Abstract
The circadian gene Timeless (TIM) provides a molecular bridge between circadian and cell cycle/DNA replication regulatory systems and has been recently involved in human cancer development and progression. However, its functional role in colorectal cancer (CRC), the third leading cause of cancer-related deaths worldwide, has not been fully clarified yet. Here, the analysis of two independent CRC patient cohorts (total 1159 samples) reveals that loss of TIM expression is an unfavorable prognostic factor significantly correlated with advanced tumor stage, metastatic spreading, and microsatellite stability status. Genome-wide expression profiling, in vitro and in vivo experiments, revealed that TIM knockdown induces the activation of the epithelial-to-mesenchymal transition (EMT) program. Accordingly, the analysis of a large set of human samples showed that TIM expression inversely correlated with a previously established gene signature of canonical EMT markers (EMT score), and its ectopic silencing promotes migration, invasion, and acquisition of stem-like phenotype in CRC cells. Mechanistically, we found that loss of TIM expression unleashes ZEB1 expression that in turn drives the EMT program and enhances the aggressive behavior of CRC cells. Besides, the deranged TIM-ZEB1 axis sets off the accumulation of DNA damage and delays DNA damage recovery. Furthermore, we show that the aggressive and genetically unstable ‘CMS4 colorectal cancer molecular subtype’ is characterized by a lower expression of TIM and that patients with the combination of low-TIM/high-ZEB1 expression have a poorer outcome. In conclusion, our results as a whole suggest the engagement of an unedited TIM-ZEB1 axis in key pathological processes driving malignant phenotype acquisition in colorectal carcinogenesis. Thus, TIM-ZEB1 expression profiling could provide a robust prognostic biomarker in CRC patients, supporting targeted therapeutic strategies with better treatment selection and patients’ outcomes.
Subject terms: Metastasis, Prognostic markers
Introduction
Colorectal cancer (CRC) is the third most common cancer worldwide [1]. Aggressive cancer frequently shows a switch to a mesenchymal phenotype, which drives tumor progression and acquisition of resistance to chemo/radiotherapy, including new targeted agents [2]. The pathological mechanisms that drive EMT in CRC have not yet been fully characterized, which hampers the identification of successful treatment protocols for refractory cancer. In such a scenario, we found that TIMELESS (TIM), a highly conserved gene, is linked to EMT in CRC. TIM in Drosophila melanogaster is a core circadian gene [3], while in human cells it takes part in the replication fork protection complex (FPC), a multiprotein complex that preserves fork integrity and genome stability during DNA replication [4–7]. Augmented levels of TIM and CLASPIN (a checkpoint adapter and DNA replication factor) were frequently observed and functionally linked to the protection of tumor cells from oncogene-induced replicative stress in a checkpoint-independent manner [8]. Recently, PARP1 was also found to physically interact with TIM in a TIPIN (i.e., TIMELESS-interacting protein) independent manner, which in turn allows PARP1 interaction with several proteins involved in DNA damage response (DDR), promoting its activation [9, 10].
TIM overexpression was associated with a pro-tumorigenic role in different human cancer types and identified as an adverse prognostic biomarker in breast cancer, cervical carcinoma, and lung cancer [11–16]. However, contrasting results were reported in breast cancer where the loss of TIM expression was associated with increased cell migration and invasion [17]. Further evidence has also suggested that loss of TIM expression may contribute to cancer development by impairing proper replication fork progression and DDR [8].
Here, we report unprecedented findings showing that loss of TIM expression in CRC favors tumor progression underpinned by induction of epithelial-to-mesenchymal transition (EMT). Furthermore, we provide evidence of an unedited TIM-ZEB1 axis in CRC pathogenesis, with several implications in the acquisition of aggressive phenotypes adversely impacting CRC prognosis. Strikingly, the loss of TIM expression occurs in a sizable fraction of human cancer types thus suggesting a “double-hedged sword” role for TIM in cancer.
Results
Pan-cancer analysis of TIM expression reveals prognostic significance in multiple tumor types including CRC
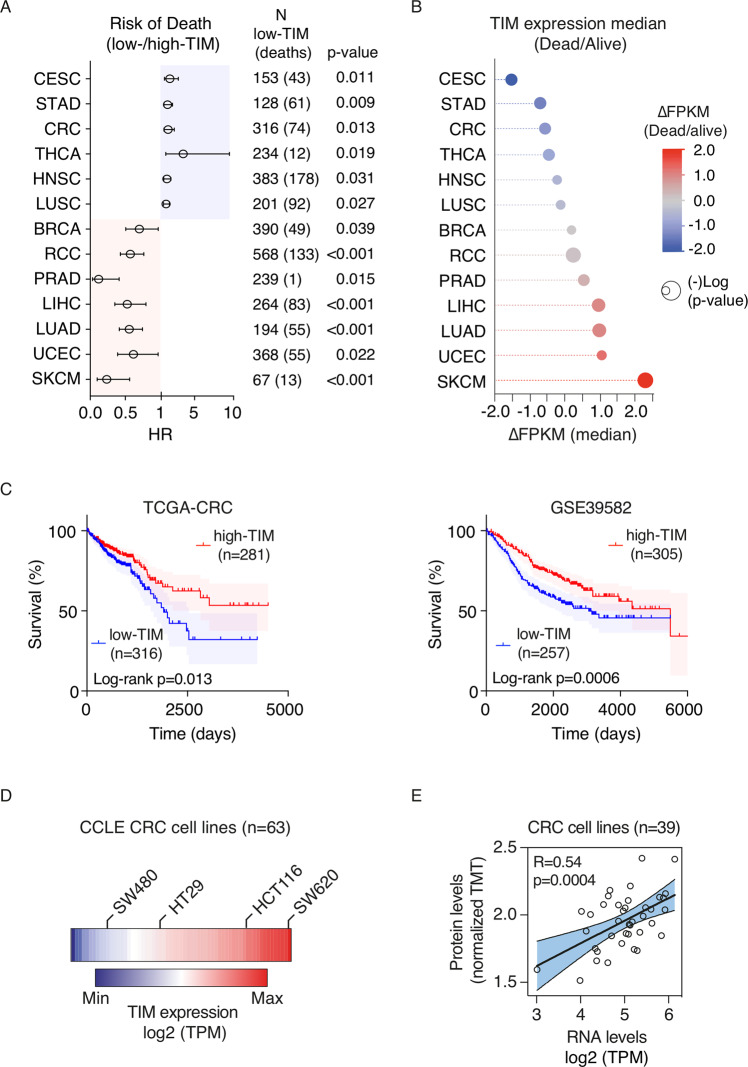
We initially investigated the pattern of TIM expression in human cancer by pan-cancer analysis using the “Human Protein Atlas” database (HPA) [18], which contains expression data of protein-coding genes from multiple tumor types (N = 21). Tumor samples were stratified into low-TIM and high-TIM expression on the basis of HPA “best-expression cut-off” (see Methods). Overall, TIM expression was prognostic in ~62% of different tumor types and subtypes (13 out of 21) (Fig. 1A and Fig. S1A, B), with ~50% of the tumor types (7 out of 13; Fig. 1A, B) showing TIM overexpression as an adverse prognostic factor while, unexpectedly, in the remaining half (6 out of 13), loss of TIM expression correlated with poor prognosis (Fig. 1A, B). In particular, loss of TIM expression was an unfavorable prognostic factor in CRC (HR = 1.559; 95% CI: 1.096–2.216; p = 0.013), in cervical squamous cell carcinoma and endocervical adenocarcinoma (HR = 1.832; 95% CI: 1.149–2.92; p = 0.011), stomach adenocarcinoma (HR = 1.539; 95% CI: 1.086–2.180; p = 0.009), thyroid carcinoma (HR = 3.541; 95% CI: 1.325–9.468; p = 0.019), head and neck squamous cell carcinoma (HR = 1.454; 95% CI: 1.068–1.979; p = 0.031) and lung squamous cell carcinoma (LUSC) (HR = 1.356; CI: 1.025–1.793; p = 0.027) (Fig. 1A, B). Furthermore, we confirmed the unfavorable prognostic value of loss of TIM expression in an additional independent cohort of 562 CRC patients (GSE39582 cohort; Fig. 1C). Lastly, loss of TIM expression correlated with advanced tumor stages, metastatic spreading (loco-regional and distant), and microsatellite stability status in CRC (Table 1).
Fig. 1. Pan-cancer screening on TIM expression reveals prognostic significance in multiple tumor types.
A Forest plot analysis of the risk of death (i.e., hazard ratios; HR) of patients with low-TIM vs. high-TIM expressing cancer. Circles represent the HR values while horizontal bars mean 95% confidence interval of HR. p-Values were calculated by log-rank test. B Bubble plot of the significant (p < 0.05) differential TIM median expression (fragments per kilobase of transcript per million mapped reads, ∆FPKM) in tumors of patients associated with an adverse prognosis (Dead) or favorable prognosis (Alive). Gene expression data were derived from the “HPA database” and were relative to TCGA cancer samples. Bubble colors are as per the legend. Bubble size represents statistical significance expressed as −Log “p-value” (log-rank test), where the larger the size the greater the significance. BRCA breast invasive carcinoma, CRC colorectal carcinoma, CESC cervical squamous cell carcinoma and endocervical adenocarcinoma, LIHC liver hepatocellular carcinoma, LUAD lung adenocarcinoma, LUSC lung squamous cell carcinoma, HNSC head, and neck squamous cell carcinoma, PRAD prostate adenocarcinoma, RCC renal cell carcinoma, STAD stomach adenocarcinoma, SKCM skin cutaneous melanoma, THCA thyroid carcinoma, UCEC uterine corpus endometrial carcinoma. C Kaplan–Meier survival curves of patients of two independent colorectal cancer cohorts [TCGA-CRC (n = 597) and GSE39582 (n = 562)] stratified based on high- or low-TIM expression (see methods). p-Values were calculated by log-rank test. D Heatmap of TIM expression profile in 63 CRC cell lines (CCLE dataset) ordered according to the TIM expression level (low to high) as per the legend. The labeled cell lines were the ones used in this study. TPM transcripts per million. E Scatter plot analysis showing TIM expression correlation in 39 colorectal cell lines where mRNA and protein expression data were available. Total protein level was measured by isobaric peptide labeling (TMT tandem mass tag) and MS3 quantification, while mRNA refers to RNA-seq expression quantification. TPM transcripts per million. p Values were determined by two-sided Pearson’s correlation test.
Table 1.
Clinicopathologic characteristics of TCGA-CRC cohort (N = 592) and GSE39582 cohort (N = 566), according to TIM level.
| TCGA-CRC cohort N = 592 | High-TIM N = 292 (49.3%) | Low-TIM N = 300 (50.7%) | p-Value | GSE39582 N = 566 | High-TIM N = 306 (54.1%) | Low-TIM N = 260 (45.9%) | p-Value | |
|---|---|---|---|---|---|---|---|---|
| Age at diagnosis [yrs] | ||||||||
| Median (Q1; Q3) | 68 (58; 76)a | 68 (57; 77)a | 67 (58; 75) | 0.57b | 68 (59; 76)d | 69 (58; 77) | 68 (60; 75)d | 0.72b |
| Min–max | 31–90 | 33–90 | 31–90 | 22–97 | 24–97 | 22–96 | ||
| Sex | ||||||||
| Male | 311 (52.5%) | 146 (50.0%) | 165 (55.0%) | 0.17c | 310 (54.8%) | 158 (51.6%) | 152 (58.5%) | 0.11c |
| Female | 279 (47.1%) | 144 (49.3%) | 135 (45.0%) | 256 (45.2%) | 148 (48.4%) | 108 (41.5%) | ||
| NA | 2 (0.3%) | 2 (0.7%) | 0 | – | – | – | ||
| Stage | ||||||||
| 0 | – | – | – | 4 (0.7%) | 1 (0.3%) | 3 (1.2%) | 0.0246c | |
| I | 103 (17.4%) | 52 (17.8%) | 51 (17.0%) | 0.08c | 33 (5.8%) | 19 (6.2%) | 14 (5.4%) | |
| II | 220 (37.2%) | 119 (40.8%) | 101 (33.7%) | 264 (46.6%) | 158 (51.6%) | 106 (40.8%) | ||
| III | 170 (28.7%) | 80 (27.4%) | 90 (30.0%) | 205 (36.2%) | 104 (34.0%) | 101 (38.8%) | ||
| IV | 85 (14.4%) | 32 (11.0%) | 53 (17.7%) | 60 (10.6%) | 24 (7.8%) | 36 (13.8%) | ||
| NA | 14 (2.4%) | 9 (3.1%) | 5 (1.7%) | – | – | – | ||
| Pathological lymph node status | ||||||||
| N0 | 340 (57.4%) | 182 (62.3%) | 158 (52.7%) | 0.0241c | 302 (53.4%) | 179 (58.5%) | 123 (47.3%) | 0.0208c |
| N+ | 249 (42.1%) | 108 (37.0%) | 141 (47.0%) | 244 (43.1%) | 119 (38.9%) | 125 (48.1%) | ||
| NX/NA | 3 (0.5%) | 2 (0.7%) | 1 (0.3%) | 20 (3.5%) | 8 (2.6%) | 12 (4.6%) | ||
| Pathological M status | ||||||||
| M0 | 439 (74.2%) | 222 (76.0%) | 217 (72.3%) | 0.06c | 482 (85.2%) | 272 (88.9%) | 210 (80.8%) | 0.0263c |
| M1 | 82 (13.9%) | 31 (10.6%) | 51 (17.0%) | 61 (10.8%) | 24 (7.8%) | 37 (14.2%) | ||
| MX/NA | 71 (12.0%) | 39 (13.4%) | 32 (10.7%) | 23 (4.1%) | 10 (3.3%) | 13 (5.0%) | ||
| Microsatellite status | ||||||||
| Instable | 177 (29.9%) | 106 (36.3%) | 71 (23.7%) | 0.0015c | 75 (13.3%) | 59 (19.3%) | 16 (6.2%) | <0.0001c |
| Stable | 410 (69.3%) | 183 (62.7%) | 227 (75.7%) | 444 (78.4%) | 215 (70.3%) | 229 (88.1%) | ||
| NA | 5 (0.8%) | 3 (1.0%) | 2 (0.7%) | 47 (8.3%) | 32 (10.5%) | 15 (5.8%) | ||
| Follow-up | N = 589 | N = 289 | N = 300 | N = 562 | N = 305 | N = 257 | ||
| Deaths | 120 (20.4%) | 51 (17.6%) | 69 (23.0%) | 191 (34.0%) | 87 (28.5%) | 104 (40.5%) | ||
| Survivors | 469 (79.6%) | 238 (82.4%) | 231 (77.0%) | 371 (66.0%) | 218 (71.5%) | 153 (59.5%) | ||
| Median (Q1; Q3) [yrs] in survivors | 1.8 (1.1–3.0) | 1.8 (1.1–3.0) | 2.0 (1.1–2.9) | 0.45b | 5.3 (3.4–7.1) | 5.2 (3.4–7.3) | 5.5 (2.9–7.1) | 0.79b |
Percentages could not add up 100 due to rounding.
