Abstract
The nitrogen assimilation control protein (NAC) from Klebsiella aerogenes or Escherichia coli (NACK or NACE, respectively) is a transcriptional regulator that is both necessary and sufficient to activate transcription of the histidine utilization (hut) operon and to repress transcription of the glutamate dehydrogenase (gdh) operon in K. aerogenes. Truncated NAC polypeptides, generated by the introduction of stop codons within the nac open reading frame, were tested for the ability to activate hut and repress gdh in vivo. Most of the NACK and NACE fragments with 100 or more amino acids (wild-type NACK and NACE both have 305 amino acids) were functional in activating hut and repressing gdh expression in vivo. Full-length NACK and NACE were isolated as chimeric proteins with the maltose-binding protein (MBP). NACK and NACE released from such chimeras were able to activate hut transcription in a purified system in vitro, as were NACK129 and NACE100 (a NACK fragment of 129 amino acids and a NACE fragment of 100 amino acids) released from comparable chimeras. A set of NACE and NACK fragments carrying nickel-binding histidine tags (his6) at their C termini were also generated. All such constructs derived from NACE were insoluble, as was NACE itself. Of the his6-tagged constructs derived from NACK, NACK100 was inactive, but NACK120 was active. Several NAC fragments were tested for dimerization. NACK120-his6 and NACK100-his6 were dimers in solution. MBP-NACK and MBP-NACK129 were monomers in solution but dimerized when the MBP was released by cleavage with factor Xa. MBP-NACE was readily cleaved by factor Xa, but the resulting NACE was also degraded by the protease. However, MBP-NACE-his6 was completely resistant to cleavage by factor Xa, suggesting an interaction between the C and N termini of this protein.
The nitrogen regulatory (Ntr) system of enteric bacteria allows Klebsiella aerogenes, Escherichia coli, and other enteric bacteria to respond rapidly and effectively to changes in the quality of the nitrogen source provided (13). Under conditions of nitrogen limitation, a complex cascade of regulatory events involving uridylylation and phosphorylation reactions leads to the accumulation of the phosphorylated (active) form of the transcriptional activator NtrC. NtrC-phosphate can activate the ς54-dependent expression of a variety of genes involved in utilization of organic and inorganic nitrogen sources. NtrC-phosphate also activates transcription of the nac gene in K. aerogenes and E. coli (6).
The nitrogen assimilation control protein (NAC) is a regulatory protein responsible for activating the transcription of operons such as hutUH, putP, and ureDABCEFG, whose products supply the cell with ammonia or glutamate from histidine, proline, and urea, respectively (2). NAC is also responsible for repressing transcription of the gdhA and gltBD operons, whose products are involved in assimilating ammonia under conditions of nitrogen excess or limitation (2). NAC is also responsible for down-regulating its own transcription (3, 7). The expression of the nac gene is entirely dependent on the Ntr system such that under conditions of nitrogen-limited growth, the Ntr system is active and nac is actively transcribed by RNA polymerase charged with the minor sigma factor ς54 (6, 12). There are no known coeffectors involved in the regulation of transcription by NAC (17), so if nac is expressed, then NAC is produced and activates transcription of hut, put, and ure by RNA polymerase charged with the major sigma factor ς70. Thus, NAC serves as a coupling factor between the ς54-dependent Ntr system and ς70-dependent genes like hut (2).
NAC is a member of the large family of LysR-type transcriptional regulators (LTTRs; 15, 16). This group of over 50 proteins are identified on the basis of amino acid sequence similarity. Most of the members of this family have between 300 and 350 amino acids and show a surprisingly high degree of sequence similarity. For example, NAC from K. aerogenes shows about 40% sequence identity with OxyR from E. coli (16). Within the LTTRs, the DNA-binding domain occurs near the N terminus of the protein, where a predicted helix-turn-helix motif (approximately amino acids 20 to 40 in NAC) is located (15). In several LTTRs, the binding site for a required coeffector has been identified. These binding sites are generally in the C-terminal third of the polypeptide, suggesting that the DNA-binding and coeffector-binding domains are physically separated (15).
Very little structural information about LTTRs is available. A fragment of CysB containing the C-terminal 233 amino acids has been crystallized, and a structure was derived from X-ray diffraction data (18). However, this fragment lacks the N-terminal 87 amino acids and thus the DNA-binding domain. As a result, this structure says little about how the DNA-protein interactions occur, are altered by the presence of inducer, or result in the activation of transcription.
Comparison of NAC from E. coli (NACE) with NAC from K. aerogenes (NACK) revealed a surprising lack of sequence similarity (14). NACE was only about 75% identical to NACK, in contrast to other E. coli and K. aerogenes regulatory proteins, which are >95% identical. Most of the sequence divergence between NACE and NACK occurs in the C-terminal two-thirds of the protein, consistent with the lack of a need to conserve a coeffector-binding site in the C-terminal domain. This raised the question of whether the C-terminal domain has any role at all, leading us to seek the smallest N-terminal fragment that would retain the biological activities associated with NAC.
Two nac mutations also suggested that a C-terminal portion of NAC might be dispensable, further encouraging us to search for a small active fragment of NAC. The nac-112 allele of K. aerogenes is an insertion of Mudlac,amp at an unknown site within nacK, which retains considerable NAC activity (12). The nac-10 allele of E. coli, an insertion of a stop codon and a drug resistance cartridge in the center of the nacE gene, was constructed as an intermediate in constructing the nac-28 null allele (14). Strains carrying nac-10 also retained considerable NAC activity. Thus, we attempted to determine whether the C-terminal domain of NAC from E. coli and K. aerogenes plays an essential role and to define the smallest N-terminal fragment of NAC that would retain the ability to function as a transcriptional regulator.
