Abstract
Purpose
To develop and evaluate domain-specific and pretrained bidirectional encoder representations from transformers (BERT) models in a transfer learning task on varying training dataset sizes to annotate a larger overall dataset.
Materials and Methods
The authors retrospectively reviewed 69 095 anonymized adult chest radiograph reports (reports dated April 2020–March 2021). From the overall cohort, 1004 reports were randomly selected and labeled for the presence or absence of each of the following devices: endotracheal tube (ETT), enterogastric tube (NGT, or Dobhoff tube), central venous catheter (CVC), and Swan-Ganz catheter (SGC). Pretrained transformer models (BERT, PubMedBERT, DistilBERT, RoBERTa, and DeBERTa) were trained, validated, and tested on 60%, 20%, and 20%, respectively, of these reports through fivefold cross-validation. Additional training involved varying dataset sizes with 5%, 10%, 15%, 20%, and 40% of the 1004 reports. The best-performing epochs were used to assess area under the receiver operating characteristic curve (AUC) and determine run time on the overall dataset.
Results
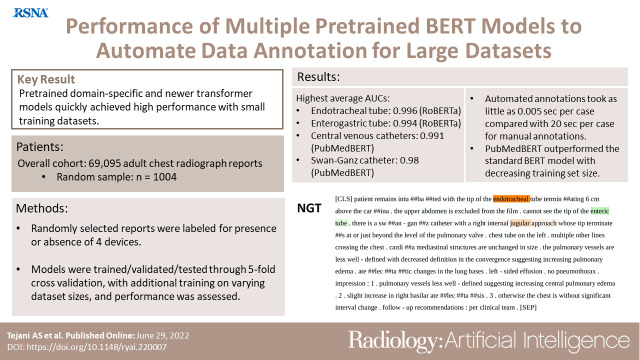
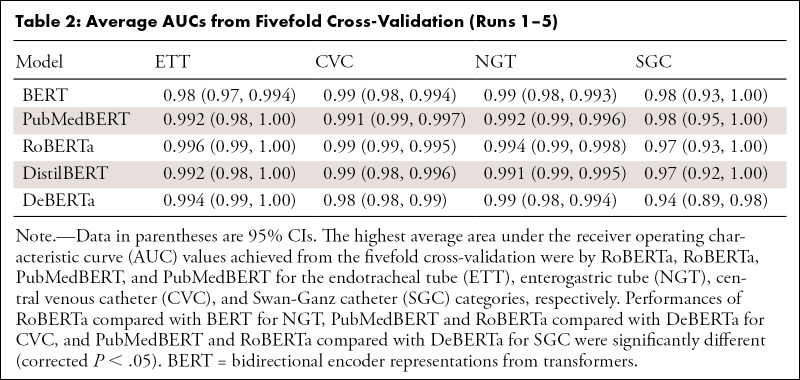
The highest average AUCs from fivefold cross-validation were 0.996 for ETT (RoBERTa), 0.994 for NGT (RoBERTa), 0.991 for CVC (PubMedBERT), and 0.98 for SGC (PubMedBERT). DeBERTa demonstrated the highest AUC for each support device trained on 5% of the training set. PubMedBERT showed a higher AUC with a decreasing training set size compared with BERT. Training and validation time was shortest for DistilBERT at 3 minutes 39 seconds on the annotated cohort.
Conclusion
Pretrained and domain-specific transformer models required small training datasets and short training times to create a highly accurate final model that expedites autonomous annotation of large datasets.
Keywords: Informatics, Named Entity Recognition, Transfer Learning
Supplemental material is available for this article.
©RSNA, 2022
See also the commentary by Zech in this issue.
Keywords: Informatics, Named Entity Recognition, Transfer Learning
Summary
Pretrained domain-specific and newer transformer models achieved high performance using small training datasets and short training times, creating a robust final model that expedited autonomous annotation of a large dataset.
Key Points
■ Domain-specific and newer transformer models achieved high area under the receiver operating characteristic curve values (>0.90) with training sets of fewer than 1000 reports in as little as 3 minutes 39 seconds with performance comparable to metrics reported for older natural language processing techniques.
■ Applying fully trained bidirectional encoder representations from transformers (BERT) models for autonomous annotation of radiology reports took as little as 0.005 second per case, compared with 20 seconds per case for manual annotations.
■ Domain-specific models (ie, PubMedBERT) consistently outperformed the standard BERT model despite progressively decreasing training set sizes (P < .05).
Introduction
Developing a consistent deep learning model that achieves high performance metrics, such as area under the receiver operating characteristic curve (AUC), for computer vision tasks relies heavily on curating a large, diverse dataset, an arduous task that can become a limiting factor in deep learning model creation (1–3). The ability of a deep learning model to produce accurate output often depends on the quality of the dataset used for model development, necessitating deliberate, time-consuming collection and annotation of representative data (1,2). The time- and labor-intensive process of manual data curation is a well-recognized issue in the creation of deep learning algorithms, and automated data curation leveraging natural language processing (NLP) has been suggested to alleviate the burden imposed by this process (2,4–6).
