Abstract
Genetic analyses have suggested that the pyrimidine moiety of thiamine can be synthesized independently of the first enzyme of de novo purine synthesis, phosphoribosylpyrophosphate amidotransferase (PurF), in Salmonella typhimurium. To obtain biochemical evidence for and to further define this proposed synthesis, stable isotope labeling experiments were performed with two compounds, [2-13C]glycine and [13C]formate. These compounds are normally incorporated into thiamine pyrophosphate (TPP) via steps in the purine pathway subsequent to PurF. Gas chromatography-mass spectrometry analyses indicated that both of these compounds were incorporated into the pyrimidine moiety of TPP in a purF mutant. This result clearly demonstrated that the pyrimidine moiety of thiamine was being synthesized in the absence of the PurF enzyme and strongly suggested that this synthesis utilized subsequent enzymes of the purine pathway. These results were consistent with an alternative route to TPP that bypassed only the first enzyme in the purine pathway. Experiments quantitating cellular thiamine monophosphate (TMP) and TPP levels suggested that the alternative route to TPP did not function at the same capacity as the characterized pathway and determined that levels of TMP and TPP in the wild-type strain were significantly altered by the presence of purines in the medium.
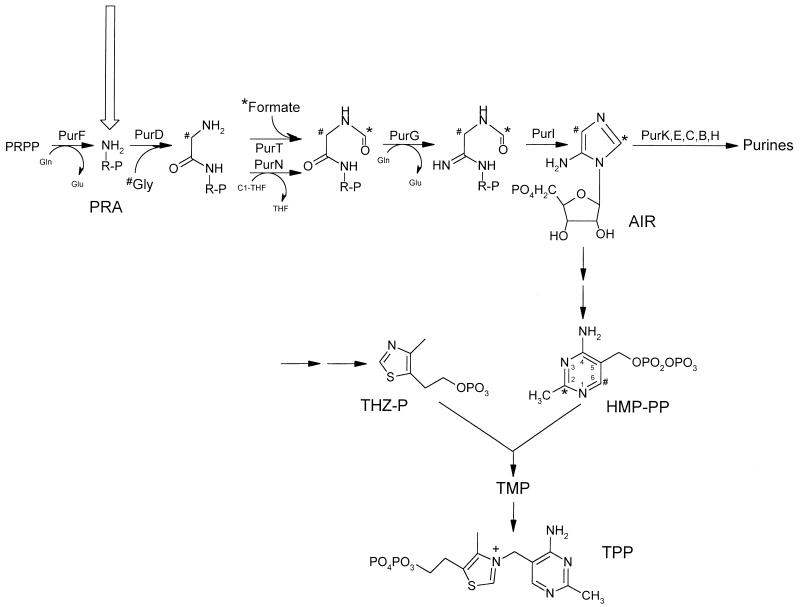
Thiamine pyrophosphate (TPP) is a cofactor for reactions involving the transfer of C2 units and is required by a number of important metabolic enzymes, including pyruvate dehydrogenase (EC 1.2.4.1), α-ketoglutarate dehydrogenase (EC 1.2.4.2), and acetolactate synthase (EC 4.1.3.18). Despite the critical role of this vitamin in cellular metabolism, its biosynthetic pathway is not well understood in any organism. Our current understanding of TPP synthesis in Salmonella typhimurium, including results presented herein, is shown in Fig. 1. Two independently synthesized precursor molecules, 4-amino-5-hydroxymethylpyrimidine pyrophosphate (HMP-PP) and 4-methyl-5-(β-hydroxyethyl)thiazole-phosphate (THZ-P), are first condensed to form thiamine monophosphate (TMP), which is subsequently phosphorylated to form TPP. The major precursor molecules for the HMP and THZ moieties have been determined by labeling studies (4–6, 13, 16, 24), but many of the enzymatic steps in these pathways have not been clearly defined. Genetic and biochemical studies have shown that HMP is derived from an intermediate in the well-characterized purine pathway, aminoimidazole ribotide (AIR) (13, 15, 22–24). The first five purine enzymes thus play a role in both purine and TPP synthesis.
FIG. 1.
Schematic representation of TPP biosynthesis. Pathways involved in TPP synthesis are shown. Purine biosynthesis is indicated with structural intermediates prior to the AIR branch point for HMP-PP synthesis. Purine gene products are indicated above the reactions that they catalyze. The carbons in HMP-PP that originate from the C-2 of glycine and the carbon in formate are indicated by the pound signs and asterisks, respectively. TPP synthesis independent of the PurF enzyme is indicated by the unfilled arrow. Enzymatic steps for the conversion of AIR to HMP-PP and for the synthesis of THZ-P have not been clearly defined. PRPP, phosphoribosylpyrophosphate.
Although the involvement of the purine pathway in HMP synthesis is clear, genetic evidence has suggested that HMP can be synthesized independently of purine enzymes in S. typhimurium. A mutant defective in the first enzyme of the purine pathway, phosphoribosylpyrophosphate amidotransferase (PurF) (EC 2.4.2.14), is able to grow in the absence of thiamine under a number of conditions, including medium containing glucose as a carbon source if exogenous pantothenate is provided (9); medium containing a number of nonglucose carbon sources, i.e., gluconate and ribose (27); or anaerobic conditions (7). This mutant lacked detectable phosphoribosylpyrophosphate amidotransferase activity (10). These results suggested that a route (or routes) independent of the PurF enzyme could synthesize HMP. As these genetic studies have progressed, it has become necessary to (i) confirm biochemically that the thiamine-independent growth of a purF mutant is due to TPP synthesis and (ii) perform biochemical experiments to address the role of subsequent purine enzymes in this synthesis.
