Abstract
The nucleotide-binding oligomerization domain protein 2 (NOD2) senses bacterial peptidoglycan to induce proinflammatory and antimicrobial responses. Dysregulation of NOD2 signaling is involved in multiple inflammatory disorders. Recently, S-palmitoylation, a novel type of post-translational modification, is reported to play a crucial role in membrane association and ligand-induced signaling of NOD2, yet its influence on the stability of NOD2 is unclear. Here we show that inhibition of S-palmitoylation facilitates the SQSTM1/p62-mediated autophagic degradation of NOD2, while S-palmitoylation of NOD2 by ZDHHC5 promotes the stability of NOD2. Furthermore, we identify a gain-of-function R444C variant of NOD2 short isoform (NOD2s-R444C) in autoinflammatory disease, which induces excessive inflammation through its high S-palmitoylation level. Mechanistically, the NOD2s-R444C variant possesses a stronger binding ability to ZDHHC5, which promotes its S-palmitoylation, and restricts its autophagic degradation by reducing its interaction with SQSTM1/p62. Taken together, our study reveals the regulatory role of S-palmitoylation in controlling NOD2 stability through the crosstalk with autophagy, and provides insights into the association between dysfunctional S-palmitoylation and the occurrence of inflammatory diseases.
Subject terms: Autophagy, Inflammation
Introduction
The innate immune system serves as the first line of host defense by detecting microbes and initiating inflammatory responses. Recognition of microbial pathogens by pattern recognition receptors (PRRs) is crucial to trigger downstream innate immune signaling cascade [1]. The nucleotide-binding oligomerization domain (NOD)-containing protein 2 (NOD2), also known as CARD15, is a well-characterized PRR of the NOD-like receptor (NLR) family and functions as an intracellular sensor of bacterial cell wall peptidoglycan (PGN) fragments [2]. NOD2 contains tandem caspase activation and recruitment domains (CARDs), a NOD domain, and multiple leucine-rich repeats regions [3]. Once activated by muramyl dipeptide (MDP), a conserved motif common to the PGN, NOD2 undergoes self-oligomerization via the central NOD domain, which in turn promotes the recruitment of the downstream receptor-interacting serine-threonine kinase 2 through CARD-CARD interactions, and subsequently triggers the activation of mitogen-activated protein kinase and nuclear factor-κB (NF-κB) signaling [4]. Ultimately, NOD2 signaling contributes to host defense via transcriptional upregulation of various proinflammatory cytokines [5].
Recent studies suggested that dysregulation of NOD2 signaling could confer susceptibility to several immunologic and inflammatory diseases, including Crohn’s disease (CD), Blau syndrome (BS), and early-onset sarcoidosis, underscoring the critical role of NOD2 activation in inflammatory homeostasis [5]. Gain-of-function NOD2 variants have been confirmed to lead to increased basal NF-κB activity in BS, while other loss-of-function NOD2 variants have been indicated to be incapable of active NOD2 signaling in CD [6–8]. TRIM22 variants that affect K63-linked NOD2 polyubiquitination, and consequent NOD2-dependent NF-κB and type I interferon signaling, are demonstrated to be associated with very-early-onset inflammatory bowel disease [9]. However, it is still poorly understood how NOD2 variants contribute to disease pathogenesis.
Accumulating evidence indicates that NOD2-mediated signaling can be modulated by multiple post-translational modifications [8]. TRIM27-induced K48-linked polyubiquitination facilities the 26S proteasome-mediated degradation of NOD2 [10], while TRIM22-mediated K63-linked polyubiquitination of NOD2 is required for the activation of NOD2 signaling, but not NOD2 degradation [9]. NOD2 can also be O-GlcNAcylated, which increases the half-life of NOD2 [11]. Recently, palmitoylation of proteins by aspartate-histidine-histidine-cysteine (DHHC) palmitoylacyltransferases has been shown to regulate protein localization, trafficking, and degradation [12, 13]. Strikingly, ZDHHC5-mediated S-palmitoylation of NOD2 is reported as a required step of its membrane recruitment for subsequent PGNs-triggered responses [14]. However, it remains unknown whether S-palmitoylation can modulate the protein stability of NOD2.
In this study, we found that S-palmitoylation of NOD2 by ZDHHC5 inhibits the SQSTM1/p62-mediated autophagic degradation of NOD2 to positively regulate NF-κB signaling. Moreover, we demonstrated that the disease-associated R444C variant in the short isoform of NOD2 (NOD2s-R444C), but not the corresponding variant in long isoform of NOD2 (NOD2l-R471C), potentiates NF-κB signaling and inflammation in a S-palmitoylation-dependent manner. Mechanistically, NOD2s-R444C variant enhances its interaction with ZDHHC5 for S-palmitoylation, which prevents NOD2 from SQSTM1/p62-mediated autophagic degradation. Our findings deepen our understanding of the function of S-palmitoylation in immune regulation and provide insights into the crosstalk between autophagy and inflammation.
Results
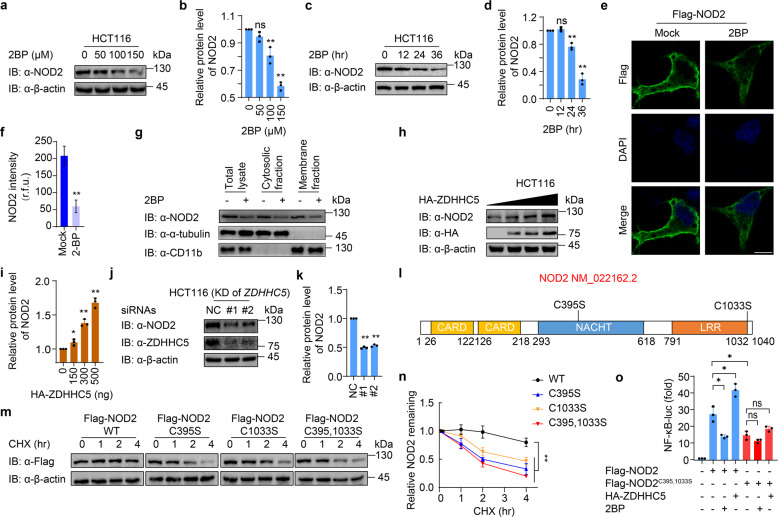
S-palmitoylation enhances the protein stability of NOD2
According to a previous study, NOD2 could be S-palmitoylated by ZDHHC5, which is a required step for membrane recruitment during pathological conditions [14]. Since S-palmitoylation might also control protein trafficking and stability [15, 16], we checked whether S-palmitoylation affects NOD2 degradation by applying 2-bromopalmitate (2BP), a general S-palmitoylation inhibitor, to reduce the S-palmitoylation in HCT116 cells (a colorectal epithelial cell line). The protein levels of NOD2 were reduced by 2BP treatment in both dose-dependent (Fig. 1a, b) and time-dependent manners (Fig. 1c, d). Yan et al. observed that treatment of 50 μM 2BP affected the redistribution of NOD2 from membranes to the cytosol, and 100 μM 2BP treatment substantially reduced the levels of NOD2 associated with membrane fraction but did not apparently affect cytosolic NOD2 protein levels in HCT116 cells [14]. These results were consistent with our study that total NOD2 protein levels remained unchanged with the treatment of 50 μM 2BP and slightly decreased with the treatment of 100 μM 2BP, as shown in Fig. 1a–d. Notably, when the concentration of 2BP was increased to 150 μM, or the duration of 2BP treatment extended to 36 h, we observed a significant decrease of NOD2 protein abundance. We further confirmed the effect of 2BP (100 μM, 24 h) on NOD2 by immunofluorescence. Indeed, the membrane localization of NOD2 was no longer obvious and the total fluorescence intensity of NOD2 protein largely decreased upon 2BP treatment (Fig. 1e, f). Such dose-dependent degradation of NOD2 by 2BP was also observed in THP-1-derived macrophages (THP-1-DMs) (Supplementary Fig. S1a). Considering the influence of the S-palmitoylation of NOD2 on its localization, we performed a subcellular fractionation assay. We found that 2BP (100 μM, 24 h) treatment significantly reduced the protein abundance of NOD2 in both cytosolic and membrane fractions, leading to a decrease of NOD2 level in total cell lysates (Fig. 1g). We, therefore, speculated that strong suppression of S-palmitoylation by 2BP could accelerate NOD2 protein degradation to eliminate the redundant NOD2 translocated from the membrane. Consistent with our hypothesis, overexpression of ZDHHC5 increased the protein level of NOD2 in a dose-dependent manner, while knockdown of ZDHHC5 significantly decreased NOD2 abundance (Fig. 1h–k). It was demonstrated that the cysteine residues in 395 and 1033 of NOD2 are the S-palmitoylation sites [14]. We constructed vectors expressing NOD2 single Cys to Ser mutants NOD2C395S or NOD2C1033S or double Cys mutant NOD2C395,1033S (Fig. 1l), and found that these mutants largely reduced the S-palmitoylation of NOD2 (Supplementary Fig. S1b). We further observed that these S-palmitoylation-deficient mutants remarkably accelerated the degradation of NOD2 (Fig. 1m, n).
