Abstract
Salmonella typhimurium in vivo-induced (ivi) genes were grouped by their coordinate behavior in response to a wide variety of environmental and genetic signals, including pH, Mg2+, Fe2+, and PhoPQ. All of the seven ivi fusions that are induced by both low pH and low Mg2+ (e.g., iviVI-A) are activated by the PhoPQ regulatory system. Iron-responsive ivi fusions include those induced under iron limitation (e.g., entF) as well as one induced by iron excess but only in the absence of PhoP (pdu). Intracellular expression studies showed that each of the pH- and Mg2+-responsive fusions is induced upon entry into and growth within three distinct mammalian cell lines: RAW 264.7 murine macrophages and two cultured human epithelial cell lines: HEp-2 and Henle-407. Each ivi fusion has a characteristic level of induction consistent within all three cell types, suggesting that this class of coordinately expressed ivi genes responds to general intracellular signals that are present both in initial and in progressive stages of infection and may reflect their responses to similar vacuolar microenvironments in these cell types. Investigation of ivi expression patterns reveals not only the inherent versatility of pathogens to express a given gene(s) at various host sites but also the ability to modify their expression within the context of different animal hosts, tissues, cell types, or subcellular compartments.
We have implemented a genetic system, termed IVET (in vivo expression technology), to identify bacterial genes that are induced during infection (26, 27). Such in vivo-induced (ivi) genes are expressed poorly on rich laboratory media (i.e., Luria broth [LB] or lactose MacConkey) but undergo elevated levels of expression in host tissues. Using this system, we previously identified over 100 different Salmonella typhimurium genes that are induced during infection of BALB/c mice and/or RAW 264.7 cultured murine macrophages (23). The functions of 25% of these ivi genes are unknown: they show no significant homology to entries in the DNA databases (8, 24). Many ivi genes reside in Salmonella-specific regions of atypical base composition that encode predicted adhesin and invasin-like functions required for full virulence. Several of these Salmonella-specific regions are inherited in a serovar-specific fashion (9).
Many virulence genes shown to be coordinately regulated in vitro are presumed to function at the same anatomical site during infection. For example, the production of cholera toxin in vitro is coordinately regulated by the same environmental (pH, temperature, and osmolarity) and genetic (e.g., ToxR) signals as production of the toxin-coregulated pilus, both of which function in the small intestine (28, 42). Thus, independent of the actual (often unknown) in vivo signals, classification of bacterial genes based on regulatory patterns in vitro may reflect coordinate expression at a given anatomical site as well as a possible functional relationship (i.e., in the same biochemical pathway or in a broader context, such as phagosome survival).
Here, we have implemented the lac fusion technology of the IVET approach to classify ivi genes isolated from different host tissues (spleen, liver, intestine) based on their coordinate expression patterns in response to environmental (pH, Mg2+, and Fe2+) and regulatory (PhoPQ) conditions known to control virulence gene expression in vitro (28–30). Low pH and low Mg2+ have been associated with Salmonella-containing vacuoles (15) and have been shown to be relevant signals for bacterial genes presumed to function in the macrophage (16, 39, 40). Low Mg2+ is the inducing signal for the activation of the PhoPQ regulatory system (16) which controls Salmonella virulence (18), including functions required for survival within (31) and during acidification of (2) the macrophage phagosome. Additionally, iron limitation is a well-characterized barrier to infection and thus may serve as an important environmental signal influencing bacterial gene expression in the animal (11, 28).
Here we demonstrate that ivi genes which are coordinately induced in response to low pH and low Mg2+ also displayed coordinate induction upon entry into cultured murine macrophages and cultured human epithelial cells. These data suggest this coordinately expressed class of ivi genes responds to general intracellular signals present during both early and late stages of infection.
MATERIALS AND METHODS
Media.
Laboratory media used in these studies included LB (10) and MOPS [3-(N-morpholino)propanesulfonic acid] minimal medium (33) with 0.4% glycerol as a carbon source to avoid catabolite repression effects. When necessary, the pH was adjusted to pH 7.6, 7.0, 6.6, or 5.5. To buffer media to pH 5.5, MOPS was replaced with MES [2-(N-morpholino)ethanesulfonic acid] at the same concentration. MOPS medium contains 10 μM iron. Medium stocks were prepared iron or Mg2+ free when required, and FeSO4 or MgCl2 was added at the desired concentration. Unless specified otherwise, final concentrations of antibiotics (Sigma) in LB and minimal medium were as follows: ampicillin, 50 and 15 μg/ml; tetracycline, 20 and 10 μg/ml; and chloramphenicol, 20 and 5 μg/ml.
Bacterial strains and phage.
All S. typhimurium strains used in this study were derived from strain ATCC 14028 (CDC 6516-60). The high-frequency generalized transducing bacteriophage P22 mutant HT 105/1, int-201 was used for all transductional crosses (36), and phage-free, phage-sensitive transductants were isolated as previously described (7). Strains used for PhoPQ regulation studies were constructed by transduction of the IVET-selected fusion into ATCC 14028 (wild type) and isogenic phoPQ derivatives MT1657 (phoP102::Tn10d-Cm [32]) and MT1658 (phoQ24 [31]), kindly provided by Karl Klose (University of Texas, San Antonio).
Cell culture.
The murine macrophage cell line RAW 264.7 and human epithelial cell lines HEp-2 (larynx carcinoma) and Henle-407 (embryonic small intestine) (ATCC TIB-71, CCL-23, and CCL-6, respectively) were obtained from the American Type Culture Collection, Rockville, Md., and maintained in minimum essential medium (MEM) supplemented with Earle’s salts, l-glutamine, and 10% heat-inactivated fetal calf serum (FCS) (Life Technologies, Rockville, Md.). Cells were grown in a humidified atmosphere of 5% carbon dioxide and 95% air at 37°C in 75-cm2 plastic flasks (Corning Glass Works, Corning, N.Y.). Cultured macrophages were harvested by scraping with a rubber policeman, or in the case of epithelial cells, by trypsinization using 0.25% trypsin–0.02% EDTA, and plated at a density of 2.5 × 105 to 5 × 105 cells/ml in 4 ml of culture medium in 35-mm-diameter, six-well dishes (Corning) and grown for 24 h to approximately 80 to 90% confluence (1 × 106 to 5 × 106 cells/well) (14).
Cultured macrophage and epithelial cell infections.
