Abstract
Purpose
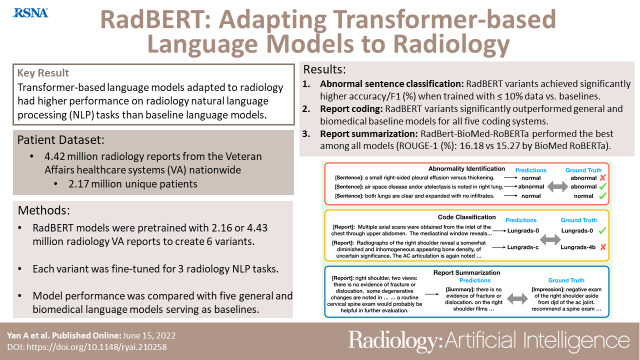
To investigate if tailoring a transformer-based language model to radiology is beneficial for radiology natural language processing (NLP) applications.
Materials and Methods
This retrospective study presents a family of bidirectional encoder representations from transformers (BERT)–based language models adapted for radiology, named RadBERT. Transformers were pretrained with either 2.16 or 4.42 million radiology reports from U.S. Department of Veterans Affairs health care systems nationwide on top of four different initializations (BERT-base, Clinical-BERT, robustly optimized BERT pretraining approach [RoBERTa], and BioMed-RoBERTa) to create six variants of RadBERT. Each variant was fine-tuned for three representative NLP tasks in radiology: (a) abnormal sentence classification: models classified sentences in radiology reports as reporting abnormal or normal findings; (b) report coding: models assigned a diagnostic code to a given radiology report for five coding systems; and (c) report summarization: given the findings section of a radiology report, models selected key sentences that summarized the findings. Model performance was compared by bootstrap resampling with five intensively studied transformer language models as baselines: BERT-base, BioBERT, Clinical-BERT, BlueBERT, and BioMed-RoBERTa.
Results
For abnormal sentence classification, all models performed well (accuracies above 97.5 and F1 scores above 95.0). RadBERT variants achieved significantly higher scores than corresponding baselines when given only 10% or less of 12 458 annotated training sentences. For report coding, all variants outperformed baselines significantly for all five coding systems. The variant RadBERT–BioMed-RoBERTa performed the best among all models for report summarization, achieving a Recall-Oriented Understudy for Gisting Evaluation–1 score of 16.18 compared with 15.27 by the corresponding baseline (BioMed-RoBERTa, P < .004).
Conclusion
Transformer-based language models tailored to radiology had improved performance of radiology NLP tasks compared with baseline transformer language models.
Keywords: Translation, Unsupervised Learning, Transfer Learning, Neural Networks, Informatics
Supplemental material is available for this article.
© RSNA, 2022
See also commentary by Wiggins and Tejani in this issue.
Keywords: Translation, Unsupervised Learning, Transfer Learning, Neural Networks, Informatics
Summary
Transformer-based language models adapted for radiology had higher performance on three radiology natural language processing tasks than five intensively studied baseline models.
Key Points
■ Bidirectional transformer-based language models tailored to radiology were superior to general domain or biomedical- and clinical-specific language models for the radiology natural language processing tasks of abnormal sentence classification, report coding, and report summarization.
■ Radiology-specialized language models were significantly better than baseline models for all five coding systems in the report coding task, outperformed baseline models for abnormal sentence classification when fine-tuned with less than 10% of training examples, and achieved Recall-Oriented Understudy for Gisting Evaluation–1 score of 16.18 versus 15.27 by the counterpart baseline model for the report summarization task.
Introduction
Creating an efficient way to extract large volumes of critical information from unstructured radiology reports has many potential applications that can benefit patients and providers alike, including improved follow-up of abnormal results, assistance in quality improvement projects, and facilitation of epidemiologic, surveillance, and cost-effectiveness investigations. The benefit is eminent in challenging health care environments where well-trained medical personnel and resources are limited.
Recently, tremendous progress has been made in natural language processing (NLP) using deep learning. One of the advances is contextualized language models based on the “transformer” architecture (1), such as bidirectional encoder representations from transformers (BERT) (2) and robustly optimized BERT pretraining approach (RoBERTa) (3). These models can effectively represent words and sentences given their document-level context; that is, words can have different representations across varied contexts. Compared with noncontextualized models such as Word2Vec (4), substantial performance improvements have been reported for a broad range of NLP tasks.
Transformer-based language models are appealing for clinical NLP because they may be used as a shared layer for transfer learning, in which pretraining them with a large amount of text data can benefit downstream tasks where annotated training data are scarce. In many applications, we may need to annotate tens of thousands of documents to effectively train a classifier. But if our classifier includes a transformer layer, then what is already learned by the transformer can be transferred to the classifier via “fine-tuning.” Such classifiers require fewer annotated training examples than training from scratch, which will greatly lower the hurdle to leveraging the latest NLP advances in clinical applications.
A few domain-specific models have been developed, such as biomedical text with BioBERT (5) and BioMed-RoBERTa (6) and clinical text with clinical-BERT (7) and BlueBERT (8). However, a model specific to radiology text has rarely been explored. Previous work trained transformers with the Medical Information Mart for Intensive Care (MIMIC-III) database (9), which includes radiology text, but they are not sufficient to fully support the radiology domain with a limited number of reports. Moreover, the duplicates present in the MIMIC-III database reported in a study by Gabriel et al (10) may degrade the effectiveness of transformers pretrained with it, as suggested in a study by Raffel et al (11) for pretraining language models.
