Abstract
Cell-based therapies have been making great advances toward clinical reality. Despite the increase in trial activity, few therapies have successfully navigated late-phase clinical trials and received market authorization. One possible explanation for this is that additional tools and technologies to enable their development have only recently become available. To support the safety evaluation of cell therapies, the Health and Environmental Sciences Institute Cell Therapy—Tracking, Circulation and Safety Committee, a multisector collaborative committee, polled the attendees of the 2017 International Society for Cell & Gene Therapy conference in London, UK, to understand the gaps and needs that cell therapy developers have encountered regarding safety evaluations in vivo. The goal of the survey was to collect information to inform stakeholders of areas of interest that can help ensure the safe use of cellular therapeutics in the clinic. This review is a response to the cellular imaging interests of those respondents. The authors offer a brief overview of available technologies and then highlight the areas of interest from the survey by describing how imaging technologies can meet those needs. The areas of interest include imaging of cells over time, sensitivity of imaging modalities, ability to quantify cells, imaging cellular survival and differentiation and safety concerns around adding imaging agents to cellular therapy protocols. The Health and Environmental Sciences Institute Cell Therapy—Tracking, Circulation and Safety Committee believes that the ability to understand therapeutic cell fate is vital for determining and understanding cell therapy efficacy and safety and offers this review to aid in those needs. An aim of this article is to share the available imaging technologies with the cell therapy community to demonstrate how these technologies can accomplish unmet needs throughout the translational process and strengthen the understanding of cellular therapeutics.
Keywords: biodistribution, cell therapy, fate, in vivo tracking, non-invasive, safety
Introduction
Cell-based therapies are making great advances toward clinical reality. This is encouraging news, as these advanced therapy medicinal products [1] offer a therapeutic opportunity for several currently incurable diseases, including blood-borne cancers [2-5], immunodeficiency disorders [6,7] and degenerative diseases [8-10].
The number of clinical trials investigating cell-based therapies is increasing, which is a good indication that there are tractable targets with promising therapeutic interventions. This increase also suggests that these putative therapies have satisfactory pre-clinical evidence of safety and efficacy. The increase in clinical trials, particularly phase 3 trials, also indicates that effective and current good manufacturing practice-compliant manufacturing processes have been developed for these complex products.
Despite the increase in clinical trial activity, very few therapies have successfully negotiated late-phase clinical trials and received marketing authorizations. Why is this the case? One possible reason is that it could simply be a matter of time, particularly in the case of adoptive immunotherapies. Although the concept has been around since the 1980s [11], the tools and technologies to enable commercialization of these therapies have only relatively recently become available. However, this is not necessarily the case for other therapies, such as stem cell-derived cellular therapies. Globally, trials for stem cell-based therapies number in the thousands (ClinicalTrials.gov) and have been pursued for decades. Scale-up manufacturing processes, although not optimal, have been available for some time, and the safety profile for some types of cell-based therapies (e.g., mesenchymal stromal cells [MSCs]) appears favorable. Indeed, autologous and allogeneic bone marrow transplants are widely used therapeutic options.
As more therapies are being developed and assessed, the Health and Environmental Sciences Institute (HESI) formed the Cell Therapy—Tracking, Circulation and Safety (CT-TRACS) Committee to combine the minds of stakeholders across the private and public sectors of academia and industry with regulators to enhance confidence in the safe use of cell therapy technologies. The mission of CT-TRACS is to improve the safety of cell-based therapies for patients by enhancing our ability to reliably apply analytical methods, devices and scientific knowledge to evaluate the distribution and fate of these cells in patients. To achieve these goals, CT-TRACS is divided into two subcommittees: (i) point of administration/biodistribution assessment and (ii) tumorigenicity assessment. Together, the two subcommittees polled the attendees of the 2017 International Society for Cell & Gene Therapy Annual Meeting to understand the gaps and needs that cell-based therapy developers encounter at the stage of safety evaluations: in vivo. The goal of this survey was to collect the information necessary to inform stakeholders of priority areas in need of development to ensure the safe use of cellular therapies in the clinic and to determine whether technologies and tools currently available in clinical settings could successfully meet those needs.
The majority of survey participants were professionals having at least 6 or more years of experience in the cell therapy field, with a mix of those from academic and pharmaceutical/biotechnology sectors. This suggests representation of cell therapies at a range of development stages, from pre-clinical to commercialization.
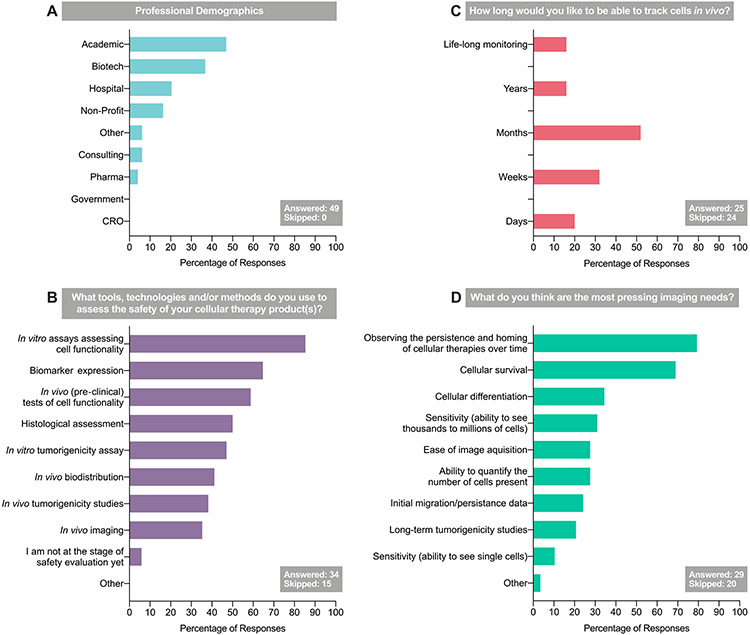
The survey results indicated that, if they could, therapy developers would like to be able to track cells from point of administration up to several months post-transplantation (Figure 1). Approximately 60% of survey respondents did not know how long the therapeutic cells in their product survived in the body. These therapeutic cells were both immune therapeutic cells and stem cell-derived. Most respondents indicated that their therapy was intended for systemic administration; as such, there is a requirement to demonstrate that the therapeutic cell has reached the disease site and is not accumulating at a site where it may cause harm. Equally, if a cell therapy is surgically implanted or delivered directly to the disease site via a catheter, it may be important to determine whether the cells remain at the site for the duration necessary to be efficacious.
Figure 1.
Results of the 2017 CT-TRACS stakeholder survey (“Assessing the needs for cell-based therapies translation”), administered at the 2017 ISCT Annual Meeting. CRO, contract research organization; ISCT, International Society for Cell & Gene Therapy; Pharma, pharmaceutical.
We also know that regulatory authorities would like therapy developers to employ cell tracking technology, if it were available, to use in clinical trials [12-17]. The authors’ survey asked therapy developers if in vivo tracking of cells was important for determining the efficacy and safety of cell therapies. The results indicated that 76% of respondents believed it was, with the majority of these indicating that both pre-clinical and clinical tracking of the cells is required.
Consequently, the HESI CT-TRACS consortium believes that the ability to fully understand therapeutic cell fate is vital for determining and understanding cell therapy efficacy and safety. Because we are concerned that the lack of efficacy and safety data could become a major block to cellular therapies becoming a mainstream therapeutic option, we set out to provide an overview of technologies for in vivo cell tracking. In this article, the authors review technologies that have the potential to track cells from point of administration up to several months post-transplantation. The authors explore how they could be used and/or adapted to address the unmet needs indicated by survey participants and regulatory authorities. The topics the authors discuss align with areas of need that were expressed by the survey respondents.
Cell Tracking Technologies: A Concise Review of Clinically Applicable Tools and Methodologies
Cellular imaging techniques tend to fall into two categories: direct and indirect labeling. Direct labeling involves the incorporation of a label or probe with the cells of interest prior to administration, and indirect labeling involves the genetic modification of cells to incorporate a reporter gene. Numerous reviews of cellular imaging technologies exist [18-22], so a brief review of emerging and clinically applicable technologies is included in the following sections. Technologies are grouped primarily by imaging modality, and a discussion of co-developed and multimodal approaches is included. These methods are further explored later, when the applications of the technologies will be discussed in more detail surrounding the survey respondents’ imaging needs and concerns.
Early means of assessment
Having an indication of the biodistribution profile of a cell therapeutic product is important, as it provides supportive information for the interpretation of pharmacology studies. Furthermore, knowledge regarding the distribution profile of the cell product could guide the design of toxicological studies. Consequently, for optimal design of the pre-clinical package, it is most favorable to start biodistribution studies at the beginning of the development phase. In these studies, methods such as quantitative polymerase chain reaction (qPCR) techniques or immunohistochemical labeling specific to the cell product are currently often utilized.
Early biodistribution assessments can help in choosing an animal model for and determining the duration of toxicology studies. Using a small number of animals at continuous study intervals will develop a basic understanding of these endpoints. Later biodistribution studies can be combined with ongoing good laboratory practice safety studies to conserve animals and contain program costs.
These good laboratory practice safety studies most commonly utilize PCR techniques or immunohistochemical labeling for cell product-specific antibodies for in vivo cell detection. The use of qPCR is labor-intensive and expensive, requiring dedicated laboratories with separate sampling processing, reagent preparation and amplification/product detection areas to minimize the possibility of cross-contamination. DNA and RNA can be isolated and purified from a variety of tissues, body fluids, cells in culture and other biologic samples. Extraction procedures must be optimized to provide the highest quality real-time qPCR results, and both quality and yield of extracted nucleic acid samples must be assessed prior to being used in the PCR assay. As the cells from a tissue are often pooled to deliver the DNA and/or RNA for the analyses, information on the spatial distribution of the cells within a tissue cannot be deduced. However, the total number of cells present in a tissue or organ could be quantified with this technique.
Immunohistochemistry is often employed as a technique for biodistribution analysis. This method requires the proper preservation of the tissue after the animal is sacrificed to make tissue slices that can be stained with specific antibodies, enabling discrimination of the (human) cell product from the tissue cells of the animal. Consequently, this method will reveal qualitative information on the spatial distribution of the cells within the slices of the respective tissue or organ; however, quantification of the total product cells present in the tissue is not possible.
Knowledge regarding cell survival of the cell product in the animal model, exposure to the product and distribution profile will thus determine the design of the toxicology program and support interpretation of pharmacology studies in animals, but it may also aid in determining the design of a potential clinical biodistribution study.
Single photon emission computed tomography: indium and technetium applications
Single photon emission computed tomography (SPECT) is one of the two main nuclear imaging modalities commonly used in both the clinic and pre-clinical research. SPECT images give three-dimensional quantitative or semi-quantitative information on the distribution of radioisotopes across either the whole body or selected anatomical regions. SPECT is commonly used with x-ray computed tomography (CT) in dedicated SPECT-CT scanners, with CT providing anatomical contrast based on tissue density to supplement the radionuclide-based SPECT image. SPECT image contrast is produced via the detection of photons (i.e., gamma rays) released during the decay of certain radioactive isotopes, allowing a three-dimensional tomographic image to be reconstructed using a specialized algorithm. This gives quantitative measurements of radioactivity biodistribution under optimal conditions and appropriate calibration. As photon energy is dependent on the isotope, simultaneous imaging of multiple isotopes is possible when their photon energy levels are sufficiently separated, allowing the biodistribution of more than one labeled cell type or molecule to be tracked with one scan.
The range of isotopes commonly used with SPECT includes metallic (indium-111 [111In] and technetium-99m [99mTc]) and non-metallic (iodine-123 [123I] and iodine-125 [125I]) isotopes, allowing labeling of cells, antibodies and small molecules, with half-lives between hours and days, suitable for tracking over a few hours to a week or more. For more than three decades, SPECT imaging has been established in the clinic for tracking autologous leukocytes to diagnose areas of inflammation, most commonly following direct labeling of cells with either 111In-oxine or 99mTc-hexamethylpropyleneamine oxime (HMPAO) prior to injection [23,24]. This labeling involves incubation of the cells with the radiolabel for approximately 20–30 min before separation of the cells and/or washing to remove unbound tracer before dosing of the cells into the patient. Although diagnostic use of direct cell labeling is established for SPECT imaging, this has rarely been used in the clinic with cell therapies because of concerns of cytotoxicity inhibiting longer-term cell survival or affecting therapeutic phenotype. With SPECT, this is due to the amounts of radioactivity needed to produce a sufficient signal, which has caused significant time-dependent toxicity in in vitro assessments of MSCs [25], hematopoietic progenitor cells [26] and lymphocytes [27] labeled with 111In. However, some examples of clinical use do exist, including tracking of primed dendritic cells for cancer therapy with 111In-oxine and 99mTc-HMPAO labeling [28], labeling of MSCs for cirrhosis therapy with 111In-oxine and tracking up to 10 days post-injection [29] and tracking of 99mTc-labeled bone marrow mononuclear cells in stroke patients for 24 h [30]. Use of SPECT isotopes with longer half-lives (68 h for 111In versus 6 h for 99mTc) has the benefit of allowing cells to be tracked for longer; however, this must be weighed against the potential for greater radioactive exposure and hence toxicity to cells. In addition to tracking cells following direct labeling, more recent work on reporter genes for SPECT has demonstrated the possibility of tracking cells for longer periods of time, although this has yet to be demonstrated with SPECT in patients (Table 1).
Table 1.
Selected examples of cell tracking with nuclear imaging.
| Imaging modality | Cell type | Information obtained |
|---|---|---|
| SPECT | MSCs (111In-oxine) | Success of delivery (% cells in target location) |
| Hematopoietic progenitor cells (111In-oxine,99mTc-HMPAO) | ||
| Longitudinal retention | ||
| Biodistribution | ||
| Leukocytes (111In-oxine, 99mTc-HMPAO) | Within-organ heterogeneity | |
| Migration/homing | ||
| Dendritic cells (111In-oxine, 99mTc-HMPAO) | Inter-patient variability | |
| PET | Islet cells (18FDG) | Success of delivery (% cells in target location) |
| Hematopoietic progenitor cells (18FDG) | ||
| Longitudinal retention | ||
| Bone marrow cells (18FDG) | Biodistribution | |
| T cells (89Zr-oxine) | Within-organ heterogeneity | |
| Natural killer cells (89Zr-oxine) | Migration/homing | |
| Inter-patient variability | ||
| MSCs (89Zr-oxine) |
Positron emission tomography: fluorodeoxyglucose and zirconium
Positron emission tomography (PET) is the other main nuclear imaging technique commonly used in research and the clinic. PET image contrast is produced via the simultaneous detection of two photons (gamma rays) of 511 keV, which are released at 180 degrees from each other following the annihilation of the positron emitted from the imaging isotope when it encounters an electron in the host tissue. Like SPECT, PET is also often commonly used together with CT in a combined PET-CT scanner, with CT providing anatomical information and soft tissue contrast. PET has the benefits of allowing a lower dose of radioisotope to be given to the patient and shorter scan times.
