Abstract
Colorectal cancer (CRC) is one of the most prevalent and deadly cancers worldwide. Despite recent improvements in treatment and prevention, most of the current therapeutic options are weighted by side effects impacting patients’ quality of life. Better patient selection towards systemic treatments represents an unmet clinical need. The recent multidisciplinary and molecular advancements in the treatment of CRC patients demand the identification of efficient biomarkers allowing to personalise patient care. Currently, core tumour biopsy specimens represent the gold-standard biological tissue to identify such biomarkers. However, technical feasibility, tumour heterogeneity and cancer evolution are major limitations of this single-snapshot approach. Genotyping circulating tumour DNA (ctDNA) has been addressed as potentially overcoming such limitations. Indeed, ctDNA has been retrospectively demonstrated capable of identifying minimal residual disease post-surgery and post-adjuvant treatment, as well as spotting druggable molecular alterations for tailoring treatments in metastatic disease. In this review, we summarise the available evidence on ctDNA applicability in CRC. Then, we review ongoing clinical trials assessing how liquid biopsy can be used interventionally to guide therapeutic choice in localised, locally advanced and metastatic CRC. Finally, we discuss how its widespread could transform CRC patients’ management, dissecting its limitations while suggesting improvement strategies.
Subject terms: Tumour biomarkers, Cancer genetics
Introduction
Colorectal cancer (CRC) is the third most common and the second most deadly cancer worldwide, representing 10.2% of new cases and 9.2% of cancer-related deaths [1]. Overall survival (OS) at 5-year from initial diagnosis spans from 87–90% in stage I–II to 68–72% in stage III, lowering to 11–14% in stage IV metastatic CRC (mCRC) [2]. Today, treatment algorithms in CRC are mainly driven by cancer staging, patients’ performance status and molecular profiling encompassing RAS, BRAF, ERBB2 and mismatch repair (MMR) status assessed on surgical or core biopsy tumour samples [3, 4]. In addition, following tumour-agnostic drug approvals, new molecular biomarkers such as NTRK1-3 translocations and high tumour mutational burden (TMB) have emerged in CRC as well as in other malignancies [5, 6]. Differently from the metastatic setting, with the exception of microsatellite instability (MSI), in stages II and III CRC there is still a lack of validated biomarkers identifying those patients more likely to benefit from adjuvant cytotoxic regimens [7]. This applies also following resections of metastatic disease, where usually post-operative treatments are administered despite the lack of prognostic biomarkers [3, 4]. To date, all molecular analyses exploited for clinical decision-making are based on core tumour biopsies, which currently represent the gold standard as per clinical guidelines [3, 4]. However, single solid tissue snapshots have several limitations such as tumour spatial and temporal heterogeneity, and technical feasibility issues [8–12]. In this clinical scenario, liquid biopsy (LB) is increasingly gaining attention as a complementary and potentially alternative non-invasive tool to bypass these limitations [13–16].
The term “liquid biopsy” defines the collection of tumour-derived biomarkers in the blood or other body fluids, such as urine, saliva, stool or cerebrospinal fluid [17–19]. Circulating tumour DNA (ctDNA), circulating tumour cells (CTCs) and exosomes are the most common tumour-related biomarkers assessed on liquid biopsy so far [8, 17]. Among these, ctDNA analysis, consisting in the isolation of DNA fragments from the bloodstream of patients, has already shown its potential in capturing the CRC molecular complexity, coupled with the technical advantages of minimal invasiveness and fast turnaround time [17, 20]. Assessment of real-time tumour-associated genomic changes and treatment selection can be guided by ctDNA monitoring in non-metastatic as well as metastatic CRC. Particularly, in mCRC the omni-comprehensiveness of ctDNA could replace solid tumour tissue biopsy as a more accurate tool in high burden diseases, potentially refining treatment tailoring such as rechallenge with anti-EGFR drugs [12, 17, 20, 21]. Given the promising retrospective data in all disease stages, validation of the interventional ctDNA role to drive treatment decisions is presently been tested to translate this tool into everyday clinical practice.
In this review, we will first discuss retrospective evidence supporting the role of ctDNA in CRC. Secondly, we will analyse currently ongoing trials testing interventional ctDNA as a tool to drive clinical decision-making in CRC focusing on initially available prospective data in this field. Finally, we will provide potential solutions to overcome current limitations for the incorporation of liquid biopsy in the management of CRC.
Circulating tumour DNA in colorectal cancer
CRC is one of the solid tumours shedding the highest amount of ctDNA in the bloodstream [22, 23]. However, since the ratio between ctDNA and circulating-free DNA (cfDNA) can greatly vary between less than 1% and more than 40%, due to many variables, including the location of the primary tumour and metastases, ctDNA detection requires highly sensitive and specific approaches [8, 12, 24]. Available assays for the detection of ctDNA have steadily increased in sensitivity and specificity. Indeed, today high-resolution PCR-based technologies, such as BEAMing, reached a sensitivity up to 0.001% with a specificity up to a single base difference, establishing the limit of detection (LOD) as 1 copy of mutant DNA/mL. It is also necessary to consider that the LOD depends on the chromosomal region (polymorphism adjacent to the hotspot could decrease the LOD) and the specificity of the probes [25, 26]. PCR-based tests, however, can detect only a few loci, while next-generation sequencing (NGS) can capture the full spectrum of genetic alterations, including Single Nucleotide Variants (SNVs), copy number variations and chromosomal rearrangements, depending on sequencing library and bioinformatic tools exploited [27]. Whole-genome sequencing (WGS) can further broaden the genomic analysis but with higher costs and lower sensitivity, given the difficulty in systematically applying barcoding systems required to increase sensitivity, especially for low-frequency ctDNA [28, 29]. In addition, if a low LOD cannot be achieved by WGS, Cristiano and colleagues developed an approach that combines the high fragmentation patterns of cell-free DNA in the genome of cancer patients with the mutation-based cell-free DNA analyses to detect 91% of patients with cancer from a healthy donor, obtaining 81% sensitivity and 95% specificity in CRC [30]. This integrated approach stood as a proof-of-concept that WGS from cfDNA could be exploited for screening, early detection and monitoring of human cancer [30]. Broadening the spectrum of genetic alterations assessable on ctDNA, methylation-specific PCR and methyl-BEAMing were found capable of detecting methylated ctDNA. However, a deeper discussion concerning the method of ctDNA detection, which is beyond the objectives of this review, is already discussed in other published reviews [8, 13, 20].
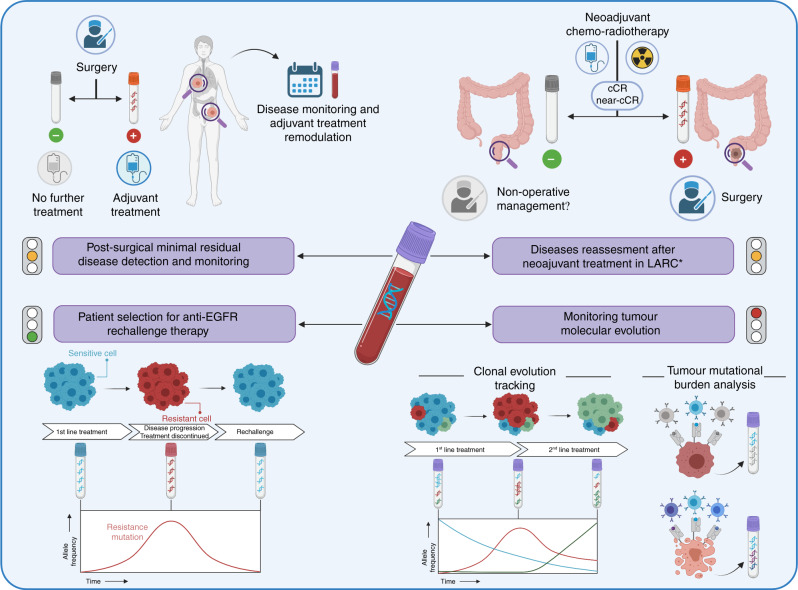
Given its potential clinical utility and improvements in technical applicability, ctDNA isolation through liquid biopsy has actively been pursued as a potential tool to refine CRC patients care (Fig. 1). Following, we discuss the clinical settings where the use of liquid biopsy can support clinical decision-making and guide therapeutic choices.
Fig. 1. Interventional liquid biopsy can orient therapeutic decision-making in CRC.
Circulating tumor DNA (ctDNA) analysis through liquid biopsy has proven to be a robust method to tailor personalised treatments for CRC) patient care. Promising results have been achieved in the post-surgical adjuvant setting and in driving treatment choice in locally advanced rectal cancer (LARC) after neoadjuvant treatment. Ongoing and future studies, exploiting ctDNA to guide anti-EGFR rechallenge therapy and treatment choices based on mutational and molecular CRC evaluation, will further expand the use of interventional liquid biopsy in CRC patients care. The “traffic lights”, close to each box where clinical strategies are defined, summarise the level of evidence supporting applicability of interventional ctDNA in the clinical practice. Figure created with BioRender.com. Keys: LARC locally advanced rectal cancer, * ctDNA negativity or positivity might be taken into account in patients treated with neoadjuvant multimodal treatment and achieving near clinical complete response (cCR) or cCR to evaluate if they might be candidate to non-operative management rather than curative surgery, respectively; cCR clinical complete response, EGFR epidermal growth factor receptor, green light = initial prospective data available, orange line = only retrospective data available, red light = only partial data available.
Colorectal cancer screening and diagnosis
The idea of exploiting cfDNA derived from cancer cells as a non-invasive screening technique is as old as the discovery of ctDNA itself [31]. It stems from the observation that CRC patients display higher levels of mutated DNA in their bloodstream compared to healthy controls [32]. The major limitation to its widespread clinical use is the suboptimal limit of detection for small invasive cancers or precancerous lesions, which characterise a relevant target of the screening campaigns [33, 34]. Moreover, since no information about the mutational profile of the cancer is available in the screening setting, panels of frequently mutated genes in CRC need to be verified to achieve an acceptable sensitivity [35], even if this approach also increases the risk of false-positive results due, for instance, to clonal haematopoiesis [36, 37]. A potential option to overcome this limitation is to associate already-known cancer protein biomarkers to mutational panels, as investigated in the CancerSEEK study, that integrated both mutations in a panel of 16 genes (in 1933 genomic positions) and 8 protein biomarkers from plasma, with a good performance in identifying 8 common cancer types [38]. Interestingly, even if the sensitivity for CRC is only around 60%, the specificity of the test is more than 99%, rendering this test a good candidate for multi-cancer screening before more invasive diagnostic procedures [38, 39].
In order to bypass limitations due to false-positive results, many approaches in this setting have focused on specific features of cancer genomes to be used as biomarkers [40, 41]. The first approach involves the study of the methylation pattern of cancer genomes compared to DNA derived from healthy cells [40, 42]. In particular, this approach may involve the evaluation of a single gene, such as the FDA-approved EpiproColon test which uses Septin9 (SEPT9) gene methylation with a sensitivity and a specificity of 68–72% and 80–82%, respectively [43–45], or the ColoSure test which evaluates vimentin (VIM) methylation state with comparable diagnostic accuracy [46]. On the other hand, the evaluation of thousands of methylation sequences using targeted bisulfite sequencing with the integration of machine learning to predict cancer-associated patterns of methylation is the approach exploited by GRAIL that, in the ambitious STRIVE study (NCT03085888), aims at identifying ctDNA from 12 tumour types including CRC with remarkable sensitivity and specificity (60–73% and >98%, respectively) in the first retrospective case-control study [47]. Another approach for the detection of cancer-specific circulating DNA relies on the analysis of the DNA fragments’ length (i.e. fragmentomics), which has proven valuable in identifying seven common cancer types including CRC [30], and is currently exploited in the DELFI study (NCT04825834). The lessons deriving from the study of ctDNA methylation and fragmentation have converged in the design of multimodality ctDNA test LUNAR-2, which has recently shown high values of sensitivity and specificity, also for stage I–II disease, of 88% and 94%, respectively [48, 49].
Finally, analysis of cfDNA derived from stool has been proposed as another means to increase the sensitivity of non-invasive faecal immunohistochemical test (FIT), particularly in early tumour stages and in precancerous lesions in which classical faecal tests display poor sensitivity [50–52]. Specifically, in a randomised clinical trial of multitarget stool DNA testing the sensitivity of detecting CRC versus FIT was improved from 74 to 92%, and the rate of detection of polyps with high-grade dysplasia was also dramatically increased from 46 to 69%, though the rate of false-positive was higher for the stool DNA test versus FIT [51]. Collectively, although the use of cfDNA for CRC screening is valuable and increasingly cheaper, it is still too early to claim its prime time in the clinic. Thus, based on available data, cfDNA analysis could only be exploited on top of clinically validated screening tools.
