Abstract
The unprecedented outbreak of the severe acute respiratory syndrome (SARS) Coronavirus-2, across the globe, triggered a worldwide uproar in the search for immediate treatment strategies. With no specific drug and not much data available, alternative approaches such as drug repurposing came to the limelight. To date, extensive research on the repositioning of drugs has led to the identification of numerous drugs against various important protein targets of the coronavirus strains, with hopes of the drugs working against the major variants of concerns (alpha, beta, gamma, delta, omicron) of the virus. Advancements in computational sciences have led to improved scope of repurposing via techniques such as structure-based approaches including molecular docking, molecular dynamic simulations and quantitative structure activity relationships, network-based approaches, and artificial intelligence-based approaches with other core machine and deep learning algorithms. This review highlights the various approaches to repurposing drugs from a computational biological perspective, with various mechanisms of action of the drugs against some of the major protein targets of SARS-CoV-2. Additionally, clinical trials data on potential COVID-19 repurposed drugs are also highlighted with stress on the major SARS-CoV-2 targets and the structural effect of variants on these targets. The interaction modelling of some important repurposed drugs has also been elucidated. Furthermore, the merits and demerits of drug repurposing are also discussed, with a focus on the scope and applications of the latest advancements in repurposing.
Keywords: SARS-CoV-2, COVID-19, Variants of concern, Drug repurposing, Protein targets, Computational sciences
Introduction
The sudden outbreak of SARS-CoV-2 in 2019 took the world by storm and despite there being vaccines, numerous other alternative treatment approaches are also being researched. Coronavirus disease-2019 (COVID-19) is a communicable disease caused by severe acute respiratory syndrome coronavirus-2 (SARS Cov-2). This disease was first detected in Wuhan, China, and has expanded its reach globally, leading to myriad deaths. According to World Health Organisation, the total number of COVID-19 confirmed cases worldwide was found to be 53,22,01,219 and total number of deaths accounted to 63,05,358 as of 10th June, 2022. Among many regions, Europe holds the top position in highest number of confirmed COVID-19 cases (22,24,17,177) then followed by America (15,89,83,746), Western Pacific (6,17,35,224), South-East Asia (5,82,17,287), Eastern Mediterranean (2,18,07,376), and Africa (90,39,645) respectively [1]. These statistics reveal that even today, the disease is still infecting several people, many of whom die due to complications. This pandemic requires quick and easily accessible knowledge for hastening the development of treatments and diagnostic tests [2]. However, the development of specific drugs for this disease takes several years, paving the way for exploring alternative approaches. One such potent strategy is repurposing drugs, for alternative utilization of already existing and approved drugs. These drugs must have already undergone all safety and clinical trials, which would further speed up the drug discovery process. The COVID-19 pandemic has ensured research to explore various alternatives, drug repurposing being one of them, as part of the disease management strategy. Several drugs such as chloroquine, hydroxychloroquine, remdesivir, tocilizumab, kaletra, and favipiravir are currently being testing in clinal trials for repurposing against COVID-19 [2].
SARS-CoV-2: structural and genomic perspectives
SARS-CoV 2 belongs to the Nidovirales family, under the beta coronaviruses genera along with other viruses such as Middle East Respiratory Syndrome Coronavirus (MERS-CoV), HCoV-HKU1, and the HCoV-OC43 (Human Coronavirus variants) [3]. The replication of SARS-CoV-2 occurs with the help of mRNAs. These viruses have the largest RNA genome among other viruses that have RNA as their genetic material. The RNA genome is approximately 30 kb in size and runs in the 5’ to 3’ direction, with single-stranded positive-sense RNA (+ssRNA). A cap at 5’ end and poly-A-tail at 3’ end at the end of the genomic RNA is a feature of the genetic material. Several open-reading frames in varying numbers are present in the genomic RNA and it encodes for 4 structural proteins, 16 non-structural proteins, and a few accessory proteins [4].
The nucleocapsid in the virus protects the viral RNA genomes from external environmental conditions. At the relaxed position, the nucleocapsid appears to be in a helical shape, but within the virus, it appears spherical. The genome of SARS-CoV-2 codes for four main structural proteins when compared to five in other coronaviruses leaving out the hemagglutinin-esterase (HE) protein. Nucleocapsid (N), membrane (M), envelope (E), and spike (S) are the four important structural proteins expressed by the SARS-CoV-2 virus [3]. On the surface of the virion, three structural proteins (M, E, and S) are present, while the nucleocapsid protein resides in the core region bound to genomic RNA. The spike proteins of the virus help in the attachment, entry, pathogenesis, and tissue tropism, whereas, the infection is carried out by the envelope protein. Ribonucleoprotein is formed with nucleocapsid protein’s assistance and the membrane proteins aid in the overall viral architecture [4]. The SARS-CoV-2 virus enters with the help of spike proteins by binding to the human angiotensin-converting enzyme 2 (hACE2) through its receptor-binding domain (RBD) and human proteases that make them proteolytically active [5].
Variants of SARS-CoV-2
Beta, gamma, delta, and omicron are the latest variants of concern for the SARS-CoV-2 virus. South Africa was first to report the beta variant of COVID-19, gamma was noted first in Brazil, the delta in India, and omicron in western Europe. Omicron which is the most recent variant of COVID-19 carried all the mutations that were present in the previous variants along with some new improved functions such as showing partial resistance to the vaccine and enhanced transmissibility compared to others [6]. The major coronavirus variants can be classified into two [7]: previously circulating variants of concern (VoC) and currently circulating variants of concern (Table 1).
Table 1.
The major variants of concern (VOCs) of SARS-CoV-2 reported worldwide
| WHO label | GISAID Code | PANGO lineage | Next strain clade | Date of VOC designation | Additional amino acid changes causing mutations |
|---|---|---|---|---|---|
| Alpha | GRY | B.1.1.7 | 20I (V1) |
VOC: 18-Dec-2020 Recent VOC: 09-Mar-2022 |
Nil |
| Beta | GH/501Y.V2 | B.1.351 | 20H (V2) |
VOC: 18-Dec-2020 Recent VOC: 09-Mar-2022 |
Nil |
| Gamma | GR/501Y.V3 | P.1 | 20 J (V3) |
VOC: 11-Jan-2021 Recent VOC: 09-Mar-2022 |
Nil |
| Delta | G/478 K.V1 | B.1.617.2 | 21A, 21I, 21 J | VOC: 11-May-2021 |
+S:K417N +S:K484K |
| Omicron | GR/484A | B.1.1.529 | 21 K | VOC: 26-Nov-2021 | +S:R346K |
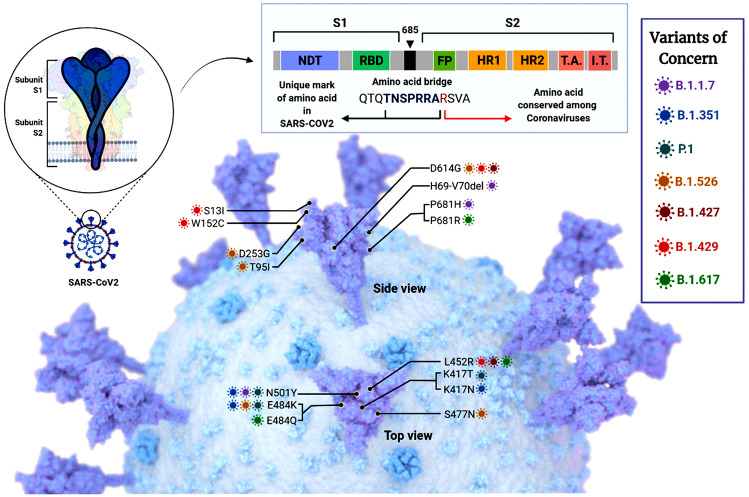
These five major variants of concerns are the current targets for research to find repurposed drugs for exploring treatment strategies. In addition to this, certain amino acids are conserved among the coronavirus viruses, while some have a unique mark in the amino acid indicating lineage mutations. The variants of concern belong to various lineages. B.1.1.7 has the mutations H69-V70del, P618H, and N501Y, all indicating alterations in the amino acids. Lineage B.1.351 has mutations K417N, N501Y, and E484K. Likewise, another variant of concern of lineage P.1 has mutations N501Y, K417T, and E484K. B.1.526 lineage is mutated at D614G, T95I, D253G, and S477N. B.1.427 has mutations at L452R and D614G. B.1.429 is mutated at sites D614G, S13I, W152C, and L452R. B.1.617 variant has variations P681R, E484Q, and L452R [8] (Fig. 1).
Fig. 1.
Various SARS-CoV-2 mutants showing the different lineages and the change in amino acids that caused the variation. The amino acid conserved among the coronaviruses is also shown. The different lineages and the mutations are also shown. B.1.1.7 has the mutations H69-V70del, P618H, and N501Y, all indicating alterations in the amino acids. Lineage B.1.351 has mutations K417N, N501Y, and E484K. Likewise, another variant of concern of lineage P.1 has mutations N501Y, K417T, and E484K. B.1.526 lineage is mutated at D614G, T95I, D253G, and S477N. B.1.427 has mutations at L452R and D614G. B.1.429 is mutated at sites D614G, S13I, W152C, and L452R. B.1.617 variant has variations P681R, E484Q, and L452R
Variants from all SARS-CoV-2 lineages
Apart from the variants identified in the most common lineages, there are approximately 1,516 SARS-CoV-2 lineages to date [4, 9]. To identify the variants in all the existing lineages and those that exist not only in the spike protein of SARS-CoV-2, this study also performed a search in https://cov-glue.cvr.gla.ac.uk/ and summarized the different types of variants (Tables 2, 3, 4, and 5). Synonymous, non-synonymous, deletions, and non-coding variations were identified, and changes in the amino acid positions, and the reference and mutated codon as screened in all 1516 lineages of the coronavirus. As per the latest data as of 6th June 2022, 131 non-synonymous mutations have been noted, 51 synonymous variants, 11 deletions, and 11 non-coding variations. These mutations are in various regions of the virus, including the spike proteins, nucleocapsid proteins, different open reading frames, and membrane and envelope proteins.
Table 2.
Non-synonymous mutations of SARS-CoV-2 from all the lineages showing the ORFs, amino acid mutation, reference codon, and the mutated codon
| Sl. No | ORF | Mutation | RefCodon | MutCodon |
|---|---|---|---|---|
| Non-Synonymous | ||||
| 1 | S | L5F | Ctt | Ttt |
| 2 | ORF1ab/nsp5A-B | L89F | Ctt | Ttt |
| 3 | ORF1ab/nsp6 | L37F | ttG | ttT |
| 4 | S | V1176F | Gtt | Ttt |
| 5 | ORF3a | G172V | gGt | gTt |
| 6 | ORF1ab/nsp14A2-B | N129D | Aat | Gat |
| 7 | N | P67S | Cct | Tct |
| 8 | S | P26S | Cct | Tct |
| 9 | ORF1ab/nsp13 | E341D | gaG | gaT |
| 10 | S | T1027I | aCt | aTt |
| 11 | ORF1ab/nsp2 | K81N | aaG | aaT |
| 12 | S | D138Y | Gat | Tat |
| 13 | S | H655Y | Cat | Tat |
| 14 | ORF1ab/nsp12 | F694Y | tTt | tAt |
| 15 | ORF1ab/nsp3 | S370L | tCa | tTa |
| 16 | ORF1ab/nsp6 | I162V | Att | Gtt |
| 17 | ORF1ab/nsp5A-B | K90R | aAg | aGg |
| 18 | ORF1ab/nsp6 | T77A | Act | Gct |
| 19 | ORF1ab/nsp14A2-B | A394V | gCt | gTt |
| 20 | ORF1ab/nsp4 | V167L | Gta | Tta |
| 21 | ORF1ab/nsp3 | P1228L | cCa | cTa |
| 22 | N | G215C | Ggt | Tgt |
| 23 | ORF1ab/nsp3 | A488S | Gct | Tct |
| 24 | ORF7b | T40I | aCt | aTt |
| 25 | ORF1ab/nsp3 | P1469S | Cct | Tct |
| 26 | S | G142D | gGt | gAt |
| 27 | N | R203K | aGG | aAA |
| 28 | N | G204R | Gga | Cga |
| 29 | S | N501Y | Aat | Tat |
| 30 | S | P681H | cCt | cAt |
| 31 | ORF1ab/nsp3 | T183I | aCt | aTt |
| 32 | S | T716I | aCa | aTa |
| 33 | S | A570D | gCt | gAt |
| 34 | S | D1118H | Gac | Cac |
| 35 | ORF1ab/nsp3 | A890D | gCt | gAt |
| 36 | ORF8 | Q27X | Caa | Taa |
| 37 | ORF8 | Y73C | tAc | tGc |
| 38 | N | S235F | tCt | tTt |
| 39 | ORF8 | R52I | aGa | aTa |
| 40 | S | S982A | Tca | Gca |
| 41 | N | D3L | GAT | CTA |
| 42 | ORF1ab/nsp3 | I1412T | aTa | aCa |
| 43 | S | T95I | aCt | aTt |
| 44 | ORF1ab/nsp3 | A1711V | gCt | gTt |
| 45 | ORF3a | Q57H | caG | caT |
| 46 | S | A222V | gCt | gTt |
| 47 | ORF8 | K68X | Aaa | Taa |
| 48 | ORF1ab/nsp2 | T85I | aCc | aTc |
| 49 | ORF1ab/nsp4 | A446V | gCt | gTt |
| 50 | ORF1ab/nsp3 | P822L | cCt | cTt |
| 51 | ORF1ab/nsp6 | V149A | gTt | gCt |
| 52 | ORF1ab/nsp13 | K460R | aAg | aGg |
| 53 | ORF1ab/nsp6 | T181I | aCt | aTt |
| 54 | S | E484K | Gaa | Aaa |
| 55 | S | L18F | Ctt | Ttt |
| 56 | ORF10 | V30L | Gta | Tta |
| 57 | N | A220V | gCt | gTt |
| 58 | ORF1ab/nsp13 | I334V | Ata | Gta |
| 59 | ORF1ab/nsp12 | P227L | cCa | cTa |
| 60 | N | P199L | cCa | cTa |
| 61 | ORF3a | E239Q | Gag | Cag |
| 62 | N | T205I | aCt | aTt |
| 63 | ORF1ab/nsp12 | F192V | Ttc | Gtc |
| 64 | ORF1ab/nsp12 | L838I | Cta | Ata |
| 65 | N | Q9L | cAg | cTg |
| 66 | ORF1ab/nsp3 | A1736V | gCa | gTa |
| 67 | ORF8 | S24L | tCa | tTa |
| 68 | S | D614G | gAt | gGt |
| 69 | ORF1ab/nsp12 | P323L | cCt | cTt |
| 70 | S | L452R | cTg | cGg |
| 71 | M | I82T | aTc | aCc |
| 72 | S | P681R | cCt | cGt |
| 73 | ORF3a | S26L | tCa | tTa |
| 74 | N | D377Y | Gat | Tat |
| 75 | ORF1ab/nsp13 | P77L | cCa | cTa |
| 76 | N | R203M | aGg | aTg |
| 77 | S | T19R | aCa | aGa |
| 78 | ORF1ab/nsp12 | G671S | Ggt | Agt |
| 79 | S | T478K | aCa | aAa |
| 80 | N | D63G | gAc | gGc |
| 81 | S | D950N | Gat | Aat |
| 82 | ORF7a | T120I | aCa | aTa |
| 83 | ORF7a | V82A | gTt | gCt |
| 84 | S | R158G | Aga | Gga |
| 85 | ORF1ab/nsp4 | T492I | aCc | aTc |
| 86 | ORF8 | E92K | Gaa | Aaa |
| 87 | ORF1ab/nsp16 | R216C | Cgc | Tgc |
| 88 | ORF1ab/nsp3 | K977Q | Aaa | Caa |
| 89 | ORF3a | S253P | Tcc | Ccc |
| 90 | N | P80R | cCa | cGa |
| 91 | S | T20N | aCc | aAc |
| 92 | S | K417T | aAg | aCg |
| 93 | S | R190S | agG | agT |
| 94 | N | S194L | tCa | tTa |
| 95 | ORF1ab/nsp2 | P129L | cCa | cTa |
| 96 | ORF1ab/nsp13 | H164Y | Cat | Tat |
| 97 | ORF1ab/nsp16 | K160R | aAg | aGg |
| 98 | ORF1ab/nsp2 | L550F | Ctc | Ttc |
| 99 | N | G204P | GGa | CCa |
| 100 | ORF1ab/nsp6 | C197F | tGc | tTc |
| 101 | ORF1ab/nsp13 | E261D | gaG | gaT |
| 102 | S | A701V | gCa | gTa |
| 103 | ORF7a | L116F | Ctc | Ttc |
| 104 | ORF3a | S171L | tCa | tTa |
| 105 | ORF8 | T11I | aCa | aTa |
| 106 | ORF7a | V71I | Gta | Ata |
| 107 | S | Y145H | Tac | Cac |
| 108 | ORF1ab/nsp2 | G339S | Ggt | Agt |
| 109 | ORF1ab/nsp3 | M1788I | atG | atA |
| 110 | ORF7a | P45L | cCa | cTa |
| 111 | ORF1ab/nsp8 | Q24R | cAg | cGg |
| 112 | ORF8 | L60F | ttG | ttT |
| 113 | ORF1ab/nsp3 | V932A | gTg | gCg |
| 114 | ORF1ab/nsp4 | L438P | cTt | cCt |
| 115 | ORF1ab/nsp13 | D260Y | Gat | Tat |
| 116 | ORF3a | W131C | tgG | tgT |
| 117 | ORF3a | P42L | cCt | cTt |
| 118 | S | S13I | aGt | aTt |
| 119 | S | V1264L | Gtg | Ttg |
| 120 | ORF1ab/nsp13 | A296S | Gct | Tct |
| 121 | S | W152C | tgG | tgT |
| 122 | ORF1ab/nsp14A2-B | P46L | cCt | cTt |
| 123 | S | W1214G | Tgg | Ggg |
| 124 | ORF1ab/nsp16 | Q238H | caG | caT |
| 125 | N | S327L | tCg | tTg |
| 126 | ORF1ab/nsp3 | H1307Y | Cat | Tat |
| 127 | N | M234I | atG | atT |
| 128 | ORF1ab/nsp4 | V94A | gTc | gCc |
| 129 | S | S477N | aGc | aAc |
| 130 | ORF1ab/nsp3 | M1441I | atG | atT |
| 131 | ORF1ab/nsp3 | D410G | gAt | gGt |
Table 3.
