Preface
The global emergence of many severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants jeopardizes the protective antiviral immunity induced following infection or vaccination. To address the public health threat caused by the increasing SARS-CoV-2 genomic diversity, the National Institute of Allergy and Infectious Diseases (NIAID) within the National Institutes of Health (NIH) established the SARS-CoV-2 Assessment of Viral Evolution (SAVE) program. This effort was designed to provide a real-time risk assessment of SARS-CoV-2 variants potentially impacting transmission, virulence, and resistance to convalescent and vaccine-induced immunity. The SAVE program serves as a critical data-generating component of the United States Government SARS-CoV-2 Interagency Group to assess implications of SARS-CoV-2 variants on diagnostics, vaccines, and therapeutics and for communicating public health risk. Here we describe the coordinated approach used to identify and curate data about emerging variants, their impact on immunity, and effects on vaccine protection using animal models. We report the development of reagents, methodologies, models, and pivotal findings facilitated by this collaborative approach and identify future challenges. This program serves as a template for the response against rapidly evolving pandemic pathogens by monitoring viral evolution in the human population to identify variants that could erode the effectiveness of countermeasures.
Introduction
SARS-CoV-2, the etiologic agent of coronavirus disease 2019 (COVID-19), has caused a devastating pandemic resulting in more than 5 million deaths worldwide (https://covid19.who.int). With continuous transmission cycles occurring around the world, SARS-CoV-2 variants have arisen with mutations throughout its genome, including the spike protein gene, the principal antigenic target of all SARS-CoV-2 vaccines currently in use1,2. The rapid emergence of variants—the latest being Omicron in November 2021— has raised concerns about how new mutations impact virus replication, infectivity, transmission, and infection- and vaccine-induced immunity. This rapid genetic evolution of SARS-CoV-2 created an immediate need to monitor and characterize variants for potential resistance to medical countermeasures.
The U.S. Department of Health and Human Services established the SARS-CoV-2 Interagency Group (SIG) to maximize coordination between the Centers for Disease Control and Prevention, NIH, Food and Drug Administration, Biomedical Advanced Research and Development Authority, and Department of Defense for the U.S. public health response to the COVID-19 pandemic3. The NIAID formed the SAVE consortium in January 2021 as a critical data-generating component for the SIG and to facilitate rapid data-sharing with global partners and the scientific community (Figure 1). The SAVE program provides a comprehensive real-time risk assessment of emerging mutations in SARS-CoV-2 strains that could impact transmissibility, virulence, and infection- or vaccine-induced immunity. SAVE is constructed as a rational, structured, and iterative risk-assessment pipeline with a goal of providing critical data to support SIG actions and ensure countermeasure effectiveness against emerging variants.
Figure 1. Overview of the SAVE program.
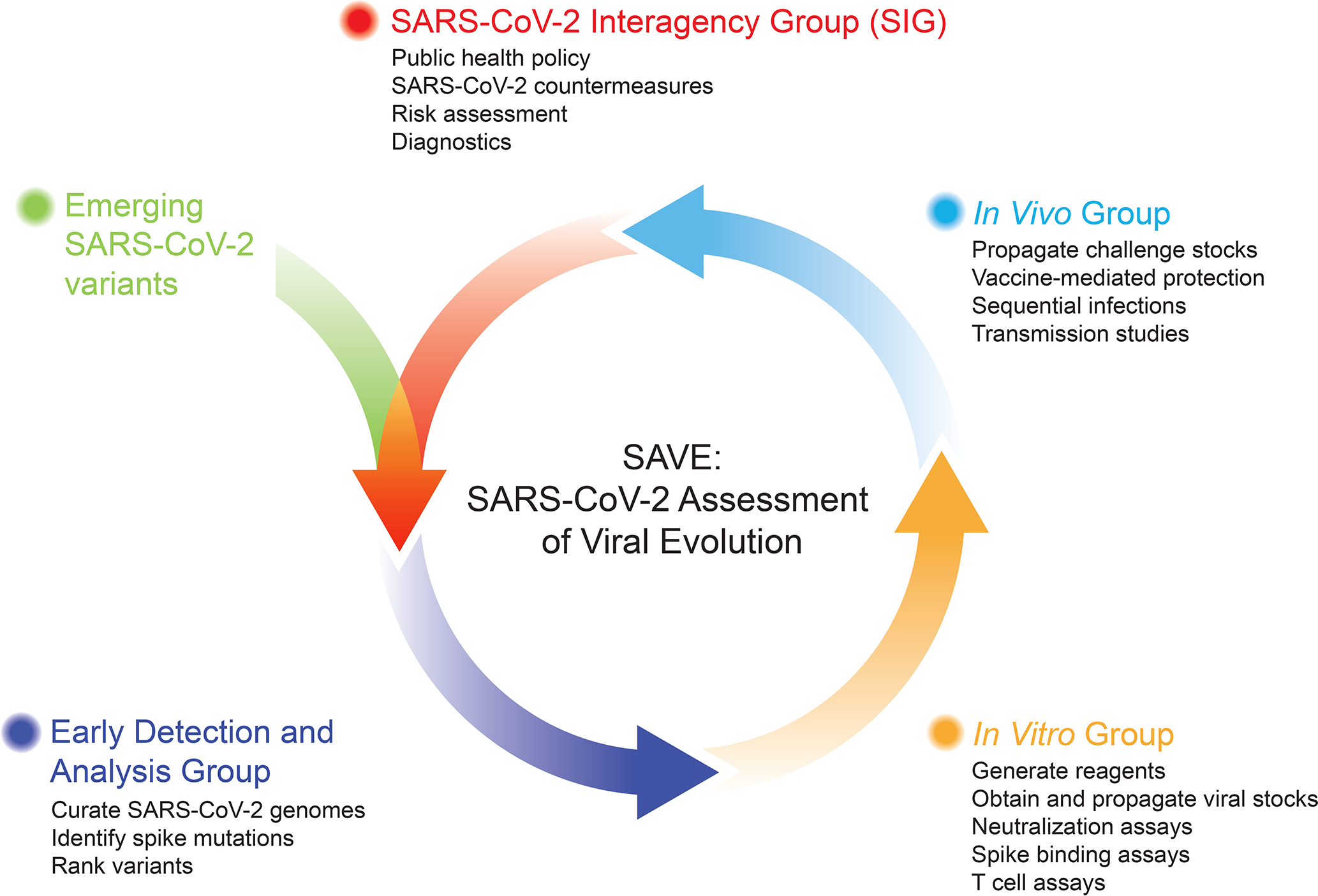
The SAVE program is divided into three working groups to provide real-time risk assessment of SARS-CoV-2 variants on infection and vaccine-induced immunity. The Early Detection and Analysis group curates and prioritizes emerging SARS-CoV-2 variants. The In Vitro group evaluates the impact of SARS-CoV-2 variants on humoral and cell-mediated immune responses. The In Vivo group uses animal models to test vaccine efficacy, transmission, and define immune mechanisms/correlates of protection. These data are fed into the SARS-CoV-2 Interagency Group (SIG), which coordinates between different U.S. government agencies to assess the impact of variants on critical SARS-CoV-2 countermeasures, including vaccines, therapeutics, and diagnostics. This iterative approach allows for information flow between the SAVE program and the SIG to continue prioritizing and testing SARS-CoV-2 variants. Created in part with BioRender.com
The SAVE program is composed of an international team of scientists with expertise in virology, immunology, vaccinology, structural biology, bioinformatics, viral genetics, and evolution. Each team member is responsible for key contributions ranging from curation of viral mutations, bioinformatics analysis, development of novel reagents, assay development and testing, in vitro characterization, and in vivo model development and countermeasure testing. The SAVE program is divided into three working groups: (1) Early Detection and Analysis group; (2) In Vitro group; and (3) In Vivo group. The Early Detection group uses public databases and analysis tools to curate and prioritize emerging SARS-CoV-2 variants. The In Vitro group evaluates the impact of SARS-CoV-2 variants on humoral and cell-mediated immune responses using in vitro assays. The In Vivo group uses small and large animal models to test vaccine efficacy, transmission, and define immune mechanisms and correlates of protection. A common theme across these subgroups is the integration of orthogonal experimental and computational approaches to validate findings and strengthen the evidence for recommendations. Collaborative efforts between the Early Detection geneticists and evolutionary biologists, and the In Vitro group virologists/immunologists allow for rapid determination of relationships between viral evolution and neutralization sensitivity. In turn, these results enable the In Vivo team to assess and evaluate vaccine protection in animal studies. The SAVE program has regularly scheduled, usually weekly, meetings that include individual subgroup meetings and an all-hands meeting, which serves as an opportunity to share key information across groups and align priorities for the most urgent experimental questions. NIAID program staff and intramural and extramural scientists share leadership responsibilities. Collaboration within and across these groups has accelerated research and discovery due to immediate and open sharing of ideas, reagents, protocols, and data4–12. The SAVE group routinely invites scientists from international sites to present a real-time assessment of SARS-CoV-2 variants and infections within their region. The SAVE group coordinates with Biodefense and Emerging Infections (BEI) Research Resources Repository, the World Reference Center for Emerging Viruses and Arboviruses (WRCEVA) and the World Health Organization (WHO) to distribute SARS-CoV-2 isolates, proteins, and plasmids. The SAVE group also has an open-face sharing policy where findings are quickly disseminated through pre-print servers while manuscripts undergo formal peer-review. The head-to-head comparison, review, and discussion of unpublished data has yielded real-time peer review that would otherwise take months to achieve.
Early Detection and Analysis group
SARS-CoV-2 genome sequencing data have been shared in public databases. As of December 2021, GISAID, the most widely used database for SARS-CoV-2, has more than 6.5 million sequences deposited with more than 150,000 sequences added weekly. This depth and rate of growth of genetic information for an emerging virus are unprecedented, providing a unique resource to track virus evolution. From late 2020, the emergence of variants of concern (VOC) with increased risk to global public health, prompted scientists to establish variant detection and tracking pipelines (e.g., Outbreak.info13; CDC SARS-CoV-2 Variant Classifications and Definitions14; BV-BRC SARS-CoV-2 Real-time Tracking and Early Warning System for Variants and Lineages of Concern (https://www.bv-brc.org)). The Early Detection and Analysis group was assembled to establish a systematic approach to identify and predict SARS-CoV-2 variants that might increase virus replication, transmission, and/or escape from immunity. The team’s main goal is to select and prioritize variants for development of key experimental reagents (e.g., spike proteins for binding assays and pseudoviruses (PSV) for neutralization assays) and viruses for challenge studies, as well as to inform the In Vitro and In Vivo groups about predicted variant properties to guide their experiments. The initial and primary focus has been on variants with mutations in the spike protein that might lead to antibody escape, with subsequent analyses considering T cell escape, infectivity, and transmission. Other important characteristics—such as replication fitness and virulence— and genomic regions outside of the spike gene are also evaluated. The process is collaborative and iterative, with seven teams using independent models and methodologies to prioritize mutations and lineages as well as rank importance for downstream testing. While the focus is on human infections, the Early Detection group also monitors variants circulating in animal populations, such as mink and deer, since they represent a potential reservoir source.
Methodology
Genomic surveillance consists of weekly downloads of SARS-CoV-2 genomes from GISAID/GENBANK, quality filtering, alignment, and the identification of variant or co-variant substitutions. The main focus has been on potential antibody escape to identify mutations in key epitopes in the receptor binding domain (RBD) and the N-terminal domain (NTD) supersite, but regions proximal to the furin cleavage site or experiencing convergent/parallel evolution are also considered. The dynamics of these spike substitutions, as a function of time and geographic spread, are evaluated considering sequence prevalence and viral population growth rate, including comparative analyses to other variants co-circulating in a given geographical location (Figure 2A). One example of recurrent substitutions with phenotypic relevance are those near the furin cleavage site, which result in enhanced spike cleavage and infectivity15,16. These mutations have been identified in different variants and in newly expanding lineages. Some teams take into account vaccine coverage when prioritizing an emerging lineage for analysis.