MATERIALS AND METHODS
Strains and plasmids.
All of the K. aerogenes strains used in this study were derived from W70 and are listed in Table 1.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| KC2668 | hutC515 Δ[bla]-2 nac+ | 10 |
| KC2725 | hutC515 Δ[bla]-2 nac-203::Tn5-131 | 10 |
| KC2972 | KC2941/pCB511 | Transformation |
| KC3220 | KC2725/pCB594 | Transformation |
| KC3555 | KC2725/pCB771 | Transformation |
| KC3568 | KC2725/pCB774 | Transformation |
| KC3572 | KC2725/pCB778 | Transformation |
| KC3578 | KC2725/pCB781 | Transformation |
| KC3580 | KC2725/pCB782 | Transformation |
| KC3591 | KC2725/pCB788 | Transformation |
| KC3615 | KC2725/pCB800/pCB648 | Transformation |
| KC3973 | KC2725/pCB941 | Transformation |
| KC3977 | KC2725/pCB529 | Transformation |
| KC3981 | KC2725/pCB606 | Transformation |
| KC4510 | KC2725/pCB547 | Transformation |
| Plasmids | ||
| pCB511 | pGB2; wild-type nacEa | This laboratory |
| pCB529 | pGB2; nac-10 allele of E. coli | This laboratory |
| pCB547 | pGB2; nac-28 allele of E. coli | This laboratory |
| pCB594 | pMalC2; MBP-NACE fusion protein | This laboratory |
| pCB606 | pMalC2; MBP-NACK fusion protein | This laboratory |
| pCB771 | pGD103; NACE-his6 | This laboratory |
| pCB774 | pGD103; wild-type NACE | This laboratory |
| pCB778 | pGD103; wild-type NACK | This laboratory |
| pCB781 | pGD103; NACK-his6 | This laboratory |
| pCB782 | pGD103; NACK-(DFGRSGHTDSL) | This laboratory |
| pCB788 | pGD103; NACK100-his6 | This laboratory |
| pCB800 | pGD103; NACK120-his6 | This laboratory |
| pCB941 | pGD103; NACE100-his6 | This laboratory |
| pGB2 | Low-copy cloning vector; insert expressed from its own promoter | 4 |
| pGD103 | Low-copy vector expressing inserted material from lacP | 5 |
| pMalC2 | MBP fusion vector expressing inserted material from tac promoter | New England BioLabs |
Vector; cloned insert or product.
Generation of C-terminal deletions of NAC.
Unidirectional exonuclease III deletions were performed to make 3′ deletions in the E. coli nac gene by using the protocol described by Henikoff (9) with minor modifications. Plasmid pCB574 contains the E. coli nac gene inserted into the EcoRI site of plasmid pGB2 (4) such that the 3′ end of the gene is proximal to the PstI site present in the multiple cloning site. Deletions were initiated by cutting with SalI and PstI to allow unidirectional deletions into nac. After digestion with exonuclease III and mung bean nuclease, the deleted DNA fragments were resuspended in 20 μl of ligation buffer (250 mM Tris-HCl [pH 7.6], 50 mM MgCl2, 1 mM dithiothreitol, 0.5 mM ATP) and 100 pmol of a dephosphorylated NheI linker (containing stop codons in three reading frames) to which T4 DNA ligase was added. The ligation mixture was used to transform E. coli DH5α to streptomycin (50 μg/ml) and spectinomycin (100 μg/ml) resistance. Deletions of interest were sequenced by using the pUC reverse primer to determine the precise endpoint of each deletion. These were brought under the control of the lac promoter by PCR amplification by using the primers CGAATTCAAACTGGAGACTCATATGAAC (forward) and AGGATCCTCACACAGGAAACAGCTATGAC (reverse) and insertion into the EcoRI and BamHI sites of pGD103 (5). Specific truncations in the K. aerogenes nac gene were generated by PCR using a forward primer (CTGGAATTCCTTACAGGAGGCA) containing an EcoRI site and reverse primers complementary to the last 18 bases prior to the desired truncation point followed by an amber stop codon and GGATCC (BamHI). PCR products were introduced into the EcoRI and BamHI sites of plasmid pGD103, and the sequences were verified by dideoxy sequencing.
Purification of MBP-NAC fusions.
The nac genes from E. coli and K. aerogenes, along with the C-terminal truncated derivatives of each, were inserted into vector pMalC2 (New England Biolabs) as a blunt-ended 5′ end of the nac gene beginning at the initiating methionine ligated to the blunt-end XmnI site within the vector. The 3′ ends were ligated into either the EcoRI or BamHI sites within the multiple cloning site. The initial construct was prepared by PCR and sequenced to verify that errors had not been introduced by PCR amplification. Later derivatives were made by replacing sequences within this clone by using internal sites (XmnI, SspI, and BglII) in the nac gene rather than reamplifying for each construct. Insertion of a blunt-ended DNA fragment into the XmnI site of the vector results in fusion of the NAC protein to the 42-kDa maltose-binding protein (MBP). The junction between the two proteins contains the recognition site for factor Xa protease (Ile Glu Gly Arg) positioned such that cleavage with this enzyme would result in separation of the intact NAC protein from the fusion.