aThree patients with missing information on age.
bWilcoxon test.
cFisher’s exact test.
dOne patient with missing information on age.
Loss of TIM expression impacts relevant cancer-related pathways
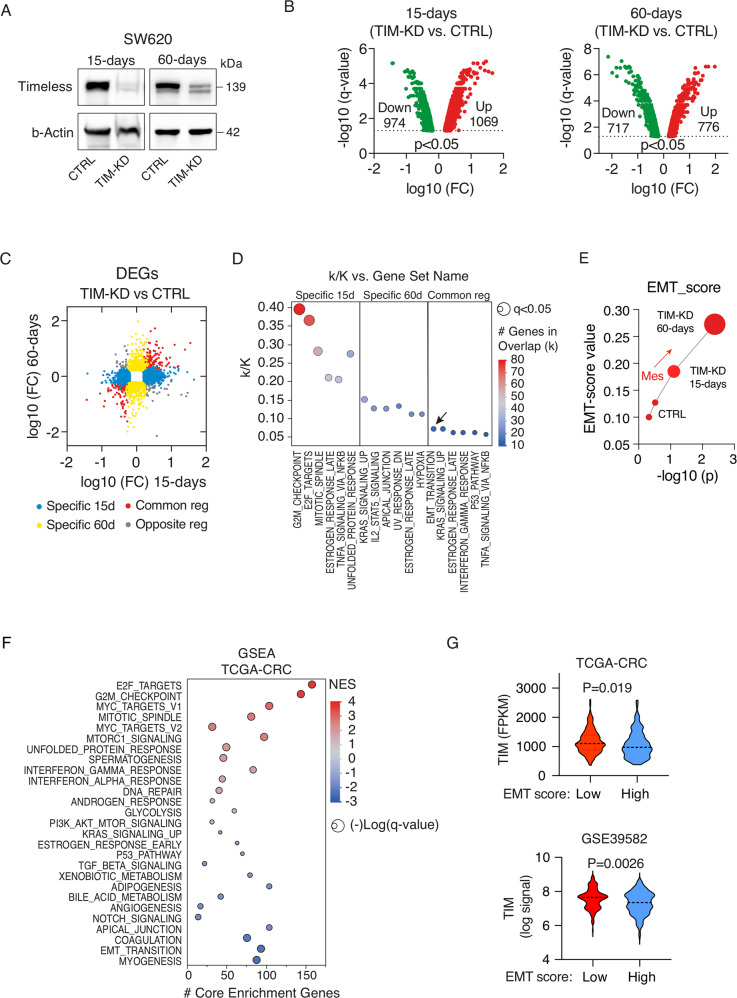
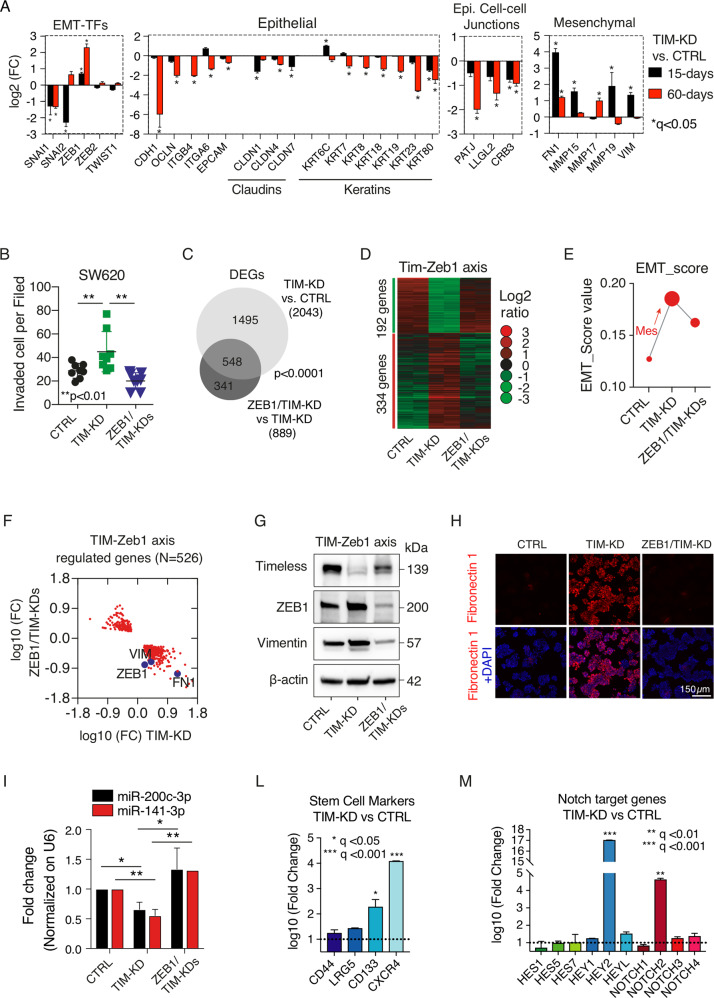
While TIM overexpression protects cancer cells from DNA replication stress and promotes proliferation [19], the link between loss of TIM expression and tumor progression lacks at present a specific mechanistic explanation. To tackle this issue, we started by analyzing the TIM expression profile in a panel of 63 CRC cell lines belonging to the cancer cell line encyclopedia (Table S1) [20]. TIM showed a wide range of expressions in these cell lines (Fig. 1D; Table S1) and, in particular, among commonly used CRC cell lines (HCT116, HT29, SW480, and SW620), which we then employed in our experimental setting (Fig. S2A–C). Notably, a positive and significant correlation was found between TIM mRNA and protein expression (R = 0.54, p = 0.0004; Fig. 1E; Table S1). We thus initially selected the SW620 (high-TIM expressing) cells to stably knockdown TIM expression using different sh-RNAs (TIM-KD cells) or an empty vector (CTRL cells) (Fig. S2D, E; see Methods). Next, we run genome-wide expression profiling of CRC cells at 15-days and at 60-days post knockdown of TIM expression (Fig. 2A; Fig. S2F) to monitor middle- (15-days) and long-term (60-days) transcriptional changes upon TIM-KD, which more resembles actual pathological conditions in CRC and is not limited only to an acute-phase KD as previously performed [8, 9, 11, 13, 15, 19]. A total of 2043 differentially expressed genes (DEG) (q-value < 0.05; log-FC > |1.5|) were found in the TIM-KD_15-days condition, while 1493 DEGs were identified in the TIM-KD_60-days condition (Fig. 2B). A total of 425 DEGs were also found overlapping in both conditions (Fig. S2G), of which 334 (~79%) were coherently regulated in TIM-KD_15-days and TIM-KD-60_days conditions (Fig. 2C, red dots; Table S2). Next, we performed Molecular Signatures Database (MSigDB) analysis (see Methods) using the set of 1618 DEGs specifically regulated in TIM-KD_15-days condition (Fig. 2C, blue dots; Fig. S2G) which revealed enrichment of the “G2M checkpoint” and “E2F targets” gene sets (q-value < 0.05) (Fig. 2D; Table S3). Yet, MSigDB analysis using the set of 1068 DEGs specifically regulated in TIM-KD_60-days (Fig. 2C, yellow dots; Fig. S2G) resulted in “KRAS signaling” and “IL2-STAT5 signaling” (relevant for metabolic reprogramming and cancer progression [21]) among the top enriched (q-value < 0.05) gene sets (Fig. 2D; Table S3). Lastly, the analysis of the set of coherently regulated 334 DEGs (Fig. 2C, red dots) revealed that the “EMT program” was the most significantly represented among coherently regulated genes in both TIM-KD_15-days and 60-days conditions (Fig. 2D; Table S3). Recent studies demonstrated that during EMT, which is a lengthy process [22], a series of intermediate phenotypic states are generated, referred to as the ‘EMT spectrum’, where cells can exhibit both epithelial and mesenchymal features [22]. In keeping with this, we explored the EMT spectrum upon TIM-KD by using a universal and quantitative EMT-score [23] (see methods). Interestingly, the EMT-score increased at TIM-KD-15-days and further augmented at TIM-KD-60-days condition (Fig. 2E), which confirmed that loss of TIM expression was able to initiate a transcriptional reprogramming characteristic of EMT. Importantly, when we analyzed the transcriptome of CRC patients by Gene Set Enrichment Analysis (GSEA; see Methods), low-TIM expressing CRCs were enriched in gene sets representing cell plasticity (i.e., EMT transition, apical junction, NOTCH signaling, and TGF-beta signaling) (Fig. 2F; Table S4). Conversely, proliferation/cell cycle-related mechanisms (i.e., E2F targets, G2M checkpoint signatures, MYC targets, and MTORC1 signaling) were enriched in high-TIM CRCs (Fig. 2F; Table S3). Remarkably, TIM expression was also found significantly lower in CRC samples with a high “EMT score” (Fig. 2G).
Fig. 2. Loss of TIM expression triggers relevant CRC pathways.
A Western blot analysis of TIM levels in SW620 cells infected with a lentiviral vector containing shRNA (TIM-KD; oligo#1; see Methods) or an empty lentiviral vector as negative control (CTRL), at 15-days and 60-days after cell selection with Puromycin. β-actin was used as a protein loading control. B Volcano plot of differentially expressed genes (microarray; q-value < 0.05; FC > |1.5|) in SW620 TIM-KD vs. CTRL cells, at 15-days or 60-days conditions. In red, upregulated genes, and, in green, downregulated genes (number of genes are indicated within the plot). FC fold change. C Scatter plot analysis of differentially expressed genes (DEGs) in SW620 TIM-KD at 15-days or TIM-KD at 60-days conditions. y-Axes, Log10FC at 60-days condition; x-axes, Log10FC at 15-days condition. Blue dots, genes exclusively regulated in 15-days condition; Yellow dots, genes exclusively regulated in 60-days condition; Red dots, commonly regulated genes with the same trend of expression; Grey dots, commonly regulated genes but with the opposite trend of regulation. D MSigDB-overlap analysis of TIM DEGs (SW620 at 15-days and 60-days) as in (C) with the Hallmark (H) gene sets (N = 50). The bubble plot shows the top six overlapping gene sets among TIM-regulated genes. Bubble size represents the statistical significance of overlap expressed as −log10(q-value), where the larger the size the greater the significance. In the x-axis, H-gene set name; in y-axes, the ratio of overlap (k/K) is shown where “k” represents the number of TIM regulated genes while “K” is the number of genes in the specific H-gene set. Bubble color reflects the number of TIM DEGs (k). The complete results are provided in Table S3. E EMT-score of SW620 TIM-KD or CTRL cells at 15-days or 60-days conditions. In y-axes, the EMT-score.; x-axes, the statistical significance of mesenchymal transition expressed as −log10(p-value) of two-sample Kolmogorov–Smirnov test. F Bubble plot of GSEA results using Hallmark (H) gene sets (N = 50) of the TCGA-CRC cohort of patients stratified accordingly to high- or low-TIM tumor expression level. y-Axes, H-gene set name; x-axes, number of core enriched (TIM-regulated) genes. Bubble size represents statistical significance −log10(q-value) of enrichment. Bubble color represents the normalized enrichment score (NES) where positive values indicate enrichment of gene sets in high-TIM expressing CRC, while negative values indicate enrichment of gene sets (Gsets) in low-TIM expressing CRC. Bubble colors are as per the legend (NES). In bold, Gsets represent mechanisms of cell plasticity. Complete results of GSEA are provided in Table S4. G Violin plots of TIM expression level in CRC samples from TCGA-CRC or GSE39582 cohorts, stratified accordingly to low- or high-EMT score (i.e., the 75th and 25th percentile rank of EMT score values were used to categorize high- or low-EMT score CRCs samples, respectively). p-Values were calculated by Mann–Whitney U-test.
