Abstract
The pygmy rabbit Brachylagus idahoensis (Merriam, 1891) is the smallest extant leporid, which naturally occurs in the Great Basin and adjacent areas in western parts of the United States of America. Its distribution is strongly associated with the sagebrush (Artemisia ssp.) vegetation. Here we present, for the first time, the complete mitochondrial genome of Brachylagus idahoensis, de novo assembled from Illumina short reads of fragmented probe-enriched DNA. The circular mitogenome is 17,021 bp in length and contains 13 protein-coding genes (PCGs), two ribosomal RNAs (16S rRNA and 12S rRNA), 22 transfer RNA genes, and a control region. The gene NAD6 and the tRNA(Gln), tRNA(Ala), tRNA(Asn), tRNA(Cys), tRNA(Tyr), tRNA(Ser), tRNA(Glu) and tRNA(Pro) are encoded on the light strand while the rest are encoded on the heavy strand. The overall nucleotide composition was 30.78% for A, 28.5% for T, 13.62% for G and 27.08% for C. The mitogenome data are available in the GenBank under the accession number OL436257.
Keywords: Mitogenome, Phylogenomics, De novo assembly, Lagomorpha, North America
Specifications Table
| Subject | Genomics |
| Specific subject area | Mitogenomics |
| Type of data | Mitogenome sequence data in FASTA file format, tables, mitogenome map in figure format (.jpg), phylogenetic tree in figure format (.jpg) Supplementary information (.zip) contains the following documents: S1: List of taxa and corresponding accession numbers used to design baits for target enrichment (.xls) S2: Assembled mitogenome sequence data in FASTA file format (.fasta) S3: Alignment file for phylogenetic analysis in PHYLIP file format (.phy) S4: Gene partition file for phylogenetic analysis in NEXUS file format (.nex) S5: Phylogenetic tree in NEWICK file format (.newick) |
| How the data were acquired | Target enrichment; Illumina NextSeq 550 high-throughput sequencing |
| Data format | Raw and analyzed |
| Description of data collection | Genomic DNA was extracted with Qiagen DNeasy Blood & Tissue Kit (Valencia, CA, USA); double-indexed and double-stranded library preparation; mitogenome enrichment with myBaits Custom 20–40 K (Daicel Arbor Biosciences, USA); sequencing: Illumina NextSeq 550 platform; mitogenome assembled de novo in NOVOPlasty v.4.3.1 and annotated in MITOS2 web server. The circular mitogenome map was drawn using OGDRAW. Phylogenetic relationships were inferred using IQ-TREE |
| Data source location | This individual was collected from Christmas Valley, Lake County, OR, USA and is preserved under the voucher number UWBM 82570 at the Burke Museum, University of Washington, WA, USA |
| Data accessibility | The mitogenome data are available in the GenBank under the accession number OL436257 (https://www.ncbi.nlm.nih.gov/nuccore/OL436257) and Mendeley data repository (http://dx.doi.org/10.17632/g799t3s5s9.1) [1]. Raw sequence data are available in Sequence Read Archive (BioProject: PRJNA839569, BioSample: SAMN28539370; http://www.ncbi.nlm.nih.gov/bioproject/839569) |
Value of the Data
-
•
The mitogenome will be useful in conservation studies of North American mammalian diversity.
-
•
The data will be useful for monitoring of potential changes in the range of the species.
-
•
The data will contribute to our knowledge on species diversification and the micro-evolution of extant leporids, in particular the North American Brachylagus–Sylvilagus complex.
-
•
The data generated will be useful in research on hybridization in Lagomorpha.
1. Data Description
The pygmy rabbit, Brachylagus idahoensis (Merriam, 1891) is the world's smallest leporid from North America, and belongs to a monotypic genus. This rabbit occurs in the Great Basin and adjacent intermontane areas from western Wyoming (easternmost), southwestern Oregon (westernmost), southwestern Montana (northernmost) to southwestern Utah (southernmost) [2,3]. The species is adapted to semiarid sagebrush habitat and it feeds mostly on the sagebrush (Artemisia ssp.), especially during winter. B. idahoensis is considered Least Concern (LC) on the IUCN Red List of Threatened Species [3]. It is the only true fossorial rabbit in North America, digging extended burrow systems [2]. The species is important for reconstruction of the lagomorph phylogeny and evolution, especially for the history of the divergence within the Leporidae family [4], as it shows an array of derived features in the skull and dentition. Furthermore, it is important species for landscape genetics [5] and climate dynamics as an indicator species. Its strong habitat dependence can be used to monitor the subtle climate changes, correlated with changes in humidity and thus, contraction or expansion of the pygmy rabbit habitats [6].
