Summary
Background
Autopsy studies have provided valuable insights into the pathophysiology of COVID-19. Controversies remain about whether the clinical presentation is due to direct organ damage by SARS-CoV-2 or secondary effects, such as overshooting immune response. SARS-CoV-2 detection in tissues by RT-qPCR and immunohistochemistry (IHC) or electron microscopy (EM) can help answer these questions, but a comprehensive evaluation of these applications is missing.
Methods
We assessed publications using IHC and EM for SARS-CoV-2 detection in autopsy tissues. We systematically evaluated commercially available antibodies against the SARS-CoV-2 proteins in cultured cell lines and COVID-19 autopsy tissues. In a multicentre study, we evaluated specificity, reproducibility, and inter-observer variability of SARS-CoV-2 IHC. We correlated RT-qPCR viral tissue loads with semiquantitative IHC scoring. We used qualitative and quantitative EM analyses to refine criteria for ultrastructural identification of SARS-CoV-2.
Findings
Publications show high variability in detection and interpretation of SARS-CoV-2 abundance in autopsy tissues by IHC or EM. We show that IHC using antibodies against SARS-CoV-2 nucleocapsid yields the highest sensitivity and specificity. We found a positive correlation between presence of viral proteins by IHC and RT-qPCR-determined SARS-CoV-2 viral RNA load (N= 35; r=-0.83, p-value <0.0001). For EM, we refined criteria for virus identification and provide recommendations for optimized sampling and analysis. 135 of 144 publications misinterpret cellular structures as virus using EM or show only insufficient data. We provide publicly accessible digitized EM sections as a reference and for training purposes.
Interpretation
Since detection of SARS-CoV-2 in human autopsy tissues by IHC and EM is difficult and frequently incorrect, we propose criteria for a re-evaluation of available data and guidance for further investigations of direct organ effects by SARS-CoV-2.
Funding
German Federal Ministry of Health, German Federal Ministry of Education and Research, Berlin University Alliance, German Research Foundation, German Center for Infectious Research.
Keywords: SARS-CoV-2, COVID-19, Autopsy, Immunohistochemistry, Electron microscopy, Pathology
Research in context.
Evidence before this study
We searched PubMed and Google scholar for all publications published between 01 April 2020 to March 2022 claiming proof of SARS-CoV-2 particles in human tissues by electron microscopy (EM) or publications discussing the challenges of this technique using the main search terms “SARS-CoV-2“, “electron microscopy” and “autopsy” or “biopsy”. In addition, we reviewed all publications on immunohistochemical (IHC) detection of SARS-CoV-2 (as of November 2021) from PubMed using the main search terms “SARS-CoV-2“, “detection” and “IHC”. Our searches were restricted to studies in English or offered an Abstract in English. Numerous studies performed in situ detection of SARS-CoV-2 using IHC and EM to define cell and organ tropism and linking presence of SARS-CoV-2 to organ damage in multiple human tissues and organs. Several correspondences and reviews discussed the challenges of virus in situ detection using EM, because a substantial number of studies misinterpreted cellular structures as virus. Nevertheless, such misinterpretations have still been published.
Added value of this study
A comprehensive evaluation of in situ methods of virus detection in human autopsy tissues is missing. We therefore systematically evaluated commercially available antibodies against SARS-CoV-2 proteins. We show in a multicentre study that IHC using antibodies against SARS-CoV-2 nucleocapsid yields the highest sensitivity and specificity. We also found a positive correlation between presence of viral proteins by IHC and RT-qPCR-determined SARS-CoV-2 viral RNA load and show that tissue compartments with low viral load are prone to false positive IHC interpretation. We show that in many publications, unreliable antibodies were used, and staining patterns apparently were misinterpreted as false positive. We used qualitative and quantitative EM analyses to refine criteria for ultrastructural identification of SARS-CoV-2 and deduce why misinterpretations continued. Of note, 135 of 144 publications misinterpret cellular structures as virus using EM or show only insufficient data. Thus, other techniques of virus detection were wrongfully validated by false positive EM data, ultimately producing a questionable basis to claim a multiorgan tropism of SARS-CoV-2. We provide recommendations for improved in situ virus detection in autopsy tissues as well as publicly accessible digitized EM sections as a reference and for training purposes.
Implications of all the available evidence
Detection of SARS-CoV-2 in human autopsy tissues by IHC and EM is prone to false positive interpretation. This needs to be considered when interpreting these data. Our proposed criteria for detecting SARS-CoV-2 using IHC and EM provide guidance for re-evaluation of available data and further investigations.
Alt-text: Unlabelled box
Introduction
In clinical routine, quantitative reverse transcriptase (RT)-qPCR and rapid antigen detection tests from nasopharyngeal swabs are robust, standardized, and validated tools for screening or diagnosing severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infections. In contrast, in situ SARS-CoV-2 detection methods such as immunohistochemistry (IHC) and electron microscopy (EM) in patient tissues are much less validated, yet autopsy studies use these methods to investigate mechanisms of organ damage and tropism of SARS-CoV-2.1,2
Detection of pathogen-specific antigens by immunohistochemical staining is a potent diagnostic tool allowing spatial correlation of pathological changes with presence of the pathogen. Early in the COVID-19 pandemic, positive controls for this newly emerging disease were not available. The urgent need to assess the distribution of SARS-CoV-2 in tissues of deceased patients with mentioned technical limitations led to inconsistencies in interpretation of SARS-CoV-2 localization and distribution by IHC and EM.1, 2, 3, 4, 5, 6
Only diagnostic EM allows direct visualization of intact SARS-CoV-2 particles.7 It thereby validates other in situ techniques, detecting viral proteins or RNA, and enables cellular/subcellular localization of virus particles.7 EM of SARS-CoV-2 in model systems expanded our understanding of structural and cellular biology, yet did not provide information on the distribution of the virus in patients.8,9 Due to complex sample processing procedures in EM and challenges in recording and interpretation of micrographs, misinterpretation of structures as SARS-CoV-2 particles in patient tissues occurred.10,11 In autopsy tissues, virus structures have to be distinguished from other structures of cells. Therefore, sufficient structural preservation and a suitable sampling strategy are needed.3,10,12 Recommendations for identifying SARS-CoV-2 by EM are mainly based on virus particles in cell cultures,11,13 which only partially reflect the situation in autopsy tissues.
Here we collated available data on in situ detection of SARS-CoV-2 in human tissue focussing on immunohistochemical detection of SARS-CoV-2 proteins and EM detection of intact SARS-CoV-2 particles. Furthermore, we determined optimally suited SARS-CoV-2 antibodies and evaluated their sensitivity and specificity in a multicentre approach. We correlated viral load as detected by RT-qPCR to presence of SARS-CoV-2 proteins and viral particles as detected by IHC or EM, respectively. Additionally, we suggest refined criteria for identifying SARS-CoV-2 by IHC and EM and provide a publicly accessible repository of digitized electron microscopical sections showing examples and pitfalls for in situ detection of SARS-CoV-2.
Methods
Ethics
Autopsies were performed after consent was given by next of kin at the Institute of Legal Medicine of the University Medical Center Hamburg-Eppendorf, Germany and Institute of Pathology Charité Universitätsmedizin Berlin, and Institute of Pathology and Electron Microscopy Facility, RWTH University of Aachen, Germany. Use of human tissue for post mortem studies after conclusion of diagnostic procedures has been reviewed and approved by the institutional review board of the independent Ethics Committee of the Hamburg Chamber of Physicians (WF-051/20; protocol-no. PV7311) and by the local Ethics Committee (Berlin: EA2/066/20) and by the Charité-BIH COVID-19 research board, and by local Ethics Committee (Aachen: EK 304/20, EK 119/20, and EK 092/20), and the study is in line with the Declaration of Helsinki.
Sample processing for immunohistochemistry
Paraffin cell blocks from SARS-CoV-2-infected and un-infected Vero cells (Supplementary Methods) were processed similar to autopsy tissues regarding fixation time and paraffin embedding. Lung and respiratory mucosa tissue from COVID-19 patients and control autopsies were formalin-fixed and paraffin-embedded (FFPE) using standard laboratory procedures from three study centres. Immunohistochemical staining with antibodies against SARS-CoV-2 proteins (Supplementary Table 1) were performed using a Ventana Benchmark XT autostainer at two centres.
Sample processing for electron microscopy
31 autopsy samples from lung, kidney, brain, heart, upper respiratory tract (olfactory mucosa and trachea) derived from 14 COVID-19 patients of the IHC cohort and 3 additional patients and SARS-CoV-2-infected Vero cells were assessed (specified in Supplementary Methods; Supplementary Table 5). For qualitative and quantitative analyses of infected cells and viral particles, autopsy lung (Supplementary Table 5, case B1) and FFPE re-embedded olfactory mucosa (case B3) with a high viral RNA load, as determined by RT-qPCR, and successful detection of particles by EM were used.
RT-qPCR
All included tissue samples for IHC and EM were analysed by RT-qPCR of the same tissue block as specified in Supplementary Methods; Supplementary Table 3 and 5).
