Abstract
The global spread of SARS-CoV-2 has proceeded at an unprecedented rate. Remarkably, characterization of the virus using modern tools in structural biology has also progressed at exceptional speed. Advances in electron-based imaging techniques, combined with decades of foundational studies on related viruses, have enabled the research community to rapidly investigate structural aspects of the novel coronavirus from the level of individual viral proteins to imaging the whole virus in a native context. Here, we provide a detailed review of the structural biology and pathobiology of SARS-CoV-2 as it relates to all facets of the viral life cycle, including cell entry, replication, and three-dimensional (3D) packaging based on insights obtained from X-ray crystallography, cryo-electron tomography, and single-particle cryo-electron microscopy. The structural comparison between SARS-CoV-2 and the related earlier viruses SARS-CoV and MERS-CoV is a common thread throughout this review. We conclude by highlighting some of the outstanding unanswered structural questions and underscore areas that are under rapid current development such as the design of effective therapeutics that block viral infection.
1. Introduction
1.1. Overview of the COVID-19 Pandemic
As early as November 2019, initial reports surfaced describing patients presenting with pneumonia-like symptoms in the Guangdong region of China, believed to be the origin of the severe acute respiratory syndrome (SARS)-associated coronavirus (SARS-CoV) in 2003.1−3 In late December 2019, the Wuhan Municipal Health Commission reported a cluster of 27 cases of pneumonia and days later identified a novel coronavirus—now named SARS-CoV-2—as the causative agent of the disease now called COVID-19 (Figure 1).4 Coronaviruses are enveloped, positive-strand RNA viruses, with SARS-CoV-2 part of the β-coronavirus genus containing SARS-CoV and MERS-CoV (the causative viruses of the 2003 SARS and 2012 MERS outbreaks, respectively).1−3,5 The first COVID-19-associated death was reported in China on January 11, 2020, and the genetic sequence of SARS-CoV-2 was published by the Global Initiative on Sharing All Influenza Data (GISAID) the following day (Figure 1).4,6 This genetic sequence revealed that SARS-CoV-2 shares 79% sequence identity with SARS-CoV.6−8 On January 13, 2020 the first case of COVID-19 outside of China was reported in Thailand, and the World Health Organization (WHO) suggested evidence of “limited human-to-human transmission” in a press briefing the following day.4,9 In early February 2020, the first COVID-19 death was reported outside of China, and countries worldwide began reporting cases.4 The WHO characterized COVID-19 as a pandemic on March 11, 2020 (Figure 1), and over the next few years, SARS-CoV-2 would spread globally, infect over 6% of the global population, mutate into more infectious variants of concern, and generally disrupt many aspects of daily life.
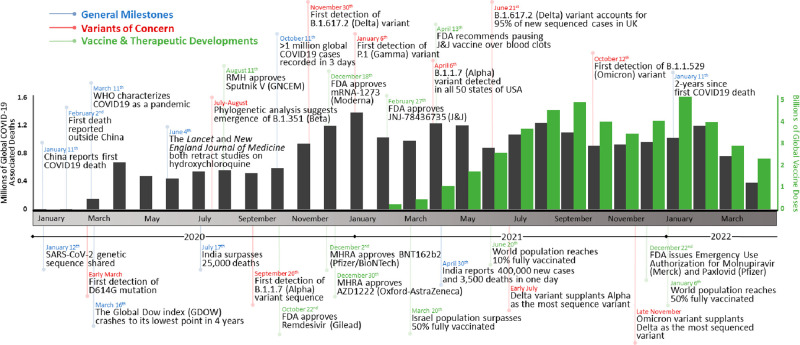
Figure 1.
Timeline of the COVID-19 pandemic. Events are divided into general milestones (blue), variants of concern (red), and vaccine and therapeutic developments (green). The number of global COVID-19-associated deaths (gray) and vaccine doses administered (green) are graphed per month over the course of the COVID-19 pandemic (ref (10)).
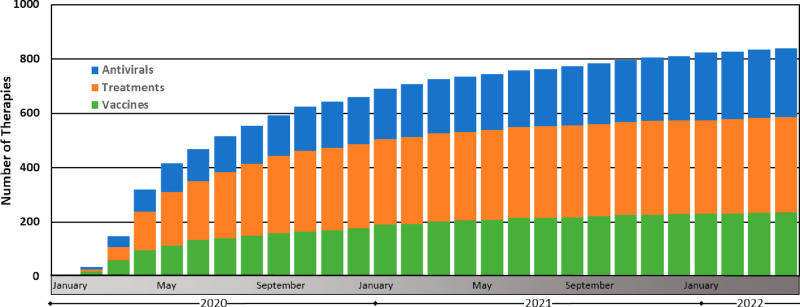
Many diverse therapeutic avenues have been employed to treat COVID-19. Small molecules, convalescent plasma, and biologics (monoclonal antibodies, human recombinant ACE2, and peptides) have all been successfully used as COVID-19 treatments.11−13 However, the majority of these therapeutic regimes are only implemented in mild-to-severe hospitalized cases as measures of treatment following SARS-CoV-2 infection. Relatively early on in the progression of this pandemic, it was clear that population-level vaccination was the most promising prophylactic approach to slow the spread of SARS-CoV-2.14−16 As such, at the time of writing, there are currently 194 and 149 vaccines in preclinical and clinical development, respectively (Figure 2).17 The rate of vaccine development and approval of SARS-CoV-2 vaccines is proceeding at an unprecedented rate, afforded by multiple factors.14,15,18 First, ongoing fundamental research on vaccine development and characterization of pathogens has provided a backdrop upon which to quickly leverage these tools and knowledge to rapidly begin vaccine development.14,15,19 Second, improvements in preclinical and clinical scientific throughput have further accelerated the development rate. Third, the overwhelming widespread need for these vaccines has pushed governmental and conglomerate regulatory agencies to speed their evaluation processes. Finally, the 10 vaccines in phase 4 clinical trials are composed of vastly different vaccine technologies, including nucleic acids, whole virus, and viral vectors, thus permitting a multiangled approach.19,20 These accelerating factors have combined to reduce the typical vaccine-development timeline from 5–10 years to under 1 year for the COVID-19 pandemic.20
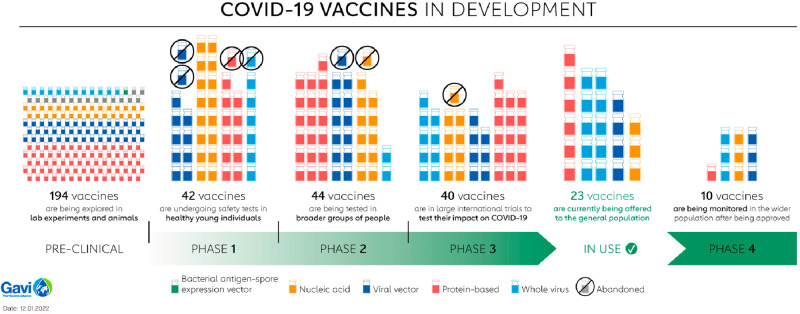
Figure 2.
Summary of SARS-CoV-2 vaccines in development. The 10 vaccines in phase 4 are the following: nucleic acids mRNA-1273 (Moderna), BNT162 (Pfizer/BioNTech), and mRNA-1273.351 (Moderna); viral vectors ChAdOx1 (AstraZeneca/Oxford), Ad5-nCoV (CanSino Biologics), and JNJ-78436735 (Johnson & Johnson); protein-based MVC-COV1901 (Medigen); whole virus CoronaVac (Sinovac), BBIBP-CorV (Beijing Institute of Biological Products), and BIBP (Sinopharm) (refs (17 and 20)). Adapted with permission from ref (20). Copyright 2022 Gavi, the Vaccine Alliance.
Genomic surveillance of SARS-CoV-2 samples during the first year of the COVID-19 pandemic revealed limited mutation.7,8,21 The D614G mutation in the spike (S) protein was the sole widespread consensus mutation, with the G614 genotype largely displacing D614 in March 2020 (Figure 3).7,8,21 In November 2020, however, the emergence of the Alpha (B.1.1.7) variant began capturing global headlines and coincided with a surge in COVID-19 cases in the United Kingdom. Within 4 months, the Alpha variant became the dominantly sequenced SARS-CoV-2 lineage worldwide (Figure 3).7,8 Emergence of the Alpha lineage was quickly proceeded by the emergence of the Beta (B.1.351), Gamma (P.1), Delta (B.1.617.2), Kappa (B.1.617.1), and Epsilon (B.1.429) variants in early 2021 (Figure 3). Most of these variants were classified as variants of concern (VoCs) by the WHO, demonstrating (a) increased transmissibility or detrimental changes in COVID-19 epidemiology, (b) increased virulence or changes in clinical disease presentation, and/or (c) decreased effectiveness of public health and social measures or available diagnostics, vaccines, and therapeutics.22 Finally, in late 2021, the Omicron (B.1.1.529) variant—which contained an unprecedented number of mutations—rapidly supplanted the Delta variant as the most sequenced variant worldwide. Given that most current SARS-CoV-2 vaccine immunogens and testing reagents are based on the original Wuhan-1 reference sequence, the mutations present in emergent VoCs warrant urgent investigation to assess their consequences on vaccine efficacy and the SARS-CoV-2 life cycle.7,8
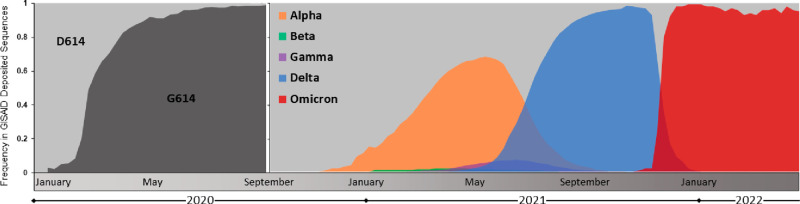
Figure 3.
Emergence and global prevalence of the D614G and variant of concern lineages of SARS-CoV-2. Sequence data was downloaded from the Global Initiative on Sharing All Influenza Data (GISAID) and graphed as weekly totals (refs (7 and 8)). D614 and G614 genotype prevalence is shown from January to September 2020, and variant of concern lineage prevalence is shown from September 2020 to April 2022.
1.2. Structural Biology Perspective of SARS-CoV-2
Scientists have never before been so readily positioned to quickly answer critical questions about the three-dimensional (3D) arrangement of emergent viruses. As highlighted in the above section, the COVID-19 pandemic has evoked unprecedented speeds of response as researchers aim to understand and combat the spread of the virus. Our cumulative knowledge and ever-advancing scientific toolbox—garnered by studying other recent viral pandemics—has fueled the swift unravelling of the SARS-CoV-2 viral life cycle. Recent epidemics such as MERS (2012–2015), Ebola (2014–2016), Zika (2015–2016), and Dengue (2019–2020) have prepared us to quickly gain an understanding of emergent viruses. Joachim Frank highlights how these pandemics have coincided with the “resolution revolution” in cryo-electron tomography (cryo-ET) and cryo-electron microscopy (cryo-EM) imaging (afforded largely by improved electron detectors), allowing researchers to more finely resolve the arrangement of these virions and the proteins that compose them.23
No single imaging technique paints a full picture of SARS-CoV-2 biology given the vast biological size scale across which viral infection, replication, and packaging takes place. Rather, the aggregation of results across the imaging size spectrum allows for a more comprehensive characterization. Cryo-ET involves the flash-freezing of a biospecimen (a virus, cell, tissue, or protein), imaging the sample through a tilt-series using an electron microscope, and finally aligning and merging the images using computational techniques to reconstruct a 3D image.24 Cryo-ET provided the first images of the SARS-CoV-2 virion and how the S and nucleocapsid (N) proteins were arranged on the viral surface and within the viral lumen, respectively. Similar to cryo-ET, single-particle cryo-EM generally involves the flash-freezing of a biospecimen (individual proteins or protein complexes), collecting images or movies of the vitrified sample using an electron microscope, and aligning and merging the images to produce a 3D image.25 Cryo-EM provided the first structures of the spike glycoprotein and actively replicating SARS-CoV-2 RNA polymerase complex.26,27 As there exists a theoretical minimum protein-size limit for high-resolution cryo-EM (∼40 kDa), under which many of the nonstructural SARS-CoV-2 proteins lie,28,29 X-ray crystallography has been used extensively to determine the structures of these small (<40 kDa) crystallizable proteins. Here, isolated protein crystals are diffracted with an incident X-ray beam, and the resulting diffraction pattern produces a 3D electron density map using Fourier transforms.30 The combination of these three structural techniques (cryo-ET, cryo-EM, and X-ray crystallography), each with their inherent strengths and limitations, has yielded us a rapid 3D understanding of SARS-CoV-2 from atomic details of viral replication machinery to visualizing entire viral particles being packaged and trafficked within a human cell.
The first structure of a SARS-CoV-2 protein, the main protease (Mpro/NSP5), was reported mere weeks after the global sharing of the viral sequence (Figure 4A).31 This initial atomic structure—in complex with an inhibitor—permitted the rational design of improved and specific protease inhibitors that may help to treat COVID-19.31−33 Additionally, the reporting of structural impacts imparted by the many mutations within VoC S proteins provides us with insights into how they may effect vaccine efficacy. These insights are particularly important given the global dominance of VoCs and the fact that the majority of vaccines in development use the S protein as their sole protein immunogen.7,8,17 The Mpro and spike proteins are just two examples of how structural biology is being employed to help researchers gain a better understanding of SARS-CoV-2.
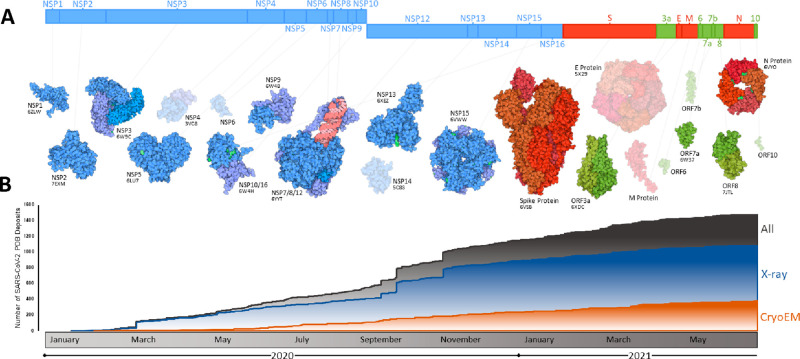
Figure 4.
