Summary
Background
Early prediction of treatment response to neoadjuvant chemotherapy (NACT) in patients with human epidermal growth factor receptor 2 (HER2)-positive breast cancer can facilitate timely adjustment of treatment regimens. We aimed to develop and validate a Siamese multi-task network (SMTN) for predicting pathological complete response (pCR) based on longitudinal ultrasound images at the early stage of NACT.
Methods
In this multicentre, retrospective cohort study, a total of 393 patients with biopsy-proven HER2-positive breast cancer were retrospectively enrolled from three hospitals in china between December 16, 2013 and March 05, 2021, and allocated into a training cohort and two external validation cohorts. Patients receiving full cycles of NACT and with surgical pathological results available were eligible for inclusion. The key exclusion criteria were missing ultrasound images and/or clinicopathological characteristics. The proposed SMTN consists of two subnetworks that could be joined at multiple layers, which allowed for the integration of multi-scale features and extraction of dynamic information from longitudinal ultrasound images before and after the first /second cycles of NACT. We constructed the clinical model as a baseline using multivariable logistic regression analysis. Then the performance of SMTN was evaluated and compared with the clinical model.
Findings
The training cohort, comprising 215 patients, were selected from Yunnan Cancer Hospital. The two independent external validation cohorts, comprising 95 and 83 patients, were selected from Guangdong Provincial People's Hospital, and Shanxi Cancer Hospital, respectively. The SMTN yielded an area under the receiver operating characteristic curve (AUC) values of 0.986 (95% CI: 0.977–0.995), 0.902 (95%CI: 0.856–0.948), and 0.957 (95%CI: 0.924–0.990) in the training cohort and two external validation cohorts, respectively, which were significantly higher than that those of the clinical model (AUC: 0.524–0.588, Pall < 0.05). The AUCs values of the SMTN within the anti-HER2 therapy subgroups were 0.833-0.972 in the two external validation cohorts. Moreover, 272 of 279 (97.5%) non-pCR patients (159 of 160 (99.4%), 53 of 54 (98.1%), and 60 of 65 (92.3%) in the training and two external validation cohorts, respectively) were successfully identified by the SMTN, suggesting that they could benefit from regime adjustment at the early-stage of NACT.
Interpretation
The SMTN was able to predict pCR in the early-stage of NACT for HER2-positive breast cancer patients, which could guide clinicians in adjusting treatment regimes.
Funding
Key-Area Research and Development Program of Guangdong Province (No.2021B0101420006); National Natural Science Foundation of China (No.82071892, 82171920); Guangdong Provincial Key Laboratory of Artificial Intelligence in Medical Image Analysis and Application (No.2022B1212010011); the National Science Foundation for Young Scientists of China (No.82102019, 82001986); Project Funded by China Postdoctoral Science Foundation (No.2020M682643); the Outstanding Youth Science Foundation of Yunnan Basic Research Project (202101AW070001); Scientific research fund project of Department of Education of Yunnan Province(2022J0249). Science and technology Projects in Guangzhou (202201020001;202201010513); High-level Hospital Construction Project (DFJH201805, DFJHBF202105).
Keywords: Early prediction, Deep learning, HER2-positive breast cancer, Pathological complete response, Neoadjuvant chemotherapy, Ultrasound, Multi-task network
Abbreviations: pCR, pathological complete response; NACT, neoadjuvant chemotherapy; HER2, human epidermal growth factor receptor 2; SMTN, Siamese multi-task network; AUC, area under the receiver operating characteristic curve; CI, confidence interval
Research in context.
Evidence before this study
We searched the observational studies through PubMed with the terms “(breast cancer OR HER2-positive breast cancer) AND (response) AND (neoadjuvant chemotherapy) AND (early OR early prediction) AND (radiomics OR deep learning)” published from database inception up to 07 April 2022 with no language restrictions. We found that previous studies were mainly based on imaging with magnetic resonance imaging (MRI). Only three studies about artificial intelligence-based early response prediction using ultrasound images were published from March 2021 to February 2022. However, these single-centre studies did not consider the therapy-induced changes and effectively extract dynamic information from longitudinal images. As the biological biomarker of each tumor can be changed during neoadjuvant chemotherapy, and an early prediction of pathological complete response (pCR) to neoadjuvant chemotherapy can assist clinicians in adjusting therapy to increase rates of pCR and avoid toxic side effects in patients with human epidermal growth factor receptor 2 (HER2)-positive breast cancer. It is crucial to capture dynamic information from longitudinal images for early pCR prediction.
Added value of this study
In this multicentre study, we developed and validated a Siamese multi-task network for early pCR prediction in patients with HER2-positive breast cancer. The Siamese multi-task network was constructed by the dynamic information from longitudinal ultrasound images at the early stage of neoadjuvant chemotherapy. The proposed strategy outperformed the clinical model and the multi-task network trained with before or after the first/second cycle of neoadjuvant chemotherapy ultrasound images.
Implications of all the available evidence
Our findings showed that pCR to neoadjuvant chemotherapy could be predicted non-invasively by the proposed Siamese multi-task network in the early stage of neoadjuvant chemotherapy for HER2-positive breast cancer. In clinical practice, the Siamese multi-task network has the potential to guide individual therapy for HER2-positive breast cancer. For patients with the potential not to achieve a pCR, it is crucial to modify the treatment regimens to increase the rates of pCR. Comparatively, patients with the potential to achieve a pCR may benefit from breast-conserving surgery.
Alt-text: Unlabelled box
Introduction
Human epidermal growth factor receptor 2 (HER2)-positive accounts for 15-20% of breast cancer cases.1,2 Neoadjuvant chemotherapy (NACT) is the standard therapeutic option for patients with HER2-positive breast cancer.2,3 Pathological complete response (pCR) to NACT serves as a surrogate marker for long-term survival.4 However, HER2-positive breast cancer is heterogeneous with variable responses to NACT, with a pCR rate ranging from 20% to 80%.3 Early assessment of responder to NACT can assist clinicians in adjusting therapy to avoid toxic side effects, increase rates of pCR, and improve disease-free survival and overall survival.5, 6, 7 Thus, early prediction of pCR to NACT is of great clinical significance.
Previous efforts have been made to explore imaging features based on a single time point to predict pCR in the early stage of NACT.8,9 However, the biomarkers of tumors can be changed during neoadjuvant chemotherapy.10 Longitudinal monitoring of the dynamic changes is crucial for early pCR prediction. Mammography and magnetic resonance imaging (MRI) have been used to monitor tumor response to NACT.11,12 However, mammography has side effects of radiation, and performing multiple repeated MRI examinations in the short-term is difficult. In contrast, ultrasound is widely utilized to evaluate the treatment response in breast cancer because of its convenience and radiation-free features. However, the conventional ultrasound method is not accurate enough.13 No clinically accepted biomarkers are available for early pCR prediction. Surgical specimens remain the gold standard for pCR assessment. There is an urgent need for more broadly accessible, cost-efficient tools to facilitate early prediction of treatment response.
