Abstract
With the rapid development of sequencing technology, research on pigs has focused on intestinal microbes. Accumulating evidence suggests that the metabolites of intestinal microbes are the key medium for interactions between microbes and the host. Amino acid metabolism is involved in the growth and immune processes of pigs. The gut microbes of pigs are heavily involved in the metabolism of amino acids in their hosts. Here, we review the latest relevant literature. Research findings show that microbial metabolites, such as indoles, short-chain fatty acids, and ammonia, play a key role in gut health. Moreover, we summarize the effects of amino acids on the structure of the gut microbial community and the metabolism of amino acids by pig gut microbes. Evidence shows that microbial amino acid metabolites act as signal molecules in the intestine and play an important role in the intestinal health of pigs.
Keywords: Pig, Amino acid, Intestinal microbe, Metabolite
1. Introduction
With the prohibition of antibiotics in the pig breeding industry, functional supplementary products such as amino acids are more widely used to improve growth performance and immunity. Intestinal microbes play crucial roles in the metabolism of amino acids in pigs and in their growth-promoting and immune functions (Xiang et al., 2020; Yin et al., 2017). Gut microbes can regulate the host's nutrition and metabolism, immune response, and even brain activity, among many physiological functions (Agus et al., 2018). The complex microbial community structure in the intestine is inextricably linked to the host's metabolism. Accumulating evidence shows that improving the dietary structure and supplementing certain nutrients can regulate intestinal microbes (Tremaroli and Bäckhed, 2012). Amino acids act as supplementary nutrients to pig diet. Through the mediation of proteins, the small intestine absorbs amino acids and transports them to various tissues. In the case of plasma amino acids, the small intestine's main role is to maintain homeostasis, whereas the main responsibility for amino acid transport lies with microbes and their metabolites in the large intestine (Bröer and Fairweather, 2018).
The dynamic changes in the metabolic products of amino acids and related microorganisms have not been sufficiently investigated. Recent studies have shown that amino acid catabolism products produced by intestinal flora are an important factor for intestinal homeostasis. Here, we review the metabolic processes of amino acids in pigs and the role of gut microbes in these processes. We discuss the role of microbial metabolism in the host and the effect of microbial metabolites' modulation mechanisms on the host's intestinal immunity (Fig. 1).
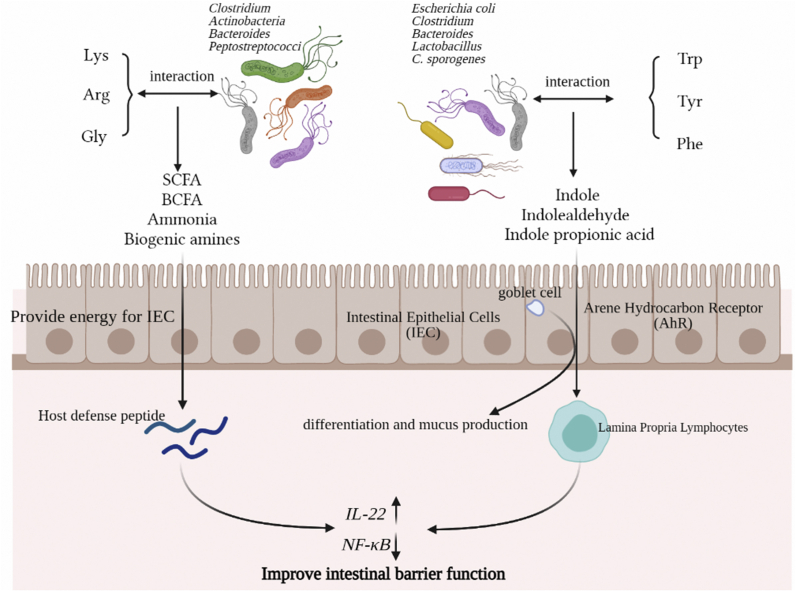
Fig. 1.
The beneficial effects of amino acids on pig intestinal health through the action of intestinal microbes. Metabolites of various types of amino acids fermented by intestinal microorganisms acting on epithelial cells to reinforce the intestinal immune barrier. SCFA = short-chain fatty acids; BCFA = branched-chain fatty acids; IL-22 = interleukin-22; NF-κB = nuclear factor-κB.
2. Bacteriala amino acid catabolism in pig intestines
The gastrointestinal tract of pigs is a complex host of diverse microbial communities. The metabolic processes of these microorganisms can improve host functions, enhance intestinal immunity, and improve intestinal nutrient absorption and health. Research has shown that intestinal microbes can metabolize and utilize amino acids. After amino acids are absorbed by the small intestine, many endogenous and exogenous nitrogen-containing compounds enter the large intestine through the cecum, where they are used by colonic microbes to produce metabolites through fermentation (Abdallah et al., 2020).