S. typhimurium was grown overnight without shaking in LB containing ampicillin. Cultured cells were inoculated with S. typhimurium at a bacterium/host cell ratio of 10:1. Fifty microliters of an overnight culture of bacteria (5 × 107) was added to 5 × 106 cultured macrophages in six-well cell culture plates (Corning); 107 bacteria were added to 106 cultured epithelial cells. The bacteria were centrifuged onto cultured monolayers at 1,000 × g for 10 min at room temperature, after which they were incubated at 37°C for 30 min in a 5% CO2 incubator. The coculture was washed twice with cell culture medium and incubated for 2 h in the presence of 100 μg of gentamicin per ml to kill extracellular bacteria (17). Cells were washed twice with cell culture medium and incubated for the time specified in cell culture medium containing 10 μg of gentamicin per ml. The coculture was then washed three times with ice-cold 1× phosphate-buffered saline (PBS), the host cells were lysed with 1% Triton X-100, and the surviving intracellular bacteria were recovered in 1× PBS. β-Galactosidase assays were performed on the recovered bacterial cells, which were also plated for single colonies to determine bacterial cell number.
β-Galactosidase assays.
β-Galactosidase activities were assayed by the method of Slauch and Silhavy (37). Unless otherwise specified, activities are given as 103 units per A600 unit per milliliter of cell suspension, where units are micromoles of o-nitrophenol (ONP) formed per minute (n = 3 trials, standard deviation < 10% of the mean).
Mg2+ and pH assays.
Bacterial strains were used to inoculate 1 ml of minimal medium at the given pH (7.6, 6.6, or 5.5) and Mg2+ concentration (10, 1, 0.05, or 0 mM). When Mg2+ was omitted from the incubation medium, sufficient growth was obtained to reach a final optical density at 600 nm (OD600) of at least 0.2 for the phoP+ strains; phoP and phoQ24 strains failed to grow under these conditions. The cultures were shaken at 37°C, and β-galactosidase assays were performed after 16 h. To assay cells grown in exponential phase, growth curves were determined for each environmental condition. Overnight cultures grown in the 12 assay conditions tested were diluted (typically 12- to 30-fold) to an OD600 of 0.1 in the same medium (cells were not tested in the absence of Mg2+ since the strains do not grow well in this medium after dilution). The subcultures were grown at 37°C and assayed at an OD600 of 0.4 to 0.6.
Iron assays.
Bacterial strains were used to inoculate 1 ml of MOPS minimal medium at the designated pH and Mg2+ concentration with the addition of the given concentration of FeSO4. Medium conditions without iron added also contained the iron-chelating agent 2,2′-dipyridyl (0.03 mM). The cultures were shaken at 37°C, and β-galactosidase assays were performed after 16 h of incubation.
Cell culture assays.
Bacterial cells were recovered from infected cultured macrophages or epithelial cells lysed with 1% Triton X-100 in 1× PBS. β-Galactosidase activities typically were assayed on 0.5 ml of the 1 ml coculture lysate. The activity was determined as described by Slauch and Silhavy (37), with the modification that the number of bacteria was determined by direct colony count; units were expressed as micromoles of ONP formed per minute per CFU. For comparison, bacteria were also grown in 1 ml of LB and cell culture medium containing 10% FCS without shaking; assays performed on the recovered bacteria (typically 0.1 ml) were determined as in the cell culture assay. β-Galactosidase activities for experiments were determined in triplicate on at least two different days.
RESULTS
pH and Mg2+ regulation of ivi genes.
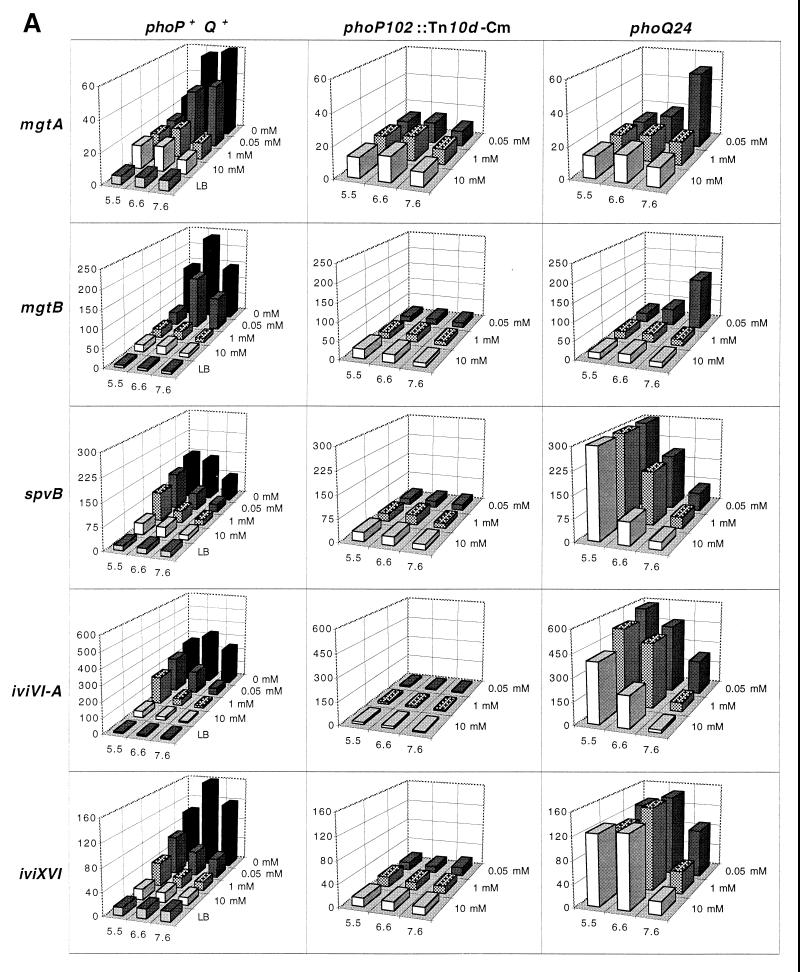
We have tested the effects of the environmental signals pH and Mg2+ on the expression patterns of 74 IVET-selected bacterial gene fusions recovered from infected BALB/c mice (intestine, spleen, and liver) and/or from cultured RAW 264.7 murine macrophages (23). These environmental signals have been associated with Salmonella-containing vacuoles (15) and are presumed to be relevant signals in the macrophage environment (16, 39, 40). The strains used in this study are shown in Table 1, and the environmental and genetic parameters governing their expression are summarized in Table 2. The strains presented in Table 2 exhibited a greater than twofold response to the signals tested. Figure 1 shows the responses of eight ivi fusions following 16 h of growth under a wide variety of Mg2+ and pH conditions. Fusions in strains that are wild type with respect to phoPQ were also assayed in exponential phase; both the actual values of β-galactosidase activity and the expression pattern of each fusion were similar to those observed after 16 h of growth (data not shown).
