Abstract
Eukaryotic cells adequately control the mass and functions of organelles in various situations. Autophagy, an intracellular degradation system, largely contributes to this organelle control by degrading the excess or defective portions of organelles. The endoplasmic reticulum (ER) is an organelle with distinct structural domains associated with specific functions. The ER dynamically changes its mass, components, and shape in response to metabolic, developmental, or proteotoxic cues to maintain or regulate its functions. Therefore, elaborate mechanisms are required for proper degradation of the ER. Here, we review our current knowledge on diverse mechanisms underlying selective autophagy of the ER, which enable efficient degradation of specific ER subdomains according to different demands of cells.
Keywords: autophagy, endoplasmic reticulum, ER‐phagy, intracellular degradation
Subject Categories: Autophagy & Cell Death
This review discusses the diverse mechanisms underlying selective autophagy of the ER, which enable efficient degradation of specific ER subdomains according to different demands of cells.

Glossary
- AIM
ATG8‐family interacting motif
- Atg11
autophagy‐related 11
- Atg39
autophagy‐related 39
- Atg40
autophagy‐related 40
- ATI1
ATG8‐interacting protein 1
- ATI2
ATG8‐interacting protein 2
- ATL3
atlastin 3
- ATZ
the Z variant of human alpha‐1 antitrypsin
- CALCOCO1
calcium‐binding and coiled‐coil domain‐containing protein 1
- CAMK2B
calcium/calmodulin‐dependent protein kinase type II subunit beta
- CCPG1
cell cycle progression gene 1
- CDK5RAP3
CDK5 regulatory subunit‐associated protein 3
- COPII
coat protein complex II
- DDRGK1
DDRGK domain containing 1
- Epr1
ER‐phagy receptor 1
- ERAD
ER‐associated degradation
- ERLAD
ER‐to‐lysosome‐associated degradation
- ERPHS
ER‐phagy sites
- ESCRT
endosomal sorting complex required for transport
- FAM134B
family with sequence similarity 134 member B
- FFAT
two phenylalanines in an acidic tract
- FIP200
focal adhesion kinase family interacting protein of 200 kDa
- GABARAP
gamma‐aminobutyric acid type A receptor‐associated protein
- HSAN I
hereditary sensory and autonomic neuropathy type I
- HSAN II
hereditary sensory and autonomic neuropathy type II
- IDR
intrinsically disordered region
- LC3
microtubule‐associated protein 1 light chain 3
- Lnp1
lunapark family member
- Lst1
lethal‐with‐sec‐thirteen 1
- LDS
LIR docking site
- LIR
LC3 interacting region
- NDV
nucleus‐derived double membrane vesicle
- NPC1
Niemann–Pick type C 1
- MIDY
mutant INS‐gene‐induced diabetes of youth
- MSBP1
membrane steroid‐binding protein 1
- PGRMC1
progesterone receptor membrane component 1
- p62
EBI3‐associated protein of 60 kDa
- RHD
reticulon homology domain
- RTN3L
the longest isoform of reticulon 3
- SEC24C
a paralog of the COPII component SEC24
- SEC62
a component of the Sec61 protein translocation machinery
- TEX264
testis expressed gene 264
- TORC1
target of rapamycin complex 1
- TRIM13
tripartite motif containing 13
- ULK1
Unc‐51 like kinase 1
- UPR
unfolded protein response
- VAP
vesicle‐associated membrane protein‐associated protein
- UDS
ubiquitin‐interacting motif docking site
- UIR
UDS‐interacting region
- UFL1
UFM1‐specific ligase 1
- UFM1
ubiquitin‐fold modifier 1
Introduction
Degradation, as well as synthesis, plays pivotal roles for cellular homeostasis in various physiological situations. Degradation of aberrant or excess components is important for the maintenance and regulation of related cellular functions. Degradation also mobilizes building blocks of macromolecules, which are used as nutrients for cells to cope with starvation. The same holds true for the degradation of organelles. Cells degrade organelles to control their mass and quality and could also obtain scarce molecules by organelle degradation. However, organelles, which are large structures bound by lipid membranes and contain different macromolecules including DNA, RNA, proteins, and sugar chains, seem difficult to be degraded. In addition, organelle degradation by cytoplasmic enzymes could result in the leakage of the contents, perturbing different cellular processes. To circumvent these difficulties, eukaryotes have evolved to utilize the system of autophagy for organelle degradation. In autophagy, organelles are sequestered within lysosomes or vacuoles, which contain different hydrolases, including lipases, proteases, nucleases, and glycosidases, and therefore can be efficiently degraded.
Autophagy mediates the degradation of a wide range of intracellular materials, including proteins, RNAs, and organelles, in lysosomes or vacuoles (Yang & Klionsky, 2010; Ohsumi, 2014; Morishita & Mizushima, 2019). Autophagy is a collective term for lysosomal/vacuolar degradation of cells' own constituents, and previous studies have revolved around three major types of autophagy: macroautophagy, microautophagy, and chaperone‐mediated autophagy (Kaushik & Cuervo, 2018; Nakatogawa, 2020; Schuck, 2020). In macroautophagy, a membrane cisterna called the isolation membrane or phagophore forms in the cytoplasm, expands, bends, and finally closes to complete a double membrane vesicle called the autophagosome (Fig 1A). During this process, cytoplasmic constituents are selectively or nonselectively sequestered within the autophagosome. Then, the outer autophagosomal membrane fuses with the endosomal/lysosomal or vacuolar membrane to allow the degradation of the inner autophagosomal membrane and the sequestered material. In selective types of macroautophagy (selective macroautophagy), proteins called macroautophagy receptors or adaptors play central roles in the selection of degradation targets (Fig 1A; Kirkin & Rogov, 2019). These proteins bind to degradation targets and interact with the scaffold protein Atg11 in yeast or FIP200 in mammals, which recruits core autophagy‐related (ATG) proteins that initiate autophagosome formation in the vicinity of the targets. Macroautophagy receptors, via a common motif called the ATG8‐family interacting motif (AIM) or LC3 interacting region (LIR) with the consensus sequence of Trp/Tyr/Phe‐X‐X‐Leu/Ile/Val, also bind to ATG8‐family proteins anchored to the isolation membrane via lipidation (conjugation to phosphatidylethanolamine) to link the targets to the membrane for their efficient sequestration into the autophagosome (Kirkin & Rogov, 2019). In microautophagy, the vacuolar or lysosomal/endosomal membrane invaginates, and membrane fission caused by the ESCRT machinery results in the release of lysosomal/endosomal membrane‐bound vesicles containing degradation targets into the lumen (Fig 1B). Microautophagy also occurs in a selective manner, and several organelles such as the nucleus, peroxisome, and the endoplasmic reticulum (ER) are known to be targets of selective microautophagy (Schuck, 2020).
Figure 1. Basic process of autophagic degradation of the ER.
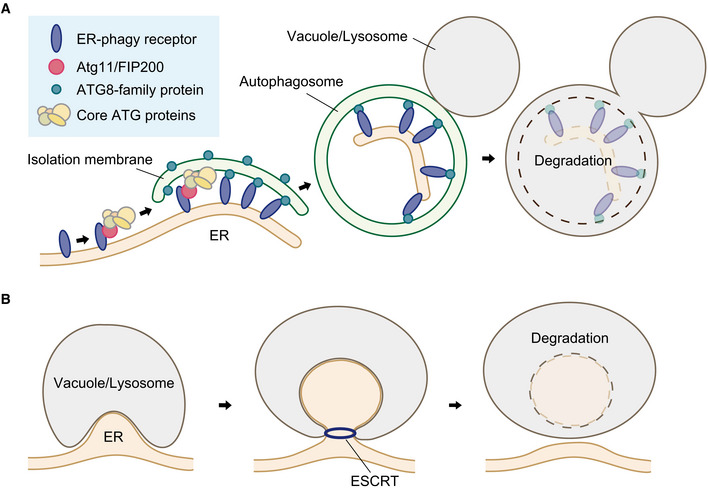
(A) In macro‐ER‐phagy, receptor proteins trigger autophagosome biogenesis in the vicinity of the ER by recruiting the Atg1 and ULK complexes via the interactions with Atg11 and FIP200 in yeast and mammalian cells, respectively. The Atg1 and ULK complexes serve as a scaffold to recruit downstream core ATG proteins, and these proteins mediate the formation and expansion of the isolation membrane (or phagophore). Receptor proteins also bind to ATG8‐family proteins anchored to the isolation membrane via conjugation to phosphatidylethanolamine to facilitate the uptake of the ER into autophagosomes following ER membrane fission and isolation membrane closure. Eventually, the outer autophagosomal membrane fuses with the vacuolar or lysosomal membrane, leading to the degradation of the inner autophagosomal membrane and sequestered ER fragments by different hydrolases within these lytic organelles. (B) In micro‐ER‐phagy, the vacuolar or lysosomal membrane directly contacts the ER, invaginates along with the ER, and causes ESCRT‐dependent fission, releasing vacuolar/lysosomal membrane‐bound vesicles containing ER fragments into the lumen.
