Abstract
After UV doses that disrupt DNA replication, the recovery of replication at replication forks in Escherichia coli requires a functional copy of the recF gene. In recF mutants, replication fails to recover and extensive degradation of the nascent DNA occurs, suggesting that recF function is needed to stabilize the disrupted replication forks and facilitate the process of recovery. We show here that the ability of recF to promote the recovery of replication requires that the disrupting lesions be removed. In the absence of excision repair, recF+ cells protect the nascent DNA at replication forks, but replication does not resume. The classical view is that recombination proteins operate in pathways that are independent from DNA repair, and therefore the functions of Rec proteins have been studied in repair-deficient cells. However, mutations in either uvr or recF result in failure to recover replication at UV doses from which wild-type cells recover efficiently, suggesting that recF and excision repair contribute to a common pathway in the recovery of replication.
The uvrA, uvrB, and uvrC genes of Escherichia coli are required for the incision and removal of UV-induced lesions from the DNA. E. coli strains mutated in any one of these genes are unable to remove these lesions and are extremely sensitive to UV irradiation (10, 39).
Other mutations which confer hypersensitivity to UV include those in the recF gene, which was originally identified as a gene required for conjugational or transductional recombination in recBC sbcBC mutants (15). In an otherwise wild-type background, however, the recF mutants are fully proficient in recombination by these assays, although, interestingly, they remain hypersensitive to UV irradiation. recO and recR mutants were identified independently and are equivalent to recF mutants in their UV sensitivity and recombinational phenotypes when tested alone or in a recF background (21, 29). Together, these genes are commonly considered to operate in the recF pathway of recombination or repair (3, 24, 46).
RecF function appears to be tightly associated with DNA replication in vivo. At the genomic level of organization, recF and recR are polycistronic with the dnaN and dnaXZ genes, respectively (9, 32). Both dnaN and dnaXZ encode core subunits of the replication holoenzyme. Additionally, a mutation in priA, a component of the primosome, has been shown to be lethal in combination with a recF mutation. Suppressors of this lethality map to the dnaC gene, which is yet another component of the replication machinery (40, 41).
A functional recF gene is implicated in several aberrant forms of replication, such as plasmid linear multimer formation, rifampin-resistant plasmid replication, stable DNA replication, and thymineless death (20, 25, 27, 28, 31). While these processes are all abnormal and nonproductive for cellular survival, they all involve extensive DNA replication.
The recovery of replication in UV-irradiated E. coli also requires a functional copy of the recF gene. In its absence, replication fails to recover and extensive degradation of the nascent DNA occurs (7). We hypothesized that the UV hypersensitivity of recF cells could be explained by a failure of these cells to recognize and resume replication from disrupted replication forks (7).
A role for RecF in the resumption of replication from disrupted replication forks could also explain how recF may promote recombination. Genetic and biochemical data suggest that RecF-mediated recombination utilizes a recombinational intermediate which mimics the structure of a disrupted replication fork. For recombination to occur in vivo, a 3′ single-stranded overhang must be paired with homologous duplex DNA (1, 18, 22, 23, 26, 33). In the case of a disrupted replication fork, this identical structure is created by the leading strand of DNA synthesis, which polymerizes an invading 3′ DNA end into a homologous duplex template (7).
The ability of RecF to promote the resumption of replication from the site of disruption in UV-irradiated cells may remain blocked by the replication-arresting lesions. If the resumption of replication requires that the arresting lesions must first be repaired, then one would predict that nucleotide excision repair should have a large effect on the resumption of replication. Indeed, the discovery of nucleotide excision repair followed from the characterization of UV-sensitive bacterial mutants in which replication did not recover (42). In order to understand the mechanism of replication recovery more clearly, we have characterized the role of excision repair in the ability of RecF to promote the recovery of replication.
MATERIALS AND METHODS
Bacterial strains.
SR108 is a thyA36 deoC2 derivative of W3110. HL946 (SR108 recF332::Tn3), HL952 (SR108 uvrA::Tn10), HL925 (SR108 uvrC::Tn10), and HL1034 (SR108 recA::Tn10) were made by P1 transduction of the recF332::Tn3, uvrA::Tn10, uvrC::Tn10, and Δ(srlR-recA)306::Tn10 markers from strains HL556, HL758, HL765, and JC10289, respectively. The recF, uvrA, uvrC, and recA phenotypes were checked by UV sensitivity.
Qualitative survival following UV irradiation.
A fresh overnight culture was evenly applied onto a Luria-Bertani medium plate with a cotton swab and incubated at 37°C for 1 h. The plate was covered by a sheet of aluminum foil and placed under a 15-W germicidal lamp (254 nm; 0.6 J/m2/s). The foil was progressively retracted following 20-J/m2 exposures. The irradiated plate was then incubated at 37°C for 8 h and photographed.
Time course of replication recovery.
Cells were grown in Davis medium supplemented with 0.4% glucose, 0.2% Casamino Acids, and 10 μg of thymine per ml (DGCthy medium) and containing 1.0 μCi of [3H]thymine per ml to an optical density at 600 nm (OD600) of 0.2 (approximately 3 × 108 cells/ml), at which point half of the culture received an incident dose of 25 J/m2 (time zero). The amount of 3H incorporated into the DNA was measured by averaging results for duplicate, 0.2-ml samples precipitated in 5% cold trichloroacetic acid and then collected on Whatman glass fiber filters.
Density labeling of replicated DNA.