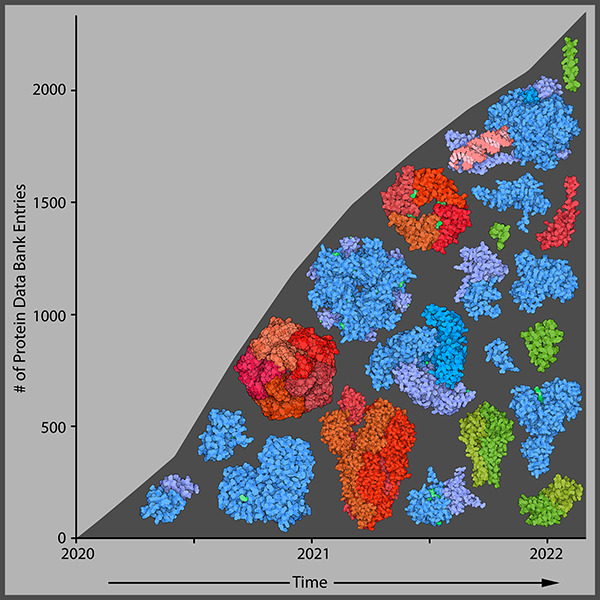
Summary of progress toward SARS-CoV-2 structural characterization. (A) Schematic of the SARS-CoV-2 genome with structurally characterized proteins indicated in full color. Proteins that have not yet been characterized are displayed through homology modeling and are shown as semitransparent images (NSP4/6/12, E protein, M protein). Protein illustrations were generated using Illustrate (ref (34)). (B) The number of X-ray crystallography, cryo-EM, and all SARS-CoV-2 protein structures deposited into the RCSB protein data bank (PDB) over the first 18 months of the COVID-19 pandemic (ref (35)).
1.3. Scope and Organization of This Review
Researchers worldwide have worked at great pace to unravel the 3D architecture of the SARS-CoV-2 virion and the atomic arrangement of its proteome. From depositing the first structure of the main protease (6LU7), just 2 weeks following the global sharing of the SARS-CoV-2 genome, to the first S protein structure deposited only 2 weeks later (6VSB), the structural characterization of SARS-CoV-2 has developed at a truly unprecedented rate.26,31 Herein, we aim to summarize these results in the context of how they inform our understanding of the SARS-CoV-2 life cycle.
The driving theme of this review is the 3D visualization of the SARS-CoV-2 life cycle, including how the arrangement of the virion, cell entry, replication, packaging, and release are orchestrated in 3D space. We strive to integrate results across the 3D visualization spectrum (X-ray crystallography, single-particle cryo-EM, cryo-ET, and molecular dynamics) and describe both a general overview and specific interesting themes throughout. Structural differences between SARS-CoV-2 and other previously emerged viruses (SARS-CoV, MERS-CoV) will additionally be highlighted. While fundamental chemical, molecular, clinical, and cellular biology have each provided crucial information in our characterization of this virus, this review will not delve deeply into these topics; rather, we will use the findings of these fields to contextualize the structural biology described herein.
This review is arranged into three main sections, each providing both a broad overview and specific insights into various processes within the SARS-CoV-2 life cycle. The initial section (Structure of the Virion) describes the 3D architecture of the viral particle and largely summarizes results obtained from cryo-ET. Specific themes in this section include the arrangement and conformations of the spike glycoprotein on the surface of the virion and how the nucleocapsids are organized within the viral particle. The next section (Entry into the Cell) examines the molecular mechanisms by which SARS-CoV-2 transits the plasma membrane to deposit its genome into host cells. Specific themes in this section include the conservation of glycosylation across coronavirus spike proteins (SARS-CoV-2, MERS-CoV, and SARS-CoV) and the spike protein–ACE2 interaction. The following section (Replication, Packaging, and Release) compiles the numerous structures of proteins encoded by the SARS-CoV-2 genome and how they come together to orchestrate replication, packaging, and release of the viral particles. Here, X-ray crystallography and cryo-EM structures of individual and complexed proteins and cryo-ET imaging of the packaging process in vivo combine to visualize these complicated final steps in the viral replication cycle. Our final section (Future Prospects) aims to highlight yet to be addressed areas of SARS-CoV-2 pathobiology that warrant continued structural investigation.
2. Structure of the Virion
2.1. Brief Overview of the SARS-CoV-2 Viral Structure
Like all coronaviruses, the SARS-CoV-2 viral particle is composed of proteins, nucleic acids, and lipids that are assembled within host cells.36 The viral envelope is derived from the membrane of the endoplasmic reticulum and is studded with membrane (M), envelope (E), and S structural proteins (Figure 5). M is the most abundant envelope protein in coronaviruses and is a critical structural component that facilitates budding and defines the shape of the viral particle.37,38 As such, the M protein is considered the central organizer of the viral envelope, as it contacts and coordinates all other structural proteins (E, S, and N).39,40 The S protein facilitates both attachment to and entry into host cells by binding the angiotensin-converting enzyme 2 (ACE2) receptor, as covered in greater detail in the next section (Entry into the Cell). E is the smallest of the structural proteins with crucial, yet currently ill-defined, mechanistic roles.41 As a viroporin, E is a hydrophobic protein that oligomerizes in the membrane of host cells, forming hydrophilic pores that precipitate membrane remodelling and viral packaging.42 Recombinant coronaviruses lacking the E protein exhibit hampered viral titers and yield propagation-incompetent progeny, demonstrating the critical nature of this structural protein in viral replication.41,43−45 These three structural proteins (M, S, and E) define the viral envelope which encapsulates an ∼30 kb viral genome. The SARS-CoV-2 genome is composed of positive-sense single-stranded RNA (ssRNA) that associates with hundreds of copies of the fourth and final structural protein, the N protein (Figure 5).46,47 The ribonucleoprotein (RNP) complex of ssRNA and N protein compresses and packages the viral genome within the virion, coordinated by interactions between the N and M proteins (covered in greater depth in section 4, Replication, Packaging, and Release).40,48−51 This brief overview of SARS-CoV-2 structural proteins leverages decades-long investigations of related coronaviruses (SARS-CoV and MERS-CoV) and is reviewed in greater depth by Schoeman and Fielding and Mariano et al.41,52 Herein, we highlight several recent studies that report in situ evidence for the overall 3D arrangement of the SARS-CoV-2 virion.
Figure 5.
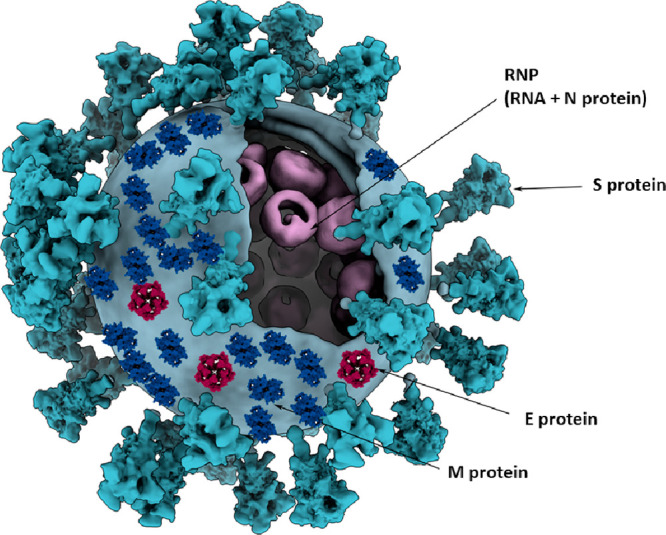
Three-dimensional model of a coronavirus particle. Membrane (M), spike (S), envelope (E), and nucleocapsid (N) structural proteins are shown. Models for E and M proteins were obtained from https://sars3d.com/ and were manually (not experimentally) arranged on the surface of a 3D model of the virion rendered using EMD-30430. Adapted with permission from ref (47). Copyright 2020 Elsevier.
2.2. Spike Protein Distribution within the Viral Envelope
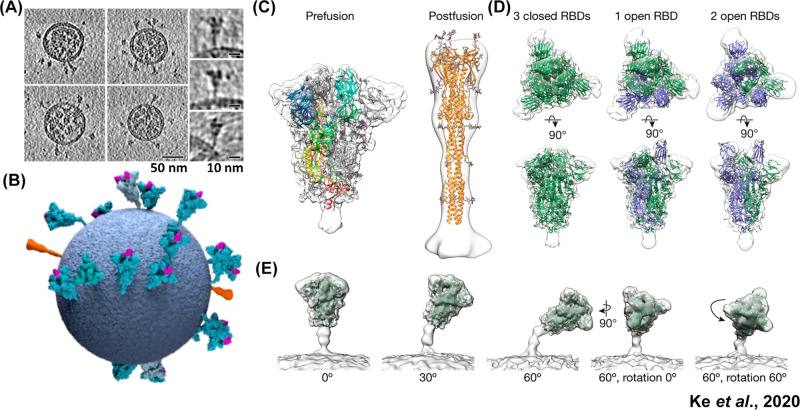
Multiple groups have leveraged cryo-ET to uncover the overall arrangement of authentic SARS-CoV-2 viral particles.47,53,54 Ke et al. reported roughly spherical particles, with an average outer diameter of 91 ± 11 nm, while Yao et al. reported both spherical and ellipsoidal shaped viruses, with dimensions of 64.8 ± 11.8, 85.9 ± 9.4, and 96.6 ± 11.8 nm for the short, medium, and the long axes of the ellipsoid envelope, respectively (Figure 6A).47,53 Both of these results are consistent with the diameter of the SARS-CoV virion (∼85 nm), which was determined in 2008 by Neuman et al., also by cryo-ET methods.40 Each SARS-CoV-2 virion contains roughly 15–40 S proteins randomly distributed across the viral surface, with the vast majority (97%) of S proteins adopting the prefusion conformation (Figure 6, parts B and C).53 Notably, SARS-CoV was previously determined to have roughly 90 S proteins per virion, with fewer S proteins potentially providing a viral fitness advantage given the immune susceptibility of this protein in viral neutralization.40 This modeling of intact SARS-CoV-2 virions approximates that there is one spike protein per 1000 nm2 of membrane surface, in contrast to approximately one hemagglutinin per 100 nm2 for the influenza A virus.55 This ∼10-fold decrease in S protein density suggests that S protein–ACE2 receptor binding may be less dependent on the avidity effects as seen in influenza A. This finding is additionally consistent with the nanomolar and millimolar affinities for the S protein–ACE2 (SARS-CoV-2) and hemagglutinin–sialic acid (influenza A) interactions, respectively.53 A further cryo-ET study by Liu et al. employed β-propiolactone-inactivated SARS-CoV-2 viruses and found that this chemical inactivation drastically shifted the spike proteins toward the postfusion conformation (∼74%), thus altering their antigenic profile.56 This finding is particularly relevant as chemical inactivation of pathogens is one of the most common vaccine strategies, with β-propiolactone used in current SARS-CoV-2 vaccine formulations.56−59
Figure 6.
Spike protein distribution, conformations, and tilt angles in authentic SARS-CoV-2 virions. (A) Tomographic slices of four representative SARS-CoV-2 virions and side projections of three individual S proteins. (B) Three-dimensional model of a single SARS-CoV-2 virion derived from subtomogram averaging. Prefusion S proteins are colored in blue with up RBDs colored pink. Postfusion S protein densities are colored in orange. (C) Prefusion and postfusion S protein trimer densities obtained by subtomogram averaging and fitted with PDBs 6VXX and 6XRA, respectively. (D) Prefusion trimer conformations as observed on intact virions. The densities corresponding to three closed, one open, and two open RBDs are fitted with PDBs 6VXX, 6VYB, and 6X2B, respectively, with protomers containing up RBDs colored in blue. (E) Averaging of trimer subsets is shown for pools centered at 0°, 30°, and 60° from the normal, as well as for two rotations of the S protein relative to the tilt direction. Adapted with permission from ref (53). Copyright 2020 Ke et al. http://creativecommons.org/licenses/by/4.0/.
These cryo-ET studies provide moderate resolution (7–8 Å) 3D reconstructions of pre- and postfusion S proteins. These resolutions enable the assignment of receptor binding domain (RBD) “open” versus “closed” states and in situ validation of higher resolution soluble ectodomain structures. Only recently was the first structure of the SARS-CoV S protein reported, which revealed the requirement for the RBD to adopt an open or “up” conformation before engaging the ACE2 receptor.60 The in situ cryo-ET classification of SARS-CoV-2 S proteins revealed three distinct states: (1) all three RBDs in the closed or “down” conformation, (2) one RBD in the open conformation, and (3) a small fraction with two RBDs in the open conformation, which has also been observed in various ectodomain structures (Figure 6D).53 Overall, these in situ S protein structures were found to be very structurally similar to soluble and recombinantly produced ectodomain structures. Three hinge points present in the stalk of the prefusion S protein afford a high degree of flexibility relative to the viral membrane, and accordingly, S proteins were found to adopt a wide range of tilt angles, with a mode of 40° from normal (Figure 6E).47,54 Interestingly, this flexibility was not observed in postfusion S proteins, and it has therefore been proposed that rigid postfusion S proteins anchor the viral particle into the cell membrane, while unbound flexible prefusion S proteins are able to “scan” and bind additional ACE2 receptors, therefore contributing to avidity effects.54
2.3. Packing of the Viral Contents
A well-defined molecular model for how coronaviruses compress ∼30 kb RNA genomes into an 80 nm diameter viral lumen remained elusive prior to the emergence of SARS-CoV-2. The N protein binds genomic RNA to form the RNP core and is the basic unit of genome packing.46,47,61,62 X-ray crystal structures of the individual structured domains of the N protein were published months after the emergence of SARS-CoV-2 and defined the location of the viral RNA’s binding site. These structures revealed relatively conserved N protein structures compared to other reported coronaviral N proteins and defined the electrostatic interactions between RNA and the N protein N-terminal domain (NTD).62 The structure of the C-terminal domain (CTD) of the N protein showed that it dimerizes in a highly conserved manner relative to SARS-CoV and tetramerizes through a conserved spacer domain.51 The synthesis of these domain-specific structural insights describes a protein that binds RNA at its NTD and oligomerizes to facilitate packing via its CTD. Notably, these in vitro studies were likely complicated given the recently demonstrated tendency of the SARS-CoV-2 N protein to undergo liquid–liquid phase separation following RNA binding.48−50,63,64
These in vitro X-ray crystallography observations were further corroborated by in situ cryo-ET studies of authentic SARS-CoV-2 viruses, wherein visualization of higher order RNP packing is possible (Figure 7A).47 A majority of RNPs were found to be membrane-proximal, which is consistent with the previously described interaction between N and M proteins.46,47,61,62 Following 3D refinement, two distinct RNP ultrastructure assemblies emerged; the first is a membrane-proximal “hexon” assembly in the shape of “eggs in a nest”, and the second is a membrane-free “tetrahedron” assembly in the shape of a “pyramid” (Figure 7B).47 A portion of the hexon-assembled RNPs simultaneously participate in tetrahedron assemblies, suggesting that the RNP pyramid is the fundamental genome packing unit in SARS-CoV-2. Backprojection of these distinct RNP assemblies onto their viral coordinates reveals a 2-fold increase in the proportion of pyramid assemblies in ellipsoid virions compared to spherical virions (Figure 7C).47 Whether differential RNP assemblies play a driving role in viral morphology, or whether virion shape simply favors different RNP assemblies, remains to be uncovered.