Positron-emitting isotopes commonly used include fluorine-18 (18F), zirconium-89 (89Zr), gallium-68 (68Ga) [31] and copper-64 (64Cu). Although no cell tracking techniques reliant on direct labeling and PET imaging are routinely used in the clinic, a couple of direct labeling options exist, including fluorodeoxyglucose ([18F]FDG) and 89Zr-oxine. The metabolic tracer [18F]FDG was initially developed for diagnosing tumors, in which uptake is high because of their increased glycolytic rate, allowing detection with PET imaging. However, glucose uptake is fairly good in most cell types, so [18F]FDG can be readily used for labeling cell therapies via a short incubation with the tracer prior to their in vivo transplantation. FDG is widely available, and its clinical and pre-clinical use is well established in oncology, which makes it a convenient agent for the alternative use of tracking therapeutic cells. The main limitation here, however, is the 110-min half-life of 18F, meaning that in practical terms the transplanted cells will only be traceable over the first few hours after transplantation, and the signal will be gone by the following day. This short half-life has the benefit of reducing radioactive dosing of patients as well as radioactive dose to the cells but does not allow the longer-term migration of cells to be followed over days or weeks.
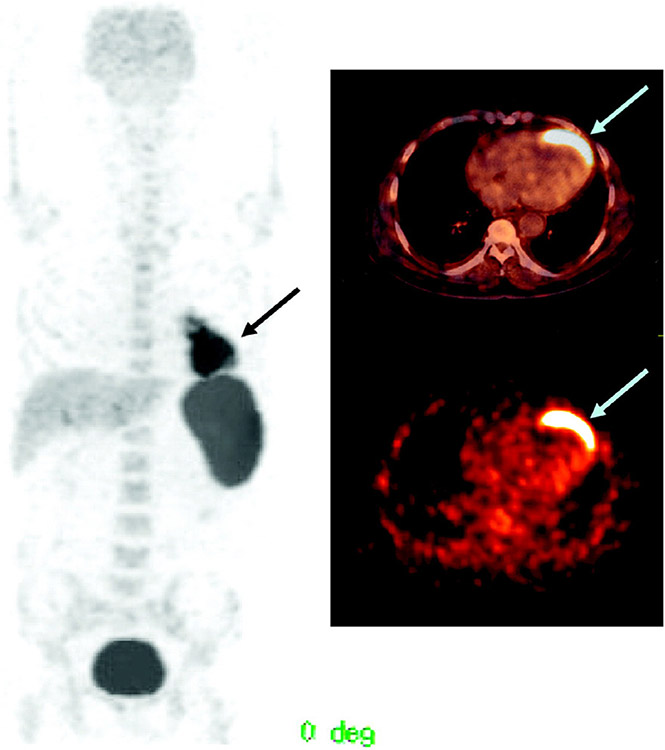
A handful of informative examples of clinical PET imaging of cell therapies with FDG exist for a range of therapies (Table 1, Figure 2) [32-36]. Although these do not show migration of cells past a few hours post-transplantation because of the short half-life of 18F, they indicate the degree of variability to be expected between patients in terms of successful delivery, heterogeneity of distribution within the target tissue, areas of off-target delivery and kinetics of retention and clearance following injection. With transplanted islet cell delivery, hot spots have been seen across the lobes of the liver, with variability in retention between lobes in each patient, consistent with embolism. In myocardial studies, retention of cells within the infarction varied over 10-fold between two populations of cells investigated [34]. This clearly illustrates the potential utility of cell tracking in investigating optimization strategies for both delivery routes and cell type at the clinical stage.
Figure 2.
PET/CT images of a 65-year-old man with a history of anterior wall infarction. After percutaneous intervention, 18F-FDG-labeled stem cells were injected via an intracoronary catheter. PET/CT images were obtained 2 h after injection. Stem cell accumulation at the myocardium is well visualized (arrow). The total amount of stem cells at the myocardium was 2.1% of the injected dose. Reproduced with permission from [35] © Society of Nuclear Medicine and Molecular Imaging (2006).
The recent development of 89Zr-oxine as a radiolabel for cell tracking with PET imaging comes out of decades of use of 111In-oxine with SPECT imaging. As with 111In-oxine and [18F]FDG labeling of cells, this relies on a short incubation (20–30 min) with the label before cells are washed/separated and injected into patients. The potential advantage of 89Zr versus 111In-based labeling strategies is that 89Zr can be imaged with PET, which has a higher sensitivity of detection than SPECT. Both 89Zr-oxine and 111In-oxine are taken up by cells because of their high lipophilicity, and they show comparable retention in cells [37]. As 89Zr has a much longer half-life than 18F (78.4 h versus 110 min), this extends the period of time over which cells can be tracked in practical terms from a few hours for [18F]FDG to a week or two for 89Zr-oxine.
Some pre-clinical examples give an idea of the relevant information that might be gained on the in vivo behavior of cell therapies using 89Zr-oxine labeling and PET imaging. Homing or co-localization to tumors has been shown with cytotoxic T cells [38], chimeric antigen receptor (CAR) T cells [39] and γδ T cells [40]. In another example of immunotherapy, adoptively transferred natural killer cells were tracked up to 7 days in rhesus macaques, showing initial delivery to the lungs and subsequent migration to the liver and spleen [41]. Although 89Zr-oxine cell tracking has not yet progressed to the clinic, the positive results obtained in a large animal study as well as the favorable comparison with the clinically established 111In-oxine make this a promising labeling method for tracking cell therapies. The half-life similar to 111In-oxine would allow tracking over days to weeks, with increased sensitivity of PET reducing the amount of radiolabel needed to label cells, suiting this better to use with therapeutic cells that might need to retain their viability and phenotype.
Magnetic resonance imaging: iron oxide and fluorine technologies
Magnetic resonance imaging (MRI) is a common non-radioactive diagnostic tool that provides sensitive non-invasive means of imaging detailed anatomy throughout the body. MRI uses a strong magnetic field to align the spins of magnetic resonance (MR)-visible non-radioactive nuclei so that they create a net magnetization. The perturbation of this alignment and the resulting relaxation and realignment produce a detectable radio frequency that can be used for imaging. Hydrogen-1 (1H) nuclei are most frequently used for MRI because of the abundance of protons within the water and fat of biological tissues. However, other MR-visible nuclei, including carbon-13 (13C), fluorine-19 (19F) and phosphorus-31 (31P), can be detected using MRI. Agents that alter the magnetic field surrounding native protons or are elements that are directly detectable by MRI have been applied to labeling and imaging cellular therapeutics [20]. Application of these techniques has been employed both clinically and pre-clinically, optimizing their utility at varying magnetic strengths and scan sequences (Box 1). Two commonly applied agents for imaging cellular therapies, iron oxides and fluorine, are discussed in the following sections.
Iron oxide has a long history of being used to label cells for detection by standard MRI (T2- and T2*-weighted 1H MRI) [42-44], creating a darkening effect or negative contrast when the iron inside the cells creates a localized disruption of the surrounding magnetic field. Iron imaging is a sensitive technique for MRI, and even nanomolar concentrations of iron are able to alter the magnetic field, providing detection of the labeled cells [45]. Iron has been utilized as dextran-coated superparamagnetic iron oxide (SPIO) nanoparticles [44,46,47] that may be applied with transfection agents or other means to boost uptake depending on a cell’s phagocytic nature [45]. SPIO has been utilized to image numerous cell types pre-clinically [48-51], with several published clinical applications [18,50,52-58]. Recently, ferumoxytol, an ultra-small SPIO (USPIO) nanoparticle, the only Food and Drug Administration-approved commercially available iron oxide nanoparticle in the United States for treating iron-deficiency anemia, has been used in clinical settings as an in vivo stem cell labeling agent in arthritis, a macrophage-imaging agent in tumors and arterial lymph node stability and a blood pool agent by MRI [59-61]. Ferumoxytol has also been further developed and utilized to label and track many stem and immune cell types, albeit off-label (Table 2) [62-69]. Using ferumoxytol-based labeling and MRI [62], homing of therapeutic natural killer cells at the tumor sites in vivo has been shown to be an important early biomarker for prediction of longitudinal tumor response, with secondary endpoint measures such as tumor volume reduction over time [65]. In clinical settings, ferumoxytol has been applied in the direct labeling of cells in a laboratory setting before injection (ClinicalTrials.gov identifier NCT01172964) [62,66] and/or in in vivo direct labeling in patients (ClinicalTrials.gov identifier NCT02893293) [63,70]. SPIO imaging sensitivity, balanced with utilizing proton MRI for both cellular location and anatomical context, has its advantages and disadvantages. Depending on the application, distinguishing between natural background, pathologies that also cause darkening of the signal (e.g., blood pooling) and labeled cells may be difficult with regard to both detection and quantification [71]. The use of 1H MRI allows for quick integration into standard clinical practice of MRI (both pre- and post-administration scans), with the potential to perform whole-body scans [45]. Iron imaging products continue to be developed and produced for standard MRI and, additionally, for visualization by magnetic particle imaging, where signals arise directly from 1H nuclei, to monitor the fate of cellular therapies [72-74].
Table 2.
Cell types tested with iron oxide and fluorine probes and respective applications.
| Cell type | Application | |
|---|---|---|
| Fluorine-labeled | Iron oxide-labeled | |
| Dendritic cells [78,79,82] | Neuronal stem cells [62,66] | On-/off-target delivery |
| T cells [83-85] | Biodistribution | |
| Hematopoietic stem cells [80] | MSCs [63,69,94] | Homing |
| Natural killer cells [64,65] | Migration | |
| Neuronal stem cells [86,87] | Immune response | |
| MSCs [89,90] | Cardiac stem cells [68] | Cell survival |
| Natural killer cells [91] | ||
| White blood cells [92] | ||
| Mixed cell populations [88] |
Non-radioactive fluorine (19F) is an alternative source of MR contrast that is currently being applied in the clinic. Perfluorocarbons that have a long history in human health as blood substitutes [75,76] are now being applied to track cellular therapeutics. Unlike iron oxide-based contrast, where the signal is derived from the relaxation effect on local water protons, the 19F signal arises from the 19F nuclei themselves. Compared with its earlier predecessors, which give a weak signal because of the inherently low sensitivity of 19F MR, the clinical compound for cellular imaging is composed of many chemically identical fluorine atoms, yielding a single major spectral resonance detectable by fluorine MRI [77]. Cells can internalize many emulsion droplets, allowing as few as thousands of cells to be detected by 19F MRI [78]. The label is taken up by simple co-incubation with the cells of interest for 4–48 h under normal culture conditions and without transfection agents [79,80]. After washing away residual label and administering the cells, they are then imaged by combining a 19F MRI scan with a 1H scan, giving a fluorine-specific image in its anatomical context that can be used to quantify the number of cells present [78,79]. The short T1/T2 (longitudinal/transverse relaxation time) of the reagent has demonstrated clinical sensitivity at 3T magnetic strength using a 9-min fluorine acquisition [81]. This compound has also been tested extensively on a number of cell types (Table 2), with a number of comprehensive data sets evaluated and published in peer-reviewed journals [78-80,82-92].
Ultrasound
Ultrasound is one of the most widely used clinical and pre-clinical imaging modalities owing to its relatively low cost, portability and rapid image acquisition. The ultrasound waves are either scattered or reflected by boundaries between tissues within the body generating image contrast. This facilitates delineation of structures such as blood, vessel walls and muscle. However, more dramatic structural differences, such as those between soft tissue and bone or air cavities, can distort and mask regions, preventing whole-body imaging.
Microbubbles (MBs) are the most frequently used contrast agents in ultrasound imaging. Gas-filled, phospholipid-coated MBs are highly echogenic owing to the mismatch in acoustic impedances between the gas and the surrounding tissue and their non-linear oscillations under pulsed ultrasound [95]. This allows tissue function and perfusion to be imaged across organs, with applications in cardiovascular, liver and renal disease as well as diagnostic cancer imaging [96] and drug delivery.
MBs have been used to directly label neural progenitor cells in vitro, allowing their detection in vivo after intravenous infusion in mice (Table 3) [97]. However, the relatively large size necessary to induce sufficient contrast and short half-life of standard MBs mean they are not ideal for cell labeling and tracking. This has led to innovative methods for initiating ultrasound contrast within grafted cells, including intracellular aggregation of silica nanoparticles within human MSCs [98]; targeting of systemically administered MBs to cell surface markers specific to grafted rat endothelial progenitor cells [99]; and genetically encoded, gas-filled, protein-shelled compartments within Halobacterium [100] and mammalian cells [101].
Table 3.
Summary of ultrasound applications of cellular imaging.
These approaches still suffer from the disadvantages of poor contrast, high background signal and lack of quantitative data. However, by combining ultrasound contrast agents with 1H or 19F MRI, nuclear, CT or photoacoustic cell imaging approaches, quantitative data on whole-body distribution and long-term cell retention can be acquired [102-105]. This multimodal approach becomes more impactful when combined with non-invasive, ultrasound-guided delivery of cells directly to the specific target organ [106,107]. Using this method, both needle position and cell injection can be assessed in real time, informing upon the success of cell delivery. The short-term nature and low specificity of ultrasound labeling methods do not detract from this approach, as live images can be acquired during injection, allowing cell-mediated changes in contrast to be immediately visualized and injection sites delineated.
Expanding upon ultrasound technologies, photoacoustic imaging involves excitation with light and detection with ultrasound, typically of certain fluorophores. The technique has recently come to clinical use, although not for cell tracking purposes [108]. Currently, the only clinically approved agent for photoacoustics, sometimes called multispectral optoacoustic tomography, is the dye Indocyanine Green (IC-Green). IC-Green has been used for cell labeling for imaging with photoacoustics [109,110]. In general, photoacoustics is an emerging technology and thus remains an open field, with a lack of clinically approved agents optimized for cell tracking and little known about sensitivity for cell tracking. The technique, however, offers several advantages, such as a lack of invasiveness and background. It is a bedside technique, with a scanner similar to ultrasound scanners. Thus, it holds a lot of potential for future application with cell imaging.
indirect imaging: reporter genes
Indirect labeling using a reporter gene imaging paradigm allows for reliable, stable visualization of cellular trafficking, persistence, proliferation and function at the target site [22]. Genetically labeled cells can be repetitively imaged for a long period of observation (months). The genetic nature of this method (i.e., stable integration into the cell genome) permits in vivo labeling of the progeny of injected cells. Using inducible genetic reporter systems, functional imaging of cell activation, cytokine signaling and pro-apoptotic events is feasible and can be performed in pre-clinical experiments and clinical settings [111] with picomolar concentrations of radiotracer administered in vivo.
Both human and non-human genes have been used as reporter genes (Table 4, Figure 3). The major impediment to the translation of virus- and bacteria-derived reporter gene imaging approaches into clinical practice is the immunogenicity of these non-human-derived reporter proteins. This is especially important when repetitive administration of a reporter gene or long-term monitoring of transgene expression is required, which could potentially be compromised by an immune reaction against this foreign protein. To circumvent this problem, several human-derived reporter genes have been proposed for nuclear imaging, including a genetically engineered prostate-specific membrane antigen (NdelPSMA) (by PET [112]), the human sodium/iodide symporter (NIS) (by SPECT [113] and PET [114]), engineered human thymidine (by PET [115]) and deoxycytidine kinases (by PET [116]), to name just a few; a comprehensive list has recently been compiled by Ashmore-Harris et al. [22]. It is expected that new human-derived reporter genes will be translated into clinical applications that require repetitive imaging for effective monitoring of various genetic and cellular therapies.
