Escherichia coli ST410, where the lineage is similar to the global pandemic ESBL E. coli lineage ST131, is emerging as new international high-risk clones.1 It is an extraintestinal pathogen associated with resistance to fluoroquinolones, third-generation cephalosporins and carbapenems. Equipped with enhanced pathogenicity, E. coli ST410 can cause severe and/or recurrent infections, including patient-to-patient transmission causing hospital outbreaks.2 While it has been reported that in Asia carbapenem-resistant E. coli ST410 was isolated as the causative pathogen of infections in patients of Sichuan hospitals in China and in Singapore,2,3 there is no report of this carbapenem-resistant E. coli ST410 arising from the natural water environment in Asia, which is an important aspect in the One Health perspective. The One Health perspective recognizes that the health of humans, domestic and wild animals, plants and the wider environment (including ecosystems) are closely linked and interdependent, and collaborative effort is needed to attain optimal health for all sectors. To this end, we have found a carbapenem resistant E. coli ST410 strain SrichA-1 with a blaNDM-5-bearing plasmid pSGNDM-5, isolated from reservoir water in Singapore (BioSample accession: SAMN18579051; SRA: SRP313016). A circular plasmid map of pSGNDM-5 was obtained by long-read next-generation sequencing (PacBio RS II) and a deep genetic investigation was applied.
E. coli strain SrichA-1 was obtained during the cephalosporin-resistant bacteria investigation from the Serangoon reservoir in Singapore in November 2018. A bottle of 800 mL of reservoir water was filtered with a 0.45 μm filter membrane and further enriched with the nutrition medium. The strain was selected on a Brilliance™ ESBL Agar (ThermoFisher, USA) and further confirmed as carbapenem resistant with microbroth dilution methods using Sensititre™ Extended Spectrum β-lactamase Plate (ThermoFisher, USA) (Table S1, available as Supplementary data at JAC-AMR Online). The SrichA-1 strain is interpreted as resistant to all the tested β-lactams including cephalosporins and carbapenems according to the CLSI M100 guideline.4
The genomic DNA of SrichA-1 was then extracted with the QIAGEN Genomic DNA Kit (Qiagen, Germany) and sequenced with the PacBio RS II system (PacBio, USA). The raw reads were assembled with SMART portal analysis software SMARTlink v2.3.0 (NovogeneAIT, Singapore). The strain was characterized with WGS analysis; all the sequencing analysis tools are listed in Table S3. The chromosome of SrichA-1 is 4 806 160 bp, and it was detected to contain 4786 genes. The MLST of SrichA-1 is ST410, in silico serotyping