Abstract
We have cloned and sequenced the 3-hydroxybutyrate dehydrogenase-encoding gene (bdhA) from Rhizobium (Sinorhizobium) meliloti. The gene has an open reading frame of 777 bp that encodes a polypeptide of 258 amino acid residues (molecular weight 27,177, pI 6.07). The R. meliloti Bdh protein exhibits features common to members of the short-chain alcohol dehydrogenase superfamily. bdhA is the first gene transcribed in an operon that also includes xdhA, encoding xanthine oxidase/dehydrogenase. Transcriptional start site analysis by primer extension identified two transcription starts. S1, a minor start site, was located 46 to 47 nucleotides upstream of the predicted ATG start codon, while S2, the major start site, was mapped 148 nucleotides from the start codon. Analysis of the sequence immediately upstream of either S1 or S2 failed to reveal the presence of any known consensus promoter sequences. Although a ς54 consensus sequence was identified in the region between S1 and S2, a corresponding transcript was not detected, and a rpoN mutant of R. meliloti was able to utilize 3-hydroxybutyrate as a sole carbon source. The R. meliloti bdhA gene is able to confer upon Escherichia coli the ability to utilize 3-hydroxybutyrate as a sole carbon source. An R. meliloti bdhA mutant accumulates poly-3-hydroxybutyrate to the same extent as the wild type and shows no symbiotic defects. Studies with a strain carrying a lacZ transcriptional fusion to bdhA demonstrated that gene expression is growth phase associated.
The soil bacterium Rhizobium (Sinorhizobium) meliloti fixes N2 in symbiotic association with leguminous plants such as alfalfa. R. meliloti cells are able to exist as two distinct entities: the free-living form competes for limiting nutrients with other soil inhabitants, while the symbiotic N2-fixing bacteroid forms an intimate association with the host plant from which a steady supply of nutrients is derived.
When excess carbon nutrient is available but a noncarbon nutrient such as N, P, or O2 is limiting for growth, many bacteria accumulate the intracellular carbon storage compound poly-3-hydroxybutyrate (PHB). Under subsequent carbon limiting conditions, the endogenous PHB stores can serve as a source of carbon and reducing energy (3). In pure culture, free-living R. meliloti is able to accumulate PHB to up to 60% of the total cellular dry weight (81). PHB deposits have also been observed to be present in bacteria within the infection thread but not in differentiated bacteroids (40, 41, 65). In contrast to the absence of PHB in R. meliloti bacteroids, PHB does accumulate in bacteroids of some other rhizobia such as Rhizobium sp. strain NGR234 (83), Rhizobium etli (11), and Bradyrhizobium japonicum (58). Thus, PHB may have important roles to play at various stages of the symbiosis.
The capacity of R. meliloti to successfully compete with other soil microorganisms for the limiting nutrient resources in soil is an important determinant for the establishment of a successful symbiosis (82, 84). The ability to synthesize and degrade PHB may influence the capability of bacterial cells to survive extended periods of starvation in the soil (77). A specific carbon source that is responsible for fueling cell growth and development during infection and nodule invasion has not been identified (28), and it has been suggested that intracellular PHB, accumulated in the rhizosphere, may be an important source of carbon and energy during infection (12). In the cases where PHB deposits are found in bacteroids, PHB may be important in fueling the N2 fixation process when the supply of plant photosynthates is reduced, for example, in darkness (9, 35), or for recovery of bacteroids after nodule senescence (48, 76). Alternatively, PHB synthesis may compete with the N2 fixation process for reducing equivalents, as was proposed to explain the observation that R. etli mutants defective in PHB synthesis induce nodules with enhanced N2 fixation capacity (11). Although it has been reported that mutants of R. meliloti unable to synthesize PHB are able to establish symbiotic association with alfalfa hosts (67), PHB may nevertheless provide carbon and energy for bacteria within the infection thread when the supply from the host is inadequate.
Biochemical studies have suggested a pathway for PHB catabolism in bacteria. Degradation is initiated with the action of a PHB depolymerase that releases the monomer 3-hydroxybutyrate. Both intracellular and extracellular PHB depolymerases have been documented (42, 43). The enzyme 3-hydroxybutyrate dehydrogenase (EC 1.1.1.30; Bdh) catalyzes the reversible oxidation of the released 3-hydroxybutyrate (HB) to acetoacetate, which is then activated to acetoacetyl coenzyme A (acetoacetyl-CoA) by a CoA transferase. A ketothiolase cleaves the acetoacetyl-CoA to yield two molecules of acetyl-CoA which are metabolized via the tricarboxylic acid cycle and glyoxylate shunt (71).
Bdh enzymes from a number of bacteria have been purified and biochemically characterized (10, 22, 46, 49, 55, 63, 78). Enzymatic studies in various bacteria have suggested that Bdh plays a key role in the control of PHB degradation because its activity is regulated by some or all of the following compounds: NAD(P)H, pyruvate, oxaloacetate, 2-oxoglutarate, and acetyl-CoA (55, 56, 71, 78). Moreover, this enzyme has been observed only in bacteria that are able to accumulate PHB (55), and in Azospirillum brasilense, the level of Bdh activity has been shown to be directly proportional to the total PHB content (77). Although R. meliloti has a single Bdh, some Rhizobium strains produce multiple forms of Bdh, the physiological significance of which remains unknown (31).
In mammals, the Bdh enzyme is involved in ketone body metabolism during periods of starvation. The mammalian enzyme is located in the matrix face of the inner mitochondrial membrane (59), whereas the bacterial enzyme is cytoplasmic. In contrast to the bacterial enzyme, the mammalian enzyme has an absolute requirement for phosphatidylcholine for activity. The primary sequence of rat Bdh places it in the short-chain alcohol dehydrogenase (SCAD) superfamily (20). An example of a bacterial Bdh sequence has not been available for comparison to establish the molecular basis of the structural and functional differences between the bacterial and mammalian Bdh enzymes.
In a recent report, we described the isolation of a R. meliloti Tn5 mutant, strain Rm11107, that is unable to metabolize HB but retains the ability to utilize acetoacetate as a sole carbon source (12). Cell extracts of strain Rm11107 cultures lack 3-hydroxybutyrate dehydrogenase activity. The mutation was mapped to megaplasmid pRmeSU47b. Here we report the phenotypic characterization of the bdhA mutant, isolation of the bdhA gene, and the complete nucleotide sequence of the gene and surrounding DNA. In addition, we show that expression of bdhA is growth phase associated.
MATERIALS AND METHODS
Bacterial strains, plasmids, transposons, and culture conditions.
A list of the bacterial strains, plasmids, and transposons used in this study is provided in Table 1. The construction of new strains is described in the text. Culture methods in LB, TY, and modified M9 minimal medium with various carbon sources, and antibiotic concentrations, were as described previously (12, 13). Thiamine (5 μM) was added to minimal medium for growth of Escherichia coli LS5218 and LS5218R. Where glycerol served as the sole carbon source, it was present at a final concentration of 1%.
TABLE 1.
