Abstract
Anaplastic lymphoma kinase (ALK)1 and the related leukocyte tyrosine kinase (LTK)2 are recently deorphanized receptor tyrosine kinases3. Together with their activating cytokines, ALKAL1 and ALKAL24–6 (also called FAM150A and FAM150B or AUGβ and AUGα, respectively), they are involved in neural development7, cancer7–9 and autoimmune diseases10. Furthermore, mammalian ALK recently emerged as a key regulator of energy expenditure and weight gain11, consistent with a metabolic role for Drosophila ALK12. Despite such functional pleiotropy and growing therapeutic relevance13, 14, structural insights into ALK and LTK and their complexes with cognate cytokines have remained scarce. Here we show that the cytokine-binding segments of human ALK and LTK comprise a novel architectural chimera of a permuted TNF-like module that braces a glycine-rich subdomain featuring a hexagonal lattice of long polyglycine type II helices. The cognate cytokines ALKAL1 and ALKAL2 are monomeric three-helix bundles, yet their binding to ALK and LTK elicits similar dimeric assemblies with two-fold symmetry, that tent a single cytokine molecule proximal to the cell membrane. We show that the membrane-proximal EGF-like domain dictates the apparent cytokine preference of ALK. Assisted by these diverse structure–function findings, we propose a structural and mechanistic blueprint for complexes of ALK family receptors, and thereby extend the repertoire of ligand-mediated dimerization mechanisms adopted by receptor tyrosine kinases.
ALK is an evolutionarily ancient receptor tyrosine kinase (RTK). The vertebrate orthologues are uniquely endowed with an amino-terminal heparin-binding domain (HBD) (Fig. 1a, Extended Data Fig. 1a, b). Gene duplication in vertebrates resulted in LTK, a second ALK-like receptor15, which evolved divergently with loss of the HBD and the MAM-LDLa-MAM module in mammals (Extended Data Fig. 1c). The common architectural hallmark in the ectodomains of ALK and LTK comprises their cytokine-binding segment, which is unique among cytokine receptors and features an array of a TNF-like (TNFL) module, a glycine-rich (GR) region, and a membrane-proximal EGF-like module (EGFL) (Fig. 1a). Whereas ALKAL1 and ALKAL2 are both strong activators of LTK4, only ALKAL2 potently activates ALK5, 6–this activation is coupled to additional regulation via glycosaminoglycan binding to its HBD16.
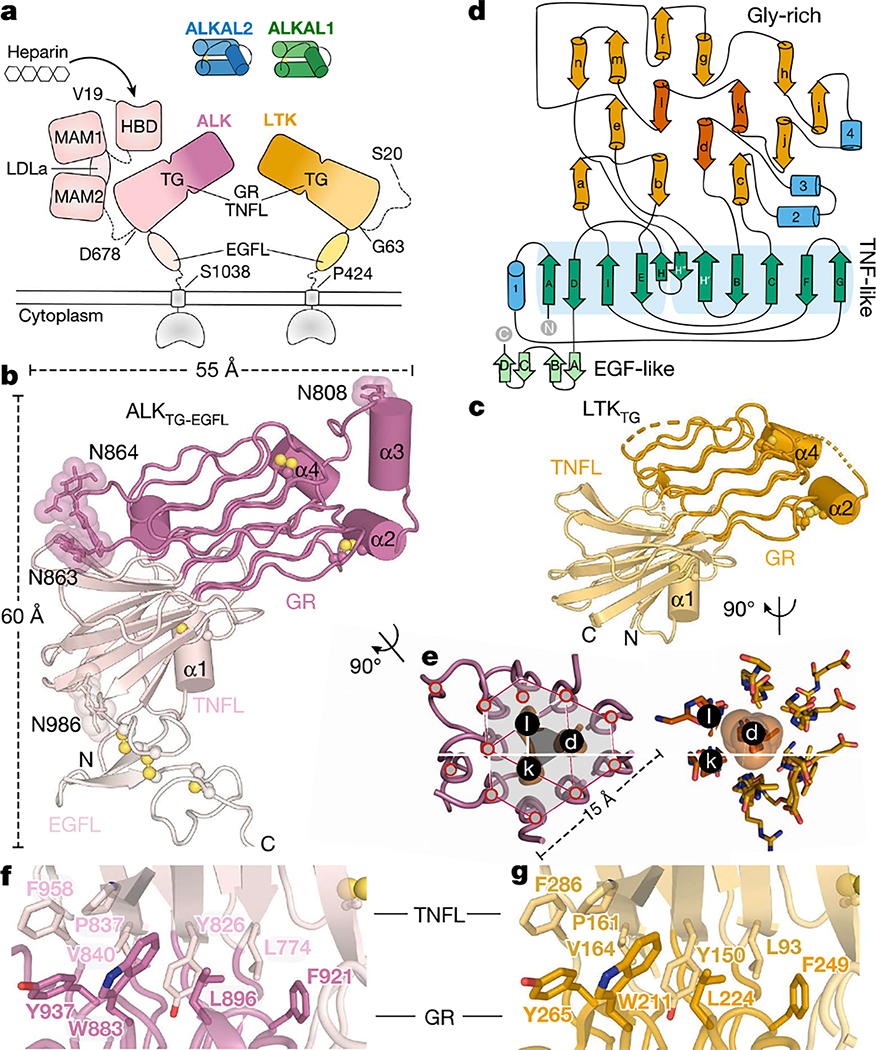
Fig. 1 |. Structure of the ALK family cytokine-binding domain.
a, Domain organization of ALK and LTK, and ligands. MAM, meprin–A5-protein–PTPμ domain; LDLa, low-density lipoprotein class A repeat; HBD, heparin-binding domain. b, c, Crystal structures of ALKTG–EGFL (b) and LTKTG (c). Disulfides (yellow spheres) and experimentally observed N-glycans in ALK (N808, N863, N864 and N986) are indicated. Helices are denoted by α. d, Topology diagram of the TG supradomain, showing β-strands (green), α-helices (blue), pGII helices (orange), central pGII helices d, k and l (vermillion). e, Cross-sections and packing of pGII helix lattices in ALK (left) and LTK (right). f, g, Hydrophobic interfaces between the TNFL and GR subdomains in the TG supradomain of ALK (f) and LTK (g).
Structure of ALK-LTK cytokine-binding domains
To characterize the cytokine-binding segments of ALK and LTK, we determined their crystal structures based on purified glycan-shaved human ALKTG–EGFL (residues 648–1038), its complex with a non-neutralizing Fab fragment17, and human LTKTG (residues 63–378) (Extended Data Fig. 2a–c, Extended Data Table 1).
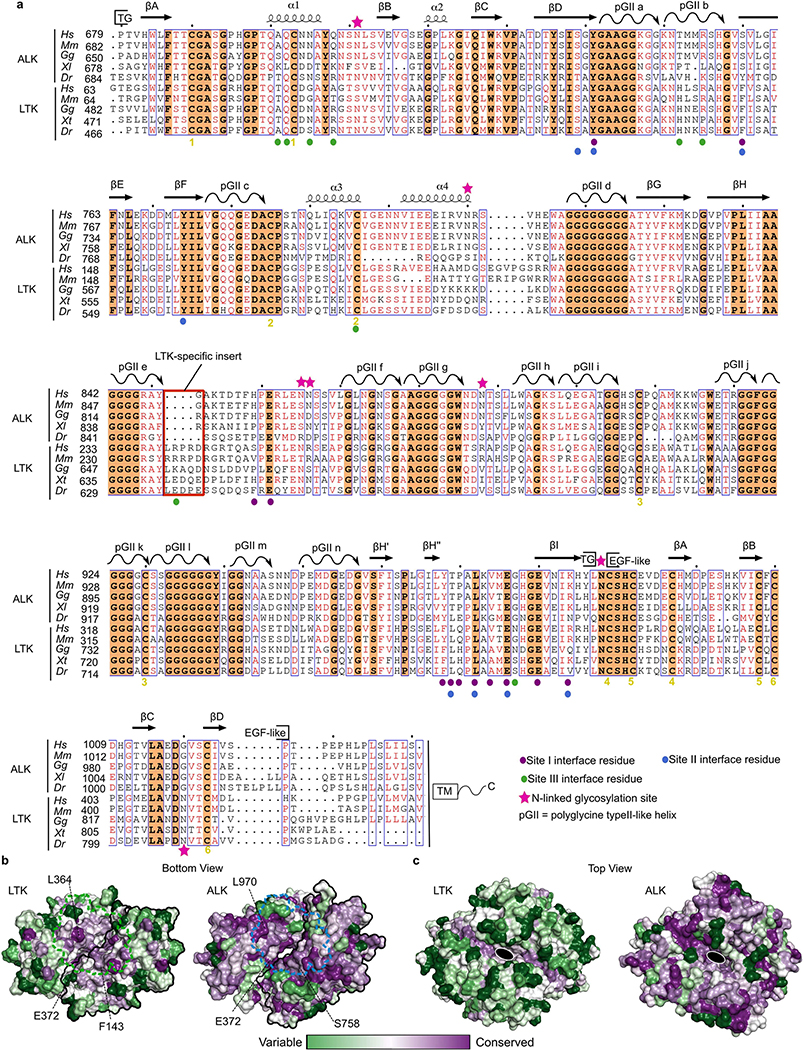
Unexpectedly, the TNFL and GR regions in ALK and LTK do not form separate domains, but rather a fully globular TNFL–GR (TG) supradomain featuring a chimeric arrangement of two interwoven subdomains with distinct secondary structure (Fig. 1b–d, Extended Data Fig. 2d–f). The TNFL subdomain is an anti-parallel β-sandwich, and the GR subdomain contains 14 long polyglycine type II (pGII) helices18 tightly packed into a honeycomb-like lattice (Fig. 1e, Extended Data Fig. 2g, h). The boundary between TNFL and GR comprises an extensive hydrophobic interface lined by conserved residues (Fig. 1f, g). The N terminus of the TG supradomain maps to the first strand of its TNFL subdomain and the C terminus is on the adjacent strand (Fig. 1d), which in ALKTG–EGFL connects to the membrane-proximal EGFL module (Cys987 to Prol025) through a short N-glycosylated linker (Fig. 1b). Additionally, the TG supradomain is decorated by four peripheral α-helices, with α1 tethered to the TNFL subdomain through a conserved disulfide, whereas the disulfide-linked α2 and α4 cluster at the tip of the GR subdomain together with α3 (Fig. 1b, c).
Notably, the GR core has three pGII helices (d, k and l) exclusively containing glycine residues, which are surrounded by six other pGII helices, establishing tight van der Waals contacts and hydrogen bonding via their main-chain amide and carbonyl groups (Fig. 1e, Extended Data Fig. 2g). Collectively, the observed interactions within the pGII helix lattices offer a rationale for the sequence conservation of the polyglycine sequences (Extended Data Fig. 2i) and for loss-of-function mutants in this region of Drosophila ALK19.
The TNFL subdomains adopt a markedly different chain topology compared with TNF/C1q-class folds, despite their spatial similarity (root mean square deviation (r.m.s.d.) = 2.8 Å against TNF/C1q, 72 Cα atoms) and the sequence homology of individual β-strands (Extended Data Fig. 2j, k). However, the shuffling of spatially equivalent β-strands between the ALKTNFL/LTKTNFL subdomain and TNF/C1q proteins goes far beyond a simple permutation and cannot be explained by a simple evolutionary path. The connected β-strands in the ALKTNFL/LTKTNFL subdomain break up the alternating sheet-to-sheet register of the TNF/C1q β-jellyroll, and instead permit the spatially contiguous sprouting of the three glycine-rich loop inserts towards the distinctive pGII helix lattice of the ALKGR/LTKGR subdomains (Extended Data Fig. 2j). The spatial coalescence of three otherwise intrinsically disordered stretches of glycine-rich insertions into a topologically tortured TNFL subdomain suggests that this subdomain has potential as a versatile scaffold for protein design20.
Assembly of ALK– and LTK–cytokine complexes
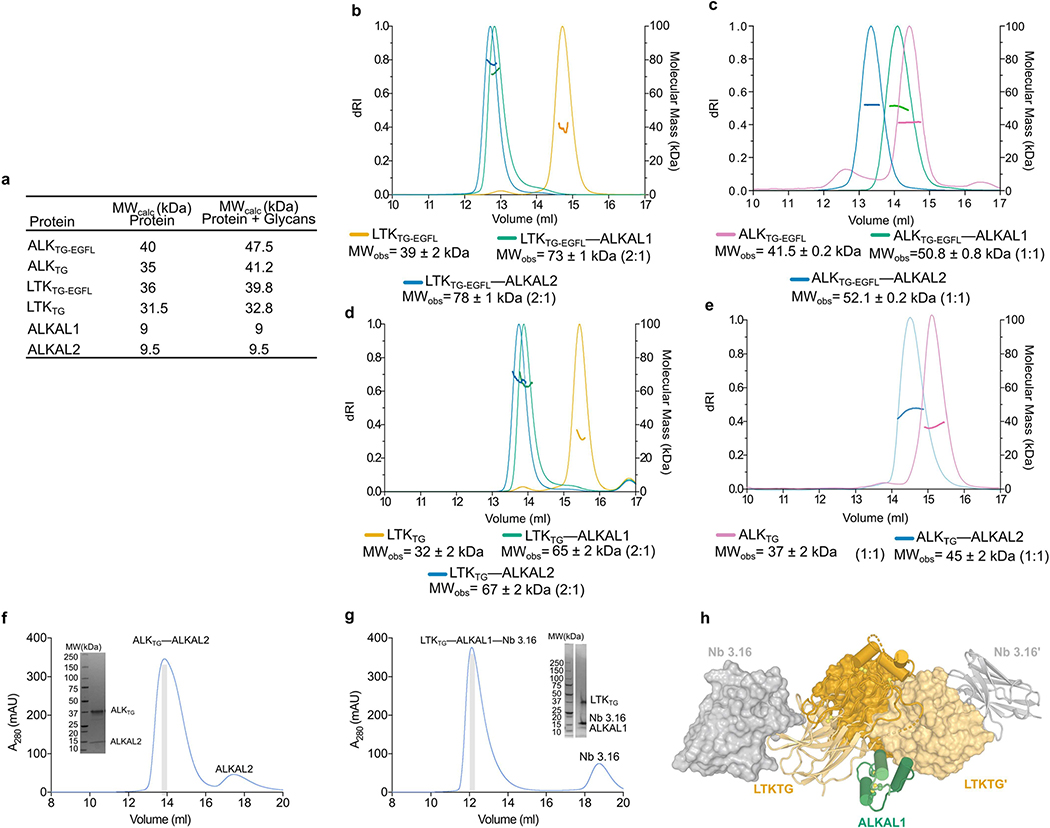
To cover the breadth of possible ALK– and LTK–cytokine complexes, we investigated structures of ALK–ALKAL2 and LTK–ALKAL1 assemblies. We first established that the conserved C-terminal domains of the cytokines (ALKAL1 and ALKAL2) and full-length ALKAL1 (ALKAL1FL) could drive ALK-dependent cellular proliferation (Extended Data Fig. 3a–c). Notably, purified complexes of ALK and LTK with cytokines displayed distinct stoichiometries. LTKTG–EGFL–ALKAL1 and LTKTG–EGFL–ALKAL2 formed 2:1 stoichiometric assemblies (2 receptors and 1 cytokine), whereas ALKTG–EGFL–ALKAL1 and ALKTG–EGFL–ALKAL2 exhibited 1:1 stoichiometries (Extended Data Fig. 4a–c). The same stoichiometric dissonance was observed for complexes of ALKTG and LTKTG lacking the membrane-proximal EGFL domain (Extended Data Fig. 4d, e). Thus, ALK/LTK–cytokine complexes deviated from the canonical 2:2 stoichiometry of RTK–cytokine complexes.
