Abstract
Rho family small GTPases are universal signaling switches in the control of cell polarity in eukaryotic cells. Their polar distribution to the cell cortex is critical for the execution of their functions, yet the mechanism for this distribution is poorly understood. Using a yeast two-hybrid method, we identified RIP1 (ROP interactive partner 1), which belongs to a family of five members of novel proteins that share a C-terminal region that interacts with ROP. When expressed in Arabidopsis pollen, green fluorescence protein GFP-tagged RIP1 was localized to the nucleus of mature pollen. When pollen grains were hydrated in germination medium, GFP-RIP1 switched from the nucleus to the cell cortex at the future pollen germination site and was maintained in the apical cortex of germinating pollen and growing pollen tubes. RIP1 was found to interact with ROP1 in pollen tubes, and the cortical RIP1 localization was influenced by the activity of ROP1. Overexpression of RIP1 induced growth depolarization in pollen tubes, a phenotype similar to that induced by ROP1 overexpression. Interestingly, RIP1 overexpression enhanced GFP-ROP1 recruitment to the plasma membrane (PM) of pollen tubes. Based on these observations, we hypothesize that RIP1 is involved in the positive feedback regulation of ROP1 localization to the PM, leading to the establishment of a polar site for pollen germination and pollen tube growth.
INTRODUCTION
Tip growth is a form of polar growth by which cell expansion is strictly restricted to the apex of the cell (Feijo et al., 2004; Hepler et al., 2001; Yang, 1998,2008). It is proposed that tip growth is controlled by a self-organizing system, termed LENS (for localization enhancing network, self-sustaining) (Cole and Fowler, 2006). Components of LENS are often localized to the apex of tip-growing cells, such as the actin cytoskeleton, calcium gradients, various signaling molecules, and vesicle trafficking machineries (Cole and Fowler, 2006; Gu et al., 2004; Hepler et al., 2001; Kost, 2008; Robinson and Messerli, 2002; Samaj et al., 2006; Staiger, 2000; Yang, 1998; Yang and Fu, 2007). Many signaling molecules that are important for the function of LENS have been identified as residents of these tip-localized signaling proteins include ROP GTPase and its associated proteins (Cheung et al., 2003; Fu et al., 2001; Gu et al., 2003,2005,2006; Jones et al., 2002; Klahre and Kost, 2006; Kost et al., 1999; Molendijk et al., 2001; Wu et al., 2001; Zhang and McCormick, 2007), protein kinases, such as calmodulin-domain like protein kinases (Yoon et al., 2006), phospholipase (Dowd et al., 2006; Helling et al., 2006), and phosphoinositide kinases (Kusano et al., 2008; Stenzel et al., 2008). How the polar localization of these signaling components is initiated and maintained remains mysterious.
The Rho family of small GTPases plays an important role in polarity establishment and polar cell growth/movement in various eukaryotic cells (Etienne-Manneville and Hall, 2002; Hall, 1998; Schmitz et al., 2000). In plants, ROP (Rho-related GTPases), a unique subfamily of Rho small GTPases, is a central player controlling tip establishment and tip growth both in root hairs and pollen tubes (Gu et al., 2004; Molendijk et al., 2004; Yang, 1998,2002,2008). Arabidopsis ROP1 controls the dynamics of the apical actin cytoskeleton and the formation of the tip-focused calcium gradient (Chen et al., 2003; Fu et al., 2001; Gu et al., 2005; Kost et al., 1999; Li et al., 1999; Lin and Yang, 1997). During pollen tube growth, ROP is preferentially localized to the apical PM of pollen tubes (Kost et al., 1999; Li et al., 1999; Lin and Yang, 1997). This polar distribution of ROPs is thought to be important for the maintenance of polarity and rapid growth of pollen tubes. Delocalization of ROP1 induced by overexpression of ROP1a pollen-specific ROP or CA-ROP1 (constitutive active ROP1) in Arabidopsis pollen tubes caused depolarization of pollen tube growth (Cheung et al., 2003; Gu et al., 2003; Kost et al., 1999; Li et al., 1999). The regulation of the apical PM localization, activation, and inactivation of ROP is an essential step for the execution of ROP functions.
In ArabidopsisROP regulators have been identified and studied to elucidate the complexities of ROP regulation. RopGAPs (ROP GTPase activating proteins) are presumed to inactivate ROP at the PM by promoting GTPase activity (Baxter-Burrell et al., 2002; Klahre and Kost, 2006; Wu et al., 2000). RopGAP1 overexpression suppressed growth depolarization induced by ROP1 overexpression (Fu et al., 2001; Hwang et al., 2005). RopGDIs (ROP guanine nucleotide disassociation inhibitors) are also important for maintaining growth polarity presumably by sequestering ROPs in the cytosol (Bischoff et al., 2000; Carol et al., 2005; Fu et al., 2001; Hwang et al., 2005; Klahre and Kost, 2006). A family of novel plant-specific RopGEFs (guanine nucleotide exchange factor) for ROP has been identified in Arabidopsis and has also been shown to be localized to the apical PM for the regulation of polar growth of pollen tubes (Berken et al., 2005; Gu et al., 2006). RopGEF12 has been shown to physically and functionally interact with AtPRK2a and this interaction seems to affect RopGEF12 localization to the apical PM (Zhang and McCormick, 2007). In spite of these important advances in our understanding of ROP1 activity regulation at the tip of pollen tubes, how ROP1 is localized to the apical PM is unknown. Activated RopGEFs could recruit ROP1 to the apical PM. Alternatively, ROP1 could be recruited to the apical PM in a manner independent of RopGEFs.
In this study, we have identified a class of novel ROP-binding proteins, termed RIPs (ROP interactive partners), which are localized to the apical region of the pollen tube cortex. Here, we used RIP1 as a representative of pollen-expressed RIPs to study the function of RIPs in pollen tube tip growth. RIP1 was also recently identified as ICR1 (interactor of constitutive active ROPs 1), which is implicated in polarized cell growth in pavement cells and root apical meristem maintenance (Lavy et al., 2007). RIP1/ICR1 was found to accumulate in a limited region of the pollen cell cortex prior to pollen germination, which marked the future site of pollen germination. RIP1/ICR1 appeared to promote ROP1 localization to the PM, while ROP1 activity was required for RIP1/ICR1 localization to the cell apex. We propose that RIP1/ICR1 may act as a key component in a positive feedback loop of ROP activation and ROP localization to the PM, which leads to the establishment of a polar site for pollen germination and tip growth.
RESULTS
Identification of RIPs Using Yeast Two-Hybrid
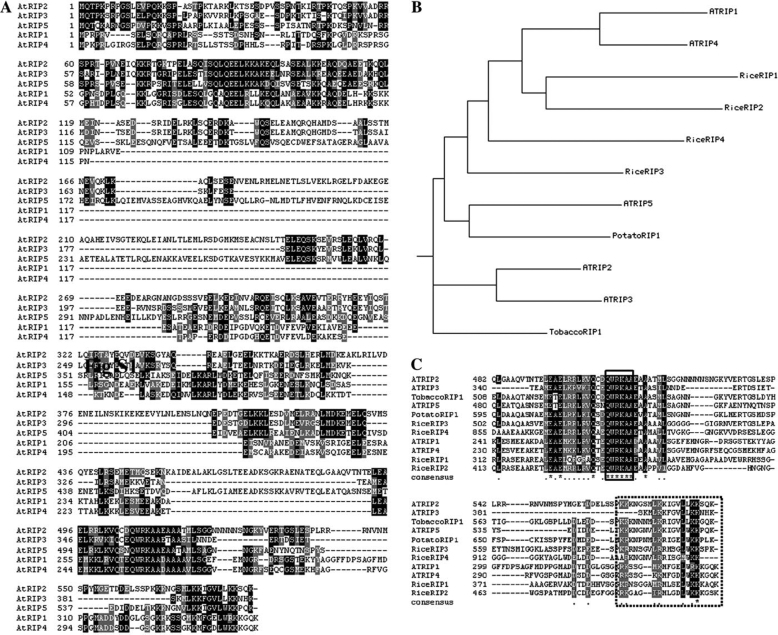
We used a yeast two-hybrid screen to identify proteins interacting with GDP-bound dominant or negative ROP1 mutant (DN-ROP1). In this screen, we recovered nine partial cDNAs encoding five proteins corresponding to genes At1g17140, At1g78430, At2g37080, At3g53350, and At5g60210. These five proteins belong to a protein family of unknown function. We named these five proteins RIP1 to 5 (ROP interactive partner) (Figure 1A). The RIPs share a relatively high identity and similarity in amino acid sequence at their N and C-termini. A Blast search using N or C-termini identified homologs of Arabidopsis RIPs in different plant species, including tobacco, potato, and rice (Figure 1B), but not in yeast and animals. Alignment of the predicted amino acids of RIPs from Arabidopsis and other plant species shows that the C-termini of RIPs are relatively conserved through evolution (Figure 1C). RIP1 was also recently identified in a two-hybrid screen for proteins interacting with the activated form of ROP10 and was named ICR1 (interactor of constitutively active ROP 1) (Lavy et al., 2007).
Figure 1.
RIPs Are Novel Plant-Specific Proteins.
(A) Alignment of the predicted amino acid sequences of Arabidopsis RIP family proteins. Black boxes represent identical residues, and gray boxes represent conserved residues.
(B) phylogentic tree of RIPs from different species.
(C) Alignment of the C-terminus of RIPs from different plant species. Black boxes represent identical residues, and gray boxes represent conserved residues. The conserved QWRKAA motif and the lysine-rich region are indicated by unfilled boxes with solid and dashed lines, respectively.