Bacterial strains, plasmids, and phage used in this studya
| Strain, plasmid, or phage | Relevant characteristics | Source, reference, or construction |
|---|---|---|
| R. meliloti | ||
| Rm1021 | SU47 str-21 | 61 |
| Rm5000 | SU47 rif-5 | 26 |
| Rm5422 | Rm1021ntrA75::Tn5 | 29 |
| Rm8501 | Rm1021lac | 37 |
| Rm11107 | Rm1021bdhA1::Tn5 | 12 |
| Rm11145 | Rm1021bdhA1::Tn5-233 | 12 |
| Rm11159 | Rm1021bdhA2::Tn5 | This study |
| Rm11191 | Rm1021bdhA3-2::Tn5-B20 | This study |
| Rm11234 | Rm1021glpK11::Tn5 | This study |
| Rm11249 | Rm5000bdhA3::ΩSmSp | This study |
| Rm11262 | Rm1021bdhA3::ΩSmSp | This study |
| E. coli | ||
| LS5218 | fadR601 atoC200 (constitutive) | CGSC 6966; reference 74 |
| LS5218R | LS5218 Rpr | This work |
| DH5α | F−endA1 hsdR17 (rK− mK−) supE44 thi-1 recA1 gyrA96 relA1 Δ(argF-lacZYA)U169 Φ80dlacZ ΔM15 λ− | BRL Inc. |
| MT607 | pro-82 thi-1 endA hsdR17 supE44 recA56 | 27 |
| MT616 | MT607(pRK600); mobilizer | 27 |
| G312 | MT607Ω5::Tn5-B20 | 24 |
| MT614 | MT607ΩTn5 | 29a |
| Plasmids | ||
| pLAFR1 | IncP cosmid cloning vector; Tcr | 33 |
| pHP45 | Ω SmrSpr; vector Apr | 68 |
| pBSKS+ | ColE1 cloning vector; Apr | Stratagene |
| pUC19 | ColE1 cloning vector; Apr | 87 |
| pSP329 | IncP cloning vector; Tcr | 66a |
| pTC344 | pLAFRI clone carrying bdhA, isolated by complementation of Rm11107 | This study |
| pTC370 | pLAFRI clone carrying bdhA, isolated by complementation of LS5218R | This study |
| pPA31 | pBSKS+ carrying bdhA on a 5.5-kb PstI fragment, derived from pTC344 | This study |
| pPA35 | pSP329 with the pTC344-derived 5.5-kb PstI fragment carrying bdhA | This study |
| pPA36 | Same as pPA35, opposite orientation | This study |
| pPA56 | pUC19 with the pTC344-derived 5.5-kb PstI fragment carrying bdhA | This study |
| pPA59 | pPA56 with Ω SmrSpr insertion in unique EcoRV site within bdhA | This study |
| pPA60 | pSP329 with the PstI fragment in pPA59 | This study |
| pPA63 | pBSKS+ carrying bdhA on a 1.2-kb XhoI-SalI fragment derived from pPA35 | This study |
| pPA67 | pSP329 carrying bdhA on a 1.2-kb XhoI-SalI fragment derived from pPA35 | This study |
| Phage | ||
| ΦM12 | Generalized transducing phage | 26 |
Genetics and molecular biology techniques.
Bacterial conjugations, ΦM12 transductions, Tn5 mutagenesis, homogenotizations, and transposon replacements were carried out as described previously (13, 14, 16). DNA manipulations were carried out by using standard methods (4). DNA sequence was determined by a combination of manual and automated (MOBIX Facility, McMaster University, Hamilton, Ontario, Canada) sequencing using both universal and custom-designed primers and an IS50-specific primer (15). Custom-designed primers were purchased from Biosource International (Keystone Labs, Camarillo, Calif.) or Life Technologies GIBCO BRL (Gaithersburg, Md.). Manual sequencing was performed with the cycle sequencing kit from Life Technologies GIBCO BRL. Automated sequencing was performed with an Applied Biosystems 373A Stretch DNA automated sequencer, using dye terminator chemistry and cycle sequencing. Sequence compilations and analyses were done with MacVector, version 6.0.1 (Oxford Molecular Group) and DNA Strider, version 1.2 (57). Multiple alignments were performed with the Clustal W program (79).
RNA isolation.
Total cellular RNA was prepared from 500 ml of TY-grown R. meliloti cultures harvested at an optical density at 600 nm (OD600) between 0.5 and 0.6 by using a modification of the hot phenol extraction method (34, 64). Prior to use, the RNA solution was treated with 160 U of RNase-free DNase I in the presence of 80 U of the RNase inhibitor RNasin (Promega, Madison, Wis.). This was followed by phenol-chloroform-isoamyl alcohol (25:24:1) extraction and ethanol precipitation of the RNA.
Primer extension analysis.
A 30-mer oligonucleotide (5′-CTCGTGGAACCCGTTATCACCGCAGTCTTG-3′) complementary to nucleotide positions 6 to 35 of the putative protein coding region was employed for mapping the transcriptional start site(s) of bdhA. The primer was end labeled with [γ-33P]ATP (Amersham, Buckinghamshire, United Kingdom) by using T4 polynucleotide kinase (Promega) at 37°C for 10 min, followed by heat inactivation at 65°C for 5 min. The labeled primer was separated from the unincorporated label by using a Microcon-3 (Amicon, Beverly, Mass.) spin column. The probe (106 cpm) was coprecipitated with 50 μg of Rm1021 RNA. Extension was performed with 40 U of Expand reverse transcriptase (Boehringer GmbH, Mannheim, Germany) for 90 min at 42°C. Half of the extension product was loaded onto a 6% polyacrylamide–7 M urea gel and run alongside a sequencing ladder generated by using the same 30-mer oligonucleotide as primer and 200 ng of plasmid pPA31 as template DNA.
Enzyme assays.
Preparation of cell extracts, protein determination, the assay for Bdh activity, and nondenaturing polyacrylamide gel electrophoresis were carried out as described previously (12). For detection of xanthine oxidase and xanthine dehydrogenase activities, cell extracts equivalent to 85 μg of total protein were electrophoresed through a nondenaturing polyacrylamide gel. Postelectrophoretic staining for xanthine oxidase (EC 1.1.3.22) and xanthine dehydrogenase (EC 1.1.1.204) activities were performed with hypoxanthine as the substrate (25, 72). Since aldehyde oxidase (EC 1.2.3.1) can also be detected by this method, a parallel control gel was stained for aldehyde oxidase activity with benzaldehyde as the substrate (25).
Generation of bdhA-lacZ fusions.
To obtain Tn5-B20 (73) insertions in bdhA, pTC344 was introduced into E. coli G312, which carries Tn5-B20 on the chromosome (24). Tn5-B20 insertions in pTC344 were identified by triparental mating of the plasmids from the donor E. coli strains into strain Rm11145 recipients by using E. coli MT616 as a mobilizer. The Smr Nmr Tcr colonies obtained were screened for the ability to grow on minimal medium with HB as the sole carbon source, and five insertions that disrupted the complementing ability of pTC344 were retained. These Tn5-B20 insertions were then tested for lacZ expression in the Lac− strain Rm8501, and three of five were Lac+. The precise locations of these insertions were determined by sequence analysis using the IS50 primer. The insertions were then homogenotized into the Rm8501 genome.
Transcriptional fusion assays.
Transcriptional assays were performed by inoculating 1.5 ml of stationary-phase cultures of strains Rm8501 and Rm11191 into 1-liter Erlenmeyer flasks containing 300 ml of TY. Samples (1.5 ml) were withdrawn every 2 h, and OD600 and β-galactosidase activities were determined as described previously (75). β-Galactosidase activity is expressed in Miller units (62).
PHB assays.
Cultures for PHB assays were obtained by growing the strains in 125-ml Erlenmeyer flasks containing 50 ml of YMB (K2HPO4, 0.5; MgSO4 · 7H2O, 0.2; NaCl, 0.1; mannitol, 10; yeast extract, 0.4 [in grams per liter]) (81) and shaking at 160 rpm for 48 h. Following a saline wash and resuspension in 50 ml of saline, PHB was extracted from a 2-ml fraction of culture and assayed by the method of Law and Slepecky (53). The remaining 48 ml of culture was used for dry weight determination after incubation of the wet pellet at 37°C until no further decrease in weight was noted.
Plant assays.
Symbiotic phenotype assays were performed with axenic alfalfa plants in covered Magenta jars with washed vermiculite containing 25 ml of Jensen’s N-free plant nutrient solution (45). Each jar, containing five 2-day-old seedlings, was inoculated with 5 ml of a 1:50 H2O dilution of a saturated TY culture. The jars were placed in a growth chamber (16 h at 25°C [day] and 8 h at 20°C [night]; light intensity, 300 μEm−2 sm−1). Shoot dry weights were determined 6 weeks after inoculation. Nodulation kinetics data were obtained by monitoring the appearance of nodules on alfalfa plants grown on 1% agar slants containing Jensen’s N-free medium.
Nucleotide sequence accession number.
The nucleotide sequence of bdhA and the surrounding region has been deposited in GenBank under accession no. AF080548.
RESULTS
Isolation, identification, and subcloning of the bdhA locus.