Loss of TIM expression induces EMT
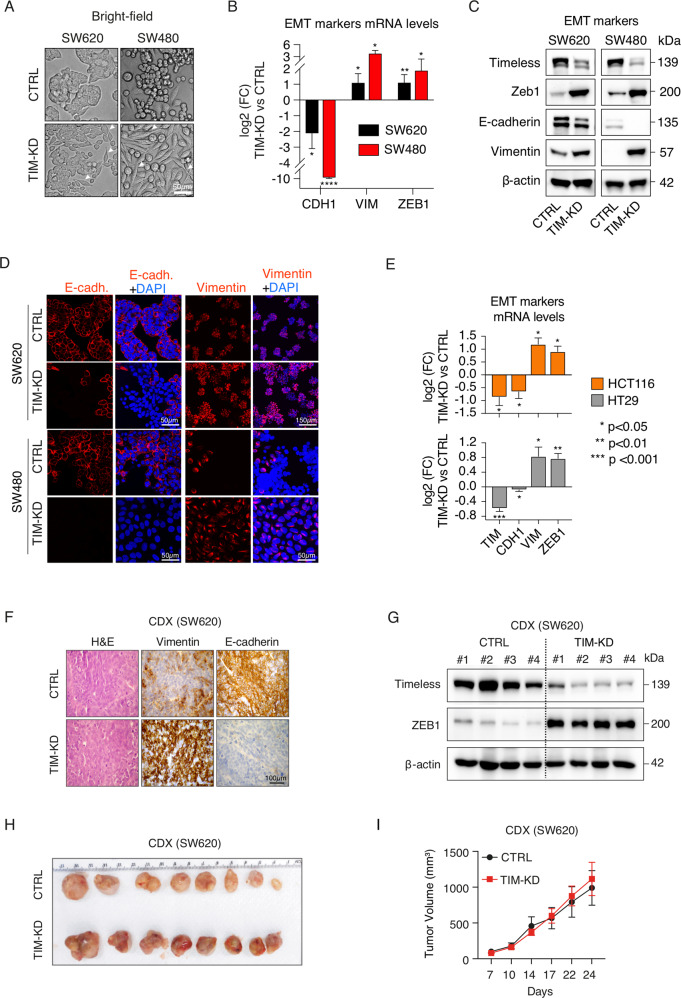
Bright-field microscopy analysis confirmed that loss of TIM expression induced a switch from cobblestone-like to spindle-like phenotype in CRC cells, which is a hallmark of EMT (Fig. 3A). Furthermore, immunofluorescence, immunoblot, and qRT-PCR analyses confirmed the increased expression of canonical EMT markers (i.e., Vimentin, VIM; Zinc finger E-box-binding homeobox 1, ZEB1) and loss of E-cadherin (CDH1) expression in TIM-KD cells (Fig. 3B-D). Such results were confirmed also in additional CRC cell lines (i.e., HCT116 and HT29) (Fig. 3E). Conversely, TIM overexpression reverted the spindle-like phenotype of TIM-KD cells to a cobblestone-like (Fig. S3A) and reduced cell invasion (Fig. S3B). Accordingly, ZEB1 expression was reduced while CDH1 expression increased (Fig. S3A–E). Next, immunohistochemistry and immunoblot analyses performed on a cell line-derived xenograft (CDX) model obtained using SW620 cells confirmed the EMT phenotype in TIM-KD tumor masses (Fig. 3F, G); tumor masses derived from TIM-KD and CTRL SW620 cells were comparable in size (Fig. 3H, I).
Fig. 3. Loss of TIM expression induces EMT in CRC cell lines and cell line-derived xenograft (CDX).
A Bright-field images of CTRL and TIM-KD CRC cells at 60-days conditions. White arrows indicate remarkable morphologic features showing fibroblast-like characteristics in TIM-KD cells. Scale bar, 50 μm. B qRT-PCR analysis of EMT markers expression in TIM-KD vs. CTRL CRC cells (SW620 and SW480) at 60-days conditions. y-Axes, −log2FC (fold change) of TIM-KD vs. CTRL. x-axes, EMT markers analyzed. The 18S ribosomal RNA was used as a housekeeping gene. Data are represented as means ± SEM of experiments run at least three times (at least three biological replicates). *p ≤ 0.05; **p ≤ 0.01; ****p ≤ 0.0001 (Student’s t-test). C Immunoblot analysis for TIM expression and EMT markers (E-cadherin, Vimentin, Zeb1) was performed in TIM-KD and CTRL CRC cells at 60-days conditions. β-actin was used as a protein loading control. D Immunofluorescence staining of Vimentin and E-cadherin used as a mesenchymal or epithelial marker, respectively, in TIM-KD and CTRL CRC cells at 60-days conditions. Scale bars are also shown. E qRT-PCR analysis of TIM and EMT markers expression in TIM-KD vs. CTRL CRC cells (i.e., HCT116 and HT29). y-axes, log2 FC (fold change) of TIM-KD vs. CTRL. x-Axes, gene symbols. The 18S ribosomal RNA was used as a housekeeping gene. Data are represented as means ± SEM of experiments run at least six times (at least six biological replicates). *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001 (Student’s t-test). F Hematoxylin and eosin (H&E) staining, E-cadherin and Vimentin immunohistochemical analysis, performed on FFPE samples of CDX (TIM-KD or CTRL condition) (Scale bar, 100 μm). G Immunoblot analysis of TIM and ZEB1 expression in CDX (TIM-KD tumors, N = 4; CTRL tumors, N = 4). β-actin was blotted as a protein loading control. H Pictures of CDX were obtained from TIM-KD and CTRL SW620 cells. Tumors were excised on day 24 after cell injection. I Tumor growth of TIM-KD and CTRL condition (Line graph). y-Axes, relative tumor volume (mm3); x-axes, days after injection. Values represent means ± SEM (N = 10). p > 0.05, statistical analysis was performed by Mann–Whitney test.
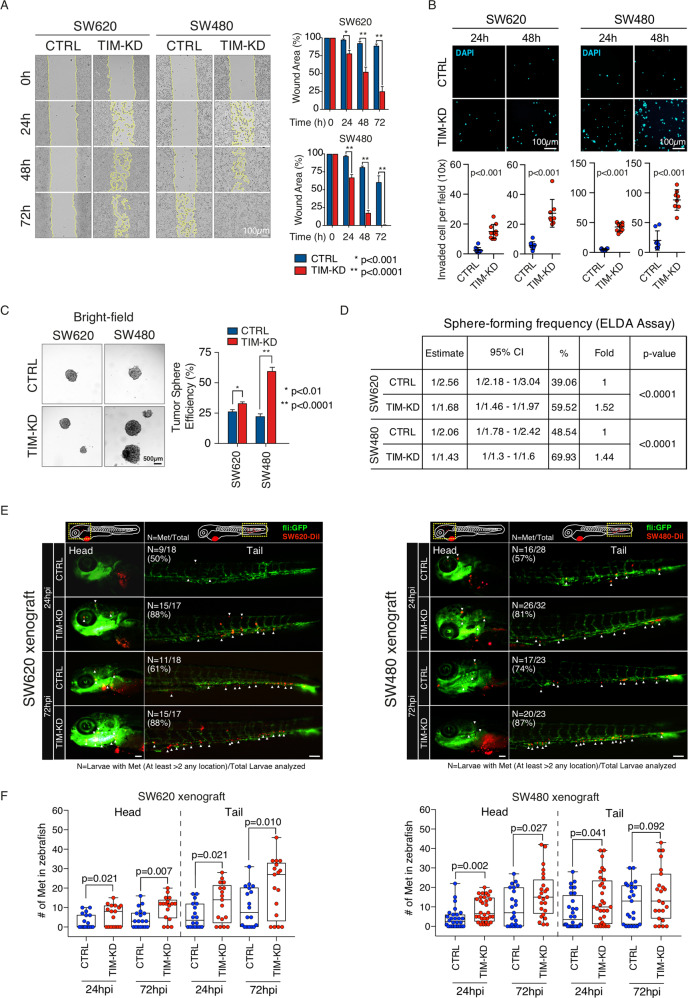
Lastly, we investigated whether the transition to a mesenchymal-like phenotype of TIM-KD CRC cells triggered increased cell aggressiveness. Wound healing assay revealed a cell migration increase in TIM-KD vs. CTRL cells (Fig. 4A). Likewise, cell invasion was increased in TIM-KD cells (Fig. 4B). We then measured the efficiency of generating tumorspheres (TSE) as a proxy for cancer stem cell (CSC) enrichment [24]. Loss of TIM expression in CRC cells correlated with a significant increase of TSE (Fig. 4C) confirmed also by extreme limiting dilution analysis (ELDA) (Fig. 4D; see Methods). Strikingly, xenografts in Zebrafish larvae (ZTX) of TIM-KD and CTRL SW620 cells and SW480 cells (see Methods) revealed that loss of TIM expression correlates with a strong and significant enhancement of CRC cells metastatic spreading (Fig. 4E, F).
Fig. 4. Loss of TIM expression in CRC cells correlates with enhanced migration, invasion, tumorsphere formation in vitro, and enhanced metastatic spreading in vivo.
A Left, representative images of wound healing assay of TIM-KD and CTRL CRC cells. The wound width was measured at 0, 24, 48, or 72 h. Right, histograms show the quantification of the percentage of area compared to the initial scratch area. Experiments were run in biological replicates (N = 4). Statistical significance (p-value) was computed by Student’s t-test. B Top, Invasion assay of TIM-KD and CTRL CRC cells. Images (10× magnification) of DAPI-staining of invaded tumor cells at 24 or 48 h post-seeding. Bottom, dot-plots are based on data as in (B), collected from two independent experiments (eight random fields). y-Axes, number of invaded cells; x-axes, experimental condition. Statistical significance (p-value) was computed by Mann–Whitney test. C Left, analysis of tumorsphere efficiency (TSE) in TIM-KD and CTRL CRC cells. Representative images of spheres. Scale bar, 500 µm. Right, bar plots represent the TSE expressed as a number of spheres over the total number of cells seeded (as a percentage). Data represent mean ± SEM of biological triplicates. Statistical significance (p-value) was computed by Student’s t-test. D TIM-KD and control cells were seeded into 96-well U-bottomed culture plates (poly-HEMA coated) at a density of 10, 5, or 1 cell per well and cultured for 7 days. The sphere-forming cell frequency was calculated by ELDA software. The data are reported as the estimated (ESTIMATE) sphere-forming frequency with relative confidence intervals (95% CI) as well as the estimated percentage of self-renewing cells (%). Fold, sphere-forming frequency of TIM-KD vs. CTRL conditions. p-Value, statistical significance calculated by the chi-squared test. E SW620-CTRL, SW620-TIMKD, SW480-CTRL, and SW480-TIMKD cells were fluorescently labeled with DiI (red) and injected into the perivitelline space (PVS) of 2 days post fertilization (dpf) Tg(fli1:eGFP) zebrafish larvae. At 24 and 72 h post-injection (hpi), ZTX was analyzed to measure the number of metastases. White arrowheads indicate the metastatic colon cancer cells (RED dot) into zebrafish head and tail. N, the number of larvae with metastases (At least >2 any location) divided by the number of total larvae analyzed (N = met/total). Scale bars, 100 μm. F Box-plots represent the number of metastatic colon cancer cells into the head and tail of Tg(fli1:eGFP) zebrafish larvae at 24 and 72 h post-injection. Each dot represents one zebrafish larvae. Results are from two independent experiments. The p-value was calculated by the longitudinal Poisson log-linear model.
Taken together, our results convincingly hint that loss of TIM expression in CRC cells induces EMT with the acquisition of aggressive features in line with the unfavorable prognosis of patients with low-TIM expressing CRC (Fig. 1C).
