Abstract
Transcription factor EB (TFEB), a member of the MiT/TFE family of basic helix-loop-helix leucine zipper transcription factors, is an established central regulator of the autophagy/lysosomal-to-nucleus signaling pathway. Originally described as an oncogene, TFEB is now widely known as a regulator of various processes, such as energy homeostasis, stress response, metabolism, and autophagy-lysosomal biogenesis because of its extensive involvement in various signaling pathways, such as mTORC1, Wnt, calcium, and AKT signaling pathways. TFEB is also implicated in various human diseases, such as lysosomal storage disorders, neurodegenerative diseases, cancers, and metabolic disorders. In this review, we present an overview of the major advances in TFEB research over the past 30 years, since its description in 1990. This review also discusses the recently discovered regulatory mechanisms of TFEB and their implications for human diseases. We also summarize the moonlighting functions of TFEB and discuss future research directions and unanswered questions in the field. Overall, this review provides insight into our understanding of TFEB as a major molecular player in human health, which will take us one step closer to promoting TFEB from basic research into clinical and regenerative applications.
Subject terms: Macroautophagy, Kinases
Facts
TFEB serves as a master regulator of autophagy-lysosomal biogenesis by promoting the ‘CLEAR’ network.
TFEB is involved in various signaling pathways, including mTOR and ERK, and is controlled by various upstream factors, including nutrient availability and stress.
Impaired autophagy-lysosomal functions consequent to defective TFEB are implicated in a plethora of diseases, such as lysosomal storage disorders, neurodegenerative diseases, cancers, and metabolic diseases.
TFEB exerts autophagy-lysosomal independent functions.
Given the important role of TFEB in human diseases, TFEB is potentially a novel therapeutic target for clinical and regenerative applications.
Open questions
TFEB: A double-edged sword with therapeutic potential. Activation of TFEB is beneficial in certain contexts, but may have negative consequences in others. It is vital to coordinate TFEB activity carefully. Is TFEB activation more dominant than its inhibition? How can TFEB modulation be segregated in specific circumstances?
Recently, it has become evident that TFEB exerts both autophagy-lysosomal dependent and independent functions. Do other crucial non-autophagy-lysosomal biogenesis functions exist and are these favorable for therapeutic applications?
Regulation by a single transcription factor, TFEB, is required for the precise control of physiological functions. How is TFEB activity controlled at the transcriptional level in a cell type-specific manner, and does this involve particular transcriptional upstream regulators of TFEB?
Introduction
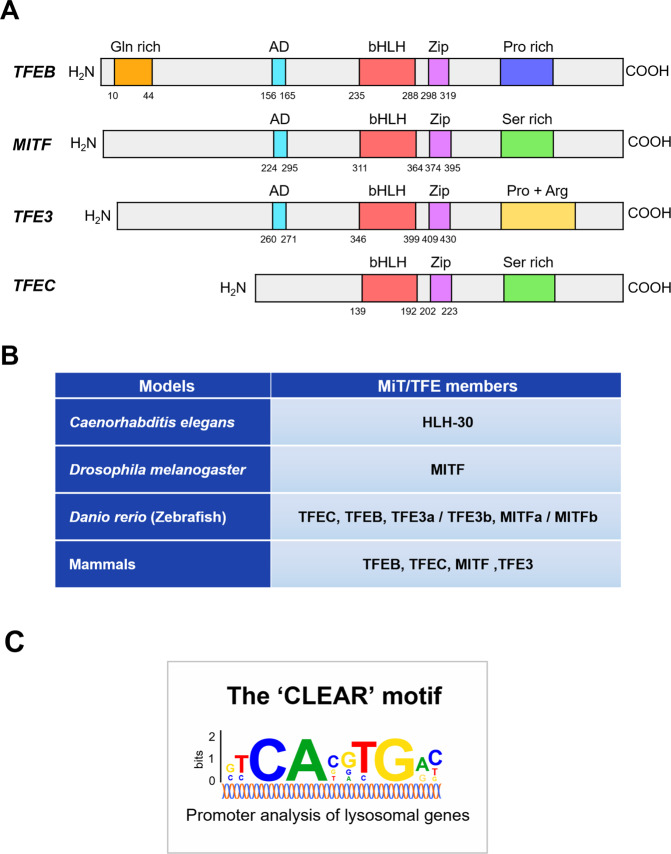
Members of the microphthalmia (MiT/TFE) family belong to the basic group of helix-loop-helix leucine zipper (bHLH-ZIP)-containing transcription factors. Originally believed to be involved in melanocyte biology, they have emerged as key players in a myriad of cellular processes, notably autophagy-lysosomal biogenesis, cellular energy homeostasis, and metabolic processes, in response to internal and external stresses. In vertebrates, the MiT/TFE family of transcription factors is composed of four evolutionarily conserved members: transcription factor EB (TFEB), microphthalmia-associated transcription factor (MITF), TFEC, and TFE3 [1] (Fig. 1A). These bHLH-ZIP transcription factors share high sequence similarities and activate target gene expression by binding to DNA through their basic domain, either in the form of homodimers or heterodimers. Structural and biochemical data suggest that all members of the MiT/TFE family share highly conserved functional domains that are required for DNA binding and homo-heterodimerization across different species, ranging from Homo sapiens, Danio rerio, and Drosophila melanogaster to Caenorhabditis elegans [2] (Fig. 1B). Their functions and regulations appear to be similar to those of TFEB [3–6]. MiT/TFE transcription factors are ubiquitously expressed and their expression levels vary considerably depending on the cell types and tissues. Extensive analysis of the promoters of numerous autophagy-lysosomal genes has revealed that MiT/TFE proteins prefer a palindromic ten base pair motif (GTCACGTGAC), known as the coordinated lysosomal expression and regulation (CLEAR) element [7, 8] (Fig. 1C).
Fig. 1. Domain structure of MiT/TFE proteins and their homologs in primitive metazoans.
A The MiT/TFE family comprises of four members: TFEB, MITF, TFE3, and TFEC, all of which share conserved basic helix-loop-helix (bHLH) and leucine zipper (Zip) domains. B Different species harbor different MiT/TFE members owing to variation in whole-genome duplication. C The CLEAR motif, which can be found in the majority of autophagy-lysosomal promoters, is recognized by MiT/TFE proteins. Abbreviations for each domain are as follows: activation domain (AD), glutamine-rich (Gln rich), proline-rich (Pro rich), serine-rich (Ser rich), and proline + arginine (Pro + Arg) domain.
In this review, we will discuss and provide detailed insights regarding the past TFEB research over the last 30 years as well as present and future perspectives.
TFEB
Introduction of TFEB
All MiT/TFE proteins, including TFEB, are evolutionarily conserved, and their homologs can be found in primitive metazoans [9, 10]. TFEB was first discovered in 1990 during the screening of a bacteriophage expression library, which uncovered a novel cDNA in the helix-loop-helix (HLH) family that encoded the TFEB protein [11]. While the function of MITF in development (such as melanosome biogenesis) and eye disorders has been long recognized, owing to the striking coat color, smaller eyes, and eye development defects in mice and rats harboring the MITF mutation [1, 12–14], the function of TFEB remained elusive until 1998, when TFEB knockout (KO) mice were found to show embryonic lethality consequent of defective placental vascularization [15].
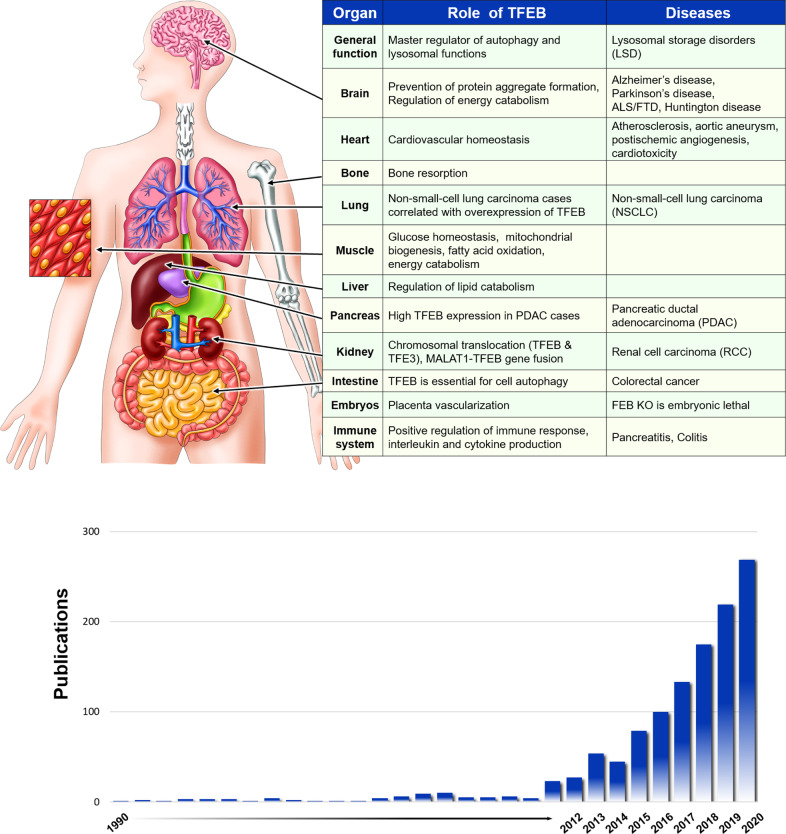
TFEB first came into the limelight and garnered public interest when it was recognized as the master regulator of lysosomal biogenesis. Systems Biology studies have also led to the identification of a gene transcriptional network involved in lysosomal biogenesis, termed the CLEAR network, with TFEB as the master regulator. In addition to lysosomal genes, TFEB also links autophagy to lysosomal biogenesis by regulating the expression of genes involved in autophagy [16]. Moreover, TFEB regulates lysosomal exocytosis [17]. Owing to the critical role of TFEB in autophagy-lysosomal biogenesis, numerous studies have exploited the benefits of TFEB in promoting cellular clearance in a variety of cellular and mouse models of human diseases related to the accumulation of un-degraded substances. Neurodegenerative diseases such as Alzheimer’s disease (AD) [18–20] and Parkinson’s disease (PD) [21–23], lysosomal storage disorders (LSDs) [17, 24–26], metabolic diseases [5], and cancers [27] are good examples of human diseases caused by the accumulation of un-degraded substances. Figure 2 shows the overall pathophysiological functions and importance of the TFEB protein, whereas Fig. 3 presents the timeline of the major key events in TFEB research over the past 30 years.
Fig. 2. Pathophysiological functions and importance of the TFEB protein.
TFEB regulates the lysosomal biogenesis & autophagy pathway, which is crucial for proper tissue-specific regulation and TFEB-mediated intracellular clearance of toxic protein aggregates (upper panel). The number of research articles has increased along with the interest in and perceived importance of the TFEB protein (bottom panel).
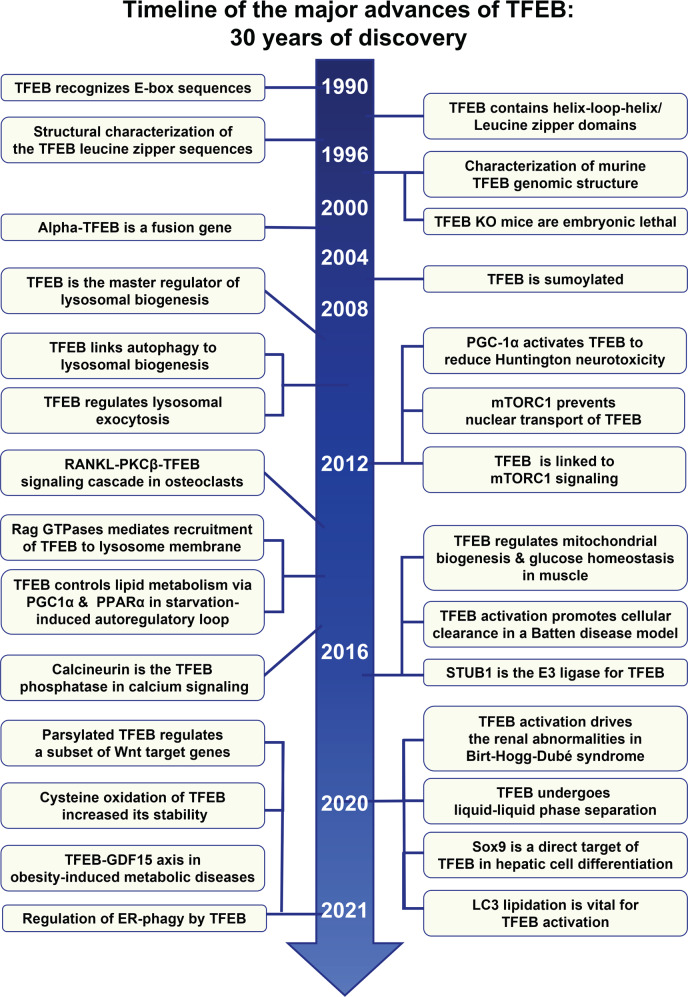
Fig. 3. Timeline of the major advances in TFEB research, in the 30 years since discovery.