Purification of the fusion protein was initiated by inducing mid-log-phase cultures (3 × 108 CFU/ml) with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG). The induced cultures (500 ml in 2-liter flasks) were shifted from 37 to 28°C and grown for an additional 2 h. Cells were chilled on ice and harvested by centrifugation. The pellets were washed twice in column buffer (100 mM Tris-HCl [pH 7.5], 250 mM NaCl, 1 mM dithiothreitol, 1 mM 2-mercaptoethanol) and resuspended in a final volume of 15 ml. Cell disruption was accomplished by passage through a French pressure cell (twice at 12,000 lb/in2). The resulting lysate was clarified by centrifugation for 30 min at 30,000 × g at 4°C. The supernatant solution containing the fusion protein was increased in volume to 30 ml with column buffer containing 50 mM freshly prepared phenylmethylsulfonyl fluoride. The crude preparation was applied to a column (2.5 cm in diameter) containing a 10-ml bed volume of amylose-agarose resin (New England Biolabs no. 800-21L) at a flow rate of 1 ml/min. Bound fusion protein was washed with 300 ml of column buffer at a 2-ml/min flow rate and eluted with column buffer supplemented with 1 M maltose at a rate of 1.5 ml/min with 3-ml fractions collected over 50 ml. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was used to determine which fractions contained MBP-NAC. The MBP-NAC fusion generally eluted over a wide peak from fractions 3 to 7. Fractions were pooled if SDS-PAGE indicated sufficient purity. Protein concentrations were determined by Bio-Rad protein assay and generally indicated values between 0.7 and 2.5 mg/ml. Diafiltration was performed by using either an Amicon 10- or 25-ml stir cell apparatus with PM30 filters to concentrate samples. Fusions were stored at −20°C following the addition of an equal volume of glycerol. If the fusion was to be cleaved or used immediately, it was kept on ice at all times. Diafiltration was also used to change the buffer in which the MBP-NAC was suspended to buffer 4 (8), which contains 100 mM NaH2PO4 (pH 7.0), 250 mM NaCl, 2.5 mM MgCl2, and 1 mM 2-mercaptoethanol. The NAC protein was not transcriptionally active as a fusion protein with MBP. Therefore, to retrieve active NAC from these fusions for in vitro transcription experiments, it was necessary to cleave NAC from the fusion by using factor Xa protease. This was accomplished by adding 1 μg of protease to 0.5 mg of the fusion protein in a 100-ml volume and then incubating it for 16 h at 22°C. Following cleavage, freshly prepared phenylmethylsulfonyl fluoride was added to 20 mM. The cleaved NAC fusion protein had to be used immediately because it degraded quickly over time.
Purification of his6-tagged NAC.
To overcome the problems associated with MBP-NAC fusions, various six-histidine (his6)-tagged constructs were built. NAC N-terminal constructs were made by cloning nac genes into the EcoRI/HindIII sites of pET28 (Novagen). The N-terminal fusions added a leader to the NAC proteins consisting of 56 amino acids which include a stretch of six histidines in a row to facilitate nickel-nitrilotriacetic acid affinity purification (Qiagen). Constructs in which carboxy-terminal his6 tags were added had the DNA sequence (CAT)6 followed by an amber stop codon designed into the primers used to amplify the nac gene. The C-terminal his6-tagged genes were inserted into a derivative of the pET28 vector in which the material between the XhoI and EcoRI sites had been removed. Histidine-tagged NAC proteins were expressed as previously described for the MBP fusions and prepared for affinity purification in a similar fashion, except that the pH of the buffer was raised to 7.8 to facilitate nickel interaction. Clarified S30 French pressate was added to a column with an internal diameter of 1.25 cm containing a 10-ml bed volume of nickel-nitrilotriacetic acid resin (Qiagen). NAC was allowed to bind the column at a flow rate of 2 ml/min. The column was washed with 150 ml of column buffer supplemented with 5 mM imidazole to eliminate nonspecific binding. NAC was eluted with elution buffer (300 mM imidazole [pH 7.5], 250 NaCl, 1 mM MgCl2, 1 mM 2-mercaptoethanol). Fractions of 3 ml were collected and screened by Bio-Rad protein assay. NAC generally eluted in a tight peak in fraction 3. Purity was monitored by SDS-PAGE.
Gel filtration analysis of purified proteins.
Gel filtration chromatography was used to determine the size and oligomeric state of purified NAC proteins. Size determinations were made by one of two methods: fast protein liquid chromatography (FPLC)-mediated gel filtration or gravity-fed chromatography. For determinations by FPLC, a Pharmacia LCC-501 FPLC apparatus was used in conjunction with a Superdex 75 HR 10/30 prepacked column. The column was equilibrated with buffer 4 (described above) and loaded with 30 μl (75 μg) of NAC. Separation was allowed to continue at a flow rate of 1 ml/min for 40 min. Detection of eluting protein was done by monitoring A280. Standardization of the column was done with gel filtration protein standards purchased from Sigma Chemical Co. resuspended in buffer 4. Generally, dimeric NAC (66-kDa dimer) eluted at a 15-ml volume at the same fraction as bovine serum albumin (66 kDa). Gravity flow gel filtration was performed by using an SR 25/100 column (25-mm internal diameter) packed with a 150-ml bed volume of Sephacryl S-100. Standardization of this column was done with the same buffers, globular weight standards, and concentrations as the FPLC, but the loading volume was increased to 1 ml. The flow rate was adjusted to 0.5 ml/min, and 150 ml was allowed to flow. Generally, dimeric NAC (66 kDa) eluted at a volume of 16 ml from this column.
In vitro transcription assays.
In vitro transcriptions were carried out essentially as described previously (8). The template used was plasmid pCB695, which carries the K. aerogenes hutUH promoter inserted into vector pTE103, similar to pAM1202, which was described previously (8). Transcription mixtures contained 0.16 pmol of supercoiled plasmid as the template and 2 to 6 pmol of RNA polymerase in a total volume of 25 μl.
Enzyme assays.