Post-surgical resection with curative intent
Surgery represents the main curative treatment of CRC patients with localised, locally advanced and also oligometastatic disease [3, 4]. In these patients the presence of ctDNA in the blood post-surgery can identify the existence of a minimal residual disease (MRD), invisible at radio-imaging and conceptually similar to the MRD in hematology [37, 53].
Post-surgical liquid biopsy in localised and locally advanced stages
In pivotal retrospective studies, a positive post-surgical liquid biopsy at a short distance after curative-intent surgery forebodes a disease recurrence within 2 years in almost all cases (Table 1 and Fig. 1). More than a decade ago, Diehl and co-workers first demonstrated that, in stage II–IV CRC patients who had undergone surgery with curative intent, median ctDNA decreased by 97% in less than a day and by 99% within 10 days. On the opposite, if supposedly curative resection was not attained, ctDNA levels decreased much less evidently or increased [54]. After this first evidence, other studies confirmed that the ctDNA was capable of predicting recurrence in CRC patients after surgical resections for localised (stage I–III) or oligometastatic disease (Table 1) [21, 55–60].
Table 1.
Main published retrospective and interventional studies suggesting the potential clinical role of circulating tumour DNA (ctDNA) assessment in colorectal cancer patients.
| Study | Disease stage | ctDNA method | N.° Pts. | Colon/rectal | Main findings |
|---|---|---|---|---|---|
| (A) Post-surgical resection with curative intent | |||||
| Tie et al. 2016 [57] | II | Safe-SeqS assay | 231 | 231/0 |
• Pts after surgery not receiving adjuvant chemotherapy: post-operative ctDNA detection correlates with higher risk of recurrence (HR 18; 95% CI 7.9–40) • Pts receiving adjuvant cytotoxic regimens: ctDNA positivity after treatment correlates with an inferior RFS (HR 11; 95% CI 1.8–68). |
| Diehl et al. 2008 [54] | II–IV | Real-time PCR | 20 | NA | • Worse RFS in ctDNA-positive pts after surgery (p = 0.006). |
| Tie et al. 2021 [59] | IV (CRLM resections) | Safe-SeqS) assay | 54 | NA |
• ctDNA post-operative positive pts had lower RFS (HR 6.3; 95% CI 2.58–15.2) and OS (HR 4.2; 95% CI 1.5–11.8). • ctDNA clearance observed in 3 pts receiving post-operative treatment, 2 of whom remained disease-free. • End-of-treatment (surgery ± adjuvant cytotoxic regimen) ctDNA positivity was associated with 0% 5-year RFS compared to 75.6% in those ctDNA-negative (HR 14.9; 95% CI 4.94–44.7). |
| Tie et al. 2019 (a) [58] | III | Safe-SeqS) assay | 96 | 96/0 |
• Positive ctDNA post-surgery correlates with an inferior RFS (HR 3.8; 95% CI, 2.4–21.0). • Positive ctDNA post-adjuvant therapy: lower 3-year RFI (HR, 6.8; 95% CI, 11.0–157.0). • Post-surgical ctDNA status is independently associated with RFI after adjusting for clinicopathologic risk factors (HR, 7.5; 95% CI, 3.5–16.1). |
| Reinert et al. 2016 [55] | I–IV | ddPCR | 11 | 5/6 | • 6/6 post-surgery positive ctDNA pts relapsed while 0/5 ctDNA-negative did (8 pts were ctDNA-positive prior to surgery but 1 was stage 1 CRC pts and 2 were stage 2 CRC pts). |
| Reinert et al. 2019 [56] | I–III | HiSeq 2500 system, Illumina Inc | 125 | 119/6 |
• Pre-operative ctDNA-positive pts were 108/122 (88.5%). • Post-operative ctDNA-positive pts were more likely to relapse (HR 7.2; 95% CI, 2.7–19.0). • Positive ctDNA pts after adjuvant cytotoxic regimens were more likely to relapse (HR 17.5; 95% CI, 5.4–56.5). • 3/10 ctDNA-positive pts were cleared by adjuvant regimens • In all multivariate analyses, ctDNA positivity was independently associated with relapse after adjusting for known clinicopathologic risk factors. |
| Parikh et al. 2021 [21] | I–IV | Guardant Reveal, Health panel | 84 | 54/30 |
• PPV in determining disease recurrence in ctDNA-positive pts: 100% • Integrating epigenomic signatures increased sensitivity by 25–36% versus genomic alterations alone. |
| Taieb et al. 2019 [60] | II–III | ddPCR | 805 | NA |
• 2-year DFS: 64% in ctDNA-positive versus 82% in those negative (HR 1.75; 95%CI 1.25–2.45). • In the multivariate analysis including age, gender, MSI, perforation, T stage, N stage and treatment arm (3 vs 6 months adjuvant treatment) ctDNA was confirmed an independent prognostic marker (HR 1.85 95% CI 1.31–2.61). |
| Kotaka et al. 2022 [62] | I–IV | Signatera bespoke multiplex-PCR NGS assay | 1564 | NA |
• Post-operative ctDNA positivity at 4 weeks after surgery was associated with an inferior DFS (HR 10.9; 95% CI 7.8–15.4). • DFS rates by ctDNA dynamics at 4 and 12 weeks postoperatively were significantly different between “positive to negative” versus “positive to positive” (HR 15.8; 95% CI 5.7–44.2). • Adjuvant treatment improved DFS in stages II, III and IV (analysed separately) among ctDNA-positive patients, while those ctDNA-negative do no benefit from medical post-operative treatment (HR 1.3; 95% CI 0.5–3.6). |
| (B) Neoadjuvant setting | |||||
| Tie et al. 2019 (b) [74] | II–III | Safe-SeqS) assay | 159 | 0/159 |
• HR for recurrence in ctDNA-negative vs ctDNA-positive after pre-operative chemotherapy: HR6.6 (p < 0.001); HR for recurrence in ctDNA-negative vs ctDNA-positive after surgery: HR 13.0 (p < 0.001). • 3-year RFS rate: 33% for the post-operative ctDNA-positive patients versus 87% for the post-operative ctDNA-negative patients, irrespective of clinicopathological risk factors (HR 6.0; p < 0.001). |
| Vidal et al. 2021 [75] | II–III | NGS (Guardant Reveal) | 72 | 0/72 |
• Detectable pre-surgery ctDNA after chemotherapy significantly associated with systemic recurrence, shorter DFS (HR 4; p = 0.033), and shorter OS (HR 23; p < 0.0001). • No significant association between ctDNA status and pathologic response. |
| McDuff et al. 2021 [76] | II–III | NGS (Guardant Reveal) | 29 | 0/29 |
• Overall margin-negative, node-negative resection rate: 88% in undetectable pre-operative ctDNA versus 44% in patients with detectable pre-operative ctDNA (p = .028). • Relapse for post-operative ctDNA-positive versus ctDNA-negative: 100% vs 13.3% (HR = 11.56; p = .007). |
| Khakoo et al. 2020 [77] | II–III | ddPCR | 47 | 0/47 |
• ctDNA status after CRT Associated with primary tumour response by mrTRG (p = 0.03). • MFS significantly shorter in ctDNA-positive patients after completing CRT (HR 7.1; p < 0.001), ctDNA-positive pre and mid-CRT (HR 3.8; p = 0.02), and ctDNA-positive pre, mid, and after CRT (HR 11.5; p < 0.001) versus ctDNA-negative or non-persistent. |
| Zhou et al. 2020 [80] | II–III | NGS (Targeted capture sequencing) | 106 | 0/106 |
• Pre-operative ctDNA-positive rate significantly lower in: patients with better pathologic tumour regression grade (ypCAP 0–1 vs ypCAP 2–3; p < 0.001), patients with pCR vs non-pCR (p = 0.02), patients with lower pT stage (ypT 0–2 vs ypT 3–4; p = 0.002). • ctDNA positivity at every timepoint after the start of neoadjuvant treatment is associated to shorter MFS (p < 0.05). |
| Wang et al. 2021 [79] | II–III | NGS (Targeted capture sequencing) | 119 | 0/119 |
• ctDNA clearance after CRT associated with a low probability of non-pCR (OR = 0.11, p = 0.04). • Incorporation of ctDNA and mrTRG after CRT exhibits high performance in predicting pCR (AUC = 0.886). • ctDNA-positive pts display a worse RFS after surgery (HR = 9.29; p < 0.001). |
| Murahashi et al. 2020 [78] | II–III | Amplicon-based deep sequencing | 85 | 0/85 |
• Change in ctDNA predicts pCR after pre-operative therapy (p = 0.0276). • Post-operative ctDNA detection predicts recurrence (p = 0.0127). |
| (C) Anti-EGFR rechallenge in the metastatic setting | |||||
| Cremolini et al. 2018 [90] | IV | ddPCR and Ion Torrent S5 XL | 28 | 22/6 |
• No RAS mutations on ctDNA in patients achieving PR • RAS wt on ctDNA patients had longer PFS than those RAS mutated on ctDNA (median PFS 4.0 vs 1.9 months; HR, 0.44; 95% CI, 0.18–0.98; p = .03). |
| Martinelli et al. 2021 [91] | IV | Idylla qPCR | 77 | 52/25 |
• RAS/BRAF wt on ctDNA patients had 17.3 months mOS (95% CI, 12.5–22.0 months) compared with 10.4 months (95% CI, 7.2–13.6 months) in those with mutated ctDNA (HR 0.49; 95% CI, 0.27–0.90; p = .02). • mPFS was 4.1 months (95% CI, 2.9–5.2 months) in RAS/BRAF wt patients compared with 3.0 months (95% CI, 2.6–3.5 months) in patients with mutated ctDNA (HR, 0.42; 95% CI, 0.23–0.75; p = .004) |
| Sartore-Bianchi et al. 2021 [92] | IV | ddPCR | 27 | 22/5 | • Among pts with RAS, BRAF and EGFR ECD wt mCRC prospectively interventionally assessed by ctDNA achieved RR 30%, DCR 63% and mPFS 16.4 weeks with panitumumab rechallenge monotherapy. |
N.° number; pts. patients, RFS relpase-free survival, OS overall survival, HR hazard ratio, ctDNA circulating tumour DNA, RFI relapse-free interval, CRC colorectal cancer, PCR polymerase chain reaction, ddPCR droplet-digital PCR, PPV predictive positive value, DFS disease free survival, DFI disease-free interval, MSI microsatellite instability, T primary tumour stage according to TNM classification, N lymph-nodes status according to TNM classification, RFS recurrence-free survival, NGS next-generation sequencing, CRT chemoradiation therapy, pCR pathological complete response, ypCAP post chemoradiation outcome according to the College of American Pathologists system, MFS metastasis-free survival, mrTRG tumour regression grade assessed by magnetic resonance, wt wild-type, AUC area under the curve, PR partial response, PFS progression-free survival, mOS median overall survival, mPFS median progression-free survival, ECD ectodomain, RR response rate, DCR disease control rate.
The prognostic role of ctDNA is specifically dramatic in high-risk stage II (T4) and stage III CRC patients. So far, due to the lack of truly predictive markers, the standard of care (SoC) for these patients is fluoropyrimidine ± oxaliplatin-based regimens despite the fact that approximately 50% of cases are cured by surgery alone, and chemotherapy is providing a relatively small net survival advantage of around 3–5% and 10–15%, respectively [7, 61]. In the last years the question has been: can ctDNA safely cherry-pick only patients that have a post-surgery MRD and thus should receive adjuvant treatment, while sparing treatment and toxicities to those already rendered disease-free by surgery alone? In untreated stage II CRC patients, ctDNA positivity indeed predicts a higher risk of recurrence with unprecedented double-digit probability statistics (HR 18; 95% CI 7.9–40) [57]. Moreover, in the same clinical context, ctDNA positivity is also associated with worse relapse-free survival (RFS) among those patients who had adjuvant treatment [57]. Later, the same authors demonstrated that ctDNA positivity was remarkably prognostic also in stage III patients, predicting disease relapse both post-surgery and post-adjuvant treatment [58]. A retrospective analysis in 805 adjuvant chemotherapy-treated stage III patients enrolled in the IDEA trial showed a worse 2-year disease-free survival (DFS) in ctDNA-positive patients if compared to negative ones (64% versus 84%, HR 1.75 95% CI 1.25–2.45) [60]. In addition, ctDNA positivity after surgery with curative intent in localised and locally advanced CRC independently correlates with a worse RFS even after adjusting for clinicopathological risk factors [55, 56, 60]. At the recent 2022 ASCO Gastrointestinal Cancer Symposium, the GALAXY study confirmed the ctDNA prognostic role in more than 1500 all stages surgically resected CRC patients [62]. Kotaka and collaborators also demonstrated that DFS rates by ctDNA dynamics 4 to 12 weeks after surgery were significantly different between “positive to negative” versus “positive to positive” (HR 15.8; 95% CI 5.7–44.2) [62]. More interestingly, they found that in ctDNA-positive patients adjuvant treatment improved 6 and 12 months DFS in stages II, III and IV, while not providing any DFS advantage in ctDNA-negative ones (HR 1.3; 95% CI 0.5–3.6) [62]. Finally, ctDNA-guided treatment in the stage II CRC adjuvant setting appears to be a cost-effective strategy aiming at reducing overtreatment in this specific setting [7, 63].