TABLE 1.
S. typhimurium strains used in this study.
| Straina |
Genotype |
|---|---|
| MT1228 | DUP3169 [Φ(mgtA′-cat+-lacZ+Y+)*pIVET8*(mgtA+)] |
| MT1865 | DUP3169 [Φ(mgtA′-cat+-lacZ+Y+)*pIVET8*(mgtA+)] phoP102::Tn10d-Cm |
| MT1866 | DUP3169 [Φ(mgtA′-cat+-lacZ+Y+)*pIVET8*(mgtA+)] phoQ24 |
| MT1695 | DUP3170 [Φ(mgtB′-cat+-lacZ+Y+)*pIVET8*(mgtB+)] |
| MT1696 | DUP3170 [Φ(mgtB′-cat+-lacZ+Y+)*pIVET8*(mgtB+)] phoP102::Tn10d-Cm |
| MT1697 | DUP3170 [Φ(mgtB′-cat+-lacZ+Y+)*pIVET8*(mgtB+)] phoQ24 |
| MT1701 | DUP3210 [Φ(spvB′-cat+-lacZ+Y+)*pIVET8*(spvB+)] |
| MT1702 | DUP3210 [Φ(spvB′-cat+-lacZ+Y+)*pIVET8*(spvB+)] phoP102::Tn10d-Cm |
| MT1703 | DUP3210 [Φ(spvB′-cat+-lacZ+Y+)*pIVET8*(spvB+)] phoQ24 |
| MT1704 | DUP3188 [Φ(iviVI-A′-cat+-lacZ+Y+)*pIVET8*(iviVI-A+)] |
| MT1705 | DUP3188 [Φ(iviVI-A′-cat+-lacZ+Y+)*pIVET8*(iviVI-A+)] phoP102::Tn10d-Cm |
| MT1706 | DUP3188 [Φ(iviVI-A′-cat+-lacZ+Y+)*pIVET8*(iviVI-A+)] phoQ24 |
| MT1818 | DUP3350 [Φ(iviXVI′-cat+-lacZ+Y+)*pIVET8*(iviXVI+)] |
| MT1898 | DUP3350 [Φ(iviXVI′-cat+-lacZ+Y+)*pIVET8*(iviXVI+)] phoP102::Tn10d-Cm |
| MT1899 | DUP3350 [Φ(iviXVI′-cat+-lacZ+Y+)*pIVET8*(iviXVI+)] phoQ24 |
| MT1371 | DUP3193 [Φ(phoP′-purA+-lacZ+Y+)*pIVET1*(phoP+)] |
| MT1681 | DUP3193 [Φ(phoP′-purA+-lacZ+Y+)*pIVET1*(phoP102::Tn10d-Cm)] |
| MT1680 | DUP3193 [Φ(phoP′-purA+-lacZ+Y+)*pIVET1*(phoP+)] phoQ24 |
| MT1698 | DUP3465 [Φ(pmrB′-cat+-lacZ+Y+)*pIVET8*(pmrB+)] |
| MT1699 | DUP3465 [Φ(pmrB′-cat+-lacZ+Y+)*pIVET8*(pmrB+)] phoP102::Tn10d-Cm |
| MT1700 | DUP3465 [Φ(pmrB′-cat+-lacZ+Y+)*pIVET8*(pmrB+)] phoQ24 |
| MT1819 | DUP3371 [Φ(iviXVII′-cat+-lacZ+Y+)*pIVET8*(iviXVII+)] |
| MT1900 | DUP3371 [Φ(iviXVII′-cat+-lacZ+Y+)*pIVET8*(iviXVII+)] phoP102::Tn10d-Cm |
| MT1901 | DUP3371 [Φ(iviXVII′-cat+-lacZ+Y+)*pIVET8*(iviXVII+)] phoQ24 |
| MT1772 | DUP3142 [Φ(fhuA′-purA+-lacZ+Y+)*pIVET1*(fhuA+)] |
| MT1833 | DUP3127 [Φ(cirA′-cat+-lacZ+Y+)*pIVET8*(cirA+)] |
| MT1801 | DUP3231 [Φ(entE′-cat+-lacZ+Y+)*pIVET8*(entF+)] |
All derived from S. typhimurium ATCC 14028 (CDC 6516-60).
TABLE 2.
Environmental and genetic regulation of S. typhimurium ivi genes
| Strain | Gene | Function | Environmental signal(s) | PhoPQ regulation | Parameters | Reference(s)a |
|---|---|---|---|---|---|---|
| MT1371 | phoP | Virulence regulator | pH + Mg2+ | pag | i.g., i.p./spleenb | 16, 23, 38, 40, this work |
| MT1698 | pmrB | Polymyxin resistance | pH + Mg2+ | pag | Cultured macrophage | 21, 23, 39, this work |
| MT1701 | spvB | Plasmid virulence | pH + Mg2+ | pag | i.p./spleen | 13, 23, this work |
| MT1228 | mgtA | Mg2+ transport | pH + Mg2+ | pag | i.p./spleen; cultured macrophage | 16, 23, 39, 40, this work |
| MT1695 | mgtB | Mg2+ transport | pH + Mg2+ | pag | i.p./spleen | 15, 16, 23, 40, 41, this work |
| MT1704 | iviVI-A | Adhesin-like | pH + Mg2+ | pag | i.p./spleen | 23, this work |
| MT1818 | iviXVI | Aldehyde dehydrogenase-like | pH + Mg2+ | pag | i.p./spleen | This work |
| MT1819 | iviXVII (pdu) | 1,2-Propanediol utilization | pH + Mg2+, iron O2, propanediol | prg | i.p./spleen | 1, 5, this work |
| MT1772 | fhuA | Iron transport | Iron | None | i.g./small intestine | 23, 41, this work |
| MT1833 | cirA | Colicin 1 receptor | Iron | None | Cultured macrophage | 23, this work |
| MT1801 | entF | Enterobactin synthesis | Iron | None | Cultured macrophage | 23, this work |
Listed are references relevant to environmental signals that affect ivi gene expression in S. typhimurium.