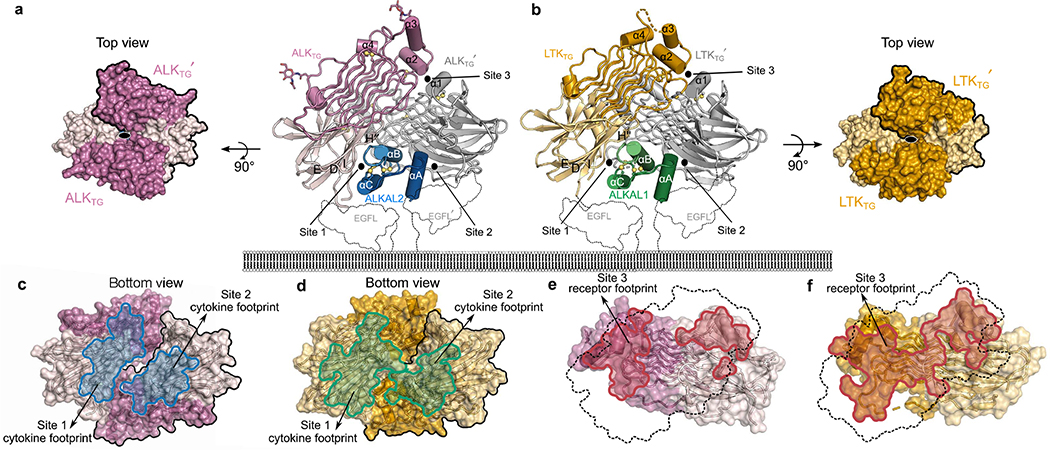
Diffraction-quality crystals could be obtained only from purified ALKTG–ALKAL2 and LTKTG–ALKAL1 stabilized by a non-neutralizing single-domain camelid antibody fragment (nanobody Nb3.16) (Extended Data Fig. 4f, g, Extended Data Table 1). Our structures revealed highly similar assemblies in which a single cytokine molecule is cradled proximal to the membrane by two receptor TG supradomains, resulting in receptor dimerization (Fig. 2a, b, Extended Data Fig. 4h). In hindsight, the inordinately high protein concentration reached during crystallization might have compensated for the low degree of cytokine-induced ALKTG dimerization in solution. ALK– and LTK–cytokine complexes feature three compact interaction interfaces. Sites 1 and 2 correspond to receptor–cytokine interactions and site 3 describes receptor–receptor contacts (Fig. 2c–f). The cytokine ligands are asymmetric three-helix bundles and use helices B and C to contact one receptor molecule via site 1, and helix A to engage the second receptor via site 2. The dimerized ALKTG and LTKTG supradomains incline their GR subdomains at approximately 45° to establish site 3. Thus, ALK family receptors establish two-fold-symmetric assemblies mediated by asymmetric cytokines, which we envisage become fully encapsulated at the cell surface, given the expected position of EGFL (Fig. 1b, Fig. 2a, b).
Fig. 2 |. Cytokine-mediated dimerization of ALK and LTK.
a, b, Structures of the ternary assembly of ALKTG with ALKAL2 (a) and LTKTG with ALKAL1 (b). The putative positions of the EGF-like domains are outlined. c, d, Interaction footprints (blue) for site 1 (755 Å2) and site 2 (630 Å2) in the ALKTG–ALKAL2 complex (c) and interaction footprints (green) for site 1 (740 Å2) and site 2 (580 Å2) in the LTKTG–ALKAL1 complex (d). e, f, Sideviews with interaction footprints for site 3 (red) in ALKTG–ALKAL2 (760 Å2) (e) and LTKTG-ALKAL1 (880 Å2) (f). The front receptor footprint is represented by a dotted outline.
Structure of ALKAL1 and ALKAL2
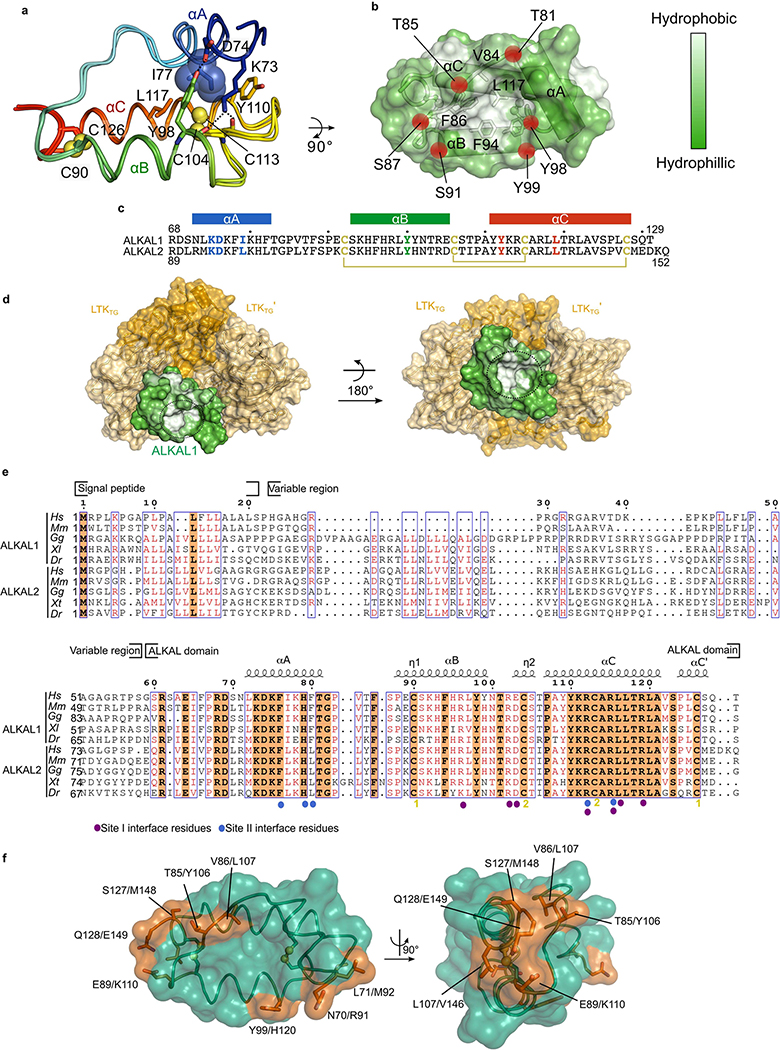
ALKAL1 and ALKAL2 adopt highly similar structures (r.m.s.d. = 0.54 Å, 56 Cα atoms) featuring a novel disulfide-stabilized three-helix bundle. (Extended Data Fig. 5a). The ALKAL1 and ALKAL2 fold is conspicuously open and lacks the classically buried hydrophobic core of helical cytokines. Rather, helix A merely buries Ile77 into a hydrophobic pocket formed by Leu117, Tyr110 and the Cys104–Cys113 disulfide in the BC loop (Extended Data Fig. 5a).
Despite their engulfment by the dimerized receptors, ALKAL1 and ALKAL2 harbour solvent-exposed hydrophobic cavities lined by conserved residues along the internal BC face (Leu117, Phe94 and Tyr98) and the AB loop (Val84 and Phe86) and hydroxyl-containing residues around the outer rim (Extended Data Fig. 5b–d). The strong conservation of all core residues suggests the universality of this fold among vertebrate ALKAL1 and ALKAL2 proteins (Extended Data Fig. 5e). The few sequence differences map to distinct patches contributed by the end of helix C and the AB loop, and might dictate cytokine specificity (Extended Data Fig. 5f).
ALK– and LTK–cytokine interactions
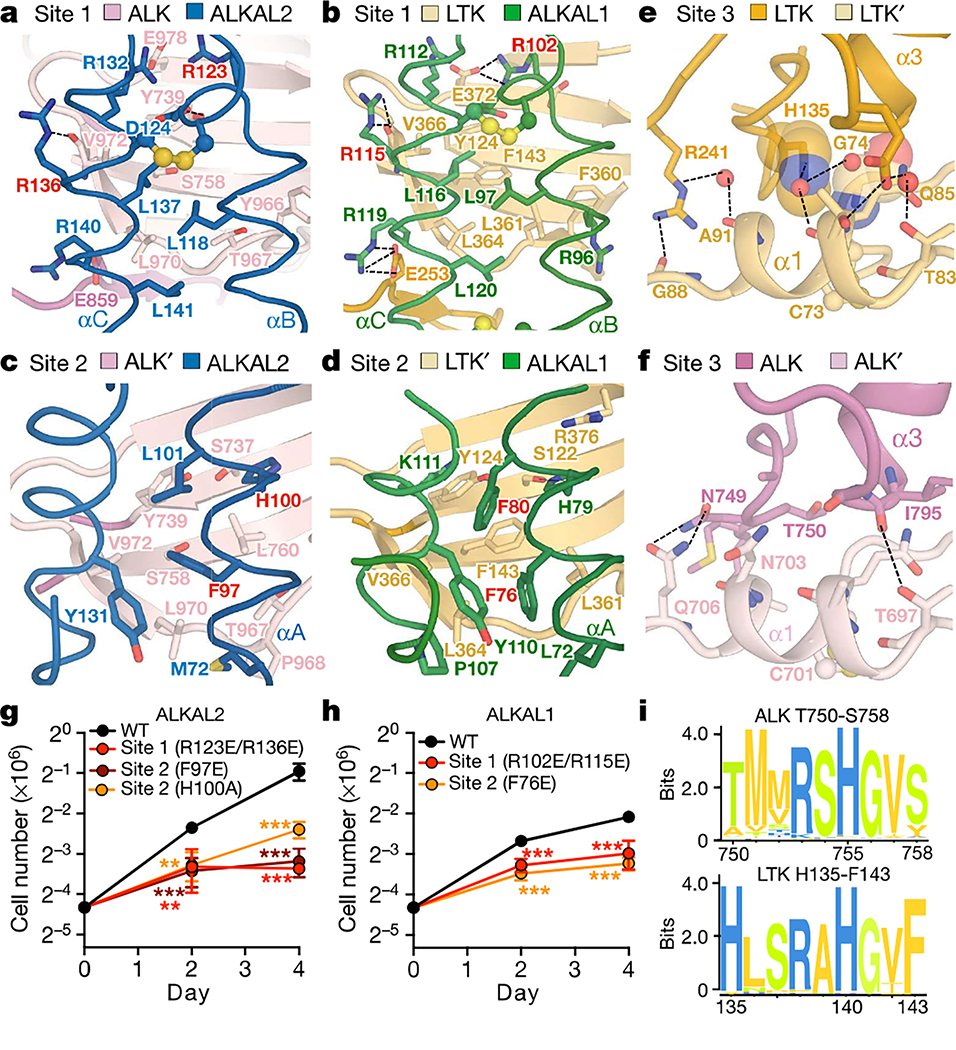
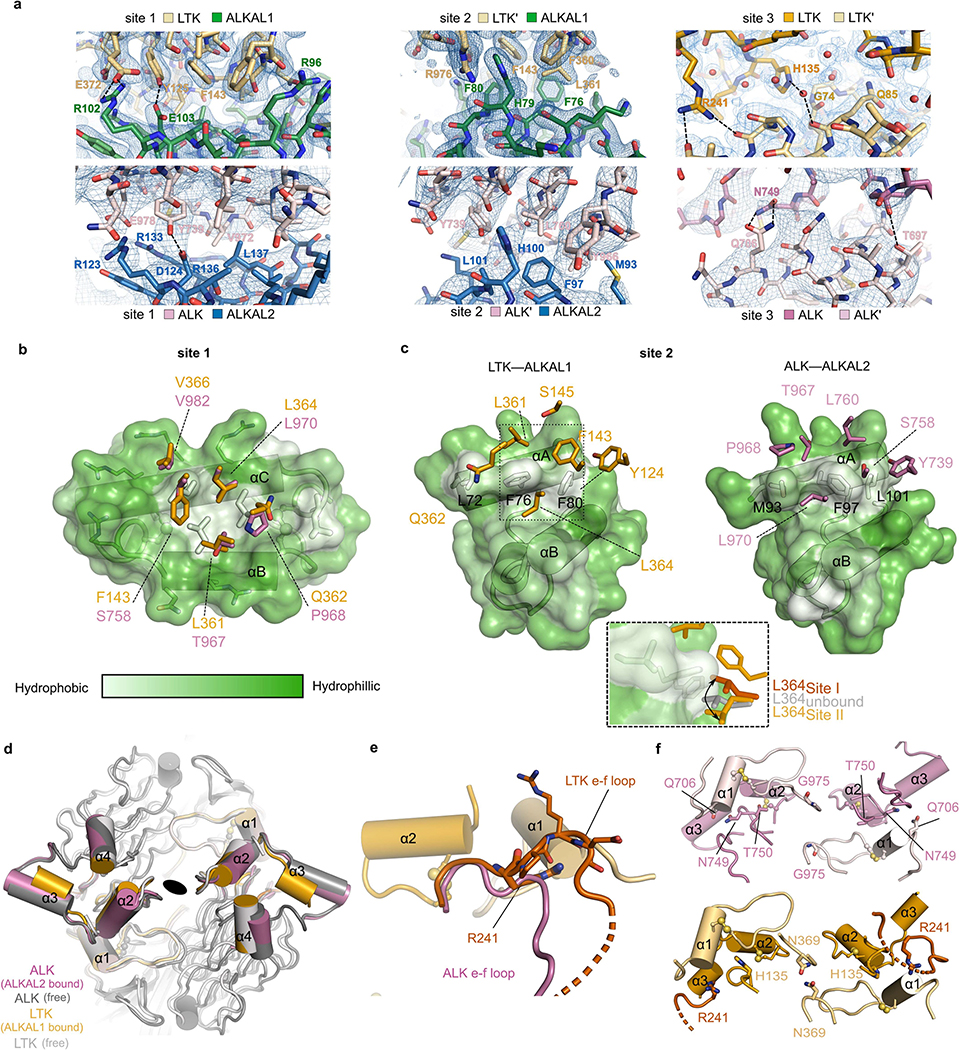
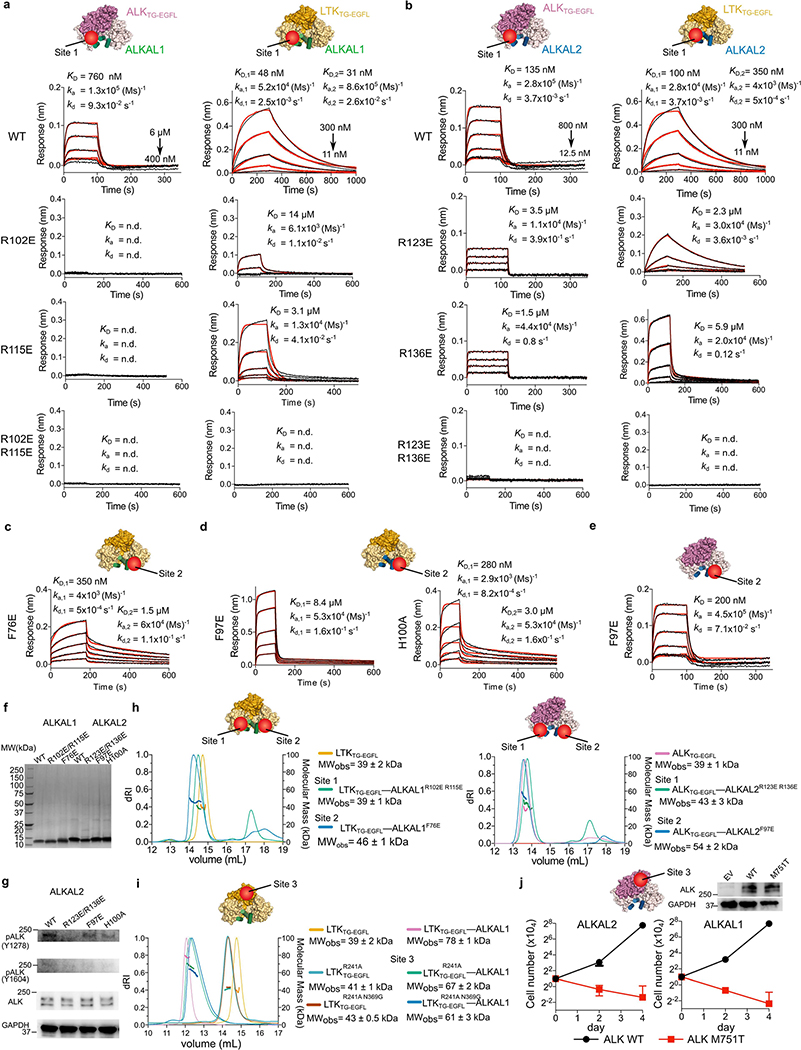
In site 1, ALKAL1 and ALKAL2 use a hydrophobic patch surrounded by arginine residues on helix B and helix C to contact one receptor molecule via its TNFL subdomain (Fig. 3a, b, Extended Data Fig. 6a, Extended Data Table 2). Here, LTKTNFL accommodates ALKAL1 via a conserved hydrophobic platform (Phe143, Leu361, Leu364 and Val366) (Fig. 3b), whereas ALKTNFL uses a rather amphipathic patch to interact with ALKAL2 (Ser758, Thr967, Leu970 and Val972) (Fig. 3a, Extended Data Fig. 6b).