The timeline summarizes 30 years of major advances and key findings in TFEB research, from 1990 through to 2021.
Molecular mechanisms of TFEB regulation
Cytoplasmic and nuclear shuttling of TFEB
Autophagy is a type of cellular adaptation to nutrient stress, as it prevents the accumulation of damaged protein aggregates and organelles in the cytosol, while allowing the recycling of essential cellular building blocks to generate energy. Therefore, autophagy activation is vital to preserving homeostasis and is effectively upregulated in response to stress conditions, such as nutrient starvation, protein misfolding, long-lived proteins, and organelle damage. Therefore, appropriate regulation of autophagosome biogenesis and endolysosomal compartments is necessary. However, the mechanism that globally governs lysosomal biogenesis remained uncharacterized until the identification of TFEB as the transcriptional master regulator of autophagy-lysosomal biogenesis [7, 16].
TFEB, initially primarily localized in the cytoplasm, is relocated from the cytoplasm to the nucleus in response to nutrient starvation. Re-feeding after starvation restores TFEB re-localization from the nucleus within minutes [16, 28, 29]. In vivo studies also showed that starvation in mice results in TFEB nuclear localization [16, 30]. Treatment with small molecules, such as rapamycin and Torin1, which regulate molecules upstream of TFEB, such as mTORC1, could also induce its nuclear localization. Even though rapamycin has been proposed as an mTORC1 inhibitor, there are several studies that appear to contradict rapamycin-induced TFEB translocation [31, 32]. This is mainly due to the partial inhibition activity of rapamycin on mTOR, as compared to that of Torin1 [33]. Instead of mTOR, transient receptor potential channel mucolipin 1 (TRPML1) is proposed as a new target of rapamycin. Rapamycin specifically and directly binds to TRPML1, thus activating its calcium ion release channel. Calcium ions then activates calcineurin, causing TFEB nuclear translocation, and promoting autophagy-lysosomal gene expression [32]. In addition, other conditions have also been found to promote the nuclear localization of TFEB, including endoplasmic reticulum (ER) stress [34], physical exercise [35], infections [36–38], mitochondrial damage [39], and inflammation [38].
TFEB signaling
mTOR-dependent/-independent TFEB regulation
TFEB activation and cellular localization are mainly controlled by protein-protein interactions and post-translational modifications (PTMs). Notably, the subcellular localization of TFEB is tightly controlled by phosphorylation (Fig. 3).
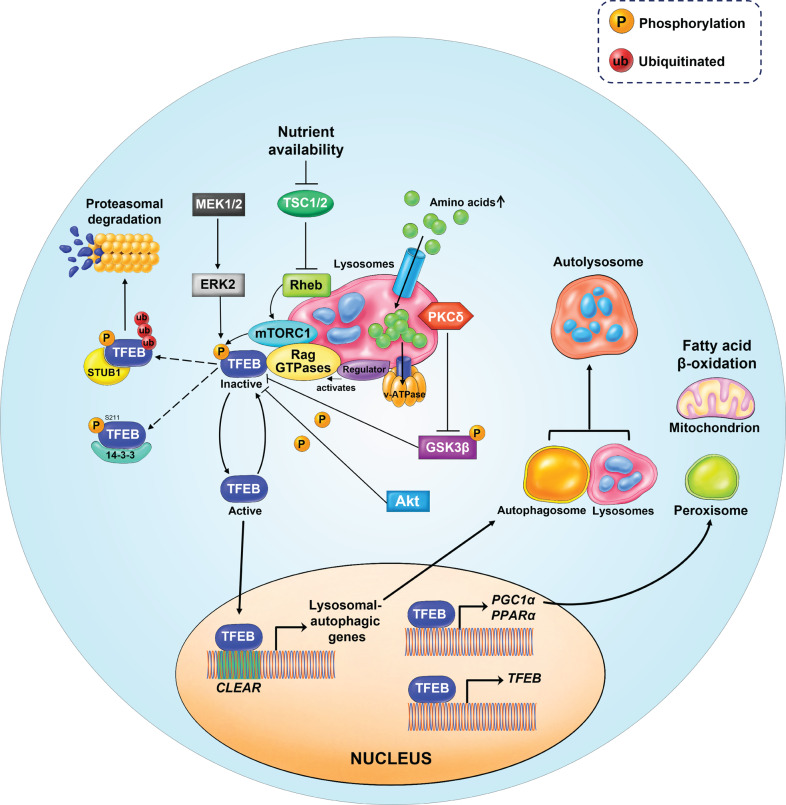
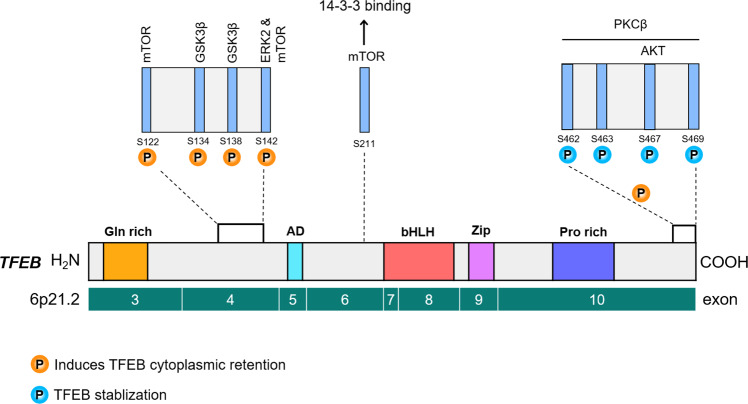
mTOR kinase is a major kinase known to phosphorylate TFEB and regulate its intracellular localization. In nutrient-rich conditions, for instance, amino stimulation, mTORC1 is recruited to the lysosomal membrane. Amino acids can freely cross the lysosomal membrane and accumulate inside the lysosomes. These amino acids are ‘sensed’ by the lysosomal lumen and signal to the Rag GTPases through the v-ATPase-Ragulator complex [40]. At the lysosomal surface, GTP-bound Ras homolog enriched in the brain (Rheb) promotes the activation of mTORC1 [40–42]. Notably, active Rag GTPases also bind to TFEB and recruit it to the lysosomal surface [43], where it is phosphorylated by the mTORC1 complex [28, 29, 31, 44] (Fig. 5) at several serine(S) residues: S122, S142, and S211. However, mTORC1-mediated phosphorylation of TFEB at S211 induces TFEB cytoplasmic retention, through binding to the cytosolic chaperone protein 14-3-3 [28, 29, 31, 44] (Fig. 4). S142 TFEB phosphorylation is mediated not only by mTORC1 but also by extracellular signal-regulated kinase 2 (ERK2) [16, 31]. Recently, STIP1 homology and U-Box containing protein 1 (STUB1), a chaperone-dependent E3 ubiquitin ligase, was found to modulate TFEB activity by targeting phosphorylated TFEB (S142 and S211) for ubiquitin-mediated proteasomal degradation [45]. This suggests that both TFEB intracellular localization and stability are dependent on its phosphorylation status [45] (Fig. 5).
Fig. 5. Schematic model of TFEB upstream regulation by mTORC1.
In nutrient-rich conditions, for instance, amino stimulation, mTORC1 is recruited to the lysosomal membrane. Amino acids can freely cross the lysosomal membrane and accumulate inside the lysosomes. These amino acids are ‘sensed’ by the lysosomal lumen and signal to the Rag GTPases through the v-ATPase-Ragulator complex. Activated Rag GTPases recruit both TFEB and mTORC1 to the lysosomal membrane. Rheb promotes the activation of membrane-localized mTORC1. Active mTORC1 phosphorylates TFEB, thus causing its inactivation and inhibiting its nuclear translocation. In addition, TFEB can also be phosphorylated by ERK2, AKT, and GSK3β. Phosphorylated TFEB is recognized by STUB1, an E3-ligase, which then targets TFEB for proteasomal degradation. However, in nutrient- deprived conditions, TFEB becomes active and free from phosphorylation. Active TFEB then translocates to the nucleus, binds to the CLEAR motif, and increases the expression of genes involved in autophagy/lysosomal biogenesis. TFEB also binds to its own promoters and upregulates its expression through a self-regulatory feedback loop. TFEB is responsible for the transcriptional activation of PGC1α, the co-activator of the nuclear receptor peroxisome proliferator-activated receptor α (PPARα) and a key regulator of lipid metabolism and mitochondrial biogenesis.
Fig. 4. Phosphorylation sites in the TFEB domain structure and their regulatory role.
TFEB can be phosphorylated at multiple serine residues by upstream kinases, such as mTOR, GSK3β, ERK2, AKT, and PKCβ. Phosphorylation of TFEB by PKCβ promotes TFEB protein stabilization. Moreover, functional studies show that TFEB localization to the lysosomal membrane is important for its inhibition by mTORC1 complex-mediated phosphorylation.
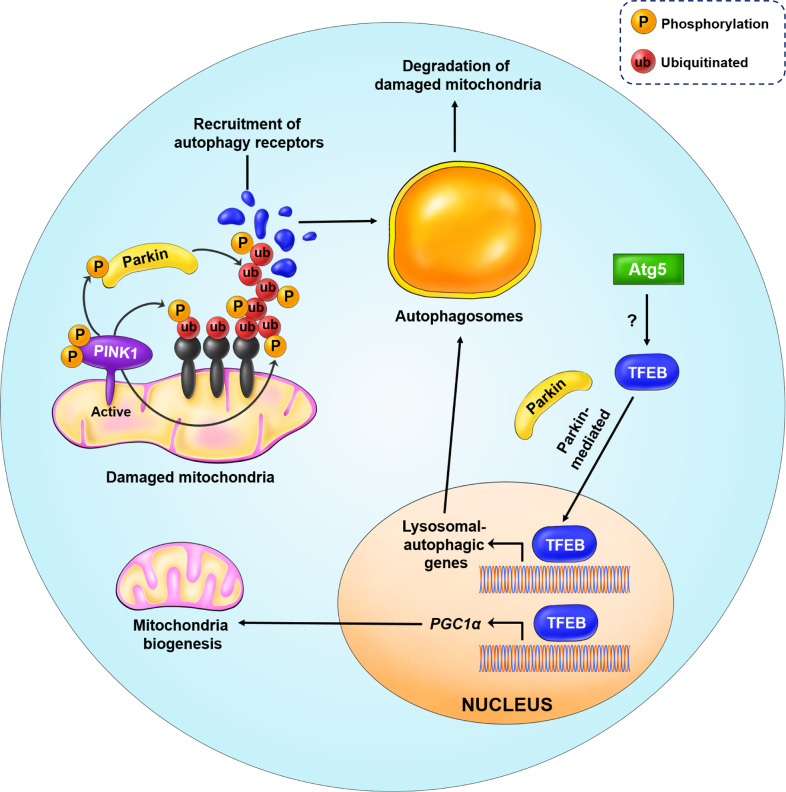
Mitophagy is a process by which damaged mitochondria are degraded through autophagy. Mitochondrial damage induces phosphatase and tensin homolog (PTEN)-induced putative kinase 1 (PINK1) to recruit Parkin, an E3 ligase, to the outer membrane of mitochondria. Parkin ubiquitinates mitochondrial proteins, and the malfunctioning and damaged mitochondria are then delivered to the autophagosome for subsequent destruction [46]. However, mitochondrial stress can activate TFEB in an mTOR-independent manner. Induction of mitophagy by either oligomycin or anti-mycin A results in TFEB nuclear localization in a PINK1- and Parkin-dependent manner. In addition, Parkin-mediated nuclear localization of TFEB upon mitophagy induction is regulated downstream of autophagy-related gene 5 (ATG5). ATG5 KO largely blocked TFEB nuclear localization in Parkin-overexpressing cells (Fig. 6) [39]. TFEB also regulates cellular lipid metabolism via a starvation-induced regulatory loop. Notably, liver-specific deletion of TFEB and rescue experiments in the liver revealed that TFEB plays a key role in regulating liver lipid metabolism [5]. TFEB induces the expression of peroxisome proliferator-activated receptor alpha (PPARα) and its co-activator, peroxisome proliferator-activated receptor gamma co-activator 1-alpha (PGC1α), a known central regulator of mitochondrial biogenesis and fatty acid oxidation (Figs. 5 and 6).
Fig. 6. Regulation of PINK1/Parkin-mediated mitophagy by TFEB.
In the case of mitochondria damage and loss of membrane potential, PINK1 is recruited to the OMM and activated through auto-phosphorylation. PINK1 triggers mitophagy initiation by phosphorylating the ubiquitin attached to the damaged and misfolded OMM proteins. Simultaneously, PINK1 also phosphorylates Parkin, an E3 ligase, causing its activation. Activated Parkin further amplifies the signal by further ubiquitinating misfolded OMM proteins. In addition, phosphorylated ubiquitin also recruits autophagy receptors (blue) to induce autophagosome formation, for degradation of damaged mitochondria. This process requires the transcriptional activity of nuclear TFEB to induce the expression of genes involved in autophagy biogenesis. Additionally, ATG5 activates TFEB via an unknown mechanism, while TFEB nuclear localization is Parkin-dependent. TFEB also induces the expression of PGC1α, a master regulator of mitochondrial biogenesis.