Table 4.
Reporter gene/reporter probe imaging studies.
| Human reporter gene/imaging modality |
Probe (half-life) | Optimal approximate probe biodistribution time |
Primary probe excretion route |
Probe overlap with normal tissue(s) |
|---|---|---|---|---|
| hdCK mutants/PET | 18F-FEAU (109 min) | 1 h | Renal | Intestine, gallbladder, kidneys, bladder |
| 124I-FIAU (4.18 days) | 1 h | Renal | Intestine, gallbladder, thyroid, stomach, kidneys, bladder | |
| hTK2 mutants/PET | 18F-L-FMAU (109 min) | 1 h | Renal | Intestine, gallbladder, kidneys, bladder |
| hNIS/PET and SPECT | 124I-iodide (4.18 days) | 2 h | Renal | Salivary gland, thyroid, stomach, kidneys, bladder |
| [18F]BF4− (109 min) | 1 h | |||
| 123I (13 h), 99mTcO4- (6 h) | 1 h | |||
| hNET/PET and SPECT | 18F-MFBG (109 min) | 4 h | Renal and hepatobiliary | Salivary gland, intestine, kidneys, bladder |
| 123I-MIBg (13 h) | 12 h | |||
| 124I-MIBG (4.18 days) | 24 h | Renal and hepatobiliary | Salivary gland, intestine, thyroid, kidneys, bladder | |
| hERL binding domain/PET | 18F-FES (109 min) | 1 h | Renal and hepatobiliary | Intestine, liver, kidneys, bladder |
| hSSTr2/PET | 68Ga-DOTATOC (68 min) | 1 h | Renal | Kidneys, bladder |
| D2R/PET | 18F-FESP (109 min) | 3 h | Renal and hepatobiliary | Intestine, liver, kidneys, bladder |
| Membrane-anchored anti-PEG antibody/PET | 124I-PEG-SHPP (4.18 days) | 24 h | Renal and hepatobiliary | Liver, thyroid |
| anti-DAbR1 fragment/PET | 86Y-AABD (15 h) | 24 h | Renal and hepatobiliary | Liver, intestine |
| CEA/PET | 124I anti-CEA antibody fragment (4.18 days) | 24 h | Renal | Thyroid, kidneys |
| PSMA/PET | 18F-DCFPyL (109 min), 68Ga-PSMA-11 (68 min) | 15–30 min | Renal | Intestine, liver, kidneys, bladder |
| Ferritin/MRI | Endogenous Fe | Iron accumulation varies | Dense, hypointense tissues | |
| Water channel AQP1/MRI | Endogenous water | |||
| OATP | Gd-EOB-DTPA/111In-EOB-DTPA (2.8 days)/ICG | 30 min to 2 h | Renal and hepatobiliary | Liver, kidneys, gallbladder, bladder, intestine |
anti-PEG, anti-polyethylene glycol; AQP1,aquaporin 1; CEA, carcinoembryonic antigen; DAbR1, DOTA antibody reporter 1; D2R, dopamine 2 receptor; Fe, iron; 68Ga, gallium-68; hdCK, human deoxycytidine kinase; hERL, human estrogen receptor alpha ligand; hNET; human norepinephrine transporter; hNIS, human NIS; hSSTr2, human somatostatin receptor subtype 2; hTK2, human thymidine kinase 2; OATP, organic anion transporting polypeptide; PSMA, prostate-specific membrane antigen.
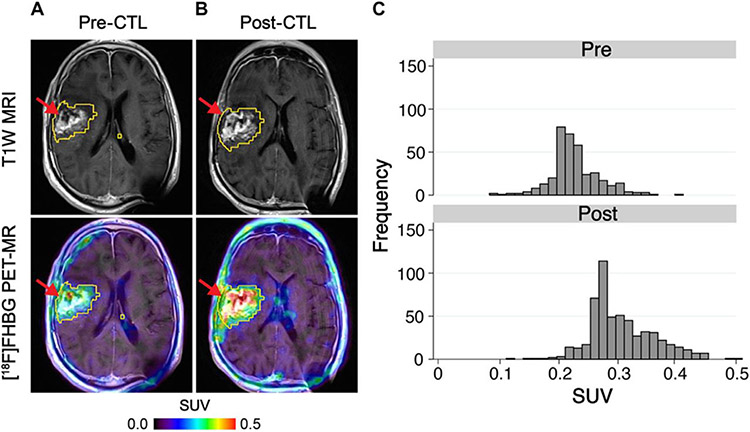
Figure 3.
HSV1-tk was fused in-frame with the hygromycin phosphotransferase gene and expressed as a fusion protein. Through imaging of HSV1-tk reporter gene expression using [18F]FHBG, which is an 18F-radiolabeled analog of the anti-herpes drug penciclovir, [18F]FHBG trapping in the brain tumor could be imaged, which corresponds to CTL accumulation. The [18F]FHBG PET imaging was performed in a patient with a recurrent right frontoparietal glioblastoma (A) before and (B) 1 week after tumor-specific HSV1tk-transduced CAR T-cell infusions. Allogeneic CAR T cells and IL-2 were injected intratumorally (red arrows). Tumor recurrence was monitored by T1W MRI (top panels). The [18F]FHBG PET images were fused with MR images (bottom panels), and 3D volumes of interest were drawn using a 50% [18F]FHBG SUVmax threshold (outlined in yellow). (C) Voxel-wise analysis of[18F]FHBG SUV in pre- and post-CTL infusion scans. Reproduced with permission from [111] © American Association for the Advancement of Science (2017). CTL, cytotoxic T lymphocyte; [18F]FHBG, 9-[4-[18F]fluoro-3-(hydroxymethyl)butyl] guanine; HSV1-tk, herpes simplex virus 1-thymidine kinase; SUV, standard uptake value; 3D, three-dimensional; T1W, T1-weighted.
Multimodal imaging: addressing limitations
Cell imaging is complex, and the imaging needs may vary within each application. For example, a small number of cells can be injected in a localized manner in deep tissue or a large number of cells can be injected systemically. No imaging modality is perfect (Table 5). Thus, in practice, multimodal imaging is used to overcome these issues. Almost all cell imaging experiments at the pre-clinical level include a fluorescent agent with other imaging agents to allow for fast and easy confirmation of the presence of label in the relevant cells using histology; thus far, the literature reports bi- and tri-modal agents [123,124]. Clinical cell tracking has used only single modality agents, the vast majority with PET or SPECT tracers [125,126], and proof-of-principle studies with 1H MRI [52] or 19F MRI [81].
Table 5.
Visual assessment of the performance of the main imaging types in different categories, such as applicability to humans and sensitivity.
| Human use | Sensitivity | Longitudinal | Quantitative | Radioactivity | Probes in parallel | Availability | Cost | Imaging time | Labels (cells) | Resolution | Depth | Patient comfort | Operator skill | Whole body | Background | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SPECT | ||||||||||||||||
| PET | ||||||||||||||||
| Ultrasound | ||||||||||||||||
| Optoacoustic | ||||||||||||||||
| 1H MRI | ||||||||||||||||
| 19F MRI | ||||||||||||||||
| Optical |
Adapted from [123,127]; note that assessment may change over time, as technologies are continually evolving. Color coding: green indicates a “good” or “high” rating, yellow “intermediate” and red “poor” or “low.” This is an approximate indication, as actual performance will depend on a combination of multiple factors, such as the application, setting and imaging parameters.
One approach that could be readily translated to the clinic involves the addition of IC-Green, a clinically approved fluorescent dye, to iron oxide nanoparticles, which would allow both fluorescence and MRI [128]. A major issue that needs to be addressed is a practical one: image co-registration when different scanners are used. Solutions include both software developments and special imaging hardware, such as subject holders [129,130].
Co-development of a multimodal imaging probe or simply employing more than one of the described modalities would remedy the shortcomings of any one imaging agent. Direct labeling approaches, for example, dilute out with cellular expansion, limiting the quantification and detection of cells with time. Combining one of these approaches with a reporter gene or other non-dilutive technology would allow for longer-term monitoring of cellular distribution. Another example, 1H MRI, although having several advantages, does not allow for the initial imaging of cells that are in the lungs after systemic administration. The challenge of detecting cells in this context is that the air-filled lungs appear dark by MRI [131-133]. When imaging the lungs in this setting, other modalities and probe detection can be applied to visualize cells [78,132,134-136]. Throughout the following sections, common research interests are examined for different probes, expanding further on the limitations and advantages of various probes. When looking at Table 5, it is important to keep in mind that none of the categories listed rank poorly across all applications. By combining more than one approach or even a probe that is multimodal, researchers could satisfy their imaging needs if they could not be achieved by one probe alone.
Common Interests in the Application of Imaging Technologies
Based on the surveys conducted at the 2017 International Society for Cell & Gene Therapy Annual Meeting in London, UK, the following topics discussed are those most desired by or of concern to the survey respondents. To address these subjects, the previously outlined methods will be highlighted.
Cellular monitoring: observing persistence and homing over time
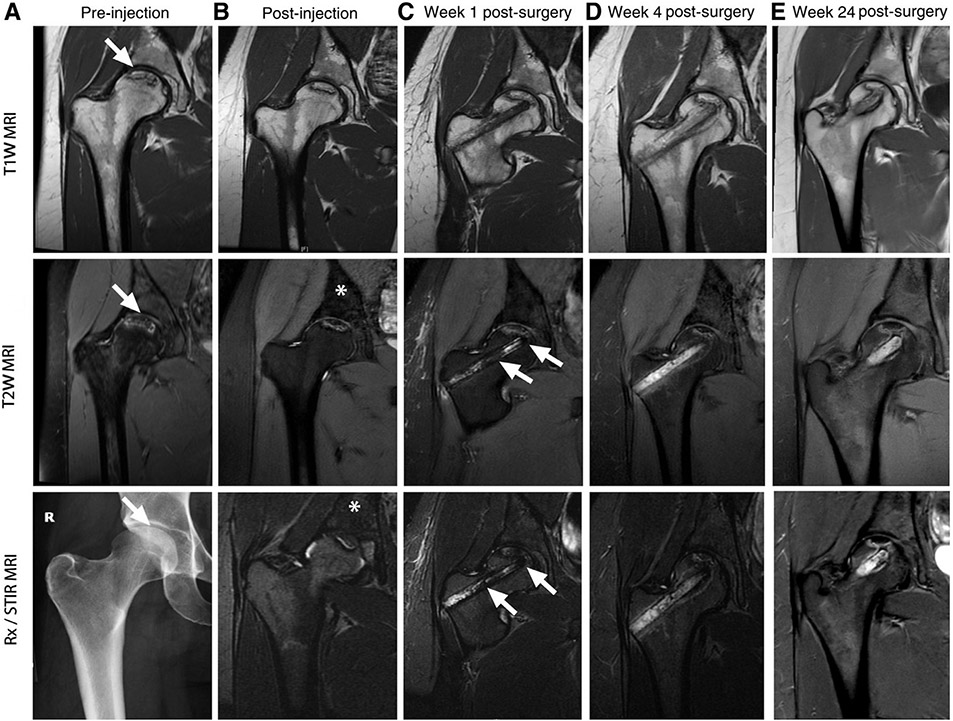
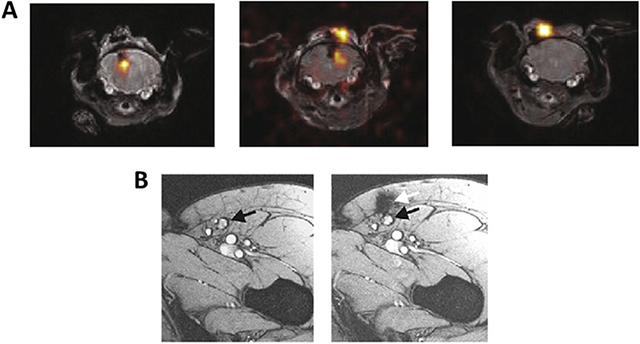
The invasive nature of classical pathology precludes the repetitive monitoring of cellular trafficking in the same living organism over time. The persistence and homing of therapeutic cells in the recipient could potentially be investigated by repetitive imaging using clinically applicable techniques, including direct ex vivo cell labeling and in vivo indirect reporter gene imaging paradigms. All categories of non-invasive cellular tracers (PET, MR, etc.) have the ability to trace cells over time to some extent. Interest in following the homing and persistence of cells by survey respondents varied between immediately after administration to days, weeks and months later. The span of applicable time adds complexity when weighing the different technologies. Some probes (FDG) have shorter half-lives, limiting the span of time in which they are effective, whereas 89Zr has the ability to be detected for a week or longer [37]. MR probes, which do not decay over time, can be detected so long as the cell population remains viable. Fluorine-labeled T cells have been observed from day of infusion to 21 days post-administration without loss of detection [85]. Iron-based agents have been detected from 7 days (Figure 4) [63] in studies of bone decompression to 6 months [50] in islet transplants depending on the activity of the cells. In further considering direct and indirect labeling approaches, cells that are directly labeled will exhibit dilution of the label between daughter cells with time. With these probes, the question of desired sensitivity and quantification also plays a role. Upon division, the cells and their probe are still detectable since the quantity of signal (especially with MR) is not diminished if the cells remain in the same region of interest; only the ability to quantify the signal would be altered upon multiple replications. With indirect labeling approaches, where cells are actively producing the means of their detection, so long as the administered probe that binds is able to find its target, cells will be able to be detected.
Figure 4.
Serial imaging of ferumoxytol-labeled bone marrow cells. (A) T1W, T2W and x-ray images of the right femur show osteonecrosis (arrows) in the femoral epiphysis with typical fat-equivalent center and serpiginous borders. (B) At 24 h after intravenous injection of ferumoxytol, the normal bone marrow in the iliac crest shows hypointense (dark) enhancement (asterisks) on T2-weighted MR images. (C) One week after core decompression and injection of iron-labeled bone marrow cells, a hypointense (dark) signal is noted in the decompression track (arrows), consistent with delivery of iron-labeled cells. (D,E) MRI follow-up at 4 weeks and 24 weeks after core decompression and transplantation of labeled cells shows decline in iron signal over time. The femoral epiphysis did not collapse during this 6-month follow-up period. Reproduced with permission from [63] © American Association for Cancer Research (2018). STIR, short tau inversion recovery; T1W, T1-weighted; T2W, T2-weighted.
Sensitivity
The sensitivity of an imaging technique is dependent on the inherent detection threshold of the label and modality and the amount of specific label taken up per cell. Sensitivity ranges in theory between an individual cell to thousands or millions of cells. Sensitivity can also refer to limits of detection as far as depth of cellular population, environment in which the cells are located or image resolution.
PET and PET probes, which offer high sensitivity, have higher sensitivity than SPECT imaging by one to two orders of magnitude. PET does not use lead collimators to achieve spatial discrimination, thereby greatly increasing the proportion of emitted photons collected by the detectors. Instead of collimators, PET achieves spatial information on probable isotope location based on the co-incident detection of the two simultaneously released photons by opposite detector panels [137]. As positron annihilation and the release of detectable photons occur at an average distance of 1–2 mm from the source of the isotope, depending on the positron-emitting isotope used, not only does effective image resolution vary between isotopes, but the resolution is also somewhat lower with SPECT, where photons are released directly from the radioisotope [137]. This is usually more than compensated for by the higher sensitivity of PET compared with SPECT.
