Abstract
Rare genomic disorders (RGDs) confer elevated risk for neurodevelopmental psychiatric disorders (NPDs). In this era of intense genomics discoveries, the landscape of RGDs is rapidly evolving. However, there has not been comparable progress to date in scalable, harmonized phenotyping methods. As a result, beyond associations with categorical diagnoses, the effects on dimensional traits remain unclear for many RGDs.
The nature and specificity of RGD effects on cognitive and behavioral traits is an area of intense investigation: RGDs are frequently associated with more than one psychiatric condition and those studied to date affect, to varying degrees, a broad range of developmental and cognitive functions. Although many RGDs have large effects, phenotypic expression is typically influenced by additional genomic and environmental factors. There is emerging evidence that using polygenic risk scores in individuals with RGDs offers opportunities to refine prediction, thus allowing for the identification of those at greatest risk of psychiatric illness. However, translation into the clinic is hindered by roadblocks, which include limited genetic testing in clinical psychiatry, and the lack of guidelines for following individuals with RGDs, who are at high risk of developing psychiatric symptoms. The Genes 2 Mental Health Network (G2MH) is a newly funded NIMH initiative that will collect, share and analyze large scale datasets combining genomics and dimensional measures of psychopathology spanning diverse populations and geography. We present here the most recent understanding of the effects of RGDs on dimensional behavioral traits and risk for psychiatric conditions. We discuss strategies that will be pursued within the G2MH network, and how expected results can be translated into clinical practice to improve patient outcomes.
Plain Language summary
Genomic methods are widely used in psychiatric research and are recommended in clinical practice to identify rare genomic variants in patients with neurodevelopmental disorders. However, we still know very little about these variants. The mission of G2MH is to collect and harmonize clinical and genomic data on large groups of individuals who carry such rare variants to understand how they affect cognition and behavior and increase the risk for psychiatric conditions.
INTRODUCTION
Rare Genomic Disorders: A Brief Overview.
Rare genomic disorders (RGDs, text box 1), including structural genomic variants such as copy number variants (CNVs), and sequence-level variants such as single nucleotide variants (SNVs), are major contributors to Neurodevelopmental Psychiatric Disorders (NPDs, Box 1, Table 1). In clinical samples, RGDs are identified in up to 14 to 37% of individuals with autism spectrum disorder (ASD) depending on ascertainment (1, 2) and 2% to 7% of individuals with schizophrenia (3). Case-control association studies have linked 102 genes and 16 CNVs with ASD (4, 5), as well as 14 CNVs with schizophrenia (SZ) (6). Among the latter, seven were also linked to ASD. However, for both disorders, a much broader landscape of rare genomic variants is implicated, as suggested by the greater burden of rare CNVs in ASD and SZ compared to controls (5–7). In fact, statistical models trained on large samples suggest that any deletion or duplication ≧1Mb encompassing coding genes would be associated with an increase in ASD risk and a decrease of cognitive functioning, albeit with a range of effect sizes (8).
Text box 1. Definitions.
RGDs:
Rare Genomic Disorders have an individual prevalence of <1/2000 but cumulatively are a major cause of morbidity, frequently involving neuropsychiatric symptoms. In the current paper we focus on RGDs with large to moderate effect sizes on cognitive and behavioral dimensions. RGDs are of clinical significance because they substantially contribute to the expression of psychiatric illness in patients who carry them.
Copy Number Variants (CNVs):
either a gain (duplication) or a loss (deletion) of a stretch of DNA > 1000 base pairs.
NPDs:
Neurodevelopmental Psychiatric Disorders refer to a broad spectrum of psychiatric, behavioral and cognitive symptoms observed in the first decades of life and interfering with daily functioning. At present, there is no consensus regarding which disorders are included under the umbrella term developmental disorders, and to what extent a distinction can be made between phenotypes related to early brain development versus those related to later brain maturation (84, 85). As such, the concept of NPDs in this paper includes, but is not limited to: developmental delay (DD), intellectual disability (ID), learning disorders, autism spectrum disorder (ASD), attention deficit hyperactivity disorder (ADHD), language and motor coordination disorders, mood and anxiety disorders, schizophrenia and related psychotic disorders.
Penetrance and effect size:
Penetrance refers to the proportion of individuals with a specific genomic variant who present with a certain categorical diagnosis. However, when phenotypes are examined as a quantitative trait, there is no clear evidence that RGDs exert effects in only a subset of carriers. Under the assumption of additive effects, genomic variants with very large effect sizes show high penetrance (e.g., trisomy 21, which causes Down syndrome, decreases IQ by 3.5 standard deviations on average, resulting in a penetrance of intellectual disability close to 100%), because symptoms are observable irrespective of genetic and environmental backgrounds. On the other hand, small effect size (low penetrance) CNVs (e.g., 15q11.2 deletion) may appear asymptomatic unless other genetic and/or environmental background factors are conducive to phenotypic expression. One important focus of research is the use of dimensional traits with the aim to replace penetrance with quantitative effect sizes. However, penetrance remains relevant in clinical practice, in which categorical diagnoses are standard, and binary decisions are required.
Additive and non-additive genetic effects:
The combined effect of several genomic variants on a quantitative trait is equal to the sum of their individual effects. The alternative possibility is that of multiplicative effects (non-additive). For complex traits such as cognition and behavior, the genetic contribution to phenotypic variance has thus far mainly been attributed to additive effects. Under this assumption, the variance of a trait is expected to be the same within a group of CNV carriers and in the general population (e.g., IQ variance is the same in 16p11.2 deletion carriers and in unselected populations). This is in line with existing research on additional familial and genetic factors that have previously been associated with the variance of cognitive and behavioral phenotypes in CNV carriers, as in the general population.
Table 1:
Recurrent CNVs frequently identified in the developmental pediatric and genetic clinic.
| Chromosome coordinates (gene) | START-STOP Mb (hg19) | Effect size IQa | Freq. of ID % | OR for ASDb | OR for SZb |
|---|---|---|---|---|---|
| 1q21.1 Del. (CHD1L) | 146.53–147.39 | −15.15c | 16.1 | 3.2d | 6.4e |
| 1q21.1 Dup. (CHD1L) | 146.53–147.39 | −25.35c | 37.8 | 5.3d | 2.9f |
| NRXN1 Del. | 50.14–51.26 | −9 | 8.1 | 7.9d | 4.7g |
| 3q29 Del. (DLG1) | 195.73–197.34 | −31.5 | 54.0 | 19d | 23g |
| 7q11.23 Dup. (ELN) | 72.7–74.1 | −13.95 | 14.2 | 32d | 16.1g |
| 15q11.2 Del. (CYFIP1) | 22.81–23.09 | −5.7 | 5.3 | 1.3h | 1.9h |
| 15q13.3, BP4-BP5 Del. (CHRNA7) | 30.92–32.51 | −21.9 | 29.5 | 15d | 18g |
| 16p13.11 Del. (MYH11) | 15.51–16.29 | −7.35 | 6.5 | 2.5d | 2.2e |
| 16p13.11 Dup. (MYH11) | 15.51–16.29 | −8.7 | 7.8 | - | 1.5f |
| 16p11.2 distal Del. (SH2B1) | 28.82–29.05 | −9.15 | 8.2 | 3.6d | 4.4g |
| 16p11.2 distal Dup. (SH2B1) | 28.82–29.05 | −3 | 3.6 | - | 1.3e |
| 16p11.2 Del. (MAPK3) | 29.65–30.2 | −26i | 39.4 | 14.3d | 1.1e |
| 16p11.2 Dup. (MAPK3) | 29.65–30.2 | −11i | 10.2 | 10.5d | 11.7g |
| 17p11.2 Dup. (RAI1) | 17–21.4 | −49.2 | 90.0 | 32d | 11.3e |
| 17q12 Del. (HNF1B) | 34.81–36.22 | −11.55 | 11 | 3.5j | 6.6f |
| 17q12 Dup. (HNF1B) | 34.81–36.22 | −6.6 | 6.0 | - | 1.9e |
| 22q11.2 Del. (TBX1) | 19.04–21.47 | −28.5 | 46.0 | 32.3d | 23k |
| 22q11.2 Dup. (TBX1) | 19.04–21.47 | −13.65 | 13.8 | 2d | 0.2g |
| General population | - | - | 2.3 | (1.9%)l | (0.7%)m |
Chromosome coordinates are provided with a well-known gene at each locus to help recognize the CNV. Values were derived from the following studies:
Derived from Huguet G et al. (22)
Derived from Sanders SJ et al. (89)
Derived from Bernier R et al. (90)
Derived from Kirov G et al. (91)
Derived from Rees E et al. (92)
Derived from Marshall CR et al. (6)
Derived from Jønch AE et al. (44)
Derived from D’Angelo D et al. (11)
Derived from Moreno-De-Luca D et al. (93)
Derived from Davies RW et al. (23)
Derived from Maenner MJ et al. (94)
Derived from McGrath J et al. (95).
Table also includes 3 CNVs with small effect sizes not considered as RGDs (15q11.2 deletion, 16p11.2 distal (SH2B1) duplication, and 17q11.2 (HNF1B) duplication.
In sharp contrast to the accelerated pace of genomic discovery, very little is known about the mechanisms by which genomic variants increase risk for NPDs. In recent years, the National Institute of Mental Health (NIMH) has invested in the study of RGDs. As such, the Genes to Mental Health Network (G2MH) is a newly established NIMH-funded initiative that collects, shares, and analyzes large-scale datasets combining genomics and dimensional measures of psychopathology. What follows is not intended as a comprehensive review but rather a perspective on the current understanding of the contribution of rare variants to measures of cognition, behavior, and risk for psychiatric conditions. To advance the field, we propose strategies that will be pursued within the G2MH network, and discuss challenges currently faced by the field and how results from such studies can be translated to the clinic with potential contributions to patient care.
