Abstract
Background
Serum starvation and hypoxia (SSH) mimics a stress condition in tumours. We have shown that intercellular adhesion molecule-1 (ICAM-1) protein is synergistically expressed in ovarian clear cell carcinoma (CCC) cells under SSH in response to an insufficient supply of fatty acids (FAs). This ICAM-1 expression is responsible for resistance against the lethal condition, thereby promoting tumour growth. However, the underlying mechanisms that link SSH-driven ICAM1 gene expression to impaired FA supply and its clinical relevance are unclear.
Methods
The underlying mechanisms of how FA deficiency induces ICAM-1 expression in cooperation with hypoxia were analysed in vitro and in vivo. Clinical significance of CCC cell-derived ICAM-1 and the mechanism associated with the transcriptional synergism were also investigated.
Results
ICAM-1 expression was mediated through lipophagy-driven lipid droplet degradation, followed by impaired FA-lipid droplet flow. Lipophagy induced ICAM1 expression through stabilisation of NFκB binding to the promoter region via Sam68 and hTERT. Analyses of clinical specimens revealed that expression of ICAM-1 and LC3B, an autophagy marker associated with lipophagy, significantly correlated with poor prognoses of CCC.
Conclusions
The lipophagy-ICAM-1 pathway induced under a tumour-like stress conditions contributes to CCC progression and is a potential therapeutic target for this aggressive cancer type.
Subject terms: Cancer metabolism, Cancer microenvironment
Background
Epithelial ovarian cancer (EOC) is one of the most lethal gynaecological disorders associated with heterogeneous histological subtypes [1]. The clear cell carcinoma (CCC) subtype is relatively frequent in Asian countries, which includes Japan, and some European countries [2]. Conversely, the serous carcinoma (SC) subtype is predominant in most Western countries [1]. CCC is highly aggressive and recurrent because it exhibits resistance against chemotherapy, which leads to a poorer prognosis than that of SC [1, 2].
Hypoxia is a common tissue condition associated with poor blood supply to tumour tissues that include ovarian cancer tissue [3]. Tumour cells are starved for molecular oxygen and other serum components, which include nutrients, growth factors, and hormones, owing to aberrant vasculature and aggressive tumour growth [3]. Cancer cells can adapt to such severe conditions for tumour progression. This involves activation of transcription mechanisms that include heterodimeric complex formation between hypoxia-inducible factor (HIF)-1α and -2α, and arylhydrocarbon receptor nuclear translocator (ARNT), followed by association with hypoxia response elements of target genes [4]. Furthermore, recent studies have shown that lipid starvation and hypoxia are lethal and adaptation to this condition is important for progression of glioblastoma [5] and clear cell renal cell carcinoma [6].
We previously reported that HIF-2α complexes with the transcription factor Sp1, thereby activating multiple target genes that include FVII and ICAM1 in CCC cells [7–9], which suggests ARNT-independent transcriptional regulation via HIFs. We further demonstrated that this Sp1-dependent transcriptional mechanism is synergistically enhanced when cells are simultaneously exposed to serum starvation and hypoxia (SSH) [7–9]. Long-chain fatty acid (LCFA) deficiency [8] and cholesterol starvation coupled with SREBP-1 activation [9] are responsible for the synergism of transcriptional activation for ICAM1 and FVII, respectively. In the case of ICAM1 expression, activation of transcription factor NFκB and mTOR and TNF-α signalling pathways contributes to the synergism in association with a decrease in the cellular lipid droplet (LD) level [8]. However, the underlying mechanisms by which LCFA deficiency causes this characteristic synergism of transcriptional activation are unclear.
ICAM1 encodes intercellular adhesion molecule-1 (ICAM-1). ICAM-1 is a transmembrane protein that belongs to the immunoglobulin superfamily. ICAM-1 is expressed in endothelial and immune cells, and contributes to tissue infiltration of leucocytes under physiological conditions [3]. Additionally, ICAM-1 is highly expressed in various cancer cell types and augments malignancy by enhancing metastatic potential [10–12], tissue vascularisation [13], and interactions with immune cells [13, 14]. Reports on the roles of ICAM-1 in EOC progression have revealed a complex overview [3]. Previous studies have demonstrated that expression of ICAM-1 suppresses the malignancy of EOC cells [15–18]. Conversely, other reports have suggested the opposite trend [19–22]. These results are based on experiments using non-CCC EOC cells. Thus, published information regarding the effect of ICAM-1 on CCC progression has been scarce. We have demonstrated that SSH is lethal for CCC cells with a reduced LD level. ICAM-1 confers resistance against SSH-driven apoptosis and facilitates CCC-tumour progression [8], although detailed mechanisms are currently unclear.
On the basis of this background information, in this study, we aimed to uncover the clinical importance of CCC cell-derived ICAM-1. We further investigated the details of how an insufficient lipid supply causes synergistic ICAM1 gene expression in cooperation with hypoxia.
Methods
Cell lines
CCC cell lines (OVSAYO and OVISE) and SC cell lines (OVSAHO and OVKATE) were cultured as described previously [8, 23]. TOV21G was purchased from the American Type Culture Collection (Manassas, VA, USA). For cell authentication, STR profiling was performed (Takara Bio, Shiga, Japan). To confirm cell line identity, STR profiles were examined by the similarity search (CLASTR) programme in a public database (Cellosaurus; https://expasy.org/cellosaurus/). We tested for mycoplasma contamination using an e-Myco™ mycoplasma PCR detection kit (iNtRON Biotechnology, Gyeonggi-do, Korea) and confirmed that the cell lines used in this study were free of mycoplasma.
Cell culture
Cells were routinely cultured in RPMI-1640 medium with 10% foetal calf serum. For hypoxic cell culture, cells were cultivated under 1% O2 using a multi-gas incubator (BL-43MD, TOSC, Tokyo, Japan). Serum-starved cell culture was performed as described previously [9]. Briefly, routinely cultured cells were washed once with serum-free medium and then cultured in the same medium.
Reagents
Fatty acid supplement (a mixture of unesterified saturated and unsaturated fatty acids not involving cholesterol or albumin; information from product website and technical support) (F-7050, Sigma-Aldrich, St. Louis, MO, USA), oleic acid (O1008, Sigma-Aldrich), palmitic acid (P0500, Sigma-Aldrich). Albumin from human serum (A3782, Sigma-Aldrich), LDL human (OPPA01439-10MG, Aviva Systems Biology, San Diego, CA, USA), water-soluble cholesterol reagent (C4951, Sigma-Aldrich), methyl-β-cyclodextrin (C4555, Sigma-Aldrich), N-acetyl-L-cysteine (NAC) (A7250, Sigma-Aldrich), and Bafilomycin A1 (B1793, Sigma-Aldrich).
siRNA transfection
siRNA reagents used in this study (See Supplementary materials) were a pool of 2–4 target sequences for each gene designed to efficiently and specifically block target mRNA expression. Transfection of siRNAs was performed using Lipofectamine RNAi MAX (Life Technologies) for OVSAYO and TOV21G cells. Transfection of OVISE cells was performed by electroporation using a Neon Transfection System (Thermo Fisher Scientific, Waltham, MA, USA). All siRNAs were used at a final concentration of 15 nM.
Quantitative RT-PCR analysis
We determined mRNA levels by real-time RT-PCR with hybridisation probes as described previously [7, 8]. All data were normalised to 18S ribosomal RNA levels.
Isolation of nuclear and cytoplasmic fractions
Nuclear and cytoplasmic fractions used for immunoblotting were prepared using a Nuclear Extract kit (Active Motif, Carlsbad, CA, USA) in accordance with the manufacturer’s protocol.
Protein quantification
Protein levels were quantified using a Micro BCA™ Protein Assay Kit (Thermo Fisher Scientific) in accordance with the manufacturer’s instructions.
Western blot analysis
Western blotting was performed as described previously [8]. In generally, whole-cell lysates were used for detection. To detect mTOR, immunoprecipitated mTOR from cell lysates prepared using RIPA buffer were used for better visibility. See Supplementary materials for primary antibodies.
Cell counting
Cells were counted by trypan blue exclusion using a Countess™ automated cell counter (Invitrogen, Waltham, MA, USA).
Immunohistochemistry (IHC) of a tissue microarray
Routinely processed formalin-fixed, paraffin-embedded specimens from 38 CCC and 50 SC patients diagnosed at Kanagawa Cancer Center Hospital (KCCH) from 2005 to 2010 were prepared for tissue microarray (TMA) production (Table S1). Additionally, whole tissue sections (WTSs) were prepared from 45 CCC patients diagnosed at KCCH from 2006 to 2017, which were not applied to the TMA (Table S2). Written consent was obtained from all patients. The study was approved by our institutional review board (approval No. 177). All tissues were reviewed and histological subtypes were diagnosed by pathologists at KCCH in accordance with the World Health Organization Classification of Tumours, Female Genital Tumours, 2020. TMA and WTS were stained with antibodies against ICAM-1 (sc-8439, Santa Cruz Biotechnology, Dallas, TX, USA; 2 µg/mL), LC3B (E7X4S, #43566, Cell Signaling Technology, 0.1 µg/mL), and LC3A (A1805a, abcepta, San Diego, CA, USA, 10 μg/ml). Immunoreactivity was visualised by the peroxidase-labelled amino acid polymer method using Histofine simple stain MAX-PO® (Nichirei Co., Tokyo, Japan) and the avidin-biotin-peroxidase complex method (LSAB+; DakoCytomation Co., Tokyo, Japan) in accordance with the manufacturers’ instructions. Sections were counterstained with hematoxylin and eosin (HE).
