Abstract
Whether adult hippocampal neurogenesis (AHN) persists in adult and aged humans continues to be extensively debated. A major question is whether the markers identified in rodents are reliable enough to reveal new neurons and the neurogenic trajectory in primates. Here, to provide a better understanding of AHN in primates and to reveal more novel markers for distinct cell types, droplet-based single-nucleus RNA sequencing (snRNA-seq) is used to investigate the cellular heterogeneity and molecular characteristics of the hippocampi in macaques across the lifespan and in aged humans. All of the major cell types in the hippocampus and their expression profiles were identified. The dynamics of the neurogenic lineage was revealed and the diversity of astrocytes and microglia was delineated. In the neurogenic lineage, the regulatory continuum from adult neural stem cells (NSCs) to immature and mature granule cells was investigated. A group of primate-specific markers were identified. We validated ETNPPL as a primate-specific NSC marker and verified STMN1 and STMN2 as immature neuron markers in primates. Furthermore, we illustrate a cluster of active astrocytes and microglia exhibiting proinflammatory responses in aged samples. The interaction analysis and the comparative investigation on published datasets and ours imply that astrocytes provide signals inducing the proliferation, quiescence and inflammation of adult NSCs at different stages and that the proinflammatory status of astrocytes probably contributes to the decrease and variability of AHN in adults and elderly individuals.
Subject terms: Neural stem cells, Ageing
Introduction
The hippocampal formation is one of the main brain regions affected in neurological diseases, such as Alzheimer’s disease, stress, and depression; the vulnerability and plasticity of hippocampal formation during aging have attracted tremendous attention due to its physiological and clinical significance.1,2 Whether adult hippocampal neurogenesis (AHN) persists in adult and aged humans continues to be extensively debated.3–10 Extensive evidence in animal models demonstrates that hippocampal neurogenesis is involved in memory processing, cognitive function, and mood regulation.11,12 There is no doubt that adult neurogenesis slows down in adults and aged animals compared to that in young animals. However, controversy still exists regarding whether AHN persists in aged humans, although remarkable efforts have been attempted, including using different antibodies, shortening the intervals of postmortem sampling, and improving the fixation conditions and procedures for immunostaining.4,7,9,10,13–15 The key concerns are whether the markers used in mice are powerful enough to reveal young neurons in humans, whether there are more robust and reliable markers in humans, and whether there are still proliferating neural stem cells (NSCs) in the dentate gyrus of aged humans.6 Recently, another round of debate was ignited.5,8 Despite the conflicting results, a consensus was reached that new technologies, such as single-cell RNA-seq, will provide new insights in this field and reveal novel markers for immature neurons and adult NSCs.
Single-cell/single-nucleus RNA sequencing (scRNA-seq/snRNA-seq) is a powerful technique to unbiasedly evaluate the expression profile, cellular diversity and heterogeneity of tissues at a single-cell resolution.16–18 Accumulating evidence has demonstrated that scRNA-seq has a great advantage in revealing cell types in the tissues examined, especially rare cell populations.19–21 Previous studies in the field of adult neurogenesis have provided insightful knowledge on cellular heterogeneity and lineage,22,23 as well as quiescent NSC maintenance and activation,24–29 in the ventricular–subventricular zone (V-SVZ) and hippocampus of mice. Regarding the controversial questions in adult human hippocampal neurogenesis, we believe that a systematic investigation of the postnatal development of hippocampi in primates will provide more promising and definitive information on the dynamic regulation of the postanal development of the hippocampus and adult neurogenesis.
Therefore, in this study, we evaluated 13 macaques across the lifespan and four human donors using droplet-based snRNA-seq. The dynamics of the landscape in primate hippocampi were depicted. In the neurogenic lineage, immature were readily revealed in macaques and humans. Several new markers in the cluster of newborn neurons, as well as new NSC markers, were revealed and verified with immunostaining in vivo. We also observed a cluster of active astrocytes and microglia exhibiting proinflammatory responses in aged samples. The interaction analysis of distinct cell types implied that inflammatory circumstances might be responsible for the decreased proliferation of NSCs and contribute to the variable hippocampal neurogenesis.
Results
Dynamic landscape of the hippocampal formation in macaques
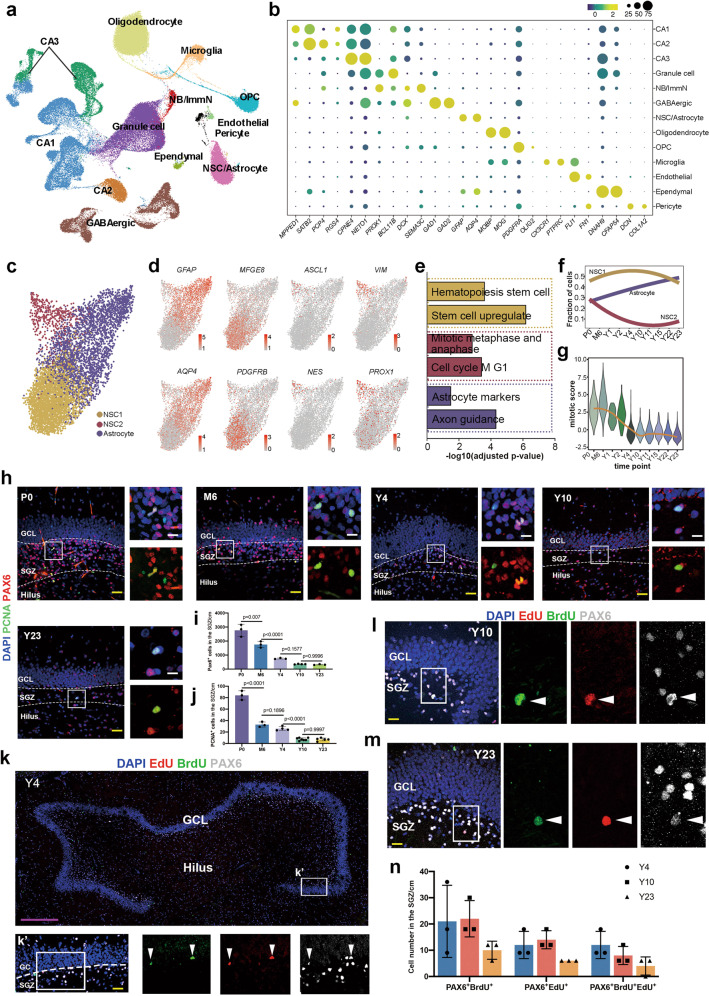
To obtain a better understanding of the postnatal development of primate hippocampi and elucidate how hippocampal neurogenesis is regulated across the lifespan, we conducted sampling and sequencing of 13 macaque hippocampi from the postnatal to aging stages (Supplementary information, Table S1). Among the 13 samples, 8 samples containing both cornu ammonis (CA) and dentate gyrus (DG) regions were sequenced; the CA and DG subregions were sequenced separately in the remaining samples (Supplementary information, Fig. S1a, b). In total, 132,524 nuclei with a median of 2203 genes detected per nucleus were collected after stringent quality control and doublet removal, and all nuclei from the 13 samples were integrated after batch effect removal to preserve biological variation (Supplementary information, Fig. S1a, b). Unsupervised clustering analysis and uniform manifold approximation and projection (UMAP) visualization were performed, and 13 major cell types were identified based on the transcriptomic differentially expressed genes (DEGs) and cell-type unique markers reported previously22,30,31 (Fig. 1a, b; Supplementary information, Fig. S1c and Table S2).
Fig. 1. Transcriptional profiles of the macaque hippocampi at single-nucleus resolution.
a Clustering of individual hippocampal cells from macaques of different ages via UMAP. Cells are colored by cluster annotation. b Dot plot showing the average expression of marker genes for the 13 main cell types in a. CA cornu ammonis, ImmN immature neuron, NSC/Astrocyte Neural stem cell/Astrocyte, OPC oligodendrocyte progenitor. c Visualization of NSC/Astrocyte subclusters using UMAP. d The expression of DEGs among the three distinct NSC/Astrocyte subclusters was projected onto UMAP. Cells are colored according to the gene expression levels (red, high; gray, low). e Bar plot of enriched GO terms in NSC/Astrocyte subclusters. f Fitted curves show the fraction of cells from three NSC/Astrocyte subclusters at different time points. g Mitotic score analysis shows that the proliferation capacity of NSCs in the SGZ of the hippocampi of macaques declined with age. h–j The number of NSCs and proliferating NSCs declined with age, as revealed by the NSC marker PAX6 and the proliferating cell marker PCNA. Sidak’s multiple comparisons test was used, and each dot represents a single experiment. Data are shown as means ± SD. Scale bar in yellow, 50 µm; scale bar in white, 20 µm. k–m Adult NSCs in the DG of the hippocampus undergo consecutive proliferation in macaques, as evidenced by EdU/BrdU/PAX6 triple staining. The arrowheads indicate EdU/BrdU/Pax6 triple-positive cells. Scale bar in magenta, 500 µm; scale bar in yellow, 50 µm. n Quantification of staining for the neural stem cell marker PAX6 with EdU and BrdU. Sidak’s multiple comparisons test was used, and each dot represents a single experiment. Data are shown as means ± SD. See also Supplementary information, Figs. S1 and S2.
The DG and CA subregions contain distinct cell types and have different physiological functions.30,32 In addition to the unique cell types, for example, pyramidal neurons (CA1–3) in the CA regions and GCs, immature neurons (ImmN) and adult NSCs in the DG subregions, other major cell types (Astrocyte, OPC, Oligodendrocyte, GABAergic interneuron and Microglia) exist in both subregions (Fig. 1a). Due to the great similarity of NSCs and astrocytes, these cells were clustered together. To delineate the developmental dynamics and specificity of NSCs in the primate hippocampi, astrocytes from the CA subregions and DG subregions should be identified correctly. To this end, we first scrutinized the NSC/Astrocyte cluster from the CA and the DG samples being sequenced separately, which possessed the spatial information. The cells from the two regions showed a region-specific expression profile. DEGs indicated that MEIS2 was a marker for CA astrocytes (Supplementary information, Fig S2a–d and Table S3). This result was confirmed with immunostaining, and we found that most GFAP positive astrocytes in the CA regions were MEIS2 positive, but this was not the case for those in the DG region (Supplementary information, Fig. S2e). Accordingly, the NSCs/Astrocytes from the DG subregions were readily isolated based on the spatial DEGs.
