosis by increasing the expression of Fas and cleaved-caspase 3 protein as well as inhibiting T lymphocyte proliferation in vitro. RIG-I-induced apoptosis may account for the mechanism of T cell lymphopenia in DM patients [200]. Furthermore, the expression of RIG-I and MDA5 are up-regulated in the perifascicular atrophy fibers in DM biopsies, suggesting that RIG-I is involved in the endogenous production of type I IFN and the formation of perifascicular atrophy within DM patient muscle tissue [201,202]. Statistical analysis performed on 44 DM patients further confirmed that RIG-I expression in perifascicular myofibers is a reliable and promising biomarker of DM [203].
In almost half of primary Sjögren's syndrome (pSS) patients, the disease activity and autoantibody presence positively correlate with the upregulated type I IFN signature. sRIG-I and MDA5 were upregulated in plasmacytoid dendritic cells and monocytes of patients with IFN-positive pSS, contributing to the pathogenic type I IFN production [204].
8. Anti-MDA5 antibody in clinical amyopathic dermatomyositis
Dermatomyositis (DM) is a systemic autoimmune disease which involves muscle, skin and organs such as joints and lungs. In the serum of patients with PM/DM, myositis-specific autoantibodies (MSAs) and myositis-associated autoantibodies (MAAs) are detected. The anti-MDA5 antibody, one of the MSAs, was first identified in a Japanese cohort by Sato et al in 2005 [205]. Anti-MDA5 antibody-positive patients are mainly seen in DM, especially in clinically amyopathic dermatomyositis (CADM), which is a subtype of DM, and its clinical manifestations are substantially different from other types of DM. Patients with CADM display only skin or predominant skin appearance, only mild proximal or no muscle weakness and elevated serum muscle enzymes. The muscle biopsies of patients with CADM are usually normal or only contain scarce inflammation and cellular infiltrates (primarily macrophages) [206].
There are only a few data concerning the pathophysiology of CADM. Nevertheless, it is emphasized that type I IFN signaling pathway is pivotal for DM pathogenesis [207]. Allenbach et al. found considerable upregulation of the expression of six ISGs in DM patients with positive anti-MDA5 antibodies [208]. Jiang et al. further proved that type I IFN signaling pathway is overexpressed in the skeletal muscles of anti-MDA5 DM patients [209]. These observations all account for the mild muscle phenotype of CADM.
The numbers and ratios of peripheral blood lymphocytes, especially CD4+ T cells and CD8+ T cells, decrease in some myositis patients with anti-MDA5 antibodies. It is associated with anaemia, thrombocytopenia, fibrinolytic abnormalities (such as increased D-dimer), liver function damage and elevated blood lipid levels. Many studies have found that the levels of TNF-α, IL-6, IL-8, IL-10, IL-18, IFN α/β/γ, IP10 and other cytokines or chemokines are increased myositis patient serum with the anti-MDA5 antibodies, and the serum ferritin concentrations were also significantly elevated. Furthermore, the levels of soluble IL-2 receptor α chain (sIL-2R or SCD25), soluble scavenger receptor CD16 (SCD163) and soluble mannose receptor CD206 (SCD206) significantly increase in serum of patients with CADM and ILD [[210], [211], [212], [213]]. These inflammatory markers and proteins are essential in activating immune cells, especially T cells and macrophages. As a result, many scholars have proposed the hypothesis that macrophage activation plays a vital role in CADM associated with RP-ILD.
The incidence rate of CADM ranges from 10% to 48% in previous studies of patients with PM/DM [214]. A link is identified between anti-MDA5 antibodies and clinical characteristics such as mechanic’s hand, skin ulcers, Gottron’s sign or papules and V rash. For this group of patients with CADM, lung injury is the most typical and prominent feature. 80% of CADM patients are associated with interstitial lung disease (ILD), and 30-60% with rapidly progressive interstitial lung disease (RP-ILD), leading to respiratory failure and mortality as high as 50% [215,216]. Lung tissue pathology revealed diffuse alveolar damage (DAD), including hyaline membrane formation, alveolar exudation, alveolar obstruction and collapse [217,218]. Patients with CADM and RP-ILD usually have very poor prognoses. In a recent meta-analysis, anti-MDA5 autoantibody showed superior sensitivity and specificity for RP-ILD, at 77% (95% CI: 64, 87%) and 86% (95% CI:79, 90%), respectively [219] in patients with anti-MDA5 antibody-positive CADM.
However, the correlation between anti-MDA5 antibody presence and the clinical manifestations of DM patients has not been determined yet. John et al. reported that no significant association exits between anti-MDA5 autoantibody and RP-ILD [220]. The frequency of positive anti-MDA5 antibodies differs among different ethnicities, as most case reports are identified in Asian populations (especially Japanese). The prevalence percentage of ILD in typical adult-onset DM patients is only about 10% in the United States but almost 40% in Japan.
The initial reduction of anti-MDA5 antibody in most DM patients, including those in fatal cases, suggests that perhaps MDA5 is not a potential biomarker for DM patients [221]. Thus, the associated clinical phenotype in different ethnicities must be further explored.
9. Development of RLRs as therapeutic targets or prognostic indicators in cancers
More and more studies have found that tumor cells can mimic viral invasion and trigger the pathogen recognition of innate immune as well as the signal transduction of type I IFN [222]. For example, it has been proved that some damaged tumor cells leak cytosolic DNA fragments and activates the STING signaling pathway to induce an anti-tumor immune response [223]. Similarly, stromal cells within the tumor tissue can release exosomes containing nucleic acids such as small nuclear RNAs (snRNAs) that can be sensed by the RIG-I/MDA5 of adjacent tumor cells. Thus, the anti-tumor process is elicited via the production of pro-inflammatory cytokines, the activation of programmed cell death as well as enhanced cytolytic activity of NK cells and increased antigen-presenting ability of antigen presenting cells (APCs), which include macrophages and dendritic cells (DCs) [80,224,225]. When co-cultured with pancreatic cancer cells, those dendritic cells with pre-activated RIG-I signaling can present tumor-derived antigens to T lymphocytes more effectively [225]. Consistent with this phenomenon, ovarian cancer cells are pre-stimulated with RIG-I performed apoptosis. They get easily phagocytosed by monocytes and DCs, further indicating that mimicking viral infection can elicit immunogenic cell death of tumor cells and enhance immune responses against the tumor through the activation of RLRs signaling pathway [226].
Lately, there is growing evidence that agonists of RIG-I and MDA5 can produce antiviral and anti-tumor effects in a type I IFN-dependent or -independent manner, leading to immunogenic tumor cell death and the regression of multiple types of tumors [227,228]. Treatments aimed at activating non-infectious RLRs signaling pathways are under pre-clinical investigation as an anticancer strategy, which functions through effectively initiating inflammatory responses against neoantigens of tumor cells and has received considerable attention. For instance, RLRs agonists that comprise poly (I:C), 5' -triphosphate dsRNA (ppp dsRNA), stem loop RNA (SLR) 14 and M8, may induce cancer cell apoptosis and immune responses against tumors. [229,[230], [231], [232], [233], [234], [235]]. Through cotreatment with RLR agonist Poly(I:C)-high molecular weight (Poly(I:C)-HMW), Sato et al. finds that the anticancer effect of ionizing radiation (IR) is enhanced, and the following upregulated Fas expression is responsive to FasL-induced apoptosis. Therefore, they propose that it is promising to use a combination of RLR agonist, IR, and FasL for treating cancers [231]. SLR14 is a RIG-I agonist which has been observed to induce robust anti-tumor responses in immunogenic or nonimmunogenic tumors and can serve as a powerful immune adjuvant of immune checkpoint blockade (ICB) therapy [232]. Das et al. designed a nanoparticle delivery of RIG-I’s natural ligand ppp- dsRNA into pancreatic cancer and successfully seized tumor progression [234].
M8, a sequence-modified RIG-I agonist, is a uridine-rich hairpin 5’pppRNA composed of 99 nucleotides and has recently shown promising potential to induce immunogenic cell death-related and damage-associated molecular patterns (ICD-DAMP) on tumor cells and synergize with other therapies to stimulate the activation of anti-tumor immune signals [235,236]. David et al. found that intertumoral administration of the RIG-I agonist SLR20 can induce immunogenic cell death and breast cancer cell microenvironment modulation, ultimately leading to the inhibition of tumor growth and metastasis [237]. RIG-I activation has been proved to take effect in resisting a large set of human tumor cells that comprise breast cancer, cervical cancer, myeloid leukemia, prostate cancer, lung cancer, pancreatic cancer and so on [230,238]. These findings further highlight the potential of therapeutic RLRs agonists in adjuvant therapy. Newly developed RLR agonists may fill the growing need for novel therapeutics against tumors in the future. However, whether these RLRs agonists are structurally stable and whether they have corresponding, potent or specific ligands in vivo is a question that needs to be addressed in future studies. Furthermore, prolonged exposure to a type I IFN environment can lead to an anti-inflammatory response, and inappropriate use of RLRs agonists may also result in an out-of-control inflammatory response. Therefore, the long-term use of RLRs agonists should be fully considered, and the periodicity and dose of administration should be further studied.
