Abstract
Aims/hypothesis
Observational studies have found an increased risk of latent autoimmune diabetes in adults (LADA) associated with low birthweight and adult overweight/obese status. We aimed to investigate whether these associations are causal, using a two-sample Mendelian randomisation (MR) design. In addition, we compared results for LADA and type 2 diabetes.
Methods
We identified 43 SNPs acting through the fetal genome as instrumental variables (IVs) for own birthweight from a genome-wide association study (GWAS) of the Early Growth Genetics Consortium (EGG) and the UK Biobank. We identified 820 SNPs as IVs for adult BMI from a GWAS of the UK Biobank and the Genetic Investigation of ANthropometric Traits consortium (GIANT). Summary statistics for the associations between IVs and LADA were extracted from the only GWAS involving 2634 cases and 5947 population controls. We used the inverse-variance weighted (IVW) estimator as our primary analysis, supplemented by a series of sensitivity analyses.
Results
Genetically determined own birthweight was inversely associated with LADA (OR per SD [~500 g] decrease in birthweight 1.68 [95% CI 1.01, 2.82]). In contrast, genetically predicted BMI in adulthood was positively associated with LADA (OR per SD [~4.8 kg/m2] increase in BMI 1.40 [95% CI 1.14, 1.71]). Robust results were obtained in a range of sensitivity analyses using other MR estimators or excluding some IVs. With respect to type 2 diabetes, the association with birthweight was not stronger than in LADA while the association with adult BMI was stronger than in LADA.
Conclusions/ interpretation
This study provides genetic support for a causal link between low birthweight, adult overweight/obese status and LADA.
Graphical abstract
Supplementary Information
The online version of this article (10.1007/s00125-022-05725-2) contains peer-reviewed but unedited supplementary material.
Keywords: Epidemiology, Genetics, Human, Weight regulation and obesity
Introduction
Latent autoimmune diabetes in adults (LADA) is a hybrid form of diabetes. Genetically, it is closely related to type 1 diabetes with a strong link to HLA genotype [1, 2], while many of its clinical features such as the metabolic syndrome are shared with type 2 diabetes [2]. LADA is characterised by pancreatic autoantibodies and its slow progression to insulin dependence, and is usually restricted to adults [2] although a similar phenomenon has been described in younger individuals (latent autoimmune diabetes in the young [LADY]) [3]. Autoantibody testing is required to separate LADA from type 2 diabetes. Around 5–14% of the adults who have been diagnosed with type 2 diabetes in Europe, North America and Asia have pancreatic autoantibodies [2].
Risk factors for type 2 diabetes have been studied extensively [4, 5] and the disease can be prevented or postponed by maintaining a healthy weight and physical activity [6]. In comparison, the evidence on environmental/lifestyle risk factors for LADA is limited [7]. However, we have previously reported an increased risk of LADA in relation to overweight/obese status [8] and low birthweight [9], in line with findings in type 2 diabetes [10, 11]. Observational studies are prone to residual confounding and reverse causation, while randomised control trials may be unfeasible for studying some risk factors of diseases. Individuals’ genotypes are randomly assigned from their parents before conception and thus are less likely to suffer from confounding or reverse causation [12]. Taking advantage of the natural experiments, Mendelian randomisation (MR) studies use genetic variants as instrumental variables (IVs) for an environmentally modifiable exposure to make causal inference about the outcome [12].
Our aim was to investigate whether low birthweight and adult adiposity are implicated in the aetiology of LADA by, for the first time, using a two-sample MR design. In addition, we wanted to compare these associations in LADA and type 2 diabetes.
Methods
Study design
This was a two-sample MR study using summary statistics from two separate genome-wide association studies (GWAS) of non-overlapping samples of the same underlying population; one provided measures of the associations between IVs and the exposure and the other on the associations between IVs and the outcome [13].
Genetic instruments
Birthweight
Birthweight can be affected by both fetal (own) SNPs and maternal SNPs (through intrauterine environment). To assess the direct association of birthweight determined by fetal SNPs with LADA, and to minimise potential bias from the intrauterine environment (pathway 2 in ESM Fig. 1), we used summary statistics (β coefficients and SEs) for the associations between fetus-only SNPs (SNPs acting only through fetal genome and not maternal genome) and own birthweight (see ESM Methods). IVs for birthweight were extracted from a meta-analysis of the Early Growth Genetics Consortium (EGG) and UK Biobank [14]. Information on birthweight had been collected by measurement at birth, obstetric records, medical registers, interviews with the mother, or self-report as adults in different included studies (ESM Table 1). The study identified 64 fetus-only SNPs associated with own birthweight at p<6.6×10−9 (genome-wide significance threshold revised by authors of the GWAS) in 298,140 European individuals. Among the 64 SNPs, we excluded nine located near genes [14] known to affect the occurrence of diabetes, insulin resistance or glucose regulation to reduce pleiotropy (pathway 3 in ESM Fig. 1; ESM Methods). We further excluded three SNPs located near imprinted genes (rs234864, rs6575803 and rs6026449) and three SNPs (rs7772579, rs8756 and rs1480470) in linkage disequilibrium (LD) (r2≥0.01) with other fetus-only SNPs (ESM Methods) [14]. Forty-three of the 49 remaining SNPs (ESM Table 2) were available in the GWAS dataset of LADA described below, explaining 0.996% of the variance in own birthweight.
