Abstract
Genetically modified (GM) crops are a major product of the global food industry. From 1996 to 2014, 357 GM crops were approved and the global value of the GM crop market reached 35% of the global commercial seed market in 2014. However, the rapid growth of the GM crop-based industry has also created controversies in many regions, including the European Union, Egypt, and Taiwan. The effective detection and regulation of GM crops/foods are necessary to reduce the impact of these controversies. In this review, the status of GM crops and the technology for their detection are discussed. As the primary gap in GM crop regulation exists in the application of detection technology to field regulation, efforts should be made to develop an integrated, standardized, and high-throughput GM crop detection system. We propose the development of an integrated GM crop detection system, to be used in combination with a standardized international database, a decision support system, high-throughput DNA analysis, and automated sample processing. By integrating these technologies, we hope that the proposed GM crop detection system will provide a method to facilitate comprehensive GM crop regulation.
Keywords: decision support system, genetically modified organism, GM crop detection, high-throughput analysis
1. Introduction
Genetically modified (GM) crops are a dominant agricultural food product worldwide owing to their superior productivity. From 1996 to 2014, 357 GM crops have been approved globally. The global value of the GM crop market was 15.7 billion US$ in 2014, representing 35% of the global commercial seed market [1]. Rapid growth of the GM crop industry also created controversies in many regions, including the European Union [2], Egypt [3,4], Japan [5], Korea [6], Brazil [7], and Taiwan [8]. To mitigate these controversies, effective regulation based on comprehensive GM crop detection is essential. DNA-based methods such as real-time quantitative PCR (qPCR) have been successfully applied to GM crop detection for the past two decades. However, the continued rapid development of new GM crop events is overwhelming the processing capacity of conventional methods. In addition, the efficacy of GM crop regulation has deteriorated further, due to the release of unauthorized GM crops/foods into the food chain [9]. To meet these challenges, it is necessary to develop a high-efficiency GM crop detection infrastructure.
2. Classification of GM crops and levels of DNA detection
The past few decades have seen significant advances in plant gene engineering. The methods for the transgenic manipulation of GM crops have also evolved, with major breakthroughs in both technology and theory. Today, GM crops can be classified into four generations according to the structure and strategy used to construct their transgenes. Therefore, the detection of GM crops/foods requires a dedicated strategy. GM crops/foods can be identified via several types of biomolecules such as specific proteins, RNA, DNA, and metabolites. Among these targets, DNA is the only molecule with advantages of being stable, abundant, and easily to amplify. Thus, detection of specific DNA sequences, especially using a PCR-based approach, is still the most effective strategy. In brief, there are four generations of GM crops and three major levels of detection.
(1) Four generations/classes of GM crops
(a) The first generation/class: single trait
Most commercial GM crops today either are of the first generation or its stacked (second generation) [9]. Most first-generation GM crops contain common transgene elements such as the cauliflower mosaic virus (CaMV), 35S promoter (CaMV35S-P), aminoglycoside 3′-phosphotransferase gene (nptII), phosphinothricin acetyltransferase gene (pat/bar), 5-enolpyruvylshikimate 3-phosphate (CP4-epsp) gene, nopaline synthase promoter (nos-P), and terminator (nos-T). In effect, because of the limited variation in high-performance transgene elements, ~90% of commercial GM crops contain one or more of the six transgene elements listed above [10].
(b) The second generation/class: stacked traits
Second-generation GM crops are usually hybrid crosses between commercialized first-generation GM crops {e.g., 59122 × MIR604 maize (DAS-59122-7 × SYN-IR604-5) [9]}. Owing to their lower developing costs, the importance and prevalence of second-generation GM crops are increasing. However, two major detection problems arose with stacked trait GM crops/foods: (1) in-depth gene analysis may require the ability to discriminate between stacked trait GM crops and unintended stacked trait GM crops, which might be produced via cross-pollination between two single GM crop events in adjacent fields and (2) the discrimination of mixed events from single stack traits was only possible by testing single seeds or plants, which prevents the technique from being used on processed GM crop products such as corn flour. The detection of second-generation GM crops is complicated by these problems, which together could pose a major threat to GM crop regulation in the near future.
(c) The third and fourth generations/classes: near-intragenics, intragenics, and cisgenics
The third generation of GM crops is comprised of so-called near-intragenics, or GM crops where the inserted transgenic elements have not been used in other (known) GM crops [9]. Near-intragenics are transgene constructs that originated from the host and have undergone minimal recombination or modification. This makes them more difficult to detect than first- or second-generation GM crops.
True intragenics and cisgenics are to be classified as the fourth generation of GM crops. The transgenic elements of fourth-generation GM crops are genuine host genes. Thus, fourth-generation GM crops/foods cannot be distinguished via their transgenic elements. The only way to identify fourth-generation GM crops/foods is to inspect the specific order and insertion loci of its transgenes.