Do Rare Variants Exert Specific or Shared Effects on Psychopathology?
The nature and specificity of RGD effects on cognitive and behavioral traits (i.e., whether a gene function or a molecular pathway leads to particular cognitive and behavioral profiles) is an area of intense investigation. Multimorbid presentation of neurodevelopmental and psychiatric conditions is common (9–12). All RGDs studied to date affect cognition to varying degrees and affect a broad range of cognitive functions, including reaction time, attention, executive functions, and social cognition (13, 14). Early studies highlighted distinctive behavioral profiles, such as social disinhibition, excessive empathy, and non-social anxiety described in 7q11.23 deletions (Williams-Beuren syndrome(15)); insatiable appetite in paternal 15q11.2-q13 deletions (Prader-Willi syndrome(16)); sleep disturbances in 17p11.2 deletions (Smith Magenis syndrome) (17); as well as childhood apraxia of speech in 16p11.2 proximal deletions (18).
Recent studies have attempted to quantify distinct and shared patterns of measurable cognitive and behavioral traits altered by specific genotypes (19, 20)(21). A study of 258 individuals with neurodevelopmental disorders, and 13 CNVs, found that 80% of carriers met criteria for one or more psychiatric disorder; but overall, the range of affected behavioral traits was broadly similar for these CNVs, and specific genotypes accounted for a low proportion of the variance (5–20%) (20). Phenotypes with impairments across all CNVs included mood, sleep, peer relationships, and sustained attention (Figure 1). Recent studies have leveraged the shared effects of RGDs on cognition by developing models that can predict the effect size of any CNV on IQ with close to 80% accuracy (22).
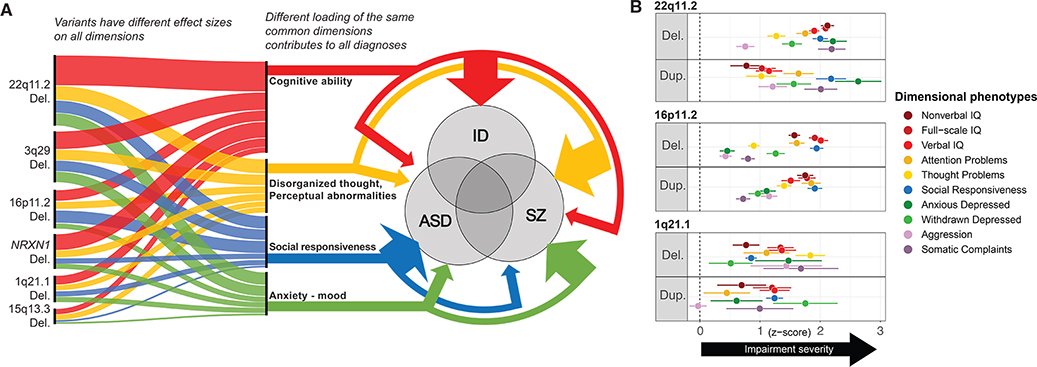
Figure 1.
Effects of CNVs on cognitive and behavioral dimensions
(A) RGDs and dimensional phenotyping, current knowledge, and hypotheses. NPD CNVs affect multiple cognitive and behavioral dimensions and increase the risk for ASD, SZ, and ID, albeit with different effect sizes. This scenario incorporates key features of the genomic architecture of psychiatric disorders including polygenicity, phenotypic variability -also referred to as pleiotropy- and genetic overlap between conditions, cognitive and behavioral dimensions. In this scenario, combinations in different proportions of common dimensions lead to different clinical manifestations classified as psychiatric diagnoses. The 4 dimensions described in figure 1A may approximately align with RDoC dimensions: Cognitive ability → Cognitive systems; Disorganized thought/perceptual abnormalities → perception (a (sub)construct of the cognitive systems domain); Social responsiveness → Systems for social processes; Anxiety-mood → Negative valence.
(B) Effects of 1q21.1, 16p11.2 and 22q11.2 deletions and duplications on neurocognitive and behavioral functioning. Measures are standardized to control mean and standard deviation. For visualization purposes, scores are converted to absolute, positive values to highlight impairments in CNV groups compared to controls (Z=0 indicated by vertical dashed line). Behavioral measures = scales from Child Behavior Checklist (CBCL). Social responsiveness was assessed by the Social Responsiveness Scale (SRS). Sample sizes: 1q21.1 Del n =11, 1q21.1 Dup n = 12 (REF); 16p11.2 Del n = 137, 16p11.2 Dup n = 127 (11); 22q11.2 Del n = 99, 22q11.2 Dup n = 34 (21); combined controls n = 214.
Phenotypic Variability
The variability of behavioral symptoms observed in individuals who carry the same RGD has been the subject of several studies (11, 23, 24). Because recurrent CNVs typically have the same breakpoints and include the same genes in unrelated individuals, they are well suited to investigate such questions. Hypotheses put forward to explain phenotypic variability may be broadly classified in 2 categories: non-additive interactions versus additive models (text box 1).
Under the former hypothesis, RGDs may interact with other variants, but non-additive effects involving variants at different loci remain elusive. In rare cases, a deletion may uncover a second variant on the remaining allele (i.e., recessive condition). For example, in 5 patients with 22q11.2 deletions, missense variants in CDC45 on the remaining allele of 22q11.2 deletion carriers were found to lead to CGS syndrome (craniosynostosis, cleft lip and palate, gastrointestinal, and genitourinary anomalies, skeletal differences and short stature) (25, 26). Likewise, RBM8 missense variants on the remaining allele of individuals with 1q21.1 deletions cause thrombocytopenia-absent-radius syndrome (25, 26). However, recent data for 22q11.2 deletion syndrome provides no evidence for such recessive effects for schizophrenia expression. Larger studies are required to investigate these challenging questions. Indeed, for traits such as cognition and behavior, the genetic contribution to phenotypic variance has mainly been attributed to additive effects (27). This is in line with studies showing that IQ variance within groups of 16p11.2 and 22q11.2 deletion carriers is similar to that in the general population, although the mean IQ is significantly lower. Therefore, variance in CNV groups is thought to be related to additive effects of genetic (and non-genetic) factors that are captured by parental IQ and common variants (11, 23, 28–30) (Figure 2).
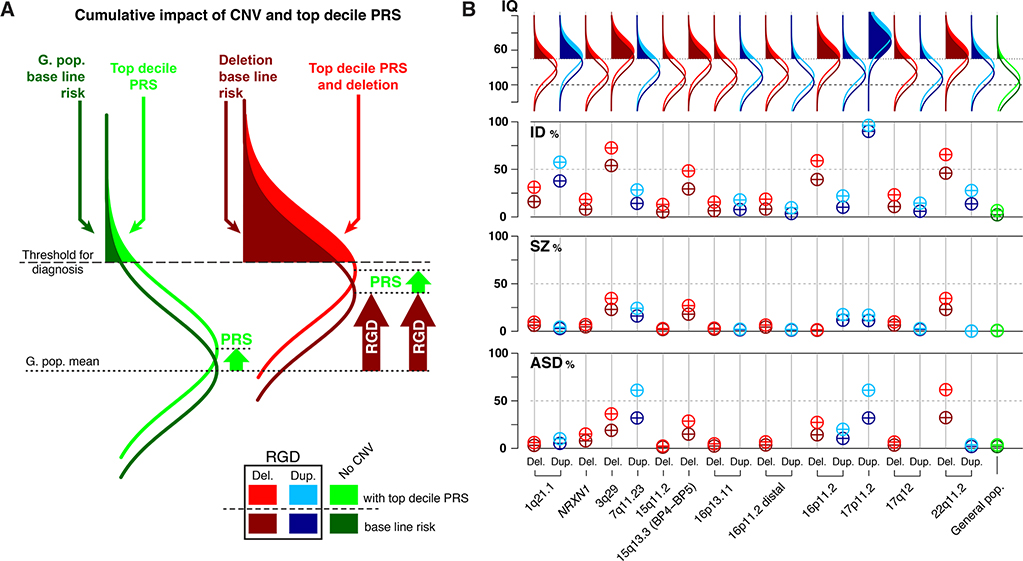
Figure 2.
Risk prediction in rare CNV carriers with and without polygenic risk (PRS) information.
(A) Schematic showing baseline risk for the general population (dark green) and a large effect size RGD (dark red). Risk for individuals of both groups with a top decile PRS score (light green and light red). Although the PRS has the same small effect size in both groups, it results in a larger increase in the penetrance of a diagnosis in the RGD group. (B) Comparing risk conferred by CNVs for carriers without PRS information (baseline risk, table 1) to those with top decile PRS values. For schizophrenia PRS, OR= 1.5 (23, 59). For Autism spectrum disorder PRS, OR = 1.91 (96). Effect size of top decile PRS cognitive ability compared to 50th percentile = 0.45 z-score (23, 97). Risk in RGD carriers with top decile PRS values is computed based on an additive model. Y-axis for IQ: IQ values. Y-axis for ID, SZ, and ASD: penetrance (from 0 to 100%) of a diagnosis in CNV carriers.
