Abstract
H-NS is a major Escherichia coli nucleoid-associated protein involved in bacterial DNA condensation and global modulation of gene expression. This protein exists in cells as at least two different isoforms separable by isoelectric focusing. Among other phenotypes, mutations in hns result in constitutive expression of the proU and fimB genes, increased fimA promoter inversion rates, and repression of the flhCD master operon required for flagellum biosynthesis. To understand the relationship between H-NS structure and function, we transformed a cloned hns gene into a mutator strain and collected a series of mutant alleles that failed to repress proU expression. Each of these isolated hns mutant alleles also failed to repress fimB expression, suggesting that H-NS-specific repression of proU and fimB occurs by similar mechanisms. Conversely, alleles encoding single amino acid substitutions in the C-terminal DNA-binding domain of H-NS resulted in significantly reduced affinity for DNA yet conferred a wild-type fimA promoter inversion frequency, indicating that the mechanism of H-NS activity in modulating promoter inversion is independent of DNA binding. Furthermore, two specific H-NS amino acid substitutions resulted in hypermotile bacteria, while C-terminal H-NS truncations exhibited reduced motility. We also analyzed H-NS isoform composition expressed by various hns mutations and found that the N-terminal 67 amino acids were sufficient to support posttranslational modification and that substitutions at positions 18 and 26 resulted in the expression of a single H-NS isoform. These results are discussed in terms of H-NS domain organization and implications for biological activity.
H-NS is a major component of the Escherichia coli nucleoid originally isolated by its ability to bind and compact chromosomal DNA (10, 34, 35, 44). This small (15.4 kDa), abundant (20,000 copies/cell), neutral (pI, ∼7.5), heat-stable protein binds avidly to double-stranded (ds) DNA with higher affinity for curved DNA substrates (30, 37, 46). H-NS is a global modulator of gene expression affecting the synthesis, both positively and negatively, of more than 50 E. coli proteins (3, 22, 47). In addition, H-NS is also involved in regulating expression of virulence-associated genes in pathogenic Shigella spp. (25, 31), Salmonella spp. (15), and enteroinvasive E. coli (5, 32).
In the best-characterized H-NS-sensitive systems, such as proU (16, 26, 40), fimB (9), bgl (6), and pap (14), gene expression is derepressed in hns insertion mutant strains. Often H-NS acts as a transcriptional repressor binding to intrinsically curved DNA sequences located near prokaryotic promoters (37). In solution, H-NS acts as a homodimer and can form higher-order oligomers at increased protein concentrations (12, 34). Multimerization has recently been shown to be critical for H-NS repressor function at both the proU and hns promoters (36, 41).
In light of the fact that H-NS is found in a cross-section of bacterial species (43) and is part of global regulatory networks involved in virulence, metabolism, and environmental stress, yet contains no known structural peptide motifs (such as zinc finger or helix-turn-helix motifs), we randomly mutagenized hns to help delineate biologically active H-NS domains. To clarify protein structure and function, we examined H-NS isoform composition and the action of H-NS in the processes of piliation and motility, as well as the more frequently studied roles of DNA binding and gene repression.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, genetic techniques, and enzyme assays.
The bacterial strains and plasmids used in this study are listed in Table 1. H-NS mutant derivatives are listed in Table 2. Media consisted of Luria-Bertani (LB) broth, LB agar, and MacConkey agar (Difco, Detroit, Mich.). When used, antibiotics were added to a final concentration of 100 μg (ampicillin) or 20 μg (tetracycline or chloramphenicol) per ml of medium. Restriction and other DNA-modifying enzymes were used as instructed by the manufacturers (Gibco-BRL, Gaithersburg, Md.; New England Biolabs, Beverly, Mass.; and Boehringer Mannheim, Indianapolis, Ind.). β-Galactosidase assays were performed as described elsewhere (27) with strains grown in LB broth.
TABLE 1.
Bacteria and plasmids used
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Bacterial strain | ||
| THK38 | thr-1 leuB thi-1 Δ(argF-lac)U169 malA1 xyl-7 ara-13 mtl-2 gal-6 rpsL fhuA2 supE44 hns+, linked to tetR | 18 |
| THK62 | THK38 except Φ(proU′-lacZYA-kan) hns2-tetR | 18 |
| ORN116 | THK38 except Φ(fimA′-lacZYA-kan) hns-1 | 29 |
| AL90 | MG1655 ΔlacZYA hns2-tetR fimB-lacZYA | 9 |
| AL29 | ORN116 except hns2-tetR | 29 |
| XL1-Red | endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac mutD5 mutS mutT Tn10(Tetr) | Stratagene |
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIqZΔM15 Tn10(Tetr)] | Stratagene |
| Plasmids | ||
| pACYC184 | Low-copy-number cloning vector, Tetr, Cmr | New England Biolabs |
| pTHK116 | pBR322 with wild-type hns gene under control of hns-1 promoter mutation, Ampr | 19 |
| pTHK116-7 | pACYC184 with 1.8-kb EcoRI-SalI hns encompassing fragment from pTHK116 | This study |
TABLE 2.