3. Level of DNA detection
(1) Element-specific
Element-specific PCR methods target individual transgenic elements (such as promoters, genes, or terminators), which may be independent of transgenic traits [9]. Due to the limited variance of transgenic elements, this is a very effective universal GM crop screening strategy, especially in multiplex form. In effect, element-specific PCR methods are the only currently available approaches to effective screening of unauthorized and unintended GM crops. The major drawbacks of element-specific PCR are its limited utility for GM crop quantification and its inability to detect intragenic and cisgenic GM crops. It should be noted that transgenic elements sharing the same name do not necessarily possess identical DNA sequences. Various sequence optimizations and variations introduced during GM crop development may decrease the specificity of element-specific PCR methods [10].
(2) Construct-specific
Construct-specific PCR targets the specific order of transgenic elements [9]. The target sequences of construct-specific PCR are usually comprised of junction(s) of two or more transgenic elements, which do not exist naturally in organisms. The resolving power of construct-specific PCR is inferior to that of event-specific PCR, because of the many GM crops that share similar transgenic construct configurations. However, the throughput of construct-specific PCR for the screening of GM crops is also constrained by its specificity to constructs but not universal transgenic elements. Thus, despite the fact that the discriminatory ability of construct-specific PCR is higher than that of element-specific PCR, construct-specific methods used in routine GM crop detection are rare. The method is simply too general for use in the identification of GM crops while being an inefficient screening method.
(3) Event-specific
As most plant transformation methods (such as Agrobacterium or Biolistic) used today are based on the random insertion of transgenic DNA, chimeric sequences comprised of host DNA and transgenic construct border sequences are present in every trait of GM crops [9]. Event-specific PCR targets these unique chimeric sequences, which are particularly suitable markers for the identification and quantification of GM crops. This form of detection is also the legal basis of GM crop authorization for commercial use as food or feed in the European Union (EU).
4. Method validation
All the GM crops/food detection methods must be validated before application to routine regulation. Specificity, sensitivity, linearity, limit of detection and limit of quantification of GM organism (GMO) detection methods are tested with intra-and interlaboratory analysis of certified reference material. An additional spike test may be needed to validate analysis method for food. The EU Database of Reference Methods for GMO Analysis (GMOMETHODS: http://gmo-crl.jrc.ec.europa.eu/gmomethods/) provides comprehensive information of fully validated GMO detection methods. Novel GMO detection methods could be validated by European Network of GMO Laboratories (ENGL) advisory group via submission.
5. Collection and processing of GM crop information
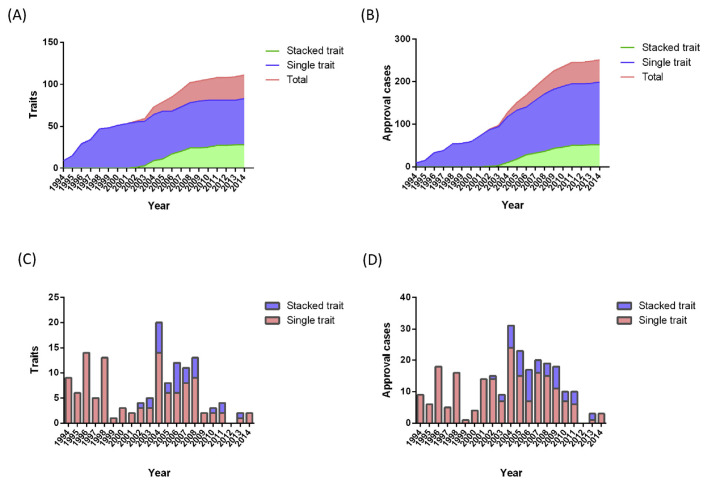
Since the first GM crop approval in 1994, the increase in the number of approved GM crops has been relatively constant over the course of the past two decades (Fig. 1A and 1C). Today, 357 GM traits in various crops such as potato, canola, maize, cotton, and soybean have been approved worldwide [1]. Besides the vast number of GM traits, the approval status (food, feed, and environment) of many GM crops varies from country to country. For example, there are four approval statuses of soybean MON-04032-6 (GTS 40-3-2) in 22 countries including food only, feed only, food/feed, and food/feed/environment (Table 1). The amount of existing GM crop-related information is now too large to process without the aid of software and databases. Thus, to deal with the numerous varieties of GM crops traded and grown worldwide, the effective regulation of GM crops requires the support of comprehensive databases. Several open-access databases such as the Center for Environmental Risk Assessment (CERA) GM crop database (http://cera-gmc.org/index.php?action=gm_crop_database) are useful repositories of GM crop-related datasets and information, including transgene constructs, plant species, traits, and approval statuses in major countries. However, information regarding a GM plant's comprehensive approval and production status in each individual country, which is necessary for the comprehensive regulation of GM crops in world trade, is not available for every country (e.g., Egypt). Furthermore, the type of information available and the structures of existing databases may not be compatible because of the varied aims and scopes of the available information sources. For example, the GM approval database of the International Service for the Acquisition of Agri-biotech Applications (http://www.isaaa.org/gmapprovaldatabase/) includes 374 traits, but there are only 158 GM crop traits listed in the CERA GM crop database [11]. Information regarding the approval status of various GM crop traits is also not consistent between these two databases, especially the coverage of countries. For example, the approval of soybean MON-04032-6 (GTS 40-3-2) of Bolivia, Chile, Costa Rica, and Indonesia are not listed in the CERA database (Tables 1 and 2). This inconsistency or incompleteness of GM crop information may compromise GM crop regulation, and even lead to conflicts in global trade. Thus, we propose the development of an international standardized GM crop database that includes information relating to transgenes, regulation status, and production status. In addition, possible unauthorized traits should be considered as information necessary for the development of GM crop regulation infrastructure.
