Abstract
The physiological function of a new gene, hereby designated merG, located between merA and merB on the broad-spectrum mer operon of Pseudomonas strain K-62 plasmid pMR26 was investigated. The 654-bp merG gene encodes a protein with a canonical leader sequence at its N terminus. The processing of the signal peptide of this protein was dose-dependently inhibited by sodium azide, a potent inhibitor of protein export. These results suggest that the mature MerG protein (ca. 20 kDa) may be located in the periplasm. Deletion of the merG gene from the broad-spectrum mer operon of pMR26 had no effect on the inorganic mercury resistance phenotype, but rendered the bacterium more sensitive to phenylmercury than its isogenic wild-type strain. Escherichia coli cells bearing pMU29, which carries a deletion of the merG gene, took up significantly more phenylmercury than the bacteria with the intact plasmid pMRA17. When the merG gene in a compatible plasmid was transformed into the E. coli strain carrying pMU29, the high uptake of and high sensitivity to phenylmercury were almost completely restored to their original levels. These results demonstrate that the merG gene is involved in phenylmercury resistance, presumably by reducing in-cell permeability to phenylmercury.
Bacterial resistance to organomercurials has mostly been shown to result from the cleavage of the carbon-mercury linkage by the organomercurial lyase enzyme, followed by the reduction of Hg2+ to Hg0 by the mercuric reductase enzyme. The genetics and biochemistry of organomercurial resistance have been extensively studied and reviewed (1, 5, 14, 19, 26, 27). These studies focused predominantly on mercury resistance (mer) operons. Most of the mer operons encoding a broad-spectrum resistance phenotype have been shown to consist of a regulatory gene (merR), an operator-promoter region, and at least four structural genes, merT, merP, merA, and merB (7, 8, 10, 11, 17, 21).
Pseudomonas strain K-62, a bacterial strain with broad-spectrum mercury resistance isolated from phenylmercury-polluted soil, has been shown to have a resistance to phenylmercury approximately a thousand times higher than that of other bacteria, such as Escherichia coli and Pseudomonas aeruginosa (30). Two separate organomercurial lyase enzymes, designated S-1 and S-2, each with somewhat different properties and substrate specificities (28, 29), were previously believed to be the elements contributing to Pseudomonas strain K-62’s resistance to phenylmercury. Genetic studies of resistance determinants, however, have shown that the mercury resistance of this soil strain had to do with pMR26 and pMR68, two of the six plasmids (8). Two mer operons in this soil strain were mapped on a 26-kb plasmid (pMR26), and one was cloned into the SacI site of the cloning vector, pBluescriptII (8). The resultant plasmid, pMRA17, expressed a typical broad-spectrum mercury resistance phenotype. DNA sequence analysis of a 6.6-kb SacI fragment, encompassing the whole region required for the broad-spectrum mercury resistance, of pMRA17 revealed that it comprised six open reading frames (ORFs), five of which were identified as merR, merT, merP, merA, and merB (10). The order and function of mer genes concerned with the mercurial resistance on the 6.6-kb fragment of pMRA17 are basically the same as those of the broad-spectrum mer operon of pDU1358, except that it has an additional ORF located between merA and merB genes (10).
The additional ORF, however, had no homology with the mer genes known to date. Deletion of this ORF rendered the bacterium more sensitive to phenylmercury than its isogenic wild-type strain, while surprisingly retaining full resistance to Hg2+ (10). The purpose of this paper is to characterize and define more precisely the roles of this ORF, hereby designated merG, in phenylmercury resistance.
MATERIALS AND METHODS
Bacterial strain, plasmids, and growth conditions.
The bacterial strain and plasmids used in this study are listed in Table 1. The E. coli strain was grown at 37°C in Luria-Bertani medium (22). When necessary, the medium was supplemented with 100 μg of ampicillin or 25 μg of kanamycin per ml.
TABLE 1.