The sensitivity of contrast-enhanced cellular 1H MRI depends on (i) the physiochemical properties of the selected contrast agent, such as chemical structure, surface chemistry and r1 and r2 relaxivities (e. g., gadolinium versus iron oxide, dextran-coated versus chitin-coated); (ii) the type and nature of the target cells, such as cell types, their uptake mechanisms and doubling time; (iii) target tissue/organ environment; and (iv) MRI signal acquisition, such as magnetic field strength, imaging sequences and parameters. A major consideration in using SPIO/USPIO labeling for MRI cellular tracking has been dilution of the contrast agent due to cellular division and the consequent decrease in the ability to detect the signal by MRI for an extended period—for example, months—depending on the nature of the cells. Additionally, since SPIO/USPIO is biodegradable and enters iron metabolic pathways, signal dilution can occur as a result [138,139]. In several studies, labeled stem cells were tracked post-administration and permitted the non-invasive visualization of cell engraftment and/or cell migration in specific locations: CD34-positive hematopoietic stem cell incorporation into tumor neovasculature [187], neural progenitor cell migration to sites of spinal cord injury [140] and brain tumors [66], natural killer cells to liver tumors [64,65], cardiac stem cells in heart repair [68] and MSCs to the orthopedic environment [67,141]. These studies showed that such labeled stem cells were visible by MRI at 1 week to 3 months after administration [64,66-68,94,140-143]. The studies also showed that as little as 4 ng/voxel can be detected by MRI. Transplanted SPIO-labeled pancreatic islets in patients have been tracked by MRI for up to 6 months post-transplantation [50].
Similar to iron, the sensitivity for detecting 19F-labeled cells depends on a number of features. The chemical structure of the 19F probe and the number of detectable, spectrally similar 19F atoms will influence sensitivity, in that the more chemically similar the fluorine atoms, the better the resolution [144,145]. Perfluoropolyether, with a single major spectral peak (i.e., chemically identical fluorine atoms), has been used clinically to image transplanted cells and will be the structure further considered here. Detection relies on the number of fluorine atoms within the cell. Fluorine uptake directly correlates with cellular size—that is, larger cells, such as dendritic cells, take up more label than smaller induced pluripotent stem cells [146]. Other factors that influence sensitivity are scan time and magnetic field strength. Clinical use of fluorine-labeled dendritic cells demonstrated that less than 3 × 104 cells could be detected per voxel using a clinical 3T scanner and 9-min image acquisition time when cells were labeled with 1 × 1012 to 1 × 1013 fluorine atoms per cell [81]. A voxel is a user-defined imaging unit that can vary from a millimeter cube to an entire organ. Smaller cells labelled with 1 × 1011 fluorine atoms per cell would require approximately 1.5 × 105 cells/voxel for detection [82]. Pre-clinical studies, where scan times and magnetic field strengths can be higher, demonstrate improved limits of detection but may be limited by milligram/kilogram dosing strategies in which the overall administered dosage will be lower in small animal studies. These studies, however, allow for the assessment of entire organs by nuclear magnetic resonance spectroscopy to quantify overall biodistribution of cells [147]. MRI probes, in general, do not suffer from limits of detection based on tissue depth, but they may require coil mapping or optimization before use depending on the type of coil/system being used. Fluorine probes, which are detected via 19F MRI, generate “hot spot” detection without the limitation posed by the natural proton signal intensity of tissues. However, like iron oxide labeling, dilution of fluorine contrast agents due to cellular division decreases the MR signal and limits the duration over which cells can be tracked.
Quantification
Quantification refers to the ability to enumerate the presence of an administered cellular product both quantitatively and qualitatively, both of which will be distinguished in this discussion. Direct labeling approaches (when cells are labeled prior to administration) provide more quantitative measurements than indirect labeling approaches, which tend to be more qualitative. Cellular populations prone to rapid expansion, especially when directly labeled in advance, will lose the ability to quantify exact cell numbers as the label is divided among daughter cells, but they retain qualitative estimations.
For PET and SPECT applications, the proportion of a radiolabeled cell population present in a tissue can be estimated using decay-corrected measurements of PET/SPECT signal in specified regions of interest for quantification. This typically gives a percentage of the administered transplanted cells that are now present in a given tissue, with the caveat that at least four sources of opposing error prevent exact quantification. First, label can leach from cells over the course of time, potentially giving an underestimation of the proportion of cells present. Second, the radiolabel may be retained in cells that have died, leading to a possible overestimation of viable cell numbers in tissue. Third, label leached out of cells can accumulate in certain tissues, including the bones or organs of excretion, leading to an overestimation of the proportion of cells in these areas. Finally, when cells proliferate, their label will be diluted out among their progeny, leading toward an underestimation of cell number in cases where cells are dividing rapidly (although this is less of a concern in non-dividing or slowly dividing cell types).
Together, these factors mean that it is not possible to quantify cell number precisely following direct labeling and nuclear imaging; however, some of these factors can be taken into account when analyzing data to improve estimates of cell number. These problems are not unique to SPECT or PET imaging of directly labeled cells and usually operate to some degree with direct labeling strategies for MRI and other modalities.
Stable genetic labeling of adoptively transferred cells with reporter genes can be used to circumvent the temporal limitations of ex vivoradiolabeling, such as a relatively low attainable level of radioactivity/concentration per cell, as well as the limited period of monitoring due to radiolabel decay, cell division and biological clearance. By contrast, stable reporter gene expression by adoptively transferred therapeutic cells allows for high temporal resolution/longitudinal monitoring and more accurate cell quantification over time. Several groups established the correlation of PET signal with cell number, and they characterized the cellular limit of detection for PET imaging using human and mouse cells transduced with different reporters, resulting in a limit of detection below 105 cells in 0.1 mL volume of region of interest [122]. This level of sensitivity enables effective assessment of cell localization at target sites and assessment of homing in vivo. Defining the levels of sensitivity of reporter gene imaging will be useful in guiding the development of novel cellular therapeutics utilizing this technology in the future.
Quantitative 1H MRI of iron oxide-labeled cells is currently performed by measuring the iron content in each cell (pg/cell) via relaxometry and inductively coupled plasma mass spectrometry, obtaining the R1, R2, R2* values (per second) at the target locations, and calculating the total number of cells using the relaxivities of the iron oxide agent (r1, r2, r2* [mM−1 s-1] at specified magnetic field strength and temperature). Additional quantitative MRI scan sequences and protocols, such as ultra-short echo time quantitative susceptibility mapping for detection of high iron concentrations in vivo, quickly and accurately minimizing interference from iron susceptibility artifact, are under development, which may further allow for in vivo quantitative cellular MRI [148-150].
Fluorine imaging by MRI, when paired with conventional proton MRI, gives highly selective detection of fluorine-labeled cells in their anatomical context [78]. Since fluorine is not native to the human body, it is also quantitative. Cells labeled ex vivo are measured for fluorine uptake by nuclear magnetic resonance. Upon administration to the patient, a fluorine reference can be placed either adjacent to the patient or within the detection coil for accurate cellular quantification [78,81]. A simple calculation of the signal from the reference compared with the signal from the cellular population, knowing the cellular labeling efficiency prior to administration, yields the number of cells in a region of interest. For cellular populations that are not rapidly dividing, accurate quantification can be maintained. Cellular populations that are rapidly dividing will still be detectable if they remain in a similar region; however, quantification loses its specificity with time. An interesting benefit of fluorine imaging is that the signal detected in the liver corresponds to the non-viable (dead) cell population [147]. Since the fluorine compound is cleared through the reticuloendothelial system and any released reagent has short blood circulation time, the signal from the dead cell population is easily detected there. This is a common phenomenon across multiple imaging modalities [151]. As a result, intentional cellular migration to the liver will not be quantitative against the non-viable population when using modalities that clear through the reticuloendothelial system.
Imaging cell survival
In vivo quantification of cell survival requires cell labeling according to one of the two fundamental principles described earlier: the cell label must (i) remain sufficiently long in the labeled cell to assess survival and (ii) reflect cellular viability. Assessing viability can theoretically be done based on label presence in a cell. However, there are several pitfalls that depend on the chosen cell labeling strategy. Direct labeling approaches by both MRI agents (iron oxide and fluorine) and PET agents (radiolabels) are fundamentally affected by label dilution and have the potential for false-positive signals—for example, when labels from non-surviving cells accumulate in macrophages at the target disease site or non-target organs, including through clearance of the dead cell fraction or of the label itself during longitudinal imaging [147,152-154]. However, recent advances in iron oxide particles and imaging protocols have enabled differential detection of live labeled cells from dead cells and/or scavenging macrophages [94,155,156]. Similarly, radiolabel may leak from both healthy and dead/dying cells and be transported to other tissues (e.g., bone in the case of 89Zr, kidney in the case of 111In, thyroid in the case of radioiodine [157,158]).
When reporter genes are employed, signal presence is used to assess cell survival in indirect cell labeling approaches, but although signal loss issues are circumvented through the generation of stable reporter-expressing cells, the approach is also not free of some pitfalls. Reporters suitable for whole-body imaging can be classified according to their contrast forming mechanism: (i) substrate conversion to a detectable tracer, (ii) tracer transport, (iii) tracer binding and (iv) traceable proteins not requiring administered tracers [22]. Class III and IV reporters can potentially be detected in live cells but also in dying cells and as part of dead cells and cell debris. The extent of this artifact is a function of intracellular and extracellular reporter degradation, both processes that remain understudied (except for fluorescent proteins, which do not have much clinical utility). Horizontal reporter gene transfer from the therapeutic cell to (other) patient cells can also not completely be ruled out, but this has so far never been reported in the context of in vivo cell tracking.
Indirect cell labeling offers many opportunities. Reporters based on substrate conversion (class I), such as luciferases, are frequently used pre-clinically for assessing cell survival in vivo. Substrate conversion by luciferases consumes cellular adenosine triphosphate (ATP), and thus contrast is formed only in live cells [159,160]. Unfortunately, luciferases are only applicable to pre-clinical studies. Analogous convertible substrate/reporter pairs for radionuclide imaging do not exist because the in vivo generation of radioactivity is impossible. However, it is possible to design radioactive tracers that become modified through reporter action, thereby, for example, affecting cellular radiotracer washout kinetics. The prime example in this context is the reporter herpes simplex virus 1-thymidine kinase, which entraps radioactive thymidine analogues through phosphorylation [161,162]. The radiotracer transport into the cells relies on endogenous transport mechanisms, whereas the herpes simplex virus 1-thymidine kinase-mediated radiotracer entrapment is ATP-dependent and thereby linked to cell viability. Reporters that are active transporters (class II) can also be linked to cell viability [163]. Transporters require intact cellular membranes in live cells for correct function. Transporters that have been exploited for cell tracking include the human NIS [164], the human norepinephrine transporter [165] and the dopamine transporter [166]. NIS, which has been most widely used, is a symporter that is highly selective for sodium ions but promiscuous for anions similar in size and structure to iodide, which is exploited for medical imaging [164]. Radiotracers that are not metabolically trapped in the thyroid are desirable for imaging and now available for both SPECT [164] and PET [167, 168] imaging. The symport is driven by the sodium gradient across plasma membranes of live cells, and the sodium gradient is maintained by sodium/potassium ion-ATPase [169]. NIS allowed imaging of live, but not dead/necrotic, cancer cells in tumors [170-172] as well as, notably, assessment of induced pluripotent stem cell-derived, hepatocyte-like cell survival after engraftment [173] and CAR T-cell tumor retention in triple-negative breast cancer models [114]. NIS has also been widely used for cell tracking in other fields. Other particularly promising host reporter genes for PET imaging in this context are a prostate-specific membrane antigen derivative, which was recently used to track CAR T cells in mouse models [112], and pyruvate kinase M2, which has been used to track cells in the brain because of the ability of its radiotracer, 1-((2-fluoro-6-(fluoro-[18F])phenyl)sulfonyl)-4-((4-methoxyphenyl)sulfonyl)piperazine, to cross the blood–brain barrier [174].
Cellular differentiation
Imaging cellular differentiation in vivo is desirable in the field of regenerative medicine. Although it is a niche use, it is difficult to achieve clinically. Classically, this has been achieved through examination of histological specimens and performed without in vivo methods. Tissue-specific differentiation of transplanted progenitors has been challenging: after delivery, adult progenitor cells often fail to transdifferentiate into target tissues. Developing technologies being applied pre-clinically to visualize differentiation include intravital microscopy and tissue- or differentiating stage-specific promoters and reporter genes. For example, embryonic stem cell differentiation into endothelial-like cells was followed using the endothelial cell lineage-specific promoter (i.e., vascular endothelial cadherin promoter), driving expression of a reporter gene that allowed for longitudinal monitoring of the fate and function of transplanted embryonic stem/endothelial cells in the heart [175]. Clinical application of lineage- and stage-specific reporter systems will allow for longitudinal non-invasive assessment of the fate and function of transplanted cells in patients undergoing treatment with stem cells and committed progenitors, but these technologies have yet to be translated into clinical reality.
Safety of probes
With regard to safety, there are two avenues to consider: the safety of the probe to cells and the safety of the probe itself to patients. The assessment of the safety of the probe to cells, which includes testing cell viability and functionality (and any characteristic deemed necessary for the therapeutic), should be addressed through in vitro studies and, if required, by in vivo animal studies. The dose and duration of labeling as well as the amount of label taken up by each cell type can vary for each application [46,79]. Varying cell types may have different sensitivities to labeling agents and different transport mechanisms for intracellular trafficking and efflux of probes and, as such, may have varying sensitivities to the labels. Considering this, the safety of the imaging agent should be evaluated for each new combination of cell type and labeling probe. These aspects are not yet covered by regulatory guidance documents for cell therapies, although these particulars can influence the establishment and interpretation of the biodistribution study.
Whether the administered cells are detected and identified by a reporter gene or by a radioactive, magnetic or other probe added by direct labeling, a probe may affect the behavior, differentiation, functionality or proliferation of the cells, and this needs to be taken into consideration. For instance, reporter genes usually entail genetic modification to incorporate a reporter gene, the expression of which may also alter the behavior of the cells. Otherwise, direct labeling of probes or the manipulations used to incorporate the probe may cause changes in function or phenotype, such as activation/deactivation of immune cells, activation of free radical defense mechanisms or impairment of chemotaxis or motility, or may impact survival and should be considered or investigated before interpretation of biodistribution study data. Awareness is present and investigation occurs, as is evidenced by articles describing the influence of a probe and labeling method on the functionality of cells. For instance, it was shown that 99mTc-HMPAO labeling of CD4-positive T cells did not affect viability and proliferation [176], whereas 99mTc-HMPAO labeling of neutrophils and eosinophils increased their activation [177,178]. Furthermore, 64Cu-pyruvaldehyde-bis(N4-methylthiosemicarbazone) labeling of CD4-positive T cells minimally affected cell viability, interferon gamma production, proliferation, DNA damage and intracellular 64Cu distribution, but the drawback was that only 14% of the radioactivity remained for 24 h [179]. In another study, labeling γδ T cells with 89Zr-oxine resulted in dose-dependent effects (in millibecquerels per cell) on proliferation, cancer cell killing, DNA damage and cell death [40]. Finally, labeling of dendritic cells with SPIO nanoparticles for tracking with MRI appeared not to affect their viability, phenotype or functionality, as assessed by T-cell stimulatory capacity, mixed lymphocyte reactions and proliferation assays [180].
