Abstract
Females and males often have markedly different mortality rates and life spans, but it is unclear why these forms of sexual dimorphism evolve. The unguarded X hypothesis contends that dimorphic life spans arise from sex differences in X or Z chromosome copy number (i.e., one copy in the “heterogametic” sex; two copies in the “homogametic” sex), which leads to a disproportionate expression of deleterious mutations by the heterogametic sex (e.g., mammalian males; avian females). Although data on adult sex ratios and sex‐specific longevity are consistent with predictions of the unguarded X hypothesis, direct experimental evidence remains scant, and alternative explanations are difficult to rule out. Using a simple population genetic model, we show that the unguarded X effect on sex differential mortality is a function of several reasonably well‐studied evolutionary parameters, including the proportion of the genome that is sex linked, the genomic deleterious mutation rate, the mean dominance of deleterious mutations, the relative rates of mutation and strengths of selection in each sex, and the average effect of mutations on survival and longevity relative to their effects on fitness. We review published estimates of these parameters, parameterize our model with them, and show that unguarded X effects are too small to explain observed sex differences in life span across species. For example, sex differences in mean life span are known to often exceed 20% (e.g., in mammals), whereas our parameterized models predict unguarded X effects of a few percent (e.g., 1–3% in Drosophila and mammals). Indeed, these predicted unguarded X effects fall below statistical thresholds of detectability in most experiments, potentially explaining why direct tests of the hypothesis have generated little support for it. Our results suggest that evolution of sexually dimorphic life spans is predominantly attributable to other mechanisms, potentially including “toxic Y” effects and sexual dimorphism for optimal investment in survival versus reproduction.
Keywords: Deleterious mutations, evolution of life span, evolutionary theory, inbreeding depression, population genetics, sex chromosomes, sex ratio, sexual dimorphism
Impact Summary
Females and males are dimorphic for a wide range of traits, including the average lengths of their life spans. Sex differences in life span are both conspicuous and variable among species. For example, in mammals, females live ∼20% longer than males (on average), whereas in birds, males live ∼10% longer than females. One leading explanation for these patterns—the unguarded X hypothesis—argues that sex differences in life span emerge from the distinct sex chromosomes that females and males inherit. For many species, one sex (e.g., female mammals; male birds) carries two copies of each X‐linked gene, whereas the other carries one. Because harmful mutations are partially recessive, the sex with only one copy of the X is more prone to expressing them, and that sex should therefore have a shorter average life span. This prediction of the unguarded X hypothesis is qualitatively consistent with observations of sex‐ratio bias in adults and sexual dimorphism for longevity (e.g., mammalian males have one copy of the X and have shorter lives than females). However, there are other possible explanations for these patterns, making it unclear how much the unguarded X explains species diversity for sex‐specific longevity. We developed a mathematical model for the contribution of unguarded X effects to sex differences in survival and life span, and used data on mutation rates and their effects on survival and fitness to quantify the importance of the unguarded X across species. The model, when combined with current data, suggests that the unguarded X hypothesis cannot explain the conspicuous sex differences in life span that are commonly reported in animal species, particularly vertebrates. Our results suggest that the unguarded X is an unlikely general explanation for the evolution of sexually dimorphic life spans, which gives weight to alternative mechanisms, including “toxic Y” effects and sex differential selection via trade‐offs between survival and reproduction.
Sex differences in life span are widely observed in nature, yet the evolutionary causes of these differences are unresolved and subject to much debate (Maklakov and Lummaa 2013; Bronikowski et al. 2022). Sexually dimorphic mortality rates might have evolved because they are adaptive and reflect sex differences in selection for traits promoting survival versus reproduction (Bonduriansky et al. 2008). Alternatively, sex differences in survival may emerge from sex differences in genetic inheritance (Maklakov and Lummaa 2013). For example, maternal inheritance of mitochondrial genes can lead to the accumulation of mutations that preferentially decrease male life span (i.e., the “mother's curse” hypothesis; Frank and Hurst 1996; Gemmell et al. 2004). Or, because sex chromosomes are asymmetrically inherited between the sexes, the deleterious mutations they harbor can differentially affect female and male fitness components, including survival (i.e., “toxic Y” and “unguarded X” hypotheses; Trivers 1985; Marais et al. 2018; Brown et al. 2020). These scenarios are not mutually exclusive and may all play some role in generating sex differences in life span.
The unguarded X hypothesis, which posits that sexual dimorphism in life span is a simple consequence of the different number of X (or Z) chromosome copies that each sex inherits, is built upon three well‐established findings from evolutionary genetics: (1) random mutations are far more likely to be harmful than beneficial; (2) these deleterious mutations are maintained at low frequencies, at an evolutionary balance between recurrent mutation and purifying selection; and (3) fitness costs of deleterious mutations tend to be partially recessive (Lynch et al. 1999; Charlesworth 2015). These conditions render haploid and inbred individuals more susceptible to expressing harmful genetic variation than outbred and diploid individuals from the same population. Consequently, the sex that is haploid for X‐ or Z‐linked genes (the “heterogametic” sex) should be more susceptible to expressing mutations that decrease survival (all else being equal) than the sex that is diploid for the X or Z chromosome (the “homogametic” sex). Unguarded X effects should, therefore, preferentially reduce male life spans in species with X chromosomes and female life spans in species with Z chromosomes.
The unguarded X hypothesis is consistent with several recently reported empirical patterns. Suggestive evidence for the hypothesis comes from field surveys of adult sex ratios (which may reflect sex differential mortality; Pipoly et al. 2015) and estimates of sex‐specific aging and longevity (Xirocostas et al. 2020; Cayuela et al. 2022). Estimates of adult sex ratios are typically skewed toward the homogametic sex (i.e., females in species with X chromosomes; males in species with a Z; Pipoly et al. 2015; Marais and Lemaitre 2022), as predicted by the unguarded X hypothesis. Similarly, estimates of sex‐specific aging and longevity indicate that the heterogametic sex has a lower mean life span than the homogametic sex (Xirocostas et al. 2020; Marais and Lemaitre 2022). These sex ratio and longevity data span a relatively small fraction of species, with terrestrial vertebrates overrepresented; further study will be required to evaluate whether the reported associations between sex chromosome system and metrics of sex‐specific survival generalize across the tree of life.
Although the patterns summarized above are broadly consistent with the unguarded X hypothesis, it is unclear whether X‐ or Z‐linkage is causal (as noted by Xirocostas et al. 2020). Indeed, taxonomic surveys (as above) can test for correlations between sex chromosome systems and sex‐specific survival, but cannot exclude other plausible contributors. For example, a strong Y‐ or W‐linked genetic basis of mortality, as suggested by recent Drosophila experiments (Brown et al. 2020; Nguyen and Bachtrog 2021), could generate taxonomic patterns that parallel those predicted by the unguarded X (Sultanova et al. 2020). Moreover, the sex determination system of a species can potentially affect the evolution of traits that are pertinent to mortality, which may lead to co‐evolved associations between the sex chromosome system and sex‐specific patterns of survival and longevity. Species with X versus Z chromosomes may, for example, be differentially prone to evolving traits that increase male mating success and decrease male survival (Reeve and Pfennig 2003; Albert and Otto 2005).
