Abstract
Context
Children with cancer undergoing chemotherapy experience a cluster of psychoneurological symptoms (PNS), including pain, fatigue, anxiety, and depressive symptoms. Metabolomics is promising to differentiate metabolic pathways associated with the PNS cluster.
Objectives
Identify metabolic pathways associated with the PNS cluster in children with cancer before and after chemotherapy.
Methods
Pain, fatigue, anxiety, and depressive symptoms were assessed using the Pediatric PROMIS scales. T-scores were computed and divided dichotomously by a cutoff point of 50; the PNS cluster was a sum of the four symptoms ranging from 0 (all T-scores <50) to 4 (all T-scores ≥50). Serum metabolites were processed using liquid chromatography mass-spectrometry untargeted metabolomics approach. Linear regression models examined metabolites associated with the PNS cluster. Metabolic pathway enrichment analysis was performed.
Results
Participant demographics (n = 40) were 55% female, 60% white, 62.5% aged 13–19 years, and 62.5% diagnoses of Hodgkin’s lymphoma and B-cell acute lymphocytic leukemia. Among 9276 unique metabolic features, 454 were associated with pain, 281 with fatigue, 596 with anxiety, 551 with depressive symptoms, and 300 with the PNS cluster across one chemotherapy cycle. Fatty acids pathways were associated with pain: de novo fatty acid biosynthesis (p < .001), fatty acid metabolism (p = .001), fatty acid activation (p = .004), and omega-3 fatty acid metabolism (p = .009). Tryptophan amino acid pathway was associated with fatigue (p < .001), anxiety (p = .015), and the PNS cluster (p = .037). Carnitine shuttle was associated with the PNS cluster (p = .015).
Conclusion
Fatty acids and amino acids pathways were associated with PNS in children undergoing chemotherapy. These findings require further investigation in a larger sample.
Keywords: cancer, chemotherapy, children, metabolomics, metabolic pathway, psychoneurological symptoms, symptom cluster
Approximately 16,000 children and adolescents are diagnosed with cancer in the United States each year (Siegel et al., 2021). Children with cancer undergoing chemotherapy report multiple psychoneurological symptoms (PNS), including pain, fatigue, anxiety, and depressive symptoms (Hooke & Linder, 2019; Linder & Hooke, 2019; Rodgers et al., 2016). These symptoms commonly co-occur as a cluster (hereafter named “the PNS cluster”), potentially due to common etiologies and biological mechanisms (Miaskowski et al., 2017). Recently, two levels of the PNS cluster (mild symptoms vs. severe symptoms) were identified in children throughout a cycle of chemotherapy (Wang et al., 2018). Inadequate treatment and management of PNS and the PNS cluster can potentially delay cancer treatment, decrease tumor response, and interfere with a child’s ability to engage in daily activities, all of which lead to lower quality of life among children with cancer (Kestler & LoBiondo-Wood, 2012; Linder & Hooke, 2019).
Due to treatment advances in recent decades, the 5-year survival rate has increased from 58% in the mid-1970s to 84% across all cancer diagnoses in children (Siegel et al., 2021). Sadly, the management of PNS across cancer treatment has not kept pace with this advance in survival (Kwekkeboom, 2016; Miaskowski et al., 2017). Literature has suggested common biological pathways underlying the development and severity of PNS in cancer, such as proinflammatory cytokines, metabolites, and neurotransmitters (Kennedy et al., 2016; Kim et al., 2012; Lyon et al., 2018). The underlying biological mechanisms of PNS are still not well investigated in children with cancer. With recent emphasis on precision medicine, understanding the biological mechanisms of PNS and the PNS cluster can advance the development of targeted therapies for children who are more prone to the development of a severe level of PNS.
Metabolomics is a systems biology approach that can be used to explore genetic and environmental influences on children’s health and disease (Baldassarre & Laforgia, 2020; Moco et al., 2013). Metabolic profiles have been assisting with characterizing markers of environmental exposures (Athersuch & Keun, 2015) and understanding health care outcomes from pre-term birth (Wilson et al., 2014) to adulthood (James et al., 2004). A variety of factors, such as age (Chiu et al., 2016; Yu et al., 2012), sex (Kochhar et al., 2006), body mass index (Jourdan et al., 2012), and diet (Holmes et al., 2008), play critical roles in determining the metabolome, including the metabolome in children (Lau et al., 2018). From a systems-level perspective, the metabolomics approach (e.g., targeted and untargeted metabolomics) has been used to identify biological mechanisms underlying cancer occurrence, therapy responses, and treatment-related toxicities (e.g., psychoneurological toxicities) (Lyon et al., 2018; Schmidt et al., 2021). However, the use of a metabolomics approach is still underexplored in children with cancer (Spiga et al., 2013).
Metabolites play functional roles in communicating with other host factors, such as the gut microbiome, to impact PNS, namely, the microbiome-gut-brain axis (MGB). The MGB proposes a bidirectional communication network between the gut and the brain to determine the level of PNS (Mayer et al., 2014; Song & Bai, 2020). Specifically, chemotherapy-induced disruptions of the gut microbiome can potentially regulate corresponding metabolic pathways in the MGB (Kennedy et al., 2016), such as short-chain fatty acids (SCFAs; van de Wouw et al., 2018), bile acids (Monteiro-Cardoso et al., 2021), and tryptophan for kynurenine pathway metabolism (Kennedy et al., 2016). These activated metabolic molecules then carry related biological signals to specific brain regions (e.g., amygdala; Cowan et al., 2017; Kennedy et al., 2016), leading to behavioral and symptomatic changes.
Specifically, SCFAs, including butyrate, acetate, and propionate, are main metabolites produced in the colon by bacterial fermentation of dietary fibers and resistant starch (Pascale et al., 2018). Alterations in SCFAs might underpin disturbances in the central nervous system, ranging from neurodevelopmental disorders (e.g., mood disorders) to neurodegenerative diseases (e.g., Alzheimer’s Disease; Silva et al., 2020). These fatty acids could influence PNS through interactions with G protein-coupled receptors or histone deacetylases, direct humoral effects, indirect hormonal and immune pathways, and neural routes (Cryan et al., 2019; Dalile et al., 2019). The regulation of tryptophan is another major pathway of PNS, with dual emphasis on the regulation of serotonin synthesis and the control of kynurenine pathway metabolism (Agus et al., 2018; Kennedy et al., 2016; Strasser et al., 2016). Tryptophan, an essential amino acid, must be obtained from dietary or microbial sources (Kennedy et al., 2015, 2016; Yanofsky, 2007). Tryptophan metabolism via kynurenine pathway is associated with regulations of neuronal function and intestinal homeostasis (Platten et al., 2019); it can influence and modulate a variety of psychiatric disorders (Berger et al., 2009), such as depression and cognitive dysfunction (Jenkins et al., 2016; Muller & Homberg, 2015; O’Mahony et al., 2015). Among patients with cancer, changes of metabolic profiles have been reported in monitoring responses towards chemotherapeutic agents (Amin et al., 2019; Debik et al., 2019; Schmidt et al., 2021) and understanding chemotherapy-related psychoneurological toxicities (Li et al., 2020; Lyon et al., 2018). Current work primarily focuses on adult cancers (e.g., breast and colorectal cancers). Exploration of metabolite contribution to chemotherapy-related psychoneurological toxicities in children with cancer is critically needed with respect to the PNS burden in pediatric oncology.
Metabolic pathways associated with PNS have been investigated in various adult cancers across cancer treatment spectrum and even during survivorship. Untargeted metabolomics analysis showed that increased pain was associated with decreased essential tryptophan and N-tryptophan, and that increased fatigue was associated with decreases in arachidonic acid, a vital fatty acid in metabolic signaling (Li et al., 2020; Lyon et al., 2018); targeted metabolomics analysis showed moderate-to-strong correlations between changes in pain and tryptophan levels and kynurenine/tryptophan ratio, as well as between changes in depressive symptoms and serotonin among women with breast cancer pre- and post-chemotherapy (Li et al., 2020; Lyon et al., 2018). Furthermore, decreased tryptophan and increased kynurenine and tryptophan/kynurenine ratio were correlated with increased PNS among adult cancer survivors (Li et al., 2020). Metabolites may provide precise targets to treat and manage PNS during cancer treatment, which has not been examined among children with cancer. The purposes of this study were to: (1) compare PNS and the PNS cluster pre-(T1) and post-(T2) one cycle of chemotherapy and (2) identify metabolic pathways associated with PNS and the PNS cluster among children with cancer receiving chemotherapy.
Materials and Methods
Study Design
This pilot sub-study was conducted in conjunction with a larger parent study to extend validation for the Patient-Reported Outcomes Measurement Information System (PROMIS) Pediatric measures while also validating a new measurement system the Pediatric Patient Reported Outcome-Common Terminology Criteria for Adverse Events (Reeve et al., 2020). As such, subjective treatment-related symptoms data were collected from children and adolescents with cancer at T1 and T2. IRB approval was obtained from Emory University prior to participant enrollment and data collection.
