Abstract
The phytopathogenic bacterium Pseudomonas syringae pv. glycinea PG4180.N9 causes bacterial blight of soybeans and preferably infects its host plant during periods of cold, humid weather conditions. To identify proteins differentially expressed at low temperatures, total cellular protein fractions derived from PG4180.N9 grown at 18 and 28°C were separated by two-dimensional gel electrophoresis. Of several proteins which appeared to be preferentially present at 18°C, a 40-kDa protein with an isoelectric point of approximately 5 revealed significant N-terminal sequence homology to morphinone reductase (MR) of Pseudomonas putida M10. The respective P. syringae gene was isolated from a genomic cosmid library of PG4180, and its nucleotide sequence was determined. It was designated ncr for NAD(P)H-dependent 2-cyclohexen-1-one reductase. Comparison of the 1,083-bp open reading frame with database entries revealed 48% identity and 52% similarity to the MR-encoding morB gene of P. putida M10. The ncr gene was overexpressed in Escherichia coli, and its gene product was used to generate polyclonal antisera. Purified recombinant Ncr protein was enzymatically characterized with NAD(P)H and various morphinone analogs as substrates. So far, only 2-cyclohexen-1-one and 3-penten-2-one were found to be substrates for Ncr. By high-pressure liquid chromatography analysis, flavin mononucleotide could be identified as the noncovalently bound prosthetic group of this enzyme. The distribution of the ncr gene in different Pseudomonas species and various strains of P. syringae was analyzed by PCR and Southern blot hybridization. The results indicated that the ncr gene is widespread among P. syringae pv. glycinea strains but not in other pathovars of P. syringae or in any of the other Pseudomonas strains tested.
Pseudomonas syringae pv. glycinea is a phytopathogenic bacterium which causes bacterial blight of soybeans [Glycine max (L.) Merrit], a foliar disease characterized by necrotic leaf spots with chlorotic halos. The symptoms of bacterial blight are most severe during periods of cold, humid weather (8). As an important virulence factor, P. syringae pv. glycinea PG4180.N9 produces the chlorosis-inducing polyketide phytotoxin coronatine (COR) in a temperature-dependent manner (2, 40). Biosynthesis of COR in P. syringae is maximal at 18°C, whereas no detectable amount of COR is produced at 28 to 30°C, a temperature range otherwise optimal for growth of this bacterium (5, 25).
Previously, synthesis of various virulence factors in plant pathogens such as Agrobacterium tumefaciens, P. syringae pv. phaseolicola, Erwinia chrysanthemi, and Erwinia carotovora had been shown to be thermoresponsive (14, 16, 17, 22, 31). Low temperatures are often associated with conditions of high humidity which, in turn, favor infections of plants by foliar pathogens. The ecological importance for this phenomenon has not been elucidated in detail. It could be speculated that a rapid response to temperature shifts enables P. syringae to take advantage of favorable conditions and to infect its host plant. Although a modified two-component regulatory system has been demonstrated to control the temperature-dependent transcription of COR biosynthesis genes (40), no putative global system for temperature sensing or any other thermoresponsive factors of P. syringae pv. glycinea have been identified so far.
The aim of a long-term project in our laboratory is the identification and characterization of proteins of P. syringae which are expressed in a temperature-dependent manner. In this context, P. syringae pv. glycinea PG4180.N9 cultures were grown at 18° and 28°C and total cellular protein fractions were separated by two-dimensional gel electrophoresis. Several protein spots which appeared to be induced or to be solely present at 18°C were N-terminally sequenced. The gene for a protein which exhibited significant N-terminal sequence homology to morphinone reductase (MR) of Pseudomonas putida M10 (10) was subcloned from a genomic library of P. syringae PG4180, overexpressed in Escherichia coli, and studied with respect to its distribution among various Pseudomonas strains. The recombinant gene product was enzymatically characterized, indicating functional similarities as well as distinct biochemical differences to MR of P. putida M10.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Tables 1 and 2. Pseudomonas strains were maintained on mannitol-glutamate medium (18) at 28°C. For liquid cultures at 18° or 28°C, bacteria were incubated in either HSC medium (25) or King’s B medium (19) as described previously (5, 15). E. coli strains were used as hosts in cloning and expression studies and were grown in Luria-Bertani (LB) broth at 37°C. Bacterial growth was monitored by measuring the optical density at 600 nm (OD600). The protein concentration in cell lysates was determined by the Bradford assay (32). The following antibiotics were added to the media when needed (values are concentrations in micrograms per milliliter): ampicillin, 50; kanamycin, 25; tetracycline, 25.
TABLE 1.
Bacterial strains used in this study and distribution of the ncr gene among Pseudomonas species
| Bacterial strain | Result of the Southern blot and PCR screeninga | Reference or source |
|---|---|---|
| E. coli DH5α | 32 | |
| E. coli M15 (pREP4) | Qiagen | |
| P. syringae | ||
| pv. glycinea PG4180 | + | R. E. Mitchell |
| pv. glycinea PG4180.N9 | + | 39 |
| pv. savastanoi GSPB2264 | − | GSPBb |
| pv. phaseolicola GSPB796 | − | GSPB |
| pv. morsprunorum Pm7 | − | A. L. Jones |
| pv. atropurpurea MAFF301313 | − | B. Völksch |
| pv. syringae C72 | − | B. Völksch |
| pv. tomato DSM50315 | − | DSMc |
| pv. maculicola GSPB2145 | − | GSPB |
| pv. pisi Psp104 | − | B. Völksch |
| pv. lachrymans GSPB82a | − | GSPB |
| P. putida GSPB1498 | − | GSPB |
| P. fluorescens GSPB1714 | − | GSPB |
| P. cichorii GSPB2097 | − | GSPB |
| P. marginalis GSPB92 | − | GSPB |
| P. viridiflava GSPB1685 | − | GSPB |
| P. fuscovaginae GSPB2309 | − | GSPB |
| P. agarici GSPB2305 | − | GSPB |
| Race typus strains P. syringae pv. glycinea | ||
| Race 0, GSPB1834 | − | GSPB |
| Race 2, GSPB1201 | + | GSPB |
| Race 4, GSPB1548 | + | GSPB |
| Race 4, GSPB1996 | + | GSPB |
| Race 5, GSPB1837 | − | GSPB |
| Race 9, GSPB2003 | + | GSPB |
| Race X, GSPB1997 | + | GSPB |
+, positive signal in both experiments; −, no signal.
GSPB, Göttinger Sammlung Phytopathogener Bakterien, Göttingen, Germany.
DSM, Deutsche Sammlung für Mikroorganismen, Braunschweig, Germany.
TABLE 2.
Plasmids used in this study
| Plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| Cosmid 5/III | Tcr, genomic cosmid library clone of PG4180, approximately 35-kb insert in pRK7813 | 15 |
| pBluescript II SK | Apr, high-copy-number cloning vector | Stratagene |
| pECos5 | Apr, 4.2-kb EcoRI fragment of cosmid 5/III in pBluescript II SK | This study |
| pMAL-c2 | Apr, expression vector for malE translational fusions | New England Biolabs |
| pQE70 | Apr, expression vector for C-terminal His tag fusions | Qiagen |
| pMAL-Ncr | Apr, contains a 1,132-bp BamHI-HindIII fragment with coding sequence of ncr downstream of the factor Xa cleavage site in pMAL-c2 | This study |
| pQE-Ncr | Apr, contains a 1,078-bp SphI-BglII fragment with the ncr gene truncated at its 3′ end in pQE70 | This study |
Determination of the N-terminal protein sequence.
