Abstract
We report the use of a fusion to the green fluorescent protein to visualize the assembly of the morphogenetic protein SpoIVA around the developing forespore during the process of sporulation in the bacterium Bacillus subtilis. Using a deconvolution algorithm to process digitally-collected optical sections, we show that SpoIVA, which is synthesized in the mother cell chamber of the sporangium, assembled into a spherical shell around the outer surface of the forespore. Time-lapse fluorescence microscopy showed that this assembly process commenced at the time of polar division and seemed to continue after engulfment of the forespore was complete. SpoIVA remained present throughout the late stages of morphogenesis and was present as a component of the fully mature spore. Evidence indicates that assembly of SpoIVA depended on the extreme C-terminal region of the protein and an additional region that directly or indirectly facilitated interaction among SpoIVA molecules. The N- and C-terminal regions of SpoIVA, including the extreme C terminus, are highly similar to the corresponding regions of the homologous protein from the distantly related endospore-forming bacterium Clostridium acetobutylicum, attesting to their importance in the function of the protein. Finally, we show that proper localization of SpoIVA required the expression of one or more genes which, like spoIVA, are under the control of the mother cell transcription factor ςE. One such gene was spoVM, whose product was required for efficient targeting of SpoIVA to the outer surface of the forespore.
Despite their small size and apparent simplicity, bacteria are able to form an impressive array of complex morphological structures, such as the flagellum, the chemoreceptor complex, and the cell division septasome (38). An underlying mechanism that is critical for the assembly of these structures is the initial subcellular localization of one or more morphogenetic proteins to a particular site within the cell. For example, in the predivisional cell of the dimorphic bacterium Caulobacter crescentus, monomers of the flagellar protein FliF are targeted to the pole of the incipient swarmer cell, where they assemble to form the MS ring (18). The MS ring then serves as the foundation for the construction of a flagellum, the locomotory apparatus that enables the swarmer cell to swim (44). In Escherichia coli, the chemoreceptor MCP (methyl-accepting chemotaxis protein) and CheW are targeted to the poles of the cell, where they form a ternary complex with CheA, a cytoplasmic histidine kinase (24). CheA transmits signals from the MCP to downstream members of the chemotaxis system, enabling the cell to respond to environmental stimuli (15, 28). Finally, in the case of the septasome, the tubulin-like protein FtsZ, which specifies the nascent division site, polymerizes into a ring at the center of the cell, where it recruits an array of other cell division proteins that mediate the process of cytokinesis (2, 3, 11, 25, 27). Here we are concerned with the subcellular localization of the sporulation protein SpoIVA (6, 29, 34, 40), a morphogenetic protein in Bacillus subtilis that is responsible for the proper assembly of the outer layers of the spore.
Sporulation in B. subtilis involves a modification of the process of cell division, in which the site of septum formation switches from the midcell position (as is characteristic of binary fission during vegetative growth) to an extreme polar position of the developing cell (the sporangium) (42). The formation of an asymmetrically-positioned septum divides the sporangium into a large compartment called the mother cell and a small compartment known as the forespore (or the prespore). Initially, the mother cell and the forespore lie side by side, but in the next stage of development, the ends of the septal membrane migrate around and thereby engulf the forespore. Eventually, the forespore is released into the mother cell cytoplasm as a free protoplast encircled by two membranes, an inner membrane that corresponds to the cytoplasmic membrane of the forespore and an outer membrane layer that originated from the enveloping mother cell membrane. During subsequent morphogenesis, a thick layer of peptidoglycan known as the cortex is formed in the space between the inner and outer membranes. Meanwhile, the outer membrane becomes encased in a thick protein shell known as the coat. The coat is composed of an electron-dense outer layer (the outer coat) and a lamellate inner layer (the inner coat). The protein components of the coat are produced in the mother cell and deposited around the outside surface of the developing forespore. When fully ripened after about 7 h of development, the mature spore is released from the sporangium by lysis of the mother cell.
The SpoIVA protein plays a central role in the proper formation of both the cortex and the coat. Sporulating cells of a spoIVA null mutant fail to synthesize a cortex, and they produce a mislocalized coat (29, 34). The mislocalized coat exhibits the electron-dense and lamellate layers characteristic of a normal coat, but the coat is misassembled as swirls within the mother cell rather than being deposited on the outside surface of the forespore. SpoIVA is synthesized in the mother cell under the direct control of the mother cell transcription factor ςE (34, 40). Previous immunoelectron and immunofluorescence microscopy experiments with antibodies against SpoIVA have shown that SpoIVA localizes to the mother cell membrane that surrounds the forespore (10, 30). SpoIVA is thus appropriately positioned both to promote formation of the cortex, which is produced just underneath the membrane, and to target assembly of the coat to the region around the forespore.
Because of the central role of SpoIVA in morphogenesis, we sought to extend our understanding of the subcellular localization and assembly of the sporulation protein and to investigate the basis for its distinctive pattern of targeting to the outer surface of the forespore. Here we report the use of a fusion to the green fluorescent protein (GFP) (5, 43) to visualize SpoIVA in living cells. In conjunction with deconvolution and time-lapse microscopy, we show that the sporulation protein assembles into a spherical structure around the forespore and that this assembly process progresses from the accumulation of SpoIVA on one side of the forespore through full encasement of the forespore by the morphogenetic protein. We also show that subcellular localization is dependent upon an amino acid sequence at the extreme C terminus of SpoIVA and upon an additional sporulation protein (SpoVM [21]) that is also produced in the mother cell under the control of ςE.
MATERIALS AND METHODS
General methods.
The construction of B. subtilis strains was carried out as described previously (8). Except as indicated, plasmid cloning and phage vectors were built according to methods outlined in Sambrook et al. (35). Drug-resistant strains were selected on Luria-Bertani agar containing chloramphenicol (5 μg/ml), neomycin (3 μl/ml), spectinomycin (100 μg/ml), kanamycin (5 μg/ml), a combination of 1 μg of erythromycin per ml and 25 μg of lincomycin per ml, or ampicillin (100 μg/ml).
Strains.
The B. subtilis strains used in this study are listed in Table 1. MO1708 and MO1433 are congenic derivatives of JH642 (9). The genetic backgrounds of 91 and 85.3 are not known. All other B. subtilis strains are congenic with the prototrophic strain PY79 (46).
TABLE 1.
Strains used in this study
| Strain | Genotype or description | Source or reference |
|---|---|---|
| 85.3 | spoVE85 trpC2 | 12 |
| 91 | spoVB91 trpC2 | 12 |
| BK332 | spoVD156 | This work |
| BSL51 | spoIVFΔAB::cat | 23 |
| EL200 | spoVMΔ::spc | 7 |
| KP73 | spoIVAΔ::neo2 | This work |
| KP163 | spoIVAΔ::neo2ΩpKP162 | This work |
| KP349 | spoIVAΔ::neo2ΩpKP343 | This work |
| KP373 | spoIVAΔ::neo2ΩpKP372 | This work |
| KP429 | amyE::gfp-spoIVA | This work |
| KP461 | amyE::gfp-spoIVA spoIVCB::Tn917ΩHU215 | This work |
| KP466 | spoIVAΔ::neo2ΩpKP464 | This work |
| KP505 | amyE::(spoIVA+ spc) spoIVAΔ::neo2ΩpKP464 spoIVCB::Tn917ΩHU215 | This work |
| KP581 | spoIVAΔ::spcΩpKP338 | This work |
| KP591 | spoIVAΔ::spcΩpKP338 spoIIIGΔ::neo | This work |
| KP592 | spoIVAΔ::spcΩpKP338 spoIIGBΔ::erm | This work |
| KP593 | spoIVAΔ::spcΩpKP338 spoIVCB::Tn917ΩHU215 | This work |
| KP598 | spoIVAΔ::spcΩpKP338 | This work |
| KP599 | spoIVAΔ::neo2ΩpKP588 | This work |
| KP612 | spoVR::kan | This work |
| KP635 | amyE::gfp-spoIVA spoVMΔ::spc | This work |
| KS287 | spoIIM::Tn917ΩHU287 | 37 |
| MO1433 | spoIIIAΔ::erm | P. Stragier |
| MO1708 | spoIIP::kan | P. Stragier |
Strain constructions.