Figure 2. Prioritization of variants by the Early Detection and Analysis group.
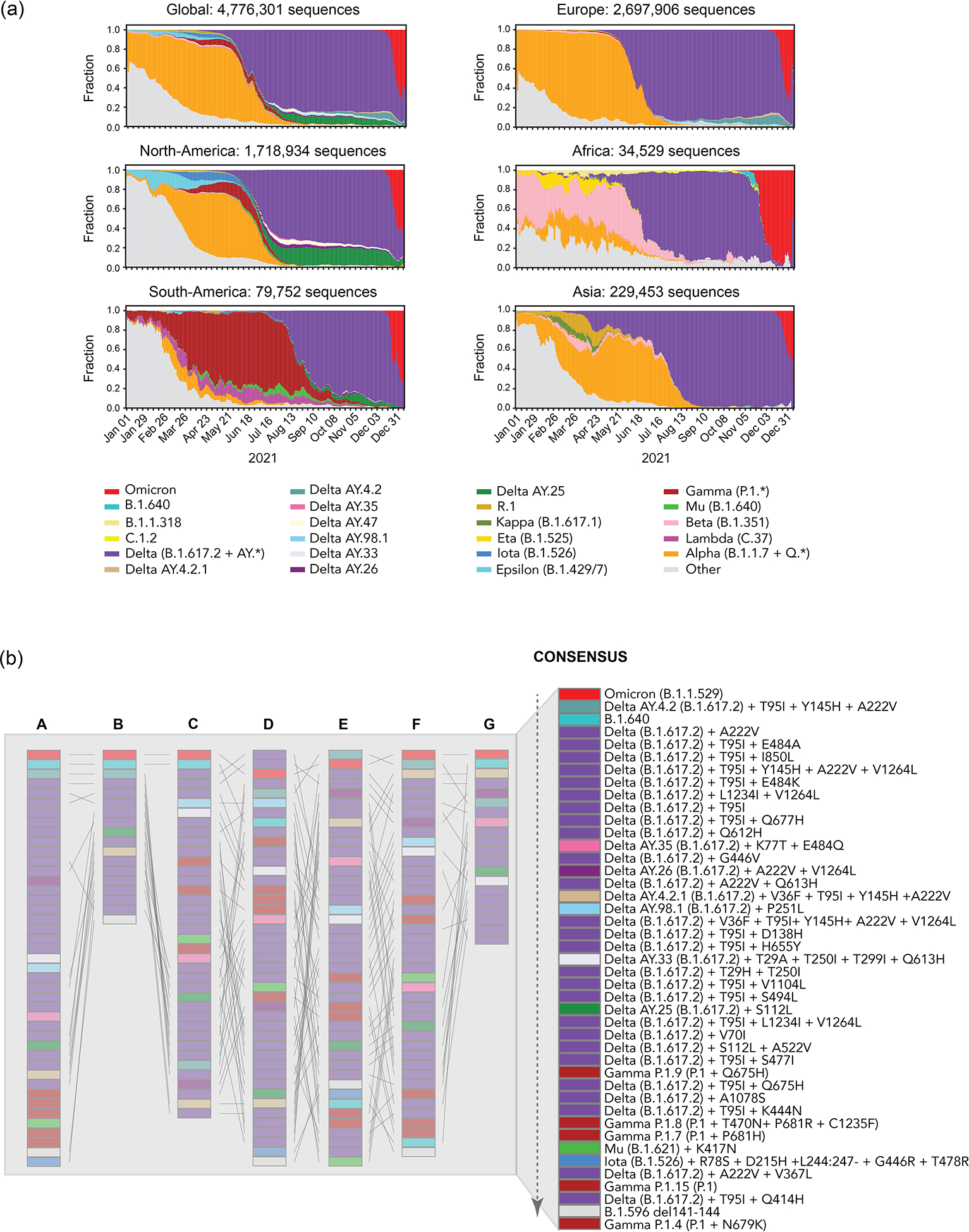
(a) The trajectory of SARS-CoV-2 variant sequence prevalence over a one-year period, Jan. 01, 2021 to Dec 31, 2021, tracking frequencies of weekly counts based on PANGO lineage designations. The data in the graphs was based on the 4.8 million SARS-CoV-2 sequences sampled in 2021 and made available through the GISAID Initiative. Updated graphs can be found at cov.lanl.gov (the tracking tool is called Embers). Top. A global summary and status on five continents is provided. Europe and North America remain the most highly sampled regions of the world, biasing the global sampling. (b) Tangle plots for comparative prioritization of circulating variants across subgroups. The list of variants to prioritize is collectively built by the whole group and prioritized by individual teams to arrive at a consensus list. Each column graph refers to the prioritization order made by each sub-team for circulating variants in December 2021 (top-highest, bottom- lowest priority): A. Cambridge University, B. LANL; C. ISMMS; D. JCVI/BV-BRC; E. UCR SOM; F. Broad Institute; G. WRAIR. The final consensus ranking of the 43 variants is produced by ordering the lineages by their mean rank across the different teams, who also have the option to defer from ranking a lineage, or to assign multiple lineages a tied ranking, and after discussion with the group to determine priority categories. The dashed arrow indicates the order of priority, top being highest priority variants for analysis, bottom means lowest priority. Colors refer to each PANGO lineage tracked but multiple blocks of the same color can also refer to different variants within a PANGO lineage. For example, besides the colored Delta AY.* sublineages indicated, Delta has 26 subvariants (purple) with different combinations of mutations that are being prioritized for analysis.
The rankings are split into two broadly distinct methodologies, each with slight variations: one is based on convergent evolution as the main signal for selection and functional impact of mutations (i.e., Cambridge and Walter Reed Army Institute of Research (WRAIR) teams); whereas the other is anchored on prevalence and growth patterns of mutations and defined lineages (i.e., Los Alamos National Lab (LANL), Icahn School of Medicine at Mount Sinai (ISMMS), J. Craig Venter Institute/Bacterial Viral Bioinformatic Resource Center (JCVI/BV-BRC), UC-Riverside, and Broad Institute teams) (Figure 2B).
Functional impact of mutations.
Cambridge prioritizes substitutions likely to cause immune escape, by looking at both experimentally determined escape from polyclonal sera and the impact of mutations on spike protein structure. Substitutions are given higher priority if they appear to be emerging and if they are in a different Barnes class17 from previously observed substitutions, and lower priority if they have already been tested experimentally. The WRAIR team tracks the prevalence of substitutions at a set of sites selected based on the strength of the interaction with known SARS-CoV-2 antibodies (using complex structures in the PDB Protein Data Bank; https://www.rcsb.org) as well as structural information or knowledge from deep mutational scanning or mutagenesis studies. Weight scores for ranking are also given for various characteristics, such as fold increase in detection over time and geographic spread or population growth in the context of high vaccination coverage.
Prevalence and growth patterns.
The ISMMS team has a similar approach where variants are ranked based on an aggregate score for sequence prevalence increase and genetic changes of concern in sites of importance associated with functional changes (ACE2 binding, antibody escape, etc.) but also assigns weight to mutations in active sites of viral enzymes. Moreover, data from surveillance cohorts in the New York City metropolitan area are used to assess lineages associated with local outbreaks and breakthrough infections after vaccination. LANL identifies emergent mutational patterns within the spike, RBD, and NTD supersite to determine global and regional sampling frequencies. Variant dynamics and global spread are tracked at multiple geographic levels using a suite of tools (available at cov.lanl.gov)5. The JCVI/BV-BRC team uses an algorithm combining sequence prevalence dynamics with functional impact predictions to rank emerging variants. Each mutation is given a sequence prevalence score, reflecting geographically localized prevalence changes, and a functional impact score, based on location within important spike protein regions and whether studies have demonstrated significant changes in either antibody or ACE2 receptor binding18–21. UC-Riverside uses relative growth in the prevalence of specific substitutions and deletions/insertions to identify the fastest growing variants and mutation combinations (https://coronavirus3d.org). For the final variant and subvariant ranking, additional criteria are included such as their potential impact on protein structure (by modeling) and the re-emergence of individual mutations in novel combinations in new variants. Finally, the team from the Broad Institute, like the UC-Riverside team, examines the accelerated growth of a variant relative to its peers, across multiple geographic regions, but fits a binomial logistic regression to each lineage’s proportion over time. Additionally, they fit hierarchical multinomial logistic regression models across geographic regions22.
Challenges (Early Detection and Analysis group)
The Early Detection and Analysis group has faced six main challenges in identifying emerging variants for functional testing: (a) the newest data are the most subject to bias and the least representative because of small numbers. The longer one waits, the more accurate the data, but the greater the delay in identification of newly emergent variants for evaluation; (b) disentanglement of epidemiological from evolutionary effects. A variant might show increased sequence prevalence within a geographic region due to founder effects, or increased incidence could be conferred by epidemiological factors rather than an evolutionary fitness advantage. An example of a founder effect is Delta AY.25 that is very common in North America but not increasing in frequency over time (Figure 2A), versus AY.4.2, which was first sampled well after Delta was increasing in the UK and that was constantly increasing in frequency in 27 countries where it was found, and, furthermore, it never significantly decreased relative to other Deltas once it emerged, suggesting positive selection; (c) selective pressures on the virus are in flux, and mutations may be transient due to a balance with requirements for retention of fitness. Pressures are exerted by the host at the level of transmission, epidemiological interventions, and immune evasion; (d) underrepresentation of variant spread and evolution in countries with limited sampling and sequencing capacity. Although some parts of the world have an abundance of sequence data (e.g. UK, US), others are underrepresented (e.g. African continent, China). There is an urgent need to increase sampling and sequencing capacity in resource-poor countries; (e) variability in data quality. The submission of consensus assemblies without underlying raw read-level data means that quality cannot be independently evaluated. Erroneous genome sequences due to technical artifacts, low coverage, or bioinformatic strategies that default to ancestral bases in regions without sequence coverage can impact the accuracy of variant amino acid calls23; and (f) the database curation quality control steps can filter based on criteria that do not apply uniformly across lineages. The B.1.621 (Mu) lineage had an unexpected stop codon in ORF3a that caused B.1.621 sequences to be flagged during automated uploads to the GISAID database, which initially led Mu to be undercounted. This can lead to a false understanding of the dynamics of a given variant lineage globally. Despite these challenges, our prioritization methods continue to evolve as more information becomes available. These efforts have allowed for the rapid generation of reagents for multiple variants before they have spread extensively in the United States and have been critical for guiding the In Vitro and In Vivo groups. A list of regularly updated prioritized variants is available at https://docs.google.com/spreadsheets/d/167uJP9LfJN07410sWaMSKU1Se-4XX687j8IgVX4MV_w/edit?usp=sharing.
In Vitro group
The In Vitro group performs antibody binding, neutralization, Fc effector, and T cell stimulation assays to understand how SARS-CoV-2 variants impact vaccine- and infection-induced immunity. The In Vitro group serves as a critical intermediary between the Early Detection and Analysis and In Vivo groups by providing valuable data to confirm variant lineage prioritization, and ranking viruses for prioritized in vivo challenge studies. The In Vitro Group was initially tasked with developing key reagents (e.g., spike and RBD antigens, and plasmids for generation of PSV) and procuring biospecimens (e.g., authentic viruses and sera/plasma from infected and vaccinated individuals). At the beginning of 2021, reagents for generating data, including variant virus isolates, recombinant infectious clones, recombinant variant spike proteins for antibody binding assays, variant-specific expression plasmids for PSV particle entry inhibition assays, and variant-specific sera were not widely available (Figure 3A). A key lesson from this process is that the streamlining of administrative procedures for reagent sharing facilitates data generation that directly informs urgent policy- and decision-making. A major and ongoing challenge requiring numerous administrative steps is to obtain authentic virus isolates from domestic and international sources. To expedite this process, we developed a pipeline between SAVE investigators to isolate, propagate, and sequence novel emerging viruses. This effort led to cataloging and isolating hundreds of SARS-CoV-2 variants representing over 40 lineages. For more difficult to obtain SARS-CoV-2, additional efforts have been made to generate infectious clones24–26. Furthermore, the Early Detection group prioritized viral variants and curated sequences to accelerate the production of recombinant variant spike proteins and expression plasmids.
