Abstract
Mono- and polyclonal antibodies directed against UMP kinase from Escherichia coli were tested with the intact protein or with fragments obtained by deletion mutagenesis. As detected in enzyme-linked immunosorbent assay tests, the carboxy-terminal quarter of UMP kinase is immunodominant. Polyclonal antibodies inhibited the enzyme activity with partial or total loss of allosteric effects exerted by UTP and GTP, respectively. These data indicate that the UTP and GTP binding sites in UMP kinase are only partially overlapping. One monoclonal antibody (44-2) recognized a linear epitope in UMP kinase between residues 171 and 180. A single substitution (D174N) in this segment of the enzyme abolished its interaction with the monoclonal antibody (44-2). Polyclonal antisera were used to identify UMP kinase in the bacterial proteome. The enzyme appears as a single spot on two-dimensional electrophoresis at a pI of 7.24 and an apparent molecular mass of 26 kDa. Immunogold labeling of UMP kinase in whole E. coli cells shows a localization of the protein near the bacterial membranes. Because the protein does not contain sequences usually required for compartmentalization, the aggregation properties of UMP kinase observed in vitro might play a role in this phenomenon. The specific localization of UMP kinase might also be related to its putative role in cell division.
Nucleoside monophosphate (NMP) kinases are present in all forms of living cells. Small and generally monomeric, they belong to the α/β class of proteins, in which a five-stranded β-sheet forming the core of the molecule is surrounded by eight or nine α-helices. The best-studied member of this family of catalysts, which has relatively well conserved primary and three-dimensional structures among different species, is adenylate kinase (AK) (3, 30). Over 60 sequences of AKs are known from either gene or protein analysis, and the crystal structures of bacterial, yeast, and mammalian AKs were deciphered at high resolution, both in the absence and in the presence of substrates (1, 9, 13, 23, 24, 35).
UMP kinase from bacteria represents a particular class of NMP kinases. Encoded by the pyrH gene, the protein, which is a hexamer, shows no sequence similarity to any other known NMP kinase and is subject to complex regulatory mechanisms (31, 33). The pyrH gene has also been described as smbA, a suppressor of the mukB null mutant, which shows defects in cell division. It was suggested that the MukB protein could be a candidate for a force-generating enzyme involved in the correct positioning of replicated chromosomes, but the relationship between UMP kinase and MukB was not completely elucidated (38). As pyrH is essential, UMP kinase might be an interesting new target for antibacterial drugs and may have functions other than catalysis. Therefore, we considered it important to design methods to detect the protein under different experimental conditions, in order to initiate a physiological study of this enzyme in Escherichia coli and to answer the question of its putative involvement in cell division. Antibodies are useful tools in characterizing proteins, especially when high-resolution three-dimensional structural data are not yet available. Monoclonal antibodies (MAbs) or polyclonal antibodies tested with the intact protein or with fragments obtained by deletion mutagenesis were used to answer a number of questions regarding the structure and catalytic properties of UMP kinase. Antibodies also served to locate the enzyme in the bacterial proteome and the intact cell. One of the most surprising results of this study is the dual localization of bacterial UMP kinase, i.e., cytosolic and close to the membranes, a result which strengthens the hypothesis of multiple functional roles of the enzyme in bacterial life.
MATERIALS AND METHODS
Bacterial strains, plasmids, growth conditions, and DNA manipulations.
General DNA manipulations were performed as described by Sambrook et al. (29). Open reading frames from the complete or truncated pyrH gene were generated by PCR and inserted into the expression vectors pET22b and pET24a (Novagen) and pET24ma (33a) (Table 1). Cloning experiments were carried out with strain NM554/pDIA17 (25, 26). The resulting plasmids were introduced into strain BL21(DE3)/pDIA17 (34) to overproduce the corresponding peptides. Recombinant strains (Table 1) were grown in 2YT medium supplemented with antibiotics to an optical density of 1 at 600 nm, and then overproduction was triggered by isopropyl-β-d-thiogalactoside induction (1 mM final concentration) for 3 h. Bacteria were harvested by centrifugation, and proteins were purified as described below.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Relevant feature(s) | Reference or source |
|---|---|---|
| Strains | ||
| NM554 | F−araD139 Δ(ara-leu)7696 galE15 galK16 Δ(lac)X74 rpsL hsdR2 mcrA mcrB1 recA13 | 26 |
| BL21(DE3) | F−ompT lon hsdSBgal dcm(DE3) | 34 |
| 14:40-42 | F−araD139 Δlac(IPOZYA)U169 rpsL thi car-14(::Mu dlac Apr) pyrH42 | 28 |
| Plasmids | ||
| pET22b | ColE1 replication origin; T7 RNA polymerase-dependent expression vector; Apr | Novagen Inc. |
| pET24a | ColE1 replication origin; T7 RNA polymerase-dependent expression vector; Kmr | Novagen Inc. |
| pET24ma | p15A replication origin; T7 RNA polymerase-dependent expression vector; Kmr | 33a |
| pDIA17 | p15A replication origin; harbors the lacI gene; Cmr | 25 |
| pSL1 | Wild-type pyrH from E. coli in pET22b | This work |
| D168N | Mutated pyrH; D168N substitution | 8 |
| D174N | Mutated pyrH; D174N substitution | 8 |
| pSL2 | Truncated UMP kinase; fragment from residue 25 to 241 included | This work |
| pSL3 | Truncated UMP kinase; fragment from residue 50 to 241 included | This work |
| pHSP233 | Truncated UMP kinase; fragment from residue 113 to 241 included | This work |
| pSL4 | Truncated UMP kinase; fragment from residue 1 to 180 included | This work |
| pSL18 | Truncated UMP kinase; fragment from residue 1 to 160 included | This work |
| pSL19 | Truncated UMP kinase; fragment from residue 1 to 140 included | This work |
| pSL8 | Truncated UMP kinase; fragment from residue 1 to 120 included | This work |
| pSL11 | Wild-type UMP kinase from S. typhimurium SL5235 in pET24ma | This work |
| pSL13 | Wild-type UMP kinase from B. subtilis 168 in pET24a | This work |
| pHSP253 | Wild-type UMP kinase from H. influenzae 102-514T in pET24a | This work |
Purification of UMP kinase and its fragments.