Adult BMI
The primary set of IVs for BMI in adulthood was obtained from a meta-analysis including 681,275 individuals of European ancestry from the UK Biobank and the Genetic Investigation of Anthropometric Traits (GIANT) consortium [15]. Information on BMI had mainly been obtained through clinical measurements (ESM Table 1). SNPs of this meta-analysis have been used as IVs for BMI in a previous MR study [16]. In the meta-analysis, independent SNPs were selected through LD clumping, in which SNPs with LD measure of r2>0.01 within a 10,000 kb window were pruned, resulting in a total of 839 independent SNPs with p<1×10−8 (genome-wide significance threshold revised by authors of the meta-analysis). Of these, 817 SNPs were available in the GWAS of LADA and proxy SNPs in LD (r2>0.8) with three of the unavailable SNPs were identified using LDlink (accessed 12 March 2021) [17]. The 820 SNPs explained 7.43% of the variance in adult BMI (ESM Table 3).
The GIANT consortium had samples overlapping with the GWAS of type 2 diabetes (described below). Therefore, we selected a secondary set of IVs (734 independent SNPs) for adult BMI exclusively from the UK Biobank study (https://gwas.mrcieu.ac.uk/, accessed 3 April 2021) when comparing the BMI–LADA association with the BMI–type 2 diabetes association. The 734 SNPs explained 7.86% of the variance in adult BMI.
GWAS of LADA
LADA was the primary outcome in the present study. We obtained summary statistics for the association between the aforementioned SNPs and LADA from the only GWAS of LADA hitherto. This study included 2634 LADA cases and 5947 population controls, comprising individuals of European ancestry from Sweden, Denmark, Germany and the UK, and the analysis was adjusted for sex and principal components (to correct for potential bias due to population structure) [1]. LADA was defined based on the following criteria: (1) adult-onset (age at diagnosis >20, 30 or 35 years); (2) the presence of diabetes-associated autoimmune autoantibodies, in particular GAD autoantibody (GADA) positivity; and (3) lack of insulin requirement for 6 months or 1 year after diagnosis [1].
GWAS of type 2 diabetes
Summary statistics for the SNP–type 2 diabetes association were obtained from the DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium, which included 26,676 type 2 diabetes cases and 132,532 controls among individuals of European ancestry [18]. Summary statistics in this study were adjusted for age, sex, and principal components [18].
Data harmonisation
We checked the effect allele, reference allele and effect allele frequency in the GWAS datasets. The β coefficient for the SNP–exposure association was ‘flipped’ if the effect allele for the SNP–exposure association was the reference allele for the SNP–outcome association. We paid attention to palindromic (A/T or C/G) SNPs and there was no ambiguity in matching effect alleles for these SNPs between GWAS datasets of the exposure and the outcome.
Statistical analysis
We measured instrument strength of each SNP using F statistic [19], which equals the square of the β coefficient for the SNP–exposure association divided by the square of the SE for the β coefficient. A larger F statistic indicated stronger instrument strength [19]. Birthweight and adult BMI were all inverse-normally transformed and analysed in an additive model in the GWAS. Therefore, the risk estimates based directly on the summary statistics were ORs and 95% CIs for LADA and type 2 diabetes per SD change in exposures.
Main analysis
The inverse-variance weighted (IVW) method was used to assess the risk of LADA in relation to birthweight based on 43 IVs and adult BMI based on 820 primary IVs. The IVW method can be fitted in weighted linear regression [20, 21]. It provides a more precise risk estimate than other methods when all the IVs are valid [22].
Sensitivity analyses
Central assumptions in an MR study are that an IV only affects the outcome through the exposure, not through a direct pathway to the outcome or via a confounder (ESM Fig. 2) [23], otherwise there will be directional pleiotropy and the IV is invalid.
The IVW estimator used in the main analyses assumes that all the IVs are valid [24]. Several sensitivity analyses using other MR estimators were conducted to test the robustness of the results based on the IVW estimator. Some of them can detect potential directional pleiotropy, and some of them do not require all IVs to be valid. First, we used the robust IVW method, which replaces the standard linear regression in the IVW method with a robust regression [25]. This method has a greater power than IVW to reject causal null hypothesis when there is balanced pleiotropy (average pleiotropic effect: 0) and the instrument strength is independent of the instrument’s direct effect (InSIDE [26] assumption) [24]. Furthermore, we used the weighted median method, which provides consistent estimates when >50% of IVs are valid and does not rely on the InSIDE assumption [27]. Egger regression of MR (MR-Egger) can give a causal estimate under the InSIDE [26] assumption even when all the IVs are invalid, with the slope coefficient of the regression model representing the logarithmic OR [26]. This method indicates overall directional pleiotropy [26] if the estimated intercept term in the regression model is non-zero [26]. Finally, the MR pleiotropy residual sum and outlier (MR-PRESSO) approach estimator was used. The MR-PRESSO approach is based on the IVW method and detects outliers (potential pleiotropic SNPs), produces corrected ORs by removing outliers, and evaluates distortion of risk estimate by outliers [28].