Setting and Sample
Forty children with cancer were enrolled in this sub-study from the AFLAC Cancer and Blood Disorder Center in Children’s Healthcare of Atlanta (CHOA) in Atlanta, Georgia. Inclusion criteria of these children were as follows: (1) 7–19 years old; (2) diagnosed with cancer; (3) undergoing chemotherapy; (4) able to read and write English for completion of self-report questionnaires; and (5) willing to provide consent/assent for study participation. Exclusion criteria included any neurological disorder or syndrome interfering with self-report.
Measures
Psychoneurological Symptoms
Computerized-adaptive testing was utilized to collect symptom reports from children and adolescents using the PROMIS measures developed by the National Institutes of Health. Pediatric PROMIS measures captured self-reports of fatigue, depressive symptoms, anxiety, and pain interference within the past 7 days recall period (Hinds et al., 2019). Higher scores on these measures indicate higher symptom burden. Pediatric PROMIS measures generate a continuous T-score (mean of 50 and standard deviation [SD] of 10) based on a reference sample (i.e., a diverse set of healthy children and children with a variety of common chronic illnesses; Irwin et al., 2010). Additional symptom measures were collected as part of the larger parent study, but not utilized in this sub-study.
Blood Draw
After obtaining consent/assent, and when other clinical labs were drawn, an extra 10 cc of non-fasting blood was collected at t two time points (T1 and T2) and placed in a red top (anticoagulant-free) tube. Specimens were placed on cold packs (∼35° environment) within 30 minutes of collection and transported to the lab for processing. Most specimens were processed and stored within 1 hour (but no more than 4 hours) from collection. In the lab, samples were centrifuged at 3000 rpm for 10 minutes in bench-top centrifuge at room temperature. Two hundred uL aliquots of serum supernatant were pipetted into clean polypropylene tubes and stored at −80°C. Sixty-five uL from one tube was assayed, with the remaining aliquots stored as back-up in the event quality control procedures indicated a need for repeated analysis.
Demographic and Clinical Variables
Demographic (e.g., sex, age, and race) and clinical variables (e.g., cancer diagnosis, chemotherapy cycles, and use of steroids) were obtained from the electronic medical record. In this study, children were enrolled at different cycles of chemotherapy, and we did not control the cycle of chemotherapy due to different types of cancer diagnoses. All children were enrolled fairly early in their treatment plan and at a time-point correlating with intense therapy (i.e., around a cycle of delayed intensification for children with acute lymphocytic leukemia). Use of steroids was collected because of its prevalence as part of children’s cancer treatment and its associations with significant complications on host metabolism and neurobehavioral toxicities (McNeer & Nachman, 2010; Mrakotsky et al., 2011).
Data Collection Procedure
All participants were recruited during their regularly scheduled clinical visits. Clinical collaborators identified eligible patients prior to their visits. The research staff obtained parents’ consent (or children’s consent if aged 18 years) in addition to assent (for those <18 years). After obtaining consent/assent, blood samples were collected at visits immediately before and 7–17 days after a cycle of chemotherapy. During the same clinical appointment as the blood draws, participants completed the PROMIS measures on a computer tablet. A study team member was allowed to read the questions, as needed, to younger children.
High-Resolution Mass Spectrometry and Data Preprocessing
After thawing, serum samples stored at −80°C were processed and analyzed based on a high-resolution, untargeted metabolomics platform described in previous literature (Chandler et al., 2016; Hoffman et al., 2014; Neujahr et al., 2014). Reference standardization was used according to concurrent analysis of pooled reference samples at predefined intervals and enables batch correction (Liu et al., 2020). For each batch of 40 samples, reference standards were used at the beginning, middle, and end. All the samples were randomized to minimize batch effects and ComBat (Johnson et al., 2006) was used to correct batch effects before further analysis.
Each sample was run with three technical replicates. Among each sample, 10 μl aliquots were injected and analyzed by liquid chromatography with Fourier transform mass spectrometry (Dionex Ultimate 3000, Q-Exactive HF, Thermo Fisher) with HILIC chromatography/positive electrospray ionization (ESI) mode and resolution of 120,000 (Soltow et al., 2013). We used the standard solution formulation through Vendors of Cambridge and Sigma. A quality control pooled reference plasma sample was included at the beginning, middle, and end of each analytical batch of 40 samples for normalization and post-hoc quantification (Go et al., 2015). Peak extraction and ion intensity quantification were performed by validated detection methods apLCMS (Yu et al., 2013) and xMSanalyzer (Uppal et al., 2013). These two methods were involved in noise filtering, feature identification, retention time (RT) correction, mass-to-charge ratio (m/z) feature alignment, and reanalysis to capture features missed due to weak signal due to the signal to noise filter (Uppal et al., 2013). The resulting data contained ions defined by m/z and RT. Triplicate injections were averaged and filtered for metabolic features less than 80% non-missing values across all samples and 80% non-missing values in at least one group (e.g., the group with pain T-score ≥50), log2 transformed, and quantile normalized. Samples were processed at the Clinical Biomarkers Laboratory at Emory University.
Metabolic Pathway Analysis and Annotation
Metabolic pathway analysis was performed for significant metabolic features (m/z) associated with PNS and the PNS cluster using the default inputs of Mummichog version 2.1.1 (Li et al., 2013). This metabolic pathway analysis provides a 2-step approach to protect against type I and type II errors in metabolomics analysis (Uppal et al., 2016). Mummichog can identify both metabolic features in conventional pathways or single-metabolite analyses and significant metabolic patterns associated with clustered metabolic reactions. In this study, we examined distinct metabolic profiles or networks associated with PNS and the PNS cluster using a raw p value of .05. Particularly, metabolic pathway enrichment analyses were conducted using significant metabolites from univariate analysis and then again with metabolites that were significant after adjustment for demographic (e.g., sex, age, and race) and clinical (e.g., steroid use) variables. Only metabolic pathways with a minimum overlap size of 3 (number of metabolites with significant discrimination between study groups within a particular metabolic pathway) were contained for analyses. Plots of intensities for selected metabolites in significant pathways were provided to compare groups of PNS and the PNS cluster.
Metabolites were annotated by matching the accurate mass m/z for adducts commonly formed under positive ESI conditions the METLIN (Guijas et al., 2018) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases (Kanehisa et al., 2015) based on a mass error threshold of 10 ppm (relative m/z error × 106). This study reported the high confidence matches metabolites after metabolic annotations.
Statistical Analysis
Descriptive statistics, including mean (SD), median (range), and frequency (%), were performed for demographic, clinical, and PNS variables. The Pediatric PROMIS measures data were computed to calculate T-score metric with mean of 50 (SD = 10). As these symptoms often occur together, a cluster variable was also created by summing the number of severe symptoms: “yes” for reporting T-score ≥50 for any one symptom, otherwise, “no.” The cluster of PNS ranged from 0 (none of the PNS with a T-score ≥50) to 4 (all 4 PNS with a T-score ≥50). This method was used to characterize the symptoms cluster (Bai et al., 2020) due to our small sample size. Paired samples T-test was used to compare PNS between T1 and T2. Mann–Whitney U-test was used to compare PNS according to age (7–12 years vs. 13–19 years), sex (male vs. female), race (White vs. Black), and steroid use (yes vs. no). Pearson’s correlation (r) was conducted for the relationships among PNS at T1 and T2, as well as between T1 and T2. These analyses were conducted by SPSS version 23 (IBM Corp, Armonk, N.Y., USA).
After the relative intensities of the metabolites were log2 transformed and quantile normalized, we used multivariable linear regression to assess metabolites associated with the four individual PNS, and the 0–4 score of the PNS cluster variable using xmsPANDA, with a p value <.05. Age, sex, race, and steroid use were controlled in linear regression models for both timepoints (T1 and T2). The Benjamini–Hochberg FDR method (Hochberg & Benjamini, 1990) was used for multiple-hypothesis correction to identify individual metabolites based on a q value of .20. As reported in previous literature (Carlson et al., 2020), the FDR correction protects against type I error but it may exclude true positive metabolites (type II error). Thus, metabolic pathways associated with PNS and the PNS cluster were determined with a raw p value of .05 (Chandler et al., 2016). These analyses were implemented by R 4.1.0.
Results
Characteristics of Participants
Participant (n = 40) demographics included the following: female (n = 22, 55%) and male (n = 18, 45%); 15 (37.5%) children aged 7–12 years and 25 (62.5%) adolescents aged 13–19 years; 24 (60%) White, 14 (35%) Black, and 2 (5%) others. Types of cancer diagnoses included the following: Hodgkin’s lymphoma (n = 13, 32.5%), B-cell acute lymphocytic leukemia (n = 12, 30%), T-cell lymphoblastic lymphoma (n = 5, 12.5%), osteosarcoma (n = 4, 10%), and others (n = 6, 15%). Fifteen (37.5%) children received steroids during treatment.
