Abstract
The Lactococcus lactis ptsH and ptsI genes, encoding the general proteins of the phosphoenolpyruvate-dependent phosphotransferase system, HPr and enzyme I, respectively, were cloned, and the regulatory role of HPr was studied by mutational analysis of its gene. A promoter sequence was identified upstream of the ptsHI operon, and the transcription start site was mapped by primer extension. The results of Northern analyses showed the presence of two glucose-inducible transcripts, one of 0.3 kb containing ptsH and a second of 2.0 kb containing both ptsH and ptsI. Disruption of the ptsH and ptsI genes in strain NZ9800 resulted in a reduced growth rate at the expense of glucose, but no growth at the expense of sucrose and fructose, confirming the dominant role of the phosphotransferase system in the uptake of these sugars in L. lactis. Complementation of the ptsH and ptsI mutants with the intact genes under the control of a regulated promoter resulted in the restoration of the wild-type phenotype. The role of HPr(Ser-P) in the recently established CcpA-mediated control of galactose metabolism as well as glycolysis was analyzed by producing an HPr mutant carrying an aspartic acid on residue 46 which mimicks a phosphorylated serine. The results of these experiments demonstrated the role of HPr(Ser-P) as corepressor in the catabolite repression of the gal operon. Furthermore, we show for the first time that HPr(Ser-P) functions as a coactivator in the CcpA-mediated catabolite activation of the pyruvate kinase and l-lactate dehydrogenase genes.
The main sugar uptake system in many bacteria is the phosphoenolpyruvate:sugar phosphotransferase system (PEP-PTS), which mediates the uptake and phosphorylation of carbohydrates (32). The PTS is a group translocation process in which the transfer of the phosphate moiety of PEP to carbohydrates is catalyzed by the general non-sugar-specific proteins enzyme I and HPr in combination with sugar-specific enzyme II (EII) proteins. After autophosphorylation of enzyme I at the expense of PEP, enzyme I catalyzes the phosphorylation of HPr at histidine 15, resulting in HPr(His-P). The phosphate group from HPr(His-P) is then transferred to the sugar substrate via a two-step phosphorylation reaction mediated by a dedicated EII protein. EII proteins can consist of one or more proteins and are composed of three domains: the EIIA and EIIB domains, which are involved in the phosphotransfer, and the membrane-located EIIC domain, which is most likely involved in the translocation of the sugar substrate (32). The genes encoding HPr and enzyme I, ptsH and ptsI, respectively, have been cloned from several bacteria and found often to be organized in an operon structure with the gene order ptsHI (13, 18, 22). Expression of the Escherichia coli and Bacillus subtilis ptsHI operons appears to be regulated at the transcriptional level, since mRNA levels were found to be higher in glucose-grown cells than in cells grown on non-PTS sugars (6, 38).
Apart from its function in the uptake of sugars, the PTS also plays a regulatory role. In E. coli and other gram-negative bacteria, the PTS regulates the concentration of cyclic AMP (cAMP) via activation of adenylate cyclase by the phosphorylated form of the glucose-specific EIIA, the concentration of which increases in the absence of PTS substrates (32). Elevated concentrations of cAMP lead to transcriptional activation of several genes via the binding of the cAMP receptor protein complexed with cAMP to operator sites located in the promoter regions of affected genes. Furthermore, the unphosphorylated form of the glucose-specific EIIA reduces the uptake of several non-PTS sugars via an interaction with the uptake protein (32). Regulatory functions for the PTS have also been described for gram-positive bacteria. The HPr(His-P)-mediated phosphorylation of two glycerol kinases results in an increased activity of both enzymes in Enterococcus spp. (4). In contrast, enzyme I/HPr(His-P)-mediated phosphorylation of the lactose permease in Streptococcus thermophilus results in a reduced permease activity leading to a decreased uptake of sugar (31). In B. subtilis, the PTS regulates the expression of the levanase operon by HPr(His-P) mediated phosphorylation of the transcriptional regulator LevR, resulting in activation of transcription (37).
Apart from phosphorylation at residue His-15, a second phosphorylation site has been identified in HPr, the function of which has been shown only in gram-positive bacteria. Phosphorylation of HPr at residue Ser-46 is catalyzed by an ATP-dependent protein kinase that is activated by fructose-1,6-diphosphate (8, 35). Recently, the genes encoding the HPr(Ser) kinase and the HPr(Ser) phosphatase were cloned from B. subtilis, and their involvement in the phosphorylation of HPr at residue Ser-46 was established (16, 33). The seryl-phosphorylated form of HPr, designated HPr(Ser-P), interacts with several PTS and non-PTS sugar permeases, and this process, termed inducer exclusion, results in a reduced uptake of sugars (35). In addition, HPr(Ser-P) allosterically activates sugar-phosphate phosphatases in Lactococcus lactis, Enterococcus faecalis, and Streptococcus pyogenes that catalyze the dephosphorylation of various phosphorylated sugars, resulting in an efflux of the sugar from the cell, a process known as inducer expulsion (44, 47). Apart from these allosteric control systems, HPr(Ser-P) can also negatively regulate the transcription of genes by an interaction with the catabolite control protein, CcpA (10, 20). It has been reported that the in vitro binding of CcpA to a cis-acting cre (catabolite-responsive element) operator site, located in the promoter region of genes controlled by CcpA, is enhanced by an interaction with HPr(Ser-P), glucose-6-P, and NADP (12, 17, 23). Deutscher et al. (9) demonstrated that expression of the gene encoding S46D HPr, which is structurally similar to HPr(Ser-P), leads to catabolite repression of the B. subtilis gluconate kinase gene even in the absence of glucose. In addition, they showed that the replacement of HPr with S46A HPr has the same effect as a mutation in the ccpA gene, rendering the gluconate kinase gene insensitive to catabolite repression.
Recently, we have demonstrated (i) the involvement of the L. lactis CcpA in the negative regulation of the expression of the genes involved in galactose metabolism and (ii) the positive control of the las operon encoding phosphofructokinase, pyruvate kinase, and l-lactate dehydrogenase (29). Here, we report the cloning and analysis of the L. lactis ptsH and ptsI genes and the involvement of HPr in the catabolite repression of galactose metabolism. Furthermore, we show for the first time the participation of HPr(Ser-P) in the CcpA-mediated transcriptional activation of the las operon.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and transformation procedure.
The L. lactis strains and plasmids used in this study are described in Table 1. The strains were cultivated without aeration at 30°C in M17 broth supplemented with different carbon sources. L. lactis was transformed by electroporation as described by Holo and Nes (22). E. coli MC1061 was used as a host for cloning experiments and grown in L-broth-based medium with aeration at 37°C. Antibiotics were used in the following concentrations: ampicillin, 50 μg/ml (for selection of pUC19-based plasmids in E. coli); chloramphenicol, 5 μg/ml (for selection of pNZ8030-based plasmids in E. coli and L. lactis); and erythromycin, 2.5 μg/ml (for selection for the integration of the erythromycin resistance [Ermr] gene into the chromosome of L. lactis). Bacterial growth was monitored spectrophotometrically at 600 nm.
TABLE 1.
