Abstract
Candida albicans is an opportunistic pathogenic yeast which frequently develops resistance to the antifungal agent fluconazole (FCZ) in patients undergoing long-term therapy. FCZ-resistant strains often display a reduced intracellular FCZ accumulation which correlates with the overexpression of the ATP-binding cassette transporters CDR1 and CDR2 or the major facilitator (MF) MDR1. We have recently cloned a C. albicans gene, named CAP1, which codes for a bZip transcription factor of the AP-1 family homologous to the Yap1 protein involved in multidrug resistance and response to oxidative stress in Saccharomyces cerevisiae. CAP1 was found to confer FCZ resistance in S. cerevisiae by transcriptionally activating FLR1, a gene coding for an MF homologous to the C. albicans MDR1 gene product (A.-M. Alarco, I. Balan, D. Talibi, N. Mainville, and M. Raymond, J. Biol. Chem. 272:19304–19313, 1997). To study the role of CAP1 in C. albicans, we constructed a CAI4-derived mutant strain carrying a homozygous deletion of the CAP1 gene (CJD21). We found that deletion of CAP1 did not affect the susceptibility of CJD21 cells to FCZ, cerulenin, brefeldin A, and diamide but caused hypersensitivity to cadmium, 4-nitroquinoline N-oxide, 1,10-phenanthroline, and hydrogen peroxide, an effect which was reverted by reintroduction of the CAP1 gene in these cells. Introduction of a hyperactive truncated allele of CAP1 (CAP1-TR) in CJD21 resulted in resistance of the cells to all of the above compounds except hydrogen peroxide. The hyperresistant phenotype displayed by the CJD21 CAP1-TR transformants was found to correlate with the overexpression of a number of potential CAP1 transcriptional targets such as MDR1, CaYCF1, CaGLR1, and CaTRR1. Taken together, our results demonstrate that CAP1 is involved in multidrug resistance and oxidative stress response in C. albicans. Finally, disruption of CAP1 in strain FR2, selected in vitro for FCZ resistance and constitutively overexpressing MDR1, did not suppress but rather increased the levels of MDR1 expression, demonstrating that CAP1 acts as a negative transcriptional regulator of the MDR1 gene in FR2 and is not responsible for MDR1 overexpression in this strain.
The opportunistic yeast Candida albicans is the leading etiologic agent of candidiasis, an infection affecting severely immunocompromised individuals. The last decade has seen an increase in the number of cases in which the immune system is chronically compromised, most notably with the onset of the AIDS pandemic, leading to a substantial increase in the incidence of Candida infections. Candidiasis is usually treated with azole antifungal agents such as ketoconazole, itraconazole, and fluconazole (FCZ), with FCZ currently the most widely used due to its high level of bioavailability and low toxicity (38). However, a growing number of studies report the occurrence of clinical failure during FCZ treatment, which correlates with increased levels of in vitro FCZ resistance in C. albicans clinical isolates (14, 30, 38).
In C. albicans and other Candida species, azole resistance has been associated with the alteration or overexpression of the azole cellular target 14α-lanosterol demethylase, encoded by the ERG11/CYP51A1 gene (41, 53, 58). In addition, a number of studies have also shown that many resistant strains display lower intracellular accumulation of FCZ involving the participation of energy-dependent transporter-mediated drug efflux mechanisms in these strains (2, 10, 22, 36, 44). Furthermore, in some C. albicans FCZ-resistant clinical and experimental isolates, azole resistance has been correlated with the overexpression of the CDR1 and CDR2 genes, encoding ATP-binding cassette (ABC) transporters, or overexpression of the MDR1 gene, encoding a major facilitator (MF) (43, 44, 58). However, the molecular mechanisms controlling the transcriptional activation of these transporter-encoding genes have not yet been elucidated.
Our studies of the molecular determinants of azole resistance led us to isolate a C. albicans gene which confers FCZ resistance when overexpressed in Saccharomyces cerevisiae (1). This gene encodes a protein highly homologous to the S. cerevisiae bZip transcription factor Yap1p and was named CAP1, for C. albicans AP-1. Yap1p has been associated with resistance to a variety of toxicants as well as with tolerance to oxidative stress induced by compounds such as H2O2 and diamide (21, 23, 26, 46, 49, 55, 56, 59). Yap1p is the prototype of a growing family of transcription factors which also includes S. cerevisiae Yap2p (7) as well as six other S. cerevisiae members identified through sequence similarity searches (15), in addition to C. albicans Cap1p (1), Schizosaccharomyces pombe Pap1p (51), Kluyveromyces lactis Klyap1p (5), and Aspergillus nidulans meaBp (37). Although related to the yeast transcription factor Gcn4 and to the mammalian AP-1 proteins Jun and Fos, Yap1p-like transcriptions factors are distinctive due to atypical residues present in their basic DNA binding domains (15). Interestingly, Yap1p, Yap2p, Cap1p, Pap1p, and Klyap1p constitute a subgroup within the YAP family of transcription factors since these proteins possess a highly conserved C-terminal region termed the cysteine-rich domain (CRD). This domain contains three invariably conserved cysteine residues and has been shown, both in Yap1p and in Pap1p, to regulate the nuclear localization of these proteins in response to oxidative stress (27, 52).
Yap1p mediates its pleiotropic phenotype through the transcriptional activation of a number of downstream target genes, including YCF1, which encodes an ABC transporter essential for cadmium resistance (56); ATR1, encoding an MF involved in 4-nitroquinoline N-oxide (4-NQO) and 3-amino-1,2,4-triazole resistance (11); TRX2, which encodes a thioredoxin conferring tolerance to oxidative stress (26); TRR1, encoding a thioredoxin reductase (32); GSH1, whose product is a γ-glutamylcysteine synthetase involved in glutathione biosynthesis (60); and GLR1, encoding a glutathione reductase (20). In addition, we have shown that YAP1 mediates FCZ resistance through the transcriptional activation of FLR1, which codes for a multidrug transporter of the MF superfamily highly homologous to the C. albicans MDR1 gene product (1). Cap1p and Yap1p are functional homologues, since CAP1 overexpression in S. cerevisiae also leads to FCZ resistance through transcriptional activation of FLR1 and expression of CAP1 in a yap1 mutant strain partially restores the ability of the cells to grow in the presence of toxic concentrations of cadmium or H2O2 (1). The high level of structural and functional similarities between Cap1p and Yap1p suggested that Cap1p could be involved in multidrug resistance (MDR) and oxidative stress response (OSR) in C. albicans. Furthermore, the CAP1/YAP1-dependent FLR1-mediated FCZ resistance phenotype in S. cerevisiae suggested that Cap1p could also mediate FCZ resistance in C. albicans through the transcriptional activation of MDR1. This paper reports the results of our investigations of Cap1p function in C. albicans.
MATERIALS AND METHODS
Yeast strains and culture conditions.
Yeast strains used in this study are listed in Table 1. The cells were routinely grown in YPD or uracil-deficient SD -Ura medium at 30°C (47).
TABLE 1.
C. albicans strains used in this study
| Strain | Parental strain | Relevant genotype | Reference |
|---|---|---|---|
| CAI4 | SC5314 | CAP1/CAP1 | 17 |
| CJD10 | CAI4 | CAP1/cap1Δ::hisG-URA3-hisG | This study |
| CJD11 | CJD10 | CAP1/cap1Δ::hisG | This study |
| CJD20 | CJD11 | cap1Δ::hisG-URA3-hisG/cap1Δ::hisG | This study |
| CJD21 | CJD20 | cap1Δ::hisG/cap1Δ::hisG | This study |
| SGY243 | A-81 | CAP1/CAP1 | 24 |
| FR2 | SGY243 | CAP1/CAP1 | 2 |
| FJD10 | FR2 | CAP1/cap1Δ::hisG-URA3-hisG | This study |
| FJD11 | FJD10 | CAP1/cap1Δ::hisG | This study |
| FJD20 | FJD11 | cap1Δ::hisG-URA3-hisG/cap1Δ::hisG | This study |
| FJD21 | FJD20 | cap1Δ::hisG/cap1Δ::hisG | This study |
Deletion of CAP1.