Here we report the first complete mitogenome of Brachylagus idahoensis, which is 17,021 bp in length (GenBank No. OL436257). The sequenced mitogenome data are summarized in Table 1. The mitogenome comprises two ribosomal RNAs (16S rRNA and 12-S rRNA), 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, and a noncoding control region (D-loop). The arrangement of these 37 genes encoded on either the heavy (H) or the light (L) strand is presented in Fig. 1. The total length of PCGs is 11,389 bp, transcribing 3796 amino acids, which accounts for 66.91% of the entire mitogenome. The gene NAD6 and the tRNA(Gln), tRNA(Ala), tRNA(Asn), tRNA(Cys), tRNA(Tyr), tRNA(Ser), tRNA(Glu) and tRNA(Pro) are encoded on the light strand while rest are encoded on the heavy strand (Fig. 1; Table 2). The overall nucleotide composition is estimated as 30.78% for A, 28.5% for T, 13.62% for G and 27.08% for C.
Table 1.
Mitogenome sequence data of Brachylagus idahoensis specimen (UWBM 82570).
| Total reads obtained | 363,630 |
| Total mapped reads | 46,294 |
| Mapped paired reads | 27,185 |
| Mean Coverage (x) | 266.0656 |
| Mean Mapping Quality | 58.98 |
Fig. 1.
Circular map of Brachylagus idahoensis mitochondrial genome. Genes encoded on the heavy (H) and light (L) strands represent the outer and inner circle, respectively. The direction of the arrows symbolises the transcription direction (both clockwise and anti-clockwise) of the genes. The inner ring (gray) indicates the GC content of the genome. The image of B. idahoensis is from Keinath & McGee [22].
Table 2.
Mitogenome features of Brachylagus idahoensis (GenBank accession number OL436257; BioProject: PRJNA839569; BioSample: SAMN28539370). Protein coding genes (PCGs) are represented in bold letters and the genes encoded on light (L) strand are italicized.
|
Position |
|||||
|---|---|---|---|---|---|
| Gene | Start | End | Size (bp) | Amino acid length | Strand |
| tRNA(Phe) | 1 | 70 | 70 | H | |
| 12S rRNA | 70 | 1029 | 960 | H | |
| tRNA(Val) | 1030 | 1095 | 66 | H | |
| 16S rRNA | 1096 | 2678 | 1583 | H | |
| tRNA(Leu) | 2678 | 2754 | 77 | H | |
| NAD1 | 2756 | 3710 | 955 | 318 | H |
| tRNA(Ile) | 3710 | 3780 | 71 | H | |
| tRNA(Gln) | 3777 | 3848 | 72 | L | |
| tRNA(Met) | 3856 | 3924 | 69 | H | |
| NAD2 | 3925 | 4968 | 1044 | 348 | H |
| tRNA(Trp) | 4975 | 5041 | 67 | H | |
| tRNA(Ala) | 5043 | 5109 | 67 | L | |
| tRNA(Asn) | 5110 | 5182 | 73 | L | |
| rep_origin | 5182 | 5224 | 43 | ||
| tRNA(Cys) | 5214 | 5281 | 68 | L | |
| tRNA(Tyr) | 5282 | 5347 | 66 | L | |
| COX1 | 5356 | 6897 | 1542 | 514 | H |
| tRNA(Ser) | 6900 | 6968 | 69 | L | |
| tRNA(Asp) | 6972 | 7040 | 69 | H | |
| COX2 | 7041 | 7724 | 684 | 228 | H |