Bacterial strain and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Strain | ||
| E. coli XL1-Blue | recA1 endA1 gyrA96 thi hsdR17 supE44 relA1 lac/[F′::Tn10 proAB+ lacIqlacZM15 traD36] | 2 |
| Plasmids | ||
| pBluescriptII SK+ | Apr ColE1 replicon, cloning vector | Stratagene |
| pSTV29 | Cmr pACYC replicon, cloning vector | 4, 25 |
| pTUE1122 | Aprtac promoter, 6xHis, expression vector | 16 |
| pMR26 | Broad-spectrum mercury resistance plasmid from Pseudomonas strain K-62 | 8, 10 |
| pMRA17 | 6.6-kb SacI fragment of pMR26 containing merRTPAGB cloned into pBluescriptII SK+ | 8, 10 |
| pMR96 | 4.7-kb BglII-EcoRV fragment of pMRA17 containing merRTPAGB cloned into pBluescriptII SK+ | This study |
| pMC89 | merB-deleted plasmid, containing merRTPAG, constructed from pMR96 | This study |
| pMU29 | merG-deleted plasmid containing merRTPAB cloned into pBluescriptII SK+ | 10 |
| pMRD141 | merAGB-deleted plasmid containing merRTP, constructed from pMRA17 | 9, 10, 31 |
| pMEV89 | 1.2-kb HindIII-BamHI fragment of pMC89 containing merG cloned into pSTV29 | This study |
| pMUE74 | 697-bp PCR products of merG cloned into pTUE1122 | This study |
Plasmid construction.
To construct a plasmid containing the broad-spectrum resistance mer operon of pMRA17, the plasmid pMRA17 was digested with BglII, and the resulting 5′ overhang was filled with deoxynucleoside triphosphates (dNTPs) by DNA polymerase I (Klenow fragment). After digestion with EcoRV, the 4.7-kb fragment was inserted into the SmaI site of the cloning vector pBluescriptII SK+. The resultant plasmid was designated pMR96. Plasmid pMC89 was constructed by digesting pMR96 with AflII-NheI, and the resulting 5′ overhang was filled with dNTPs by DNA polymerase I (Klenow fragment). The 7.2-kb fragment was religated with a DNA ligation kit.
pMEV89 containing the merG gene was constructed by cloning the 1.2-kb HindIII-BamHI fragment of pMC89 into the HindIII-BamHI site of a cloning vector, pSTV29. pMUE74 carrying the merG gene of pMRA17 was constructed as follows. A 697-bp fragment containing the merG gene was PCR amplified with primer 1 (5′3325GCTCTAGAGGGCACCAAAACAGGG3348 3′) and primer 2 (5′3998GAAGATCTCTTTGTGCCTTGCCGG4021 3′) containing restriction sites for XbaI and BglII, respectively. pMRA17 was used as the template. After digestion with XbaI and BglII, the fragment was cloned into the XbaI-BglII site of plasmid pTUE1122, which contains DNA sequence encoding six consecutive histidine residues immediately posterior to the BglII site (Table 1). The structures of the relevant genes and restriction sites in the constructed plasmid used in this study are schematically illustrated in Fig. 1.
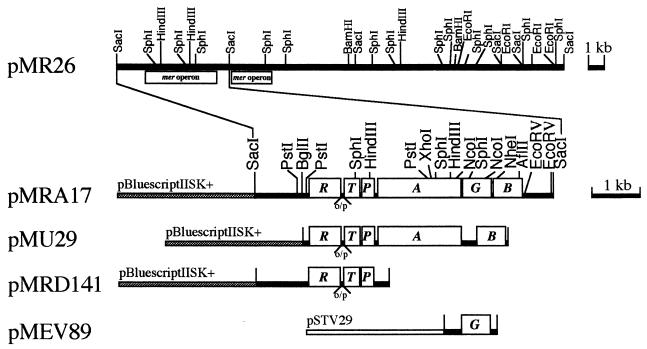
FIG. 1.
Structure of the relevant mer genes and restriction endonuclease sites in plasmid pMRA17 and its derivatives. R, merR (regulatory gene); T, merT (encodes mercury transport protein); P, merP (encodes periplasmic mercury-binding protein); A, merA (encodes the enzyme mercuric reductase); G, merG (new gene for phenylmercury resistance); B, merB (encodes the enzyme organomercurial lyase). o/p, operator-promoter region.
DNA sequencing.
The complete sequence of the 654-bp merG gene was determined from both strands by using the dideoxy chain-termination DNA sequencing method of Sanger et al. (24). Sequence analysis was also performed with a DNA autosequencer (model 373A; Applied Biosystems, Inc., Foster City, Calif.) with a Taq dye terminator cycle sequencing kit.
Maxicell analysis.
