SUMMARY
Nuclear clearance of the RNA-binding protein TDP-43 is a hallmark of neurodegeneration and an important therapeutic target. Our current understanding of TDP-43 nucleocytoplasmic transport does not fully explain its predominantly nuclear localization or mislocalization in disease. Here, we show that TDP-43 exits nuclei by passive diffusion, independent of facilitated mRNA export. RNA polymerase II blockade and RNase treatment induce TDP-43 nuclear efflux, suggesting that nuclear RNAs sequester TDP-43 in nuclei and limit its availability for passive export. Induction of TDP-43 nuclear efflux by short, GU-rich oligomers (presumably by outcompeting TDP-43 binding to endogenous nuclear RNAs), and nuclear retention conferred by splicing inhibition, demonstrate that nuclear TDP-43 localization depends on binding to GU-rich nuclear RNAs. Indeed, RNA-binding domain mutations markedly reduce TDP-43 nuclear localization and abolish transcription blockade-induced nuclear efflux. Thus, the nuclear abundance of GU-RNAs, dictated by the balance of transcription, pre-mRNA processing, and RNA export, regulates TDP-43 nuclear localization.
Graphical Abstract
In brief
Duan et al. demonstrate that TDP-43 nuclear export occurs by passive diffusion through nuclear pore channels and is restricted by nuclear GU-rich RNA binding. Processes that modulate nuclear RNA abundance or TDP-43-RNA binding—such as transcription, splicing, and mRNA export—regulate TDP-43 nuclear localization and availability for export.
INTRODUCTION
Transactive response DNA binding protein 43 kDa (TDP-43) is an essential DNA/RNA-binding protein that plays a major role in RNA processing and stability (reviewed in Prasad et al., 2019; François-Moutal et al., 2019). TDP-43 nuclear clearance and cytoplasmic aggregation are pathological hallmarks of multiple neurodegenerative diseases, including amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) (Neumann et al., 2006; Arai et al., 2006). Substantial evidence links TDP-43 disruption to ALS and FTD pathogenesis via loss of nuclear splicing regulation (Polymenidou et al., 2011; Tollervey et al., 2011; Ling et al., 2015) and the toxic effects of cytoplasmic aggregates (reviewed in Vanden Broeck et al., 2014; Prasad et al., 2019). Rare TDP-43 mutations in families with inherited ALS (Kabashi et al., 2008; Sreedharan et al., 2008; Deerlin et al., 2008; Gitcho et al., 2008) and FTD (Borroni et al., 2009; Kovacs et al., 2009) cause neurodegeneration in disease models (reviewed in Buratti, 2015), supporting a role for TDP-43 disruption in disease. However, most ALS cases with TDP-43 cytoplasmic mislocalization are not associated with TDP-43 mutations, indicating that diverse genetic, environmental, or age-related causes—possibly all involved in the same underlying process-drive TDP-43 nuclear clearance. However, the initiating cause of TDP-43 mislocalization in disease remains unknown. Moreover, the regulation of TDP-43 nuclear localization in healthy cells is incompletely understood.
TDP-43 is a member of the heterogeneous ribonucleoprotein (hnRNP) family of RNA-binding proteins (RBPs) that contains an N-terminal nuclear localization signal (NLS), two RNA recognition motifs (RRM1 and RRM2), and a C-terminal intrinsically disordered region (IDR) (reviewed in François-Moutal et al., 2019). RNA crosslinking and immunoprecipitation-sequencing (CLIP-seq) studies show that TDP-43 preferentially binds GU-rich RNA motifs, particularly in introns (Polymenidou et al., 2011; Tollervey et al., 2011), consistent with its role in regulating alternative splicing and repression of cryptic exons (Ling et al., 2015). Like many hnRNPs, TDP-43 shuttles between the nucleus and cytoplasm (Ayala et al., 2008). Ran-regulated active TDP-43 nuclear import occurs via NLS binding to importins α and β (Ayala et al., 2008; Nishimura et al., 2010). Modest passive import also likely occurs, based on size and the low level of persistent nuclear entry of TDP-43-ΔNLS (Ayala et al., 2008; Winton et al., 2008; TDP-43 nuclear export was initially thought to occur via the exportin-1 (XPO1) receptor and a putative nuclear export signal (NES) in RRM2 (Winton et al., 2008). However, NES deletions do not disrupt TDP-43 export, nor does XPO1 knockdown or selective inhibitors of nuclear export (Ederle et al., 2018; Pinarbasi et al., 2018; Archbold et al., 2018). Adding 54- to 119-kDa tags to TDP-43 strongly slowed its export, as expected for passive export rather than an energy-dependent, transport receptor-facilitated process (Pinarbasi et al., 2018; Ederle et al., 2018). Meanwhile, transcriptional blockade (Ayala et al., 2008; Ederle et al., 2018) and overexpression of the mRNA export receptor NXF1 (Archbold et al., 2018) promote TDP-43 nuclear efflux, suggesting a role for nuclear RNA in mediating TDP-43 nuclear localization and raising the possibility of TDP-43 co-export with mRNA, perhaps via the TREX (TRanscription/EXport) pathway (discussed in Ederle and Dormann, 2017). Arguing against RNA co-export, small interfering RNA (siRNA) knockdown of AlyREF, a TREX complex protein that recruits NXF1, did not promote TDP-43 nuclear accumulation (Archbold et al., 2018; Ederle et al., 2018).
Here, we aimed to elucidate the mechanism of TDP-43 nuclear export and test the hypothesis that TDP-43 nuclear localization depends on its binding to nuclear RNAs. We found that RNase-mediated nuclear RNA degradation in permeabilized cells markedly disrupted TDP-43 nuclear sequestration, allowing TDP-43 to diffuse from the nucleus in low-ATP conditions. Moreover, in permeabilized and live cells, the introduction of GU-rich oligomers induced TDP-43 nuclear efflux, likely by competitive dissociation from endogenous nuclear RNAs. Splicing inhibition caused dose-dependent TDP-43 nuclear accumulation and abolished transcriptional blockade-induced nuclear exit, further demonstrating that nuclear localization of TDP-43 depends on association with pre-mRNAs. Together, our results indicate that binding to GU-rich RNAs retains TDP-43 within nuclei and controls its availability for passive nuclear export.
RESULTS
NVP2-induced RNA pol II inhibition promotes TDP-43 nuclear export
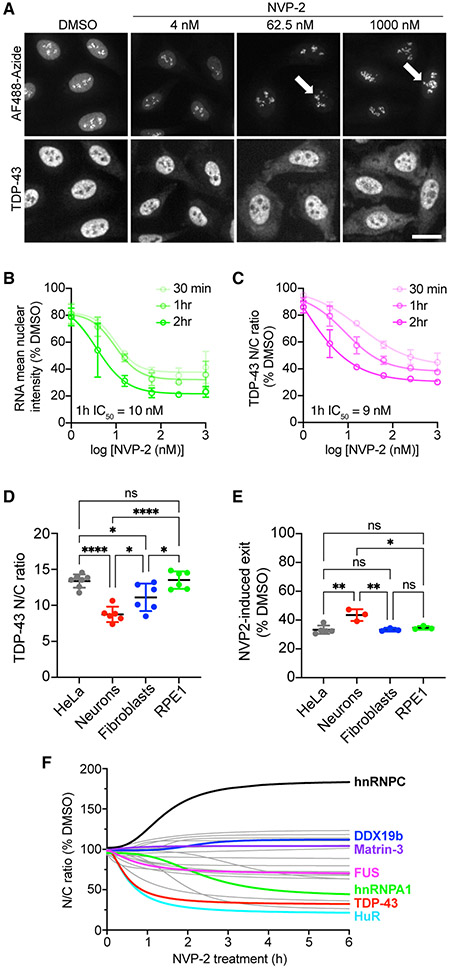
Actinomycin D (ActD), a DNA-intercalator and pan-transcriptional inhibitor, induces the nuclear efflux of nuclear RBPs including hnRNPA1 (Piñol-Roma and Dreyfuss, 1992), SR proteins (Cáceres et al., 1998), and TDP-43 (Ayala et al., 2008; Ederle et al., 2018). To confirm that TDP-43 nuclear localization is sensitive to inhibition of mRNA synthesis and avoid off-target effects of DNA intercalation, we treated HeLa cells with NVP2, an inhibitor of CDK9-dependent RNA pol II activation (Olson et al., 2018), and monitored TDP-43 localization by immunofluorescence (IF) (Figures 1A-1C). RNA synthesis was analyzed via a 30-min pulse of 5-ethynyl-uridine (5-EU) before fixation, followed by click-chemistry labeling of nascent RNAs (Jao and Salic, 2008). Cells were imaged with an automated high-content confocal microscope, and a translocation algorithm was used to quantify the background-corrected fluorescence intensity in the nuclear and cytoplasmic compartments (Hayes et al., 2020). Like ActD (Figures S1A-S1C), NVP2 caused progressive, dose-dependent inhibition of RNA synthesis (Figures 1A and 1B) but spared nucleolar ribosomal RNA (rRNA) puncta resulting from RNA pol I activity (Figure 1A, arrows) (Sharifi and Bierhoff, 2018). In parallel, NVP2 induced progressive TDP-43 nuclear efflux, as measured by a decrease in the nuclear to cytoplasmic (N/C) ratio (Figures 1A and 1C). Importantly, the NVP2- and ActD-induced drop in the TDP-43 N/C ratio corresponded to decreased nuclear and increased cytoplasmic intensity (Figures S1D and S1E), consistent with N/C translocation. In addition to HeLa cells, NVP2-induced TDP-43 nuclear efflux was also observed in mouse primary cortical neurons, HFF1 human primary fibroblasts, and immortalized human retinal pigment epithelial cells (RPE1) (Figures 1D, 1E, and S2), demonstrating conservation across cell types. Despite the lower steady-state TDP-43 N/C in neurons (Figure 1D), NVP2 still induced marked neuronal TDP-43 nuclear efflux (Figures 1E and S2).
Figure 1. NVP2-induced RNA pol II inhibition promotes TDP-43 nuclear export.
(A) Nascent RNA labeled with AF488-picolyl azide via 5-EU/click chemistry (top) and TDP-43 IF (bottom) in HeLa cells treated with NVP2 for 1 h. Arrows indicate rRNA puncta unaffected by NVP2. Scale bar, 25 μm.
(B and C) Nascent RNA (AF488) mean nuclear intensity (B) and TDP-43 nuclear/cytoplasmic ratio (N/C) (C) expressed as percentage of DMSO control. Mean ± SD is shown for >2,000 cells/group in each of three biological replicates. IC50 at 1 h was calculated by non-linear regression. See also Figure S1.
(D) Steady-state TDP-43 N/C in HeLa cells, mouse primary cortical neurons, human HFF1 fibroblasts, and human retinal pigment epithelial (RPE1) cells fixed and immunostained with mouse monoclonal anti-TDP-43. Mean ± SD is shown for 6–7 biological replicates. See also Figure S2.
(E) NVP2-induced shift in TDP-43 N/C, shown as percentage of DMSO control, following 3 h with 250 nM NVP2. Mean ± SD is shown for 3–5 biological replicates.
(F) RBP N/C in HeLa cells treated with 250 nM NVP2 for 30 min to 6 h. Curves were fit by non-linear regression using the mean of 3–4 biological replicates per RBP. See also Figure S3.
In (D), (E), and (F), >2,000 cells/replicate for cell lines and 60–80 cells/replicate for neurons. ns, not significant; *p < 0.05, **p < 0.01, ****p < 0.0001 by one-way ANOVA with Tukey’s test.
Next, we compared the effect of NVP2 on the localization of TDP-43 versus a panel of RBPs, including hnRNPs, RNA export, exon-junction complex (EJC), and splicing-associated proteins (Figures 1D and S3). HeLa cells were treated with NVP2 for 30 min to 6 h, fixed, and RBP localization analyzed by IF and high-content analysis. RBP responses were diverse, including rapid nuclear efflux (TDP-43, HuR), slow nuclear efflux (FUS, hnRNPA1), and no change (Matrin-3). DDX19b, an ATP-dependent RNA helicase in the mRNA export pathway that is tethered to the nuclear pore complex (NPC) (Napetschnig et al., 2009; Hodge et al., 2011), was also unchanged. hnRNPC, which does not exhibit N/C shuttling (Piñol-Roma and Dreyfuss, 1992; Ederle et al., 2018), showed nuclear accumulation. Interestingly, TDP-43 was among the most highly responsive RBPs to transcriptional blockade, suggesting strict dependence on nascent nuclear RNA levels.
TDP-43 exits the nucleus by passive diffusion
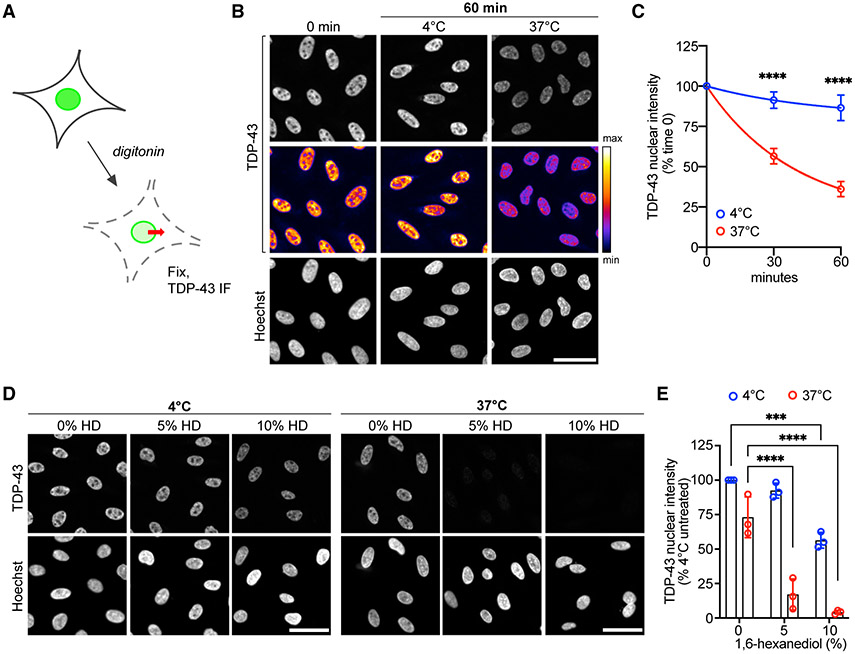
Recent studies showed that TDP-43 nuclear export is independent of XPO1 (Archbold et al., 2018; Pinarbasi et al., 2018; Ederle et al., 2018) and strongly size limited, as expected for a passive export cargo (Pinarbasi et al., 2018; Ederle et al., 2018). Consistent with this, we found that HeLa cells transiently transfected with TDP43-YFP (+27 kDa) showed a marked increase in the steady-state TDP-43 N/C ratio, and the YFP tag strongly inhibited NVP2-induced TDP-43 nuclear efflux (Figure S4A). To investigate the passive versus active mechanism of TDP-43 nuclear export, we next developed a permeabilized cell TDP-43 export assay (Figure 2A). Permeabilized cell assays are widely used to study N/C transport and permit a broad range of experimental perturbations (Adam et al., 1990; Cassany and Gerace, 2009). Digitonin is used to selectively perforate the plasma membrane (Colbeau et al., 1971; Adam et al., 1990), releasing the cytoplasm and leaving the nuclear membrane and NPCs intact for passive or energy-dependent transport (Hayes et al., 2020). Following permeabilization, HeLa cells were incubated in an energy-free buffer and fixed at regular intervals to analyze the localization of endogenous TDP-43 by IF (Figure 2B). Nuclear envelope and NPC integrity were monitored by verifying nuclear restriction of a 70-kDa dextran (Figure S4B), relying on the progressive exclusion of cargoes >30–60 kDa by the NPC permeability barrier (Mohr et al., 2009; Timney et al., 2016). Luciferase reporter-based ATP measurements showed that ATP content dropped by 97% immediately after permeabilization (Figure S4C), confirming low ATP levels in the permeabilized cell system.
Figure 2. TDP-43 exits the nucleus by passive diffusion in permeabilized cells.
(A) Permeabilized cell TDP-43 passive export assay. See also Figure S4.
(B) TDP-43 IF (top) and Hoechst (bottom) in HeLa cells fixed immediately (time 0) or 60 min post permeabilization, following incubation at 4°C or 37°C. The intensity histogram for TDP-43 images was normalized to time 0, prior to pseudo-color look-up table (LUT) (middle). Scale bar, 50 μm.
(C) TDP-43 integrated nuclear intensity in permeabilized HeLa cells normalized to time 0. Mean ± SD is shown for >2,000 cells/well in five biological replicates.
(D) TDP-43 IF (top) and Hoechst (bottom) in permeabilized HeLa cells incubated for 30 min at 4°C versus 37°C with 1,6-HD. The intensity histogram for TDP-43 images was normalized to untreated cells at 4°C. Scale bar, 50 μm.
(E) TDP-43 integrated nuclear intensity in permeabilized HeLa cells incubated for 30 min at 4°C versus 37°C with 1,6-HD, normalized to untreated cells at 4°C. Mean ± SD is shown for >2,000 cells/well in three biological replicates.
In (C) and (E), ***p < 0.001, ****p < 0.0001 by two-way ANOVA with Tukey’s test.