Previous studies have identified macroautophagy receptors for the degradation of almost all the membrane‐bound organelles, that is, mitochondria, peroxisomes, lysosomes, the nucleus, the Golgi apparatus, and the ER (Gatica et al, 2018; Nthiga et al, 2021; Li & Nakatogawa, 2022). The ER, which has versatile functions, including protein and lipid synthesis, calcium homeostasis, and cell death, forms tubular and sheet‐like structures, which connect with each other and establish a network widely spreading throughout the cytoplasm (English & Voeltz, 2013). Given that the ER has distinct functional and structural domains and responds to different signals or stresses within or without cells, multiple mechanisms would be required for the proper execution of selective autophagy of the ER (ER‐phagy) to maintain the homeostasis of this organelle. A specific mechanism by which ER tubules or sheets are efficiently packed into autophagosomes would also be required. Indeed, as overviewed in this article, recent studies have revealed that diverse mechanisms involving multiple receptors work for the spatiotemporal regulation of ER‐phagy in different organisms.
ER‐phagy in yeast
Macro‐ER‐phagy in budding yeast
Macro‐ER‐phagy defined with specific receptor proteins was first reported in the budding yeast Saccharomyces cerevisiae and mammals (see below; Mochida et al, 2015; Khaminets et al, 2015). In S. cerevisiae, searching for macroautophagy receptors among Atg8‐binding proteins identified Atg39 and Atg40 as receptors responsible for the degradation of the ER membrane protein Sec63 (a subunit of the Sec61 protein translocation channel; Mochida et al, 2015). Sec63 degradation was partially impaired by deletion of either ATG39 or ATG40 and completely blocked by deletion of both. In yeast cells, the ER consists of three distinct subdomains, the perinuclear ER, the cortical (cell periphery) ER, and the cytoplasmic ER (Friedman & Voeltz, 2011; Fig 2A). While Sec63 resides in all of these ER subdomains, Atg39 and Atg40 had a clear preference in their localization, that is, the perinuclear ER and the cortical and cytoplasmic ER, respectively. Consistent with this observation, the disruption of ATG39 and ATG40 preferentially impaired the degradation of the perinuclear ER protein Hmg1 and the cortical and cytoplasmic ER protein Rtn1, respectively. Thus, Atg39 and Atg40 were found to be macro‐ER‐phagy receptors specific to these ER subdomains, rather than those redundantly acting in degradation of the entire ER (Mochida et al, 2015). However, in S. cerevisiae cells, the perinuclear ER scarcely develops and is therefore practically equivalent to the nuclear envelope. In addition, nucleus‐derived double membrane vesicles (NDVs) of ~200 nm, which form through the budding of the nuclear envelope and contain intranuclear components, are sequestered within autophagosomes depending on Atg39 (Fig 2B). Cells lacking Atg39 or expressing Atg39 mutants defective in the interaction with Atg11 and/or Atg8 exhibit abnormal nuclear morphology and cause cell death under nitrogen starvation, suggesting that Atg39‐mediated macroautophagy is important for nuclear homeostasis under the condition (Mochida et al, 2015). In addition, a recent work suggests that Atg39 is involved in the selection of nuclear proteins for autophagic degradation (also see below; Chandra et al, 2021; Mochida et al, 2022). By contrast, there is no evidence for a role of Atg39 in the maintenance or regulation of perinuclear ER function and mass. Therefore, currently, it may be more adequate to regard Atg39‐mediated autophagy as macronucleophagy rather than macro‐ER‐phagy.
Figure 2. Macro‐ER‐phagy receptors in yeasts.
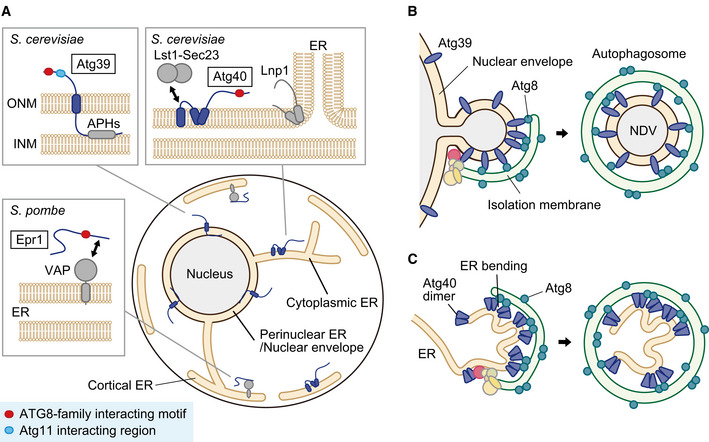
(A) The sub‐ER localization and domain architecture of ER‐phagy receptors in S. cerevisiae and S. pombe. Atg39 binds to the outer and inner nuclear membranes (ONM and INM) via the transmembrane domain and amphipathic helices (APHs), respectively. Atg40 localizes to a site for ER‐phagy depending on Lnp1 and cooperates with the Lst1‐Sec23 complex. Epr1 interacts with VAPs to localize to the ER. (B) Atg39 localizes to the outer nuclear membrane/perinuclear ER and recruits core Atg proteins to initiate autophagosome formation. It also deforms the nuclear envelope to form a nucleus‐derived double membrane vesicle (NDV) and binds to Atg8 on the isolation membrane to load the vesicle into the autophagosome. (C) Atg40 dimers mainly localize to the cortical and cytoplasmic ER, form a super‐assembly at ER contacts with the isolation membrane via multivalent interactions between Atg8, and bend the ER by the reticulon‐like function for efficient ER packing into the autophagosome.
Macro‐ER‐phagy in S. cerevisiae is induced by nitrogen starvation or rapamycin, suggesting that inactivation of TORC1 is a key to trigger macro‐ER‐phagy (Hamasaki et al, 2005; Mochida et al, 2015). In addition, ER stress, such as cell treatment with the reducing reagent dithiothreitol or the N‐linked glycosylation inhibitor tunicamycin, also induces macro‐ER‐phagy (Mizuno et al, 2020). The expression of ATG39 and ATG40 is both upregulated in response to TORC1 inactivation or ER stress (Mochida et al, 2015; Vevea et al, 2015; Mizuno et al, 2020). The transcription of ATG40 is repressed under normal conditions by the Rpd3‐Pho23 histone deacetylase complex, and this repression is thought to be canceled upon TORC1 inactivation or ER stress (Cui et al, 2019). In the transcriptional regulation of ATG39, the AMP‐activated protein kinase Snf1 phosphorylates the transcription repressors Mig1 and Mig2 to dissociate them from the ATG39 promoter in response to ER stress but not nitrogen starvation (Mizuno et al, 2020). In addition, protein kinase A and the transcription factors Msn2 and Msn4 are involved in ATG39 upregulation upon both ER stress and nitrogen starvation (Mizuno & Irie, 2021). Overexpression of Atg39 and Atg40 is not sufficient for macro‐ER‐phagy/nucleophagy induction (Mochida et al, 2015), which is therefore likely to require stress‐induced expression or posttranslational modification of related factors. For the latter, Atg39 and Atg40 may undergo phosphorylation to interact with Atg11 as with other macroautophagy receptors in yeast (Kanki et al, 2013; Mochida et al, 2014; Pfaffenwimmer et al, 2014; Tanaka et al, 2014).
Atg40 is an integral membrane protein with two hydrophobic segments and has the AIM in the C‐terminal cytoplasmic tail (the Atg11‐interacting region in Atg40 has not been determined; Fig 2A; Mochida et al, 2015). The second hydrophobic segment has a structural similarity to the reticulon homology domain (RHD), which is shared by reticulon‐family proteins and adopts a wedge‐like conformation (Voeltz et al, 2006). These proteins insert the RHD into the ER membrane and generate membrane curvature to shape ER tubules and the edge of ER sheets (Voeltz et al, 2006; Shibata et al, 2010). Atg40 also has an ability to generate membrane curvature in the ER (Mochida et al, 2020). Atg40 forms a dimer, and Atg40 dimers are further assembled via the interaction with Atg8 at contact sites between the ER and the forming autophagosomal membrane, and locally concentrated Atg40 exerts reticulon‐like activity high enough to fold the ER for its efficient packing into the autophagosome (Fig 2C). Vps13, which transfers lipids at contacts between the ER and several different organelles, was also reported to be important for ER packaging into autophagosomes (Chen et al, 2020). However, how Vps13 is involved in this process awaits further analysis.