Fig. 1. S-palmitoylation of NOD2 by ZDHHC5 enhances the protein stability of NOD2.
a Immunoblot analysis showing NOD2 degradation in HCT116 cells treated with 2BP at the indicated concentrations (0, 50, 100, and 150 μM) for 24 h. b Quantification of the protein level of NOD2 after 2BP treatment in (a). c Immunoblot analysis showing NOD2 degradation in HCT116 cells treated with 100 μM 2BP for 0, 12, 24, and 36 h. d Quantification of the protein level of NOD2 after 2BP treatment in (c). e Immunofluorescence staining of Flag-NOD2 in HEK293T cells with or without 2-BP (100 μM, 24 h) treatment. The nucleus is stained with 4′,6-diamidino-2-phenylindole (DAPI). Scale bars, 10 μm. f Quantification of the NOD2 intensity (30 cells per sample). r.f.u. relative fluorescence unit. g Cytosolic, and membrane fractions of THP-1-derived macrophages (THP-1-DM) pretreated with DMSO or 2BP (100 μM) for 24 h. THP-1 cells were differentiated with PMA (100 ng/ml) overnight. h Immunoblot analysis showing stabilization of NOD2 in HCT116 cells transfected with 0-500 ng ZDHHC5 for 24 h. i Quantification of the protein level of NOD2 after overexpression of ZDHHC5 in (h). j Immunoblot analysis showing NOD2 degradation in HCT116 cells after knockdown of ZDHHC5 for 36 h. k Quantification of the protein level of NOD2 with overexpression of ZDHHC5 in (j). l Schematic view of NOD2 (full-length) and the location of the S-palmitoylation-deficient NOD2 variants. m Immunoblot analysis showing the stability of wild-type (WT) or S-palmitoylation-deficient NOD2 mutants. Cells were treated with CHX (100 μg/ml) for 0, 1, 2 and 4 h, and then collected and lysed for IB. n Quantification of WT or S-palmitoylation-deficient NOD2 mutants in (m). o NF-κB-dependent luciferase reporter gene activities in HEK293T cells expressing Flag-NOD2 or Flag-NOD2C395,1033S, together with HA-empty vector or HA-ZDHHC5, treated with DMSO or 2BP (100 μM, 24 h). NF-κB promoter-driven luciferase activity was measured and normalized to the renilla luciferase activity. In b, d, f, i, k, n, and o, all error bars, mean values ± SEM, P values were determined by unpaired two-tailed Student’s t-test of n = 3 independent biological experiments. *P < 0.05; **P < 0.01; ns not significant. For a, c, e, g-h, j, and m, similar results are obtained from three independent biological experiments.
To further investigate the influence of such S-palmitoylation-mediated NOD2 stabilization on its downstream signaling, we examined the NOD2-mediated NF-κB activation by manipulating NOD2 S-palmitoylation. We found that NOD2-induced NF-κB activation was decreased by 2BP treatment or mutation of S-palmitoylation sites of NOD2, while ZDHHC5 overexpression promoted the activation of NOD2 signaling. Moreover, neither ZDHHC5 overexpression nor 2BP treatment could significantly change the NOD2-mediated NF-κB activation when NOD2 is deficient in S-palmitoylation (Fig. 1o). Collectively, these results indicate that S-palmitoylation of NOD2 inhibits its degradation and strengthens the NOD2-mediated NF-κB signaling.
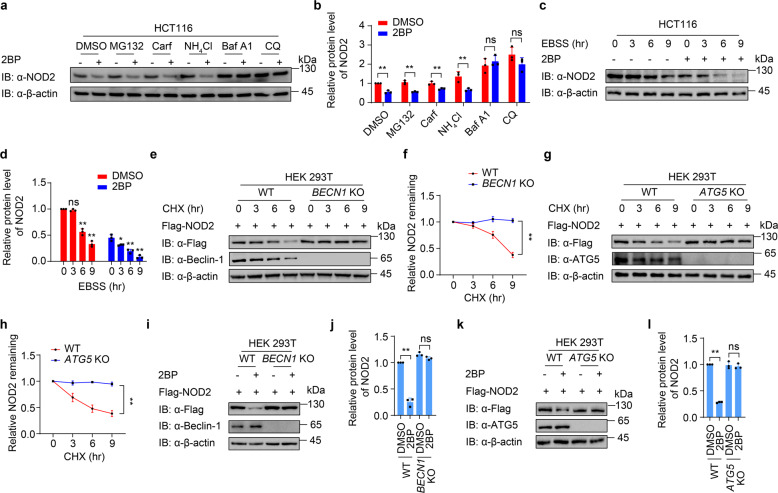
S-palmitoylation attenuates the autophagic degradation of NOD2
To figure out which degradation pathway of NOD2 is dominantly regulated by S-palmitoylation, we determined whether the degradation of NOD2 under 2BP treatment could be abrogated by the inhibitors of proteasome or the autophagy degradation pathway. Our results showed that the degradation of NOD2 by 2BP treatment could be blocked by autophagy and autolysosome inhibitors bafilomycin A1 (Baf A1) and chloroquine (CQ), but not the proteasome inhibitor MG132 and carfilzomib, indicating that suppressed S-palmitoylation might result in the autophagic degradation of both ectopically expressed NOD2 in HEK293T cells and endogenous NOD2 in HCT116 cells (Supplementary Fig. S2a, b and Fig. 2a, b). It is worth noting that the protein levels of ectopic NOD2 in the cells treated with the proteasome inhibitor MG132 and carfilzomib were significantly elevated when compared to that with DMSO treatment no matter 2BP was added or not (Supplementary Fig. S2a, b), indicating that the overexpressed NOD2 undergoes proteasomal degradation, which is consistent with the observations by Zurek et al. [10]. We next observed that autophagy-initiated by Earle’s balanced salt solution (EBSS) treatment remarkably degraded NOD2, and 2BP further facilitated EBSS-induced NOD2 degradation (Fig. 2c, d). To further demonstrate that NOD2 degrades through the autophagy pathway, we detected the protein degradation of NOD2 in BECN1 or ATG5 knockout (KO) cells, in which autophagy was largely impaired. The degradation of NOD2 was significantly abolished in these autophagy-deficient cells (Fig. 2e–h). In addition, the degradation of NOD2 by 2BP treatment was mostly abrogated in BECN1 or ATG5 KO cells (Fig. 2i–l). We next observed that the degradation of NOD2 by ZDHHC5 depletion was restored by CQ treatment in THP-1 DMs (Supplementary Fig. S3). Taken together, these data indicate that S-palmitoylation prevents NOD2 from autophagic degradation.
Fig. 2. Palmitoylation attenuates the autophagic degradation of NOD2.
a Immunoblot analysis showing NOD2 protein level in HCT116 cells treated with proteasome inhibitors MG132 (10 μM), carfilzomib (100 nM), or autolysosome inhibitor ammonium chloride (NH4Cl, 20 mM), bafilomycin A1 (Baf A1, 0.2 μM), or chloroquine (CQ, 50 μM) for 6 h. b Quantification of relative protein level of NOD2 in (a). c Immunoblot analysis showing NOD2 degradation in HCT116 cells treated with DMSO or 2BP (100 μM, 24 h) upon starvation-induced autophagy activation under Earle’s balanced salt solution (EBSS)-cultured condition for 0, 3, 6 or 9 h. d Quantification of relative protein level of NOD2 in (c). e, h Immunoblot analysis showing protein level of Flag-NOD2 in wild-type (WT) and BECN1 (e) or ATG5 (g) knockout (KO) HEK293T cells. Cells were transfected with Flag-NOD2 for 24 h, and then treated with CHX (100 μg/ml) for 0, 3, 6 and 9 h. f, h Quantification of relative NOD2 remaining in (e and g). i–l Immunoblot analysis showing protein level of Flag-NOD2 in WT and BECN1 (i, j) or ATG5 (k, l) KO HEK293T cells treated with DMSO or 2BP (100 μM, 24 h). j, l Quantification of relative protein level of NOD2 in (i and k). In b, d, f, j, and l, all error bars, mean values ± SEM, P values were determined by unpaired two-tailed Student’s t-test of n = 3 independent biological experiments. *P < 0.05; **P < 0.01; ns not significant. For a, c–e, g, i, k, similar results are obtained from three independent biological experiments.