Synonymous mutations of SARS-CoV-2 from all the lineages showing the ORFs, amino acid mutation, reference codon, and the mutated codon
| Sl. No | ORF | Mutation | RefCodon | MutCodon |
|---|---|---|---|---|
| Synonymous | ||||
| 1 | ORF1ab/nsp3 | F106F | ttC | ttT |
| 2 | ORF1ab/nsp4 | D144D | gaC | gaT |
| 3 | ORF1ab/nsp6 | V120V | gtA | gtG |
| 4 | S | I68I | atA | atC |
| 5 | ORF1ab/nsp12 | P412P | ccC | ccT |
| 6 | ORF1ab/nsp12 | T912T | acT | acC |
| 7 | ORF1ab/nsp12 | H613H | caC | caT |
| 8 | ORF1ab/nsp3 | F1089F | ttC | ttT |
| 9 | ORF1ab/nsp2 | S36S | tcC | tcT |
| 10 | ORF1ab/nsp13 | N268N | aaT | aaC |
| 11 | ORF1ab/nsp9 | L112L | Cta | Tta |
| 12 | ORF1ab/nsp12 | N552N | aaT | aaC |
| 13 | ORF1ab/nsp3 | T955T | acA | acG |
| 14 | ORF1ab/nsp3 | F1107F | ttC | ttT |
| 15 | ORF1ab/nsp3 | T1189T | acC | acT |
| 16 | ORF1ab/nsp2 | N435N | aaC | aaT |
| 17 | ORF1ab/nsp1 | V60V | gtT | gtC |
| 18 | ORF1ab/nsp16 | A199A | gcG | gcC |
| 19 | M | L93L | ctC | ctG |
| 20 | S | I882I | atC | atT |
| 21 | ORF7a | G38G | ggA | ggC |
| 22 | N | S202S | AGt | TCt |
| 23 | ORF1ab/nsp4 | Y335Y | taC | taT |
| 24 | ORF1ab/nsp15A1-B | N73N | aaT | aaC |
| 25 | N | F363F | ttC | ttT |
| 26 | ORF1ab/nsp13 | Y217Y | taC | taT |
| 27 | S | I410I | atC | atT |
| 28 | ORF3a | L106L | ctC | ctT |
| 29 | ORF1ab/nsp3 | V1298V | gtA | gtG |
| 30 | ORF1ab/nsp1 | D156D | gaT | gaC |
| 31 | ORF8 | H17H | caC | caT |
| 32 | ORF1ab/nsp3 | P1200P | ccA | ccG |
| 33 | ORF1ab/nsp3 | D10D | gaC | gaT |
| 34 | ORF1ab/nsp15A1-B | L214L | ttA | ttG |
| 35 | M | F53F | ttC | ttT |
| 36 | ORF1ab/nsp15A1-B | L216L | ttA | ttG |
| 37 | ORF8 | F120F | ttC | ttT |
| 38 | ORF1ab/nsp14A2-B | L495L | ctC | ctT |
| 39 | ORF1ab/nsp12 | N600N | aaC | aaT |
| 40 | ORF1ab/nsp2 | G154G | ggC | ggT |
| 41 | M | Y71Y | taC | taT |
| 42 | S | D1259D | gaC | gaT |
| 43 | ORF1ab/nsp12 | Y455Y | taC | taT |
| 44 | S | D574D | gaT | gaC |
| 45 | ORF1ab/nsp3 | Y840Y | taC | taT |
| 46 | ORF1ab/nsp3 | D1075D | gaC | gaT |
| 47 | ORF1ab/nsp12 | D760D | gaC | gaT |
| 48 | ORF3a | S74S | tcC | tcT |
| 49 | ORF6 | D30D | gaT | gaC |
| 50 | ORF1ab/nsp3 | N506N | aaT | aaC |
| 51 | ORF1ab/nsp9 | Y87Y | taT | taC |
| 52 | S | N856N | aaC | aaT |
| 53 | S | V1122V | gtG | gtT |
| 54 | ORF1ab/nsp14A2-B | Y235Y | taC | taT |
| 55 | ORF1ab/nsp14A2-B | L280L | Cta | Tta |
| 56 | ORF1ab/nsp12 | D140D | gaC | gaT |
| 57 | N | S412S | agC | agT |
| 58 | N | R259R | cgG | cgA |
| 59 | ORF1ab/nsp9 | Y31Y | taC | taT |
Table 4.
Deletions in SARS-CoV-2 from all the lineages showing the ORFs, amino acid mutation, reference codon, and the mutated codon
| Sl. No | ORF | Mutation | RefCodon | MutCodon |
|---|---|---|---|---|
| Deleterious | ||||
| 1 | ORF1ab/nsp6 | F108del | TTT | – |
| 2 | S | H69del | CAT | – |
| 3 | S | V70del | GTC | – |
| 4 | S | Y144del | TAT | – |
| 5 | ORF1ab/nsp1 | V84del | GTT | – |
| 6 | ORF1ab/nsp6 | G107del | GGT | – |
| 7 | ORF8 | D119del | GAT | – |
| 8 | ORF8 | F120del | TTC | – |
| 9 | S | F157del | TTC | – |
| 10 | S | E156del | GAG | – |
| 11 | ORF1ab/nsp6 | S106del | TCT | – |
Table 5.
Non-coding mutations of SARS-CoV-2 from all the lineages showing the ORFs, amino acid mutation, reference codon, and the mutated codon
| Sl. No | ORF | Mutation | RefCodon | MutCodon |
|---|---|---|---|---|
| Non-coding | ||||
| 1 | noncoding | A28272T | A | T |
| 2 | noncoding | C29870A | C | A |
| 3 | noncoding | T29834A | T | A |
| 4 | noncoding | C222T | C | T |
| 5 | noncoding | G174T | G | T |
| 6 | noncoding | G204T | G | T |
| 7 | noncoding | C241T | C | T |
| 8 | noncoding | A28271- | A | - |
| 9 | noncoding | G210T | G | T |
| 10 | noncoding | G29742T | G | T |
| 11 | noncoding | A29700G | A | G |
Additionally, some mutations are characteristic of specific regions, while some others are present in all the regions in SARS-CoV-2. For instance, in Asia, three mutations in QLA46612 (isolated from South Korea) were observed within the same sequence [10]. This particular study stressed the fact that binding sites of the ligands and potential drugs change if the structure of the spike protein is damaged. Moreover, the study also tried to comprehend if more than one mutation affects the binding of the drug, thereby altering the potentially identified drug binding sites. It was later revealed that drug binding sites were greatly affected in structures with multiple variations [10]. Thus, the SARS-CoV-2 variants may have different binding sites making them identifying a suitable drug challenging. However, this arena will pave the way for prospective vaccine trials.
Limitations of the existing COVID-19 therapies
It has been reported that traditional Chinese medical therapies have increased the clinical manifestation of the virus among patients, along with enhanced the severity of the disease [11]. Major concerns of the recent vaccines which were developed during the pandemic were their safety and efficacy, the emergence of new variants, challenges that occur due to vaccine distribution among a larger population, and many more. Recently, an mRNA-based vaccine (BNT162b2) was reported to have adverse side effects such as fatigue, headaches, and immense pain at the site of injection. Variants are concerns among the present therapies due to severe mutations that occur in the spike protein as well as the increased transmission rate when compared to its predecessor. Distribution of the vaccines to the greater population becomes highly challenging to the lack of storage facilities in different geographical locations as well as the stability of the vaccines which highly depends on the storage conditions such as temperature [12]. These limitations warrant the use of alternative approaches, with drug repurposing being the most effective strategy that is currently being researched.
Introduction and mechanism of drug repurposing
The concept of drug repurposing holds promises of swift clinical trials and a reduced cost than drug development de novo. Drug repurposing, also known as drug repositioning, is the process of using approved and existing drugs for treating diseases other than the intended. Drug repurposing is pragmatic and alluring, given the considerable time and cost requirements for typical drug development [13]. Additionally, a great number of potential drugs do not reach clinical trials, with lesser than 15% of the compounds receiving approval [14]. Some examples of approved repositioned drugs include aspirin, the cyclooxygenase inhibitor for coronary heart disease, antibiotic erythromycin for gastric motility, and sildenafil, a phosphodiesterase inhibitor for erectile dysfunction [15]. Other drugs that have disconcerting side effects also merit repurposing as corroborated by the successful implementation of thalidomide, an antiemetic against multiple myeloma [16].
Currently, there is a huge interest in the repurposing of drugs to speed up the identification of drugs that can prevent or treat COVID-19. The chief importance of repositioning is to accelerate the traditional drug discovery process by the detection of novel, safe, and effective clinal-use drugs in humans. The primary rationale for this is the utilization of drugs that have the same molecular pathways but are involved in different diseases [17]. The mechanism of repurposing depends on the type of targets that the drugs are being used against. One of the most valuable drivers of repurposing has been the chance discovery of the pharmacological activity of new drug targets, implying a newer indication of drug use [18]. For instance, drugs lopinavir and ritonavir have been used for the treatment of Human Immunodeficiency Virus-1 and 2 (HIV) [19]. These drugs were also the first drugs to be repurposed for the treatment of COVID-19. The main reason for this is due to the resemblance of the SARS-CoV and MERS-CoV (beta coronavirus) genome sequences to SARS-CoV-2 [20, 21]. Thus, the drugs being utilized for the treatment of MERS-CoV and SARS-CoV-2 could be essentially used for COVID-19 treatment, implying that the mechanism of action of repositioned drugs against their targets should be similar to or resemble its family target members [22, 23]. Successful administration to patients with moderate COVID-19 symptoms has been previously observed, confirming the hypothesis of drug repurposing [24, 25]. From all this, it can be elucidated that the concept of drug repurposing relies on two major scientific bases: (1) the idea of pleiotropic drugs, (2) the discovery of drugs via human genome interpretation that certain diseases share the common targets.