Recombinant wild-type UMP kinase and two modified forms (D168N and D174N) were purified from overproducing bacteria as previously described (8, 31). The activity of the wild-type enzyme under standard conditions (i.e., 1 mM ATP, 0.3 mM UMP, 30°C, and pH 7.4) was 70 U/mg (1 U corresponds to 1 μmol of UDP formed in 1 min). UMP kinase fragments were overproduced as inclusion bodies. They were obtained after ultrasound disruption of bacteria and solubilization with 8 M urea.
MAbs and polyclonal antibodies.
Biozzi mice were immunized with 20 μg of antigen at 12-day intervals. After four injections and a last intraperitoneal booster injection, splenic lymphocyte fusions were performed as described by Köhler and Milstein (17). Cell culture supernatants were screened for antibody production by enzyme-linked immunosorbent assay (ELISA). Ascitic fluid from positive clones was obtained by intraperitoneal injection into BALB/c mice. Antibodies from ascitic fluid were purified by two steps of ammonium sulfate precipitation (40 and 45%).
Anti-UMP kinase sera were also obtained by immunizing rabbits with 250 μg of purified recombinant protein, as described for mice. To improve the signal-to-noise ratio, immune sera were adsorbed against a bacterial sonicate from strain 14:40-42 (28).
Immunochemical methods.
Antibody preparations were analyzed by ELISA, as described by Voller et al. (37), with slight modifications. Microtiter plates (Nunc) were incubated with antigen preparations (1 μg/ml diluted in phosphate-buffered saline [PBS]), saturated with PBS containing 0.1% Tween 20 supplemented with 0.5% gelatin, and further incubated with serial dilutions of antibodies or peroxidase-conjugated anti-mouse immunoglobulins in the same buffer. Plate washes were performed with PBS containing 0.1% Tween 20. Peroxidase activity was revealed with ortho-phenylenediamine substrate and optical density measurement at 490 nm. Subclasses of MAbs were determined by ELISA with anti-mouse immunoglobulins specific for light chains (κ or λ), immunoglobulin M (IgM), or various types of IgGs. The affinities of MAbs for various immunogens were determined by ELISA as described by Friguet et al. (11). Western blotting was performed as follows. Protein samples were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred to a nitrocellulose membrane which was further treated as described for ELISA, except that alkaline phosphatase-conjugated anti-mouse (or anti-rabbit) immunoglobulins were used. Alkaline phosphatase activity was revealed by using the nitroblue tetrazolium–5-bromo-4-chloro-3-indolylphosphate dye system.
Two-dimensional gel electrophoresis.
Bacteria were grown in 2YT medium to an optical density of 0.5 at 600 nm, harvested by centrifugation, and then sonicated in 50 mM Tris-HCl (pH 8) containing DNase and RNase (final concentrations of 1 and 0.5 mg/ml, respectively). Insoluble material was removed by centrifugation, and supernatants were boiled for 5 min with 0.3% SDS and 50 mM dithiothreitol. Extracts were quickly frozen in liquid nitrogen, lyophilized, and then resuspended in 9.95 M urea–4% Nonidet P-40–2% ampholytes–100 mM dithiothreitol and stored at −20°C until used. The electrophoresis procedure was as previously described (12, 19), with some modifications. Ten-microliter samples (50 μg of protein) were loaded onto an isoelectric focusing gel (Millipore Inc.; ampholytes, pH range 3 to 10) and focused for 20,000 V × h, and the second-dimension electrophoresis was performed on 10% slab gels. Detection of proteins was performed by silver nitrate staining as described by Morrissey (22). Molecular masses, isoelectric points (pIs), and spot quantifications were determined by using Melanie II software and a GS-700 densitometer (Bio-Rad).
Immunoelectron microscopy.
Bacteria were fixed with 4% formaldehyde (freshly made from paraformaldehyde) and 0.2% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.4) for 1 h at 4°C. The cell pellets were rinsed with cacodylate buffer and then treated with 0.5% aqueous uranyl acetate solution (4), followed by a final rinse in distilled water. Bacteria were embedded in 2% agarose (type IX; Sigma). Small blocks were embedded in Unicryl by the PLT method and modified procedure, as described by Gounon and Rolland (15). Ultrathin sections were collected on Formvar-carbon-coated nickel grids. Sections were then incubated in the following solutions: PBS containing 50 mM NH4Cl (10 min), PBS containing 1% bovine serum albumin (BSA) and 1% normal goat serum (7) (10 min), and rabbit polyclonal anti-UMP kinase antiserum (1/100 dilution) or mouse monoclonal anti-CMP kinase antibodies (100 μg/ml) (1 h). Two washes (5 min each) in PBS containing 0.1% BSA and then one wash in PBS were performed. Incubations were for 45 min in a solution containing gold-conjugated anti-rabbit or anti-mouse immunoglobulin (5- or 10-nm particles; British Biocell Laboratories, Cardiff, United Kingdom) diluted 1/20 in PBS containing 0.01% fish skin gelatin (Sigma). Sections were washed once in PBS and three times in distilled water, fixed for 2 min with 1% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.4), and finally rinsed with distilled water and dried. Optional counterstaining was performed by treating the sections with 2% aqueous uranyl acetate solution for 40 min, followed by a 3-min incubation in Millonig’s lead tartrate solution (21). Specimens were examined with a Philips CM12 electron microscope under standard conditions.
Other analytical procedures.