Histidase and glutamate dehydrogenase (GDH) activities were measured as described previously (12). Specific activities are reported as nanomoles of urocanate formed (histidase) or NADPH2 oxidized (GDH) per minute per milligram of cell protein. Cell protein was determined by the method of Lowry et al. (11).
Gel mobility shift assays.
Gel mobility shift assays were performed by using purified NACE or NACK or fragments thereof. The ability of NAC to bind DNA was determined by using the 330-bp EcoRI-to-HindIII fragment of the ureD promoter, which contains a strong NAC-binding site. Binding reaction mixtures consisted of 1 μl of DNA (0.05 pmol), 1 μl of poly[d(I·C) (50 ng/ml), 4 μl of deionized distilled H2O, and 1 μl of a NAC dilution (0.35 to 1.7 pmol) in 50 mM NaH2PO4 (pH 7)–125 mM NaCl–0.5 mM MgCl2–0.1 mM β-mercaptoethanol–50% glycerol, 1-mg/ml bovine serum albumin. NAC was incubated with the DNA for 30 min before addition of 1.5 μl of loading buffer (40 mM Tris [pH 8.4], 4 mM EDTA, 0.2% bromthymol blue, 0.2% xylene cyanol, 15% Ficoll) and applied to a 2% Tris-borate-EDTA agarose gel. Electrophoresis was carried out at room temperature at 15 V/cm for approximately 1 h. Gels were stained with ethidium bromide, and the fragments were visualized by UV fluorescence.
RESULTS
Residual NAC activity in nac-10 mutants.
The nac-10 allele of the E. coli nac gene has a stop codon and a kanamycin resistance gene inserted into the middle of nacE after codon 165. The resulting NAC polypeptide has four amino acids encoded by the polylinker fused to the 165th amino acid of NAC; the C-terminal 140 amino acids of NAC are not present. The nac gene of E. coli was replaced with nac-10 as described elsewhere (14), and the replacement was confirmed by Southern blotting (data not shown). To test for NAC activity, we measured the ability of a strain carrying nac-10 to activate the NAC-dependent expression of the hut operon in response to nitrogen-limited growth. The nac-10 mutant showed about half as much histidase as the wild type under NAC-dependent conditions (data not shown), suggesting that the NAC protein encoded by nac-10 retained considerable activity. However, NAC-dependent activation of histidase expression in E. coli is weak, making it dangerous to draw strong conclusions based on small differences.
NAC-dependent activation of histidase expression is stronger in K. aerogenes, and repression of glutamate dehydrogenase formation by NAC serves as another measure of NAC activity. Therefore, we studied the effect of the nac-10 allele in K. aerogenes. The nac-10 allele was cloned in low-copy expression vector pGD103 and tested for the ability to complement a nac-203 mutation in K. aerogenes. The wild-type nac gene from E. coli was able to complement nacK-203 fully, showing 15-fold activation of histidase formation and 7-fold repression of GDH expression in response to nitrogen starvation. This is similar to the effect seen in wild-type (nac+) K. aerogenes (Table 2, cf. KC2668 and KC2972). In contrast, a known E. coli null allele, nacE-28, failed to complement nacK-203 with strain KC4510 (nac-28/nac-203), showing no more activation of histidase or repression of GDH expression than the nac-203 mutant, KC2725. Strain KC3977 (nac-10/nac-203) showed about sevenfold activation of histidase formation and threefold repression of GDH formation in response to nitrogen starvation. Thus, the nac-10 mutation, with an insertion of a stop codon and a kanamycin resistance cassette after amino acid 165, retained about half as much ability to activate and repress transcription as wild-type nac+ from E. coli, despite the absence of the entire C-terminal half of NAC.
TABLE 2.
Residual activity of nac-10 mutationa
| Strain | Chromosomal allele | Plasmid allele | Presence of N in medium | Activityb of:
|
|
|---|---|---|---|---|---|
| Histidase | GDH | ||||
| KC2668 | nac+ | None | + | 28 | 410 |
| − | 389 | 65 | |||
| KC2725 | nac-203 | None | + | 31 | 413 |
| − | 54 | 436 | |||
| KC2972 | nac-203 | nac+ | + | 31 | 292 |
| − | 460 | 41 | |||
| KC3977 | nac-203 | nac-10 | + | 35 | 470 |
| − | 265 | 141 | |||
| KC4510 | nac-203 | nac-28 | + | 33 | 397 |
| − | 59 | 425 | |||
Strains were grown to mid-log phase in glucose minimal medium with 0.2% (wt/vol) glutamine as the sole nitrogen source (−) or with a combination of 0.2% glutamine and 0.2% ammonium sulfate as the nitrogen source (+) and assayed for histidase and GDH (12).
Specific activity is reported as nanomoles per minute per milligram of cell protein.
This residual activity is particularly surprising compared to the nacK-203 mutation of K. aerogenes. nac-203 is an insertion of Tn5 (or Tn5-131) after the 199th codon of nacK and has one of the tightest Nac− phenotypes of all of the characterized K. aerogenes nac mutants (12). Therefore, we began a systematic search for the smallest NAC fragment with biological activity.
The N-terminal 100 amino acids of NAC are sufficient to activate and repress transcription.
A collection of mutants with deletions of portions of the C-terminal part of the K. aerogenes and E. coli nac genes (nacK and nacE, respectively) was generated by exonuclease III digestion as described in Materials and Methods. These truncated nac genes were cloned into pGD103, where nac expression was under the control of the lacZ promoter. K. aerogenes nac-203 mutant KC2725 was transformed with the resulting plasmids, and histidase and glutamate dehydrogenase activities were determined as a measure of NAC-dependent activation and repression. For convenience, the truncated proteins will be named by the number of the last remaining amino acid. Thus, wild-type E. coli NAC is NACE305 and a K. aerogenes NAC lacking the last five amino acids is NACK300.