Beyond the role in treating cancer, RLRs also function as prognostic indicators for specific cancers. A monocentric study by Dominik et al. prove that high RIG-I expression is associated with ovarian cancer (OC) poor outcome, including disease progression and recurrence after remission. This correlation is much more significant in p53-mutated Type-II OC. Survival analysis further disclose RIG-I as a new biomarker for OC survival with independent prognostic usages [239]. Conversely, a low level of RIG-I suggests poor prognosis in melanoma patients. As previous studies have reported, IFN-α plays a crucial role in inducing cancer cell apoptosis and performing antitumor immune responses [240,241].
Melanoma tumor-repopulating cells (TRCs) are a subpopulation of highly malignant and tumorigenic cancer cells. To counteract the IFN-α-induced cell apoptosis and achieve immune evasion, TRCs downregulate RIG-I expression and hamper STAT1 activation via the integrin β3 (ITGB3)-c-SRC-STAT3 signaling pathway [241]. A combination treatment of STAT3 inhibitor stattic and IFN-α could improve the therapeutic effect of melanoma, and this discovery may provide new ideas for melanoma immunotherapy. As for neuroblastoma (NB), high cytoplasmic LGP2 expression in tumor tissues is found to strongly correlate with the differentiated histological grade and higher survival rates for NB patients, suggesting that LGP2 may exert an antitumor effect and serve as a prognostic marker in NB [242]. The mechanism of LGP2’s role as a tumor suppressor may be that LGP2 can enhance the sensitivity to poly (I:C)-induced apoptotic cell death and increase the sub-G1 phase of NB cells. In short, RLRs serve as crucial participants of innate immune and can influence the anti-tumor immune response of hosts through many different mechanisms, playing a potential role in tumor suppression and opening up a new way for tumor pathogenesis and immunotherapies.
10. Summary and outlook
The RLRs family and its downstream IFN signaling pathway, as one of the critical cytoplasmic RNA sensors for pathogen recognition, play an indispensable part in mediating an effective immune response of the host. However, just as the coin has both sides, the role of the RLRS family in diseases can be a double-edged sword. Moderate and controlled activation of RLRs protects the host from invading pathogens, whereas excessive and uncontrolled RLR activation may lead to unexpected pathological damage and cytokine storms, and may even cause autoimmune diseases or autoinflammatory diseases. Furthermore, the SNPs and functional activity of RLRs are associated with many monogenic inherited diseases and tumors. Therefore, understanding the role of RLRs in diseases and how to utilize it effectively has become an urgent clinical problem. Fortunately, there are currently many RLR agonists or antagonists developed and used as antiviral agents, vaccine adjuvants and antitumor drugs. With better understanding of the molecular biology of RLRs and their regulatory mechanisms, these therapies may benefit patients with a variety of treatment options in the future.
Declaration of Competing Interest
We declare that we do not have any commercial or associative interest that represents a conflict of interest in connection with the work submitted.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (NSFC) (grant numbers 82172146, 82070018).
We thank Xianzhong Xiao (recipient of NSFC grant no. 82172146) for supervision and advice.
We declare no conflicts of interest.
References
- 1.Loo Y.M., Gale M.J. Immune signaling by RIG-I-like receptors. Immunity. 2011;34(5):680–692. doi: 10.1016/j.immuni.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shekarian T., et al. Pattern recognition receptors: immune targets to enhance cancer immunotherapy. Ann Oncol. 2017;28(8):1756–1766. doi: 10.1093/annonc/mdx179. [DOI] [PubMed] [Google Scholar]
- 3.Aleynick M., et al. Pathogen molecular pattern receptor agonists: treating cancer by mimicking infection. Clin Cancer Res. 2019;25(21):6283–6294. doi: 10.1158/1078-0432.CCR-18-1800. [DOI] [PubMed] [Google Scholar]
- 4.Li X., et al. Structural basis of double-stranded RNA recognition by the RIG-I like receptor MDA5. Arch Biochem Biophys. 2009;488(1):23–33. doi: 10.1016/j.abb.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 5.Fairman-Williams M.E., Guenther U.P., Jankowsky E. SF1 and SF2 helicases: family matters. Curr Opin Struct Biol. 2010;20(3):313–324. doi: 10.1016/j.sbi.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schweibenz B.D., et al. The intrinsically disordered CARDs-Helicase linker in RIG-I is a molecular gate for RNA proofreading. EMBO J. 2022;41(10) doi: 10.15252/embj.2021109782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kawai T., et al. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6(10):981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 8.Bruns A.M., Horvath C.M. Activation of RIG-I-like receptor signal transduction. Crit Rev Biochem Mol Biol. 2012;47(2):194–206. doi: 10.3109/10409238.2011.630974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duic I., et al. Viral RNA recognition by LGP2 and MDA5, and activation of signaling through step-by-step conformational changes. Nucleic Acids Res. 2020;48(20):11664–11674. doi: 10.1093/nar/gkaa935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duic I., et al. Viral RNA recognition by LGP2 and MDA5, and activation of signaling through step-by-step conformational changes. Nucleic Acids Res. 2020;48(20):11664–11674. doi: 10.1093/nar/gkaa935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Komuro A., Horvath C.M. RNA- and virus-independent inhibition of antiviral signaling by RNA helicase LGP2. J Virol. 2006;80(24):12332–12342. doi: 10.1128/JVI.01325-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kang D.C., et al. Expression analysis and genomic characterization of human melanoma differentiation associated gene-5, mda-5: a novel type I interferon-responsive apoptosis-inducing gene. Oncogene. 2004;23(9):1789–1800. doi: 10.1038/sj.onc.1207300. [DOI] [PubMed] [Google Scholar]
- 13.Yoneyama M., et al. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J Immunol. 2005;175(5):2851–2858. doi: 10.4049/jimmunol.175.5.2851. [DOI] [PubMed] [Google Scholar]
- 14.Bruns A.M., Horvath C.M. LGP2 synergy with MDA5 in RLR-mediated RNA recognition and antiviral signaling. Cytokine. 2015;74(2):198–206. doi: 10.1016/j.cyto.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Satoh T., et al. LGP2 is a positive regulator of RIG-I- and MDA5-mediated antiviral responses. Proc Natl Acad Sci U S A. 2010;107(4):1512–1517. doi: 10.1073/pnas.0912986107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Venkataraman T., et al. Loss of DExD/H box RNA helicase LGP2 manifests disparate antiviral responses. J Immunol. 2007;178(10):6444–6455. doi: 10.4049/jimmunol.178.10.6444. [DOI] [PubMed] [Google Scholar]
- 17.Bruns A.M., et al. The innate immune sensor LGP2 activates antiviral signaling by regulating MDA5-RNA interaction and filament assembly. Mol Cell. 2014;55(5):771–781. doi: 10.1016/j.molcel.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miyamoto M., Komuro A. PACT is required for MDA5-mediated immunoresponses triggered by Cardiovirus infection via interaction with LGP2. Biochem Biophys Res Commun. 2017;494(1-2):227–233. doi: 10.1016/j.bbrc.2017.10.048. [DOI] [PubMed] [Google Scholar]
- 19.Bruns A.M., et al. The innate immune sensor LGP2 activates antiviral signaling by regulating MDA5-RNA interaction and filament assembly. Mol Cell. 2014;55(5):771–781. doi: 10.1016/j.molcel.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sanchez D.R., et al. LGP2 binds to PACT to regulate RIG-I- and MDA5-mediated antiviral responses. Sci Signal. 2019;12(601) doi: 10.1126/scisignal.aar3993. [DOI] [PubMed] [Google Scholar]