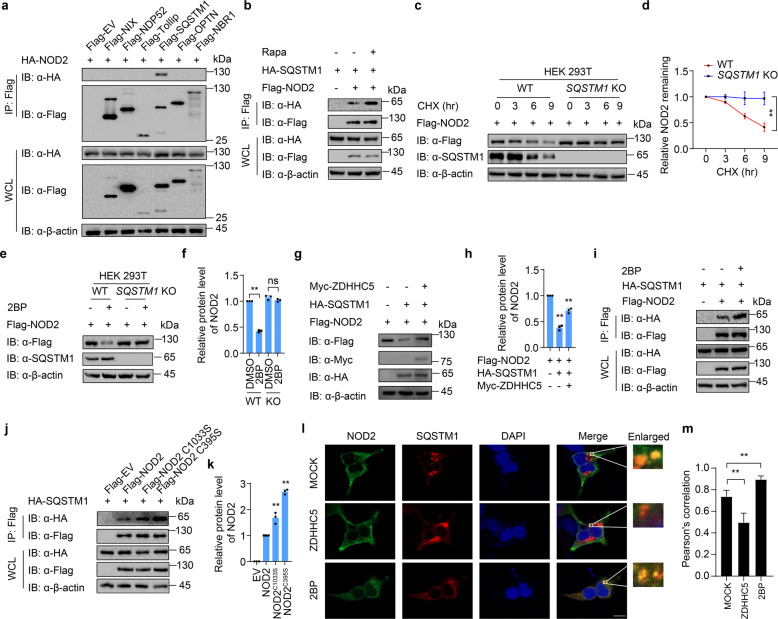
Cargo receptor SQSTM1/p62 is required for NOD2 degradation
During the autophagy process, cargo receptors are responsible to deliver substrates to the autophagosome for selective degradation [17]. To investigate which cargo receptor mediates the autophagic degradation of NOD2, we detected the interaction of NOD2 and several reported cargo receptors, including NIX, NDP52, Tollip, SQSTM1/p62, OPTN, and NBR1. Co-immunoprecipitation (Co-IP) assays showed that NOD2 mainly interacted with SQSTM1/p62 rather than other cargo receptors (Fig. 3a). Moreover, rapamycin (Rapa)-induced autophagy enhanced the interaction of NOD2 and SQSTM1/p62 (Fig. 3b). CHX assays indicated that the degradation of NOD2 was almost disappeared in SQSTM1 KO cells (Fig. 3c, d). Thus, our results suggest that the autophagic degradation of NOD2 is mediated by cargo receptor SQSTM1/p62.
Fig. 3. Palmitoylation attenuates the interaction between NOD2 and cargo receptor SQSTM1/p62.
a HEK293T cells were transfected with HA-NOD2 and indicated Flag-tagged cargo receptors. Cell lysates were co-immunoprecipitated with anti-Flag beads and immunoblotted with anti-HA antibody. b HEK293T cells were transfected with HA-NOD2 and Flag-tagged SQSTM1, followed by rapamycin (250 nM) treatment for 24 h together with Baf A1 (0.2 μM). Cell lysates were co-immunoprecipitated with anti-Flag beads and immunoblotted with anti-HA antibody. c Immunoblot analysis showing protein level of Flag-NOD2 in wild-type (WT) and SQSTM1 knockout (KO) HEK293T cells, treated with CHX (100 μg/ml) for indicated time points. d Quantification of relative NOD2 remaining in (c). e Immunoblot analysis showing protein level of Flag-NOD2 in WT and SQSTM1 KO HEK293T cells transfected with Flag-NOD2 for 24 h, followed by treatment of DMSO or 2BP (100 μM, 24 h). f Quantification of relative protein level of NOD2 in (e). g Immunoblot analysis showing protein level of Flag-NOD2 in HEK293T cells transfected with Flag-NOD2, together with HA-SQSTM1 or Myc-ZDHHC5 for 24 h. h Quantification of relative protein level of NOD2 in (g). i HEK293T cells were transfected with Flag-NOD2 and HA-SQSTM1, with or without 2BP treatment, followed by rapamycin (250 nM) treatment together with Baf A1 (0.2 μM). Cell lysates were co-immunoprecipitated with anti-Flag beads and immunoblotted with anti-HA antibody. j HEK293T cells were transfected with Flag-tagged WT or S-palmitoylation-deficient NOD2, together with HA-SQSTM1 and Myc-ZDHHC5. Cell lysates were co-immunoprecipitated with anti-Flag beads and immunoblotted with anti-HA antibody. k Quantification of relative protein level of NOD2 in (k). l Immunofluorescence staining of Flag-NOD2 (green) and HA-SQSTM1 (red) in HeLa cells with or without 2BP (100 μM) treatment, together with EBSS and CQ treatment. The nucleus is stained with DAPI. Scale bars, 10 μm. m Statistics of colocalization of NOD2 and SQSTM1/p62 indicated by Pearson’s correlation (30 cells per sample). In d, f, h, k, and m, all error bars, mean values ± SEM, P values were determined by unpaired two-tailed Student’s t-test of n = 3 independent biological experiments. *P < 0.05; **P < 0.01; ns not significant. For a–c, e, g, i, and j, similar results are obtained from three independent biological experiments.
To further confirm that S-palmitoylation restrains SQSTM1/p62-mediated autophagic degradation of NOD2, we applied 2BP to inhibit the S-palmitoylation in wild-type (WT) and SQSTM1 KO cells. The protein levels of NOD2 were decreased with the treatment of 2BP in WT cells, while no NOD2 degradation was observed in SQSTM1 KO cells (Fig. 3e, f). We also found that overexpression of SQSTM1/p62 led to NOD2 degradation, but this phenomenon was partially rescued in the presence of ectopic ZDHHC5 (Fig. 3g, h). Moreover, S-palmitoylation of NOD2 by ZDHHC5 restricted the interaction between NOD2 and SQSTM1/p62 (Supplementary Fig. S4). In contrast, inhibition of NOD2 S-palmitoylation by 2BP or mutation of S-palmitoylation sites enhanced NOD2-SQSTM1/p62 interaction (Fig. 3i–k). Confocal microscopy also revealed that the colocalization of NOD2 and SQSTM1/p62 was significantly reduced by ZDHHC5 overexpression, while 2BP treatment markedly enhanced NOD2-SQSTM1/p62 colocalization (Fig. 3l, m). Since SQSTM1/p62 was reported to bind to ubiquitinated protein by its ubiquitin-associated (UBA) domain, we compared the ubiquitination level of WT and S-palmitoylation-deficient NOD2, and found that S-palmitoylation-deficiency in both two main NOD2 isoforms (the canonical NOD2l, 1040 bp; short form NOD2s, 1013 bp; NOD2s differs from NOD2l by deficiency of the first 27 amino acids) showed the same level of ubiquitination (Supplementary Fig. S5a). We next determined the binding of NOD2 and SQSTM1/p62 or SQSTM1/p62 ΔUBA, and found that the interaction between NOD2 and SQSTM1/p62 did not require the UBA domain of p62 (Supplementary Fig. S5b). These results excluded the possibility that S-palmitoylation affects the ubiquitination of NOD2 to influence the binding of SQSTM1/p62 and NOD2. Collectively, our results show that S-palmitoylation decreased the interaction between NOD2 and SQSTM1/p62, thus inhibiting NOD2 delivery to the autophagosome for selective degradation.
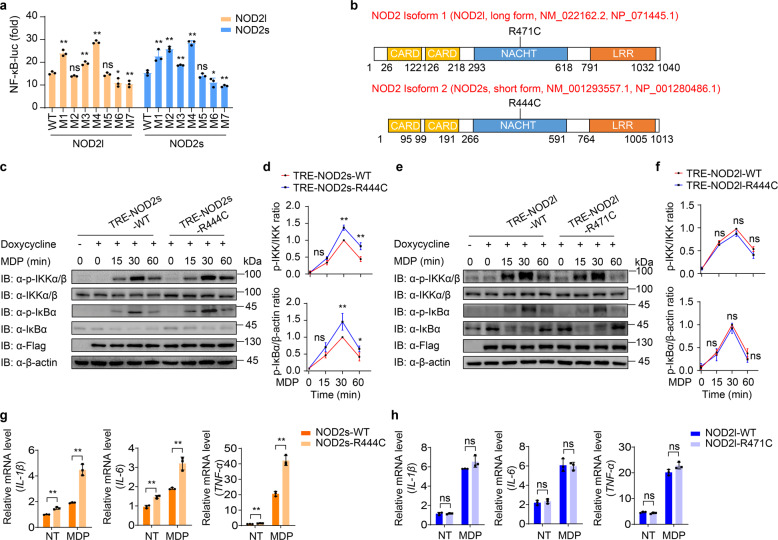
NOD2s-R444C variant causes increased NF-κB activation and inflammation
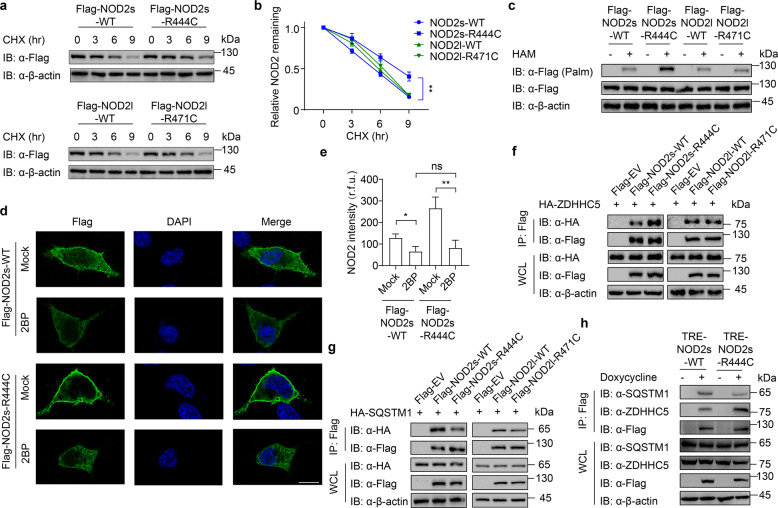
Many dysfunctional NOD2 variants are closely related to the occurrence of inflammatory diseases, but the exact molecular mechanisms by which they cause disease remain unclear [5, 18]. Considering that there are two main NOD2 isoforms, we examined NF-κB activation of several reported NOD2 variants in both NOD2l and NOD2s isoforms (R391W, R471C, G481D, C495Y, R587C, A755V and R702W in NOD2l, and corresponding variants in NOD2s). Among them, we found the NOD2l R471C variant (NOD2l-R471C) did not affect the NOD2-mediated NF-κB activation, but the NOD2s R444C variant (NOD2s-R444C, which has the same amino acid mutation) could significantly enhance NF-κB activation, compared with that of WT NOD2 (Fig. 4a). The NOD2l-R471C or NOD2s-R444C variant is encoded by rs1078327 and has been reported in patients suffering from IBD, BS, acute myeloid leukemia or aggressive periodontitis [19–22]. The allele frequency for this site is 0.01591 in gnomAD East Asian cohort, and 0.001485 in the entire gnomAD exomes cohort. Sequence alignment analysis revealed that NOD2l-R471 or NOD2s-R444 site was located at the NACHT domain of NOD2 (Fig. 4b).