Results of stable isotope labeling experiments presented in this report demonstrated that HMP synthesis occurred in the absence of the PurF enzyme in S. typhimurium and strongly suggested that, under the growth conditions tested, this synthesis utilized the remaining purine enzymes involved in the formation of AIR. Experiments quantitating cellular TMP and TPP levels suggested that PurF-independent TPP synthesis did not function at the same capacity as that involving the PurF enzyme. In addition, these measurements determined that levels of TMP and TPP in a wild-type strain were decreased when purines were present in the growth medium, suggesting the biochemical basis for previously described adenine-sensitive phenotypes.
MATERIALS AND METHODS
Bacterial strains, media, and chemicals.
The two strains used in this study were S. typhimurium LT2 (wild type) and DM1936, a derivative of LT2 that contains a deletion of the purF gene (purF2085) (9). DM1936 lacks detectable phosphoribosylpyrophosphate amidotransferase activity and any sequences that hybridize to the purF gene (9, 10). The NCE medium of Berkowitz et al. (2) was used as minimal medium. Carbon sources (glucose, gluconate, and ribose) were added to a final concentration of 16 mM. Difco nutrient broth (8 g/liter) with NaCl (5 g/liter) added was used as rich medium. When present in the culture media and unless otherwise indicated, the compounds were used at the indicated final concentrations: adenine, 0.4 mM; pantothenate, 100 μM; formate, 20 mM. Stable isotopic forms of glycine (99 atom% 2-13C) and sodium formate (99 atom% 13C) were obtained from Aldrich Chemical Company (Milwaukee, Wis.) and added at final concentrations of 4.4 and 4.8 mM, respectively. All other chemicals were purchased from Sigma Chemical Company, St. Louis, Mo.
Quantitation of TMP and TPP. (i) Bacterial growth.
To quantitate TMP and TPP levels over the growth of a bacterial culture, a 1-liter culture of strain LT2 was grown in glucose minimal medium. Aliquots (125 ml) were removed and pelleted, and A650 was measured at the indicated time points. For quantitation of TMP-TPP under different growth conditions, a 200-ml culture of either LT2 or DM1936 (purF2085) was grown in minimal medium with the indicated supplements to 100 Klett units and pelleted.
(ii) TMP and TPP extraction and derivatization.
TMP and TPP were extracted and assayed by a modification of CNBr thiochrome derivatization (19). Briefly, pelleted cells were resuspended in 1 ml of double-distilled H2O and divided into two aliquots. One aliquot was used for dry weight determination. A 0.1-ml quantity of 1 M HCl was added to the other aliquot followed by incubation on ice for 30 min with intermittent vortexing. During the course of experiments measuring TMP and TPP, it was found that the standard protocol for extracting thiamine (20) involving boiling cells in 0.1 M HCl resulted in significant breakdown of TPP to TMP; performing the acid extraction at 0°C reduced this degradation. After extraction, the sample was centrifuged at 30,000 × g for 20 min to remove cell debris. For derivatization, 12.5 μl of 0.3 M CNBr (or double-distilled H2O as a control) was added to 100 μl of extract. After vortexing, 12.5 μl of 1 M NaOH was added to neutralize each sample. This derivatized extract (2.5 to 10 μl) was analyzed by high-pressure liquid chromatography (HPLC).
(iii) Thiochrome HPLC analysis.
The thiochromes in the derivatized assay supernatant were separated by normal-phase HPLC with a Lichrosorb-NH2 10-μm column (Altech, Deerfield, Ill.) as has been described elsewhere (19, 25). The mobile phase used was acetonitrile–90 mM potassium phosphate (pH 8.4) in a 60:40 ratio with a flow rate of 1.4 ml/min. Under these conditions, thiochrome standards of thiamine, TMP, and TPP eluted at 2.5, 5.3, and 7.1 min, respectively. Thiochromes were detected with a Waters 990 spectrofluorimeter detector set at 375 nm (excitation) and 432 nm (emission). Waters Millennium 2000 software was used to determine the area under the eluted peak. Concentrations were calculated by using a standard curve with known TMP and TPP concentrations.
Isolation of the pyrimidine moiety of thiamine.
A 1.5-liter culture of strain LT2 or DM1936 (purF2085) was grown in minimal medium with the indicated supplements to an A650 of approximately 0.8. Cells were pelleted, and a previously described procedure for derivatization and purification of the HMP moiety of thiamine (35) was applied without modification. Briefly, TPP was extracted from cells by boiling them in 0.1 M HCl for 20 min. After removal of cell debris by centrifugation, TPP in the crude extract was cleaved by ethanethiol to thiazole diphosphate and 2-methyl-4-amino-5-[(ethyl-thio)methyl]pyrimidine (ETMP). ETMP was then purified by repeated extractions with dichloromethane. The extractions were reduced to dryness with a stream of nitrogen, and the residue containing ETMP was resuspended in 30 μl of ethyl acetate for gas chromatography-mass spectrometry (GC-MS) analysis.
GC-MS.
GC-MS analysis was performed with a Shimadzu GC-17A gas chromatograph and a Shimadzu QP-5000 mass spectrometer. The GC-MS spectrometer was equipped with a 30-m by 0.25-mm SGE BPX5 column packed with 5% phenyl polysilphenylene-siloxane. Conditions for the analysis of ETMP were as follows: column, 140°C; injector, 200°C; and interface, 220°C. All spectra were recorded with the detector at 1.2 keV. Under these conditions, ETMP had a retention time of 28 min. The mass spectra of 10 to 12 successive scans of the ETMP peak were averaged with Shimadzu GC-MS Class-5000 software, and this average was used for calculating the ion intensities of the main fragment (percentage of total).
RESULTS
Stable isotope labeling of HMP in a mutant lacking the PurF enzyme.
