Abstract
Introduction
Small cell lung cancer (SCLC) is an aggressive malignancy with no established biomarkers. Schlafen 11(SLFN11), a DNA/RNA helicase that sensitises cancer cells to DNA-damaging agents, has emerged as a promising predictive biomarker for several drug classes including platinum and PARP inhibitors. Detection of SLFN11 in circulating tumour cells (CTCs) may provide a valuable alternative to tissue sampling.
Methods
SLFN11 expression was evaluated in tumour samples and characterised in circulating tumour cells (CTC) longitudinally to determine its potential role as a biomarker of response.
Results
Among 196 SCLC tumours, 51% expressed SLFN11 by IHC. In addition, 20/29 extra-thoracic high-grade neuroendocrine tumours expressed SLFN11 expression. In 64 blood samples from 42 SCLC patients, 83% (53/64) of samples had detectable CTCs, and SLFN11-positive CTCs were detected in 55% (29/53). Patients actively receiving platinum treatment had the lowest number of CTCs and a lower percentage of SLFN11-positive CTCs (p = 0.014). Analysis from patients with longitudinal samples suggest a decrease in CTC number and in SLFN11 expression that correlates with clinical response.
Conclusions
SLFN11 levels can be monitored in CTCs from SCLC patients using non-invasive liquid biopsies. The ability to detect SLFN11 in CTCs from SCLC patients adds a valuable tool for the detection and longitudinal monitoring of this promising biomarker.
Subject terms: Small-cell lung cancer, Predictive markers
Introduction
Small cell lung cancer (SCLC) is an aggressive high-grade neuroendocrine carcinoma. Its clinical course is notable for early metastases and rapid relapse, despite initial response to frontline treatments. To date, there are no validated predictive biomarkers in SCLC to guide treatment selection at presentation or relapse. In recent years, Schlafen 11 (SLFN11), a DNA/RNA helicase that blocks replication at stressed replication forks, which lead to cell death, has emerged as a promising predictive biomarker for several therapeutics used in SCLC, including platinum, topoisomerase inhibitors (topotecan), poly (ADP-ribose) polymerase inhibitors (PARPi), and lurbinectedin [1–3]. In a phase II trial of relapsed SCLC treated with the combination of temozolomide and the PARPi veliparib or a placebo, patients with SLFN11-positive tumours by immunohistochemistry (IHC) had significantly longer progression free survival (PFS) (5.7 vs. 3.6 mo, p = 0.009) and overall survival (OS) (12.2 vs.7.5 mo, p = 0.014) compared to patients with SLFN11-negative tumours in the temozolomide/veliparib combination group [4]. In another phase II trial studying veliparib in combination with carboplatin/etoposide in patients with treatment naive extensive-stage SCLC patients, SLFN11 positivity by IHC was found to have a trend towards improved PFS in patients received veliparib versus placebo (HR, 0.6; 80% CI, 0.36–0.97) [5]. Additionally, SLFN11 was found to be an important biomarker in other cancer types such as Ewing sarcoma, prostate cancer and ovarian cancer [6–8].
Patients with SCLC are typically diagnosed at an advanced stage and rarely undergo surgical resection. As a result, tissue samples (commonly from fine needle aspirations or small biopsies) are often scarce and inadequate in size and quality for biomarker testing. For example, in the analysis of the SLFN11 expression by IHC in a randomised phase II trial, analysis was limited by the fact that adequate archival tissue was available for only 45% of the patients [4]. This lack of tissue has posed a major challenge in studying SCLC biology and treatment response. In addition to practical limitations posed by small biopsies, several groups including our own have found that the molecular profile of SCLC can change over time due to the selective evolutionary pressures of treatment [9–12]. This is especially true for SLFN11, where pre-clinical studies showed SLFN11 to be downregulated as early as 72 h following treatment with platinum chemotherapy in cell lines [1]. Furthermore, PDX models treatment with an epigenetic modifier EZH2 for 7 days re-induced SLFN11 expression [11, 13]. Thus, archival tissue collected at the time of diagnosis may not accurately reflect the current SLFN11 biomarker status for patients with relapsed SCLC. However, a repeat biopsy may not be clinically feasible due to potential procedure risks or the need to avoid treatment delays.
Given the limited tissue and the need for repeat biopsies for biomarker testing, an attractive alternative would be using minimally invasive, blood-based biomarker approaches to assess a patient’s current biomarker status, especially in patients with relapsed SCLC. Blood-based biomarkers may be especially well suited to SCLC, since patients with SCLC tend to have an abundance of circulating tumour cells (CTCs), consistent with its early metastatic potential through circulation [14, 15]. This presents a unique opportunity to study CTC-based biomarkers in SCLC. In contrast to non-small cell lung cancer, SCLC generally lacks driver mutations and fusions that are more suitable for detection in the circulating tumour DNA, thus assessing protein biomarkers in real-time within CTCs presents a considerable opportunity. Of note, SLFN11 expression is highly prevalent - previously shown to be present in about 50% all cancer cell lines and in 50% of tumour samples from SCLC patients [3, 4, 16] which makes it a suitable candidate biomarker to be tested in CTCs.
To further characterise the prevalence, heterogeneity and dynamic expression of SLFN11 in SCLC, we developed a novel CTC assay utilising a non-enrichment platform (Epic Sciences), in addition to the previously validated SLFN11 IHC tissue assay. This CTC platform had been used successfully to develop a variety of prognostic and predictive tools in prostate cancer [8, 17–22], one of which, the CTC AR-V7 assay, has generated sufficient clinical utility data and credentialing to receive reimbursement from Centers for Medicare & Medicaid Services and is listed in the National Comprehensive Cancer Network guidelines for men with progressing castrate-resistant prostate cancer considering a second generation androgen receptor targeted agent [17, 19, 20]. Here, we demonstrate that this novel technology can detect CTCs in patients with SCLC, and can monitor the dynamic expression of SLFN11 in CTCs over time in relation to patients’ treatment and clinical status.
Methods
Patient selection
Written consents were obtained from patients. Patients diagnosed with SCLC at the University of Texas M.D. Anderson Cancer Center were selected based on extensive-stage disease irrespective of age, gender or other clinical criteria. Patients underwent informed consent to Institutional Review Board (IRB)-approved protocol LAB10-0442 (“Evaluation of blood-based test for the detection of circulating tumour cells and circulating proteins and microRNAs and molecular analysis for polymorphisms and mutations”) and blood was collected.