Sequence analysis of hns mutations
| H-NS derivativea | Codon change or alteration | No. of isolates |
|---|---|---|
| M1I | ATG→ATA | 1 |
| A18E | GCA→GAA | 2 |
| L26P | CTG→CCG | 2 |
| L65P | CTG→CCG | 1 |
| R93H | CGT→CAT | 1 |
| T108I | ACC→ATC | 1 |
| G111S | GGC→AGC | 2 |
| N67 | 1-bp insertion causing frameshift and new sequence encoding 64MLIA67→64NADR67 before early stop codon | 1 |
| N82 | AAA→TAA stop codon | 1 |
| N93 | CCG→TGA stop codon | 1 |
| N123 | 2-bp deletion causing frameshift and new sequence encoding 122AM→122DG before early stop codon | 1 |
| N124 | 1-bp insertion causing frameshift and new sequence encoding 122AMD→122SNG before early stop codon | 3 |
Plasmid construction and hns mutagenesis.
Plasmid pTHK116-7 was generated to serve as the hns template for the mutagenesis as follows. Plasmid pTHK116 was linearized by EcoRI digestion, blunt-ended with T4 DNA polymerase, and ligated to BamHI linkers (Stratagene, La Jolla, Calif.). The 1.8-kb hns fragment was released by BamHI-SalI digestion, gel purified (Qiagen, Chatsworth, Calif.), and cloned into the BamHI-SalI sites of pACYC184. The ligation reaction was transformed into DH5α, and colonies were selected on chloramphenicol plates. DNA from Cmr Tets transformants was subsequently verified by PCR screening and restriction digestion.
The hns gene was randomly mutated by transforming and propagating pTHK116-7 in the mutD mutS mutT strain XL1-Red as suggested by the manufacturer (Stratagene). Plasmid DNA was purified (Qiagen) from Cmr transformants, retransformed into the indicator strain THK62, and plated on lactose MacConkey medium. Plasmid DNA from these strains was verified to contain hns by HpaI digestion and sequenced to determine the mutations with forward 5′ CAG TCC TGC TCG CTT CGC 3′ and reverse 5′ GGT GTT ATC CAC GAA ACG GC 3′ primers.
Motility, gel shift assays, and protein purifications.
Motility was assayed by measuring swarm diameters as previously described (8) except measurements were taken at only one time point. Gel shift assays and H-NS purifications were done as previously described (9) except the mutant protein lysate (T108I) was washed from the ds-DNA-cellulose column with a 125 mM NaCl solution.
Two-dimensional gel electrophoresis.
Whole-cell lysates were prepared by using a French pressure cell as previously described (9). Samples were dissolved in O’Farrell’s solution (28) containing 1.5% pH 5 to 7 and 0.5% pH 3 to 10 ampholytes (Bio-Rad, Hercules, Calif.) and focused in the first dimension in tube gels according to the manufacturer’s instructions (Hoefer Scientific Instruments, San Francisco, Calif.). After isoelectric focusing (IEF), proteins in the extracted tubes were separated in the second dimension by sodium dodecyl sulfate–15% polyacrylamide gel electrophoresis, transferred to nitrocellulose, probed with H-NS antiserum, and detected as described previously (9). Some filters were probed with H-NS antiserum preadsorbed against an hns2 tetR strain in order to eliminate cross-reactivity to non-H-NS proteins.
RESULTS
Isolation of hns mutations.
H-NS is a global regulator of a variety of unlinked and unrelated genes in E. coli and Salmonella typhimurium (2, 43). Mutations in hns are pleiotropic, affecting the synthesis of a vast array of gene products involved in numerous biological processes (3, 22, 47). In an attempt to define and correlate separate H-NS domains with different biological activities, we subjected hns to a random mutagenesis scheme followed by an in vivo genetic screen to isolate a series of hns mutants.
We constructed plasmid pTHK116-7 (Table 1) to serve as the low-level hns-expressing template for the mutagenesis in order to avoid the potentially deleterious effects of overexpressing mutant hns protein products in the cell (35). This clone carried the wild-type hns gene driven by the down-regulated hns-1 promoter mutation (19) in the low-copy-number vector pACYC184. Plasmid pTHK116-7 was transformed into a triple-repair-deficient E. coli mutator strain (XL1-Red) to generate spontaneous, random mutations. Approximately 100 antibiotic-resistant colonies were picked, and plasmid DNA was purified from each of these transformants and individually retransformed into strain THK62 carrying a chromosomal hns2-tetR insertion mutation and proU′-lacZ transcriptional fusion (Table 1). H-NS is a direct transcriptional repressor of proU (16, 23, 40), and expression is phenotypically detectable in strains with proU-lacZ fusions on lactose MacConkey indicator medium. Thus, our strategy was to select for mutations that were unable to complement the hns2-tetR mutation and therefore represented the derepression of proU expression (Lac+, red colony phenotype). Seventy-one red THK62-based transformants from lactose MacConkey plates were chosen as putative hns mutant candidates.
To ensure that these potential hns mutations encoded stable proteins and to study point mutations rather than large gene deletions, we probed mutant strains with anti-H-NS antiserum in Western blots (data not shown). H-NS from 29 of 71 strains reacted with anti-H-NS antibody with 25 isolates producing apparently full-length protein on SDS-PAGE. We transformed plasmids representing 17 hns isolates into a clean background strain (XL1-Blue) and sequenced the DNA. We found seven independent missense and five nonsense mutations, which all maintained the original pTHK116-7 promoter sequence. The remaining five isolates expressed duplicate hns mutations. Each missense mutation encoded inferred single amino acid substitutions while the nonsense mutations resulted in C-terminally truncated forms of H-NS (Table 2).