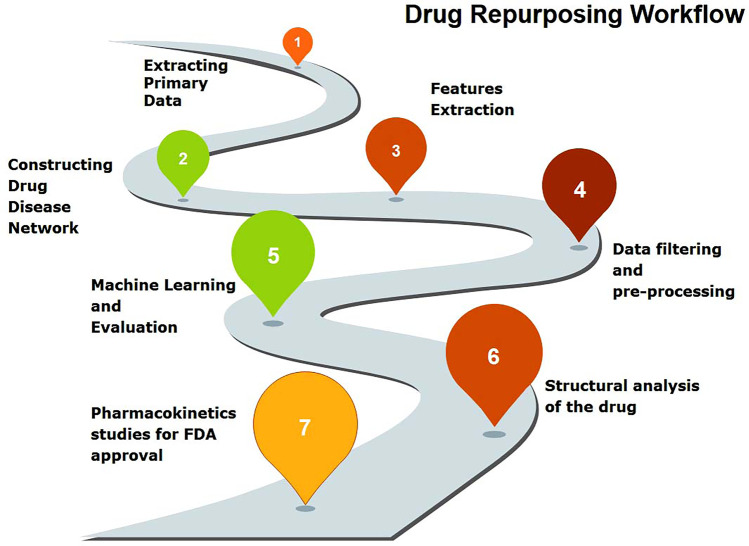
It begins with the extraction of primary data on the drug, disease, and target. A drug-disease network is eventually constructed and its features of it are extracted to filter the data and pre-process it thoroughly. A computational-based evaluation is carried out via methods detailed in the following sections, including advanced approaches such as machine learning that entails the structural analysis of the drug candidates. The pharmacokinetic studies ensue, which are essential for obtaining an FDA approval for use by the public. This general workflow and roadmap to drug repurposing are detailed in Fig. 2.
Fig. 2.
The roadmap to drug repurposing. It begins with the extraction of primary data on the drug, disease, and target. A drug-disease network is eventually constructed and its features of it are extracted to filter the data and pre-process it thoroughly. A computational-based evaluation is carried out via methods detailed in the following sections, including advanced approaches such as machine learning that entails the structural analysis of the drug candidates. The pharmacokinetic studies ensue, which are essential for obtaining an FDA approval for use by the public
Scope of drug repurposing
Drug repurposing is known to decrease the costs needed for the development of new drugs, with additional savings in pre-clinical phases I and II [18]. Aside from this, the scope of drug repurposing expands to the following, with instances in COVID-19 research, as discussed in the below sections:
Precision medicine: This area is an emerging strategy that studies the variability of individual genes, the lifestyle, and the environment for each person to decide upon the correct treatment approach [26]. Disorders with traits that are common are characterized as a single condition and are part of a spectrum of illnesses. Safer and more effective medications can be found if the drugs could be tailored to the variations in the genome of the individual, their proteome, transcriptome, or metabolome. This approach would help target the specific types of a general condition, with repositioning paving the way for precision medicine. Recent studies have implicated the use of computational techniques such as machine learning algorithms as a precision medicine tool for drug repurposing for the treatment of COVID-19 [27]. The decision tree algorithms used in the study identify patients with longer survival rates and better oxygen support when treatment was provided and suggest corticosteroids and remdesivir as effective COVID-19 repurposed drugs.
Rapid therapeutic options: With the advent of the COVID-19 pandemic and reduced therapeutic options for the disease, repurposing offers rapid therapeutic strategies to combat the disease. El Bairi et al. [28] reviewed various anticancer drugs for the management of COVID-19. Likewise, another study has elucidated and suggested the use of interleukin inhibitors, Janus-kinase pathway inhibitors, interferons, vascular endothelial growth factor (VEGF) inhibitors, immune check-point inhibitors, Bruton tyrosine kinase inhibitors (BTK), and XPO-1 (a selective inhibitor of nuclear export) as some of the alternative drugs that could be repurposed for rapid COVID-19 treatment [29].
Identification of multiple purposes for existing drugs: The latest applications of already approved drugs well beyond the scope of their existing medical aid in the identification of multiple purposes for such drugs in just four major steps–identification of compound, acquisition of the compound, development of the compound, and FDA approval after monitoring of its safety. This reduces the time taken for traditional drug discovery, lowers developmental cost as well as risk, and also reveals novel pathways and targets for already approved drugs, allowing them to have higher functionality [30, 31]. For instance, a recent study has repurposed antiviral protease inhibitors such as chloroquine, remdesivir, and lopinavir, which have already shown efficient inhibition of the SARS-CoV-2 virus in in vitro studies, using the extra-cellular vesicles, to make it more robust against the virus [32].
Identification of direct-acting and host-targeting antivirals: Drug repurposing also has applications in the identification of existing antivirals that are either direct-acting or host-targeting. Studies have shown that anti-hepatitis C combinations act as potential direct-acting main protease inhibitors of SARS-CoV-2 [33]. Similarly, another study suggested p38 MAPK (mitogen-activated protein kinase), as a vital host target for inhibiting the replication of coronaviruses, after studies proved that Ralimetiniib mesylate was found to be active in both Calu-3 and HeLa-ACE2 cells [34, 35].
Thus, drug repositioning for COVID-19 research has now taken the front seat and has a huge scope for tackling the SARS-CoV-2 virus.
Need for repurposed molecules and the latest developments in computational medicinal chemistry
Drugs are small molecules that affect the molecular pathways of important protein targets that are most likely responsible for causing a specific disease. These molecules have a lower molecular weight and are typically designed using rational drug design or structure-based drug design. With increasing incidences of emerging diseases such as COVID-19, for which immediate treatments were not available, the need for drugs to treat the disease became indispensable. With the SARS-CoV virus, mutating at a fast pace and affecting millions of people globally, there was a pressing need to address this issue by using all alternative approaches to discovering a drug [36, 37]. In this light, drug repurposing came into the picture. Comprehending detailed molecular mechanisms between the drugs and their targets is critical in the discovery of drugs. Even though several biochemical and biophysical methods can explicate the molecular interactions of the small molecules with their proteins, the most powerful tool is structural and computational biology which can aid in interpreting the mechanism of action of the interaction complexes [38]. Bearing this in mind, drug repurposing is the alternative approach meant to identify the most suitable drug candidates from among the existing libraries, to combat specific diseases.
Various approaches in computational medicinal chemistry for drug repurposing
Currently, several approaches exist that pave the way for drug repurposing. Advancements in computational chemistry and chem-bioinformatics pave the way for exploring more accurate predictive repurposing.
Technological advancements in repurposing
Regarding developments in multiplex assays, cell-based screening, mining of data, chem-informatics and bioinformatics approaches, and their databases, pharmaceutical industries have now shown augmented keenness in digging up molecules that had once failed due to several reasons [39]. Generally, there are three important steps before repositioning drugs: (a) drug candidate identification for the specific disease, (b) assessment of the selected drug candidate in pre-clinical models, (c) safety evaluation in phase II clinical trials [18].
The approach of repositioning itself can be subdivided into experimental and computational approaches. Using these approaches in combination yields better outcomes than using them individually. Computational strategies are driven by a huge amount of data that entails the utilization of genetic expressions, the structure of the chemical compounds, and the proteomic drug candidate data [40]. Several computational medicinal approaches add value to the drug repurposing process.
Structure-based approaches
Structure-based approaches such as virtual screening help in the identification of small molecules from an existing library, that can be used for repurposing. Previous studies have most commonly employed molecular docking for virtual screening against targets of SARS-CoV-2, such as the main protease, with possible drugs for repurposing such as digitoxigenin, beta-eudesmol, crocin, rutin, ritonavir, emetine, hesperidin, and indinavir [41, 42]. Likewise, other virtual screening approaches used previously for COVID-19 drug repurposing include combinations of docking with molecular dynamic (MD) simulations and calculation of free energy using MM-GBSA studies [43] and two-way docking with in silico ADMET studies [44]. The following strategies fall under structure-based approaches for drug repurposing.
Molecular docking
Molecular docking is a chem-informatics strategy to predict the conformation of the docking site/binding site of a drug candidate to the protein target [45]. The prediction of the best conformation where the drug and protein form an interaction complex, that will stop the molecular pathway of the target, in turn affecting the occurrence of the disease is the idea of docking [46]. Novel interactions that will help in understanding the potential drug candidates for repurposing can be identified using docking studies. The advantage of this strategy is that it can be followed by other computational validation techniques such as molecular dynamics simulations and Quantitative Structure–Activity Relationship studies (QSAR), to understand the complex stability and thereby increase the drug repurposing opportunity. The authors’ previous studies have used this computational technique against various targets of SARS-CoV-2 [47, 48]. The disadvantage is that not all proteins of interest are available, due to a lack of macromolecular structure. Applying more than one computational medicine approach will provide further authenticity to the predicted results.
MD simulations
The static binding energies between the drugs and targets obtained via molecular docking studies are further simulated dynamically in real-time to comprehend the stability of the interaction complex in biological environments. Molecular dynamic simulations have proved to be one of the most reliant strategies in drug repurposing. Elfiky [49, 50] proved via MD simulations that Setrobuvir, YAK, and ID-184 had the best antiviral activity against RNRP (RNA-dependent RNA polymerase) of SARS-CoV-2, after screening out 31 compounds that were in clinical trials or had known antiviral activity. Similarly, MD simulations identified the stability of lopinavir complexed with SARS-CoV-2 main protease as the most stable in biological conditions [51, 52], along with other identified antivirals such as carfilzomib, valrubicin, elbasvir, and eravacycline.
QSAR
Quantitative structure–activity relationship acts as a tool to identify the relationship between the structure and the activity of compounds. With regard to COVID-19, this approach combined with docking and simulations has brought immense benefits to the design and repurposing of drugs for SARS-CoV-2. A study has previously created a QSAR model founded on machine learning strategies utilizing several hundred inhibitor molecules of the SARS-CoV-2 main protease. This model was employed for virtual screening and docking, followed by shortlisting of the 20 best candidates for MD simulation validation [53]. Similarly, another recent study designed a binary-QSAR model for the virtual screening of several drugs that are FDA-approved and are in clinical studies against the main protease of SARS-CoV-2 [54]. This QSAR protocol resulted in cefuroxime pivoxetil as a potential drug repositioning molecule against SARS-CoV-2.
Network-based approach
Network-based approaches are commonly employed in repurposing due to the capability of amalgamating and integrating several sources of data [20, 21]. Advancements in high-throughput technology and computational approaches allow the modeling of biological systems by the network which facilitates diagnostic and pharmaceutical research that is structure-guided, with the possibility of identification of novel targets [55, 56]. Studies have stated the importance of drug-drug networks, drug-target networks, and drug-disease networks in drug repurposing [57]. The two-most common types of network-based strategies in computational biology include network-based propagation and network-based clustering approaches [58]. King et al. [59] have previously studied PPI network-clustering technique for drug repurposing, while Vanunu et al. [60] have used the disease-gene network propagation technique to accurately predict the disease-gene relationships.
Artificial intelligence-based approaches
Recently, the use of AI technology for drug repositioning has surfaced with several studies focusing on repurposing for COVID-19 research. A deep-learning-based approach to screen a huge number of molecules having allocated datasets to seek compounds possessing SARS-CoV-2-inhibiting activities was carried out previously by Ke et al. [61]. To validate the designed model and verify the antiviral activity of the compound that the model predicted, an in vitro cell line for feline coronavirus replication was also set up in the study. Likewise, Beck et al. [62] utilized an already trained deep-learning prediction model for drug-target interactions known as Molecule Transformer-Drug Target Interaction (MT-DTI), to find antiviral drugs that are commercially available and which could perturb SARS-CoV-2 components including RNA-dependent RNA polymerase, proteinase, and helicase. Thus, AI-based techniques are providing the scientific communities a chance to fight COVID-19 in a much faster way, owing to the accessible web servers, tools, and models that reduce the time required for a drug to come into the market.
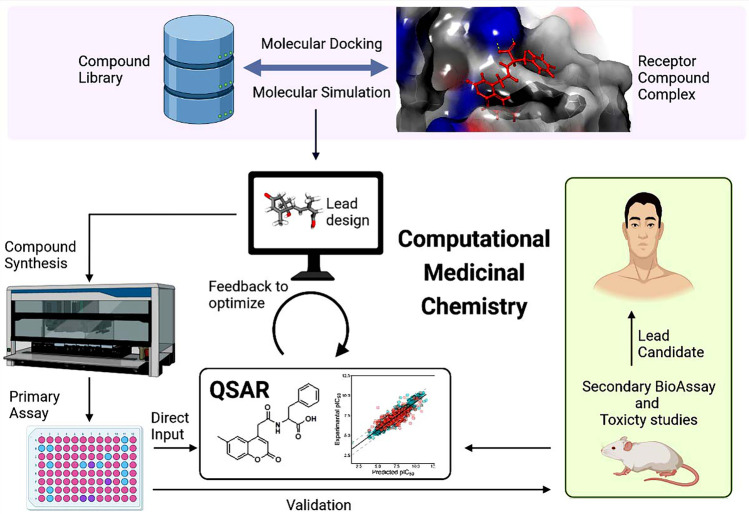
The scope and developments of computational medicinal chemistry toward COVID-19 treatment strategies are summarized in Fig. 3.
Fig. 3.
The latest developments and scope of computational medicinal chemistry in the treatment of COVID-19. These include molecular docking and simulations that form receptor-drug complexes. The simulation then leads to the lead design, followed by synthesis of the compound if required, primary in vitro assays, and QSAR studies. Validation of the repurposed drug can be performed via secondary bioassays and toxicity studies, using in vitro, in vivo, and in silico platforms
Drug repurposing construct: a computational chem-bioinformatic perspectives
Several computational approaches for repurposing have been used over the years to enhance the efficiency of the predictive outcomes and decrease the drawbacks. Currently, advancements in the field of computer science have led to the use of machine learning methods, network-based techniques, and matrix decomposition methods, along with other algorithms such as tensor decomposition for repurposing [63]. This technique has been performing better than several basic algorithms such as Network-based Random Walk with Restart (NRWRH), SNScore, and Collective Matrix Factorization (CMF). This technique solves the difficulty of negative sample training in cases where deep learning methods are used such as multilayer perceptron, auto encoders, and deep belief networks [64]. The tensor decomposition technique involves the construction and decomposition of 3D tensors that enable the associations among protein targets, drugs, and diseases [63].
Topological data analysis (TDA) is then taken into consideration to examine the drug properties, diseases, and disease targets to be able to recover the missing drug associations, diseases, and their targets, and to also predict the new repositioning candidate [40]. Moreover, drug repositioning can be performed on the drug candidates that arise from two clusters with robust connections, instead of assuming only the new-drug associations. To define these clusters, the diseases with similar molecular mechanisms are more inclined to appear within the same cluster. This approach, therefore, offers huge potential to explore predictive drug repurposing research.