Several conclusions can be drawn from the body of literature on RGDs: 1) Although many RGDs have large effects, in most instances, phenotypic expression of NPDs occurs in the context of additional genomic and environmental factors; 2) similar to studies focused only on common single nucleotide polymorphisms (SNPs), as sample sizes increase, an ever-growing number of rare variants are associated with intellectual disability (ID), ASD, SZ, and other NPDs; 3) RGDs are invariably associated with more than one psychiatric condition; 4) Some RGDs show preferential association with SZ or ASD such as 22q11.2 deletions and duplications, respectively (6). Conceptual models may be put forward to link these observations (Figure 1a). For example, in a “common dimensionality” scenario, we posit that each RGDs alter (with different effect sizes and proportions) the same dimensions of cognition and psychopathology. Variable alterations across each dimension may lead to distinct clinical manifestations classified as overt neurodevelopmental psychiatric diagnoses (Figure 1). A systematic and quantitative, standardized phenotyping approach across RGDs would help decompose the contribution of these dimensions to psychiatric diagnoses.
KNOWLEDGE GAPS
An Elusive Phenome: Closing the Gap Between the Tidal Wave of Gene Discovery and Phenotypes
In this era of rapidly evolving gene discovery, many genomic variants conferring varying degrees of risk for neurodevelopmental and psychiatric conditions have been identified. However, there has not been comparable progress in scalable, harmonized phenotyping measures and methods, which generally remain more expensive and labor-intensive than genotyping. As a result, genomic associations have largely been established for categorical diagnoses but the effects of variants on multiple phenotypes and dimensional traits has not been investigated on a large scale. Dimensional phenotyping across diagnostic categories is needed to identify the underlying mechanisms that may mediate some of the causal pathways between RGDs and diagnostic psychiatric outcomes (31). A barrier to achieving this has been a lack of cohorts providing consistent dimensional assessments across a broad spectrum of RGDs irrespective of psychiatric diagnoses. The IMAGINE-ID cohort in the UK has been one of the few studies of RGDs to employ a multi-dimensional deep phenotyping approach across a range of loci (20). It is critical that multidimensional phenotyping methods that capture the full breadth of the neuropsychiatric phenome are developed that are scalable and culturally appropriate for large international multi-site studies. Previous publicly available resource efforts, such as the Simons Simplex Collection(32), have focused on specific diagnoses, providing dimensional phenotyping and exome sequencing in probands with ASD. The latter allowed researchers to establish general relationships between RGDs, IQ, gross motor, phonological memory, and language skills (33, 34). Although these resources have been instrumental in advancing our knowledge, they include a limited set of RGDs and many measures used are either disorder-specific (e.g., the Autism Diagnostic Observation Schedule -ADOS-, developed for ASD), or are only developmentally appropriate for a particular age range, making cross-study harmonization of measures a challenge.
The G2MH strategy aims to capture dimensional phenotypes consistently assessed across a range of clinically ascertained and unselected populations worldwide. This approach is optimally suited for the Research Domain Criteria (RDoC) framework (35) because genetic effects are likely to manifest along dimensions of behavior, and individuals who harbor RGDs can be assessed irrespective of whether they present subthreshold symptoms or meet full criteria for psychiatric diagnoses.
RDoC is a neuroscience-based framework to guide psychiatric research. It was designed to examine and link the distribution of key behavioral domains to genetics and brain circuitry, thereby advancing mechanistic understanding of psychiatric conditions (36). Within the RDoC framework, RGDs provide a unique opportunity for genome-phenome association. Such efforts may require adaptations of RDoC measures to pediatric populations, as most NPDs present in childhood. The establishment of normative charts for development will contribute to early identification of risk and protective parameters and advance precision psychiatry across the lifespan.
Historically, an important question has been to investigate whether clusters of dimensional measures associated with RGDs are representative of behaviorally defined conditions such as ASD and SZ diagnosed in individuals without any identified RGD (Figure 1). A study comparing individuals with different CNVs with autism to individuals with ‘idiopathic’ autism found evidence of a range of profile differences, however, these tended to be subtle(24). Another recent study showed that CNVs including genes intolerant to haploinsufficiency identified in ASD cohorts were not associated with any differences in ASD severity (34). Individuals with de novo SNVs (as a group) in ASD cohorts are more likely to present motor delay and a less severe symptom profile with respect to social communication and language deficits, relative to those with ASD without de novo variants (33). Studies of 22q11.2 deletion syndrome indicate representativeness of NPDs by specific RGD may vary by phenotype. For example, the SZ clinical phenotype in 22q11.2 deletion syndrome has similar phenomenology to that of idiopathic SZ (37, 38). In contrast, current evidence suggests that ADHD in 22q11.2 Deletion Syndrome has a more specific profile characterized predominantly by inattention symptoms rather than hyperactivity symptoms (39). Overall, these findings suggest that Considering idiopathic cases as most phenotypically representative of a condition may be questionable. There may or may not be a clear distinction between the phenotypic presentation of individuals with and without a known RGD who meet standard diagnostic criteria for a particular NPD.
Furthermore, as RGDs can be identified early in development, there are novel opportunities to conduct studies using accelerated longitudinal designs to uncover the early developmental correlates of adult dimensional traits. Pioneering work in 22q11.2 deletion syndrome has identified longitudinal dimensional antecedents of psychotic phenomenology, including verbal IQ decline (40), executive functions, spatial working memory, sustained attention and early language measures (41, 42).
Biases and Challenges in the Field
1). Ascertainment methods
As for all research involving human subjects, several sources of ascertainment bias may affect studies of RGDs. Such studies have mainly been conducted in clinically ascertained individuals (i.e., referred clinically for genetic testing), which is expected to favor inclusion of more severely affected cases. This is particularly problematic for genomic variants that have effects that are disproportionately milder than typical expression required for clinical referral. The 15q11.2 deletion is a prime example because it has mild effects and is relatively frequent in the general population and therefore frequently identified in neurodevelopmental disorder and genetic clinics. The literature is extensive and confusing with clinical series reporting cases with mild to severe neurodevelopmental symptoms, as well as malformations leading authors to delineate a microdeletion syndrome with considerable incomplete penetrance (43). Yet studies in unselected populations and meta-analysis of case-control studies have demonstrated that in fact this deletion, on average, confers very small risk for major disease (OR<2 for ASD, SZ, ID and congenital heart disease) and has small effects on cognitive ability (a 4 point drop in IQ) (44, 45).
Methods to limit certain ascertainment biases, especially clinical severity biases, include family studies, unselected populations and general population cohorts. Family studies evaluating first degree relatives who do not carry the variant of interest allow adjustment for genetic and environmental background factors. Such approaches have provided robust estimates of the effects of RGDs on cognitive and behavioral traits (11, 28). Unselected population studies have been successfully used to accurately estimate effect sizes of RGDs on cognitive traits (22, 45–47). As sample sizes rapidly increase, this approach will become relevant even for very rare variants (48). However, such studies introduce other biases because “unselected” cohorts tend to select against the inclusion of the most affected individuals.
Genomic studies in general population cohorts (epidemiological samples representative of the entire populations) remain uncommon, and would theoretically represent a gold standard for unbiased estimates. The iPSYCH study has examined 30,000 randomly sampled controls and 57,377 cases born from 1981 to 2005 identified within the Danish health system to have a diagnosis of ADHD, Major Depressive Disorder, SZ, ASD or Bipolar Disorder (49). Population prevalence was 1:3672 and 1:1606 for deletions and duplications and 60% of deletion carriers were missed by clinical ascertainment as would be expected for a cohort born before the routine use of Chromosomal Microarray Analysis in diagnostics. By age 32 years, 10% of these CNV carriers had a recorded diagnosis of ADHD, ASD or ID. These estimates suggest that previously reported ASD and SZ prevalence in clinically ascertained samples may have been overestimated (37). However, epidemiological studies such as iPSYCH can potentially introduce other biases such as the lack of systematic standardized assessment methods leading to underestimation of diagnoses, symptoms and disabilities. Further, expression of both cognitive and behavioral phenotypes varies widely across the lifespan (50). The size and number of genomic studies in unselected population cohorts have significantly increased in the past decade. Because CNVs can be identified using data from genotyping arrays, such studies have informed on the effects of CNVs on cognition and behavior in individuals who were not selected for a particular condition. However, such studies tend to recruit individuals who are on average healthier than the general population and therefore carriers of RGDS can be significantly underrepresented (e.g., the frequency of 16p11.2 and 22q11.2 deletion carriers in the UKBB was 1/4000 and 1/30000 respectively - 2 and 6-fold lower than expected) (22, 51, 52). Mortality effects of certain CNVs (53, 54) may also play a role.
2). Cultural biases
Improving the representation in mental health and genomic research of diverse populations (i.e., cultural, geographical background and ancestry) is an urgent scientific priority. However, the majority of assessment tools for cognition, psychiatric and developmental disorders have been developed in high resource contexts with a large proportion of validation samples being of anglophone and European ancestry. Studies adapting or validating assessment tools to account for different environments, languages and cultural backgrounds remain scarce. As a result, the overall validity of phenotypic data outside of these populations can be questioned. In particular, the interpretation of reported or observed symptoms and comparability of data from different contexts and populations remains a challenge (55, 56).
3). Subjective methods of assessments - diagnostic criteria
Inherent to all studies in this field is the challenge stemming from the fact that the DSM-5 and ICD remain almost entirely descriptive and agnostic to etiology. Consequently, even though based on objective criteria describing symptoms and functional impact, psychiatric nosology does not necessarily coincide with biologically valid distinctions between phenotypic classes (57). While this challenge is not exclusive for psychiatry, – in other fields of medicine there are also diagnoses without established biological correlates, – this challenge is substantially more prominent in psychiatry. Where for many non-psychiatric conditions a variety of lab examinations are available to confirm a diagnosis, the adequate identification of psychopathology depends on the reporting ability (or willingness) of the patient and/or proxy and the observational skills of the examiner. Subsequently, the obtained behavioral data require interpretation, adding another level of variability. For example, difficulty with eye contact may be interpreted as social anxiety or indicative of communication deficits such as ASD. Ascertainment may also vary across clinics where patients with RGDs are followed (psychiatry, psychology, developmental pediatrics, neurology, clinical genetics). These difficulties are pertinent to any study of psychiatric diagnoses, and as a result of these challenges, rates of psychiatric diagnoses and symptoms in patients with RGDs are not definitively established.