Psychoneurological Symptoms
Table 1 presents PNS and the comparisons of PNS between T1 and T2. Mean T-scores across the 4 PNS ranged from 41.8 (SD = 9.2) to 46.0 (SD = 2.0) at T1 and from 41.5 (SD = 9.0) to 43.9 (SD = 10.5) at T2. These mean PNS scores were below the average score (T-score = 50) for the PROMIS measures’ reference population. From the highest to lowest average T scores (T1 and T2), children experienced depressive symptoms, fatigue, anxiety, and pain. Paired-samples T test showed no significant differences for any of the PNS between T1 and T2 (all p > .05). At T1 (before a cycle of chemotherapy), children reported the PNS cluster with greater frequency than at T2 (p < .0001). Mann–Whitney U test showed that children aged 7–12 years reported lower fatigue than those aged 13–19 years in T1 (p = .032) and T2 (p = .028); children receiving steroids had more pain than those not receiving steroids (p = 027) at T2. Gender and race were not associated with the PNS scores. Age, gender, race, and steroid use were not associated with the PNS cluster (all p > .05).
Table 1.
Comparisons of Psychoneurological Symptoms Pre-(T1) and Post-(T2) Chemotherapy Cycle.
| Variable | T1 (n = 40) | T2 (n = 39) d | p-value |
|---|---|---|---|
| Pain (mean [SD]) | 41.8 (9.2) | 41.5 (9.0) | .80 a |
| Fatigue (mean [SD]) | 44.9 (13.8) | 43.7 (14.3) | .53 a |
| Anxiety (mean [SD]) | 43.9 (11.0) | 42.4 (11.8) | .26 a |
| Depressive symptoms (mean [SD]) | 46.0 (12.0) | 43.9 (10.5) | .11 a |
| Pain group (T score ≥50 vs.<50), n (%) | 6 (15) versus 34 (85) | 8 (20.5) versus 31 (79.5) | .63 b |
| Fatigue group (T score ≥50 vs.<50), n (%) | 14 (35) versus 26 (65) | 15 (38.5) versus 24 (61.5) | .99 b |
| Anxiety group (T score ≥50 vs. <50), n (%) | 11 (27.5) versus 29 (72.5) | 8 (20.5) versus 31 (79.5) | .45 b |
| Depressive symptoms group (T score ≥50 vs. <50), n (%) | 13 (32.5) versus 27 (67.5) | 12 (30.8) versus 27 (69.2) | .99 b |
| PNS cluster across groups, n (%) | <0.0001 c | ||
| 0 | 23 (29.1) | 18 (46.2) | |
| 1 | 2 (2.5) | 9 (23.1) | |
| 2 | 8 (10.1) | 5 (12.8) | |
| 3 | 4 (5.1) | 4 (10.3) | |
| 4 | 3 (3.8) | 3 (7.7) |
Note. T1, pre-chemotherapy; T2, post-chemotherapy. PNS, psychoneurological symptoms; SD, standard deviation.
aPaired t-test.
bMcNemar test.
cChi-Square test.
d One patient with missing data of PNS post-chemotherapy, thus, our analyses were conducted for 39 patients.
Table 2 presents the correlations of PNS with each other and between T1 and T2. All the 4 PNS were positively and significantly correlated with each other at T1 (r ranged from 0.36 to 0.88, all p < .05), T2 (r ranged from 0.35 to 0.77, all p < .05), as well as between T1 and T2 (r ranged from 0.35 to 0.75, all p < .05) except that depressive symptoms at T1 showed a marginal correlation with pain at T2 (r = 0.30, p = .06).
Table 2.
Correlations between Psychoneurological Symptoms Pre-(T1) and Post-(T2) Chemotherapy Cycle.
| Variable | PainT1 | FatigueT1 | AnxietyT1 | Depressive symptomsT1 | PainT2 | FatigueT2 | AnxietyT2 | Depressive symptomsT2 |
|---|---|---|---|---|---|---|---|---|
| PainT1 | 1.0 | |||||||
| FatigueT1 | .54*** | 1.0 | ||||||
| AnxietyT1 | .42** | .58*** | 1.0 | |||||
| Depressive symptomsT1 | .36* | .69*** | .88*** | 1.0 | ||||
| PainT2 | .65* | .62*** | .41** | .30 | 1.0 | |||
| FatigueT2 | .35* | .68*** | .48** | .51*** | .63*** | 1.0 | ||
| AnxietyT2 | .42** | .53*** | .75*** | .62*** | .48** | .68*** | 1.0 | |
| Depressive symptomsT2 | .37* | .64*** | .70*** | .75*** | .35* | .67*** | .77*** | 1.0 |
Note. T1, pre-chemotherapy; T2, post-chemotherapy.
*p < .05, ** p < .01, *** p < .001.
Metabolic Pathways of PNS and the PNS Cluster
A total of 9276 unique metabolomic features were identified after filtering, including 127 laboratory confirmed metabolites, 801 metabolic features with high confidence annotations, 1581 medium confidence annotations, 3099 low confidence annotations, and 3668 unknown metabolic features. Table 3 shows the number of significant metabolic features associated with PNS and the PNS cluster at T1, T2, and from T1 to T2. Anxiety (n = 596) and depressive symptoms (n = 551) showed the largest number of associated metabolic features compared with pain (n = 454) and fatigue (n = 281). These identified metabolites in Table 3 were used for pathway enrichment analysis.
Table 3.
Number of Metabolic Features Associated with PNS and the PNS Cluster Pre-(T1), Post-(T2) Chemotherapy Cycle, and from T1 to T2.
| Variable | T1 | T2 | T1 to T2 |
|---|---|---|---|
| Pain | 330 | 409 | 454 |
| Fatigue | 410 | 353 | 281 |
| Anxiety | 575 | 641 | 596 |
| Depressive symptoms | 916 | 673 | 551 |
| PNS cluster | 473 | 436 | 300 |
Note. T1, pre-chemotherapy; T2, post-chemotherapy; PNS, psychoneurological symptoms.
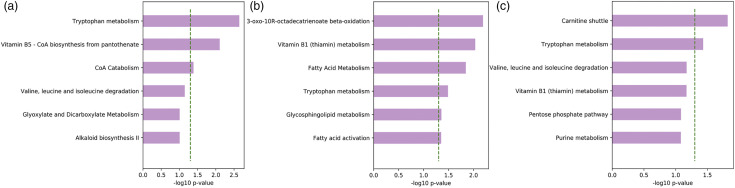
Supplemetal Tables S1 and S2 (shown in the supplementary material) demonstrate the metabolic pathways associated with PNS and the PNS cluster at T1 and T2, respectively. Metabolomic pathway enrichment analysis using Mummichog showed that fatty acid metabolic pathways were associated with pain, including de novo fatty acid biosynthesis (p = .013), fatty acid metabolism (p = .005), and fatty acid activation (p = .005), and depressive symptoms, such as octadecatrienoate beta-oxidation (p = .015) at T1. At T1, amino acid metabolic pathways, particularly tryptophan, were associated with multiple symptoms: tryptophan with anxiety (p = .034), depressive symptoms (p = .044), and the PNS cluster (p = .002). Figure 1A describes metabolic pathways significantly differentiating different levels of the PNS cluster at T1.
Figure 1.
Metabolic Pathways Associated with psychoneurological symptoms (PNS). (a) Pre-chemotherapy; (b) Post-chemotherapy; and (c) from pre-chemotherapy to post-chemotherpay. Dotted lines indicate p < .05
Among children post-chemotherapy, Supplemental Table S2 shows that vitamin pathways and fatty acid metabolism were primarily associated with multiple symptoms: vitamin B3 was associated with pain (p = .049), anxiety (p = .038), and depressive symptoms (p = .033); vitamin B6 was associated with fatigue (p = .005). Tryptophan was associated with fatigue (p = .012) and anxiety (p = .013). The PNS cluster was associated with fatty acid metabolism (p = .014), fatty acid activation (p = .006), glycosphingolipid metabolism (p = .044), 3-oxo-10R-octadecatrienoate beta-oxidation (p = .016), and tryptophan (p = .033). Figure 1B describes metabolic pathways significantly differentiating different levels of the PNS cluster at T2.
Repeated measures linear regression was used to analyze the longitudinal associations between metabolic pathways and individual PNS or the PNS cluster by considering both between-subjects and within-subjects effects. Table 4 shows the metabolic enrichment pathways and examples of significant metabolites identified in each pathway associated with PNS and the PNS cluster based on our data from T1 to T2. We found that fatty acid pathways were associated with pain: de novo fatty acid biosynthesis (p < .001), fatty acid metabolism (p = .001), and fatty acid activation (p = .004), omega-3 fatty acid metabolism (p = .009), as well as bile acid biosynthesis (p = .005). Tryptophan pathway was associated with fatigue (p = .004) and anxiety (p = .015). The PNS cluster was associated with carnitine shuttle (p = .015) and tryptophan (p = .037). Figure 1C describes metabolic pathways significantly differentiating different levels of the PNS cluster from T1 to T2.
Table 4.