Scope and applications of computational biology and chem-bioinformatics in COVID-19 research
There are several applications of computational biology and chem-bioinformatic approaches for drug discovery in COVID-19 research. A recent study has developed a multi-model deep learning-based algorithm for drug repurposing to aid in COVID-19 research. This work utilized the multimodel-restricted Boltzmann machine approach (MM-RBM), which can associate data about multiple modalities, to amalgamate two data types, inclusive of the chemical structures of small drug molecules and genes expressed differentially [65]. The results from this study showed clusters that were employed to identify the drugs that were notably similar to the proposed and existing COVID-19 treatment medication. This work also demonstrated results in predicting drugs with minimum side effects and high efficiency.
Another recent study proposed a mechanism-driven neural network technique called DeepCE, that employs a graphical network to model the gene–gene associations and the chemical structure-gene associations to predict the gene expression profile [64]. This work also proposed a data augmentation approach that extricates only beneficial data from a set of variable datasets, to screen novel drug candidates for COVID-19. A recent application of signature mapping on COVID-19 research demonstrated the use of Connectivity Map databases [66] for generating drug perturbation profiles [67]. A modified version of this study was implemented by Jia et al. [68] where the input used was the expression profile data from healthy and infected individuals to generate a pathway-based drug repurposing framework. Co-clusters were identified before performing reverse signature matching [68].
Another recent study designed and developed Deep Docking, which uses QSAR (Quantitative Structure–Activity Relationship) models to predict the docking scores of drug candidates that target the SARS-CoV-2 3CL protein. Deep Docking docked all the compounds and provided a list of potential drug hits, that could be used to repurpose for COVID-19 [69]. Several studies have implemented random-walk algorithms to identify drug repurposing candidates for COVID-19 [70–73]. The pathogenic mechanisms instigated by the spike protein using the information from all related coronaviruses were also explored and random walk algorithms were implemented on the molecular networks to recognize the most relevant COVID-19 drug targets [74]. Thus, these computational perspectives on drug repurposing provide a unique outlook on the alternative treatment options for the ongoing COVID-19 disease.
Proposed mechanisms of action and their targets of major repurposed drugs available for COVID-19
Drugs such as lopinavir, ritonavir, boceprevir, carmofur, and doxycycline act on the 3-chymotrypsin-like proteins/SARS-CoV-2 main protease that play an important role in cleaving and releasing the 16 non-structural proteins [4]. Fostamatinib disodium and platycodin target the Nsp3 proteins which play a major role in the infection and pathogenesis. The RNA-dependent RNA polymerases (RdRp, Nsp 12) that are primarily involved in important activities such as transcription and replication of the viral RNA genome are targeted by drugs such as remdesivir, favipiravir, and ribavirin [4]. For the helicase (Nsp13) of SARS-CoV-2, a multifunctional protein that helps in the proliferation and replication of the virus, drugs such as vapreotide, atazanavir, daclatasvir, and bismuth potassium citrate commonly block this function of the target. Likewise, dinucleoside 123 and raltegravir affect the pathways of methyltransferases (guanine-N7-methyltransferase, 2’-O-methyltransferase), while gliclazide and memantine affect the SARS-CoV-2 E protein, which is the envelope protein of the virus. Arbidol and chloroquine target the spike protein that helps in the viral entry, along with ACE2 protein in the host, thereby affecting the spike protein pathway [4] (Table 6).
Table 6.
FDA-approved drugs for repurposing, their molecular targets and functions in SARS-CoV-2
| Repurposed drug | Molecular target | Reference |
|---|---|---|
| Paritaprevir | 3-chymotrypsin-like proteins/SARS-CoV 2 Main protease – play an important role in cleaving as well as releasing 16 non-structural proteins (Nsps), ACE2-SARS-CoV-2-RBD complex: Spike protein which helps in the viral entry along with the help of ACE2 protein in host organism, RNA-dependent RNA polymerase (RdRp, Nsp12) – involved in important activities such as transcription, replication of viral RNA genome | |
| Ledipasvir | ACE2-SARS-CoV-2-RBD complex: Spike protein which helps in the viral entry along with the help of ACE2 protein in host organism | |
| Vancomycin | ACE2-SARS-CoV-2-RBD complex: Spike protein which helps in the viral entry along with the help of ACE2 protein in host organism, RNA-dependent RNA polymerase (RdRp, Nsp12)–involved in important activities such as transcription, replication of viral RNA genome | [75, 76] |
| Sirolimus | ACE2-SARS-CoV-2-RBD complex: Spike protein which helps in the viral entry along with the help of ACE2 protein in host organism | |
| Nilotinib | 3-chymotrypsin-like proteins/SARS-CoV 2 Main protease – play an important role in cleaving as well as releasing 16 non-structural proteins (Nsps), ACE2-SARS-CoV-2-RBD complex: Spike protein which helps in the viral entry along with the help of ACE2 protein in host organism | |
| Irinotecan | CD147, member of the immunoglobulin superfamily (IgSF) interacts with spike protein of SARS-CoV-2 virus | |
| Abemaciclib | CD147, member of the immunoglobulin superfamily (IgSF) interacts with spike protein of SARS-CoV-2 virus | |
| Estradiol benzoate | CD147, member of the immunoglobulin superfamily (IgSF) interacts with spike protein of SARS-CoV-2 virus | |
| Capmatinib | CD147, member of the immunoglobulin superfamily (IgSF) interacts with spike protein of SARS-CoV-2 virus | |
| Olaparib | CD147, member of the immunoglobulin superfamily (IgSF) interacts with spike protein of SARS-CoV-2 virus | |
| Lumacaftor | CD147, member of the immunoglobulin superfamily (IgSF) interacts with spike protein of SARS-CoV-2 virus | |
| Pazopanib | CD147, member of the immunoglobulin superfamily (IgSF) interacts with spike protein of SARS-CoV-2 virus | |
| Rifabutin | RNA-dependent RNA polymerase (RdRp, Nsp12)–involved in important activities such as transcription, replication of viral RNA genome | |
| Dactinomycin | RNA-dependent RNA polymerase (RdRp, Nsp12)–involved in important activities such as transcription, replication of viral RNA genome | |
| Dihydroergotamine | 3-chymotrypsin-like proteins/SARS-CoV 2 Main protease–play an important role in cleaving as well as releasing 16 non-structural proteins (Nsps) | |
| Bromocriptine | 3-chymotrypsin-like proteins/SARS-CoV 2 Main protease–play an important role in cleaving as well as releasing 16 non-structural proteins (Nsps) | |
| Entrectinib | 3-chymotrypsin-like proteins/SARS-CoV 2 Main protease–play an important role in cleaving as well as releasing 16 non-structural proteins (Nsps) | |
| Selinexor | 3-chymotrypsin-like proteins/SARS-CoV 2 Main protease–play an important role in cleaving as well as releasing 16 non-structural proteins (Nsps) | |
| Elbasvir | 3-chymotrypsin-like proteins/SARS-CoV 2 Main protease–play an important role in cleaving as well as releasing 16 non-structural proteins (Nsps) | |
| Quinupristin | 3-chymotrypsin-like proteins/SARS-CoV 2 Main protease–play an important role in cleaving as well as releasing 16 non-structural proteins (Nsps) | |
| Rifapentine | 3-chymotrypsin-like proteins/SARS-CoV 2 Main protease–play an important role in cleaving as well as releasing 16 non-structural proteins (Nsps) | |
| Rutin | 3-chymotrypsin-like proteins/SARS-CoV 2 Main protease–play an important role in cleaving as well as releasing 16 non-structural proteins (Nsps) | |
| Diosmin | 3-chymotrypsin-like proteins/SARS-CoV 2 Main protease–play an important role in cleaving as well as releasing 16 non-structural proteins (Nsps) | |
| Digitoxin | 3-chymotrypsin-like proteins/SARS-CoV 2 Main protease–play an important role in cleaving as well as releasing 16 non-structural proteins (Nsps) | |
| Antrafenine | 3-chymotrypsin-like proteins/SARS-CoV 2 Main protease–play an important role in cleaving as well as releasing 16 non-structural proteins (Nsps) |
The ACE receptors are blocked via losartan and captopril, transmembrane protease serine 2 (TMPRSS2) is blocked by camostat mesylate and with anyone, and cathepsin L is inhibited by hydroxychloroquine, which and teicoplanin. The viral neuraminidases and the DPP4 cell receptor pathways are hindered due to the action of oseltamivir and sitagliptin, while pemirolast, nitrofurantoin, isoniazid pyruvate, eridictoyl, hypericin, cepharanthine, and ergoloid block the human ACE2 and the S protein of SARS-CoV-2 [4]. The most important Janus kinase pathway is hindered by baricitinib, RdRp, helicase, exo-nuclease, and methyltransferase of SARS-CoV 2 by Efavirenz and Poly [ADP-ribose] polymerase 1 (PARP-1) by Mefuparib hydrochloride. Poly [ADP-ribose] polymerase 1 (PARP-1), glycogen synthase kinase 3 (GSK-3), and also biological pathways such as NF-kappa B signaling are all affected by the use of repurposed drugs toremifene, sirolimus, mercaptopurine, irbesartan, emodin, dactinomycin, and melatonin [4]. Other common drugs such as nitazoxanide, ivermectin, and azithromycin tend to symptoms of the virus, and play a role in inhibiting the role of SARS-CoV-2.
Despite these drugs being implicated as a potential treatment for COVID-19, ethical issues arise regarding the use of these drugs without proper clinical phase trials and FDA (Food and Drug Administration) approval. Therefore, a list of the FDA-approved drugs for the treatment of COVID-19 is provided in Table 6. Mahdian et al. [75] and Yuce et al. [76] state that Paritaprevir is important against 3-chymotrypsin-like proteins/SARS-CoV-2 Main protease and plays an important role in cleaving as well as releasing 16 non-structural proteins (Nsps). It also affects the ACE2-SARS-CoV-2-RBD complex. Likewise, ledipasvir, vancomycin, and sirolimus target the ACE2-SARS-CoV-2-RBD complex, a spike protein that helps in the viral entry along with the help of ACE2 protein in the host organism. Similarly, FDA-approved irinotecan, abemaciclib, estradiol benzoate, capmatinib, olaparib, lumacaftor, and pazopanib target CD147, a member of the immunoglobulin superfamily (IgSF) interacts with spike protein of SARS-CoV-2 virus [75, 76]. Other drugs such as nilotinib, rifabutin, dactinomycin, dihydroergotamine, bromocriptine, entrectinib, selinexor, elbasvir, quinupristin, rifapentine, rutin, digitoxin, diosmin, and antrafenine target 3-chymotrypsin-like proteins/SARS-CoV-2 Main protease that plays an important role in cleaving as well as releasing 16 non-structural proteins (Nsps).
Clinical trials data on potential COVID-19 repurposed drugs
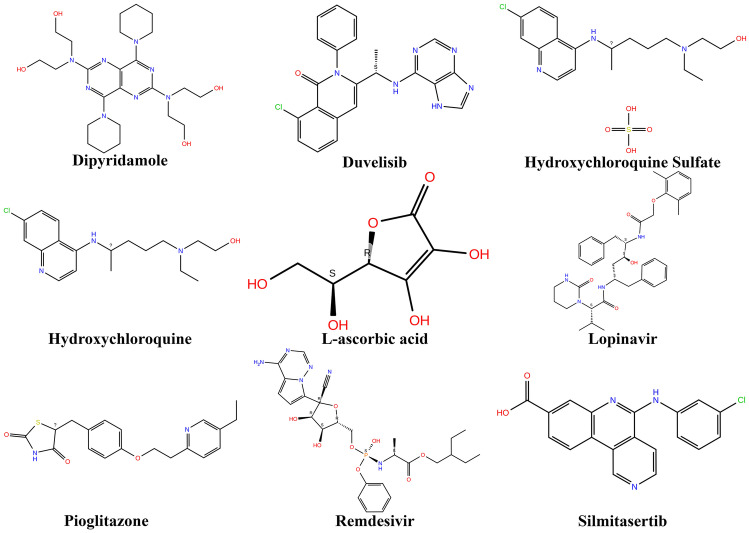
According to the latest updates from https://clinicaltrials.gov, phase 3 trials are completed for remdesivir, lopinavir/ritonavir, interferon beta-1A, hydroxychloroquine, and AZD7442 (clinicaltrials.gov registration number: NCT04315948). Trials for Silmitasertib are active and in phase 2, while for Pioglitazone, phase 4 trials are completed (clinicaltrials.gov registration number: NCT04473274). Phase 2 clinical trials are completed for dipyridamole 100 mg (NCT04391179), and phase 2 is completed for Leronlimab 700 mg (NCT04678830). L-ascorbic acid has phases 1 and 2 completed (NCT04357782), and phase 1 for aerolized hydroxychloroquine sulfate completed (NCT04461353). Similarly, drugs duvelisib (NCT04487886), ruconest (NCT04530136), and infliximab (NCT04425538) have phase 2 clinical trials completed. This data is provided in detail in Table 7 and the 2D structures of some of these drugs are shown in Fig. 4.
Table 7.
Clinical trials phase data for repurposed drugs that are in different phases of clinical trials
Fig. 4.
The 2D structures of some important repurposed drugs that are FDA approved and which are in clinical phase trials for COVID-19
When remdesivir was administered intravenously to the first patient diagnosed with SARS-CoV-2 in the USA, no adverse effects were noted with some clinical benefits in the patients [77]. Evidence also showed an accelerated recovery rate of 4 days when remdesivir was employed [78]. However, when randomized, double-blind, placebo-controlled trials were conducted by other researchers, it was found that remdesivir was not associated with significant improvement statistically [51, 52]. Additionally, due to some adverse effects early on, remdesivir was then discontinued, and WHO has now provided a conditional recommendation for the use of remdesivir against COVID-19 [79]. Likewise, real-world usage of lopinavir/ritonavir revealed no beneficial response against COVID-19 hospitalized patients in a randomized, controlled, trial [80]. Overall, the available data show an unfavourable response of patients to lopinavir/ritonavir, and hence the use of this drug has to be monitored and controlled vigilantly. Despite there being studies on hydroxychloroquine, an analogue of chloroquine, where improvements have been noted in patients suffering from COVID-19 in a study conducted in Wuhan, there still seems to be very limited data on the reliability of the drug [79]. Interferon-beta-1A has been used in hospitalized patients for treating COVID-19 at Shaid Behesti University of Medical Sciences, in combination with other drugs. However, despite it showing promising outcomes, more clinical data is required to corroborate the reliability of the drug [79]. Therefore, the clinical data presented provide evidence of the efficacy of the use of these drugs,however, more research is indispensable for the proper application and employability of these drugs for SARS-CoV-2 treatment.