- 21.Yin X., et al. MDA5 Governs the Innate Immune Response to SARS-CoV-2 in Lung Epithelial Cells. Cell Rep. 2021;34(2) doi: 10.1016/j.celrep.2020.108628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loo Y.M., et al. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J Virol. 2008;82(1):335–345. doi: 10.1128/JVI.01080-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hornung V., et al. 5’-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314(5801):994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 24.Dias J.A., Sampaio N.G., Rehwinkel J. A Balancing Act: MDA5 in Antiviral Immunity and Autoinflammation. Trends Microbiol. 2019;27(1):75–85. doi: 10.1016/j.tim.2018.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goubau D., et al. Antiviral immunity via RIG-I-mediated recognition of RNA bearing 5'-diphosphates. Nature. 2014;514(7522):372–375. doi: 10.1038/nature13590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kato H., et al. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med. 2008;205(7):1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hornung V., et al. 5’-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314(5801):994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 28.Pichlmair A., et al. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5'-phosphates. Science. 2006;314(5801):997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 29.Goubau D., et al. Antiviral immunity via RIG-I-mediated recognition of RNA bearing 5'-diphosphates. Nature. 2014;514(7522):372–375. doi: 10.1038/nature13590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rehwinkel J., Gack M.U. RIG-I-like receptors: their regulation and roles in RNA sensing. Nat Rev Immunol. 2020;20(9):537–551. doi: 10.1038/s41577-020-0288-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davis W.G., et al. The 3' untranslated regions of influenza genomic sequences are 5'PPP-independent ligands for RIG-I. PLoS One. 2012;7(3) doi: 10.1371/journal.pone.0032661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schnell G., et al. Uridine composition of the poly-U/UC tract of HCV RNA defines non-self recognition by RIG-I. PLoS Pathog. 2012;8(8) doi: 10.1371/journal.ppat.1002839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hafner A.M., Corthesy B., Merkle H.P. Particulate formulations for the delivery of poly(I:C) as vaccine adjuvant. Adv Drug Deliv Rev. 2013;65(10):1386–1399. doi: 10.1016/j.addr.2013.05.013. [DOI] [PubMed] [Google Scholar]
- 34.Kato H., et al. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med. 2008;205(7):1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dhir A., et al. Mitochondrial double-stranded RNA triggers antiviral signalling in humans. Nature. 2018;560(7717):238–242. doi: 10.1038/s41586-018-0363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCartney S.A., et al. MDA-5 recognition of a murine norovirus. PLoS Pathog. 2008;4(7) doi: 10.1371/journal.ppat.1000108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kato H., et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441(7089):101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 38.Abe Y., et al. The toll-like receptor 3-mediated antiviral response is important for protection against poliovirus infection in poliovirus receptor transgenic mice. J Virol. 2012;86(1):185–194. doi: 10.1128/JVI.05245-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee H.C., Chathuranga K., Lee J.S. Intracellular sensing of viral genomes and viral evasion. Exp Mol Med. 2019;51(12):1–13. doi: 10.1038/s12276-019-0299-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Francisco E., et al. Cell-type specificity and functional redundancy of RIG-I-like receptors in innate immune sensing of Coxsackievirus B3 and encephalomyocarditis virus. Virology. 2019;528:7–18. doi: 10.1016/j.virol.2018.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Slater L., et al. Co-ordinated role of TLR3, RIG-I and MDA5 in the innate response to rhinovirus in bronchial epithelium. PLoS Pathog. 2010;6(11) doi: 10.1371/journal.ppat.1001178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chalubinski M., et al. Human rhinovirus 16 induces antiviral and inflammatory response in the human vascular endothelium. APMIS. 2021;129(3):143–151. doi: 10.1111/apm.13103. [DOI] [PubMed] [Google Scholar]
- 43.Fredericksen B.L., et al. Establishment and maintenance of the innate antiviral response to West Nile Virus involves both RIG-I and MDA5 signaling through IPS-1. J Virol. 2008;82(2):609–616. doi: 10.1128/JVI.01305-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Loo Y.M., et al. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J Virol. 2008;82(1):335–345. doi: 10.1128/JVI.01080-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saito T., et al. Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proc Natl Acad Sci U S A. 2007;104(2):582–587. doi: 10.1073/pnas.0606699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schlee M., et al. Recognition of 5' triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity. 2009;31(1):25–34. doi: 10.1016/j.immuni.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cao X., et al. MDA5 plays a critical role in interferon response during hepatitis C virus infection. J Hepatol. 2015;62(4):771–778. doi: 10.1016/j.jhep.2014.11.007. [DOI] [PubMed] [Google Scholar]
- 48.Zalinger Z.B., et al. MDA5 Is Critical to Host Defense during Infection with Murine Coronavirus. J Virol. 2015;89(24):12330–12340. doi: 10.1128/JVI.01470-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kato H., Takahasi K., Fujita T. RIG-I-like receptors: cytoplasmic sensors for non-self RNA. Immunol Rev. 2011;243(1):91–98. doi: 10.1111/j.1600-065X.2011.01052.x. [DOI] [PubMed] [Google Scholar]
- 50.Thorne L.G., et al. SARS-CoV-2 sensing by RIG-I and MDA5 links epithelial infection to macrophage inflammation. EMBO J. 2021;40(15) doi: 10.15252/embj.2021107826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hertzog J., et al. Infection with a Brazilian isolate of Zika virus generates RIG-I stimulatory RNA and the viral NS5 protein blocks type I IFN induction and signaling. Eur J Immunol. 2018;48(7):1120–1136. doi: 10.1002/eji.201847483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Akhrymuk I., Frolov I., Frolova E.I. Both RIG-I and MDA5 detect alphavirus replication in concentration-dependent mode. Virology. 2016;487:230–241. doi: 10.1016/j.virol.2015.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kato H., et al. Cell type-specific involvement of RIG-I in antiviral response. Immunity. 2005;23(1):19–28. doi: 10.1016/j.immuni.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 54.Habjan M., et al. Processing of genome 5' termini as a strategy of negative-strand RNA viruses to avoid RIG-I-dependent interferon induction. PLoS One. 2008;3(4) doi: 10.1371/journal.pone.0002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Luthra P., et al. Activation of IFN-β expression by a viral mRNA through RNase L and MDA5. Proc Natl Acad Sci U S A. 2011;108(5):2118–2123. doi: 10.1073/pnas.1012409108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou S., et al. Induction and inhibition of type I interferon responses by distinct components of lymphocytic choriomeningitis virus. J Virol. 2010;84(18):9452–9462. doi: 10.1128/JVI.00155-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.He F., et al. Ebolavirus protein VP24 interferes with innate immune responses by inhibiting interferon-lambda1 gene expression. Virology. 2017;509:23–34. doi: 10.1016/j.virol.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 58.Faul E.J., et al. Rabies virus infection induces type I interferon production in an IPS-1 dependent manner while dendritic cell activation relies on IFNAR signaling. PLoS Pathog. 2010;6(7) doi: 10.1371/journal.ppat.1001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Linder A., et al. Defective Interfering Genomes and the Full-Length Viral Genome Trigger RIG-I After Infection With Vesicular Stomatitis Virus in a Replication Dependent Manner. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.595390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu B., et al. Structural basis for dsRNA recognition, filament formation, and antiviral signal activation by MDA5. Cell. 2013;152(1–2):276–289. doi: 10.1016/j.cell.2012.11.048. [DOI] [PubMed] [Google Scholar]
- 61.Gitlin L., et al. Melanoma differentiation-associated gene 5 (MDA5) is involved in the innate immune response to Paramyxoviridae infection in vivo. PLoS Pathog. 2010;6(1) doi: 10.1371/journal.ppat.1000734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ikegame S., et al. Both RIG-I and MDA5 RNA helicases contribute to the induction of alpha/beta interferon in measles virus-infected human cells. J Virol. 2010;84(1):372–379. doi: 10.1128/JVI.01690-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cuellar T.L., et al. Silencing of retrotransposons by SETDB1 inhibits the interferon response in acute myeloid leukemia. J Cell Biol. 2017;216(11):3535–3549. doi: 10.1083/jcb.201612160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Oosenbrug T., et al. Induction of Robust Type I Interferon Levels by Oncolytic Reovirus Requires Both Viral Replication and Interferon-alpha/beta Receptor Signaling. Hum Gene Ther. 2021;32(19-20):1171–1185. doi: 10.1089/hum.2021.140. [DOI] [PubMed] [Google Scholar]