In the present study, we further excluded outliers detected by MR-PRESSO from the IVs, and then re-analysed the data using all different MR estimators including the IVW estimator.
We did other sensitivity analyses based on the IVW method by excluding some SNPs from the 43 IVs for birthweight and 820 primary IVs for adult BMI. These sensitivity analyses included several conservative analyses to minimise the possibility that the IVs affect the outcome through a pathway outside the exposure, and a leave-one-out analysis to investigate whether the association in the main analysis would disappear by excluding any one of the IVs. In the analysis of birthweight, conservative analysis 1 excluded SNPs associated with diabetes-related traits at nominal significance level (Bonferroni-corrected), to minimise the possibility that IVs affect LADA directly. We further excluded SNPs associated with any trait (except birthweight) at genome-wide significance in conservative analysis 2. In the analysis of adult BMI, we excluded SNPs associated with diabetes-related traits at nominal significance level (Bonferroni-corrected) or any trait (except adult body size) at genome-wide significance (conservative analysis 1), SNPs associated with lifestyle factors at nominal significance level (Bonferroni-corrected, conservative analysis 2), and SNPs associated with birthweight at nominal significance level (Bonferroni-corrected, conservative analysis 3). Further details about conservative analyses are provided in ESM Fig. 3 and ESM Table 4.
We additionally did a sensitivity analysis by using summary statistics adjusted for maternal genotypes for birthweight based on the 43 fetus-only SNPs [14].
Comparison between LADA and type 2 diabetes
For comparison between LADA and type 2 diabetes, we used the same 43 IVs for birthweight and 734 secondary IVs for adult BMI in the analyses, to ensure the comparability of results. The associations of birthweight and adult BMI with type 2 diabetes were assessed using all the different MR estimators described above. Outlier-corrected ORs (95% CIs) from MR-PRESSO were used for comparison between LADA and type 2 diabetes if MR-PRESSO detected outliers, otherwise the results of IVW were used for comparison.
MR analysis was conducted using MendelianRandomization and MR-PRESSO package in R 4.0.4 [29]. All statistical tests were two-sided, with p<0.05 indicating statistical significance.
Ethical approval
No ethical permit was required as we used only GWAS summary statistics and no individual-level data.
Results
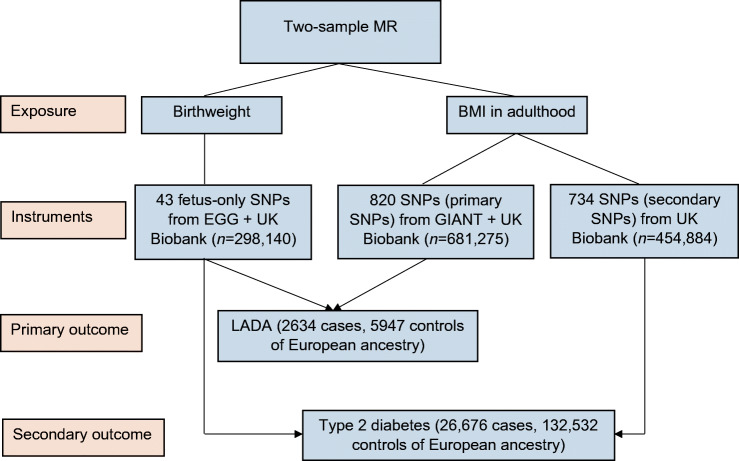
A flow chart of the study design is provided in Fig. 1. The SD of birthweight and BMI in adulthood was approximately 500 g and 4.8 kg/m2, respectively (ESM Table 1). F statistics for IVs were all above 10 in the present study (ESM Tables 2, 3).
Fig. 1.
Flow chart of the design of the present study
Birthweight and LADA
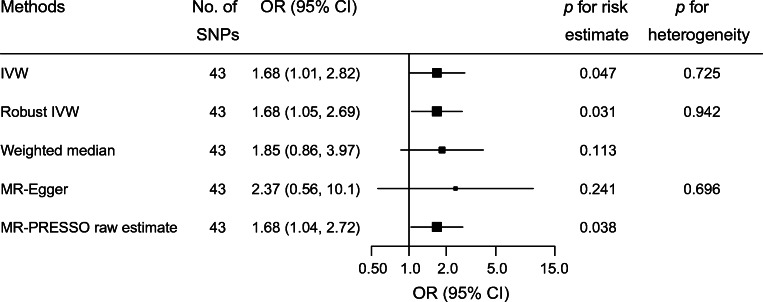
Genetically determined birthweight based on 43 IVs was inversely associated with LADA (ESM Fig. 4). In the main analysis, the OR of LADA was 1.68 (95% CI 1.01, 2.82) for each SD decrease in birthweight (Fig. 2).
Fig. 2.
The risk of LADA in relation to one SD (500 g) decrease in birthweight. MR-Egger intercept −0.009, p for directional pleiotropy = 0.618. MR-PRESSO detected no outliers (p for global test of pleiotropy = 0.736) and the raw estimate is presented
The inverse association between birthweight and LADA was also observed using other MR estimators (Fig. 2). MR-Egger indicated no directional pleiotropy (intercept −0.009, p for directional pleiotropy = 0.618). MR-PRESSO detected no outliers and the raw estimate was 1.68 (95% CI 1.04, 2.72).