Fig. 1.
Annually introduced genetically modified (GM) crop traits and approval cases for food/feed use. (A) Accumulated GM crop traits. (B) Accumulated GM crop approval cases. (C) New GM crop traits introduced each year. (D) New approval cases introduced each year.
Table 1.
A summary of the legal approval status of soybean MON-04032-6 (GTS 40-3-2) in the Center for Environmental Risk Assessment genetically modified crop database [8].
| Country | Environment | Food &/or feed | Food | Feed | Marketing |
|---|---|---|---|---|---|
| Argentina | 1996 | — | 1996 | 1996 | — |
| Australia | — | — | 2000 | — | — |
| Brazil | 1998 | — | 1998 | 1998 | — |
| Canada | 1995 | — | 1996 | 1995 | — |
| China | — | 2004 | — | — | — |
| Colombia | — | 2005 | — | — | — |
| Czech Republic | — | — | 2001 | 2001 | 2001 |
| European Union | — | 2005 | — | — | 1996 |
| Japan | 1996 | — | 1996 | 1996 | — |
| Korea | — | — | 2000 | 2004 | — |
| Mexico | 1998 | — | 1998 | 1998 | — |
| Paraguay | 2004 | 2004 | 1996 | — | — |
| Philippines | — | — | 2000 | 2003 | — |
| Russia | — | — | 1998 | — | 1999 |
| South Africa | 2001 | — | 1996 | 2001 | — |
| Switzerland | — | — | 1996 | 1996 | — |
| Taiwan | — | — | 2002 | — | — |
| United Kingdom | — | — | 2001 | 1996 | — |
| United States | 1994 | 1994 | — | — | — |
| Uruguay | 1997 | — | 1996 | 1997 | — |
CERA = Center for Environmental Risk Assessment.
Table 2.
A summary of the regulatory approvals of soybean MON-04032-6 (GTS 40-3-2) in the genetically modified approval database of the International Service for the Acquisition of Agri-biotech Applications (http://www.isaaa.org/gmapprovaldatabase/).
| Country | Food | Feed | Cultivation |
|---|---|---|---|
| Argentina | 1996 | 1996 | 1996 |
| Australia | 2000 | — | — |
| Bolivia | 2005 | 2005 | 2005 |
| Brazil | 1998 | 1998 | 1998 |
| Canada | 1996 | 1995 | 1995 |
| Chile | — | — | 2007 |
| China | 2002 | 2002 | — |
| Colombia | 2005 | 2007 | — |
| Costa Rica | — | — | 2001 |
| European Union | 2005 | 2005 | — |
| Indonesia | 2011 | — | 2012 |
| Japan | 2001 | 2003 | 2005 |
| Malaysia | 2010 | 2010 | — |
| Mexico | 1996 | — | 1996 |
| New Zealand | 2000 | — | — |
| Paraguay | 2004 | 2004 | 2004 |
| Philippines | 2003 | 2003 | — |
| Russian Federation | 2007 | 2008 | — |
| South Africa | — | — | 2001 |
| South Korea | 2002 | 2004 | — |
| Switzerland | 1996 | 1996 | — |
| Taiwan | 2002 | — | — |
| Turkey | — | 2011 | — |
| United States of America | 1995 | 1995 | 1993 |
| Uruguay | 1996 | 1996 | 1996 |
6. Screening of transgenic elements
The preliminary screening of transgenic elements (element-specific) is an efficient approach to both authorized and unauthorized GM crop detection. Although several PCR-free detection methods such as direct detection by DNA microarray [12] and magnetic capture with fluorescence cross-correlation spectroscopy [13] have been described, PCR-based methods are still the method of choice due to their versatility, sensitivity, and high-throughput potential. Preliminary screening by PCR is usually arranged in a multiplex or other equivalent form, to increase its screening efficiency. Theoretically, all the GM crops could be detected by incorporating a large number (>18) of primer sets into a multiplex PCR [10]. However, the robustness of detection declines following the addition of primer sets to multiplex PCR. Besides, the addition of more than six primer sets will only marginally contribute to GM crop detection coverage [10]. Thus, the number of primer sets used for qPCR [14,15] and conventional PCR [10] assays have been limited to under six, in order to achieve a balance between reasonable trait coverage and robustness. Despite the fact that, in practice, methods of screening for transgenic elements cannot cover all existing GM crop traits, it is still a valuable technique for the preliminary screening of GM crops owing to its higher throughput potential. Besides, screening for transgenic elements may be the only viable approach to the detection and classification of unauthorized GM crops. Therefore, we suggest the screening of transgenic elements as a primary method of unauthorized GM crop screening, and an auxiliary approach for the identification of authorized GM crops. However, it should not be used as a primary screening method. The identification and classification of unauthorized GM crop will be discussed later in this article.