Plasmid pBC-CAP1 was generated by inserting a 3.2-kb KpnI-HindIII CAP1 fragment excised from plasmid YEp352-CAP1 (1) into plasmid pBC (Stratagene, PDI Bioscience, Aurora, Ontario, Canada) cut with KpnI and HindIII. The CAP1 open reading frame (ORF) in pBC-CAP1 was removed by replacing a 1.35-kb ClaI-BsaBI CAP1 fragment with a 4-kb BamHI-BglII hisG-URA3-hisG cassette excised from plasmid pMB7 (17), generating plasmid pBC/cap1Δ::hisG-URA3-hisG. A linear 5.8-kb cap1Δ::hisG-URA3-hisG fragment was released from pBC/cap1Δ::hisG-URA3-hisG by digestion with SalI and used to transform C. albicans CAI4 and FR2 to Ura+ prototrophy. Transformations were performed by the lithium acetate method (18). Counterselection of the URA3 gene was carried out on plates containing 5-fluoroorotic acid (5-FOA; 1 mg/ml; Toronto Research Chemicals Inc., North York, Ontario, Canada) as described by Boeke (6) except that uracil was replaced with uridine (25 μg/ml).
Genomic DNA isolation and Southern blot analyses.
C. albicans genomic DNA was prepared essentially as described for S. cerevisiae (39) except that Zymolyase-100T (ICN Biomedicals, Costa Mesa, Calif.) was added to a final concentration of 0.4 mg/ml. Genomic DNAs (2 μg) were digested to completion with BglII, electrophoresed in triplicate on 1% agarose gels, and transferred to a nylon membrane (Hybond-N; Amersham Corp., Arlington Heights, Ill.). Prehybridization was performed for 2 h at 65°C in 6× SSC (0.9 M NaCl, 0.9 M sodium citrate)–5× Denhardt’s solution (0.1% bovine serum albumin, 0.1% Ficoll, 1% polyvinylpyrrolidone)–0.5% sodium dodecyl sulfate (SDS) in the presence of 20 μg of denatured salmon sperm DNA per ml of prehybridization solution. Hybridization was carried out for 16 h at 65°C with 106 cpm of radiolabeled probe per ml of hybridization solution, using a 3.2-kb KpnI/HindIII fragment containing the entire CAP1 gene, a 341-bp AccI DNA fragment (positions +276 to +617 relative to the start codon) internal to the CAP1 ORF (1), or a 900-bp BamHI/BglII hisG fragment isolated from plasmid pMB7 (17). The blots were washed at high stringency (0.1× SSC–0.1% SDS at 65°C) and exposed at −80°C with two intensifying screens.
Plasmid construction.
Plasmids YEp352-CAP1 and YEp352-CAP1TR (1) were digested with PstI and SmaI to generate a 3.2-kb CAP1 and a 3-kb CAP1-TR (truncated form) fragment, respectively. Both DNA fragments were blunt ended and cloned into plasmid pMK22 (29) (obtained from R. Rachubinski, University of Alberta) linearized with ScaI, generating plasmids PMK-CAP1 and PMK-CAP1TR, respectively. Plasmids PMK22, PMK-CAP1, and PMK-CAP1TR were transformed in CAI4 and CJD21 cells by the lithium acetate method (18). Single Ura+ colonies were picked and restreaked on SD −Ura medium. Individual clones for CJD21/PMK-CAP1 and CJD21/PMK-CAP1TR transformants were selected based on wild-type levels of CAP1 expression, as determined by Northern blot analysis (data not shown).
Drug resistance assays.
Stock solutions of FCZ (10 mg/ml) and cadmium sulfate (100 mM) were prepared in water. Stock solutions of 4-NQO (5 mM), and diamide (1 M) were prepared in dimethyl sulfoxide. Stock solutions of cerulenin (1 mg/ml), brefeldin A (1 mg/ml), and 1,10-phenanthroline (10 mg/ml) were prepared in ethanol. Dilutions of 30% H2O2 were prepared in water. FCZ was obtained from Pfizer Canada Inc. (Arnprior, Ontario, Canada), cadmium sulfate was obtained from MAT Laboratory (Beauport, Québec, Canada), and the other compounds were obtained from Sigma (Mississauga, Ontario, Canada). Drug resistance was determined by spot assay. PMK22 transformants (CAI4/PMK, CJD21/PMK, CJD21/PMK-CAP1, and CJD21/PMK-CAP1TR) were grown overnight on SD −Ura plates. Cells were then suspended in YPD to an A600 of 0.1. Three microliters of fivefold serial dilutions of each yeast culture were spotted onto YPD plates supplemented or not with a specific antifungal compound. Qualitative growth differences among the yeast transformants were recorded following incubation of the plates at 30°C for 2 days. Growth was not affected by the presence of ethanol (1%) or dimethyl sulfoxide (0.15%).
RNA preparation and Northern blotting.
Total RNA from cells grown in YPD or in SD −Ura medium (PMK22 transformants) was prepared by using the glass beads extraction protocol (9). RNA samples (20 μg) were electrophoresed on a 7.5% formaldehyde–1% agarose gel and transferred by capillarity to a Zeta-Probe nylon membrane (Bio-Rad Laboratories, Mississauga, Ontario, Canada). Detection of specific RNAs was performed by hybridization with 32P-labeled DNA probes as previously described (1). The CAP1 probe was a 614-bp XbaI-HincII DNA fragment (positions +47 to +661 relative to the translation initiation codon [1]). An MDR1 DNA fragment was generated by PCR with primers 5′-ACTCTTGCTGATGATACA and 5′-TATATGGATGTACGACCA and overlaps region +135 to +559 of the MDR1 gene (positions are relative to the translation initiation codon [16]). This PCR fragment was subsequently gel purified and cloned blunt into the pCRII vector (TA cloning kit; Invitrogen). The MDR1 DNA fragment used to generate this probe was then obtained following EcoRI digestion of the PCR-MDR1 plasmid. The C. albicans CaYCF1, CaGLR1, and CaTRR1 probes were obtained by PCR using oligonucleotides derived from sequences available from the C. albicans information pages on the Alces server (http://alces.med.umn.edu/Candida.html). A 220-bp CaYCF1 fragment was amplified with primers 5′-TATCAATATTGATGGTATAG and 5′-CCTCGGTAGTCTCCCTC; a 256-bp CaGLR1 fragment was generated with primers 5′-TCCATCAGTGATTTTCTC and 5′-GCAACACCAAAACCTTG; and a 286-bp CaTRR1 fragment was obtained with primers 5′-TCTGAAGTATCATGGAAC and 5′-CAGCAGAAGTGGTGGCTT. All DNA fragments were gel purified before random labeling with [α-32P]ATP (40). The membranes were also hybridized with an ACT1 probe (kindly provided by B. Magee, University of Minnesota), confirming that the RNA samples had been equally loaded and transferred to the membranes.
Preparation of antisera.
A glutathione S-transferase (GST)-CAP1 in-frame gene fusion was constructed by inserting a 341-bp AccI CAP1 fragment blunt ended with T4 DNA polymerase and inserted into the SmaI site of vector pGEX-4T-1 (Pharmacia), generating plasmid pGEX-CAP350. The resulting fusion protein contains 114 amino acids of the Cap1p protein (amino acid positions 93 to 207 [1]). Escherichia coli DH5α cells transformed with pGEX-CAP350 were treated with isopropyl-β-d-thiogalactoside (0.1 mM) for 4 h at 30°C to induce the expression of the fusion protein. The GST-Cap1p fusion protein was soluble and was thus purified from a crude bacterial lysate by affinity chromatography on immobilized glutathione (48). The purified fusion protein was used to raise polyclonal antibodies in two New Zealand White rabbits, yielding two anti-Cap1p-350 antisera of high titers (C1 and C2). The C1 antiserum was used without any further purification for the detection of Cap1p in our experiments.
Protein preparation and Western blot analysis.