R. meliloti Rm11107, the previously reported bdhA1::Tn5 mutant, is unable to utilize HB as a sole carbon source and lacks Bdh activity in cell extracts but is able to utilize acetoacetate as a sole carbon source (12). A pLAFR1 genomic library of the wild-type strain Rm1021 (33) was introduced into Rm11107 in a triparental mating. Clones that confer upon Rm11107 the ability to utilize HB were selected by plating on minimal medium containing HB (M9-HB) as a sole carbon source. Colonies were streak purified twice before the complementing cosmid clones were transferred by conjugation into E. coli MT607. A representative complementing cosmid clone was retained for further study and designated pTC344. The complementing ability of pTC344 was verified by reintroduction into Rm11107 followed by testing for HB utilization. A 5.5-kb PstI fragment of pTC344 was isolated by subcloning into pBSKS+ to give pPA31 and subsequently transferred into pSP329 in both orientations. Plasmids pPA35 and pPA36 thus generated were able to complement Rm11107 for HB utilization. Further subcloning of pPA35 identified a 1.2-kb XhoI-SalI fragment by first cloning into pBSKS+, to give pPA63, and then cloning into pSP329 as a KpnI-HindIII fragment (by using convenient flanking restriction sites in the multiple cloning site). The resulting plasmid, pPA67, was able to complement the HB utilization phenotype of Rm11107 (Fig. 1).
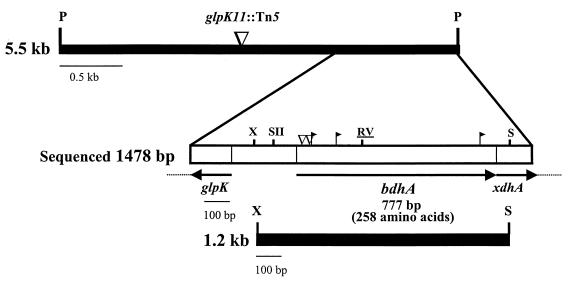
FIG. 1.
Physical and genetic map of the 5.5-kb PstI complementing fragment containing the entire bdhA gene and flanking regions. The relevant restriction sites shown are PstI (P), EcoRV (RV), XhoI (X), SalI (S), and SacII (SII) (underline indicates unique restriction site). The location of insertion pPA35glpK11::Tn5 is marked (▿). The open box delineates the 1,478-bp region sequenced. The positions of the Tn5 (▿) and Tn5-B20 (▸|) insertions that abolish complementation of HB utilization are also indicated. The orientations of the arrows above the gene designations indicate directions of transcription. The lower closed box represents the 1.2-kb XhoI-SalI fragment that retains complementing ability.
Complementation of E. coli LS5218R for HB utilization.
E. coli K-12 derivatives do not metabolize HB since they lack 3-hydroxybutyrate dehydrogenase activity, but they are able to utilize acetoacetate as a sole carbon source. The K-12 derivative strain LS5218 constitutively expresses the genes for acetoacetate utilization (44, 74). We reasoned that heterologous expression of the R. meliloti bdhA gene should confer upon LS5218 the ability to utilize HB as a growth substrate. Accordingly, the R. meliloti clone bank was also introduced into LS5218R (a spontaneous Rpr derivative of LS5218) as an independent attempt to isolate bdhA, and complemented transconjugants were selected on M9-HB. A complementing clone designated pTC370 was thus isolated and subjected to Tn5 mutagenesis. Plasmid pTC370bdhA2::Tn5, which is unable to complement LS5218R for growth on HB, was isolated and recombined into the wild-type genome to generate strain Rm11159, which was unable to utilize HB as a sole carbon source. Consistent with this result, cell extracts of Rm11159 strain lacked Bdh activity (data not shown). Also, both pTC370 and pTC344 were able to complement both Rm11159 and Rm11107 for HB utilization. To compare the genomic locations of the Tn5 insertions in Rm11107 and Rm11159, the insertion in Rm11145 (Rm1021bdhA1::Tn5-233) was transduced into strain Rm11159. All 89 transductants tested were Nms, establishing that the bdhA1 and bdhA2 insertions are tightly linked in transduction. The precise location of the Tn5 insertion in pTC370bdhA2::Tn5 was determined by sequencing the region flanking the Tn5 insertion and is shown in Fig. 2.
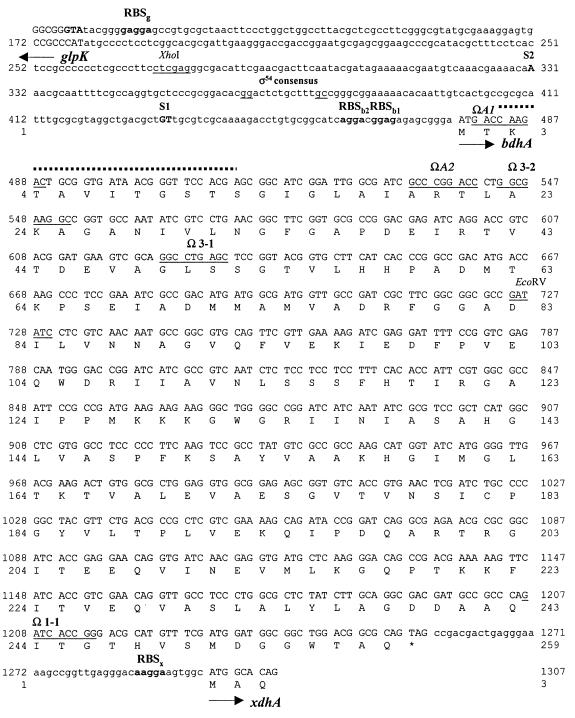
FIG. 2.
Nucleotide and deduced amino acid sequences of a 1,136-bp region comprising bdhA and portions of the flanking ORFs. Additional sequence information on the ORFs flanking bdhA beyond what is included in this figure is available under GenBank accession no. AF080548. Arrows indicate directions of transcription. Putative ribosome binding sites (RBS) for bdhA, glpK, and xdhA are in boldtype, followed by the first letter of the respective gene. The −12 and −24 regions of the putative ς54 consensus sequence are underlined. The position of the primer used for primer extension analysis is overlined (broken lines), and the transcriptional start sites, S1 and S2, are shown in uppercase, boldfaced letters. The locations of the Tn5 insertions in Rm11107 (bdhA1::Tn5) and Rm11159 (bdhA2::Tn5) and the Tn5-B20 insertions on pTC344 are indicated by the underlined 9-bp duplicated sequence at the site of the insertion. The bdhA stop codon is marked by an asterisk. The recognition sequences of the restriction enzymes XhoI and EcoRV are underlined.
Nucleotide sequence of bdhA and surrounding region.
A 1,478-bp region, encompassing the entire complementing region as determined by the subcloning and Tn5 mutagenesis experiments was sequenced (Fig. 1). The precise location of bdhA1::Tn5 was determined by sequence analysis of the PCR amplification product obtained with Rm11107 genomic DNA as the template, the IS50-specific forward primer, and a reverse primer complementary to sequence between nucleotide positions 1304 and 1322. A major open reading frame (ORF) comprising 777 bp which was predicted to encode a polypeptide of 258 amino acid residues was detected (Fig. 2). The computed molecular mass of the predicted protein is 27,177 Da with an estimated pI of 6.07. The G+C content of the R. meliloti bdhA gene is 61%, which agrees well with the G+C values observed for other genes from this organism. An ORF immediately downstream of bdhA exhibits homology to xanthine oxidases and dehydrogenases. The deduced partial primary sequence has 43% identity (61% similarity) with the chicken (Gallus gallus) Xdh (70) sequence over an aligned length of 62 amino acid residues. Another ORF upstream of bdhA shows homology to glpK, the glycerol kinase-encoding gene and, based on sequence orientation, is predicted to be transcribed in the opposite direction. The putative partial polypeptide sequence has 61% identity (74% similarity) with the sequence for GlpK of Enterococcus casseliflavus (18) over an aligned length of 61 amino acid residues. glpK mutants of various organisms are unable to utilize glycerol as a sole carbon source (54). Additionally, the ability to utilize glycerol as a sole carbon source is abolished when a Tn5 insertion on plasmid pPA35, located in the glpK region (Fig. 1), is recombined into the Rm1021 genome to give strain Rm11234.