Next, we identified conditions permissive of TDP-43 nuclear export. After plasma membrane permeabilization, with no added transport receptors or ATP, we saw a progressive decrease in nuclear TDP-43 intensity, to 36% at 1 h, in cells heated to 37°C (Figures 2B and 2C). There was little change in nuclear TDP-43 intensity over the same time when cells were permeabilized and kept at 4°C. Thus, while TDP-43 can passively leave the nucleus in low-ATP conditions at 37°C, temperature-sensitive mechanism(s) hinder its nuclear exit at 4°C, beyond the expected minor temperature effects on free diffusion (Soh et al., 2010). The behavior of TDP-43 was in contrast to that of DEK, a chromatin-bound EJC protein (Waldmann et al., 2004). DEK showed no shift in localization after transcriptional blockade (Figure S3) and remained localized to the nucleus in permeabilized cells even after 1 h at 37°C (Figures S4D and S4E).
To further test the passive nature of TDP-43 export, we added 1,6-hexanediol (1,6-HD), an aliphatic alcohol that reversibly disrupts the phenylalanine-glycine (FG) permeability barrier lining the central channel of the NPC (Ribbeck and Görlich, 2002). 1,6-HD caused a marked, dose-dependent acceleration of TDP-43 nuclear efflux, suggesting that NPC permeability is rate limiting for TDP-43 export (Figures 2D and 2E). Interestingly, 1,6-HD accelerated TDP-43 nuclear exit at 37°C but also at 4°C, suggesting that the FG barrier is temperature sensitive and contributes, at least in part, to the hindrance of TDP-43 passive export at 4°C. 1,6-HD also reversibly inhibits TDP-43 liquid-liquid phase separation (LLPS) (Gopal et al., 2017; Mann et al., 2019). If LLPS affects TDP-43 availability for diffusion across the NPC, disruption of nuclear TDP-43 condensates may also contribute to 1,6-HD-induced acceleration of TDP-43 export. Next, we tested the effect of adding ATP and saw no change in TDP-43 export at concentrations far above those that support active nuclear transport in permeabilized cells (Cassany and Gerace, 2009; Hayes et al., 2020, 2021) (Figure S4F). To confirm that the data are not confounded by TDP-43 re-import into the nucleus, we tested the ability of permeabilized cells to support active import. Although no import receptors or Ran cycle proteins were added to the assay, residual importins remain at NPCs upon cell permeabilization and could conceivably bind and import cargo (Kapinos et al., 2017). We analyzed the capacity for nuclear import of Rango, a Förster resonance energy transfer sensor and importin β cargo (Kalab et al., 2006; Hayes et al., 2020), and saw no import in our lysate-free conditions, with or without added ATP (Figure S4G). Therefore, our findings are unlikely to be confounded by the nuclear re-entry of TDP-43. These data demonstrate that TDP-43 nuclear efflux in permeabilized cells is accelerated by heat and 1,6-HD, but not ATP, consistent with passive diffusion from the nucleus.
TDP-43 export is independent of NXF1-mediated mRNA export
The observation that TDP-43 exits permeabilized cell nuclei under passive conditions suggests that energy-dependent RNA export pathways are unlikely to be required for TDP-43 export. These include the exportin family of receptors, which rely on Ran-GTPase (e.g., XPO1, XPOT, and XPO5), and the TREX bulk mRNA export pathway, which is independent of Ran but requires ATP-dependent RNA helicases (including UAP56 and DDX19b) for mRNP export complex assembly and disassembly (Okamura et al., 2015). siRNA knockdown of XPO1, XPO5, XPOT, NXF1, and AlyREF did not promote TDP-43 nuclear accumulation in previous studies, further suggesting that canonical RNA export pathways are not involved in TDP-43 nuclear export (Archbold et al., 2018; Ederle et al., 2018). Similarly, TDP-43 RRM mutations did not prevent nuclear export in the heterokaryon assay (Ederle et al., 2018). However, XPO1, XPO7, and NXF1 overexpression promoted TDP-43 cytoplasmic mislocalization (Archbold et al., 2018), and NXF1 was identified as a potential TDP-43 interactor (Freibaum et al., 2010).
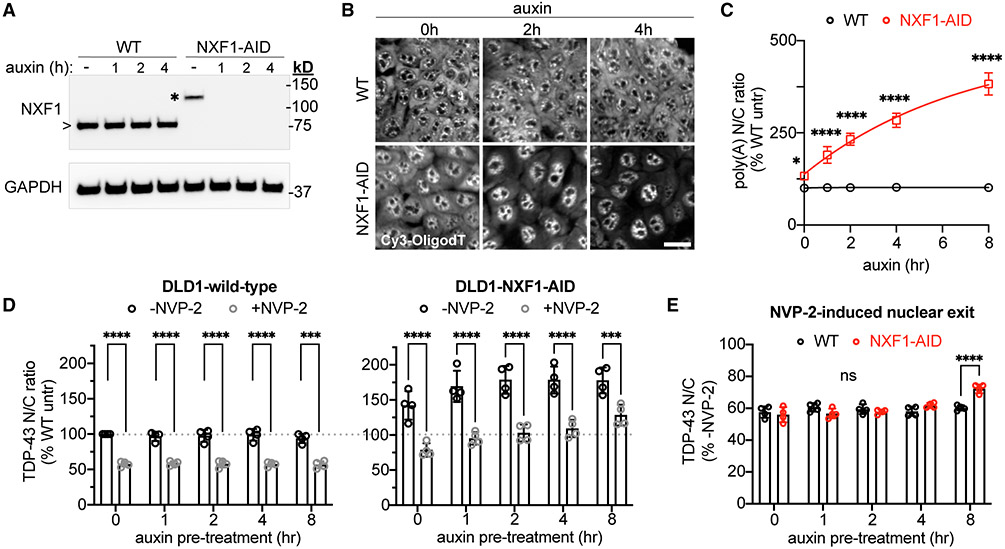
To further investigate the role of the NXF1/TREX pathway in TDP-43 nuclear export, we utilized a DLD1 cell line containing an auxin-inducible degron (AID) tag integrated at the endogenous NXF1 locus (Aksenova et al., 2020). TIR1 ligase, which drives ubiquitination of AID-tagged proteins upon auxin-mediated recruitment, was inserted at the C terminus of the nuclear protein RCC1 via a self-cleavable P2A sequence. Rapid degradation of the NXF1 protein occurs in this cell line within 1 h of auxin treatment (Figure 3A). Due to TIR1 leakage, DLD1-NXF1-AID cells showed mildly reduced NXF1 expression even before auxin treatment (Figure 3A, asterisk). To examine the effect of NXF1 ablation on mRNA export, DLD1-wild-type and -NXF1-AID cells were treated with auxin for 0–8 h and fixed, and polyA-RNA N/C localization was analyzed by fluorescence in situ hybridization (FISH) (Figures 3B and 3C). DLD1-NXF1-AID cells showed a ~30% increase in the polyA-RNA N/C ratio even without auxin (p < 0.05 versus DLD1-wild-type), consistent with the reduced NXF1 expression. Auxin induced further, progressive accumulation of nuclear polyA-RNA to >300% of baseline by 8 h, consistent with mRNA export inhibition (Figures 3B and 3C), together with a dose-dependent increase in the steady-state TDP-43 N/C ratio, presumably due to TDP-43 binding to the nuclear RNAs marked by the polyA-FISH signal (Figure 3D). To determine the effect of NXF1 ablation on transcriptional blockade-induced TDP-43 export, we treated cells with auxin for 0–8 h, followed by 2 h ± NVP2 before fixation and TDP-43 IF (Figures 3D and 3E). NVP2 still induced robust TDP-43 nuclear export in DLD1-NXF1-AID cells at all time points. When adjusted for the increased steady-state TDP-43 N/C ratio, there was no alteration in NVP2-induced TDP-43 export until 8 h, when subtle slowing was observed (Figure 3E). Thus, inhibiting mRNA export promotes TDP-43 nuclear accumulation, but the NXF1 receptor does not mediate TDP-43 export across the NPC.
Figure 3. Acute NXF1 ablation does not alter NVP2-induced TDP-43 nuclear export.
(A) NXF1 immunoblot in DLD1-wild-type versus NXF1-AID cells treated with 0.5 mM auxin for 0–4 h. Note the increased molecular weight for AID-tagged (*)versus endogenous NXF1 (>).
(B) PolyA-FISH (Cy3-OligodT(45)) in DLD1-wild-type versus NXF1-AID cells treated with 0.5 mM auxin. Scale bar, 25 μm.
(C) PolyA-RNA N/C shown as percentage of wild-type untreated cells.
(D) TDP-43 N/C in DLD1-wild-type cells (left) and NXF1-AID cells (right) pretreated with 0.5 mM auxin for 0–8 h, followed by 2 h auxin ± 250 nM NVP2. Data in both panels are normalized to untreated DLD1-wild-type cells (dotted line).
(E) TDP-43 N/C in NVP2 treated versus untreated cells in DLD1-wild-type (black) versus NXF1-AID (red) cells. These are the same data as (D), adjusted for differences in the steady-state TDP-43 N/C ratio to permit comparison of NVP2-induced TDP-43 nuclear exit.
In (C), (D), and (E), mean ± SD is shown for >3,000 cells/well in four biological replicates. ns, not significant; ***p < 0.001, ****p < 0.0001 by two-way ANOVA with Sidak’s test.
TDP-43 nuclear localization depends on binding to GU-rich RNAs
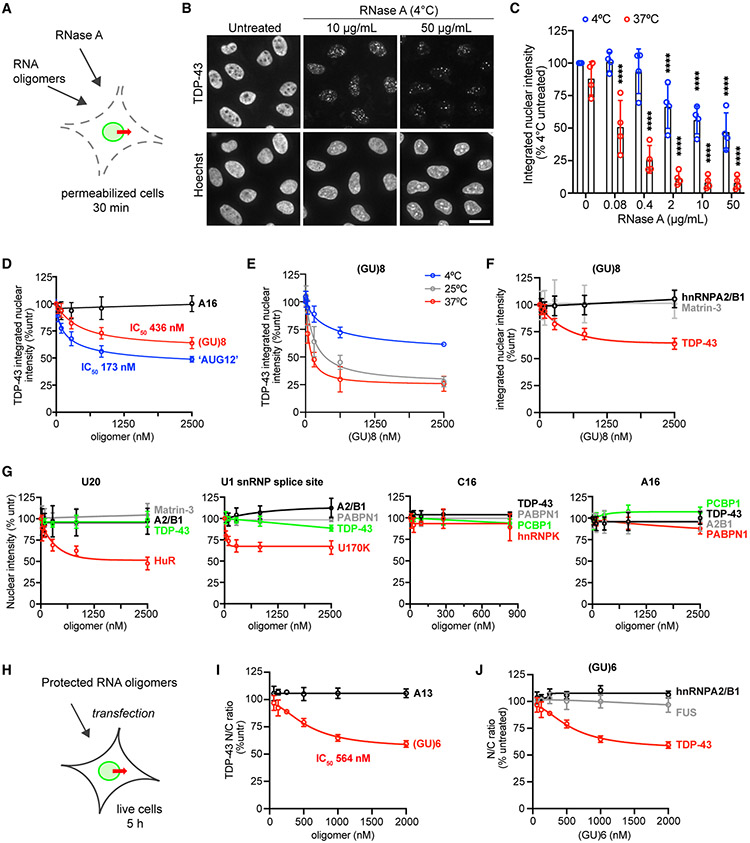
Because TDP-43 is primarily nuclear, despite its ability to diffuse through the NPC, we reasoned that TDP-43 nuclear localization must depend on intranuclear sequestration, perhaps by binding to RNA given its transcription-dependent and mRNA export-dependent shifts in localization. To test this idea, we treated permeabilized cells with RNase A and monitored the effect on TDP-43 nuclear localization (Figures 4A-4C). RNase caused rapid TDP-43 nuclear efflux over 30 min even at 4°C, suggesting that RNA tethers TDP-43 in the nucleus. The residual TDP-43 in RNase-treated nuclei formed puncta (Figure 4B), as previously observed in live-cell nuclei microinjected with RNase (Maharana et al., 2018). To assess nuclear integrity, we also analyzed the effect of RNase on the nuclear localization of DDX19b (54 kDa) and Nup50 (50 kDa), two proteins of similar size localized to the NPC and nucleoplasm (Figure S5A). The nuclear intensities of DDX19b and Nup50 were unaffected by RNase at 4°C and modestly decreased at 37°C but remarkably stable compared with TDP-43. The observation of the crucial role of RNA in TDP-43 nuclear localization led us to test whether RNA degradation contributes to TDP-43 nuclear efflux in our permeabilized cell assay (Figure S5B). We saw no difference in the rate of TDP-43 nuclear efflux in permeabilized cells kept at 37°C for 30–60 min, with or without RNase inhibitor, suggesting that RNA degradation does not contribute to passive export in this system. Nevertheless, RNase inhibitor was added to the transport buffer for all permeabilized cell assays as a precaution.
Figure 4. RNase and GU-rich oligomers induce TDP-43 nuclear efflux.
(A) Addition of RNase A or RNA oligomers to the permeabilized cell TDP-43 passive export assay.
(B) TDP-43 IF (top) and Hoechst (bottom) in permeabilized HeLa cells incubated for 30 min at 4°C ± RNase A. Scale bar, 25 μm.
(C) TDP-43 integrated nuclear intensity in permeabilized HeLa cells incubated for 30 min at 4°C versus 37°C ± RNase A. Data are normalized to untreated cells kept at 4°C. ****p < 0.0001 versus untreated by two-way ANOVA with Tukey’s test. See also Figure S5.
(D) TDP-43 integrated nuclear intensity (percent untreated) in permeabilized HeLa cells incubated for 30 min at 4°C with A16, (GU)8, or “AUG12.”
(E) TDP-43 integrated nuclear intensity (percent untreated) in permeabilized HeLa cells incubated at 4°C, 25°C, or 37°C for 30 min with (GU)8.
(F) Integrated nuclear intensity of nuclear RBPs (percent untreated) in permeabilized HeLa cells incubated for 30 min at 4°C with (GU)8. Note that TDP-43 data are the same as in (D).
(G) Integrated nuclear intensity of nuclear RBPs (percent untreated) in permeabilized HeLa cells incubated for 30 min at 4°C with the indicated oligomers.
(H) Transfection of live HeLa cells with protected RNA oligomers.
(I) TDP-43 N/C (percent untreated) 5 h post transfection with protected A13 or (GU)6.
(J) RBP N/C (percent untreated) 5 h post transfection with protected (GU)6. Note that TDP-43 data are the same as the (GU)6 data in (I).
In (F), (G), and (J), RBP in red is the most closely predicted binding partner for that motif, green is moderately similar, and gray and black denote no or low predicted binding. In (C) to (J), mean ± SD is shown for >2,000 cells/well in 3–4 biological replicates. IC50 was calculated by non-linear regression. See Table S1 for oligomer sequences.
CLIP-seq studies show that TDP-43 preferentially binds GU-rich motifs within introns (Polymenidou et al., 2011; Tollervey et al., 2011), and the RRM domains optimally recognize an “AUG12” (GUGUGAAUGAAU) motif (Lukavsky et al., 2013). To investigate the specificity of the RNA-mediated nuclear tethering of TDP-43, we added synthetic GU-RNA “decoy” oligomers to the permeabilized cell passive export assay (Figures 4D-4F). Following permeabilization, (GU)8, “AUG12,” or A16 was added, and cells were incubated at 4°C for 30 min (Figure 4D). Both “AUG12” and (GU)8 induced dose-dependent nuclear efflux of TDP-43, presumably by competitive dissociation of TDP-43 from endogenous nuclear RNAs, freeing TDP-43 to passively exit the nucleus. A16 had no effect. Incubating permeabilized cells with (GU)8 at increasing temperatures (25°C and 37°C) markedly accelerated (GU)8-induced TDP-43 export (Figure 4E), consistent with the expected temperature sensitivity of NPC permeability, TDP-43 dissociation from nuclear RNAs, and free diffusion kinetics.
Next, we evaluated the selectivity of oligomer-induced nuclear efflux by comparing different RNA motifs and RBPs. (GU)8 induced the nuclear efflux of TDP-43 but not hnRNPA2/B1 (AGGACUGC consensus motif) (Wu et al., 2018) or Matrin-3 (UUUCUXUUU consensus motif) (Uemura et al., 2017) (Figure 4F). PolyU (U20) induced the nuclear efflux of HuR, which binds AU-rich elements and other U-rich motifs (e.g., UUGGUUU) (Ripin et al., 2019; de Silanes et al., 2004), but did not affect TDP-43 or other RBPs (Figure 4G). An oligomer based on the U1 small nuclear ribonucleoprotein (snRNP) splice site (CAAAGGUAAGUUGGA) (Kondo et al., 2015) selectively induced the nuclear export of the U1 small nuclear ribonucleoprotein 70 kDa (U1-70K or snRNP70), with no significant effect on the localization of TDP-43, hnRNPA2/B1, or PABPN1. C16 and A16 did not induce the export of any RBPs tested, even those predicted to bind (i.e., hnRNPK and PABPN1, respectively) (Choi et al., 2009; Goss and Kleiman, 2013). These results in permeabilized cells suggest that the nuclear localization of a subset of RBPs, including TDP-43, depends on selective binding to preferred motifs within nuclear RNAs.
