Abstract
Dissemination of antimicrobial resistance (AMR) genes by horizontal gene transfer (HGT) mediated through plasmids is a major global concern. Genomic epidemiology studies have shown varying success of different AMR plasmids during outbreaks, but the underlying reasons for these differences are unclear. Here, we investigated two Shigella plasmids (pKSR100 and pAPR100) that circulated in the same transmission network but had starkly contrasting epidemiological outcomes to identify plasmid features that may have contributed to the differences. We used plasmid comparative genomics to reveal divergence between the two plasmids in genes encoding AMR, SOS response alleviation and conjugation. Experimental analyses revealed that these genomic differences corresponded with reduced conjugation efficiencies for the epidemiologically successful pKSR100, but more extensive AMR, reduced fitness costs, and a reduced SOS response in the presence of antimicrobials, compared with the less successful pAPR100. The discrepant phenotypes between the two plasmids are consistent with the hypothesis that plasmid-associated phenotypes contribute to determining the epidemiological outcome of AMR HGT and suggest that phenotypes relevant in responding to antimicrobial pressure and fitness impact may be more important than those around conjugation in this setting. Plasmid phenotypes could thus be valuable tools in conjunction with genomic epidemiology for predicting AMR dissemination.
Keywords: conjugation, plasmid fitness cost, antimicrobial resistance, bacterial SOS response
1. Introduction
Antimicrobial resistance (AMR) is a pressing global public health crisis. Bacterial pathogens become resistant to antimicrobials through either chromosomal mutations or by acquiring new AMR determinants through horizontal gene transfer (HGT) of mobile genetic elements (MGEs), such as plasmids [1,2]. HGT plays an important part in the dissemination of AMR genes, evidenced by multiple reports of emergence of similar AMR genes from different locations around the world. For example, plasmid-mediated colistin resistance conferred by the mcr-1 gene was first reported in 2015 in an isolate from 2011 in China, and was subsequently identified across five continents [3,4] in multiple species of Enterobacteriaceae by virtue of its being carried on a plasmid capable of inhabiting multiple hosts after it was first mobilized by a composite transposon [5]. Similarly, Klebsiella pneumoniae carbapenemases (KPCs), which were originally observed in the USA in 1996 [6,7], and CTX-M extended spectrum beta lactamases (ESBL), which were thought to be mobilized from the chromosome of Kluyvera spp., have been reported in multiple geographical regions as a result of AMR plasmid dissemination [8–11]. Owing to the rapid and extensive dissemination of AMR genes through HGT, it is critical to understand how different plasmid ‘vehicles’ affect the behaviour and spread of AMR genes.
Headway has been made towards understanding AMR epidemiology with reference to the specific plasmids that carry AMR determinants for a number of pathogen–plasmid combinations, largely from hospital settings [12–16]. In vitro work as well as modelling studies—in some cases supported by epidemiological data—suggest that the phenotypes of plasmids, such as plasmid fitness costs, resistance gene profile and other plasmid-conferred traits, may have a role in driving transmission and persistence of AMR across bacterial host populations [17–22]. Recent studies have attempted to associate non-AMR phenotypes such as conjugation with pathogen and plasmid epidemiology, including in clinical settings [12,13,23]. However, the drivers of AMR-HGT in non-hospital associated pathogens are likely to be distinct owing to the environmental differences (e.g. in antimicrobial pressures and transmission routes) and the comparatively under-observed bacterial populations.
Tracking plasmid-mediated AMR emergence in the community therefore requires a well-surveyed and characterized pathogen population. The Gram-negative diarrhoeal pathogen Shigella provides a highly observable community infection because infection is almost always symptomatic, the disease is reportable and there is no substantial animal or environmental reservoir (with the exception of sporadically reported cases in non-human primates [24,25]). Using genomic epidemiology, we recently investigated the emergence of AMR in the relatively closed transmission network of Shigella infections among men who have sex with men (MSM) in the United Kingdom [26,27]. The selective pressure caused by antimicrobial treatment of sexually transmitted infections (STIs) such as syphilis and gonorrhoea (common co-infections among shigellosis affected MSM [26,28]) led to the global emergence of a sublineage of Shigella flexneri 3a following the acquisition of plasmid pKSR100, encoding azithromycin resistance [26]. This sustained selection pressure subsequently led to the convergent acquisition of multiple azithromycin resistance plasmids in multiple other Shigella sublineages, directly triggering and intensifying epidemic waves of shigellosis [27,29]. The acquisition of different azithromycin resistance plasmids by these different sublineages provides us with an epidemiologically grounded model to test why some plasmids proliferate in a population, and why some do not.
The two IncFII plasmids under study here derive from a cross-sectional analysis of UK Shigella strains detected during routine surveillance between 2008 and 2014. During this period, we demonstrated that S. flexneri 2a emerged within the MSM community (during a cross-sectional study of 179 isolates) as two azithromycin resistant sublineages with markedly different epidemiology [27]. The ‘minor’ sublineage was estimated to have emerged earlier (most recent common ancestor (MRCA) in 1996) but only seven cases were observed in the cross section during the study period, whereas the comparatively recent (MRCA 2011) persistent or ‘major’ sublineage, caused 49 cases in the cross section during the study period, and disseminated internationally [27,29,30]. Although both sublineages were azithromycin resistant, the plasmids conferring azithromycin resistance varied. The successful ‘major’ sublineage carried pKSR100 [27], a plasmid which has continued to spread globally throughout shigellae, while the other ‘minor’ sublineage carried azithromycin resistance on a different plasmid (herein termed pAPR100) which has failed to mirror the epidemiological success of pKSR100. Furthermore, pKSR100 has continued to acquire novel AMR genes including a multidrug resistance (MDR) integron [27] and, more recently, ESBL genes [31]. As both pKSR100 and pAPR100 plasmids conferred the critical azithromycin resistance phenotype, we hypothesized that non-azithromycin resistance related plasmid phenotypes of these co-circulating plasmids may have contributed to their disparate epidemiological outcomes.
Thus here, we use these two plasmids to examine which phenotypes conferred on the bacterial host by the plasmid associate with the emergence of AMR in a community-transmitting obligate pathogen. We extend the genomic epidemiological evidence for the respective global distribution of the plasmids and use a comparative genomics-guided approach to determine the discrepant phenotypes between these two co-circulating plasmids with markedly different epidemiological outcomes.
2. Material and methods
(a) . Plasmid comparison and comparative genomics
Full plasmid sequences were extracted from genome sequences from a previous study [27]. Specifically, for pKSR100 (now deposited in NCBI under accession CP090161) the isolate corresponding to accession number ERR1364116 was used and for pAPR100 (now deposited in NCBI under accession CP090162) the contiguous sequence 24 from the isolate corresponding with accession number ERR1364014 was used.
A detailed version of the comparative genomics methods is available in the electronic supplementary material. Briefly, extracted genomic sequences of the plasmids were annotated using RAST server [32] and regions of similarity were identified as BLAST percentage identity cut-off of 95% or above. Roary v. 3.11.2 [33] was then used to examine the unique regions of the two plasmids (electronic supplementary material, file S2). NCBI non-redundant database and 661 K COBS data structure were used to assess the global distribution of the two plasmids.
