Abstract
In gram-positive bacteria, HPr, a protein of the phosphoenolpyruvate:sugar phosphotransferase system, is phosphorylated on a serine residue at position 46 by an ATP-dependent protein kinase. The HPr(Ser) kinase of Streptococcus salivarius ATCC 25975 was purified, and the encoding gene (hprK) was cloned by using a nucleotide probe designed from the N-terminal amino acid sequence. The predicted amino acid sequence of the S. salivarius enzyme showed 45% identity with the Bacillus subtilis enzyme, the conserved residues being located mainly in the C-terminal half of the protein. The predicted hprK gene product has a molecular mass of 34,440 Da and a pI of 5.6. These values agree well with those found experimentally by polyacrylamide gel electrophoresis in the presence of sodium dodecyl sulfate, molecular sieve chromatography in the presence of guanidine hydrochloride, and chromatofocusing using the purified protein. The native protein migrates on a Superdex 200 HR column as a 330,000-Da protein, suggesting that the HPr(Ser) kinase is a decamer. The enzyme requires Mg2+ for activity and functions optimally at pH 7.5. Unlike the enzyme from other gram-positive bacteria, the HPr(Ser) kinase from S. salivarius is not stimulated by FDP or other glycolytic intermediates. The enzyme is inhibited by inorganic phosphate, and its Kms for HPr and ATP are 31 μM and 1 mM, respectively.
Streptococci have a major impact on a wide variety of human activities in their roles as adjuncts in food production (30), pathogens (38), and probiotic agents (17). These lactic acid bacteria are nutritionally very demanding and obtain their energy mainly by oxidizing sugars that are taken up, in several cases, by the phosphoenolpyruvate:sugar phosphotransferase system (PTS). This multienzymatic system, composed of the general energy-coupling proteins enzyme I (EI) and HPr and the multidomain sugar-specific proteins called EII, catalyzes the transport and phosphorylation of mono- and disaccharides and also plays a key role in the control of sugar metabolism by regulating the expression of catabolic genes, modulating the activity of key metabolic enzymes, and controlling the activity of sugar transport systems (20, 32, 33, 36). The regulatory functions of the PTS in gram-positive bacteria are associated with a posttranslational modified form of HPr, HPr(Ser-P) generated by phosphorylation of the protein at a serine residue at position 46 by an ATP-dependent HPr kinase (4, 10, 22, 25, 37, 43). The HPr(Ser) kinases of several organisms, including Bacillus subtilis and Enterococcus faecalis, are activated by cellular metabolites—the most potent being FDP—cellular concentrations of which vary in response to environmental conditions (22, 37, 43). These HPr kinase-activating metabolites are thus cellular indicators of external energy or carbon availability and are the first elements of a teleonomic transmission signal pathway that enables the cell to modify its physiology when faced with changes in the external milieu in order to ensure optimal growth.
The regulatory functions of HPr(Ser-P) in gram-positive bacteria include control of sugar permeases (47, 48) and regulation of gene transcription (4, 7, 27, 31). The latter function is accomplished in conjunction with a DNA binding protein called CcpA that recognizes a specific DNA sequence called CRE (catabolite-responsive element) located in the promoter regions of target operons. The association of CcpA with a number of CRE sequences is promoted by HPr(Ser-P) (6, 10, 18) and results in the activation or inhibition of gene transcription, depending on whether the CRE sequence is located upstream or downstream from the promoter sequence (23).
Although no direct evidence for the involvement of HPr(Ser-P) in the regulation of sugar transport or gene transcription in oral streptococci has been provided, the fact that these bacteria produce HPr(Ser-P) (44) and possess a CcpA-like (28) protein and CRE sequences (23) suggests that HPr(Ser-P)-dependent regulatory mechanisms described for other gram-positive bacteria such as bacillus and lactobacilli also occur in oral streptococci. Recently, a study was conducted with a mutant of Streptococcus salivarius harboring a mutation in the ptsH gene that replaces the isoleucine at position 47 by a threonine. The results established the involvement of HPr in the control of gene expression in this organism and showed that the preferential metabolism of PTS sugars over non-PTS sugars by streptococci is dependent on HPr (15). However, the role of CcpA in catabolite repression in streptococci remains equivocal. Simpson and Russell (41) recently identified in Streptococcus mutans a gene called regM that showed 53% identity with ccpA from B. subtilis. Inactivation of regM did not abolish diauxic growth in S. mutans and did not relieve catabolite repression of α-galactosidase, mannitol-1-P dehydrogenase, or P-β-galactosidase. In contrast, glucose caused a more severe repression of these enzymes in a regM-minus background.
Here we report the purification and characterization of the ATP-dependent HPr(Ser) kinase of S. salivarius ATCC 25975. Using a nucleotide probe designed from the N-terminal amino acid sequence, we cloned and sequenced the gene coding for the HPr-kinase (hprK). The protein encoded by hprK was then expressed in Escherichia coli and shown to phosphorylate HPr at the expense of ATP.
MATERIALS AND METHODS
Chemicals.
Restriction enzymes were purchased from Gibco-BRL Life Technologies (Burlington, Ontario, Canada) or from New England Biolabs (Missisauga, Ontario, Canada). Glycolytic intermediates and dimethyldichlorosilane were obtained from Sigma Chemical Company (St. Louis, Mo.). Radiolabeled materials were purchased from Amersham Life Science Inc. (Toronto, Ontario, Canada). Low-range protein standards were from BioRad Laboratories (Canada) Ltd. (Missisauga, Ontario, Canada). HPr was purified from S. salivarius ATCC 25975 (45).
Bacterial strains, plasmids, and culture conditions.
Strains and plasmids used in this study are described in Table 1. S. salivarius was grown at 37°C in Trypticase yeast extract (TYE) broth containing 10 g of tryptone, 5 g of yeast extract, 2.5 g of NaCl, and 2.5 g of disodium phosphate per liter. Glucose was sterilized by filtration (Millipore filter; 0.22-μm pore size) and added aseptically to the medium to a final concentration of 0.5%. E. coli was grown with aeration at 37°C in Luria-Bertani (LB) medium. When necessary, 50 μg of ampicillin per ml, 10 μg of tetracycline per ml, or 50 μg of kanamycin per ml was added.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or characteristics | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| XL-1 Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac[F′ proAB lacIqZΔM15 Tn10 (Tetr)] | Stratagene |
| TOP10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 deoR recA1 araD139 Δ(ara-leu)7697 galU rpsL endA1 nupG | Invitrogen |
| S. salivarius ATCC 25975 | Wild type | 19 |
| Plasmids | ||
| pUC18 | Cloning vector; AprlacZ′ (lacZα) | 46 |
| pCR-Blunt | Cloning vector | Invitrogen |
| pHPK20 | Contains 2-kbp HindIII fragment from S. salivarius cloned into pUC18; carries the last 36 nucleotides of hprK | This work |
| pHPK221 | Contains a 2.9-kbp StuI fragment from S. salivarius cloned into pCR-Blunt in a transcriptional orientation opposite that of the lacZ promoter; carries hprK | This work |
| pHPK229 | Contains the same fragment as pHPK221 cloned in the opposite orientation | This work |
| pHPK224 | pCR-Blunt containing an uncharacterized 2.9-kb StuI DNA fragment from S. salivarius without hprK | This work |
Determination of HPr(Ser) kinase activity.