Cells were grown in DGCthy medium containing 0.2 μCi of [14C]thymine per ml to an OD600 of between 0.3 and 0.4 before being harvested by filtration and resuspended in DGC medium containing 10 μg of 5-bromodeoxyuridine per ml. Half of the culture received 25 J/m2, and each half received 0.5 μCi of [3H]thymine per ml and was then incubated for 1 h. Ten-milliliter samples were placed in an equal volume of ice-cold NET buffer (100 mM NaCl, 10 mM Tris [pH 8.0], 10 mM EDTA), pelleted, and lysed in 0.4 ml of 0.5 M K3PO4 (pH 12.5) containing 40 μl of 10% sarcosyl. The solution was then subjected to isopycnic alkaline CsCl gradient sedimentation as described previously (45). Thirty fractions were collected on Whatman no. 17 paper. The amounts of 14C and 3H in each fraction were determined by scintillation counting.
Measurement of global DNA repair.
Cells were grown in DGCthy medium containing 1.0 μCi of [3H]thymine per ml to an OD600 of 0.4, at which point cells were irradiated with a dose of 25 J/m2 in the defined medium and returned to the shaking, 37°C water bath. Ten-milliliter samples were removed at each time point and mixed with 2 volumes of ice-cold NET. Cells were pelleted, resuspended in 0.5 ml of NET and 100 μg of RNase per ml, and lysed by sonication in a Branson Sonifier. Ten microliters of 10-mg/ml proteinase K and 10 μl of 10% sarcosyl were added to the lysate and incubated for 1 h at 65°C. The DNA was extracted with phenol-chloroform and precipitated in 2.5 M ammonium acetate and 2 volumes of ethanol. Purified DNA was resuspended in NET. The concentration of each sample was determined by fluorometry with Hoechst 33258 dye (2). The removal of cyclobutane pyrimidine dimers (CPDs) and 6-4 photoproducts (6-4PPs) from the DNA was measured by using an immunoassay (19). Following denaturation by boiling, 200 ng (CPDs) or 1 μg (6-4PPs) of each DNA sample was loaded in triplicate onto a Hybond N+ membrane, using a slot blot apparatus. The membrane was incubated for 2 h in the presence of a mouse antibody against either CPDs (TDM-2) or 6-4PPs (64M-2) diluted 1:2,000 in phosphate-buffered saline (PBS) (antibodies were a generous gift from Toshio Mori [30]). Horseradish peroxidase-conjugated secondary antibodies were used at a dilution of 1:5,000 and detected with enhanced chemiluminescence (Amersham) and subsequent phosphorimager (Bio-Rad) analysis. Following detection, the amount of 3H-labeled DNA loaded in each slot was confirmed by scintillation counting.
DNA degradation following UV irradiation.
Cells were grown in DGCthy medium containing 0.2 μCi of [14C]thymine per ml to an OD600 of between 0.3 and 0.4. Ten seconds before harvesting by filtration, 1 μCi of [3H]thymine per ml was added to the culture. Cells were resuspended in nonradioactive DGCthy medium and irradiated with a dose of 25 J/m2 unless otherwise indicated. Approximately 10 and 20 s elapsed between resuspension and irradiation. The amounts of 14C and 3H remaining in the DNA were measured as before (see above).
RESULTS
Replication recovery is inhibited in excision repair mutants and in recF mutants.
The recF gene is generally considered to function independently of the uvr genes. However, the survival following moderate doses of UV requires that both genes be functional (Fig. 1) (35). Previous studies revealed a dose-dependent inhibition of replication in both excision-deficient mutants and recF mutants (35, 37, 42). To assess the contributions of excision repair and recF in the normal recovery process, we compared the recovery of replication in uvr and recF mutants to that in wild-type cells.
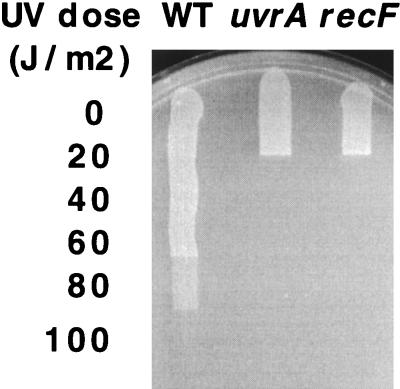
FIG. 1.
Survival of wild-type (WT), uvrA, and recF strains following UV irradiation with the indicated dose.
Using the incorporation of [3H]thymine to quantitate replication, we found that following UV irradiation with 25 J/m2, the wild-type cells exhibited a brief arrest of DNA synthesis before replication resumed at a rate comparable to that in unirradiated cells. However, when recF or uvr mutants were examined, the recovery of replication either was significantly delayed or did not occur (Fig. 2A).
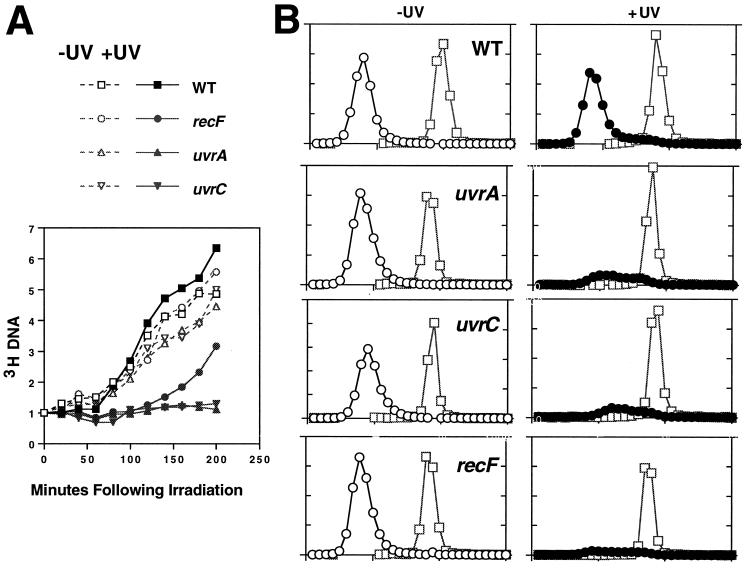
FIG. 2.