(b) . Bacterial conjugation
All bacterial strains used in conjugation experiments were grown in TSB overnight and diluted 1 : 100 into fresh media and grown for 3 h before preparing the conjugation mixture. Donor and recipient strains were mixed 1 : 1 in a final volume in 500 µl for the conjugation experiments in liquid media and incubated shaking at 215 r.p.m. at 37°C for 75 min. Conjugation mixtures were serially diluted and plated on selective media to distinguish between donors, recipients and transconjugants every 15 min and the resulting colony forming units were counted to enumerate each. Conjugation mixtures were prepared as described above for the conjugation experiments on solid media and 10 µl of the conjugation mixture was spotted onto a sterile nitrocellulose filter paper placed on an agar plate and incubated at 37°C without shaking for 5 h. Conjugation mixtures present on the filter papers were submerged in 500 µl of sterile PBS, agitated by vortexing, serially diluted and plated as above every hour for CFU measurements. Conjugation efficiencies (CEs) for four biological replicates were calculated as previously described [34] using the following equation:
where η = conjugation efficiency (CE) measured in ml cell−1 h−1, T = transconjugants, D = donors, R = recipients and Δt = total conjugation time.
(c) . SOS response measurements during conjugation and ciprofloxacin exposure
SOS induction during both conjugation and exposure to ciprofloxacin was measured by using GFP expression as a proxy from a Pint-gfp fusion reporter plasmid p9092 (kindly gifted by Mazel and co-workers [35]) where the expression of the integrase promoter Pint is a strong signal of SOS induction [35]. Briefly, the strains of interest were transformed with p9092 where the GFP expressing cells were counted using a Bio-Rad ZE5 Cell Analyzer flow cytometer under the relevant test conditions and the proportion of GFP expressing cells were calculated for each instance. Specific details for each of the experiments are available in the electronic supplementary material.
(d) . Bacterial growth curves and fitness
Transconjugant Escherichia coli strains (with constitutive GFP expression) or Shigella sonnei 216 strains, carrying pKSR100 or pAPR100 from conjugation experiments were frozen in 25% glycerol and used as the source freezer stock for the growth curves. Pre-cultures for the bacterial growth curves were grown in M9 medium without azithromycin selection overnight incubating at 37°C with 215 r.p.m. shaking and diluted 1 : 100 into fresh M9 media adjusting all the cultures to the lowest OD600 value. The cultures were then distributed into a sterile 96-well plate (Greiner Bio One, UK) with a moisture barrier seal (4titude, UK) and incubated at 37°C with shaking in a Synergy H1 multi-mode plate reader taking optical density readings at 600 nm every 15 min. The resulting values were plotted using the R package Growthcurver [36] to determine the area under the curve (AUC) for each growth curve. Average values of the technical replicates for AUC of each of the plasmid carrying strains were normalized using the average AUC value of the technical replicates of plasmid-free E. coli and S. sonnei 216 strains to obtain relative fitness of each of the transconjugant E. coli and S. sonnei 216 strains for three biological replicates.
(e) . Minimum inhibitory concentration measurements
Minimum inhibitory concentration (MIC) measurements were carried out using Liofilchem MIC test strips (Liofilchem, Italy) following manufacturer's guidelines. Bacterial inocula for the MIC testing were prepared following the EUCAST guidelines for broth microdilution testing breakpoint table (https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_11.0_Breakpoint_Tables.pdf) and were spread on Mueller Hinton Agar plates (Bio-Rad, France) using sterile cotton swabs after which the MIC test strip was applied, and plates were incubated at 37°C for 18 h before the readings were recorded.
3. Results
(a) . pKSR100 has greater epidemiological success than pAPR100
As pKSR100 and pAPR100 had markedly different epidemiological success in the original study (conducted in the UK and France from 2008 to 2016) [27], we investigated whether the broader scientific research base and subsequent public health surveillance activity supported this discrepancy. In the original study, the pKSR100 carrying sublineage of S. flexneri 2a rapidly emerged as a dominant sublineage in a short space of time compared to the pAPR100 carrying sublineage even though the latter was circulating in the population prior to the acquisition of pKSR100 [27].
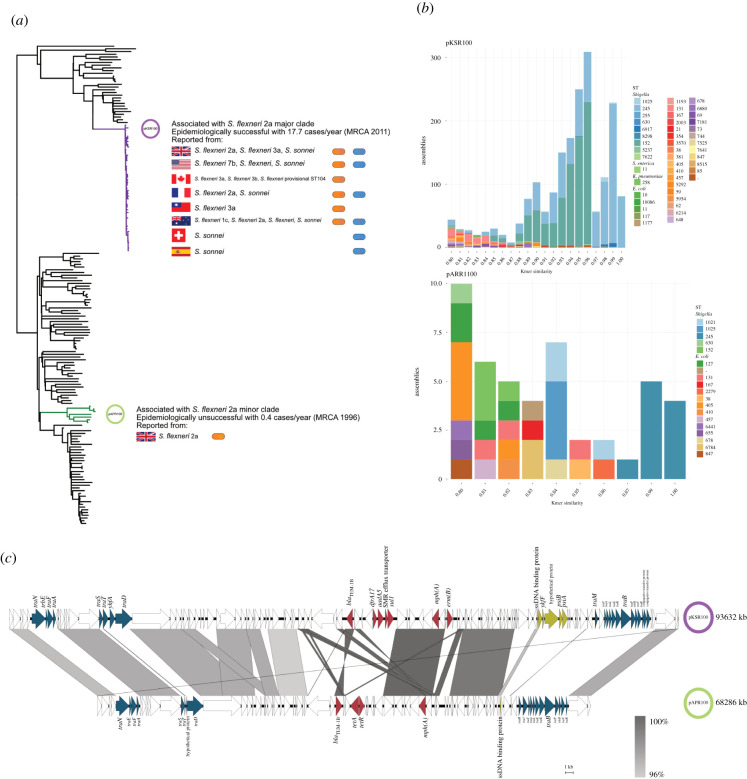
Since the original reporting of pKSR100 in Australia, Canada, France and the UK, further pKSR100 and pKSR100-like plasmids have been reported and/or deposited in the National Center for Biotechnology Information (NCBI) non-redundant database. Specifically, a BLASTn search returned 15 hits with over 95% query coverage and 95% identity from greater than or equal to 7 Shigella serotypes across four continents, continuing to drive national and global shigellosis dynamics [29,30,37–40] (figure 1a). Contrasting with the widespread dissemination of pKSR100, no BLAST hits with more than 87% query coverage for pAPR100 were returned, suggesting no distribution beyond its original detection in S. flexneri 2a in the UK. To extend the investigation of plasmid dissemination to unassembled genomes (such as those from routine public health surveillance activity), we screened both plasmids against the 661 K COBS data structure, which provides a snapshot of all bacterial genome data in the ENA as of November 2018 [41]. This search detected kmer similarity matches of over 0.80 of pKSR100 in 1926 publicly available bacterial genomes compared with only 46 bacterial genomes for pAPR100, further confirming the extensive dissemination of pKSR100. The genomes containing similarity matches for pKSR100 belonged to diverse bacterial hosts comprising 9 sequence types of Shigella, and 34 sequence types of E. coli and two other species (a K. pneumoniae ST258 and a Salmonella enterica ST11), while pAPR100 hits only came from 5 sequence types of Shigella and 13 sequence types of E. coli (figure 1b). Having established that pKSR100 had ongoing epidemiological success compared with pAPR100 through these analyses, we then set out to compare phenotypes of the two plasmids.
Figure 1.