Although reporter gene-expressing cells are less likely to be affected by the radiotracers used to probe them, as radioactivity per cell is typically much less and radionuclides with shorter half-lives are used, some testing has been performed. Sharif-Paghaleh et al. [181] showed that regulatory T-cell lines derived from CD4+CD25+FoxP3+ cells retrovirally transduced with a reporter gene construct encoding the human NIS and the fluorescent protein mCherry and radiolabeled in vitro with 99mTc-pertechnetate (NIS substrate) did not affect cell viability, phenotype or function. Notably, Volpe et al.[114] very recently demonstrated that neither 99mTc-pertechnetate nor 18F-tetrafluoroborate negatively impacted CAR T-cell viability or function over a period of several days, with no significant DNA damage being detected at cellular radiotracer concentrations comparable to those observed during in vivo cell tracking.
Charoenphun et al. [37] studied the status of cancer cells labeled with a 89Zr-oxine radiolabel, making use of a reporter gene. A myeloma cell line expressing enhanced green fluorescent protein (GFP) also labeled with a 89Zr-oxine radiolabel to track them in mice using PET imaging was used to measure in vivo survival and retention of the radiolabel over a 7-day period [37]. After imaging over the 7-day period, cells from the distribution target organs and GFP-postive and GFP-negative cells were sorted, and radioactivity was measured with a gamma counter. GFP-positive cells remained radioactive, whereas GFP-negative cells did not, confirming that 89Zr-oxine-labeled cells followed with PET imaging represented a live cell population.
For the clinical safety evaluation, four factors help assess the amount of exposure a patient will have to an imaging agent and thus its potential for adverse effects [182]: (i) the mass dose of the agent used, (ii) the route of administration, (iii) the frequency of use and (iv) the agent’s biological, physical and effective half-life. The safety margin should be 100- or 25-fold toward the no-observed-adverse-effect level determined in single-dose toxicity or repeated-dose toxicity studies, respectively. Investigating genotoxicity pre-clinically is also encouraged before moving into humans. Imaging agents that are biologic in nature require both pre-clinical evidence of safety and thorough patient monitoring after use to establish clinical safety [182]. For the majority of the technologies mentioned in this article (namely, the ones being used clinically), the initial safety studies to permit their use in humans have already been completed.
In general, factors taken into consideration with regard to an agent’s use include reagent formulation; potential for adverse effects to the cells and the patients, as established through the results of non-clinical studies; initial clinical experience; and performance. Key factors determining performance are as follows: (i) ability of the imaging technology to image non-invasively, (ii) detection specific to the labeled cell and (iii) not altering the cellular characteristics of the therapeutic [21]. When considering an imaging agent, the cellular characteristics should be examined for each different cell type [51,79,183-185]. With all of these factors considered, it comes down to what each developer wants to be able to image and what method fits best into their cellular production without disrupting the functionality of the cellular product (see the modality summaries in previous sections and Table 5).
Regulatory context
As cellular imaging for cell therapy is relatively new, the regulatory requirements for both the imaging probe and the labeled cells have not yet been clearly defined (Box 2). Although cell-based therapies are generally classified as pharmaceutical products, the status of imaging probes alone or for labeled cells is less clear because this depends on their function and the geographical area. In the European Union, diagnostic probes are classified as pharmaceutical products; however, for imaging probes used for monitoring cellular therapies, classification is less evident and could potentially be approached as a pharmaceutical product, a starting material (for a pharmaceutical product) or a medical device. Further, if an imaging probe is used only in combination with therapeutic cells, it is unclear whether a separate classification and resulting set of regulatory requirements for the probe alone are needed. This issue is particularly relevant if the imaging probe is intended to be combined with different cellular therapies. Although a separate set of requirements for the imaging probe could facilitate the combination of the probe with various cell products, this does not take into account the potential effect of the interaction between the probe and the cell type. In the absence of regulatory guidance on the use of cellular imaging probes, the requirements for their use, in terms of quality and performance, should be determined on a case-by-case basis. Ultimately, the data required for a certain probe will depend on various factors, such as the combination of the probe and the cell type, previous experience and development phase of the cell product in which the probe is used. For development and use of new in vivo labeling probes, some guidance for the medical imaging agent may be found in a guideline for probes administered in vivo [182], whereas guidance for cell-based therapies might be relevant for imaging probes added ex vivo to the cell therapeutic prouduct [14,16]. Over time, when more experience has been gained, the regulatory requirements will become clearer.
Discussion
Cell-based therapies are making great strides toward becoming a standard therapeutic option for the treatment of disease. Recent approvals of cellular immunotherapies and MSC therapies highlight the progress that is being made. As the industry achieves marketing authorization of cellular therapeutics and new applications (e.g., solid tumors) enter clinical trials, a greater understanding of the safety profile of therapeutic cells is required. This includes survival, persistence, proliferation, distribution, differentiation and integration into the host physiology. These parameters can be evaluated by utilizing in vivo imaging techniques, such as reporter genes, dyes and particles. Although one technology may not answer every question, there are many available that will help answer these safety questions while additional technologies are moving toward the clinic.
Frangioni and Hajjar [21] introduced eight characteristics of an ideal imaging technology for tracking cellular therapeutics in the clinic: biocompatible, safe and nontoxic; no genetic modification or perturbation to the cell; single-cell detection at any anatomic location; quantification of cell number; minimal or no dilution with cell division; minimal or no transfer of contrast agent to other cells; non-invasive imaging in the living subject over months to years; and no requirement for injectable contrast agents. With the advancement of imaging technologies, some of these points have changed, but the overarching thesis remains: imaging technologies applied to imaging cellular therapeutics have to be detectable, quantitative and unique to the cell and cannot harm or alter the therapeutic. With these points in mind, the goal is to assess the fate of therapeutic cells in vivo. Knowing the location, migration, persistence and quantity of cells at a given location would provide improved understanding and thus added value to cellular therapy applications. However, what information (added value) is being missed when imaging is not performed? Imaging addresses more than simply determining where the cells went, whether they are alive and whether they reached the target; imaging technologies allow for insight to optimize and speed along the success of cellular therapeutics.
Only a limited number of clinical trials have incorporated cellular imaging into the analysis of their therapeutics [50,52-58,81,186]. Hesitance to adopt cellular imaging as part of a clinical trial is driven, in part, by the belief that inclusion of imaging is not required to further mitigate patient risk. In addition, there is a perception that imaging results could lead to findings that could be detrimental to the therapy’s progress to clinical authorization. In fact, in vivo imaging can add valuable safety data and the ability to overcome what, without imaging, researchers would not know to address (e.g., redosing to overcome a non-response or adjusting the route of administration to allow for more robust delivery or response). The acquisition of information regarding on-/off-target delivery, correct delivery to the target site and optimal dosing (i.e., location and quantity of dose) is valuable as the field of cellular therapy races toward tackling solid tumors. These measures could also result in an early biomarker of clinical efficacy: if cells reach the target coordinates with clinical success, this could signal early efficacy or, in its absence, the need for alternative therapy, co-therapy or redosing of the cellular therapeutic. Imaging data would facilitate this type of discussion about the cellular therapy and enable the appropriate exploration. Therefore, rather than relying on clinical endpoints alone, imaging offers an opportunity for early confidence in safety and important insights into the mode of action in vivo.
Although investigators need to consider the availability and suitability of imaging services associated with the chosen clinical trial delivery center, they need not have concerns over the practicality of including imaging as part of clinical trials because the use of in vivo imaging to track cells is familiar to radiology departments across the globe. Indeed, the base technologies (SPECT, PET, MRI, ultrasound, etc.) are used routinely for diagnostic procedures numerous times per day.
Cell tracking by imaging at the whole-body level has been part of the clinical routine for 30 years as it relates to tracking autologous leukocytes to detect sites of infection/inflammation. Thus far, among the imaging modalities described elsewhere in this article, only radionuclide imaging with gamma-emitting radionuclides (scintigraphy) has become part of this clinical routine. The standard radiolabeling methodology has been non-specific assimilation of lipophilic, metastable complexes of 111In (with oxine, tropolone and occasionally other bidentate chelators) and, later, 99mTc. Regenerative medicine and cell-based therapies and new insights into immunology are creating new roles for tracking stem cells and CAR-expressing T cells and for imaging the migration of individual immune cell types (e.g., eosinophils and neutrophils, T cells and dendritic cells) in cancer, atherosclerosis, stroke, transplantation and asthma. Conventional labeling methods have been applied in some of these areas; however, for clinical use, some of these new applications will require resolution, quantification and sensitivity (e.g., detecting small numbers of cells) beyond that achievable with available tracers, creating a need for improved labels, providing better sensitivity, quantification and resolution of the cellular product.
As reviewed earlier, different methods have different advantages, and with the advancement of technology over time, there may be an imaging solution for the broadest of needs. However, there are a plethora of currently available imaging techniques and probes that can allow researchers to label and track cells in most clinical and pre-clinical settings. The aim of this review is to raise awareness of these techniques and allow researchers to identify a potential imaging technique that could lead to generation of valuable safety and mode-of-action data. Probably of more importance, the authors hope to enable connection of therapy developers to imaging expertise. This will be essential, as any therapy developer wishing to benefit from in vivo tracking of cells will require a multidisciplinary team to match and validate a technique to provide a fit-for-purpose imaging solution.
When selecting the imaging technique, most probes have solutions for overcoming probe-related toxicities (duration of incubation time, reduced concentration, etc.), and different methods may overcome the shortcomings of other technologies. The community within the HESI CT-TRACS consortium has a wealth of understanding to both minimize the impact of an imaging technique on the cellular product and deliver data that will enhance confidence in the therapy’s safety profile. Furthermore, with the changing landscape of cellular therapy, co-development of imaging strategies remains an appealing option. This would allow for customization of the imaging solution to meet a developer’s needs. Although it is likely there will be a combination of probe and modality to meet these needs, this community is well placed to develop bespoke solutions or multimodal applications should they be required. Such a situation where an immediate solution would need to be developed is tracking of long-lived, rapidly dividing cells where repeat imaging is desired. However, most of the concerns from survey respondents could be addressed with the application of existing imaging techniques.
Through this article and the activities within CT-TRACS, the authors hope to have introduced and enhanced the understanding of the capabilities capabilities of in vivo imaging to advance the development of safe and efficacious cellular therapies. The authors also hope we have increased understanding to the point that investigators feel able to include imaging as part of the evidence gathered to demonstrate the value of their therapy. Throughout the preparation of this article, and reflected in its authorship, the authors have involved therapy developers, tool and reagent suppliers and, critically, regulatory authorities, demonstrating a cross-industry desire for broadening the type and availability of safety data.
Box 1.
Administration and misadministration. (A) 19F-labeled NK cells were injected intracranially to treat medulloblastoma. Images demonstrate successful administration, partial success and delivery failure of labeled NK cells (left to right respectively). Reproduced with permission from [93] © Springer Nature (2019). (B) Imaging the administration of dendritic cells. MRI before dendritic cell vaccination (left), with the inguinal lymph node to be injected, with cells identified by black arrow. MRI after injection (right) showing that dendritic cells were not accurately delivered into the lymph node (black arrow) but into the subcutaneous fat adjacent to the lymph node (white arrow). Reproduced with permission from [52] © Springer Nature (2015). NK, natural killer.
Box 2.
Can imaging add knowledge and experience to the development of cellular therapeutics? (A) In the discovery of small molecules, there are decades of experience and knowledge to draw upon, and these data have shaped the drug discovery pathway, from the idea for a therapeutic target to a licensed product. Many of the in vitro and pre-clinical tests to determine drug safety are also applicable to large molecules and biologics; however, there are fewer established methods for the field of cellular therapies. Arguably, some pre-clinical testing may be relevant, but options are limited for autologous and allogeneic cellular therapies. By and large, the established tools for safety assessment do not transfer well to cellular therapies, but in vivo imaging can play a major part in enabling the gathering of biodistribution data. This will be essential as the field moves to treating solid tumors, where evidence to prove that the cells reach their target will be required, and it is difficult to apply pre-clinical animal data to meet this need for cellular therapeutics. (B) In the development of cellular therapeutics, imaging can help proof-of-concept studies demonstrate mechanism of action. Additionally, imaging stands to play a large role in safety and efficacy in clinical trials. These approaches may help optimize delivery dose and route of administration, demonstrate efficacy of delivery or migration, show how/why some people respond better than others (i.e., responders versus non-responders) and lay the groundwork for co-therapeutics. ADME, absorption, distribution, metabolism and excretion; DMPK, drug metabolism and pharmacokinetics.
Acknowledgments
The authors thank all members (volunteers) of the HESI CT-TRACS Committee for their valuable time and expert input in the discussions leading to this article. The authors also thank Ms Christina West for manuscript review and editorial support.
Funding
This work was supported by the HESI CT-TRACS Committee. HESI is a non-profit scientific organization that facilitates public and private partnerships in human and environmental health. MP’s contribution was partly funded by the European Union’s Horizon 2020 research and innovation program under the nTRACK project (grant no. 761031).
Footnotes
Declaration of Competing Interest
BMH, SJ, MS, MST and WDS are employees of companies developing cellular therapies, providing testing or enabling technologies for the manufacture of cellular therapies.