Large-scale electron microscopy and transmission electron microscopy
To screen for SARS-CoV-2 particles, sections prepared from four resin blocks of two autopsy lung samples (case B1) and sections from two resin blocks of SARS-CoV-2-infected Vero cells were completely digitized at 3-4 nm pixel size as described14 (Supplementary Material). Large-scale screening datasets were generated via TrakEM2 for stitching and nip2 for export to high-resolution tif files14 for analysis using QuPath. Infected cells in autopsy lung were annotated and digitized at 1 nm pixel size for quantitative analysis. To assess the heterogeneity of coronavirus (CoV) particles in infected cells, four types of particles were specified (Figure 5d-s). These types were counted manually in QuPath, and intracellular CoV particles per µm2 cytoplasmic area were determined. Also, particle diameter (largest diameter without spikes), ribonucleoprotein (RNP) diameter (smallest diameter), and diameter of tubular structures were measured. In FFPE re-embedded olfactory mucosa (case B3), we restricted quantitative analysis to measurement of particle diameter via Fiji, based on TEM images, due to limited preservation. We compared viral RNA load with virus particle number per section and cell to estimate the likelihood of finding CoV particles via thin section EM (Supplementary Methods).
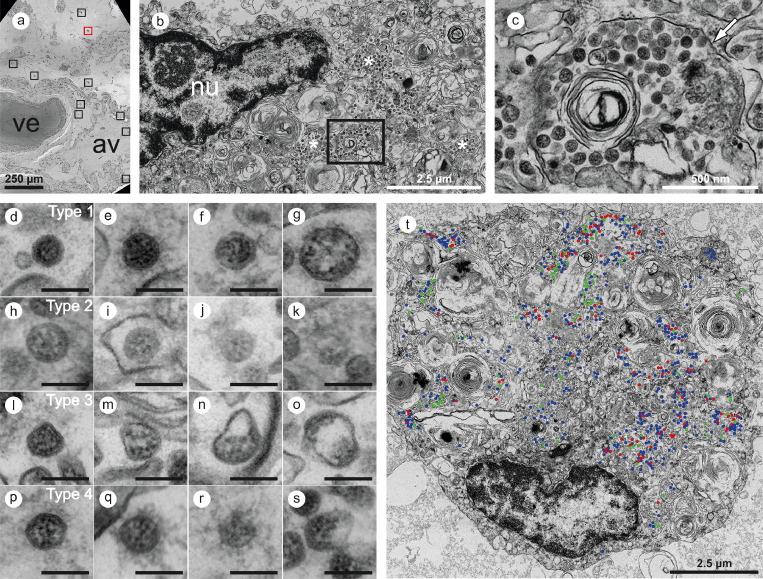
Figure 5.
Large-scale electron microscopy of SARS-CoV-2 in human autopsy lung tissue. Four entire sections of B1 were automatically digitized (dataset 1 is depicted here) at 3-4 nm pixel size to screen for SARS-CoV-2 infected cells (a-c). In the overview (a), a large vessel (ve) and an alveolus (av) are readily detectable. Preserved microanatomy allows to precisely locate regions of interest at the nanometer scale within the histological context at the millimeter scale. The red box in (a) indicates the position of the infected cell shown in (B; cell 6), the nine black boxes indicate locations of other infected cells. Numerous coronavirus (CoV) particles are located in the region next to the nucleus (nu), note several well-preserved membrane compartments with numerous CoV particles (white asterisks in b, and box in b that is further magnified and marked by the white arrow in c). The screening resolution is sufficient to detect typical CoV morphology (c) with prominent ribonucleoprotein (RNP) within the interior of particles. All infected cells were digitized at a very high resolution of 1 nm pixel size (d-s; T, cell 7) to resolve CoV substructure with improved image quality for validation and quantitative analysis. (d-s) Ultrastructural types of coronavirus particles; for quantitative analysis of CoV particles, different morphological types were defined. (d-g) Type 1 CoV particles are electron dense, corresponding to a large particle volume within the section and are relatively well preserved with a round to oval shape. Different representative appearances are shown here; small particle (d), a standard-sized particle with evenly distributed granular-appearing RNP (e), partly elongated-appearing RNP (f), a larger particle with slightly irregular interior (g). (h-k) Less electron dense type 2 CoV particles, corresponding to a smaller particle volume within the section, while being also relatively well preserved; relatively electron dense particle with well-recognizable biomembrane (h), less electron dense particles with also less distinct biomembrane (i-k). Note that the partly granular and partly elongated RNP profiles are still visible. (l-o) Type 3 CoV particles have the electron density of type 1 or type 2 CoV particles, but show more bizarre shapes. (p-s) Type 4 CoV particles were defined as all extracellular particles next to infected cells; well preserved dark particle (p), some particles showed prominent “fuzzy” coats (q), less electron dense (r) and deformed (s) particles. (t) Visualization of different particle types in QuPath; type 1 (blue), type 2 (red) and type 3 (green). See also http://www.nanotomy.org/OA/Krasemann2022eBioMedicine/index.html for internet browser-based open access pan-and-zoom analysis of the full resolution datasets and for our Supplementary Video demonstrating how large-scale electron microscopy facilitates ultrastructural analysis and visual pattern recognition.
Search and analysis of publications demonstrating immunohistochemical and/or ultrastructural detection of SARS-CoV-2
We defined specific search strategies to find scientific publications using immunohistochemical datasets from autopsy studies with antibodies to SARS-CoV-2 proteins. Additionally, we searched for all scientific publications claiming ultrastructural proof of SARS-CoV-2 particles in infected human tissues (Supplementary Methods).
Statistics
For the multicentre study evaluating IHC to SARS-CoV-2 nucleocapsid, we used antibody N#9. Cases were selected form the autopsy cohorts of the university medical centers of Hamburg, Berlin and Aachen and selected to represent the spectrum of different viral loads (high, low and medium) and different post-mortem interval to mirror the heterogeneity of routine diagnostic cases. Inclusion criteria were cause of death COVID-19 and SARS-CoV-2 qPCR positive in autopsy lung tissue. Exclusion criteria was unavailability of tissue for further analysis. As controls, we selected ARDS and Influenza cases as well as normal appearing autopsy lung tissue as specified in Supplementary Table 3a-c. Staining specificity and intensity were evaluated by eight independent pathologists/neuropathologists at four different centres (Pathology Aachen, Neuropathology Berlin, Neuropathology Hamburg, Pathology Paris). Observers received instructions and teaching slides and were asked to categorize slides in a blinded fashion, using a published four-tiered semiquantitative approach (none (0), slight (+), moderate (++), and severe (+++))15 (Figure 3). In brief, to achieve a (+) single-positive cells <10 cells / total section were required, for (++) single-positive cells and clusters of cells (>10 but below 50) were required, and for (+++) multiple positive cells (>50) and/or several clusters of positive cells were required, all cell types were included.
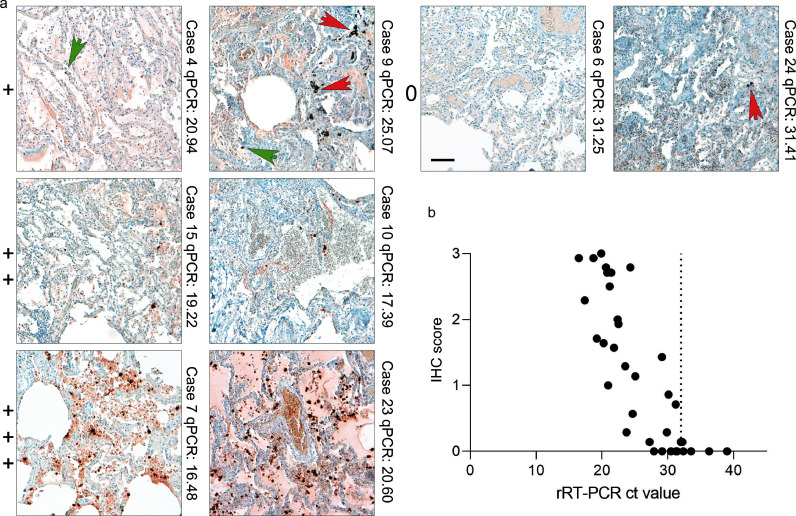
Figure 3.
IHC for SARS-CoV-2 nucleocapsid correlates with SARS-CoV-2 viral RNA load determined by RT-qPCR. (a) Lung tissues of COVID-19 patients and respective controls were stained with an anti-nucleocapsid-antibody (#9) and scored in a blinded fashion by pathologists from four different centres (patient details in Supplementary Tables 3a-c; scoring results in Supplementary Table 4). We defined four categories for the scoring of nucleocapsid abundance: 0= no detection; += detection of single and/or regionally separated positive cells; ++= several positive cells and/or cluster of cells in a regionally restricted manner; +++= high abundance of positive cells and/or several highly positive cluster. Representative images of lung tissues for all four scoring categories are shown together with the case number and the RT-qPCR value. Green arrows point towards single positive cells; red arrows mark typical pigmentation due to formalin or anthracosis as further pitfalls for IHC interpretation in lung tissue. Scale bar: 100 µm. (b) SARS-CoV-2 viral RNA load was determined by RT-qPCR of consecutive tissue block paraffin sections. Viral RNA loads correlate with detection of SARS-CoV-2 nucleocapsid by IHC (r= -0.83, p-value <0.0001; 35 pairs). Of note, widespread detection (score+++) of nucleocapsid in lung tissue is only associated with high RNA loads/low ct values, whereas at high ct values (low RNA) detection of positive cells is comparatively low.