Relative epidemiological success and comparative genomics of pKSR100 and pAPR100. The disparate epidemiological success of pKSR100- and pAPR100- bearing clades in a cross section of 179 Shigella isolates from UK surveillance data between 2008 and 2014 [26]. The pKSR100-bearing major clade is highlighted in purple and the pARP100-bearing minor clade is highlighted in green (a). The pKSR100 associated major clade had a higher case rate despite a more recent MRCA (most recent common ancestor) compared to the lower case rate and older MRCA of the minor clade, highlighting the more rapid spread of the major clade through the UK. The information alongside the clades depict the disparate global and species distribution of other pKSR100 bacterial hosts being reported in multiple countries across multiple Shigella subtypes (as determined by BLASTn against the NCBI non-redundant database and literature review). pKSR100 and pKSR100-like plasmids have been detected in and reported from eight countries and in multiple Shigella subtypes. By contrast, pAPR100 has only been detected in S. flexneri 2a in the UK. (b) Species and sequence type distribution of genomes sharing kmer similarities of greater than 0.80 with pKSR100 or pAPR100 from across greater than 600 000 publicly available bacterial genomes (in the 661 K COBS data structure). (c) Comparison of the genetic content of both plasmids. Areas of synteny (using a cut-off of 95% BLAST identity) are shown intervening grey bars coloured according to the inlaid legend. Regions with variation between the two plasmids are broadly categorized into three main groups; (i) conjugation machinery related (blue), (ii) SOS response alleviation related (green) and (iii) AMR related (red). (Online version in colour.)
(b) . pKSR100 and pAPR100 broadly differ across three gene categories
To guide our phenotypic experiments, we first performed comparative genomic analysis of the plasmid sequences to identify areas of differing genetic content. The plasmids had a high degree of conservation between their genetic content (figure 1c, electronic supplementary material, file S2) with a total of 49 genes shared between both plasmids and 122 genes unique to each. However, the variances we observed with the gene content on both plasmids could be categorized into three broad functional categories; namely (i) conjugation machinery related genes, implying different conjugative abilities between pKSR100 and pAPR100 (ii) genes related to SOS response alleviation, indicating that perhaps pKSR100 conferred a fitness advantage related to stress responses and (iii) AMR genes, suggesting that perhaps the advantage of pKSR100 lay in conferring greater or broader resistance to antimicrobials (figure 1c). Thus, we then proceeded to measure and compare the phenotypic differences between these two plasmids with disparate epidemiological outcomes. The two sequences of the pKSR100 and pAPR100 variants used in this study have been uploaded to the NCBI under accession numbers CP090161 and CP090162, respectively.
(c) . pKSR100 is less conjugative than pAPR100 from its native donor
Although both plasmids carried the conjugation related tra genes and trb genes, we observed variations in the genes between the two plasmids (figure 1c). These variations at the nucleotide level were observed at less than 95% BLAST similarity between the two plasmid sequences. Given these discrepancies in conjugation related genes and the differential dissemination of the plasmids at an epidemiological level, we investigated the CE of pKSR100 and pAPR100 in shigellae and in model strain E. coli MG1655.
Both pKSR100 and pAPR100 carried T4SS genes required for conjugation and were conjugative from S. flexneri to, and between, E. coli MG1655 and a clinical isolate of S. sonnei (figure 2). The S. sonnei clinical isolate we used, S. sonnei 216, from an MSM-associated sublineage that bore neither plasmid, but from the same collection of isolates as the native hosts of pKSR100 and pAPR100 [27] so represented a natural and relevant recipient. We measured CE across a number of variables including: from various donors (native and isogenic); to various recipients (E. coli and S. sonnei 216); in/on liquid and solid media; and across several time points (figure 2; electronic supplementary material, figure S2A).
Figure 2.
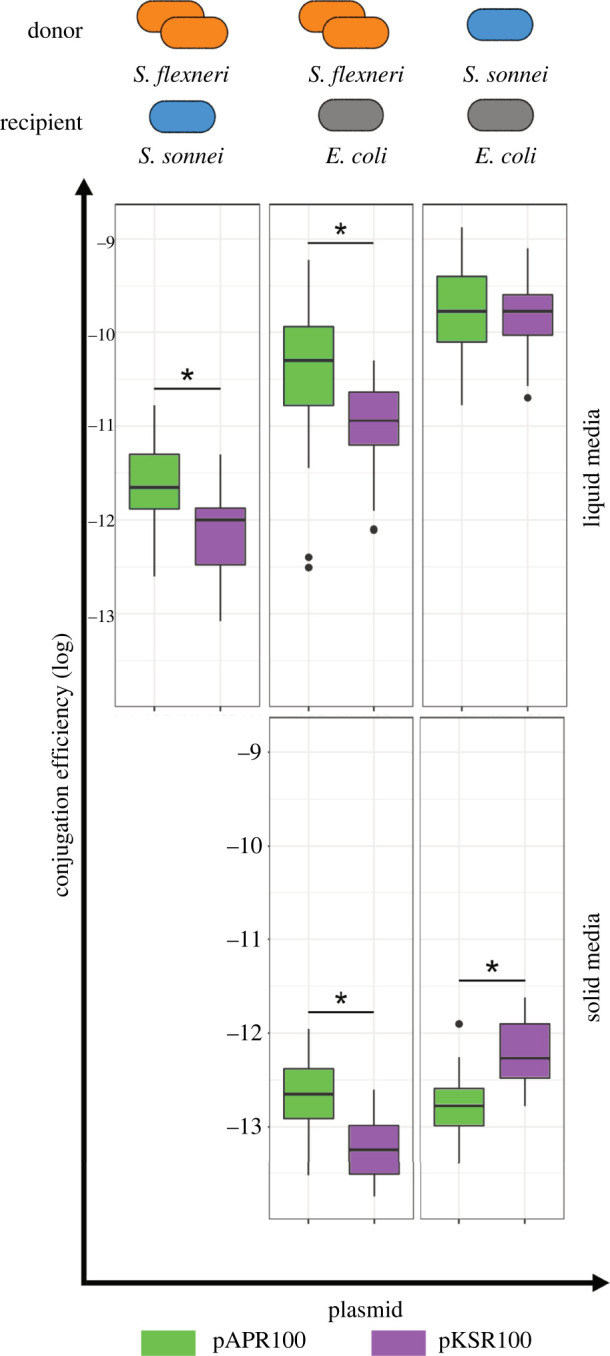
Conjugation efficiencies of pKSR100 and pAPR100 among different donors, recipients and media conditions. Different donor strains included native S. flexneri hosts (orange) and clinical isolate S. sonnei 216 as an isogenic donor (blue) while the recipient strains were either E. coli MG1655 (in grey) or S. sonnei 216 (in blue). Results for each plasmid are coloured according to the inlaid key. Results from liquid media are shown upper while solid is shown lower. Each box plot represents the combined results of all the time points used in LM models, where there are 12 replicates (four biological, three technical) for each time point. The asterisks denote significance as determined LMs. The p-values for each of the panels from left to right are as follows: top row, p = 0.043, p < 0.000; bottom row, p = 0.026, p = 0.025. Note: conjugations between S. flexneri donors and S. sonnei recipients are not shown here as no transconjugants were recovered until 3 h after mating, making it difficult to control for the growth rates of strains involved. Therefore, our main claims are based on liquid media conjugations where first transconjugants were recovered within 15 min of mating. (Online version in colour.)
Generally, pAPR100 had a better CE compared to pKSR100 from their native donors (i.e. the major and minor sublineages of S. flexneri 2a [27]) into either S. sonnei 216 or E. coli MG1655, regardless of media type (post hoc Tukey's tests on linear model (LM), p < 0.05 for all comparisons) (figure 2; electronic supplementary material, tables S5 and S4). However, these patterns varied across donors (liquid media LM hostpair : plasmid interaction F2,87 = 3.7, p < 0.029; solid media host : plasmid interaction F1,28 = 18.3, p < 0.001), such that in experiments with isogenic donors (S. sonnei 216 donation to E. coli) we detected no difference between the plasmids in liquid media (post hoc Tukey's test p = 1) and increased CE of pKSR100 versus pAPR100 on solid media (p = 0.025). This indicates that the CE differences between the plasmids observed with the native host donors may be due to the different donor backgrounds (electronic supplementary material, table S4). In support of the importance of donor effects, the isogenic S. sonnei donor facilitated a greater CE than the native S. flexneri donors (all relevant post hoc comparison p < 0.05) (electronic supplementary material, table S4). Recipient effects were also important as the E. coli recipient facilitated more efficient conjugation than S. sonnei (all relevant post hoc comparison p < 0.05) (electronic supplementary material, table S4).