GFP-RIP1/ICR1 Developmentally Controlled Polar Localization during Arabidopsis Pollen Germination and Pollen Tube Growth
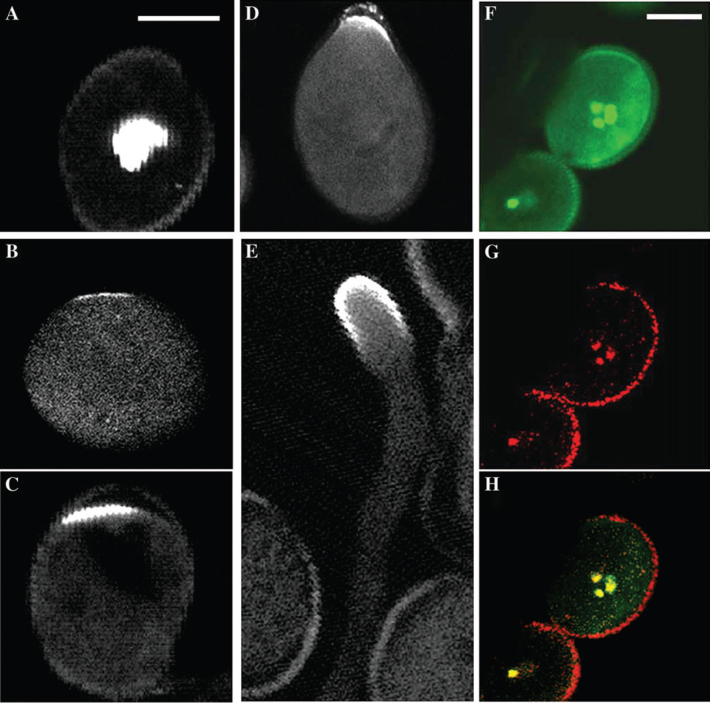
Since ROP1 was preferentially expressed in pollen and controls pollen tip growth (Li et al., 1998,1999), we were interested in investigating the function of RIPs in Arabidopsis pollen tip growth. RIP1/ICR1, which represented the highest number of clones recovered in the yeast two-hybrid screen, was chosen for further studies. A LAT52::GFP-RIP1/ICR1 construct was stably transformed into Arabidopsis (Col-0) and GFP-RIP1/ICR1 localization was examined in mature, activated, and germinating pollen. In mature pollen prior to activation, GFP-RIP1/ICR1 was localized in the nuclei (Figure 2A and 2F–2H). About 30 min after mature pollen was incubated in germination medium, GFP-RIP1/ICR1 was shifted to a cortical region, which predicted the germination pore (Figure 2B–2D). At this time, there was no visible sign of germination. Therefore, GFP-RIC1/ICR1 clearly marks the site of germination. Time lapse imaging revealed that GFP-RIP1/ICR1 was initially accumulated to a very restricted site of the cortex and was rapidly amplified so that the GFP intensity was very intense and spread to a broader region of the cortex, which presumably determined the width of emerging tubes (Supplemental Video). GFP-RIP1/ICR1 was maintained at the germination site and then at the apical cortex of growing pollen tubes (Figure 2E). This apical localization is similar to the localization of ROP1 and RopGEFs (Lin et al., 1996; Gu et al., 2005).
Figure 2.
GFP-RIP1/ICR1 Localization in Arabidopsis Pollen and Pollen Tubes.
LAT52::GFP-RIP1/ICR1 was transformed into Arabidopsis (Col-0) and pollen grains were collected and germinated on semi-solid medium (1% agar).
(A) GFP-RIP1/ICR1 localization in a pollen grain before germination.
(B–D) GFP-RIP1/ICR1 localization during pollen germination.
(E) GFP-RIP1/ICR1 localization in a growing pollen tube.
(F) GFP-RIP1/ICR1 localization in a pollen grain before germination.
(G) DAPI staining of the pollen grain in (F).
(H) Overlay of (F) and (G). Bar (5 μm) in (A) is for (A–E). Bar (5 μm) in (F) is for (F–H).
RIP1/ICR1 Interacts with ROP In Vivo
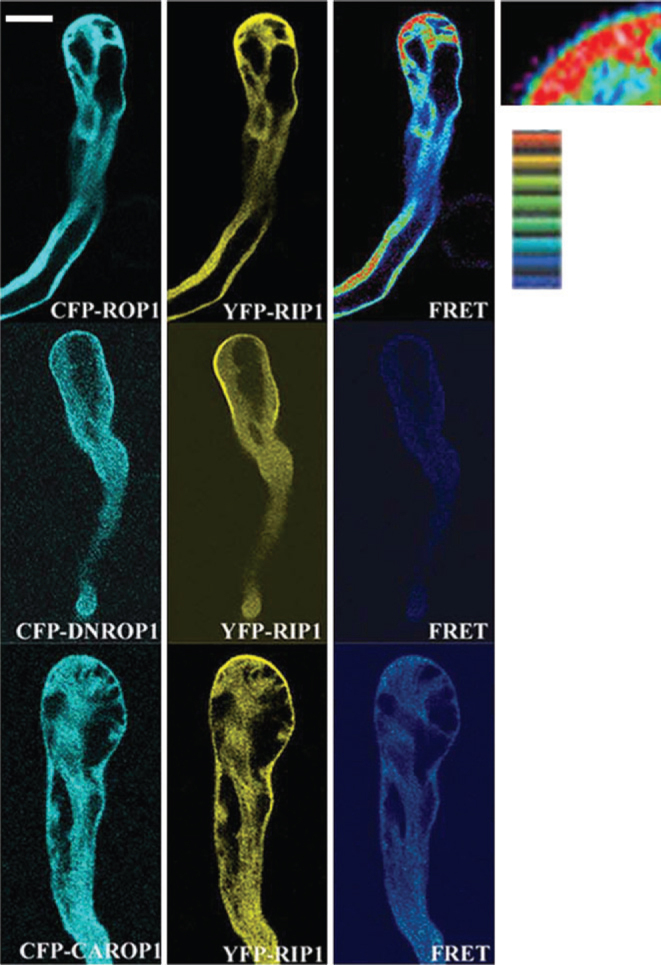
To confirm the interaction between RIP1/ICR1 and ROP1 in vivoFRET analysis was conducted in tobacco pollen tubes (Figure 3). We co-expressed LAT52::YFP-RIP1/ICR1 with different forms of ROP1 tagged with CFP. YFP-RIP1/ICR1 was localized to the apical PM of the tube similar to GFP-RIP1 in Arabidopsis pollen tubes. However, CFP-ROP1 had a weak apical PM localization. A strong FRET signal was detected in a region of the cortical region close to the apical PM and a weak FRET signal was detected on the PM. A weak FRET signal was detected between YFP-RIP1/ICR1 and CFP-DN-ROP1 in the tobacco pollen tube cytoplasm, even though the localization of both YFP-RIP1/ICR1 and CFP-DN-ROP1 was primarily in the PM. CFP-CA-ROP1 interaction with YFP-RIP1/ICR1 was stronger than with CFP-DN-ROP1.
Figure 3.
FRET Analysis of RIP1/ICR1 Interaction with ROP1 in Tobacco Pollen Tubes.
Equal amounts (1 μg) of LAT52::YFP-RIP1/ICR1 and LAT52::CFP-ROP1 or LAT52::CFP-DNROP1 or LAT52::CFP-CAROP1 were mixed and used in each transformation. Tobacco pollen germinated and grew in liquid medium for 5 h before observation. All images are single sections through the middle plane of the pollen tube. Bar indicates the intensity of the FRET signal: red represents a strong signal and blue a weak signal. Bar is 15 μm.
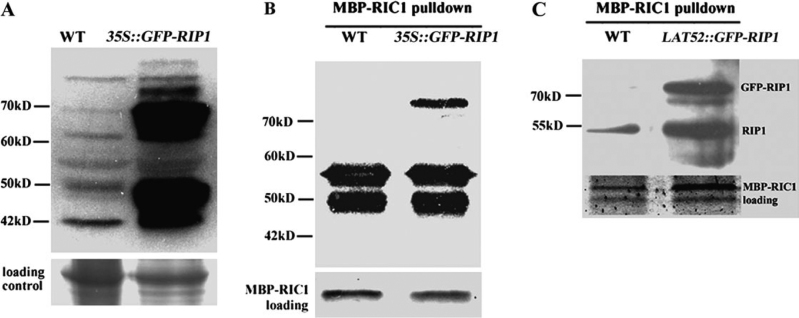
We developed a RIP1/ICR1-specific polyclonal antibody using His-tagged recombinant RIP1/ICR1 (full-length). Purified RIP1/ICR1 antibodies did not react with RIP2 and RIP3 recombinant proteins, confirming that these antibodies were RIP1-specific (data not shown). Purified RIP1/ICR1 antibodies recognized a protein (42-kD) of the expected molecular weight of unmodified RIP1/ICR1 from 10-day-old wild-type seedlings in Arabidopsis (Figure 4A). In addition, RIP1/ICR1 antibodies recognized several proteins larger than predicted RIP1/ICR1 (50, 55, 65, 70, and 90 kD). To assess whether these larger proteins were non-specific or represented modified forms of RIP1, RIP1/ICR1 antibodies were also used to analyze GFP-RIP1/ICR1 from 35S::GFP-RIP1/ICR1 transgenic plants. A major band about 70 kD corresponding to GFP-RIP1/ICR1 was detected as well as some bands larger than GFP-RIP1/ICR1 (75, 90, and 100 kD). Furthermore, RIP1 antibodies did not detect these larger proteins from total proteins isolated from GFP-RIP2 and GFP-RIP3 overexpressing plants (data not shown). We conclude that the anti-RIP1 reactive proteins that were larger than RIP1/ICR1 most likely represent modified RIP1/ICR1 proteins.
Figure 4.
RIP1/ICR1 and GFP-RIP1/ICR1 Interact with GTP-Bound ROP in Arabidopsis.
(A) A Western-blotting analysis with RIP1/ICR1 polyclonal antibodies shows the expression of GFP-RIP1/ICR1 in the 35S::GFP-RIP1/ICR1transgenic plants. About 10 μg of total proteins extracted from 10-day-old seedlings were loaded in each lane.
(B) A Western-blotting analysis using RIP1/ICR1 polyclonal antibodies shows that RIP1/ICR1 and GFP-RIP1/ICR1 interact with GTP-bound ROP pulled down by MBP-RIC1, a ROP effector that specifically interacts with GTP-bound ROP.
(C) A Western-blotting analysis shows that GFP-RIP1/ICR1 interacts with ROPs in Arabidopsis pollen grain or pollen tubes. To purify active ROP complex from Arabidopsis7-day-old seedlings or open flowers (1 g) were ground in liquid nitrogen hydrated extraction buffer. The supernatant was mixed with MBP-RIC1 containing beads (about 5 μg MBP-RIC1). The pellet was washed three times and the liquid fraction was completely removed by a syringe and then the MBP-RIC1-bound active ROP complex was eluted by maltose. Proteins were fractionated by 12% SDS–PAGE and transferred to nitrocellulose membrane.