L. lactis strains and plasmids used in this study
| Strain or plasmid | Relevant properties | Reference |
|---|---|---|
| Strains | ||
| NZ9800 | 26 | |
| NZ9880 | NZ9800 derivative; ΔptsH | This work |
| NZ9881 | NZ9800 derivative: ΔptsI | This work |
| Plasmids | ||
| pUC19E | pUC19 derivative containing an Ermr gene | 28 |
| pNZ8030 | Lactococcal cloning and expression vector | 7 |
| pNZ9279 | pGEM-T containing a 0.8-kb PCR product derived from the L. lactis ptsHI operon | This work |
| pNZ9280 | pUC19 containing a 4.5-kb SstI-ClaI fragment carrying the ptsH gene and part of the ptsI gene | This work |
| pNZ9281 | pUC19 containing a 3.5-kb EcoRV-BamHI fragment carrying the ptsH and ptsI genes | This work |
| pNZ9282 | pNZ8030 containing the L. lactis ptsH gene translationally fused to the nisA promoter | This work |
| pNZ9283 | pNZ8030 containing the B. subtilis ptsH gene translationally fused to the nisA promoter | This work |
| pNZ9284 | pNZ8030 containing the S46A ptsH gene translationally fused to the nisA promoter | This work |
| pNZ9285 | pNZ8030 containing the S46D ptsH gene translationally fused to the nisA promoter | This work |
| pNZ9286 | pNZ8030 containing the ptsH and ptsI genes translationally fused to the nisA promoter | This work |
| pNZ9287 | pNZ9286 derivative containing the ptsI gene transcriptionally fused to the nisA promoter | This work |
| pNZ9288 | pNZ8030 containing S46A ptsH and ptsI translationally fused to the nisA promoter | This work |
| pNZ9289 | pNZ8030 containing S46D ptsH and ptsI translationally fused to the nisA promoter | This work |
| pNZ9290 | ptsH disruption construct | This work |
| pNZ9291 | pUC19E derivative containing an internal ClaI-PstI fragment of ptsI | This work |
DNA techniques and DNA sequence analysis.
All manipulations with recombinant DNA were carried out according to standard procedures (36) and as specified by the enzyme manufacturer (Gibco/BRL Life Technologies [Breda, The Netherlands] or United States Biochemical Corp. [Cleveland, Ohio]). Plasmid and chromosomal DNA of L. lactis was isolated as described previously (41). The nucleotide sequence of the ptsHI genes was determined on both strands, using an ALF DNA sequencer (Pharmacia LKB Biotechnology, Uppsala, Sweden). PCR was performed in a total volume of 50 μl containing 10 mM Tris-HCl (pH 8.8), 50 mM NaCl, 2 mM MgCl2, 10 μg of gelatin, 200 μM each deoxynucleoside triphosphate, 1 U of Taq polymerase (Gibco/BRL), 10 pmol of each primer, and 10 to 100 ng of template DNA. A small volume of mineral oil was added to prevent evaporation. PCR amplifications were performed in 25 cycles, each cycle consisting of a denaturation step at 95°C for 1 min, a primer annealing step at the appropriate temperature for 1 min, and a primer extension step at 72°C for 2.5 min, carried out in a DNA Thermocycler 480 (Perkin Elmer, Norwalk, Conn.).
Construction of plasmids.
Primers PTS1 (5′-AACWGGWATTCATGCWMGWCCWGC-3′) and PTS2 (5′-GGTACCWCCAATATTWGTWACAAAWGC-3′) were used in a PCR with chromosomal DNA from L. lactis NZ9800 to amplify part of the ptsHI operon. The PCR product was cloned in the pGEM-T vector (Promega, Madison, Wis.), yielding pNZ9279. The insert from pNZ9279 was used as a probe to clone a 4.5-kb SstI-ClaI fragment from the chromosomal DNA of L. lactis into SstI-AccI-digested pUC19 (43), resulting in pNZ9280. The same fragment was used as a probe to clone a 3.5-kb EcoRV-BamHI fragment from the chromosomal DNA in SmaI-BamHI-digested pUC19, resulting in pNZ9281.
By the use of oligonucleotides HPRNCOI (5′-GAACCATAACCATGGCATCTAAAG-3′) and HPRHINDIII (5′-GACAAGCAAGCTTGCCTTAGCTACTG-3′) the ptsH gene was amplified by PCR with chromosomal DNA from L. lactis NZ9800 as the template. Bases that were changed to introduce the NcoI and HindIII restriction sites are underlined. These sites were used to clone the PCR product in NcoI-HindIII-digested pNZ8030, resulting in plasmid pNZ9282.
The B. subtilis ptsH gene (18) was amplified by PCR with oligonucleotides BSHPRNCO (5′-GGAGAATGATAACCATGGCACAAAAAAC-3′) and BSHPRHIND (5′-GCAGAGATGTTAAGCTTTTCAACC-3′) and chromosomal DNA from B. subtilis 168 as the template. The PCR product was digested with NcoI and HindIII and cloned into NcoI-HindIII-digested pNZ8030, yielding pNZ9283.
Residue serine 46 of L. lactis HPr was changed to an alanine and an aspartic acid by PCR-mediated megaprimer mutagenesis of the ptsH gene, as described previously (25), with the mutagenic primers HPR46A (3′-GTAAACCTTAAAGCAATCATGGGTGTTATGTC-3′) and HPR46D (5′-GTAAACCTTAAAGATATCATGGGTGTTATGTC-3′), respectively. The substitutions needed to change codon 46 are underlined. Two amplification rounds were used: the first with the mutagenic primers and the antiparallel primer PTSHINDIII; the second with the purified PCR products from the first round as megaprimer, together with primer PTSHNCOI. The PCR products were digested with NcoI and HindIII and cloned into NcoI-HindIII-digested pNZ8030, yielding pNZ9284 and pNZ9285, carrying the genes encoding S46A HPr and S46D HPr, respectively.
For the construction of a translational fusion between the nisA promoter and the entire ptsHI operon encoding wild-type, S46A, or S46D HPr together with enzyme I, a 3.2-kb DraIII-BamHI fragment was isolated from pNZ9281. After treatment with Klenow DNA polymerase, this fragment was cloned into pNZ9282, pNZ9284, or pNZ9285 digested with DraIII and HindIII. The latter site was made blunt by treatment with Klenow DNA polymerase, and the resulting plasmids were designated pNZ9286, pNZ9288, and pNZ9289, respectively.
Plasmid pNZ9287 was constructed by digesting plasmid pNZ9286 with NcoI and DraIII to remove the 5′ end of the ptsH gene and then treating the ends with Klenow enzyme and T4 DNA polymerase, followed by self-ligation of the resulting DNA. To disrupt the ptsH gene, a BamHI site was introduced in the ptsH open reading frame by PCR using primers HPRBAM2 (5′-GTGTTGGATCCCTCGGTGTTGGTCAAG-3′), corresponding to nucleotides 403 to 419, and PTS6 (5′-GATGGATTGTAAGGTTGATA-3′), complementary to nucleotides 1947 to 1967 of the ptsHI genes. The PCR product was digested with BamHI and HindIII, and the 1.0-kb fragment was cloned into BamHI-HindIII-digested pUC19. The resulting plasmid was digested with SstI and BamHI, and a 1.2-kb Sau3A fragment from plasmid pIL253 carrying an Ermr gene and a 3.5-kb SstI-BclI fragment carrying the upstream region of the ptsH gene from plasmid pNZ9280 were cloned into these sites. The expected constructs were verified by restriction endonuclease analysis, one product, containing the Ermr gene in the same orientation as the ptsI open reading frame and designated pNZ9290, was selected for further study. Plasmid pNZ9291 was constructed by cloning a 0.9-kb ClaI-PstI fragment from plasmid pNZ9281, the ClaI site of which was made blunt by Klenow DNA polymerase treatment, into PstI-BamHI-digested pUC19E, the BamHI site of which was filled in by use of Klenow DNA polymerase. The nucleotide sequences of all DNA fragments obtained via PCR were verified and found to be correct.