In this paper, we present RadBERT, a family of transformer-based language models adapted to radiology. RadBERT variants were pretrained with millions of radiology reports from the U.S. Department of Veterans Affairs (VA) health care system nationwide on top of a variety of language models as the initialization to investigate if tailoring a transformer-based language model to radiology is beneficial for radiology NLP applications.
Materials and Methods
VA Radiology Report Corpus
Figure 1 shows an overview of our study design. The study team had institutional review board approval for exemption from informed consent to use 150 million radiology reports from 130 VA facilities nationwide from the past 20+ years to develop various artificial intelligence applications in radiology. Among these reports, 4.42 million reports with 2.17 million unique patients, 466 million tokens, and 2.6 GB in size were retrieved from the clinical data warehouse of the VA, de-identified, and de-duplicated for the study. Appendix E1 (supplement) describes the details of our preprocessing steps. An estimation of the distributions of modalities and body parts covered in these reports is given in Appendix E2 (supplement).
Figure 1:
Overview of our study design, which includes pretraining and fine-tuning of RadBERT. (A) In pretraining, different weight initializations were considered to create variants of RadBERT. (B) The variants were fine-tuned for three important radiology natural language processing (NLP) tasks: abnormal sentence classification, report coding, and report summarization. The performance of RadBERT variants for these tasks was compared with a set of intensively studied transformer-based language models as baselines. (C) Examples of each task and how performance was measured. In the abnormality identification task, a sentence in a radiology report was considered “abnormal” if it reported an abnormal finding and “normal” otherwise. A human-annotated abnormality was considered ground truth to evaluate the performance of an NLP model. In the code classification task, models were expected to output diagnostic codes (eg, abdominal aortic aneurysm, Breast Imaging Reporting and Data System [BI-RADS], and Lung Imaging Reporting and Data System [Lung-RADS]) that match the codes given by human providers as the ground truth for a given radiology report. During report summarization, the models generated a short summary given the findings in a radiology report. Summary quality was measured by how similar it was to the impression section of the input report. AAA = abdominal aortic aneurysm, BERT = bidirectional encoder representations from transformers, RadBERT = BERT-based language model adapted for radiology, RoBERTa = robustly optimized BERT pretraining approach.
Pretraining of Transformers
We followed the standard pretraining procedures as used in BERT (3) for all RadBERT models. Input texts were tokenized with WordPiece (12) as subword tokens and fed into the model. The training objective was the masked language model introduced in the study by Devlin et al (2). Liu et al (3) reported that skipping the next sentence prediction loss slightly improved downstream task performance while substantially simplifying pretraining compared with the original pretraining algorithm used to pretrain BERT. We followed their suggestion when pretraining RadBERT models. We used the original vocabulary of previous BERT-base (2) language models, which allows weights pretrained on general domain corpora to be reused.
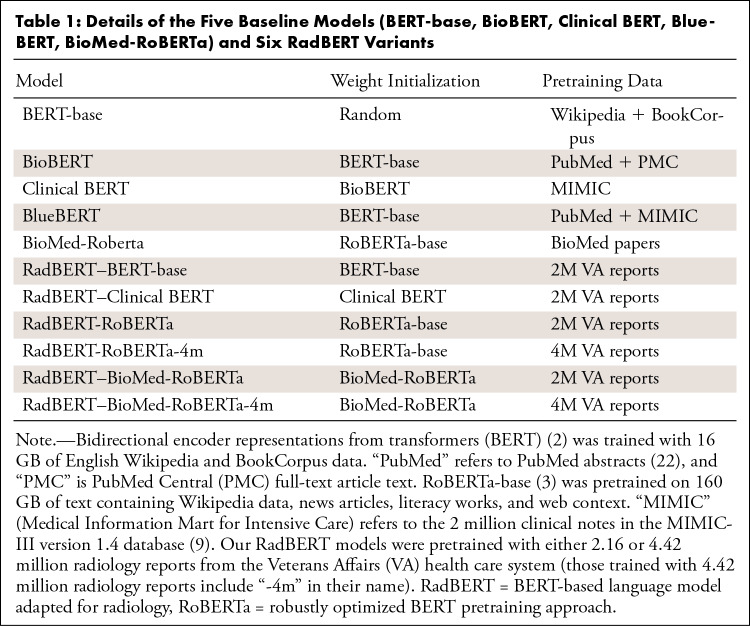
The details of five baseline models and six RadBERT variants are shown in Table 1. Appendix E3 (supplement) provides more implementation details for pretraining and the three tasks.
Table 1:
Details of the Five Baseline Models (BERT-base, BioBERT, Clinical BERT, BlueBERT, BioMed-RoBERTa) and Six RadBERT Variants
Task 1: Abnormal Sentence Classification
The task consisted of identifying sentence-level abnormal findings in a radiology report by classifying if a sentence reports normal or abnormal conditions. Following the common practice for fine-tuning transformers for sentence classification, we fed the output representation of the first token from the transformer into a single linear layer to classify the input sentence (Fig 1B). We used the labeled dataset from Harzig et al (13), which is a subset of the Open-I chest radiograph radiology report dataset (14) available in the public domain, to fine-tune a classifier on top of each transformer. The dataset was annotated previously by labeling whether a sentence describes abnormality and was readily split into 12 458, 1557, and 1558 sentences for training, validation, and testing, respectively. We measured the classification performance by using F1 score and accuracy, given seven different percentages of training sentences (from 1% to 100%). To ensure that a reported result was not an outlier due to a specific random seed initialization, mean and SD results of five runs with different random seed initializations were reported.