SLFN11 IHC assay
The SLFN11 IHC assay has been standardised and validated in the MD Anderson Cancer Center (MDACC) clinical IHC laboratory using standard approaches for clinical test validation. This assay was used in the two clinical trials mentioned above [4, 5], in 29 extra-thoracic high-grade neuroendocrine tumour samples from patients at MDACC, and in xenograft models derived from CTCs. The same assay was later certified by Clinical Laboratory Improvement Amendments (CLIA) provisions and used in an ongoing clinical trial (NCT04334941). The anti-human SLFN11 antibody used in this IHC assay is an unconjugated polyclonal antibody (IgG Isotype) manufactured by Millipore-Sigma (HPA023030). The SLFN11 IHC slides were reviewed and scored by a thoracic pathologist. SLFN11 positivity is defined as H-score (percentage of tumoral labelling × intensity score) of ≥1.
SCLC CTC Identification
Blood samples from adult patients with a confirmed histological diagnosis of SCLC were collected in 10 mL cell-free preservative blood tubes (StreckTM, Omaha, Nebraska), and shipped overnight at room temperature to Epic Sciences for processing using the method previously described [23]. Briefly, red blood cells were lysed, and nucleated cells were dispensed on up to 12 glass slides at a density of 3M cells/slide and stored at −80 °C [23]. Two slides, or approximately 6 million total cells from each patient were evaluated by immunofluorescence (IF) assay using antibodies targeting pan-cytokeratin (CK) and Cluster of Differentiation 45 (CD45) and stained with 4’,6-diamidino-2-phenylindole (DAPI). Each nucleated cell was imaged individually, and CTC candidates identified by a proprietary digital pathology algorithm. The results were reviewed by trained technicians. CK+/CD45− cells with intact DAPI-stained nuclei exhibiting tumour-associated morphologies were classified as CTCs [24]. Using the WHO guidelines for small-cell diagnosis in tissue as reference, CTCs were also sub-classified using an equivalent set of single-cell CTC criteria for defining a CTC with small-cell histology: a small and circular CD45−/CK+ cell with high nuclear to cytoplasmic (N/C) ratio lacking detectable nucleoli and harbouring salt and pepper chromatin [25–27]. We confirmed SLFN11 levels in xenograft models derived from patient CTCs using western blots using sc-347339 (Santa Cruz Biotechnology).
SLFN11 expression in CTCs and immune cells
In addition to CK and CD45, SLFN11 (Cell Signaling Technologies D8W1B) was assessed by IF. The analytical threshold for categorising SLFN11 expression into “positive” (SLFN11+) and “negative” (SLFN11−) was established using laboratory-derived samples (LDS). LDS consisted of 6 healthy donor blood samples spiked with cell lines comprising varying levels of SLFN11 expression to mimic physiologically appropriate ranges as previously described [8]. SLFN11 expression was also determined in immune cell populations, specifically cells that were CK−/CD45+/SLFN11+ or CK−/CD45−/SLFN11+ that were imaged alongside traditional CK+ CD45− CTCs.
Statistical analyses
ANOVA was used to compare the SLFN11 biomarker signal in CTCs among patients with different platinum-treatment and clinical status. The pairwise comparisons among patient status was conducted using Tukey’s HSD (honestly significant difference) test. The CTC ratios were analysed using the Kruskal–Wallis rank sum test, and the Dunn’s test was performed on multiple pairwise comparisons after the Kruskal–Wallis test. All statistical analyses were performed using R version 4.0.1 [28].
Results
SLFN11 expression in SCLC and extra-thoracic clinical tumour samples
To survey the prevalence of SLFN11-positive tumours in SCLC patients, we examined SLFN11 IHC data from 196 SCLC patient tumour samples tested at MDACC from 2 clinical trials. In addition, we performed SLFN11 IHC in 29 extra-thoracic high-grade neuroendocrine tumour samples at MDACC, with majority of them being high-grade neuroendocrine tumours of the cervix (n = 20) and small cell carcinoma of the prostate (SCPC, n = 8). The SLFN11 IHC assay was validated and standardised under the direction of a clinical pathologist in the laboratory at MDACC and quantified by H-score, a weighted function of percentage of positive cells and the intensity of the cell stained (1+, 2 +, 3+). H-score of > =1 is considered positive for SLFN11 expression [6]. In aggregate, SLFN11 IHC expression was detected in 51% (100/196) of total tumour samples from patients with SCLC, and in 69% of the tumour samples from patients with extra-thoracic high-grade neuroendocrine cancers (Tables 1, 2). Interestingly, none of the tumour samples from primary SCPC were positive, whereas the two tumour samples from metastatic site of small cell transformation of metastatic prostate adenocarcinoma were both positive for SLFN11. In contrast, the majority (86%) of the cervical/vaginal neuroendocrine tumour samples were positive for SLFN11.
Table 1.
SLFN11 expression by IHC in SCLC patient tumour samples.
Table 2.
SLFN11 expression by IHC in extra-thoracic high-grade neuroendocrine tumour samples.
| Extra-thoracic tumour samples (n) | SLFN11 positive % (n) | Average H-score in SLFN11-positive samples (range) |
|---|---|---|
| Cervical/vaginal neuroendocrine or mixed histology (21)a | 86% (18) | 25.7 (1–153.1) |
| Prostate cancer with small cell or mixed histology (6) | 0% (0) | n/a |
| Metastatic sites with small cell transformation of prostate adenocarcinoma (2)b | 100% (2) | 25 (1.5–48.4) |
aAll tumours were from cervical primary except 1 from vaginal primary.
bSamples were taken from (1). lymph node, (2). paravertebral region.