Fig. 3 |. ALK– and LTK–cytokine complexes harbour three distinct interaction sites.
a–f Site 1 interactions in ALK–ALKAL2 (a) and LTK–ALKAL1 (b) complexes. Site 2 interactions in ALK–ALKAL2 (c) and LTK–ALKAL1 (d) complexes. Site 3 interactions in the LTK–ALKAL1 (e) and ALK–ALKAL2 (f) complexes. Residues investigated by mutagenesis are shown in red. g, h, Proliferation of Ba/F3 cells expressing wild-type (WT) ALK with wild-type or R123E/R136E, F97E or H100A mutants of ALKAL2 (g), or with wild-type or R102E/R115E or F76E mutants of ALKAL1 (h). n = 3 biologically independent experiments; mean ± s.d.; two-way analysis of variance (ANOVA) with Tukey’s multiple comparison test compared with the wild type. *P < 0.05, **P < 0.01 and ***p< 0.001; Exact P-values are provided in Source Data, i, Sequence motif analysis of ALKT750–S758 and LTKH135–F143 derived from 248 and 224 vertebrate sequences.
In site 2, the short helix A in ALKAL1 and ALKAL2 and the tip of the BC hairpin engage the second receptor molecule (Fig. 3c, d). In the LTKTG–ALKAL1 complex, Phe80 and Phe76 protrude from helix A in ALKAL1 into a conserved hydrophobic pocket on LTK (Tyr124, Phe143 and Val366). However, site 2 in the ALKTG–ALKAL2 complex is substantially less hydrophobic because ALKTG projects Ser758 instead of a phenylalanine (Extended Data Fig. 6c). Thus, although the locations of sites 1 and 2 on LTK and ALK are identical, these sites are less hydrophobic in ALK.
Cytokine-mediated dimerization of ALKTG and LTKTG results in receptor–receptor contacts via site 3 by the locking of α1 of the TNFL subdomain in one receptor with α2 and α3 at the distal end of the GR subdomain in the second receptor (Fig. 2e, f, Extended Data Fig. 6d). However, the site 3 interface in ALK is markedly less extensive (Fig. 3e, f, Extended Data Fig. 6d–f).
Site 1 drives ALK– and LTK–cytokine complexes
To obtain insights into the contribution of the three interaction interfaces, we interrogated polar interactions in site 1 and the hydrophobic pocket in site 2, and receptor–receptor contacts in site 3.
Following identification of ALKAL2 as the high-affinity cytokine for ALK and ALKAL1 for LTK, we inserted charge-reversal mutations of two conserved polar contacts in the cytokines at site 1 (ALKAL1(R102E), ALKAL1(R115E), ALKAL1(R102E/R115E), ALKAL2(R123E), ALKAL2(R136E) and ALKAL2(R123E/R136E)), which markedly reduced the affinity for both receptors (Extended Data Fig. 7a, b). The effect of site 2 mutations ALKAL1(F76E), ALKAL2(F97E) and ALKAL2(H100A) on binding to LTK was less pronounced, although still substantial (Extended Data Fig. 7c, d). By contrast, ALKAL2(F97E) bound to ALK in a similar manner to wild-type ALKAL2 (Extended Data Fig. 7e). Nevertheless, all mutants markedly impaired Ba/F3 cell proliferation via ALK (Fig. 3g, h, Extended Data Fig. 7f, g). Of note, site 2 mutants retained their ability to form 1:1 stoichiometric complexes with both ALK and LTK, whereas site 1 mutants did not (Extended Data Fig. 7h), indicating that site 1 engagement might drive ALK/LTK–cytokine encounter complexes.
Site 3 mutants in LTK (LTK(R241A) and LTK(R241A/N369G)) did not undergo ALKALl-mediated dimerization (Extended Data Fig. 7i). To probe site 3 in ALK, we expressed ALK(M751T) in Ba/F3 cells, hypothesizing that this mutation would introduce an N-glycosylation site at Asn749. We found that ALK(M751T) did not drive cytokine-dependent cellular proliferation, although it was expressed at similar levels to wild-type ALK (Extended Data Fig. 7j).
ALK appears less well-equipped to fully harness contributions from sites 2 and 3 for its dimerization. Most notably, it lacks equivalents of Phe143 and His135 in LTK (Fig. 3i, Extended Data Fig. 8a) and a critical insert in the e–f loop (Extended Data Fig. 6e, f). Nevertheless, the conservation of interfacing residues at the three interaction sites supports a common binding mode among all vertebrate ALK family orthologues (Extended Data Figs. 5a, 8a, b), consistent with reported species cross-reactivity of zebrafish ALKAL2a and human ALKALs21. Finally, amino acids on the opposite side of the ALK cytokine-binding interfaces are well conserved, in contrast to those in LTK, suggesting their relevance for interactions with the N-terminal domains, which are absent in LTK (Extended Data Fig. 8c).
Mapping of somatic mutations in ALK
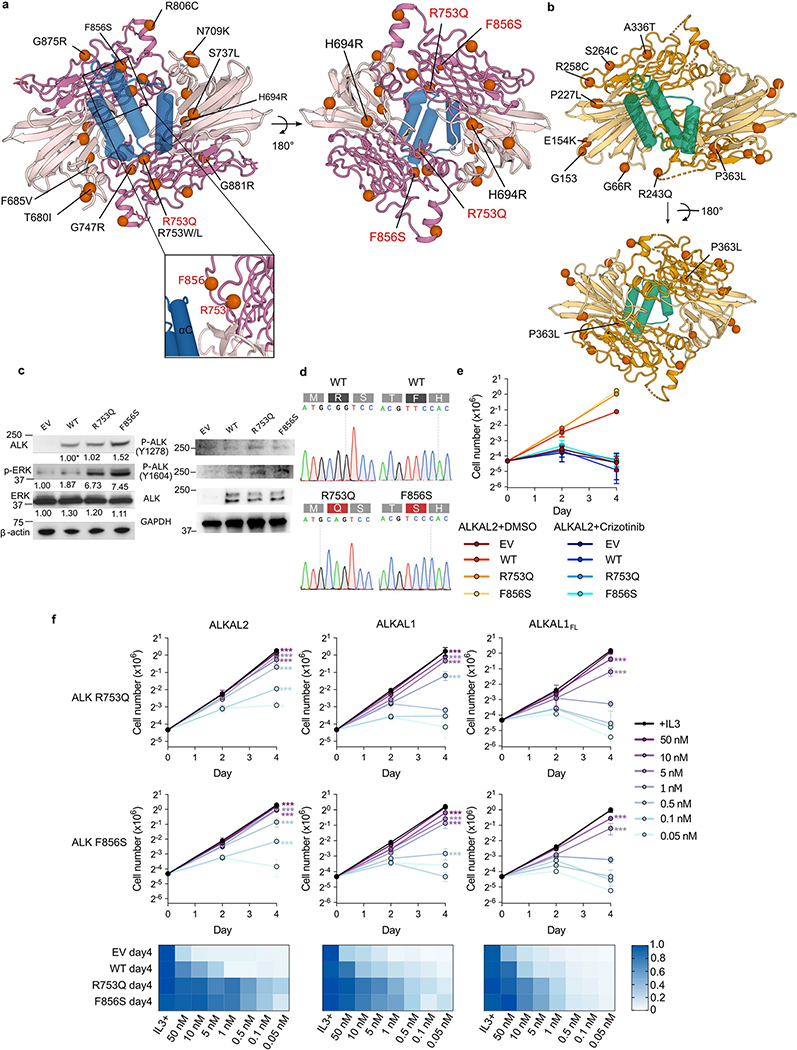
To broaden structure–function insights and to contextualize possible disease-relevant ALK and LTK variants, we structurally mapped mutations in ALKTG and LTKTG, combining documented oncogenic potential and frequently occurring missense single-nucleotide polymorphisms (Extended Data Fig. 9a, b). Mutations leading to constitutive receptor activation, enhancement of receptor–receptor contacts or stabilization of active receptor states are widely studied to evaluate oncogenic potential. LTK variant R243Q and ALK variants G685R, G747R and H694R are localized at positions that would be compatible with such roles. Notably, H694R is a gain-of-function mutation in lung adenocarcinoma leading to constitutive activation of ALK22. Mutations that might affect cytokine binding–for example, by increasing affinity–are less common. Here we identify two such candidate mutations: LTK variant P363L and ALK variant S737L, which map to interaction sites 2 and 1, respectively.
ALK(F856S), a gain-of-function mutation linked to acute myeloid leukemia23, and ALK(R753Q) identified in histiocytic neoplasms24, are roughly equidistant from the start of helix C of bound ALKAL2 (Extended Data Fig. 9a). Expression of ALK(F856S) and ALK(R753Q) in Ba/F3 cells resulted in markedly increased cytokine-dependent cell proliferation (Extended Data Fig. 9c–f). In contrast to an earlier study23, we found that ALKF856S was not constitutively active. Thus, these mutations might facilitate reorganization of key regions of the cytokine–receptor interfaces.
EGFL dictates ALK cytokine specificity
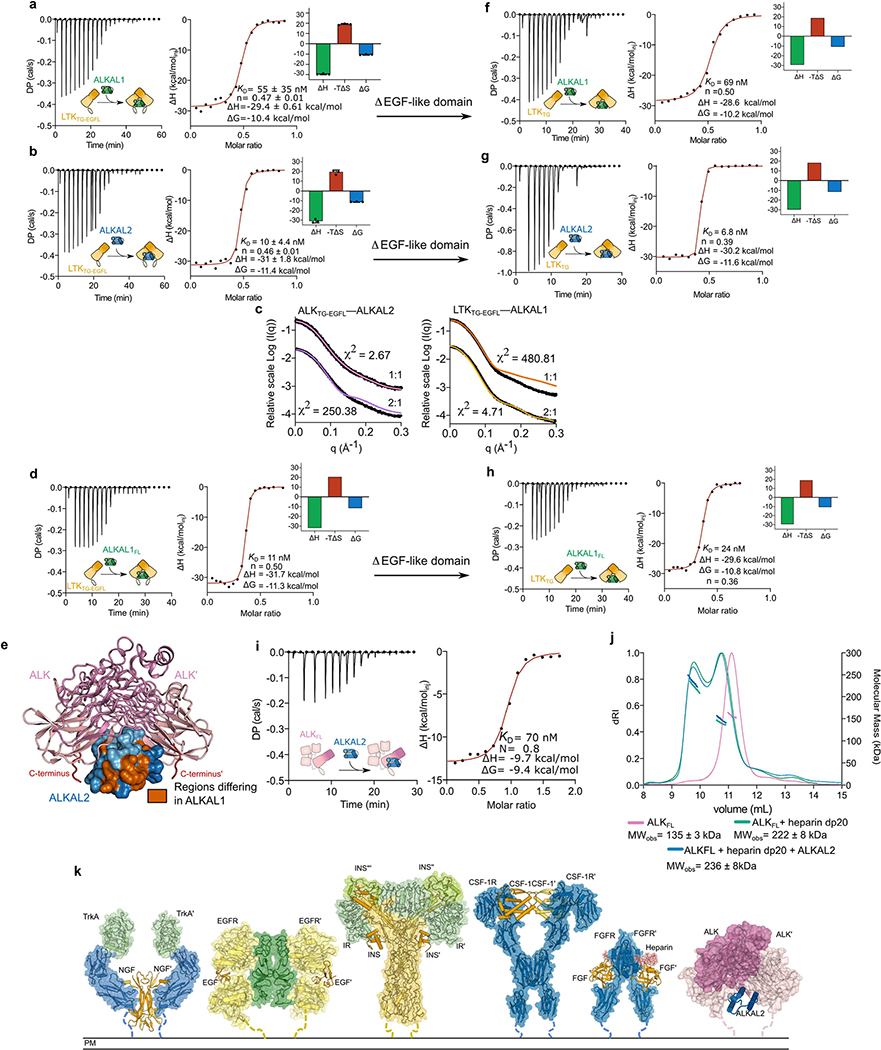
Given the structural similarity of ALKTC– and LTKTG–cytokine assemblies and the ligand-binding capacities of EGFL modules25, we hypothesized that the membrane-proximal EGFL domain might underlie the preference of ALK for ALKAL2 over ALKAL1. Benchmarking of binding thermodynamics showed that even at micromolar concentrations, ALKTG–EGFL formed enthalpically driven binary complexes with either cytokine characterized by a markedly higher affinity for ALKAL2 (dissociation constant (KD) = 40 nM) than for ALKAL1 (KD = 600 nM) (Fig. 4a, b). By contrast, LTKTG–EGFL bound with high affinity and 2:1 stoichiometry to both cytokines (KD,ALKAL1 = 10 nM, KD,ALKAL2 = 55 nM) (Extended Data Fig. 10a, b) in agreement with stoichiometries derived from small-angle X-ray scattering (Extended Data Fig. 10c). The affinity of ALKAL1FL for LTKTG–EGEL was only modestly higher than that of the truncated form (Extended Data Fig. 10d), indicating that the conserved C-terminal part of ALKAL1 determines receptor engagement.