Next, we analyzed the NSCs/Astrocytes residing in the DG subregions. Three subclusters with distinct gene expression profiles were identified (Fig. 1c, d; Supplementary information, Table S4). According to the DEGs and Gene Ontology (GO) enrichment analysis, the clusters were annotated as Astrocyte, NSC1, and NSC2. Mature astrocytic markers, such as GFAP, AQP4, CD44 and ITGB4, were preferentially expressed in the Astrocyte cluster. The NSC1 subpopulation was delineated by the dominant expression of the well-documented quiescent NSC markers MFGE8 and PDGFRB, while the NSC2 subcluster prominently expressed ASCL1, PROX1, NES, and VIM, which represented active NSCs (Fig. 1d, e). The distribution of the three subpopulations in each sample showed that the proportion of active NSC2 decreased, indicating decreased proliferation in the adult and aged brains (Fig. 1f). Additionally, the mitotic score, which represents the mitotic capacity of the NSCs, sharply declined from 2-year-old (Y2) onward and then remained stable until Y23 (Fig. 1g). This conclusion was verified with the immunofluorescent results of the NSC marker PAX6 and the proliferative marker PCNA, which showed that the number of proliferating cells and the number of adult stem cells declined from the neonatal to the aged samples (Fig. 1h–j). However, compared to previous studies that rarely observed proliferative NSCs located in the subgranular zone (SGZ),2 we did observe nearly 10 cells coexpressing both PAX6 and PCNA in each section in Y23 macaques. Furthermore, we traced the proliferation of NSCs and adult neurogenesis by performing consecutive EdU and BrdU labeling in adult and aging Macaca mullata (Y4, Y10, and Y23). EdU, BrdU and PAX6 were used to evaluate the continuous proliferation of adult NSCs. We detected NSCs expressing PAX6 that incorporated both EdU and BrdU, even in 23-year-old macaques (Fig. 1k–n), indicating that these adult NSCs entered the S-phase twice in the experimental time window. Overall, we found that the proliferation of NSCs declined with age but still existed even in aged macaques.
Divergent characteristics of adult NSCs in macaques and rodents
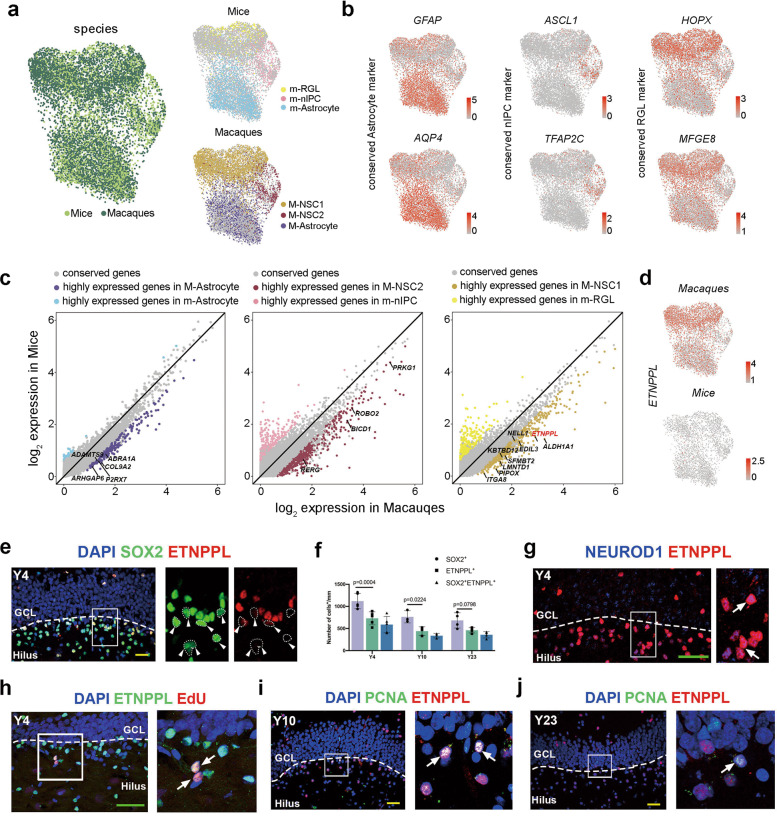
Next, we addressed whether primate NSCs/Astrocytes exhibited shared and distinct features compared with those of the well-studied rodent models. We compared our dataset with the dataset containing information on postnatal stages in mice.22 Cells in the Astrocyte, NSC1, and NSC2 clusters matched perfectly with mouse astrocytes, Radial glial-like cells (RGLs) and neural intermediate progenitor cells (nIPCs), respectively (Fig. 2a, b). The divergent and conserved molecular signatures of distinct cell types were present (Fig. 2c). Then, we focused on the expressional features of macaque NSCs and mouse RGLs. Most expressed genes were conserved between macaques and mice (9470/10300), 307 genes were highly enriched in mice, and 523 genes were prominently abundant in macaques (Fig. 2c; Supplementary information, Table S5). By assessing the expression of the 523 genes in the Allen Brain RNA-ISH Database (https://mouse.brain-map.org), we found that 29 of these genes were undetectable in the DG region of the mouse hippocampus (Supplementary information, Table S5), which provided a reservoir of macaque-specific NSC signatures. For instance, the expression of ETNPPL (ethanol phosphate phospholyase, an enzyme involved in lipid metabolism) was markedly detected in the NSCs of macaques but was not expressed in mice at either the RNA or protein levels (Fig. 2d; Supplementary information, Fig. S3a, b). The expression of ETNPPL was readily observed in the SGZ of adult and aged macaques (Fig. 2e–j, Supplementary information, Fig. S3c). Coexpression analysis with the traditional NSC marker SOX2 showed that ETNPPL was expressed in a subset of SOX2-positive cells (Fig. 2e), and the significance of the difference decreased with age (Fig. 2f). Moreover, a portion of ETNPPL also colocalized with the committed neurogenic progenitor marker NEUROD118,33 but rarely with the astrocytic marker Vim and did not colocalize with the microglial marker IBA1 (Fig. 2g; Supplementary information, Fig. S3d–f), suggesting the neurogenic nature of ETNPPL+ NSCs. Then, the colocalization of ETNPPL with S-phase or M-phase markers, such as EdU and PCNA, indicated that some of the ETNPPL-expressing cells were mitotically active (Fig. 2h–j; Supplementary information, Fig. S3c). In general, we identified a group of primate-specific NSC markers and verified the reliability of ETNPPL at both the RNA and protein levels.
Fig. 2. ETNPPL is identified as a primate-specific NSC marker.
a UMAP showing the integration of macaques and mouse cells (left). UMAP showing the distribution of mouse cells (right-upper panel) and macaque cells (right-lower panel) in the integrated dataset. b The expression of conserved genes in mice and macaques among the three distinct NSC/Astrocyte subclusters. Cells are colored according to the gene expression levels (red, high; gray, low). c Scatter plot showing species-enriched genes in mice and macaques among the three distinct NSC/Astrocyte subclusters. d Featureplot showing ETNPPL expression in macaques (upper panel) but not in mice (lower panel). e, f Colocalization analysis of SOX2 and ETNPPL showed that most ETNPPL+ cells coexpressed SOX2, and the arrowheads indicate SOX2+ETNPPL− cells. g A portion of ETNPPL-expressing cells coexpressing the committed neurogenic progenitor marker NEUROD1; the arrows indicate ETNPPL/NEUROD1 double-positive cells. h Immunostaining showing ETNPPL colocalization with EdU in 4-year-old brain tissue. The arrows indicate ETNPPL/EdU double-positive cells. ETNPPL+ PCNA+ cells in the SGZ of the hippocampi of 10-year-old (i) and 23-year-old (j) macaques. Scale bar in green, 80 µm; scale bar in yellow, 50 µm. The arrows indicate ETNPPL/PCNA double-positive cells. See also Supplementary information, Fig. S3.
AHN continues in the adult and aged macaques
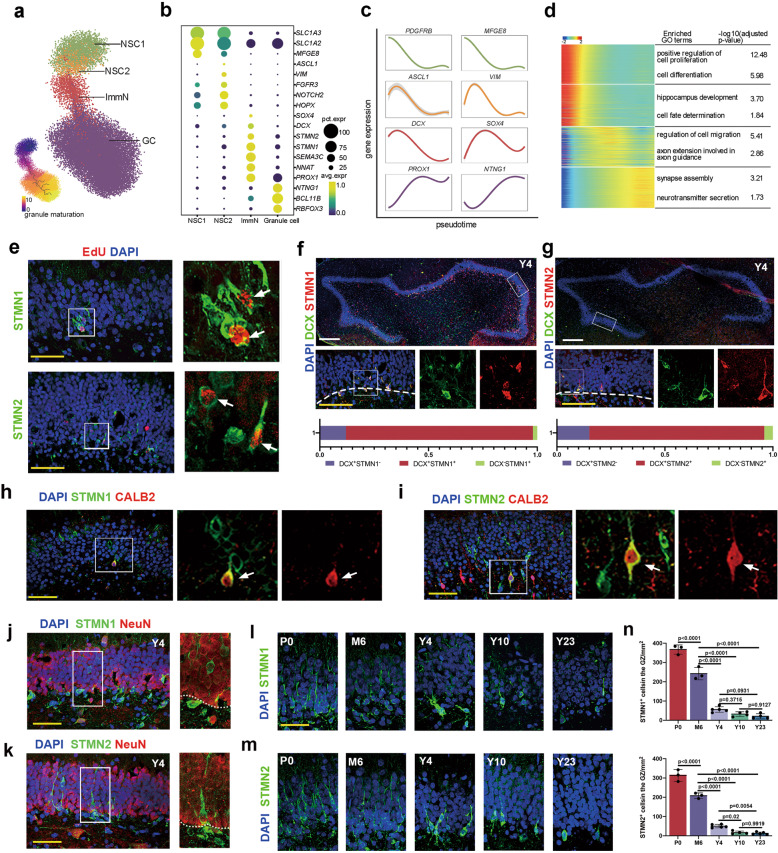
With the identified NSCs, we next investigated the developmental process and regulatory continuum of postnatal and adult neurogenesis in macaques. The neurogenic lineage-related clusters (NSC1, NSC2, ImmN, and GC) were chosen for further analysis (Fig. 3a). Representative markers for each cell type are highlighted (Fig. 3b). Cells of ImmN cluster highly expressed classical immature neuronal markers, such as SOX4, SEMA3C, NNAT, and PROX1 (Supplementary information, Fig. S4a), which was supposed to contain both migrating neuroblasts (also named early immature GCs) and late immature GCs.11,31,34 To investigate the subtypes of immature neurons, ImmN cluster were divided into two distinct subclusters, including a group of neuroblast-like(NB-like)cells expressing genes ELAVL2 and DCX and a group of late immature GCs (Late-ImmGC) expressing genes RASGRF2 and TENM1 (Supplementary information, Fig. S4b, c), as reported in mouse immature neuron.22 The trajectory inference with monocle3 showed that NSC1 transitioned to NSC2, followed by ImmN, and then differentiated into mature GC (Fig. 3a lower panel). The expression of DEGs along the pseudotime plot exactly recapitulated the regulatory continuum of the process of adult neurogenesis (Fig. 3c). An analysis of enriched GO terms validated the biological process and function of each cell type, such as ‘cell proliferation’ for NSC, ‘cell migration’ and ‘axon extension’ for ImmN, and ‘neurotransmitter secretion’ and ‘synapse assembly’ for mature GC (Fig. 3d).