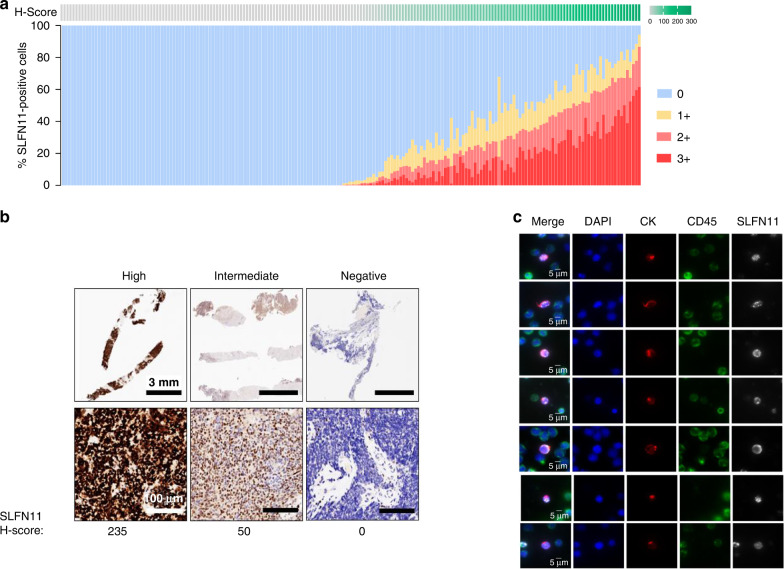
In SLFN11-positive SCLC tumours, significant intra-tumoral heterogeneity of SLFN11 expression was detected, as depicted by the wide H-score ranges in the positive samples (Table 1, Fig. 1a). The H-score in the SLFN11- positive samples (H-score >1) across the two SCLC cohorts ranged from 1.5 to 235.0. Representative images of tumours with high (H-score: 235), intermediate (H-score: 50), or no SLFN11 expression were shown in Fig. 1b.
Fig. 1. SLFN11 levels in SCLC patient samples.
a SLFN11 IHC H-score and % positive cells for 196 SCLC patient tumour samples. b Representative IHC images for SLFN11-high, -intermediate and -negative tumours. c SCLC patient CTCs identified by CK-positivity, CD45-negativity and screened for SLFN11 expression. Scale bar = 3 mm, 100 µm (b), 5 µm (c).
SCLC patients have abundant CTCs
64 blood samples from 42 patients were collected for CTC analyses. Fifteen patients had more than one sample available collected at different timepoints. All patients had extensive-stage or recurrent SCLC, and they were further stratified by their platinum-treatment status at the time of sample collection. CTCs were detected using a non-enrichment based platform to analyse all nucleated cells by IF and detects rare cells using automated digital pathology [19, 21, 29]. For each sample 6 million nucleated cells, consisting of both CTCs and immune cells, corresponding to analysis of approximately 1 mL of blood were analysed.
Fifty-three (83%) of samples had detectable CTCs defined as any CK+/CD45− cell, ranging from 1 to 316 per sample (0.6 to 181 normalised per 1 ml of blood) (Table 3). The median number of CTCs was 4.4 per 1 mL when normalised to the blood volume tested. Patients were stratified into three groups at the time of collection: newly diagnosed treatment-naive patients (or “platinum naive”) (n = 14), patients actively receiving platinum-based frontline treatment at the time of blood collection were designated as “on-platinum”(n = 8), and patients who relapsed after platinum-based treatment as “platinum relapsed” (n = 42). We calculated the percentage of patients with CTCs stratified by the three groups and found that CTCs were detected most frequently in platinum-relapsed samples (90%), followed by platinum-naive (79%), and on-platinum (50%) (Table 3).
Table 3.
CTCs in patient samples stratified by platinum treatment status.
| Platinum status (no. of patient samples) | % (n) of patient samples with CTCs | Range of CTC number per ml | % (n) of patient samples with SLFN11-positive CTCs |
|---|---|---|---|
| Platinum naive (14) | 79% (11) | 0–181 | 71% (10) |
| On-platinum (8) | 50% (4) | 0–38.2 | 25% (2) |
| Platinum relapsed (42) | 90% (38) | 0–64.6 | 40% (17) |
| All (64) | 83% (53) | 0–181 | 45% (29) |
SLFN11 expression, and small-cell pathology criteria can be detected in CTCs
Given the challenge associated with tumour tissue paucity in SCLC, we developed an IF assay to detect SLFN11 expression in CTCs within the blood. The anti-SLFN11 antibody clone (D8W1B) was selected for IF assay development. Six cell-lines were initially used to develop and test the CTC based assay; SHP-77 (negative control), MDA-MB-231 (negative control), MCF-7 (low expressing), MDA-MBA-436 (medium expressing), PC3 (high expressing), DU-145 (high expressing). Using these results a signal-to-noise ratio of 6 was selected for defining whether a given CTC was positive or negative for SLFN11 (dotted line Supplementary Fig. 1).
The SLFN11 assay was then used to analyse the 64 collected SCLC patient blood samples. Overall, SLFN11-positive CTCs were detected in 45% (29) of all 64 patient samples, or 54.7% of the 53 samples with detectable CTCs (Table 3). SLFN11-positive CTCs comprised 55% of the total number of CTCs detected. This is in concordance with the approximately 50% SLFN11-positivity observed in clinical tumour tissues by IHC (Table 1). SLFN11 expression was detected almost exclusively within the nucleus of the CTCs, as was observed in other reported studies(Fig. 1c) [1, 30].
A subset of CTCs detected were re-imaged at higher magnification and morphologic criteria for diagnosing small-cell or neuroendocrine transformation as defined by the WHO were extracted from CTC images. These features included cell size (<90 μm2), nuclear-to-cytoplasm (NC) ratio (>0.8), lack of visible nucleoli, and salt-and-pepper like or textured chromatin. Additionally, we extracted whether the CTC had dot-like or speckled CK indicative of transformation away from epithelial lineage. This latter feature is observed in a subset of small-cell transformed tumours [31, 32]. (Supplementary Table 1, Supplementary Fig. 2).