2.1. Microbial catabolism of nonaromatic amino acids in pig intestines
Amino acids are absorbed and first metabolized by porcine small intestinal epithelial cells. Mucosal cells of the small intestine play a major role in metabolizing lysine. Research has shown that lysine can also be metabolized by the intestinal microbiota of piglets (Stoll et al., 1998). Moreover, growing evidence shows that other amino acids, such as methionine and phenylalanine, can also be used by small intestine microorganisms (Davila et al., 2013). Most intestinal microorganisms preferentially use amino acids and ammonia as high-quality nitrogen sources. To some extent, some amino acids are absorbed by intestinal microorganisms as substrates and transported to bacterial cells as protein structures for catabolism (Neis et al., 2015). Clostridium clusters, Proteobacteria, and Bacillus, Lactobacillus, and Streptococcus are the most common strains known to participate in amino acid fermentation in the small intestine (Dai et al., 2011). In vitro experiments have shown that lysine, arginine, threonine, and glutamic acid are the amino acids most commonly used by small intestine microorganisms. The metabolism of these amino acids depends on the type of microorganism and the gut compartment (Dai et al., 2010, 2012). The main microorganisms involved in amino acid metabolism in the large intestine are Clostridium and Peptostreptococcus (Davila et al., 2013). These bacteria can metabolize lysine, arginine, glycine, and other amino acids into complex mixtures of ammonia, short-chain fatty acids (SCFA), and branched-chain fatty acids (BCFA) (Dai et al., 2011). Metabolites such as SCFA can promote the production of aryl hydrocarbon receptors and hypoxia-inducible factor-1α (HIF-1α), increasing the expression of interleukin-22 (IL-22) and ultimately improving intestinal immunity (Yang et al., 2020). Biogenic amines are other substances produced by microorganisms through amino acid decarboxylation. Biogenic amines derived from animal intestinal metabolism mainly include cadaverine, a decarboxylation product of lysine, and agmatine, a product of arginine (Sánchez-Jiménez et al., 2013). Lactobacillus rhamnosus can ferment histidine as a substrate and decarboxylate it to produce histamine, a microbial metabolite (Frei et al., 2013). Biogenic amines in appropriate amounts are crucial for intestinal health, as they can maintain intestinal function stability and improve intestinal immunity (Fan et al., 2017). However, excessive amounts can cause poisoning, headaches, and digestive disorders (Özogul and Hamed, 2018). Fusobacteria in the intestine can use cysteine to produce hydrogen sulfide through the action of intestinal epithelial cells. Although hydrogen sulfide is known to be a toxic gas, recent studies have shown that low concentrations of hydrogen sulfide produced by endogenous metabolism can maintain the integrity of the intestinal mucus layer and reduce inflammation of the intestinal mucosa (Blachier et al., 2019).
2.2. Microbial catabolism of aromatic amino acids in pig intestines
Many studies have examined the metabolic processes of aromatic amino acids. As early as the 18th century, it was discovered that tryptophan could be metabolized into indole by Escherichia coli and Vibrio cholera. Consequently, indole was once used as a metabolite marker to distinguish E. coli from other intestinal microorganisms (Lee and Lee, 2010). The metabolism of tryptophan to indole is mediated by tryptophanase, which is found in the expression of many intestinal microorganisms, including E. coli, Clostridium, and Bacteroides (Devlin et al., 2016; Smith and Macfarlane, 1996). It has also been found that tryptophan can be used by other microorganisms to produce indole derivatives through various metabolic pathways. For example, Lactobacillus spp. can use aromatic amino acid aminotransferase and indolelactic acid dehydrogenase to metabolize tryptophan into indole-3-aldehyde and indolelactic acid (Zelante et al., 2013). Moreover, tryptophan can be metabolized into indole propionic acid by various intestinal microorganisms, such as Clostridium sporogenes and Clostridium botulinum (Wikoff et al., 2009). Indole propionic acid can improve the intestinal barrier function through pregnane X receptor and Toll-like receptor protein (Venkatesh et al., 2014). The metabolic pathways of other aromatic amino acids have also been extensively studied. Through the action of Streptomyces maritimus, phenylalanine can be converted to cinnamic acid and ammonia through nonoxidative deamination by phenylalanine ammonia lyase (Xiang and Moore, 2005). Studies have shown that these processes of amino acid metabolism are determined by key genes of the involved microorganisms. The fldH, fldC, and acdA genes play an essential role in the reduction metabolism of threonine, phenylalanine, and tyrosine, whereas porA is involved in the reduction metabolism of phenylalanine and tyrosine.
3. Role of dietary amino acids in the regulation of intestinal microbes in pigs
Intestinal microbes have received increasing attention and have shown intricate relationships with animal growth, development, and health. The intestinal flora exhibits considerable plasticity and can undergo structural changes under the influence of the environment (such as diet changes and acute stress) (Conlon and Bird, 2014). Proteins and decomposed amino acids in the diet are important substrate sources of intestinal microbial fermentation in pigs. Amino acids can also be used as a source of nitrogen, promoting the growth of gut microbiota and the host's growth and development (Wu et al., 2011). The addition of amino acids to pigs' diet can significantly adjust the structure and functional characteristics of the intestinal microbial community.
Several mechanisms through which amino acids act on gut microbes have been recognized. The intake of amino acids promotes the secretion of intestinal β-defensin, endogenous cationic peptides, and other antibacterial substances in the intestines, effectively inhibiting the growth of harmful bacteria (Sherman et al., 2006). Amino acids can also promote the contraction of the gallbladder, affecting the production of cholecystokinin and changing the metabolic processes of intestinal microbes (Steinert et al., 2015). Furthermore, porcine gut microbes can catabolize amino acids to produce metabolites, such as kynurenine and indole, which contribute to the host's feedback regulation (Sun et al., 2020). In the following subsections, we discuss the interactions between pig gut microbes and amino acids. Table 1 describes the regulatory relationship between amino acids and gut microbes.
Table 1.