Macro‐ER‐phagy also requires proteins involved in the deformation or shaping of the ER membrane. In S. cerevisiae, the COPII protein Sec24, which forms a complex with Sec23 and plays pivotal roles for cargo selection and COPII vesicle formation, has two paralogs, Iss1 and Lst1. A recent study discovered that the Lst1–Sec23 complex associates with Atg40 via Lst1 to organize ER‐phagy sites (ERPHS) in the ER, which are distinct from conventional ER exit sites (ERES) where the Sec24–Sec23 complex acts to form COPII vesicles (Fig 2A; Cui et al, 2019). At ERPHS, the Lst1–Sec23 complex is thought to be involved in ER deformation and/or selection of ER proteins or domains to be degraded by Atg40‐mediated macro‐ER‐phagy. Macro‐ER‐phagy in mammalian cells also requires the Lst1 homolog SEC24C but no other SEC24 isoforms (Cui et al, 2019). The finding of ERPHS was linked to the previous observation that Lnp1, which localizes to three‐way junctions formed by ER tubules and stabilizes the ER network, is important for macro‐ER‐phagy (Chen et al, 2018); Lnp1‐mediated ER remodeling is required for Atg40 recruitment to ERPHS (Fig 2A). Macro‐ER‐phagy also requires Ypt1 (RAB1 homolog), which acts in COPII vesicle targeting to the Golgi in the secretory pathway (Lipatova et al, 2013). In the context of autophagy, this GTPase is required for the recruitment of the core Atg proteins that mediate autophagosome formation as well as that of COPII vesicles which contribute to autophagosomal membranes (Wang et al, 2013, 2015; Davis et al, 2016; Shima et al, 2019). Thus, Ypt1 is likely to be involved in autophagosome formation rather than ER deformation during macro‐ER‐phagy. In COPII vesicle recruitment, Ypt1 activates Hrr25 (casein kinase 1δ), which phosphorylates Sec24, enhancing its binding to Atg9 residing in another membrane source called Atg9 vesicles (Yamamoto et al, 2012; Davis et al, 2016). Hrr25 also phosphorylates a number of macroautophagy receptors to enhance their interactions with Atg11, stimulating selective macroautophagy pathways mediated by these receptors (Mochida et al, 2014; Pfaffenwimmer et al, 2014; Tanaka et al, 2014). Thus, there is another possibility that Ypt1 activates macro‐ER‐phagy by stimulating Hrr25‐mediated phosphorylation of Atg39 and Atg40.
Atg39 is a single membrane‐spanning protein and contains the AIM and Atg11‐binding sequence in its N‐terminal region in the cytoplasm (Fig 2A; Mochida et al, 2022, Mochida et al, 2015; Chandra et al, 2021). While Atg39 is anchored to the outer nuclear membrane via the transmembrane segment, it also binds to the inner nuclear membrane via amphipathic helices in the perinuclear space region, linking these membranes and preventing Atg39 from leaking into the ER which is continuous with the nuclear envelope (Chandra et al, 2021; Mochida et al, 2022). Overexpression of Atg39 leads to the formation of protrusions or blebs from the nuclear envelope, which are likely to represent intermediate structures in the formation of NDVs to be sequestered within autophagosomes (Chandra et al, 2021; Mochida et al, 2022). Since amphipathic helices are known to deform membranes by inserting into one side of the lipid bilayer (Drin & Antonny, 2010), the amphipathic helices in the perinuclear space region of Atg39 are likely to play a central role for nuclear envelope deformation during macronucleophagy (Fig 2B).
Previous studies also described the physiological impact of macro‐ER‐phagy in yeast. The expression of ATG40 is upregulated by overexpression of ATZ, an aggregation‐prone protein that induces the unfolded protein response (UPR) and can be subjected to ER‐associated degradation (ERAD) when expressed in yeast cells (Cui et al, 2019). In addition, aggregates of ATZ accumulate in cells lacking Atg40 or Lst1. Moreover, ATG40 expression is upregulated in cells defective in the UPR, and the UPR is more strongly induced in macro‐ER‐phagy‐deficient cells in the presence or even absence of ER stress. These results suggest that Atg40‐mediated macro‐ER‐phagy, in cooperation with ERAD, degrades aberrant ER proteins to maintain proteostasis in the ER. A recent study also showed that expression of Atg40 is upregulated during meiosis and Atg40‐mediated macro‐ER‐phagy degrades the collapsed cortical ER for structural remodeling of the ER (Otto et al, 2021).
It was reported that when plasma membrane proteins are overexpressed in yeast cells, these proteins are stacked in the ER and, along with a number of endogenous ER proteins, targeted to degradation by macro‐ER‐phagy in the presence of nutrients, which depends on Atg11 but not Atg39 and Atg40 (Lipatova et al, 2013, 2020; Lipatova & Segev, 2015). There may exist an unknown receptor that mediates this type of ER‐phagy.
Macro‐ER‐phagy in fission yeast
While the homologs of Atg39 and Atg40 are not found in the fission yeast Schizosaccharomyces pombe, a recent study identified the soluble Atg8‐binding protein Epr1 as a receptor for macro‐ER‐phagy induced by ER stress (dithiothreitol treatment) in this yeast (Zhao et al, 2020). In response to ER stress, EPR1 expression is upregulated depending on the UPR sensor Ire1. VAMP‐associated proteins (VAPs) interact with proteins containing the two phenylalanines in an acidic tract (FFAT) motif to tether these proteins to the ER or establish inter‐organelle contacts (Murphy & Levine, 2016). Epr1 localizes to the ER by interacting with VAPs (Scs2 and Scs22) via the FFAT motif (Fig 2A) and targets the ER to forming autophagosomal membranes by binding to Atg8. Intriguingly, although VAPs do not contribute to autophagosome biogenesis in S. pombe unlike mammals (Zhao et al, 2018), their role for the maintenance of contacts between the ER and the plasma membrane is important for macro‐ER‐phagy. The possibility was discussed that the morphology and/or lipid composition of the ER maintained by ER‐plasma membrane contacts are important for ER fragmentation for its sequestration into autophagosomes (Zhao et al, 2020). EPR1 knockout sensitized S. pombe cells to dithiothreitol, suggesting that macro‐ER‐phagy is crucial for cellular homeostasis under ER stress. Importantly, Epr1 is dispensable for macro‐ER‐phagy induced by nitrogen starvation, implying the existence of another receptor acting under starvation conditions.
Micro‐ER‐phagy in budding yeast
Saccharomyces cerevisiae induces not only macro‐ER‐phagy but also micro‐ER‐phagy under ER stress. As a strategy to cope with accumulation of aberrant proteins in the ER, S. cerevisiae cells expand the volume of the ER via the UPR (Bernales et al, 2006; Schuck et al, 2009). Electron microscopy analyses demonstrated that stacks of expanded ER cisternae establish tight contacts with the vacuolar membrane, and then bend into whorls during their uptake into the vacuole through vacuolar membrane invagination (Bernales et al, 2006; Schuck et al, 2014; Schäfer et al, 2020). Subsequently, vacuolar membrane fission depending on the ESCRT machinery releases vacuolar membrane‐bound vesicles containing these ER whorls into the vacuolar lumen for degradation. This micro‐ER‐phagy pathway proceeds independently of macro‐ER‐phagy factors such as the core Atg proteins, Atg39, and Atg40. ER expansion without ER stress (accumulation of misfolded proteins) by activating lipid biosynthesis also led to the formation of ER whorls and their degradation via micro‐ER‐phagy, suggesting that the physiological significance of micro‐ER‐phagy under ER stress is to control the ER volume.
Micro‐ER‐phagy in fission yeast has not been described so far.
ER‐phagy in mammals
In mammals, eleven ER‐phagy receptors, FAM134B, FAM134A, FAM134C, RTN3L, CCPG1, SEC62, TEX264, ATL3, CALCOCO1, p62, and CDK5RAP3, have been identified to date—SEC62 was first reported as a receptor for macro‐ER‐phagy but recently turned out to mediate micro‐ER‐phagy as described in the later section. These receptors are diverse in terms of their molecular characteristics and functions while they share the AIM/LIR for the interaction with ATG8 family proteins and induce degradation of distinct ER subdomains in different physiological or pathological contexts (Fig 3A). In addition, some of the receptors were found to mediate the degradation of specific ER proteins. Thus, mammals seem to have evolved more elaborate mechanisms for ER‐phagy control, implicating the significance of ER‐phagy in the maintenance or regulation of ER functions in higher eukaryotes.
Figure 3. Macro‐ER‐phagy receptors in mammals.
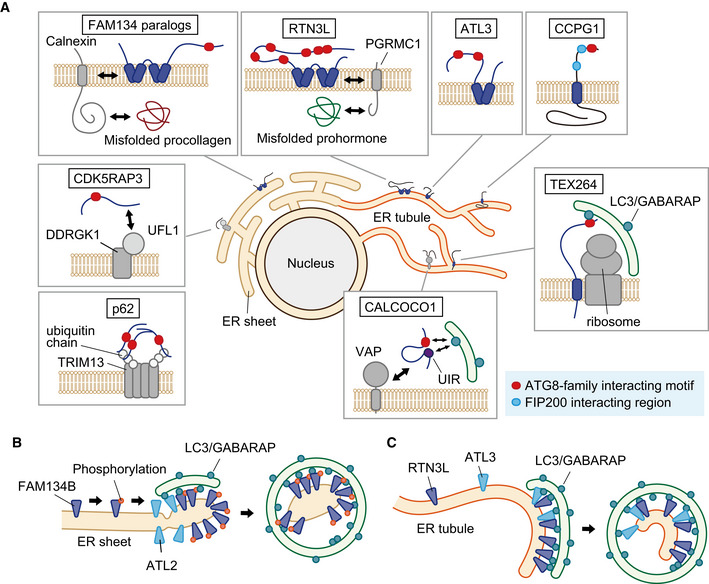
(A) The ER subdomain localization and domain architecture of mammalian ER‐phagy receptors. FAM134 paralogs and RTN3L interact with calnexin and PGRMC1 for selective degradation of misfolded procollagens and prohormones, respectively. CALCOCO1, p62, and CDK5RAP3 localize to the ER by binding to VAPs, ubiquitylated TRIM13, and the UFL1–DDRGK1 complex, respectively. The long intrinsically disordered region of TEX264 allows its LIR to bind to ATG8‐family proteins on the isolation membrane across ribosomes associated with the ER. (B) FAM134B oligomerization, which is important for its membrane fission activity, is promoted by CAMK2B‐mediated phosphorylation. FAM134B clusters at the edge of ER sheets by sensing high membrane curvature in these regions. Membrane fission activity of FAM134B, a pulling force generated by the interaction of FAM134B with ATG8‐family proteins on the isolation membrane, and ATL2 is thought to cooperatively work to separate these ER regions for sequestration into autophagosomes. (C) RTN3L forms oligomers, localizes to the tubular ER, and exerts membrane fission activity to sequester ER tubules into autophagosomes in cooperation with ATL3.