Because a range of environmental or evolutionary variables may systematically differ between species carrying an X versus a Z chromosome, direct experimental tests for unguarded X effects are essential for evaluating the hypothesis. One experimentally testable prediction of the unguarded X is that inbreeding depression should have a weaker effect on the heterogametic sex (which cannot be inbred for the X or Z) compared to the homogametic sex (which can). Although Vega‐Trejo et al. (2022) recently showed (using meta‐analysis) that inbreeding depression is generally higher in females than males (also see Carazo et al. 2016; Sultanova et al. 2018), this pattern is independent of the sex chromosome system. Moreover, as the authors note, there are other reasons aside from sex linkage why inbreeding depression may differentially affect the sexes, including intrinsic sex differences in the effects of mutations on fitness components (e.g., Whitlock and Agrawal 2009). Direct experimental tests of the unguarded X hypothesis—carried out in Drosophila where manipulative experiments of the effect of X‐linked homozygosity on fitness components are feasible—have likewise failed to identify even marginal unguarded X effects (see Eanes et al. 1985; Brengdahl et al. 2018; Narayan et al. 2022).
Here, we argue that there is a more fundamental reason to doubt that unguarded X effects contribute substantially to sexually dimorphic life spans. Building upon prior theory (e.g., Werren 1993; Charlesworth and Charlesworth 1999; Pipoly et al. 2015), we present a simple model that shows that the magnitude of the unguarded X effect depends on several evolutionary parameters that previous studies have estimated, including the genomic deleterious mutation rate, the fraction of the haploid genome that is X‐ or Z‐linked, the average dominance of deleterious mutations, sex biases in mutation and selection, and the average effects of mutations on individual fitness components (e.g., survival and longevity) relative to their effects on total fitness. All of these parameters have been estimated in Drosophila, where we predict that unguarded X effects generate sex differences in life span on the order of 3% or less. Furthermore, available data on sex chromosome sizes, mutation rates, and fitness effects, which we review, imply that unguarded X effects in other taxa should generally be of similar magnitude to those predicted for Drosophila. The magnitude of our estimates is too small to explain the large sex differences in life span that are often documented in animals (e.g., mammals, where females live on average ∼20% longer than males; Lemaître et al 2020). Hence, although unguarded X effects should contribute to sex differences in life span, other factors are likely to be more important in explaining conspicuous patterns of sexual dimorphism for aspects of survival.
A Model of the Unguarded X
We consider a simple model for the evolution of unguarded X effects arising from a large number of loci maintained at mutation‐selection balance. The focus of the unguarded X hypothesis is on fitness components relevant to mortality in each sex, including survival to reproductive maturity and longevity, and we therefore focus on these fitness components in our model. We present explicit results for the case of male heterogamety assuming that the Y chromosome is degenerate (i.e., XX/XY or XX/XO sex chromosome systems, where males are haploid for X‐linked genes) and the X chromosome is functionally diploid in females (e.g., in mammals with random X inactivation; Deng et al. 2014). Our results are easily reframed to cases of female heterogamety (Z chromosome systems with a degenerate W), by simply reversing the sex labels in our models.
Following previous theory (e.g., Haldane 1937; Connallon 2010), and assuming strong selection relative to mutation (, where and represent the female and male mutation rates for the ith locus, sf,i and sm,i are the selection coefficients, and hi is the dominance coefficient; Table 1), the equilibrium frequency of a deleterious a allele at the ith X‐linked locus is
| (1) |
Table 1.
Effect of the ith X‐linked locus on sex‐specific survival and overall fitness
| X‐Linked Genotype | |||
|---|---|---|---|
| AA, A | Aa, – | aa, a | |
| Female fitness | 1 | 1 – hisf,i | 1 – sf,i |
| Male fitness | 1 | – | 1 – sm,i |
| Female survival | 1 | 1 – hisf,iαf,i | 1 – sf,iαf,i |
| Male survival | 1 | – | 1 – sm,iαm,i |
Parameters include female and male selection coefficients (sf,i , sm,i ), the dominance coefficient (hi ), and the effect of the mutation on survival relative to its effect on overall fitness (αf,i , αm,i , where, e.g., αf,i = 1 under pure viability selection and αf,i = 0 under pure fecundity selection).
The contribution of the locus to mean survival or longevity of females and males, respectively, will be and , where represents the effect of the deleterious mutation on the fitness component of the jth sex relative to its effect on overall fitness (see Charlesworth 2015; Table 1).
Following Charlesworth and Charlesworth (1999), we assume that loci have multiplicative effects on fitness and fitness components. With nX X‐linked loci at mutation‐selection balance, the mean value of the female fitness component with respect to the X‐linked loci will be
| (2) |
and the mean value of the male fitness component will be
| (3) |
The unguarded X effect can be quantified by taking the ratio of equations (2) and (3):
| (4) |
Unguarded X effects contribute to lower survival or longevity in the heterogametic sex when equation (4) exceeds unity (), which requires that the summation is positive.
BASELINE PREDICTIONS FOR THE UNGUARDED X
If we assume that the rates and effects of mutations are equal between the sexes (i.e., sf,i = sm,i and αf,i = αm,i ), and that hi , , and are independently distributed across X‐linked loci, then equation (4) simplifies to
| (5) |
which is derived using a Taylor series expansion, to second order in , where is the mean and var(h) is the variance of the dominance coefficients of deleterious mutations; is the average effect of mutations on the fitness component relative to overall fitness; and is the total deleterious mutation rate per X chromosome.
If we further assume that deleterious mutations are generally partially recessive (0 < hi < 0.5), which is consistent with mutation‐accumulation data (Manna et al. 2011; Charlesworth 2015), then terms of var(h) will be small enough to neglect (see the Supporting Information), and equation (5) simplifies to
| (6) |
Thus, the magnitude of the unguarded X effect should increase with the X‐linked deleterious mutation rate (UX ) and the mean effect of mutations on the fitness component relative to their effects overall fitness (). The unguarded X effect should decrease with mean dominance of deleterious mutations (). For the extreme case in which mutations are completely recessive (hi = 0), and purifying selection is entirely based on mutational effects on viability (, corresponding to the unlikely scenario where mutations do not affect reproductive fitness components), equation (6) simplifies to , which represents an upper limit for the unguarded X effect that was previously derived by Pipoly et al. (2015).