BK332 was constructed by the congression of chromosomal DNA from 1S45 (Bacillus Genetic Stock Center) into strain PY78 (36). To construct KP73, pSR54 was linearized and introduced into PY79 by transformation by selecting for neomycin resistance and screening for sensitivity to chloramphenicol, indicating that the plasmid had integrated via double-crossover (marker replacement) recombination. (pSR54, a plasmid containing a neomycin resistance marker flanked by sequences from upstream and downstream of the spoIVA open reading frame, was constructed by first digesting pSR28 [34] with Tth111I and AccI and subsequently rendering the ends flush with T4 DNA polymerase. The 1,362-bp SmaI-SmaI fragment of pBEST501 [16] was then cloned into pSR28 such that the orientation of the gene conferring resistance to neomycin was opposite that of the spoIVA transcriptional unit.)
KP163 was constructed in a series of steps. First, the BamHI-KpnI fragment harboring the erm gene of pUC18-ERM (19) was rendered flush and cloned into filled EcoO109I-digested pLACZ1 (17) to yield pKP49. Next, the HindIII-AgeI fragment of pSR28 was rendered flush and cloned into filled EcoRI-digested pKP49 to generate pKP58. The PstI-DraI fragment of pLACZ3 (17) was ligated to the DraI-PstI-digested backbone of pKP58 to generate pKP111. Subsequently, the 841-bp SphI-KpnI fragment of pKP58 was cloned into similarly-digested M13mp18 (45) to generate KPRF1. The Amersham oligonucleotide-mediated site-directed mutagenesis system with OL339 (5′-ATCATCCTGTTATACCGGAAT-3′) was then used to convert the middle nucleotide of the spoIVA stop codon from an adenosine to a thymidine residue, generating KPRF2. The NruI-KpnI fragment of KPRF2 was cloned into similarly-digested pKP111, yielding KP126. Next, the PmeI-PstI fragment of pKP126 was ligated to the PstI-SmaI-digested backbone of pCW44 (harboring gfp) (32) to create pKP154. The HindIII-SpeI fragment of pKP154 was then ligated to the HindIII-XbaI backbone of pER19 (33) to generate pKP156. Finally, pKP156 was digested with PstI, trimmed, and religated. The resulting plasmid, pKP162, which contains spoIVA fused at its 3′ terminus to gfp via a polylinker domain, was integrated into KP73 by single-crossover (Campbell-type) reciprocal recombination to generate KP163.
KP349 was created by first subcloning the HindIII-XbaI fragment of pSR28 (34) into similarly-digested pER19 to generate pKP297. OL137 (5′-TGCGCCAGGATTTAAGCG-3′) and OL487 (5′-GTACCGGTTTATAAGCCGCCAGAGCCTTCGTT-3′) were used to PCR amplify pSR28. Adenosine residues were added to the ends of the resulting approximately-850-bp fragment before it was ligated to pCR2.1 (Invitrogen), a TA cloning vector, to generate pKP328. The BlpI-AgeI fragment of pKP328 was then cloned into the AgeI-BlpI backbone of pKP297 to create pKP343, a plasmid containing spoIVA missing five codons from the 3′ terminus of its open reading frame. pKP343 was integrated via Campbell-type reciprocal recombination into KP73 to create KP349.
Using the Amersham oligo-mediated site-directed mutagenesis kit and OL377 (5′-AATATCGACCTTTTCACTAGTATGCATCAAGTGATCCCCTC-3′), we inserted a 12-bp fragment containing the NsiI and SpeI restriction enzyme sites immediately downstream of the spoIVA start codon harbored in SRRF49, yielding KPRF3. The Tth111I-ApaI fragment of KPRF3 was cloned into similarly-digested pKP126 to create pKP160. The PmeI-SacII fragment of pKP160 was then cloned into the SacII-PmeI backbone of pKP297, yielding pKP333. OL527 (5′-ATGCATATGAGTAAAGGAGAAGAACTTTTCACTG-3′) and OL528 (5′-ACTAGTGCAGCCGGATCCTTTGTATAG-3′), containing NsiI and SpeI sites, respectively, were used to PCR amplify a fragment containing gfp from pCW44. Adenosine residues were added to the ends of this fragment, which was subsequently cloned into pCR2.1 to generate pKP357. The NsiI-SpeI fragment of pKP357 containing gfp was ligated into similarly-digested pKP333 to create pKP372, a plasmid containing gfp fused to the N terminus of spoIVA (gfp-spoIVA). pKP372 was integrated into KP73 via Campbell-type reciprocal recombination to generate KP373.
To construct KP429, the HindIII-BamHI fragment of pKP372 was cloned into the HindIII-BamHI backbone of pDG364 (8) to generate pKP427, a plasmid containing gfp-spoIVA at amyE. pKP427 was linearized with PstI and BglII and used to transform PY79 to chloramphenicol resistance.
To generate KP461, KS215 (37) chromosomal DNA was transformed into KP429 by selecting for MLS resistance.
The HindIII-SphI fragment pKP372, containing gfp, was cloned into the SphI-HindIII-digested backbone of pKP343 to create pKP464, a plasmid harboring gfp-spoIVA missing five codons from the 3′ terminus of the spoIVA open reading frame. pKP464 was integrated into KP73 via Campbell-type reciprocal recombination to create KP466.
To create KP505, the BamHI-HindIII fragment of pSR28 was first cloned into HindIII-BamHI-digested pLD30 (13). The resulting plasmid, pKP477, containing a wild-type copy of spoIVA and a spectinomycin resistance marker at amyE, was linearized and transformed into PY79 via double-crossover reciprocal recombination to create KP479. KP479 chromosomal DNA was used to transform KP466 to spectinomycin resistance, yielding the strain KP494. KP505 was created by transforming KS215 chromosomal DNA into KP494 and selecting for MLS resistance.
SDMOL1 (5′-GCTGGCGAAAGGGGGATGTG-3′) and OL508 (5′-ATGCATGTCAGTAACTTCCACAGTAGTTCA-3′) were used to PCR amplify an approximately-340-bp fragment from pSM79-1, a pBluescript (Stratagene)-derived vector harboring spoVG. Adenosine residues were added to the ends of this fragment before it was cloned into pCR2.1 to create pKP332. Next the HindIII-NsiI fragment of pKP332 was cloned into NsiI-HindIII-digested pKP333 to generate pKP338, a plasmid containing spoIVA under the control of the spoVG promoter. The EcoRI-SpeI fragment of pJL74 (20) was trimmed and cloned into Tth111I-AgeI-digested pKP297 to generate pKP551, which contains a spectinomycin resistance marker flanked by sequences from upstream and downstream of spoIVA. pKP551 was then linearized and used to transform KP73 to spectinomycin resistance, yielding KP559. This strain was sensitive to neomycin, indicating that integration had occurred via double-crossover reciprocal recombination. To create KP581, pKP338 was used to transform KP559 to chloramphenicol resistance via a Campbell-type reciprocal recombination event.
Chromosomal DNA from strains PM357 (26), EU8701 (19), KS215, and KJP324 (31) was used to transform KP581 to drug resistance, yielding the strains KP591, KP592, KP593, and KP598, respectively. (KJP324 was constructed by transforming MO2235 [spoIIAC::kan] chromosomal DNA into PY79. PM357 is a strain in which the internal HindIII-BamHI fragment of spoIIIG, encoding ςG, has been replaced with the HindIII-BamHI fragment of pBEST502 [16].)
Oligonucleotide site-directed mutagenesis with OL626 (5′-AAGGATCCGGCTGCCTGCAGACTAGTGAAAAGGT-3′) and OL627 (5′-ACCTTTTCACTAGTCTGCAGGCAGCCGGATCCTT-3′) was used to insert a PstI site immediately downstream of the SpeI site of pKP372. This procedure led to the generation of KP507. pKP507 was then digested with PstI and NgoMI and trimmed and religated to create pKP588, a plasmid containing gfp-spoIVA missing the first 64 codons of the spoIVA open reading frame. pKP588 was used to transform KP73 to chloramphenicol resistance via Campbell-type reciprocal recombination, yielding KP599.
KP612 was constructed by transforming chromosomal DNA from AH832 (1) into PY79 and selecting for kanamycin resistance.
To construct KP635, EL200 chromosomal DNA was used to transform KP429 to spectinomycin resistance.
Growth conditions.
All strains harboring a gfp fusion were inoculated as a single colony into 1 ml of liquid Difco sporulation media (DSM) (8) and sporulated overnight at 37°C. KP599 was additionally sporulated overnight in DSM at 25°C. KP461 was sporulated by the resuspension method (39) to obtain cells for time-lapse imaging. Cells used for immunofluorescence imaging and Western blot analysis were sporulated by the resuspension method, and samples of these cells were taken at hour 4 of sporulation.