NLP represents a subfield of artificial intelligence that analyzes natural language data, serving as an intersection between computer science and linguistics (7–9). NLP has proven to be effective in extracting information from radiology reports for tasks, including detection of critical findings, quality assessment, and annotation and generation of datasets. However, these functions often use relatively older techniques featuring simple machine learning algorithms (“term frequency–inverse document frequency”) or convolutional neural networks (“word embedding” or “Word2vec”) (3,8,10–12). Bidirectional encoder representations from transformers (BERT) is a recently developed language representation model based on the transformer architecture that has been shown to improve outcomes in NLP tasks and maintain stability against varying training dataset sizes and class imbalance, potentially offering a means of rapid and accurate curation of text-based datasets (3,7,9,13). The ability of BERT and other transformer models to outperform more traditional NLP models derives in part from an attention mechanism and nonsequential processing of input text that can capture long-range dependencies (ie, connecting a fact in the first sentence of the first paragraph to the last sentence of the last paragraph just as well as the second sentence of the first paragraph). This represented an improvement over previous recurrent neural network and unidirectional models (9,13).
The other major improvement derives from pretraining on large unlabeled datasets with subsequent fine-tuning for specific tasks. For example, one such model, BioBERT, represents a BERT model pretrained on biomedical text, including 21.3 billion words from text on platforms such as PubMed and PMC (3). This domain-specific BERT model for biomedical language representation demonstrated substantial improvements in several performance markers over BERT for biomedical text mining tasks (3). However, the initial creation of domain-specific BERT models, such as BioBERT, can require multiple weeks to complete because of large training dataset sizes (3). Fortunately, pretrained BERT models are publicly available for further transfer learning tasks, reducing the time required for training. For example, CheXbert, a biomedically pretrained BERT model tasked with chest radiograph report labeling for 14 different findings, required 30 minutes for training on ground truth data (14). Accordingly, an advantage of using domain-specific models is the substantially less time required to fine-tune a pretrained model for a specific language modeling task.
Though BERT has been shown to achieve high levels of accuracy in radiology-specific language representation tasks, prior studies have required large training datasets containing millions of studies (9). Furthermore, even reported pretrained models for radiology-specific tasks have required large training sets in the order of hundreds of thousands of reports. For example, development of RadBERT-CL, a biomedically pretrained BERT model tasked with chest radiograph report classification that demonstrated improvement over CheXbert, required 301 688 reports for training (15). Similarly, 155 000 reports from mammography, US, MRI, and image-guided biopsies were used to train BI-RADS BERT, a biomedically pretrained BERT model tasked with classification of reports per information in the Breast Imaging Reporting and Data System (ie, BI-RADS) (16). The purpose of this study was to develop and evaluate pretrained and newer BERT models further trained on varying training dataset sizes of chest radiograph reports to annotate a large dataset that can be used to train subsequent deep learning models for downstream computer vision tasks.
Materials and Methods
Study Sample
This study was approved by the institutional review board and designated “exempt” status from full board review with waived informed consent on the basis of minimal risk and adequate provisions for maintaining confidentiality. This study complied with the Health Information Portability and Accountability Act.
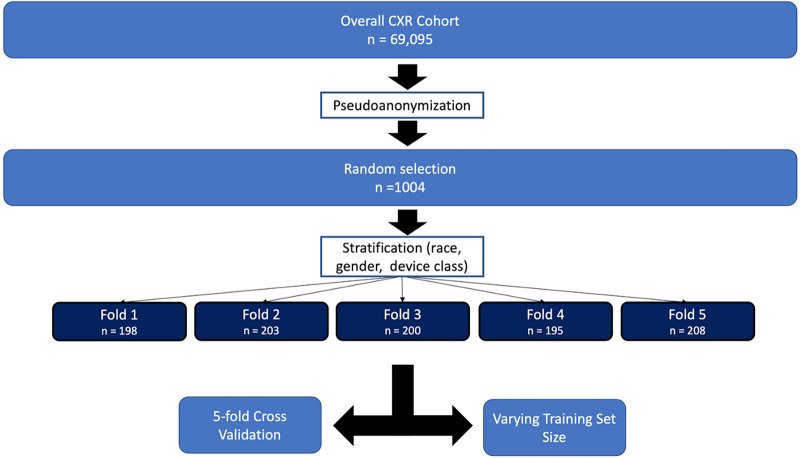
The initial phase of this study involved the retrospective review of 69 095 anonymized chest radiograph reports dated from April 1, 2020, through March 31, 2021, from an academic tertiary care center. The dataset consisted of 69 095 adult patients (>18 years old), with 31 875 (46%) women, 37 188 (54%) men, and 32 (0.05%) who had a gender categorization of unknown, declined, or nonbinary. With regard to self-reported race, the dataset included 43 081 (62%) White, 14 857 (22%) African American, 8635 (12%) mixed or unknown, 2261 (3%) Asian, 167 (0.2%) American Indian or Alaska Native, and 94 (0.1%) Hawaiian or Pacific Islander patients. Study reports were de-identified and underwent a process of pseudoanonymization, which involved replacing patient health information with unique identifiers and encrypting the original data. From the overall cohort, 1004 reports were randomly selected for the training, validation, and testing sets and featured 475 (47%) women and 528 (53%) men. This smaller cohort also featured 613 (61%) White, 234 (23%) African American, 83 (8.3%) mixed or unknown, 26 (2.6%) Asian, two (0.2%) Hawaiian or Pacific Islander, and one (0.01%) American Indian or Alaska Native patients. The remaining 68 061 reports were used only to evaluate inference time to determine the resulting algorithms’ total run time.
