Abstract
Vertebrate embryogenesis involves a conserved and fundamental process, called the maternal-to-zygotic transition (MZT), which marks the switch from a maternal factors-dominated state to a zygotic factors-driven state. Yet the precise mechanism underlying MZT remains largely unknown. Here we report that the RNA helicase Ddx3xb in zebrafish undergoes liquid–liquid phase separation (LLPS) via its N-terminal intrinsically disordered region (IDR), and an increase in ATP content promotes the condensation of Ddx3xb during MZT. Mutant form of Ddx3xb losing LLPS ability fails to rescue the developmental defect of Ddx3xb-deficient embryos. Interestingly, the IDR of either FUS or hnRNPA1 can functionally replace the N-terminal IDR in Ddx3xb. Phase separation of Ddx3xb facilitates the unwinding of 5’ UTR structures of maternal mRNAs to enhance their translation. Our study reveals an unprecedent mechanism whereby the Ddx3xb phase separation regulates MZT by promoting maternal mRNA translation.
Subject terms: Developmental biology, Cell biology
Introduction
The maternal-to-zygotic transition (MZT), involving maternal mRNA clearance and zygotic genome activation (ZGA), is a conserved and fundamental process during vertebrate embryogenesis.1–3 During this critical developmental time, the embryo undergoes dramatic reprogramming by switching from a maternal factor-dominated state to a zygotic factor-driven state. Due to the transcriptional quiescence of the zygotic genome during the initial developmental stages, selective maintenance and translational control of maternal mRNAs is essential.1,3–6 Recent studies have illustrated that RNA 5-methylcytosine (m5C) modification7 and RNA structure8 can selectively protect the stability of maternal mRNAs, while other factors are involved in the clearance of maternal mRNAs, such as miR-430,9 codon usage,10 N6-methyladenosine (m6A),11 and uridylation.12 In addition, selective translation of maternal factors, including Nanog, Pou5f1, and SoxB1, is responsible for initiating zygotic gene activation in zebrafish.4 Cytoplasmic mRNA polyadenylation of maternal mRNAs is also reported to coincide with translational activation during MZT.13 Nonetheless, how fertilization mechanistically leads to MZT is not understood yet.
Liquid–liquid phase separation (LLPS) is an important mechanism that regulates cellular processing by driving the formation of biomolecular condensates.14,15 These membraneless compartments induced by LLPS concentrate proteins, nucleic acids, and other biomolecules to accurately control diverse biological processes, including heterochromatin maintenance,16 germ cell specification,17 synaptic complex formation,18 cytoskeleton assembly,19 and autophagic degradation.20 RNA helicases of the DEAD box family are highly conserved and required for RNA metabolism, such as transcription, splicing, transport, translation, and degradation.21 Many intrinsically disordered regions (IDRs), especially those involved in RNA metabolism, are known to mediate LLPS,22 and at least one-third of DEAD-box ATPase proteins contain IDRs. Therefore, phase separation may be important for the functions of DEAD-box ATPase.23,24 Here, we show that an increase in ATP concentration upon fertilization promotes LLPS of an RNA helicase called Ddx3xb. Such event promotes the enzymatic activity of the RNA helicase, which leads to unwinding of maternal mRNAs. This specificity of Ddx3xb selectively regulates translation and facilitates MZT during embryogenesis. Our study shows that fertilization leads to LLPS, which selectively regulates translation during a critical period of early development. Our data reveal a completely novel and highly spatially and temporally controlled mechanism that is essential for embryogenesis.
Results
Zebrafish Ddx3xb features that contribute to condensates formation
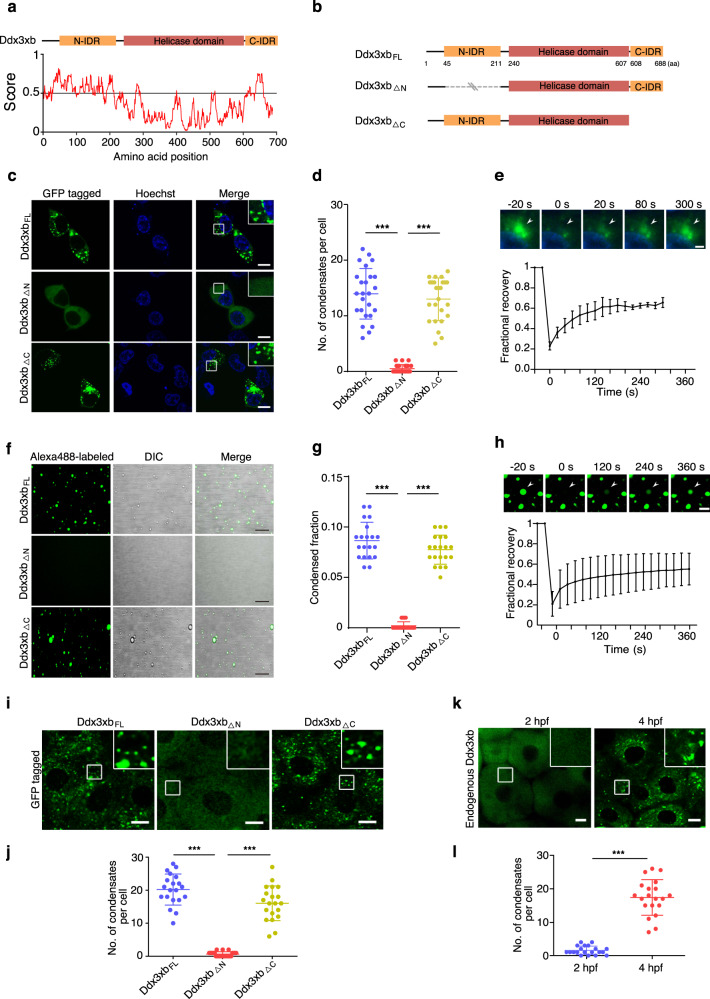
Family members of RNA-dependent DEAD-box (DDX) helicases are regulators of membraneless ribonucleoprotein (RNP) organelles in living cells,23 and LLPS has been shown to be an important organizing principle for RNPs.14 We therefore speculated that DDX proteins might be involved in LLPS and play a role during MZT. To test this hypothesis, we mainly focused on the RNA helicase Ddx3xb based on our findings that it showed ubiquitous and high expression during early zebrafish embryogenesis determined by quantitative-PCR (qPCR) and whole-mount in situ hybridization (Supplementary information, Fig. S1a, b). In particular, Ddx3xb showed an obvious increase in protein level at 2 hpf (Supplementary information, Fig. S1c). The human and yeast orthologs of the zebrafish Ddx3xb have been reported to control translation.25–27 The abundant expression of Ddx3xb prompted us to explore the regulatory role of Ddx3xb during zebrafish early embryogenesis. We analyzed the functional domains present in the zebrafish Ddx3xb protein and found that besides a helicase domain, Ddx3xb also contains N-terminal and C-terminal IDR domains (Fig. 1a; Supplementary information, Fig. S2a). To investigate whether these IDRs could facilitate condensation of Ddx3xb, we generated a series of full-length or truncated versions of N-terminally GFP-tagged Ddx3xb, namely Ddx3xbFL (full-length Ddx3xb), Ddx3xb△N (Ddx3xb lacking N-terminal IDR domain), and Ddx3xb△C (Ddx3xb lacking C-terminal IDR domain) (Fig. 1b). We expressed these versions in HEK293 cells (Supplementary information, Fig. S2b) and evaluated subcellular localization of the proteins using confocal microscopy. Accordingly, we found that only Ddx3xb containing N-terminal IDR can form droplets (Fig. 1c, d), indicating that the N-terminal IDR is required for droplet formation in HEK293 cells. Furthermore, fluorescence recovery after photobleaching (FRAP) analysis showed that GFP-tagged Ddx3xb droplets were associated with fluorescence recovery (Fig. 1e; Supplementary information, Video S1). Of note, pairs of droplets slowly fused and relaxed into larger droplets (Supplementary information, Fig. S2c), suggesting that Ddx3xb forms condensates through LLPS in living cells. To test whether Ddx3xb proteins form condensates in vitro, we performed phase separation assays using Alexa-488-labelled purified Ddx3xbFL, Ddx3xb△N, and Ddx3xb△C proteins. 10 μM Ddx3xb variant proteins were incubated in buffer containing 50 mM HEPES-KOH, pH 7.0, 100 mM KCl at room temperature. The results showed that the N-terminal IDR is essential for Ddx3xb phase separation in vitro (Fig. 1f, g). FRAP also demonstrates the formation of Ddx3xb condensates through LLPS in vitro (Fig. 1h; Supplementary information, Video S2), which is consistent with our previous observations in HEK293 cells (Fig. 1e; Supplementary information, Video S1). To test whether this also occurs in vivo, we injected GFP-tagged ddx3xbFL, ddx3xb△N, and ddx3xb△C mRNAs into zebrafish embryos at 1-cell stage (Supplementary information, Fig. S2d). In consistent with the in vitro results (Fig. 1f, g), defective LLPS formation was only observed under deletion of the N-terminal IDR (Fig. 1i, j), illustrating that Ddx3xb phase separation depends on the presence of its N-terminal IDR.
Fig. 1. Zebrafish Ddx3xb forms liquid–liquid phase separation via its N-terminal IDR.
a Bottom, predictions of intrinsic disorder tendency of Ddx3xb by IUPred2A (https://iupred2a.elte.hu/). Scores above 0.5 indicate disorder. Top, schematic diagram showing different truncated forms of zebrafish Ddx3xb. b Schematic diagram showing different truncated forms of zebrafish Ddx3xb. c Expression pattern of N-terminally GFP-tagged Ddx3xb variants (Ddx3xbFL, Ddx3xb△N and Ddx3xb△C) in HEK293 cells. Scale bars, 10 μm. d Quantification of the number of droplets per cell in fluorescence images. n = 25 cells per condition. Error bars, means ± SD. P values were determined by the two-tailed Student’s t-test. ***P < 0.001. e Bottom, plot of fluorescence intensity of GFP-Ddx3xb droplets in HEK293 cells after photobleaching over time. The black curve represents the mean of the fluorescence intensity in the photobleached region in distinct droplets (n = 5); error bars, means ± SD. Top, representative images of fluorescence recovery. Scale bars, 2 μm. f In vitro phase separation of Alexa-488-labeled Ddx3xb variants: Ddx3xbFL, Ddx3xb△N or Ddx3xb△C. Scale bars, 10 μm. g Quantification of condensed phase fraction of Ddx3xb variants as observed in experiments shown in f. The ImageJ software was used for distinguishing the dilute phase and condensed phase by fluorescence intensity difference and condensed phase fraction was measured by calculation of condensed phase area/the given field area. n = 20 fields per condition. Error bars, means ± SD. P values were determined by the two-tailed Student’s t-test. ***P < 0.001. h Top, representative images of fluorescence recovery. Scale bars, 2 μm. Bottom, changes in the fluorescence intensity of Alexa-488-labeled Ddx3xb droplets after photobleaching were plotted over time. The black curve represents the mean of the fluorescence intensity of photobleached regions in distinct droplets (n = 5). Error bars, means ± SD. i Representative images of zebrafish embryos expressing variants of N-terminally GFP-tagged Ddx3xb at 4 hpf. Scale bars, 10 μm. j Quantification of the number of droplets per cell in images obtained from experiments shown in i. n = 20 cells per condition. Error bars, means ± SD. P values were determined by the two-tailed Student’s t-test. ***P < 0.001. k Immunofluorescence staining showing the expression pattern of endogenous Ddx3xb at 2 hpf and 4 hpf. Scale bars, 10 μm. l Quantification of the number of droplets per cell in images obtained from experiments shown in k. n = 20 cells per condition. Error bars, means ± SD. P values were determined by the two-tailed Student’s t-test. ***P < 0.001.