IHC image acquisition, scoring, and survival analysis
Images (×20 objective lens) of tumour tissues stained by IHC were acquired under a BM53 microscope (Olympus, Tokyo, Japan). Three images with a representative staining pattern were selected and quantified using ImageJ software (http://rsb.info.nih.gov/ij/). Briefly, the stromal region in images was selected and discarded using the freehand selections tool in this software. Then, the stained tumour cell area was quantified. Expression scores were obtained by normalising the stained area to the total examined tumour cell area. Mean scores of the images were used for survival analysis. Kaplan–Meier analysis of the relationship between the expression scores and overall survival (OS) periods of patients and multivariable analysis by the Cox regression method were performed using SPSS statistics 19 software (IBM, Chicago, IL, USA).
Fluorescence microscopy of BODIPY and Filipin III staining
Cellular LDs and free cholesterols were stained using BODIPY™ (D3922, Invitrogen, Eugene, OR, USA) and Filipin III (sc-205323A, Santa Cruz Biotechnology), respectively. Images were acquired under a BZ-9000 fluorescence microscope (Keyence, Osaka, Japan). Quantitative analysis of images, which included the fluorescence area and number of nuclei, was performed using BZ-Analyzer software (Keyence) and ImageJ software.
Immunofluorescence microscopy
For immunocytochemistry of LC3A/B, cells were seeded in a four-well chamber polystyrene vessel (#354114, Corning, Big Flats, NY, USA) and then cultured under the indicated conditions. Cells were fixed with methanol in accordance with the protocol for the primary antibody [#12741, LC3A/B (D3U4C) XP rabbit mAb]. In the case of xenografted tumour tissues, samples were briefly washed and fixed with 4% paraformaldehyde in PBS for 10 min. Fixed cells were washed twice with PBS and permeabilized with 0.1% Triton X-100 (Sigma). Cells were incubated with primary antibodies. After washing the cells with PBS, they were incubated for 1 h with anti-mouse (A11029 and A11031, Molecular Probes, Eugene, OR, USA) and anti-rabbit (A11008, Molecular Probes and DI-1794, Vector Laboratories, Burlingame, CA, USA) secondary antibodies conjugated with Alexa Fluor 488, 568, or 594 diluted with blocking reagent. Labelled cells were washed and mounted using VECTASHIELD HardSet™ Mounting Medium with DAPI (Vector Laboratories). All steps were carried out at ambient temperature. Cellular localisation of target proteins was visualized under the BZ-9000 fluorescence microscope. Primary antibodies against ICAM-1, pimonidazole-adducts (hypoxyprobe) have been described previously [8]. The antibody against LC3B was the same as that in IHC.
Chromatin immunoprecipitation analysis
Chromatin immunoprecipitation (ChIP) analyses of the ICAM1 promoter region were performed as described previously [8, 9]. See Supplementary materials for antibodies used.
Xenografted tumour establishment
The Institutional Review Board at Kanagawa Cancer Center Research Institute approved this study. OVISE cells were implanted as described previously [8]. Briefly, cells were subcutaneously injected into two 6-week-old female NOD-SCID mice. Two representative tumors isolated from different mice were used for ChIP as described previously [9]. Alternatively, tumours fixed using neutral formalin [8] were used for histochemical analysis.
Detection of reactive oxygen species
Detection of reactive oxygen species (ROS) was performed using a CellROX green flow cytometry assay kit (C10492, Life Technologies) in accordance with the manufacturer’s protocol.
Statistics
Statistical significance was evaluated for two data sets using SPSS statistics 19 software. Parametric (Student’s and Welch’s (no assumption of equal variance between data sets) t-tests) methods were used. Non-parametric method (Mann–Whitney U-test) was also used based on non-normal distribution determined with the Shapiro–Wilk test. p < 0.05 was considered statistically significant.
Results
ICAM-1 is associated with poor prognosis of CCC patients
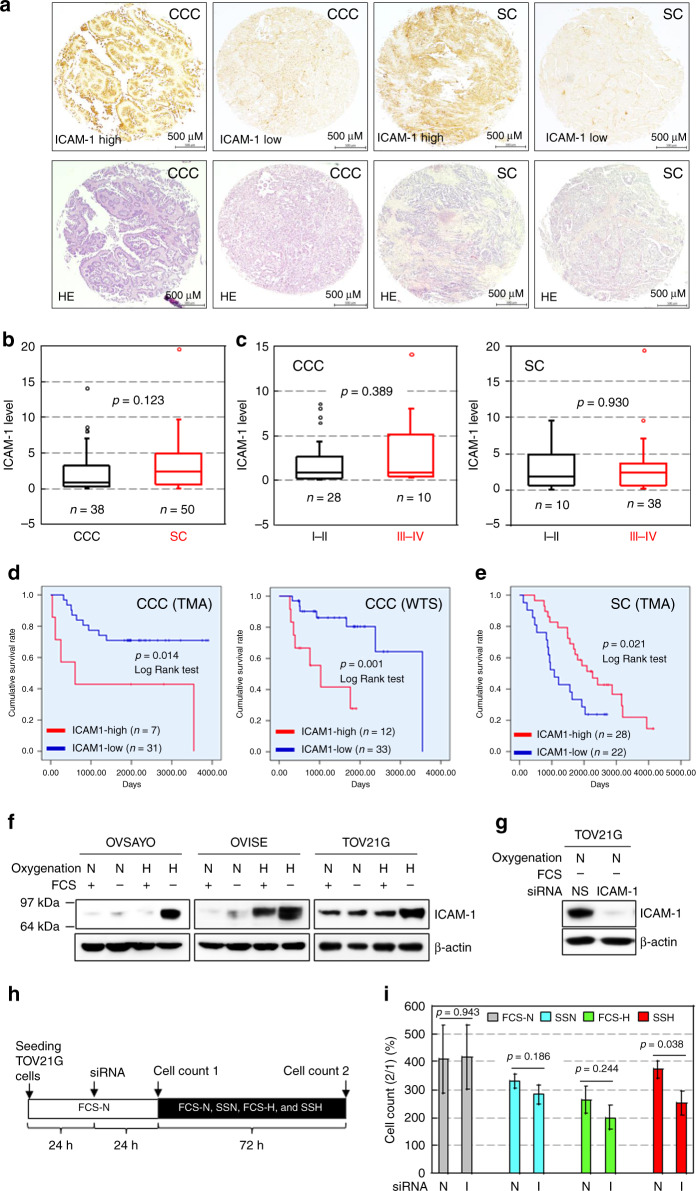
Previous studies have shown that ICAM-1 expression in non-CCC EOC contributes to suppression of malignant phenotypes [15–18]. This is consistent with information in a public database, which shows that ICAM-1 transcript levels in tumour cells of 373 patients based on The Cancer Genome Atlas RNA-seq data significantly correlate with favourable prognoses of SC patients (see the website: Human Protein Atlas; https://www.proteinatlas.org). Thus, we examined the expression levels of ICAM-1 in CCC tissues surgically removed from 38 Japanese patients and compared them with those of SC tissues from 50 Japanese patients (Table S1). IHC of ICAM-1 using a TMA (Fig. 1a), followed by scoring of expression levels revealed that ICAM-1 was expressed similarly in CCC and SC tissues (Figs. 1a, b, and S1). Additionally, ICAM-1 levels were not different between disease (FIGO) stages for both histological subtypes (Fig. 1c). Kaplan–Meier analysis revealed that a high ICAM-1 level correlated significantly with a poor OS rate of CCC patients (Fig. 1d, left). Additional IHC using WTSs with a different diagnosis period (Table S2), followed by Kaplan–Meier analysis revealed the same trend (Fig. 1d, right). Multivariable analysis showed that ICAM-1 was a prognostic factor for CCC with a risk ratio of 4.183 (TMA) and 4.653 (WTS) (Table 1). Conversely, a high ICAM-1 level in SC tissues was significantly related to a better OS rate (Fig. 1e), which was consistent with published data showing that ICAM-1 transcript levels are associated with better prognoses of EOC patients.