A GenBank BLASTP search (2) with the deduced primary sequence of R. meliloti Bdh revealed similarity to many members of the SCAD superfamily. The highest score was obtained with a putative oxidoreductase in the PEPT-KATB region of the Bacillus subtilis genome, designated YxjF, with 40% identity (62% similarity) in a 259-amino-acid residue overlap. On the basis of sequence similarity alone, yxjF has been determined to encode gluconate 5-dehydrogenase (51). Our analysis shows that YxjF has only 33% identity (52% similarity) in a 266-residue overlap with gluconate 5-dehydrogenase from Gluconobacter oxydans (47), compared to 40% identity (62% similarity) in a 259-amino-acid overlap with R. meliloti BdhA. Based on similarity scores with bdhA, we propose that yxjF may encode a 3-hydroxybutyrate dehydrogenase.
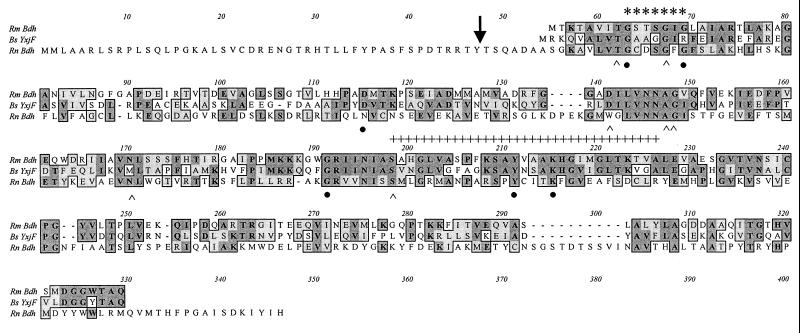
R. meliloti BdhA exhibited only 24% identity (43% similarity) in a 347-amino-acid overlap with the Rat Bdh sequence (Fig. 3). However, Clustal W alignment demonstrated that except for the first 47 amino acids that are involved in targeting the protein to its mitochondrial location, the Rat Bdh and the R. meliloti BdhA do show a certain degree of alignment along most of the length of the polypeptide, particularly in regions found to be highly conserved among members of SCAD (Fig. 3). The R. meliloti BdhA and the B. subtilis YxjF do not contain the C-terminal domain found in the mammalian Bdh which has been shown to be involved in binding phosphatidylcholine and which is essential for enzyme activity (1, 38, 52). This is consistent with the observation that the bacterial Bdh does not demonstrate a requirement for phosphatidylcholine for activity.
FIG. 3.
Alignment of the deduced primary sequences of R. meliloti BdhA, the putative oxidoreductase, YxjF from B. subtilis (51), and Bdh from Rattus norvegicus (20). The position of cleavage of the mitochondrial targeting sequence of the Rat Bdh is marked by an arrow. The amino acid residues involved in coenzyme binding are indicated by asterisks. The amino acid residues corresponding to the proposed consensus for SCAD are indicated by a broken hatched overline. The six strictly conserved residues are indicated by solid circles below the residues. The seven other conserved residues common to most members of the SCAD superfamily are highlighted by carets under the residues. Alignment was performed by using the ClustalW program.
Analysis of the sequence immediately upstream of bdhA.
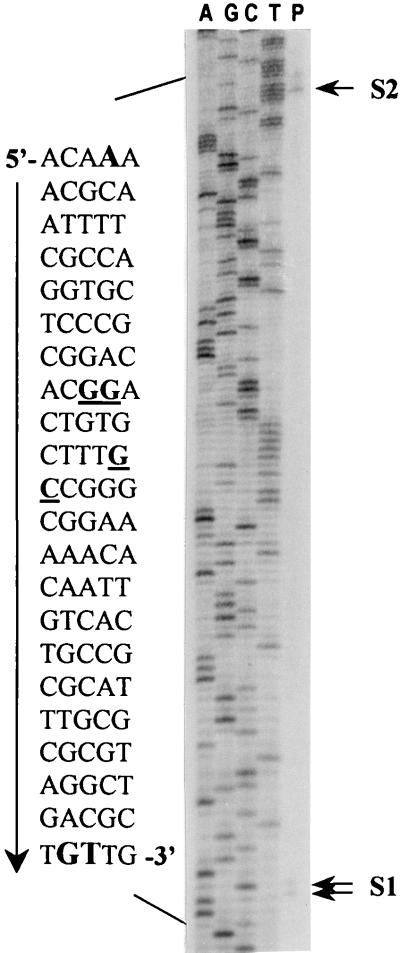
The ability of the 1.2-kb XhoI-SalI fragment to complement for HB utilization indicated that the entire ORF is present within this region. A putative ATG start codon was assigned on the basis of locations of insertions that disrupt the BdhA phenotype. Moreover, we located two possible ribosome binding site sequences 10 and 15 nucleotides preceding the assigned ATG start codon (Fig. 2). Transcription start site determination by primer extension analysis identified two transcripts that correspond to positions S1 and S2 (Fig. 4). Assuming that the bands are not a result of premature termination of the extension reaction, S1 and S2 are 46 to 47 and 148 bases upstream of the putative ATG start codon, respectively (Fig. 2 and 4). Examination of the sequence around S1 and S2 did not reveal the presence of any known consensus promoter sequences. We did, however, identify a ς54 consensus sequence (80) within the stretch of sequence between S1 and S2 (Fig. 2 and 4). To determine whether ς54 (encoded by the rpoN gene) is required for HB utilization, we compared the growth on M9-HB of an rpoN mutant, strain Rm5422, with that of the wild-type strain Rm1021. The growth of both these strains was comparable, indicating that ς54 is not required for HB utilization in free-living R. meliloti.
FIG. 4.
Determination of the transcription start site(s) of bdhA by primer extension analysis. The products of the sequencing reactions, using the same primer, are shown on the left of the extension product (P). The sequence surrounding the extension products is shown on the left of the ladder. The putative ς54 binding consensus sequence is underlined. The positions of the observed extension products are marked by arrows labeled S1 and S2 and the corresponding nucleotides are boldfaced and capitalized in the adjoining sequence ladder.
BdhA is a member of the SCAD superfamily.
As shown in Fig. 3, the deduced BdhA primary sequence contains the PROSITE consensus pattern for SCADs, [LIVSPADNK]-x (12)-Y-[PSTAGNCV]-[STAGNQCIVM]-[STAGC]-K-{PC}-[SAGFR]-[LIVMSTAGD]-x(2)-[LIVMFYW]-x(3)-[LIVMFY WGAPTHQ]-[GSACQRHM] (5). A general feature of SCAD that is also exhibited by R. meliloti BdhA is an N-terminally located glycine-rich GXXXGXG signature sequence for coenzyme binding (Fig. 3). According to Persson et al. (66), there are six strictly conserved residues in SCADs. These include three glycine residues at positions 14, 19, and 132 and the three polar residues D-64, Y-152, and K-156 (numbers assigned in accordance with the Drosophila melanogaster alcohol dehydrogenase); the R. meliloti BdhA sequence exhibits all six of them (Fig. 3). In addition, there are seven other residues conserved in most but not all members of this family, i.e., T-13, G-17, D-87, A-93, G-94, N-114, and S-139 (numbers assigned in accordance with the D. melanogaster alcohol dehydrogenase) (66); these seven other residues are present in the R. meliloti BdhA sequence (Fig. 3). The conserved polar residues S-139, Y-152, and K-156, predicted to be of functional importance (66), are also among the residues contained in the R. meliloti BdhA sequence (Fig. 3). The R. meliloti BdhA sequence, however, does not display the active-site S-T motif found in most but not all SCAD family members. The S-T pattern refers to the presence of a serine or threonine residue just after the tyrosine and/or before the lysine residue. Where there are exceptions to the S-T pattern, distant serine or threonine residues have been suggested to compensate (50), and this may also be the case for R. meliloti BdhA.
The bdhA transcript is multicistronic.