To explore whether Ddx3xb phase separation is involved in zebrafish embryonic development, we analyzed the endogenous Ddx3xb protein expression pattern by using specific anti-Ddx3xb antibody. Intriguingly, we observed that endogenous Ddx3xb formed condensates through LLPS only at 4 hpf, but not at 2 hpf (Fig. 1k, l). Ddx3xb protein levels did not change notably between 2 hpf and 4 hpf (Supplementary information, Fig. S1c).
An increase in ATP concentration after fertilization promotes LLPS of Ddx3xb
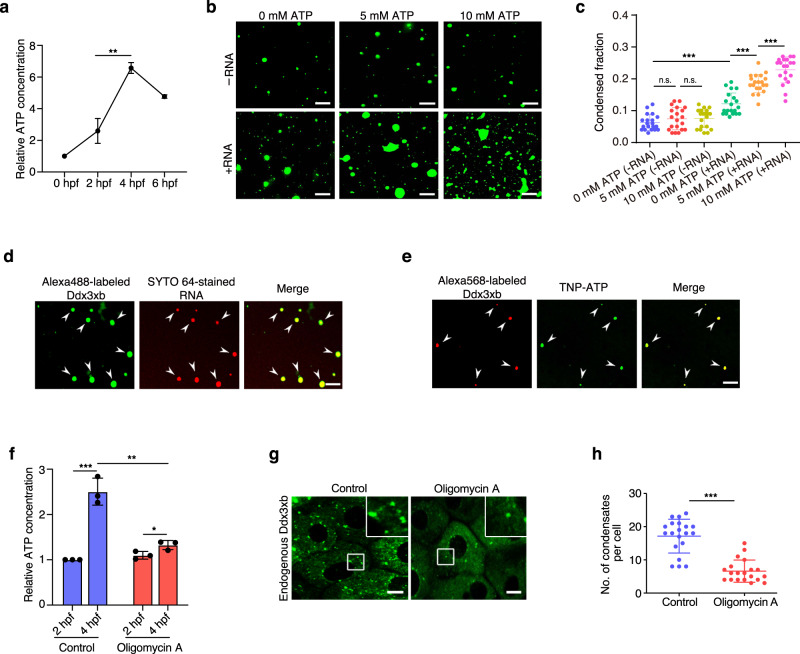
Previous studies have demonstrated that ATP concentration increases following fertilization.28,29 Moreover, ATP-binding state can promote phase separation of DDXs proteins.23 Since Ddx3xb is a DDX protein, we wanted to test if ATP can indeed affect Ddx3xb LLPS. Therefore, we monitored the ATP concentration during zebrafish embryogenesis. We found that ATP concentration significantly increased from 2 hpf to 4 hpf (Fig. 2a; Supplementary information, Fig. S3a). Next, purified Ddx3xb protein labelled with Alexa-488 was used to test whether the RNA and ATP influence Ddx3xb condensation. Ddx3xbFL protein at a final concentration of 4 μM was incubated in 50 mM HEPES-KOH, pH 7.0, 100 mM KCl, with gradient ATP concentrations (0, 5, 10 mM) with or without 100 ng/µL zebrafish mRNA. The result showed that mRNA significantly promotes the phase separation of Ddx3xb. In addition, an increase in ATP concentration significantly promotes Ddx3xb phase separation in the presence of RNA, while ATP alone failed to show this effect (Fig. 2b, c; Supplementary information, Fig. S3b). Then, we performed phase separation assay by using Alexa-488-labelled Ddx3xb and SYTO 64-stained RNA, where the SYTO 64 prefers to stain double-stranded RNA. We found that the SYTO 64-stained RNA was enriched in the droplets of Ddx3xb (Fig. 2d), and further associated with fluorescence recovery (Supplementary information, Fig. S3c), indicating that RNA could dynamically be shuttled in and outside the droplets. To further investigate whether ATP is enriched in Ddx3xb droplets, we used a fluorescence-labeled ATP analog, TNP-ATP, together with Alexa-568-labeled Ddx3xb, to perform phase separation assay in vitro. The result showed that TNP-ATP was enriched in the droplets of Alexa-568-labeled Ddx3xb (Fig. 2e). We also used the AMP-PNP (a non-hydrolyzable ATP analog) to test whether non-hydrolyzable ATP analog still promotes LLPS. We found that AMP-PNP can promote Ddx3xb phase separation in the presence of RNA (Supplementary information, Fig. S3d, e), indicating that non-hydrolyzable ATP analog still kept the promoting effect on Ddx3xb phase separation. Overall, these results indicate that RNA facilitates Ddx3xb condensation, and that ATP promotes Ddx3xb phase separation only in the presence of RNA. To further validate this finding, an ATP synthase inhibitor, Oligomycin A, was used to treat zebrafish embryos at 2 hpf. As expected, Oligomycin A treatment significantly decreased ATP concentration in zebrafish embryos at 4 hpf (Fig. 2f). Accordingly, we observed fewer endogenous Ddx3xb droplets in zebrafish embryos at 4 hpf (Fig. 2g, h). Similar results were obtained when performing phase separation assay in HEK293 cells (Supplementary information, Fig. S3f, g). We also performed ATP injection in Oligomycin A-treated embryos at 3 hpf, and observed ATP injection leading to Ddx3xb granules number rescue (Supplementary information, Fig. S3h, i).
Fig. 2. An increase in ATP concentration promotes both exogenous and endogenous phase separation of zebrafish Ddx3xb.
a Relative ATP concentration during zebrafish embryogenesis, normalized by the ATP concentration at 0 hpf. n = 3 replicates per stage. Error bars, means ± SD. P values were determined by the two-tailed Student’s t-test. **P < 0.01. b Phase separation analysis of full-length Ddx3xb with and without RNA (100 ng/μL) in response to changes in ATP concentration. Scale bars, 10 μm. c Quantification of the phase separation analysis from assays as shown in b. n = 20 fields per condition. Error bars, means ± SD. P values were determined by the two-tailed Student’s t-test. n.s., not significant; ***P < 0.001. d Double-stranded RNA is enriched in Ddx3xb phase separation droplets. Scale bar, 10 μm. e TNP-ATP is enriched in the droplets of Alexa-568-labeled Ddx3xb. Scale bars, 10 μm. f Relative ATP concentration in untreated and Oligomycin A-treated groups, normalized by the ATP concentration of the untreated group at 2 hpf. n = 3 replicates per stage. Error bars, means ± SD. P values were determined by the two-tailed Student’s t-test. *P < 0.05; **P < 0.01; ***P < 0.001. g Immunofluorescence staining showing the expression pattern of endogenous Ddx3xb in control and Oligomycin A treatment groups. Scale bars, 10 μm. h Quantification of the number of droplets per cell in images obtained from assays shown in g. n = 20 cells per condition. Error bars, means ± SD. P values were determined by the two-tailed Student’s t-test. ***P < 0.001.
Overall, we discovered that the phase separation of zebrafish DDx3xb is regulated via its N-terminal IDR domain. In addition, increased ATP promotes Ddx3xb phase separation during MZT. These data may hint at a role of Ddx3xb in regulating MZT through LLPS activity.
Ddx3xb phase separation is essential for zebrafish embryogenesis
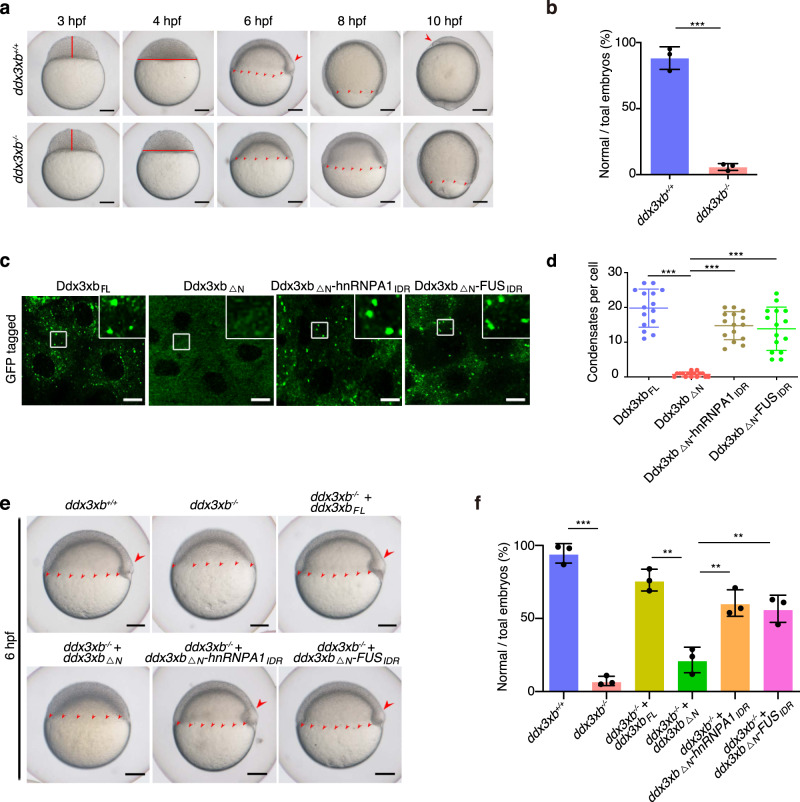
To determine whether Ddx3xb is functionally required for zebrafish embryogenesis, we generated a ddx3xb-null frameshift mutant by CRISPR/Cas9 (Supplementary information, Fig. S4a–d). Both the wild-type and mutant embryos could reach the 1k-cell stage at 3 hpf. However, subsequent developmental processes, including gastrulation, were obviously delayed in mutant embryos (Fig. 3a, b; Supplementary information, Table S1). As an additional control, we knocked down ddx3xb by ddx3xb morpholino (MO) injection (Supplementary information, Fig. S5a) as verified by western blot (Supplementary information, Fig. S5b). Relative to control, ddx3xb morphants (MO-injected embryos) exhibited obvious developmental delays starting from 4 hpf (Supplementary information, Fig. S5c, d and Table S1). Of note, the development was normal in Ddx3xb-deficient embryos before the mid-blastula transition (about 3 hpf), suggesting that Ddx3xb mainly regulates the developmental process after the rapid synchronous cell division period. Therefore, our findings demonstrate that Ddx3xb deficiency causes a developmental defect.