| tRNA(Lys) | 7727 | 7799 | 73 | H | |
| ATP8 | 7800 | 8006 | 207 | 69 | H |
| ATP6 | 7961 | 8641 | 681 | 227 | H |
| COX3 | 8641 | 9424 | 784 | 261 | H |
| tRNA(Gly) | 9425 | 9495 | 71 | H | |
| NAD3 | 9495 | 9841 | 347 | 116 | H |
| tRNA(Arg) | 9842 | 9908 | 67 | H | |
| NAD4-L | 9910 | 10,206 | 297 | 99 | H |
| NAD4 | 10,200 | 11,577 | 1378 | 459 | H |
| tRNA(His) | 11,578 | 11,646 | 69 | H | |
| tRNA(Ser) | 11,647 | 11,705 | 59 | H | |
| tRNA(Leu) | 11,706 | 11,775 | 70 | H | |
| NAD5 | 11,776 | 13,584 | 1809 | 603 | H |
| NAD6 | 13,581 | 14,101 | 521 | 174 | L |
| tRNA(Glu) | 14,102 | 14,169 | 68 | L | |
| COB | 14,173 | 15,312 | 1140 | 380 | H |
| tRNA(Thr) | 15,312 | 15,377 | 66 | H | |
| tRNA(Pro) | 15,378 | 15,443 | 66 | L | |
| Control region | 15,444 | 17,021 | 1578 | ||
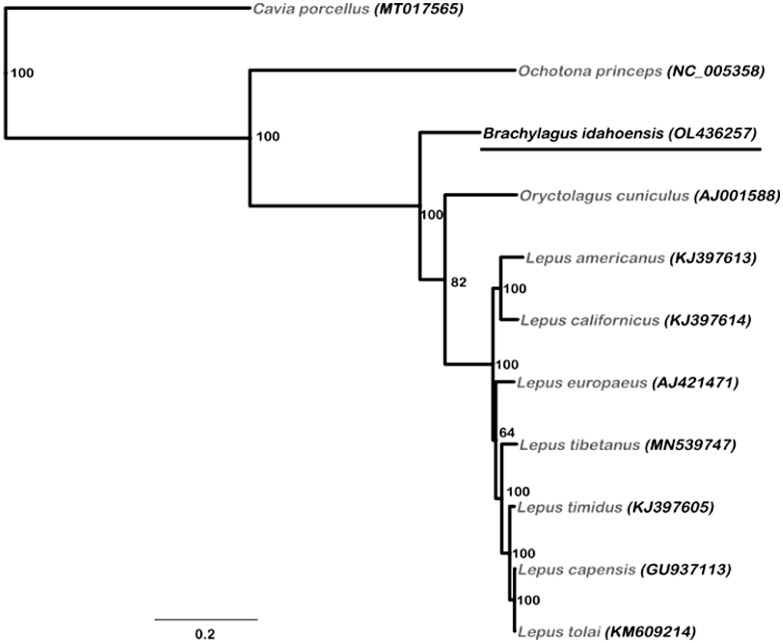
The phylogenetic inference is limited by the incomplete taxon sampling due to the unavailability of mitogenomes of other leporid species. Although the phylogenetic position of B. idahoensis appears resolved in the tree (Fig. 2; bootstrap value=100), the precise relationship within the Leporidae cannot be ascertained due to the many species lacking. However, the description of this novel mitogenome is a crucial input to lagomorph evolutionary studies.
Fig. 2.
Phylogeny of Brachylagus idahoensis constructed with nine lagomorph species (eight leporid and one ochotonid used as an outgroup) complete mitochondrial genomes using the maximum-likelihood (ML) method. Second outgroup is the guinea pig, a caviomorph rodent. Numbers to the right side of each node demonstrate the ML bootstrap support. The corresponding GenBank accession numbers are given after each taxon name. The GenBank accession number for the underlined taxon is generated in this study.
2. Experimental Design, Materials and Methods
2.1. Biological Sample
An individual of Brachylagus idahoensis was collected on August 23, 2011 in Christmas Valley, Lake County, OR, USA. It was preserved in 100% ethanol at the collection of the Burke Museum, University of Washington, WA, USA (UWBM 82570; tissue number JEB1781; https://www.burkemuseum.org/collections-and-research/biology/mammalogy/collections-database/search.php). A sample of muscle tissue was obtained by ZM and UO from the museum collection and transferred to AS for genomic analyzes. Total genomic DNA was extracted from the tissue with Qiagen DNeasy Blood & Tissue Kit (Cat#69-504; Valencia, CA, USA) following the manufacturer's protocol. The extracted DNA was subjected to spectrophotometric quantification, followed by fragmentation in M220 ultrasonicator (Covaris) and size-selection with magnetic beads (1.5X) to remove fragments <300 bp.