To evaluate the ability of GU-RNA oligomers to alter TDP-43 nuclear localization in live cells, we next transfected HeLa cells with (GU)6 or A13 oligomers with 2′-O-methyl groups and phosphorothioate bonds to protect against cellular RNases (Figure 4H). To confirm that the transfected oligomers enter the nucleus, we transfected cells with biotin-tagged (GU)6 and labeled it with AF568-conjugated streptavidin (SAV) following cell fixation (Figures S5C and S5D). Transfected (GU)6-biotin oligomers readily entered the nucleus in a time- and dose-dependent manner. As in permeabilized cells, (GU)6 but not A13 induced dose-dependent TDP-43 nuclear efflux (Figure 4I). Again, this was specific to TDP-43, as there was no change in the N/C ratio of RBPs that recognize other motifs (Figure 4J).
To verify that the transfected GU-RNA oligomers directly interact with TDP-43 to induce its nuclear exit, we performed biochemical pull-downs with SAV-conjugated microbeads from lysates of cells transfected with (GU)6- or A12-biotin (Figures S5E and S5F). Endogenous TDP-43 strongly bound to SAV-captured (GU)6 but not A12. The TDP-43·(GU)6 association was disrupted by RNase A, demonstrating dependence on RNA binding. Pull-downs from cells that were UV crosslinked before lysis additionally showed the formation of RNase-resistant higher-molecular-weight forms of TDP-43 (> in Figure S5F), indicative of covalently crosslinked RNA-protein complexes. These studies confirm that in live cells, binding to endogenous GU-rich nuclear RNAs opposes the tendency of TDP-43 to passively leave the nucleus.
Inhibition of pre-mRNA splicing promotes TDP-43 nuclear accumulation
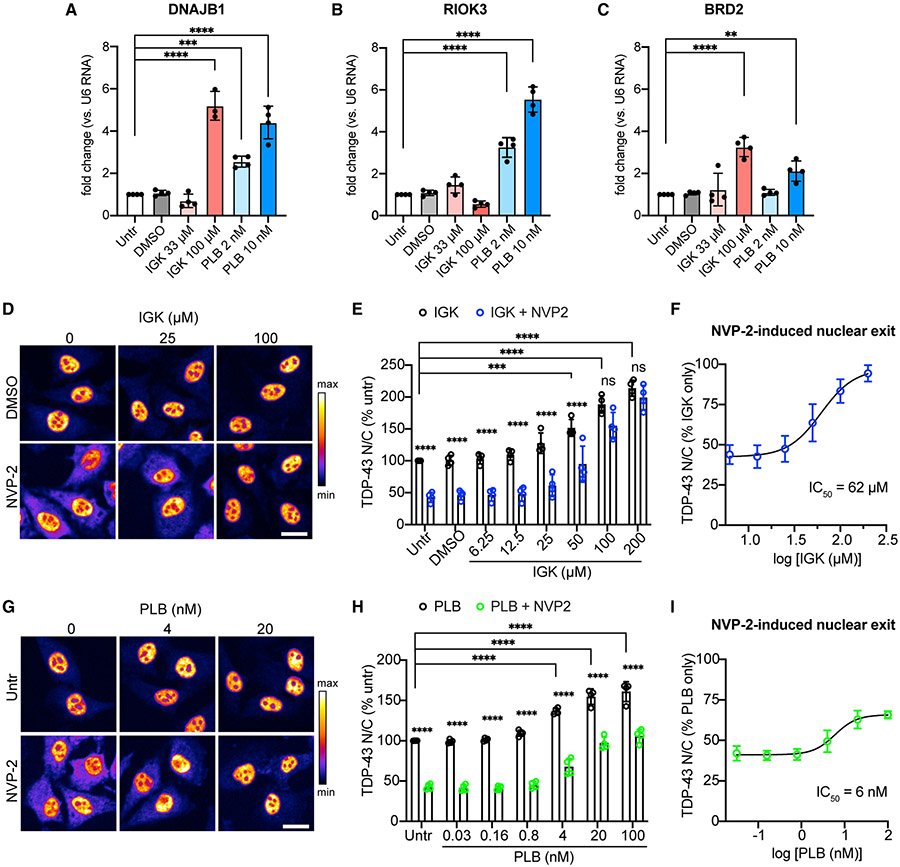
Since GU-rich RNA binding appears critical for TDP-43 nuclear retention, we hypothesized that binding to GU-rich intronic sequences in newly transcribed pre-mRNAs supports TDP-43 nuclear localization. As inhibition of pre-mRNA splicing stalls intron excision and leads to nuclear accumulation of unspliced pre-mRNAs (Carvalho et al., 2017), we used two splicing inhibitors, isoginkgetin (IGK) (O’Brien et al., 2008) and pladienolide B (PLB) (Sato et al., 2014), to test whether nuclear intron accumulation reduces TDP-43 nuclear efflux. PLB binds and inhibits SF3B1 in the U2 snRNP in the first step of spliceosome assembly (Kotake et al., 2007). The precise target of IGK is not known, but it blocks the complex A-to-B transition during spliceosome assembly (O’Brien et al., 2008). After 4 h, both IGK and PLB induced accumulation of introns by qRT-PCR, normalized to the expression of U6 small nuclear RNA, an unspliced transcript of RNA pol III (O’Brien et al., 2008) (Figures 5A-5C). Over the same period there was a dose-dependent increase in the steady-state TDP-43 N/C ratio (Figures 5D, 5E, 5G, and 5H). Remarkably, 4 h of treatment with IGK or PLB also induced dose-dependent resistance to NVP2-induced TDP-43 nuclear efflux (Figures 5F and 5I), even after adjusting for the steady-state nuclear accumulation of TDP-43. IGK was more potent than PLB at promoting TDP-43 nuclear retention, albeit at higher doses (IGK IC50 = 62 μM, PLB IC50 = 6 nM), for reasons that are unclear based on the similar mechanism of splicing disruption of these compounds. However, IGK also inhibits transcription elongation, potentially further increasing nuclear pre-mRNA accumulation (Boswell et al., 2017). Along with the effects of GU-rich oligomers, these data strongly suggest that TDP-43 nuclear localization depends on the abundance of nuclear GU-rich RNA-binding sites.
Figure 5. Inhibition of pre-mRNA splicing promotes TDP-43 nuclear accumulation.
(A–C) qRT-PCR quantification of DNAJB1 (A), RIOK3 (B), and BRD2 (C) introns, normalized to U6 small nuclear RNA in HeLa cells treated for 4 h with IGK or PLB. Mean ± SD is shown for four biological replicates. A single outlier was removed from IGK at 100 μM (fold change 18.78) via Grubbs test (α = 0.05). **p < 0.01, ***p < 0.001, ****p < 0.0001 by one-way ANOVA with Dunnett’s test.
(D) TDP-43 IF in HeLa cells treated with IGK for 4 h, followed by 2 h ± 250 nM NVP2. Scale bar, 25 μm.
(E) TDP-43 N/C (percent untreated) in IGK or IGK + NVP2-treated cells.
(F) TDP-43 N/C (same data as B) shown as percentage of IGK only to permit comparison of NVP2-induced nuclear exit. IC50 Was calculated by non-linear regression.
(G) TDP-43 IF in HeLa cells treated with PLB for 4 h, followed by 2 h PLB ± 250 nM NVP2. Scale bar, 25 μm.
(H) TDP-43 N/C (percent untreated) in PLB or PLB + NVP2-treated cells.
(I) TDP-43 N/C (same data as E) shown as percentage of PLB only, to permit comparison of NVP2-induced nuclear exit. IC50 was calculated by non-linear regression.
In (D) and (G), the intensity histogram for each image was independently maximized across the full range and a pseudo-color LUT was applied. In (E), (F), (H), and (I), mean ± SD of >2,000 cells/well in four biological replicates. ns, not significant; ***p < 0.001, ****p < 0.0001 by two-way ANOVA with Tukey’s test. See also Figure S6.
To explore whether related RBPs also show splicing-dependent localization, we analyzed the effect of splicing inhibition on hnRNPA1. Like TDP-43, hnRNPA1 displays N/C shuttling but is predominantly nuclear and acts as a splicing regulator (Clarke et al., 2021). hnRNPA1 also shows ActD-induced (Piñol-Roma and Dreyfuss, 1992) and NVP2-induced nuclear efflux (Figures 1D and S3). Like TDP-43, IGK and PLB induced a dose-dependent increase in steady-state hnRNPA1 nuclear localization and conferred resistance to NVP2-induced nuclear efflux (IGK IC50 = 69 μM, PLB IC50 = 3 nM) (Figures S6A-S6D). In contrast, the predominantly nuclear localization of Matrin-3, a DNA- and RNA-binding protein that functions in RNA stability, export, and gene regulation (Salton et al., 2011; Iradi et al., 2018; Cha et al., 2021), was not affected by the inhibition of transcription (Figures 1D and S3) or pre-mRNA splicing by IGK or PLB (Figures S6E-S6H). These data suggest that the nuclear localization of a selective subset of proteins involved in pre-mRNA splicing, including TDP-43, depends on pre-mRNA splicing activity.
TDP-43 RRM domains confer RNA-dependent TDP-43 nuclear localization
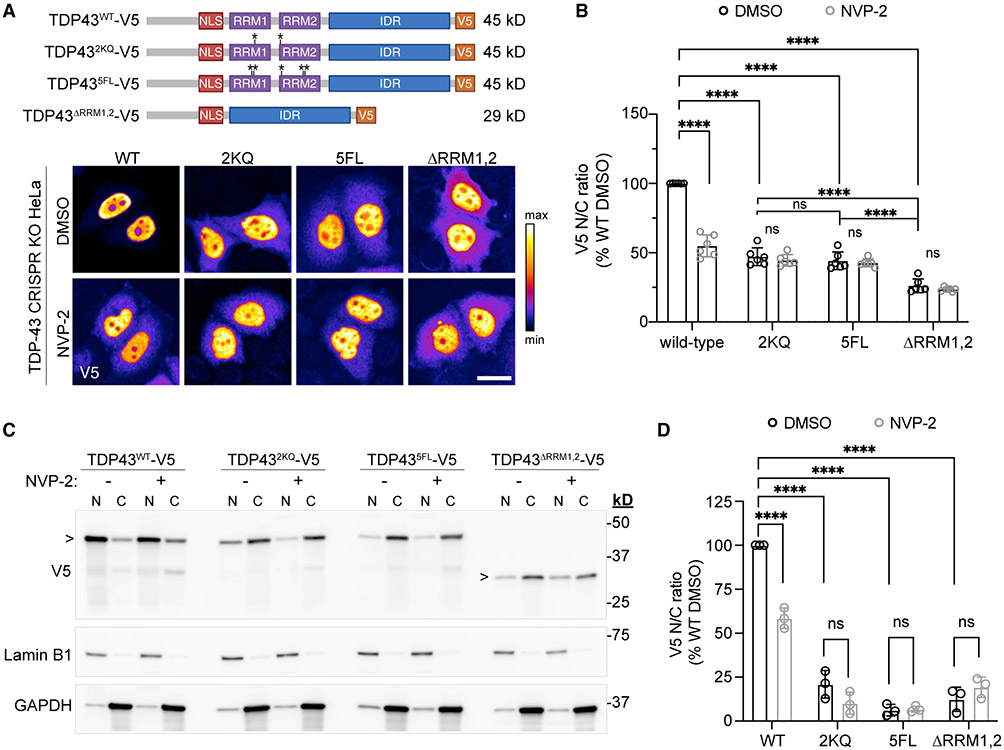
The TDP-43 RRM-domain RNA-binding properties have been extensively characterized (Buratti and Baralle, 2001; Lukavsky et al., 2013; Cohen et al., 2015; Flores et al., 2019). Although RRM1,2 mutation or deletion does not abolish TDP-43 nuclear localization (Elden et al., 2010; Wang et al., 2020), these observations arise from fluorescently tagged TDP-43 constructs, whose higher molecular weight markedly inhibits TDP-43 export (Figure S4A) (Ederle et al., 2018; Pinarbasi et al., 2018). However, TDP-43 RRM mutants with a small (~1 kDa) FLAG tag showed increased cytoplasmic localization by N/C fractionation and immunoblotting (Ayala et al., 2008). To further assess the role of the RRM domains in TDP-43 nucleocytoplasmic localization, we generated V5-tagged (+1.4 kDa) wild-type and RRM mutant TDP-43 constructs (Figure 6A and Table S2), including point mutations of phenylalanine residues (5F → L) and acetylation sites (2K → Q) critical for RNA binding (Buratti and Baralle, 2001; Cohen et al., 2015), and a full RRM deletion (ΔRRM1-2). Constructs were transiently transfected into a stable HeLa cell line depleted of TDP-43 by CRISPR (Figures S7A and S7B) (provided by S. Ferguson [Roczniak-Ferguson and Ferguson, 2019]) to avoid potential confounds of hetero-oligomerization with endogenous TDP-43. High-content analysis showed a significant drop in the steady-state N/C ratio of all three RRM mutants, likely exaggerated for ΔRRM1-2 due to its small size (29 kDa) compared with the point mutants (~45 kDa, p < 0.0001) (Figures 6A and 6B). In addition to reduced steady-state nuclear localization of the RRM mutants, NVP2-induced nuclear efflux was abolished. Of note, nuclear puncta of V5-tagged wild-type and TDP-43 RRM mutants were only rarely observed, compared with frequent nuclear puncta observed in cells transfected with YFP-tagged wild-type TDP-43 and TDP-43-5F → L (Figures S7C-S7E) (Elden et al., 2010; Wang et al., 2020; Yu et al., 2020). Since 27-kDa-YFP contains three residues (F223, L221, A206) that cause dimerization of GFP (Day and Davidson, 2009), the presumed nuclear LLPS of TDP-43 RRM mutants could be exacerbated by size or dimerization propensity (Snapp et al., 2003; Falcón-Pérez et al., 2005). Increased nuclear TDP-43-YFP protein concentration as a consequence of delayed nuclear export may also contribute (Figure S4A).
Figure 6. Mutation or deletion of TDP-43 RRM domains disrupts TDP-43 nuclear localization.
(A) V5-tagged TDP-43 RRM mutant constructs (top) and V5 IF (bottom) in transiently transfected TDP-43 CRISPR KO HeLa cells, after 1 h with 250 nM NVP2. The intensity histogram for each image was independently maximized across the full range and a pseudo-color LUT was applied. Scale bar, 25 μm. For plasmids see Table S2. See also Figure S7.
(B) V5 N/C ratio of TDP-43WT versus RRM mutants in transiently transfected TDP-43 CRISPR KO HeLa cells, following 3 h with 250 nM NVP2. Data are normalized to DMSO-treated TDP43WT cells. Mean ± SD is shown of >1,000 cells/well in six biological replicates.
(C) N/C fractionation and immunoblotting of TDP-43 CRISPR KO HeLa cells, transiently transfected with V5-tagged TDP-43 constructs and treated for 3 h with 250 nM NVP2. Lamin B1, nuclear marker; GAPDH, cytoplasmic marker.
(D) V5 N/C normalized to DMSO-treated TDP43WT cells. Mean ± SD is shown for three biological replicates.
In (B) and (D), ns denotes not significant; ****p < 0.0001 by two-way ANOVA with Tukey’s test.
Finally, we confirmed our imaging findings with N/C fractionation and immunoblotting (Figures 6C and 6D), which also showed a marked reduction in the N/C ratio of all three TDP-43 RRM mutants and no response to NVP2. Thus, TDP-43 RRM domains are required for nuclear TDP-43 localization and transcription-dependent shuttling.