Total protein extracts of P. syringae cells grown at 18 and 28°C were separated by two-dimensional gel electrophoresis according to the method of O’Farrell (24). The sodium dodecyl sulfate (SDS)-polyacrylamide gels were stained with 0.1% Coomassie blue R250 and destained with 40% methanol and 10% acetic acid. Subsequently, gels were washed with water. Protein spots were cut out of the gel, and the N-terminal sequence was determined by standard procedures (41).
Isolation of the ncr gene of P. syringae.
Oligonucleotide primers derived from conserved regions of the morB gene of P. putida M10 and homologous genes (10) were used to amplify a 550-bp fragment from total genomic DNA of P. syringae pv. glycinea PG4180.N9 by PCR. The respective primers morF (5′-GACGAATTCATG GCGCCGCTGACCCGC-3′) and morR (5′-ATAGAATTCGAAGCGCGCCCGGTTCTC-3′) contained EcoRI restriction sites (underlined) at their 5′ ends. The 550-bp PCR product was used as the DNA probe in a Southern hybridization screening of a genomic cosmid library of P. syringae PG4180 (15). Two positive cosmids were characterized by restriction endonuclease mapping and Southern blot analysis. A 4.2-kb EcoRI DNA fragment containing the full-length ncr gene was isolated from a cosmid designated 5/III and subcloned into pBluescript II SK to generate pECos5.
Standard genetic procedures.
Genomic DNA was isolated from P. syringae by established procedures (38). Agarose gel electrophoresis, restriction digests, purification of DNA fragments from agarose gels, electroporations, PCR, and small-scale plasmid DNA preparations were performed by standard techniques (32). Southern blot hybridizations were carried out with a nonradioactive nucleotide labeling and detection kit (Boehringer, Mannheim, Germany). Subclones were generated in pBluescript II SK (Stratagene, Heidelberg, Germany). Nested deletion clones were constructed with the Erase-a-Base system (Promega, Mannheim, Germany). Large-scale preparations of plasmid DNA from E. coli were carried out by alkaline lysis and purified with the Nucleobond AX 100 kit (Macherey-Nagel, Düren, Germany).
Nucleotide sequencing and analysis.
Nucleotide sequencing reactions were performed by the dideoxynucleotide method (32) with the Thermo Sequenase fluorescent labeled primer cycle sequencing kit (Amersham-Buchler, Braunschweig, Germany) and Cy5-labeled T3/T7 oligonucleotide primers (Pharmacia, Freiburg, Germany). Automated DNA sequencing was accomplished with an ALF Express sequencing apparatus (Pharmacia). Sequence data were aligned and processed with the Lasergene version 4.1 software package (DNASTAR, Madison, Wis.). DNA and protein sequence homology searches of the GenBank, EMBL, PIR, and SwissProt databases were performed with the University of Wisconsin Genetics Computer Group programs BLASTX, BLASTN, FASTEMBL, and BESTFIT.
Cloning of ncr into the expression vectors pMAL-c2 and pQE70.
The ncr gene was amplified by PCR, with plasmid pECos5 serving as the template DNA. For cloning in pMAL-c2 (New England Biolabs, Schwalbach, Germany), flanking oligonucleotides with either a BamHI or a HindIII recognition site (underlined) were used for PCR amplification (Ncr-BamHI, 5′-CGCGGATCCATGCCGACTCTTTTCGAC-3′ and Ncr-HindIII, 5′-CCCAAGCTTATATTGAGGTGCGCAGCC-3′). For cloning in pQE70 (Qiagen, Hilden, Germany), flanking oligonucleotides with either an SphI or a BglII restriction enzyme site (underlined) were used (Ncr-SphI, 5′-AACCCCCGCATGCCGACT-3′, and Ncr-BglI, 5′-GGAAGATCTTTGGTCAGCGGTGGGGTA-3′). The appropriate restriction sites were used to clone the amplification products into the expression vectors.
High-level expression and purification of MBP-Ncr fusion protein.
Overexpression of the ncr gene as a translational fusion to the malE gene coding for maltose binding protein (MBP) was carried out with the expression vector pMAL-c2 in E. coli DH5α. Transformants with the correct insert size were screened for expression of MBP-Ncr by SDS-polyacrylamide gel electrophoresis (PAGE). A clone carrying plasmid pMAL-Ncr showed high levels of expression and was chosen for large-scale protein purification. LB medium (400 ml) was inoculated (1:100) with an overnight culture of E. coli DH5α (pMAL-Ncr). At an OD600 of 0.5, the culture was induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and subsequently incubated for an additional 4 h. The cells were then pelleted and resuspended in protein extraction buffer (50 mM Tris, 200 mM NaCl, 1 mM EDTA, 5 mM dithiothreitol). Bacterial cells were broken by four passages through a French pressure cell. MBP-Ncr fusion protein was purified from crude cell lysate by affinity chromatography as recommended by the manufacturer (New England Biolabs). The purified protein was then cleaved with protease factor Xa, and Ncr was isolated following SDS-PAGE. Purified Ncr was used to raise antibodies in rabbits (BioGenes, Berlin, Germany).
Immunodetection of Ncr.
Total protein extracts were isolated from cell pellets of bacterial cultures (1.5 ml). Proteins were diluted, and equal amounts (15 μg/lane for crude extracts and 1.5 μg/lane for purified protein) were separated by SDS–10% PAGE. Electroblotting and hybridizations on nitrocellulose membranes were conducted according to standard procedures (32). The specificity of the Ncr antiserum at a dilution of 1:4,000 was evaluated with recombinant Ncr protein from E. coli and crude protein extracts of P. syringae PG4180.N9. For signal detection, secondary anti-rabbit immunoglobulin G antibodies conjugated to alkaline phosphatase (Sigma, Darmstadt, Germany) were used at a concentration of 1:4,000, and the reaction was visualized by using 5-bromo-4-chloro-3-indolyl phosphate and nitroblue tetrazolium salt.
Overexpression of Ncr as a His tag fusion and purification.
To construct plasmid pQE-Ncr, the ncr gene was PCR amplified with oligonucleotide primers Ncr-SphI and Ncr-BglII. The SphI- and BglII-cleaved PCR product was cloned into the expression vector pQE70, carrying a sequence coding for six consecutive histidine residues located at the 3′ end of its multiple cloning site. Positive clones were transformed into E. coli M15 (pREP4) and screened for high-level expression of the ncr-His6 gene fusion to yield plasmid pQE-Ncr. Four hundred milliliters of LB medium was inoculated (1:100) with an overnight culture of E. coli M15 (pREP4/pQE-Ncr) grown at 37°C. At an OD600 of 0.5 the culture was treated with 0.1 mM IPTG and subsequently incubated at 10°C and 280 rpm for 40 h. Bacterial cells were harvested and disrupted as described above. Purification of Ncr-His6 was performed with Ni-nitrilotriacetic acid (Ni-NTA) agarose according to the manufacturer’s recommendations (Qiagen).
Assay for enzymatic activity.