Macro‐ER‐phagy mediated by canonical receptors
FAM134 paralogs
In mammalian cells, macro‐ER‐phagy constitutively occurs at a basal level and is enhanced upon nutrient withdrawal (Khaminets et al, 2015; Liang et al, 2018). FAM134B is the first discovered macro‐ER‐phagy receptor in mammals (Khaminets et al, 2015). Its genetic perturbation impairs both constitutive and starvation‐enhanced macro‐ER‐phagy, results in abnormal ER expansion, and renders cells sensitive to different stress such as ER stress and nutrient starvation (Khaminets et al, 2015). Mutations in FAM134B can cause hereditary sensory and autonomic neuropathy type II (HSAN II), and the disease‐associated mutations were shown to decrease macro‐ER‐phagy activity (Kurth et al, 2009; Khaminets et al, 2015). FAM134B knockout mice manifest phenotypes similar to sensory neuropathy. Thus, ER turnover via FAM134B‐mediated macro‐ER‐phagy is critical for ER homeostasis in mammalian cells, especially peripheral neurons.
Similar to yeast Atg40, the N‐terminal transmembrane domains of FAM134B adopt the RHD with membrane bending (liposome/ER fragmenting) activity (Khaminets et al, 2015; Bhaskara et al, 2019; Jiang et al, 2020). However, whereas Atg40 prefers the tubular ER, FAM134B localizes to the sheet region of the ER (Fig 3A; Khaminets et al, 2015). A molecular dynamics simulation‐based study suggested that the RHD of FAM134B favors positively curved membranes, leading to the enrichment of FAM134B in the edge of ER sheets and deformation of these ER regions (Fig 3B; Bhaskara et al, 2019). It was also proposed that the interaction of FAM134B with ATG8‐family proteins on the growing isolation membrane provides a pulling force, promoting the deformation and fission of the ER regions. Molecular dynamics simulation also suggested that FAM134B is able to generate asymmetric lipid bilayer through the formation of membrane bud, which might be important for ER remodeling during macro‐ER‐phagy (Siggel et al, 2021). Atlastins are integral membrane GTPases involved in ER morphogenesis, among which ATL2 was shown to colocalize with FAM134B, and play a role in macro‐ER‐phagy. ATL2 may participate in ER remodeling to separate FAM134B‐enriched ER regions for their sequestration into autophagosomes (Fig 3B; Liang et al, 2018). FAM134B, as well as Atg40 and other reticulon family proteins, forms oligomers (Voeltz et al, 2006; Shibata et al, 2008; Jiang et al, 2020; Mochida et al, 2020). This oligomerization, which involves the RHD, is important for ER fission activity of FAM134B (Bhaskara et al, 2019; Jiang et al, 2020). Interestingly, ER stress stimulates phosphorylation in the FAM134B RHD by the calcium/calmodulin‐dependent protein kinase CAMK2B, facilitating the oligomerization of FAM134B (Jiang et al, 2020). In the same study, the authors found that one of the HSAN II‐associated mutations (G216R) enhances the oligomerization and ER fission activity of FAM134B, resulting in excessive macro‐ER‐phagy and cell death in mouse sensory neurons. This finding suggests that abnormally unregulated macro‐ER‐phagy can also be a cause of human diseases. This notion is consistent with the previous finding that excessive FAM134B‐dependent macro‐ER‐phagy causes ER stress and cell death in Hela cells (Liao et al, 2019). FAM134B and RTN3L (see below) were also suggested to be involved in the suppression of viruses, including Ebola, Dengue, and Zika; these viruses utilize the ER for their proliferation (Wu et al, 2014; Chiramel et al, 2016; Lennemann & Coyne, 2017). A recent study showed that the mTORC1‐TFEB/TFE3 axis is responsible for the transcriptional regulation of FAM134B in response to nutrient starvation (Cinque et al, 2020). In addition, this study showed that fibroblast growth factor signaling also activates macro‐ER‐phagy by upregulating FAM134B transcription via the same axis in chondrocytes. FAM134B‐mediated macro‐ER‐phagy was suggested to be important for skeletogenesis, expanding the role for macro‐ER‐phagy to tissue development.
FAM134B paralogs, FAM134A and FAM134C, also contain the LIR motif in the C‐terminal tail, which is important for ER sequestration into the autophagosome (Reggio et al, 2021). FAM134A and FAM134C contribute to ER turnover under starvation and ER stress. Stress‐responsive activation of these paralogs is thought to involve posttranslational modifications or other related factors induced by these stimuli.
FAM134B was also shown to be involved in degradation of a mutant form of NPC1, a multispanning membrane protein related to neurodegenerative Niemann–Pick type C (NPC) disease (Pallottini & Pfrieger, 2020). NPC1 is synthesized in the ER, and correctly folded NPC1 is transported to late endosomes/lysosomes, where it mediates cholesterol trafficking (Meng et al, 2020). Loss‐of‐function mutations in NPC1 cause cholesterol accumulation in late endosomes/lysosomes, leading to NPC disease. The most common, misfolding–prone mutant of NPC1 (I1061T) accumulates in the ER and is degraded via two independent pathways, ERAD and autophagy. Knockdown of BECN1 (a component of the phosphatidylinositol 3‐kinase complex essential for autophagosome formation) or FAM134B caused the accumulation of the NPC1 I1061T mutant protein, whereas it did not affect the level of wild‐type NPC1, suggesting that misfolded NPC1 is selectively degraded by FAM134B‐mediated macro‐ER‐phagy. Misfolded NPC1 may be recognized by an ER chaperone complexed with FAM134B as with misfolded procollagens as described in the later section (the I1061T mutation is located within the ER lumenal loop of NPC1).
As described in the previous section, ATZ expressed in yeast cells is cleared by Atg40‐mediated macro‐ER‐phagy (Cui et al, 2019). In mammalian cells, FAM134B and the LC3 lipidation machinery were found to be involved in lysosomal degradation of ATZ in a quite unique manner (Kamimoto et al, 2006; Fregno et al, 2018). In this pathway named ER‐to‐lysosome‐associated degradation (ERLAD), ATZ aggregates are recognized by a complex of FAM134B and the ER chaperone calnexin and loaded into ER‐derived single membrane vesicles, whose formation requires FAM134B. Subsequently, these vesicles are targeted to endolysosomes via the interaction between FAM134B and LC3 lipidated on endolysosomal membranes, followed by membrane fusion involving the ER‐resident SNARE STX17 and the endolysosomal SNARE VAMP8, which are also responsible for autophagosome–endolysosome fusion in macroautophagy pathways. Intriguingly, it was further shown that a folding‐defective mutant of type II procollagen is also degraded via ERLAD and that N‐glycan processing (deglucosylation/reglucosylation cycle) of ATZ and the procollagen mutant is important for their persistent association with calnexin and therefore for targeting ERAD‐resistant aggregates of these proteins to ERLAD (Fregno et al, 2021). The FAM134B–calnexin complex also mediates selective degradation of misfolded procollagens via macro‐ER‐phagy as described in the later section.
RTN3L and ATL3
Whereas the reticulon family protein RTN3 is involved in ER morphogenesis, its longest splicing variant (RTN3L), which contains six LIR motifs in the cytoplasmic N‐terminal region specific to this variant, was found to act as a macro‐ER‐phagy receptor under nutrient‐deprived conditions (Fig 3A; Grumati et al, 2017). In contrast to FAM134B, RTN3L localizes to the tubular ER and triggers autophagic degradation of this ER subdomain. The overexpression of RTN3L expanded the tubular ER network under nutrient‐rich conditions, and induced ER fragmentation upon nutrient starvation depending on its LIR motifs and autophagosome biogenesis. In addition, the forcible oligomerization of RTN3L caused ER fragmentation without starvation. These results suggest that RTN3L forms clusters at ER‐isolation membrane contacts via the interaction with ATG8‐family proteins on the isolation membrane, and these RTN3L clusters cause ER fission and sequester ER fragments into autophagosomes (Fig 3C; Grumati et al, 2017). A recent study also reported that RTN3L cooperates with the ER‐enriched small GTPase Rab32 for autophagic degradation of proteins enriched in mitochondria‐ER contacts (Herrera‐Cruz et al, 2021).