Total protein extracts were prepared from strains CAI4, CJD21, CJD21/PMK, CJD21/PMK-CAP1, and CJD21/PMK-CAP1TR as described previously (3). Protein concentration was determined by the Bradford method (8), using bovine serum albumin as the standard. Total protein extracts (50 μg) were suspended in Laemmli sample buffer, boiled for 10 min, and separated by electrophoresis on an SDS–12% polyacrylamide gel. Proteins were transferred to a nitrocellulose membrane and analyzed with the anti-Cap1p-350 polyclonal antibody at a dilution of 1:5,000, using a chemiluminescence detection system under conditions recommended by the manufacturer (Boehringer Mannheim).
RESULTS
Chromosomal deletion of the CAP1 gene in C. albicans CAI4.
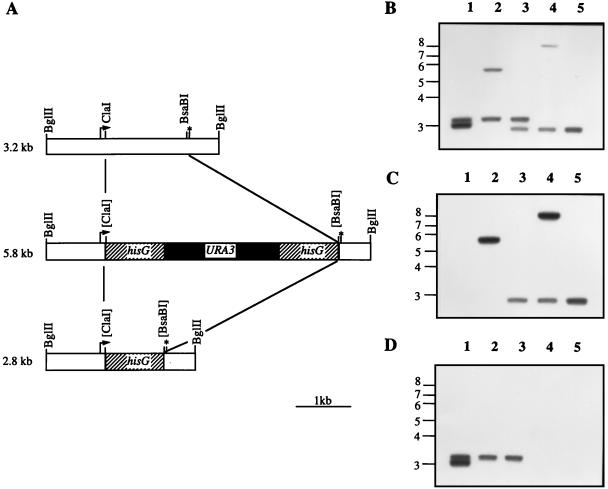
To investigate the biological function of Cap1p in C. albicans, both copies of the CAP1 gene were deleted in the FCZ-sensitive C. albicans strain CAI4, using the Ura-blaster strategy (17). A linear 5.8-kb cap1Δ::hisG-URA3-hisG deletion fragment was constructed as described in Materials and Methods (Fig. 1A) and used to transform CAI4. In CAI4, the CAP1 locus is polymorphic: the two CAP1 alleles are located on a 3.0- and a 3.2-kb BglII genomic fragment (Fig. 1B, lane 1). Deletion of the first CAP1 allele occurred in strain CJD10 at the 3.0-kb BglII CAP1 locus, as seen from the disappearance of the 3.0-kb BglII fragment and the appearance of a new 5.8-kb BglII fragment hybridizing with the 3.2-kb CAP1 probe (Fig. 1B, lane 2). After counterselection on 5-FOA, a 2.8-kb BglII fragment resulting from the recombination between the two hisG direct repeats was detected in strain CJD11 (Fig. 1B, lane 3). This strain was used for a second round of transformation with the cap1Δ fragment. Deletion of the remaining 3.2-kb BglII CAP1 allele was confirmed in strain CJD20 by the appearance of a new fragment of about 9 kb which hybridizes with the CAP1 probe (the size of this fragment suggests that two cap1Δ fragments were inserted during the recombination process) (Fig. 1B, lane 4). Proper loop-out of the URA3 marker in strain CJD21 was confirmed by the appearance of a second 2.8-kb fragment hybridizing with the CAP1 probe (Fig. 1B, lane 5). Southern blotting analysis was also performed with a hisG and a CAP1 internal probe, confirming the correct genotypes of the different strains (Fig. 1C and D). CJD21 cells are Ura− and thus suitable for transformation with a URA3-based vector for the expression of CAP1 in a null mutant background.
FIG. 1.
Chromosomal deletion of CAP1 in CAI4. (A) Schematic representation of the disruption strategy. The CAP1 locus is contained within a 3.2-kb BglII fragment (top). The start (arrow) and stop (asterisk) codons of the CAP1 ORF are indicated. The disruption cassette (middle) was generated by replacing a 1.35-kb ClaI-BsaBI CAP1 fragment by the 4-kb hisG-URA3-hisG cassette. After counterselection on 5-FOA, recombination between the two hisG direct repeats should generate a 2.8-kb BglII fragment (bottom). Southern blot analysis was used to characterize the different steps of the disruption (B to D). Genomic DNA was extracted from strains CAI4 CAP1/CAP1 (lanes 1), CJD10 CAP1/cap1Δ::hisG-URA3-hisG (lanes 2), CJD11 CAP1/cap1Δ::hisG (lanes 3), CJD20 cap1Δ::hisG-URA3-hisG/cap1Δ::hisG (lanes 4), and CJD21 cap1Δ::hisG/cap1Δ::hisG (lanes 5). DNA samples (2 μg) were digested in triplicate with BglII, separated by electrophoresis on agarose gels, and transferred to nylon membranes. The blots were then probed with either the 3.2-kb BglII fragment comprising the entire wild-type CAP1 gene (B), a 0.9-kb BamHI-BglII hisG fragment (C), or a 0.6-kb XbaI-HincII CAP1 internal fragment deleted in the cap1Δ::hisG-URA3-hisG allele (D). Positions of molecular size markers (in kilobases) are indicated on the left. Membranes were exposed for 6 h at −80°C with two intensifying screens.
Involvement of CAP1 in C. albicans MDR and OSR.
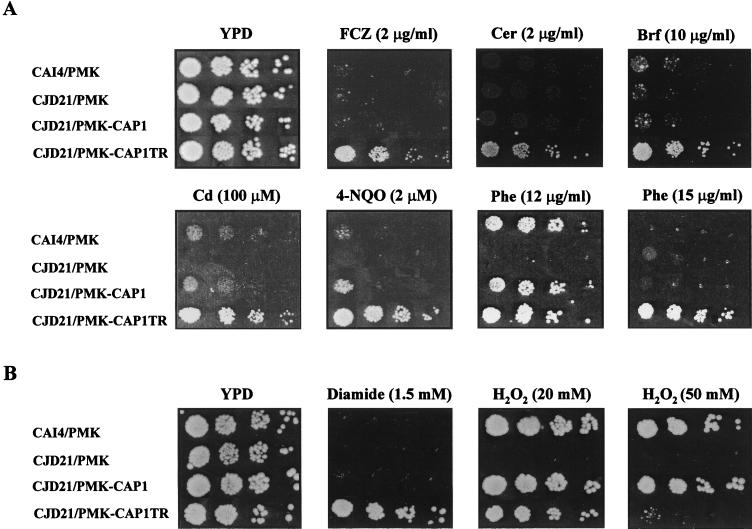
Our previous findings that CAP1 mediates FCZ resistance when overexpressed in S. cerevisiae led us to investigate the role of CAP1 in C. albicans FCZ resistance. First, we compared the levels of FCZ susceptibility of the CAI4 and CJD21 strains, using a spot assay. This experiment showed that deleting CAP1 does not change the ability of the cells to grow on FCZ, as both strains displayed an FCZ MIC of 1 μg/ml in this assay (data not shown). We then analyzed the FCZ resistance profile of CJD21 cells transformed with plasmid pMK22 carrying either the full-length CAP1 gene or a hyperactive allele of CAP1 (CAP1-TR). This allele, identified previously during our isolation of the CAP1 gene (1), codes for a truncated Cap1p protein which lacks 165 amino acids at its C terminus but is otherwise identical to the full-length protein and which is hyperactive in conferring FCZ resistance in S. cerevisiae (Fig. 2A) (1). Total protein extracts from CJD21 cells transformed with plasmids PMK22, PMK-CAP1, and PMK-CAP1TR were analyzed by Western blotting using an anti-Cap1p polyclonal antibody (Fig. 2B). This antibody was raised against a region of the Cap1p protein (Cap1-350) located downstream of the bZip domain and thus recognizes both the full-length and the truncated forms of Cap1 (Fig. 2A). This experiment showed that the anti-Cap1p antibody detects a 65-kDa protein in CAI4 which is absent in the CJD21 cells, thus demonstrating that this protein is Cap1p and confirming the complete deletion of the CAP1 gene in strain CJD21. We noted a difference between the apparent molecular mass of 65 kDa for the Cap1p protein deduced from its mobility relative to the migration of the molecular mass markers and its predicted molecular weight of 55,000 based on the deduced amino acid sequence of Cap1p (1). The reasons for this difference are not known, but a similar discrepancy has been reported for the Yap1p protein (33, 50). The CJD21/PMK22-CAP1 transformants produce a 65-kDa protein similar in size and abundance to that detected in CAI4, thus confirming the proper expression of Cap1p from the plasmid-borne copy of the CAP1 gene (Fig. 2B; compare CJD21/PMK-CAP1 with CAI4). In the CJD21/PMK-CAP1TR transformants, the anti-Cap1p antibody detects a protein with an apparent molecular mass of 45 kDa, which is in close agreement with the calculated molecular weight of 38,000 for the truncated protein encoded by the CAP1-TR allele (1). The truncated Cap1p appears to be less abundant than the full-length protein, most likely due to a decreased stability as previously reported for C-terminal deletion versions of Yap1p (57). Analysis of the FCZ resistance profile of the CJD21 transformants confirmed that the deletion of CAP1 does not affect the level of FCZ susceptibility of the CJD21 cells and that reintroduction of CAP1 in CJD21 has, as anticipated, no effect on their FCZ resistance profile when analyzed on various concentrations of FCZ (Fig. 3A). However, expression of the truncated Cap1p did confer FCZ resistance, as judged from the ability of the CJD21/PMK-CAP1TR cells to grow on FCZ at concentrations of up to 2 μg/ml, compared to cells expressing the full-length Cap1p protein, whose growth is completely abrogated at this FCZ concentration (Fig. 3A). These results are consistent with our previous finding that the truncated Cap1p protein elicits high levels of FCZ resistance in S. cerevisiae (1) and also establish the involvement of CAP1 in C. albicans FCZ resistance.