Figure 7.
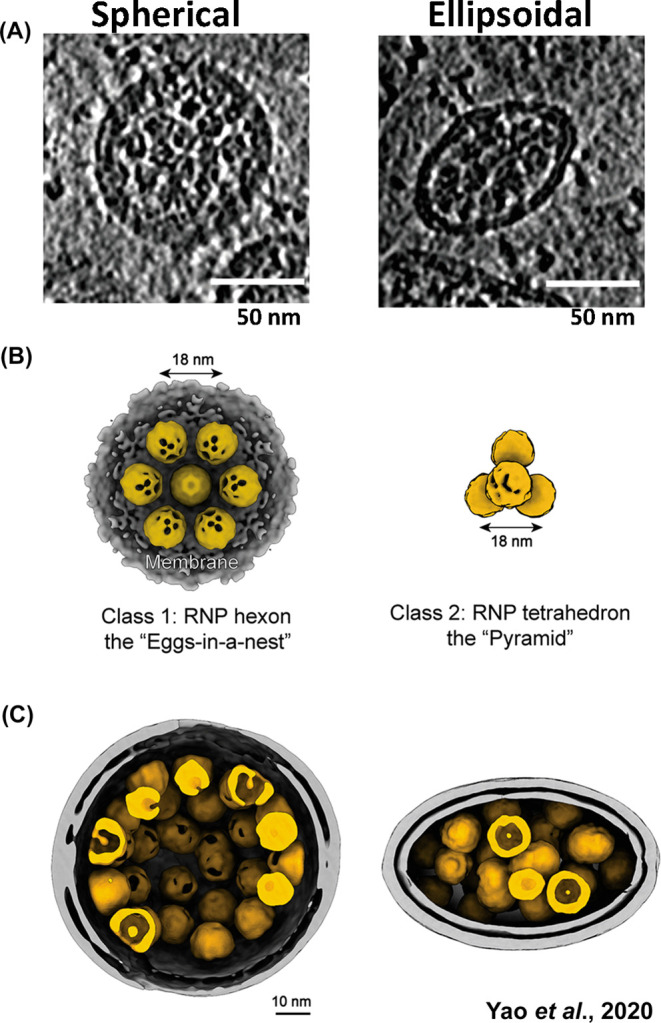
Packing of the ribonucleoprotein (RNP) complex within spherical and ellipsoidal SARS-CoV-2 particles. (A) Representative tomogram slices (5 Å thick) of spherical and ellipsoid viral particles. RNPs are visible as granular densities within the viral lumen. (B) Hexon and pyramid in situ ultrastructure reconstructions of the RNP. There was an approximately 2-fold increase in pyramid RNP reconstructions in ellipsoid viruses compared to spherical viruses. (C) Representative RNP packing arrangements in spherical and ellipsoid SARS-CoV-2 virions. Adapted with permission from ref (47). Copyright 2020 Elsevier.
3. Entry into the Cell
3.1. Overview of SARS-CoV-2 Cell Entry
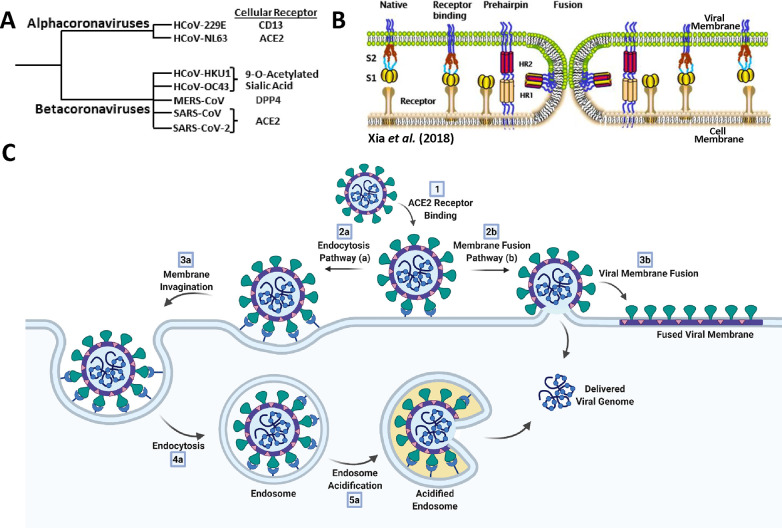
Provided a viral particle has evaded innate and adaptive immune responses, the first step in viral infection is attachment and subsequent entry into host cells. Cellular attachment by the SARS-CoV-2 viral particle exploits the same receptor, ACE2, as the related coronaviruses SARS-CoV and HCoV-NL63 (Figure 8A).65−67 The viral particle binds the ACE2 receptor via the RBD of its S protein, with the first structures of this interaction reported by Shang et al. and Lan et al. in March 2020.68−70 The S protein is composed of an N-terminal S1 subunit that mediates cell attachment and a C-terminal S2 subunit which facilitates fusion of the viral and host membranes (Figure 8B).69,71,72 During expression and processing of the S protein, it is cleaved at the S1/S2 boundary, but the domains remain associated through noncovalent interactions. Upon binding ACE2, the SARS-CoV-2 viral genome may transit the cellular plasma membrane by two distinct entry mechanisms: (1) the membrane fusion pathway (early pathway) or (2) the endocytosis pathway (late pathway) (Figure 8C; comprehensively reviewed by Tang et al. and others).73,74 The membrane fusion pathway involves cleavage of the S protein at its S2′ boundary by the TMPRSS2 host protease, followed by dissociation of the S1 subunit, leaving S2 exposed. Through a highly conserved (across the coronavirus family and in the HIV gp41 protein), yet structurally uncharacterized, mechanism the S2 domain unfolds to adopt its postfusion conformation and extends into the host-cell plasma membrane.71,75,76 The postfusion S protein then ratchets the two membranes together, again by a structurally uncharacterized process that results in membrane fusion (Figure 8B). In contrast, the late endocytosis pathway does not reply upon TMPRSS2 cleavage and exploits the host cells’ innate endocytosis process to transit the plasma membrane (Figure 8C). Prolonged attachment of the virus at the exterior of the cell triggers receptor-mediated endocytosis of the viral particle. The particle is invaginated into an endosome, wherein acidification activates cathepsin L and other host proteases, which cleave at the S protein S2′ site, resulting in endosome–viral membrane fusion.74 These two distinct cell entry pathways both result in deposition of the SARS-CoV-2 genome into the cytosol to precipitate viral replication.
Figure 8.
Overview of coronavirus cell-entry mechanisms. (A) Members of the α- and β-coronavirus genera and their major associated cellular receptors. (B) Model of coronavirus receptor-mediated membrane-fusion mechanism between viral and cellular membranes. Adapted from ref (72). Copyright 2018 Xia et al. http://creativecommons.org/licenses/by/4.0/. (C) Endocytosis (a) and membrane fusion (b) pathways of coronavirus cell entry. Created with BioRender.
3.2. S Protein–ACE2 Interaction
As described above, both the membrane fusion and endocytosis pathways of SARS-CoV-2 cellular infection critically rely upon association of the S protein with the ACE2 receptor. This interaction was first predicted in January 2020 by Wan et al., wherein the authors leveraged decade-long structural studies of SARS-CoV.77 Using homology modeling to gain insights into the yet to be validated SARS-CoV-2 S protein–ACE2 interaction, the authors emphasized that the N501 residue was not ideal for binding human ACE2 and that “2019-nCoV [SARS-CoV-2] evolution in patients should be closely monitored for the emergence of novel mutations at the 501 position”. Therefore, early structural biology insights enabled the prediction of a mutation—N501Y—that would replace N501 as the dominantly sequenced genotype over a year later (January 2021, see the Introduction, Figure 3).7,8 Following this prediction, experimental evidence by Hoffmann et al. and others confirmed that SARS-CoV-2 cell entry is dependent upon ACE2.78−80 Additionally, recent reports have found overexpression of specific lectins (DC-/L-SIGN and SIGLEC1) to enhance SARS-CoV-2 infectivity, potentially implicating the heavily glycosylated S protein NTD in viral particle attachment.81,82
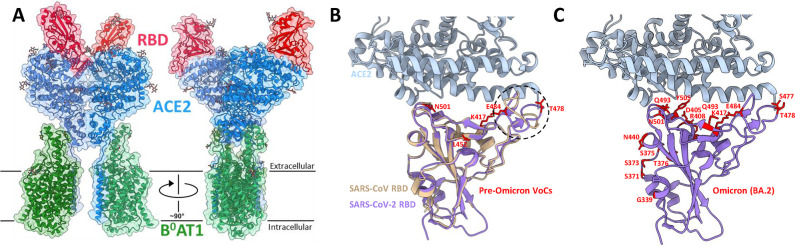
Following the structurally predicted and experimentally validated SARS-CoV-2 S protein–ACE2 interaction, Lan et al. and Yan et al. solved structures of the ACE2 receptor in complex with the S protein RBD by X-ray crystallography and cryo-EM methods, respectively.69,70 The structure by Yan et al. included the ACE2-associated B0AT1 protein and suggested that two S protein trimers are able to simultaneously bind the ACE2 homodimer (Figure 9A).69 Both studies discussed the SARS-CoV-2 RBD–ACE2 interaction in the context of the SARS-CoV interaction and concluded that the interaction is structurally similar; however, several small sequence and conformational variations are present in the respective ACE2 interfaces. The higher resolution X-ray structure allowed for the conclusion that there are subtle rearrangements within the SARS-CoV-2 receptor binding motif (RBM; the portion of the RBD that forms the interface with ACE2) that cause the RBM ridge to become more compact and form better contacts with the N-terminal helix of ACE2 (circled in Figure 9B).70 The synthesis of structural and biochemical data reported by these groups and others revealed that the SARS-CoV-2 RBD recognizes and binds ACE2 better than the SARS-CoV RBD.26,68−70,80 The interplay of these two structures, the X-ray structure illuminating subtle rearrangements at the ACE2-RBM interface and the cryo-EM structure yielding a more global picture of S protein binding relative to the membrane plane, combined to provide an early understanding of how SARS-CoV-2 particles attach to our cells. These structures additionally provided the basis for structure-based rational design of neutralizing binders with enhanced affinities to either ACE2 or the S protein.
Figure 9.
Structural insights into the SARS-CoV-2 S protein–ACE2 interaction. (A) Cryo-EM structure of the SARS-CoV-2 RBD–ACE2–B0AT1 protein complex reported by Yan et al. (6M17). The complex is shown as a colorized ribbon model and molecular surface with the RBD, ACE2, and B0AT1 shown in red, blue, and green, respectively. (B) Superposition of ACE2-complexed SARS-CoV (2AJF, brown) and SARS-CoV-2 (6M0J, purple) RBDs aligned by the RBD (refs (70 and 93)). The ACE2 structure for the SARS-CoV-2 complex is shown alone to simplify the RBD–ACE2 interface. The major structural discrepancy between the SARS-CoV and SARS-CoV-2 RBDs is circled with a black dotted line. The side chains of residues mutated in variants of concern (prior to the Omicron variant) are shown and labeled in red. (C) The same as in panel B, but mutated residues are shown for the Omicron BA.2 variant.
Mutations within the S protein have been the major focus of the structural characterization of emergent SARS-CoV-2 variants, with several RBD mutations shared between multiple VoCs (Alpha, Beta, Gamma, Delta, and Omicron). Figure 9B demonstrates that these VoC mutations localize to the RBD–ACE2 interface and therefore may elicit effects on ACE2 binding affinity. Indeed, the combination of these mutations (N501Y, E484 K/Q, L452R, T478 K) have been biochemically implicated in increasing ACE2 binding affinity.83−88 The N501Y mutation was structurally demonstrated to insert into a cavity at the ACE2 binding interface and form a perpendicular π–π stacking interaction with Y41.83 This additional interaction likely underlies the increased ACE2 affinity afforded by the N501Y mutation and rationalizes its presence in the Alpha, Beta, Gamma, and Omicron VoCs. In contrast to these mutations that enhance ACE2 affinity, the ambiguous mutation of residue K417 to either T or N (K417T/N) uniquely decreases the ACE2 binding affinity.84,85,89 Accordingly, the K417N/T mutations are not significantly prevalent in the absence of N501 or E484 mutations, which likely compensate for the loss in ACE2 affinity.7,8 The structural rationale for this decreased ACE2 affinity by K417N/T is the loss of the K417–D30 salt bridge that spans the ACE2–S protein complex. K417N potently escapes neutralizing antibodies, justifying its inclusion in these variants, despite the imparted penalty on ACE2 affinity. The recently emerged Omicron variant contains over 3 times the number of S protein mutations relative to any other previously emerged VoC, with many mutations localizing to the RBD–ACE2 interface (Figure 9C).90 These numerous mutations again balance ACE2 binding affinity and afford unprecedented escape from convalescent and vaccine-induced antibodies, likely rationalizing the rapid replacement of the Delta variant by Omicron in late 2021.90
Mutations elsewhere within the S protein, such as the omnipresent D614G mutation and the A570D and S982A mutations in the Alpha variant, have also been implicated in increasing ACE2 affinity through allosteric mechanisms including influencing the propensity of the RBD to occupy the up or open conformation.91,92 Additionally, these variants are defined by mutations within other viral proteins which have been superficially structurally characterized relative to the S protein mutations. The antigenic dominance of the S protein and its inclusion as the sole protein antigenic component of all approved COVID-19 vaccines rationalizes this hyper focus on S protein mutation.