RNA analysis.
RNA was isolated from L. lactis cultures as described previously (26). RNA was denatured and size fractionated on a 1% agarose gel containing formaldehyde according to standard procedures (36). The RNA was stained by adding ethidium bromide to the sample buffer. As molecular weight markers, a 0.24- to 9.5-kb RNA ladder from Bethesda Research Laboratories was used. The gel was blotted to a nylon membrane (Gene Screen Plus nylon membranes; NEN Research Products, Wilmington, Del.) as recommended by the manufacturer. Blots were hybridized with oligonucleotides PEPTSH (for ptsH) and PTS6 (for ptsI). Following autoradiography, bands were cut out and total radioactivity was determined in a liquid scintillation counter (Beckman LKS 7500).
Primer extension analysis.
The oligonucleotide used for priming cDNA synthesis was PEPTSH (5′-CTGCAACGATGTGGAATTCTTTAG-3′), complementary to nucleotides 254 to 278 in the coding strand of the ptsH gene. Primer extension reactions were performed by annealing 2 ng of oligonucleotide to 100 μg of total RNA as described elsewhere (26).
Enzyme assays.
Pyruvate kinase and l-lactate dehydrogenase activities were determined according to standard methods (5, 21). Protein concentrations were assayed by the method of Bradford (3), with bovine serum albumin as the standard.
Nucleotide sequence accession number.
The nucleotide sequence data reported in this paper will appear in the EMBL, GenBank, and DDBJ nucleotide sequence databases under accession no. Z97203.
RESULTS
Cloning of the ptsHI operon.
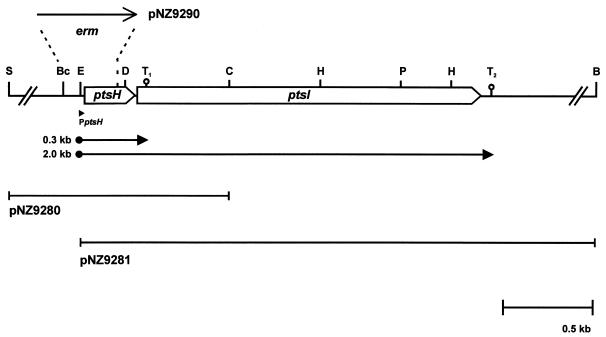
Sequences of HPr proteins from different gram-positive bacteria were aligned. Oligonucleotide PTS1 was designed based on the conserved region TGIHARP, encompassing the catalytic phosphorylation site His-15 in several HPr sequences. The reported active-site sequence of the L. lactis enzyme I (1) was used to design an antisense oligonucleotide, PTS2. Assuming that the gene order in L. lactis would be identical to that found in other bacteria (ptsHI), primers PTS1 and PTS2 were used in a PCR under nonstringent conditions with chromosomal DNA from L. lactis NZ9800 as the template. The PCR product had the expected size of 0.8 kb. The nucleotide sequence of this fragment was analyzed and found to contain two partial open reading frames encoding proteins with high sequence homology to HPr and enzyme I sequences. This fragment was used as a probe and hybridized to a single DNA fragment of the L. lactis chromosome that was cloned as two overlapping fragments, resulting in plasmids pNZ9280 and pNZ9281.
Characterization of the ptsH and ptsI genes.
Analyses of the nucleotide sequences in the inserts of plasmids pNZ9280 and pNZ9281 revealed the presence of two complete open reading frames. The first, designated ptsH (see below), could encode a protein of 88 amino acids that shows high sequence homology to HPr proteins from different bacteria, with up to 91% identical residues compared to the S. salivarius HPr protein (13). A sequence (5′-AAGGAG-3′) resembling a typical lactococcal ribosome binding site was identified 10 nucleotides upstream of the ATG start codon (11). The second open reading frame, designated ptsI (see below), could encode a protein of 575 amino acids with high sequence similarity to enzyme I proteins of various bacteria, i.e., 83% identity compared to the S. salivarius enzyme I (14). The putative ATG start codon of the ptsI gene, which is located eight nucleotides downstream of the ptsH gene, is preceded by a putative lactococcal ribosome binding site (5′-AAGGA-3′) with an unusual spacing of 14 bp. Two stem-loop structures were identified in the ptsHI operon: one with a ΔG value of −6.4 kcal/mol located downstream of the ptsH gene (terminator T1), and a second with a ΔG value of −9.0 kcal/mol followed by an AT-rich stretch of nucleotides downstream of the ptsI gene (T2) (Fig. 1).
FIG. 1.
Transcriptional organization of the ptsHI operon of L. lactis NZ9800. The genes (open arrows) connected by intergenic regions (thin lines) are shown with their products. The orientation of the Ermr gene (erm) in plasmid pNZ9290 is indicated. The arrows below the genes denote the promoter and transcripts mapped with primer extension and Northern analysis, respectively. The putative terminators downstream of ptsH and ptsI, (T1 and T2, respectively) are indicated. Relevant restriction sites: S, SstI; Bc, BclI; E, EcoRV; D, DraIII; C, ClaI; H, HindIII; P, PstI; B, BamHI.
Transcriptional analysis of the ptsHI operon.
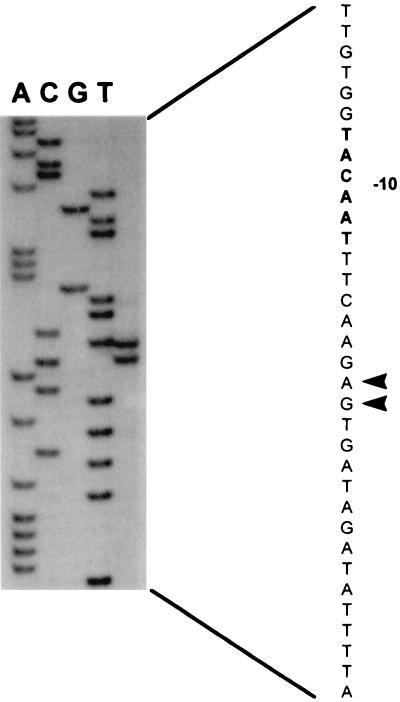
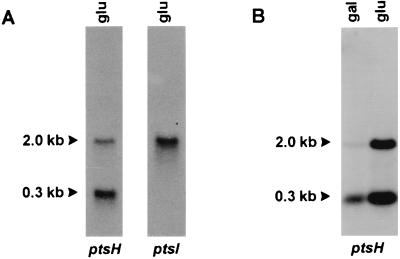
Primer extension analysis was performed with total RNA isolated from strain NZ9800 grown on glucose. Two equally intense labeled primer extension products were obtained, indicating that transcription starts at two adjacent sites (Fig. 2). Upstream of these start sites, a sequence corresponding to consensus L. lactis promoter sequences was identified (11, 40). Northern analysis of the ptsHI genes after hybridization with a ptsH-specific probe revealed the presence of two transcripts, one of 0.3 kb and a second of 2.0 kb (Fig. 3A). When the same blot was stripped and rehybridized with a ptsI-specific probe, only the 2.0-kb transcript hybridized (Fig. 3A). RNA isolated from cells grown on galactose and glucose was hybridized with a ptsH-specific probe. The results showed that the transcription of the ptsHI operon varies in response to the carbon source, since transcription levels were at least 10-fold higher in glucose-grown cells than in galactose-grown cells, as determined by scintillation counting (Fig. 3B). This finding demonstrates that expression of the L. lactis ptsHI operon is regulated at the transcriptional level in response to the carbon source.