recF and uvr mutants show a delay in the recovery of DNA synthesis following UV irradiation. (A) Cells were prelabeled with [3H]thymine. At time zero, half of the culture was removed and given a dose of 25 J/m2 (closed symbols), while the other half was left unirradiated (open symbols). The relative increase of DNA as measured by 3H incorporation is plotted. The initial 3H level was between 1,000 and 1,500 cpm for all strains. (B) The amount of replication occurring within 1 h postirradiation was analyzed with alkaline CsCl density gradients. Cells prelabeled with [14C]thymine were irradiated or not, filtered, and grown in medium containing 5-bromodeoxyuridine and [3H]thymine for 1 h to density label replication occurring this time period. □, 14C-prelabeled DNA, ○, 3H-labeled replicated DNA in unirradiated cultures; •, 3H-labeled, replicated DNA in irradiated cultures. The range of the peak fraction of 3H in unirradiated cultures was 58,000 to 91,000 cpm for all strains. The range of the peak fraction of 14C was 900 to 2,100 cpm in all cases. The ratio of the maximum value between the 3H axis and 14C axis is held constant in all graphs.
The inhibition of replication in recF and uvr mutants can also be observed by density labeling the DNA with 5-bromouracil to quantitate the amount of DNA replicated during the first hour after irradiation. Cultures receiving either 25 J/m2 or no irradiation were incubated in medium containing 5-bromouracil (in place of thymine) for a period of 1 h, so that any DNA replicated during this period would be of a greater density than the DNA synthesized before the time of irradiation. The denser, replicated DNA in each culture was separated from the rest of the DNA by centrifugation in an isopycnic alkaline CsCl gradient and quantitated. By this assay, irradiated wild-type cells had replicated nearly as much DNA as the unirradiated control. However, neither the recF nor the uvr mutants appeared to replicate significant amounts of DNA within this period of time (Fig. 2B). In contrast, while recBC mutants are just as sensitive to UV as recF mutants, they recover replication normally following UV irradiation, suggesting that the failure to recover replication is not related to increased cell death in these populations (7).
The loss of replication recovery in either the recF or uvr mutants, at doses from which wild-type cells completely recover, suggests that functional copies of the recF, uvrA, and uvrC genes are required for the efficient recovery of replication. The results also suggest that in a wild-type cell, recF function in replication recovery is greatly enhanced by the presence of excision repair.
recF mutants do not recover replication despite the repair of the UV lesions.
recF mutants have been reported to have an altered induction of the SOS response (48). SOS induction has been demonstrated to enhance the excision repair rate of the primary UV photoproducts (8). Thus, the lack of recovery in recF mutants could be due to a failure to repair DNA lesions efficiently. However, Rothman and Clark found that the ability of UV-irradiated phage lambda to infect and form plaques was not significantly impaired in recF cells, implying that excision repair was functional (35). Rothman then demonstrated by thin-layer chromotography that dimers were excised in recF cells (34). To confirm this, we examined the rate of removal of the two primary DNA lesions produced by UV, the 6-4PP and the CPD, using monoclonal antibodies directed against each lesion.
In agreement with the results of Rothman and Clark (35), we found that recF cells removed both lesions with rates comparable to those of wild-type cells (Fig. 3). uvrA mutants, as expected, did not remove significant amounts of either lesion. Although no difference between the rates of 6-4PP removal in wild-type and recF cells could be detected, we observed a slight reduction in the rate of removal of CPDs in recF mutants, which may be a consequence of the delayed induction of the SOS response. However, repair was nearly complete within an hour in both the wild type and recF mutants, suggesting that the lack of replication recovery in recF cells is not due to a failure to remove lesions from the template.
FIG. 3.
recF cells remove UV lesions with kinetics that are comparable to those of wild-type cells. Monoclonal antibodies specific for CPDs (A) and 6-4PPs (B) were used to assay lesions in DNA isolated at the indicated times following irradiation with 25 J/m2. Points represent the averages from two independent experiments, each slotted in triplicate. A representative time course for each strain is shown next to each graph.
Nascent-strand degradation at the replication fork occurs following replication disruption.
The failure to recover replication in UV-irradiated recF mutants is associated with the extensive loss of nascent DNA made just prior to irradiation. Since replication also fails to recover in uvr mutants, we examined the degradation pattern in these mutants to determine whether their phenotype was similar to that of the recF mutants. Exponentially growing, [14C]thymine-prelabeled cultures were pulse-labeled with [3H]thymine for 10 s to label the DNA at replication forks and then transferred to nonradioactive medium just prior to irradiation. The 14C prelabel allowed us to compare the degradation occurring in the overall genome to that in the 3H-labeled DNA made at replication forks just prior to UV irradiation.
Wild-type cells degraded very little of their overall genomic DNA following irradiation. However, the nascent DNA exhibited moderate degradation at times prior to the recovery of replication, as determined above. The increase in 3H after 60 min is probably due to intracellular pools of [3H]thymine incorporated following recovery which we were unable to wash out (data not shown). In contrast to wild-type cells, the recF mutant degraded approximately half of the nascent DNA. Similar to the case for wild-type cells, however, the degradation in recF cells was localized primarily to the replication fork DNA, and very little degradation of the genome overall was detected (Fig. 4A).