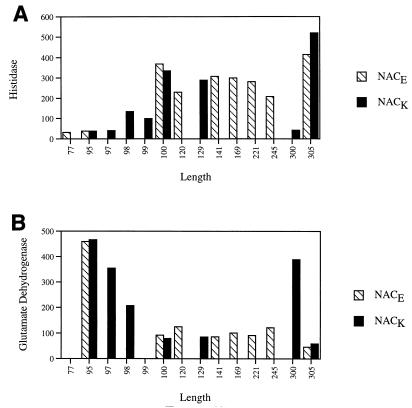
Most of the truncated genes that retained 100 or more codons at the amino terminus were able to complement the nacK-203 allele for activation of histidase formation and repression of GDH formation (Fig. 1). The magnitude of the effect varied from about 50 to 90% as much activation or repression as the wild type, and the mutants retaining only 100 amino acids of NAC (NACK100 and NACE100) were among the most active of the truncations in vivo. This result was as true of the E. coli nac gene as of the K. aerogenes nac gene and appears to be a general property of NAC rather than a species-specific anomaly.
FIG. 1.
hut activation (A) and gdh repression (B) by N-terminal fragments of NAC. K. aerogenes KC2725 (nac-203::Tn5-131) was transformed with derivatives of pGD103 containing an insert that coded for either full-length NAC or truncated versions of NAC. These transformants were assayed for NAC-dependent activation of histidase formation and repression of GDH formation. The values on the x axis represent the number of amino acids remaining in the NAC fragment. Thus, 305 represents the wild type and 100 represents the N-terminal 100 amino acids. Histidase and GDH values are reported as specific activities.
The single exception in this set was NACK300, which lacks only the five C-terminal amino acids of NACK and was totally inactive in complementation. Western blot assays of extracts prepared from cells carrying wild-type NACK or NACK300 were carried out by using anti-NAC serum. The wild type was readily seen, but NACK300 was not (data not shown), suggesting that the NACK300 product is unstable and degraded rapidly.
Although NACK100 has nearly full activity in vivo, NACK98 complements nacK-203 less well, and truncations with 77, 95, or 97 amino acids remaining failed to complement nacK-203. We have not determined whether the failure of these very short NAC truncation products resulted from rapid degradation or an inactive protein. However, it is clear that a mutant nac gene that encodes only the first 100 of the 305 amino acids of NAC is functional in activation and repression of transcription, at least in vivo.
Activity of N-terminal fragments of NAC purified using MBP tags.
Fragments of NAC containing only the N-terminal 100, 120, or 129 amino acids were nearly as active as wild-type NAC in vivo. In order to prove that these fragments were directly responsible for the activity, we attempted to demonstrate activation of transcription in vitro by using purified components. To purify these fragments, we fused the MBP to the N terminus via a peptide linker that could be cleaved with factor Xa protease, leaving an N-terminal methionine, as is seen in wild-type NAC. MBP-tagged variants of wild-type NAC (NACK305 and NACE305) and two N-terminal fragments (NACK129 and NACE100) were constructed. All of these fusion proteins were inactive in vivo.
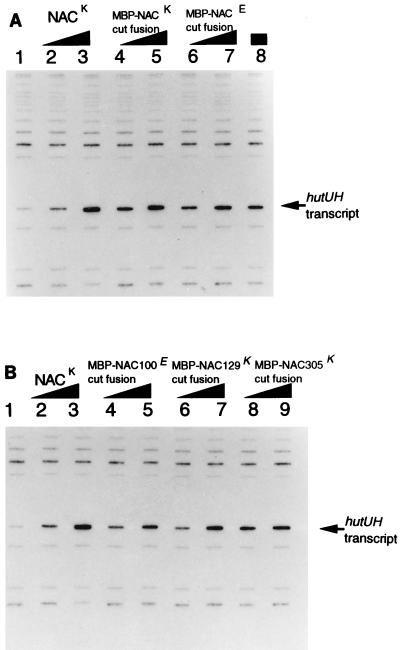
NAC fragments released from MBP-NAC fusions were unstable in the presence of factor Xa (or thrombin when a thrombin-sensitive linker was used); therefore, the factor Xa-mediated cleavage products of MBP-NAC were added immediately to an in vitro transcription reaction mixture without further purification. As expected, cleaved MBP-NACK305 was able to activate hut transcription (Fig. 2A). Cleaved MBP-NACE305 was also able to activate hut transcription, consistent with the observation that the nacE gene was able to complement a nacK mutant in vivo. This also provided the first direct evidence that NACE was active in vitro as well as in vivo, since we had been unable purify NACE because of its insolubility under all of the conditions tested. Cleaved MBP-NACK129 and MBP-NACE100 were also able to activate hut transcription almost as well as cleaved MBP-NACK305 (Fig. 2B). Thus, NACK129 and NACE100 were able to activate hut operon expression in vitro as well as in vivo.
FIG. 2.