The regulatory relationship between amino acids and gut microbes.
| Amino acids | Gut microbiota | Relation | References |
|---|---|---|---|
| Glycine | Clostridium | ↑ | |
| Tyrosine | Lactobacillus algidus | ↑ | Säde et al. (2020) |
| Rikenellaceae | ↑ | Zhang et al. (2019) | |
| Aspartic | Clostridium | ↑ | Li et al. (2019) |
| Intestinibacter | ↑ | ||
| Prevotella | ↑ | Zhou et al. (2016) | |
| Lysine | Streptococcu | ↑ | Yin et al. (2018) |
| Bacteroides | ↑ | ||
| Bacillus | ↑ | ||
| Clostridium | ↑ | ||
| Actinobacteria | ↑ | Yin et al. (2017) | |
| Saccharibacteria | ↑ | ||
| Synergistetes | ↑ | ||
| Tryptophan | Lactobacillus | ↑ | Liang et al. (2018) |
| Clostridium | ↑ | ||
| Streptococcus | ↓ | ||
| Escherichia coli | ↓ | Messori et al. (2013) | |
| Methionine | Phascolarctobacterium | ↑ | Azad et al. (2018) |
| Bacteroides | ↑ | ||
| Threonine | Escherichia coli | ↓ | Trevisi et al. (2015) |
3.1. Interactions between nonessential amino acids and pig gut microbes
Nonessential amino acids can be synthesized by pigs themselves or obtained through the ingestion and transformation of other amino acids. However, the body content also affects the growth and immune functions of pigs and exerts a profound regulatory effect on the structure of the intestinal microbial community (Dai et al., 2013). Low glycine levels can lead to disorders of the metabolic system. Rom et al. found that this phenomenon can significantly increase the abundance of Clostridium spp. (Rom et al., 2020). Another study found that the serum tyrosine content of obese pigs was significantly higher than that of lean pigs, leading to changes in microbial metabolites (trimethylamine-N-oxide) related to obesity (He et al., 2012). Furthermore, an experiment conducted to investigate the growth performance of pigs showed that sufficient aspartic acid intake can increase daily weight gain, reduce the abundance of Actinobacteria and Bacteroides in the intestinal tract, and increase the levels of Clostridium and Intestinibacter. The levels of these nonessential amino acids in the body are mainly related to the growth and metabolism of pigs. They are mostly involved in the regulation of the microbial community involved in membrane transport and metabolism (Li et al., 2019).
3.2. Interactions between essential amino acids and pig gut microbes
The 11 essential amino acids for pigs are lysine, tryptophan, methionine, cystine, arginine, histidine, leucine, isoleucine, threonine, phenylalanine, and valine. Pigs either cannot synthesize them or cannot synthesize them in sufficient amounts to meet their needs. Therefore, these amino acids must be added to pigs' diet to maintain their nitrogen balance, meet their growth needs, and promote immunity (Chen et al., 2009). Lysine supplements have been found to increase the feed intake of piglets and reduce the abundance of Streptococcus, Bacteroides, Bacillus, Pasteurella, Clostridium sensu stricto, Faecalibacterium, Paucisalibacillus, and Lachnoclostridium. Most of these microbial communities are related to host metabolism, especially amino acid metabolism (Yin et al., 2018). Tryptophan is mostly added to diets for weaned piglets. It can reduce the abundance of Clostridium sensu stricto and Streptococcus and increase the abundance of Lactobacillus and Clostridium XI, which are related to tryptophan metabolism in the jejunum, ultimately improving the intestinal barrier function (Liang et al., 2018). Tryptophan can boost the resistance of weaned piglets to F4 enterotoxigenic E. coli colonization and increase the diversity of intestinal microbes (Messori et al., 2013). Furthermore, trials have shown that adding 0.48% methionine to lactating sows' diet can significantly increase their antioxidant capacity and increase the abundance of Phascolarctobacterium and Bacteroides, contributing to maintaining piglet health (Azad et al., 2018). The addition of arginine to feed can alleviate the effects of weaning stress in piglets and counteract stress-induced metabolic disturbances. However, it cannot restore disturbed intestinal flora (He et al., 2011). Threonine can produce mucin and immunoglobulin, protecting weaned piglets from E. coli K88ac infection, which can affect their growth performance (Trevisi et al., 2015). Most of these essential amino acids are related to piglets’ growth performance and immunity. Therefore, they are used in the swine industry to increase productivity and reduce stress and disease during growth.
4. Microbial amino acid metabolites as signaling molecules
Most research on amino acid microbial metabolites as signal molecules has focused on aromatic amino acids, whereas the role of other amino acids has not been extensively explored. Indole, the main metabolite of tryptophan, is well known as an intercellular molecular signal (Roager and Licht, 2018). E. coli has been shown to produce 600 μmol/L of indole in suspension culture species (Domka et al., 2006), and the concentration of indole detected in human feces ranges from 250 to 1,100 μmol/L (Bansal et al., 2010). Indole can inhibit the production of spores and biofilms, compromise the stability of plasmids, and produce virulence factors, thereby regulating the structure of intestinal microbes (Lee and Lee, 2010). Indole derivatives in the intestine can also affect the integrity of the intestinal barrier through the aryl hydrocarbon receptor (AhR)/IL-22 axis. Indole-3-aldehyde can use AhR to activate lamina propria lymphocytes and produce IL-22, which further promotes the proliferation of intestinal epithelial cells and ensures the integrity of the intestinal barrier structure (Hou et al., 2018). Moreover, indole-3-aldehyde and indolelactic acid can activate AhR in CD4+ T cells through Lactobacillus reuteri and program them into immune regulatory T cells (Cervantes-Barragan et al., 2017). It seems that the microbial metabolites of aromatic amino acids and their ligand AhR play a decisive role in intestinal barrier immunity. In vitro experiments have shown that tryptophan metabolites indole-3-ethanol, indole-3-pyruvate, and indole-3-aldehyde inhibit tumor necrosis factor-α (TNF-α)-induced increased epithelial permeability in human Caco-2 IEC cell lines (Hou et al., 2018). Indolepropionic acid (IPA), another microbial-derived metabolite of indole, can also inhibit the expression of TNF-α in intestinal epithelial cells and at the same time enhance intestinal immunity through the mediation of TLR4 (Venkatesh et al., 2014) and IL-10R1 (Alexeev et al., 2018). Arginine can increase the expression of pBD2 and pBD3 in ileum tissue, promote the secretion of host defense peptides, and protect the integrity of the intestinal barrier function (Osei-Boadi et al., 2013). Nitric oxide, the microbial metabolite of arginine, is also essential in this process, as it can prevent intestinal mucosal damage and inhibit inflammation (Leitão et al., 2011). Isoleucine, leucine, and valine seem to increase the expression of β-defensin in porcine intestinal epithelial cells by activating the sirtuin-1-ERK-90-kDa ribosomal S6 kinase pathway, thereby enhancing intestinal immunity (Ren et al., 2016).