SEX DIFFERENCES IN SELECTION AND THE UNGUARDED X
Our baseline model (eq. 6) should provide reasonable predictions for unguarded X effects provided mutations have similar effects on fitness and the fitness components of each sex. We can think of three plausible ways that this assumption may be violated. First, at least some deleterious mutations have sex‐limited fitness effects (e.g., mutations underlying sterility are typically sex limited; Connallon and Clark 2011). Second, stronger sexual selection on one sex (e.g., males) can give rise to systematic sex differences in the strength of purifying selection against deleterious mutations (Whitlock and Agrawal 2009; Grieshop et al. 2021). Third, several studies argue that a lack of dosage compensation, which pertains to some taxa with strongly heteromorphic sex chromosomes, might dampen the phenotypic effects of mutations in the heterogametic (e.g., Charlesworth et al. 1987; Hitchcock and Gardner 2020; Rayner et al. 2021). As we will show, each of these factors tend, if anything, to dampen unguarded X effects, particularly in male‐heterogametic species.
First, consider the case of sex‐limited selection against deleterious mutations. Because we have no reason to expect sex differences in the number of loci affecting viability, we consider the unrealistic but illuminating case where half of X‐linked loci have female‐limited fitness effects and the other half have male‐limited effects (current data, nevertheless, imply that most mutations decrease fitness of both sexes; e.g., Mallet et al. 2011; Sharp and Agrawal 2013). In this idealized case, equations ((2), (3), (4)) simplify to , , and . The latter expression suggests that unguarded X effects will vanish, regardless of the dominance of deleterious mutations, when . Although unguarded X effects could persist when viability‐related fitness components are more important (relative to total fitness) for males than for females (i.e., when ), stronger sexual selection in males should, if anything, tend to disproportionately decrease the relative contribution of viability‐related fitness components to male fitness. If so, we expect a higher X‐linked load for the survival or longevity in females compared to males (i.e., when ), thereby reversing the unguarded X effect.
Second, consider the more plausible case of sex‐differential purifying selection, where mutations similarly affect viability in each sex ( in our models), but sexual selection leads to stronger purifying selection in males (sm,i > sf,i ). Following the same approach that we used to derive equation (6) and assuming, for simplicity, that that the ratios β = sm,i /sf,i = αf,i /αm,i are constant across loci, the unguarded X effect becomes
| (7) |
which, by definition, must have a smaller magnitude than our baseline model (eq. 6) whenever purifying selection is stronger in males (i.e., β > 1). To the extent that sexual selection increases β, unguarded X effects on survival and longevity should be dampened. Overall, we expect sexual selection to decrease the magnitude of the unguarded X effect in species with XX/XY (or XX/XO) sex chromosome systems. However, the reverse may be true in species with Z chromosomes, in which loci with sex‐limited mutational effects and stronger purifying selection in males than females may accentuate “unguarded Z” effects relative to our baseline model (eq. 6).
Third, consider a scenario in which a lack of dosage compensation systematically reduces the hemizygous fitness effects of X‐linked mutations relative to their effects in homozygous state, as some previous theoretical models have assumed (e.g., Charlesworth et al. 1987; Hitchcock and Gardner 2020). Although mutation accumulation data are needed to validate this assumption, evaluating how it should affect predictions about the direction and magnitude of the unguarded X effect is straightforward (see the Supporting Information, including Fig. S1). Under such conditions, we show that the range of dominance coefficients that will generate an unguarded X effect becomes more restrictive in species lacking dosage compensation relative to those with dosage compensation. Moreover, the magnitude of the unguarded X effect, when it does occur, is dampened in species that lack dosage compensation, except in the unlikely scenario where all deleterious mutations are completely recessive.
Unguarded X Effects Predicted from Theory and Mutation Data
All of the key parameters in our unguarded X model (UX , , , , β) have been estimated in Drosophila, and a subset have also been estimated in other taxa. We first review what we currently know about these parameters (we have greatly benefitted from relatively recent reviews that focus on one or more of these parameters; see Manna et al. 2011; Charlesworth 2015). We subsequently use these estimates to predict the likely contribution of the unguarded X to observed sex differences and life span.
X‐LINKED AND Z‐LINKED DELETERIOUS MUTATION RATES
The total X‐linked or Z‐linked deleterious mutation rate, UX , depends on three factors that vary among species: (1) the haploid genomic deleterious rate (UH ), which is most accurately estimated using modern genomic approaches (see below); (2) the proportion of the haploid genome that is X‐linked (PX ) or Z‐linked (PZ ), which has been estimated from karyotype data and annotated, whole‐genome sequences (e.g., Ross et al. 2005; Stiglec et al. 2007; Sultanova et al. 2020); and (3) the ratio of male to female mutation rates (), which can be estimated from the sequences of parent‐offspring trios or from molecular evolution data (e.g., Ellegren 2007; Hedrick 2007). Taking these factors into account, the total X‐linked deleterious mutation is
| (8) |
and the Z‐linked deleterious mutation rate is
| (9) |
In species (like Drosophila) where sex biases in mutation are trivial, equations (8) and (9) reduce to and . In species with strongly male‐biased mutation rates, UX and UZ approach the following limits: and .
Genomic deleterious mutation rate estimates are primarily based on autosomal sequences, where the mutation rates of each sex receive equal weighting. For example, with n functional sites on the autosomes, the haploid genomic deleterious mutation rate will be , where is the mean mutation rate per nucleotide site (Keightley 2012; Lynch et al. 2016). UH can be estimated using genome sequence data (Kondrashov and Crow 1993; Haag‐Liautard et al. 2007), in which n is based on estimates of genome size and the proportion of sites that are evolutionarily conserved, and is estimated by sequencing mutation‐accumulation lines or pedigrees. Current estimates of UH range from ∼0.5 in Caenorhabditis elegans, Drosophila, and mice (Denver et al. 2004; Haag‐Liautard et al. 2007; Uchimura et al. 2015) to UH ∼ 1.1 in humans (Keightley 2012; Dukler et al. 2021). Recent work suggests that UH is likely to be higher in species with relatively small historical effective population sizes (Lynch et al. 2016; e.g., humans and many other terrestrial vertebrates), and the human estimates may thus be tentatively regarded as near the upper end of UH among species. Nevertheless, it is currently difficult to generalize about the distribution of UH among species, and further empirical attention to the issue is warranted.
Most species fall somewhere along a spectrum of modest‐to‐strong male‐biased mutation rates (; Fig. 1), in which and , so that male‐biased mutation should somewhat dampen X‐linked mutation rates and elevate Z‐linked rates. We compiled estimates of male‐biased mutation rate from peer‐reviewed articles and preprints up to February 9, 2022, by searching Google Scholar and the Web of Science using key phrases: “sex‐biased mutation,” “male‐biased mutation,” “mutation rate,” “substitution rate,” and “male‐driven evolution.” Where available, we recorded point estimates of , upper and lower bounds for the estimate (e.g., confidence intervals), the species or lineage associated with the estimate, and the method and primary source from which estimates were derived (the entire dataset is provided in the Supporting Information). We excluded studies that were unlikely to be informative for genome‐wide sex biases in mutation (i.e., estimates calculated exclusively using disease‐affecting loci or using organelle genomes) and those whose estimates were later corrected (e.g., Labuda et al. 2010; Lohmueller et al. 2010). Overall, we collected 206 estimates, from 48 studies and 147 species or lineages. The vast majority of estimates were from mammalian (46.3% of all estimates) and avian taxa (44.4%), and were calculated using the molecular evolutionary method (91.2%) proposed by Miyata et al. (1987). The data (Fig. 1) suggest that, typically, for mammals and in birds. Although fewer estimates are available for other taxa, the full dataset indicated that most species should fall within the range, and for Drosophila species.