Maxicells were used to specifically label plasmid-encoded polypeptides. The transformants of E. coli CSR603 carrying the plasmids of interest were incubated aerobically at 37°C in K medium (M9 medium supplemented with 1% Casamino Acids and 0.1 μg of thiamine per ml). After 50 s of UV irradiation (50 J/m2), the cells were treated with d-cycloserine (150 μg/ml), and the maxicell proteins were then labeled with [35S]methionine (1,000 Ci/mmol; New England Nuclear) according to the original protocol (23). For induction of the mer gene-encoded polypeptides, 1 μM HgCl2 was added to the cells during the 1-h period of labeling with [35S]methionine. Gel electrophoresis was performed by the method of Laemmli (12), and sodium salicylate was used for detection of the 35S-labeled polypeptide (3).
Protein purification and analysis.
An E. coli XL1-Blue strain carrying pMUE74 was cultured in Luria-Bertani medium supplemented with 100 μg of ampicillin per ml at 37°C for 2 h, and 1 mM isopropyl-β-o-thiogalactopyranoside (IPTG) was added for a further 30 min of cultivation. After incubation with IPTG, the cells were incubated at 37°C in the absence or presence of various concentrations of sodium azide, an inhibition of protein transport (18). After 2 h, the cells were harvested by centrifugation at 10,000 × g for 1 min and resuspended in 1% sodium dodecyl sulfate (SDS) sonication buffer (1% SDS, 100 mM phosphate buffer [pH 7.8], 300 mM NaCl). The cells were disrupted by boiling for 5 min, and the cell debris was removed by centrifugation at 10,000 × g for 5 min at 4°C. The resultant supernatant was gently mixed with Ni-nitrilotriacetic acid (NTA) resin (Qiagen) at room temperature for 16 h. The mixture was centrifuged, and the pellet was washed three times with 1% SDS sonication buffer. The pellet was then mixed with SDS-polyacrylamide gel electrophoresis (PAGE) sample buffer (2% SDS, 60 mM Tris-HCl [pH 6.8], 5% mercaptoethanol, 5% sucrose, 0.002% bromophenol blue) containing 100 mM EDTA. After incubation for 5 min at 100°C, the supernatant obtained by centrifugation was resolved by SDS-PAGE (17.5% polyacrylamide gel). Gel electrophoresis was performed by the method of Laemmli (12), and Coomassie brilliant blue was used for protein staining. After purification, the N-terminal amino acid sequences of the 25- and 21-kDa proteins were determined, respectively, with an HP G1005A protein sequencing system (Hewlett-Packard).
Mercury resistance assay.
Resistance of bacteria to HgCl2 and phenylmercuric acetate (PMA) was determined on petri dishes according to the method of Foster et al. (6). The zones of inhibition of growth around the disks were measured after incubation at 37°C for 16 h.
Mercury volatilization assay.
A qualitative detection of nonradioactive mercury volatilization by the cells was done according to the X-ray film method described by Nakamura and Nakahara (15). Cells were grown, induced with 1 μM Hg2+, and harvested. Cells were resuspended in 0.15 ml of 70 mM phosphate buffer (pH 7.0) containing 0.5 mM EDTA, 0.2 mM magnesium acetate, 5 mM sodium thioglycolate, and 184 μM HgCl2 or 149 μM PMA in a microplate. The plate was covered with X-ray film in the darkroom. The foggy area on the film was the result of the reduction of Ag+ emulsion by the mercury vapor formed from HgCl2 and PMA.
Mercury uptake assay.
Bacterial cells were grown in Luria-Bertani medium in the absence or presence of 0.5 μM HgCl2 or PMA. The late-log-phase cells were harvested and resuspended in Luria-Bertani medium containing 100 μg of chloramphenicol per ml, 100 μM EDTA, and 5 μM 203Hg2+ (136 mCi/mmol) or 50 μM 14C6H5Hg+ (30 mCi/mmol). After incubation at 37°C, aliquots (0.5 ml) were removed periodically and filtered on a Whatman GF/B glass microfiber filter (0.45-μm pore diameter). The filters were washed three times with 5 ml of Luria-Bertani medium, and the radioactivity on the filter was measured with a Beckman gamma scintillation counter (GAMMA-5500) or liquid scintillation spectrometer (Aloka LSC-3500). The standard deviations of measurement were less than 10%.
Nucleotide sequence accession number.
The nucleotide sequence of merG has been deposited in the EMBL/GenBank/DDBJ Nucleotide Sequence Database under accession no. D83080.