Enzymatic activity of Ncr-His6 was determined spectrophotometrically by monitoring the decrease in absorbance at 366 nm concomitant with the disappearance of NAD(P)H (ɛ366nm = 3.3 mM−1 cm−1) essentially as described previously (11). The reaction was performed at 30°C and started by addition of purified Ncr-His6 (50 μg/ml) to 2-cyclohexen-1-one (0.01 to 0.4 mM) or respective other compounds and NAD(P)H (0.2 to 0.5 mM) in sodium phosphate buffer (50 mM, pH 7.0). No absorbance at 366 nm could be detected for either cyclohexanone or 2-cyclohexen-1-one at 0.1 M, a concentration much higher than that used in the assays. The background rate of NAD(P)H oxidation in the absence of 2-cyclohexen-1-one was determined and subtracted. One unit of enzyme activity was defined as the amount of enzyme required for the turnover of 1 μmol of substrate per min at 30°C. Specific activities were expressed as units per milligram of protein.
Determination of the prosthetic group of Ncr.
For the analysis of noncovalently bound flavins, purified Ncr-His6 (0.5 mg in 1 ml; 12.5 μM) was denatured by adding trichloroacetic acid (TCA) to a final concentration of 5% and incubation on ice for 15 min. Precipitated protein was collected by centrifugation and washed once with 50 μl of 5% TCA. Both supernatants were combined and adjusted to pH 6.5 with 2 M K2HPO4. For the identification of the flavin prosthetic group, 1 μl of supernatant was applied to a 125 by 4 mm Spherisorb C18 reverse-phase column (Sykam, Fürstenfeldbruck, Germany) by high-pressure liquid chromatography (HPLC). Flavins were eluted with the following solvent system at a flow rate of 1 ml/min: 25% (vol/vol) methanol-100 mM ammonium formate (pH 3.7). Detection was carried out at 370 nm with a S3200 UV/VIS detector (Sykam). The standards flavin adenine dinucleotide (FAD), flavin mononucleotide (FMN), and riboflavin (RF) were applied at a concentration of 5 μM (1 μl). For the detection of covalently bound flavin prosthetic groups, the TCA-pelleted protein (0.5 mg) was resuspended in 0.5 ml of 100 mM Tris-HCl (pH 8.0) containing 50 μg of proteinase K and incubated for 12 h at 37°C to digest the protein. The clear solution was then adjusted to pH 6.5, and the fluorescence at 525 nm was monitored following an excitation at 475 nm with a Shimadzu RF 540 spectrofluorometer.
Gas chromatography.
Cyclohexanone, 2-cyclohexen-1-one, and 2-cyclohexen-1-ol were quantitated with a Carlo Erba 8000 series gas chromatograph (CE Instruments, Milan, Italy) equipped with a 30 m by 0.32 mm CDB-FAAD capillary and a flame ionization detector. The injector temperature was set to 70°C, and the detector and oven temperatures were 220 and 100°C, respectively. Hydrogen was used as carrier gas at a flow rate of 55 kPa.
Nucleotide sequence accession number.
The nucleotide sequence reported in this study was deposited with GenBank and EMBL under accession no. AF093246.
RESULTS
Isolation of proteins differentially expressed depending on temperature.
P. syringae pv. glycinea PG4180.N9 cultures were grown in HSC medium at 18 and 28°C. Cells were harvested in the stationary phase, and total cellular protein extracts were separated by two-dimensional gel electrophoresis. Use of the Coomassie staining method for protein detection was intended to identify only differentially expressed proteins which occurred in relatively large quantities. Several protein spots which appeared to be induced or solely present at 18°C were N-terminally sequenced. Of those, the N-terminal sequence of a 40-kDa protein with an isoelectric point of approximately 5 revealed significant similarities to the amino acid sequence of the enzyme morphinone reductase (MR) of P. putida M10 (10) (Fig. 1). MR is encoded by the morB gene and converts morphinone to hydromorphone. It is involved in the degradation of morphine by P. putida M10, a strain which was isolated from sewage water of an opiate-producing pharmaceutical factory (4). The 40-kDa protein was chosen for further characterization because it showed the most obvious temperature dependence of all proteins investigated. Moreover, its potential capability to use plant-borne alkaloids such as morphinone seemed to be in line with the scope of identification of proteins involved in plant-microbe interaction.
FIG. 1.
Alignment of the determined N-terminal amino acid sequence of the isolated thermoresponsive protein from P. syringae pv. glycinea PG4180.N9 (Ncr) with the MR sequence of P. putida M10 (10).
Cloning of the P. syringae pv. glycinea gene homologous to morB.
According to a previously published alignment of the morB gene of P. putida M10 and several closely related oxidoreductase genes (10), oligonucleotide primers were designed to amplify a predicted 550-bp PCR product from genomic DNA of P. syringae pv. glycinea PG4180.N9. The 550-bp fragment was used for Southern hybridization analysis of a genomic cosmid library of PG4180. Two individual cosmid clones contained a common 4.2-kb EcoRI fragment which hybridized with the PCR probe and which was subcloned into pBluescript II SK to yield plasmid pECos5.
Nucleotide sequence analysis of the genetic locus for the MR homolog.
To obtain subclones of pECos5 for nucleotide sequencing, nested deletions from both sides of the insert were generated. The nucleotide sequence of the entire insert DNA was determined, and one open reading frame (ORF) of 1,083 or 1,107 bp with respect to two potential translational start sites was obtained (Fig. 2). This ORF was designated ncr for NAD(P)H-dependent 2-cyclohexen-1-one reductase (Ncr) due to results of its enzymatic characterization (see below). This ORF contained binding sites for the oligonucleotide primers morF and morR, indicating that it harbored the DNA of interest. The DNA regions up- and downstream of ncr contained putative ORFs of shorter length or incomplete ORFs. Of those, only one incomplete ORF of more than 1,400 bp showed significant sequence similarities to database entries, namely to chemotaxis transducer proteins (data not shown). This incomplete ORF had its putative translational start site more than 600 bp upstream of the ncr gene and was oriented in the opposite direction to ncr. Compared with GenBank, EMBL, SwissProt, and PIR database entries, ncr showed significant sequence homology (48% identity and 52% similarity) to the morB gene of P. putida M10 and to genes for pentaerythritol tetranitrate reductase from Enterobacter cloacae, N-ethylmaleimide reductase of E. coli, and glycerol trinitrate reductase of Agrobacterium radiobacter (10, 12, 23, 37). The highest degree of similarity to enzymes of eukaryotic origin was found for 12-oxophytodienoate (OPDA) reductase from Arabidopsis thaliana (34) (41% identity and 49% similarity). To a lesser extent, similarities to a number of additional genes coding for NAD(P)H-dependent flavin oxidoreductases from various organisms were found (40 to 48% similarities) (9, 35). The predicted ncr gene product was 360 amino acids in length and had a molecular mass of 39.4 kDa. In order to obtain information about the potential promoter region of ncr, the nucleotide sequence located upstream of the ncr translational start was examined for regulatory sequence motifs common to gram-negative bacteria as well as hrp boxes, which are specific to pathogenicity-associated genes in plant-pathogenic bacteria (42). None of those were found, indicating a novel type of transcriptional regulation.
FIG. 2.
Nucleotide sequence and predicted translation product of the ncr gene from P. syringae pv. glycinea PG4180.N9. Nucleotides and amino acid residues are numbered on the left and right, respectively. Horizontal arrows below the nucleotide sequence indicate putative transcriptional terminator sequences, whereas horizontal arrows above the sequence represent binding sites for oligonucleotide primers used in this study. Shaded boxes mark the conserved regions and amino acid residues found in 15 sequences of NAD(P)H-dependent oxidoreductases from various organisms (10).