SLFN11 expression differs by platinum treatment status
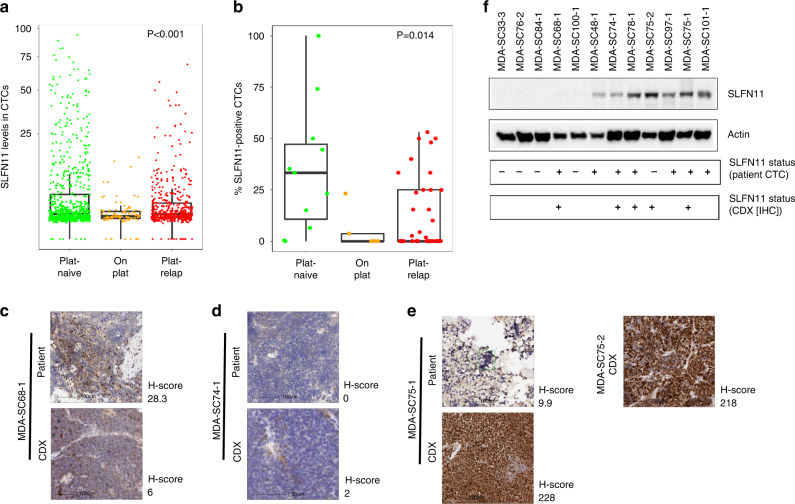
When stratified by platinum-based treatment status, SLFN11-positive CTCs were found to be highest in platinum-naive patient samples, followed by platinum-relapsed, and lowest in patients while on platinum treatment (p < 0.001 by ANOVA) (Table 3, Fig. 2a, b). Among samples with SLFN11-positive CTCs, the concentration of SLFN11-positive CTCs range from 0.4 CTCs/ml to 131 CTCs/ml, and the median concentration was highest in patients with platinum-naive status (3.75 CTCs/ml), followed by on-platinum treatment status (2.1 CTCs/ml) and platinum-relapsed status (1.57 CTCs/ml).
Fig. 2. SLFN11 levels in patient CTCs and tumours, and CTC-derived xenograft (CDX) models.
a SLFN11 levels in patient CTCs stratified by treatment status (SLFN11 analytical cut-off = 6). b Percentage of SLFN11-positive CTCs per patient stratified by treatment status. c–e SLFN11 IHC in patients and their CDX models. F western blot analysis of SLFN11 levels in CDXs and the corresponding positivity in patient IHC and CTCs. Scale bar = 100 µm (c–e).
SLFN11 expression was also observed frequently on CD45+ WBCs [median 199,484 cells/mL, range = 1929/mL to 896,304/mL]. Intriguingly, the percentage of SLFN11+ WBCs also appeared to be lowest when patient was undergoing platinum-containing treatment (Supplementary Fig. 3).
SLFN11 expression in CTC-derived patient xenografts (CDXs)
We then compared SLFN11 expression in CDX models generated from patient CTCs collected at the same time as the CTCs analysed for IF. As expected, liquid biopsies from patients with the highest CTC numbers were most likely to generate CDX models (MDA-SC68-1, MDA-SC75-1) [11]. SLFN11 IHC demonstrated a range of expression in the CDXs. For three patient samples (MDA-SC68-1, MDA-SC74-1, MDA-SC75-1), we additionally performed SLFN11 IHC on the available original diagnostic core needle biopsy from patients (MDA-SC68-1, MDA-SC74-1, MDA-SC75-1) (Fig. 2c–e).
We found concordance between SLFN11 levels in patient CTCs and CDXs in most cases (Fig. 2f). However, in this small cohort, variations can be seen across SLFN11 detection methods. For example, MDA-SC68-1 had negative SLFN11 expression by western blot from the CDX model, but positive expression by IHC from CDX, as well as in CTCs and in patient’s tumour biopsy. While for MDA-SC75-2, SLFN11 status was negative in CTC, but positive by Western and IHC from the CDX model.
Dynamic changes of CTC and SLFN11 expression were correlated with patients’ clinical response
For patients with more than one blood sample available at different time points, we also investigated whether the change in SLFN11 expression correlated with change in clinical status.
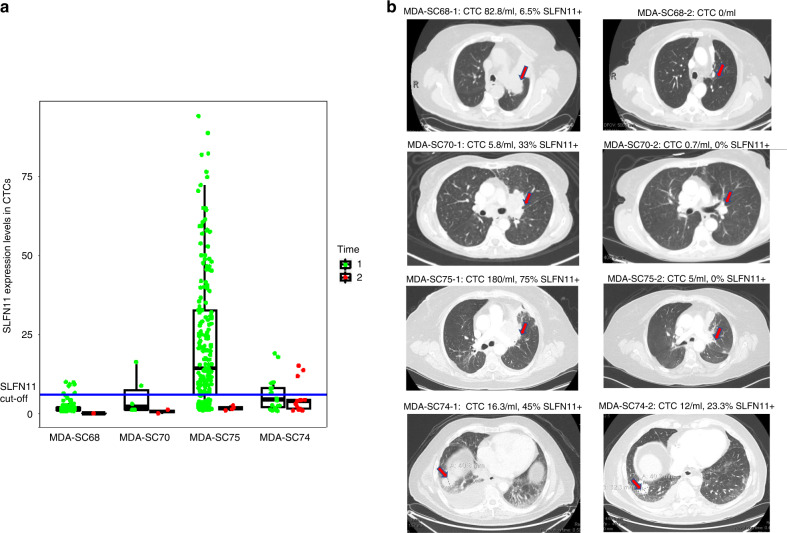
We found that in three patients with serial draws at the time of new diagnosis (time 1) and while on platinum-based treatment (time 2) and having clinical response (MDA-SC68, MDA-SC70 and MDA-SC75), both the number of CTCs and of SLFN11-positive CTCs dropped considerably (Fig. 3a). For example, patient MDA-SC75 initially presented with ES-SCLC with liver and bone metastases, CTC detection from blood draw at the time of diagnosis (Time 1) demonstrated a high number of CTCs (180 CTCs/mL), with 75% of them being SLFN11-positive. The patient started platinum-based chemotherapy treatment. Two months later, while still on treatment and having a partial clinical response as assessed by radiographic findings (Fig. 3b, not by strict Response Evaluation Criteria in Solid Tumours [RECIST] criteria), analyses showed total CTC number went down to 5 CTCs per ml and none were SLFN11-positive (Time 2). Similarly, patient MDA-SC68 was diagnosed with ES-SCLC with bony metastases. Time 1 sample was collected before treatment with 82.8 CTCs/ml, and 6.5% of them were SLFN11-positive. Three months later, while on platinum-based treatment and having a complete response in the bone (partial response in the chest, Fig. 3b), CTC numbers were undetectable (Time 2). In contrast, CTCs in patient MDA-SC74 remained abundant at Time 2 with no significant radiographic improvement (Fig. 3a, b). In relapsed patients on subsequent lines of treatment, persistent CTCs with variable SLFN11% changes correlated with progressive disease (Supplementary Fig. 4). Overall, these data demonstrate dynamic changes of CTC numbers and SLFN11 levels, measured in longitudinal liquid biopsies, may have both prognostic and predictive values in assessing clinical status and treatment response for SCLC patients.