RESULTS
Evidence that merG codes for phenylmercury resistance.
Plasmid pMRA17 carries a 6.6-kb SacI-fragment of the Pseudomonas strain K-62 plasmid pMR26, which encodes a typical broad-spectrum mercurial resistance based on the degradation of organomercury and reduction of the resultant Hg2+ to Hg0 (10). A new gene, merG, which specified a protein with a deduced molecular mass of 20 kDa, was found in the broad-spectrum mer operon of pMR26.
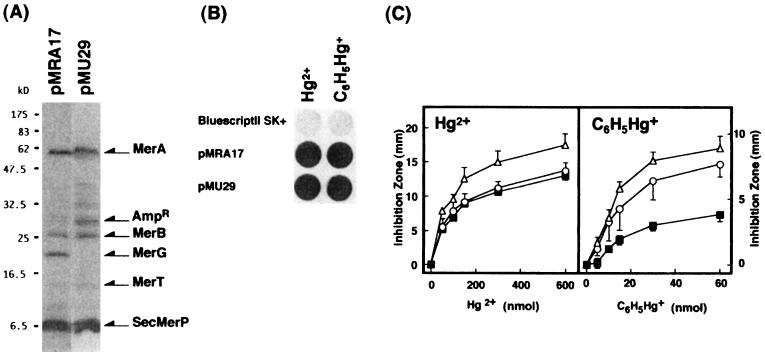
To define the roles of merG, a series of deletion plasmids were constructed (Fig. 1 and Table 1). As shown in Fig. 2A, the 20-kDa protein was not detected in maxicells carrying merG-deleted plasmid pMU29. pMU29, however, still expressed its ability to volatilize mercury, not only from Hg2+, but also from C6H5Hg+ (Fig. 2B). Deletion mutation in the merG region did not impair Hg2+ resistance but significantly decreased phenylmercury resistance (Fig. 2C). These results suggest that merG may be involved in phenylmercury resistance.
FIG. 2.
mer polypeptides (A), mercury volatilization (B), and mercury resistance encoded by deletion plasmid pMU29. (A) E. coli CSR603 harboring pMRA17 or pMU29 was cultured, induced with 1 μM Hg2+, UV irradiated, and labeled with [35S]methionine. Labeled cells were collected and lysed, and proteins were analyzed on SDS-polyacrylamide gels. Molecular mass markers are indicated to the left. The arrows marked MerA, MerB, MerG, MerT, and SecMerP indicate the polypeptides of the respective genes. (B) Cells were grown, induced with 1 μM Hg2+, and harvested. Volatilization of mercury from mercurials was detected by the X-ray film method as described in Materials and Methods. (C) Resistance of E. coli XL1-Blue with pMRA17 (■), pMU29 (○), and pBluescriptII SK+ (▵) to mercuric chloride and phenylmercuric acetate was determined by measuring the diameter of the inhibition zone (minus the 6.5-mm disk diameter) on petri dishes after overnight growth at 37°C. All values are the means of triplicate experiments.
DNA sequence and properties of merG.
The nucleotide sequence of the 950-bp NcoI-NheI fragment of pMEV89 which contained the entire merG gene was carefully redetermined and is identical to that previously reported for the merG region (10). (Note that an error was made at position 277 of the merG gene region. There should have been a “c” at this position, but it was inadvertently omitted in the previous report [10].) There is a 654-bp merG gene, preceded by a potential ribosome binding site. From the DNA sequence data, the protein encoded by this gene has been predicted to have molecular mass of 23.8 kDa, which is about 4 kDa larger than that detected in maxicell analysis (Fig. 2A).