Immunofluorescence microscopy.
Samples were prepared for immunofluorescence microscopy as described previously (30) with a few modifications. Cells were fixed in 2.7% paraformaldehyde and 0.008% glutaraldehyde. The fixed and washed cells were subjected to lysozyme treatment for 4 min. They were not treated with methanol or acetone. Cells were treated overnight with a 1:2,500 dilution of anti-SpoIVA antibodies and then treated for 2 h with a 1:100 dilution of anti-rabbit immunoglobulin G fluorescein-conjugated secondary antibodies.
Microscopy and photography.
Cells were observed and photographed by using an Olympus BX 60 microscope equipped with a Micromax (Princeton Instruments, Trenton, N.J.)-cooled charge-coupled device camera driven by the METAMORPH software package (version 3.0; Universal Imaging, Media, Pa.). All images were taken by using the UPlan Fluorite phase-contrast objective (magnification, ×100; numerical aperture, 1.3). The images presented in Fig. 1 and 3 were obtained by overlapping two separate images; either phase-contrast and GFP or propidium iodide (PI)- and fluorescein-stained images were combined by using the ARITHMETIC command of METAMORPH. Phase-contrast images and images of GFP were taken with an excitation cube unit (U-MWIB; Olympus) with a wide-band-pass (460- to 490-nm) excitation filter and a long-pass (515-nm) barrier filter. PI staining and fluorescein staining were visualized as described previously (14).
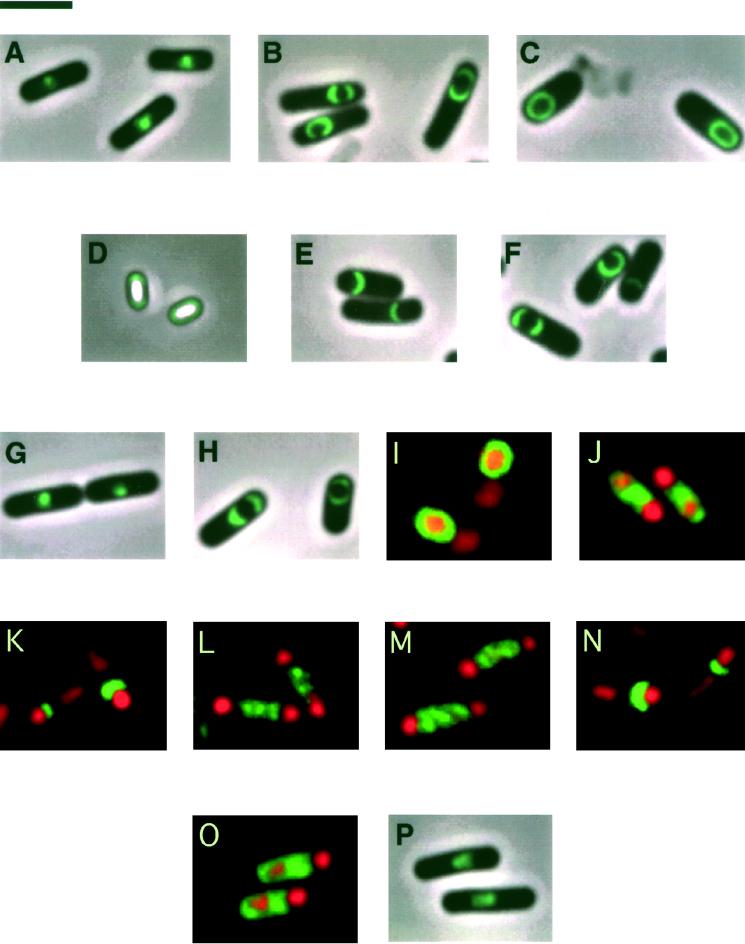
FIG. 1.
Visualizing SpoIVA and SpoIVA fused to GFP by fluorescence microscopy. (A) Sporangia of strain KP163 synthesizing GFP fused to the C terminus of SpoIVA (SpoIVA-GFP). SpoIVA-GFP is mislocalized as a bright spot in the mother cell cytoplasm. (B and C) Cells harboring a fusion of GFP to the N terminus of SpoIVA (GFP-SpoIVA) in the absence of wild-type SpoIVA (strain KP373) (B) and the presence of wild-type SpoIVA (strain KP461) (C). Note that the pattern of GFP-SpoIVA localization improves in the presence of a wild-type copy of SpoIVA. (D) Visualization of GFP-SpoIVA near the periphery of spores produced by a strain that synthesizes both GFP-SpoIVA and wild-type SpoIVA (strain KP429). (E and F) Sporangia of strain KP599, which synthesizes GFP-SpoIVA65–492, that had been sporulated at 37°C (E) or 25°C (F). (G and H) Sporangia from strains that synthesize GFP-SpoIVA1–487 in the absence (strain KP466) (G) or presence (strain KP505) (H) of wild-type SpoIVA. The fusion protein mislocalizes when synthesized in the absence of wild-type SpoIVA but assembles as caps around the forespore in the presence of wild-type SpoIVA. (I to O) Immunofluorescence microscopy of cells at 4 h of sporulation stained to visualize SpoIVA (green) and the mother cell and forespore chromosomes (red). (I) Localization of SpoIVA around the forespore in wild-type sporangia of strain PY79. (J) Failure of SpoIVA1–487 to localize to the forespore in sporangia of strain KP349; fluorescence is instead distributed throughout the mother cell cytoplasm. (K to N) Sporangia producing SpoIVA exclusively under the control of the spoVG promoter in a strain that is otherwise wild type (strain KP581) (K) or mutant for the transcription factors ςF (strain KP598) (L), ςE (strain KP592) (M), or ςG (strain KP591) (N). SpoIVA partially envelopes the forespore in the sporangia of K and N but not those of L and M. (O and P) spoVM mutant strains. (O) Mislocalization of SpoIVA throughout the mother cell cytoplasm in sporangia from a spoVM mutant (strain KP626). (P) Mislocalization of GFP-SpoIVA as a bright spot in the mother cell cytoplasm in sporangia from a spoVM mutant that synthesizes both GFP-SpoIVA and wild-type SpoIVA (strain KP635). Bar, 3 μm.
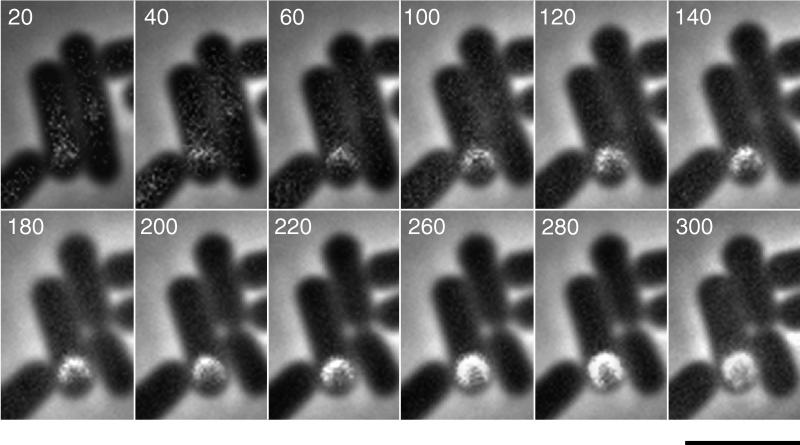
FIG. 3.
Time-lapse photography of GFP-SpoIVA assembly. A sample of cells was placed on agarose 2 h after suspension in Sterlini-Mandelstam (39) medium, and images were acquired beginning shortly thereafter at intervals of 20 min. Numbers in the upper left of each panel represent the time in minutes after the first image was acquired. Bar, 3 μm.
For three-dimensional imaging, a piezoelectric device (Physik Instruments, Auburn, Mass.) was used to photograph a series of sections that were spaced at 0.05-μm intervals along the z axis of a cell of strain KP461. The images were then subjected to deconvolution by using an EXHAUSTIVE PHOTON REASSIGNMENT program (Scanalytics, Billerica, Mass.). METAMORPH was used to obtain a three-dimensional reconstruction of the image stack.
To perform time-lapse microscopy, cells that had been sporulating for 2 h in resuspension media were placed under a coverslip on a bed of resuspension medium containing 1% agarose. To keep the sample hydrated, liquid resuspension medium was added approximately every half an hour to small pieces of tissue paper that were placed on either side of the agarose bed. Phase-contrast and fluorescence images of the sample were taken every 20 min over the course of 5 h by using the ACQUIRE TIMELAPSE command of METAMORPH.