Post-surgical liquid biopsy in oligometastatic disease
In selected stage IV oligometastatic patients, surgical resection of metastasis can be pursued with curative intent [3, 4, 64]. In this setting as well, ctDNA provided striking results comparable to those described above for non-metastatic disease (Table 1). Diehl and co-workers first reported the prognostic potential of ctDNA detection in 20 patients with the liver-limited disease treated by partial hepatectomy. Of these, 16 were ctDNA-positive after surgery and all but one relapsed while those ctDNA-negative did not [54]. More recently, Tie and co-workers demonstrated that patients with liver-only metastases undergoing surgical resection had a lower RFS and survived less in the case of ctDNA positivity [59]. Of note, ctDNA clearance was achieved in 2 out of 3 patients receiving post-operative treatment supporting the effectiveness of post-operative treatments in this setting [59].
Despite this exciting amount of retrospective evidence suggesting that ctDNA is a potential predictive marker of disease recurrence in radically resected stage I–IV CRC patients, the actual clinical benefit is yet to be proven in prospective interventional trials. We reviewed (using ClinicalTrial.gov) ongoing trials aiming to verify if the assessment of ctDNA on plasma might be exploited interventionally in treating CRC patients. This search was performed in October 2021 and the Medical Subject Headings terms used were (“Colo-rectal Cancer” as condition/disease) and (“circulating tumor dna” as other terms) (Table 2). Particularly, several ctDNA-guided clinical trials are ongoing in the post-surgical setting exploiting different approaches and designs (Table 2). As an example, in the ongoing PEGASUS trial 140 high-risk stage II and stage III MSS patients are being ctDNA screened within a month from surgery and the intensity of their adjuvant treatment is modulated by the results (NCT04259944). ctDNA-negative patients receive capecitabine monotherapy while positive receive CAPOX for 3 months. In the LB-negative arm a series of shortly spaced ctDNA assessments are used to minimise the risk of false-negative, allowing to rapidly escalate to CAPOX whenever this is necessary. In the LB positive arm patients are re-biopsied for ctDNA at the end of the third month of capecitabine plus oxaliplatin (CAPOX regimen) and, if negative, they are de-escalated to a maintenance period with capecitabine, while if still positive they are switched to 5-fluorouracil plus irinotecan (FOLFIRI regimen) that is the SoC combination treatment in the setting of resistance to adjuvant treatment [3, 4]. In addition, the ALTAIR (NCT04457297) trial is investigating the role of trifluridine/tipiracil versus placebo in the case of stage II–IV resected ctDNA-positive CRC patients, while the VEGA (jRCT1031200006) study is assessing if surgery alone is non-inferior to adjuvant capecitabine plus oxaliplatin (CAPOX) in ctDNA-negative patients [65]. Other trials such as the MEDOCC-CrEATE [66], DYNAMIC-II and COBRA are focusing on stage II CRC patients to assess ctDNA role in driving treatment decision-making (Table 2). In stage III CRC instead, the DYNAMIC-III (ACTRN12617001566325) trial is aiming at assessing whether a treatment escalation strategy based on ctDNA might be superior to the standard of care in terms of RFS (Table 2). Overall, several studies are currently ongoing trying to prospectively assess the interventional role of ctDNA in stages I–III CRC through different experimental strategies and designs (Table 2). As depicted in the upper left box of Fig. 1, ctDNA might dramatically impact future post-operative treatment algorithms on top of main clinicopathological variables such as tumour staging and resection margins status. Indeed, radically resected CRC patients with no ctDNA in their plasma might be spared from receiving adjuvant treatments, while those positive for ctDNA will be candidates to receive adjuvant or an even more intensive treatment regimen. Furthermore, when results from these studies will be available, meta-analyses encompassing the range of different studies in this setting will be warranted to derive robust and potentially practice-changing conclusions.
Table 2.
Main ongoing clinical trials (N = 21) investigating the role of interventional circulating tumour DNA (ctDNA) to drive treatment decision-making in colorectal cancer (CRC) patients according to different clinical settings, suggesting potential future applications and developments.
| Study (Code Identifiers) Location |
Trial design Status |
Estimated enrolment (N pts) ctDNA analysis |
Main characteristics and inclusion criteria |
|---|---|---|---|
| Post-surgical resection with curative intent | |||
|
PEGASUS Italy–Spain |
Phase IIa Recruiting |
140 LUNAR1 panel |
• Resected stage III or T4N0 stage II colon cancer • ctDNA-guided adjuvant treatment: initially those ctDNA-positive will receive CAPOX while those negative capecitabine monotherapy; following treatment will be tailored on following ctDNA reassessment |
|
OPTIMIZE Denmark |
Randomised Phase II Not yet recruiting |
350 NA |
• Radical intended treatment for metastatic CRC with no evidence of further disease • Clinically eligible for adjuvant chemotherapy • ctDNA-guided post-surgical treatment |
|
DYNAMIC-II (ACTRN12615000381 583) Australia |
Phase III Recruiting |
450 NA |
• Resected stage II CRC • Pts will be randomly assigned to ctDNA treatment-guided group or not, and to those ctDNA-positive 5-FU will be given while to ctDNA-negative will be followed up |
|
DYNAMIC-III trial (ACTRN12617001566325) Australia |
Randomised Phase II/III |
1000 NA |
• Resected stage III colon cancer • ctDNA-negative pts in experimental arm will be de-escalated adjuvant treatment strategy and those ctDNA-positive will be escalated adjuvant treatment strategy; control will be treated as per SoC |
|
MEDOCC-CrEATE (NL6281/NTR6455) Netherlands |
Randomised TwiCs design Recruiting |
1320 NGS PGDx elio panel |
• Stage II colon cancer pts without indication for adjuvant treatment according to current guidelines • ctDNA-positive pts will be offered 8 cycles of adjuvant capecitabine plus oxaliplatin while ctDNA-negative pts and control group will be followed up |
|
COBRA (NCT04068103 and NRG-GI005) USA |
Phase II/III Recruiting |
1408 LUNAR panel |
• Stage IIA resected CRC • Pts in experimental arm II will receive adjuvant treatment (at investigator choice) if ctDNA-positive and surveillance if ctDNA-negative |
|
IMPROVE-IT Denmark |
Phase II randomised Recruiting |
64 NA |
• Stage I or II disease radically resected • Detectable ctDNA in post-operative plasma sample • No indication for adjuvant chemotherapy according to DCCG guidelines but standard adjuvant chemotherapy administered if ctDNA-positive |
|
USA |
Phase Ib/II Recruiting |
74 NA |
• Pts with detectable ctDNA following resection of all known liver metastases will receive treatment with an anti-PD-L1/TGFbetaRII Fusion Protein M7824 • Resected MSS metastatic CRC |
|
ALTAIR Japan |
Phase III Recruiting |
240 Signatera panel |
• Pts who undergone radical curative resection of the primary and metastatic tumours • Pts tested positive for ctDNA but with no evidence of disease at imaging will receive TAS-102 or placebo |
|
VEGA (jRCT1031200006) Japan |
Phase III Recruiting |
1240 NA |
• High‐risk stage II or low‐risk stage III (T1‐3 and N1) CRC, and ctDNA‐negative status at week 4 after surgery • Randomisation between surgery alone versus adjuvant CAPOX |
|
BESPOKE USA |
NA Recruiting |
2000 Signatera panel |
• Resected stage II or III colorectal cancer (CRC) • Pts may be recommended for adjuvant treatment or observation by their treating clinician |
| Neoadjuvant setting | |||
|
SYNCOPE Finland |
Randomised Not yet recruiting |
93 NA |
• LARC randomised to receive TNT using capecitabine/oxaliplatin and SCRT vs long course CRT using capecitabine • ctDNA and organoid-guided adjuvant therapy as experimental arm compared to SoC • Assessment of MRD after surgery and correlation with prognosis |
| Metastatic unresectable disease | |||
|
USA |
Phase II Recruiting |
100 NA |
• Pts clinically eligible for either regorafenib or trifluridin-tipiracil • Pts will continue treatment beyond 1st cycle depending on ctDNA results |
|
China |
Phase II Not yet recruiting |
100 NA |
• Pts must have failed after first-line treatment containing cetuximab • Individualised second-line targeted therapy based on ctDNA analysis |
|
FOLICOLOR International |
NA Recruiting |
60 NPY Methylation |
• Unresectable metastatic disease • Identification of PD by NPY Methylation in liquid biopsies • To assess response and progression to first-line FOLFOX/FOLFIRI treatment on liquid biopsy |
|
China |
Phase III Not yet recruiting |
50 NA |
• RAS wt on ctDNA • Non-resectable liver metastases candidate to anti-EGFR rechallenge based on ctDNA results |
|
LIBImAb Italy |
Phase III Not yet recruiting |
280 KRAS, NRAS and in BRAFV600 status assessment using the Idylla system (Biocartis) |
• RAS/BRAF wt on solid tumour biopsy but with RAS mutant at liquid biopsy • To compare di efficacy of FOLFIRI + Cetuximab or Bevacizumab in tissue wt but liquid mutant RAS mCRC |
|
China |
Phase II Not yet recruiting |
35 RAS/BRAF status assessment |
• First-line therapy of FOLFOX/FOLFIRI/FOLFOXIRI + Cetuximab effectively and the PFS is not less than 6 months • ≥4 months after the last time treated with Cetuximab • RAS/BRAF wt on ctDNA |
|
PARERE Italy |
Phase II Recruiting |
214 IdyllaTM ctKRAS-NRAS-BRAF Mutation Test |
• RAS and BRAF wt status of primary CRC or related metastasis • RAS and BRAF wt ctDNA at the time of screening • Previous first-line anti-EGFR-containing therapy with at least a PR or SD ≥ 6 months; ≥4 months elapsed between the end of first-line anti-EGFR administration and screening; ≥1 line of therapy between the end of first-line anti-EGFR administration and screening |
|
Saudi Arabia |
Phase II Recruiting |
60 RAS status assessment |
• Baseline must be RAS/BRAF wt on solid tumour tissue • RAS wt on ctDNA • Tumour burden with <4 organ involvement |
|
USA |
Phase II Recruiting |
120 Guardant360 assay |
• RAS and BRAF wt on tumour tissue taken from primary or metastatic site • PD after treatment with an anti-EGFR monoclonal antibody for at least 4 months • ≥ 90 days from the last anti-EGFR treatment • BRAF, EGFR, ERBB2, RAS, MET wt highest allele frequency reported for any gene mutation <2% |
These studies were retrieved through an extensive search performed on ClinicalTrial.gov in October 2021. The Medical Subject Headings terms used were (“Colo-rectal Cancer” as condition/disease) and (“circulating tumor dna” as other terms).
ctDNA circulating tumour DNA, N number, pts patients, NA not available, CRC colorectal cancer, CAPOX capecitabine plus oxaliplatin, NGS next-generation sequencing, 5-FU 5-fluorouracil, SoC standard of care, DCCG Dutch Colorectal Cancer Group, LARC locally advanced rectal cancer, SCRT short course radiotherapy, CRT chemoradiotherapy, MRD minimal residual disease, FOLFOX 5-fluorouracil plus oxaliplatin, TNT total neoadjuvant treatment, cCR clinical complete response, PD progressive disease, MSS microsatellite stable, NPY Neuropeptide Y, wt wild-type.
aMatched historical control 1:3 with TOSCA trial patients.
Neoadjuvant setting in locally advanced rectal cancer (LARC)
The current consensus on the management of locally advanced rectal cancer (LARC) below the peritoneal reflection consists of a multimodality treatment of neoadjuvant chemoradiotherapy (CRT) [67]. The pre-operative treatment of LARC, requiring a detailed pre- and post-treatment disease staging [68, 69], has contributed to decreasing the risk of local and distant relapse over time but has not disruptively changed the chance of survival [70].