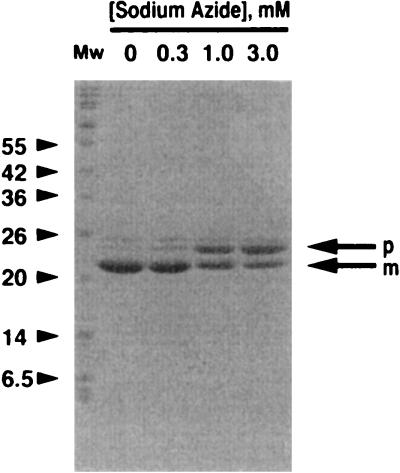
The fate of MerG protein fused on its C terminus of eight amino acid residues (RSHHHHHH; 1 kDa) in cells with pMUE74 was studied. As shown in Fig. 3, a protein with molecular mass of 21 kDa was predominantly detected in the cells with pMUE74. When the cells were treated with sodium azide, an inhibitor of protein export (18), an additional protein with a molecular mass of 25 kDa was detected in the cells. The appearance of the 25-kDa protein was dose-dependently increased by sodium azide, along with the decrease of the appearance of the 21-kDa protein (Fig. 3). The N-terminal amino acid sequences of the 25- and 21-kDa proteins overlapped completely with those predicted from translation of the DNA sequence at positions 1 to 5 (MFCDI) and 35 to 39 (AYDFS). These results suggest that the 21-kDa protein is the exported product of the 25-kDa protein encoded by pMUE74.
FIG. 3.
Effect of sodium azide on the translocation of MerG. E. coli XL1-Blue carrying pMUE74 was cultured in Luria-Bertani medium and induced with 1 mM IPTG for 30 min. After a 2-h treatment with sodium azide, the cells were harvested and disrupted by boiling for 5 min, and the resultant supernatant was mixed with Ni-NTA resin (Qiagen) at room temperature for 16 h. After centrifugation, the pellet was washed, mixed with SDS-PAGE sample buffer containing 100 mM EDTA, and boiled for 5 min. The proteins in the sample were resolved by SDS-PAGE, and Coomassie brilliant blue was used for protein staining as described in Materials and Methods. p, the precursor MerG; m, the mature MerG. Mw, molecular mass markers (kilodaltons).
Role of merG in phenylmercury resistance.
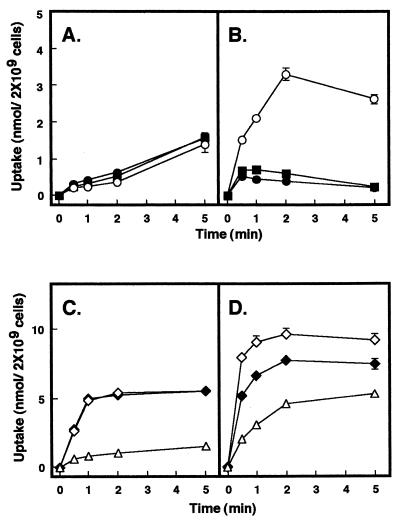
To study the function of merG in mercury resistance, uptake of radioactive mercury by the cells with the merG-deleted plasmid pMU29 was examined. No significant difference in the uptake of 203Hg2+ was found between the cells with pMU29 and pMRA17 (Fig. 4A). However, the cells with pMU29 took up appreciably more 14C6H5Hg+ than the cells with pMRA17 (Fig. 4B). Transformation of plasmid pMEV89 containing the merG gene into the pMU29-bearing cells had no effect on the uptake of 203Hg2+, but caused a significant reduction in the uptake of 14C6H5Hg+ (Fig. 4A and B).
FIG. 4.
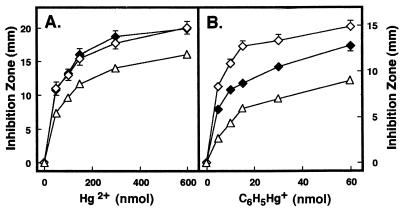
Effect of merG on bacterial uptake of 203Hg2+ (A and C) and 14C6H5Hg+ (B and D). E. coli XL1-Blue cells with pMRA17 (■), pMU29 (○), pMU29 and pMEV89 (•), pMRD141 (◊), pMRD141 and pMEV89 (⧫) and pBluescriptII SK+ (▵) were grown, induced, and harvested as described in Materials and Methods. After addition of the radioactive mercurials, aliquots were removed periodically, filtered, and washed. All values are the means of triplicate determinations from three experiments.
The effect of merG on bacterial uptake of 14C6H5Hg+ was next studied in the absence of functional MerB and MerA proteins. pMRD141, containing the intact mercury transport genes (merT and merP), but not the detoxifying genes (merA and merB), conferred bacterial hyperaccumulation of both 203Hg2+ and 14C6H5Hg+ (Fig. 4C and D). This finding is in agreement with our earlier results (9, 31). Interestingly, when pMEV89 was transformed into the pMRD141-bearing cells, the hyperaccumulation of 14C6H5Hg+ in these cells was significantly reduced, while the hyperaccumulation of 203Hg2+ was not (Fig. 4C and D). Figure 5 shows that bacteria carrying pMRD141 consistently showed hypersensitivity, not only to Hg2+, but also to C6H5Hg+. When pMEV89 was transferred into the bacterial strain harboring pMRD141, the bacterium was more resistant to C6H5Hg+ than was the strain carrying pMRD141 only. However, transformation of pMEV89 into the strain had no effect on the phenotype of hypersensitivity to Hg2+ (Fig. 5).