as H9, fimH24. Besides, 14 antimicrobial resistance genes were detected using CGE ResFinder 4.0 (https://cge.food.dtu.dk/services/ResFinder/), among which 12 were determined to be located on an IncF-type plasmid [F-:A1: B49],5 except mdf(A) and blaCMY-2, which were found on the chromosome. Based on both core genome (cg) MLST and cg SNP (performed via BacWGSdb; http://bacdb.cn/BacWGSTdb/analysis_single.php), the closest genome is strain SIEC197 (accession: SAMN11399743) from Thailand, which carries four of the same β-lactamase genes as our isolate.
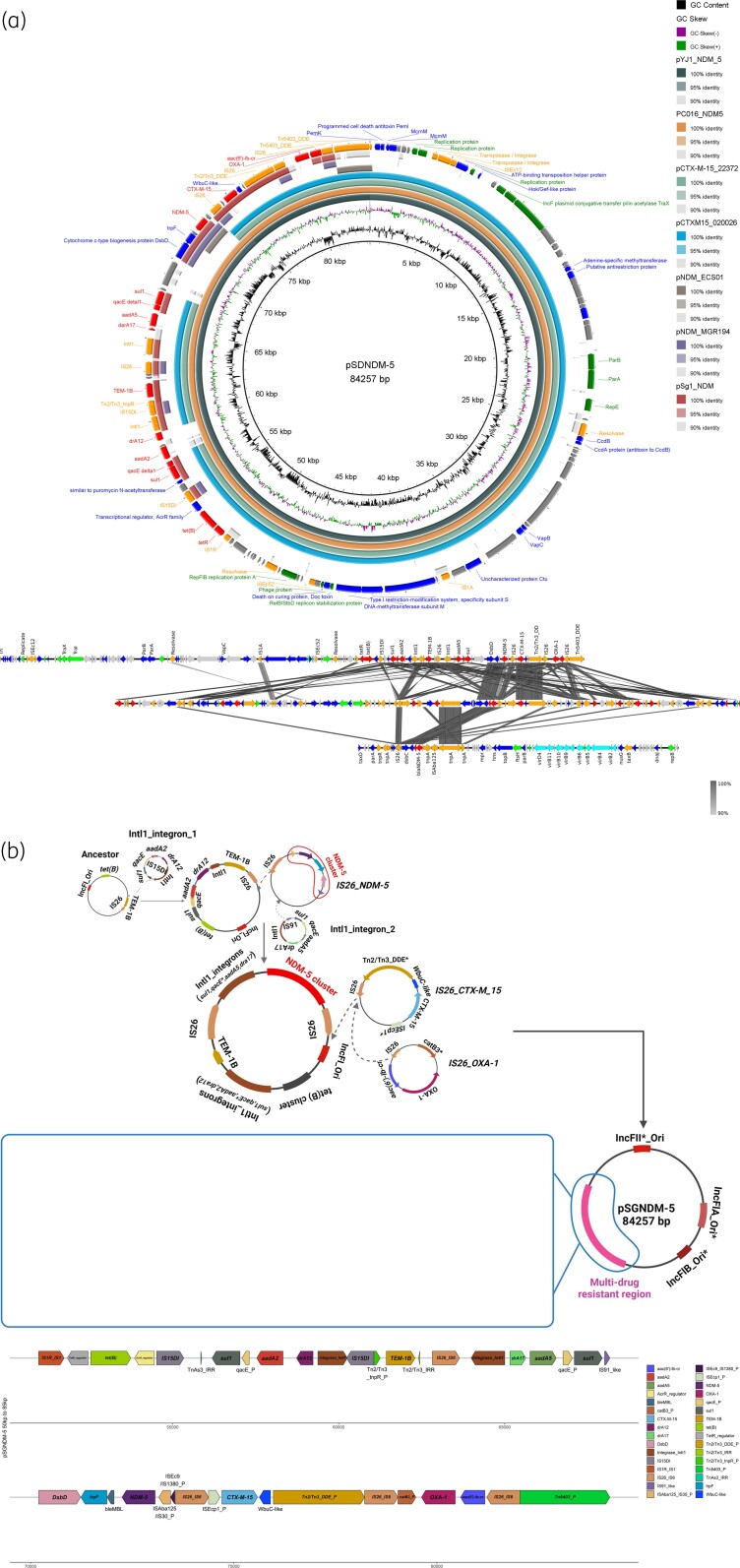
The SrichA-1 strain contains two plasmids: the one harbouring blaNDM-5 is 84 257 bp and named pSGNDM-5. Four close neighbours of pSGNDM-5 from other Asian countries have been found. The pSGNDM-5 plasmid is highly similar to pYJ1-NDM-5 and pC016-NDM5 reported from Myanmar and Thailand. Besides, pSGNDM-5 shared a similar backbone with pCTX-M15_020026 and pCTX-M15_22372 from China and India. Compared to pSGNDM-5, pCTX-M15_020026 and pCTX-M15_22372 are lacking the integron (intI1-drA12-aadA2) and blaNDM-5 (ISAba125-blaNDM-5-bleMBL-trpF-dsbD) cluster. Another circular blaNDM-5-bearing plasmid from Singapore was also chosen for comparison.6 However, their backbones are quite different; only the antimicrobial resistance region shared some conserved clusters (blaCTX-M-15, blaNDM-5, blaOXA-1, blaTEM-1B). Two normally used reference plasmids in Singapore, pNDM_ECS01 and pNDM-MGR194, are also included in the alignment, but their structures are completely different from that of pSGNDM-5 (Figure 1a and Table S2).7
Figure 1.
pSGNDM-5 genetic environment comparison analysis with other blaNDM-bearing plasmids and potential formation history. (a) pSGNDM-5 is used as reference and backbone. Seven plasmids from other Asian countries and one plasmid from Singapore (pSg-NDM) were chosen for comparison. The details of plasmids are shown in Table S2. The circle alignment was plotted with Brig (java package) and genes under different function catalogs are indicated with different colours. Genes were annotated with the RAST server. The pairwise comparison was performed with Blastn and visualized with Easyfig (java package). DNA fragments with length over 50 bp and identity over 90% are linked with grey lines. (b) The potential formation history of pSGNDM-5. The original ancestor plasmid may consequently have obtained an IS26-bounded transposon and intI1 integron harbouring resistance genes and finally formed the MDR plasmid. _P, partial, a fragment of the reference gene without full length. A higher resolution version of this figure is available in the Supplementary data.