Unless otherwise mentioned, HPr kinase activity was determined in a mixture containing 100 mM Tris-acetate (pH 7.5), 5 mM MgCl2, 12.5 mM NaF, 10 μM HPr, 1 mM [γ-32P]ATP (0.1 μCi/nmol), and HPr(Ser) kinase in a final volume of 20 or 50 μl. Incubation was at 37°C for 2 to 30 min. The reaction was stopped by the addition of 10 or 25 μl of denaturing buffer (187.5 mM Tris-HCl [pH 6.8], 6% sodium dodecyl sulfate [SDS], 15% 2-mercaptoethanol, 30% glycerol, 0.003% bromophenol blue) followed by heating at 100°C for 5 min. The product, HPr(Ser-32P), was isolated by SDS-polyacrylamide gel electrophoresis (PAGE) (29) with a 15% resolving gel. Electrophoresis was at 200 V at room temperature until the bromophenol blue reached the bottom of the gel. HPr(Ser-32P) was located by autoradiography, and the labeled gel band was excised and counted in 5 ml of Cytosint ES (ICN) in a liquid scintillation counter.
Purification of HPr(Ser) kinase.
Results of preliminary experiments had indicated that the HPr(Ser) kinase readily adhered to plastic and glass. To prevent loss of activity during purification, material that came into contact with the kinase such as tubes, Eppendorf tips, and Pasteur pipettes was siliconized by leaving it for 30 min under vacuum in dimethyldichlorosilane vapor. The material was then rinsed thoroughly with water and autoclaved at 110°C for 15 min (9). Cells were grown with gentle stirring in six 20-liter Pyrex bottles, each containing 12 liters of TYE medium with 0.5% glucose. Each bottle was inoculated with a 1-liter overnight culture. Cells were harvested in the stationary growth phase and washed once with 50 mM Tris-HCl (pH 7.0). Cells (173 g, wet weight) were lysed by pulsed sonication (three 5-min bursts, 60% duty cycle) at maximum energy level with a Heat System-Ultrasonic model W350 sonicator. The sonication was carried out on a mixture containing 2 to 3 g of cells, 0.5 g of alumina, 50 mM Tris-HCl (pH 7.5), 0.1 mM phenylmethylsulfonyl fluoride (PMSF), 0.1 μM leupeptin, and 0.1 μM pepstatin per ml. During sonication, the tube containing the cells was kept in a mixture of 80% ethanol and dry ice. All remaining steps except the fast protein liquid chromatography (FPLC) were performed at 4°C. Alumina was removed by centrifugation for 5 min at 3,000 × g, and cell debris and unbroken cells were sedimented at 20,000 × g for 20 min. The supernatant was collected, and membrane fragments were sedimented at 145,000 × g for 16 to 18 h. All enzyme activity was associated with the membranes, which were homogenized in 50 mM Tris-HCl (pH 7.5) and left at 4°C for 2 h. The solution was then centrifuged at 150,000 × g for 4 h. The supernatant (323 ml, 1.63 g of protein), containing 50% of the HPr(Ser) kinase activity, was separated into three fractions that were manipulated identically but separately. The supernatant was applied to a Hiload Q-Sepharose 16/10 (Fast Flow) column equilibrated with 20 mM Tris-HCl (pH 7.5). The column was washed with 50 ml of equilibration buffer. The proteins were eluted with a 0 to 0.5 M KCl gradient (500 ml) at a flow rate of 2.5 ml/min, and 5-ml fractions were collected. HPr(Ser) kinase eluted at approximately 0.3 M KCl. Active fractions were pooled, concentrated by ultrafiltration in an Amicon cell with a YM3 membrane (Mr cutoff = 3,000), and dialyzed against 2 liters of 20 mM Tris-acetate (pH 5.0) for 16 h. The dialyzed solution was centrifuged at 12,000 × g for 20 min to collect the insoluble material, which contained 97 to 99% of the HPr(Ser) kinase activity. The pellet was solubilized in 2 ml of 20 mM Tris-HCl (pH 7.5) containing 0.1 M KCl (TK buffer) and loaded on a Sephacryl S200 HR column (2.6 by 64 cm) equilibrated with TK buffer. The column was eluted with 400 ml of the same buffer at 1 ml/min, and 2-ml fractions were collected. The fractions containing activity were pooled, diluted twofold with TK buffer, and concentrated to approximately 1 ml on a Mono Q anion-exchange column. A Superdex 75 HR column equilibrated with TK buffer was loaded with 200-μl fractions. The column was eluted with 25 ml of buffer TK at 0.5 ml/min, and 0.5-ml fractions were collected. The HPr(Ser) kinase obtained at this step was used for enzyme characterization.
The following procedure was used to obtain HPr(Ser) kinase suitably pure for N-terminal amino acid sequence determination. Five milliliters of partially purified HPr(Ser) kinase obtained following chromatography on Superdex 75 HR was mixed with trichloroacetic acid to give a final acid concentration of 10%. The solution was left on ice for 45 min to permit protein precipitation. The mixture was then centrifuged at 10,000 × g for 5 min, and the pellet was washed twice with 0.1 ml of cold acetone. The pellet was dissolved in 50 μl of buffer containing 62.6 mM Tris-HCl (pH 6.8), 2% SDS, 5% 2-mercaptoethanol, and 10% glycerol. The sample was heated at 100°C for 3 min, then loaded onto a 10% polyacrylamide gel, and subjected to SDS-PAGE using currents of 15 and 25 mA/gel during migration in the stacking and resolving gels, respectively. The proteins were then electrophoretically transferred to a polyvinylidene difluoride membrane (0.45-μm pore size), using 10 mM CAPS (3-[cyclohexylamino]-1-propanesulfonic acid) buffer (pH 11.0) at 55 V for 80 min. The membrane was stained for 5 min with Coomassie blue (0.1% in 50% methanol) and destained for 15 min in a solution of 50% methanol and 10% acetic acid. The portion of the membrane containing the HPr(Ser) kinase was excised and used for amino acid sequencing.
The recombinant S. salivarius HPr(Ser) kinase was purified from crude extracts of E. coli One Shot TOP10[pHPK229]. Cells were cultured at 37°C with aeration in 400 ml of LB medium containing 50 μg of kanamycin per ml, harvested by centrifugation when the optical density at 600 nm reached 0.65, and washed twice with 100 ml of 50 mM Tris-HCl, pH 7.5 (TH50 buffer). Cells were then suspended in 10 ml of TH50 buffer containing 0.1 mM PMSF, 0.1 μM leupeptin, and 0.1 μM pepstatin and ruptured by sonication (three 15-s bursts, energy level 7) with a Heat System-Ultrasonic model W350 sonicator. The cellular extract was centrifuged at 20,000 × g for 20 min at 4°C. The supernatant was collected and centrifuged at 200,000 × g for 6 h. The pellet, which contained the HPr(Ser) kinase activity, was suspended in 10 ml of TH50 buffer and gently stirred for 2 h at 4°C. The mixture was then centrifuged at 130,000 × g for 4 h. The supernatant (total volume of approximately 10 ml, 2.3 mg of protein/ml), which contained approximately 50% of the HPr(Ser) kinase, was loaded onto a Mono Q HR 5/5 column equilibrated with 20 mM Tris-HCl (pH 7.5). The column was washed with 5 ml of equilibration buffer and then with a 0 to 0.5 M KCl gradient (20 ml) at 1 ml/min, and 1-ml fractions were collected. The enzyme eluted at 0.25 M KCl. The recombinant HPr(Ser) kinase was further purified by chromatography on a Superdex 200 HR column equilibrated with 20 mM Tris-HCl (pH 7.5) containing 0.1 M KCl.
N-terminal amino acid sequencing.