7. Event identification and quantification
Event-specific PCR, especially event-specific qPCR, is the gold standard of GM crop detection methods. Event-specific detection is necessary for authorized GM crop screening and identification in the EU [16]. Event-specific PCR methods are rarely used to screen GM crops because the number of GM crop traits far exceeds the capabilities of single multiplex PCR/qPCR. With this method, a large number of tests are needed to analyze unknown samples such as single seeds for event-specific detection. However, event-specific detection is the only approach that can specifically identify/quantify all GM crops at the trait level. Thus, despite its low efficiency, event-specific detection is the only comprehensive solution for authorized GM crop screening. With the help of high-throughput and automated technology, event-specific detection is recommended for the screening of authorized GM crops.
8. Unidentifiable and undetectable GM crops
Unidentifiable GM crops are those that are detectable by either element-specific or event-specific methods, but whose traits cannot be determined via simple genetic analysis. These unidentifiable GM crops may include: (1) unauthorized GM crops with detectable transgenic elements introduced by genetic manipulation; (2) unintended GM crops produced via cross pollination between GM and wild-type crops; (3) unintended stacked traits produced by unintended crosses between two GM crops; and (4) genuine stacked-trait GM crops. For the first type, unidentifiable (unauthorized) GM crops, identification is impossible without information about their genetic modifications and sources. The identification of unauthorized GM crops relies entirely on the traceability of food trade chains. Thus, comprehensive traceability and real-time information exchange is critical for unauthorized GM crop identification.
The second type of unidentifiable GM crop (the unintended GM crops) is virtually indistinguishable from GM donor crops when single-seed samples are not available. Even when a single-seed sample is available, multidisciplinary analysis may be required to discriminate between the traits of most crops. It may difficult to discriminate between crop traits using seed samples alone, and discrimination of crop traits in the processed food is usually impossible. Thus the regulation of the second type of identifiable GM crop may rely on the comprehensive traceability of crop production chain rather than detection.
Unintended stacked-trait (the 3rd type) GM crops originate from unintended cross-pollination between two adjacent GM crops in the field. There is no straightforward approach to discriminate between unintended stacked-trait and genuine stacked-trait GM crops, except for those trait combinations that are unauthorized. For angiosperm crops such as maize, the polyploid nature of the endosperm makes the genetic background of angiosperm plants traceable via the quantification of parental- and maternal-specific sequences [17]. Thus, the unintended stacked traits of GM angiosperm plants may be distinguished from their genuine stacked-trait GM counterparts by the ratio of transgenic sequences of the individual parental traits.
Genuine (Type 4 unidentifiable) GM crops are identified via a combination of several methods targeting individual parental traits, rather than a single method specific to stacked-trait plants. Thus, it is difficult to distinguish genuine stacked-trait GM crops from a mixture of parental GM crops without a single-seed test. This test is not only essential for the identification of genuine stacked-trait GM crops but is also highly recommended for the analysis of unauthorized (Type 1) and unintended stacked-trait (Type 3) GM crops. For processed food, the analysis of stacked-trait GM crops may be complicated by the presence of two or more GM crops in the ingredients. Besides DNA or protein-based analysis, various attempts have been made to identify a specific plant variety or component by metabolomics-scale analysis [17–20]. However, the reproducibility of metabolite profile was limited due to variations of individual, climate, and agricultural management, especially for the processed foods. Thus the identification of stacked-trait GM crops in the processed food may very difficult, which emphasized the importance of a comprehensive regulation at crop (raw material) level.
Screening of common transgenic elements is currently the best strategy to detect unintended or unauthorized GM crops [10,21,22]. However, there are certain unauthorized GM crops that are undetectable because they have neither common transgenic elements nor a pre-existing event-specific detection method. Although the unauthorized GM crops (including intended release) would cause a high risk to food safety, events of these unauthorized GM crops during laboratory-phase trials are virtually untraceable. As with other unauthorized GM crops, the control of undetectable GM crops entirely relies on the regulation of laboratory-phase GM crops, which is beyond the scope of this review.
9. Automation and high-throughput technologies for GM crop screening
The emergence of automated nucleic acid extraction and handling systems in conjunction with the development of high-throughput analysis technologies has significantly improved the capabilities of modern nucleic acid analysis. As the vast number of GM crop traits has become a major burden on GM crop detection, automated high-throughput technologies are necessary for future GM crop detection.
Because most GM crop detection is based on DNA sequence analysis, the first step of GM crop detection is the preparation of sample DNA. DNA extraction is a labor-intensive process because of the significant number of liquid transfer and centrifugation steps required for the analysis of each individual sample. Thus, manual sample DNA preparation has become the main bottleneck in the GM crop detection process. According to our experience, the rate of DNA extraction is usually <100 samples/d/technician, with ready-to-use DNA extraction kits. The labor burden of DNA extraction is further exacerbated by the fact that GM crop samples are usually seeds, which need to be ground into flour prior to extraction. Thus, for large-scale GM crop screening, the aid of automated sample processers such as a tissue homogenizer and DNA extraction system is essential.