Figure 3-. In Vitro group.
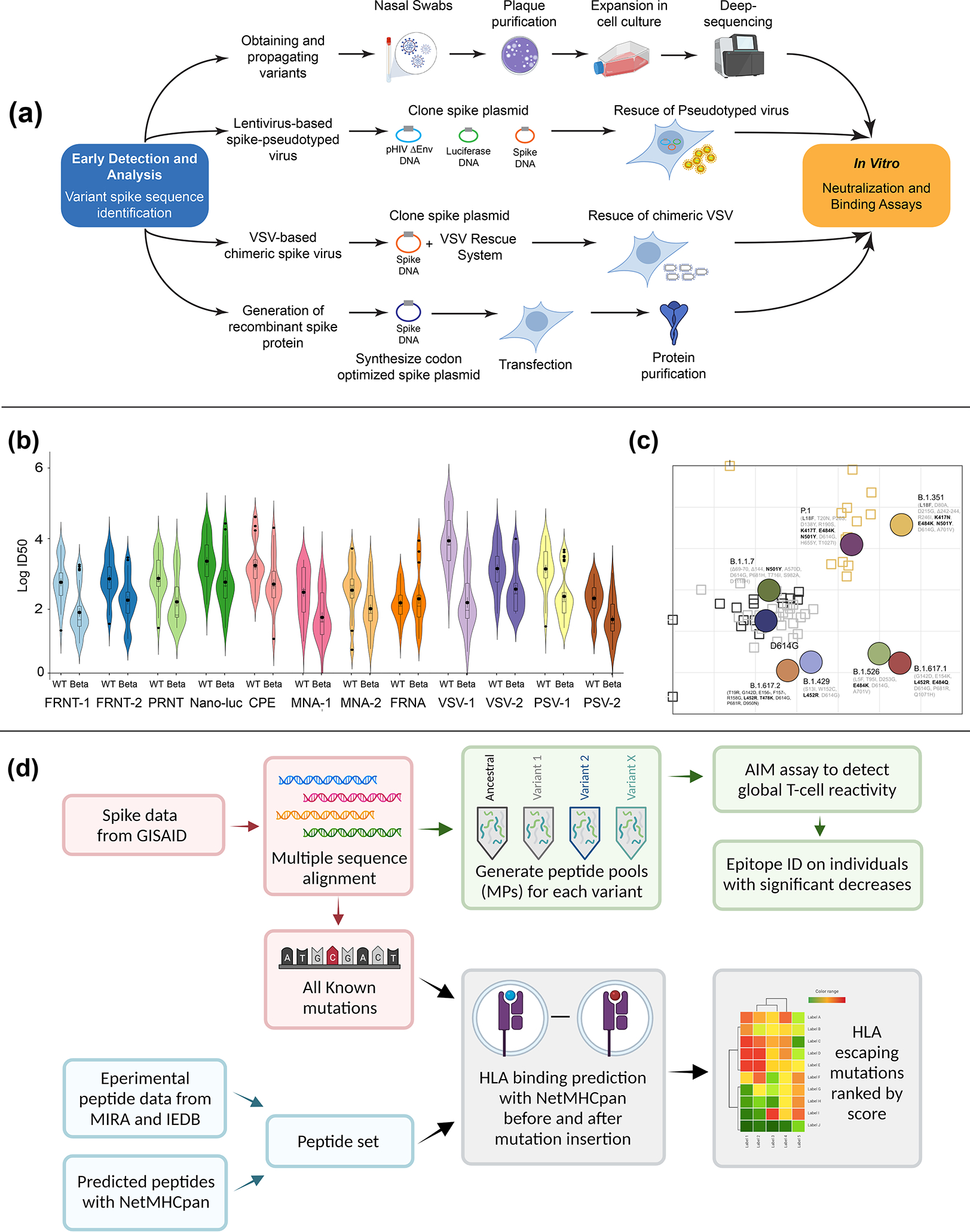
(a) Live virus- nasal swabs in viral transport media or seed stocks are obtained followed by plaque purification and deep-sequencing. Pseudotyped virus- plasmids encoding the variant spike sequence are synthesized to generate pseudotyped lentivirus stocks. Vesicular Stomatitis virus (VSV) chimeric virus- the glycoprotein gene (G) is replaced with the spike protein of SARS-CoV-2 (VSV-eGFP-SARS-CoV-2) and a GFP reporter gene33. (b) The In Vitro group conducted performance testing between 12 neutralization assays involving live authentic virus consisting of focus-reduction neutralization test (FRNT-1), recombinant SARS-CoV-2 reporter virus FRNT (FRNT-2), plaque reduction neutralization test (PRNT), recombinant SARS-CoV-2 expressing nano-luc (Nano-luc), cytopathic effect assay (CPE), microneutralization assay (MNA), and focus-reduction neutralization assay (FRNA) and lentivirus and VSV pseudotyped neutralization assays, and VSV chimeric assays. Example dataset between wild-type (WT) and Beta virus is presented. (c) Antigenic cartography. ID50 neutralization titers in a lentivirus-based pseudovirus assay were determined against a panel of SARS-CoV-2 variants and serum from Moderna vaccinated or infected individuals. Distance between serum to an antigen corresponds to the titer of that serum for the antigen37. Grid lines represent 2-fold dilution of antiserum. Vertical and horizontal axes represent antigenic distance. Antigens- circle; Sera-squares. (d) SARS-CoV-2 variant T cell responses. Sequence data are curated for coding mutations (pink boxes). Curated mutations are tested on convalescent T cell responses using functional assays (Activation Induced Marker- AIM assays; green boxes). Immune Epitope Database (IEDB) and the Immunocode Multiplex Identification of T cell Receptor Antigen Specificity dataset (MIRA) are analyzed to generate curated peptide sets of immunodominant epitopes (blue boxes). Data are integrated to produce a ranked score list of variant epitope changes weighted by their likelihood to disrupt epitope binding and the relative size of the affected population (gray boxes). Created in part with BioRender.com.
At the start of the pandemic, the correlates of immune protection were unknown for COVID-19. Multiple teams within the In Vitro group conducted assessments of vaccine-induced serum neutralization using parallel but independent methods across laboratories. Studies with clinical samples show neutralizing antibody titers are a strong predictor of protection against severe disease27. As such, a major undertaking of the In Vitro group has been to use neutralization assays to assess the impact of spike mutations on the inhibitory activity of clinically-approved monoclonal antibodies and serum/plasma from vaccinated or infected individuals. One of the strengths of the In Vitro group is the use of orthogonal SARS-CoV-2 neutralization assays based on authentic live viruses, PSV, and chimeric viruses. An initial task of this group was to compare neutralization assay platforms across 12 independent laboratories using a defined serum panel from both Pfizer and Moderna vaccinated individuals. Using either the ancestral wild-type virus (Wuhan-1) or more recent variants (e.g., Beta variant shown in Figure 3B), team members performed neutralization assays that varied based on live virus assay readouts (foci, plaques, cytopathic effect, luciferase, fluorescence), target cells, and expression of ACE2 and/or TMPRSS2 on target cells28–35. This type of performance testing has highlighted differences between assay platforms, cell targets, and readouts that can impact neutralization potency. Nonetheless, in most cases, there was remarkable congruence across platforms. Another area of emphasis is using variant-infected serum\plasma samples to visualize the antigenic evolution of spike through a process called antigenic cartography (Figure 3C)36,37. This two-dimensional map provides a landscape of how spike mutations drive loss in neutralizing activity.
For many viruses, the affinity and magnitude of antibody binding to viral glycoproteins associates with virus neutralizing activity, and a strong correlation has been shown for SARS-CoV-238–41. Investigating the correlation between neutralizing and binding activity of vaccine-induced antibodies showed that spike mutations alter this slope, and virus neutralization is often more affected than antibody binding42,43. This has been confirmed through different platforms to measure changes in binding to either native spike proteins or the receptor binding domain, including ELISA44 and multiplexed spike antigen detection platforms7. One potential explanation for this is that many more binding than neutralizing epitopes exist on the spike protein. Some antibodies that have neutralizing activity against the wild-type virus may lose activity to variants, yet overall binding is still maintained – a phenomenon observed for other viruses (e.g., influenza virus45).
Binding antibodies can still have considerable protective effect, irrespective of neutralizing activity due to Fc effector functions, as seen with influenza virus or Ebola virus46–48. The humoral immune response restricts microbes via the coordinated effort of the Fab (antigen-binding) and Fc (constant) domains49. Following infection or vaccination, polyclonal antibodies are induced that target pathogens at multiple sites via their Fab domains. Fab domains that directly or indirectly hinder virus entry are neutralizing; however, the remaining “non-neutralizing” antibodies, can bind and opsonize the pathogen to form immune complexes, or bind spike proteins on the surface of infected cells. Once complexed, the Fc-domains act as molecular beacons that draw in immune cells via Fc-gamma receptors (FcγRs), providing instructions on how the immune system should destroy the antibody-opsonized material. Fc-effector functions of antibodies are linked to natural resolution of COVID-1950–53, correlate with vaccine mediated protection from infection in animal models54–56, and are associated with protection following passive convalescent serum or monoclonal antibody transfer57–60. While emerging variants of SARS-CoV-2 can escape from neutralizing antibodies, their substitutions alter a limited fraction of the overall humoral immune response to the SARS-CoV-2 spike56,61. Thus, Fc-effector functions have more resilience in the face of variation across the spike, for both mRNA and the adenoviral 26 (Ad26) vaccines, offering mechanisms by which antibodies may continue to confer protection despite escape from neutralization.
Growing evidence from animal models and human studies indicate that CD4+ and CD8+ T cells have protective roles in preventing severe disease and death from SARS-CoV-2 infection6,62–64. T cells are an attractive target for intervention as they are less susceptible to viral escape than antibodies6,65. This is largely for two reasons: (1) in convalescent individuals, T cells can target peptides derived from the entire proteome, not just surface-exposed epitopes, and (2) HLA-restriction and diversity creates interpersonal variation in the repertoire of targets, limiting the immunological pressure on any one epitope. Given the presumed role of T cells in limiting severe disease and their potential for sustaining protection against variant mutation, the SAVE In Vitro group included assessment of T cell responses. The goal was to determine empirical drift from vaccination and infection-induced immunity, and to develop tools to predict the impact of variant-associated mutations on immunodominant T cell responses.
The T cell investigations follow two parallel approaches to assess the impact of variant mutations on T cell reactivity and a broad range of different variants (Figure 3D). The first entails measuring overall reactivity against the entire spike protein (in the case of vaccination) or the entire proteome (in the case of infection) and expressing the results as fold difference relative to ancestral sequences. A parallel approach characterizes the mutational impact on specific single epitopes, and monitors if individuals with decreased T cell reactivity have responses that selectively recognize certain epitopes in the context of particular HLA types. Regarding the first approach, at the general population level, the results thus far have detected limited impact of mutations within the spike after natural infection or mRNA vaccination6 against the most concerning variants at the time the study was performed (B.1.1.7, B.1.351, P.1, and B.1.427/429). These findings were corroborated66 and expanded to adenoviral vector-based vaccination67. However, in a minority of individuals, 2-3-fold decreases in the CD8+ T cell responses against B.1.351 (Beta) and B.1.427/429 (Epsilon) variants were noted6. These findings suggest that a more in-depth characterization at the single-epitope level is required to understand the mechanisms behind the reduced CD8+ T cell response in specific subjects. Additionally, it is critical to monitor and predict the effect of emerging circulating variants on T cell reactivity, particularly regarding to-date the most concerning B.1.617.2 (Delta) variant (including AY.* sub-lineages) and B.1.1.529 (Omicron) variant. The experimental data will be used to confirm and improve the bioinformatics analysis and infer the impact of current and upcoming variants on SARS-CoV-2 specific T cell responses.