DISCUSSION
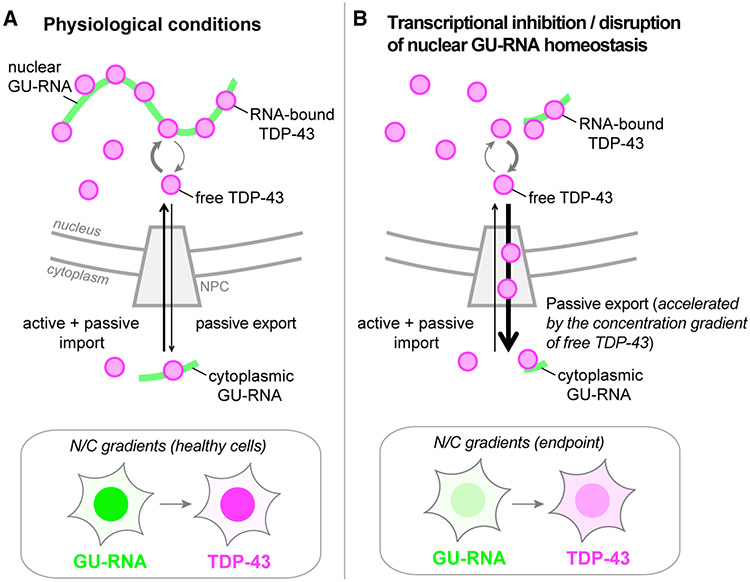
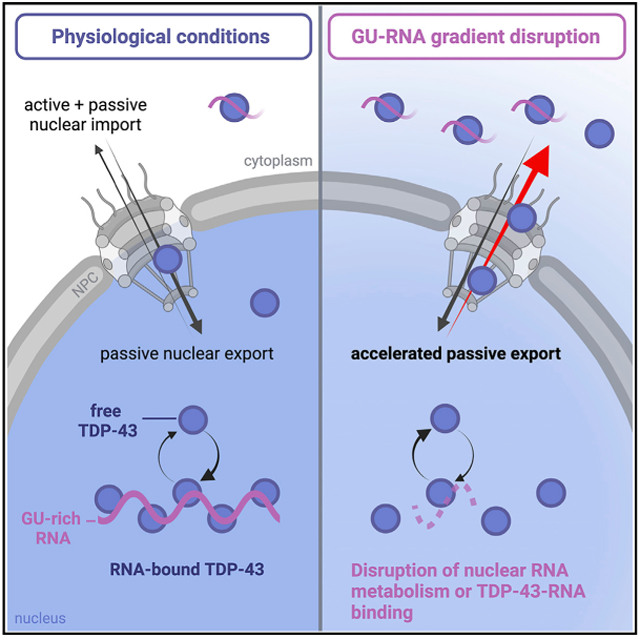
Here, we investigated the mechanism of TDP-43 nuclear export and the regulatory role of RNA. TDP-43 readily exited permeabilized cell nuclei in low-ATP conditions, consistent with passive diffusion through NPCs. Acute NXF1 depletion did not alter TDP-43 export in live cells, further excluding active co-export with mRNA. Three lines of evidence indicate that binding to GU-rich nuclear RNAs sequesters TDP-43 in nuclei and controls its availability for passive export. (1) RNA pol II inhibition in live cells and RNase treatment in permeabilized cells induced rapid TDP-43 nuclear efflux. (2) Short GU-rich oligomers induced nuclear TDP-43 exit, likely by competitive displacement of TDP-43 from endogenous nuclear RNAs. Splicing inhibitors also promoted TDP-43 nuclear accumulation and resistance to transcriptional blockade-induced nuclear efflux, suggesting that binding to unspliced introns promotes TDP-43 nuclear localization. (3) Mutation of the TDP-43 RRM domains reduced TDP-43 nuclear localization and abolished its transcription-dependent shuttling. These findings support a model (Figure 7) in which TDP-43 N/C distribution results from a balance between active and passive TDP-43 nuclear import, nuclear sequestration by binding to GU-rich RNAs, and passive nuclear export. In this model, TDP-43 moves in and out of the nucleus via a reaction-diffusion-controlled mechanism (Bastiaens et al., 2006; Soh et al., 2010), whereby transient formation of TDP-43 complexes with nuclear RNAs sequesters TDP-43 and opposes the availability of free TDP-43 for passive diffusion through the NPC. Because inhibition of nuclear RNA synthesis and competitive displacement by GU oligomers both induced TDP-43 nuclear exit in live cells, active import of TDP-43 is insufficient to balance passive export resulting from the loss of nuclear RNA-binding sites. According to this model, a decrease in TDP-43 nuclear RNA-binding sites or reduced TDP-43 RNA-binding activity will increase the fraction of TDP-43 that can passively leave the nucleus. By maintaining the nuclear abundance of GU-rich RNAs, the dynamic and interlinked balance of transcription, pre-mRNA processing, and nuclear RNA export may be the primary factor determining TDP-43 nuclear localization under physiological conditions.
Figure 7. Mechanisms coupling TDP-43 localization with nuclear GU-rich pre-mRNA binding and abundance.
(A) Under physiological conditions, nuclear TDP-43 is highly bound to GU-rich nuclear RNAs, limiting the pool of free TDP-43 capable of exiting nuclei by free diffusion through NPCs.
(B) Perturbations that deplete nuclear GU-rich RNAs or impair TDP-43 RNA binding (e.g., transcriptional blockade, RNase, RRM mutations, or putative disruption of GU-RNA homeostasis in disease) cause an initial increase in nuclear free TDP-43, accelerating its passive export until reaching steady state at a lower N/C ratio.
Insets depict the cellular GU-RNA N/C gradients that dictate TDP-43 localization.
Transcriptional dependence of RBP nuclear enrichment
Consistent with previous studies (Piñol-Roma and Dreyfuss, 1992; Cáceres et al., 1998; Ayala et al., 2008; Ederle et al., 2018), we observed that transcriptional blockade altered the N/C partitioning of numerous RBPs, ranging from prominent cytoplasmic accumulation (TDP-43, HuR) to a marked nuclear increase (hnRNPC). Cytoplasmic-shifting RBPs included those with distinct RNA-binding preferences, such as TDP-43 (Polymenidou et al., 2011,Tollervey et al., 2011), HuR (Lebedeva et al., 2011), and hnRNPA1 (Bruun et al., 2016; Jain et al., 2017), as well as NXF1, which binds RNA in a sequence-independent manner (Tuck and Tollervey, 2013; Baejen et al., 2014). Similarly, RBPs retained or enriched in nuclei included sequence-specific binders such as U2AF65 (Wu et al., 1999; Zorio and Blumenthal, 1999; Merendino et al., 1999) and the DEAD-box helicase DDX19b, which has no sequence requirement (Lin et al., 2018). Thus, the diverse response of individual RBPs to transcriptional blockade appears to be defined by RBP-specific protein-RNA and protein-protein interactions rather than by a particular RNA motif. The mechanism of NVP2-induced nuclear enrichment of hnRNPC is unclear. hnRNPC forms a tetrameric nuclear complex to measure pol II transcript length and sort pre-mRNAs for nuclear export (McCloskey et al., 2012). Unlike TDP-43 and other shuttling RBPs, hnRNPC contains a nuclear retention sequence that prevents its nuclear export (Nakielny and Dreyfuss, 1996). Whether RNA pol II inhibition may disrupt hnRNPC turnover or trigger an autoregulatory increase in expression is unknown.
Our understanding of RNA-dependent RBP shuttling, including factors that mediate RBP nuclear export or retention, is incomplete (Nakielny and Dreyfuss, 1999). Several RBP-specific domains have been identified to mediate N/C shuttling, including the HuR HNS domain (Fan and Steitz, 1998), the KNS motif of hnRNPK (Michael et al., 1997), the M9 signal for transportin-mediated import and transcriptional blockade-induced export of hnRNPA1 (Michael et al., 1995), the nuclear retention signal of hnRNPC (Nakielny and Dreyfuss, 1996), and the SR domain of SF2 (SRSF1), which confers ActD-dependent shuttling to other proteins (Cáceres et al., 1998). Here, we demonstrate that the TDP-43 RRM domains mediate its transcriptional blockade-induced nuclear efflux and act as a nuclear retention domain. Interestingly, depletion of cytoplasmic RNA via activation of cytoplasmic Xrn1 exonuclease induced the nuclear translocation of many RBPs, including the cytoplasmic polyA-binding protein PABPC (Gilbertson et al., 2018). In a spatial inverse to our model of TDP-43 shuttling, the findings of Gilbertson et al. suggest that the cytoplasmic abundance of polyA-RNA may be responsible for PABPC localization. N/C gradients of RNA-binding motifs may represent a broadly relevant paradigm for regulating RBP localization, which warrants further investigation.
Passive diffusion of TDP-43 across the NPC
The FG permeability barrier of the NPC does not have a strict size cutoff. Instead, diffusion of cargoes through NPC channels is increasingly restricted from 30 to 60 kDa (Mohr et al., 2009; Timney et al., 2016). Thus, 43-kDa TDP-43 monomers are predicted to diffuse across the NPC with moderate efficiency. Indeed, as expected for passive diffusion from the nucleus, we observed that TDP-43 exited nuclei of permeabilized cells in low-ATP conditions. NPC permeabilization further accelerated the passive TDP-43 nuclear efflux. These data corroborate predictions from live-cell assays showing marked size restriction of TDP-43 nuclear export, including slowed TDP43-tdTomato (+54 kDa) export in the heterokaryon assay (Pinarbasi et al., 2018), lack of TDP43-GCR2-EGFP2 (+119 kDa) cytoplasmic recovery in a dexamethasone-induced shuttling assay (Ederle et al., 2018), and failure of TDP43-YFP (+27 kDa) nuclear efflux following transcriptional blockade (Figure S4A). Delay in active nuclear import has been demonstrated for very large cargoes (Paci et al., 2020), and the rate of active mRNP export is modestly size dependent (Grünwald et al., 2011). However, the exquisite size limitation of TDP-43 nuclear export in live-cell assays is most consistent with passive diffusion from the nucleus, as our permeabilized cell export assay confirms.
In addition to size, cargo surface properties also critically dictate NPC passage, including hydrophobic residues, which augment NPC transport (Frey et al., 2018). Besides the glycine-rich C-terminal IDR, hydrophobic patches are present throughout the TDP-43 sequence (https://web.expasy.org/protscale/) (Gasteiger et al., 2005), including in the N-terminal domain, RRM1, and RRM2; however, their roles in NPC passage are not known. A recent study showed that intrinsically disordered proteins up to ~63 kDa could passively transit through NPC channels (Junod et al., 2020), suggesting that the disordered state of the TDP-43 IDR could potentially contribute to its NPC translocation, which remains to be examined.
N/C GU-RNA gradients and TDP-43 localization
CLIP-seq studies from mouse brain showed that 94% of TDP-43 binding sites reside in GU-rich intronic sequences (Polymenidou et al., 2011). A high preference for introns (74%) was also detected in human embryonic stem cells and SH-SY5Y cells (Tollervey et al., 2011). A lower frequency of intronic binding sites (58%) was seen in human postmortem brain, which may be due to the reduced stability of intronic RNAs. On average, introns are much longer than exons in human pre-mRNAs (7.5 kb versus 320 b) (Lee and Rio, 2015). Some introns are also retained in exported mRNAs due to alternative splicing or incomplete debranching and are detectable in the cytoplasm as stable intron lariats (Talhouarne and Gall, 2018; Saini et al., 2019). However, a substantial proportion of nuclear introns are likely short lived due to the tight coupling of transcription with pre-mRNA splicing, pre-mRNA quality surveillance, and rapid degradation via the nuclear exosome (Lee and Rio, 2015; Kilchert et al., 2016; Bresson and Tollervey, 2018). CLIP-seq also identified TDP-43 binding to GU-rich motifs in exonic 3′ UTRs, long non-coding RNAs (lncRNAs), and intergenic sites (Polymenidou et al., 2011; Tollervey et al., 2011). Active transcription could thus potentially give rise to the N/C concentration gradient of TDP-43 binding sites in several classes of GU-rich RNAs. The evident nuclear accumulation of newly synthesized 5-EU-labeled RNAs in HeLa cells, neurons, fibroblasts, and RPE1 cells (Figures 1A, S1, and S2) demonstrates a steep N/C gradient of U-rich nascent RNA across multiple cell types. Recent advances in CLIP-seq and related methods (Wheeler et al., 2018; Hafner et al., 2021) have yielded detailed insights into the RNA-RBP interactions and their dynamics (Nostrand et al., 2020). Nevertheless, available computational methods are not well suited for the global quantification of particular sequence motifs (e.g., GU repeats). Thus, direct quantification of the putative N/C gradient of TDP-43 GU-RNA-binding sites might require the development of new methodological approaches.
Biophysical regulation of TDP-43-RNA binding
We observed a drastic reduction in TDP-43 passive nuclear exit from permeabilized cell nuclei at 4°C (Figures 2B and 2C), far beyond the ~2-fold decrease in the passive import of ERK2-GFP at 4°C in a similar permeabilized cell system (Whitehurst et al., 2002), Thus, temperature dependence of free diffusion alone is unlikely to fully account for the slowing we observed at 4°C. The ability of 1,6-HD to elicit TDP-43 efflux at 4°C (Figures 2D and 2E) suggests that the FG permeability barrier is temperature sensitive and likely contributes to the hindrance of TDP-43 passive export at 4°C. Interestingly, TDP-43 nuclear efflux induced by RNase or GU-rich oligomers was also delayed at 4°C, suggesting temperature sensitivity of TDP-43-RNA binding, which remains to be verified. RNA-protein interactions generally involve dynamic rearrangements of both binding partners and their stabilization in the complex (Corley et al., 2020), suggesting that increased complex mobility at higher temperatures may promote TDP-43-RNA dissociation. Consistent with this, van der Waals forces support intramolecular RRM1-RRM2 interactions, which strongly influence RNA affinity (Lukavsky et al., 2013) and most TDP-43-RNA interactions (Sun et al., 2021). As van der Waals forces steeply decline with the distance of the interacting atoms, a temperature-dependent increase in molecular fluctuation could stochastically promote TDP-43 unbinding from nuclear RNAs at 37°C.
The TDP-43 GU-rich RNA-binding partners identified by CLIP-seq are structurally diverse (Polymenidou et al., 2011; Tollervey et al., 2011) and include GU-rich RNAs with wide-ranging dissociation constants (~3–3,000 nM), with affinity increasing with the number of perfect GU repeats (Bhardwaj et al., 2013). The existence of multiple low-affinity binding sites might have a physiological role in TDP-43 autoregulation (Avendaño-Vázquez et al., 2012; Bhardwaj et al., 2013) and could explain the ability of short GU-rich oligomers, which bind TDP-43 with low-nM affinity (Bhardwaj et al., 2013; French et al., 2019), to displace TDP-43 from endogenous RNAs (Figures 4D and 4I). Although RNase caused a near-complete evacuation of TDP-43 from nuclei in permeabilized cells (Figures 4B and 4C), TDP-43 efflux induced by transcriptional blockade (Figures 1C, S1, and S2) and GU-rich “decoy” oligomers (Figure 4I) showed a time- and dose-dependent plateau. In addition to ongoing active nuclear import, the existence of an “export-resistant” TDP-43 pool may result from binding to other nuclear RNAs such as lncRNAs (Polymenidou et al., 2011; Tollervey et al., 2011), which play a role in TDP-43 nuclear LLPS (NEAT1) (Wang et al., 2020) and nuclear localization (Malat1) (Nguyen et al., 2019). “Export-resistant” TDP-43 may also represent TDP-43 bound to chromatin or nuclear matrix constituents, as supported by TDP-43 chromatin fractionation (Ayala et al., 2008). Since TDP-43 preferentially binds single-stranded DNA (Buratti and Baralle, 2001), direct binding to genomic DNA is uncertain, but TDP-43 could be indirectly associated with chromatin via protein-protein interactions with histones or other chromatin-associated proteins (Freibaum et al., 2010). Indeed, TDP-43 was recently identified among chromatin-associated RBPs (Rafiee et al., 2021). Nevertheless, a large fraction of endogenous TDP-43 exhibited nuclear RNA concentration-dependent shuttling in our assays.
Other physiological and pathological factors that may regulate TDP-43 shuttling
Together with active nuclear import, our results show that TDP-43 RRM-dependent RNA binding plays a critical role in establishing the concentration gradient of TDP-43 across the nuclear envelope (Figure 6). Multiple additional processes likely function in parallel to modulate TDP-43 steady-state localization and availability for active or passive transport. These include: oligomerization (Mompeán et al., 2017; Afroz et al., 2017; French et al., 2019); LLPS (Molliex et al., 2015; Conicella et al., 2016, 2020; Zacco et al., 2018; Mann et al., 2019; Carter et al., 2021); alternative splicing to truncated isoforms (Weskamp et al., 2020); post-translational modifications, such as phosphorylation, ubiquitination, SUMOylation, acetylation, and C-terminal cleavage (Prasad et al., 2019; François-Moutal et al., 2019); and pathological aggregation (Arai et al., 2006; Neumann et al., 2006). Aberrant maturation of TDP-43 LLPS condensates induced by seeding (Gasset-Rosa et al., 2019) or optogenetic clustering (Mann et al., 2019) diverts soluble TDP-43 into insoluble, high-molecular-weight cytoplasmic aggregates that become unavailable for nuclear transport. Interestingly, GU-RNA oligomers attenuate recombinant TDP-43 aggregation (French et al., 2019) and optogenetically induced cytoplasmic TDP-43 aggregation in live cells (Mann et al., 2019), consistent with the notion that access to GU-rich RNAs promotes TDP-43 solubility and availability for N/C transport.
Disruption of RNA metabolism in ALS and FTD
The observation that GU-rich nuclear RNAs regulate TDP-43 nuclear localization suggests that disruption of nuclear RNA metabolism could contribute to TDP-43 nuclear clearance in ALS and FTD. Indeed, disruption of RNA processing, stability and localization are of growing interest in neurodegenerative diseases (Nussbacher et al., 2019; Butti and Patten, 2019; Zaepfe and Rothstein, 2021). The status of the GU-rich RNA gradient in TDP-43-mislocalized cells in ALS and FTD is unknown, and few studies to date have examined factors predicted to alter the availability of nuclear GU-rich RNA-binding sites. However, analysis of genome-wide RNA stability in fibroblasts and induced pluripotent stem cells by metabolic labeling (Bru-seq) demonstrated RNA destabilization in cells derived from ALS patients (Tank et al., 2018). Spliceosomal machinery has also appeared as a common denominator in knockout and interactome analyses of RBPs implicated in familial ALS and FTD (Chi et al., 2018a; 2018b). Further investigation of these factors is warranted as well as analysis of N/C transcriptome compartmentalization, which has shown changes in nuclear RNA retention in neuropsychiatric disorders (Price et al., 2019) and may provide insights into the perturbation of RNA gradients in ALS and FTD. Based on the conservation of transcriptional blockade-induced TDP-43 nuclear efflux across multiple cell lines and neurons, RNA-based regulation of TDP-43 nuclear localization appears to be conserved across somatic and neuronal cells. It is tempting to speculate that cell-specific modifications of RNA processing and metabolism (Mauger et al., 2016; Hermey et al., 2017; Jaffrey and Wilkinson, 2018; Furlanis et al., 2019; Tyssowski and Gray, 2019; Ling et al., 2020) may contribute to TDP-43 mislocalization in disease.