Fig. 1. ICAM-1 level in cancer cells is associated with poor prognoses of CCC patients.
a Staining pattern of ICAM-1 in EOC tissues evaluated by IHC. H&E staining is also shown for comparison. b Comparison of ICAM-1 levels between CCC and SC tissues. c Comparison of ICAM-1 levels between FIGO stages in EOC tissues. d,e Kaplan–Meier analysis of the correlation between the ICAM-1 level and OS of CCC (d) and SC (e) patients. f Western blot analysis of ICAM-1 expression in the indicated cells exposed to the indicated culture conditions for 48 h. FCS: 10% foetal calf serum, N and H indicate normoxia (ambient air) and hypoxia (1% O2), respectively. β-Actin was examined as a protein loading control. g Example of ICAM-1 knockdown (KD) in TOV21G cells. Cells were cultured under the SSN condition for 48 h after the indicated siRNA transfection. NS: non-specific. h Scheme of the cell growth assay. The effect of ICAM-1-KD on proliferation of TOV21G cells was examined under various culture conditions. i Viability of TOV21G cells under the indicated culture conditions. Data are the mean (N = 3) ± SD. Statistical significance was evaluated by the two-sided t-test. N non-specific, I ICAM-1.
Table 1.
Multivariable (with ICAM-1) analysis of OS rate of CCC patients.
| TMA | WTSs | |||||
|---|---|---|---|---|---|---|
| Variable | HR | 95% CI | p value | HR | 95% CI | p value |
| ICAM-1 | 4.183 | 1.075–16.279 | 0.039 | 4.653 | 1.369–15.816 | 0.014 |
| Age (year) | 0.934 | 0.869–1.005 | 0.068 | 1.015 | 0.956–1.078 | 0.619 |
| Disease stage (FIGO I, II vs III, IV) | 10.824 | 3.028–38.691 | <0.001 | 2.367 | 0.748–7.485 | 0.142 |
HR hazard ratio, CI confidence interval.
We previously reported that ICAM-1 expression in CCC cell lines, OVSAYO and OVISE is very low under normoxia with serum (FCS-N), but dramatically increases in response to serum starvation and hypoxia (SSH) (H with FCS—in Fig. 1f), thereby increasing cell viability under this harsh condition [8]. In this study, we found that ICAM-1 was expressed in an additional CCC cell line, TOV21G, even under the FCS-N condition (Fig. 1f) and the expression was highest under SSH similarly to OVSAYO and OVISE cell lines (Fig. 1f). ICAM-1 knockdown (KD) (Fig. 1g), followed by elucidation of cell viability under culture conditions, which included serum starvation and normoxia (SSN) and hypoxia with serum (FCS-H) (Fig. 1h), revealed that the number of live cells was significantly decreased by ICAM-1-KD only when the cells were cultured under SSH (Fig. 1i). We also examined ICAM-1 expression in two additional SC cell lines [23]. We found that OVKATE, (Fig. S2a) but not OVSAHO (Fig. S2b), cells expressed ICAM-1. SSH-induced ICAM-1 expression was not observed in both cell lines (Fig. S2a, b). Furthermore, ICAM-1-KD (Fig. S2c), followed by the cell viability assay as shown in Fig. 1h revealed that ICAM-1 tended to suppress (p < 0.1) growth of OVKATE cells under SSH (Fig. S2d). Thus, ICAM-1 conferred a survival advantage specifically to multiple CCC cell lines exposed to SSH, which may contribute to a poor survival rate of CCC patients.
Loss of fatty acid-LD flow under SSH induces ICAM-1 expression
Our previous study showed that fatty acid (FA) uptake, followed by LD biogenesis is important to alleviate SSH stress [8]. Similarly, a recent study revealed that an unsaturated FA (oleic acid) derived from neutral LD hydrolysis alleviates saturated FA-driven cytotoxicity in cells exposed to limited serum and O2 supply, because this condition reduces both uptake and unsaturation of FAs [6].
Low-density lipoprotein (LDL) is a major plasma lipid and source of LD biogenesis because it supplies multiple esterified FAs and cholesterol [24, 25]. Cholesterol facilitates progression of cancer via multiple mechanisms [26–28] and its deficiency, followed by SREBP-1 activation, can be critical for progression of glioblastoma under hypoxia [5]. However, the effect of FA saturation and cholesterol on CCC cell survival under SSH is currently unclear. Thus, we next examined this issue to better understand the relationship between cell viability, ICAM-1 expression, the LD level, and LD components.
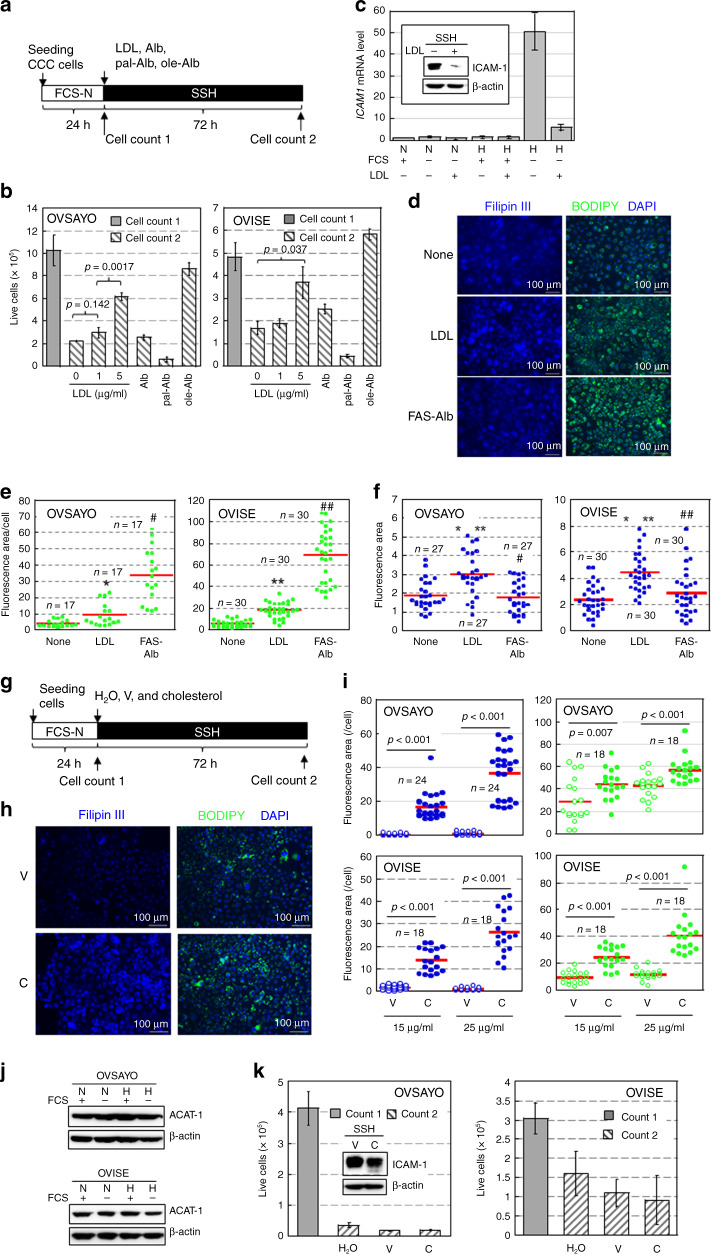
We examined the effect of LDL treatment on the viability of OVSAYO and OVISE cells cultured under SSH for 72 h (Fig. 2a). We found that LDL rescued the impaired cell viability (cell count 2) compared with the viability of initial live cells (cell count 1) (Fig. 2a, b) similarly to oleic acid-Alb (fatty acid-free albumin) (Fig. 2b) and 0.3% fatty acid supplement (FAS, a mixture of unsaturated and saturated FAs, see “Methods”)-Alb (Fig. S3a) treatments because these treatments cancelled the SSH condition. However, saturated palmitic acid-Alb treatment impaired cell viability (Fig. 2b).
Fig. 2. Impairment of FA-LD flow is associated with synergistic ICAM-1 expression and decreased cell viability under SSH.