Task 2: Report Coding
The task classified reports into different diagnostic codes (see Appendix E4 [supplement]). Unlike abnormal sentence classification, report coding is a multiclass classification task on a report level.
The fine-tuning procedure was similar to that used for the abnormal sentence classification (Fig 1B). Average accuracy and macro average of F1 scores of multiclass classification for each coding system were reported to evaluate the performance of each model.
Task 3: Summarization
We applied an extractive summarization method that was based on a transformer-based language model (15) for this task. We randomly chose 1000 reports and their corresponding impressions as evaluation data. Note that the evaluated samples were reserved from the radiology corpus and not included in the pretraining. The length distributions for the reports and impressions are shown in Figure 2. The extracted summaries were scored by Recall-Oriented Understudy for Gisting Evaluation (ROUGE)–1, ROUGE-2, and ROUGE-L (16), the standard performance metrics of automated summarization and machine translation in NLP. ROUGE-1 and ROUGE-2 compute the overlap of n grams (n consecutive tokens) with n = 1 and 2, respectively, between a predicted summary and the ground truth, while ROUGE-L measures the overlap between summaries on the basis of the longest common subsequences appearing in the pair of summaries to be compared. Intuitively, ROUGE-1 and ROUGE-2 measure if a predicted summary captures information contents similar to the ground truth, while ROUGE-L measures fluency and coherence. The range of ROUGE lies from 0 to 1 (0%–100%). A higher score indicates better quality. A typical ROUGE number for summarization tasks could fall between 0 and 0.3, depending on the model and the difficulty of the task.
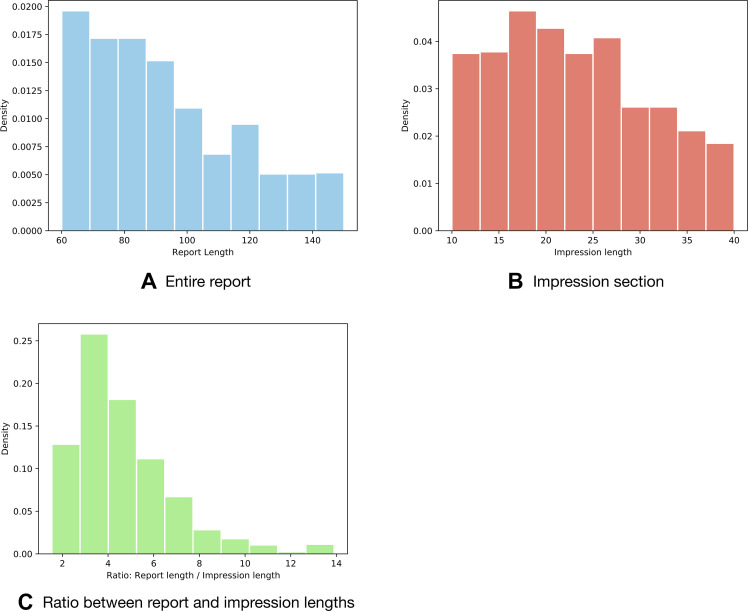
Figure 2:
Distribution histograms of (A) the length of the entire 1000 radiology reports evaluated in task 3 and (B) their impression sections. Density is the proportion of reports or impressions (ie, documents) with a certain length. Length is the number of words in a report or impression (words per document). (C) The ratio between one report's length and its corresponding impression mostly lies in the range from 2 to 6. The average number of words in a sentence in these reports is 11. The histograms may be used to estimate the number of sentences that need to be extracted from a report as a summary to match the impressions.
Statistical Analysis
We conducted bootstrap resampling as the statistical significance test for our results, following the methods described in the study by Smith et al (17). For task 1, given 1558 predictions from each model on the test set, we randomly sampled 1558 data with replacements from these predictions. The accuracy and F1 score were computed from the samples. We conducted 10 000 repetitive trials. For a pair of models, A and B, the proportion of times in these 10 000 trials that the F1 score of model A exceeded the F1 score of model B was noted. We labeled such pairs statistically significant if this proportion was greater than 95% for the predetermined significance level at .05.
We considered five pairs of models with different training data proportions (10%, 5%, 2%, and 1%) to confirm the significance in performance improvement between a RadBERT variant and their corresponding baseline models: (a) BERT-base and RadBERT–BERT-base, (b) Clinical BERT and RadBERT–Clinical BERT, (c) BlueBERT and RadBERT–Clinical BERT, (d) BioMed-RoBERTa and RadBERT-RoBERTa, and (e) BioMed-RoBERTa and RadBERT–BioMed-RoBERTa (see Table 2).