(d) . pKSR100 has little fitness cost and alleviates the SOS response
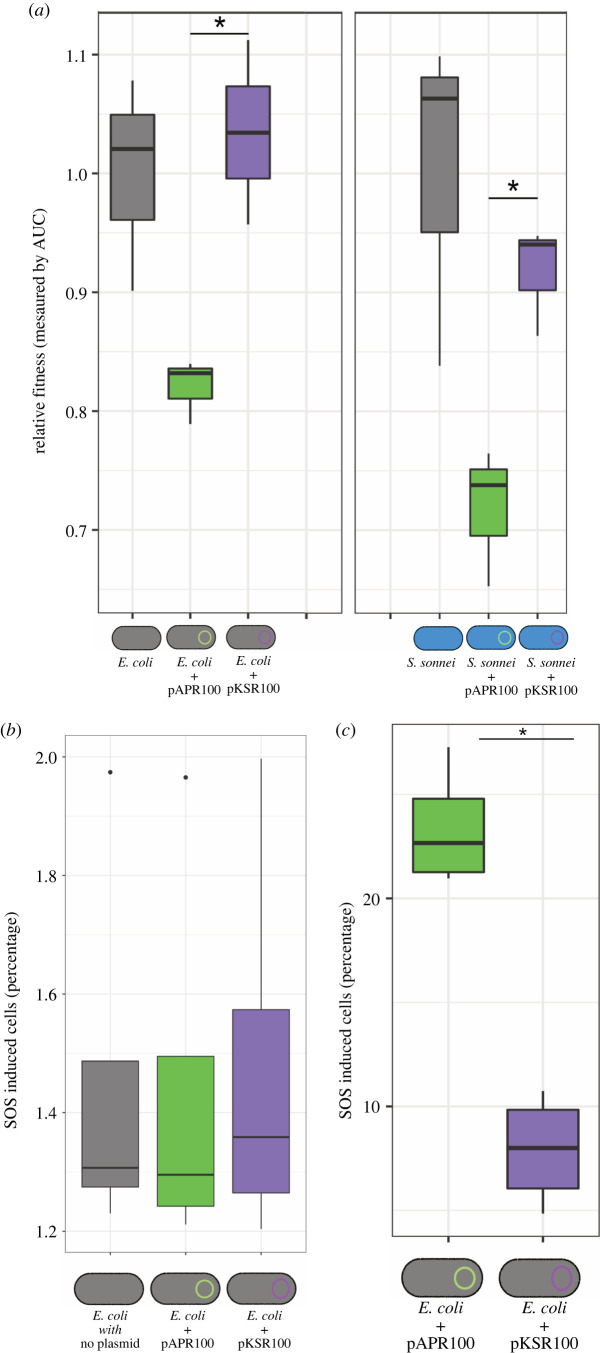
As plasmid burden on bacterial fitness is commonly investigated as a factor contributing to plasmid population dynamics, we investigated the fitness cost of plasmid carriage during growth. Comparing the growth of plasmid-bearing and plasmid-free strains revealed that pAPR100 imposed approximately 20% fitness cost in E. coli MG1655, whereas pKSR100 did not have a significant fitness cost based on the AUC measurements (figure 3a). Similar patterns were observed in S. sonnei 216, though unlike in E. coli we observed a small (approx. 6%) cost of pKSR100 in S. sonnei, suggesting that host factors also contribute to the overall relative fitness impact of plasmid carriage (figure 3a). Collectively, pAPR100 imposed an increased fitness cost relative to pKSR100 across the hosts.
Figure 3.
Relative fitness cost and SOS response induction during conjugation and antimicrobial exposure of pKSR100 and pAPR100. (a) Relative fitness of E. coli MG1655 (grey icons) and S. sonnei 216 (blue icons) carrying either pKSR100 (purple) or pAPR100 (green) compared to plasmid-free wild-type (grey bars in the graph). Asterisks denote significance where p = 0.01071 for E. coli strains and p = 0.009971 for S. sonnei strains as determined by two sample t-test. (b,c) SOS response levels by plasmid as a proportion of cells in which SOS is induced (measured by GFP expression using a reporter plasmid, see methods) during conjugation (b) and following a 2 h exposure to sub-inhibitory concentrations of ciprofloxacin (c). Individual box plots represent the median, range and IQR of four independent biological replicate data points adjusted to a negative control for each replicate (see methods, electronic supplementary material, figure S3). No statistically significant difference was observed between the two plasmids during conjugation (p = 0.7864), but there was a marked level of SOS response alleviation in cells carrying pKSR100 (p = 0.000237) as determined by a two-sample t-test. (Online version in colour.)
The comparative genomic analysis revealed that pKSR100 contained a cluster of five genes including single stranded DNA binding protein gene, plasmid SOS inhibition A (psiA) and B (psiB) genes which were absent in pAPR100 (figure 1c; electronic supplementary material, figure S1A) and are known to alleviate the bacterial SOS response in E. coli [42]. A gene encoding for a hypothetical protein and an uncharacterized protein family (UPF) 0401 protein ykfF gene was also found in the same cluster though their roles in SOS response alleviation, if any, is unclear. The SOS response is often induced during conjugation and by environmental stressors and is known to be costly [43]. Given the disparate fitness costs between pKSR100 and pAPR100, we investigated whether there were differences in SOS response and alleviation attributable to the plasmids. Firstly, because of the differences between conjugation genes and the known activation of the SOS response during conjugation [35], we measured SOS response induction during conjugation anticipating that pAPR100 conjugation might elicit a greater SOS response in the recipients due to the absence of the gene cluster-containing psiB, and that pKSR100 (which has the psiB gene cluster) would alleviate the SOS response. However, we observed no statistically significant difference among SOS induction between the plasmids during conjugation when 100 000 events were counted (figure 3b) or when 1 million events were counted (data not shown), though both showed a slight increase in SOS relative to the negative control (figure 3b).
Because these plasmids derive from a patient community with high levels of antimicrobial use, we then sought to determine if either plasmid alleviated the SOS induction in response to sublethal concentrations of antibiotics. Specifically, we investigated the fluoroquinolone antibiotic ciprofloxacin, which is known to induce bacterial SOS response as a result of DNA mutagenesis stimulation in E. coli [44]; the recommended, and overwhelmingly administered, treatment for shigellosis in this patient community [45,46]; and an antimicrobial to which neither pKSR100 or pAPR100 confer resistance (see below and table 1). Our subsequent measuring of SOS induction among cells (carrying either pKSR100 or pAPR100) after a 2 h exposure to sub-inhibitory concentrations of ciprofloxacin revealed that cells harbouring pKSR100 were significantly better at alleviating the SOS induction than cells carrying pAPR100 (figure 3c). Thus cells harbouring pKSR100 were less likely to activate the costly SOS response following exposure of sublethal concentrations of an antimicrobial for which the plasmid conferred no resistance.
Table 1.