Fig. 3. Hippocampal neurogenesis analysis across the lifespan of macaques.
a The developmental trajectory of the neurogenic lineage is visualized using UMAP. The pseudotime analysis of individual cells is also shown (bottom left panel). b Dot plot showing the average expression of DEGs among different cell types in a. c Fitted curve showing the expression of representative genes over pseudotime. d Heatmap illustrating the expression of genes that covary across pseudotime. The enriched GO terms are also shown. e The colocalization of STMN1 and STMN2 with EdU indicates that these cells are generated via mitosis. The arrows indicate STMN1/EdU- or STMN2/EdU-positive cells. Scale bar in white, 500 µm; scale bar in yellow, 50 µm. f, g Immunostaining analysis showing STMN1/DCX (86%) and STMN2/DCX (81%) double-positive cells. h–k Colocalization analysis of STMN1 (h) and STMN2 (i) with CALB2; the arrows indicate colocalization. Colocalization analysis of STMN1 (j) and STMN2 (k) with NeuN. i–n Time-course analysis and quantification of STMN1 and STMN2 expression in the DG of the hippocampus indicating decreased but persistent neurogenesis in the aged brains. Sidak’s multiple comparisons test was used, and each dot represents a single experiment. Data are shown as means ± SD. Scale bar in white, 500 µm; scale bar in yellow, 50 µm. See also Supplementary information, Figs. S4–S6.
To assess adult hippocampal neurogenesis in adult and aged macaques, we applied the widely used young neuron markers DCX and PSA-NCAM to hippocampal tissues in the Y4, Y10 and Y23 samples. Abundant DCX and PSA-NCAM double-positive young neurons exhibiting typical immature GC morphology were found in the DG of the hippocampus in 4-year-old macaques. In the 10- and 23-year-old samples, DCX+ PSN-NCAM+ cells were also readily detected; however, their total number was considerably decreased compared to those in 4-year-old macaques (Supplementary information, Fig. S5a, b).
To consolidate these DCX-positive immature neurons were produced from NSCs via mitosis, an EdU labeling assay was conducted. Briefly, EdU (5 mg/kg body weight) was administered to macaques intravenously twice a week for 10 weeks. The animals were sacrificed after 4 months for further analysis (see the details in the Materials and Methods). In all EdU-injected monkeys, most cells with EdU-labeled nuclei were observed in the DG. By costaining with DCX, a large number of EdU+ DCX+ double-positive cells were observed in the SGZ and GCL of the hippocampal formations in the 4-, 10- and 23-year-old macaques (Supplementary information, Fig. S5c–f). We also observed that some EdU-positive cells were colabeled with the mature GC marker NeuN (Supplementary information, Fig. S5g). These results indicated that there were newly generated neurons in the DG of adult and aged hippocampal formations via mitosis. Thus, we detected a group of immature GCs in the adult and aged hippocampal DG in macaques with widely used markers for young neurons and EdU-labeling assays.
STMN1 and STMN2 are new markers for immature neurons in macaques
Among the genes enriched in the ImmN cluster, STMN1 and STMN2, two members of the Stathmin family, were specifically and predominantly expressed together with the classic markers DCX and PROX1 (Fig. 3b). STMN1 or STMN2 are reportedly expressed in the neural progenitor cells of the hippocampus in mice,28 as well as mature neurons in the neocortex of humans.35 In our datasets, both genes were heavily expressed in immature GCs at the transcriptional level, implying distinct functions in different species and cellular contexts. Then, the expression of STMN1 or STMN2 was validated by immunostaining. In the newborn samples, abundant STMN1- and STMN2-positive cells were detected, which colocalized with DCX (Supplementary information, Fig. S6a). We precisely identified the characteristics of these two markers by further verifying their expression in samples from 4-year-old animals. First, some STMN1+ or STMN2+ cells were also EdU positive (Fig. 3e), which suggested that they were generated via the mitosis of NSCs. Next, the colocalization analysis showed that STMN1 or STMN2 exhibited over 80% overlap with DCX and PSA-NCAM, which implied the reliability of revealing immature GCs (Fig. 3f, g; Supplementary information, Fig. S6b). We also detected that some STMN1+ or STMN2+ cells were colabeled with Calb2, a transiently-expressing newborn neuron marker36 (Fig. 3h, i), but were less positive for either SOX2 or NEUROD1 (Supplementary information, Fig. S6c–g). Furthermore, the colabeling of STMN1 or STMN2 with NeuN, a late expressed neuronal marker, showed that 88% of STMN1+ cells and 52% of STMN2+ cells were NeuN-negative (Fig. 3j, k), suggesting that these two genes were selectively expressed by nascent neurons. Finally, the time-course analysis revealed a substantial decrease in the number of STMN1- and STMN2-positive immature GCs in aged samples (Fig. 3l–n). In summary, we determined the specific developmental continuum of adult hippocampal neurogenesis at the single-cell level and ascertained that adult neurogenesis continues in aged macaques. We also detected a reservoir of macaque-specific signatures in NSCs and validated STMN1 and STMN2 as reliable immature neuronal markers.
Neuroinflammation contributes to the decreased AHN in aged macaques
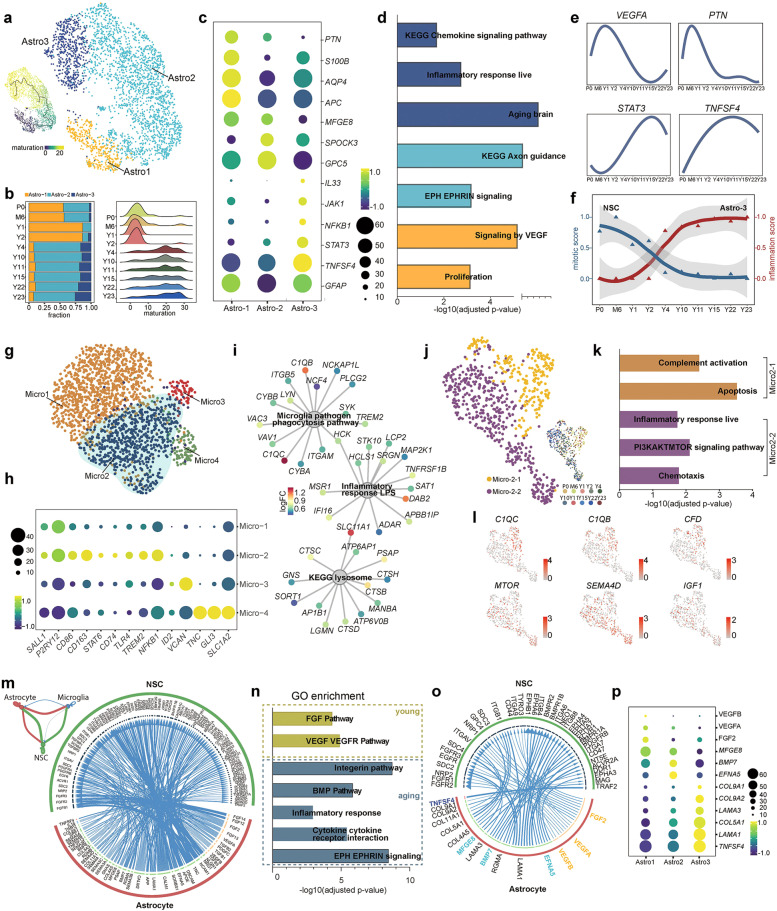
A broad body of evidence indicates that adult neurogenesis and the quiescence of adult NSCs are sophistically orchestrated by both intrinsic and extrinsic factors.37,38 Niche cells provide the major source of extrinsic regulators. Among them, astrocytes play fundamental roles in the proliferation and maintenance of adult NSCs.39–42 We subgrouped astrocytes into 3 distinct clusters to investigate the transcriptome dynamics and putative function of astrocytes in the DG of the hippocampus (Fig. 4a). Young samples were predominantly enriched in Astro1 and Astro2, while adult and aged samples were abundant in Astro2 and Astro3 (Fig. 4b). The maturation analysis indicated that astrocytes from young stages (P0–Y2) were less mature than those from older stages (Y4–Y23) (Fig. 4b). DEGs and GO term enrichment analyses indicated that Astro1 was prominently associated with VEGF signaling and proliferation function, and Astro2 entailed mature astrocyte functions, such as axonal guidance. Astro3 was an active astrocyte population that was responsible for chemokine signal transduction and the inflammatory response, expressing NFKB1, JAK1, STAT3, IL33, etc. (Fig. 4c, d; Supplementary information, Fig. S7a). Representative gene expression also confirmed that proliferation-related factors decreased with age, whereas inflammation-related gene expression increased (Fig. 4e).
Fig. 4. Transcriptional heterogeneity of cells derived from macaque astrocytes and microglia.
a UMAP showing the clustering of astrocytes with cells colored by astrocyte subclusters (right panel) and pseudotime (left panel). b The fraction of cells in three distinct astrocyte subclusters at different time points is shown in the bar plot (left panel); the distribution of pseudotime values at each time point is visualized in a ridge plot (right panel). c Dot plot displaying the average expression of DEGs among the three astrocyte subclusters. d GO enrichment analysis shows the cell properties of three astrocyte subclusters. e Smoothed relative expression of the proliferation regulation-related genes VEGFA and PTN and the inflammatory-related genes STAT3 and TNFSF4 over pseudotime. f Graph showing the smoothed mitotic score and inflammation score at different time points for NSCs and Astro3 cells. g UMAP showing the clustering of microglial cells. h Dot plot showing the average expression of DEGs among 4 different microglial clusters. i Network plot of enriched GO terms in Micro2. Nodes for genes were colored by log2fold change computed as the ratio of gene expression between Micro2 and the rest of human microglia (Micro1 and Micro3) derived from analysis of DEGs. j Subclusters of activated Micro2 microglia. Cells are colored by cell type (left panel) and age (right panel). k Enriched GO terms in Micro2-1 and Micro2-2. l UMAP showing the expression of genes related to complement-dependent synaptic pruning and the inflammatory response. m Network plot showing the ligand–receptor interaction strength between each of the two different cell types (upper left panel). Circos plot illustrating the ligand–receptor interactions between astrocytes and NSCs. n Bar plot showing the GO enriched terms of ligands and receptors identified in o. o Circos plot showing the interactions between ligands and receptors that were present in the enriched GO terms. p Dot plot showing the average expression of identified ligands across three astrocyte subclusters. See also Supplementary information, Figs. S6–S8.
To verify the results obtained in snRNA-seq analysis, we examined the proinflammatory state of astrocytes with an antibody against p-STAT3, a key effector of proinflammation. The results showed that p-STAT3 was specifically expressed in the astrocytes of the DG in aged samples but was barely detected in young samples (Supplementary information, Fig. S7b–d). Next, we wondered whether astrocyte inflammation is detrimental to the proliferation of adult NSCs. By analyzing the mitotic score for adult NSCs and the inflammation score for astrocytes, we observed a concurrence of increased inflammation in astrocytes and decreased proliferation in adult NSCs (Fig. 4f). This result implies that niche inflammation may exert an important function in slowing down the mitotic activity of NSCs in primates, as reported in the V-SVZ of mice.43
Microglia are another niche cell that play fundamental roles in the inflammatory response in the brain. A subset of microglial cells residing in the DG subregion but not in the CA subregion was isolated (Supplementary information, Fig. S8a–c). Then, the microglia in the DG were subclustered into 4 populations with distinct gene expression profiles (Fig. 4g; Supplementary information, Fig. S8d). Micro1 represented a resting state of microglia expressing P2RY12, SALL1, etc. Micro2 was a group of active microglia expressing CD74, CD86, CD163, TLR4, and NFKB1 at high levels. Micro3 and Micro4 were two small populations expressing some astrocyte- or OPC-related markers. We proposed that these microglia might engulf some debris from apoptotic cells (such as astrocytes and OPCs), which were captured during sample preparation as previously reported44 (Fig. 4h). The analysis of enriched GO terms showed that Micro2 was associated with the phagocytosis pathway, inflammatory response and lysosome pathways (Fig. 4i). Active microglia exert beneficial functions via phagocytosis or detrimental functions by secreting cytotoxic cytokines.45–49 To detail the nature of Micro2, it was divided into two subclusters, Micro2-1 and Micro2-2 (Fig. 4j). The Micro2-1 subcluster, mainly found in young samples, exhibited anti-inflammatory signatures, such as complement activation and apoptosis, which might be associated with synaptic pruning and the phagocytosis of debris from apoptotic cells during early postnatal stages. The Micro2-2 subcluster, found in aged samples, was involved in the inflammatory response and PI3K/AKT/mTOR pathway activation, which is reportedly related to aging-associated microglial activation50 (Fig. 4k, l). Therefore, our data suggested that active microglia might exert different functions at different stages: at young stages, they positively regulate the development of the hippocampus via the phagocytic pathway, whereas at aged stages, active microglia mainly play a proinflammatory role.