More recently, several randomised trials have shown that pre-operative chemotherapy intensification as part of a total neoadjuvant treatment (TNT) strategy doubles the pathological complete response (pCR) rate achieved by conventional neoadjuvant chemoradiation (25 vs 12%) [71]. The doubling in pCR rate suggests that through TNT surgery might be avoided in a higher proportion of cases, paving the way towards a safer surgery-free “watch-and-wait” approach [72]. This expanding complexity in the management of LARC, poses pressing clinical questions including patients selection for different pre-operative treatments and early disease reassessment but, given the predictive importance of pCR for a non-surgical strategy, perhaps the most pivotal question relates to the timing and methodology for assessing the clinical complete response (cCR) after the completion of the neoadjuvant treatment. Tracking cancer in blood more than any other biological classifier, including the immunoscore [73], has the potential to fill the gaps of the unmet clinical needs in LARC [37], as suggested by the retrospective studies mentioned above. In the studies looking at pre-surgery or post-surgery liquid biopsy, a ctDNA-positive test was associated with an extremely high risk of recurrence and shortened survival reaching impressively results in double-digit hazard ratios for most studies (Table 1). Tie and colleagues, found a recurrence rate significantly higher in 13 and 19 of 159 patients with detectable ctDNA both after pre-operative treatment and after surgery, with a hazard ratio (HR) of 6.6 and 13, respectively [74]. Similarly, in a cohort of 72 patients undergoing TNT in the GEMCAD 1402 trial, pre-surgery ctDNA detected MRD in 15% and was significantly associated with shorter DFS (HR 4; P = 0.033) and OS (HR 23; P < 0.0001) [75]. In another study, the overall margin-negative, node-negative resection rate significantly doubled in 17 patients with undetectable versus 9 patients with pre-operative detectable ctDNA (88 vs 44%; P = 0.007) [76].
Notably, other studies have instead focused on the role of LB in monitoring early response during neoadjuvant therapy, using sequential samplings before, during and after the end of the pre-operative treatment. In the study by Khakoo and colleagues, ctDNA detection after pre-operative CRT was associated with primary tumour regression by magnetic resonance tumour regression grade (mrTRG) [77]. In the same work, the detection of ctDNA at any timepoint (pre-CRT, mid-CRT, or after CRT) was associated with shorter metastasis-free survival (MFS), fostering the way for an early prognostic evaluation during neoadjuvant treatment. Other studies found correlations between pre-operative ctDNA-positive rate after CRT and pathologic features after surgical resection such as pathologic ypT stage, tumour regression grade (TRG), and pathologic complete response (pCR) rate [78–80]. On the other hand, no correlation was identified between pre-operative ctDNA and pathologic response in patients receiving TNT, possibly reflecting the higher sensitivity of the method used in this work [75]. In addition, a currently ongoing trial (NCT04842006) is prospectively investigating the role of ctDNA in defining adjuvant approach after TNT in LARC (Table 2). As depicted in the upper right box of Fig. 1, ctDNA might play a key role when deciding for curative surgery versus non-operative management in patients achieving near clinical complete response (cCR) or cCR after neoadjuvant multimodal CRT, as assessed by imaging and endoscopy.
Non-resectable advanced disease
The majority of studies investigated the use of LB in unresectable metastatic CRC patients. The first important finding in this setting is that the molecular landscape of tumours analysed using ctDNA or tissue samples is concordant in the vast majority of cases [9, 20, 81]. This was initially reported in a cohort of 106 mCRC patients, where ctDNA analysis through allele-specific quantitative PCR achieved 100% specificity and sensitivity in capturing BRAFV600E mutations and specificity and sensitivity of 98% and 92%, respectively, in detecting six KRAS point mutation tested (G12A, G12C, G12D, G12S, G12V, G13D) [82]. Similarly, in a population of 100 mCRC patients the mutational status of KRAS, BRAF and NRAS in plasma samples achieved a 97% concordance in capturing “RAS pathway mutations” between solid tissue and blood was evaluated [9]. In both studies and others, discordant samples were linked to real biological differences secondary to intra-tumour inter-lesion heterogeneity, previous treatments and/or low burden of disease [9, 81, 82]. A liquid biopsy has the added advantage that ctDNA captures alterations occurring in multiple genes, specifically EGFR, ERBB2, PIK3CA or MAP2K1, unshadowing new potential targets for treatment as well as putative mechanisms of resistance to SoC targeted therapies such as anti-EGFR, anti-BRAF and anti-HER2 agents [9, 10, 12, 24, 24, 81, 83, 84]. In addition, and more recently, in a cohort of 232 CRC patients both solid tumour tissue and ctDNA were genotyped and an overall high concordance (84.9–100.0%) increased to near 100% (97.0–100.0%) when considering only clonal alterations (Fig. 1) [23]. Finally, through the GI-SCREEN network, the same authors demonstrated that ctDNA genotyping significantly shortens biomarker evaluation turnaround time (3 days versus 11 in standard pathological assessment) and increases screening efficiency for targeted agents trial enrolment (9.5% enrolment versus 4.1%) [23]. Similarly, the TARGET study conducted in different solid tumour including CRC patients showed that ctDNA can enhance patients enrolment into early phase clinical trials [83, 85].
Of note, even though CTCs isolation and analysis might offer the chance to study the CRC genome in its integrity rather than small fragments of DNA with potential further biological insights, currently ctDNA analysis represents the most effective strategy to assess mCRC molecular alterations in the advanced stage of disease [86]. Indeed, directly comparing the amount of ctDNA and CTCs in mCRC patients, we found that the median number of CTCs was 0 (ranging from 0 to 73) while the median amount ctDNA was 732,573 genome equivalent (GE, being the total number of fragments of cfDNA/mL)/mL (ranging from 174,774 to 174,078,615 GE/mL) [86]. Similarly, other studies confirmed the paucity of CTC in the blood of CRC patients and/or complexities of CTCs isolation in this setting [86–88].
Disease monitoring and the Darwinian evolution model of CRC clones
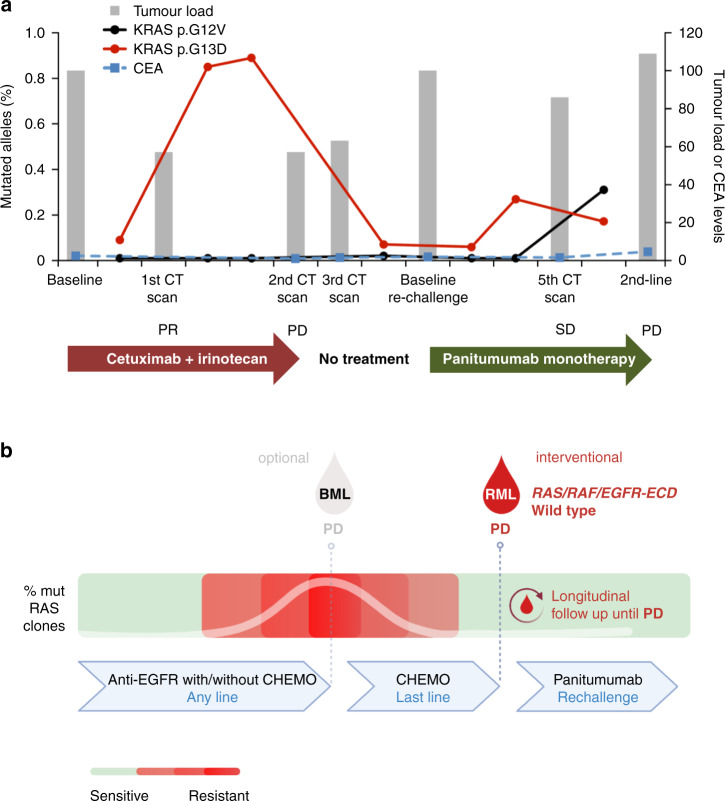
In metastatic CRC, ctDNA was investigated as a tool to dynamically monitor the molecular evolution of CRC over time, under the pressure of different courses of treatment (Fig. 1) [9, 11, 20]. Indeed, we found that the amount of mutations conferring resistance to approved anti-EGFR agents is reflected by quantitative fluctuation and qualitative molecular landscapes change in ctDNA (Fig. 2a), revealing molecular evolution of CRCs which would have been impossible to assess by tissue biopsy [9, 24]. This pulsatile behaviour of tumour-specific mutant clones, identified through mutation monitoring over time on ctDNA, provided a scientific rational for the strategy of retreatment with anti-EGFR, previously attempted on clinical empiricism (Fig. 2b) [89]. Two studies showed retrospectively that mCRC patients harbouring RAS mutations on ctDNA achieved a significantly inferior response rate and progression-free survival when rechallenged with anti-EGFR agents if compared to those RAS wild-type on ctDNA (Table 1) [90, 91]. Both studies suggested that RAS assessment in ctDNA could improve the clinically-based selection of patients to be rechallenged with anti-EGFR retreatment [90, 91]. More recently, the CHRONOS trial prospectively confirmed this hypothesis (Fig. 2b) [92]. In CHRONOS, CRC patients approaching third or later line of treatment were assessed for RAS, BRAF and EGFR ectodomain status in ctDNA and rechallenged with anti-EGFR treatment only if a mutation-negative status was found [92]. Interestingly, by using this strategy a 30% response rate and a 63% disease control rate were achieved [92]. These figures favourably compare with those achieved by anti-EGFR rechallenge trials selecting patients empirically, and also by current SoC chemotherapies for the late disease space in mCRC [89–94]. Furthermore, CHRONOS supports the concept that a timely RAS assessment on ctDNA might be more reliable to select patients for anti-EGFR rechallenge than previously proposed mathematical models [92, 95]. Further albeit, retrospective correlative data on the role of ctDNA in the rechallenge setting are expected from an ongoing randomised phase III trial comparing third line standard of care versus anti-EGFR rechallenge strategy (AIO-KRK-0114; NCT02934529). In conclusion, liquid biopsy-driven rechallenge with anti-EGFR antibody monotherapy led to objective responses in one-third of mCRC patients. These results showed for the first time prospectively that genotyping tumour DNA in the blood of CRC patients can be used to direct therapy and can be effectively incorporated in the management of advanced CRC patients. As supported by initial prospective data and depicted in the lower-left box of Fig. 1, anti-EGFR rechallenge represents the real-world clinical scenario which will likely be impacted sooner by the introduction of interventional ctDNA assessment.
Fig. 2. Tumour clones change consequently to drug-selective pressure.
Specifically, mutated RAS mutant clones dynamically evolve in response to pulsatile EGFR-specific antibody administration in metastatic colorectal cancer (mCRC) patients. In the upper panel, the dynamic of RAS altered clones retrospectively monitored through circulating tumour DNA (ctDNA) of a mCRC patient ONCG-CRC69 (a)—adapted from Siravegna et al. Nat Med, 2015. Each treatment received by this patient are indicated. Grēy bars represent tumour load change during treatments. Tumour load was calculated as percentage change based on measurable disease at baseline assumed as 100%. Dotted blue line indicates changes in CEA values (ng/ml). Treatment outcome are reported according to RECIST criteria. Red lines indicate the frequency of RAS mutation (percentage of alleles) detected in circulating free DNA. In the bottom panel, a schematic representation CHRONOS clinical trial design (b). The CHRONOS trial is the first phase II trial prospectively aiming to assess the role of interventional ctDNA assessment to molecularly select mCRC patients towards rechallenge with anti-EGFR monotherapy. In this trial, mCRC patients RAS, BRAF and EGFR ectodomain wild-type on ctDNA received panitumumab monotherapy up to disease progression or toxicity. Finally, in this trial all enrolled patients are periodically and prospectively followed up for ctDNA collection to be retrospectively analysed to derive further exploratory translational data. Keys: CEA carcinoembryonic antigen.
Liquid biopsies for immunotherapy and beyond
All the applications discussed so far in CRC are focused either on the correlation between ctDNA presence and tumour burden (e.g. in the detection of MRD) or the identification of molecular alterations that predict response or resistance to targeted agents. However, recent developments in our understanding of the cancer genome and the increasing availability of sequencing technologies at progressively lower prices are paving the way for new biomarkers analysis also in ctDNA [14, 96].