Overexpression and purification of the ncr gene product.
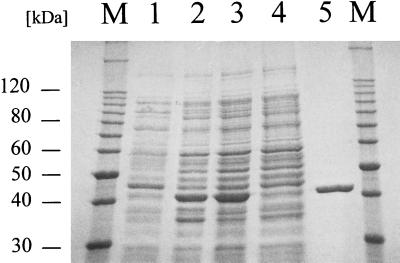
Two different methods for overexpression of the Ncr protein were tested simultaneously. Following the overexpression of the fusion protein MBP-Ncr derived from E. coli DH5α (pMAL-Ncr), the proteolytically cleaved-off Ncr protein was used to generate polyclonal antibodies (see below) but unfortunately turned out to be enzymatically inactive (data not shown). To test the enzymatic activity of Ncr, expression plasmid pQE-Ncr was constructed containing an in-frame fusion of the ncr gene with a sequence of pQE70 which codes for six histidine residues. Ncr-His6 could be detected in the soluble protein fraction of E. coli M15 (pREP4/pQE-Ncr) (Fig. 3). The Ncr-His6 fusion was purified by affinity chromatography and showed a bright yellow color, which indicated the presence of a flavin prosthetic group. This result suggested that Ncr-His6 might have undergone correct protein folding (13) and therefore might be enzymatically active.
FIG. 3.
SDS-PAGE analysis of protein fractions obtained during purification of Ncr-His6. Lanes: 1, homogenate from E. coli M15 (pREP4/pQE-Ncr); 2, E. coli M15 (pREP4/pQE-Ncr) after induction with 0.1 mM IPTG; 3, crude extract after cell lysis; 4, soluble proteins passed through the Ni-NTA column; 5, Ncr-His6 after elution from the Ni-NTA column with 250 mM imidazole; M, molecular size marker.
Immunodetection of the ncr gene product.
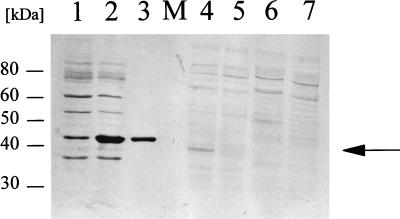
In preliminary Western blotting experiments the Ncr antiserum reacted with an approximately 40-kDa protein in cellular extracts from PG4180.N9 grown at 18°C. In order to monitor Ncr abundance in response to growth temperature, PG4180.N9 was incubated at 18, 22, and 28°C in HSC minimal medium, and total cellular proteins were extracted in the stationary phase. Samples from 22°C-grown cultures were included in this assay because P. syringae pv. glycinea PG4180 is also virulent at this temperature. Analysis by Western blotting indicated that the signal for Ncr was significantly increased in protein extracts from 18°C- and 22°C-incubated PG4180.N9 cultures compared to that in samples from 28°C cultures (Fig. 4 and data not shown), confirming the initial finding that this protein was preferentially expressed at lower temperatures. The signal was absent in protein extracts obtained from representatives of P. syringae pv. glycinea races 0 and 5, confirming data on the distribution of the ncr gene among P. syringae strains reported below. We repeated the Western blot analysis with protein extracts from PG4180.N9 cells grown in complex King’s B medium at 18, 22, and 28°C (data not shown). However, signals were not visible in any of those protein samples, indicating a possible lack of Ncr expression in complex medium.
FIG. 4.
Western blot analysis of overexpressed Ncr-His6 protein in E. coli and total cellular protein fractions of P. syringae. Pseudomonas cells were grown to stationary phase (OD600 = 5.0) at 18, 22, and 28°C and then subjected to total protein extraction. Western blotting was performed with polyclonal antibodies raised against Ncr. The specific signal is marked with an arrow. Similar signals were detected in protein samples from cultures grown at 22°C (data not shown). Lanes: 1, crude extract of E. coli M15 (pREP4/pQE-Ncr); 2, crude extract of E. coli M15 (pREP4/pQE-Ncr) after induction with 0.1 mM IPTG; 3, purified Ncr-His6 protein; 4, P. syringae pv. glycinea PG4180.N9 grown at 18°C; 5, PG4180.N9 grown at 28°C; 6, P. syringae pv. glycinea race 0 grown at 18°C; 7, P. syringae pv. glycinea race 5 grown at 18°C; M, molecular size marker.
Identification of the prosthetic group of the Ncr-His6 protein.
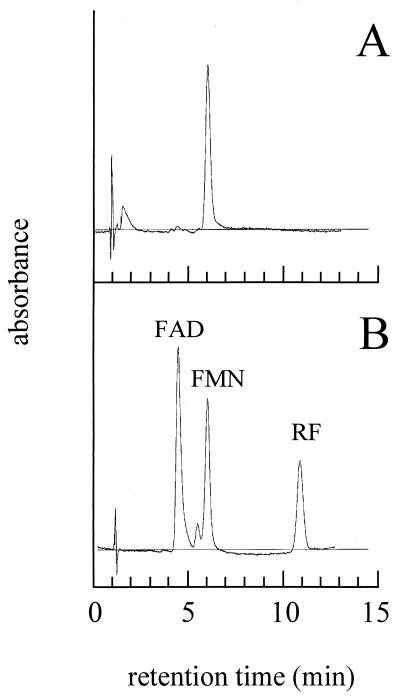
The predicted amino acid sequence of Ncr showed significant homologies to sequences from within the protein family of class I α/β-barrel flavoprotein oxidoreductases (35), suggesting that Ncr might possess a flavin prosthetic group. To test this, purified Ncr-His6 protein was monitored spectrophotometrically. The UV/visible spectrum showed characteristic flavin peaks at 275, 385, and 470 nm (data not shown). The flavin compound could be liberated from the protein following its denaturation with 5% TCA, indicating that the flavin prosthetic group was not covalently bound. In support of this, no flavin could be detected in the TCA-precipitated protein fraction. The liberated flavin was analyzed by HPLC with FAD, FMN, and riboflavin as standards (Fig. 5). Chromatography performed with the three standards resulted in clearly distinguishable peaks for FAD (retention time = 4.5 min), FMN (retention time = 6.1 min), and riboflavin (retention time = 11.1 min) (Fig. 5B). The chromatogram representing the flavin compound released from Ncr-His6 showed one distinct peak at 6.1 min (Fig. 5A), indicating that FMN was the noncovalently bound prosthetic group of the Ncr protein.
FIG. 5.
HPLC analysis for the determination of the prosthetic group of Ncr. (A) One microliter of supernatant resulting from TCA precipitation of Ncr-His6. (B) One microliter of the flavin standards FAD, FMN, and RF, each at a concentration of 5 μM. The standards gave rise to three clearly distinguishable peaks at 4.5 (FAD), 6.1 (FMN), and 11.1 (RF) min.
Enzymatic characterization of the Ncr protein.