Fig. 4 |. The EGF-like domain dictates cytokine specificity in ALK.
a–d, Isothermal titration calorimetry (ITC) thermograms for the titration of ALKTC–EGFL (10 μM) with ALKAL2 (100 μM) (a), ALKAL1 (25 μM) with ALKTC–ECFL (210 μM) (b), ALKAL2 (33 μM) with ALKTC (330 μM) (c), ALKAL1 (33 μM) with ALKTG (330 μM) (d). Data are mean ± s.d. from three measurements. DP, differential electrical power, e, Proposed assembly mechanism of ligand-mediated complexes in the ALK receptor family.
Sequence differences in ALKAL1 and ALKAL2 map to distinct patches proximal to EGFL (Extended Data Figs. 5b, 10e). Notably, removal of EGFL from ALK reduced the receptor’s affinity for ALKAL2 by 30-fold, but reduced the affinity for ALKAL1 by only 4-fold (Fig. 4d, c); by contrast, the LTKTG domain retained its high affinity to both ALKAL1 and ALKAL2 (Extended Data Fig. 10f–h). Thus, the EGFL domain of ALK drives cytokine specificity and might orchestrate the orientation of the membrane-proximal domains and their connected transmembrane helices to fine tune signal transduction.
Finally, we measured the binding affinity of full-length ALK ectodomain (ALKFL) for ALKAL2. We obtained a similar affinity and 1:1 stoichiometry to the ALKTG–EGFL–ALKAL2 complex, indicating that the four N-terminal domains of ALK are not important for cytokine binding (Extended Data Fig. 10i). Nevertheless, consistent with the interaction of canine ALK with heparin16, heparin induced dimerization of a large fraction of human ALKFL in the presence or absence of bound cytokine (Extended Data Fig. 10j).
Mechanistic considerations
Despite the substantial structural similarity of ALK– and LTK–cytokine complexes, their assembly mechanism appears to be distinct and critically dependent on initial receptor–cytokine interactions via site 1 and the extent of receptor–receptor contacts. LTK–cytokine complexes are fully cytokine-driven, whereas ALK–cytokine assemblies might require the synergy of glycosaminoglycan binding4 and the reduced dimensionality of membrane-proximal engagement (Fig. 4e). The currently known modes of ligand-induced activation of RTKs range from receptor dimerization exclusively driven by the ligand (for example, TRKA–NGF)26 to complexes mediated fully via receptor contacts (for example, EGFR–EGF)27. However, several cytokine–RTK assemblies feature a combination of ligand- and receptor-mediated contributions (for example, CSF-1–CSF-1R)28, including the involvement of accessory molecules (for example, FGF–FGFR)29 or co-receptors, and the use of multiple cytokine copies (for example, insulin receptor)30. The use of a single monomeric cytokine by ALK family receptors to dimerize with two-fold receptor symmetry introduces a novel cytokine-driven assembly mechanism among RTKs (Extended Data Fig. 10k). Our findings are poised to facilitate interrogation of ALK and LTK signalling in physiology and disease, and in the therapeutic targeting of ALK and LTK ectodomains14 and their cognate cytokines13.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41586-021-03959-5.
Methods
No statistical methods were used to predetermine sample size. The experiments were not randomized and the investigators were not blinded to allocation during experiments and outcome assessment. All cell lines used have been tested for mycoplasma contamination.
Uncropped images of protein gel electrophoresis and western blot analyses reported in Extended Data Figs. 2, 4, 7, 9 are provided in Supplementary Fig. 1.
Protein expression constructs for recombinant protein production in mammalian cells
Sequence-optimized DNA for full-length wild-type ALK (Uniprot ID Q9UM73), LTK (Uniprot ID P29376), ALKAL1 (Uniprot ID Q6UXT8) and ALKAL2 (Uniprot ID Q6UX46) were purchased from Genscript. DNA encoding different human ALK constructs comprising amino acids 19–1038 (ALKFL), 648–1038 (ALKTG–EGFL) and 648–985 (ALKTG), and human LTK constructs comprising amino acids 63–420 (LTKTG–EGFL) and 63–378 (LTKTG) were cloned in the pHLsec vector31 in frame with a N-terminal chicken RTPμ-like signal peptide sequence and a C-terminal caspase-3-cleavabIe Fc–His6 tag at the C-terminus.
Sequences encoding ALKAL1FL (residues 28–129), ALKAL1 (residues 57–129) and ALKAL2 (residues 78–152) were cloned in the pHLsec vector in frame with a N-terminal chicken RTPμ-like signal peptide sequence followed by a caspase-3-cleavable Sumostar-tag at the C-terminus. Sequence-optimized DNA encoding the light and heavy chains of Fab324 were purchased from IDT as GBlocks. The N-terminal signal peptide sequences were exchanged for a chicken RTPμ-like signal peptide sequence. The heavy chain was cloned in frame with a C-terminal caspase-3 site followed by an Avi–His6 tag, while the light chain was cloned without a purification tag.
Protein expression in HEK 293 cells and purification from conditioned medium
Production of all ALKTG–EGFL and ALKTG constructs was performed in adherently grown HEK 293 MGAT1−/− cells32 (obtained from P. Reeves, University of Essex, UK) maintained in DMEM supplemented with 10% FCS. Cells with a confluence of 80% were transiently transfected using branched polyethylenimine 25 kDa as transfection reagent in DMEM with 3.6 mM valproic acid without FCS.
Expression of the Fab fragment was achieved in adherent HEK 293T cells (obtained from N. Callewaert, VIB-Medical Biotechnology Center) using the same method. For the heterodimeric Fab fragment, plasmids encoding each chain were co-transfected in a 1:1 ratio.
Protein production for ALKFL, ALKAL1, ALKAL2 and LTK constructs was performed in HEK 293S (obtained from N. Callewaert) cells grown in suspension and maintained in a mixture of 50% Freestyle (Thermofisher), 50% Ex-Cell (Sigma-Aldrich) growth medium. Transient transfection was performed with linear polyethylenimine 25 kDa (Polysciences) as transfection reagent. One day after transfection valproic acid was added33 to a final concentration of 1.5 mM.
For expression in suspension cells, conditioned medium was collected after 5 days and subsequently clarified by centrifugation for 12 min at 8,000g while medium from adherently grown cells was collected after 6 days and centrifuged for 15 min at 6,000g. After centrifugation media were filtered through a 0.22-μm filter prior to chromatographic purification steps.
ALK and LTK proteins were captured via their Fc tag on a protein A column (HiTrap Protein AHP, Cytiva) followed by on-column digestion with caspase-3 for 1 h at 37 °C and an additional 2-h incubation at room temperature, and were finally eluted with HBS (20mM HEPES, pH 7.4, 150 mM NaCl). The tag-free proteins were concentrated and injected onto a HiLoad 16/600 SD200 (Cytiva) size-exclusion chromatography column pre-equilibrated with HBS. Purified proteins were stored at −80 °C until further use.
ALKAL-containing medium was diluted fourfold with 20mM HEPES pH 7.4 before loading on a cation-exchange column packed with SP Sepharose Fast Flow resin (Cytiva) equilibrated in 20 mM HEPES, pH7.4, 50 mM NaCl. ALKALs were eluted using a NaCl gradient from 50 mM–750 mM for 20 min. Fractions containing ALKALs were immediately diluted with 20 mM HEPES pH7.4 to a NaCl concentration of 200 mM and further supplemented with 0.1% (w/v) CHAPS. The C-terminal Sumo-tag was cleaved with caspase-3 overnight at 20 °C. To remove undigested protein as well as the cleaved Sumo tag, the digestion mixture was loaded on a MonoQ 5/50 GL column (Cytiva). Flowthrough containing the ALKALs was concentrated and injected on a Superdex 75 Increase 10/300 GL equilibrated in HBS supplemented with 0.1% (w/v) CHAPS. Purified ALKALs were stored at −80 °C at a concentration of 1 mg ml−1 until further use.
For ALKALs used in BaF/3 assays, endotoxin levels were measured using a Endosafe PTS limulus amoebocyte lysate assay (Charles River) and were below 5 EU mg−1.
Fab fragments were captured using complete His-tag purification resin (Roche) and eluted using HBS supplemented with 250 mM imidazole followed by buffer exchange to HBS on a HIPrep 26/10 desalting column. Caspase-3 was added to the purified Fab fragment to remove the Avi-His6 tag of the heavy chain by overnight digestion at 20 °C. The sample was loaded on an immobilized metal affinity column (IMAC) to remove the enzyme and undigested protein. The flow-through containing tagless Fab fragments was concentrated and injected on a Superdex 200 Increase 10/300 GL (Cytiva) column pre-equilibrated in HBS.
Production of a non-neutralizing single domain camelid nanobody against LTK
Single domain camelid nanobodies against LTK were raised by immunizing llamas with LTKTG–EGFL and were selected for specific binding to LTKTG–EGFL via ELISA and BLI. Epitope binning via biolayer interferometry (BLI) led to the identification of candidate nanobodies with non-neutralizing behaviour with respect to cytokine binding.
The sequence of non-neutralizing nanobody Nb 3.16 was cloned in a MoClo-derived yeast expression vector in frame with an N-terminal preproMF secretory leader sequence followed by the N-terminal His6 tag and a caspase cleavage site. Komagataella phaffii cells were transformed by electroporation and grown on yeast extract peptone dextrose agar with sorbitol, containing 500 μg ml−1 zeocin. One colony was picked to inoculate 500 ml of buffered glycerol-complex medium supplemented with 100 μg ml−1 zeocin and grown at 28 °C for 24 h. Next, cells were pelleted by centrifugation at 500g for 7 min and resuspended in 500 ml buffered methanol-complex medium without zeocin and incubated overnight at 28 °C. Expression was further induced by adding 2.5 ml of 50% methanol, followed by addition of the same volume of methanol after 8 h and 24 h. The cells were subsequently incubated for another 8 h at 28 °C and conditioned medium was collected by centrifugation for 10 min at 6,000g.
His-tagged camelid single-domain nanobodies were captured by addition of 2 ml cOmplete resin (Roche) to 500 ml conditioned medium followed by overnight incubation at 4 °C while shaking. Nanobodies were eluted in HBS supplemented with 250 mM imidazole and buffer exchanged to HBS on a HiPrep 26/10 desalting column (Cytiva). The N-terminal His6 tag was removed by an overnight digestion with caspase-3 at 20 °C. Undigested protein and the enzyme were removed by IMAC. The flowthrough containing tagless nanobody was concentrated and injected on an SD75 Increase 10/300 GL column (Cytiva) pre-equilibrated in HBS. Purified nanobody was concentrated to a concentration of 4 mg ml−1 and flash-frozen and stored at −80 °C.
Crystallization and crystal structure determination
For ALKTG–EGFL–Fab324 crystals, a 1.5 molar excess of Fab324 was added to ALKTG–EGFL and N-linked glycans were digested overnight with EndoH. The complex was purified by size-exclusion chromatography (SEC) on a Superdex 200 Increase 10/300 GL (Cytiva) and concentrated to 13.5 mg ml−1. Commercial sparse-matrix crystallization screens were set up using a Mosquito liquid handling robot (TTP Labtech) in sitting-drop format using 100 nl protein mixed with 100 nl mother liquor in SwissSci 96-well triple drop plates incubated at 287 K. A first hit in the Morpheus II screen34 was further optimized to a condition consisting of (40 mM polyamines, 0.1 M Gly–Gly/2-amino-2-methyl-1,3-propanediol pH 8.5, 11% w/v PEG4000, 19 % w/v 1,2,6-hexanetrioI) in sitting-drop format with a 100 nl protein mixed with 200 nl mother liquor geometry. Crystals were cryoprotected in mother liquor containing 25% w/v 1,2,6-hexanetriol prior to being cryocooled in liquid nitrogen. Diffraction data was collected at 100 K at the ID23-2 beam line at ESRF, Grenoble. The datasets were processed using XDS35. Initial phases were calculated by molecular replacement with PHASER36 using the coordinates of a Fab fragment exhibiting the highest sequence identity (PDB: 5NUZ, chain A) followed by rigid body refinement in Buster37. A partial polyalanine model was built into the visible electron density in Coot38 followed by density modification via Resolve39. The density-modified map showed density for several aromatic side chains allowing for tracing of the correct ALK sequence. Additional refinement steps were carried out in PHENIX40 using individual B-factor refinement in combination with TLS, XYZ refinement, optimizing the X-ray/geometry weights as well as local torsion angle non-crystallographic symmetry restraints.
For crystals of ALKTG–EGFL, glycans were trimmed by overnight enzymatic digestion with EndoH in HBS. The complex was polished via SEC and concentrated to 10.5 mg ml−1. Commercial sparse-matrix crystallization screens were set up as described above. One hit was obtained in the Morpheus II screen and optimized to 0.5 mM manganese (II) chloride tetrahydrate, 0.5 mM cobalt (II) chloride hexahydrate, 0.5 mM nickel (II) chloride hexahydrate, 0.5 mM zinc acetate dihydrate, 13% w/v PEG 3000, 28% v/v 1,2,4-butanetriol, 1% w/v non-detergent sulfobetaine (NDSB) 256 in hanging-drop format. Crystals were cryoprotected in mother liquor containing 25% 1,2,4-butanetriol prior to being flash frozen in liquid nitrogen. Diffraction data were collected at 100 K at the P14 microfocus beam line at PETRA III, Hamburg and integrated using XDS with standard parameters except for the BEAM_DIVERGENCE parameter which was doubled. Initial phases were obtained using maximum likelihood molecular replacement in Phaser using the structure of the ALKTG domain. The structure was refined using Phenix.refine followed by manual building in COOT. The EGF-like domain was manually built into the electron density. Refinement in phenix followed the same protocol as for the ALKTG–EGFL–Fab324 structure except for the absence of non-crystallographic symmetry restraints and implementation of additional reference restraints based on the structure of ALKTG in complex with Fab324.
For crystals of LTKTG, purified LTKTG was concentrated to 10 mg ml−1 and used to set up sparse matrix screens as previously described. Crystals appeared in a condition of the Morpheus II screen with the final optimized condition (MOPSO/Bis-Tris pH 6.3, 12% PEG 20000, 26% trimethyl propane 1% w/v NDSB 195 5 mM yttrium (III) chloride hexahydrate, 5 mM erbium (III) chloride hexahydrate, 5 mM terbium (III) chloride hexahydrate, 5 mM ytterbium (III) 1 chloride hexahydrate) set up in sitting-drop format with a 100 nl protein mixed with 200 nl mother liquor geometry. For data collection, single crystals were cryoprotected in mother liquor and cryo-cooled in liquid nitrogen. Diffraction data were collected at 100 K at the P13 microfocus beam line at PETRAIII, Hamburg and processed using XDS as previously described. Phases were obtained by single-wavelength anomalous dispersion making use of the anomalous signal from lanthanide atoms. Determination of the lanthanide substructure for four sites was performed by the hybrid substructure search as implemented in Phenix. Phases were calculated using Phaser-EP. The experimental electron density was readily interpretable, and a model was manually built in Coot and further refined in Phenix implementing an anisotropic individual B-factor model.