One explanation for the thiamine-independent growth of a purF mutant was that an alternative enzyme (or enzymes) synthesized 5-phosphoribosylamine (PRA), the product of the PurF-catalyzed reaction. In such a scenario, the subsequent four purine enzymes would be required for HMP synthesis. Because this model did not simply explain all of our genetic results, we sought to demonstrate HMP synthesis in the absence of the PurF enzyme and define the role of subsequent purine enzymes in this synthesis. To do this, we performed stable isotope labeling of HMP with glycine and formate in wild-type and purF mutant strains. Glycine and formate are incorporated into the pyrimidine moiety of TPP in wild-type strains via enzymes in the purine pathway (Fig. 1). The nitrogen, C-1, and C-2 of glycine are incorporated into the N-1, C-4, and C-6 of HMP, respectively, by glycinamide ribotide (GAR) synthetase (PurD) (14, 35), while the carbon in formate is incorporated into the C-2 of the pyrimidine ring by GAR transformylase T (PurT) (20).
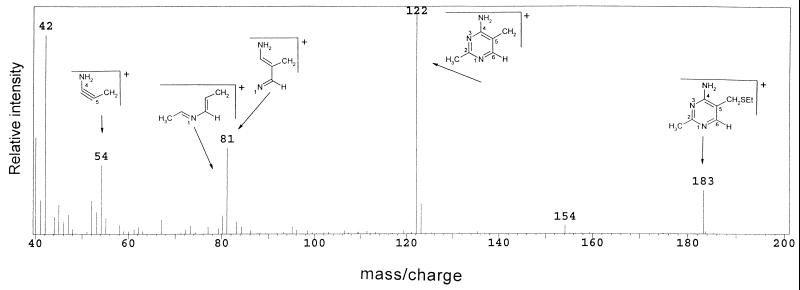
Labeling studies with the wild-type (LT2) and purF mutant (DM1936) strains were performed with cells grown on glucose adenine pantothenate medium and gluconate adenine medium. Previous genetic data suggested that PurF-independent TPP synthesis might occur by two different mechanisms under these two growth conditions (9, 27, 36). TPP from labeled cultures was cleaved by ethanethiol, and the pyrimidine moiety, in the form of ETMP, was subsequently purified and analyzed by GC-MS. The mass spectrum of unlabeled ETMP purified from strain LT2 grown on gluconate adenine medium is shown in Fig. 2 and detailed in Table 1, row 1. This spectrum was identical in its significant composition to that previously reported for Escherichia coli (35). The fragmentation pattern that has been previously reported is also shown.
FIG. 2.
Mass spectrum of unlabeled ETMP derived from strain LT2 grown in gluconate adenine medium. The fragmentation pattern shown was expected (35). The ETMP molecular ion (M+) is shown and has an m/z value of 183.
TABLE 1.
Incorporation of [2-13C]glycine and [13C]formate into the pyrimidine moiety of thiamine in a wild-type strain and purF mutanta
| Genotype | Growth medium | Label | Ions observed | Ion intensities of main fragment (% of total) for m/z:
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 122 | 123 | 124 | 125 | |||||||
| Wild type | Glc Ade | None | 54 | 81 | 122 | 183 | 88 | 12 | ||
| Wild type | Glc Ade | [2-13C]glycine | 54 | 82 | 124 | 185 | 6 | 7 | 79 | 8 |
| Wild type | Glu Ade Pan | [2-13C]glycine | 54 | 82 | 124 | 185 | 7 | 9 | 76 | 8 |
| Wild type | Glc Ade formate | [2-13C]glycine | 54 | 82 | 123*/124 | 184 | 5 | 67 | 27 | 1 |
| purF2085 | Glc Ade | [2-13C]glycine | 54 | 82 | 124 | 185 | 3 | 13 | 77 | 7 |
| purF2085 | Glu Ade Pan | [2-13C]glycine | 54 | 82 | 124 | 185 | 3 | 6 | 83 | 8 |
| purF2085 | Glc Ade formate | [2-13C]glycine | 54 | 82 | 123*/124 | 184 | 3 | 66 | 29 | 2 |
| Wild type | Glc Ade | [13C]formate | 54 | 81 | 122/123* | 184 | 24 | 70 | 6 | |
| Wild type | Glu Ade Pan | [13C]formate | 54 | 81 | 122/123* | 184 | 27 | 66 | 7 | |
| purF2085 | Glc Ade | [13C]formate | 54 | 81 | 122/123* | 184 | 24 | 72 | 4 | |
| purF2085 | Glu Ade Pan | [13C]formate | 54 | 81 | 122/123* | 184 | 25 | 70 | 5 | |
Strains were grown in the indicated media and labeled with the indicated stable isotope. The pyrimidine moiety of thiamine in the form of ETMP was isolated from these cultures and analyzed by GC-MS as described in Materials and Methods. The isotopic composition of the main fragment was calculated by averaging 10 to 12 successive scans from the mass spectrum. The asterisk indicates the major peak observed. Abbreviations: Glc, gluconate; Glu, glucose; Ade, adenine; Pan, pantothenate.
(i) Labeling studies with [2-13C]glycine.