Limitations of the study
This study tests the hypothesis that the N/C gradient of GU-RNAs regulates TDP-43 localization, owing to its preferential binding of GU-rich motifs. Although transcriptional dependence of TDP-43 localization was confirmed in primary neurons and fibroblasts most data are from in vitro studies in immortalized cell lines. It remains to be seen whether nuclear GU-RNAs regulate TDP-43 localization in human neurons and in vivo, which is particularly important to the future application of these results in disease models. Results obtained from transfection with synthetic RNA oligomers may be confounded by limited control over the intracellular concentration. In addition, although studies were performed with both unprotected and 2′-O-methyl- and phosphorothioate-protected oligomers, we have not systematically explored the effects of the protective groups on binding affinity and RBP selectivity. Finally, our results indicate that TDP-43 can passively diffuse through NPC channels when either unbound from RNA (post-RNase digestion) or bound to short synthetic RNAs. However the molecular state of TDP-43 passively traversing the NPC in living cells is unknown, including whether TDP-43 exits in a monomeric or oligomerized form or when bound to RNA or other ligands. Moreover, it remains to be tested how the availability o TDP-43 for passive export is affected by the interplay between its RNA binding, nuclear oligomerization, and LLPS.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Lindsey Hayes (lhayes@jhmi.edu).
Materials availability
Plasmids generated in this study will be shared by the lead contact upon completion of a material transfer agreement.
Data and code availability
All data reported in this paper will be shared by the lead contact upon request. Original Western blot images are included in the supplement (Data S1).
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell lines
A single cell-derived clone of HeLa cells (ATCC; originally female-derived) was maintained in OptiMEM (Gibco/ThermoFisher) with 4% FBS and penicillin-streptomycin. HEK293T cells (ATCC; originally female-derived) and a monoclonal TDP-43 CRISPR-depleted HeLa cell line (a generous gift from Shawn Ferguson (Roczniak-Ferguson and Ferguson, 2019)), were maintained in DMEM (Gibco/ThermoFisher) with 10% FBS and penicillin-streptomycin. DLD1-wildtype cells (ATCC; originally male-derived) and DLD1-NXF1-AID cells (Aksenova et al., 2020) were maintained in DMEM (Gibco/ThermoFisher) with 10% FBS and penicillin-streptomycin. HFF1 human fibroblasts (ATCC; originally male-derived) were maintained in DMEM (Gibco/ThermoFisher) with 10% FBS. hTERT-immortalized retinal pigment epithelial (RPE1) cells (ATCC; originally female-derived) were maintained in DMEM/F12 (Gibco/ThermoFisher) with 10% FBS. All cells were grown at 37°C in humidified air containing 5% CO2. All cell lines were validated by STR profiling (ATCC), routinely verified to be mycoplasma negative (Genlantis), and frequently refreshed from frozen stocks.
Mouse primary cortical neuron culture
All animal procedures were approved by the Johns Hopkins Animal Care and Use Committee. Timed pregnant C57BL/6J female mice (Jackson Laboratory) were sacrificed by cervical dislocation at E16, cortex dissociated, and cells (of mixed male/female gender) were plated at 50,000/well on poly-D-lysine/laminin-coated, optical glass-bottom 96-well plates as described (Hayes et al., 2020, 2021). The growth medium consisted of Neurobasal supplemented with B27, Glutamax, and penicillin/streptomycin (Gibco/ThermoFisher). Cells were grown at 37°C in humidified air containing 5% CO2.
METHOD DETAILS
RNA oligonucleotides
Desalted lyophilized synthetic RNA oligonucleotides (Table S1) were obtained from IDT. Upon reconstitution in sterile, RNAse- and DNAse-free water, single-use aliquots were stored at −70°C.
Cloning of recombinant constructs
Plasmids for the expression of wild-type (WT) human TDP-43 and its variants in tissue culture cells (Table S2) were prepared by Twist Biosciences via gene synthesis between the HindIII and NheI sites in the pTwistEF1α expression vector. The expected sequences of TDP-43 open reading frames were verified by Sanger sequencing.
Permeabilized cell TDP-43 export assays
HeLa cells were plated on Matrigel-coated optical glass-bottom 96 well plates (CellVis) at 12,000–15,000 cells per well, targeting 80–90% confluence at 24 h. To permeabilize, cells were rinsed for 2 min in ice-cold PBS, and permeabilized on ice for 10 min in 25–35 ug/mL digitonin (Calbiochem) in transport buffer (TRB, 20 mM HEPES, 110 mM KOAc, 2 mM Mg(OAc)2, 5 mM NaOAc, 0.5 mM EGTA, 250 mM sucrose, pH 7.4, with freshly added Halt protease inhibitor (1:100, Promega)). The optimal digitonin concentration varied by cell density and passage number and was optimized before each assay such that the majority of plasma membranes were permeabilized while maintaining the nuclear exclusion of a 70 kD fluorescent dextran (ThermoFisher). Before initiating the export assay, the permeabilized cells were washed twice in ice-cold TRB.
Assay components, including 1,6-HD (Sigma-Aldrich), ATP (pH 7.5, Sigma-Aldrich), RNase A (ThermoFisher), or RNA oligomers (Table S1, synthesized by IDT or Sigma-Aldrich) were premixed in TRB in 96-well plates and equilibrated at the appropriate temperature (4°C, 25°C, or 37°C) before cell permeabilization. Except for RNase A-containing assays, TRB was routinely supplemented with RNasin Ribonuclease Inhibitor (1000 U/mL, Promega). Following permeabilization, the assay mix was transferred onto permeabilized cells via a multichannel pipette, and export was allowed to proceed at the temperature (4°C, 25°C, or 37°C) and for the time (30–60 min) indicated in the figure legends before fixation in 4% paraformaldehyde/PBS (Electron Microscopy Sciences) for 15 min. An additional plate of cells designated as ‘time 0’ was fixed immediately post-permeabilization for each replicate to enable data normalization across independent biological replicates.
Permeabilized cell Rango nuclear import assay
The recombinant Rango sensor was prepared as used in nuclear import assays as recently described (Hayes et al., 2020, 2021). Briefly, following HeLa cell permeabilization as above, Rango nuclear import assays were performed with or without 2.5 mM ATP (pH 7.5, Sigma-Aldrich) and HEK whole cell lysate (2.5 mg/mL in TRB) at 37°C for 30 min. Cells were fixed for 15 min in 4% paraformaldehyde/PBS, washed 2x with PBS containing Hoechst 33342, and transferred to 50% glycerol/PBS for immediate imaging. A subset of assays included recombinant WT human importin β and RanGTP-resistant importin β (71-876) (Drutovic et al., 2020).
Live cell TDP-43 localization assays
HeLa and DLD1 cells were plated in uncoated, optical glass-bottom 96-well plates (CellVis) to achieve ~75% confluence at the time of shuttling assays. For transcriptional inhibition, cells were treated with ActD (Sigma-Aldrich, in DMSO) or NVP2 (Tocris Bioscience, in DMSO) at doses/times indicated in the figure legends. For splicing inhibition, cells were pretreated for 4 h with escalating doses of isoginkgetin (IGK, Millipore-Sigma, in DMSO) or pladienolide B (PLB, Cayman Chemicals, in DMSO) before the addition of 250 nM NVP2. For NXF1 ablation experiments, DLD1-wildtype and DLD1-NXF1-AID cells were pretreated for 0–8 h with 0.5 mM synthetic auxin (3-indoleacetic acid, Sigma, in ethanol) before the addition of 250 nM NVP2. Synthetic RNA oligonucleotides were transfected using Lipofectamine RNAiMax (ThermoFisher) according to the manufacturers’ instructions. Serial dilutions of RNA oligomers were prepared in Opti-MEM (Gibco/ThermoFisher) and mixed 1:1 with diluted RNAiMAX (1.5 μL per 25 μL Opti-MEM). After a 5 min incubation at room temperature, transfection mixes were added to culture media (10 μL transfection mix per 100 μL media in 96-well format). RNA oligonucleotide concentrations listed in the figures denote the final concentration after diluting the transfection mix into culture media. Cells treated with RNAiMAX alone were used as a mock transfection control. At the conclusion of the experiments, cells were fixed in 4% paraformaldehyde/PBS (Electron Microscopy Sciences) for 15 min before immunostaining.
Immunofluorescence
Paraformaldehyde-fixed cells were rinsed with PBS and simultaneously permeabilized and blocked with 0.1% Triton-X 100 and 10% normal goat serum (NGS, Vector Labs) in PBS for 30 min at room temperature. Primary antibodies in 10% NGS/PBS were added to the cells and incubated for 60–90 min at room temperature or overnight at 4°C. Cells were rinsed twice with PBS and Alexa Fluor (AF)-labeled secondary antibodies (ThermoFisher) were added in 10% NGS/PBS and incubated for 1 h at room temperature. In experiments with biotinylated oligomers, Streptavidin-AF568 (1:500, ThermoFisher) was added together with the secondary antibody. Cells were rinsed with PBS containing Hoechst 33342 and transferred to 50% glycerol/PBS for imaging.
RNA labeling
For polyA fluorescence in situ hybridization (FISH), cells were fixed in 10% molecular grade-formaldehyde (Sigma-Aldrich) in PBS for 20 min, permeabilized in 0.1% Triton-X 100/PBS for 10 min, and washed 3 times in 1x PBS and 2 times in 2x SSC buffer (Sigma-Aldrich) for five min each. Cells were prehybridized for 1 h in hybridization buffer at 42°C (Thermo) and then incubated in Cy3-oligo-dT(45) probe (IDT, 100 nM in hybridization buffer) overnight at 42°C. Cells were washed with decreasing concentrations of SSC buffer at 42°C (2x, 0.5x, and 0.1x in PBS, for 20 min each), with Hoechst 33342 nuclear counterstain included in the final wash.
For click-chemistry labeling of nascent RNA synthesis, cells were treated with 200 μM 5-ethynyl-uridine in culture media for 30 min, fixed with 4% paraformaldehyde/PBS for 15 min, permeabilized and blocked in 2% BSA/0.1%TX-100 in PBS for 30 min, and labeled with 0.5 μM AF488-Picolyl Azide using the Click-&-Go Cell Reaction Buffer kit according to the manufacturers’ instructions (all reagents from Click Chemistry Tools). Cells were subsequently immunolabeled for TDP-43 as described above.
qRT-PCR
Cells were rinsed in PBS and lysed in TRIzol (ThermoFisher), and total RNA was isolated following the manufacturer’s protocol and resuspended in nuclease-free water. Subsequently, first-strand cDNA synthesis was performed with 1mg total RNA using the High-Capacity cDNA Reverse Transcription Kit (ThermoFisher). qRT-PCR was performed using PowerSYBR Green PCR Master Mix (ThermoFisher) with 10ng cDNA per well and intron primers (500nM) or with TaqMan Fast Advanced Master Mix (ThermoFisher) with 12.5ng of cDNA per well using TaqMan probes. All qRT-PCR assays were performed on a QuantStudio 3 Real-Time PCR System.
N/C fractionation and immunoblotting
24–48 h post-transfection with V5-tagged TDP-43 and RRM mutants, TDP-43 CRISPR KO HeLa cells were lysed for N/C fractionation and SDS-PAGE with the NE-PER kit (ThermoFisher) according to the manufacturer’s instructions. Total protein concentration was measured using the DC Protein Assay kit (Bio-Rad). Nuclear (5 μg total protein) and cytoplasmic (10 μg total protein) fractions were boiled in Laemmli buffer (Bio-Rad), run on Criterion 4–20% Gels (Bio-Rad), and transferred to nitrocellulose membranes using a TransBlot Turbo system (BioRad). Membranes were blocked with 5% non-fat milk in TBS-Tween and probed by sequential incubation in primary antibody overnight at 4°C. Detection was via HRP-conjugated secondary antibodies/chemiluminescence using an ImageQuant LAS 4000 system (GE). Band intensities were measured using ImageQuant software.
Pulldowns with biotinylated RNA oligomers
HeLa cells were transfected as described above with 500 nM biotinylated-A12, -(GU)6, or RNAiMAX alone (mock transfection). 5 h post-transfection, cells were rinsed with DPBS (no Ca2+ or Mg2+), and half were UV crosslinked (λ = 254 nm, 1.5 J/cm2 on ice). Cells were harvested by trypsinization, spun at 300g for 2 min, washed twice with DPBS, and resuspended in lysis buffer (1% NP40 in PBS, pH 7.4 with protease inhibitor, and 2.5% v/v RNase inhibitor). After 10s sonication, lysates were incubated on ice for 5 min and clarified by centrifugation (21,000g, 5 min, 4°C). After protein concentration measurement (Bio-Rad Protein Assay Kit I), samples were diluted with lysis buffer to 4 μg/μL. 70 μL of lysate was then added to DPBS-washed magnetic SAV-conjugated beads (ThermoFisher, 15 μL beads/sample) and rotated at 4°C for 1.5 h. Supernatants were collected using a magnetic stand and beads were washed 5 times with ice-cold RIPA buffer (50mM Tris-HCl, pH 8.0, 5mM EDTA, 1% Triton X-100, 0.5% Sodium Deoxycholate, 0.1% SDS, 150mM NaCl). Beads were rinsed once with water, resuspended in 15 μL RNase elution buffer (100mM triethylammonium bicarbonate, 1% SDS, 0.2 μg/μL RNase A) (Villanueva et al., 2020), kept overnight at 4°C, and then incubated in a thermomixer at 37°C at 1500 rpm for 1.5h. After collecting the RNase eluate on a magnetic stand, beads were rinsed with an additional 6 μL RNase elution buffer that was combined with the eluate. Finally, samples were re-eluted with Laemmli sample buffer (Bio-Rad, 2X with 5% β-mercaptoethanol) for 5 min at 100°C to elute any remaining bead-bound proteins. Samples were separated by SDS-PAGE, transferred to PVDF membranes, and immunoblots developed as described above.
ATP quantification
Permeabilized and live cells in TRB were lysed and ATP levels quantified using the CellTiter-Glo luminescence assay (Promega) according to the manufacturers’ instructions. Luminescence was measured using a SpectraMax M3 microplate reader (Molecular Devices).
Image processing for figures
IF images were cropped and minimally processed for figures using Adobe Photoshop 2021(v22.4.3) as follows and as indicated in the figure legends. For raw intensity comparisons, including the nascent RNA signal (Figures 1 and S1) and the TDP-43 nuclear signal in permeabilized cells (Figures 2 and 4), the intensity histogram of the designated control was maximized based on the brightest and dimmest pixels, and those parameters were subsequently applied to all other images. For comparisons of shifts in the N/C ratio (all other figures), the intensity histogram was independently maximized according to the brightest and dimmest pixels in each image to visualize the nuclear and cytoplasmic compartments. The fire pseudo-color LUT was applied in selected images using FIJI/ImageJ (v2.1.0/1.53c) to aid data visualization. The quantitative/linear map is provided in the images. All adjustments were linear (no gamma changes) and applied equally to the entire image. Immunoblots were cropped for space, and no other processing was applied.
QUANTIFICATION AND STATISTICAL ANALYSIS
High content imaging and analysis
Automated cell imaging was carried out using an ImageXpress Micro Confocal high content microscope with MetaXpress software (Molecular Devices) as previously described (Hayes et al., 2020, 2021). Briefly, nine non-overlapping fields per well were imaged at 20x (immortalized cells) or 40x (neurons) magnification in spinning disc confocal mode with 60 μm pinhole, with exposures targeting half-maximal saturation (33,000 / 65,536 relative fluorescent units (RFU) in unbinned, 16-bit images). For HeLa, DLD1, RPE1, and fibroblast cell lines, the background-corrected mean and integrated nuclear and cytoplasmic intensities and the nuclear/cytoplasmic (N/C) ratio were calculated using the MetaXpress translocation-enhanced module as described (Hayes et al., 2020, 2021). Nuclear puncta (Figure S7D) were quantified using the granularity module with liberal cutoff values to identify puncta from 0.5 to 6 μM in size (1–18 pixels at 20x magnification), with intensity ≥ 7500 RFU above the local background. Hoechst was used to identify the nucleus, and nuclear and cytoplasmic compartments were set several pixels inside and outside the nuclear envelope to avoid edge effects. All image analysis was carried out on raw, unaltered images. Raw data were uniformly filtered to exclude errors of cell identification (probe intensity = 0) and non-physiologic N/C ratios (N/C < 0.1 or >100). For transient transfections, data were filtered by mean intensity to exclude untransfected cells (<1000 RFU) and highly expressing cells (>45,000 RFU). As indicated in the figure legends, the resulting data were normalized across technical and biological replicates as % untreated/vehicle-treated controls or percent time 0. The approximate number of cells/well across replicates is also provided.