Advanced computational tools for assessment of SARS-CoV-2 genome mutations on HLA binding have enabled prediction of the effect of mutations within a VOC on T-cell reactivity. Due to the broad diversity of HLA genotypes, T-cell escape on a population level is not likely, as demonstrated for multiple VOC6. However, previous work on HIV and influenza virus has identified associations between specific HLA class I alleles, disease severity68,69 and vaccine efficacy70. We anticipate that as SARS-CoV-2 continues to spread globally, T cell immunity will eventually drive viral evolution. In these situations, specific HLA alleles may become associated with reduced ability to mount responses against dominant T cell epitopes, which may affect clinical outcomes. The T cell subgroup has developed a computational pipeline to assess the effects of specific mutations on HLA binding by also ranking all individual mutations on a T cell escape score, based on experimentally verified and predicted T cell responses (Figure 3D). This ranking will provide early identification of specific mutations associated with T cell escape, particularly CD8+ T cells, and testable hypotheses for T cell experiments. In our preliminary analyses of VOCs, the B.1.617.2 variant was identified as the first in which mutations were associated with reduced HLA binding at a population level. These data suggest that T cell cross-reactivity to B.1.617.2 may be reduced in some individuals. Due to the extensive number of SARS-CoV-2 viral genomes, and large-scale clinical cohorts that are being studied, the T cell SAVE group plans to assemble a database linking HLA genotypes with clinical outcome and viral genomes, which may provide a unique opportunity to study HLA associations with clinical disease and viral evolution at a resolution never attempted previously.
Challenges (In Vitro group)
Work by the In Vitro group has focused mostly on characterizing neutralizing antibody responses to the spike protein with some analysis of the impact of variants on T-cell responses as well. With the recent surge of Omicron infections in vaccinated and unvaccinated individuals, a challenge moving forward will be to disentangle vaccine- and infection-induced immunity, breakthrough infections, waning immunity and other covariates associated with increased risk of symptomatic infection (immunocompromised, age, obesity, diabetes, etc.). Many other key aspects of SARS-CoV-2 and its variants remain unexplored. While neutralizing antibodies correlate with protection from SARS-CoV-2, neutralization is not the only function of antibodies. In fact, non-neutralizing antibodies can afford substantial protection against influenza virus46,71–73 and similar mechanisms remain to be explored for SARS-CoV-2. In addition, differences between wild-type and variant viruses in ACE2 binding, fusion, impact of mutations on spike processing by proteases, and potentially fusion at the cell membrane and cell-to-cell fusion, remain poorly understood15. Furthermore, the spike protein is just one of many SARS-CoV-2 proteins. The impact of mutations in non-spike proteins on immunity and viral fitness, including transmission, virus-host interaction, and polymerase fidelity has not yet been assessed. The use of reverse genetics systems and PSV can be leveraged to understand the contribution of individual mutations on viral fitness and evasion of antibody responses24–26,74. We acknowledge that differences between the ancestral and variant viruses also may impact neutralization assays in different cell lines. While we have seen some of these cell-line specific effects in in vitro neutralization, we do not yet understand their underlying mechanisms. Further, we need to increase the use of reference standards in binding and neutralization assays, such as the WHO International Standard and International Reference Panel for anti-SARS-CoV-2 immunoglobulin75, to calibrate assays and provide a means to compare serological findings. Rare and volume-limited variant-specific sera/plasma are difficult to obtain and share across borders, and\or between academic institutions, and the process is often slowed by administrative hurdles. On occasion, SARS-CoV-2 variant sample sharing has not been possible within the needed timeframe, impeding the research response to this public health emergency. While access to virus isolates outside the United States and variant-specific human sera remains limited, the In Vitro group has created an extensive network of collaborations to overcome these hurdles. Finally, much remains to be explored for both antibody and T cell responses about emerging variants like B.1.617.2, B.1.617.2 subvariants, B.1.1.529 and other new VOC/VOI.
In Vivo group
SARS-CoV-2 animal models have been critical for the development and testing of vaccines and antiviral therapeutics76–84. Initial countermeasures targeted the spike protein from the SARS-CoV-2 strain circulating during the early phase of the pandemic in 2020 and focused on efficacy testing against homologous strains. However, the emergence of variants and their possible effects on transmission, pathogenesis, and infection- or vaccine-mediated immunity required rapid adaptation of animal models to confirm vaccine efficacy against VOC. The In Vivo group was assembled to develop animal models, standardize reagents and assays, and examine the impact of SARS-CoV-2 variants on protection elicited by vaccine- or infection-induced immunity and transmission. The identification of variants for investigation by the Early Detection and Analysis group that are validated in the In Vitro group are then forwarded to the In Vivo group. The In Vivo group studies protection against SARS-CoV-2 variants using an array of animal models, including mice, hamsters, and non-human primates (NHPs). This has led to a collaborative process in which transmission, pathogenesis, and protection data are shared to develop consensus on the impact of emerging variants on protective immunity.
Animal model development
One of the first tasks of the In Vivo group was to standardize viral challenge stocks, routes and doses of infection, and vaccination strategies across each of the animal models. To minimize variability and adventitious mutations associated with virus propagation in different cell types, Vero-TMPRSS2 cells were used to generate challenge stocks and distributed among team members85. As new variants emerge, viruses are tested in small animal models to determine infectivity and pathogenicity. For vaccination studies, the group focused on evaluating both approved (including Emergency Use Authorization (EUA)) vaccines and those undergoing advanced clinical testing in humans, including mRNA vaccines (Pfizer BNT162b and Moderna mRNA-1273), protein-based vaccines (Novavax NVX-CoV2373) and virus-vectored vaccines (J&J Ad26.COV2.S). For these studies, vaccine doses were optimized to model magnitude and durability of vaccine-induced immunity across the animal models. For each vaccination experiment, many parameters are studied including neutralizing antibody potency and kinetics, pathogenesis of ancestral and variant viruses, as well as levels of virus in various respiratory tract tissues.
Mouse models
The ancestral SARS-CoV-2 strain does not replicate in conventional laboratory mice as the spike protein inefficiently binds to murine ACE286,87. To overcome this obstacle, several mouse models were developed, including human ACE2 (hACE2) transgenic mice (e.g., K18-hACE288, originally developed for studies of SARS-CoV88) and mice that express hACE2 transiently after transduction with viral vectors (e.g., Adenovirus)89,90. The K18-hACE2 transgenic mice are highly permissive for most SARS-CoV-2 strains and variants, and infection typically results in weight loss, nasal turbinate and lung infection, pneumonia, and death60,91,92. Lungs from SARS-CoV-2 infected mice show denuding bronchiolitis, mixed inflammatory infiltrate, alveolar edema, and alveolitis92–94. Some mice, especially young K18-hACE2 mice, develop infection in the brain and encephalitis, which may confound interpretation of clinical disease95. The spike mutation at position N501Y, which is found in mouse-adapted strains and several emerging variants (Alpha, Beta, Gamma, Mu and Omicron variants), increases the affinity of SARS-CoV-2 spike protein for the murine ACE2 receptor and allows for direct infection of inbred mice86. Thus, in addition to the K18-hACE2 mouse model, challenge of conventional laboratory mice with mouse-adapted virus or SARS-CoV-2 variants containing an N501Y substitution within the spike protein cause pneumonia in BALB/c, 129S2, and C57BL/6 mice in an age-dependent manner89,96–98. The initial characterization of each variant in a variety of mouse strains at different doses allowed for vaccination studies to be conducted with validated stocks and consistent phenotypes. The In Vivo Group has utilized both inbred mouse strains (e.g., BALB/c, C57BL/6, 129S1, 129S2) and K18-hACE2 transgenic mice for iterative infection and vaccination studies.
As part of the experimental design, the In Vivo team uses a ‘high’ and ‘low’ vaccine dose strategy with Ad26.COV2.S, NVX-CoV2373, BNT162b, and mRNA-1273 to study the impact of variant mutations on protection. Mice inoculated with a high vaccine dose are useful for evaluating antibody-mediated protection of the upper and lower respiratory tract after challenge. Mice inoculated with the lower vaccine dose serve as a model for suboptimal immunity (as might be seen in the elderly or immunocompromised) and breakthrough infection. Through this effort, the group has demonstrated that low doses of mRNA vaccines (BNT162b and mRNA-1273), or protein vaccine (NVX-CoV2373) show reduced protection as compared to high vaccine doses (Figure 4) against B.1.351 and B.1.617.2 variants99. These experiments are being extended to study additional variants and a range of vaccination doses.
Figure 4-. In Vivo group.
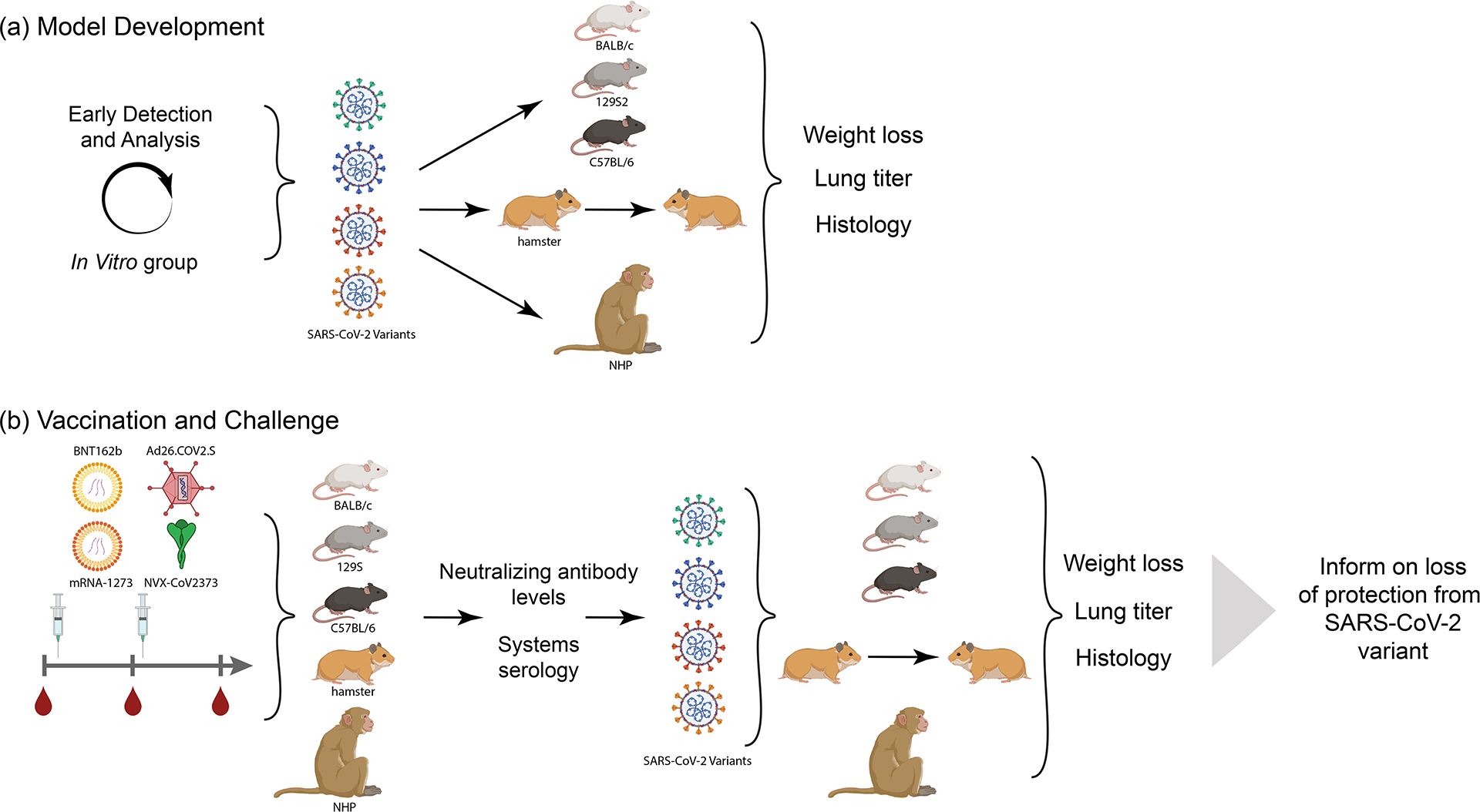
(a) Animal model development. After selection of variants to analysis by the Early Detection and In Vitro Analysis Group, isolates are grown, validated by next-generation sequencing, and analyzed in each animal model at different doses to determine pathogenicity, viral kinetics and transmission (in hamsters). Weight loss, lung titer and lung pathology as assessed to have benchmarks for vaccine studies. (b) Vaccine Challenge. Each animal model is immunized with selected vaccines. Animal serum is examined after vaccination for neutralizing antibody levels and across a systems serology analysis before viral challenge with the chosen variants. Protection against infection and disease in each model is analyzed to determine the protective capability of each vaccine and variant. Data on protection of each animal model with each vaccine platform and challenged with variant viruses is shared with the SAVE consortium and SIG. Created with BioRender.com
Another goal of the SAVE team is to assess the ability of prior infection to protect against secondary challenge with SARS-CoV-2. To model this, middle aged C57BL/6 mice are inoculated with SARS-CoV-2 (B.1.1.7 (Alpha), B.1.351 (Beta) or P.1 (Gamma) variants) followed by a homologous or heterologous challenge up to 120 days later with either variants or mouse-adapted SARS-CoV-2. Mice infected with any of the three variants remain asymptomatic but develop a neutralizing antibody response that is measurable at 21 days post infection. However, neutralizing antibody cannot be detected in most mice at 3 months post infection, yet mice are still partly protected from variant virus challenge or from a dose of mouse-adapted SARS-CoV-2 that is lethal to mice without pre-existing immunity. Part of the goal of this project is to measure T and B cell memory responses to understand why the mice remain protected.