Fig. 3. Longitudinal assessment of CTCs and SLFN11 positivity in CTCs with corresponding radiographic images.
a CTCs and SLFN11 expression levels at Time 1 (in green, at the time of diagnosis, treatment naive) and Time 2 (in red, on frontline platinum-based chemotherapy treatment). b Representative radiographic images at Time 1 (left panel) and Time 2 (right panel).
Discussion
SCLC is a recalcitrant cancer with poor patient outcomes and in dire need of more effective therapies. Research in SCLC is particularly challenging due to the scarcity of tissue samples and the lack of validated clinical biomarkers. In this study, we demonstrate that SLFN11, a promising predictive biomarker for DNA damaging agents (e.g. platinum), PARPi and lurbinectedin in SCLC, can be reliably detected by IHC in about 50% of clinical SCLC tumour samples. In addition, non-invasive CTC-based detection represents an important complementary method to tissue-based techniques such as IHC and has a unique advantage of monitoring longitudinally.
Our data demonstrate that SCLC patients exhibit high numbers of CTCs at the time of diagnosis and relapse, and the number of CTCs were suppressed while on treatment. Previous reports showed CTCs were detected in 60-85% of SCLC patients [14, 33], which is similar to what we have reported here. The total numbers of CTCs in our study was lower than previous reports, and this may be due to reduced blood volume used for the analysis (~1 ml, versus 7.5 ml in the other studies) [14, 33]. Additionally, CTCs here were identified by pan-cytokeratin expression, which would exclude any CTCs undergoing epithelial to mesenchymal transition. Longitudinal monitoring of CTCs could potentially be used to monitor SCLC disease status, as reported in other tumour types such as non-small cell lung cancer and melanoma [34, 35]. The same CTC detection technology used in this paper was recently validated prospectively in prostate cancer patient samples [21, 22]. Similarly, we have validated the detection of SLFN11 on CTCs, and demonstrated that the percentage of SLFN11-positive CTCs was lowest in SCLC patients on platinum-based treatment. We further found that in longitudinal patient samples, the number of CTCs and SLFN11-positive CTCs reflect patient response - CTC numbers were reduced in patients having a clinical response, and SLFN11 levels were reduced following treatment. Conversely, CTCs persist in patients with clinical progression on their restaging scans. These findings suggest the dynamic nature of SLFN11 expression and are consistent with preclinical studies demonstrating downregulation of SLFN11 after treatment [11, 13]. Furthermore, this raises serious concerns of whether archival tissue accurately captures the current status of SLFN11 in order to predict response to further lines of treatment. Instead, longitudinal assessment may indicate the SLFN11 biomarker status more accurately compared to the status in pre-treatment biopsy tissue. Additionally, in our CTC cohort, most patients do not have archival tumour samples available for SLFN11 analyses, except for MDA-SC68, MDA-SC74, and MDA-SC75 (Fig. 2c–f). Although future studies are needed, and an ongoing study (PRIO—NCT04728230) is investigating the correlation of SLFN11 status in tumour IHC and in CTCs at matching time points longitudinally, our data also highlights the challenges of obtaining repeat biopsies from SCLC patients. Thereforeiquid biopsy methodology could be essential for longitudinal monitoring and guide treatment selection. Furthermore, this methodology can be extended to other biomarkers of interest in SCLC, such as EZH2 and PD-L1 expression.
Recently, Gay and colleagues described four subtypes of small cell lung cancer, defined by expression of specific transcription factors; ASCL1(SCLC-A), NEUROD1 (SCLC-N), POU2F3 (SCLC-P), and a newly defined subtype which lacks expression of the transcription factors but has enriched immune cell infiltration (SCLC-I) [9, 36]. Each of these subtypes has unique therapeutic vulnerabilities, which opens the possibility of matching patients to personalised treatment. Interestingly, within subtype SCLC-A, distinct SLFN11 high and SLFN11 low groups can be found (based on SLFN11 bimodality), and stark differences in sensitivity to cisplatin and PARPi olaparib in these two groups [9]. Another recent published paper by Qu and colleagues used IHC to assess SLFN11 expression in 146 primary SCLC tumours, and found SLFN11 to be present in about 60% of the tumours [37]. Notably, SLFN11 expression was absent in tumours with higher CD8+ T cell infiltration and lacking all four markers (ASCL1, NEUROD1, POU2F3 and YAP1), which highlights the heterogeneity of SLFN11 expression in subtypes of SCLC [37]. Despite uncertain clinical evidence so far [4, 5], SLFN11 as a predictive biomarker clearly warrants further validation. Currently, SLFN11 positivity by IHC is being used in our CLIA lab for prospective patient selection in an ongoing clinical trial (NCT04334941), which tests talazoparib in addition to atezolizumab in the maintenance phase of the frontline treatment of patients with SLFN11-positive extensive-stage SCLC. This is the first trial enriching for SLFN11 prospectively as a biomarker in SCLC. Additionally, in an investigator-initiated phase I/II trial (PRIO—NCT04728230) evaluating the promising strategy of durvalumab in combination with PARP inhibitor olaparib and thoracic radiation following frontline carboplatin, etoposide durvalumab for ES-SCLC, SLFN11 expression will be analysed in longitudinal blood and biopsy samples to evaluate its role as a predictive biomarker of response and survival.