- 65.Dou Y., et al. The innate immune receptor MDA5 limits rotavirus infection but promotes cell death and pancreatic inflammation. EMBO J. 2017;36(18):2742–2757. doi: 10.15252/embj.201696273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Samanta M., et al. EB virus-encoded RNAs are recognized by RIG-I and activate signaling to induce type I IFN. EMBO J. 2006;25(18):4207–4214. doi: 10.1038/sj.emboj.7601314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang F., et al. RIG-I mediates the co-induction of tumor necrosis factor and type I interferon elicited by myxoma virus in primary human macrophages. PLoS Pathog. 2008;4(7) doi: 10.1371/journal.ppat.1000099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang N., et al. Lung type II alveolar epithelial cells collaborate with CCR2(+) inflammatory monocytes in host defense against poxvirus infection. Nat Commun. 2022;13(1):1671. doi: 10.1038/s41467-022-29308-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rasmussen S.B., et al. Herpes simplex virus infection is sensed by both Toll-like receptors and retinoic acid-inducible gene- like receptors, which synergize to induce type I interferon production. J Gen Virol. 2009;90(Pt 1):74–78. doi: 10.1099/vir.0.005389-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Melchjorsen J., et al. Early innate recognition of herpes simplex virus in human primary macrophages is mediated via the MDA5/MAVS-dependent and MDA5/MAVS/RNA polymerase III-independent pathways. J Virol. 2010;84(21):11350–11358. doi: 10.1128/JVI.01106-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sato S., et al. The RNA sensor RIG-I dually functions as an innate sensor and direct antiviral factor for hepatitis B virus. Immunity. 2015;42(1):123–132. doi: 10.1016/j.immuni.2014.12.016. [DOI] [PubMed] [Google Scholar]
- 72.Zao X., et al. NFATc3 inhibits hepatocarcinogenesis and HBV replication via positively regulating RIG-I-mediated interferon transcription. Oncoimmunology. 2021;10(1):1869388. doi: 10.1080/2162402X.2020.1869388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lu H.L., Liao F. Melanoma differentiation-associated gene 5 senses hepatitis B virus and activates innate immune signaling to suppress virus replication. J Immunol. 2013;191(6):3264–3276. doi: 10.4049/jimmunol.1300512. [DOI] [PubMed] [Google Scholar]
- 74.Louber J., et al. Kinetic discrimination of self/non-self RNA by the ATPase activity of RIG-I and MDA5. BMC Biol. 2015;13:54. doi: 10.1186/s12915-015-0166-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Song B., et al. Ordered assembly of the cytosolic RNA-sensing MDA5-MAVS signaling complex via binding to unanchored K63-linked poly-ubiquitin chains. Immunity. 2021;54(10):2218–2230.e5. doi: 10.1016/j.immuni.2021.09.008. [DOI] [PubMed] [Google Scholar]
- 76.Yu Q., Qu K., Modis Y. Cryo-EM Structures of MDA5-dsRNA Filaments at Different Stages of ATP Hydrolysis. Mol Cell. 2018;72(6):999–1012.e6. doi: 10.1016/j.molcel.2018.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Seth R.B., et al. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122(5):669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 78.Shi Y., et al. An autoinhibitory mechanism modulates MAVS activity in antiviral innate immune response. Nat Commun. 2015;6:7811. doi: 10.1038/ncomms8811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nombel A., Fabien N., Coutant F. Dermatomyositis with Anti-MDA5 antibodies: bioclinical features, pathogenesis and emerging therapies. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.773352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wu Y., et al. The anticancer functions of RIG-I-like receptors, RIG-I and MDA5, and their applications in cancer therapy. Transl Res. 2017;190:51–60. doi: 10.1016/j.trsl.2017.08.004. [DOI] [PubMed] [Google Scholar]
- 81.Zhang W., et al. Lactate is a natural suppressor of RLR signaling by targeting MAVS. Cell. 2019;178(1):176–189.e15. doi: 10.1016/j.cell.2019.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liu J., et al. Application and prospect of targeting innate immune sensors in the treatment of autoimmune diseases. Cell Biosci. 2022;12(1):68. doi: 10.1186/s13578-022-00810-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.van der Veen A.G., et al. The RIG-I-like receptor LGP2 inhibits Dicer-dependent processing of long double-stranded RNA and blocks RNA interference in mammalian cells. EMBO J. 2018;37(4) doi: 10.15252/embj.201797479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bruns A.M., Horvath C.M. LGP2 synergy with MDA5 in RLR-mediated RNA recognition and antiviral signaling. Cytokine. 2015;74(2):198–206. doi: 10.1016/j.cyto.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Goulet M.L., et al. Systems analysis of a RIG-I agonist inducing broad spectrum inhibition of virus infectivity. PLoS Pathog. 2013;9(4) doi: 10.1371/journal.ppat.1003298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schoggins J.W., et al. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472(7344):481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hou P., et al. A novel selective autophagy receptor, CCDC50, delivers K63 polyubiquitination-activated RIG-I/MDA5 for degradation during viral infection. Cell Res. 2021;31(1):62–79. doi: 10.1038/s41422-020-0362-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kok K.H., et al. The double-stranded RNA-binding protein PACT functions as a cellular activator of RIG-I to facilitate innate antiviral response. Cell Host Microbe. 2011;9(4):299–309. doi: 10.1016/j.chom.2011.03.007. [DOI] [PubMed] [Google Scholar]
- 89.Kok K.H., et al. The double-stranded RNA-binding protein PACT functions as a cellular activator of RIG-I to facilitate innate antiviral response. Cell Host Microbe. 2011;9(4):299–309. doi: 10.1016/j.chom.2011.03.007. [DOI] [PubMed] [Google Scholar]
- 90.Lui P.Y., et al. PACT Facilitates RNA-Induced Activation of MDA5 by Promoting MDA5 Oligomerization. J Immunol. 2017;199(5):1846–1855. doi: 10.4049/jimmunol.1601493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sanchez D.R., et al. LGP2 binds to PACT to regulate RIG-I- and MDA5-mediated antiviral responses. Sci Signal. 2019;12(601) doi: 10.1126/scisignal.aar3993. [DOI] [PubMed] [Google Scholar]
- 92.Vaughn L.S., Chukwurah E., Patel R.C. Opposite actions of two dsRNA-binding proteins PACT and TRBP on RIG-I mediated signaling. Biochem J. 2021;478(3):493–510. doi: 10.1042/BCJ20200987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lian H., et al. The Zinc-Finger Protein ZCCHC3 Binds RNA and Facilitates Viral RNA Sensing and Activation of the RIG-I-like Receptors. Immunity. 2018;49(3):438–448.e5. doi: 10.1016/j.immuni.2018.08.014. [DOI] [PubMed] [Google Scholar]
- 94.Wu X.M., et al. NOD1 Promotes Antiviral Signaling by Binding Viral RNA and Regulating the Interaction of MDA5 and MAVS. J Immunol. 2020;204(8):2216–2231. doi: 10.4049/jimmunol.1900667. [DOI] [PubMed] [Google Scholar]
- 95.Chen H., et al. RAVER1 is a coactivator of MDA5-mediated cellular antiviral response. J Mol Cell Biol. 2013;5(2):111–119. doi: 10.1093/jmcb/mjt006. [DOI] [PubMed] [Google Scholar]
- 96.Zhu Q., et al. DHX29 functions as an RNA co-sensor for MDA5-mediated EMCV-specific antiviral immunity. PLoS Pathog. 2018;14(2) doi: 10.1371/journal.ppat.1006886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhong X., et al. ZFYVE1 negatively regulates MDA5- but not RIG-I-mediated innate antiviral response. PLoS Pathog. 2020;16(4) doi: 10.1371/journal.ppat.1008457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kitai Y., et al. Negative regulation of melanoma differentiation-associated gene 5 (MDA5)-dependent antiviral innate immune responses by Arf-like protein 5B. J Biol Chem. 2015;290(2):1269–1280. doi: 10.1074/jbc.M114.611053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cadena C., et al. Ubiquitin-Dependent and -Independent Roles of E3 Ligase RIPLET in Innate Immunity. Cell. 2019;177(5):1187–1200.e16. doi: 10.1016/j.cell.2019.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lin J.P., Fan Y.K., Liu H.M. The 14-3-3eta chaperone protein promotes antiviral innate immunity via facilitating MDA5 oligomerization and intracellular redistribution. PLoS Pathog. 2019;15(2) doi: 10.1371/journal.ppat.1007582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gack M.U., et al. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446(7138):916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- 102.Yan J., et al. TRIM4 modulates type I interferon induction and cellular antiviral response by targeting RIG-I for K63-linked ubiquitination. J Mol Cell Biol. 2014;6(2):154–163. doi: 10.1093/jmcb/mju005. [DOI] [PubMed] [Google Scholar]