To confirm the in-vivo interaction between RIP1/ICR1 and ROP1, we performed a co-immunoprecipitation assay. We used MBP-RIC1 (a ROP effector that specifically interacts with GTP-bound active ROP) to pull down a protein complex containing GTP-bound active ROP from 10-day-old seedlings of wild-type and 35S::GFP-RIP1/ICR1 Arabidopsis (Figure 4B). Using RIP1/ICR1 antibodies, we detected two species of proteins (50 and 55 kD) that were co-purified with GTP-bound active ROP from wild-type seedlings and an extra band (about 75 kD) from 35S::GFP-RIP1/ICR1 seedlings. These proteins were larger than expected unmodified forms of RIP1/RIC1 and GFP-RIP1/ICR, respectively, suggesting that they could be modified forms of the corresponding proteins. To confirm that the 50 and 55-kD bands that co-purified with GTP-bound ROP were indeed RIP1/ICR1 proteins, the 55-kD band was isolated for nano-liquid chromatography/tandem mass spectrometry analysis (nano-LC MS/MS). RIP1/ICR1 was found to be a major protein in this band (data not shown). These results suggest that the active form of ROPs specifically interacts with the modified 50 and 55-kD RIP1/ICR1 proteins, whereas the total ROPs may interact with the unmodified 42-kD RIP1/ICR1 and several other modified RIP1/ICR1 proteins.
To test whether GFP-RIP1/ICR1 can interact with ROP in Arabidopsis pollen or pollen tubes, we used MBP-RIC1 to pull down GTP-bound ROP extracted from open flowers of wild-type and LAT52::GFP-RIP1/ICR1 plants (Figure 4C). Unlike the result obtained from the seedlings, only one band (55-kD) of RIP1/ICR1 co-purified with active ROP from wild-type flowers. The 75-kD band, which corresponds to the presumably modified GFP-RIP1/ICR1, was co-purified with active ROP from transgenic LAT52::GFP-RIP1/ICR1 flowers. Since the LAT52 promoter is preferentially expressed in mature pollen grains and pollen tubes, this result suggests that active ROP1 indeed interacts with the modified 55-kD RIP1/ICR1. These results also suggest that pollen RIP1/ICR1 exhibits different modification patterns from the vegetative tissues. Taken together, our results show that RIP1/ICR1 interaction with ROP1 is complex and does not follow the typical interaction between an active Rho GTPase and its effector, which raises the possibility that RIP1/ICR1 may not just simply be a ROP effector.
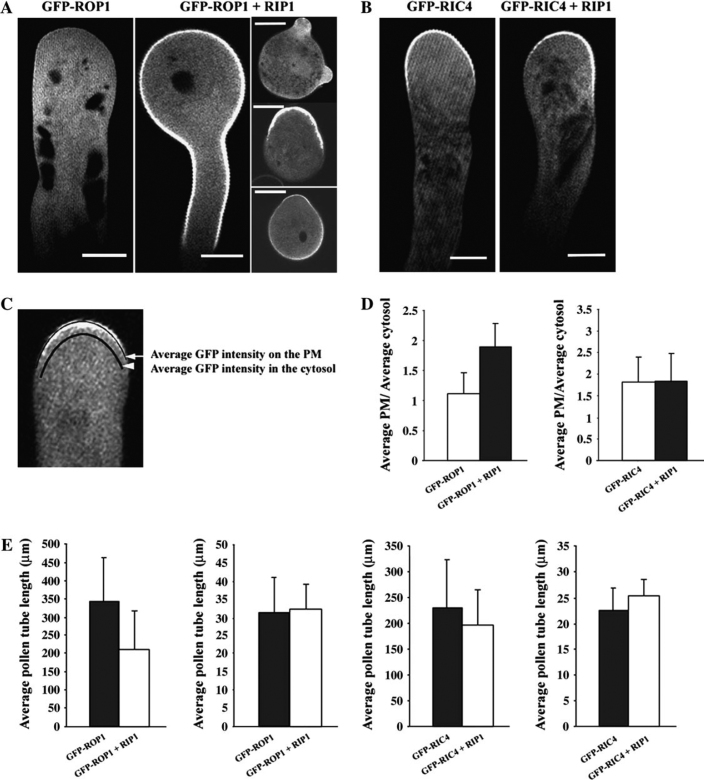
RIP1/ICR1 Promotes GFP-ROP1 Association with the Cell Cortex of the Tobacco Pollen Tube
Overexpression of GFP-RIP1/ICR1 or YFP-RIP1/ICR1 caused severe growth depolarization in tobacco pollen tubes (i.e. a ballooned tip). Similar phenotype was also induced by transient expression of LAT52::RIP1/ICR1 (data not shown). This result implies a positive effect of RIP1/ICR1 on ROP signaling. However, we could not detect any guanine nucleotide exchange activity on recombinant RIP1/ICR1 proteins using a fluorescent-based GEF assay towards ROP1 (data not shown) (Gu et al., 2005). We next tested whether RIP1/ICR1 induced growth depolarization by altering the PM localization of ROP1. Transient overexpression of GFP-ROP1 in tobacco pollen tubes produces depolarization of pollen tube growth (Fu et al., 2001; Gu et al., 2003). GFP-ROP1 was primarily distributed in the cytosol, while its PM localization was quite weak (Figure 5A). Co-overexpression of GFP-ROP1 and RIP1/ICR1 in tobacco pollen tubes induced a dramatic shift of GFP-ROP1 to the PM. PM-associated GFP-ROP1 was greatly increased in intensity and was extended to the flank of the tube. We compared the average GFP-ROP1 intensity at the apical PM to the GFP-ROP1 intensity in the cytosol close to the apical PM (Figure 5C–5E). RIP1/ICR1 greatly increased the GFP-ROP1 intensity on the apical PM. Interestingly, half of the pollen grains overexpressing GFP-ROP1 and RIP1/ICR1 did not have substantial pollen tube growth, even though they germinated (Figure 5A). GFP-ROP1 localization in these pollen grains was either randomly localized to the PM or localized to a broad region on the PM at the germination site. By the measurement of the width of the tobacco pollen tube tip (excluding those that do not have substantial tube growth), we found that co-overexpression of GFP-ROP1 and RIP1/ICR1 in the pollen tube resulted in a similar degree of growth depolarization, as seen in transient expression of GFP-ROP1 alone (Figure 5E). However, the length of the pollen tube was dramatically decreased in tubes co-expressing GFP-ROP1 and RIP1/ICR1 compared to those overexpressing GFP-ROP1 alone. Based on these results, we propose that RIP1/ICR1 promotes the stabilization of ROP1 on the PM but not the activation of ROP1.
Figure 5.
Overexpression of RIP1/ICR1 Alters the Localization of GFP-ROP1 but not GFP-RIC4.
(A) GFP-ROP1 localization in tobacco pollen tubes with and without co-expressed RIP1/ICR1.
(B) GFP-RIC4 localization in tobacco pollen tubes with and without co-expressed RIP1/ICR1.
(C) Schematic view of the measurements of the average GFP-ROP1 or GFP-RIC4 intensity on the PM and in the cytosol.
(D) The change in GFP-ROP1 and GFP-RIC4 localization caused by RIP1/ICR1 overexpression indicated by the ratio of the average GFP intensity at the PM and in the cytosol. For each measurement, 40 tubes were used. GFP-ROP1 or GFP-RIC4 was used as control. Error bars indicate S.D.
(E) Measurements of tobacco pollen tube lengths and widths 6 h after transient expression. The pollen tube length and width measurements were from 40 pollen tubes for each sample. GFP-ROP1 or GFP-RIC4 was used as control. Error bars indicated S.D. LAT52::GFP-ROP1 or LAT52::GFP-RIC4 plasmid DNA (1 μg) was used in each transient experiment. For co-transient expression, equal amounts (1 μg) of LAT52::GFP-ROP1 or LAT52::GFP-RIC4 and LAT52::RIP1/ICR1 were mixed and transformed into tobacco pollen grains. Pollen grains germinated and grew in liquid medium and images of GFP-ROP1 localization were recorded 6 h after transient expression. All images were single sections through the middle plane of the pollen tube. Bar = 15 μm.
To test whether RIP1/ICR1 overexpression increases ROP1 activity, we transiently co-overexpressed RIP1/ICR1 with GFP-RIC4, a marker for active ROP1 (Hwang et al., 2005). ROP1 activation is indicated by RIC4 localization to the apical PM (Figure 5B). By comparing the average GFP-RIC4 intensity at the apical PM to the average GFP-RIC4 intensity in the cytosol near the apical PM, we found that RIP1/ICR1 overexpression did not promote the association of GFP-RIC4 to the PM (Figure 5D and 5E). Measurements of the pollen tube tip width also indicated that RIP1/ICR1 only slightly enhanced depolarization of tobacco pollen tubes induced by GFP-RIC4 overexpression, consistent with its limited effect of RIC4 localization to the apex PM. Increased ROP1 localization to the PM did not result in an increase in ROP1 activation, suggesting that the ROP1 activator was rate-limiting in these tubes. RIP1/ICR1 overexpression also reduced the length of tobacco pollen tubes expressing GFP-RIC4 but the degree of this reduction was not as dramatic as that induced by co-expression of RIP1/ICR1 and GFP-ROP1. The reduction in pollen tube length could be explained by RIP1/ICR1 sequestering ROP1 to the flank of pollen tubes, reducing the amount of ROP1 that could be targeted to the tip to promote tip growth. Alternatively, cycling between the PM and the cytosol may be critical for the function of ROP1 in promoting tip growth (Klahre et al., 2006).