Fig. 3. Ddx3xb phase separation is essential for zebrafish early embryogenesis.
a, b Time-matched images (a) and statistics (b) of zebrafish embryogenesis showing a developmental delay after 3 hpf in the ddx3xb-null frameshift mutant. The red straight lines denote the height (3 hpf) and width (4 hpf) of blastula. The continuous small red arrowheads denote the edge of epiboly. The big red arrowheads denote the embryonic shield (6 hpf) and polster (10 hpf). Scale bars, 100 μm. n = 3 biological replicates. Error bars, means ± SD. P values were calculated by the two-tailed Student’ s t-test. ***P < 0.001. c Images of zebrafish embryos expressing N-terminally GFP-tagged Ddx3xb variants, including Ddx3xbFL, Ddx3xb△N, Ddx3xb△N-hnRNPA1IDR, and Ddx3xb△N-FUSIDR, at 4 hpf. Scale bars, 10 μm. d Quantification of the number of droplets per cell in images obtained from assays shown in c. n = 20 cells per condition. Error bars, means ± SD. P values were determined by the two-tailed Student’s t-test. ***P < 0.001. e, f Bright-field images of representative zebrafish embryos under different Ddx3xb backgrounds (e), and histograms quantifying the phenotype (f). The continuous small red arrowheads denote the edge of epiboly. The big red arrowheads denote the embryonic shield. Scale bars, 100 μm. n = 3 biological replicates. Error bars, means ± SD. P values were calculated by the two-tailed Student’ s t-test. **P < 0.01; ***P < 0.001.
To test whether the phase separation behavior of Ddx3xb is essential for zebrafish early embryogenesis, we generated two chimeric proteins, Ddx3xb△N-FUSIDR and Ddx3xb△N-hnRNPA1IDR, with the N-terminal IDR of Ddx3xb replaced by the IDR from FUS30 and hnRNPA1,31 respectively (Supplementary information, Fig. S6a). FUS and hnRNPA1 are well-known RNA binding proteins (RBPs) that are involved in phase separation through their IDRs. We also replaced the N-terminal IDR of Ddx3xb with an non-RNA-binding IDR from YAP protein whose phase separation can be initiated by coiled-coil domain-mediated hydrophobic interaction and formed by weak interactions of short IDR.32 We found that chimeric Ddx3xb proteins containing IDRs from other proteins can also fulfill phase separation in both zebrafish embryos and HEK293 cells (Fig. 3c, d; Supplementary information, Fig. S6b–f). We then injected ddx3xbFL or ddx3xb△N mRNA into ddx3xb mutant embryos at 1-cell stage. We found that the developmental delay caused by Ddx3xb deficiency can be rescued by injecting ddx3xbFL mRNA, but not ddx3xb△N mRNA (Fig. 3e, f). We also observed that such developmental delay can be rescued when injecting chimeric ddx3xb mRNAs, including ddx3xb△N-FUSIDR and ddx3xb△N-hnRNPA1IDR (Fig. 3e, f; Supplementary information, Table S1). We also co-injected ddx3xb MO with ddx3xb variant mRNA together into 1-cell stage embryos. The sequence in translation initiation region of these chimeric constructs has been modified for not being targeted by the ddx3xb MO. Similar observations were obtained by injecting full-length ddx3xb, truncated ddx3xb, or chimeric ddx3xb mRNA into ddx3xb morphants (Supplementary information, Fig. S7a, b and Table S1). Together, these results suggest that loss of Ddx3xb induces developmental delay, and LLPS ability of Ddx3xb is essential for zebrafish embryogenesis.
Ddx3xb phase separation facilitates zebrafish MZT
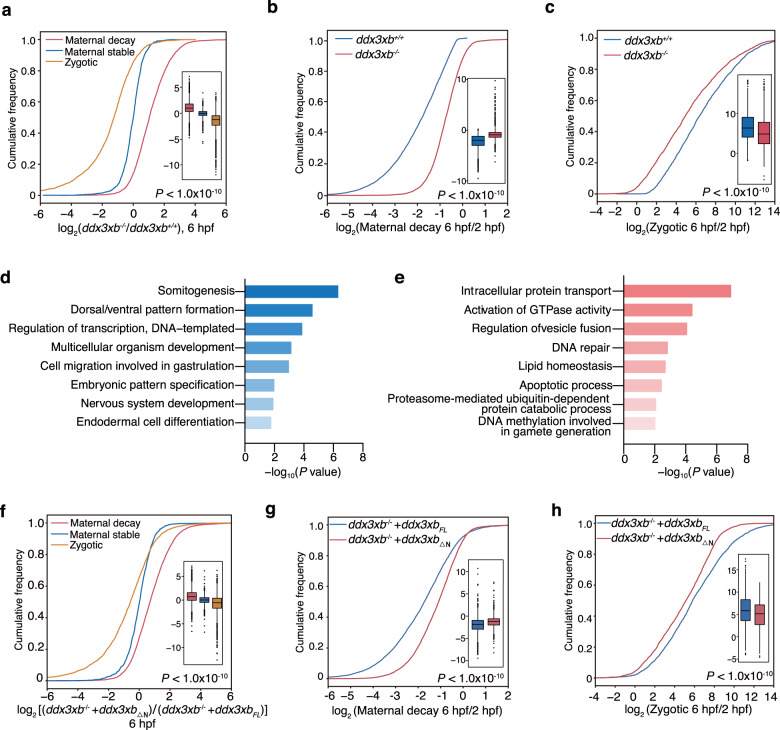
MZT process is an essential embryonic developmental period, proceeding from maternally to zygotically controlled state. To illustrate the potential impact of Ddx3xb phase separation on MZT, we performed RNA-seq to define the effect of Ddx3xb at a transcriptome-wide scale at 2 hpf and 6 hpf of wild-type and ddx3xb mutant embryos (Supplementary information, Fig. S8a). We compared gene expression levels of wild-type embryos between 2 hpf and 6 hpf, and grouped genes according to the following criteria: (1) maternal decay genes which express RNAs with an RPKM > 1 at 2 hpf, log2(fold change) > log2(1.2), and P value < 0.05; (2) maternal stable genes which express RNAs with an RPKM > 1 at 2 hpf, log2(fold change) < log2(1.2); and (3) zygotic genes which express RNAs with an RPKM < 1 at 2 hpf and RPKM > 1 at 6 hpf (Supplementary information, Table S2). Ddx3xb deficiency resulted in the upregulation of maternal decay mRNAs as well as downregulation of zygotic mRNAs at 6 hpf (Fig. 4a). Further analysis demonstrated that Ddx3xb deficiency results in a delay in both maternal mRNA degradation (Fig. 4b) and zygotic gene activation (Fig. 4c). RNA-seq analysis of ddx3xb morphants also revealed a similar observation to that of ddx3xb mutant embryos (Supplementary information, Fig. S8b–e). To determine whether the delayed clearance of maternal RNA and zygotic RNA transcription upon Ddx3xb deficiency is directly caused by functional loss of Ddx3xb or indirectly by a general delay of embryo development, we performed RNA-seq using stage-matched wild-type (6 hpf) and ddx3xb mutant (7.2 hpf) embryos at the shield stage (Supplementary information, Fig. S8f). The results demonstrated that Ddx3xb deficiency caused a delayed clearance of maternal RNA and zygotic RNA transcription (Supplementary information, Fig. S8g–i), indicating that the delayed MZT in ddx3xb mutant embryos is directly caused by loss of Ddx3xb function. Gene Ontology (GO) analysis showed that downregulated zygotic genes are enriched in processes of somitogenesis, dorsal/ventral pattern formation, and regulation of transcription (Fig. 4d; Supplementary information, Table S3). Upregulated maternal decay genes are enriched in processes, such as intracellular protein transport, activation of GTPase activity, and regulation of vesicle fusion (Fig. 4e; Supplementary information, Table S3). Moreover, to determine whether the effect of Ddx3xb deficiency on MZT is dependent on phase separation, we performed RNA-seq in ddx3xbFL or ddx3xb△N mRNA-injected ddx3xb mutant embryos at 2 hpf and 6 hpf (Supplementary information, Fig. S9a). The results showed the upregulation of maternal decay mRNAs and downregulation of zygotic mRNAs in ddx3xb△N group, compared to ddx3xbFL group, at 6 hpf (Fig. 4f). In addition, a delay in both maternal mRNA degradation (Fig. 4g) and zygotic gene activation was found (Fig. 4h). We also performed RT-qPCR-based rescue assays using the chimeric protein (Ddx3xb△N-FUSIDR and Ddx3xb△N-hnRNPA1IDR). The results showed that the delayed maternal gene decay (Supplementary information, Fig. S9b) and zygotic gene activation (Supplementary information, Fig. S9c) caused by Ddx3xb deficiency could be rescued by injecting ddx3xb△N-FUSIDR or ddx3xb△N-hnRNPA1IDR mRNAs, further supporting the importance of LLPS ability of Ddx3xb in zebrafish MZT. Taken together, these results suggest that the phase separation activity of Ddx3xb is critical for MZT.
Fig. 4. Ddx3xb phase separation facilitates zebrafish MZT.
a Cumulative distribution and boxplots (inside) of the log2 fold changes of RNA levels of ddx3xb mutant versus wild-type embryos in maternal decay, maternal stable, and zygotic groups at 6 hpf. P values were calculated by the Kruskal Wallis test. b Cumulative distribution and boxplots (inside) of the log2 fold changes of RNA levels of maternal decay group in ddx3xb wild-type and mutant embryos during the period of 2 hpf to 6 hpf. P values were calculated by the Kruskal Wallis test. c Cumulative distribution of the log2 fold changes of RNA levels of zygotic group in ddx3xb wild-type and mutant embryos during the period of 2 hpf to 6 hpf. P values were calculated by the Kruskal Wallis test. d Gene set enrichment analysis of downregulated genes in the zygotic group compared between ddx3xb mutant and wild-type embryos at 6 hpf. e Gene set enrichment analysis of upregulated genes in the maternal decay group compared between ddx3xb mutant and wild-type embryos at 6 hpf. f Cumulative distribution and boxplots (inside) of the log2 fold changes of RNA levels between ddx3xbFL and ddx3xb△N mRNAs-injected ddx3xb mutant embryos in maternal decay, maternal stable, and zygotic groups at 6 hpf. P values were calculated by the Kruskal Wallis test. g Cumulative distribution and boxplots (inside) of the log2 fold changes of RNA levels of maternal decay group in ddx3xbFL and ddx3xb△N mRNAs-injected ddx3xb mutant embryos during the period of 2 hpf to 6 hpf. P values were calculated by the Kruskal Wallis test. h Cumulative distribution and boxplots (inside) of the log2 fold changes of RNA levels of zygotic group in ddx3xbFL and ddx3xb△N mRNAs-injected ddx3xb mutant embryos during the period of 2 hpf to 6 hpf. P values were calculated by the Kruskal Wallis test.