N-terminal amino acid sequences were determined by Edman degradation using a model 473A pulsed liquid-phase sequencer from Applied Biosystems. The sample was applied to a trifluoroacetic acid-treated cartridge filter coated with 1.5 mg of Polybrene and 0.1 mg of NaCl. The phenylthiohydantoin amino acid derivatives were identified by comparison with standards (PTH Analyzer standards; Applied Biosystems) analyzed on-line prior to the sequence analysis.
Identification of the phosphorylated residue.
HPr (11 μg) was phosphorylated with [γ-32P]ATP as described for determination of HPr(Ser) kinase activity. The reaction was performed at 37°C for 30 min. The proteins were then precipitated with trichloroacetic acid for 30 min at 4°C. After centrifugation, the pellet was dissolved in 6 N HCl, and the solution was incubated under vacuum for 2 h at 110°C. The acid was eliminated by evaporation, and the sample was resuspended in 20 μl of water. The HPr amino acid residue phosphorylated by the purified ATP-dependent HPr kinase was identified by electrophoresis on a thin-layer cellulose plate followed by ascending chromatography as described by Duclos et al. (8).
Molecular mass determination.
The molecular mass of the HPr(Ser) kinase was determined by using the enzyme purified from S. salivarius and with the recombinant HPr(Ser) kinase purified from E. coli One Shot TOP10[pHPK229]. The molecular mass of the enzyme was also determined with E. coli One Shot TOP10[pHPK229] cellular extracts obtained using three different rupture procedures. In the first procedure, the cells were lysed in lysozyme-EDTA at pH 8.0. Bacteria from a 500-ml culture were harvested in mid-exponential growth and washed twice at 4°C with 10 mM Tris-HCl buffer (pH 8.0). After centrifugation, the pellet was resuspended in 80 ml of 30 mM Tris-HCl buffer (pH 8.0) containing 10 mM EDTA and 0.5 mg of lysozyme per ml. The solution was gently stirred at room temperature for 30 min. Then, 0.5 mg of DNase I per ml, 0.1 mM PMSF, and 10 mM MgCl2 were added, and the solution was kept at 4°C for 1 h. Whole cells and cell debris were removed by centrifugation at 4°C (23,000 × g for 1 h). The second rupture procedure also involved lysozyme, but lysis was performed at pH 10. Bacteria from a 30-ml culture were harvested in mid-exponential growth and washed twice at 4°C with 10 mM Tris-HCl buffer (pH 8.0). The cells were resuspended in 20 ml of 30 mM Tris-HCl buffer (pH 8.0) containing 10 mM EDTA, 0.5 mg of lysozyme per ml, and 20% (wt/vol) sucrose. They were left at room temperature for 30 min and then harvested by centrifugation. The pellet was resuspended in CAPS buffer (pH 10), and the solution was gently stirred at room temperature for 30 min. Whole cells and cell debris were eliminated by centrifugation at 4°C (23,000 × g for 1 h). The molecular mass determination of HPr(Ser) kinase was also carried out with cellular extracts obtained from cells ruptured by ultrasonic disintegration. Cells resuspended in 100 mM Tris-acetate (pH 7.5) containing 12.5 mM NaF, 0.1 mM PMSF, 0.1 μM leupeptin, and 0.1 μM pepstatin were lysed by pulsed sonication (three 10-s bursts, 50% duty cycle, energy level 6) with a Heat System-Ultrasonic model W350 sonicator.
The native molecular mass was determined by molecular sieve chromatography on a Superdex 200 HR column (cross-linked agarose and dextran) operated with a Pharmacia FPLC system using a 20 mM Tris-HCl buffer (pH 7.5) containing 0.1 M or 0.3 M KCl. The column was equilibrated with seven protein markers of different molecular masses: thyroglobulin (669 kDa), apoferritin (443 kDa), amylase (200 kDa), alcohol dehydrogenase (150 kDa), transferrin, (81 kDa), ovalbumin (43 kDa), and myoglobin (17.6 kDa). Elution of the protein markers was monitored by light absorption at 280 nm, whereas elution of the HPr(Ser) kinase was determined by measurement of enzyme activity. This experiment was also performed on a Sephacryl S200 HR column (dextran cross-linked with N,N′-methylenebisacrylamide) and an Ultrogel AcA-34 column (acrylamide and agarose).
The molecular mass of the HPr(Ser) kinase was also estimated by molecular sieve chromatography in the presence of guanidine hydrochloride on a Superdex 200 HR column. The protein was dissolved in 50 mM Tris-HCl buffer (pH 7.5) containing 6 M guanidine hydrochloride and 5 mM dithiothreitol. The column was eluted at 0.5 ml/min, and 0.5-ml fractions were collected. The column was equilibrated with five protein markers of different molecular masses: bovine serum albumin (68 kDa), ovalbumin (43 kDa), carbonic anhydrase (29 kDa), chymotrypsinogen (25 kDa), and myoglobin (17 kDa). Elution of the protein markers was monitored by light absorption at 280 nm. HPr(Ser) kinase purified from S. salivarius was detected by SDS-PAGE after protein precipitation with 90% ethanol at −20°C, whereas recombinant HPr(Ser) kinase was detected by light absorption at 280 nm. The molecular mass of the enzyme determined by molecular sieve chromatography was calculated from the plot of the logarithm of molecular mass versus the partition coefficient (Kav) of the marker proteins.
The molecular mass of the denatured protein was also determined by disc gel electrophoresis in the presence of SDS (29).
DNA purification and manipulations.
Chromosomal DNA was isolated from S. salivarius as described previously (14). Unless otherwise mentioned, DNA manipulations were performed by standard procedures (1). E. coli XL-1 Blue cells were made competent and transformed with plasmid DNA by electroporation (39). Unless otherwise mentioned, DNA fragments used for subcloning and probes were isolated from agarose gels with an Elu-Quik DNA purification kit (Schleicher & Schuell).
Cloning and sequencing of the hprK gene.
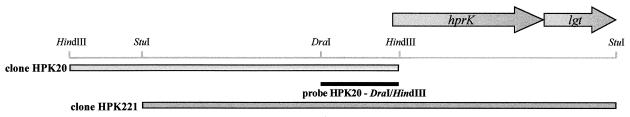
Two degenerate oligonucleotides, db1 (5′AARCCWATYCAAGGWGCWGAYATYACYCG) and db2 (5′ACYGTWAARATGYTIGTWGAYATYACWCG), were designed on the basis of the N-terminal amino acid sequence of the HPr(Ser) kinase. These oligonucleotides were radiolabeled with [γ-32P]ATP and T4 polynucleotide kinase. Restriction endonuclease digests of chromosomal DNA from S. salivarius ATCC 25975 were prepared by using EcoRI, HindIII, Sau3AI, XbaI, BglII, PstI, and StyI. Southern blot analyses of the generated fragments using the aforementioned oligonucleotides as probes allowed the identification of a 2-kb HindIII DNA fragment that hybridized strongly with oligonucleotide db2. Chromosomal DNA from S. salivarius was thus digested with HindIII, the digest was separated on an agarose gel, and approximately 2-kb fragments were isolated by using a Prep-a-Gene (Bio-Rad) kit. The DNA fragments were cloned into the pUC18 HindIII/BAP vector (Pharmacia). Ligation mixtures were used to transform E. coli XL-1 Blue by electroporation. Colony screening with oligonucleotide db2 resulted in the isolation of a clone, designated HPK20, containing the first 36 nucleotides of the 5′ end of hprK at one end (Fig. 1). The cloned HindIII fragment was digested with DraI to give a 475-bp DNA fragment, which was labeled with digoxigenin by random priming. DNA digested with various restriction enzymes was separated into fragments by electrophoresis. Southern blot analyses with probe HPK20-DraI/HindIII revealed the presence of a 2.9-kb StuI fragment that contained the entire hprK gene. StuI-digested DNA was separated by agarose electrophoresis, and DNA fragments of approximately 2.9 kb were recovered from the gel and cloned into the pCRblunt vector. Recombinant plasmids were used to transform E. coli One Shot TOP10 (Zeroblunt PCR cloning kit; Invitrogen) (hereafter referred to as simply E. coli TOP10). Screening with probe HPK20-DraI/HindIII resulted in the isolation of four clones possessing the entire HPr(Ser) kinase gene. Clones HPK221 and HPK229 were selected for further study (Fig. 1).