FIG. 5.
Mercurial resistance. E. coli XL1-Blue cells with pMRD141 (◊), pMRD141 and pMEV89 (⧫), and pBluescript II SK+ (▵) with mercuric chloride (A) and phenylmercuric acetate (B) were grown at 37°C. After 16 h of incubation, the diameter of the inhibition zone (minus the 6.5-mm disk diameter) on petri dishes was measured. All values are the means of triplicate experiments.
DISCUSSION
From a Pseudomonas strain K-62 plasmid, pMR26, we had previously uncovered a new gene which had no homology with any published mer genes (10). The new gene was found to be located between the merA and merB genes on the broad-spectrum mer operon of pMR26. In this paper, we have further characterized and defined the physiology of this new gene. The new gene is hereby designated merG.
In agreement with our previous results (10), deletion of this new gene resulted in a decrease in phenylmercury resistance, but not in a total loss of the resistance or of volatilization activity (Fig. 2B and C). These results clearly demonstrate that merG plays a role in the expression of phenylmercury resistance.
An independent determination of the DNA sequence agreed with what was previously reported. There is, however, an error in the DNA sequence in the previous report (10). There should have been a “c” at position 3639, but it was inadvertently omitted. The merG gene is 654-bp long, encoding a 217-amino-acid polypeptide with a deduced molecular mass of 23.8 kDa; however, no 23.8-kDa protein was detected in the maxicell analysis (Fig. 2A). There is no doubt that the 20-kDa protein detected by maxicell analysis is encoded by merG, because this protein was no longer detected coincidentally along with the gene deleted from the mer operon (Fig. 2A). It was this discrepancy that prompted us to characterize the merG gene further.
The amino acid sequence of the gene product, predicted from the DNA sequence of merG, has a good leader sequence which contains a short positively charged region at the N terminus followed by a hydrophobic region and a signal peptide cleavage site (ALAA) at position 32 to 35. This characteristic N-terminal sequence is homologous to the “leader sequences” of known periplasmic proteins (13). The formation of the 21-kDa protein was significantly inhibited by sodium azide in a dose-dependent manner (Fig. 3). In addition, the 21-kDa protein was predominantly detected in the periplasmic fraction, suggesting that the MerG protein may be located in the periplasm.
It is interesting to note that deletion of the merG gene resulted in a significant decrease in the phenylmercury resistance, but not in a total loss of the resistance (Fig. 2C). Deletion of merG did not impair the enzymatic activities encoded by merA and merB, because the bacteria carrying pMRA17 and pMU29 still volatilized mercury from the C6H5Hg+ taken up by the cells (Fig. 2B), and no difference in the rate of 14C6H5Hg+ disappearance was found between the cells with pMRA17 and those with pMU29 (data not shown). In addition, the radioactivity in the culture medium was constant during the culture of bacteria carrying plasmid pMRD141 and pMEV89 in the presence of 14C6H5Hg+ (data not shown). These results suggest that the MerG protein-specified phenylmercury resistance seems to be due to the efflux mechanism rather than mercury biotransformation.
Reduced uptake of mercury has been put forward as one of the mechanisms for bacterial resistance to mercurials (20). Several lines of evidence obtained in this study strongly suggest that merG is involved in the reduction of cellular permeability to C6H5Hg+. (i) Although deletion of the merG gene had no apparent effect on the accumulation of Hg2+, it caused an increase in the accumulation of 14C6H5Hg+ compared to the level in the strain with pMRA17 (Fig. 4A and B). (ii) The high accumulation of 14C6H5Hg+ was completely abolished when pMEV89 was introduced into the bacterial strain with pMU29 (Fig. 4B). (iii) Transformation of pMEV89 into the bacteria carrying pMRD141 caused a significant reduction in the uptake of 14C6H5Hg+, but not that of 203Hg2+ (Fig. 4C and D). And finally, (iv) the hypersensitivity to phenylmercury in the bacteria with pMRD141 was markedly reduced (Fig. 5). Tonomura et al. (30) previously reported that approximately 70% of the phenylmercury taken up by Pseudomonas strain K-62 was detected mainly on the cell surface, and they speculated that this soil strain might be less permeable to mercurials than other mercurial-sensitive bacteria. Our results confirmed their speculation.