Every case was rated by seven investigators. Cases with ≥70% rater-agreement (≥5 of 7 raters) were classified as positive (“+” or more) or negative. Cases with unclear classification into positive or negative due to inconsistent ratings (less than 60% agreement) were defined as non-classifiable (n=2; one control / one COVID-19). To measure a linear dependence between viral load and immunohistochemical scores pearson correlation calculation was done with Graphpad Prism (GraphPad, La Jolla, USA). Here non classifiable cases (n=2) were excluded. Sensitivity was calculated as Sensitivity = True positive/True positive + False negative. Specificity was calculated as Specificity = True negative/True negative + False positive (Supplementary Table 4). Sample size determination was not calculated given the explorative character of the study.
Role of funders
The funders had no role in study design, data collection, data analyses, interpretation, writing or submission of the publication.
Results
Assessing commercially available antibodies against SARS-CoV-2 proteins
We curated papers with immunohistochemical datasets and testing of different antibodies.16, 17, 18, 19, 20, 21, 22, 23, 24 We found that the majority of studies neither validated the antibodies nor reported adequate positive and negative controls. Detection of SARS-CoV-2 proteins by IHC in autopsy tissues showed differences regarding protein amounts and interpretation of data. However, lack of controls or usage of non-validated antibodies does not mean that the results are invalid per se, but if this comes along with low viral RT-qPCR load, findings should be critically re-evaluated.
Thirteen commercially available anti-SARS-CoV-2 protein antibodies were evaluated in this study. Three target spike protein, two non-structural protein 3 (Nsp3), seven nucleocapsid protein, and one targets double-strand RNA (dsRNA) (Supplementary Table 1).
Firstly, we tested antibodies on FFPE SARS-CoV-2 infected and un-infected Vero cells (Supplementary Figure 1) simultaneously processed to avoid batch effects. Of the three different anti-spike-antibodies, two gave a specific signal, but one also produced mild nonspecific staining. One widely used antibody (#3) did not produce a signal at all. Both anti-Nsp3-antibodies generated high background staining in un-infected cells. An antibody against dsRNA generated a faint signal in infected cells. Of the seven nucleocapsid-antibodies, five produced specific staining on infected cells with minimal background in un-infected cells. One widely used antibody showed high nonspecific background in un-infected cells (#12), while another did not produce any signal (#13) (Supplementary Figure 1).
To further investigate the specificity of widely used antibodies that did not perform in our FFPE cell-based evaluation, we double-stained those (spike #3, Nsp3 #5, dsRNA #6, and nucleocapsid #12, Supplementary Figure 2) in formalin-fixed, but non-embedded SARS-CoV-2-infected Vero cells together with one well-working anti-nucleocapsid-antibody (#7 or #9). Un-infected cells served as control. While antibodies #5, #6, and #12 produced specific staining, antibody #3 did not produce a detectable signal again (Supplementary Figure 2). Antibodies #4 and #13 did not produce specific signals in FFPE cell blocks or fixed cells and were not further considered.
To assess the performance of anti-SARS-CoV-2 antibodies in COVID-19 autopsy tissues, we tested nine of the antibodies on FFPE autopsy tissues from COVID-19 patients and controls. We chose lung and respiratory mucosa as these harbour high SARS-CoV-2 viral loads and stained consecutive sections (Figure 1; Supplementary Figure 3). Two anti-spike-antibodies produced specific staining, while one gave no signal (#3). Of the six different nucleocapsid antibodies, five showed specific and robust staining. However, background staining in COVID-19-positive tissues was higher in the polyclonal (#7) and one monoclonal (#11) antibody compared cell blocks. Staining caused by SARS-CoV-2 proteins in cellular debris or abundance of tissue artefacts was higher in lung tissue when compared to respiratory mucosa (Figure 1; Supplementary Figure 3). Interestingly, signals obtained with anti-nucleocapsid-antibodies were more abundant when compared to anti-spike-antibodies (Figure 1; Supplementary Figure 3). To evaluate this we performed immunofluorescence double-staining on Vero cells, lung, and respiratory mucosa and consistently observed more nucleocapsid-positive cells than spike-positive cells. Moreover, nucleocapsid signals were evenly distributed and more abundant than spike-protein within double-positive cells (Figure 2). This effect was observed with several anti-spike antibodies targeting spike S1 and receptor-binding domain (RBD) (Supplementary Figure 4).
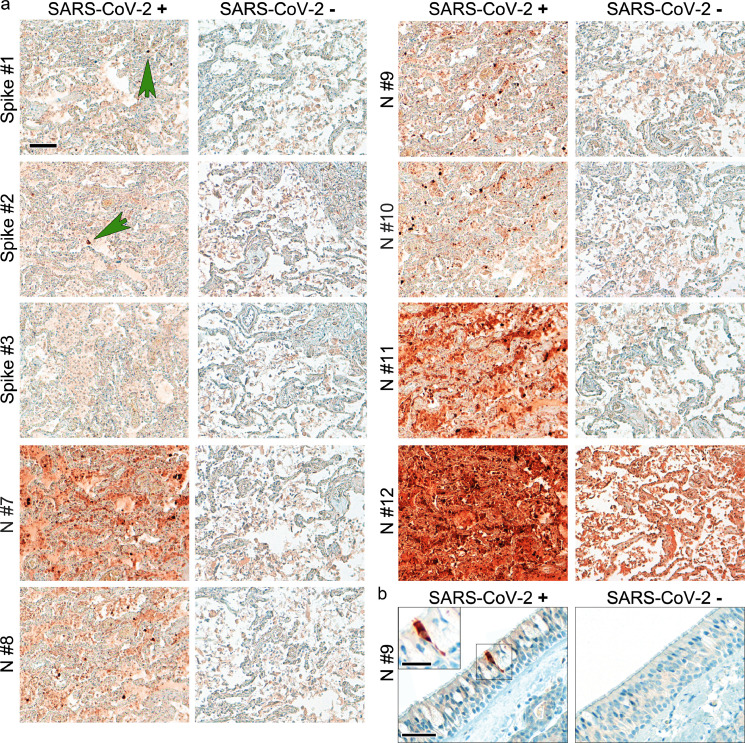
Figure 1.
Suitable antibodies for SARS-CoV-2 protein detection show an optimal signal to background ratio in human autopsy tissues. (a) Formalin-fixed paraffin-embedded lung tissues of COVID-19 (Case Hamburg12; pm time 72 h) and non-COVID-19-control were stained with different anti-spike (spike) and anti-nucleocapsid (N) antibodies (see Supplementary Table 1). Consecutive sections were used to perform the staining to enable comparability of the different antibodies. Green arrows point to single spike-protein positive cells. One widely used antibody (Spike #3) did not detect spike protein in SARS-CoV-2 positive lung tissue. Moreover, one other widely used antibody (N#12) produced very high background staining in non-COVID-19-control lung tissue. Of note, abundance of spike protein is much lower than that of nucleocapsid. Representative images are shown; scale bar: 100 µm. (b) Formalin-fixed paraffin-embedded respiratory mucosa from two COVID-19 patients were stained for SARS-CoV-2 nucleocapsid (antibody #9). The SARS-CoV-2-positive cell is evenly stained by the antibody and does not show single dots; scale bar: 50 µm, close-up: 25 µm.
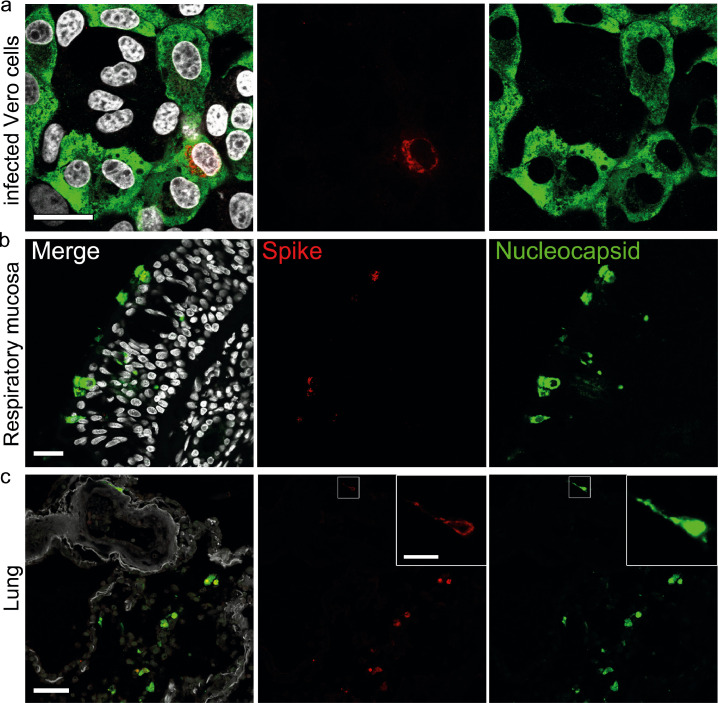
Figure 2.