FIG. 1.
Restriction and genetic map of the S. salivarius DNA region coding for hprK. Clone HPK20 was isolated by using two degenerated oligonucleotides designed on the basis of the N-terminal amino acid sequence of the HPr(Ser) kinase as probes. The DNA fragment HPK20-DraI/HindIII was used as a probe to clone a 2.9-kb StuI fragment that contained the entire hprK gene. Arrows indicate names of genes and directions of transcription. The region upstream of hprK was not sequenced.
The DNA sequences were determined on both strands by the dideoxy chain termination method of Sanger et al. (40). The primers used for sequencing the HPK20-HindIII DNA fragment were the reverse and forward lacZ universal primers; those for sequencing the HPK221-StuI DNA fragment were the lacZ primers and three specific primers complementary to previously determined sequences: 5′-TATGATTGGCCCTGGTGCTA-3′, 5′-GGTAAGAGTGAAACAGGG-3′, and 5′-CCAAGACGATCAAAGAGG-3′.
Computer analysis.
Sequence analyses were performed with the BestFit, Pileup, Pretty, Map, and Peptidestructure programs in the Wisconsin Package software (version 9.0) of the Genetics Computer Group (16).
RESULTS
Purification of the HPr(Ser) kinase.
Purification of the HPr(Ser) kinase from E. faecalis and Streptococcus pyogenes has been reported previously (3, 35). We thus first tried to purify the HPr(Ser) kinase from S. salivarius by using these published procedures, but without success. Results of several attempts indicated that the HPr(Ser) kinase of S. salivarius (i) was associated with the membrane, (ii) adhered readily to plastic and glass, (iii) was present in very low amounts, and (iv) did not possess a molecular mass of 65,000 Da as reported earlier (3, 35). Taking into account these factors, we developed the purification procedure described in Materials and Methods. It was not possible to obtain a homogeneous preparation. When the enzyme solution was chromatographed on a Superdex 75 HR FPLC column, the activity was eluted as a sharp peak. By comparing the protein profile of each fractions revealed by SDS-PAGE to the activity profile, a protein with a molecular mass of 34,000 Da was shown to be the HPr(Ser) kinase. Several experiments of this type were performed, and in all cases the activity was associated with the 34-kDa protein. Attempts to purify the enzyme to homogeneity by other chromatographic procedures (phenyl-Superose, butyl-TSK, hydroxylapatite, dye ligand chromatography, ATP-agarose, and phosphocellulose) did not result in improved purification of the enzyme.
Nature of the phosphorylated residue.
The phosphoryl-bond form of HPr resulting from the incubation of HPr with the purified kinase and ATP was found to be heat stable and resistant to acid treatment (not shown). These properties are characteristic of phosphoserine, phosphothreonine, and phosphotyrosine (8). Hydrolysis of the phosphoyl-bound form with HCl followed by two-dimensional separation of the products (not shown) revealed that the phosphorylated residue was a serine, confirming that the purified protein was an HPr(Ser) kinase.
N-terminal amino acid sequence.
The sequence of the first 35 N-terminal amino acids of the protein identified as the HPr(Ser) kinase was (G,M,S)V(T,C,Q)VKMLVDKLKLKVIYGNEKLLSKPIQGADITRP. At the time when these results were obtained, this sequence showed no significant similarity with proteins and translated DNAs in current databases (GenBank version 103.0, EMBL version 52.0, and Swiss-Prot version 34.0).
Sequence analysis of the gene coding for the HPr(Ser) kinase.
The gene coding for the HPr(Ser) kinase (hprK) was cloned as described in Materials and Methods. hprK was located upstream of lgt, a gene coding for a putative prolipoprotein, diacylglyceryl transferase (Fig. 1). The same gene organization was found in B. subtilis (12, 34). The predicted hprK gene product contains 309 amino acids (Fig. 2). Amino acids at positions 3 through 28 of the N terminus of the peptide inferred by hprK were identical to those of the N-terminal amino acid sequence determined from the purified protein. A putative ribosome binding (Shine-Dalgarno) site (CGGAGG) was identified 7 bp upstream from the hprK ATG start codon. A putative promoter region with characteristic features such as AT-rich regions and two likely −10 and −35 regions separated by 19 bp was also found. The estimated size of the translated protein was 34,440 Da, with a pI of 5.6. Sequence analysis of the deduced hprK product identified an ATP/GTP binding motif (GX4GKS) at residues 153 through 160, which is consistent with the function of the protein. The sequence indicated that the protein contained no cysteine. The sequence of the N-terminal end up to residue 26 was particularly remarkable as three amino acids—lysine (six residues), leucine (five residues), and valine (four residues)—account for over half of the amino acids in this region. Moreover, the 6 lysine residues in this region represent almost one-third of the total of 19 lysines in the protein, giving the N terminus region a pI of 10.7. Secondary structure prediction using the methods of Chou and Fasman (2) and Garnier and Robson (13) suggested that the N-terminal half of the protein (up to residue 150) is composed mainly of β strands possessing only three small helical structures, the longest helix being composed of approximately 10 residues (positions 96 to 105). The C-terminal end is characterized by the presence of a long amphiphatic α helix of approximately 25 amino acids initiated by a cluster of five helix-promoting amino acid residues (EAAAM) and possibly C capped by an asparagine (residues 276 to 300). The Kyte-Doolittle hydropathy profile generated with a scanning window size of nine amino acids (not shown) suggested that the HPr(Ser) kinase is a soluble protein with five major but small hydrophobic regions, most corresponding to β-strand regions (residues 1 to 20, 83 to 95, 135 to 151, 205 to 217, and 271 to 280).
FIG. 2.
Nucleotide sequence and predicted translation product of hprK. Underlined sequences correspond to the −35 and −10 boxes of the putative promoter region, the ribosome binding Shine-Dalgarno [SD] site, and the start codon. The region corresponding to the ATP/GTP binding motif is shown white on black.
Sequence comparison with other HPr(Ser) kinases.
The B. subtilis gene coding for the HPr(Ser) kinase has recently been cloned and sequenced (12, 34), and genes coding for HPr(Ser) kinase-like proteins have been identified in genome sequencing projects. Figure 3 shows a multiple alignment of the predicted amino acid sequence of S. salivarius HPr(Ser) kinase with those of other gram-positive bacteria and two mycoplasmas. This analysis complements that of Reizer et al. (34) and Gallinier et al. (12), who recently reported the cloning of hprK from B. subtilis. Conserved residues among HPr(Ser) kinases were much more abundant in the C-terminal half of the protein (from residue 140 to the end) than in the N-terminal region. The sequence is particularly well conserved from residues 140 through 180, a stretch that includes the ATP/GTP binding motif. Reizer et al. (34) recently proposed a signature sequence for the HPr kinase family by comparing the sequence of six enzymes, including two proteins from gram-negative bacteria. This putative signature sequence (E[LIVM]RG[LIVM]G[LIVM]2[NDEQ][LIVMF]) was found in the HPr(Ser) kinase of S. salivarius at positions 202 through 211 (EIRGVGIIDV) (numbers refer to positions of the amino acids in the S. salivarius protein and correspond to psoitions 204 through 213 in Fig. 3). The penultimate amino acid of this sequence (D) is conserved in the proteins of all streptococci and of Clostridium acetobutylicum but not in the protein from B. subtilis, where it is replaced by an asparagine. Interestingly, analysis of the protein kinase signatures reported in PROSITE revealed that D is a strictly conserved active-site residue in two signature sequences of eucaryotic protein kinases that resemble the signature sequence of the HPr(Ser) kinase. The S. salivarius HPr(Ser) kinase revealed the greatest identity with the proteins from S. pneumoniae (81% identity; this sequence, however, is incomplete) and S. pyogenes (72% identity). The weakest identity was with the mycoplasma enzyme (28% identity). The percentages of similarity and identity with the HPr(Ser) kinase of B. subtilis were 57 and 45, respectively.