3.3. Glycosylation of the S Protein
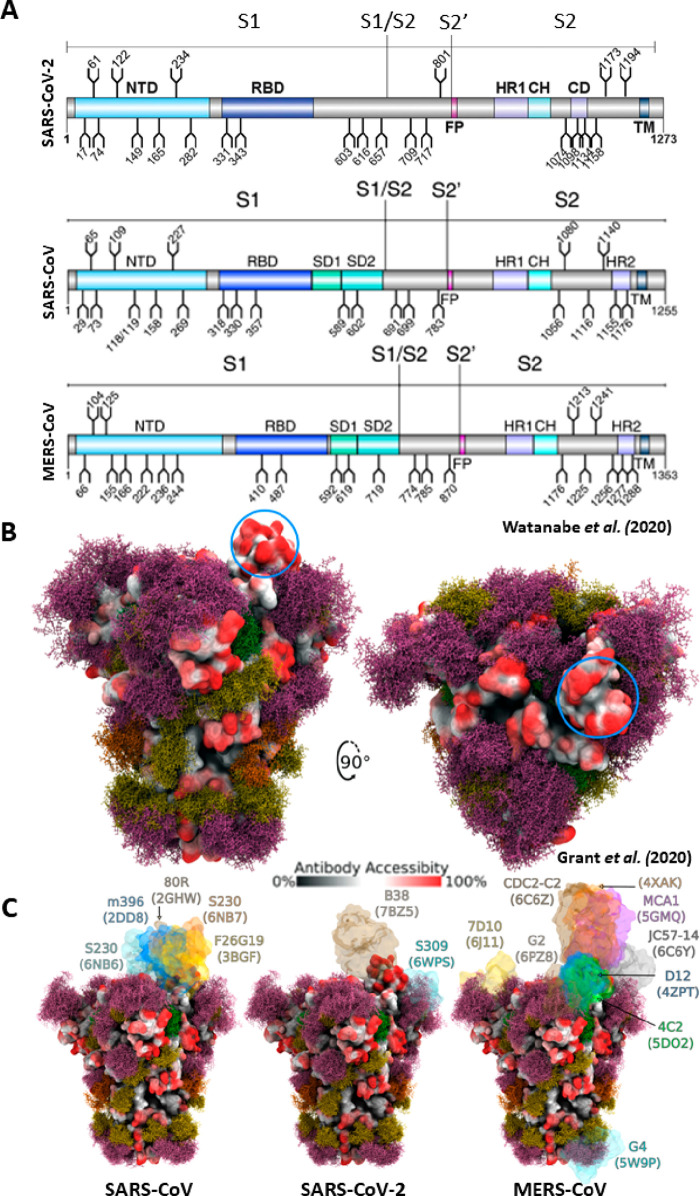
Complementary structural and mass spectrometry analyses of the SARS-CoV-2 S protein confirmed that, like the related proteins from SARS-CoV and MERS-CoV viruses, the SARS-CoV-2 S protein is also extensively glycosylated.94,95 Glycosylation has a myriad of roles in viral pathobiology including shielding vulnerable neutralizing epitopes, shaping viral tropism, and mediating S protein folding and stability.94−100Figure 10A shows that there are 22, 22, and 23 glycosylation sites in the SARS-CoV-2, SARS-CoV, and MERS-CoV S proteins, respectively, with glycosylation preferentially localizing to the NTD, S1/S2 boundary, and stem helix of the S2 fusion domain.94,95 Using the first reported structure of the SARS-CoV-2 spike protein (6VSB), Grant et al. generated 3D structures of the S protein glycoforms and subjected them to molecular dynamic (MD) simulations to determine the antibody-accessible surface area.101 Despite only accounting for 17% of the molecular weight of the S trimer, Grant et al. found that the glycans shield approximately 40% of the S protein surface (Figure 10B). The most exposed protein epitope comprises the ACE2 receptor site of the RBD in the up or open conformation (indicated by the blue circle in Figure 10B). The RBD has been demonstrated to present the antigen against which the vast majority (∼90%) of patient-derived neutralizing antibodies bind.102 Therefore, the exposure of the RBD in the up position due to lack of glycan shielding is likely a vulnerability necessitated by the crucial requirement for the S protein to bind the ACE2 receptor. Taking glycan microheterogeneity into account, the authors further conclude that variations in glycan identity may affect local structural fluctuation at either the protein or glycan level, which may influence S protein function and stability.101
Figure 10.
Glycosylation of the SARS-CoV-2, SARS-CoV, and MERS-CoV spike proteins. (A) Schematic representation of the SARS-CoV-2, SARS-CoV, and MERS-CoV protein open reading frames with glycosylation sites indicated (refs (94 and 95)). Adapted with permission from ref (95). Copyright 2020 Watanabe et al. http://creativecommons.org/licenses/by/4.0/. (B) Moss surface representation of SARS-CoV-2 S protein glycosylation from molecular dynamic simulations performed by Grant et al. (ref (101)). Glycans are shown in ball-and-stick representations and colorized accordingly: M9, green; M5, dark yellow; hybrid, orange; complex, pink. The S protein surface (6VSB) is colored according to antibody accessibility from black (least accessible) to red (most accessible). The RBD in the up conformation is circled in blue. Adapted with permission from ref (101). Copyright 2020 Grant et al. http://creativecommons.org/licenses/by/4.0/. (C) The same as in panel B, but for SARS-CoV, SARS-CoV-2, and MERS-CoV S proteins and with available S protein–antibody structures overlapped.
Given the high degree of structural similarity between the SARS-CoV, MERS-CoV, and SARS-CoV-2 S proteins, comparison between their respective glycan shields, and therefore their impact on antibody accessibility, was possible. Figure 10C presents a side-by-side comparison of the glycan shield and antibody-accessible area of these three viral S proteins, with neutralizing antibody co-complexes superimposed. From this analysis and various reports mapping the epitopes of patient-derived neutralizing antibodies, it can be concluded that the majority (∼90%) of neutralizing antibodies map to the RBD of the SARS-CoV-2 S protein.102−104 Additionally, these simulated structures show a remarkable degree of epitope conservation among the SARS-CoV, MERS-CoV, and SARS-CoV-2 coronavirus S proteins. The strong correlation between the predicted gaps in the S protein glycan shield and the observed antibody binding sites highlights the importance of these epitopes in the elicitation of neutralizing antibodies by therapeutics such as vaccines. This point may underlie the hampered (decades-long) efforts to develop successful vaccines incorporating the even more densely glycosylated HIV-1 Env and gp120 proteins.101,105
4. Replication, Packaging, and Release
4.1. Overview
Our understanding of the SARS-CoV-2 life cycle after it enters a host cell has greatly benefited from the ability to “peer” into infected cells using numerous 2D and 3D imaging technologies. Collectively, these snapshots highlight the extensive spatial and temporal coordination employed during replication, packaging, and release of newly formed viral particles, where each step takes place in discrete but highly coordinated cytoplasmic compartments.
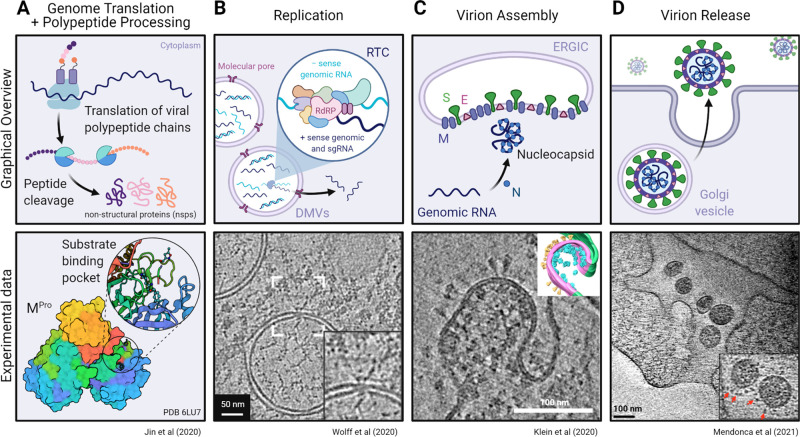
Initial translation of the ∼30 kb long positive-sense SARS-CoV-2 genome by host ribosomes occurs in the cytoplasm (Figure 11A).106−108 Beginning at a single ribosome entry site and exploiting ribosomal frame-shifting, two large polyproteins (pp1a and pp1ab) are translated from the first two-thirds of the genome.108−112 These polyproteins encode 16 individual nonstructural proteins (NSPs), many of which function as components of the replication–transcription complex (RTC) responsible for viral RNA synthesis.111,113−116 Overall, the SARS-CoV-2 NSPs share 86% sequence identity with the NSPs produced by SARS-CoV.117 The first four NSPs (NSPs 1–4) are cleaved by the viral protease encoded by NSP3 (PLpro).118−120 The remaining NSPs (NSPs 5–16) are cleaved by the main viral protease, NSP5 (Mpro, also called 3CLpro).31 Mpro undergoes autolytic cleavage from the polyprotein, and then assembles as an asymmetric dimer (Figure 11A, bottom panel) before cleaving the remaining downstream NSPs.31 The Mpro substrate-binding pocket is highly conserved in all β-coronaviruses and contains a Cys-His catalytic diad located in a cleft formed between two adjacent protein domains.31 Since the cleavage recognition sequence for Mpro is distinct from that of human proteases, and given its high conservation and functional importance in coronavirus replication, Mpro has become a prominent target for therapeutic development.11,31,33,121−124
Figure 11.
Overview of RNA translation and replication, viral packaging, and release of the SARS-CoV-2 virion. Schematic representations (top) and experimental data (bottom) of the cellular machinery and viral proteins involved in (A) genome translation and initial polypeptide processing, (B) replication of genomic and subgenomic RNA, (C) assembly of the virion at the ER–Golgi intermediate compartment (ERGIC), and (D) final egress of the viral particle into the extracellular environment. (A, bottom) Mpro dimer surface model (6LU7) (ref (31)) colored by chain and the substrate binding pocket (inset) depicting the bound Mpro inhibitor N3 (sticks). Experimental data from panels B–D show tomographic slices from cryo-ET studies of (B) murine hepatitis virus (MHV) or (C and D) SARS-CoV-2-infected cells highlighting the transport of RNA through a molecular DMV pore, budding of a SARS-CoV-2 virion, and a viral exit tunnel, respectively. Panel B was adapted with permission from ref (138). Copyright 2020 Wolff et al. http://creativecommons.org/licenses/by/4.0/. Panel C was adapted with permission from ref (125). Copyright 2020 Klein et al. http://creativecommons.org/licenses/by/4.0/. Panel D was adapted with permission from ref (139). Copyright 2021 Mendonça et al. http://creativecommons.org/licenses/by/4.0/. Created with BioRender.
The remaining stages of replication require extensive spatial reorganization of the cytoplasm to sequester RNA synthesis within viral replication organelles (vROs). These vROs have been extensively studied using transmission electron microscopy (TEM) of stained plastic sections, serial cryo-focused ion beam (FIB)/scanning electron microscopy (SEM), and cryo-ET, which all reveal a perinuclear network of interconnected membrane compartments created by reorganization of the rough endoplasmic reticulum (ER) (Figure 11B).54,56,125−132 This network is predominantly made up of double-membrane vesicles (DMVs) and is induced by the combined action of NSP3, NSP4, NSP6, and various host factors.126,133,134 This spatial segregation is thought to have several beneficial effects for viral replication, as sequestering RNA synthesis into DMVs may not only concentrate RNA replication machinery but also provide cover from host innate immune sensors that detect the double-stranded RNA replication intermediates produced by this process.135,136 However, early cellular studies on coronavirus infection raised the issue of how newly made genomic RNA and subgenomic mRNAs could be transported out of fully enclosed DMVs to the site of viral assembly.137 High-resolution electron microscopy analysis has shed light onto this conundrum with the identification of a 3 MDa molecular pore complex that spans both DMV membranes (Figure 11B, bottom panel).56,138 This pore contains NSP3 as a structural component and likely serves as the export channel for viral mRNA and genomic RNA back into the cytoplasm.138
4.2. Viral Replication–Transcription Complex (RTC)
Within the lumen of SARS-CoV-2 DMVs, the viral genome is transcribed by the multiprotein RTC (Figures 11B and 12). This complex is not only responsible for transcription of the entire 30 kb genome but also the numerous subgenomic mRNAs encoded within the final one-third of the genome required for structural protein synthesis (i.e., N, M, E, and S proteins). In fact, genomic RNA only accounts for a small fraction of the total RNA produced during replication.137 The RTC uses a discontinuous transcription mechanism that relies on complementary transcription regulatory sequences throughout the genome to produce these subgenomic mRNAs (reviewed by Sawicki et al.).140 These mRNAs are then exported through the DMV molecular pore to the cytoplasm and translocated to the ER/Golgi for protein production.
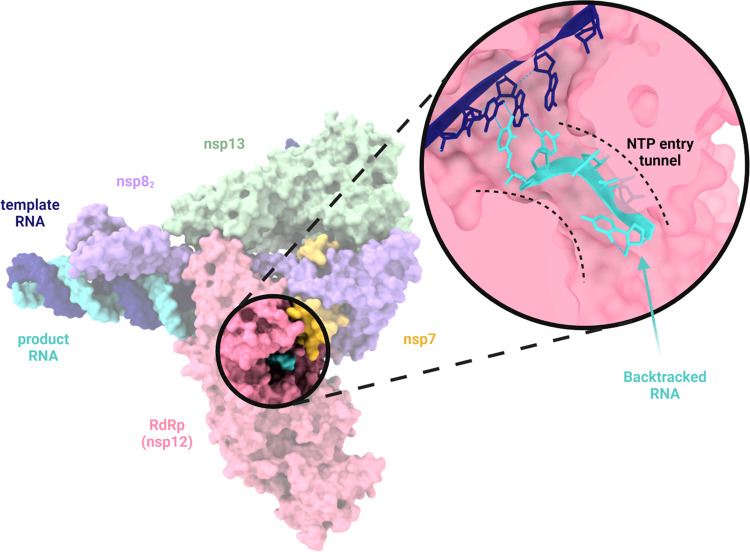
Figure 12.
Structure of the SARS-CoV-2 multiprotein replication–transcription complex (RTC). Surface representation (PDB 7KRN) (ref (143)) of the RTC highlighting the relative positions of the RNA-dependent RNA polymerase (RdRp, NSP12), processivity cofactors (NSP7 and NSP8), and the viral helicase (NSP13) as determined by single-particle cryo-EM. The NTP entry tunnel (inset) plays a critical role in the backtracking/proofreading function of the RTC, as erroneously incorporated ribonucleotides are frayed into the entry tunnel where they can then be removed by the 3′-5′ exonuclease, NSP14, to ensure high-fidelity replication of the viral genome. Created with BioRender.