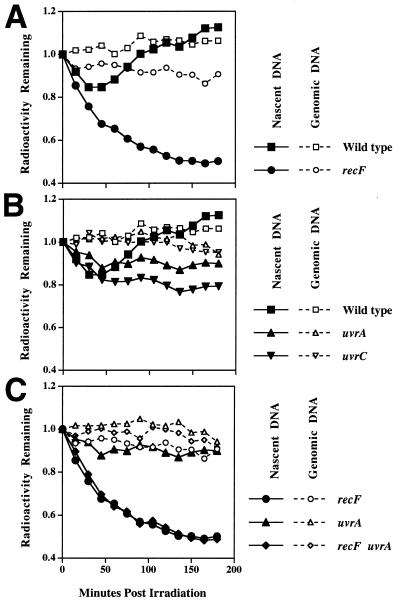
FIG. 4.
Following irradiation, increased degradation occurs at the growing fork in recF mutants but not uvr mutants. [3H]thymine was added to [14C]thymine-prelabeled cells for 10 to 15 s immediately before the cells were filtered and irradiated with 25 J/m2 in nonlabeled medium. The fraction of the radioactivity remaining in the DNA is plotted against time. The loss of 14C genomic DNA (open symbols) can be compared to the loss of the 3H DNA synthesized at the growing fork just prior to irradiation (closed symbols). The range of the initial 14C level was 900 to 1,200 cpm, and that of the initial 3H level was 5,800 to 10,000 cpm in all cases.
In contrast to the case for the recF mutants, the nascent-strand degradation in the uvr mutants was limited to approximately the extent and duration seen in wild-type cells (Fig. 4B). This result is interesting, because although neither uvr nor recF mutants recovered replication, the uvr mutants did not display the extensive nascent-strand degradation associated with recF deficiency. The absence of the uvr proteins, however, did not seem to prevent the disruption of replication, since degradation still occurred in the uvr mutants. In addition, a recF uvrA double mutant exhibited the same extensive nascent-DNA loss as did the recF single mutant (Fig. 4C).
Thus, while replication disruption appears to occur in wild-type, uvr, and recF cells as evidenced by the loss of nascent-strand DNA following UV irradiation, only recF cells fail to recognize the nascent strands and to protect them from extensive degradation. The results suggest that the failure to recover replication in uvr mutants is not due to a failure to recognize and protect the nascent strands of the disrupted DNA fork.
Replication is only partially inhibited at low UV doses.
Previous studies that have focused on the recombination pathways of E. coli have examined postirradiation replication in either recF or uvr mutants at lower doses of UV (11–13, 37, 38). Since we found that replication is significantly inhibited following UV irradiation, we examined replication in these mutants at the lower doses used in other studies.
The amount of replication occurring postirradiation was quantitated as before by incubating irradiated cultures in 5bromouracil to density label any DNA replicated within a 1-h incubation period. The denser, newly replicated DNA was then separated in an isopycnic alkaline CsCl gradient, and the amounts replicated after various doses were compared.
Consistent with the results of previous studies (36, 37, 42), we found that replication was only partially inhibited at the lower doses. In either mutant, the inhibition of replication increased as the UV dose increased, and the levels of inhibition for the uvrA and recF mutants were roughly comparable at a given dose (Fig. 5A and B). However, the fact that wild-type cells completely recover replication at doses which totally inhibit recovery in either mutant suggests that the resumption of DNA synthesis in wild-type cells is dependent on both gene products (Fig. 2B).
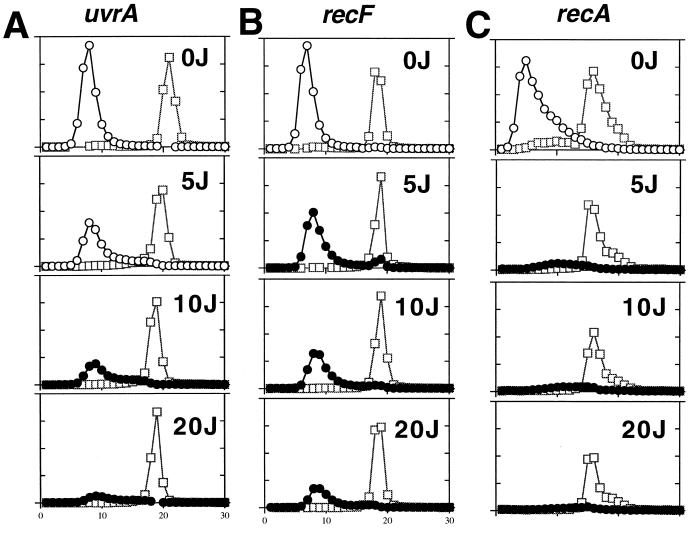
FIG. 5.
Replication is only partially inhibited after low doses of UV irradiation. The amount of replication occurring within 1 h postirradiation at various doses was analyzed with alkaline CsCl density gradients. A single, [14C]thymine-prelabeled culture was filtered and placed in medium containing 5-bromodeoxyuracil and [3H]thymine. Ten-milliliter aliquots were immediately irradiated with the indicated dose. Cells were allowed to recover in a 37°C shaking water bath for 1 h density label any replication occurring after irradiation. □, 14C-prelabeled DNA; ○, 3H-labeled replicated DNA in unirradiated cultures; •, 3H-labeled replicated DNA in irradiated cultures. The range of the peak fraction of 3H in unirradiated cultures was 16,000 to 47,000 cpm for all strains. The range of the peak fraction of 14C was 900 to 4,100 cpm in all cases. The range of the peak fraction of 14C was 900 to 2,100 cpm in all cases. The ratio of the maximum value between the 3H axis and 14C axis is held constant in all graphs.