Machine learning has been extensively applied to the diagnosis, therapeutic evaluation, and prognosis prediction in various types of cancer.14, 15, 16 This promising technology has the potential to reveal tumor characteristics that failed to be detected by naked eyes.17 Nevertheless, previous researches have mainly concentrated on hand-crafted features. With the development of artificial intelligence, deep learning algorithms can automatically extract imaging features,18 which has demonstrated higher efficiency and reproducibility than hand-crafted features.19 Previous studies have evaluated tumor response in breast cancer based on before and mid-treatment ultrasound images using the deep learning method.20, 21, 22 However, these single-centre studies did not consider the therapy-induced changes and effectively extract dynamic information from longitudinal images. Siamese multi-task network (SMTN) was introduced to capturing dynamic information from longitudinal image.23 Furthermore, multi-task network could achieve positive feedbacks among related tasks to improve the performance of the overall model.24 Therefore, we, in the present study, developed SMTN that combined tumor segmentation and pCR prediction using dynamic change information from ultrasound images before and after the first/second cycle of NACT for HER2-positive breast cancer.
The present study aimed to develop and validate SMTN to predict early treatment response for HER2-positive breast cancer. Using the proposed SMTN, early adjusted therapy can be performed for non-pCR patients to increase rates of pCR and avoid toxic side effects.
Methods
Study design
The overall design of this study is shown in Figure 1. In the present multicentre, retrospective cohort study, we analysed clinicopathological characteristics and ultrasound imaging data of HER2-positive breast cancer patients who received NACT from three institutions. This study was approved by the ethics committees of Yunnan Cancer Hospital (YNCH), Guangdong Provincial People's Hospital (GPPH), and Shanxi Cancer Hospital (SCH), and was conducted according to the Declaration of Helsinki. A waiver of informed consent was granted due to the retrospective nature of the study. All patient-relevant information was anonymous and de-identified. We conducted the study following the Standards for Reporting of Diagnostic Accuracy (STARD) guidelines.
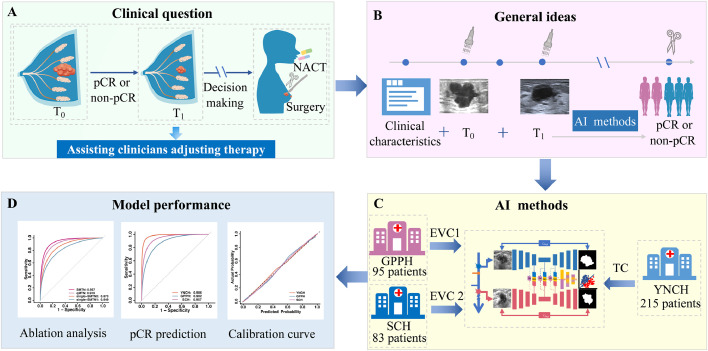
Figure 1.
The overall design of the study. (A) Early prediction of pCR in breast cancer can assist clinicians in adjusting therapy. (B) We constructed a deep learning-based model for early prediction of pCR combined clinical characteristics and longitudinal ultrasound images features. (C) Patients enrolled from YNCH were used as the training cohort, while others recruited from GPPH and SCH were used as two independent external validation cohorts. (D) Model performance was assessed using AUC and calibration curve. Abbreviations: pCR: pathological complete response; NACT: Neoadjuvant chemotherapy; T0: before neoadjuvant chemotherapy; T1: after the first/second cycle of neoadjuvant chemotherapy; AI: artificial intelligence; YNCH: Yunnan Cancer Hospital; GPPH: Guangdong Provincial People's Hospital; SCH: Shanxi Cancer Hospital; TC: training cohort; EVC: external validation cohort; AUC: area under the curve.
Patients
Data of patients with HER2-positive breast cancer who received NACT between December 16, 2013, and March 05, 2021, were collected from three hospitals in different regions of China. Patients enrolled from YNCH were used as the training cohort (TC), while others recruited from GPPH (external validation cohort 1, EVC1) and SCH (external validation cohort 2, EVC2) were used as two independent external validation cohorts (Supplementary Figure S1). Details of inclusion and exclusion criteria were shown in the Supplementary materials.
Conventional ultrasound protocol
Ultrasound examinations were performed by one of ten sonographers in the TC, one of three sonographers in the EVC1, and one of five sonographers in the EVC2. Each of whom had over 10 years of experience in breast ultrasound. The collected breast B-mode ultrasound images were converted into digital imaging and communications in medicine (DICOM) format in the TC and EVC 2, while those were converted into joint photographic experts group (JPEG) format in the EVC 1. Ultrasound examinations were conducted before (T0) and after the first/second cycle of NACT (T1). The breast ultrasound images were acquired by twelve different devices (Supplementary Table S1).
Image processing
In this study, per-image for per-lesion at T0 and T1 was used for analysis. To improve the learning efficiency of the SMTN, it is necessary to define a rectangular region of interest (ROI) by the coordinates in the ultrasound images to eliminate the interference caused by irrelevant data, such as text and icons. The X and Y coordinates in the ITK-SNAP (www.itksnap.org) were used to label the ROI with a rectangular bounding box. After that, the ROIs were cropped with the target lesion and the surrounding tissues (Supplementary Figure S2). All the images were addressed by data augmentation and normalization to reduce differences caused by different imaging protocols and intra/inter-operator variability. Data augmentation process was applied for improving the robustness (Supplementary materials).
Histopathology analysis
Ultrasound-guided needle biopsies were performed using 12- to 18-gauge needles for the diagnosis of the target breast tumor within 2 weeks before NACT. The status of estrogen receptor (ER), progesterone receptor (PR), HER2, and Ki-67 index was determined by immunohistochemistry (IHC). The cut-off level of Ki-67 was 30%.25 The status of ER and PR was regarded as positive if the tumor showed at least 1% of positive cells on nuclear staining.26 HER2-positive was defined as IHC 3+ or IHC 2+ and amplified by fluorescence in situ hybridization (FISH). HER2-negative was defined as IHC 0 or IHC 1+ or IHC 2+ and FISH-negative.2 The molecular subtypes were classified as hormone receptor (HR)-positive and HR-negative.
The status of pCR for each target tumor was determined by surgical-pathological results within 1 month after NACT. pCR was defined as a complete absence of invasive tumor cells both in the breast and axillary lymph nodes regardless of the presence of residual ductal carcinoma in situ (ypT0/isypN0).27
Clinicopathological data
Clinicopathological data were obtained from patient's medical records. Clinical data included age, menopausal status, clinical T stage, NACT regimen, and NACT cycles. Histopathological results of the breast cancer included tumor type, ER status, PR status, HER2 status, Ki-67 proliferation index, and pCR status.
