Summary
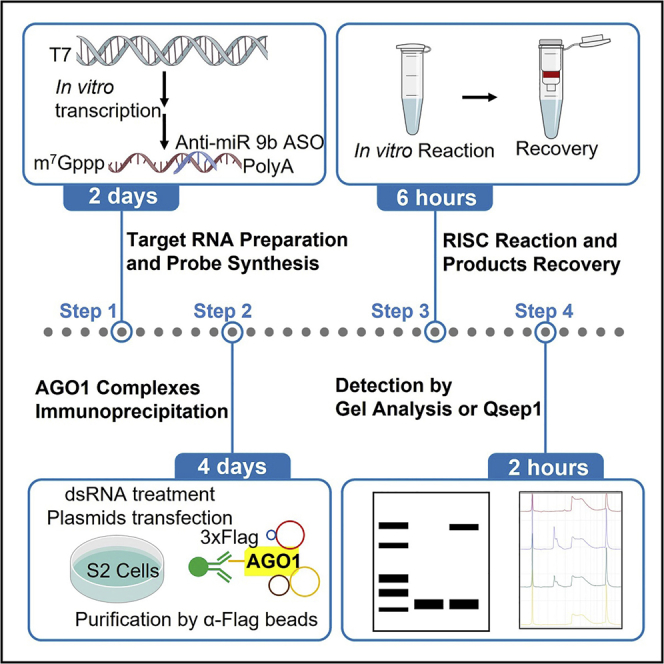
Here, we provide an optimized RNA-induced silencing complex (RISC) assembly and cleavage protocol in vitro without using radiolabeled RNA. The protocol is useful to characterize the biochemical properties of the RISC. We describe the preparation of RNA probes, the target RNA, and Drosophila cell lysates for RISC assembly assay. We then detail AGO1 complexes immunoprecipitation for RISC cleavage assay. This protocol can detect RISC assembly and cleavage products within 5 days. Moreover, it can detect 5′- and 3′-cleavage products simultaneously.
For complete details on the use and execution of this protocol, please refer to Gao et al. (2022).
Subject areas: Cell-based Assays, Molecular Biology, Gene Expression, Molecular/Chemical Probes, Protein Biochemistry
Graphical abstract
Highlights
-
•
In vitro RISC assembly and cleavage without radiolabeled RNA
-
•
Steps to prepare RNA probes, target RNA, and cell lysates for RISC assembly
-
•
AGO1 complexes immunoprecipitation for RISC cleavage assay
-
•
Detection of RISC assembly and cleavage products within five days
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Here, we provide an optimized RNA-induced silencing complex (RISC) assembly and cleavage protocol in vitro without using radiolabeled RNA. The protocol is useful to characterize the biochemical properties of the RISC. We describe the preparation of RNA probes, the target RNA, and Drosophila cell lysates for RISC assembly assay. We then detail AGO1 complexes immunoprecipitation for RISC cleavage assay. This protocol can detect RISC assembly and cleavage products within 5 days. Moreover, it can detect 5′- and 3′-cleavage products simultaneously.
Before you begin
Small interfering RNAs (siRNAs) and microRNAs (miRNAs) mediated gene silencing are a major process of gene expression at the transcriptional and post-transcriptional levels (Iwakawa and Tomari, 2022; Treiber et al., 2019). Small RNAs bind to Argonaute (AGO) proteins to form RISC to silence their target mRNA by RNA cleavage or by preventing protein translation (Bartel, 2018; Duchaine and Fabian, 2019). AGOs, as the catalytic core of RISC, play essential roles in mediating sequence-specific target gene silencing.
To achieve their function for target gene silencing. siRNAs and miRNAs must form RISC with AGO proteins in a similar manner. RISC assembly is divided into two steps: the loading step and the maturation step. In the loading step, a small RNA duplex is loaded into the AGO protein to form the pre-RISC (Iwakawa and Tomari, 2022). In the maturation step, the duplex is separated, and only the guide strands reside in the AGO protein to form the mature RISC via wedging and passenger ejection (Kwak and Tomari, 2012). Mature siRNA-RISC induces the perfectly complementary target mRNA decay via cleavage activity, and mature miRNA-RISC generally causes the translational repression of partially complementary target mRNA.
Further applications into therapeutics of RISC arise from the fact that siRNAs and miRNAs can be designed to target mRNA for silencing, and many factors can fine-tune the core silence activity of RISC. In vitro RISC assembly and cleavage assay provides powerful biochemical readouts to assess the activity and stability of RISC. Agarose native gel electrophoresis has been used to detect in vitro AGO-RISC assembly (Kawamata et al., 2009) and cleavage (Meister et al., 2004; Miyoshi et al., 2005) to investigate the molecular mechanism of RISC biogenesis in Drosophila AGO1 or human AGO2 pathways. However, previous protocols depended on radiolabeled RNA (Kawamata and Tomari, 2011) and required labeling and gel assay steps to be performed in an isotope lab. Here, we describe a similar agarose native gel electrophoresis system to analyze mature AGO-RISC assembly and cleavage without using radiolabeled RNA probes.
Preparation of the ASO for AGO1-RISC assembly
Timing: Approximately 2 days
-
1.Anti-miR-9b antisense oligonucleotide (ASO) design and synthesis.
-
a.Design anti-miR-9b ASO with 5′-FAM and 3′-2-O-methylated modification, complementary to miR-9b, except for one mismatch in the central region (Figure 1).
-
a.
Note: This ASO is designed to have a central bulge to prevent endo-nucleolytic cleavage by AGO1-RISC.
Alternatives: Other fluorescence modifications such as Cy3 or Cy5 would also work.
CRITICAL: The RNA probe is very sensitive to the RNases. All the steps are conducted by using fresh RNase-Free water and RNase-Free pipet tips.
Figure 1.