In contrast to the case for recF and uvrA mutants, the inhibition of replication occurs at much lower doses in recA cells (Fig. 5C). In addition, the recA cells degrade 80 to 90% of both the nascent and genomic DNAs following even a low UV dose (7). The DNA degradation which occurs in recA cells has been shown to progress back from disrupted replication forks and does not occur in nonreplicating cultures (16). Since replication is disrupted at these low fluences in recA cells, it is unlikely that the partial inhibition seen in recF and uvrA mutants is due to a nonuniform exposure of the cell population to UV.
DISCUSSION
Following UV irradiation, recF and uvr mutants fail to recover replication at doses from which wild-type cells recover efficiently. In recF cells, the DNA lesions are removed but the nascent strands of the disrupted replication fork are not protected and undergo more extensive degradation. In uvr mutants, the nascent strands are recognized and protected, but the recovery of replication remains blocked because the UV lesions are not removed. The data strongly suggest that in wild-type cells, both RecF and excision repair operate in a common pathway of replication recovery.
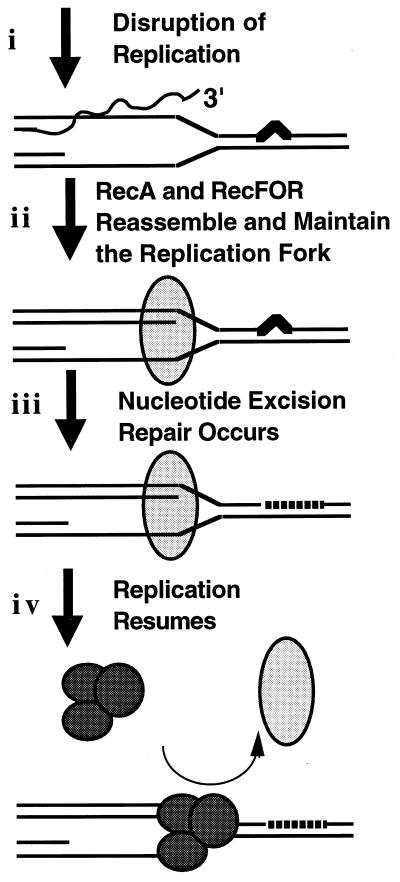
We believe that the data are most consistent with the idea that following the disruption of replication by UV irradiation, recF function is required for the resumption of DNA synthesis from the disrupted replication forks following the removal of the UV lesions by excision repair (Fig. 6). The disruption of replication as evidenced by the transient arrest of DNA synthesis and loss of nascent DNA presumably allows both the time and accessibility required for excision repair to occur.
FIG. 6.
Model of replicational recovery following UV irradiation. Replication is disrupted by a UV lesion in the DNA (i). Because the replication fork has been disrupted, resumption of replication requires that strand-pairing and -exchange proteins (RecA and RecFOR) are present to reassemble and maintain the strands of the replication fork (ii) until the nucleotide excision repair proteins can remove the blocking lesions from the parental DNA template (iii) and the replication can resume (iv). In such a model, the recombination activities of the RecA and RecF proteins function exactly as they have been characterized biochemically. In vivo, however, it suggests that these enzymes are required to re-pair the strands of the replication fork as they were before the disruption event occurred, rather than pairing them with other homologous strands.
Because recombination proteins are usually considered to function independently from the process of nucleotide excision repair, previous models of RecF function have focused on how replication deals with lesions that arrest replication but which cannot be repaired. A large body of work on uvr mutants has demonstrated that following UV irradiation, the limited replication that occurs in the absence of excision repair is accompanied by significant amounts of recF-dependent strand exchange (11, 13, 37, 38). It has been proposed that in this case DNA replication can resume downstream of DNA lesions, creating single-strand gaps which are later repaired through recF-dependent strand exchanges with sister chromosomes, a process termed postreplication recombinational repair or daughter strand gap repair (14).
Both the model presented in Fig. 6 and classical postreplication repair models suggest that replication is disrupted and then resumes upon encounters with UV lesions. The data presented here suggest that excision repair plays a large role in the ability to resume replication in wild-type cells. As presented in Fig. 6, if excision repair occurs following disruption, replication may simply resume from the site of disruption rather than reinitiating from a new site downstream. The lack of replication recovery in UV-irradiated recF mutants despite the proficient overall repair of the genome suggests that following disruption, replication does not efficiently resume downstream of disrupting lesions. If it did, one might expect recF mutants to have wild-type levels of replication recovery but simply leave a gap(s) at the site(s) of disruption. Further, the fact that the nascent DNA is accessible to nucleases indicates that the region is not hidden by a stalled replication complex and implies that the region may also be accessible to repair enzymes. However, we cannot exclude more complex models in which replication reinitiates downstream of the lesion but then arrests again until the required steps of both recombination and repair have been completed.
The partial recovery which occurs in uvr and recF mutants following low doses of UV may highlight the conditions which promote recombination. However, it may not represent the predominant mechanism of recovery in wild-type cells, since the wild-type cells remain unaffected under these conditions while significant reductions in both replication recovery and cell survival occur in either mutant. The fact that replication is not completely inhibited at low doses in these mutants could suggest that a class of lesions (such as those on the lagging-strand template) do not disrupt replication or that these mutants retain a limited ability to bypass lesions.