The images were prepared for final publication by using Adobe Systems (Mountain View, Calif.) PHOTOSHOP 3.0 or PHOTOSHOP 4.0, CANVAS 5.0 (Deneba, Miami, Fla.), and a Tektronix Phaser 450 printer.
Western blot analysis.
Whole cell extracts were prepared from 1-ml samples. The cells were lysed in 48 μl of lysis buffer (40 mM Tris-HCl [pH 8.0], 10 mM EDTA, 8% sucrose) containing 12 μl of 7.5 mg of lysozyme per ml. After incubation at 37°C for 20 min, 20 μl of 4× sodium dodecyl sulfate (SDS) sample buffer and 8 μl of 1 M dithiothreitol were added to each sample. The samples were then boiled for 5 min, vortexed, and subsequently boiled again for 5 min before being loaded onto an SDS–8% polyacrylamide gel. After separation by electrophoresis, the extracts were electroblotted to an Immobilon-P membrane (Millipore). The blot was subsequently blocked in TBST (10 mM Tris-HCl [pH 8.0], 150 mM NaCl, 0.05% Tween 20) and 1.5% bovine serum albumin overnight at room temperature, with shaking. Immunodetection of SpoIVA was achieved by first incubating the blot in a 1:5,000 dilution of anti-SpoIVA antibodies in TBST and then using a colorimetric detection system (PhotoBlot Western blot AP system; Promega).
RESULTS
Use of a fusion to GFP to visualize SpoIVA.
Previous work by immunofluorescence microscopy and immunoelectron microscopy showed that SpoIVA localizes as a ring at the mother cell membrane that surrounds the forespore (10, 30). To visualize SpoIVA in living cells, we created a fusion to GFP. Initially, we joined GFP at the C terminus of SpoIVA (SpoIVA-GFP), but such a fusion protein did not localize properly. Instead, it coalesced as a bright spot next to the forespore in the mother cell cytoplasm (Fig. 1A). Next, we constructed a fusion of GFP to the amino terminus of SpoIVA by cloning the gene (gfp) for GFP in frame between the first and second codons of spoIVA. The resulting gene fusion contained the spoIVA promoter and ribosome-binding site, followed by a fusion of gfp near the beginning of the spoIVA open reading frame. Cells harboring gfp-spoIVA in place of the wild-type gene reached the stage of engulfment but did not progress further into sporulation, indicating that the fusion protein was not fully functional. Nevertheless, the fusion protein retained the capacity to localize to the forespore. Consistent with results obtained by immunoelectron and immunofluorescence microscopy (10, 30), GFP-SpoIVA localized to the region around the mother cell membrane that surrounds the forespore, preferentially accumulating as caps on either side of the forespore (Fig. 1B).
The pattern of subcellular localization was sharpened significantly by the use of merodiploid cells that harbored gfp-spoIVA as well as a wild-type copy of spoIVA. Because cells producing GFP-SpoIVA as well as unmodified SpoIVA were sporulation proficient, we carried out this analysis with a merodiploid strain (KP461) that additionally harbored a mutation in the sporulation gene spoIVCB (41). The purpose of the spoIVCB mutation was to block sporulation at the stage (IV) of cortex formation and thereby prevent the maturing forespores from becoming refractile, which interferes with the visualization of GFP. In cells producing both GFP-SpoIVA and unmodified SpoIVA, the fusion protein formed a strikingly sharp, contoured ring around the forespore (Fig. 1C). The simplest interpretation of these results is that the assembly of SpoIVA involves (direct or indirect) interactions between molecules of the sporulation protein and that the presence of the wild-type protein in merodiploid cells facilitated the localization and assembly of the fusion protein. See the cover of this issue for a fluorescence image of a field of sporangia from a GFP-SpoIVA-producing merodiploid strain.
Because SpoIVA plays a central role in coat assembly during sporulation, we wondered whether the morphogenetic protein persists during the late stages of spore maturation and is present in the fully ripened spore. In previous work, Driks et al. (10) failed to detect SpoIVA in mature spores by immunoelectron microscopy, but this technique is relatively insensitive. Indeed, using fluorescence microscopy and the GFP-SpoIVA fusion, we observed a distinct halo of green fluorescence at the periphery of spores from a merodiploid strain (KP429) that harbored both gfp-spoIVA and a wild-type copy of spoIVA (Fig. 1D). A caveat in the interpretation of this experiment is that SpoIVA may not ordinarily be present in mature spores and that either the presence of an N-terminal extension caused the fusion protein to persist aberrantly during morphogenesis or the GFP moiety of the fusion protein remained intact and visible after SpoIVA had been degraded.
Use of deconvolution microscopy to visualize GFP-SpoIVA in three dimensions.
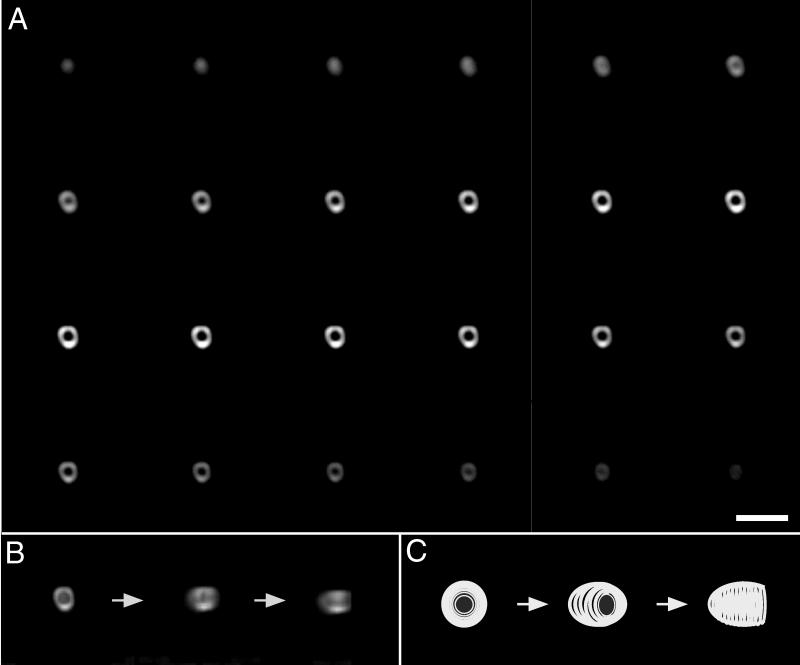
Conventional fluorescence microscopy allowed us to visualize SpoIVA as a two-dimensional ring around the mother cell membrane that surrounds the forespore. Presumably, however, SpoIVA forms a shell around the forespore and thus should be arranged in a spherical shape. To attempt to visualize the localization of SpoIVA in three dimensions, we collected a series of optical sections through a sporangium of the GFP-SpoIVA-producing, merodiploid strain KP461. Digital images were collected in increments of 0.05 μm beginning at a distance of approximately 1.8 μm above the focal point of the sporangium and proceeding for a total distance of 3 μm. We then processed the images with a deconvolution algorithm to improve the resolution by removing out-of-focus light and reassigning it to its correct points of origin (4, 25) (Fig. 2A). The first image of the series is of a disc, representing a plane at the top of the forespore. In subsequent images of slices from the middle of the stack, SpoIVA appears as a ring. At the bottom of the stack, SpoIVA can be visualized once again as a disc. We combined the images to construct a three-dimensional view of SpoIVA that could be rotated by using imaging software (Fig. 2B). Because more images were obtained on one side of the central plane than on the other in the series of Fig. 2A, the stack appears as a cup. In addition, the shape of the stack resembles a football more than a sphere, due to z axis elongation, a phenomenon that inherently extends the width of optical sections. As the stack is being rotated in the images in Fig. 2B, initially the cup appears on its side, with its rim facing the viewer. After an approximately-45°-counterclockwise rotation, the rim of the cup faces slightly to the right of the viewer. A further 45°-counterclockwise rotation brings the rim around such that it is facing to the viewer’s right. We conclude from these findings that SpoIVA assembles into a sphere-like shell around the forespore.
FIG. 2.