Hamster models
Syrian golden hamsters are highly susceptible to SARS-CoV-2 infection and disease without any species-specific adaptation of the virus, and have a disease phenotype that resembles mild disease observed in human COVID-19 cases. Loss of 10%–20% of the initial body weight is seen at 6–7 days after infection depending on the age and sex of the animal and the variant and dose of the virus100–102. Virus replication is confined to the upper and lower respiratory tract, which peaks at 3 days post infection and then wanes to undetectable amounts by 10 days after infection. Imaging of the lungs of infected hamsters shows abnormalities during the course of infection that do not directly resolve even after virus clearance100.
Pathological changes in the lungs of the hamsters are comparable to those in some humans and are characterized by widespread, moderate to severe broncho-interstitial pneumonia100,103. Lung lesions are comprised of focal extensive areas of pulmonary edema and consolidation with evidence of interstitial pneumonia. Histopathological lesions include fibrin deposits and edema in alveolar spaces, influx of neutrophils and macrophages into alveolar spaces, presence of syncytial cells and prominent type II pneumocyte hyperplasia. Secondary bacterial infections are often detected in the lungs. Despite a robust infection, SARS-CoV-2 is not lethal in healthy hamsters and infected animals recover.
Since the utility of the hamster model for SARS-CoV-2 infection was established using an early isolate of SARS-CoV-2, infection studies have been performed with several variants. Hamsters are largely agnostic to the variants, demonstrating little differences in viral replication and shedding kinetics between different variants including D614G, B.1.1.7 and B.1.351104. While competition infections are more sensitive to revealing small effects of SARS-CoV-2 mutations on fitness for airway infection and transmission25,105, recent studies indicate that B.1.1.529 (Omicron) is attenuated in hamsters with less infection in the lung106.
Given their general susceptibility to SARS-CoV-2, hamsters are an excellent model to study vaccine-induced immunity against variants. Although immunological reagents are less widely available for hamsters, antibody responses induced by vaccination can be measured in neutralization assays and ELISAs by using hamster-specific IgG, IgA, and IgM secondary antibodies. Cohorts of animals receive two immunizations with either the Pfizer BNT162b2 and Moderna mRNA-1273 vaccine given three and four weeks apart, respectively. The vaccine dose in these studies has generally been one-third of the dose given to humans (i.e., 10 μg of the Pfizer vaccine or 35 μg of the Moderna vaccine), and an additional freeze-thaw of the Pfizer and Janssen vaccines does not decrease immunogenicity. IgG antibody titers against the SARS-CoV-2 spike can be detected after the first vaccination but are more robust three weeks after the second vaccination, similar to data in human studies. Since long-term vaccine immunity is a key question in SARS-CoV-2 research, vaccinated animals are held for extended periods of time before challenge with new emerging variants.
Transmission studies are well established in hamsters, as SARS-CoV-2 can transmit efficiently through the aerosol route from an infected to naive hamster. Increase in transmission potential of the D614G and the B.1.1.7 variants have been observed compared to other isolates of SARS-CoV-2. Using a direct contact transmission model, intramuscular or intranasal vaccinated ChadOx1/AZD1222 hamsters were protected from disease but not upper respiratory tract infection107,108. This suggests the hamster transmission model is a useful tool to study vaccine efficacy in the context of natural exposure.
Non-human primate model
Vaccinated non-human primates (NHP) are an important experimental model for demonstrating immunogenicity and protective efficacy against SARS-CoV-230,109–112. NHPs have several advantages for clinical translation. First, NHP are outbred, and their innate immune responses and B and T cell repertoires have greater similarity to humans than those of rodents. Because of the diversity in class I and II MHC, NHPs also support study of the breadth of T cell responses induced by vaccines. Second, NHP allows use of clinically relevant vaccine doses and are an excellent model to study the durability of immune responses. Third, following intranasal and intratracheal administration, viral replication occurs rapidly in the upper and lower airways, respectively, with similar kinetics to humans109,110. For most SARS-CoV-2 strains, infection is cleared by 7–10 days post-challenge. Inflammation and pathology in the lung are consistent with mild infection as described in humans. The NHP model has been used to show immunogenicity and protection following vaccination with mRNA77, ChAdOx-178, Ad26-Spike84, protein/adjuvant79 or inactivated whole virus,113,114 which have all been approved for use in humans. The NHP model also has been used to understand immune correlates and mechanisms of protection. The SAVE investigators are currently studying homologous and heterologous prime/boost vaccinations using EUA or approved vaccines for their ability to induce humoral and cellular immunity, longevity of immune responses and the mechanisms associated with induction of long-lived immunity and protection in the upper and lower airway.
Challenges (In Vivo group)
SARS-CoV-2 animal models provide an opportunity to understand mechanisms of infection, inflammation, pathogenesis, and transmission across species and against different vaccine platforms. Similar to the concerns raised with the In Vitro group, an initial challenge for the In Vivo group was obtaining authentic viruses without cell culture-adaptive mutations for challenge studies. To overcome this hurdle, a parallel pipeline for propagating and sequencing challenge stocks was developed, not only to ensure presence of lineage-defining mutations but also absence of mutations associated with propagation in tissue culture, and which included standardizing virus dose and routes of inoculation and distributing the same stocks to all team members. SARS-CoV-2 strains are constantly evolving, which challenges the decision as to which strains are most relevant for in vivo study. Each animal model has unique opportunities and limitations that are considered when evaluating protective immunity against a variant. Mice are a tractable system with an array of immunological tools, assays, and genetic knockout strains that allow for experimental rigor and mechanistic analysis However, inbred mouse strains are limited to SARS-CoV-2 variants that possess an N501Y mutation in the spike protein, and the genetic background (e.g., BALB/c, 129S2, C57BL/6) can impact virus replication and pulmonary pathology. Most variants appear to infect hamsters at similar levels with comparable lung inflammation and pathology, with the apparent exception of B.1.1.529106. However, infection in hamsters causes a mild to moderate disease and there are limited immunological reagents to probe the response to infection and vaccination. The transmission models are exquisitely sensitive and further studies are needed to understand experimental parameters (e.g., airflow, contact time, relative humidity, temperature) that modulate transmission efficiency and standardize experimental systems. The NHP model has been used in conjunction with the small animal models using the same viral stocks to provide a more comprehensive analysis across species for how the vaccines are mediating protection. As NHP are more limited in their availability, the rodent models can inform the best use of this model. As the SARS-CoV-2 pandemic continues, animal models will need to be adapted to reflect the immune status of the population (e.g., natural infection, vaccination, and booster shots). These large and small animal models will be essential for further testing of next-generation vaccines, boosters formulated with variant spikes, and immunological imprinting.
Summary
Collaborative science and open sharing of results in near real-time have defined the SAVE program. This cross-fertilization has allowed for efficient and rapid analysis of the impact of emerging variants on infection and vaccine-induced immunity. The emergence of the B.1.1.529 (Omicron) variant, which contains more than 30 mutations in the spike protein, threatens clinically approved monoclonal antibodies and infection- and vaccine-induced immunity. The SAVE group rapidly responded by generating plasmids and spike protein, isolating, propagating, and distributing authentic Omicron viral stocks, submitting reagents to public repositories, performing binding and neutralization assays and evaluating virus infection across different animal models106,115–122. The data from these studies were rapidly shared with government agencies and submitted as manuscripts on pre-print servers.
Over the past two decades, we have witnessed the emergence\re-emergence of several RNA viruses, including West Nile virus, H1N1 influenza virus, Chikungunya virus, Zika virus, SARS-CoV-1, MERS-CoV, and Ebola virus, that have threatened global public health. Developing collaborative programs between academic, industry and commercial partners is essential to respond to rapidly evolving viruses. This progressive approach combined with open communication and coordination by NIAID/NIH has facilitated rapid prioritization, reagent development, testing, and assessment of SARS-CoV-2 variants. The mutual relationship between the SAVE group and the SIG has provided feed-forward and feedback loops to aid with key decision matters involving risk assessment, SARS-CoV-2 countermeasures, diagnostics and public health policy. In addition to the SAVE group, other national\international networks have been developed for assessing the risk of SARS-CoV-2 mutations on immunity. This includes the ‘G2P-UK’ National Virology Consortium, the Genotype to Phenotype (G2P)-Japan Consortium, the NIH Accelerating COVID-19 therapeutic interventions and vaccines (ACTIV) Tracking resistance and coronavirus evolution (Trace) consortium, the World Health Organization R&D Blueprint, and the National Cancer Institute Serological Sciences Network (SeroNet). These partnerships must continue to increasingly include scientists across the world to ensure that variants are rapidly identified and characterized to determine their impact on transmission, infection, replication, and immune evasion. This SAVE program serves as a template to develop reagents, models, assays, and diagnostics and test therapeutics and vaccines in pre-clinical models against rapidly evolving pathogens.