Protein concentrations were measured as described by Bradford (6). SDS-PAGE was performed as described by Laemmli (18). UMP kinase activity was determined at 30°C and 334 nm in a coupled spectrophotometric assay (0.5-ml final volume) with an Eppendorf ECOM6122 photometer (5). The reaction medium contained 50 mM Tris-HCl (pH 7.4), 50 mM KCl, 1 mM phosphoenolpyruvate, 0.2 mM NADH, 1 mM ATP, 0.3 mM UMP, and 2 U each of pyruvate kinase, nucleoside diphosphate kinase, and lactate dehydrogenase. The reaction was started either with UMP kinase or with UMP.
RESULTS
Purification of UMP kinase and its fragments.
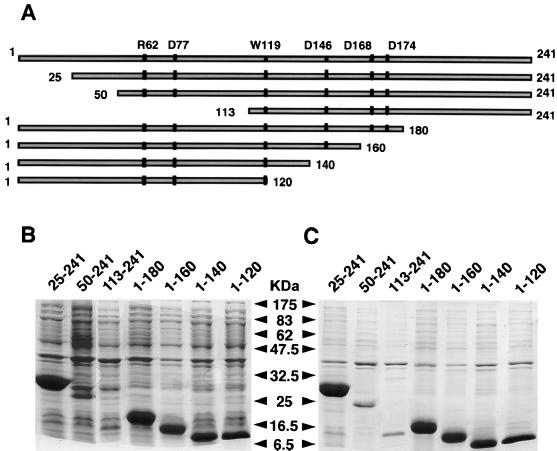
The wild-type UMP kinase and two modified forms (D168N and D174N) were purified as previously described (8). The samples were stored at 4°C in 50 mM Tris-HCl (pH 7.4) as insoluble proteins. After solubilization with 0.1 M borate (pH 9) or with 1 mM UTP in 50 mM Tris-HCl (pH 7.4), the enzymes were fully active. To identify the immunodominant segment(s) in UMP kinase, several amino- or carboxy-terminal deletion forms of the enzyme were generated by mutagenesis (Fig. 1A). The overproduced peptides represented between 20 and 30% of total E. coli proteins, except the fragment encompassing amino acids 50 to 241 (fragment 50-241) and fragment 113-241, whose levels were significantly lower (Fig. 1B). They appeared in the pellet after centrifugation of the bacterial extract at 10,000 × g (Fig. 1C). After several washings of sonicated bacterial pellet with 50 mM Tris-HCl (pH 7.4), UMP kinase fragments were solubilized with 8 M urea in the same buffer. None of the fragments displayed UMP kinase activity.
FIG. 1.
Schematic representation of UMP kinase and of its fragments generated by deletion mutagenesis and SDS-PAGE analysis of peptides overproduced in E. coli. (A) Key residues involved in substrate binding, catalysis, or regulation by nucleotides are indicated by vertical bars. (B) Total bacterial extract after sonication (50 μg of protein). (C) Pellet equivalent to 50 μg of total extract after centrifugation for 5 min at 10,000 × g. The molecular mass markers are from New England Biolabs.
Interaction of polyclonal antibodies with UMP kinase and its fragments.
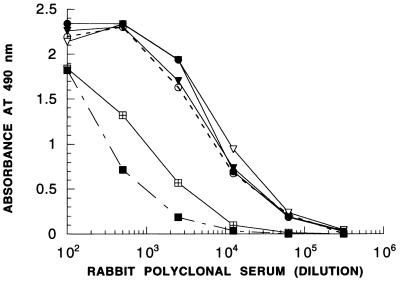
Rabbit polyclonal antisera obtained by using wild-type UMP kinase as the immunogen yielded high absorbance values in ELISA tests in the dilution range of 103 to 104, indicating reasonably high immunogenicity of the protein (Fig. 2). ELISA tests with unrelated proteins demonstrated high specificities and good signal-to-noise ratios of anti-UMP kinase sera. The carboxy-terminally truncated forms of the protein (i.e., fragments 1-120 and 1-180) were detected with approximately 10-fold-lower sensitivity than the complete molecule (Fig. 2).
FIG. 2.
Determination of immunodominant segments of UMP kinase. UMP kinase (○) and various fragments were detected by ELISA with rabbit polyclonal antiserum as described in Materials and Methods. ▾, 25-241 fragment; ▿, 50-241 fragment; •, 113-241 fragment; ⊞, 1-180 fragment; ■, 1-120 fragment.
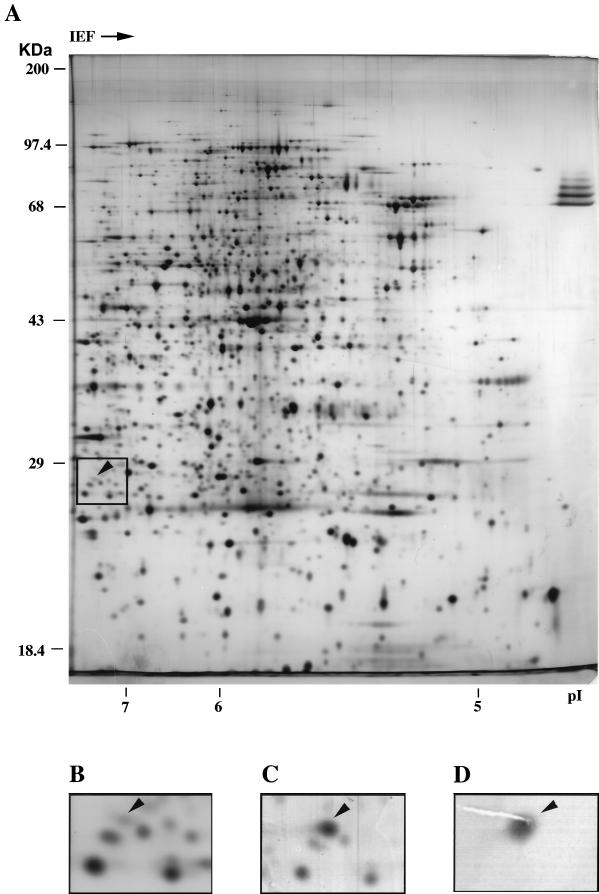
The various UMP kinase fragments in crude extracts or in partially purified forms were also identified by Western blot analysis (not shown). UMP kinase represents approximately 0.05% of the total proteins in E. coli, a value close to the ratio between the enzyme specific activity in crude extracts and the pure form (32). The enzyme was also identified in the E. coli proteome (Fig. 3A and B). Crude extract from a strain harboring the pyrH gene on a multicopy plasmid was subjected to two-dimensional gel electrophoresis and silver stained, and the protein pattern was compared to that from the recipient strain alone (Fig. 3C). In other experiments, crude extract from the wild-type strain was subjected to gel electrophoresis, and then proteins were transferred onto a nitrocellulose membrane and UMP kinase was immunodetected with rabbit polyclonal antiserum (Fig. 3D). A single spot of protein with an apparent molecular mass of 26 kDa and a pI of 7.24 was identified.