Route of delivery (intragastric [i.g.] or intraperitoneal [i.p.])/host tissue from which bacteria were recovered.
FIG. 1.
Expression profiles of ivi fusions in response to pH and Mg2+ concentration. pH values are represented on the x axis, β-galactosidase activities [(micromoles of ONP formed per minute per A600 unit per milliliter of cell suspension] [103]) are plotted on the y axis, and Mg2+ concentrations are plotted on the z axis. Values represent the averages of three independent cultures with a standard deviation of <10% of the mean. (A) Seven PhoP-activated ivi fusions. (B) PhoP-repressed fusion iviXVII (pdu). In panels A and B, the columns represent, from left to right, expression profiles in a PhoP+ background, in a PhoP− (phoP102::Tn10d-Cm) background, and in a phoQ24 background. A very similar induction profile was obtained for exponentially growing cells of the wild type under all 15 conditions tested (data not shown). The pH of each wild-type culture was quantitated under the 15 conditions tested before and after incubation. After 16 h of growth, the pH in MOPS minimal medium typically decreased 1 to 1.8 pH units; the pH in MOPS-buffered LB increased 1.0 to 1.5 pH units. The pH of exponentially grown cultures in MOPS minimal medium typically decreased 0.4 to 0.6 pH units in MOPS minimal medium or remained within 0.4 pH units in MOPS-buffered LB.
Figure 1 shows a set of eight ivi fusions that respond to pH and Mg2+ in the media; each fusion displays a characteristic response to these signals. Examples include five known ivi genes and three unknown ivi genes, some of which were shown previously to respond to these environmental conditions (referenced in Table 2). Known genes include mgtA and mgtB which encode two high-affinity Mg2+ transport systems; mgtB, part of an operon with a gene (mgtC) residing on Salmonella pathogenicity island 3 and required for intramacrophage survival (4); spvB, which resides on the Salmonella virulence plasmid and facilitates growth at systemic sites of infection (19); and phoP and pmrAB, which encode two-component regulatory systems that affect the expression of Salmonella virulence genes (18). Unknown genes include iviVI-A (23), which encodes a predicted product that shows extensive sequence similarity to enterotoxigenic Escherichia coli Tia, an outer membrane protein involved in attachment to and invasion of gut epithelial cells (14). We have mapped iviXVI to between min 30 and 37 on the Salmonella chromosome (35), and its predicted amino acid sequence (using 262 bp from the fusion joint) shows similarity (57 to 62% identity) to several aldehyde dehydrogenases of E. coli. We have recently identified iviXVII as a member of the pdu operon (9), involved in vitamin B12-dependent 1,2-propanediol utilization (34).
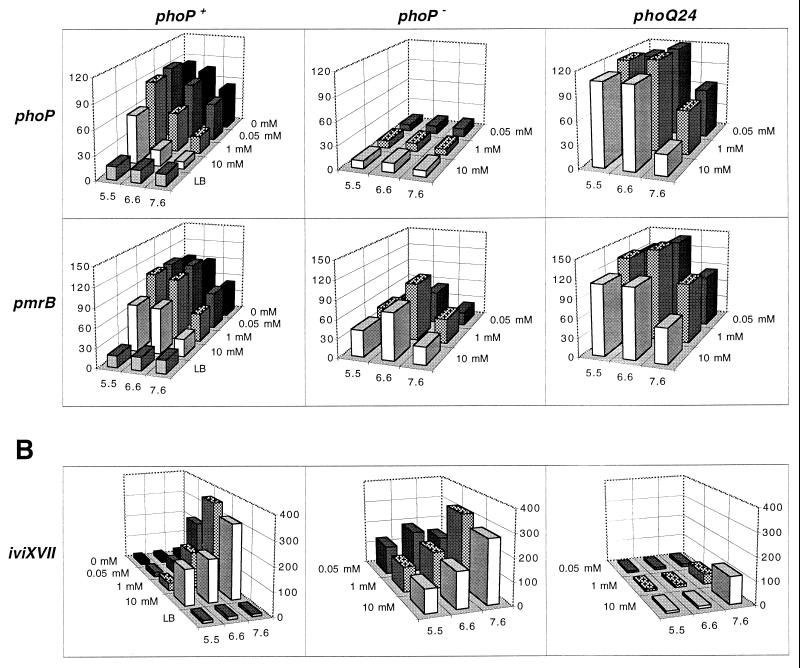
Figure 1A shows that seven of eight ivi fusions recovered from the spleen and/or cultured macrophages are induced by both low pH and low Mg2+, an environment likely to be found within Salmonella-containing vacuoles (15). However, although iviXVII (pdu) was also recovered from the spleen, Fig. 1B shows that it exhibits a reciprocal pattern of expression with regard to pH and Mg2+: it is induced when grown under high-Mg2+ and mild alkaline conditions but repressed when grown under low-pH and low-Mg2+ conditions. Thus, although pdu is capable of responding to signals in a different fashion than the other ivi fusions, the data in Fig. 1B and 2 show that under conditions presumed to exist in vacuolar environments, pdu expression may have been sufficient for IVET selection in the spleen.
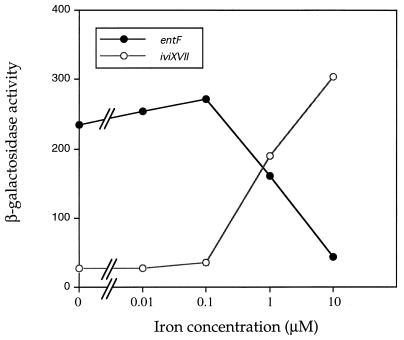
FIG. 2.
iviXVII (pdu) and entF expression as a function of iron concentration in the medium. Cultures were grown overnight in iron-free MOPS minimal medium at pH 7.0. The cultures were diluted to OD600 of 0.1 in the same medium grown to an OD600 of 0.4 (mid-log phase). FeSO4 was then added at the given concentration, and β-galactosidase activities were determined after 5 h. Values represent the averages of three independent cultures with a standard deviation of <10% of the mean.
Regulation of ivi genes by PhoPQ.