The impact of pKSR100 and pAPR100 on antimicrobial resistance phenotypes.
| MIC values by plasmid and hosta |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| antimicrobial resistance |
gene present in: |
neither plasmid |
with pKSR100 |
with pAPR100 |
||||||||
| class | gene | antibiotic | pKSR100 | pAPR100 | E. coli | S. sonnei 216 | S. flexneri (native) | E. coli + pKSR100 | S. sonnei 216 + pKSR100 | S. flexneri (native) | E. coli + pAPR100 | S. sonnei 216 + pAPR100 |
| macrolide | mph(A) | azithromycin | √ | √ | 4 | 4 | >256 | >256 | >256 | 48 | 64/32 | 64 |
| macrolide | erm(B) | clindamycin | √ | >256 | 48 | >256 | >256 | >256 | >256 | >256 | >256 | |
| sulfonamide | sul1 | sulfamethoxazole | √ | 3 | 8 | >1024 | 12 | >1024 | 4 | 3 | 8 | |
| trimethoprim | dfrA17 | trimethoprim | √ | 0.125 | >32 | >32 | >32 | >32 | >32 | 0.125 | >32 | |
| penicillins | blaTEM1b/1a | ampicillin | √ | √ | 2 | 4 | >256 | >256 | >256 | >256 | >256 | >256 |
| aminoglycoside | aadA5 | streptomycin | √ | 1 | 16 | 48/64 | 4 | 24 | 128 | 1 | 16 | |
| tetracycline | tetA | tetracycline | √ | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 64 | 48 | 32 | |
| fluoroquinolone | ciprofloxacin | 0.006 | 0.004 | 0.006 | 0.006 | 0.004 | 0.004 | 0.006 | 0.006 | |||
| cephalosporin | cefotaxime | 0.016 | <0.016 | 0.023 | 0.016 | <0.016 | 0.016 | 0.016 | <0.016 | |||
aMIC values in bold represent a change of four-fold or more in MIC when the relevant plasmid is present.
(e) . pKSR100 offers a greater range and magnitude of AMR
Owing to the importance of AMR in driving emergence of shigellosis among MSM populations, we also investigated the AMR phenotypes conferred by the two plasmids. Predictions of AMR genes identified six AMR genes in pKSR100, which accounted for 50% (6 of 12) of the total AMR genes in the host strain of S. flexneri 2a, while pAPR100 only had three AMR genes contributing 27% (3 of 11) to the total AMR gene content of its host S. flexneri 2a strain (electronic supplementary material, tables S2 and S3). Analysis of AMR genes present on the plasmids using ResFinder predicted that pKSR100 encoded resistance to sulphonamides, macrolides, trimethoprim, beta-lactams and aminoglycosides and that pAPR100 encoded resistance to macrolides, beta-lactams and tetracycline showing that the AMR genes of the plasmids differed both in the number and variety of drug classes they provided resistance against (table 1). Thus, both resistance plasmids conferred MDR and contained resistance genes relevant to transmission among MSM, but pKSR100 was predicted to confer broader-spectrum resistance.
To test the genotypic predictions of increased AMR conferred by the plasmids, we correlated our genotypic information with MIC phenotypes. The antimicrobials tested were based on the genotypic AMR predictions, and to differentiate plasmid and host factors, MICs were determined for the native hosts, and in both E. coli and S. sonnei 216 transconjugants of both plasmids compared to plasmid-free wild-types (table 1). The two plasmid-free hosts carried intrinsic resistance to three antibiotics (i.e. E. coli was resistant against clindamycin and S. sonnei 216 to trimethoprim and streptomycin). Collectively, however, the results demonstrated general agreement between the AMR genotype carried on plasmids and phenotype with a minimum fourfold increase in the MIC values compared to the controls when the AMR genes were present (table 1). Notably, however, pKSR100 conferred a higher level of resistance to azithromycin, a phenotype critical for driving circulation of shigellosis in this community at that time, by virtue of encoding the additional macrolide resistance gene ermB (table 1) [26,27].
In addition to those antimicrobials where resistance was anticipated to change based on genotypic prediction, we tested whether the plasmids conferred AMR against other clinically relevant antimicrobials; ciprofloxacin and cefotaxime. This was to ensure we were not missing relevant AMR conferred by potentially novel genes, or potentiating genes, and to ensure the robustness of our SOS induction results. As expected, neither plasmid altered the MIC values against these antimicrobials.
Collectively, these AMR results show that pKSR100 confers resistance to a more extensive range of antimicrobials, and a higher level of resistance to azithromycin, than pAPR100.
4. Discussion
Here, we investigated two AMR plasmids with different epidemiological outcomes from the same clinical niche to identify putative plasmid phenotypes associated with epidemiological success, using comparative genomics as a guide. And since the epidemiological fate of these plasmids had already been documented with regard to azithromycin resistance carried on these plasmids [26], our objective was to investigate the potential other factors that helped shape the observed epidemiological outcome of these plasmids beyond the fact that pKSR100 carried more AMR genes and conferred more AMR phenotypes. The spread and persistence of plasmids may depend on several plasmid and host associated factors such as the plasmid type, host range, conjugative capacity and fitness cost as well as the environments they are in [47–52]. Understanding the contributions of each of these factors is important in determining the fate of AMR plasmids found in clinical settings and predicting evolutionary trajectories of those plasmids [17]. In summary, our results identified that the epidemiologically successful pKSR100 conferred increased AMR, reduced fitness cost and reduced SOS response in the presence of sub-inhibitory concentrations of antibiotics, compared with the less successful pAPR100, despite pKSR100 having a lower CE in native hosts.
The epidemiologically prominent plasmid, pKSR100, carried more AMR genes than pAPR100. In addition to the AMR genes carried on the plasmid, the bacterial host also carried AMR genes on the chromosome (electronic supplementary material, tables S2 and S3). However, there were few discrepancies between the non-plasmid AMR gene content of the native hosts (only a dfrA1 and additional blaTEM gene), and these did not result in phenotypic differences between the native hosts against the cognate antimicrobials (trimethoprim and ampicillin, respectively; table 1). Thus, it is unlikely that there was a relationship between the AMR genes present on the respective bacterial chromosomes (i.e. the S. flexneri 2a major and minor sublineages) and the spread of the plasmids. This is further evidenced by the remarkable onward global spread of pKSR100 from the setting in which it was originally described (figure 1). Thus, it is possible that the more extensive range of AMR and higher azithromycin resistance conferred by pKSR100 relative to pAPR100 was a key determining factor in the resulting epidemiological success and widespread dissemination of pKSR100. However, given that AMR genes frequently recombine between plasmids, and both plasmids already clearly encoded MDR, we are still faced with the question of why it was pKSR100 that acquired the broader AMR profile and went on to globally disseminate despite the comparative longevity of circulation of pAPR100 in the community. To address this, we considered further non-AMR factors guided by the comparative genomics, namely CE and fitness cost.
Conjugation is an efficient mechanism for plasmid dissemination and has been widely studied [1]. Theoretically, a higher conjugative capacity might result in a greater spread of plasmids. However, in our case, we observed that the globally widespread pKSR100 had a lower CE than pAPR100 from the native hosts. The observed differences in CE between pSKR100 and pAPR100 were mostly in the range of one order of magnitude (figure 2), but significant differences were observed for various combinations of donors and recipients, as well as various media conditions, indicating that, though modest, this effect is real. Furthermore, similar results were observed with different CE calculations (electronic supplementary material, figure S2B) adding to the persistency of our results. While the conjugative ability of plasmids can depend on donor or recipient strains' background as well as abiotic environmental factors, the lack of apparent importance of the CE exclusively for broader spread is consistent with a previous study examining the contribution of conjugation and other factors such as fitness effects towards plasmid spread in hospital settings [13]. However, we also observed that the differences of CE between the two plasmids diminished from an isogenic donor background, suggesting that donor factors are influencing CE in our model. This is not unexpected as the effect of donor and recipient backgrounds are known to affect the conjugative capacity of plasmids [53]. Hence, our results provide strong indications that the CE is dependent on donor, recipient and environmental conditions and that CE alone might not be an accurate measure of successful dissemination of a plasmid among bacterial populations in real world settings.