Since members of the enzyme family of α/β-barrel flavin oxidoreductases catalyze the reduction of 2-cyclohexen-1-one (Fig. 7, inset), this compound was used in our study as a substrate in standard assays. A clear Michaelis-Menten kinetic was observed when the purified Ncr-His6 protein (50 μg/ml) was incubated in phosphate buffer (pH 7.0) with 0.5 mM NAD(P)H and various concentrations of 2-cyclohexen-1-one. Following assays with various 2-cyclohexen-1-one concentrations and 0.5 mM NADPH or 0.5 mM NADH, the double-reciprocal Lineweaver-Burk plot of the rates of NAD(P)H oxidation versus the 2-cyclohexen-1-one concentration resulted in apparent Km values for 2-cyclohexen-1-one of 0.32 mM (Vmax = 3.9 U/mg) and 0.062 mM (Vmax = 0.7 U/mg), respectively. As controls, crude protein extracts of E. coli M15 (pREP4/pQE70) were assayed under identical conditions but showed no detectable reductase activity with 2-cyclohexen-1-one as the substrate. Furthermore, incubation of Ncr-His6 with 0.1 M cyclohexanone and 10 mM NAD(P)+ in 0.2 M Tris-HCl buffer (pH 8.0) did not result in measurable enzymatic activity at a wavelength of 340 nm [specific absorption maximum for NAD(P)H], indicating that the enzyme could not catalyze the reverse reaction. In the standard assay, addition of 0.1 M cyclohexanone did not inactivate the enzyme. Additionally, a more sensitive method to test the reverse reaction was carried out by directly oxidizing nascent NAD(P)H to NAD(P)+ and therefore immediately removing it from the reaction equilibrium. This was achieved by a coupled reaction with 80 μM 2-(p-iodophenyl)-3-(p-nitrophenyl)-5-phenyl-tetrazoliumchloride (INT) and 80 μM 8-dimethylamino-2,3-benzophen-oxazine (Meldola’s blue) and by measuring the absorbance at 492 nm. The reverse reaction was still not observed, confirming the results reported above.
FIG. 7.
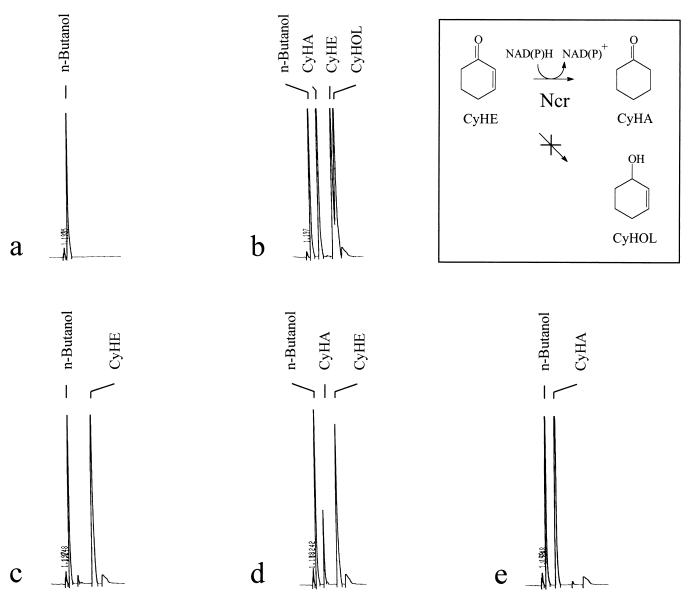
Gas chromatographic analysis of the product of 2-cyclohexen-1-one reduction by the Ncr-His6 protein. Reaction mixtures containing 0.2 mM 2-cyclohexen-1-one, 0.5 mM NAD(P)H, and 0.1 mg of purified Ncr-His6 in sodium phosphate buffer, pH 7.0, were incubated at 30°C for 30 min. Aliquots of 1 μl were removed and separated on a gas chromatographic capillary. (a) n-Butanol as internal standard; (b) standards of cyclohexanone (CyHA), 2-cyclohexen-1-one (CyHE), and 2-cyclohexen-1-ol (CyHOL); (c) reaction mixture at t0 (0 min); (d) reaction mixture at t1 (1 min); (e) reaction mixture at t2 (5 min).
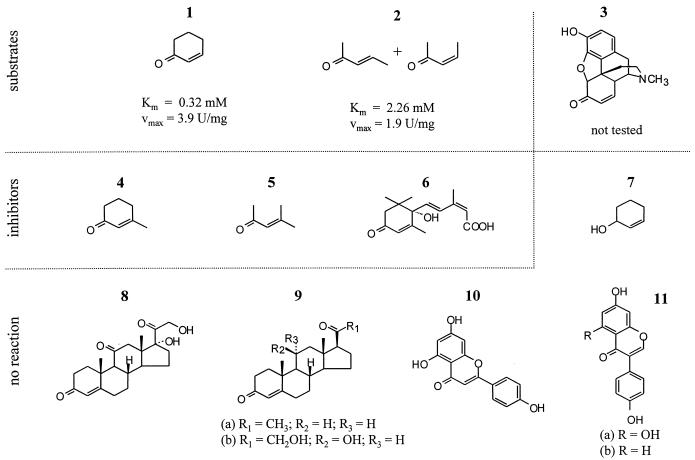
Several substances with structural similarities to morphinone and 2-cyclohexen-1-one were tested as potential substrates for Ncr-His6 (Fig. 6). Neither plant-associated compounds, such as apigenin, genistein, and daidzein, nor steroid compounds, such as progesterone, cortisone, and corticosterone, could be identified as substrates or competitive inhibitors for this enzyme. Likewise, 2-cyclohexen-1-ol did not function as a substrate or inhibitor, demonstrating the importance of the keto group of 2-cyclohexen-1-one for the enzymatic activity of Ncr. Interestingly, 3-penten-2-one, another nonsubstituted α,β-unsaturated ketone, functioned as a substrate for Ncr (Fig. 6). In contrast, compounds such as 3-methyl-2-cyclohexen-1-one, 4-methyl-3-penten-2-one, and abscisic acid, which all possess a substituted carbon-carbon double bond, significantly inhibited the enzymatic activity of Ncr but did not function as substrates.
FIG. 6.
Structures of the following chemical compounds tested as potential substrates or inhibitors of Ncr activity: 1, 2-cyclohexen-1-one; 2, 3-penten-2-one; 3, morphinone; 4, 3-methyl-2-cyclohexen-1-one; 5, 4-methyl-3-penten-2-one; 6, (+/−)-2-cis-4-trans-abscisic acid; 7, 2-cyclohexen-1-ol; 8, cortisone-21-acetate; 9a, progesterone; 9b, corticosterone; 10, apigenin; 11a, genistein; and 11b, daidzein. Compounds 1 and 2 functioned as Ncr substrates, with Km and Vmax values given below the diagrams of the structures. Presence of compounds 4 to 6 inhibited Ncr activity, whereas compounds 7 to 11b neither functioned as substrates nor inhibited Ncr activity.
Identification of cyclohexanone as the reaction end product.
Theoretically, two possible end products could be generated from 2-cyclohexen-1-one by the Ncr protein, cyclohexanone or 2-cyclohexen-1-ol (Fig. 7, inset). To identify the actual end product, aliquots of the enzyme assays were subjected to gas chromatographic analysis. Purified Ncr-His6 (0.1 mg) was incubated with 0.2 mM 2-cyclohexen-1-one and 0.5 mM NAD(P)H at 30°C. At several time points, aliquots were removed from the assay mixture and separated by gas chromatography. As controls, cyclohexanone, 2-cyclohexen-1-one, and 2-cyclohexen-1-ol were also subjected to gas chromatographic analysis. As shown in Fig. 7, only 2-cyclohexen-1-one was present in the mixture when the reaction was started (time zero [t0] = 0 min). Aliquots taken after 1 min already showed appearance of the end product cyclohexanone. After 5 min, 2-cyclohexen-1-one had completely been transformed into cyclohexanone (Fig. 7e). 2-Cyclohexen-1-ol was not detected during this procedure, suggesting that this compound was not an end product of the reaction catalyzed by Ncr.