PRECISION PSYCHIATRY, TRANSLATING KNOWLEDGE into the CLINICAL SETTING
Predictive Testing
Given the substantial heritability of many psychiatric disorders (Figure 3), there is increasing interest in using genetic variants to identify individuals at high risk of developing psychiatric symptoms. To date, polygenic risk scores (PRS), which are additive models aggregating the effect of thousands of common variants, remain poor predictors of psychiatric disease in general medical settings (58). For example, individuals with schizophrenia-PRS values in the top decile have approximately a 2% risk of developing SZ compared to 1% in the rest of the population (23, 59). While PRS will continue to improve as sample sizes increase for GWAS, we are still a long way from filling in the current “missing heritability” gap of psychiatric conditions and cognitive traits (60). In addition, the GWAS used to compute PRS were performed in individuals of European descent and do not generalize to individuals with other ancestries.
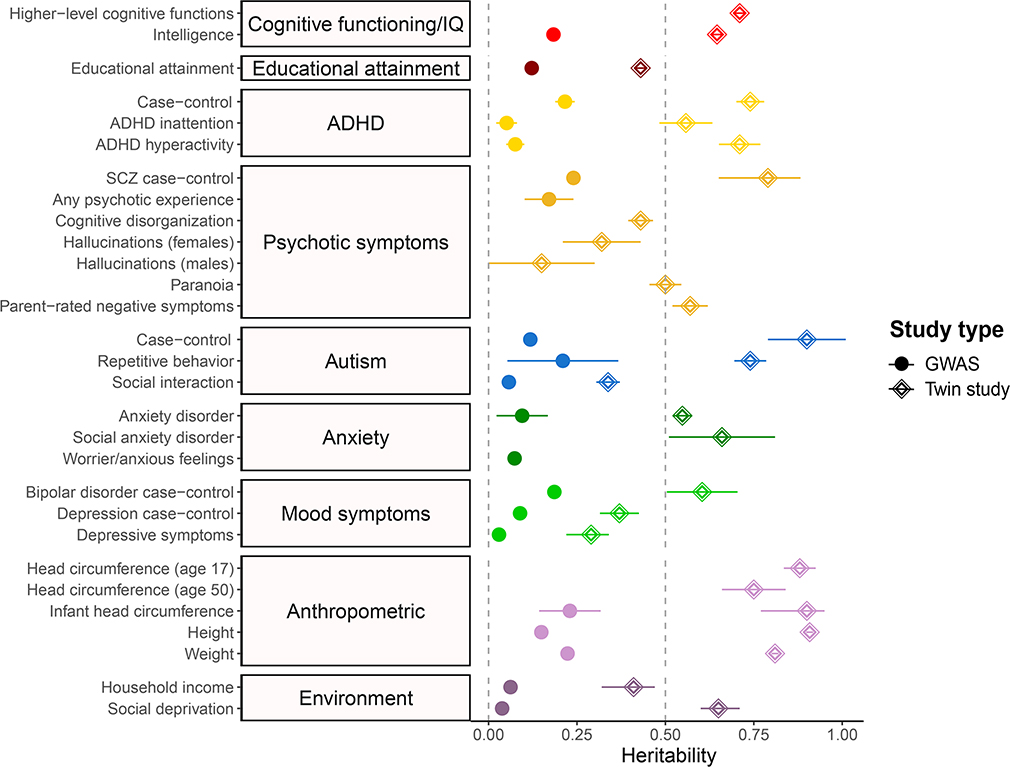
Figure 3.
Heritability / SNP heritability for cognitive and behavioral dimensions relevant to RGDs
Comparison between SNP-based (GWAS) heritabilities and twin-based heritabilities for phenotypes of interest. Heritability estimates and 95% CI are shown. Heritability estimates were derived from studies listed in supplemental table 1. Heritability estimates can differ depending on age (e.g., head circumference) and sex (e.g., hallucinations).
On the other hand, while NPD RGDs only explain a small proportion of population-based liability for psychiatric risk, their large effect sizes substantially increase baseline risk in carriers. Using PRS in the context of a RGD offers additional opportunities to refine prediction. A recent study has shown that applying SZ- and IQ-PRS information to high risk individuals with 22q11.2 deletions provided positive predictive values that approach levels of clinical utility (23). The baseline prevalence of schizophrenia and ID in 22q11.2 deletion carriers was 23% and 41%, respectively. However, individuals in the highest and lowest PRS-SZ risk decile had schizophrenia in 33% and 9% respectively. Similarly, 63% of individuals in the lowest PRS-IQ decile had ID versus 24% in the highest decile (23).
These results in individuals with 22q11.2 deletions appear consistent with additive models, as shown in Figure 2a–b, where we estimate positive predictive values by combining the previously published effect sizes of 19 CNVs on cognitive ability and psychiatric risk for SZ and ASD as well as effects of PRS-IQ, PRS-ASD and PRS-SZ. If our assumptions are correct, this approach could eventually provide clinically meaningful stratification for CNVs with large effect sizes in the range of that observed for 22q11.2 deletions (61). Furthermore, modest increases in PRS performance - which we expect in the near future - could have a significant impact on psychiatric risk stratification for some RGDs. On the other hand, under additive model assumptions, RGDs with smaller effect sizes are unlikely to benefit as much from PRS unless the field uncovers evidence of non-additive interactions. In addition, CNVs with extremely large effect sizes (e.g., the impact of trisomy 21 on IQ, Figure 2b.) may not benefit from PRS stratification because most of the risk is already conferred by the RGD itself.
Preventive Care and Interventions
Guidelines for follow-up of individuals who carry specific RGDs have been published for a few variants (62–64). More general guidelines for the assessment and management of those with RGDs have also been published (65). These include multi-system assessment beyond the initial reason for referral and covering biological, psychological and social factors, recognition of the potential need for co-ordinated multidisciplinary care, and that interventions take account of any relevant comorbidities (62–64). Individuals with RGDs often have needs that transcend single specialties and can provide challenges to existing services (62–64)(66). Many of the highly penetrant risk variants for neurodevelopmental disorders also seem to confer considerable risk for mood and anxiety disorders (50, 52, 67), which reinforces the need for psychiatric services.
Predictive testing for neuropsychiatric disorders occurs routinely as a by-product of chromosomal microarray and whole exome or genome sequencing prescribed in the prenatal(68), neonatology and developmental pediatric clinics (although diagnostic practices vary widely across health systems). Although these tests are typically ordered to diagnose malformations or non-specific - and often transient - motor symptoms such as neonatal hypotonia, in many cases the diagnosed RGD provides knowledge about increased risk for psychiatric symptoms and disorders that may manifest years or even decades later, and about other treatable medical conditions. Currently, however, patients are only seen by psychiatric services if they are experiencing symptoms.
The positive predictive value of RGDs, especially when combined with additional genetic factors PRS - as discussed above - has the potential to provide great clinical value and represent opportunities to investigate the efficacy of preventive interventions. Information about underlying genetic causes can also be beneficial for the planning of appropriate educational support (62–64). Early involvement and close collaboration of primary care, clinical genetics, and developmental pediatrics with child, adolescent and adult psychiatry may be beneficial for providing clinical care that can optimize outcomes.
Lack of routine genetic testing in psychiatric clinics (text box 2)
Text box 2. Clinical vignettes.
Patient 1.
Related to her participation in a research project, a 41-year-old woman diagnosed with idiopathic epilepsy, learning disabilities, and an unspecified psychotic disorder was found to have a 15q13.3 deletion. This recurrent deletion has been well-described in the literature and is associated with variable NPD phenotypes that include epilepsy, schizophrenia, and intellectual disability. On reviewing her genetics laboratory report with a genetic counselor, the woman expressed profound relief at finally having a tangible, medical explanation for her history. She described the stigma of living with learning and psychiatric disabilities, and she perceived that extended family members, and even some doctors, blamed her past drug use and lifestyle choices for these conditions. In recent years, the woman had become disillusioned with medical specialists and was inconsistent in showing up for doctor appointments. Subsequent communication with her primary care and specialty providers about her genetic diagnosis led to scheduling of long overdue follow-up care. In addition, two of the woman’s adolescent children were also found to have the 15q13.3 deletion through cascade testing. One 16-year-old daughter was receiving special education services for ASD, while her 19-year-old son had low average intelligence, inactive epilepsy, and a diagnosis of ADHD which was untreated. Two younger children were developing typically and were not genetically tested. A medical plan was developed for the woman’s son and daughter, and the family followed through with neurological and developmental medicine appointments. During a follow-up genetic counseling visit, the woman and her husband expressed high satisfaction at finally understanding a unifying genetic explanation for the family’s history of NPD. The family’s strong interest in their genetic diagnosis has led to a re-engagement, after many years without medical follow-up, with specialty providers who can now provide care informed by current and future knowledge about the 15q13.3 deletion, including the potential for targeted treatments(86–88).
Patient 2.