Development of the deep learning-based model
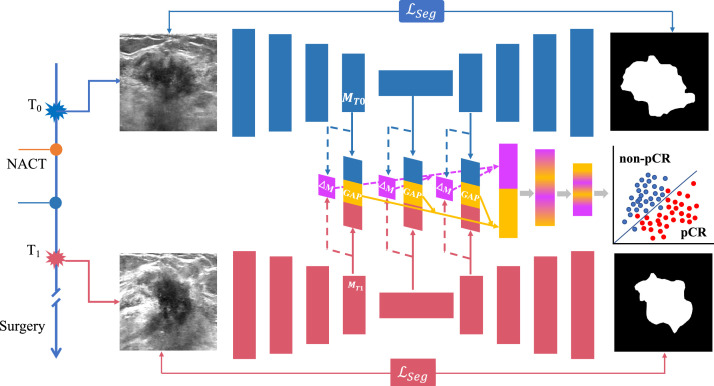
To simultaneously annotate tumor and capture dynamics imaging data from T0 and T1, we proposed a SMTN for breast tumor segmentation and pCR prediction. The SMTN was composed of two subnetworks (Figure 2): tumor segmentation networks and pCR prediction networks. The tumor segmentation network consists of two same U-nets to perform ultrasound image segmentation at T0 and T1 time points. To capture the dynamic change information of the tumor, the prediction network extracts different scale features from the layers of encoder, bottom and decoder of the two U-nets. The different scale features and dynamic change information were integrated through global average pooling and fully-connection, and finally used for pCR prediction.
Figure 2.
Details of the SMTN architecture. Our proposed SMTN contains two subnetworks: one for tumor segmentation consists of two Unets (upper and lower, represents by blue and red color), and the other (middle) for pCR prediction integrates the features from tumor segmentation subnetwork. The SMTN takes T0 and T1 images as inputs (image size: 256 × 256). Abbreviations: T0: before neoadjuvant chemotherapy; T1: after the first/second cycle of neoadjuvant chemotherapy; pCR: pathological complete response; NACT: neoadjuvant chemotherapy. GAP: global average pooling; ℒseg: segmentation loss. MT0, MT1: feature maps generated from T0 and T1 images via SMTN. ΔM: change values between MT0 and MT1. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Class imbalance is an important challenge for accurately predicting treatment response in our study. The number of non-pCR patients was twice more than that of pCR. To address this issue, we utilized a modified weighted focal loss to serve as an early pCR prediction loss function to mitigate the imbalance during training.28 The imbalance between the foreground and background of ultrasound was another challenge. To account for this, a combination of binary cross-entropy and Dice loss was utilized to train the segmentation subnetwork. Finally, the total loss was linearly combined with the pCR prediction and segmentation loss as a multi-task loss.
To alleviate the overfitting of the model, data augmentation was implemented using three transformations: 1) rotation, 2) flip, and 3) change lighting conditions (e.g., brightness, contrast, and saturation). In addition, strategies, such as dropout, batch normalization, and early stopping were employed in the network design to mitigate overfitting. The proposed SMTN was implemented using the Keras framework with TensorFlow backend on NVIDIA GeForce RTX 3090. Further details were described in the Supplementary Materials.
Ablation analysis for SMTN
To explore whether different components of the proposed network are required to accurately predict pCR, we conducted a set of ablation experiments29 by modifying the network structure. First, we removed half of the network and dynamic information change capture branch, while remained a multi-task network for simultaneous tumor segmentation and pCR prediction (denoted as single-MTN) (Supplementary Figure S3). Second, the dynamic information changes capture branch of SMTN was removed, and the remaining structure was used to predict pCR based on longitudinal ultrasound images (denoted as qMTN, Supplementary Figure S4). Third, we removed the decoders of SMTN (denoted as RD-SMTN), which perform pCR prediction only, but no segmentation (Supplementary Figure S5). In addition, we removed the decoders and the dynamic information changes capture branch of SMTN (denoted as RDD-SMTN). Again, this is a single-task learning designed for pCR prediction only (Supplementary Figure S6). In addition, we substituted the SMTN architecture with Siamese Densenet12120 (denote as S-Densenet121) and concatenated features from the fully connection layer for the T0 and T1 images. This network was used for pCR prediction only.
Integration with clinicopathological characteristics
Univariable analysis was performed to identify covariates associated with pCR. Multivariable logistic regression analysis was conducted to construct a clinical model (Supplementary Figure S7A). Then, we further developed an integration model with both an image-based model and clinicopathological characteristics.
Model performance evaluation metrics
First, to evaluate SMTN segmentation performance, we compared it against manual delineation. Second, to confirm the importance of integrating longitudinal information in multi-task networks, we compared the performance between SMTN and the single-MTN model. Third, to estimate the importance of capturing the dynamic information from longitudinal ultrasound images, we compared the performance between SMTN and qMTN. Lastly, to investigate whether tumor segmentation was truly necessary to achieve good performance, we compared the performance of SMTN with RD-SMTN and RDD-SMTN.
Then, the model performance for early pCR prediction was assessed using the receiver operating characteristic (ROC) curve, and the area under the curve (AUC) was calculated. We defined pCR as a positive event. False positive event was defined as patients with pCR predicted by SMTN who were non-pCR in the surgery. We also calculated the sensitivity, specificity, accuracy, positive-predictive value (PPV), and negative-predictive value (NPV) for pCR prediction. For tumor segmentation, the dice coefficient (Dice) was calculated.
Finally, to ascertain the decision information captured by SMTN in predicting pCR, we utilized gradient-weighted class activation mapping (Grad-CAM) to visualize the location and distribution of decision making on ultrasound images.30
Statistical analysis
Descriptive statistics were summarized as mean ± standard deviation (SD). Comparisons between groups were made using the Student's t-test or the Mann-Whiney U test for continuous variables and the Chi-square or the Fisher's exact test for categorical variables. The AUC was used to estimate the probability of the correct prediction of pCR. Differences between various AUCs were compared by the DeLong test.31 All statistical tests were two-sided, and P < 0.05 was considered statistically significant. All statistical analyses were performed using R 3.4.1 and SPSS 23.0 (IBM, Armonk, NY, USA) software.
Role of the funding source
The funder of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report. All authors had full access to all the data and approved the final manuscript for submission.
Results
Baseline characteristics
A total of 393 patients (215 patients in the TC, 95 patients in the EVC 1, and 83 patients in the EVC 2) were consecutively enrolled for further analysis (Supplementary materials, Supplementary Figure S1). All patients underwent anthracycline-based and/or taxane-based regimens. The details of neoadjuvant chemotherapy regimens were shown in the Supplementary Table S2. Anti-HER2 agents were used in 41.9% (90/215) of patients in the TC (24.7% patients treated with trastuzumab, 17.2% treated with trastuzumab plus pertuzumab), 89.5% (85/95) of patients in the EVC 1(72.6% patients treated with trastuzumab, 16.9% treated with trastuzumab plus pertuzumab), and 32.5% (27/83) of patients in the EVC 2(19.2% patients treated with trastuzumab, 13.3% treated with trastuzumab plus pertuzumab) (Table 1).
Table 1.