To determine the potential interaction between niche cells and adult NSCs, we investigated the ligand–receptor interaction with the iTALK package.51 We compared the interaction strength among microglia, astrocytes and NSCs, and the results showed that astrocytes and NSCs had a stronger interaction (Fig. 4m). Thus, we further delineated the potential interactions between astrocytes and NSCs (Fig. 4m). The analysis of representative GO terms showed that VEGF–VEGFR signaling and the FGF pathway were enriched in the young samples (P0–Y2), and the integrin pathway, BMP pathway, Eph–Ephrin signaling, and cytokine–receptor interaction were enriched in the old samples (Y4–Y23) (Fig. 4n, o). Our results were consistent with previous findings that VEGF and FGF are involved in the mechanism by which astrocytes promote adult NSC proliferation;37,42,52 the integrin pathway,53,54 BMP signaling,55,56 and Eph–Ephrin signaling41,57 have consistently been shown to be involved in regulating NSC quiescence by niche cells.
We next investigated the expression levels of the ligands in the subclusters of astrocytes. VEGFA, VEGFB, and FGF2 were expressed at high levels in Astro1. Genes involved in the quiescence of adult NSCs (e.g., MFGE8, BMP7, and EFNA5) were abundantly enriched in Astro2. In Astro3, the cytokine TNFSF4 might lead to an inflammatory effect on NSCs. Interestingly, we also found that extracellular matrix-related genes, which play an important role in modulating the local inflammatory milieu, as well as the homeostasis of NSCs, were enriched in Astro3 (Fig. 4p). Overall, we provided evidence that distinct astrocyte subtypes exerted different effects on modulating adult NSCs, including increasing proliferation (Astro1), maintaining quiescence (Astro2), and inducing an inflammatory response (Astro3). The decreased function of Astro1 and increased functions of Astro2 and Astro3 may contribute to the decline in adult neurogenesis during aging.
AHN in aged humans
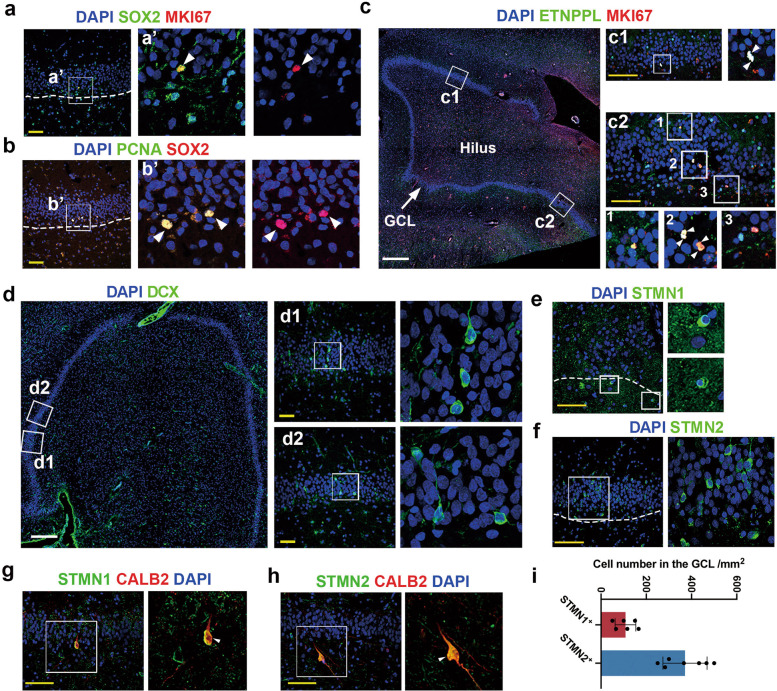
Whether new neurons are continuously generated and whether NSCs exist in the DG of adult and aged hippocampi of humans are still being extensively debated. We evaluated whether adult NSCs exist and proliferate in the hippocampi of aged humans (Supplementary information, Table S6) indicated by the expression of canonical NSC marker SOX2 or the new NSC marker ETNPPL with the cell cycle-related marker MKI67 or PCNA (Fig. 5a–d). First, we found the colocalization of SOX2 and ETNPPL, as we observed in macaques, indicating that ETNPPL is a conserved NSC marker in primates (Supplementary information, Fig. S9a, b). Next, MKI67+ SOX2+ double-positive and PCNA+ SOX2+ double-positive cells indicated that even in the hippocampi of elderly individuals, adult NSCs were still in active mitosis (Fig. 5a, b). Additionally, MKI67 and ETNPPL double-positive cells were observed in the DG of the hippocampi in aged humans (Fig. 5c). Among these MKI67 and ETNPPL double-positive cells, we detected several pairs of compacted nuclei that were juxtaposed, resembling two daughter cells from a mother cell (Fig. 5c). In summary, we observed some cells expressing NSC markers and proliferating markers in the GCL of the hippocampus, which implied that NSCs still exist and were proliferating in aged humans.
Fig. 5. Proliferating NSCs and immature GCs are readily detected in aged human hippocampi.
a, b Expression of the NSC marker SOX2 and the cell cycle markers MKI67 (a) and PCNA (b) in the DG of the hippocampus of aged humans. The arrowheads indicate SOX2/MKI67-positive cells in a and SOX2/PCNA-positive cells in b. c Proliferating NSCs were detected by the coexpression of the primate-specific NSC marker ETNPPL with MKI67 in the DG of the hippocampi of aged humans. d–f The arrowheads in c2-2 indicate two MKI67/ETNPPL double-positive cells juxtaposed. Immature neurons were detected with classic (DCX in d) and new markers (STMN1 in e, STMN2 in f) in the DG of the hippocampi of aged brains. Scale bar in white: 500 µm, Scale bar in yellow, 50 µm. g, h STMN1 and STMN2 are colabeled with the newborn granule cell marker CALB2 in the DG of aged human hippocampi. Scale bar in yellow, 50 µm. i Quantification of STMN1- and STMN2-positive cells in the DG in aged humans. See also Supplementary information, Fig. S9.
The next question we want to address is whether there is still adult neurogenesis in adult and aged humans. Therefore, we examined several human samples (aged 52, 67, 78, and 85 years, Supplementary information, Table S6) using the classic immature neuronal marker DCX and an optimized staining protocol. Fewer DCX-positive cells (Y52 and Y78, Supplementary information, Fig. S9c, e, f) or more DCX-positive cells (Y85 in Fig. 5d; Y67 in Supplementary information, Fig. S9d, f) were detected in each sample, implying remarkable individual variation in AHN. We applied the immature markers revealed in macaques, and STMN1- or STMN2-positive cells with GC morphology were readily detected in aged human samples (Fig. 5e, f, i). We also detected the colocalization of STMN1 or STMN2 with CALB2 but not with NEUROD1, indicating the reliability of both markers to reveal immature neurons (Fig. 5g, h; Supplementary information, Fig. S9g, h).
Transcription dynamics of hippocampal neurogenesis in aged humans
To further investigate adult neurogenesis and the regulatory continuum in aged human hippocampi, four human samples from donors (Y67, Y85, Y87, and Y92) were collected and analyzed using snRNA-seq (Supplementary information, Table S6). In total, 22,119 nuclei were obtained and categorized into 13 cell populations. A cluster of ImmN was revealed in our dataset (Fig. 6a, b; Supplementary information, Fig. S10a–c and Table S7). Adult NSCs were subgrouped from the NSC/Astrocyte cluster (Supplementary information, Fig. S10d–f and Table S8). Although most NSCs are in quiescent state in aged humans, we also observed a small group of NSCs that highly and specifically expressed the active NSC markers, such as ASCL1, and HES6, suggesting they might be active NSCs (Supplementary information, Fig. S11a, b). Lineage trajectory inference showed that the ImmN cluster in humans was positioned between the NSC and GC clusters, which expressed immature neuron markers similar to those in macaques at high levels, including DCX, STMN1, STMN2, PROX1, NNAT, and SOX4. (Fig. 6c, e). ImmN cluster in humans also contained two subclusters, NB-like cluster and Late-ImmGC cluster by expressing conserved markers as in macauqes and rodents (Supplementray Fig. S11c, d).The correlation analysis showed that the human ImmM (H-ImmN) cluster was strongly correlated with the macaque ImmN cluster, indicating evolutionarily conserved transcriptome profiles in primates (Fig. 6c, d). The expression of DEGs along the pseudotime plot and GO term enrichment analysis in the neurogenic lineage confirmed that adult neurogenesis continued in the aged human hippocampi (Fig. 6f, g).
Fig. 6. Persistent adult hippocampal neurogenesis was identified in aged humans.
a UMAP visualization of human hippocampal cells. Cells are colored by the 13 annotated clusters. b Dot plot showing the mean expression of marker genes for the 13 major cell types. c Developmental trajectory inference of the neurogenic lineage is visualized with UMAP (left panel); the pseudotime analysis of individual cells is also presented (right panel). d Correlation analysis of NSC, ImmN, and GC clusters between humans and macaques. e UMAP showing the expression of representative markers for each cluster shown in c. f Fitted curve showing the expression of representative genes along the human adult neurogenic trajectory. g Heatmap illustrating the expression of genes that covary across pseudotime (left panel). The enriched GO terms are also shown (right panel). h Integrated analysis of human hippocampal cells from the present study and CA and DG cells from Franjic’s study. i UMAP showing the clustering of astrocytes in the human dentate gyrus. j Visualization of the expression of differentially expressed genes across astrocyte subclusters in a dot plot. k UMAP showing the clustering of microglia in the human dentate gyrus. l Dot plot showing the expression of DEGs across microglial subclusters. m Circos plot illustrating the ligand–receptor interactions between human astrocytes and NSCs. n Dot plot showing that the expression of inflammation-related key effectors is highly expressed in astrocytes from Franjic’s dataset compared to those from the present dataset. o Comparison of the inflammation score in astrocytes across two datasets. See also Supplementary information, Figs. S10–S14.