Schematic of the Anti-miR-9b antisense oligonucleotide
Preparation of the dsRNA
Timing: Approximately 1 day
-
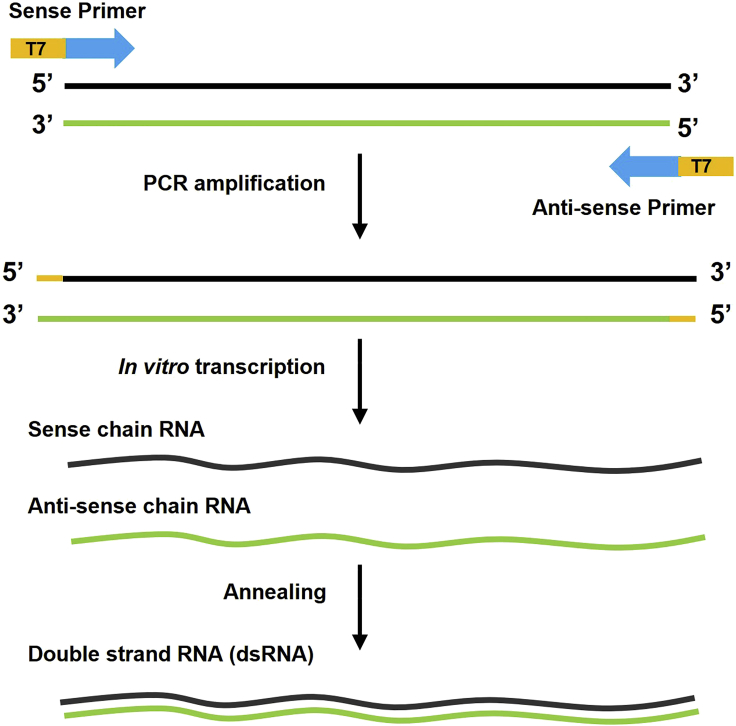
2.dsRNA synthesis (Figure 2).
-
a.Preparation of the in vitro transcriptional template by PCR. Prepare DNA templates of dsRNAs from Drosophila cDNA with gene specific primers harboring the T7 promoter sequence. Gene specific primers used to prepare the templates are listed in key resources table.
-
b.In vitro transcription reaction. Because the DNA templates used for synthesizing dsRNA contain the T7 promoter sequence at both 5′ and 3′ sides, the templates can be used to synthesize both sense and antisense RNA sequences by the T7 RiboMAX Express Large-Scale RNA Production Kit (Promega) according to the standard protocol.
-
c.Sense and antisense RNAs are annealed to generate dsRNAs. Briefly microfuge the tube and collect the RNA solution at the bottom of the tube. Use the PCR program as follows to perform the annealing.
Steps Temperature Time Cycles Denaturation 94°C 2 min 1 Annealing 94°C–22°C 0.1°C/s 1 Incubation 22°C 5 min 1 -
d.dsRNA recovery and quantitation. Before the recovery procedure, perform DNase I treatment to remove the template DNA. Purify the dsRNAs following instructions of the T7 RiboMAX Express Large-Scale RNA Production Kit. These procedures remove the nucleotides, short oligonucleotides, proteins, and salts from dsRNAs. Quantitation can be determined by Nanodrop (Thermo Fisher Scientific).
-
e.Store the dsRNA at −20°C or −80°C for up to one year.
-
a.
Figure 2.
Schematic of dsRNA synthesis
Preparation of the agarose gel
Timing: Approximately 4 h
-
3.Vertical agarose gel preparation.
-
a.Clean and dry the glass plates to avoid the formation of air bubbles while pouring the gel.
-
b.Assemble the glass plates and set them up in a standing (vertical) position with the gel casting equipment (Tanon).
-
c.For a 102 × 85 × 1.5 mm plate, use 10 mL of TBE-agarose. Add 0.3 g agarose to 20 mL 0.5 × TBE solution in a 150 mL conical flask.
-
d.Melt the agarose in a microwave oven until it is completely dissolved.
-
e.Carefully and slowly pour the agarose into the glass plates, and immediately insert a 1.5 mm, 15-well comb between the glass plates.
-
f.When the gel has solidified, carefully remove the comb. These gels should be stored at 4°C.
-
a.
Preparation of substrate RNAs for the cleavage assay
Timing: Approximately 8 h
-
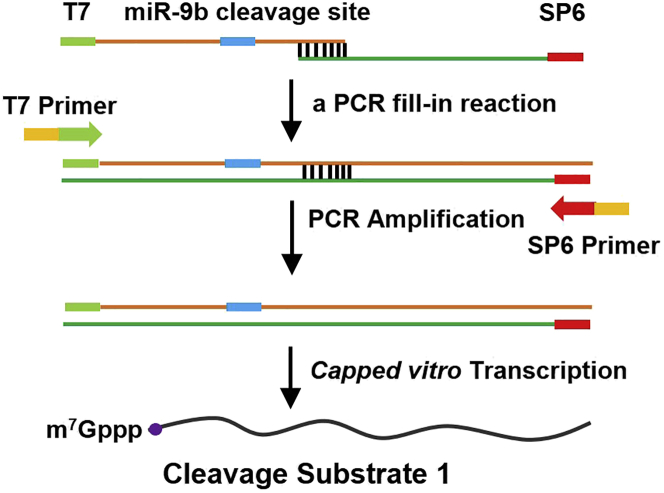
4.Substrate RNAs preparation (Figure 3). See troubleshooting 2 and 4.
-
a.Preparation of the transcriptional template by PCR.
-
i.Synthesize the partially complementary oligonucleotides containing the miR-9b cleavage site as follows.miR-9b-S:5′-GAACAATTGCTTTTACAGATGCACATATCGAGGTGAACATCACGTACGCATACAGCTAAAATCACCAAAGATCGGTTGGCAGAAGCTAT-3′.miR-9b-AS:5′-GGCATAAAGAATTGAAGAGAGTTTTCACTGCATACGACGATTCTGTGATTTGTATTCAGCCCATATCGTTTCATAGCTTCTGCCAACCGA-3′.
-
ii.Use the miR-9b-S and miR-9b-AS oligonucleotides for a PCR fill-in reaction via the PCR program.
Reagent Amount 10 μM miR-9b-S 1 μL 10 μM miR-9b-AS 1 μL 2 × Phanta Mixture 25 μL ddH2O N/A Total 46 μL -
iii.PCR program: 60 s at 95°C, 5 min at 72°C, then cooling to 4°C.
-
iv.For the second PCR amplification reaction, directly add 2 μL of T7 and SP6 primer (10 μM) to the PCR mixture. Set up the following PCR program.
Steps Temperature Time Cycles Initial denaturation 95°C 2 min 1 Denaturation 95°C 15 s 30 cycles Annealing 56°C 15 s Extension 72°C 30 s Final extension 72°C 3 min 1 Hold 4°C forever -
v.Purify the PCR products after separation on a 1.5% agarose gel via gel DNA extraction according to the instructions.
-
vi.Ligate the PCR fragment into the pEASY cloning vector using the pEASY-Blunt Zero Cloning Kit.