Major targets of SARS-CoV-2 for repurposed drugs
According to literature, coronavirus papain-like proteases (PLpro), main protease (3CLpro), helicase, and RNA-dependent RNA polymerase (RdRp) are the considered to be the potential molecular targets and are involved in important functional activities such as protein synthesis, RNA transcription, translation, replication, processing, and infection respectively. All these proteins come under non-structural proteins (nsps) for coronaviruses [81]. The below sections discuss the important protein targets from a structural perspective.
Papain-like proteases (PLpro)
The papain-like proteases have a “thumb-palm-fingers” structure architecture that houses domains and subdomains. The active site residues/binding pockets present in these domains are His272, Cys111, and Asp286 which form the catalytic triad of the same protease/protein. These papain-like proteases (PLpro) share their structure with the ubiquitin-specific proteases (USPs) which have two main domains, ubiquitin-like (Ubl) domain, an N-terminal domain along with the catalytic triad which forms thumb-palm-fingers architecture [82].
RNA-dependent DNA polymerase (RdRp)
RNA-dependent DNA polymerase is mainly involved in the replication and transcription of the virus. Since it plays an important role in viral infection and hence it is considered a potential molecular target of the SARS-CoV-2 virus, human telomerase reverse transcriptase (hTERT) was first thought to be RNA-dependent DNA polymerase but it is DNA polymerase that shares structural similarities with RdRp. Single-stranded RNA (ssRNA) encodes for RdRp proteins whose structure has a right-handed closed polymerase-like structure with the thumb, finger, and palm domains. At N-terminal, there is the presence of beta-hairpin nidovirus RdRp-associated nucleotidyltransferase (NiRAN) domain (residues ranging from D60 to R249) and C-terminal/polymerase domain (residues ranging from S367 to F920) which houses thumb-fingers-palm subdomains. There are seven motifs present in the RdRp protein of SARS-CoV 2 ranging from A to G, where five are said to be present in the most conserved domain, i.e., the thumb domain [83–85].
Main protease (3CLpro)
Non-structural protein 5 coding gene encodes the main protease (3CLpro) which is mainly involved in the proteolytic activity of viral polyproteins and replicates. They share structural similarities between the 3C proteases of picornaviruses in terms of core homology as well as substrate specificities. The size of the protein is approximately 30 kDa which has three domains rich in cysteine amino acids. The protease comprises chymotrypsin-like folds along with a catalytic triad and a domain that is inundated with cysteine [86].
Nucleoprotein (N)
The nucleoprotein (N) of SARS CoV-2 binds to the genomic RNA (gRNA) to form the ribonucleoprotein (RNP) complex, a major virion core of the coronavirus. Nucleoprotein along with other proteins plays an important role replication and transcription of the virus. The N-terminal and the C-terminal domain of the nucleocapsid protein share similarities with CoVs N proteins. The N-terminal domain is a right-handed fist-shaped structure that has the topology of five beta strands sandwiched between two alpha-helices. The C-terminal domain is a homodimer with two protomers resembling a rectangular slab-like structure. Each protomer comprises two 3–10 helices, two beta-strands, and five alpha-helices respectively. The beta hairpin-like structure connects two protomers by inserting itself into their cavities [87].
Enveloped protein (E)
There are three domains in the enveloped protein of the SARS-CoV-2 virus which comprise of N-terminal domain, C-terminal domain, and transmembrane domain. The overall size of the E protein is about 8.4–12 kDa and forms the integral membrane protein of the virus comprising 76–109 amino acids. There are about 7–12 amino acids at the N-terminal which is short and makes it hydrophilic in nature. The transmembrane domain has about 25 amino acids, out of which two are non-polar and neutral making it hydrophobic in nature. The C-terminal domain forms the majority of the protein structure [88].
Spike glycoproteins
The genome of the SARS-CoV-2 encodes for the ectodomain trimeric protein which projects out of mature virions and plays an important role in the initial attachment process and entry into the host organism. The spike glycoproteins in SARS-CoV-2 are strongly glycosylated and are trimeric in structure. The protomers present in the spike glycoprotein structure contain approximately 1260 amino acids. The first and foremost part of the spike glycoprotein is the surface subunit (S1) which comprises four domains (two SD1, SD2 subdomains, N-terminal domain, and C-terminal domain which is also the receptor-binding domain of the protein) with 672 amino acids in length. The transmembrane subunit S2 is made up of 588 amino acids and contains a transmembrane domain, cytoplasmic tail, and two heptad repeats [89].
Non-structural proteins
The non-structural proteins of the SARS-CoV-2 are encoded by the open reading frame 1a and ab (ORF1ab) which is present in the downstream region of the 5’ end of the ssRNA (single-stranded ribonucleic acid) in the SARS-CoV-2 virus. They are polyproteins pp1a and pp1ab. Nsp 1 to 11 is categorized under pp1 polyprotein, while Nsp 12 to 16 is classified under polyprotein pp1ab [90].
These structural details are of utmost essential when the proteins interact with the drug candidates, to understand the precise molecular mechanism of action of the small molecule.
Effect of variants on the SARS-CoV-2 targets and immune escape
Mutations in the spike received early attention and some of the residues in spike variations influenced the polyclonal antibody recognition, thereby causing immune escape. Studies have shown that among all the receptor-binding domains (RBD) residues for which mutations in the form of substitutions affected the recognition by certain convalescent serums, E484 was identified by deep mutation scanning as having importance, with changes to amino acids K, P, or Q [91]. Moreover, the mutation E484K is recognized as an escape mutation, that occurs when exposure to monoclonal antibodies C144 and C121 occurs [92]. Moreover, E484K was the only variable that was found to reduce the neutralization capacity of MAbs REGN10934 and REGN10989 [93]. It was also studied that mutations K444E, L452R, G446V, and F490S also escaped some of the convalescent sera, thereby making these variants of SARS-Cov-2 stronger and more robust for survival in the human body [94]. Similarly, the substitution N439K enhances the SARS-CoV-2 ACE2 receptor affinity [95]. This mutation is also identified to decrease the potential of neutralization of the plasma. However, other deep mutation scanning studies pose contradictory results, stating that N439K did not affect the impact of the polyclonal antibodies used [96]. This discrepancy could be because immune escape caused by this mutation is via ACE2 affinity than by direct antibody epitope identification, making it a variant that should compel more studies.
Studies have also identified that the n-terminal domain (NTD) mutations in spike also cause immune escape [97]. Studies have shown that mutations K150R, K150T, K150E, S151P, K150Q, and N148S were escape mutations [92]. Moreover, deletions in the NTD have been noted to be the cause of SARS-CoV-2 evolution repeatedly, by altering the antigenicity of the NTDs [98]. Another study noted that Δ140 mutant in the spike eventually acquired the E484K variation, thus directly resulting in escaping the antibody responses [99].
Our previous studies show that mutations in the spike glycoprotein demonstrated enhanced stability in the protein, and the residues that were undergoing mutations were also part of the hydroxychloroquine drug-binding active pockets [100]. The mutations that occurred in other prioritized proteins of interest evidenced a decrease in the stability of the protein itself. These mutations were also part of the drug-binding active sites of remdesivir, favipiravir, and dexamethasone. The study also concluded that the presence of mutations in various regions reduced the effect of remdesivir on the targets, thereby allowing the potential drugs to not work on the targets of interest [100]. This shows the presence and effect the variants have on the drug-binding sites.
All these studies prove that there is distinct evidence as to the altering of the SARS-CoV-2 proteins’ structural antigenicity, causing them to evade the immune system and become more robust. As of now, evidence suggests the changes that mutations cause in the structure of spike proteins, thereby influencing the binding of small molecules. More research is required to assess the precise mechanism of how various structural and non-structural proteins are being affected by the variants. Amino acid deletions and substitutions in the spike affect the antibodies which may also lead to an increase in the global-viral resistance. A greater understanding of the viral genome sequence data and the impact of variants on the drug-binding sites will be essential to better obtain repurposed drugs.
Interaction modeling of the major repurposed drugs
Currently, as we have seen from various evidences, there are several repurposed drugs reported for use against COVID-19, some already available on the market. The potential binding of several FDA-approved drugs has been studied previously; however, not much data is revealed on the precise molecular basis of these interactions against SARS-CoV-2 targets. In the previous section, evidence of how the variants alter the drug binding efficacy and how the virus escapes the immune system is provided. In our previous study, however, various targets of SARS-CoV-2 were analyzed at the molecular level to comprehend the changes it undergoes during the binding event of some of the important FDA-approved drugs for COVID-19. Favipiravir, lopinavir, chloroquine, hydroxychloroquine, lopinavir, ritonavir, and remdesivir were studied. These drugs have also been mentioned in other computational biological studies as well [101]. Though there are limited experimental data to highlight the spike glycoprotein of SARS-CoV-2 act as the target for repurposed drugs, however, several computational biology prediction revelated the scope of utilising spike glycoproteins as the putative molecular target for repurposed drugs. A recent study stated that drugs ritonavir and eriodictoyl bind to the active sites of spike protein of SARS-CoV-2 with potential binding energy and stability [102]. Both these drugs interacted with the binding energy of −7.6 kcal/mol, indicating stable binding. Likewise, another study showed that repurposed drugs such as pralatrexate, carumonam, aclerasteride, and granotapide interacted with spike protein of SARS-CoV-2 and acted as potential binders [103]. Additionally, another recent study has reported Remdesivir and ivermectin, potential repurposed drugs interacted with the spike protein of SARS-CoV-2 [104]. The study stated that ivermectin bound effectively to the spike protein of the virus, and inhibited the entry of SARS-CoV-2 into the host. Remdesivir was also suggested to be involved in stable interaction with the spike target, apart from the other targets. These studies demonstrate that repurposed drugs were suggested to be targeted the spike protein at various binding pockets and interacted with the spike glycoprotein. To understand further on the interaction modelling of some important repurposed drugs, molecular docking and molecular dynamic studies were performed in our previous study and the results showed ritonavir and lopinavir with better binding energy, followed by remdesivir against the prioritized targets [105].
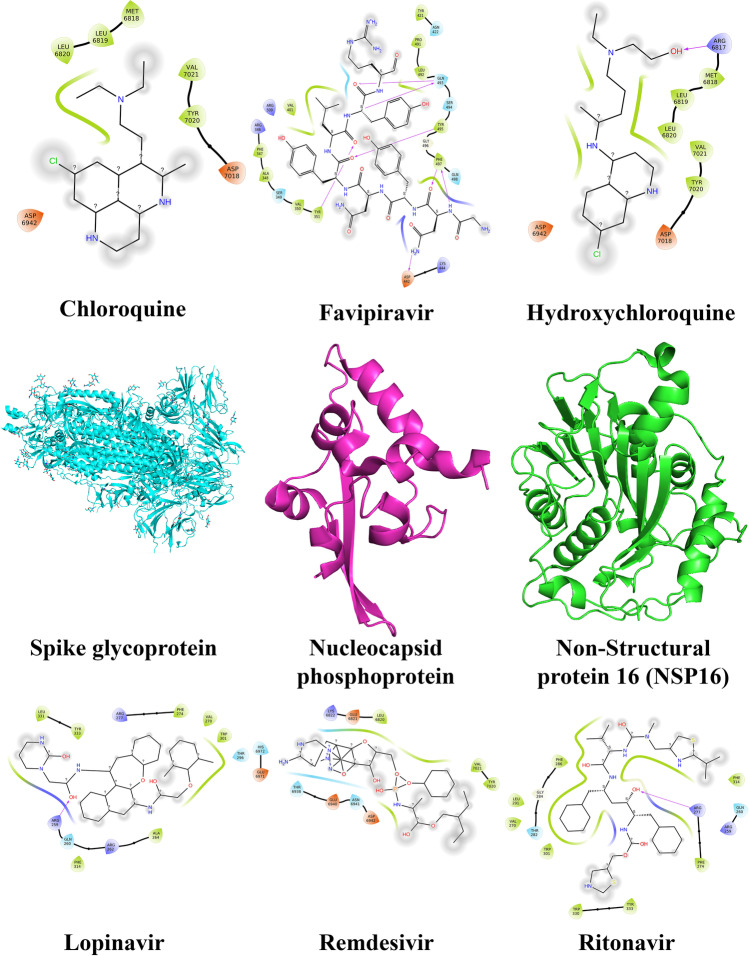
At a molecular level, Fig. 5 showed that chloroquine interacted with amino acids Leu6820, Leu6819, Met6818, Val7021, Tyr7020, Asp7018, and Asp6942 of the spike glycoprotein. Leu331, Tyr333, Arg277, Phe274, Val270, Trp301, Thr296, Ala264, Arg262, Phe314, Gln260, and Arg259 were the major amino acids involved when spike glycoprotein interacted with Lopinavir [105]. Likewise, Tyr421, Asn422, Pro491, Leu492, Gln493, Ser494, Tyr495, Gly496, Phe497, Gln498, Lys444, Asp442, Tyr351, Val350, Ser349, Ala348, Phe347, Arg346, Arg509, and Val401 were involved in complex favipiravir-nucleocapsid phosphoprotein. Lys6822, Glu6821, Leu6820, Val7021, Tyr7020, Asp6942, Asn6941, Glu6940, and Thr6938 were observed for complex remdesivir-nucleocapsid phosphoprotein. NSP16 interacted with hydroxychloroquine at residues Arg6817, Met6818, Leu6819, Leu6820, Tyr7020, Val7021, Asp7018, and Asp6942 and with ritonavir at residues Phe314, Gln260, Arg259, Arg277, Phe274, Tyr333, Trp330, Trp301, Thr282, Gly284, Phe286, Leu291, and Val270 [105] (Fig. 5).
Fig. 5.
Amino acid interactions between repurposed drug candidates and the protein targets of SARS-CoV-2 from our previous studies [105]. The targets shown are spike glycoprotein, nucleocapsid phosphoprotein, and NSP16 (middle row). The repurposed drugs that interacted with it included remdesivir, lopinavir, hydroxychloroquine, favipiravir, ritonavir, and chloroquine (top and bottom rows)
Bearing in mind the changes that the drug binding sites undergo if variations occur as already evidenced in the previous sections, it may also be hinted that the residues at the interaction sites also change, rendering a different molecular basis in case of variants. More comprehensive studies are essential to further confirm this.
Implementation of repurposed drugs and translation from in silico to in vivo platform
The major repurposed drugs displayed in Table 3 dhow that research is ongoing and always improving for tackling the problem of COVID-19. Drugs such as lopinavir, ritonavir, remdesivir, ribavirin, and favipiravir are some repurposed drugs. Lopinavir/ritonavir was generally used against HIV [106], remdesivir against major SARS viruses [107], and favipiravir which was previously used as a general viral pro-drug [108]. With the advent of research techniques to identify more repurposed drugs, several other candidates have come up, evidencing the successful implementation of repositioning.