Figure 1.
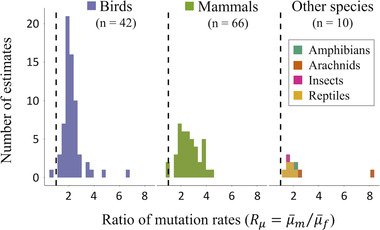
Curated published estimates of species‐specific sex‐biased mutation rates, expressed as the ratio of male to female rates (). Results were compiled by surveying the primary literature, including preprints (for details, see the main text and the Supporting Information). We focused on the subset of estimates in which sex‐biased mutation rates could be calculated for individual species or species lineages rather than groups of related species. In cases where there were multiple estimates for a species, we took the average (we excluded a single outlier of from humans; Wilson Sayres et al. 2011). This process yielded a total of 118 species‐specific estimates (numbers per species group are shown in the figure) from 29 studies (17 mammal, six bird, six other). The vertical broken lines denote equal rates of mutation between the sexes (); values exceeding one are male biased .
EFFECTS OF DELETERIOUS MUTATIONS ON VIABILITY, LONGEVITY, AND FITNESS
Representative data on the dominance coefficients of deleterious mutations have been obtained from mutation‐accumulation experiments using model organisms (e.g., Drosophila, C. elegans, and Saccharomyces cerevisiae). Although there are many case studies of dominance associated with segregating polymorphisms, these are generally unfit for purpose because they represent a biased sample: such loci have been filtered by natural selection and are undoubtedly enriched for alleles with unusually large (and, thus, empirically discernible) phenotypic effects. Focusing on mutation‐accumulation results, the Drosophila data suggest a mean dominance coefficient of (Charlesworth 2015). A meta‐analysis of estimates spanning four decades, and including data from Drosophila, C. elegans, and S. cerevisiae, yields a similar conclusion ( with a plausible range of ; see Manna et al. 2011). Moreover, these results conform closely to theoretical predictions for the dominance of deleterious mutations arising in populations that have evolved to an optimum (Manna et al. 2011).
Estimates of for several fitness components have been obtained from Drosophila mutation‐accumulation experiments (reviewed in table S8 of Charlesworth 2015), including estimates for juvenile viability (i.e., egg‐to‐adult survival, where ) and longevity (where ). Estimates of are typically much lower than one‐half, consistent with studies of standing genetic variation that show substantially weaker genotypic effects on viability components relative to total fitness (e.g., Wilton and Sved 1979; Eanes et al. 1985). Although we lack estimates of from vertebrates, the Drosophila estimates for viability and longevity may very well exceed those of vertebrate taxa; these longer lived species are likely to have lower rates of preadult mortality relative to Drosophila yet high variances in reproductive success among adults, which should cause to be smaller for vertebrate viability.
Finally, sex differences in the overall strength of purifying selection (which affects β in eq. 7) have been estimated in several insect studies, although the most powerful tests have focused on Drosophila. Their results suggest that new deleterious mutations tend to harm both sexes, with the strength of purifying selection likely to range from equal between the sexes to roughly 50% stronger in males than females (; Mallet et al. 2011; Sharp and Agrawal 2013; Singh and Agrawal 2022). Recent work further suggests that sex biases in purifying selection are influenced by both the mating system and extrinsic environmental factors that may vary over time, space, or among taxa (Long et al. 2009; Yun et al. 2018).
PREDICTING THE MAGNITUDE OF UNGUARDED X EFFECTS IN Drosophila AND OTHER SPECIES
In Drosophila melanogaster, the X represents upward of 20% of the genome (PX ∼ 0.2) and mutation rates are approximately equal between the sexes (), which (given UH ∼ 0.5; Haag‐Liautard et al. 2007) suggests an X‐linked deleterious mutation rate of UX = 0.1. Assuming , and using (Charlesworth 2015) and β = 1 (the approximate lower bound for sex‐specific purifying selection; see above), we estimate that the unguarded X should lead to a 3% elevation in female relative to male viability or life span (; Fig. 2). Parameterization using lower values of and higher values of β decreases the magnitude of the unguarded X effect for D. melanogaster (e.g., given and β = 1.5, which are plausible, , corresponding to a sex difference of less than 1%). These small predicted unguarded X effects will become even smaller if a substantial fraction of mutations have sex‐limited effects on life span because sex‐limited loci tend to equalize the contribution of X‐linked mutations to sex differences in mortality (see above).
Figure 2.
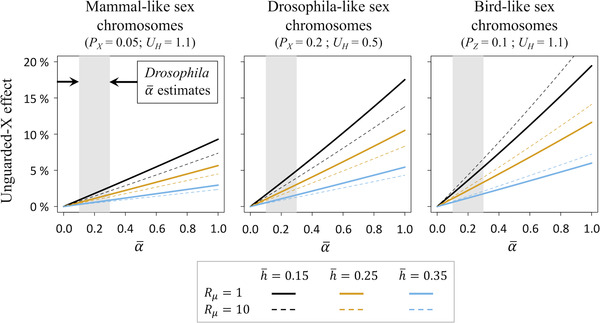
Unguarded X effects in species with mammalian‐like, Drosophila‐like, and bird‐like sex chromosomes. Results are shown for plausible values of the haploid genomic deleterious mutation rate (UH = 1.1 for vertebrates, based on a relatively high estimate from humans: Keightley 2012; Dukler et al. 2021; UH = 0.5 for Drosophila: Haag‐Liautard et al. 2007), with typical sex chromosome sizes for mammals, flies, and birds (bird results are based on the upper end of range of Z chromosome sizes: 0.07 < PZ < 0.1; Stiglec et al. 2007; Sultanova et al. 2020), and equally strong purifying selection in each sex (β = 1). The gray‐shaded regions correspond to the range of estimates obtained from Drosophila studies (; Charlesworth 2015), and the orange lines correspond to dominance values consistent with both theory and mutation accumulation studies from model organisms (; see Manna et al. 2011).