Various high-throughput nucleic acid analysis methods such as DNA microarrays [23], optical thin-film biosensor chips [24], capillary electrophoresis [25,26], microdroplet PCR [26], multiwall carbon nanotube-doped polypyrrole DNA biosensors [27], and loop-mediated isothermal amplification (LAMP) [28] have been successfully applied to GM crop detection. However, the large-scale application of most of these technologies to GM crop detection is limited because they require specialized instruments and reagents. Of these technologies, only the DNA microarray and LAMP are currently commercially available for GM crop detection. DNA microarrays are quickly becoming standard tools in molecular biology [29]. Various microarrays have been developed for GMO screening, including the DualChip GMO microarray [30]. The major drawback of microarray analysis is the complex and laborious sample/microarray preparation process, which includes PCR, hybridization, multiple wash/liquid transfers, and the development of fluorescence. As a result, specialized instruments such as liquid handling stations and microarray scanners are required to guarantee reproducibility [29]. The high cost of microarray chips and its peripheral instruments greatly reduces the benefits of the application of microarray technology to routine GM crop screening.
LAMP is an emerging DNA amplification technique for sequence detection, with the potential to be an alternative to PCR [28]. LAMP techniques have the following advantages: (1) a moderate incubation temperature leads to simple instrument requirements; (2) high amplification product yields, which can be detected either visually or using a simple detector; (3) highly robust because of superior tolerance of substances that typically inhibit PCR; (4) high specificity and sensitivity compared to PCR; and (5) rapid detection—typical LAMP reaction time is 10–20 minutes [30,31]. In addition, LAMP is highly versatile because of its compatibility with a wide range of popular analysis methods, including electrophoresis, colorimetry, fluorescence, and real-time PCR. Simplicity, low instrument costs, and robustness make LAMP a high-throughput method with great potential for application to large-scale GM crop screening.
10. Decision-supported system and GM crop detection workflow
Current routine GM crop detection methods are not efficient enough to screen every crop sample. Most current GM crop screening at customs and border control is conducted by taking random samples of a fixed percentage of cargo, leading to significant gaps in GM crop regulation. Thus, efficiency of the GM crop-detection workflow is the limiting factor in comprehensive GM crop regulation.
The decision-supported system (DSS) is a computer-assist matrix that aids decision-making. It greatly facilitates decision-making processes by extracting and processing information from large and complex data sets. DSS has a great potential to facilitate the validation of GM crop approval, the prediction of analysis results, the screening of possible unknown GM crops, and the selection of detection and analysis methods. To deal with an increasingly large and complex GM crop database (Fig. 1), several DSSs have been tested in previous studies on GM crop detection and identification [10,25,32–34]. The JRC GMO-Matrix [33] integrated GM crop database with detection method database to form a cross-index search system, which was a significant progress of DSS to routine GM crop regulation. However, the integration of databases with various instruments and computer software programs is still elusive.
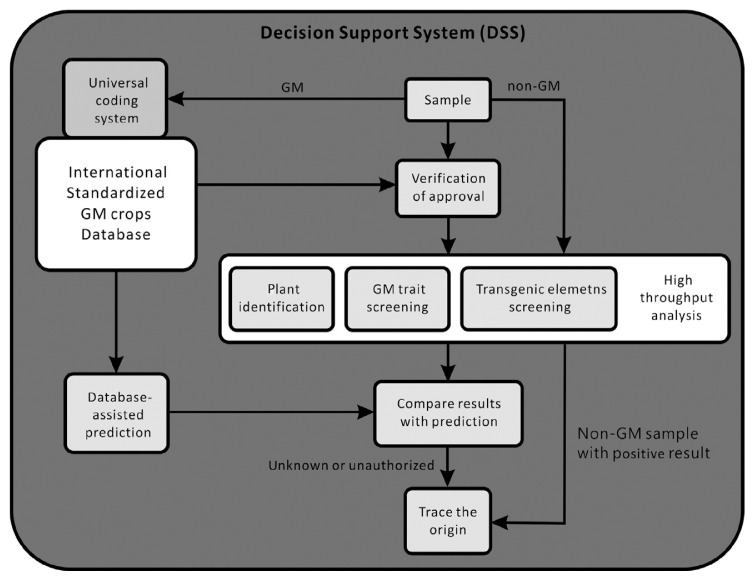
Thus, to maximize the benefits of DSS application, we propose the development of an integrated GM crop detection workflow incorporating high-throughput DNA analysis and complete DSS support (Fig. 2). The major steps in the GM crop detection workflow include: (1) sample legitimacy validation; (2) method validation and result prediction; (3) detection and identification of GM crops; (4) verification of the results of analyses; and (5) the origin of a GM crop, if necessary. Besides DSS, a simple, standardized, systematic GM crop coding system is also required to facilitate the exchange of information and workflow control (Fig. 2). The aim of the proposed workflow is to increase the efficiency of GM crop regulation at the field level such as at customs and border control.
Fig. 2.
Proposed workflow of the integrated lab system for genetically modified crop detection.