FIG. 2.
Expression of the Cap1p and Cap1p-TR proteins in C. albicans. (A) Schematic representation of the full-length (Cap1p) and the truncated (Cap1p-TR) proteins. Positions of the bZip domain (grey) and the CRD (stippled) are indicated. The region used to generate a GST-Cap1p fusion protein (Cap1-350) is indicated. This fusion protein was used to raise an anti-Cap1p-350 polyclonal antibody. (B) Western blot analysis of Cap1p and Cap1p-TR expression. Total proteins were extracted from CAI4, CJD21, CJD21/PMK, CJD21/PMK-CAP1, and CJD21/PMK-CAP1TR cells. Protein samples (50 μg) were separated by electrophoreses on an SDS–12% polyacrylamide gel, transferred to a nitrocellulose membrane, and analyzed with the anti-Cap1p-350 polyclonal antibody. Arrows on the right indicate positions of the full-length (Cap1p) and truncated (Cap1p-TR) proteins. The position of a nonspecific cross-reacting protein is also indicated (★). Positions of molecular size standards are shown on the left.
FIG. 3.
Involvement of CAP1 in C. albicans MDR and OSR. Strains CAI4/PMK, CJD21/PMK, CJD21/PMK-CAP1, and CJD21/PMK-CAP1TR were analyzed by spot assay (as described in Materials and Methods) for the ability to grow on YPD plates containing different antifungal (A) and prooxidant (B) agents. Growth differences were monitored after 2 days at 30°C. A representative plate of control growth on YPD medium is shown for each set of experiments (YPD). Abbreviations for substances are FCZ (fluconazole), Cer (cerulenin), Brf (brefeldin A), Cd (cadmium), 4-NQO (4-nitroquinoline N-oxide), and Phe (1,10-phenanthroline).
We have shown that in S. cerevisiae, overexpression of CAP1 mediates resistance not only to FCZ but also to cycloheximide and 4-NQO (1). These results, together with the fact that S. cerevisiae YAP1 mediates resistance to a wide variety of antifungal agents (21, 23, 45, 56) and prooxidants (26, 46), prompted us to characterize the levels of susceptibility of our different transformants to a series of compounds displaying antifungal activity. As seen for FCZ, deletion of the CAP1 gene did not affect the ability of the CJD21/PMK transformants to grow on cerulenin and brefeldin A when their growth was compared to that of the CAI4/PMK transformants, both transformants displaying similar cerulenin and brefeldin A MICs (1 and 6 μg/ml, respectively [data not shown]). Reintroduction of the full-length CAP1 gene had no effect on cell growth in the presence of these drugs, whereas expression of the truncated Cap1p protein in strain CJD21 resulted in increased resistance of the cells to these two compounds (Fig. 3A). On the other hand, we found that the deletion of CAP1 led to hypersensitivity of the cells to cadmium, 4-NQO, and 1,10-phenanthroline (Fig. 3A; compare CAI4/PMK and CJD21/PMK) which was compensated for by the reintroduction of CAP1 in strain CJD21. Finally, expression of the truncated form of Cap1p in CJD21 led to a hyperresistance of the cells to these three compounds (Fig. 3A). Taken together, these results indicate that CAP1 is a determinant of MDR in C. albicans.
YAP1 has been shown to be involved in OSR in S. cerevisiae by regulating the expression of a number of effector genes (20, 26, 32, 60). In addition, we have shown that CAP1 complements a yap1 deletion mutant for its ability to grow on H2O2 (1). Therefore, we were interested in determining if CAP1 also participates in OSR in C. albicans. To this end, CAI4 and CJD21 transformants were grown on plates containing different concentrations of either diamide, a thiol oxidant (25), or H2O2, an oxidizing agent whose effects include lipid peroxidation, protein oxidation, and DNA damage (12). We found that deletion of CAP1 did not affect the susceptibility of the cells to diamide, since both CAI4/PMK and CJD21/PMK cells display a diamide MIC of 1 mM (data not shown). However, the truncated Cap1p was found to confer resistance to this compound, as CJD21/PMK-CAP1TR transformants were able to grow at diamide concentrations inhibitory for the growth of the CAI4/PMK and CJD21/PMK-CAP1 transformants (Fig. 3B). On the other hand, the deletion of CAP1 resulted in hypersensitivity of the cells to H2O2, as the growth of CJD21/PMK cells was completely abrogated on 20 mM H2O2, whereas growth of CAI4/PMK cells was unaffected at this H2O2 concentration (Fig. 3B). These results indicate that CAP1 is essential for H2O2 tolerance in C. albicans. Reintroduction in the CJD21 cells of either the full-length or the truncated CAP1 gene restored normal levels of tolerance to H2O2 at a concentration of 20 mM (Fig. 3B). However, only the full-length CAP1 gene was able to restore wild-type growth in CJD21 at 50 mM H2O2. Taken together, these results clearly establish that CAP1, in addition to MDR, is involved in OSR in C. albicans. Furthermore, the different behavior of the CJD21/PMK-CAP1TR transformants when exposed to diamide or H2O2 suggests that complex regulatory mechanisms control the activity of Cap1p in response to these two oxidants.
Identification of potential CAP1 transcriptional targets.