For LTKTG–ALKAL1–Nb3.16 crystals, a threefold molar excess of Nb3.16 was added to the LTKTG–ALKAL1 complex, concentrated and injected into a Superdex 200 Increase 10/300 GL (Cytiva) equilibrated in HBS. Eluted fractions were concentrated to 13.5 mg ml−1 and used to set up sparse-matrix crystallization screens in sitting-drop format as described above. Crystals were obtained in a condition consisting of MOPSO/Bis-Tris pH 6.3, 13% PEG 8000, 22% w/v 1,5-pentanediol, 5 mM sodium chromate tetrahydrate, 5 mM sodium molybdate tetrahydrate, 5 mM sodium tungstate tetrahydrate, 5 mM sodium orthovanadate tetrahydrate. Crystals were cryoprotected in mother liquor containing 25% 1,2,4-butanetriol and were cryo-cooled in liquid nitrogen. Diffraction experiments were performed at 100 K Proxima 2 (Soleil Synchrotron). Initial phases were obtained by maximum-likelihood molecular replacement using Phaser with the previously obtained LTKTG structure as a search model. A model for Nb3.16 was automatically built using Arp Warp41, 42 followed by manual building of ALKAL1 in Coot. Refinement was performed in Phenix with individual anisotropic ADP parameters with a TLS model.
For ALKTG–ALKAL2 crystals a threefold molar excess of ALKAL2 was added to ALKTG and subjected to an overnight EndoH digest, concentrated and injected into a Superdex 200 Increase 10/300 GL (Cytiva) equilibrated in HBS. Eluted fractions were concentrated to 8 mg ml−1 and used to set up sparse-matrix crystallization screens in sitting-drop format as described above. Initial hits were obtained in a condition consisting of 0.1 M MES pH 6.5, 15% w/v PEG 6000, 5% w/v MPD. A single crystal was used to prepare a seed stock43 using the PTFE seed bead (Hampton Research). The best diffracting crystals were obtained by seeding with a 1:1,000 dilution of the seed stock into the optimization screen. Crystals were cryoprotected in 78% (v/v) mother liquor supplemented with 22% (v/v) ethylene glycol before flash freezing in liquid nitrogen. Diffraction data were collected at 100 K at the Proxima 2 microfocus beam line at the Soleil synchrotron. Data as processed as described above with the difference that anisotropy correction was implemented by the UCLA diffraction anisotropy server44. Initial phases were obtained by molecular replacement in Phaser using the previously determined ALKTG and ALKAL1 structures. Initial refinement cycles were performed in Buster followed by iterative refinement using Coot and Phenix. B-factors were refined using two isotropic atomic displacement parameters complemented by TLS. During refinement structures of ALKTG and ALKAL1 provided reference model restraints. For Fab324 crystals, tag-free Fab324 was concentrated to 18.5 mg ml−1 and a pH versus (NH4)2SO4 concentration screen was set up in sitting drop format resulting in crystals in a condition consisting of (1.5 M (NH4)2SO4,40 mM glycine pH 9.5). The crystal was cryoprotected using a saturated (NH4)2SO4 solution. Diffraction experiments were performed at the P13 beamline at PetraIII, Hamburg and data were processed using XDS. Phases were obtained by molecular replacement using our structure of Fab324 obtained from the ALKTG–EGFL–Fab324 complex. The structure was refined using Coot and Phenix.
All display items containing structures were generated using the PyMOL Molecular Graphics System, version 2.0.5 (Schrödinger).
Isothermal titration calorimetry
Experiments were performed using a MicroCal PEAQ-ITC instrument at 310 K. Proteins used in ITC experiments were expressed in HEK 293S cells grown in suspension. As a final purification step all proteins were buffer exchanged to the same HBS buffer via size-exclusion chromatography. Titrations were preceded by an initial injection of 0.5 μl. The injection spacing was optimized per experiment to allow for the signal to get back to a stable baseline. Throughout the titration the sample was stirred at a speed of 750 rpm. Data were analysed using PEAQ-ITC analysis software (version 1.1.0.1262, Malvern) and fit using a ‘one set of sites’ model.
Multi-angle laser light scattering
Protein samples of 100 μl at approximately 1.0 mg ml−1 were injected onto a Superdex 200 increase 10/300 GL column (Cytiva) connected in line to a UV-detector (Shimadzu), a miniDawnTReos (Wyatt) Multi-angle laser light scattering (MALLS) detector and an Optilab T-Rex (Wyatt) refractometer. The refractive increment value (dn/dc) was 0.185 ml g−1. Band broadening was corrected for using reference measurements of BSA (Pierce). Data analysis was carried out using the Astra 6.1.6 software and standard deviations were calculated using Prism8 (Graphpad Software).
Biolayer interferometry
Benchmarking of binding kinetics and screening of mutant ALKALs were performed by immobilizing wild-type and mutant ALKAL1/2 variants. To this end, wild-type ALKAL1 (residues 57–129) and mutant variants (R102E, R115E, R102E/R115E, F76E and F80E) as well as wild-type ALKAL2 (residues 78–152) and interface mutants (R123E, R136E, R123E/R136E, F97E and H100A) were cloned in the pHLsec vector in frame with a C-terminal Avi tag. All constructs were transiently co-transfected in suspension-grown HEK 293S cells together with a BirA expression plasmid (pDisplayBirA-ER45) as previously described and supplemented with 100 μM biotin upon transfection. After 4 days of expression, excess biotin was removed by desalting the conditioned media to HBS on a HIPrep 26/10 desalting column (Cytiva).
All measurements of binding kinetics and dissociation constants were performed using an Octet Red 96 (Forté Bio) in assay buffer (20 mM HEPES pH 7.4, 150 mM NaCl, 0.02% (w/v) BSA, 0.002% (v/v) Tween 20) at 298 K. ALKALs were immobilized to a level of 0.5 nm on streptavidin-coated biosensors (Forté Bio). To verify that no nonspecific binding was present during the assay, non-functionalized biosensors were used as a control by measuring in parallel all ligand concentrations as well as running buffer. For all mutants a two fold dilution series from 6.4 μM–400 nM was employed. Data analysis was performed using Data Analysis software 9.0.0.14 (Forté Bio) and binding curves were exported to Prism8 (Graphpad Software) for plotting of curves.
Small angle X-ray scattering
SEC–small angle X-ray scattering (SAXS) data were collected at the SWING beamline at Soleil (France) using an integrated online HPLC set-up. Purified samples of ALKTG–EGFL–ALKAL2 (18.5 mg ml−1) and LTKTG–EGFL–ALKAL1 (19 mg ml−1) expressed in HEK 293S MGAT1−/− cells were injected on a Biosec-3 column (Agilent) with HBS as a running buffer. The scattering data were collected in continuous flow mode with a flow speed of 0.3 ml min−1 and a 1-s exposure time per frame. Buffer and sample frames were selected and subtracted using the program RAW46. Theoretical scattering curves and fitting to experimental scattering data was performed with AllosMod-FoXS.
In brief, a model for the C-terminal EGFL domain of LTK was generated by homology modelling starting from the crystal structure of the ALK EGF-like domain using the SWISS-MODEL server47. This model was manually placed and connected to the C terminus of the LTKTG domain in Pymol based on the ALKTG–EGFL structure. Missing regions in the ALK and LTK structures were added using MODELER48. The resulting models were subsequently energy-minimized using Rosetta-Relax, and used as an input for AllosMod to add N-linked glycans at positions Asn709, Asn808, Asn886 and Asn986 for ALK and Asn380 and Asn412 for LTK, and the resulting model energy landscapes were calculated. The output of AllosMod was then used in AllosMOD-FoXS to calculate fits with theoretical scattering curves during fast AllosMod simulations at 300 K.
Cell culture and retroviral transduction
Ba/F3 (mouse pro-B cell line) cells were cultured in RPMI/10% FCS supplemented with 1 ng ml−1 mouse interleukin-3 (IL-3). Ba/F3 cell line was not listed in the database of commonly misidentified cell lines maintained by ICLAC and NCBI Biosample. Ba/F3 cells were transduced with murine stem cell virus (MSCV) supernatant from MSCV expressing ALK(WT), ALK(R753Q), ALK(F856S) or EV-IRES-GFP (where EV is empty vector) for 2 days in RPMI, 10% FCS supplemented with mouse IL-3 as previously described49. GFP-sorted cells were used for the cell growth assays and western blots. After removal of IL-3 from the medium, the cells were stringently washed with PBS for three times. The cell growth curves and heat maps were made using GraphPad Prism 9 software as mean values, with error bars representing standard deviation.
Reagents
For western blotting, the following antibodies were used: ALK (Purchased from Cell Signaling Technologies; catalogue no. 3633; dilution: 1:1,000), phospho-ALK (Tyr1278) (Cell Signaling Technologies; 6941; 1:250), phospho-ALK (Tyr1604) (Cell Signaling Technologies; 3341; 1:250), phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) (Cell Signaling Technologies; 4370; 1:2,000), p44/42 MAPK (Erk1/2) (Cell Signaling Technologies; 9102; 1:1,000), β-actin (Sigma-Aldrich; A-5441; 1:2,000), GAPDH (GENETEX; GTX100118; 1:2000). For the cell growth assay, Crizotinib (Sigma-Aldrich; PZ0191-5MG), DMSO (Sigma-Aldrich; D8418-100ML), and IL-3 (Peprotech; AF-213-13) were used.
Statistics and reproducibility
Statistical significance was determined by two-way ANOVA followed by Tukey’s multiple comparison test where multiple comparisons should be adjusted. Data were plotted using GraphPad Prism 9 software as mean values, with error bars representing s.d. Heat maps were also made using GraphPad Prism 9 based on mean values. *P < 0.05, **P < 0.01 and ***P < 0.001, unless otherwise specified.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this paper.
Data availability
Coordinates and structure factors for the complexes have been deposited in the Protein Data Bank (PDB) under accessions 7NWZ (ALKTG–ALKAL2), 7NX0 (LTKTG–ALKAL1–Nb3.16), 7NX1 (LTKTG), 7NX2 (unbound Fab324), 7NX3 (ALKTG–EGFL–Fab324), 7NX4 (ALKTG–EGFL). Most common single-nucleotide polymorphisms were obtained from the COSMIC database (https://cancer.sanger.ac.uk/cosmic) and gnomAD database (https://gnomad.broadinstitute.org). The Ba/F3 cell lines expressing ALK(WT), ALK(R753Q) or ALK(F856S) can be obtained from the authors upon request. Source data are provided with this paper.
Extended Data
Extended Data Fig. 1 |. Phylogenetic analysis and evolution of ALK and LTK.
a, Phylogenetic analysis of ALK (a). Multiple sequence alignment was performed with MAFFT 7 using the E-INS-I algorithm for refinement. The phylogenetic tree was visualized using Phyl.io tool as implemented in MAFFT. ALK is an evolutionarily ancient RTK in C. elegans and D. melanogaster, where it is activated by FIEN-150 and Jeb51, 52. b, Sequence alignment of ALK sequences focusing on the heparin binding motif. Positively charged residues are highlighted in blue. The multiple sequence alignment was performed with MAFFT 7 using the E-INS-I algorithm for refinement. c, Phylogenetic analysis of LTK performed as for ALK. In mammals the LTK ectodomain consists of the TG and EGF-like domains while other vertebrates contain the additional MAM-LDLa-MAM domains at the N-terminus.
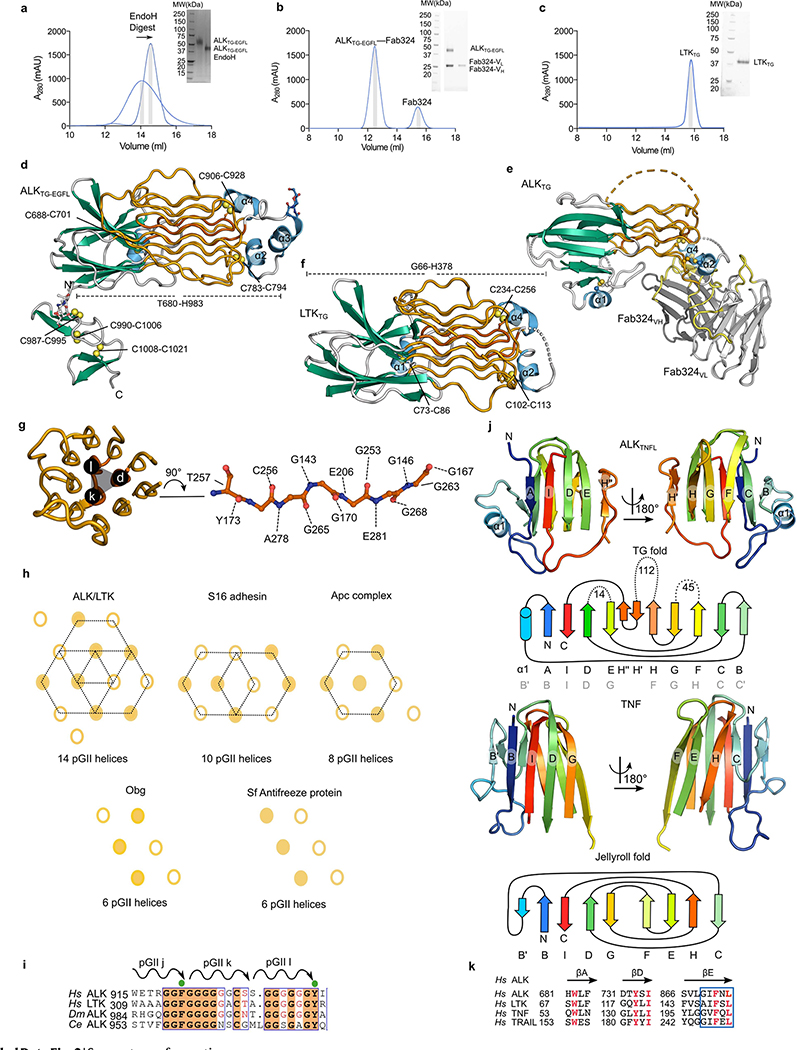
Extended Data Fig. 2 |. Purification of ALKTG-EGEL and LTKTG and structural details of the TNFL and GR subdomains of the novel TG supradomain fold.