- 103.Kuniyoshi K., et al. Pivotal role of RNA-binding E3 ubiquitin ligase MEX3C in RIG-I-mediated antiviral innate immunity. Proc Natl Acad Sci U S A. 2014;111(15):5646–5651. doi: 10.1073/pnas.1401674111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wang W., et al. RNF122 suppresses antiviral type I interferon production by targeting RIG-I CARDs to mediate RIG-I degradation. Proc Natl Acad Sci U S A. 2016;113(34):9581–9586. doi: 10.1073/pnas.1604277113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Arimoto K., et al. Negative regulation of the RIG-I signaling by the ubiquitin ligase RNF125. Proc Natl Acad Sci U S A. 2007;104(18):7500–7505. doi: 10.1073/pnas.0611551104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhao C., et al. The E3 ubiquitin ligase TRIM40 attenuates antiviral immune responses by targeting MDA5 and RIG-I. Cell Rep. 2017;21(6):1613–1623. doi: 10.1016/j.celrep.2017.10.020. [DOI] [PubMed] [Google Scholar]
- 107.Zhao K., et al. Cytoplasmic STAT4 promotes antiviral type I IFN production by blocking CHIP-mediated degradation of RIG-I. J Immunol. 2016;196(3):1209–1217. doi: 10.4049/jimmunol.1501224. [DOI] [PubMed] [Google Scholar]
- 108.Lang X., et al. TRIM65-catalized ubiquitination is essential for MDA5-mediated antiviral innate immunity. J Exp Med. 2017;214(2):459–473. doi: 10.1084/jem.20160592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lin R., et al. Negative regulation of the retinoic acid-inducible gene I-induced antiviral state by the ubiquitin-editing protein A20. J Biol Chem. 2006;281(4):2095–2103. doi: 10.1074/jbc.M510326200. [DOI] [PubMed] [Google Scholar]
- 110.Friedman C.S., et al. The tumour suppressor CYLD is a negative regulator of RIG-I-mediated antiviral response. EMBO Rep. 2008;9(9):930–936. doi: 10.1038/embor.2008.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Nguyen N.T., et al. Ubiquitin-like modifier FAT10 attenuates RIG-I mediated antiviral signaling by segregating activated RIG-I from its signaling platform. Sci Rep. 2016;6:23377. doi: 10.1038/srep23377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kim M.J., et al. Negative feedback regulation of RIG-I-mediated antiviral signaling by interferon-induced ISG15 conjugation. J Virol. 2008;82(3):1474–1483. doi: 10.1128/JVI.01650-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Mirzalieva O., et al. ISG15 and ISGylation in human diseases. Cells. 2022;11(3) doi: 10.3390/cells11030538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhang D., Zhang D.E. Interferon-stimulated gene 15 and the protein ISGylation system. J Interferon Cytokine Res. 2011;31(1):119–130. doi: 10.1089/jir.2010.0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Liu G., et al. ISG15-dependent activation of the sensor MDA5 is antagonized by the SARS-CoV-2 papain-like protease to evade host innate immunity. Nature Microbiology. 2021;6(4):467–478. doi: 10.1038/s41564-021-00884-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Maharaj N.P., et al. Conventional protein kinase C-alpha (PKC-alpha) and PKC-beta negatively regulate RIG-I antiviral signal transduction. J Virol. 2012;86(3):1358–1371. doi: 10.1128/JVI.06543-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gack M.U., et al. Phosphorylation-mediated negative regulation of RIG-I antiviral activity. J Virol. 2010;84(7):3220–3229. doi: 10.1128/JVI.02241-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Nistal-Villan E., et al. Negative role of RIG-I serine 8 phosphorylation in the regulation of interferon-beta production. J Biol Chem. 2010;285(26):20252–20261. doi: 10.1074/jbc.M109.089912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sun Z., et al. Phosphorylation of RIG-I by casein kinase II inhibits its antiviral response. J Virol. 2011;85(2):1036–1047. doi: 10.1128/JVI.01734-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Takashima K., et al. RIOK3-mediated phosphorylation of MDA5 interferes with its assembly and attenuates the innate immune response. Cell Rep. 2015;11(2):192–200. doi: 10.1016/j.celrep.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 121.Wies E., et al. Dephosphorylation of the RNA sensors RIG-I and MDA5 by the phosphatase PP1 is essential for innate immune signaling. Immunity. 2013;38(3):437–449. doi: 10.1016/j.immuni.2012.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Fan Y., et al. SUMOylation in viral replication and antiviral defense. Adv Sci (Weinh) 2022;9(7) doi: 10.1002/advs.202104126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hu M.M., et al. Innate immunity to RNA virus is regulated by temporal and reversible sumoylation of RIG-I and MDA5. J Exp Med. 2017;214(4):973–989. doi: 10.1084/jem.20161015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Fu J., et al. MDA5 is SUMOylated by PIAS2beta in the upregulation of type I interferon signaling. Mol Immunol. 2011;48(4):415–422. doi: 10.1016/j.molimm.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Choi S.J., et al. HDAC6 regulates cellular viral RNA sensing by deacetylation of RIG-I. EMBO J. 2016;35(4):429–442. doi: 10.15252/embj.201592586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Liu H.M., et al. Regulation of retinoic acid inducible gene-I (RIG-I) activation by the histone deacetylase 6. EBioMedicine. 2016;9:195–206. doi: 10.1016/j.ebiom.2016.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Gao D., et al. REUL is a novel E3 ubiquitin ligase and stimulator of retinoic-acid-inducible gene-I. PLoS One. 2009;4(6) doi: 10.1371/journal.pone.0005760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Oshiumi H., et al. Riplet/RNF135, a RING finger protein, ubiquitinates RIG-I to promote interferon-beta induction during the early phase of viral infection. J Biol Chem. 2009;284(2):807–817. doi: 10.1074/jbc.M804259200. [DOI] [PubMed] [Google Scholar]
- 129.Chen W., et al. Induction of Siglec-G by RNA viruses inhibits the innate immune response by promoting RIG-I degradation. Cell. 2013;152(3):467–478. doi: 10.1016/j.cell.2013.01.011. [DOI] [PubMed] [Google Scholar]
- 130.Narayan K., et al. TRIM13 is a negative regulator of MDA5-mediated type I interferon production. J Virol. 2014;88(18):10748–10757. doi: 10.1128/JVI.02593-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Cui J., et al. USP3 inhibits type I interferon signaling by deubiquitinating RIG-I-like receptors. Cell Res. 2014;24(4):400–416. doi: 10.1038/cr.2013.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Fan Y., et al. USP21 negatively regulates antiviral response by acting as a RIG-I deubiquitinase. J Exp Med. 2014;211(2):313–328. doi: 10.1084/jem.20122844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zhong H., et al. Ubiquitin-specific proteases 25 negatively regulates virus-induced type I interferon signaling. PLoS One. 2013;8(11) doi: 10.1371/journal.pone.0080976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wang L., et al. USP4 positively regulates RIG-I-mediated antiviral response through deubiquitination and stabilization of RIG-I. J Virol. 2013;87(8):4507–4515. doi: 10.1128/JVI.00031-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Pauli E.K., et al. The ubiquitin-specific protease USP15 promotes RIG-I-mediated antiviral signaling by deubiquitylating TRIM25. Sci Signal. 2014;7(307) doi: 10.1126/scisignal.2004577. (p. ra3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Chen R., et al. The ubiquitin-specific protease 17 is involved in virus-triggered type I IFN signaling. Cell Res. 2010;20(7):802–811. doi: 10.1038/cr.2010.41. [DOI] [PubMed] [Google Scholar]