Fig. 4. NOD2s-R444C variant specifically promotes NF-κB activation and inflammation.
a Luciferase activity in 293T cells transfected with the NF-κB luciferase reporter, pRL-TK, together with a vector encoding wild-type (WT) NOD2l, NOD2s, or indicated NOD2 variants (NOD2l: M1, R391W; M2, R471C; M3, G481D; M4, C495Y; M5, R587C; M6, A755V; M7, R702W. NOD2s: M1, R364W; M2, R444C; M3, G454D; M4, C468Y; M5, R560C; M6, A728V; M7, R675W). b Schematic view of NOD2 (full-length/long isoform, NOD2l; short isoform, NOD2s) and the location of the NOD2l-R471C and NOD2s-R444C variants. c–f Immunoblot analysis of muramyl dipeptide (MDP, 10 μg/ml)-induced phosphorylation of IKK and IκBα in NOD2s-WT/R444C (c–d) or NOD2l-WT/R471C (e–f) THP-1-DM cells. Quantitative comparison of signaling activation between WT and NOD2s-R444C (d) or NOD2l-R471C (f) variant cells by density scanning of the immunoblots in three independent experiments. g, h Real-time PCR for IL-1β, IL-6, and TNFα transcription in MDP (10 μg/ml)-induced NOD2s-WT/R444C (g) or NOD2l-WT/R471C (h) THP-1-DM cells. Cells in (c–h) were differentiated with PMA (100 ng/ml) overnight and subsequently treated with Dox (100 ng/ml) for 10 h. In a, d, f, g, and h, all error bars, mean values ± SEM, P values were determined by unpaired two-tailed Student’s t-test of n = 3 independent biological experiments. *P < 0.05; **P < 0.01; ns not significant. For c and e, similar results are obtained from three independent biological experiments.
To explicitly demonstrate the excessive NF-κB activation induced by NOD2s-R444C variant, we generated doxycycline (Dox)-inducible NOD2l-WT, NOD2l-R471C, NOD2s-WT, and NOD2s-R444C macrophages using a human monocytic THP-1 cell line. After doxycycline treatment, these THP-1 cell lines could induce the expression of Flag-tagged NOD2 (Supplementary Fig. S6a, b). We then differentiated these THP-1 cells into macrophages, and found that NOD2s-R444C cells showed enhanced MDP-induced phosphorylation of IKK and IκBα compared to that of NOD2s-WT cells (Fig. 4c, d). However, similar p-IKK and p-IκBα level were observed in NOD2l-WT and NOD2l-R471C THP-1 DMs (Fig. 4e, f). Consistently, MDP stimulation resulted in higher expression levels of pro-inflammatory cytokines IL-1β, IL-6 and TNF-α in NOD2s-R444C cells, compared to those in NOD2s-WT cells (Fig. 4g), while the same amount of these proinflammatory cytokines were expressed in NOD2l-WT and NOD2l-R471C THP-1 DMs (Fig. 4h). These results confirm that NOD2s-R444C variant markedly promotes NOD2s-mediated NF-κB activation, but NOD2l-R471C variant has no significant effect on NOD2l-mediated NF-κB activation in human macrophages.
R444C mutation promotes NOD2s S-palmitoylation to enhance its stability
Next, we attempted to explore the molecular mechanisms that underlie the enhanced activation of NOD2s-R444C. CHX-chase assay revealed that NOD2s-R444C variant increased NOD2s stability, but NOD2l-R471C showed the same degradation rate as NOD2l-WT (Fig. 5a, b). Since we found that S-palmitoylation attenuated the SQSTM1/p62-mediated autophagic degradation of NOD2, we next checked whether the NOD2s-R444C variant keeps NOD2s stability by affecting its S-palmitoylation. Indeed, acyl-biotin exchange (ABE) assays indicated that R444C mutation increased the S-palmitoylation of NOD2s, while there was no obvious change of S-palmitoylation in NOD2l-R471C variant (Fig. 5c).
Fig. 5. R444C variant promotes NOD2s palmitoylation through enhancing its binding ability with ZDHHHC5.
a Immunoblot analysis showing degradation of NOD2s-WT/R444C or NOD2l-WT/R471C, under CHX (100 μg/ml) treatment for 0, 3, 6, and 9 h. b Quantification of NOD2s-WT/R444C or NOD2l-WT/R471C in (a). c Streptavidin blot detection of palmitoylated NOD2s-WT/R444C or NOD2l-WT/R471C in HEK293T cells transfected with Flag-tagged NOD2s-WT/R444C or NOD2l-WT/R471C by acyl-biotin exchange (ABE) assay with or without hydroxylamine (HAM, 1 M). The S-palmitoylated NOD2 in immunoprecipitated samples were detected using anti-Flag antibody. d Immunofluorescence staining of Flag-NOD2s-WT or Flag-NOD2s-R444C in HEK293T cells transfected with the same amount of Flag-NOD2s-WT or Flag-NOD2s-R444C plasmids, with or without 2-BP (100 μM, 24 h) treatment. The nucleus is stained blue with DAPI. Scale bars, 10 μm. e Quantification of the NOD2 intensity (30 cells per sample). r.f.u. relative fluorescence unit. f, g Co-immunoprecipitation and immunoblot analysis showing the interaction of NOD2 and ZDHHC5 (f) or SQSTM1 (g) in HEK293T cells transfected with Flag-tagged NOD2s-WT/R444C or NOD2l-WT/R471C and HA-ZDHHC5 (f) or HA-SQSTM1 (g) for 24 h. h Immunoblot analysis showing the interaction of NOD2 and ZDHHC5 or SQSTM1 in MDP (10 μg/ml)-induced NOD2s-WT/R444C THP-1-DM cells. Cells were differentiated with PMA (100 ng/ml) overnight and subsequently treated with Dox for 10 h. In a–c and f–h, we adjusted the amount of transfected plasmids or Dox to make the protein level of NOD2 the same. In b and e, all error bars, mean values ± SEM, P values were determined by unpaired two-tailed Student’s t-test of n = 3 independent biological experiments. *P < 0.05; **P < 0.01; ns not significant. For a, c, d, and f–h, similar results are obtained from three independent biological experiments.
Previously, we used endogenous NOD2l, the most canonical isoform of NOD2, to study the mechanism that S-palmitoylation by ZDHHC5 inhibits autophagic degradation of NOD2. We further confirmed that NOD2s also underwent autophagic degradation by 2BP treatment, which could be restored by autolysosome inhibitor CQ (Supplementary Fig. S7a). Moreover, EBSS-induced autophagy also led to NOD2s degradation, which could be accelerated by 2BP (Supplementary Fig. S7b). In addition, both NOD2s and NOD2l exhibited an equivalent binding ability to ZDHHC5 (Supplementary Fig. S7c).