a,b,c Representative chromatograms and SDS-PAGE gels for the purification of ALKTG-ECFL (a), ALKTG–GFL–Fab324 (b) and LTKTG (c). The arrow indicates the shift in elution volume after EndoH digest of ALKTG-ECFL. Each protein was purified several times, chromatograms and SDS-PAGE analysis of each sample are representative for different protein batches. Uncropped gels are included in source data, d, ALKTG-ECFL structure colored according to secondary structure elements. α-Helices (blue), β-strands (green), pGII-helices (orange), loops (grey), e, Structure of the ALKTG-ECFL–Fab324 complex with ALK coloured according to its secondary structure elements. CDR loops of Fab324 are coloured yellow. The constant domains of Fab324 are omitted for clarity. f, LTKTG structure colored according to secondary structure elements. g, Hexagonal pGII-helix arrangement surrounding pGII-helix d in LTK. Vermillion pGII-helices consist exclusively of glycine residues. pGII-helix d shown as sticks, hydrogen bonds to other residues in LTK are indicated as dotted lines. The glycine-rich segment has complicated detection of a globular fold but has led to its sequence-based classification as Glycine-rich PFAM domain PF1281053. h, Schematic representations of pGII-helix arrangements in reported structures. Drastically less extensive pGII-helix arrangements than the one displayed in the GR subdomain of ALK and LTK have been observed in synthetic polyglycines20 and four functionally diverse proteins54–57. Full circles indicate pGII-helices coming out of the plane of the page while empty circles indicate helices going into the plane of the page. S16 adhesin (pdb: 6F45) Ape complex (pdb: 5L9W) obg (pdb: 5M04) Sf antifreeze protein (pdb: 3BOI). i, Sequence alignment performed with Clustal Omega of human ALK, human LTK, C. elegans ALK (SCD-2) and D. melanogaster ALK covering pGII-helices j,k and l. Residues conserved across all four species are indicated with an orange background. Conserved hydrophobic residues involved in the hydrophobic groove between the TNF-like and glycine rich region are indicated by a green sphere, j, The β-sheet subregions of ALK and a trimmed view of TNF (pdb: 1TNF) are coloured in a N-(blue) to C-terminus (red) gradient and shown side by side after structural superposition. Topology diagram for the TNFL domain of ALK and the jelly-roll fold of TNF follow the same colour scheme. Jelly-roll fold nomenclature starts with strand B according to convention. For ALKTNFL the nomenclature in black is according to the TG domain notation used in this study while the nomenclature according to the TNF convention (first β-strand labeled B) is shown in grey. Structural queries63 using the TFNL subdomains retrieved TNF/C1q-class folds (r.m.s.d =2.8 Å against C1q and TNF, 72 Cα atoms). Topology-independent searches58 covered an additional −20 residues in the canonical TNF fold, and structure-based sequence alignments clarified the sequence homology between the A, D and E β-strands in ALK/LTKTNFL and β-strands B, E and F in TNF or TRAIL. The distinctly connected β-strands in the ALKTNFL/LTKTNFL subdomain break up the alternating sheet-to-sheet register of the TNF/C1q β-jellyroll, and instead permit the spatially contiguous sprouting of the three glycine-rich loop inserts (between β-strands D and E, F and G, and H and H’) towards the distinctive pGII-helix lattice of the ALKGR/LTKGR subdomain. The sequential B to 1 β-strands of the TNF/C1q β-jelly roll smoothly sew together the two β-sheets (that feature characteristic B’BIDG and FEHC faces) whereas the ALK/LTKTNFL subdomain has AIDEH” and H’HGFCB faces (primed small caps denote additional, edge β-strands). k, Annotated alignment of selected β-strands of human ALK, LTK, TNF and TRAIL. Conserved hydrophobic residues are indicated in red.
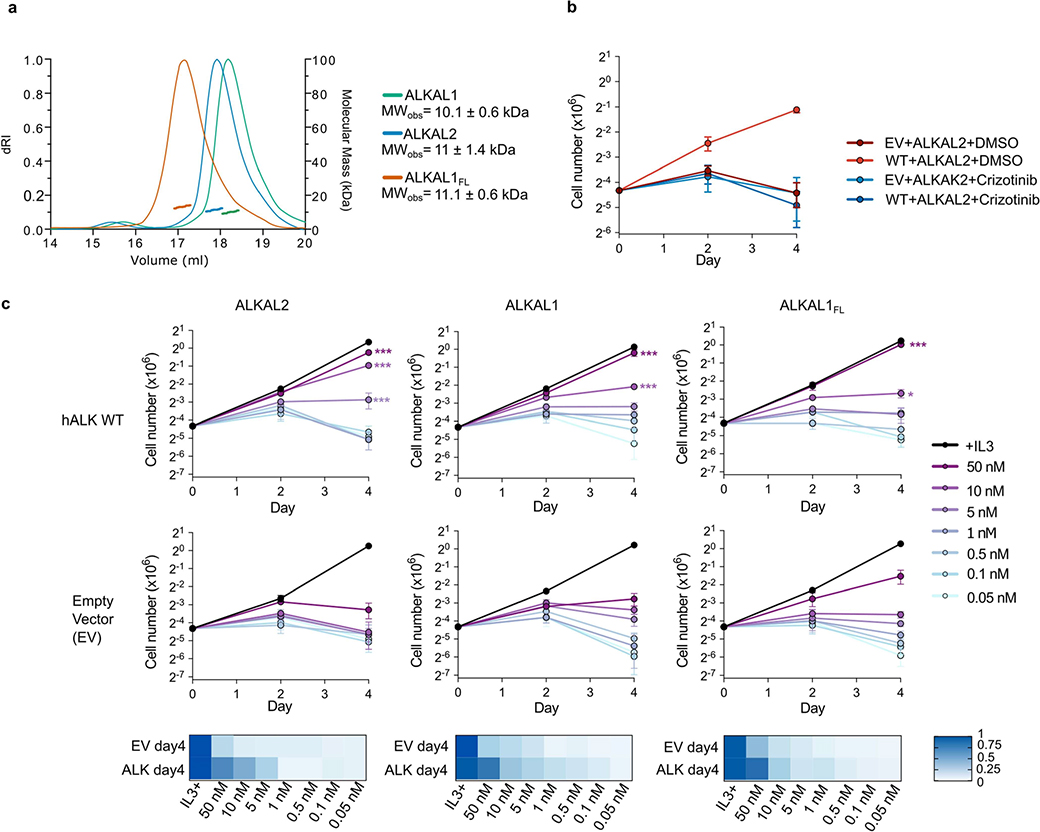
Extended Data Fig. 3 |. ALKAL1- and ALKAL2-dependent proliferation of Ba/F3 cells expressing ALK.
a, Molecular mass determination of ALKAL2, ALKAL1 and ALKAL1FL by size-exclusion chromatography and in-line multi-angle laser light scattering (SEC-MALLS). The differential refractive index (left vertical axis) is plotted against the determined molecular weight (right vertical axis). ALKAL2 (blue trace), ALKAL1 (green trace) and ALKAL1FL (vermillion trace). b, Cell proliferation of ALKWT or EV (empty vector) expressing Ba/F3 cells treated with 10 nM ALKAL2 with and without addition of crizotinib (n = 3 biologically independent experiments; mean ± s.d). c, Cell proliferation of Ba/F3 cells expressing ALKWT or EV upon stimulation by a concentration series of ALKAL2, ALKAL1 or ALKAL1FL at indicated concentrations. (n = 3 biologically independent experiments; mean ± s.d.; two-way ANOVA with Tukey’s multiple comparison test compared with EV; *P < 0.05, **P < 0.01 and ***P < 0.001; Exact P-values are provided in source data. The ratio of the observed ALKAL-induced cell growth with the IL-3 induced cell growth is shown in a heatmap representation for the measurements on day 4). Volume (ml)
Extended Data Fig. 4 |. Biophysical characterization and purification of ALKAL-mediated complexes of ALK and LTK.
a, Calculated molecular masses for the ALK, LTK and ALKAL constructs under study. b-e, Experimental molecular mass determination of ALKAL-mediated complexes with LTKTG-EGFL (b), ALKTG-EGFL (deglycosylated) (c), LTKTG (d), ALKTG (deglycosylated) (e) by SEC-MALLS. The differential refractive index (left vertical axis) is plotted against the experimentally measured molecular mass (right vertical axis). Unbound LTK and ALK are in orange and pink respectively. Complexes with ALKAL1 or ALKAL2 are in green and blue respectively. The reported molecular mass represents the average molecular mass ± s.d. across the elution peak. f,g, Representative SEC elution profiles and SDS-PAGE analysis for the purification of the ALKTG-EGFL–ALKAL2 complex (f) and the LTKTG-EGFL–ALKAL1–Nb3.16 complex (g). Each protein was purified several times, chromatograms and SDS-PAGE analysis of each sample are representative for different protein batches. Uncropped gels are included in source data, h, Surface representation of the LTKTG-EGFL–ALKAL1–Nb3.16 complex illustrating the location of the binding sites of the non-neutralizing Nb3.16 to LTKTG far from any cytokine–receptor or receptor–receptor binding site.
Extended Data Fig. 5 |. Structure of ALKAL1 and ALKAL2.
a, Structural superposition of the conserved C-terminal segments of ALKAL1 and ALKAL2 (coloured in an N- (blue) to C-terminus (red) gradient) as observed in their complexes with cognate receptors. ALKAL1 and ALKAL2 share high sequence identity (66%) in their C-terminal domain, but have variable N-terminal regions. ALKAL1 residues in the interface between helix A and helices B and C are shown as sticks. αA connects via a conserved short loop to a helical hairpin constructed from αB and αC, which in turn are tethered by two conserved disulfides. A conserved stretch of −10 residues preceding αA was not ordered in the reported structures, suggesting they might help to stabilize the soluble forms of these cytokines rather than contribute to direct receptor engagement or might help reduce the entropic cost of binding, b, Surface representation of ALKAL1 coloured according to the Eisenberg hydrophobicity scale. Hydroxyl groups surrounding the central cavity are shown as red spheres, c, Sequence alignment by ClustalOmega of human ALKAL1 with ALKAL2. Residues in the interface between helix A and helices B and C are coloured according to their position. Cysteines are coloured yellow and disulfide bonds are shown as yellow lines, d, Surface representation of the LTKTG–ALKAL1 complex with LTK in ALKAL1 coloured according to the Eisenberg hydrophobicity scale (white is more hydrophobic). The black circle denotes the ALKAL1 hydrophobic cavity, e, Multiple sequence alignments of various vertebrate ALKAL1 and ALKAL2 sequences using the ESPripT server (http://espript.ibcp.fr/ESPript/ESPript/) and structural annotation according to secondary structure elements. Symbols indicate residues participating in interaction sites 1 and 2 according to the graphical legend. Hs (Homo sapiens), Mm (Mus musculus), Gg (Gallus gallus) Xl (Xenopus laevis), Xt (Xenopus tropicalis) Dr (Danio rerio). f, ALKAL1 shown in green transparent surface and ribbon representation. Residues differing with ALKAL2 are colored Vermillion and labeled with the ALKAL1/ALKAL2 numbering.
Extended Data Fig. 6 |. Structural details of receptor–cytokine and receptor–receptor interactions in ALK/LTK–cytokine complexes.
a, 2Fo-Fc electron density maps contoured at +1 r.m.s.d. showing details of site 1,2 and 3 of the LTK–ALKAL1 and ALK–ALKAL2 complexes, b, Transparent surface of ALKAL1 according to the Eisenberg hydrophobicity scale illustrating similarities and differences in site 1 of LTK/ALK–cytokine complexes. Shown is the central conserved hydrophobic patch formed by leucines (L97, L116 and L120) and the interacting residues of LTK (orange). The equivalent ALK residues (pink) are shown after alignment with LTK. c, View of Site 2 in the LTK–ALKAL1 and ALK–ALKAL2 complexes. ALKALs are coloured according to the Eisenberg hydrophobicity scale. Receptor residues surrounding the hydrophobic triad of helix A (L72, F76, F80 in ALKAL1 and M93, F97, L101 in ALKAL2) are shown as sticks for LTK (orange) and ALK (pink). d, Superposition of unbound ALK (dark gray), unbound LTK (light gray), bound ALK (pink, only helices shown) and bound LTK (orange, only helices shown), e, Superposition of the ALK–ALKAL2 and LTK–ALKAL1 complexes, zoomed in on the region around the e-f loop. f, View of the site 3 groove of ALK (top) and LTK (bottom). In LTK, site 3 centers on His153, which stacks against Gly74 and Arg241 on an LTK-specific loop with Asn369 residues hydrogen bonding across the twofold axis of the complex.
Extended Data Fig. 7 |. Functional interrogation of site 1, site 2, and site 3 interfaces in ALK/LTK–cytokine complexes.
a,b Representative response curves as measured by biolayer-interferometry (BLI) for the interaction of wild type ALKAL1 and ALKAL1 mutants (containing charge-reversal mutations of residues involved in site 1) (a) and WT ALKAL2 and ALKAL2 mutants (b) with ALKTG–EGFL and LTKTG–EGFL. For wild type ALKALs LTK curves were fitted with a 2:1 binding model (red) while for ALK a 1:1 model was used. Start and end concentrations of the 2-foid dilution series used for the WT measurements is shown as an inset while for all mutants a 2-fold dilution series from 6.4 μM-400nM was used, c, BLI response curves for the interaction of the site 2 ALKAL1F76E mutant with LTKTG–EGFL. d, BLI response curves for the interaction of the site 2 ALKAL2F97E and ALKAL2H100A with LTKTG–EGFL. e, BLI response curves for the interaction of the site 2 ALKAL2F97E with ALKTG–EGFL. f, SDS-PAGE analysis of purified ALKAL1 and ALKAL2 mutants used in Ba/F3 and SEC-MALLS assays. Each protein was purified several times, SDS-PAGE analysis of each sample are representative for different protein batches. Uncropped gels are included in source data, g, Western blot analysis of phosphorylated ALK (Y1278 and Y1604) after stimulation with ALKAL2WT, ALKAL2R123E/R136E, ALKAL2F97E and ALKAL2H100A. Uncropped western blot scans are provided in source data. h, Capacity of ALKAL1 and ALKAL2 mutants to form complexes with LTKTG-EGFL and ALK TG-EGFL respectively as characterized by SEC-MALLS. Differential refractive index (left axis) is plotted against the determined molecular weight (right axis). LTKTG-EGFL (orange trace), LTKTG-EGFL–ALKAL1R102E/R115E (green trace) and LTKTG-EGFL–ALKAL1F76E (blue trace). ALKTG-EGFL (pink trace), ALKTGEGFL–ALKAL2R123E/R136E (green trace) and ALKTG-EGFL-ALKAL2F97E (blue trace). The ALKAL1 site 1 mutant is unable to form a complex with ALK while the site 2 mutant still forms a binary complex. The reported molecular mass represents the average molecular mass ± s.d. across the elution peak, i, Capacity of the LTKTG-EGFLR241A LTKTG-EGFLR241A/N369G site 3 mutants to form complexes with ALKAL1 as characterized by SEC-MALLS. LTKTG-EGFLR241A (red trace), LTKTG-EGFLR241A/N369G (cyan trace), LTKTG-EGFLR241A–ALKAL1 (green trace), LTKTG-EGFLR241A/N369G–ALKAL1 (blue trace). LTKTG-EGFL (orange trace) and LTKTG-EGFL–ALKAL1 (pink trace) are shown for comparison. The reported molecular mass represents the average molecular mass ± s.d. across the elution peak, j, Cell proliferation of Ba/F3 cells expressing ALKWT or ALKM7S1T upon stimulation with 50 nM ALKAL1 or 50 nM ALKAL2. Western blot analysis of ALKWT or ALKM7SIT expression is a representative of three biologically independent experiments with similar results. Uncropped western blot scans are provided in source data.