Metabolites Associated with Psychoneurological Symptoms from Pre-(T1) and Post-(T2) Chemotherapy Cycle.
| Variable | Metabolic pathways | Overlap size | Pathway size | p-value | Metabolites |
|---|---|---|---|---|---|
| Pain | de novo fatty acid biosynthesis | 8 | 35 | <.001 | Hexadecanoate (n-C16:0); Linoelaidic acid (all trans C18:2); Octadecadienoate (n-C18:2); (9Z,12Z,15Z)-Octadecatrienoic acid; alpha-Linolenic acid; 9,12,15-Octadecatrienoic acid; Linolenate; (6Z,9Z,12Z)-Octadecatrienoic acid; 6,9,12-Octadecatrienoic acid; Gamolenic acid |
| Fatty Acid metabolism | 7 | 29 | .001 | Linoelaidic acid (all trans C18:2); Octadecadienoate (n-C18:2); Hexadecanoate (n-C16:0); linoleic acid (all cis C18:2) n-6; Dodecanoate (n-C12:0) | |
| Linoleate metabolism | 9 | 48 | .002 | (9Z,12Z,15Z)-Octadecatrienoic acid; alpha-Linolenic acid; 9,12,15-Octadecatrienoic acid; Linolenate; alpha-Linolenate; (6Z,9Z,12Z)-Octadecatrienoic acid; 6,9,12-Octadecatrienoic acid; gamma-Linolenic acid; Gamolenic acid | |
| Fatty acid activation | 9 | 53 | .004 | linoelaidic acid (all trans C18:2); Octadecadienoate (n-C18:2); Hexadecanoate (n-C16:0); Butanoic acid; Linoleic acid (all cis C18:2) n-6 | |
| Bile acid biosythesis | 5 | 21 | .005 | 3alpha,7alpha-Dihydroxy-5beta-cholestane; 5beta-Cholestane-3alpha,7alpha-diol; 3alpha,7alpha,12alpha-trihydroxy-5beta-cholestan-27-al; 27-Hydroxycholesterol; 7alpha-Hydroxy-5beta-cholestan-3-one; 27-Hydroxycholesterol; 20alpha-Hydroxycholesterol | |
| Omega-3 fatty acid metabolism | 4 | 16 | .009 | (9Z,12Z,15Z)-Octadecatrienoic acid; alpha-Linolenic acid; 9,12,15-Octadecatrienoic acid; Linolenate; alpha-Linolenate; (6Z,9Z,12Z)-Octadecatrienoic acid; 6,9,12-Octadecatrienoic acid; gamma-Linolenic acid; Gamolenic acid | |
| Glycerophospholipid metabolism | 9 | 61 | .009 | 9,12,15-Octadecatrienoic acid; Linolenate; alpha-Linolenate; (6Z,9Z,12Z)-Octadecatrienoic acid; 6,9,12-Octadecatrienoic acid; gamma-Linolenic acid; Gamolenic acid; Lysophosphatidylcholine; 1-Alkyl-2-lyso-sn-glycero-3-phosphocholine | |
| Fatigue | Tryptophan metabolism | 7 | 74 | .004 | Indole-3-acetaldehyde; N-Methyltryptamine; N-Methylindoleethylamine; 1-methyl-2-(3-indolyl)ethylamine; 6-Hydroxymelatonin; L-Kynurenine; Formyl-5-hydroxykynurenamine |
| Drug metabolism - cytochrome P450 | 3 | 24 | .018 | 5-Phenyl-1,3-oxazinane-2,4-dione; 5-Hydroxyindoleacetate; Monoethylglycinexylidide; 3-Carbamoyl-2-phenylpropionic acid | |
| Anxiety | Glycerophospholipid metabolism | 9 | 61 | .011 | Tetrahydropteridine; 5,6,7,8-Tetrahydropteridine; Glycerol 3-phosphate; Platelet-activating factor; sn-Glycero-3-phosphoethanolamine |
| Tryptophan metabolism | 10 | 74 | .015 | Indolepyruvate; Indolepyruvic acid; (Indol-3-yl)pyruvate; Indole-3-pyruvate; 3-(Indol-3-yl)pyruvate; Indole-3-acetaldehyde; L-Kynurenine Formyl-5-hydroxykynurenamine | |
| Phosphatidylinositol phosphate metabolism | 4 | 18 | .015 | D-myo-Inositol 1,2-cyclic phosphate; 1D-myo-Inositol 1,2-cyclic phosphate; N-Acetyl-D-mannosamine; 2-Acetamido-2-deoxy-D-mannose N-Acetylgalactosamine; N-Acetylchondrosamine; 2-Acetamido-2-deoxygalactose; GalNAc N-Acetyl-D-glucosamine N-Acetyl-D-galactosamine | |
| Sialic acid metabolism | 3 | 15 | .042 | N-Acetyl-D-mannosamine; 2-Acetamido-2-deoxy-D-mannose N-Acetylgalactosamine; N-Acetylchondrosamine; 2-Acetamido-2-deoxygalactose; GalNAc N-Acetyl-D-glucosamine N-Acetyl-D-galactosamine | |
| Depressive symptoms | Limonene and pinene degradation | 3 | 12 | .013 | (+)-trans-Carveol; (4S,6 R)-trans-Carveol; Perillyl alcohol; (−)-Perillyl alcohol; p-Mentha-1,8-dien-7-ol; (−)-Perillylalcohol; (−)-trans-Carveol; Perillyl aldehyde; Perillaldehyde |
| Phosphatidylinositol phosphate metabolism | 3 | 18 | .038 | 1D-myo-Inositol 1,4,5,6-tetrakisphosphate; 1D-myo-Inositol 1,3,4,5,6-pentakisphosphate; Diacylglycerol; Diglyceride | |
| Ordinal PNS cluster | Carnitine shuttle | 4 | 31 | .015 | Stearoylcarnitine; dihomo-gamma-linolenyl carnitine; Linoleyl carnitine; Vaccenyl carnitine; Elaidic carnitine; Octadecenoyl carnitine |
| Tryptophan metabolism | 6 | 74 | .037 | Indole-3-acetaldehyde; Nicotinate; L-Kynurenine; Formyl-5-hydroxykynurenamine; N-Methyltryptamine; N-Methylindoleethylamine; 1-methyl-2-(3-indolyl)ethylamine |
Note. T1, pre-chemotherapy; T2, post-chemotherapy.
Overlap size means significant metabolite hits associated with the outcomes; metabolic pathways with an overlap size <3 were removed from our findings. Pathway size means total number of KEGG metabolites in the pathways. Metabolites represent some examples of identified metabolites that are associated with PNS and the PNS cluster.
Discussion
As the first of its kind, this study examined the associations of metabolic pathways and metabolites with individual PNS and the PNS cluster among children with cancer receiving chemotherapy. The PNS showed positive correlations among each other, while no significant changes in mean symptom scores were found between pre- and post-chemotherapy throughout one cycle of chemotherapy. We found that fatty acid, bile acid, and tryptophan pathways were primarily associated with PNS in children with cancer receiving chemotherapy.
Children with cancer experience a cluster of co-occurring PNS during chemotherapy (Rodgers et al., 2016; Wang et al., 2018). Multiple data-driven methods have been widely used to identify the symptom clusters among children with cancer, such as principal component analysis, exploratory factor analysis, and hierarchical agglomerative cluster analysis (Linder & Hooke, 2019). Due to the exploratory nature of this study, the PNS cluster was a sum of the four symptoms ranging from 0 to 4. Although different clusters of symptoms were identified in children with cancer, the PNS cluster is widely reported in this population (Rodgers et al., 2016; Wang et al., 2018). Consistent with previous studies (Collins et al., 2000; Hooke & Linder, 2019), our results demonstrated that these symptoms showed significant correlations with each other at two timepoints during a single cycle of chemotherapy, providing further validation that these four symptoms co-occur. This cluster of PNS reflects the prevalence, severity, and disturbance of the symptom experiences in children with cancer across one cycle of chemotherapy.
Previous work suggests significant changes of PNS across treatment trajectories (Collins et al., 2000; Hooke & Linder, 2019). Hockenberry et al. examined fatigue in children aged 3–15 years undergoing acute lymphoblastic leukemia and found a decrease of fatigue over the course of treatment (Hockenberry et al., 2014). Prevalence of pain (Dupuis et al., 2016; Hockenberry et al., 2014), anxiety (Dupuis et al., 2016), depressive symptoms (Baggott et al., 2010; Hockenberry et al., 2014) was reported to improve over cancer treatments. No significant difference of the PNS was found between two study timepoints (pre- vs. post-chemotherapy) in this study. This may be due to the short period of time between immediately before and 7–17 days after a cycle of chemotherapy, and the heterogeneities of cancer types. The PNS cluster varied in composition across cancer treatments (Linder & Hooke, 2019). In this study, children immediately prior to receiving a cycle of chemotherapy showed a higher prevalence of 2 PNS as a cluster while children post-chemotherapy indicated a higher prevalence of single PNS symptom. All children were recruited across one cycle of chemotherapy and their previous experience of chemotherapy may enhance their report of PNS at the beginning of one cycle of chemotherapy. On the other hand, the recovery from cancer treatment-related procedures and acute chemotherapy toxicities, and discharge from the hospital after the completion of chemotherapy may lead to report lower PNS.