The potential of in silico studies has been documented for ages, especially in the field of drug discovery. In this case, repurposing beings with computational bioinformatics work, with much exploration into chem-informatics, chemo-genomics, and computer science aspects such as machine learning, deep learning, artificial intelligence, computational biology, structural biology, and genomics/proteomics. A thorough analysis of the chemical structures of drug candidates helps in the identification of its working mechanism against specific targets. Using computational biology and computer science algorithms will help in predicting the possible and potent drug candidates in silico, a work that might otherwise take years without computational studies. The best drug candidates working against the protein targets of interest causing a specific disease will then be tested experimentally in vitro to validate the predictions. Accurate laboratory confirmation narrows down the best working drug that could be taken forward for in vivo animal model studies, before clinical trials. For example, remdesivir a broad-spectrum antiviral came into the market to be repurposed as a COVID-19 drug after in vitro studies revealed that this drug inhibited the SARS-CoV and MERS-CoV in the human respiratory epithelial cells [109] initially, followed by inhibiting the epithelial cells in SARS-CoV-2 [110], the pathogenic strain of COVID-19 disease. Likewise, the in vivo study of remdesivir when infected with SARS-CoV-2 infected rhesus monkey demonstrated an improvement in the pulmonary lesions and decreased the viral load after 7 days of treatment with the drug. No coronaviral disease was observed after the successful use of this drug [111]. Thus, repurposed drugs follow the path of in silico to in vitro to the in vivo platform before they are successfully used.
Repurposed drugs, their original targets, and SARs-CoV-2 targets
There are several FDA-approved drugs/drugs in the clinical phase, their original targets, and the repurposed targets (Table 8). Remdesivir was originally meant for treating the Ebola virus against the Ebola RdRp target, and now with potential repurposing capabilities against the RdRp and NSP12 of SARS-Cov-2 [75, 76]. Lopinavir/ritonavir is a drug for the HIV-1 protease, that now evidences potential for the main protease of SARS-CoV-2 [112]. Silmitasertib has its original target as casein kinase 2, which inhibits the casein kinase 2 pathway in Sonic hedgehog (SHH). This drug is also known to inhibit the casein kinase 2 pathway in SARS-CoV-2 [113]. Likewise, dipyridamole 100 mg is an inhibitor of phosphodiesterase originally and works against the main protease of SARS-CoV-2 and stops replication [114]. Leronlimab 700 mg works against the C–C Chemokine receptor type 5 in HIV and now shows potential against the same target in SARS-CoV-2 [115, 116]. Ruconest targets the c1 esterase and inhibits angioedema, while it targets C1, C5a, and C3a esterases in SARS-CoV-2 [117]. Infliximab attacks the tumor necrosis factor in SARS-CoV-2 as well as in its original target in Rheumatoid arthritis, Crohn’s disease, and ankylosing spondylitis [118]. This data is also available at https://go.drugbank.com/drugs/ [119] and the same is elucidated in Table 8.
Table 8.
Repurposed drugs, their original targets, and SARS-Cov-2 targets
| Drug | Original target | SARS-CoV-2 target | Reference |
|---|---|---|---|
| Remdesivir | RdRp, Ebola virus | RdRp, Nsp12 | https://go.drugbank.com/drugs/DB14761 [75, 76] |
| Lopinavir/ritonavir | HIV-1 protease inhibitor | Main protease | https://go.drugbank.com/drugs/DB01601 [112] |
| Interferon Beta-1A | Type 1 interferon receptor activity (multiple sclerosis) | Prohibits replication (3CLprotease) | https://go.drugbank.com/drugs/DB00060 [123] |
| Hydroxychloroquine | Transmembrane signaling receptor activity and inhibits toll-like receptors (Prophylaxis of malaria) | Inhibits terminal glycosylation of ACE2 | https://go.drugbank.com/drugs/DB01611 [124] |
| AZD7442 | Spike protein (SARS-CoV-2) | Spike protein | https://go.drugbank.com/drugs/DB15787 [125] |
| Silmitasertib | Casein kinase 2 inhibitor (Sonic hedgehog (SHH) medulloblastoma) | Inhibits casein kinase 2 | https://go.drugbank.com/drugs/DB15408 [113] |
| Dipyridamole 100 Milligram(mg) | Phosphodiesterase inhibitor (Postoperative thromboembolic) | Main protease of SARS-CoV-2 virus and suppressed the replication | https://go.drugbank.com/drugs/DB00975 [114] |
| Leronlimab (700 mg) | C–C Chemokine receptor type 5, HIV and Cancer | C–C Chemokine receptor type 5, SARS-CoV-2 | https://go.drugbank.com/drugs/DB05941 [115, 116] |
| Aerolized Hydroxychloroquine Sulfate | Transmembrane signaling receptor activity and inhibits toll-like receptors (Malaria, rheumatoid arthritis, chronic discoid lupus erythematosus and systemic lupus erythematosus) | Inhibits terminal glycosylation of ACE2 | https://go.drugbank.com/drugs/DB01611 [124] |
| Duvelisib | Inhibits phosphatidylinositol 3-kinase delta and gamma (Chronic lymphocytic leukemia or small lymphocytic lymphoma) | Reduce the levels of cytokines | https://go.drugbank.com/drugs/DB11952 [126] |
| Ruconest | Targets c1 esterase and inhibits (Angioedema) | Targets C1, C5a and C3a esterases | [117] |
| Infliximab | Attacks tumor necrosis factor alpha (Rheumatoid arthritis, Crohn’s disease and ankylosing spondylitis) | Attacks TNF-alpha there by controlling the cytokine storm | [118] |
Merits and applications of drug repurposing
With all the important approaches and scope discussed for repurposing drugs, some important advantages can be classified into three main focuses:
Translational: drug repurposing directly benefits translational research by fostering major collaborations, essential for scientists to identify important repurposed drugs. Bridging this gap is one major advantage of drug repositioning research.
Target: drug repurposing research aids in understanding thoroughly the pathways associated with specific targets and how the same can be exploited to benefit patients who must be treated. The comprehension of the complete metabolic pathways for specific targets opens up avenues for remaining drug discovery research.
Disease: drug repurposing has huge merits in discovering drugs for new, emerging diseases when there are no other treatment approaches yet discovered. The emergence of SARS-CoV-2 has illuminated the purpose and benefits of repositioning drugs to provide emergency treatments.
Other advantages of repurposed drugs include:
Lower cost of production
Lesser time for identifying useful, effective, and safe drugs for quick use
Research can be carried out using the existing, easily accessible bioinformatics and chem-informatics data and resources available for specific diseases, as was the case in COVID-19 cases
Applications of chem-bioinformatic approaches to drug repurposing
Molecular docking: The emergence and prevalence of COVID-19 disease have paved the way for molecular docking studies. Previously, molecular docking has been used to determine the potential anti-virals for curbing the spread of the disease. Poly-ADP-ribose polymerase-1, CVL218, a potent inhibitor that binds to the N-terminal of the nucleocapsid protein of the SARS-CoV-2 was identified [120].
Signature matching: Drug-drug comparisons and drug-disease comparisons can be carried out using the transcriptomic signature patterns. This technique has been previously used for repositioning, however, for lymphoid malignancies [121]. The use of signature matching for COVID-19 remains an unexplored avenue.
Pathway mapping: Identification of potential drugs using this technique has been previously carried out to analyze the network of drugs and diseases. Respiratory viral infections have been previously studied for repurposing candidates and a phosphodiesterase inhibitor was identified in the process [122].
Genome-wide association studies: Studying the small variations (SNPs) of the genome of interest helps in identifying traits associated with specific diseases. Reports have stated the use of GWAS in combination with other computational approaches for target prediction in drug repositioning [18, 40].
Thus, despite there being several advantages of repurposing, there are some limitations as elucidated in the following sections.
Limitations of repurposed drugs
With so many repurposed drugs in the market today, and several more upcoming owing to the intensity and acceleration of COVID-19 research, there are more advantages than disadvantages to the use of such drugs. However, some limitations to the drug repositioning, include:
Weak intellectual property rights protection for repurposed drugs due to reduced rate of return
The molecular docking technique used for identifying drugs for repurposing cannot be applied to all existing drugs due to the larger computational operations
Some network-based approaches used for drug repurposing do not always yield promising candidates
These approaches are data-dependent, due to which the results may be inaccurate due to the lower volume of existing COVID-19 data
Lack of strong, solid evidence for the mechanism of action of repurposed drugs on COVID-19 19
Experimental evidence is required for validating the predictive outcomes
Overall low success rates in new disease settings
Extensive data to be studied before repurposing
Dosing and safety issues, i.e., approved range of dosage and delivery capabilities are not very strong
Accurate, robust toxicological studies of repurposed drugs essential before delivery and testing
Rare to find unique and novel drug-protein interactions all within the restraints of the approved therapeutic window [17].
Future perspectives
With drug repurposing emerging as one of the most important research strategies for treating the occurrence of sudden, new diseases, there arrives the question of its efficacy when compared to existing viral vaccines or other small molecules, both considered standard treatment options. However, using high-throughput screening techniques and various other predictive approaches for identifying drugs for repositioning will help maintain the genomic integrity, thereby resolving the issue of efficacy. These approaches when amalgamated with advanced computational techniques such as machine learning will pave the way for producing better-repurposed drugs. In the wake of the emergence of highly mutated and resistant SARS-CoV-2 strains, the question of how already repurposed drugs work comes into the picture. But with improvements in technologies that can help repurposed drugs function better against such mutations, the future for drug repurposing looks bright.
Another perspective is the use of drug repurposing as a therapy for novel diseases for which specific drugs are not yet available, COVID-19 being one such important example. Repositioning played a major role in this case and is still in the research arena where different drugs are being explored using advanced computational techniques. Other advanced techniques are also being researched that will provide accurate predictive drug repositioning results that are more reliable and can be used for a variety of diseases. Although this review comprehensively lists all the current and latest drugs repurposed for SARS-CoV-2 and the existing computational approaches that in turn improve the process of repurposing, further advancements in the software and techniques such as using ensemble machine learning will improve the predictive process of repositioning.
A dimension that still requires improvement is the patent protection issues that still do not provide full protection to the repurposed drugs. If these avenues are improved, then the future of drug repurposing appears bright.
Conclusion
With high mortality rates, COVID-19 has become one of the most prevailing diseases globally. With the CDC and WHO declaring several recent variants of concern for SARS-CoV-2, there is an urgent need to explore alternative options of treatment when there are no specific drugs targeting the disease. One such alternative is drug repurposing, where the use of existing, FDA-approved drugs with similar functions and mechanisms of action against the same family of viruses can be employed. This concept relies heavily on robust preliminary in silico work using advanced and latest techniques in computational biology and chem-bioinformatics. The use of the machine and deep learning algorithms are also a way to obtain a better prediction of repurposed drugs. This review sheds light on the various existing drugs that have been repurposed for use against SARS-CoV-2 and also illuminates the important drug targets against which these drugs are used: spike glycoproteins, RDRP, main protease, papain-like protease, nucleoproteins, non-structural proteins, and enveloped proteins of SARS-CoV-2. The most important advantage of repositioning drugs against COVID-19 is the reduced cost and time of development and the drawback is the absence of safety/toxicity profiles via in vivo trials for the disease. Despite this, repurposing has found its niche with advancements in all related interdisciplinary fields of science and acts as a major research avenue in the current pandemic.
Author contribution
VN and ASS collected the data and drafted the manuscript. CK and AU involved the preparation of the manuscript especially the figures and tables. KMK involved in the preparation and review of the manuscript. SS supervised, reviewed, edited, and finalized the manuscript. All the authors have verified the manuscript and approved for submission.
Declarations
Conflict of interest
The authors declare no competing interests.