References
- [1].European Medicines Agency. Advanced therapy medicinal products: overview. https://www.ema.europa.eu/en/human-regulatory/overview/advanced-therapy-medicinal-products-overview; 2020. [accessed 20 March 2020]. [Google Scholar]
- [2].Boyiadzis MM, Dhodapkar MV, Brentjens RJ, Kochenderfer JN, Neelapu SS, Maus MV, et al. Chimeric antigen receptor (CAR) T therapies for the treatment of hematologic malignancies: clinical perspective and significance. J Immunother Cancer 2018;6:137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].June CH, O'Connor RS, Kawalekar OU, Ghassemi S, Milone MC. CAR T cell immunotherapy for human cancer. Science 2018;359:1361–5. [DOI] [PubMed] [Google Scholar]
- [4].Maude SL, Laetsch TW, Buechner J, Rives S, Boyer M, Bittencourt H, et al. Tisagenlecleucel in children and young adults with B-cell lymphoblastic leukemia. N Engl J Med 2018;378:439–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Kochenderfer JN, Rosenberg SA. Treating B-cell cancer with T cells expressing anti-CD19 chimeric antigen receptors. Nat Rev Clin Oncol 2013;10:267–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Carbonaro DA, Jin X, Wang X, Yu XJ, Rozengurt N, Kaufman ML, et al. Gene therapy/bone marrow transplantation in ADA-deficient mice: roles of enzyme-replacement therapy and cytoreduction. Blood 2012;120:3677–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Candotti F, Shaw KL, Muul L, Carbonaro D, Sokolic R, Choi C, et al. Gene therapy for adenosine deaminase-deficient severe combined immune deficiency: clinical comparison of retroviral vectors and treatment plans. Blood 2012;120:3635–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Fadini GP, Agostini C, Avogaro A. Autologous stem cell therapy for peripheral arterial disease meta-analysis and systematic review of the literature. Atherosclerosis 2010;209:10–7. [DOI] [PubMed] [Google Scholar]
- [9].Kim SU, de Vellis J. Stem cell-based cell therapy in neurological diseases: a review. J Neurosci Res 2009;87:2183–200. [DOI] [PubMed] [Google Scholar]
- [10].Klinker MW, Wei CH. Mesenchymal stem cells in the treatment of inflammatory and autoimmune diseases in experimental animal models. World J Stem Cells 2015;7:556–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Gross G, Waks T, Eshhar Z. Expression of immunoglobulin-T-cell receptor chimeric molecules as functional receptors with antibody-type specificity. Proc Natl Acad Sci U S A 1989;86:10024–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Food and Drug Administration. Guidance for industry: Preclinical Assessment of Investigational Cellular and Gene Therapy Products, November 2013. https://www.fda.gov/media/87564/download; 2013 [accessed 20 March 2020]. [Google Scholar]
- [13].Food and Drug Administration. Draft Guidance for Industry: Considerations for the Design of Early-Phase Clinical Trials of Cellular and Gene Therapy Products; Availability. 78 FR 39736. Fed Regis 2013;78. FR 39736:39736–7. [Google Scholar]
- [14].Food and Drug Administration. Guidance for Industry: Considerations for the Design of Early-Phase Clinical Trials of Cellular and Gene Therapy Products, June 2015. http://fda.gov/regulatory-information/search-fda-guidance-documents/considerations-design-early-phase-clinical-trials-cellular-and-gene-therapy-products; 2015 [accessed 20 March 2020]. [Google Scholar]
- [15].European Medicines Agency. CHMP Guideline on Human Cell-Based Medicinal Products, May 2008. https://www.ema.europa.eu/en/documents/scientific-guide-line/guideline-human-cell-based-medicinal-products_en.pdf; 2008 [accessed 20 March 2020]. [Google Scholar]
- [16].European Medicines Agency. Reflection paper on stem cell-based medicinal products, January 2011. https://www.ema.europa.eu/en/documents/scientific-guideline/reflection-paper-stem-cell-based-medicinal-products_en.pdf; 2011 [accessed 20 March 2020]. [Google Scholar]
- [17].Health Canada. Draft Guidance Document: Guidance for Sponsors: Preparation of Clinical Trial Applications for use of Cell Therapy Products in Humans, October 2014. https://www.canada.ca/en/health-canada/services/drugs-health-products/drug-products/applications-submissions/guidance-documents/clinical-trials/guidance-document-preparation-clinical-trial-applications-use-cell-therapy-products-humans.html; 2014 [accessed 20 March 2020]. [Google Scholar]
- [18].Bulte JWM, Daldrup-Link HE. Clinical tracking of cell transfer and cell transplantation: trials and tribulations. Radiology 2018;289:604–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Chapelin F, Capitini CM, Ahrens ET. Fluorine-19 MRI for detection and quantification of immune cell therapy for cancer. J Immunother Cancer 2018;6. 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Ahrens ET & Bulte JW. Tracking immune cells in vivo using magnetic resonance imaging. Nat Rev Immunol 2013;13:755–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Frangioni JV, Hajjar RJ. In vivo tracking of stem cells for clinical trials in cardiovascular disease. Circulation 2004;110:3378–83. [DOI] [PubMed] [Google Scholar]
- [22].Ashmore-Harris C, Iafrate M, Saleem A, Fruhwirth GO. Non-invasive reporter gene imaging of cell therapies, including T cells and stem cells. Mol Ther 2020;28:1392–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Roca M, de Vries EF, Jamar F, Israel O, Signore A. Guidelines for the labelling of leucocytes with (111)In-oxine. Inflammation/Infection Taskgroup of the European Association of Nuclear Medicine. Eur J Nucl Med Mol Imaging 2010;37:835–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].de Vries EF, Roca M, Jamar F, Israel O, Signore A. Guidelines for the labelling of leucocytes with (99m)Tc-HMPAO. Inflammation/Infection Taskgroup of the European Association of Nuclear Medicine. Eur J Nucl Med Mol Imaging 2010;37:842–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Gholamrezanezhad A, Mirpour S, Ardekani JM, Bagheri M, Alimoghadam K, Yarmand S, et al. Cytotoxicity of 111In-oxine on mesenchymal stem cells: a time-dependent adverse effect. Nucl Med Commun 2009;30:210–6. [DOI] [PubMed] [Google Scholar]
- [26].Nowak B, Weber C, Schober A, Zeiffer U, Liehn EA, von Hundelshausen P, et al. Indium-111 oxine labelling affects the cellular integrity of haematopoietic progenitor cells. Eur J Nucl Med Mol Imaging 2007;34:715–21. [DOI] [PubMed] [Google Scholar]
- [27].Balaban EP, Simon TR, Frenkel EP. Toxicity of indium-111 on the radiolabeled lymphocyte. J Nucl Med 1987;28:229–33. [PubMed] [Google Scholar]
- [28].Blocklet D, Toungouz M, Kiss R, Lambermont M, Velu T, Duriau D, et al. 111In-oxine and 99mTc-HMPAO labelling of antigen-loaded dendritic cells: in vivo imaging and influence on motility and actin content. Eur J Nucl Med Mol Imaging 2003;30:440–7. [DOI] [PubMed] [Google Scholar]
- [29].Gholamrezanezhad A, Mirpour S, Bagheri M, Mohamadnejad M, Alimoghaddam K, Abdolahzadeh L, et al. In vivo tracking of 111In-oxine labeled mesenchymal stem cells following infusion in patients with advanced cirrhosis. Nucl Med Biol 2011;38:961–7. [DOI] [PubMed] [Google Scholar]
- [30].Rosado-de-Castro PH, Schmidt Fda R, Battistella V, Lopes de Souza SA, Gutfilen B, Goldenberg RC, et al. Biodistribution of bone marrow mononuclear cells after intra-arterial or intravenous transplantation in subacute stroke patients. Regen Med 2013;8:145–55. [DOI] [PubMed] [Google Scholar]
- [31].Jung KO, Kim TJ, Yu JH, Rhee S, Zhao W, Ha B, et al. Whole-body tracking of single cells via positron emission tomography. Nat Biomed Eng 2020;4:835–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Eich T, Eriksson O, Lundgren T. Visualization of early engraftment in clinical islet transplantation by positron-emission tomography. N Engl J Med 2007;356: 2754–5. [DOI] [PubMed] [Google Scholar]
- [33].Eriksson O, Eich T, Sundin A, Tibell A, Tufveson G, Andersson H, et al. Positron emission tomography in clinical islet transplantation. Am J Transplant 2009;9:2816–24. [DOI] [PubMed] [Google Scholar]
- [34].Hofmann M, Wollert KC, Meyer GP, Menke A, Arseniev L, Hertenstein B, et al. Monitoring of bone marrow cell homing into the infarcted human myocardium. Circulation 2005;111:2198–202. [DOI] [PubMed] [Google Scholar]
- [35].Kang WJ, Kang HJ, Kim HS, Chung JK, Lee MC, Lee DS. Tissue distribution of 18F-FDG-labeled peripheral hematopoietic stem cells after intracoronary administration in patients with myocardial infarction.J Nucl Med 2006;47:1295–301. [PubMed] [Google Scholar]
- [36].Blocklet D, Toungouz M, Berkenboom G, Lambermont M, Unger P, Preumont N, et al. Myocardial homing of nonmobilized peripheral-blood CD34+ cells after intracoronary injection. Stem Cells 2006;24:333–6. [DOI] [PubMed] [Google Scholar]
- [37].Charoenphun P, Meszaros LK, Chuamsaamarkkee K, Sharif-Paghaleh E, Ballinger JR, Ferris TJ, et al. [(89)Zr]oxinate4 for long-term in vivo cell tracking by positron emission tomography. Eur J Nucl Med Mol Imaging 2015;42:278–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Sato N, Wu H, Asiedu KO, Szajek LP, Griffiths GL, Choyke PL. (89)Zr-oxine complex PET cell imaging in monitoring cell-based therapies. Radiology 2015;275: 490–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Weist MR, Starr R, Aguilar B, Chea J, Miles JK, Poku E, et al. PET of adoptively transferred chimeric antigen receptor T cells with (89)Zr-oxine. J Nucl Med 2018;59:1531–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Man F, Lim L, Volpe A, Gabizon A, Shmeeda H, Draper B, et al. In vivo PET tracking of (89)Zr-labeled Vgamma9Vdelta2 t cells to mouse xenograft breast tumors activated with liposomal alendronate. Mol Ther 2019;27:219–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Davidson-Moncada J, Sato N, Hoyt RF, Reger RN, Thomas M, Clevenger R, et al. A novel method to study the in vivo trafficking and homing of adoptively transferred NK cells in rhesus macaques and humans. Blood 2014;124:659. [Google Scholar]
- [42].Hamm B, Taupitz M, Hussmann P, Wagner S, Wolf KJ. MR lymphography with iron oxide particles: dose-response studies and pulse sequence optimization in rabbits.AJR Am J Roentgenol 1992;158:183–90. [DOI] [PubMed] [Google Scholar]
- [43].White DL, Aicher KP, Tzika AA, Kucharczyk J, Engelstad BL, Moseley ME. Iron-dextran as a magnetic susceptibility contrast agent: flow-related contrast effects in the T2-weighted spin-echo. Magn Reson Med 1992;24:14–28. [DOI] [PubMed] [Google Scholar]
- [44].Arbab AS, Wilson LB, Ashari P, Jordan EK, Lewis BK, Frank JA. A model of lysosomal metabolism of dextran coated superparamagnetic iron oxide (SPIO) nanoparticles: implications for cellular magnetic resonance imaging. NMR Biomed 2005;18:383–9. [DOI] [PubMed] [Google Scholar]
- [45].Liu W, Frank JA. Detection and quantification of magnetically labeled cells by cellular MRI. Eur J Radiol 2009;70:258–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Oude Engberink RD, van der Pol SM, Dopp EA, de Vries HE, Blezer EL. Comparison of SPIO and USPIO for in vitro labeling of human monocytes: MR detection and cell function. Radiology 2007;243:467–74. [DOI] [PubMed] [Google Scholar]
- [47].Bulte JW, Kraitchman DL. Iron oxide MR contrast agents for molecular and cellular imaging. NMR Biomed 2004;17:484–99. [DOI] [PubMed] [Google Scholar]
- [48].Walczak P, Zhang J, Gilad AA, Kedziorek DA, Ruiz-Cabello J, Young RG, et al. Dual-modality monitoring of targeted intraarterial delivery of mesenchymal stem cells after transient ischemia. Stroke 2008;39:1569–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Wu YL, Ye Q, Foley LM, Hitchens TK, Sato K, Williams JB, et al. In situ labeling of immune cells with iron oxide particles: an approach to detect organ rejection by cellular MRI. Proc Natl Acad Sci U S A 2006;103:1852–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Toso C, Vallee JP, Morel P, Ris F, Demuylder-Mischler S, Lepetit-Coiffe M, et al. Clinical magnetic resonance imaging of pancreatic islet grafts after iron nanoparticle labeling. Am J Transplant 2008;8:701–6. [DOI] [PubMed] [Google Scholar]
- [51].Balakumaran A, Pawelczyk E, Ren J, Sworder B, Chaudhry A, Sabatino M, et al. Superparamagnetic iron oxide nanoparticles labeling of bone marrow stromal (mesenchymal) cells does not affect their “stemness.”. PLoS One 2010;5. e11462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].de Vries IJ, Lesterhuis WJ, Barentsz JO, Verdijk P, van Krieken JH, Boerman OC, et al. Magnetic resonance tracking of dendritic cells in melanoma patients for monitoring of cellular therapy. Nat Biotechnol 2005;23:1407–13. [DOI] [PubMed] [Google Scholar]
- [53].Zhu J, Zhou L, XingWu F. Tracking neural stem cells in patients with brain trauma. N Engl J Med 2006;355:2376–8. [DOI] [PubMed] [Google Scholar]
- [54].Callera F, de Melo CM. Magnetic resonance tracking of magnetically labeled autologous bone marrow CD34+ cells transplanted into the spinal cord via lumbar puncture technique in patients with chronic spinal cord injury: CD34+ cells' migration into the injured site. Stem Cells Dev 2007;16:461–6. [DOI] [PubMed] [Google Scholar]
- [55].Saudek F, Jirak D, Girman P, Herynek V, Dezortova M, Kriz J, et al. Magnetic resonance imaging of pancreatic islets transplanted into the liver in humans. Transplantation 2010;90:1602–6. [DOI] [PubMed] [Google Scholar]
- [56].Karussis D, Karageorgiou C, Vaknin-Dembinsky A, Gowda-Kurkalli B, Gomori JM, Kassis I, et al. Safety and immunological effects of mesenchymal stem cell transplantation in patients with multiple sclerosis and amyotrophic lateral sclerosis. Arch Neurol 2010;67:1187–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Jozwiak S, Habich A, Kotulska K, Sarnowska A, Kropiwnicki T, Janowski M, et al. Intracerebroventricular transplantation of cord blood-derived neural progenitors in a child with severe global brain ischemic injury. Cell Med 2010;1:71–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Richards JMJ, Shaw CA, Lang NN, Williams MC, Semple SIK, MacGillivray TJ, et al. In vivo mononuclear cell tracking using superparamagnetic particles of iron oxide: feasibility and safety in humans. Circ Cardiovasc Imaging 2012;5:509–17. [DOI] [PubMed] [Google Scholar]
- [59].Neuwelt EA, Hamilton BE, Varallyay CG, Rooney WR, Edelman RD, Jacobs PM, et al. Ultrasmall superparamagnetic iron oxides (USPIOs): a future alternative magnetic resonance (MR) contrast agent for patients at risk for nephrogenic systemic fibrosis (NSF)? Kidney Int 2009;75:465–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Bashir MR, Bhatti L, Marin D, Nelson RC. Emerging applications for ferumoxytol as a contrast agent in MRI. J Magn Reson Imaging 2015;41:884–98. [DOI] [PubMed] [Google Scholar]
- [61].McCullough BJ, Kolokythas O, Maki JH, Green DE. Ferumoxytol in clinical practice: implications for MRI.J Magn Reson Imaging 2013;37:1476–9. [DOI] [PubMed] [Google Scholar]