Two integrases, intI, leading gene cassettes were detected upstream and downstream of blaTEM-1B, respectively. The downstream integron (intI1-dfrA17-aadA5) and the conserved blaNDM-5 cluster with a partial IS, ISAba125, are led by an IS26. Potentially, these two clusters are co-located on an IS26-bounded transposon during spreading.8 The blaCTX-M-15 cluster (ISEcp1-blaCTX-M-15-wubC-like), which has been reported to be chromosomal in other E. coli isolates, was also found on this plasmid. But the first 1375 bp of ISEcp1 was also replaced by IS26. Besides, the IS26 is also associated with the blaOXA-1 cluster (aac(6′)-lb-cr-blaOXA-1-catB3) on this plasmid. IS26 has been widely reported to mediate resistance transfer particularly of blaNDM-5 in other Asian countries like China.9 The high presence of IS26 in pSGNDM-5 may be a consequence of several recombinations. Perhaps during its evolution and spread, the ancestor plasmid subsequently acquired resistance genetic clusters mediated by IS26, which resulted in a 30 kb MDR region on pSGNDM-5 (Figure 1b). The genetic environment investigation of this plasmid reflects the important role of IS26 in forming MDR genomic regions.
To our knowledge, this is the first complete chromosome and circular plasmid sequence report of the environmental E. coli ST410 strain carrying blaNDM-5 from Southeast Asia, where the antimicrobial resistance rate is high. There is one report of an environmental E. coli ST410 strain, which was cephalosporin resistant but carbapenem susceptible, from Southeast Asia.10 From a One Health perspective, the finding of E. coli ST410 whose plasmid carries blaNDM-5 from the environment may suggest this resistant clone was in existence within Singapore long ago, and could potentially cause severe infections.
Supplementary Material
Contributor Information
Yang Zhong, Department of Pharmacy, Singapore General Hospital, Singapore, Singapore; Nanyang Technological University Food Technology Centre (NAFTEC), 62 Nanyang Drive, Singapore 637459, Singapore; School of Chemical and Biomedical Engineering, Nanyang Technological University, 62 Nanyang Drive, Singapore 637459, Singapore.
Siyao Guo, Nanyang Technological University Food Technology Centre (NAFTEC), 62 Nanyang Drive, Singapore 637459, Singapore; School of Chemical and Biomedical Engineering, Nanyang Technological University, 62 Nanyang Drive, Singapore 637459, Singapore.
Joergen Schlundt, Nanyang Technological University Food Technology Centre (NAFTEC), 62 Nanyang Drive, Singapore 637459, Singapore; School of Chemical and Biomedical Engineering, Nanyang Technological University, 62 Nanyang Drive, Singapore 637459, Singapore.
Andrea L Kwa, Department of Pharmacy, Singapore General Hospital, Singapore, Singapore; Emerging Infectious Diseases, Duke-NUS Medical School, Singapore, Singapore; Singhealth Duke-NUS Medicine Academic Clinical Programme, Singapore, Singapore.
Funding
The research work was supported by Nanyang Technological University and Singapore General Hospital.
Transparency declarations
None to declare.
Supplementary data
Tables S1 to S3 and a higher resolution version of Figure 1 are available as Supplementary data at JAC-AMR Online.
References
- 1. Roer L, Overballe-Petersen S, Hansen Fet al. Escherichia coli sequence type 410 is causing new international high-risk clones. mSphere 2018; 3: e00337-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Feng Y, Liu L, Lin Jet al. Key evolutionary events in the emergence of a globally disseminated, carbapenem resistant clone in the Escherichia coli ST410 lineage. Commun Biol 2019; 2: 322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Koh TH, Cao D, Tee NWet al. Escherichia coli with blaIMP-8 in Singapore. Antimicrob Agents Chemother 2014; 58: 617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. CLSI . Performance Standards for Antimicrobial Susceptibility Testing—Thirty-Second Edition: M100. 2022.
- 5. Kleinheinz KA, Joensen KG, Larsen MV. Applying the ResFinder and VirulenceFinder web-services for easy identification of acquired antibiotic resistance and E. coli virulence genes in bacteriophage and prophage nucleotide sequences. Bacteriophage 2014; 4: e27943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Khong WX, Marimuthu K, Teo Jet al. Tracking inter-institutional spread of NDM and identification of a novel NDM-positive plasmid, pSg1-NDM, using next-generation sequencing approaches. J Antimicrob Chemother 2016; 71: 3081–9. [DOI] [PubMed] [Google Scholar]
- 7. Khong WX, Xia E, Marimuthu Ket al. Local transmission and global dissemination of New Delhi metallo-β-lactamase (NDM): a whole genome analysis. BMC Genomics 2016; 17: 452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Harmer CJ, Hall RM. IS26-mediated formation of transposons carrying antibiotic resistance genes. mSphere 2016; 1: e00038-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liu Z, Xiao X, Liu Yet al. Recombination of NDM-5-producing plasmids mediated by IS26 among Escherichia coli. Int J Antimicrob Agents 2020; 55: 105815. [DOI] [PubMed] [Google Scholar]
- 10. Nadimpalli ML, de Lauzanne A, Phe Tet al. Escherichia coli ST410 among humans and the environment in Southeast Asia. Int J Antimicrob Agents 2019; 54: 228–32. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.