The general view that the recombination function is independent from excision repair derives from early studies demonstrating that a recA uvrA double mutant was more sensitive to UV irradiation than either single mutant (17). However, wild-type cells survive irradiations producing thousands of lesions per genome, whereas a mutation in either uvrA or recA reduces the lethal dose to fewer than fifty lesions per genome, with more than 99.9% of cells losing viability before any cell death can be detected in wild-type cells (17). The extreme hypersensitivity of either a uvrA or recA mutant suggests that the majority of the survival and recovery occurring in wild-type cells requires that both genes be functional. Similar to mutations inactivating recA, recF mutations also increase the sensitivity of uvr strains (35). However, as is the case with recA, the increase in hypersensitivity due to the addition of a recF mutation represents an almost insignificant portion of the lethality observed in either recF or uvr mutants when compared to the survival of wild-type cells (Fig. 1).
Other studies have also suggested a link between recombination genes and excision repair. Studies of the phenomenon termed long-patch excision repair documented a similar dependency on both the uvr proteins and recF (4–6). Following UV irradiation, the size distribution of the DNA repair patches was found to be bimodal. At early times, short patches representing normal excision repair were the predominant species generated. However at the time that replication was seen to recover, longer patches of 1,500 and >9,000 bp in length were found. These patches, which correspond in both size and ratio to those predicted for lagging- and leading-strand DNA synthesis, respectively, have been shown by two-dimensional gel analysis to be localized at DNA replication forks (4). It is tempting to speculate that these uvr- and recF-dependent patches may in fact represent the resumption of chromosomal replication following removal of the disrupting lesions.
The biochemical activity of RecF in the initiation of replication remains unknown. RecF may serve a largely structural role. This possibility is supported by observations that RecA filaments dissociate upon encountering DNA ends. In vitro, combinations of the RecFOR proteins function by stabilizing RecA filaments at DNA ends and limit the length of filaments extending into duplex DNA (43, 44, 47). The biochemical reaction of reassembling the replication fork structure is identical, in principle, to the mechanism by which RecA is thought to promote homologous strand pairing. The RecFOR proteins may function through stabilizing the RecA filaments which maintain the replication fork structure following disruption. Alternatively, the RecF protein may play a more active role in the reestablishment of the replication machinery at these sites. The latter possibility is attractive considering the genomic organization and genetic associations of the recF pathway with replication proteins as outlined in the introduction. It would be interesting if these associations extended to direct biochemical interactions between the DNA and replication proteins as well.
Although the cellular role of recombination proteins is tightly associated with the replication of the chromosome, recombination proteins are generally studied independently from the process of replication. Replication is able to duplicate the genome in a semiconservative fashion, without alteration, generation after generation. The fact that many of the rec mutants of E. coli appear to be compromised in this ability suggests that these proteins contribute to the semiconservative duplication of the chromosome. Recombination events, i.e., strand exchanges, occur at a very high cost to the organism, and in higher organisms they are intimately associated with genomic instability and a progression towards cancer. The requirement of strand-pairing activities for accurate resumption of replication from disrupted replication forks may be the reason that cells endure this cost. Strand exchange may be a minor, perhaps inappropriate resolution of the strand reassembly process that is required following disruption. Genetic analysis, however, whether for scoring cancer in humans or an auxotrophic marker in E. coli, reflects only the exchanges rather than the normal events that maintain the integrity of the genome.
ACKNOWLEDGMENTS
We thank Ann Ganesan for many fruitful discussions and for critically reading the manuscript.
This research is supported by grant CA44349 from the National Cancer Institute. JC is supported by a traineeship from the National Cancer Institute (DHHS no. CA09302).
Footnotes
This paper is dedicated to the memory of Tokio Kogoma. His work, comments, and insights have significantly contributed to the present work and will be missed in the future.
REFERENCES
- 1.Anderson D G, Kowalczykowski S C. The recombination hot spot chi is a regulatory element that switches the polarity of DNA degradation by the RecBCD enzyme. Genes Dev. 1997;11:571–581. doi: 10.1101/gad.11.5.571. [DOI] [PubMed] [Google Scholar]
- 2.Brunk C F, Jones K C, James T W. Assay for nanogram quantities of DNA in cellular homogenates. Anal Biochem. 1979;92:497–500. doi: 10.1016/0003-2697(79)90690-0. [DOI] [PubMed] [Google Scholar]