Distribution of the ncr gene among different Pseudomonas strains.
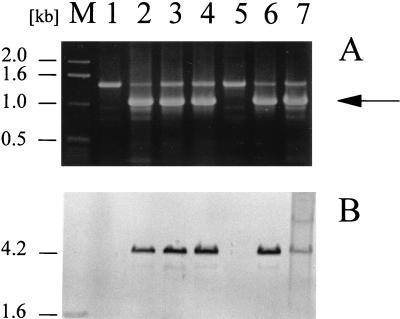
Since P. syringae pv. glycinea PG4180.N9 contains five indigenous plasmids ranging in size from 35 to 95 kb (3), we initially tested whether the ncr gene was located on the chromosome or on one of the plasmids. When preparations of plasmid DNA versus chromosomal DNA were transferred onto a nylon membrane and hybridized with an ncr probe, signals appeared only with chromosomal DNA fragments, indicating that the gene was not located on plasmid DNA. To define the number of chromosomal copies of ncr, PG4180.N9 genomic DNA was individually cleaved with six different restriction enzymes and subjected to Southern blot analysis. Results indicated that only a single copy of ncr was present on the chromosome (data not shown). Furthermore, genomic DNA was prepared from strains of six different races of P. syringae pv. glycinea, 10 different pathovars of P. syringae, and seven other Pseudomonas species. The genomic DNA preparations were treated with restriction endonuclease EcoRI, electrophoretically separated, and analyzed for DNA-DNA hybridization to an ncr probe. Under diverse stringency conditions (60 to 68°C), positive hybridization signals were detected in representatives of P. syringae pv. glycinea races 2, 4, 9, and X, but not in the two strains of races 0 and 5 or in any of the other strains tested (Table 1 and Fig. 8B). In addition, the genomic DNA was used as a template in PCRs performed with oligonucleotide primers Ncr-SphI and Ncr-BglII. Following a PCR under low-stringency conditions (annealing temperature of 56°C), products were analyzed by agarose gel electrophoresis (Table 1 and Fig. 8A). The results confirmed data from the Southern blot analysis; only genomic DNA from strains hybridizing with the ncr probe gave rise to PCR products of the correct molecular mass. These data indicated that the ncr gene was present in a number of different P. syringae pv. glycinea strains but not in representatives of other pathovars of this bacterial species or in other distantly related strains.
FIG. 8.
PCR and Southern blot analyses of the distribution of the ncr gene among various P. syringae pv. glycinea strains. (A) PCR carried out with primers Ncr-Sph and Ncr-Bgl on total genomic DNA of strains representing six different races of P. syringae pv. glycinea. The arrow indicates the specific amplification product of 1.1 kb. (B) Southern blot analysis of genomic DNA cleaved with EcoRI and probed with ncr. Lanes: M, molecular size marker; 1, P. syringae pv. glycinea, race 0; 2, race 2; 3 and 4, two different strains of race 4; 5, race 5; 6, race 9; 7, race X.
DISCUSSION
From a plant pathogen, P. syringae pv. glycinea, we have isolated a novel gene coding for an NAD(P)H-dependent flavoprotein which reduces the olefinic bond of the α,β-unsaturated ketone 2-cyclohexen-1-one and which was predominately synthesized in minimal medium at 18°C. Sequence comparisons revealed that the Ncr protein belongs to the growing family of class I flavin-dependent α/β-barrel oxidoreductases (28, 35). Not surprisingly, the best alignments were obtained when the deduced amino acid sequence of Ncr was compared with those of oxidoreductases from gram-negative bacteria, although the respective substrates were remarkably different (10, 12, 23, 37). The most significant similarities to eukaryotic enzymes of this family were to OPDA reductase from A. thaliana and old yellow enzyme from yeast (9, 34).
Ncr exhibited the highest degree of sequence similarity to MR of P. putida M10, which catalyzes the reduction of morphinone and codeinone to hydromorphone and hydrocodone, respectively (11). MR also reduces 2-cyclohexen-1-one to cylohexanone, a reaction used to identify members of the family of NAD(P)H-dependent flavin oxidoreductases. Consequently, 2-cyclohexen-1-one could be identified as a substrate for Ncr. For MR, enzymatic activity was additionally demonstrated with morphinone, codeinone, and neopinone as substrates (11). None of these compounds was available during the course of this study. However, by testing the influence of some previously reported MR inhibitors on Ncr activity, we aimed at defining a potential substrate spectrum for Ncr. Neither progesterone, cortisone, nor corticosterone significantly inhibited Ncr activity.
The prosthetic group of the overproduced Ncr-His6 protein was found to be FMN. A large number of enzymes, mostly catalyzing redox reactions, use flavins as prosthetic groups. During protein purification and enzymatic analysis, it was demonstrated that the flavin prosthetic group was tightly but not covalently bound to Ncr. This result is in line with data for most other flavin-dependent enzymes (36).
Both NADPH and NADH were found to be suitable electron donors for the Ncr enzyme in the reaction with 2-cyclohexen-1-one. In comparison, MR of P. putida mainly utilized NADH as electron donor, whereas the OPDA reductase from A. thaliana showed a preference for NADPH (11, 33). The apparent Km of Ncr for 2-cyclohexen-1-one at 0.4 mM NADH was found to be approximately 55-fold lower than that reported for MR (11).
Analysis of the end product of the Ncr-catalyzed reaction revealed that cyclohexanone but not 2-cyclohexen-1-ol was synthesized, confirming findings for flavin-dependent reductases, dehydrogenases, and electron transferases, which frequently use α,β-unsaturated ketones as substrates (36). In this context, incubation of Ncr-His6 with 2-cyclohexen-1-ol did not lead to any NAD(P)H turnover, suggesting that the keto group of 2-cyclohexen-1-one located next to the double bond was essential for Ncr activity.
Despite our current lack of knowledge about the actual substrate for Ncr, experiments carried out in this study somewhat delimited the number of possible substrates. Compounds with nonsubstituted olefinic bonds of α,β-unsaturated ketones were reduced by Ncr. In contrast, structurally related molecules with respective substitutions inhibited the enzymatic activity of Ncr. Larger compounds which also carried substitutions at the olefinic bond, such as the soybean isoflavones, genistein or daidzein, were not substrates and had no inhibitory effect, possibly due to their sizes or due to incompatible spatial arrangements.
Taking into account the diversity of origins for the enzymes most closely related to Ncr and for P. syringae pv. glycinea PG4180 as an isolate from soybean leaves, we speculate that the substrate spectra may differ remarkably. As demonstrated by Craig and coworkers (6), MR of P. putida M10 could have branched off the class I flavin-dependent α/β-barrel oxidoreductases relatively late in evolution. Our results suggested that enzymes related to MR can also be found in phytopathogens but that their distribution within plant-pathogenic bacteria seemed to be closely restricted.