Almost all NSPs produced by cleavage of the viral polyproteins play a role in the structure and function of the RTC. NSPs 2–11 provide RTC supporting functions, while the core enzymatic functions of RNA synthesis, RNA proofreading, and RNA modification are carried out by NSPs 12–16.52 The main component of the RTC is the RNA-dependent RNA polymerase (RdRp) NSP12. The structure of the RdRp has been likened to a “right hand” wrapped around the replicating RNA, with the conserved polymerase motifs (A–G) located in the “palm” domain and the NSP7/NSP8 cofactor binding sites located in the “thumb” and “fingers” (Figure 12).27,141,142 NSP7 and NSP8 act as processivity factors for the RdRp and bind the NSP12 thumb as a heterodimer.27,141,142 An additional copy of NSP8 also occupies the NSP12 fingers domain.27,141,142 While many structural features of the RdRp are consistent with prior information obtained from SARS-CoV, the SARS-CoV-2 RdRp structure highlights a previously unresolved N-terminal β hairpin that is predicted to stabilize the overall structure.142 The positively charged RNA template and NTP entry tunnels located at the back of the RdRp join together in a central hydrophilic cavity in the palm where template-directed RNA synthesis occurs (Figure 12, inset).142 Specificity of the polymerase for RNA over DNA synthesis is likely conferred through recognition of the 2′-OH group of the NTP by residues N691, S682, and D623 in the RdRp palm.27,141 After incorporation of the nucleotide into the nascent RNA strand, the double-stranded RNA intermediate exits through a tunnel located at the front side of the polymerase.142
4.3. RNA Proofreading and Modification
One of the most intriguing qualities of the coronavirus RTC is its exceptionally high fidelity during replication. The high mutation rates of typical RNA viruses promote genetic diversity and viral adaptation; however, coronaviruses have a mutation rate that is an order of magnitude lower compared to most other RNA viruses.124,144 The driving force of this high-fidelity replication is the ability of the SARS-CoV-2 RTC to backtrack along the nascent RNA strand and remove erroneously incorporated ribonucleotides. This backtracking and proofreading function is driven by the viral helicase, NSP13, and the 3′-5′ exonuclease, NSP14 (also known as ExoN).143,145 On the basis of single-particle cryo-EM structures of backtracked complexes and molecular dynamics analysis of the RTC, it is suggested that, when a ribonucleotide is misincorporated into the growing product RNA, the 5′-3′ helicase activity of NSP13 pushes the mismatched duplex RNA backward into the RdRp.143,145 In doing so, the template RNA and product RNA are separated by a structural motif of the RdRp that frays the misincorporated ribonucleotide into the more favorable environment of the NTP entry tunnel (Figure 12, inset).143,145 Exposure of the erroneously incorporated ribonucleotide in the entry tunnel allows NSP14 (along with its stabilizing cofactor NSP10) to remove it. It is this backtracking and proofreading function that limits the efficacy of nucleotide analogue drugs (e.g., remdesivir) for the treatment of COVID-19.
Final processing of the viral mRNAs involves addition of a 5′ cap and polyadenylation of the 3′ end that together aid in viral mRNA stability, translation initiation, and escape from the cellular innate immune system. Synthesis of the 5′ cap is facilitated by the nucleotide triphosphatase activity of NSP13, C-terminal N7-methyltransferase activity of NSP14 (ExoN), and 2′-O methylation by NSP16.124,146 To easily facilitate capping, these enzymes are positioned in close proximity to the newly synthesized viral mRNA as part of the large RTC.
4.4. Viral Packaging
Formation of the complete SARS-CoV-2 virion requires all components of the viral genome and envelope to assemble at the same time and place. Numerous high-resolution electron microscopy imaging studies of SARS-CoV-2-infected cells have shown that virions bud into the lumen of the ER–Golgi intermediate compartment (ERGIC) (Figure 11C).125,147 The membrane-associated structural proteins M, E, and S are all translated from viral mRNA and inserted into the ER membrane. These virus assembly sites are frequently observed in close proximity to DMV molecular pores, thereby aiding in the spatiotemporal coordination of the packaging process.125,139
Given the large size of the SARS-CoV-2 genome, it must be extensively condensed prior to encapsulation by the viral envelope. This packaging is mediated by RNA binding and dimerization of the N protein which coats the genomic RNA to form a tightly packed RNP complex.47,125 Since the N protein is required to bind variable regions all along the genome, interactions between its N-terminal RNA-binding domain and RNA are likely nonspecific and largely electrostatic.61 However, this nonspecificity gives rise to the issue of how genomic RNA is specifically packaged over the other abundant RNAs produced during the process of viral replication. To date, no specific packaging signal has been conclusively identified within the SARS-CoV-2 genome that drives selective packaging by the N protein. However, Syed et al. carried out a series of truncations of the SARS-CoV-2 genome—guided by reported packaging sequences for related viruses (murine hepatitis virus and SARS-CoV)—and found that deletion of a region termed “T20” (nucleotides 20080–22222) resulted in significant impairment of viral infectivity.148 Additionally, it has been suggested that phase separation of the N protein, driven by its three intrinsically disordered regions, may play a role in recruiting the nascent viral RNA.46,49,50,63,64,149 Once condensed, the RNP complex is retained at the cytoplasmic side of the ERGIC through interactions with the highly abundant M protein.125
Oligomerization and association of the M protein with the viral RNP complex and E and S proteins drives assembly, membrane curvature, and eventual budding of the viral particles into the ERGIC.150 These interactions are mediated through its soluble C-terminal domain that extends into the viral lumen.46 Trimeric prefusion spike proteins are produced in the Golgi/ER network and carried by small transport vesicles to the viral assembly site.151 Here, spike proteins cluster exclusively in association with RNP complexes, likely through a bridged interaction with the M protein.139 As previously mentioned, the precise role of the E protein in SARS-CoV-2 assembly remains unclear, as deletion of the E protein in other related β-coronaviruses does not impact viral assembly but instead attenuates the viral particles.43,44,152 The M2 protein of influenza viruses is also a viroporin and has been demonstrated to alter membrane cholesterol levels, inducing membrane scission and viral particle budding; however, further experiments are needed to determine whether the E protein plays a similar mechanistic role in SARS-CoV-2 viral packaging.153,154 Nevertheless, the M protein-driven clustering of the E protein with all other viral components induces positive membrane curvature of the ERGIC and eventual fusion of the viral envelope to produce a fully encapsulated and infectious SARS-CoV-2 virion.
4.5. Virion Release
Once all components of the virion have been assembled, the final phase of the SARS-CoV-2 replication cycle is release of the viral particles into the extracellular environment. It was initially assumed that β-coronaviruses use vesicles from the biosynthetic pathway for cellular egress similar to other enveloped RNA viruses that assemble and bud directly from the plasma membrane.155−157 In contrast, assembled SARS-CoV-2 virions in the ERGIC are trafficked to the plasma membrane via lysosomal exocytosis.125,158,159 This pathway poses a unique challenge for cellular egress of the viral particles as the typical acidification of lysosomes would result in particle inactivation and degradation. To circumvent this, β-coronaviruses have been shown to significantly disrupt lysosomal acidification and inactivate lysosomal proteases.150 While the exact mechanism of this deacidification is currently unknown, ORF3a of SARS-CoV-2 and other related β-coronaviruses has been implicated in this process.150,158 As a consequence of this deacidification, antigen presentation on SARS-CoV-2-infected cells is limited and could potentially serve as an additional path for immune evasion.158
Given that many of the SARS-CoV-2 structural proteins require post-translational modifications, such as phosphorylation, glycosylation, and/or cleavage, single-membrane vesicles (SMVs) containing single or multiple virions are first trafficked to the Golgi and trans-Golgi network where these post-translational modifications can be made.150 From here, lysosomes carry the viral particles to the cell periphery. Cryo-FIB/SEM and cryo-ET imaging analyses reveal that final egress of the virions into the extracellular space is mediated through exit tunnels connected to the cell membrane (Figure 11D), likely formed by fusion of the SMVs with the plasma membrane.125,139
5. Future Prospects
Recent advancements in electron microscopy have made viral proteins and processes that were intractable only years previously now structurally tractable. Here, we have reviewed SARS-CoV-2 structural insights and placed them into the context of greater viral pathobiology. What follows is a discussion of the limitations to our current understanding of the SARS-CoV2 viral life cycle and examples of how structural results are being used to inform therapeutic design.
Our understanding of the 3D rearrangement of viral biology stems from the synthesis of results across imaging techniques. However, due to technology specialization, there is a tendency for structural fields to compare results exclusively within their own imaging field (i.e., cryo-EM structures compared with other cryo-EM structures) and a lack of integration of results across imaging techniques. An excellent example of the implementation of multitechnique structural investigation was conducted by Cortese et al., wherein the authors describe the effects of viral infection on cellular structure, visualizing fixed samples by cryo-ET and FIB/SEM and live cells by confocal and super-resolution (STED) light microscopy.147 This integrative imaging approach revealed in situ remodeling of internal cellular membrane systems upon SARS-CoV-2 infection and provided a functional link by demonstrating the pharmacological inhibition of cytoskeleton remodeling to restrict viral replication.
While broad 3D characterization of the viral infection cycle has been achieved, there still exist a few structural unknowns. High-resolution details for the M and E structural proteins remain elusive, with pan-coronavirus implications given their high degree of conservation. An additional structural unknown is the molecular mechanism by which the SARS-CoV-2 S protein transitions from the pre- to postfusion state during cellular infection. This mechanism is presumed to be highly conserved across many viruses (coronaviruses and HIV-1), and therefore the in-depth experimental characterization of this process may inform broad therapeutic targeting.71,75,76
We would like to finally highlight how some of these structural results have aided in the development of several SARS-CoV-2 therapies. Overall, therapeutic discovery has steadily advanced throughout the COVID-19 pandemic (Figure 13). A specific example of structure-based therapeutic design was reported by Hunt et al., wherein high-affinity small proteins (“minibinders”) were designed to bind and geometrically complement the ACE2 binding site on the SARS-CoV-2 S protein.161 These in silico and structurally designed minibinders were further demonstrated to potently neutralize SARS-CoV-2 VoCs, with protection provided in human ACE2-expressing transgenic mice (both prophylactically and therapeutically). Foundational work by Pallesen et al. and Hsieh et al. demonstrating structure-based design of prefusion-stabilized MERS-CoV, SARS-CoV, and SARS-CoV-2 stabilized spike proteins has undoubtedly informed the formulation of the BNT162b2 (Pfizer, Inc.) and mRNA-1273 (Moderna, Inc.) mRNA vaccines.162,163 Finally, the development of the recently announced oral pills Molnupiravir (Merck & Co.) and Paxlovid (Pfizer Inc.) have likely benefited from the high-resolution structural data available for their viral targets (the RTC and 3CL protease, respectively). The interdisciplinary combination of structural biology, immunology, virology, and continual scientific technological advancements has proven to represent our best approach to understanding and mitigating the COVID-19 pandemic.
Figure 13.
Therapeutic discovery during the COVID-19 pandemic. These data were compiled by the Biotechnology Innovation Organization and are presented here as the cumulative monthly number of antiviral, treatment, and vaccine therapies in development (ref (160)). Antivirals are defined here as drugs that interact directly with the virus or disrupt its ability to replicate. Treatments are defined here as drugs that treat various COVID-19-associated illnesses resulting from SARS-CoV-2 viral infection. Vaccines are defined here as prophylactic therapeutics that stimulate immunity against SARS-CoV-2.
Acknowledgments
We thank Katharine Tuttle for creating the graphical abstract associated with this publication. Sriram Subramaniam is funded by a Canada Excellence Research Chair Award; the VGH Foundation; Genome BC, Canada; and the Tai Hung Fai Charitable Foundation. J.W.S. is supported by a CIHR Frederick Banting and Charles Best Canada Graduate Scholarships Doctoral Award (CGS-D) and a University of British Columbia President’s Academic Excellence Initiative PhD Award.
Biographies
James Saville is a Ph.D. candidate in the department of Biochemistry and Molecular Biology at the University of British Columbia. James received his B.Sc. in Biochemistry and Microbiology from the University of Victoria. His current research involves the characterization of variant SARS-CoV-2 spike proteins by biochemical and structural methods.
Dr. Alison Berezuk is the electron microscopy facility manager for the Program in Cryo-EM Guided Drug Design at the University of British Columbia. Previously, Alison received a Ph.D. in Molecular and Cellular Biology from the University of Guelph where she studied the role of proteins involved in bacterial cell division and their potential as targets for the development of novel antibiotics. She went on to complete postdoctoral training at the University of British Columbia where she helped establish a high-end cryo-EM facility used to visualize a vast array of proteins and protein complexes, including drug- or antibody-bound proteins.
Dr. Shanti Swaroop Srivastava is an assistant professor at the Indian Institute of Science Education and Research (IISER) in Berhampur, India. He received his Ph.D. from the Centre for Cellular and Molecular Biology in Hyderabad, India before completing postdoctoral training at the National Cancer Institute in Maryland, U.S.A. and the University of British Columbia in Vancouver, Canada. Shanti’s research has spanned from the use of X-ray crystallography to uncover the mechanism of metal binding in microbial βγ-crystallins to cryo-EM to characterize emerging SARS-CoV-2 variants of concern.
Dr. Sriram Subramaniam is the Gobind Khorana Canada Excellence Research Chair in Cancer Drug design at the University of British Columbia and also the founder and CEO of Gandeeva Therapeutics, a drug discovery company based in Vancouver, Canada. Prior to his arrival in Vancouver, Sriram was a Senior Investigator at the National Institutes of Health where he also founded and directed the U.S. National Cryo-EM Facility. He received his Ph.D. in physical chemistry from Stanford University and completed postdoctoral training in the Departments of Chemistry and Biology at the Massachusetts Institute of Technology. His work showed for the first time that the structures of proteins and bound drugs, as well as a variety of dynamic protein complexes, could be visualized using cryo-EM methods at near-atomic resolution.
The authors declare the following competing financial interest(s): S.S. is the founder and CEO of Gandeeva Therapeutics Inc.
References
- Xu R.-H.; He J.-F.; Evans M. R.; Peng G.-W.; Field H. E.; Yu D.-W.; Lee C.-K.; Luo H.-M.; Lin W.-S.; Lin P.; et al. Epidemiologic Clues to SARS Origin in China. Emerging Infectious Diseases 2004, 10, 1030–1037. 10.3201/eid1006.030852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong N.; Zheng B.; Li Y.; Poon L.; Xie Z.; Chan K.; Li P.; Tan S.; Chang Q.; Xie J.; et al. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People’s Republic of China, in February, 2003. Lancet 2003, 362, 1353–1358. 10.1016/S0140-6736(03)14630-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heymann D. L.; Mackenzie J. S.; Peiris M. SARS legacy: outbreak reporting is expected and respected. Lancet 2013, 381, 779–781. 10.1016/S0140-6736(13)60185-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Listings of WHO’s Response to COVID-19. World Health Organization, June 29, 2020. https://www.who.int/news/item/29-06-2020-covidtimeline (accessed 2021-09-07).