To determine whether the enhanced stability of NOD2s-R444C variant comes from its low level of S-palmitoylation, we treated NOD2s-R444C or NOD2s-WT cells with 2BP, and found that the fluorescence intensity of NOD2s-R444C decreased to the same level as that of NOD2s-WT (Fig. 5d, e). We further performed a Co-IP assay and found that the interaction of ZDHHC5 and NOD2s-R444C was much stronger than that of NOD2-WT (Fig. 5f). In addition, NOD2s-R444C variant showed the weaker binding ability to SQSTM1/p62 compared to that of NOD2s-WT (Fig. 5g). However, NOD2l-R471 and NOD2l-WT exhibited the same binding abilities with both ZDHHC5 and SQSTM1/p62. The structural basis underlying NOD2-ZDHHC5 interactions has not been resolved yet. We got the predicted structure of human NOD2 and ZDHHC5 produced by AlphaFold [23]. Computational analysis based on the structure prediction by PyMOL for NOD2-ZDHHC5 complex suggested that the nearest amino acid residue of R471 site of NOD2l and R444 site of NOD2s in ZDHHC5 is 139N (hydrogen bond distance: 2.36, 4.49, and 5.43), and the nearest amino acid residue of C471 site of NOD2l in ZDHHC5 is A107 and T108 (hydrogen bond distance: 2.99, 3.86, and 5.38), while the nearest amino acid residue of C444 site of NOD2s in ZDHHC5 is 151L (hydrogen bond distance: 1.91, 4.1, and 4.06). It is likely that the absence of these 27 amino acids leads to an increased association with ZDHHC5 due to the change of spatial structure and the increase of surface binding force (Supplementary Fig. S8). The enhanced binding ability with ZDHHC5 and attenuated binding ability with SQSTM1/p62 by NOD2s-R444C variant were further confirmed in THP-1-DMs (Fig. 5h). To address whether the reduced binding of SQSTM1/p62 for the R444C variant is caused by altered localization of NOD2, we determined the binding of SQSTM1/p62 and NOD2 in cytosolic fractions. We still found that R444C variant, but not R471C variant, decreased the interaction of NOD2 and SQSTM1/p62 (Supplementary Fig. S9a). To figure out if this decrease is due to altered ubiquitination of NOD2, we detected ubiquitination of NOD2l/s and their variants accordingly, and found that all the indicated variants of NOD2 showed an equal level of ubiquitination as WT NOD2 did (Supplementary Fig. S9b). Finally, we examined whether it is caused by a binding competition between ZDHHC5 and SQSTM1/p62 to NOD2, and our results showed that ZDHHC5 and SQSTM1/p62 competitively interacted with NOD2 (Supplementary Fig. S9c). In Supplementary Fig. S5b, our results indicated that the interaction between NOD2 and SQSTM1/p62 did not require UBA domain of p62 protein. It is possible that the binding of ZDHHC5 as well as the palmitoyl group on NOD2 mediated by ZDHHC5 could restrict the binding of SQSTM1/p62 to NOD2 and inhibit the autophagic degradation of NOD2. In summary, these results suggest that the NOD2s-R444C variant strengthens its interaction with ZDHHC5 to facilitate its S-palmitoylation, therewith restraining its binding ability with the cargo receptor SQSTM1/p62, leading to the inhibition of autophagic degradation of NOD2.
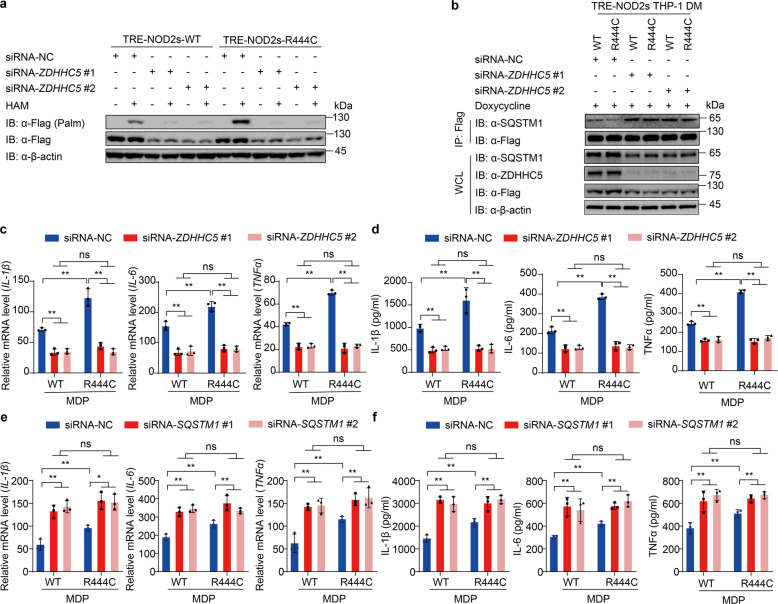
Inhibition of NOD2s-R444C S-palmitoylation impairs the excessive NOD2-induced inflammation
Since enhanced S-palmitoylation by ZDHHC5 is accounted for the promoted stability and excessive inflammation activation of NOD2s-R444C variant, we next investigated whether knockdown of ZDHHC5 by siRNAs could impair the excessive activation of NOD2s-R444C variant. Our results showed that knockdown of ZDHHC5 in THP-1-DMs markedly decreased the S-palmitoylation of both NOD2s-WT and NOD2s-R444C variant (Fig. 6a), and increased their interaction with SQSTM1/p62 (Fig. 6b). As expected, the excessive mRNA expression and secretion of IL-1β, IL-6, and TNF-α by NOD2s-R444C-mediated inflammation activation were completely eliminated by ZDHHC5 deficiency (Fig. 6c, d). We also found that knockdown of SQSTM1 increased the mRNA expression and secretion of IL-1β, IL-6, and TNF-α in NOD2s-WT and NOD2s-R444C THP-1-DMs to the same level (Supplementary Fig. S10 and Fig. 6e, f). Together, these data suggest that the excessive activation of NOD2s-R444C is due to its high level of ZDHHC5-mediated S-palmitoylation, which impeded SQSTM1/p62-dependent autophagic degradation of NOD2.
Fig. 6. Inhibition of NOD2s-R444C S-palmitoylation reverses NOD2s-induced excessive inflammatory responses.
a NOD2s-WT/R444C THP-1 cells were differentiated with PMA (100 ng/ml) overnight. After replacement of fresh medium supplemented with Dox (NOD2s-WT, 140 ng/ml; NOD2s- R444C, 100 ng/ml) for 12 h, cells were transfected with siRNA-NC or ZDHHC5 siRNAs for 36 h. Streptavidin blot detection of palmitoylated NOD2s-WT/R444C by ABE assay with or without HAM (1 M). b Immunoblot analysis showing the interaction of SQSTM1 and NOD2-WT/R444C in THP-1-DM cells supplemented with Dox (100 ng/ml) for 12 h. c, d Real-time PCR for IL-1β, IL-6 and TNFα transcription (c) and ELISA for IL-1β, IL-6, and TNFα secretion (d) in MDP (10 μg/ml)-induced NOD2s-WT/R444C THP-1-DM cells with knockdown of ZDHHC5. e, f Real-time PCR for IL-1β, IL-6, and TNFα transcription (e) and ELISA for IL-1β, IL-6 and TNFα secretion (f) in MDP (10 μg/ml)-induced NOD2s-WT/R444C THP-1-DM cells with knockdown of SQSTM1. In b–f, Dox was used at 100 ng/ml for both NOD2s-WT and NOD2s-R444C THP-1-derived macrophages. In c–f, all error bars, mean values ± SEM, P values were determined by unpaired two-tailed Student’s t-test of n = 3 independent biological experiments. *P < 0.05; **P < 0.01; ns not significant. For a, b, similar results are obtained from three independent biological experiments.
Discussion
Autophagy is a highly conserved cytoplasmic homeostasis process for the recycling and degradation of cytoplasmic constituents, and involves many human diseases, especially neurodegenerative, cancer, and inflammatory disorders [24–26]. Recently, autophagy is also reported to modulate the antimicrobial immune response by directly decreasing the levels of PAMPs or selectively degrading several PRRs, such as DDX58/RIG-I and NLRP3 [27, 28]. Selectivity of autophagy is achieved by specific cargo receptors that harbor both ubiquitin-binding domains and LC3-interacting regions, such as SQSTM1/p62, OPTN and NDP52 [25, 29]. These cargo receptors take charge of cargoes’ recognition and bridge ATG8-family proteins to ubiquitin chains or other selective signals on cargoes [17, 30]. However, the substrates and mechanisms of selective autophagy still remain largely unclear. Although the autophagic cargo receptor SQSTM1/p62 mainly recognizes ubiquitinated cargoes, more and more studies showed that SQSTM1/p62 can also bind to the substrates, such as IKKβ, RIG-I and Sindbis virus (SINV) capsid, in a ubiquitination-independent manner for autophagic degradation [31–34]. In our study, we demonstrated that NOD2, an important PRR, undergoes SQSTM1/p62-mediated autophagic degradation in a ubiquitination-independent manner. Since NOD2 S-palmitoylation by ZDHHC5 could attenuate the interaction between NOD2 and cargo receptor SQSTM1/p62, the autophagic degradation of NOD2 could be regulated by its S-palmitoylation state.
According to Cooney et al.’s report, under MDP stimulation, NOD2 mediates autophagy for both bacterial handling and generation of MHC class II antigen-specific CD4+ T cell responses in DCs [35]. SQSTM1/p62 does not play a role in such NOD2-mediated autophagy flux, but could act as a cargo receptor to promote the autophagic degradation of NOD2, which is crucial for avoiding excessive NOD2-mediated response. Ultimately, SQSTM1/p62 provides a negative feedback mechanism for NOD2 signaling and ensures appropriate response during host defense.