To determine if the environmental regulation was mediated through the PhoPQ regulatory system, the expression patterns of ivi fusions found in the wild type were compared to those observed in a PhoP− (phoP102::Tn10d-Cm) genetic background. Figure 1A (second column) shows that all of the eight ivi genes that respond to both pH and Mg2+ are also PhoP-regulated genes (the responses are summarized in Table 2). Seven of these (mgtA, mgtB, spvB, phoP, pmrB, iviVI-A, and iviXVI) are PhoP-activated genes (pags [16, 21, 23, 38–40]), since their expression is dependent on the presence of a functional PhoP regulatory protein. Note that there is suboptimal expression of pmrB in the absence of PhoP under several environmental conditions (e.g., pH 6.6 and 1 mM Mg2+). Conversely, Fig. 1B shows that the expression levels of iviXVII (pdu) are increased under several conditions in the absence of PhoP, and thus iviXVII is defined as a PhoP-repressed gene (prg).
The conformation of the PhoQ sensor protein has been shown to respond to levels of Mg2+ in the medium (16). The phoQ24 mutation renders the PhoQ protein less sensitive to its known inhibitory signal, Mg2+ (16), resulting in enhanced phosphorylation of PhoP (20). Such enhanced PhoP phosphorylation causes increased expression of pags and reduced expression of prgs (31). As expected, the effect of phoQ24 is to amplify the expression of the PhoP-activated genes spvB, phoP, pmrB, iviVI-A, and iviXVI compared to that found in the wild type over a wide range of conditions (Fig. 1A, third column). Figure 1B shows that phoQ24 abolishes the expression of iviXVII (pdu) under most conditions tested, as expected for a prg.
Iron regulation of ivi genes.
The 74 IVET-selected fusions that were tested for their response to Mg2+ and pH were also screened for their response to iron. Three ivi fusions, fhuA, cirA, and entF, are induced 5-, 8-, and 22-fold, respectively, by iron limitation (no iron added, 0.03 mM 2,2′-dipyridyl) compared to growth in iron excess (0.01 mM iron), as shown previously for E. coli (reference 11 and data not shown). Each of these ivi genes is presumed to play a role in iron transport: fhuA encodes a siderophore uptake system; cirA encodes the colicin 1 receptor, which also shows extensive homology to siderophore uptake systems; and entF encodes enterobactin synthetase component F (11). Conversely, iviXVII (pdu) expression increased 13-fold when cells were grown under high versus low iron conditions in minimal medium but not in the high-iron conditions present in LB (10 μM [reference 17 and data not shown]). Thus, all iron-responsive ivi fusions are repressed when grown in LB but must be expressed in the animal in response to the IVET selection.
To resolve the apparent paradox that reciprocally regulated fusions (with respect to iron) are repressed in LB and expressed in the animal (and in cultured mammalian cells [see below]), the effect of iron concentration on entF and iviXVII expression was quantitated. Figure 2 shows that in 1 μM iron (the concentration indirectly estimated for Salmonella-containing vacuoles [15]), both entF and iviXVII are expressed at high levels, presumably sufficient for both to respond to the IVET selection.
Table 3 shows the effects of phoPQ, medium composition (pH and Mg2+), and iron on expression of iviXVII (pdu) in vitro. The data indicate that the ability of iviXVII to respond to high iron is dependent on the absence of the PhoP regulatory protein. This can be achieved by either growth condition or mutation. For example, iviXVII is induced by iron under growth conditions where PhoP is not expressed (pH 7.0 to 7.6 and 0.5 to 10 mM Mg2+ [Fig. 1B]) or in a PhoP− background.
TABLE 3.
Effects of phoPQ, medium composition, and iron on expression of a iviXVII::lac (pdu) fusion
| iviXVII::lac derivative | β-Galactosidase activity (U)a
|
|||||
|---|---|---|---|---|---|---|
| pH 5.5, 0.05 mM Mg2+
|
pH 7.0, 0.05 mM Mg2+
|
pH 7.6, 10 mM Mg2+
|
||||
| −Iron | +Iron | −Iron | +Iron | −Iron | +Iron | |
| phoP+ | 22 | 25 | 27 | 288 | 27 | 322 |
| phoP102::Tn10d-Cm | 13 | 216 | 15 | 361 | 13 | 344 |
| phoQ24 | 11 | 10 | 12 | 15 | 13 | 247 |
Determined in cultures grown for 16 h in MOPS minimal medium at the given pH and Mg2+ concentration in the presence or absence of 50 μM iron. Medium without iron added also contained the iron-chelating agent 2,2′-dipyridyl (0.03 mM). Data represent averages of three independent cultures with a standard deviation of <10% of the mean.
Intracellular expression of ivi genes.
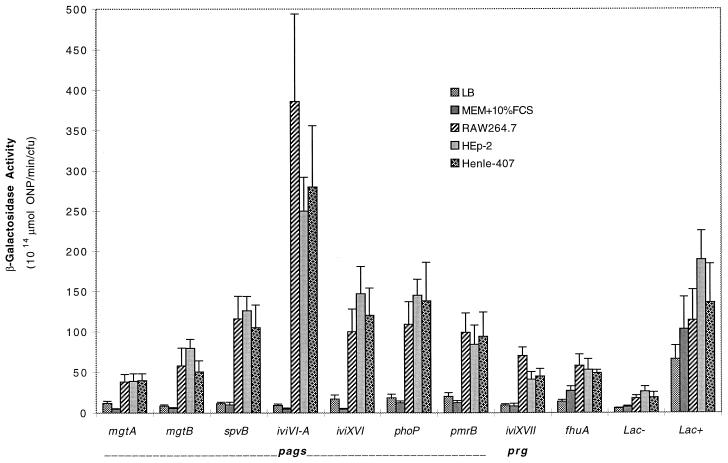
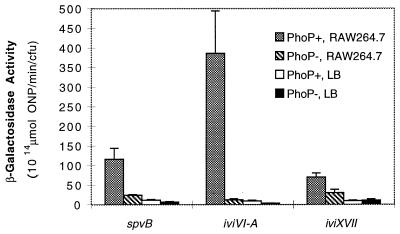
The eight ivi genes that respond to pH, Mg2+, and PhoP in vitro were tested for expression upon entry into and growth within three cultured host cell lines: RAW 264.7 (murine macrophage) and HEp-2 and Henle-407 (human epithelium). Preselected Lac− and Lac+ strains were obtained from the initial nonselected pool of integrated IVET fusions. Figure 3 indicates that these strains showed no significant intracellular induction relative to the PhoP-regulated ivi genes which were induced at 4 h following infection of all three cell lines. While each ivi fusion has a characteristic level of intracellular induction, that level is relatively consistent within all three cell lines tested, as shown previously for spvB in several cell lines (13).
FIG. 3.