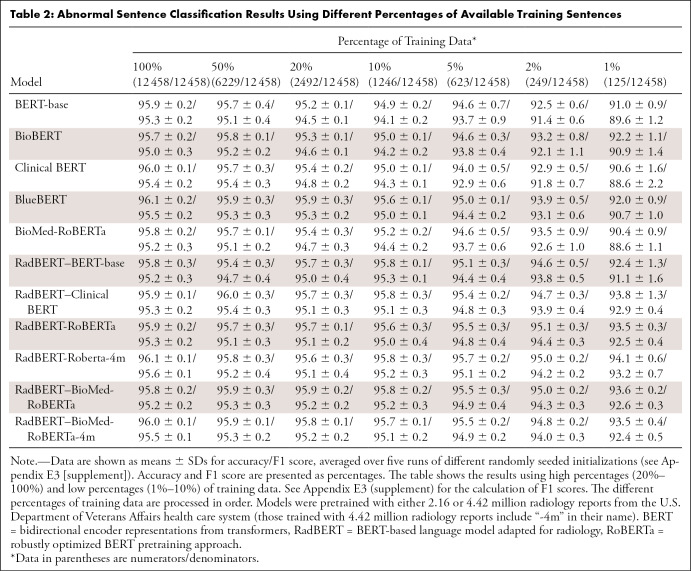
Table 2:
Abnormal Sentence Classification Results Using Different Percentages of Available Training Sentences
For task 2, the same pairwise comparisons as described above were used to verify statistical significance of the results for all five coding systems. For task 3, we compared the variant RadBERT–BioMed-RoBERTa with five baselines over the sum of ROUGE scores. Bonferroni correction was used to accommodate multiple (five) tests.
Model Availability
RadBERT models can be released upon request with a data usage agreement.
Results
Task 1: Abnormal Sentence Classification
Results for abnormal sentence classification are reported in Table 2. In general, the performance improved with more training data used for all models. When fine-tuned with sufficient annotated data (eg, thousands of sentences or more), general BERT models and those pretrained on the biomedical or clinical domain could achieve strong results, showing the effectiveness of pretraining. However, when the data for fine-tuning became scarce—that is, the number of training data were less than a thousand (<5% of training data)—adapting the language model to the specific domain was necessary.
One of the best-performing RadBERT variants was RadBERT-RoBERTa-4m, which was initialized with the RoBERTa model and then pretrained with 4.42 million radiology reports. When fine-tuned with 5% of available training data, RadBERT-RoBERTa-4m performed as well as BioBERT fine-tuned with 100% of available training data. This is particularly valuable, as annotating datasets by experts (ie, radiologists) is expensive and a major bottleneck in developing deep learning NLP applications in radiology.
Statistical tests on RadBERT variants compared with their corresponding baseline models under low percentages of training data (10% and lower) showed that for all pairs of models evaluated, the performance of the RadBERT variant was statistically significantly better than its counterpart baseline model, with P less than .05 given for all four different percentages of training data. P values were less than .0063 when only 5% or less of training data were given, suggesting that the differences are statistically significant when accommodating for testing five pairs with Bonferroni-adjusted significance level of .01. Given 10% of training data, the differences are still significant (P ≤ .0066), except for the pair between BlueBERT and RadBERT–Clinical BERT (P = .026).
Task 2: Report Coding
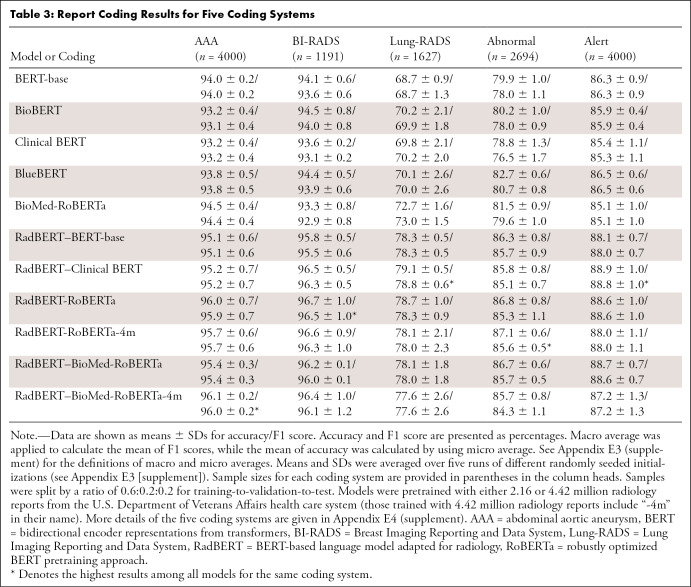
Table 3 shows the results of the code classification task. We found that all variants of RadBERT models significantly outperformed general BERT models or biomedical BERT models by a large margin, which further demonstrates the benefit of tailoring radiology-specific language models. For all coding systems and all pairs of RadBERT variants and their corresponding baseline models, P values were close to zero from our statistical tests. The best-overall-performing RadBERT variant is RadBERT-RoBERTa, achieving the best average accuracy and F1 score for all five coding systems, while the worst-overall performer among RadBERT variants (RadBERT–BioMed-RoBERTa-4m) still performed better than the best-overall baseline models, which are BlueBERT in terms of average accuracy over all five coding systems and BioMed-RoBERTa in terms of F1 score. In fact, the worst-performing RadBERT variant outperformed the best-performing baseline for each of the five coding systems in terms of both accuracy and F1 score.
Table 3:
Report Coding Results for Five Coding Systems
We also observed that, first, by comparing RadBERT variants, models initialized from general, biomedical, or clinical domains showed similar performance. The variance of accuracy or F1 scores among RadBERT variants is less than 0.4% for any coding system, while the variances for the baselines can be as high as 2.28% in accuracy and 2.63% in F1 score for the coding system “abnormal” (see Appendix E4 [supplement]). Overall, the variances are 0.08% in accuracy and 0.06% in F1 score among RadBERT variants, suggesting that regardless of weight initialization, after pretraining with a large corpus of radiology reports, the resulting RadBERT language models could reach a similar level of performance improvement for a radiology report coding task, with slight differences.