Recently, it has been uncovered that Yes-associated protein (YAP)/TFEB signaling is responsible for the autophagic cell death and development of cardiomyopathy in LSDs [47]. YAP, which is an oncogene, physically interacts with TFEB to promote excessive accumulation of autophagosomes in a mouse model of LSD with cardiomyopathy. Conditional KO of Rag A/B induces nuclear accumulation of YAP in the mouse hearts. Nuclear endogenous YAP physically interacts with TFEB to promote the transcription of TFEB target genes, which are involved in autophagosome formation. This causes the accumulation of undigested autophagosomes, resulting in hypertrophic cardiomyopathy in LSDs [47].
Upstream regulators of TFEB
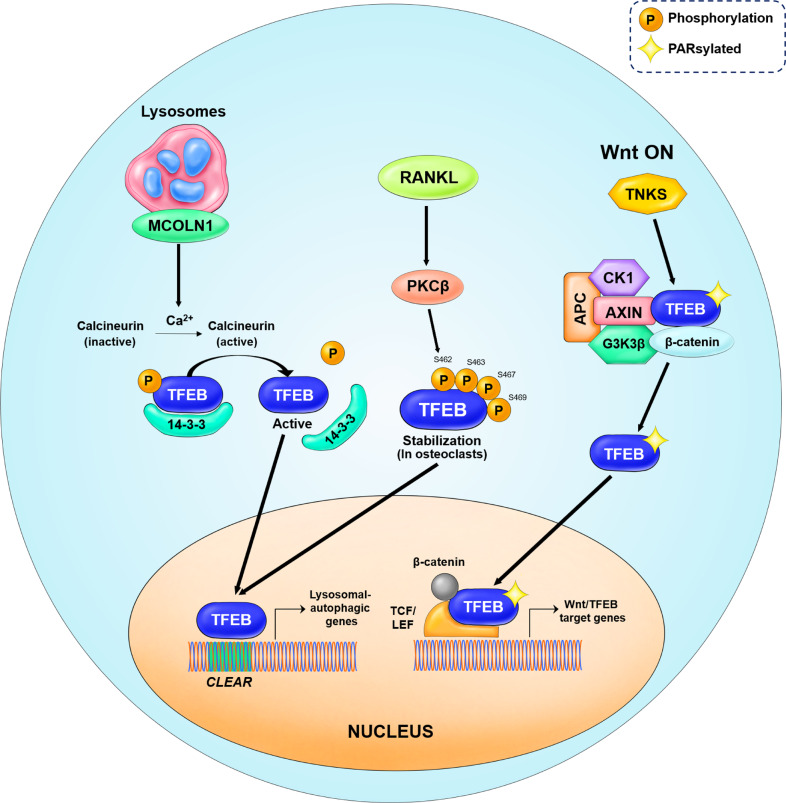
After activation of PKCβ, one of the numerous PKC isoforms, by receptor activator of nuclear factor κB ligand (RANKL), PKCβ phosphorylates multiple serine residues (S462, S463, S467, and S469) located in C-terminal region of the TFEB protein, which resulted in increased TFEB nuclear localization [48, 49] (Fig. 7). In turn, the PKC isoforms PKCα and PKCδ inhibit the phosphorylation of TFEB by GSK3β, to induce TFEB activation and nuclear translocation [50] (Fig. 5).
Fig. 7. Schematic model of the upstream regulators of TFEB.
In starvation conditions, calcium is released from lysosomes through the mucolipin 1 (MCOLN1) channel protein. Calcium ions induce the activation of calcineurin, a phosphatase, resulting in TFEB dephosphorylation. Dephosphorylated TFEB detaches from the 14-3-3 chaperone proteins and enters the nucleus to induce autophagy-lysosomal biogenesis. PKCβ activation is mediated by RANKL. The active PKCβ phosphorylates multiple serine residues in the C-terminal region of the TFEB protein. Such phosphorylation leads to TFEB nuclear translocation, inducing lysosomal biogenesis and secretion of lysosomal hydrolases in osteoclasts. In the Wnt ON condition, TFEB undergoes the PARsylation post-translational modification. The PARsylated TFEB translocates to the nucleus to form a transcription factor complex with β-catenin-TCF/LEF1. This complex regulates a subset of genes that are distinct from the canonical β-catenin target genes, known as Wnt/TFEB target genes.
ERK2, a master regulator of cellular growth, is also the main protein kinase that phosphorylates TFEB in nutrient-rich conditions in most cell types [16, 28, 29, 31]. Similar to mTORC1, ERK2 also localizes to subcellular compartments, including the lysosome surface [51]. Phosphorylation of TFEB by ERK2 at S142 promotes TFEB cytosolic retention, whereas inhibition of ERK2, either through its depletion or inhibitor treatment, promotes nuclear localization of TFEB [16] (Fig. 5).
The results of in vitro kinase assays and mutation of modification sites indicate that TFEB can also be phosphorylated by GSK3β at S134 and S138, leading to cytoplasmic retention [50] (Figs. 4 and 5). TFEB phosphorylation at these sites appears to direct it to the lysosomal surface, where it interacts with mTORC1. Double mutation of S134 and S138 to alanine not only decreases TFEB lysosomal localization, but also reduces its interaction with 14-3-3 chaperone proteins, thus promoting TFEB nuclear localization.
Akt phosphorylates TFEB at S467 and inhibits its nuclear translocation [52] (Figs. 4 and 5). Consistent with this, treatment with an Akt inhibitor induces TFEB nuclear localization. Mutation of the Akt phosphorylation site S467 to alanine induces TFEB nuclear translocation under nutrient-rich conditions, with this effect appearing to be independent of mTOR-dependent TFEB phosphorylation. Thus, it will be crucial to parse out the individual contributions of each of the multiple phosphorylation sites in the serine-rich region of TFEB, to obtain a more complete understanding of TFEB regulation.
According to the TFEB nuclear translocation model, TFEB needs to undergo dephosphorylation to be active. Calcineurin is a serine/threonine phosphatase that connects calcium signaling and the phosphorylation states of numerous substrates, such as transcription factors and proteins associated with mitochondria and microtubules. Calcineurin is usually activated by an increase in intracellular calcium concentrations [53, 54]. Under nutrient deprived conditions, calcium is released from the lysosomes through mucolipin 1 (MCOLN1), to induce the activation of calcineurin, which then dephosphorylates TFEB at phosphorylated serine residues. Dephosphorylated TFEB, i.e., the active form, enters the nucleus and induces autophagy-lysosomal biogenesis [55] (Fig. 7). A schematic of TFEB signaling is presented in Figs. 5, 6, and 7.
Folliculin (FLCN), a well-known tumor suppressor gene for Birt-Hogg-Dubé (BHD) syndrome, is an emerging mechanism of TFEB control [56, 57]. FLCN was first identified as the gene responsible for BHD syndrome, which affects the skin, kidneys, and lungs [57]. FLCN often forms a complex with folliculin-interacting proteins 1 and 2 (FNIP1/2) and this FLCN/FNIP complex acts as a GTPase-activating protein (GAP) for Rag GTPases (Rag A, Rag B, Rag C, and Rag D), which are small GTPases [42]. RagGTPase complex is composed of a formation of two pairs, Rag A/B with Rag C/D. In the inactive state, Rag A/B bind to GDP and Rag C/D bind to GTP. However upon activation, for instance by amino acid-stimulation or growth factors, the FLCN/FNIP complex elicits its GAP activity toward Rag C/D, resulting in Rag C/D binding to GDP and Rag A/B binding to GTP, thus activating Rags [42]. FLCN is required for the recruitment of mTORC1 to the lysosomal membrane and its subsequent activation by the Rheb protein. Active mTORC1 then phosphorylates and inactivates TFEB, thus inhibiting its nuclear localization [58–61].
Exogenous and endogenous factors such as drugs, pathogens, crystals, and silicas can induce lysosomal membrane permeabilization (LMP)/rupture. Recently, TFEB has been proposed as a key mediator for lysosomal homeostasis after lysosomal damage [62]. TFEB is activated during lysosomal damage and its activation is dependent on LC3 lipidation, which is mediated by the ATG conjugation system. During lysosomal damage, LC3 is recruited to the lysosomal surface and undergoes lipidation. Lipidated LC3 interacts with TRPML1, which is a calcium channel, triggering large amount of calcium efflux essential for TFEB activation. Moreover, cells lacking autophagy initiation factors such ATG3, ATG7, and ATG5 and LC3 lipidation factors (LC3A, LC3B, LC3C, and GABARAP) show TFEB inactivation during lysosomal damage [62]. TFEB activation by lipidated LC3 is essential to prevent the progression of lysosomal damage.
Acetylation is a key PTM that acts by controlling protein properties such as stability, solubility, enzymatic function, and subcellular localization [63]. Lysine acetyltransferases (KATs), such as E1A binding protein p300 (also known as KAT3B) encoded by the EP300 gene, acetylate proteins at the ε-amino group of lysine residues [64]. p300 is mainly localized in the nucleus, but can be moved to the cytoplasm depending on intracellular signaling. p300 appears to acetylate many ATG proteins that control autophagy at multiple levels. p300 knockdown can induce autophagy, whereas its overexpression inhibits autophagy [64]. Activated mTORC1 interacts with p300 and phosphorylates it, resulting in its activation. As a result, starvation-induced autophagy is inhibited, while lipogenesis is promoted [65]. Interestingly, p300 promotes TFEB acetylation [66–68], suggesting that the mTOR pathway modulates p300 activity to control TFEB subcellular localization.
TFEB in disease
Lysosomal storage disorders
LSDs are usually caused by defects in genes encoding enzymes that are involved in various aspects of lysosomal homeostasis and are responsible for the degradation of macromolecules [69]. The molecular mechanisms by which TFEB affects LSDs, such as multiple sulfatase deficiency (MSD), Pompe disease, and Batten disease are discussed in the following section.
MSD is caused by mutation of the sulfatase-modifying factor-1 gene (SUMF1) [70, 71]. SUMF1 post-translationally modifies the catalytic site of sulfatases, which is essential for activation of sulfatases. Deficiency of active SUMF1 leads to the simultaneous loss of function of 17 sulfatases, most of which are lysosomal [72]. The main feature of MSD is the accumulation of un-degraded sulfated glycosaminoglycans (GAGs), which subsequently cause neurodegeneration in humans. Systemic adenovirus-mediated overexpression of TFEB in adult MSD mice resulted in reduction of GAGs in the liver and skeletal muscles, tissue inflammation, and cell death [17]. TFEB-induced GAG clearance involves lysosomal exocytosis [17].
Pompe disease is caused by deficiency of the lysosomal enzyme acid α-glucosidase (GAA; acid maltase), which catabolizes glycogen to glucose in acidic lysosomes. The near-complete lack of GAA activity results in the accumulation of glycogen, enlargement of lysosomes, and presence of aberrant autophagic debris in skeletal muscle fibers. The disease symptoms include muscle weakness, feeding difficulties, hypertrophic cardiomyopathy, and respiratory infections [73]. The pathogenesis of muscle damage in Pompe disease has been studied using a GAA KO mouse model (GFP-LC3:GAA−/−) [25]. Adenovirus-mediated overexpression of TFEB in Pompe muscle reduced autophagic build-up and cleared the load of glycogen [25]. In addition, TFEB treatment stimulated an increase in lysosomal number; moreover, a glycogen clearance effect was mainly mediated by lysosomal exocytosis.
Batten disease is a disorder comprising a family of 13 fatal diseases that constitute inherited neurodegenerative disorders of genetically distinct origins. Each of the 13 diseases is abbreviated as CLN succeeded by a numeral. At the cellular level, neuronal accumulation of autofluorescent ceroid lipopigments is observed in patients with Batten disease [74, 75]. However, the biochemical function of CLN3 remains unknown. Increasing endogenous TFEB levels by oral administration of trehalose to Cln3Δex7/8 mice, a well-recognized model of Batten disease, showed therapeutic benefits [52]. Trehalose treatment also cleared the electron-dense material typical of Batten pathology in cortical neurons. Biochemically, trehalose caused Akt inhibition, thereby enhancing TFEB nuclear translocation independently of mTORC1. Nevertheless, the mechanism underlying how TFEB activation reduces the lysosomal storage of ceroid lipopigment in CLN3 KO neurons and the biochemical connection between CLN3 and the accumulation of ceroid lipopigment requires further study.
Neurodegenerative diseases
AD is an age-associated neurodegenerative disease characterized by pathological hallmarks of extracellular Aβ plaques and intracellular Tau tangles [76]. Results from a comprehensive study of the autophagic process from the initial to late stages of AD in the human hippocampus suggested that autophagy is upregulated throughout the AD process. Notably, higher activation of TFEB was observed in glia than in neurons [77]. However, several studies have also reported reduced TFEB activity in AD. The microRNA (miR)-128 targets TFEB mRNA, and thus, reduces its expression [7]. Patients with AD exhibit increased hippocampal expression of miR-128 [78], which decreases TFEB expression and Aβ clearance [79]. A separate relationship between TFEB function and AD pathology was suggested by results showing that PSEN deficiency inhibits the activation of TFEB. Consistent with this, neuron-specific TFEB deletion resulted in Aβ and p-Tau accumulation in the mouse brain [80]. Another group also reported that TFEB overexpression markedly reduced paired-helical filament Tau levels in the P301S tauopathy model mice [81]. The TFEB signaling mechanisms in AD are summarized in Table 1.