The inverse association between birthweight and LADA was also observed in a series of conservative analyses by excluding SNPs associated with diabetes-related traits at p<0.05/43, and by further excluding SNPs associated with any other trait at p<5×10−8 (ESM Table 5). Leaving out one of the 43 SNPs each time did not change the direction of association, although some 95% CIs crossed 1.00 (ESM Fig. 5). Among the 43 SNPs, rs4144829 was in LD with a SNP acting through both fetal and maternal genome (rs2174633, not used as an IV in this study). The leave-one-out analysis showed that there was no major change in point estimate (1.67 [95% CI 0.99, 2.83]) after leaving rs4144829 out. The OR (1.58 [95% CI 0.96, 2.48]) estimated by using summary statistics adjusted for maternal genotypes was similar to that of the main analysis.
Adult BMI and LADA
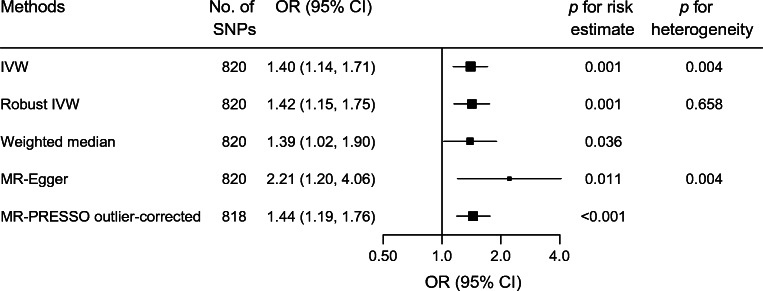
Genetically determined adult BMI based on the 820 primary IVs was positively associated with LADA (ESM Fig. 6). One SD increase in adult BMI was associated with an OR of 1.40 (95% CI 1.14, 1.71) for LADA using the IVW method (Fig. 3).
Fig. 3.
The risk of LADA in relation to one SD (4.8 kg/m2) increase in adult BMI. MR-PRESSO identified two outliers: rs11066188 and rs10840606. Outliers were excluded from the outlier-corrected estimate (p for distortion of estimate 0.777)
MR-Egger indicated no directional pleiotropy (intercept −0.007, p for directional pleiotropy = 0.119), while MR-PRESSO detected two outliers (rs11066188 and rs10840606). The outliers did not distort the results (p for distortion test = 0.777) and the outlier-corrected OR was similar to the OR obtained with IVW. Robust IVW and weighted median also showed results comparable with those estimated by IVW (Fig. 3). After excluding rs11066188 and rs10840606 from the IVs, no major change in results from different MR estimators was observed (ESM Fig. 7).
All the three conservative analyses (ESM Table 5) and the leave-one-out analysis (data not shown) showed similar results to the main analysis.
Comparison between LADA and type 2 diabetes
When assessing the association between birthweight and type 2 diabetes, MR-PRESSO detected three outliers and indicated no distortion by outliers (p for distortion = 0.533; ESM Table 6). After correction for outliers, the OR for type 2 diabetes in relation to one SD decrease in genetically determined birthweight was 1.64 (95% CI 1.32, 2.04), which was similar to the magnitude of the birthweight–LADA association (Table 1). The leave-one-out analysis (ESM Fig. 8) and the sensitivity analysis using summary statistics adjusted for maternal genotypes (OR 1.47 [95% CI 1.11, 1.95]) also showed an inverse association between birthweight and type 2 diabetes.
Table 1.
Comparison between LADA and type 2 diabetes
| Exposures | Outcomes | No. of initial SNPs | No. of outliersa | GWAS dataset for SNP–exposure association | Methods | OR (95% CI)b | p for risk estimate |
|---|---|---|---|---|---|---|---|
| Birthweight | LADA | 43 | 0 | EGG+UK Biobank | IVWc | 1.68 (1.01, 2.82) | 0.047 |
| Birthweight | Type 2 diabetes | 43 | 3 | EGG+UK Biobank | MR-PRESSO outlier-corrected | 1.64 (1.32, 2.04) | <0.001 |
| BMI in adulthood | LADA | 734 | 3 | UK Biobank | MR-PRESSO outlier-corrected | 1.42 (1.18, 1.72) | <0.001 |
| BMI in adulthood | Type 2 diabetes | 734 | 9 | UK Biobank | MR-PRESSO outlier-corrected | 2.33 (2.16, 2.51) | <0.001 |
aOutliers were excluded from the outlier-corrected estimates
bOR (95% CI) for LADA or type 2 diabetes per SD (500 g) decrease in birthweight or per SD (4.8 kg/m2) increase in BMI in adulthood
cMR-PRESSO detected no outliers and therefore the result of the IVW method was presented
MR-PRESSO detected nine outliers when assessing the association between adult BMI and type 2 diabetes based on the 734 secondary IVs (ESM Table 7). The OR of type 2 diabetes was 2.33 (95% CI 2.16, 2.51) for each SD increase in adult BMI after outlier removal (Table 1 and ESM Table 7). For LADA, the same IVs yielded an outlier-corrected OR of 1.42 (95% CI 1.18, 1.72) (Table 1 and ESM Table 7), which was similar to the association between LADA and adult BMI observed based on the 820 primary IVs.