What about other species? Relative to D. melanogaster, the proportion of the genome that is X‐ or Z‐linked is smaller in most other taxa (Tree of Sex Consortium 2014; Sultanova et al. 2020). Because there is little reason to expect major differences in UH , , and between Drosophila and other insect species, the smaller sex chromosomes of most insect species (e.g., PZ ∼ 0.02–0.05 in Lepidoptera: Presgraves 2002; Fraïsse et al. 2017; PX ∼ 0.05 in fireflies [Coleoptera]: Lower et al. 2017; PX < 0.08 in cockroaches [Blattodea]: Meisel et al. 2019; PX < 0.1 is generally implied by the high chromosome numbers of most insects orders: Blackmon et al. 2017; Sylvester et al. 2020) should lead to even smaller unguarded X (or Z) effects than predicted for D. melanogaster. Important exceptions include the subset of Drosophila species with larger sex chromosomes than D. melanogaster (i.e., PX ∼ 0.4; see Turelli and Begun 1997) and haplo‐diploid species (e.g., Hymenoptera, where PX = 1).
Although vertebrates also have smaller sex chromosomes than Drosophila, they may typically have higher genomic deleterious mutation rates (given the negative relation between UH and population size, in which the latter is known to be smaller in vertebrates; see Lynch et al. 2016; Buffalo 2021). If we take humans as a representative case for mammals (thus, UH = 1.1 and PX = 0.05; Ross et al. 2005; Keightley 2012), and assume that and , then unguarded X effects are predicted to be even smaller than those of Drosophila (i.e., or 1.7% longer life span for females; Fig. 2); male‐biased mutation and purifying selection further reduce the predicted effect.
Among vertebrates, our model predicts that birds should have higher potential for unguarded X effects because their sex chromosomes are somewhat large compared to mammals and their genomic deleterious mutation rates may be comparable to those of mammals (Fig. 2; but note that UH may be substantially smaller in birds compared to humans; Smeds et al. 2016). Male‐biased mutation rates (, as is typical for birds; Fig. 1) should further elevate Z‐linked deleterious mutation rates in avian taxa (i.e., based on eq. 9). Using a relatively high value of PZ = 0.1 for the Z‐linked proportion of the genome (0.07 < PZ < 0.1 in birds; Stiglec et al. 2007; Sultanova et al. 2020), a high genomic deleterious mutation rate (UH = 1.1, which is comparable to humans), strongly male‐biased mutation (), and the remaining parameters based on Drosophila estimates (, , β = 1.5), the unguarded X is predicted to increase male longevity by 5.2% relative to females (recall that females are haploid). The prediction declines to ∼3.3% in the absence of systematic sex differences in mutation or purifying selection, and further declines with lower values for PZ and UH (e.g., PZ = 0.07 and UH < 1, both of which are plausible).
Discussion
Large sex differences in survival, ageing, and longevity have been reported in surveys of adult sex ratios and sex‐specific life spans, and these differences often qualitatively agree with the unguarded X hypothesis. However, our population genetic models of the unguarded X effect, along with current parameter estimates for the rates and effects of deleterious mutations, fall considerably short of explaining the relatively large differences in mortality that are observed in nature (Pipoly et al. 2015 reached a similar conclusion concerning adult sex ratios, using a specific case of our model; see eq. 6 and surrounding text). For example, our predictions for mammals (∼1–2% higher mean female survival) fall far below the 20% elevation in mean female life span that has been reported in empirical surveys of mammalian longevity (Lemaître et al 2020; Xirocostas et al. 2020). Predictions for birds (∼3–5% higher male survival) similarly fall short of the average sex difference in longevity (∼10%) based on avian data included in Xirocostas et al. (2020). Predictions for D. melanogaster (∼1–3% higher female survival) are insufficiently large to explain the extent of female‐biased longevity in laboratory populations of D. melanogaster (i.e., data compiled in Ziehm et al. 2015 suggest that female life span is typically >10% longer than male life span; see the Supporting Information), and may further help explain the negligible estimates of X‐linked inbreeding depression in females with respect to viability and longevity (Eanes et al. 1985; Brengdahl et al. 2018; Narayan et al. 2022). Although these experiments were by no means small, they were underpowered to detect levels of X‐linked inbreeding depression predicted by our model (Eanes et al. 1985 could only reject effects exceeding 5%).
Two additional observations imply that the unguarded X is unlikely to provide a general explanation for sex differences in longevity. First, extensive sex differences for ageing and life span are observed in species with largely homomorphic sex chromosomes, including higher mortality rates in the heterogametic sex (Cayuela et al. 2022), which cannot be explained by unguarded X effects. Second, although birds are predicted (by unguarded X theory) to exhibit stronger sex differences in life span than mammals, data support the opposite pattern (the sex difference in life span in birds is roughly half the average difference observed in mammals; Lemaître et al. 2020; Xirocostas et al. 2020).
Does this mean that the unguarded X hypothesis is dead? Not entirely. Although current theory and data suggest the hypothesis is unlikely to explain broad taxonomic patterns of sex‐specific longevity, several factors might elevate its importance for some species. First, species with exceptionally large X chromosomes, including haplodiploids, could experience moderate or even large unguarded X effects. For example, with mutation parameters similar to those of Drosophila (e.g., , , , ), our model predicts a ∼15% reduction in male relative to female survival in haplodiploid species, with similar reductions in viability expected for inbred females. Although such effect sizes should be statistically discernible in reasonably sized experiments, most tests for inbreeding depression in haplodiploid species suggest that its effect on survival and longevity is typically weak (Saito et al. 2000; Mori et al. 2005; Tien et al. 2015; for an exception, see Henter 2003). Second, synergistic epistasis between deleterious mutations (which our models do not consider) can amplify inbreeding depression and genetic loads expressed by haploid (or heterogametic) individuals relative to diploid individuals, particularly if mutations are strongly recessive (; Kondrashov and Crow 1991). Although it is unclear how widespread synergistic epistasis is (Charlesworth and Charlesworth 1999; de Visser and Elena 2007; Domínguez‐García et al. 2019), epistatic effects on viability could substantially elevate unguarded X effects in species where sex chromosomes are large (and, hence, UX is also large; Kondrashov and Crow 1991; Charlesworth 1998). Finally, estimates of and especially are primarily derived from Drosophila studies (Manna et al. 2011; Charlesworth 2015), and it is unclear how much these parameters vary among species. Although there is no obvious reason for to differ among taxa, there is good reason to expect variation in , owing to among‐species variability in the contribution of preadult mortality to total fitness variance. In plants and broadcast spawning marine organisms, juvenile mortality will often be exceptionally high, and in such cases, viability selection may represent the dominant component of overall selection, causing to be larger than the Drosophila estimates imply. Those species that also carry heteromorphic sex chromosomes (e.g., Silene latifolia: Charlesworth 2016; Paracentrotus lividus: Lipani et al. 1996) are potential candidates for substantial unguarded X effects.