Our observations of the nucleotide sequence indicated the probability of xdhA being transcribed from the same promoter(s) as bdhA. To verify whether bdhA and xdhA are organized as an operon and to determine whether the bdhA1::Tn5 insertion in Rm11107 exerts a polar effect on xdhA expression, we assayed for xanthine oxidase and xanthine dehydrogenase activities in cell extracts of Rm11107. Since Tn5 insertions do not always generate polar mutations (8, 21), a strain in which the bdhA gene was disrupted by a ΩSmSp interposon cassette (68) was constructed. The interposon has strong transcriptional and translational stop signals and therefore invariably generates polar mutations (32). The interposon insertion was constructed by subcloning the 5.5-kb PstI fragment containing a single EcoRV site (within the bdhA coding sequence) to pUC19, resulting in plasmid pPA56. The blunt-ended 2.0-kb interposon was then ligated to EcoRV-digested pPA56 to give pPA59. The resulting 7.5-kb PstI fragment in pPA59 was cloned into the IncP vector pSP329, resulting in pPA60, which when introduced into Rm11107 was unable to complement for HB utilization. The insertion, bdhA3::ΩSmSp, was recombined into the Rm5000 genome to give strain Rm11249 and then transduced into the Rm1021 background, which resulted in strain Rm11262, which was unable to utilize HB as a sole carbon source.
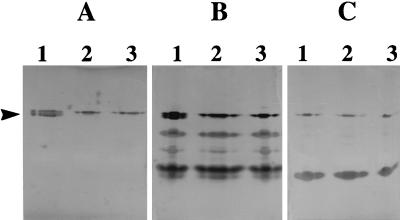
Cell extracts of Rm1021, Rm11107, and Rm11262 were separated by native polyacrylamide gel electrophoresis and stained for xanthine oxidase, xanthine dehydrogenase, and aldehyde oxidase activities. Cell extracts of the bdhA mutants and the wild type revealed the presence of the unidentified dehydrogenases and aldehyde oxidase bands of equal intensity in Fig. 5. In gels stained for xanthine oxidase (Fig. 5A) and xanthine dehydrogenase (Fig. 5B) activities, the lower band of the doublet present in the wild-type cell extracts (Fig. 5A and B, lanes 1) is absent in cell extracts of both of the bdhA mutant strains (Fig. 5A and B, lanes 2 and 3). This confirms that xdhA lacks an independent promoter that is active under the culture conditions examined and that xdhA is probably transcribed from a bdhA promoter(s).
FIG. 5.
Staining for xanthine oxidase (A), xanthine dehydrogenase (B), and aldehyde oxidase (C) activities on native polyacrylamide gels. Lane 1, Rm1021 (wild type); lane 2, Rm11107 (bdhA1::Tn5); lane 3, Rm11262 (bdhA3::ΩSmSp). Cell extracts, equivalent to 85 μg of total protein, were loaded in each lane.
PHB accumulation and bdhA gene expression.
To determine whether a genetic lesion in the PHB degradation pathway results in excessive PHB accumulation, the total PHB content of YMB-grown cultures of the bdhA mutant strain Rm11107 and the wild-type strain Rm1021 was determined. The proportion of the total cellular dry weight contributed by PHB was not significantly different between the wild-type and the bdhA mutant under the growth conditions tested (data not presented).
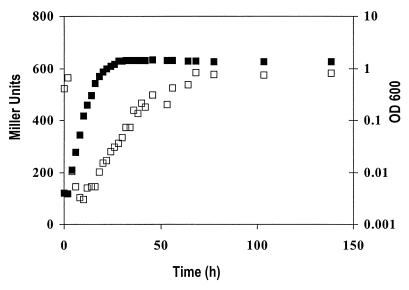
The insertion on plasmid pTC344bdhA3-2::Tn5-B20 was homogenotized into Rm8501 (Lac−), resulting in Rm11191. When grown on complex TY medium, the specific β-galactosidase activity over time was found to be growth phase dependent. Activity was lowest in the lag phase, then increased steadily in log phase, and finally leveled off in stationary phase. Due to the stability of β-galactosidase, whether gene expression in the stationary phase is actually repressed cannot be concluded from this study (Fig. 6).
FIG. 6.
Growth (■) and expression (□) of bdhA-lacZ fusion in strain Rm11191 grown in TY complex medium.
Symbiotic phenotype of the bdhA mutant.
Symbiotic assays with alfalfa host plants showed no significant differences in shoot dry weight per plant inoculated with either the bdhA mutant strain Rm11107 or the wild-type strain Rm1021 (data not presented). The inoculated plants appeared healthy and green, whereas the uninoculated plants were stunted and yellow in appearance. Nodulation kinetic data also did not reveal any differences between the mutant and the wild type either in terms of numbers of nodules induced or the time course of appearance of the nodules (data not presented).
DISCUSSION
We have cloned and sequenced the bdhA gene of R. meliloti, which encodes 3-hydroxybutyrate dehydrogenase. To our knowledge, this is the first example of genetic and molecular characterization of a nonmammalian 3-hydroxybutyrate dehydrogenase-encoding gene. Analysis of the predicted polypeptide indicates that R. meliloti BdhA is a member of the SCAD family of proteins. A survey of Genpept96 (39) identified at least 350 members belonging to the SCAD superfamily of oxidoreductases. Members of this group are 250 to 300 amino acids long and are either dimeric or tetrameric. There is no striking overall similarity between the mammalian and bacterial Bdh primary sequences other than what is typically observed between most members of this family. A detailed examination of these differences may provide a better understanding of the structure-function relationship of these functionally similar proteins that have only a minimal degree of identity at the primary sequence level. The low level of sequence homology between the bacterial and mammalian Bdh proteins suggests that they may be of independent evolutionary origins.
Most of the conserved residues in members of the SCAD superfamily are glycine residues, which are believed to be involved in coenzyme binding and contribute to the maintenance of the tertiary structure by allowing bend formation. Conserved residues other than the glycines are believed to have functional importance, particularly Y-152, K-156, and S-139, which are found at the active site and therefore considered to be strong candidates for residues involved in catalysis (66). Ghosh et al. (36) described the active site as a deep cleft located around the center of the tetramer in 3α.20β-hydroxysteroid dehydrogenase, which is also a member of the SCAD superfamily. Both the bacterial and mammalian Bdh proteins have also been described as homotetramers (60, 63, 78).
The recently completed sequence of the B. subtilis genome (51) shows the bdhA homologue yxjF positioned beside two loci designated yxjE and yxjD, arranged in the order yxjDEF. On the basis of sequence similarity analyses, yxjE and yxjD have been proposed to encode the two subunits of 3-oxoadipate CoA transferase, an enzyme that is able to catalyze the activation of acetoacetate to acetoacetyl-CoA. We also noted that yxjF and yxjE are interrupted by a mere 16 nucleotides, making the existence of an independent promoter in this region highly unlikely. Taken together, these observations suggest the presence of a PHB degradation operon in B. subtilis. In R. meliloti, the only organism in which studies on the genetics of PHB degradation has been addressed, loci involved in PHB degradation appear to be scattered on the genome, with two loci on the chromosome and two on one of the megaplasmids, pRmeSU47b (12).
The R. meliloti megaplasmid pRmeSU47b carries a number of catabolic genes, including those encoding enzymes for utilization of carbon sources such as C4-dicarboxylates, dulcitol, lactose, melibiose, HB, and acetoacetate (12, 13, 17, 86, 88), as well as genes encoding enzymes for phosphate utilization (6). In this report, we have shown that pRmeSU47b also carries glpK, a gene required for utilization of glycerol as a sole carbon source, and xdhA, which in B. subtilis is involved in the utilization of purines as a sole nitrogen source (30). This further reinforces the significant catabolic capacity of megaplasmid pRmeSU47b.
The relevance of the coexistence of xdhA and bdhA in the same operon is not clear. Research on hypoxanthine salvage and hypoxanthine catabolism in Streptomyces spp. (85) and B. subtilis (19) has shown that both of these pathways are subject to nitrogen control. When nitrogen is in excess, catabolism is repressed, whereas when nitrogen is limiting, purine salvage is repressed and catabolism is induced. Furthermore, enhanced Xdh activity in a glnA mutant background has been reported, suggesting the involvement of glutamine synthetase (encoded by the glnA gene)-mediated nitrogen catabolite repression (19). This leads us to consider whether the organization of these two genes, bdhA and xdhA, the products of which have roles to play during carbon and nitrogen starvation, respectively, reflect a physiological relationship between these two different starvation responses.