FIG. 3.
Amino acid sequence alignment of HPr(Ser) kinases of gram-positive bacteria and mycoplasma. Residues conserved in all sequences are indicated by gray boxes, and those found in at least four proteins are included in the consensus sequence. The sequences of S. pyogenes, S. pneumoniae, and E. faecalis are incomplete. Shown are sequences for kinases derived from M. genitalium (Swiss-Prot accession no. P47331); M. pneumoniae (Swiss-Prot accession no. P75548); S. pneumoniae (TIGR microbial database, contig stp_4147); S. pyogenes (TIGR microbial database, contig 210); S. salivarius (GenBank accession no. AF069743); B. subtilis (GenBank accession no. AF017113); C. acetobutylicum (http://www.cric.com/genesequences/clostridium/clospage.html, contig538_34042077_C3_74); E. faecalis (Swiss-Prot accession no. O07664).
Expression of S. salivarius hprK in E. coli.
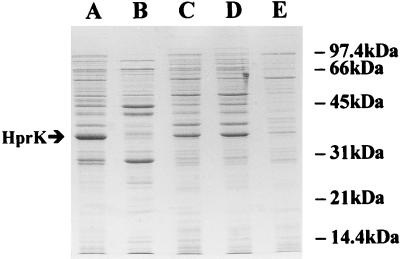
E. coli TOP10[pHPK229] produced a 34-kDa component not found in E. coli TOP10 or E. coli TOP10 transformed with a plasmid in which an S. salivarius DNA fragment that did not contain hprK was cloned (strain E. coli TOP10[pHPK224]) (not shown). Determination of HPr(Ser) kinase activity revealed that the cellular extract of E. coli TOP10[pHPK229] was able to phosphorylate purified S. salivarius HPr at the expense of ATP, whereas cellular extracts prepared from cells without plasmids or transformed with pHPK224 had no activity (not shown). To determine where the enzyme is located in recombinant E. coli, we ultracentrifuged a crude cellular extract from E. coli TOP10[pHPK229] to separate soluble proteins from membrane fragments and membrane-associated proteins. The pellet was suspended in 10 ml of TH50 buffer and gently stirred for 2 h at 4°C. The mixture was then centrifuged at 130,000 × g for 4 h to separate proteins that were loosely associated with the membrane from membrane fragments. Determination of HPr(Ser) kinase activity indicated that most of the enzyme remained associated with the membrane after the first ultracentrifugation and was partially (approximately 50%) released following incubation of the membranes in TH50 buffer (not shown). Consistent with these results, Coomassie blue-stained SDS-polyacrylamide gels showed that the 34-kDa component was not found in the cytoplasmic fraction (Fig. 4, lane B) but was present in the supernatant of the second ultracentrifugation and in the membrane fraction (lanes C and D). The use of different procedures to rupture the cells (alumina grinding and osmotic lysis) as well as treatment of the crude cellular extract with DNase I gave similar results (not shown). The recombinant HPr(Ser) kinase could be purified to near homogeneity by chromatography on a Mono Q HR 5/5 column followed by a Superdex 200 HR column. A yield of approximately 2 mg of purified protein, with a purity of approximately 98%, was obtained from a 400-ml culture of uninduced cells (without isopropyl-β-d-thiogalactopyranoside). The identity of the purified HPr(Ser) kinase was confirmed by measuring its ability to phosphorylate HPr at the expense of ATP.
FIG. 4.
Cellular localization of HPr(Ser) kinase in E. coli TOP10[pHPK229]. Cells were ruptured by sonication and centrifuged at 20,000 × g for 20 min (crude cellular extract). The supernatant was collected and ultracentrifuged at 200,000 × g for 6 h. The supernatant (cytoplasmic fraction) was collected, and the pellet (membrane fraction) was suspended in 50 mM Tris-HCl (pH 7.5) for 2 h at 4°C. The membrane suspension was then ultracentrifuged at 130,000 × g for 4 h to separate proteins that were loosely associated with the membranes (membrane extract) from membrane fragments. The protein content of the various fractions was revealed by SDS-PAGE. The gel was stained with Coomassie blue. Lane A, crude cellular extract (5 μg of protein); lane B, cytoplasmic fraction (5 μg of protein); lane C, membrane fraction (10 μg of protein); lane D, membrane extract (5 μg of protein); lane E, membrane fragments (10 μg of protein). The position of HPr(Ser) kinase is indicated by the arrow.
Estimation of pI and molecular mass.
The pI of the native enzyme as estimated by chromatofocusing was 5.6 (not shown), identical to the pI calculated from the amino acid sequence. As mentioned, the HPr(Ser) kinase in its reduced state migrated on SDS-PAGE as a protein with a molecular mass of 34,000 Da. Molecular sieve chromatography of the enzyme purified from S. salivarius and of the recombinant protein in the presence of guanidine hydrochloride gave the same result (not shown).
Molecular sieve chromatography of the native protein was performed as described in Materials and Methods. The experiment was conducted with the enzyme purified from S. salivarius, with the recombinant kinase purified from E. coli, and with cellular extracts prepared from E. coli TOP10[pHPK221] ruptured by sonication or treatment with lysosyme at two different pHs. An experiment was also carried out with the purified recombinant enzyme in the presence of 0.3 M KCl. In all cases, the activity eluted from the Superdex 200 HR column as a sharp peak at a position corresponding to a protein with a molecular mass of approximately 330,000 Da (not shown). Examination of the elution profile revealed that the width of the peak corresponding to the HPr(Ser) kinase was similar to the width of the peaks generated by the marker proteins, suggesting that the HPr(Ser) kinase did not form a heterogeneous population of oligomers resulting from nonspecific aggregation in solution. On the other columns tested (Sephacryl S200 HR and Ultragel AcA-34), the activity eluted at a position close to the void volume, corresponding to a protein with a molecular mass larger than 250,000 Da.
Enzymatic properties of the HPr(Ser) kinase purified from S. salivarius.