Discussion
Main findings
This study provides genetic evidence to show that low birthweight and adult adiposity confer an increased risk of LADA. We also confirm findings of previous MR studies showing that lower birthweight [30–32] and higher BMI [33–40] in adulthood are associated with increased risk of type 2 diabetes. To the best of our knowledge, this is the first MR study to explore the role of environmental/lifestyle factors in the aetiology of LADA.
Main findings in relation to previous studies
The results regarding birthweight are in line with those of our previous observational study indicating a twofold increased risk of LADA in individuals with a birthweight <3 kg compared with ≥4 kg [9]. Notably, the association with birthweight was not weaker for LADA than for type 2 diabetes, also in line with previous observational data [9]. The mechanism linking birthweight to LADA and type 2 diabetes remains unclear. The Barker hypothesis proposes that an adverse intrauterine environment leads to both lower birthweight and higher risk of future cardiometabolic risk [41]. This hypothesis was not tested in our study since such analysis would require data on mother–child pairs for both the exposure and the outcome [41]. However, we restricted the IVs to SNPs with fetus-only effects; this implies that the inverse association is not explained by pleiotropy introduced by the intrauterine environment, although we cannot fully rule out the possibility that some of the fetus-only SNPs might be found to have both fetal and maternal components by larger GWAS in the future. Notably, results of our sensitivity analysis where we adjusted for maternal genotypes were also compatible with inverse associations between birthweight and LADA/type 2 diabetes. The fetal insulin hypothesis proposes that genetically determined insulin resistance in the fetus results in impaired insulin-mediated fetal growth as well as insulin resistance in adult life [42]. However, our findings do not support this hypothesis since the association between birthweight and LADA was not attenuated after excluding SNPs currently known to be associated with diabetes-related traits (including insulin resistance) at nominal significance level (Bonferroni-corrected). Our findings suggest that lower birthweight determined by own SNPs might have a causal effect on the risk of LADA. The results have public health implication in identifying individuals (those with lower birthweight) susceptible to LADA. These individuals might need to adopt a healthier lifestyle to alleviate the risk of LADA since our previous findings suggest that the combination of low birthweight and being overweight as an adult may be particularly detrimental [9]. However, we acknowledge that it is complicated to study birthweight as an exposure in an MR framework and the underlying mechanism for the suggested inverse association remains to be explored. With the accumulation of GWAS data, future MR studies based on mother–child pairs [41] are needed to gain deeper insights into the potential mechanisms linking perinatal factors to adult-onset diseases such as LADA.
Findings regarding LADA and adult BMI were also in line with previous observational data [8]. Overweight and obese status is strongly associated with development of insulin resistance [43] and this may explain a causal link between adult adiposity and LADA. In support hereof, a positive association between BMI and insulin resistance was observed in individuals with LADA [8]. The association with BMI was stronger for type 2 diabetes than LADA, which is in line with previous findings [8]. This is to be expected since insulin resistance tends to be less pronounced in LADA compared with type 2 diabetes [2, 8, 44]. A previous MR study also found support for a link between childhood adiposity and type 1 diabetes [45]. This implies that overweight/obese status is implicated in the promotion of all major types of diabetes and emphasises that it is crucial to prevent overweight status in order to reduce the incidence of diabetes.
Assessment of MR assumptions
A major concern in MR studies is the violation of IV assumptions. These assumptions cannot be fully tested, although we used several approaches to minimise this potential bias. First, we applied different MR estimators, some of which detected and corrected for potential directional pleiotropy from a statistical perspective. There was no major change in ORs after excluding outliers, indicating the robustness of the results. Second, we excluded some SNPs in several conservative analyses, in which the associations of birthweight and adult BMI with LADA persisted. The risk of LADA is linked to genes in the HLA region, primarily susceptibility within HLA-DRB1 and HLA-DQB1 haplotypes [1, 46, 47]. Among the 43 SNPs for birthweight, no SNPs are located within 300 kb windows of HLA-related genes [14]. It should be noted that excluding SNPs in the conservative analyses does not prove that they are in fact invalid SNPs.
Strengths and limitations
There are several strengths in the present study. First, the two-sample MR design using genetic variants as unbiased proxy minimises confounding and reverse causation. Second, the application of different MR estimators and a series of conservative analyses reduces the risk of bias caused by directional pleiotropy. Third, we confirmed findings from previous studies on type 2 diabetes and used type 2 diabetes as a ‘positive control’ to show that the instruments and methods used for LADA in the present study are reliable. Further, this provides a good opportunity to compare the aetiology of LADA and type 2 diabetes. There are also some limitations. First, the lack of individual data rules out the possibility of exploring potential non-linear association between exposures and outcomes; this is a common limitation in MR studies based on summary statistics. The linear assumption is less likely to be violated in our MR study. For LADA, there is no evidence with sufficient statistical power to support a non-linear association [8, 9]. Adult BMI seemed to be linearly (positively) associated with type 2 diabetes [35, 48]. Observational studies found that the linear (inverse) association between birthweight and type 2 diabetes held when birthweight was <4.0 kg [49] or <4.5 kg [50]. The range of birthweight (study-specific, 2.5–4.5 kg, or within the range of mean ± 5SD) in the GWAS [14] used by the present MR analysis is generally in the range of linear association. Moreover, deviation from the linear assumption is likely to reduce the statistical power in risk estimate, rather than generating spurious associations [51]. Second, findings from the present study are only applicable to the European population since the only GWAS on LADA was conducted in individuals of European ancestry. It is unclear to what extent the findings are generalisable to other populations.