The first step of the proposed workflow is to generate a unique code for every sample. This code includes the cargo serial number, GM crop trait/plant identity, usage (food/feed), sample type (seed/flour), and test/replicate number. At the same time, a complete sample information file including the country of origin and shipping information is established. The legitimacy of a sample is checked prior to DNA analysis. Samples designated as illegitimate, such as illegal imports, samples designated for illegal intended usages, or samples that are illegal to grow in their country of origin, will be rejected at this step without further analysis. Following the legitimacy check, the DSS will generate a prediction of the DNA analysis results, according to the information in the sample file and the GM crop database. Meanwhile, the sample is sent for high-throughput DNA analysis, for identification, GM trait screening (event-specific detection), and transgenic element screening. The results of the DNA analysis are then verified against the DSS prediction. Unmatched samples will be rejected and sent for further investigation. By contrast, samples declared as non-GM crop are directly tested with high-throughput analysis instrument. Non-GM crops with any positive GM reaction will be halted and sent for further investigation.
The efficiency of GM crop detection relies not only on a comprehensive workflow but also on a fully integrated instrument system. An ideal GM crop detection analyzer requires the integration of data processing/DSS, a nucleic acid extraction unit, and a high-throughput DNA analysis unit. The tissue homogenizer/nucleic acid extraction unit and high-throughput DNA analysis unit cooperate with the DSS to complete the GM crop detection workflow with full automation [35]. We also suggest the use of the fluorescent-LAMP based method in high-density array form for GM crop detection because of its simple instrument requirements and the rapid development of the LAMP method as a tool for GM crop detection [28,35–41].
11. Conclusion
As food becomes increasingly important worldwide, it is worthwhile to mitigate the controversies surrounding GM crop trading by working on a comprehensive GM crop regulation system. The major gap in GM crop regulation is at the application of detection technologies to field regulation practices; thus, efforts should be made to develop an integrated, standardized, and high-throughput GM crop detection system. In this review, we proposed an integrated GM crop detection system combining an international standardized database, a decision support system, high-throughput DNA analysis, and automated sample processing. A high-density LAMP reaction array including the detection of complete event-specific and transgenic elements is included for authorized and unauthorized GM crop screening, respectively. By integrating these available technologies, we hope that the proposed GM crop detection system will be the solution to the problems currently impeding comprehensive GM crop regulation.
Footnotes
Conflicts of interest
The authors declare no conflicts of interest.
References
- 1.James C. Global Status of Commercialized Biotech/GM Crops. Ithaca: ISAAA; 2014. Brief No 49 ISAAA. [Google Scholar]
- 2. Kuntz M, Davison J, Ricroch AE. What the French ban of Bt MON810 maize means for science-based risk assessment. Nat Biotechnol. 2013;31:498–500. doi: 10.1038/nbt.2613. [DOI] [PubMed] [Google Scholar]
- 3. Adenle AA. Response to issues on GM agriculture in Africa: are transgenic crops safe? BMC Res Notes. 2011;4:388–93. doi: 10.1186/1756-0500-4-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sarant L. Egypt's legal battle to regulate Monsanto's GMOs. [accessed January 29, 01, 14]; Egypt Independent. 2012 Available at: http://www.egyptindependent.com/news/egypt-s-legal-battle-regulate-monsanto-s-gmos-0. [Google Scholar]
- 5. Morris B. Rogue GMO wheat found in Oregon, Japan bans import, EU will test. [accessed March 29, 03, 15]; Politics in the Zeros. 2013 Available at: http://polizeros.com/2013/06/03/rogue-gmo-wheat-found-in-oregon-japan-bans-import-eu-will-test/ [Google Scholar]
- 6. Choi SJ. South Korean institute discovers ‘mystery plants’ from imported GMOs. [accessed 29, 03, 15]; Sustanible Pulse. 2015 Available at: http://sustainablepulse.com/2015/01/24/south-korean-institute-discovers-mystery-plants-imported-gmos/#.VRfEP_mUfIY. [Google Scholar]
- 7.Da Página do MST. Brazilian farmers occupy and cancel approval meeting for GMO trees. Movimento dos Trabalhadores Sem Terra. 2015. [accessed 29, 03, 15]. Available at: http://www.mst.org.br/2015/03/05/apos-ocupacao-na-suzano-outros-300-camponeses-ocupam-predio-da-ctnbio-em-bsb.html.