Funding:
AA is funded by HHSN272201700060C, GA is funded by the Ragon Institute of MGH, MIT, and Harvard, the Massachusetts Consortium on Pathogen Readiness (MassCPR), the National Institutes of Health (NIH) (3R37AI080289-11S1, R01AI146785, U19AI42790-01, U19AI135995-02, U19AI42790-01, 1U01CA260476-01), the Gates Foundation Global Health Vaccine Accelerator Platform funding (OPP1146996 and INV-001650), and the Musk Foundation, DCM and XS are supported in part by the National Institute of Allergy and Infectious Diseases (NIAID)/NIH Collaborative Influenza Vaccine Innovation Centers (CIVIC) under contract 75N93019C00050- Duke University, AE, AGor, JMC, FK, and VS are funded in part by NIH/NIAID CIVIC under contract 75N93019C00051-Icahn School of Medicine at Mt. Sinai, GA, BL, VAM, SSC, and PGT are funded in part by the NIH/NIAID Center for Influenza Vaccine Research for High-Risk Populations (CIVR-HRP) CIVIC under contract 75N93019C0052- University of Georgia, MSD, VS, LBT, HVB, LAV, RAMF, AGS, MBF, RMJ, JPL, SMW, BLH, DDH, YH, YL, MSN, PW, MW, DJS, AN, EBL, SLJ, STüre, SW, STurn, JMC, FK, BMW, BR and BY are supported by the NIH/NIAID Centers of Excellence for Influenza Research and Response (CEIRR) under contract 75N93021C00014-Icahn School of Medicine at Mt. Sinai, TPF, GF, JF, TJ, LK, BL, VAM, SSC, AS, ST, PGT, EK, LC-L, TH, SSac and RJW are funded in part by the NIH\NIAID CEIRR under contract 75N9302100016-St Jude Children’s Research Hospital, RRA, VVE, MDG and MSS is funded in part by the NIH/NIAID CEIRR under contract 75N93021C00017-Emory University, AE, TPF, JF, TJ, LK, BL, VAM, SSC, and ST SPJW, ZL, L-MB, PWR, MCP, Sstu, AG, AS and RJW are funded in part by the NIH/NIAID Centers of Excellence for Influenza Research and Surveillance (CEIRS) under contract HHSN272201400006C-St Jude Children’s Research Hospital, AP is supported in part by the NIH/NIAID CEIRS under contract HHS N2772201400007C- Johns Hopkins University, AE, MBF, RMJ, AGS, JPL, SMW, PJH, YK, JMC, FK, MSc, and VS are funded in part by the NIH/NIAID CEIRS under contract HHSN272201400008C- Icahn School of Medicine at Mt. Sinai, RRA is funded by 3R01AI148378-01S1, JDB is supported in part by R01AI141707 and is an Investigator of the Howard Hughes Medical Institute, ACMB is funded by U01AI151810, EB is supported by an Intramural NIH Research Program project #ZIA AI005156-02, DLB is supported by NIH/NIAID under contract 75N93019D0025 to CAMRIS International, EG and VJM are supported by the Intramural Research Program of the NIAID, NIH, BLD and MR are supported by a cooperative agreement between The Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc. and the U.S. Department of the Army (W81XWH-18-2-0040), MSD is supported in part by R01AI157155, MD is supported by 1R01AI143639, NADR, DCD, ABM, NJS, RAK, SO,SDS, and RAS are supported by the Intramural Research Program of the Vaccine Research Center, NIAID and NIH, CD, BAM, and TCJ are funded in part by the German Ministry of research under project codes DZIF, MolTrax and PREPARED, AE is supported by NIAID grants U01AI141990 and 1U01AI150747, VVE, MDG and MSS are supported in part by the NIH\NIAID (P51OD011132 and 3U19AI057266-17S1), Emory Executive Vice President for Health Affairs Synergy Fund award, and Woodruff Health Sciences Center 2020 COVID-19 CURE Award, Agod, LJ, MSe are supported by HHSN272201700060C, AGor is supported by R01AI20997, MRH is supported by NIAID contract HHSN272201800013C, MPGK is supported by VEO - Versatile emerging infectious disease observatory: Forecasting, nowcasting and tracking in a changing world. VEO has received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement No. 874735, COVID-19 vaccination in kidney patients (RECOVAC) - ZonMw # 10430072010002 Monitoring the evolution, spread and transmission of SARS-CoV-2 through whole genome sequencing to enable fast genotype to phenotype prediction’ funded by ZonMw - project# 10150062010005, APW 202645827; To support the structured evaluation of SARS-CoV-2 evolution funded by WHO APW 202605269; To gain in-depth understanding of the evolution, spread and transmission of SARS-CoV-2 during the coming phase of the pandemic funded by WHO, BK, JT and HY are funded by the Los Alamos National Laboratory and the Gates Foundation OPP1169339, RAK is funded by Intramural NIH funding, MJM is supported by UM1AI068618, VDM is supported by R01AI153602, SMP is supported by P01AI060699 and R01AI129269, PYS is supported by NIH grants AI134907 and awards from the Sealy Smith Foundation, the Kleberg Foundation, the John S. Dunn Foundation, the Amon G. Carter Foundation, the Gillson Longenbaugh Foundation, and the Summerfield Robert Foundation, VS is supported by the NIH/NCI: Serological Sciences Network (SeroNet) 75N91019D00024, PGT is supported by U01AI144616, U01AI150747, R01AI136514, R01AI154470, ZSW, RHS, AMN, MSh are supported by NIH, NIAID Contract 75N93019C00076, WW and CDW are supported by US Food and Drug Administration institutional research funds. SCW is supported by NIH grant R24 AI120942 and by the Sealy and Smith Foundation.
Footnotes
Conflicts of Interest:
DHB is a co-inventor on provisional vaccine patents (63/121,482; 63/133,969; 63/135,182), JDB consults for Moderna and Flagship Labs 77 on topics related to viral evolution, and is an inventor on Fred Hutch licensed patents related to viral deep mutational scanning, The Boon laboratory has received unrelated funding support in sponsored research agreements from AI Therapeutics, GreenLight Biosciences Inc., and Nano targeting & Therapy Biopharma Inc. The Boon laboratory has received funding support from AbbVie Inc., for the commercial development of SARS-CoV-2 mAb. A.C.M.B. was a recipient of a licensing agreement with Abbvie Inc., for commercial development of SARS-CoV-2 mAb, M.S.D. is a consultant for Inbios, Vir Biotechnology, Senda Biosciences, and Carnival Corporation, and on the Scientific Advisory Boards of Moderna and Immunome. The Diamond laboratory has received unrelated funding support in sponsored research agreements from Vir Biotechnology, Moderna, and Emergent BioSolutions, The Ellebedy laboratory received funding under sponsored research agreements that are unrelated to the data presented in the current study from Emergent BioSolutions and from AbbVie. A.H.E. has received consulting fees from InBios International, Inc, Fimbrion Therapeutics, Mubadala Investment Company and Goldman Sachs and is the founder of ImmuneBio Consulting, MBF has funding from Novavax which is outside the scope of this research. They had no role in the funded research from the SAVES consortium, the Garcia-Sastre laboratory has received research support from Pfizer, Senhwa Biosciences, Kenall Manufacturing, Avimex, Johnson & Johnson, Dynavax, 7Hills Pharma, Pharmamar, ImmunityBio, Accurius, Nanocomposix, Hexamer, N-fold LLC, Model Medicines and Merck, outside of the reported work. A.G.-S. has consulting agreements for the following companies involving cash and/or stock: Vivaldi Biosciences, Contrafect, 7Hills Pharma, Avimex, Vaxalto, Pagoda, Accurius, Esperovax, Farmak, Applied Biological Laboratories, Pharmamar and Pfizer, outside of the reported work. A.G.-S. is inventor on patents and patent applications on the use of antivirals and vaccines for the treatment and prevention of virus infections and cancer, owned by the Icahn School of Medicine at Mount Sinai, New York, outside of the reported work, Aubree Gordon serves on a scientific advisory board for Janssen, Erasmus MC has a Proprietary IP on MERS, BK is part of provisional patent applications for strategies for next generation SARS-CoV-2 vaccines that address diversity, The Icahn School of Medicine at Mount Sinai has filed patent applications relating to SARS-CoV-2 serological assays which lists Viviana Simon as a co-inventor and NDV-based SARS-CoV-2 vaccines which list Florian Krammer as co-inventor, Mount Sinai has spun out a company, Kantaro, to market serological tests for SARS-CoV-2. Florian Krammer has consulted for Merck and Pfizer (before 2020), and is currently consulting for Pfizer, Seqirus, 3rd Rock Ventures and Avimex. The Krammer laboratory is also collaborating with Pfizer on animal models of SARS-CoV-2, VDM have filed a patent on the reverse genetic system and reporter SARS-CoV-2, DCM receives funding from Moderna to perform blinded assessments of vaccine-elicited neutralizing antibody responses in clinical studies of their COVID-19 vaccines, A.S. is a consultant for Gritstone Bio, Flow Pharma, Arcturus Therapeutics, ImmunoScape, CellCarta, Avalia, Moderna, Fortress and Repertoire., PYS laboratory has received funding support in sponsored research agreements from GSK, Pfizer, Gilead, Novartis, Merck, IGM Biosciences, and Atea Pharmaceuticals. P.Y.S. is a member of the Scientific Advisory Boards of AbImmune and is Founder of FlaviTech, M.S.S serves on the advisory board for Moderna and Ocugen, PGT serves on the scientific advisory board for Immunoscape and Cytoagents and has consulted for Johnson and Johnson. PGT has received travel support and honoraria from Illumina and 10X Genomics. PGT has patents related to viral infection treatment and T cell receptor biology. S.P.J.W. and Z.L. have filed a patent with Washington University for VSV- SARS-CoV-2 mutants to characterize antibody panels. S.P.J.W has received unrelated funding support in sponsored research agreements with Vir Biotechnology, AbbVie, and sAB therapeutics. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the US Centers for Disease Control and Prevention. Use of trade names is for identification only and does not imply endorsement by the US Centers for Disease Control and Prevention or the US Department of Health and Human Services.
References
- 1.Krammer F SARS-CoV-2 vaccines in development. Nature 586, 516–527, doi: 10.1038/s41586-020-2798-3 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Plante JA et al. The variant gambit: COVID-19’s next move. Cell Host Microbe 29, 508–515, doi: 10.1016/j.chom.2021.02.020 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walensky RP, Walke HT & Fauci AS SARS-CoV-2 Variants of Concern in the United States-Challenges and Opportunities. JAMA 325, 1037–1038, doi: 10.1001/jama.2021.2294 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fischer W et al. HIV-1 and SARS-CoV-2: Patterns in the evolution of two pandemic pathogens. Cell Host Microbe 29, 1093–1110, doi: 10.1016/j.chom.2021.05.012 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Korber B et al. Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell 182, 812–827 e819, doi: 10.1016/j.cell.2020.06.043 (2020). *This was the first study showing that a newly emerging mutation in the Spike protein could come to rapidly dominate across the globe, indicating a fitness advantage.
- 6.Tarke A et al. Impact of SARS-CoV-2 variants on the total CD4(+) and CD8(+) T cell reactivity in infected or vaccinated individuals. Cell Rep Med 2, 100355, doi: 10.1016/j.xcrm.2021.100355 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pegu A et al. Durability of mRNA-1273 vaccine-induced antibodies against SARS-CoV-2 variants. Science 373, 1372–1377, doi: 10.1126/science.abj4176 (2021). ***This study describes the impact of several SARS-CoV-2 variants on antibody binding and neutralization on mRNA-1273 vaccine sera between 1 to 6 months post-2nd dose.