Table 1.
TFEB relevant studies in Alzheimer’s disease.
| TFEB Signaling mechanism | Tissue | Phenotype | Reference |
|---|---|---|---|
| Reduced TFEB expression resulted in lysosomal deficits | AD patient lymphocytes and monocytes | [79] | |
| Selective nuclear TFEB exclusion | AD patient brain samples | Negative correlation between pathology & nuclear TFEB levels | [129] |
| TFEB is retained in cytosol hence lysosomal activity is reduced in AD patient fibroblasts & iPSC-derived human | Double presenilin KO cells, AD patient fibroblasts & iPSC-derived human AD neurons | [80] | |
| APOE ɛ4 may compete with TFEB for binding to CLEAR elements in the lysosomal gene promoters & inhibition of the CLEAR network genes | APOE ɛ4/ɛ4 AD patients. | [130] | |
| Increased TFEB expression | AD patient fibroblasts carrying the familial presenilin-1 A246E mutation | [131] | |
| TFEB levels are unchanged however, upregulation of a subset of CLEAR network genes | presenilin-1 conditional KO mice | [132] | |
| Transcriptome upregulation of TFEB targets | 5x-FAD mice | [133] | |
| Enhanced levels of TFEB and TFE3 resulted in elevated expression of autophagy target genes | Hippocampus of AD subjects | [77] | |
| GSK inhibitor VIII induces TFEB nuclear translocation | reduced APP levels in vitro | [134] | |
| CNS delivery of TFEB selectively cleared p-Tau species | rTg4510 mouse tauopathy model | Rescue of behavioral phenotypes, synaptic deficits, & neurodegeneration | [18] |
| Neuron specific TFEB overexpression resulted in decline of toxic p-Tau & lipofuscin levels in the cortex and hippocampus | P301S tauopathy mouse model | Improved performance in memory & learning tests | [81] |
| Astrocyte-specific delivery of TFEB promoted Aβ uptake & clearance | hippocampus of of APP/PS1 transgenic mice | Reduced Aβ levels & hippocampal plaque load | [19] |
| SIRT1 causes the deacetylation of TFEB increased lysosomal biogenesis & microglial degradation of fibrillary Aβ | ex vivo brain slices of APP/PS1 mice | Reduction of amyloid plaque | [135] |
| GCN5 causes TFEB acetylation leading to inhibition of autophagy and lysosome biogenesis | hTau Drosophila model | hTau aggregates & neurotoxicity | [136] |
| Curcumin analog C1 activated TFEB, increased autophagy and lysosomal activity | 5xFAD, P301S and 3xTg-AD mice | Improved synaptic & cognitive function | [137] |
PD is caused by the neurodegeneration of dopamine neurons in the substantia nigra, and is characterized by the accumulation of Lewy bodies [82]. PD-associated molecules, such as α-synuclein (A53T and A30P mutants), Parkin, and PINK1 have been reported to regulate autophagy pathway [82]. Postmortem examination of brain samples from patients with PD showed accumulation of autophagosomes (AP) accompanied by autophagic cell death in dopaminergic neurons [83]. TFEB is excluded from the nucleus of dopaminergic neurons in patients with PD [22]. Genetic activation of lysosomal biogenesis through TFEB overexpression inhibits lysosomal depletion and reverses induced cell death.
Phosphorylation of TFEB increases the association of TFEB with YWHA/14-3-3 proteins, leading to cytosolic retention of TFEB [29]. Moreover, Decressac et al. showed that both α-synuclein and 14-3-3 interacted with TFEB in α-synuclein-overexpressing midbrain tissue [22]. α-Synuclein overexpression leads to the cytoplasmic retention of TFEB, inhibiting the autophagy-lysosomal pathway and, thereby, its own clearance. Notably, TFEB is also involved in a positive transcriptional feedback loop with PGC1α, as PARIS (parkin interacting substrate)-mediated suppression of PGC1α also reduces the expression of TFEB [84]. The TFEB signaling mechanisms in PD are summarized in Table 2.
Table 2.
TFEB relevant studies in neurodegenerative diseases.
| Disease | TFEB Signaling mechanism | Tissue | Phenotype | Reference |
|---|---|---|---|---|
| Parkinson’s disease | α-synuclein interacts with TFEB, sequestering it away from its canonical 14-3-3 partners and preventing its transactivation & nuclear import | AAV–α-syn rat model, postmortem human brains | Accumulation of α-syn oligomers | [22] |
| Elevated PARIS levels and reduction of PGC1α-TFEB signaling | Parkin Q311X mutant mice | Mitochondrial impairment & neuronal cell loss | [138] | |
| Viral TFEB overexpression rescued lysosomal breakdown and autophagic vacuole accumulation | MPTP-induced dopaminergic neurotoxicity in mice | Reduction of α–synuclein aggregates & neuroprotection | [21] | |
| 2-Hydroxypropyl-β-cyclodextrin is a TFEB activator which promote autophagic clearance of α-synuclein | H4/α-syn-GFP cells | [23] | ||
| CCI-79 is a mTOR inhibitor resulting in nuclear translocation of TFEB | AAV–α-syn injected rats | Absence of motor impairments | [22] | |
| ALS/FTD | TFEB promotes autophagy by enhancing the expression of Beclin-1 | Spinal cord of SOD1-G39A transgenic mice, SOD1-G93A NSC-34 cell model | [139] | |
| Low nuclear TFEB levels | ALS patient brain samples | [129] | ||
| Reduced mTOR activity resulted in increase of TFEB and loss of C9orf72 enhances autophagic flux, | brain homogenates of C9orf72 KO mice | C9orf72 KO mice died prematurely | [140] | |
| Loss of TDP‐43 enhanced the nuclear translocation of TFEB | TDP‐43 depleted Drosophila model | Severe neurodegenerative phenotype | [141] | |
| Trehalose-induced TFEB activation is regulated by PPP3 | immortalized motor neurons expressing GFP-SOD1A4V or GFP-SOD1G93A | Clearance of insoluble GFP-SOD1 mutant protein | [142] | |
| Ibudilast enhances TFEB levels & increased autophagic flux to clear the pathogenic aggregates | TDP-43 NSC-34 cells | Protection of motor neurons from cell death induced by TDP-43 aggregations | [143] | |
| Impaired TFEB nuclear import by the C9orf72-hexanucleotide repeat expansion | C9-ALS human motor cortex. | [144] | ||
| Huntington disease | Inhibition of PGC1α-mediated transcription impaired TFEB levels and function | N171-82Q transgenic mice | [84] | |
| TFEB overexpression promotes efficient clearance of polyQ-Htt | striatum of zQ175 HD mice | Lowered mHTT levels | [86] |
HD is an autosomal-dominant neurodegenerative disease caused by abnormal expansion of cytosine-adenine-guanine (CAG) repeats in the huntingtin (HTT) gene [85]. The beneficial effects of TFEB have been demonstrated in in vitro models of HD. TFEB overexpression has been shown to reduce the accumulation of polyglutamine (polyQ)-expanded HTT in the rat HD43 striatal cell model [7]. A follow-up study revealed that PGC-1α is a regulator of TFEB in the clearance of mutant HTT aggregates [84]. Increased PGC-1α induced the expression of TFEB, which cleared the mutant HTT protein aggregates and restored mitochondrial function [84]. Moreover, exogenous expression of TFEB in the striatum of zQ175 HD-mutant mice induced autophagy and lysosome activity as well as efficient clearance of mutant HTT protein aggregates [86]. The TFEB signaling mechanisms in HD are summarized in Table 2.
Cancer
Based on emerging evidence, it is believed that cancer cells exploit TFEB-mediated transcriptional activation of autophagy-lysosomal biogenesis (ALB) for their oncogenic survival [87]. Chromosomal translocations involving the MiT/TFE family, resulting in TFEB, and TFE3 gene fusion, cause a particular type of renal cell carcinoma (RCC), referred to as translocation RCC. TFE3, and less frequently TFEB gene fusions, consistently preserve the TFEB/TFE3 open reading frame and DNA binding domains [88]. The TFEB and TFE3 chromosomal translocations have also been reported in pediatric renal carcinoma [88–90] and alveolar soft part sarcoma [91].
The TFEB fusion partner MALAT1 has also been implicated in a subset of pediatric renal neoplasms. 6p21/11q13 chromosomal translocation, an extremely rare phenomenon, results in the fusion of the regulatory region of the non-coding MALAT1 gene with the coding region of the TFEB gene (MALAT1-TFEB) [88, 92]. Augmentation of the intrinsic pro-oncogenic potential of TFEB is a major consequence of gene fusion, as the TFEB oncogene becomes highly expressed, owing to the introduction of a new active promoter [93]. Moreover, renal cancer may also be induced through TFEB amplification. In particular, a group of aggressive RCCs that share morphological features exhibits genomic amplification in the 6p21.1 region, which includes the proximal TFEB and VEGFA genes that are thereby co-amplified [94]. In addition, kidney-specific TFEB overexpression in transgenic mice caused renal clear cells, severe cystic pathology, renal cysts, and papillary carcinomas with liver metastases [95].
Altered TFEB expression is also associated with pancreatic ductal adenocarcinoma (PDAC) tumorigenesis and pancreatic cancer cell proliferation [87, 96]. MITF, TFE3, and TFEB remain constitutively active and are localized in the nucleus of PDAC cells regardless of nutrient status, owing to their interaction with the nucleocytoplasmic transporters importin 8 (IPO8) and importin 7 (IPO7), as suggested by the decreased nuclear levels of these proteins in PDAC cell lines with IPO7 and IPO8 knockdown [87]. Nuclear localization of TFEB in primary PDAC cell lines was also observed under starvation conditions [97].
TFEB depletion has been associated with reduced migratory and invasive phenotypes of oral squamous cell carcinoma (OSCC) and non-small cell lung cancer (NSCLC) cell lines, suggesting that it may regulate cellular migration [98, 99]. Tumors with high TFEB expression and ALB display greater regrowth capability and metastasis [98]. In another study, p53, a tumor suppressor, was found to regulate the TFEB-dependent autophagy-lysosomal pathway in lung cancer cells (Table 3). Specifically, p53 inhibition increased TFEB nuclear translocation and TFEB-mediated autophagic activity [100].
Table 3.
TFEB relevant studies in cancer.
| Disease | TFEB Signaling mechanism | Tissue | Phenotype | Reference |
|---|---|---|---|---|
| Pancreatic cancer | Overexpression of TFEB in PDAC | Human PDAC samples | [97] | |
| TFEB levels are negatively correlated to KRAS expression | Primary PDAC cell lines | [97] | ||
| Lung cancer | TFEB overexpression induced lysosomal biogenesis | NSCLC samples | Enhanced metastatic and regrowth capability | [98] |
| Inhibition of p53 increased TFEB nuclear transport and p53 controlled autophagy-lysosome pathway by TFEB | Lung cancer cell lines (A549 & H1299) | [100] | ||
| Breast cancer | TFEB levels are associated with LAMP2A, CTSD, LC3A, and HIF2-α expression | T1-stage samples | [145] | |
| Glioblastoma | PI3K inhibitor GDC-0941, increased the expression of TFEB via inhibition of mTORC1 and stimulates lysosomal expansion and maturation | Glioblastoma U87MG & T98G cell lines | GDC-0941 causes apoptosis in glioblastoma cells | [146] |
| Oral squamous cell carcinomas (OSCC) | TFEB depletion reduced the invasiveness and aggressiveness, but elevated the adhesion ability of OSCC cell lines | Human OSCC cell lines | TFEB induces OSCC progression and metastasis | [99] |
| Colorectal cancer (CRC) | TFEB controls autophagy by enhancing Beclin1 transcription | CRC cell lines (HCT-116 & LoVo) | TFEB expression correlated with rapid progression, deep mucosal infiltration, lymphatic propagation, & aggressive clinical aspects of CRC | [147] |
NSCLC non-small cell lung carcinoma, PDAC pancreatic ductal adenocarcinoma.
BHD is an inherited disorder caused by loss-of-function mutations in the mTORC1 regulator, FLCN, that is defined by kidney cancer [101]. Recently, Ballabio and colleagues reported that TFEB overexpression in humans and mice induced kidney cysts and renal cancer, and this phenotype is markedly identical to that caused by the loss of FLCN expression [102]. This implied that TFEB activation is a key driver of the renal phenotype and mTORC1 hyperactivity is associated with BHD syndrome. In the normal condition, amino-acid-mediated activation of RagC and RagD GTPases induces the phosphorylation of S6K, 4E-BP1, and TFEB and inhibits nuclear translocation. FLCN is a positive regulator of RagC/D, which is critical for mTORC1-mediated phosphorylation of TFEB. In BHD syndrome, depletion of FLCN function leads to the nuclear localization of TFEB and hyperactivation of mTORC1 [102].