a Scheme of the assay. Effect of LDL (0, 1, and 5 μg/ml), Alb (44 μM [8]) alone, and palmitic acid (pal)- and oleic acid (ole)-Alb complexes (50 μg/ml) on cell viability, LD levels, and cholesterol levels were examined. b Results of a. Data are the mean (N = 3) ± SD. Statistical significance was evaluated by the two-sided t-test. c LDL (5 μg/ml) suppresses ICAM-1 mRNA expression in OVSAYO cells exposed to SSH for 16 h (N = 3) ± SD. Inset: western blot analysis of ICAM-1 expression in OVSAYO cells exposed to SSH for 48 h. Symbols are described in Fig. 1. d Effect of LDL on cholesterol (Filipin III) and LD (BODIPY) levels in OVSAYO cells cultured under SSH for 16 h. Cellular lipids were stained with the indicated dyes and detected by fluorescence microscopy. Nuclei were counterstained with DAPI. e LD levels were quantified by ImageJ software. The stained area was evaluated for the indicated number of images acquired from three independent replicates and normalised to the number of cells (nucleus) in each image. Statistical significance was evaluated by the Mann–Whitney U-test. *p = 0.005 (compared with none), #p < 0.001 (compared with LDL), **p < 0.001 (compared with none), and ##p < 0.001 (compared with LDL). f Cholesterol levels were quantified as described in (e). Statistical significance was evaluated by the Mann–Whitney U-test. *p < 0.001 (compared with none), **p < 0.001 (compared with FAS-Alb), and #p = 0.514 (compared with none), ##p = 0.213 (compared with none). The stained area was evaluated for the indicated number of images acquired from three independent replicates. g Scheme of the cell growth assay. The effects of cholesterol on cellular LD, cholesterol, and cell viability were examined. h Image of cholesterol and LD in OVSAYO cells treated with a cholesterol supplement under SSH for 16 h. C: 25 μg/ml cholesterol; V: corresponding amount of vehicle (methyl-β-cyclodextrin). i Cholesterol (blue) and LD (green) levels were quantified as described in e and f. C cholesterol, V vehicle (methyl-β-cyclodextrin). The stained area was evaluated for the indicated number of images acquired from three independent replicates. j Western blot analysis of ACAT-1 in CCC cells cultured under the indicated conditions for 48 h. Symbols are described in Fig. 1f. k Effect of cholesterol treatment on viability of CCC cells cultured as described in (g). Data are the mean (N = 3) ± SD. C: 25 μg/ml cholesterol; V: corresponding amount of vehicle (methyl-β-cyclodextrin). Inset: effect of cholesterol treatment on ICAM-1 expression. Western blotting was performed using cells exposed to SSH for 48 h.
Real-time RT-PCR and western blot analyses showed that ICAM1 expression in OVSAYO cells was largely decreased (approximately one-tenth) by treatments with 5 μg/ml LDL (Fig. 2c) and 0.3% FAS-Alb (Fig. S3b).
We next examined effect of LDL treatment on cellular LD and cholesterol levels. BODIPY staining of OVSAYO and OVISE cells (Fig. 2d and S3c) treated with 5 μg/ml LDL revealed that the LD level was increased by LDL treatment (Fig. 2d, e). However, this was significantly lower than that induced by FAS-Alb treatment (Fig. 2d, e), although both treatments similarly suppressed ICAM-1 mRNA expression (Fig. 2c and S3b) and improved cell viability (Fig. 2b and S3a). Filipin III staining revealed that the cellular free cholesterol level was significantly increased only in LDL-treated cells (Fig. 2f) with a plasma membrane staining pattern (Fig. S3d). This indicated that FAS-Alb treatment efficiently increased the LD level and cell viability without a cholesterol supply.
We further examined the effect of cholesterol treatment on the LD level and viability of CCC cells cultured under SSH (Fig. 2g). OVSAYO and OVISE cells were cultured under SSH with 15 and 25 μg/ml cholesterol (cholesterol supplement, see “Methods”). LD levels in CCC cells were increased by cholesterol treatment (Fig. 2h, i). However, they were quite low considering the robust increase of free cholesterol levels compared with negative controls (Fig. 2h, i). It is unlikely that this low cholesterol-LD generation was due to low esterification potency because acyl-CoA cholesterol acyltransferase (ACAT-1) [26], which is responsible for cellular cholesterol esterification, followed by LD biogenesis were abundant in CCC cells (Fig. 2j). We found that cholesterol treatment did not rescue the impaired viability of cells under SSH with sustained ICAM-1 expression (Fig. 2k). Considering that LDL treatment improved cell viability with a similar LD level as cholesterol treatment (Fig. 2e, i), LDL alleviated SSH stress through unsaturated FA-mediated LD biogenesis and the effect of cholesterol was dispensable. Collectively, our data showed that impairment of FA-LD flow, which involved incorporation of oleic acid, was critical for SSH-driven cell death and concomitant ICAM-1 expression.
Lipophagy contributes to SSH-driven expression of ICAM-1 in CCC cells
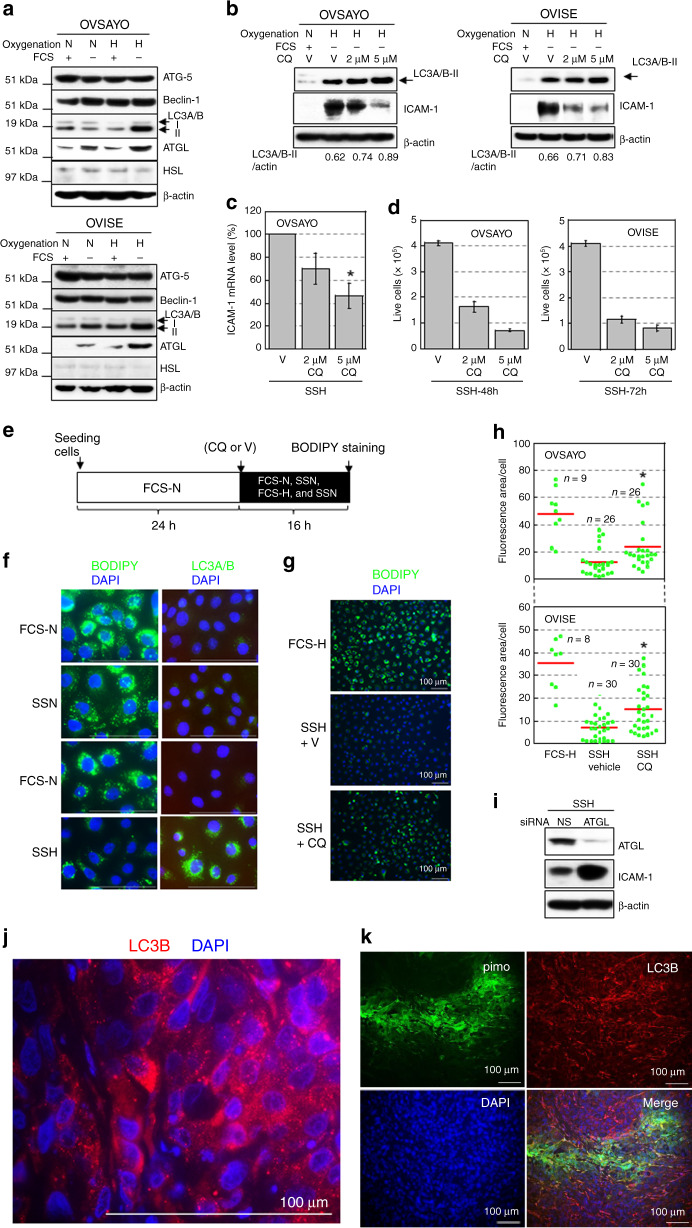
Our data demonstrated that unsaturated FA uptake, followed by LD biosynthesis are important for survival of CCC cells. However, this pathway was impaired in response to SSH with ICAM-1 expression. Thus, we next investigated the mechanisms of these events. In our metabolomics analysis [9], the total amino acid level in OVSAYO cells cultured under SSH was increased compared with that in cells cultured under FCS-H (Fig. S4), which suggested activation of a cellular protein degradation process. Autophagy was a candidate mechanism because it promotes cancer progression (Autophagy to Disease; http://auto2disease.nwsuaflmz.com). Thus, we next assessed whether autophagy contributed to SSH-driven ICAM1 expression.
Western blot analysis showed that autophagy executor proteins ATG-5 and beclin-1 (ATG-6) [29, 30] were expressed equally under all four culture conditions in OVSAYO and OVISE cells (Fig. 3a). However, LC3A/B-II levels were increased when OVSAYO and OVISE cells were cultured under SSH (Fig. 3a), which suggested accumulation of autophagosomes [31] in CCC cells under this condition. LC3A/B-II levels were further increased by treatment with chloroquine (CQ) that inhibits fusion between autophagosomes and lysosomes [31, 32] (Fig. 3b). Therefore, we concluded that autophagy was induced in response to SSH. We further found that CQ treatment cancelled SSH-driven ICAM-1 expression at protein (Fig. 3b) and mRNA (Fig. 3c) levels in CCC cells with impairment of cell viability (Fig. 3d). These results suggest that autophagy contributed to resistance against SSH via ICAM-1 expression.
Fig. 3. Lipophagy is responsible for SSH-driven ICAM-1 expression and associated with an LD decrease in CCC cells.
a Western blot analysis of the indicated proteins in CCC cells. Cells were cultured under the indicated conditions for 16 h. N and H are described in Fig. 1f. β-Actin was probed as a protein-loading control. b Effect of CQ on LC3A/B-II and ICAM-1 levels in CCC cells. Cells were cultured under the indicated conditions for 16 h (LC3A/B-II) and 48 h (ICAM-1). V: Vehicle. c Effect of CQ on ICAM-1 mRNA levels in OVSAYO cells exposed to SSH for 16 h. V: Vehicle. Data are the mean (N = 3) ± SD. Statistical significance was evaluated by the two-sided t-test. *p = 0.02 (compared with vehicle). d Effect of CQ on the viability of CCC cells exposed to SSH for the indicated periods. Data are the mean (N = 3) ± SD. e Scheme of the assay. Cellular LD level under the indicated conditions and effect of CQ treatment on LD level were examined. f LD and LC3A/B levels in OVSAYO cells cultured under the indicated conditions for 16 h. Bars: 100 μm. g Effect of CQ on the LD level in OVSAYO cells cultured under SSH. h LD levels in CCC cells were quantified by ImageJ software. The stained area was evaluated for the indicated number of images acquired from three independent replicates and normalised to the number of cells (nucleus) in each image. Statistical significance was evaluated by the Mann–Whitney U-test. *p < 0.001 (compared with SSH vehicle). i Effect of ATGL-KD on expression of ICAM-1 in OVSAYO cells cultured under SSH for 48 h. j, k Immunofluorescence staining of grafted OVISE tumour tissue for the indicated markers. j is a magnified version of Fig. S7.