Selective targeting and disposal of proteins in eukaryotic cells usually occur by proteasomes and autophagy [36]. Zurek et al. reported that ectopically expressed NOD2 could be ubiquitinated followed by proteasomal degradation [10]. This is consistent with our results that the protein levels of ectopic NOD2 in the cells treated with the proteasome inhibitor MG132 and carfilzomib were significantly elevated when compared to that with DMSO treatment no matter 2BP is added or not. We also found that the degradation of endogenous NOD2 in HCT116 cells by 2BP treatment could be blocked by autophagy and autolysosome inhibitors, but not the proteasome inhibitors. Hence, it was proposed that degradation of NOD2 through the ubiquitin-proteasome pathway may occur only when NOD2 was activated.
Palmitoylation has emerged as a crucial posttranslational modification that regulates protein–membrane association and protein–protein interactions [37]. Although close to 3000 proteins are known to undergo palmitoylation, how this modification modulates inflammation and innate immune responses remain unclear [37]. A recent study unraveled that NOD2 is a palmitoylated protein, and NOD2 S-palmitoylation is required for membrane recruitment and immune signaling. They also identified ZDHHC5 as a palmitoyltransferase responsible for NOD2 S-palmitoylation [14]. Here, we found that S-palmitoylation of NOD2 by ZDHHC5 restricted its interaction with the cargo receptor SQSTM1/p62, thus inhibiting its delivery to autophagosomes for degradation and promoting NOD2-mediated inflammation. Thus, S-palmitoylation facilitates NOD2 signaling in two distinct ways: (1) direct redistribution of NOD2 to membranes, which is required for NOD2 signaling; (2) enhancement of the stability of NOD2 by preventing its autophagic degradation. Collectively, the dual functions of NOD2 S-pamitoylation orchestrate the NOD2-mediated host defense.
NOD2 signaling to bacterial PGN has long been recognized to be vital to immune homeostasis. Numerous studies showed that NOD2 loss-of-function or gain-of-function variants are associated with many diseases, including CD, BS, and early-onset sarcoidosis [8]. However, little is known about the exact molecular mechanisms by which they cause these diseases. NACHT domain displays ATPase activity and is vital for NOD2 activation and oligomerization. We found a previously ignored NOD2 mutation (NOD2s-R444C) in NACHT domain that caused increased NF-κB activation and proinflammatory cytokine production.
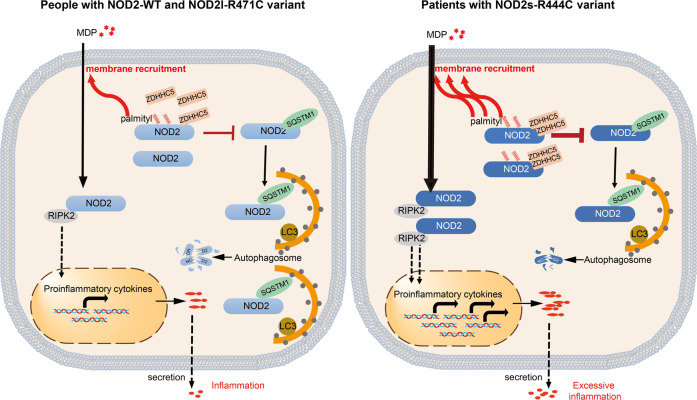
Growing evidence showed that different isoforms of a protein may be involved in diverse regulation of protein function, although they are only partially different. For instance, MyD88 is an adapter protein that is involved in Toll-like receptor (TLR)- and interleukin-1 receptor (IL-1R)-induced activation of NF-κB. However, a lipopolysaccharide-inducible isoform of MyD88, MyD88S, is not able to activate NF-κB, and in contrast, specifically prevents the recruitment of IRAK-4 into the IL-1R complex and functions as a dominant-negative inhibitor of TLR/IL-1R-induced NF-κB activation [38]. Previously, we also demonstrated that TRIM9s, but not TRIM9l, is a positive regulator of type I interferon signaling in response to both DNA and RNA viruses by recruiting GSK3β to TBK1 [39]. There are three NOD2 isoforms, and all of them can activate NF-κB signaling. Interestingly, although we and other groups found that NOD2l-R471C variant exhibited no effect on NF-κB signaling and cytokine secretion, compared to the NODl-WT [21], the corresponding variant of NOD2s-R444C markedly promoted inflammatory response. Mechanistically, NOD2s-R444C variant possessed stronger ability to interact with ZDHHC5, making it more easily to be S-palmitoylated. Enhanced S-palmitoylation of NOD2s-R444C impaired its interaction with the cargo receptor SQSTM1/p62, leading to less SQSTM1/p62-mediated autophagic degradation of NOD2. Thus, the patients with NOD2s-R444C variant would more easily overrespond to bacterial PGN fragments, and trigger excessive inflammation (Fig. 7). Knockdown of ZDHHC5 diminished NOD2 abundance and excessive inflammatory response, suggesting a strategy of targeting ZDHHC5 for anti-inflammatory therapy of these patients.
Fig. 7. A proposed working model to illustrate how the NOD2s-R444C variant contributes to excessive inflammation.
Upon MDP stimulation, NOD2 undergoes self-oligomerization and recruitment of the downstream adaptor molecule RIPK2 via homophilic CARD-CARD interaction, leading to NF-κB translocation to the nucleus and the transcription of proinflammatory genes. Normally, palmitoylation of NOD2 by ZDHHC5 tightly regulates NOD2 signaling in two ways by promoting the membrane recruitment of NOD2 and restricting the SQSTM1/p62-mediated autophagic degradation of NOD2. In the patients with NOD2s-R444C variant, palmitoylation of NOD2s by ZDHHC5 is largely increased through their enhanced protein interactions. Enhanced NOD2 palmitoylation might reduce the threshold of NOD2 activation, giving rise to excessive NOD2-mediated inflammation and the risk of developing inflammatory diseases.
In summary, we demonstrate that S-palmitoylation of NOD2 by ZDHHC5 reduces its binding ability to cargo receptor SQSTM1/p62 and restrains it from autophagic degradation, ultimately promoting NOD2-mediated inflammatory response. We further identify a gain-of-function NOD2s-R444C variant, which is associated with excessive inflammation through enhanced S-palmitoylation level. This study highlights that S-palmitoylation-regulated autophagic degradation of NOD2 is critical in the occurrence of inflammatory diseases, and would facilitate diagnosis and precision treatment for patients with NOD2-associated AIDs.
Materials and methods
Cell lines
Human embryonic kidney cells (HEK293T) and HCT116 from ATCC were cultured in DMEM (Corning) with 10% fetal bovine serum (GenStar) and 1% L-glutamine (Gibco). THP-1 cells from ATCC were maintained in RPMI-1640 medium (Gibco) with 10% fetal bovine serum and 1% L-glutamine (Gibco). THP-1 cells were differentiated into macrophages by treatment with 100 ng/ml phorbol-12-myristate-13-acetate (PMA; Millipore Sigma) for 12 h.
Plasmids and siRNA transfection
HA-, Flag-, or Myc-tagged NOD2l/NOD2s, ZDHHC5, or SQSTM1/p62 constructs were cloned into the pcDNA3.1 vector. Site-directed mutagenesis was performed with the QuickChange Lightning Kit (210519-5; Agilent Technologies) according to the manufacturer’s instructions. Chemically synthesized 21-nt siRNA duplexes were obtained from TranSheepBio and transfected using Lipofectamine RNAiMAX (13778150; Invitrogen) according to the manufacturer’s instructions.
The following RNA oligonucleotides were used in this study:
Control siRNA (siRNA-NC) forward: 5′-UUCUCCGAACGUGUCACGUTT-3′;
Reverse: 5′-ACGUGACACGUUCGGAGAATT-3′;
ZDHHC5 siRNA #1 forward: 5′-UCAGAUGAUUCAAAGAGAUTT-3′;
Reverse: 5′-AUCUCUUUGAAUCAUCUGAGG-3′;
ZDHHC5 siRNA #2 forward: 5′-AUGUCUUACAGCAGCCAAATT-3′;
Reverse: 5′-UUUGGCUGCUGUAAGACAUCG-3′;
SQSTM1 siRNA #1 forward: 5′-CGGAUAACUGUUCAGGAGGTT-3′;
Reverse: 5′-CCUCCUGAACAGUUAUCCGTT-3′;
SQSTM1 siRNA #2 forward: 5′- UCUUCCGAAUCUACAUUAATT -3′;
Reverse: 5′- UUAAUGUAGAUUCGGAAGATT-3′.
Reagents and antibodies
Doxycycline (Dox; D9891), the palmitoyl acyltransferase inhibitor, 2-bromohexadecanoic acid (2BP, 238422), MG132 (C2211), rapamycin (Rapa, 37094), chloroquine phosphate (PHR1258), NH4Cl (A9434), horseradish peroxidase (HRP)-anti-FLAG (M2) (A8592), and anti-β-actin (A1978) were purchased from Merck. Bafilomycin A1 (S1413) and carfilzomib (S2853) were purchased from Selleck. Goat anti-rabbit (sc-2004), goat anti-mouse (sc-2005), and anti-NOD2 (sc-56168) were purchased from Santa Cruz Biotechnology. Anti-c-Myc-HRP (11814150001) and anti-HA-peroxidase (high-affinity from rat immunoglobulin G1) (12013819001) were purchased from Roche Applied Science. Anti-ZDHHC5 polyclonal antibody (21324-1-AP) was purchased from Proteintech.