- 8.Carreno JM et al. Evidence for retained spike-binding and neutralizing activity against emerging SARS-CoV-2 variants in serum of COVID-19 mRNA vaccine recipients. EBioMedicine 73, 103626, doi: 10.1016/j.ebiom.2021.103626 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jansen JE & Retpen JA Acute spontaneous streptococcal myositis. Case report. Acta Chir Scand 154, 323–324 (1988). [PubMed] [Google Scholar]
- 10.Lusvarghi S et al. Key substitutions in the spike protein of SARS-CoV-2 variants can predict resistance to monoclonal antibodies, but other substitutions can modify the effects. J Virol, JVI0111021, doi: 10.1128/JVI.01110-21 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neerukonda SN et al. Establishment of a well-characterized SARS-CoV-2 lentiviral pseudovirus neutralization assay using 293T cells with stable expression of ACE2 and TMPRSS2. PLoS One 16, e0248348, doi: 10.1371/journal.pone.0248348 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pino M et al. A yeast expressed RBD-based SARS-CoV-2 vaccine formulated with 3M-052-alum adjuvant promotes protective efficacy in non-human primates. Sci Immunol 6, doi: 10.1126/sciimmunol.abh3634 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mullen Julia L., Alaa Abdel Latif GT, Alkuzweny Manar, Cano Marco, Haag Emily, Zhou Jerry, Zeller Mark, Hufbauer Emory, Matteson Nate, Andersen Kristian G., Wu Chunlei, Su Andrew I., Gangavarapu Karthik, Hughes Laura D., and the Center for Viral Systems Biology. outbreak.info, < https://outbreak.info/ > (2020). [Google Scholar]
- 14.Centers for Disease Control and Prevention SARS-CoV-2 Variant Classifications and Definitions. (2021).
- 15.Escalera A et al. Mutations in SARS-CoV-2 variants of concern link to increased spike cleavage and virus transmission. Cell Host Microbe, doi: 10.1016/j.chom.2022.01.006 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Y et al. Delta spike P681R mutation enhances SARS-CoV-2 fitness over Alpha variant. bioRxiv, doi: 10.1101/2021.08.12.456173 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barnes CO et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature 588, 682–687, doi: 10.1038/s41586-020-2852-1 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Starr TN, Greaney AJ, Dingens AS & Bloom JD Complete map of SARS-CoV-2 RBD mutations that escape the monoclonal antibody LY-CoV555 and its cocktail with LY-CoV016. Cell Rep Med 2, 100255, doi: 10.1016/j.xcrm.2021.100255 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greaney AJ et al. Comprehensive mapping of mutations in the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human plasma antibodies. Cell Host Microbe 29, 463–476 e466, doi: 10.1016/j.chom.2021.02.003 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Starr TN et al. Prospective mapping of viral mutations that escape antibodies used to treat COVID-19. Science 371, 850–854, doi: 10.1126/science.abf9302 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Starr TN et al. Deep Mutational Scanning of SARS-CoV-2 Receptor Binding Domain Reveals Constraints on Folding and ACE2 Binding. Cell 182, 1295–1310 e1220, doi: 10.1016/j.cell.2020.08.012 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Obermeyer F et al. Analysis of 2.1 million SARS-CoV-2 genomes identifies mutations associated with transmissibility. 2021.2009.2007.21263228, doi: 10.1101/2021.09.07.21263228%J medRxiv (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davis JJ et al. Analysis of the ARTIC Version 3 and Version 4 SARS-CoV-2 Primers and Their Impact on the Detection of the G142D Amino Acid Substitution in the Spike Protein. Microbiol Spectr 9, e0180321, doi: 10.1128/Spectrum.01803-21 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xie X et al. An Infectious cDNA Clone of SARS-CoV-2. Cell Host Microbe 27, 841–848 e843, doi: 10.1016/j.chom.2020.04.004(2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu Y et al. The N501Y spike substitution enhances SARS-CoV-2 infection and transmission. Nature, doi: 10.1038/s41586-021-04245-0 (2021). **This study tested all spike mutations identified in the first variant of concern, B.1.1.7 (Alpha), and identified N501Y as being important for enhanced transmission and an adaptive mutation of concern.
- 26.Xie X et al. Neutralization of SARS-CoV-2 spike 69/70 deletion, E484K and N501Y variants by BNT162b2 vaccine-elicited sera. Nat Med 27, 620–621, doi: 10.1038/s41591-021-01270-4 (2021). [DOI] [PubMed] [Google Scholar]
- 27.Khoury DS et al. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat Med 27, 1205–1211, doi: 10.1038/s41591-021-01377-8 (2021). [DOI] [PubMed] [Google Scholar]
- 28.Vanderheiden A et al. Development of a Rapid Focus Reduction Neutralization Test Assay for Measuring SARS-CoV-2 Neutralizing Antibodies. Curr Protoc Immunol 131, e116, doi: 10.1002/cpim.116 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang P et al. Increased resistance of SARS-CoV-2 variant P.1 to antibody neutralization. Cell Host Microbe 29, 747–751 e744, doi: 10.1016/j.chom.2021.04.007 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Corbett KS et al. Protection against SARS-CoV-2 beta variant in mRNA-1273 vaccine-boosted nonhuman primates. Science, eabl8912, doi: 10.1126/science.abl8912 (2021). [DOI] [PubMed] [Google Scholar]
- 31.Chen RE et al. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat Med 27, 717–726, doi: 10.1038/s41591-021-01294-w (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hou YJ et al. SARS-CoV-2 Reverse Genetics Reveals a Variable Infection Gradient in the Respiratory Tract. Cell 182, 429–446 e414, doi: 10.1016/j.cell.2020.05.042 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Case JB et al. Neutralizing Antibody and Soluble ACE2 Inhibition of a Replication-Competent VSV-SARS-CoV-2 and a Clinical Isolate of SARS-CoV-2. Cell Host Microbe 28, 475–485 e475, doi: 10.1016/j.chom.2020.06.021 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saadat S et al. Binding and Neutralization Antibody Titers After a Single Vaccine Dose in Health Care Workers Previously Infected With SARS-CoV-2. JAMA 325, 1467–1469, doi: 10.1001/jama.2021.3341 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muruato AE et al. A high-throughput neutralizing antibody assay for COVID-19 diagnosis and vaccine evaluation. Nat Commun 11, 4059, doi: 10.1038/s41467-020-17892-0 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fonville JM et al. Antibody landscapes after influenza virus infection or vaccination. Science 346, 996–1000, doi: 10.1126/science.1256427 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith DJ et al. Mapping the antigenic and genetic evolution of influenza virus. Science 305, 371–376, doi: 10.1126/science.1097211 (2004). [DOI] [PubMed] [Google Scholar]
- 38.Wajnberg A et al. Robust neutralizing antibodies to SARS-CoV-2 infection persist for months. Science 370, 1227–1230, doi: 10.1126/science.abd7728 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Amanat F et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat Med 26, 1033–1036, doi: 10.1038/s41591-020-0913-5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suthar MS et al. Rapid Generation of Neutralizing Antibody Responses in COVID-19 Patients. Cell Rep Med 1, 100040, doi: 10.1016/j.xcrm.2020.100040 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cohen KW et al. Longitudinal analysis shows durable and broad immune memory after SARS-CoV-2 infection with persisting antibody responses and memory B and T cells. Cell Rep Med 2, 100354, doi: 10.1016/j.xcrm.2021.100354 (2021). ***This study describes the durability of humoral and T cell mediated immunity after SARS-CoV-2 infection.
- 42.Carreño JM et al. Reduced neutralizing activity of post-SARS-CoV-2 vaccination serum against variants B.1.617.2, B.1.351, B.1.1.7+E484K and a sub-variant of C.37. 2021.2007.2021.21260961, doi: 10.1101/2021.07.21.21260961 %J medRxiv (2021). [DOI] [Google Scholar]
- 43.Amanat F et al. SARS-CoV-2 mRNA vaccination induces functionally diverse antibodies to NTD, RBD, and S2. Cell 184, 3936–3948 e3910, doi: 10.1016/j.cell.2021.06.005 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stadlbauer D et al. SARS-CoV-2 Seroconversion in Humans: A Detailed Protocol for a Serological Assay, Antigen Production, and Test Setup. Curr Protoc Microbiol 57, e100, doi: 10.1002/cpmc.100 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roubidoux EK et al. Mutations in the Hemagglutinin Stalk Domain Do Not Permit Escape from a Protective, Stalk-Based Vaccine-Induced Immune Response in the Mouse Model. mBio 12, doi: 10.1128/mBio.03617-20 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Asthagiri Arunkumar G et al. Broadly Cross-Reactive, Nonneutralizing Antibodies against Influenza B Virus Hemagglutinin Demonstrate Effector Function-Dependent Protection against Lethal Viral Challenge in Mice. J Virol 93, doi: 10.1128/JVI.01696-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saphire EO et al. Systematic Analysis of Monoclonal Antibodies against Ebola Virus GP Defines Features that Contribute to Protection. Cell 174, 938–952 e913, doi: 10.1016/j.cell.2018.07.033 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.DiLillo DJ, Tan GS, Palese P & Ravetch JV Broadly neutralizing hemagglutinin stalk-specific antibodies require FcgammaR interactions for protection against influenza virus in vivo. Nat Med 20, 143–151, doi: 10.1038/nm.3443 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lu LL, Suscovich TJ, Fortune SM & Alter G Beyond binding: antibody effector functions in infectious diseases. Nat Rev Immunol 18, 46–61, doi: 10.1038/nri.2017.106 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zohar T et al. Compromised Humoral Functional Evolution Tracks with SARS-CoV-2 Mortality. Cell 183, 1508–1519 e1512, doi: 10.1016/j.cell.2020.10.052 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Atyeo C et al. Distinct Early Serological Signatures Track with SARS-CoV-2 Survival. Immunity 53, 524–532 e524, doi: 10.1016/j.immuni.2020.07.020 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Anand SP et al. Longitudinal analysis of humoral immunity against SARS-CoV-2 Spike in convalescent individuals up to 8 months post-symptom onset. Cell Rep Med 2, 100290, doi: 10.1016/j.xcrm.2021.100290 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tso FY et al. Presence of antibody-dependent cellular cytotoxicity (ADCC) against SARS-CoV-2 in COVID-19 plasma. PLoS One 16, e0247640, doi: 10.1371/journal.pone.0247640 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McMahan K et al. Correlates of protection against SARS-CoV-2 in rhesus macaques. Nature 590, 630–634, doi: 10.1038/s41586-020-03041-6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mercado NB et al. Single-shot Ad26 vaccine protects against SARS-CoV-2 in rhesus macaques. Nature 586, 583–588, doi: 10.1038/s41586-020-2607-z (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gorman MJ et al. Fab and Fc contribute to maximal protection against SARS-CoV-2 following NVX-CoV2373 subunit vaccine with Matrix-M vaccination. Cell Rep Med 2, 100405, doi: 10.1016/j.xcrm.2021.100405 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Begin P et al. Convalescent plasma for hospitalized patients with COVID-19: an open-label, randomized controlled trial. Nat Med 27, 2012–2024, doi: 10.1038/s41591-021-01488-2 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Winkler ES et al. Human neutralizing antibodies against SARS-CoV-2 require intact Fc effector functions for optimal therapeutic protection. Cell 184, 1804–1820 e1816, doi: 10.1016/j.cell.2021.02.026 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schafer A et al. Antibody potency, effector function, and combinations in protection and therapy for SARS-CoV-2 infection in vivo. J Exp Med 218, doi: 10.1084/jem.20201993 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yamin R et al. Fc-engineered antibody therapeutics with improved anti-SARS-CoV-2 efficacy. Nature 599, 465–470, doi: 10.1038/s41586-021-04017-w (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kaplonek P et al. Subtle immunological differences in mRNA-1273 and BNT162b2 COVID-19 vaccine induced Fc-functional profiles. bioRxiv, doi: 10.1101/2021.08.31.458247 (2021). [DOI] [Google Scholar]
- 62.Mateus J et al. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science 370, 89–94, doi: 10.1126/science.abd3871 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Grifoni A et al. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell 181, 1489–1501 e1415, doi: 10.1016/j.cell.2020.05.015 (2020). ***This study described the SARS-CoV-2-specific CD4+ and CD8+ T cell responses in COVID-19 patients and in unexposed individuals.
- 64. Dan JM et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science 371, doi: 10.1126/science.abf4063 (2021). ***This study assessed the durability of immune responses against SARS-CoV-2 variants in individuals that received the mRNA vaccine.