Extended Data Fig. 8 |. Conservation of TG supradomains in ALK and LTK.
a, Structurally annotated multiple sequence alignments of the TG and EGFL domains of ALK and LTK using the ESPripT server (http://espript.ibcp.fr/ESPript/ESPript/). Hs (Homo sapiens), Mm (Mus musculus), Gg (Gallus gallus) Xl (Xenopus laevis), Xt (Xenopus tropicalis) Dr (Danio rerio). Symbols indicate residues participating in the different interfaces according to the graphical legend. b, Bottom view of dimerized LTK (left) and ALK (right) in surface representation coloured according to residue conservation. The ALKAL binding sites are shown as an outline of ALKAL1 (green) and ALKAL2 (blue) are shown as dashed lines. Conservation analysis was performed using the Consurf server (https://consurf.tau.ac.il) based on an alignment of 248 vertebrate sequences for ALK and 225 for LTK by COBALT. c, Top view of dimerized LTK (left) and ALK (right) coloured according to sequence conservation levels.
Extended Data Fig. 9 |. Mapping of missense mutations on the structures of the ALK–ALKAL2 and LTK–ALKAL1 complexes.
a, Mapping of most frequent SNPs (GnomAD) to the ALKAL2–ALKTG complex shown in top view. SNPs also found in COSMIC database (https://cancer.sanger.ac.uk/cosmic) are also indicated on the bottom view. Mutations further characterized in this study are coloured red. Inset shows the detailed position of R753 in the ALKTG-ALKAL2 complex. b, Mapping of most frequent SNPs (GnomAD) to the ALKAL1–LTKTG complex shown in top view. SNPs also found in COSMIC database (https://cancer.sanger.ac.uk/cosmic) are also indicated on the bottom view, c, Western blot analysis of the expression levels of ALKWT and the ALKR753Qand ALKF856S mutants and their ERK phosphorylation (left). On the right-side western blot analysis of phosphorylated ALKWT, ALKR753Qand ALKF856S is shown. Representative results from three biologically independent experiments with similar results. Uncropped western blot scans are provided in source data, d, Sanger sequencing of cDNA showing WT or mutant ALK expression in isogenic Ba/F3 cells. e, Cell proliferation of ALK expressing Ba/F3 cells treated with 10 nM ALKAL2 with and without addition of crizotinib for ALK R753Q and F856S mutants. Data for EV and WT ALK are repeated from Extended Data Fig. 3 for direct comparison. Crizotinib is also able to inhibit ALKAL2 induced proliferation for mutant ALK, indicating ALK dependent signalling. (n = 3 biologically independent experiments; mean ± s.d.; two-way ANOVA with Tukey’s multiple comparison test compared with DMSO control). f, Proliferation of Ba/F3 cells expressing ALK carrying the R7S3Q or F856S mutation upon stimulation by a concentration series of ALKAL2, ALKAL1 or ALKAL1FL at indicated concentration (n=3 biologically independent experiments; mean ± s.d.; two-way ANOVA with Tukey’s multiple comparison test compared with ALKWT. ALKAL-induced cell growth relative to that of cells cultured with IL-3 is shown in a heatmap representation. E V and ALKWT controls are included for comparison. *P < 0.05, **P < 0.01 and ***P <0.001. Exact P-values are provided in source data).
Extended Data Fig. 10 |. Mechanistic insights into the assembly of ALK/LTK–cytokine complexes derived from microcalorimetry, SAXS, and SEC-MALLS.
a,b ITC experiments for the titration of LTKTG-EGFL (5 μM) with ALKAL1 (56 μM). (a) Titration of LTKTG-EGEL (12 μM) with ALKAL2 (56 μM). (b). ITC titration curves, the left panel shows the raw data with the differential electrical power (DP) plotted against time. The right panel represents the binding isotherm obtained from the integration of the raw data and fitted to a one-site model. mean ± s.d were calculated based on 3 measurements, c, Small-angle X-ray scattering analysis and calculated FoXS fits of the binary ALKTG-EGFL–ALKAL2 (pink), ternary ALKTG-EGFL–ALKAL2 (purple), binary LTKTG-EGFE–ALKAL1 (orange) and ternary LTKTG-EGEL–ALKAL1 (light orange) to experimental SAXS data (black curves), d, ITC experiments for the titration of LTKTG-EGFL (7.2 μM) with ALKAL1FE (55 μM) Data representative of 2 independent experiments, e, Ternary ALKTG:ALKAL2 complex with regions differing with ALKAL1 coloured Vermillion. C-termini of the TG domains leading towards the EGF-like domains are coloured red. f, ITC experiments for the titration of LTKTC (10 μM) with ALKAL1 (70 μM) Data representative of 2 independent experiments, g, ITC experiments for the titration of LTKTG (10 μM) with ALKAL2 (100 μM) Data representative of 2 independent experiments, h, ITC experiments for the titration of LTKTG (10 μM) with ALKAL1FL (40 μM) Data representative of 2 independent experiments, i, ITC experiments for the titration of ALKFL (8 μM) with ALKAL2 (82 μM) Data representative of 2 independent experiments, j, Characterization of heparin-induced ALKFL dimerization by SEC-MALLS. Differential refractive index (left vertical axis) is plotted against the determined molecular weight (right vertical axis). ALKFL (pink), ALKFL complexes with heparin dp20 (green) and ALKFF complexed with heparin dp20 and ALKAL2 (blue trace). The reported molecular mass represents the average molecular mass ± s.d. across the elution peak, k, Overview of the different ligand-mediated extracellular assemblies across RTKs. Trka–NGF (PDB: 21FG), EGFR–EGF (PDB: 1IVO), INSR–INS (PDB: 6PXW), CSF1R–CSF1 (PDB: 4WRM), FGFR–heparin–FGF (PDB: 1FQ9), ALKTC–ALKAL2 (PDB: 7NWZ).
Extended Data Table 1 |.
Crystallographic data and refinement statistics
| ALKTG-EGFL | ALKTG-EGFL—Fab324 | LTKTG | ALKTG—ALKAL2 | LTKTG—ALKAL1—Nb3.16 | Fab324 | |
|---|---|---|---|---|---|---|
|
| ||||||
| Crystallization conditions | 0.1M Gly-Gly/AMPD pH 8.5 15% w/v PEG 3000 20% v/v 1,2,4-Butanetriol 1% w/v NDSB 256 5mM NiCl2.6H2O 5mM CoC12.6H2O 5mM ZnCl2.6H2O 5mM MnCl2.6H2O |
40mMPolyamines, 0.1M Gly-Gly/AMPD pH8.5 12.5% w/v PEG4000 20% w/v 1,2,6-Hexanetriol |
MOPSO/Bis-Tris pH 6.3 12% PEG 20000 25% Trimethyl propane 2% w/v NDSB 195 5mM YC13.6H2O 5mM ErCl3.6H2O 5mM TbCl3.6H2O 5mM YbCl3.6H2O |
0.1M MES pH 6.5 15% w/v PEG 6000 5% w/v MPD |
MOPSO/BIS-TRIS pH 6.5 13% PEG 8000 22% w/v 1,5-pentanediol 5mM Na2CrO4.4H2O 5mM Na2MoO4.4H2O 5mM Na2WO4.4H2O 5mM Na2VO4.4H2O |
1.5M (NH4)2SO4 40 mM Glycine pH 9.5 |
| Cryoprotectant | 25% v/v 1,2,4-Butanetriol | 25% w/v 1,2,6-Hexanetriol | 25% Trimethyl propane | 22% ethylene glycol | 25% 1,5-pentanediol | Saturated (NH4)2SO4 |
| Data collection | ||||||
| X-ray source (beamline) | PETRAIII (P14) | ESRF (ID-23-2) | PETRAIII (P13) | SOLEIL (Proxima 2A) | SOLEIL (Proxima 2A) | PETRAIII (P13) |
| Wavelength (Å) | 1.033 | 0.8731 | 0.9763 | 0.9800 | 0.9800 | 0.9762 |
| Space group | I41 | P 21 21 21 | P21 | P 43 21 2 | P 61 | P 21 21 21 |
| Cell dimensions | ||||||
| a, b, c (Å) | 84.93 84.93 148.67 | 109.7 137.47 144.26 | 55.04 54.46 55.01 | 97.57 97.57 355.35 | 129.11 129.11 109.75 | 81.24 87.09 126.6 |
| α,β,γ (°) | 90.00, 90.00, 90.00 | 90.00, 90.00, 90.00 | 90 111.284 90 | 90.00, 90.00, 90.00 | 90.00 90.00 120.00 | 90.00, 90.00,, 90.00 |
| Resolution (Å) | 60.05 – 3.0 (3.11 – 3.0) | 48.04 – 2.81 (2.91 – 2.81) | 51.26 – 1.3 (1.38 – 1.3) | 48.96 – 4.18 (4.32 – 4.18) | 42.3 – 1.95 (2.02 – 1.95) | 71.76 – 1.47 (1.52 – 1.47) |
| Rmeas (%) | 18.9 (114.9) | 25.4 (120.1) | 6.9 (125.5) | 20.6 (168.1) | 7.6 (147.1) | 4.2 (35.8) |
| <I/σ> | 10.15 (1.26) | 11.11 (1.91) | 11.5 (1.0) | 13.9 (2.0) | 26.6 (1.9) | 17.32 (2.4) |
| CC ½ (%) | 100 (75.8) | 99.9 (34.1) | 99.9 (54.6) | 99.9 (89.2) | 99.9 (88.5) | 99.7 (42.1) |
| Completeness (%) | 99.90 (99.90) | 99.19 (93.06) | 98.90 (98.5) | 99.50 (94.5) | 99.7 (96.3) | 96.20 (73.3) |
| Redundancy | 14.3 (14.4) | 5.8 (4.5) | 3.4 (3.2) | 25.6 (24.6) | 20.8 (19.8) | 3.8 (3.2) |
| Wilson B factor (Å2) | 104.20 | 39.46 | 14.23 | 105.89 | 38.80 | 16.20 |
| Refinement | ||||||
| Resolution (Å) | 60.05 – 3.0 | 48.04 – 2.81 | 51.26 – 1.3 | 48.78 – 4.18 | 42.3 – 1.95 | 71.76 – 1.47 |
| No. reflections | 10550 | 53379 | 145183 | 13500 | 75501 | 146924 |
| Rwork / Rfree (%) | 23.4/26.0 | 22.6 / 26.3 | 16.2 / 18.1 | 25.3 / 26.8 | 16.9 / 19.6 | 16.9 / 20.5 |
| No. atoms | ||||||
| Protein | 2496 | 10653 | 2098 | 9763 | 6502 | 6486 |
| Ligand/ion | 31 | - | 46 | 98 | 4 | - |
| Water | - | - | 232 | - | 691 | 917 |
| B-factors (Å2) | ||||||
| Protein | 103.45 | 52.21 | 21.08 | 162.0 | 53.25 | 19.60 |
| Ligand/ion | 116.68 | - | 25.67 | 195.3 | 116.64 | 24.54 |
| Water | - | - | 36.65 | - | 57.86 | 30.49 |
| R.m.s. deviations | ||||||
| Bond lengths (Å) | 0.002 | 0.005 | 0.007 | 0.002 | 0.006 | 0.008 |
| Bond angles (°) | 0.53 | 0.87 | 1.09 | 0.50 | 0.90 | 1.03 |
| Pdb id | 7NX4 | 7NX3 | 7NX1 | 7NWZ | 7NX0 | 7NX2 |
Each data set was collected from a single crystal. Values in parentheses correspond to the highest resolution shell.
Extended Data Table 2 |.