Condensation of Ddx3xb promotes maternal mRNA translation through unwinding 5’ UTR structures
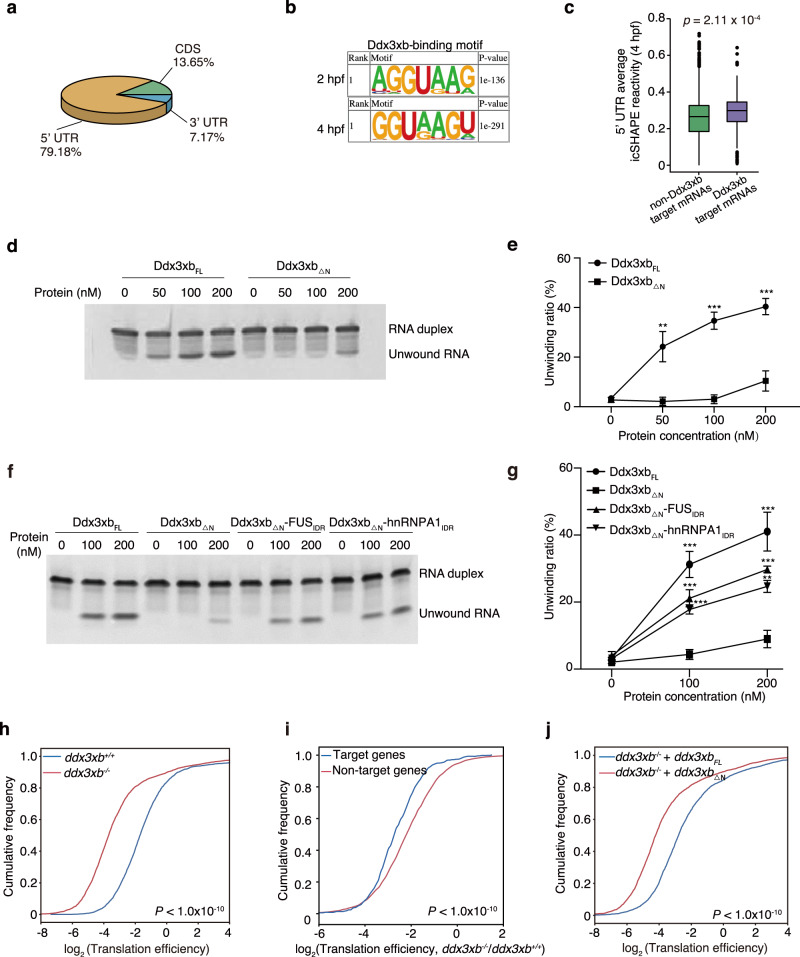
To further explore the regulatory mechanism of Ddx3xb phase separation in MZT, we preformed Flag-Ddx3xb RIP-seq to identify RNA targets of Ddx3xb in zebrafish embryos at 2 hpf or 4 hpf (Supplementary information, Fig. S10a). The results showed that Ddx3xb predominantly binds to 5’ UTRs via a GGUAAG motif (Fig. 5a, b). We thus focused on 5’ UTRs of the Ddx3xb target mRNAs for subsequent analysis. Ddx3xb displayed a higher binding affinity on 5’ UTR at 4 hpf than at 2 hpf (Supplementary information, Fig. S10b, c and Table S4). In addition, Ddx3xb target genes are highly enriched in processes related to chordate embryonic development, transcription, cell cycle, and regulation of cell shape (Supplementary information, Fig. S10d and Table S5). As human and yeast orthologs of the zebrafish Ddx3xb had been reported to be involved in RNA processing regulation by unwinding RNA structures,25–27 we therefore set to evaluate whether Ddx3xb could function similarly on its mRNA targets. Previously we reported the landscape of RNA structures during MZT using in vivo click selective 2’-hydroxyl acylation and profiling experiment (icSHAPE).8 Here we combined these icSHAPE data with our RIP-seq data to examine the effect of Ddx3xb on RNA structures in common Ddx3xb target mRNAs detected at 2 hpf and 4 hpf. Surprisingly, we discovered that 5’ UTR structures of Ddx3xb target mRNAs were significantly more open than those of non-Ddx3xb targets at 4 hpf (Fig. 5c). Such difference was not observed in 2 hpf embryos (Supplementary information, Fig. S10e). This analysis suggests that Ddx3xb unwinds 5’ UTR structures at 4 hpf. Furthermore, cnot6a and lin28a (Ddx3xb target RNAs) as well as gapdh (nontarget RNA) from RIP-seq data were chosen for synthesizing Cy3-labelled mRNAs by in vitro transcription. Then, these Cy3-labelled mRNAs were injected into zebrafish embryos and analyzed for the fluorescent probe localization. The results showed that both Ddx3xb target RNAs, but not the nontarget-gapdh, were enriched in Ddx3xb condensates (Supplementary information, Fig. S10f). Given that Ddx3xb phase separation occurs at 4 hpf (Fig. 1k, l), we speculate that phase separation of Ddx3xb may influence its helicase function.
Fig. 5. Condensation of Ddx3xb regulates maternal mRNA translation via unwinding RNA structures.
a Pie chart depicting relative fraction of Ddx3xb-binding reads that mapped to 5’ UTR, CDS and 3’ UTR regions of mRNAs identified by RIP-seq. mRNA target regions were subdivided into three transcript segments. b Sequence motif identified within the binding region of Ddx3xb at 5’ UTR by HOMER. c Boxplot showing the average icSHAPE reactivity in 5’ UTR of non-Ddx3xb target mRNAs and Ddx3xb target mRNAs at 4 hpf (a high icSHAPE reactivity implies a single-stranded conformation, a low icSHAPE reactivity implies a double-stranded conformation). P values were calculated by the Kruskal Wallis test. d, e In vitro helicase assays using different doses (0 nM, 50 nM, 100 nM, and 200 nM) of purified Ddx3xbFL and Ddx3xb△N proteins. The unwinding ratio was calculated by (unwound RNA)/((unwound RNA) + (RNA duplex)). Error bars, means ± SD (n = 3). P values were determined by the two-tailed Student’s t-test. **P < 0.01, ***P < 0.001. f, g In vitro helicase assays using different doses (0 nM, 100 nM, and 200 nM) of purified Ddx3xb△N, Ddx3xb△N-hnRNPA1IDR, and Ddx3xb△N-FUSIDR proteins. The unwinding ratio was calculated by (unwound RNA)/((unwound RNA) + (RNA duplex)). Error bars, means ± SD (n = 3). P values were determined by the two-tailed Student’s t-test. **P < 0.01; ***P < 0.001. h Cumulative distribution of the translation efficiency of Ddx3xb mutant and wild-type embryos at 4 hpf. P values were calculated by the Kruskal Wallis test. i Cumulative distribution of the log2 fold changes of translation efficiency of Ddx3 target genes and Ddx3 non-target genes between Ddx3xb mutant and wild-type embryos at 4 hpf. P values were calculated by the Kruskal Wallis test. j Cumulative distribution of the translation efficiency of ddx3xbFL and ddx3xb△N mRNAs-injected ddx3xb mutant embryos at 4 hpf. P values were calculated by the Kruskal Wallis test.
To test whether the LLPS contributes to the helicase activity of Ddx3xb, in vitro helicase assays were performed using full-length and truncation versions of Ddx3xb (Ddx3xbFL and Ddx3xb△N). We found that the RNA helicase ability of Ddx3xb△N was significantly decreased compared with Ddx3xbFL (Fig. 5d, e; Supplementary information, Fig. S11a), suggesting that IDR is necessary for Ddx3xb helicase activity. To substantiate our findings that protein phase separation and Ddx3xb helicase activity are interlinked, we generated three chimeric proteins, Ddx3xb△N-FUSIDR, Ddx3xb△N-hnRNPA1IDR and Ddx3xb△N-YAPIDR. The results showed that all these chimeric proteins exhibit a significant higher helicase activity as compared with Ddx3xb△N with the N-terminal IDR deleted (Fig. 5f, g; Supplementary information, Fig. S11b–e). To further investigate the role of phase separation in helicase activity of Ddx3xb, we performed the helicase assay by using the dilute phase and condensed phase of Ddx3xb. Firstly, we performed a sedimentation assay to separate the condensed liquid phase from the bulk Ddx3xb aqueous solutions (2 μM) by centrifugation.18 The top 90% of the bulk Ddx3xb aqueous solution was defined as dilute phase, the bottom 10% as the condensed phase. By using Alexa-488-labeled Ddx3xb to test the effiency of sedimentation assay, we found that the condensed phase fraction of bottom layer (condensed phase) was much higher than that of supernatant (dilute phase) (Supplementary information, Fig. S11f, g). We then used the same volume of these two separated phases for the helicase assay, and the condensed phase showed a much higher level of helicase activity than the dilute phase (Supplementary information, Fig. S11h, i). We found that the protein concentration of condensed phase was much higher than that of dilute phase (Supplementary information, Fig. S11j). It is likely that the difference in protein amount per volume results in different catalytic efficiencies of these two phases. To investigate the role of phase separation in helicase activity of Ddx3xb more accurately, the condensed phase was diluted to achieve the same protein amount (1 × 10−2 nmol) as dilute phase in the same reaction volume (25 μL) for the helicase assay. In this case, the consensed phase group was named as condensed phase (diluted). The condensed phase fraction and helicase activity were measured respectively after 20 min of reaction. By using Alexa-488-labeled Ddx3xb, we found that the condensed phase fraction of the condensed phase (diluted) group was still higher than that of dilute phase group (Supplementary information, Fig. S11k, l). The results of helicase assay showed that the helicase activity of condensed phase (diluted) group was higher than that of dilute phase (Supplementary information, Fig. S11m, n). Of note, a considerable amount of Ddx3xb protein in the condensed phase (diluted) group remained in a condensed state at the end of the helicase assay (Supplementary information, Fig. S11o, p). Thus, our findings demonstrate that phase separation, mediated by the N-terminal IDR in Ddx3xb, facilitates the Ddx3xb helicase activity, and the N-terminal IDR in Ddx3xb can be functionally replaced by the IDR of FUS, hnRNPA1 or YAP. These results, combined with other evidence (Fig. 2d; Supplementary information, Fig. S3c), strongly support that Ddx3xb is able to enrich dsRNAs and open their structures in the droplets, and then unwound RNAs are released. The human and yeast orthologs of the zebrafish Ddx3xb had been reported to control translation through unwinding RNA structures in 5' UTRs.25–27 We further performed ribosome profiling assays to determine whether Ddx3xb affects mRNA translation in zebrafish embryogenesis (Supplementary information, Fig. S12a, b). When comparing translation efficiency (TE) between wild-type and ddx3xb mutant embryos at 4 hpf, we found that the TE was decreased in ddx3xb mutant embryos (Fig. 5h; Supplementary information, Table S6). Based on the Flag-Ddx3xb RIP-seq data, all genes were categorized into two groups: Ddx3xb target and non-target gene groups and the difference in TE between these two group genes were compared. We found that TE of Ddx3xb target genes decreased much more than that of non-target genes upon Ddx3xb deficiency (Fig. 5i), supporting the function of Ddx3xb in translation control during zebrafish embryogenesis. We further performed ribosome profiling in ddx3xbFL or ddx3xb△N mRNA-injected ddx3xb mutant embryos at 4 hpf (Supplementary information, Fig. S12c, d), and found that TE was lower in ddx3xb△N group than that in ddx3xbFL group (Fig. 5j; Supplementary information, Table S6), supporting the function of Ddx3xb phase separation in translation control during zebrafish embryogenesis (Supplementary information, Fig. S12e). Moreover, at 4 hpf, more than 90% of transcripts detected by ribosome profiling overlapped with the maternal stable and maternal decay genes (Supplementary information, Fig. S12f). This result indicates that transcripts of maternal factors still dominate during this time period. Taken together, these results suggest that Ddx3xb facilitates MZT by promoting maternal mRNA translation through unwinding their 5’ UTR structures in a phase separation-dependent fashion.