- [62].Thu MS, Bryant LH, Coppola T, Jordan EK, Budde MD, Lewis BK, et al. Self-assembling nanocomplexes by combining ferumoxytol, heparin and protamine for cell tracking by magnetic resonance imaging. Nat Med 2012;18:463–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Theruvath AJ, Nejadnik H, Muehe AM, Gassert F, Lacayo NJ, Goodman SB, et al. Tracking cell transplants in femoral osteonecrosis with magnetic resonance imaging: a proof-of-concept study in patients. Clin Cancer Res 2018;24:6223–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Li K, Gordon AC, Zheng L, Li W, Guo Y, Sun J, et al. Clinically applicable magnetic-labeling of natural killer cells for MRI of transcatheter delivery to liver tumors: preclinical validation for clinical translation. Nanomedicine (Lond) 2015;10: 1761–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Su Z, Wang X, Zheng L, Lyu T, Figini M, Wang B, et al. MRI-guided interventional natural killer cell delivery for liver tumor treatment. Cancer Med 2018;7:1860–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Gutova M, Frank JA, D'Apuzzo M, Khankaldyyan V, Gilchrist MM, Annala AJ, et al. Magnetic resonance imaging tracking of ferumoxytol-labeled human neural stem cells: studies leading to clinical use. Stem Cells Transl Med 2013;2:766–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Khurana A, Nejadnik H, Chapelin F, Lenkov O, Gawande R, Lee S, et al. Ferumoxytol: a new, clinically applicable label for stem-cell tracking in arthritic joints with MRI. Nanomedicine (Lond) 2013;8:1969–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Vandergriff AC, Hensley TM, Henry ET, Shen D, Anthony S, Zhang J, et al. Magnetic targeting of cardiosphere-derived stem cells with ferumoxytol nanoparticles for treating rats with myocardial infarction. Biomaterials 2014;35:8528–39. [DOI] [PubMed] [Google Scholar]
- [69].Yin Y, Zhou X, Guan X, Liu Y, Jiang CB, Liu J. In vivo tracking of human adipose-derived stem cells labeled with ferumoxytol in rats with middle cerebral artery occlusion by magnetic resonance imaging. Neural Regen Res 2015;10:909–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Aghighi M, Theruvath AJ, Pareek A, Pisani LL, Alford R, Muehe AM, et al. Magnetic resonance imaging of tumor-associated macrophages: clinical translation. Clin Cancer Res 2018;24:4110–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Hitchens TK, Ye Q, Eytan DF, Janjic JM, Ahrens ET, Ho C. 19F MRI detection of acute allograft rejection with in vivo perfluorocarbon labeling of immune cells. Magn Reson Med 2011;65:1144–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Daldrup-Link HE. Ten things you might not know about iron oxide nanoparticles. Radiology 2017;284:616–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Zhou XY, Jeffris KE, Yu EY, Zheng B, Goodwill PW, Nahid P, et al. First in vivo magnetic particle imaging of lung perfusion in rats. Phys Med Biol 2017;62:3510–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Wang YX. Superparamagnetic iron oxide based MRI contrast agents: current status of clinical application. Quant Imaging Med Surg 2011;1:35–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Flaim SF. Pharmacokinetics and side effects of perfluorocarbon-based blood substitutes. Artif Cells Blood Substit Immobil Biotechnol 1994;22:1043–54. [DOI] [PubMed] [Google Scholar]
- [76].Krafft MP. Fluorocarbons and fluorinated amphiphiles in drug delivery and biomedical research. Adv Drug Deliv Rev 2001;47:209–28. [DOI] [PubMed] [Google Scholar]
- [77].Janjic JM, Srinivas M, Kadayakkara DK, Ahrens ET. Self-delivering nanoemulsions for dual fluorine-19 MRI and fluorescence detection. J Am Chem Soc 2008;130:2832–41. [DOI] [PubMed] [Google Scholar]
- [78].Ahrens ET, Flores R, Xu HY, Morel PA. In vivo imaging platform for tracking immunotherapeutic cells. Nat Biotechnol 2005;23:983–7. [DOI] [PubMed] [Google Scholar]
- [79].Helfer BM, Balducci A, Nelson AD, Janjic JM, Gil RR, Kalinski P, et al. Functional assessment of human dendritic cells labeled for in vivo (19)F magnetic resonance imaging cell tracking. Cytotherapy 2010;12:238–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Helfer BM, Balducci A, Sadeghi Z, O'Hanlon C, Hijaz A, Flask CA, et al. (19)F MRI tracer preserves in vitro and in vivo properties of hematopoietic stem cells. Cell Transplantation 2013;22:87–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Ahrens ET, Helfer BM, O'Hanlon CF, Schirda C. Clinical cell therapy imaging using a perfluorocarbon tracer and fluorine-19 MRI. Magn Reson Med 2014;72:1696–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Bonetto F, Srinivas M, Heerschap A, Mailliard R, Ahrens ET, Figdor CG. A novel (19)F agent for detection and quantification of human dendritic cells using magnetic resonance imaging. Int J Cancer 2011;129:365–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Kadayakkara DK, Beatty PL, Turner MS, Janjic JM, Ahrens ET, Finn OJ. Inflammation driven by overexpression of the hypoglycosylated abnormal mucin 1 (MUC1) links inflammatory bowel disease and pancreatitis. Pancreas 2010;39:510–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Srinivas M, Morel PA, Ernst LA, Laidlaw DH, Ahrens ET. Fluorine-19 MRI for visualization and quantification of cell migration in a diabetes model. Magn Reson Med 2007;58:725–34. [DOI] [PubMed] [Google Scholar]
- [85].Srinivas M, Turner MS, Janjic JM, Morel PA, Laidlaw DH, Ahrens ET. In vivo cytometry of antigen-specific T cells using F-19 MRI. Magn Reson Med 2009;62:747–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Bible E, Dell'acqua F, Solanky B, Balducci A, Crapo PM, Badylak SF, et al. Non-invasive imaging of transplanted human neural stem cells and ECM scaffold remodeling in the stroke-damaged rat brain by (19)F- and diffusion-MRI. Biomaterials 2012;33:2858–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Boehm-Sturm P, Mengler L, Wecker S, Hoehn M, Kallur T. In vivo tracking of human neural stem cells with 19F magnetic resonance imaging. PLoS One 2011;6:e29040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Rose LC, Kadayakkara DK, Wang G, Bar-Shir A, Helfer BM, O'Hanlon CF, et al. Fluorine-19 labeling of stromal vascular fraction cells for clinical imaging applications. Stem Cells Transl Med 2015;4:1472–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Muhammad G, Jablonska A, Rose L, Walczak P, Janowski M. Effect of MRI tags: SPIO nanoparticles and 19F nanoemulsion on various populations of mouse mesenchymal stem cells. Acta Neurobiol Exp (Wars) 2015;75:144–59. [PMC free article] [PubMed] [Google Scholar]
- [90].Ribot EJ, Gaudet JM, Chen Y, Gilbert KM, Foster PJ. In vivo MR detection of fluorine-labeled human MSC using the bSSFP sequence. Int J Nanomedicine 2014;9:1731–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Bouchlaka MN, Ludwig KD, Gordon JW, Kutz MP, Bednarz BP, Fain SB, et al. 19F-MRI for monitoring human NK cells in vivo. Oncoimmunology 2016;5: e1143996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Fink C, Gaudet JM, Fox MS, Bhatt S, Viswanathan S, Smith M, et al. (19)F-perfluorocarbon-labeled human peripheral blood mononuclear cells can be detected in vivo using clinical MRI parameters in a therapeutic cell setting. Sci Rep 2018;8:590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Kennis BA, Michel KA, Brugmann WB, Laureano A, Tao RH, Somanchi SS, et al. Monitoring of intracerebellarly-administered natural killer cells with fluorine-19 MRI. J Neurooncol 2019;142:395–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Khurana A, Chapelin F, Beck G, Lenkov OD, Donig J, Nejadnik H, et al. Iron administration before stem cell harvest enables MR imaging tracking after transplantation. Radiology 2013;269:186–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Ferrara K, Pollard R, Borden M. Ultrasound microbubble contrast agents: fundamentals and application to gene and drug delivery. Annu Rev Biomed Eng 2007;9:415–47. [DOI] [PubMed] [Google Scholar]
- [96].Blomley MJ, Cooke JC, Unger EC, Monaghan MJ, Cosgrove DO. Microbubble contrast agents: a new era in ultrasound. BMJ 2001;322:1222–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Cui W, Tavri S, Benchimol MJ, Itani M, Olson ES, Zhang H, et al. Neural progenitor cells labeling with microbubble contrast agent for ultrasound imaging in vivo. Biomaterials 2013;34:4926–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Jokerst JV, Khademi C, Gambhir SS. Intracellular aggregation of multimodal silica nanoparticles for ultrasound-guided stem cell implantation. Sci Transl Med 2013;5. 177ra35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Kuliszewski MA, Fujii H, Liao C, Smith AH, Xie A, Lindner JR, et al. Molecular imaging of endothelial progenitor cell engraftment using contrast-enhanced ultrasound and targeted microbubbles. Cardiovasc Res 2009;83:653–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Shapiro MG, Goodwill PW, Neogy A, Yin M, Foster FS, Schaffer DV, et al. Biogenic gas nanostructures as ultrasonic molecular reporters. Nat Nanotechnol 2014;9:311–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Farhadi A, Ho GH, Sawyer DP, Bourdeau RW, Shapiro MG. Ultrasound imaging of gene expression in mammalian cells. Science 2019;365:1469–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Kempen PJ, Greasley S, Parker KA, Campbell JL, Chang HY, Jones JR, et al. Theranostic mesoporous silica nanoparticles biodegrade after pro-survival drug delivery and ultrasound/magnetic resonance imaging of stem cells. Theranostics 2015;5:631–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Nam SY, Ricles LM, Suggs LJ, Emelianov SY. In vivo ultrasound and photoacoustic monitoring of mesenchymal stem cells labeled with gold nanotracers. PLoS One 2012;7:e37267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Xu S, Yang F, Zhou X, Zhuang Y, Liu B, Mu Y, et al. Uniform PEGylated PLGA microcapsules with embedded Fe3O4 nanoparticles for US/MR dual-modality imaging. ACS Appl Mater Interfaces 2015;7:20460–8. [DOI] [PubMed] [Google Scholar]
- [105].Barnett BP, Ruiz-Cabello J, Hota P, Ouwerkerk R, Shamblott MJ, Lauzon C, et al. Use of perfluorocarbon nanoparticles for non-invasive multimodal cell tracking of human pancreatic islets. Contrast Media Mol Imaging 2011;6:251–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Rodriguez-Porcel M, Gheysens O, Chen IY, Wu JC, Gambhir SS. Image-guided cardiac cell delivery using high-resolution small-animal ultrasound. Mol Ther 2005;12:1142–7. [DOI] [PubMed] [Google Scholar]
- [107].Patrick PS, Bear JC, Fitzke HE, Zaw-Thin M, Parkin IP, Lythgoe MF, et al. Radiometal cross-linking of alginate hydrogels for non-invasive in vivo imaging. Biomaterials 2020;243:119930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Steinberg I, Huland DM, Vermesh O, Frostig HE, Tummers WS, Gambhir SS. Photoacoustic clinical imaging. Photoacoustics 2019;14:77–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Swider E, Daoudi K, Staal AHJ, Koshkina O, van Riessen NK, van Dinther E, et al. Clinically-applicable perfluorocarbon-loaded nanoparticles for in vivo photoacoustic, (19)F magnetic resonance and fluorescent imaging. Nanotheranostics 2018;2:258–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Sharkey J, Scarfe L, Santeramo I, Garcia-Finana M, Park BK, Poptani H, et al. Imaging technologies for monitoring the safety, efficacy and mechanisms of action of cell-based regenerative medicine therapies in models of kidney disease. Eur J Pharmacol 2016;790:74–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Keu KV, Witney TH, Yaghoubi S, Rosenberg J, Kurien A, Magnusson R, et al. Reporter gene imaging of targeted T cell immunotherapy in recurrent glioma. Sci Transl Med 2017;9. eaag2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Minn I, Huss DJ, Ahn HH, Chinn TM, Park A, Jones J, et al. Imaging CAR T cell therapy with PSMA-targeted positron emission tomography. Sci Adv 2019;5: eaaw5096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Emami-Shahri N, Foster J, Kashani R, Gazinska P, Cook C, Sosabowski J, et al. Clinically compliant spatial and temporal imaging of chimeric antigen receptor T-cells. Nat Commun 2018;14:1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Volpe A, Lang C, Lim L, Man F, Kurtys E, Ashmore-Harris C, et al. Spatiotemporal PET Imaging Reveals Differences in CAR-T Tumor Retention in Triple-Negative Breast Cancer Models. Mol Ther 2020;28:2271–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Ponomarev V, Doubrovin M, Shavrin A, Serganova I, Beresten T, Ageyeva L, et al. A human-derived reporter gene for noninvasive imaging in humans: mitochondrial thymidine kinase type 2. J Nucl Med 2007;48:819–26. [DOI] [PubMed] [Google Scholar]
- [116].Likar Y, Zurita J, Dobrenkov K, Shenker L, Cai S, Neschadim A, et al. A new pyrimidine-specific reporter gene: a mutated human deoxycytidine kinase suitable for PET during treatment with acycloguanosine-based cytotoxic drugs. J Nucl Med 2010;51:1395–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Cohen B, Dafni H, Meir G, Harmelin A, Neeman M. Ferritin as an endogenous MRI reporter for noninvasive imaging of gene expression in C6 glioma tumors. Neoplasia 2005;7:109–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Deans AE, Wadghiri YZ, Bernas LM, Yu X, Rutt BK, Turnbull DH. Cellular MRI contrast via coexpression of transferrin receptor and ferritin. Magn Reson Med 2006;56:51–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Genove G, DeMarco U, Xu H, Goins WF, Ahrens ET. A new transgene reporter for in vivo magnetic resonance imaging. Nat Med 2005;11:450–4. [DOI] [PubMed] [Google Scholar]
- [120].Iordanova B, Robison CS, Ahrens ET. Design and characterization of a chimeric ferritin with enhanced iron loading and transverse NMR relaxation rate. J Biol Inorg Chem 2010;15:957–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Drexler HG, Gaedicke G, Minowada J. Isoenzyme studies in human leukemia-lymphoma cell lines—1. Carboxylic esterase. Leuk Res 1985;9:209–29. [DOI] [PubMed] [Google Scholar]
- [122].Moroz MA, Zhang H, Lee J, Moroz E, Zurita J, Shenker L, et al. Comparative analysis of T cell imaging with human nuclear reporter genes. J Nucl Med 2015;56:1055–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Srinivas M, Melero I, Kaempgen E, Figdor CG, de Vries IJ. Cell tracking using multimodal imaging. Contrast Media Mol Imaging 2013;8:432–8. [DOI] [PubMed] [Google Scholar]
- [124].Lee HW, Gangadaran P, Kalimuthu S, Ahn BC. Advances in molecular imaging strategies for in vivo tracking of immune cells. Biomed Res Int 2016. 2016:1946585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Gangadaran P, Ahn BC. Molecular imaging: a useful tool for the development of natural killer cell-based immunotherapies. Front Immunol 2017;8:1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Lesterhuis WJ, de Vries IJ, Schreibelt G, Lambeck AJ, Aarntzen EH, Jacobs JF, et al. Route of administration modulates the induction of dendritic cell vaccine-induced antigen-specific T cells in advanced melanoma patients. Clin Cancer Res 2011;17:5725–35. [DOI] [PubMed] [Google Scholar]