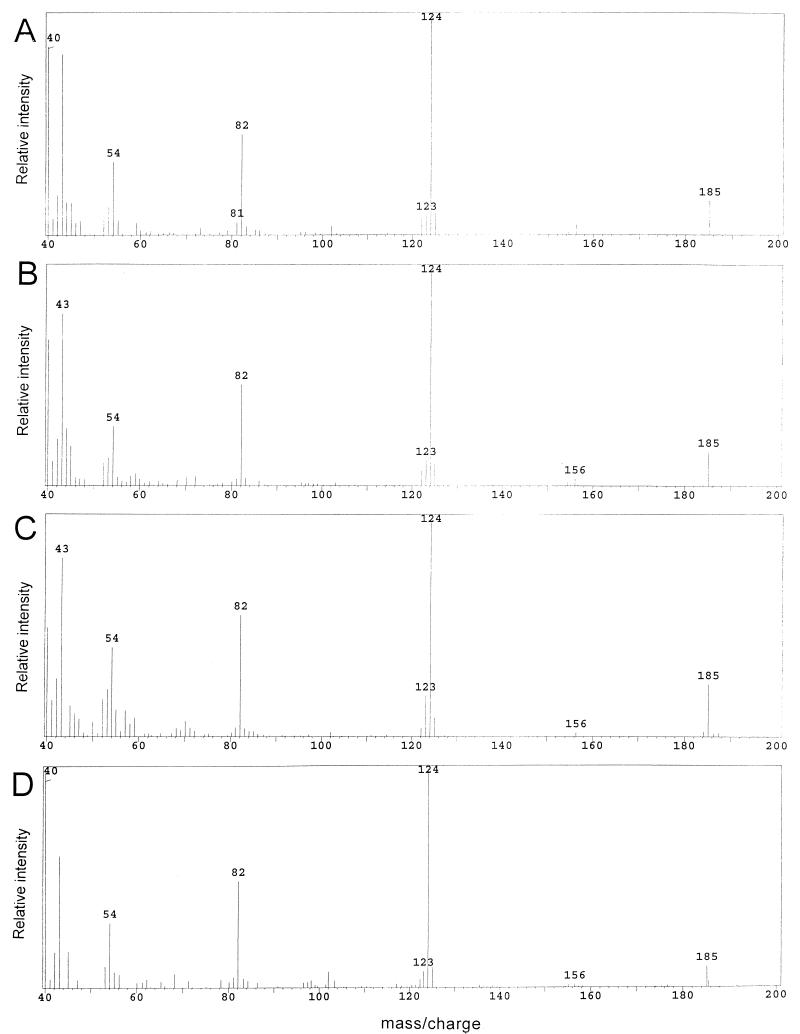
The results of labeling studies with [2-13C]glycine are shown in Fig. 3 and Table 1, rows 2 to 7. These mass spectra clearly indicated that [2-13C]glycine labeled the pyrimidine moiety of TPP in a purF mutant twice when this strain was grown in either gluconate adenine or glucose adenine pantothenate medium (Fig. 3C and D, respectively). Significantly, a similar labeling pattern was observed with the wild-type strain under the same conditions (Fig. 3A and B, respectively). Under both growth conditions, the m/z values of the molecular ion (M+, m/z 183) and the main fragment [(M-SC2H5)+, m/z 122] were shifted by 2 U (to 185 and 124, respectively). Calculations from the intensities of the main fragment showed that the percentage of molecules that had an m/z of 124 ranged from 76 to 83% in both LT2 and DM1936 (Table 1). The ions observed in each spectrum were consistent with one of the [2-13C]glycine labels being incorporated into the C-6 of the pyrimidine ring as previously described (through the activity of GAR synthetase, PurD) (35) and the other being incorporated into the C-2 of the pyrimidine ring (see below). Incorporation of label into both C-2 and C-6 of the pyrimidine ring (Fig. 2) would result in a shift of the 80, 122, and 183 ions by 2 mass units (to 82, 124, and 185, respectively), a 1-mass unit shift in the 81 ion (to 82, due to incorporation at C-6), and no shift in the 54 ion. As shown in Fig. 3, this was the observed result with both the purF mutant and wild-type strains.
FIG. 3.
Mass spectra of ETMP derived from the following cultures labeled with [2-13C]glycine: strain LT2 grown in gluconate adenine medium (A), strain LT2 grown in glucose adenine pantothenate medium (B), DM1936 (purF2085) grown in gluconate adenine medium (C), and DM1936 grown in glucose adenine pantothenate medium (D).
Control labeling experiments performed in the presence of excess formate (Table 1, rows 4 and 7) demonstrated that [2-13C]glycine was most likely incorporated into the C-2 of the pyrimidine ring through folate metabolism. Under these conditions, mass 124 and 185 decreased to 123 and 184, respectively, but mass 82 remained. Thus, according to Fig. 2, excess formate interfered with incorporation of [2-13C]glycine into the C-2 of the pyrmidine ring. The C-2 of glycine is known to contribute to the cellular pool of formyl tetrahydrofolate by the glycine cleavage system (29). We propose that such labeled C1 units were then incorporated into TPP by GAR transformylase N (PurN [Fig. 1]). Formate would thus dilute this incorporation through the activity of GAR transformylase T (PurT [Fig. 1]). The high efficiency of incorporation of [2-13C]glycine into the C-2 of the pyrimidine ring was likely due to the presence of purines in the growth medium (30) and reduced levels of formate due to exogenous glycine (26).
The incorporation of [2-13C]glycine into the pyrimidine moiety of TPP in a purF mutant demonstrated unequivocally that HMP was being synthesized independently of the PurF enzyme in S. typhimurium. Since [2-13C]glycine was incorporated into HMP in a purF mutant in a manner identical to that of the wild-type strain (where incorporation has been shown previously to be due to function of the purine pathway), these results suggested that the subsequent purine enzymes involved in the formation of AIR were utilized in this synthesis.
(ii) Labeling studies with [13C]formate.