Next, an input of ∼200 ng size-selected DNA was used as a template for the preparation of double indexed, double-stranded DNA library based on the protocol of Meyer and Kircher [7]. The library (∼150 ng) was further enriched for the mitogenome following the manufacturer's protocol, using a total of 28,756 unique custom designed RNA probes with 3 × tilling that were based on mitogenomes of 71 species representing all five extant orders of Euarchontoglires, obtained from GenBank (Supplementary file S1.xls; list of species, taxonomic classification and corresponding accession numbers). The custom-designed probes were included in a myBaits Custom 20–40 K kit (Cat#300-248.V5 201119–92; Daicel Arbor Biosciences, USA). Libraries were pooled and sequenced on Illumina NextSeq 550 using 2 × 75 bp mode.
2.2. Complete Mitogenome Generation
Raw reads were demultiplexed using bcl2fastq (Illumina), adapters and low-quality bases were trimmed and overlapping reads were collapsed using AdapterRemoval v.2 [8]. The mitogenome was de novo assembled in NOVOPlasty v.4.3.1 [9] with default parameters and kmer value of 23 to reproduce the candidate mitogenome. The sequence assembled by NOVOPlasty was used as a reference for mapping using BWA-MEM [10]. Duplicates and reads with low mapping quality (mapQ<30) were removed using samtools [11]. Number of unique mapped reads, mean coverage and mapping quality of the assembled genome was estimated by Qualimap 2 [12], followed by a manual checkinTablet software [13]. The mitogenome was annotated on the MITOS2 web server ([14]; http://mitos2.bioinf.uni-leipzig.de/index.py). The annotations for individual PCG's start and stop codons were manually confirmed by nucleotide BLAST analysis. The circular mitogenome map was drawn using OGDRAW [15].
To reconstruct the mitogenomic phylogeny of available leporid species we used an alignment of eight closely related Leporidae taxa and two outgroup taxa (Cavia porcellus MT017565, a rodent, and the ochotonid lagomorph Ochotona princeps NC_005358, aligned with MAFFT [16] and subsequently removed ambiguously aligned sites in GBLOCKS [17,18]; http://molevol.cmima.csic.es/castresana/Gblocks_server.html). A Maximum-Likelihood (ML) based phylogeny (Fig. 2) was inferred from the resulting alignment (15,999 bp) on IQ-TREE web server ([19]; http://iqtree.cibiv.univie.ac.at/), simultaneously using the model selection (auto) algorithm [20] and ultrafast bootstrap method [21] for 1000 iterations.
Ethics Statement
This study is based on non-living animal individuals, the only tissue sample has been collected from a museum specimen. Therefore, no ethic statement is required.
CRediT authorship contribution statement
Anwesha Saha: Methodology, Software, Investigation, Formal analysis, Writing – original draft. Mateusz Baca: Methodology, Software, Writing – review & editing, Project administration. Danijela Popović: Methodology, Data curation. Zeinolabedin Mohammadi: Resources, Validation, Writing – review & editing. Urban Olsson: Resources, Writing – review & editing. Łucja Fostowicz-Frelik: Conceptualization, Visualization, Supervision, Funding acquisition, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work is supported by National Science Center (Cracow, Poland) Grant No. 2015/18/E/NZ8/00637. We are grateful to Sharon Birks and Jeff Bradley at the Burke Museum, University of Washington, WA, USA for granting access to the sample of tissue of B. idahoensis and to Sanjukta Chakravorti, Indian Statistical Institute, Kolkata, India for help with Fig. 1.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.dib.2022.108314.
Appendix. Supplementary materials
Data Availability
Brachylagus idahoensis complete mitogenome sequence (Original data) (GenBank).