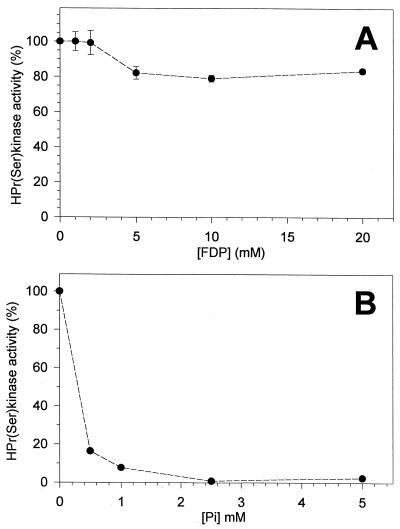
The enzyme was active over a pH range of 5.5 to 9.5, with a maximum at pH 7.5 and midpoints at pH 6.5 and 8.7 (not shown). At pH 5.5, the enzyme exhibited less than 15% of its optimal activity. We evaluated the pH stability of the enzyme by incubating the HPr(Ser) kinase at different pHs for 30, 60, and 90 min prior to measuring its activity. Stability was optimum at pH 8.0, decreasing two- to threefold at pH 7.0 and below and at pH 9.0 and above (not shown). The presence of a divalent cation is essential for the activity of the HPr(Ser) kinase of S. salivarius. Activity was observed in the presence of Mg2+, Mn2+, and Co2+ but not in the presence of Ca2+ and Cu2+ (not shown). Magnesium was the preferred cation and gave maximum activity at a concentration of 2 mM in the presence of 1 mM ATP. The effects of several cellular metabolites on the activity of the HPr(Ser) kinase were investigated. The following intermediates, tested at concentrations found in growing streptococcal cells (24), had no effect: 0.5 mM glucose-6-phosphate, 0.05 mM fructose-6-phosphate, 1 mM dihydroxyacetone phosphate, 0.2 mM glyceraldehyde phosphate, 0.25 mM 2-phosphoglycerate, 0.5 mM phosphoenolpyruvate, 1 mM pyruvate, 0.1 mM NADH, 1 mM NAD+, and 0.5 mM ADP. 3-Phosphoglycerate at a concentration of 2.0 mM and 2,3-diphosphoglycerate at a concentration of 5 mM reduced the activity of the HPr(Ser) kinase by approximately 50%. The effect of FDP was investigated in more detail, as this intermediate has been reported to stimulate the activity of the HPr(Ser) kinases of other gram-positive bacteria (3, 34, 35). Figure 5A illustrates the effect of FDP on the activity of the S. salivarius HPr(Ser) kinase. As can be seen, FDP at concentrations up to 20 mM had virtually no effect on activity. These experiments were conducted with 0.15 ng of HPr(Ser) kinase in the reaction mixtures. To test whether the effect of FDP was dependent on enzyme concentration, we performed experiments in the presence of 5 mM FDP with increased amounts of enzyme in the reaction mixtures (5 and 10 ng). In no case was FDP found to activate de HPr(Ser) kinase (not shown). Pi, on the other hand, was found to be a strong inhibitor (Fig. 5B).
FIG. 5.
Effects of FDP (A) and Pi (B) on HPr(Ser) kinase activity. Activity was measured as described in Materials and Methods; the concentration of ATP was 1 mM in all cases.
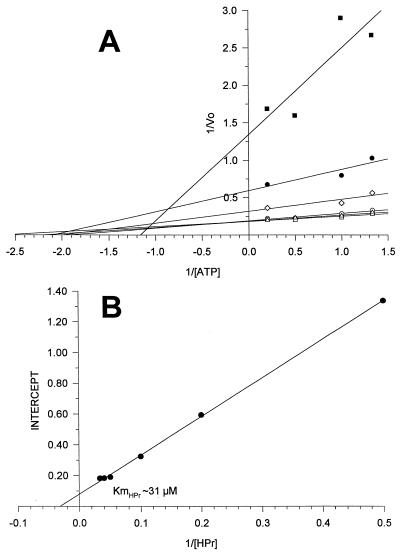
The Michaelis-Menten constant of the kinase for HPr and ATP was determined. Figure 6A shows the results obtained when the reciprocal of the initial velocity of HPr(Ser-P) formation was plotted as of function of the reciprocal of ATP concentration for various HPr concentrations. A family of straight lines with different slopes, characteristic of a ternary complex mechanism, was obtained. By plotting the intercept of each curve as a function of the reciprocal of HPr concentration (Fig. 6B), we found a Km for HPr of 31 μM. Using the same approach, we found a Km of approximately 1 mM for ATP. The addition of FDP at different concentrations did not modify the affinity of the enzyme for its substrates (not shown).
FIG. 6.
Determination of the Km of HPr(Ser) kinase for HPr. (A) Double-reciprocal plots of 1/V0 versus 1/ATP at various constant concentrations of HPr (2, 5, 10, 20, 25, and 30 μM). (B) The intercepts were used to obtain the Km of HPr (B).
DISCUSSION
We have reported in this communication the purification and characterization of the HPr(Ser) kinase as well as the cloning of the encoding gene, hprK. Sequence comparison of translated S. salivarius hprK demonstrated that the amino acid sequence of the HPr(Ser) kinase was highly conserved among streptococci. The similarity with proteins from other gram-positive bacteria was, however, lower. Interestingly, we observed that the C-terminal half of the protein contained more conserved residues than the N-terminal region. This dichotomous distribution of conserved amino acids suggests that the C-terminal portion of the HPr(Ser) kinase is vital for the phosphorylation of HPr at the expense of ATP, whereas the N-terminal region would be less important in catalytic functions. The sequence of the N-terminal region may have diverged during evolution, conferring on the enzyme idiosyncratic properties that are compatible with the physiology of species living in different ecological habitats. In S. salivarius, the N-terminal end of the HPr(Ser) kinase contains a high proportion of lysine, giving the amino-terminal end of the protein a basic isoelectric point. It seems likely that the high positive charge of the N terminus is involved in the attachment of the HPr(Ser) kinase to the inner surface of the cytoplasmic membrane by electrostatic interaction with negatively charged groups. This hypothesis is consistent with the observation that the HPr(Ser) kinase remained attached to the membrane after cell lysis and centrifugation of cellular extracts from S. salivarius and recombinant E. coli.
The molecular mass of the S. salivarius HPr(Ser) kinase calculated from the deduced amino acid sequence was 34,440 Da, a value consistent with the molecular mass determined under denaturing conditions either by SDS-PAGE or by gel filtration in the presence of guanidine hydrochoride (34,000 Da). The molecular mass of the denatured S. salivarius protein is similar to that of the B. subtilis HPr(Ser) kinase calculated from the nucleotide sequence of the encoding gene (34,668 Da) (12, 34) or determined by mass ion spray spectrometry (34,529 and 35,313 Da) (34). Determination of the molecular mass by gel filtration chromatography under nondenaturing conditions indicated that the size of the native protein was approximately 330,000 Da, suggesting that the enzyme is a decamer. Several results support the conclusion that this result was not the consequence of protein aggregation or nonspecific interactions between the HPr(Ser) kinase and the chromatographic support: (i) the activity was eluted from the column as a single sharp peak in a very reproducible manner; (ii) using 0.3 M KCl in the elution buffer did not change the elution pattern of the enzyme; (iii) the migration was not affected by the chemical composition of the chromatographic support; and (iv) the activity was eluted at the same position whether the experiment was carried out with a crude cellular extract, a partially purified enzyme, or a purified recombinant protein. Whether the decameric structure is required for activity remains to be determined.
Kinetic analyses of HPr phosphorylation by the HPr(Ser) kinase indicated that the process occurs via sequential reactions rather than a ping-pong mechanism. This finding is consistent with the observation that no phosphorylated derivative of the enzyme was observed. The Michaelis-Menten constant of the S. salivarius HPr(Ser) kinase for HPr was estimated at 31 μM, which is comparable to the value found for the S. pyogenes enzyme (35) but twofold lower than the Km reported for the B. subtilis enzyme (34). As the cellular concentration of HPr in S. salivarius is approximately 1 mM (15), it seems clear that the rate of HPr(Ser-P) synthesis in the cell is not limited by HPr concentration. On the other hand, the affinity of the enzyme for ATP was estimated as 1 mM. The concentration of ATP in actively growing streptococcal cells is approximately 4 mM and may drop to as low as 0.2 mM in starved cells (11). It is thus likely that the amount of cellular ATP is a rate-limiting factor for the phosphorylation of HPr at Ser46. The HPr kinase activity and stability decreased rapidly under acidic conditions. These properties are of importance considering the fact that oral streptococci are unable to maintain an intracellular pH above 7.0 in response to external acidic conditions (21, 26). Thus, acidification of the external milieu caused by sugar fermentation may modify the intracellular concentration of HPr(Ser-P) if the organic acids produced are not rapidly neutralized.