Although there are several reports demonstrating that Bdh activity is negatively regulated by NAD(P)H, adenosine nucleotides, pyruvate, oxaloacetate, and 2-ketoglutarate (55, 71, 78), regulation of Bdh at the level of gene expression was not examined. The growth phase-associated gene expression may reflect cellular PHB levels, and this will be the subject of further study. The relationship of bdhA gene expression to the pattern of cellular PHB accumulation is a topic that should be addressed, especially considering the proposed key role of Bdh in the control of PHB degradation.
From our PHB assay data, we are able to conclude that a mutation in bdhA has no effect on the ability of R. meliloti to accumulate PHB under the culture conditions tested. For a more complete assessment of the effect of the bdhA mutation on cellular PHB levels, the ability to utilize endogenous PHB stores in a carbon-poor medium needs to be determined.
The nature of the promoters corresponding to transcription start sites S1 and S2 is not apparent, as no familiar promoter consensus sequences were recognized immediately upstream of either of these sites. It will be important to delineate the upstream regions that are required for expression, especially since relatively little is known about promoters of nonsymbiotic genes in R. meliloti (64).
The presence of a ς54 consensus sequence between S1 and S2 is intriguing, especially considering the importance of ς54 in the regulation of symbiotic functions (69, 83). Primer extension analysis with bacteroid RNA may help clarify the involvement, if any, of ς54-dependent expression of bdhA-xdhA. The absence of consensus sequences for binding of any of the known ς54-associated transcriptional activators raises the possibility that as-yet-undiscovered transcriptional factors are involved in regulation of bdhA-xdhA gene expression.
Since the bdhA mutant shows no symbiotic defects, we conclude that Bdh-dependent PHB degradation is not essential for bacteroid development or function. The behavior of the mutant in the soil and rhizosphere environments remains to be investigated. Consideration of the biology of the free-living organism will potentially be as enlightening as the symbiotic studies.
ACKNOWLEDGMENTS
We are grateful for strains received from Mary Berlyn (E. coli Genetic Stock Center) and Turlough M. Finan. We thank Sylvie Bardin and Kirk Bartholomew for advice on the RNA isolation and primer extension analysis.
This work was supported by operating grants to T.C.C. from the Natural Sciences and Engineering Research Council of Canada and Fonds pour la formation de chercheurs et l’aide à la recherche (Québec).
REFERENCES
- 1.Adami P, Duncan T M, McIntyre J O, Carter C E, Fu C, Melin M, Latruffe N, Fleischer S. Monoclonal antibodies for the structure-function studies of (R)-3-hydroxybutyrate dehydrogenase, a lipid-dependent membrane bound enzyme. Biochem J. 1993;292:863–872. doi: 10.1042/bj2920863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schäffer A A, Zhang Z, Miller W, Lipman D. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson A J, Dawes E A. Occurrence, metabolism, metabolic role, and industrial uses of bacterial polyhydroxyalkanoates. Microbiol Rev. 1990;54:450–472. doi: 10.1128/mr.54.4.450-472.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1997. [Google Scholar]
- 5.Bairoch A, Bucher P, Hofman K. The PROSITE database, its status in 1997. Nucleic Acids Res. 1997;25:217–221. doi: 10.1093/nar/25.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bardin S, Dan S, Østerås M, Finan T M. A phosphate transport system is required for symbiotic nitrogen fixation by Rhizobium meliloti. J Bacteriol. 1996;178:4540–4547. doi: 10.1128/jb.178.15.4540-4547.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berg C M, Berg D E. Uses of transposable elements and maps of known insertions. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Vol. 2. Washington, D.C: American Society for Microbiology; 1987. pp. 1071–1109. [Google Scholar]
- 8.Berg D E, Weiss A, Crossland L. Polarity of Tn5 insertion mutations in Escherichia coli. J Bacteriol. 1980;142:439–446. doi: 10.1128/jb.142.2.439-446.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bergersen F J, Peoples M B, Turner G L. A role for poly-β-hydroxybutyrate in bacteroids of soybean root nodules. Proc R Soc Lond Ser B. 1991;245:59–64. [Google Scholar]
- 10.Bergmeyer H U, Gawehn K, Klotzsch H, Krebs H A, Williamson D H. Purification and properties of crystalline 3-hydroxybutyrate dehydrogenase from Rhodopseudomonas spheroides. Biochem J. 1967;102:423–431. doi: 10.1042/bj1020423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cevallos M A, Encarnación S, Leija A, Mora Y, Mora J. Genetic and physiological characterization of a Rhizobium etli mutant strain unable to synthesize poly-β-hydroxybutyrate. J Bacteriol. 1996;178:1646–1654. doi: 10.1128/jb.178.6.1646-1654.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Charles T C, Cai G Q, Aneja P. Megaplasmid and chromosomal loci for the PHB degradation pathway in Rhizobium (Sinorhizobium) meliloti. Genetics. 1997;146:1211–1220. doi: 10.1093/genetics/146.4.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Charles T C, Finan T M. Analysis of a 1600-kilobase Rhizobium meliloti megaplasmid using defined deletions generated in vivo. Genetics. 1991;127:5–20. doi: 10.1093/genetics/127.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Charles T C, Finan T M. Genetic map of Rhizobium meliloti megaplasmid pRmeSU47b. J Bacteriol. 1990;172:2469–2476. doi: 10.1128/jb.172.5.2469-2476.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Charles T C, Nester E W. A chromosomally encoded two-component sensory transduction system is required for virulence of Agrobacterium tumefaciens. J Bacteriol. 1993;175:6614–6625. doi: 10.1128/jb.175.20.6614-6625.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Charles T C, Newcomb W, Finan T M. ndvF, a novel locus located on megaplasmid pRmeSU47b (pEXO) of Rhizobium meliloti, is required for normal nodule development. J Bacteriol. 1991;173:3981–3992. doi: 10.1128/jb.173.13.3981-3992.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Charles T C, Singh R S, Finan T M. Lactose utilization and enzymes encoded by megaplasmids in Rhizobium meliloti SU47: implications for population studies. J Gen Microbiol. 1990;136:2497–2502. [Google Scholar]
- 18.Charrier V, Buckley E, Parsonage D, Galinier A, Darbon E, Jaquinod M, Forest E, Deutscher J, Clairborne A. Cloning and sequencing of two enterococcal glpK genes and regulation of the encoded glycerol kinases by phosphoenolpyruvate-dependent phosphotransferase system-catalysed phosphorylation of a single histidyl residue. J Biol Chem. 1997;272:14166–14174. doi: 10.1074/jbc.272.22.14166. [DOI] [PubMed] [Google Scholar]
- 19.Christiansen L C, Schou S, Nygaard P, Saxild H H. Xanthine metabolism in Bacillus subtilis: characterization of the xpt-pbuX operon and evidence for purine- and nitrogen-controlled expression of genes involved in xanthine salvage and catabolism. J Bacteriol. 1997;179:2540–2550. doi: 10.1128/jb.179.8.2540-2550.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Churchill P, Hempel J, Romovacek H. Primary structure of rat liver D-beta-hydroxybutyrate dehydrogenase from cDNA and protein analyses: a short-chain alcohol dehydrogenase. Biochemistry. 1992;31:3793–3799. doi: 10.1021/bi00130a009. [DOI] [PubMed] [Google Scholar]
- 21.Clark A, Satin L, Chu C C. Transcription of the Escherichia coli recE gene from a promoter in Tn5 and IS50. J Bacteriol. 1994;176:7024–7031. doi: 10.1128/jb.176.22.7024-7031.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Delafield F P, Cooksey K E, Doudoroff M. β-Hydroxybutyric dehydrogenase and dimer hydrolase of Pseudomonas lemoignei. J Biol Chem. 1965;240:4023–4028. [PubMed] [Google Scholar]
- 23.De Vos G F, Walker G C, Signer E R. Genetic manipulations in Rhizobium meliloti utilizing two new transposon Tn5 derivatives. Mol Gen Genet. 1986;204:485–491. doi: 10.1007/BF00331029. [DOI] [PubMed] [Google Scholar]
- 24.Driscoll B T, Finan T M. NADP+-dependent malic enzyme of Rhizobium meliloti. J Bacteriol. 1996;178:2224–2231. doi: 10.1128/jb.178.8.2224-2231.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feinstein R N, Lindahl R. Detection of oxidases on polyacrylamide gels. Anal Biochem. 1973;56:353. doi: 10.1016/0003-2697(73)90201-7. [DOI] [PubMed] [Google Scholar]
- 26.Finan T M, Hartwieg E K, LeMieux K, Bergman K, Walker G C, Signer E R. General transduction in Rhizobium meliloti. J Bacteriol. 1984;159:120–124. doi: 10.1128/jb.159.1.120-124.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Finan T M, Kunkel B, DeVos G F, Signer E R. Second symbiotic megaplasmid in Rhizobium meliloti carrying exopolysaccharide and thiamine synthesis genes. J Bacteriol. 1986;167:66–72. doi: 10.1128/jb.167.1.66-72.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Finan T M, McWhinnie E, Driscoll B, Watson R J. Complex symbiotic phenotypes result from gluconeogenic mutations in Rhizobium meliloti. Mol Plant Microbe Interact. 1991;4:386–392. [Google Scholar]
- 29.Finan T M, Oresnik I, Bottacin A. Mutants of Rhizobium meliloti defective in succinate metabolism. J Bacteriol. 1988;170:3396–3403. doi: 10.1128/jb.170.8.3396-3403.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29a.Finan, T. M. Unpublished results.