- [127].Srinivas M, Aarntzen EH, Bulte JW, Oyen WJ, Heerschap A, de Vries IJ, et al. Imaging of cellular therapies. Adv Drug Deliv Rev 2010;62:1080–93. [DOI] [PubMed] [Google Scholar]
- [128].Park HS, Kim J, Cho MY, Lee H, Nam SH, Suh YD, et al. Convenient and effective ICGylation of magnetic nanoparticles for biomedical applications. Sci Rep 2017;7:8831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Song G, Han J, Zhao Y, Wang Z, Du H. A review on medical image registration as an optimization problem. Curr Med Imaging Rev 2017;13:274–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Chehade M, Srivastava AK, Bulte JW. Co-registration of bioluminescence tomography, computed tomography, and magnetic resonance imaging for multimodal in vivo stem cell tracking. Tomography 2016;2:159–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Makela AV, Murrell DH, Parkins KM, Kara J, Gaudet JM, Foster PJ. Cellular imaging with MRI. Top Magn Reson Imaging 2016;25:177–86. [DOI] [PubMed] [Google Scholar]
- [132].Bulte JW. Superparamagnetic iron oxides as MPI tracers: a primer and review of early applications. Adv Drug Deliv Rev 2019;138:293–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Sehl OC, Makela AV, Hamilton AM, Foster PJ. Trimodal Cell Tracking In Vivo: Combining Iron- and Fluorine-Based Magnetic Resonance Imaging with Magnetic Particle Imaging to Monitor the Delivery of Mesenchymal Stem Cells and the Ensuing Inflammation. Tomography 2019;5:367–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Zheng B, von See MP, Yu E, Gunel B, Lu K, Vazin T, et al. Quantitative Magnetic Particle Imaging Monitors the Transplantation, Biodistribution, and Clearance of Stem Cells In Vivo. Theranostics 2016;6:291–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Kraitchman D, Tatsumi M, Gilson WD, Ishimori T, Kedziorek D, Walczak P, et al. Dynamic imaging of allogeneic mesenchymal stem cells trafficking to myocardial infarction. Circulation 2005;112:1451–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Ebner B, Behm P, Jacoby C, Burghoff S, French BA, Schrader J, et al. Early assessment of pulmonary inflammation by 19F MRI in vivo. Circ Cardiovasc Imaging 2010;3:202–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Rahmim A, Zaidi H. PET versus SPECT: strengths, limitations and challenges. Nucl Med Commun 2008;29:193–207. [DOI] [PubMed] [Google Scholar]
- [138].Arbab AS, Bashaw LA, Miller BR, Jordan EK, Lewis BK, Kalish H, et al. Characterization of biophysical and metabolic properties of cells labeled with superparamagnetic iron oxide nanoparticles and transfection agent for cellular MR imaging. Radiology 2003;229:838–46. [DOI] [PubMed] [Google Scholar]
- [139].Pawelczyk E, Arbab AS, Pandit S, Hu E, Frank JA. Expression of transferrin receptor and ferritin following ferumoxides-protamine sulfate labeling of cells: implications for cellular magnetic resonance imaging. NMR Biomed 2006;19:581–92. [DOI] [PubMed] [Google Scholar]
- [140].Sykova E, Jendelova P. Magnetic resonance tracking of transplanted stem cells in rat brain and spinal cord. Neurodegener Dis 2006;3:62–7. [DOI] [PubMed] [Google Scholar]
- [141].Jendelova P, Herynek V, DeCroos J, Glogarova K, Andersson B, Hajek M, et al. Imaging the fate of implanted bone marrow stromal cells labeled with superparamagnetic nanoparticles. Magn Reson Med 2003;50:767–76. [DOI] [PubMed] [Google Scholar]
- [142].Arbab AS, Jordan EK, Wilson LB, Yocum GT, Lewis BK, Frank JA. In vivo trafficking and targeted delivery of magnetically labeled stem cells. Hum Gene Ther 2004;15:351–60. [DOI] [PubMed] [Google Scholar]
- [143].Sykova E, Jendelova P. Migration, fate and in vivo imaging of adult stem cells in the CNS. Cell Death Differ 2007;14:1336–42. [DOI] [PubMed] [Google Scholar]
- [144].Partlow KC, Chen J, Brant JA, Neubauer AM, Meyerrose TE, Creer MH, et al. 19F magnetic resonance imaging for stem/progenitor cell tracking with multiple unique perfluorocarbon nanobeacons. Faseb j 2007;21:1647–54. [DOI] [PubMed] [Google Scholar]
- [145].Janjic JM, Ahrens ET. Fluorine-containing nanoemulsions for MRI cell tracking. Wiley Interdiscip Rev Nanomed Nanobiotechnol 2009;1:492–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Helfer BM, Balducci A, Sadeghi Z, O'Hanlon C, Hijaz A, Flask CA, et al. (1)(9)F MRI tracer preserves in vitro and in vivo properties of hematopoietic stem cells. Cell Transplant 2013;22:87–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Chapelin F, Gao S, Okada H, Weber TG, Messer K, Ahrens ET. Fluorine-19 nuclear magnetic resonance of chimeric antigen receptor T cell biodistribution in murine cancer model. Sci Rep 2017;7:17748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Lu X, Jang H, Ma Y, Jerban S, Chang EY, Du J. Ultrashort echo time quantitative susceptibility mapping (UTE-QSM) of highly concentrated magnetic nanoparticles: a comparison study about different sampling strategies. Molecules 2019;24:1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Lu X, Ma Y, Chang EY, He Q, Searleman A, von Drygalski A, et al. Simultaneous quantitative susceptibility mapping (QSM) and R2* for high iron concentration quantification with 3D ultrashort echo time sequences: an echo dependence study. Magn Reson Med 2018;79:2315–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Ma YJ, Shao H, Du J, Chang EY. Ultrashort echo time magnetization transfer (UTE-MT) imaging and modeling: magic angle independent biomarkers of tissue properties. NMR Biomed 2016;29:1546–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Yanagisawa K, Moriyasu F, Miyahara T, Yuki M, Iijima H. Phagocytosis of ultrasound contrast agent microbubbles by Kupffer cells. Ultrasound Med Biol 2007;33:318–25. [DOI] [PubMed] [Google Scholar]
- [152].Terrovitis J, Stuber M, Youssef A, Preece S, Leppo M, Kizana E, et al. Magnetic resonance imaging overestimates ferumoxide-labeled stem cell survival after transplantation in the heart. Circulation 2008;117:1555–62. [DOI] [PubMed] [Google Scholar]
- [153].Gaudet JM, Ribot EJ, Chen Y, Gilbert KM, Foster PJ. Tracking the fate of stem cell implants with fluorine-19 MRI. PLoS One 2015;10. e0118544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Ahrens ET, Zhong J. In vivo MRI cell tracking using perfluorocarbon probes and fluorine-19 detection. NMR Biomed 2013;26:860–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Berman SC, Galpoththawela C, Gilad AA, Bulte JW, Walczak P. Long-term MR cell tracking of neural stem cells grafted in immunocompetent versus immunodeficient mice reveals distinct differences in contrast between live and dead cells. Magn Reson Med 2011;65:564–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Nejadnik H, Lenkov O, Gassert F, Fretwell D, Lam I. Daldrup-Link HE. Macrophage phagocytosis alters the MRI signal of ferumoxytol-labeled mesenchymal stromal cells in cartilage defects. Sci Rep 2016;6:25897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Moroz MA, Zanzonico P, Lee JT, Ponomarev V. Ex vivo radiolabeling and in vivo PET imaging of T cells expressing nuclear reporter genes. Methods Mol Biol 2018;1790:153–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Koehne G, Doubrovin M, Doubrovina E, Zanzonico P, Gallardo HF, Ivanova A, et al. Serial in vivo imaging of the targeted migration of human HSV-TK-transduced antigen-specific lymphocytes. Nat Biotechnol 2003;21:405–13. [DOI] [PubMed] [Google Scholar]
- [159].Xu T, Close D, Handagama W, Marr E, Sayler G, Ripp S. The expanding toolbox of in vivo bioluminescent imaging. Front Oncol 2016;6:150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Mezzanotte L, van 't Root M, Karatas H, Goun EA, Lowik C. In vivo molecular bioluminescence imaging: new tools and applications. Trends Biotechnol 2017;35:640–52. [DOI] [PubMed] [Google Scholar]
- [161].Blasberg RG, Tjuvajev JG. Herpes simplex virus thymidine kinase as a marker/reporter gene for PET imaging of gene therapy. Q J Nucl Med 1999;43:163–9. [PubMed] [Google Scholar]
- [162].Yaghoubi SS, Couto MA, Chen CC, Polavaram L, Cui G, Sen L, et al. Preclinical safety evaluation of 18F-FHBG: a PET reporter probe for imaging herpes simplex virus type 1 thymidine kinase (HSV1-tk) or mutant HSV1-sr39tk's expression. J Nucl Med 2006;47:706–15. [PubMed] [Google Scholar]
- [163].Brader P, Serganova I, Blasberg RG. Noninvasive molecular imaging using reporter genes.J Nucl Med 2013;54:167–72. [DOI] [PubMed] [Google Scholar]
- [164].Portulano C, Paroder-Belenitsky M, Carrasco N. The Na+/I− symporter (NIS): mechanism and medical impact. Endocr Rev 2014;35:106–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Zhang H, Huang R, Lewis J, Blasberg R. Imaging human norepinephrine transporter (hNET) expressing reporter cells and tumors with 4-18F-fluorobenzylguanidine. J Nucl Med 2012;53:1584. [Google Scholar]
- [166].Pule M, Badar A, Kiru L, Lythgoe M, Peters A, Detecting a Therapeutic Cell, US patent 20170056534, Aug 25, 2015. https://uspto.report/patent/grant/10,286,092.
- [167].Jauregui-Osoro M, Sunassee K, Weeks AJ, Berry DJ, Paul RL, Cleij M, et al. Synthesis and biological evaluation of [18 F] tetrafluoroborate: a PET imaging agent for thyroid disease and reporter gene imaging of the sodium/iodide symporter. Eur J Nucl Med Mol Imaging 2010;37:2108–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Khoshnevisan A, Jauregui-Osoro M, Shaw K, Torres JB, Young JD, Ramakrishnan NK, et al. [(18)F]tetrafluoroborate as a PET tracer for the sodium/iodide symporter: the importance of specific activity. EJNMMI Res 2016;6:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Dohan O, De la Vieja A, Paroder V, Riedel C, Artani M, Reed M, et al. The sodium/iodide Symporter (NIS): characterization, regulation, and medical significance. Endocr Rev 2003;24:48–77. [DOI] [PubMed] [Google Scholar]
- [170].Fruhwirth GO, Diocou S, Blower PJ, Ng T, Mullen GE. A whole-body dual-modality radionuclide optical strategy for preclinical imaging of metastasis and heterogeneous treatment response in different microenvironments. J Nucl Med 2014;55:686–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Diocou S, Volpe A, Jauregui-Osoro M, Boudjemeline M, Chuamsaamarkkee K, Man F, et al. [(18)F]tetrafluoroborate-PET/CT enables sensitive tumor and metastasis in vivo imaging in a sodium iodide symporter-expressing tumor model. Sci Rep 2017;7:946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Volpe A, Man F, Lim L, Khoshnevisan A, Blower J, Blower PJ, et al. Radionuclide-fluorescence reporter gene imaging to track tumor progression in rodent tumor models. J Vis Exp 2018;133:57088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Ashmore-Harris C, Blackford SJ, Grimsdell B, Kurtys E, Glatz MC, Rashid TS, et al. Reporter gene-engineering of human induced pluripotent stem cells during differentiation renders in vivo traceable hepatocyte-like cells accessible. Stem Cell Res 2019;41:101599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Haywood T, Beinat C, Gowrishankar G, Patel CB, Alam IS, Murty S, et al. Positron emission tomography reporter gene strategy for use in the central nervous system. Proc Natl Acad Sci U S A 2019;116:11402–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Li Z, Wu JC, Sheikh AY, Kraft D, Cao F, Xie X, et al. Differentiation, survival, and function of embryonic stem cell derived endothelial cells for ischemic heart disease. Circulation 2007;116:I46–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Sharif-Paghaleh E, Leech J, Sunassee K, Ali N, Sagoo P, Lechler RI, et al. Monitoring the efficacy of dendritic cell vaccination by early detection of (99m) Tc-HMPAO-labelled CD4(+)T cells. Eur J Immunol 2014;44:2188–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Lukawska JJ, Livieratos L, Sawyer BM, Lee T, O'Doherty M, Blower PJ, et al. Imaging inflammation in asthma: real time, differential tracking of human neutrophil and eosinophil migration in allergen challenged, atopic asthmatics in vivo. EBio-Medicine 2014;1:173–80. 10.1016/j.ebiom.2014.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Lukawska JJ, Livieratos L, Sawyer BM, Lee T, O'Doherty M, Blower PJ, et al. Realtime differential tracking of human neutrophil and eosinophil migration in vivo. J Allergy Clin Immunol 2014;133. 233–9.e1. [DOI] [PubMed] [Google Scholar]
- [179].Griessinger CM, Kehlbach R, Bukala D, Wiehr S, Bantleon R, Cay F, et al. In vivo tracking of Th1 cells by PET reveals quantitative and temporal distribution and specific homing in lymphatic tissue. J Nucl Med 2014;55:301–7. [DOI] [PubMed] [Google Scholar]
- [180].Tavare R, Sagoo P, Varama G, Tanriver Y, Warely A, Diebold SS, et al. Monitoring of in vivo function of superparamagnetic iron oxide labelled murine dendritic cells during anti-tumour vaccination. PLoS One 2011;6. e19662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Sharif-Paghaleh E, Sunassee K, Tavare R, Ratnasothy K, Koers A, Ali N, et al. In vivo SPECT reporter gene imaging of regulatory T cells. PLoS One 2011;6:e25857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Food and Drug Administration. Guidance for Industry: Developing Medical Imaging Drug and Biological Products, Part 1: Conducting Safety Assessments. Maryland: FDA; 2004. [Google Scholar]
- [183].Bansal A, Pandey MK, Demirhan YE, Nesbitt JJ, Crespo-Diaz RJ, Terzic A, et al. Novel (89)Zr cell labeling approach for PET-based cell trafficking studies. EJNMMI Res 2015;5:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Crich SG, Biancone L, Cantaluppi V, Duo D, Esposito G, Russo S, et al. Improved route for the visualization of stem cells labeled with a Gd-/Eu-chelate as dual (MRI and fluorescence) agent. Magn Reson Med 2004;51:938–44. [DOI] [PubMed] [Google Scholar]
- [185].Gildehaus FJ, Haasters F, Drosse I, Wagner E, Zach C, Mutschler W, et al. Impact of indium-111 oxine labelling on viability of human mesenchymal stem cells in vitro, and 3D cell-tracking using SPECT/CT in vivo. Mol Imaging Biol 2011;13:1204–14. [DOI] [PubMed] [Google Scholar]
- [186].Janowski M, Walczak P, Kropiwnicki T, Jurkiewicz E, Domanska-Janik K, Bulte JW, et al. Long-term MRI cell tracking after intraventricular delivery in a patient with global cerebral ischemia and prospects for magnetic navigation of stem cells within the CSF. PLoS One 2014;9:e97631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Arbab Ali S, Frenkel Victor, Pandit Sunil D, Anderson Stasia A, Yocum Gene T, Bur Monica, et al. Magnetic Resonance Imaging and Confocal Microscopy Studies of Magnetically Labeled Endothelial Progenitor Cells Trafficking to Sites of Tumor Angiogenesis. STEM CELLS 2006;24:671–8. 10.1634/stem-cells.2005-0017. [DOI] [PubMed] [Google Scholar]