-
vii.Perform sequencing using the M13 forward and reverse primers to verify positive colonies.
-
viii.Set up another PCR mixture using the template from a sequence-verified positive colony and use the PCR program from step (iv) above. The PCR products from this step will be used as the template for in vitro transcription.
Reagent Amount 10 μM T7 Primer 2 μL 10 μM SP6 Primer 2 μL 2 × Phanta Mixture 25 μL ddH2O N/A Total 50 μL
-
i.
-
b.Capped transcription reaction for cleavage substrate 1 preparation.
-
i.Assemble the capped in vitro transcription mixture at room temperature (22°C–28°C) according to the instruction of the mMESSAGE mMACHINE kit as follows.
Component Amount 2 × NTP/CAP 10 μL 10 × Reaction buffer 2 μL Template DNA ∼ 200 ng Nuclease-free H2O N/A Total 20 μL -
ii.Mix thoroughly by gently pipetting the mixture up and down, and then briefly microfuge the tube and collect the solution at the bottom of the tube.
-
iii.Incubate at 37°C for 4 h or overnight (4–12 h).Note: The mMESSAGE mMACHINE Kit is designed for optimal function with transcription templates in the 300–1,000 base range. In general, maximum yield can be achieved after a 2 h incubation. The second hour of incubation is necessary for transcription of < 300 base products.
-
iv.Add 1 μL TURBO DNase, mix well and incubate at 37°C for 15 min.
-
v.Recover the RNA by lithium chloride precipitation according to the standard protocol. The RNA resulting from capped in vitro transcription provides the cleavage substrate 1 for the RISC cleavage assay.
-
i.
-
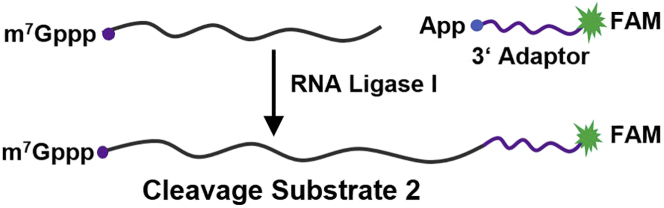
c.Cleavage substrate 2 preparation.
-
i.Synthesize the 3′-RNA adaptor designed with both 5′-phosphate and 3′-6-FAM modification. 5′-phosphate-GrUrGrCrUrCrGrArGrUrCrGrCrGrGrCrCrGrCrArArGrGrArArCrArUrUrCrGrGrC-3′-6-FAM.
-
ii.Ligate the 3′-RNA adaptor to cleavage substrate 1 by T4 RNA ligase 1 following the manufacturer’s instructions as follows (Figure 4).
Component Amount Cleavage substrate 1 20 pmol 3′RNA adaptor 40 pmol 10 × T4 RNA ligase buffer 2 μL 10 mM ATP 2 μL 50% PEG 8000 7 μL RNase inhibitor (40 U/μL) 1 μL T4 RNA ligase I 1 μL Nuclease-free H2O N/A Total 20 μL -
iii.Incubate at 25°C for 4 h.
-
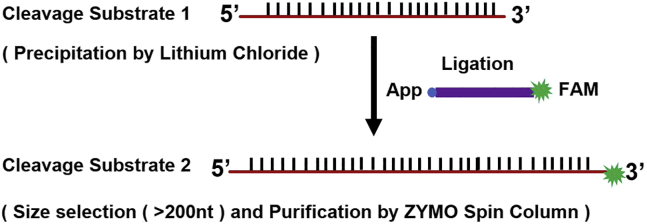
iv.Purify the ligated RNA by size selection (> 200 nucleotides) using the Separated Fraction of the ZYMO-Spin Column, and use the purified ligated RNA for cleavage substrate 2 (Figure 5).
 CRITICAL: Avoid multiple freezes and thaw cycles for the RNA.
CRITICAL: Avoid multiple freezes and thaw cycles for the RNA.
-
i.
-
a.
Figure 3.
Schematic of cleavage substrate 1 preparation
Figure 4.
Schematic of cleavage substrate 2 preparation
Figure 5.
Purification and size selection of the cleavage substrate
Preparation of buffers and solutions
Timing: Approximately 1 day
-
5.
Preparation of lysis buffer for RISC assembly lysate preparation.
Note: Refer to Materials and equipment.
-
6.
Preparation of the 5 × TBE solution.
Note: Refer to Materials and equipment.
-
7.
Preparation of the RISC assembly reaction mixture.
Note: Refer to Materials and equipment.
-
8.
Preparation of immunoprecipitation (IP) lysis buffer.
Note: Refer to Materials and equipment.
-
9.
Preparation of IP washing buffer.
Note: Refer to Materials and equipment.
-
10.
Preparation of cleavage reaction buffer.
Note: Refer to Materials and equipment.
Design qPCR primers for RNAi efficiency detection
Timing: Approximately 20 min
-
11.