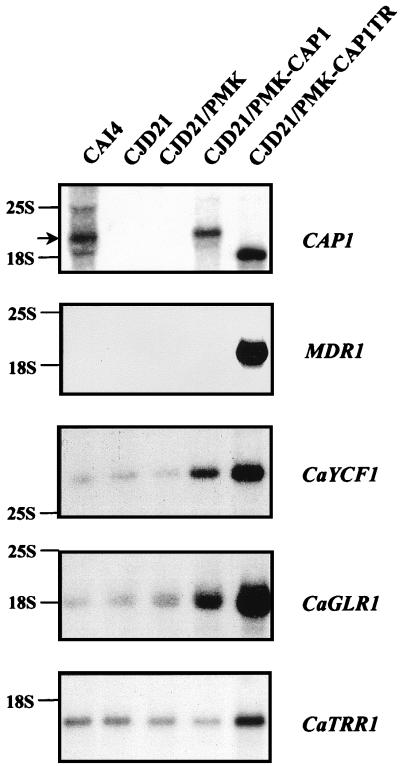
Yap1p has been shown to transcriptionally activate a number of downstream target genes, including FLR1, YCF1, ATR1, TRX2, TRR1, and GLR1 (1, 11, 20, 26, 32, 56). We were thus interested in investigating if Cap1p regulates similar targets in C. albicans and whether the CAP1-mediated phenotypes observed in C. albidans correlate with the overexpression of such targets. To this end, total RNAs prepared from CAI4 and CJD21 cells as well as from the CJD21/PMK, CJD21/PMK-CAP1, and CJD21/PMK-CAP1TR transformants were analyzed by Northern blotting, a technique which assess steady-state levels of RNA expression, using probes for the MDR1, CaYCF1, CaGLR1, and CaTRR1 genes. First, hybridization with a CAP1 probe identified a major 2.2-kb transcript in the wild-type CAI4 strain (Fig. 4, top panel). This transcript is absent in the CJD21 and CJD21/PMK cells (Fig. 4, top panel), as expected from the complete deletion of CAP1 in this strain. Restoration of CAP1 expression in the CJD21/PMK-CAP1 cells was confirmed by the detection of the CAP1-specific transcript (Fig. 4, top panel). The introduction of the truncated CAP1 gene in CJD21 cells led to the appearance in the CJD21/PMK-CAP1TR cells of a shorter CAP1 transcript of approximately 1.7 kb which codes for the truncated Cap1p protein observed in Fig. 2B (Fig. 4, top panel, lane CJD21/PMK-CAP1TR). The high level of CAP1-TR mRNA detected in the CJD21/PMK-CAP1TR cells by Northern blotting indicates that the lower amount of truncated Cap1p as compared to full-length Cap1p observed in the Western blot (Fig. 2B) is not due to a lower level of expression of the truncated CAP1 allele mRNA. This experiment showed that deletion of the CAP1 gene has no effect on the basal transcription of any of the genes analyzed (Fig. 4; compare lanes CAI4 and CJD21). Analysis of the expression of the MDR1 gene showed that the MDR1 transcript is undetectable in all strains but is massively overexpressed in the CJD21/PMK-CAP1TR transformants (Fig. 4, MDR1 panel). This result suggests that the FCZ-resistant phenotype of the CJD21/PMK-CAP1TR transformants may be due to the overexpression of the MDR1 gene and demonstrates that CAP1 modulates MDR1 expression. Since MDR1 expression in S. cerevisiae has also been shown to confer resistance to cerulenin, brefeldin A, 4-NQO, and 1,10-phenanthroline (43), it is likely that MDR1 overexpression in CJD21/PMK-CAP1TR transformants is also responsible for the resistance of the cells to these compounds. As seen for MDR1, expression of the truncated Cap1p protein resulted in strong overexpression of the CaYCF1, CaGLR1, and CaTRR1 genes. Given that the sequence homology between the C. albicans genes CaYCF1, CaGLR1, and CaTRR1 genes and their S. cerevisiae counterparts translates into functional homology, increased expression of these genes upon expression of the Cap1p-TR protein could explain the cadmium and diamide resistance phenotypes of the CJD21/PMK-CAP1TR cells (Fig. 3). We also found that reintroduction of the full-length CAP1 gene in CJD21 resulted in a small increase in CaYCF1 and CaGLR1 expression detected in the CJD21/PMK-CAP1 cells (Fig. 4, CaYCF1 and CaGLR1 panels). However, this increase did not translate into increased resistance to cadmium or diamide, respectively (Fig. 3). Finally, hybridization with a CDR1 probe showed that CDR1 expression is not modulated by Cap1p in these different strains (data not shown). Taken together, our results suggest that CAP1 acts as a molecular determinant of MDR and OSR in C. albicans through the transcriptional control of a number of specific downstream target genes.
FIG. 4.
Northern blot analysis of CAP1 transcriptional targets in C. albicans. Total RNA was extracted from strains CAI4, CJD21, CJD21/PMK, CJD21/PMK-CAP1, and CJD21/PMK-CAP1TR. RNA samples (20 μg) were separated by electrophoresis on a 1% agarose gel, transferred onto nylon membranes, and probed with a CAP1, MDR1, CaYCF1, CaGLR1, or CaTRR1 probe. Positions of rRNAs are indicated on the left; the position of the major CAP1 transcript is indicated by an arrow (top panel). The membranes were exposed for either 24 h (CAP1), 3 days (CaYCF1, CaGLR1, and CaTRR1), or 12 days (MDR1) at −80°C with two intensifying screens.
Chromosomal deletion of the CAP1 gene in the C. albicans FCZ-resistant FR2 strain.
A number of C. albicans FCZ-resistant clinical isolates have been shown to overexpress the MDR1 gene (44, 58). In addition, the FCZ-resistant strain FR2, which was derived in vitro from the FCZ-susceptible strain SGY243 (24) through serial passages in medium containing increasing concentrations of FCZ, also overexpresses the MDR1 gene (2). FCZ resistance in these strains is a stable phenotype (2, 44), suggesting that it could be caused by genetic alterations including dominant activating mutations in a trans-acting factor controlling MDR1 expression. Such dominant hyperactive mutations have been identified in the transcription factors Pdr1p and Pdr3p and have been shown to elicit MDR in S. cerevisiae by causing the overexpression of a number of drug efflux transporters of the ABC family (reviewed in reference 4). It was thus of interest to evaluate the potential contribution of CAP1 in the constitutive MDR1 overexpression displayed by these FCZ-resistant isolates. Unlike clinical isolates, FR2 is auxotrophic for uracil and thus amenable to gene disruption with the Ura-blaster strategy (17). We thus deleted both alleles of the CAP1 gene in strain FR2 and determined the consequence of this deletion on the levels of MDR1 expression. Disruption of both CAP1 alleles was performed as described for CAI4 except that the CAP1 alleles are both located on a 3.2-kb BglII fragment in FR2 (data not shown). To confirm that the CAP1 gene was completely deleted, we performed a Northern blot analysis (Fig. 5A) using total RNA extracted from the FCZ-sensitive SGY243 strain, its FCZ-resistant derivative FR2, the disruption intermediate FJD11 (CAP1/cap1Δ::hisG), and the homozygous disruptant FJD21 (cap1Δ::hisG/cap1Δ::hisG). Using an internal CAP1 fragment as a probe, two CAP1 RNA transcripts were detected in strains SGY243 and FR2 (Fig. 5A), showing that there are no quantitative or qualitative differences in CAP1 RNA expression between the FCZ-sensitive strain SGY243 and the FCZ-resistant strain FR2. Disruption of one of the CAP1 alleles resulted in a weaker signal with the CAP1 probe in strain FJD11 (Fig. 5A). This signal was completely absent in the double disruptant FJD21, thus confirming the complete deletion of CAP1 in FR2 (Fig. 5A). Northern blot analysis of the same strains with the MDR1 probe confirmed that the FR2 strain overexpresses MDR1 compared to its parental strain SGY243 (Fig. 5B) (2). If CAP1 was involved in MDR1 overexpression in FR2, it would be expected that deletion of CAP1 would result in the downregulation of MDR1 expression in the resulting cap1Δ/cap1Δ mutant strain. However, we found that the disruption of CAP1 correlates with increased levels of MDR1 expression: the heterozygous FJD11 CAP1/cap1 cells have higher levels of MDR1 RNA than their parental FR2 strain, and MDR1 expression is further increased in the FJD21 cap1/cap1 homozygous strain (Fig. 5B). These results demonstrate that CAP1 acts as a negative transcriptional regulator of the MDR1 gene in FR2 and is not the determinant of the stable MDR1 overexpression in this strain.
FIG. 5.
Northern blot analysis of CAP1 and MDR1 in FR2 cap1 disruptants. Total RNA was extracted from strains SGY243, FR2, FJD11 (CAP1/cap1Δ::hisG), and FJD21 (cap1Δ::hisG/cap1Δ::hisG). RNA samples (10 μg) were separated by electrophoresis in duplicate on a 1% agarose gel and transferred onto nylon membranes. The membranes were probed with a CAP1 (A) or MDR1 (B) probe. Both blots were subsequently hybridized with an ACT1 probe as a control for RNA loading and transfer. Membranes were exposed for 10 days (MDR1 and CAP1) or 24 h (ACT1) at −80°C with two intensifying screens. Positions of rRNAs are indicated on the left.