N-terminal IDR of Ddx3xb is necessary for maternal mRNA translation in zebrafish early embryogenesis
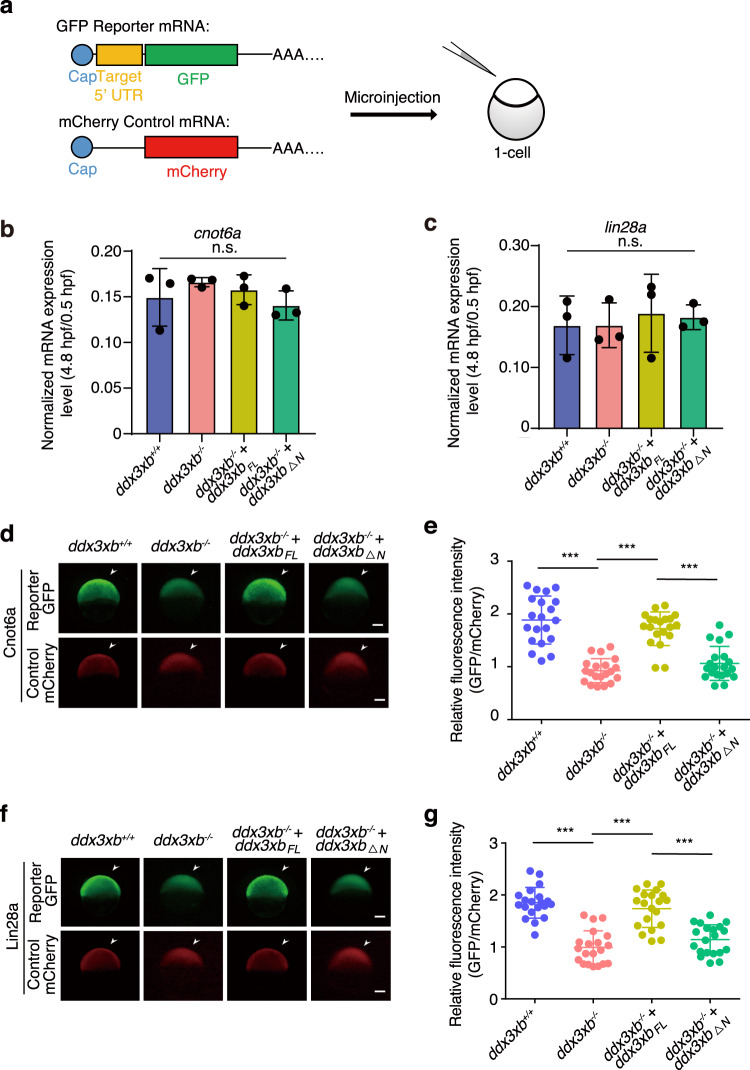
To explore whether Ddx3xb phase separation is able to influence maternal mRNA translation efficiency, we generated three GFP reporter mRNA constructs. These were derived from endogenous 5’ UTRs of Ddx3xb target maternal mRNAs, including cnot6a, lin28a, and smarcad1a. mCherry mRNA was used as control (Fig. 6a). These GFP reporter mRNAs and mCherry control mRNA were co-injected into wild-type, ddx3xb mutant, ddx3xbFL rescue or ddx3xb△N rescue embryos at 1-cell stage. The abundances of these reporter mRNAs were examined by reverse-transcription quantitative PCR (RT-qPCR). The results showed that the expression levels of reporter mRNAs are not influenced by ddx3xb deficiency (Fig. 6b, c; Supplementary information, Fig. S13a). GFP protein expression was significantly higher in wild-type compared to ddx3xb mutant, indicating that the Ddx3xb defect leads to the translation block (Fig. 6d–g; Supplementary information, Fig. S13b, c). Intriguingly, the translation block can be rescued by ddx3xbFL, but not by ddx3xb△N (Fig. 6d–g; Supplementary information, Fig. S13b, c).
Fig. 6. Ddx3xb regulates maternal mRNA translation in a condensation-dependent manner.
a Design of reporter mRNAs. 5’ UTR sequences of Ddx3xb targets were fused with mRNA of GFP. b, c Relative mRNA level (4.8 hpf vs 0.5 hpf) of GFP reporter under the control of 5’ UTR of cnot6a (b) and lin28a (c). n = 3 for each group. Error bars, means ± SD. P values were determined by the two-tailed Student’s t-test. ns, not significant. d–g Protein levels of 5’ UTR (Cnot6a)-GFP reporter (d, e) and 5’ UTR (Lin28a)-GFP reporter (f, g) and mCherry control at 4.8 hpf in different embryos analyzed by fluorescence intensities. Representative images of embryos are shown in d, f. Scale bars, 100 μm. Relative fluorescence intensities (GFP/mCherry) are shown in e, g. n = 20 for each group. Error bars, means ± SD. P values were determined by the two-tailed Student’s t-test. ***P < 0.001.
Overall, our study demonstrates that Ddx3xb condensation, which is mediated by its N-terminal IDR, is promoted by an increase in ATP concentration. During MZT, Ddx3xb phase separation modulates maternal mRNA translation via opening 5’ UTR RNA structures, and this translation control effect is essential for zebrafish early development (Fig. 7).
Fig. 7. Schematic model showing that Ddx3xb regulates maternal mRNA translation efficiency in a condensation-dependent manner.
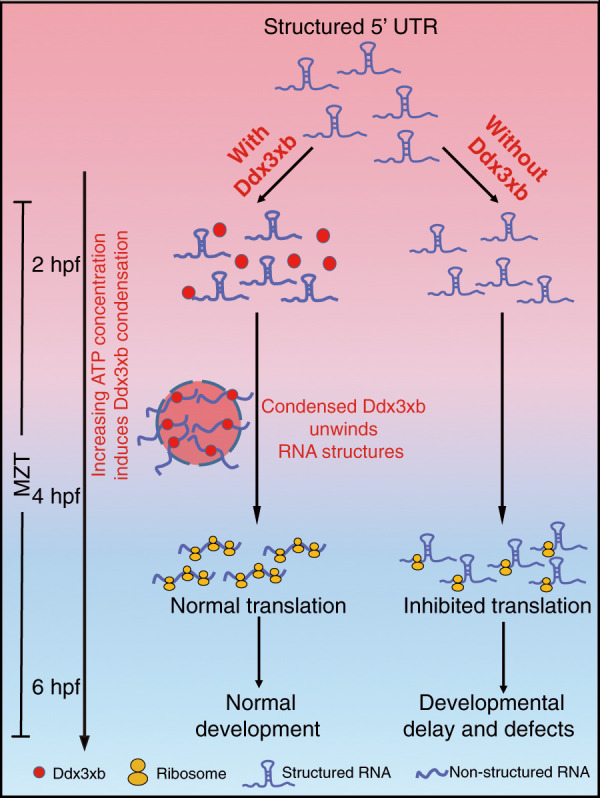
Condensation of Ddx3xb promoted by increased ATP concentration at 4 hpf unwinds the structured 5’ UTRs to promote translation of maternal mRNAs. Without Ddx3xb, structured 5’ UTRs cannot be opened, which results in translation inhibition, further leading to developmental delay and defects during zebrafish embryogenesis.
Discussion
In this study, we show that fertilization leads to phase separation of Ddx3xb, which selectively regulates translation during a critical period of early development. Our data reveal a completely novel and temporally controlled mechanism that is essential for embryogenesis. Specifically, we show that (1) a gradual increase in ATP concentration, stimulated by fertilization, promotes phase separation of RNA helicase Ddx3xb; (2) the LLPS induction was mediated by Ddx3xb N-terminal IDR which could be functionally replaced by IDR of FUS or hnRNPA1, and this LLPS mechanism enhances the Ddx3xb RNA helicase activity; (3) Ddx3xb RNA helicase specifically unwinds maternal mRNAs, driving maternal mRNA translation required for MZT and embryogenesis. Overall, our findings demonstrate that condensation of the Ddx3xb helicase represents a completely novel mechanism for translational control, which is critical during early embryogenesis. This mechanism is tightly controlled, and occurs only at a specific time. Furthermore, Ddx3xb seems to be a molecular sensor for differential ATP concentrations, which allows it to regulate MZT of early embryogenesis.
In vertebrates, early embryogenesis is characterized by the conversion of a fertilized egg into a multicellular complex organism via a series of programmed genetic and epigenetic events.33 The MZT is one of the most critical events during this process. Because of the transcriptional quiescence of the zygotic genome, the orchestrated regulation of maternal mRNAs is particularly vital for ZGA and maternal clearance. Recent studies have illustrated that m5C-modified maternal mRNAs are stabilized by Ybx1 and Pabpc1a during zebrafish MZT,7 and RBP Elavl1a regulates maternal mRNA stability in an RNA structure-dependent fashion.8 In addition, the selective translation of maternal factors, including Nanog, Pou5f1, and SoxB1, is crucial for the ZGA.4 The gradual elongation of poly(A) tails also coincides with translation efficiency.12,13 However, how maternal mRNAs are spatiotemporally regulated, thereby allowing accurate execution of zygote fate transition remains largely unknown. Our findings provide a new perspective that phase separation-mediated translational control facilitates MZT and is a critical regulator for early embryogenesis.
Biomolecular condensates are involved in diverse biochemical processes including RNA metabolism, ribosome biogenesis, the DNA damage response, and signal transduction.14 DDX3 proteins, such as Ddx3xb, have a conservative structure that is defined by a core of helicase domains flanked by two IDRs of diverse length and sequence composition.23 However, the specific regions that are sufficient for the formation of condensates are diverse among different species.27,34 Previous studies have demonstrated that an N-terminal IDR of LAF-1, a DDX3 RNA helicase found in the Caenorhabditis elegans, is necessary and sufficient for phase separation.34 The condensation of its yeast ortholog Ded1p is greatly affected by the C-terminal IDR under heat stress.27 In our study, by in vitro and in vivo phase separation assays, we show that the N-terminal IDR is essential for coalescence of zebrafish Ddx3xb, which can be functionally replaced by IDR of FUS or hnRNP1 protein. However, results from the phase separation assays showed that the average number of chimeric protein condensates was lower than that of wild-type Ddx3xb, suggesting that the IDRs from FUS and hnRNPA1 could not fully replace the function of the N-terminal IDR of Ddx3xb. These results indicate that functional IDRs in DDX3 proteins are diverse in different species or conditions.