A woman in her early 30s was admitted to an inpatient psychiatric service for first episode psychosis. She experienced a gradual onset of psychotic symptoms over 2 years, culminating in delusions, paranoid ideation, auditory hallucinations and inability to function. Pertinent medical history was noted for normal pregnancy and an episode of cyanosis shortly after birth revealed a ventricular septal defect that spontaneously closed. At age 8 she was diagnosed and treated for hypothyroidism. Chronic otitis media since childhood required tubes and eventuated with hearing loss requiring hearing aids. She has been treated for thrombocytopenia and iron deficiency anemia diagnosed at age 8. At age 16 she developed seizures, well controlled on medications. Developmental history is noted for slight delay in achieving milestones. She was a good student in lower school, yet math was always a challenge. She had accommodations in high school and IQ assessed at age 18 was average. She graduated college with support and attained a part time job that she maintained for years. Psychiatric history is noted for anxiety diagnosed at age 8, which improved with treatment of hypothyroidism. Following inpatient care, psychotic symptoms improved significantly. She has not been able to return to her previous job, but does volunteer work regularly. Examination and a full review of history when she presented to the outpatient psychosis program suggested that 22q11.2 deletion syndrome may underlie the range of disorders manifested. She had never been genetically evaluated and agreed to be tested; the diagnosis was confirmed. The relief experienced by her and the family was immense. The realization that all the multiple issues and challenges she faced were not due to their failure of upbringing but of a rare genetic disorder literally “took a rock off the chest.” They expressed their wish that they had known the genetic diagnosis much earlier, when they could have connected with support groups and learned from the experience of others with this condition.
Guidelines have been established for clinical genetic testing in patients with developmental delay, intellectual disabilities, ASD, and complex learning disabilities, in addition to major congenital anomalies (69–71). Genetic testing for CNVs and SNVs is primarily carried out in genetic clinics, prenatal clinics, and some developmental pediatric and neonatal clinics. However, knowledge about clinical applications of genetics is modest, and diagnostic genetic testing is still a rare practice in child and adult psychiatry and in many resource-limited settings (72). This has led to lost opportunities for patient care and training for practitioners who may follow many patients with unrecognized RGDs (73).
Key challenges to implementation of diagnostic genetic testing recommendations include inadequate medical genetics training by psychiatrists, particularly related to practical issues of test selection, results interpretation, and genetic counseling. A recent study showed that the likelihood for child psychiatrists to order genetic testing was related to the clinician’s own perceived knowledge about these aspects (74). Another challenge is the lack of a unified diagnostic approach for patients with RGDs, whereby psychiatric symptoms and genetic etiology are diagnosed and managed as two entirely separate clinical realities, despite their obvious connection (75), except where specialty clinics exist (62–64)(66).
While inconsistencies remain in recommendations for clinical genetic testing and medical education, depending on the professional group cited and the recency of its published statement, initiatives for harmonization are starting to occur. For example, the Residency Education Committee of the International Society of Psychiatric Genetics (ISPG) published recommendations for the medical training of psychiatrists and endorsed consideration of genetic testing (76). Both the American College of Medical Genetics and Genomics (ACMG) and European College of Medical Genetics recommend chromosomal microarray analyses as a first tier evaluation of all children with ASD, ID, neurodevelopmental disorders, or congenital anomalies (71, 77). The UK’s National Institute for Health and Care Excellence however suggests deferring to evaluation by Medical Genetics specialists for decisions on genetic testing. A separate multidisciplinary consensus statement that included a meta-analysis by an expert consensus group cites evidence for high diagnostic yields and recommends exome sequencing as a first-tier clinical test in individuals with ID and ASD (78). Individual research groups have also endorsed genetic testing for patients with schizophrenia, based on evidence for clinically relevant CNVs in this population (79, 80). However, practice guidelines from the American Psychiatric Association vaguely suggest “genetic testing” in the context of psychotic symptoms and mention 22q11.2 deletions without further clarification, while the Canadian Psychiatric Association recommends “genetic testing based on the history and physical examination of the patient, especially at the time of the first episode of psychosis.” A position statement by the IISPG opines that diagnostic genetic testing may have value for patients with ASD and ID, while offering no specific recommendations on schizophrenia.
Lack of testing in the psychiatric clinic also has significant impacts on scientific advances limiting the number of clinically initiated projects and reports. As an example, there are very few large-scale reports on the yield of genetic testing for schizophrenia in a clinical setting. Increasing the implementation of genetic testing in psychiatry will require the integration of medical genetics in clinical training.
G2MH ROADMAP
Given the promise of genomics approaches to neurodevelopmental psychiatric research, but recognizing the need to tackle the knowledge gaps and challenges discussed throughout this review, the “Genes to Mental Health” (G2MH) network (https://genes2mentalhealth.com) was established in 2019 with funding from the National Institute of Mental Health (NIMH) and the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD). The network currently includes researchers representing 14 institutions in North America, Europe, and Africa, all with a shared goal of creating a robust phenotypic pipeline to standardize and accelerate the clinical characterization of RGDs. Conceptually, while few clinical conclusions can be drawn by studying a single individual with a specific RGD, aggregated data on standardized measures from a substantial sample of individuals with the same genetic disorder would allow meaningful analyses, fuel hypotheses, and drive discovery. As such, the G2MH network espouses a strong commitment to data-sharing as an essential strategy to reach sample sizes required to power analyses. Its initial focus is on establishing processes for the collection, sharing, and harmonization of quantitative data from measures of cognition and behavior (Figure 3) across multiple genomic variants that confer increased risk of adverse developmental and psychiatric outcomes. Currently, the five main activities of the network include:
New Data Collection
Cognitive and behavioral measures investigating the dimensions detailed in Figure 1 will be collected in 2300 probands who carry RGDs at 5 target loci (1q21.1, 15q13.3, 16p11.2, 22q11.2, and CHD8) and their family members, as well as in a cohort of 1000 individuals with ASD and related neurodevelopmental disorders and their parents and a matched number of unaffected community control children. Recruitment and assessments will take place in very different health systems across North America, Europe and Africa. Additional genetic factors present in probands and family members will be characterized via genotyping and whole genome sequencing. Phenomena such as dynastic effects and assortative mating can induce correlations between genotypes and phenotypes. By using family data, we will investigate how familial (genetic and environmental) factors may have affected the estimated effect-sizes of genomic variants on cognitive and behavioral traits (81).
Leveraging Archival Data
There are many opportunities to identify rare CNVs in large unselected and clinical populations, with cognitive and behavioral assessments as well as data from genotyping arrays. Therefore, G2MH will also coalesce data on close to 800,000 individuals across 30 datasets including individuals from 5 to 80 years of age. CNVs will be identified using genotyping array data. The effect of recurrent CNVs as well as non-recurrent CNVs will be consistently measured across cognitive and behavioral dimensions discussed above (Figure 1). Many of these population cohorts will provide much more information on smaller CNVs than pathogenic large-effect-size CNVs that are the focus of this review. However the latter are typically multigenic and inherently complex (e.g., 22q11.2 deletions include 50 genes, 10 of them being intolerant to haploinsufficiency). Several studies have shown that models trained on observations of small, non-pathogenic variants in the general population can predict the effects of large multigenic pathogenic variants(22, 34, 47). Therefore, studying all recurrent and non-recurrent CNVs regardless of their effect-size is a strategy to help decipher the diversity of mechanisms that are at play in large pathogenic CNVs.
Phenotypic Harmonization Strategy
New phenotypic data collection and archival data will require harmonization for joint analyses. Cohort specific phenotypic data (diagnostic instruments, rating scales, self-reports, and cognitive assessments) will be mapped to the key dimensions in Figure 1 and integrated into a normalized dataset in a central database. Integrative analyses will be used where factor models estimated in disparate studies are transformed to minimize measurement invariance.
New Satellite Projects
Currently, G2MH focuses on a small proportion of NPD-associated RGDs. Ongoing and future studies targeting other genomic loci are encouraged to join the network, including studies of monogenic disorders or CNVs. G2MH now includes investigators studying additional loci such as 3q29, 16p12.1, and 17q12. The harmonization strategies described above will be applied in order to perform analyses across as many genomic loci as possible.
Data Sharing
Data will be made available to the broader research community. The central hub for data sharing will be the National Institute of Mental Health Data Archive (NDA). Individual projects will make regular deposits of genomic data. Access to data derived from human samples will be shared using a controlled access mechanism that complies with regulatory requirements and governance policies regarding protection of personal information.
The projects currently included in G2MH are limited to prospective data collection on the most frequent recurrent CNVs, as well as data available from CNV carriers identified in unselected populations. Therefore, a broad spectrum of RGDs will remain undocumented. When fully operational, an expanding number of G2MH member sites will continuously contribute to its central database, harmonized phenotype and genotype data from consented families who carry RGDs at additional genomic loci. Data will be derived from a variety of sources, including prospectively-collected and archival RGD research. Additionally, some G2MH sites have established processes to capture research-grade data generated as a byproduct of clinical care of consented individuals with RGDs, a cost-effective strategy that is not utilized by other large-scale phenotyping efforts. This unique “learning healthcare” approach (82) capitalizes on freely available clinical information, redirecting valuable research dollars away from data generation toward analysis and discovery. Ultimately, the G2MH aims to fuel breakthroughs in treatment by creating a cost-effective and continuously expanding data resource with actively engaged, well-characterized families who would be potentially interested in participating in future drug development and non-pharmacological interventions. G2MH is complementary to other major psychiatric genomics efforts, such as the PsychENCODE Consortium (https://www.nimhgenetics.org/resources/psychencode), which focuses on non-coding functional genomic elements in human brain, with the goal of elucidating their role in the molecular pathophysiology of NPDs. Preclinical models (both animal and in vitro/cellular models) of these genetic defects offer ‘bottom-up’ insights into neurobiological mechanisms underlying these human conditions (83).