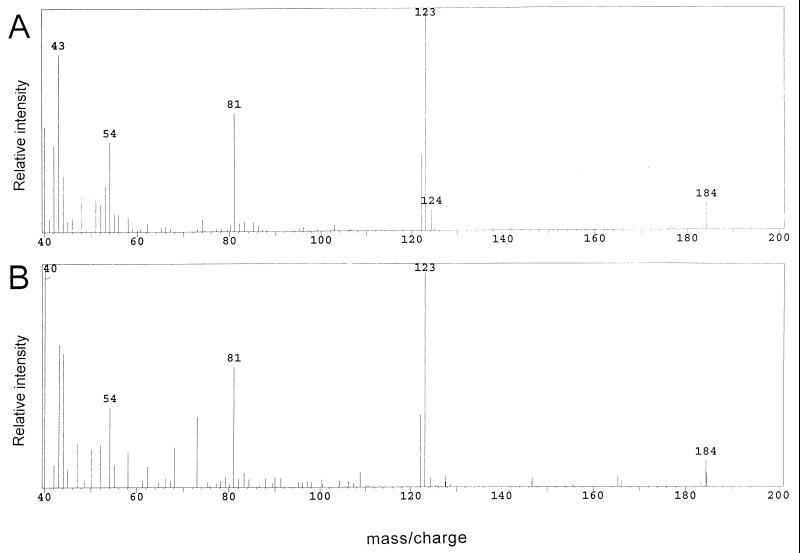
To minimize the possibility that the above labeling results mistakenly implicated purine enzymes in PurF-independent TPP synthesis, additional labeling studies were performed with [13C]formate. The results are shown in Fig. 4 and Table 1, rows 8 to 11. These data clearly demonstrated that [13C]formate labeled the pyrimidine moiety of TPP in a purF mutant in a manner identical to that of the wild-type strain. The m/z values of the molecular ion (M+, m/z 183) and the main fragment [(M-SC2H5)+, m/z 122] were shifted by 1 U, but the values of the 54 and 81 ions remained the same, consistent with previous data indicating that formate was incorporated into the C-2 of the pyrimidine ring (20). [13C]formate was incorporated into the pyrimidine moiety of TPP whether strains were grown in gluconate adenine or in glucose adenine pantothenate medium. Ion intensity calculations from the main fragment showed that 66 to 72% of the molecules from both LT2 and DM1936 had an m/z of 123, while 24 to 27% of the molecules had an m/z of 122 (unlabeled) (Table 1). The dilution of label was most likely due to the fact that the C-2 of the pyrimidine moiety of TPP is incorporated by two enzymes, only one of which incorporates formate directly (Fig. 1). The results obtained from these labeling studies demonstrated that formate was incorporated into the pyrimidine moiety of TPP independently of the PurF enzyme and provided further evidence that, under both growth conditions tested, enzymes of the purine pathway were utilized for this TPP synthesis.
FIG. 4.
Mass spectra of ETMP derived from the following cultures labeled with [13C]formate: strain LT2 grown in gluconate adenine medium (A) and DM1936 (purF2085) grown in gluconate adenine medium (B).
Comparison of cellular TMP and TPP levels in wild-type and purF mutant strains under different growth conditions.
The above results clearly demonstrated that TPP synthesis could occur independently of the PurF enzyme in S. typhimurium. We therefore sought to quantitate the TPP accumulation in strains that were dependent on PurF-independent TPP synthesis for growth. TMP and TPP levels were measured in strains grown under three different conditions that allowed thiamine-independent growth of a purF mutant. The results are shown in Table 2. Several points can be made from these data. (i) purF mutants accumulated significant levels of TPP (8.1 to 23.8 pmol/mg [dry weight]), consistent with the demonstrated synthesis above. (ii) TPP levels in the purF mutant were lower than those found under the same conditions in the wild-type strain (22.5 to 41.4 pmol/mg [dry weight]). (iii) The purF mutant accumulated the highest levels of TPP when ribose was used as a carbon source. Under this condition, TPP levels in the purF mutant approached the levels measured in the wild-type strain. This result was consistent with previous genetic data indicating that function of the alternative route(s) to TPP was dependent on levels of ribose-5-P (11, 27). (iv) There was a striking effect of exogenous purines (adenine) on both the levels and the ratios of TPP and TMP in the wild-type strain. The TPP/TMP ratio in the wild-type strain grown on each of the three carbon sources increased four- to fivefold when adenine was included in the medium. Although adenine appeared to have a slight effect on TPP levels (20 to 45% reduction), the major effect was on TMP levels, which decreased seven- to eightfold. This result demonstrated a physiological effect of exogenous adenine not quantitated previously and provided a possible biochemical basis for thiamine-correctable adenine-sensitive phenotypes of some E. coli and S. typhimurium mutants (1, 3, 8, 18, 31).
TABLE 2.
TPP and TMP content in a wild-type strain and a purF mutant grown in different mediaa
| Content | Genotype | Value (pmol/mg [dry wt]) for growth medium:
|
||||||
|---|---|---|---|---|---|---|---|---|
| Glucose | Glucose adenine | Glucose adenine Pan | Gluconate | Gluconate adenine | Ribose | Ribose adenine | ||
| TPP | Wild type | 41.1 ± 1.3 | 23.7 ± 2.3 | 22.5 ± 1.5 | 50.7 ± 4.3 | 41.4 ± 2.1 | 43.1 ± 2.4 | 34.0 ± 1.3 |
| purF2085 | NG | NG | 8.1 ± 1.1 | NG | 15.4 ± 1.5 | NG | 23.8 ± 0.3 | |
| TMP | Wild type | 6.6 ± 1.4 | 1.0 ± 0.4 | 0.7 ± 0.2 | 13.9 ± 0.5 | 2.5 ± 0.7 | 6.6 ± 2.2 | 0.9 ± 0.1 |
| purF2085 | NG | NG | 0.5 ± 0.1 | NG | 0.6 ± 0.3 | NG | 1.2 ± 0.1 | |
A wild-type strain (LT2) and a purF mutant (DM1936, purF2085) were grown in the indicated media to 100 Klett units. Cells were pelleted and assayed for TPP and TMP content as described in Materials and Methods. Abbreviations: Pan, pantothenate; NG, no growth.
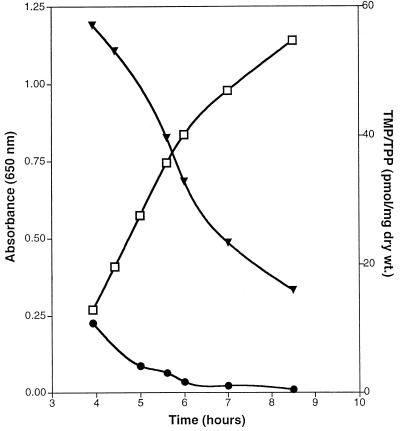
TPP and TMP levels change during cell growth.