Symptoms within the PNS cluster may share a common etiology. Literature has summarized that molecular indicators, such as release of proinflammatory cytokines (e.g., interleukin [IL]-1, IL-6, and tumor necrosis factor-alpha [TNF-α]) (Ji et al., 2017; Oliveira Miranda et al., 2014), hypothalamic-pituitary-adrenal (HPA) axis, and monoamine neurotransmission system (e.g., serotonin), may be associated with the development of PNS (Kim et al., 2012). Specifically, the signals of proinflammatory cytokines in response to tumor tissue cells and chemotherapy-related damaged cells can be transmitted to specific brain regions (Quan & Banks, 2007), and then lead to PNS for children with cancer. In addition, chemotherapy can result in dysregulations of the HPA axis, an essential pattern to maintain homeostasis. An altered HPA axis may cause some homeostatic imbalances associated with various acute and chronic symptoms (Kim et al., 2012; Oh et al., 2019). Importantly, the proinflammatory cytokines and the HPA axis seem to interact with neurotransmitters to adjust central nervous system pathologies contributing to the development and severity of PNS.
The metabolomics approach is promising to elucidate biological mechanisms associated with the development and severity of PNS. Recently, the role of metabolites in PNS has been explored in patients with cancer (Li et al., 2020; Lyon et al., 2018). Our study found that metabolic pathways of fatty acids, bile acids, and amino acids were associated with the PNS in children with chemotherapy. These results were consistent with previous work from adult cancers (Li et al., 2020; Lyon et al., 2018). Specifically, moderate-to-strong correlations were found between changes in pain and tryptophan concentration and kynurenine/tryptophan ratio, as well as between changes in depression and serotonin level in women with breast cancer (Lyon et al., 2018). Additionally, increases in pain from pre-to post-chemotherapy were associated with increased concentrations of betaine (an n-trimethylated amino acid); decreases of multiple bile-related compounds were significantly associated with increases in fatigue and fatty acids (e.g., arachidonic acid, n-malate, citramalate, and n-citramalate) were associated with fatigue post-chemotherapy; and nicotinamide (a water-soluble form of vitamin B3) was associated with depression in this population (Lyon et al., 2018). We found that fatty acid metabolic pathways (e.g., de novo fatty acid biosynthesis, fatty acid metabolism, omega-3 fatty acid metabolism, and fatty acid activation) and bile acid biosynthesis were associated with pain; and tryptophan metabolism was associated with fatigue, anxiety, and the PNS cluster across one cycle of chemotherapy.
Fatty acids pathways are crucial molecules that determine human brain’s integrity and ability to perform (Chang et al., 2009). Some essential fatty acids, particularly the omega-3 fatty acids, are found to decrease the symptoms of fatigue and pain in patients during chemotherapy, possibly due to weight maintenance and reduced inflammatory status (Freitas & Campos, 2019). Bile acids facilitate excretion, absorption, and transportation of fat and sterols in the liver and intestines and play a role in cholesterol homeostasis and microbiome signaling (Kelly et al., 2015). Similar findings of decreased bile acid synthesis were reported in patients with chronic fatigue syndrome (Naviaux et al., 2016).
Tryptophan, an essential amino acid, is required for structural and functional processes of protein biosynthesis and immunoregulation (Ball et al., 2014). The depletion of tryptophan and formation of kynurenine pathway metabolites modulates the activity of the mammalian immune, reproductive, and central nervous systems. A reduced tryptophan and increased kynurenine/tryptophan ratio were associated with an increased burden of pain, fatigue, anxiety, depression (Capuron et al., 2002; Lanser et al., 2020), as well as a lower quality of life (Huang et al., 2002; Schroecksnadel et al., 2007). Consistent with our findings in children with cancer, these findings revealed significant relationship of altered tryptophan and symptom burden from PNS during chemotherapy.
Moreover, cancer and its treatment (e.g., chemotherapy) can disturb the gut microbiome profiles, which regulate specific metabolic pathways and metabolites (e.g., SCFAs) associated with PNS based on the MGB (Cryan et al., 2019; Kennedy et al., 2016; Song & Bai, 2020). Until now, specific gut microbes and microbial associated metabolic signatures have been identified to contribute to PNS in patients with head and neck, breast, and colorectal cancers (Bai et al., 2020; González-Mercado et al., 2021; Lyon et al., 2018). For example, patients with a high level of PNS cluster were more likely to have higher abundances in phylum Bacteroidetes and genera Ruminiclostridium\, Tyzzerella, Eubacterium_fissicatena, and DTU089, while those with the low PNS cluster had higher abundances in genera Lactococcus, Phascolarctobacterium, and Desulfovibrio. This dysbiosis of the gut microbiome may regulate the PNS cluster through lipid metabolism and nervous system (Bai et al., 2020). More work is needed to confirm our findings in patients with cancer, including children. Further understanding the role of metabolites and associated molecular indicators in PNS can help develop precise interventions to relieve children’s suffering from PNS.
Several limitations must be noted in this study. Most importantly, is the pilot nature of the study. As a sub-study, the heterogeneities of cancer types and limitations to address potential confounders (e.g., chemotherapeutic agents) may limit the generalization of the findings. Due to the small sample size, the PNS cluster was categorized based on the cutoff point of T-scores rather than using widely reported modeling techniques (Kim et al., 2013). Second, this study used the untargeted metabolomics approach, providing a broader range of metabolites associated with PNS and the PNS cluster. Although the untargeted metabolomics approach is a more comprehensive method to study the metabolome, future work should confirm our findings in a large sample and further understand these associations using the targeted metabolomics approach via focusing on the most salient pathways. Third, the metabolomics data cannot be collected during fasting due to the special needs of children with cancer. Last, this study only focused on one snapshot of a specific cycle of chemotherapy (before and 7–17 days after a chemotherapy cycle) in a group of children with several types of cancer. Future work should examine the impacts of specific chemotherapy agents and multiple cycles of chemotherapy on the metabolome profiles in children with cancer.
In conclusion, fatty acid, bile acid, and amino acid metabolic pathways seemed to contribute to individual PNS and the PNS cluster in children undergoing chemotherapy. These findings require further investigation in a large sample size over longitudinal cycle of chemotherapy.
Supplemental Material
sj-pdf-1-brn-10.1177_10998004211069619 for Metabolic Pathways Associated With Psychoneurological Symptoms in Children With Cancer Receiving Chemotherapy by Jinbing Bai, Janice Withycombe and Ronald C. Eldridge in Biological Research For Nursing
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by theInstitutional Research Grant of American Cancer Society (PI: Withycombe); National Institute of Health/National Institute of Nursing Research (1K99NR017897-01, 4R00NR017897-03, PI: Bai); National Institute of Health/National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) (U19AR069522, PIs: Reeve and Schanberg); National Institutes of Health/National Cancer Institute (R01CA175759, PIs: Hinds and Reeve); 2018 and 2019 Transdisciplinary Research in Energetics and Cancer (TREC) Training Workshop (R25CA203650, PI: Irwin). Georgia Clinical and Translational Science Alliance (UL1TR002378, KL2TR002381, PI: Eldridge).
Supplemental Material: Supplemental material for this article is available online.