Terminology
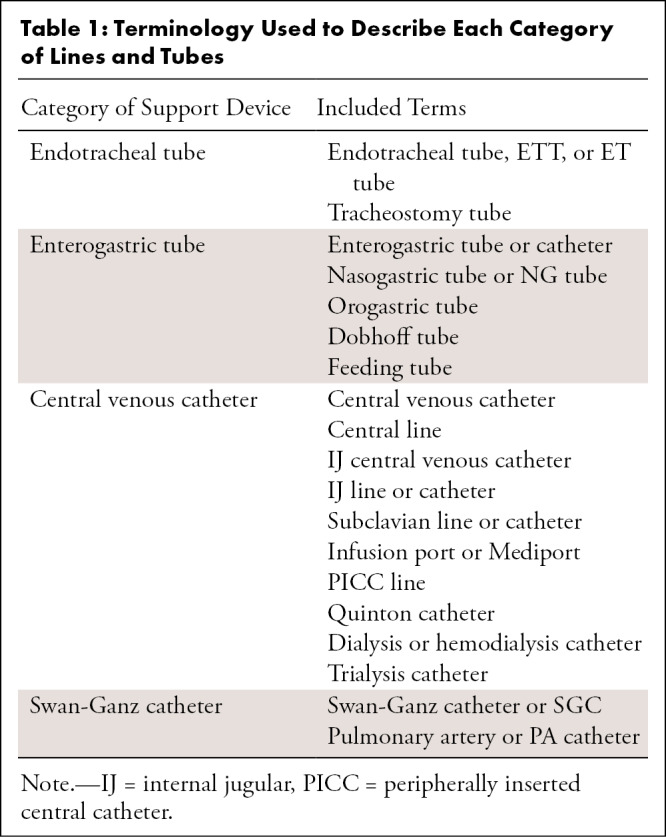
Four primary groups of lines and tubes were designated for this study: endotracheal tubes (ETTs), enterogastric tubes (NGTs), central venous catheters (CVCs), and Swan-Ganz catheters (SGCs), on the basis of categories assigned in a large, publicly available chest radiograph dataset (17). Table 1 summarizes specific terms in each of the four categories for the purpose of training and testing in this study. Reports demonstrated the absence of all devices versus a combination of one or more of the designated devices.
Table 1:
Terminology Used to Describe Each Category of Lines and Tubes
Ground Truth Labeling
A radiology resident (A.S.T.) with experience in identifying tubes and lines on plain radiographs manually annotated 1004 reports selected randomly from the overall cohort, specifically noting the presence or absence of an ETT, NGT, CVC, or SGC as determined by the provided lexicon described in Table 2. The resident noted the presence or absence of each device as denoted on the radiology reports, as the specific task in this study required annotation of textual data contained in radiology reports. Each radiology report was unique to a given study instance and/or accession, without any repeated reports in this cohort. The randomly selected reports included both structured and unstructured reports. Average time to manually annotate each case was calculated by dividing the total number of reports (1004) by the total amount of time required to annotate all cases.
Table 2:
Average AUCs from Fivefold Cross-Validation (Runs 1–5)
Software and Hardware Information
All model development and testing were performed with Py-Torch Lightning version 1.3.8 using PyTorch version 1.8.1 (18). All model development, training, and testing were performed on a standard workstation with an Nvidia GTX 1080ti GPU.
Model Development
PubMedBERT is a domain-specific BERT model trained specifically on biomedical text from PubMed abstracts (19). DistilBERT features a smaller, faster BERT model trained using the concept of knowledge distillation (20). RoBERTa is a transformer model that was also pretrained on English-language text data from sources such as Wikipedia, though using a dynamic variation of masked language modeling intended for fine-tuning on downstream tasks (21). DeBERTa is a recent transformer model that has been shown to outperform RoBERTa, with improvements attributed to using disentangled attention and an enhanced mask decoder (22).
The architecture of these pretrained BERT models, including structural alterations or training differences from BERT models trained on general text, is documented in cited literature. The pretrained BERT-derived models were fine-tuned on the institution-specific, manually annotated chest radiograph reports for the purposes of this study. Training, validation, and testing of the models required determining the presence versus absence of each of the devices (ETT, NGT, CVC, SGC) as four independent tasks.