Primers used in the protocol are listed in the key resources table.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-dmAGO1. Working dilution: 1:2000 | Abcam | Cat# ab5070; RRID: AB_2277644 |
| Rabbit polyclonal anti-Flag. Working dilution: 1:3000 | Sigma-Aldrich | Cat# SAB1306078; RRID: N/D |
| Mouse monoclonal anti-β-Tubulin. Working dilution: 1:1000 | Cowin | Cat# CW0098M; RRID: AB_2814800 |
| Bacterial and virus strains | ||
| E. coli DH5α | AlpaLife | Cat# KTSM101L |
| Chemicals, peptides, and recombinant proteins | ||
| 2 × Phanta Max Master Mix | Vazyme | Cat# P525-01 |
| Protein A/G agarose beads | Thermo Fisher Scientific | Cat# 21186 |
| Lipofectamine 2000 | Thermo Fisher Scientific | Cat# 11668019 |
| HEPES | Thermo Fisher Scientific | Cat# 11344041 |
| KOH | Aladdin | Cat# P11287-500g |
| KAC | Aladdin | Cat# P108329-500g |
| Nonidet P-40 | Thermo Fisher Scientific | Cat# 28324 |
| Glycerol | Thermo Fisher Scientific | Cat# 17904 |
| RNase Free H2O | Solarbio | Cat# R1600 |
| cOmplete Protease Inhibitor Cocktail | Roche | Cat# 4693116001 |
| Triton X-100 | Sigma | Cat# X-100 |
| Tween 20 | Sigma | Cat# P1379 |
| PageRuler Prestained Protein Ladder, 10–180 kDa | Thermo Fisher Scientific | Cat# 26616 |
| Agarose | Invitrogen | Cat# 16500500 |
| EDTA (Ethylenediaminetetraacetic acid) | Sigma | Cat# 798681 |
| Trizol | TIANGEN | Cat# DP424 |
| SYBR qPCR Master Mix | Vazyme | Cat# Q711 |
| T4 RNA ligase I | NEB | Cat# M0437M |
| RNase inhibitor | NEB | Cat# M0314L |
| RNA Loading Dye | NEB | Cat# B0363S |
| ssRNA Ladder | NEB | Cat# N0364S |
| Schneider’s Drosophila Medium | Gibco | Cat# 11720-067 |
| Opti-MEM | Gibco | Cat# 31985088 |
| Tris Base | Sigma-Aldrich | Cat# 11814273001 |
| Boric Acid | Sigma-Aldrich | Cat# B0394 |
| PBS | Thermo Fisher Scientific | Cat# 14190144 |
| DTT | Thermo Fisher Scientific | Cat# P2325 |
| Creatine Phosphate | Sigma-Aldrich | Cat# 237911 |
| Creatine phosphokinase | Sigma-Aldrich | Cat# C3755 |
| ATP | Thermo Fisher Scientific | Cat# R0441 |
| GTP | Thermo Fisher Scientific | Cat# R0461 |
| ddH2O | N/A | N/A |
| Liquid nitrogen | N/A | N/A |
| Critical commercial assays | ||
| FasePure Gel DNA Extraction Kit | Vazyme | Cat# DC301-01 |
| pEASY-Blunt Zero Cloning Kit | TransGen Biotech | Cat# CB501-01 |
| T7 RiboMAX Express Large-Scale RNA Production Kit | Promega | Cat# P1700 |
| HiScript III Reverse Transcriptase | Vazyme | Cat#R302 |
| mMESSAGE mMACHINE kit | Thermo Fisher Scientific | Cat# AM1344 |
| RNA clean and Concentrator-5 | Zymo Research | Cat# R1013 |
| RNase, RNA and DNA Remover | Vazyme | Cat# R504 |
| Deposited data | ||
| Raw and analyzed data | Mendeley | https://data.mendeley.com/datasets/4gmsw3t82z/1 |
| Experimental models: Cell lines | ||
| D. melanogaster cell line S2 | (Xia et al., 2010) | N/A |
| Oligonucleotides | ||
| T7 primer | (Gao et al., 2022) | 5′-TAATACGACTCACTATAGA ACAATTGCTTTTACAG-3′ |
| SP6 primer | (Gao et al., 2022) | 5′-ATTTAGGTGACACTATAGGC ATAAAGAATTGAAGA-3′ |
| dsRNA(gfp) Forward Primer | (Gao et al., 2022) | 5′-CTCACTATAGGGAGA atggtgagcaagggc-3′ |
| dsRNA(gfp) Reverse Primer | (Gao et al., 2022) | 5′-CTCACTATAGGGAGA ggtgcgctcctggac-3′ |
| dsRNA(ago1-3′UTR) Forward Primer | (Gao et al., 2022) | 5′-CTCACTATAGGGAGA aaagtatcgcccctccc-3′ |
| dsRNA(ago1-3′UTR) Reverse Primer | (Gao et al., 2022) | 5′-CTCACTATAGGGAGA tttcctatttgctttcaatt-3′ |
| dsRNA(ago2) Forward Primer | (Gao et al., 2022) | 5′-CTCACTATAGGGAGA agccaaggccaataccaa-3′ |
| dsRNA(ago2) Reverse Primer | (Gao et al., 2022) | 5′-CTCACTATAGGGAGA cagaccgaccagggc-3′ |
| dsRNA(dcr-2) Forward Primer | (Gao et al., 2022) | 5′-CTCACTATAGGGAGA ctacgcagcttccatagc-3′ |
| dsRNA(dcr-2) Reverse Primer | (Gao et al., 2022) | 5′-CTCACTATAGGGAGA ggcattaccgtcccga-3′ |
| dsRNA(r2d2) Forward Primer | (Gao et al., 2022) | 5′-CTCACTATAGGGAGA atggataacaagtcagccgt-3′ |
| dsRNA(r2d2) Reverse Primer | (Gao et al., 2022) | 5′-CTCACTATAGGGAGA ttcaatggccgctcgc-3′ |
| RT-rp49 Forward Primer | (Gao et al., 2022) | 5′-ATGACCATCCGCCCAGCATAC-3′ |
| RT- rp49 Reverse Primer | (Gao et al., 2022) | 5′-CTGCATGAGCAGGACCTCCAG-3′ |
| RT-Ago2 Forward Primer | (Gao et al., 2022) | 5′-TCCAGGGCACGGCCAAGCCA-3′ |
| RT-Ago2 Reverse Primer | (Gao et al., 2022) | 5′-CGATTGCAACGAGGGAACAT-3′ |
| RT-r2d2 Forward Primer | (Gao et al., 2022) | 5′-AGGCATTGCGCAGAAAGAAA-3′ |
| RT-r2d2 Reverse Primer | (Gao et al., 2022) | 5′-GCAAGGGAACCAACGATGAA-3′ |
| RT-Ago1 Forward Primer | (Gao et al., 2022) | 5′-GGAGATCAAGGGTTTGAAGATCG-3′ |
| RT-Ago1 Reverse Primer | (Gao et al., 2022) | 5′-AGTGGGAATGATTGCATCTGAG-3′ |
| RT-dcr2 Forward Primer | (Gao et al., 2022) | 5′-TCTAGCCTTGTGGCGAGAAA-3′ |
| RT-dcr2 Reverse Primer | (Gao et al., 2022) | 5′-GCCTCAAGGGTATCGGCTAT-3′ |
| 3′-RNA adaptor | (Gao et al., 2022) | 5′-phosphate-GrUrGrCrUrCrGrArGr UrCrGrCrGrGrCrCrGrCrArArGrGrAr ArCrArUrUrCrGrGrC-3′-6-FAM. |
| miR-9b-S | (Gao et al., 2022) | 5′-GAACAATTGCTTTTACAGATGCA CATATCGAGGTGAACATCACGTAC GCATACAGCTAAAATCACCAAAGA TCGGTTGGCAGAAGCTAT-3′ |