Higher abundance of nucleocapsid in comparison to spike in infected cells and tissues from COVID-19 patients. (a) Vero cells were infected with SARS-CoV-2 (MOI 0.5; 24 h), fixated in formalin, and directly stained. Representative staining of SARS-CoV-2 spike (red; antibody spike#1) and nucleocapsid (green; antibody N#7); nuclei/DAPI in white; scale bar: 25 µm. (b) FFPE sections of respiratory mucosa (Case Hamburg14); scale bar: 25 µm or (c) lung tissue of a COVID-19 patient (Case Hamburg12) were stained as above for spike (red), nucleocapsid (green), nuclei/DAPI in white; scale bar: 50 µm, close-up: 10 µm. Note that the signal strength and abundance of spike protein and spike-positive cells is always less than that for nucleocapsid. Thus, we recommend using antibodies against nucleocapsid for the detection of SARS-CoV-2 protein positive cells in human autopsy tissues.
Of note, two widely used and published antibodies did not perform well in our analyses. One antibody (anti-N, #12) produced high background staining even in control tissues (Figure 1; Supplementary Figures 1,3), while another (anti-spike, #3) did not result in measurable staining in SARS-CoV-2-infected Vero cells or COVID-19 tissues (Figure 1; Supplementary Figures 1-3). In summary, six anti-SARS-CoV-2 protein antibodies reliably work on FFPE autopsy tissues from COVID-19 patients, with anti-nucleocapsid-antibodies providing better results. Of note, detection of nucleocapsid in virus-infected cells was also conserved in the novel variant of concern (VOC) Omicron (B.1.1.529) when we stained trachea from a patient deceased after infection (Supplementary Figure 5a). In contrast, several anti-spike-antibodies that worked with SARS-CoV-2 WT did not detect B.1.1.529-infected cells (Supplementary Figure 5a, b). Based on our thorough evaluation we suggest recommendations for the use of commercial antibodies for the detection of SARS-CoV-2 in human autopsy tissues by IHC (Table 1, Supplementary Table 2).
Table 1.
Recommendations for detection of SARS-CoV-2 proteins by IHC in formalin-fixed paraffin-embedded (FFPE) human autopsy tissues.
| 1 | Determination of virus load by RT-qPCR may give an overestimated picture of the tissue burden; SARS-CoV-2 positive cells might be comparatively rare. |
| 2 | IHC is not suited to detect low virus amounts in cells due to unfavourable signal to background ratio in human autopsy tissues; positive cells in tissues carry a rather high virus protein load (see Figures 1,3; Supplementary Figures 3,5). |
| 3 | Always use positive/negative controls (e.g. infected versus un-infected FFPE cells (see Supplementary Methods for generation of cell blocks)). If using autopsy tissues, control for fixation time and degree of inflammation. |
| 4 | Some antibodies are not recommended for detection of SARS-CoV-2 in human autopsy tissues. They either produce high background in control tissue or do not stain specifically for SARS-CoV-2 proteins (compare Figure 1; Supplementary Figures 1,3). |
| 5 | Home-made or novel antibodies need to be evaluated using FFPE cell blocks of SARS-CoV-2 infected versus un-infected cells and appropriate positive and negative control tissues. To improve comparability of different COVID-19 datasets, we recommend using one of the established antibodies in addition to the potential new one. |
| 6 | Consult an experienced pathologist to avoid misinterpretation of typical tissue artefacts (e.g. lipofuscin in neurons; carbon deposition in lung; formalin-induced artefacts). Use polarized light to identify formalin-induced artefacts in FFPE tissues (e.g. punctate or dark precipitates). |
| 7 | Nucleocapsid has a higher abundance in virus-protein positive cells, thus, usage of anti-nucleocapsid antibodies is recommended to increase the sensitivity of detection (see Figure 2; Supplementary Figure 4). Epitopes of nucleocapsid might show higher conservation in novel VOC and thus, produce more reliable detection in the future. Anti-spike antibodies are not recommended for detection, but are suitable as additional tools to confirm specificity in double stainings. |
| 8 | COVID-19 tissues often present with high inflammatory changes which are prone to produce higher background staining. Keep in mind that more (nonspecific) signal in tissues could be infection specific, but might not be a SARS-CoV-2 virus protein staining. It is highly recommended to employ a secondary antibody only control or isotype control in IHC. |
| 9 | We recommend to evaluate staining results on a microscope (possibility to focus in z-plane) and not on a scanned image to avoid misinterpretation of nonspecific staining artefacts on the tissue surface. |
| 10 | Some autopsy tissues provide exceptional high background staining such as kidney or placenta and should only be validated together with RT-qPCR results and comparable positive and negative tissues. |
| 11 | Nucleocapsid staining is planar and intracellular, but does not produce single punctuate dots. |
| 12 | Fluorescence microscopy is more prone to background signal than chromogenic IHC due to autofluorescence in human autopsy tissues. Check tissues in several channels to exclude autofluorescent “dots”. |
A multicentre study assessing SARS-CoV-2 immunohistochemistry
Using one optimally performing anti-nucleocapsid-antibody (#9) (Figure 1a,b; Supplementary Figures 1,3), we performed a blinded multicentre study aiming to (1) assess, whether IHC staining in human lung autopsy tissue by experienced pathologists is a suitable method to detect SARS-CoV-2 protein and (2) investigate the correlation between SARS-CoV-2 load as measured by RT-qPCR and IHC (Figure 3). Three centres contributed lung tissues from COVID-19 deceased individuals. Abundance of SARS-CoV-2 nucleocapsid in lung tissue in COVID-19 patients was highly variable and present in a clustered, inhomogeneous pattern (Figure 3; Supplementary Figure 6). However, the specificity of the antibody #9 to detect nucleocapsid was not altered by tissue fixation time (Supplementary Figure 7). Viral loads were determined by RT-qPCR of consecutive sections of the same paraffin tissue blocks as the staining (Supplementary Table 4). Lung tissues from control patients without pathological lung changes, from patients dying of non-COVID-19-related Acute Respiratory Distress Syndrome (ARDS), and those dying with Influenza infections were included (Figure 3; Supplementary Figure 6). Anonymized patient details are summarized in Supplementary Tables 3a-c. Slides were evaluated blinded by pathologists from four different centres and scored in a semiquantitative manner (see Figure 3 for overview and examples; Supplementary Table 4).
We assessed false positive and false negative ratings in lung tissues. The utilized monoclonal antibody (#9) had a sensitivity of 0.72 (Supplementary Table 4). Extensive tissue damage seen in COVID-19 lungs presenting with pre-necrotic epithelial cells and hyaline membranes obscured interpretation of signals. The specificity for antibody #9 was high at 0.95 (41 True negative/ 41 True negative + 2 False positive; see Supplementary Table 4). A correlation between RT-qPCR-defined viral loads and presence of immunohistochemically detected SARS-CoV-2 nucleocapsid protein was seen in tissues with high SARS-CoV-2 viral loads (Figure 3). COVID-19 tissues with low viral RNA loads were often rated false negative (n=7 with a mean ct value of 28.7) which could be true negative as, in these cases, low RNA signal translates to negligible SARS-CoV-2 protein, however, in our calculations these figure as false negative. Only 2/74 cases were not classifiable with an interrater agreement of less than 60%. The overall interrater reliability documented as interrater agreement frequency was 61% and reliability increased up to 81% if only trained raters with experience in SARS-CoV-2 IHC were included (Supplementary Table 4). The cases with interrater discrepancy (all raters) were controls (n=14) and COVID-19 (n=15) samples with only single positive cells. In contrast, COVID-19 cases with a high viral RNA load and a high IHC score were always classified correctly.
Thus, our multicentre study showed that detection of SARS-CoV-2 proteins in human autopsy tissues is feasible yet pitfalls such as discrimination of true signal from the background and limited sensitivity of the method in tissues with low viral RNA load cause interpretational difficulties, highlighting the importance of proper controls in autopsy studies and training of evaluators.
Ultrastructural analysis of SARS-CoV-2 in human tissues
Guidance for identifying SARS-CoV-2 has been provided, albeit without detailed consideration of the specific challenges of autopsy tissues.11,13 We searched for SARS-CoV-2 viral particles (Figure 4) in the lung, olfactory mucosa, medulla oblongata, kidney, trachea, and myocardium in autopsy tissues of 17 COVID-19 patients (n=14 from the above-mentioned cohort). Virus particles were found in lung (case B1) and olfactory mucosa (case B3). Viral particle detection correlated well with RT-qPCR ct values and immunohistochemical staining scores (Supplementary Table 5). Also, in situ hybridization of case B1 (olfactory mucosa) was positive, and published previously.3
Figure 4.