Clinicopathological characteristics of patients in the training cohort and external validation cohorts.
| characteristics | Training cohort (n = 215) |
P | External validation cohort 1 (n = 95) |
P | External validation cohort 2 (n = 83) |
P | |||
|---|---|---|---|---|---|---|---|---|---|
| pCR | non-pCR | pCR | non-pCR | pCR | non-pCR | ||||
| Age (mean ± SD) | 49.1±8.0 | 47.1±8.6 | 0.522 | 50.8±8.3 | 49.4±9.2 | 0.539 | 49.8±12.8 | 50.9±9.1 | 0.070 |
| Age group | 0.215 | 0.103 | 0.790 | ||||||
| <40 | 7 | 27 | 6 | 8 | 4 | 10 | |||
| 40-50 | 12 | 50 | 10 | 24 | 5 | 20 | |||
| ≥50 | 36 | 83 | 25 | 22 | 9 | 35 | |||
| menopausal status | 0.419 | 0.315 | 0.601 | ||||||
| premenopausal | 36 | 114 | 17 | 28 | 9 | 37 | |||
| Postmenopausal | 19 | 46 | 24 | 26 | 9 | 28 | |||
| Clinical T stage | 0.513 | 0.071 | 0.769 | ||||||
| T1 | 0 | 4 | 8 | 3 | 5 | 12 | |||
| T2 | 40 | 111 | 29 | 40 | 12 | 46 | |||
| T3 | 8 | 30 | 1 | 7 | 1 | 6 | |||
| T4 | 7 | 15 | 3 | 4 | 0 | 1 | |||
| ER | 0.148 | < 0.01 | 0.155 | ||||||
| Negative | 26 | 58 | 24 | 13 | 10 | 24 | |||
| Positive | 29 | 102 | 17 | 41 | 8 | 41 | |||
| PR | 0.962 | 0.002 | 0.529 | ||||||
| Negative | 17 | 50 | 29 | 21 | 12 | 38 | |||
| Positive | 38 | 110 | 12 | 33 | 6 | 27 | |||
| Ki-67 | 0.018 | 0.635 | 0.477 | ||||||
| Negative | 12 | 63 | 14 | 21 | 3 | 16 | |||
| Positive | 43 | 97 | 27 | 33 | 15 | 49 | |||
| NACT regimen | 0.402 | 0.403 | 0.032 | ||||||
| Anthracycline-based | 2 | 7 | 0 | 0 | 0 | 1 | |||
| Taxane-based | 5 | 26 | 27 | 31 | 7 | 8 | |||
| Anthracycline and Taxane-based |
48 | 127 | 14 | 23 | 11 | 56 | |||
| Anthracycline | 0.192 | 0.403 | 0.017 | ||||||
| No | 5 | 26 | 27 | 31 | 7 | 9 | |||
| Yes | 50 | 134 | 14 | 23 | 11 | 56 | |||
| Anti-HER2 therapy | 0.065 | 0.246 | 0.001 | ||||||
| No | 27 | 98 | 4 | 6 | 8 | 48 | |||
| H | 20 | 33 | 33 | 36 | 3 | 13 | |||
| HP | 8 | 29 | 4 | 12 | 7 | 4 | |||
| Molecular subtype | 0.669 | 0.003 | 0.130 | ||||||
| HR-negative | 15 | 39 | 22 | 13 | 9 | 20 | |||
| HR-positive | 40 | 121 | 19 | 41 | 9 | 45 | |||
Data were presented as number of patients, with the exception of age (mean ± SD).
Abbreviations: pCR: pathological complete response; SD: standard deviation; NACT: neoadjuvant chemotherapy; H: trastuzumab; HP: trastuzumab plus pertuzumab; ER: estrogen receptor; PR: progesterone receptor; HER2: human epidermal growth factor receptor; HR: hormone receptor.
The pCR rates ranged from 21.7% to 43.2% in the three cohorts. The baseline clinicopathological characteristics of the three cohorts are summarized in Table 1. No significant difference was detected between patients with pCR and non-pCR in terms of age, menopausal status, and clinical T stage (P > 0.05) among the three cohorts. pCR was significantly associated with Ki-67 index only in the TC (P < 0.05). ER, PR, and molecular subtypes showed a significant association with pCR only in the EVC 1. NACT regimen, anthracycline-containing, and anti-HER2 therapy were significantly associated with pCR status only in the EVC 2. Multivariable logistic regression analysis revealed that Ki-67 index was a significant predictor of pCR status in the TC (Supplementary Figure S7A), and it was used to construct the clinical model as a baseline. Consistent with previous studies,32,33 the performance of clinical models in predicting pCR was unsatisfactory, with AUC values of 0.588 (95% confidence interval (CI): 0.521, 0.655), 0.524 (95%CI: 0.425, 0.622), and 0.540 (95%CI: 0.437, 0.643) in the three cohorts. (Supplementary Table S3).
Model performance
To evaluate the segmentation performance of the SMTN, we compared the segmentation results with manual delineation. The SMTN achieved promising segmentation performances in two external cohorts (DICEmean > 0.764 (range: 0.764-0.793, 0.793 ± 0.098 in the TC, 0.764 ± 0.096 in the EVC1, 0.782 ± 0.100 in the EVC2, respectively, Supplementary Figure S8)).
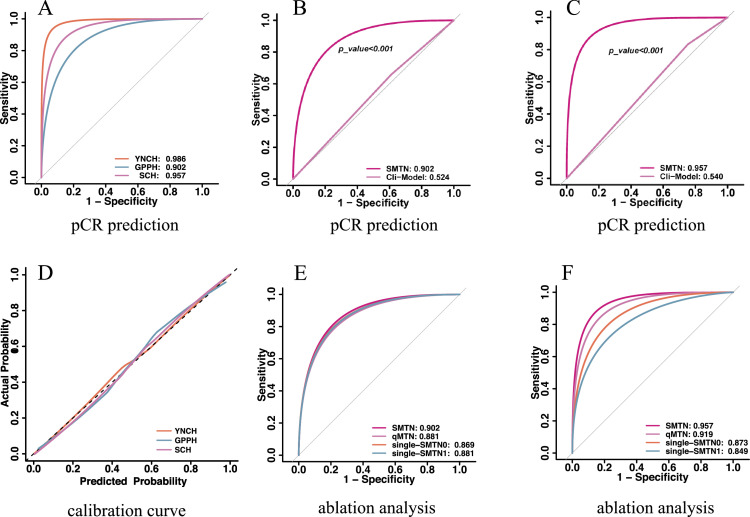
The proposed SMTN obtained a high accuracy with an AUC of 0. 986 (95%CI: 0.977-0.995) in the TC. Furthermore, the SMTN achieved consistently a high accuracy with AUC values of 0.902 (95%CI: 0.856-0.948) and 0.957 (95%CI: 0.924-0.990) in two EVCs, respectively (Table 2). Notably, the sensitivity and specificity slightly decreased, whereas were still satisfactory, achieving 0.863 (95%CI: 0.794-0.932), and 0.874 (95%CI: 0.807-0.940) in the EVC 1, and 0.940 (95%CI: 0.889-0.991) and 0.904 (95%CI: 0.840-0.967) in the EVC 2, respectively. Additionally, the SMTN performed remarkably well in terms of PPV and NPV, ranging from 0.872 (95%CI: 0.805, 0.940) to 0.907 (95%CI: 0.846, 0.968), and 0.865 (95%CI: 0.796-0.933) to 0.938 (95%CI: 0.884-0.991) in two EVCs (Supplementary Table S4, Figure 3A). Moreover, 272 of 279 (97.5%) patients with non-pCR (159 of 160 (99.4%), 53 of 54 (98.1%), and 60 of 65 (92.3%) patients in the TC and two EVCs, respectively) were successfully identified by SMTN. Meanwhile, 94 of 114 (82.5%) patients with pCR (49 of 55 (89.1%), 30 of 41 (73.2%), 15 of 18 (83.3%) patients in the TC and two EVCs, respectively) were successfully identified by the SMTN (Supplementary Figure S9).