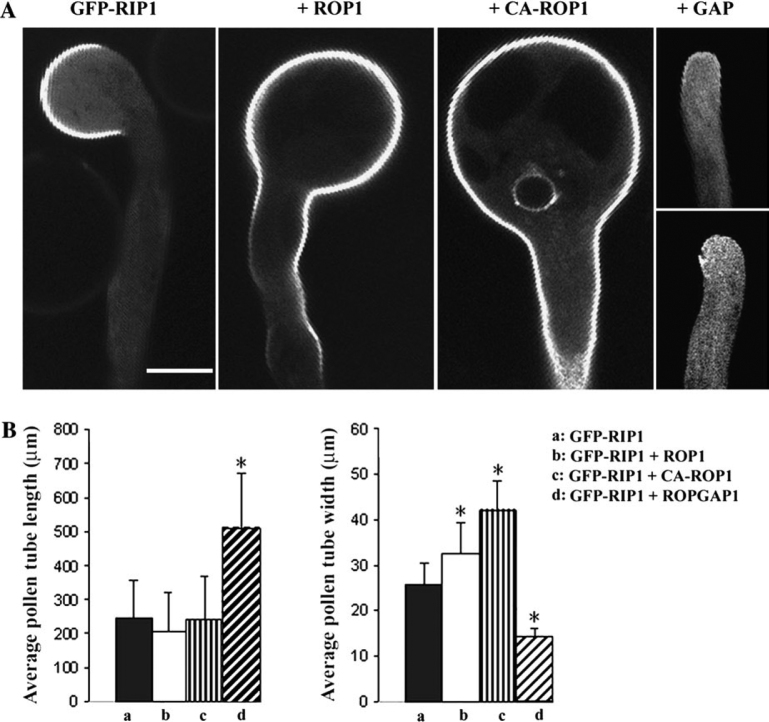
The Apical PM Localization of GFP-RIP1/ICR1 Is Dependent on ROP Activity
We next tested whether the RIP1/ICR1 localization to the apical PM is dependent on ROP activity. GFP-RIP1/ICR1 was co-transiently overexpressed with different components in ROP signaling (Figure 6A). ROP1 overexpression caused GFP-RIP1/ICR1 to localize to a broader region of the apical PM compared to tubes expressing GFP-RIP1/ICR1 alone. This might be due to a strong physical interaction between ROP1 and GFP-RIP1/ICR1 on the PM. However, this was unlikely, because the FRET signal between YFP-RIP1/ICR1 and CFP-ROP1 was more restricted to the tip (Figure 3). Alternatively, ROP1 overexpression may change the properties of the apical PM, which may promote a strong association of GFP-RIP1/ICR1 to PM. To test this possibility, GFP-RIP1/ICR1 was co-expressed with CA-ROP1. CA-ROP1 was mainly localized to the cytosol as indicated by CFP-ROP1 (Figure 3). However, the association of GFP-RIP1/ICR1 with the whole PM was promoted of the tobacco pollen tube when CA-ROP1 was co-expressed, suggesting that the strong association of GFP-RIP1/ICR1 to the PM was not due to its direct interaction with CA-ROP1. We then determined whether ROP1 activation was necessary for GFP-RIP1/ICR1 association with the PM. GFP-RIP1/ICR1 was co-overexpressed with RopGAP1, which inactivated ROP1 by promoting its GTP hydrolysis. GFP-RIP1/ICR1 localization to the PM was diminished in most of the tobacco pollen tubes. In addition, GFP-RIP1/ICR1-induced pollen tube depolarization was abolished by RopGAP1 (Figure 6A and 6B).
Figure 6.
GFP-RIP1/ICR1 Localization at the Apex of the Tobacco Pollen Tube Is ROP Activity-Dependent.
(A) Localization of GFP-RIP1/ICR1 in tobacco pollen tubes and the effects of the ROP signaling components on GFP-RIP1/ICR1 localization. LAT52::GFP-RIP1/ICR1 plasmid DNA (1 μg) was introduced into tobacco pollen grains via microprojectile bombardment. For co-expression, equal amounts (1 μg) of LAT52::GFP-RIP1/ICR1 and LAT52::CA-ROP1 or LAT52:: ROP1 or LAT52:: RopGAP1 was used in each transformation. Pollen was germinated and cultured in liquid germination medium for 6 h before observation under a confocal microscope. All images are single sections through the middle plane of the pollen tube. Bar = 20 μm.
(B) Phenotypic analysis of the effects of ROP signaling components on GFP-RIP1/ICR1 by measurement of pollen tube length and width. The measurements were from 60 pollen tubes after 6 h germination. Asterisks indicate a significant difference from control at the same data point (P < 0.01; t-test). Error bars indicate S.D. GFP-RIP1 was used as control.
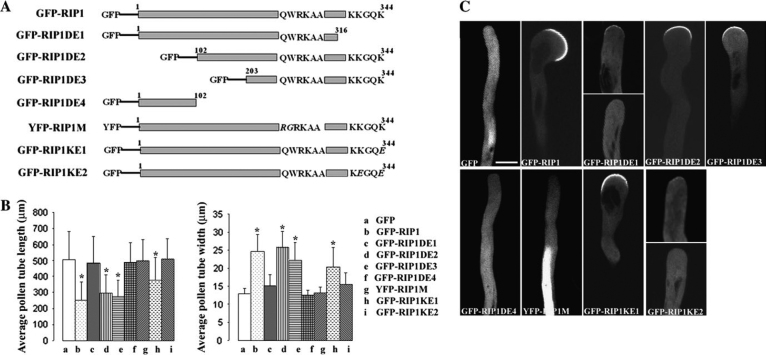
The C-Terminal Region of RIP1/ICR1 Is Necessary and Sufficient for the RIP1/ICR1 Localization to the Apical PM
To better understand the mechanisms for RIP1/ICR1 promotion of ROP1 association with the PM and for ROP1 regulation of RIP1/ICR1 localization to the PM, we dissected domains of RIP1/ICR1 required for ROP1 binding and PM localization. From the yeast two-hybrid results, all of the recovered partial cDNA clones of RIPs encoded their C-terminal region. We speculated that a ROP binding domain(s) may be located in this region. As shown in Figure 7Atwo N-terminal deletion mutants, RIP1/ICR1DE2 and RIP1/ICR1DE3, were made and tagged with GFP at their N-termini (Figure 7A). GFP-tagged deletion mutants were transiently expressed in tobacco pollen tubes and the localization patterns and pollen tube phenotypes were examined (Figure 7B and 7C). We found that both N-terminal deletions have little impact on the PM localization of RIP1/ICR1 and the depolarized pollen tube growth phenotype. Therefore, the C-terminal part of RIP1/ICR1 was sufficient for PM localization and the induction of pollen tube growth depolarization.
Figure 7.
Analysis of RIP1/ICR1 Mutations in Tobacco Pollen Tubes.
(A) Schematics of GFP or YFP-tagged RIP mutants.
(B) Tobacco pollen tube phenotypes caused by transient overexpression of GFP-or YFP-tagged RIP1/ICR1 mutants. Measurements of pollen tube length and width were performed 6 h after pollen transformation with the GFP-tagged mutants. Asterisks indicate a significant difference from control at the same data point (P < 0.01; t-test). GFP was used as control. Error bars indicate S.D. For each transient expression, 1 μg of plasmid DNA was used.
(C) Localization of GFP or YFP-tagged RIP1/ICR1 mutants in tobacco pollen tubes. All images are single sections through the middle plane of a tobacco pollen tube. Bar = 20 μm.
An alignment of the predicted amino acids of RIPs from Arabidopsis and other plant species showed that the C-termini of RIPs are relatively conserved. The conserved regions include a conserved box (QWRKAA) located near the C-terminus and a lysine-rich region at the very C-terminus (Figure 1C). To test the importance of those two motifs, two C-terminal deletion mutants (RIP1/ICR1DE1 and RIP1/ICR1DE4) were made and fused with GFP at their N-terminus (Figure 7A). GFP-tagged C-terminal deletion mutants were transiently overexpressed in tobacco pollen tubes (Figure 7B and 7C). When the lysine-rich region was deleted, the localization of RIP1/ICR1 was shifted to the cytosol in the majority of pollen tubes observed, with only a few showing weak localization to the apical PM. The growth depolarization phenotype was also reduced dramatically. When both the lysine-rich region and the QWRKAA motif were deleted, both PM localization and growth depolarization were completely eliminated (Figure 7A–7C).
Since the QWRKAA motif was highly conserved, we speculated that this motif might be involved in ROP binding or PM targeting. To test this hypothesis, we made a rip1/icr1 mutant (RIP1/ICR1M) with two amino acids QW in this motif replaced with RG (Figure 7A). YFP-RIP1/ICR1M was not localized to the PM, but instead was found in the nucleus (Figure 7C). Overexpression of YFP-RIP1/ICR1M did not induce pollen tube depolarization. A RIP1/ICR1 mutant (RIP1/ICR1DE) with deletion of the QWRKAADAAA also lost PM localization and effects on pollen tube polarity (data not shown). We concluded that at least the QWKRAA is critical for PM targeting.
Deletion of the lysine-rich region of the C-terminus of RIP1/ICR1 also dramatically decreased the PM localization. All the RIPs except the rice RIP4 had the KKXXK at their C-termini. In order to test whether these lysine residues were important for the PM localization of RIP1/ICR1, we generated two point mutations, RIP1/ICR1KE1 and RIP1/ICR1KE2, in which the last K and the last two Ks were mutated to Es, respectively (Figure 7A). GFP-RIP1/ICR1KE1 was still localized to the apical PM and induced depolarization of tobacco pollen tube growth (Figure 7C). However, the degree of depolarization induced by GFP-RIP1/ICR1KE1 was reduced compared to by GFP-RIP1/ICR1 (Figure 7B). GFP-RIP1/ICR1KE2 localization was greatly shifted from the PM to the cytosol, and its effect on growth depolarization was also dramatically reduced similar to the effect of deleting the whole lysine-rich domain (Figure 7B and 7C). We conclude that both the QWRKAA motif and the lysine-rich region are required for PM targeting.
We next tested whether the QWRKAA motif and the lysine-rich region were required for ROP1 binding. His-tagged RIP1/ICR1 mutant proteins were purified from E. coli (Supplemental Figure 1A) and an in-vitro ROP1 binding assay was conducted (Supplemental Figure 1B). RIP1/ICR1 interacted with all forms of ROP1. RIP1/ICR1M lost its ROP1 binding ability. Surprisingly, RIP1/ICR1DE1 interacted strongly with all forms of ROP1. We conclude that the QWRKAA motif, but not the lysine-rich region, is required for ROP binding. Similarly, this motif was also found to be critical for the interaction of ICR1 with other ROPs and for the function of ICR1 in Arabidopsis pavement cells (Lavy et al., 2007). All together, our results suggest that ROP binding and PM targeting are two separable functions of RIP1/ICR1.