- Boni M. F.; Lemey P.; Jiang X.; Lam T. T.-Y.; Perry B. W.; Castoe T. A.; Rambaut A.; Robertson D. L. Evolutionary origins of the SARS-CoV-2 sarbecovirus lineage responsible for the COVID-19 pandemic. Nature Microbiology 2020, 5, 1408–1417. 10.1038/s41564-020-0771-4. [DOI] [PubMed] [Google Scholar]
- Lu R.; Zhao X.; Li J.; Niu P.; Yang B.; Wu H.; Wang W.; Song H.; Huang B.; Zhu N.; et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbe S.; Buckland-Merrett G. Data, disease and diplomacy: GISAID’s innovative contribution to global health. Global Challenges 2017, 1, 33–46. 10.1002/gch2.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Y.; McCauley J. GISAID: Global initiative on sharing all influenza data – from vision to reality. EuroSurveillance 2017, 22, 30494. 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edrada E. M.; Lopez E. B.; Villarama J. B.; Salva Villarama E. P.; Dagoc B. F.; Smith C.; Sayo A. R.; Verona J. A.; Trifalgar-Arches J.; Lazaro J.; et al. First COVID-19 infections in the Philippines: a case report. Trop. Med. Health 2020, 48, 21. 10.1186/s41182-020-00203-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roser M.; Ortiz-Ospina E.. Global Education. Our World in Data, 2016. https://ourworldindata.org/global-education (accessed 2021-11-27).
- Chen P.-L.; Lee N.-Y.; Cia C.-T.; Ko W.-C.; Hsueh P.-R. A Review of Treatment of Coronavirus Disease 2019 (COVID-19): Therapeutic Repurposing and Unmet Clinical Needs. Front. Pharmacol. 2020, 11, 584956. 10.3389/fphar.2020.584956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welte T.; Ambrose L. J.; Sibbring G. C.; Sheikh S.; Müllerová H.; Sabir I. Current evidence for COVID-19 therapies: a systematic literature review. Eur. Respir. Rev. 2021, 30, 200384. 10.1183/16000617.0384-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y.; Shamsuddin A.; Campbell H.; Theodoratou E. Current COVID-19 treatments: Rapid review of the literature. J. Global Health. 2021, 11, 10003. 10.7189/jogh.11.10003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amanat F.; Krammer F. SARS-CoV-2 Vaccines: Status Report. Immunity 2020, 52, 583–589. 10.1016/j.immuni.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krammer F. SARS-CoV-2 vaccines in development. Nature 2020, 586, 516–527. 10.1038/s41586-020-2798-3. [DOI] [PubMed] [Google Scholar]
- Dong Y.; Dai T.; Wei Y.; Zhang L.; Zheng M.; Zhou F. A systematic review of SARS-CoV-2 vaccine candidates. Signal Transduction Targeted Ther. 2020, 5, 237. 10.1038/s41392-020-00352-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- COVID-19 vaccine tracker and landscape. World Health Organization. https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines (accessed 2021-09-07).
- Jones I.; Roy P. Sputnik V COVID-19 vaccine candidate appears safe and effective. Lancet 2021, 397, 642–643. 10.1016/S0140-6736(21)00191-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W.-H.; Strych U.; Hotez P. J.; Bottazzi M. E. The SARS-CoV-2 Vaccine Pipeline: an Overview. Current Tropical Medicine Reports 2020, 7, 61–64. 10.1007/s40475-020-00201-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The COVID-19 vaccine race – weekly update. Gavi. https://www.gavi.org/vaccineswork/covid-19-vaccine-race (accessed 2021-09-07).
- Korber B.; Fischer W. M.; Gnanakaran S.; Yoon H.; Theiler J.; Abfalterer W.; Hengartner N.; Giorgi E. E.; Bhattacharya T.; Foley B.; et al. Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell 2020, 182, 812–827. 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tracking SARS-CoV-2 variants. World Health Organization. https://www.who.int/activities/tracking-SARS-CoV-2-variants (accessed 2021-10-15).
- Frank J. Just in Time”: The Role of Cryo-Electron Microscopy in Combating Recent Pandemics. Biochemistry 2021, 60, 3449–3451. 10.1021/acs.biochem.1c00318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerr A. Cryo-electron tomography. Nat. Methods 2017, 14, 34–34. 10.1038/nmeth.4115. [DOI] [Google Scholar]
- Ognjenović J.; Grisshammer R.; Subramaniam S. Frontiers in Cryo Electron Microscopy of Complex Macromolecular Assemblies. Annu. Rev. Biomed. Eng. 2019, 21, 395–415. 10.1146/annurev-bioeng-060418-052453. [DOI] [PubMed] [Google Scholar]
- Wrapp D.; Wang N.; Corbett K. S.; Goldsmith J. A.; Hsieh C.-L.; Abiona O.; Graham B. S.; McLellan J. S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020, 367, 1260–1263. 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillen H. S.; Kokic G.; Farnung L.; Dienemann C.; Tegunov D.; Cramer P. Structure of replicating SARS-CoV-2 polymerase. Nature 2020, 584, 154–156. 10.1038/s41586-020-2368-8. [DOI] [PubMed] [Google Scholar]
- Herzik M. A.; Wu M.; Lander G. C. High-resolution structure determination of sub-100 kDa complexes using conventional cryo-EM. Nat. Commun. 2019, 10, 1032. 10.1038/s41467-019-08991-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X.; Wang J.; Zhang X.; Yang Z.; Zhang J.-C.; Zhao L.; Peng H.-L.; Lei J.; Wang H.-W. Single particle cryo-EM reconstruction of 52 kDa streptavidin at 3.2 Angstrom resolution.. Nat. Commun. 2019, 10, 2386. 10.1038/s41467-019-10368-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes G.Crystallography Made Crystal Clear; Complementary Science Series; Academic Press: Cambridge, MA, 2006; pp 29–41. [Google Scholar]
- Jin Z.; Du X.; Xu Y.; Deng Y.; Liu M.; Zhao Y.; Zhang B.; Li X.; Zhang L.; Peng C.; et al. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature 2020, 582, 289–293. 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- Mengist H. M.; Dilnessa T.; Jin T. Structural Basis of Potential Inhibitors Targeting SARS-CoV-2 Main Protease. Front. Chem. 2021, 9, 622898. 10.3389/fchem.2021.622898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacco M. D.; Ma C.; Lagarias P.; Gao A.; Townsend J. A.; Meng X.; Dube P.; Zhang X.; Hu Y.; Kitamura N.; et al. Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against Mpro and cathepsin L. Sci. Adv. 2020, 6, 0751. 10.1126/sciadv.abe0751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodsell D. S.; Autin L.; Olson A. J. Illustrate: Software for Biomolecular Illustration. Structure 2019, 27, 1716–1720. 10.1016/j.str.2019.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman H. M. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M.-Y.; Zhao R.; Gao L.-J.; Gao X.-F.; Wang D.-P.; Cao J.-M. SARS-CoV-2: Structure, Biology, and Structure-Based Therapeutics Development. Front. Cell. Infect. Microbiol. 2020, 10, 587269. 10.3389/fcimb.2020.587269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrier A.; Bonnin A.; Desmarets L.; Danneels A.; Goffard A.; Rouillé Y.; Dubuisson J.; Belouzard S. The C-terminal domain of the MERS coronavirus M protein contains a trans-Golgi network localization signal. J. Biol. Chem. 2019, 294, 14406–14421. 10.1074/jbc.RA119.008964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ujike M.; Taguchi F. Incorporation of Spike and Membrane Glycoproteins into Coronavirus Virions. Viruses 2015, 7, 1700–1725. 10.3390/v7041700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng Y.-T.; Chang C.-H.; Wang S.-M.; Huang K.-J.; Wang C.-T. Identifying SARS-CoV Membrane Protein Amino Acid Residues Linked to Virus-Like Particle Assembly. PLoS One 2013, 8, e64013. 10.1371/journal.pone.0064013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman B. W.; Kiss G.; Kunding A. H.; Bhella D.; Baksh M. F.; Connelly S.; Droese B.; Klaus J. P.; Makino S.; Sawicki S. G.; et al. A structural analysis of M protein in coronavirus assembly and morphology. J. Struct. Biol. 2011, 174, 11–22. 10.1016/j.jsb.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeman D.; Fielding B. C. Coronavirus envelope protein: current knowledge. Virol. J. 2019, 16, 69. 10.1186/s12985-019-1182-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieva J. L.; Madan V.; Carrasco L. Viroporins: structure and biological functions. Nature Reviews Microbiology 2012, 10, 563–574. 10.1038/nrmicro2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeDiego M. L.; Álvarez E.; Almazán F.; Rejas M. T.; Lamirande E.; Roberts A.; Shieh W.-J.; Zaki S. R.; Subbarao K.; Enjuanes L. A Severe Acute Respiratory Syndrome Coronavirus That Lacks the E Gene Is Attenuated In Vitro and In Vivo. Journal of Virology 2007, 81, 1701–1713. 10.1128/JVI.01467-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo L.; Masters P. S. The Small Envelope Protein E Is Not Essential for Murine Coronavirus Replication. Journal of Virology 2003, 77, 4597–4608. 10.1128/JVI.77.8.4597-4608.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortego J.; Escors D.; Laude H.; Enjuanes L. Generation of a Replication-Competent, Propagation-Deficient Virus Vector Based on the Transmissible Gastroenteritis Coronavirus Genome. Journal of Virology 2002, 76, 11518–11529. 10.1128/JVI.76.22.11518-11529.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S.; Ye Q.; Singh D.; Cao Y.; Diedrich J. K.; Yates J. R.; Villa E.; Cleveland D. W.; Corbett K. D. The SARS-CoV-2 nucleocapsid phosphoprotein forms mutually exclusive condensates with RNA and the membrane-associated M protein. Nat. Commun. 2021, 12, 502. 10.1038/s41467-020-20768-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao H.; Song Y.; Chen Y.; Wu N.; Xu J.; Sun C.; Zhang J.; Weng T.; Zhang Z.; Wu Z.; et al. Molecular Architecture of the SARS-CoV-2 Virus. Cell 2020, 183, 730–738. 10.1016/j.cell.2020.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson C. R.; Asfaha J. B.; Ghent C. M.; Howard C. J.; Hartooni N.; Safari M.; Frankel A. D.; Morgan D. O. Phosphoregulation of Phase Separation by the SARS-CoV-2 N Protein Suggests a Biophysical Basis for its Dual Functions. Mol. Cell 2020, 80, 1092–1103. 10.1016/j.molcel.2020.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.; Cui Y.; Han X.; Hu W.; Sun M.; Zhang Y.; Wang P.-H.; Song G.; Chen W.; Lou J. Liquid-liquid phase separation by SARS-CoV-2 nucleocapsid protein and RNA. Cell research 2020, 30, 1143–1145. 10.1038/s41422-020-00408-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubuk J.; Alston J. J.; Incicco J. J.; Singh S.; Stuchell-Brereton M. D.; Ward M. D.; Zimmerman M. I.; Vithani N.; Griffith D.; Wagoner J. A.; et al. The SARS-CoV-2 nucleocapsid protein is dynamic, disordered, and phase separates with RNA. Nat. Commun. 2021, 12, 1936. 10.1038/s41467-021-21953-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Q.; West A. M. V.; Silletti S.; Corbett K. D. Architecture and self-assembly of the SARS-CoV-2 nucleocapsid protein. Protein Sci. 2020, 29, 1890–1901. 10.1002/pro.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariano G.; Farthing R. J.; Lale-Farjat S. L. M.; Bergeron J. R. C. Structural Characterization of SARS-CoV-2: Where We Are, and Where We Need to Be. Front. Mol. Biosci. 2020, 7, 605236. 10.3389/fmolb.2020.605236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke Z.; Oton J.; Qu K.; Cortese M.; Zila V.; McKeane L.; Nakane T.; Zivanov J.; Neufeldt C. J.; Cerikan B.; et al. Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Nature 2020, 588, 498–502. 10.1038/s41586-020-2665-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turoňová B.; Sikora M.; Schürmann C.; Hagen W. J. H.; Welsch S.; Blanc F. E. C.; von Bülow S.; Gecht M.; Bagola K.; Hörner C.; et al. In situ structural analysis of SARS-CoV-2 spike reveals flexibility mediated by three hinges. Science 2020, 370, 203–208. 10.1126/science.abd5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong X.; Coombs P. J.; Martin S. R.; Liu J.; Xiao H.; McCauley J. W.; Locher K.; Walker P. A.; Collins P. J.; Kawaoka Y.; et al. Receptor binding by a ferret-transmissible H5 avian influenza virus. Nature 2013, 497, 392–396. 10.1038/nature12144. [DOI] [PubMed] [Google Scholar]
- Liu C.; Mendonça L.; Yang Y.; Gao Y.; Shen C.; Liu J.; Ni T.; Ju B.; Liu C.; Tang X.; et al. The Architecture of Inactivated SARS-CoV-2 with Postfusion Spikes Revealed by Cryo-EM and Cryo-ET. Structure 2020, 28, 1218–1224. 10.1016/j.str.2020.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q.; Bao L.; Mao H.; Wang L.; Xu K.; Yang M.; Li Y.; Zhu L.; Wang N.; Lv Z.; et al. Development of an inactivated vaccine candidate for SARS-CoV-2. Science 2020, 369, 77–81. 10.1126/science.abc1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A.; Lamirande E. W.; Vogel L.; Baras B.; Goossens G.; Knott I.; Chen J.; Ward J. M.; Vassilev V.; Subbarao K. Immunogenicity and Protective Efficacy in Mice and Hamsters of a β-Propiolactone Inactivated Whole Virus SARS-CoV Vaccine. Viral Immunology 2010, 23, 509–519. 10.1089/vim.2010.0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta D.; Parthasarathy H.; Sah V.; Tandel D.; Vedagiri D.; Reddy S.; Harshan K. H. Inactivation of SARS-CoV-2 by β-propiolactone Causes Aggregation of Viral Particles and Loss of Antigenic Potential. Virus Res. 2021, 305, 198555. 10.1016/j.virusres.2021.198555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui M.; Song W.; Zhou H.; Xu J.; Chen S.; Xiang Y.; Wang X. Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding. Cell Research 2017, 27, 119–129. 10.1038/cr.2016.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chechetkin V. R.; Lobzin V. V. Ribonucleocapsid assembly/packaging signals in the genomes of the coronaviruses SARS-CoV and SARS-CoV-2: detection, comparison and implications for therapeutic targeting. J. Biomol. Struct. Dyn. 2022, 40, 508–522. 10.1080/07391102.2020.1815581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S.; Yang M.; Hong Z.; Zhang L.; Huang Z.; Chen X.; He S.; Zhou Z.; Zhou Z.; Chen Q.; et al. Crystal structure of SARS-CoV-2 nucleocapsid protein RNA binding domain reveals potential unique drug targeting sites. Acta Pharmaceutica Sinica B 2020, 10, 1228–1238. 10.1016/j.apsb.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.; Dai T.; Qin Z.; Pan T.; Chu F.; Lou L.; Zhang L.; Yang B.; Huang H.; Lu H.; et al. Targeting liquid–liquid phase separation of SARS-CoV-2 nucleocapsid protein promotes innate antiviral immunity by elevating MAVS activity. Nat. Cell Biol. 2021, 23, 718–732. 10.1038/s41556-021-00710-0. [DOI] [PubMed] [Google Scholar]