FIG. 2.
Determination of the transcription start sites of the pts promoter. The transcription start sites are indicated with arrowheads. The putative −10 region in the coding strand is in bold. RNA was isolated from strain NZ9800 grown on glucose.
FIG. 3.
(A) Northern analysis of ptsH and ptsI transcription in L. lactis NZ9800 grown at the expense of glucose; (B) transcriptional analysis of the ptsH gene in L. lactis strain NZ9800 grown at the expense of galactose and glucose. Estimated sizes of the transcripts are indicated with arrowheads.
Disruption of the ptsH and ptsI genes.
To analyze the functional and regulatory roles of the L. lactis ptsH and ptsI genes, both genes were disrupted by replacement recombination. The ptsH gene was disrupted by chromosomal integration of plasmid pNZ9290, which contains an Ermr cassette flanked by upstream and downstream regions of the ptsH gene. Integrants were obtained and analyzed via PCR and Southern hybridization. We selected one strain in which the integration had occurred via two simultaneous crossover events resulting in the replacement of the ptsH promoter and the region encoding residues His-15 and Ser-46 by the Ermr cassette. This strain, designated NZ9880, had the Ermr cassette integrated in the same orientation as the ptsI gene. Disruption of the ptsI gene was achieved by the integration of plasmid pNZ9291 into the chromosome of strain NZ9800, resulting in a strain, named NZ9881, that contains two disrupted copies of the ptsI gene.
Growth characteristics of the ptsH-disrupted strain.
Disruption of the ptsH or ptsI gene resulted in a severe reduction of the growth rate on all sugars tested (Table 2). No growth at the expense of sucrose and fructose was observed, while growth rate at the expense of glucose was reduced to approximately 1/10 of that observed in the wild-type strain. Remarkably, the growth at the expense of galactose and maltose was also affected by these mutations; the growth rate on both carbon sources was reduced to half of that observed for the wild-type strain (Table 2). Results for disruption of the ptsI gene were identical to those for the ptsH disruption, confirming the important role of the PTS in the utilization of these sugars.
TABLE 2.
Maximal growth rates of strains used in this study
| Strains | Nisin A induction (μg/ml) | Mean maximal growth rate (h−1) ± SEa
|
||||
|---|---|---|---|---|---|---|
| Glucose | Sucrose | Fructose | Galactose | Maltose | ||
| NZ9800 (wtb) | 1.40 ± 0.06 | 1.16 ± 0.04 | 0.91 ± 0.02 | 0.59 ± 0.1 | 0.50 ± 0.04 | |
| NZ9880 (ΔptsH) | 0.13 ± 0.03 | NG | NG | 0.30 ± 0.04 | 0.24 ± 0.06 | |
| NZ9880 + pNZ9282 (L. lactis ptsH) | 10−3 | ND | 0.09 ± 0.03 | ND | 0.35 ± 0.02 | ND |
| NZ9880 + pNZ9283 (B. subtilis ptsH) | 10−3 | ND | 0.10 ± 0.06 | ND | 0.39 ± 0.09 | ND |
| NZ9880 + pNZ9286 (wt ptsHI) | 10−5 | 1.39 ± 0.09 | 1.09 ± 0.06 | 0.90 ± 0.09 | 0.61 ± 0.04 | 0.48 ± 0.03 |
| NZ9880 + pNZ9288 (S46A ptsHI) | 10−5 | 1.17 ± 0.08 | 0.99 ± 0.01 | 0.80 ± 0.05 | 0.48 ± 0.03 | 0.39 ± 0.03 |
| NZ9880 + pNZ9289 (S46D ptsHI) | 10−5 | 0.12 ± 0.04 | NG | NG | 0.21 ± 0.04 | 0.20 ± 0.04 |
| NZ9881 (ΔptsI) | 0.16 ± 0.05 | NG | NG | 0.25 ± 0.08 | 0.26 ± 0.06 | |
| NZ9881 + pNZ9286 (wt ptsHI) | 10−5 | 1.35 ± 0.09 | 1.18 ± 0.09 | 0.84 ± 0.06 | 0.63 ± 0.08 | 0.50 ± 0.07 |
| NZ9881 + pNZ9287 (wt ptsI) | 10−3 | 1.20 ± 0.03 | 1.02 ± 0.03 | 0.89 ± 0.04 | 0.55 ± 0.06 | 0.43 ± 0.07 |
| NZ9800 + pNZ9284 (S46A ptsH) | 10−4 | 1.40 ± 0.11 | 1.18 ± 0.10 | ND | 0.61 ± 0.09 | ND |
| NZ9800 + pNZ9285 (S46D ptsH) | 10−4 | 1.35 ± 0.02 | 1.02 ± 0.07 | ND | 0.09 ± 0.02 | ND |
NG, no growth; ND, not determined.
wt, wild type.
Complementation of the ptsH and ptsI mutations.
Since the pIL253-derived Ermr cassette used to construct the disruption mutants does not contain a terminator structure (27), readthrough of the Ermr promoter leads to expression of the ptsI gene in strain NZ9880 (ΔptsH), albeit not to the wild-type level (data not shown). Strain NZ9880 was transformed with plasmid pNZ9282 or pNZ9283, containing the L. lactis or B. subtilis ptsH gene, respectively, under the control of the regulated nisA promoter. The nisA promoter allows inducible expression in response to the amount of nisin A added to the growth medium (7). Induction of the expression of ptsH of either L. lactis or B. subtilis by the addition of inducing concentrations nisin A resulted in partial complementation of the observed growth defect of strain NZ9880 grown at the expense of sucrose and galactose (Table 2). Comparable results were obtained with glucose, fructose, and maltose. Without nisin A induction, strain NZ9880 harboring either pNZ9282 or pNZ9283 did not grow at the expense of sucrose. The addition of a relatively high concentration of nisin A (10−3 μg/ml) restored the ability to grow on sucrose, although the maximal specific growth rate was approximately 1/10 of that observed for the wild-type strain, probably because of insufficiently high expression of ptsI. Transformation with plasmid pNZ9286, which contains the entire L. lactis ptsHI operon under control of the nisA promoter, almost completely restored the observed growth defect of strain NZ9880 after induction by the addition of a low concentration of nisin A (10−5 μg/ml). Similarly, the ptsI mutation in strain NZ9881 could be complemented with plasmid pNZ9286 (containing the entire ptsHI operon) or plasmid pNZ9287 (containing the ptsI gene transcriptionally fused to the nisA promoter), although the latter strain required higher concentrations of nisin A to reach optimal induction of ptsI (Table 2).