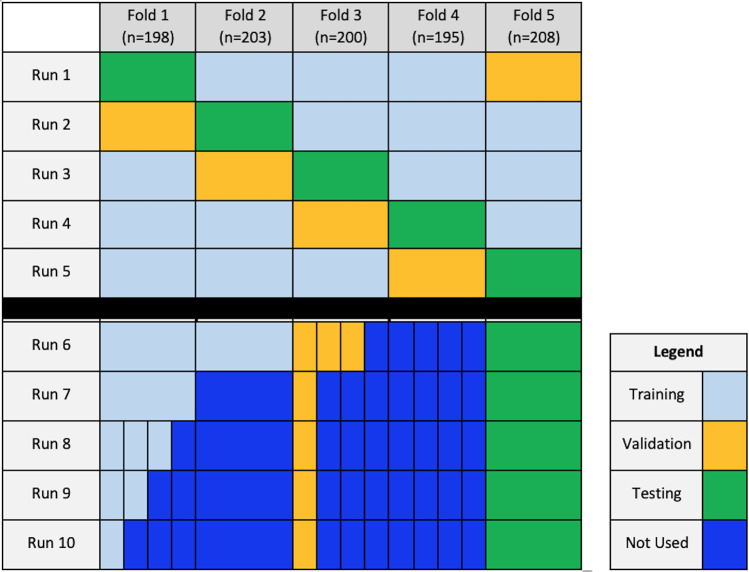
The 1004 reports were divided into five folds (n = approximately 200 per fold), and the distribution of tube presence, gender, race, study class (immediate [ie, stat], inpatient, outpatient), and procedure type (two view vs one view) were maintained; fold groups were selected by their pseudoanonymized medical record number. In this way, no single patient was in two different folds. At this specific institution, study class designates the ordering providers’ desired examination priority, with associated report turnaround goals of 1 hour for immediate, 24 hours for inpatient, and 72 hours for outpatient studies.
The pretrained BERT model was initially trained, validated, and tested on 60%, 20%, and 20% of the cohort, respectively, with validation and testing sets designated independently of the training dataset through fivefold cross-validation. This represents runs 1–5, and the average AUC of five folds is reported for each tube class, which represents outputs from model performance on only the test set. The validation set was used to monitor model performance during training, and weights from the best-performing iteration of each model were locked prior to the testing step.
Additional training involved varying training and validation dataset sizes, labeled as runs 6–10, selected from the original 1004 annotated cases, while the testing dataset size used for run 5 (208 of 1004, 21%) was maintained for all runs. Varying training set sizes represented 40% (401 of 1004, run 6), 20% (198 of 1004, run 7), 15% (149 of 1004, run 8), 10% (99 of 1004, run 9), and 5% (50 of 1004, run 10) of the 1004 reports. Training and validation set sizes collectively represented 55% (551 of 1004, run 6), 25% (248 of 1004, run 7), 20% (199 of 1004, run 8), 15% (149 of 1004, run 9), and 10% (100 of 1004, run 10) of the 1004 reports compared with 79% (796 of 1004) for run 5. Detailed illustrations of the data splitting scheme are shown in Figures 1 and 2.
Figure 1:
Flowchart of case selection prior to training and validation. Please refer to Figure 2 for details on varying training size sets. CXR = chest radiograph.
Figure 2:
Schematic of data splitting for each run. Runs 1–5 featured fivefold cross-validation, alternating folds used for training, validation, and testing. Runs 6–10 represent additional runs with varying training dataset size.
Each training run consisted of 12 epochs, with each epoch representing one pass through the entire training set, and performance was then validated on the validation set chosen. The model state was saved at the end of each epoch if the validation loss (binary cross-entropy with loss logits) was better than previous epochs—that is, weights from the epoch with the lowest validation loss were selected for each run. All model parameters were eligible for fine-tuning without the restriction of optimization to a subset of transformer parameters. The selected epoch model state was then run on the test set, and the AUC values were calculated to assess performance, specifically indicating the models’ abilities to detect presence versus absence of each of the four devices on the presented radiology reports. The model was also applied in inference to the entire cohort (n = 69 095) to assess time-to-run performance. Specifically, model weights were locked, and data from the entire cohort were provided as input to the model to obtain corresponding outputs without any additional training.
The models were optimized using an AdamW optimizer with standard initialization and learning rate initialized at 5e-6. A learning rate scheduler composed of cosine annealing with gradual warmup was employed, such that the learning rate ramped up by 10× to 5e-5 by the second epoch and annealed to 1e-6 by the 12th epoch.
Statistical Analysis
AUCs from runs 1–5 were averaged from performance on the testing set while reporting performance on the full dataset, and 95% CIs were calculated for the average AUCs. When comparing to the reduced training and validation datasets (runs 6–10), only the AUC from run 5 was used, as it was based on the same testing dataset. The DeLong test was used to test the statistical significance of the difference between the AUC of each model (23). Holm test for multiple comparisons was applied to correct initial P values. All statistical analysis was performed with R version 4.0.2 (R Core Team). For all analysis, P less than .05 indicated statistical significance.
Results
Sample Characteristics
Among the 1004 randomly selected reports, 358 contained at least one of the designated lines or tubes; specifically, the reports included 133 ETT (37.2%), 133 NGT (37.2%), 273 CVC (76.2%), and 39 SGC (10.9%). Folds 1–5 were composed of 198, 203, 200, 195, and 208 cases, respectively. Manual annotation by the radiology resident took an average of 20 seconds per report.