| miR-9b-AS | (Gao et al., 2022) | 5′-GGCATAAAGAATTGAAGAGAGT TTTCACTGCATACGACGATTCTGTG ATTTGTATTCAGCCCATATCGTTTCA TAGCTTCTGCCAACCGA-3′ |
| Recombinant DNA | ||
| pAc5.1-Flag-AGO1 | (Gao et al., 2022) | https://doi.org/10.1016/j.molcel.2022.02.035 |
| pAc5.1-Flag-EGFP | (Gao et al., 2022) | https://doi.org/10.1016/j.molcel.2022.02.035 |
| pAc5.1-miR 9b | (Gao et al., 2022) | https://doi.org/10.1016/j.molcel.2022.02.035 |
| Software and algorithms | ||
| ImageJ | (Schneider et al., 2012) | https://imagej.nih.gov/ij/download.html |
| Snap gene | SnapGene | https://www.snapgene.com/try-snapgene/ |
| Other | ||
| Dounce homogenizer (with tight pestle) | WHEATON | Cat# 1984-10002 |
| Eppendorf centrifuge | Eppendorf | Cat# 5427R |
| Eppendorf centrifuge | Eppendorf | Cat# 5910R |
| Cold room | N/A | N/A |
| Membrane filter, pore size 0.22 m | Millipore | Cat# GSWP04700 |
| Casting Unit | Tanon | Cat# 180-1600&1808 |
| NanoDrop | Thermo Scientific | Cat# ND-ONE-W |
| LightCycler®480 Real-time PCR | Roche | N/A |
| Qsep1-lite | BiOptic | Cat# C100001-L |
| Tanon-6600 imaging workstation | Tanon | N/A |
Materials and equipment
Lysis buffer for RISC assembly lysates
| Reagent | Final concentration | Amount |
|---|---|---|
| HEPES-KOH (pH 7.4) | 30 mM | 1.4298 g |
| KAC | 100 mM | 1.9628 g |
| Mg(AC)2 | 5 mM | 0.1424 g |
| ddH2O | N/A | N/A |
| Total | N/A | 200 mL |
Store at 4°C for 6 months. Filtered buffer can be stored at 4°C for up to one year.
5 × TBE solution
| Reagent | Final concentration | Amount |
|---|---|---|
| Tris base | 445 mM | 56.3 g |
| Boric acid | 445 mM | 27.6 g |
| EDTA | 10 mM | 3.7 g |
| ddH2O | N/A | N/A |
| Total | N/A | 1000 mL |
Store at room temperature (22°C–28°C) for up to 6 months. Filter buffer and avoid RNase contamination.
RISC assembly reaction mixture
| Reagent | Final concentration | Amount |
|---|---|---|
| KAC | 133.3 mM | 16 μL of 1 M stock |
| ATP | 3.33 mM | 4 μL of 100 mM stock |
| DTT | 30 mM | 2 μL of 1 M stock |
| Creatine monophosphate | 83.3 mM | 20 μL of 500 mM stock |
| Creatine phosphokinase | 0.1 U/μL | 6 μL of 2 U/μL stock |
| RNase Free H2O | N/A | 72 μL |
| Total | N/A | 120 μL |
Store aliquots at −80°C for up to one year.
CRITICAL: DTT is a hazardous chemical. Carefully work under a chemical hood, and must wear gloves and a lab coat when handing. DTT waste needs to be collected and disposed of according to institute regulations.
IP lysis buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| Tris-HCl (pH 7.4) | 50 mM | 3.028 g |
| KCl | 300 mM | 11.183g |
| EDTA | 2 mM | 0.372 g |
| Nonidet P-40 | 0.5% | 5 mL |
| Glycerol | 10% | 50 mL |
| ddH2O | N/A | N/A |
| Total | N/A | 500 mL |
Store at 4°C for 6 months, and add DTT to a final concentration of 1 mM immediately before use.
IP washing buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| Tris-HCl (pH 7.4) | 50 mM | 3.028 g |
| NaCl | 300 mM | 8.766 g |
| MgCl2 | 5 mM | 0.238 g |
| Nonidet P-40 | 0.05% | 0.5 mL |
| ddH2O | N/A | N/A |
| Total | N/A | 500 mL |
Store at 4°C for 6 months.
Cleavage reaction buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| HEPES | 150 mM | 18 μL of 1 M stock |
| KCl | 200 mM | 24 μL of 1 M stock |
| ATP | 3.33 mM | 4 μL of 100 mM stock |
| DTT | 10 mM | 1.2 μL of 1 M stock |
| MgCl2 | 25 mM | 6 μL of 0.5 M stock |
| RNase Free H2O | N/A | 66.8 μL |
| Total | N/A | 120 μL |
Store aliquots at−80°C for up to one year.
Alternatives: Distilled H2O treated with 0.1% diethylpyrocarbonate (DEPC) can be used instead of commercial RNase Free H2O.
Step-by-step method details
Production of S2 lysates for RISC assembly
Timing: 4 days
This first part of the protocol is modified from a previous publication (Kawamata et al., 2009). This section describes how to prepare the cell lysates that will be used for RISC assembly assay.
-
1.Preparation of Drosophila S2 cells.
-
a.Culture Drosophila S2 cells in insect cell culture medium (Gibco) supplemented with penicillin-streptomycin and 10% FBS at 27°C.Note: To keep a healthy S2 cell culture going, the passage numbers of S2 cell is limited in 10–15.
-
b.Dilute confluent cells (∼1 × 107 cells/mL) and seed at a density of 5 × 105 cells/mL.
-
a.
-
2.dsRNA treatment.
-
a.Seed S2 cells into a 10 cm dish with volumes of 10 mL at a density of 1 × 105 cells/mL and prepare for RNAi.
-
b.Thaw the frozen dsRNA reagents and place them on ice.
-
c.Add 8 μg dsRNA for targetting each gene(dcr-2, r2d2, ago2, or ago1 (3′-UTR)) to the cell culture medium and shake gently.
-
d.Transfer 1 mL of cells from the 10 cm dish to a 12-well plate to detect the efficiency of RNAi knockdown. See troubleshooting 1.
-
a.
-
3.Plasmid transfection.
-
a.After 24 h of dsRNA treatment, prepare for plasmid transfection.
-
b.Preheat the Lipofectamine 2000 and Opti-MEM at room temperature (22°C–28°C).
-
c.Add 8 μg pAc5.1-Flag-AGO1 and 5 μg of pAc5.1-miR-9b plasmids to 500 μL Opti-MEM, and vortex the mixture.Note: The transfection of the miRNA is not essential. It depends on the expression levels of miRNA in the cell line.