The newly identified TIM-ZEB1 axis
Since EMT is tightly controlled by activation of EMT-inducing transcription factors (EMT-TFs) such as SNAI1, SNAI2, TWIST1, ZEB1, and ZEB2 [25, 26], we analyzed EMT-TFs expression profile in TIM-KD and CTRL cells. ZEB1 expression was increased, while SNAI1 and SNAI2 diminished (Fig. 5A). Yet, the expression landscape of epithelial and mesenchymal markers confirmed the switch to a mesenchymal phenotype of TIM-KD cells (Fig. 5A). Therefore, we asked whether ZEB1 overexpression in TIM-KD cells was functionally related to EMT and increased in malignant phenotype. To tackle this issue, we stably knocked down ZEB1 in SW620 cells (i.e., ZEB1-KD cells) (Fig. S3F; see Methods) followed by TIM knockdown (i.e., ZEB1/TIM-KDs cells; Fig. S3G; see Methods), with TIM-KD only as further experimental control (TIM-KD cells). The invasive potential of TIM-KD cells was confirmed to be increased when compared to CTRL cells (Fig. 5B). Contrariwise, the absence of ZEB1 in ZEB1/TIM-KDs cells resulted in a significant reduction of cell invasion (Fig. 5B). Furthermore, genome-wide expression profiling revealed 889 DEGs in ZEB1/TIM-KDs vs. TIM-KD cells (q-value < 0.05; log-fold change (FC) > |1.5|; Fig. 5C). Of these, 548 DEGs (~62%) were overlapping with the 2043 DEGs in TIM-KD cells vs. CTRL cells (Fig. 5C). Importantly, 526 (~96%) out of 548 DEGs showed an opposite trend of regulation in ZEB1/TIM-KDs cells vs. TIM-KD cells (Fig. 5D; Table S5). Likewise, the EMT-score was reduced in ZEB1/TIM-KDs cells (Fig. 5E) as well as the expression profile of several mesenchymal markers including Fibronectin 1 (FN1), which, on the contrary, were upregulated in TIM-KD vs CTRL analysis (Fig. 5F–H). These results revealed the existence of a TIM-ZEB1 axis functional to EMT and to the acquisition of an aggressive phenotype of CRC cells. Recently, the miR-200(cluster)-Zeb1 axis has been revealed as having a central role in controlling EMT, tumor invasion [27], and stemness [28]. Therefore, we assayed the expression levels of miR-200 family members (miR-141-3p and miR-200c-3p) [29]; the analysis by qRT-PCR revealed a marked expression reduction of both miR-141-3p and miR-200c-3p in TIM-KD cells when compared to CTRL cells (Fig. 5I; see Methods). However, in ZEB1/TIM-KDs cells the expression of miR-141-3p and miR-200c-3p reverted to comparable levels as observed in CTRL cells (Fig. 5I). Notably, miR-200 members target the Notch pathway components [30] and Bmi-1 [31] which control core cellular processes including proliferation, differentiation, survival, and stemness features acquisition. Indeed, we found several stem-related markers and Notch signaling targets to be upregulated upon TIM-KD (Fig. 5L, M) which is in line with the observed significant enrichment of CSC compartment confirmed both by TSE analysis and ELDA in TIM-KD cells (Fig. 4C, D).
Fig. 5. TIM-ZEB1 axis regulates EMT in CRC.
A Expression profile (microarray) of EMT-TFs, epithelial-, mesenchymal-, and epithelial cell–cell junction markers in SW620 cells infected with a lentiviral vector containing shRNA (TIM-KD; oligo#1; see methods) or an empty lentiviral vector as negative control (CTRL), at 15-days and 60-days after cell selection with Puromycin. y-Axes, the expression level of EMT-TFs (log2 FC). Statistical significance is reported as FDR adjusted p-value (or q-value). The analyses were performed in two independent biological experiments, and gene expression data were Log2 transformed before analyses. B Invasion assay of CTRL, TIM-KD, ZEB1-KD, or ZEB1/TIM-KDs cells. y-Axes, number of invaded cells; x-axes, relative experimental condition. Eight or ten random fields in two independent experiments were selected for statistical analysis. Statistical analysis was performed by Mann–Whitney test. C Venn diagram representing the overlap of significantly regulated genes (FC > | 1.5 | ; q-value < 0.05) in TIM-KD vs. CTRL cells (light grey circle) or TIM-KD vs. ZEB1/TIM-KDs (dark grey circle). D Heat-map of DEGs in TIM-KD vs. CTRL cells whose expression profile is reverted upon ZEB1-KD. The colors are as per the legend. Each column represents mean expression data from biological replicates (N = 2). E EMT-score value (y-axes) of CTRL, TIM-KD, or ZEB1/TIM-KD cells (x-axes). F Scatter plot analysis of differentially expressed genes (DEGs) in ZEB1/TIM-KD vs. CTRL cells (y-axes), and in TIM-KD vs. CTRL (x-axes). Relevant EMT-related genes are highlighted in the picture (Violet circle). G Immunoblot analysis for the indicated protein performed CTRL, TIM-KD, and ZEB1/TIM-KD cells. β-actin was used as a loading control. H Immunofluorescence analysis of Fibronectin 1 (Red) in CTRL, TIM-KD, and ZEB1/TIM-KD cells. Nuclei were stained with DAPI (blue). The scale bar is also shown. I Expression profile analysis of miR-200c-3p and miR-141-3p by qRT-PCR. Data were normalized using U6 snRNA (see also Table S7). Y-axes, fold chance expression difference. x-Axes, different experimental conditions are shown as per the label. Data are represented as means ± SEM of biological replicates (N = 5) run in triplicate (technical). p-Value: *p ≤ 0.05; **p ≤ 0.01 (Student’s t-test). L Expression profile (microarray) of genes involved in stemness in TIM-KD vs. CTRL cells. y-Axes, the expression level of selected genes (log10 FC). Statistical significance is explained by an FDR-adjusted p-value (or q-value). M Expression profile (microarray) of Notch target genes in TIM-KD vs. CTRL cells. y-Axes, the expression level of selected genes (log10 FC). Statistical significance is explained by FDR adjusted p-value (or q-value).
Loss of TIM and ZEB1 expression induces DSBs
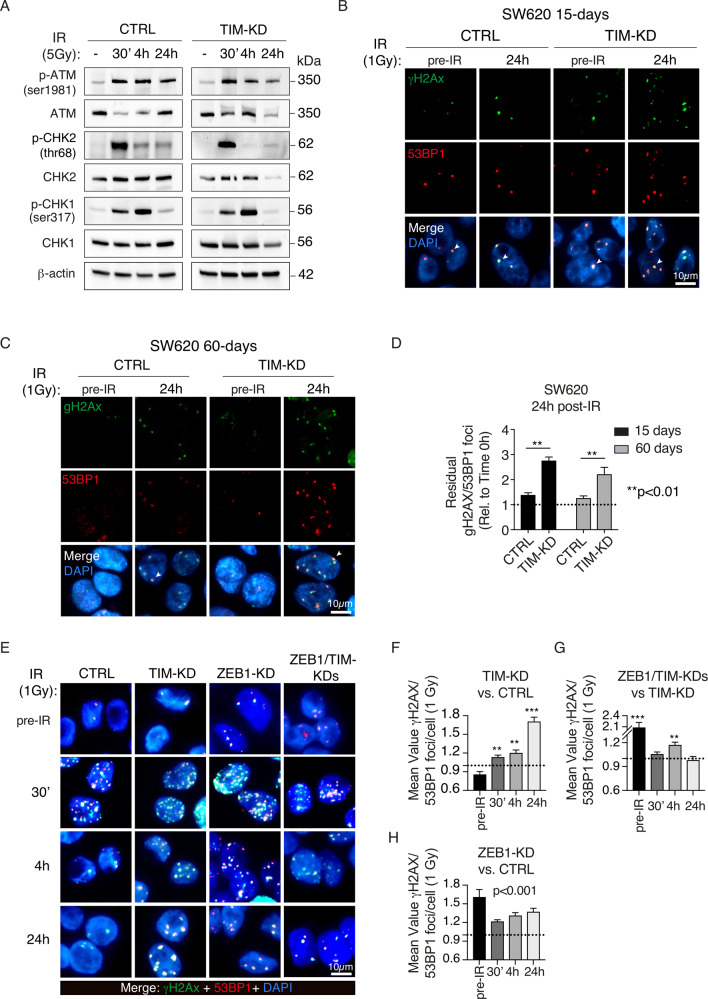
Previous evidence showed that loss of TIM leads to impairment of FPC with proper fork progression and DDR [8–10, 19]. Yet, ZEB1 has also been recently described to take part in DDR [32]. We, therefore, asked if TIM-ZEB1 axis may play a role for DDR in CRC cells. CTRL and TIM-KD cells were exposed to γ-radiation (IR; 5 Gγ) and incubated for 30’, 4 h, and 24 h. Immunoblot analysis revealed an increase in pATM, p-CHK1, and p-CHK2 at different time points (i.e., at 30’ and 4 h) post-IR in both CTRL and TIM-KD cells, consistently with induction of DNA double-strand breaks (DSB; [33] (Fig. 6A). Immunofluorescence analysis of p53 binding protein 1 (53BP1) and γ-H2AX localization, a well-recognized marker of DSBs [34–36], showed higher co-localizing nuclear foci (canonical DSBs hallmark) in TIM-KD cells compared to CTRLs at 24 h post-IR (1 Gy; see Methods) (Fig. 6B–D). Notably, the number of γ-H2AX/53BP1 foci in irradiated CTRL cells was comparable to non-irradiated cells, thus confirming an efficient repair of DSBs (Fig. 6B–D). Time-course analysis revealed a less efficient DSBs repair in TIM-KD cells at the different time points (Fig. 6E, F). Interestingly, we observed that the number of γ-H2AX/53BP1 foci was significantly higher in ZEB1/TIM-KD cells vs. TIM-KD cells at 4 h post-IR condition (Fig. 6G), thus suggesting a delay in the DNA damage recovery upon the TIM-ZEB1 axis impairment. Remarkably, ZEB1-KD cells showed a significantly higher number of γ-H2AX/53BP1foci in pre-IR conditions compared to CTRL cells (Fig. 6H). This difference was appreciable even in post-IR conditions (Fig. 6H) in keeping with the described role of ZEB1 in DDR [32].
Fig. 6. Loss of TIM expression induces accumulation of DSBs.
A Immunoblot for DNA damage markers in CTRL and TIM-KD cells before (−) and after γ-ionizing radiation treatment (30’−24 h) with a dose of 5 Gy. β-actin was used as a protein loading control. B Immunofluorescence analysis of γH2AX (green) and 53BP1 (red) foci in TIM-KD and CTRL cells (15-days condition). Cells were analyzed before (pre-IR) and after irradiation (24 h; 1 Gy). DNA was counterstained with DAPI (blue), and images were merged to determine the γH2AX/53BP foci (i.e., white arrows indicate representative foci) (scale bar, 10 μm). C Immunofluorescence analysis of γH2AX (green) and 53BP1 (red) foci in TIM-KD and CTRL cells (60-days condition). Cells were analyzed before (pre-IR) and after irradiation (24 h; 1 Gy), compared with unirradiated controls. DNA was counterstained with DAPI (blue) and images were merged to determine the γH2AX/53BP foci (i.e., white arrows indicate representative foci) (scale bar, 10 μm). D Quantitative analysis of the γ-H2AX/53BP1 foci in the same experimental setting as in (B, C). Data shown are means ± SE of more than 50 nuclei analyzed in two independent experiments. p-Value (p) was calculated using the Mann–Whitney U-test. E Immunofluorescence analysis of the γH2AX (green) and the 53BP1 (red) foci in CTRL, TIM-KD, ZEB1-KD, and ZEB1/TIM-KD cells before (pre-IR) and after γ-ionizing radiation treatment (30’−24 h; 1 Gy). DNA was counterstained with DAPI (blue) and images were merged to determine the γH2AX/53BP foci (scale bar, 10 μm). F–H Quantitative analysis of the γ-H2AX/53BP1 foci in the same experimental setting as in (E). Data shown are means ± SD of more than 50 nuclei analyzed in two independent experiments. Statistical significance was calculated using the Mann–Whitney U-test. **p ≤ 0.01; ***p ≤ 0.01.