Intracellular expression of ivi genes in cultured macrophages and cultured epithelial cells. Individual ivi fusions were incubated with cultured RAW 264.7 murine macrophages and cultured human epithelial cell lines HEp-2 (larynx carcinoma) and Henle-407 (embryonic small intestine). The coculture was incubated for 30 min, washed, treated with gentamicin to kill extracellular bacteria, washed, and incubated for 4 h. The cultured mammalian cells were lysed with Triton X-100, and β-galactosidase assays were performed on the recovered intracellular bacteria. For comparison, β-galactosidase assays were also performed on individual ivi fusions grown for 4 h in LB and in MEM supplemented with 10% FCS. Preselected Lac− and Lac+ strains were obtained from the initial nonselected pool of integrated IVET fusions. Error bars represent 1 standard deviation of the measured value.
Effects of PhoPQ on intracellular ivi gene expression.
To determine whether the regulatory behavior of ivi genes mediated by PhoPQ in vitro mimics what is observed intracellularly, we assayed the effect of PhoPQ on strains containing either PhoP-activated or PhoP-repressed ivi genes in cultured macrophages. Figure 4 shows that introduction of a PhoP mutation into either pag strain (iviVI-A and spvB), resulted in reduced intracellular expression in cultured RAW 264.7 macrophages, indicating that PhoP is a major regulatory protein involved in the expression of these ivi genes in vitro and in cultured cells. In contrast, the intracellular expression of the PhoP-repressed gene iviXVII (pdu) was not enhanced after introduction of a PhoP mutation. This finding suggests that intracellular PhoP is not phosphorylated sufficiently to repress the low-level intracellular expression of iviXVII and/or other factors contribute to iviXVII expression 4 h postinfection. Additionally, phoP and iviXVII show significant levels of expression under some common conditions in vitro (Fig. 1) and during growth within cultured mammalian cells (Fig. 3). Taken together, these data suggest that although pags and prgs are capable of responding differently to a similar set of signals, this does not preclude their concomitant expression within the same cell type (see Discussion).
FIG. 4.
Intracellular expression of ivi genes in PhoP+ and PhoP− (phoP102::Tn10d-Cm) backgrounds in cultured macrophages. Individual ivi fusions were incubated with cultured RAW 264.7 murine macrophages. The coculture was incubated for 30 min, washed, treated with gentamicin to kill extracellular bacteria, washed, and incubated for 4 h. The cultured macrophages were lysed with Triton X-100. β-Galactosidase assays were performed on the recovered intracellular bacteria and also on individual ivi fusions grown for 4 h in LB. Error bars represent 1 standard deviation of the measured value.
DISCUSSION
Here we have grouped several ivi genes based on their coordinate behavior in response to environmental and genetic signals known to control virulence gene expression in vitro. Such in vitro expression was shown to parallel their intracellular induction both in cultured murine macrophages and in human epithelial cells. Moreover, the ivi fusions are each induced to similar levels in both cell types, suggesting that they respond to environmental and genetic signals that are present at both early and late stages of infection. These coordinately expressed ivi genes reside in three different types of DNA sequences, including native chromosomal sequence (mgtA), plasmid sequence (spvB), and acquired sequence of atypical base composition (iviVI-A). Thus, acquired sequences either have evolved to respond to the regulatory circuitry of the recipient Salmonella genome or were responsive to similar regulatory controls in the donor organism.
All known ivi genes analyzed in this study have been shown to be required for, or have been implicated in, virulence (9, 23). Additionally, two genes, iviVI-A and iviXVII (pdu), reside in regions of atypical base composition whose removal confers virulence defects (9). There is a strong correlation between the expression of ivi genes in response to a given set of environmental and genetic signals in vitro and their intracellular expression in mammalian cells. All ivi fusions that were induced by low pH and low Mg2+ were activated by PhoPQ. These genes were also induced upon entry into and growth within mammalian cells; moreover, spvB and iviVI-A were also shown to be activated by PhoPQ in cultured macrophages. Not all pags require PhoP for their induction under all environmental conditions. Notably, pmrB showed significant expression in the absence of PhoP (e.g., pH 6.6 and 1 mM Mg2+), which may be important under conditions found in the host. Coordinate intracellular expression of genes that respond similarly in vitro (e.g., low pH and low Mg2+) may reflect their response to similar vacuolar environments in these cell types. Defining the bacterial genes that are expressed in Salmonella-containing vacuoles in macrophages and/or epithelial cells will identify both general and specific functions that may be required for survival in these intracellular microenvironments as has been shown recently (6).
It is not implied that ivi genes used in this study are expressed only in the cell types tested or that other ivi genes are not expressed in the same cell types or subcellular compartments. For example, previously we have shown that phoP is induced after both intragastric and intraperitoneal inoculation (23), indicating that phoP is expressed at early and late stages of infection presumably in several cell types. Indeed, it has been recently shown that PhoPQ is a component of the acid tolerance response (3), which is presumed to contribute to bacterial survival at many anatomical sites. Additionally, most ivi genes isolated from the same infected tissue (e.g., spleen) do not respond to any of the signals tested here (e.g., iron, Mg2+, pH, and PhoP), suggesting that other signals may govern their expression in these tissues.
PhoP-activated and PhoP-repressed genes are capable of responding to the same set of signals in diverse and robust fashion. However, both classes show significant levels of expression under the same conditions in vitro (Fig. 1) and during growth within cultured mammalian cells (Fig. 3). Thus, pags and prgs may be induced within the same cell type and subcellular compartment (e.g., macrophage phagosome). The low level induction of iviXVII (pdu) observed within cultured macrophages may make a significant contribution to the intracellular fitness of the bacterium within the host. Indeed, the extent to which an individual gene is induced relative to another gene is not necessarily an indication of how important a particular gene is to a particular downstream event; it is simply a measurable factor. Correspondingly, removal of the pdu region confers a defect in systemic survival (9).
The expression pattern of spvB presented here differs from results of previous studies using alternatively constructed fusions. SpvB translational fusions from a low-copy-number plasmid containing S. dublin sequences have been reported to be induced in a PhoP-independent manner (12); moreover, the fusion protein and native mRNA levels are induced in stationary phase (12, 25). The spvB transcriptional fusion in this study resides at its native site on the S. typhimurium virulence plasmid, is expressed in LB at very low levels, is activated by PhoP, and is subject to coordinate environmental and genetic regulation with other PhoP-activated genes both in vitro and in cultured mammalian cells. One explanation for this disparity may lie in the structure of the spvB ivi fusion construct. Although the integrated spvB::lac fusion contains all spv wild-type sequences and their native control regions, two types of messages are produced since the promoter region is duplicated in this construct (covering 3 kb of sequence upstream of the spvB fusion join). Thus, the spvB::lac fusion-bearing message does not contain sequences that may be relevant to production and/or stability of full-length message in stationary phase, allowing the observation of subtle but significant aspects of spv regulation whose biological role is yet to be determined.