DISCUSSION
A central question in cell polarity is how a polarizing cue initiates localized signaling and how localized signaling is maintained to develop cell polarity. In plants, ROP GTPases play a critical role in the regulation of cell polarity and polarized cell growth (Gu et al., 2004; Krichevsky et al., 2007; Yang, 2002,2008; Yang and Fu, 2007). In pollen tubes, ROP is localized to or ROP signaling is detected at the tip of pollen tubes to promote tip growth and to maintain growth polarity (Cheung et al., 2003; Fu et al., 2001; Hwang et al., 2005; Kost et al., 1999; Li et al., 1999). In the current study, we have identified a ROP-interacting protein, RIP1/ICR1, which marks the site of pollen germination and regulates polarized pollen tube growth. We show that RIP1/ICR1 promotes or stabilizes ROP1 on the PM, but RIP1/ICR1 localization to the tip requires ROP1 activation. We hypothesize that a circuitry involving RIP1/ICR1-ROP1 interaction participates in the establishment of cell polarity during pollen germination and pollen tube tip growth.
RIP1/ICR1 Marks Pollen Germination Sites
In Arabidopsisthe constitutive active (CA) form of ROP1 promotes in-vitro pollen germination (Li et al., 1999). However, how is the germination site selected and initiated is still a mystery. RIP1/ICR1 is the first identified protein that predicts future germination sites in pollen grains. During pollen tube tip initiation, ROP needs to be targeted to the pollen tube initiation site. Interestingly, RIP1/ICR1 are localized to the nucleus of mature pollen grain before germination and this localization shifts to the cytosol and the PM at the pollen tube initiation site. The localization of RIP1/ICR1 in the nucleus may prevent it from targeting ROP to the PM of the tube initiation site. When RIP1/ICR1 moves out of the nucleus, it can interact with ROP in the cytosol and then target ROP to the PM of the germination sites. This nucleus-to-PM pattern of RIP1/ICR1 is similar to that of Cdc24p (Cdc42p guanine-nucleotide-exchange factor). Cdc24p shuttles from the nucleus to the cytosol/budding site to initiate budding or mating tube formation in budding yeast (Nern and Arkowitz, 2000; Toenjes et al., 1999). This nucleus to cytosol targeting is critical for the establishment of localized Cdc42p activity at site of cell polarization. It is possible that pollen germination also adopts a similar nucleo-cytoplasmic shuttling mechanism for components that are required for the initial establishment of polar germination sites. In the budding yeast, a stable polarity axis can be spontaneously established without any extracellular cue. This cell polarization is dependent on the activity of Cdc42p (the counterpart of ROP1 in budding yeast) and actin polymerization (Wedlich-Soldner et al., 2003). Detailed study revealed that this polarity initiation required a rapid exchage of Cdc42p between polar caps and the cytosol (Wedlich-Soldner et al., 2004). A similar rapid exchange of ROP between the initiation site and the cytosol may also be required for a successful pollen tube initiation. Interestingly, GFP-RIP1/ICR1 is not just amplified on the germinating sites, but also in oscillates (Supplemental Movie 1). This GFP-RIP1/ICR1 dynamic pattern may reflect a rapid exchange of ROP activity/localization at the putative germination sites. RIP1/ICR1 was also found to interact with Arabidopsis SEC3A, a subunit of the exocyst complex (Lavy et al., 2007). Interestingly, Arabidopsis SEC8, a putative exocyst 70 subunit, is required for pollen germination and pollen tube growth (Cole et al., 2005). It would be interesting to determine whether RIP1/ICR1 is involved in the recruitment of exocyst to the site of pollen germination.
RIP1/ICR1 Is a Component Involved in Polar ROP PM Localization
Our results show that overexpression of a functional fusion protein of GFP-RIP1/ICR1 in the tobacco pollen tube leads to depolarization of tube growth similar to the effect of ROP1 overexpression or overactivation. This implies that RIP1/ICR1 is a component of ROP-mediated signaling. Our observation that RIP1/ICR1 dramatically increased the GFP-ROP1 PM targeting in the tobacco pollen tube suggests that RIP1/ICR1 was involved in the polar ROP PM localization. This efficient ROP PM targeting could be a result of either RIP1/ICR1 facilitation of the GFP-ROP1 transport to the PM or RIP1/ICR1 stabilization of the PM association of GFP-ROP1. Our FRET analysis indicated that YFP-RIP1/ICR1 selectively interacted with CFP-ROP1 in the cytosol. This implies that RIP1/ICR1 forms a complex with ROP1 in the cytosol and the complex somehow allows ROP1 to be stabilized or recruited to the PM. It is known that ROP1 is prenylated and that the prenylation is important for ROP1 association with the PM (Li et al., 1999; Lin et al., 1996). However, the prenylation alone is probably not sufficient for the stable association of ROP1 with a specific domain of the PM. Evidence suggested that acylation of the active form of ROP6 stabilized its PM association, which could be a mechanism for localization of ROPs to a specific PM domain when ROPs were locally activated (Sorek et al., 2007). We propose that ROP association with RIP1/ICR1 provides another mechanism for localized distribution of ROPs to the PM. We have shown that the C-terminal poly-lysine motif of RIP1/ICR1 is required for its PM association but not for its binding to ROP1. RIP1/ICR1 association with the PM requires ROP activation, suggesting that localized ROP signaling may generate a specialized PM domain (by altering either the PM lipid composition or the PM-associated protein) (see below), which can recruit RIP1/ICR1-ROP complex, allowing ROP1 to be recruited or stabilized to this specific PM domain.
The Regulation of RIP1/ICR1 in Pollen Tube Tip Growth
In the budding yeast, Cdc42p, a distant homologue of ROP, is subjected to a positive feedback mechanism regulating its activity during bud initiation and budding (Butty et al., 2002; Wedlich-Soldner et al., 2003,2004). This positive feedback mechanism requires the actin cytoskeleton and secretion. In plants, a positive feedback mechanism involving calcium signaling and ROS is required for the establishment of polar root hairs in Arabidopsis (Takeda et al., 2008). A positive feedback mechanism also exists to regulate ROP activity in tip growth of the pollen tube (Hwang et al., 2005). The targeting of ROP to the PM could be a critical step in this mechanism. Our observation indicates that at least a positive feedback mechanism exists at the level of ROP PM localization. First, RIP1/ICR1 dramatically increases the PM localization of GFP-ROP1 in the tobacco pollen tube tip. The increased PM localization of GFP-ROP1 does not induce additional pollen tube depolarization compared to GFP-ROP1 alone. This indicates that RIP1/ICR1 only promotes the PM localization of GFP-ROP1 but not the activation event. Second, the PM localization of GFP-RIP1/ICR1 depends on ROP activity. ROP activity-induced PM localization of GFP-RIP1/ICR1 could be the result of either a tight interaction between GFP-RIP1/ICR1 and active ROP or a change of PM properties by ROP activity. Our data generated from the deletion and mutation study of RIP1/ICR1 favor the hypothesis that ROP activity changes the properties of the PM, then subsequently affects GFP-RIP1/ICR1 localization. The deletion of the lysine-rich region at the end of the C-terminus of RIP1/ICR1 (RIP1/ICR1DE1) greatly reduces the RIP1/ICR1 PM localization. However, RIP1/ICR1DE1 does not lose its in-vitro ROP binding ability. It is likely that the interaction between RIP1/ICR1 and ROP1 in the cytosol is a prerequisite for the PM targeting of RIP1/ICR1. This interaction may confer RIP1/ICR1’s PM localization via two mutually non-exclusive mechanisms. The formation of the RIP1/ICR1-ROP1 complex may facilitate the transport of this complex to the PM. Alternatively or simultaneously, the binding of ROP1 to RIP1/ICR1 may change the conformation of RIP1/ICR1’s C-terminus, allowing RIP1/ICR1 to associate with the PM.
ROP Dynamics Are Critical for Rapid Tip Growth and Polarity Maintenance of the Pollen Tube
It has been observed in different species that rapid pollen tube growth is associated with a growth rate oscillation (Feijo et al., 2001,2004; Hwang et al., 2005; Messerli et al., 2000; Robinson and Messerli, 2002). This growth oscillation has been linked to the oscillation of a steep tip-focused calcium gradient in the growing pollen tube (Li et al., 1998; Messerli et al., 2000), dynamic and oscillating tip F-actin, and the dynamic and oscillating tip-focused ROP activity in the pollen tube (Fu et al., 2001; Kost et al., 1999; Li et al., 1999). Among these oscillators, ROP1 activity seems to be the leader (Hwang et al., 2005). ROP1 activation is required for the formation of calcium gradients and F-actin at the tip. Stabilization of ROP1 activity by overexpression or overactivation of ROP1 or its effector proteins causes tip growth arrest (Hwang et al., 2005). Interestingly, we find that GFP-ROP1 loses its normal oscillation when RIP1/ICR1 is overexpressed in the tobacco pollen tube. Co-overexpression of GFP-ROP1 and RIP1/ICR1 in tobacco pollens causes pollen tube arrest, even though GFP-ROP1 is dramatically promoted to the PM. In severe cases, pollen grains overexpressing GFP-ROP1 and RIP1/ICR1 do not have substantial tube growth after germination. This arrested pollen tube growth phenotype is less severe in tobacco pollen tubes overexpressing GFP-ROP1 or RIP1/ICR1 alone. We reason that the pollen tube tip has a delicate mechanism to balance both the activity and the amount of ROP in a temporal and spatial distribution. This balanced ROP activity distribution may be necessary for the oscillation of many signaling components that support rapid pollen tube tip growth. Disruption of this balanced temporal and spatial ROP activity distribution through adding exogenous ROP and stabilizing ROP on the PM by RIP1/ICR1 may surpass the buffering capability to sustain normal oscillation of the pollen tube.