Neuron linescan analysis
Neuronal nuclear and cytoplasmic intensities were manually analyzed in Image J (FIJI v2.1.0/1.53c) using a linescan method, as the asymmetry of neuronal cytoplasm precluded accurate automated analysis using circular regions of interest around the nucleus. A 35-pixel line was drawn from the center of the nucleus into the cytoplasm of 60–80 cells per group, and the mean nuclear and cytoplasmic intensity was calculated from the plotted intensity profile. Mean background intensity was sampled from three randomly selected cell-free regions of each image and subtracted from the nuclear and cytoplasmic intensities before calculation of the N/C ratio.
Statistical analysis
Prism v9.2.0-v9.3.1 (Graphpad) was used for all statistical analyses. Dose-response curves were fit by non-linear regression, using raw or log10-transformed concentrations. As reported in the figure legends, statistical differences among means were analyzed by two-tailed unpaired t-test, one-way ANOVA, or two-way ANOVA. Test assumptions, including Gaussian distribution and equal variance were carefully considered along with sample size and experimental design in statistical tests. The number of biological replicates (independent cell passages and experiments) was used as the N for statistical analysis for all assays. Post-hoc tests for multiple comparisons were selected based on the type of ANOVA and the comparisons being performed (i.e., comparison with a single control or comparison of all values), according to Prism default settings. Outliers were detected with the extreme Studentized deviate (Grubbs) method to detect a single outlier (α = 0.05). This resulted in one outlier being removed (Figure 5A). ANOVA tables and exact p values for all comparisons are provided in the supplement (Data S2).
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti-TDP43 monoclonal (WB: 1:2000, IF: 1:2000-1:5000) | Abcam [3H8] | Cat# ab104223; RRID:AB_10710019 |
| Rabbit anti-TDP43 polyclonal (WB (RNA-IP) 1:2600) | Proteintech | Cat# 10782-2-AP; RRID:AB_615042 |
| Rabbit anti-hnRNP A1 polyclonal (IF: 1:2000) | Proteintech | Cat# 11176-1-AP; RRID:AB_2117177 |
| Mouse anti-hnRNP A2/B1 monoclonal (IF: 1:50) | Santa Cruz Biotechnology | Cat# sc-53531; RRID:AB_2248245 |
| Rabbit anti-hnRNP A2/B1 polyclonal (IF: 1:1000) | Thermo Fisher Scientific | Cat# PA5-30061; RRID:AB_2547535 |
| Mouse anti-hnRNP C1/C2 monoclonal (IF: 1:100) | Santa Cruz Biotechnology | Cat# sc-32308; RRID:AB_627731 |
| Rabbit anti-hnRNP F polyclonal (IF 1:500) | Thermo Fisher Scientific | Cat# PA5-22341; RRID:AB_11155780 |
| Mouse anti-hnRNP K monoclonal (IF: 1:100) | Santa Cruz Biotechnology | Cat# sc-28380; RRID:AB_627734 |
| Rabbit anti-hnRNP M polyclonal (IF: 1:500) | Thermo Fisher Scientific | Cat# PA5-30247; RRID:AB_2547721 |
| Rabbit anti-FUS polyclonal (IF: 1:3000) | Bethyl | Cat# A300-302A; RRID:AB_309445 |
| Rabbit anti-PABPN1 monoclonal (IF: 1:1000) | Thermo Fisher Scientific | Cat# MA5-32621; RRID:AB_2809898 |
| Rabbit anti-PCBP1 polyclonal (IF: 1:500) | Thermo Fisher Scientific | Cat# PA5-65369; RRID:AB_2663337 |
| Rabbit anti-Matrin 3 monoclonal (IF 1:1000) | Abcam | Cat# ab151714; RRID:AB_2491618 |
| Mouse anti-AlyREF monoclonal (IF: 1:500) | Santa Cruz Biotechnology | Cat# sc-32311; RRID:AB_626667 |
| Rabbit anti-Nup50 monoclonal (IF: 1:250) | Abcam | Cat# ab137092; RRID:AB_2921286 |
| Rabbit anti-DDX19b polyclonal (IF: 1:3000) | Abcam | Cat# ab151478; RRID:AB_2921287 |
| Rabbit anti-NXF1 monoclonal (IF 1:2000, WB: 1:2000) | Abcam | Cat# ab129160; RRID:AB_11142853 |
| Rabbit anti-ELAVL1/HuR monoclonal (IF: 1:100) | Cell Signaling Technology | Cat# 12582; RRID:AB_2797964 |
| Mouse anti-DEK monoclonal (IF: 1:100) | Santa Cruz Biotechnology | Cat# sc-136222; RRID:AB_2245948 |
| Mouse anti-Y14 monoclonal (IF 1:100) | Santa Cruz Biotechnology | Cat# sc-32312; RRID:AB_2178827 |
| Mouse anti-sc35 monoclonal (IF 1:2000) | Sigma-Aldrich | Cat# S4045; RRID:AB_477511 |
| Mouse anti-U1-70K monoclonal (IF 1:500) | Millipore | Cat# 05-1588; RRID:AB_10805959 |
| Mouse anti-U2AF65 (IF 1:100) | Santa Cruz Biotechnology | Cat# sc-53942; RRID:AB_831787 |
| Mouse anti-UAP56 (BAT1) monoclonal (IF 1:50) | Santa Cruz Biotechnology | Cat# sc-271395; RRID:AB_10609494 |
| Rabbit anti-DHX15 polyclonal (IF 1:200) | Thermo Fisher Scientific | Cat# PA5-61413; RRID:AB_2640582 |
| Rabbit anti-GAPDH monoclonal (WB: 1:3000) | Cell Signaling Technology | Cat# 5174; RRID:AB_10622025 |
| Rabbit anti-V5 monoclonal (WB: 1:1000, IF 1:2000) | Cell Signaling Technology | Cat# 13202; RRID:AB_2687461 |
| Rabbit anti-Lamin B1 polyclonal (WB: 1:1000) | Abcam | Cat# ab16048; RRID:AB_443298 |
| Goat anti-mouse IgG AF488 polyclonal (IF: 1:1000) | Thermo Fisher Scientific | Cat# A32723; RRID:AB_2633275 |
| Goat anti-rabbit IgG AF488 polyclonal (IF: 1:1000) | Thermo Fisher Scientific | Cat# A32731; RRID:AB_2633280 |
| Goat anti-mouse IgG AF647 polyclonal (IF: 1:1000) | Thermo Fisher Scientific | Cat# A32728; RRID:AB_2633277 |
| Horse anti-mouse IgG HRP-conjugated polyclonal (WB: 1:5000) | Cell Signaling Technology | Cat# 7076; RRID:AB_330924 |
| Goat anti-rabbit IgG HRP-conjugated polyclonal (WB: 1:5000) | Cell Signaling Technology | Cat# 7074; RRID:AB_2099233 |
| Bacterial and virus strains | ||
| NEB 5-alpha E.coli (High Efficiency) | New England Biolabs | Cat# C2987H |
| BL21(DE3) Competent E. coli | New England Biolabs | Cat# C2527H |
| Chemicals, peptides, and recombinant proteins | ||
| Recombinant RNasin Ribonuclease Inhibitor | Promega | Cat# N2515 |
| RNase A | Thermo Fisher Scientific | Cat# EN0531 |
| Digitonin, high purity | Calbiochem | Cat# 300410 |
| 1,6-Hexanediol | Sigma-Aldrich | Cat# 240117 |
| ATP disodium salt hydrate | Sigma-Aldrich | Cat# A6419 |
| Actinomycin D | Sigma-Aldrich | Cat# A9415 |
| NVP-2 | Tocris Bioscience | Cat# 6535 |
| 5-ethynyl-uridine | Click Chemistry Tools | Cat# 1261 |
| Isoginkgetin | Millipore Sigma | Cat# 416154 |
| Pladienolide B | Cayman Chemicals | Cat# 13568 |
| Auxin (3-indoleacetic acid) | Sigma-Aldrich | Cat# I2886 |
| Dynabeads, MyOne Streptavidin C1 | Thermo Fisher Scientific | Cat# 65001 |
| Streptavidin, Alexa Fluor-568 conjugate | Thermo Fisher Scientific | Cat# S11226 |
| ULTRAhyb-Oligo hybrididzation buffer | Invitrogen | Cat# AM8663 |
| SSC Buffer 20X Concentrate | Sigma-Aldrich | Cat# S6639 |
| Lipofectamine RNAiMAX | Thermo Fisher Scientific | Cat# 13778030 |
| Triethylammonium bicarbonate | Sigma-Aldrich | Cat# 18597-100ML |
| SDS, 20% Solution, RNase-free | Thermo Fisher Scientific | Cat# AM9820 |
| Critical commercial assays | ||
| Click-&-Go Cell Reaction Buffer kit | Click Chemistry Tools | Cat# 1263 |
| CellTiter-Glo 2.0 assay kit | Promega | Cat# G9242 |
| NE-PER Nuclear and Cytoplasmic Extraction Reagents | Thermo Fisher Scientific | Cat# 78833 |
| TDP-43 Taqman assay | Thermo Fisher Scientific | Cat# Hs00429203_gH |
| GAPDH Taqman assay | Thermo Fisher Scientific | Cat# Hs02786624_g1 |
| PowerSYBR Green PCR Master Mix | Thermo Fisher Scientific | Cat# 4368577 |
| TaqMan Fast Advanced Master Mix | Thermo Fisher Scientific | Cat# 4444556 |
| MycoScope PCR Mycoplasma Detection Kit | Genlantis | Cat# MY01100 |
| Experimental models: Cell lines | ||
| HeLa cells (human) | ATCC and (Kalab et al., 2006) | HeLa clone 61 |
| TDP-43 CRISPR KO HeLa cells (human) | (Roczniak-Ferguson and Ferguson, 2019) | N/A |
| HEK293T cells (human) | ATCC | Cat# CRL-3216 |
| DLD1-wildtype cells (human) | ATCC and (Aksenova et al., 2020) | Cat# CCL-221 |
| DLD1-NXF1-AID cells (human) | (Aksenova et al., 2020) | N/A |
| Mouse primary cortical neurons | C57BL/6J E16 timed pregnant mice (Jackson laboratory) | Cat# 000664 |
| HFF1 human fibroblasts | ATCC | Cat# SCRC-1041 |
| hTERT RPE1 cells | ATCC | Cat# CRL-4000 |
| Experimental models: Organisms/strains | ||
| C57BL/6J timed pregnant female mice | Jackson Laboratory | Cat# 000664 |
| Oligonucleotides | ||
| RNA oligonucleotides | This study | See Table S1 |
| Cy3-oligo-dT(45) | IDT | N/A |
| U6 forward primer: CGCTTCGGCAGCACATATAC | (O’Brien et al., 2008) | N/A |
| U6 reverse primer: GAATTTGCGTGTCATCCTTG | (O’Brien et al., 2008) | N/A |
| DNAJB1 Intron 3 Forward GGCCTGATGGGTCTTATCTATGG | (Kotake et al., 2007) | N/A |
| DNAJB1 Intron 3 Reverse TTAGATGGAAGCTGGCTCAAGAG | (Kotake et al., 2007) | N/A |
| RIOK3 Intron 3 Forward TCAATGGAGATAGCAAAGGTATTATAAC | (Kotake et al., 2007) | N/A |
| RIOK3 Intron 3 Reverse AGATTTACTTAGGAGCACATTATGAGTG | (Kotake et al., 2007) | N/A |
| BRD2 Intron 3 Forward AGGTAATGTCACAGGATGGGAAGT | (Kotake et al., 2007) | N/A |
| BRD2 Intron 3 Reverse CCCTGCTGCCTTTCTCTAACC | (Kotake et al., 2007) | N/A |
| Recombinant DNA | ||
| V5-tagged TDP-43 expression constructs | This study | See Table S2 |
| pcDNA3.2 TDP-43 YFP | (Elden et al., 2010) via Addgene | Cat#84911 |
| pcDNA3.2 TDP-43 5F-L YFP | (Elden et al., 2010) via Addgene | Cat#84914 |
| pRSET Rango-2/α1+linkers | (Hayes et al., 2020) | pK44 |
| Importin β (71-876) | (Drutovic et al., 2020) | pKW488 |
| Software and algorithms | ||
| MetaXpress (v6.1.0.2071) | Molecular Devices | https://www.moleculardevices.com/products/cellular-imaging-systems/acquisition-and-analysis-software/metaxpress#gref |
| MetaXpress N/C translocation-enhanced algorithm | (Hayes et al., 2020, 2021) | N/A |
| MetaXpress granularity algorithm | This study | N/A |
| ImageQuant TL (v7.0) | GE Healthcare | https://imagequanttl.software.informer.com/7.0/ |
| Prism (v9.2.0–9.3.1) | Graphpad | https://www.graphpad.com/ |
| FIJI/Image J (v2.1.0/1.53c) | NIH | https://imagej.net/software/fiji/ |
| Photoshop 2021 (v22.4.3) | Adobe | https://www.adobe.com/products/photoshop.html |
| Other | ||
| ImageXpress Micro Confocal System | Molecular Devices | N/A |
| LSM980 Confocal Microscope | Zeiss | N/A |
| Trans-Blot Turbo Transfer System | Bio-Rad | Cat#1704150 |
| Nano-drop 1000 | Thermo Scientific | N/A |
| QuantStudio 3 Real-Time PCR System | ThermoFisher | N/A |
| ImageQuant LAS 4000 System | GE Healthcare | N/A |
| Spectramax M3 plate reader | Molecular Devices | N/A |
| Thermomixer F1.5 | Eppendorf | Cat#5384000020 |
| UV Stratalinker 1800 | Stratagene | N/A |
Highlights.