Previous studies demonstrate that yeast RNA helicase Dhh1 promotes phase separation in its ATP-bound form,23 whereas ATP hydrolysis induces compartment turnover and RNA release. Fertilization increases ATP levels with the Ca2+ transient in mice,28 and ATP concentration in zebrafish embryos also shows the similar pattern.29 In support, we observed that an increase in ATP concentration promotes condensation of zebrafish Ddx3xb at 4 hpf, and blocking ATP synthase from zebrafish 2 hpf leads to the failure of Ddx3xb phase separation formation, suggesting that Ddx3xb phase separation is regulated by ATP concentration during MZT. However, because of the complexity of in vivo environment, it could not be excluded that other factors might also be involved in Ddx3xb condensation during this period.
Condensates substantially increase local concentration of biomolecules. As such, condensates can increase reaction kinetics and specificity.14 Previous study has shown that the IDR is essential for Drosophila Otu enzymatic activity by inducing coalescence of Otu.35 In our study, deletion of the N-terminal IDR domain results in a decrease in the Ddx3xb helicase activity. Our domain replacement experiments also show that IDRs from other RBPs are able to mediate the formation of the chimeric Ddx3xb condensates. These results indicate that the N-terminal IDR is necessary for zebrafish Ddx3xb helicase activity. Previous studies indicate that the human and yeast orthologs of the zebrafish Ddx3xb influence translation through directly and preferentially binding 5’ UTRs of its targets.25–27 By combining icSHAPE8 with RIP-seq data, we show that Ddx3xb prefers to bind to 5’ UTRs of mRNAs. We further observed fewer mRNA 5’ UTR structures formed in Ddx3xb-bound mRNAs in comparison to non-Ddx3xb-bound mRNAs at 4 hpf. In contrast, no significant difference was observed at 2 hpf. These results reveal that the regulation of Ddx3xb helicase activity depends on its LLPS state in early embryogenesis, supporting an ‘on-and-off’ switch model in which a protein’s enzymatic activity is dynamically regulated by IDR-mediated LLPS.
In conclusion, our study demonstrates that Ddx3xb condensation is promoted by an increase in ATP concentration. This selectively modulates maternal mRNA translation via opening 5’ UTR RNA structures during MZT. This selective translational regulation by phase separation spatiotemporally coordinates zebrafish early development. We reasonably speculate that the phase separation-dependent translation control mechanism probably coordinates similar developmental processes in other vertebrates as well.
Materials and methods
Animal models and chemical treatment
Zebrafish wild-type strains AB and Tubingen were raised in system water at 28.5 °C under standard condition. Zebrafish embryos were obtained by natural spawning. In Oligomycin A treatment experiments, embryos were treated with 5 μM Oligomycin A (Selleck, S1478) from 2 hpf to the stages for detection. For ATP treatment assay, 1 nL ATP solution (25 mM) was injected into zebrafish embryos at 3 hpf. All zebrafish experiments were approved and carried out in accordance with the Animal Care Committee at the Institute of Zoology, Chinese Academy of Sciences, China.
Replication, size estimation, and randomization
At least two biological replicates were performed for all experiments. No sample size estimation was performed. Animals for all experiments were randomly selected. All animals have been included in statistical analyses, and no exclusions have been done. Images were randomized before quantification.
Cell lines
HEK293 cells (Homo sapiens, RRID: CVCL_0045) were originally purchased from ATCC, routinely confirmed to be free of mycoplasma. Cells were maintained in standard DMEM (Gibco) supplemented with 10% fetal bovine serum (Gibco) and 1× penicillin/streptomycin (Invitrogen) in humidified 5% CO2, 37 °C cell culture incubator.
MOs, vector construction, mRNA synthesis and injection
MOs used in this study, including ddx3xb MO (5ʹ-TCTACGGCCACATGACTCATAACTA-3ʹ) and Control MO (5ʹ-CCTCTTACCTCAGTTACAATTTATA-3ʹ), were purchased from GeneTools. MOs (6 ng for ddx3xb MO, 6 ng for Control MO) were injected into embryos at 1-cell stage. For truncated or chimeric versions of ddx3xb mRNA overexpression experiments, truncated or chimeric versions of ddx3xb were cloned into a pCS2+ vector. mRNA was transcribed using the mMESSAGE mMACHINE™ SP6 kit (Invitrogen, AM1340) and injected into zebrafish embryos at 1-cell stage. For mRNA colocalization assay, the T7 promoter sequence fused with PCR products of the 5’ UTR and CDS region (cnot6a, lin28a and gapdh) were used as templates for in vitro transcription. Cy3-labeled mRNAs were generated using the MEGAscript™ T7 Transcription Kit (Invitrogen, AM1333) with Cy3-UTP (Enzo Life Sciences). For mRNA translation reporter experiments, N-terminally GFP-tagged reporters and mCherry mRNA control were cloned into pCS2+ vectors. Capped mRNAs were generated using the mMACHINE™ SP6 kit (Invitrogen, AM1340) according to the manufacturer’s protocol. 30 pg of GFP-tagged reporter mRNA and mCherry mRNA were co-injected into zebrafish embryos at 1-cell stage.
Mutant generation by CRISPR/Cas9
The ddx3xb mutant was generated using CRISPR/Cas9, following a method for Cas9 mRNA and guide RNA synthesis as described previously.36 pXT7-Cas9 was used for cas9 mRNA transcription by the T7 mMessage Machine kit (Invitrogen, AM1344). Cas9 mRNA was purified using the RNA clean Kit (TIANGEN, DP412). The ddx3xb gRNA, which targeted 5ʹ-GGACCAGGAGAAGCTCTGC-3ʹ sequence, was generated by in vitro transcription using T7 RNA polymerase (Promega, P2075). The mutant was identified by DNA sequencing analysis.
Microscopy imaging
High-resolution fluorescent images were taken with a Nikon A1 confocal microscope. Microscopic observation and photography were carried out as previously described.37 The condensed phase fraction was analyzed using ImageJ, and the P values were calculated by using GraphPad Prism. Embryos were observed with a Nikon SMZ1500 microscope and imaged with a Nikon digital camera in bright field or for fluorescence detection. The fluorescence intensities of reporters were analyzed using ImageJ, and the P values were calculated using GraphPad Prism.
Phase separation assay
In vitro phase separation assay was performed as previously described.23 Briefly, proteins were labeled with Alexa Fluor 488 NHS ester (Invitrogen, A20000) or Alexa Fluor 568 NHS ester (Invitrogen, A20003) at the final labeling ratio of 5%. For Fig. 1f, 10 μM Ddx3xb variant proteins were incubated in 50 mM HEPES-KOH, pH 7.0, 100 mM KCl at room temperature. For Fig. 2b, d, e and Supplementary information, Fig. S3b, d, 4 μM Ddx3xbFL proteins were incubated in 50 mM HEPES-KOH, pH 7.0, 100 mM KCl, with 100 ng/µL zebrafish mRNA and different concentrations of ATP at room temperature. 2 μM SYTO-64 (Invitrogen, S11346) was used to stain RNA in Fig. 2d. TNP-ATP (Tocris, 2464) or AMP-PNP (Roche, 10102547001) was used in Fig. 2e or Supplementary information, Fig. S3d. For the sedimentation assay, 2 μM Ddx3xb proteins was subjected to centrifugation at 8000× g for 5 min at room temperature. Dilute phase and condensed phase were separated into two tubes immediately after centrifugation. Samples were examined under Nikon A1 confocal microscopy.
FRAP assay
FRAP experiments were carried out on a Nikon A1 confocal microscope. GFP-tagged or Alexa-488-labeled Ddx3xb proteins were bleached using a 488-nm laser beam at 50% power. SYTO 64-stained RNA was bleached using a 561-nm laser beam at 50% power. To analyze the condensates in HEK293 cells, a circular region was selected for FRAP assay. To analyze condensates in solution, a circular region containing one droplet was assayed. For quantitative analysis, the average fluorescence intensity of frames before photobleaching was normalized to 100%. Data were analyzed using GraphPad Prism.
ATP concentration measurement
ATP levels of zebrafish embryos were determined using an Enhanced ATP Assay Kit (Beyotime, China). Twenty embryos were collected for each stage and lysed in lysis buffer. Lysed embryos were centrifuged for 5 min at 12,000× g, 4 °C, and the supernatant was collected. Detecting solution was added to a 96-well plate and incubated at room temperature for 5 min. The supernatant was then added to the plate, mixed quickly, and the signals were detected by BioTek Synergy4 (Gene Company) within 30 min.
Whole-mount in situ hybridization
Whole-mount in situ hybridization was carried out with a Digoxigenin-uridine-5’-triphosphate (Roche)-labeled RNA probe for ddx3xb. For probe synthesis, PCR products from primers (ddx3xb F:5’-GGAGGATTCGGAAACTTCTACAG-3’, ddx3xb R:5’-GCGTCGCGTCTCCAGAGTAAAG-3’) were cloned into the pGEM-T vector (Promega). The RNA probe was transcribed with T7 RNA polymerase (Promega). After hybridization, RNA probes were detected by AP-conjugated anti-DIG antibody (Roche) using BM purple (Roche) as substrate.
Western blot and immunofluorescence
Western blot was performed as previously reported38 using the following antibodies: anti-zebrafish Ddx3xb antibody (custom-made by company, Abclonal), anti-β-actin antibody (Cell Signaling Technology, 4967, RRID:AB_330288), anti-Flag antibody (Sigma-Aldrich, F7425). Immunofluorescence experiments were performed as previously reported39 using the following antibodies: anti-zebrafish Ddx3xb (custom-made by company, Abclonal).
RIP experiments
Flag-ddx3xb mRNA-injected embryos were collected at 2 hpf or 4 hpf and lysed in NETN lysis buffer (150 mM NaCl, 0.5% NP-40, 50 mM Tris-HCl, pH 7.4). Ddx3xb RIP was carried out following a published protocol,40 but with some modifications. Lysate was incubated with anti-FLAG M2 Magnetic Beads (Merck, M8823) for 4 h at 4 °C. RNA was extracted and subjected to library construction using the SMARTer smRNA-Seq kit (Clontech, 635031).
RNA-seq
Total RNA was isolated from zebrafish embryos at different time points with TRIzol reagent and mRNA was purified using the Dynabeads mRNA purification kit (Ambion, 61006). Fragmented mRNA was used for library construction using the KAPA Stranded mRNA-Seq Kit (KAPA, K8401), according to the manufacturer’s protocol.