Conclusions
These findings provide genetic support for a causal link between low birthweight, adult overweight/obese status and LADA. The results persisted in a series of sensitivity analyses. Measures should be taken to reduce the prevalence of adult overweight/obese status for the prevention of diabetes with and without an autoimmune component. The mechanism linking low birthweight to diabetes remains to be explored.
Supplementary information
(PDF 1.97 mb)
Acknowledgments
Authors’ relationships and activities
The authors declare that there are no relationships or activities that might bias, or be perceived to bias, their work.
Contribution statement
YW and SC conceived and designed the study. YW collected the summary data, analysed data and wrote the first draft of the manuscript. YZ contributed to methodological issues. All authors critically revised the manuscript for important intellectual content and made substantial contributions to the interpretation of data. All authors reviewed and approved the final manuscript. YW is the guarantor of this work.
Abbreviations
- DIAGRAM
DIAbetes Genetics Replication And Meta-analysis Consortium
- EGG
Early Growth Genetics Consortium
- GIANT
Genetic Investigation of Anthropometric Traits Consortium
- GWAS
Genome-wide association studies
- InSIDE
Instrument Strength Independent of Direct Effect
- IV
Instrumental variable
- IVW
Inverse-variance weighted
- LADA
Latent autoimmune diabetes in adults
- LADY
Latent autoimmune diabetes in the young
- MR
Mendelian randomisation
- MR-Egger
Egger regression of MR
- MR-PRESSO
MR pleiotropy residual sum and outlier approach
Funding
Open access funding is provided by the Karolinska Institute. The study was supported by the Swedish Research Council (2018–03035), Research Council for Health, Working Life and Welfare (FORTE, 2018–00337) and Novo Nordisk Foundation (NNF19OC0057274). YW received a scholarship from the China Scholarship Council (student no. 202006010041). The sponsors had no role in the study design, data collection, data analysis and interpretation, writing of the report or the decision to submit the article for publication.
Data availability
This study only used summary data and these data are publicly available.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yuxia Wei, Email: yuxia.wei@ki.se.
Yiqiang Zhan, Email: zhanyq8@mail.sysu.edu.cn.
References
- 1.Cousminer DL, Ahlqvist E, Mishra R, et al. First genome-wide association study of latent autoimmune diabetes in adults reveals novel insights linking immune and metabolic diabetes. Diabetes Care. 2018;41(11):2396–2403. doi: 10.2337/dc18-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tuomi T, Santoro N, Caprio S, Cai M, Weng J, Groop L. The many faces of diabetes: a disease with increasing heterogeneity. Lancet. 2014;383(9922):1084–1094. doi: 10.1016/S0140-6736(13)62219-9. [DOI] [PubMed] [Google Scholar]
- 3.Bering B, Devendra D. Latent autoimmune diabetes in the young. Clin Med (Lond) 2009;9(1):93. doi: 10.7861/clinmedicine.9-1-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yuan S, Larsson SC. An atlas on risk factors for type 2 diabetes: a wide-angled Mendelian randomisation study. Diabetologia. 2020;63(11):2359–2371. doi: 10.1007/s00125-020-05253-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bellou V, Belbasis L, Tzoulaki I, Evangelou E. Risk factors for type 2 diabetes mellitus: an exposure-wide umbrella review of meta-analyses. PLoS One. 2018;13(3):e0194127. doi: 10.1371/journal.pone.0194127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tuomilehto J, Lindström J, Eriksson JG, et al. Prevention of type 2 diabetes mellitus by changes in lifestyle among subjects with impaired glucose tolerance. N Engl J Med. 2001;344(18):1343–1350. doi: 10.1056/nejm200105033441801. [DOI] [PubMed] [Google Scholar]
- 7.Carlsson S. Etiology and pathogenesis of latent autoimmune diabetes in adults (LADA) compared to type 2 diabetes. Front Physiol. 2019;10:320. doi: 10.3389/fphys.2019.00320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hjort R, Ahlqvist E, Carlsson PO, et al. Overweight, obesity and the risk of LADA: results from a Swedish case-control study and the Norwegian HUNT study. Diabetologia. 2018;61(6):1333–1343. doi: 10.1007/s00125-018-4596-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hjort R, Alfredsson L, Carlsson PO, et al. Low birthweight is associated with an increased risk of LADA and type 2 diabetes: results from a Swedish case-control study. Diabetologia. 2015;58(11):2525–2532. doi: 10.1007/s00125-015-3711-8. [DOI] [PubMed] [Google Scholar]
- 10.Li Y, Ley SH, Tobias DK, et al. Birth weight and later life adherence to unhealthy lifestyles in predicting type 2 diabetes: prospective cohort study. BMJ. 2015;351:h3672. doi: 10.1136/bmj.h3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abdullah A, Peeters A, de Courten M, Stoelwinder J. The magnitude of association between overweight and obesity and the risk of diabetes: a meta-analysis of prospective cohort studies. Diabetes Res Clin Pract. 2010;89(3):309–319. doi: 10.1016/j.diabres.2010.04.012. [DOI] [PubMed] [Google Scholar]