Footnotes
The original online version of this article was revised: The affiliation number of the authors are wrongly mentioned in the published version of the manuscript.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
8/31/2022
A Correction to this paper has been published: 10.1007/s11224-022-02045-4
References
- 1.World Health Organisation (2021) WHO Official COVID-19 info - World Health Organization. https://covid19.who.int/. Accessed 14 Apr 2022
- 2.Sadegh S, Matschinske J, Blumenthal DB, Galindez G, Kacprowski T, List M, Baumbach J. Exploring the SARS-CoV-2 virus-host-drug interactome for drug repurposing. Nat Commun. 2020;11(1):1–9. doi: 10.1038/s41467-020-17189-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rabaan AA, Al-Ahmed SH, Haque S, Sah R, Tiwari R, Malik YS, Rodriguez-Morales AJ. SARS-CoV-2, SARS-CoV, and MERS-COV: a comparative overview. Infez Med. 2020;28(2):174–184. [PubMed] [Google Scholar]
- 4.Senger MR, Evangelista TCS, Dantas RF, Santana MVDS, Gonçalves LCS, de Souza Neto LR, Silva-Junior FP. COVID-19: molecular targets, drug repurposing and new avenues for drug discovery. Mem Inst Oswaldo Cruz. 2020;115:e200254. doi: 10.1590/0074-02760200254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shang J, Wan Y, Luo C, Ye G, Geng Q, Auerbach A, Li F. Cell entry mechanisms of SARS-CoV-2. Proc Natl Acad Sci. 2020;117(21):11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Torjesen I. Covid-19: Omicron may be more transmissible than other variants and partly resistant to existing vaccines, scientists fear. BMJ. 2021;2021:375. doi: 10.1136/bmj.n2943. [DOI] [PubMed] [Google Scholar]
- 7.World Health Organisation (2021) Tracking SARS-CoV-2 variants. https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/. Accessed 14 Apr 2022
- 8.Pango Lineages: Latest epidemiological lineages of SARS-CoV-2. https://cov-lineages.org/. Accessed 14 Apr 2022
- 9.CoV-GLUE: enabled by data from GISAID. https://covglue.cvr.gla.ac.uk/. Accessed on Accessed 3 Jun 2022
- 10.Aktas E. Bioinformatics analysis unveils certain mutations implicated in spike structure damage and ligand-binding site of severe acute respiratory syndrome coronavirus 2. Bioinform Biol Insights. 2021;15:11779322211018200. doi: 10.1177/11779322211018200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rauf A, Abu-Izneid T, Olatunde A, Ahmed Khalil A, Alhumaydhi FA, Tufail T, Rengasamy KR. COVID-19 pandemic: epidemiology, etiology, conventional and non-conventional therapies. Int J Environ Res Public Health. 2020;17(21):8155. doi: 10.3390/ijerph17218155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yan Y, Pang Y, Lyu Z, Wang R, Wu X, You C, Zhao H, Manickam S, Lester E, Wu T, Pang CH (2021) The COVID-19 Vaccines: Recent Development, Challenges and Prospects. Vaccines (Basel) 9(4):349. 10.3390/vaccines9040349 [DOI] [PMC free article] [PubMed]
- 13.Nosengo N. Can you teach old drugs new tricks? Nature. 2016;534(7607):314–316. doi: 10.1038/534314a. [DOI] [PubMed] [Google Scholar]
- 14.Hay M, Thomas DW, Craighead JL, Economides C, Rosenthal J. Clinical development success rates for investigational drugs. Nat Biotechnol. 2014;32(1):40–51. doi: 10.1038/nbt.2786. [DOI] [PubMed] [Google Scholar]
- 15.Goldstein I, Lue TF, Padma-Nathan H, Rosen RC, Steers WD, Wicker PA. Oral sildenafil in the treatment of erectile dysfunction. N Engl J Med. 1998;338(20):1397–1404. doi: 10.1056/NEJM199805143382001. [DOI] [PubMed] [Google Scholar]
- 16.Palumbo A, Facon T, Sonneveld P, Blade J, Offidani M, Gay F, Harousseau JL. Thalidomide for treatment of multiple myeloma: 10 years later. Blood J Am Soc Hematol. 2008;111(8):3968–3977. doi: 10.1182/blood-2007-10-117457. [DOI] [PubMed] [Google Scholar]
- 17.Oprea TI, Bauman JE, Bologa CG, Buranda T, Chigaev A, Edwards BS, Sklar LA. Drug repurposing from an academic perspective. Drug Discov Today Ther Strateg. 2011;8(3–4):61–69. doi: 10.1016/j.ddstr.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pushpakom S, Iorio F, Eyers PA, Escott KJ, Hopper S, Wells A, Doig A, Guilliams T, Latimer J, McNamee C, Norris A, Sanseau P, Cavalla D, Pirmohamed M (2019) Drug repurposing: progress, challenges and recommendations. Nat Rev Drug Discov. 18(1):41–58. 10.1038/nrd.2018.168 [DOI] [PubMed]
- 19.Wishart DS, Feunang YD, Guo AC, Lo EJ, Marcu A, Grant JR, Wilson M. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46(D1):D1074–D1082. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Shi ZL. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou Y, Hou Y, Shen J, Huang Y, Martin W, Cheng F. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. 2020;6(1):1–18. doi: 10.1038/s41421-020-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yao TT, Qian JD, Zhu WY, Wang Y, Wang GQ. A systematic review of lopinavir therapy for SARS coronavirus and MERS coronavirus–A possible reference for coronavirus disease-19 treatment option. J Med Virol. 2020;92(6):556–563. doi: 10.1002/jmv.25729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yao X, Ye F, Zhang M, Cui C, Huang B, Niu P, Liu D. In vitro antiviral activity and projection of optimized dosing design of hydroxychloroquine for the treatment of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Clin Infect Dis. 2020;71(15):732–739. doi: 10.1093/cid/ciaa237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harrison C. Coronavirus puts drug repurposing on the fast track. Nat Biotechnol. 2020;38(4):379–381. doi: 10.1038/d41587-020-00003-1. [DOI] [PubMed] [Google Scholar]
- 25.Siddiqi HK, Mehra MR. COVID-19 illness in native and immunosuppressed states: a clinical–therapeutic staging proposal. J Heart Lung Transplant. 2020;39(5):405–407. doi: 10.1016/j.healun.2020.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garrido P, Aldaz A, Vera R, Calleja MA, De Alava E, Martín M, Palacios J. Proposal for the creation of a national strategy for precision medicine in cancer: a position statement of SEOM, SEAP, and SEFH. Clin Transl Oncol. 2018;20(4):443–447. doi: 10.1007/s12094-017-1740-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lam C, Siefkas A, Zelin NS, Barnes G, Dellinger RP, Vincent JL, Das R. Machine learning as a precision-medicine approach to prescribing covid-19 pharmacotherapy with remdesivir or corticosteroids. Clin Ther. 2021;43(5):871–885. doi: 10.1016/j.clinthera.2021.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.El Bairi K, Trapani D, Petrillo A, Le Page C, Zbakh H, Daniele B, Afqir S. Repurposing anticancer drugs for the management of COVID-19. Eur J Cancer. 2020;141:40–61. doi: 10.1016/j.ejca.2020.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Borcherding N, Jethava Y, Vikas P. Repurposing anti-cancer drugs for COVID-19 treatment. Drug Des Dev Ther. 2020;14:5045. doi: 10.2147/DDDT.S282252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Papapetropoulos A, Szabo C. Inventing new therapies without reinventing the wheel: the power of drug repurposing. Br J Pharmacol. 2018;175(2):165. doi: 10.1111/bph.14081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xue H, Li J, Xie H, Wang Y. Review of drug repositioning approaches and resources. Int J Biol Sci. 2018;14(10):1232. doi: 10.7150/ijbs.24612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumar S, Zhi K, Mukherji A, Gerth K. Repurposing antiviral protease inhibitors using extracellular vesicles for potential therapy of COVID-19. Viruses. 2020;12(5):486. doi: 10.3390/v12050486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hamed M, El-Hasab M, Mansour FR. Direct acting anti-hepatitis C combinations as potential COVID-19 protease inhibitors. VirusDisease. 2021;32(2):279–285. doi: 10.1007/s13337-021-00691-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bakowski MA, Beutler N, Wolff KC, Kirkpatrick MG, Chen E, Nguyen TTH, Rogers TF. Drug repurposing screens identify chemical entities for the development of COVID-19 interventions. Nat Commun. 2021;12(1):1–14. doi: 10.1038/s41467-021-23328-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dong W, Xie W, Liu Y, Sui B, Zhang H, Liu L, Peng G. Receptor tyrosine kinase inhibitors block proliferation of TGEV mainly through p38 mitogen-activated protein kinase pathways. Antiviral Res. 2020;173:104651. doi: 10.1016/j.antiviral.2019.104651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCall B. COVID-19 and artificial intelligence: protecting health-care workers and curbing the spread. Lancet Digit Health. 2020;2(4):e166–e167. doi: 10.1016/S2589-7500(20)30054-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sapkota N, Karwowski W, Davahli MR, Al-Juaid A, Taiar R, Murata A, Marek T. The chaotic behavior of the spread of infection during the COVID-19 pandemic in the United States and globally. IEEE Access. 2021;9:80692–80702. doi: 10.1109/ACCESS.2021.3085240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Q, Kang C. Mechanisms of action for small molecules revealed by structural biology in drug discovery. Int J Mol Sci. 2020;21(15):5262. doi: 10.3390/ijms21155262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sleigh SH, Barton CL. Repurposing strategies for therapeutics. Pharm Med. 2010;24(3):151–159. doi: 10.1007/BF03256811. [DOI] [Google Scholar]
- 40.Low ZY, Farouk IA, Lal SK. Drug repositioning: new approaches and future prospects for life-debilitating diseases and the COVID-19 pandemic outbreak. Viruses. 2020;12(9):1058. doi: 10.3390/v12091058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aanouz I, Belhassan A, El-Khatabi K, Lakhlifi T, El-Ldrissi M, Bouachrine M. Moroccan medicinal plants as inhibitors against SARS-CoV-2 main protease: computational investigations. J Biomol Struct Dyn. 2021;39(8):2971–2979. doi: 10.1080/07391102.2020.1758790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Das S, Sarmah S, Lyndem S, Singha Roy A. An investigation into the identification of potential inhibitors of SARS-CoV-2 main protease using molecular docking study. J Biomol Struct Dyn. 2021;39(9):3347–3357. doi: 10.1080/07391102.2020.1763201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Elmezayen AD, Al-Obaidi A, Şahin AT, Yelekçi K. Drug repurposing for coronavirus (COVID-19): in silico screening of known drugs against coronavirus 3CL hydrolase and protease enzymes. J Biomol Struct Dyn. 2021;39(8):2980–2992. doi: 10.1080/07391102.2020.1758791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Islam R, Parves MR, Paul AS, Uddin N, Rahman MS, Mamun AA, Halim MA. A molecular modeling approach to identify effective antiviral phytochemicals against the main protease of SARS-CoV-2. J Biomol Struct Dyn. 2021;39(9):3213–3224. doi: 10.1080/07391102.2020.1761883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alghamdi HA, Attique SA, Yan W, Arooj A, Albulym O, Zhu D, Nawaz MZ. Repurposing the inhibitors of COVID-19 key proteins through molecular docking approach. Process Biochem. 2021;110:216–222. doi: 10.1016/j.procbio.2021.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meng XY, Zhang HX, Mezei M, Cui M. Molecular docking: a powerful approach for structure-based drug discovery. Curr Comput Aided Drug Des. 2011;7(2):146–157. doi: 10.2174/157340911795677602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Skariyachan S, Gopal D, Deshpande D, Joshi A, Uttarkar A, Niranjan V. Carbon fullerene and nanotube are probable binders to multiple targets of SARS-CoV-2: Insights from computational modeling and molecular dynamic simulation studies. Infect Genet Evol. 2021;96:105155. doi: 10.1016/j.meegid.2021.105155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Skariyachan S, Gopal D, Muddebihalkar AG, Uttarkar A, Niranjan V. Structural insights on the interaction potential of natural leads against major protein targets of SARS-CoV-2: molecular modelling, docking and dynamic simulation studies. Comput Biol Med. 2021;132:104325. doi: 10.1016/j.compbiomed.2021.104325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Elfiky AA. Anti-HCV, nucleotide inhibitors, repurposing against COVID-19. Life Sci. 2020;248:117477. doi: 10.1016/j.lfs.2020.117477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Elfiky AA. Ribavirin, Remdesivir, sofosbuvir, galidesivir, and tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): a molecular docking study. Life Sci. 2020;253:117592. doi: 10.1016/j.lfs.2020.117592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang M, Cao R, Zhang L, Yang X, Liu J, Xu M, et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) In Vitro. Cell Res. 2020;30(3):269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang M, Cao R, Zhang L, Yang X, Liu J, Xu M, Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30(3):269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tejera E, Munteanu CR, López-Cortés A, Cabrera-Andrade A, Pérez-Castillo Y. Drugs repurposing using QSAR, docking and molecular dynamics for possible inhibitors of the SARS-CoV-2 Mpro protease. Molecules. 2020;25(21):5172. doi: 10.3390/molecules25215172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oktay L, Erdemoğlu E, İlayda TOLU, Yumak Y, Özcan A, Elif ACAR, Durdaği S. Binary-QSAR guided virtual screening of FDA approved drugs and compounds in clinical investigation against SARS-CoV-2 main protease. Turk J Biol. 2021;45(4):459. doi: 10.3906/biy-2106-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen H, Zhang H, Zhang Z, Cao Y, Tang W. Network-based inference methods for drug repositioning. Comput Math Methods Med. 2015;2015:130620. doi: 10.1155/2015/130620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fakhraei S, Huang B, Raschid L, Getoor L. Network-based drug-target interaction prediction with probabilistic soft logic. IEEE/ACM Trans Comput Biol Bioinf. 2014;11(5):775–787. doi: 10.1109/TCBB.2014.2325031. [DOI] [PubMed] [Google Scholar]
- 57.Li J, Lu Z. Pathway-based drug repositioning using causal inference. BMC Bioinform. 2013;14(16):1–10. doi: 10.1186/1471-2105-14-S16-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tu YF, Chien CS, Yarmishyn AA, Lin YY, Luo YH, Lin YT, Chiou SH. A review of SARS-CoV-2 and the ongoing clinical trials. Int J Mol Sci. 2020;21(7):2657. doi: 10.3390/ijms21072657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.King AD, Pržulj N, Jurisica I. Bacterial Molecular Networks. New York, NY: Springer; 2012. Protein complex prediction with RNSC; pp. 297–312. [DOI] [PubMed] [Google Scholar]