Protein purification in mammalian cells
HEK293 cells were transfected with plasmids encoding Flag-tagged Ddx3xb variants using the PEI transfection reagent (MKbio). After 48 h, cells were lysed with lysis buffer (50 mM HEPES-KOH, pH 7.4, 1 M KCl, 1% NP-40, 1 mM DTT, 2 mM EDTA, Protease inhibitor cocktail, benzonase) and sonicated (10% output, 10 s pulse-on, 20 s pulse-off) for 1 min by a Sonic Dismembrator (Thermo Fisher). After removing cell debris through centrifugation at 13,300 rpm for 20 min, the lysates were incubated with the anti-Flag M2 Affinity Gel (Sigma-Aldrich, A2220) for 4 h, at 4 °C. Samples were then washed five times with lysis buffer and twice with TBS buffer. The gel-bound proteins were eluted with 4 mg/mL 3× Flag peptide (Sigma-Aldrich, F4799) by incubating the mixture for 1 h, at 4 °C. The eluate containing purified protein was concentrated using VIVASPIN 500 (Sartorius Stadium Biotech) and quantified by Coomassie brilliant blue staining and Western blot analysis.
Helicase assay
The RNA helicase assay was performed as previously reported.41 Briefly, we used unlabeled RNA oligomers (5ʹ-GCGUCUUUACGGUGCUUAAAACAAAACAAAACAAAACAAAA-3ʹ) and the FAM-labeled ones (5ʹ- AGCACCGUAAAGACGC-3ʹ). These oligomers were annealed to form RNA duplexes in the presence of different Ddx3xb variant proteins, 100 μM ATP, RNA inhibitor Rnasin (Promega) in reaction buffer containing 50 mM Tris, pH 7.0, 2 mM MgCl2, 3% glycerol, and 1 mM DTT at room temperature. Reactions were stopped with stop solution containing proteinase K (1 mg/mL) and SDS (0.6%) after a 30-min incubation. Reactions were mixed with 5 μL 5× Hi-Density TBE sample buffer and separated on a 6% TBE gel on ice for 30 min at 80 V. The gel was scanned by a Typhoon 9500 (GE Healthcare) imager.
Ribosome profiling experiments
Ribosome profiling experiments were performed as previously reported,42 with some modifications. Zebrafish embryos were incubated in 100 mg/mL cycloheximide in E3 buffer (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, 0.33 mM MgSO4) for 5 min at room temperature. Embryos were then transferred into 600 μL of ice-cold lysis buffer. After triturating the cells ten times through a 26-G needle, the lysate was clarified by centrifugation for 10 min at 20,000× g at 4 °C to recover the soluble supernatant. 3 μL of RNase I (100 U/µL) was added to 600 μL of lysate. After a 45-min incubation at room temperature with gentle mixing, 10 µL of SUPERase·In (Invitrogen, AM2694) RNase inhibitor was added to stop the nuclease digestion. MicroSpin S-400 HR columns (GE Healthcare, 27-5140-01) were equilibrated with 3 mL of mammalian polysome buffer by gravity flow and emptied by centrifugation at 600× g for 4 min. Immediately 100 µL of the digested lysate was loaded on the column and eluted by centrifugation at 600× g for 2 min. RNA was extracted from the flow-through (approximately 125 µL) using Trizol LS (Life Technologies, 10296-010). Ribosomal RNA fragments were removed using the RiboZero Kit (Illumina, MRZH11124) and were separated on a 17% denaturing urea-PAGE gel (National Diagnostics, EC-829). RNA fragments sized 27 nt to 35 nt were cut out of the gel, and were subjected to library preparation using the Smarter smRNA-Seq kit (Clontech Laboratories, Inc.).
RT-qPCR
RT-qPCR was carried out to examine the relative abundance of target RNA. 0.1 µg of total RNA was used for cDNA synthesis using the RevertAid™ First Strand cDNA Synthesis Kit (Thermo). Experiments were performed with the Takara SYBR Premix Ex Taq (Takara) according to the manufacturer’s instructions and examined by a CFX96 Real-Time PCR System (Bio-Rad). The primers used for RT-qPCR in this study are listed as follows:
ddx3xa-F: 5ʹ-GCCCATGTGTAGTGTACGGT-3ʹ;
ddx3xa-R: 5ʹ-GTCACGGGCCAGAATCTGAA-3ʹ
ddx3xb-F: 5ʹ-CGCCGAAACACCGAAATCAA-3ʹ
ddx3xb-R: 5ʹ-GGAATGTAGCGCCTTCCAGT-3ʹ
gfp-F: 5ʹ-CGTAAACGGCCACAAGTTCAG-3ʹ
gfp-R: 5ʹ-GCGGCTGAAGCACTGCACGCCG-3ʹ.
stt3b-F: 5ʹ-GGGGTGGTTTTCCTTACGGT-3ʹ
stt3b-R: 5ʹ-ATGGCATACAGAGCCACGAA-3ʹ
hsp90b1-F: 5ʹ-GTGGGCTTCTACTCCGCTTT-3ʹ
hsp90b1-R: 5ʹ-TCTACGGTCTCGGTCTTGCT-3ʹ
desi2-F: 5ʹ-GCCACAGAACTCGGAGAGAC-3ʹ
desi2-R: 5ʹ-TCTCGACCACAAAGGATCTCAG-3ʹ
uck2a-F: 5ʹ-AGGAGTTCTGTCTGCCAACAA-3ʹ
uck2a-R: 5ʹ-CAATCCATCCATCCGTCCGT-3ʹ
gdf3-F: 5ʹ-CGTAATCCGAAGGGGAAGGT-3ʹ
gdf3-R: 5ʹ-TTTACCTCCGCACAGTGTCT-3ʹ
nanog-F: 5ʹ-CACCAGCTTCTCAATCTCAGTTTC-3ʹ
nanog-R: 5ʹ-CTGATGGGGTTGGGGAACTC-3ʹ
sumf2-F: 5ʹ-TTGCTCTGTTCTGTTGCTGTTC-3ʹ
sumf2-R: 5ʹ-ACACCATCTCATCATCTGCTGC-3ʹ
bahd1-F: 5ʹ-AGTGTTCATCGTGGCTGAGG-3ʹ
bahd1-R: 5ʹ-CTGAGTGTGTTCTGGGCGAT-3ʹ
kctd15a-F: 5ʹ-TGTCCTCTGTTATTCGGGCTG-3ʹ
kctd15a-R: 5ʹ-GACATACTCCTTCCCTCCTTGG-3ʹ
ephb3a-F: 5ʹ-GGAGAGTTCGGGGAGGTTTG-3ʹ
ephb3a-R: 5ʹ-TGCCATCGTTCAGTCTGAGG-3ʹ
plekhg4-F: 5ʹ-TCTGTCTGGCAGTGGCTCTA-3ʹ
plekhg4-R: 5ʹ-AGCATCCCCCGCTGAGA-3ʹ
abca1a-F: 5ʹ-AGCCGCTTCAGTCTTGAGTC-3ʹ
abca1a-R: 5ʹ-ACCGAGGTGGTCATTAGTGC-3ʹ
RNA-seq data processing and analysis
Raw sequencing reads were processed using FastQC (http://www.bioinfor-matics.babraham.ac.uk/projects/fastqc/). Low-quality bases were trimmed and filtered by cutadapt (v1.13)43 and Trimmomatic (v0.36).44 Processed reads were mapped to the zebrafish genome (Zv9) from the Ensemble annotation using hisat2 (v2.0.5)45 with default parameters (Supplementary information, Table S7). After quality filtering (≥ 20) using SAM tools (v1.9),46 read counts and corresponding RPKMs were calculated. The final RPKM of each transcript is the average of RPKMs from replicates.
The fold change in transcriptional expression levels between samples was calculated using the DEseq2 (v1.26.0)47 package. Based on comparison of expression levels between 2 hpf and 6 hpf, we defined three groups: (i) genes with RPKM > 1 at 2 hpf, log2(FC) > log2(1.2), and FDR < 0.05 were defined as maternal decay genes; (ii) genes with RPKM > 1 at 2 hpf, log2(FC) < log2(1.2) were defined as maternal stable genes; (iii) genes with RPKM < 1 at 2 hpf and RPKM > 1 at 6 hpf were defined as zygotic genes. GO analysis was performed using the DAVID website (https://david.ncifcrf.gov/)48,49 and using all genes as background.
Ribosome profiling RNA-seq data processing and analysis
Single read sequencing data were used to obtain ribosome protected fragments RNA-seq data. The quality of raw sequencing reads was processed using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Low-quality bases were trimmed and filtered by cutadapt (v1.13)43 and Trimmomatic (v0.36).44 Processed reads were mapped to the zebrafish rRNA transcriptome using Bowtie50 and unmapped reads were retained. The remaining reads were mapped to zebrafish genome (Zv9) using TopHat (v2.1.1)51 with “–bowtie1” (Supplementary information, Table S7). The expression levels of mRNAs were evaluated by Cufflinks v2.2.152 with “-p x -u -G”. Translation efficiency of each mRNA was calculated by the ratio of RPFs and the mean expression of input mRNA replicates.
RIP-seq data processing and analysis
Preprocessing and peak calling
The quality of raw sequencing reads was evaluated using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/), and low-quality bases were trimmed and filtered by cutadapt (V 1.13)43 and Trimmomatic (V 0.36).44 Processed reads were mapped to the zebrafish genome (Zv9) using TopHat (v2.1.1)51 with “–bowtie1” (Supplementary information, Table S7). The binding read proportions of Ddx3xb were normalized by the length of 5’UTR, CDS region and 3’UTR. The target binding regions of Ddx3xb were identified using the MACS2 software (v2.0.10)53 with options ‘–nomodel’ –keep-dup and –gsize = 1.4e9, and using RNA-seq data as input. A stringent cutoff threshold with a P value of 0.01 was used to obtain target genes of Ddx3xb. Target genes were annotated based on Ensembl (release 79) gene annotation information, using BEDTools’ intersectBed (v2.16.2).54
Binding motif identification
Sites annotated within 5ʹ UTR regions were used to identify motif regions. Enriched motif regions within Ddx3xb-binding sites were identified by HOMER (v4.7).55 Motif length was restricted to 7 nucleotides.
Quantitation and statistical analysis
Data in Figs. 1d, e, g, h, j, l; 2a, c, f, h; 3b, d, f; 5e, g; 6b, c, e, g; Supplementary information, Figs. S1a, c; S2b, d; S3a, c, e, g, i; S4c; S5d; S6b, c, e; S7b; S9b, c; S11e, g, i, j, l, n, p; S13a, c were analyzed using Prism (GraphPad). Statistical significance was determined by the two-sided Student’s t-test, as indicated in figure legends. P value of less than 0.05 was considered significant. *P < 0.05, **P < 0.01, and ***P < 0.001. Information about sample sizes and biologically independent replicates are included in figure legends. Data in Figs. 4a–c, f–h; 5c, h–j; Supplementary information, Figs. S8c–e, g–i; S10c, e; S12e were analyzed using Python (3.6.4). Statistical significance was determined by Kruskal Wallis test with the scipy package, as indicated in figure legends. Data in Fig. 5b were analyzed using HOMER. Statistical significance was determined by binomial test, as indicated in figure legends.