- 12.Lawlor DA, Harbord RM, Sterne JA, Timpson N, Davey Smith G. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat Med. 2008;27(8):1133–1163. doi: 10.1002/sim.3034. [DOI] [PubMed] [Google Scholar]
- 13.Burgess S, Scott RA, Timpson NJ, Davey Smith G, Thompson SG. Using published data in Mendelian randomization: a blueprint for efficient identification of causal risk factors. Eur J Epidemiol. 2015;30(7):543–552. doi: 10.1007/s10654-015-0011-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Warrington NM, Beaumont RN, Horikoshi M, et al. Maternal and fetal genetic effects on birth weight and their relevance to cardio-metabolic risk factors. Nat Genet. 2019;51(5):804–814. doi: 10.1038/s41588-019-0403-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yengo L, Sidorenko J, Kemper KE, et al. Meta-analysis of genome-wide association studies for height and body mass index in approximately 700000 individuals of European ancestry. Hum Mol Genet. 2018;27(20):3641–3649. doi: 10.1093/hmg/ddy271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou W, Liu G, Hung RJ, et al. Causal relationships between body mass index, smoking and lung cancer: Univariable and multivariable Mendelian randomization. Int J Cancer. 2021;148(5):1077–1086. doi: 10.1002/ijc.33292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Machiela MJ, Chanock SJ. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics. 2015;31(21):3555–3557. doi: 10.1093/bioinformatics/btv402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scott RA, Scott LJ, Mägi R, et al. An expanded genome-wide association study of type 2 diabetes in Europeans. Diabetes. 2017;66(11):2888–2902. doi: 10.2337/db16-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burgess S, Thompson SG. Mendelian randomization: methods for using genetic variants in causal estimation. Florida, USA: CRC Press; 2015. [Google Scholar]
- 20.Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol. 2013;37(7):658–665. doi: 10.1002/gepi.21758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Slob EAW, Burgess S. A comparison of robust Mendelian randomization methods using summary data. Genet Epidemiol. 2020;44(4):313–329. doi: 10.1002/gepi.22295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burgess S, Foley CN, Allara E, Staley JR, Howson JMM. A robust and efficient method for Mendelian randomization with hundreds of genetic variants. Nat Commun. 2020;11(1):376. doi: 10.1038/s41467-019-14156-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burgess S, Bowden J, Fall T, Ingelsson E, Thompson SG. Sensitivity analyses for robust causal inference from Mendelian randomization analyses with multiple genetic variants. Epidemiology. 2017;28(1):30–42. doi: 10.1097/ede.0000000000000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burgess S, Bowden J, Dudbridge F, Thompson SG (2016) Robust instrumental variable methods using multiple candidate instruments with application to Mendelian randomization. arXiv: 1606.03729
- 25.Koller M, Stahel WA. Sharpening Wald-type inference in robust regression for small samples. Comput Stat Data Anal. 2011;55(8):2504–2515. doi: 10.1016/j.csda.2011.02.014. [DOI] [Google Scholar]
- 26.Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through egger regression. Int J Epidemiol. 2015;44(2):512–525. doi: 10.1093/ije/dyv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. 2016;40(4):304–314. doi: 10.1002/gepi.21965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Verbanck M, Chen CY, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. 2018;50(5):693–698. doi: 10.1038/s41588-018-0099-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.R Core Team (2021). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL https://www.R-project.org/
- 30.Huang T, Wang T, Zheng Y, et al. Association of birth weight with type 2 diabetes and glycemic traits: a Mendelian randomization study. JAMA Netw Open. 2019;2(9):e1910915. doi: 10.1001/jamanetworkopen.2019.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zanetti D, Tikkanen E, Gustafsson S, Priest JR, Burgess S, Ingelsson E. Birthweight, type 2 diabetes mellitus, and cardiovascular disease: addressing the barker hypothesis with Mendelian randomization. Circ Genom Precis Med. 2018;11(6):e002054. doi: 10.1161/circgen.117.002054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang T, Huang T, Li Y, et al. Low birthweight and risk of type 2 diabetes: a Mendelian randomisation study. Diabetologia. 