Second, models pretrained with a larger dataset of 4 million reports did not yield a higher average performance than models pretrained with 2 million reports, though the differences are below 0.5% in either accuracy or F1 score.
We visualized confusion matrices to compare BERT-base and RadBERT-RoBERTa as examples, shown in Figure 3. The comparison between Figure 3A and 3B demonstrated that the BERT-base model was confused by the categories “suspicious nodule-b” and “prior lung cancer,” while our RadBERT-RoBERTa model was able to classify the two categories with a lower error rate. Similar results were observed in Figure 3C and 3D, where the BERT-base model confused the class “major abnormality, no attn needed” with other classes often, while our pretrained RadBERT-RoBERTa model more accurately distinguished between these classes.
Figure 3:
Confusion matrices for report coding with two language models (BERT-base and RadBERT-RoBERTa) fine-tuned to assign diagnostic codes in two coding systems (Lung Imaging Reporting and Data System [Lung-RADS] and abnormal) (see Appendix E4 [supplement]). (A, B) The Lung-RADS dataset consisted of six categories: “incomplete,” “benign nodule appearance or behavior,” “probably benign nodule,” “suspicious nodule-a,” “suspicious nodule-b,” and “prior lung cancer,” denoted as numbers 1 to 6 in the figure. (C, D) The abnormal dataset also consisted of six categories: “major abnormality,” “no attn needed,” “major abnormality, physician aware,” “minor abnormality,” “possible malignancy,” “significant abnormality, attn needed,” and “normal.” The figures show that RadBERT-RoBERTa improved from BERT-base by better distinguishing code numbers 5 and 6 for Lung-RADS and making fewer errors for code number 1 of the abnormal dataset. BERT = bidirectional encoder representations from transformers, RadBERT = BERT-based language model adapted for radiology, RoBERTa = robustly optimized BERT pretraining approach.
Task 3: Summarization
In this task, we extracted sentence embeddings from the models directly, without fine-tuning. Because extractive text summarization depends on how well a transformer represents sentences, the results quantitatively measure whether pretraining offers contextual sentence embeddings that reflect the semantics of the sentences. In addition to its potential use in radiology reporting, the task provides a useful test to investigate if pretraining on the radiology domain is helpful for learning better contextual word embeddings.
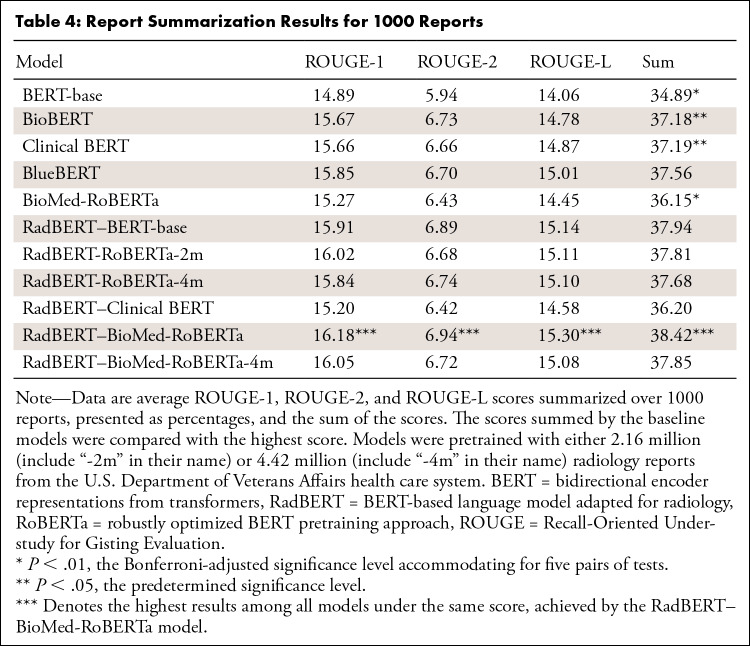
Table 4 reports the average ROUGE scores. There was a clear performance gain between RadBERT models and general BERT models (the relative improvement between each pair of models, eg, BERT-base and RadBERT-base in terms of all three ROUGE scores). The best-performing RadBERT variant is RadBERT–BioMed-RoBERTa, which outperformed all baseline models. The differences are statistically significant compared with BioBERT and Clinical-BERT (P < .05) and with BERT-base and BioMed-RoBERTa (P < .004). Other RadBERT variants also outperformed all baseline models, except RadBERT–Clinical BERT.