Supplementary information
Author contributions
Y.-G.Y. and F.L. conceived this project, supervised the study; B.S., J.H. and Y.Y. performed the experiments; J.-Y.Z., B.S. and W.-Y.Z. performed bioinformatics analysis; Y.-G.Y., F.L., M.J.K., P.L., Y.-L.Z., Y.Y., J.-Y.Z., J.H. and B.S. discussed and integrated the data, wrote the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by grants from the National Natural Science Foundation of China (32030058, 32121001, 31625016, 32030032), the Strategic Priority Research Program of the Chinese Academy of Sciences, China (XDA16010207, XDA16010501, XDPB2004), the National Key R&D Program of China (2018YFA0800200, 2019YFA0110901), the Youth Innovation Promotion Association of Chinese Academy of Sciences (2018133), Shanghai Municipal Science and Technology Major Project (2017SHZDZX01), and Research Unit of Medical Neurobiology, Chinese Academy of Medical Sciences (2019RU003).
Data availability
The RNA-seq, RIP, and ribosome profiling data supporting the conclusions of this article have been deposited in the Gene Expression Omnibus database (GEO: GSE169169), and also the Genome Sequence Archive (GSA: CRA003999 linked to the BioProject with accession Number PRJCA004602).
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Boyang Shi, Jian Heng, Jia-Yi Zhou, Ying Yang.
Contributor Information
Feng Liu, Email: liuf@ioz.ac.cn.
Yun-Gui Yang, Email: ygyang@big.ac.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41422-022-00655-5.
References
- 1.Abrams EW, Mullins MC. Early zebrafish development: it’s in the maternal genes. Curr. Opin. Genet. Dev. 2009;19:396–403. doi: 10.1016/j.gde.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vastenhouw NL, Cao WX, Lipshitz HD. The maternal-to-zygotic transition revisited. Development. 2019;146:11. doi: 10.1242/dev.161471. [DOI] [PubMed] [Google Scholar]
- 3.Lee MT, Bonneau AR, Giraldez AJ. Zygotic genome activation during the maternal-to-zygotic transition. Annu. Rev. Cell Dev. Biol. 2014;30:581–613. doi: 10.1146/annurev-cellbio-100913-013027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee MT, et al. Nanog, Pou5f1 and SoxB1 activate zygotic gene expression during the maternal-to-zygotic transition. Nature. 2013;503:360–364. doi: 10.1038/nature12632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun J, Yan L, Shen W, Meng A. Maternal Ybx1 safeguards zebrafish oocyte maturation and maternal-to-zygotic transition by repressing global translation. Development. 2018;145:19. doi: 10.1242/dev.166587. [DOI] [PubMed] [Google Scholar]
- 6.Yan L, et al. Maternal Huluwa dictates the embryonic body axis through β-catenin in vertebrates. Science. 2018;362:6417. doi: 10.1126/science.aat1045. [DOI] [PubMed] [Google Scholar]
- 7.Yang Y, et al. RNA 5-methylcytosine facilitates the maternal-to-zygotic transition by preventing maternal mRNA decay. Mol. Cell. 2019;75:1188–1202. doi: 10.1016/j.molcel.2019.06.033. [DOI] [PubMed] [Google Scholar]
- 8.Shi B, et al. RNA structural dynamics regulate early embryogenesis through controlling transcriptome fate and function. Genome Biol. 2020;21:120. doi: 10.1186/s13059-020-02022-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giraldez AJ, et al. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- 10.Mishima Y, Tomari Y. Codon usage and 3’ UTR length determine maternal mRNA stability in Zebrafish. Mol. Cell. 2016;61:874–885. doi: 10.1016/j.molcel.2016.02.027. [DOI] [PubMed] [Google Scholar]
- 11.Zhao BS, et al. m6A-dependent maternal mRNA clearance facilitates zebrafish maternal-to-zygotic transition. Nature. 2017;542:475–478. doi: 10.1038/nature21355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang H, et al. Terminal uridylyltransferases execute programmed clearance of maternal transcriptome in vertebrate embryos. Mol. Cell. 2018;70:72–82.e7. doi: 10.1016/j.molcel.2018.03.004. [DOI] [PubMed] [Google Scholar]
- 13.Subtelny AO, Eichhorn SW, Chen GR, Sive H, Bartel DP. Poly(A)-tail profiling reveals an embryonic switch in translational control. Nature. 2014;508:66–71. doi: 10.1038/nature13007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Banani SF, Lee HO, Hyman AA, Rosen MK. Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 2017;18:285–298. doi: 10.1038/nrm.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roden C, Gladfelter AS. RNA contributions to the form and function of biomolecular condensates. Nat. Rev. Mol. Cell Biol. 2021;22:183–195. doi: 10.1038/s41580-020-0264-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li CH, et al. MeCP2 links heterochromatin condensates and neurodevelopmental disease. Nature. 2020;586:440–444. doi: 10.1038/s41586-020-2574-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brangwynne CP, et al. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 2009;324:1729–1732. doi: 10.1126/science.1172046. [DOI] [PubMed] [Google Scholar]
- 18.Zeng M, et al. Phase transition in postsynaptic densities underlies formation of synaptic complexes and synaptic plasticity. Cell. 2016;166:1163–1175.e12. doi: 10.1016/j.cell.2016.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li P, et al. Phase transitions in the assembly of multivalent signalling proteins. Nature. 2012;483:336–340. doi: 10.1038/nature10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang G, Wang Z, Du Z, Zhang H. mTOR regulates phase separation of PGL granules to modulate their autophagic degradation. Cell. 2018;174:1492–1506.e22. doi: 10.1016/j.cell.2018.08.006. [DOI] [PubMed] [Google Scholar]
- 21.Linder P, Jankowsky E. From unwinding to clamping - the DEAD box RNA helicase family. Rev. Mol. Cell Biol. 2011;12:505–516. doi: 10.1038/nrm3154. [DOI] [PubMed] [Google Scholar]
- 22.Alberti S, Gladfelter A, Mittag T. Considerations and challenges in studying liquid-liquid phase separation and biomolecular condensates. Cell. 2019;176:419–434. doi: 10.1016/j.cell.2018.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hondele M, et al. DEAD-box ATPases are global regulators of phase-separated organelles. Nature. 2019;573:144–148. doi: 10.1038/s41586-019-1502-y. [DOI] [PubMed] [Google Scholar]
- 24.Nott TJ, et al. Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol. Cell. 2015;57:936–947. doi: 10.1016/j.molcel.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oh S, et al. Medulloblastoma-associated DDX3 variant selectively alters the translational response to stress. Oncotarget. 2016;7:28169–28182. doi: 10.18632/oncotarget.8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guenther UP, et al. The helicase Ded1p controls use of near-cognate translation initiation codons in 5’ UTRs. Nature. 2018;559:130–134. doi: 10.1038/s41586-018-0258-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iserman C, et al. Condensation of Ded1p promotes a translational switch from housekeeping to stress protein production. Cell. 2020;181:818–831.e19. doi: 10.1016/j.cell.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Campbell K, Swann K. Ca2+ oscillations stimulate an ATP increase during fertilization of mouse eggs. Dev. Biol. 2006;298:225–233. doi: 10.1016/j.ydbio.2006.06.032. [DOI] [PubMed] [Google Scholar]
- 29.Dutta A, Sinha DK. Zebrafish lipid droplets regulate embryonic ATP homeostasis to power early development. Open Biol. 2017;7:170063. doi: 10.1098/rsob.170063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murray DT, et al. Structure of FUS protein fibrils and its relevance to self-assembly and phase separation of low-complexity domains. Cell. 2017;171:615–627.e16. doi: 10.1016/j.cell.2017.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Molliex A, et al. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell. 2015;163:123–133. doi: 10.1016/j.cell.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu M, et al. Interferon-γ induces tumor resistance to anti-PD-1 immunotherapy by promoting YAP phase separation. Mol. Cell. 2021;81:1216–1230.e9. doi: 10.1016/j.molcel.2021.01.010. [DOI] [PubMed] [Google Scholar]
- 33.Eckersley-Maslin MA, Alda-Catalinas C, Reik W. Dynamics of the epigenetic landscape during the maternal-to-zygotic transition. Nat. Rev. Mol. Cell Biol. 2018;19:436–450. doi: 10.1038/s41580-018-0008-z. [DOI] [PubMed] [Google Scholar]
- 34.Elbaum-Garfinkle S, et al. The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc. Natl. Acad. Sci. USA. 2015;112:7189–7194. doi: 10.1073/pnas.1504822112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ji S, et al. LC domain-mediated coalescence is essential for otu enzymatic activity to extend drosophila lifespan. Mol. Cell. 2019;74:363–377.e5. doi: 10.1016/j.molcel.2019.02.004. [DOI] [PubMed] [Google Scholar]
- 36.Chang N, et al. Genome editing with RNA-guided Cas9 nuclease in zebrafish embryos. Cell Res. 2013;23:465–472. doi: 10.1038/cr.2013.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Renaud O, Herbomel P, Kissa K. Studying cell behavior in whole zebrafish embryos by confocal live imaging: application to hematopoietic stem cells. Nat. Protoc. 2011;6:1897–1904. doi: 10.1038/nprot.2011.408. [DOI] [PubMed] [Google Scholar]
- 38.Heng J, et al. Rab5c-mediated endocytic trafficking regulates hematopoietic stem and progenitor cell development via Notch and AKT signaling. PLoS Biol. 2020;18:e3000696. doi: 10.1371/journal.pbio.3000696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murphey RD, Stern HM, Straub CT, Zon LI. A chemical genetic screen for cell cycle inhibitors in zebrafish embryos. Chem. Biol. Drug Des. 2006;68:213–219. doi: 10.1111/j.1747-0285.2006.00439.x. [DOI] [PubMed] [Google Scholar]
- 40.Zhang C, et al. m(6)A modulates haematopoietic stem and progenitor cell specification. Nature. 2017;549:273–276. doi: 10.1038/nature23883. [DOI] [PubMed] [Google Scholar]
- 41.Bol GM, et al. Targeting DDX3 with a small molecule inhibitor for lung cancer therapy. EMBO Mol. Med. 2015;7:648–669. doi: 10.15252/emmm.201404368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Calviello L, et al. Detecting actively translated open reading frames in ribosome profiling data. Nat. Methods. 2016;13:165–170. doi: 10.1038/nmeth.3688. [DOI] [PubMed] [Google Scholar]
- 43.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. journal. 2011;17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 44.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 50.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Duttke SH, Chang MW, Heinz S, Benner C. Identification and dynamic quantification of regulatory elements using total RNA. Genome Res. 2019;29:1836–1846. doi: 10.1101/gr.253492.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq, RIP, and ribosome profiling data supporting the conclusions of this article have been deposited in the Gene Expression Omnibus database (GEO: GSE169169), and also the Genome Sequence Archive (GSA: CRA003999 linked to the BioProject with accession Number PRJCA004602).