Activation of hutUH transcription in vitro by NAC and N-terminal fragments of NAC. In vitro transcription was carried out by using purified components as described in Materials and Methods. The supercoiled template, pCB695, was similar to pAM1202 (8) and contained the hutUH promoter region cloned upstream of a strong transcriptional terminator such that transcription from this promoter would yield a transcript with the mobility on SDS-PAGE indicated at the right. All reactions contained RNA polymerase, nucleotides (including radioactive UTP), and buffers. NAC and NAC fragments were added as indicated. (A) Addition of full-length NAC. Lanes: 1, no NAC; 2 and 3, 0.04 and 0.4 μg of NAC (0.06 and 0.6 pmol of dimers) purified from K. aerogenes by salt precipitation (8); 4 and 5, 0.1 and 1 μg of MBP-NACK (1.3 and 13.3 pmol of MBP-NACK monomers) treated with factor Xa for 30 min immediately before addition to the transcription mixture; 6 and 7, 0.1 and 1 μg of MBP-NACE (1.3 and 13.3 pmol of monomers) treated with factor Xa for 30 min immediately before addition to the transcription mixture; 8, 3 μg of MBP-NACE (40 pmol of monomers) treated with factor Xa for 16 h before addition to the transcription mixture. (B) Addition of NAC fragments. Lanes: 1, no NAC; 2 and 3, 0.07 and 0.7 μg of NAC (1.1 and 10.5 pmol of dimers) purified from K. aerogenes by salt precipitation; 4 and 5, 0.1 and 1 μg of MBP-NACE100 (1.9 and 19 pmol of monomers) treated with factor Xa for 30 min immediately before addition to the transcription mixture; 6 and 7, 0.13 and 1.3 μg of MBP-NACK129 (1.9 and 19 pmol of monomers) treated with factor Xa for 30 min immediately before addition to the transcription mixture; 8 and 9, 0.14 and 1.4 μg of MBP-NACK305 (1.9 and 19 pmol of monomers) treated with factor Xa for 30 min immediately before addition to the transcription mixture.
The instability of the NAC fragments in the presence of factor Xa precluded any attempt to quantify NAC in these reaction mixtures, as well as any attempt to isolate large quantities of NAC fragments. Thus, we attempted to characterize NAC fragments with a nickel-binding his6 tag at the C terminus.
Activity of NAC fragments with his6 tags.
Full-length NACE305 and NACK305 with the nickel-binding his6 tag at the C terminus were active in vivo. In fact, the activation of histidase and repression of GDH formation were even stronger with NAC305-his6 that with the corresponding unmodified NAC305 (Table 3). This increased activation and repression appears not to be specific to the nature of the C-terminal tag, since a cloning artifact that fused an 11-amino-acid peptide (DFGRSGHTDSL) to the C terminus of NACK305 also resulted in more activation and repression than NACK305 (Table 3). Although the modified E. coli NAC (NACE305-his6) was active in vivo, the modification did not prove useful as a means of purifying NACE, since NACE305-his6 was insoluble in all of the buffers tested, just like NACE305.
TABLE 3.
Activation of histidase formation and repression of GDH formation in vivo by modified forms of NAC
| NAC source and straina | Plasmidb | NAC productc | Sp actd of:
|
|
|---|---|---|---|---|
| Histidase | GDH | |||
| E. coli | ||||
| KC2725 | None | None | 31 | 413 |
| KC3568 | pCB774 | NACE | 415 | 46 |
| KC3210 | pCB594 | MBP-NACE | 34 | 435 |
| KC3555 | pCB771 | NACE-his6 | 890 | 21 |
| KC3973 | pCB941 | NACE100-his6 | 42 | 354 |
| K. aerogenes | ||||
| KC3572 | pCB778 | NACK | 520 | 59 |
| KC3981 | pCB606 | MBP-NACK | 37 | 417 |
| KC3578 | pCB781 | NACK-his6 | 650 | 35 |
| KC3580 | pCB782 | NACK-DFGRSGHDTSL | 720 | 32 |
| KC3615 | pCB800 | NACK120-his6 | 295 | NDe |
| KC3591 | pCB788 | NACK100-his6 | 45 | 335 |
All strains are derived from K. aerogenes KC2725 (nac-203::Tn5-131), grown in glucose minimal medium to mid-log phase, and assayed as previously described (12). IPTG (0.5 mM) was included for strain KC3981 to guarantee expression from the pMalC2 vector.
All plasmids except pCB594 and PCB606 were derived from pGD103 as described in Materials and Methods. Plasmids pCB594 and pCB606 were derived from pMalC2.
The code is described in detail in the text. Subscripts indicate derivation from E. coli (E) or K. aerogenes (K). MBP indicates MBP fused to the N terminus of NAC. his6 indicates six histidine residues added to the C terminus. DFGRSGHTDSL indicates fusion of 11 amino acids to the C terminus of NAC (amino acids are indicated by the one-letter code). NAC100 and NAC120 indicate that only the first 100 or 120 amino acids of NAC were present.
Specific activity is reported as nanomoles per minute per milligram of cell protein.
ND, not determined.
Three N-terminal fragments with the his6 tag at the C terminus were constructed as described in Materials and Methods: NACE100-his6, NACK100-his6, and NACK120-his6. In contrast to their unmodified counterparts, both NACK100-his6 and NACE100-his6 were inactive in vivo (Table 3). Thus, we focused our attention on NACK120-his6. NACK120-his6 was purified by Ni affinity chromatography and shown to be able to activate hut operon transcription by purified RNA polymerase (data not shown). Unfortunately, we have had difficulty purifying NACK120-his6 from strains carrying either high-copy or low-copy expression vectors on a reliable basis. We have no explanation for this difficulty, although it may represent either instability of the polypeptide or toxicity of the fragment.
In a final attempt to isolate an active form of NACE for comparison to NACK in vitro, we constructed a gene encoding NACE that was modified at the N terminus with MBP and at the C terminus with his6 (MBP-NACE-his6). We hoped to use the his6 tag to separate the NACE fragment from factor Xa after cleavage of MBP-NACE-his6. However, MBP-NACE-his6 was completely resistant to cleavage by factor Xa, in marked contrast to MBP-NACE. Only the addition of strong denaturants allowed MBP to be removed, and this was accompanied by very rapid disappearance of NACE. No active NACE was recovered after attempts to renature the material. Apparently, the C-terminal his6 tag had altered the factor Xa cleavage site in MBP-NACE-his6 so as to make it inaccessible to cleavage by factor Xa, suggesting that the C terminus of the folded protein is close to the N terminus. Thus, the C-terminal his6 tag would be able to prevent the protease from reaching the cleavage site at the N terminus of the NAC polypeptide.