Binding to nuclear GU-rich RNAs restricts TDP-43 availability for nuclear export
RNA-binding domain mutations disrupt TDP-43 nuclear localization
Perturbations of RNA transcription, splicing, or export alter TDP-43 localization
TDP-43 unbound from RNA exits nuclei by passive diffusion via nuclear pore channels
ACKNOWLEDGMENTS
We thank Lin Xue, Svetlana Vidensky, and Lin Jin for technical assistance, Pei-Hsun Wu for imaging assistance, Nae-Yuh Wang from the JHU Biostatistics Center for statistical consultation, and Lyudmila Mamedova and Barbara Smith for administrative support. The graphical abstract was created with BioRender.com. This research was supported by NIH K08NS104273 (L.R.H.), NIH R01NS123538 (L.R.H.), NIH R03NS127011 (P.K., L.R.H.), DOD/CDRMP W81XWH1910209 (J.D.R., L.R.H.), the Robert Packard Center for ALS Research (L.R.H., P.K.), the Guy McKhann Scholar Award (L.R.H.), NICHD Division of Intramural Research Funds ZIA-HD008954 (M.D.), and T32GM007445 (B.L.Z.).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.111106.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Adam SA, Marr RS, and Gerace L (1990). Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J. Cell Biol 111, 807–816. 10.1083/jcb.111.3.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afroz T, Hock E-M, Ernst P, Foglieni C, Jambeau M, Gilhespy LAB, Laferrière F, Maniecka Z, Plückthun A, Mittl P, et al. (2017). Functional and dynamic polymerization of the ALS-linked protein TDP-43 antagonizes its pathologic aggregation. Nat. Commun 8, 45. 10.1038/s41467-017-00062-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aksenova V, Smith A, Lee H, Bhat P, Esnault C, Chen S, Iben J, Kaufhold R, Yau KC, Echeverria C, et al. (2020). Nucleoporin TPR is an integral component of the TREX-2 mRNA export pathway. Nat. Commun 11, 4577. 10.1038/s41467-020-18266-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai T, Hasegawa M, Akiyama H, Ikeda K, Nonaka T, Mori H, Mann D, Tsuchiya K, Yoshida M, Hashizume Y, and Oda T (2006). TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem. Biophys. Res. Commun 351, 602–611. 10.1016/j.bbrc.2006.10.093. [DOI] [PubMed] [Google Scholar]
- Archbold HC, Jackson KL, Arora A, Weskamp K, Tank EMH, Li X, Miguez R, Dayton RD, Tamir S, Klein RL, and Barmada SJ (2018). TDP43 nuclear export and neurodegeneration in models of amyotrophic lateral sclerosis and frontotemporal dementia. Sci. Rep 8, 4606. 10.1038/s41598-018-22858-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avendaño-Vázquez SE, Dhir A, Bembich S, Buratti E, Proudfoot N, and Baralle FE (2012). Autoregulation of TDP-43 mRNA levels involves interplay between transcription, splicing, and alternative polyA site selection. Genes Dev. 26, 1679–1684. 10.1101/gad.194829.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala YM, Zago P, D’Ambrogio A, Xu Y-F, Petrucelli L, Buratti E, and Baralle FE (2008). Structural determinants of the cellular localization and shuttling of TDP-43. J. Cell Sci 121, 3778–3785. 10.1242/jcs.038950. [DOI] [PubMed] [Google Scholar]
- Baejen C, Torkler P, Gressel S, Essig K, Söding J, and Cramer P (2014). Transcriptome maps of mRNP biogenesis factors define pre-mRNA recognition. Mol. Cell 55, 745–757. 10.1016/j.molcel.2014.08.005. [DOI] [PubMed] [Google Scholar]
- Bastiaens P, Caudron M, Niethammer P, and Karsenti E (2006). Gradients in the self-organization of the mitotic spindle. Trends Cell Biol. 16, 125–134. 10.1016/j.tcb.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Bhardwaj A, Myers MP, Buratti E, and Baralle FE (2013). Characterizing TDP-43 interaction with its RNA targets. Nucleic Acids Res. 41, 5062–5074. 10.1093/nar/gkt189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borroni B, Bonvicini C, Alberici A, Buratti E, Agosti C, Archetti S, Papetti A, Stuani C, Di Luca M, Gennarelli M, and Padovani A (2009). Mutation within TARDBP leads to Frontotemporal Dementia without motor neuron disease. Hum. Mutat 30, E974–E983. 10.1002/humu.21100. [DOI] [PubMed] [Google Scholar]
- Boswell SA, Snavely A, Landry HM, Churchman LS, Gray JM, and Springer M (2017). Total RNA-seq to identify pharmacological effects on specific stages of mRNA synthesis. Nat. Chem. Biol 13, 501–507. 10.1038/nchembio.2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresson S, and Tollervey D (2018). Surveillance-ready transcription: nuclear RNA decay as a default fate. Open Biol. 8, 170270. 10.1098/rsob.170270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanden Broeck L, Callaerts P, and Dermaut B (2014). TDP-43-mediated neurodegeneration: towards a loss-of-function hypothesis? Trends Mol. Med 20, 66–71. 10.1016/j.molmed.2013.11.003. [DOI] [PubMed] [Google Scholar]
- Bruun GH, Doktor TK, Borch-Jensen J, Masuda A, Krainer AR, Ohno K, and Andresen BS (2016). Global identification of hnRNP A1 binding sites for SSO-based splicing modulation. BMC Biol. 14, 54. 10.1186/s12915-016-0279-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratti E, and Baralle FE (2001). Characterization and functional implications of the RNA binding properties of nuclear factor TDP-43, a novel splicing regulator of CFTR exon 9. J. Biol. Chem 276, 36337–36343. 10.1074/jbc.m104236200. [DOI] [PubMed] [Google Scholar]
- Buratti E (2015). Functional significance of TDP-43 mutations in disease. Adv. Genet 91, 1–53. 10.1016/bs.adgen.2015.07.001. [DOI] [PubMed] [Google Scholar]
- Butti Z, and Patten SA (2019). RNA dysregulation in amyotrophic lateral sclerosis. Front. Genet 9, 712. 10.3389/fgene.2018.00712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cáceres JF, Screaton GR, and Krainer AR (1998).A specific subset of SR proteins shuttles continuously between the nucleus and the cytoplasm. Genes Dev. 12, 55–66. 10.1101/gad.12.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter GC, Hsiung C-H, Simpson L, Yang H, and Zhang X (2021). N-Terminal domain of TDP43 enhances liquid-liquid phase separation of globular proteins. J. Mol. Biol 433, 166948. 10.1016/j.jmb.2021.166948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho T, Martins S, Rino J, Marinho S, and Carmo-Fonseca M (2017). Pharmacological inhibition of the spliceosome subunit SF3b triggers exon junction complex-independent nonsense-mediated decay. J. Cell Sci 130, 1519–1531. 10.1242/jcs.202200. [DOI] [PubMed] [Google Scholar]
- Cassany A, and Gerace L (2009). Reconstitution of nuclear import in permeabilized cells. Methods Mol. Biol 464, 181–205. 10.1007/978-1-60327-461-6_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha HJ, Uyan Ö, Kai Y, Liu T, Zhu Q, Tothova Z, Botten GA, Xu J, Yuan G-C, Dekker J, and Orkin SH (2021). Inner nuclear protein Matrin-3 coordinates cell differentiation by stabilizing chromatin architecture. Nat. Commun 12, 6241. 10.1038/s41467-021-26574-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi B, O’Connell JD, Iocolano AD, Coady JA, Yu Y, Gangopadhyay J, Gygi SP, and Reed R (2018a). The neurodegenerative diseases ALS and SMA are linked at the molecular level via the ASC-1 complex. Nucleic Acids Res. 46, 11939–11951. 10.1093/nar/gky1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi B, O’Connell JD, Yamazaki T, Gangopadhyay J, Gygi SP, and Reed R (2018b). Interactome analyses revealed that the U1 snRNP machinery overlaps extensively with the RNAP II machinery and contains multiple ALS/SMA-causative proteins. Sci. Rep 8, 8755. 10.1038/s41598-018-27136-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HS, Hwang CK, Song KY, Law P-Y, Wei L-N, and Loh HH (2009). Poly(C)-binding proteins as transcriptional regulators of gene expression. Biochem. Biophys. Res. Commun 380, 431–436. 10.1016/j.bbrc.2009.01.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke JP, Thibault PA, Salapa HE, and Levin MC (2021). A comprehensive analysis of the role of hnRNP A1 function and dysfunction in the pathogenesis of neurodegenerative disease. Front. Mol. Biosci 8, 659610. 10.3389/fmolb.2021.659610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen TJ, Hwang AW, Restrepo CR, Yuan C-X, Trojanowski JQ, and Lee VMY (2015). An acetylation switch controls TDP-43 function and aggregation propensity. Nat. Commun 6, 5845. 10.1038/ncomms6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colbeau A, Nachbaur J, and Vignais PM (1971). Enzymic characterization and lipid composition of rat liver subcellular membranes. Biochim. Biophys. Acta 249, 462–492. 10.1016/0005-2736(71)90123-4. [DOI] [PubMed] [Google Scholar]
- Conicella AE, Dignon GL, Zerze GH, Schmidt HB, D’Ordine AM, Kim YC, Rohatgi R, Ayala YM, Mittal J, and Fawzi NL (2020). TDP-43 α-helical structure tunes liquid–liquid phase separation and function. Proc. Natl. Acad. Sci. USA 117, 5883–5894. 10.1073/pnas.1912055117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conicella A, Zerze G, Mittal J, and Fawzi N (2016). ALS mutations disrupt phase separation mediated by α-helical structure in the TDP-43 low-complexity C-terminal domain. Structure 24, 1537–1549. 10.1016/j.str.2016.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corley M, Burns MC, and Yeo GW (2020). How RNA-binding proteins interact with RNA: molecules and mechanisms. Mol. Cell 78, 9–29. 10.1016/j.molcel.2020.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day RN, and Davidson MW (2009). The fluorescent protein palette: tools for cellular imaging. Chem. Soc. Rev 38, 2887–2921. 10.1039/b901966a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Deerlin VM, Leverenz JB, Bekris LM, Bird TD, Yuan W, Elman LB, Clay D, Wood EM, Chen-Plotkin AS, Martinez-Lage M, et al. (2008). TARDBP mutations in amyotrophic lateral sclerosis with TDP-43 neuropathology: a genetic and histopathological analysis. Lancet Neurol. 7, 409–416. 10.1016/s1474-4422(08)70071-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drutovic D, Duan X, Li R, Kalab P, and Solc P (2020). RanGTP and importin β regulate meiosis I spindle assembly and function in mouse oocytes. EMBO J. 39, e101689. 10.15252/embj.2019101689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ederle H, and Dormann D (2017). TDP-43 and FUS en route from the nucleus to the cytoplasm. FEBS Lett. 591, 1489–1507. 10.1002/1873-3468.12646. [DOI] [PubMed] [Google Scholar]
- Ederle H, Funk C, Abou-Ajram C, Hutten S, Funk EBE, Kehlenbach RH, Bailer SM, and Dormann D (2018). Nuclear egress of TDP-43 and FUS occurs independently of Exportin-1/CRM1. Sci. Rep 8, 7084. 10.1038/s41598-018-25007-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elden AC, Kim H-J, Hart MP, Chen-Plotkin AS, Johnson BS, Fang X, Armakola M, Geser F, Greene R, Lu MM, et al. (2010). Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature 466, 1069–1075. 10.1038/nature09320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcón-Pérez JM, Nazarian R, Sabatti C, and Dell’Angelica EC (2005). Distribution and dynamics of Lamp1-containing endocytic organelles in fibroblasts deficient in BLOC-3. J. Cell Sci 118, 5243–5255. 10.1242/jcs.02633. [DOI] [PubMed] [Google Scholar]
- Fan XC, and Steitz JA (1998). HNS, a nuclear-cytoplasmic shuttling sequence in HuR. Proc. Natl. Acad. Sci. USA 95, 15293–15298. 10.1073/pnas.95.26.15293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores BN, Li X, Malik AM, Martinez J, Beg AA, and Barmada SJ (2019). An intramolecular salt bridge linking TDP43 RNA binding, protein stability, and TDP43-dependent neurodegeneration. Cell Rep. 27, 1133–1150.e8. 10.1016/j.celrep.2019.03.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- François-Moutal L, Perez-Miller S, Scott DD, Miranda VG, Mollasalehi N, and Khanna M (2019). Structural insights into TDP-43 and effects of post-translational modifications. Front. Mol. Neurosci 12, 301. 10.3389/fnmol.2019.00301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freibaum BD, Chitta RK, High AA, and Taylor JP (2010). Global analysis of TDP-43 interacting proteins reveals strong association with RNA splicing and translation machinery. J. Proteome Res 9, 1104–1120. 10.1021/pr901076y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French RL, Grese ZR, Aligireddy H, Dhavale DD, Reeb AN, Kedia N, Kotzbauer PT, Bieschke J, and Ayala YM (2019). Detection of TAR DNA-binding protein 43 (TDP-43) oligomers as initial intermediate species during aggregate formation. J. Biol. Chem 294, 6696–6709. 10.1074/jbc.ra118.005889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey S, Rees R, Schünemann J, Ng SC, Fünfgeld K, Huyton T, and Görlich D (2018). Surface properties determining passage rates of proteins through nuclear pores. Cell 174, 202–217.e9. 10.1016/j.cell.2018.05.045. [DOI] [PubMed] [Google Scholar]
- Furlanis E, Traunmüller L, Fucile G, and Scheiffele P (2019). Landscape of ribosome-engaged transcript isoforms reveals extensive neuronal-cell-class-specific alternative splicing programs. Nat. Neurosci 22, 1709–1717. 10.1038/s41593-019-0465-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasset-Rosa F, Lu S, Yu H, Chen C, Melamed Z, Guo L, Shorter J, Da Cruz S, and Cleveland DW (2019). Cytoplasmic TDP-43 de-mixing independent of stress granules drives inhibition of nuclear import, loss of nuclear TDP-43, and cell death. Neuron 102, 339–357.e7. 10.1016/j.neuron.2019.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, and Bairoch A (2005). The Proteomics Protocols Handbook, pp. 571–607. [Google Scholar]
- Gilbertson S, Federspiel JD, Hartenian E, Cristea IM, and Glaunsinger B (2018). Changes in mRNA abundance drive shuttling of RNA binding proteins, linking cytoplasmic RNA degradation to transcription. Elife 7, e37663. 10.7554/elife.37663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitcho MA, Baloh RH, Chakraverty S, Mayo K, Norton JB, Levitch D, Hatanpaa KJ, White CL, Bigio EH, Caselli R, et al. (2008). TDP-43 A315T mutation in familial motor neuron disease. Ann. Neurol 63, 535–538. 10.1002/ana.21344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopal PP, Nirschl JJ, Klinman E, and Holzbaur ELF (2017). Amyotrophic lateral sclerosis-linked mutations increase the viscosity of liquid-like TDP-43 RNP granules in neurons. Proc. Natl. Acad. Sci. USA 114, E2466–E2475. 10.1073/pnas.1614462114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goss DJ, and Kleiman FE (2013). Poly(A) binding proteins: are they all created equal? Wiley Interdiscip. Rev. RNA 4, 167–179. 10.1002/wrna.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grünwald D, Singer RH, and Rout M (2011). Nuclear export dynamics of RNA-protein complexes. Nature 475, 333–341. 10.1038/nature10318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Katsantoni M, Köster T, Marks J, Mukherjee J, Staiger D, Ule J, and Zavolan M (2021). CLIP and complementary methods. Nat. Rev. Methods Primers 1, 20. 10.1038/s43586-021-00018-1. [DOI] [Google Scholar]
- Hayes LR, Duan L, Bowen K, Kalab P, and Rothstein JD (2020). C9orf72 arginine-rich dipeptide repeat proteins disrupt karyopherin-mediated nuclear import. Elife 9, e51685. 10.7554/elife.51685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes LR, Duan L, Vidensky S, and Kalab P (2021). Nuclear transport assays in permeabilized mouse cortical neurons. J. Vis. Exp 10.3791/62710. [DOI] [PubMed] [Google Scholar]
- Hermey G, Blüthgen N, and Kuhl D (2017). Neuronal activity-regulated alternative mRNA splicing. Int. J. Biochem. Cell Biol 91, 184–193. 10.1016/j.biocel.2017.06.002. [DOI] [PubMed] [Google Scholar]
- Hodge CA, Tran EJ, Noble KN, Alcazar-Roman AR, Ben-Yishay R, Scarcelli JJ, Folkmann AW, Shav-Tal Y, Wente SR, and Cole CN (2011). The Dbp5 cycle at the nuclear pore complex during mRNA export I: dbp5 mutants with defects in RNA binding and ATP hydrolysis define key steps for Nup159 and Gle1. Genes Dev. 25, 1052–1064. 10.1101/gad.2041611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iradi MCG, Triplett JC, Thomas JD, Davila R, Crown AM, Brown H, Lewis J, Swanson MS, Xu G, Rodriguez-Lebron E, and Borchelt DR (2018). Characterization of gene regulation and protein interaction networks for Matrin 3 encoding mutations linked to amyotrophic lateral sclerosis and myopathy. Sci. Rep 8, 4049. 10.1038/s41598-018-21371-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffrey SR, and Wilkinson MF (2018). Nonsense-mediated RNA decay in the brain: emerging modulator of neural development and disease. Nat. Rev. Neurosci 19, 715–728. 10.1038/s41583-018-0079-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain N, Lin H-C, Morgan CE, Harris ME, and Tolbert BS (2017). Rules of RNA specificity of hnRNP A1 revealed by global and quantitative analysis of its affinity distribution. Proc. Natl. Acad. Sci. USA 114, 2206–2211. 10.1073/pnas.1616371114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jao CY, and Salic A (2008). Exploring RNA transcription and turnover in vivo by using click chemistry. Proc. Natl. Acad. Sci. USA 105, 15779–15784. 10.1073/pnas.0808480105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junod SL, Kelich JM, Ma J, and Yang W (2020). Nucleocytoplasmic transport of intrinsically disordered proteins studied by high-speed super-resolution microscopy. Protein Sci. 29, 1459–1472. 10.1002/pro.3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabashi E, Valdmanis PN, Dion P, Spiegelman D, McConkey BJ, Velde CV, Bouchard J-P, Lacomblez L, Pochigaeva K, Salachas F, et al. (2008). TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat. Genet 40, 572–574. 10.1038/ng.132. [DOI] [PubMed] [Google Scholar]
- Kaláb P, Pralle A, Isacoff EY, Heald R, and Weis K (2006). Analysis of a RanGTP-regulated gradient in mitotic somatic cells. Nature 440, 697–701. 10.1038/nature04589. [DOI] [PubMed] [Google Scholar]
- Kapinos LE, Huang B, Rencurel C, and Lim RY (2017). Karyopherins regulate nuclear pore complex barrier and transport function. J. Cell Biol 216, 3609–3624. 10.1083/jcb.201702092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilchert C, Wittmann S, and Vasiljeva L (2016). The regulation and functions of the nuclear RNA exosome complex. Nat. Rev. Mol. Cell Biol 17, 227–239. 10.1038/nrm.2015.15. [DOI] [PubMed] [Google Scholar]