Determining Model Attention
Figure 3 shows an example word importance map of input text from the PubMedBERT model with respect to an individual classification output task, serving as a heat map–equivalent visualization tool for this particular NLP task. Specifically, the figure demonstrates the relative importance of each word on a report provided to the pretrained BERT algorithm in classifying the presence or absence of a given device (24). Highlighted annotation of certain words in Figure 3 indicates that the given model provided attention over a certain threshold to those words before providing an output as dictated by the specified task. Review of word importance shows the ability of PubMedBERT to derive from its domain-specific library, including terms such as “musculoskeletal, pneumothorax, mediastinal, pulmonary,” and others. Figure E1 (supplement) demonstrates an example word importance map when one of the models, in this case PubMedBERT, evaluates for presence or absence of all four devices as four separate tasks.
Figure 3:
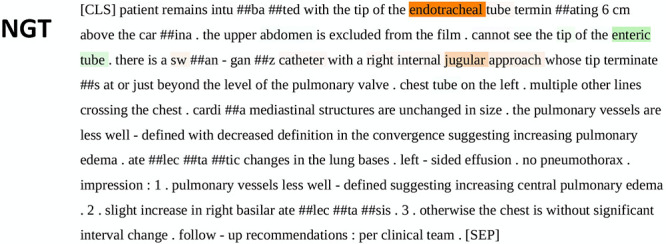
Example of “sequence classification explainer” from PubMedBERT. This figure demonstrates the process of “tokenization,” an automatic process that occurs after unprocessed text is introduced to the model. Words represented by the pretrained model’s “vocabulary” are maintained in their entirety, while those not in the model vocabulary are broken down into fragments that do exist in the vocabulary. These fragments are annotated with preceding “##.” Though the fragments do not hold real meaning, the model learns what combinations of the fragments mean through training in context of the text despite absence of these terms in the model’s vocabulary. Words are highlighted and color-coded per their positive (green), neutral (white), or negative (red) impact on the given task. The resulting saliency map provides insight and context for the model’s probability output. Highlighted annotation of certain words indicates that the model provided attention over a certain threshold to those words before providing an output. BERT = bidirectional encoder representations from transformers, NGT = enterogastric tube.
Model Performance
Performance metrics from the fivefold cross-validation of the pretrained BERT models are summarized in Table 2. Of 1004 reports used in this study, 358 of 1004 (35.6%) had at least one type of tube presented; specifically, 72 of 198 (36.4%), 65 of 203 (32.0%), 65 of 200 (32.5%), 78 of 195 (40.0%), and 77 of 208 (37.0%), respectively, on folds 1–5. The highest average AUCs achieved from the fivefold cross-validation were 0.996 (RoBERTa) for ETT, 0.994 (RoBERTa) for NGT, 0.991 (PubMedBERT) for CVC, and 0.98 (PubMedBERT) for SGC. Results of the DeLong test showed that the performances of RoBERTa compared with BERT for NGT, PubMedBERT and RoBERTa compared with DeBERTa for CVC, and PubMedBERT and RoBERTa compared with DeBERTa for SGC were significantly different (corrected P < .05). Table E1 (supplement) shows pairwise corrected P values between model performance for device classification.
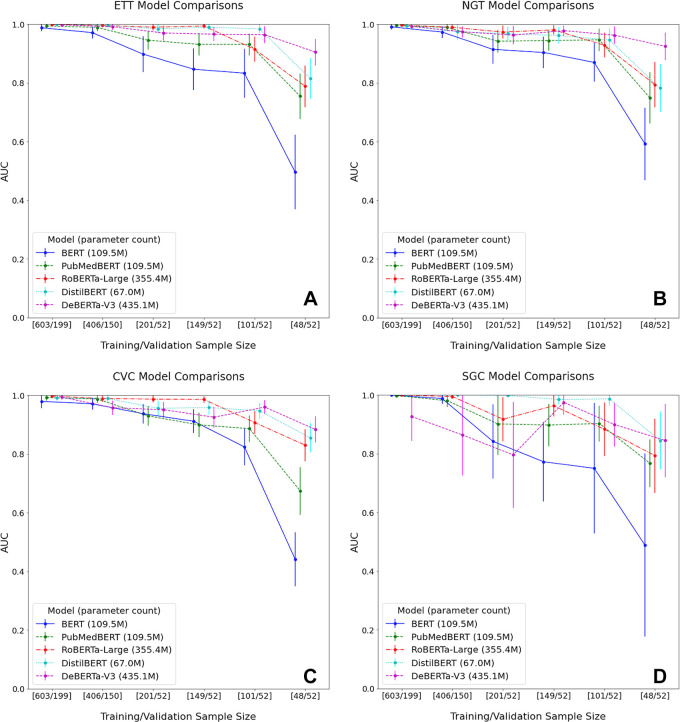
Performance metrics from varying training set sizes are shown in Figure 4, demonstrating the relationship between AUC versus training and validation set sizes. Notably, while training on just 5% (n = approximately 50) of the overall dataset, DeBERTa (matched by DistilBERT for SGC) achieved the highest AUCs of 0.91, 0.93, 0.88, and 0.85 on ETT, NGT, CVC, and SGC, respectively; this model outperformed all others, with the exception of identifying SGC, per the DeLong test (corrected P < .05). Additional results from the DeLong test indicated that each of the newer, pretrained transformer models demonstrated improved performance compared with BERT across all runs, with a progressively smaller training set size (P < .05). However, the presence of statistical differences varied when comparing the newer transformer models. Table E2 (supplement) shows pairwise corrected P values between model performance from runs 6–10.