TIM-ZEB1 axis derangement is a hallmark of CRC progression
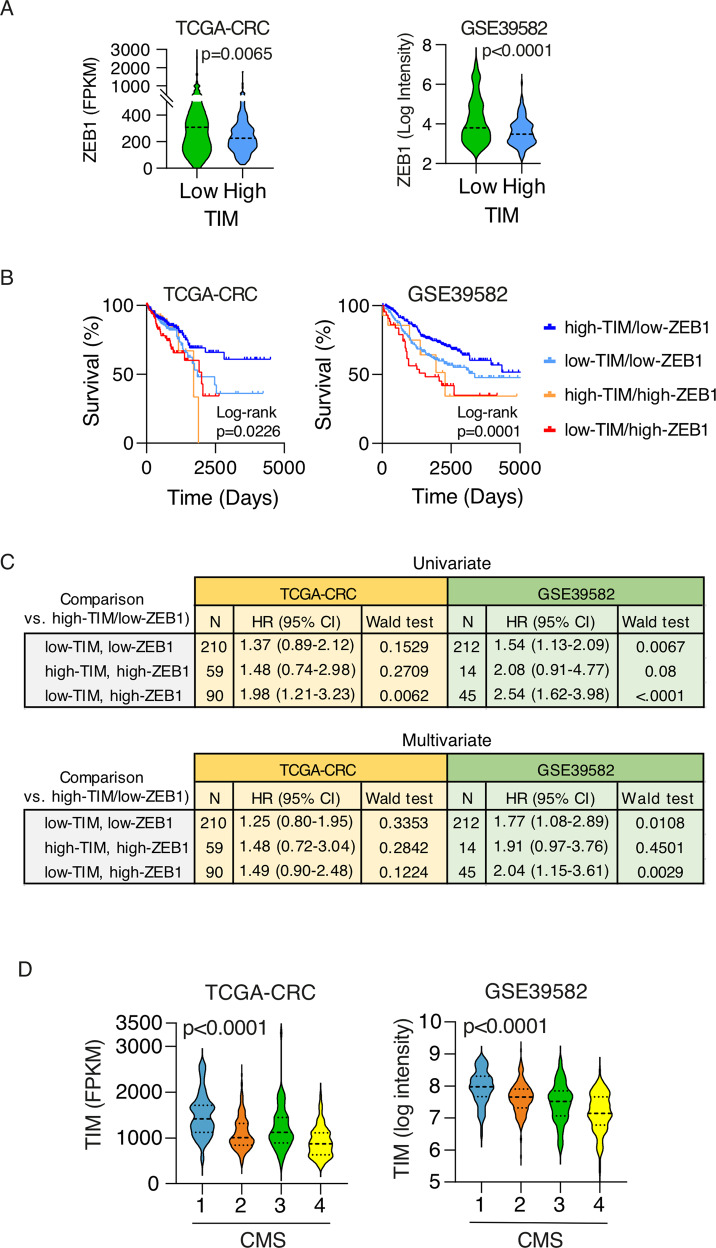
Finally, we explored the relevance of this unedited TIM-ZEB1 axis in human CRCs. Low-TIM expressing CRCs showed indeed a significantly increased expression of ZEB1 in both TCGA-CRC and GSE39582 datasets cohorts (Fig. 7A). Patients with low-TIM/high-ZEB1 expressing CRCs showed a poorer prognosis like the patients with high-TIM/high-ZEB1 expressing CRCs in the TCGA-CRC cohort (Fig. 7B). However, univariate COX analysis revealed that the low-TIM/high-ZEB1 group was associated with the higher risk of death (HR = 1.98, p-value = 0.0062, TCGA-CRC; HR = 2.54, p-value < 0.0001, GSE39562; Fig. 7C). In the multivariate analysis (corrected for age, stage and sex), patients with low-TIM/high-ZEB1 CRC still remained at higher risk of death in the GSE39562 cohort (HR = 2.04, p-value = 0.0029) and also in the TCGA-CRC cohort though with borderline significance (HR = 1.49, p-value = 0.12; Fig. 7C; Table S6).
Fig. 7. TIM-ZEB1 axis is prognostic in CRC.
A Violin plots of ZEB1 expression level in CRC samples (TCGA CRC and GSE39582 cohorts) stratified into low- and high-TIM expressing CRC. The p-value is calculated by Mann–Whitney U-test. B Kaplan–Meier survival plots of patients stratified on the basis of TIM and ZEB1 tumor expression as per the legend. Two independent CRC cohorts of patients were considered (TCGA CRC, N = 597; GSE39582, N = 562). p-Values were calculated by log-rank test. C Univariate and multivariate survival risk analysis of CRC patients stratified based on TIM and ZEB1 tumor expression. Hazard-Ratios (HR) are shown with a relative 95% CI. Multivariate analysis was corrected for age, stage, and sex. p-Values were calculated by Wald test. D Violin plots of TIM expression in CRC samples, from TCGA CRC and GSE39582 datasets, stratified according to the consensus molecular subtype (CMS1-4). The p-Value is calculated by Mann–Whitney U-test.
Importantly, when we analyzed TIM expression across the intrinsic molecular subtypes of CRC by using the Consensus Molecular Subtypes of Colorectal Cancer [37] (CMS), we found that the CSM4 group of tumors had a significant lower TIM expression (Fig. 7D). CMS4 is a hallmark of aggressive CRC [37], is associated with loss of genome integrity [38], and is characterized by upregulation of genes implicated in the EMT process (including ZEB1), in transforming growth factor-β (TGFβ) signaling, angiogenesis, complement-dependent inflammatory system, and in downregulation of miR-200 family members, in line with most of the molecular and phenotypic features of TIM-KD CRC cells we herein characterized.
Discussion
In the last decade, numerous studies have identified TIM as a prognostic biomarker in a variety of human cancers [3, 8]. Timeless is a gene conserved in different living organisms, coding for a protein involved in many cellular processes, such as the circadian circuitry, embryonic development, replication termination, and cell cycle progression [3]. Deranged TIM expression greatly impacts these critical processes as well as the prognosis of cancer patients. We found that TIM expression regulation in cancer correlates with specific cell/tissue context and, possibly, genetic background. As a matter of fact, in non-small cell lung cancer (NSCLC), TIM was over-expressed in aggressive lung adenocarcinoma (LUAD) while downregulated in aggressive LUSC (Fig. 2A). LUAD and LUSC are two NSCLC subtypes originated from different cell types [39] (i.e., the alveolar type II cell for adenocarcinoma, preferentially; the basal/club cells for squamous cell carcinoma) and characterized by distinct oncogenic lesions (e.g., NKX2-1 amplification in lung adenocarcinoma; SOX2 gain and PTEN loss in squamous cell carcinoma) [40, 41] and molecular profile [42, 43].
Intriguingly, TIM expression regulation showed prominent but opposite prognostic significance when evaluated in the various cancer types comprised in the HPA database [18]. In particular, low-TIM expression is associated with advanced stages, metastatic spreading, and unfavorable prognosis in patients suffering from aggressive CRC which accounts for ~1,9 million new cases and ~900,000 deaths in 2020, worldwide [1]. This evidence seemed at odds with the protective role of TIM against oncogene-induced DNA replication stress, especially when overexpressed [8]. DNA replication stress is crucial to support cancer onset and progression [44], but at a higher level can induce mitotic catastrophe which kills cancer cells [44–46].
Our in vitro and in vivo experiments reconcile the apparent paradox of loss TIM expression correlating with CRC progression by showing, for the first time, that loss of TIM expression triggers additional mechanisms which determine aggressive cell phenotypes and fate. Notably, we designed our experimental setting to analyze middle-/long-term transcriptional changes induced by TIM-KD, which could better recapitulate actual pathological processes going on in human CRC. Previous studies analyzed TIM-related phenotypes only at an acute phase (i.e., ~24–72 h [8, 9, 11, 13, 15, 19]); which, therefore, were not appropriate to explore the role of TIM in determining EMT transition, known to be a lengthy process [22]. Indeed, when we performed TIM-KD in acute phase condition the EMT markers expression was relatively unchanged (Fig. S3H). Importantly, we showed that loss of TIM expression in CRC cells triggers a specific and strong induction of ZEB1, an EMT-inducing zinc finger transcription factor that reprograms the transcriptional profile of several coding and non-coding genes (including the miR-200 family). EMT and its reverse process named mesenchymal-to-epithelial transition are fundamental steps during embryogenesis that determine migration and plasticity of cells thus favoring differentiation in diverse cell and tissue types [22]. In cancer, EMT is aberrantly re-activated during progression in order to enhance cell migration, invasion, and stemness phenotypes. Indeed, ZEB1 overexpression in low-TIM expressing CRCs does correlate with the EMT process and metastatic spreading. Our results also suggest that ZEB1 overexpression in the TIM-KD condition is accompanied by downregulation of miR-141-3p and miR-200c-3p, two canonical miRNAs involved in a negative feedback loop with ZEB1 [47]. In our experimental setting, miR-141/200c loss of expression seems to result from the perturbation of this feedback, which we found to be further regulated by TIM. Notably, Notch pathway elements [30] and Bmi-1 [31], which manage crucial cellular processes such as proliferation, differentiation, survival, and stemness phenotype, are targeted by miR-200 family members. As a matter of fact, TIM loss of expression correlated with increased tumor sphere-forming efficiency (Fig. 4C), i.e., a proxy for increased stemness [24], and upregulation of several stemness-related markers and Notch signaling targets (Fig. 5L). Yet, we observed that decreased expression of miR-200 family members (i.e., miR-141-3p and miR-200c-3p) in TIM-KD condition was reverted by silencing ZEB1 thus suggesting possible reciprocal feedback. Further studies are therefore warranted to explore the epigenetic and transcriptional control mechanisms impinging on this new TIM-ZEB1 axis and relevant for CRC progression.
ZEB1 was found also to stabilize CHK1 during DDR [32], which is particularly relevant when transformed cells acquire genomic instability. Loss of TIM expression was largely associated to the impairment of FPC and DDR response [8, 19]; indeed, we found an increase in γ-H2AX and 53BP1 foci in TIM-KD cells that is a hallmark of augmented DNA damage. It is tempting to speculate that ZEB1 overexpression could prevent, at least in part, the accumulation of toxic DNA damage due to defective FPC and DDR upon loss of TIM expression [8, 10, 19]. Nevertheless, we showed that loss of TIM expression causes DNA replication stress and enhanced expression of ZEB1, ultimately leading to EMT and the acquisition of metastatic traits of CRC cells, thus potentially representing another alternative mechanism for TIM-dependent ZEB1 regulation besides the miR-200 family regulation. It is worth noting that the aggressive and genetically unstable CRC-CMS4 subtype [37] is characterized by lower expression of TIM (Fig. 7D), in agreement with adverse outcome hallmarking patients with low-TIM and high-ZEB1 expressing CRCs.
In a recent publication, the TIM protein expression profile was analyzed by immunohistochemistry (IHC) in a cohort of 114 CRC patients [48]. In univariate analysis, high TIM protein expression was found to positively correlate with the patient’s adverse outcome [48], which is in stark contrast with the results we obtained by analyzing two sizeable independent cohorts of CRC patients (N = 1159), i.e., the TCGA CRC and GSE39582 cohorts (Fig. 1C). Furthermore, Cao et al. described the interaction and positive expression correlation of β-catenin and Myosin IIA (aka Myosin-9) with Timeless which would synergize to favor EMT. In our case, we did not observe any positive correlation either at the mRNA level using the TCGA-CRC dataset (i.e., TIM-MYH9 pair, Spearman correlation coefficient ρ = 0.022, p-value = 0.58; TIM-CTNNB1, Spearman correlation coefficient ρ = −0.09, p-value = 0.03) (Fig. S4A) or when β-catenin and Myosin IIA protein expression profile was investigated (i.e., TIM-Myosin IIA pair, Spearman correlation coefficient ρ = 0.048, p-value = 0.49; TIM-Myosin IIA_pS1943 pair, Spearman correlation coefficient ρ = 0.042, p-value = 0.55; TIM-β-catenin pair, Spearman correlation coefficient ρ = 0.002; p-value = 0.98) in a subset of TCGA-CRC samples with proteomic data available (i.e., the TCPA dataset, N = 204; [49]) (Fig. S4B).
In conclusion, our results suggest the engagement of an unedited TIM-ZEB1 axis in key pathological processes in colorectal carcinogenesis. Besides, TIM-ZEB1 expression profiling could provide a robust prognostic biomarker in CRC patients, supporting targeted therapeutic strategies with superior treatment selection and outcome.
Materials and methods
Cell culture
Human CRC cell lines, SW480, SW620, HT29, and HCT116 were obtained from the American type culture collection (ATCC, Manassas, VA, USA). SW480 and HT29 were grown in DMEM Media—GlutaMAX™-I (Gibco; ThermoFisher Scientific, Waltham, MA, USA) with 10% FBS, 1% penicillin/streptomycin (Gibco; Thermo Fisher Scientific, Waltham, MA, USA); SW620 and HCT116 were cultured in RPMI 1640 (Gibco; Thermo Fisher Scientific, Waltham, MA, USA), with 10% FBS and 1% penicillin/streptomycin. Cells were maintained at 37 °C and 5% CO2 in a humidified incubator. Cells were routinely verified as free of mycoplasma contamination.