-
d.Add 20 μL Lipofectamine 2000 to 500 μL Opti-MEM, and vortex the mixture.
-
e.Add diluted plasmids to the diluted Lipofectamine 2000 mixture.
-
f.Vortex the mixture, and incubate at room temperature (22°C–28°C) for 15 min.
-
g.Add the plasmid-lipid complex dropwise to the cell medium of the 10 cm dish and shake gently to mix.
-
a.
-
4.qRT-PCR to confirm the efficiency of RNAi knockdown.
-
a.Design primers to perform the qPCR. Primers used for qPCR are listed in the key resources table.
-
b.After 72 h of dsRNA treatment, collect cells for subsequent real-time PCR.
-
c.Extract total RNA from the S2 cells treated with dsRNAs by the Trizol method.
-
d.Prepare cDNA with the HiScript III Reverse Kit (Vazyme) according to the instructions and dilute to a volume of 100 μL.
-
e.Set up the qRT-PCR reaction system.
Reagent Amount 2 × SYBR Green Master Mix 10 μL 100 μM RT-Forward Primer 0.1 μL 100 μM RT-Reverse Primer 0.1 μL cDNA template 9.8 μL Total 20 μL -
f.Run the real-time PCR instrument.
Steps Temperature Time Cycles Initial denaturation 95°C 30 s 1 Denaturation 95°C 10 s 40 cycles Annealing/ Extension 60°C 30 s Melting curve 95°C 5 s 1 65°C 60 s 97°C 5 s Cooling 40 30 s 1 -
g.Calculate the efficiency of RNAi knockdown. The relative mRNA expression is determined using the formula: 2-Ct method (Schmittgen and Livak, 2008) (Figure 6). See troubleshooting 1.Alternatives: The TaqMan probes can be used instead of SYBR Green mixture and oligonucleotides.
-
a.
-
5.Preparation of cell lysates.
-
a.After 48 h of transfection, collect the transfected S2 cells by centrifugation at 500 g for 5 min at 4°C, and then wash cells with cold PBS twice.Note: The waste of cell culture medium and the PBS needs to be collected and thoroughly mixed with an appropriate amount of 84 disinfectants, then disposed of according to institute regulations.
-
b.Resuspend cells in 1 mL lysis buffer (freshly supplemented with 2 mM DTT and protease inhibitor cocktail) and transfer to a pre-chilled Dounce homogenizer.Note: Dissolve one Complete EDTA-free protease inhibitor tablet in 500 μL lysis buffer as 100× stock solutions. Prepare freshly before use.
-
c.Prepare homogenates by performing 50 strokes using the tight pestle.
 CRITICAL: This step should be performed on ice.
CRITICAL: This step should be performed on ice. CRITICAL: Between samples, clean the Douncer and pestle with lysis buffer 3 times to avoid cross-contamination between different samples.
CRITICAL: Between samples, clean the Douncer and pestle with lysis buffer 3 times to avoid cross-contamination between different samples. -
d.Clear the lysates by centrifugation at 15,000 g for 15 min at 4°C.
-
e.Transfer the supernatant and aliquot 50 μL/tube.
-
f.Freeze the lysates using liquid nitrogen and store them at −80°C.Note: We recommend immediately starting the AGO-RISC assembly reaction using fresh lysates.Alternatives: Instead of the lysates prepared from S2 cells overexpressing AGO1, and in which dcr-2, r2d2, ago2, and ago1 are knocked down, the lysates prepared from the embryo of dcr-2 mutant flies can be used (Lee et al., 2004).
 CRITICAL: Avoid multiple freezes and thaw cycles.
CRITICAL: Avoid multiple freezes and thaw cycles. Pause point: The lysates can be stored at −80°C for up to one month.
Pause point: The lysates can be stored at −80°C for up to one month.
-
a.
Figure 6.
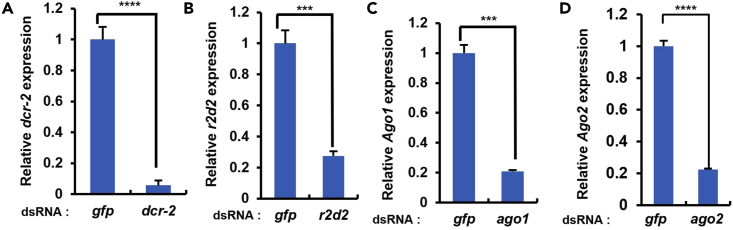
Relative expression levels of indicated genes in S2 cells treated with dsRNA measured by qRT-PCR
(A–D) S2 cells were treated with dsRNAs against dcr-2 (A), r2d2 (B), ago1 (C) or ago2 (D). Three days post dsRNA treatment, expressions of indicated genes were analyzed by qRT-PCR analysis. The two-tailed Student's t test was used to analyze statistical variance. Error bars indicate mean ± s.d (n=3). ∗∗∗, p<0.001, ∗∗∗∗, p<0.0001.
Native gel analysis of AGO1-RISC assembly
Timing: 4 h
This section describes how to perform the in vitro RISC assembly.
-
6.
Before beginning the in vitro RISC assembly, set up the agarose gel in the electrophoresis tank and fill it with pre-chilled 0.5 × TBE buffer.
Note: Cooling the electrophoresis buffer is critical to detect complexes in the AGO1-RISC assembly. It is optimal to perform the electrophoresis at 4°C.
-
7.
Prepare the in vitro RISC assembly as follows.
| Reagent | Amount |
|---|---|
| RISC Assembly mixture | 3 μL |
| S2 cell lysates | 4 μL |
| 50 nM miR-9b ASO | 1 μL |
| RNase inhibitor | 0.5 μL |
| RNase-free H2O | 1.5 μL |
| Total | 10 μL |
-
8.
Incubate the mixture at 27°C for 40 min. See troubleshooting 3.
-
9.
Load 3 μL of the assembly products into the 1.5% agarose native gel. See troubleshooting 5.
CRITICAL: Washing the gel and the gel wells with 0.5 × TBE buffer before running is essential for a good resolution.
-
10.
Load 3 μL of 2 × RNA loading dye and perform electrophoresis at 300 V in cold 0.5 × TBE buffer for 20 min.
CRITICAL: We have noticed that the RISC assembly sample will not rapidly load to the bottom of the wells, likely due to the RISC assembly mixture not containing glycerol. We load the sample using long pipet pips to sink the sample to the bottom of the wells.