Accumulating evidence suggests that autophagy contributes to LD degradation via lipophagy [29, 30, 33]. Alternatively, LD can be catabolized through lipolysis by neutral cytosolic lipases such as adipocyte triglyceride lipase (ATGL) [29] and hormone-sensitive lipase (HSL) [6]. We found that the ATGL, but not HSL, level was increased and showed the highest expression under SSH in OVSAYO and OVISE cells (Fig. 3a), which suggested that neutral lipolysis by ATGL is also activated under SSH. Thus, we next examined whether lipophagy was involved in SSH-driven ICAM-1 expression. Cultivation of OVSAYO cells under the indicated conditions for 16 h (Fig. 3e), followed by fluorescence microscopy demonstrated that LC3A/B were obviously increased in cells with a robust decrease in the LD level (Fig. 3f). This diminished LD level in OVSAYO and OVISE cells was restored by CQ treatment prior to SSH culture (Fig. 3g, h).
It has been reported that ATGL activates lipophagy in the liver [34]. Interestingly, western blotting showed that the ICAM-1 level increased in response to SSH was further increased by ATGL-KD in OVSAYO cells (Fig. 3i), presumably because the equilibrium between lipophagy and neutral lipolysis shifted to the former. Thus, lipophagy but not cytosolic lipolysis induced by ATGL, likely mediated SSH-driven ICAM-1 expression in CCC cells.
Our metabolomics analysis [9] showed that a reduced glutathione/oxidised glutathione (GSH/GSSG) ratio was decreased in OVSAYO cells cultured under SSH compared with those cultured under FCS-H (Fig. S5a), which implied that the ROS level was higher under SSH. Thus, we determined whether ROS contributes to SSH-driven ICAM-1 expression. Western blot analysis demonstrated that ICAM-1 induced under SSH was not decreased by NAC treatment (Fig. S5b), although a positive control experiment with a ROS-inducing drug, tert-butyl-hydroperoxide (TBHP), showed that the same concentration of NAC substantially decreased ROS generation in OVSAYO cells (Fig. S5c).
We further addressed whether lipophagy occurred in hypoxic tumour regions in vivo. OVISE cells were used for this purpose because of favourable engraftment efficacy in immunocompromised mice. Mice with grafted OVISE tumours were treated with hypoxia probe pimonidazole and then the hypoxic region, ICAM-1 expression, and LC3B expression in tumour tissues were examined by IHC. As expected, ICAM-1 was strongly expressed in the severe hypoxic tumour area (Fig. S6). We found that autophagosomes stained for LC3B (Fig. 3j and S7), a predominant marker of autophagy [31], were unevenly distributed in tumour tissue (Fig. S7) that included the pimonidazole-positive region (Fig. 3k and S8). Our previous study of frozen tumour sections revealed that the LD level in the blood vessel-poor (hypoxic) grafted tumour area was significantly lower than that in the vessel-rich area [8]. Thus, the hypoxia-lipophagy-driven LD catabolism-ICAM-1 expression axis was likely active in vivo.
Lipophagy contributes to SSH-driven ICAM-1 expression via NFκB binding
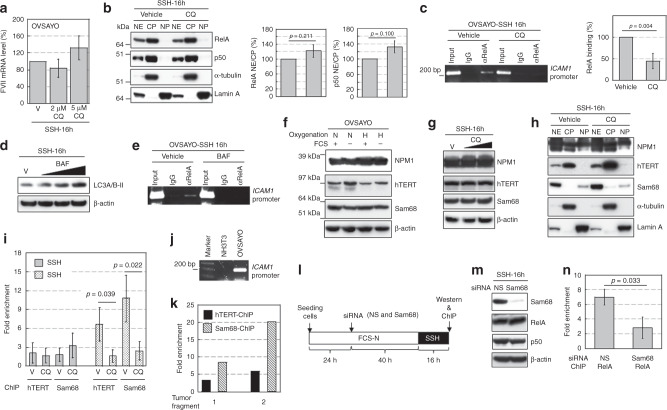
We found that lipophagy and hypoxia collaborated to induce robust ICAM-1 expression in CCC cells at the gene expression level. We next investigated how lipophagy contributed to this synergistic transcriptional activation. Our previous data showed that phosphorylation of RelA, a component of transcription factor NFκB, followed by binding to the ICAM1 promoter is enhanced in response to SSH [8]. This NFκB activation is associated with mTOR and TNF-α pathways [8]. In this study, we found that SSH-driven expression of the FVII gene, which is independent of NFκB [9], was not inhibited by CQ treatment (Fig. 4a). Thus, we surmised that the NFκB pathway was responsible for the lipophagy-mediated ICAM1 expression. To test this hypothesis, we first examined the effect of CQ treatment on SSH-driven phosphorylation of RelA in OVSAYO cells. Western blotting revealed that CQ did not reduce the phospho-RelA level (Fig. S9a). Additionally, expression of proteins associated with NFκB activation (TNF-α) [8], dimerisation (p50) [8, 35], and suppression (GILZ [9] and IκB-α [35]) were not changed by CQ treatment (Fig. S9a, b). Phosphorylation of mTOR was also not altered by CQ treatment (Fig. S9c). Thus, CQ did not interfere with the activation process of NFκB under SSH.
Fig. 4. hTERT and Sam68 contribute to NFκB binding to ICAM1 under SSH.
a Real-time RT-PCR analysis of FVII mRNA expression in cells exposed to CQ under the indicated conditions. b Western blot analysis of RelA and p50 expression in cytoplasmic and nuclear fractions of OVSAYO cells. NE, CP, and NP are the nuclear extract, cytoplasmic extract, and nuclear pellet, respectively. Cytoplasmic (α-tubulin) and nuclear (lamin A) markers are shown. Right panels show quantitation of the NE/CP ratio. Data are the mean (N = 3) ± SD. Statistical significance was evaluated by the two-sided t-test. c Effect of CQ treatment on RelA binding to the ICAM1 promoter region under SSH was analysed by ChIP. Gel electrophoresis (2%) was performed. Right: Binding levels were determined by quantitative PCR. Data are the mean (N = 3) ± SD. Statistical significance was evaluated by the two-sided t-test. d Effect of BAF treatment (5, 15, and 30 nM) on LC3A/B-II levels in OVSAYO cells cultured under the indicated conditions was evaluated by western blotting. e Effect of BAF treatment on binding of RelA to the ICAM1 region in cells cultured under the indicated conditions was analyzed by ChIP. f Western blot analysis of NPM1, hTERT, and Sam68 in cells cultured under the indicated conditions for 48 h. Symbols are described in Fig. 1f. g Effect of CQ treatment (2 and 5 μM) on expression of the indicated proteins in OVSAYO cells cultured under SSH. h Western blot analysis of NPM1, hTERT, and Sam68 in cytoplasmic and nuclear fractions prepared from OVSAYO cells cultured under the indicated conditions as described in b. i ChIP analysis with real-time PCR of hTERT and Sam68 bindings to the ICAM1 promoter region. The binding level is shown as fold enrichment of DNA fragments relative to control immunoprecipitation with normal IgG. Data are the mean (N = 3 for SSN, N = 4 for SSH) ± SD. Statistical significance was evaluated by the two-sided t-test. j PCR primers used for ChIP assay detected the human, but not murine, ICAM1 region as revealed by 2% gel electrophoresis. k Quantitative ChIP assay of grafted OVISE tumours. l Scheme of the Sam68-driven RelA binding analysis. Effect of Sam68 on RelA binding to the ICAM1 region was examined under SSH. m Effect of Sam68-KD on expression of RelA and p50 in OVSAYO cells cultured under SSH. n ChIP analysis revealed the effect of Sam68-KD on RelA binding to the ICAM1 region under SSH. Data are the mean (N = 3) ± SD. Statistical significance was evaluated by the two-sided t-test.
HSC70 is involved in chaperone-mediated autophagy in association with lipophagy [29]. HSC70 also binds to RelA and activates NFκB-targeted genes [36]. Thus, we further investigated whether HSC70 was involved in SSH-driven expression of ICAM-1. HSC70 protein was expressed equally in OVSAYO cells cultured under the four conditions (Fig. S9d). Western blotting showed that the ICAM-1 level under SSH was not reduced by HSC70-KD (Fig. S9e), which suggested that HSC70 was not involved in ICAM1 expression.