As mentioned above, an earlier study reported the involvement of atlastins in ER remodeling during macro‐ER‐phagy (Liang et al, 2018). It was later on found that ATL3 but no other paralogs also functions as an macro‐ER‐phagy receptor (Fig 3A; Chen et al, 2019). Among mammalian ATG8 homologs, ATL3 specifically interacts with GABARAP via two GABARAP interacting motifs, and this interaction is required for ATL3‐mediated macro‐ER‐phagy. ATL3 also interacts with RTN3L to cooperatively promote autophagic degradation of the tubular ER under nutrient starvation. Specific ATL3 binding to GABARAP may be important for the cooperation with RTN3L to avoid their competition for ATG8 homologs on forming autophagosomal membranes. It is interesting to know how these two macro‐ER‐phagy receptors, which both possess membrane remodeling activity, collaborate to sequester ER fragments into autophagosomes. Importantly, ATL3 is a responsible factor for HSAN I and ATL3 mutations linked to this disease impair the ATL3–GABARAP interaction and macro‐ER‐phagy, highlighting the importance of macro‐ER‐phagy in neuronal homeostasis along with the results of analysis of FAM134B.
CCPG1
CCPG1 is a single‐spanning ER membrane protein containing the LIR in its N‐terminal region. CCPG1 is upregulated at a transcription level upon ER stress and triggers macroautophagic degradation of the tubular ER (Smith et al, 2018; Fig 3A). Pancreatic cells in Ccpg1‐deficient mice accumulate SDS‐insoluble aggregates of ER chaperones and secretory enzymes such as amylase and carboxypeptidase, resulting in the chronic induction of the UPR and ER swelling. These mice also exhibit tissue injury associated with inflammatory infiltration in the exocrine pancreas. These results suggest that CCPG1‐dependent macro‐ER‐phagy plays an important role for the maintenance of proteostasis in the ER in exocrine pancreatic cells, which synthesize large amounts of secretory proteins and suffer from protein folding burden in the ER.
Autophagosome formation is initiated by the ULK1 and Atg1 protein kinase complexes in mammalian and yeast cells, respectively (Nakatogawa, 2020). CCPG1 interacts with the ULK1 complex component FIP200, a functional counterpart of Atg11 in yeast, via two FIP200 interacting regions (Smith et al, 2018; Fig 3A). These regions contain amino acid sequences similar to the Atg11‐binding motif in yeast macroautophagy receptors (Kirkin & Rogov, 2019). CCPG1 is likely to induce autophagosome formation in the vicinity of the ER by recruiting the ULK1 complex via the interaction with FIP200. In this regard, whether and how other ER‐phagy receptors initiate autophagosome formation remains to be clarified.
In contrast to FAM134 paralogs, RTN3L, and ATL3, CCPG1 and ER‐phagy receptors described in the following sections do not seem to have an ability to deform the ER membrane given their amino acid sequences or structural features. Therefore, the cooperation with other proteins with membrane remodeling activity would be required for ER fission during macro‐ER‐phagy involving these receptors.
TEX264
TEX264 is a single transmembrane protein with the LIR‐containing C‐terminal region exposed to the cytoplasm (Fig 3A; Chino et al, 2019; An et al, 2019). This protein has a long intrinsically disordered region (IDR) between a gyrase inhibitor‐like domain following the transmembrane domain and the LIR motif, which is important for LIR motif's passing through a high density of ribosomes associated with the ER and reaching ATG8‐family proteins on the isolation membrane (Fig 3A; Chino et al, 2019). Viewed in this light, other macro‐ER‐phagy receptors also contain IDRs between the transmembrane domain and the LIR motif, and these regions may play a similar role in linking the ER to the isolation membrane (Chino et al, 2019). TEX264 is expressed in various tissues of mice and binds to ATG8‐family proteins more efficiently than other macro‐ER‐phagy receptors (Chino et al, 2019). In addition, knockdown/knockout analyses suggested that among macro‐ER‐phagy receptors known at the moment, TEX264 contributes most to autophagic degradation of the ER under both basal and nutrient starvation conditions in HeLa cells (Chino et al, 2019). Moreover, mass spectrometry estimated that about 50% of autophagic degradation of ER proteins under amino acid depletion depends on TEX264 in 293T cells (An et al, 2019). These results suggest that TEX264 acts as a major macro‐ER‐phagy receptor in these conditions. However, the physiological significance of TEX264‐dependent macro‐ER‐phagy awaits further investigation. Interestingly, TEX264 localizes to three‐way junctions in the tubular ER network, and a cohort of ER resident proteins decrease during nutrient starvation in a TEX264‐dependent manner (An et al, 2019; Chino et al, 2019). These results suggest that TEX264 mediates the degradation of a spectrum of ER proteins different from those degraded via macro‐ER‐phagy initiated by other receptors. A recent study also showed that serine residues in the LIR of TEX264 are phosphorylated by casein kinase 2, enhancing its binding to ATG8‐family proteins and thereby stimulating TEX264‐mediated macro‐ER‐phagy (Chino et al, 2022). Another study reported that a subset of TEX264 localizes to the inner nuclear membrane and participates in the regulation of DNA replication by forming a complex with topoisomerase 1, SPRTN, and p97 (Fielden et al, 2020). ER‐phagy might be involved in this process via degradation of TEX264.
CALCOCO1
Similar to Epr1 in S. pombe, CALCOCO1 itself is a soluble protein and binds to VAPs (VAPA and VAPB) via the FFAT‐like motif to localize to the ER (Fig 3A). CALCOCO1 interacts with ATG8‐family proteins in a unique manner: it has an atypical LIR and a ubiquitin‐interacting motif docking site (UDS)‐interacting region (UIR), which bind to the LIR docking site (LDS) and the UDS on ATG8‐family proteins, respectively (CALCOCO1 preferentially interacts with GABARAP subfamily proteins; Nthiga et al, 2020). CALCOCO1 contributes to the degradation of the tubular ER under nutrient starvation or proteotoxic stress (cell treatment with the proteasome inhibitor MG132), but the physiological impact of CALCOCO1‐mediated macro‐ER‐phagy needs further analysis. It is noteworthy that CALCOCO1 is also involved in Golgiphagy (selective autophagy of the Golgi apparatus) during starvation, in which CALCOCO1 localizes to the Golgi by interacting with the Golgi‐resident palmitoyltransferase ZDHHC17, instead of VAPs, via the ZDHHC‐AR‐binding motif (Nthiga et al, 2021). It is tempting to speculate that CALCOCO1 changes its localization between the ER and the Golgi to balance the functions or mass of these organelles via autophagic degradation.
p62—N‐degron‐mediated macro‐ER‐phagy
In the N‐degron pathway, specific amino acid residues at protein N termini are recognized by proteins called N‐recognins and accordingly degraded via the ubiquitin‐proteasome system (Varshavsky, 2019). In some cases, arginine is enzymatically attached to the N termini of target proteins, and this N‐terminal arginine residue is recognized by N‐recognins. This pathway was shown to be involved in macro‐ER‐phagy in association with the ubiquitin‐binding macroautophagy receptor/adaptor p62 (or Sequestome‐1), which mediates autophagic degradation of different cellular components, including ubiquitylated proteins, Keap1 (a component of the Keap1‐Nrf2 oxidative stress response system), and peroxisomes (Lamark et al, 2017; Sánchez‐Martín & Komatsu, 2018; Ji et al, 2019). In N‐degron‐mediated macro‐ER‐phagy, p62 serves as an N‐recognin and, via the ZZ domain, binds to ER lumenal proteins, such as BiP and PDI, which are N‐terminally arginylated in the cytoplasm following retrotranslocation, leading to the oligomerization of p62 (Cha‐Molstad et al, 2015; Ji et al, 2019). These p62 oligomers are recruited to the ER by interacting with the ER‐resident E3 enzyme TRIM13, and this recruitment is promoted by the K63‐linked self‐ubiquitylation of TRIM13 (Fig 3A). Clusters of these proteins are likely to recruit core ATG proteins and initiate autophagosome formation and ER deformation. N‐degron‐mediated macro‐ER‐phagy is stimulated by the accumulation of misfolded proteins such as ATZ in the ER lumen, which promotes the retrotranslocation of ER lumenal proteins and their N‐terminal arginylation, and gets rid of the misfolded proteins from the ER.
CDK5RAP3—UFM1‐mediated macro‐ER‐phagy
Recent studies have revealed the involvement of the ubiquitin‐like protein UFM1 in mammalian and plant macro‐ER‐phagy (a UFM1 homolog does not exist in yeast; Liang et al, 2020; Stephani et al, 2020). Similar to ubiquitin, UFM1 is covalently attached to target proteins via a series of reactions catalyzed by UBA5 (E1), UFC1 (E2), and UFL1 (E3) (Komatsu et al, 2004; Tatsumi et al, 2010). A genome‐wide screen in mammalian cells identified factors associated with protein UFMylation as required for autophagic degradation of ER sheets (but not ER tubules) under nutrient starvation, including DDRGK1 (or UFBP1), an integral ER membrane protein that interacts with UFL1 (Tatsumi et al, 2010; Liang et al, 2020). UFL1 is recruited to the ER by binding to DDRGK1 and UFMylates ribosomal proteins such as RPL26 and the oligosaccharyltransferase complex subunit RPN1 (Walczak et al, 2019; Liang et al, 2020; Wang et al, 2020). Although whether the UFMylation of these proteins is important for macro‐ER‐phagy remains to be examined, it can be speculated that protein UFMylation serves as a marker for the recruitment of proteins involved in autophagosome biogenesis, similar to protein ubiquitylation of mitochondrial proteins by the PINK1‐Parkin system in selective autophagy of depolarized mitochondria (Narendra et al, 2008; Quinn et al, 2020). In the same study, proteins involved in mitochondrial oxidative phosphorylation were also identified to be important for ER‐phagy, but the underlying mechanism is still unknown.