Future studies need to validate whether SLFN11 status may be different in biopsies from primary site versus metastatic site in SCLC. In a recent study, SLFN11 expression was found to be higher in metastatic sites than primary ones in breast cancer PDXs [38]. Intriguingly, in our data SLFN11 expression was absent from primary prostate tumour biopsy with small cell histology but present from the metastatic sites with small cell transformation of prostate adenocarcinoma. In addition, preclinical studies suggest that SLFN11 expression is associated with the activation of innate immune response via type I IFNs, and SLFN11 may regulate IFNγ (type II interferon) -mediated cytotoxic T cell killing [1, 39, 40] In our CTC analyses, we found dynamic changes in CD45+ leukocyte SLFN11 expression (Supplementary Fig. 2). It is not surprising given that SLFN11 is inducible by IFNs and associated with immune activation. In a recently published study, SLFN11 was found to be expressed by many types of immune cells infiltrating high-grade serous ovarian cancer (HGSOC), including CD4+, CD8+ T cells, B cells, macrophages and NK cells. The authors found that SLFN11 in cancer and non-cancer cells independently predicted response to platinum-based CT in HGSOC [7]. This raises an interesting possibility whether the SLFN11 expression levels in immune cells could serve as a predictive biomarker to treatment, particularly in relation to immunotherapy. This could be further correlated with the SLFN11 expression in CTCs from the same sample, and with SLFN11 expression by IHC on immune cells in order to elucidate the biomarker value of SLFN11 on immune cells.
In summary, our findings highlight the feasibility of detecting SLFN11 expression by IHC and by liquid biopsy as a predictive biomarker in SCLC. Given the challenge of obtaining adequate SCLC tumour tissue, liquid biopsy should be considered as an important tool in the study and treatment of SCLC. The ability to monitor for CTC number and SLFN11 levels longitudinally is particularly valuable as this may have prognostic and predictive value in patients undergoing treatment for SCLC. Given SLFN11 has been implicated as a potential biomarker across several cancer types [6], this approach may also be relevant for the study of other cancers beyond SCLC.
Supplementary information
Acknowledgements
We would like to acknowledge the patients who participated in this study and volunteered for blood draws (under protocol LAB10-0442) to help further our understanding of SCLC. We would also like to thank Patrice Hartsfield and Heather Napoleon from the protocol blood collection team. This work was supported by: The NIH/NCI CCSG P30-CA016672 (Bioinformatics Shared Resource); NIH/NCI R01-CA207295; NIH/NCI U01-CA213273; NIH/NCI R50-CA243698; The University of Texas-Southwestern and MD Anderson Lung SPORE (5 P50 CA070907); NIH T32 training grant (T32-CA009666); and CPRIT Early Clinical Investigator Award. and through generous philanthropic contributions to The University of Texas MD Anderson Lung Cancer Moon Shot Program and to the Byers Lab; The Andrew Sabin Family Fellowship; The Abell Hangar Foundation; the LUNGevity Foundation Career Development Award, and the Rexanna Foundation for Fighting Lung Cancer.
Author contributions
BZ, CAS, RJC, LAB and CMG: conceptualised this study. BZ, CAS, QW, PR, JF, LS, RW, LF, AJ, CG, JDS, JB and JJ contributed to methodology development, data curation and analyses of the study. VN, PA, LH, HT, AA, MN, JZ, WW and IW provided administration and resources support. JW, RW, LAB and CMG supervised and provided funding for the study. All authors contributed to the writing, editing and review of the manuscript.
Funding
This work was supported by NIH T32 CA009666 (BZ); NIH/NCI R50-CA243698 (CS); University of Texas-Southwestern and MD Anderson Lung SPORE (5 P50 CA070907), The University of Texas MD Anderson Lung Cancer Moon Shot Program, The Andrew Sabin Family Fellowship, The Abell Hangar Foundation; the LUNGevity Foundation Career Development Award, the Rexanna Foundation for Fighting Lung Cancer, and CPRIT Early Clinical Investigator Award (CG); NIH/NCI U01-CA213273 and NIH R01-CA207295 (LB).
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Competing interests
IW received honorarium from Genentech/Roche, Bayer, Bristol-Myers Squibb, AstraZeneca, Pfizer, HTG Molecular, Asuragen, Merck, GlaxoSmithKline, Guardant Health, Flame, Novartis, Sanofi, Daiichi Sankyo, Amgen, Oncocyte, and MSD; and received research support from Genentech, HTG Molecular, DepArray, Merck, Bristol-Myers Squibb, Medimmune, Adaptive, Adaptimmune, EMD Serono, Pfizer, Takeda, Amgen, Karus, Johnson & Johnson, Bayer, Iovance, 4D, Novartis, and Akoya. JZ served on advisory board for AstraZeneca, Novartis, Johnson and Johnson, Geneplus and received speaker’s fees from BMS, OrigMed, Innovent, grants from Merck, Novartis, Johnson and Johnson from outside the submitted work JS received stock options from Epic Sciences at the time of this work. MN reports research funding to institution: Mirati, Novartis, Checkmate, Ziopharm, AstraZeneca, Pfizer, Genentech; consulted for Mirati, Merk and served on advisory board for MSD; meal expenses from Ziopharm. LB received research funding from AstraZeneca, GenMab, Sierra Oncology, ToleroPharmaceuticals; served as an advisor/consultant for AstraZeneca, GenMab, Sierra Oncology, PharmaMar, AbbVie, Bristol-Myers Squibb, Alethia, Merck, Pfizer, Jazz Pharmaceuticals, Genentech, Debiopharm Group. CG received research funding from AstraZeneca; served as an adviser for Bristol-Myers Squibb, Jazz Pharmaceuticals, AstraZeneca, Kisoji; and served on the Speaker’s Bureau for AstraZeneca, Beigene.
Ethics approval and consent to participate
Participants underwent informed written consent to Institutional Review Board (IRB)-approved protocol LAB10-0442 (“Evaluation of blood-based test for the detection of circulating tumour cells and circulating proteins and microRNAs and molecular analysis for polymorphisms and mutations”). The study was performed in accordance with the Declaration of Helsinki.
Consent to publish
Not applicable.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Bingnan Zhang, C. Allison Stewart.
Contributor Information
Lauren A. Byers, Email: Lbyers@mdanderson.org
Carl M. Gay, Email: Cgay@mdanderson.org
Supplementary information
The online version contains supplementary material available at 10.1038/s41416-022-01811-9.