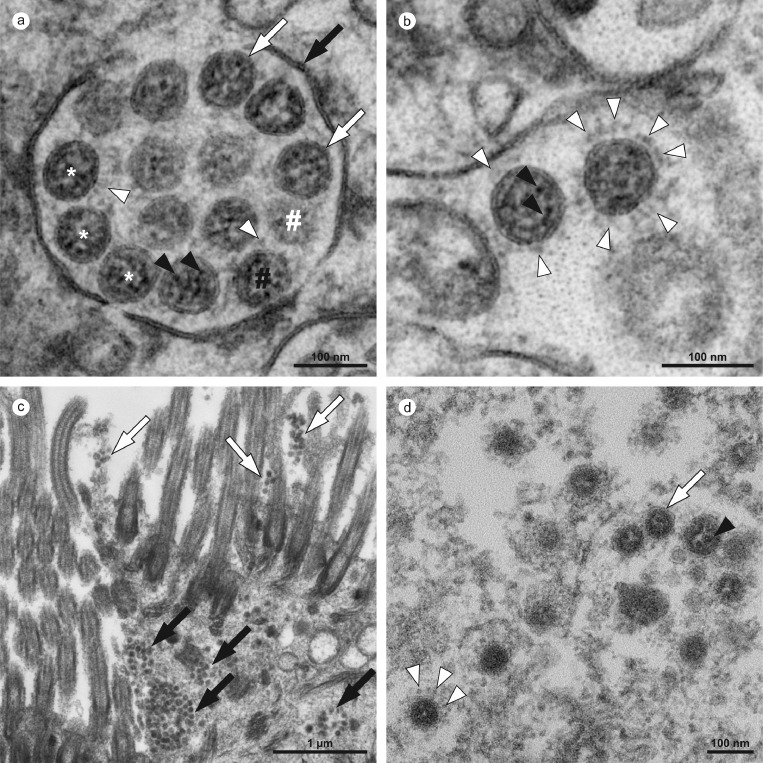
Ultrastructural characteristics of coronavirus particles in different types of autopsy samples. (a,b) Autopsy lung tissue, (c,d) FFPE-re-embedded olfactory mucosa. (a) Well-preserved intracellular coronavirus (CoV) particles (some indicated by white asterisks) in autopsy lung tissue, located within a membrane compartment (black arrow) and showing a distinct biomembrane (white arrows) and, some of them also, faint surface projections (white arrowheads). Note the granular and relatively fine dispersed ribonucleoprotein (RNP; black arrowheads). The appearance of a different electron density between the individual particles (black vs. white #) is a result of different particle volumes (about 90 nm in diameter) captured within the section volume (about 60 nm thickness); a larger particle volume within the section results in higher electron density, a phenomenon that is typical for spherical particles with an electron dense interior (as compared to e.g. empty vesicles). (b) Very few particles demonstrate prominent (well-visible) surface projections, e.g. the right CoV particle (white arrowheads) as compared to the left CoV particle. The two particles show the heterogeneous nature of the RNP in such preparations, with distinct granular and some elongated profiles (black arrowheads) in the left particle and a more granular luminal matrix with few distinct profiles in the right particle. (c) Infected cells within the olfactory mucosa also show membrane compartments (black arrows) with numerous coronavirus particles and also grouped extracellular particles that typically adhere to kinocilia and microvilli (white arrows), but individual particles are less well-preserved and more difficult to identify than particles in a and b. Virus particles appear as groups of electron dense particles of rather uniform size. (d) Virus particles at higher magnification (another region as shown in c). The particles are surrounded by a biomembrane (white arrow) which only rarely show surface projections (white arrowheads; note the globular shape of their peripheral part and also the relatively low electron density as compared to e.g. the RNP). The granular luminal matrix, formed by the RNP, is only visible in a few particles (black arrowhead). Individual images were acquired manually at high resolution by scanning transmission electron microscopy (STEM; a,b) and transmission electron microscopy (TEM; c,d).
Overall, virus particles could be identified in 15 cells within lung tissue of one patient (case B1), and all cells were found in two of the four digitized sections. Virus particles were well-preserved with distinct substructures (Figure 4a,b). Virus-containing cells could be identified as type 2 pneumocytes and alveolar macrophages (Supplementary Table 6). Potential viral mimics such as swollen mitochondria, vesicles of rough endoplasmic reticulum (rER), and coated vesicles could be distinguished from viral particles (Supplementary Figure 8).13 We recorded all 15 infected cells in the lung at very high resolution (Figure 5; datasets on www.nanotomy.org) for morphometric analyses and found 1557 intracellular and 144 extracellular viral particles. Infected cells showed virus loads ranging from 4 to 620 intracellular particles per sectioned cell profile (or 0.07 to 5.44 intracellular particles per µm2 cytoplasmic area, Supplementary Figure 9). Intracellular particles showed a mean diameter of 87 nm (±13 nm; n=1369; 55 to 177 nm). The RNP showed a mean diameter of 7.2 nm (± 1.6 nm; n=433; 3.6 to 13 nm). Different morphological types of CoV were observed (Figure 5). According to our quantitative analysis and calculations (Supplementary Methods), cells may contain thousands (up to 40,000; cell 7) of CoV particles. Based on a rough approximation from viral RNA load, we can expect 0.006 (case B9) or 500 (case B1) CoV particles/ mm2 section area for low and high RNA load, respectively, which implies a huge difference in the likelihood of particle detection (Supplementary Methods; calculations). No definitive typical replication compartments such as double-membrane vesicles (DMV) were found. However, four cells of the autopsy lung demonstrated peculiar tubular structures which are possibly associated with CoV infection (Supplementary Figure 10).25
Virus particles could also be identified in several ciliated cells within olfactory mucosa of case B3 (Supplementary Table 5; Figure 4c,d). However, virus particles were less well-preserved due to FFPE-re-embedding. Intracellular virus particles appeared more condensed than particles after standard preparation with a mean diameter of 73 nm (± 7 nm; n=175; 58 to 108 nm).
Of note, no virus particles were found in 28 samples of 15 patients in different tissues (Supplementary Table 5). Interestingly, we did not find virus particles in autopsy lung of two cases (B8 and Hamburg case 10) using FFPE-re-embedded material, guided by positive IHC signals, and with a high viral RNA load as determined by RT-qPCR.
Based on reliable reference publications3,9,10,12,25 (see also Supplementary Methods), and the results presented here, we developed refined criteria for identification of CoV particles in autopsy samples (Table 2).
Table 2.
Recommendations for detection of intact SARS-CoV-2 particles using electron microscopy in human autopsy tissues.
| Criteria for ultrastructural identification | ||
|---|---|---|
| General considerations | It is sufficient if all of these criteria are met by a group of closely associated and similar particles within one individual cell, but individual particles of different cells should not be combined. Identification of cell types in autopsy tissues is challenging and often not possible, complicated also by pathological and virus-induced alterations that may mimick e.g. lamellar bodies of type 2 pneumocytes. | |
| 1 | Shape | Round to oval. |
| 2 | Size | 50-180 nm (mean = 87 ± 13 nm; without spikes), with smaller particles in re-embedded FFPE material (mean = 73 ± 7 nm, 58-108 nm). In the range also described for cell culture9 and autopsy lung.11 |
| 3 | Membrane | At least partially visible around the particle. |
| 4 | Surface projections | Thin stalk and a globular component (in total about 20 nm long9), at least the globular component needs to be discernible with some distance from the biomembrane. The electron density is considerably lower than surface structures of e.g. coated vesicles and the particle surface usually is not entirely covered.9 Note that the visibility of surface projections may be heterogeneous within the sample and also depends on additives applied during tissue embedding such as en bloc treatment with tannic acid and uranyl acetate.9,25 |
| 5 | Interior structure | Inhomogeneous granular (never empty or homogeneous at low electron density), ribonucleoprotein (RNP) profiles are round/aggregated or oval/longitudinal structures. Based on our findings, the RNP profile diameter is generally between 3.6-13 nm (mean = 7.2 nm ± 1.6 nm), as published.11,13 |
| 6 | Number | Particles must be present at higher number and should often occur in groups within cells.3,12 |
| 7 | Location | Extracellular: individual particles or small groups, sometimes attaching to outer surface of membranes. Intracellular: within small compartments with e.g. 1 particle up to very large compartments with dozens of particles, sometimes attaching to the inner surface of the membranes, but compartments with more than 1 particle should be identifiable as different structures such as swollen mitochondria may produce a “one-particle within a membrane compartment” appearance. |
| Recommendations for sampling and analysis | ||
|---|---|---|
| General considerations | Prioritize analysis of few, carefully selected samples with a high viral load. Controls are not required for virus identification, because EM allows a direct (label-free) proof of the respective particles. | |
| 1 | Sampling | Analyse multiple blocks of the most strongly RT-qPCR-positive samples (see 3), facultatively at different levels to locate infection foci, even in apparently suboptimal samples, such as samples with a relatively long post mortem interval, or samples that have been frozen or paraffin embedded. Identify virus particles in the best specimen to get an internal “positive control”. |
| 2 | Correlation: IHC/ISH | Identify infection foci by using IHC or ISH and process corresponding FFPE regions via a punch biopsy or paraffin sections for EM as previously described.3,12 Serial paraffin sections may be processed to stain virus antigens and cell type markers and detect intact virus particles of the same cell. |
| 3 | Correlation: RT-qPCR | Try to roughly estimate the likelihood of finding virus particles based on RT-qPCR data as shown in our work with 0.006 or 500 expected particles per mm2 section area for low or high viral load, respectively. |
| 4 | Structural preservation/ image quality | Adjustment of fixation, embedding, sectioning and staining may be required for sufficient preservation of virus.7,43 Use adequate magnification to clearly resolve all relevant details of possible virus particles (0.2-1 nm pixel size) and adjust the respective EM settings correctly.43 |
| 5 | Labelling techniques | Pre-embedding or post-embedding techniques should be used and interpreted with caution, as structural preservation usually is negatively affected. Morphological features of virus particles should still be identifiable and adequate controls should be used. Additional conventional EM for virus detection should be used. |
| 6 | Screening | Screen individual sections systematically at a medium resolution at which groups of viruses are easily detected (as can be tested by our large-scale datasets), and in some regions also at higher resolution to avoid missing of single virus particles present in the cells. |
| 7 | Pattern recognition | Learn the visual pattern of virus particles in tissue samples by using correctly identified virus particles (see large-scale repository datasets: http://www.nanotomy.org/OA/Krasemann2022eBioMedicine/index.html). |
| 8 | Figures | Ensure adequate image size, resolution, colour profile, brightness and contrast when publishing figures. |
Analysis of publications demonstrating ultrastructural evidence for SARS-CoV-2 in human samples
We surveyed publications (published April 2020 to March 2022) using ultrastructural findings as proof of the presence of virus particles in human samples (Supplementary Tables 7,8) and re-evaluated the data using our refined criteria for virus identification. Nine publications presented sufficient structural evidence to prove the presence of virus particles, while 135 publications misinterpreted different cellular structures as virus or showed only insufficient structural evidence for the presence of virus particles (Supplementary Figure 11). In these 135 publications, in total, only 67 of 337 electron micrographs (20%) showed sufficient structural preservation and image quality. In two of the 135 publications,26, 41 images of putative CoV are shown which did not allow a clear decision whether the particles represent CoV particles or not (see also comments on borderline cases in Supplementary Table 7).