The activity of the S. salivarius HPr(Ser) kinase was not activated by FDP or any other glycolytic intermediates as opposed to the HPr(Ser) kinases of other gram-positive bacteria. Moreover, the presence of FDP had no effect on the affinity of the enzyme for its substrates. These results suggest that the intracellular levels of HPr(Ser-P) in oral streptococci are independent of FDP levels. Nonetheless, the relative proportions of the different forms of HPr vary in the cell with respect to growth conditions, HPr(Ser-P) and HPr(Ser-P)(His∼P) being the predominant forms in rapidly growing cells, while slowly growing or stationary-phase cells contain mostly free HPr and HPr(His∼P) (42, 44). We have observed that Pi is a strong inhibitor of S. salivarius HPr(Ser) kinase. Other studies have indicated that Pi activates HPr(Ser-P) phosphatase (5). Taking into account the effect of ATP on the rate of HPr phosphorylation as discussed above, it is proposed that the intracellular concentrations of the different forms of HPr in oral streptococci are governed by the relative cellular concentrations of ATP and Pi, which are indicators of the energy status of the cells.
ACKNOWLEDGMENTS
This research was supported by operating grant MT-6979 from the Medical Research Council of Canada.
We thank Michel Frenette and Louis André Lortie for stimulating discussions and technical support, and we thank Gene Bourgeau for editorial assistance. We thank Josef Deutscher and Jonathan Reizer for providing data on the HPr(Ser) kinase from B. subtilis prior to publication.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Greene Publishing and Wiley Interscience; 1997. [Google Scholar]
- 2.Chou P W, Fasman G D. Prediction of the secondary structure of proteins from their amino acid sequence. Adv Enzymol. 1978;47:45–148. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- 3.Deutscher J, Engelmann R. Purification and characterization of an ATP-dependent protein kinase from Streptococcus faecalis. FEMS Microbiol Lett. 1984;23:157–162. [Google Scholar]
- 4.Deutscher J, Saier M H., Jr ATP-dependent protein kinase-catalyzed phosphorylation of a seryl residue in HPr, a phosphate carrier protein of the phosphotransferase system in Streptococcus pyogenes. Proc Natl Acad Sci USA. 1983;80:6790–6794. doi: 10.1073/pnas.80.22.6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deutscher J, Kessler U, Hengstenberg W. Streptococcal phosphoenolpyruvate:sugar phosphotransferase system: purification and characterization of a phosphoprotein phosphatase which hydrolyzes the phosphoryl bond in seryl-phosphorylated histidine-containing protein J. Bacteriol. 1985;163:1203–1209. doi: 10.1128/jb.163.3.1203-1209.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deutscher J, Küster E, Bergstedt U, Charrier V, Hillen W. Protein kinase-dependent HPr/CcpA interaction links glycolytic activity to carbon catabolite repression in gram-positive bacteria. Mol Microbiol. 1995;15:1049–1053. doi: 10.1111/j.1365-2958.1995.tb02280.x. [DOI] [PubMed] [Google Scholar]
- 7.Deutscher J, Reizer J, Fischer C, Galinier A, Saier M H, Jr, Steinmetz M J. Loss of protein kinase-catalyzed phosphorylation of HPr, a phosphocarrier protein of the phosphotransferase system, by mutation of the ptsH gene confers catabolite repression resistance to several catabolic genes of Bacillus subtilis. J Bacteriol. 1994;176:3336–3344. doi: 10.1128/jb.176.11.3336-3344.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duclos B, Marcandier S, Cozzone A J. Chemical properties and separation of phosphoamino acids by thin-layer chromatography and/or electrophoresis. Methods Enzymol. 1991;201:10–21. doi: 10.1016/0076-6879(91)01004-l. [DOI] [PubMed] [Google Scholar]
- 9.Ferrari S, Thomas G. Micro- and macropurification methods for protein kinases. Methods Enzymol. 1991;200:159–169. doi: 10.1016/0076-6879(91)00136-k. [DOI] [PubMed] [Google Scholar]
- 10.Fujita Y, Miwa Y, Galinier A, Deutscher J. Specific recognition of the Bacillus subtilis gnt cis-acting catabolite-responsive element by a protein complex formed between CcpA and seryl-phosphorylated HPr. Mol Microbiol. 1995;17:953–960. doi: 10.1111/j.1365-2958.1995.mmi_17050953.x. [DOI] [PubMed] [Google Scholar]
- 11.Fukui K, Kato K, Kodama T, Ohta H, Shimamoto T, Shimono T. Kinetic study of a change in intracellular ATP level associated with aerobic catabolism of ethanol by Streptococcus mutans. J Bacteriol. 1988;170:4589–4593. doi: 10.1128/jb.170.10.4589-4593.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galinier A, Kravanja M, Engelman R, Hengstenberg W, Kilhoffer M C, Deutscher J, Haiech J. New protein kinase and protein phosphatase families mediate signal transduction in bacterial catabolite repression. Proc Natl Acad Sci USA. 1998;95:1823–1828. doi: 10.1073/pnas.95.4.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garnier J, Robson B. In: Prediction of protein structure and the principles of protein conformation. Fasman G D, editor. New York, N.Y: Plenum; 1989. pp. 417–466. [Google Scholar]
- 14.Gauthier L, Thomas S, Gagnon G, Frenette M, Trahan L, Vadeboncoeur C. Positive selection for resistance to 2-deoxyglucose gives rise, in Streptococcus salivarius, to seven classes of pleiotropic mutants, including ptsH and ptsI missense mutants. Mol Microbiol. 1994;13:1101–1109. doi: 10.1111/j.1365-2958.1994.tb00501.x. [DOI] [PubMed] [Google Scholar]
- 15.Gauthier M, Brochu D, Eltis L D, Thomas S, Vadeboncoeur C. Replacement of isoleucine-47 by threonine in the HPr protein of Streptococcus salivarius abrogates the preferential metabolism of glucose and fructose over lactose and melibiose but does not prevent the phosphorylation of HPr on serine-46. Mol Microbiol. 1997;25:695–705. doi: 10.1046/j.1365-2958.1997.4981870.x. [DOI] [PubMed] [Google Scholar]
- 16.Genetics Computer Group Inc. Wisconsin Package, version 9.0. Madison, Wis: Genetics Computer Group, Inc.; 1996. [Google Scholar]
- 17.Gorbach S L. Lactic acid bacteria and human health. Ann Med. 1990;22:37–41. doi: 10.3109/07853899009147239. [DOI] [PubMed] [Google Scholar]
- 18.Gösseringer R, Küster E, Galinier A, Deutscher J, Hillen W. Cooperative and non-cooperative DNA binding modes of catabolite control protein CcpA from Bacillus megaterium result from sensing two different signals. J Mol Biol. 1997;266:665–676. doi: 10.1006/jmbi.1996.0820. [DOI] [PubMed] [Google Scholar]
- 19.Hamilton I R. Synthesis and degradation of intracellular polyglucose in Streptococcus salivarius. Can J Microbiol. 1968;14:65–77. doi: 10.1139/m68-011. [DOI] [PubMed] [Google Scholar]