To confirm the results observed in snRNA-seq, we also performed scATAC-seq on aged human samples to examine the existence of immature neurons and neural stem cells in aged human hippocampi. In total, 8680 nuclei were obtained and grouped into 8 clusters (Supplementary information, Fig. S12a). Broad cell types were annotated based on the gene activity scores of the representative marker genes (Supplementary information, Fig. S12b). To identify cell types at a higher resolution, we conducted Seurat integrated analysis to map human scATAC-seq data onto human snRNA-seq data and transfer cell-type labels from snRNA-seq to scATAC-seq. After the integrated analysis, populations of NSCs, ImmNs and GCs were identified (Supplementary information, Fig. S12c). Next, we computed differentially accessible peaks among these three cell types and profiled the pile-up of the ATAC-seq signal for each cell type with the top 1000 cluster-enriched peaks to confirm the cell identity in the scATAC-seq dataset. Then, significantly enriched transcription factor motifs for each cell type-specific peak set were computed and motifs for cell type representative transcription factors were enriched in the corresponding cell types, including sites for SOX2 (NSC); NEUROG2 (ImmN) and NEUROD1 (GC). Furthermore, we conducted GO analysis and found that genes located closest to the cell-type-specific peaks were enriched for biological processes associated with cell-type identities (Supplementary information, Fig. S12d). Therefore, based on snRNA-seq and sc-ATACseq, we identified adult NSCs and immature neurons in aged humans.
Recently, Franjic et al. failed to reveal clear evidence of adult neurogenesis in human tissue samples by using snRNA-seq.58 In our dataset, we observed a cohort of cells expressing immature neuron markers (Fig. 6c, e). To nail down the divergence of the two datasets, we integrated our dataset with theirs by using the CCA algorithm, and we identified 12 clusters using unsupervised clustering (Fig. 6h). All other major cell types except ImmNs in the two studies were integrated perfectly. The ImmN cluster (Cluster 12) from our dataset was located adjacent to the integrated GC cluster (Cluster 1) on the UMAP, indicating a close relationship with GCs (Fig. 6h). Additionally, we observed that the ImmNs expressed neuroblast markers that were not detected in Franjic’s dataset (Fig. 6c, e). Therefore, we ascertained that newborn neurons existed and adult hippocampal neurogenesis persisted in the samples that we evaluated.
Next, we examined the NSC niche in aged humans since we observed a close negative correlation between neurogenesis and inflammation in macaques. The astrocytes in humans were subgrouped into two populations, H-Astro1 and H-Astro2 (Fig. 6i). H-Astro1 was enriched in genes related to the inflammatory response, similar to those in macaques (e.g., the JAK/STAT pathway, NFKB1, and TNFSF4), which strengthened the hypothesis that a group of astrocytes was activated during brain aging (Fig. 6j). H-Astro2 expressed genes related to regulating NSC quiescence (e.g., PDGFA/B/C, MFGE8, and the Ephrin family). We integrated the two datasets to investigate whether the regulation of active astrocytes was conserved between humans and macaques. The river plot showed that active astrocytes (M-Astro3 and H-Astro1) were conserved between macaques and humans (Supplementary information, Fig. S13a, b). The integration analysis with Disease-associated astrocytes (DAA) from an Alzheimer’s disease mouse model59 also indicated that M-Astro3 and H-Astro1 processed a pro-inflammatory signature (Supplementary information, Fig. S13c–i). Likewise, the analysis of microglia in aged humans also revealed a cluster of active microglia that correlated strongly with the active microglia in macaques and expressed CD74, IFNGR1, TLR2, and TNFRSF1B (Fig. 6k, l; Supplementary information, Fig. S13j, k). The interaction of astrocytes and adult NSCs revealed conserved pathways between humans and macaques, such as the EPH–EPHRIN pathway, the BMP pathway, and the inflammatory signaling pathway. However, in the Eph–Ephrin pathway, EFNA5 was the only ligand enriched in macaques, while EFNB1, EFNB2, and EFNA1 were also significantly expressed in humans, indicating conserved pathways but distinct preferred ligand–receptor counterparts between humans and macaques (Fig. 6m).
Next, we wondered whether the inflammatory state of astrocytes was different between the two datasets. The expression levels of some key effectors of neuroinflammatory signals, such as NFKB1, STAT3, and STAT5B, as well as the inflammation score, were significantly higher in astrocytes in Franjic’s dataset than in those in the present dataset (Fig. 6n, o), which implied that the severe proinflammatory state of the samples might hinder hippocampal neurogenesis. To further validate this observation, we also analyzed the anti-inflammation score with the gene set of ‘negative regulation of inflammation’ (GO:0050728). The result clearly showed that the anti-inflammation score was lower in the astrocytes of our dataset than that in Franjic’s dataset (Supplementary information, Fig. S14a). We next calculated the mitotic score and the inflammation score in each sample in our dataset. We observed a negative correlation between mitotic score and inflammation score in each sample as we observed in macaques (Supplementary information, Fig. S14b, c). Given the controversial conclusions drawn in different laboratories with immunostaining assays, we proposed that proinflammatory conditions might contribute to the variety of adult hippocampal neurogenesis in humans.
Discussion
Whether adult hippocampal neurogenesis exists and whether the proliferation of adult neural stem cells continues in adult humans have been controversial recently.3,4,6,7,9,10,13,34,60 In the adult or elderly stages, newborn neurons are scarce and not evenly distributed in the hippocampal formation.6,34,61–63 Moreover, these newborn neurons are very vulnerable to experimental procedures, such as fixatives, postmortem intervals, different batches of antibodies, and methods for antigen retrieval. Sc/snRNA-seq provides a robust and sensitive tool that can bypass the limitations of traditional methods, such as RNA in situ hybridization and immunohistochemistry, to reveal rare cell types.19–21 Additionally, snRNA-seq will provide more molecular details about the expression of each cell type that might provide more primate-specific markers for NSCs and immature GCs. Therefore, in this study, we systematically evaluated the molecular and cellular dynamics of the hippocampi in macaques across the lifespan and in aged humans using high-throughput snRNA-seq.
In the neurogenic lineage, immature GCs were readily revealed with the unsupervised cluster method in both macaques and aged humans. The developmental and expressional continuum from adult NSCs to immature and mature GCs was delineated. Whether there are more robust and reliable markers to reveal NSCs and immature neurons in the primate hippocampus is a fundamental question. Integrated cross-species analysis between mice and monkeys highlighted a trail of primate-specific NSC signatures. ETNPPL was validated as a primate-specific marker for NSCs in both humans and macaques. We also verified STMN1 and STMN2 as new markers to detect immature GCs in the hippocampi of primates. Therefore, we provide strong evidence that hippocampal NSCs and adult neurogenesis persist in adult humans and macaques, albeit the number of immature neurons and the number of NSCs decline with age.
Adult neurogenesis and the proliferation of adult NSCs are modulated by both intrinsic and extrinsic factors in mice.37,38 Regarding extrinsic factors, environmental enrichment, proper exercise, stress, and even diet remarkably impact adult hippocampal neurogenesis.64–69 Another key modulator of hippocampal function and adult neurogenesis is neuroinflammation, which is induced by the invasion of pathogens, brain injury, neurological diseases and brain aging.70–73 In our study, subclustering analysis showed that both active microglia and active astrocytes exhibited an inflammatory signature. However, active astrocytes were present only in the aged samples, whereas active microglia were detected in both young and aged samples. The GO terms and DEG analysis indicated that microglia remove extra synapses and apoptotic debris at young stages, which is beneficial for proper circuit formation. At aged stages, both microglia and astrocytes contribute to the neuroinflammatory response, which is considered a detrimental effect on brain function.
It is an interesting question how active astrocytes and active microglia affect other cell types. In the present study, the interaction strength among microglia, astrocytes, and NSCs showed that astrocytes and NSCs have a stronger interaction, implying that active astrocytes have direct effects on NSCs. The transcriptome dynamics of astrocytes and the analysis of the interaction of astrocytes with NSCs infer that distinct astrocyte subtypes exert distinct functions by providing distinct signals to adult NSCs. Previous studies in mice also indicate that active astrocytes induce the death of neurons and oligodendrocytes.74 Normal aging-induced upregulation of reactive genes by astrocytes could contribute to cognitive decline in vulnerable brain regions, such as the hippocampus and striatum, and to the greater vulnerability of the aged brain to injury.75 In AD human samples, scientists also reported the upregulation of active astrocyte signatures in AD-pathology-associated astrocyte subpopulations using snRNA sequencing.76 Altogether, all of the data, including our data in aged humans and macaques, manifest a key role of active astrocytes in the regulation of brain function and homeostasis. The correlation of decreased neurogenesis with increased neuroinflammation implies that manipulating the neuroinflammatory status of the brain might be a feasible approach to restore adult hippocampal neurogenesis in aged individuals and individuals with disease.
Materials and methods
Macaque sample collection
Macaque monkeys were raised and received cares in the non-human primate center of the Institute of Biophysics, Chinese Academy of Sciences (IBP, CAS; Supplementary information, Table S1). All procedures were conducted in accordance with the Principles for the Ethical Treatment of Non-Human Primates and were approved by the Institutional Animal Care and Use Committee of the IBP, CAS (N-W-20131104).
Human sample collection
Postmortem human brain samples (Supplementary information, Table S6) from individuals without neurological disorders were provided by the National Human Brain Bank for Development and Function, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China. The informed consent form was signed by the donors before the collection. All sample collection and experimental procedures were followed national laws and international ethical and technical guidelines.The whole procedure was examined and approved by the Ethic Committee of Peking Union Medical College (009-2014) and Institute of Biophysics, CAS (H-W-20131104R1).
We collected tissue blocks from samples with a shorter post-mortem interval. Blocks were either frozen in liquid nitrogen or fixed overnight in 4% paraformaldehyde (PFA) at 4 °C. Frozen blocks were used for snRNA-seq. Fixed blocks were cryoprotected using a 30% sucrose solution and then embedded in OCT. Embedded tissue was cut on a Leica CMl950 very low temperature freezer into 20 μm coronal sections, and mounted on glass slides for immunohistochemistry analysis.
Consecutive EdU and BrdU labeling
To trace the proliferation of NSCs and adult neurogenesis, EdU (Thermo Fisher Scientific, A10044) and BrdU (Thermo Fisher Scientific, B23151) labeling was conducted. Briefly, EdU labeling (5 mg/kg body weight) was administered to macaques intravenously twice a week for 10 weeks. With a two-month-interval, BrdU was intravenously administered (10 mg/kg) twice a week for 10 weeks. Then, the animals were sacrificed 2 months post administration.
For macaque brain sample preparation, adult and senile macaque monkeys were anesthetized with Ketamine (10 mg/kg body weight) intramuscularly and perfused with cold artificial cortico-spinal fluid (ACSF). The whole brains were rapidly dissected on ice. For snRNA-seq, the hippocampal formations and other brain regions in the right hemisphere were isolated and quick frozen in liquid nitrogen. For the immunostaining, the left part of each brain was cut coronally into around 1cm-thick slabs and fixed in 4% PFA for 12 h, following being cryoprotected in a sucrose gradient from 10%, 20% to 30%, and then was embedded with OCT.