-
11.
End the electrophoresis, and keep the gel attached to the glass plate.
-
12.
Perform phosphor imaging to detect the mature AGO1-RISC.
Immunoprecipitation of AGO1 complexes for cleavage assay
Timing: 8–12 h
This section details the associated procedures of immunoprecipitation of AGO1 complexes. This part of the protocol is critical since the immunoprecipitation of AGO1 complexes will be used for the RISC cleavage assay.
-
13.
Perform the cell culture as described under production of S2 lysates for RISC assembly.
-
14.Preparation of the cell lysates.
-
a.Harvest S2 cells by centrifugation at 500 g for 5 min.
-
b.Wash cells two times with cold PBS.
-
c.Add 1 mL of lysis buffer per 10 cm dish, resuspend the cell pellet by pipetting to ensure cells are fully lysed, and incubate on ice for 10 min.
-
d.Centrifuge at 12,000 g for 15 min at 4°C.
-
e.Carefully transfer the supernatant to new tubes and aliquot a 30 μL sample as the input fraction for western blot analysis.
-
a.
Note: When we transfer supernatant, do not disturb the pellets.
CRITICAL: Buffers should be pre-chilled on ice, and the centrifuge should be pre-cooled to 4°C.
-
15.Immunoprecipitation.
-
a.For immunoprecipitation of endogenous AGO1, incubate the supernatants with anti-AGO1 antibody or control IgG by rotation at 4°C for 5 h or overnight (6–12 h).
-
b.Use 20 μL of protein A/G agarose beads per sample.
-
c.Wash the beads once with 1 mL lysis buffer.
-
d.Add the washed beads to the lysates and incubate at 4°C for 3 h on a rotator.
-
e.Wash the samples three times with washing buffer.
-
f.Transfer the beads to a fresh tube and take an aliquot of 5 μL beads as the immunoprecipitation fraction for western blot analysis.
-
a.
CRITICAL: All steps should be performed on ice or at 4°C.
RISC cleavage assay
Timing: 5 h
The section of this protocol is modified from a previous publication (Miyoshi et al., 2005), and this section describes how to perform the RISC cleavage assay in vitro.
-
16.
Remove the supernatant.
-
17.
Prepare the in vitro RISC cleavage reaction mixture as follows.
| Reagent | Amount |
|---|---|
| Cleavage reaction mixture | 5 μL |
| Protein A/G Beads +/- Immunoprecipitated AGO1 | 10 μL |
| Cleavage substrate 1 or 2 | 100 ng |
| RNase inhibitor | 1 μL |
| RNase-free H2O | N/A |
| Total | 25 μL |
-
18.
Incubate at 27°C for 2 h.
-
19.
Add 1 mL of Trizol reagent to the cleavage RNA sample and pipet the sample up and down several times to homogenize.
-
20.
Incubate for 10 min to allow complete dissociation of the AGO1-RNA complex.
-
21.
Add 0.2 mL of chloroform to 1 mL of Trizol, cap the tube and mix thoroughly by shaking.
-
22.
Incubate at room temperature (22°C–28°C) for 5 min.
CRITICAL: Trizol reagent and chloroform are hazardous solutions. Carefully work under a fume hood, must wear gloves and a lab coat. Trizol and chloroform waste must be collected and disposed of according to institute regulations.
-
23.
Centrifuge the sample for 15 min at 13,000 g at 4°C.
-
24.
Transfer 0.5 mL of the aqueous phase to a new tube.
Note: After centrifugation, three phases will be obtained. An upper aqueous phase contains RNA, interphase contains DNA, and a lower phase contains proteins. Carefully pipet the aqueous phase and avoid transferring the interphase into the pipette.
-
25.
Add 0.5 mL of 100% ethanol to the aqueous phase and mix well.
-
26.
Proceed with the RNA Clean-up protocol of the ZYMO-Spin column.
-
27.
Subject the cleaved fragment of substrate 1 to the Qsep1 for size distribution analysis, and the cleaved fragment of substrate 2 to gel electrophoresis analysis.
Alternatives: Agilent bioanalyzer (Agilent 2100 Bioanalyzer) with the Agilent RNA Pico Kit (Cat # 5067-1513RUO) can be used instead of Qsep1.
Expected outcomes
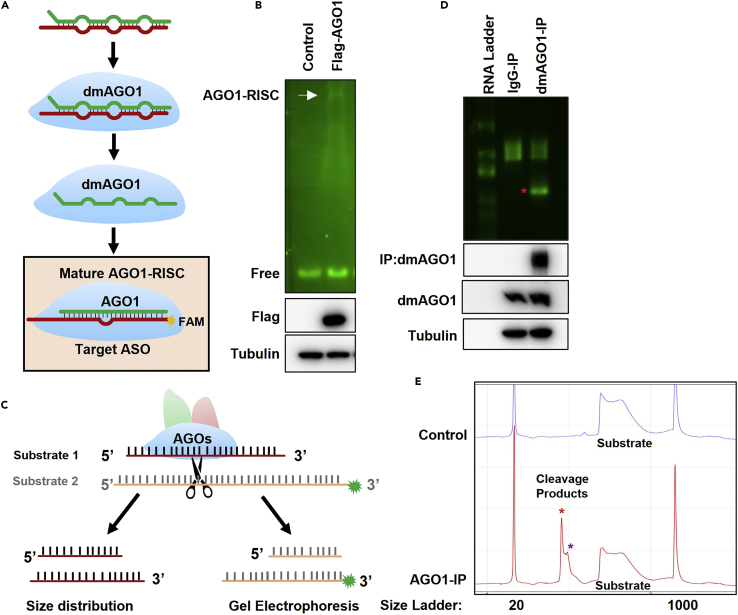
Here, we provide an optional protocol to detect AGO1-RISC assembly and cleavage by fluorescent labeling of RNA instead of radiolabeling. We use a FAM-ASO that is complementary to the miR-9b guide strand to detect the mature AGO1-RISC (Figures 7A and 7B). Moreover, we prepare FAM-labeled RNA substrate by T4 RNA ligation and perform the cleavage assay (Figures 7C–7E).
Figure 7.
In vitro RISC assembly and cleavage assay
(A) Schematic of AGO-RNA-induced silencing complex assembly.