We next determined whether CQ interfered with translocation of NFκB from the cytoplasm to nucleus. Western blotting of RelA and its binding partner p50 revealed that their relative expression levels in the nucleus under SSH were not decreased by CQ treatment (Fig. 4b), which suggested that CQ did not inhibit the translocation process.
Some cofactors, such as NPM1 [37], human telomerase reverse transcriptase (hTERT) [38], and Src-associated substrate during mitosis of 68 kDa (Sam68) [39], stabilise NFκB binding to target gene regions to enhance transcription, which indicates that NFκB activity is controlled at its binding level. Moreover, there have been no reports on the roles of these cofactors in ICAM1 expression. Thus, we further examined whether CQ interfered with NFκB binding to the ICAM1 promoter region. Chromatin immunoprecipitation (ChIP) analysis of RelA revealed that the binding of NFκB under SSH was significantly impaired in CQ-treated CCC cells (Fig. 4c and S10a). This finding was also observed for bafilomycin A1, another autophagy inhibitor (Fig. 4d) with a distinct chemical structure (Fig. 4e). Thus, it was unlikely that CQ directly inhibited NFκB binding.
Western blotting of NPM1, hTERT, and Sam68 in whole OVSAYO and OVISE cell lysates revealed that these proteins were expressed under the indicated conditions (Fig. 4f and S10b). The expression levels of these proteins in cells cultured under SSH were not altered by CQ treatment (Fig. 4g and S10c). Additionally, nuclear localisation of these proteins under SSH was not decreased by CQ treatment (Fig. 4h), which suggested that CQ did not affect both the expression or nuclear localisation of these proteins at least within the tested treatment period.
Binding of the NFκB complex to target genes is enhanced by direct association to hTERT and Sam68 [38, 39]. Thus, we next examined whether these proteins bound to the ICAM1 promoter region in OVSAYO cells. ChIP assays with real-time PCR revealed that the binding level of hTERT and Sam68 was relatively low under SSN in OVSAYO cells. However, binding of these proteins was enhanced by culturing the cells under SSH (Fig. 4i). We also found that such protein binding under SSH was inhibited by CQ treatment in CCC cells (Fig. 4i and S10d). ChIP analysis of grafted OVISE tumours revealed that both proteins considerably bound to the human ICAM1 region in vivo (Fig. 4j, k) and the binding was predominant for Sam68 (Fig. 4k).
CQ potentially blocks NFκB binding by inhibition of hTERT and Sam68. To test this possibility, we further examined whether Sam68-KD under SSH decreases the association of NFκB with the promoter region (Fig. 4l). Sam68-KD under SSH did not affect the expression levels of RelA and p50 in OVSAYO cells (Fig. 4m). However, RelA binding was significantly diminished by Sam68-KD (Fig. 4n), which suggests that Sam68 promoted NFκB binding under SSH. We also examined hTERT, but the hTERT level was only partially reduced even when CCC cells were treated with siRNA for a longer period probably, owing to high protein stability (data not shown). Therefore, we did not reach the same conclusion.
LC3B expression in cancer cells significantly correlates with poor prognoses of CCC patients
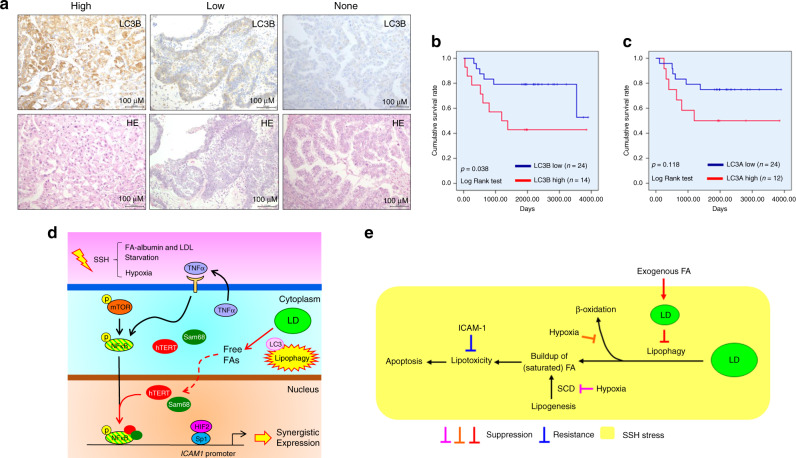
It has been shown that LC3B better reflects the autophagy status than LC3A [40]. Additionally, it has been reported that LC3A expression in tumour tissues is associated with poor prognoses of CCC patients [41]. We found that lipophagy associated with increased LC3A/B level induced synergistic ICAM-1 expression in CCC cells in response to SSH. Thus, we further examined the correlation between tissue LC3 levels and the prognosis of CCC patients. IHC of LC3A and LC3B using the TMA, followed by scoring of expression levels revealed that the LC3B level was considerably heterogeneous among CCC cases (Fig. 5a). Kaplan–Meier analysis revealed that the LC3B level was significantly associated with a poor OS rate (Fig. 5b). Multivariable analysis showed that LC3B tended to be a prognostic factor for CCC (p < 0.1) with a risk ratio of 2.634 (Table S3), although the LC3A level did not (Figs. S11 and 5c), which may be consistent with the notion that LC3B better reflects the autophagy status. Thus, the lipophagy-ICAM-1 pathway contributed to the aggressiveness of CCC.
Fig. 5. Lipophagy-ICAM-1 pathway is involved in aggressiveness of CCC through resistance against cell death.
a Staining pattern of LC3B in CCC cells of tumour tissue evaluated by IHC. H&E staining of corresponding regions is also shown. b and c Kaplan–Meier analysis of the correlation between LC3B (b) and LC3A (c) levels and the OS rate of CCC patients. d Model of lipophagy-driven expression of the ICAM1 gene in CCC cells on the basis of results of present and former [8] studies. Pathways indicated by black and red arrows represent mechanistic routes revealed in former [8] and the present studies, respectively. Dashed arrow indicates involvement of unknown mechanisms. “p” with a yellow circle indicates protein phosphorylation. Yellow symbols with a red margin are shown to highlight important events. e Model of SSH-driven cell death (apoptosis) and its blockade by LD and ICAM-1. In this model, CCC cells undergo apoptosis owing to lipophagy-driven buildup of fatty acids. Inhibition of β-oxidation and FA unsaturation (orange and magenta T-bars) may assist lipotoxicity. This cytotoxicity is cancelled by inhibition of lipophagy owing to exogenous FA-mediated LD synthesis (red symbols). When exogenous FA is unavailable, ICAM-1 confers resistance via unknown mechanisms. SCD: stearoyl-CoA desaturase.
Discussion
The present study demonstrated that SSH-driven activation of the ICAM1 gene in CCC cells was mediated via lipophagy, a lipid catabolism induced in association with autophagy. It is well known that autophagy is induced in cancer cells, which include ovarian cancer cells, in response to nutrient deprivation [33] and hypoxia [42]. The presented mechanism is characteristic because it occurs when availabilities of oxygen and fatty acids (FA-albumin complex and lipoproteins such as LDL) are simultaneously limited (Fig. 5d). Additionally, we demonstrated that this mechanism involves stabilisation of NFκB binding to the ICAM1 promoter region (Fig. 5d). Thus, the present study provides a greater understanding of how ICAM-1 is produced in CCC cells, thereby augmenting malignancy of CCC tissues.
We found that ICAM-1 levels were not significantly different between disease stages, which implied that malignant CCC cases tend to highly express ICAM-1 even at the onset of the disease. So far, the reported roles of ICAM-1 in EOC cell biology have been controversial as described in the Introduction. We first demonstrated that ICAM-1 expression is related to poor prognoses of CCC patients although there are limitations owing to relatively small sample size. This is in agreement with data showing that ICAM-1 confers a survival advantage to CCC cells under SSH [8] and that generation of autophagosomes in association with ICAM-1 expression can correlate with a poor prognosis. Considering that ICAM-1 and autophagy contribute to cancer progression and can be therapeutically targeted by specific antibodies [14] and CQ [43], respectively, CCC may also be treated with these drugs.
We showed that FA, but not cholesterol, uptake is predominantly associated with LD biogenesis in CCC cells, thereby increasing cell viability under SSH. To date, some studies have suggested mechanisms by which LD confers survival advantages to cancer cells under hypoxia, such as maintenance of endoplasmic reticulum homoeostasis [44] and detoxification of ROS [45]. Thus, the effect of LD generation on cell survival under SSH might involve these reported molecular mechanisms. However, these functions of LD may not replace those of ICAM-1 because SSH-driven ER-stress [8] and potential ROS generation (Fig. S5) are not responsible for the ICAM-1 expression.