5. Effects of microbial amino acid metabolites on intestinal health
Metabolites of amino acids produced through intestinal microbial fermentation are closely related to intestinal health (Lallès, 2016). Indole, the most abundant microbial metabolite of aromatic amino acids, exerts strong intestinal anti-inflammatory activity, and its anti-inflammatory mechanism has been thoroughly studied (Jansson et al., 2009; Santoru et al., 2017). Indole exhibits an immune-enhancing effect on the colon. It can inhibit the activation of nuclear factor-κB (NF-κB), increase intestinal transmembrane resistance, and improve the intestinal barrier function (Bansal et al., 2010). Moreover, experiments have demonstrated that indoleacrylic acid can mediate goblet cell differentiation and mucus production through AhR, thereby improving intestinal immunity and repairing intestinal epithelial barrier damage (Wlodarska et al., 2017). The content of indole derivatives in the intestine, including indole-3-aldehyde and indole acetic acid, is significantly reduced under intestinal inflammation. This metabolic disorder in the intestine leads to damage to the intestinal barrier and increases the risk of infection and inflammation (Lamas et al., 2016). Indole metabolized by intestinal microbes can affect the time of action potentials fired by colonic L cells and promote the production of glucagon-like peptide-1 (Chimerel et al., 2014), which participates in the regulation of appetite and plays an important role in fat loss and metabolic regulation (de Mello et al., 2017).
Although most SCFA in the intestine are produced through the fermentation of dietary fibers, less than 1% of intestinal microbes can use amino acids for fermentation to form propionic acid and butyric acid (Dai et al., 2011). For example, Intestinimonas AF211 can ferment lysine to form butyric acid in several ways (Bui et al., 2015). Clostridium can enter the pyruvate cycle through coenzyme B12-dependent glutamate mutase. Pyruvate is disproportionated to butyric acid, acetic acid, and propionic acid (Buckel, 2001). Short-chain fatty acids can mediate the expression of host defense peptides in pig intestines (Wu et al., 2020). Due to the metabolism of intestinal microorganisms, the SCFA content in the colon is the highest. In addition to providing energy to intestinal epithelial cells, they can also promote the secretion of host defense peptides (Hamer et al., 2008). Many experiments have shown that butyrate can promote the expression of anti-inflammatory factors in intestinal epithelial cells while stabilizing the structure of the intestinal microbial community, increasing metabolic activity, stabilizing intestinal development, and improving immunity (Chung et al., 2012; Kamada et al., 2013; Lee and Mazmanian, 2010). Specifically, butyrate can downregulate the expression of the NF-κB and histone deacetylase 1 (HDAC1) genes in colonic epithelial cells to reduce inflammation (Xu et al., 2016). Propionate has been shown to change the microbial structure, increase the abundance of probiotics, and affect the expression of pro-inflammatory cytokines, thereby regulating colon health in fistula pig models (Li et al., 2020).
6. Conclusion
This review of recent research provides a thorough understanding of the effects of amino acids on the growth and immune processes of pigs. Extensive evidence demonstrates the effects of various amino acids on changes in the structure of pig intestinal microbial communities, as well as the processes of microbial metabolism and decomposition of amino acids. The growing understanding of these processes has led to investigations of the role of microbial amino acid metabolites as signaling molecules in the intestine. The findings have demonstrated the importance of microbial amino acid metabolites for intestinal health. This provides a solid research basis for more efficient pig breeding and new options for antibiotic-free production. However, many aspects remain underexplored. For example, most studies on microbial amino acid metabolites have focused on functional amino acids (arginine and tryptophan), whereas few studies have examined other types of microbial amino acid metabolites. Therefore, further research is needed to gain a deeper understanding of the microbial metabolism of other amino acids and its effects on pig intestinal health.
Author contributions
Yong Ma: Writing – original draft preparation, revision, and investigation. Xuebing Han: Revision. Jun Fang: Supervision – oversight and leadership responsibility for the research activity planning and execution, including mentorship external to the core team. Hongmei Jang: Validation.
Declaration of competing interest
We declare that we have no financial and personal relationships with other people or organizations that can inappropriately influence our work, and there is no professional or other personal interest of any nature or kind in any product, service and/or company that could be construed as influencing the content of this paper.