- Perdikari T. M.; Murthy A. C.; Ryan V. H.; Watters S.; Naik M. T.; Fawzi N. L. SARS-CoV-2 nucleocapsid protein phase-separates with RNA and with human hnRNPs. EMBO J. 2020, 39, e106478. 10.15252/embj.2020106478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G.; Deering C.; Macke M.; Shao J.; Burns R.; Blau D. M.; Holmes K. V.; Davidson B. L.; Perlman S.; McCray P. B. Human Coronavirus 229E Infects Polarized Airway Epithelia from the Apical Surface. Journal of Virology 2000, 74, 9234–9239. 10.1128/JVI.74.19.9234-9239.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X.; Liu J. Q. L.; Fung T. S.. Human Coronavirus-229E, -OC43, -NL63, and -HKU1 (Coronaviridae). In Encyclopedia of Virology, 4th ed.; Bamford D. H., Zuckerman M., Eds.; Academic Press: Cambridge, MA, 2021; pp 428–440. [Google Scholar]
- Bačenková D.; Trebuňová M.; Špakovská T.; Schnitzer M.; Bednarčíková L.; Živčák J. Comparison of Selected Characteristics of SARS-CoV-2, SARS-CoV, and HCoV-NL63. Applied Sciences 2021, 11, 1497. 10.3390/app11041497. [DOI] [Google Scholar]
- Shang J.; Ye G.; Shi K.; Wan Y.; Luo C.; Aihara H.; Geng Q.; Auerbach A.; Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature 2020, 581, 221–224. 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R.; Zhang Y.; Li Y.; Xia L.; Guo Y.; Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 2020, 367, 1444–1448. 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan J.; Ge J.; Yu J.; Shan S.; Zhou H.; Fan S.; Zhang Q.; Shi X.; Wang Q.; Zhang L.; et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020, 581, 215–220. 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Fan X.; Cao D.; Kong L.; Zhang X. Cryo-EM analysis of the post-fusion structure of the SARS-CoV spike glycoprotein. Nat. Commun. 2020, 11, 3618. 10.1038/s41467-020-17371-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S.; Xu W.; Wang Q.; Wang C.; Hua C.; Li W.; Lu L.; Jiang S. Peptide-Based Membrane Fusion Inhibitors Targeting HCoV-229E Spike Protein HR1 and HR2 Domains. Int. J. Mol. Sci. 2018, 19, 487. 10.3390/ijms19020487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu G.; Wang Q.; Gao G. F. Bat-to-human: spike features determining ‘host jump’ of coronaviruses SARS-CoV, MERS-CoV, and beyond. Trends in Microbiology 2015, 23, 468–478. 10.1016/j.tim.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang T.; Bidon M.; Jaimes J. A.; Whittaker G. R.; Daniel S. Coronavirus membrane fusion mechanism offers a potential target for antiviral development. Antiviral Res. 2020, 178, 104792. 10.1016/j.antiviral.2020.104792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J.; Wan Y.; Luo C.; Ye G.; Geng Q.; Auerbach A.; Li F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. U. S. A. 2020, 117, 11727–11734. 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Zhang X.; Leng Yap Y. Structural similarity between HIV-1 gp41 and SARS-CoV S2 proteins suggests an analogous membrane fusion mechanism. Journal of Molecular Structure: THEOCHEM 2004, 677, 73–76. 10.1016/j.theochem.2004.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y.; Shang J.; Graham R.; Baric R. S.; Li F. Receptor Recognition by the Novel Coronavirus from Wuhan: an Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. J. Virol. 2020, 94, e00127. 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M.; Kleine-Weber H.; Schroeder S.; Krüger N.; Herrler T.; Erichsen S.; Schiergens T. S.; Herrler G.; Wu N.-H.; Nitsche A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280. 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letko M.; Marzi A.; Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nature Microbiology 2020, 5, 562–569. 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A. C.; Park Y.-J.; Tortorici M. A.; Wall A.; McGuire A. T.; Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181, 281–292. 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lempp F. A.; Soriaga L.; Montiel-Ruiz M.; Benigni F.; Noack J.; Park Y.-J.; Bianchi S.; Walls A. C.; Bowen J. E.; Zhou J.; et al. Lectins enhance SARS-CoV-2 infection and influence neutralizing antibodies. Nature 2021, 598, 342–347. 10.1038/s41586-021-03925-1. [DOI] [PubMed] [Google Scholar]
- Trbojević-Akmačić I.; Petrović T.; Lauc G. SARS-CoV-2 S glycoprotein binding to multiple host receptors enables cell entry and infection. Glycoconjugate Journal 2021, 38, 611–623. 10.1007/s10719-021-10021-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X.; Mannar D.; Srivastava S. S.; Berezuk A. M.; Demers J.-P.; Saville J. W.; Leopold K.; Li W.; Dimitrov D. S.; Tuttle K. S.; et al. Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies. PLoS Biol. 2021, 19, e3001237. 10.1371/journal.pbio.3001237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S.; Nelson G.; Olson C. A.; Buzko O.; Higashide W.; Shin A.; Gonzalez M.; Taft J.; Patel R.; Buta S.; et al. An ACE2 Triple Decoy that neutralizes SARS-CoV-2 shows enhanced affinity for virus variants. Sci. Rep. 2021, 11, 12740. 10.1038/s41598-021-91809-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laffeber C.; de Koning K.; Kanaar R.; Lebbink J. H. G. Experimental Evidence for Enhanced Receptor Binding by Rapidly Spreading SARS-CoV-2 Variants. J. Mol. Biol. 2021, 433, 167058. 10.1016/j.jmb.2021.167058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian F.; Tong B.; Sun L.; Shi S.; Zheng B.; Wang Z.; Dong X.; Zheng P. N501Y mutation of spike protein in SARS-CoV-2 strengthens its binding to receptor ACE2. eLife 2021, 10, 69091. 10.7554/eLife.69091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H.; Zhang Q.; Wei P.; Chen Z.; Aviszus K.; Yang J.; Downing W.; Jiang C.; Liang B.; Reynoso L.; et al. The basis of a more contagious 501Y.V1 variant of SARS-CoV-2. Cell Res. 2021, 31, 720–722. 10.1038/s41422-021-00496-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saville J. W.; Mannar D.; Zhu X.; Srivastava S. S.; Berezuk A. M.; Demers J.-P.; Zhou S.; Tuttle K. S.; Sekirov I.; Kim A.; et al. Structural and biochemical rationale for enhanced spike protein fitness in delta and kappa SARS-CoV-2 variants. Nat. Commun. 2022, 13, 742. 10.1038/s41467-022-28324-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannar D.; Saville J. W.; Zhu X.; Srivastava S. S.; Berezuk A. M.; Zhou S.; Tuttle K. S.; Kim A.; Li W.; Dimitrov D. S.; et al. Structural analysis of receptor binding domain mutations in SARS-CoV-2 variants of concern that modulate ACE2 and antibody binding. Cell Rep. 2021, 37, 110156. 10.1016/j.celrep.2021.110156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannar D.; Saville J. W.; Zhu X.; Srivastava S. S.; Berezuk A. M.; Tuttle K. S.; Marquez A. C.; Sekirov I.; Subramaniam S. SARS-CoV-2 Omicron variant: Antibody evasion and cryo-EM structure of spike protein–ACE2 complex. Science 2022, 375, 760–764. 10.1126/science.abn7760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T.-J.; Yu P.-Y.; Chang Y.-C.; Liang K.-H.; Tso H.-C.; Ho M.-R.; Chen W.-Y.; Lin H.-T.; Wu H.-C.; Hsu S.-T. D. Effect of SARS-CoV-2 B.1.1.7 mutations on spike protein structure and function. Nature Structural & Molecular Biology 2021, 28, 731–739. 10.1038/s41594-021-00652-z. [DOI] [PubMed] [Google Scholar]
- Gobeil S. M. C.; Janowska K.; McDowell S.; Mansouri K.; Parks R.; Stalls V.; Kopp M. F.; Manne K.; Li D.; Wiehe K.; et al. Effect of natural mutations of SARS-CoV-2 on spike structure, conformation, and antigenicity. Science 2021, 373, eabi6226. 10.1126/science.abi6226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F.; Li W.; Farzan M.; Harrison S. C. Structure of SARS Coronavirus Spike Receptor-Binding Domain Complexed with Receptor. Science 2005, 309, 1864–1868. 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- Watanabe Y.; Berndsen Z. T.; Raghwani J.; Seabright G. E.; Allen J. D.; Pybus O. G.; McLellan J. S.; Wilson I. A.; Bowden T. A.; Ward A. B.; et al. Vulnerabilities in coronavirus glycan shields despite extensive glycosylation. Nat. Commun. 2020, 11, 2688. 10.1038/s41467-020-16567-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y.; Allen J. D.; Wrapp D.; McLellan J. S.; Crispin M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science 2020, 369, 330–333. 10.1126/science.abb9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y.; Bowden T. A.; Wilson I. A.; Crispin M. Exploitation of glycosylation in enveloped virus pathobiology. Biochimica et Biophysica Acta (BBA) - General Subjects 2019, 1863, 1480–1497. 10.1016/j.bbagen.2019.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigerust D. J.; Shepherd V. L. Virus glycosylation: role in virulence and immune interactions. Trends in Microbiology 2007, 15, 211–218. 10.1016/j.tim.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagdonaite I.; Wandall H. H. Global aspects of viral glycosylation. Glycobiology 2018, 28, 443–467. 10.1093/glycob/cwy021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagdonaite I.; Thompson A. J.; Wang X.; Søgaard M.; Fougeroux C.; Frank M.; Diedrich J. K.; Yates J. R.; Salanti A.; Vakhrushev S. Y.; et al. Site-Specific O-Glycosylation Analysis of SARS-CoV-2 Spike Protein Produced in Insect and Human Cells. Viruses 2021, 13, 551. 10.3390/v13040551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oommen A.; Cunningham S.; Joshi L. Transcriptomic Analysis of Respiratory Tissue and Cell Line Models to Examine Glycosylation Machinery during SARS-CoV-2 Infection. Viruses 2021, 13, 82. 10.3390/v13010082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant O. C.; Montgomery D.; Ito K.; Woods R. J. Analysis of the SARS-CoV-2 spike protein glycan shield reveals implications for immune recognition. Sci. Rep. 2020, 10, 14991. 10.1038/s41598-020-71748-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum M.; De Marco A.; Lempp F. A.; Tortorici M. A.; Pinto D.; Walls A. C.; Beltramello M.; Chen A.; Liu Z.; Zatta F.; et al. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell 2021, 184, 2332–2347. 10.1016/j.cell.2021.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaney A. J.; Loes A. N.; Crawford K. H. D.; Starr T. N.; Malone K. D.; Chu H. Y.; Bloom J. D. Comprehensive mapping of mutations in the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human plasma antibodies. Cell Host Microbe 2021, 29, 463–476. 10.1016/j.chom.2021.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes C. O.; West A. P.; Huey-Tubman K. E.; Hoffmann M. A. G.; Sharaf N. G.; Hoffman P. R.; Koranda N.; Gristick H. B.; Gaebler C.; Muecksch F.; et al. Structures of Human Antibodies Bound to SARS-CoV-2 Spike Reveal Common Epitopes and Recurrent Features of Antibodies. Cell 2020, 182, 828–842. 10.1016/j.cell.2020.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seabright G. E.; Doores K. J.; Burton D. R.; Crispin M. Protein and Glycan Mimicry in HIV Vaccine Design. J. Mol. Biol. 2019, 431, 2223–2247. 10.1016/j.jmb.2019.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.; Kim S.; Park J.; Chang H. R.; Chang J.; Ahn J.; Park H.; Park J.; Son N.; Kang G.; et al. A high-resolution temporal atlas of the SARS-CoV-2 translatome and transcriptome. Nat. Commun. 2021, 12, 5120. 10.1038/s41467-021-25361-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Breyne S.; Vindry C.; Guillin O.; Condé L.; Mure F.; Gruffat H.; Chavatte L.; Ohlmann T. Translational control of coronaviruses. Nucleic Acids Res. 2020, 48, 12502–12522. 10.1093/nar/gkaa1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel Y.; Mizrahi O.; Nachshon A.; Weingarten-Gabbay S.; Morgenstern D.; Yahalom-Ronen Y.; Tamir H.; Achdout H.; Stein D.; Israeli O.; et al. The coding capacity of SARS-CoV-2. Nature 2021, 589, 125–130. 10.1038/s41586-020-2739-1. [DOI] [PubMed] [Google Scholar]