In conclusion, the merG gene is involved in the bacterial expression of phenylmercury resistance. It is plausible that the resistance is the result of a reduction in the cellular permeability to phenylmercury.
ACKNOWLEDGMENTS
We are grateful to K. Watabe and H. Takamatsu of Setsunan University for the gift of plasmid pTUE1122. We also thank Y. Uno and M. Nakayama for technical assistance and C. Lee for critical reading of the manuscript.
This work was supported in part by a Grant-in-Aid for Encouragement of Young Scientists (no. 09780512) to M.K. and a Grant-in-Aid for Scientific Research (C) (no. 10680555) to H.P.-H. from the Ministry of Education, Science and Culture, Japan, and a grant from Sumitomo Foundation to H.P.-H.
REFERENCES
- 1.Brown N L. Bacterial resistance to mercury—reductio ad absurdum? Trends Biochem Sci. 1985;10:400–403. [Google Scholar]
- 2.Bullock W O, Fernandez J M, Short J M. XL1-Blue: a high efficiency plasmid transforming recA Escherichia coli strain with β-galactosidase selection. BioTechniques. 1987;5:376–379. [Google Scholar]
- 3.Chamberlain J P. Fluorographic detection of radioactivity in polyacrylamide gels with the water-soluble fluor, sodium salicylate. Anal Biochem. 1979;98:132–135. doi: 10.1016/0003-2697(79)90716-4. [DOI] [PubMed] [Google Scholar]
- 4.Chang A C Y, Cohen S N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Foster T J. The genetics and biochemistry of mercury resistance. Crit Rev Microbiol. 1987;15:117–140. doi: 10.3109/10408418709104455. [DOI] [PubMed] [Google Scholar]
- 6.Foster T J, Nakahara H, Weiss A A, Silver S. Transposon A-generated mutations in the mercuric resistance genes of plasmid R100-1. J Bacteriol. 1979;140:167–181. doi: 10.1128/jb.140.1.167-181.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Griffin H G, Foster T J, Silver S, Misra T K. Cloning and DNA sequence of the mercuric- and organomercurial-resistance determinants of plasmid pDU1358. Proc Natl Acad Sci USA. 1987;84:3112–3116. doi: 10.1073/pnas.84.10.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kiyono M, Omura T, Fujimori H, Pan-Hou H. Organomercurial resistance determinants in Pseudomonas K-62 are present on two plasmids. Arch Microbiol. 1995;163:242–247. doi: 10.1007/BF00393375. [DOI] [PubMed] [Google Scholar]
- 9.Kiyono M, Omura T, Fujimori H, Pan-Hou H. Lack of involvement of merT and merP in methylmercury transport in mercury resistant Pseudomonas K-62. FEMS Microbiol Lett. 1995;128:301–306. doi: 10.1111/j.1574-6968.1995.tb07540.x. [DOI] [PubMed] [Google Scholar]
- 10.Kiyono M, Omura T, Inuzuka M, Fujimori H, Pan-Hou H. Nucleotide sequence and expression of the organomercurial-resistance determinants from a Pseudomonas K-62 plasmid, pMR26. Gene. 1997;189:151–157. doi: 10.1016/s0378-1119(96)00741-x. [DOI] [PubMed] [Google Scholar]
- 11.Laddaga R A, Chu L, Misra T K, Silver S. Nucleotide sequence and expression of the mercurial-resistance operon from Staphylococcus aureus plasmid pI258. Proc Natl Acad Sci USA. 1987;84:5106–5110. doi: 10.1073/pnas.84.15.5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 13.Michaelis S, Beckwith J. Mechanism of incorporation of cell envelope proteins in Escherichia coli. Annu Rev Microbiol. 1982;36:435–465. doi: 10.1146/annurev.mi.36.100182.002251. [DOI] [PubMed] [Google Scholar]
- 14.Misra T K. Bacterial resistance to inorganic mercury salts and organomercurials. Plasmid. 1992;27:4–16. doi: 10.1016/0147-619x(92)90002-r. [DOI] [PubMed] [Google Scholar]