Three-dimensional reconstruction of GFP-SpoIVA assembled around the forespore. (A) Fluorescence images were collected at regularly spaced intervals (0.05 μm) along the z axis for a sporangium (strain KP461) in which GFP-SpoIVA was localized around the forespore. In the upper left of the panel, a section from the top of the stack shows GFP-SpoIVA as a disc. Depicted across the figure from left to right and top to bottom are serial images taken as the plane of focus was moved down through the sporangium. GFP-SpoIVA appears as a ring in slices close to the middle of the stack. At the bottom of the stack, GFP-SpoIVA appears again as a disc. Note that more sections were collected above the central plane than below. (B) Rotation of a three-dimensional reconstruction of localized GFP-SpoIVA. The optical sections depicted in panel A were subjected to deconvolution and combined to generate a three-dimensional image of GFP-SpoIVA as a football-shaped cup. In the first of three images, the cup of GFP-SpoIVA can be seen with its rim facing the viewer. To obtain the second image, the cup was rotated on its z axis 45° counterclockwise. Finally, to generate the third image, the cup was rotated on its z axis an additional 45° counterclockwise such that the rim of the cup faced to the viewer’s right. (C) Cartoon of events depicted in panel B. Bar, 3 μm.
Use of time-lapse microscopy to visualize the assembly of GFP-SpoIVA.
Next, we used time-lapse photography to visualize the assembly of SpoIVA around the forespore during the course of sporulation. For this experiment, cells of the merodiploid strain harboring spoIVCB were suspended in sporulation medium at 37°C (39). After 2 h, the cells were placed on a bed of agarose containing sporulation medium, covered with a coverslip, and incubated at room temperature (∼23°C). As sporulation continued, images were collected of a single cell at intervals of 20 min for 5 h. A subset of those images is presented in Fig. 3. Twenty minutes after placement on agarose, SpoIVA was visualized as a region of weak fluorescence next to the forespore. During the next 40 min the fluorescence became brighter and appeared as a crescent on the mother cell side of the forespore. From 60 min to about 280 min the crescent became progressively more defined as SpoIVA migrated around the forespore. Finally, by 300 min, the assembly process was completed and SpoIVA could be seen completely encircling the forespore. For comparison, under the same conditions most cells undergoing sporulation had completed engulfment by 120 min of incubation on agarose (data not shown).
Identification of a region of SpoIVA important for its localization.
To identify regions of SpoIVA that are important for its localization, we constructed two gfp-spoIVA fusions truncated at the 5′ or 3′ end of spoIVA open reading frame. One such deletion mutant specified a truncated fusion protein, GFP-SpoIVA65–492, lacking the first 64 amino acids at the N terminus of SpoIVA. In cells lacking a wild-type copy of spoIVA (and hence blocked after the stage of engulfment), GFP-SpoIVA65–492 localized to the forespore, appearing most often as a crescent on one side of the forespore when sporulation was carried out at 37°C (Fig. 1E). When sporulation was carried out at 25°C, GFP-SpoIVA65–492 showed improved localization, frequently appearing as caps on both sides of the forespore or as an almost complete circle around the forespore (Fig. 1F). Thus, SpoIVA truncated at its N terminus exhibited a modest localization defect, one that could be partially corrected by carrying out sporulation at low temperature. Western blot analysis showed that at 37°C GFP-SpoIVA65–492 accumulated at approximately half the level of the full-length fusion protein (data not shown), a finding that provides a possible explanation for the modest localization defect exhibited by the truncated fusion protein.
In contrast to GFP-SpoIVA65–492, a fusion protein (GFP-SpoIVA1–487) lacking five amino acids from the C terminus of GFP-SpoIVA exhibited a severe localization defect. In a very small number of sporangia, GFP-SpoIVA1–487 appeared to form a crescent next to the forespore (data not shown). Generally, however, in cells sporulating at 37°C and lacking a wild-type copy of spoIVA, GFP-SpoIVA1–487 formed a blotchy spot in the mother cell cytoplasm near the forespore (Fig. 1G). A similarly severe defect in localization was observed at 25°C. The localization defect of GFP-SpoIVA1–487 could not be attributed to reduced levels of the truncated fusion protein, because Western blot analysis showed that GFP-SpoIVA1–487 and GFP-SpoIVA65–492 accumulated to similar levels during sporulation (data not shown).
A stringent requirement for the C-terminal five amino acids in proper localization was also demonstrated with a truncated SpoIVA protein that had not been fused to GFP at its N terminus. In this case, immunostaining was used to visualize the truncated protein (green) and PI to stain the mother cell and forespore chromosomes (red). In wild-type cells that have undergone engulfment as shown previously by Pogliano et al. (30), SpoIVA appeared as a ring around the forespore (Fig. 1I). In contrast, SpoIVA1–487 did not localize to the forespore. Unlike GFP-SpoIVA1–487, however, which formed a blotchy spot in the mother cell cytoplasm, SpoIVA1–487 exhibited a uniform pattern of staining throughout the mother cell cytoplasm (Fig. 1J). Evidently, the presence of the GFP moiety caused the truncated sporulation protein to coalesce into a clump within the mother cell. In any event, as judged both by the use of a GFP fusion and by immunostaining, the C-terminal five amino acids of SpoIVA appear to play a crucial role in proper localization. The importance of the extreme C terminus provides an explanation for the failure of the SpoIVA-GFP fusion (in which GFP was joined at the C terminus) to properly localize, as indicated above.
Localization may depend on oligomerization as well as targeting.
That the presence of wild-type SpoIVA can improve the localization of GFP-SpoIVA had suggested (see above) that SpoIVA molecules may interact with each other, although this interaction may be indirect. This suggests a model in which localization of SpoIVA is mediated by two domains of the protein. One domain(s) is responsible for targeting the sporulation protein to a receptor or other landmark on the outer surface of the forespore, and the other domain(s) allows SpoIVA molecules to interact (directly or indirectly) with each other (i.e., oligomerize). To investigate this possibility, we asked whether the presence of unmodified SpoIVA would restore proper localization to GFP-SpoIVA1–487. The results show that in the presence of SpoIVA, the truncated fusion protein was able to localize, often appearing as caps on either side of the engulfed forespore (Fig. 1H). (As in the meridiploid strain producing full-length GFP-SpoIVA, the strain producing both the truncated fusion protein and wild-type SpoIVA contained a spoIVCB mutation to block the formation of phase-bright spores.) We conclude that the C-terminal five amino acids play a key role in targeting SpoIVA to the surface of the forespore and that SpoIVA molecules (directly or indirectly) interact with each other through a region outside of the extreme C terminus.
A gene under ςE control is important for SpoIVA localization.
What feature of the forespore is responsible for targeting SpoIVA? One possibility is that the product of a gene expressed during sporulation directly or indirectly enables SpoIVA to recognize and assemble around the forespore, perhaps serving as a receptor on the outer surface of the mother cell membrane that surrounds the forespore. Such a gene might be expressed in the predivisional sporangium, perhaps under the control of the sporulation sigma factor ςH, which directs gene expression at the onset of sporulation. (See reference 42 for a review of sigma factors that govern sporulation.) Alternatively, the hypothetical gene might be expressed in the forespore under the control of the forespore-specific sigma factor ςF. Perhaps the product of such a ςF-controlled gene is displayed on the surface of the forespore, where it recruits SpoIVA molecules made in the mother cell. Yet a third alternative is that the gene in question is expressed in the mother cell under the control of the mother cell sigma factor ςE. Because spoIVA is transcribed under the control of ςE and because the activation of ςE depends on the prior action of ςH and ςF, it has heretofore not been possible to study the dependence of SpoIVA localization on ςH-, ςF-, or ςE-directed gene expression.
To circumvent this problem and to help distinguish among these possibilities, we freed spoIVA transcription from the control of ςE by replacing the spoIVA promoter with the promoter for spoVG (47), a sporulation gene whose transcription is induced in the predivisional sporangium under the control of ςH. We then substituted the PspoVG-spoIVA fusion for the normal gene in otherwise wild-type cells and in mutants of ςF, ςE, and ςG. In these experiments, we used immunofluorescence in conjunction with antibodies against SpoIVA to visualize the sporulation protein in cells that had been sporulating for 4 h and thus presumably had undergone engulfment. The results show that SpoIVA localized as a crescent on one side of the forespore in otherwise wild-type cells in which its synthesis had been brought under the direct control of ςH (Fig. 1K). Evidently, localization to the forespore does not stringently depend upon the time at which SpoIVA synthesis commences. (SpoIVA produced under the control of ςH did not, however, fully encircle the forespore, probably because the level of the sporulation protein produced from PspoVG-spoIVA was somewhat lower [as judged by Western blot analysis; data not shown] than that synthesized from the wild-type gene.)