Immunohistofluorescence
Frozen sections were equilibrated to room temperature (RT) and rinsed with PBST buffer (0.3% Triton-X in PBS) for 10 min. To reduce the remaining aldehyde groups, the sections were incubated with a 0.5% sodium triacetoxyborohydride (NaBH(OAc)3; Merck, 316393) solution for 30 min at RT. Next, the samples were boiled in a sodium citrate buffer (10 mM sodium citrate tribasic dihydrate, pH 6.0) for 20 min, and cooled down to RT. For PSA-NCAM antibody staining, we conducted antigen retrieval in Tris-EDTA buffer (50 mM Tris, 1 mM EDTA, pH 9.0). After antigen retrieval, samples were washed with PBST for 10 min and then blocked with a 5% normal donkey serum solution (NDS) (JACKSON, 017-000-121T) for 1 h at RT. Subsequently, samples were incubated for three to five days at 4 °C with 5% NDS containing the primary antibodies (Supplementary information, Table S9). The secondary antibody incubation and DAPI nuclear staining were performed at RT for at least 2 h. Autofluorescence Eliminator reagent (0.1% Sudan Black in 70% ethanol) was applied to the aged samples to eliminate lipofuscin autofluorescence. After three washes with PBST, sections were mounted with Fluoromount-G (SouthernBiotech, 0100-01) and stored at 4 °C in the dark.
To visualize EdU-labeled cells, a Click-iT EdU Imaging Kit (Invitrogen, C10639) was used according to manufacturer’s guidelines. For BrdU staining, the tissue was incubated with 2 M HCl at RT for 20 min, followed by sequential incubation with 5% NDS, the anti-BrdU antibody and secondary antibody, respectively.
Imaging
Serial coronal sections of the hippocampus were collected from macaques and humans. Images were acquired using an Olympus FV3000 confocal microscope with a 30× oil immersion objective (NA 1.30). Tiled images were acquired and stitched using Olympus FluoView software. Image processing and analysis were conducted using FLUOVIEW FV3000 and Fiji/ImageJ software and compiled with Adobe Photoshop 2020.
Single nuclei isolation, 10× Genomics Chromium, and sequencing
Nuclei were isolated from fresh-frozen tissue as previously described with some modifications and improvements. Briefly, tissue samples were minced into pieces <5 mm and then homogenized using a glass dounce tissue grinder (Sigma, D8938) in 2 mL of Nuclei EZ lysis buffer (Sigma, NUC-101) on ice. After two incubations on ice (with 4 mL of lysis buffer and 5 min each time), the homogenate was filtered through a 70 µm strainer, and then we used Debris Removal Solution (Miltenyi Biotech, 130-109-398) to perform density gradient centrifugation to clean the nuclear suspension according to the manufacturer’s protocol. Isolated nuclei were resuspended and washed with nuclei suspension buffer (NSB, consisting of 1× PBS, 0.1% BSA and 0.4 U/µL Ambion™ RNase inhibitor (Thermo Fisher, AM2684)) and filtered through a 35 µm cell strainer. Nuclei were counted using a hemocytometer and diluted to 1000 nuclei/μL for optimal 10× loading. Approximately 8000 nuclei were targeted and captured for each reaction. Steps from Chip B loading to cDNA library construction was carried out with Chromium Single Cell 3’ Reagent Kits v3 according to the 10× official instructions.
scATAC library preparation and high-throughput sequencing
Human hippocampal frozen tissue was homogenized using a glass tissue grinder (Sigma, D8938) in 500 µL chilled 0.1× lysis buffer and incubated for 10 min on ice. Then, 500 µL chilled wash buffer was added. The homogenate was filtered through a 40 µm strainer, centrifuged at 500 × g for 5 min at 4 °C, resuspended the nuclei pellet with chilled PBS. After that, Debris Removal Solution (Miltenyi Biotech, 130-109-398) was used to perform density gradient centrifugation to clean the nuclei suspension following the manufacture’s protocol. Isolated nuclei were resuspended with chilled Diluted Nuclei Buffer at the proper concentration to generate ATAC library with Chromium Single Cell ATAC Reagent Kits v1.0 following the default process (www.10xgenomics.com). Finally, the library was processed on the Illumina NovaSeq platform for sequencing with 50 bp paired-end reads.
Processing of macaque snRNA-seq data
A cell-by-gene count matrix was generated following sequence alignment with Cell Ranger software (https://support.10xgenomics.com/). The raw count matrix from each sample was loaded to the scrublet pipeline individually, with the parameter expected_doublet_ratio set to 0.06 to identify possible doublets.77 A total of 32,049/182,036 cells with a doublet score greater than the doublet score threshold was considered as doublets and thus discarded from subsequent analysis. The filtered cell-by-gene count matrix was then loaded into the Seurat pipeline78 for downstream analysis. Cells that did not meet the following criteria were omitted: 1) cells with a number of detected genes ranging from 800 to 7500; 2) cells with nUMI greater than 1000; and 3) cells with a percentage of mitochondrial counts less than 1%. Overall, 132,524 cells were retained for downstream analysis. Next, the filtered cell-by-gene matrix was imported to function CreateSeuratObject to create a Seurat object followed by log-normalization of the gene expression matrix with function NormalizeData. Then, we identified variable genes that exhibited high cell-to-cell variations using the function FindVariableFeatures and performed a dimension reduction analysis in the space of those variable features with the functions RunPCA and RunUMAP. We removed batch effects and preserved the biological variation present in our dataset by conducting a canonical correction analysis among individual samples with the functions FindIntegrationAnchors and IntegrateData.78 Clustering analysis was performed with the functions FindNeighbors and FindClusters.
Distinguishing DG and CA cells based on regional transcriptional identities
We first computed DEGs between cells obtained from individual dissection of CA and DG for the each of the following individual cell types to distinguish DG and CA cells from whole hippocampal samples: Astrocytes, Microglia, OPCs, Oligodendrocytes and GABAergic neurons. Then, we performed principal component analysis of each cell type in whole hippocampal samples based on the identified regional DEGs. Next, we fitted the resulting principal components to the k-means algorithm for clustering analysis. Subsequently, we determined the regional characteristics of the identified clusters by calculating the enrichment score for genes enriched in CA and DG across all clusters using the AUCell package.79 A cluster with a higher enrichment score for DG-enriched genes was considered as a DG-derived group, and a cluster with a higher enrichment score for CA-enriched genes was identified as a CA-derived group.
Construction of the macaque adult neurogenic trajectory
We first extracted NSCs, ImmNs and granule from the whole dataset and performed the dimension reduction analysis using those cells to infer the adult neurogenic trajectory in macaque hippocampi. Then, for trajectory construction, we converted the Seurat object to a SingleCellExperiment object and performed trajectory graph learning analysis and pseudotime measurement with the functions learn_graph and order_cells from monocle3,80–82 respectively.
Comparison of transcriptional profiles between macaque and mouse astrocytes and NSC
We first assembled cells from our dataset with those from a published scRNA-seq dataset of the mouse dentate gyrus22 based on orthologues identified between the macaque and mouse genomes with the R package biomaRt.83 Next, we subdivided astrocytes or NSCs from the merged dataset and computed DEGs between macaque and mouse astrocyte or NSCs with the Seurat function FindMarkers.
Identification of cell–cell communication
We studied cell–cell communication using iTALK (https://github.com/Coolgenome/iTALK), a package designed to study cell interactions. Briefly, the rawParse function was applied to retain the top 50% highly expressed genes as input for the FindLR function to identify ligand–receptor pairs between cell types. We mainly focused on the ligand–receptor pairs where ligands were obtained from astrocytes and receptors were expressed in NSCs. The identified interactions were visualized using the NetView and LRPlot functions.
Processing of human snRNA-seq data
Reads were aligned to the human reference genome hg19 with Cell Ranger software (https://support.10xgenomics.com/), and a cell-by-gene count matrix was then generated. Then, candidate doublets were identified by individually importing the raw count matrix from each human sample into the scrublet software by setting the parameter expected_doublet_ratio to 0.06.77 A total of 2287/26,865 cells with a doublet score exceeding the setting threshold were then omitted from the subsequent analysis. The filtered cell-by-gene count matrix was then loaded into the Seurat pipeline. Then, cells that did not meet the following criteria were discarded: 1) cells with more than 200 detected genes and 2) cells with fewer than 10,000 detected genes. After removing low-quality cells, 22,119 cells were retained for the subsequent analysis. The filtered count matrix was then log-normalized with the NormalizeData function. Next, variable features were computed with the function FindVariableFeatures, and dimension reduction analysis was performed on these variable features with the functions RunPCA and RunUMAP. Batch effects derived from nonbiological variations were removed by conducting a canonical correction analysis among individual samples with the functions FindIntegrationAnchors and IntegrateData. The clustering analysis was carried out with the functions FindNeighbors and FindClusters.
Construction of the human adult neurogenic trajectory
Similar to the trajectory inference analysis conducted for the macaque dataset, we first extracted NSCs, ImmNs and GCs from the whole human dataset and then performed the dimension reduction analysis on those cells. Next, we used the UMAP embedding and clusters generated by Seurat as input for the trajectory learning analysis and pseudotime measurement analysis with the functions learn_graph and order_cells from monocle3.
Comparison of human hippocampal cells across datasets
To compare the constitutions and characteristics of adult human hippocampi cells from our study and those from Franjic’s study, we integrated the adult human hippocampi cells from both studies following the Seurat integration analysis. In order to do so, we first subtracted DG and CA cells from Franjic’s study and then selected genes that are repeatedly variable across two datasets with function SelectIntegrationFeatures. Subsequently, we identified cross-dataset cell-pairs that are in a matched biological state using function FindIntegrationAnchors followed by integrating these two datasets with function IntegrateData. Next, we performed dimension reduction analysis on the integrated datasets with function RunPCA and RunUMAP. The clustering analysis was accomplished with functions FindNeighbors and FindClusters. The correspondences of cell types across two datasets were illustrated with riverplot conducted with R package riverplot.
Estimation of mitotic capacity and inflammation status of cells
To estimate the mitotic capacity and inflammation status of cells, we computed the inflammatory score and mitotic score by following the method adapted from the previous publication.35 Briefly, we first collected cell cycle- and inflammation response-related genes from the molecular signature database MSigDB and identified genes to be included in the subsequent analysis by computing the intersection between the collected genes and genes detected in our dataset. Next, to calculate inflammation score and mitotic score, we performed principal component analysis in the space of the inflammation response-related genes and cell cycle-related genes, respectively. To infer inflammation and mitotic capacity of cells across postnatal development time points, we plotted inflammation score or mitotic score along time points and computed the fitted curve with locally weighted regression (loess).
Processing of human snATAC-seq data
Reads filtering, alignment and transposase cut sites identification were performed with the Cell Ranger software. The cell-by-peak count matrix was then binarized and passed to Signac for subsequent dimension reduction and clustering analysis. Briefly, the binarized count matrix was first frequency-inverse document frequency (TF-IDF) transformed by dividing each index by the cells-wise sum of the matrix multiplying with the inverse document frequency computed as log(1+ncol(matrix)/rowSums(matrix)). Singular value decomposition (SVD) was then conducted on the TD-IDF matrix for dimension reduction. Clustering was done with function FindNeighbors and FindClusters.