By contrast, the lack of a fitness cost imposed by pKSR100 may have been an important factor in perpetuating its epidemiological success. The fitness costs imposed by plasmids depend on the plasmid and host combination as well as their environmental factors along with gene conflicts [47,54] and, previous work has demonstrated that fitness cost measured in a laboratory setting might not translate to success in the natural environment [23]. We addressed host variability by measuring the fitness cost imposed by our plasmids in multiple backgrounds, and the broader database analysis (figure 1; electronic supplementary material, figure S1B) indicated a similar host range for the two plasmids and their close relatives. While our results may not relate exactly to the fitness cost imposed by these plasmids on their natural hosts, it helps us understand the effects that fitness costs may have on transmission through other residents in their natural gut microbe community. Our observation of the unsuccessful pAPR100 having a greater fitness cost is consistent with fitness costs affecting plasmid fates in microbial populations, and suggests that fundamental plasmid properties can contribute to epidemiological outcomes, specifically in a community-transmitting pathogen in our case.
Given the difference in SOS alleviation gene content between the two plasmids, we also examined the impact of the two plasmids on the induction of the costly SOS response. Since plasmids move as single stranded DNA molecules to recipient cells during conjugation, the possible induction of the SOS response may act as an influencing factor for the dissemination of plasmids. The pKSR100-related plasmid R100 had been shown to induce the SOS response when conjugated into Vibrio cholerae recipient (alleviated by the psiB gene cluster [55,56]). Hence, finding no difference in SOS induction during conjugation with the psiB cluster-containing pKSR100 and pAPR100 (which does not contain the cluster) in E. coli, indicates that SOS inductions by plasmids could be host dependent. However, as sub-inhibitory concentrations of ciprofloxacin have been demonstrated to induce the SOS response [57] and this is a highly relevant exposure for our system (as ciprofloxacin is a recommended treatment for shigellosis and other STIs circulating among MSM [45]) our finding that pKSR100 protected against the induction of the SOS response during exposure to sub-inhibitory concentrations of a clinically relevant antimicrobial supports the notion that an SOS response alleviation phenotype may contribute to AMR emergence in an epidemiological setting of high antimicrobial use.
The results of our study are consistent with the theory that, alongside AMR, plasmid-associated non-AMR phenotypes may play a crucial role in facilitating the dissemination of AMR plasmids. Although our study used only two plasmids from a single model system, the plasmids we used were important examples drawn from an established pathogen and real world epidemiological scenario (capturing complex networks in their natural habitats). Thus, our findings may help guide future studies towards the development of universal ground truths regarding important plasmid phenotypes for AMR emergence and establishment within the dynamic and problematic Enterobacteriaceae. We also mitigated the possibility that phenotypic findings may have been dependent on non-plasmid factors by varying the host and environmental conditions, which will vary in the natural habitat. However, our study raises further questions, particularly concerning the molecular mechanisms behind the disparate epidemiological outcomes of the two plasmids. Interestingly, though we could generate strains carrying both pKSR100 and pAPR100 in the laboratory (electronic supplementary material), this pairing has never been detected in any of our collection of approximately 179 epidemiological isolates. The separation of these two plasmids could be due to specific evolutionary conflicts and deleterious interactions of the two plasmids in their natural hosts. Further investigations into the evolutionary consequences of harbouring both plasmids is another subject for future research. In addition to that, our observations on SOS response alleviation after exposure to ciprofloxacin in the presence of pKSR100 could also be taken further to evaluate the long-term survival of ciprofloxacin exposure as another future research objective. Together, our data demonstrate that working backwards from an established epidemiological scenario is a valuable approach for identifying plasmid-associated phenotypes that contribute to the epidemiological trajectory of AMR, and perhaps one day could be turned prospectively to aid prediction and prevention of AMR emergence in future surveillance.
Data accessibility
Plasmid sequences used are publicly available in NCBI database and the accession numbers are provided in the main text. Raw data for used for the linear modelling and electronic supplementary material, file S2 are deposited in the Dryad Digital Repository (doi:10.5061/dryad.sxksn0363) [58].
Electronic supplementary material is available online [59].
Authors' contributions
P.M.D.S.: formal analysis, investigation, methodology, visualization, writing—original draft, writing—review and editing; G.E.S.: formal analysis, methodology, software, visualization, writing—review and editing; G.A.B.: formal analysis, methodology, software, visualization, writing—review and editing; R.J.B.: formal analysis, methodology, software, writing—review and editing; C.J.: conceptualization, resources, supervision, writing—review and editing; J.P.J.H.: conceptualization, formal analysis, methodology, resources, software, supervision, visualization, writing—review and editing; K.S.B.: conceptualization, formal analysis, funding acquisition, methodology, resources, software, supervision, visualization, writing—original draft, writing—review and editing.
All authors gave final approval for publication and agreed to be held accountable for the work performed therein.
Conflict of interest declaration
Authors declare no competing interests
Funding
This work was supported by an Academy of Medical Sciences Springboard award (SBF002\1114). K.S.B. was supported by a Wellcome Trust Clinical Research Career Development Award (106690/A/14/Z). G.E.S. is supported by a studentship from the MRC Discovery Medicine North (DiMeN) Doctoral Training Partnership (MR/N013840/1). P.M.D.S. is supported by a Biotechnology and Biological Sciences Research Council project grant no. BB/V009184/1. R.J.B. is supported by a Medical Research Council and UK Department for International Development (DFID) project grant no. MR/R020787/1. K.S.B. is affiliated to the National Institute for Health Research Health Protection Research Unit (NIHR HPRU) in Gastrointestinal Infections at University of Liverpool in partnership with UK Health Security Agency (UKHSA), in collaboration with University of Warwick. The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR, the Department of Health and Social Care or UKHSA. We would like to thank Michael Bottery (University of Manchester) for the kind gift of GFP and mCherry tagged E. coli MG1655 strains and Didier Mazel for his kind gift of the Pint-gfp fusion reporter plasmid p9092.