Table 2.
The performance of models.
| Models | AUC (95%CI) |
||
|---|---|---|---|
| Training cohort | External validation cohort 1 | External validation cohort 2 | |
| SMTN | 0.986 (0.977,0.995) | 0.902 (0.856, 0.948) | 0.957 (0.924, 0.990) |
| Clinical model | 0.588 (0.521, 0.655) | 0.524 (0.425, 0.622) | 0.540 (0.437,0.643) |
| Clinical + SMTN | 0.989 (0.982, 0.996) | 0.904 (0.861, 0.948) | 0.952 (0.920, 0.983) |
Abbreviations: AUC: area under the receiver operating characteristics curve; SMTN: Siamese multi-task network.
Figure 3.
Performances for pCR prediction. A: AUCs of the SMTN in the training cohort and two validation cohorts; B: AUCs of the SMTN and clinical model in the external validation cohort 1; C: AUCs of the SMTN and clinical model in the external validation cohort 2; D: The calibration curve of the SMTN in the training cohort and two validation cohorts; E: AUC of the SMTN, qMTN, single-MTN0, and single-MTN1 in the external validation cohort 1; F: AUC of the SMTN, qMTN, single-MTN0, and single-MTN1 in the external validation cohort 2. pCR: pathological complete response; ROC: receiver operating characteristics; AUC: area under the receiver operating characteristics curve; YNCH: Yunnan Cancer Hospital (training cohort); GPPH: Guangdong Provincial People's Hospital (external validation cohort 1); SCH: Shanxi Cancer Hospital (external validation cohort 2); SMTN: Siamese multi-task network; Clin-model: clinical model; single-MTN0: a multi-task network trained with ultrasound image before neoadjuvant chemotherapy; single-MTN1: a multi-task network trained with ultrasound image after the first/second cycle of neoadjuvant chemotherapy, qMTN: the dynamic information changes capture branch of SMTN was removed, and the remaining structure was used to predict pCR based on longitudinal ultrasound images.
Finally, we evaluated the calibration of the SMTN in pCR prediction. The predicted probabilities of the model were close to the observed probabilities and showed a good calibration (Hosmer-Lemeshow test: P-value ranged from 0.103 to 0.442 in the three cohorts, Figure 3D).
Superiority verification of SMTN
The SMTN, integrating temporal and spatial information from longitudinal ultrasound images, showed a superior performance in evaluating treatment response compared with the traditional clinical model and single ultrasound information.
Compared with the clinical model, the predictive performance of the SMTN was better than that of the clinical model among the three cohorts (Figure 3B-C, Table 2, Supplementary Tables S3-S4). We further integrated the clinical model into the SMTN to achieve better predictive performance. However, it did not improve the predictive performance of the SMTN (Table 2, Supplementary Tables S4-S5). We found that none of the clinical factors were significant variables by analyzing their relative contribution (Supplementary Figure S7B).
To illustrate the advantages of longitudinal ultrasound in the early prediction of pCR, we trained single-MTN with ultrasound images at T0 or T1 time points, respectively. Unsurprisingly, the SMTN achieved a better predictive performance than the single-MTN (AUC: 0.986 vs. 0.858 vs. 0.916 in the TC (P<0.001), 0.902 vs. 0.869 vs. 0.881 in the EVC 1 (P > 0.05), 0.957 vs. 0.873 vs. 0.849 (P <0.05) in the EVC 2 (P<0.05) (Supplementary Table S4 and Tables S6-7, Figure 3E-F).
To estimate the importance of capturing the dynamic information from longitudinal ultrasound images, we simultaneously trained the SMTN and qMTN on longitudinal ultrasound images and compared their performance. There was a significant improvement in prediction accuracy after adding dynamic information in the TC (AUC: 0.918 to 0.986, DeLong test, P < 0.05). Although differences did not reach statistical significance, there was also an increase in predictive performance in the EVCs (AUC: 0.881 to 0.902, 0.919 to 0.957, respectively, DeLong test, P > 0.05) (Tables S4 and S8, and Figure 3E-F).
To investigate whether tumor segmentation is truly necessary for accurate pCR prediction, we then compared the performance between SMTN and single-task learning. The proposed SMTN achieved higher performance than RD-SMTN and RDD-SMTN (AUC: 0.986 vs.0.890 vs .0.832 in the TC (PSMTN VS. RD-SMTN < 0.001, PSMTN VS. RDD-SMTN < 0.001), 0.902 vs. 0.876 vs. 0.811 in the EVC 1 (PSMTN VS. RD-SMTN =0.451, PSMTN VS. RDD-SMTN =0.019), 0.957 vs. 0.913 vs.0.843 (PSMTN VS. RD-SMTN =0.118, PSMTN VS. RDD-SMTN =0.001) in the EVC 2 (Supplementary Table S4 and Tables S9-10, Supplementary Figure S10).
The proposed SMTN also achieved better performance for pCR prediction compared with S- Densenet121 (AUC: 0.902 vs. 0.841 in the EVC1(P =0.097), 0.957 vs. 0.842 in the EVC2 (P=0.001), respectively).
Subgroup analysis of the SMTN
The SMTN achieved a good performance among anti-HER2 therapy groups, with AUC values of 0.940 (no anti-HER2 therapy), 0.903 (trastuzumab), and 0.895 (trastuzumab and pertuzumab) in the EVC 1, and 0.972 (no anti-HER2 therapy), 0.833 (trastuzumab) and 0.959 (trastuzumab and pertuzumab) in the EVC 2 (Supplementary Table S11).
We conducted a subgroup analysis based on HR. Prediction within HR- and HR+ subgroups achieved a good performance, with AUC values of 0.869 (HR-) and 0.953 (HR+) in the EVC 1, and 0.940 (HR-) and 0.988 (HR+) in the EVC 2 (Supplementary Table S12).
We also performed subgroup analysis based on the different ultrasound machines. The performances of SMTN in the subgroup of the same and different ultrasound machines used at T0 and T1 were similar in the whole study population (supplementary Table S13). We then performed subgroup analysis based on the ultrasound machines in patients who underwent ultrasound examination at T0 and T1 using the same machine, the performances were better in patients underwent ultrasound examination using Hitachi (supplementary Table S14). This may be because of the higher amount of data in the Hitachi group.