Immunoblot and immunoprecipitation analysis
For immunoprecipitation, whole-cell extracts were prepared after transfection or treatment, followed by incubation overnight with anti-Flag, anti-HA beads (Millipore Sigma) or Protein A/G beads (Pierce). Beads were washed five times with lysis buffer, and immunoprecipitates were eluted for SDS-PAGE. Proteins were transferred to polyvinylidene fluoride membranes (Bio-Rad), and ECL Western Blotting Substrate (Pierce) was used for protein detection.
Immunofluorescence staining analysis
For immunofluorescence staining analysis, cells were cultured on Glass Bottom culture dishes (Nest Scientific, 801002). After transfection or treatment for the indicated time, cells were fixed with 4% paraformaldehyde for 10 min, and then permeabilized in methyl alcohol for 10 min at –20 °C. After washing with PBS for three times, cells were blocked in 10% fetal goat serum (Boster Biological, AR1009) for 1 h. After washing with PBS, cells were successively incubated with primary antibodies and fluorescently labeled secondary antibody. Confocal images were examined using a microscope (LSM800; Carl Zeiss). The images were processed for gamma adjustments using Zen 3.3 or ImageJ software (National Institutes of Health).
Subcellular fractionations assays
Cells cultured on 10 cm culture dish were washed twice with cold PBS and harvested using a cell scraper. Collected cells by centrifugation (500–600 × g, 5 min). Cytosolic and membrane fractions of the collected cells were further isolated using Invent MinuteTM Plasma Membrane Protein Isolation and Cell Fractionation Kit (Invent, SM-005), according to the manufacturer’s protocols.
Luciferase reporter assays
Cells were plated in 24-well plates and transfected with plasmids encoding the NF-κB luciferase reporter (firefly luciferase; 80 ng) and pRL-TK (Renilla luciferase plasmid; 8 ng), together with different plasmids (100 ng). Cells were harvested in a passive lysis buffer (Promega, E1941). Enzyme activity was normalized by the efficiency of transfection on the basis of Renilla luciferase activity levels. Fold induction relative to the basal level was measured in cells. The values were means ± SEM of three independent transfections performed in parallel.
Generation knockout (KO) cell lines by the CRISPR cas9 system
For the BECN1, ATG5, and SQSTM1 KO HEK293T cells, single guide RNAs (sgRNAs) for each gene were annealed and cloned into pLentiCRISPRv2 (Addgene #52961) by cutting with BsmB I, and then cotransfected with Δ8.9 and VSVG into HEK293T cells. Then the transfected cells were cultured with puromycin for selection. The sequences of target-related gene are as follows:
BECN1-sgRNA: 5′-ATTTATTGAAACTCCTCGCC-3′;
ATG5-sgRNA: 5′-GTGCTTCGAGATGTGTGGTT-3′;
SQSTM1-sgRNA: 5′- TCAGGAGGCGCCCCGCAACA -3′.
Generation of doxycycline-inducible NOD2l-WT, NOD2l-R471C, NOD2s-WT, and NOD2s-R444C THP-1 cell lines
For NOD2-inducible expression, lentiviral particles were produced by transfecting HEK293T cells with pL-Teton3G-iZ-NOD2l-WT/NOD2l-R471C/NOD2s-WT/NOD2s-R444C and the Δ8.9 and VSVG lentivirus expression system. THP-1 cells were infected by incubation with lentivirus-containing supernatant for 48 h. Cells were treated with doxycycline (100–200 ng/ml) to induce expression of Flag-tagged NOD2l-WT, NOD2l-R471C, NOD2s-WT or NOD2s-R444C.
Quantitative RT-PCR assay
Total RNAs were extracted using the Trizol reagent (Invitrogen) and cDNA was generated with HiScript® II Q RT SuperMix for qPCR (+gDNA wiper) (Vazyme, R223-01), according to the manufacturer’s instructions. RT-PCR analysis assay was performed using the 2×Taq PCR StarMix (GenStar). The following primers were used:
IL-1β-F: 5′-AGCTACGAATCTCCGACCAC-3′;
IL-1β-R: 5′-CGTTATCCCATGTGTCGAAGAA-3′;
IL-6-F: 5′-ACTCACCTCTTCAGAACGAATTG-3′;
IL-6-R: 5′-CCATCTTTGGAAGGTTCAGGTTG-3′;
TNFα-F: 5′-CCTCTCTCTAATCAGCCCTCTG-3′;
TNFα-R: 5′-GAGGACCTGGGAGTAGATGAG-3′;
18S-F: 5′-CAGCCACCCGAGATTGAGCA-3′;
18S-R: 5′-TAGTAGCGACGGGCGGTGTG-3′;
β-actin-F: 5′-ACCATGTACCCTGGCATTGC-3′;
β-actin-R: 5′-CGGACTCGTCATACTCCTGC-3′.
Cytokine quantification from culture supernatants
Human IL-1β, IL-6, and TNF-α concentrations were determined by ELISA using BD OptEIA™ Human IL-1β ELISA Kit II, BD OptEIA™ Human IL-6 ELISA Kit II and BD OptEIA™ Human TNF ELISA Kit II (BD Biosciences), respectively, according to the manufacturer’s protocols.
Acyl-biotin exchange (ABE) assay
The ABE assay was performed as previously described with minor modifications [40]. In brief, HEK293T cells transiently expressing Flag-tagged NOD2 or doxycycline-inducible NOD2 THP-1 cell lines were harvested and washed with cold PBS. Prepare fresh N-ethylmaleimide (Millipore Sigma, 04260, 50 mM), protease inhibitor (Bimake, B14012, 50 mM), and phosphatase inhibitor (Roche, 04906837001, 50 mM) containing lysis buffer (LB, 50 mM Tris-HCl, 150 mM NaCl, 1 mM MgCl2, 1% NP-40, 10% glycerol) of pH 7.5. Cells were then suspended in the above buffer for 1 h at 4 °C and the cell lysates (supernatants) were incubated with anti-flag beads at 4 °C overnight. Then the beads were washed five times with LB of pH 7.5 and then three times with LB of pH 7.2. Then, the beads were incubated with hydroxylamine (HAM, Millipore Sigma, 467804, 1 M), a protease inhibitor (50 mM), and phosphatase inhibitor (50 mM) containing LB of pH 7.2 at room temperature for 1 h. Each sample was divided into two parts, one omitting the HAM cleavage step (–HAM), and one including the HAM step (+HAM). After being washed four times with LB of pH 7.2 and once with LB of pH 6.2, beads were treated with a thiol-reactive biotin molecule, Biotin-BMCC (Sangon Biotech, C100222-0050) in Lysis Buffer of pH 6.2 at 4 °C for 1 h. Gently wash all samples once in LB of pH 6.2, and three times in LB of pH 7.5. The immunoprecipitate samples were analyzed by immunoblot using anti-Flag antibody.
Statistical analysis
Data are represented as mean ± SEM unless otherwise indicated, and Student’s t-test was used for all statistical analyses with the GraphPad Prism 8 software. P values were determined by unpaired two-tailed Student’s t-test of n = 3 independent biological experiments. Differences between the two groups were considered significant when P value was less than 0.05.
Supplementary information
Author contributions
JC, SJ, and HZ conceived and directed the project. JC, LZ, SJ, and HZ drafted the manuscript. LZ, XH, LW, PW, ZC, SZ, SJ, and HZ performed the experiments and analyzed the data. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (92042303, 31870862, 32000544, 81873869, 31970700, and 32170876), Guangdong Basic and Applied Basic Research Foundation (2020B1515120090), the Natural Science Foundation of Guangdong Province (No. 2020A1515110193), and Guangzhou Women and Children’s Medical Centre project grant (0160001).
Data availability
Data supporting the present study are available from the corresponding author upon reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Edited by H Zhang
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Lingli Zhou, Xing He.
Contributor Information
Shouheng Jin, Email: jinshh3@mail.sysu.edu.cn.
Huasong Zeng, Email: zenghuasong@gwcmc.org.
Jun Cui, Email: cuij5@mail.sysu.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41418-022-00942-z.