Effects of hns mutations on gene expression.
We measured β-galactosidase activity (27) from THK62 carrying each hns mutant allele to quantitate the effects of the mutations on proU expression (Fig. 1A). As expected, proU-lacZ expression increased significantly in all hns mutant strains tested, albeit to various degrees. We also examined the effects of these mutations on fimB expression by using a fimB-lacZ fusion strain (AL90). We have recently shown that H-NS acts as a repressor of fimB expression by directly binding to the promoter region and inhibiting transcription (9). As with proU, fimB-lacZ expression was 6- to 30-fold higher in the hns mutant strains relative to that of the wild-type hns-expressing strain (Fig. 1B). Similar trends develop if one compares the level of derepression of both promoters over the course of each hns mutation. That is, each hns mutation affected proU and fimB expression in a similar fashion. For example, hnsL26P greatly increased expression whereas hnsT108I and hnsA18E resulted in the least severe mutant phenotypes at both promoter fusions. We concluded that (i) all of the hns mutations isolated induced proU and fimB expression, (ii) these mutants were defective in their repression ability, and (iii) H-NS probably employs similar repression mechanisms at each promoter.
FIG. 1.
Effects of hns mutant alleles on gene expression. All strains containing either proU-lacZ (THK62) (A) fimB-lacZ (AL90) (B) fusions and the indicated hns plasmids were grown to mid-log phase at 30°C and assayed for β-galactosidase activity (27). Data represent the averages of three to four independent experiments in duplicate. Error bars, standard errors of the means.
Effects of hns mutations on fimA promoter inversion.
In order to sort our series of hns mutants into separate phenotypic classes we tested their effects on fimA promoter inversion. Type 1 pilus expression is controlled transcriptionally by the inversion of a DNA element encompassing the fimA promoter (1, 11). Piliation is phase variable (on ⇔ off), and changes in expression may be monitored phenotypically on lactose MacConkey agar (29). Individual colonies from wild-type strains carrying a fimA′-lacZ fusion are either red (Lac+, fimA promoter on) or white (Lac−, fimA promoter off). Our laboratory has previously shown that hns mutations cause a 100-fold increase in the fimA promoter inversion rate, yielding pink colonies consisting of approximately equal numbers of on and off promoter-oriented cells (18, 19).
The hns mutations fell into two classes in regards to fimA promoter inversion (Table 3). The point mutations in the C-terminal third of the protein did not affect inversion, whereas the remaining four point mutations and three C-terminal truncations all resulted in the mutant, rapid promoter-inversion phenotype. It has been well documented that the DNA-binding domain is located in the C terminus of H-NS (33, 42). Thus, our results suggest that H-NS regulated type 1 pilus promoter inversion by some non-DNA-binding mechanism. We concluded that the ability of H-NS to control promoter flipping resides in the N-terminal half of the protein and that the C terminus may be needed only for some undefined role.
TABLE 3.
Effects of hns mutations on fimA promoter inversiona
| H-NS derivative | Inversion phenotype |
|---|---|
| pTHK116-7 (wild-type hns) | Red and white (normal) |
| pACYC184 (vector) | Pink (rapid) |
| M1I | Pink |
| A18E | Pink |
| L26P | Pink |
| L65P | Pink |
| R93H | Red and white |
| T108I | Red and white |
| G111S | Red and white |
| N67 | Pink |
| N93 | Pink |
| N123 | Pink |
All plasmids were transformed into AL29 (fimA′lacZYA-kan, hns2-tetR) and grown at 37°C on lactose MacConkey indicator medium. Individual colonies were picked and restruck to verify phenotypes.
Effects of hns mutations on motility.
H-NS is a positive regulator of flhCD, the master operon which controls the expression of all flagellar genes required for motility (24). Mutations in hns render bacteria nonmotile due to the reduced expression of flhCD and subsequent lack of flagellar biogenesis (4, 17, 20). Strains harboring each of the hns plasmids were inoculated onto semisolid agar plates and incubated at 30°C for 13 to 17 h. Motility was determined by measuring the diameter of the bacterial swarms and calculating a relative rate (Table 4). Most of the mutations had no major effect on motility with swarm rates close to 1. However the two hns point mutations A18E and T108I exhibited a unique hypermotile phenotype. We independently analyzed the basis for this hypermotile phenotype and determined that A18E and T108I affected flagellar rotational speed (8). Three other mutants were severely defective in the ability to move through the soft agar. The initial M1I mutant was completely nonmotile and two truncations, N93 and N124, displayed two- to threefold decreases in motility rates, respectively. These results suggested that the carboxy-terminal portion of H-NS may be involved in some aspect of bacterial motility that does not necessarily include DNA binding.
TABLE 4.
Effects of hns mutations on motility
| H-NS derivative | Relative motility ratea |
|---|---|
| pACYC184 (vector) | 0.20 ± .05 |
| M1I | 0.17 ± .03 |
| A18E | 2.50 ± .17 |
| L26P | 0.79 ± .17 |
| L65P | 0.80 ± .11 |
| R93H | 0.99 ± .15 |
| T108I | 2.10 ± .21 |
| G111S | 0.94 ± .20 |
| N67 | 0.60 ± .13 |
| N93 | 0.43 ± .06 |
| N124 | 0.32 ± .08 |
Motility rates were determined by comparing the swarm diameters of the hns2-tetR strains expressing the mutant hns plasmids relative to those of the same strains, on the same plates, expressing the wild-type hns plasmid. A swarm rate of 1 indicates identical swarm sizes between the two strains. Data represent the means ± standard errors of the means for five to seven individual experiments.