TMB is presently being debated in CRC and other solid tumours given its correlation with response to immunotherapy and the recent Food and Drugs Administration (FDA) approval as an agnostic biomarker to access cancer immunotherapy with pembrolizumab or dostarlimab [97, 98]. TMB is defined as the number of mutations per megabase of DNA (Mut/Mb), and in CRC it is typically increased in case of microsatellite instability (MSI) or pathogenic mutations occurring in the proofreading domains of the DNA polymerases POLE and POLD, leading to a consequential increase of tumour neoantigens (TNA) likely driving response to immune checkpoint blockade [99]. It is noteworthy that a minority of TMB-high (≥20 Mut/Mb) cases occur also in MSS and POLE/POLD wild-type gastrointestinal cancers, mainly associated with mutations in other DNA damage response genes [100], although the effective response to immunotherapy for these tumours is yet to be prospectively evaluated and has shown inconsistent results across different retrospective analyses [101]. The gold standard for TMB evaluation is tumour tissue specimens [102] even if intra-tumour heterogeneity constitutes a relevant limit to its exact estimation, thus supporting the role of a ctDNA-based evaluation, as it already achieved in non-small cell lung cancer (NSCLC) [103]. Moreover, as for any other genetic or genomic biomarker, TMB can change under treatment with standard cytotoxic agents in CRC [104]. Thus, ctDNA-based evaluation of TMB as a criterion to predict response to immunotherapy after cytotoxic priming with temozolomide in O-6-Methylguanine-DNA Methyltransferase (MGMT) methylated mCRC, is currently being investigated in the ARETHUSA trial (NCT03519412) (Table 2 and Fig. 1). Importantly, even though this biomarker is promising, the chromosomal regions to be considered for its calculation, as well as the fractional abundance of supported mutations and its cut-off values are far from being standardised, despite many ongoing international efforts to harmonise the way TMB is analysed and reported [105].
On the other hand, MSI is currently the most relevant biomarker for immunotherapy sensitivity in CRC, typically assessed on solid tissue specimens [3, 4]. However, similarly to TMB, MSI status is subjected to both spatial and temporal heterogeneity [106], making its monitoring through LB therapeutically valuable.
Further potential exploitation of LB in CRC is the analysis of methylation biomarkers, which is rapidly emerging as a powerful methodology for early diagnosis and prognosis [20]. However, until epigenetic drugs reach the clinical setting in CRC, clear ctDNA interventional applications in this setting will be lacking [107]. In addition, another intriguing frontier in the analysis of ctDNA, although speculative at present time, is the study of mutational signatures as a proxy to identify cancer evolution and as predictive factors for treatment and/or the onset of resistance [108]. In summary, despite being of high potential translational value as depicted in the lower right box of Fig. 1, these ctDNA applications need further refinements before they can be deployed clinically.
Overcoming limitations to interventional use of liquid biopsy in colorectal cancer
Despite the wealth of clinical opportunities offered by the analyses of ctDNA in CRC, logistical and biological limitations still limit its extensive application (Supplementary Table 1). The main logistical reason hampering the use of ctDNA-based analyses consists in their feasibility outside academic or comprehensive cancer centres [109]. One way to overcome this might be the centralisation of ctDNA analyses in selected referral centres, but this would require large cost-effectiveness studies and initiatives that need national and/or international public support. Conversely, using LB as a companion diagnostic could be particularly useful in those cases for which the result predicts response to medical treatment (targeted or immunotherapy) [110]. These two approaches could be complementary for the different settings, even though they would both require a simplification and standardisation of sample acquisition, in order to decrease pre-analytical biases as much as possible [111]. A second relevant logistical limitation for ctDNA analysis relies on the availability of different assays with different technical features in terms of sensitivity, LOD and reproducibility, whose accurate description is beyond the scope of this review since thoroughly discussed elsewhere [112]. Moreover, even if the industry has been a driving force in the development of the available LB platforms, we lack head-to-head comparisons of the different techniques for the same setting, making its cost-effectiveness evaluation difficult [113]. However, although no absolute preference can be given for one specific technology over the other, a balance between sensitivity required by the clinical question and widespread availability of the technique is advisable. For instance, less sensitive and/or automated platforms have shown good clinical performance in some settings [91, 114], while more sensitive and/or customised platforms, even based on a combination of different approaches, might be better for MRD detection [30, 115].
Biological limitations hampering ctDNA applicability into the clinic, must be analysed by considering other relevant CRC pathological, clinical and biological features (Supplementary Table 1). Tumour DNA shedding constitutes the first and more relevant of these limitations, and is known to be variable across stages, ranging between 73 and 100%, and primary tumour location in CRC [22, 116]. Moreover, DNA shedding not only correlates with tumour burden, similarly to other serum biomarkers such as CEA and CA19-9 [117], but also with the localisation of tumour metastases [118, 119]. In particular, the results from several studies investigating ctDNA-based analysis of KRAS mutations in mCRC have shown that the absence of liver metastases (e.g. in nodal or peritoneal-limited advanced disease) translates to reduced concordance with tissue analysis, that can be as low as 56% [120–122]. In a recent study, Bando and colleagues reported that patients with lung-only and peritoneum-only metastasis had significantly lower variant allele frequencies (VAFs) and lower numbers of detected variants, suggesting lower DNA release of subclonal variants in the blood [119]. Specifically, ctDNA was detectable in patients with lung-only metastases only in case of ≥20 mm of longest diameter and/or more than 20 lesions, and more than 20 mm of longest diameter in patients with peritoneum-only disease [119]. Collectively, these results evidence how low DNA shedding from lung- or peritoneal-limited disease could be considered another limitation to the application of liquid biopsies in patients with mCRC.
Moreover, the histopathological context of the cancer—that takes into account growth rate, stromal and inflammatory component, the extent of tumour cell death and necrosis—represents another determinant of cfDNA shedding that is intrinsically variable across CRC patients [123]. While the problem of DNA shedding affects mainly the sensitivity of LB, the presence of genetic aberrations in cfDNA originating from not cancerous tissues is a limitation to the specificity and sensitivity of circulating DNA analyses. Clonal Hematopoiesis (CH) is defined as the age-related accumulation of somatic mutations in hematopoietic stem cells which leads to clonal expansion of mutations in blood cells, and this is a primary source of false-positive results from ctDNA analysis [124]. CH is a relevant phenomenon that is reported in more than 10% of tumour-free patients over the age of 70 [125]. In a large dataset of more than 17,000 advanced cancer patients, it was shown that 5% of the patients would have at least 1 CH-associated mutation misattributed as tumour-derived in the absence of matched germline DNA sequencing [126]. The most common mutations derived from CH involve genes implicated in haematological tumorigenesis, such as DNMT3A, TET2, ASXL1 and JAK2, but also genes frequently mutated in solid tumours such as TP53, KRAS, PIK3CA and EGFR can be often reported, potentially leading to misinterpretation of the actionability of cancer [81, 124]. A recent work by Huang and co-workers identified KRAS mutations in three mCRC patients pre-treated with chemotherapy to be CH-derived by paired peripheral blood cells (PBCs) sequencing, even if the fractional abundance of all these mutations were reported <5% [127]. The impact of CH in the detection of MRD was also investigated by Chan and colleagues, whereby 17% of the pre-operative cfDNA mutations were CH-related and recurrently detected after surgery or completion of adjuvant chemotherapy [128]. Collectively, these results indicate that paired peripheral blood marrow cells (PBCs) or solid tumour tissue sample (either from the primary tumour or a metastatic site) sequencing should be taken into consideration whenever ctDNA results influence therapeutic choices to rule out between CH and CRC specific alterations (Supplementary Table 1). Moreover, this limitation might be overtaken by exploiting barcoded DNA sequencing methods and integrating different approaches for ctDNA detection (i.e. using fragmentomics) to increase the capability of discriminating the CH-driven molecular background from cancer mutations derived from colorectal tumours.
At the present time, another limitation of performing extended ctDNA molecular panels in CRC patients is the lack of clear evidence that therapeutic intervention can be driven by liquid biopsy findings. Indeed, and differently from NSCLC [129], apart from the recently completed CHRONOS trial, there is a lack of evidence indicating the activity of a specific agent based on ctDNA analyses in CRC.
Conclusions
Liquid biopsy is increasingly gaining traction in the clinical management of CRC patients in several clinical settings (Fig. 1). Retrospective data indicate that ctDNA can identify CRC patients requiring adjuvant treatments or conversely, not needing surgery after neoadjuvant treatment for LARC. Accordingly, once confirmed prospectively, the use of LB to detect MRD post-surgery with curative intent will likely be widely used in the management of early-stage CRC. Recently, the CHRONOS clinical trial demonstrated that ctDNA-based anti-EGFR rechallenge treatments can improve the therapeutic index of this therapeutic regimen. Presently, this is the only prospective and interventional evidence supporting the use ctDNA in CRC patients’ management, and accordingly anti-EGFR rechallenge is the setting in which ctDNA appears closer to clinical application. However as discussed above, ctDNA is likely to play a role also in selecting CRC potentially benefitting from other targeted therapies and immunotherapy, given its potential capability of capturing TMB and MSI features. In summary, even in presence of several biological and logistical limitations, LB will likely become central to rationally guiding CRC management. Ultimately, the accumulation of data from an ongoing perspective and randomised trials will determine the impact of ctDNA assessment for CRC patients’ care.
Supplementary information
Acknowledgements
We also thank members of the Molecular Oncology Laboratory at Candiolo Cancer Institute for scientific support and critical reading of the manuscript. GM is a PhD student within the European School of Molecular Medicine (SEMM). Figure 1 was created using Biorender.com.
Author contributions
GM, PPV and AB conceived and wrote the manuscript and tables. AS designed the figures and critically reviewed the manuscript. AS-B, GC, SM and SS critically reviewed and edited the manuscript.
Funding
This work was supported, in part, by Fondazione AIRC under 5 per Mille 2018-ID. 21091 program-P.I. AB, G.L. SS; AIRC under IG 2018-ID. 21923 project—P.I. AB; AIRC IG [no. 20685] to SS; BiLiGeCT—Progetto PON ARS01_00492 (AB); Terapia Molecolare Tumori by Fondazione Oncologia Niguarda Onlus to AS-B and SS; International Accelerator Award, ACRCelerate, jointly funded by Cancer Research UK (A26825 and A28223), FC AECC (GEACC18004TAB) and AIRC (22795) to AB; Ministero Salute, RC 2020 to AB; the project leading to this application has received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement No 101020342).
Data availability
Not applicable.
Competing interests
SS is advisory board member for Amgen, Bayer, BMS, CheckmAb, Daiichi-Sankyo, Guardant Health, Merck, Novartis, Roche-Genentech, and Seattle Genetics. AS-B is advisory board member for Amgen, Bayer, Sanofi and Servier. AB is a member of the scientific advisory board of NeoPhore, Illumina and Inivata, and a shareholder of NeoPhore. PPV has acted as a consultant of Biocartis and speaker for Merck. The remaining authors declare no competing interests.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Gianluca Mauri and Pietro Paolo Vitiello
Supplementary information
The online version contains supplementary material available at 10.1038/s41416-022-01769-8.