Tissue-specific function
TFEB functions in liver lipid metabolism
Lipophagy is a selective autophagy process where the lipid droplets accumulated in the cytoplasm are shuttled to the lysosome to be degraded by acid lipase, thus generating free fatty acids and free cholesterol [103]. The redistribution and recycle process of intracytoplasmic lipid droplets via lipophagy is essential for the maintenance of cellular lipid homeostasis.
Pioneer work by Settembre et al. showed that in starvation, TFEB is upregulated through a self-regulatory feedback loop [5]. TFEB is also responsible for the transcriptional activation of PGC1α, a co-activator of the nuclear PPARα and main cellular regulator of lipid degradation [5]. TFEB-mediated stimulation of the PGC1α-PPARα program induces genes essential for free fatty acid oxidation in the mitochondria and peroxisomes. The free fatty acids released from the lysosome are broken down by enzymes in the peroxisomes and mitochondria, to generate ATP [5]. Livers from HFD-fed TFEB-injected mice showed decreased lipid content, and normal weight, indicating that TFEB overexpression ameliorated the deleterious effects of the HFD by accelerating lipid breakdown [5]. Notably, when C. elegans are exposed to starvation, TFEB (HLH-30) replaces the repressor Max-like 3 (MXL-3) and increases lysosomal lipase expression [4].
TFEB is enriched in the promoter of the cholesterol 7α-hydroxylase (CYP7A1) gene in human and mouse hepatocytes. As a result, TFEB induces hepatic bile acid synthesis, leading to the inhibition of cholesterol accumulation and metabolic disorders in Western diet-fed mice [104]. Moreover, TFEB activation was induced in diet-induced atherogenesis modeled using APOE KO mice with myeloid cell-specific sorting nexin 10 (SNX10) deficiency, which caused the promotion of lysosomal acid lipase and mitochondrial fatty acid oxidation [105]. Recently, Lee et al. found that TFEB is also induced by organelle stress. Deficient accumulation of the lysosomal digestion fragment, lipofuscin, in adipose tissue macrophages (ATMs) of obese mice or humans caused the activation of TFEB, indicative of obesity-dependent lysosomal stress in ATMs [106].
Recently, it has been discovered that the fasting-induced hormone Fgf21-TFEB axis integrates extracellular hormonal signaling and lysosome homeostasis to orchestrate lipid metabolism during fasting [30]. Fgf21 depletion inhibits hepatic lysosomal function and enhances lipid accumulation in the liver tissue, by blocking TFEB nuclear shuttling. This results in the suppression of TFEB target genes involved in lipid metabolism, which include lipid oxidation and intracellular lipid degradation [30].
TFEB in skeletal muscle function
The AMPK-PGC1α pathway is known to coordinate contraction-mediated adaptive responses; however, TFEB has emerged as a novel metabolic coordinator [107]. Using TFEB muscle-specific gain-and loss-of-function approaches, Mansueto et al. found that TFEB is a key mediator of metabolic flexibility in the muscle during exercise [35]. During exercise, TFEB translocate into the nucleus and increases the expression of genes related to glucose metabolism and mitochondrial homeostasis, which are necessary for muscle function and endurance. Whether TFEB levels are reduced in patients with metabolic disorders should also be evaluated.
The recent identification of FGF/TFEB/FAM134B signaling event represents the first study that implicates the role of TFEB in ER-phagy and its physiological relevance during skeletal development in vivo [108]. Nuclear TFEB enhances the transcription of lysosomal genes and FAM134B, an ER-phagy receptor and newly identified target gene of TFEB.
Role of TFEB in the centrality and dynamics of lysosomes
TFEB in autophagic fluxes, lysosomal exocytosis, and lysosomal positioning
With respect to intracellular lysosome pool, it has been proposed that aberrant activation of TFEB increases the lysosome pool to the proximity of the plasma membrane and promotes lysosome docking and fusion with the plasma membrane [17].
By transcriptionally regulating the genes involved in both autophagy and lysosomal biogenesis, TFEB mediates the autophagy/lysosomal-mediated degradation pathway which includes autophagosome and autolysosome formation. In addition, serine–threonine kinase RIP1 (RIPK1), a regulator of cell survival and cell death, negatively regulates autophagic flux via TFEB, to control sensitivity to cell death [109]. TFEB also transcriptionally regulates lysosomal exocytosis [17]. TFEB overexpression enhances trafficking and plasma membrane proximity of lysosomes. TFEB triggers Ca2+ elevation through activating its target gene, MCOLN1. Activation of MCOLN1 increases the lysosome pools/population in the proximity of the plasma membrane [17].
Recently, it has been found that lysosomal positioning may influence lysosomal functions and this is mediated by the TFEB/TMEM55B/JIP4 pathway, which coordinates lysosome movement in response to stress conditions [110]. In brief, TFEB dynamically modulates the lysosomal positioning in response to nutrient deprivation, by directly regulating the expression of levels of TMEM55B, which in turn recruits JIP4 to the lysosomes [110].
TFEB in autophagy and lysosome-dependent cell death (LDCD)
Cell death mediated by components of autophagy machinery is termed as autophagy-dependent cell death (ADCD) [111–113]. The activation of TFEB shows anticancer effects by induction of ADCD in proliferating cells of melanoma and multiple myeloma [114, 115]. Reduction of NAD+ intracellular stores enhances ADCD in multiple myeloma cells, by promoting nuclear translocation of TFEB and transcription of autophagy-related genes [114]. Mammalian metabolite, Dendrogenin A, binds to ligand-dependent transcription factor, NR1H2, to induce TFEB gene expression, which further activates ADCD and shows anticancer effects in mice [115]. In the nucleus, YAP interacts with TFEB and promotes ADCD, leading to cardiomyopathy in RagA/B KO mice [47].
In breast cancer, the interaction of signal transducer and activator of transcription 3 (STAT3) with nuclear TFEB increases LDCD, thus acting as an antitumor response [116]. L-leucyl-L-leucine methyl ester (LLME) causes the release of lysosomal enzymes, leading to LMP, followed by LDCD. BHLHE40/41 are TFEB target genes. Specifically in LLME-induced LDCD, TFEB protects the cells, whereas BHLHE40/41 sensitizes the cells [117]. DNA damage response causes inhibition of mTORC1, resulting in TFEB/TFE3 activation as an early response. TFE3/TFEB stimulates genes associated with LMP and apoptosis, upon prolonged DNA damage [118].
TFEB in lysophagy
Generally, Leu-Leu-OMe is used to induce lysosomal damage resulting in the accumulation of Galectin-3 puncta on damaged lysosomes. The critical step in the beginning of autophagy response is initiated by cytosolic lectins, Galectin-3, which recognize β-galactoside located on the luminal surface of organelle and recruit autophagic receptors such as TRIM16 [119]. The tripartite motif protein, TRIM16 regulates autophagy by interacting with ATG16L1, ULK1, and Beclin-1. In Galectin-3 KO cells exposed to lysosomal damage, there was an enhancement of TFEB nuclear translocation. Lysosomal membrane damage has been reported to stimulate nuclear translocation of TFEB and promote lysosomal biogenesis [119, 120]. The function of TRIM16 in mTOR inhibition and its effects on TFEB during autophagy [119] all add to lysosome replacement.
Recently Nakamura and colleagues proposed that lipidated LC3 is necessary for TFEB activation in response to lysosomal damage [62, 121]. The lysosomal damage in mammalian cells causes leakage of calcium from lysosomes. The released calcium induces LC3 lipidation (LC-3II) and localizes on lysosomes. Subsequently, the lipidated LC3 associates with, MCOLN1, and expedites the release of substantial quantities of calcium necessary for TFEB activation into the cytoplasm [62].
Autophagy-lysosomal-independent role of TFEB
Notably, in triple-negative breast cancer (TNBC), TFEB has a novel lysosomal independent function in response to DNA damage [122]. TFEB depletion reduces the viability of TNBC cells and promotes caspase-3-dependent apoptosis. However, TFEB retains the ability to protect TNBC cells from apoptosis in the presence of lysosomal inhibitors [122]. Similarly, high basal levels of autophagy in pancreatic cancer cells are not related to TFEB activation, as TFEB knockdown does not affect autophagic flux under normal conditions [123].
Activation of Wnt signaling induces the PARsylation of TFEB. However, PARsylated TFEB does not activate TFEB-mediated lysosomal target genes [124] (Fig. 7). Additionally, TFEB also regulates the pluripotency transcriptional network in mouse embryonic stem cells (mESCs), independent of ALB. Modulation of TFEB levels inmESCs did not affect the expression of TFEB lysosome-autophagy target genes [125].
In another study, TFEB appeared to induce the differentiation of murine liver stem/progenitor cells into progenitor/cholangiocyte lineage, while inhibiting hepatocyte differentiation in an autophagy-lysosomeal-independent manner. TFEB directly activates Sox9, a precursor and biliary cell marker [126]. In addition to regulating liver cell fate, TFEB also protects against obesity and insulin resistance in an autophagy-independent manner. Specifically, TFEB induction of GDF15 expression is crucial for regulating obesity-induced metabolic diseases [106].
Future perspective
In this review article, we focused on the major advances in TFEB research over the past 30 years, along with its pathophysiological functions and importance. These advances have provided new insights and biological relevance regarding the importance of TFEB transcriptional mechanisms, thus highlighting TFEB as a powerful potential tool for modulating both health and disease.
However, when trying to modify TFEB activity in order to heal diseases, we must keep in mind that the same perturbation of TFEB activity (either activating or inhibiting) can elicit distinct biological outcomes depending on the input conditions or time points of a potential feedback loop. Similar findings were recently published by Pavel et al., who found that inhibiting autophagy activity can result in opposite Hippo signaling output depending on the level of α-catenin: when autophagy activity is inhibited, Hippo signaling is ON or OFF, depending on whether the cells have high or low levels of α-catenin [127].
Cell heterogeneity in TFEB signaling states was also observed between different cell types [128]. One such example is that mTOR could induce TFEB cytoplasmic retention maximally in HeLa cells, while only doing a fraction of that in MCF7 cells. It is suggested that differences of homeostatic mechanisms in different cell types contribute to the heterogeneity of TFEB signaling states.
Activation of TFEB is beneficial in neurological and lysosomal disorders; however, in the case of cancer, constitutively enhanced TFEB may promote tumorigenesis. Therefore, it is important to coordinate TFEB activity carefully and in considerable detail. Activating TFEB activity for a certain period or enhancing specific subsets of TFEB may be suitable alternatives and should be taken into consideration when developing drugs targeting TFEB. Overall, continued efforts and progress toward elucidating the therapeutic potential of TFEB will take us one step closer to advancing TFEB from basic research into clinical and regenerative applications.
Supplementary information
Author contributions
AT and RP planned and wrote the manuscript. CL designed and drew the figures. EJ directed, edited, and finalized the manuscript. All authors have read and approved the final version of the manuscript.
Funding
The authors’ work was supported by grants from the National Research Foundation of Korea (NRF- 2016R1A5A1010764 and 2020R1A2C3013746) to E.J.
Competing interests
The authors declare no competing interests.
Footnotes
Edited by D Rubinsztein
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Anderson Tan, Renuka Prasad.
Supplementary information
The online version contains supplementary material available at 10.1038/s41418-022-01028-6.