Another group identified the conserved cytoplasmic protein CDK5RAP3 (or C53) as an ATG8‐binding protein (Stephani et al, 2020). CDK5RAP3 localizes to the ER by forming a complex with DDRGK1 and UFL1, and acts as a macro‐ER‐phagy receptor under ER stress in plant and mammalian cells (Wu et al, 2010; Walczak et al, 2019; Stephani et al, 2020). CDK5RAP3 consists of the N‐ and C‐terminal α‐helical domains and the central IDR connecting these domains, and binds to Atg8 via a noncanonical AIM named the shuffled AIM (Ile‐Asp‐Trp‐Gly) within the IDR. The group further showed that UFM1 and Atg8 competitively interact with CDK5RAP3 and that ER stress weakens UFM1 binding to CDK5RAP3 while enhancing Atg8 binding to CDK5RAP3. It was proposed that UFM1 transfer from CDK5RAP3 to other proteins upon ER stress triggers macro‐ER‐phagy. Loss of the UFMylation machinery or CDK5RAP3 induces ER stress in mammalian cells and renders plant cells sensitive to ER stress (Lemaire et al, 2011; Liang et al, 2020; Stephani et al, 2020), suggesting that CDK5RAP3‐mediated macro‐ER‐phagy is crucial for ER stress tolerance.
Interestingly, the UFMylation of the ribosomal subunit RPL26 is stimulated when protein translation is stalled during co‐translational protein translocation into the ER, and CDK5RAP3 colocalizes with stalled nascent ER proteins in the ER (Stephani et al, 2020; Wang et al, 2020). CDK5RAP3 was shown to be important for the lysosomal transport of these nascent ER proteins (Stephani et al, 2020). It is possible that macro‐ER‐phagy involving CDK5RAP3 and protein UFMylation mediates this transport to eliminate stalled nascent proteins from the ER.
SEC62‐mediated micro‐ER‐phagy
Cells cope with ER stress by upregulating proteins such as molecular chaperones and ERAD‐related proteins and increasing the volume of the ER (Hetz et al, 2020). Meanwhile, it is also important for cells to restore these changes in the ER after the resolution of ER stress. SEC62 is a subunit of the machinery for protein translocation across the ER membrane. This protein was found to mediate autophagic degradation of the ER during the recovery from ER stress, named recov‐ER‐phagy (Fumagalli et al, 2016). In the first report, given the results that SEC62 interacted with LC3 via the LIR motif and that ER degradation depended on ATG5 and ATG7, which are essential for LC3 lipidation and therefore important for autophagosome formation, it was concluded that recov‐ER‐phagy is a type of macroautophagy. However, a continuing work with ultrastructural analysis revealed that recov‐ER‐phagy occurs in a microautophagy manner, in which ER‐derived vesicles are directly engulfed by endolysosomes depending on SEC62, the LC3 lipidation system, and the ESCRT machinery, but not other core ATG proteins (Loi & Raimondi, 2019). Overexpression of SEC62 alone or depletion of another subunit of the protein translocation machinery induced SEC62 LIR‐dependent ER‐phagy (Fumagalli et al, 2016). It is likely that recov‐ER‐phagy initiates with SEC62 dissociation from the protein translocation machinery, and then it is recognized by LC3 lipidated in the endolysosomal membrane, followed by membrane invagination and closure involving the ESCRT machinery.
STING‐mediated macro‐ER‐phagy
A bacterial infection was also shown to induce autophagic degradation of the ER (Moretti et al, 2017). In infection with Gram‐positive bacteria, cyclic‐di‐adenosine monophosphate (c‐di‐AMP) serves as a pathogen‐associated molecular patterns (PAMP) (Kieser & Kagan, 2017), which is recognized by the ER‐resident pattern recognition receptor STING in phagocytes, leading to the upregulation of autophagy through the activation of the ER stress response and the subsequent inactivation of mTORC1, a negative regulator of autophagy (Moretti et al, 2017). Quantitative proteomics of autophagosome‐enriched fractions suggested that ER proteins including STING were sequestered within autophagosomes in response to bacterial stimulation of macrophages. Blocking autophagy in macrophages results in sustained ER stress and increases cell death following bacterial infection. Thus, this macro‐ER‐phagy pathway induced by STING is thought to be important for cell survival during infection by resolving ER stress. A recent study reported that STING directly interacts with LC3 via its LIR motifs, raising the possibility that STING acts as an ER‐phagy receptor in this pathway (Liu et al, 2019).
Selective degradation of specific ER proteins by ER‐phagy
As described in the previous section, ER‐phagy receptors localize to specific ER subdomains, suggesting that proteins enriched in these ER subdomains are preferentially degraded via ER‐phagy triggered by the corresponding receptors. In addition, ATZ and mutant NPC1 are efficiently degraded by FAM134B‐mediated ER‐phagy (Fregno et al, 2018; Schultz et al, 2018), suggesting the existence of the mechanism that recognizes these misfolded proteins. Moreover, recent studies have discovered the mechanisms by which procollagens or prohormones misfolded in the ER lumen are recognized and targeted to macro‐ER‐phagy.
Procollagens
Collagens are major components of the extracellular matrix in vertebrates. Procollagen is formed via the assembly of three procollagen α chains into a long triple helix in the ER lumen prior to secretion out of the cell through the Golgi apparatus. The triple helix formation of procollagens requires the collagen‐specific ER chaperone HSP47, and mutations in procollagens or HSP47 cause the accumulation of misfolded procollagens, leading to diseases such as osteogenesis imperfecta (Ito & Nagata, 2021). An earlier work reported that ERAD‐resistant aggregates of misfolded procollagens are subjected to autophagic degradation (Ishida et al, 2009). It was also reported that autophagy is important for the homeostasis of type II procollagen in the ER in growth‐plate chondrocytes (Cinque et al, 2015). A recent study revealed that these procollagens with abnormal conformation are degraded via FAM134B‐dependent macro‐ER‐phagy (Forrester et al, 2019). Although FAM134B does not have a region exposed to the ER lumen, it interacts with the transmembrane chaperone calnexin, which can bind to procollagens in the ER lumen. Cells deficient for FAM134B, calnexin, or core ATG proteins are unable to transport procollagens to lysosomes. Thus, FAM134B collaborates with calnexin to selectively degrade misfolded procollagens within the ER via macro‐ER‐phagy (Fig 3A). FAM134A and FAM134C were also shown to interact with calnexin for selective degradation of procollagen I (Reggio et al, 2021). However, FAM134A lacking the LIR could support this procollagen degradation as efficiently as the wild‐type protein, suggesting the importance of the nonreceptor function of FAM134A (e.g., ER deformation) and the presence of a coreceptor for macro‐ER‐phagy of procollagen I.
Micro‐ER‐phagy was also reported to participate in degradation of procollagen I (Omari et al, 2018). A subset of procollagen I accumulates in ERES, where COPII vesicles form in the secretory pathway, and these parts of the ER are directly sequestered within lysosomes. A misfold–prone mutant of procollagen I was degraded more efficiently than the wild‐type protein, suggesting that this pathway works as part of the quality control system for procollagen I in the ER. Intriguingly, in addition to COPII proteins, core ATG proteins, ubiquitin, and p62 accumulate on the cytoplasmic side of these ERES. However, it remains unknown whether these macroautophagy‐related factors are involved in this micro‐ER‐phagy pathway.
Prohormones
Recent studies have discovered macro‐ER‐phagy of prohormones misfolded in the ER and begun to unravel the molecular mechanism. In pancreatic β cells, insulin is synthesized as preproinsulin and translocated into the ER, where its signal peptide is cleaved to release proinsulin. Subsequently, correctly folded proinsulin is selected and transported to the Golgi, where it matures into insulin following the removal of its C‐peptide. Insulin gene mutations have been identified to cause a syndrome called mutant INS‐gene‐induced diabetes of youth (MIDY). In cells depleted for core ATG proteins including ATG5 and BECN1 or RTN3 but not for other ER‐phagy receptors, a MIDY mutant form of proinsulin was shown to form aggregates, suggesting that RTN3 (RTN3L)‐dependent macro‐ER‐phagy contributes to the alleviation of MIDY by degrading the mutant proinsulin (Cunningham et al, 2019). Remarkably, the same study showed that RTN3 was also involved in the removal of mutants of other prohormones such as pro‐opiomelanocortin and pro‐arginine‐vasopressin. Since RTN3, as well as FAM134B, does not possess an ER lumenal region, another protein that directly recognized these mutant prohormones in the ER lumen was assumed to cooperate with RTN3. Indeed, a proteomic approach in a continuing work identified the single‐spanning membrane protein PGRMC1 as an RTN3 interactor (Chen et al, 2021). Knockdown of PGRMC1, as well as that of RTN3, resulted in the accumulation of aggregates of mutant prohormones in the ER lumen. PGRMC1 was shown to interact with both RTN3 and mutant prohormones through the transmembrane domain and the C‐terminal ER lumenal region, respectively. Thus, PGRMC1 acts as a cargo receptor in RTN3‐dependent macro‐ER‐phagy (Fig 3A). Intriguingly, among seven proinsulin mutants analyzed, knockdown of PGRMC1 differently affected the degradation of these mutants, although that of RTN3 blocked degradation of all the mutants. Proinsulin mutants whose degradation involved PGRMC1 shared the feature of relatively small aggregation size (< 150 kDa; the size of monomeric proinsulin is ~10 kDa), and a mutation that decreases the aggregation size rendered degradation of a PGRMC1‐independent proinsulin mutant dependent on PGRMC1. These results suggest that PGRMC1 can recognize the size of mutant proinsulin aggregates in the ER. Future studies will reveal the mechanism of this size recognition by PGRMC1 as well as how RTN3 recognizes large aggregates of proinsulin mutants independently of PGRMC1. Proinsulin mutants entangle the wild‐type protein and thereby inhibit its secretion in a dominant‐negative manner. RTN3 overexpression and PGRMC1 knockdown were both shown to increase the secretion of wild‐type proinsulin in the presence of mutant proinsulin, providing a new insight into the development of treatments for MIDY (Cunningham et al, 2019; Chen et al, 2021).