A crescent of SpoIVA was similarly observed in cells mutant for ςG, a forespore-specific sigma factor that comes into play after the engulfment stage of sporulation and after the induction of the spoIVA promoter (Fig. 1N). In contrast, SpoIVA produced from the PspoVG-spoIVA fusion in cells mutant for ςF and ςE was substantially mislocalized, the pattern of immunolabeling being diffuse fluorescence throughout the mother cell cytoplasm (Fig. 1L and M). (As can be seen in the figure, mutants of ςF and ςE produce disporic sporangia, in which forespores are formed at both poles of the sporangium [42].) Because activation of ςE depends on the prior action of ςF (42), the simplest interpretation of these results is that the localization of SpoIVA requires the product of a gene(s) which, like spoIVA, is under the control of ςE. It is entirely possible, however, that localization additionally requires a ςF- or ςH-controlled gene.
Proper localization depends on the ςE-controlled gene spoVM.
In addition to spoIVA itself, 12 spo genes are known whose transcription is under ςE control (42). Localization of SpoIVA was known not to be dependent upon spoIID, spoIIID, and spoVID (10). We have now extended this analysis to the remaining nine genes by immunofluorescence microscopy: spoIIM, spoIIP, spoIIA, spoIVF, spoVB, spoVD, spoVE, spoVM, and spoVR. In mutants of eight of the genes, localization of SpoIVA was normal. In the case of a spoVM mutant, however, localization was substantially impaired. Using immunofluorescence microscopy, we observed that in a spoVM mutant, fluorescence from SpoIVA was spread throughout the mother cell cytoplasm (Fig. 1O). Moreover, when a spoVM mutation was introduced into cells harboring the gfp-spoIVA fusion and the wild-type spoIVA gene, GFP-SpoIVA did not localize as a sharp green ring around the forespore, as observed in cells of the parent strain. Instead, GFP-SpoIVA was seen as a clump close to, but not localized around, the forespore (Fig. 1P). The case of spoVM shows, once again, that SpoIVA and GFP-SpoIVA behave differently when they fail to localize: unlocalized SpoIVA spreads out in the mother cell cytoplasm (Fig. 1L), whereas unlocalized GFP-SpoIVA and SpoIVA-GFP coalesce into a clump near the center of the sporangium (Fig. 1A and 1P) (22).
DISCUSSION
Exploiting the capacity of GFP to tag proteins fluorescently in living cells in combination with the techniques of optical sectioning, deconvolution, and time-lapse microscopy, we have obtained a four-dimensional view of the localization and assembly of a key morphogenetic protein in the sporulation process. Central to our experiments was the finding that a fusion of GFP at the N terminus of SpoIVA in combination with the presence of the unmodified, wild-type protein made possible the visualization of the sporulation protein at high resolution without interfering with the normal course of morphogenesis. Using the GFP-SpoIVA fusion in fluorescence microscopy experiments in which images of optical sections were collected in increments of 0.05 μm and processed by using a deconvolution algorithm (4, 25), we have observed that SpoIVA assembles into a spherical shell around the forespore. Thus, in confirmation and extension of previous results based on two-dimensional analysis (10, 30), we conclude that SpoIVA coats the outer surface of the mother cell membrane that surrounds the forespore. Therefore, SpoIVA is advantageously positioned both to trigger biosynthesis of the cortex underneath the engulfment membrane and to recruit from the mother cell cytoplasm the components of the protein coat that will envelop the mature spore. Furthermore, it appears that SpoIVA remains present in the spore after maturation is complete, although we cannot exclude the possibility that the presence of the GFP moiety caused the fusion protein to persist aberrantly after morphogenesis was complete.
The use of GFP-SpoIVA in time-lapse experiments enabled us to visualize the assembly of the morphogenetic protein over the course of sporulation. In cells growing on sporulation medium at room temperature, GFP-SpoIVA initially collected in the region of the mother cell closest to the forespore, where it assembled into a crescent. Over time the crescent expanded, eventually spreading around and fully encircling the forespore. Assembly and accretion of additional fusion protein around the forespore continued for several hours. We cannot exclude the possibility that the repeated excitation of the sporangia during the course of the time-lapse experiment artificially slowed morphogenesis to some extent. Nevertheless, the accretion of SpoIVA around the forespore evidently continued after the process of engulfment was complete.
Together with previous results indicating that SpoIVA assembly commences at the stage (II) of polar division (10), we propose the following model for the formation of the SpoIVA shell around the forespore. Initially, some SpoIVA begins to accumulate on the mother cell face of the polar septum. Next, during engulfment SpoIVA remains associated with the septal membrane as it migrates around the forespore. Importantly, however, SpoIVA molecules (presumably from ongoing synthesis) continue to accumulate around the forespore well after the engulfment process is complete. During this accumulation process SpoIVA preferentially collects on the mother cell side of the engulfed forespore, but eventually SpoIVA is deposited equally all around the forespore.
What features of SpoIVA are responsible for directing the protein to the outside surface of the forespore? The generation of truncated forms of GFP-SpoIVA indicated that the extreme C-terminal region of SpoIVA plays a critical role in proper localization. A truncated form of the fusion protein lacking 64 residues at the N terminus (GFP-SpoIVA65–492) was capable of localizing as a crescent on the forespore, whereas GFP-SpoIVA lacking five amino acids from the C terminus (GFP-SpoIVA1–487) exhibited little or no recognition of the forespore. Proper localization of the truncated fusion protein was, however, restored by the presence of unmodified SpoIVA. We take this as evidence that the C-terminal truncation did not cause global misfolding of the fusion protein. Also, Western blot analysis showed that the C-terminal truncation mutant GFP-SpoIVA1–487 was no more subject to proteolysis than GFP-SpoIVA65–492. We therefore favor the possibility that the C-terminal region plays a direct role in targeting SpoIVA to a hypothetical receptor or other feature on the outside surface of the mother cell membrane around the forespore. Whatever this feature is (see below), it must be present at the polar septum, because localization of SpoIVA commences prior to the stage of engulfment (10, 30).
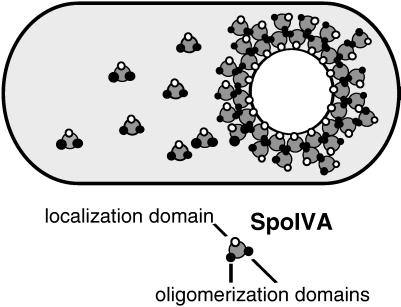
That wild-type SpoIVA can improve the capacity of GFP-SpoIVA to localize and restore the capacity of the C-terminal truncation mutant to assemble around the forespore implies that SpoIVA molecules interact with each other, either directly or indirectly, by means of one or more (unknown) intervening molecules. Assuming for the sake of simplicity that this interaction is direct, we infer that SpoIVA contains a domain(s) that enables it to oligomerize and that this domain lies between the extreme C-terminal region of the protein and the N-terminal 64 amino acids. Figure 4 summarizes our model for the localization of SpoIVA in which we posit that SpoIVA contains both an oligomerization domain and a separate targeting domain, which corresponds, at least in part, to the C-terminal five amino acids of the protein. An example of a morphogenetic protein that both oligomerizes and localizes to a specific region of the cell is the cell division protein FtsZ, which assembles into a cytokinetic ring at the midcell position and is responsible for the formation of the septasome (25).
FIG. 4.
Model of SpoIVA assembly. SpoIVA has a targeting domain that enables the protein to recognize the forespore and one or more oligomerization domains that allow the protein to interact with other identical molecules. Thus, when fully assembled, SpoIVA forms a multilayered shell around the forespore.