Subunit structures.
We next used the tagged NAC polypeptides to search for the dimerization site on NAC. These data (along with a summary of the in vitro activation data and DNA-binding data) are presented in Table 4. NACE and NACK with an N-terminal MBP tag eluted as monomers. If factor Xa was added, the elution profile was complex and included a peak of the size expected for MBP-NAC (uncleaved, monomer), MBP (monomer), NAC (dimer), and an unknown peak with an apparent size of 8.5 kDa (presumed to be a degradation product of the 33-kDa NAC monomer).
TABLE 4.
In vitro properties of modified forms of NACa
| Construct | Apparent molecular size in solution (kDa) | Formb | Gel mobility shiftc | Transcription activationc |
|---|---|---|---|---|
| NACE | I | |||
| MBP-NACE | 75 (66) | M (D) | Y (Y) | (Y) |
| 56his6-NACE | Y (Y) | N (N) | ||
| NACE-his6 | I | |||
| NACE100-his6 | I | |||
| MBP-NACE100 | (Y) | |||
| NACK | 66 | D | Y | Y |
| MBP-NACK | 70 (66) | M (D) | Y (Y) | N (Y) |
| 56his6-NACK | 77 | D | Y | |
| NACK-his6 | Y | Y | ||
| MBP-NACK129 | 56 (28) | M (D) | (Y) | |
| NACK120-his6 | 27 | D | Y | Y |
| NACK100-his6 | 22 | D | Y |
NACE, NACK, and several modified forms thereof were purified as described in Materials and Methods. The apparent molecular size in solution was determined by gel filtration, and this size was used to infer whether the product was monomeric or dimeric in solution. The ability to bind DNA was determined in a gel mobility shift assay using the urease operon promoter (ureDp) as a target. The ability to activate transcription from the hutUH promoter (hutUp) was tested as described in the legend to Fig. 2. The values in parentheses were determined after cleavage with Factor Xa or thrombin.
M, monomer; I, insoluble; D, dimer.
Y, yes; N, no.
A smaller N-terminal modification was also tested by fusing a 56-amino-acid peptide that included a his6 tag in its midst to the N terminus of NACK and NACE. Both of these fusions were inactive in vivo (data not shown). The N-terminally his6-tagged NACE and NACK were purified by Ni affinity chromatography. The tagged NACK was found to be dimeric in solution even before cleavage (NACE was not tested). However, neither the NACK nor the NACE fusion protein was able to activate transcription in vitro (Table 4). Cleavage of the tagged NACE with thrombin removed the his6 domain but left an 18-amino-acid peptide fused to the N terminus of NACE. This cleavage product was also unable to activate transcription in vitro (Table 4). In summary, fusions to the N terminus of NACE increase its solubility, but even small additions at the N terminus block its ability to activate transcription. Large additions (MBP) block the ability to dimerize in solution (Table 4).
Somewhat surprisingly, the C-terminally his6-tagged NAC fragments of NACK (NACK100-his6 and NACK120-his6), as well as the product released from MBP-NACK129 by (factor Xa) cleavage, were all dimers in solution as measured by gel filtration. Thus, the N-terminal domain of 100 amino acids also appears to contain the signals required for dimer formation, as well as for activation of transcription.
DISCUSSION
Perhaps the most surprising finding in these experiments is that the N-terminal 100 amino acids of NAC contain the determinants for all of the known functions of NAC. NAC100 was able to activate histidase formation in vivo and hutUp transcription in vitro. NAC100 was able to repress GDH formation in vivo. NACK100-his6 was able to bind a DNA fragment with a NAC-binding site, and NACK100-his6 was present as a dimer in solution. In short, NACK100 (or NACK100-his6) displayed all four known properties of NAC: activation of transcription, repression of transcription, site-specific DNA binding, and dimerization.
All of the truncations of NAC isolated in these experiments, except NACK300, showed considerable NAC activity. However, several tight nac mutants have been isolated before by using selection and screening. One of these, nac-203::Tn5, has been sequenced and is an insertion of Tn5 after the 199th codon of nacK. The data in Fig. 1 suggest that this mutation should leave NAC partially active, but nac-203 is one of the tightest nac mutants in our collection (although even nac-203 retains some activity, particularly on the ure operon promoter). Thus, it seems likely that NACK300 and NACK199 are inactive because of instability or misfolding rather than because of any lack of informative sequence.
The data presented here also provide some information about the function of the N and C termini of NAC. Additions to the N terminus block the ability of NAC to activate transcription, and small additions to the C terminus increase this activity, at least in vivo. The effect of the N-terminal additions appears to be intrinsic to the protein, since it is also seen in vitro with purified protein. Moreover, the ability to activate transcription can be recovered (in the case of MBP-NAC) by removing the N-terminal addition. The effect of the C-terminal additions may likewise be intrinsic to the protein, but it may also reflect an increased stability of the C-terminally modified NAC. We have been unable to raise a high-titer antibody with sufficient specificity for NAC to address this question directly, but we have previously suggested that wild-type NAC is rapidly turned over in vivo (14).
Two lines of evidence led us to suspect an interaction between the N and C termini of NAC and perhaps other LTTRs. First, MBP-NAC-his6 was completely resistant to cleavage by factor Xa, while MBP-NAC was readily cleaved. Second, other work in this laboratory (1) has shown that replacement of the amino-terminal 20 amino acids of NAC with the corresponding amino acids from OxyR results in an inactive NAC unless the C-terminal domain of this NAC-OxyR chimera is also replaced with the C-terminal domain from OxyR.