RIP1/ICR1 May Act as Both ROP1 Effector and Regulator
In ArabidopsisRICs have been identified as plant-specific ROP effectors and they specifically interact with the active form of ROPs (Gu et al., 2005; Wu et al., 2001). RIP1/ICR1 was also identified as an effector of ROP10 or ROP6 in a yeast two-hybrid screen, since it preferentially interacted with both wild-type and constitutively active (CA) form of ROP10, but not with the dominant negative form of ROP10 (Lavy et al., 2007). We also show that RIP1/ICR1 interacts with the active form of ROP1. It will be interesting to determine whether RIP1/ICR1 indeed acts as a ROP1 effector and interacts with SEC3 to recruit exocyst to the cortex of growth sites (Lavy et al., 2007). However, our data also support a possible role for RIP1/ICR1 as ROP1 regulator in pollen and pollen tubes. First, partial cDNAs of all five RIPs were found to interact with DN-ROP1 in the yeast two-hybrid assay. Second, His-tagged RIP1 interacted with GTP-bound, GDP-bound and wild-type ROP1 equally well in an in-vitro binding assay. Furthermore, our FRET analysis in tobacco pollen tube demonstrated a strong interaction between YFP-RIP1/ICR1 and CFP-ROP1 in the cytosol of pollen tube tip. This is in contrast to our previous finding that active ROP1 interacts with its effector RIC4 at the PM (Gu et al., 2005; Hwang et al., 2005). Last, our co-immunoprecipitation experiments also demonstrated that active ROPs did not interact with all different forms of RIP1/ICR1, supporting a complex functional relationship between ROPs and RIP1/ICR1. Nonetheless, further biochemical study will be necessary to elucidate properties of different forms of RIP1/ICR1 and their functions in ROP signaling.
METHODS
Yeast Two-Hybrid Screens
A dominant negative ROP1 (D121A) and isoprenylation-defective mutant (C188S) were generated by site-directed mutagenesis as described previously (Li et al., 1999). The D121A/C188S double mutant (DN2S) was fused to the GAL4 DNA binding domain in bait vector PAS2. The PAS2-DN2S was introduced into Y190 by an electroporation method. One transformant was designated as Y190-pAS2-DN2S and used for the transformation with 10 μg of the Arabidopsis flower cDNA library (Hu et al., 2003). All transformants were plated on SC-Trp-Leu-His + 100 mM 3-aminotriazole (3-AT, sigma) plates. About 3 million transformants were obtained. A filter assay was performed to test the β-galactosidase activity on all transformants. Putative positives were traced back to plates using filter papers and grown in fresh selective medium (SC-Trp-Leu-His) for 3 d at 30°C. A total of 72 putative positives were recovered and confirmed by their ability to grow on SC-Trp-Leu-His medium and for β-galactosidase activity using the filter assay for a second time. These putative positives were grown on SC-Leu medium containing 2.5 g ml−1 cycloheximide to select for cells only containing prey vector. DNA was isolated from yeast cells and transformed into E. coli. Only 41 of the 72 putative clones were recovered. All plasmid DNAs were sequenced using T7 primer. A total of nine partial cDNA clones were recovered for the RIP protein family. Four clones represent AtRIP1/ICR1 (At1g17140); two clones represent AtRIP2 (At2g37080); and single clones represent AtRIP3 (At3g53350), AtRIP4 (At1g78430), and AtRIP5 (At5g60210).
Database Search, Sequence Alignment, and Phylogenetic Analysis
DNA sequences obtained were used to identify Arabidopsis genes using BLAST-N against both NCBI (www.ncbi.nlm.nih.gov/BLAST) and TAIR database (www.arabidopsis.org/Blast). The GenBank accession numbers for each RIP other than Arabidopsis RIPs are: Rice RIP1/ICR1 (AAU03160), Rice RIP1/ICR1 (AAO72652), Rice RIP3 (AAS72367), Rice RIP4 (BAB92927), Tobacco Rice RIP1/ICR1 (CAC84774), and Potato RIP1/ICR1 (AAT39303). Protein sequences were aligned using ClustalW (www.ebi.ac.uk/clustalW). Data generated from alignments were loaded onto a Phylodendron Phylogenetic tree Printer (http://iubio.bio.indiana.edu/treeapp/treeprint-form.html) to reconstruct the rooted neighbor-joining phylogenetic tree. Aligned sequences were processed using Boxshade (www.ch.embnet.org/software/BOX_form.html) in a fraction of 0.7 and output as new RTF format.
DNA Manipulation and Plasmid Construction
PCR primers 5′RIP1/ICR1-BglII (5′-ccagatctatgccaagaccaagagtt-3’) and 3′RIP1/ICR1-KpnI (5′-gcggtacctcacttttgccctttcttcct-3’) were designed according to the predicted coding sequence of RIP1/ICR1. The coding sequence for AtRIP1/ICR1 was amplified by PCR from a flower cDNA library. Amplified RIP1/ICR1 fragments were ligated into pGEM T-EASY vector (Promega) to create pGEM::RIP1/ICR1 and then sequenced with T7 and SP6 primers. To create LAT52::GFP-RIP1/ICR1 constructs, the RIP1/ICR1 fragment was cut from pGEM::RIP1/ICR1 with BglII/KpnI digestion, and was cloned into pC1300LAT52::GFP with BglII/KpnI to create a translational fusion. For the construction of LAT52::YFP-RIP1/ICR1the RIP1/ICR1 fragment obtained from digestion of pGEM::RIP1/ICR1 with BglII/SacI was ligated into pLAT52::YFP to generate a translational fusion (Gu et al., 2005). To create 35S::GFP-RIP1/ICR1 RIP1/ICR1 (BglII/KpnI) was inserted into pC130035S::GFP (BglII/KpnI) to create a translational fusion.
Primers were designed for the generation of RIP1/ICR1 deletion mutants; RIP1/ICR1DE1: 5′RIP1/ICR1-BglII and 3′RIP1/ICR1DE1-KpnI (5′-gcggtaccttaatcatcagccattgccggtg-3’); RIP1/ICR1DE2: 5′RIP1/ICR1DE2-BglII (5′-ccagatctcttcataagaagtccaagaaacc-3’) and 3′RIP1/ICR1-KpnI; RIP1/ICR1DE3: 5′RIP1/ICR1DE3-BglII (5′-ccagatctgcttcagagatttctaatgtgaaa-3’); RIP1/ICR1DE4: 5′RIP1/ICR1-BglII and 3′RIP1/ICR1DE4-KpnI (5′-gcggtaccttacttcttatgaagctcatcttga-3’). All the deletion fragments were amplified from pGEM::RIP1/ICR1 as the template and were subsequently cloned into pGEM-T EASY vector. The resulting plasmids were confirmed by DNA sequencing; then the RIP1/ICR1 mutant fragments obtained using BglII/KpnI digestion were cloned into digested pC1300LAT52::GFP (BglII/KpnI).
Site-Directed Mutagenesis of RIP1/ICR1
We used PCR-based site-directed mutagenesis to generate RIP1/ICR1KE1 RIP1/ICR1KE2and RIP1/ICR1M. Primers were designed to amplify RIP1/ICR1KE1 (5′RIP1/ICR1-BglII and 3′RIP1/ICR1KE1-KpnI: 5′-gcggtacctcactcttgccctttcttcctcca-3’) and RIP1/ICR1KE2 (5′RIP1/ICR1-BglII and 3′RIP1/ICR1KE2-KpnI: 5′-gcggtacctcacttttgcccttcctccctcca-3’) from the template, pGEM::RIP1/ICR1. The amplified RIP1/ICR1KE1 and RIP1/ICR1KE2 fragments were cloned into pGEM-T EASY vector and the mutations were confirmed by DNA sequencing. Then, the RIP1/ICR1KE1 and RIP1/ICR1KE2 fragments were subcloned into pC1300LAT52::GFP using BglII/KpnI enzyme sites. To create point mutations of RIP1/ICR1Mthe primer 5′RIP1/ICR1-BglII and a mutant primer, 3′RIP1/ICR1M-NotI (5′-cgcggccgcgctcggtttgaaccctgaacttc-3’) containing Q265R and W266G mutations, were used to amplify mutant RIP1/ICR1 fragment A from pGEM::RIP1/ICR1. RIP1/ICR1 fragment A was ligated into pGEM-T EASY vector and was sequenced to confirm the proper mutation. The RIP1/ICR1 fragment B was amplified using primers 5′RIP1/ICR1M-NotI (5′-gcgcggccgcaaggcagcggatgctgcagcag-3’) and 3′RIP1/ICR1-SacI and then ligated into pGEM-T EASY followed by DNA sequencing to confirm the proper mutation. The pLAT52::YFP-RIP1/ICR1M plasmid was constructed by ligating the RIP1/ICR1 fragment A into pLAT52::YFP plasmid using enzyme sites BglII/NotI and followed by the ligation of fragment B using NotI/SacI enzyme site.
Expression of Recombinant Proteins
Coding sequences of RIP1/ICR1 RIP1/ICR1DE1and RIP1/ICR1M fragments were cut from pGEM RIP1/ICR1 pPGEM RIP1/ICR1DE1and pGEM RIP1/ICR1Mrespectively. These fragments were cloned into BamHI/EcoRI-digested pET30a vectors (Novagen, Madison, WI). The cDNAs were expressed in Escherichia coli BL21 (DE3) and each recombinant protein was incorporated a 6 His tag on the N-terminus. Bacterial cultures were grown in LB medium supplemented with 50 μg mL−1 of Kanamycin at 37°C until the culture reached a density of 0.6 measured at 600 nm. His-tagged protein was induced by the addition of 1 mM IPTG for 4 h. The bacteria were pelleted and re-suspended in protein extraction buffer (50 mM NaH2PO4pH 8.0, 300 mM NaCl, and 10 mM imidazole) containing protease inhibitors (1 mM Phenylmethylsulfonyl fluoride (PMSF), 10 mg mL−1 of leupeptin, and 10 mg mL−1 of pepstatin) and sonicated. The bacterial lysates were centrifuged at 10 000 g for 30 min and the supernatant was applied to columns containing Ni-NTA agarose resin (Qiagen). The columns were washed three times with protein extraction buffer containing 25 mM imidazole. The bound proteins were eluted with 500 mM imidazole and dialyzed overnight against distilled water.
Plant Growth Conditions and Plant Transformation
Arabidopsis (ecotype Columbia-0) was grown at 22°C with a 12-h light/12-h dark cycle in an environmentally controlled growth chamber. Tobacco (Nicotiana tabacum) plants were grown in growth chambers at 22°C under a light regimen of 12 h of darkness and 12 h of light. The pC1300LAT52::GFP-RIP1/ICR1 and pC130035S::GFP-RIP1/ICR1 constructs were transformed into Arabidopsis (Col-0) by floral dipping and transformants selected on half-strength MS agar medium containing 30 μg mL−1 hygromycin and 100 μg mL−1 carbenicillin (Sigma). Homozygous lines were determined via the resistance to hygromycin.