References
- 1.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer Statistics, 2021. CA Cancer J Clin. 2021;71:7–33. doi: 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- 2.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 3.Yoshino T, Arnold D, Taniguchi H, Pentheroudakis G, Yamazaki K, Xu R-H, et al. Pan-Asian adapted ESMO consensus guidelines for the management of patients with metastatic colorectal cancer: a JSMO-ESMO initiative endorsed by CSCO, KACO, MOS, SSO and TOS. Ann Oncol. 2018;29:44–70. doi: 10.1093/annonc/mdx738. [DOI] [PubMed] [Google Scholar]
- 4.Benson AB, Venook AP, Al-Hawary MM, Arain MA, Chen Y-J, Ciombor KK, et al. Colon Cancer, Version 2.2021, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw. 2021;19:329–59. doi: 10.6004/jnccn.2021.0012. [DOI] [PubMed] [Google Scholar]
- 5.Amatu A, Sartore-Bianchi A, Bencardino K, Pizzutilo EG, Tosi F, Siena S. Tropomyosin receptor kinase (TRK) biology and the role of NTRK gene fusions in cancer. Ann Oncol. 2019;30:viii5–15. doi: 10.1093/annonc/mdz383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marabelle A, Fakih M, Lopez J, Shah M, Shapira-Frommer R, Nakagawa K, et al. Association of tumour mutational burden with outcomes in patients with advanced solid tumours treated with pembrolizumab: prospective biomarker analysis of the multicohort, open-label, phase 2 KEYNOTE-158 study. Lancet Oncol. 2020;21:1353–65. doi: 10.1016/S1470-2045(20)30445-9. [DOI] [PubMed] [Google Scholar]
- 7.Argilés G, Tabernero J, Labianca R, Hochhauser D, Salazar R, Iveson T, et al. Localised colon cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2020;31:1291–305. doi: 10.1016/j.annonc.2020.06.022. [DOI] [PubMed] [Google Scholar]
- 8.Diaz LA, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol. 2014;32:579–86. doi: 10.1200/JCO.2012.45.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Siravegna G, Mussolin B, Buscarino M, Corti G, Cassingena A, Crisafulli G, et al. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat Med. 2015;21:827. doi: 10.1038/nm0715-827b. [DOI] [PubMed] [Google Scholar]
- 10.Russo M, Siravegna G, Blaszkowsky LS, Corti G, Crisafulli G, Ahronian LG, et al. Tumor heterogeneity and lesion-specific response to targeted therapy in colorectal cancer. Cancer Discov. 2016;6:147–53. doi: 10.1158/2159-8290.CD-15-1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Siena S, Sartore-Bianchi A, Garcia-Carbonero R, Karthaus M, Smith D, Tabernero J, et al. Dynamic molecular analysis and clinical correlates of tumor evolution within a phase II trial of panitumumab-based therapy in metastatic colorectal cancer. Ann Oncol. 2018;29:119–26. doi: 10.1093/annonc/mdx504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parikh AR, Leshchiner I, Elagina L, Goyal L, Levovitz C, Siravegna G, et al. Liquid versus tissue biopsy for detecting acquired resistance and tumor heterogeneity in gastrointestinal cancers. Nat Med. 2019;25:1415–21. doi: 10.1038/s41591-019-0561-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Patelli G, Vaghi C, Tosi F, Mauri G, Amatu A, Massihnia D, et al. Liquid biopsy for prognosis and treatment in metastatic colorectal cancer: circulating tumor cells vs circulating tumor DNA. Target Oncol. 2021;16:309–24. doi: 10.1007/s11523-021-00795-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nagayama S, Low S-K, Kiyotani K, Nakamura Y. Precision medicine for colorectal cancer with liquid biopsy and immunotherapy. Cancers (Basel) 2021;13:4803. doi: 10.3390/cancers13194803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mazouji O, Ouhajjou A, Incitti R, Mansour H. Updates on clinical use of liquid biopsy in colorectal cancer screening, diagnosis, follow-up, and treatment guidance. Front Cell Dev Biol. 2021;9:660924. doi: 10.3389/fcell.2021.660924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heidrich I, Abdalla TSA, Reeh M, Pantel K. Clinical applications of circulating tumor cells and circulating tumor DNA as a liquid biopsy marker in colorectal cancer. Cancers (Basel) 2021;13:4500. doi: 10.3390/cancers13184500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Siravegna G, Marsoni S, Siena S, Bardelli A. Integrating liquid biopsies into the management of cancer. Nat Rev Clin Oncol. 2017;14:531–48. doi: 10.1038/nrclinonc.2017.14. [DOI] [PubMed] [Google Scholar]
- 18.Crisafulli G, Mussolin B, Cassingena A, Montone M, Bartolini A, Barault L, et al. Whole exome sequencing analysis of urine trans-renal tumour DNA in metastatic colorectal cancer patients. ESMO Open. 2019;4:e000572. [DOI] [PMC free article] [PubMed]
- 19.Jiang J, Gao J, Wang G, Lv J, Chen W, Ben J, et al. Case Report: Vemurafenib treatment in brain metastases of BRAFS365L -mutant lung papillary cancer by genetic sequencing of cerebrospinal fluid circulating tumor DNA detection. Front Oncol. 2021;11:688200. doi: 10.3389/fonc.2021.688200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Di Nicolantonio F, Vitiello PP, Marsoni S, Siena S, Tabernero J, Trusolino L, et al. Precision oncology in metastatic colorectal cancer - from biology to medicine. Nat Rev Clin Oncol. 2021;18:506–25. doi: 10.1038/s41571-021-00495-z. [DOI] [PubMed] [Google Scholar]
- 21.Parikh AR, Van Seventer EE, Siravegna G, Hartwig AV, Jaimovich A, He Y, et al. Minimal residual disease detection using a plasma-only circulating tumor DNA assay in patients with colorectal cancer. Clin Cancer Res. 2021;27:5586–94. [DOI] [PMC free article] [PubMed]
- 22.Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6:224ra24. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakamura Y, Taniguchi H, Ikeda M, Bando H, Kato K, Morizane C, et al. Clinical utility of circulating tumor DNA sequencing in advanced gastrointestinal cancer: SCRUM-Japan GI-SCREEN and GOZILA studies. Nat Med. 2020;26:1859–64. doi: 10.1038/s41591-020-1063-5. [DOI] [PubMed] [Google Scholar]
- 24.Siravegna G, Lazzari L, Crisafulli G, Sartore-Bianchi A, Mussolin B, Cassingena A, et al. Radiologic and genomic evolution of individual metastases during HER2 blockade in colorectal cancer. Cancer Cell. 2018;34:148–62. doi: 10.1016/j.ccell.2018.06.004. [DOI] [PubMed] [Google Scholar]
- 25.Diehl F, Li M, He Y, Kinzler KW, Vogelstein B, Dressman D. BEAMing: single-molecule PCR on microparticles in water-in-oil emulsions. Nat Methods. 2006;3:551–9. doi: 10.1038/nmeth898. [DOI] [PubMed] [Google Scholar]
- 26.Sanmamed MF, Fernández-Landázuri S, Rodríguez C, Zárate R, Lozano MD, Zubiri L, et al. Quantitative cell-free circulating BRAFV600E mutation analysis by use of droplet digital PCR in the follow-up of patients with melanoma being treated with BRAF inhibitors. Clin Chem. 2015;61:297–304. doi: 10.1373/clinchem.2014.230235. [DOI] [PubMed] [Google Scholar]
- 27.Heitzer E, Haque IS, Roberts CES, Speicher MR. Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat Rev Genet. 2019;20:71–88. doi: 10.1038/s41576-018-0071-5. [DOI] [PubMed] [Google Scholar]
- 28.Liu MC, Oxnard GR, Klein EA, Swanton C, Seiden MV, Liu MC, et al. Sensitive and specific multi-cancer detection and localization using methylation signatures in cell-free DNA. Ann Oncol. 2020;31:745–59. doi: 10.1016/j.annonc.2020.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rheinbay E, Nielsen MM, Abascal F, Wala JA, Shapira O, Tiao G, et al. Analyses of non-coding somatic drivers in 2,658 cancer whole genomes. Nature. 2020;578:102–11. doi: 10.1038/s41586-020-1965-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cristiano S, Leal A, Phallen J, Fiksel J, Adleff V, Bruhm DC, et al. Genome-wide cell-free DNA fragmentation in patients with cancer. Nature. 2019;570:385–9. doi: 10.1038/s41586-019-1272-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sidransky D. Nucleic acid-based methods for the detection of cancer. Science. 1997;278:1054–9. doi: 10.1126/science.278.5340.1054. [DOI] [PubMed] [Google Scholar]
- 32.Vymetalkova V, Cervena K, Bartu L, Vodicka P. Circulating cell-free DNA and colorectal cancer: a systematic review. Int J Mol Sci. 2018;19:E3356. doi: 10.3390/ijms19113356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Issa IA, Noureddine M. Colorectal cancer screening: an updated review of the available options. World J Gastroenterol. 2017;23:5086–96. doi: 10.3748/wjg.v23.i28.5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kanth P, Inadomi JM. Screening and prevention of colorectal cancer. BMJ. 2021;374:n1855. doi: 10.1136/bmj.n1855. [DOI] [PubMed] [Google Scholar]
- 35.Fleshner P, Braunstein GD, Ovsepyan G, Tonozzi TR, Kammesheidt A. Tumor-associated DNA mutation detection in individuals undergoing colonoscopy. Cancer Med. 2018;7:167–74. doi: 10.1002/cam4.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu Y, Ulrich BC, Supplee J, Kuang Y, Lizotte PH, Feeney NB, et al. False-positive plasma genotyping due to clonal hematopoiesis. Clin Cancer Res. 2018;24:4437–43. doi: 10.1158/1078-0432.CCR-18-0143. [DOI] [PubMed] [Google Scholar]
- 37.Dasari A, Morris VK, Allegra CJ, Atreya C, Benson AB, Boland P, et al. ctDNA applications and integration in colorectal cancer: an NCI Colon and Rectal–Anal Task Forces whitepaper. Nat Rev Clin Oncol. 2020;17:757–70. doi: 10.1038/s41571-020-0392-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cohen JD, Li L, Wang Y, Thoburn C, Afsari B, Danilova L, et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science. 2018;359:926–30. doi: 10.1126/science.aar3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Killock D. Diagnosis: CancerSEEK and destroy - a blood test for early cancer detection. Nat Rev Clin Oncol. 2018;15:133. doi: 10.1038/nrclinonc.2018.21. [DOI] [PubMed] [Google Scholar]
- 40.Petit J, Carroll G, Gould T, Pockney P, Dun M, Scott RJ. Cell-free DNA as a diagnostic blood-based biomarker for colorectal cancer: a systematic review. J Surg Res. 2019;236:184–97. doi: 10.1016/j.jss.2018.11.029. [DOI] [PubMed] [Google Scholar]
- 41.Ferrari A, Neefs I, Hoeck S, Peeters M, Van Hal G. Towards novel non-invasive colorectal cancer screening methods: a comprehensive review. Cancers (Basel) 2021;13:1820. doi: 10.3390/cancers13081820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jung G, Hernández-Illán E, Moreira L, Balaguer F, Goel A. Epigenetics of colorectal cancer: biomarker and therapeutic potential. Nat Rev Gastroenterol Hepatol. 2020;17:111–30. doi: 10.1038/s41575-019-0230-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.deVos T, Tetzner R, Model F, Weiss G, Schuster M, Distler J, et al. Circulating methylated SEPT9 DNA in plasma is a biomarker for colorectal cancer. Clin Chem. 2009;55:1337–46. doi: 10.1373/clinchem.2008.115808. [DOI] [PubMed] [Google Scholar]
- 44.Johnson DA, Barclay RL, Mergener K, Weiss G, König T, Beck J, et al. Plasma Septin9 versus fecal immunochemical testing for colorectal cancer screening: a prospective multicenter study. PLoS ONE. 2014;9:e98238. doi: 10.1371/journal.pone.0098238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liles EG, Coronado GD, Perrin N, Harte AH, Nungesser R, Quigley N, et al. Uptake of a colorectal cancer screening blood test is higher than of a fecal test offered in clinic: a randomized trial. Cancer Treat Res Commun. 2017;10:27–31. doi: 10.1016/j.ctarc.2016.12.004. [DOI] [Google Scholar]
- 46.Ned RM, Melillo S, Marrone M. Fecal DNA testing for Colorectal Cancer Screening: the ColoSureTM test. PLoS Curr. 2011;3:RRN1220. doi: 10.1371/currents.RRN1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu MC, Oxnard GR, Klein EA, Swanton C, Seiden MV. CCGA Consortium. Sensitive and specific multi-cancer detection and localization using methylation signatures in cell-free DNA. Ann Oncol. 2020;31:745–59. doi: 10.1016/j.annonc.2020.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee J, Kim HC, Kim ST, He Y, Sample P, Nakamura Y, et al. Multimodal circulating tumor DNA (ctDNA) colorectal neoplasia detection assay for asymptomatic and early-stage colorectal cancer (CRC). 2021. https://meetinglibrary.asco.org/record/196511/abstract
- 49.Victoria M Raymond LH, Guardant Health I, Guardant Health RC. Evaluation of the ctDNA LUNAR-2 Test In an Average Patient Screening Episode (ECLIPSE). 2021. https://meetinglibrary.asco.org/record/194200/abstract
- 50.Sidransky D, Tokino T, Hamilton SR, Kinzler KW, Levin B, Frost P, et al. Identification of ras oncogene mutations in the stool of patients with curable colorectal tumors. Science. 1992;256:102–5. doi: 10.1126/science.1566048. [DOI] [PubMed] [Google Scholar]
- 51.Imperiale TF, Ransohoff DF, Itzkowitz SH, Levin TR, Lavin P, Lidgard GP, et al. Multitarget stool DNA testing for colorectal-cancer screening. N Engl J Med. 2014;370:1287–97. doi: 10.1056/NEJMoa1311194. [DOI] [PubMed] [Google Scholar]
- 52.Niedermaier T, Balavarca Y, Brenner H. Stage-specific sensitivity of fecal immunochemical tests for detecting colorectal cancer: systematic review and meta-analysis. Am J Gastroenterol. 2020;115:56–69. doi: 10.14309/ajg.0000000000000465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Siravegna G, Mussolin B, Venesio T, Marsoni S, Seoane J, Dive C, et al. How liquid biopsies can change clinical practice in oncology. Ann Oncol. 2019;30:1580–90. doi: 10.1093/annonc/mdz227. [DOI] [PubMed] [Google Scholar]
- 54.Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14:985–90. doi: 10.1038/nm.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reinert T, Schøler LV, Thomsen R, Tobiasen H, Vang S, Nordentoft I, et al. Analysis of circulating tumour DNA to monitor disease burden following colorectal cancer surgery. Gut. 2016;65:625–34. doi: 10.1136/gutjnl-2014-308859. [DOI] [PubMed] [Google Scholar]
- 56.Reinert T, Henriksen TV, Christensen E, Sharma S, Salari R, Sethi H, et al. Analysis of plasma cell-free DNA by ultradeep sequencing in patients with stages I to III colorectal cancer. JAMA Oncol. 2019;5:1124–31. doi: 10.1001/jamaoncol.2019.0528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tie J, Wang Y, Tomasetti C, Li L, Springer S, Kinde I, et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci Transl Med. 2016;8:346ra92. doi: 10.1126/scitranslmed.aaf6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tie J, Cohen JD, Wang Y, Christie M, Simons K, Lee M, et al. Circulating tumor DNA analyses as markers of recurrence risk and benefit of adjuvant therapy for stage III colon cancer. JAMA Oncol. 2019;5:1710–7. doi: 10.1001/jamaoncol.2019.3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tie J, Wang Y, Cohen J, Li L, Hong W, Christie M, et al. Circulating tumor DNA dynamics and recurrence risk in patients undergoing curative intent resection of colorectal cancer liver metastases: a prospective cohort study. PLoS Med. 2021;18:e1003620. doi: 10.1371/journal.pmed.1003620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Taieb J, Taly V, Vernerey D, Bourreau C, Bennouna J, Faroux R, et al. Analysis of circulating tumour DNA (ctDNA) from patients enrolled in the IDEA-FRANCE phase III trial: prognostic and predictive value for adjuvant treatment duration. Ann Oncol. 2019;30:v867. doi: 10.1093/annonc/mdz394.019. [DOI] [Google Scholar]
- 61.André T, Boni C, Mounedji-Boudiaf L, Navarro M, Tabernero J, Hickish T, et al. Oxaliplatin, fluorouracil, and leucovorin as adjuvant treatment for colon cancer. N Engl J Med. 2004;350:2343–51. doi: 10.1056/NEJMoa032709. [DOI] [PubMed] [Google Scholar]
- 62.Kotaka M, Shirasu H, Watanabe J, Yamazaki K, Hirata K, Akazawa N, et al. Association of circulating tumor DNA dynamics with clinical outcomes in the adjuvant setting for patients with colorectal cancer from an observational GALAXY study in CIRCULATE-Japan. J Clin Oncol. 2022;40:9. https://meetinglibrary.asco.org/record/204528/abstract.