- 30.Fisher S H. Utilization of amino acids and other nitrogen-containing compounds. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. Washington D.C: American Society for Microbiology; 1993. pp. 221–228. [Google Scholar]
- 31.Fottrell P F, O’Hora A. Multiple forms of d(−)3-hydroxybutyrate dehydrogenase in Rhizobium. J Gen Microbiol. 1969;57:287–292. doi: 10.1099/00221287-57-3-287. [DOI] [PubMed] [Google Scholar]
- 32.Frey J, Krisch H M. Ω mutagenesis in gram-negative bacteria: a selectable interposon which is strongly polar in a wide range of bacterial species. Gene. 1985;36:143–150. doi: 10.1016/0378-1119(85)90078-2. [DOI] [PubMed] [Google Scholar]
- 33.Friedman A M, Long S R, Brown S E, Buikema W J, Ausubel F M. Construction of a broad host range cloning vector and its use in the genetic analysis of Rhizobium mutants. Gene. 1982;18:289–296. doi: 10.1016/0378-1119(82)90167-6. [DOI] [PubMed] [Google Scholar]
- 34.Gary J X, Djordjevic M A, Rolfe B G. Two genes that regulate exopolysaccharide production in Rhizobium sp. strain NGR234: DNA sequences and resultant phenotypes. J Bacteriol. 1990;172:193–203. doi: 10.1128/jb.172.1.193-203.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gerson T, Patel J J, Wong M N. The effects of age, darkness and nitrate on poly-β-hydroxybutyrate levels and nitrogen-fixing ability of Rhizobium in Lupinus angustifolius. Physiol Plant. 1978;42:420–424. [Google Scholar]
- 36.Ghosh D, Weeks C M, Grochulski P, Daux W L, Erman M, Rimsay R L, Orr J C. Three-dimensional structure of holo 3α.20β-hydroxysteroid dehydrogenase: a member of a short-chain dehydrogenase family. Proc Natl Acad Sci USA. 1991;88:10064–10068. doi: 10.1073/pnas.88.22.10064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Glazebrook J, Walker G C. Genetic techniques in Rhizobium meliloti. Methods Enzymol. 1991;204:398–418. doi: 10.1016/0076-6879(91)04021-f. [DOI] [PubMed] [Google Scholar]
- 38.Green D, Marks A R, Fleischer S, McIntyre J O. Wild type and mutant human heart (R)-3-hydroxybutyrate dehydrogenase expressed in insect cells. Biochemistry. 1996;35:8158–8165. doi: 10.1021/bi952807n. [DOI] [PubMed] [Google Scholar]
- 39.Grundy W N, Bailey T L, Elkan C P, Baker M E. Hidden Markov model analysis of motifs in steroid dehydrogenases and their homologs. Biochem Biophys Res Commun. 1997;231:760–766. doi: 10.1006/bbrc.1997.6193. [DOI] [PubMed] [Google Scholar]
- 40.Hirsch A M, Bang M, Ausubel F M. Ultrastructural analysis of ineffective alfalfa nodules formed by nif::Tn5 mutants of Rhizobium meliloti. J Bacteriol. 1983;155:367–380. doi: 10.1128/jb.155.1.367-380.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hirsch A M, Long S R, Bang M, Haskins N, Ausubel F M. Structural studies of alfalfa roots infected with nodulation mutants of Rhizobium meliloti. J Bacteriol. 1982;151:411–419. doi: 10.1128/jb.151.1.411-419.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huisman G W, Wonink E, Meima R, Kazemier B, Terpstra P, Witholt B. Metabolism of poly(3-hydroxyalkanoates) by Pseudomonas oleovorans: identification and sequences of genes and function of the encoded proteins in the synthesis and degradation of PHA. J Biol Chem. 1991;266:2191–2198. [PubMed] [Google Scholar]
- 43.Jendrossek D, Frisse A, Behrends A, Andermann M, Kratzin H D, Stanislawski T, Schlegel H G. Biochemical and molecular characterization of the Pseudomonas lemoignei polyhydroxyalkanoate depolymerase system. J Bacteriol. 1995;177:596–607. doi: 10.1128/jb.177.3.596-607.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jenkins L S, Nunn W D. Genetic and molecular characterization of the genes involved in short-chain fatty acid degradation in Escherichia coli: the ato system. J Bacteriol. 1987;169:42–52. doi: 10.1128/jb.169.1.42-52.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jensen H L. Nitrogen fixation in leguminous plants. I. General characters of root-nodule bacteria isolated from species of Medicago and Trifolium in Australia. Proc Linn Soc N S W. 1942;66:98–108. [Google Scholar]
- 46.Jurtshuk P, Manning S, Barrera C R. Isolation and purification of the d(−)-β-hydroxybutyric dehydrogenase of Azotobacter vinelandii. Can J Microbiol. 1968;14:775–783. doi: 10.1139/m68-129. [DOI] [PubMed] [Google Scholar]
- 47.Klasen R, Bringer-Meyer S, Sahm H. Biochemical characterization and sequence analysis of the gluconate:NADP 5-oxidoreductase gene from Gluconobacter oxydans. J Bacteriol. 1995;177:2637–2643. doi: 10.1128/jb.177.10.2637-2643.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Klucas R V. Studies on soybean nodule senescence. Plant Physiol. 1975;54:612–616. doi: 10.1104/pp.54.4.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kovar J, Matyskova I, Motyska L. Kinetics of d-3-hydroxybutyrate dehydrogenase from Paracoccus denitrificans. Biochim Biophys Acta. 1986;871:302–309. [Google Scholar]
- 50.Krozowski Z. The short-chain alcohol dehydrogenase superfamily—variations on a common theme. J Steroid Biochem Mol Biol. 1994;51:125–130. doi: 10.1016/0960-0760(94)90084-1. [DOI] [PubMed] [Google Scholar]
- 51.Kunst F, et al. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 52.Langston H P, Jones L, Churchill S, Churchill P F. Purification and characterization of a (R)-3-hydroxybutyrate dehydrogenase deletion mutant—evidence for C-terminal involvement in enzyme activation by lecithin. Arch Biochem Biophys. 1996;327:45–52. doi: 10.1006/abbi.1996.0091. [DOI] [PubMed] [Google Scholar]
- 53.Law J H, Slepecky R A. Assay of poly-β-hydroxybutyric acid. J Bacteriol. 1961;82:33–36. doi: 10.1128/jb.82.1.33-36.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lin E C C. Glycerol dissimilation and its regulation in bacteria. Annu Rev Microbiol. 1976;30:535–578. doi: 10.1146/annurev.mi.30.100176.002535. [DOI] [PubMed] [Google Scholar]
- 55.Lötter L H, Dubery I A. Metabolic regulation of β-hydroxybutyrate dehydrogenase in Acinetobacter calcoaceticus var lwoffi. Water S A (Pretoria) 1989;15:65–70. [Google Scholar]
- 56.Manchak J, Page W. Control of polyhydroxyalkanoate synthesis in Azotobacter vinelandii strain UWD. Microbiology. 1994;140:953–963. [Google Scholar]
- 57.Marck C. DNA Strider: a “C” program for the fast analysis of DNA and protein sequences on the Apple Macintosh family of computers. Nucleic Acids Res. 1988;16:1829–1836. doi: 10.1093/nar/16.5.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McDermott T R, Griffith S M, Vance C P, Graham P H. Carbon metabolism in Bradyrhizobium japonicum bacteroids. FEMS Microbiol Rev. 1989;63:327–340. [Google Scholar]