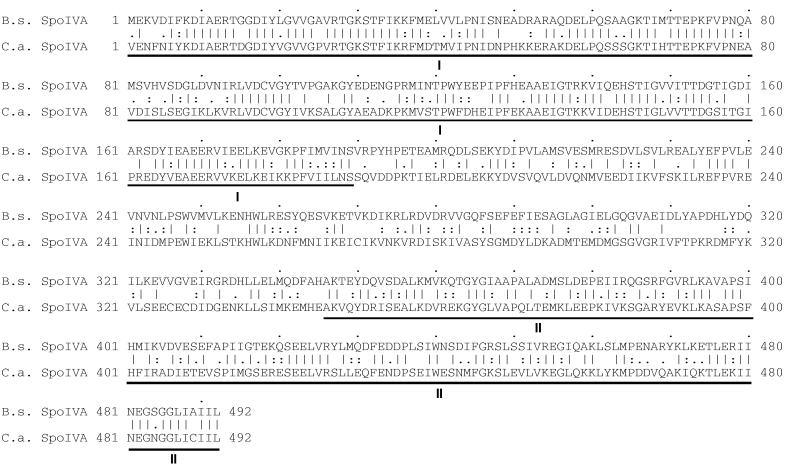
An alignment of the amino acid sequence of the B. subtilis SpoIVA protein and its homolog in the distantly related endospore-forming bacterium Clostridium acetobutylicum provides further insight into the functional anatomy of the sporulation protein (Fig. 5). The two SpoIVA proteins, which are both 492 residues in length, exhibit 49% overall identity in amino acid sequence. The similarity is most extensive in two large regions at the N terminus and the C terminus, which extend 189 and 147 amino acids, respectively. Identity across the N-terminal and C-terminal regions is 63 and 54%, respectively. Consistent with the assignment of the extreme C terminus in proper localization, the B. subtilis and C. acetobutylicum proteins are identical in four of five residues at the C terminus. We know from our truncation analysis that the N-terminal 64 amino acids of SpoIVA are functionally important although largely dispensable for proper localization. That the N terminus lies in a region of high conservation is consistent with the idea that this region is important for some aspect of SpoIVA function, such as coat assembly or cortex formation or both.
FIG. 5.
Alignment of the amino acid sequences of SpoIVA from B. subtilis and C. acetobutylicum. The alignment was generated by the GAP program of the Wisconsin Genetics Computer Group sequence analysis package (version 9.0). Identical residues are indicated by lines, whereas highly and moderately similar residues are denoted by double dots and single dots, respectively. The database accession number corresponding to the B. subtilis SpoIVA sequence is M80926. The C. acetobutylicum SpoIVA sequence is accessible on the Genome Therapeutics Corporation web page: http://www.genomecorp.com/genesequences/clostridium/clospage.html. Regions of highest identity are denoted below the sequences by roman numbers I and II.
What feature of the outer surface of the forespore is responsible for recruiting SpoIVA? An important clue comes from our finding that proper localization depends on the expression of another ςE-controlled gene. In a survey of spo genes known to be under the control of ςE, only a mutant of spoVM caused a pronounced effect on SpoIVA localization. The product of spoVM is remarkably small, being only 26 residues in length, and exhibits no significant similarity to other proteins in the databases (21). Little is known about the function of SpoVM, but genetic and biochemical analysis by Cutting et al. (7) indicate that it is a substrate for FtsH protease, a member of the AAA (ATPases associated with diverse cellular activities) family of ATPases. The work of Cutting et al. (7) also indicated that the interaction of SpoVM with FtsH is important for engulfment. Our present results raise the question of whether the SpoVM peptide is itself targeted to the outside surface of the forespore and whether it serves in part as a receptor for SpoIVA, conceivably interacting with the C-terminal region of the morphogenetic protein.
The dependence of SpoIVA localization on SpoVM does, however, raise an apparent paradox. A spoVM null mutation is known to block cortex formation, but, unlike a spoIVA null mutation, a spoVM null mutation allows for the assembly of a thin but detectable layer of coat protein around the forespore. In other words, the phenotype of a spoVM null mutant is distinct from and less severe with respect to coat assembly than a spoIVA mutation, which causes the coat to misassemble as a swirl in the mother cell cytoplasm. One way to reconcile these apparently contradictory findings is to suppose that SpoIVA is able to localize to a limited extent (below our capacity to detect) in the absence of SpoVM and that this low level of localization is capable of supporting the formation of a thin layer of properly positioned coat. If so, then SpoVM may not be the only factor involved in recruiting SpoIVA to the forespore.
In summary, we have shown that fluorescence microscopy can be used to visualize morphogenesis in living bacterial cells with respect to both space and time. We have shown that the sporulation protein SpoIVA assembles into a spherical shell around the forespore and that accretion of SpoIVA molecules around the forespore seems to continue after the time at which the forespore is wholly engulfed within the mother cell. We have also shown that targeting of SpoIVA to the forespore critically depends on the extreme C-terminal region of the protein, and we presented evidence for a model for morphogenesis in which assembly is determined by both oligomerization and targeting domains in SpoIVA. Finally, we have presented evidence showing that proper targeting of SpoIVA depends on ςE-controlled gene expression in the mother cell and that the product of one such ςE-controlled gene (spoVM) plays a key role in the recruitment of SpoIVA to the outer surface of the forespore.
ACKNOWLEDGMENTS
This work was supported by NIH grant GM181568 and the Office of Naval Research.
We thank E. Angert, A. Teleman, and K. Pogliano for advice and assistance with three-dimensional imaging and deconvolution, P. Fawcett for assistance with image processing, P. Stragier for strains MO1708 and MO1433, and A. Driks and K. Pogliano for critical reading of the manuscript.
REFERENCES
- 1.Beall B, Moran C P., Jr Cloning and characterization of spoVR, a gene from Bacillus subtilis involved in spore cortex formation. J Bacteriol. 1994;176:2003–2012. doi: 10.1128/jb.176.7.2003-2012.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bi E, Lutkenhaus J. FtsZ ring structure associated with division in Escherichia coli. Nature. 1991;354:161–164. doi: 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- 3.Bramhill D. Bacterial cell division. Annu Rev Cell Dev Biol. 1997;13:395–424. doi: 10.1146/annurev.cellbio.13.1.395. [DOI] [PubMed] [Google Scholar]
- 4.Carter K C, Bowman D, Carrington W, Fogarty K, McNeil J A, Fay F S, Lawrence J B. A three-dimensional view of precursor messenger RNA metabolism within the mammalian nucleus. Science. 1993;259:1330–1335. doi: 10.1126/science.8446902. [DOI] [PubMed] [Google Scholar]
- 5.Chalfie M, Tu Y, Euskirchen G, Ward W W, Prasher D C. Green Fluorescent Protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 6.Coote J G. Sporulation in Bacillus subtilis. Characterization of oligosporogenous mutants and comparison of their phenotypes with those of asporogenous mutants. J Gen Microbiol. 1972;71:1–15. doi: 10.1099/00221287-71-1-1. [DOI] [PubMed] [Google Scholar]
- 7.Cutting S, Anderson M, Lysenko E, Page A, Tomoyasu T, Tatematsu K, Tatsuta T, Kroos L, Ogura T. SpoVM, a small protein essential to development in Bacillus subtilis, interacts with the ATP-dependent protease FtsH. J Bacteriol. 1997;179:5534–5542. doi: 10.1128/jb.179.17.5534-5542.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cutting S M, Vander Horn P B. Genetic analysis. In: Harwood C R, Cutting S M, editors. Molecular biological methods for Bacillus. Chichester, United Kingdom: John Wiley & Sons Ltd.; 1990. pp. 27–74. [Google Scholar]
- 9.Dean D R, Hoch J A, Aronson A I. Alteration of the Bacillus subtilis glutamine synthetase results in overproduction of the enzyme. J Bacteriol. 1977;131:981–987. doi: 10.1128/jb.131.3.981-987.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Driks A, Roels S, Beall B, Moran C P, Jr, Losick R. Subcellular localization of proteins involved in the assembly of the spore coat of Bacillus subtilis. Genes Dev. 1994;8:234–244. doi: 10.1101/gad.8.2.234. [DOI] [PubMed] [Google Scholar]
- 11.Erickson H P, Taylor D W, Taylor K A, Bramhill D. Bacterial cell division protein FtsZ assembles into protofilament sheets and minirings, structural homologs of tubulin polymers. Proc Natl Acad Sci USA. 1996;93:519–523. doi: 10.1073/pnas.93.1.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Errington J, Mandelstam J. Use of a lacZ gene fusion to determine the dependence pattern of sporulation operon spoIIA in spo mutants of Bacillus subtilis. J Gen Microbiol. 1986;132:2967–2976. doi: 10.1099/00221287-132-11-2967. [DOI] [PubMed] [Google Scholar]