Our main conclusion—that the unguarded X hypothesis is unlikely to explain diverse and often conspicuous patterns of sexual dimorphism for life span among species—gives weight to prominent alternative hypotheses for the evolutionary origins of sexually dimorphic survival and longevity. One such alternative is the toxic Y hypothesis, which posits that nonrecombining Y and W chromosomes serve as reservoirs for accumulation of mutations, which (owing to male‐ and female‐limited inheritance of the Y and W, respectively) solely decrease survival of the heterogametic sex. Although this hypothesis is relatively young, it is supported by experimental evidence from Drosophila (Brown et al. 2020; Nguyen and Bachtrog 2021), and there is suggestive evidence that it might apply broadly to other taxa (Sultanova et al. 2020; Peona et al. 2021). The toxic Y effect has yet to be formally modeled, which would help identify biological conditions mediating its magnitude and taxa where it is likely to be particularly important. A second major hypothesis is that sexually dimorphic longevity reflects a fundamental sex difference in the optimal rate of investment in survival versus reproduction (Maklakov and Lummaa 2013). Although it is widely thought that such sexually dimorphic optima often favor the evolution of sex‐differential mortality, it remains an open question whether such scenarios are also likely to play out differently in species with distinct sex determination systems. There are plausible theoretical arguments that could explain why the sex chromosome system of a species might influence the evolution of sexual dimorphism for traits correlated with susceptibility to pathogens, predators, and other sources of mortality (Hastings 1994; Andrés and Morrow 2003; Reeve and Pfennig 2003; Kirkpatrick and Hall 2004; Albert and Otto 2005; Brom et al. 2018). Such scenarios should be taken seriously as potential factors contributing to predictable differences in life span between the heterogametic and homogametic sex.
AUTHOR CONTRIBUTIONS
TC, IB, YM, and FR conceptualized the study. TC developed the model. IB and YM compiled the sex‐biased mutation estimates. TC, IB, YM, and FR reviewed the relevant scientific literature. TC wrote the manuscript with extensive input from IB, YM, and FR.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
DATA ARCHIVING
The compiled dataset of sex‐biased mutation estimates is provided in full in the Supporting Information. Data and code used to create Figure 1 can be found in the GitHub repository: https://github.com/IJbeasley/unguarded_x_journal_club.
Supporting information
Supplementary Information
Supplementary Information
ACKNOWLEDGMENTS
We wish to thank the associate editor and two anonymous reviewers for suggestions that helped improve this article. This work was supported by funds from the Australian Research Council and the School of Biological Sciences at Monash University.
LITERATURE CITED
- Albert, A.Y.K. & Otto, S.P. (2005) Sexual selection can resolve sex‐linked sexual antagonism. Science, 310, 119–121. [DOI] [PubMed] [Google Scholar]
- Andrés, J.A. & Morrow, E.H. (2003) The origin of interlocus sexual conflict: is sex‐linkage important?. J. Evol. Biol, 16, 219–223. [DOI] [PubMed] [Google Scholar]
- Blackmon, H. , Ross, L. & Bachtrog, D. (2017) Sex determination, sex chromosomes, and karyotype evolution in insects. J. Hered, 108, 78–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonduriansky, R. , Maklakov, A. , Zajitschek, F. & Brooks, R. (2008) Sexual selection, sexual conflict and the evolution of ageing and life span. Funct. Ecol, 22, 443–453. [Google Scholar]
- Brengdahl, M. , Kimber, C.M. , Maguire‐Baxter, J. & Friberg, U. (2018) Sex differences in life span: females homozygous for the X chromosome do not suffer the shorter life span predicted by the unguarded X hypothesis. Evolution, 72, 568–577. [DOI] [PubMed] [Google Scholar]
- Brom, T. , Massot, M. & Laloi, D. (2018) The sex chromosome system can influence the evolution of sex‐biased dispersal. J. Evol. Biol, 31, 1377–1385. [DOI] [PubMed] [Google Scholar]
- Bronikowski, A.M. , Meisel, R.P. , Biga, P.R. , Walters, J.R. , Mank, J.E. , Larschan, E. , et al. (2022) Sex‐specific aging in animals: perspective and future directions. Aging Cell, 21:e13542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, E.J. , Nguyen, A.H. & Bachtrog, D. (2020) The Y chromosome may contribute to sex‐specific ageing in Drosophila . Nat. Ecol. Evol, 4, 853–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffalo, V. (2021) Quantifying the relationship between genetic diversity and population size suggests natural selection cannot explain Lewontin's Paradox. Elife, 10:e67509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carazo, P. , Green, J. , Sepil, I. , Pizzari, T. & Wigby, S. (2016) Inbreeding removes sex differences in lifespan in a population of Drosophila melanogaster . Biol. Lett, 12:20160337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayuela, H. , Lemaître, J.‐F. , Léna, J.‐P. , Ronget, V. , Martínez‐Solano, I. , Muths, E. , et al. (2022) Sex‐related differences in aging rate are associated with sex chromosome system in amphibians. Evolution, 76, 346–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth, B. (1998) The effect of synergistic epistasis on the inbreeding load. Genet. Res, 71, 85–89. [DOI] [PubMed] [Google Scholar]
- Charlesworth, B. (2015) Causes of natural variation in fitness: evidence from studies of Drosophila populations. Proc. Natl. Acad. Sci. USA, 112, 1662–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth, B. & Charlesworth, D. (1999) The genetic basis of inbreeding depression. Genet. Res, 74, 329–340. [DOI] [PubMed] [Google Scholar]
- Charlesworth, B. , Coyne, J.A. & Barton, N.H. (1987) The relative rates of evolution of sex chromosomes and autosomes. Am. Nat, 130, 113–146. [Google Scholar]
- Charlesworth, D. (2016) Plant sex chromosomes. Annu. Rev. Plant Biol, 67, 397–420. [DOI] [PubMed] [Google Scholar]
- Connallon, T. (2010) Genic capture, sex linkage, and the heritability of fitness. Am. Nat, 175, 564–576. [DOI] [PubMed] [Google Scholar]
- Connallon, T. & Clark, A.G. (2011) Association between sex‐biased gene expression and mutations with sex‐specific phenotypic consequences in Drosophila . Genome. Biol. Evol, 3, 151–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, X. , Berletch, J.B. , Nguyen, D.K. & Disteche, C.M. (2014) X chromosome regulation: diverse patterns in development, tissues and disease. Nat. Rev. Genet, 15, 367–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denver, D.R. , Morris, K. , Lynch, M. & Thomas, W.K. (2004) High mutation rate and predominance of insertions in the Caenorhabditis elegans nuclear genome. Nature, 430, 679–682. [DOI] [PubMed] [Google Scholar]