(B) The lysates from transfected S2 cells treated with dsRNA targeting dcr-2, ago2, r2d2, and ago1 (Figure 6) were incubated with 5′-FAM-labeled and 3′-2′-O-methylated anti-miR-9b ASO complementary to the guide strand of miR-9b for 40 min. Then the complexes were analyzed by native gel electrophoresis.
(C) Schematic of the cleavage assay procedure. The AGO complexes purified by immunoprecipitation cleaved the RNA substrate at a defined position.
(D and E) The AGO1 was immunoprecipitated by anti-AGO1 protein A/G beads from S2 cell lysates and incubated with cleavage substrate 1 or 2 containing a sequence complementary to miR-9b. The cleaved fragment was detected by gel electrophoresis (D) and Qsep1 (E).∗Indicates the signal corresponding to the cleaved substrate.
Limitations
Because this method labels the target RNA of RISC in the in vitro RISC assembly reaction, only the mature AGO1-RISC is detected. This protocol is not applicable to analyzing the dynamic formation of AGO1-RISC complexes. The sensitivity of fluorescent labeling is lower than that of radiolabeling.
Troubleshooting
Problem 1
Low RNAi knockdown efficiency (steps 2 and 4).
Potential solution
Increase the amount of dsRNA used for RNAi. Seed the S2 cells at a lower density. After dsRNA treatment, treat the S2 cells by starvation using a medium without serum for 30 min.
Problem 2
Low yield of the cleavage RNA substrate (step of preparation of substrate RNAs for the cleavage assay).
Potential solution
Increase the reaction time to 4 h or overnight (6–12 h). Increase the amount of template DNA.
Problem 3
There is no signal for AGO1-RISC present in the agarose gel (step 8).
Potential solution
Check the amount and quality of the miR-9b ASO on an agarose gel to ensure it is intact and of the expected size. Extend the reaction time. The reaction temperature can also be adjusted. The temperature is critical for the formation and stability of mature AGO1-RISC complexes.
Problem 4
RNA degradation (step of preparation of substrate RNAs for the cleavage assay).
Potential solution
RNA is highly sensitive to RNases. To avoid RNase contamination, the pipettes, pipette boxes and tube racks are treated with RNase, RNA and DNA Remover (Vazyme); Use the RNase-free 1.5 mL tubes, pipette tips, and H2O even though the buffer or reagent contains RNase inhibitors, the probability of degradation increases with time and temperature. To preserve RNA quality and avoid degradation, keep everything cold and spend no more than 60 min for RNA recovery.
Problem 5
Images of the RISC assembly contain high background (step 9).
Potential solution
The time of in vitro RISC assembly and the loading volumes of the products should be tested. The background might be reduced by a different loading volume or a longer reaction time.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr. Dahua Chen (chendh@ynu.edu.cn).
Materials availability
All unique materials generated from this study are available from the lead contact with a complete Materials Transfer Agreement.
Acknowledgments
This work is supported by Basic Science Center Program of NSFC (grant #31988101) and the National Key R&D Program of China (2018YFC1003300 and 2021YFA0805800).
Author contributions
Y.G., Y.Z., Q.S., and D.C. designed experiments. Y.G. and Y.Z. performed experiments. Y.G., Y.Z., Q.S., and D.C. performed data analysis. Y.G., Y.Z., Q.S., and D.C. wrote the manuscript. All authors approved the submission.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Yajie Gao, Email: gaoyajie@ynu.edu.cn.
Qinmiao Sun, Email: qinmiaosun@ioz.ac.cn.
Dahua Chen, Email: chendh@ynu.edu.cn.
Data and code availability
Original data have been deposited to Mendeley Data: https://data.mendeley.com/datasets/4gmsw3t82z/1.
References
- Bartel D.P. Metazoan MicroRNAs. Cell. 2018;173:20–51. doi: 10.1016/j.cell.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchaine T.F., Fabian M.R. Mechanistic insights into MicroRNA-mediated gene silencing. Cold Spring Harb. Perspect. Biol. 2019;11:a032771. doi: 10.1101/cshperspect.a032771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Zhu Y., Wang H., Cheng Y., Zhao D., Sun Q., Chen D. Lipid-mediated phase separation of AGO proteins on the ER controls nascent-peptide ubiquitination. Mol. Cell. 2022;82:1313–1328.e8. doi: 10.1016/j.molcel.2022.02.035. [DOI] [PubMed] [Google Scholar]
- Iwakawa H.-O., Tomari Y. Life of RISC: formation, action, and degradation of RNA-induced silencing complex. Mol. Cell. 2022;82:30–43. doi: 10.1016/j.molcel.2021.11.026. [DOI] [PubMed] [Google Scholar]
- Kawamata T., Tomari Y. In: Argonaute Proteins. Hobman T.C., Duchaine T.F., editors. Humana Press; 2011. Native gel analysis for RISC assembly; pp. 91–105. [Google Scholar]
- Kawamata T., Seitz H., Tomari Y. Structural determinants of miRNAs for RISC loading and slicer-independent unwinding. Nat. Struct. Mol. Biol. 2009;16:953–960. doi: 10.1038/nsmb.1630. [DOI] [PubMed] [Google Scholar]
- Kwak P.B., Tomari Y. The N domain of Argonaute drives duplex unwinding during RISC assembly. Nat. Struct. Mol. Biol. 2012;19:145–151. doi: 10.1038/nsmb.2232. [DOI] [PubMed] [Google Scholar]
- Lee Y.S., Nakahara K., Pham J.W., Kim K., He Z., Sontheimer E.J., Carthew R.W. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117:69–81. doi: 10.1016/s0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- Meister G., Landthaler M., Patkaniowska A., Dorsett Y., Teng G., Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Miyoshi K., Tsukumo H., Nagami T., Siomi H., Siomi M.C. Slicer function of Drosophila Argonautes and its involvement in RISC formation. Genes Dev. 2005;19:2837–2848. doi: 10.1101/gad.1370605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen T.D., Livak K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treiber T., Treiber N., Meister G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat. Rev. Mol. Cell Biol. 2019;20:5–20. doi: 10.1038/s41580-018-0059-1. [DOI] [PubMed] [Google Scholar]
- Xia L., Jia S., Huang S., Wang H., Zhu Y., Mu Y., Kan L., Zheng W., Wu D., Li X., et al. The fused/smurf complex controls the fate of Drosophila germline stem cells by generating a gradient BMP response. Cell. 2010;143:978–990. doi: 10.1016/j.cell.2010.11.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Original data have been deposited to Mendeley Data: https://data.mendeley.com/datasets/4gmsw3t82z/1.