DISCUSSION
To unravel the biological function of Cap1p in C. albicans, we deleted the CAP1 gene in strain CAI4 and analyzed the consequence of this deletion on (i) the level of expression of Cap1p transcriptional targets and (ii) the susceptibility of the cells to a variety of toxic compounds. On the one hand, we found that deletion of CAP1 has no effect on the basal transcription of the genes analyzed, although these genes were found to be Cap1p transcriptional targets, as shown by their upregulation in the CAP1-TR transformants (Fig. 4). This observation is consistent with what has been found in S. cerevisiae for Yap1p targets such as YCF1, GSH1, and TRX2, whose expression is not reduced in a yap1 deletion strain under noninducing conditions (50, 56). Rather, Yap1p has been shown to be required for stress-induced transcriptional activation of its targets (50), a situation mimicked by elevated gene dosage (56) or by expression of Yap1p derivatives carrying an altered CRD (see below). On the other hand, we found that deletion of CAP1 in CAI4 elicits hypersensitivity to 4-NQO, 1,10-phenanthroline, cadmium, and H2O2, which could be reverted by reintroduction of a plasmid-borne copy of the CAP1 gene in the cap1 deletion strain (Fig. 3). These results clearly establish that Cap1p is a key determinant of cellular tolerance to different drugs and prooxidants in C. albicans. The hypersensitivity of the CJD21/PMK cells to 4-NQO and 1,10-phenanthroline could result from a defective upregulation of MDR1 in the cap1 mutant since overexpression of MDR1 in S. cerevisiae has been shown to mediate resistance to 4-NQO and 1,10-phenanthroline (43) and since deletion of the MDR1 gene in C. albicans results in hypersensitivity of the cells to 4-NQO (19, 42). Similarly, the hypersensitivity of CJD21/PMK cells to cadmium (Fig. 3A) could reflect the absence of CaYCF1 induction upon cap1 deletion. Alternatively, other CAP1 transcriptional targets may be responsible for the cadmium hypersensitivity of CJD21/PMK cells, such as the recently characterized CIP2 gene (35). Expression of this gene is massively induced upon exposure of cells to cadmium or diamide. Moreover, CIP2 possesses a putative Yap1 response element in its promoter region (35), suggesting that cadmium and diamide induction of the CIP2 gene expression could be CAP1 dependent. Finally, the H2O2-hypersensitive phenotype of CJD21/PMK cells (Fig. 3B) could be due to a reduced induction of CaGLR1 and/or CaTRX2 transcripts in the absence of Cap1p, since induction of these genes by H2O2 in S. cerevisiae has been shown to be dramatically reduced in a yap1 mutant (20, 50); alternatively, it could be due to the involvement of other, yet unidentified CAP1 target genes involved in the OSR.
The essential role of CAP1 in cellular tolerance to the reactive oxygen species (ROS) H2O2 is of particular interest since it suggests that Cap1p may be involved in C. albicans response to the ROS produced by the host immune system. The production of ROS such as H2O2, hydroxyl radicals, and superoxide anions by human phagocytic neutrophils is a major line of defense against fungal infections (34) since it has been shown to correlate directly with fungicidal activity (13), while antioxidants impair killing of C. albicans (54). These findings emphasize the need for C. albicans to adapt to the presence of ROS in order to counteract the host immune response. CAP1 could thus be part of C. albicans defense mechanisms against ROS produced by the host immune system, through the transcriptional regulation of target genes coding for proteins with antioxidant activities. Experiments are currently under way to address the role of CAP1 in tolerance to ROS and virulence of C. albicans.
We have previously shown that expression of the truncated CAP1 gene in S. cerevisiae confers high levels of drug resistance compared to its full-length counterpart (1), thus behaving as a gain-of-function allele. A similar behavior was observed in C. albicans, since CJD21 cells expressing CAP1-TR are resistant to FCZ, cerulenin, brefeldin A, cadmium, 1,10-phenanthroline, and diamide compared to cells expressing the full-length CAP1 (Fig. 3). CJD21 cells expressing CAP1-TR do not display increased resistance to drugs such as ketoconazole and fluphenazine, two substrates of the Cdr1p transporter, demonstrating that the resistance phenotype conferred by the truncated CAP1 allele is specific for certain drugs and, by extension, for CAP1 transcriptional targets (data not shown). Expression of the truncated CAP1 allele in CAI4 was also found to cause drug resistance, demonstrating that this hyperactive allele of CAP1 is dominant (data not shown). The hyperactivity of the CAP1 truncated allele most likely reflects the absence of the highly conserved CRD, the effect of such a truncation being well characterized in the case of Yap1p and Pap1p (7, 51, 57). Upon imposition of an oxidative stress, Yap1p and Pap1p relocalize from the cytoplasm to the nucleus by a regulated protein export mechanism which involves the Crm1p/Xpo1p nuclear export factor (27, 28, 52). It has been shown that Crm1p interacts with the CRD and that this interaction is redox regulated via the conserved cysteine residues present in the CRD (28, 52). Thus, Yap1p and Pap1p are exported out of the nucleus in the absence of stress but remain in the nucleus upon imposition of an oxidative stress, due to the disruption of the Crm1p-CRD interactions. Interestingly, expression of the truncated Cap1p leads to resistance to diamide but not to H2O2 (Fig. 3B). This difference in behavior toward these two oxidative stress-inducing agents is reminiscent of what has been reported for Yap1p:YAP1 mutants carrying mutations in the CRD are able to grow in the presence of diamide but not of H2O2, probably due to their inability to activate transcription of the TRX2 gene (27, 55). It thus seems that the region deleted in the Cap1-TR protein, although not essential for the ability of Cap1p to activate its transcriptional targets involved in resistance to FCZ, cerulenin, brefeldin A, cadmium, 1,10-phenanthroline, and diamide, is essential for Cap1p to activate its target(s) conferring resistance to H2O2. We found that resistance of the CJD21/pMK-CAP1TR transformants was accompanied by the overexpression of MDR1, CaYCF1, CaGLR1, and CaTRR1 (Fig. 4), demonstrating that these genes are transcriptional targets of CAP1 and suggesting that they may be involved in the resistant phenotype conferred by the expression of Cap1p-TR in CJD21.
Our previous data suggested that the overexpression of MDR1 in FR2 and in a number of FCZ-resistant clinical isolates could be due to a gain-of-function mutation in a transcriptional regulator of MDR1 such as CAP1. However, we observed that the deletion of CAP1 in the FR2 strain did not abolish MDR1 expression (Fig. 5), showing that Cap1p is not responsible for the constitutive overexpression of MDR1 in FR2. This finding suggests that FR2 probably contains a mutation in another MDR1 transcriptional regulator or in the MDR1 promoter itself which would be responsible for the stable MDR1 overexpression observed in this strain. Moreover, we found that the deletion of CAP1 in FR2 resulted in MDR1 upregulation (Fig. 5), indicating that Cap1p behaves as a negative transcriptional regulator of MDR1 in this strain. This finding was unexpected, in light of our previous demonstration that Cap1p (and Yap1p) behaves as a positive transcriptional regulator of FLR1 in S. cerevisiae (1) and that deletion of CAP1 does not result in MDR1 upregulation in C. albicans CAI4 (Fig. 4). It is possible that in FR2, the genetic alteration leading to MDR1 overexpression also modifies the activity of Cap1p, which, in this mutated context, would function as a transcriptional repressor rather than as an activator. Nevertheless, our data showing that Cap1p is not responsible for the constitutive overexpression of MDR1 in FR2 suggests, by extension, that CAP1 is probably not responsible for the constitutive overexpression of MDR1 observed in a number of FCZ-resistant clinical isolates. Identifying the molecular mechanisms leading to MDR1 upregulation is becoming particularly important given recent data showing that overexpression of MDR1 constitutes a very early and widespread event in the emergence of FCZ resistance in C. albicans and related species (31, 58). The molecular dissection of the cis- and trans-acting regulatory elements controlling the expression of MDR1 will help answer these questions.