References
- 1.Allison Stewart C, Tong P, Cardnell RJ, Sen T, Li L, Gay CM, et al. Dynamic variations in epithelial-to-mesenchymal transition (EMT), ATM, and SLFN11 govern response to PARP inhibitors and cisplatin in small cell lung cancer. Oncotarget. 2017;8:28575–87. doi: 10.18632/oncotarget.15338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sousa FG, Matuo R, Tang SW, Rajapakse VN, Luna A, Sander C, et al. Alterations of DNA repair genes in the NCI-60 cell lines and their predictive value for anticancer drug activity. DNA Repair (Amst) 2015;28:107–15. doi: 10.1016/j.dnarep.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zoppoli G, Regairaz M, Leo E, Reinhold WC, Varma S, Ballestrero A, et al. Putative DNA/RNA helicase Schlafen-11 (SLFN11) sensitizes cancer cells to DNA-damaging agents. Proc Natl Acad Sci USA. 2012;109:15030–5. doi: 10.1073/pnas.1205943109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pietanza MC, Waqar SN, Krug LM, Dowlati A, Hann CL, Chiappori A, et al. Randomized, double-blind, phase II study of temozolomide in combination with either veliparib or placebo in patients with relapsed-sensitive or refractory small-cell lung cancer. J Clin Oncol. 2018;36:2386–94. doi: 10.1200/JCO.2018.77.7672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Byers, LA, Bentsion, D, Gans, S, Penkov, K, Son, C, Sibille, A et al. Veliparib in combination with carboplatin and etoposide in patients with treatment-naive extensive-stage small cell lung cancer: a phase 2 randomized study. Clin Cancer Res. 2021; 10.1158/1078-0432.CCR-20-4259. [DOI] [PubMed]
- 6.Zhang, B, Ramkumar, K, Cardnell, RJ, Gay, CM, Stewart, CA, Wang, WL et al. A wake-up call for cancer DNA damage: the role of Schlafen 11 (SLFN11) across multiple cancers. Br J Cancer. 2021; 10.1038/s41416-021-01476-w. [DOI] [PMC free article] [PubMed]
- 7.Winkler, C, King, M, Berthe, J, Ferraioli, D, Garuti, A, Grillo, F et al. SLFN11 captures cancer-immunity interactions associated with platinum sensitivity in high-grade serous ovarian cancer. JCI Insight 2021;6:e146098. [DOI] [PMC free article] [PubMed]
- 8.Conteduca V, Ku SY, Puca L, Slade M, Fernandez L, Hess J, et al. SLFN11 Expression in advanced prostate cancer and response to platinum-based chemotherapy. Mol Cancer Ther. 2020;19:1157–64. doi: 10.1158/1535-7163.MCT-19-0926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gay CM, Stewart CA, Park EM, Diao L, Groves SM, Heeke S, et al. Patterns of transcription factor programs and immune pathway activation define four major subtypes of SCLC with distinct therapeutic vulnerabilities. Cancer Cell. 2021;39:346–360.e347. doi: 10.1016/j.ccell.2020.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ireland AS, Micinski AM, Kastner DW, Guo B, Wait SJ, Spainhower KB, et al. MYC Drives temporal evolution of small cell lung cancer subtypes by reprogramming neuroendocrine fate. Cancer Cell. 2020;38:60–78.e12. doi: 10.1016/j.ccell.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stewart CA, Gay CM, Xi Y, Sivajothi S, Sivakamasundari V, Fujimoto J, et al. Single-cell analyses reveal increased intratumoral heterogeneity after the onset of therapy resistance in small-cell lung cancer. Nat Cancer. 2020;1:423–36. doi: 10.1038/s43018-019-0020-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simpson KL, Stoney R, Frese KK, Simms N, Rowe W, Pearce SP, et al. A biobank of small cell lung cancer CDX models elucidates inter- and intratumoral phenotypic heterogeneity. Nat Cancer. 2020;1:437–51. doi: 10.1038/s43018-020-0046-2. [DOI] [PubMed] [Google Scholar]
- 13.Gardner EE, Lok BH, Schneeberger VE, Desmeules P, Miles LA, Arnold PK, et al. Chemosensitive relapse in small cell lung cancer proceeds through an EZH2-SLFN11 Axis. Cancer Cell. 2017;31:286–99. doi: 10.1016/j.ccell.2017.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hou JM, Krebs MG, Lancashire L, Sloane R, Backen A, Swain RK, et al. Clinical significance and molecular characteristics of circulating tumor cells and circulating tumor microemboli in patients with small-cell lung cancer. J Clin Oncol. 2012;30:525–32. doi: 10.1200/JCO.2010.33.3716. [DOI] [PubMed] [Google Scholar]
- 15.Hodgkinson CL, Morrow CJ, Li Y, Metcalf RL, Rothwell DG, Trapani F, et al. Tumorigenicity and genetic profiling of circulating tumor cells in small-cell lung cancer. Nat Med. 2014;20:897–903. doi: 10.1038/nm.3600. [DOI] [PubMed] [Google Scholar]
- 16.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–7. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Armstrong AJ, Halabi S, Luo J, Nanus DM, Giannakakou P, Szmulewitz RZ, et al. Prospective multicenter validation of androgen receptor splice variant 7 and hormone therapy resistance in high-risk castration-resistant prostate cancer: The PROPHECY Study. J Clin Oncol. 2019;37:1120–9. doi: 10.1200/JCO.18.01731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Puca, L, Gavyert, K, Sailer, V, Conteduca, V, Dardenne, E, Sigouros, M et al. Delta-like protein 3 expression and therapeutic targeting in neuroendocrine prostate cancer. Sci Transl Med. 2019;11:eaav0891. [DOI] [PMC free article] [PubMed]