- Kondo Y, Oubridge C, van Roon AMM, and Nagai K (2015). Crystal structure of human U1 snRNP, a small nuclear ribonucleoprotein particle, reveals the mechanism of 5′ splice site recognition. Elife 4, e04986. 10.7554/elife.04986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotake Y, Sagane K, Owa T, Mimori-Kiyosue Y, Shimizu H, Uesugi M, Ishihama Y, Iwata M, and Mizui Y (2007). Splicing factor SF3b as a target of the antitumor natural product pladienolide. Nat. Chem. Biol 3, 570–575. 10.1038/nchembio.2007.16. [DOI] [PubMed] [Google Scholar]
- Kovacs GG, Murrell JR, Horvath S, Haraszti L, Majtenyi K, Molnar MJ, Budka H, Ghetti B, and Spina S (2009). TARDBP variation associated with frontotemporal dementia, supranuclear gaze palsy, and chorea. Movement Disord. 24, 1842–1847. 10.1002/mds.22697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebedeva S, Jens M, Theil K, Schwanhäusser B, Selbach M, Landthaler M, and Rajewsky N (2011). Transcriptome-wide analysis of regulatory interactions of the RNA-binding protein HuR. Mol. Cell 43, 340–352. 10.1016/j.molcel.2011.06.008. [DOI] [PubMed] [Google Scholar]
- Lee Y, and Rio DC (2015). Mechanisms and regulation of alternative pre-mRNA splicing. Annu. Rev. Biochem 84, 291–323. 10.1146/annurev-biochem-060614-034316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin DH, Correia AR, Cai SW, Huber FM, Jette CA, and Hoelz A (2018). Structural and functional analysis of mRNA export regulation by the nuclear pore complex. Nat. Commun 9, 2319. 10.1038/s41467-018-04459-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling JP, Pletnikova O, Troncoso JC, and Wong PC (2015). TDP-43 repression of nonconserved cryptic exons is compromised in ALS-FTD. Science 349, 650–655. 10.1126/science.aab0983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling JP, Wilks C, Charles R, Leavey PJ, Ghosh D, Jiang L, Santiago CP, Pang B, Venkataraman A, Clark BS, et al. (2020). ASCOT identifies key regulators of neuronal subtype-specific splicing. Nat. Commun 11, 137. 10.1038/s41467-019-14020-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukavsky PJ, Daujotyte D, Tollervey JR, Ule J, Stuani C, Buratti E, Baralle FE, Damberger FF, and Allain FH-T (2013). Molecular basis of UG-rich RNA recognition by the human splicing factor TDP-43. Nat. Struct. Mol. Biol 20, 1443–1449. 10.1038/nsmb.2698. [DOI] [PubMed] [Google Scholar]
- Maharana S, Wang J, Papadopoulos DK, Richter D, Pozniakovsky A, Poser I, Bickle M, Rizk S, Guillén-Boixet J, Franzmann TM, et al. (2018). RNA buffers the phase separation behavior of prion-like RNA binding proteins. Science 360, 918–921. 10.1126/science.aar7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann JR, Gleixner AM, Mauna JC, Gomes E, DeChellis-Marks MR, Needham PG, Copley KE, Hurtle B, Portz B, Pyles NJ, et al. (2019). RNA binding antagonizes neurotoxic phase transitions of TDP-43. Neuron 102, 321–338.e8. 10.1016/j.neuron.2019.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauger O, Lemoine F, and Scheiffele P (2016). Targeted intron retention and excision for rapid gene regulation in response to neuronal activity. Neuron 92, 1266–1278. 10.1016/j.neuron.2016.11.032. [DOI] [PubMed] [Google Scholar]
- McCloskey A, Taniguchi I, Shinmyozu K, and Ohno M (2012). hnRNP C tetramer measures RNA length to classify RNA polymerase II transcripts for export. Science 335, 1643–1646. 10.1126/science.1218469. [DOI] [PubMed] [Google Scholar]
- Merendino L, Guth S, Bilbao D, Martínez C, and Valcárcel J (1999). Inhibition of msl-2 splicing by Sex-lethal reveals interaction between U2AF35 and the 3′ splice site AG. Nature 402, 838–841. 10.1038/45602. [DOI] [PubMed] [Google Scholar]
- Michael WM, Choi M, and Dreyfuss G (1995). A nuclear export signal in hnRNP A1: a signal-mediated, temperature-dependent nuclear protein export pathway. Cell 83, 415–422. 10.1016/0092-8674(95)90119-1. [DOI] [PubMed] [Google Scholar]
- Michael WM, Eder PS, and Dreyfuss G (1997). The K nuclear shuttling domain: a novel signal for nuclear import and nuclear export in the hnRNP K protein. EMBO J. 16, 3587–3598. 10.1093/emboj/16.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr D, Frey S, Fischer T, Güttler T, and Görlich D (2009). Characterisation of the passive permeability barrier of nuclear pore complexes. EMBO J. 28, 2541–2553. 10.1038/emboj.2009.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj A, Kim H, Mittag T, and Taylor J (2015). Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163, 123–133. 10.1016/j.cell.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mompeán M, Romano V, Pantoja-Uceda D, Stuani C, Baralle FE, Buratti E, and Laurents DV (2017). Point mutations in the N-terminal domain of transactive response DNA-binding protein 43 kDa (TDP-43) compromise its stability, dimerization, and functions. J. Biol. Chem 292, 11992–12006. 10.1074/jbc.m117.775965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakielny S, and Dreyfuss G (1996). The hnRNP C proteins contain a nuclear retention sequence that can override nuclear export signals. J. Cell Biol 134, 1365–1373. 10.1083/jcb.134.6.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakielny S, and Dreyfuss G (1999). Transport of proteins and RNAs in and out of the nucleus. Cell 99, 677–690. 10.1016/s0092-8674(00)81666-9. [DOI] [PubMed] [Google Scholar]
- Napetschnig J, Kassube SA, Debler EW, Wong RW, Blobel G, and Hoelz A (2009). Structural and functional analysis of the interaction between the nucleoporin Nup214 and the DEAD-box helicase Ddx19. Proc. Natl. Acad. Sci. USA 106, 3089–3094. 10.1073/pnas.0813267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM, et al. (2006). Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314, 130–133. 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- Nguyen TM, Kabotyanski EB, Reineke LC, Shao J, Xiong F, Lee J-H, Dubrulle J, Johnson H, Stossi F, Tsoi PS, et al. (2019). The SINEB1 element in the long non-coding RNA Malat1 is necessary for TDP-43 proteostasis. Nucleic Acids Res. 48, 2621–2642. 10.1093/nar/gkz1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura AL, Župunski V, Troakes C, Kathe C, Fratta P, Howell M, Gallo J, Hortobágyi T, Shaw CE, and Rogelj B (2010). Nuclear import impairment causes cytoplasmic trans-activation response DNA-binding protein accumulation and is associated with frontotemporal lobar degeneration. Brain 133, 1763–1771. 10.1093/brain/awq111. [DOI] [PubMed] [Google Scholar]
- Nostrand ELV, Freese P, Pratt GA, Wang X, Wei X, Xiao R, Blue SM, Chen J-Y, Cody NAL, Dominguez D, et al. (2020). A large-scale binding and functional map of human RNA-binding proteins. Nature 583, 711–719. 10.1038/s41586-020-2077-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussbacher JK, Tabet R, Yeo GW, and Lagier-Tourenne C (2019). Disruption of RNA metabolism in neurological diseases and emerging therapeutic interventions. Neuron 102, 294–320. 10.1016/j.neuron.2019.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien K, Matlin AJ, Lowell AM, and Moore MJ (2008). The biflavonoid isoginkgetin is a general inhibitor of pre-mRNA splicing. J. Biol. Chem 283, 33147–33154. 10.1074/jbc.m805556200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura M, Inose H, and Masuda S (2015). RNA export through the NPC in eukaryotes. Genes(Basel) 6, 124–149. 10.3390/genes6010124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson CM, Jiang B, Erb MA, Liang Y, Doctor ZM, Zhang Z, Zhang T, Kwiatkowski N, Boukhali M, Green JL, et al. (2018). Pharmacological perturbation of CDK9 using selective CDK9 inhibition or degradation. Nat. Chem. Biol 14, 163–170. 10.1038/nchembio.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paci G, Zheng T, Caria J, Zilman A, and Lemke EA (2020). Molecular determinants of large cargo transport into the nucleus. Elife 9, e55963. 10.7554/elife.55963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinarbasi ES, Cağatay T, Fung HYJ, Li YC, Chook YM, and Thomas PJ (2018). Active nuclear import and passive nuclear export are the primary determinants of TDP-43 localization. Sci. Rep 8, 7083. 10.1038/s41598-018-25008-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piñol-Roma S, and Dreyfuss G (1992). Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature 355, 730–732. 10.1038/355730a0. [DOI] [PubMed] [Google Scholar]
- Polymenidou M, Lagier-Tourenne C, Hutt KR, Huelga SC, Moran J, Liang TY, Ling S-C, Sun E, Wancewicz E, Mazur C, et al. (2011). Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat. Neurosci 14, 459–468. 10.1038/nn.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad A, Bharathi V, Sivalingam V, Girdhar A, and Patel BK (2019). Molecular mechanisms of TDP-43 misfolding and pathology in amyotrophic lateral sclerosis. Front. Mol. Neurosci 12, 25. 10.3389/fnmol.2019.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AJ, Hwang T, Tao R, Burke EE, Rajpurohit A, Shin JH, Hyde TM, Kleinman JE, Jaffe AE, and Weinberger DR (2019). Characterizing the nuclear and cytoplasmic transcriptomes in developing and mature human cortex uncovers new insight into psychiatric disease gene regulation. Genome Res. 30, 1–11. 10.1101/gr.250217.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafiee M-R, Zagalak JA, Sidorov S, Steinhauser S, Davey K, Ule J, and Luscombe NM (2021). Chromatin-contact atlas reveals disorder-mediated protein interactions and moonlighting chromatin-associated RBPs. Preprint at bioRxiv. 10.1101/2020.07.13.200212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribbeck K, and Görlich D (2002). The permeability barrier of nuclear pore complexes appears to operate via hydrophobic exclusion. EMBO J. 21, 2664–2671. 10.1093/emboj/21.11.2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripin N, Boudet J, Duszczyk MM, Hinniger A, Faller M, Krepl M, Gadi A, Schneider RJ, Šponer J, Meisner-Kober NC, and Allain FHT (2019). Molecular basis for AU-rich element recognition and dimerization by the HuR C-terminal RRM. Proc. Natl. Acad. Sci. USA 116, 2935–2944. 10.1073/pnas.1808696116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roczniak-Ferguson A, and Ferguson SM (2019). Pleiotropic requirements for human TDP-43 in the regulation of cell and organelle homeostasis. Life Sci. Alliance 2, e201900358. 10.26508/lsa.201900358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini H, Bicknell AA, Eddy SR, and Moore MJ (2019). Free circular introns with an unusual branchpoint in neuronal projections. Elife 8, e47809. 10.7554/elife.47809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salton M, Elkon R, Borodina T, Davydov A, Yaspo M-L, Halperin E, and Shiloh Y (2011). Matrin 3 binds and stabilizes mRNA. PLoS One 6, e23882. 10.1371/journal.pone.0023882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, Muguruma N, Nakagawa T, Okamoto K, Kimura T, Kitamura S, Yano H, Sannomiya K, Goji T, Miyamoto H, et al. (2014). High antitumor activity of pladienolide B and its derivative in gastric cancer. Cancer Sci. 105, 110–116. 10.1111/cas.12317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharifi S, and Bierhoff H (2018). Regulation of RNA polymerase I transcription in development, disease, and aging. Annu. Rev. Biochem 87, 51–73. 10.1146/annurev-biochem-062917-012612. [DOI] [PubMed] [Google Scholar]
- de Silanes IL, Zhan M, Lal A, Yang X, and Gorospe M (2004). Identification of a target RNA motif for RNA-binding protein HuR. Proc. Natl. Acad. Sci. USA 101, 2987–2992. 10.1073/pnas.0306453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snapp EL, Hegde RS, Francolini M, Lombardo F, Colombo S, Pedrazzini E, Borgese N, and Lippincott-Schwartz J (2003). Formation of stacked ER cisternae by low affinity protein interactions. J. Cell Biol 163, 257–269. 10.1083/jcb.200306020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soh S, Byrska M, Kandere-Grzybowska K, and Grzybowski B (2010). Reaction-diffusion systems in intracellular molecular transport and control. Angew. Chem. Int. Ed. Engl 49, 4170–4198. 10.1002/anie.200905513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreedharan J, Blair IP, Tripathi VB, Hu X, Vance C, Rogelj B, Ackerley S, Durnall JC, Williams KL, Buratti E, et al. (2008). TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science 319, 1668–1672. 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Chen W, Chen L, and Zheng W (2021). Exploring the molecular basis of UG-rich RNA recognition by the human splicing factor TDP-43 using molecular dynamics simulation and free energy calculation. J. Comput. Chem 42, 1670–1680. 10.1002/jcc.26704. [DOI] [PubMed] [Google Scholar]
- Talhouarne GJS, and Gall JG (2018). Lariat intronic RNAs in the cytoplasm of vertebrate cells. Proc. Natl. Acad. Sci. USA 115, E7970–E7977. 10.1073/pnas.1808816115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tank EM, Figueroa-Romero C, Hinder LM, Bedi K, Archbold HC, Li X, Weskamp K, Safren N, Paez-Colasante X, Pacut C, et al. (2018). Abnormal RNA stability in amyotrophic lateral sclerosis. Nat. Commun 9, 2845. 10.1038/s41467-018-05049-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timney BL, Raveh B, Mironska R, Trivedi JM, Kim SJ, Russel D, Wente SR, Sali A, and Rout MP (2016). Simple rules for passive diffusion through the nuclear pore complex. J. Cell Biol 215, 57–76. 10.1083/jcb.201601004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey JR, Curk T, Rogelj B, Briese M, Cereda M, Kayikci M, König J, Hortobágyi T, Nishimura AL, Župunski V, et al. (2011). Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat. Neurosci 14, 452–458. 10.1038/nn.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuck A, and Tollervey D (2013).A transcriptome-wide atlas of RNP composition reveals diverse classes of mRNAs and lncRNAs. Cell 154, 996–1009. 10.1016/j.cell.2013.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyssowski KM, and Gray JM (2019). The neuronal stimulation–transcription coupling map. Curr. Opin. Neurobiol 59, 87–94. 10.1016/j.conb.2019.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura Y, Oshima T, Yamamoto M, Reyes CJ, Costa Cruz PH, Shibuya T, and Kawahara Y (2017). Matrin3 binds directly to intronic pyrimidine-rich sequences and controls alternative splicing. Genes Cells 22, 785–798. 10.1111/gtc.12512. [DOI] [PubMed] [Google Scholar]
- Villanueva E, Smith T, Queiroz RML, Monti M, Pizzinga M, Elzek M, Dezi V, Harvey RF, Ramakrishna M, Willis AE, and Lilley KS (2020). Efficient recovery of the RNA-bound proteome and protein-bound transcriptome using phase separation (OOPS). Nat. Protoc 15, 2568–2588. 10.1038/s41596-020-0344-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldmann T, Scholten I, Kappes F, Hu HG, and Knippers R (2004). The DEK protein—an abundant and ubiquitous constituent of mammalian chromatin. Gene 343, 1–9. 10.1016/j.gene.2004.08.029. [DOI] [PubMed] [Google Scholar]
- Wang C, Duan Y, Duan G, Wang Q, Zhang K, Deng X, Qian B, Gu J, Ma Z, Zhang S, et al. (2020). Stress induces dynamic, cytotoxicity-antagonizing TDP-43 nuclear bodies via paraspeckle LncRNA NEAT1-mediated liquid-liquid phase separation. Mol. Cell 79, 443–458.e7. 10.1016/j.molcel.2020.06.019. [DOI] [PubMed] [Google Scholar]
- Weskamp K, Tank EM, Miguez R, McBride JP, Gómez NB, White M, Lin Z, Gonzalez CM, Serio A, Sreedharan J, and Barmada SJ (2020). Shortened TDP43 isoforms upregulated by neuronal hyperactivity drive TDP43 pathology in ALS. J. Clin. Invest 130, 1139–1155. 10.1172/jci130988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler EC, Nostrand ELV, and Yeo GW (2018). Advances and challenges in the detection of transcriptome-wide protein–RNA interactions. Wiley Interdiscip. Rev. RNA 9, e1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehurst AW, Wilsbacher JL, You Y, Luby-Phelps K, Moore MS, and Cobb MH (2002). ERK2 enters the nucleus by a carrier-independent mechanism. Proc. Natl. Acad. Sci. USA 99, 7496–7501. 10.1073/pnas.112495999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winton MJ, Igaz LM, Wong MM, Kwong LK, Trojanowski JQ, and Lee VMY (2008). Disturbance of nuclear and cytoplasmic TAR DNA-binding protein (TDP-43) induces disease-like redistribution, sequestration, and aggregate formation. J. Biol. Chem 283, 13302–13309. 10.1074/jbc.m800342200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Romfo CM, Nilsen TW, and Green MR (1999). Functional recognition of the 3′ splice site AG by the splicing factor U2AF35. Nature 402, 832–835. 10.1038/45590. [DOI] [PubMed] [Google Scholar]
- Wu B, Su S, Patil DP, Liu H, Gan J, Jaffrey SR, and Ma J (2018). Molecular basis for the specific and multivariant recognitions of RNA substrates by human hnRNP A2/B1. Nat. Commun 9, 420. 10.1038/s41467-017-02770-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Lu S, Gasior K, Singh D, Vazquez-Sanchez S, Tapia O, Toprani B, Beccari MS, Yates JR, Da Cruz S, et al. (2020). HSP70 chaperones RNA-free TDP-43 into anisotropic intranuclear liquid spherical shells. Science 371, eabb4309. 10.1126/science.abb4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacco E, Martin SR, Thorogate R, and Pastore A (2018). The RNA-recognition motifs of TAR DNA-binding protein 43 may play a role in the aberrant self-assembly of the protein. Front. Mol. Neurosci 11, 372. 10.3389/fnmol.2018.00372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaepfel BL, and Rothstein JD (2021). RNA is a double-edged sword in ALS pathogenesis. Front. Cell. Neurosci 15, 708181. 10.3389/fncel.2021.708181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorio DAR, and Blumenthal T (1999). Both subunits of U2AF recognize the 3′ splice site in Caenorhabditis elegans. Nature 402, 835–838. 10.1038/45597. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data reported in this paper will be shared by the lead contact upon request. Original Western blot images are included in the supplement (Data S1).
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.