- 3.Clark A J, Sandler S J. Homologous genetic recombination: the pieces begin to fall into place. Crit Rev Microbiol. 1994;20:125–142. doi: 10.3109/10408419409113552. [DOI] [PubMed] [Google Scholar]
- 4.Cooper P K. Carcinogenic DNA damage. Department of Energy Project Summary. Berkeley, Calif: University of California Lawrence Berkeley Laboratory; 1990. [Google Scholar]
- 5.Cooper P K. Characterization of long patch excision repair of DNA in ultraviolet-irradiated Escherichia coli: an inducible function under rec-lex control. Mol Gen Genet. 1982;185:189–197. doi: 10.1007/BF00330785. [DOI] [PubMed] [Google Scholar]
- 6.Cooper P K, Hanawalt P C. Heterogeneity of patch size in repair replicated DNA in Escherichia coli. J Mol Biol. 1972;67:1–10. doi: 10.1016/0022-2836(72)90381-6. [DOI] [PubMed] [Google Scholar]
- 7.Courcelle J, Carswell-Crumpton C, Hanawalt P C. recF and recR are required for the resumption of replication at DNA replication forks in Escherichia coli. Proc Natl Acad Sci USA. 1997;94:3714–3719. doi: 10.1073/pnas.94.8.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crowley D J, Hanawalt P C. Induction of the SOS response increases the efficiency of global nucleotide excision repair of cyclobutane pyrimidine dimers, but not 6-4 photoproducts, in UV-irradiated Escherichia coli. J Bacteriol. 1998;180:3345–3352. doi: 10.1128/jb.180.13.3345-3352.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flower A M, McHenry C S. Transcriptional organization of the Escherichia coli dnaX gene. J Mol Biol. 1991;220:649–658. doi: 10.1016/0022-2836(91)90107-h. [DOI] [PubMed] [Google Scholar]
- 10.Friedberg E C, Graham C W, Siede W. DNA repair and mutagenesis. Washington, D.C: ASM Press; 1995. [Google Scholar]
- 11.Ganesan A K. Persistence of pyrimidine dimers during post-replication repair in ultraviolet light-irradiated Escherichia coli. J Mol Biol. 1974;87:103–119. doi: 10.1016/0022-2836(74)90563-4. [DOI] [PubMed] [Google Scholar]
- 12.Ganesan A K, Seawell P C. The effect of lexA and recF mutations on post-replication repair and DNA synthesis in Escherichia coli K-12. Mol Gen Genet. 1975;141:189–205. doi: 10.1007/BF00341799. [DOI] [PubMed] [Google Scholar]
- 13.Ganesan A K, Smith K C. The duration of recovery and repair in excision-deficient derivatives of Escherichia coli K-12 after ultraviolet light irradiation. Mol Gen Genet. 1971;113:285–296. doi: 10.1007/BF00272328. [DOI] [PubMed] [Google Scholar]
- 14.Hanawalt P C, Cooper P K, Ganesan A K, Smith C A. DNA repair in bacteria and mammalian cells. Annu Rev Biochem. 1979;48:783–836. doi: 10.1146/annurev.bi.48.070179.004031. [DOI] [PubMed] [Google Scholar]
- 15.Horii Z, Clark A J. Genetic analysis of the recF pathway to genetic recombination in Escherichia coli K12: isolation and characterization of mutants. J Mol Biol. 1973;80:327–344. doi: 10.1016/0022-2836(73)90176-9. [DOI] [PubMed] [Google Scholar]
- 16.Horii Z, Suzuki K. Degradation of the DNA of Escherichia coli K12 rec- (JC1569b) after irradiation with ultraviolet light. Photochem Photobiol. 1968;8:93–105. doi: 10.1111/j.1751-1097.1970.tb05976.x. [DOI] [PubMed] [Google Scholar]
- 17.Howard-Flanders P, Theriot L, Stedeford J B. Some properties of excision-defective recombination-deficient mutants of Escherichia coli K-12. J Bacteriol. 1969;97:1134–1141. doi: 10.1128/jb.97.3.1134-1141.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Joseph J W, Kolodner R. Exonuclease VIII of Escherichia coli. I. Purification and physical properties. J Biol Chem. 1983;258:10411–10417. [PubMed] [Google Scholar]
- 19.Koehler D R, Courcelle J, Hanawalt P C. Kinetics of pyrimidine(6-4)pyrimidone photoproduct repair in Escherichia coli. J Bacteriol. 1996;178:1347–1350. doi: 10.1128/jb.178.5.1347-1350.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kogoma T, Lark K G. DNA replication in Escherichia coli: replication in absence of protein synthesis after replication inhibition. J Mol Biol. 1970;52:143–164. doi: 10.1016/0022-2836(70)90022-7. [DOI] [PubMed] [Google Scholar]
- 21.Kolodner R, Fishel R A, Howard M. Genetic recombination of bacterial plasmid DNA: effect of RecF pathway mutations on plasmid recombination in Escherichia coli. J Bacteriol. 1985;163:1060–1066. doi: 10.1128/jb.163.3.1060-1066.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Konforti B B, Davis R W. 3′ homologous free ends are required for stable joint molecule formation by the RecA and single-stranded binding proteins of Escherichia coli. Proc Natl Acad Sci USA. 1987;84:690–694. doi: 10.1073/pnas.84.3.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Konforti B B, Davis R W. The preference for a 3′ homologous end is intrinsic to RecA-promoted strand exchange. J Biol Chem. 1990;265:6916–6920. [PubMed] [Google Scholar]
- 24.Kowalczykowski S C, Dixon D A, Eggleston A K, Lauder S D, Rehrauer W M. Biochemistry of homologous recombination in Escherichia coli. Microbiol Rev. 1994;58:401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kusano K, Nakayama K, Nakayama H. Plasmid-mediated lethality and plasmid multimer formation in an Escherichia coli recBC sbcBC mutant. Involvement of RecF recombination pathway genes. J Mol Biol. 1989;209:623–634. doi: 10.1016/0022-2836(89)90000-4. [DOI] [PubMed] [Google Scholar]