- 63.To YH, Degeling K, Kosmider S, Wong R, Lee M, Dunn C, et al. Circulating tumour DNA as a potential cost-effective biomarker to reduce adjuvant chemotherapy overtreatment in stage II colorectal cancer. Pharmacoeconomics. 2021;39:953–64. doi: 10.1007/s40273-021-01047-0. [DOI] [PubMed] [Google Scholar]
- 64.Adam R, De Gramont A, Figueras J, Guthrie A, Kokudo N, Kunstlinger F, et al. The oncosurgery approach to managing liver metastases from colorectal cancer: a multidisciplinary international consensus. Oncologist. 2012;17:1225–39. doi: 10.1634/theoncologist.2012-0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Taniguchi H, Nakamura Y, Kotani D, Yukami H, Mishima S, Sawada K, et al. CIRCULATE‐Japan: Circulating tumor DNA–guided adaptive platform trials to refine adjuvant therapy for colorectal cancer. Cancer Sci. 2021;112:2915–20. doi: 10.1111/cas.14926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schraa SJ, van Rooijen KL, van der Kruijssen DEW, Rubio Alarcón C, Phallen J, Sausen M, et al. Circulating tumor DNA guided adjuvant chemotherapy in stage II colon cancer (MEDOCC-CrEATE): study protocol for a trial within a cohort study. BMC Cancer. 2020;20:790. doi: 10.1186/s12885-020-07252-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Glynne-Jones R, Wyrwicz L, Tiret E, Brown G, Rödel C, Cervantes A, et al. Rectal cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2018;29:iv263. doi: 10.1093/annonc/mdy161. [DOI] [PubMed] [Google Scholar]
- 68.Taylor FGM, Quirke P, Heald RJ, Moran BJ, Blomqvist L, Swift IR, et al. Preoperative magnetic resonance imaging assessment of circumferential resection margin predicts disease-free survival and local recurrence: 5-year follow-up results of the MERCURY study. J Clin Oncol. 2014;32:34–43. doi: 10.1200/JCO.2012.45.3258. [DOI] [PubMed] [Google Scholar]
- 69.Bhoday J, Balyasnikova S, Wale A, Brown G. How should imaging direct/orient management of rectal cancer? Clin Colon Rectal Surg. 2017;30:297–312. doi: 10.1055/s-0037-1606107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Papaccio F, Roselló S, Huerta M, Gambardella V, Tarazona N, Fleitas T, et al. Neoadjuvant chemotherapy in locally advanced rectal cancer. Cancers. 2020;12:3611. doi: 10.3390/cancers12123611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bahadoer RR, Dijkstra EA, van Etten B, Marijnen CAM, Putter H, Kranenbarg EM-K, et al. Short-course radiotherapy followed by chemotherapy before total mesorectal excision (TME) versus preoperative chemoradiotherapy, TME, and optional adjuvant chemotherapy in locally advanced rectal cancer (RAPIDO): a randomised, open-label, phase 3 trial. Lancet Oncol. 2021;22:29–42. doi: 10.1016/S1470-2045(20)30555-6. [DOI] [PubMed] [Google Scholar]
- 72.Giunta EF, Bregni G, Pretta A, Deleporte A, Liberale G, Bali AM, et al. Total neoadjuvant therapy for rectal cancer: making sense of the results from the RAPIDO and PRODIGE 23 trials. Cancer Treat Rev. 2021;96:102177. doi: 10.1016/j.ctrv.2021.102177. [DOI] [PubMed] [Google Scholar]
- 73.Bruni D, Angell HK, Galon J. The immune contexture and Immunoscore in cancer prognosis and therapeutic efficacy. Nat Rev Cancer. 2020;20:662–80. doi: 10.1038/s41568-020-0285-7. [DOI] [PubMed] [Google Scholar]
- 74.Tie J, Cohen JD, Wang Y, Li L, Christie M, Simons K, et al. Serial circulating tumour DNA analysis during multimodality treatment of locally advanced rectal cancer: a prospective biomarker study. Gut. 2019;68:663–71. doi: 10.1136/gutjnl-2017-315852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vidal J, Casadevall D, Bellosillo B, Pericay C, Garcia-Carbonero R, Losa F, et al. Clinical impact of presurgery circulating tumor DNA after total neoadjuvant treatment in locally advanced rectal cancer: a biomarker study from the GEMCAD 1402 trial. Clin Cancer Res. 2021;27:2890–8. doi: 10.1158/1078-0432.CCR-20-4769. [DOI] [PubMed] [Google Scholar]
- 76.McDuff SGR, Hardiman KM, Ulintz PJ, Parikh AR, Zheng H, Kim DW, et al. Circulating tumor DNA predicts pathologic and clinical outcomes following neoadjuvant chemoradiation and surgery for patients with locally advanced rectal cancer. JCO Precis Oncol. 2021;5:123–32. [DOI] [PMC free article] [PubMed]
- 77.Khakoo S, Carter PD, Brown G, Valeri N, Picchia S, Bali MA, et al. MRI tumor regression grade and circulating tumor DNA as complementary tools to assess response and guide therapy adaptation in rectal cancer. Clin Cancer Res. 2020;26:183–92. doi: 10.1158/1078-0432.CCR-19-1996. [DOI] [PubMed] [Google Scholar]
- 78.Murahashi S, Akiyoshi T, Sano T, Fukunaga Y, Noda T, Ueno M, et al. Serial circulating tumour DNA analysis for locally advanced rectal cancer treated with preoperative therapy: prediction of pathological response and postoperative recurrence. Br J Cancer. 2020;123:803–10. doi: 10.1038/s41416-020-0941-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang Y, Yang L, Bao H, Fan X, Xia F, Wan J, et al. Utility of ctDNA in predicting response to neoadjuvant chemoradiotherapy and prognosis assessment in locally advanced rectal cancer: a prospective cohort study. PLoS Med. 2021;18:e1003741. doi: 10.1371/journal.pmed.1003741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhou J, Wang C, Lin G, Xiao Y, Jia W, Xiao G, et al. Serial circulating tumor DNA in predicting and monitoring the effect of neoadjuvant chemoradiotherapy in patients with rectal cancer: a prospective multicenter study. Clin Cancer Res. 2021;27:301–10. doi: 10.1158/1078-0432.CCR-20-2299. [DOI] [PubMed] [Google Scholar]
- 81.Strickler JH, Loree JM, Ahronian LG, Parikh AR, Niedzwiecki D, Pereira AAL, et al. Genomic landscape of cell-free DNA in patients with colorectal cancer. Cancer Discov. 2018;8:164–73. doi: 10.1158/2159-8290.CD-17-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Thierry AR, Mouliere F, El Messaoudi S, Mollevi C, Lopez-Crapez E, Rolet F, et al. Clinical validation of the detection of KRAS and BRAF mutations from circulating tumor DNA. Nat Med. 2014;20:430–5. doi: 10.1038/nm.3511. [DOI] [PubMed] [Google Scholar]
- 83.Nakamura Y, Okamoto W, Kato T, Esaki T, Kato K, Komatsu Y, et al. Circulating tumor DNA-guided treatment with pertuzumab plus trastuzumab for HER2-amplified metastatic colorectal cancer: a phase 2 trial. Nat Med. 2021;27:1899–903. doi: 10.1038/s41591-021-01553-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Siena S, Raghav K, Masuishi T, Yamaguchi K, Nishina T, Elez E, et al. 386O Exploratory biomarker analysis of DESTINY-CRC01, a phase II, multicenter, open-label study of trastuzumab deruxtecan (T-DXd, DS-8201) in patients (pts) with HER2-expressing metastatic colorectal cancer (mCRC) Ann Oncol. 2021;32:S532. doi: 10.1016/j.annonc.2021.08.908. [DOI] [PubMed] [Google Scholar]
- 85.Rothwell DG, Ayub M, Cook N, Thistlethwaite F, Carter L, Dean E, et al. Utility of ctDNA to support patient selection for early phase clinical trials: the TARGET study. Nat Med. 2019;25:738–43. doi: 10.1038/s41591-019-0380-z. [DOI] [PubMed] [Google Scholar]
- 86.Germano G, Mauri G, Siravegna G, Dive C, Pierce J, Di Nicolantonio F, et al. Parallel evaluation of circulating tumor DNA and circulating tumor cells in metastatic colorectal cancer. Clin Colorectal Cancer. 2018;17:80–3. doi: 10.1016/j.clcc.2017.10.017. [DOI] [PubMed] [Google Scholar]
- 87.Cohen SJ, Alpaugh RK, Gross S, O’Hara SM, Smirnov DA, Terstappen LWMM, et al. Isolation and characterization of circulating tumor cells in patients with metastatic colorectal cancer. Clin Colorectal Cancer. 2006;6:125–32. doi: 10.3816/CCC.2006.n.029. [DOI] [PubMed] [Google Scholar]
- 88.Denève E, Riethdorf S, Ramos J, Nocca D, Coffy A, Daurès J-P, et al. Capture of viable circulating tumor cells in the liver of colorectal cancer patients. Clin Chem. 2013;59:1384–92. doi: 10.1373/clinchem.2013.202846. [DOI] [PubMed] [Google Scholar]
- 89.Mauri G, Pizzutilo EG, Amatu A, Bencardino K, Palmeri L, Bonazzina EF, et al. Retreatment with anti-EGFR monoclonal antibodies in metastatic colorectal cancer: Systematic review of different strategies. Cancer Treat Rev. 2019;73:41–53. doi: 10.1016/j.ctrv.2018.12.006. [DOI] [PubMed] [Google Scholar]
- 90.Cremolini C, Rossini D, Dell’Aquila E, Lonardi S, Conca E, Del Re M, et al. Rechallenge for patients with RAS and BRAF wild-type metastatic colorectal cancer with acquired resistance to first-line cetuximab and irinotecan: a phase 2 single-arm clinical trial. JAMA Oncol. 2019;5:343–50. [DOI] [PMC free article] [PubMed]
- 91.Martinelli E, Martini G, Famiglietti V, Troiani T, Napolitano S, Pietrantonio F, et al. Cetuximab rechallenge plus avelumab in pretreated patients with RAS wild-type metastatic colorectal cancer: the phase 2 single-arm clinical CAVE trial. JAMA Oncol. 2021;7:1529–35. [DOI] [PMC free article] [PubMed]
- 92.Sartore-Bianchi A, Pietrantonio F, Lonardi S, Mussolin B, Rua F, Fenocchio E, et al. Phase II study of anti-EGFR rechallenge therapy with panitumumab driven by circulating tumor DNA molecular selection in metastatic colorectal cancer: The CHRONOS trial. J Clin Oncol. 2021;39:3506. https://meetinglibrary.asco.org/record/195971/abstract.