FIG. 3.
Identification of UMP kinase in the E. coli proteome by two-dimensional gel electrophoresis. (A) Fifty micrograms of soluble protein from strain BL21(DE3) was silver stained after two-dimensional gel electrophoresis. The molecular mass markers are indicated on the left of the gel, and the isoelectric point scale is on the bottom. (B) Enlarged portion of panel A, in which the position of UMP kinase (arrowhead) and surrounding protein spots can be easily recognized. (C) A multicopy plasmid harboring the pyrH gene was introduced into strain BL21(DE3), and a 10-μg sample of soluble protein was analyzed as for panel A. An enlarged portion is shown, as in panel B. (D) One hundred micrograms of soluble protein from strain NM554 was separated by two-dimensional gel electrophoresis, and UMP kinase was immunodetected with polyclonal rabbit antiserum as described in Materials and Methods. An enlarged portion is shown, with the same magnification as in panels B and C.
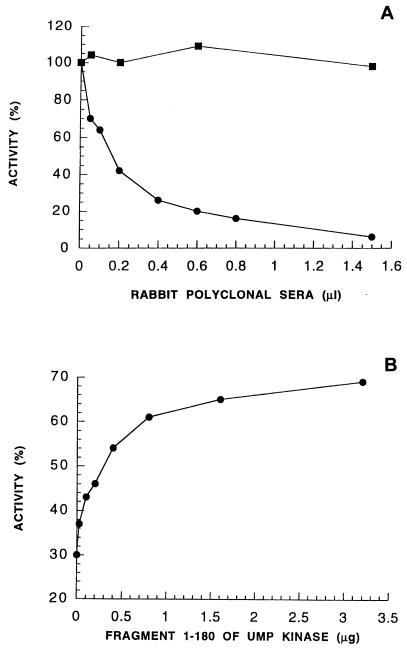
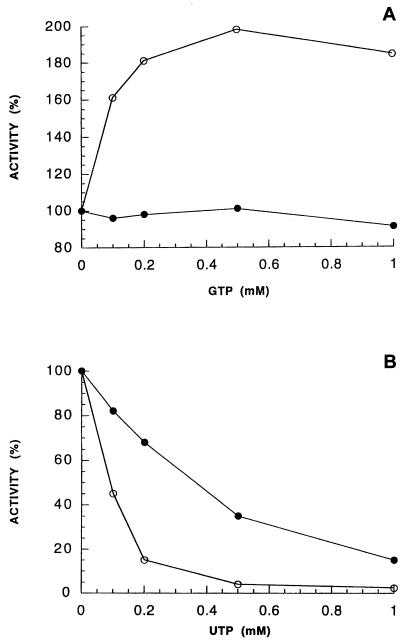
Incubation of UMP kinase with polyclonal antiserum resulted in a concentration-dependent loss of activity. Rabbit antiserum against an unrelated protein had no effect on UMP kinase activity (Fig. 4A). Fragment 1-180 in stoichiometric ratio with UMP kinase alleviated the inhibitory effect (Fig. 4B). Polyclonal antibodies at concentrations where the enzyme was half inhibited completely reversed the activation by GTP (Fig. 5A) but affected the inhibition by UTP to a lesser extent (Fig. 5B).
FIG. 4.
Effect of rabbit polyclonal antiserum on UMP kinase activity. (A) UMP kinase (0.035 U, corresponding to 0.5 μg of protein) in 100 μl of PBS was incubated for 15 min with various amounts of polyclonal anti-UMP kinase antiserum as indicated on the abscissa, and then the residual activity was measured as described in Materials and Methods, with 1 mM ATP and 0.3 mM UMP as substrates (•). Equivalent volumes of unrelated rabbit polyclonal antiserum (■) were used as a control. (B) Antiserum at a concentration calculated to achieve 70% inhibition of enzymatic activity was incubated in 40 μl with fragment 1-180 in the indicated amounts. UMP kinase (0.04 U, corresponding to 0.7 μg of protein) was then added to this mixture, and after 1 min of incubation, the residual activity was measured with 1 mM ATP and 0.3 mM UTP as substrates. One hundred percent represents the UMP kinase activity that would be obtained in the absence of both the polyclonal antiserum and fragment 1-180.
FIG. 5.
Effect of rabbit polyclonal antiserum on GTP activation and UTP inhibition. UMP kinase was incubated as described for Fig. 4A with a polyclonal antiserum solution calculated to yield 50% inhibition. This resulting activity (•), in the absence of GTP or UTP, was considered 100% in this set of experiments. Enzyme activity was then assayed with 1 mM ATP, 1 mM UMP, and various concentrations of GTP (A) or with 1 mM ATP, 0.1 mM UMP, and various concentrations of UTP (B). Results of control experiments in the absence of polyclonal antiserum (○) are also indicated.