In the course of experiments investigating levels of TMP and TPP, we monitored the levels of these compounds during the growth of a bacterial culture. The results, shown in Fig. 5, indicated that both TMP and TPP varied significantly during the course of bacterial growth. The TPP levels decreased approximately 3.6-fold, from 57 pmol/mg (dry weight) during early log phase (optical density at 650 nm, 0.27) to 16 pmol/mg (dry weight) in early stationary phase (optical density at 650 nm, 1.14). Variation of the TMP levels was more dramatic, decreasing 22-fold, from 11 pmol/mg (dry weight) in early log phase to 0.5 pmol/mg (dry weight) in early stationary phase. After 24 h of growth, TPP and TMP levels were 9 and 0.7 pmol/mg (dry weight), respectively.
FIG. 5.
TPP and TMP levels change significantly during growth. Strain LT2 was grown in minimal glucose medium, and aliquots were removed at the indicated times. Aliquots were assayed for TMP and TPP content as described in Materials and Methods. Growth of the culture was monitored by A650 (empty squares). TPP and TMP levels (picomoles per milligram [dry weight]) are indicated by the filled inverted triangles and filled circles, respectively. The data shown are representative of at least three independent experiments.
A number of experiments were performed to address whether the observed variations in TPP and TMP levels over growth were due to experimental procedure. To investigate whether the high initial levels of TPP and TMP were due to high levels of these compounds in the inoculum, the culture shown in Fig. 5 was subcultured from a culture grown to full density in identical minimal medium. Previous experiments indicated that TPP and TMP levels in full-density LT2 cultures grown under these conditions were approximately 12 and 3 pmol/mg (dry weight), respectively, i.e., levels of these compounds in the inoculum were low. Additional experiments to address whether the decrease in TPP and TMP levels was due to altered aeration of the culture (from removal of aliquots during the experiment) were performed. Results from these experiments, which involved measuring TPP and TMP levels from independently grown cultures stopped at various points during growth, did not support this idea (data not shown). Cultures grown in different media, i.e., utilizing gluconate as a carbon source, or different strains, i.e., DM1936 (purF2085), also showed a similar pattern (data not shown). These results suggested that the decrease in cellular TMP and TPP content was of a general nature in S. typhimurium and not specific for a strain or a growth condition. Preliminary experiments to address whether the variations in TPP and TMP observed during growth were due to regulation of thiamine biosynthetic genes at the transcriptional level were performed with a thiH::Mud-lacZ gene fusion. The thiH gene is in an operon (thiCEFSGH) known to be regulated by TPP in S. typhimurium (33). These experiments failed to detect parallel regulation (34). The physiological significance of the apparent decrease in TMP-TPP levels is not obvious but may represent a regulatory effect on TPP metabolism not previously explored.
DISCUSSION
This study was initiated to biochemically demonstrate HMP synthesis in the absence of the PurF enzyme and to determine the involvement of the purine pathway in this alternative route to HMP. Stable isotope labeling experiments demonstrated that atoms in glycine and formate, two compounds incorporated into HMP via steps in the purine pathway, were incorporated into the pyrimidine moiety of TPP in a purF mutant. Furthermore, the pattern of incorporation was identical to that of the wild-type strain under the same conditions. This result demonstrated unequivocally that thiamine-independent growth of a purF mutant under the tested conditions was due to HMP synthesis, not to an altered TPP requirement. This result was significant since (i) there is evidence that some growth conditions allow thiamine-independent growth because of a reduction in the TPP requirement (12) and (ii) it validated conclusions previously supported only by genetic data.
The labeling studies performed strongly suggested that the remaining purine enzymes involved in the synthesis of AIR were required for PurF-independent TPP synthesis. [2-13C]glycine and [13C]formate labeled the atoms of HMP predicted if the purine enzymes, PurD and PurT, were being utilized for HMP synthesis. While it would be formally possible for this incorporation to occur completely independently of the purine pathway, this possibility seems highly unlikely. These data are in agreement with previous genetic data indicating that insertion mutations in purine genes required for the synthesis of AIR result in thiamine auxotrophy under the conditions tested here (9, 27).
Significantly, glycine and formate were incorporated into HMP in a purF mutant whether strains were grown on glucose adenine pantothenate or on gluconate adenine medium. Previous genetic data had led to a model that these two growth conditions resulted in distinct mechanisms of PurF-independent TPP synthesis. The isolation of mutations (apbA) that specifically eliminated PurF-independent TPP synthesis on gluconate adenine medium (27) and the existence of purG and purI point mutations that resulted in a Pur− Thi+ phenotype (36) had been integral in this model. The recent demonstration that apbA encodes a pantothenate biosynthetic enzyme (PanE) has provided an explanation for the distinct phenotypes caused by this mutation (17). Recent results have also suggested that the Pur− Thi+ phenotype caused by some purG and purI point mutations is due to low levels of enzyme (37). Thus, all of these data are consistent with labeling results presented herein suggesting that PurF-independent TPP synthesis occurs by the same mechanism whether strains are grown on glucose adenine pantothenate or on gluconate adenine medium; results suggest that this alternative route bypasses the PurF enzyme but requires PurD, PurN/T, PurG, and PurI, i.e., all of the enzymes needed for the synthesis of AIR. Significant questions remain in defining how S. typhimurium compensates for the lack of the PurF enzyme in the synthesis of HMP and what role pantothenate has in this process. Attempts to isolate an alternative PRA-forming activity both biochemically and genetically have thus far been unsuccessful.
Physiological significance of TPP levels in purF mutant and wild-type strains.
Quantitation of TPP levels demonstrated that significant levels of TPP accumulated in the purF mutant, although these levels were reduced in comparison to those observed in the wild-type (purF+) strain. We postulate that the PurF-independent route to TPP does not function to the same capacity as the de novo pathway, at least under the conditions tested. This result is consistent with previous genetic data indicating that the alternative pathway is able to bypass the requirement for the PurF enzyme in TPP synthesis but not purine biosynthesis. It is possible that the enzyme(s) that bypasses PurF is not dedicated to TPP biosynthesis and/or that TPP synthesis independent of PurF exists to supplement the de novo pathway only under certain physiological conditions. One possible physiological explanation for the development of a PurF-independent mechanism for TPP synthesis would be to allow ample TPP synthesis in the presence of exogenous purines, a condition known to restrict PurF activity (21, 28).