- 20.Hamilton I R. Effect of changing environment on sugar transport and metabolism by oral bacteria. In: Reizer J, Peterkofsky A, editors. Sugar transport and metabolism by Gram-positive bacteria. Chichester, England: Ellis Horwood; 1987. pp. 94–133. [Google Scholar]
- 21.Hamilton I R. Maintenance of proton motive force by Streptococcus mutans and Streptococcus sobrinus during growth in continuous culture. Oral Microbiol Immunol. 1990;5:280–287. doi: 10.1111/j.1399-302x.1990.tb00426.x. [DOI] [PubMed] [Google Scholar]
- 22.Hueck C J, Hillen W. Catabolite repression in Bacillus subtilis: a global regulatory mechanism for the Gram-positive bacteria? Mol Microbiol. 1995;15:395–401. doi: 10.1111/j.1365-2958.1995.tb02252.x. [DOI] [PubMed] [Google Scholar]
- 23.Hueck C J, Hillen W, Saier M H., Jr Analysis of a cis-active sequence mediating catabolite repression in Gram-positive bacteria. Res Microbiol. 1994;145:503–518. doi: 10.1016/0923-2508(94)90028-0. [DOI] [PubMed] [Google Scholar]
- 24.Iwami Y, Yamada T, Araya S. Glycolytic intermediates in Streptococcus mutans PK1. Arch Oral Biol. 1975;20:695–697. doi: 10.1016/0003-9969(75)90140-5. [DOI] [PubMed] [Google Scholar]
- 25.Jones B E, Dossonnet V, Küster E, Hillen W, Deutscher J, Klevit R E. Binding of the catabolite repressor protein CcpA to its DNA target is regulated by phosphorylation of its corepressor HPr. J Biol Chem. 1997;272:26530–26535. doi: 10.1074/jbc.272.42.26530. [DOI] [PubMed] [Google Scholar]
- 26.Kashket S, Kashket E R. Dissipation of the proton motive force in oral streptococci by fluoride. Infect Immun. 1985;48:19–22. doi: 10.1128/iai.48.1.19-22.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krüger S, Gertz S, Hecker M. Transcriptional analysis of bglH expression in Bacillus subtilis: evidence for two distinct pathways mediating carbon catabolite repression. J Bacteriol. 1996;178:2637–2644. doi: 10.1128/jb.178.9.2637-2644.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Küster E, Luesink E J, de Vos W M, Hillen W. Immunological crossreactivity to the catabolite control protein CcpA from Bacillus megaterium is found in many Gram-positive bacteria. FEMS Microbiol Lett. 1996;139:109–115. doi: 10.1111/j.1574-6968.1996.tb08188.x. [DOI] [PubMed] [Google Scholar]
- 29.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 30.Marshall V M. Lactic acid bacteria: starters for flavour. FEMS Microbiol Rev. 1987;46:327–336. [Google Scholar]
- 31.Martin-Verstraete I, Stülke J, Klier A, Rapoport G. Two different mechanisms mediate catabolite repression of the Bacillus subtilis levanase operon. J Bacteriol. 1995;177:6919–6927. doi: 10.1128/jb.177.23.6919-6927.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meadow N D, Fox D K, Roseman S. The bacterial phosphoenolpyruvate:glycose phosphotransferase system. Annu Rev Biochem. 1990;59:497–542. doi: 10.1146/annurev.bi.59.070190.002433. [DOI] [PubMed] [Google Scholar]
- 33.Postma P W, Lengeler J W, Jacobson G R. Phosphoenolpyruvate:carbohydrate phosphotransferase system of bacteria. Microbiol Rev. 1993;57:543–594. doi: 10.1128/mr.57.3.543-594.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reizer J, Hoischen C, Titgemeyer F, Rivolta C, Rabus R, Stülke J, Karamata D, Saier M H, Jr, Hillen W. A novel protein kinase that controls carbon catabolite repression in bacteria. Mol Microbiol. 1998;27:1157–1169. doi: 10.1046/j.1365-2958.1998.00747.x. [DOI] [PubMed] [Google Scholar]
- 35.Reizer J, Novotny M J, Hengstenberg W, Saier M H., Jr Properties of ATP-dependent protein kinase from Streptococcus pyogenes that phosphorylates a seryl residue in HPr, a phosphocarrier protein of the phosphotransferase system. J Bacteriol. 1984;160:333–340. doi: 10.1128/jb.160.1.333-340.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saier M H, Jr, Reizer J. Proposed uniform nomenclature for the proteins and protein domains of the bacterial phosphoenolpyruvate:sugar phosphotransferase system. J Bacteriol. 1992;174:1433–1438. doi: 10.1128/jb.174.5.1433-1438.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saier M H, Jr, Chauvaux S, Cook G M, Deutscher J, Paulsen I T, Reizer J, Ye J J. Catabolite repression and inducer control in Gram-positive bacteria. Microbiology. 1996;142:217–230. doi: 10.1099/13500872-142-2-217. [DOI] [PubMed] [Google Scholar]
- 38.Salyers A A, Whitt D D. Bacterial pathogenesis: a molecular approach. Washington, D.C: ASM Press; 1994. [Google Scholar]
- 39.Sambrook J E, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 40.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simpson C L, Russell R B B. Identification of a homolog of CcpA catabolite repressor protein in Streptococcus mutans. Infect Immun. 1998;66:2085–2092. doi: 10.1128/iai.66.5.2085-2092.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thevenot T, Brochu D, Vadeboncoeur C, Hamilton I R. Regulation of ATP-dependent P-(Ser)-HPr formation in Streptococcus mutans and Streptococcus salivarius. J Bacteriol. 1995;177:2751–2759. doi: 10.1128/jb.177.10.2751-2759.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vadeboncoeur C, Pelletier M. The phosphoenolpyruvate:sugar phosphotransferase system of oral streptococci and its role in the control of sugar metabolism. FEMS Microbiol Rev. 1997;19:187–207. doi: 10.1111/j.1574-6976.1997.tb00297.x. [DOI] [PubMed] [Google Scholar]
- 44.Vadeboncoeur C, Brochu D, Reizer J. Quantitative determination of the intracellular concentration of the various forms of HPr, a phosphocarrier protein of the phosphoenolpyruvate:sugar phosphotransferase system in growing cells of oral streptococci. Anal Biochem. 1991;196:24–30. doi: 10.1016/0003-2697(91)90112-7. [DOI] [PubMed] [Google Scholar]
- 45.Rodrigue L, Lacoste L, Trahan L, Vadeboncoeur C. Effect of nutritional constraint on the biosynthesis of the components of the phosphoeolpyruate:sugar phosphotransferase system in a fresh isolate of Streptococcus mutans. Infect Immun. 1988;56:518–522. doi: 10.1128/iai.56.2.518-522.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vector and host strains: nucleotide sequences of the m13mp18 and pUC19 vector. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 47.Ye J J, Reizer J, Saier M H., Jr Regulation of 2-deoxyglucose phosphate accumulation in Lactococcus lactis vesicles by metabolite-activated, ATP-dependent phosphorylation of serine-46 in HPr of the phosphotransferase system. Microbiology. 1994;140:3421–3429. doi: 10.1099/13500872-140-12-3421. [DOI] [PubMed] [Google Scholar]
- 48.Ye J J, Reizer J, Cui X, Saier M H., Jr ATP-dependent phosphorylation of serine in HPr regulates lactose: H+ symport in Lactobacillus brevis. Proc Natl Acad Sci USA. 1994;91:3102–3106. doi: 10.1073/pnas.91.8.3102. [DOI] [PMC free article] [PubMed] [Google Scholar]