DNA-binding activity of altered H-NS protein.
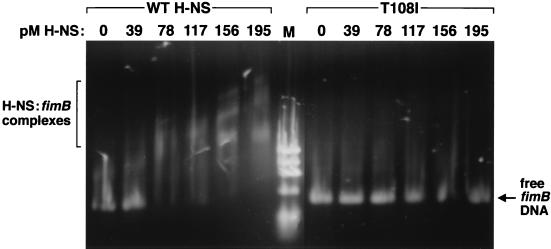
One of the most extensively studied properties of H-NS is its ability to bind DNA, since this interaction is often coupled to gene regulation (9, 38, 40). Combined genetic and biochemical efforts have isolated the DNA-binding region of H-NS to the C-terminal third of the protein (33, 42). We performed gel shift assays to compare the binding affinities of wild-type H-NS to protein encoded by hns with a mutation in the proposed DNA-binding fragment (T108I). Equal and increasing amounts of each purified protein were added to a known H-NS target, the fimB promoter (9) (Fig. 2).
FIG. 2.
Gel shift assay comparing the DNA-binding abilities of wild-type H-NS and altered T108I H-NS. Each lane contains 1.6 pM fimB promoter DNA and the indicated amount of either wild-type or T108I H-NS. Reaction mixtures were incubated, electrophoresed on a 1% agarose gel, and stained with ethidium bromide (9). M, ΦX174 marker.
In agreement with our previous study on H-NS–fimB interactions (9), we determined that (i) wild-type H-NS avidly bound the fimB promoter fragment, (ii) binding occurred at a low protein-to-DNA molar ratio, indicating a high-affinity binding capacity, and (iii) there were multiple H-NS binding sites within the fimB promoter region. In contrast, H-NS encoded by hnsT108I was unable to bind and retard fimB DNA. Even at the maximum protein concentrations added, complexes were undetectable even though the intensity of free DNA slightly decreased. These results demonstrated that H-NST108I is defective in its ability to bind relevant fimB DNA and corroborates evidence that the H-NS C terminus is essential for this function.
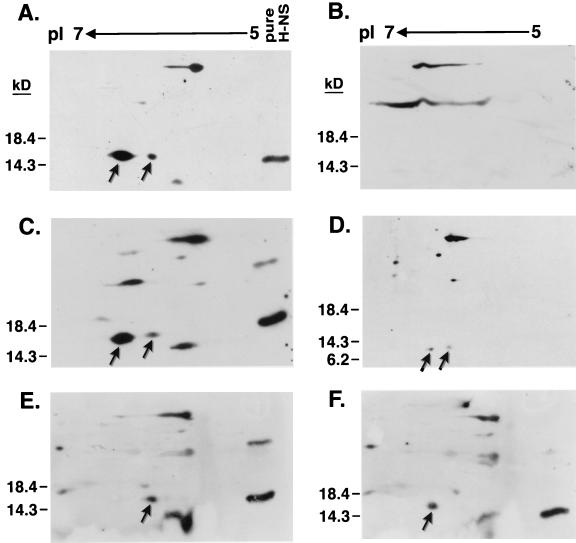
H-NS isoforms.
In the cell, H-NS exists as two or three isoforms differing only in their pI values (21, 34, 43). Presumably, H-NS undergoes a posttranslational modification, yet the type and site of modification and biological function of each form is still unknown. To begin uncovering these details we subjected whole-cell lysates of the hns mutant strains to two-dimensional electrophoresis followed by H-NS identification via Western blot analysis (Fig. 3). We consistently observed two isoforms present at 15.4 kDa that reacted with anti-H-NS antiserum in the wild-type hns strain (Fig. 3A) but absent in the vector-only control (Fig. 3B). The same two polypeptides were also synthesized from the hnsT108I mutant allele (Fig. 3C). Two low-intensity H-NS specific spots were also visible with the N67 derivative (Fig. 3D). This mutation encodes a C-terminally truncated form of H-NS that is approximately 7 kDa. However, each of the N-terminal point mutations, A18E and L26P, yielded only one H-NS isoform (Fig. 3E and F, respectively). Thus, these results suggested that the H-NS modification site is located within the amino half of the protein, potentially near residues 18 to 26.
FIG. 3.
Identification of H-NS isoforms by two-dimensional gel electrophoresis. Shown are Western blots of H-NS lysates separated by pI in the first dimension and by size in the second dimension and probed with anti-H-NS antiserum. All blots are in the same orientation with the same pI scale and protein standards indicated. Arrows indicate H-NS isoforms. For some panels e.g., panel A, purified H-NS (pure H-NS) was included as an internal marker. The H-NS protein and derivatives used were wild-type (A), vector (B), T108I (C), N67 (D), A18E (E), and L26P (F).