- 15.Nakamura K, Nakamura H. Simplified X-ray film method for detection of bacterial volatilization of mercury chloride by Escherichia coli. Appl Environ Microbiol. 1988;54:2871–2873. doi: 10.1128/aem.54.11.2871-2873.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakane A, Takamatsu H, Oguro A, Sadaie Y, Nakamura K, Yamane K. Acquisition of azide-resistance by elevated SecA ATPase activity confers azide-resistance upon cell growth and protein translocation in Bacillus subtilis. Microbiology. 1995;141:113–121. doi: 10.1099/00221287-141-1-113. [DOI] [PubMed] [Google Scholar]
- 17.Nucifora G, Chu L, Silver S, Misra T K. Mercury operon regulation by the merR gene of the organomercurial resistance system of plasmid pDU1358. J Bacteriol. 1989;171:4241–4247. doi: 10.1128/jb.171.8.4241-4247.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oliver D B, Cabelli R J, Dolan K M, Jarosik G P. Azide-resistant mutants of Escherichia coli alter the SecA protein, an azide-sensitive component of the protein export machinery. Proc Natl Acad Sci USA. 1990;87:8227–8231. doi: 10.1073/pnas.87.21.8227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Osborn A M, Bruce K D, Strike P, Ritchie D A. Distribution, diversity and evolution of the bacterial mercury resistance (mer) operon. FEMS Microbiol Rev. 1997;19:239–262. doi: 10.1111/j.1574-6976.1997.tb00300.x. [DOI] [PubMed] [Google Scholar]
- 20.Pan-Hou H, Nishimoto M, Imura N. Possible role of membrane proteins in mercury resistance of Enterobacter aerogenes. Arch Microbiol. 1981;130:93–95. doi: 10.1007/BF00411057. [DOI] [PubMed] [Google Scholar]
- 21.Reniero D, Galli E, Barbieri P. Cloning and comparison of mercury- and organomercurial-resistance determinants from Pseudomonas stutzeri plasmid. Gene. 1995;166:77–82. doi: 10.1016/0378-1119(95)00546-4. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook, J., E. F. Fritsch, and T. Maniatis. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 23.Sancer A, Hack A M, Rupp W D. Simple method for identification of plasmid-coded proteins. J Bacteriol. 1979;137:692–693. doi: 10.1128/jb.137.1.692-693.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Selzer G, Som T, Itoh T, Tomizawa J. The origin of replication of plasmid p15A and comparative studies on the nucleotide sequences around the origin of related plasmids. Cell. 1983;32:119–129. doi: 10.1016/0092-8674(83)90502-0. [DOI] [PubMed] [Google Scholar]
- 26.Silver S, Phung L T. Bacterial heavy metal resistance. Annu Rev Microbiol. 1996;50:753–789. doi: 10.1146/annurev.micro.50.1.753. [DOI] [PubMed] [Google Scholar]
- 27.Summers A O. Organisation, expression and evolution of genes for mercury resistance. Annu Rev Microbiol. 1986;40:607–634. doi: 10.1146/annurev.mi.40.100186.003135. [DOI] [PubMed] [Google Scholar]
- 28.Tezuka T, Tonomura K. Purification and properties of an enzyme catalyzing the splitting of carbon-mercury linkages from mercury-resistant Pseudomonas K-62 strain. J Biochem. 1976;80:79–87. doi: 10.1093/oxfordjournals.jbchem.a131261. [DOI] [PubMed] [Google Scholar]
- 29.Tezuka T, Tonomura K. Purification and properties of a second enzyme catalyzing the splitting of carbon-mercury linkages from mercury-resistant Pseudomonas K-62. J Bacteriol. 1978;135:138–143. doi: 10.1128/jb.135.1.138-143.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tonomura K, Maeda K, Futai F, Nakagami T, Yamada M. Stimulative vaporization of phenylmercuric acetate by mercury-resistant bacteria. Nature. 1968;217:644–646. doi: 10.1038/217644b0. [DOI] [PubMed] [Google Scholar]
- 31.Uno Y, Kiyono M, Tezuka T, Pan-Hou H. Phenylmercury transport mediated by merT-merP genes of Pseudomonas K-62 plasmid pMR26. Biol Pharm Bull. 1997;20:107–109. doi: 10.1248/bpb.20.107. [DOI] [PubMed] [Google Scholar]