The occurrence of the ncr gene in various representatives of P. syringae pv. glycinea, its absence in related plant pathogens, and its differential expression at virulence-promoting low temperatures encouraged us to set up the following working hypothesis. Ncr might be of special importance under low-temperature conditions during the interaction of the bacterial pathogen with its soybean host plant. This protein could be required for certain as yet unknown processes of the bacterial secondary metabolism. In support of this, the use of NADPH as an electron donor is rather typical for biosynthetic enzymes. Alternatively, the ability to specifically reduce the olefinic bond of α,β-unsaturated ketones such as 2-cyclohexen-1-one could indicate that plant-borne defense compounds with such a target structure may be substrates for Ncr. Many of those mediate antimicrobial effects and eventually need to be detoxified by invading pathogens (7, 21, 26). At least one other possibility could involve the functional analogy of Ncr to OPDA reductase, an enzyme involved in octadecanoid biosynthesis leading to jasmonic acid, which is a plant signal transducer associated with pathogen defense (33). OPDA reductase is a key enzyme of octadecanoid synthesis because it converts 12-oxophytodienoate into a readily available intermediate for β-oxidation and therefore triggers jasmonate synthesis. Interestingly, the P. syringae phytotoxin COR, which is synthesized under conditions similar to those used for Ncr, is known to mimic jasmonate at the molecular level (43). Experiments to further define the actual substrate spectrum for Ncr are under way in our laboratory.
It remains to be elucidated why the Ncr protein occurred predominantly in crude extracts of P. syringae cells grown at 18 and 22°C but not in samples grown at 28°C. This could be due to increased transcription of the ncr gene or increased protein stability at the lower temperatures. A similar phenomenon has been described for at least two different proteins involved in the temperature-dependent biosynthesis of COR, the polyketide phytotoxin synthesized by P. syringae pv. glycinea (5, 27, 30). It is very unlikely that Ncr functions in COR biosynthesis, because the ncr gene was found to be chromosomally located, whereas all necessary functions for heterologous COR biosynthesis are encoded on the 95-kb conjugative plasmid p4180A in strain PG4180 (1, 44). In this study, copies of the ncr gene were found in phytotoxin-negative strains of P. syringae pv. glycinea but not in all COR-producing strains tested (data not shown). At the moment, we cannot rule out the possibility that the temperature-dependent occurrences of Ncr and COR biosynthetic enzymes are somehow linked at the regulatory level. The preferential expression of Ncr in minimal medium was in line with many other virulence-associated processes in plant-pathogenic bacteria (16, 22, 25, 38). Examples for a global regulation of various virulence factors in P. syringae have been demonstrated previously (20, 29).
Molecular tools to investigate the transcriptional activation of the ncr gene as well as the stability of its respective messenger and gene product are now available and will be used for respective studies in our laboratory. Furthermore, by generating a knockout mutant and testing it in plant inoculation studies, we hope to identify possible functions of Ncr in P. syringae virulence.
ACKNOWLEDGMENTS
This study was financed by the Max-Planck-Gesellschaft and by the Deutsche Forschungsgemeinschaft.
We are grateful to Holger Berk, Steffen Heim, and Hans Scholten for their very helpful contributions during the initial stages of this work and to Antonio Pierik for critical review of the manuscript and for his help with the Meldola’s blue assay.
REFERENCES
- 1.Bender C L, Liyanage H, Palmer D, Ullrich M, Young S, Mitchell R. Characterization of the genes controlling biosynthesis of the polyketide phytotoxin coronatine including conjugation between coronafacic and coronamic acid. Gene. 1993;133:31–38. doi: 10.1016/0378-1119(93)90221-n. [DOI] [PubMed] [Google Scholar]
- 2.Bender C L, Palmer D A, Penaloza-Vazquez A, Rangaswamy V, Ullrich M. Biosynthesis of coronatine, a thermoregulated phytotoxin produced by the phytopathogen Pseudomonas syringae. Arch Microbiol. 1996;166:71–75. [Google Scholar]
- 3.Bender C L, Young S A, Mitchell R E. Conservation of plasmid DNA sequences in coronatine-producing pathovars of Pseudomonas syringae. Appl Environ Microbiol. 1991;57:993–999. doi: 10.1128/aem.57.4.993-999.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruce N C, Wilmot C J, Jordan K N, Trebilcock A E, Gray Stephens L D, Lowe C R. Microbial degradation of the morphine alkaloids: identification of morphinone as an intermediate in the metabolism of morphine by Pseudomonas putida M10. Arch Microbiol. 1990;154:465–470. doi: 10.1007/BF00245229. [DOI] [PubMed] [Google Scholar]
- 5.Budde I P, Rohde B H, Bender C L, Ullrich M S. Growth phase and temperature influence promoter activity, transcript abundance, and protein stability during biosynthesis of the Pseudomonas syringae phytotoxin coronatine. J Bacteriol. 1998;180:1360–1367. doi: 10.1128/jb.180.6.1360-1367.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Craig D H, Moody P C E, Bruce N C, Scrutton N S. Reductive and oxidative half-reactions of morphinone reductase from Pseudomonas putida M10: a kinetic and thermodynamic analysis. Biochemistry. 1998;37:7598–7607. doi: 10.1021/bi980345i. [DOI] [PubMed] [Google Scholar]
- 7.Dempsey D A, Silva H, Klessig D F. Engineering disease and pest resistance in plants. Trends Microbiol. 1998;6:54–61. doi: 10.1016/S0966-842X(97)01186-4. [DOI] [PubMed] [Google Scholar]
- 8.Dunleavy J M. Bacterial, fungal, and viral diseases affecting soybean leaves. St. Paul, Minn: American Phytopathological Society; 1988. [Google Scholar]
- 9.Fox K M, Karplus P A. Old yellow enzyme at 2 Å resolution: overall structure, ligand binding, and comparison with related flavoproteins. Structure. 1994;2:1089–1105. [PubMed] [Google Scholar]
- 10.French C E, Bruce N C. Bacterial morphinone reductase is related to Old Yellow Enzyme. Biochem J. 1995;312:671–678. doi: 10.1042/bj3120671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.French C E, Bruce N C. Purification and characterization of morphinone reductase from Pseudomonas putida M10. Biochem J. 1994;301:97–103. doi: 10.1042/bj3010097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.French C E, Nicklin S, Bruce N C. Sequence and properties of pentaerythritol tetranitrate reductase from Enterobacter cloacae. J Bacteriol. 1996;178:6623–6627. doi: 10.1128/jb.178.22.6623-6627.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gondry M, Diep Le K H, Manson F D, Chapman S K, Mathews F S, Reid G A, Lederer F. On the lack of coordination between protein folding and flavin insertion in Escherichia coli for flavocytochrome b2 mutant forms Y254L and D282N. Protein Sci. 1995;4:925–935. doi: 10.1002/pro.5560040512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goss R W. The relation of temperature to common halo blight of beans. Phytopathology. 1970;30:258–264. [Google Scholar]