DISCUSSION
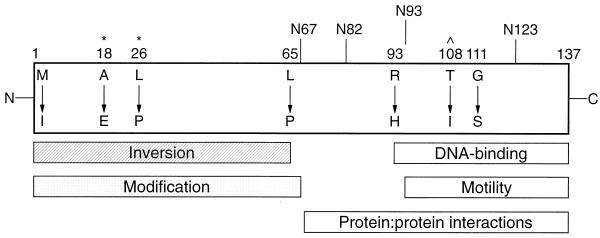
In this study, we combined genetic and biochemical approaches to identify key H-NS features necessary for a number of different functions. Transforming an hns-bearing plasmid into an E. coli mutator strain provided a simple method to obtain a set of random hns mutations. This technique, as opposed to chemical mutagenesis, had the advantage of being truly random and unbiased. We isolated hns mutant alleles with base pair insertions, deletions, transitions, and transversions which resulted in the alteration of different individual amino acid residues (Table 2). The locations of these amino acid substitutions and their effects on various H-NS activities allowed us to construct a model of H-NS domain organization (Fig. 4).
FIG. 4.
Schematic representation of H-NS. Altered residues are indicated by position number and amino acid substitution. Truncations are denoted by vertical lines. Rectangles represent domains involved in the indicated H-NS functions based on this study. The DNA-binding domain is as defined by Ueguchi et al. (42). Caret denotes residue implicated in DNA binding in this study; asterisks represent potential sites of H-NS modification.
As anticipated, all of the hns mutations isolated were defective in their ability to repress the expression of both proU- and fimB-lacZ promoter fusions (Fig. 1). The magnitude of induction varied greatly between mutations with the major difference between test promoters being that proU expression was ∼10 times more repressed by wild-type H-NS than fimB. Moreover, individual hns mutations had equivalent effects at both promoters. This observation led us to postulate that the mode of operation of repression is probably the same for genes at which H-NS has been shown to be a direct transcriptional inhibitor such as fimB (9), proU (40), and rrnB (38). Other investigators have suggested alternate mechanisms for H-NS repression of the semi-synthetic gal promoter (45) and the bgl operon (42).
Since its discovery (34, 44), the ability to bind DNA has been a major focus of H-NS research. Based on the binding properties of an H-NS truncation containing only the C-terminal 47 amino acids of the protein (33) and analysis of H-NS mutants (42), it has been established that the C-terminal third of H-NS encompasses the DNA-binding domain. Two lines of evidence generated here with our hnsT108I mutation confirmed these data. First, during purification H-NS T108I did not bind as tightly as wild-type H-NS to a ds-DNA-cellulose column (7), dissociating at lower salt concentrations (data not shown). Secondly, in gel shift assays T108I did not bind a fimB promoter fragment at a protein-to-DNA molar ratio upwards of 120:1 (Fig. 2). This result is in contrast to the strong binding exhibited by a low 45:1 molar ratio of wild-type H-NS to the same piece of DNA. Thus, our work on T108I complemented previous intrinsic fluorescence H-NS studies (13, 39) that suggested that the proximal tryptophan-109 closely interacted with DNA. The caveat of in vitro binding assays is that other factors that may be present in the cell and necessary for efficient protein binding may not be included. However, this problem is minimized when comparing the binding affinities of two proteins, such as wild-type and T108I H-NS, under the exact same conditions. Also, as previously suggested (42), a defect in DNA binding is only partially responsible for maximal gene derepression. This observation is exemplified by the fact that H-NS encoded by hnsT108I was unable to bind fimB DNA (Fig. 2) yet this mutation caused the smallest increase in fimB-lacZ expression (Fig. 1B).
The molecular mechanism by which H-NS influences promoter inversion and type 1 piliation is unknown. By inversion assays (Table 3) we showed that the residues within the N-terminal half of H-NS were important for controlling inversion since changing them resulted in a mutant inversion phenotype. In contrast, inversion regulation was not dependent on DNA binding since strains with altered proposed H-NS–DNA contact sites (R93H, T108I, and G111S) retained the wild-type inversion frequency. In particular, substitutions at Thr-108 (Fig. 2) and Gly-111 (42) have specifically been shown to cause drastic reductions in the ability of H-NS to bind DNA yet neither alteration adversely affected fimA promoter inversion frequency. Interestingly, even though residues within the DNA-binding domain of H-NS were not required for proper fimA promoter flipping, the entire C-terminal half of the protein was needed in some capacity, as evidenced by the mutant inversion phenotype exhibited by the H-NS truncations. We postulate that H-NS regulates inversion without binding DNA, possibly through protein-protein interactions with other molecules present at the fimA invertible element, and that the C-terminal domain may be important for overall structural integrity and protein stabilization. Data presented here also corroborated our previous conclusion that the regulation of FimB recombinase levels in the cell by H-NS is not the sole cause of the rapid inversion witnessed in hns mutant backgrounds (9). Consistent with this hypothesis, H-NS derivatives R93H, T108I, and G111S caused 6- to 25-fold increases in fimB expression (Fig. 1B) while maintaining a wild-type inversion phenotype (Table 3).
As with inversion, the motility results (Table 4) implicated the nonregulatory role of the H-NS carboxy terminus. The negative effects of hns mutations on bacterial motility were mainly clustered to the H-NS truncations, arguing that the conformation of this domain, rather than individual residues, is relevant to this motility phenotype. Rather than acting solely as a transcriptional activator of flagellar gene expression (4) the effect of H-NS on motility may be determined by protein interactions between H-NS and flagellar rotor components, as we have recently demonstrated with FliG (8).