Given the intracellular induction of a subset of ivi genes, we now can ask how these genes contribute to bacterial survival during infection. One intracellularly expressed fusion, iviVI-A, resembles an enterotoxigenic E. coli adhesin and the Opa adherence and invasion proteins of Neisseria gonorrhoeae. The intracellular expression of an adhesin/invasin-like protein may seem puzzling. However, bacterial proteins often play roles other than those for which they were first defined. Indeed, the Opa adherence proteins have an additional function once the gonococci are intracellular: these outer membrane proteins bind host pyruvate kinase at the bacterial surface, leading to an environment encompassing the bacterium that is rich in pyruvate, which is one of few known carbon sources utilized by N. gonorrhoeae in vitro. (43). Thus, the adhesin-like IviVI-A protein may contribute other capabilities to S. typhimurium within host vacuoles.
Determination of the genetic and environmental factors that regulate ivi expression and the host site(s) in which they are expressed provides clues to both the intracellular environment and possible functions of ivi genes at these specific host sites. This complex and overlapping regulatory circuitry offers a pathogen tremendous flexibility to express different sets of genes at various sites. The functions of some of these genes may change dependent on the context of the animal (chicken versus cow), tissue (intestine versus spleen), cell type (macrophage versus hepatocyte), or subcellular compartment (phagosome versus phagolysosome).
ACKNOWLEDGMENTS
We thank Bob Sinsheimer for critically reading the manuscript.
This work was supported by NIH grant AI36373, Beckman Young Investigator Award, and University of California-Lawrence Livermore National Laboratory Collaboration (M.J.M.) and by NIH grant AI08649, ACS grant IRG 158K, and the Duke University Medical Center (P.C.H.).
REFERENCES
- 1.Ailion M, Bobik T A, Roth J R. Two global regulatory systems (Crp and Arc) control the cobalamin/propanediol regulon of Salmonella typhimurium. J Bacteriol. 1993;175:7200–7208. doi: 10.1128/jb.175.22.7200-7208.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alpuche-Aranda C M, Swanson J A, Loomis W P, Miller S I. Salmonella typhimurium activates virulence gene transcription within acidified macrophage phagosomes. Proc Natl Acad Sci USA. 1992;89:10079–10083. doi: 10.1073/pnas.89.21.10079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bearson B, Wilson L L, Foster J W. A low pH-inducible PhoPQ-dependent acid tolerance response protects Salmonella typhimurium against inorganic acid stress. J Bacteriol. 1998;180:2409–2417. doi: 10.1128/jb.180.9.2409-2417.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blanc-Potard A B, Groisman E A. The Salmonella selC locus contains a pathogenicity island mediating intramacrophage survival. EMBO J. 1997;16:5376–5385. doi: 10.1093/emboj/16.17.5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bobik T A, Ailion M, Roth J R. A single regulatory gene integrates control of vitamin B12 synthesis and propanediol degradation. J Bacteriol. 1992;174:2253–2266. doi: 10.1128/jb.174.7.2253-2266.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burns-Keliher L, Nickerson C A, Morrow B J, Curtiss R C., III Cell-specific proteins synthesized by Salmonella typhimurium. Infect Immun. 1998;66:856–861. doi: 10.1128/iai.66.2.856-861.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan R K, Botstein D, Watanabe T, Ogata Y. Specialized transduction of tetracycline resistance by phage P22 in Salmonella typhimurium. II. Properties of a high-frequency-transducing lysate. Virology. 1972;50:883–898. doi: 10.1016/0042-6822(72)90442-4. [DOI] [PubMed] [Google Scholar]
- 8.Connor C P, Heithoff D M, Mahan M J. In vivo gene expression: contributions to infection, virulence, and pathogenesis. Curr Top Microbiol Immunol. 1998;225:1–12. doi: 10.1007/978-3-642-80451-9_1. [DOI] [PubMed] [Google Scholar]
- 9.Connor C P, Heithoff D M, Julio S M, Sinsheimer R L, Mahan M J. Differential patterns of acquired virulence genes distinguish Salmonella strains. Proc Natl Acad Sci USA. 1998;95:4641–4645. doi: 10.1073/pnas.95.8.4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davis R W, Botstein D, Roth J R. Advanced bacterial genetics. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1980. [Google Scholar]
- 11.Earhart C F. Uptake and metabolism of iron and molybdenum. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 1075–1090. [Google Scholar]
- 12.Fang F C, Krause M, Roudier C, Fierer J, Guiney D G. Growth regulation of a Salmonella plasmid gene essential for virulence. J Bacteriol. 1991;173:6783–6789. doi: 10.1128/jb.173.21.6783-6789.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fierer J, Eckmann L, Fang F, Pfeifer C, Finlay B B, Guiney D. Expression of the Salmonella virulence plasmid gene spvB in cultured macrophages and nonphagocytic cells. Infect Immun. 1993;61:5231–5236. doi: 10.1128/iai.61.12.5231-5236.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fleckenstein J M, Kopecko D J, Warren R L, Elsinghorst E A. Molecular characterization of the tia invasion locus from enterotoxigenic Escherichia coli. Infect Immun. 1996;64:2256–2265. doi: 10.1128/iai.64.6.2256-2265.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia-del Portillo F, Foster J W, Maguire M E, Finlay B B. Characterization of the micro-environment of Salmonella typhimurium-containing vacuoles within MDCK epithelial cells. Mol Microbiol. 1992;6:3289–3297. doi: 10.1111/j.1365-2958.1992.tb02197.x. [DOI] [PubMed] [Google Scholar]
- 16.Garcia Vescovi E, Soncini F C, Groisman E A. Mg2+ as an extracellular signal: environmental regulation of Salmonella virulence. Cell. 1996;84:165–174. doi: 10.1016/s0092-8674(00)81003-x. [DOI] [PubMed] [Google Scholar]
- 17.Goldberg M B, DiRita V J, Calderwood S B. Identification of an iron-regulated virulence determinant in Vibrio cholerae, using TnphoA mutagenesis. Infect Immun. 1990;58:55–60. doi: 10.1128/iai.58.1.55-60.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Groisman E A, Heffron F. Regulation of Salmonella virulence by two-component regulatory systems. In: Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C: ASM Press; 1995. pp. 319–332. [Google Scholar]