Structures misinterpreted as virus particles were coated vesicles, vesicles of rough endoplasmic reticulum, multivesicular bodies, and autolytic mitochondria. Thirty-eight publications discussed the challenges of SARS-CoV-2 identification by EM (Supplementary Table 9).
Discussion
One key question in COVID-19 is whether organ damage is due to direct organ targeting of SARS-CoV-2 or downstream effects such as an overshooting immune response. During the COVID-19 pandemic, autopsy-driven research, using multimodal approaches, attempted at defining viral organ tropism and organ-specific pathomechanisms,3,4,15,27 yet a critical and systematic study investigating the limitations of in situ detection of SARS-CoV-2 in autopsy tissues has not been performed. Methods directly detecting viral RNA are superior to IHC and EM in terms of sensitivity and ISH may also provide a reasonable spatial resolution if performed correctly including adequate positive and negative controls.28 However, they may be prone to contaminations by RNA from nearby sources, which cannot be excluded during autopsies. Moreover, RNA might be more prone to degradation upon inadequate handling of tissue sections e.g. long storage of cut sections. Accordingly, the minute amounts of virus found in some autopsy studies18,29,30 could be caused by droplet spread during organ/tissue extraction or viral RNA in blood. Thus, because of the patchy infection pattern of SARS-CoV-2,31 RT-PCR should be combined with IHC and/or ISH as well as EM on the same tissue specimen to achieve optimal reliability.
The discrepancy of IHC results from studies using autopsy material to determine organ tropism and identification of organ-specific target cell types of SARS-CoV-2 led to conflicting results hampering research progress.2, 3, 4, 5,32 For instance, published data show direct infection of neurons by SARS-CoV-2 with substantial neuroinvasion,33 single infected cells in a subset of patients,15 but also absence of virus and COVID-19-specific alterations.34 Validations are complicated by the use of various, often not well-evaluated antibodies in different studies and the lack of appropriate controls. Furthermore, multiple studies reported a possible renal tropism,1,35,36 but EM data on kidney tissue in all 21 publications included in our literature research showed false-positive results. Importantly, multiple studies also reported that they did not find virus using IHC, ISH, and EM in kidney samples,6,37 and the specificity of rare, equivocal ISH signals were discussed.6 It was also questioned that a low abundance of virus, as detected by RT-qPCR, would be sufficient for the observed pathologic changes, therefore arguing for indirect mechanisms of kidney damage in COVID-19.6 The significance of low abundance of virus RNA and/or protein in e.g. kidney tissue,4 present as virus particles or virus fragments, therefore remains to be elucidated. One may speculate that a low abundance of virus RNA and/or protein (or even intact particles) in tissues such as kidney and brain is of pathophysiological significance, but this needs to be critically discussed and differentiated from a bona fide viral tropism. Our study provides novel insight concerning the use of in situ SARS-CoV-2 detection methods which should be considered when assessing multiorgan tropism of SARS-CoV-2.
We determined the limitations of SARS-CoV-2 detection by IHC and EM using a defined set of control tissues including FFPE cell blocks and autopsy tissues with high SARS-CoV-2 viral load. We performed a multicentre study assessing how well IHC performs in detecting SARS-CoV-2 proteins in autopsy tissues. However, studies at an even bigger scale, including further tissue and more methods, like ISH, need to be conducted, preferentially initiated by pathology societies, to define consensus criteria for a reliable in situ detection of SARS-CoV-2 in human autopsy tissues.
Assessment of a wide range of commercially available anti-SARS-CoV-2 antibodies showed that only a subset can be reliably used in autopsy tissues (Figure 1, Supplementary Figure 3). Interestingly, anti- nucleocapsid-antibodies showed highest sensitivity. This may be since, of all SARS-CoV-2 proteins, nucleocapsid is produced at the highest levels during the SARS-CoV-2 lifecycle in cells.38 Of note, nucleocapsid-antibodies also performed well in detecting infected cells by the novel VOC Omicron B.1.1.529, while several anti-spike antibodies that detected SARS-CoV-2 WT spike protein failed to detect Omicron. The targeted epitopes of the nucleocapsid may be more structurally conserved and therefore result in more robust detection. Correspondingly, we found much less spike protein than nucleocapsid, based on both, the amount per cell and the general abundance in affected tissues. This should be considered when interpreting studies showing multiorgan tropism and target cell types, especially when using anti-spike-antibodies.1,2,33,39 In fact, in one study the unspecific nature of the used anti-spike antibody (our #3) was noticed between the preprint and the final publications5 and the erroneous interpretation could thus be corrected.
We found a discrepancy between RT-qPCR-determined viral RNA loads and detection of SARS-CoV-2 proteins by immunohistochemistry. In tissues with low viral RNA loads, immunohistochemistry is not a reliable method to determine organ tropism or target cells as interpretation of rare immunosignals is difficult and nonspecific staining may be falsely interpreted as a positive signal. Interestingly interpretation of ISH is faced with similar problems.23,40 In agreement with this, we found low inter-observer reliability in tissues with low viral RNA loads (Supplementary Table 4). This may be due to uneven distribution of virus, even in highly affected organ systems such as lungs (Figure 3). The absence of viral proteins should not be used as an argument for the absence of SARS-CoV-2-related tissue pathology, as autopsy tissues can only provide an incomplete snapshot of what has occurred in the sometimes very long clinical phase of the disease.32 Recent studies have addressed this by studying autopsy tissues at different stages of COVID-19.32 We show that it is crucial to choose suitable control tissues for immunohistochemistry not only based on high viral RNA loads but also on tissue integrity. Lung, for example, the tissue with the highest viral RNA loads cannot be considered an optimal control tissue as it tends to produce false-positive signals and difficult-to-interpret staining patterns, probably related to extensive tissue damage with pre-necrotic epithelial cells and hyaline membranes giving rise to false-positive signals. Based on the data shown in this manuscript, our recommendations for the usage of antibodies for the detection of anti-SARS-CoV-2 proteins in human autopsy tissues are summarized in Table 1 and Supplementary Table 2.
Using EM to detect intact SARS-CoV-2 particles in autopsy tissues holds specific challenges such as a relatively low sensitivity as compared to e.g. RT-qPCR, limitations in cell-type identification, and high inter-observer variability depending on EM expertise. We found virus particles only in a minor fraction of patient samples (with comparatively high viral RNA load, high IHC scores, and positive ISH,3 and virus particles were spatially highly confined. In fact, individual cells may contain up to tens of thousands CoV particles and hundreds of thousands of RNA copies. Thus, even in samples with a high SARS-CoV-2 load, few infected cells (∼20 or less per 10,000 cells), possibly also mobile cells, could make up a significant fraction of the total viral load. This result aligns with our IHC findings suggesting a focal infection and argues for the complementarity of both methods so as not to misguide research.10,12 The importance of careful implementation of multiple techniques is also supported by our negative results of virus particles in two samples with a high virus RNA load and guided by positive IHC. Absence of identifiable virus particles in these cases could variably be attributable to insufficient structural preservation, but could also be due to presence of mainly virus fragments (RNA and/or proteins) instead of intact particles. Here, virus fragments could be the result of virus particle degradation, but could also be due to incomplete virus assembly in infected cells. Moreover, presence of intact intracellular particles in e.g. macrophages may variably be a result of phagocytosis and not infection, thus indicating a need for further research on this correlation.
We expanded and detailed previously published criteria11,13 primarily based on cell culture data and provide recommendations on a suitable strategy for identifying virus-infected cells in Table 2.