- 26.Lovett S T, Kolodner R D. Identification and purification of a single-stranded-DNA-specific exonuclease encoded by the recJ gene of Escherichia coli. Proc Natl Acad Sci USA. 1989;86:2627–2631. doi: 10.1073/pnas.86.8.2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maaloe O, Hanawalt P C. Thymine deficiency and the normal DNA replication cycle. I. J Mol Biol. 1961;3:144–155. doi: 10.1016/s0022-2836(61)80041-7. [DOI] [PubMed] [Google Scholar]
- 28.Magee T R, Kogoma T. Rifampin-resistant replication of pBR322 derivatives in Escherichia coli cells induced for the SOS response. J Bacteriol. 1991;173:4736–4741. doi: 10.1128/jb.173.15.4736-4741.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mahdi A A, Lloyd R G. Identification of the recR locus of Escherichia coli K-12 and analysis of its role in recombination and DNA repair. Mol Gen Genet. 1989;216:503–510. doi: 10.1007/BF00334397. [DOI] [PubMed] [Google Scholar]
- 30.Mori T, Nakane M, Hattori T, Matsunaga T, Ihara M, Nikaido O. Simultaneous establishment of monoclonal antibodies specific for either cyclobutane pyrimidine dimer or (6-4)photoproduct from the same mouse immunized with ultraviolet-irradiated DNA. Photochem Photobiol. 1991;54:225–232. doi: 10.1111/j.1751-1097.1991.tb02010.x. [DOI] [PubMed] [Google Scholar]
- 31.Nakayama K, Shiota S, Nakayama H. Thymineless death in Escherichia coli mutants deficient in the RecF recombination pathway. Can J Microbiol. 1988;34:905–907. doi: 10.1139/m88-157. [DOI] [PubMed] [Google Scholar]
- 32.Perez-Roger I, Garcia-Sogo M, Navarro-Avino J P, Lopez-Acedo C, Macian F, Armengod M E. Positive and negative regulatory elements in the dnaA-dnaN-recF operon of Escherichia coli. Biochimie. 1991;73:329–334. doi: 10.1016/0300-9084(91)90220-u. [DOI] [PubMed] [Google Scholar]
- 33.Phillips G J, Prasher D C, Kushner S R. Physical and biochemical characterization of cloned sbcB and xonA mutations from Escherichia coli K-12. J Bacteriol. 1988;170:2089–2094. doi: 10.1128/jb.170.5.2089-2094.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rothman R H. Dimer excision and repair replication patch size in a recL152 mutant of Escherichia coli K-12. J Bacteriol. 1978;136:444–448. doi: 10.1128/jb.136.1.444-448.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rothman R H, Clark A J. The dependence of postreplication repair on uvrB in a recF mutant of Escherichia coli K-12. Mol Gen Genet. 1977;155:279–286. doi: 10.1007/BF00272806. [DOI] [PubMed] [Google Scholar]
- 36.Rothman R H, Kato T, Clark A J. The beginning of an investigation of the role of recF in the pathways of metabolism of ultraviolet-irradiated DNA in Escherichia coli. Basic Life Sci. 1975;5A:283–291. doi: 10.1007/978-1-4684-2895-7_37. [DOI] [PubMed] [Google Scholar]
- 37.Rupp W D, Howard-Flanders P. Discontinuities in the DNA synthesized in an excision-defective strain of Escherichia coli following ultraviolet irradiation. J Mol Biol. 1968;31:291–304. doi: 10.1016/0022-2836(68)90445-2. [DOI] [PubMed] [Google Scholar]
- 38.Rupp W D, Wilde C E I, Reno D L, Howard-Flanders P. Exchanges between DNA strands in ultraviolet-irradiated Escherichia coli. J Mol Biol. 1971;61:25–44. doi: 10.1016/0022-2836(71)90204-x. [DOI] [PubMed] [Google Scholar]
- 39.Sancar A. DNA excision repair. Annu Rev Biochem. 1996;65:43–81. doi: 10.1146/annurev.bi.65.070196.000355. [DOI] [PubMed] [Google Scholar]
- 40.Sandler S J. Overlapping functions for recF and priA in cell viability and UV-inducible SOS expression are distinguished by dnaC809 in Escherichia coli K-12. Mol Microbiol. 1996;19:871–880. doi: 10.1046/j.1365-2958.1996.429959.x. [DOI] [PubMed] [Google Scholar]
- 41.Sandler S J, Samra H S, Clark A J. Differential suppression of priA2::kan phenotypes in Escherichia coli K-12 by mutations in priA, lexA, and dnaC. Genetics. 1996;143:5–13. doi: 10.1093/genetics/143.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Setlow R B, Swenson P A, Carrier W L. Thymine dimers and inhibition of DNA synthesis by ultraviolet irradiation of cells. Science. 1963;142:1464–1466. doi: 10.1126/science.142.3598.1464. [DOI] [PubMed] [Google Scholar]
- 43.Shan Q, Bork J M, Webb B L, Inman R B, Cox M M. RecA protein filaments: end-dependent dissociation from ssDNA stabilization by RecO and RecR proteins. J Mol Biol. 1997;265:519–540. doi: 10.1006/jmbi.1996.0748. [DOI] [PubMed] [Google Scholar]
- 44.Shan Q, Cox M M. RecA filament dynamics during DNA strand exchange reactions. J Biol Chem. 1997;272:11063–11073. doi: 10.1074/jbc.272.17.11063. [DOI] [PubMed] [Google Scholar]
- 45.Smith C A, Cooper P K, Hanawalt P C. Measurement of replication by equilibrium sedimentation. In: Friedberg E C, Hanawalt P C, editors. DNA repair. A manual of research procedures. 1b. New York, N.Y: Marcel Dekker Inc.; 1981. pp. 289–305. [Google Scholar]
- 46.Smith G R. Homologous recombination in prokaryotes. Microbiol Rev. 1988;52:1–28. doi: 10.1128/mr.52.1.1-28.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Webb B L, Cox M M, Inman R B. Recombinational DNA repair: the RecF and RecR proteins limit the extension of RecA filaments beyond single-strand DNA gaps. Cell. 1997;91:347–356. doi: 10.1016/s0092-8674(00)80418-3. [DOI] [PubMed] [Google Scholar]
- 48.Whitby M C, Lloyd R G. Altered SOS induction associated with mutations in recF, recO, and recR. Mol Gen Genet. 1995;246:174–179. doi: 10.1007/BF00294680. [DOI] [PubMed] [Google Scholar]