Acknowledgements
This research was supported by National Natural Science Foundation of China (No. 31772642, 31672457, 41807135), Hunan Provincial Science and Technology Department (2019TP2004, 2020NK2004, 2020ZL2004), China Postdoctoral Science Foundation (2018M632963, 2019T120705), Scientific Research Fund of Hunan Provincial Education Department (2020JGYB112, 18B107), Double First-class Construction Project of Hunan Agricultural University (SYL201802003, YB2018007, CX20190497), Natural Science Foundation of Hunan province, China (No. 2019JJ50220), and Undergraduate on Innovation and Entrepreneurship Training Program (No. S202010537084).
Footnotes
Peer review under responsibility of Chinese Association of Animal Science and Veterinary Medicine.
Contributor Information
Jun Fang, Email: fangjun1973@hunau.edu.cn.
Hongmei Jiang, Email: jhmndcn@hunau.edu.cn.
References
- Abdallah A., Elemba E., Zhong Q., Sun Z. Gastrointestinal interaction between dietary amino acids and gut microbiota: with special emphasis on host nutrition. Curr Protein Pept Sci. 2020;21:785–798. doi: 10.2174/1389203721666200212095503. [DOI] [PubMed] [Google Scholar]
- Agus A., Planchais J., Sokol H. Gut microbiota regulation of tryptophan metabolism in health and disease. Cell Host Microbe. 2018;23:716–724. doi: 10.1016/j.chom.2018.05.003. [DOI] [PubMed] [Google Scholar]
- Alexeev E.E., Lanis J.M., Kao D.J., Campbell E.L., Kelly C.J., Battista K.D., Gerich M.E., Jenkins B.R., Walk S.T., Kominsky D.J., et al. Microbiota-derived indole metabolites promote human and murine intestinal homeostasis through regulation of interleukin-10 receptor. Am J Pathol. 2018;188:1183–1194. doi: 10.1016/j.ajpath.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azad M.A.K., Bin P., Liu G., Fang J., Li T., Yin Y. Effects of different methionine levels on offspring piglets during late gestation and lactation. Food & function. 2018;9:5843–5854. doi: 10.1039/c8fo01343h. [DOI] [PubMed] [Google Scholar]
- Bansal T., Alaniz R.C., Wood T.K., Jayaraman A. The bacterial signal indole increases epithelial-cell tight-junction resistance and attenuates indicators of inflammation. Proc Natl Acad Sci USA. 2010;107:228–233. doi: 10.1073/pnas.0906112107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blachier F., Beaumont M., Kim E. Cysteine-derived hydrogen sulfide and gut health: a matter of endogenous or bacterial origin. Curr Opin Clin Nutr Metab Care. 2019;22:68–75. doi: 10.1097/mco.0000000000000526. [DOI] [PubMed] [Google Scholar]
- Bröer S., Fairweather S.J. Amino acid transport across the mammalian intestine. Compr Physiol. 2018;9:343–373. doi: 10.1002/cphy.c170041. [DOI] [PubMed] [Google Scholar]
- Buckel W. Unusual enzymes involved in five pathways of glutamate fermentation. Appl Microbiol Biotechnol. 2001;57:263–273. doi: 10.1007/s002530100773. [DOI] [PubMed] [Google Scholar]
- Bui T.P., Ritari J., Boeren S., de Waard P., Plugge C.M., de Vos W.M. Production of butyrate from lysine and the amadori product fructoselysine by a human gut commensal. Nat Commun. 2015;6:10062. doi: 10.1038/ncomms10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes-Barragan L., Chai J.N., Tianero M.D., Di Luccia B., Ahern P.P., Merriman J., Cortez V.S., Caparon M.G., Donia M.S., Gilfillan S., et al. Lactobacillus reuteri induces gut intraepithelial cd4(+)cd8αα(+) t cells. Science (New York, NY) 2017;357:806–810. doi: 10.1126/science.aah5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Li P., Wang J., Li X., Gao H., Yin Y., Hou Y., Wu G. Catabolism of nutritionally essential amino acids in developing porcine enterocytes. Amino Acids. 2009;37:143–152. doi: 10.1007/s00726-009-0268-1. [DOI] [PubMed] [Google Scholar]
- Chimerel C., Emery E., Summers D.K., Keyser U., Gribble F.M., Reimann F. Bacterial metabolite indole modulates incretin secretion from intestinal enteroendocrine l cells. Cell Rep. 2014;9:1202–1208. doi: 10.1016/j.celrep.2014.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung H., Pamp S.J., Hill J.A., Surana N.K., Edelman S.M., Troy E.B., Reading N.C., Villablanca E.J., Wang S., Mora J.R., et al. Gut immune maturation depends on colonization with a host-specific microbiota. Cell. 2012;149:1578–1593. doi: 10.1016/j.cell.2012.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlon M.A., Bird A.R. The impact of diet and lifestyle on gut microbiota and human health. Nutrients. 2014;7:17–44. doi: 10.3390/nu7010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Z.L., Li X.L., Xi P.B., Zhang J., Wu G., Zhu W.Y. Metabolism of select amino acids in bacteria from the pig small intestine. Amino Acids. 2012;42:1597–1608. doi: 10.1007/s00726-011-0846-x. [DOI] [PubMed] [Google Scholar]
- Dai Z.L., Li X.L., Xi P.B., Zhang J., Wu G., Zhu W.Y. L-glutamine regulates amino acid utilization by intestinal bacteria. Amino Acids. 2013;45:501–512. doi: 10.1007/s00726-012-1264-4. [DOI] [PubMed] [Google Scholar]