- de Visser, J. & Elena, S.F. (2007) The evolution of sex: empirical insights into the roles of epistasis and drift. Nat. Rev. Genet, 8, 139–149. [DOI] [PubMed] [Google Scholar]
- Domínguez‐García, S. , García, C. , Quesada, H. & Caballero, A. (2019) Accelerated inbreeding depression suggests synergistic epistasis for deleterious mutations in Drosophila melanogaster . Heredity, 123, 709–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dukler, N. , Mughal, M.R. , Ramani, R. , Huang, Y.F. & Siepel, A. (2021) Extreme purifying selection against point mutations in the human genome. bioRxiv, 10.1101/2021.08.23.457339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eanes, W.F. , Hey, J. & Houle, D. (1985) Homozygous and hemizygous viability variation on the X chromosome of Drosophila melanogaster . Genetics, 111, 831–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegren, H. (2007) Characteristics, causes and evolutionary consequences of male‐biased mutation. Proc. R. Soc. B Biol. Sci, 274, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraïsse, C. , Picard, M.A.L. & Vicoso, B. (2017) The deep conservation of the Lepidoptera Z chromosome suggests a non‐canonical origin of the W. Nat. Commun, 8, 1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank, S.A. & Hurst, L.D. (1996) Mitochondria and male disease. Nature, 383, 224. [DOI] [PubMed] [Google Scholar]
- Gemmell, N.J. , Metcalf, V.J. & Allendorf, F.W. (2004) Mother's curse: the effect of mtDNA on individual fitness and population viability. Trends Ecol. Evol, 19, 238–244. [DOI] [PubMed] [Google Scholar]
- Grieshop, K. , Maurizio, P.L. , Arnqvist, G. & Berger, D. (2021) Selection in males purges the mutation load on female fitness. Evol. Lett, 5, 328–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag‐Liautard, C. , Dorris, M. , Maside, X. , Macaskill, S. , Halligan, D.L. , Charlesworth, B. , et al. (2007) Direct estimation of per nucleotide and genomic deleterious mutation rates in Drosophila . Nature, 445, 82–85. [DOI] [PubMed] [Google Scholar]
- Haldane, J.B.S. (1937) The effect of variation on fitness. Am. Nat, 71, 337–349. [Google Scholar]
- Hastings, I.M. (1994) Manifestations of sexual selection may depend on the genetic basis of sex determination. Proc. R. Soc. B Biol. Sci, 258, 83–87. [DOI] [PubMed] [Google Scholar]
- Hedrick, P.W. (2007) Sex: differences in mutation, recombination, selection, gene flow, and genetic drift. Evolution, 61, 2750–2771. [DOI] [PubMed] [Google Scholar]
- Henter, H.J. (2003) Inbreeding depression and haplodiploidy: experimental measures in a parasitoid and comparisons across diploid and haplodiploid insect taxa. Evolution, 57, 1793–1803. [DOI] [PubMed] [Google Scholar]
- Hitchcock, T.J. & Gardner, A. (2020) A gene's‐eye view of sexual antagonism. Proc. R. Soc. B Biol. Sci, 287:20201633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley, P.D. (2012) Rates and fitness consequences of new mutations in humans. Genetics, 190, 295–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick, M. & Hall, D.W. (2004) Sexual selection and sex linkage. Evolution, 58, 683–691. [DOI] [PubMed] [Google Scholar]
- Kondrashov, A.S. & Crow, J.F. (1991) Haploidy or diploidy: which is better? Nature, 351, 314–315. [DOI] [PubMed] [Google Scholar]
- ———. (1993) A molecular approach to estimating the human deleterious mutation rate. Hum. Mutat, 2, 229–234. [DOI] [PubMed] [Google Scholar]
- Labuda, D. , Lefebvre, J.F. , Nadeau, P. & Roy‐Gagnon, M.H. (2010) Female‐to‐male breeding ratio in modern humans‐an analysis based on historical recombinations. Am. J. Hum. Genet, 86, 353–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaître, J.F. , Ronget, V. , Tidière, M. , Allainé, D. , Berger, V. , Cohas, A. , et al. (2020) Sex differences in adult lifespan and aging rates of mortality across wild mammals. Proc. Natl. Acad. Sci. USA, 117, 8546–8553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipani, C. , Vitturi, R. , Sconzo, G. & Barbata, G. (1996) Karyotype analysis of the sea urchin Paracentrotus lividus (Echinodermata): evidence for a heteromorphic chromosome sex mechanism. Mar. Biol, 127, 67–72. [Google Scholar]
- Lohmueller, K.E. , Degenhardt, J.D. & Keinan, A. (2010) Sex‐averaged recombination and mutation rates on the X chromosome: a comment on Labuda et al. Am. J. Hum. Genet, 86, 978–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, T.A.F. , Pischedda, A. , Stewart, A.D. & Rice, W.R. (2009) A cost of sexual attractiveness to high‐fitness females. PLoS Biol, 7:e1000254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lower, S.S. , Johnston, J.S. , Stanger‐Hall, K.F. , Hjelmen, C.E. , Hanrahan, S.J. , Korunes, K. , et al. (2017) Genome size in north american fireflies: substantial variation likely driven by neutral processes. Genome. Biol. Evol, 9, 1499–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch, M. , Blanchard, J. , Houle, D. , Kibota, T. , Schultz, S. , Vassilieva, L. , et al. (1999) Perspective: spontaneous deleterious mutation. Evolution, 53, 645–663. [DOI] [PubMed] [Google Scholar]
- Lynch, M. , Ackerman, M.S. , Gout, J.F. , Long, H. , Sung, W. , Thomas, W.K. , et al. (2016) Genetic drift, selection and the evolution of the mutation rate. Nat. Rev. Genet, 17, 704–714. [DOI] [PubMed] [Google Scholar]
- Maklakov, A.A. & Lummaa, V. (2013) Evolution of sex differences in lifespan and aging: causes and constraints. Bioessays, 35, 717–724. [DOI] [PubMed] [Google Scholar]
- Mallet, M.A. , Bouchard, J.M. , Kimber, C.M. & Chippindale, A.K. (2011) Experimental mutation‐accumulation on the X chromosome of Drosophila melanogaster reveals stronger selection on males than females. BMC Evol. Biol, 11, 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manna, F. , Martin, G. & Lenormand, T. (2011) Fitness landscapes: an alternative theory for the dominance of mutation. Genetics, 189, 923–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marais, G.A.B. & Lemaître, J.F. (2022) Sex chromosomes, sex ratios and sex gaps in longevity in plants. Philos. Trans. R. Soc. B Biol. Sci, 377:20210219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marais, G.A.B. , Gaillard, J.M. , Vieira, C. , Plotton, I. , Sanlaville, D. , Gueyffier, F. , et al. (2018) Sex gap in aging and longevity: can sex chromosomes play a role? Biol. Sex Differ, 9, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisel, R.P. , Delclos, P.J. & Wexler, J.R. (2019) The X chromosome of the German cockroach, Blattella germanica, is homologous to a fly X chromosome despite 400 million years divergence. BMC Biol, 17, 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata, T. , Hayashida, H. , Kuma, K. , Mitsuyasu, K. & Yasunaga, T. (1987) Male‐driven molecular evolution: a model and nucleotide sequence analysis. Cold Spring Harb. Symp. Quant. Biol, 52, 863–867. [DOI] [PubMed] [Google Scholar]