RNA interference and plasmid transfection
For small interfering RNA (siRNA)-mediated knockdown of TIM, SW620 cells were seeded in 6-well plates and transfected with 10 nM of the targeting or control siRNA (Sigma-Aldrich) using Lipofactamine RNAiMax (Life Technologies). Cells were incubated at 37 °C and experiments were performed 48 or 72 h after transfection. The siRNA sequences used were as follows: CTR siRNA pool (Oligo 8017542333-000050 sequence 5′-UUCUCCGAACGUGUCACGU[dT][dT]-3′ and Oligo 8017542333-000060 sequence 5′-ACGUGACACGUUCGGAGAA[dT][dT]-3′) and hTIM siRNA pool (Oligo 8017542333-000020 sequence 5′-GUAGCUUAGUCCUUUCAAA[dT][dT]-3′ and Oligo 8017542333-000030 sequence 5′-UUUGAAAGGACUAAGCUAC[dT][dT]-3′).
To generate stably overexpression of Timeless, the SW620-KDTIM cells (60 days) were transfected with pcDNA4-Flag-TIMELESS (wild-type ORF) (Addgene plasmid # 22887) or pcDNA™4/myc-His as control (Invitrogen; V86320). The cells were then grown into a selection medium containing 100 μg/mL of Zeocin and selection was continued for 2 weeks.
Lentivirus vector production and transduction
Lentiviral constructs functionally knocking-down TIM (p.LKO.1#1—TRCN0000153760 and p.LKO.1#2—TRCN0000153090, Sigma-Aldrich, St. Louis, MO, USA) and knocking-down ZEB1 (p.LKO.1#1—TRCN0000369267 and p.LKO.1#2—TRCN0000017565, Sigma-Aldrich), along with the corresponding control (p.LKO.1-puro (SHC002), Sigma-Aldrich) were transduced and packaged in 293 T cells. The “p.LKO.1#1—TRCN0000153760” (i.e., TIM-KD) and the “p.LKO.1#2—TRCN0000017565” (i.e., ZEB1-KD) were then preferentially used. 293 T cells were seeded into cell-culture dishes of 10 cm in diameter at a density of 3.5 × 106 cells per plate and grown in DMEM Media - GlutaMAX™-I supplemented with 10% FBS, 1% penicillin/streptomycin (Gibco; ThermoFisher Scientific, Waltham, MA, USA). Lipofectamin 3000 Transfection Reagent (Invitrogen; Thermo Fisher Scientific, Waltham, MA, USA) was used for lentiviral constructs transfection (10 µg of backbone p.LKO.1 plasmid, 5 µg of packaging psPAX2 plasmid, and 2.5 µg of pMD2.G envelope plasmid) in 293 T cells according to the manufacturer’s instructions.
Stable knockdown of TIM and/or ZEB1 (Fig. S2A, B, D; Fig. 5F) in the different CRC cell lines were generated via lentiviral transduction in the presence of polybrene (8 μg/ml) (Sigma-Aldrich, St. Louis, MO, USA) and selected with puromycin (Sigma-Aldrich, St. Louis, MO, USA). All experiments were performed at least three times.
Ionizing radiation treatment
The SW620 cells infected with various lentiviral constructs were irradiated during the exponential phase of growth, at room temperature (RT), with an X-ray dose of 1 or 5 Gy depending on the analyzed endpoint (e.g., a lower dose of IR is recommended in IF experiments to allow better discrimination of the foci), using an MGL 300/6‐D X‐ray apparatus (Gilardoni S.P.A., Mandello del Lario (LC), Italy; 250 kV, 6 mA, Cu filter) operating at 0.53 Gy/min dose rate (University Roma Tre X-rays facility). Cells were harvested after irradiation according to the experimental plan.
RNA and protein extraction
Total RNA was extracted using RNeasy Mini Kit (Qiagen, Hilden, Germany), treated with DNase-RNase free (Qiagen, Hilden, Germany), and quantified by Nanodrop (Thermo Fisher Scientific).
The whole protein extracts were prepared using an appropriate volume of RIPA buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 1% NP-40, 1% Na-deoxycholate, 1 mM EDTA, 0.1% SDS) supplemented with PhosSTOP Phosphatase Inhibitor Cocktail Tablets (Roche) and complete Mini Protease Inhibitor Cocktail Tablets (Roche). The whole protein extracts were prepared using an appropriate volume of RIPA buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 1% NP-40, 1% Na-deoxycholate, 1 mM EDTA, 0.1% SDS) supplemented with PhosSTOP Phosphatase Inhibitor Cocktail Tablets (Roche, Basel, Switzerland) and complete Mini Protease Inhibitor Cocktail Tablets (Roche).
Genome-wide expression profiling
The gene expression profiling was performed using the GeneChip® Human Clarion S Array (Thermo Fisher Scientific) including more than 210,000 distinct probes representative of >20,000 well-annotated genes (hg19; Genome Reference Consortium Human Build 37 (GRCh37)). RNA samples were amplified, fragmented, and labeled for array hybridization according to the manufacturer’s instructions. Samples were then hybridized ON, washed, stained, and scanned using the GeneChip Hybridization Oven 640, Fluidic Station 450, and Scanner 3000 7 G (Thermo Fisher Scientific) to generate the raw data files (.CEL files). The quality control and the normalization of CEL files were performed using the Transcriptome Analysis Console (TAC) software v4.0 (Thermo Fisher Scientific) by performing the “Gene level SST-RMA” summarization method with the human genome version hg38. The analyses were performed in two independent biological experiments, and gene expression data were Log2 transformed before analyses. Differentially expressed genes were defined as those with a fold-change (FC) difference of at least 1.5 (i.e., TIM-KD vs CTRL cells) and a false-discovery rate (FDR) less than 5% (adjusted p-value based on Benjamini-Hochberg Step-Up FDR-controlling Procedure). Raw and normalized gene expression data are available in the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/; GEO Database accession #: GSE169576.
Reverse transcription and quantitative real-time PCR (qRT-PCR)
Reverse transcription was performed using a High-Capacity cDNA Reverse Transcription Kit (ThermoFisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. qRT-PCR was performed using the QuantStudio 12 K Flex Real-Time PCR System (ThermoFisher Scientific, Waltham, MA, USA). TaqMan-based real-time PCR reactions were performed using TaqMan Universal PCR Master Mix and TaqMan probes (ThermoFisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions, with the following program: i) 95 °C for 10 minutes; ii) 95 °C for 15 seconds and 60 °C for 1 minute, repeated for 40 cycles. A list of genes with the relative primers code is reported in Table S7. Each cDNA sample was tested in triplicate (technical replicates) and the 18 S ribosomal RNA was used as housekeeping. The delta-delta Ct method was used to calculate log2 normalized ratios of gene expression. Statistical analyses were performed using triplicate experiments (biological replicates).
miRNAs detection by quantitative real time-PCR (qRT-PCR)
Hsa-miR-200c-3p, hsa-miR-141-3p, and U6 snRNA (see details in Table S7) were reverse transcribed using a TaqMan MicroRNA Reverse Transcription Kit (ThermoFisher Scientific, Waltham, MA, USA) starting from 10 ng of total RNA. 2.5 uL of RT product were pre-amplified (14 cycles of PCR) with TaqMan PreAmp Master Mix (2X) and Custom PreAmp Primers Pool (ThermoFisher Scientific), according to the manufacturer’s instructions. qRT-PCRs were conducted on 1 uL of diluted pre-amplification reaction according to the manufacturer’s instructions. qRT-PCR was carried out on a QuantStudio 12k Flex thermocycler (ThermoFisher Scientific) using the manufacturer’s recommended cycling conditions.
Western blotting
Cell pellets were incubated for 30 minutes on ice. Lysates were cleared by centrifugation at 17000 × g for 30 minutes at 4 °C. Protein concentration was determined by the Pierce BCA Protein Assay kit (ThermoFisher Scientific) according to the manufacturer’s instructions. Thirty micrograms of whole protein lysate were loaded onto Mini-protean TGX gels (Bio-Rad, Hercules, CA, USA) and then transferred on a polyvinylidene fluoride (PVDF) membrane (Bio-Rad) using Trans-blot Turbo (Bio-Rad). Membranes were blocked for 1 h at RT with either 3% BSA/0.1% Tween-20/TBS/(w/v/v) or with 5% non-fat dry milk/0.1% Tween-20/TBS (w/v/v), and then incubated overnight (ON) at 4 °C with primary antibodies. The list of antibodies is reported in Table S8. Membranes were then incubated for 1 h at RT with goat anti-mouse IgG or goat anti-rabbit IgG secondary antibodies (Promega, Madison, WI, USA). Proteins were visualized using ClarityTM or Clarity MaxTM Western ECL substrates (Promega, Madison, WI, USA). Images were acquired using the ChemiDoc™ Imaging system (Bio-Rad). Protein levels were quantified using the Image Lab software (version 2.1.0.35.deb, Bio-Rad). Experiments were repeated at least three times.
Immunofluorescence staining
As many as 9000 cells were seeded on µ-Slide 8 Well (IBIDI) and incubated in appropriate medium. When cells reached the desired confluency, they were washed 2 times with PBS pH7.4 with Ca2+/Mg2+ and then fixed using 4% paraformaldehyde at RT for 10 min. After 3 washes in PBS, cells are permeabilized for 5 min at RT with 0.5% triton/PBS. The cells were incubated with Blocking Buffer (2% BSA + 0.05% Triton+5% Donkey Serum in PBS) for 1 h at RT, and primary antibodies, listed in Table S8, diluted in Blocking Buffer, were added and incubated overnight at 4 °C. After washing, Alexa Fluor 555-conjugated secondary antibodies (see Table S8) were added for 1 h at RT. Nuclei were counterstained with DAPI (1 ng/ml working solution; Sigma-Aldrich, San Luis, MO). Pictures were taken using a confocal laser microscope Leica SP5 X Laser Scanning Confocal Microscope (Leica Microsystems GmbH, Wetzlar, Germany). For DNA repair analysis, cells were grown on glass coverslips, irradiated, and fixed with 4% paraformaldehyde by maintaining cells for 10 min on ice. After permeabilization with PBS/0.2% Triton X-100 (v/v), cells were blocked in 3% BSA/PBS (w/v). γ-H2AX and 53BP1 localizations were detected by co-staining slides ON at 4 °C with 5 mg/mL anti-γ-H2AX and 10 mg/mL anti-53BP1 primary antibodies (see Table S8). Slides were then incubated for 1 h at 37 °C with 10 mg/mL of Alexa Fluor 488- and Alexa Fluor 610-secondary antibodies (Immunological Sciences, Rome, Italy). DNA was counterstained with 0.2 mg/mL DAPI (Sigma-Aldrich) and slides were mounted with Vectashield anti-fade solution (Vector Laboratories, Burlingame, CA, USA) [35, 36, 50]. The Axio Imager M1 fluorescent microscope (Zeiss, Oberkochen, Germany) equipped with a charged coupled device camera was used to analyze samples. Quantitative analysis was carried out by counting foci in at least 50 cells per experiment. All the immunofluorescence experiments were repeated two times.
Wound healing assay
The cells were seeded in a 6-well plate at a density of 106 cells/well and when the cells reached confluency, a linear scratch wound was created in the middle of the confluent monolayer using a sterile P-200 pipette tip. The culture medium was then removed and replaced with a fresh medium. The edges of the initial scratch are indicated with a yellow line in the figure and the wound closure values refer to this initial position. The scratched area was captured at 0-, 24-, 48-, and 72 h after scratching using an Eclipse TE300 Microscope (Nikon, Melville, NY, USA) with a 10X phase objective. The cell-free area (percentage of control at 0 h) was then measured by Fiji software (ImageJ; https://imagej.nih.gov/ij/). All experiments were performed at least three times in triplicates.
Cell invasion assay
Transwell assay: 1 × 105 cells were resuspended in serum-free cell culture media and seeded onto Matrigel-coated Transwell filters (8-µm pore size) (Costar, Corning Inc., Corning, NY, USA) coated with 200 μl of Corning® Matrigel® matrix (final concentration of 250 μg/mL) according to the manufacturer protocol. The outer chamber was filled with 750 μl of medium containing 20% FBS and incubated at 37 °C for 24 h or 48 h. Non-invading cells on the upper surface of the insert were removed with a cotton swab, while those on the lower surface (invasive cells) were fixed with 4% paraformaldehyde and stained with DAPI (1 ng/ml working solution; Sigma-Aldrich). The number of invading cells from 8 fields, each of two separate experiments, was counted under the Eclipse TE300 Microscope using a 10× objective and fields pictures analyzed by using the Cell Count tool of the Fiji software (ImageJ).
Tumor spheres formation assay
Cells in serum-free RPMI were passed through a 50μm strainer and a total of 500 cells were admixed to Matrigel (growth factor reduced; Corning, NY, USA, Lot 4132005) in a 1:1 ratio and left to solidify for 1 h at 37 °C in a humidified incubator with 5% CO2. Spheroids were grown in a DMEM-F12 (Sigma Aldrich) supplemented with 2 mM Glu, 100 U/ml, 5 μg/ml insulin, 0.5 μg/ml hydrocortisone, 2% B27 (Invitrogen; Thermo Fisher Scientific), 20 ng/ml EGF and human b-FGF, and 4 μg/ml heparin. After Matrigel solidification, 1 ml of pre-warmed (37 °C) spheroid media was slowly added in the center of the well to avoid Matrigel disruption. After 15 days, spheres were counted and images were acquired at 4× magnification using the Eclipse TE300 Microscope (Nikon, Melville, NY, USA). Images were analyzed using the Fiji software (ImageJ; https://imagej.nih.gov/ij/) to measure the sphere diameter by setting the cut-off at 50 μm.