Using these refined criteria for SARS-CoV-2 identification, 135 of 144 publications do not sufficiently prove the presence of viral particles in various human tissues. Of these 135 publications, two are borderline cases with particles that may be CV particles. In Birkhead et al.26 and in Qadir et al.41 ( lower image), particles fulfil most criteria, but the RNP substructure appears untypically granular26 and the image quality is not optimal41 (lower image). In such borderline cases, additional techniques for virus detection and more EM data can help. The problem of misinterpretation has already been discussed11,13 and resulted in specific recommendations for the correct detection of CoV particles. However, a general loss of diagnostic EM expertise occurred,10,42 and most EM facilities have no experience with in situ detection of viruses.13 Both probably complicate transfer of recommendations into practice, as also indicated by the lack of quality standards43 of many published EM data. This is further illustrated by the fact that also during the SARS-CoV pandemic, detection of the virus by EM was tainted with technical and interpretational difficulties, and non-viral particles in different organs (e.g. kidney and lung44,45) were used to propose a multiorgan tropism of SARS-CoV. These misinterpretations were then perpetuated early on in the SARS-CoV-2 pandemic.46 Importantly, misinterpretations also occurred in non-human samples i.e. in cell culture,47 and brain organoids.33
For a robust reliable work-up, a multistep approach of analyzing samples based on the same tissue is advisable. RT-PCR allows for screening of tissues with sufficient RNA load. Then, IHC/ISH allows fast and easy in situ detection of virus thus guiding EM analysis. Although throughput and sensitivity of EM are limited it still plays an important role in validating, the other faster, cheaper and more sensitive techniques for virus detection.11
Our large-scale datasets, corresponding to approximately 130,000 conventional electron micrographs, may help in acquiring visual pattern recognition skills, which cannot be acquired using small sets of preselected conventionally published electron micrographs lacking cellular and microanatomical context. As demonstrated by our Supplementary Video, virus particles can be found by spanning the scales from millimeter to nanometer. This technique14 also provides a promising approach for fast and precise ultrastructural in silico analysis for future pandemics, especially in light of innovative high-throughput EM imaging approaches.
The main limitation of our study is its limited scope regarding assessed tissues and anti-SARS-CoV-2 antibodies. Also, autolysis and a limited number of cases with successful identification of virus particles by EM did not allow to perform a systematic analysis of infected cells.48 Similarly, we did not find definitive DMVs, most probably because of the limited structural preservation as recently, these could be seen in well-preserved autopsy lung.49
In summary, usage of autopsy tissues with in situ detection of SARS-CoV-2 is valuable if interpreted within the limits of all applied methods and tissues. In the early phase of the COVID-19 pandemic, researchers have not fully abided to this, leading to controversies regarding SARS-CoV-2 multiorgan tropism. Key aspects to consider are that due to the heterogeneous distribution of virus-containing cells, IHC is not optimally suited to detect low amounts of SARS-CoV-2 in autopsy tissue and usage of anti-nucleocapsid antibodies increases the detection sensitivity. For EM identification of SARS-CoV-2 particles in autopsy tissue, RT-PCR and IHC/ISH should be used to select samples and only tissue with good structural preservation should be used. Virus particles can be recognized if key features such as round to oval shape, correct size (50-180 nm), characteristic surface projections and interior structures are present.
Contributors
Autor's contributions were as follows: conceptualisation of the study: SK, CD, HR, ML, MG. Autopsy sample collection: SvS,JeM, FH, TADA, JR, AO, SS, JI, FD, VA, JM, BO, MO, WS, FLH, PB, HR, ML, MG. Generated and/or analysed IHC data: SK, SvS, JeM, RvM, JR, AO, SE, DH, JM, DS, HAB, FLH, PB, HR, MG. Analysed EM data: CD, RvM, TADA, EMB, ST, SB, PB, ML. Analyzed PCR data: VC, SSB, GG. Infected culture cells: SP. Paper writing figure preparation: SK, CD, PB, HR, ML, MG. Approved final version of the manuscript: SK, CD, SvS, JeM, FH, KH, SP, ET, RvM, TADA, JR, AO, SS, EMB, JI, FD, VA, SE, DH, AH, ST, SB, VC, HHG, JM, SSB, GG, DS, HAB, BO, MO, WS, FLH, PB, HR, ML, MG. SK, HR, and MG are responsible for the validity of the underlying data.
Data sharing statement
Large-scale EM datasets of SARS-CoV-2 infected autopsy tissues and cell culture are openly available at http://www.nanotomy.org/OA/Krasemann2022ebiomedicine/index.html. More anonymized data of the present study are available upon reasonable request to the authors.
Declaration of interests
All authors declare that they have no conflict of interests regarding this manuscript.
Acknowledgments
SK (head) and KH (technician) are running the Core Facility for Experimental Pathology of the UKE (“Mouse Patho”). We thank Annette Gries, Hanna Jania and Gudrun Holland for excellent technical assistance and the UMIF/UKE for using their microscopes. Our condolences to the families and all those who have lost their loved ones during the ongoing pandemic. We thank all relatives who made the difficult decision to give their permission for autopsy and research.
This work was supported by the German Registry of COVID-19 Autopsies (www.DeRegCOVID.ukaachen.de), funded Federal Ministry of Health (ZMVI1-2520COR201), by the Federal Ministry of Education and Research within the framework of the network of university medicine (DEFEAT PANDEMICs, 01KX2021 and NATON, 01KX2121). ACH was supported by Berlin University Alliance GC2 Global Health (Corona Virus Pre-Exploration Project), BMBF (RAPID and Organo-Strat 01KX2021) as well as DFG (SFB-TR 84, B6 / Z1a), HR by DFG (RA 2491/1-1), SB by DFG SFB 1365 C04, S01 and NIH 2R01DK05149-19A1, subaward 1016678, while SK was funded by the German Center for Infectious Research (TTU.01.929).
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2022.104193.
Appendix. Supplementary materials
References
- 1.Bradley BT, Maioli H, Johnston R, et al. Histopathology and ultrastructural findings of fatal COVID-19 infections in Washington State: a case series. Lancet. 2020;396(10247):320–332. doi: 10.1016/S0140-6736(20)31305-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kanczkowski W, Evert K, Stadtmüller M, et al. COVID-19 targets human adrenal glands. The Lancet Diabetes Endocrinol. 2021;10(1):13–16. doi: 10.1016/S2213-8587(21)00291-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meinhardt J, Radke J, Dittmayer C, et al. Olfactory transmucosal SARS-CoV-2 invasion as a port of central nervous system entry in individuals with COVID-19. Nat Neurosci. 2021;24(2):168–175. doi: 10.1038/s41593-020-00758-5. [DOI] [PubMed] [Google Scholar]
- 4.Puelles VG, Lutgehetmann M, Lindenmeyer MT, et al. Multiorgan and renal tropism of SARS-CoV-2. N Engl J Med. 2020;383(6):590–592. doi: 10.1056/NEJMc2011400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang AC, Kern F, Losada PM, et al. Dysregulation of brain and choroid plexus cell types in severe COVID-19. Nature. 2021;595(7868):565–571. doi: 10.1038/s41586-021-03710-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kudose S, Batal I, Santoriello D, et al. Kidney Biopsy Findings in Patients with COVID-19. J Am Soc Nephrol. 2020;31(9):1959–1968. doi: 10.1681/ASN.2020060802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goldsmith CS, Miller SE. Modern uses of electron microscopy for detection of viruses. Clin Microbiol Rev. 2009;22(4):552–563. doi: 10.1128/CMR.00027-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turonova B, Sikora M, Schurmann C, et al. In situ structural analysis of SARS-CoV-2 spike reveals flexibility mediated by three hinges. Science. 2020;370(6513):203–208. doi: 10.1126/science.abd5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laue M, Kauter A, Hoffmann T, Moller L, Michel J, Nitsche A. Morphometry of SARS-CoV and SARS-CoV-2 particles in ultrathin plastic sections of infected Vero cell cultures. Sci Rep. 2021;11(1):3515. doi: 10.1038/s41598-021-82852-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dittmayer C, Meinhardt J, Radbruch H, et al. Why misinterpretation of electron micrographs in SARS-CoV-2-infected tissue goes viral. Lancet. 2020;396(10260):e64–e65. doi: 10.1016/S0140-6736(20)32079-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bullock HA, Goldsmith CS, Zaki SR, Martines RB, Miller SE. Difficulties in differentiating coronaviruses from subcellular structures in human tissues by electron microscopy. Emerg Infect Dis. 2021;27(4):1023–1031. doi: 10.3201/eid2704.204337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martines RB, Ritter JM, Matkovic E, et al. Pathology and pathogenesis of SARS-CoV-2 associated with fatal coronavirus disease, United States. Emerg Infect Dis. 2020;26(9):2005–2015. doi: 10.3201/eid2609.202095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hopfer H, Herzig MC, Gosert R, et al. Hunting coronavirus by transmission electron microscopy - a guide to SARS-CoV-2-associated ultrastructural pathology in COVID-19 tissues. Histopathology. 2021;78(3):358–370. doi: 10.1111/his.14264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dittmayer C, Goebel HH, Heppner FL, Stenzel W, Bachmann S. Preparation of samples for large-scale automated electron microscopy of tissue and cell ultrastructure. Microsc Microanal. 2021;27(4):815–827. doi: 10.1017/S1431927621011958. [DOI] [PubMed] [Google Scholar]