- 8. Hsiao A. Ninety percent of soybeans genetically modified: experts. [accessed January 29, 01, 14]; Taipei Times. 2013 Available at: http://www.taipeitimes.com/News/taiwan/archives/2013/03/31/2003558445. [Google Scholar]
- 9. Holst-Jensen A, Bertheau Y, de Loose M, Grohmann L, Hamels S, Hougs L, Morisset D, Pecoraro S, Pla M, Van den Bulcke M, Wulff D. Detecting un-authorized genetically modified organisms (GMOs) and derived materials. Biotechnol Adv. 2012;30:1318–35. doi: 10.1016/j.biotechadv.2012.01.024. [DOI] [PubMed] [Google Scholar]
- 10. Lu IJ, Lin CH, Pan TM. Establishment of a system based on universal multiplex-PCR for screening genetically modified crops. Anal Bioanal Chem. 2010;396:2055–64. doi: 10.1007/s00216-009-3214-x. [DOI] [PubMed] [Google Scholar]
- 11.CERA Center for Environmental Risk Assessment (CERA) GM Crop Database. Washington: ILSI Research Foundation; 2012. [accessed 02, 06, 15]. Available at: http://cera-gmc.org/gmcropdatabase. [Google Scholar]
- 12. Von Gotz F. See what you eat—broad GMO screening with microarrays. Anal Bioanal Chem. 2010;396:1961–7. doi: 10.1007/s00216-009-3204-z. [DOI] [PubMed] [Google Scholar]
- 13. Zhou X, Xing D, Tang Y, Chen WR. PCR-free detection of genetically modified organisms using magnetic capture technology and fluorescence cross-correlation spectroscopy. PLoS One. 2009;4:e8074. doi: 10.1371/journal.pone.0008074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Huber I, Block A, Sebah D, Debode F, Morisset D, Grohmann L, Berben G, Stebih D, Milavec M, Zel J, Busch U. Development and validation of duplex, triplex and pentaplex real-time PCR screening assays for the detection of genetically modified organisms in food and feed. J Agric Food Chem. 2013;61:10293–301. doi: 10.1021/jf402448y. [DOI] [PubMed] [Google Scholar]
- 15. Dorries HH, Remus I, Grönewald A, Grönewald C, Berghof-Jäger K. Development of qualitative, multiplex real-time PCR kit for screening of genetically modified organisms (GMOs) Anal Bioanal Chem. 2010;396:2043–54. doi: 10.1007/s00216-009-3149-2. [DOI] [PubMed] [Google Scholar]
- 16. European Commission. Regulation (EC) No 1829/2003 of the European Parliament and of the Council of 22 September 2003 on genetically modified food and feed. Off J Eur Union. 2003;L268:1–23. [Google Scholar]
- 17. Dan W, Jirui H, Yueming J, Bao Y. Quality analysis of Polygala tenuifolia root by ultrahigh performance liquid chromatography–tandem mass spectrometry and gas chromatography–mass spectrometry. J Food Drug Anal. 2014;23:144–51. doi: 10.1016/j.jfda.2014.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yang IC, Tsai CY, Hsieh KW, Yang CW, Ouyang F, Lo YM, Chen S. Integration of SIMCA and near-infrared spectroscopy for rapid and precise identification of herbal medicines. J Food Drug Anal. 2014;21:268–78. [Google Scholar]
- 19. Li YS, Church JS. Raman spectroscopy in the analysis of food and pharmaceutical nanomaterials. J Food Drug Anal. 2014;22:29–48. doi: 10.1016/j.jfda.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chuang YK, Yang IC, Lo YM, Chao-Yin Tsai, Chen S. Integration of independent component analysis with near-infrared spectroscopy for analysis of bioactive components in the medicinal plant Gentiana scabra Bunge. J Food Drug Anal. 2014;22:336–44. doi: 10.1016/j.jfda.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Candela H, Hake S. The art and design of genetic screens: maize. Nat Rev Genet. 2008;9:192–203. doi: 10.1038/nrg2291. [DOI] [PubMed] [Google Scholar]
- 22. Mano J, Oguchi T, Akiyama H, Teshima R, Hino A, Furui S, Kitta K. Simultaneous detection of recombinant DNA segments introduced into genetically modified crops with multiplex ligase chain reaction coupled with multiplex polymerase chain reaction. J Agric Food Chem. 2009;57:2640–6. doi: 10.1021/jf803361a. [DOI] [PubMed] [Google Scholar]
- 23. Shao N, Jiang SM, Zhang M, Wang J, Guo SJ, Jiang HW, Liu CX, Ling Z, Zhang DB, Yang LT, Tao SC. MACRO: a combined microchip-PCR and microarray system for high-throughput monitoring of genetically modified organisms. Anal Chem. 2014;86:1269–76. doi: 10.1021/ac403630a. [DOI] [PubMed] [Google Scholar]
- 24. Bai SL, Zhang J, Li SC, Chen HD, Terzaghi W, Zhang X, Chi XR, Tian J, Luo HX, Huang WS, Chen Y, Zhang YC. Detection of six genetically modified maize lines using optical thin-film biosensor chips. J Agric Food Chem. 2010;58:8490–4. doi: 10.1021/jf100598k. [DOI] [PubMed] [Google Scholar]
- 25. Ruttink T, Morisset D, Van Droogenbroeck B, Lavrac N, Van Den Eede GLM, Zel J, De Loose M. Knowledge-technology-based discovery of unauthorized genetically modified organisms. Anal Bioanal Chem. 2010;396:1951–9. doi: 10.1007/s00216-009-3218-6. [DOI] [PubMed] [Google Scholar]