Peak calling for each cluster was performed with fragments from cells in the cluster using MACS2 with parameters ‘–nomodel –shift 75 –extsize 150 –qval 5e-2 -B -f BED –nolambda –keep-dup all –call-summits’ callpeak with MACS2 software (v 2.2.6). Peaks from each cluster were then assembled and a binarized cell-by-peak matrix was generated by converting non-zero counts to 1.
The created cell-by-peak matrix was then loaded into Signac for downstream analysis. Cells that pass the following quality criterion were kept: (1) the number of fragments in peaks between 2000 and 15,000; (2) the fraction of fragments in peaks greater than 10%; (3) transcriptional start site (TSS) enrichment score greater than 2; (4) ratio reads in genomic blacklist regions smaller than 0.05; (5) nucleosomal signal strength smaller than 4.5. Overall, 8680 cells passed the quality control and retained for subsequent analyses. Next, we passed the filtered cell-by-peak matrix to Signac function RunTFIDF and RunSVD for dimension reduction and function FindNeighbors and FindClusters for cluster identification. FindNeighbors was performed with reduced dimension components 1:15. FindClusters was performed by setting resolution to 0.3. Then, we computed gene activity matrix by quantifying the activity of each gene in the genome by accessing gene associated chromatin accessibility with function GeneActivity. Clusters were then annotated based on gene activity score profiles of the representative cell-type markers (SATB2 for CA1, CPNE4 for CA3, PROX1 and CALB2 for GCs, SLC32A1 for GABAergic neurons, GFAP and AQP4 for astrocytes, MOG for oligodendrocytes, OLIG2 for OPCs and CSF2R for microglia).
To identify cell types at a higher resolution and anchor transcriptional expression and chromatin state profiles of cells, we integrated snRNA-seq and snATAC-seq datasets by following the Seurat integration analysis. Briefly, anchors between two datasets were first computed with function FindTransferAnchors by correlating patterns in gene activity matrix from snATAC-seq dataset and gene expression profiles of snRNA-seq dataset. Then, the computed anchors were used to transfer cell type labels derived from the snRNA-seq dataset and an imputed gene expression matrix was generated with function TransferData. Then, the co-embedding of snRNA-seq and snATAC-seq datasets was created by conducting dimensional reduction analysis with function RunPCA and RunUMAP with the imputed gene expression matrix.
Motif enrichment
Motif enrichment was performed with FIMO from the MEME suites to identify enriched binding motif in the genes’ regulatory regions. Homer function annotatePeaks.pl was then used for motif binding site annotation.
Quantification and statistical analysis
All data obtained from macaques and humans at each age were collected from one sample with at least three independent experiments. The error bars represent the SD. Statistical analyses were performed by unpaired two-tailed Student’s t-tests and the multiple comparison test after ANOVA using GraphPad Prism software. Sample size and P values are given in the Figure legends.
Supplementary information
Acknowledgements
This work was supported by the National Key R&D Program of China (2020YFA0112200, 2019YFA0110100), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB32010100, XDA16020601), the National Natural Science Foundation of China (NSFC) (32122037, 32192411 and 81891001), CAS Project for Young Scientists in Basic Research (YSBR-013), the China Brain Project (2021ZD0200102), BUAA-CCMU Big Data and Precision Medicine Advanced Innovation Center Project (BHME-2019001), Collaborative Research Fund of Chinese Institute for Brain Research, Beijing (2020-NKX-PT-02 and 2020-NKX-PT-03). Human tissues were provided by the National Human Brain Bank for Development and Function, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China. This study was supported by the Institute of Basic Medical Sciences, Chinese Academy of Medical Sciences, Neuroscience Center, and the China Human Brain Banking Consortium.
Author contributions
W.W., X.W., Q.W., and W.J. conceived the project. W.W., and X.W., designed the experiments. M. Y. and W.W. performed immunostaining and imaging. M.W., B.Z., X.J., and J.Z. analyzed the snRNA-seq data and scATAC-seq data. W.W., C.Y., and B. W. prepared the macaque samples. Q.M., Y.G., Y.L., and Q.Z. performed snRNA-seq and scATAC-seq experiments. W.Q., and C.M. prepared and provided the human samples. W.W, M.W., Q.W. and X.W. wrote the manuscript. All authors edited and proofread the manuscript.
Data availability
The single-nucleus RNA-seq data used in this study has been deposited in the Gene Expression Omnibus (GEO) under the accession number GSE163737. The scATAC-seq data has been deposited in the Genome Sequence Archive (HRA002222).
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Wei Wang, Mengdi Wang, Meng Yang, Bo Zeng, Wenying Qiu.
Contributor Information
Qian Wu, Email: qianwu@bnu.edu.cn.
Chao Ma, Email: machao@ibms.cams.cn.
Xiaoqun Wang, Email: xiaoqunwang@bnu.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41422-022-00678-y.
References
- 1.Navarro Negredo P, Yeo RW, Brunet A. Aging and rejuvenation of neural stem cells and their niches. Cell Stem Cell. 2020;27:202–223. doi: 10.1016/j.stem.2020.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartsch T, Wulff P. The hippocampus in aging and disease: from plasticity to vulnerability. Neuroscience. 2015;309:1–16. doi: 10.1016/j.neuroscience.2015.07.084. [DOI] [PubMed] [Google Scholar]
- 3.Paredes MF, et al. Does adult neurogenesis persist in the human hippocampus? Cell Stem Cell. 2018;23:780–781. doi: 10.1016/j.stem.2018.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sorrells SF, et al. Human hippocampal neurogenesis drops sharply in children to undetectable levels in adults. Nature. 2018;555:377–381. doi: 10.1038/nature25975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sorrells SF, et al. Positive controls in adults and children support that very few, if any, new neurons are born in the adult human Hippocampus. J. Neurosci. 2021;41:2554–2565. doi: 10.1523/JNEUROSCI.0676-20.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kempermann G, et al. Human adult neurogenesis: evidence and remaining questions. Cell Stem Cell. 2018;23:25–30. doi: 10.1016/j.stem.2018.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moreno-Jimenez EP, et al. Adult hippocampal neurogenesis is abundant in neurologically healthy subjects and drops sharply in patients with Alzheimer’s disease. Nat. Med. 2019;25:554–560. doi: 10.1038/s41591-019-0375-9. [DOI] [PubMed] [Google Scholar]
- 8.Moreno-Jimenez EP, Terreros-Roncal J, Flor-Garcia M, Rabano A, Llorens-Martin M. Evidences for adult Hippocampal neurogenesis in humans. J. Neurosci. 2021;41:2541–2553. doi: 10.1523/JNEUROSCI.0675-20.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boldrini M, et al. Human hippocampal neurogenesis persists throughout aging. Cell Stem Cell. 2018;22:589–599. doi: 10.1016/j.stem.2018.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tobin MK, et al. Human hippocampal neurogenesis persists in aged adults and Alzheimer’s disease patients. Cell Stem Cell. 2019;24:974–982. doi: 10.1016/j.stem.2019.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Toda T, Gage FH. Review: adult neurogenesis contributes to hippocampal plasticity. Cell Tissue Res. 2018;373:693–709. doi: 10.1007/s00441-017-2735-4. [DOI] [PubMed] [Google Scholar]
- 12.Abbott LC, Nigussie F. Adult neurogenesis in the mammalian dentate gyrus. Anat. Histol. Embryol. 2020;49:3–16. doi: 10.1111/ahe.12496. [DOI] [PubMed] [Google Scholar]
- 13.Cipriani S, et al. Hippocampal radial glial subtypes and their neurogenic potential in human fetuses and healthy and Alzheimer’s disease adults. Cereb. Cortex. 2018;28:2458–2478. doi: 10.1093/cercor/bhy096. [DOI] [PubMed] [Google Scholar]
- 14.Dennis C, Suh L, Rodriguez M, Kril J, Sutherland G. Human adult neurogenesis across the ages: an immunohistochemical study. Neuropathol. Appl. Neurobiol. 2016;42:621–638. doi: 10.1111/nan.12337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flor-Garcia M, et al. Unraveling human adult hippocampal neurogenesis. Nat. Protoc. 2020;15:668–693. doi: 10.1038/s41596-019-0267-y. [DOI] [PubMed] [Google Scholar]
- 16.Pollen AA, et al. Molecular identity of human outer radial glia during cortical development. Cell. 2015;163:55–67. doi: 10.1016/j.cell.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhong S, et al. A single-cell RNA-seq survey of the developmental landscape of the human prefrontal cortex. Nature. 2018;555:524–528. doi: 10.1038/nature25980. [DOI] [PubMed] [Google Scholar]
- 18.Zhong S, et al. Decoding the development of the human hippocampus. Nature. 2020;577:531–536. doi: 10.1038/s41586-019-1917-5. [DOI] [PubMed] [Google Scholar]
- 19.Aizarani N, et al. A human liver cell atlas reveals heterogeneity and epithelial progenitors. Nature. 2019;572:199–204. doi: 10.1038/s41586-019-1373-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jindal A, Gupta P, Jayadeva, Sengupta D. Discovery of rare cells from voluminous single cell expression data. Nat. Commun. 2018;9:4719. doi: 10.1038/s41467-018-07234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Montoro DT, et al. A revised airway epithelial hierarchy includes CFTR-expressing ionocytes. Nature. 2018;560:319–324. doi: 10.1038/s41586-018-0393-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hochgerner H, Zeisel A, Lonnerberg P, Linnarsson S. Conserved properties of dentate gyrus neurogenesis across postnatal development revealed by single-cell RNA sequencing. Nat. Neurosci. 2018;21:290–299. doi: 10.1038/s41593-017-0056-2. [DOI] [PubMed] [Google Scholar]
- 23.Mizrak D, et al. Single-cell analysis of regional differences in adult V-SVZ neural stem cell lineages. Cell Rep. 2019;26:394–406. doi: 10.1016/j.celrep.2018.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Basak O, et al. Troy+ brain stem cells cycle through quiescence and regulate their number by sensing niche occupancy. Proc. Natl. Acad. Sci. USA. 2018;115:E610–E619. doi: 10.1073/pnas.1715911114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dulken BW, Leeman DS, Boutet SC, Hebestreit K, Brunet A. Single-cell transcriptomic analysis defines heterogeneity and transcriptional dynamics in the adult neural stem cell lineage. Cell Rep. 2017;18:777–790. doi: 10.1016/j.celrep.2016.12.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Llorens-Bobadilla E, et al. Single-cell transcriptomics reveals a population of dormant neural stem cells that become activated upon brain injury. Cell Stem Cell. 2015;17:329–340. doi: 10.1016/j.stem.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 27.Luo Y, et al. Single-cell transcriptome analyses reveal signals to activate dormant neural stem cells. Cell. 2015;161:1175–1186. doi: 10.1016/j.cell.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shin J, et al. Single-cell RNA-seq with waterfall reveals molecular cascades underlying adult neurogenesis. Cell Stem Cell. 2015;17:360–372. doi: 10.1016/j.stem.2015.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Artegiani B, et al. A single-cell RNA sequencing study reveals cellular and molecular dynamics of the Hippocampal neurogenic niche. Cell Rep. 2017;21:3271–3284. doi: 10.1016/j.celrep.2017.11.050. [DOI] [PubMed] [Google Scholar]