Analysis of the effects of overproduction of altered HPr proteins on galactose utilization.
Two mutated ptsH genes that encode proteins affected in the phosphorylation on residue serine 46 were constructed. One mutated ptsH gene encodes S46D HPr, where residue serine 46 was changed to an aspartic acid that mimics a phosphate group on the seryl residue; another encodes S46A HPr, carrying an alanine on residue 46 that can no longer be phosphorylated. Both mutated genes were tested for the ability to complement the ptsH mutation in strain NZ9880 in comparison to the wild-type ptsH gene (Table 2). This was done by expressing either the wild-type or the mutated gene together with the wild-type ptsI gene, using plasmid pNZ9288 and pNZ9289, respectively. Plasmid pNZ9288 (coding for S46A HPr) but not plasmid pNZ9289 (coding for S46D HPr) was, after induction with low concentrations of nisin A, able to restore the growth defect in the ptsH mutant strain. The observed maximal growth rate after complementation with pNZ9288 was approximately 80% of that observed after complementation with pNZ9286 carrying the wild-type ptsHI genes.
Recently, we showed that CcpA is involved in the catabolite repression of the galactose operon in L. lactis, most likely through an interaction with a cre site located in the promoter region of the gal operon (29). To investigate the role of HPr(Ser-P) in this regulatory process, the mutant ptsH genes were also overexpressed without the coexpression of ptsI in the wild-type strain NZ9800. Moderate overexpression of mutant ptsH genes encoding S46A HPr and S46D HPr, by the addition of 10−5 μg of nisin A/ml, had no effect on the growth rate of the cells (data not shown). However, high overexpression of the mutant gene encoding S46D HPr, by the addition of 10−3 μg of nisin A/ml, resulted in a severe reduction of the growth on galactose, whereas the high overexpression of the gene encoding S46A HPr did not affect growth on galactose. The high overexpression of genes encoding S46A HPr or S46D HPr did not influence the rates of growth on glucose and sucrose (Table 2).
Analysis of the role of HPr(Ser-P) in regulation of the las operon.
Previously, we showed that the activities of pyruvate kinase and l-lactate dehydrogenase vary in response to the carbon source provided (29). Cells grown on glucose show higher activities of both enzymes than cells grown on galactose (Table 3). Furthermore, we found that the disruption of the L. lactis ccpA gene resulted in a reduction of the expression of the las operon, which encodes phosphofructokinase, pyruvate kinase, and l-lactate dehydrogenase, when the cells were grown on glucose. These results indicated that CcpA acts as an activator of the expression of the las operon most likely by an interaction with a cre site located in the promoter region of the las operon (29). Expression of the pNZ9285-borne gene encoding S46D HPr in galactose-grown cells resulted in pyruvate kinase and l-lactate dehydrogenase activities higher than those in wild-type cells (Table 3). In contrast, the overproduction of S46A HPr did not affect both activities. When either of the mutant ptsH genes was expressed in cells grown on glucose, the activities of pyruvate kinase and l-lactate dehydrogenase were not altered. Higher levels of expression of the mutant genes achieved by the addition of higher amounts of nisin A to the growth medium resulted in lower pyruvate kinase and l-lactate dehydrogenase activities (data not shown). The disruption of the ptsH gene in strain NZ9880 resulted in a reduction of both pyruvate kinase and l-lactate dehydrogenase activities to 70% of the values measured in wild-type cells. Higher pyruvate kinase and l-lactate dehydrogenase activities were measured in strain NZ9880 (ΔptsH) grown on glucose than in cells grown on galactose, suggesting that regulation of the las operon expression is not dependent on the presence of the ptsH gene. To assess the role of HPr(Ser-P) in the CcpA-mediated activation of the las operon, we expressed the S46D HPr gene in strain NZ9870, which carries a disrupted copy of the ccpA gene. Since strain NZ9870 does not grow on galactose, the effects of the overproduction of S46D HPr could be analyzed only in cells grown on glucose. Overexpression of the S46D HPr gene in strain NZ9870 (ΔccpA) did not result in an increase of the activities to the levels measured in the wild-type strain NZ9800 grown on glucose (Table 3). The results of these experiments indicate the requirement of an intact CcpA for the observed HPr(Ser-P)-mediated activation of pyruvate kinase and l-lactate dehydrogenase.
TABLE 3.
l-Lactate dehydrogenase and pyruvate kinase activities of strains used in this study
| Strain | Nisin A induction (μg/ml) | Carbon source | Activity (mean ± SE)
|
|
|---|---|---|---|---|
| Pyruvate kinase (μmol of PEP formed/mg/min) | l-Lactate dehydrogenase (μmol of NADH formed/mg/min) | |||
| NZ9800 (wild type) | Glucose | 3.20 ± 0.57 | 14.2 ± 0.20 | |
| Galactose | 1.67 ± 0.48 | 9.05 ± 0.31 | ||
| NZ9800/pNZ9284 (S46A ptsH) | 10−5 | Glucose | 3.19 ± 0.43 | 13.84 ± 0.39 |
| 10−5 | Galactose | 1.84 ± 0.06 | 9.12 ± 0.65 | |
| NZ9800/pNZ9285 (S46D ptsH) | 10−5 | Glucose | 2.95 ± 0.09 | 12.38 ± 0.23 |
| 10−5 | Galactose | 2.85 ± 0.16 | 12.43 ± 0.90 | |
| NZ9870 (ΔccpA) | Glucose | 0.79 ± 0.78 | 6.32 ± 0.55 | |
| NZ9870/pNZ9285 (S46D ptsH) | 10−3 | Glucose | 0.75 ± 0.26 | 6.42 ± 0.54 |
| NZ9880 (ΔptsH) | Glucose | 2.14 ± 0.53 | 9.87 ± 0.65 | |
| Galactose | 1.45 ± 0.56 | 8.31 ± 0.32 | ||
DISCUSSION
Using inactivation, mutation, and overexpression studies, we have cloned and analyzed the L. lactis ptsH and ptsI genes in order to assess their roles in the phosphorylation cascade of the PTS and the global regulation of carbohydrate metabolism. The genes are located in an operon structure with the gene order ptsHI. Two equally strong, adjacent transcriptional start sites were mapped and found to be located downstream of a typical lactococcal promoter sequence (11, 40). The results of the transcriptional analysis revealed that the ptsH gene was located on two transcripts: a small transcript of approximately 0.3 kb and a larger transcript of 2 kb. Probing with a ptsI-specific probe indicated that this gene is located only on the 2-kb transcript. These results indicate that the ptsHI genes are transcribed as a single mRNA species of 2 kb that terminates at the rho-independent terminator structure (T2) located downstream of ptsI and that the stem-loop structure (T1) located downstream of ptsH functions as a limited transcriptional terminator. This transcriptional organization most likely results in a higher level of expression of the ptsH gene, in accordance with the observation that higher amounts of HPr protein than of enzyme I are found in several bacteria, including Staphylococcus carnosus (24).