- 26. Guo JC, Yang L, Chen LT, Morisset D, Li X, Pan LW, Zhang DB. MPIC: a high-throughput analytical method for multiple DNA targets. Anal Chem. 2011;83:1579–86. doi: 10.1021/ac103266w. [DOI] [PubMed] [Google Scholar]
- 27. Lien TTN, Lam TD, An VTH, Hoang TV, Quang DT, Khieu DQ, Tsukahara T, Lee YH, Kim JS. Multi-wall carbon nanotubes (MWCNTs)-doped polypyrrole DNA biosensor for label-free detection of genetically modified organisms by QCM and EIS. Talanta. 2010;80:1164–9. doi: 10.1016/j.talanta.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 28. Randhawa GJ, Singh M, Morisset D, Sood P, Zel J. Loop-mediated isothermal amplification: rapid visual and real-time methods for detection of genetically modified crops. J Agric Food Chem. 2013;61:11338–46. doi: 10.1021/jf4030085. [DOI] [PubMed] [Google Scholar]
- 29. Berndt ML, Yauk CL. Review of the literature examining the correlation among DNA microarray technologies. Environ Mol Mutagen. 2007;48:380–94. doi: 10.1002/em.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hamels S, Leimanis S, Mazzara M, Bellocchi G, Foti N, Moens W, Remacle J, Van de Eede G. EUR22935 EN. Luxembourg: JRC Scientific and Technical Reports; 2007. Microarray method for the screening of EU approved GMOs by identification of their genetic elements. [Google Scholar]
- 31. Ahmad F, Hashsham SA. Miniaturized nucleic acid amplification system for rapid and point-of-care diagnostics: a review. Anal Chim Acta. 2012;733:1–15. doi: 10.1016/j.aca.2012.04.031. [DOI] [PubMed] [Google Scholar]
- 32. Block A, Debode F, Grohmann L, Hulin J, Taverniers I, Kluga L, Barbau-Piednoir E, Broeders S, Huber I, Van den Gruden M, Heinze P, Berben G, Busch U, Roosens N, Janssen E, Žel J, Gruden K, Morisset D. The GMO seek matrix: a decision support tool for optimizing the detection of genetically modified plants. BMC Bioinformatics. 2013;14:256. doi: 10.1186/1471-2105-14-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vanden Bulcke, Leivens A, Barbau-Piednoir E, MbongoloMbella G, Rossens N, Sneyers M, Casi AL. A theoretical introduction to “Combinatory SYBR Green qPCR Screening”, a matrix-based approach for the detection of materials derived from genetically modified plants. Anal Bioanal Chem. 2010;396:2113–23. doi: 10.1007/s00216-009-3286-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Angers-Loustau A, Petrillo M, Bonfini L, Gatto F, Rosa S, Patak A, Kreysa J. JRC GMO-Matrix: a web application to support Genetically Modified Organisms detection strategies. BMC Bioinformatics. 2014;15:417. doi: 10.1186/s12859-014-0417-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang M, Liu Y, Chen L, Quan S, Jiang S, Zhang D, Yang L. One simple DNA extraction device and its combination with modified visual loop-mediated isothermal amplification for rapid on-field detection of genetically modified organisms. Anal Chem. 2013;85:75–82. doi: 10.1021/ac301640p. [DOI] [PubMed] [Google Scholar]
- 36. Kiddle G, Hardinge P, Buttigieg N, Gandelman O, Pereira C, McElgunn CJ, Rizzoli M, Jackson R, Appleton N, Moore C, Tisi LC, Murray JA. GMO detection using a bioluminescent real time reporter (BART) of loop mediated isothermal amplification (LAMP) suitable for field use. BMC Biotechnol. 2012;12:15. doi: 10.1186/1472-6750-12-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chen L, Guo J, Wang Q, Kai G, Yang L. Development of the visual loop-mediated isothermal amplification assays for seven genetically modified maize events and their application in practical samples analysis. J Agric Food Chem. 2011;59:5914–8. doi: 10.1021/jf200459s. [DOI] [PubMed] [Google Scholar]
- 38. Chen X, Wang X, Jin N, Zhou Y, Huang S, Miao Q, Zhu Q, Xu J. Endpoint visual detection of three genetically modified rice events by loop-mediated isothermal amplification. Int J Mol Sci. 2012;13:14421–33. doi: 10.3390/ijms131114421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Huang X, Chen L, Xu J, Ji HF, Zhu S, Chen H. Rapid visual detection of phytase gene in genetically modified maize using loop-mediated isothermal amplification method. Food Chem. 2014;156:184–9. doi: 10.1016/j.foodchem.2014.01.102. [DOI] [PubMed] [Google Scholar]
- 40. Zhang F, Wang L, Wang R, Ying Y, Wu J. Simple screening strategy with only water bath needed for the identification of insect-resistant genetically modified rice. Anal Chem. 2015;87:1523–6. doi: 10.1021/ac504384p. [DOI] [PubMed] [Google Scholar]
- 41. Feng J, Tang S, Liu L, Kuang X, Wang X, Hu S, You S. Development of a loop-mediated isothermal amplification (LAMP) assay for rapid and specific detection of common genetically modified organisms (GMOs) Int J Food Sci Nutr. 2015:1–11. doi: 10.3109/09637486.2014.979318. [DOI] [PubMed] [Google Scholar]