- Dai Z.L., Wu G., Zhu W.Y. Amino acid metabolism in intestinal bacteria: links between gut ecology and host health. Front Biosci. 2011;16:1768–1786. doi: 10.2741/3820. [DOI] [PubMed] [Google Scholar]
- Dai Z.L., Zhang J., Wu G., Zhu W.Y. Utilization of amino acids by bacteria from the pig small intestine. Amino Acids. 2010;39:1201–1215. doi: 10.1007/s00726-010-0556-9. [DOI] [PubMed] [Google Scholar]
- Davila A.M., Blachier F., Gotteland M., Andriamihaja M., Benetti P.H., Sanz Y., Tomé D. Re-print of "intestinal luminal nitrogen metabolism: role of the gut microbiota and consequences for the host. Pharmacol Res. 2013;69:114–126. doi: 10.1016/j.phrs.2013.01.003. [DOI] [PubMed] [Google Scholar]
- de Mello V.D., Paananen J., Lindström J., Lankinen M.A., Shi L., Kuusisto J., Pihlajamäki J., Auriola S., Lehtonen M., Rolandsson O., et al. Indolepropionic acid and novel lipid metabolites are associated with a lower risk of type 2 diabetes in the Finnish diabetes prevention study. Sci Rep. 2017;7:46337. doi: 10.1038/srep46337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin A.S., Marcobal A., Dodd D., Nayfach S., Plummer N., Meyer T., Pollard K.S., Sonnenburg J.L., Fischbach M.A. Modulation of a circulating uremic solute via rational genetic manipulation of the gut microbiota. Cell Host Microbe. 2016;20:709–715. doi: 10.1016/j.chom.2016.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domka J., Lee J., Wood T.K. Ylih (bssr) and ycep (bsss) regulate escherichia coli k-12 biofilm formation by influencing cell signaling. Appl Environ Microbiol. 2006;72:2449–2459. doi: 10.1128/aem.72.4.2449-2459.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan P., Song P., Li L., Huang C., Chen J., Yang W., Qiao S., Wu G., Zhang G., Ma X. Roles of biogenic amines in intestinal signaling. Curr Protein Pept Sci. 2017;18:532–540. doi: 10.2174/1389203717666160627073048. [DOI] [PubMed] [Google Scholar]
- Frei R., Ferstl R., Konieczna P., Ziegler M., Simon T., Rugeles T.M., Mailand S., Watanabe T., Lauener R., Akdis C.A., et al. Histamine receptor 2 modifies dendritic cell responses to microbial ligands. J Allergy Clin Immunol. 2013;132:194–204. doi: 10.1016/j.jaci.2013.01.013. [DOI] [PubMed] [Google Scholar]
- Hamer H.M., Jonkers D., Venema K., Vanhoutvin S., Troost F.J., Brummer R.J. Review article: the role of butyrate on colonic function. Aliment Pharmacol Ther. 2008;27:104–119. doi: 10.1111/j.1365-2036.2007.03562.x. [DOI] [PubMed] [Google Scholar]
- He Q., Ren P., Kong X., Wu Y., Wu G., Li P., Hao F., Tang H., Blachier F., Yin Y. Comparison of serum metabolite compositions between obese and lean growing pigs using an nmr-based metabonomic approach. J Nutr Biochem. 2012;23:133–139. doi: 10.1016/j.jnutbio.2010.11.007. [DOI] [PubMed] [Google Scholar]
- He Q., Tang H., Ren P., Kong X., Wu G., Yin Y., Wang Y. Dietary supplementation with l-arginine partially counteracts serum metabonome induced by weaning stress in piglets. J Proteome Res. 2011;10:5214–5221. doi: 10.1021/pr200688u. [DOI] [PubMed] [Google Scholar]
- Hou Q., Ye L., Liu H., Huang L., Yang Q., Turner J.R., Yu Q. Lactobacillus accelerates iscs regeneration to protect the integrity of intestinal mucosa through activation of stat3 signaling pathway induced by lpls secretion of il-22. Cell Death Differ. 2018;25:1657–1670. doi: 10.1038/s41418-018-0070-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansson J., Willing B., Lucio M., Fekete A., Dicksved J., Halfvarson J., Tysk C., Schmitt-Kopplin P. Metabolomics reveals metabolic biomarkers of crohn's disease. PloS one. 2009;4 doi: 10.1371/journal.pone.0006386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada N., Seo S.U., Chen G.Y., Núñez G. Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol. 2013;13:321–335. doi: 10.1038/nri3430. [DOI] [PubMed] [Google Scholar]
- Lallès J.P. Microbiota-host interplay at the gut epithelial level, health and nutrition. J Anim Sci Biotechnol. 2016;7:66. doi: 10.1186/s40104-016-0123-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamas B., Richard M.L., Leducq V., Pham H.P., Michel M.L., Da Costa G., Bridonneau C., Jegou S., Hoffmann T.W., Natividad J.M., et al. Card9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat Med. 2016;22:598–605. doi: 10.1038/nm.4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.H., Lee J. Indole as an intercellular signal in microbial communities. FEMS Microbiol Rev. 2010;34:426–444. doi: 10.1111/j.1574-6976.2009.00204.x. [DOI] [PubMed] [Google Scholar]