- 15.Matschke J, Lutgehetmann M, Hagel C, et al. Neuropathology of patients with COVID-19 in Germany: a post-mortem case series. Lancet Neurol. 2020;19(11):919–929. doi: 10.1016/S1474-4422(20)30308-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Best Rocha A, Stroberg E, Barton LM, et al. Detection of SARS-CoV-2 in formalin-fixed paraffin-embedded tissue sections using commercially available reagents. Lab Invest. 2020;100(11):1485–1489. doi: 10.1038/s41374-020-0464-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Borczuk AC, Salvatore SP, Seshan SV, et al. COVID-19 pulmonary pathology: a multi-institutional autopsy cohort from Italy and New York City. Mod Pathol. 2020;33(11):2156–2168. doi: 10.1038/s41379-020-00661-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duarte-Neto AN, Caldini EG, Gomes-Gouvea MS, et al. An autopsy study of the spectrum of severe COVID-19 in children: from SARS to different phenotypes of MIS-C. EClinicalMedicine. 2021;35 doi: 10.1016/j.eclinm.2021.100850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.El Jamal SM, Pujadas E, Ramos I, et al. Tissue-based SARS-CoV-2 detection in fatal COVID-19 infections: sustained direct viral-induced damage is not necessary to drive disease progression. Hum Pathol. 2021;114:110–119. doi: 10.1016/j.humpath.2021.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ko CJ, Harigopal M, Gehlhausen JR, Bosenberg M, McNiff JM, Damsky W. Discordant anti-SARS-CoV-2 spike protein and RNA staining in cutaneous perniotic lesions suggests endothelial deposition of cleaved spike protein. J Cutan Pathol. 2021;48(1):47–52. doi: 10.1111/cup.13866. [DOI] [PubMed] [Google Scholar]
- 21.McMullen P, Pytel P, Snyder A, et al. A series of COVID-19 autopsies with clinical and pathologic comparisons to both seasonal and pandemic influenza. J Pathol Clin Res. 2021;5:459–470. doi: 10.1002/cjp2.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ray A, Jain D, Goel A, et al. Clinico-pathological features in fatal COVID-19 infection: a preliminary experience of a tertiary care center in North India using postmortem minimally invasive tissue sampling. Expert Rev Respir Med. 2021;10:1367–1375. doi: 10.1080/17476348.2021.1951708. [DOI] [PubMed] [Google Scholar]
- 23.Roden AC, Vrana JA, Koepplin JW, et al. Comparison of in situ hybridization, immunohistochemistry, and reverse transcription-droplet digital polymerase chain reaction for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) testing in tissue. Arch Pathol Lab Med. 2021;145(7):785–796. doi: 10.5858/arpa.2021-0008-SA. [DOI] [PubMed] [Google Scholar]
- 24.Szabolcs M, Sauter JL, Frosina D, et al. Identification of immunohistochemical reagents for in situ protein expression analysis of coronavirus-associated changes in human tissues. Appl Immunohistochem Mol Morphol. 2021;29(1):5–12. doi: 10.1097/PAI.0000000000000878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goldsmith CS, Tatti KM, Ksiazek TG, et al. Ultrastructural characterization of SARS coronavirus. Emerg Infect Dis. 2004;10(2):320–326. doi: 10.3201/eid1002.030913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Birkhead M, Glass AJ, Allan-Gould H, Goossens C, Wright CA. Ultrastructural evidence for vertical transmission of SARS-CoV-2. Int J Infect Dis. 2021;111:10–11. doi: 10.1016/j.ijid.2021.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wenzel J, Lampe J, Muller-Fielitz H, et al. The SARS-CoV-2 main protease M(pro) causes microvascular brain pathology by cleaving NEMO in brain endothelial cells. Nat Neurosci. 2021;24(11):1522–1533. doi: 10.1038/s41593-021-00926-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.von Stillfried S, Boor P. Detection methods for SARS-CoV-2 in tissue. Pathologe. 2021;(suppl 1):81–88. doi: 10.1007/s00292-021-00920-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dolhnikoff M, Ferreira Ferranti J, de Almeida Monteiro RA, et al. SARS-CoV-2 in cardiac tissue of a child with COVID-19-related multisystem inflammatory syndrome. Lancet Child Adolesc Health. 2020;4(10):790–794. doi: 10.1016/S2352-4642(20)30257-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Valdespino-Vazquez MY, Helguera-Repetto CA, Leon-Juarez M, et al. Fetal and placental infection with SARS-CoV-2 in early pregnancy. J Med Virol. 2021;93(7):4480–4487. doi: 10.1002/jmv.26965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Desai N, Neyaz A, Szabolcs A, et al. Temporal and spatial heterogeneity of host response to SARS-CoV-2 pulmonary infection. Nat Commun. 2020;11(1):6319. doi: 10.1038/s41467-020-20139-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khan M, Yoo SJ, Clijsters M, et al. Visualizing in deceased COVID-19 patients how SARS-CoV-2 attacks the respiratory and olfactory mucosae but spares the olfactory bulb. Cell. 2021;184(24):5932–5949. doi: 10.1016/j.cell.2021.10.027. e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Song E, Zhang C, Israelow B, et al. Neuroinvasion of SARS-CoV-2 in human and mouse brain. J Exp Med. 2021;218(3) doi: 10.1084/jem.20202135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deigendesch N, Sironi L, Kutza M, et al. Correlates of critical illness-related encephalopathy predominate postmortem COVID-19 neuropathology. Acta Neuropathol. 2020;140(4):583–586. doi: 10.1007/s00401-020-02213-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Su H, Yang M, Wan C, et al. Renal histopathological analysis of 26 postmortem findings of patients with COVID-19 in China. Kidney Int. 2020;98(1):219–227. doi: 10.1016/j.kint.2020.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Varga Z, Flammer AJ, Steiger P, et al. Electron microscopy of SARS-CoV-2: a challenging task - authors' reply. Lancet. 2020;395(10238):e100. doi: 10.1016/S0140-6736(20)31185-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Daniel E, Sekulic M, Kudose S, et al. Kidney allograft biopsy findings after COVID-19. Am J Transplant. 2021;21(12):4032–4042. doi: 10.1111/ajt.16804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.V'Kovski P, Kratzel A, Steiner S, Stalder H, Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat Rev Microbiol. 2021;19(3):155–170. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schurink B, Roos E, Radonic T, et al. Viral presence and immunopathology in patients with lethal COVID-19: a prospective autopsy cohort study. Lancet Microbe. 2020;1(7):e290–e299. doi: 10.1016/S2666-5247(20)30144-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Massoth LR, Desai N, Szabolcs A, et al. Comparison of RNA in situ hybridization and immunohistochemistry techniques for the detection and localization of SARS-CoV-2 in human tissues. Am J Surg Pathol. 2021;45(1):14–24. doi: 10.1097/PAS.0000000000001563. [DOI] [PubMed] [Google Scholar]
- 41.Qadir MMF, Bhondeley M, Beatty W, et al. SARS-CoV-2 infection of the pancreas promotes thrombofibrosis and is associated with new-onset diabetes. JCI Insight. 2021;6(16) doi: 10.1172/jci.insight.151551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Haro T, Furness P. Current and future delivery of diagnostic electron microscopy in the UK: results of a national survey. J Clin Pathol. 2012;65(4):357–361. doi: 10.1136/jclinpath-2011-200601. [DOI] [PubMed] [Google Scholar]
- 43.Stirling JW, Curry A. Quality standards for diagnostic electron microscopy. Ultrastruct Pathol. 2007;31(5):365–367. doi: 10.1080/01913120701638660. [DOI] [PubMed] [Google Scholar]
- 44.Gu J, Gong E, Zhang B, et al. Multiple organ infection and the pathogenesis of SARS. J Exp Med. 2005;202(3):415–424. doi: 10.1084/jem.20050828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ding Y, He L, Zhang Q, et al. Organ distribution of severe acute respiratory syndrome (SARS) associated coronavirus (SARS-CoV) in SARS patients: implications for pathogenesis and virus transmission pathways. J Pathol. 2004;203(2):622–630. doi: 10.1002/path.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li YC, Bai WZ, Hashikawa T. The neuroinvasive potential of SARS-CoV2 may play a role in the respiratory failure of COVID-19 patients. J Med Virol. 2020;92(6):552–555. doi: 10.1002/jmv.25728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xiao K, Zhai J, Feng Y, et al. Isolation of SARS-CoV-2-related coronavirus from Malayan pangolins. Nature. 2020;583(7815):286–289. doi: 10.1038/s41586-020-2313-x. [DOI] [PubMed] [Google Scholar]
- 48.Eymieux S, Rouille Y, Terrier O, et al. Ultrastructural modifications induced by SARS-CoV-2 in Vero cells: a kinetic analysis of viral factory formation, viral particle morphogenesis and virion release. Cell Mol Life Sci. 2021;78(7):3565–3576. doi: 10.1007/s00018-020-03745-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kataoka M, Tsukamoto T, Tajima Y, et al. Coronavirus hunted in human pneumocytes and alveolar macrophages: a case report. Histopathology. 2022;7:1130–1134. doi: 10.1111/his.14637. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.