ORCID iDs
Jinbing Bai https://orcid.org/0000-0001-6726-5714
Janice Withycombe https://orcid.org/0000-0002-3042-9049
References
- Agus A., Planchais J., Sokol H. (2018. Gut microbiota regulation of tryptophan metabolism in health and disease. Cell Host & Microbe, 23(6), 716–724. 10.1016/j.chom.2018.05.003 [DOI] [PubMed] [Google Scholar]
- Amin S., Rattner J., Keramati M. R., Farshidfar F., McNamara M. G., Knox J. J., Kopciuk K., Vogel H. J., Bathe O. F. (2019). A strategy for early detection of response to chemotherapy drugs based on treatment-related changes in the metabolome. PLoS One, 14(4), Article e0213942. 10.1371/journal.pone.0213942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athersuch T. J., Keun H. C. (2015). Metabolic profiling in human exposome studies. Mutagenesis, 30(6), 755–762. 10.1093/mutage/gev060 [DOI] [PubMed] [Google Scholar]
- Baggott C., Dodd M., Kennedy C., Marina N., Matthay K. K., Cooper B. A., Miaskowski C. (2010. Changes in children’s reports of symptom occurrence and severity during a course of myelosuppressive chemotherapy. Journal of Pediatric Oncology Nursing: Official Journal of the Association of Pediatric Oncology Nurses, 27(6), 307–315. 10.1177/1043454210377619 [DOI] [PubMed] [Google Scholar]
- Bai J., Bruner D. W., Fedirko V., Beitler J. J., Zhou C., Gu J., Zhao H., Lin I. H., Chico C. E., Higgins K. A., Shin D. M., Saba N. F., Miller A. H., Xiao C. (2020). Gut microbiome associated with the psychoneurological symptom cluster in patients with head and neck cancers. Cancers (Basel), 12(9), 2531. 10.3390/cancers12092531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldassarre M. E., Laforgia N. (2020). Metabolomics applications in children: A right way to go. Metabolites, 10(9), 364. 10.3390/metabo10090364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball H. J., Jusof F. F., Bakmiwewa S. M., Hunt N. H., Yuasa H. J. (2014). Tryptophan-Catabolizing Enzymes – Party of Three [Review]. Frontiers in Immunology, 5, 485. 10.3389/fimmu.2014.00485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger M., Gray J. A., Roth B. L. (2009). The expanded biology of serotonin. Annual Review of Medicine, 60(1), 355–366. 10.1146/annurev.med.60.042307.110802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capuron L., Ravaud A., Neveu P. J., Miller A. H., Maes M., Dantzer R. (2002). Association between decreased serum tryptophan concentrations and depressive symptoms in cancer patients undergoing cytokine therapy. Molecular Psychiatry, 7(5), 468–473. 10.1038/sj.mp.4000995 [DOI] [PubMed] [Google Scholar]
- Carlson N. S., Frediani J. K., Corwin E. J., Dunlop A., Jones D. (2020). Metabolic Pathways Associated With Term Labor Induction Course in African American Women. Biological Research for Nursing, 22(2), 157–168. 10.1177/1099800419899730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler J. D., Hu X., Ko E. J., Park S., Lee Y. T., Orr M., Fernandes J., Uppal K., Kang S. M., Jones D. P., Go Y. M. (2016). Metabolic pathways of lung inflammation revealed by high-resolution metabolomics (HRM) of H1N1 influenza virus infection in mice. American Journal of Physiology. Regulatory, Integrative and Comparative Physiology, 311(5), R906–r916. 10.1152/ajpregu.00298.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C. Y., Ke D. S., Chen J. Y. (2009). Essential fatty acids and human brain. Acta Neurologica Taiwanica, 18(4), 231–241. 10.1016/b978-1-4160-4417-8.50040-1 [DOI] [PubMed] [Google Scholar]
- Chiu C.-Y., Yeh K.-W., Lin G., Chiang M.-H., Yang S.-C., Chao W.-J., Yao T.-C., Tsai M.-H., Hua M.-C., Liao S.-L., Lai S.-H., Cheng M.-L., Huang J.-L. (2016). Metabolomics reveals dynamic metabolic changes associated with age in early childhood. PLoS One, 11(2), Article e0149823. 10.1371/journal.pone.0149823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins J. J., Byrnes M. E., Dunkel I. J., Lapin J., Nadel T., Thaler H. T., Polyak T., Rapkin B., Portenoy R. K. (2000). 5The Measurement of Symptoms in Children with Cancer. Journal of Pain and Symptom Management, 19(5), 363–377. 10.1016/S0885-3924(00)00127-5. [DOI] [PubMed] [Google Scholar]
- Cowan C. S. M., Hoban A. E., Ventura-Silva A. P., Dinan T. G., Clarke G., Cryan J. F. (2017). Gutsy Moves: The Amygdala as a Critical Node in Microbiota to Brain Signaling. Bioessays: News and Reviews in Molecular, Cellular and Developmental Biology, 40, 1700172. 10.1002/bies.201700172 [DOI] [PubMed] [Google Scholar]
- Cryan J. F., O'Riordan K. J., Cowan C. S. M., Sandhu K. V., Bastiaanssen T. F. S., Boehme M., Codagnone M. G., Cussotto S., Fulling C., Golubeva A. V., Guzzetta K. E., Jaggar M., Long-Smith C. M., Lyte J. M., Martin J. A., Molinero-Perez A., Moloney G., Morelli E., Morillas E., Dinan T. G. (2019). The microbiota-gut-brain axis. Physiological Reviews, 99(4), 1877–2013. 10.1152/physrev.00018.2018 [DOI] [PubMed] [Google Scholar]
- Dalile B., Van Oudenhove L., Vervliet B., Verbeke K. (2019). The role of short-chain fatty acids in microbiota-gut-brain communication. Nature Reviews. Gastroenterology & Hepatology, 16(Suppl 1), 1. 10.1038/s41575-019-0157-3 [DOI] [PubMed] [Google Scholar]
- Debik J., Euceda L. R., Lundgren S., Gythfeldt H. v. d. L., Garred Ø., Borgen E., Engebraaten O., Bathen T. F., Giskeødegård G. F. (2019). Assessing treatment response and prognosis by serum and tissue metabolomics in breast cancer patients. Journal of Proteome Research, 18(10), 3649–3660. 10.1021/acs.jproteome.9b00316 [DOI] [PubMed] [Google Scholar]
- Dupuis L. L., Lu X., Mitchell H. R., Sung L., Devidas M., Mattano L. A., Jr., Carroll W. L., Winick N., Hunger S. P., Maloney K. W., Kadan-Lottick N. S. (2016). Anxiety, pain, and nausea during the treatment of standard-risk childhood acute lymphoblastic leukemia: A prospective, longitudinal study from the children’s oncology group. Cancer, 122(7), 1116–1125. 10.1002/cncr.29876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitas R. D. S., Campos M. M. (2019). Protective effects of omega-3 fatty acids in cancer-related complications. Nutrients, 11(5), 945. 10.3390/nu11050945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Mercado V. J., Henderson W. A., Sarkar A., Lim J., Saligan L. N., Berk L., Dishaw L., McMillan S., Groer M., Sepehri F., Melkus G. D. (2021). Changes in gut microbiome associated with co-occurring symptoms development during chemo-radiation for rectal cancer: A proof of concept study. Biological Research for Nursing, 23(1), 31–41. 10.1177/1099800420942830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Go Y.-M., Walker D. I., Liang Y., Uppal K., Soltow Q. A., Tran V., Strobel F., Quyyumi A. A., Ziegler T. R., Pennell K. D., Miller G. W., Jones D. P. (2015). Reference standardization for mass spectrometry and high-resolution metabolomics applications to exposome research. Toxicological Sciences: an Official Journal of the Society of Toxicology, 148(2), 531–543. 10.1093/toxsci/kfv198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guijas C., Montenegro-Burke J. R., Domingo-Almenara X., Palermo A., Warth B., Hermann G., Koellensperger G., Huan T., Uritboonthai W., Aisporna A. E., Wolan D. W., Spilker M. E., Benton H. P., Siuzdak G. (2018). METLIN: A Technology Platform for Identifying Knowns and Unknowns. Analytical Chemistry, 90(5), 3156–3164. 10.1021/acs.analchem.7b04424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinds P. S., Wang J., Cheng Y. I., Stern E., Waldron M., Gross H., DeWalt D. A., Jacobs S. S. (2019). PROMIS pediatric measures validated in a longitudinal study design in pediatric oncology. Pediatric Blood & Cancer, 66(5), Article e27606. 10.1002/pbc.27606 [DOI] [PubMed] [Google Scholar]
- Hochberg Y., Benjamini Y. (1990). More powerful procedures for multiple significance testing. Statistics in Medicine, 9(7), 811–818. 10.1002/sim.4780090710 [DOI] [PubMed] [Google Scholar]