Figure 4:
Model performance with decreasing training and validation sample size for the four devices: (A) endotracheal tube (ETT), (B) enterogastric tube (NGT), (C) central venous catheter (CVC), and (D) Swan-Ganz catheter (SGC). For each device, PubMedBERT and newer BERT models outperformed BERT as sample size decreased. DeBERTa demonstrated the best performance for each device at 5% of the training set size. Relatively high performance was achieved for all models except BERT, with as little as 10% of the original training set size. Results from the DeLong test indicated that each of the newer, pretrained transformer models demonstrated improved performance compared with BERT across all runs, with a progressively smaller training set size (P < .05). Note: Data points in each line plot appear slightly offset toward the right along the x-axis relative to corresponding axis labels to allow for ease of visualization. Actual training and validation set sizes are designated on the x-axis as discrete values without continuity between labels. AUC = area under the receiver operating characteristic curve, BERT = bidirectional encoder representations from transformers.
Training and validation time as represented by the average of the first five runs across five folds was shortest for DistilBERT at 3 minutes 39 seconds on the annotated cohort, with automated annotation of all 69 095 cases taking 6 minutes 15 seconds, in inference. DeBERTa was the slowest model, taking 22 minutes 48 seconds for training on the annotated cohort and 43 minutes 10 seconds in inference to annotate the larger overall cohort. PubMedBERT took 5 minutes 48 seconds to train and validate, compared with 6 minutes 20 seconds for BERT. Run times are fully detailed in Table E3 (supplement). As suggested in the table, model training times demonstrated a positive correlation with the number of training parameters, contributing to increased training time for RoBERTa and DeBERTa compared with the other models.
Discussion
Curation of a large dataset may often become the rate-limiting step in the creation of a computer vision tool (1,2). The results of this study demonstrate the role of pretrained BERT models further trained on task-specific data to rapidly and autonomously curate large amounts of data. If we extrapolate from the manual human performance time taken to label each of the original 1004 studies (20 seconds per case), the total time required for the overall cohort of 69 095 studies would be approximately 384 hours. In contrast, applying the fastest fully trained transformer model took 6 minutes 15 seconds (0.005 second per case). Differences between transformer models were modest, as the highest-performing model in each device category from fivefold cross-validation did not perform differently than three of the other four models (P < .05). Furthermore, statistical significance of the poorest-performing model varied without a consistent pattern between device categories. The fivefold cross-validation results show that the domain-specific BERT, PubMedBERT, outperformed the BERT model for every support device, though differences in performance were not substantial for a majority of the runs. In fact, select comparisons highlight better performance of a newer transformer model (RoBERTa) compared with BERT and a domain-specific BERT (PubMedBERT) compared with newer transformer models (DeBERTa). However, PubMedBERT outperformed BERT at all levels of progressively smaller training and validation set sizes, with substantial differences between AUCs for most runs.
Varying training set sizes in this context allowed us to discern a clear relationship with performance metrics of the model, namely the AUC. As expected, model performance improved with greater amounts of data included in the training set. However, a higher AUC than expected (approximately 0.88–0.91) was achieved for three of the four targets (ETT, NGT, and CVC) with a training dataset as small as 50 cases, demonstrating the potency of a pretrained, domain-specific algorithm, as well as the more recently developed transformer models. RoBERTa, DistilBERT, and DeBERTa consistently demonstrated statistically significant higher AUC with decreasing training data compared with BERT and PubMedBERT. Specifically, DeBERTa demonstrated the highest AUC when 5% (n = 48) of the fully annotated dataset was used for training, statistically significant for each support device except SGC, in which case DeBERTa and DistilBERT exhibited similar performance. However, the AUC achieved by DeBERTa with smaller training datasets represented lower performance than other transformer models trained on fivefold cross-validation with a larger training set size. Furthermore, DeBERTa demonstrated relatively weaker performance on fivefold cross-validation compared with three of the other four models, including BERT, for identification of SGC and two of the other four models for identification of CVC.
AUC increased steadily with more training data, and the use of a pretrained model enabled minimization of the time required to train the final task-specific BERT model. The models in this particular study required anywhere from 3 minutes 39 seconds to 22 minutes 48 seconds to train and validate on our standard hardware, as opposed to the time period of weeks reported in other studies (3). The relatively shorter model training time in this study may derive from fine-tuning a pretrained model, leveraging established model weights for similar tasks, compared with training a given transformer model from scratch. Of note, PubMedBERT had a training and validation time comparable to BERT, but improved performance in several experiments. Our findings suggest that the use of a pretrained, domain-specific model or newer transformer models through transfer learning may be an effective alternative to developing a model de novo in resource-constrained environments with a paucity of data.
Creating pretrained and newer transformer models requires consideration of available resources, specifically, available GPUs and the associated financial cost. For instance, reported training resources for a larger version of DeBERTa required 16 v100 GPUs and a training duration of 30 days (18). With these requirements, the cost of one training run amounts to tens to hundreds of thousands of dollars if renting such hardware from available cloud compute instances, before even considering the added cost necessary for hyperparameter tuning with multiple runs (22). Accordingly, transfer learning using established models allows for considerable cost savings.