Variation of TMP and TPP levels under different growth conditions.
In addition to increasing our understanding of HMP synthesis independent of the PurF enzyme, this study has provided the basis for further analysis of physiological conditions affecting TPP synthesis. Significant differences in TMP and TPP levels were observed during growth of the bacterial culture. Additional experiments to address the mechanisms regulating TMP and TPP levels during growth will be important in determining a possible physiological role for this phenomenon. Another factor that had a major effect on TPP and especially TMP levels was the presence of purines (adenine) in the medium. One explanation for this result is that adenine negatively affects synthesis of TMP and TPP. Adenine is known to inhibit growth of a number of S. typhimurium strains, and this inhibition is reversed by the addition of thiamine (1, 3, 8, 18, 31). Our data thus biochemically demonstrate an effect of adenine on TMP and TPP synthesis that has been suggested by previous genetic studies and, as such, provide the basis for further studies to understand this phenomenon. Neither the mechanism nor the target of adenine inhibition is clear. Feedback inhibition of the PurF enzyme (21) and/or repression of purine enzyme transcription by purines (28) has been proposed previously. Other explanations such as effects on the conversion of AIR to HMP or the synthesis of the thiazole moiety (Fig. 1) need to be explored. The dramatic effect of adenine on the TMP/TPP ratio suggests that the cell is additionally regulating the conversion of TMP to TPP via thiamine monophosphate kinase (ThiL). This is an attractive hypothesis since previously we identified mutations in thiL that resulted in a defect in thi gene regulation (32).
This study has provided critical biochemical data about PurF-independent TPP synthesis in S. typhimurium. Our results demonstrated that thiamine-independent growth of purF mutants was due to synthesis of HMP and strongly suggested that this synthesis required the four subsequent purine enzymes under the growth conditions tested. These results have further defined PurF-independent thiamine synthesis and have predicted an alternative PRA-generating activity. Data presented here have also raised new questions regarding TPP synthesis and/or metabolism. Results suggest that bacterial growth and exogenous adenine significantly affect levels of thiamine phosphoesters. These results lay the groundwork for future studies investigating the mechanisms of this regulation and the components involved.
ACKNOWLEDGMENTS
This work was supported by NIH grant GM47296 to D.M.D. J.L.E. was supported by the Pfizer Fellowship in Microbial Physiology.
We thank Dan Sykes and Martha Vestling for help with the GC-MS analyses and Jorge Escalante-Semerena for critical reading of the manuscript.
REFERENCES
- 1.Beck B J, Downs D M. The apbE gene encodes a lipoprotein involved in thiamine biosynthesis in Salmonella typhimurium. J Bacteriol. 1998;180:885–891. doi: 10.1128/jb.180.4.885-891.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berkowitz D, Hushon J M, Whitfield H J, Roth J, Ames B N. Procedure for identifying nonsense mutations. J Bacteriol. 1968;96:215–220. doi: 10.1128/jb.96.1.215-220.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dalal F R, Gots R E, Gots J E. Mechanism of adenine inhibition in adenine-sensitive mutants of Salmonella typhimurium. J Bacteriol. 1966;91:507–513. doi: 10.1128/jb.91.2.507-513.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.David S, Estramareix B, Fischer J C, Therisod M. The biosynthesis of thiamine. Synthesis of [1,1,1,5-2H4]-1-deoxy-d-threo-2-pentulose and incorporation of this sugar in biosynthesis of thiazole by Escherichia coli cells. J Chem Soc Perkin Trans. 1982;1:2131–2137. [Google Scholar]
- 5.David S, Estramareix J-C, Therisod M. 1-Deoxy-d-threo-2-pentulose: the precursor of the five-carbon chain of the thiazole of thiamine. J Am Chem Soc. 1981;103:7341–7342. [Google Scholar]
- 6.DeMoll E, Shive W. Determination of the metabolic origin of the sulfur atom in thiamine of Escherichia coli by mass spectrometry. Biochem Biophys Res Commun. 1985;132:217–222. doi: 10.1016/0006-291x(85)91010-1. [DOI] [PubMed] [Google Scholar]
- 7.Downs D M. Evidence for a new, oxygen-regulated biosynthetic pathway for the pyrimidine moiety of thiamine in Salmonella typhimurium. J Bacteriol. 1992;174:1515–1521. doi: 10.1128/jb.174.5.1515-1521.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Downs D M, Petersen L. apbA, a new genetic locus involved in thiamine biosynthesis in Salmonella typhimurium. J Bacteriol. 1994;176:4858–4864. doi: 10.1128/jb.176.16.4858-4864.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Downs D M, Roth J R. Synthesis of thiamine in Salmonella typhimurium independent of the purF function. J Bacteriol. 1991;173:6597–6604. doi: 10.1128/jb.173.20.6597-6604.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Enos-Berlage J L. Characterization of thiamine-pyrophosphate synthesis independent of the PurF enzyme (phosphoribosylpyrophosphate amidotransferase) in Salmonella typhimurium. Ph.D. thesis. Madison: University of Wisconsin—Madison; 1998. [Google Scholar]