- Lee Y.K., Mazmanian S.K. Has the microbiota played a critical role in the evolution of the adaptive immune system? Science (New York, NY) 2010;330:1768–1773. doi: 10.1126/science.1195568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitão R.F., Brito G.A., Oriá R.B., Braga-Neto M.B., Bellaguarda E.A., Silva J.V., Gomes A.S., Lima-Júnior R.C., Siqueira F.J., Freire R.S., et al. Role of inducible nitric oxide synthase pathway on methotrexate-induced intestinal mucositis in rodents. BMC Gastroenterol. 2011;11:90. doi: 10.1186/1471-230x-11-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H.Y., Zhang H.L., Zhao F.J., Wang S.Q., Wang Z.X., Wei Z.Y. Modulation of gut microbiota, short-chain fatty acid production, and inflammatory cytokine expression in the cecum of porcine deltacoronavirus-infected chicks. Front Microbiol. 2020;11:897. doi: 10.3389/fmicb.2020.00897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Han H., Yin J., He X., Tang Z., Li T., Yao K., Yin Y. D- and l-aspartate regulates growth performance, inflammation and intestinal microbial community in young pigs. Food & function. 2019;10:1028–1037. doi: 10.1039/c8fo01410h. [DOI] [PubMed] [Google Scholar]
- Liang H., Dai Z., Kou J., Sun K., Chen J., Yang Y., Wu G., Wu Z. Dietary l-tryptophan supplementation enhances the intestinal mucosal barrier function in weaned piglets: implication of tryptophan-metabolizing microbiota. Int J Mol Sci. 2018;20 doi: 10.3390/ijms20010020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messori S., Trevisi P., Simongiovanni A., Priori D., Bosi P. Effect of susceptibility to enterotoxigenic escherichia coli f4 and of dietary tryptophan on gut microbiota diversity observed in healthy young pigs. Vet Microbiol. 2013;162:173–179. doi: 10.1016/j.vetmic.2012.09.001. [DOI] [PubMed] [Google Scholar]
- Neis E.P., Dejong C.H., Rensen S.S. The role of microbial amino acid metabolism in host metabolism. Nutrients. 2015;7:2930–2946. doi: 10.3390/nu7042930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osei-Boadi K., Gordon E., Melgarejo T. Select amino acids induced expression of human beta-defensin in caco-2 cells. Faseb J. 2013 [Google Scholar]
- Özogul F., Hamed I. The importance of lactic acid bacteria for the prevention of bacterial growth and their biogenic amines formation: a review. Crit Rev Food Sci Nutr. 2018;58:1660–1670. doi: 10.1080/10408398.2016.1277972. [DOI] [PubMed] [Google Scholar]
- Ren M., Zhang S., Liu X., Li S., Mao X., Zeng X., Qiao S. Different lipopolysaccharide branched-chain amino acids modulate porcine intestinal endogenous β-defensin expression through the sirt1/erk/90rsk pathway. J Agric Food Chem. 2016;64:3371–3379. doi: 10.1021/acs.jafc.6b00968. [DOI] [PubMed] [Google Scholar]
- Roager H.M., Licht T.R. Microbial tryptophan catabolites in health and disease. Nat Commun. 2018;9:3294. doi: 10.1038/s41467-018-05470-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rom O., Liu Y., Liu Z., Zhao Y., Wu J., Ghrayeb A., Villacorta L., Fan Y., Chang L., Wang L., et al. Glycine-based treatment ameliorates nafld by modulating fatty acid oxidation, glutathione synthesis, and the gut microbiome. Sci Transl Med. 2020;12 doi: 10.1126/scitranslmed.aaz2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Säde E., Johansson P., Heinonen T., Hultman J., Björkroth J. Growth and metabolic characteristics of fastidious meat-derived lactobacillus algidus strains. Int J Food Microbiol. 2020;313:108379. doi: 10.1016/j.ijfoodmicro.2019.108379. [DOI] [PubMed] [Google Scholar]
- Sánchez-Jiménez F., Ruiz-Pérez M.V., Urdiales J.L., Medina M.A. Pharmacological potential of biogenic amine-polyamine interactions beyond neurotransmission. Br J Pharmacol. 2013;170:4–16. doi: 10.1111/bph.12109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoru M.L., Piras C., Murgia A., Palmas V., Camboni T., Liggi S., Ibba I., Lai M.A., Orrù S., Blois S., et al. Cross sectional evaluation of the gut-microbiome metabolome axis in an Italian cohort of ibd patients. Sci Rep. 2017;7:9523. doi: 10.1038/s41598-017-10034-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman H., Chapnik N., Froy O. Albumin and amino acids upregulate the expression of human beta-defensin 1. Mol Immunol. 2006;43:1617–1623. doi: 10.1016/j.molimm.2005.09.013. [DOI] [PubMed] [Google Scholar]
- Smith E.A., Macfarlane G.T. Enumeration of human colonic bacteria producing phenolic and indolic compounds: effects of ph, carbohydrate availability and retention time on dissimilatory aromatic amino acid metabolism. J Appl Bacteriol. 1996;81:288–302. doi: 10.1111/j.1365-2672.1996.tb04331.x. [DOI] [PubMed] [Google Scholar]
- Steinert R.E., Landrock M.F., Ullrich S.S., Standfield S., Otto B., Horowitz M., Feinle-Bisset C. Effects of intraduodenal infusion of the branched-chain amino acid leucine on ad libitum eating, gut motor and hormone functions, and glycemia in healthy men. Am J Clin Nutr. 2015;102:820–827. doi: 10.3945/ajcn.115.114488. [DOI] [PubMed] [Google Scholar]
- Stoll B., Henry J., Reeds P.J., Yu H., Jahoor F., Burrin D.G. Catabolism dominates the first-pass intestinal metabolism of dietary essential amino acids in milk protein-fed piglets. J Nutr. 1998;128:606–614. doi: 10.1093/jn/128.3.606. [DOI] [PubMed] [Google Scholar]