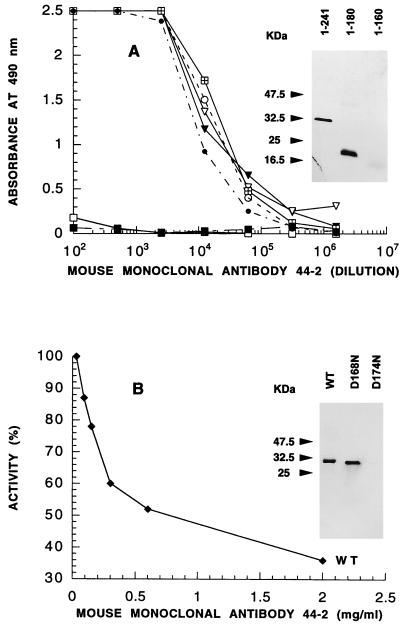
Characterization of a MAb interacting with the native UMP kinase and with a truncated form (fragment 1-180) of the enzyme.
Attempts to obtain murine MAbs by using the native hexameric enzyme as the immunogen failed. As the polyclonal response was biased towards the hydrophilic carboxy-terminal end of the molecule, and as the amino-terminal part alleviated the antiserum inhibition, we used fragment 1-180 of UMP kinase for immunization of mice. Overproduced as inclusion bodies and requiring chaotropic agents for solubilization, this fragment induced a significant polyclonal response and allowed a set of MAbs to be obtained. One MAb (44-2), which is an IgG2a, recognized the native protein as well as the immunogen, with a Kd in the 10−9 M range. Using various UMP kinase fragments in ELISA and Western blotting, we located the epitope first between amino acids 161 and 180 (Fig. 6A) and then between residues 171 and 180 (data not shown). A modified UMP kinase from E. coli (D174N) resulting from site-directed mutagenesis (8) was not recognized by the 44-2 MAb, whereas the vicinally modified variant (D168N) was detected in Western blot analysis with a sensitivity similar to that for the wild-type protein (Fig. 6B, inset). On the other hand, when equivalent amounts (0.1 μg) of UMP kinases from Haemophilus influenzae, Bacillus subtilis, and Salmonella typhimurium were tested on Western blots, only the enzyme from S. typhimurium was recognized by the 44-2 MAb (data not shown). Incubation of the MAb with the wild-type UMP kinase from E. coli resulted in loss of enzymatic activity in a dose-dependent manner, with the maximal inhibition being 60% (Fig. 6B). As expected, the activity of the D174N mutant of UMP kinase was not affected by the MAb.
FIG. 6.
Epitope mapping of MAb 44-2 and its inhibitory effect on UMP kinase. (A) ELISA was performed with UMP kinase (○) or its fragments (▾, 25-241; ▿, 50-241; •, 113-241; ⊞, 1-180; ■, 1-120; □, BSA) as described in Materials and Methods. The inset shows Western blot analysis with 100 ng of protein, performed as described in Materials and Methods. 1-241 corresponds to the complete UMP kinase. (B) UMP kinase inhibition by MAb 44-2 was performed as described for Fig. 4A. The inset shows Western blot analysis with 100 ng of wild-type (WT) or mutated variants of UMP kinase.
Immunoelectron microscopy of UMP kinase of E. coli.
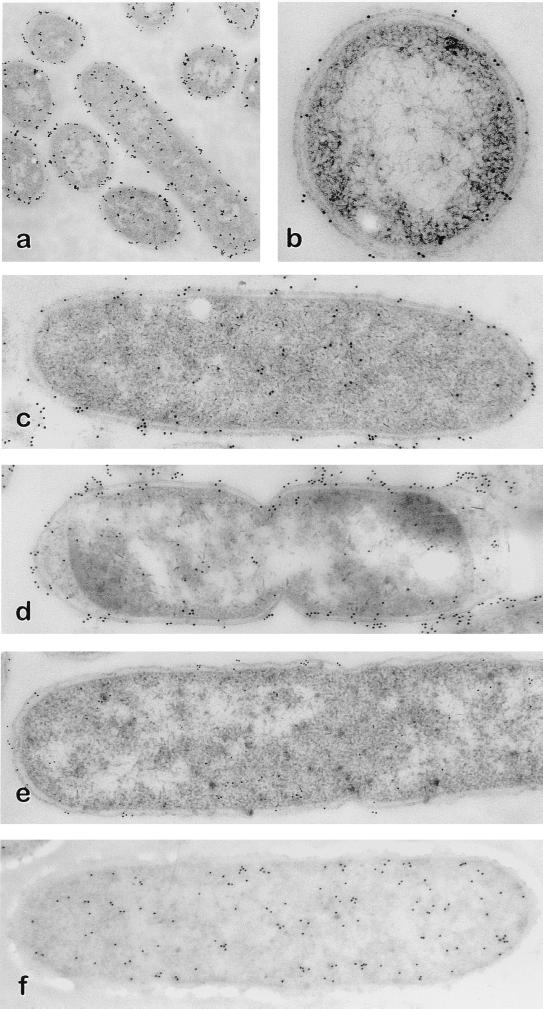
Various E. coli strains were subjected to transmission immunoelectron microscopy analysis after immunogold labeling of UMP kinase (Fig. 7). The labeling patterns showed a localization close to the membranes in strain NM554, which accounted for about 70% of the total gold particles when bacterial samples were harvested at late stationary phase (Fig. 7a to d). Strain 14:40-42, which is defective in UMP kinase activity, was considerably less immunostained than strain NM554 (Fig. 7e). Immunogold labeling of CMP kinase with a MAb (18a) yielded a uniform pattern of particles in the E. coli cytoplasm (Fig. 7f), indicating that the localization of UMP kinase near the membranes is a specific feature of this enzyme. Due to the total size of the immunochemical detection system, which consists of a primary antibody revealed by a secondary antibody linked to a gold particle, the distance between the epitope and the gold particle is about 30 nm. Gold particles will therefore display a statistical scattering centered upon the actual position of the target protein. It is thus not possible to assign an accurate subcellular localization of UMP kinase on the basis of these observations alone, despite the fact that substructures such as the inner membrane, outer membrane, and periplasmic space are clearly visible.