2016;59(9):1920–1927. doi: 10.1007/s00125-016-4019-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Richardson TG, Sanderson E, Elsworth B, Tilling K, Davey Smith G. Use of genetic variation to separate the effects of early and later life adiposity on disease risk: mendelian randomisation study. BMJ. 2020;369:m1203. doi: 10.1136/bmj.m1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Q, Pan J, Berzuini C, Rutter MK, Guo H. Integrative analysis of Mendelian randomization and Bayesian colocalization highlights four genes with putative BMI-mediated causal pathways to diabetes. Sci Rep. 2020;10(1):7476. doi: 10.1038/s41598-020-64493-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wainberg M, Mahajan A, Kundaje A, et al. Homogeneity in the association of body mass index with type 2 diabetes across the UK biobank: a Mendelian randomization study. PLoS Med. 2019;16(12):e1002982. doi: 10.1371/journal.pmed.1002982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shu X, Wu L, Khankari NK, et al. Associations of obesity and circulating insulin and glucose with breast cancer risk: a Mendelian randomization analysis. Int J Epidemiol. 2019;48(3):795–806. doi: 10.1093/ije/dyy201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu Z, Zheng Z, Zhang F, et al. Causal associations between risk factors and common diseases inferred from GWAS summary data. Nat Commun. 2018;9(1):224. doi: 10.1038/s41467-017-02317-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lyall DM, Celis-Morales C, Ward J, et al. Association of body mass index with cardiometabolic disease in the UK biobank: a Mendelian randomization study. JAMA Cardiol. 2017;2(8):882–889. doi: 10.1001/jamacardio.2016.5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Corbin LJ, Richmond RC, Wade KH, et al. BMI as a modifiable risk factor for type 2 diabetes: refining and understanding causal estimates using Mendelian randomization. Diabetes. 2016;65(10):3002–3007. doi: 10.2337/db16-0418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Holmes MV, Lange LA, Palmer T, et al. Causal effects of body mass index on cardiometabolic traits and events: a Mendelian randomization analysis. Am J Hum Genet. 2014;94(2):198–208. doi: 10.1016/j.ajhg.2013.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.D'Urso S, Wang G, Hwang LD, Moen GH, Warrington NM, Evans DM. A cautionary note on using Mendelian randomization to examine the barker hypothesis and developmental origins of health and disease (DOHaD) J Dev Orig Health Dis. 2021;12(5):688–693. doi: 10.1017/s2040174420001105. [DOI] [PubMed] [Google Scholar]
- 42.Hattersley AT, Tooke JE. The fetal insulin hypothesis: an alternative explanation of the association of low birthweight with diabetes and vascular disease. Lancet. 1999;353(9166):1789–1792. doi: 10.1016/s0140-6736(98)07546-1. [DOI] [PubMed] [Google Scholar]
- 43.Barazzoni R, Gortan Cappellari G, Ragni M, Nisoli E. Insulin resistance in obesity: an overview of fundamental alterations. Eat Weight Disord. 2018;23(2):149–157. doi: 10.1007/s40519-018-0481-6. [DOI] [PubMed] [Google Scholar]
- 44.Maddaloni E, Coleman RL, Agbaje O, Buzzetti R, Holman RR. Time-varying risk of microvascular complications in latent autoimmune diabetes of adulthood compared with type 2 diabetes in adults: a post-hoc analysis of the UK prospective diabetes study 30-year follow-up data (UKPDS 86) Lancet Diabetes Endocrinol. 2020;8(3):206–215. doi: 10.1016/s2213-8587(20)30003-6. [DOI] [PubMed] [Google Scholar]
- 45.Censin JC, Nowak C, Cooper N, Bergsten P, Todd JA, Fall T. Childhood adiposity and risk of type 1 diabetes: a Mendelian randomization study. PLoS Med. 2017;14(8):e1002362. doi: 10.1371/journal.pmed.1002362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mishra R, Åkerlund M, Cousminer DL, et al. Genetic discrimination between LADA and childhood-onset type 1 diabetes within the MHC. Diabetes Care. 2020;43(2):418–425. doi: 10.2337/dc19-0986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Andersen MK. New insights into the genetics of latent autoimmune diabetes in adults. Curr Diab Rep. 2020;20(9):43. doi: 10.1007/s11892-020-01330-y. [DOI] [PubMed] [Google Scholar]
- 48.Narayan KM, Boyle JP, Thompson TJ, Gregg EW, Williamson DF. Effect of BMI on lifetime risk for diabetes in the U.S. Diabetes Care. 2007;30(6):1562–1566. doi: 10.2337/dc06-2544. [DOI] [PubMed] [Google Scholar]
- 49.Harder T, Rodekamp E, Schellong K, Dudenhausen JW, Plagemann A. Birth weight and subsequent risk of type 2 diabetes: a meta-analysis. Am J Epidemiol. 2007;165(8):849–857. doi: 10.1093/aje/kwk071. [DOI] [PubMed] [Google Scholar]
- 50.Knop MR, Geng TT, Gorny AW, et al. Birth weight and risk of type 2 diabetes mellitus, cardiovascular disease, and hypertension in adults: a meta-analysis of 7 646 267 participants from 135 studies. J Am Heart Assoc. 2018;7(23):e008870. doi: 10.1161/jaha.118.008870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carreras-Torres R, Johansson M, Gaborieau V, et al. The role of obesity, type 2 diabetes, and metabolic factors in pancreatic Cancer: a Mendelian randomization study. J Natl Cancer Inst. 2017;109(9):djx012. doi: 10.1093/jnci/djx012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 1.97 mb)
Data Availability Statement
This study only used summary data and these data are publicly available.