References
- 1.Frost LS, Leplae R, Summers AO, Toussaint A. 2005. Mobile genetic elements: the agents of open source evolution. Nat. Rev. Microbiol. 3, 722-732. ( 10.1038/nrmicro1235) [DOI] [PubMed] [Google Scholar]
- 2.Norman A, Hansen LH, Sorensen SJ. 2009. Conjugative plasmids: vessels of the communal gene pool. Phil. Trans. R. Soc. B 364, 2275-2289. ( 10.1098/rstb.2009.0037) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang R, et al. 2018. The global distribution and spread of the mobilized colistin resistance gene mcr-1. Nat. Commun. 9, 1179. ( 10.1038/s41467-018-03205-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu Y-Y, et al. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161-168. ( 10.1016/S1473-3099(15)00424-7) [DOI] [PubMed] [Google Scholar]
- 5.Snesrud E, He S, Chandler M, Dekker JP, Hickman AB, Mcgann P, Dyda F. 2016. A model for transposition of the colistin resistance gene mcr-1 by ISApl1. Antimicrob. Agents Chemother. 60, 6973-6976. ( 10.1128/AAC.01457-16) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Munoz-Price LS, et al. 2013. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect. Dis. 13, 785-796. ( 10.1016/S1473-3099(13)70190-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee CR, Lee JH, Park KS, Kim YB, Jeong BC, Lee SH. 2016. Global dissemination of carbapenemase-producing Klebsiella pneumoniae: epidemiology, genetic context, treatment options, and detection methods. Front. Microbiol. 7, 895. ( 10.3389/fmicb.2016.00895) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pitout JD, Laupland KB. 2008. Extended-spectrum beta-lactamase-producing Enterobacteriaceae: an emerging public-health concern. Lancet Infect. Dis. 8, 159-166. ( 10.1016/S1473-3099(08)70041-0) [DOI] [PubMed] [Google Scholar]
- 9.Canton R, Coque TM. 2006. The CTX-M beta-lactamase pandemic. Curr. Opin. Microbiol. 9, 466-475. ( 10.1016/j.mib.2006.08.011) [DOI] [PubMed] [Google Scholar]
- 10.Canton R, Gonzalez-Alba JM, Galan JC. 2012. CTX-M enzymes: origin and diffusion. Front. Microbiol. 3, 110. ( 10.3389/fmicb.2012.00110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Livermore DM, et al. 2007. CTX-M: changing the face of ESBLs in Europe. J. Antimicrob. Chemother. 59, 165-174. ( 10.1093/jac/dkl483) [DOI] [PubMed] [Google Scholar]
- 12.Arredondo-Alonso S, et al. 2020. Plasmids shaped the recent emergence of the major nosocomial pathogen Enterococcus faecium. mBio 11, 03284-19. ( 10.1128/mBio.03284-19) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leon-Sampedro R, et al. 2021. Pervasive transmission of a carbapenem resistance plasmid in the gut microbiota of hospitalized patients. Nat. Microbiol. 6, 606-616. ( 10.1038/s41564-021-00879-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gumpert H, et al. 2017. Transfer and persistence of a multi-drug resistance plasmid in situ of the infant gut microbiota in the absence of antibiotic treatment. Front. Microbiol. 8, 1852. ( 10.3389/fmicb.2017.01852) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Conlan S, et al. 2014. Single-molecule sequencing to track plasmid diversity of hospital-associated carbapenemase-producing Enterobacteriaceae. Sci. Transl. Med. 6, 254ra126. ( 10.1126/scitranslmed.3009845) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hegstad K, Mikalsen T, Coque TM, Werner G, Sundsfjord A. 2010. Mobile genetic elements and their contribution to the emergence of antimicrobial resistant Enterococcus faecalis and Enterococcus faecium. Clin. Microbiol. Infect. 16, 541-554. ( 10.1111/j.1469-0691.2010.03226.x) [DOI] [PubMed] [Google Scholar]
- 17.San Millan A. 2018. Evolution of plasmid-mediated antibiotic resistance in the clinical context. Trends Microbiol. 26, 978-985. ( 10.1016/j.tim.2018.06.007) [DOI] [PubMed] [Google Scholar]
- 18.Lopatkin AJ, Meredith HR, Srimani JK, Pfeiffer C, Durrett R, You L. 2017. Persistence and reversal of plasmid-mediated antibiotic resistance. Nat. Commun. 8, 1689. ( 10.1038/s41467-017-01532-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sorensen SJ, Bailey M, Hansen LH, Kroer N, Wuertz S. 2005. Studying plasmid horizontal transfer in situ: a critical review. Nat. Rev. Microbiol. 3, 700-710. ( 10.1038/nrmicro1232) [DOI] [PubMed] [Google Scholar]
- 20.Millan AS, Toll-Riera M, Qi Q, Maclean RC. 2015. Interactions between horizontally acquired genes create a fitness cost in Pseudomonas aeruginosa. Nat. Commun. 6, 6845. ( 10.1038/ncomms7845) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vogwill T, Maclean RC. 2015. The genetic basis of the fitness costs of antimicrobial resistance: a meta-analysis approach. Evol. Appl. 8, 284-295. ( 10.1111/eva.12202) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maclean RC, San Millan A. 2015. Microbial evolution: towards resolving the plasmid paradox. Curr. Biol. 25, R764-R767. ( 10.1016/j.cub.2015.07.006) [DOI] [PubMed] [Google Scholar]
- 23.Alonso-del Valle A, León-Sampedro R, Rodríguez-Beltrán J, DelaFuente J, Hernández-García M, Ruiz-Garbajosa P, Cantón R, Peña-Miller R, San Millán A. 2021. Variability of plasmid fitness effects contributes to plasmid persistence in bacterial communities. Nat. Commun. 12, 2653. ( 10.1038/s41467-021-22849-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lederer I, Much P, Allerberger F, Voracek T, Vielgrader H. 2005. Outbreak of shigellosis in the Vienna Zoo affecting human and non-human primates. Int. J. Infect. Dis. 9, 290-291. ( 10.1016/j.ijid.2004.11.003) [DOI] [PubMed] [Google Scholar]
- 25.Kennedy FM AJ, Needham JR, Cheasty T. 1993. Shigellosis due to occupational contact with non-human primates. Epidemiol. Infect. 110, 247-251. ( 10.1017/S0950268800068163) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baker KS, et al. 2015. Intercontinental dissemination of azithromycin-resistant shigellosis through sexual transmission: a cross-sectional study. Lancet Infect. Dis. 15, 913-921. ( 10.1016/S1473-3099(15)00002-X) [DOI] [PubMed] [Google Scholar]
- 27.Baker KS, et al. 2018. Horizontal antimicrobial resistance transfer drives epidemics of multiple Shigella species. Nat. Commun. 9, 1462. ( 10.1038/s41467-018-03949-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gilbart VL, Simms I, Jenkins C, Furegato M, Gobin M, Oliver I, Hart G, Gill ON, Hughes G. 2015. Sex, drugs and smart phone applications: findings from semistructured interviews with men who have sex with men diagnosed with Shigella flexneri 3a in England and Wales. Sex. Transm. Infect. 91, 598-602. ( 10.1136/sextrans-2015-052014) [DOI] [PubMed] [Google Scholar]
- 29.Ingle DJ, et al. 2019. Co-circulation of multidrug-resistant Shigella among men who have sex with men in Australia. Clin. Infect. Dis. 69, 1535-1544. ( 10.1093/cid/ciz005) [DOI] [PubMed] [Google Scholar]
- 30.Van Den Beld MJC, et al. 2020. A multifactorial approach for surveillance of Shigella spp. and entero-invasive Escherichia coli is important for detecting (inter)national clusters. Front. Microbiol. 11, 564103. ( 10.3389/fmicb.2020.564103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Locke RK, Greig DR, Jenkins C, Dallman TJ, Cowley LA. 2021. Acquisition and loss of CTX-M plasmids in Shigella species associated with MSM transmission in the UK. Microb. Genom. 7, 000644. ( 10.1099/mgen.0.000644) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aziz RK, et al. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genom. 9, 75. ( 10.1186/1471-2164-9-75) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Page AJ, et al. 2015. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31, 3691-3693. ( 10.1093/bioinformatics/btv421) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sysoeva TAKY, Rodriguez J, Lopatkin AJ, You L. 2020. Growth-stage-dependent regulation of conjugation. AlChE J. 66, 16848. ( 10.1002/aic.16848) [DOI] [Google Scholar]