FIG. 7.
Transmission immunoelectron microscopy of various E. coli strains. Strain NM554 in log phase was treated with anti-UMP kinase antiserum, as described in Materials and Methods, and immunogold stained with (a) or without (b) (cross section) silver amplification staining. Strain NM554, either in log phase (c) or in stationary phase (d), was treated with anti-UMP kinase antiserum and immunogold stained. Strains 14:40-42 (e) and NM554 (f) in log phase were treated with anti-UMP kinase antiserum (e) or with anti-CMP kinase MAbs (f) and immunogold stained.
DISCUSSION
One challenging issue regarding NMP kinases is whether they are involved in processes other than mere nucleotide metabolism. Recent data suggest that these enzymes have more than simple housekeeping functions and that AMP kinase, TMP kinase, and UMP kinase are essential for cell life (10, 14, 16, 20, 27, 38, 39).
UMP kinase is the most intriguing member of the bacterial NMP kinase family. The protein exhibits limited sequence similarity with aspartokinase (residues 145 to 194) and phosphoglycerate kinase (residues 35 to 78), has an oligomeric structure, and is subject to complex regulation. On the other hand, UMP kinase was shown to participate in transcription regulation of the carAB operon and most probably is involved in cell division (16, 38). In the absence of three-dimensional structural data, immunochemical methods might elucidate the role of an enzyme whose function in the bacterial cell is not well understood.
(i) Structure-function analysis.
Previous experiments suggested that amino acids between residues 62 and 77 and between residues 146 and 174 are involved in regulation of UMP kinase and in catalysis, respectively (8). The carboxy-terminal quarter of UMP kinase was predicted to be immunodominant, due to its hydrophilicity. This indeed seems to be the case, as polyclonal antiserum raised against the whole protein exhibited a pronounced detection bias in favor of the carboxy terminus. Although only approximately 10% of the total immune response was targeted to the domains of the molecule relevant for catalysis, the polyclonal antibodies strongly inhibited the UMP kinase activity. At antiserum dilutions where the enzyme still retained 50% of its activity, the activation by GTP was completely reversed. Under identical conditions, the inhibition by UTP, although decreased, was not abolished. Assuming that these effects are due to perturbations in nucleotide binding, this suggests that the GTP and UTP binding sites in the protein are not completely overlapping, in agreement with fluorescence analyses and chemical modification studies (31).
MAbs are better suited for analyzing discrete effects upon binding to a target protein. The fragment situated between residues 1 and 180 generated a MAb displaying high affinity and specificity towards native UMP kinase. It recognized a linear and surface-exposed epitope, between amino acids 171 and 180, having the sequence 171FTADPAKDPT180. A single conservative amino acid substitution (D174N) abolished the interaction of this MAb with UMP kinase. The fact that H. influenzae and B. subtilis UMP kinases were not recognized by this antibody was not surprising, as only 5 of 10 amino acids were conserved in the above-mentioned sequence. Conversely, the S. typhimurium enzyme was detected by the MAb, in agreement with the fact that nine residues of this segment are common to E. coli and S. typhimurium (sequencing data are unpublished). Although a detailed analysis of the stoichiometry of the UMP kinase-MAb interaction was not undertaken, the inhibition of enzyme activity is not complete at saturating concentrations of the MAb. This suggests a particular distribution of UMP kinase subunits in the hexamer. Further physicochemical and electron microscopic studies will clarify this point (Asp174 has been demonstrated to play a role in binding of UMP to the enzyme) (8).
(ii) Localization of UMP kinase in E. coli.
The E. coli proteome, defined as the total protein complement of a genome, has been widely investigated over the past years by two-dimensional electrophoresis (36). Until now, among the one-quarter of the gene products identified, adenylate kinase and CMP kinase were the only members of the NMP kinase family that had been localized in the two-dimensional reference map. The combination of two-dimensional gel electrophoresis and specific immunolabeling allowed the detection of a third NMP kinase (i.e., UMP kinase), whose molecular mass and isoelectric point were in agreement with those calculated from the protein sequence. Electron microscopy of immunolabeled E. coli samples showed that UMP kinase was located predominantly near the membranes. This was an unexpected result, because the protein does not display features suggesting compartmentalization, such as membrane-spanning segments or a typical leader peptide. The biological relevance of this observation is under study, and a detailed analysis of the interaction between UMP kinase and the bacterial membranes will help clarify this point. As a major concern with immunolocalization techniques is antibody specificity, care was taken to detect any cross-reacting species. UMP kinase was the single spot appearing after dye revelation. Overexposure of the nitrocellulose membranes revealed a weak spot corresponding to DnaK. On the other hand, strain 14:40-42 of E. coli, which is defective in UMP kinase activity, was considerably less immunolabeled with polyclonal antibodies, in agreement with its low reactivity in Western blots. We hypothesize that the structural factor which might contribute to UMP kinase stacking on the bacterial membranes is the strong tendency of the hexameric molecule to aggregate at physiological pH values (32). The concentration of UMP kinase in E. coli is approximately 0.1 mg/ml (4 μM in terms of the monomer), a value which corresponds to the solubility limit of the protein in vitro. The aggregate forms planar structures, as suggested by our preliminary studies by electron microscopy. It is known that complex aggregates such as S-layers in bacterial envelopes also form planar structures (2). The unique topology of UMP kinase compared to the other members of NMP kinase family might also be related to its putative role in cell division. The gene encoding UMP kinase (pyrH) is identical to the smbA gene, which was originally recognized as being involved in proper chromosome partitioning during cell division. Two smbA mutations (R62H and D201N) were responsible for the altered morphological phenotype under nonpermissive conditions, suggesting defects in specific membrane sites. The absence of normal UMP kinase function presumably causes defects in the septation site, and smbA mutants are SDS sensitive (38). The specific localization of UMP kinase might thus result from interactions of this enzyme with integral membrane proteins or other proteins known for their localization near the membranes, which remain to be identified.