Interpretability of the SMTN
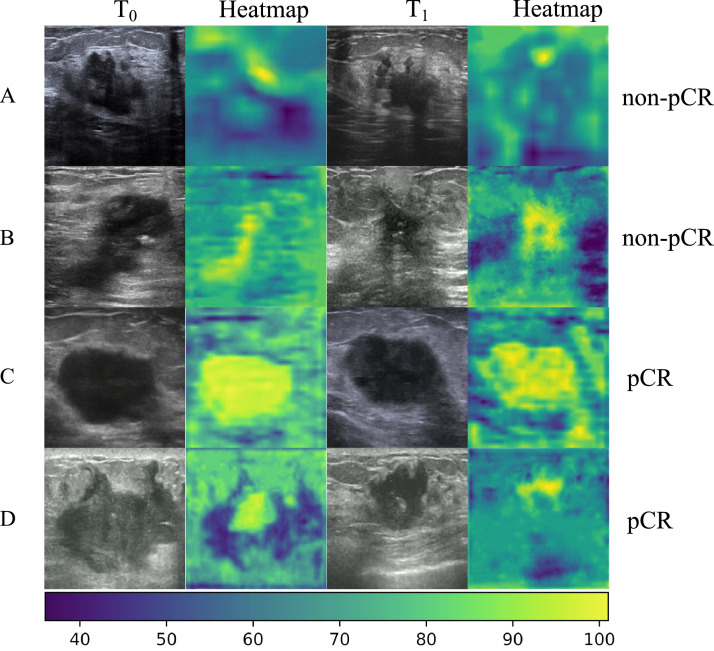
For investigating the interpretability of the SMTN in early pCR prediction, the heatmaps were generated in key channels at the bottom of the SMTN (Figure 4). There were two locations valuable for early pCR prediction on ultrasound images (namely, the peritumor region and intratumor region). To better understand the decision made by the SMTN, we selected four patients in the external validation cohorts, two with pCR and two with non-pCR (Figure 4). For patients with non-pCR, the peritumor region was highlighted, while the intratumor region was not in T0 and T1 ultrasound images (Figure 4A-B). For patients with pCR, the intratumor region was highlighted, whereas the peritumor region was not in T0 and T1 ultrasound images (Figure 4C-D). In addition, we also selected four patients who failed to be correctly predicted (Supplementary Figure S11). For patients who were wrongly predicted, the SMTN cannot focus on the lesions. In addition, we demonstrated that more than half of the 256 feature map channels in T1 were inactive (feature map active rate <50%) compared to T0 for patient with pCR. What's more, the proportion of active regions in feature map was reduced by 3%. By contrast, more than half of the feature map channels were activated at T1 for patient with non-pCR, and the proportion of activated regions increased by 5.7% (Supplementary Figure S12).
Figure 4.
SMTN visualization and interpretation. Color-code heatmaps overlaid with the corresponding ultrasound images at T0 and T1 time points for four patients who were accurately predicted. A, B: The heatmaps highlighted the peritumor tissue in patients with non-pCR, indicating that information exploited from the peritumor region of the tumor contributed to the prediction of non-pCR by the SMTN; C, D: The heatmaps highlighted the intratumor region of the tumor in patients with pCR, indicating that information exploited from the intratumor region of the tumor contributed to the prediction of pCR by the SMTN. T0: before neoadjuvant chemotherapy; T1: after the first/second cycle of neoadjuvant chemotherapy; pCR: pathological complete response; SMTN: Siamese multi-task network. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Discussion
In the present multicentre study, we developed and validated a deep learning-based model for early pCR prediction using longitudinal ultrasound images at the early stage of NACT in patients with HER2-positive breast cancer. The SMTN achieved favorable results in terms of AUC, sensitivity, specificity, and calibration curve. Moreover, the performance of the SMTN was not influenced by HR molecular subtypes and anti-HER2 therapy regimens. Our study presented a precise and robust model, which could assist clinicians in the early adjusting treatment regimens for patients with non-pCR, thereby increasing the rates of pCR and avoiding toxic effects.
The biomarkers of each tumor can be changed during NACT,10 including tumor size and tumor cellular density.34 These changes can be reflected on longitudinal ultrasound images.34,35 Thus, it is crucial to integrate longitudinal ultrasound images for early pCR prediction. To capture the dynamic changes induced by NACT, we developed the SMTN using two Siamese subnetworks based on ultrasound images at T0 and T1 time points. As expected, the SMTN achieved an increased performance compared with single-MTN, qMTN, RD-SMTN, and RDD-SMTN (AUC: 0.902 vs. 0.869 vs. 0.881 vs. 0.881 vs.0.876 vs.0.811 in the EVC1, and 0.957 vs. 0.873 vs. 0.849 vs.0.919 vs. 0.913 vs. 0.843 in the EVC2). Previous studies have evaluated early treatment response using deep learning methods based on before and mid-treatment ultrasound images, with AUC values of 0.797-0.94.20,21 However, they did not take therapy-induced changes into account. Moreover, the clinical applicability remained unclear as these studies had a small sample size and were not validated in the external validations, resulting in the possibility of over-fitting. Radiomics has been used to predict pCR based on pre- and mid-treatment ultrasound images of breast cancer patients with an AUC of 0.866 in the internal validation cohort,36 which had a lower prediction performance than our method. The reason for this discrepancy may be because our model not only captured the intratumor information but also peritumor information for early pCR prediction, which was evident from heatmaps (Figure 4). We also surveyed the current literature on pCR prediction for breast cancer based on MRI or positron emission tomography (PET). Our results showed that SMTN was superior to other prediction models based on MRI and PET findings.14,37, 38, 39 This promising result presents the ability to early predict pCR using less complex, less costly, and more accessible ultrasound images compared with MRI or PET, which has enormous potential clinical and economic benefits.
The proposed SMTN provided an automatic segmentation approach that did not require any handcrafted features. Tumor segmentation was extremely important for pCR prediction. Previous studies mainly used manual segmentation for pCR prediction,20,40 which is mainly subjective and time-consuming. In the present study, we developed a multi-task network that could automatically segment tumors. The performance of algorithms for automated segmentation was highly consistent with manual delineation (DICEmean > 0.764 (range: 0.764 − 0793)). The proposed networks saved time for manual delineation and reduced inter/intra-observer variability even among senior sonographers.
Regarding the clinical applicability, the SMTN has the potential to guide individual therapy for HER2-positive breast cancer. For patients without a potential to achieve a pCR, it is necessary to modify the treatment regimens to reduce the toxic effects and increase the rates of pCR. In this study, 272 of 279 (97.5%) non-pCR patients were successfully identified by the SMTN who may benefit from adjusting treatment regimes. Comparatively, for patients with a potential to achieve a pCR, it is worthwhile to administer NACT with the same chemotherapy regimens. Of the 114 patients with pCR in our study, 94 patients were predicted accurately who may benefit from breast-conserving surgery and omission of axillary node dissection.
The heatmap enabled clinicians to visualize the basis of the SMTN prediction, which could guide their clinical decision. The heatmaps highlighted the intratumor region in patients with pCR and peritumor tissue in patients with non-pCR. Importantly, pCR was associated with changes in tumor echogenicity.34 In contrast, tumor microenvironment in peritumor tissue (for example, lymph vascular invasion) was associated with non-pCR.33,41 Importantly, we observed substantial decreases in features form T0 and T1 ultrasound images for patients with pCR, while there were increases in features in patients with non-pCR. This further indicates that information about treatment response is mainly contained in the change of ultrasound imaging phenotypes at T0 and T1, confirming the effectiveness of our approach.
To investigate whether anti-HER2 therapy regimes could affect the performance of the SMTN, stratification analysis in subgroups was performed. We found that the SMTN achieved a promising performance in no anti-HER2 therapy, trastuzumab (H), and trastuzumab plus pertuzumab (HP) groups, with AUCs values of 0.833-0.972. The SMTN could guide clinicians in identifying subpopulations of HER2-positive patients who would benefit from more/less therapy and spare some patients from unnecessary exposure to ineffective treatments.