- 13.Garsin D A, Paskowitz D M, Duncan L, Losick R. Evidence for common sites of contact between the antisigma factor SpoIIAB and its partners SpoIIAA and the developmental transcription factor ςF in Bacillus subtilis. J Mol Biol. 1998;284:557–568. doi: 10.1006/jmbi.1998.2201. [DOI] [PubMed] [Google Scholar]
- 14.Harry E J, Pogliano K, Losick R. Use of immunofluorescence to visualize cell-specific gene expression during sporulation in Bacillus subtilis. J Bacteriol. 1995;177:3386–3393. doi: 10.1128/jb.177.12.3386-3393.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hess J F, Oosawa K, Kaplan N, Simon M I. Phosphorylation of three proteins in the signalling pathway of bacterial chemotaxis. Cell. 1988;53:79–87. doi: 10.1016/0092-8674(88)90489-8. [DOI] [PubMed] [Google Scholar]
- 16.Itaya M, Kondo K, Tanaka T. A neomycin resistance cassette selectable in a single copy state in the Bacillus subtilis chromosome. Nucleic Acids Res. 1989;17:4410. doi: 10.1093/nar/17.11.4410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jain C. New improved lacZ fusion vectors. Gene. 1993;133:99–102. doi: 10.1016/0378-1119(93)90231-q. [DOI] [PubMed] [Google Scholar]
- 18.Jenal U, Shapiro L. Cell cycle-controlled proteolysis of a flagellar motor protein that is asymmetrically distributed in the Caulobacter predivisional cell. EMBO J. 1996;15:2393–2406. [PMC free article] [PubMed] [Google Scholar]
- 19.Kenney T J, Moran C P., Jr Organization and regulation of an operon that encodes a sporulation-essential sigma factor in Bacillus subtilis. J Bacteriol. 1987;169:3329–3339. doi: 10.1128/jb.169.7.3329-3339.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.LeDeaux J R, Grossman A D. Isolation and characterization of kinC, a gene that encodes a sensor kinase homologous to the sporulation sensor kinases KinA and KinB in Bacillus subtilis. J Bacteriol. 1995;177:166–175. doi: 10.1128/jb.177.1.166-175.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levin P A, Fan N, Ricca E, Driks A, Losick R, Cutting S. An unusually small gene required for sporulation by Bacillus subtilis. Mol Microbiol. 1993;9:761–771. doi: 10.1111/j.1365-2958.1993.tb01736.x. [DOI] [PubMed] [Google Scholar]
- 22.Lewis P J, Errington J. Use of Green Fluorescent Protein for detection of cell-specific gene expression and subcellular protein localization during sporulation in Bacillus subtilis. Microbiology. 1996;142:733–740. doi: 10.1099/00221287-142-4-733. [DOI] [PubMed] [Google Scholar]
- 23.Lu S, Kroos L. Overproducing the Bacillus subtilis mother cell sigma factor precursor, pro-ςK, uncouples ςK-dependent gene expression from dependence on intercompartmental communication. J Bacteriol. 1994;176:3936–3943. doi: 10.1128/jb.176.13.3936-3943.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maddock J, Shapiro L. Polar location of the chemoreceptor complex in the Escherichia coli cell. Science. 1993;259:1717–1723. doi: 10.1126/science.8456299. [DOI] [PubMed] [Google Scholar]
- 25.Margolin W. A green light for the bacterial cytoskeleton. Trends Microbiol. 1998;6:233–238. doi: 10.1016/s0966-842x(98)01280-3. [DOI] [PubMed] [Google Scholar]
- 26.Margolis P, Driks A, Losick R. Establishment of cell type by compartmentalized activation of a transcription factor. Science. 1991;254:562–565. doi: 10.1126/science.1948031. [DOI] [PubMed] [Google Scholar]
- 27.Mukherjee A, Lutkenhaus J. Guanine nucleotide-dependent assembly of FtsZ into filaments. J Bacteriol. 1994;176:2754–2758. doi: 10.1128/jb.176.9.2754-2758.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oosawa K, Hess J F, Simon M I. Mutants defective in bacterial chemotaxis show modified protein phosphorylation. Cell. 1988;53:89–96. doi: 10.1016/0092-8674(88)90490-4. [DOI] [PubMed] [Google Scholar]
- 29.Piggot P J. Mapping of asporogenous mutations of Bacillus subtilis: a minimum estimate of the number of sporulation operons. J Bacteriol. 1973;114:1241–1253. doi: 10.1128/jb.114.3.1241-1253.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pogliano K, Harry E, Losick R. Visualization of the subcellular location of sporulation proteins in Bacillus subtilis using immunofluorescence microscopy. Mol Microbiol. 1995;18:459–470. doi: 10.1111/j.1365-2958.1995.mmi_18030459.x. [DOI] [PubMed] [Google Scholar]
- 31.Pogliano K, Hofmeister A E M, Losick R. Disappearance of the ςE transcription factor from the forespore and the SpoIIE phosphatase from the mother cell contributes to establishment of cell-specific gene expression during sporulation in Bacillus subtilis. J Bacteriol. 1997;179:3331–3341. doi: 10.1128/jb.179.10.3331-3341.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Resnekov O, Alper S, Losick R. Subcellular localization of proteins governing the proteolytic activation of a developmental transcription factor in Bacillus subtilis. Genes Cells. 1996;1:529–542. doi: 10.1046/j.1365-2443.1996.d01-262.x. [DOI] [PubMed] [Google Scholar]
- 33.Ricca E, Cutting S, Losick R. Characterization of bofA, a gene involved in intercompartmental regulation of pro-ςK processing during sporulation in Bacillus subtilis. J Bacteriol. 1992;174:3177–3184. doi: 10.1128/jb.174.10.3177-3184.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roels S, Driks A, Losick R. Characterization of spoIVA, a sporulation gene involved in coat morphogenesis in Bacillus subtilis. J Bacteriol. 1992;174:575–585. doi: 10.1128/jb.174.2.575-585.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 36.Sandman K, Kroos L, Cutting S, Youngman P, Losick R. Identification of the promoter for a spore coat protein gene in Bacillus subtilis and studies on the regulation of its induction at a late stage of sporulation. J Mol Biol. 1988;200:461–473. doi: 10.1016/0022-2836(88)90536-0. [DOI] [PubMed] [Google Scholar]
- 37.Sandman K, Losick R, Youngman P. Genetic analysis of Bacillus subtilis spo mutations generated by Tn917-mediated insertional mutagenesis. Genetics. 1987;117:603–617. doi: 10.1093/genetics/117.4.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shapiro L, Losick R. Protein localization and cell fate in bacteria. Science. 1997;276:712–718. doi: 10.1126/science.276.5313.712. [DOI] [PubMed] [Google Scholar]
- 39.Sterlini J M, Mandelstam J. Commitment to sporulation in Bacillus subtilis and its relationship to development of actinomycin resistance. Biochem J. 1969;113:29–37. doi: 10.1042/bj1130029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stevens C M, Daniel R, Illing N, Errington J. Characterization of a sporulation gene, spoIVA, involved in spore coat morphogenesis in Bacillus subtilis. J Bacteriol. 1992;174:586–594. doi: 10.1128/jb.174.2.586-594.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stragier P, Kunkel B, Kroos L, Losick R. Chromosomal rearrangement generating a composite gene for a developmental transcription factor. Science. 1989;243:507–512. doi: 10.1126/science.2536191. [DOI] [PubMed] [Google Scholar]
- 42.Stragier P, Losick R. Molecular genetics of sporulation in Bacillus subtilis. Annu Rev Genet. 1996;30:297–341. doi: 10.1146/annurev.genet.30.1.297. [DOI] [PubMed] [Google Scholar]
- 43.Webb C D, Decatur A, Teleman A, Losick R. Use of green fluorescent protein for visualization of cell-specific gene expression and subcellular protein localization during sporulation in Bacillus subtilis. J Bacteriol. 1995;177:5906–5911. doi: 10.1128/jb.177.20.5906-5911.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu J, Newton A. Regulation of the Caulobacter flagellar gene hierarchy; not just for motility. Mol Microbiol. 1997;24:233–239. doi: 10.1046/j.1365-2958.1997.3281691.x. [DOI] [PubMed] [Google Scholar]
- 45.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 46.Youngman P, Perkins J B, Losick R. Construction of a cloning site near one end of Tn917 into which foreign DNA may be inserted without affecting transposition in Bacillus subtilis or expression of the transposon-borne erm gene. Plasmid. 1984;12:1–9. doi: 10.1016/0147-619x(84)90061-1. [DOI] [PubMed] [Google Scholar]
- 47.Zuber P, Losick R. Use of a lacZ fusion to study the role of the spoO genes of Bacillus subtilis in developmental regulation. Cell. 1983;35:275–283. doi: 10.1016/0092-8674(83)90230-1. [DOI] [PubMed] [Google Scholar]