ACKNOWLEDGMENTS
This work was supported by grants from the Centre National de la Recherche Scientifique (URA 1129) and the Institut Pasteur.
We thank J. Neuhard for many inspiring discussions, N. Glansdorff for the kind gift of E. coli 14:40-42, D. Sourdive for the kind gift of plasmid pET24ma, O. Jeannequin for skillful technical help, and M. Ferrand for excellent secretarial assistance.
REFERENCES
- 1.Abele U, Schulz G E. High-resolution structures of adenylate kinase from yeast ligated with inhibitor Ap5A, showing the pathway of phosphoryl transfer. Protein Sci. 1995;4:1262–1271. doi: 10.1002/pro.5560040702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bahl H, Scholz H, Bayan N, Chami M, Leblon G, Gulik-Krzywicki T, Shechter E, Fouet A, Mesnage S, Tosi-Couture E, Gounon P, Mock M, Conway de Macario E, Macario A J, Fernandez-Herrero L A, Olabarria G, Berenguer J, Blaser M J, Kuen B, Lubitz W, Sara M, Pouwels P H, Kolen C P, Boot H J, Resch S. Molecular biology of S-layers. FEMS Microbiol Rev. 1997;20:47–98. doi: 10.1111/j.1574-6976.1997.tb00304.x. [DOI] [PubMed] [Google Scholar]
- 3.Bârzu O, Gilles A-M. Relation structure-fonction chez les enzymes ATP-dépendants, vue à travers la plus petite phosphotransférase, l’adényl kinase. Ann Inst Pasteur/Actualités. 1993;4:121–132. [Google Scholar]
- 4.Benichou J C, Frehel C, Ryter A. Improved sectioning and ultrastructure of bacteria and animal cells embedded in Lowicryl. J Electron Microsc Technique. 1990;14:289–297. doi: 10.1002/jemt.1060140402. [DOI] [PubMed] [Google Scholar]
- 5.Blondin C, Serina L, Wiesmüller L, Gilles A-M, Bârzu O. Improved spectrophotometric assay of nucleoside monophosphate kinase activity using the pyruvate kinase/lactate dehydrogenase coupling system. Anal Biochem. 1994;220:219–221. doi: 10.1006/abio.1994.1326. [DOI] [PubMed] [Google Scholar]
- 6.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 7.Brorson S H. Bovine serum albumin (BSA) as a reagent against non-specific immunogold labeling on LR-White and epoxy resin. Micron. 1997;28:189–195. doi: 10.1016/s0968-4328(97)00030-9. [DOI] [PubMed] [Google Scholar]
- 8.Bucurenci N, Serina L, Zaharia C, Landais S, Danchin A, Bârzu O. Mutational analysis of UMP kinase from Escherichia coli. J Bacteriol. 1998;180:473–477. doi: 10.1128/jb.180.3.473-477.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dreusicke D, Karplus P A, Schulz G E. Refined structure porcine cytosolic adenylate kinase at 2.1 Å resolution. J Mol Biol. 1988;199:359–371. doi: 10.1016/0022-2836(88)90319-1. [DOI] [PubMed] [Google Scholar]
- 10.Fricke J, Neuhard J, Kelln R A, Pedersen S. The cmk gene encoding cytidine monophosphate kinase is located in the rpsA operon and is required for normal replication rate in Escherichia coli. J Bacteriol. 1995;177:517–523. doi: 10.1128/jb.177.3.517-523.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Friguet B, Chaffotte A F, Djavadi-Ohaniance L, Goldberg M E. Measurements of the true-affinity constant in solution of antigen-antibody complexes by enzyme-linked immunosorbent assay. J Immunol Methods. 1985;77:305–319. doi: 10.1016/0022-1759(85)90044-4. [DOI] [PubMed] [Google Scholar]
- 12.Garrels J I. Quantitative two-dimensional electrophoresis of proteins. Methods Enzymol. 1983;100:411–423. doi: 10.1016/0076-6879(83)00070-1. [DOI] [PubMed] [Google Scholar]
- 13.Gerstein M, Schulz G, Chothia C. Domain closure in adenylate kinase joints on either side of two helices close like neighboring fingers. J Mol Biol. 1993;229:494–501. doi: 10.1006/jmbi.1993.1048. [DOI] [PubMed] [Google Scholar]
- 14.Goelz S E, Cronan J E., Jr Adenylate kinase of Escherichia coli: evidence for a functional interaction in phospholipid synthesis. Biochemistry. 1982;21:189–195. doi: 10.1021/bi00530a032. [DOI] [PubMed] [Google Scholar]
- 15.Gounon P, Rolland J-P. Modification of unicryl composition for rapid polymerization at low temperature without alteration of immunocytochemical sensitivity. Micron. 1998;29:293–296. doi: 10.1016/s0968-4328(98)00013-4. [DOI] [PubMed] [Google Scholar]
- 16.Kholti A, Charlier D, Gigot D, Huysveld N, Roovers M, Glansdorff N. pyrH-encoded UMP-kinase directly participates in pyrimidine-specific modulation of promoter activity in Escherichia coli. J Mol Biol. 1998;280:571–582. doi: 10.1006/jmbi.1998.1910. [DOI] [PubMed] [Google Scholar]
- 17.Köhler G, Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature. 1975;256:495–497. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- 18.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 18a.Landais, S., et al. Unpublished data.