CONCLUSION
With the increasing implementation of genetic testing into clinical care, RGDs are being more routinely identified. While resolution, scope and accuracy of genetic testing methods have advanced substantially over the past two decades, our ability to describe associated neurodevelopmental and psychiatric phenotypes, both categorically and dimensionally in a consistent and unified way, is lagging behind. RGDs represent unique opportunities to elucidate mechanisms underlying risk for NPDs. In order to fully exploit these opportunities, comprehensive and harmonized phenotyping strategies are required. G2MH will provide large scale data to investigate the effects of RGDs on dimensional phenotypes and additional genetic and environmental factors that modulate these effects. These results will help refine clinical outcome predictions and design future interventions that can be implemented in the clinic to improve patient outcomes.
Supplementary Material
ACKNOWLEDGMENTS
This paper offers a synthesis of the ideas and aims of the Genome to Mental Health Network - G2MH- a new initiative funded under the RFA ‘Rare Genetic Disorders as a Window into the Genetic Architecture of Mental Disorders by the National Institute of Mental Health (NIMH) and the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD).
FUNDING
This work is supported by NIMH R01MH085953, 1U01MH119736, 1U01MH119736-01:S1 (Dr. C.E. Bearden). NIMH U01MH119738, U01MH119738:S1, U01MH119746 (Dr. J. Sebat). Binational Science Foundation 2017369 (Dr. R.E. Gur). NIMH U01MH119759 (Dr. A. Swillen). NIMH 1U01MH119690, 1U01MH119739, Canadian Institutes of Health Research (CIHR) 159734, 173280 (Dr. S. Jacquemont). NIMH 1U01MH119738 (Dr. M.B.M. van den Bree). Wellcome Trust ISSF Fellowship 204824/Z/16/Z (Dr. S.J.R.A. Chawner). NIMH U01 MH119690, U01 MH119689, U01 MH119690 02S1, Tommy Fuss Center for Neuropsychiatric Disease Research (Dr. D.C. Glahn). NIMH U01 MH119737-02, U01-MH119737-02S1, PO1-HD070454, RO1- GM125757 (Dr. D. McDonald-McGinn). NIMH U01MH119705, R01MH107431 (Dr. C.L. Martin / Dr. D. H. Ledbetter). Dutch Research Council Rubicon fellowship 45219212 (Dr. M. Klein). NIMH U01MH119741-01, Canadian Institutes of Health Research (CIHR) 162323 (Dr. J.A.S. Vorstman) NIMH U01MH119741-01, Canadian Institutes of Health Research (CIHR) MOP-313331, Dalglish Family Chair in 22q11.2 Deletion Syndrome (Dr. A.S. Bassett).
Footnotes
Members of the G2MH network: Kaitlyn Ahlers Ph.D. (University of Washington), Laura Almasy Ph.D. (Children’s Hospital of Philadelphia), Dustin Baldridge M.D., Ph.D. (Washington University in St. Louis), Anne Bassett M.D., FRCPC (University of Toronto; Centre for Addiction and Mental Health; University Health Network), Carrie Bearden Ph.D. (University of California, Los Angeles) Raphael Bernier Ph.D. (University of Washington), Meghan Bradley (The Hospital for Sick Children), Tonia Brown (University of Washington), Matthew Brown Ph.D. (Geisinger Health System), Christopher Chabris Ph.D. (Geisinger Health System), Elaine Chang (The Hospital for Sick Children), Samuel Chawner Ph.D. (Cardiff University), John Constantino M.D. (Washington University in St. Louis), Anthony Curtis Eayrs (University of Washington), Olga Del Rosario M.D. (Washington University in St. Louis), Kirsty Donald MBChB, Ph.D. (University of Cape Town), Rachel Earl Ph.D. (University of Washington), Evan Eichler Ph.D. (University of Washington & Howard Hughes Medical Institute), Beverly Emanuel Ph.D. (Children’s Hospital of Philadelphia and University of Pennsylvania), Ania M. Fiksinski Ph.D. (Maastricht University), Brenda Finucane M.S., LGC (Geisinger Health System), Sean Gallagher M.S. (University of Pennsylvania), Liliane Geyskens (Katholieke Universiteit Leuven), Madelyn Gillentine Ph.D. (University of Washington), Santhosh Girirajan MBBS, Ph.D. (Pennsylvania State University), David Glahn Ph.D. (Boston Children’s Hospital), Vardui Grigoryan B.A. (University of California, Los Angeles), Sinan Guloksuz Ph.D. (Maastricht University), Raquel Gur M.D., Ph.D. (University of Pennsylvania), Ruben Gur Ph.D. (University of Pennsylvania), Jessica Hall Ph.D. (Cardiff University), Janet Harwood Ph.D. (Cardiff University, Alexis Heidlebaugh Sc.M. (Geisinger Health System), Karahlyn Holdren B.S. (Geisinger Health System), Peter Holmans Ph.D. (Cardiff University), Guillaume Huguet Ph.D. (University of Montreal), Sebastien Jacquemont M.D. (University of Montreal), Jathishinie Jegathisawaran (The Hospital for Sick Children), Amina Kambic (Washington University in St. Louis), Claire Karlen Ph.D. (Washington University in St. Louis), Lauren Kasparson Walsh M.A. (Geisinger Health System), Marieke Klein Ph.D. (Radboud University Nijmegen), Eva Kurtz-Nelson Ph.D. (University of Washington), Leila Kushan M.Sc (University of California, Los Angeles), Virginia Lanzotti M.S. (Washington University in St. Louis), Hannah Lazarus (Children’s Hospital of Philadelphia), David Ledbetter Ph.D. (Dascena), Rene Lepore (Broad Institute), David Linden M.D., Ph.D. (Maastricht University), Christa Martin Ph.D., FACMG (Geisinger Health System), Donna McDonald-McGinn M.S., CGC (Children’s Hospital of Philadelphia), Hayley Moss (Cardiff University), Jennifer Mulle M.H.S., Ph.D. (Emory University), Scott Myers M.D. (Geisinger Health System), Jennifer Nunamaker (Geisinger Health System), Anne O’Donnell-Luria M.D., Ph.D. (Broad Institute), Matthew Oetjens Ph.D. (Geisinger Health System), Michael Owen Ph.D., M.B., FRCPsych, FMedSci, FLSW (Cardiff University), Katrina Palad M.Ed. (The Hospital for Sick Children), Megan Panther (Washington University in St. Louis), Alana Peters-Whiting B.S. (University of Washington), Laura Peyras (University of Montreal), Hannah Rea Ph.D. (University of Washington), David Roalf Ph.D. (University of Pennsylvania), Elise Robinson Sc.D. (Broad Institute), Kosha Ruparel M.S.E. (University of Pennsylvania), Tara Rutter M.S. (University of Washington), Christina Sargent B.A., B.S. (University of Washington), Stephen Scherer Ph.D., DSc, FRSC (The Hospital for Sick Children), Laura Schultz Ph.D. (Children’s Hospital of Philadelphia), Jonathan Sebat Ph.D. (University of California, San Diego), Autumn Swan M.S. (Geisinger Health System), Ann Swillen Ph.D. (Katholieke Universiteit Leuven), Cora Taylor Ph.D. (Geisinger Health System), Scott Troyan (University of Pennsylvania), Therese van Amelsvoort M.D., Ph.D. (Maastricht University), Kris Van de Woestyne (University Hospital Gasthuisberg), Marianne van den Bree Ph.D. (Cardiff University), Celia Van Der Merwe Ph.D., M.Sc. (University of Cape Town), Mieke van Haelst M.D. (Amsterdam UMC), Jente Verbesselt Ph.D. (Katholieke Universiteit Leuven), Claudia Vingerhoets Ph.D. (Maastricht University), Jacob Vorstman M.D., Ph.D. (The Hospital for Sick Children), Tianyun Wang Ph.D. (University of Washington), Lauren White Ph.D. (Children’s Hospital of Philadelphia), Nigel Williams Ph.D. (Cardiff University), Elaine Zackai M.D. (Children’s Hospital of Philadelphia and University of Pennsylvania), Stephanie Zettlemoyer (Geisinger Health System), Janneke Zinkstok Ph.D. (UMC Utrecht).