- 19.Scher HI, Armstrong AJ, Schonhoft JD, Gill A, Zhao JL, Barnett E, et al. Development and validation of circulating tumour cell enumeration (Epic Sciences) as a prognostic biomarker in men with metastatic castration-resistant prostate cancer. Eur J Cancer. 2021;150:83–94. doi: 10.1016/j.ejca.2021.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scher HI, Lu D, Schreiber NA, Louw J, Graf RP, Vargas HA, et al. Association of AR-V7 on circulating tumor cells as a treatment-specific biomarker with outcomes and survival in castration-resistant prostate cancer. JAMA Oncol. 2016;2:1441–9. doi: 10.1001/jamaoncol.2016.1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schonhoft JD, Zhao JL, Jendrisak A, Carbone EA, Barnett ES, Hullings MA, et al. Morphology-predicted large-scale transition number in circulating tumor cells identifies a chromosomal instability biomarker associated with poor outcome in castration-resistant prostate cancer. Cancer Res. 2020;80:4892–903. doi: 10.1158/0008-5472.CAN-20-1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brown, LC, Halabi, S, Schonhoft, JD, Yang, Q, Luo, J, Nanus, DM et al. Circulating tumor cell chromosomal instability and neuroendocrine phenotype by immunomorphology and poor outcomes in men with mCRPC treated with abiraterone or enzalutamide. Clin Cancer Res. 2021; 10.1158/1078-0432.CCR-20-3471. [DOI] [PMC free article] [PubMed]
- 23.Werner SL, Graf RP, Landers M, Valenta DT, Schroeder M, Greene SB, et al. Analytical validation and capabilities of the epic CTC platform: enrichment-free circulating tumour cell detection and characterization. J Circ Biomark. 2015;4:3. doi: 10.5772/60725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beltran H, Jendrisak A, Landers M, Mosquera JM, Kossai M, Louw J, et al. The initial detection and partial characterization of circulating tumor cells in neuroendocrine prostate cancer. Clin Cancer Res. 2016;22:1510–9. doi: 10.1158/1078-0432.CCR-15-0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dorantes-Heredia R, Ruiz-Morales JM, Cano-Garcia F. Histopathological transformation to small-cell lung carcinoma in non-small cell lung carcinoma tumors. Transl Lung Cancer Res. 2016;5:401–12. doi: 10.21037/tlcr.2016.07.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Travis WD, Brambilla E, Burke AP, Marx A, Nicholson AG. Introduction to The 2015 World Health Organization Classification of Tumors of the Lung, Pleura, Thymus, and Heart. J Thorac Oncol. 2015;10:1240–2. doi: 10.1097/JTO.0000000000000663. [DOI] [PubMed] [Google Scholar]
- 27.Travis WD, Brambilla E, Nicholson AG, Yatabe Y, Austin JHM, Beasley MB, et al. The 2015 World Health Organization Classification of Lung Tumors: Impact of Genetic, Clinical and Radiologic Advances Since the 2004 Classification. J Thorac Oncol. 2015;10:1243–60. doi: 10.1097/JTO.0000000000000630. [DOI] [PubMed] [Google Scholar]
- 28.R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, V., Austria. 2020; https://www.R-project.org/.
- 29.Scher HI, Graf RP, Schreiber NA, Jayaram A, Winquist E, McLaughlin B, et al. Assessment of the validity of nuclear-localized androgen receptor splice variant 7 in circulating tumor cells as a predictive biomarker for castration-resistant prostate cancer. JAMA Oncol. 2018;4:1179–86. doi: 10.1001/jamaoncol.2018.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murai J, Feng Y, Yu GK, Ru Y, Tang SW, Shen Y, et al. Resistance to PARP inhibitors by SLFN11 inactivation can be overcome by ATR inhibition. Oncotarget. 2016;7:76534–50. doi: 10.18632/oncotarget.12266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Travis WD. Update on small cell carcinoma and its differentiation from squamous cell carcinoma and other non-small cell carcinomas. Mod Pathol. 2012;25:S18–30. doi: 10.1038/modpathol.2011.150. [DOI] [PubMed] [Google Scholar]
- 32.Thunnissen E, Borczuk AC, Flieder DB, Witte B, Beasley MB, Chung JH, et al. The use of immunohistochemistry improves the diagnosis of small cell lung cancer and its differential diagnosis. An International Reproducibility Study in a Demanding Set of Cases. J Thorac Oncol. 2017;12:334–46. doi: 10.1016/j.jtho.2016.12.004. [DOI] [PubMed] [Google Scholar]
- 33.Tay RY, Fernandez-Gutierrez F, Foy V, Burns K, Pierce J, Morris K, et al. Prognostic value of circulating tumour cells in limited-stage small-cell lung cancer: analysis of the concurrent once-daily versus twice-daily radiotherapy (CONVERT) randomised controlled trial. Ann Oncol. 2019;30:1114–20. doi: 10.1093/annonc/mdz122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shishido SN, Carlsson A, Nieva J, Bethel K, Hicks JB, Bazhenova L, et al. Circulating tumor cells as a response monitor in stage IV non-small cell lung cancer. J Transl Med. 2019;17:294. doi: 10.1186/s12967-019-2035-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kiniwa Y, Nakamura K, Mikoshiba A, Ashida A, Akiyama Y, Morimoto A, et al. Usefulness of monitoring circulating tumor cells as a therapeutic biomarker in melanoma with BRAF mutation. BMC Cancer. 2021;21:287. doi: 10.1186/s12885-021-08016-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frese KK, Simpson KL, Dive C. Small cell lung cancer enters the era of precision medicine. Cancer Cell. 2021;39:297–9. doi: 10.1016/j.ccell.2021.02.002. [DOI] [PubMed] [Google Scholar]
- 37.Qu, S, Fetsch, P, Thomas, A, Pommier, Y, Schrump, DS, Miettinen, MM et al. Molecular Subtypes of Primary SCLC Tumors and Their Associations With Neuroendocrine and Therapeutic Markers. J Thorac Oncol. 2021; 10.1016/j.jtho.2021.08.763. [DOI] [PMC free article] [PubMed]
- 38.Winkler C, Armenia J, Jones GN, Tobalina L, Sale MJ, Petreus T, et al. SLFN11 informs on standard of care and novel treatments in a wide range of cancer models. Br J Cancer. 2021;124:951–62. doi: 10.1038/s41416-020-01199-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mezzadra R, de Bruijn M, Jae LT, Gomez-Eerland R, Duursma A, Scheeren FA, et al. SLFN11 can sensitize tumor cells towards IFN-gamma-mediated T cell killing. PLoS ONE. 2019;14:e0212053. doi: 10.1371/journal.pone.0212053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mavrommatis E, Fish EN, Platanias LC. The schlafen family of proteins and their regulation by interferons. J Interferon Cytokine Res. 2013;33:206–10. doi: 10.1089/jir.2012.0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.