References
- 1.Steingrímsson E, Copeland NG, Jenkins NA. Melanocytes and the microphthalmia transcription factor network. Annu Rev Genet. 2004;38:365–411. doi: 10.1146/annurev.genet.38.072902.092717. [DOI] [PubMed] [Google Scholar]
- 2.La Spina M, Contreras PS, Rissone A, Meena NK, Jeong E, Martina JA. MiT/TFE family of transcription factors: an evolutionary perspective. Front Cell Dev Biol. 2020;8:609683. [DOI] [PMC free article] [PubMed]
- 3.Lapierre LR, De Magalhaes Filho CD, McQuary PR, Chu C-C, Visvikis O, Chang JT, et al. The TFEB orthologue HLH-30 regulates autophagy and modulates longevity in Caenorhabditis elegans. Nat Commun. 2013;4:1–8. doi: 10.1038/ncomms3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O’Rourke EJ, Ruvkun G. MXL-3 and HLH-30 transcriptionally link lipolysis and autophagy to nutrient availability. Nat Cell Biol. 2013;15:668–76. doi: 10.1038/ncb2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Settembre C, De Cegli R, Mansueto G, Saha PK, Vetrini F, Visvikis O, et al. TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nat Cell Biol. 2013;15:647–58. doi: 10.1038/ncb2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tognon E, Kobia F, Busi I, Fumagalli A, De Masi F, Vaccari T. Control of lysosomal biogenesis and Notch-dependent tissue patterning by components of the TFEB-V-ATPase axis in Drosophila melanogaster. Autophagy. 2016;12:499–514. doi: 10.1080/15548627.2015.1134080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sardiello M, Palmieri M, Di Ronza A, Medina DL, Valenza M, Gennarino VA, et al. A gene network regulating lysosomal biogenesis and function. Science. 2009;325:473–7. doi: 10.1126/science.1174447. [DOI] [PubMed] [Google Scholar]
- 8.Palmieri M, Impey S, Kang H, di Ronza A, Pelz C, Sardiello M, et al. Characterization of the CLEAR network reveals an integrated control of cellular clearance pathways. Hum Mol Genet. 2011;20:3852–66. doi: 10.1093/hmg/ddr306. [DOI] [PubMed] [Google Scholar]
- 9.Simionato E, Ledent V, Richards G, Thomas-Chollier M, Kerner P, Coornaert D, et al. Origin and diversification of the basic helix-loop-helix gene family in metazoans: insights from comparative genomics. BMC Evolut Biol. 2007;7:1–18. doi: 10.1186/1471-2148-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gyoja F. A genome-wide survey of bHLH transcription factors in the Placozoan Trichoplax adhaerens reveals the ancient repertoire of this gene family in metazoan. Gene. 2014;542:29–37. doi: 10.1016/j.gene.2014.03.024. [DOI] [PubMed] [Google Scholar]
- 11.Carr CS, Sharp PA. A helix-loop-helix protein related to the immunoglobulin E box-binding proteins. Mol Cell Biol. 1990;10:4384–8. doi: 10.1128/mcb.10.8.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hodgkinson CA, Moore KJ, Nakayama A, Steingrímsson E, Copeland NG, Jenkins NA, et al. Mutations at the mouse microphthalmia locus are associated with defects in a gene encoding a novel basic-helix-loop-helix-zipper protein. Cell. 1993;74:395–404. doi: 10.1016/0092-8674(93)90429-t. [DOI] [PubMed] [Google Scholar]
- 13.Steingrímsson E, Moore KJ, Lamoreux ML, Ferré-D’Amaré AR, Burley SK, Zimring DCS, et al. Molecular basis of mouse microphthalmia (mi) mutations helps explain their developmental and phenotypic consequences. Nat Genet. 1994;8:256–63. doi: 10.1038/ng1194-256. [DOI] [PubMed] [Google Scholar]
- 14.Opdecamp K, Nakayama A, Nguyen M, Hodgkinson CA, Pavan WJ, Arnheiter H. Melanocyte development in vivo and in neural crest cell cultures: crucial dependence on the Mitf basic-helix-loop-helix-zipper transcription factor. Development. 1997;124:2377–86. doi: 10.1242/dev.124.12.2377. [DOI] [PubMed] [Google Scholar]
- 15.Steingrímsson E, Tessarollo L, Reid SW, Jenkins NA, Copeland NG. The bHLH-Zip transcription factor Tfeb is essential for placental vascularization. Development. 1998;125:4607–16. doi: 10.1242/dev.125.23.4607. [DOI] [PubMed] [Google Scholar]
- 16.Settembre C, Di Malta C, Polito VA, Arencibia MG, Vetrini F, Erdin S, et al. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332:1429–33. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Medina DL, Fraldi A, Bouche V, Annunziata F, Mansueto G, Spampanato C, et al. Transcriptional activation of lysosomal exocytosis promotes cellular clearance. Developmental Cell. 2011;21:421–30. doi: 10.1016/j.devcel.2011.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Polito VA, Li H, Martini‐Stoica H, Wang B, Yang L, Xu Y, et al. Selective clearance of aberrant tau proteins and rescue of neurotoxicity by transcription factor EB. EMBO Mol Med. 2014;6:1142–60. doi: 10.15252/emmm.201303671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiao Q, Yan P, Ma X, Liu H, Perez R, Zhu A, et al. Enhancing astrocytic lysosome biogenesis facilitates Aβ clearance and attenuates amyloid plaque pathogenesis. J Neurosci. 2014;34:9607–20. doi: 10.1523/JNEUROSCI.3788-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chauhan S, Ahmed Z, Bradfute SB, Arko-Mensah J, Mandell MA, Choi SW, et al. Pharmaceutical screen identifies novel target processes for activation of autophagy with a broad translational potential. Nat Commun. 2015;6:1–15. doi: 10.1038/ncomms9620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dehay B, Bové J, Rodríguez-Muela N, Perier C, Recasens A, Boya P, et al. Pathogenic lysosomal depletion in Parkinson’s disease. J Neurosci. 2010;30:12535–44. doi: 10.1523/JNEUROSCI.1920-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Decressac M, Mattsson B, Weikop P, Lundblad M, Jakobsson J, Björklund A. TFEB-mediated autophagy rescues midbrain dopamine neurons from α-synuclein toxicity. Proc Natl Acad Sci. 2013;110:E1817–26. doi: 10.1073/pnas.1305623110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kilpatrick K, Zeng Y, Hancock T, Segatori L. Genetic and chemical activation of TFEB mediates clearance of aggregated α-synuclein. PloS One. 2015;10:e0120819. doi: 10.1371/journal.pone.0120819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song W, Wang F, Savini M, Ake A, Di Ronza A, Sardiello M, et al. TFEB regulates lysosomal proteostasis. Hum Mol Genet. 2013;22:1994–2009. doi: 10.1093/hmg/ddt052. [DOI] [PubMed] [Google Scholar]
- 25.Spampanato C, Feeney E, Li L, Cardone M, Lim JA, Annunziata F, et al. Transcription factor EB (TFEB) is a new therapeutic target for Pompe disease. EMBO Mol Med. 2013;5:691–706. doi: 10.1002/emmm.201202176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rega LR, Polishchuk E, Montefusco S, Napolitano G, Tozzi G, Zhang J, et al. Activation of the transcription factor EB rescues lysosomal abnormalities in cystinotic kidney cells. Kidney Int. 2016;89:862–73. doi: 10.1016/j.kint.2015.12.045. [DOI] [PubMed] [Google Scholar]
- 27.Bahrami A, Bianconi V, Pirro M, Orafai HM, Sahebkar A. The role of TFEB in tumor cell autophagy: Diagnostic and therapeutic opportunities. Life Sci. 2020;244:117341. doi: 10.1016/j.lfs.2020.117341. [DOI] [PubMed] [Google Scholar]
- 28.Roczniak-Ferguson A, Petit CS, Froehlich F, Qian S, Ky J, Angarola B, et al. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci Signal. 2012;5:ra42–ra. doi: 10.1126/scisignal.2002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martina JA, Chen Y, Gucek M, Puertollano R. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy. 2012;8:903–14. doi: 10.4161/auto.19653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen L, Wang K, Long A, Jia L, Zhang Y, Deng H, et al. Fasting-induced hormonal regulation of lysosomal function. Cell Res. 2017;27:748–63. doi: 10.1038/cr.2017.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Settembre C, Zoncu R, Medina DL, Vetrini F, Erdin S, Erdin S, et al. A lysosome‐to‐nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 2012;31:1095–108. doi: 10.1038/emboj.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang X, Chen W, Gao Q, Yang J, Yan X, Zhao H, et al. Rapamycin directly activates lysosomal mucolipin TRP channels independent of mTOR. PLoS Biol. 2019;17:e3000252. doi: 10.1371/journal.pbio.3000252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y, et al. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem. 2009;284:8023–32. doi: 10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martina JA, Diab HI, Brady OA, Puertollano R. TFEB and TFE 3 are novel components of the integrated stress response. EMBO J. 2016;35:479–95. doi: 10.15252/embj.201593428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mansueto G, Armani A, Viscomi C, D’Orsi L, De Cegli R, Polishchuk EV, et al. Transcription factor EB controls metabolic flexibility during exercise. Cell Metab. 2017;25:182–96. doi: 10.1016/j.cmet.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Visvikis O, Ihuegbu N, Labed SA, Luhachack LG, Alves A-MF, Wollenberg AC, et al. Innate host defense requires TFEB-mediated transcription of cytoprotective and antimicrobial genes. Immunity. 2014;40:896–909. doi: 10.1016/j.immuni.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Campbell GR, Rawat P, Bruckman RS, Spector SA. Human immunodeficiency virus type 1 Nef inhibits autophagy through transcription factor EB sequestration. PLoS Pathog. 2015;11:e1005018. doi: 10.1371/journal.ppat.1005018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pastore N, Brady OA, Diab HI, Martina JA, Sun L, Huynh T, et al. TFEB and TFE3 cooperate in the regulation of the innate immune response in activated macrophages. Autophagy. 2016;12:1240–58. doi: 10.1080/15548627.2016.1179405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nezich CL, Wang C, Fogel AI, Youle RJ. MiT/TFE transcription factors are activated during mitophagy downstream of Parkin and Atg5. J Cell Biol. 2015;210:435–50. doi: 10.1083/jcb.201501002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zoncu R, Bar-Peled L, Efeyan A, Wang S, Sancak Y, Sabatini DM. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H+-ATPase. Science. 2011;334:678–83. doi: 10.1126/science.1207056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, et al. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martina JA, Puertollano R. Rag GTPases mediate amino acid–dependent recruitment of TFEB and MITF to lysosomes. J Cell Biol. 2013;200:475–91. doi: 10.1083/jcb.201209135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vega-Rubin-de-Celis S, Peña-Llopis S, Konda M, Brugarolas J. Multistep regulation of TFEB by MTORC1. Autophagy. 2017;13:464–72. doi: 10.1080/15548627.2016.1271514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sha Y, Rao L, Settembre C, Ballabio A, Eissa NT. STUB 1 regulates TFEB‐induced autophagy–lysosome pathway. EMBO J. 2017;36:2544–52. doi: 10.15252/embj.201796699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Narendra D, Kane LA, Hauser DN, Fearnley IM, Youle RJ. p62/SQSTM1 is required for Parkin-induced mitochondrial clustering but not mitophagy; VDAC1 is dispensable for both. Autophagy. 2010;6:1090–106. doi: 10.4161/auto.6.8.13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ikeda S, Nah J, Shirakabe A, Zhai P, Oka S-I, Sciarretta S, et al. YAP plays a crucial role in the development of cardiomyopathy in lysosomal storage diseases. J Clin Invest. 2021;131:e143173. [DOI] [PMC free article] [PubMed]
- 48.Ferron M, Settembre C, Shimazu J, Lacombe J, Kato S, Rawlings DJ, et al. A RANKL–PKCβ–TFEB signaling cascade is necessary for lysosomal biogenesis in osteoclasts. Genes Dev. 2013;27:955–69. doi: 10.1101/gad.213827.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peña‐Llopis S, Vega‐Rubin‐de‐Celis S, Schwartz JC, Wolff NC, Tran TAT, Zou L, et al. Regulation of TFEB and V‐ATPases by mTORC1. EMBO J. 2011;30:3242–58. doi: 10.1038/emboj.2011.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li Y, Xu M, Ding X, Yan C, Song Z, Chen L, et al. Protein kinase C controls lysosome biogenesis independently of mTORC1. Nat Cell Biol. 2016;18:1065–77. doi: 10.1038/ncb3407. [DOI] [PubMed] [Google Scholar]