- 137.Maharaj N.P., et al. Conventional protein kinase C-alpha (PKC-alpha) and PKC-beta negatively regulate RIG-I antiviral signal transduction. J Virol. 2012;86(3):1358–1371. doi: 10.1128/JVI.06543-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Nistal-Villan E., et al. Negative role of RIG-I serine 8 phosphorylation in the regulation of interferon-beta production. J Biol Chem. 2010;285(26):20252–20261. doi: 10.1074/jbc.M109.089912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Gack M.U., et al. Phosphorylation-mediated negative regulation of RIG-I antiviral activity. J Virol. 2010;84(7):3220–3229. doi: 10.1128/JVI.02241-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Sun Z., et al. Phosphorylation of RIG-I by casein kinase II inhibits its antiviral response. J Virol. 2011;85(2):1036–1047. doi: 10.1128/JVI.01734-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wies E., et al. Dephosphorylation of the RNA sensors RIG-I and MDA5 by the phosphatase PP1 is essential for innate immune signaling. Immunity. 2013;38(3):437–449. doi: 10.1016/j.immuni.2012.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Fu J., et al. MDA5 is SUMOylated by PIAS2beta in the upregulation of type I interferon signaling. Mol Immunol. 2011;48(4):415–422. doi: 10.1016/j.molimm.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Nguyen N.T., et al. Ubiquitin-like modifier FAT10 attenuates RIG-I mediated antiviral signaling by segregating activated RIG-I from its signaling platform. Sci Rep. 2016;6:23377. doi: 10.1038/srep23377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Hou J., et al. MicroRNA-146a feedback inhibits RIG-I-dependent Type I IFN production in macrophages by targeting TRAF6, IRAK1, and IRAK2. J Immunol. 2009;183(3):2150–2158. doi: 10.4049/jimmunol.0900707. [DOI] [PubMed] [Google Scholar]
- 145.Sheng Y., et al. MicroRNA-92a inhibits macrophage antiviral response by targeting retinoic acid inducible gene-I. Microbiol Immunol. 2018;62(9):585–593. doi: 10.1111/1348-0421.12640. [DOI] [PubMed] [Google Scholar]
- 146.Qiu Y., et al. MicroRNA-218 inhibits type I interferon production and facilitates virus immune evasion via targeting RIG-I. Biotechnol Appl Biochem. 2020;67(3):396–403. doi: 10.1002/bab.1882. [DOI] [PubMed] [Google Scholar]
- 147.Ingle H., et al. The microRNA miR-485 targets host and influenza virus transcripts to regulate antiviral immunity and restrict viral replication. Sci Signal. 2015;8(406) doi: 10.1126/scisignal.aab3183. (p. ra126) [DOI] [PubMed] [Google Scholar]
- 148.Zhao L., et al. Identification of cellular microRNA-136 as a dual regulator of RIG-I-mediated innate immunity that antagonizes H5N1 IAV replication in A549 cells. Sci Rep. 2015;5:14991. doi: 10.1038/srep14991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Yeh Y.J., et al. Dual effects of Let-7b in the early stage of Hepatitis C virus infection. J Virol. 2021;95(4) doi: 10.1128/JVI.01800-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Karlsen T.A., Brinchmann J.E. Liposome delivery of microRNA-145 to mesenchymal stem cells leads to immunological off-target effects mediated by RIG-I. Mol Ther. 2013;21(6):1169–1181. doi: 10.1038/mt.2013.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Zhang C., et al. Hsa_circ_0012919 regulates expression of MDA5 by miR-125a-3p in CD4+ T cells of systemic lupus erythematous. Lupus. 2020;29(7):727–734. doi: 10.1177/0961203320920706. [DOI] [PubMed] [Google Scholar]
- 152.Chu Q., et al. Characterization of MDA5 and microRNA-203 negatively regulates the RLR signaling pathway via targeting MDA5 in miiuy croaker. Dev Comp Immunol. 2022;126 doi: 10.1016/j.dci.2021.104259. [DOI] [PubMed] [Google Scholar]
- 153.Han J., et al. microRNA-145 regulates the RLR signaling pathway in miiuy croaker after poly(I:C) stimulation via targeting MDA5. Dev Comp Immunol. 2017;68:79–86. doi: 10.1016/j.dci.2016.11.021. [DOI] [PubMed] [Google Scholar]
- 154.Li Z., et al. MiR-34b-5p Suppresses Melanoma Differentiation-Associated Gene 5 (MDA5) Signaling Pathway to Promote Avian Leukosis Virus Subgroup J (ALV-J)-Infected Cells Proliferaction and ALV-J Replication. Front Cell Infect Microbiol. 2017;7:17. doi: 10.3389/fcimb.2017.00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Liu D., et al. MicroRNA-33/33* inhibit the activation of MAVS through AMPK in antiviral innate immunity. Cell Mol Immunol. 2021;18(6):1450–1462. doi: 10.1038/s41423-019-0326-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Amado-Rodriguez L., et al. Effects of IFIH1 rs1990760 variants on systemic inflammation and outcome in critically ill COVID-19 patients in an observational translational study. Elife. 2022;11 doi: 10.7554/eLife.73012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Loske J., et al. Pre-activated antiviral innate immunity in the upper airways controls early SARS-CoV-2 infection in children. Nat Biotechnol. 2022;40(3):319–324. doi: 10.1038/s41587-021-01037-9. [DOI] [PubMed] [Google Scholar]
- 158.Liehl P., et al. Host-cell sensors for Plasmodium activate innate immunity against liver-stage infection. Nat Med. 2014;20(1):47–53. doi: 10.1038/nm.3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Wu J., et al. Strain-specific innate immune signaling pathways determine malaria parasitemia dynamics and host mortality. Proc Natl Acad Sci U S A. 2014;111(4):E511–E520. doi: 10.1073/pnas.1316467111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Ye W., et al. Microvesicles from malaria-infected red blood cells activate natural killer cells via MDA5 pathway. PLoS Pathog. 2018;14(10) doi: 10.1371/journal.ppat.1007298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Ofir-Birin Y., et al. Malaria parasites both repress host CXCL10 and use it as a cue for growth acceleration. Nat Commun. 2021;12(1):4851. doi: 10.1038/s41467-021-24997-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Pagliuso A., et al. An RNA-binding protein secreted by a bacterial pathogen modulates RIG-I signaling. Cell Host Microbe. 2019;26(6):823–835.e11. doi: 10.1016/j.chom.2019.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Schmolke M., et al. RIG-I detects mRNA of intracellular Salmonella enterica serovar Typhimurium during bacterial infection. mBio. 2014;5(2):e01006–e01014. doi: 10.1128/mBio.01006-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Cheng Y., Schorey J.S. Mycobacterium tuberculosis-induced IFN-beta production requires cytosolic DNA and RNA sensing pathways. J Exp Med. 2018;215(11):2919–2935. doi: 10.1084/jem.20180508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Yong H.Y., Luo D. RIG-I-Like receptors as novel targets for pan-antivirals and vaccine adjuvants against emerging and re-emerging viral infections. Front Immunol. 2018;9:1379. doi: 10.3389/fimmu.2018.01379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Elion D.L., et al. Therapeutically active RIG-I agonist induces immunogenic tumor cell killing in breast cancers. Cancer Res. 2018;78(21):6183–6195. doi: 10.1158/0008-5472.CAN-18-0730. [DOI] [PubMed] [Google Scholar]
- 167.Heidegger S., et al. RIG-I activating immunostimulatory RNA boosts the efficacy of anticancer vaccines and synergizes with immune checkpoint blockade. EBioMedicine. 2019;41:146–155. doi: 10.1016/j.ebiom.2019.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Nejentsev S., et al. Rare variants of IFIH1, a gene implicated in antiviral responses, protect against type 1 diabetes. Science. 2009;324(5925):387–389. doi: 10.1126/science.1167728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Sutherland A., et al. Genomic polymorphism at the interferon-induced helicase (IFIH1) locus contributes to Graves’ disease susceptibility. J Clin Endocrinol Metab. 2007;92(8):3338–3341. doi: 10.1210/jc.2007-0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Amy Strange F.C.C.C. A genome-wide association study identifies new psoriasis susceptibility loci and an interaction between HLA-C and ERAP1. Nature Genetics. 2010;42(11):985–990. doi: 10.1038/ng.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Sheng Y., et al. Sequencing-based approach identified three new susceptibility loci for psoriasis. Nat Commun. 2014;5:4331. doi: 10.1038/ncomms5331. [DOI] [PubMed] [Google Scholar]
- 172.Kato H., Fujita T. RIG-I-like receptors and autoimmune diseases. Curr Opin Immunol. 2015;37:40–45. doi: 10.1016/j.coi.2015.10.002. [DOI] [PubMed] [Google Scholar]