- 60.Vanunu O, Magger O, Ruppin E, Shlomi T, Sharan R. Associating genes and protein complexes with disease via network propagation. PLoS Comput Biol. 2010;6(1):e1000641. doi: 10.1371/journal.pcbi.1000641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ke YY, Peng TT, Yeh TK, Huang WZ, Chang SE, Wu SH, Chen CT. Artificial intelligence approach fighting COVID-19 with repurposing drugs. Biomed J. 2020;43(4):355–362. doi: 10.1016/j.bj.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Beck BR, Shin B, Choi Y, Park S, Kang K. Predicting commercially available antiviral drugs that may act on the novel coronavirus (SARS-CoV-2) through a drug-target interaction deep learning model. Comput Struct Biotechnol J. 2020;18:784–790. doi: 10.1016/j.csbj.2020.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang R, Li S, Cheng L, Wong M, Leung K. Predicting associations among drugs, targets and diseases by tensor decomposition for drug repositioning. BMC Bioinform. 2019;20:628. doi: 10.1186/s12859-019-3283-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pham TH, Qiu Y, Zeng J, Xie L, Zhang P. A deep learning framework for high-throughput mechanism-driven phenotype compound screening and its application to COVID-19 drug repurposing. Nat Mach Intell. 2021;3(3):247–257. doi: 10.1038/s42256-020-00285-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hooshmand SA, Zarei Ghobadi M, Hooshmand SE, Azimzadeh Jamalkandi S, Alavi SM, Masoudi-Nejad A. A multimodal deep learning-based drug repurposing approach for treatment of COVID-19. Mol Diversity. 2021;25(3):1717–1730. doi: 10.1007/s11030-020-10144-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Subramanian A, Narayan R, Corsello SM, Peck DD, Natoli TE, Lu X, Golub TR. A next generation connectivity map: L1000 platform and the first 1,000,000 profiles. Cell. 2017;171(6):1437–1452. doi: 10.1016/j.cell.2017.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Loganathan T, Ramachandran S, Shankaran P, Nagarajan D. Host transcriptome-guided drug repurposing for COVID-19 treatment: a meta-analysis based approach. PeerJ. 2020;8:e9357. doi: 10.7717/peerj.9357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jia Z, Song X, Shi J, Wang W, He K. Transcriptome-based drug repositioning for coronavirus disease 2019 (COVID-19) Pathog Dis. 2020;78(4):ftaa036. doi: 10.1093/femspd/ftaa036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gentile F, Agrawal V, Hsing M, Ton AT, Ban F, Norinder U, Cherkasov A. Deep docking: a deep learning platform for augmentation of structure-based drug discovery. ACS Cent Sci. 2020;6(6):939–949. doi: 10.1021/acscentsci.0c00229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Galindez G, Matschinske J, Rose TD, Sadegh S, Salgado-Albarrán M, Späth J, Pauling JK. Lessons from the COVID-19 pandemic for advancing computational drug repurposing strategies. Nat Comput Sci. 2021;1(1):33–41. doi: 10.1038/s43588-020-00007-6. [DOI] [PubMed] [Google Scholar]
- 71.Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hoffmann M, Schroeder S, Kleine-Weber H, Müller MA, Drosten C, Pöhlmann S. Nafamostat mesylate blocks activation of SARS-CoV-2: new treatment option for COVID-19. Antimicrob Agents Chemother. 2020;64(6):e00754–e820. doi: 10.1128/AAC.00754-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Law JN, Akers K, Tasnina N, Della Santina CM, Kshirsagar M, Klein-Seetharaman J, Murali TM (2020) Identifying human interactors of SARS-CoV-2 proteins and drug targets for COVID-19 using network-based label propagation. arXiv preprint. http://arxiv.org/abs/2006.01968
- 74.Messina F, Giombini E, Agrati C, Vairo F, Ascoli Bartoli T, Al Moghazi S, Ippolito G. COVID-19: viral–host interactome analyzed by network based-approach model to study pathogenesis of SARS-CoV-2 infection. J Transl Med. 2020;18(1):1–10. doi: 10.1186/s12967-020-02405-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mahdian S, Zarrabi M, Panahi Y, Dabbagh S. Repurposing FDA-approved drugs to fight COVID-19 using in silico methods: targeting SARS-CoV-2 RdRp enzyme and host cell receptors (ACE2, CD147) through virtual screening and molecular dynamic simulations. Inform Med Unlocked. 2021;23:100541. doi: 10.1016/j.imu.2021.100541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yuce M, Cicek E, Inan T, Dag AB, Kurkcuoglu O, Sungur FA. Repurposing of FDA-approved drugs against active site and potential allosteric drug-binding sites of COVID-19 main protease. Proteins Struct Funct Bioinform. 2021;89(11):1425–1441. doi: 10.1002/prot.26164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Holshue ML, DeBolt C, Lindquist S, Lofy KH, Wiesman J, Bruce H, et al. First case of 2019 novel coronavirus in the United States. N Engl J Med. 2020;382(10):929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jorgensen SCJ, Kebriaei R, Dresser LD. Remdesivir: review of pharmacology, pre-clinical data, and emerging clinical experience for COVID-19. Pharmacotherapy. 2020;40(7):659–671. doi: 10.1002/phar.2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zou H, Yang Y, Dai H, Xiong Y, Wang JQ, Lin L, Chen ZS. Recent updates in experimental research and clinical evaluation on drugs for COVID-19 treatment. Front Pharmacol. 2021;12:732403. doi: 10.3389/fphar.2021.732403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cao B, Wang Y, Wen D, Liu W, Wang J, Fan G, et al. A trial of lopinavir-ritonavir in adults hospitalized with severe Covid-19. N Engl J Med. 2020;382(19):1787–1799. doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Muhammed Y. Molecular targets for COVID-19 drug development: enlightening Nigerians about the pandemic and future treatment. Biosaf Health. 2020;2(04):210–216. doi: 10.1016/j.bsheal.2020.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Osipiuk J, Azizi SA, Dvorkin S, Endres M, Jedrzejczak R, Jones KA, Joachimiak A. Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors. Nat Commun. 2021;12(1):1–9. doi: 10.1038/s41467-021-21060-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gao Y, Yan L, Huang Y, Liu F, Zhao Y, Cao L, Rao Z. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020;368(6492):779–782. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Machitani M, Yasukawa M, Nakashima J, Furuichi Y, Masutomi K. RNA-dependent RNA polymerase, RdRP, a promising therapeutic target for cancer and potentially COVID-19. Cancer Sci. 2020;111(11):3976–3984. doi: 10.1111/cas.14618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhu W, Chen CZ, Gorshkov K, Xu M, Lo DC, Zheng W. RNA-dependent RNA polymerase as a target for COVID-19 drug discovery. SLAS DISCOV Adv Sci Drug Discov. 2020;25(10):1141–1151. doi: 10.1177/2472555220942123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Roe MK, Junod NA, Young AR, Beachboard DC, Stobart CC. Targeting novel structural and functional features of coronavirus protease nsp5 (3CLpro, Mpro) in the age of COVID-19. J Gen Virol. 2021;102(3):001558. doi: 10.1099/jgv.0.001558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bai Z, Cao Y, Liu W, Li J. The SARS-CoV-2 nucleocapsid protein and its role in viral structure, biological functions, and a potential target for drug or vaccine mitigation. Viruses. 2021;13(6):1115. doi: 10.3390/v13061115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Schoeman D, Fielding BC. Coronavirus envelope protein: current knowledge. Virol J. 2019;16(1):1–22. doi: 10.1186/s12985-019-1182-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Duan L, Zheng Q, Zhang H, Niu Y, Lou Y, Wang H. The SARS-CoV-2 spike glycoprotein biosynthesis, structure, function, and antigenicity: implications for the design of spike-based vaccine immunogens. Front Immunol. 2020;11:2593. doi: 10.3389/fimmu.2020.576622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yadav R, Chaudhary JK, Jain N, Chaudhary PK, Khanra S, Dhamija P, Handu S. Role of structural and non-structural proteins and therapeutic targets of SARS-CoV-2 for COVID-19. Cells. 2021;10(4):821. doi: 10.3390/cells10040821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Greaney AJ, Loes AN, Crawford KH, Starr TN, Malone KD, Chu HY, Bloom JD. Comprehensive mapping of mutations in the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human plasma antibodies. Cell Host Microbe. 2021;29(3):463–476. doi: 10.1016/j.chom.2021.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Weisblum Y, Schmidt F, Zhang F, DaSilva J, Poston D, Lorenzi JC, Bieniasz PD. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. Elife. 2020;9:e61312. doi: 10.7554/eLife.61312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Baum A, et al. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science. 2020;369:1014–1018. doi: 10.1126/science.abd0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Liu Z, et al. Identification of SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. Cell Host Microbe. 2021;29:477–488. doi: 10.1016/j.chom.2021.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Starr TN, et al. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell. 2020;182:1295–1310.e1220. doi: 10.1016/j.cell.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Thomson EC, et al. Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity. Cell. 2021;184:1171–1187. doi: 10.1016/j.cell.2021.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Harvey WT, Carabelli AM, Jackson B, Gupta RK, Thomson EC, Harrison EM, Robertson DL. SARS-CoV-2 variants, spike mutations and immune escape. Nat Rev Microbiol. 2021;19(7):409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.McCallum M, et al. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell. 2021 doi: 10.1016/j.cell.2021.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.McCarthy KR, et al. Recurrent deletions in the SARS-CoV-2 spike glycoprotein drive antibody escape. Science. 2021;371:1139–1142. doi: 10.1126/science.abf6950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Uttarkar A, Niranjan V, Pandit S, Subash S. Study of SARS-nCoV2 Indian isolates gaining insights into mutation frequencies, protein stability and prospective effect on pathogenicity. Coronaviruses. 2021;2(10):e170821190411. doi: 10.2174/2666796702666210118155636. [DOI] [Google Scholar]
- 101.Lavanya C, Upadhyaya A, Neogi AG, Niranjan V. Identification of novel regulatory pathways across normal human bronchial epithelial cell lines (NHBEs) and peripheral blood mononuclear cell lines (PBMCs) in COVID-19 patients using transcriptome analysis. Inform Med Unlocked. 2022;31:100979. doi: 10.1016/j.imu.2022.100979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Deshpande RR, Tiwari AP, Nyayanit N, Modak M. In silico molecular docking analysis for repurposing therapeutics against multiple proteins from SARS-CoV-2. Eur J Pharmacol. 2020;886:173430. doi: 10.1016/j.ejphar.2020.173430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cavasotto CN, Di Filippo JI. In silico drug repurposing for COVID-19: Targeting SARS-CoV-2 proteins through docking and consensus ranking. Mol Inf. 2021;40(1):2000115. doi: 10.1002/minf.202000115. [DOI] [PubMed] [Google Scholar]
- 104.Eweas AF, Alhossary AA, Abdel-Moneim AS (2021) Molecular docking reveals ivermectin and remdesivir as potential repurposed drugs against SARS-CoV-2. Front Microbiol 3602 [DOI] [PMC free article] [PubMed]
- 105.Skariyachan S, Gopal D, Chakrabarti S, Kempanna P, Uttarkar A, Muddebihalkar AG, Niranjan V. Structural and molecular basis of the interaction mechanism of selected drugs towards multiple targets of SARS-CoV-2 by molecular docking and dynamic simulation studies-deciphering the scope of repurposed drugs. Comput Biol Med. 2020;126:104054. doi: 10.1016/j.compbiomed.2020.104054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cvetkovic RS, Goa KL. Lopinavir/ritonavir. Drugs. 2003;63(8):769–802. doi: 10.2165/00003495-200363080-00004. [DOI] [PubMed] [Google Scholar]
- 107.Nhean S, Varela ME, Nguyen YN, Juarez A, Huynh T, Udeh D, Tseng AL (2021) COVID-19: a review of potential treatments (corticosteroids, Remdesivir, tocilizumab, bamlanivimab/etesevimab, and casirivimab/imdevimab) and pharmacological considerations. J Pharm Pract 08971900211048139 [DOI] [PMC free article] [PubMed]
- 108.Vora A, Tiwaskar M. Favipiravir. J Assoc Phys India. 2020;68:91–92. [PubMed] [Google Scholar]
- 109.Sheahan TP, Sims AC, Graham RL, et al. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci Transl Med. 2017;9(396):eaal3653. doi: 10.1126/scitranslmed.aal3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Brown AJ, Won JJ, Graham RL, et al. Broad spectrum antiviral remdesivir inhibits human endemic and zoonotic deltacoronaviruses with a highly divergent RNA dependent RNA polymerase. Antivir Res. 2019;169:104541. doi: 10.1016/j.antiviral.2019.104541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Williamson B, Feldmann F, Schwarz B, et al. Clinical benefit of remdesivir in rhesus macaques infected with SARS-CoV-2. Nature. 2020 doi: 10.1038/s41586-020-2423-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kumar GN, Jayanti V, Lee RD, Whittern DN, Uchic J, Thomas S, Denissen JF. In vitro metabolism of the HIV-1 protease inhibitor ABT-378: species comparison and metabolite identification. Drug Metab Dispos. 1999;27(1):86–91. [PubMed] [Google Scholar]
- 113.Thompson EM, Ashley D, Landi D. Current medulloblastoma subgroup specific clinical trials. Transl Pediatr. 2020;9(2):157. doi: 10.21037/tp.2020.03.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Liu X, Li Z, Liu S, Sun J, Chen Z, Jiang M, Luo HB. Potential therapeutic effects of dipyridamole in the severely ill patients with COVID-19. Acta Pharm Sin B. 2020;10(7):1205–1215. doi: 10.1016/j.apsb.2020.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Patterson BK, Seethamraju H, Dhody K, Corley MJ, Kazempour K, Lalezari J, Sacha JB. CCR5 inhibition in critical COVID-19 patients decreases inflammatory cytokines, increases CD8 T-cells, and decreases SARS-CoV2 RNA in plasma by day 14. Int J Infect Dis. 2021;103:25–32. doi: 10.1016/j.ijid.2020.10.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Trkola A, Ketas TJ, Nagashima KA, Zhao L, Cilliers T, Morris L, Olson WC. Potent, broad-spectrum inhibition of human immunodeficiency virus type 1 by the CCR5 monoclonal antibody PRO 140. J Virol. 2001;75(2):579–588. doi: 10.1128/JVI.75.2.579-588.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Song WC, FitzGerald GA. COVID-19, microangiopathy, hemostatic activation, and complement. J Clin Investig. 2020;130(8):3950–3953. doi: 10.1172/JCI140183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Pum A, Ennemoser M, Adage T, Kungl AJ. Cytokines and chemokines in SARS-CoV-2 infections–therapeutic strategies targeting cytokine storm. Biomolecules. 2021;11(1):91. doi: 10.3390/biom11010091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Drug Bank Online. Building the foundation for better health outcomes. https://go.drugbank.com/. Accessed 5 Jun 2022
- 120.Ge Y, Tian T, Huang S, Wan F, Li J, Li S, Zeng J. A data-driven drug repositioning framework discovered a potential therapeutic agent targeting COVID-19. BioRxiv; 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wei G, Twomey D, Lamb J, Schlis K, Agarwal J, Stam RW, Armstrong SA. Gene expression-based chemical genomics identifies rapamycin as a modulator of MCL1 and glucocorticoid resistance. Cancer Cell. 2006;10(4):331–342. doi: 10.1016/j.ccr.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 122.Smith SB, Dampier W, Tozeren A, Brown JR, Magid-Slav M. Identification of common biological pathways and drug targets across multiple respiratory viruses based on human host gene expression analysis. PLoS ONE. 2012;7(3):e33174. doi: 10.1371/journal.pone.0033174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hensley LE, Fritz EA, Jahrling PB, Karp C, Huggins JW, Geisbert TW. Interferon-β 1a and SARS coronavirus replication. Emerg Infect Dis. 2004;10(2):317. doi: 10.3201/eid1002.030482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lim HS, Im JS, Cho JY, Bae KS, Klein TA, Yeom JS, Park JW. Pharmacokinetics of hydroxychloroquine and its clinical implications in chemoprophylaxis against malaria caused by Plasmodium vivax. Antimicrob Agents Chemother. 2009;53(4):1468–1475. doi: 10.1128/AAC.00339-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Levin MJ, Ustianowski A, De Wit S, Launay O, Avila M, Templeton A, Esser MT (2022) Intramuscular AZD7442 (Tixagevimab–CILGAVIMAB) for prevention of COVID-19. N Engl J Med [DOI] [PMC free article] [PubMed]
- 126.Boyle DL, Kim HR, Topolewski K, Bartok B, Firestein GS. Novel dual phosphoinositide 3-kinase-delta, gamma inhibitor: Potent anti-inflammatory effects and joint protection in models of rheumatoid arthritis. J Pharmacol Exp Ther. 2013;348(2):271–280. doi: 10.1124/jpet.113.205955. [DOI] [PubMed] [Google Scholar]