ELDA assay
TIM-KD and control cells were seeded into 96-well U-bottomed culture plates, previously coated (48 h) with poly-HEMA (1.2 mg/well, Sigma Aldrich), at a density of 10, 5, or 1 cell per well and cultured for 7 days. Spheroids were grown in a DMEM-F12 (Sigma Aldrich) supplemented with 2 mM Glu, 100 U/ml, 5 μg/ml insulin, 0.5 μg/ml hydrocortisone, 2% B27 (Invitrogen; Thermo Fisher Scientific), 20 ng/ml EGF and human b-FGF, and 4 μg/ml heparin. Scoring was performed 7 days after seeding and the images were analyzed using Fiji software (ImageJ; https://imagej.nih.gov/ij/) to measure sphere diameter by setting the cut-off at 50 μm. The sphere-forming cell frequency was calculated by ELDA software (https://bioinf.wehi.edu.au/software/elda/) [51]. The data are reported as the estimated sphere-forming frequency confidence intervals, as well as the estimated percentage of self-renewing cells. Statistical analysis of differences between groups was performed using the chi-squared test.
A xenograft mouse model for in vivo studies
Approximately, 3 × 106 SW620 CTRL and KD-TIM cell lines were subcutaneously transplanted into two ventral sites of 5 female athymic nude mice (6–8-weeks-old; Charles River, Calco, Lecco, Italy; N = 10 tumor lesions which were considered sufficient for statistical analysis according to the standard operating procedure of Plaisant animal facility (Rome, Italy)). No randomization was used. No blinding was done. All procedures were performed in accordance with the Guidelines for Animal Care and Use of the National Institutes of Health (Ethical committee for animal experimentation Prot. N 088/2016-PR for Plaisant Animal Facility, Roma, Italy). Athymic nude mice were maintained at the Plaisant animal facility (Rome, Italy), and tumor volumes were monitored twice a week by caliper measurement. On day 24, the animals were subjected to gaseous anesthesia (2–3% isoflurane and 1 lt/min O2) and sacrificed by cervical dislocation. Tumor masses were evaluated and excised for further analysis. Immunoblots and immunohistochemistry analysis were performed to evaluate the TIMELESS downregulation in TIM-KD cells and to assess the well-known EMT markers.
Histological and immunohistochemistry data
IHC analysis was performed on mouse xenografted tissues fixed in 10% formalin after surgical resection and embedded in paraffin. Serial 4-μm sections were mounted on poly-l-lysine–coated slides and automatically processed in the Ventana BenchMark XT instrument (Roche Tissue Diagnostics, Arizona, USA). The primary monoclonal antibodies used were the anti-Vimentin (Roche) and anti-CDH1 (Roche) provided with the instrument and dispended to each slide at a prefixed dilution in a 100 µl final volume. Each experiment was performed in duplicate. Primary antibodies were omitted in negative controls. The histopathologic H&E analysis was performed on three sections (2 µm each). Image acquisition and analysis were performed on the Olympus CX41 Microscope (Olympus Corporation, Japan).
Zebrafish xenograft (ZTX) model of human cancer metastasis
Cell culture and labeling
Before injection in zebrafish (Danio rerio), SW480 and SW620 cell lines were labeled with red cell tracker CM-DiI (cat. n. C7000, Thermo Fisher Scientific, Waltham, Massachusetts, USA) according to the manufacturer’s instructions. Then the cells were trypsinized, washed, and resuspended in PBS to obtain a cell suspension for zebrafish xenotransplantation.
Zebrafish husbandry and xenotransplantation
Animal experiments were performed in accordance with the European Council Directive 2010/63/EU and approved by Biogem s.c.ar.l. internal Ethics Committee (OPBA). Tg(fli1:EGFP) zebrafish line, with green fluorescent vessels, was raised, maintained, and paired under standard conditions. Zebrafish eggs were obtained from natural spawnings and maintained in an incubator at 28 °C for 48 hours in E3 medium (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, 0.33 mM MgSO4). Two days post-fertilization (dpf) embryos were dechorionated and anesthetized with 0.04% of tricaine (Sigma Aldrich, St. Louis, Missouri, USA) before cell microinjection. Approximately 100 cells/embryo of SW480 cell line and 300 cells/embryo of SW620 cell line, were injected in the perivitelline space of each embryo (using a pneumatic PicoPump PV830 injector (World Precision Instruments, Sarasota, FL, USA) equipped with an injection borosilicate glass needle (Sutter Instruments, Novato, CA, USA). At least 50 embryos per group were injected according to standard operating procedure. Following transplantation (0 hours post-injection), larvae with correct engraftment in the yolk sac were selected under Leica M205 FA fluorescence stereomicroscope (Leica, Microsystems, Wetzlar, Germany) for further analysis and kept in an incubator at 34 °C for 72 h. A total of N = 17 (TIM) and N = 18 (CTRL) embryos were considered for the SW620 cells engraftment experiment, while N = 32 (TIM) and N = 28 (CTRL) embryos were considered for the SW480 experiment. No randomization was used. No blinding was done. Imaging. Zebrafish larvae were anesthetized (as previously described) and evaluated at 24 and 72 h post-injection by fluorescence stereomicroscope. Different filters were selected for fluorescence imaging and captured with a Leica DFC450 C camera. Images of embryos at different stages of each experimental group were analyzed with ImageJ software (National Institutes of Health, Bethesda, MD, USA). The analyses were performed in two independent biological experiments.
External datasets analyses
Cancer patients database
Survival curves of Timeless expression in human cancer transcriptome were done using the ‘Human Protein Atlas’ database (HPA) which contains expression data of protein-coding genes from multiple tumor types (TCGA data; N = 21 tumors) (https://www.proteinatlas.org/ENSG00000111602-TIMELESS/pathology). Tumor samples were stratified into low-TIM and high-TIM expression on the basis of HPA “best-expression cut-off”. The TIM best expression cut-off was selected on the basis of survival analysis. In detail, the best expression cut-off refers to the TIM—fragments per kilobase of transcript per million mapped reads; RNA seq-data or normalized log-signal (microarray data) value that yields maximal difference with regard to survival between the two groups at the lowest log-rank P-value. Gene expression analysis of timeless in cohorts of CRC patients was performed by using the following publicly available data sets: GSE39582 series (included 566 Colon cancer patients; Gene Expression Omnibus (GEO) database; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi) and the TCGA-CRC series (594 Colorectal cancers patients; TCGA, PanCancer Atlas; [52]). The survival curves were obtained by stratifying the patients according to the best Timeless expression cut-off. The consensus molecular subtyping (CMS) of colorectal cancer was downloaded from the Colorectal Cancer Subtyping Consortium (CRCSC) (https://www.synapse.org/#!Synapse:syn2623706) website. The Cancer Proteome Atlas (TCPA) of colorectal cancer was downloaded from https://tcpaportal.org/tcpa/.
Cell lines Database
Cancer Cell Line Encyclopedia dataset was obtained from the Depmap portal (https://depmap.org/ [20, 53]) (18Q2 release). From the original file, we extracted only the timeless level (RNA and protein) in colon cancer cell lines, resulting in a total of 63 cell lines (see Table S1). The protein abundance data in 50 colorectal cancer cell lines were derived from the work by Roumeliotis et al. [54].
EMT score calculation
The EMT score was computed using the two-sample Kolmogorov–Smirnov test on a specific gene expression signature, as previously reported [23]. This method compares cumulative distribution functions of epithelial and mesenchymal signatures. The EMT score ranges from −1.0 (fully epithelial) to +1.0 (fully mesenchymal) and the computation of the EMT score was requested through http://www.csi.nus.edu.sg/bioinfo/index.php (available online). The ranked EMT score levels in CRC samples from TCGA CRC and GSE39582 cohorts are shown in Fig. S4C, D.
GSEA and MSigDB analysis
We performed the GSEA (https://www.gsea-msigdb.org/gsea/index.jsp) comparing TIM-Low vs TIM-high CRC patients in the TCGA dataset. The GSEA was performed using the Signal2Noise metric, 1000 random gene sets permutation and median gene expression values for class comparison. The molecular signature database (MSigDB v7.0; UC San Diego, CA, USA, and Broad Institute, Boston, MA, USA [55]) was interrogated to compute overlapping analysis of TIM- and TIM-ZEB1 regulated genes, in microarray screening and in independent datasets analysis, with the Hallmark gene sets (N = 50) which represent “specific well-defined biological states or processes and display coherent expression” [55].
Statistical analyses
Hierarchical clustering and heatmap analyses were performed using Cluster 3.0 for Mac OS X (C Clustering Library 1.56; http://bonsai.hgc.jp/~mdehoon/software/cluster/software.htm) and Java Tree View (Version 1.1.6r4; http://jtreeview.sourceforge.net). The uncentered correlation and centroid linkage clustering method was used. Bubble plot analysis was performed using the JMP 15 software (SAS Institute Inc., Cary, NC, USA), while all other plots (Histograms, Forest plot, volcano plots, scatter plot, Box-plot, violin-plot, and Kaplan–Meir) were generated using GraphPad Prism 7 (GraphPad Software Inc., San Diego, CA, USA). All statistical analyses were performed using GraphPad Prism 7 (GraphPad Software Inc.) and JMP 15 (SAS Institute Inc.). Association between experiment condition (TIM-KD vs. CTRL) and a number of metastases in zebrafish xenografts was evaluated through longitudinal Poisson log-linear model, taking into account the within-individual correlation among repeated measures at a different time (24 and 72 hpi) and location (head and tail). The type of statistical test used in the various analyses is indicated in the relative figure legend. All tests were two-sided. A p-value less than 0.05 was considered for statistical significance.
Supplementary information
Supplementary Figures and Tables legends
Acknowledgements
We would like to thank Cristiana Tiberio for technical support and Chiara Di Giorgio for English editing and paper proofreading. The Grant of Excellence Departments, MIUR (ARTICOLO 1, COMMI 314–337 LEGGE 232/2016) to the Department of Science, University Roma TRE is also gratefully acknowledged.
Author contributions
FB, TC, and GM conceived and designed the study; JA, GB, TC, RC, AC, NF, FM, AdM, TN, OP, and LS performed the experiments and acquired data; FB, TC, and ED performed the statistical and bioinformatics analyses; FB, TC, and GM wrote the paper; AdM, FB, TC, VC, ED, GM, and LS reviewed the scientific literature and critically revised the paper; all authors approved the final version of the paper and agreed to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work were appropriately investigated and resolved.
Funding
This study was supported by Associazione Italiana Ricerca sul Cancro [IG-22827 to F.B.], the Italian Ministry of Health [GR-2016-02363975 and CLEARLY to FB; GR-2019-12370460 to TC; Ricerca Corrente 2021 to FB and GM]. This study was supported in part by Ateneo Roma Tre and Fondo di Finanziamento per le Attività base di Ricerca (FFABR) to AdM. TC was a recipient of a fellowship from Associazione Italiana Ricerca sul Cancro (#19548) and of a fellowship from Fondazione Umberto Veronesi. RC was a recipient of a fellowship from Fondazione Umberto Veronesi and of a fellowship from Fondazione Pezcoller. The study funders had no role in the design of the study, the collection, analysis, and interpretation of the data, the writing of the paper, and the decision to submit the paper for publication.
Data availability
All datasets on which the conclusions of the paper rely are available to readers. The raw data for the Gene expression data generated in this study can be accessed through the NCBI Gene Expression Omnibus (GEO accession number GSE169576).
Ethics approval
Please refer to mouse model establishment paragraphs in “Materials and methods.”
Competing interests
The authors declare no competing interests.
Footnotes
Edited by A. Oberst
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Lina Sabatino, Fabrizio Bianchi, Gianluigi Mazzoccoli.
Contributor Information
Fabrizio Bianchi, Email: f.bianchi@operapadrepio.it.
Gianluigi Mazzoccoli, Email: g.mazzoccoli@operapadrepio.it.
Supplementary information
The online version contains supplementary material available at 10.1038/s41418-022-00935-y.