In-Vitro Arabidopsis Pollen Culture
Arabidopsis flowers were collected and pollen was dehydrated at 23°C for 2 h and then dipped onto semi-solid agar medium containing 1 mM MgSO42.5 mM Ca(NO3)22.5 mM CaCl20.01% boric acid, 18% sucrose, and 0.7% selected agar. To visualize the GFP fusion protein before germination, pollen samples were immediately checked after the dipping onto the medium. After 2 h of germination, pollen samples were used to visualize the GFP fusion protein during germination. To visualize the GFP fusion proteins during pollen tube growth, 6 h of germination were needed. For DAPI and GFP-RIP1/ICR1 co-localization experiments, the Arabidopsis pollen germination medium was supplemented with 1 μg mL−1 of DAPI.
In-Vitro ROP1 Binding Assay
We used RIP1/ICR1 and RIP1/ICR1 mutants fused with His at their N-terminus and ROPs fused with glutathione S-transferase (GST) for pull-down assays, as described previously (Wu et al., 2000). Approximately 10 μg of GST-ROP1 fusion proteins were bound to glutathione-conjugated agarose beads, and similar amounts of His-tagged RIP1/ICR1 or His-tagged RIP1/ICR1 mutant proteins were used in each assay. His-tagged proteins were detected using His probes and visualized with enhanced chemiluminescence (Sigma).
RIP1/ICR1 Antibody Production and Immunoblot Analysis
Purified His-RIP1/ICR1 protein was used to raise polyclonal antibodies in rabbits (Cocalico Biologicals, Inc., PA). The RIP1/ICR1 polyclonal antibody was affinity-purified using the recombinant MBP-RIC1 protein. To test the specificity of RIP1/ICR1 antibody, total protein was extracted using protein sample buffer (50 mM Tris-HCl, pH 6.8, 50 mM DTT, 4% SDS, 0.05% bromphenol blue, 10% glycerol) from 7-day-old Arabidopsis seedlings of wild-type and 35S::GFP-RIP1/ICR1 and fractionated on 10% SDS–PAGE and transferred to nitrocellulose membrane. Immunoblots incubated with RIP1/ICR1 antibody (1:5000), followed by horseradish peroxidase-conjugated anti-rabbit secondary antibody (1:10 000). Signals were developed on x-ray film (Hyperfilm, Amersham) with enhanced chemiluminescence kit (Sigma).
Isolation of the RIP1/ICR1-Containing Active ROP Complex by MBP-RIC1 Pull-Down
The MBP (maltose-binding protein) fused RIC1 protein was overexpressed in E. coli and purified (Wu et al., 2001). To purify active ROP complex from Arabidopsis7-day-old seedlings or open flowers (1g) were ground in liquid nitrogen, hydrated in 2 ml of extraction buffer (25 mM Hepes (pH 7.4), 10 mM MgCl2100 mM NaCl, 5 mM Sodium fluoride, 1 M PMSF, 10 μg mL−1 aprotinin (Sigma), 10 μg mL−1 leupeptin (Sigma), 1% (v/v) Triton X-100) and centrifuged at 13 000 g for10 min at 4°C. 10 μl of supernatant was conserved for ROP input control and the rest mixed with 2 ml of extraction buffer lacking Triton X-100 and MBP-RIC1 containing beads (about 5 μg MBP-RIC1), gently shaken for 1 h at 4°C and centrifuged at 1000 g for 3 min. The pellet was washed three times with 3 ml washing buffer (25 mM Hepes (pH 7.4), 1 mM EDTA, 5 mM MgCl21 mM DTT, 0.5% (v/v) Triton X-100). The liquid fraction was completely removed by a syringe and then the MBP-RIC1-bound active ROP complex was eluted by maltose. Proteins were fractionated by 12% SDS–PAGE and transferred to nitrocellulose membrane and blotted using the same conditions as described above.
Particle Bombardment-Mediated Transient Expression in Tobacco Pollen and Microscopy
Tobacco pollen grains were collected and used for transient expression using a procedure described previously (Fu et al., 2001). All plasmid DNAs were purified using Qiagen Plasmid Kits according to the manufacturer's instructions (Qiagen, Calencia, CA). For each bombardment, 1 μg of plasmid DNA was used to coat 0.5 mg gold particles. For co-transient expression, 1 μg of plasmid DNA of each construct was mixed and coated on 0.5 mg of gold particles. Bombarded pollen grains were washed into Petri dishes with 0.7 ml of tobacco pollen germination medium (1 mM MgSO45 mM Ca(NO3)25 mM CaCl2 boric acid, 18% sucrose). The pollen grains were incubated for 6 h before observation under a laser scanning confocal microscope with a Nikon Optiphot upright microscope equipped with a MRC 600 confocal laser scanning device (Bio-Rad, Hercules, CA). One-micrometer optical sections were scanned and captured using Comos software (Bio-Rad). For pollen tubes used for the measurement of tube length and width, pollen tube pictures were taken within a 30-min time period after 6 h incubation in germination medium. Confocal images were analyzed using MetaMorph version 4.5 and processed using Photoshop version 5.0 (Adobe Systems, Mountain View, CA). For the GFP-RIP1/ICR1M and DAPI co-localization experiment, tobacco pollen germination was supplemented with 1 μg mL−1 of DAPI.
FRET Analysis
FRET analysis was performed using a Leica confocal TCS SP2 microscope equipped with a He-Cd laser (specially used for CFP excitation at 442-nm wavelength) and an Argon laser (provides excitation at 514 nm for YFP). Filters for these experiments were: CFP-ROP1/CFP-DN-ROP1/CFP-CA-ROP1 (excitation 442 nm, emission 450–490 nm); YFP-RIP1/ICR1 (excitation 514 nm, emission 525–600 nm); FRET (excitation 422 nm, emission 560–638 nm). CFP and YFP images were acquired simultaneously and FRET images were acquired separately at the same focal section. Only cells expressing similar levels of CFP-ROP and YFP-RIP1/ICR1 were selected for FRET analysis.
SUPPLEMENTARY DATA
Supplementary Data are available at Molecular Plant Online.
FUNDING
This work is supported by grants from Department of Energy (DE-FG02-04ER15555) and National Science Foundation (MCB0111082) to Z.Y.
ACKNOWLEDGMENTS
We thank members of the Yang lab and Lord lab for their stimulating discussion. No conflict of interest declared.
Footnotes
Published by the Molecular Plant Shanghai Editorial Office in association with Oxford University Press on behalf of CSPP and IPPE, SIBS, CAS.
Supplementary Material
Supplementary Movie
Supplementary Figure
REFERENCES
- Baxter-Burrell A., Yang Z.B., Springer P.S., Bailey-Serres J. RopGAP4-dependent Rop GTPase rheostat control of Arabidopsis oxygen deprivation tolerance. Science. 2002;296:2026–2028. doi: 10.1126/science.1071505. [DOI] [PubMed] [Google Scholar]
- Berken A., Thomas C., Wittinghofer A. A new family of RhoGEFs activates the Rop molecular switch in plants. Nature. 2005;436:1176–1180. doi: 10.1038/nature03883. [DOI] [PubMed] [Google Scholar]
- Bischoff F., Vahlkamp L., Molendijk A., Palme K. Localization of AtROP4 and AtROP6 and interaction with the guanine nucleotide dissociation inhibitor AtRhoGDI1 from Arabidopsis. Plant Mol. Biol. 2000;42:515–530. doi: 10.1023/a:1006341210147. [DOI] [PubMed] [Google Scholar]
- Butty A.C., et al. A positive feedback loop stabilizes the guanine-nucleotide exchange factor Cdc24 at sites of polarization. EMBO J. 2002;21:1565–1576. doi: 10.1093/emboj/21.7.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carol R.J., et al. A RhoGDP dissociation inhibitor spatially regulates growth in root hair cells. Nature. 2005;438:1013–1016. doi: 10.1038/nature04198. [DOI] [PubMed] [Google Scholar]
- Chen C.Y., Cheung A.Y., Wu H.M. Actin-depolymerizing factor mediates Rac/Rop GTPase-regulated pollen tube growth. Plant Cell. 2003;15:237–249. doi: 10.1105/tpc.007153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung A.Y., Chen C.Y.H., Tao L.Z., Andreyeva T., Twell D., Wu H.M. Regulation of pollen tube growth by Rac-like GTPases. J. Exp. Bot. 2003;54:73–81. doi: 10.1093/jxb/erg044. [DOI] [PubMed] [Google Scholar]
- Cole R.A., Fowler J.E. Polarized growth: maintaining focus on the tip. Curr. Opin. Plant Biol. 2006;9:579–588. doi: 10.1016/j.pbi.2006.09.014. [DOI] [PubMed] [Google Scholar]
- Cole R.A., Synek L., Zarsky V., Fowler J.E. SEC8, a subunit of the putative Arabidopsis exocyst complex, facilitates pollen germination and competitive pollen tube growth. Plant Physiol. 2005;138:2005–2018. doi: 10.1104/pp.105.062273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowd P.E., Coursol S., Skirpan A.L., Kao T.H., Gilroy S. Petunia phospholipase c1 is involved in pollen tube growth. Plant Cell. 2006;18:1438–1453. doi: 10.1105/tpc.106.041582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etienne-Manneville S., Hall A. Rho GTPases in cell biology. Nature. 2002;420:629–635. doi: 10.1038/nature01148. [DOI] [PubMed] [Google Scholar]
- Feijo J.A., Costa S.S., Prado A.M., Becker J.D., Certal A.C. Signalling by tips. Curr. Opin. Plant Biol. 2004;7:589–598. doi: 10.1016/j.pbi.2004.07.014. [DOI] [PubMed] [Google Scholar]
- Feijo J.A., Sainhas J., Holdaway-Clarke T., Cordeiro M.S., Kunkel J.G., Hepler P.K. Cellular oscillations and the regulation of growth: the pollen tube paradigm. Bioessays. 2001;23:86–94. doi: 10.1002/1521-1878(200101)23:1<86::AID-BIES1011>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Fu Y., Wu G., Yang Z. Rop GTPase-dependent dynamics of tip-localized F-actin controls tip growth in pollen tubes. J. Cell Biol. 2001;152:1019–1032. doi: 10.1083/jcb.152.5.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y., et al. A Rho family GTPase controls actin dynamics and tip growth via two counteracting downstream pathways in pollen tubes. J. Cell Biol. 2005;169:127–138. doi: 10.1083/jcb.200409140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y., Li S., Lord E.M., Yang Z. Members of a novel class of Arabidopsis Rho guanine nucleotide exchange factors control Rho GTPase-dependent polar growth. Plant Cell. 2006;18:366–381. doi: 10.1105/tpc.105.036434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y., Vernoud V., Fu Y., Yang Z.B. ROP GTPase regulation of pollen tube growth through the dynamics of tip-localized F-actin. J. Exp. Bot. 2003;54:93–101. doi: 10.1093/jxb/erg035. [DOI] [PubMed] [Google Scholar]