- 15.Hettwer U, Jaeckel F R, Boch J, Meyer M, Rudolph K, Ullrich M S. Cloning, nucleotide sequencing and expression in Escherichia coli of levansucrase genes from the plant pathogens Pseudomonas syringae pv. glycinea and P. syringae pv. phaseolicola. Appl Environ Microbiol. 1998;64:3180–3187. doi: 10.1128/aem.64.9.3180-3187.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hugouvieux-Cotte-Pattat N, Dominguez H, Robert-Baudouy J. Environmental conditions affect transcription of the pectinase genes of Erwinia chrysanthemi 3937. J Bacteriol. 1992;174:7807–7818. doi: 10.1128/jb.174.23.7807-7818.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin S, Song Y-N, Deng W-Y, Gordon M P, Nester E W. The regulatory VirA protein of Agrobacterium tumefaciens does not function at elevated temperatures. J Bacteriol. 1993;175:6830–6835. doi: 10.1128/jb.175.21.6830-6835.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keane P J, Kerr A, New P B. Crown gall of stone fruit. II. Identification and nomenclature of Agrobacterium isolates. Aust J Biol Sci. 1970;23:585–595. [Google Scholar]
- 19.King E O, Ward M K, Raney D E. Two simple media for the demonstration of pyocyanin and fluorescein. J Lab Clin Med. 1954;44:301–307. [PubMed] [Google Scholar]
- 20.Kitten T, Kinscherf T G, McEvoy J L, Willis D K. A newly identified regulator is required for virulence and toxin production in Pseudomonas syringae. Mol Microbiol. 1998;28:917–929. doi: 10.1046/j.1365-2958.1998.00842.x. [DOI] [PubMed] [Google Scholar]
- 21.Lamb C J, Ryals J A, Ward E R, Dixon R A. Emerging strategies for enhancing crop resistance to microbial pathogens. Biotechnology. 1992;10:1436–1445. doi: 10.1038/nbt1192-1436. [DOI] [PubMed] [Google Scholar]
- 22.Lanham P G, McIlravey K I, Perombelon M C M. Production of the cell wall dissolving enzymes by Erwinia carotovora subsp. atroseptica in vitro at 27°C and 30.5°C. J Appl Bacteriol. 1991;70:20–24. [Google Scholar]
- 23.Miura K, Tomioka Y, Suzuki H, Yonezawa M, Hishinuma T, Mizugaki M. Molecular cloning of the nemA gene encoding N-ethylmaleimide reductase from Escherichia coli. Biol Pharm Bull. 1997;20:110–112. doi: 10.1248/bpb.20.110. [DOI] [PubMed] [Google Scholar]
- 24.O’Farrell P H. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975;250:4007–4020. [PMC free article] [PubMed] [Google Scholar]
- 25.Palmer D A, Bender C L. Effects of environmental and nutritional factors on production of the polyketide phytotoxin coronatine by Pseudomonas syringae pv. glycinea. Appl Environ Microbiol. 1993;59:1619–1623. doi: 10.1128/aem.59.5.1619-1626.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Phillips D A, Kapulnik Y. Plant isoflavonoids, pathogens and symbionts. Trends Microbiol. 1995;3:58–64. doi: 10.1016/s0966-842x(00)88876-9. [DOI] [PubMed] [Google Scholar]
- 27.Rangaswamy V, Ullrich M, Jones W, Mitchell R, Parry R, Reynolds P, Bender C L. Expression and analysis of coronafacate ligase, a thermoregulated gene required for production of the phytotoxin coronatine in Pseudomonas syringae. FEBS Lett. 1997;154:65–72. doi: 10.1111/j.1574-6968.1997.tb12625.x. [DOI] [PubMed] [Google Scholar]
- 28.Rice P A, Goldman A, Steitz T A. A helix-turn-strand structural motif common in alpha-beta proteins. Proteins. 1990;8:334–340. doi: 10.1002/prot.340080407. [DOI] [PubMed] [Google Scholar]
- 29.Rich J J, Kinscherf T G, Kitten T, Willis D K. Genetic evidence that the gacA gene encodes the cognate response regulator for the lemA sensor in Pseudomonas syringae. J Bacteriol. 1994;176:7468–7475. doi: 10.1128/jb.176.24.7468-7475.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rohde B H, Pohlack B, Ullrich M S. Occurrence of thermoregulation of genes involved in coronatine biosynthesis among various Pseudomonas syringae strains. J Basic Microbiol. 1998;38:41–50. [PubMed] [Google Scholar]
- 31.Rowley K B, Clements D E, Mandel M, Humphreys T, Patil S S. Multiple copies of a DNA sequence from Pseudomonas syringae pathovar phaseolicola abolish thermoregulation of phaseolotoxin production. Mol Microbiol. 1993;8:625–635. doi: 10.1111/j.1365-2958.1993.tb01606.x. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 33.Schaller F, Weiler E W. Enzymes of octadecanoid biosynthesis in plants: 12-oxo-phytodienoate 10,11-reductase. Eur J Biochem. 1997;245:294–299. doi: 10.1111/j.1432-1033.1997.t01-1-00294.x. [DOI] [PubMed] [Google Scholar]
- 34.Schaller F, Weiler E W. Molecular cloning and characterization of 12-oxophytodienoate reductase, an enzyme of the octadecanoid signaling pathway from Arabidopsis thaliana. J Biol Chem. 1997;272:28066–28072. doi: 10.1074/jbc.272.44.28066. [DOI] [PubMed] [Google Scholar]
- 35.Scrutton N S. Alpha/beta barrel evolution and the modular assembly of enzymes: emerging trends in the flavin oxidase/dehydrogenase family. Bioessays. 1994;16:115–122. doi: 10.1002/bies.950160208. [DOI] [PubMed] [Google Scholar]
- 36.Sinnott M. Comprehensive biological catalysis: a mechanistic reference. Vol. 3. London, England: Academic Press; 1998. [Google Scholar]
- 37.Snape J R, Walkley N A, Morby A P, Nicklin S, White G F. Purification, properties, and sequence of glycerol trinitrate reductase from Agrobacterium radiobacter. J Bacteriol. 1997;179:7796–7802. doi: 10.1128/jb.179.24.7796-7802.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Staskawicz B J, Dahlbeck D, Keen N T. Cloned avirulence gene of Pseudomonas syringae pv. glycinea determines race-specific incompatibility on Glycine max (L.) Merr. Proc Natl Acad Sci USA. 1984;81:6024–6028. doi: 10.1073/pnas.81.19.6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ullrich M, Guenzi A C, Mitchell R E, Bender C L. Cloning and expression of genes required for coronamic acid (2-ethyl-1-aminocyclopropane 1-carboxylic acid), an intermediate in the biosynthesis of the phytotoxin coronatine. Appl Environ Microbiol. 1994;60:2890–2897. doi: 10.1128/aem.60.8.2890-2897.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ullrich M, Penaloza-Vazquez A, Bailey A M, Bender C L. A modified two-component regulatory system is involved in temperature-dependent biosynthesis of the Pseudomonas syringae phytotoxin coronatine. J Bacteriol. 1995;177:6160–6169. doi: 10.1128/jb.177.21.6160-6169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Völker U, Engelmann S, Maul B, Riethdorf S, Volker A, Schmid R, Mach H, Hecker M. Analysis of the induction of general stress proteins of Bacillus subtilis. Microbiology. 1994;140:741–752. doi: 10.1099/00221287-140-4-741. [DOI] [PubMed] [Google Scholar]
- 42.Wei Z B, Beer S V. HrpL activates Erwinia amylovora hrp gene transcription and is a member of the ECF subfamily of sigma factors. J Bacteriol. 1995;177:6201–6210. doi: 10.1128/jb.177.21.6201-6210.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weiler E W, Kutchan T M, Gorba T, Brodschelm W, Niesel U, Bublitz F. The Pseudomonas phytotoxin coronatine mimics octadecanoid signalling molecules of higher plants. FEBS Lett. 1994;345:9–13. doi: 10.1016/0014-5793(94)00411-0. [DOI] [PubMed] [Google Scholar]
- 44.Young S A, Park S K, Rodgers C, Mitchell R E, Bender C L. Physical and functional characterization of the gene cluster encoding the polyketide phytotoxin coronatine in Pseudomonas syringae pv. glycinea. J Bacteriol. 1992;174:1837–1843. doi: 10.1128/jb.174.6.1837-1843.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]