- Hockenberry M. J., Taylor O. A., Pasvogel A., Rodgers C., McCarthy K., Gundy P., Montgomery D. W., Ribbeck P., Scheurer M. E., Moore I. M. (2014). The influence of oxidative stress on symptom occurrence, severity, and distress during childhood leukemia treatment. Oncology Nursing Forum, 41(4), E238–E247. 10.1188/14.ONF.E238-E247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman J. M., Soltow Q. A., Li S., Sidik A., Jones D. P., Promislow D. E. (2014). Effects of age, sex, and genotype on high-sensitivity metabolomic profiles in the fruit fly, Drosophila melanogaster. Aging Cell, 13(4), 596–604. 10.1111/acel.12215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes E., Loo R. L., Stamler J., Bictash M., Yap I. K., Chan Q., Ebbels T., De Iorio M., Brown I. J., Veselkov K. A., Daviglus M. L., Kesteloot H., Ueshima H., Zhao L., Nicholson J. K., Elliott P. (2008). Human metabolic phenotype diversity and its association with diet and blood pressure. Nature, 453(7193), 396–400. 10.1038/nature06882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooke M. C., Linder L. A. (2019). Symptoms in children receiving treatment for cancer-part i: Fatigue, sleep disturbance, and nausea/vomiting. Journal of Pediatric Oncology Nursing: Official Journal of the Association of Pediatric Oncology Nurses, 36(4), 244–261. 10.1177/1043454219849576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang A., Fuchs D., Widner B., Glover C., Henderson D. C., Allen-Mersh T. G. (2002). Jun 5)Serum tryptophan decrease correlates with immune activation and impaired quality of life in colorectal cancer. British Journal of Cancer, 86(11), 1691–1696. 10.1038/sj.bjc.6600336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin D. E., Stucky B. D., Thissen D., Dewitt E. M., Lai J. S., Yeatts K., Varni J. W., DeWalt D. A. (2010)Sampling plan and patient characteristics of the PROMIS pediatrics large-scale survey. Quality of Life Research: an International Journal of Quality of Life Aspects of Treatment, Care and Rehabilitation, 19(4), 585–594. 10.1007/s11136-010-9618-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James S. J., Cutler P., Melnyk S., Jernigan S., Janak L., Gaylor D. W., Neubrander J. A. (2004). Metabolic biomarkers of increased oxidative stress and impaired methylation capacity in children with autism. The American Journal of Clinical Nutrition, 80(6), 1611–1617. 10.1093/ajcn/80.6.1611 [DOI] [PubMed] [Google Scholar]
- Jenkins T. A., Nguyen J. C., Polglaze K. E., Bertrand P. P. (2016). Influence of tryptophan and serotonin on mood and cognition with a possible role of the gut-brain axis. Nutrients, 8(1), 56. 10.3390/nu8010056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y.-B., Bo C.-L., Xue X.-J., Weng E.-M., Gao G.-C., Dai B.-B., Ding K.-W., Xu C.-P. (2017). Association of inflammatory cytokines with the symptom cluster of pain, fatigue, depression, and sleep disturbance in chinese patients with cancer. Journal of Pain and Symptom Management, 54(6), 843–852. 10.1016/j.jpainsymman.2017.05.003 [DOI] [PubMed] [Google Scholar]
- Johnson W. E., Li C., Rabinovic A. (2006). Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics, 8(1), 118–127. 10.1093/biostatistics/kxj037 [DOI] [PubMed] [Google Scholar]
- Jourdan C., Petersen A.-K., Gieger C., Döring A., Illig T., Wang-Sattler R., Meisinger C., Peters A., Adamski J., Prehn C., Suhre K., Altmaier E., Kastenmüller G., Römisch-Margl W., Theis F. J., Krumsiek J., Wichmann H. E., Linseisen J. (2012). Body fat free mass is associated with the serum metabolite profile in a population-based study. PLoS One, 7(6), Article e40009. 10.1371/journal.pone.0040009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Sato Y., Kawashima M., Furumichi M., Tanabe M. (2015). KEGG as a reference resource for gene and protein annotation. Nucleic Acids Research, 44(D1), D457–D462. 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly J. R., Kennedy P. J., Cryan J. F., Dinan T. G., Clarke G., Hyland N. P. (2015). Breaking down the barriers: The gut microbiome, intestinal permeability and stress-related psychiatric disorders. Front Cell Neurosci, 9, 392. 10.3389/fncel.2015.00392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy P. J., Allen A. P., O'Neill A., Quigley E. M., Cryan J. F., Dinan T. G., Clarke G. (2015Acute tryptophan depletion reduces kynurenine levels: Implications for treatment of impaired visuospatial memory performance in irritable bowel syndrome. Psychopharmacology, 232(8), 1357–1371. 10.1007/s00213-014-3767-z [DOI] [PubMed] [Google Scholar]
- Kennedy P. J., Cryan J. F., Dinan T. G., Clarke G. (2016). Kynurenine pathway metabolism and the microbiota-gut-brain axis. Neuropharmacology, 112(Pt B), 399–412. 10.1016/j.neuropharm.2016.07.002 [DOI] [PubMed] [Google Scholar]
- Kestler S. A., LoBiondo-Wood G. (2012)Review of symptom experiences in children and adolescents with cancer. Cancer Nursing, 35(2), E31–E49. 10.1097/NCC.0b013e3182207a2a [DOI] [PubMed] [Google Scholar]
- Kim H. J., Abraham I., Malone P. S. (2013). Analytical methods and issues for symptom cluster research in oncology. Current Opinion in Supportive and Palliative Care, 7(1), 45–53. 10.1097/SPC.0b013e32835bf28b [DOI] [PubMed] [Google Scholar]
- Kim H. J., Barsevick A. M., Fang C. Y., Miaskowski C. (2012. Common biological pathways underlying the psychoneurological symptom cluster in cancer patients. Cancer Nursing, 35(6), E1–E20. 10.1097/NCC.0b013e318233a811 [DOI] [PubMed] [Google Scholar]
- Kochhar S., Jacobs D. M., Ramadan Z., Berruex F., Fuerholz A., Fay L. B. (2006). Probing gender-specific metabolism differences in humans by nuclear magnetic resonance-based metabonomics. Analytical Biochemistry, 352(2), 274–281. 10.1016/j.ab.2006.02.033 [DOI] [PubMed] [Google Scholar]
- Kwekkeboom K. L. (2016. Cancer symptom cluster management. Seminars in Oncology Nursing, 32(4), 373–382. 10.1016/j.soncn.2016.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanser L., Kink P., Egger E. M., Willenbacher W., Fuchs D., Weiss G., Kurz K. (2020). Inflammation-induced tryptophan breakdown is related with anemia, fatigue, and depression in cancer [review]. Frontiers in Immunology, 11, 249. 10.3389/fimmu.2020.00249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau C.-H. E., Siskos A. P., Maitre L., Robinson O., Athersuch T. J., Want E. J., Urquiza J., Casas M., Vafeiadi M., Roumeliotaki T., McEachan R. R. C., Azad R., Haug L. S., Meltzer H. M., Andrusaityte S., Petraviciene I., Grazuleviciene R., Thomsen C., Coen M. (2018). Determinants of the urinary and serum metabolome in children from six European populations. BMC Medicine, 16(1), 202. 10.1186/s12916-018-1190-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Liu T., Heinsberg L. W., Lockwood M. B., Wainwright D. A., Jang M. K., Doorenbos A. Z. (2020). Systematic review of the kynurenine pathway and psychoneurological symptoms among adult cancer survivors. Biological Research for Nursing, 22(4), 472–484. 10.1177/1099800420938141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder L. A., Hooke M. C. (2019. Symptoms in children receiving treatment for cancer-part II: Pain, sadness, and symptom clusters. Journal of Pediatric Oncology Nursing: Official Journal of the Association of Pediatric Oncology Nurses, 36(4), 262–279. 10.1177/1043454219849578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Park Y., Duraisingham S., Strobel F. H., Khan N., Soltow Q. A., Jones D. P., Pulendran B. (2013). Predicting network activity from high throughput metabolomics. PLoS Computational Biology, 9(7), Article e1003123. 10.1371/journal.pcbi.1003123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K. H., Nellis M., Uppal K., Ma C., Tran V., Liang Y., Walker D. I., Jones D. P. (2020). Reference standardization for quantification and harmonization of large-scale metabolomics. Analytical Chemistry, 92(13), 8836–8844. 10.1021/acs.analchem.0c00338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon D. E., Starkweather A., Yao Y., Garrett T., Kelly D. L., Menzies V., Dereziński P., Datta S., Kumar S., Jackson-Cook C. (2018). Pilot study of metabolomics and psychoneurological symptoms in women with early stage breast cancer. Biological Research for Nursing, 20(2), 227–236. 10.1177/1099800417747411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer E. A., Knight R., Mazmanian S. K., Cryan J. F., Tillisch K. (2014). Gut microbes and the brain: Paradigm shift in neuroscience. The Journal of Neuroscience: the Official Journal of the Society for Neuroscience, 34(46), 15490–15496. 10.1523/JNEUROSCI.3299-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeer J. L., Nachman J. B. (2010). The optimal use of steroids in paediatric acute lymphoblastic leukaemia: No easy answers. British Journal of Haematology, 149(5), 638–652. 10.1111/j.1365-2141.2010.08192.x [DOI] [PubMed] [Google Scholar]
- Miaskowski C., Barsevick A., Berger A., Casagrande R., Grady P. A., Jacobsen P., Kutner J., Patrick D., Zimmerman L., Xiao C., Matocha M., Marden S. (2017). Advancing symptom science through symptom cluster research: Expert Panel Proceedings and recommendations. JNCI: Journal of the National Cancer Institute, 109(4), Article djw253. 10.1093/jnci/djw253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moco S., Collino S., Rezzi S., Martin F.-P. (2013). Metabolomics perspectives in pediatric research. Pediatric Research, 73(4 Pt 2), 570–576. 10.1038/pr.2013.1 [DOI] [PubMed] [Google Scholar]