Some limitations of the present study should be pointed out. Firstly, some patients with HER2-positive breast cancer did not receive anti-HER2 therapy. Because China is a developing country, some patients may not be able to afford anti-HER2 therapy, especially patients in Yunnan and Shanxi provinces. Secondly, a limited number of patients underwent ultrasound at the first and second cycles of NACT, thus, we did not analyse prediction results in the first and second cycles separately, which should be studied in the future research. Last but not least, this is a retrospective study, and more evidence from prospective studies is required to validate the SMTN before its clinical application in the future.
In summary, the proposed SMTN could be a promising strategy for early pCR prediction in HER2-positive breast cancer following NACT. Future prospective studies on a large group of patients are warranted.
Contributors
Conceptualization: Y.L., L.W., Y.W.
Methodology: Y.L., L.W., YX.W., Y.X., XT.Y.
Accessing and verifying the underlying data: Y.L., L.W., ZY.L., YF.C.
Investigation: Y.L., YF.C., L.W., D.C., CH.L.
Visualization: YF.C., SW.F., WQ.S., X.C.
Supervision: Y.X., MX.Y., BJ.Q., GQ.D.
Writing—original draft: Y.L., YX.W., L.W., Y.X., Y.W.
Writing—review & editing: CH.L., ZY.L., ZH.L., XT.Y., L.W.
Data sharing statement
Due to the privacy of patients, the data related to patients cannot be available for public access but can be obtained from the corresponding author (https://liangchanghong@gdph.org.cn) on reasonable request approved by the institutional review board of all enrolled centres.
Declaration of interests
All authors declare no competing interests.
Acknowledgments
This work was funded by the Key-Area Research and Development Program of Guangdong Province (No.2021B0101420006); National Natural Science Foundation of China (No.82071892, 82171920); Guangdong Provincial Key Laboratory of Artificial Intelligence in Medical Image Analysis and Application (No.2022B1212010011); the National Science Foundation for Young Scientists of China (No.82102019, 82001986); Project Funded by China Postdoctoral Science Foundation (No.2020M682643); the Outstanding Youth Science Foundation of Yunnan Basic Research Project (202101AW070001); Scientific research fund project of Department of Education of Yunnan Province(2022J0249). Science and technology Projects in Guangzhou (202201020001; 202201010513); High-level Hospital Construction Project (DFJH201805, DFJHBF202105).
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.eclinm.2022.101562.
Contributor Information
Zhenhui Li, Email: lizhenhui621@qq.com.
Xiaotang Yang, Email: yangxt210@126.com.
Changhong Liang, Email: liangchanghong@gdph.org.cn.
Lei Wu, Email: wl858998458@163.com.
Appendix. Supplementary materials
References
- 1.Zhao B, Zhao H. Impact of clinicopathological characteristics on the efficacy of neoadjuvant therapy in patients with human epidermal growth factor receptor-2-positive breast cancer. Int J Cancer. 2018;142(4):844–853. doi: 10.1002/ijc.31097. [DOI] [PubMed] [Google Scholar]
- 2.Loibl S, Gianni L. HER2-positive breast cancer. Lancet. 2017;389(10087):2415–2429. doi: 10.1016/S0140-6736(16)32417-5. [DOI] [PubMed] [Google Scholar]
- 3.Pusztai L, Foldi J, Dhawan A, et al. Changing frameworks in treatment sequencing of triple-negative and HER2-positive, early-stage breast cancers. Lancet Oncol. 2019;20(7):e390–e396. doi: 10.1016/S1470-2045(19)30158-5. [DOI] [PubMed] [Google Scholar]
- 4.Spring LM, Fell G, Arfe A, et al. Pathologic complete response after neoadjuvant chemotherapy and impact on breast cancer recurrence and survival: a comprehensive meta-analysis. Clin Cancer Res. 2020;26(12):2838–2848. doi: 10.1158/1078-0432.CCR-19-3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fatayer H, Sharma N, Manuel D, et al. Serial MRI scans help in assessing early response to neoadjuvant chemotherapy and tailoring breast cancer treatment. Eur J Surg Oncol. 2016;42(7):965–972. doi: 10.1016/j.ejso.2016.03.019. [DOI] [PubMed] [Google Scholar]
- 6.Coudert B, Pierga JY, Mouret-Reynier MA, et al. Use of [(18)F]-FDG PET to predict response to neoadjuvant trastuzumab and docetaxel in patients with HER2-positive breast cancer, and addition of bevacizumab to neoadjuvant trastuzumab and docetaxel in [(18)F]-FDG PET-predicted non-responders (AVATAXHER): an open-label, randomised phase 2 trial. Lancet Oncol. 2014;15(13):1493–1502. doi: 10.1016/S1470-2045(14)70475-9. [DOI] [PubMed] [Google Scholar]
- 7.von Minckwitz G, Blohmer JU, Costa SD, et al. Response-guided neoadjuvant chemotherapy for breast cancer. J Clin Oncol. 2013;31(29):3623–3630. doi: 10.1200/JCO.2012.45.0940. [DOI] [PubMed] [Google Scholar]
- 8.Chen JH, Bahri S, Mehta RS, et al. Breast cancer: evaluation of response to neoadjuvant chemotherapy with 3.0-T MR imaging. Radiology. 2011;261(3):735–743. doi: 10.1148/radiol.11110814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bae MS, Shin SU, Ryu HS, et al. Pretreatment MR imaging features of triple-negative breast cancer: association with response to neoadjuvant chemotherapy and recurrence-free survival. Radiology. 2016;281(2):392–400. doi: 10.1148/radiol.2016152331. [DOI] [PubMed] [Google Scholar]
- 10.Park YH, Lal S, Lee JE, et al. Chemotherapy induces dynamic immune responses in breast cancers that impact treatment outcome. Nat Commun. 2020;11(1):6175. doi: 10.1038/s41467-020-19933-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Di Cosimo S, Campbell C, Azim HA, Jr., et al. The use of breast imaging for predicting response to neoadjuvant lapatinib, trastuzumab and their combination in HER2-positive breast cancer: results from Neo-ALTTO. Eur J Cancer. 2018;89:42–48. doi: 10.1016/j.ejca.2017.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hylton NM, Blume JD, Bernreuter WK, et al. Locally advanced breast cancer: MR imaging for prediction of response to neoadjuvant chemotherapy–results from ACRIN 6657/I-SPY TRIAL. Radiology. 2012;263(3):663–672. doi: 10.1148/radiol.12110748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marinovich ML, Houssami N, Macaskill P, et al. Accuracy of ultrasound for predicting pathologic response during neoadjuvant therapy for breast cancer. Int J Cancer. 2015;136(11):2730–2737. doi: 10.1002/ijc.29323. [DOI] [PubMed] [Google Scholar]
- 14.Liu Z, Li Z, Qu J, et al. Radiomics of multiparametric MRI for pretreatment prediction of pathologic complete response to neoadjuvant chemotherapy in breast cancer: a multicenter study. Clin Cancer Res. 2019;25(12):3538–3547. doi: 10.1158/1078-0432.CCR-18-3190. [DOI] [PubMed] [Google Scholar]
- 15.Huang YQ, Liang CH, He L, et al. Development and validation of a radiomics nomogram for preoperative prediction of lymph node metastasis in colorectal cancer. J Clin Oncol. 2016;34(18):2157–2164. doi: 10.1200/JCO.2015.65.9128. [DOI] [PubMed] [Google Scholar]