- 173.Cananzi M., et al. IFIH1 loss-of-function variants contribute to very early-onset inflammatory bowel disease. Hum Genet. 2021;140(9):1299–1312. doi: 10.1007/s00439-021-02300-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Martinez A., et al. Association of the IFIH1-GCA-KCNH7 chromosomal region with rheumatoid arthritis. Ann Rheum Dis. 2008;67(1):137–138. doi: 10.1136/ard.2007.073213. [DOI] [PubMed] [Google Scholar]
- 175.Jin Y., et al. Genome-wide association analyses identify 13 new susceptibility loci for generalized vitiligo. Nat Genet. 2012;44(6):676–680. doi: 10.1038/ng.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Enevold C., et al. Multiple sclerosis and polymorphisms of innate pattern recognition receptors TLR1-10, NOD1-2, DDX58, and IFIH1. J Neuroimmunol. 2009;212(1–2):125–131. doi: 10.1016/j.jneuroim.2009.04.008. [DOI] [PubMed] [Google Scholar]
- 177.Robinson T., et al. Autoimmune disease risk variant of IFIH1 is associated with increased sensitivity to IFN-alpha and serologic autoimmunity in lupus patients. J Immunol. 2011;187(3):1298–1303. doi: 10.4049/jimmunol.1100857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Zurawek M., et al. Cumulative effect of IFIH1 variants and increased gene expression associated with type 1 diabetes. Diabetes Res Clin Pract. 2015;107(2):259–266. doi: 10.1016/j.diabres.2014.11.008. [DOI] [PubMed] [Google Scholar]
- 179.Van Eyck L., et al. Brief Report: IFIH1 mutation causes systemic lupus erythematosus with selective IgA deficiency. Arthritis Rheumatol. 2015;67(6):1592–1597. doi: 10.1002/art.39110. [DOI] [PubMed] [Google Scholar]
- 180.Rice G.I., et al. Gain-of-function mutations in IFIH1 cause a spectrum of human disease phenotypes associated with upregulated type I interferon signaling. Nature Genetics. 2014;46(5):503–509. doi: 10.1038/ng.2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Nejentsev S., et al. Rare variants of IFIH1, a gene implicated in antiviral responses, protect against type 1 diabetes. Science. 2009;324(5925):387–389. doi: 10.1126/science.1167728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Jaidane H., Hober D. Role of coxsackievirus B4 in the pathogenesis of type 1 diabetes. Diabetes Metab. 2008;34(6 Pt 1):537–548. doi: 10.1016/j.diabet.2008.05.008. [DOI] [PubMed] [Google Scholar]
- 183.Cen H., et al. Association of IFIH1 rs1990760 polymorphism with susceptibility to autoimmune diseases: a meta-analysis. Autoimmunity. 2013;46(7):455–462. doi: 10.3109/08916934.2013.796937. [DOI] [PubMed] [Google Scholar]
- 184.Blum S.I., Tse H.M. Innate Viral Sensor MDA5 and Coxsackievirus Interplay in Type 1 Diabetes Development. Microorganisms. 2020;8(7) doi: 10.3390/microorganisms8070993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Oikarinen S., et al. Characterisation of enterovirus RNA detected in the pancreas and other specimens of live patients with newly diagnosed type 1 diabetes in the DiViD study. Diabetologia. 2021;64(11):2491–2501. doi: 10.1007/s00125-021-05525-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Geravandi S., et al. Localization of enteroviral RNA within the pancreas in donors with T1D and T1D-associated autoantibodies. Cell Rep Med. 2021;2(8) doi: 10.1016/j.xcrm.2021.100371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Cananzi M., et al. IFIH1 loss-of-function variants contribute to very early-onset inflammatory bowel disease. Hum Genet. 2021;140(9):1299–1312. doi: 10.1007/s00439-021-02300-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Funabiki M., et al. Autoimmune disorders associated with gain of function of the intracellular sensor MDA5. Immunity. 2014;40(2):199–212. doi: 10.1016/j.immuni.2013.12.014. [DOI] [PubMed] [Google Scholar]
- 189.Crampton S.P., et al. Ifih1 gene dose effect reveals MDA5-mediated chronic type I IFN gene signature, viral resistance, and accelerated autoimmunity. J Immunol. 2012;188(3):1451–1459. doi: 10.4049/jimmunol.1102705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Xiao W., et al. Case Report: Aicardi-Goutieres Syndrome and Singleton-Merten Syndrome Caused by a Gain-of-Function Mutation in IFIH1. Front Genet. 2021;12 doi: 10.3389/fgene.2021.660953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Xiao W., et al. Case Report: Aicardi-Goutieres Syndrome and Singleton-Merten Syndrome Caused by a Gain-of-Function Mutation in IFIH1. Front Genet. 2021;12 doi: 10.3389/fgene.2021.660953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Oda H., et al. Aicardi-Goutieres syndrome is caused by IFIH1 mutations. Am J Hum Genet. 2014;95(1):121–125. doi: 10.1016/j.ajhg.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Ahmad S., et al. Breaching Self-Tolerance to Alu Duplex RNA Underlies MDA5-Mediated Inflammation. Cell. 2018;172(4):797–810.e13. doi: 10.1016/j.cell.2017.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Lamborn I.T., et al. Recurrent rhinovirus infections in a child with inherited MDA5 deficiency. J Exp Med. 2017;214(7):1949–1972. doi: 10.1084/jem.20161759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Zaki M., et al. Recurrent and Prolonged Infections in a Child with a Homozygous IFIH1 Nonsense Mutation. Front Genet. 2017;8:130. doi: 10.3389/fgene.2017.00130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Asgari S., et al. Severe viral respiratory infections in children with IFIH1 loss-of-function mutations. Proc Natl Acad Sci U S A. 2017;114(31):8342–8347. doi: 10.1073/pnas.1704259114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Lu C., MacDougall M. RIG-I-Like Receptor Signaling in Singleton-Merten Syndrome. Front Genet. 2017;8:118. doi: 10.3389/fgene.2017.00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Jang M.A., et al. Mutations in DDX58, which encodes RIG-I, cause atypical Singleton-Merten syndrome. Am J Hum Genet. 2015;96(2):266–274. doi: 10.1016/j.ajhg.2014.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Rutsch F., et al. A specific IFIH1 gain-of-function mutation causes Singleton-Merten syndrome. Am J Hum Genet. 2015;96(2):275–282. doi: 10.1016/j.ajhg.2014.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Zhang L., et al. The RIG-I pathway is involved in peripheral T cell lymphopenia in patients with dermatomyositis. Arthritis Res Ther. 2019;21(1):131. doi: 10.1186/s13075-019-1905-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Suarez-Calvet X., et al. Altered RIG-I/DDX58-mediated innate immunity in dermatomyositis. J Pathol. 2014;233(3):258–268. doi: 10.1002/path.4346. [DOI] [PubMed] [Google Scholar]
- 202.Li L., et al. Role of Toll-like receptors and retinoic acid inducible gene I in endogenous production of type I interferon in dermatomyositis. J Neuroimmunol. 2015;285:161–168. doi: 10.1016/j.jneuroim.2015.06.009. [DOI] [PubMed] [Google Scholar]
- 203.Suarez-Calvet X., et al. RIG-I expression in perifascicular myofibers is a reliable biomarker of dermatomyositis. Arthritis Res Ther. 2017;19(1):174. doi: 10.1186/s13075-017-1383-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Maria N.I., et al. Contrasting expression pattern of RNA-sensing receptors TLR7, RIG-I and MDA5 in interferon-positive and interferon-negative patients with primary Sjogren's syndrome. Ann Rheum Dis. 2017;76(4):721–730. doi: 10.1136/annrheumdis-2016-209589. [DOI] [PubMed] [Google Scholar]
- 205.Sato S., et al. Autoantibodies to a 140-kd polypeptide, CADM-140, in Japanese patients with clinically amyopathic dermatomyositis. Arthritis Rheum. 2005;52(5):1571–1576. doi: 10.1002/art.21023. [DOI] [PubMed] [Google Scholar]
- 206.Nombel A., Fabien N., Coutant F. Dermatomyositis with Anti-MDA5 antibodies: bioclinical features, pathogenesis and emerging therapies. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.773352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Salajegheh M., et al. Interferon-stimulated gene 15 (ISG15) conjugates proteins in dermatomyositis muscle with perifascicular atrophy. Ann Neurol. 2010;67(1):53–63. doi: 10.1002/ana.21805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Allenbach Y., et al. Dermatomyositis with or without anti-melanoma differentiation-associated gene 5 antibodies: common interferon signature but distinct NOS2 EXPRESSION. Am J Pathol. 2016;186(3):691–700. doi: 10.1016/j.ajpath.2015.11.010. [DOI] [PubMed] [Google Scholar]