References
- 1.Liu J, Qian C, Cao XT. Post-translational modification control of innate immunity. Immunity. 2016;45:15–30. doi: 10.1016/j.immuni.2016.06.020. [DOI] [PubMed] [Google Scholar]
- 2.Caruso R, Warner N, Inohara N, Nunez G. NOD1 and NOD2: signaling, host defense, and inflammatory disease. Immunity. 2014;41:898–908. doi: 10.1016/j.immuni.2014.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maekawa S, Ohto U, Shibata T, Miyake K, Shimizu T. Crystal structure of NOD2 and its implications in human disease. Nat Commun. 2016;7:1–11. doi: 10.1038/ncomms11813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–20. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 5.Philpott DJ, Sorbara MT, Robertson SJ, Croitoru K, Girardin SE. NOD proteins: regulators of inflammation in health and disease. Nat Rev Immunol. 2014;14:9–23. doi: 10.1038/nri3565. [DOI] [PubMed] [Google Scholar]
- 6.Nakamura N, Lill JR, Phung Q, Jiang ZS, Bakalarski C, de Maziere A, et al. Endosomes are specialized platforms for bacterial sensing and NOD2 signalling. Nature. 2014;509:240–4. doi: 10.1038/nature13133. [DOI] [PubMed] [Google Scholar]
- 7.Maeda S, Hsu LC, Liu HJ, Bankston LA, Iimura M, Kagnoff MF, et al. Nod2 mutation in Crohn’s disease potentiates NF-kappa B activity and IL-10 processing. Science. 2005;307:734–8. doi: 10.1126/science.1103685. [DOI] [PubMed] [Google Scholar]
- 8.Baker PJ, De Nardo D, Moghaddas F, Tran LS, Bachem A, Nguyen T, et al. Posttranslational modification as a critical determinant of cytoplasmic innate immune recognition. Physiol Rev. 2017;97:1165–209. doi: 10.1152/physrev.00026.2016. [DOI] [PubMed] [Google Scholar]
- 9.Li Q, Lee CH, Peters LA, Mastropaolo LA, Thoeni C, Elkadri A, et al. Variants in TRIM22 that affect NOD2 signaling are associated with very-early-onset inflammatory bowel disease. Gastroenterology. 2016;150:1196–207. doi: 10.1053/j.gastro.2016.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zurek B, Schoultz I, Neerincx A, Napolitano LM, Birkner K, Bennek E, et al. TRIM27 negatively regulates NOD2 by ubiquitination and proteasomal degradation. PloS One. 2012;7:e41255. doi: 10.1371/journal.pone.0041255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hou C-W, Mohanan V, Zachara NE, Grimes CL. Identification and biological consequences of the O-GlcNAc modification of the human innate immune receptor, Nod2. Glycobiology. 2016;26:13–18. doi: 10.1093/glycob/cwv076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang H, Zhang X, Chen X, Aramsangtienchai P, Tong Z, Lin H. Protein lipidation: occurrence, mechanisms, biological functions, and enabling technologies. Chem Rev. 2018;118:919–88. doi: 10.1021/acs.chemrev.6b00750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ko PJ, Dixon SJ. Protein palmitoylation and cancer. Embo Rep. 2018;19:e46666. doi: 10.15252/embr.201846666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu Y, Zheng YP, Coyaud E, Zhang C, Selvabaskaran A, Yu YY, et al. Palmitoylation of NOD1 and NOD2 is required for bacterial sensing. Science. 2019;366:460–7. doi: 10.1126/science.aau6391. [DOI] [PubMed] [Google Scholar]
- 15.Rossin A, Durivault J, Chakhtoura-Feghali T, Lounnas N, Gagnoux-Palacios L, Hueber A. Fas palmitoylation by the palmitoyl acyltransferase DHHC7 regulates Fas stability. Cell Death Differ. 2015;22:643–53. doi: 10.1038/cdd.2014.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yi L, Zheng CF. The emerging roles of ZDHHCs-mediated protein palmitoylation in the antiviral innate immune responses. Crit Rev Microbiol. 2021;47:34–43. doi: 10.1080/1040841X.2020.1835821. [DOI] [PubMed] [Google Scholar]
- 17.Gatica D, Lahiri V, Klionsky DJ. Cargo recognition and degradation by selective autophagy. Nat Cell Biol. 2018;20:233–42. doi: 10.1038/s41556-018-0037-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coretti L, Natale A, Cuomo M, Florio E, Keller S, Lembo F, et al. The interplay between defensins and microbiota in Crohn’s disease. Mediators Inflamm. 2017;2017:8392523. doi: 10.1155/2017/8392523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mizuno N, Kume K, Nagatani Y, Matsuda S, Iwata T, Ouhara K, et al. Aggressive periodontitis and NOD2 variants. J Hum Genet. 2020;65:841–6. doi: 10.1038/s10038-020-0777-z. [DOI] [PubMed] [Google Scholar]
- 20.Li C, Zhang J, Li S, Han T, Kuang W, Zhou Y, et al. Gene mutations and clinical phenotypes in Chinese children with Blau syndrome. Sci China Life Sci. 2017;60:758–62. doi: 10.1007/s11427-017-9090-6. [DOI] [PubMed] [Google Scholar]
- 21.Parkhouse R, Boyle JP, Monie TP. Blau syndrome polymorphisms in NOD2 identify nucleotide hydrolysis and helical domain 1 as signalling regulators. FEBS Lett. 2014;588:3382–9. doi: 10.1016/j.febslet.2014.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vasseur E, Boniotto M, Patin E, Laval G, Quach H, Manry J, et al. The evolutionary landscape of cytosolic microbial sensors in humans. Am J Hum Genet. 2012;91:27–37. doi: 10.1016/j.ajhg.2012.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–9. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paludan SR, Pradeu T, Masters SL, Mogensen TH. Constitutive immune mechanisms: mediators of host defence and immune regulation. Nat Rev Immunol. 2021;21:137–50. doi: 10.1038/s41577-020-0391-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clarke AJ, Simon AK. Autophagy in the renewal, differentiation and homeostasis of immune cells. Nat Rev Immunol. 2019;19:170–83. doi: 10.1038/s41577-018-0095-2. [DOI] [PubMed] [Google Scholar]
- 26.Paludan SR, Pradeu T, Masters SL, Mogensen TH. Constitutive immune mechanisms: mediators of host defence and immune regulation. Nat Rev Immunol. 2020;21:137–150. doi: 10.1038/s41577-020-0391-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saitoh T, Akira S. Regulation of inflammasomes by autophagy. J Allergy Clin Immun. 2016;138:28–36. doi: 10.1016/j.jaci.2016.05.009. [DOI] [PubMed] [Google Scholar]
- 28.Xian H, Yang S, Jin S, Zhang Y, Cui J. LRRC59 modulates type I interferon signaling by restraining the SQSTM1/p62-mediated autophagic degradation of pattern recognition receptor DDX58/RIG-I. Autophagy. 2020;16:408–18. doi: 10.1080/15548627.2019.1615303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wurzer B, Zaffagnini G, Fracchiolla D, Turco E, Abert C, Romanov J, et al. Oligomerization of p62 allows for selection of ubiquitinated cargo and isolation membrane during selective autophagy. Elife. 2015;4:e08941. doi: 10.7554/eLife.08941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xia H, Green DR, Zou W. Autophagy in tumour immunity and therapy. Nat Rev Cancer. 2021;21:281–97. doi: 10.1038/s41568-021-00344-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fan Q-W, Yan X-H. Mechanisms of selective autophagy. In: Xie Z, editor. Autophagy: biology and diseases: technology and methodology. Springer Singapore: Singapore; 2021, p. 79–98.
- 32.Liu K, Zhang L, Zhao Q, Zhao Z, Zhi F, Qin Y, et al. SKP2 attenuates NF-κB signaling by mediating IKKβ degradation through autophagy. J Mol Cell Biol. 2018;10:205–15. doi: 10.1093/jmcb/mjy012. [DOI] [PubMed] [Google Scholar]
- 33.Du Y, Duan T, Feng Y, Liu Q, Lin M, Cui J, et al. LRRC25 inhibits type I IFN signaling by targeting ISG15‐associated RIG‐I for autophagic degradation. EMBO J. 2018;37:351–66. doi: 10.15252/embj.201796781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Orvedahl A, MacPherson S, Sumpter R, Jr, Tallóczy Z, Zou Z, Levine B. Autophagy protects against Sindbis virus infection of the central nervous system. Cell Host Microbe. 2010;7:115–27. doi: 10.1016/j.chom.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cooney R, Baker J, Brain O, Danis B, Pichulik T, Allan P, et al. NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nat Med. 2010;16:90–97. doi: 10.1038/nm.2069. [DOI] [PubMed] [Google Scholar]
- 36.Lu KF, den Brave F, Jentsch S. Pathway choice between proteasomal and autophagic degradation. Autophagy. 2017;13:1799–1800. doi: 10.1080/15548627.2017.1358851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Niu JX, Sun Y, Chen BE, Zheng BH, Jarugumilli GK, Walker SR, et al. Fatty acids and cancer-amplified ZDHHC19 promote STAT3 activation through S-palmitoylation. Nature. 2019;573:139–43. doi: 10.1038/s41586-019-1511-x. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 38.Burns K, Janssens S, Brissoni B, Olivos N, Beyaert R, Tschopp J. Inhibition of interleukin 1 receptor/toll-like receptor signaling through the alternatively spliced, short form of MyD88 is due to its failure to recruit IRAK-4. J Exp Med. 2003;197:263–8. doi: 10.1084/jem.20021790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qin YF, Liu QX, Tian S, Xie WH, Cui J, Wang RF. TRIM9 short isoform preferentially promotes DNA and RNA virus-induced production of type I interferon by recruiting GSK3 beta to TBK1. Cell Res. 2016;26:613–28. doi: 10.1038/cr.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brigidi GS, Bamji SX. Detection of protein palmitoylation in cultured hippocampal neurons by immunoprecipitation and acyl-biotin exchange (ABE) J Vis Exp. 2013;18:50031. doi: 10.3791/50031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data supporting the present study are available from the corresponding author upon reasonable request.