Table 4:
Report Summarization Results for 1000 Reports
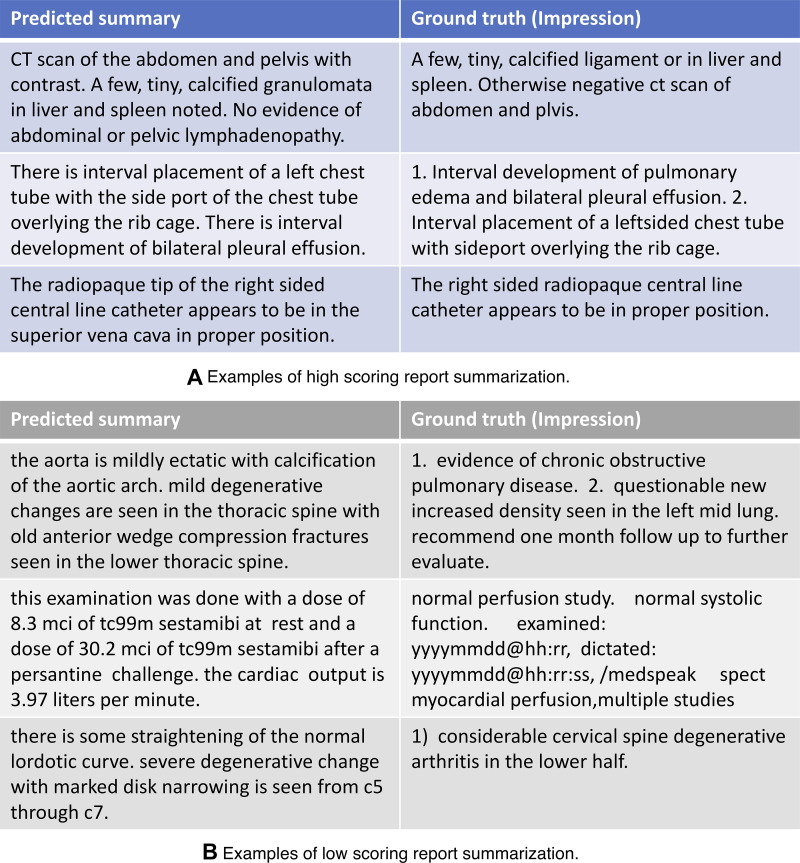
Figure 4 shows three high- and three low-scoring examples of predicted summarization by the best RadBERT model, along with their ground truth.
Figure 4:
Examples of (A) three high-scoring predicted summarizations and (B) three low-scoring predicted summarizations by RadBERT–BioMed-RoBERTa, the best-performing RadBERT model, and their corresponding ground truths (the impression section of input reports). BERT = bidirectional encoder representations from transformers, RadBERT = BERT-based language model adapted for radiology, RoBERTa = robustly optimized BERT pretraining approach.
Discussion
This paper reports our use of 4 million radiology reports from the VA nationwide to develop RadBERT, a family of language models tailored to facilitate the development of radiology NLP applications. RadBERT is based on the transformer architecture, a breakthrough in NLP. On top of RadBERT, one can apply fine-tuning to develop new site-specialized radiology NLP applications.
BERT (2) and RoBERTa (3) are language models that use pretraining objectives that are based on a “masked language model” to train a transformer deep neural network architecture (1), enabling the model to learn bidirectional representations and scale up with large training corpora. We chose BERT- and RoBERTa-based models for adaptation to the radiology domain because they provide a strong baseline and were used to train previous domain-specific models (eg, BioMed-RoBERTa [6], Clinical BERT [7]), making them suitable for fair evaluation and comparison of RadBERT performance.
From the results of the three radiology NLP tasks, we found strong experimental evidence with statistical significance that RadBERT models are superior to the baseline general domain or clinical-specific language models for radiology NLP tasks, suggesting that pretraining on radiology corpora is crucial when applying transformer-based language models to this specific domain. We also observed that RadBERT variants pretrained with different weight initializations from either the general domain (ie, models are initialized with BERT-base or RoBERTa weights) or biomedical domain (ie, models are initialized with Clinical BERT or BioMed-RoBERTa weights) performed similarly, suggesting that the performance gains were achieved mainly from adapting these models by pretraining on a large radiology report corpus.
Applying NLP to extract critical information from large volumes of radiology reports offers many opportunities to advance radiology and improve quality of care. One of the most clinically significant uses of radiology NLP is to identify radiographic examinations that require follow-up through automated identification of relevant abnormal findings and requests for follow-up in the radiology reports (eg, see a review in a study by Chen et al [18]). Every year, abnormal findings and subsequent recommendations in radiology reports are not acted upon. Inadequate follow-up can result in a combination of patient morbidity, patient mortality, and expensive litigation. One percent missed follow-ups over 10 000 cases amounts to 100 patients’ health. In some cases, their lives could be on the line if the abnormalities are life-threatening.
Effective radiology NLP may provide a solution of automated follow-up tracking by reliably minimizing missed follow-ups. However, the need for a large number of manually annotated training examples has long been a costly hurdle to applying deep learning to exploring radiology report documents, because the annotations would require substantial medical expertise. As a result, NLP still faces considerable challenges to being explored at scale and having its potential unleashed. However, RadBERT is particularly helpful when human-annotated data are scarce. Our results showed that with less than 5% of training data available, the performance gains between RadBERT and baseline models were greater than the results from training on 100% data, demonstrating the effectiveness in reducing the need for expensive human annotation by applying RadBERT. An effective radiology-specialized language model will expedite the development of deep learning–based radiology NLP applications that are cost affordable for medium-size nonresearch health care facilities, because a new application can be created by domain experts annotating a small number of radiology reports as training examples to fine-tune the pretrained radiology-specialized language model. Annotation and fine-tuning can be integrated into an easy-to-use tool to streamline the whole development process without programming. Such tools have been developed and commercialized for other domains (eg, Prodigy [19]). Adapting such tools to radiology will require enhanced data security and regulatory compliance.