Determining the type and location of the modified group(s) of H-NS is valuable for devising a mechanistic view of multiple H-NS activities. It is possible that separate H-NS isoforms possess differential functions. Using two-dimensional gel analysis (Fig. 3), we showed for the first time that multiple isoforms were present when an hns clone encoding only residues 1 through 67 was the sole source of H-NS expression. Further H-NS analysis revealed that a potential modification site may lie within or near a nine-residue span from A18 to L26. Thus, alterations in these specific amino acids may directly interfere with the addition of a modified group or cause local structural rearrangements in proximal residues.
Utilizing a random mutagenesis scheme followed by multiple phenotypic analyses, we have been able to aid in defining the relationship between H-NS structure and function. Several general themes have arisen from this study. The gene expression experiments in conjunction with the H-NS–DNA binding assays have confirmed that the ability to bind DNA is only one of the duties H-NS has in modulating gene expression. Taken together, the effects of hns mutations and deletions on fimA promoter inversion and bacterial motility suggested that the H-NS C-terminal domain may have multiple functions including involvement in protein-protein interactions as well as interactions with DNA. In agreement with this concept, we have recently shown that alterations in H-NS at residue 108 affect binding to FliG (8) and Spurio et al. (36) have shown that changes and deletions at Pro-116 impair H-NS oligomerization. We also speculate that the N and C termini may interact with each other, and we await determination of the three-dimensional crystal structure of H-NS to test this hypothesis. Although some H-NS functions may be isolated to particular domains, it seems likely that all portions of the protein work coordinately and in concert with each other to exert full H-NS activity.
ACKNOWLEDGMENTS
We thank Ian Blomfield for the fimB-lacZYA strain, Bob Bourret for advice on bacterial motility, and members of the Kawula lab for their comments on the manuscript. We gratefully acknowledge the technical assistance of Martin Schuster and the sequence provided by the UNC Automated DNA Sequence Facility.
This work was supported by grant R01 AI34176 from the National Institutes of Health.
REFERENCES
- 1.Abraham J M, Freitag C S, Clements J R, Eisenstein B I. An invertible element of DNA controls phase variation of type 1 fimbriae of Escherichia coli. Proc Natl Acad Sci USA. 1985;82:5724–5727. doi: 10.1073/pnas.82.17.5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atlung T, Ingmer H. H-NS: a modulator of environmentally regulated gene expression. Mol Microbiol. 1997;24:7–17. doi: 10.1046/j.1365-2958.1997.3151679.x. [DOI] [PubMed] [Google Scholar]
- 3.Bertin P, Lejeune P, Laurent-Winter C, Danchin A. Mutations in bglY, the structural gene for the DNA-binding protein H1, affect expression of several Escherichia coli genes. Biochimie. 1990;72:889–891. doi: 10.1016/0300-9084(90)90008-5. [DOI] [PubMed] [Google Scholar]
- 4.Bertin P, Terao E, Lee E H, Lejeune P, Colson C, Danchin A, Collatz E. The H-NS protein is involved in the biogenesis of flagella in Escherichia coli. J Bacteriol. 1994;176:5537–5540. doi: 10.1128/jb.176.17.5537-5540.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colonna B, Casalino M, Fradiani P A, Zagaglia C, Naitza S, Leoni L, Prosseda G, Coppo A, Ghelardini P, Nicoletti M. H-NS regulation of virulence gene expression in enteroinvasive Escherichia coli harboring the virulence plasmid integrated into the host chromosome. J Bacteriol. 1995;177:4703–4712. doi: 10.1128/jb.177.16.4703-4712.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeFez R, DeFelice M. Cryptic operon for β-glucosidase metabolism in Escherichia coli K-12: genetic evidence for a regulatory protein. Genetics. 1981;97:11–25. doi: 10.1093/genetics/97.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dersch P, Schmidt K, Bremer E. Synthesis of the Escherichia coli K-12 nucleoid-associated DNA-binding protein H-NS is subjected to growth-phase control and autoregulation. Mol Microbiol. 1993;8:875–889. doi: 10.1111/j.1365-2958.1993.tb01634.x. [DOI] [PubMed] [Google Scholar]
- 8.Donato, G. M., and T. H. Kawula. Enhanced binding of altered H-NS protein to flagellar rotor protein FliG causes increased flagellar rotational speed and hypermotility in Escherichia coli. J. Biol. Chem., in press. [DOI] [PubMed]
- 9.Donato G M, Lelivelt M J, Kawula T H. Promoter-specific repression of fimB expression by the Escherichia coli nucleoid-associated protein H-NS. J Bacteriol. 1997;179:6618–6625. doi: 10.1128/jb.179.21.6618-6625.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Durrenberger M L A, Citro G, Venanzi F, Gualerzi C O, Pon C L. Escherichia coli DNA-binding protein H-NS is localized in the nucleoid. Res Microbiol. 1991;142:373–380. doi: 10.1016/0923-2508(91)90106-k. [DOI] [PubMed] [Google Scholar]