- Gu Y., Wang Z., Yang Z. ROP/RAC GTPase: an old new master regulator for plant signaling. Curr. Opin. Plant Biol. 2004;7:527–536. doi: 10.1016/j.pbi.2004.07.006. [DOI] [PubMed] [Google Scholar]
- Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- Helling D., Possart A., Cottier S., Klahre U., Kost B. Pollen tube tip growth depends on plasma membrane polarization mediated by tobacco PLC3 activity and endocytic membrane recycling. Plant Cell. 2006;18:3519–3534. doi: 10.1105/tpc.106.047373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepler P.K., Vidali L., Cheung A.Y. Polarized cell growth in higher plants. Annu. Rev. Cell Dev. Biol. 2001;17:159–187. doi: 10.1146/annurev.cellbio.17.1.159. [DOI] [PubMed] [Google Scholar]
- Hu W., Wang Y., Bowers C., Ma H. Isolation, sequence analysis, and expression studies of florally expressed cDNAs in Arabidopsis. Plant Mol. Biol. 2003;53:545–563. doi: 10.1023/B:PLAN.0000019063.18097.62. [DOI] [PubMed] [Google Scholar]
- Hwang J.U., Gu Y., Lee Y.J., Yang Z. Oscillatory ROP GTPase activation leads the oscillatory polarized growth of pollen tubes. Mol. Biol. Cell. 2005;16:5385–5399. doi: 10.1091/mbc.E05-05-0409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M.A., Shen J.J., Fu Y., Li H., Yang Z., Grierson C.S. The Arabidopsis Rop2 GTPase is a positive regulator of both root hair initiation and tip growth. Plant Cell. 2002;14:763–776. doi: 10.1105/tpc.010359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klahre U., Kost B. Tobacco RhoGTPase ACTIVATING PROTEIN1 spatially restricts signaling of RAC/Rop to the apex of pollen tubes. Plant Cell. 2006;18:3033–3046. doi: 10.1105/tpc.106.045336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klahre U., Becker C., Schmitt A.C., Kost B. Nt-RhoGDI2 regulates Rac/Rop signaling and polar cell growth in tobacco pollen tubes. Plant J. 2006;46:1018–1031. doi: 10.1111/j.1365-313X.2006.02757.x. [DOI] [PubMed] [Google Scholar]
- Kost B. Spatial control of Rho (Rac-Rop) signaling in tip-growing plant cells. Trends Cell Biol. 2008;18:119–127. doi: 10.1016/j.tcb.2008.01.003. [DOI] [PubMed] [Google Scholar]
- Kost B., et al. Rac homologues and compartmentalized phosphatidylinositol 4, 5-bisphosphate act in a common pathway to regulate polar pollen tube growth. J. Cell Biol. 1999;145:317–330. doi: 10.1083/jcb.145.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krichevsky A., Kozlovsky S.V., Tian G.W., Chen M.H., Zaltsman A., Citovsky V. How pollen tubes grow. Dev. Biol. 2007;303:405–420. doi: 10.1016/j.ydbio.2006.12.003. [DOI] [PubMed] [Google Scholar]
- Kusano H., et al. The Arabidopsis phosphatidylinositol phosphate 5-kinase PIP5K3 is a key regulator of root hair tip growth. Plant Cell. 2008;15:15. doi: 10.1105/tpc.107.056119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavy M., et al. A novel ROP/RAC effector links cell polarity, root-meristem maintenance, and vesicle trafficking. Curr. Biol. 2007;17:947–952. doi: 10.1016/j.cub.2007.04.038. [DOI] [PubMed] [Google Scholar]
- Li H., Lin Y., Heath R.M., Zhu M.X., Yang Z. Control of pollen tube tip growth by a Rop GTPase-dependent pathway that leads to tip-localized calcium influx. Plant Cell. 1999;11:1731–1742. doi: 10.1105/tpc.11.9.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Wu G., Ware D., Davis K.R., Yang Z. Arabidopsis Rho-related GTPases: differential gene expression in pollen and polar localization in fission yeast. Plant Physiol. 1998;118:407–417. doi: 10.1104/pp.118.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y., Yang Z. Inhibition of pollen tube elongation by microinjected anti-Rop1Ps antibodies suggests a crucial role for Rho-Type GTPases in the control of tip growth. Plant Cell. 1997;9:1647–1659. doi: 10.1105/tpc.9.9.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y., Wang Y., Zhu J.K., Yang Z. Localization of a Rho GTPase implies a role in tip growth and movement of the generative cell in pollen tubes. Plant Cell. 1996;8:293–303. doi: 10.1105/tpc.8.2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messerli M.A., Creton R., Jaffe L.F., Robinson K.R. Periodic increases in elongation rate precede increases in cytosolic Ca2+ during pollen tube growth. Dev. Biol. 2000;222:84–98. doi: 10.1006/dbio.2000.9709. [DOI] [PubMed] [Google Scholar]
- Molendijk A.J., et al. Arabidopsis thaliana Rop GTPases are localized to tips of root hairs and control polar growth. EMBO J. 2001;20:2779–2788. doi: 10.1093/emboj/20.11.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molendijk A.J., Ruperti B., Palme K. Small GTPases in vesicle trafficking. Curr. Opin. Plant Biol. 2004;7:694–700. doi: 10.1016/j.pbi.2004.09.014. [DOI] [PubMed] [Google Scholar]
- Nern A., Arkowitz R.A. Nucleocytoplasmic shuttling of the Cdc42p exchange factor Cdc24p. J. Cell Biol. 2000;148:1115–1122. doi: 10.1083/jcb.148.6.1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson K.R., Messerli M.A. Pulsating ion fluxes and growth at the pollen tube tip. Sci. STKE. 2002;2002:PE51. doi: 10.1126/stke.2002.162.pe51. [DOI] [PubMed] [Google Scholar]
- Samaj J., Muller J., Beck M., Bohm N., Menzel D. Vesicular trafficking, cytoskeleton and signalling in root hairs and pollen tubes. Trends Plant Sci. 2006;11:594–600. doi: 10.1016/j.tplants.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Schmitz A.A.P., Govek E.E., Bottner B., Van Aelst L. Rho GTPases: signaling, migration, and invasion. Exp. Cell Res. 2000;261:1–12. doi: 10.1006/excr.2000.5049. [DOI] [PubMed] [Google Scholar]
- Sorek N., Poraty L., Sternberg H., Bar E., Lewinsohn E., Yalovsky S. Activation status-coupled transient S acylation determines membrane partitioning of a plant Rho-related GTPase. Mol. Cell Biol. 2007;27:2144–2154. doi: 10.1128/MCB.02347-06. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Staiger C.J. Signaling to the actin cytoskeleton in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2000;51:257–288. doi: 10.1146/annurev.arplant.51.1.257. [DOI] [PubMed] [Google Scholar]
- Stenzel I., et al. The type B phosphatidylinositol-4-phosphate 5-kinase 3 is essential for root hair formation in Arabidopsis thaliana. Plant Cell. 2008;20:124–141. doi: 10.1105/tpc.107.052852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda S., Gapper C., Kaya H., Bell E., Kuchitsu K., Dolan L. Local positive feedback regulation determines cell shape in root hair cells. Science. 2008;319:1241–1244. doi: 10.1126/science.1152505. [DOI] [PubMed] [Google Scholar]
- Toenjes K.A., Sawyer M.M., Johnson D.I. The guanine-nucleotide-exchange factor Cdc24p is targeted to the nucleus and polarized growth sites. Curr. Biol. 1999;9:1183–1186. doi: 10.1016/S0960-9822(00)80022-6. [DOI] [PubMed] [Google Scholar]
- Wedlich-Soldner R., Altschuler S., Wu L., Li R. Spontaneous cell polarization through actomyosin-based delivery of the Cdc42 GTPase. Science. 2003;299:1231–1235. doi: 10.1126/science.1080944. [DOI] [PubMed] [Google Scholar]
- Wedlich-Soldner R., Wai S.C., Schmidt T., Li R. Robust cell polarity is a dynamic state established by coupling transport and GTPase signaling. J. Cell Biol. 2004;166:889–900. doi: 10.1083/jcb.200405061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Gu Y., Li S.D., Yang Z.B. A genome-wide analysis of Arabidopsis Rop-interactive CRIB motif-containing proteins that act as Rop GTPase targets. Plant Cell. 2001;13:2841–2856. doi: 10.1105/tpc.010218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Li H., Yang Z.B. Arabidopsis RopGAPs are a novel family of Rho GTPase-activating proteins that require the Cdc42/Rac-interactive binding motif for Rop-specific GTPase stimulation. Plant Physiol. 2000;124:1625–1636. doi: 10.1104/pp.124.4.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. Signaling tip growth in plants. Curr. Opin. Plant Biol. 1998;1:525–530. doi: 10.1016/s1369-5266(98)80046-0. [DOI] [PubMed] [Google Scholar]
- Yang Z. Small GTPases: versatile signaling switches in plants. Plant Cell. 2002;14:S375–S388. doi: 10.1105/tpc.001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. Cell polarity signaling in Arabidopsis. Annu. Rev. Cell Dev. Biol. 2008 doi: 10.1146/annurev.cellbio.23.090506.123233. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Fu Y. ROP/RAC GTPase signaling. Curr. Opin. Plant Biol. 2007;10:490–494. doi: 10.1016/j.pbi.2007.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon G.M., Dowd P.E., Gilroy S., McCubbin A.G. Calcium-dependent protein kinase isoforms in Petunia have distinct functions in pollen tube growth, including regulating polarity. Plant Cell. 2006;18:867–878. doi: 10.1105/tpc.105.037135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., McCormick S. A distinct mechanism regulating a pollen-specific guanine nucleotide exchange factor for the small GTPase Rop in Arabidopsis thaliana. Proc. Natl Acad. Sci. U S A. 2007;104:18830–18835. doi: 10.1073/pnas.0705874104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Movie
Supplementary Figure