The helix-turn-helix region thought to be the DNA-binding site is located very close to the N terminus of LTTRs (about amino acids 20 to 40 for NAC). The binding site for coeffectors of LTTRs has generally been found in the C-terminal domain, between amino acids 100 and 200 (15). Thus, an effect of the C-terminal domain on the N-terminal domain must exist. The specificity requirement in the OxyR-NAC chimeras (1) and the ability of the C-terminal his6 tag to prevent cleavage of MBP from the N terminus of MBP-NAC-his6 seem more consistent with a hypothesis of direct interaction than with transmission of a signal through the middle part of the protein. However, in the absence of experiments to test a direct interaction, such arguments remain speculative at best.
It was exciting to discover that the four known regulatory properties of NAC are encoded within a NAC fragment containing only the N-terminal 100 amino acids, NAC100. It was disappointing to discover that a fifth property of NAC, the insolubility of NACE, was also encoded within this fragment. Most of the amino acid differences between NACE and NACK lie in the C-terminal domain; the N-terminal domains are more similar (14). Nevertheless, even NACE100-his6 is much more insoluble than NACK100-his6. Thus, future studies will continue to focus on NACK for in vitro studies.
ACKNOWLEDGMENT
This work was supported by Public Health Service grant GM47156 from the National Institutes of Health to R.A.B.
REFERENCES
- 1.Adams, G. M., and R. A. Bender. Unpublished observations.
- 2.Bender R A. The role of the NAC protein in the nitrogen regulation of Klebsiella aerogenes. Mol Microbiol. 1991;5:2575–2580. doi: 10.1111/j.1365-2958.1991.tb01965.x. [DOI] [PubMed] [Google Scholar]
- 3.Best E A, Bender R A. Cloning of the Klebsiella aerogenes nac gene which encodes a factor required for nitrogen regulation of the histidine utilization (hut) operons in Salmonella typhimurium. J Bacteriol. 1990;172:7043–7048. doi: 10.1128/jb.172.12.7043-7048.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Churchward G, Belin D, Nagamine Y. A pSC101-derived plasmid which shows no sequence homology to other commonly used cloning vectors. Gene. 1984;31:165–171. doi: 10.1016/0378-1119(84)90207-5. [DOI] [PubMed] [Google Scholar]
- 5.Deich R A, Metcalf B J, Finn C W, Farley J E, Green B A. Cloning of genes encoding a 15,000-dalton peptidoglycan-associated outer membrane lipoprotein and an antigenically related 15,000-dalton protein from Haemophilus influenzae. J Bacteriol. 1988;170:489–498. doi: 10.1128/jb.170.2.489-498.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feng J, Goss T J, Bender R A, Ninfa A J. Activation of transcription initiation from the nac promoter of Klebsiella aerogenes. J Bacteriol. 1995;177:5523–5534. doi: 10.1128/jb.177.19.5523-5534.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feng J, Goss T J, Bender R A, Ninfa A J. Repression of the nac promoter of Klebsiella aerogenes. J Bacteriol. 1995;177:5535–5538. doi: 10.1128/jb.177.19.5535-5538.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goss T J, Bender R A. The nitrogen assimilation control protein, NAC, is a DNA binding transcription activator in Klebsiella aerogenes. J Bacteriol. 1995;177:3546–3555. doi: 10.1128/jb.177.12.3546-3555.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henikoff S. Unidirectional digestion with exonuclease III creates targeted break points for DNA sequencing. Gene. 1984;28:351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- 10.Janes B K, Bender R A. Alanine catabolism in Klebsiella aerogenes: molecular characterization of the dadAB operon and its regulation by the nitrogen assimilation control protein. J Bacteriol. 1998;180:563–570. doi: 10.1128/jb.180.3.563-570.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 12.Macaluso A, Best E A, Bender R A. Role of the nac gene product in the nitrogen regulation of some NTR-regulated operons of Klebsiella aerogenes. J Bacteriol. 1990;172:7249–7255. doi: 10.1128/jb.172.12.7249-7255.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Magasanik B. Regulation of nitrogen utilization. In: Neidhardt F C, Curtis III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C: American Society for Microbiology; 1996. pp. 1344–1356. [Google Scholar]
- 14.Muse W B, Bender R A. The nac (nitrogen assimilation control) gene from Escherichia coli. J Bacteriol. 1998;180:1166–1173. doi: 10.1128/jb.180.5.1166-1173.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schell M A. Molecular biology of the LysR family of transcriptional regulators. Annu Rev Microbiol. 1993;47:597–626. doi: 10.1146/annurev.mi.47.100193.003121. [DOI] [PubMed] [Google Scholar]
- 16.Schwacha A, Bender R A. The nac (nitrogen assimilation control) gene from Klebsiella aerogenes. J Bacteriol. 1993;175:2107–2115. doi: 10.1128/jb.175.7.2107-2115.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwacha A, Bender R A. The product of the Klebsiella aerogenes nac (nitrogen assimilation control) gene is sufficient for activation of the hut operons and repression of the gdh operon. J Bacteriol. 1993;175:2116–2124. doi: 10.1128/jb.175.7.2116-2124.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tyrrell R, Verschueren K H G, Dodson E J, Murshudov G N, Addy C, Wilkinson A J. The structure of the cofactor-binding fragment of the LysR family member, CysB: a familiar fold with a surprising subunit arrangement. Structure. 1997;5:1017–1032. doi: 10.1016/s0969-2126(97)00254-2. [DOI] [PubMed] [Google Scholar]