- 30.Cembrowski MS, Wang L, Sugino K, Shields BC, Spruston N. Hipposeq: a comprehensive RNA-seq database of gene expression in hippocampal principal neurons. Elife. 2016;5:e14997. doi: 10.7554/eLife.14997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller JA, et al. Conserved molecular signatures of neurogenesis in the hippocampal subgranular zone of rodents and primates. Development. 2013;140:4633–4644. doi: 10.1242/dev.097212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bond AM, Ming GL, Song H. Adult mammalian neural stem cells and neurogenesis: five decades later. Cell Stem Cell. 2015;17:385–395. doi: 10.1016/j.stem.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuwabara T, et al. Wnt-mediated activation of NeuroD1 and retro-elements during adult neurogenesis. Nat. Neurosci. 2009;12:1097–1105. doi: 10.1038/nn.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuhn HG, Toda T, Gage FH. Adult hippocampal neurogenesis: a coming-of-age story. J. Neurosci. 2018;38:10401–10410. doi: 10.1523/JNEUROSCI.2144-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nowakowski TJ, et al. Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science. 2017;358:1318–1323. doi: 10.1126/science.aap8809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brandt MD, et al. Transient calretinin expression defines early postmitotic step of neuronal differentiation in adult hippocampal neurogenesis of mice. Mol. Cell Neurosci. 2003;24:603–613. doi: 10.1016/S1044-7431(03)00207-0. [DOI] [PubMed] [Google Scholar]
- 37.Vicidomini C, Guo N, Sahay A. Communication, cross talk, and signal integration in the adult hippocampal neurogenic niche. Neuron. 2020;105:220–235. doi: 10.1016/j.neuron.2019.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Urban N, Blomfield IM, Guillemot F. Quiescence of adult mammalian neural stem cells: a highly regulated rest. Neuron. 2019;104:834–848. doi: 10.1016/j.neuron.2019.09.026. [DOI] [PubMed] [Google Scholar]
- 39.Garber C, et al. Astrocytes decrease adult neurogenesis during virus-induced memory dysfunction via IL-1. Nat. Immunol. 2018;19:151–161. doi: 10.1038/s41590-017-0021-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Casse F, Richetin K, Toni N. Astrocytes’ contribution to adult neurogenesis in physiology and Alzheimer’s disease. Front. Cell Neurosci. 2018;12:432. doi: 10.3389/fncel.2018.00432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ashton RS, et al. Astrocytes regulate adult hippocampal neurogenesis through ephrin-B signaling. Nat. Neurosci. 2012;15:1399–1406. doi: 10.1038/nn.3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Song H, Stevens CF, Gage FH. Astroglia induce neurogenesis from adult neural stem cells. Nature. 2002;417:39–44. doi: 10.1038/417039a. [DOI] [PubMed] [Google Scholar]
- 43.Kalamakis G, et al. Quiescence modulates stem cell maintenance and regenerative capacity in the aging brain. Cell. 2019;176:1407–1419. doi: 10.1016/j.cell.2019.01.040. [DOI] [PubMed] [Google Scholar]
- 44.Li Q, et al. Developmental heterogeneity of microglia and brain myeloid cells revealed by deep single-cell RNA sequencing. Neuron. 2019;101:207–223. doi: 10.1016/j.neuron.2018.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Masuda T, Sankowski R, Staszewski O, Prinz M. Microglia heterogeneity in the single-cell era. Cell Rep. 2020;30:1271–1281. doi: 10.1016/j.celrep.2020.01.010. [DOI] [PubMed] [Google Scholar]
- 46.Paolicelli RC, et al. Synaptic pruning by microglia is necessary for normal brain development. Science. 2011;333:1456–1458. doi: 10.1126/science.1202529. [DOI] [PubMed] [Google Scholar]
- 47.Sato K. Effects of microglia on neurogenesis. Glia. 2015;63:1394–1405. doi: 10.1002/glia.22858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ekdahl CT. Microglial activation–tuning and pruning adult neurogenesis. Front. Pharmacol. 2012;3:41. doi: 10.3389/fphar.2012.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sierra A, et al. Microglia shape adult hippocampal neurogenesis through apoptosis-coupled phagocytosis. Cell Stem Cell. 2010;7:483–495. doi: 10.1016/j.stem.2010.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chu E, Mychasiuk R, Hibbs ML, Semple BD. Dysregulated phosphoinositide 3-kinase signaling in microglia: shaping chronic neuroinflammation. J. Neuroinflammation. 2021;18:276. doi: 10.1186/s12974-021-02325-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang, Y. et al. iTALK: an R package to characterize and illustrate intercellular communication. bioRxiv507871, 10.1101/507871 (2019).
- 52.Shetty AK, Hattiangady B, Shetty GA. Stem/progenitor cell proliferation factors FGF‐2, IGF‐1, and VEGF exhibit early decline during the course of aging in the hippocampus: role of astrocytes. Glia. 2005;51:173–186. doi: 10.1002/glia.20187. [DOI] [PubMed] [Google Scholar]
- 53.Zhou Y, et al. Autocrine Mfge8 signaling prevents developmental exhaustion of the adult neural stem cell pool. Cell Stem Cell. 2018;23:444–452. doi: 10.1016/j.stem.2018.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Morgner J, et al. Integrin-linked kinase regulates the niche of quiescent epidermal stem cells. Nat. Commun. 2015;6:8198. doi: 10.1038/ncomms9198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sachdeva R, et al. BMP signaling mediates glioma stem cell quiescence and confers treatment resistance in glioblastoma. Sci. Rep. 2019;9:14569. doi: 10.1038/s41598-019-51270-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mira H, et al. Signaling through BMPR-IA regulates quiescence and long-term activity of neural stem cells in the adult hippocampus. Cell Stem Cell. 2010;7:78–89. doi: 10.1016/j.stem.2010.04.016. [DOI] [PubMed] [Google Scholar]
- 57.Dong J, et al. A neuronal molecular switch through cell-cell contact that regulates quiescent neural stem cells. Sci. Adv. 2019;5:eaav4416. doi: 10.1126/sciadv.aav4416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Franjic D, et al. Transcriptomic taxonomy and neurogenic trajectories of adult human, macaque, and pig hippocampal and entorhinal cells. Neuron. 2022;110:452–469. doi: 10.1016/j.neuron.2021.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Habib N, et al. Disease-associated astrocytes in Alzheimer’s disease and aging. Nat. Neurosci. 2020;23:701–706. doi: 10.1038/s41593-020-0624-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Petrik D, Encinas JM. Perspective: of mice and men - how widespread is adult neurogenesis? Front. Neurosci. 2019;13:923. doi: 10.3389/fnins.2019.00923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Eriksson PS, et al. Neurogenesis in the adult human hippocampus. Nat. Med. 1998;4:1313–1317. doi: 10.1038/3305. [DOI] [PubMed] [Google Scholar]
- 62.Bonfanti L. Adult neurogenesis 50 years later: limits and opportunities in mammals. Front. Neurosci. 2016;10:44. doi: 10.3389/fnins.2016.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Goncalves JT, Schafer ST, Gage FH. Adult neurogenesis in the hippocampus: from stem cells to behavior. Cell. 2016;167:897–914. doi: 10.1016/j.cell.2016.10.021. [DOI] [PubMed] [Google Scholar]
- 64.Kempermann G, Kuhn HG, Gage FH. More hippocampal neurons in adult mice living in an enriched environment. Nature. 1997;386:493–495. doi: 10.1038/386493a0. [DOI] [PubMed] [Google Scholar]
- 65.van Praag H, Kempermann G, Gage FH. Running increases cell proliferation and neurogenesis in the adult mouse dentate gyrus. Nat. Neurosci. 1999;2:266–270. doi: 10.1038/6368. [DOI] [PubMed] [Google Scholar]
- 66.Lemaire V, Koehl M, Le Moal M, Abrous DN. Prenatal stress produces learning deficits associated with an inhibition of neurogenesis in the hippocampus. Proc. Natl. Acad. Sci. USA. 2000;97:11032–11037. doi: 10.1073/pnas.97.20.11032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Coe CL, et al. Prenatal stress diminishes neurogenesis in the dentate gyrus of juvenile rhesus monkeys. Biol. Psychiatry. 2003;54:1025–1034. doi: 10.1016/S0006-3223(03)00698-X. [DOI] [PubMed] [Google Scholar]
- 68.Mirescu C, Peters JD, Gould E. Early life experience alters response of adult neurogenesis to stress. Nat. Neurosci. 2004;7:841–846. doi: 10.1038/nn1290. [DOI] [PubMed] [Google Scholar]
- 69.Dranovsky A, et al. Experience dictates stem cell fate in the adult hippocampus. Neuron. 2011;70:908–923. doi: 10.1016/j.neuron.2011.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hou Y, et al. Aging as a risk factor for neurodegenerative disease. Nat. Rev. Neurol. 2019;15:565–581. doi: 10.1038/s41582-019-0244-7. [DOI] [PubMed] [Google Scholar]
- 71.Lyman M, Lloyd DG, Ji X, Vizcaychipi MP, Ma D. Neuroinflammation: the role and consequences. Neurosci Res. 2014;79:1–12. doi: 10.1016/j.neures.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 72.Shabab T, Khanabdali R, Moghadamtousi SZ, Kadir HA, Mohan G. Neuroinflammation pathways: a general review. Int. J. Neurosci. 2017;127:624–633. doi: 10.1080/00207454.2016.1212854. [DOI] [PubMed] [Google Scholar]
- 73.Villeda SA, et al. The aging systemic milieu negatively regulates neurogenesis and cognitive function. Nature. 2011;477:90–94. doi: 10.1038/nature10357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liddelow SA, et al. Neurotoxic reactive astrocytes are induced by activated microglia. Nature. 2017;541:481–487. doi: 10.1038/nature21029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Clarke LE, et al. Normal aging induces A1-like astrocyte reactivity. Proc. Natl. Acad. Sci. USA. 2018;115:E1896–E1905. doi: 10.1073/pnas.1800165115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mathys H, et al. Single-cell transcriptomic analysis of Alzheimer’s disease. Nature. 2019;570:332–337. doi: 10.1038/s41586-019-1195-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wolock SL, Lopez R, Klein AM. Scrublet: computational identification of cell doublets in single-cell transcriptomic data. Cell Syst. 2019;8:281–291. doi: 10.1016/j.cels.2018.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Stuart T, et al. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Aibar S, et al. SCENIC: single-cell regulatory network inference and clustering. Nat. Methods. 2017;14:1083–1086. doi: 10.1038/nmeth.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cao J, et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature. 2019;566:496–502. doi: 10.1038/s41586-019-0969-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Qiu X, et al. Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods. 2017;14:979–982. doi: 10.1038/nmeth.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Trapnell C, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 2014;32:381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Durinck S, et al. BioMart and Bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics. 2005;21:3439–3440. doi: 10.1093/bioinformatics/bti525. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The single-nucleus RNA-seq data used in this study has been deposited in the Gene Expression Omnibus (GEO) under the accession number GSE163737. The scATAC-seq data has been deposited in the Genome Sequence Archive (HRA002222).