There were still limitations to this study. First, the study did not exhaustively compare all possible transformer weight initializations. Because of the restrictions of our computational resources, we only trained BERT-base models with 110 million parameters. The BERT-large model with 340 million parameters was not tested, which could have potentially led to better performance with a larger architecture when trained with more data. Second, though we pretrained RadBERT variants with corpora containing either 2 million or 4 million radiology reports, the study did not reveal how many radiology reports are sufficient or whether there is an optimal amount for specialization pretraining, rather than “the more the better.” It is also interesting to investigate more fine-grained pretraining within the radiology domain, for example, focusing on specific modalities or body parts.
We presented RadBERT and demonstrated its effectiveness compared with five existing general or biomedical domain language models on performing three radiology NLP application tasks. These applications can save substantial time, ease the workload of radiologists and clinicians, and benefit patients. Abnormal sentence classification can help identify missed follow-ups. Report coding can standardize documentation for disease tracking and surveillance. Report summarization helps reduce radiologist workload and burnout. There are other radiology NLP tasks that may become feasible to explore in the future with RadBERT, for example, automated radiology report generation (eg, see studies by Ni et al [20] and Yan et al [21]).
Supported by award no. W81XWH–20–1–0693, log no. DM190543, Accelerating Innovation in Military Medicine Program, Office of the Assistant Secretary of Defense for Health Affairs, Department of Defense; and Career Award no. 1750063, National Science Foundation.
Disclosures of conflicts of interest: A.Y. No relevant relationships. J.M. No relevant relationships. X.L. No relevant relationships. J.D. No relevant relationships. E.Y.C. No relevant relationships. A.G. Department of Defense grant paid to UCSD (covers a small percentage of the author's salary). C.N.H. No relevant relationships.
Abbreviations:
- BERT
- bidirectional encoder representations from transformers
- MIMIC
- Medical Information Mart for Intensive Care
- NLP
- natural language processing
- RadBERT
- BERT-based language model adapted for radiology
- RoBERTa
- robustly optimized BERT pretraining approach
- ROUGE
- Recall-Oriented Understudy for Gisting Evaluation
- VA
- U.S. Department of Veterans Affairs
References
- 1. Vaswani A , Shazeer N , Parmar N , et al . Attention is all you need . In: Advances in Neural Information Processing Systems 30 (NIPS 2017) , 2017. ; 5998 – 6008 . https://papers.nips.cc/paper/2017/hash/3f5ee243547dee91fbd053c1c4a845aa-Abstract.html .
- 2. Devlin J , Chang MW , Lee K , Toutanova K . BERT: Pretraining of deep bidirectional transformers for language understanding . arXiv:1810.04805 [preprint] https://arxiv.org/abs/1810.04805. Posted October 11, 2018. Accessed June 7, 2022 .
- 3. Liu Y , Ott M , Goyal N , et al . RoBERTa: A robustly optimized BERT pretraining approach . arXiv:1907.11692 [preprint] https://arxiv.org/abs/1907.11692. Posted July 26, 2019. Accessed June 7, 2022 .
- 4. Mikolov T , Chen K , Corrado G , Dean J . Efficient estimation of word representations in vector space . arXiv:1301.3781 [preprint] https://arxiv.org/abs/1301.3781. Posted January 16, 2013. Accessed June 7, 2022 .
- 5. Lee J , Yoon W , Kim S , et al . BioBERT: a pre-trained biomedical language representation model for biomedical text mining . Bioinformatics 2020. ; 36 ( 4 ): 1234 – 1240 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gururangan S , Marasović A , Swayamdipta S , et al . Don't stop pretraining: Adapt language models to domains and tasks . arXiv:2004.10964 [preprint] https://arxiv.org/abs/2004.10964. Posted April 23, 2020. Accessed June 7, 2022 .
- 7. Alsentzer E , Murphy JR , Boag W , et al . Publicly available clinical BERT embeddings . arXiv:1904.03323 [preprint] https://arxiv.org/abs/1904.03323. Posted April 6, 2019. Accessed June 7, 2022 .
- 8. Peng Y , Yan S , Lu Z . Transfer learning in biomedical natural language processing: An evaluation of BERT and ELMo on ten benchmarking datasets . arXiv:1906.05474 [preprint] https://arxiv.org/abs/1906.05474. Posted June 13, 2019. Accessed June 7, 2022 .
- 9. Johnson AEW , Pollard TJ , Shen L , et al . MIMIC-III, a freely accessible critical care database . Sci Data 2016. ; 3 : 160035 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gabriel RA , Kuo TT , McAuley J , Hsu CN . Identifying and characterizing highly similar notes in big clinical note datasets . J Biomed Inform 2018. ; 82 : 63 – 69 . [DOI] [PubMed] [Google Scholar]
- 11. Raffel C , Shazeer N , Roberts A , et al . Exploring the limits of transfer learning with a unified text-to-text transformer . J Mach Learn Res 2020. ; 21 ( 140 ): 1 – 67 . https://jmlr.org/papers/v21/20-074.html . 34305477 [Google Scholar]
- 12. Wu Y , Schuster M , Chen Z , et al . Google's neural machine translation system: Bridging the gap between human and machine translation . arXiv:1609.08144 [preprint] https://arxiv.org/abs/1609.08144. Posted September 26, 2016. Accessed June 7, 2022 .
- 13. Harzig P , Chen YY , Chen F , Lienhart R . Addressing data bias problems for chest x-ray image report generation . arXiv:1908.02123 [preprint] https://arxiv.org/abs/1908.02123. Posted August 6, 2019. Accessed June 7, 2022 .