Expression of the L. lactis ptsHI operon is regulated at the transcriptional level, since mRNA levels, as determined by scintillation counting, were 10-fold higher in glucose-grown cells than in galactose-grown cells. Induction of ptsHI gene expression by glucose has also been reported for E. coli and B. subtilis (6, 34, 38). The induction by glucose of the B. subtilis ptsGHI operon, which also contains the gene encoding the glucose-specific EII (ptsG), is mediated via the antiterminator protein GlcT (38). No cis-acting sequences like cre sites (42), involved in catabolite repression, or ribonucleic antiterminator sequences (2), which play a role in antitermination mediated by proteins of the BglG/SacY family, were detected at relevant positions in the DNA sequence of the L. lactis ptsHI operon.
The disruption of both the ptsH and ptsI genes resulted in strains that were severely affected in growth at the expense of sucrose and fructose, indicating that these sugars are taken up exclusively via the PTS. Growth at the expense of glucose was affected but not completely abolished, in accordance with a previous report which indicated that L. lactis can take up glucose via two transport systems, a permease and a PTS (39). Surprisingly, growth at the expense of galactose and maltose was also affected by the ptsH and ptsI mutations since the growth rates were reduced to half of those observed with the wild-type strain. Both sugars are presumed to be taken up via non-PTS permeases in L. lactis (19, 27). Our results suggest that a functional PTS is required for the efficient utilization of galactose and maltose in L. lactis. An activating role for the PTS component HPr(His-P) has been reported before in the case of the B. subtilis levanase regulator LevR (37) and the enterococcal GlpK proteins (4). HPr(His-P) is absent in both NZ9880 (ΔptsH) and NZ9881 (ΔptsI) since phosphorylation at His-15 requires the presence of both HPr and enzyme I. A role for either HPr(Ser-P) or enzyme I in the observed reduction of the growth rate on non-PTS sugars in the PTS-negative strains can be ruled out since these proteins are still present in strains NZ9881 (ΔptsI) and NZ9880 (ΔptsH), respectively. This finding suggests that HPr(His-P), the concentration of which increases in the absence of a PTS substrate, stimulates the utilization of galactose and maltose in L. lactis.
Further evidence for the functionality of the ptsH gene was obtained by the complementation of the ptsH mutation in strain NZ9880 by the expression of either the L. lactis or the B. subtilis ptsH gene under the control of the inducible nisA promoter. Induction of the nisA promoter with increasing concentrations of nisin A resulted in only a partial restoration of the growth defect observed in the ptsH mutant strain when grown on the PTS sugar sucrose, while growth at the expense of galactose was not affected. The readthrough from the promoter of the Ermr gene was most likely not sufficient to allow full functional expression of the ptsI gene, since coexpression of the ptsH and ptsI genes did result in complete complementation of the growth defect. This result also demonstrates the dependence of ptsI expression on the promoter located upstream of the ptsH gene.
The ptsI mutation in strain NZ9881 was complemented by the expression of either the entire ptsHI operon or the ptsI gene alone, transcriptionally fused to the nisA promoter on plasmid pNZ9287. These results primarily show the functionality of the ptsI gene but also establish the functionality of the ribosome binding site located upstream of the ptsI gene, despite its unusual spacing (14 bp) relative to the putative ATG start codon of the ptsI open reading frame.
To analyze the regulatory role of HPr in the metabolism of L. lactis, we constructed in the L. lactis ptsH gene site-specific mutations encoding HPr proteins that were affected in the phosphorylation of residue Ser-46. Replacement of residue Ser-46 with alanine (S46A HPr) has been shown to inactivate HPr as a regulatory molecule (9), while the introduction of aspartate at residue 46 (S46D HPr) results in a permanently negatively charged residue 46 that resembles a phosphorylated serine. Production of S46D HPr in the wild-type strain NZ9800 resulted in a strong reduction of the growth rate on galactose, whereas no effects were observed when S46A HPr was overproduced. This can be explained by an enhanced repression of gal gene expression by CcpA, in the presence of S46D HPr, but not of S46A HPr. S46DHPr has been shown to stimulate the binding of CcpA to cre sites, thus mediating catabolite repression (12, 17). The role of CcpA in this mechanism could not be further substantiated since a ccpA-negative strain is impaired in the utilization of galactose (29). Moreover, the reduced growth on galactose after overexpression of the S46D HPr gene could be explained by the S46D HPr-mediated activation of the inducer exclusion and expulsion mechanisms. These mechanisms, which result in a reduction of inducer concentration, have been described for nongrowing L. lactis, and their activities are stimulated by the introduction of B. subtilis S46D HPr protein in L. lactis vesicles via electroporation (45, 46). The production of L. lactis HPr mutants in growing cells is a nonartificial system to study the inducer exclusion and expulsion mechanisms. Production of S46D HPr could result in a reduced uptake of galactose via an interaction with the galactose permease. In addition, S46D HPr-mediated activation of the recently purified sugar-phosphate phosphatase (47) could lead to an increased dephosphorylation of galactose intermediates, resulting in a reduced growth rate. In conclusion, we have shown that varying the expression level of HPr mutants influences the growth rate, gal gene expression, and pyruvate kinase and lactate dehydrogenase activities. Moreover, we show that L. lactis regulates the expression level of ptsH as a function of the carbon source used (Fig. 3B), which stresses the physiological importance of the regulation.
Since it has been reported that CcpA can form a complex with HPr phosphorylated at residue serine 46 and that this complex enhances the binding of CcpA to cre sites (12, 17, 20), we were interested in determining if this complex also plays a role in the regulation of the las operon, known to be transcriptionally activated by CcpA (29). Production of S46D HPr but not S46A HPr in wild-type cells grown on galactose resulted in pyruvate kinase and l-lactate dehydrogenase activities which were elevated to the same level as found in the wild-type grown on glucose, suggesting that HPr(Ser-P) acts as a signal molecule that modulates the expression level of the pyruvate kinase and l-lactate dehydrogenase genes in response to the carbon source provided. To establish the role of CcpA in this mechanism, the gene encoding S46D HPr was also expressed in strain NZ9870 (ΔccpA). Due to the inactivation of the ccpA gene, the activities of both pyruvate kinase and l-lactate dehydrogenase were no longer activated by CcpA and therefore lower than those measured in the wild-type strain (29). The fact that these no longer responded to the production of S46D HPr indicated that CcpA is required for the observed S46D HPr-mediated activation of the activities in the wild-type strain. These results indicate that HPr(Ser-P) acts as a signal molecule involved in the CcpA-mediated transcriptional activation of the las operon, providing an activation mechanism that has not been described before for a gram-positive microorganism. Remarkably, disruption of the ptsH gene did not result in a strong reduction of the activities of pyruvate kinase and l-lactate dehydrogenase, nor did it affect carbon source-dependent regulation. This finding suggests that additional signal molecules may be involved in the CcpA-dependent regulation of the las operon. Possible candidates are glucose-6-P and NADP, which have been shown to enhance the binding of CcpA to cre sites (17, 23), or a lactococcal analogue of Crh, an HPr-like protein that was recently identified in B. subtilis and that functions as a signal molecule in the catabolite repression of several genes (15).
The results presented here show the predominant role of the PTS in the uptake of various sugars in L. lactis. Furthermore, they show the regulatory functions of HPr in establishing a hierarchy in the preference for different sugars in L. lactis. Finally, we provide experimental evidence for the involvement of HPr(Ser-P) in the CcpA-mediated transcriptional activation of genes encoding glycolytic enzymes in L. lactis.