References
- 1.Saha A., Baca M., Popović D., Mohammadi Z., Olsson U., Fostowicz-Frelik Ł. The first complete mitochondrial genome data of the pygmy rabbit Brachylagus idahoensis, the world's smallest leporid. Mendeley Data. 2021;1 doi: 10.17632/g799t3s5s9.1. [data] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Green J.S., Flinders J.T. Brachylagus idahoensis. Mamm. Species. 1980:1–4. https://academic.oup.com/mspecies/article/doi/10.2307/3503856/2600225 [Google Scholar]
- 3.IUCN: Brachylagus idahoensis: J. Rachlow, P.A. Becker, L. Shipley. The IUCN Red List of Threatened Species (2016): e.T2963A45176206. doi: 10.2305/IUCN.UK.2016-3.RLTS.T2963A45176206.en. Accessed December 9, 2021. [DOI]
- 4.Robinson T.J., Matthee C.A. Phylogeny and evolutionary origins of the Leporidae: a review of cytogenetics, molecular analyses and a supermatrix analysis. Mamm. Rev. 2005;35:231–247. doi: 10.1111/j.1365-2907.2005.00073.x. [DOI] [Google Scholar]
- 5.Byer N.W., Dilts T.E., Larrucea E.S., Crowell M.M., Shoemaker K.T., Weisberg P.J., Matocq M.D. Holocene-era landscape conditions affect genetic connectivity in a sagebrush obligate species, the pygmy rabbit (Brachylagus idahoensis) Landsc. Ecol. 2021;36:3575–3590. doi: 10.1007/s10980-021-01328-1. [DOI] [Google Scholar]
- 6.Riddle B.R., Jezkova T., Hornsby A.D., Matocq M.D. Assembling the modern Great Basin mammal biota: insights from molecular biogeography and the fossil record. J. Mammal. 2014;95:1107–1127. doi: 10.1644/14-MAMM-S-064. [DOI] [Google Scholar]
- 7.Meyer M., Kircher M. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb. Protoc. 2010 doi: 10.1101/pdb.prot5448. [DOI] [PubMed] [Google Scholar]
- 8.Schubert M., Lindgreen S., Orlando L. AdapterRemoval v2: rapid adapter trimming, identification, and read merging. BMC Res. Notes. 2016;9:1–7. doi: 10.1186/s13104-016-1900-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dierckxsens N., Mardulyn P., Smits G. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2017;45:e18. doi: 10.1093/nar/gkw955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li H., Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. 1000 Genome project data processing subgroup, the sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Okonechnikov K., Conesa A., García-Alcalde F. Qualimap 2: advanced multi-sample quality control for high-throughput sequencing data. Bioinformatics. 2015;32:1–3. doi: 10.1093/bioinformatics/btv566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Milne I., Bayer M., Cardle L., Shaw P., Stephen G., Wright F., Marshall D. Tablet-next generation sequence assembly visualization. Bioinformatics. 2009;26:401–402. doi: 10.1093/bioinformatics/btp666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Donath A., Jühling F., Al-Arab M., Bernhart S.H., Reinhardt F., Stadler P.F., Middendorf M., Bernt M. Improved annotation of protein-coding genes boundaries in metazoan mitochondrial genomes. Nucleic Acids Res. 2019;47:10543–10552. doi: 10.1093/nar/gkz833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greiner S., Lehwark P., Bock R. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019;47:W59–W64. doi: 10.1093/nar/gkz238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakamura T., Yamada K.D., Tomii K., Katoh K. Parallelization of MAFFT for large-scale multiple sequence alignments. Bioinformatics. 2018;34:2490–2492. doi: 10.1093/bioinformatics/bty121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 18.Talavera G., Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007;56:564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- 19.Trifinopoulos J., Nguyen L.T., von Haeseler A., Minh B.Q. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016;44:232–235. doi: 10.1093/nar/gkw256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kalyaanamoorthy S., Minh B.Q., Wong T.K.F., Von Haeseler A., Jermiin L.S. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoang D.T., Chernomor O., Von Haeseler A., Minh B.Q., Vinh L.S. UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2018;35:518–522. doi: 10.1093/molbev/msx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keinath D.A., McGee M. United States Department of the Interior Bureau of Land Management Wyoming State Office Cheyenne; Wyoming: 2004. Species Assessment for Pygmy Rabbit (Brachylagus Idahoensis) in Wyoming.https://www.uwyo.edu/wyndd/_files/docs/reports/speciesassessments/pygmyrabbit-mar2004.pdf. Accessed December 9, 2021. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Brachylagus idahoensis complete mitogenome sequence (Original data) (GenBank).