- Mori, K. , Saito, Y. , Sakagami, T. & Sahara, K. (2005) Inbreeding depression of female fecundity by genetic factors retained in natural populations of a male‐haploid social mite (Acari: Tetranychidae). Exp. Appl. Acarol, 36, 15–23. [DOI] [PubMed] [Google Scholar]
- Narayan, V.P. , Wilson, A.J. & Chenoweth, S.F. (2022) Genetic and social contributions to sex differences in lifespan in Drosophila serrata . J. Evol. Biol, 35, 657–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, A.H. & Bachtrog, D. (2021) Toxic Y chromosome: increased repeat expression and age‐associated heterochromatin loss in male Drosophila with a young Y chromosome. PLoS Genet, 17:e1009438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peona, V. , Palacios‐Gimenez, O.M. , Blommaert, J. , Liu, J. , Haryoko, T. , Jønsson, K.A. , et al. (2021) The avian W chromosome is a refugium for endogenous retrovirusus with likely effects on female‐biased mutational load and genetic incompatibilities. Philos. Trans. R. Soc. B Biol. Sci, 376:20200186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pipoly, I. , Bókony, V. , Kirkpatrick, M. , Donald, P.F. , Székely, T. & Liker, A. (2015) The genetic sex‐determination system predicts adult sex ratios in tetrapods. Nature, 527, 91–94. [DOI] [PubMed] [Google Scholar]
- Presgraves, D.C. (2002) Patterns of postzygotic isolation in Lepidoptera. Evolution, 56, 1168–1183. [DOI] [PubMed] [Google Scholar]
- Rayner, J.G. , Hitchcock, T.J. & Bailey, N.W. (2021) Variable dosage compensation is associated with female consequences of an X‐linked, male‐beneficial mutation. Proc. R. Soc. B Biol. Sci, 288:20210355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeve, H.K. & Pfennig, D.W. (2003) Genetic biases for showy males: are some genetic systems especially conducive to sexual selection? Proc. Natl. Acad. Sci. USA, 100, 1089–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross, M.T. , Grafham, D.V. , Coffey, A.J. , Scherer, S. , McLay, K. , Muzny, D. , et al. (2005) The DNA sequence of the human X chromosome. Nature, 434, 325–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito, Y. , Sahara, K. & Mori, K. (2000) Inbreeding depression by recessive deleterious genes affecting female fecundity of a haplo‐diploid mite. J. Evol. Biol, 13, 668–678. [Google Scholar]
- Sharp, N.P. & Agrawal, A.F. (2013) Male‐biased fitness effects of spontaneous mutations in Drosophila melanogaster . Evolution, 67, 1189–1195. [DOI] [PubMed] [Google Scholar]
- Singh, A. & Agrawal, A.F. (2022) Sex‐specific variance in fitness and the efficacy of selection. Am. Nat, 199, 587–602. [DOI] [PubMed] [Google Scholar]
- Smeds, L. , Qvarnström, A. & Ellegren, H. (2016) Direct estimate of the rate of germline mutation in a bird. Genome Res, 26, 1211–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiglec, R. , Ezaz, T. & Graves, J.A.M. (2007) A new look at the evolution of avian sex chromosomes. Cytogenet. Genome Res, 117, 103–109. [DOI] [PubMed] [Google Scholar]
- Sultanova, Z. , Andic, M. & Carazo, P. (2018) The “unguarded‐X” and the genetic architecture of lifespan: inbreeding results in a potentially maladaptive sex‐specific reduction of female lifespan in Drosophila melanogaster . Evolution, 27, 540–552. [DOI] [PubMed] [Google Scholar]
- Sultanova, Z. , Downing, P.A. & Carazo, P. (2020) Genetic sex determination and sex‐specific lifespan in tetrapods—evidence of a toxic Y effect. bioRxiv, 10.1101/2020.03.09.983700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sylvester, T. , Hjelmen, C.E. , Hanrahan, S.J. , Lenhart, P.A. , Johnston, J.S. & Blackmon, H. (2020) Lineage‐specific patterns of chromosome evolution are the rule not the exception in Polyneoptera insects. Proc. R. Soc. B Biol. Sci, 287:20201388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tien, N.S.H. , Sabelis, M.W. & Egas, M. (2015) Inbreeding depression and purging in a haplodiploid: gender‐related effects. Heredity, 114, 327–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tree of Sex Consortium . (2014) Tree of Sex: a database of sexual systems. Sci. Data, 1:140015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivers, R. (1985) Social evolution. The Benjamin/Cummings Publishing Company, Menlo Park, CA. [Google Scholar]
- Turelli, M. & Begun, D.J. (1997) Haldane's rule and X‐chromosome size in Drosophila . Genetics, 147, 1799–1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchimura, A. , Higuchi, M. , Minakuchi, Y. , Ohno, M. , Toyoda, A. , Fujiyama, A. , et al. (2015) Germline mutation rates and the long‐term phenotypic effects of mutation accumulation in wild‐type laboratory mice and mutator mice. Genome Res, 25, 1125–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega‐Trejo, R. , de Boer, R.A. , Fitzptrick, J.L. & Kotrschal, A. (2022) Sex‐specific inbreeding depression: a meta‐analysis. Ecol. Lett, 25, 1009–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werren, J.H. (1993) The evolution of inbreeding in haplodiploid organisms. Pp. 42–59 in Thornhill, N.W. , ed. The natural history of inbreeding and outbreeding: theoretical and empirical perspectives. University of Chicago Press, Chicago. [Google Scholar]
- Whitlock, M.C. & Agrawal, A.F. (2009) Purging the genome with sexual selection: reducing mutation load through selection on males. Evolution, 63, 569–582. [DOI] [PubMed] [Google Scholar]
- Wilson Sayres, M.A. , Venditti, C. , Pagel, M. & Makova, K.D. (2011) Do variations in substitution rates and male mutation bias correlate with life‐history traits? A study of 32 mammalian genomes. Evolution, 65, 2800–2815. [DOI] [PubMed] [Google Scholar]
- Wilton, A.N. & Sved, J.A. (1979) X‐chromosomal heterosis in Drosophila melanogaster . Genet. Res, 34, 303–315. [DOI] [PubMed] [Google Scholar]
- Xirocostas, Z.A. , Everingham, S.E. & Moles, A.T. 2020. . The sex with the reduced sex chromosome dies earlier: a comparison across the tree of life. Biol. Lett, 16:20190867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun, L. , Chen, P.J. , Kwok, K.E. , Angell, C.S. , Rundle, H.D. & Agrawal, A.F. (2018) Competition for mates and the improvement of nonsexual fitness. Proc. Natl. Acad. Sci. USA, 115, 6762–6767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziehm, M. , Ivanov, D.K. , Bhat, A. , Partridge, L. & Thornton, J.M. (2015) SurvCurv database and online survival analysis platform update. Bioinformatics, 31, 3878–3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Supplementary Information
Data Availability Statement
The compiled dataset of sex‐biased mutation estimates is provided in full in the Supporting Information. Data and code used to create Figure 1 can be found in the GitHub repository: https://github.com/IJbeasley/unguarded_x_journal_club.