REFERENCES
- 1.Tammimies K, Marshall CR, Walker S, et al. : Molecular Diagnostic Yield of Chromosomal Microarray Analysis and Whole-Exome Sequencing in Children With Autism Spectrum Disorder. JAMA 2015; 314:895–903 [DOI] [PubMed] [Google Scholar]
- 2.Gaugler T, Klei L, Sanders SJ, et al. : Most genetic risk for autism resides with common variation. Nat Genet 2014; 46:881–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Costain G, Lionel AC, Merico D, et al. : Pathogenic rare copy number variants in community-based schizophrenia suggest a potential role for clinical microarrays. Hum Mol Genet 2013; 22:4485–4501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Satterstrom FK, Kosmicki JA, Wang J, et al. : Large-Scale Exome Sequencing Study Implicates Both Developmental and Functional Changes in the Neurobiology of Autism. Cell 2020; 180:568–584.e23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sanders SJ, He X, Willsey AJ, et al. : Insights into Autism Spectrum Disorder Genomic Architecture and Biology from 71 Risk Loci. Neuron 2015; 87:1215–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marshall CR, Howrigan DP, Merico D, et al. : Contribution of copy number variants to schizophrenia from a genome-wide study of 41,321 subjects. Nat Genet 2017; 49:27–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coe BP, Witherspoon K, Rosenfeld JA, et al. : Refining analyses of copy number variation identifies specific genes associated with developmental delay. Nat Genet 2014; 46:1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Douard E, Zeribi A, Schramm C, et al. : Effects-sizes of deletions and duplications on autism risk across the genome [Internet]. bioRxiv 2020; 2020.03.09.979815[cited 2020 Mar 19] Available from: https://www.biorxiv.org/content/10.1101/2020.03.09.979815v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vorstman JAS, Ophoff RA: Genetic causes of developmental disorders. Curr Opin Neurol 2013; 26:128–136 [DOI] [PubMed] [Google Scholar]
- 10.Niarchou M, Zammit S, van Goozen SHM, et al. : Psychopathology and cognition in children with 22q11.2 deletion syndrome. Br J Psychiatry 2014; 204:46–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.D’Angelo D, Lebon S, Chen Q, et al. : Defining the Effect of the 16p11.2 Duplication on Cognition, Behavior, and Medical Comorbidities. JAMA Psychiatry 2016; 73:20–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bassett AS, Scherer SW, Brzustowicz LM: Copy number variations in schizophrenia: critical review and new perspectives on concepts of genetics and disease. Am J Psychiatry 2010; 167:899–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moberg PJ, Richman MJ, Roalf DR, et al. : Neurocognitive Functioning in Patients with 22q11.2 Deletion Syndrome: A Meta-Analytic Review. Behav Genet 2018; 48:259–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gur RE, Yi JJ, McDonald-McGinn DM, et al. : Neurocognitive development in 22q11.2 deletion syndrome: comparison with youth having developmental delay and medical comorbidities. Mol Psychiatry 2014; 19:1205–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mervis CB, Robinson BF, Bertrand J, et al. : The Williams syndrome cognitive profile. Brain Cogn 2000; 44:604–628 [DOI] [PubMed] [Google Scholar]
- 16.Vogels A, De Hert M, Descheemaeker MJ, et al. : Psychotic disorders in Prader-Willi syndrome. Am J Med Genet A 2004; 127A:238–243 [DOI] [PubMed] [Google Scholar]
- 17.Shayota BJ, Elsea SH: Behavior and sleep disturbance in Smith-Magenis syndrome. Curr Opin Psychiatry 2019; 32:73–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fedorenko E, Morgan A, Murray E, et al. : A highly penetrant form of childhood apraxia of speech due to deletion of 16p11.2. Eur J Hum Genet 2016; 24:302–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bruining H, Eijkemans MJ, Kas MJ, et al. : Behavioral signatures related to genetic disorders in autism. Mol Autism 2014; 5:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chawner SJRA, Owen MJ, Holmans P, et al. : Genotype–phenotype associations in children with copy number variants associated with high neuropsychiatric risk in the UK (IMAGINE-ID): a case-control cohort study. The Lancet Psychiatry 2019; 6:493–505 [DOI] [PubMed] [Google Scholar]
- 21.Lin A, Vajdi A, Kushan-Wells L, et al. : Reciprocal Copy Number Variations at 22q11.2 Produce Distinct and Convergent Neurobehavioral Impairments Relevant for Schizophrenia and Autism Spectrum Disorder. Biol Psychiatry 2020; 88:260–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huguet G, Schramm C, Douard E, et al. : Genome-wide analysis of gene dosage in 24,092 individuals estimates that 10,000 genes modulate cognitive ability [Internet]. Mol Psychiatry 2021; Available from: 10.1038/s41380-020-00985-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davies RW, Fiksinski AM, Breetvelt EJ, et al. : Using common genetic variation to examine phenotypic expression and risk prediction in 22q11.2 deletion syndrome [Internet]. Nat Med 2020; Available from: 10.1038/s41591-020-1103-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chawner SJRA, Doherty JL, Anney RJL, et al. : A Genetics-First Approach to Dissecting the Heterogeneity of Autism: Phenotypic Comparison of Autism Risk Copy Number Variants. Am J Psychiatry 2021; 178:77–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Unolt M, Kammoun M, Nowakowska B, et al. : Pathogenic variants in CDC45 on the remaining allele in patients with a chromosome 22q11.2 deletion result in a novel autosomal recessive condition. Genet Med 2020; 22:326–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Albers CA, Paul DS, Schulze H, et al. : Compound inheritance of a low-frequency regulatory SNP and a rare null mutation in exon-junction complex subunit RBM8A causes TAR syndrome. Nat Genet 2012; 44:435–9, S1–2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wray NR, Wijmenga C, Sullivan PF, et al. : Common Disease Is More Complex Than Implied by the Core Gene Omnigenic Model. Cell 2018; 173:1573–1580 [DOI] [PubMed] [Google Scholar]
- 28.Moreno-De-Luca A, Evans DW, Boomer KB, et al. : The role of parental cognitive, behavioral, and motor profiles in clinical variability in individuals with chromosome 16p11.2 deletions. JAMA Psychiatry 2015; 72:119–126 [DOI] [PubMed] [Google Scholar]
- 29.Cleynen I, Engchuan W, Hestand MS, et al. : Genetic contributors to risk of schizophrenia in the presence of a 22q11.2 deletion [Internet]. Mol Psychiatry 2020; Available from: 10.1038/s41380-020-0654-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fiksinski AM, Heung T, Corral M, et al. : Within-family influences on dimensional neurobehavioral traits in a high-risk genetic model. Psychol Med 2021; 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fiksinski AM, Schneider M, Murphy CM, et al. : Understanding the pediatric psychiatric phenotype of 22q11.2 deletion syndrome. Am J Med Genet A 2018; 176:2182–2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fischbach GD, Lord C: The Simons Simplex Collection: a resource for identification of autism genetic risk factors. Neuron 2010; 68:192–195 [DOI] [PubMed] [Google Scholar]
- 33.Bishop SL, Farmer C, Bal V, et al. : Identification of Developmental and Behavioral Markers Associated With Genetic Abnormalities in Autism Spectrum Disorder. Am J Psychiatry 2017; 174:576–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Douard E, Zeribi A, Schramm C, et al. : Effect Sizes of Deletions and Duplications on Autism Risk Across the Genome. Am J Psychiatry 2020; appiajp202019080834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cuthbert BN, Insel TR: Toward the future of psychiatric diagnosis: the seven pillars of RDoC. BMC Med 2013; 11:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Insel T, Cuthbert B, Garvey M, et al. : Research domain criteria (RDoC): toward a new classification framework for research on mental disorders. Am J Psychiatry 2010; 167:748–751 [DOI] [PubMed] [Google Scholar]
- 37.Van L, Boot E, Bassett AS: Update on the 22q11.2 deletion syndrome and its relevance to schizophrenia. Curr Opin Psychiatry 2017; 30:191–196 [DOI] [PubMed] [Google Scholar]
- 38.Monks S, Niarchou M, Davies AR, et al. : Further evidence for high rates of schizophrenia in 22q11.2 deletion syndrome. Schizophr Res 2014; 153:231–236 [DOI] [PubMed] [Google Scholar]
- 39.Niarchou M, Martin J, Thapar A, et al. : The clinical presentation of attention deficit-hyperactivity disorder (ADHD) in children with 22q11.2 deletion syndrome. Am J Med Genet B Neuropsychiatr Genet 2015; 168:730–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vorstman JAS, Breetvelt EJ, Duijff SN, et al. : Cognitive decline preceding the onset of psychosis in patients with 22q11.2 deletion syndrome. JAMA Psychiatry 2015; 72:377–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chawner SJRA, Niarchou M, Doherty JL, et al. : The emergence of psychotic experiences in the early adolescence of 22q11.2 Deletion Syndrome. J Psychiatr Res 2019; 109:10–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morrison S, Chawner SJRA, van Amelsvoort TAMJ, et al. : Cognitive deficits in childhood, adolescence and adulthood in 22q11.2 deletion syndrome and association with psychopathology. Transl Psychiatry 2020; 10:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cox DM, Butler MG: The 15q11.2 BP1-BP2 microdeletion syndrome: a review. Int J Mol Sci 2015; 16:4068–4082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jønch AE, Douard E, Moreau C, et al. : Estimating the effect size of the 15Q11.2 BP1-BP2 deletion and its contribution to neurodevelopmental symptoms: recommendations for practice. J Med Genet 2019; 56:701–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stefansson H, Meyer-Lindenberg A, Steinberg S, et al. : CNVs conferring risk of autism or schizophrenia affect cognition in controls. Nature 2014; 505:361–366 [DOI] [PubMed] [Google Scholar]