- 11.Eisenstein B I. Phase variation of type 1 fimbriae in Escherichia coli is under transcriptional control. Science. 1981;214:337–339. doi: 10.1126/science.6116279. [DOI] [PubMed] [Google Scholar]
- 12.Falconi M, Gualtieri M T, LaTeana A, Losso M A, Pon C L. Proteins from the prokaryotic nucleoid: primary and quaternary structure of the 15-kD Escherichia coli DNA-binding protein H-NS. Mol Microbiol. 1988;2:323–329. doi: 10.1111/j.1365-2958.1988.tb00035.x. [DOI] [PubMed] [Google Scholar]
- 13.Friedrich K, Gualerzi C O, Lammi M, Losso M A, Pon C L. Proteins from the prokaryotic nucleoid: interaction of nucleic acids with the 15 kDa Escherichia coli histone-like protein H-NS. FEBS Lett. 1988;229:197–202. doi: 10.1016/0014-5793(88)80826-3. [DOI] [PubMed] [Google Scholar]
- 14.Goransson M, Sonden B, Nilsson P, Dagberg B, Forsman D, Emanuelsson K, Uhlin B E. Transcriptional silencing and thermoregulation of gene expression in Escherichia coli. Nature (London) 1990;344:682–685. doi: 10.1038/344682a0. [DOI] [PubMed] [Google Scholar]
- 15.Harrison J A, Pickard D, Higgins C F, Khan A, Chatfield S N, Ali T, Dorman C J, Hormaeche C E, Dougan G. Role of hns in the virulence phenotype of pathogenic salmonellae. Mol Microbiol. 1994;13:133–140. doi: 10.1111/j.1365-2958.1994.tb00408.x. [DOI] [PubMed] [Google Scholar]
- 16.Higgins C F, Dorman C J, Stirling D A, Waddell L, Booth L R, May G, Bremer E. A physiological role for DNA supercoiling in the osmotic regulation of gene expression in S. typhimurium and E. coli. Cell. 1988;52:569–584. doi: 10.1016/0092-8674(88)90470-9. [DOI] [PubMed] [Google Scholar]
- 17.Hinton J C, Santos D S, Seirafi A, Hulton C S, Pavitt G D, Higgins C F. Expression and mutational analysis of the nucleoid-associated protein H-NS of Salmonella typhimurium. Mol Microbiol. 1992;6:2327–2337. doi: 10.1111/j.1365-2958.1992.tb01408.x. [DOI] [PubMed] [Google Scholar]
- 18.Kawula T H, Lelivelt M J. Mutations in a gene encoding a new Hsp70 suppress rapid DNA inversion and bgl activation, but not proU derepression, in hns-1 mutant Escherichia coli. J Bacteriol. 1994;176:610–619. doi: 10.1128/jb.176.3.610-619.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kawula T H, Orndorff P E. Rapid site-specific DNA inversion in Escherichia coli mutants lacking the histonelike protein H-NS. J Bacteriol. 1991;173:4116–4123. doi: 10.1128/jb.173.13.4116-4123.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kutsukake K. Autogenous and global control of the flagellar master operon, flhD, in Salmonella typhimurium. Mol Gen Genet. 1997;254:440–448. doi: 10.1007/s004380050437. [DOI] [PubMed] [Google Scholar]
- 21.Laurent-Winter C, Lejeune P, Danchin A. The Escherichia coli DNA-binding protein H-NS is one of the first proteins to be synthesized after a nutritional upshift. Res Microbiol. 1995;146:5–16. doi: 10.1016/0923-2508(96)80266-x. [DOI] [PubMed] [Google Scholar]
- 22.Laurent-Winter C, Ngo S, Danchin A, Bertin P. Role of Escherichia coli histone-like nucleoid-structuring protein in bacterial metabolism and stress response—identification of targets by two-dimensional electrophoresis. Eur J Biochem. 1997;244:767–773. doi: 10.1111/j.1432-1033.1997.00767.x. [DOI] [PubMed] [Google Scholar]
- 23.Lucht J M, Dersch P, Kempf B, Bremer E. Interactions of the nucleoid-associated DNA-binding protein H-NS with the regulatory region of the osmotically controlled proU operon of Escherichia coli. J Biol Chem. 1994;269:6578–6586. [PubMed] [Google Scholar]
- 24.Macnab R M. Genetics and biogenesis of bacterial flagella. Annu Rev Genet. 1992;26:131–158. doi: 10.1146/annurev.ge.26.120192.001023. [DOI] [PubMed] [Google Scholar]
- 25.Maurelli A T, Sansonetti P J. Identification of a chromosomal gene controlling temperature-regulated expression of Shigella virulence. Proc Natl Acad Sci USA. 1988;85:2820–2824. doi: 10.1073/pnas.85.8.2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.May G, Dersch P, Haardt M, Middendorf A, Bremer E. The osmZ (bglY) gene encodes the DNA-binding protein H-NS (H1a), a component of the Escherichia coli K12 nucleoid. Mol Gen Genet. 1990;224:81–90. doi: 10.1007/BF00259454. [DOI] [PubMed] [Google Scholar]
- 27.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 28.O’Farrell P H. High-resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- 29.Orndorff P E, Spears P A, Schauer D, Falkow S. Two modes of control of pilA, the gene encoding type 1 pilin in Escherichia coli. J Bacteriol. 1985;164:321–330. doi: 10.1128/jb.164.1.321-330.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Owen-Hughes T A, Pavitt G D, Santos D S, Sidebotham J M, Hulton C S J, Hinton J C D, Higgins C F. The chromatin-associated protein H-NS interacts with curved DNA to influence DNA topology and gene expression. Cell. 1992;71:255–265. doi: 10.1016/0092-8674(92)90354-f. [DOI] [PubMed] [Google Scholar]