ACKNOWLEDGMENTS
We are grateful to Marke Beerthuyzen and Hans Paalman for technical assistance and to Michiel Kleerebezem, Roland Siezen, and Jeroen Hugenholtz for critically reading the manuscript.
This work was partly supported by the BIOTECH Programme of the European Community (contract BIO2-CT92-0137).
REFERENCES
- 1.Alpert C A, Doerschung M, Saffen D, Deutscher J, Hengstenberg W. The bacterial phosphoenolpyruvate-dependent phosphotransferase system. Isolation of active site peptides by reversed-phase high-performance liquid chromatography and determination of their primary structure. J Chromatogr. 1985;326:363–371. doi: 10.1016/s0021-9673(01)87462-8. [DOI] [PubMed] [Google Scholar]
- 2.Aymerich S, Steinmetz M. Specificity determinants and structural features in the RNA target of the bacterial antiterminator proteins of the BglG/SacY family. Proc Natl Acad Sci USA. 1992;89:10410–10414. doi: 10.1073/pnas.89.21.10410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principal of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 4.Charrier V, Buckley E, Parsonage D, Galinier A, Darbon E, Jaquinod M, Forest E, Deutscher J, Claiborne A. Cloning and sequencing of two enterococcal glpK genes and regulation of the encoded glycerol kinases by phosphoenolpyruvate-dependent, phosphotransferase system-catalyzed phosphorylation of a single histidyl residue. J Biol Chem. 1997;272:14166–14174. doi: 10.1074/jbc.272.22.14166. [DOI] [PubMed] [Google Scholar]
- 5.Collins L B, Thomas T D. Pyruvate kinase of Streptococcus lactis. J Bacteriol. 1974;120:52–58. doi: 10.1128/jb.120.1.52-58.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Reuse H, Kolb A, Danchin A. Positive regulation of the expression of the Escherichia coli pts operon. Identification of the regulatory regions. J Mol Biol. 1992;226:623–635. doi: 10.1016/0022-2836(92)90620-y. [DOI] [PubMed] [Google Scholar]
- 7.de Ruyter P G G A, Kuipers O P, de Vos W M. Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl Environ Microbiol. 1996;62:3662–3667. doi: 10.1128/aem.62.10.3662-3667.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deutscher J, Saier M H., Jr ATP-dependent protein kinase-catalyzed phosphorylation of a seryl residue in HPr, a phosphate carrier protein of the phosphotransferase system in Streptococcus pyogenes. Proc Natl Acad Sci USA. 1983;80:6790–6794. doi: 10.1073/pnas.80.22.6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deutscher J, Reizer J, Fischer C, Galinier A, Saier M H, Jr, Steinmetz M. Loss of protein kinase-catalyzed phosphorylation of HPr, a phosphocarrier protein of the phosphotransferase system, by mutation of the ptsH gene confers catabolite repression resistance to several catabolic genes of Bacillus subtilis. J Bacteriol. 1994;176:3336–3344. doi: 10.1128/jb.176.11.3336-3344.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deutscher J, Kuester E, Bergstedt U, Charrier V, Hillen W. Protein kinase-dependent HPr/CcpA interaction links glycolytic activity to carbon catabolite repression in Gram-positive bacteria. Mol Microbiol. 1995;15:1049–1053. doi: 10.1111/j.1365-2958.1995.tb02280.x. [DOI] [PubMed] [Google Scholar]
- 11.de Vos W M, Simons G. Gene cloning and expression systems in lactococci. In: Gasson M J, de Vos W M, editors. Genetics and biotechnology of lactic acid bacteria. London, England: Chapman and Hall; 1994. pp. 52–105. [Google Scholar]
- 12.Fujita Y, Miwa Y, Galinier A, Deutscher J. Specific recognition of the Bacillus subtilis gnt cis-acting catabolite-responsive element by a protein complex formed between CcpA and seryl-phosphorylated HPr. Mol Microbiol. 1995;17:953–960. doi: 10.1111/j.1365-2958.1995.mmi_17050953.x. [DOI] [PubMed] [Google Scholar]
- 13.Gagnon G, Vadeboncoeur C, Frenette M. Phosphotransferase system of Streptococcus salivarius: characterization of the ptsH gene and its product. Gene. 1993;136:27–34. doi: 10.1016/0378-1119(93)90443-7. [DOI] [PubMed] [Google Scholar]
- 14.Gagnon G, Vadeboncoeur C, Gauthier L, Frenette M. Regulation of ptsH and ptsI gene expression in Streptococcus salivarius ATCC 25975. Mol Microbiol. 1995;16:1111–1121. doi: 10.1111/j.1365-2958.1995.tb02336.x. [DOI] [PubMed] [Google Scholar]
- 15.Galinier A, Haiech J, Kilhoffer M-C, Jaquinod M, Stuelke J, Deutscher J, Martin-Verstraete I. The Bacillus subtilis crh gene encodes a HPr-like protein involved in carbon catabolite repression. Proc Natl Acad Sci USA. 1997;94:8439–8444. doi: 10.1073/pnas.94.16.8439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galinier A, Kravanja M, Engelmann R, Hengstenberg W, Kilhoffer M-C, Deutscher J, Haiech J. New protein kinase and protein phosphatase families mediate signal transduction in bacterial catabolite repression. Proc Natl Acad Sci USA. 1998;95:1823–1828. doi: 10.1073/pnas.95.4.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goesseringer R, Kuester E, Galinier A, Deutscher J, Hillen W. Cooperative and non-cooperative DNA binding modes of catabolite control protein CcpA from Bacillus megaterium result from sensing two different signals. J Mol Biol. 1997;266:665–676. doi: 10.1006/jmbi.1996.0820. [DOI] [PubMed] [Google Scholar]
- 18.Gonzy-Treboul G, Zagorec M, Rain-Guion M C, Steinmetz M. Phosphoenolpyruvate:sugar phosphotransferase system of Bacillus subtilis: nucleotide sequence of ptsX, ptsH and the 5′-end of ptsI and evidence for a ptsHI operon. Mol Microbiol. 1989;3:103–112. doi: 10.1111/j.1365-2958.1989.tb00109.x. [DOI] [PubMed] [Google Scholar]
- 19.Grossiord B P, Vaughan E E, Luesink E J, de Vos W M. Genetics of galactose utilization in lactic acid bacteria. Lait. 1998;78:77–84. [Google Scholar]
- 20.Jones B E, Dossonnet V, Kuester E, Hillen W, Deutscher J, Klevit R E. Binding of the catabolite control protein CcpA to its DNA target is regulated by phosphorylation of its corepressor HPr. J Biol Chem. 1998;272:26530–26535. doi: 10.1074/jbc.272.42.26530. [DOI] [PubMed] [Google Scholar]
- 21.Hillier A J, Jago G R. L-lactate dehydrogenase, FDP-activated, from Streptococcus cremoris. Methods Enzymol. 1982;89:362–367. doi: 10.1016/s0076-6879(82)89065-4. [DOI] [PubMed] [Google Scholar]