- 14. Demner-Fushman D , Kohli MD , Rosenman MB , et al . Preparing a collection of radiology examinations for distribution and retrieval . J Am Med Inform Assoc 2016. ; 23 ( 2 ): 304 – 310 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Miller D . Leveraging BERT for extractive text summarization on lectures . arXiv:1906.04165 [preprint] https://arxiv.org/abs/1906.04165. Posted June 7, 2019. Accessed June 7, 2022 .
- 16. Lin CY . ROUGE: A package for automatic evaluation of summaries . In: Text Summarization Branches Out , Barcelona, Spain: , July 2004 . Association for Computational Linguistics; , 2004. ; 74 – 81 . [Google Scholar]
- 17. Smith L , Tanabe LK , Ando RJ , et al . Overview of BioCreative II gene mention recognition . Genome Biol 2008. ; 9 ( Suppl 2 ): S2 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen MC , Ball RL , Yang L , et al . Deep learning to classify radiology free-text reports . Radiology 2018. ; 286 ( 3 ): 845 – 852 . [DOI] [PubMed] [Google Scholar]
- 19. Prodigy Web site . https://prodi.gy/. Published 2017. Accessed October 8, 2021 .
- 20. Ni J , Hsu CN , Gentili A , McAuley J . Learning visual-semantic embeddings for reporting abnormal findings on chest X-rays . arXiv:2010.02467 [preprint] https://arxiv.org/abs/2010.02467. Posted October 6, 2020. Accessed June 7, 2022 .
- 21. Yan A , He Z , Lu X , et al . Weakly supervised contrastive learning for chest x-ray report generation . arXiv:2109.12242 [preprint] https://arxiv.org/abs/2109.12242. Posted September 25, 2021. Accessed June 7, 2022 .
- 22. Fiorini N , Leaman R , Lipman DJ , Lu Z . How user intelligence is improving PubMed . Nat Biotechnol 2018. ; 36 ( 10 ): 937 – 945 . [DOI] [PubMed] [Google Scholar]
- 23. Neamatullah I , Douglass MM , Lehman LW , et al . Automated de-identification of free-text medical records . BMC Med Inform Decis Mak 2008. ; 8 ( 1 ): 32 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Goldberger AL , Amaral LA , Glass L , et al . PhysioBank, PhysioToolkit, and PhysioNet: components of a new research resource for complex physiologic signals . Circulation 2000. ; 101 ( 23 ): E215 – E220 . [DOI] [PubMed] [Google Scholar]
- 25. VA Office of Information and Technology . VA technical reference model v 22.5 . https://www.oit.va.gov/Services/TRM/TRMHomePage.aspx. Published 2021. Accessed June 23, 2022 .
- 26. Kingma DP , Ba J . Adam: A method for stochastic optimization . arXiv:1412.6980 [preprint] https://arxiv.org/abs/1412.6980. Posted December 22, 2014. Accessed June 7, 2022 .



![Overview of our study design, which includes pretraining and fine-tuning of RadBERT. (A) In pretraining, different weight initializations were considered to create variants of RadBERT. (B) The variants were fine-tuned for three important radiology natural language processing (NLP) tasks: abnormal sentence classification, report coding, and report summarization. The performance of RadBERT variants for these tasks was compared with a set of intensively studied transformer-based language models as baselines. (C) Examples of each task and how performance was measured. In the abnormality identification task, a sentence in a radiology report was considered “abnormal” if it reported an abnormal finding and “normal” otherwise. A human-annotated abnormality was considered ground truth to evaluate the performance of an NLP model. In the code classification task, models were expected to output diagnostic codes (eg, abdominal aortic aneurysm, Breast Imaging Reporting and Data System [BI-RADS], and Lung Imaging Reporting and Data System [Lung-RADS]) that match the codes given by human providers as the ground truth for a given radiology report. During report summarization, the models generated a short summary given the findings in a radiology report. Summary quality was measured by how similar it was to the impression section of the input report. AAA = abdominal aortic aneurysm, BERT = bidirectional encoder representations from transformers, RadBERT = BERT-based language model adapted for radiology, RoBERTa = robustly optimized BERT pretraining approach.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/9343/9344353/59525b12c91e/ryai.210258.fig1.jpg)
![Confusion matrices for report coding with two language models (BERT-base and RadBERT-RoBERTa) fine-tuned to assign diagnostic codes in two coding systems (Lung Imaging Reporting and Data System [Lung-RADS] and abnormal) (see Appendix E4 [supplement]). (A, B) The Lung-RADS dataset consisted of six categories: “incomplete,” “benign nodule appearance or behavior,” “probably benign nodule,” “suspicious nodule-a,” “suspicious nodule-b,” and “prior lung cancer,” denoted as numbers 1 to 6 in the figure. (C, D) The abnormal dataset also consisted of six categories: “major abnormality,” “no attn needed,” “major abnormality, physician aware,” “minor abnormality,” “possible malignancy,” “significant abnormality, attn needed,” and “normal.” The figures show that RadBERT-RoBERTa improved from BERT-base by better distinguishing code numbers 5 and 6 for Lung-RADS and making fewer errors for code number 1 of the abnormal dataset. BERT = bidirectional encoder representations from transformers, RadBERT = BERT-based language model adapted for radiology, RoBERTa = robustly optimized BERT pretraining approach.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/9343/9344353/84e245f2aade/ryai.210258.fig3.jpg)