- Wu A.; Peng Y.; Huang B.; Ding X.; Wang X.; Niu P.; Meng J.; Zhu Z.; Zhang Z.; Wang J.; et al. Genome Composition and Divergence of the Novel Coronavirus (2019-nCoV) Originating in China. Cell Host & Microbe 2020, 27, 325–328. 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M. T.; Irfan M.; Ahsan H.; Ahmed A.; Kaushik A. C.; Khan A. S.; Chinnasamy S.; Ali A.; Wei D.-Q. Structures of SARS-CoV-2 RNA-Binding Proteins and Therapeutic Targets. Intervirology 2021, 64, 55–68. 10.1159/000513686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J.; Li F.; Shi Z.-L. Origin and evolution of pathogenic coronaviruses. Nature Reviews Microbiology 2019, 17, 181–192. 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moustaqil M.; Ollivier E.; Chiu H.-P.; Van Tol S.; Rudolffi-Soto P.; Stevens C.; Bhumkar A.; Hunter D. J. B.; Freiberg A. N.; Jacques D.; et al. SARS-CoV-2 proteases PLpro and 3CLpro cleave IRF3 and critical modulators of inflammatory pathways (NLRP12 and TAB1): implications for disease presentation across species. Emerging Microbes & Infections 2021, 10, 178–195. 10.1080/22221751.2020.1870414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Báez-Santos Y. M.; St. John S. E.; Mesecar A. D. The SARS-coronavirus papain-like protease: Structure, function and inhibition by designed antiviral compounds. Antiviral Res. 2015, 115, 21–38. 10.1016/j.antiviral.2014.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan S.; Wu G. Spatial and temporal roles of SARS-CoV PL pro —A snapshot. FASEB J. 2021, 35, e21197. 10.1096/fj.202002271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav R.; Chaudhary J. K.; Jain N.; Chaudhary P. K.; Khanra S.; Dhamija P.; Sharma A.; Kumar A.; Handu S. Role of Structural and Non-Structural Proteins and Therapeutic Targets of SARS-CoV-2 for COVID-19. Cells 2021, 10, 821. 10.3390/cells10040821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harcourt B. H.; Jukneliene D.; Kanjanahaluethai A.; Bechill J.; Severson K. M.; Smith C. M.; Rota P. A.; Baker S. C. Identification of Severe Acute Respiratory Syndrome Coronavirus Replicase Products and Characterization of Papain-Like Protease Activity. Journal of Virology 2004, 78, 13600–13612. 10.1128/JVI.78.24.13600-13612.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manfredonia I.; Nithin C.; Ponce-Salvatierra A.; Ghosh P.; Wirecki T. K.; Marinus T.; Ogando N. S.; Snijder E. J.; Martijn J.; Bujnicki J. M.; et al. Genome-wide mapping of SARS-CoV-2 RNA structures identifies therapeutically-relevant elements. Nucleic Acids Res. 2020, 48, 12436–12452. 10.1093/nar/gkaa1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm T.; Ebert G.; Calleja D. J.; Allison C. C.; Richardson L. W.; Bernardini J. P.; Lu B. G.; Kuchel N. W.; Grohmann C.; Shibata Y.; et al. Mechanism and inhibition of the papain-like protease, PLpro, of SARS-CoV-2. EMBO J. 2020, 39, e106275. 10.15252/embj.2020106275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClain C. B.; Vabret N. SARS-CoV-2: the many pros of targeting PLpro. Signal Transduction Targeted Ther. 2020, 5, 223. 10.1038/s41392-020-00335-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osipiuk J.; Azizi S.-A.; Dvorkin S.; Endres M.; Jedrzejczak R.; Jones K. A.; Kang S.; Kathayat R. S.; Kim Y.; Lisnyak V. G.; et al. Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors. Nat. Commun. 2021, 12, 743. 10.1038/s41467-021-21060-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L.; Lin D.; Sun X.; Curth U.; Drosten C.; Sauerhering L.; Becker S.; Rox K.; Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science 2020, 368, 409–412. 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao J.; Li Y.-S.; Zeng R.; Liu F.-L.; Luo R.-H.; Huang C.; Wang Y.-F.; Zhang J.; Quan B.; Shen C.; et al. SARS-CoV-2 Mpro inhibitors with antiviral activity in a transgenic mouse model. Science 2021, 371, 1374–1378. 10.1126/science.abf1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidman D.; Gehrtz P.; Filep M.; Fearon D.; Gabizon R.; Douangamath A.; Prilusky J.; Duberstein S.; Cohen G.; Owen C. D.; et al. An automatic pipeline for the design of irreversible derivatives identifies a potent SARS-CoV-2 Mpro inhibitor. Cell Chemical Biology 2021, 28, 1795–1806. 10.1016/j.chembiol.2021.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartenian E.; Nandakumar D.; Lari A.; Ly M.; Tucker J. M.; Glaunsinger B. A. The molecular virology of coronaviruses. J. Biol. Chem. 2020, 295, 12910–12934. 10.1074/jbc.REV120.013930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein S.; Cortese M.; Winter S. L.; Wachsmuth-Melm M.; Neufeldt C. J.; Cerikan B.; Stanifer M. L.; Boulant S.; Bartenschlager R.; Chlanda P. SARS-CoV-2 structure and replication characterized by in situ cryo-electron tomography. Nat. Commun. 2020, 11, 5885. 10.1038/s41467-020-19619-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.; Lan Y.; Sanyal S. Membrane heist: Coronavirus host membrane remodeling during replication. Biochimie 2020, 179, 229–236. 10.1016/j.biochi.2020.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoops K.; Kikkert M.; Worm S. H. E. V. D.; Zevenhoven-Dobbe J. C.; Van Der Meer Y.; Koster A. J.; Mommaas A. M.; Snijder E. J. SARS-Coronavirus Replication Is Supported by a Reticulovesicular Network of Modified Endoplasmic Reticulum. PLoS Biol. 2008, 6, e226. 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Wilde A. H.; Raj V. S.; Oudshoorn D.; Bestebroer T. M.; Van Nieuwkoop S.; Limpens R. W. A. L.; Posthuma C. C.; Van Der Meer Y.; Bárcena M.; Haagmans B. L.; et al. MERS-coronavirus replication induces severe in vitro cytopathology and is strongly inhibited by cyclosporin A or interferon-α treatment. Journal of General Virology 2013, 94, 1749–1760. 10.1099/vir.0.052910-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana J.; López-Montero N.; Elliott R. M.; Fernández J. J.; Risco C. The unique architecture of Bunyamwera virus factories around the Golgi complex. Cellular Microbiology 2008, 10, 2012–2028. 10.1111/j.1462-5822.2008.01184.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenorio R.; Fernández de Castro I.; Knowlton J. J.; Zamora P. F.; Lee C. H.; Mainou B. A.; Dermody T. S.; Risco C. Reovirus σNS and μNS Proteins Remodel the Endoplasmic Reticulum to Build Replication Neo-Organelles. mBio 2018, 9, 01253. 10.1128/mBio.01253-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Brey I.; Merz A.; Chiramel A.; Lee J.-Y.; Chlanda P.; Haselman U.; Santarella-Mellwig R.; Habermann A.; Hoppe S.; Kallis S.; et al. Three-Dimensional Architecture and Biogenesis of Membrane Structures Associated with Hepatitis C Virus Replication. PLoS Pathog. 2012, 8, e1003056. 10.1371/journal.ppat.1003056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul D.; Hoppe S.; Saher G.; Krijnse-Locker J.; Bartenschlager R. Morphological and Biochemical Characterization of the Membranous Hepatitis C Virus Replication Compartment. Journal of Virology 2013, 87, 10612–10627. 10.1128/JVI.01370-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelini M. M.; Akhlaghpour M.; Neuman B. W.; Buchmeier M. J. Severe Acute Respiratory Syndrome Coronavirus Nonstructural Proteins 3, 4, and 6 Induce Double-Membrane Vesicles. mBio 2013, 4, 00534. 10.1128/mBio.00524-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oudshoorn D.; Rijs K.; Limpens R. W. A. L.; Groen K.; Koster A. J.; Snijder E. J.; Kikkert M.; Bárcena M. Expression and Cleavage of Middle East Respiratory Syndrome Coronavirus nsp3–4 Polyprotein Induce the Formation of Double-Membrane Vesicles That Mimic Those Associated with Coronaviral RNA Replication. mBio 2017, 8, 01658. 10.1128/mBio.01658-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M.; Carmichael G. G. Antisense RNA: Function and Fate of Duplex RNA in Cells of Higher Eukaryotes. Microbiology and Molecular Biology Reviews 1998, 62, 1415–1434. 10.1128/MMBR.62.4.1415-1434.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur S. Double-Stranded RNA Sensors and Modulators in Innate Immunity. Annu. Rev. Immunol. 2019, 37, 349–375. 10.1146/annurev-immunol-042718-041356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E. J.; Limpens R. W. A. L.; De Wilde A. H.; De Jong A. W. M.; Zevenhoven-Dobbe J. C.; Maier H. J.; Faas F. F. G. A.; Koster A. J.; Bárcena M. A unifying structural and functional model of the coronavirus replication organelle: Tracking down RNA synthesis. PLoS Biol. 2020, 18, e3000715. 10.1371/journal.pbio.3000715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff G.; Limpens R. W. A. L.; Zevenhoven-Dobbe J. C.; Laugks U.; Zheng S.; De Jong A. W. M.; Koning R. I.; Agard D. A.; Grünewald K.; Koster A. J.; et al. A molecular pore spans the double membrane of the coronavirus replication organelle. Science 2020, 369, 1395–1398. 10.1126/science.abd3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendonça L.; Howe A.; Gilchrist J. B.; Sheng Y.; Sun D.; Knight M. L.; Zanetti-Domingues L. C.; Bateman B.; Krebs A.-S.; Chen L.; et al. Correlative multi-scale cryo-imaging unveils SARS-CoV-2 assembly and egress. Nat. Commun. 2021, 12, 4629. 10.1038/s41467-021-24887-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicki S. G.; Sawicki D. L.; Siddell S. G. A Contemporary View of Coronavirus Transcription. Journal of Virology 2007, 81, 20–29. 10.1128/JVI.01358-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillen H. S. Structure and function of SARS-CoV-2 polymerase. Current Opinion in Virology 2021, 48, 82–90. 10.1016/j.coviro.2021.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y.; Yan L.; Huang Y.; Liu F.; Zhao Y.; Cao L.; Wang T.; Sun Q.; Ming Z.; Zhang L.; et al. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science 2020, 368, 779–782. 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone B.; Chen J.; Wang Q.; Llewellyn E.; Choi Y. J.; Olinares P. D. B.; Cao X.; Hernandez C.; Eng E. T.; Chait B. T.; et al. Structural basis for backtracking by the SARS-CoV-2 replication–transcription complex. Proc. Natl. Acad. Sci. U. S. A. 2021, 118, e2102516118. 10.1073/pnas.2102516118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjuán R.; Nebot M. R.; Chirico N.; Mansky L. M.; Belshaw R. Viral Mutation Rates. Journal of Virology 2010, 84, 9733–9748. 10.1128/JVI.00694-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S.; Chen H.; Chen Z.; Yang F.; Ye F.; Zheng Y.; Yang J.; Lin X.; Sun H.; Wang L.; et al. Crystal structure of SARS-CoV-2 nsp10 bound to nsp14-ExoN domain reveals an exoribonuclease with both structural and functional integrity. Nucleic Acids Res. 2021, 49, 5382–5392. 10.1093/nar/gkab320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano M.; Ruggiero A.; Squeglia F.; Maga G.; Berisio R. A Structural View of SARS-CoV-2 RNA Replication Machinery: RNA Synthesis, Proofreading and Final Capping. Cells 2020, 9, 1267. 10.3390/cells9051267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortese M.; Lee J.-Y.; Cerikan B.; Neufeldt C. J.; Oorschot V. M. J.; Köhrer S.; Hennies J.; Schieber N. L.; Ronchi P.; Mizzon G.; et al. Integrative Imaging Reveals SARS-CoV-2-Induced Reshaping of Subcellular Morphologies. Cell Host & Microbe 2020, 28, 853–866. 10.1016/j.chom.2020.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed A. M.; Taha T. Y.; Tabata T.; Chen I. P.; Ciling A.; Khalid M. M.; Sreekumar B.; Chen P.-Y.; Hayashi J. M.; Soczek K. M.; et al. Rapid assessment of SARS-CoV-2–evolved variants using virus-like particles. Science 2021, 374, 1626–1632. 10.1126/science.abl6184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iserman C.; Roden C.; Boerneke M.; Sealfon R.; McLaughlin G.; Jungreis I.; Park C.; Boppana A.; Fritch E.; Hou Y. J.; et al. Specific viral RNA drives the SARS CoV-2 nucleocapsid to phase separate. bioRxiv, June 12, 2020. 10.1101/2020.06.11.147199 (accessed November 6, 2021) [DOI] [Google Scholar]
- Ghosh S.; Dellibovi-Ragheb T. A.; Kerviel A.; Pak E.; Qiu Q.; Fisher M.; Takvorian P. M.; Bleck C.; Hsu V. W.; Fehr A. R.; et al. β-Coronaviruses Use Lysosomes for Egress Instead of the Biosynthetic Secretory Pathway. Cell 2020, 183, 1520–1535. 10.1016/j.cell.2020.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan L.; Zheng Q.; Zhang H.; Niu Y.; Lou Y.; Wang H. The SARS-CoV-2 Spike Glycoprotein Biosynthesis, Structure, Function, and Antigenicity: Implications for the Design of Spike-Based Vaccine Immunogens. Front. Immunol. 2020, 11, 576622. 10.3389/fimmu.2020.576622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar M.; Saha S. Structural insight into the role of novel SARS-CoV-2 E protein: A potential target for vaccine development and other therapeutic strategies. PLoS One 2020, 15, e0237300. 10.1371/journal.pone.0237300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracquemond D.; Muriaux D. Betacoronavirus Assembly: Clues and Perspectives for Elucidating SARS-CoV-2 Particle Formation and Egress. mBio 2021, 12, 02371. 10.1128/mBio.02371-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossman J. S.; Jing X.; Leser G. P.; Lamb R. A. Influenza Virus M2 Protein Mediates ESCRT-Independent Membrane Scission. Cell 2010, 142, 902–913. 10.1016/j.cell.2010.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rheinemann L.; Sundquist W. I.. Virus Budding. In Encycloperia of Virology, 4th ed.; Bamford D. H., Zuckerman M., Eds.; Academic Press: Cambridge, MA, 2021; pp 519–528. [Google Scholar]
- Welsch S.; Müller B.; Kräusslich H.-G. More than one door - Budding of enveloped viruses through cellular membranes. FEBS Lett. 2007, 581, 2089–2097. 10.1016/j.febslet.2007.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pornillos O.; Garrus J. E.; Sundquist W. I. Mechanisms of enveloped RNA virus budding. Trends in Cell Biology 2002, 12, 569–579. 10.1016/S0962-8924(02)02402-9. [DOI] [PubMed] [Google Scholar]
- Chen D.; Zheng Q.; Sun L.; Ji M.; Li Y.; Deng H.; Zhang H. ORF3a of SARS-CoV-2 promotes lysosomal exocytosis-mediated viral egress. Developmental Cell 2021, 56, 3250–3263. 10.1016/j.devcel.2021.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorshkov K.; Chen C. Z.; Bostwick R.; Rasmussen L.; Tran B. N.; Cheng Y.-S.; Xu M.; Pradhan M.; Henderson M.; Zhu W.; et al. The SARS-CoV-2 Cytopathic Effect Is Blocked by Lysosome Alkalizing Small Molecules. ACS Infectious Diseases 2021, 7, 1389–1408. 10.1021/acsinfecdis.0c00349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BIO COVID-19 Therapeutic Development Tracker. Biotechnology Innovation Organization. https://www.bio.org/policy/human-health/vaccines-biodefense/coronavirus/pipeline-tracker (accessed 2021-12-22).
- Hunt A. C.; Case J. B.; Park Y.-J.; Cao L.; Wu K.; Walls A. C.; Liu Z.; Bowen J. E.; Yeh H.-W.; Saini S. Multivalent designed proteins neutralize SARS-CoV-2 variants of concern and confer protection against infection in mice. Sci. Transl. Med. 2022, 14, 1252. 10.1126/scitranslmed.abn1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh C.-L.; Goldsmith J. A.; Schaub J. M.; Divenere A. M.; Kuo H.-C.; Javanmardi K.; Le K. C.; Wrapp D.; Lee A. G.; Liu Y.; et al. Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science 2020, 369, 1501–1505. 10.1126/science.abd0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallesen J.; Wang N.; Corbett K. S.; Wrapp D.; Kirchdoerfer R. N.; Turner H. L.; Cottrell C. A.; Becker M. M.; Wang L.; Shi W.; et al. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc. Natl. Acad. Sci. U. S. A. 2017, 114, E7348–E7357. 10.1073/pnas.1707304114. [DOI] [PMC free article] [PubMed] [Google Scholar]