ACKNOWLEDGMENTS
We are grateful to R. Cannon for providing strain FR2. R. Rachubinski for the pMK22 vector, and B. Magee for the ACT1 plasmid. We also thank François Comte for technical assistance.
This work was supported by a joint research grant to M.R. from the Medical Research Council (MRC) of Canada and Pfizer Canada Inc. M.R. is supported by a scholarship from MRC.
REFERENCES
- 1.Alarco A-M, Balan I, Talibi D, Mainville N, Raymond M. AP1-mediated multidrug resistance in Saccharomyces cerevisiae requires FLR1 encoding a transporter of the major facilitator superfamily. J Biol Chem. 1997;272:19304–19313. doi: 10.1074/jbc.272.31.19304. [DOI] [PubMed] [Google Scholar]
- 2.Albertson G D, Niimi M, Cannon R D, Jenkinson H F. Multiple efflux mechanisms are involved in Candida albicans fluconazole resistance. Antimicrob Agents Chemother. 1996;40:2835–2841. doi: 10.1128/aac.40.12.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balan I, Alarco A-M, Raymond M. The Candida albicans CDR3 gene codes for an opaque-phase ABC transporter. J Bacteriol. 1997;179:7210–7218. doi: 10.1128/jb.179.23.7210-7218.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balzi E, Goffeau A. Yeast multidrug resistance: the PDR network. J Bioenerg Biomembr. 1995;27:71–76. doi: 10.1007/BF02110333. [DOI] [PubMed] [Google Scholar]
- 5.Billard P, Dumond H, Bolotin-Fukuhara M. Characterization of an AP-1-like transcription factor that mediates an oxidative stress response in Kluyveromyces lactis. Mol Gen Genet. 1997;257:62–70. doi: 10.1007/s004380050624. [DOI] [PubMed] [Google Scholar]
- 6.Boeke J D, LaCroute F, Fink G R. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- 7.Bossier P, Fernandes L, Rocha D, Rodrigues-Pousada C. Overexpression of YAP2, coding for a new yAP protein, and YAP1 in Saccharomyces cerevisiae alleviates growth inhibition caused by 1,10-phenanthroline. J Biol Chem. 1993;268:23640–23645. [PubMed] [Google Scholar]
- 8.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 9.Carlson M, Botstein D. Two differentially regulated mRNAs with different 5′ ends encode secreted intracellular forms of yeast invertase. Cell. 1982;28:145–154. doi: 10.1016/0092-8674(82)90384-1. [DOI] [PubMed] [Google Scholar]
- 10.Clark F S, Parkinson T, Hitchcock C A, Gow N A R. Correlation between rhodamine 123 accumulation and azole sensitivity in Candida species: possible role for drug efflux in drug resistance. Antimicrob Agents Chemother. 1996;40:419–425. doi: 10.1128/aac.40.2.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coleman S T, Tseng E, Moye-Rowley W S. Saccharomyces cerevisiae basic region-leucine zipper protein regulatory networks converge at the ATR1 structural gene. J Biol Chem. 1997;272:23224–23230. doi: 10.1074/jbc.272.37.23224. [DOI] [PubMed] [Google Scholar]
- 12.Collinson L P, Dawes I W. Inducibility of the response of yeast cells to peroxide stress. J Gen Microbiol. 1992;138:329–335. doi: 10.1099/00221287-138-2-329. [DOI] [PubMed] [Google Scholar]
- 13.Diamond R D, Lyman C A, Wysong D R. Disparate effects of interferon-gamma and tumor necrosis factor-alpha on early neutrophil respiratory burst and fungicidal responses to Candida albicans hyphae in vitro. J Clin Investig. 1991;87:711–720. doi: 10.1172/JCI115050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dupont B F, Dromer F, Improvisi L. The problem of azole resistance in Candida. J Mycol Med. 1996;6:12–19. [Google Scholar]
- 15.Fernandes L, Rodrigues-Pousada C, Struhl K. Yap, a novel family of eight bZIP proteins in Saccharomyces cerevisiae with distinct biological functions. Mol Cell Biol. 1997;17:6982–6993. doi: 10.1128/mcb.17.12.6982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fling M E, Kopf J, Tamarkin A, Gorman J A, Smith H A, Koltin Y. Analysis of a Candida albicans gene that encodes a novel mechanism for resistance to benomyl and methotrexate. Mol Gen Genet. 1991;227:318–329. doi: 10.1007/BF00259685. [DOI] [PubMed] [Google Scholar]
- 17.Fonzi W A, Irwin M Y. Isogenic strain construction and gene mapping in Candida albicans. Genetics. 1993;134:717–728. doi: 10.1093/genetics/134.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gietz R D, Schiestl R H, Willems A R, Woods R A. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- 19.Goldway M, Teff D, Schmidt R, Oppenheim A B, Koltin Y. Multidrug resistance in Candida albicans: disruption of the BENr gene. Antimicrob Agents Chemother. 1995;39:422–426. doi: 10.1128/aac.39.2.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grant C M, Collinson L P, Roe J-H, Dawes I W. Yeast glutathione reductase is required for protection against oxidative stress and is a target gene for yAP-1 transcriptional regulation. Mol Microbiol. 1996;21:171–179. doi: 10.1046/j.1365-2958.1996.6351340.x. [DOI] [PubMed] [Google Scholar]
- 21.Hertle K, Haase E, Brendel M. The SNQ3 gene of Saccharomyces cerevisiae confers hyper-resistance to several functionally unrelated chemicals. Curr Genet. 1991;19:429–433. doi: 10.1007/BF00312733. [DOI] [PubMed] [Google Scholar]
- 22.Hitchcock C A. Resistance of Candida albicans to azole antifungal agents. Biochem Soc Trans. 1993;21:1039–1047. doi: 10.1042/bst0211039. [DOI] [PubMed] [Google Scholar]
- 23.Hussain M, Lenard J. Characterization of PDR4, a Saccharomyces cerevisiae gene that confers pleiotropic drug resistance in high-copy number: identity with YAP1, encoding a transcriptional activator. Gene. 1991;101:149–152. doi: 10.1016/0378-1119(91)90238-7. [DOI] [PubMed] [Google Scholar]
- 24.Kelly R, Miller S M, Kurtz M B, Kirsch D R. Directed mutagenesis in Candida albicans: one-step gene disruption to isolate ura3 mutants. Mol Cell Biol. 1987;7:199–208. doi: 10.1128/mcb.7.1.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kosower N S, Kosower E M. Formation of disulfides with diamide. Methods Enzymol. 1987;143:264–270. doi: 10.1016/0076-6879(87)43050-4. [DOI] [PubMed] [Google Scholar]
- 26.Kuge S, Jones N. YAP1 dependent activation of TRX2 is essential for the response of Saccharomyces cerevisiae to oxidative stress by hydroperoxides. EMBO J. 1994;13:655–664. doi: 10.1002/j.1460-2075.1994.tb06304.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuge S, Jones N, Nomoto A. Regulation of yAP-1 nuclear localization in response to oxidative stress. EMBO J. 1997;16:1710–1720. doi: 10.1093/emboj/16.7.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuge S, Toda T, Iizuka N, Nomoto A. Crm1 (XpoI) dependent nuclear export of the budding yeast transcription factor yAP-1 is sensitive to oxidative stress. Genes Cells. 1998;3:521–532. doi: 10.1046/j.1365-2443.1998.00209.x. [DOI] [PubMed] [Google Scholar]
- 29.Kurtz M B, Cortelyou M W, Miller S M, Lai M, Kirsch D R. Development of autonomously replicating plasmids for Candida albicans. Mol Cell Biol. 1987;7:209–217. doi: 10.1128/mcb.7.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maenza J R, Merz W G, Romagnoli M J, Keruly J C, Moore R D, Gallant J E. Infection due to fluconazole-resistant Candida in patients with AIDS: prevalence and microbiology. Clin Infect Dis. 1997;24:28–34. doi: 10.1093/clinids/24.1.28. [DOI] [PubMed] [Google Scholar]