Notably, high levels of performance were achieved with a smaller dataset in our study than used in prior studies examining BERT in radiology-specific tasks (25,26). Additionally, these models demonstrated high performance on a cohort that represented a blend of both structured and unstructured reports, obviating differentiating between structured and unstructured dictations. These results underscore the potential role of a pretrained advanced NLP model such as BERT to rapidly annotate large datasets, saving substantial time in an otherwise labor-intensive process.
Despite overall high performance, there were certain instances that led to algorithm failure. Specifically, though PubMedBERT featured a model pretrained on a domain-specific lexicon, it demonstrated instances of algorithm failure regarding device context. Regarding ETT, several cases demonstrated failure to differentiate the terms “gastric, enteric, and thoracostomy” from “endotracheal” in front of the word “tube.” For all devices, several cases demonstrated that each of the algorithms in this study failed to recognize the words “removal” or “removal of” in the context of the report.
In addition to the noted cases of algorithm failure, there were several other limitations worth noting. The ability to train the pretrained BERT model required determination of the spectrum of terms used in reports to describe common lines and tubes. Accordingly, the terms selected to further train this specific model were limited to those used in the randomly selected 1004 reports, which likely do not contain every possible phrasing in the overall cohort and therefore capture a representative sample. Additionally, the reports used in this study were retrieved from an academic tertiary care center, which serves a different demographic than those at other centers, such as affiliated county hospitals. Consideration of the demographic of patients with lines and tubes warrants attention in this setting, as studies have shown racial and sex disparities in catheter use (27). Furthermore, reporting of lines and tubes is possibly heterogeneous across institutions. Accordingly, future studies should examine model performance at external institutions on datasets obtained from varying patient demographics to determine model generalizability versus degradation with these changing factors. As demonstrated, the overall cohort in this study featured a near-even balance with regard to sex. Of note, 64.4% of the reports in this study did not have any lines or tubes, raising concern for potential “negative set” bias. However, the stratified composition of each training fold ensured balance between the number of positive and negative cases, as the difference between prevalence in each fold was not statistically significant. Finally, the heterogeneity of reports used to train a given transformer model can impact the accuracy of the final model’s output. Including inconsistent, inaccurate reports in training datasets risks poor dataset annotation because of erroneous labels from the resulting transformer model. Special mention is warranted regarding the relative paucity of SGC-positive cases in this cohort. The small amount of data regarding SGC with potential negative set bias may explain the relatively poor performance observed in this arm of the study. Furthermore, relatively low performance with regard to SGC on the test dataset may reflect a component of overfitting to the training dataset.
In conclusion, use of a pretrained, domain-specific or newer transformer model such as PubMedBERT in a transfer learning task requires relatively smaller-sized training datasets and a short amount of time to create a robust final classifier model that can expedite autonomous annotation of large datasets.
Authors declared no funding for this work.
Disclosures of conflicts of interest: A.S.T. No relevant relationships. Y.S.N. No relevant relationships. Y.X. No relevant relationships. J.R.F. No relevant relationships. T.G.B. Consulting fees from Change Healthcare. J.C.R. No relevant relationships.
Abbreviations:
- AUC
- area under the receiver operating characteristic curve
- BERT
- bidirectional encoder representations from transformers
- CVC
- central venous catheter
- ETT
- endotracheal tube
- NGT
- enterogastric tube
- NLP
- natural language processing
- SGC
- Swan-Ganz catheter
References
- 1. Willemink MJ , Koszek WA , Hardell C , et al . Preparing medical imaging data for machine learning . Radiology 2020. ; 295 ( 1 ): 4 – 15 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lee K , Famiglietti ML , McMahon A , et al . Scaling up data curation using deep learning: An application to literature triage in genomic variation resources . PLOS Comput Biol 2018. ; 14 ( 8 ): e1006390 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lee J , Yoon W , Kim S , et al . BioBERT: a pre-trained biomedical language representation model for biomedical text mining . Bioinformatics 2020. ; 36 ( 4 ): 1234 – 1240 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hripcsak G , Austin JH , Alderson PO , Friedman C . Use of natural language processing to translate clinical information from a database of 889,921 chest radiographic reports . Radiology 2002. ; 224 ( 1 ): 157 – 163 . [DOI] [PubMed] [Google Scholar]