- 16.Park H, Lim Y, Ko ES, et al. Radiomics signature on magnetic resonance imaging: association with disease-free survival in patients with invasive breast cancer. Clin Cancer Res. 2018;24(19):4705–4714. doi: 10.1158/1078-0432.CCR-17-3783. [DOI] [PubMed] [Google Scholar]
- 17.Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology. 2016;278(2):563–577. doi: 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu Y, Hosny A, Zeleznik R, et al. Deep learning predicts lung cancer treatment response from serial medical imaging. Clin Cancer Res. 2019;25(11):3266–3275. doi: 10.1158/1078-0432.CCR-18-2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Russakovsky O, Deng J, Su H, et al. Imagenet large scale visual recognition challenge. Int J Comput Vis. 2015;115(3):211–252. [Google Scholar]
- 20.Gu J, Tong T, He C, et al. Deep learning radiomics of ultrasonography can predict response to neoadjuvant chemotherapy in breast cancer at an early stage of treatment: a prospective study. Eur Radiol. 2022;32(3):2099–2109. doi: 10.1007/s00330-021-08293-y. [DOI] [PubMed] [Google Scholar]
- 21.Byra M, Dobruch-Sobczak K, Klimonda Z, et al. Early prediction of response to neoadjuvant chemotherapy in breast cancer sonography using siamese convolutional neural networks. IEEE J Biomed Health Inform. 2021;25(3):797–805. doi: 10.1109/JBHI.2020.3008040. [DOI] [PubMed] [Google Scholar]
- 22.Xie J, Shi H, Du C, et al. Dual-branch convolutional neural network based on ultrasound imaging in the early prediction of neoadjuvant chemotherapy response in patients with locally advanced breast cancer. Front Oncol. 2022;12 doi: 10.3389/fonc.2022.812463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jin C, Yu H, Ke J, et al. Predicting treatment response from longitudinal images using multi-task deep learning. Nat Commun. 2021;12(1):1851. doi: 10.1038/s41467-021-22188-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fu S, Lai H, Huang M, et al. Multi-task deep learning network to predict future macrovascular invasion in hepatocellular carcinoma. EClinicalMedicine. 2021;42 doi: 10.1016/j.eclinm.2021.101201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nielsen TO, Leung SCY, Rimm DL, et al. Assessment of Ki67 in breast cancer: updated recommendations from the international Ki67 in Breast Cancer Working Group. J Natl Cancer Inst. 2021;113(7):808–819. doi: 10.1093/jnci/djaa201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Allison KH, Hammond MEH, Dowsett M, et al. Estrogen and progesterone receptor testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists Guideline Update. Arch Pathol Lab Med. 2020;144(5):545–563. doi: 10.5858/arpa.2019-0904-SA. [DOI] [PubMed] [Google Scholar]
- 27.von Minckwitz G, Untch M, Blohmer JU, et al. Definition and impact of pathologic complete response on prognosis after neoadjuvant chemotherapy in various intrinsic breast cancer subtypes. J Clin Oncol. 2012;30(15):1796–1804. doi: 10.1200/JCO.2011.38.8595. [DOI] [PubMed] [Google Scholar]
- 28.Lin TY, Goyal P, Girshick R, et al. 2017 IEEE International Conference on Computer Vision (ICCV) 2017. Focal loss for dense object detection. [Google Scholar]
- 29.Vishnusai Y, Kulakarni TR, Sowmya Nag K. In: Innovative Data Communication Technologies and Application. ICIDCA 2019. Lecture Notes on Data Engineering and Communications Technologies. Raj J., Bashar A., Ramson S., editors. Springer International Publishing; Cham: 2020. Ablation of artificial neural networks; pp. 453–460. [Google Scholar]
- 30.Selvaraju RR, Cogswell M, Das A, et al. Grad-CAM: visual explanations from deep networks via gradient-based localization. Int J Comput Vis. 2019;128(2):336–359. [Google Scholar]
- 31.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44(3):837–845. [PubMed] [Google Scholar]
- 32.Qi TH, Hian OH, Kumaran AM, et al. Multi-center evaluation of artificial intelligent imaging and clinical models for predicting neoadjuvant chemotherapy response in breast cancer. Breast Cancer Res Treat. 2022;193(1):121–138. doi: 10.1007/s10549-022-06521-7. [DOI] [PubMed] [Google Scholar]
- 33.Zhang J, Xiao L, Pu S, et al. Can we reliably identify the pathological outcomes of neoadjuvant chemotherapy in patients with breast cancer? Development and validation of a logistic regression nomogram based on preoperative factors. Ann Surg Oncol. 2021;28(5):2632–2645. doi: 10.1245/s10434-020-09214-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ochi T, Tsunoda H, Matsuda N, et al. Accuracy of morphologic change measurements by ultrasound in predicting pathological response to neoadjuvant chemotherapy in triple-negative and HER2-positive breast cancer. Breast Cancer. 2021;28(4):838–847. doi: 10.1007/s12282-021-01220-5. [DOI] [PubMed] [Google Scholar]
- 35.Adrada BE, Candelaria R, Moulder S, et al. Early ultrasound evaluation identifies excellent responders to neoadjuvant systemic therapy among patients with triple-negative breast cancer. Cancer. 2021;127(16):2880–2887. doi: 10.1002/cncr.33604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang M, Liu H, Dai Q, et al. Treatment response prediction using ultrasound-based pre-, post-early, and delta radiomics in neoadjuvant chemotherapy in breast cancer. Front Oncol. 2022;12 doi: 10.3389/fonc.2022.748008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Antunovic L, De Sanctis R, Cozzi L, et al. PET/CT radiomics in breast cancer: promising tool for prediction of pathological response to neoadjuvant chemotherapy. Eur J Nucl Med Mol Imaging. 2019;46(7):1468–1477. doi: 10.1007/s00259-019-04313-8. [DOI] [PubMed] [Google Scholar]
- 38.Fan M, Chen H, You C, et al. Radiomics of tumor heterogeneity in longitudinal dynamic contrast-enhanced magnetic resonance imaging for predicting response to neoadjuvant chemotherapy in breast cancer. Front Mol Biosci. 2021;8 doi: 10.3389/fmolb.2021.622219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lo Gullo R, Eskreis-Winkler S, Morris EA, et al. Machine learning with multiparametric magnetic resonance imaging of the breast for early prediction of response to neoadjuvant chemotherapy. Breast. 2020;49:115–122. doi: 10.1016/j.breast.2019.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang M, Li CL, Luo XM, et al. Ultrasound-based deep learning radiomics in the assessment of pathological complete response to neoadjuvant chemotherapy in locally advanced breast cancer. Eur J Cancer. 2021;147:95–105. doi: 10.1016/j.ejca.2021.01.028. [DOI] [PubMed] [Google Scholar]
- 41.Braman N, Prasanna P, Whitney J, et al. Association of peritumoral radiomics with tumor biology and pathologic response to preoperative targeted therapy for HER2 (ERBB2)-positive breast cancer. JAMA Netw Open. 2019;2(4) doi: 10.1001/jamanetworkopen.2019.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.