- 31.Moran G P, Sanglard D, Donnelly S M, Shanley D B, Sullivan D J, Coleman D C. Identification and expression of multidrug transporters responsible for fluconazole resistance in Candida dubliniensis. Antimicrob Agents Chemother. 1998;42:1819–1830. doi: 10.1128/aac.42.7.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morgan B A, Banks G R, Toone W M, Raitt D, Kuge S, Johnston L H. The Skn7 response regulator controls gene expression in the oxidative stress response of the budding yeast Saccharomyces cerevisiae. EMBO J. 1997;16:1035–1044. doi: 10.1093/emboj/16.5.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moye-Rowley W S, Harshman K D, Parker C S. Yeast YAP1 encodes a novel form of the jun family of transcriptional activator proteins. Genes Dev. 1989;3:283–292. doi: 10.1101/gad.3.3.283. [DOI] [PubMed] [Google Scholar]
- 34.Murphy J W. Mechanisms of natural resistance to human pathogenic fungi. Annu Rev Microbiol. 1991;45:509–538. doi: 10.1146/annurev.mi.45.100191.002453. [DOI] [PubMed] [Google Scholar]
- 35.Park K S, Kwon J, Choi S Y. Cloning, characterization, and expression of the CIP2 gene induced under cadmium stress in Candida sp. FEMS Microbiol Lett. 1998;162:325–330. doi: 10.1111/j.1574-6968.1998.tb13016.x. [DOI] [PubMed] [Google Scholar]
- 36.Parkinson T, Falconer D J, Hitchcock C A. Fluconazole resistance due to energy-dependent drug efflux in Candida glabrata. Antimicrob Agents Chemother. 1995;39:1696–1699. doi: 10.1128/aac.39.8.1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Polley S D, Caddick M X. Molecular characterisation of meaB, a novel gene affecting nitrogen metabolite repression in Aspergillus nidulans. FEBS Lett. 1996;388:200–205. doi: 10.1016/0014-5793(96)00541-8. [DOI] [PubMed] [Google Scholar]
- 38.Rex J H, Rinaldi M G, Pfaller M A. Resistance of Candida species to fluconazole. Antimicrob Agents Chemother. 1995;39:1–8. doi: 10.1128/aac.39.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rose M D, Winston F, Hieter P. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- 40.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 41.Sanglard D, Ischer F, Koymans L, Bille J. Amino acid substitutions in the cytochrome P-450 lanosterol 14-α-demethylase (CYP51A1) from azole-resistant Candida albicans clinical isolates contribute to resistance to azole antifungal agents. Antimicrob Agents Chemother. 1998;42:241–253. doi: 10.1128/aac.42.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sanglard D, Ischer F, Monod M, Bille J. Susceptibilities of Candida albicans multidrug transporter mutants to various antifungal agents and other metabolic inhibitors. Antimicrob Agents Chemother. 1996;40:2300–2305. doi: 10.1128/aac.40.10.2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sanglard D, Ischer F, Monod M, Bille J. Cloning of Candida albicans genes conferring resistance to azole antifungal agents: characterization of CDR2, a new multidrug ABC transporter gene. Microbiology. 1997;143:405–416. doi: 10.1099/00221287-143-2-405. [DOI] [PubMed] [Google Scholar]
- 44.Sanglard D, Kuchler K, Ischer F, Pagani J L, Monod M, Bille J. Mechanisms of resistance to azole antifungal agents in Candida albicans isolates from AIDS patients involve specific multidrug transporters. Antimicrob Agents Chemother. 1995;39:2378–2386. doi: 10.1128/aac.39.11.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schnell N, Entian K D. Identification and characterization of a Saccharomyces cerevisiae gene (PAR1) conferring resistance to iron chelators. Eur J Biochem. 1991;200:487–493. doi: 10.1111/j.1432-1033.1991.tb16209.x. [DOI] [PubMed] [Google Scholar]
- 46.Schnell N, Krems B, Entian K D. The PAR1 (YAP1/SNQ3) gene of Saccharomyces cerevisiae, a c-jun homologue, is involved in oxygen metabolism. Curr Genet. 1992;21:269–273. doi: 10.1007/BF00351681. [DOI] [PubMed] [Google Scholar]
- 47.Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- 48.Smith D B, Johnson K S. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 49.Stephen D W, Rivers S L, Jamieson D J. The role of the YAP1 and YAP2 genes in the regulation of the adaptive oxidative stress responses of Saccharomyces cerevisiae. Mol Microbiol. 1995;16:415–423. doi: 10.1111/j.1365-2958.1995.tb02407.x. [DOI] [PubMed] [Google Scholar]
- 50.Takeuchi T, Miyahara K, Hirata D, Miyakawa T. Mutational analysis of Yap1 protein, an AP-1-like transcriptional activator of Saccharomyces cerevisiae. FEBS Lett. 1997;416:339–343. doi: 10.1016/s0014-5793(97)01233-7. [DOI] [PubMed] [Google Scholar]
- 51.Toda T, Shimanuki M, Yanagida M. Fission yeast genes that confer resistance to staurosporine encode an AP-1-like transcription factor and a protein kinase related to the mammalian ERK1/MAP2 and budding yeast FUS3 and KSS1 kinases. Genes Dev. 1991;5:60–73. doi: 10.1101/gad.5.1.60. [DOI] [PubMed] [Google Scholar]
- 52.Toone W M, Kuge S, Samuels M, Morgan B A, Toda T, Jones N. Regulation of the fission yeast transcription factor PAP1 by oxidative stress: requirement for the nuclear export factor Crm1 (exportin) and the stress-activated map kinase Sty1/Spc1. Genes Dev. 1998;12:1453–1463. doi: 10.1101/gad.12.10.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vanden Bossche H, Marichal P, Odds F C, Le Jeune L, Coene M C. Characterization of an azole-resistant Candida glabrata isolate. Antimicrob Agents Chemother. 1992;36:2602–2610. doi: 10.1128/aac.36.12.2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wagner D K, Collins-Lech C, Sohnle P G. Inhibition of neutrophil killing of Candida albicans pseudohyphae by substances which quench hypochlorous acid and chloramines. Infect Immun. 1986;51:731–735. doi: 10.1128/iai.51.3.731-735.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wemmie J A, Steggerda S M, Moye-Rowley W S. The Saccharomyces cerevisiae AP-1 protein discriminates between oxidative stress elicited by the oxidants H2O2 and diamide. J Biol Chem. 1997;272:7908–7914. doi: 10.1074/jbc.272.12.7908. [DOI] [PubMed] [Google Scholar]
- 56.Wemmie J A, Szczypka M S, Thiele D J, Moye-Rowley W S. Cadmium tolerance mediated by the yeast AP-1 protein requires the presence of an ATP-binding cassette transporter-encoding gene, YCF1. J Biol Chem. 1994;269:32592–32597. [PubMed] [Google Scholar]
- 57.Wemmie J A, Wu A L, Harshman K D, Parker C S, Moye-Rowley W S. Transcriptional activation mediated by the yeast AP-1 protein is required for normal cadmium tolerance. J Biol Chem. 1994;269:14690–14697. [PubMed] [Google Scholar]
- 58.White T C. Increased mRNA levels of ERG16, CDR, and MDR1 correlate with increases in azole resistance in Candida albicans isolates from a patient infected with human immunodeficiency virus. Antimicrob Agents Chemother. 1997;41:1482–1487. doi: 10.1128/aac.41.7.1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu A, Wemmie J A, Edgington N P, Goebl M, Guevara J L, Moye-Rowley W S. Yeast bZip proteins mediate pleiotropic drug and metal resistance. J Biol Chem. 1993;268:18850–18858. [PubMed] [Google Scholar]
- 60.Wu A L, Moye-Rowley W S. GSH1, which encodes γ-glutamylcysteine synthetase, is a target gene for yAP-1 transcriptional regulation. Mol Cell Biol. 1994;14:5832–5839. doi: 10.1128/mcb.14.9.5832. [DOI] [PMC free article] [PubMed] [Google Scholar]