The involvement of RTN3, which mediates tubular ER degradation, in ER‐phagy of mutant prohormones suggested the importance of retaining these prohormones in the tubular ER. A recent study showed that LNPK knockout or CLIMP63 overexpression, which increased the sheet ER, allowed mutant proinsulin to form larger liquid droplet‐like condensates in the sheet region of the ER, and that these condensates decreased in size when the tubular ER was increased by overexpression of RTN4 (Parashar et al, 2021). Condensate growth is likely to be restricted within narrow ER tubules. RTN4 overexpression in LNPK knockout cells stimulated lysosomal transport of mutant proinsulin. These results suggest that small proinsulin condensates in the tubular ER are degraded by RTN3 (RTN3L)‐mediated macro‐ER‐phagy, whereas the mutant forms large condensates (or aggregates) in the sheet ER when its degradation retards. It was also shown that the COPII subunit SEC24C (see the previous section Macro‐ER‐phagy in budding yeast) is involved in this degradation of mutant proinsulin (Cui et al, 2019; Parashar et al, 2021).
ER‐phagy in plants
In Arabidopsis thaliana cells, macro‐ER‐phagy is induced by CDK5RAP3 and UFM1 under ER stress as described in the previous section (Fig 4; Stephani et al, 2020). An earlier study reported that ER stress‐induced macro‐ER‐phagy in A. thaliana is under the control of the UPR sensor IRE1b but not its splicing target bZIP60 (Liu et al, 2012). The ER‐phagy receptor functions of SEC62 and reticulons (RTN1/RTN2) under ER stress were also suggested in Arabidopsis and maize cells, respectively (Fig 4; Hu et al, 2020; Zhang et al, 2020). The plant‐specific, Atg8‐binding transmembrane proteins ATI1 and ATI2 were found to trigger macro‐ER‐phagy upon dark‐induced carbon starvation in A. thaliana (Fig 4; Honig et al, 2012; Wu et al, 2021). These ATI proteins localize to the ER and associate with ER‐derived bodies (ER ATI bodies) under carbon starvation to sequester these bodies into autophagosomes for degradation in the vacuole. In addition, the ATI proteins interact with the membrane steroid‐binding protein MSBP1 and thereby facilitate its degradation via macro‐ER‐phagy following carbon starvation (Wu et al, 2021). The ATI proteins also interact with the argonaute protein AGO1 associated with the ER and induce its vacuolar degradation following the expression of the viral suppressor of RNA silencing protein P0 (Michaeli et al, 2019). It is noteworthy that the ATI proteins were also reported to be involved in macroautophagic degradation of plastid‐derived bodies under carbon starvation and during senescence (Michaeli et al, 2014).
Figure 4. Macro‐ER‐phagy receptors in plants.

CDK5RAP3 is targeted to the ER via the interaction with the UFL1–DDRGK1 complex and acts as a macro‐ER‐phagy receptor in A. thaliana cells in a manner similar to that in mammalian cells. ATI1 and ATI2 interact with Atg8 to sequester ER‐derived bodies into autophagosomes upon dark‐induced carbon starvation in A. thaliana. MSBP1 interacts with these ATI proteins to be efficiently degraded via this macro‐ER‐phagy pathway. SEC62 and RTN1/RTN2 were also suggested to be macro‐ER‐phagy receptors in A. thaliana and maize, respectively.
Conclusions
Eukaryotes, especially mammals, have evolved diverse mechanisms to regulate different modes of ER‐phagy: macro‐ER‐phagy, micro‐ER‐phagy, and direct fusion of ER‐derived vesicles to endolysosomes. Although ER‐phagy was discovered more recently than selective autophagy of other organelles such as peroxisomes and mitochondria, the largest number of receptors have been identified for ER‐phagy. These receptors mediate the degradation of specific ER subdomains in different situations, including nutrient starvation, ER stress, and the recovery phase from ER stress, as well as in the context of development, immunity, and disease pathogenesis. In addition, ER‐phagy receptors have acquired unique molecular functions and characteristics for efficiently packing portions of the ER, which widely spreads throughout the cytoplasm and is associated with a high density of ribosomes. Moreover, some ER‐phagy receptors were found to capture misfolded proteins in the ER lumen via cargo receptors. Unlike core ATG proteins, most ER‐phagy receptors are not evolutionarily conserved, suggesting that these receptors have emerged independently during evolution so as to fit the requirements of each organism. Meanwhile, it is an interesting fact that different ER‐phagy receptors share functional modules, such as the AIM/LIR, FFAT motif, and RHD. The diversity in molecules and mechanisms emphasizes the significance of ER‐phagy in the maintenance of ER homeostasis and the regulation of ER functions in different organisms.
As overviewed in this article, recent studies have largely advanced our understanding of the mechanisms and physiological roles of ER‐phagy. However, there remain fundamental issues to be addressed. Many ER‐phagy receptors were found to mediate subdomain‐specific ER degradation, but its physiological significance remains unknown. Whereas protein ubiquitylation plays pivotal roles in the initiation of selective autophagy of mitochondria, peroxisomes, and damaged lysosomes in mammalian cells (Gatica et al, 2018), its involvement in ER‐phagy is still poorly understood, although N‐degron‐mediated ER‐phagy was reported recently (Ji et al, 2019). Further analysis may discover new ubiquitin‐dependent ER‐phagy pathways. Alternatively, since the ubiquitin–proteasome system (ERAD) has a dominant role for the quality control of ER proteins, ubiquitin‐independent mechanisms might have been employed for ER‐phagy to avoid undesirable crosstalk. Nonetheless, a collaboration between ERAD and ER‐phagy in ER protein quality control is an important issue, which awaits deeper analysis. The cooperation of ER‐phagy receptors and the interplay between different ER‐phagy pathways are also interesting issues to be investigated. Future studies will answer these questions and further reveal the unexpected roles and elaborate mechanisms of ER‐phagy.
In need of answers.
How do ER‐phagy receptors trigger ER‐phagy?
How do different ER‐phagy receptors cooperate in the initiation of ER‐phagy, ER deformation/fission, and ER sequestration into the autophagosome?
Is there an interplay between different ER‐phagy pathways?
What is the physiological significance of subdomain‐specific ER degradation by ER‐phagy?
How significantly is protein ubiquitylation involved in ER‐phagy?
How do ER‐phagy pathways collaborate with ERAD in ER protein quality control?
Author contributions
Keisuke Mochida: Writing – original draft; writing – review and editing. Hitoshi Nakatogawa: Writing – original draft; writing – review and editing.
Disclosure and competing interests statement
The authors declare that they have no conflict of interest.
Acknowledgments
Research in the authors' group is supported in part by KAKENHI Grants‐in‐Aid for Scientific Research JP17H01430 and JP19H05708 (to H.N.) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan; AMED Grant Number JP21gm1410004 (to H.N.); STAR Grant funded by the Tokyo Tech Fund (to H.N.).
EMBO reports (2022) 23: e55192
See the Glossary for abbreviations used in this article.