- 51.Nada S, Hondo A, Kasai A, Koike M, Saito K, Uchiyama Y, et al. The novel lipid raft adaptor p18 controls endosome dynamics by anchoring the MEK–ERK pathway to late endosomes. EMBO J. 2009;28:477–89. doi: 10.1038/emboj.2008.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Palmieri M, Pal R, Nelvagal HR, Lotfi P, Stinnett GR, Seymour ML, et al. mTORC1-independent TFEB activation via Akt inhibition promotes cellular clearance in neurodegenerative storage diseases. Nat Commun. 2017;8:1–19. doi: 10.1038/ncomms14338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hogan PG, Li H. Calcineurin. Curr Biol. 2005;15:R442–R3. doi: 10.1016/j.cub.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 54.Creamer TP. Calcineurin. Cell Commun Signal. 2020;18:1–12. doi: 10.1186/s12964-020-00636-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Medina DL, Di Paola S, Peluso I, Armani A, De Stefani D, Venditti R, et al. Lysosomal calcium signalling regulates autophagy through calcineurin and TFEB. Nat Cell Biol. 2015;17:288–99. doi: 10.1038/ncb3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Birt AR, Hogg GR, Dubé WJ. Hereditary multiple fibrofolliculomas with trichodiscomas and acrochordons. Arch Dermatol. 1977;113:1674–7. [PubMed] [Google Scholar]
- 57.Nickerson ML, Warren MB, Toro JR, Matrosova V, Glenn G, Turner ML, et al. Mutations in a novel gene lead to kidney tumors, lung wall defects, and benign tumors of the hair follicle in patients with the Birt-Hogg-Dube syndrome. Cancer Cell. 2002;2:157–64. doi: 10.1016/s1535-6108(02)00104-6. [DOI] [PubMed] [Google Scholar]
- 58.Ramírez-Reyes JM, Cuesta R, Pause A. Folliculin: a regulator of transcription through AMPK and mTOR signaling pathways. Front Cell Developmental Biol. 2021;9:961. doi: 10.3389/fcell.2021.667311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang S, Tsun Z-Y, Wolfson RL, Shen K, Wyant GA, Plovanich ME, et al. Lysosomal amino acid transporter SLC38A9 signals arginine sufficiency to mTORC1. Science. 2015;347:188–94. doi: 10.1126/science.1257132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lawrence RE, Fromm SA, Fu Y, Yokom AL, Kim DJ, Thelen AM, et al. Structural mechanism of a Rag GTPase activation checkpoint by the lysosomal folliculin complex. Science. 2019;366:971–7. doi: 10.1126/science.aax0364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fromm SA, Lawrence RE, Hurley JH. Structural mechanism for amino acid-dependent Rag GTPase nucleotide state switching by SLC38A9. Nat Struct Mol Biol. 2020;27:1017–23. doi: 10.1038/s41594-020-0490-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nakamura S, Shigeyama S, Minami S, Shima T, Akayama S, Matsuda T, et al. LC3 lipidation is essential for TFEB activation during the lysosomal damage response to kidney injury. Nat Cell Biol. 2020;22:1252–63. doi: 10.1038/s41556-020-00583-9. [DOI] [PubMed] [Google Scholar]
- 63.Drazic A, Myklebust LM, Ree R, Arnesen T. The world of protein acetylation. Biochimica et Biophysica Acta (BBA)-Proteins Proteom. 2016;1864:1372–401. doi: 10.1016/j.bbapap.2016.06.007. [DOI] [PubMed] [Google Scholar]
- 64.Son SM, Park SJ, Fernandez-Estevez M, Rubinsztein DC. Autophagy regulation by acetylation—implications for neurodegenerative diseases. Exp Mol Med. 2021;53:30–41. doi: 10.1038/s12276-021-00556-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wan W, You Z, Xu Y, Zhou L, Guan Z, Peng C, et al. mTORC1 phosphorylates acetyltransferase p300 to regulate autophagy and lipogenesis. Mol Cell. 2017;68:323–35. doi: 10.1016/j.molcel.2017.09.020. [DOI] [PubMed] [Google Scholar]
- 66.Louphrasitthiphol P, Siddaway R, Loffreda A, Pogenberg V, Friedrichsen H, Schepsky A, et al. Tuning transcription factor availability through acetylation-mediated genomic redistribution. Mol Cell. 2020;79:472–87. doi: 10.1016/j.molcel.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Price ER, Horstmann MA, Wells AG, Weilbaecher KN, Takemoto CM, Landis MW, et al. α-Melanocyte-stimulating hormone signaling regulates expression of microphthalmia, a gene deficient in Waardenburg syndrome. J Biol Chem. 1998;273:33042–7. doi: 10.1074/jbc.273.49.33042. [DOI] [PubMed] [Google Scholar]
- 68.Sato S, Roberts K, Gambino G, Cook A, Kouzarides T, Goding C. CBP/p300 as a co-factor for the Microphthalmia transcription factor. Oncogene. 1997;14:3083–92. doi: 10.1038/sj.onc.1201298. [DOI] [PubMed] [Google Scholar]
- 69.Parenti G, Andria G, Ballabio A. Lysosomal storage diseases: from pathophysiology to therapy. Annu Rev Med. 2015;66:471–86. doi: 10.1146/annurev-med-122313-085916. [DOI] [PubMed] [Google Scholar]
- 70.Cosma MP, Pepe S, Annunziata I, Newbold RF, Grompe M, Parenti G, et al. The multiple sulfatase deficiency gene encodes an essential and limiting factor for the activity of sulfatases. Cell. 2003;113:445–56. doi: 10.1016/s0092-8674(03)00348-9. [DOI] [PubMed] [Google Scholar]
- 71.Dierks T, Schmidt B, Borissenko LV, Peng J, Preusser A, Mariappan M, et al. Multiple sulfatase deficiency is caused by mutations in the gene encoding the human Cα-formylglycine generating enzyme. Cell. 2003;113:435–44. doi: 10.1016/s0092-8674(03)00347-7. [DOI] [PubMed] [Google Scholar]
- 72.Diez-Roux G, Ballabio A. Sulfatases and human disease. Annu Rev Genomics Hum Genet. 2005;6:355–79. doi: 10.1146/annurev.genom.6.080604.162334. [DOI] [PubMed] [Google Scholar]
- 73.Kishnani PS, Hwu W-L, Mandel H, Nicolino M, Yong F, Corzo D, et al. A retrospective, multinational, multicenter study on the natural history of infantile-onset Pompe disease. J Pediatrics. 2006;148:671–6. doi: 10.1016/j.jpeds.2005.11.033. [DOI] [PubMed] [Google Scholar]
- 74.Palmer DN, Fearnley IM, Walker JE, Hall NA, Lake BD, Wolfe LS, et al. Mitochondrial ATP synthase subunit c storage in the ceroid‐lipofuscinoses (Batten disease) Am J Med Genet. 1992;42:561–7. doi: 10.1002/ajmg.1320420428. [DOI] [PubMed] [Google Scholar]
- 75.Anderson GW, Goebel HH, Simonati A. Human pathology in NCL. Biochimica et Biophysica Acta (BBA)-Mol Basis Dis. 2013;1832:1807–26. doi: 10.1016/j.bbadis.2012.11.014. [DOI] [PubMed] [Google Scholar]
- 76.Bertram L, Tanzi RE. Thirty years of Alzheimer’s disease genetics: the implications of systematic meta-analyses. Nat Rev Neurosci. 2008;9:768–78. doi: 10.1038/nrn2494. [DOI] [PubMed] [Google Scholar]
- 77.Bordi M, Berg MJ, Mohan PS, Peterhoff CM, Alldred MJ, Che S, et al. Autophagy flux in CA1 neurons of Alzheimer hippocampus: Increased induction overburdens failing lysosomes to propel neuritic dystrophy. Autophagy. 2016;12:2467–83. doi: 10.1080/15548627.2016.1239003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lukiw WJ. Micro-RNA speciation in fetal, adult and Alzheimer’s disease hippocampus. Neuroreport. 2007;18:297–300. doi: 10.1097/WNR.0b013e3280148e8b. [DOI] [PubMed] [Google Scholar]
- 79.Tiribuzi R, Crispoltoni L, Porcellati S, Di Lullo M, Florenzano F, Pirro M, et al. miR128 up-regulation correlates with impaired amyloid β (1-42) degradation in monocytes from patients with sporadic Alzheimer’s disease. Neurobiol Aging. 2014;35:345–56. doi: 10.1016/j.neurobiolaging.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 80.Reddy K, Cusack CL, Nnah IC, Khayati K, Saqcena C, Huynh TB, et al. Dysregulation of nutrient sensing and CLEARance in presenilin deficiency. Cell Rep. 2016;14:2166–79. doi: 10.1016/j.celrep.2016.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang H, Wang R, Carrera I, Xu S, Lakshmana MK. TFEB overexpression in the P301S model of tauopathy mitigates increased PHF1 levels and lipofuscin puncta and rescues memory deficits. Eneuro. 2016;3:1–18. [DOI] [PMC free article] [PubMed]
- 82.Kalia LV, Lang AE. Evolving basic, pathological and clinical concepts in PD. Nat Rev Neurol. 2016;12:65–6. doi: 10.1038/nrneurol.2015.249. [DOI] [PubMed] [Google Scholar]
- 83.Anglade P, Vyas S, Javoy-Agid F, Herrero M, Michel P, Marquez J, et al. Apoptosis and autophagy in nigral neurons of patients with Parkinson’s disease. Histol Histopathol. 1997;12:25–31. [PubMed]
- 84.Tsunemi T, Ashe TD, Morrison BE, Soriano KR, Au J, Roque RAV, et al. PGC-1α rescues Huntington’s disease proteotoxicity by preventing oxidative stress and promoting TFEB function. Sci Transl Med. 2012;4:142ra97–ra97. doi: 10.1126/scitranslmed.3003799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bar-Yosef T, Damri O, Agam G. Dual role of autophagy in diseases of the central nervous system. Front Cell Neurosci. 2019;13:196. doi: 10.3389/fncel.2019.00196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Vodicka P, Chase K, Iuliano M, Tousley A, Valentine DT, Sapp E, et al. Autophagy activation by Transcription Factor EB (TFEB) in striatum of HD Q175/Q7 Mice. J Huntington’s Dis. 2016;5:249–60. doi: 10.3233/JHD-160211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Perera RM, Stoykova S, Nicolay BN, Ross KN, Fitamant J, Boukhali M, et al. Transcriptional control of autophagy–lysosome function drives pancreatic cancer metabolism. Nature. 2015;524:361–5. doi: 10.1038/nature14587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kuiper RP, Schepens M, Thijssen J, van Asseldonk M, van den Berg E, Bridge J, et al. Upregulation of the transcription factor TFEB in t (6; 11)(p21; q13)-positive renal cell carcinomas due to promoter substitution. Hum Mol Genet. 2003;12:1661–9. doi: 10.1093/hmg/ddg178. [DOI] [PubMed] [Google Scholar]
- 89.Argani P, Lui MY, Couturier J, Bouvier R, Fournet J-C, Ladanyi M. A novel CLTC-TFE3 gene fusion in pediatric renal adenocarcinoma with t (X; 17)(p11. 2; q23) Oncogene. 2003;22:5374–8. doi: 10.1038/sj.onc.1206686. [DOI] [PubMed] [Google Scholar]
- 90.Clark J, Lu Y-J, Sidhar SK, Parker C, Gill S, Smedley D, et al. Fusion of splicing factor genes PSF and NonO (p54 nrb) to the TFE3 gene in papillary renal cell carcinoma. Oncogene. 1997;15:2233–9. doi: 10.1038/sj.onc.1201394. [DOI] [PubMed] [Google Scholar]
- 91.Ladanyi M, Lui MY, Antonescu CR, Krause-Boehm A, Meindl A, Argani P, et al. The der (17) t (X; 17)(p11; q25) of human alveolar soft part sarcoma fuses the TFE3 transcription factor gene to ASPL, a novel gene at 17q25. Oncogene. 2001;20:48–57. doi: 10.1038/sj.onc.1204074. [DOI] [PubMed] [Google Scholar]
- 92.Davis IJ, Hsi B-L, Arroyo JD, Vargas SO, Yeh YA, Motyckova G, et al. Cloning of an Alpha-TFEB fusion in renal tumors harboring the t (6; 11)(p21; q13) chromosome translocation. Proc Natl Acad Sci. 2003;100:6051–6. doi: 10.1073/pnas.0931430100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kauffman EC, Ricketts CJ, Rais-Bahrami S, Yang Y, Merino MJ, Bottaro DP, et al. Molecular genetics and cellular features of TFE3 and TFEB fusion kidney cancers. Nat Rev Urol. 2014;11:465. doi: 10.1038/nrurol.2014.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gupta S, Johnson SH, Vasmatzis G, Porath B, Rustin JG, Rao P, et al. TFEB-VEGFA (6p21. 1) co-amplified renal cell carcinoma: a distinct entity with potential implications for clinical management. Mod Pathol. 2017;30:998–1012. doi: 10.1038/modpathol.2017.24. [DOI] [PubMed] [Google Scholar]
- 95.Calcagnì A, Verschuren E, De Cegli R, Zampelli N, Nusco E, Confalonieri S, et al. Modelling TFE renal cell carcinoma in mice reveals a critical role of WNT signaling. Elife. 2016;5:e17047. doi: 10.7554/eLife.17047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Marchand B, Arsenault D, Raymond-Fleury A, Boisvert F-M, Boucher M-J. Glycogen synthase kinase-3 (GSK3) inhibition induces prosurvival autophagic signals in human pancreatic cancer cells. J Biol Chem. 2015;290:5592–605. doi: 10.1074/jbc.M114.616714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Klein K, Werner K, Teske C, Schenk M, Giese T, Weitz J, et al. Role of TFEB-driven autophagy regulation in pancreatic cancer treatment. Int J Oncol. 2016;49:164–72. doi: 10.3892/ijo.2016.3505. [DOI] [PubMed] [Google Scholar]
- 98.Giatromanolaki A, Kalamida D, Sivridis E, Karagounis IV, Gatter KC, Harris AL, et al. Increased expression of transcription factor EB (TFEB) is associated with autophagy, migratory phenotype and poor prognosis in non-small cell lung cancer. Lung Cancer. 2015;90:98–105. doi: 10.1016/j.lungcan.2015.07.008. [DOI] [PubMed] [Google Scholar]
- 99.Sakamoto H, Yamashita K, Okamoto K, Kadowaki T, Sakai E, Umeda M, et al. Transcription factor EB influences invasion and migration in oral squamous cell carcinomas. Oral Dis. 2018;24:741–8. doi: 10.1111/odi.12826. [DOI] [PubMed] [Google Scholar]
- 100.Zhang Z, Wang H, Ding Q, Xing Y, Xu D, Xu Z, et al. The tumor suppressor p53 regulates autophagosomal and lysosomal biogenesis in lung cancer cells by targeting transcription factor EB. Biomedicine Pharmacother. 2017;89:1055–60. doi: 10.1016/j.biopha.2017.02.103. [DOI] [PubMed] [Google Scholar]