- 19.Gulig P A, Danbara H, Guiney D G, Lax A J, Norel F, Rhen M. Molecular analysis of the spv virulence genes of the Salmonella virulence plasmids. Mol Microbiol. 1993;7:825–830. doi: 10.1111/j.1365-2958.1993.tb01172.x. [DOI] [PubMed] [Google Scholar]
- 20.Gunn J S, Hohman E L, Miller S I. Transcriptional regulation of Salmonella virulence: a PhoQ periplasmic domain mutation results in increased net phosphotransfer to PhoP. J Bacteriol. 1996;178:6369–6373. doi: 10.1128/jb.178.21.6369-6373.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gunn J S, Miller S I. PhoP-PhoQ activates transcription of pmrAB, encoding a two-component regulatory systems involved in Salmonella typhimurium antimicrobial peptide resistance. J Bacteriol. 1996;178:6857–6864. doi: 10.1128/jb.178.23.6857-6864.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hall H K, Foster J W. The role of Fur in the acid tolerance response of Salmonella typhimurium is physiologically and genetically separable from its role in iron acquisition. J Bacteriol. 1996;178:5683–5691. doi: 10.1128/jb.178.19.5683-5691.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heithoff D M, Conner C P, Hanna P C, Julio S M, Hentschel U, Mahan M J. Bacterial infection as assessed by in vivo gene expression. Proc Natl Acad Sci USA. 1997;94:934–939. doi: 10.1073/pnas.94.3.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heithoff D M, Conner C P, Mahan M J. Dissecting the biology of a pathogen during infection. Trends Microbiol. 1997;5:509–513. doi: 10.1016/S0966-842X(97)01153-0. [DOI] [PubMed] [Google Scholar]
- 25.Krause M, Fang F, Guiney D G. Regulation of plasmid virulence gene expression in Salmonella dublin involves an unusual operon structure. J Bacteriol. 1992;174:4482–4489. doi: 10.1128/jb.174.13.4482-4489.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mahan M J, Slauch J M, Mekalanos J J. Selection of bacterial virulence genes that are specifically induced in host tissues. Science. 1993;259:686–688. doi: 10.1126/science.8430319. [DOI] [PubMed] [Google Scholar]
- 27.Mahan M J, Tobias J W, Slauch J M, Hanna P C, Collier R J, Mekalanos J J. Antibiotic-based selection for bacterial genes that are specifically induced during infection of a host. Proc Natl Acad Sci USA. 1995;92:669–673. doi: 10.1073/pnas.92.3.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mahan M J, Slauch J M, Mekalanos J J. Environmental regulation of virulence gene expression in Escherichia, Salmonella, and Shigella spp. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 1075–1090. [Google Scholar]
- 29.Mekalanos J J. Environmental signals controlling the expression of virulence determinants in bacteria. J Bacteriol. 1992;174:1–7. doi: 10.1128/jb.174.1.1-7.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller J F, Mekalanos J J, Falkow S. Coordinate regulation and sensory transduction in the control of bacterial virulence genes. Science. 1989;243:916–922. doi: 10.1126/science.2537530. [DOI] [PubMed] [Google Scholar]
- 31.Miller S I, Mekalanos J J. Constitutive expression of the PhoP regulon attenuates Salmonella virulence and survival within macrophages. J Bacteriol. 1990;172:2485–2490. doi: 10.1128/jb.172.5.2485-2490.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller S I, Kukral A M, Mekalanos J J. A two component regulatory system (phoP and phoQ) controls Salmonella typhimurium virulence. Proc Natl Acad Sci USA. 1989;86:5054–5058. doi: 10.1073/pnas.86.13.5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neidhardt F C, Bloch P L, Smith D F. Culture medium for enterobacteria. J Bacteriol. 1974;119:736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roth J R, Lawrence J G, Rubenfield M, Kieffer-Higgins S, Church G M. Characterization of the cobalamin (vitamin B12) biosynthetic genes of Salmonella typhimurium. J Bacteriol. 1993;175:3303–3316. doi: 10.1128/jb.175.11.3303-3316.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanderson K E, Hessel A, Liu S-L, Rudd K E. The genetic map of Salmonella typhimurium, edition VIII. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 1903–1999. [Google Scholar]
- 36.Schmieger H. Phage P22-mutants with increased or decreased transduction abilities. Mol Gen Genet. 1972;119:75–88. doi: 10.1007/BF00270447. [DOI] [PubMed] [Google Scholar]
- 37.Slauch J M, Silhavy T. cis-acting ompF mutations that result in OmpR-dependent constitutive expression. J Bacteriol. 1991;173:4039–4048. doi: 10.1128/jb.173.13.4039-4048.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Soncini F C, Garcia Vescovi E, Groisman E A. Transcriptional autoregulation of the Salmonella typhimurium phoPQ operon. J Bacteriol. 1995;177:4364–4371. doi: 10.1128/jb.177.15.4364-4371.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soncini F C, Groisman E A. Two-component regulatory systems can interact to process multiple environmental signals. J Bacteriol. 1996;178:6796–6801. doi: 10.1128/jb.178.23.6796-6801.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soncini F C, Garcia Vescovi E, Solomon F, Groisman E A. Molecular basis of the magnesium deprivation response in Salmonella typhimurium: identification of PhoP-regulated genes. J Bacteriol. 1996;178:5092–5099. doi: 10.1128/jb.178.17.5092-5099.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsolis R M, Baumler A J, Stojiljkovic I, Heffron F. Fur regulon of Salmonella typhimurium: identification of new iron-regulated genes. J Bacteriol. 1995;177:4628–4637. doi: 10.1128/jb.177.16.4628-4637.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Waldor M K, Mekalanos J J. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science. 1996;272:1910–1914. doi: 10.1126/science.272.5270.1910. [DOI] [PubMed] [Google Scholar]
- 43.Williams J M, Chen G, Zhu L, Rest R F. Using the yeast two-hybrid system to identify human epithelial cell proteins that bind gonococcal Opa proteins: intracellular gonococci bind pyruvate kinase via their Opa proteins and require host pyruvate for growth. Mol Microbiol. 1998;27:171–186. doi: 10.1046/j.1365-2958.1998.00670.x. [DOI] [PubMed] [Google Scholar]