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J Clin. 2021; 10.3322/caac.21660. [DOI] [PubMed]
- 2.Shibue T, Weinberg RA. EMT, CSCs, and drug resistance: the mechanistic link and clinical implications. Nat Rev Clin Oncol. 2017;14:611–29. doi: 10.1038/nrclinonc.2017.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mazzoccoli G, Laukkanen MO, Vinciguerra M, Colangelo T, Colantuoni V. A Timeless link between circadian patterns and disease. Trends Mol Med. 2016;22:68–81. doi: 10.1016/j.molmed.2015.11.007. [DOI] [PubMed] [Google Scholar]
- 4.Baretić D, Jenkyn-Bedford M, Aria V, Cannone G, Skehel M, Yeeles JTP. Cryo-EM structure of the fork protection complex bound to CMG at a replication fork. Mol Cell. 2020;78:926–940.e13. doi: 10.1016/j.molcel.2020.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leman AR, Noguchi C, Lee CY, Noguchi E. Human Timeless and Tipin stabilize replication forks and facilitate sister-chromatid cohesion. J Cell Sci. 2010;123:660–70. doi: 10.1242/jcs.057984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kemp MG, Akan Z, Yilmaz S, Grillo M, Smith-Roe SL, Kang T-H, et al. Tipin-replication protein A interaction mediates Chk1 phosphorylation by ATR in response to genotoxic stress. J Biol Chem. 2010;285:16562–71. doi: 10.1074/jbc.M110.110304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Unsal-Kaçmaz K, Mullen TE, Kaufmann WK, Sancar A. Coupling of human circadian and cell cycles by the Timeless protein. Mol Cell Biol. 2005;25:3109–16. doi: 10.1128/MCB.25.8.3109-3116.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bianco JN, Bergoglio V, Lin Y-L, Pillaire M-J, Schmitz A-L, Gilhodes J, et al. Overexpression of Claspin and Timeless protects cancer cells from replication stress in a checkpoint-independent manner. Nat Commun. 2019;10:910. doi: 10.1038/s41467-019-08886-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Young LM, Marzio A, Perez-Duran P, Reid DA, Meredith DN, Roberti D, et al. TIMELESS forms a complex with PARP1 distinct from its complex with TIPIN and plays a role in the DNA damage response. Cell Rep. 2015;13:451–9. doi: 10.1016/j.celrep.2015.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xie S, Mortusewicz O, Ma HT, Herr P, Poon RYC, Poon RRY, et al. Timeless Interacts with PARP-1 to promote homologous recombination repair. Mol Cell. 2015;60:163–76. doi: 10.1016/j.molcel.2015.07.031. [DOI] [PubMed] [Google Scholar]
- 11.Chi L, Zou Y, Qin L, Ma W, Hao Y, Tang Y, et al. TIMELESS contributes to the progression of breast cancer through activation of MYC. Breast Cancer Res. 2017;19:53. doi: 10.1186/s13058-017-0838-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang W, He W, Shi Y, Zhao J, Liu S, Zhang F, et al. Aberrant TIMELESS expression is associated with poor clinical survival and lymph node metastasis in early-stage cervical carcinoma. Int J Oncol. 2017;50:173–84. doi: 10.3892/ijo.2016.3784. [DOI] [PubMed] [Google Scholar]
- 13.Yoshida K, Sato M, Hase T, Elshazley M, Yamashita R, Usami N, et al. TIMELESS is overexpressed in lung cancer and its expression correlates with poor patient survival. Cancer Sci. 2013;104:171–7. doi: 10.1111/cas.12068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu S-L, Lin H-X, Lin C-Y, Sun X-Q, Ye L-P, Qiu F, et al. TIMELESS confers cisplatin resistance in nasopharyngeal carcinoma by activating the Wnt/β-catenin signaling pathway and promoting the epithelial mesenchymal transition. Cancer Lett. 2017;402:117–30. doi: 10.1016/j.canlet.2017.05.022. [DOI] [PubMed] [Google Scholar]
- 15.Neilsen BK, Frodyma DE, McCall JL, Fisher KW, Lewis RE. ERK-mediated TIMELESS expression suppresses G2/M arrest in colon cancer cells. PLoS ONE. 2019;14:e0209224. doi: 10.1371/journal.pone.0209224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elgohary N, Pellegrino R, Neuman O, Elzawahry HM, Saber MM, Zeneldin AA, et al. Protumorigenic role of Timeless in hepatocellular carcinoma. Int J Oncol. 2015;46:597–606. doi: 10.3892/ijo.2014.2751. [DOI] [PubMed] [Google Scholar]
- 17.Li B, Mu L, Li Y, Xia K, Yang Y, Aman S, et al. TIMELESS inhibits breast cancer cell invasion and metastasis by down-regulating the expression of MMP9. Cancer Cell Int. 2021;21:38. doi: 10.1186/s12935-021-01752-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uhlen M, Zhang C, Lee S, Sjöstedt E, Fagerberg L, Bidkhori G, et al. A pathology atlas of the human cancer transcriptome. Science. 2017;357:eaan2507. doi: 10.1126/science.aan2507. [DOI] [PubMed] [Google Scholar]
- 19.Rageul J, Park JJ, Zeng PP, Lee E-A, Yang J, Hwang S, et al. SDE2 integrates into the TIMELESS-TIPIN complex to protect stalled replication forks. Nat Commun. 2020;11:5495. doi: 10.1038/s41467-020-19162-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghandi M, Huang FW, Jané-Valbuena J, Kryukov GV, Lo CC, McDonald ER, et al. Next-generation characterization of the cancer cell line encyclopedia. Nature. 2019;569:503–8. doi: 10.1038/s41586-019-1186-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dey P, Li J, Zhang J, Chaurasiya S, Strom A, Wang H, et al. Oncogenic KRAS-driven metabolic reprogramming in pancreatic cancer cells utilizes cytokines from the tumor microenvironment. Cancer Discov. 2020;10:608–25. doi: 10.1158/2159-8290.CD-19-0297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, Weinberg RA. Epithelial-to-mesenchymal transition in cancer: complexity and opportunities. Front Med. 2018;12:361–73. doi: 10.1007/s11684-018-0656-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tan TZ, Miow QH, Miki Y, Noda T, Mori S, Huang RY, et al. Epithelial‐mesenchymal transition spectrum quantification and its efficacy in deciphering survival and drug responses of cancer patients. EMBO Mol Med. 2014;6:1279–93. doi: 10.15252/emmm.201404208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dontu G. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253–70. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Puisieux A, Brabletz T, Caramel J. Oncogenic roles of EMT-inducing transcription factors. Nat Cell Biol. 2014;16:488–94. doi: 10.1038/ncb2976. [DOI] [PubMed] [Google Scholar]
- 26.Dongre A, Weinberg RA. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat Rev Mol Cell Biol. 2019;20:69–84. doi: 10.1038/s41580-018-0080-4. [DOI] [PubMed] [Google Scholar]
- 27.Title AC, Hong S-J, Pires ND, Hasenöhrl L, Godbersen S, Stokar-Regenscheit N, et al. Genetic dissection of the miR-200–Zeb1 axis reveals its importance in tumor differentiation and invasion. Nat Commun. 2018;9:4671. doi: 10.1038/s41467-018-07130-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilson MM, Weinberg RA, Lees JA, Guen VJ. Emerging mechanisms by which EMT programs control stemness. Trends Cancer. 2020;6:775–80. doi: 10.1016/j.trecan.2020.03.011. [DOI] [PubMed] [Google Scholar]
- 29.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 30.Brabletz S, Bajdak K, Meidhof S, Burk U, Niedermann G, Firat E, et al. The ZEB1/miR-200 feedback loop controls Notch signalling in cancer cells: ZEB1 activates Notch signalling. EMBO J. 2011;30:770–82. doi: 10.1038/emboj.2010.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wellner U, Schubert J, Burk UC, Schmalhofer O, Zhu F, Sonntag A, et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol. 2009;11:1487–95. doi: 10.1038/ncb1998. [DOI] [PubMed] [Google Scholar]
- 32.Zhang P, Wei Y, Wang L, Debeb BG, Yuan Y, Zhang J, et al. ATM-mediated stabilization of ZEB1 promotes DNA damage response and radioresistance through CHK1. Nat Cell Biol. 2014;16:864–75. doi: 10.1038/ncb3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bartek J, Lukas J. Chk1 and Chk2 kinases in checkpoint control and cancer. Cancer Cell. 2003;3:421–9. doi: 10.1016/s1535-6108(03)00110-7. [DOI] [PubMed] [Google Scholar]
- 34.Panier S, Boulton SJ. Double-strand break repair: 53BP1 comes into focus. Nat Rev Mol Cell Biol. 2014;15:7–18. doi: 10.1038/nrm3719. [DOI] [PubMed] [Google Scholar]
- 35.di Masi A, Cilli D, Berardinelli F, Talarico A, Pallavicini I, Pennisi R, et al. PML nuclear body disruption impairs DNA double-strand break sensing and repair in APL. Cell Death Dis. 2016;7:e2308. doi: 10.1038/cddis.2016.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pennisi R, Antoccia A, Leone S, Ascenzi P, di Masi A. Hsp90α regulates ATM and NBN functions in sensing and repair of DNA double-strand breaks. FEBS J. 2017;284:2378–95. doi: 10.1111/febs.14145. [DOI] [PubMed] [Google Scholar]
- 37.Guinney J, Dienstmann R, Wang X, de Reyniès A, Schlicker A, Soneson C, et al. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015;21:1350–6. doi: 10.1038/nm.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alonso MH, Aussó S, Lopez-Doriga A, Cordero D, Guinó E, Solé X, et al. Comprehensive analysis of copy number aberrations in microsatellite stable colon cancer in view of stromal component. Br J Cancer. 2017;117:421–31. doi: 10.1038/bjc.2017.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheung WKC, Nguyen DX. Lineage factors and differentiation states in lung cancer progression. Oncogene. 2015;34:5771–80. doi: 10.1038/onc.2015.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ferone G, Song J-Y, Sutherland KD, Bhaskaran R, Monkhorst K, Lambooij J-P, et al. SOX2 is the determining oncogenic switch in promoting lung squamous cell carcinoma from different cells of origin. Cancer Cell. 2016;30:519–32. doi: 10.1016/j.ccell.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yatabe Y, Mitsudomi T, Takahashi T. TTF-1 expression in pulmonary adenocarcinomas. Am J Surg Pathol. 2002;26:767–73. doi: 10.1097/00000478-200206000-00010. [DOI] [PubMed] [Google Scholar]
- 42.Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–50. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cancer Genome Atlas Research N. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–25. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Macheret M, Halazonetis TD. DNA replication stress as a hallmark of cancer. Annu Rev Pathol Mech Dis. 2015;10:425–48. doi: 10.1146/annurev-pathol-012414-040424. [DOI] [PubMed] [Google Scholar]
- 45.Baillie KE, Stirling PC. Beyond kinases: targeting replication stress proteins in cancer therapy. Trends Cancer 2020; S2405803320302843. [DOI] [PubMed]
- 46.Liu H, Zhang H, Wu X, Ma D, Wu J, Wang L, et al. Nuclear cGAS suppresses DNA repair and promotes tumorigenesis. Nature. 2018;563:131–6. doi: 10.1038/s41586-018-0629-6. [DOI] [PubMed] [Google Scholar]
- 47.Brabletz S, Brabletz T. The ZEB/miR‐200 feedback loop—a motor of cellular plasticity in development and cancer? EMBO Rep. 2010;11:670–7. doi: 10.1038/embor.2010.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cao M, Wang Y, Xiao Y, Zheng D, Zhi C, Xia X, et al. Activation of the clock gene TIMELESS by H3k27 acetylation promotes colorectal cancer tumorigenesis by binding to Myosin-9. J Exp Clin Cancer Res. 2021;40:162. doi: 10.1186/s13046-021-01936-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li J, Lu Y, Akbani R, Ju Z, Roebuck PL, Liu W, et al. TCPA: a resource for cancer functional proteomics data. Nat Methods. 2013;10:1046–7. doi: 10.1038/nmeth.2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mendez G, Cilli D, Berardinelli F, Viganotti M, Ascenzi P, Tanzarella C, et al. Cleavage of the BRCT tandem domains of nibrin by the 657del5 mutation affects the DNA damage response less than the Arg215Trp mutation. IUBMB Life. 2012;64:853–61. doi: 10.1002/iub.1077. [DOI] [PubMed] [Google Scholar]
- 51.Hu Y, Smyth GK. ELDA: Extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods. 2009;347:70–78. doi: 10.1016/j.jim.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 52.ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium. Pan-cancer analysis of whole genomes. Nature. 2020;578:82–93. doi: 10.1038/s41586-020-1969-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–7. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roumeliotis TI, Williams SP, Gonçalves E, Alsinet C, Del Castillo Velasco-Herrera M, Aben N, et al. Genomic determinants of protein abundance variation in colorectal cancer cells. Cell Rep. 2017;20:2201–14. doi: 10.1016/j.celrep.2017.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The molecular signatures database hallmark gene set collection. Cell Syst. 2015;1:417–25. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures and Tables legends
Data Availability Statement
All datasets on which the conclusions of the paper rely are available to readers. The raw data for the Gene expression data generated in this study can be accessed through the NCBI Gene Expression Omnibus (GEO accession number GSE169576).