- 59.McIntyre J O, Bock H G, Fleischer S. The orientation of D-beta-hydroxybutyrate dehydrogenase in the mitochondrial inner membrane. Biochim Biophys Acta. 1978;513:255–267. doi: 10.1016/0005-2736(78)90178-5. [DOI] [PubMed] [Google Scholar]
- 60.McIntyre J O, Churchill P, Maurer A, Berenski C J, Jung C Y, Fleischer S. Target size of D-beta-hydroxybutyrate dehydrogenase. Functional and structural molecular weight based on radiation inactivation. J Biol Chem. 1983;258:953–959. [PubMed] [Google Scholar]
- 61.Meade H M, Long S R, Ruvkun G B, Brown S E, Ausubel F M. Physical and genetic characterization of symbiotic and auxotrophic mutants of Rhizobium meliloti induced by transposon mutagenesis. J Bacteriol. 1982;149:114–122. doi: 10.1128/jb.149.1.114-122.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 63.Nakada T, Fukui T, Saito T, Miki K, Oji C, Matsuda S, Ushijima A, Tomita K. Purification and properties of d-beta-hydroxybutyrate dehydrogenase from Zoogloea ramigera I-16-M. J Biochem. 1981;89:625–635. doi: 10.1093/oxfordjournals.jbchem.a133239. [DOI] [PubMed] [Google Scholar]
- 64.Østerås M, Driscoll B T, Finan T M. Molecular and expression analysis of the Rhizobium meliloti phosphoenolpyruvate carboxykinase (pckA) gene. J Bacteriol. 1995;177:1452–1460. doi: 10.1128/jb.177.6.1452-1460.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Paau A S, Bloch C B, Brill W J. Developmental fate of Rhizobium meliloti bacteroids in alfalfa nodules. J Bacteriol. 1980;143:1480–1490. doi: 10.1128/jb.143.3.1480-1490.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Persson B, Krook M, Jörnvall H. Characteristics of short-chain alcohol dehydrogenases and related enzymes. Eur J Biochem. 1991;200:537–543. doi: 10.1111/j.1432-1033.1991.tb16215.x. [DOI] [PubMed] [Google Scholar]
- 66a.Porter, S. Unpublished results.
- 67.Povolo S, Tombolini R, Morea A, Anderson A J, Casella S, Nuti M P. Isolation and characterization of mutants of Rhizobium meliloti unable to synthesize poly-β-hydroxybutyrate (PHB) Can J Microbiol. 1994;40:823–829. [Google Scholar]
- 68.Prentki P, Krisch H M. In vitro insertional mutagenesis with a selectable DNA fragment. Gene. 1984;29:303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- 69.Ronson C W, Nixon B T, Albright L M, Ausubel F M. Rhizobium meliloti ntrA (rpoN) gene is required for diverse metabolic functions. J Bacteriol. 1987;169:2424–2431. doi: 10.1128/jb.169.6.2424-2431.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sato A, Nishino T, Noda K, Amaya Y, Nishino T. The structure of chicken liver xanthine dehydrogenase: cDNA cloning and the domain structure. J Biol Chem. 1995;270:2818–2826. doi: 10.1074/jbc.270.6.2818. [DOI] [PubMed] [Google Scholar]
- 71.Senior P J, Dawes E A. The regulation of poly-β-hydroxybutyrate metabolism in Azotobacter beijerinckii. Biochem J. 1973;134:225–248. doi: 10.1042/bj1340225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shaw C R, Prasad R. Starch gel electrophoresis of enzymes: a compilation of recipes. Biochem Genet. 1970;4:297. doi: 10.1007/BF00485780. [DOI] [PubMed] [Google Scholar]
- 73.Simon R, Quandt J, Klipp W. New derivatives of transposon Tn5 suitable for mobilization of replicons, generation of operon fusions, and induction of genes in Gram-negative bacteria. Gene. 1989;80:161–169. doi: 10.1016/0378-1119(89)90262-x. [DOI] [PubMed] [Google Scholar]
- 74.Spratt S K, Ginsburgh C L, Nunn W D. Isolation and genetic characterization of Escherichia coli mutants defective in propionate metabolism. J Bacteriol. 1981;146:1166–1169. doi: 10.1128/jb.146.3.1166-1169.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stachel S E, An G, Flores C, Nester E W. A Tn3 lacZ transposon for the random generation of beta-galactosidase gene fusions: application to the analysis of gene expression in Agrobacterium. EMBO J. 1985;4:891–898. doi: 10.1002/j.1460-2075.1985.tb03715.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sutton W D. Nodule development and senescence. In: Broughton W J, editor. Nitrogen fixation. 3. Legumes. Oxford, United Kingdom: Oxford University Press; 1983. pp. 144–212. [Google Scholar]
- 77.Tal S, Okon Y. Production of the reserve material poly-β-hydroxybutyrate and its function in Azospirillum brasilense Cd. Can J Microbiol. 1985;31:608–613. [Google Scholar]
- 78.Tal S, Smirnoff P, Okon Y. Purification and characterization of d-(−)-β-hydroxybutyrate dehydrogenase from Azospirillum brasilense Cd. J Gen Microbiol. 1990;136:645–649. [Google Scholar]
- 79.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Thöny B, Hennecke H. The −24/−12 promoter comes of age. FEMS Microbiol Rev. 1989;63:341–357. doi: 10.1016/0168-6445(89)90028-4. [DOI] [PubMed] [Google Scholar]
- 81.Tombolini R, Nuti M P. Poly (β-hydroxyalkanoate) biosynthesis and accumulation by different Rhizobium species. FEMS Microbiol Lett. 1989;60:299–304. [Google Scholar]
- 82.Toro N. Nodulation competitiveness in the Rhizobium-legume symbiosis. World J Microbiol Biotechnol. 1996;12:157–162. doi: 10.1007/BF00364680. [DOI] [PubMed] [Google Scholar]
- 83.van Slooten J C, Cervantes E, Broughton W J, Wong C H, Stanley J. Sequence and analysis of the rpoN sigma factor gene of Rhizobium sp. strain NGR234, a primary coregulator of symbiosis. J Bacteriol. 1990;172:5563–5574. doi: 10.1128/jb.172.10.5563-5574.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Vlassak K M, Vanderleyden J. Factors influencing nodule occupancy by inoculant rhizobia. Crit Rev Plant Sci. 1997;16:163–229. [Google Scholar]
- 85.Watanabe Y, Ohe T, Tsujisaka Y. Changes in the metabolic pathways of hypoxanthine in Streptomyces. J Gen Appl Microbiol. 1976;22:13–23. [Google Scholar]
- 86.Watson R J, Chan Y-K, Wheatcroft R, Yang A-F, Han S. Rhizobium meliloti genes required for C4-dicarboxylate transport and symbiotic nitrogen fixation are located on a megaplasmid. J Bacteriol. 1988;170:927–934. doi: 10.1128/jb.170.2.927-934.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 88.Yarosh O K, Charles T C, Finan T M. Analysis of C4-dicarboxylate transport genes in Rhizobium meliloti. Mol Microbiol. 1989;3:813–823. doi: 10.1111/j.1365-2958.1989.tb00230.x. [DOI] [PubMed] [Google Scholar]