- 65.Goel RR et al. mRNA vaccines induce durable immune memory to SARS-CoV-2 and variants of concern. Science, eabm0829, doi: 10.1126/science.abm0829 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Geers D et al. SARS-CoV-2 variants of concern partially escape humoral but not T-cell responses in COVID-19 convalescent donors and vaccinees. Sci Immunol 6, doi: 10.1126/sciimmunol.abj1750 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Alter G et al. Immunogenicity of Ad26.COV2.S vaccine against SARS-CoV-2 variants in humans. Nature 596, 268–272, doi: 10.1038/s41586-021-03681-2 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Moore CB et al. Evidence of HIV-1 adaptation to HLA-restricted immune responses at a population level. Science 296, 1439–1443, doi: 10.1126/science.1069660 (2002). [DOI] [PubMed] [Google Scholar]
- 69.Hertz T et al. HLA targeting efficiency correlates with human T-cell response magnitude and with mortality from influenza A infection. Proc Natl Acad Sci U S A 110, 13492–13497, doi: 10.1073/pnas.1221555110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gartland AJ et al. Analysis of HLA A*02 association with vaccine efficacy in the RV144 HIV-1 vaccine trial. J Virol 88, 8242–8255, doi: 10.1128/JVI.01164-14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Henry Dunand CJ et al. Both Neutralizing and Non-Neutralizing Human H7N9 Influenza Vaccine-Induced Monoclonal Antibodies Confer Protection. Cell Host Microbe 19, 800–813, doi: 10.1016/j.chom.2016.05.014 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tan GS et al. Broadly-Reactive Neutralizing and Non-neutralizing Antibodies Directed against the H7 Influenza Virus Hemagglutinin Reveal Divergent Mechanisms of Protection. PLoS Pathog 12, e1005578, doi: 10.1371/journal.ppat.1005578 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jegaskanda S, Weinfurter JT, Friedrich TC & Kent SJ Antibody-dependent cellular cytotoxicity is associated with control of pandemic H1N1 influenza virus infection of macaques. J Virol 87, 5512–5522, doi: 10.1128/JVI.03030-12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Plante JA et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature, doi: 10.1038/s41586-020-2895-3 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Knezevic I et al. WHO International Standard for evaluation of the antibody response to COVID-19 vaccines: call for urgent action by the scientific community. Lancet Microbe, doi: 10.1016/S2666-5247(21)00266-4 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Corbett KS et al. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature 586, 567–571, doi: 10.1038/s41586-020-2622-0 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vogel AB et al. BNT162b vaccines protect rhesus macaques from SARS-CoV-2. Nature 592, 283–289, doi: 10.1038/s41586-021-03275-y (2021). [DOI] [PubMed] [Google Scholar]
- 78.van Doremalen N et al. ChAdOx1 nCoV-19 vaccine prevents SARS-CoV-2 pneumonia in rhesus macaques. Nature 586, 578–582, doi: 10.1038/s41586-020-2608-y (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Guebre-Xabier M et al. NVX-CoV2373 vaccine protects cynomolgus macaque upper and lower airways against SARS-CoV-2 challenge. Vaccine 38, 7892–7896, doi: 10.1016/j.vaccine.2020.10.064 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tian JH et al. SARS-CoV-2 spike glycoprotein vaccine candidate NVX-CoV2373 immunogenicity in baboons and protection in mice. Nat Commun 12, 372, doi: 10.1038/s41467-020-20653-8 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pruijssers AJ et al. Remdesivir Inhibits SARS-CoV-2 in Human Lung Cells and Chimeric SARS-CoV Expressing the SARS-CoV-2 RNA Polymerase in Mice. Cell Rep 32, 107940, doi: 10.1016/j.celrep.2020.107940 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hansen J et al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science 369, 1010–1014, doi: 10.1126/science.abd0827 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Boras B et al. Preclinical characterization of an intravenous coronavirus 3CL protease inhibitor for the potential treatment of COVID19. Nat Commun 12, 6055, doi: 10.1038/s41467-021-26239-2 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.He X et al. Low-dose Ad26.COV2.S protection against SARS-CoV-2 challenge in rhesus macaques. Cell 184, 3467–3473 e3411, doi: 10.1016/j.cell.2021.05.040 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hirose R et al. A Cytopathic Effect-Based Tissue Culture Method for HCoV-OC43 Titration Using TMPRSS2-Expressing VeroE6 Cells. mSphere 6, doi: 10.1128/mSphere.00159-21 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Munoz-Fontela C et al. Animal models for COVID-19. Nature 586, 509–515, doi: 10.1038/s41586-020-2787-6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ozono S et al. SARS-CoV-2 D614G spike mutation increases entry efficiency with enhanced ACE2-binding affinity. Nat Commun 12, 848, doi: 10.1038/s41467-021-21118-2 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.McCray PB Jr. et al. Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus. J Virol 81, 813–821, doi: 10.1128/JVI.02012-06 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sun J et al. Generation of a Broadly Useful Model for COVID-19 Pathogenesis, Vaccination, and Treatment. Cell 182, 734–743 e735, doi: 10.1016/j.cell.2020.06.010 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sun CP et al. Rapid generation of mouse model for emerging infectious disease with the case of severe COVID-19. PLoS Pathog 17, e1009758, doi: 10.1371/journal.ppat.1009758 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Oladunni FS et al. Lethality of SARS-CoV-2 infection in K18 human angiotensin-converting enzyme 2 transgenic mice. Nat Commun 11, 6122, doi: 10.1038/s41467-020-19891-7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Winkler ES et al. SARS-CoV-2 infection of human ACE2-transgenic mice causes severe lung inflammation and impaired function. Nat Immunol 21, 1327–1335, doi: 10.1038/s41590-020-0778-2 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Golden JW et al. Human angiotensin-converting enzyme 2 transgenic mice infected with SARS-CoV-2 develop severe and fatal respiratory disease. JCI Insight 5, doi: 10.1172/jci.insight.142032 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Moreau GB et al. Evaluation of K18-hACE2 Mice as a Model of SARS-CoV-2 Infection. Am J Trop Med Hyg 103, 1215–1219, doi: 10.4269/ajtmh.20-0762 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zheng J et al. COVID-19 treatments and pathogenesis including anosmia in K18-hACE2 mice. Nature 589, 603–607, doi: 10.1038/s41586-020-2943-z (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Vanderheiden A et al. CCR2 Signaling Restricts SARS-CoV-2 Infection. mBio, e0274921, doi: 10.1128/mBio.02749-21 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Muruato A et al. Mouse-adapted SARS-CoV-2 protects animals from lethal SARS-CoV challenge. PLoS Biol 19, e3001284, doi: 10.1371/journal.pbio.3001284 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rathnasinghe R et al. The N501Y mutation in SARS-CoV-2 spike leads to morbidity in obese and aged mice and is neutralized by convalescent and post-vaccination human sera. medRxiv, doi: 10.1101/2021.01.19.21249592 (2021). [DOI] [Google Scholar]
- 99.Ying B et al. Protective activity of mRNA vaccines against ancestral and variant SARS-CoV-2 strains. Sci Transl Med, eabm3302 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Imai M et al. Syrian hamsters as a small animal model for SARS-CoV-2 infection and countermeasure development. Proc Natl Acad Sci U S A 117, 16587–16595, doi: 10.1073/pnas.2009799117 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bricker TL et al. A single intranasal or intramuscular immunization with chimpanzee adenovirus-vectored SARS-CoV-2 vaccine protects against pneumonia in hamsters. Cell Rep 36, 109400, doi: 10.1016/j.celrep.2021.109400 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sia SF et al. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature 583, 834–838, doi: 10.1038/s41586-020-2342-5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Imai M et al. Characterization of a new SARS-CoV-2 variant that emerged in Brazil. Proc Natl Acad Sci U S A 118, doi: 10.1073/pnas.2106535118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Schmitz AJ et al. A vaccine-induced public antibody protects against SARS-CoV-2 and emerging variants. Immunity 54, 2159–2166 e2156, doi: 10.1016/j.immuni.2021.08.013 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Plante JA et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature 592, 116–121, doi: 10.1038/s41586-020-2895-3 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Halfmann PJ et al. SARS-CoV-2 Omicron virus causes attenuated disease in mice and hamsters. Nature, doi: 10.1038/s41586-022-04441-6 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.van Doremalen N et al. Intranasal ChAdOx1 nCoV-19/AZD1222 vaccination reduces viral shedding after SARS-CoV-2 D614G challenge in preclinical models. Sci Transl Med 13, doi: 10.1126/scitranslmed.abh0755 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Fischer RJ et al. ChAdOx1 nCoV-19 (AZD1222) protects Syrian hamsters against SARS-CoV-2 B.1.351 and B.1.1.7. Nat Commun 12, 5868, doi: 10.1038/s41467-021-26178-y (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Francica JR et al. Protective antibodies elicited by SARS-CoV-2 spike protein vaccination are boosted in the lung after challenge in nonhuman primates. Sci Transl Med 13, doi: 10.1126/scitranslmed.abi4547 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Corbett KS et al. Immune correlates of protection by mRNA-1273 vaccine against SARS-CoV-2 in nonhuman primates. Science 373, eabj0299, doi: 10.1126/science.abj0299 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Corbett KS et al. mRNA-1273 protects against SARS-CoV-2 beta infection in nonhuman primates. Nat Immunol 22, 1306–1315, doi: 10.1038/s41590-021-01021-0 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gagne M et al. Protection from SARS-CoV-2 Delta one year after mRNA-1273 vaccination in rhesus macaques coincides with anamnestic antibody response in the lung. Cell 185, 113–130 e115, doi: 10.1016/j.cell.2021.12.002 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bewley KR et al. Immunological and pathological outcomes of SARS-CoV-2 challenge following formalin-inactivated vaccine in ferrets and rhesus macaques. Sci Adv 7, eabg7996, doi: 10.1126/sciadv.abg7996 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhou J et al. Immunogenicity, safety, and protective efficacy of an inactivated SARS-associated coronavirus vaccine in rhesus monkeys. Vaccine 23, 3202–3209, doi: 10.1016/j.vaccine.2004.11.075 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Cameroni E et al. Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. Nature, doi: 10.1038/s41586-021-04386-2 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Pajon R et al. SARS-CoV-2 Omicron Variant Neutralization after mRNA-1273 Booster Vaccination. N Engl J Med, doi: 10.1056/NEJMc2119912 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Edara V-V et al. mRNA-1273 and BNT162b2 mRNA vaccines have reduced neutralizing activity against the SARS-CoV-2 omicron variant. Cell Reports Medicine 3, doi: 10.1016/j.xcrm.2022.100529 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Liu L et al. Striking Antibody Evasion Manifested by the Omicron Variant of SARS-CoV-2. Nature, doi: 10.1038/s41586-021-04388-0 (2021). [DOI] [PubMed] [Google Scholar]
- 119.Tarke A et al. SARS-CoV-2 vaccination induces immunological T cell memory able to cross-recognize variants from Alpha to Omicron. Cell, doi: 10.1016/j.cell.2022.01.015 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Valanparambil RM et al. Antibody response to SARS-CoV-2 mRNA vaccine in lung cancer patients: Reactivity to vaccine antigen and variants of concern. medRxiv, doi: 10.1101/2022.01.03.22268599 (2022). [DOI] [Google Scholar]
- 121.Bartsch Y et al. Preserved Omicron Spike specific antibody binding and Fc-recognition across COVID-19 vaccine platforms. medRxiv, doi: 10.1101/2021.12.24.21268378 (2021). [DOI] [Google Scholar]
- 122.Carreno JM et al. Activity of convalescent and vaccine serum against SARS-CoV-2 Omicron. Nature, doi: 10.1038/s41586-022-04399-5 (2021). [DOI] [PubMed] [Google Scholar]