- 11.Enos-Berlage J L, Downs D M. Involvement of the oxidative pentose phosphate pathway in thiamine biosynthesis in Salmonella typhimurium. J Bacteriol. 1996;178:1476–1479. doi: 10.1128/jb.178.5.1476-1479.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Enos-Berlage J L, Downs D M. Mutations in sdh (succinate dehydrogenase genes) alter the thiamine requirement of Salmonella typhimurium. J Bacteriol. 1997;179:3989–3996. doi: 10.1128/jb.179.12.3989-3996.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Estramareix B, David S. Conversion of 5-aminoimidazole ribotide to the pyrimidine of thiamine in enterobacteria: study of the pathway with specifically labeled samples of riboside. Biochim Biophys Acta. 1990;1035:154–160. doi: 10.1016/0304-4165(90)90110-i. [DOI] [PubMed] [Google Scholar]
- 14.Estramareix B, Lesieur M. Biosynthesis of the pyrimidine moiety of thiamine: origin of carbons in positions 2 and 4 in Salmonella typhimurium. Biochim Biophys Acta. 1969;192:375–377. [PubMed] [Google Scholar]
- 15.Estramareix B, Therisod M. Biosynthesis of thiamine: 5-aminoimidazole ribotide as the precursor of all the carbon atoms of the pyrimidine moiety. J Am Chem Soc. 1984;106:3857–3860. [Google Scholar]
- 16.Estramareix B, Therisod M. Tyrosine as a factor in biosynthesis of the thiazole moiety of thiamine in Escherichia coli. Biochim Biophys Acta. 1972;273:275–282. [PubMed] [Google Scholar]
- 17.Frodyma M E, Downs D M. The panE gene, encoding ketopantoate reductase, maps at 10 minutes and is allelic to apbA in Salmonella typhimurium. J Bacteriol. 1998;180:4757–4759. doi: 10.1128/jb.180.17.4757-4759.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gots J S. Regulation of purine and pyrimidine metabolism. In: Vogel H J, editor. Metabolic regulation. V. New York, N.Y: Academic Press; 1971. pp. 225–254. [Google Scholar]
- 19.Kawasaki T. Determination of thiamine and its phosphate esters by high-performance liquid chromatography. In: Chytil F, McCormick D B, editors. Vitamins and coenzymes, part G. Vol. 122. New York, N.Y: Academic Press, Inc.; 1986. pp. 15–29. [DOI] [PubMed] [Google Scholar]
- 20.Kumaoka H, Brown G. Biosynthesis of thiamine: incorporation of formate into carbon atom two of the pyrimidine moiety of thiamine. Arch Biochem Biophys. 1967;122:378–384. doi: 10.1016/0003-9861(67)90208-1. [DOI] [PubMed] [Google Scholar]
- 21.Messenger L J, Zalkin H. Glutamine phosphoribosyl pyrophosphate amidotransferase from Escherichia coli. J Biol Chem. 1979;254:3382–3392. [PubMed] [Google Scholar]
- 22.Newell P C, Tucker R G. Biosynthesis of the pyrimidine moiety of thiamine. Biochem J. 1968;106:279–287. doi: 10.1042/bj1060279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Newell P C, Tucker R G. New pyrimidine pathway involved in the biosynthesis of the pyrimidine of thiamine. Nature (London) 1967;215:1384–1385. doi: 10.1038/2151384a0. [DOI] [PubMed] [Google Scholar]
- 24.Newell P C, Tucker R G. Precursors of the pyrimidine moiety of thiamine. Biochem J. 1968;106:271–277. doi: 10.1042/bj1060271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nosaka K, Kaneko Y, Nishimura H, Iwashima A. Isolation and characterization of a thiamine pyrophosphokinase gene, thi80, from Saccharomyces cerevisiae. J Biol Chem. 1993;268:17440–17447. [PubMed] [Google Scholar]
- 26.Nygaard P, Smith J M. Evidence for a novel glycinamide ribonucleotide transformylase in Escherichia coli. J Bacteriol. 1993;175:3591–3597. doi: 10.1128/jb.175.11.3591-3597.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Petersen L A, Enos-Berlage J L, Downs D M. Genetic analysis of metabolic crosstalk and its impact on thiamine synthesis in Salmonella typhimurium. Genetics. 1996;143:37–44. doi: 10.1093/genetics/143.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rolfes R J, Zalkin H. Escherichia coli gene purR encoding a repressor protein for purine nucleotide synthesis. J Biol Chem. 1988;263:19653–19661. [PubMed] [Google Scholar]
- 29.Stauffer G V. Biosynthesis of serine, glycine, and one-carbon units. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C: ASM Press; 1996. pp. 506–513. [Google Scholar]
- 30.Steiert J G, Rolfes R J, Zalkin H, Stauffer G V. Regulation of Escherichia coli glyA gene by the purR gene product. J Bacteriol. 1990;172:3799–3830. doi: 10.1128/jb.172.7.3799-3803.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Dyk T K, LaRossa R A. Prevention of endogenous 2-ketobutyrate toxicity in Salmonella typhimurium. In: Barak Z, Chipman D M, Schloss J V, editors. Biosynthesis of branched amino acids. Weinheim, Germany: VCH and Balglan Publishers; 1990. pp. 123–130. [Google Scholar]
- 32.Webb E, Downs D M. Characterization of thiL, encoding thiamine monophosphate kinase, in Salmonella typhimurium. J Biol Chem. 1997;252:15702–15707. doi: 10.1074/jbc.272.25.15702. [DOI] [PubMed] [Google Scholar]
- 33.Webb E, Febres F, Downs D M. Thiamine pyrophosphate negatively regulates transcription of some thi genes of Salmonella typhimurium. J Bacteriol. 1996;178:2533–2538. doi: 10.1128/jb.178.9.2533-2538.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weber, S., and D. M. Downs. 1998. Unpublished results.
- 35.White R H, Rudolph F B. Biosynthesis of the pyrimidine moiety of thiamine in Escherichia coli: incorporation of stable isotope-labeled glycines. Biochemistry. 1979;18:2632–2636. doi: 10.1021/bi00579a031. [DOI] [PubMed] [Google Scholar]
- 36.Zilles J L, Downs D M. A novel involvement of the PurG and PurI proteins in thiamine synthesis via the alternative pyrimidine biosynthetic (APB) pathway in Salmonella typhimurium. Genetics. 1996;144:883–892. doi: 10.1093/genetics/144.3.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zilles, J. L., and D. M. Downs. 1998. Unpublished data.