- 209.Jiang Y., et al. Mitochondrial morphology and MAVS-IFN1 signaling pathway in muscles of anti-MDA5 dermatomyositis. Ann Clin Transl Neurol. 2021;8(3):677–686. doi: 10.1002/acn3.51311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Peng Q.L., et al. Elevated serum levels of soluble CD163 in polymyositis and dermatomyositis: associated with macrophage infiltration in muscle tissue. J Rheumatol. 2015;42(6):979–987. doi: 10.3899/jrheum.141307. [DOI] [PubMed] [Google Scholar]
- 211.Enomoto Y., et al. Clinical significance of soluble CD163 in polymyositis-related or dermatomyositis-related interstitial lung disease. Arthritis Res Ther. 2017;19(1):9. doi: 10.1186/s13075-016-1214-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Horiike Y., et al. Successful classification of macrophage-mannose receptor CD206 in severity of anti-MDA5 antibody positive dermatomyositis associated ILD. Rheumatology (Oxford) 2019;58(12):2143–2152. doi: 10.1093/rheumatology/kez185. [DOI] [PubMed] [Google Scholar]
- 213.Nishioka A., et al. Serum neopterin as well as ferritin, soluble interleukin-2 receptor, KL-6 and anti-MDA5 antibody titer provide markers of the response to therapy in patients with interstitial lung disease complicating anti-MDA5 antibody-positive dermatomyositis. Mod Rheumatol. 2019;29(5):814–820. doi: 10.1080/14397595.2018.1548918. [DOI] [PubMed] [Google Scholar]
- 214.Fujimoto M., et al. Recent advances in dermatomyositis-specific autoantibodies. Curr Opin Rheumatol. 2016;28(6):636–644. doi: 10.1097/BOR.0000000000000329. [DOI] [PubMed] [Google Scholar]
- 215.Cao H., et al. Clinical manifestations of dermatomyositis and clinically amyopathic dermatomyositis patients with positive expression of anti-melanoma differentiation-associated gene 5 antibody. Arthritis Care Res (Hoboken) 2012;64(10):1602–1610. doi: 10.1002/acr.21728. [DOI] [PubMed] [Google Scholar]
- 216.Li J., et al. Associations between anti-melanoma differentiation-associated gene 5 antibody and demographics, clinical characteristics and laboratory results of patients with dermatomyositis: A systematic meta-analysis. J Dermatol. 2018;45(1):46–52. doi: 10.1111/1346-8138.14092. [DOI] [PubMed] [Google Scholar]
- 217.Yoshida N., et al. An autopsy case of anti-melanoma differentiation-associated gene-5 antibody-positive clinical amyopathic dermatomyositis complicated by rapidly progressive interstitial lung disease. Intern Med. 2016;55(12):1653–1659. doi: 10.2169/internalmedicine.55.6055. [DOI] [PubMed] [Google Scholar]
- 218.Watanabe T., et al. Fatal and extensive multiorgan hemorrhages in anti-melanoma differentiation-associated gene 5 antibody-positive dermatomyositis: An autopsy case report. Medicine (Baltimore) 2020;99(3) doi: 10.1097/MD.0000000000018600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Chen Z., et al. Utility of anti-melanoma differentiation-associated gene 5 antibody measurement in identifying patients with dermatomyositis and a high risk for developing rapidly progressive interstitial lung disease: a review of the literature and a meta-analysis. Arthritis Care Res (Hoboken) 2013;65(8):1316–1324. doi: 10.1002/acr.21985. [DOI] [PubMed] [Google Scholar]
- 220.Hall J.C., et al. Anti-melanoma differentiation-associated protein 5-associated dermatomyositis: expanding the clinical spectrum. Arthritis Care Res (Hoboken) 2013;65(8):1307–1315. doi: 10.1002/acr.21992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Abe Y., et al. Clinical characteristics and change in the antibody titres of patients with anti-MDA5 antibody-positive inflammatory myositis. Rheumatology (Oxford) 2017;56(9):1492–1497. doi: 10.1093/rheumatology/kex188. [DOI] [PubMed] [Google Scholar]
- 222.Elion D.L., et al. Therapeutically Active RIG-I Agonist Induces Immunogenic Tumor Cell Killing in Breast Cancers. Cancer Res. 2018;78(21):6183–6195. doi: 10.1158/0008-5472.CAN-18-0730. [DOI] [PubMed] [Google Scholar]
- 223.Flood B.A., et al. STING pathway agonism as a cancer therapeutic. Immunol Rev. 2019;290(1):24–38. doi: 10.1111/imr.12765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Boelens M.C., et al. Exosome transfer from stromal to breast cancer cells regulates therapy resistance pathways. Cell. 2014;159(3):499–513. doi: 10.1016/j.cell.2014.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Elion D.L., Cook R.S. Harnessing RIG-I and intrinsic immunity in the tumor microenvironment for therapeutic cancer treatment. Oncotarget. 2018;9(48):29007–29017. doi: 10.18632/oncotarget.25626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Kubler K., et al. Targeted activation of RNA helicase retinoic acid-inducible gene-I induces proimmunogenic apoptosis of human ovarian cancer cells. Cancer Res. 2010;70(13):5293–5304. doi: 10.1158/0008-5472.CAN-10-0825. [DOI] [PubMed] [Google Scholar]
- 227.Sprooten J., Agostinis P., Garg A.D. Type I interferons and dendritic cells in cancer immunotherapy. Int Rev Cell Mol Biol. 2019;348:217–262. doi: 10.1016/bs.ircmb.2019.06.001. [DOI] [PubMed] [Google Scholar]
- 228.Iurescia S., Fioretti D., Rinaldi M. The innate immune signalling pathways: turning RIG-I sensor activation against cancer. Cancers (Basel) 2020;12(11) doi: 10.3390/cancers12113158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Wu Y., et al. The anticancer functions of RIG-I-like receptors, RIG-I and MDA5, and their applications in cancer therapy. Transl Res. 2017;190:51–60. doi: 10.1016/j.trsl.2017.08.004. [DOI] [PubMed] [Google Scholar]
- 230.Jiang X., et al. Intratumoral delivery of RIG-I agonist SLR14 induces robust antitumor responses. J Exp Med. 2019;216(12):2854–2868. doi: 10.1084/jem.20190801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Sato Y., et al. Fas Ligand enhances apoptosis of human lung cancer cells cotreated with RIG-I-like receptor agonist and radiation. Curr Cancer Drug Targets. 2020;20(5):372–381. doi: 10.2174/1568009620666200115161717. [DOI] [PubMed] [Google Scholar]
- 232.Jiang X., et al. Intratumoral delivery of RIG-I agonist SLR14 induces robust antitumor responses. J Exp Med. 2019;216(12):2854–2868. doi: 10.1084/jem.20190801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Iurescia S., Fioretti D., Rinaldi M. The innate immune signalling pathways: turning RIG-I sensor activation against cancer. Cancers (Basel) 2020;12(11) doi: 10.3390/cancers12113158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Das M., et al. Nanoparticle delivery of RIG-I agonist enables effective and safe adjuvant therapy in pancreatic cancer. Mol Ther. 2019;27(3):507–517. doi: 10.1016/j.ymthe.2018.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Castiello L., et al. An optimized retinoic acid-inducible gene I agonist M8 induces immunogenic cell death markers in human cancer cells and dendritic cell activation. Cancer Immunol Immunother. 2019;68(9):1479–1492. doi: 10.1007/s00262-019-02380-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Chiang C., et al. Sequence-specific modifications enhance the broad-spectrum antiviral response activated by RIG-I Agonists. J Virol. 2015;89(15):8011–8025. doi: 10.1128/JVI.00845-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Elion D.L., et al. Therapeutically active RIG-I agonist induces immunogenic tumor cell killing in breast cancers. Cancer Res. 2018;78(21):6183–6195. doi: 10.1158/0008-5472.CAN-18-0730. [DOI] [PubMed] [Google Scholar]
- 238.Iurescia S., Fioretti D., Rinaldi M. The innate immune signalling pathways: turning RIG-I sensor activation against cancer. Cancers (Basel) 2020;12(11) doi: 10.3390/cancers12113158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Wolf D., et al. High RIG-I expression in ovarian cancer associates with an immune-escape signature and poor clinical outcome. Int J Cancer. 2020;146(7):2007–2018. doi: 10.1002/ijc.32818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Lazear H.M., Schoggins J.W., Diamond M.S. Shared and distinct functions of Type I and type III interferons. Immunity. 2019;50(4):907–923. doi: 10.1016/j.immuni.2019.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Hotz C., et al. Local delivery of mRNA-encoded cytokines promotes antitumor immunity and tumor eradication across multiple preclinical tumor models. Sci Transl Med. 2021;13(610) doi: 10.1126/scitranslmed.abc7804. (p. eabc7804) [DOI] [PubMed] [Google Scholar]
- 242.Lin L.L., et al. Innate immune sensor laboratory of genetics and physiology 2 suppresses tumor cell growth and functions as a prognostic marker in neuroblastoma. Cancer Sci. 2018;109(11):3494–3502. doi: 10.1111/cas.13790. [DOI] [PMC free article] [PubMed] [Google Scholar]