- 22.Holo H, Nes I F. High-frequency transformation of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl Environ Microbiol. 1989;55:3119–3123. doi: 10.1128/aem.55.12.3119-3123.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim J H, Voskuil M I, Chambliss G H. NADP, corepressor for the Bacillus subtilis catabolite control protein CcpA. Proc Natl Acad Sci USA. 1998;95:9590–9595. doi: 10.1073/pnas.95.16.9590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kohlbrecher D, Eiserman R, Hengstenberg W. Staphylococcal phosphoenolpyruvate-dependent phosphotransferase system: molecular cloning and nucleotide sequence of the Staphylococcus carnosus ptsI gene: expression and complementation studies of the product. J Bacteriol. 1992;174:2208–2214. doi: 10.1128/jb.174.7.2208-2214.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuipers O P, Boot H J, de Vos W M. Improved site-directed mutagenesis method using PCR. Nucleic Acids Res. 1991;19:4558. doi: 10.1093/nar/19.16.4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuipers O P, Beerthuyzen M M, Siezen R J, de Vos W M. Characterization of the nisin gene cluster nisABTCIPR of Lactococcus lactis. Requirement of expression of the nisA and nisI genes for the development of immunity. Eur J Biochem. 1993;216:281–291. doi: 10.1111/j.1432-1033.1993.tb18143.x. [DOI] [PubMed] [Google Scholar]
- 27.Law J, Buist G, Haandrikman A, Kok J, Venema G, Leenhouts K. A system to generate chromosomal mutations in Lactococcus lactis which allows fast analysis of targeted genes. J Bacteriol. 1995;177:7011–7018. doi: 10.1128/jb.177.24.7011-7018.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leenhouts K J, Kok J, Venema G. Stability of integrated plasmids in the chromosome of Lactococcus lactis. Appl Environ Microbiol. 1990;56:2726–2735. doi: 10.1128/aem.56.9.2726-2735.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luesink E J, van Herpen R E M A, Grossiord B P, Kuipers O P, de Vos W M. Transcriptional activation of the glycolytic las operon and catabolite repression of the gal operon in Lactococcus lactis are mediated by the catabolite control protein CcpA. Mol Microbiol. 1998;30:789–798. doi: 10.1046/j.1365-2958.1998.01111.x. [DOI] [PubMed] [Google Scholar]
- 30.Martin B, Alloing G, Mejean V, Claverys J P. Constitutive expression of the erythromycin resistance mediated by the ermAM determinant of plasmid pAMbeta 1 results from deletion of the 5′ leader sequences. Plasmid. 1987;18:250–253. doi: 10.1016/0147-619x(87)90068-0. [DOI] [PubMed] [Google Scholar]
- 31.Poolman B, Knol J, Mollet B, Nieuwenhuis B, Sulter G. Regulation of the bacterial sugar-H+ symport by phosphoenolpyruvate-dependent enzyme I/HPr-mediated phosphorylation. Proc Natl Acad Sci USA. 1995;92:778–782. doi: 10.1073/pnas.92.3.778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Postma P W, Lengeler J W, Jacobson G R. Phosphoenopyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol Rev. 1993;57:543–594. doi: 10.1128/mr.57.3.543-594.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reizer J, Hoischen C, Titgemeyer F, Rivolta C, Rabus R, Stuelke J, Karamata D, Saier M H, Jr, Hillen W. A novel protein kinase that controls carbon catabolite repression in bacteria. Mol Microbiol. 1998;27:1157–1169. doi: 10.1046/j.1365-2958.1998.00747.x. [DOI] [PubMed] [Google Scholar]
- 34.Ryu S, Ramseier T M, Michotey V, Saier M H, Jr, Garges S. Effect of the FruR regulator on the transcription of the pts operon in Escherichia coli. J Biol Chem. 1995;270:2489–2496. doi: 10.1074/jbc.270.6.2489. [DOI] [PubMed] [Google Scholar]
- 35.Saier M H, Jr, Chauvaux S, Cook G M, Deutscher J, Paulsen I T, Reizer J, Ye J J. Catabolite repression and inducer control in Gram-positive bacteria. Microbiology. 1996;142:217–230. doi: 10.1099/13500872-142-2-217. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 37.Stuelke J, Martin-Verstraete I, Charrier V, Klier A, Deutscher J, Rapoport G. The HPr protein of the phosphotransferase system links induction and catabolite repression of the Bacillus subtilis levanase operon. J Bacteriol. 1995;177:6928–6936. doi: 10.1128/jb.177.23.6928-6936.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stuelke J, Martin-Verstraete I, Zagorec M, Rose M, Klier A, Rapoport G. Induction of the Bacillus subtilis ptsGHI operon by glucose is controlled by a novel antiterminator, GlcT. Mol Microbiol. 1997;25:65–78. doi: 10.1046/j.1365-2958.1997.4351797.x. [DOI] [PubMed] [Google Scholar]
- 39.Thompson J, Chassy B M, Egan W. Lactose metabolism in Streptococcus lactis: studies with a mutant lacking glucokinase and mannose-phosphotransferase activities. J Bacteriol. 1985;162:217–223. doi: 10.1128/jb.162.1.217-223.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van der Vossen J M, van Lelie D, Venema G. Isolation and characterization of Streptococcus cremoris Wg2-specific promoters. Appl Environ Microbiol. 1987;53:2452–2457. doi: 10.1128/aem.53.10.2452-2457.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vos P, van Asseldonk M, van Jeveren F, Siezen R, Simons G, de Vos W M. A maturation protein is essential for the production of active forms of Lactococcus lactis SK11 serine proteinase located in or secreted from the cell envelope. J Bacteriol. 1989;171:2795–2802. doi: 10.1128/jb.171.5.2795-2802.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weickert M J, Chambliss G H. Site directed mutagenesis of a catabolite repression operator sequence in Bacillus subtilis. Proc Natl Acad Sci USA. 1990;87:6238–6242. doi: 10.1073/pnas.87.16.6238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 44.Ye J J, Minarcik J, Saier M H., Jr Inducer expulsion and the occurrence of an HPr(Ser-P)-activated sugar-phosphate phosphatase in Enterococcus faecaelis and Streptococcus pyogenes. Microbiology. 1996;142:585–592. doi: 10.1099/13500872-142-3-585. [DOI] [PubMed] [Google Scholar]
- 45.Ye J J, Reizer J, Cui X, Saier M H. Inhibition of the phosphoenolpyruvate:lactose phosphotransferase system and activation of a cytoplasmic sugar-phosphate phosphatase in Lactococcus lactis by ATP-dependent metabolite-activated phosphorylation of serine 46 in the phosphocarrier protein HPr. J Biol Chem. 1994;269:11837–11844. [PubMed] [Google Scholar]
- 46.Ye J J, Reizer J, Saier M H., Jr Regulation of 2-deoxyglucose phosphate accumulation in Lactococcus lactis vesicles by metabolite-activated, ATP-dependent phosphorylation of serine-46 in HPr of the phosphotransferase system. Microbiology. 1994;140:3421–3429. doi: 10.1099/13500872-140-12-3421. [DOI] [PubMed] [Google Scholar]
- 47.Ye J J, Saier M H. Purification and characterization of a small membrane-associated sugar phosphate phosphatase that is allosterically activated by HPr(Ser(P)) of the phosphotransferase system in Lactococcus lactis. J Biol Chem. 1995;270:16740–16744. doi: 10.1074/jbc.270.28.16740. [DOI] [PubMed] [Google Scholar]