- 101.Schmidt LS, Linehan WM. FLCN: the causative gene for Birt-Hogg-Dubé syndrome. Gene. 2018;640:28–42. doi: 10.1016/j.gene.2017.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Napolitano G, Di Malta C, Esposito A, de Araujo ME, Pece S, Bertalot G, et al. A substrate-specific mTORC1 pathway underlies Birt–Hogg–Dubé syndrome. Nature. 2020;585:597–602. doi: 10.1038/s41586-020-2444-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Khawar MB, Abbasi MH, Rafiq M, Naz N, Mehmood R, Sheikh N. A decade of mighty lipophagy: what we know and what facts we need to know? Oxid Med Cell Longev. 2021;2021:5539161. [DOI] [PMC free article] [PubMed]
- 104.Wang Y, Gunewardena S, Li F, Matye DJ, Chen C, Chao X, et al. An FGF15/19-TFEB regulatory loop controls hepatic cholesterol and bile acid homeostasis. Nat Commun. 2020;11:1–16. doi: 10.1038/s41467-020-17363-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.You Y, Bao W-L, Zhang S-L, Li H-D, Li H, Dang W-Z, et al. Sorting nexin 10 mediates metabolic reprogramming of macrophages in atherosclerosis through the Lyn-Dependent TFEB signaling pathway. Circulation Res. 2020;127:534–49. doi: 10.1161/CIRCRESAHA.119.315516. [DOI] [PubMed] [Google Scholar]
- 106.Kim J, Kim SH, Kang H, Lee S, Park S-Y, Cho Y, et al. TFEB–GDF15 axis protects against obesity and insulin resistance as a lysosomal stress response. Nat Metab. 2021;3:410–27. doi: 10.1038/s42255-021-00368-w. [DOI] [PubMed] [Google Scholar]
- 107.Markby GR, Sakamoto K. Transcription factor EB and TFE3: new metabolic coordinators mediating adaptive responses to exercise in skeletal muscle? Am J Physiol-Endocrinol Metab. 2020;319:E763–8. doi: 10.1152/ajpendo.00339.2020. [DOI] [PubMed] [Google Scholar]
- 108.Cinque L, De Leonibus C, Iavazzo M, Krahmer N, Intartaglia D, Salierno FG, et al. MiT/TFE factors control ER‐phagy via transcriptional regulation of FAM 134B. EMBO J. 2020;39:e105696. doi: 10.15252/embj.2020105696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yonekawa T, Gamez G, Kim J, Tan AC, Thorburn J, Gump J, et al. RIP 1 negatively regulates basal autophagic flux through TFEB to control sensitivity to apoptosis. EMBO Rep. 2015;16:700–8. doi: 10.15252/embr.201439496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Willett R, Martina JA, Zewe JP, Wills R, Hammond GR, Puertollano R. TFEB regulates lysosomal positioning by modulating TMEM55B expression and JIP4 recruitment to lysosomes. Nat Commun. 2017;8:1–17. doi: 10.1038/s41467-017-01871-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Galluzzi L, Vitale I, Aaronson SA, Abrams JM, Adam D, Agostinis P, et al. Molecular mechanisms of cell death: recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. 2018;25:486–541. doi: 10.1038/s41418-017-0012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Green DR, Galluzzi L, Kroemer G. Metabolic control of cell death. Science. 2014;345:1250256. [DOI] [PMC free article] [PubMed]
- 113.Kriel J, Loos B. The good, the bad and the autophagosome: exploring unanswered questions of autophagy-dependent cell death. Cell Death Differ. 2019;26:640–52. doi: 10.1038/s41418-018-0267-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Cea M, Cagnetta A, Patrone F, Nencioni A, Gobbi M, Anderson KC. Intracellular NAD+ depletion induces autophagic death in multiple myeloma cells. Autophagy. 2013;9:410–2. doi: 10.4161/auto.22866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Silvente-Poirot S, Segala G, Poirot MC, Poirot M. Ligand-dependent transcriptional induction of lethal autophagy: A new perspective for cancer treatment. Autophagy. 2018;14:555–7. doi: 10.1080/15548627.2018.1425059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Li L, Sun B, Gao Y, Niu H, Yuan H, Lou H. STAT3 contributes to lysosomal-mediated cell death in a novel derivative of riccardin D-treated breast cancer cells in association with TFEB. Biochemical Pharmacol. 2018;150:267–79. doi: 10.1016/j.bcp.2018.02.026. [DOI] [PubMed] [Google Scholar]
- 117.Carey KL, Paulus GL, Wang L, Balce DR, Luo JW, Bergman P, et al. TFEB transcriptional responses reveal negative feedback by BHLHE40 and BHLHE41. Cell Rep. 2020;33:108371. doi: 10.1016/j.celrep.2020.108371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Brady OA, Jeong E, Martina JA, Pirooznia M, Tunc I, Puertollano R. The transcription factors TFE3 and TFEB amplify p53 dependent transcriptional programs in response to DNA damage. Elife. 2018;7:e40856. doi: 10.7554/eLife.40856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chauhan S, Kumar S, Jain A, Ponpuak M, Mudd MH, Kimura T, et al. TRIMs and galectins globally cooperate and TRIM16 and galectin-3 co-direct autophagy in endomembrane damage homeostasis. Developmental Cell. 2016;39:13–27. doi: 10.1016/j.devcel.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.San Min Leow SXS, Chua GV, Liang Shen LL. Sub-lethal oxidative stress induces lysosome biogenesis via a lysosomal membrane permeabilization-cathepsin-caspase 3-transcription factor EB-dependent pathway. Oncotarget. 2017;8:16170. doi: 10.18632/oncotarget.14016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Nakamura S, Akayama S, Yoshimori T. Autophagy-independent function of lipidated LC3 essential for TFEB activation during the lysosomal damage responses. Autophagy. 2021;17:581–3. doi: 10.1080/15548627.2020.1846292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Slade L, Biswas D, Ihionu F, El Hiani Y, Kienesberger PC, Pulinilkunnil T. A lysosome independent role for TFEB in activating DNA repair and inhibiting apoptosis in breast cancer cells. Biochemical J. 2020;477:137–60. doi: 10.1042/BCJ20190596. [DOI] [PubMed] [Google Scholar]
- 123.Kim JH, Lee J, Cho Y-R, Lee S-Y, Sung G-J, Shin D-M, et al. TFEB supports pancreatic cancer growth through the transcriptional regulation of glutaminase. Cancers. 2021;13:483. doi: 10.3390/cancers13030483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kim S, Song G, Lee T, Kim M, Kim J, Kwon H, et al. PARsylated transcription factor EB (TFEB) regulates the expression of a subset of Wnt target genes by forming a complex with β-catenin-TCF/LEF1. Cell Death Differ. 2021;28:2555–70. [DOI] [PMC free article] [PubMed]
- 125.Tan A, Prasad R, Jho E-H. TFEB regulates pluripotency transcriptional network in mouse embryonic stem cells independent of autophagy–lysosomal biogenesis. Cell Death Dis. 2021;12:1–14. doi: 10.1038/s41419-021-03632-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Pastore N, Huynh T, Herz NJ, Klisch TJ, Brunetti L, Kim KH, et al. TFEB regulates murine liver cell fate during development and regeneration. Nat Commun. 2020;11:1–14. doi: 10.1038/s41467-020-16300-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Pavel M, Park SJ, Frake RA, Son SM, Manni MM, Bento CF, et al. alpha-Catenin levels determine direction of YAP/TAZ response to autophagy perturbation. Nat Commun. 2021;12:1703.. doi: 10.1038/s41467-021-21882-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zapata PAM, Beese CJ, Jünger A, Dalmasso G, Brady NR, Hamacher-Brady A. Time course decomposition of cell heterogeneity in TFEB signaling states reveals homeostatic mechanisms restricting the magnitude and duration of TFEB responses to mTOR activity modulation. BMC Cancer. 2016;16:1–19. doi: 10.1186/s12885-016-2388-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wang H, Wang R, Xu S, Lakshmana MK Transcription factor EB is selectively reduced in the nuclear fractions of Alzheimer’s and amyotrophic lateral sclerosis brains. Neurosci J. 2016;2016:4732837. [DOI] [PMC free article] [PubMed]
- 130.Parcon PA, Balasubramaniam M, Ayyadevara S, Jones RA, Liu L, Reis RJS, et al. Apolipoprotein E4 inhibits autophagy gene products through direct, specific binding to CLEAR motifs. Alzheimer’s Dement. 2018;14:230–42. doi: 10.1016/j.jalz.2017.07.754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Coffey EE, Beckel JM, Laties AM, Mitchell CH. Lysosomal alkalization and dysfunction in human fibroblasts with the Alzheimer’s disease-linked presenilin 1 A246E mutation can be reversed with cAMP. Neuroscience. 2014;263:111–24. doi: 10.1016/j.neuroscience.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zhang X, Garbett K, Veeraraghavalu K, Wilburn B, Gilmore R, Mirnics K, et al. A role for presenilins in autophagy revisited: normal acidification of lysosomes in cells lacking PSEN1 and PSEN2. J Neurosci. 2012;32:8633–48. doi: 10.1523/JNEUROSCI.0556-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Landel V, Baranger K, Virard I, Loriod B, Khrestchatisky M, Rivera S, et al. Temporal gene profiling of the 5XFAD transgenic mouse model highlights the importance of microglial activation in Alzheimer’s disease. Mol Neurodegeneration. 2014;9:1–18. doi: 10.1186/1750-1326-9-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Parr C, Carzaniga R, Gentleman SM, Van Leuven F, Walter J, Sastre M. Glycogen synthase kinase 3 inhibition promotes lysosomal biogenesis and autophagic degradation of the amyloid-β precursor protein. Mol Cell Biol. 2012;32:4410–8. doi: 10.1128/MCB.00930-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Bao J, Zheng L, Zhang Q, Li X, Zhang X, Li Z, et al. Deacetylation of TFEB promotes fibrillar Aβ degradation by upregulating lysosomal biogenesis in microglia. Protein Cell. 2016;7:417–33. doi: 10.1007/s13238-016-0269-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wang Y, Huang Y, Liu J, Zhang J, Xu M, You Z, et al. Acetyltransferase GCN5 regulates autophagy and lysosome biogenesis by targeting TFEB. EMBO Rep. 2020;21:e48335. doi: 10.15252/embr.201948335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Song JX, Malampati S, Zeng Y, Durairajan SSK, Yang CB, Tong BCK, et al. A small molecule transcription factor EB activator ameliorates beta‐amyloid precursor protein and Tau pathology in Alzheimer’s disease models. Aging Cell. 2020;19:e13069. doi: 10.1111/acel.13069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Siddiqui A, Bhaumik D, Chinta SJ, Rane A, Rajagopalan S, Lieu CA, et al. Mitochondrial quality control via the PGC1α-TFEB signaling pathway is compromised by parkin Q311X mutation but independently restored by rapamycin. J Neurosci. 2015;35:12833–44. doi: 10.1523/JNEUROSCI.0109-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Chen Y, Liu H, Guan Y, Wang Q, Zhou F, Jie L, et al. The altered autophagy mediated by TFEB in animal and cell models of amyotrophic lateral sclerosis. Am J Transl Res. 2015;7:1574. [PMC free article] [PubMed] [Google Scholar]
- 140.Ugolino J, Ji YJ, Conchina K, Chu J, Nirujogi RS, Pandey A, et al. Loss of C9orf72 enhances autophagic activity via deregulated mTOR and TFEB signaling. PLoS Genet. 2016;12:e1006443. doi: 10.1371/journal.pgen.1006443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Xia Q, Wang H, Hao Z, Fu C, Hu Q, Gao F, et al. TDP‐43 loss of function increases TFEB activity and blocks autophagosome–lysosome fusion. EMBO J. 2016;35:121–42. doi: 10.15252/embj.201591998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Rusmini P, Cortese K, Crippa V, Cristofani R, Cicardi ME, Ferrari V, et al. Trehalose induces autophagy via lysosomal-mediated TFEB activation in models of motoneuron degeneration. Autophagy. 2019;15:631–51. doi: 10.1080/15548627.2018.1535292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Chen Y, Wang H, Ying Z, Gao Q. Ibudilast enhances the clearance of SOD1 and TDP-43 aggregates through TFEB-mediated autophagy and lysosomal biogenesis: The new molecular mechanism of ibudilast and its implication for neuroprotective therapy. Biochem Biophys Res Commun. 2020;526:231–8. doi: 10.1016/j.bbrc.2020.03.051. [DOI] [PubMed] [Google Scholar]
- 144.Cunningham KM, Maulding K, Ruan K, Senturk M, Grima JC, Sung H, et al. TFEB/Mitf links impaired nuclear import to autophagolysosomal dysfunction in C9-ALS. Elife. 2020;9:e59419. doi: 10.7554/eLife.59419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Giatromanolaki A, Sivridis E, Kalamida D, Koukourakis MI. Transcription factor EB expression in early breast cancer relates to lysosomal/autophagosomal markers and prognosis. Clin Breast Cancer. 2017;17:e119–e25. doi: 10.1016/j.clbc.2016.11.006. [DOI] [PubMed] [Google Scholar]
- 146.Enzenmüller S, Gonzalez P, Karpel-Massler G, Debatin K-M, Fulda S. GDC-0941 enhances the lysosomal compartment via TFEB and primes glioblastoma cells to lysosomal membrane permeabilization and cell death. Cancer Lett. 2013;329:27–36. doi: 10.1016/j.canlet.2012.09.007. [DOI] [PubMed] [Google Scholar]
- 147.Liang J, Jia X, Wang K, Zhao N. High expression of TFEB is associated with aggressive clinical features in colorectal cancer. OncoTargets Ther. 2018;11:8089. doi: 10.2147/OTT.S180112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.