Interaction interface analysis of ALK—ALKAL2 and LTK—ALKAL1 complexes
a
|
|
|
||||||
|
| ||||||||
| LTK—ALKAL1 site 1 | LTK—ALKAL1 site 2 | LTK—ALKAL1 site 3 | ||||||
|
| ||||||||
| ALKAL1 Chain A | LTK Chain B | dist (Å) | ALKAL1 Chain A | LTK Chain C | dist (Å) | LTK Chain B | LTK Chain C | dist (Å) |
|
| ||||||||
| Hydrogen-bonds and salt bridges | Hydrogen-bonds and salt bridges | Hydrogen-bonds and salt bridges | ||||||
| GLU 103[OE2] | TYR 124[OH] | 2.57 | HIS 79[O] | ARG 376[NH2] | 3.43 | ARG 241[NE] | GLY 88[O] | 3.57 |
| ARG 102[NH2] | TYR 124[OH] | 3.51 | HIS 79[NE2] | SER 122[OG] | 2.8 | ARG 241[NH2] | GLY 88[O] | 3.46 |
| ARG 119[NH2] | SER 251[O] | 3.28 | LYS 111[NZ] | TYR 124[OH] | 3.0 | ARG 241[NH2] | ALA 89[O] | 2.86 |
| ARG 119[NH2] | GLU 253[OE1] | 3.09 | ARG 112[NH2] | GLU 368[OE1] | 3.06 | ARG 241[NH2] | ALA 91[O] | 2.62 |
| ARG 119[NE] | GLU 253[OE2] | 2.80 | van der Waals contacts | GLN 85[NE2] | CYS 179[O] | 3.37 | ||
| HIS 93[ND1] | GLN 362[OE1] | 3.46 | THR 84[OG1] | GLU 182[OE2] | 2.77 | |||
| ARG 115[NH2] | VAL 366[O] | 2.89 | PHE 80 | TYR 124 | ARG 138[NH1] | ASN 369[OD1] | 3.51 | |
| ARG 115[NE] | VAL 366[O] | 2.84 | PHE 143 | ASN 369[ND2] | ASN 369[OD1] | 3.22 | ||
| ARG 112[NH1] | GLU 372[OE1] | 3.59 | PRO 107 | LEU 364 | GLY 88[O] | ARG 241[NH2] | 3.16 | |
| ARG 102[NH1] | GLU 372[OE2] | 3.28 | VAL 366 | ALA 89[O] | ARG 241[NH2] | 2.94 | ||
| ARG 102[NH2] | GLU 372[OE2] | 3.20 | ALA 108 | ALA 365 | ALA 91[O] | ARG 241[NH2] | 2.89 | |
| ASN 100[O] | ARG 376[NH1] | 3.61 | PHE 76 | LEU 361 | ALA 91[O] | ARG 241[NH1] | 2.86 | |
| PHE 143 | CYS 179[O] | GLN 85[NE2] | 3.31 | |||||
| van der Waals contacts | GLN 362 | ASN 369[OD1] | ARG 138[NH1] | 3.14 | ||||
| ARG 96 | PHE 360 | LEU 364 | ASN 369[OD1] | ASN 369[ND2] | 3.17 | |||
| LEU 97 | LEU 361 | TYR 110 | LEU 364 | van der Waals contacts | ||||
| SER 123 | GLN 362 | HIS 79 | LEU120 | |||||
| GLU 103 | TYR 124 | PHE143 | HIS 135 | CYS 73 | ||||
| LEU116 | VAL 266 | ARG376 | GLY 74 | |||||
| LEU 364 | THR 84 | |||||||
| PHE 143 | LEU 180 | GLN 85 | ||||||
| LEU 120 | LEU 364 | GLU 182 | GLY 92 | |||||
| GLN 362 | ARG 243 | HIS 135 | ||||||
| CYS 73 | ||||||||
| GLY 74 | ||||||||
| THR 84 | LEU 180 | |||||||
| GLN 85 | GLU 182 | |||||||
| GLY 92 | ARG 243 | |||||||
|
| ||||||||
b
|
|
|
||||||
|
| ||||||||
| ALK—ALKAL2 site 1 | ALK—ALKAL2 site 2 | ALK—ALKAL2 site 3 | ||||||
|
| ||||||||
| ALKAL2 Chain C | ALK Chain A | dist (Å) | ALKAL2 Chain C | ALK Chain B | dist (Å) | ALK Chain A | ALK Chain B | dist (Å) |
|
| ||||||||
| Hydrogen-bonds and salt bridges | Hydrogen-bonds and salt bridges | Hydrogen-bonds and salt bridges | ||||||
| TYR 739[OH] | ASP 134[OE2] | 2.8 | HIS 100[O] | LYS 982[NZ] | 3.53 | THR 697[OG1] | ILE 795[O] | 3.54 |
| HIS 857[O] | ARG 140[NH2] | 3.90 | LYS 96[NZ] | TYR 966[O] | 3.30 | GLN 700[N] | ILE 795[O] | 3.83 |
| HIS 857[O] | ARG 140[NH2] | 3.77 | HIS 100[NE2] | SER 737[OG] | 3.49 | ASN 749[ND2] | GLN 706[OE1] | 3.20 |
| GLU 859[OE2] | ARG 140[NE] | 2.96 | LYS 132[NZ] | GLU 978[OE1] | 2.91 | ILE 795[O] | THR 697[OG1] | 3.38 |
| TYR 966[OH] | ASN 121[ND1] | 3.02 | CYS 794[O] | GLN 700[NE2] | 3.84 | |||
| VAL 972[O] | ARG 136[NH2] | 3.23 | ASN 749[OD1] | GLN 706[NE2] | 3.31 | |||
| GLU 974[OE2] | ARG 136[NE] | 3.08 | ||||||
| GLU 978[OE1] | ARG 133[NH1] | 2.8 | ||||||
| van der Waals contacts | van der Waals contacts | van der Waals contacts | ||||||
| ARG 117 | TYR 966 | MET 93 | THR 967 | GLY 689 | THR 750 | |||
| LEU 118 | LEU 970 | PRO 968 | THR 750 | GLY 689 | ||||
| LEU116 | THR 967 | LEU 970 | ILE 795 | GLN 706 | ||||
| LEU 970 | LEU 101 | TYR 739 | ||||||
| VAL 972 | PHE 97 | THR 967 | ||||||
| LEU 120 | LEU 970 | LEU 970 | ||||||
| LYS 132 | VAL 972 | |||||||
| MET 973 | ||||||||
List of contacting amino acids at interaction sites 1, 2, and 3 of the ALKAL2—ALKTG and ALKAL1—LTKTG complexes. Hydrogen bonding residues were determined using the PISA server at EBI (https://www.ebi.ac.uk/msd-srv/prot_int/pistart.html) and confirmed by analysis in ChimeraX. van der Waals contacts were analyzed using the ‘find contacts’ function in ChimeraX. The molecular components involved in each described interface are shown above each column.
Supplementary Material
Acknowledgements
We thank the staff of beamlines P13 and P14 (Petra III, Deutsches Elektronen-Synchrotron), Proxima 2A (SOLEIL) and ID23-2 (ESRF) and SWING (SOLEIL) for their technical support and beamtime allocation. S.D.M. was supported by a predoctoral fellowship from the Flanders Agency for Innovation and Entrepreneurship (VLAIO-Flanders, Belgium). Y.B. is a post-doctoral research fellow of Research Foundation Flanders (FWO). A.Y. acknowledges support from the Japan Society for the Promotion of Science (JSPS) Home-Returning Researcher Development Research (grant number 19K24691), KAKENHI (grant number 21H04828) and National Cancer Center Research and Development Funds (2020-A-2). S.N.S. acknowledges research support from the FWO (grant number G0B4918N), Ghent University (grant number BOF17-GOA-028), the Hercules Foundation (no. AUGE-11-029) and the Flanders Institute for Biotechnology (VIB).
Footnotes
Competing interests The authors declare no competing interests.
Additional information
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41586-021-03959-5.
References
- 1.Morris SW et al. Fusion of a kinase gene, ALK, to a nucleolar protein gene, NPM, in non-Hodgkin’s lymphoma. Science 263, 1281–1284 (1994). [DOI] [PubMed] [Google Scholar]
- 2.Ben-Neriah Y & Bauskin AR Leukocytes express a novel gene encoding a putative transmembrane protein-kinase devoid of an extracellular domain. Nature 333, 672–676 (1988). [DOI] [PubMed] [Google Scholar]
- 3.Zhang H et al. Deorphanization of the human leukocyte tyrosine kinase (LTK) receptor by a signaling screen of the extracellular proteome. Proc. Natl Acad. Sci. USA 111, 15741–15745(2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reshetnyak AV et al. Augmentor α and β (FAM150) are ligands of the receptor tyrosine kinases ALK and LTK: Hierarchy and specificity of ligand-receptor interactions. Proc. Natl Acad. Sci. USA 112, 15862–15867 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guan J et al. FAM150A and FAM150B are activating ligands for anaplastic lymphoma kinase. eLife 4, e09811 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reshetnyak AV et al. Identification of a biologically active fragment of ALK and LTK-ligand 2 (augmentor-α). Proc. Natl Acad. Sci. USA 115, 8340–8345 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hallberg B & Palmer RH Mechanistic insight into ALK receptor tyrosine kinase in human cancer biology. Nat. Rev. Cancer 13, 685–700 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Janostiak R, Malvi P & Wajapeyee N Anaplastic lymphoma kinase confers resistance to BRAF kinase inhibitors in melanoma. iScience 16, 453–467 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Javanmardi N et al. Analysis of ALK, MYCN, and the ALK Ligand ALKAL2 (FAM150B/AUGa) in neuroblastoma patient samples with chromosome arm 2p rearrangements. Genes Chromosomes Cancer 59, 50–57 (2020). [DOI] [PubMed] [Google Scholar]
- 10.Li N et al. Gain-of-function polymorphism in mouse and human Ltk: implications for the pathogenesis of systemic lupus erythematosus. Hum. Mol. Genet. 13, 171–179 (2004). [DOI] [PubMed] [Google Scholar]
- 11.Orthofer M et al. Identification of ALK in thinness. Cell 181, 1246–1262.e22 (2020). [DOI] [PubMed] [Google Scholar]
- 12.Pospisilik JA et al. Drosophila genome-wide obesity screen reveals Hedgehog as a determinant of brown versus white adipose cell fate. Cell 140, 148–160 (2010). [DOI] [PubMed] [Google Scholar]
- 13.Borenäs M et al. ALK ligand ALKAL2 potentiates MYCN-driven neuroblastoma in the absence of ALK mutation. EMBO J. 40, e105784 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sano R et al. An antibody-drug conjugate directed to the ALK receptor demonstrates efficacy in preclinical models of neuroblastoma. Sci. Transl. Med. 11, eaau9732 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dornburg A et al. Comparative genomics within and across bilaterians illuminates the evolutionary history of ALK and LTK proto-oncogene origination and diversification. Genome Biol. Evol. 13, evaa228 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murray PB et al. Heparin is an activating ligand of the orphan receptor tyrosine kinase ALK. Sci. Signal. 8, ra6 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Alvarado D et al. Anti-ALK antibodies and methods for use thereof. US patent 15/755421 (2021).
- 18.Crick FHC & Rich A Structure of polyglycine II. Nature 176,780–781 (1955). [DOI] [PubMed] [Google Scholar]
- 19.Lorén CE et al. A crucial role for the anaplastic lymphoma kinase receptor tyrosine kinase in gut development in Drosophila melanogaster. EMBO Rep. 4, 781–786 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kolodny R Searching protein space for ancient sub-domain segments. Curr. Opin. Struct. Biol. 68,105–112 (2021). [DOI] [PubMed] [Google Scholar]
- 21.Fadeev A et al. ALKALs are in vivo ligands for ALK family receptor tyrosine kinases in the neural crest and derived cells. Proc. Natl Acad. Sci. USA 115, E630–E638 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang YW et al. Identification of oncogenic point mutations and hyperphosphorylation of anaplastic lymphoma kinase in Lung cancer. Neoplasia 13, 704–715 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maxson JE et al. Therapeutically targetable ALK mutations in leukemia. Cancer Res. 75, 2146–2150 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Durham BH et al. Activating mutations in CSF1R and additional receptor tyrosine kinases in histiocytic neoplasms. Nat. Med. 25, 1839–1842 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malinauskas T, Aricescu AR, Lu W, Siebold C & Jones EY Modular mechanism of Wnt signaling inhibition by Wnt inhibitory factor 1. Nat. Struct. Mol. Biol. 18, 886–893 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wehrman T et al. Structural and mechanistic insights into nerve growth factor interactions with the TrkA and p75 receptors. Neuron 53, 25–38 (2007). [DOI] [PubMed] [Google Scholar]
- 27.Ogiso H et al. Crystal structure of the complex of human epidermal growth factor and receptor extracellular domains. Cell 110, 775–787 (2002). [DOI] [PubMed] [Google Scholar]
- 28.Elegheert J et al. Extracellular complexes of the hematopoietic human and mouse CSF-1 receptor are driven by common assembly principles. Structure 19, 1762–1772 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schlessinger J et al. Crystal structure of a ternary FGF–FGFR–heparin complex reveals a dual role for heparin in FGFR binding and dimerization. Mol. Cell 6, 743–750 (2000). [DOI] [PubMed] [Google Scholar]
- 30.Uchikawa E, Choi E, Shang G, Yu H & Xiao-Chen B Activation mechanism of the insulin receptor revealed by cryo-EM structure of the fully liganded receptor–ligand complex. eLife 8, e48630 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aricescu AR, Lu W & Jones EY A time- and cost-efficient system for high-level protein production in mammalian cells. Acta Crystallogr. D 62,1243–1250 (2006). [DOI] [PubMed] [Google Scholar]
- 32.Reeves PJ, Callewaert N, Contreras R & Khorana HG Structure and function in rhodopsin: high-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline-inducible N-acetylglucosaminyltransferase I-negative HEK 293S stable mammalian cell line. Proc. Natl Acad Sci. USA 99,13419–13424 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Backliwal G et al. Valproic acid: A viable alternative to sodium butyrate for enhancing protein expression in mammalian cell cultures. Biotechnoi Bioeng. 101,182–189 (2008). [DOI] [PubMed] [Google Scholar]
- 34.Gorrec F The MORPHEUS II protein crystallization screen. Acta Crystallogr. F 71, 831–837 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kabsch W XDS. Acta Crystallogr. D. 66,125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCoy AJ et al. Phaser crystallographic software. J. Appl. Crystallogr. 40,658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bricogne G, et al. BUSTER 2.11.2 (United Kingdom Global Phasing Ltd, 2017). [Google Scholar]
- 38.Emsley P, Lohkamp B, Scott WG & Cowtan K Features and development of Coot. Acta Crystallogr. D 66,486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Terwilliger TC Maximum-likelihood density modification. Acta Crystallogr. D 56, 965–972(2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liebschner D et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. Sect. D 75,861–877 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Langer G, Cohen SX, Lamzin VS & Perrakis A Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat. Protoc. 3,1171–1179 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murshudov GN et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D 67,355–367 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.D’Arcy A, Villard F & Marsh M An automated microseed matrix-screening method for protein crystallization. Acta Crystallogr. D 63,550–554 (2007). [DOI] [PubMed] [Google Scholar]
- 44.Strong M et al. Toward the structural genomics of complexes: crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc. Natl Acad Sci. USA 103, 8060–8065 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Howarth M & Ting AY Imaging proteins in Live mammalian cells with biotin ligase and monovalent streptavidin. Nat. Protoc. 3, 534–545 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hopkins JB, Gillilan RE & Skou S BioXTAS RAW: Improvements to a free open-source program for small-angle X-ray scattering data reduction and analysis. J. Appl. Crystallogr. 50, 1545–1553(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Biasini M et al. SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 42, W252 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Webb B & Sali A Comparative protein structure modeling using MODELLER. Curr. Protoc. Bioinforma. 2016, 5.6.1–5.6.37(2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yoshimi A et al. Coordinated alterations in RNA splicing and epigenetic regulation drive leukaemogenesis. Nature 574, 273–277 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ishihara T et al. HEN-1, a secretory protein with an LDL receptor motif, regulates sensory integration and learning in Caenorhabditis elegans. Cell 109, 639–649 (2002). [DOI] [PubMed] [Google Scholar]
- 51.Englund C et al. Jeb signals through the Aik receptor tyrosine kinase to drive visceral muscle fusion. Nature 425, 512–516 (2003). [DOI] [PubMed] [Google Scholar]
- 52.Lee HH, Norris A, Weiss JB & Frasch M Jelly belly protein activates the receptor tyrosine kinase Aik to specify visceral muscle pioneers. Nature 425, 507–512 (2003). [DOI] [PubMed] [Google Scholar]
- 53.Blum M et al. The InterPro protein families and domains database: 20 years on. Nucleic Acids Res. 48, D344–D354 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pentelute BL et al. X-ray structure of snow flea antifreeze protein determined by racemic crystallization of synthetic protein enantiomers. J. Am. Chem. Soc. 130, 9695–9701 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Buglino J, Shen V, Hakimian P & Lima CD Structural and biochemical analysis of the Obg GTP binding protein. Structure 10, 1581–1592 (2002). [DOI] [PubMed] [Google Scholar]
- 56.Weidenweber S et al. Structure of the acetophenone carboxylase core complex: Prototype of a new class of ATP-dependent carboxylases/hydrolases. Sci. Rep. 7, 1–10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dunne M et al. Salmonella phage S16 tail fiber adhesin features a rare polyglycine rich domain for host recognition. Structure 26, 1573–1582.e4 (2018). [DOI] [PubMed] [Google Scholar]
- 58.Krissinel E & Henrick K Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr., Sect. D: Biol. Crystallogr. 60, 2256–2268 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Coordinates and structure factors for the complexes have been deposited in the Protein Data Bank (PDB) under accessions 7NWZ (ALKTG–ALKAL2), 7NX0 (LTKTG–ALKAL1–Nb3.16), 7NX1 (LTKTG), 7NX2 (unbound Fab324), 7NX3 (ALKTG–EGFL–Fab324), 7NX4 (ALKTG–EGFL). Most common single-nucleotide polymorphisms were obtained from the COSMIC database (https://cancer.sanger.ac.uk/cosmic) and gnomAD database (https://gnomad.broadinstitute.org). The Ba/F3 cell lines expressing ALK(WT), ALK(R753Q) or ALK(F856S) can be obtained from the authors upon request. Source data are provided with this paper.