Additionally, it is important to consider why CCC cells undergo apoptosis under SSH to understand how LD confers a survival advantage to CCC cells. Cells store excess FA as a component of triglyceride, thereby protecting them from lipotoxicity [6]. Mild lipophagy in CCC cells under mild O2 and lipid deprivation likely generates FA from LD to compensate for impaired exogenous lipid supply and FA unsaturation [6]. However, aggressive SSH-driven lipophagy may produce excess unsaturated and saturated FAs, which is toxic rather than beneficial (Fig. 5e). Inhibition of β-oxidation [46] may accelerate this process (Fig. 5e, orange T-bar). Additionally, inhibition of fatty acid unsaturation under hypoxia may promote a buildup of toxic saturated FAs [6] (Fig. 5e, magenta T-bar). Our data suggest that FA uptake under hypoxia prevents SSH-driven cytotoxicity via LD biogenesis (Fig. 5e, red symbols). Oleate incorporated into LD may play roles in suppression of saturated FA-driven lipotoxicity [6]. Our data showing that unsaturated oleic acid rescues whereas saturated palmitic acid promotes SSH-driven cell death, respectively (Fig. 2b), support these arguments. When exogenous FAs are unavailable, ICAM-1 synthesised in response to SSH might replace impaired LD functions and confer resistance against apoptosis via currently unknown mechanisms (Fig. 5e, blue T-bar).
Our results suggest that lipophagy contributes to stabilisation of NFκB binding to the ICAM1 promoter region, presumably through association of hTERT and Sam68 with the NFκB complex (Fig. 5d). However, lipophagy does not contribute to the activation process of NFκB, which includes mTOR and TNF-α (Fig. 5d). This is in contrast to a previous report showing that cytokine-induced autophagy activates NFκB, but this is not associated with ICAM1 expression in endothelial cells [47]. Activities of hTERT and Sam68 are regulated by phosphorylation of their amino acid residues [39, 48]. Peroxisome proliferator-activated receptors (PPARs) are cellular receptors of free FAs and involved in lipid metabolism [25]. The PPARγ-hTERT axis contributes to prevention of endothelial cell apoptosis [49]. Thus, phosphorylation of hTERT and Sam68 through FAs released by lipophagy-driven degradation of LD might contribute to SSH-driven ICAM-1 expression. Future studies are necessary to uncover how lipophagy facilitates the association between NFκB and these coactivators (Fig. 5d, red-dashed arrow).
In summary, the present study revealed that lipophagy connects SSH-driven ICAM1 gene expression and fatty acid deficiency. Considering that expression of ICAM-1 is associated with poor survival of CCC patients, suppression of the lipophagy-ICAM-1 pathway may be a promising therapeutic strategy. Currently, the mechanisms of how ICAM-1 confers a survival advantage to CCC cells are unclear. Future elucidation of this topic together with the application of existing drugs may result in effective therapeutic options for ovarian cancer patients.
Supplementary information
Acknowledgements
We thank Mitchell Arico from Edanz (https://jp.edanz.com/ac) for editing a draft of this manuscript.
Author contributions
SK designed the study, performed experiments, wrote the manuscript, and supervised the project. TT performed experiments. YN and MY contributed to IHC. YO contributed to animal experiments. SS, HT, and EM contributed to data interpretation. TY diagnosed cancer patients. HK contributed to TMA preparation. YM contributed to drafting the manuscript and supervised the project. All authors approved the final version of the manuscript.
Funding
This study was partially supported by JSPS KAKENHI Grant Number 26430135.
Data availability
All data generated in this study are included in this manuscript.
Competing interests
The authors declare no competing interests.
Ethics and consent to participate
For human samples, written consent was obtained from all patients. The study was approved by our institutional review board (Kanagawa Cancer Center Research Institute) (approval No. 177). Animal studies were also reviewed and approved by our institutional review board.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Shiro Koizume, Email: skoizume@gancen.asahi.yokohama.jp.
Yohei Miyagi, Email: miyagi@gancen.asahi.yokohama.jp.
Supplementary information
The online version contains supplementary material available at 10.1038/s41416-022-01808-4.
References
- 1.Cho KR, Shih L-M. Ovarian cancer. Annu Rev Pathol. 2009;4:287–313. doi: 10.1146/annurev.pathol.4.110807.092246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kato N. Pathology of clear cell carcinoma of the ovary: a basic view based on cultured cells and modern view from comprehensive approaches. Pathol Int. 2020;70:591–601. doi: 10.1111/pin.13001. [DOI] [PubMed] [Google Scholar]
- 3.Koizume S, Miyagi Y. Potential coagulation factor-driven pro-inflammatory responses in ovarian cancer tissues associated with insufficient O2 and plasma supply. Int J Mol Sci. 2017;18:809. doi: 10.3390/ijms18040809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee P, Chandel NS, Simon MC. Cellular adaptation to hypoxia through hypoxia inducible factors and beyond. Nat Rev Mol Cell Biol. 2020;21:268–83. doi: 10.1038/s41580-020-0227-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lewis CA, Brault C, Peck B, Bensaad K, Griffith B, Mitter R, et al. SREBP maintains lipid biosynthesis and viability of cancer cells under lipid- and oxygen-deprived conditions and defines a gene signature associated with poor survival in glioblastoma multiforme. Oncogene. 2015;34:5128–40. doi: 10.1038/onc.2014.439. [DOI] [PubMed] [Google Scholar]
- 6.Ackerman D, Tumanov S, Qiu B, Michalopoulou E, Spata M, Azzam A, et al. Triglycerides promote lipid homeostasis during hypoxic stress by balancing fatty acid saturation. Cell Rep. 2018;24:2596–605. doi: 10.1016/j.celrep.2018.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koizume S, Ito S, Miyagi E, Hirahara F, Nakamura Y, Sakuma Y, et al. HIF2α-Sp1 interaction mediates a deacetylation-dependent FVII-gene activation under hypoxic conditions in ovarian cancer cells. Nucleic Acids Res. 2012;40:5389–401. doi: 10.1093/nar/gks201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koizume S, Ito S, Nakamura Y, Yoshihara M, Furuya M, Yamada R, et al. Lipid starvation and hypoxia synergistically activates ICAM1 and multiple genes in an Sp1-dependent manner to promote the growth of ovarian cancer. Mol Cancer. 2015;14:77. doi: 10.1186/s12943-015-0351-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koizume S, Takahashi T, Yoshihara M, Nakamura Y, Ruf W, Takenaka K, et al. Cholesterol starvation and hypoxia activate the FVII gene via the SREBP1-GILZ pathway in ovarian cancer cells to produce procoagulant microvesicles. Thromb Haemost. 2019;119:1058–1071. doi: 10.1055/s-0039-1687876. [DOI] [PubMed] [Google Scholar]
- 10.Rosette C, Roth RB, Oeth P, Braun A, Kammerer S, Ekblom J, et al. Role of ICAM1 in invasion of human breast cancer cells. Carcinogenesis. 2005;26:943–50. doi: 10.1093/carcin/bgi070. [DOI] [PubMed] [Google Scholar]
- 11.Maruo Y, Gochi A, Kaihara A, Shimamura H, Yamada T, Tanaka N, et al. ICAM-1 expression and the soluble ICAM-1 level for evaluating the metastatic potential of gastric cancer. Int J Cancer. 2002;100:486–90. doi: 10.1002/ijc.10514. [DOI] [PubMed] [Google Scholar]
- 12.Yang S-F, Chen M-K, Hsieh Y-S, Chung T-T, Hsieh Y-H, Lin C-W, et al. Prostaglandin E2/EP1 signaling pathway enhances intercellular adhesion molecule 1(ICAM-1) expression and cell motility in oral cancer cells. J Biol Chem. 2010;285:29808–16. doi: 10.1074/jbc.M110.108183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roland CL, Dineen SP, Toombs JE, Carbon J,G, Smith C, W., Brekken RA, et al. Tumor-derived intercellular adhesion molecule-1 mediates tumor-associated leukocyte infiltration in orthtopic pancreatic xenografts. Exp Biol Med. 2010;235:263–9. doi: 10.1258/ebm.2009.009215. [DOI] [PubMed] [Google Scholar]
- 14.Veitonmäki N, Hansson M, Zhan F, Sundberg A, Löfstedt T, Ljungars A, et al. A human ICAM-1 antibody isolated by a function-first approach has potent macrophage-dependent antimyeloma activity in vivo. Cancer Cell. 2013;23:502–15. doi: 10.1016/j.ccr.2013.02.026. [DOI] [PubMed] [Google Scholar]
- 15.Arnold JM, Cummings M, Purdie D, Chenevix-Trench G. Reduced expression of intercellular adhesion molecule-1 in ovarian adenocarcinomas. Br J Cancer. 2001;85:1351–8. doi: 10.1054/bjoc.2001.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Groote ML, Kazemier HG, Huisman C, van der Gun B,TF, Faas MM, Rots MG. Upregulation of endogenous ICAM-1 reduces ovarian cancer cell growth in the absence of immune cells. Int J Cancer. 2014;134:280–90. doi: 10.1002/ijc.28375. [DOI] [PubMed] [Google Scholar]
- 17.Srivastava P, Paluch BE, Matsuzaki J, James SR, Collamat-Lai G, Taverna P. Immunomodulatory action of the DNA methyltransferase inhibitor SGI-110 in epithelial ovarian cancer cells and xenografts. Epigenetics. 2015;10:237–46. doi: 10.1080/15592294.2015.1017198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bu S, Li B, Wang Q, Gu T, Dong Q, Miao X, et al. Epithelial ovarian cancer stem-like cells are resistant to the cellular lysis of cytokine-induced killer cells via HIF1A-mediated down regulation of ICAM-1. Int J Oncol. 2019;55:179–90. doi: 10.3892/ijo.2019.4794. [DOI] [PubMed] [Google Scholar]
- 19.Egan K, Crowley D, Smyth P, O’Toole S, Spillane C, Martin C, et al. Platelet adhesion and degranulation induce pro-survival and pro-angiogenic signaling in ovarian cancer cells. PLoS ONE. 2011;6:e26125. doi: 10.1371/journal.pone.0026125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matte I, Lane D, Laplante C, Rancourt C, Piché A. Profiling of cytokines in human epithelial ovarian cancer ascites. Am J Cancer Res. 2012;2:566–80. [PMC free article] [PubMed] [Google Scholar]
- 21.Pei H, Yang Y, Cui L, Yang J, Li X, Yang Y, et al. Bisdemethoxycurcumin inhibits ovarian cancer via deducing oxidative stress mediated MMPs expressions. Sci Rep. 2016;6:28773. doi: 10.1038/srep28773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yin M, Li X, Tan S, Zhou HJ, Ji W, bellone S, et al. Tumor-associated macrophages drive spheroid formation during early transcoelomic metastasis of ovarian cancer. J Clin Investig. 2016;126:4157–73. doi: 10.1172/JCI87252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koizume S, Shin Ito, Yoshioka Y, Kanayama T, Nakamura Y, Yoshihara M, et al. High-level secretion of tissue factor-rich extracellular vesicles from ovarian cancer cells mediated by filamin-A and protease-activated receptors. Thromb Haemost. 2016;115:299–310. doi: 10.1160/th15-03-0213. [DOI] [PubMed] [Google Scholar]
- 24.Heeren J, Beisiegel U. Receptor-mediated endocytosis and intracellular trafficking of lipoproteins. In Ehnholm C ed. Cellular Lipid Metabolism. Berlin Heidelberg: Springer-Veralg; 2009. p. 213–35.