- 46.Männik K, Mägi R, Macé A, et al. : Copy number variations and cognitive phenotypes in unselected populations. JAMA 2015; 313:2044–2054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huguet G, Schramm C, Douard E, et al. : Measuring and Estimating the Effect Sizes of Copy Number Variants on General Intelligence in Community-Based Samples. JAMA Psychiatry 2018; 75:447–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martin CL, Wain KE, Oetjens MT, et al. : Identification of Neuropsychiatric Copy Number Variants in a Health Care System Population [Internet]. JAMA Psychiatry 2020; Available from: 10.1001/jamapsychiatry.2020.2159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Olsen L, Sparsø T, Weinsheimer SM, et al. : Prevalence of rearrangements in the 22q11.2 region and population-based risk of neuropsychiatric and developmental disorders in a Danish population: a case-cohort study. Lancet Psychiatry 2018; 5:573–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schneider M, Debbané M, Bassett AS, et al. : Psychiatric disorders from childhood to adulthood in 22q11.2 deletion syndrome: results from the International Consortium on Brain and Behavior in 22q11.2 Deletion Syndrome. Am J Psychiatry 2014; 171:627–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kendall KM, Bracher-Smith M, Fitzpatrick H, et al. : Cognitive performance and functional outcomes of carriers of pathogenic copy number variants: analysis of the UK Biobank. Br J Psychiatry 2019; 214:297–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kendall KM, Rees E, Bracher-Smith M, et al. : Association of Rare Copy Number Variants With Risk of Depression [Internet]. JAMA Psychiatry 2019; Available from: 10.1001/jamapsychiatry.2019.0566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Van L, Heung T, Graffi J, et al. : All-cause mortality and survival in adults with 22q11.2 deletion syndrome. Genet Med 2019; 21:2328–2335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Crawford K, Bracher-Smith M, Owen D, et al. : Medical consequences of pathogenic CNVs in adults: analysis of the UK Biobank. J Med Genet 2019; 56:131–138 [DOI] [PubMed] [Google Scholar]
- 55.Uysal-Bozkir Ö, Parlevliet JL, de Rooij SE: Insufficient cross-cultural adaptations and psychometric properties for many translated health assessment scales: a systematic review. J Clin Epidemiol 2013; 66:608–618 [DOI] [PubMed] [Google Scholar]
- 56.Casaletto KB, Heaton RK: Neuropsychological Assessment: Past and Future. J Int Neuropsychol Soc 2017; 23:778–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kapur S, Phillips AG, Insel TR: Why has it taken so long for biological psychiatry to develop clinical tests and what to do about it? Mol Psychiatry 2012; 17:1174–1179 [DOI] [PubMed] [Google Scholar]
- 58.Torkamani A, Wineinger NE, Topol EJ: The personal and clinical utility of polygenic risk scores. Nat Rev Genet 2018; 19:581–590 [DOI] [PubMed] [Google Scholar]
- 59.Zheutlin AB, Dennis J, Karlsson Linnér R, et al. : Penetrance and Pleiotropy of Polygenic Risk Scores for Schizophrenia in 106,160 Patients Across Four Health Care Systems. Am J Psychiatry 2019; 176:846–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Plomin R, von Stumm S: The new genetics of intelligence. Nat Rev Genet 2018; 19:148–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huguet G, Schramm C, Douard E, et al. : Genome wide analysis of gene dosage in 24,092 individuals shows that 10,000 genes modulate cognitive ability [Internet]. Cold Spring Harbor Laboratory 2020; 2020.04.03.024554[cited 2020 Oct 22] Available from: https://www.biorxiv.org/content/10.1101/2020.04.03.024554v3 [Google Scholar]
- 62.Fung WLA, Butcher NJ, Costain G, et al. : Practical guidelines for managing adults with 22q11.2 deletion syndrome. Genet Med 2015; 17:599–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bassett AS, McDonald-McGinn DM, Devriendt K, et al. : Practical guidelines for managing patients with 22q11.2 deletion syndrome. J Pediatr 2011; 159:332–9.e1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sanchez Russo R, Gambello MJ, Murphy MM, et al. : Deep phenotyping in 3q29 deletion syndrome: recommendations for clinical care [Internet]. Genet Med 2021; Available from: 10.1038/s41436-020-01053-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chawner SJ, Watson CJ, Owen MJ: Clinical evaluation of patients with a neuropsychiatric risk copy number variant. Curr Opin Genet Dev 2021; 68:26–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sommer IE, Bearden CE, van Dellen E, et al. : Early interventions in risk groups for schizophrenia: what are we waiting for? NPJ Schizophr 2016; 2:16003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Linden SC, Watson CJ, Smith J, et al. : The psychiatric phenotypes of 1q21 distal deletion and duplication. Transl Psychiatry 2021; 11:105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Costain G, McDonald-McGinn DM, Bassett AS: Prenatal genetic testing with chromosomal microarray analysis identifies major risk variants for schizophrenia and other later-onset disorders. Am J Psychiatry 2013; 170:1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Moeschler JB, Shevell M, Committee on Genetics: Comprehensive evaluation of the child with intellectual disability or global developmental delays. Pediatrics 2014; 134:e903–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hyman SL, Levy SE, Myers SM, et al. : Identification, Evaluation, and Management of Children With Autism Spectrum Disorder [Internet]. Pediatrics 2020; 145Available from: 10.1542/peds.2019-3447 [DOI] [PubMed] [Google Scholar]
- 71.Miller DT, Adam MP, Aradhya S, et al. : Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet 2010; 86:749–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Moreno-De-Luca D, Kavanaugh BC, Best CR, et al. : Clinical Genetic Testing in Autism Spectrum Disorder in a Large Community-Based Population Sample [Internet]. JAMA Psychiatry 2020; Available from: 10.1001/jamapsychiatry.2020.0950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tchaconas A, Adesman A: Diagnostic Evaluation of Children with Autism Spectrum Disorders: Clinician Compliance with Published Guidelines. J Dev Behav Pediatr 2017; 38:29–38 [DOI] [PubMed] [Google Scholar]
- 74.Soda T, Pereira S, Small BJ, et al. : Child and Adolescent Psychiatrists’ Perceptions of Utility and Self-rated Knowledge of Genetic Testing Predict Usage for Autism Spectrum Disorder [Internet]. J Am Acad Child Adolesc Psychiatry 2021; Available from: 10.1016/j.jaac.2021.01.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vorstman J, Scherer SW: What a finding of gene copy number variation can add to the diagnosis of developmental neuropsychiatric disorders. Curr Opin Genet Dev 2021; 68:18–25 [DOI] [PubMed] [Google Scholar]
- 76.Nurnberger JI Jr, Austin J, Berrettini WH, et al. : What Should a Psychiatrist Know About Genetics? Review and Recommendations From the Residency Education Committee of the International Society of Psychiatric Genetics [Internet]. J Clin Psychiatry 2018; 80Available from: 10.4088/JCP.17nr12046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Silva M, de Leeuw N, Mann K, et al. : European guidelines for constitutional cytogenomic analysis. Eur J Hum Genet 2019; 27:1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Srivastava S, Love-Nichols JA, Dies KA, et al. : Meta-analysis and multidisciplinary consensus statement: exome sequencing is a first-tier clinical diagnostic test for individuals with neurodevelopmental disorders. Genet Med 2019; 21:2413–2421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Foley C, Heron EA, Harold D, et al. : Identifying schizophrenia patients who carry pathogenic genetic copy number variants using standard clinical assessment: retrospective cohort study. Br J Psychiatry 2020; 216:275–279 [DOI] [PubMed] [Google Scholar]
- 80.Baker K, Costain G, Fung WLA, et al. : Chromosomal microarray analysis-a routine clinical genetic test for patients with schizophrenia. Lancet Psychiatry 2014; 1:329–331 [DOI] [PubMed] [Google Scholar]
- 81.Morris TT, Davies NM, Hemani G, et al. : Population phenomena inflate genetic associations of complex social traits. Science Advances 2020; 6:eaay0328[cited 2020 Apr 21] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Roundtable on Translating Genomic-Based Research for Health, Board on Health Sciences Policy, Institute of Medicine: Genomics-Enabled Learning Health Care Systems: Gathering and Using Genomic Information to Improve Patient Care and Research: Workshop Summary. Washington (DC), National Academies Press (US), 2015 [PubMed] [Google Scholar]
- 83.Khan TA, Revah O, Gordon A, et al. : Neuronal defects in a human cellular model of 22q11.2 deletion syndrome. Nat Med 2020; 26:1888–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schaaf CP, Betancur C, Yuen RKC, et al. : A framework for an evidence-based gene list relevant to autism spectrum disorder. Nat Rev Genet 2020; 21:367–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Myers SM, Challman TD, Bernier R, et al. : Insufficient Evidence for “Autism-Specific” Genes. Am J Hum Genet 2020; 106:587–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sullivan PF, Agrawal A, Bulik CM, et al. : Psychiatric Genomics: An Update and an Agenda. Am J Psychiatry 2018; 175:15–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cubells JF, Deoreo EH, Harvey PD, et al. : Pharmaco-genetically guided treatment of recurrent rage outbursts in an adult male with 15q13.3 deletion syndrome. Am J Med Genet A 2011; 155A:805–810 [DOI] [PubMed] [Google Scholar]
- 88.Gillentine MA, Yin J, Bajic A, et al. : Functional Consequences of CHRNA7 Copy-Number Alterations in Induced Pluripotent Stem Cells and Neural Progenitor Cells. Am J Hum Genet 2017; 101:874–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sanders SJ, Sahin M, Hostyk J, et al. : A framework for the investigation of rare genetic disorders in neuropsychiatry. Nat Med 2019; 25:1477–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bernier R, Steinman KJ, Reilly B, et al. : Clinical phenotype of the recurrent 1q21.1 copy-number variant. Genet Med 2016; 18:341–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kirov G, Rees E, Walters JTR, et al. : The penetrance of copy number variations for schizophrenia and developmental delay. Biol Psychiatry 2014; 75:378–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rees E, Walters JTR, Georgieva L, et al. : Analysis of copy number variations at 15 schizophrenia-associated loci. Br J Psychiatry 2014; 204:108–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Moreno-De-Luca D, Sanders SJ, Willsey AJ, et al. : Using large clinical data sets to infer pathogenicity for rare copy number variants in autism cohorts. Mol Psychiatry 2013; 18:1090–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Maenner MJ, Shaw KA, Baio J, et al. : Prevalence of Autism Spectrum Disorder Among Children Aged 8 Years - Autism and Developmental Disabilities Monitoring Network, 11 Sites, United States, 2016. MMWR Surveill Summ 2020; 69:1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.McGrath J, Saha S, Chant D, et al. : Schizophrenia: a concise overview of incidence, prevalence, and mortality. Epidemiol Rev 2008; 30:67–76 [DOI] [PubMed] [Google Scholar]
- 96.Grove J, Ripke S, Als TD, et al. : Identification of common genetic risk variants for autism spectrum disorder. Nat Genet 2019; 51:431–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Richards AL, Pardiñas AF, Frizzati A, et al. : The Relationship Between Polygenic Risk Scores and Cognition in Schizophrenia. Schizophr Bull 2020; 46:336–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.