- Monteiro-Cardoso V. F., Corlianò M., Singaraja R. R. (2021). Bile acids: A communication channel in the gut-brain axis. NeuroMolecular Medicine, 23(1), 99–117. 10.1007/s12017-020-08625-z [DOI] [PubMed] [Google Scholar]
- Mrakotsky C. M., Silverman L. B., Dahlberg S. E., Alyman M. C. A., Sands S. A., Queally J. T., Miller T. P., Cranston A., Neuberg D. S., Sallan S. E., Waber D. P. (2011). Neurobehavioral side effects of corticosteroids during active treatment for acute lymphoblastic leukemia in children are age-dependent: Report from dana-farber cancer institute ALL consortium protocol 00-01. Pediatric Blood & Cancer, 57(3), 492–498. 10.1002/pbc.23060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller C. P., Homberg J. R. (2015). Serotonin revisited. Behavioural Brain Research, 277, 1-2. 10.1016/j.bbr.2014.10.008. [DOI] [PubMed] [Google Scholar]
- Naviaux R. K., Naviaux J. C., Li K., Bright A. T., Alaynick W. A., Wang L., Baxter A., Nathan N., Anderson W., Gordon E. (2016). Metabolic features of chronic fatigue syndrome. Proc Natl Acad Sci U S A, 113(37), E5472–E5480. 10.1073/pnas.1607571113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neujahr D. C., Uppal K., Force S. D., Fernandez F., Lawrence C., Pickens A., Bag R., Lockard C., Kirk A. D., Tran V., Lee K., Jones D. P., Park Y. (2014). Bile acid aspiration associated with lung chemical profile linked to other biomarkers of injury after lung transplantation. American Journal of Transplantation: Official Journal of the American Society of Transplant Surgeons, 14(4), 841–848. 10.1111/ajt.12631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh I. J., Kim K. S., Kim Y. C., Park J. Y., Yoo K. Y., Do S. H., Ahn R. S. (2019). Altered hypothalamus-pituitary-adrenal axis function: A potential underlying biological pathway for multiple concurrent symptoms in patients with advanced lung cancer. Psychosomatic Medicine, 81(1), 41–50. 10.1097/psy.0000000000000648 [DOI] [PubMed] [Google Scholar]
- Oliveira Miranda D., Soares de Lima T. A., Ribeiro Azevedo L., Feres O., Ribeiro da Rocha J. J., Pereira-da-Silva G. (2014). Proinflammatory Cytokines correlate with depression and anxiety in colorectal cancer patients. Biomed Research International, 2014(5), 739650. 10.1155/2014/739650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Mahony S. M., Clarke G., Borre Y. E., Dinan T. G., Cryan J. F. (2015). Serotonin, tryptophan metabolism and the brain-gut-microbiome axis. Behavioural Brain Research, 277, 32-48. 10.1016/j.bbr.2014.07.027. [DOI] [PubMed] [Google Scholar]
- Pascale A., Marchesi N., Marelli C., Coppola A., Luzi L., Govoni S., Giustina A., Gazzaruso C. (2018). Microbiota and metabolic diseases. Endocrine, 61(3), 357–371. 10.1007/s12020-018-1605-5 [DOI] [PubMed] [Google Scholar]
- Platten M., Nollen E. A. A., Röhrig U. F., Fallarino F., Opitz C. A. (2019). Tryptophan metabolism as a common therapeutic target in cancer, neurodegeneration and beyond. Nature Reviews Drug Discovery, 18(5), 379–401. 10.1038/s41573-019-0016-5 [DOI] [PubMed] [Google Scholar]
- Quan N., Banks W. A. (2007). Brain-immune communication pathways. Brain, Behavior, and Immunity, 21(6), 727–735. 10.1016/j.bbi.2007.05.005 [DOI] [PubMed] [Google Scholar]
- Reeve B. B., McFatrich M., Mack J. W., Maurer S. H., Jacobs S. S., Freyer D. R., Withycombe J. S., Baker J. N., Castellino S. M., Lin L., Lucas N. R., Hinds P. S. (2020). Validity and reliability of the pediatric patient-reported outcomes version of the common terminology criteria for adverse events. JNCI: Journal of the National Cancer Institute, 112(11), 1143–1152. 10.1093/jnci/djaa016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers C., Hooke M. C., Ward J., Linder L. A. (2016). Symptom Clusters in Children and Adolescents with Cancer. Seminars in Oncology Nursing, 32(4), 394–404. 10.1016/j.soncn.2016.08.005 [DOI] [PubMed] [Google Scholar]
- Schmidt D. R., Patel R., Kirsch D. G., Lewis C. A., Vander Heiden M. G., Locasale J. W. (2021). Metabolomics in cancer research and emerging applications in clinical oncology. Ca: a Cancer Journal for Clinicians, 71(4), 333–358. 10.3322/caac.21670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroecksnadel K., Fiegl M., Prassl K., Winkler C., Denz H. A., Fuchs D. (2007). Diminished quality of life in patients with cancer correlates with tryptophan degradation. J Cancer Res Clin Oncol, 133(7), 477–485. 10.1007/s00432-007-0191-3 [DOI] [PubMed] [Google Scholar]
- Siegel R. L., Miller K. D., Fuchs H. E., Jemal A. (2021). Cancer Statistics, 2021. Ca: a Cancer Journal for Clinicians, 71(1), 7–33. 10.3322/caac.21654 [DOI] [PubMed] [Google Scholar]
- Silva Y. P., Bernardi A., Frozza R. L. (2020). The role of short-chain fatty acids from gut microbiota in gut-brain communication [review]. Frontiers in Endocrinology, 11, 25. 10.3389/fendo.2020.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltow Q. A., Strobel F. H., Mansfield K. G., Wachtman L., Park Y., Jones D. P. (2013). High-performance metabolic profiling with dual chromatography-Fourier-transform mass spectrometry (DC-FTMS) for study of the exposome. Metabolomics, 9(1), S132–S143. 10.1007/s11306-011-0332-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song B. C., Bai J. (2020. Microbiome-gut-brain axis in cancer treatment-related psychoneurological toxicities and symptoms: A systematic review. Supportive Care in Cancer: Official Journal of the Multinational Association of Supportive Care in Cancer, 29(10), 605–617. 10.1007/s00520-020-05739-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiga L., Atzori L., Noto A., Moretti C., Mussap M., Masile A., Lussu M., Fanos V. (2013). Metabolomics in paediatric oncology: A potential still to be exploited. The Journal of Maternal-Fetal & Neonatal Medicine, 26(Suppl 2), 20–23. 10.3109/14767058.2013.832062 [DOI] [PubMed] [Google Scholar]
- Strasser B., Becker K., Fuchs D., Gostner J. M. (2016. Kynurenine pathway metabolism and immune activation: Peripheral measurements in psychiatric and co-morbid conditions. Neuropharmacology, 112(Pt B), 286–296. 10.1016/j.neuropharm.2016.02.030 [DOI] [PubMed] [Google Scholar]
- Uppal K., Soltow Q. A., Strobel F. H., Pittard W. S., Gernert K. M., Yu T., Jones D. P. (2013). xMSanalyzer: Automated pipeline for improved feature detection and downstream analysis of large-scale, non-targeted metabolomics data. BMC Bioinformatics, 14(1), 15. 10.1186/1471-2105-14-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uppal K, Walker DI, Liu K, Li S, Go YM, Jones DP. (2016, Dec 19). Computational Metabolomics: A Framework for the Million Metabolome. Chemical Research in Toxicology, 29(12), 1956–1975. 10.1021/acs.chemrestox.6b00179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Wouw M., Boehme M., Lyte J. M., Wiley N., Strain C., O'Sullivan O., Clarke G., Stanton C., Dinan T. G., Cryan J. F. (2018). Short-chain fatty acids: Microbial metabolites that alleviate stress-induced brain-gut axis alterations. J Physiol, 596(20), 4923–4944. 10.1113/jp276431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Jacobs S., Dewalt D. A., Stern E., Gross H., Hinds P. S. (2018). A Longitudinal study of PROMIS pediatric symptom clusters in children undergoing chemotherapy. Journal of Pain and Symptom Management, 55(2), 359–367. 10.1016/j.jpainsymman.2017.08.021 [DOI] [PubMed] [Google Scholar]
- Wilson K., Hawken S., Ducharme R., Potter B. K., Little J., Thébaud B., Chakraborty P. (2014). Metabolomics of prematurity: Analysis of patterns of amino acids, enzymes, and endocrine markers by categories of gestational age. Pediatric Research, 75(2), 367–373. 10.1038/pr.2013.212 [DOI] [PubMed] [Google Scholar]
- Yanofsky C. (2007. RNA-based regulation of genes of tryptophan synthesis and degradation, in bacteria. RNA, 13(8), 1141–1154. 10.1261/rna.620507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu T., Park Y., Li S., Jones D. P. (2013). Hybrid feature detection and information accumulation using high-resolution LC–MS metabolomics data. Journal of Proteome Research, 12(3), 1419–1427. 10.1021/pr301053d [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z., Zhai G., Singmann P., He Y., Xu T., Prehn C., Römisch‐Margl W., Lattka E., Gieger C., Soranzo N., Heinrich J., Standl M., Thiering E., Mittelstraß K., Wichmann H. E., Peters A., Suhre K., Li Y., Adamski J., Wang-Sattler R. (2012). Human serum metabolic profiles are age dependent. Aging Cell, 11(6), 960–967. 10.1111/j.1474-9726.2012.00865.x [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
sj-pdf-1-brn-10.1177_10998004211069619 for Metabolic Pathways Associated With Psychoneurological Symptoms in Children With Cancer Receiving Chemotherapy by Jinbing Bai, Janice Withycombe and Ronald C. Eldridge in Biological Research For Nursing