- 31.Porter M E, Dorman C J. A role for H-NS in the thermo-osmotic regulation of virulence gene expression in Shigella flexneri. J Bacteriol. 1994;176:4187–4191. doi: 10.1128/jb.176.13.4187-4191.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prosseda G, Fradiani P A, DiLorenzo M, Falconi M, Micheli G, Casalino M, Nicoletti M, Colonna B. A role for H-NS in the regulation of the virF gene of Shigella and enteroinvasive Escherichia coli. Res Microbiol. 1998;149:15–25. doi: 10.1016/s0923-2508(97)83619-4. [DOI] [PubMed] [Google Scholar]
- 33.Shindo H, Iwaki T, Ieda R, Kurumizaka H, Ueguchi C, Mizuno T, Morikawa S, Nakamura H, Kuboniwa H. Solution structure of the DNA binding domain of a nucleoid-associated protein, H-NS, from Escherichia coli. FEBS Lett. 1995;360:125–131. doi: 10.1016/0014-5793(95)00079-o. [DOI] [PubMed] [Google Scholar]
- 34.Spassky A, Rimsky S, Garreau H, Buc H. H1a, an E. coli DNA-binding protein which accumulates in stationary phase, strongly compacts DNA in vitro. Nucleic Acids Res. 1984;12:5321–5340. doi: 10.1093/nar/12.13.5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spurio R, Durrenberger M, Falconi M, La Teana A, Pon C L, Gualerzi C O. Lethal overproduction of the Escherichia coli nucleoid protein H-NS: ultramicroscopic and molecular autopsy. Mol Gen Genet. 1992;231:201–211. doi: 10.1007/BF00279792. [DOI] [PubMed] [Google Scholar]
- 36.Spurio R, Falconi M, Brandi A, Pon C L, Gualerzi C O. The oligomeric structure of nucleoid protein H-NS is necessary for recognition of intrinsically curved DNA and for DNA bending. EMBO J. 1997;16:1795–1805. doi: 10.1093/emboj/16.7.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tanaka K, Muramatsu S, Yamada H, Mizuno T. Systematic characterization of curved DNA segments randomly cloned from Escherichia coli and their functional significance. Mol Gen Genet. 1991;226:367–376. doi: 10.1007/BF00260648. [DOI] [PubMed] [Google Scholar]
- 38.Tippner D, Afflerbach H, Bradaczek C, Wagner R. Evidence for a regulatory function of the histone-like Escherichia coli protein H-NS in ribosomal RNA synthesis. Mol Microbiol. 1994;11:589–604. doi: 10.1111/j.1365-2958.1994.tb00339.x. [DOI] [PubMed] [Google Scholar]
- 39.Tippner D, Wagner R. Fluorescence analysis of the Escherichia coli transcription regulator H-NS reveals two distinguishable complexes dependent on binding to specific or nonspecific DNA sites. J Biol Chem. 1995;270:22243–22247. doi: 10.1074/jbc.270.38.22243. [DOI] [PubMed] [Google Scholar]
- 40.Ueguchi C, Mizuno T. The Escherichia coli nucleoid protein H-NS functions directly as a transcriptional repressor. EMBO J. 1993;12:1039–1046. doi: 10.1002/j.1460-2075.1993.tb05745.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ueguchi C, Seto C, Suzuki T, Mizuno T. Clarification of the dimerization domain and its functional significance for the Escherichia coli nucleoid protein H-NS. J Mol Biol. 1997;274:145–151. doi: 10.1006/jmbi.1997.1381. [DOI] [PubMed] [Google Scholar]
- 42.Ueguchi C, Suzuki T, Yoshida T, Tanaka K, Mizuno T. Systematic mutational analysis revealing the functional domain organization of Escherichia coli nucleoid protein H-NS. J Mol Biol. 1996;263:149–162. doi: 10.1006/jmbi.1996.0566. [DOI] [PubMed] [Google Scholar]
- 43.Ussery D W, Hinton J C D, Jordi B J A M, Granum P E, Seirafi A, Stephen R J, Tupper A E, Berridge G, Sidebotham J M, Higgins C F. The chromatin-associated protein H-NS. Biochimie. 1994;76:968–980. doi: 10.1016/0300-9084(94)90022-1. [DOI] [PubMed] [Google Scholar]
- 44.Varshavsky A J, Nedospasov S A, Bakayev V V, Bakayeva T G, Georgiev G P. Histone-like proteins in the purified Escherichia coli deoxyribonucleoprotein. Nucleic Acids Res. 1977;4:2725–2745. doi: 10.1093/nar/4.8.2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Williams R M, Rimsky S, Buc H. Probing the structure, function, and interactions of the Escherichia coli H-NS and StpA proteins by using dominant negative derivatives. J Bacteriol. 1996;178:4335–4343. doi: 10.1128/jb.178.15.4335-4343.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamada H, Muramatsu S, Mizuno T. An Escherichia coli protein that preferentially binds to sharply curved DNA. J Biochem. 1990;108:420–425. doi: 10.1093/oxfordjournals.jbchem.a123216. [DOI] [PubMed] [Google Scholar]
- 47.Yamada H, Yoshida T, Tanaka K, Sasakawa C, Mizuno T. Molecular analysis of the Escherichia coli hns gene encoding a DNA-binding protein, which preferentially recognizes curved DNA sequences. Mol Gen Genet. 1991;230:332–336. doi: 10.1007/BF00290685. [DOI] [PubMed] [Google Scholar]