- 19.Laurent-Winter C, Ngo S, Danchin A, Bertin P. Role of Escherichia coli histone-like nucleoid-structuring protein in bacterial metabolism and stress response. Identification of targets by two-dimensional electrophoresis. Eur J Biochem. 1997;244:767–773. doi: 10.1111/j.1432-1033.1997.00767.x. [DOI] [PubMed] [Google Scholar]
- 20.Lu Q, Inouye M. Adenylate kinase complements nucleoside diphosphate kinase deficiency in nucleotide metabolism. Proc Natl Acad Sci USA. 1996;93:5720–5725. doi: 10.1073/pnas.93.12.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Millonig G. A modified procedure for leading staining of thin sections. J Biophys Biochem Cytol. 1961;11:736–739. doi: 10.1083/jcb.11.3.736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morrissey J H. Silver stain for proteins in polyacrylamide gels: a modified procedure with enhanced uniform sensitivity. Anal Biochem. 1981;117:307–310. doi: 10.1016/0003-2697(81)90783-1. [DOI] [PubMed] [Google Scholar]
- 23.Müller C W, Schulz G E. Structure of the complex between adenylate kinase from Escherichia coli and the inhibitor Ap5A refined at 1.9 Å resolution. A model for a catalytic transition state. J Mol Biol. 1992;224:159–177. doi: 10.1016/0022-2836(92)90582-5. [DOI] [PubMed] [Google Scholar]
- 24.Müller-Dieckmann H-J, Schultz G E. The structure of uridylate kinase with its substrates, showing the transition state geometry. J Mol Biol. 1994;236:361–367. doi: 10.1006/jmbi.1994.1140. [DOI] [PubMed] [Google Scholar]
- 25.Munier H, Gilles A-M, Glaser P, Krin E, Danchin A, Sarfati R S, Bârzu O. Isolation and characterization of catalytic and calmodulin-binding domains of Bordetella pertussis adenylate cyclase. Eur J Biochem. 1991;196:469–474. doi: 10.1111/j.1432-1033.1991.tb15838.x. [DOI] [PubMed] [Google Scholar]
- 26.Raleigh E A, Murray N E, Revel H, Blumenthal R M, Westaway D, Reith A D, Rigby P W J, Elhai J, Hanahan D. McrA and McrB restriction phenotypes of some E. coli strains and implications for gene cloning. Nucleic Acids Res. 1988;16:1563–1575. doi: 10.1093/nar/16.4.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reynes J-P, Tiraby M, Baron M, Drocourt D, Tiraby G. Escherichia coli thymidilate kinase: molecular cloning, nucleotide sequence, and genetic organization of the corresponding tmk locus. J Bacteriol. 1996;178:2804–2812. doi: 10.1128/jb.178.10.2804-2812.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roovers M, Charlier D, Feller A, Gigot D, Holemans F, Lissens W, Piérard A, Glansdorff N. carP, a novel gene regulating the transcription of the carbamoylphosphate synthetase operon of Escherichia coli. J Mol Biol. 1988;204:857–865. doi: 10.1016/0022-2836(88)90046-0. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 30.Schultz G E, Schiltz E, Tomasselli A G, Frank R, Brune M, Wittinghofer A, Schirmer R H. Structural relationships in the adenylate kinase family. Eur J Biochem. 1986;161:127–132. doi: 10.1111/j.1432-1033.1986.tb10132.x. [DOI] [PubMed] [Google Scholar]
- 31.Serina L, Blondin C, Krin E, Sismeiro O, Danchin A, Sakamoto H, Gilles A-M, Bârzu O. Escherichia coli UMP-kinase, a member of the aspartokinase family, is a hexamer regulated by guanine nucleotides and UTP. Biochemistry. 1995;34:5066–5074. doi: 10.1021/bi00015a018. [DOI] [PubMed] [Google Scholar]
- 32.Serina L, Bucurenci N, Gilles A-M, Surewicz W K, Fabian H, Mantsch H H, Takahashi M, Petrescu I, Batelier G, Bârzu O. Structural properties of UMP-kinase from Escherichia coli: modulation of protein solubility by pH and UTP. Biochemistry. 1996;35:7003–7011. doi: 10.1021/bi960062v. [DOI] [PubMed] [Google Scholar]
- 33.Smallshaw J, Kelln R A. Cloning, nucleotide sequence and expression of the Escherichia coli K-12 pyrH gene encoding UMP kinase. Genetics (Life Sci Adv) 1992;11:59–65. [Google Scholar]
- 33a.Sourdive, D. Unpublished data.
- 34.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 35.Tsai M-D, Yan H. Mechanism of adenylate kinase: site-directed mutagenesis versus X-ray and NMR. Biochemistry. 1991;30:6806–6818. doi: 10.1021/bi00242a002. [DOI] [PubMed] [Google Scholar]
- 36.VanBogelen R A, Abshire K Z, Pertsemlidis A, Clark R L, Neidhardt F C. Gene-protein database of Escherichia coli K-12, edition 6. In: Neidhardt F C, Curtis III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molec ular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 2067–2117. [Google Scholar]
- 37.Voller A, Bidwell D, Huldt G, Engvall E. A microplate method of enzyme-linked immunosorbent assay and its application to malaria. Bull W H O. 1974;51:209–211. [PMC free article] [PubMed] [Google Scholar]
- 38.Yamanaka K, Ogura T, Niki H, Hiraga S. Identification and characterization of the smbA gene, a suppressor of the mukB null mutant of Escherichia coli. J Bacteriol. 1992;174:7517–7526. doi: 10.1128/jb.174.23.7517-7526.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamanaka K, Ogura T, Koonin E V, Niki H, Hiraga S. Multicopy suppressors, mssA and mssB, of an smbA mutation of Escherichia coli. Mol Gen Genet. 1994;243:9–16. doi: 10.1007/BF00283870. [DOI] [PubMed] [Google Scholar]