- Sun M., Ma N., He T., Johnston L.J., Ma X. Tryptophan (trp) modulates gut homeostasis via aryl hydrocarbon receptor (ahr) Crit Rev Food Sci Nutr. 2020;60:1760–1768. doi: 10.1080/10408398.2019.1598334. [DOI] [PubMed] [Google Scholar]
- Tremaroli V., Bäckhed F. Functional interactions between the gut microbiota and host metabolism. Nature. 2012;489:242–249. doi: 10.1038/nature11552. [DOI] [PubMed] [Google Scholar]
- Trevisi P., Corrent E., Mazzoni M., Messori S., Priori D., Gherpelli Y., Simongiovanni A., Bosi P. Effect of added dietary threonine on growth performance, health, immunity and gastrointestinal function of weaning pigs with differing genetic susceptibility to escherichia coli infection and challenged with e. Coli k88ac. J Anim Physiol Anim Nutr. 2015;99:511–520. doi: 10.1111/jpn.12216. [DOI] [PubMed] [Google Scholar]
- Venkatesh M., Mukherjee S., Wang H., Li H., Sun K., Benechet A.P., Qiu Z., Maher L., Redinbo M.R., Phillips R.S., et al. Symbiotic bacterial metabolites regulate gastrointestinal barrier function via the xenobiotic sensor pxr and toll-like receptor 4. Immunity. 2014;41:296–310. doi: 10.1016/j.immuni.2014.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikoff W.R., Anfora A.T., Liu J., Schultz P.G., Lesley S.A., Peters E.C., Siuzdak G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci USA. 2009;106:3698–3703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wlodarska M., Luo C., Kolde R., d'Hennezel E., Annand J.W., Heim C.E., Krastel P., Schmitt E.K., Omar A.S., Creasey E.A., et al. Indoleacrylic acid produced by commensal peptostreptococcus species suppresses inflammation. Cell Host Microbe. 2017;22:25–37. doi: 10.1016/j.chom.2017.06.007. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G.D., Chen J., Hoffmann C., Bittinger K., Chen Y.Y., Keilbaugh S.A., Bewtra M., Knights D., Walters W.A., Knight R., et al. Linking long-term dietary patterns with gut microbial enterotypes. Science (New York, NY) 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Ma N., Johnston L.J., Ma X. Dietary nutrients mediate intestinal host defense peptide expression. Advances in nutrition (Bethesda, Md) 2020;11:92–102. doi: 10.1093/advances/nmz057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang L., Moore B.S. Biochemical characterization of a prokaryotic phenylalanine ammonia lyase. J Bacteriol. 2005;187:4286–4289. doi: 10.1128/jb.187.12.4286-4289.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Q., Wu X., Pan Y., Wang L., Cui C., Guo Y., Zhu L., Peng J., Wei H. Early-life intervention using fecal microbiota combined with probiotics promotes gut microbiota maturation, regulates immune system development, and alleviates weaning stress in piglets. Int J Mol Sci. 2020;21 doi: 10.3390/ijms21020503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Chen X., Yu S., Su Y., Zhu W. Effects of early intervention with sodium butyrate on gut microbiota and the expression of inflammatory cytokines in neonatal piglets. PloS one. 2016;11 doi: 10.1371/journal.pone.0162461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W., Yu T., Huang X., Bilotta A.J., Xu L., Lu Y., Sun J., Pan F., Zhou J., Zhang W., et al. Intestinal microbiota-derived short-chain fatty acids regulation of immune cell il-22 production and gut immunity. Nat Commun. 2020;11:4457. doi: 10.1038/s41467-020-18262-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J., Han H., Li Y., Liu Z., Zhao Y., Fang R., Huang X., Zheng J., Ren W., Wu F., et al. Lysine restriction affects feed intake and amino acid metabolism via gut microbiome in piglets. Cell Physiol Biochem : international journal of experimental cellular physiology, biochemistry, and pharmacology. 2017;44:1749–1761. doi: 10.1159/000485782. [DOI] [PubMed] [Google Scholar]
- Yin J., Li Y., Han H., Liu Z., Zeng X., Li T., Yin Y. Long-term effects of lysine concentration on growth performance, intestinal microbiome, and metabolic profiles in a pig model. Food & function. 2018;9:4153–4163. doi: 10.1039/c8fo00973b. [DOI] [PubMed] [Google Scholar]
- Zelante T., Iannitti R.G., Cunha C., De Luca A., Giovannini G., Pieraccini G., Zecchi R., D'Angelo C., Massi-Benedetti C., Fallarino F., et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity. 2013;39:372–385. doi: 10.1016/j.immuni.2013.08.003. [DOI] [PubMed] [Google Scholar]
- Zhang F., Zheng W., Xue Y., Yao W. Suhuai suckling piglet hindgut microbiome-metabolome responses to different dietary copper levels. Appl Microbiol Biotechnol. 2019;103:853–868. doi: 10.1007/s00253-018-9533-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Fang L., Sun Y., Su Y., Zhu W. Effects of the dietary protein level on the microbial composition and metabolomic profile in the hindgut of the pig. Anaerobe. 2016;38:61–69. doi: 10.1016/j.anaerobe.2015.12.009. [DOI] [PubMed] [Google Scholar]