- 93.Grothey A, Van Cutsem E, Sobrero A, Siena S, Falcone A, Ychou M, et al. Regorafenib monotherapy for previously treated metastatic colorectal cancer (CORRECT): an international, multicentre, randomised, placebo-controlled, phase 3 trial. Lancet. 2013;381:303–12. doi: 10.1016/S0140-6736(12)61900-X. [DOI] [PubMed] [Google Scholar]
- 94.Mayer RJ, Van Cutsem E, Falcone A, Yoshino T, Garcia-Carbonero R, Mizunuma N, et al. Randomized trial of TAS-102 for refractory metastatic colorectal cancer. N Engl J Med. 2015;372:1909–19. doi: 10.1056/NEJMoa1414325. [DOI] [PubMed] [Google Scholar]
- 95.Parseghian CM, Loree JM, Morris VK, Liu X, Clifton KK, Napolitano S, et al. Anti-EGFR-resistant clones decay exponentially after progression: implications for anti-EGFR re-challenge. Ann Oncol. 2019;30:243–9. doi: 10.1093/annonc/mdy509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cescon DW, Bratman SV, Chan SM, Siu LL. Circulating tumor DNA and liquid biopsy in oncology. Nat Cancer. 2020;1:276–90. doi: 10.1038/s43018-020-0043-5. [DOI] [PubMed] [Google Scholar]
- 97.Passaro A, Stenzinger A, Peters S. Tumor mutational burden as a pan-cancer biomarker for immunotherapy: the limits and potential for convergence. Cancer Cell. 2020;38:624–5. doi: 10.1016/j.ccell.2020.10.019. [DOI] [PubMed] [Google Scholar]
- 98.Sha D, Jin Z, Budzcies J, Kluck K, Stenzinger A, Sinicrope FA. Tumor mutational burden (TMB) as a predictive biomarker in solid tumors. Cancer Discov. 2020;10:1808–25. doi: 10.1158/2159-8290.CD-20-0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Amodio V, Mauri G, Reilly NM, Sartore-Bianchi A, Siena S, Bardelli A, et al. Mechanisms of immune escape and resistance to checkpoint inhibitor therapies in mismatch repair deficient metastatic colorectal cancers. Cancers (Basel). 2021;13:2638. doi: 10.3390/cancers13112638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Parikh AR, He Y, Hong TS, Corcoran RB, Clark JW, Ryan DP, et al. Analysis of DNA damage response gene alterations and tumor mutational burden across 17,486 tubular gastrointestinal carcinomas: implications for therapy. Oncologist. 2019;24:1340–7. doi: 10.1634/theoncologist.2019-0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rousseau B, Foote MB, Maron SB, Diplas BH, Lu S, Argilés G, et al. The spectrum of benefit from checkpoint blockade in hypermutated tumors. N Engl J Med. 2021;384:1168–70. doi: 10.1056/NEJMc2031965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Schrock AB, Ouyang C, Sandhu J, Sokol E, Jin D, Ross JS, et al. Tumor mutational burden is predictive of response to immune checkpoint inhibitors in MSI-high metastatic colorectal cancer. Ann Oncol. 2019;30:1096–103. doi: 10.1093/annonc/mdz134. [DOI] [PubMed] [Google Scholar]
- 103.Gandara DR, Paul SM, Kowanetz M, Schleifman E, Zou W, Li Y, et al. Blood-based tumor mutational burden as a predictor of clinical benefit in non-small-cell lung cancer patients treated with atezolizumab. Nat Med. 2018;24:1441–8. doi: 10.1038/s41591-018-0134-3. [DOI] [PubMed] [Google Scholar]
- 104.Pich O, Muiños F, Lolkema MP, Steeghs N, Gonzalez-Perez A, Lopez-Bigas N. The mutational footprints of cancer therapies. Nat Genet. 2019;51:1732–40. doi: 10.1038/s41588-019-0525-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Stenzinger A, Allen JD, Maas J, Stewart MD, Merino DM, Wempe MM, et al. Tumor mutational burden standardization initiatives: recommendations for consistent tumor mutational burden assessment in clinical samples to guide immunotherapy treatment decisions. Genes Chromosomes Cancer. 2019;58:578–88. doi: 10.1002/gcc.22733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.He W-Z, Hu W-M, Wang F, Rong Y-M, Yang L, Xie Q-K, et al. Comparison of mismatch repair status between primary and matched metastatic sites in patients with colorectal cancer. J Natl Compr Canc Netw. 2019;17:1174–83. doi: 10.6004/jnccn.2019.7308. [DOI] [PubMed] [Google Scholar]
- 107.Rodriguez-Casanova A, Costa-Fraga N, Bao-Caamano A, López-López R, Muinelo-Romay L, Diaz-Lagares A. Epigenetic landscape of liquid biopsy in colorectal cancer. Front Cell Dev Biol. 2021;9:37. doi: 10.3389/fcell.2021.622459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Woolston A, Barber LJ, Griffiths B, Pich O, Lopez-Bigas N, Matthews N, et al. Mutational signatures impact the evolution of anti-EGFR antibody resistance in colorectal cancer. Nat Ecol Evol. 2021;5:1024–32. doi: 10.1038/s41559-021-01470-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Downing A, Morris EJ, Corrigan N, Sebag-Montefiore D, Finan PJ, Thomas JD, et al. High hospital research participation and improved colorectal cancer survival outcomes: a population-based study. Gut. 2017;66:89–96. doi: 10.1136/gutjnl-2015-311308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ignatiadis M, Sledge GW, Jeffrey SS. Liquid biopsy enters the clinic—implementation issues and future challenges. Nat Rev Clin Oncol. 2021;18:297–312. doi: 10.1038/s41571-020-00457-x. [DOI] [PubMed] [Google Scholar]
- 111.Vacante M, Ciuni R, Basile F, Biondi A. The liquid biopsy in the management of colorectal cancer: an overview. Biomedicines. 2020;8:308. doi: 10.3390/biomedicines8090308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Siravegna G, Marsoni S, Siena S, Bardelli A. Integrating liquid biopsies into the management of cancer. Nat Rev Clin Oncol. 2017;14:531–48. doi: 10.1038/nrclinonc.2017.14. [DOI] [PubMed] [Google Scholar]
- 113.Antoniotti C, Pietrantonio F, Corallo S, De Braud F, Falcone A, Cremolini C. Circulating tumor DNA analysis in colorectal cancer: from dream to reality. JCO Precis Oncol. 2019;3:1–14. [DOI] [PubMed]
- 114.Vitiello PP, De Falco V, Giunta EF, Ciardiello D, Cardone C, Vitale P, et al. Clinical practice use of liquid biopsy to identify RAS/BRAF mutations in patients with metastatic colorectal cancer (mCRC): a single institution experience. Cancers (Basel). 2019;11:1504. [DOI] [PMC free article] [PubMed]
- 115.Wan JCM, Heider K, Gale D, Murphy S, Fisher E, Mouliere F, et al. ctDNA monitoring using patient-specific sequencing and integration of variant reads. Sci Transl Med. 2020;12:eaaz8084. doi: 10.1126/scitranslmed.aaz8084. [DOI] [PubMed] [Google Scholar]
- 116.Massihnia D, Pizzutilo EG, Amatu A, Tosi F, Ghezzi S, Bencardino K, et al. Liquid biopsy for rectal cancer: a systematic review. Cancer Treat Rev. 2019;79:101893. doi: 10.1016/j.ctrv.2019.101893. [DOI] [PubMed] [Google Scholar]
- 117.Sefrioui D, Beaussire L, Gillibert A, Blanchard F, Toure E, Bazille C, et al. CEA, CA19-9, circulating DNA and circulating tumour cell kinetics in patients treated for metastatic colorectal cancer (mCRC) Br J Cancer. 2021;125:725–33. doi: 10.1038/s41416-021-01431-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ou S-HI, Nagasaka M, Zhu VW. Liquid biopsy to identify actionable genomic alterations. Am Soc Clin Oncol Educ Book. 2018;38:978–97. doi: 10.1200/EDBK_199765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bando H, Nakamura Y, Taniguchi H, Shiozawa M, Yasui H, Esaki T, et al. Impact of a metastatic site on circulating tumor DNA (ctDNA) analysis in patients (pts) with metastatic colorectal cancer (mCRC) J Clin Oncol. 2021;39:3554. doi: 10.1200/JCO.2021.39.15_suppl.3554. [DOI] [Google Scholar]
- 120.Grasselli J, Elez E, Caratù G, Matito J, Santos C, Macarulla T, et al. Concordance of blood- and tumor-based detection of RAS mutations to guide anti-EGFR therapy in metastatic colorectal cancer. Ann Oncol. 2017;28:1294–301. doi: 10.1093/annonc/mdx112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Vidal J, Muinelo L, Dalmases A, Jones F, Edelstein D, Iglesias M, et al. Plasma ctDNA RAS mutation analysis for the diagnosis and treatment monitoring of metastatic colorectal cancer patients. Ann Oncol. 2017;28:1325–32. doi: 10.1093/annonc/mdx125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bachet JB, Bouché O, Taieb J, Dubreuil O, Garcia ML, Meurisse A, et al. RAS mutation analysis in circulating tumor DNA from patients with metastatic colorectal cancer: the AGEO RASANC prospective multicenter study. Ann Oncol. 2018;29:1211–9. doi: 10.1093/annonc/mdy061. [DOI] [PubMed] [Google Scholar]
- 123.Méhes G. Liquid biopsy for predictive mutational profiling of solid cancer: the pathologist’s perspective. J Biotechnol. 2019;297:66–70. doi: 10.1016/j.jbiotec.2019.04.002. [DOI] [PubMed] [Google Scholar]
- 124.Chan HT, Chin YM, Nakamura Y, Low S-K. Clonal Hematopoiesis in Liquid Biopsy: From Biological Noise to Valuable Clinical Implications. Cancers. 2020;12:2277. doi: 10.3390/cancers12082277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Jaiswal S, Fontanillas P, Flannick J, Manning A, Grauman PV, Mar BG, et al. Age-Related Clonal Hematopoiesis Associated with Adverse Outcomes [Internet]. Massachusetts Medical Society; 2014. http://www.nejm.org/doi/10.1056/NEJMoa1408617 [DOI] [PMC free article] [PubMed]
- 126.Ptashkin RN, Mandelker DL, Coombs CC, Bolton K, Yelskaya Z, Hyman DM, et al. Prevalence of clonal hematopoiesis mutations in tumor-only clinical genomic profiling of solid tumors. JAMA Oncol. 2018;4:1589–93. doi: 10.1001/jamaoncol.2018.2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Huang F, Yang Y, Chen X, Jiang H, Wang H, Shen M, et al. Chemotherapy-associated clonal hematopoiesis mutations should be taken seriously in plasma cell-free DNA KRAS/NRAS/BRAF genotyping for metastatic colorectal cancer. Clin Biochem. 2021;92:46–53. doi: 10.1016/j.clinbiochem.2021.03.005. [DOI] [PubMed] [Google Scholar]
- 128.Chan HT, Nagayama S, Chin YM, Otaki M, Hayashi R, Kiyotani K, et al. Clinical significance of clonal hematopoiesis in the interpretation of blood liquid biopsy. Mol Oncol. 2020;14:1719–30. doi: 10.1002/1878-0261.12727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Remon J, Caramella C, Jovelet C, Lacroix L, Lawson A, Smalley S, et al. Osimertinib benefit in EGFR-mutant NSCLC patients with T790M-mutation detected by circulating tumour DNA. Ann Oncol. 2017;28:784–90. doi: 10.1093/annonc/mdx017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.