- 5. Zhou Y , Amundson PK , Yu F , Kessler MM , Benzinger TL , Wippold FJ . Automated classification of radiology reports to facilitate retrospective study in radiology . J Digit Imaging 2014. ; 27 ( 6 ): 730 – 736 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Li AY , Elliot N . Natural language processing to identify ureteric stones in radiology reports . J Med Imaging Radiat Oncol 2019. ; 63 ( 3 ): 307 – 310 . [DOI] [PubMed] [Google Scholar]
- 7. Wiggins WF , Kitamura F , Santos I , Prevedello LM . Natural language processing of radiology text reports: interactive text classification . Radiol Artif Intell 2021. ; 3 ( 4 ): e210035 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cai T , Giannopoulos AA , Yu S , et al . Natural language processing technologies in radiology research and clinical applications . RadioGraphics 2016. ; 36 ( 1 ): 176 – 191 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bressem KK , Adams LC , Gaudin RA , et al . Highly accurate classification of chest radiographic reports using a deep learning natural language model pre-trained on 3.8 million text reports . Bioinformatics 2021. ; 36 ( 21 ): 5255 – 5261 . [DOI] [PubMed] [Google Scholar]
- 10. Chen MC , Ball RL , Yang L , et al . Deep learning to classify radiology free-text reports . Radiology 2018. ; 286 ( 3 ): 845 – 852 . [DOI] [PubMed] [Google Scholar]
- 11. Yu S , Kumamaru KK , George E , et al . Classification of CT pulmonary angiography reports by presence, chronicity, and location of pulmonary embolism with natural language processing . J Biomed Inform 2014. ; 52 : 386 – 393 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chapman BE , Lee S , Kang HP , Chapman WW . Document-level classification of CT pulmonary angiography reports based on an extension of the ConText algorithm . J Biomed Inform 2011. ; 44 ( 5 ): 728 – 737 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Devlin J , Chang M , Lee K , Toutanova K . BERT: Pre-training of deep bidirectional transformers for language understanding . arXiv:1810.04805 [preprint] https://arxiv.org/abs/1810.04805. Posted October 11, 2018 Accessed December 15, 2021 . [Google Scholar]
- 14. Smit A , Jain S , Rajpurkar P , Pareek A , Ng A , Lungren MP . CheXbert: combining automatic labelers and expert annotations for accurate radiology report labeling using BERT . arXiv:2004.09167 [preprint] https://arxiv.org/abs/2004.09167 Posted April 20, 2020. Accessed December 15, 2021 . [Google Scholar]
- 15. Jaiswal A , Yang L , Ghosh M , Rousseau J , Peng YL , Ding Y . RadBERT-CL: factually-aware contrastive learning for radiology report classification . arXiv:2110.15426 [preprint] https://arxiv.org/abs/2110.15426. Posted October 28, 2021. Accessed December 15, 2021 . [PMC free article] [PubMed] [Google Scholar]
- 16. Kuling G , Curpen B , Martel A . BI-RADS BERT & using section tokenization to understand radiology reports . arXiv:2110.07552 [preprint] https://arxiv.org/abs/2110.07552. Posted October 14, 2021 Accessed December 15, 2021 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tang JSN , Seah JCY , Zia A , et al . CLiP, catheter and line position dataset . Sci Data 2021. ; 8 ( 1 ): 285 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Paszke A , Gross S , Massa F , et al . PyTorch: an imperative style, high-performance deep learning library . Red Hook, NY: : Curran Associates; , 2019. . [Google Scholar]
- 19. Gu Y , Tinn R , Cheng H , et al . Domain-Specific Language Model Pretraining for Biomedical Natural Language Processing . arXiv:2007.15779 [preprint] https://arxiv.org/abs/2007.15779. Posted July 31, 2020. Accessed December 15, 2021 . [Google Scholar]
- 20. Sanh V DL , Chaumond J , et al . DistilBERT, a distilled version of BERT: smaller, faster, cheaper, and lighter . arXiv:1910.01108 [preprint] https://arxiv.org/abs/1910.01108. Posted October 2, 2019. Accessed December 15, 2021 . [Google Scholar]
- 21. Liu Y OM , Goyal N , et al . RoBERTa: a robustly optimized {BERT} pretraining approach . arXiv:1907.11692 [preprint] https://arxiv.org/abs/1907.11692. Posted July 26, 2019. Accessed December 15, 2021 . [Google Scholar]
- 22. He P GJ , Chen W . DeBERTaV3: Improving DeBERTa using ELECTRA-Style Pre-Training with Gradient-Disentangled Embedding Sharing . arXiv:2111.09543 [preprint] https://arxiv.org/abs/2111.09543. Posted November 18, 2021. Accessed December 15, 2021 . [Google Scholar]
- 23. DeLong ER , DeLong DM , Clarke-Pearson DL . Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach . Biometrics 1988. ; 44 ( 3 ): 837 – 845 . [PubMed] [Google Scholar]
- 24. Pierse C . Transformers Interpret . (Version 0.5.2) [Computer Software]. https://github.com/cdpierse/transformers-interpret. Published 2021. Accessed December 15, 2021.
- 25. Olthof AW , Shouche P , Fennema EM , et al . Machine learning based natural language processing of radiology reports in orthopaedic trauma . Comput Methods Programs Biomed 2021. ; 208 : 106304 . [DOI] [PubMed] [Google Scholar]
- 26. Zaman S , Petri C , Vimalesvaran K , et al . Automatic diagnosis labeling of cardiovascular MRI by using semisupervised natural language processing of text reports . Radiol Artif Intell 2021. ; 4 ( 1 ): e210085 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Arya S , Melanson TA , George EL , et al . Racial and sex disparities in catheter use and dialysis access in the united states medicare population . J Am Soc Nephrol 2020. ; 31 ( 3 ): 625 – 636 . [DOI] [PMC free article] [PubMed] [Google Scholar]