- 25.Koizume S, Miyagi Y. Lipid droplets: a key cellular organelle associated with cancer cell survival under normoxia and hypoxia. Int J Mol Sci. 2016;17:1430. doi: 10.3390/ijms17091430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ayyagari VN, Wang X, Diaz-Sylvester PL, Groesch K, Brard L. Assessment of acyl-CoA cholesterol acyltransferase (ACAT-1) role in ovarian cancer progression-An in vitro study. PLoS ONE. 2020;15:e0228024. doi: 10.1371/journal.pone.0228024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gabitova L, Restifo D, Gorin A, Manocha K, Handorf E, Yang D-H, et al. Endogenous sterol metabolites regulate growth of EGFR/KRAS-dependent tumors via LXR. Cell Rep. 2015;38:1927–38. doi: 10.1016/j.celrep.2015.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Riscal R, Skuli N, Simon MC. Even cancer cells watch their cholesterol! Mol Cell. 2019;76:220–31. doi: 10.1016/j.molcel.2019.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ward C, Martinez-Lopezm N, Ottenm EG, Carroll B, Maetzel D, Singh R. Autophagy, lipophagy and lysosomal lipid storage disorders. Biochim Biophys Acta. 2016;1861:269–84. doi: 10.1016/j.bbalip.2016.01.006. [DOI] [PubMed] [Google Scholar]
- 30.Klionsky DJ, Abdel-Aziz AK, Abdelfatah S, Abdellatif M, Abdoli A, Abel S, et al. Guideline for the use and interpretation of assays for monitoring autophagy (4th edition) Autophagy. 2021;17:1–382. doi: 10.1080/15548627.2020.1797280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mizushima N, Yoshimori T. How to interpret LC3 immunoblotting. Autophagy. 2007;3:542–5. doi: 10.4161/auto.4600. [DOI] [PubMed] [Google Scholar]
- 32.Mauthe M, Orhon I, Rocchi C, Zhou X, Luhr M, Hijkema K-J, et al. Chloroquine inhibits autophagic flux by decreasing autophagosome-lysosome fusion. Autophagy. 2018;14:1435–55. doi: 10.1080/15548627.2018.1474314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roy D, Mondal S, Khurana A, Jung D-B, Hoffmann R, He X, et al. Loss of HSulf-1: the missing link between autophagy and lipid droplets in ovarian cancer. Sci Rep. 2017;7:41977. doi: 10.1038/srep41977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sathyanarayan A, Mashek MT, Mashek DG. ATGL promotes autophagy/lipophagy via SIRT1 to control hepatic lipid droplet catabolism. Cell Rep. 2017;19:1–9. doi: 10.1016/j.celrep.2017.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Q, Lenardo MJ, Baltimore D. 30 years of NF-κB: a blossoming of relevance to human pathology. Cell. 2017;168:37–57. doi: 10.1016/j.cell.2016.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klenke C, Wildera D, Engelen T, Müller J, Noll T, Niehaus K, et al. Hsc70 is a novel interactor of NF-kappaB p65 in living Hippocampal neurons. PLoS ONE. 2013;8:e65280. doi: 10.1371/journal.pone.0065280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin J, Kato M, Nagata K, Okuwaki M. Efficient DNA binding of NF-κB requires the chaperone-like function of NPM1. Nucleic Acids Res. 2017;45:3707–23. doi: 10.1093/nar/gkw1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ghosh A, Saginc G, Leow SC, Khattar E, Shin EM, Yan TD, et al. Telomerase directly regulates NF-κB-dependent transcription. Nat Cell Biol. 2012;14:1270–81. doi: 10.1038/ncb2621. [DOI] [PubMed] [Google Scholar]
- 39.Fu K, Sun X, Zheng W, Weir EM, Hodgson A, Tran DQ, et al. Sam68 modulates the promoter specificity of NF-κB and mediates expression of CD25 in activated T cells. Nat Commun. 2013;4:1909. doi: 10.1038/ncomms2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barth S, Glick D, Macleod KF. Autophagy: assays and artifacts. J Pathol. 2010;221:117–24. doi: 10.1002/path.2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spowart JE, Townsend KN, Huwait H, Eshragh S, West NR, Ries JN, et al. The autophagy protein LC3A correlates with hypoxia and is a prognostic marker of patient survival in clear cell ovarian cancer. J Pathol. 2012;228:437–47. doi: 10.1002/path.4090. [DOI] [PubMed] [Google Scholar]
- 42.DeVorkin L, Hattersley M, Kim P, Ries J, Spowart J, Anglesio MS, et al. Autophagy inhibition enhances sunitinib efficacy in clear cell ovarian carcinoma. Mol Cancer Res. 2017;15:250–8. doi: 10.1158/1541-7786.MCR-16-0132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kinsey CG, Camolotto SA, Boespflug AM, Guillen KP, Foth M, Truong A, et al. Protective autophagy elicited by RAF→MEK→ERK inhibition suggests a treatment strategy for RAS-driven cancers. Nat Med. 2019;25:620–7. doi: 10.1038/s41591-019-0367-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qiu B, Ackerman D, Sanchez DJ, Li B, Ochocki JD, Grazioli A, et al. HIF2α-dependent lipid storage promotes endoplasmic reticulum homeostasis in clear-cell renal cell carcinoma. Cancer Discov. 2015;5:652–67. doi: 10.1158/2159-8290.CD-14-1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bensaad K, Favaro E, Lewis CA, Peck B, Lord S, Collins JM. Fatty acid uptake and lipid storage induced by HIF-1α contribute to cell growth and survival after hypoxia-reoxygenation. Cell Rep. 2014;9:349–65. doi: 10.1016/j.celrep.2014.08.056. [DOI] [PubMed] [Google Scholar]
- 46.Munir R, Lisec J, Swinnen JV, Zaidi N. Lipid metabolism in cancer cells under metabolic stress. Br J Cancer. 2019;120:1090–8. doi: 10.1038/s41416-019-0451-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chu L-Y, Hsueh Y-C, Cheng H-L, Wu KK. Cytokine-induced autophagy promotes long-term VCAM-1 but not ICAM-1 expression by degrading late-phase IκBα. Sci Rep. 2017;7:12472. doi: 10.1038/s41598-017-12641-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jie M-M, Chang X, Zeng S, Liu C, Liao G-B, Wu Y-R, et al. Diverse regulatory manners of human telomerase reverse transcriptase. Cell Commun Signal. 2019;17:63. doi: 10.1186/s12964-019-0372-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Werner C, Gensch C, Pöss J, Haendeler J, Böhm M, Laufs U. Pioglitazone activates aortic telomerase and prevents stress-induced endothelial apoptosis. Atherosclerosis. 2011;216:23–24. doi: 10.1016/j.atherosclerosis.2011.02.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated in this study are included in this manuscript.