- 35.Baharoglu Z, Krin E, Mazel D. 2012. Connecting environment and genome plasticity in the characterization of transformation-induced SOS regulation and carbon catabolite control of the Vibrio cholerae integron integrase. J. Bacteriol. 194, 1659-1667. ( 10.1128/JB.05982-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sprouffske K, Wagner A. 2016. Growthcurver: an R package for obtaining interpretable metrics from microbial growth curves. BMC Bioinf. 17, 172. ( 10.1186/s12859-016-1016-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Worley JN, et al. 2021. Genomic drivers of multidrug-resistant Shigella affecting vulnerable patient populations in the United States and Abroad. mBio 12, 03188-20. ( 10.1128/mBio.03188-20) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liao YS, Liu YY, Lo YC, Chiou CS. 2016. Azithromycin-nonsusceptible Shigella flexneri 3a in men who have sex with men, Taiwan, 2015–2016. Emerg. Infect. Dis. 23, 345-346. ( 10.3201/eid2302.161260) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moreno-Mingorance A, et al. 2021. Circulation of multi-drug-resistant Shigella sonnei and Shigella flexneri among men who have sex with men in Barcelona, Spain, 2015–2019. Int. J. Antimicrob. Agents. 58, 106378. ( 10.1016/j.ijantimicag.2021.106378) [DOI] [PubMed] [Google Scholar]
- 40.Hinic V, Seth-Smith H, Stockle M, Goldenberger D, Egli A. 2018. First report of sexually transmitted multi-drug resistant Shigella sonnei infections in Switzerland, investigated by whole genome sequencing. Swiss. Med. Wkly. 148, w14645. [DOI] [PubMed] [Google Scholar]
- 41.Blackwell GA, Hunt M, Malone KM, Lima L, Horesh G, Alako BTF, Thomson NR, Iqbal Z. 2021. Exploring bacterial diversity via a curated and searchable snapshot of archived DNA sequences. PLoS Biol. 19, e3001421. ( 10.1371/journal.pbio.3001421) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Petrova V, Chitteni-Pattu S, Drees JC, Inman RB, Cox MM. 2009. An SOS inhibitor that binds to free RecA protein: the PsiB protein. Mol. Cell 36, 121-130. ( 10.1016/j.molcel.2009.07.026) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baharoglu Z, Mazel D. 2014. SOS, the formidable strategy of bacteria against aggressions. FEMS Microbiol. Rev. 38, 1126-1145. ( 10.1111/1574-6976.12077) [DOI] [PubMed] [Google Scholar]
- 44.Qin TT, Kang HQ, Ma P, Li PP, Huang LY, Gu B. 2015. SOS response and its regulation on the fluoroquinolone resistance. Ann. Transl. Med. 3, 358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Williams PCM, Berkley JA. 2018. Guidelines for the treatment of dysentery (shigellosis): a systematic review of the evidence. Paediatr. Int. Child Health 38(sup1), S50-S65. ( 10.1080/20469047.2017.1409454) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murray K, et al. 2017. Increasing antibiotic resistance in Shigella spp. from infected New York City residents, New York, USA. Emerg. Infect. Dis. 23, 332-335. ( 10.3201/eid2302.161203) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.San Millan A, Maclean RC. 2017. Fitness costs of plasmids: a limit to plasmid transmission. Microbiol. Spectr. 5. ( 10.1128/microbiolspec.MTBP-0016-2017) [DOI] [PubMed] [Google Scholar]
- 48.Humphrey B, Thomson NR, Thomas CM, Brooks K, Sanders M, Delsol AA, Roe JM, Bennett PM, Enne VI. 2012. Fitness of Escherichia coli strains carrying expressed and partially silent IncN and IncP1 plasmids. BMC Microbiol. 12, 53. ( 10.1186/1471-2180-12-53) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Redondo-Salvo S, Fernández-López R, Ruiz R, Vielva L, de Toro M, Rocha EP, Garcillán-Barcia MP, de la Cruz F. 2020. Pathways for horizontal gene transfer in bacteria revealed by a global map of their plasmids. Nat. Commun. 11, 3602. ( 10.1038/s41467-020-17278-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hall JP, Wood AJ, Harrison E, Brockhurst MA. 2016. Source-sink plasmid transfer dynamics maintain gene mobility in soil bacterial communities. Proc. Natl Acad. Sci. USA 113, 8260-8265. ( 10.1073/pnas.1600974113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baltrus DA. 2013. Exploring the costs of horizontal gene transfer. Trends Ecol. Evol. 28, 489-495. ( 10.1016/j.tree.2013.04.002) [DOI] [PubMed] [Google Scholar]
- 52.Hall JPJ, Brockhurst MA, Dytham C, Harrison E. 2017. The evolution of plasmid stability: are infectious transmission and compensatory evolution competing evolutionary trajectories? Plasmid 91, 90-95. ( 10.1016/j.plasmid.2017.04.003) [DOI] [PubMed] [Google Scholar]
- 53.Benz F, et al. 2021. Plasmid- and strain-specific factors drive variation in ESBL-plasmid spread in vitro and in vivo. ISME J. 15, 862-878. ( 10.1038/s41396-020-00819-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hall JPJ, Wright RCT, Harrison E, Muddiman KJ, Wood AJ, Paterson S, Brockhurst MA. 2021. Plasmid fitness costs are caused by specific genetic conflicts enabling resolution by compensatory mutation. PLoS Biol. 19, e3001225. ( 10.1371/journal.pbio.3001225) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Baharoglu Z, Bikard D, Mazel D. 2010. Conjugative DNA transfer induces the bacterial SOS response and promotes antibiotic resistance development through integron activation. PLoS Genet. 6, e1001165. ( 10.1371/journal.pgen.1001165) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Golub E, Bailone A, Devoret R. 1988. A gene encoding an SOS inhibitor is present in different conjugative plasmids. J. Bacteriol. 170, 4392-4394. ( 10.1128/jb.170.9.4392-4394.1988) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smirnova GV, Tyulenev AV, Muzyka NG, Peters MA, Oktyabrsky ON. 2017. Ciprofloxacin provokes SOS-dependent changes in respiration and membrane potential and causes alterations in the redox status of Escherichia coli. Res. Microbiol. 168, 64-73. ( 10.1016/j.resmic.2016.07.008) [DOI] [PubMed] [Google Scholar]
- 58.Malaka De Silva P, Stenhouse GE, Blackwell GA, Bengtsson RJ, Jenkins C, Hall JPJ, Baker KS. 2022. Data from: A tale of two plasmids: contributions of plasmid associated phenotypes to epidemiological success among Shigella. Dryad Digital Repository. ( 10.5061/dryad.sxksn0363) [DOI] [PMC free article] [PubMed]
- 59.Malaka De Silva P, Stenhouse GE, Blackwell GA, Bengtsson RJ, Jenkins C, Hall JPJ, Baker KS. 2022. A tale of two plasmids: contributions of plasmid associated phenotypes to epidemiological success among Shigella. Figshare. ( 10.6084/m9.figshare.c.6114878) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Malaka De Silva P, Stenhouse GE, Blackwell GA, Bengtsson RJ, Jenkins C, Hall JPJ, Baker KS. 2022. Data from: A tale of two plasmids: contributions of plasmid associated phenotypes to epidemiological success among Shigella. Dryad Digital Repository. ( 10.5061/dryad.sxksn0363) [DOI] [PMC free article] [PubMed]
- Malaka De Silva P, Stenhouse GE, Blackwell GA, Bengtsson RJ, Jenkins C, Hall JPJ, Baker KS. 2022. A tale of two plasmids: contributions of plasmid associated phenotypes to epidemiological success among Shigella. Figshare. ( 10.6084/m9.figshare.c.6114878) [DOI] [PMC free article] [PubMed]
Data Availability Statement
Plasmid sequences used are publicly available in NCBI database and the accession numbers are provided in the main text. Raw data for used for the linear modelling and electronic supplementary material, file S2 are deposited in the Dryad Digital Repository (doi:10.5061/dryad.sxksn0363) [58].
Electronic supplementary material is available online [59].