References
- An H, Ordureau A, Paulo JA, Shoemaker CJ, Denic V, Harper JW (2019) TEX264 is an endoplasmic reticulum‐resident ATG8‐interacting protein critical for ER remodeling during nutrient stress. Mol Cell 74: 891–908.e10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernales S, McDonald KL, Walter P (2006) Autophagy counterbalances endoplasmic reticulum expansion during the unfolded protein response. PLoS Biol 4: e423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskara RM, Grumati P, Garcia‐Pardo J, Kalayil S, Covarrubias‐Pinto A, Chen W, Kudryashev M, Dikic I, Hummer G (2019) Curvature induction and membrane remodeling by FAM134B reticulon homology domain assist selective ER‐phagy. Nat Commun 10: 2370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha‐Molstad H, Sung KS, Hwang J, Kim KA, Yu JE, Yoo YD, Jang JM, Han DH, Molstad M, Kim JG et al (2015) Amino‐terminal arginylation targets endoplasmic reticulum chaperone BiP for autophagy through p62 binding. Nat Cell Biol 17: 917–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra S, Mannino PJ, Thaller DJ, Ader NR, King MC, Melia TJ, Lusk CP (2021) Atg39 selectively captures inner nuclear membrane into lumenal vesicles for delivery to the autophagosome. J Cell Biol 220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Cui Y, Parashar S, Novick PJ, Ferro‐Novick S (2018) ER‐phagy requires Lnp1, a protein that stabilizes rearrangements of the ER network. Proc Natl Acad Sci U S A 115: E6237–E6244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y‐J, Knupp J, Arunagiri A, Haataja L, Arvan P, Tsai B (2021) PGRMC1 acts as a size‐selective cargo receptor to drive ER‐phagic clearance of mutant prohormones. Nat Commun 12: 5991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Mari M, Parashar S, Liu D, Cui Y, Reggiori F, Novick PJ, Ferro‐Novick S (2020) Vps13 is required for the packaging of the ER into autophagosomes during ER‐phagy. Proc Natl Acad Sci U S A 117: 18530–18539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Xiao Y, Chai P, Zheng P, Teng J, Chen J (2019) ATL3 is a tubular ER‐Phagy receptor for GABARAP‐mediated selective autophagy. Curr Biol 29: 846–855.e6 [DOI] [PubMed] [Google Scholar]
- Chino H, Hatta T, Natsume T, Mizushima N (2019) Intrinsically disordered protein TEX264 mediates ER‐phagy. Mol Cell 74: 909–921.e6 [DOI] [PubMed] [Google Scholar]
- Chino H, Yamasaki A, Ode KL, Ueda HR, Noda NN, Mizushima N (2022) Phosphorylation by casein kinase 2 enhances the interaction between ER‐phagy receptor TEX264 and ATG8 proteins. EMBO Rep 23: e54801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiramel AI, Dougherty JD, Nair V, Robertson SJ, Best SM (2016) FAM134B, the selective autophagy receptor for endoplasmic reticulum turnover, inhibits replication of Ebola virus strains Makona and Mayinga. J Infect Dis 214: S319–S325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinque L, Forrester A, Bartolomeo R, Svelto M, Venditti R, Montefusco S, Polishchuk E, Nusco E, Rossi A, Medina DL et al (2015) FGF signalling regulates bone growth through autophagy. Nature 528: 272–275 [DOI] [PubMed] [Google Scholar]
- Cinque L, Leonibus C, Iavazzo M, Krahmer N, Intartaglia D, Salierno FG, De Cegli R, Di Malta C, Svelto M, Lanzara C et al (2020) MiT/ TFE factors control ER ‐phagy via transcriptional regulation of FAM 134B. EMBO J 39: e105696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y, Parashar S, Zahoor M, Needham PG, Mari M, Zhu M, Chen S, Ho H‐C, Reggiori F, Farhan H et al (2019) A COPII subunit acts with an autophagy receptor to target endoplasmic reticulum for degradation. Science 365: 53–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham CN, Williams JM, Knupp J, Arunagiri A, Arvan P, Tsai B (2019) Cells deploy a two‐pronged strategy to rectify misfolded proinsulin aggregates. Mol Cell 75: 442–456.e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis S, Wang J, Zhu M, Stahmer K, Lakshminarayan R, Ghassemian M, Jiang Y, Miller EA, Ferro‐Novick S (2016) Sec24 phosphorylation regulates autophagosome abundance during nutrient deprivation. eLife 5: e21167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drin G, Antonny B (2010) Amphipathic helices and membrane curvature. FEBS Lett 584: 1840–1847 [DOI] [PubMed] [Google Scholar]
- English AR, Voeltz GK (2013) Endoplasmic reticulum structure and interconnections with other organelles. Cold Spring Harb Perspect Biol 5: a013227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fielden J, Wiseman K, Torrecilla I, Li S, Hume S, Chiang S‐C, Ruggiano A, Narayan Singh A, Freire R, Hassanieh S et al (2020) TEX264 coordinates p97‐ and SPRTN‐mediated resolution of topoisomerase 1‐DNA adducts. Nat Commun 11: 1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrester A, De Leonibus C, Grumati P, Fasana E, Piemontese M, Staiano L, Fregno I, Raimondi A, Marazza A, Bruno G et al (2019) A selective ER‐phagy exerts procollagen quality control via a calnexin‐FAM134B complex. EMBO J 38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fregno I, Fasana E, Bergmann TJ, Raimondi A, Loi M, Soldà T, Galli C, D'Antuono R, Morone D, Danieli A et al (2018) ER ‐to‐lysosome‐associated degradation of proteasome‐resistant ATZ polymers occurs via receptor‐mediated vesicular transport. EMBO J 37: 1–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fregno I, Fasana E, Soldà T, Galli C, Molinari M (2021) N‐glycan processing selects ERAD‐resistant misfolded proteins for ER‐to‐lysosome‐associated degradation. EMBO J 40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman JR, Voeltz GK (2011) The ER in 3D: A multifunctional dynamic membrane network. Trends Cell Biol 21: 709–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli F, Noack J, Bergmann TJ, Cebollero E, Pisoni GB, Fasana E, Fregno I, Galli C, Loi M, Soldà T et al (2016) Translocon component Sec62 acts in endoplasmic reticulum turnover during stress recovery. Nat Cell Biol 18: 1173–1184 [DOI] [PubMed] [Google Scholar]
- Gatica D, Lahiri V, Klionsky DJ (2018) Cargo recognition and degradation by selective autophagy. Nat Cell Biol 20: 233–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grumati P, Morozzi G, Hölper S, Mari M, Harwardt M‐LI, Yan R, Müller S, Reggiori F, Heilemann M, Dikic I (2017) Full length RTN3 regulates turnover of tubular endoplasmic reticulum via selective autophagy. eLife 6: e25555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamasaki M, Noda T, Baba M, Ohsumi Y (2005) Starvation triggers the delivery of the endoplasmic reticulum to the vacuole via autophagy in yeast. Traffic 6: 56–65 [DOI] [PubMed] [Google Scholar]
- Herrera‐Cruz MS, Yap MC, Tahbaz N, Phillips K, Thomas L, Thomas G, Simmen T (2021) Rab32 uses its effector reticulon 3L to trigger autophagic degradation of mitochondria‐associated membrane (MAM) proteins. Biol Direct 16: 22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetz C, Zhang K, Kaufman RJ (2020) Mechanisms, regulation and functions of the unfolded protein response. Nat Rev Mol Cell Biol 21: 421–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honig A, Avin‐Wittenberg T, Ufaz S, Galili G (2012) A new type of compartment, defined by plant‐specific Atg8‐interacting proteins, is induced upon exposure of Arabidopsis plants to carbon starvation. Plant Cell 24: 288–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S, Ye H, Cui Y, Jiang L (2020) AtSec62 is critical for plant development and is involved in ER‐phagy in Arabidopsis thaliana. J Integr Plant Biol 62: 181–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida Y, Yamamoto A, Kitamura A, Lamandé SR, Yoshimori T, Bateman JF, Kubota H, Nagata K (2009) Autophagic elimination of misfolded procollagen aggregates in the endoplasmic reticulum as a means of cell protection. Mol Biol Cell 20: 2744–2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Nagata K (2021) Quality control of procollagen in cells. Annu Rev Biochem 90: 631–658 [DOI] [PubMed] [Google Scholar]
- Ji CH, Kim HY, Heo AJ, Lee SH, Lee MJ, Bin KS, Srinivasrao G, Mun SR, Cha‐Molstad H, Ciechanover A et al (2019) The N‐degron pathway mediates ER‐phagy. Mol Cell 75: 1058–1072.e9 [DOI] [PubMed] [Google Scholar]
- Jiang X, Wang X, Ding X, Du M, Li B, Weng X, Zhang J, Li L, Tian R, Zhu Q et al (2020) FAM134B oligomerization drives endoplasmic reticulum membrane scission for ER‐phagy. EMBO J 39: e102608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamimoto T, Shoji S, Hidvegi T, Mizushima N, Umebayashi K, Perlmutter DH, Yoshimori T (2006) Intracellular inclusions containing mutant alpha1‐antitrypsin Z are propagated in the absence of autophagic activity. J Biol Chem 281: 4467–4476 [DOI] [PubMed] [Google Scholar]
- Kanki T, Kurihara Y, Jin X, Goda T, Ono Y, Aihara M, Hirota Y, Saigusa T, Aoki Y, Uchiumi T et al (2013) Casein kinase 2 is essential for mitophagy. EMBO Rep 14: 788–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik S, Cuervo AM (2018) The coming of age of chaperone‐mediated autophagy. Nat Rev Mol Cell Biol 19: 365–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaminets A, Heinrich T, Mari M, Grumati P, Huebner AK, Akutsu M, Liebmann L, Stolz A, Nietzsche S, Koch N et al (2015) Regulation of endoplasmic reticulum turnover by selective autophagy. Nature 522: 354–358 [DOI] [PubMed] [Google Scholar]
- Kieser KJ, Kagan JC (2017) Multi‐receptor detection of individual bacterial products by the innate immune system. Nat Rev Immunol 17: 376–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkin V, Rogov VV (2019) A diversity of selective autophagy receptors determines the specificity of the autophagy pathway. Mol Cell 76: 268–285 [DOI] [PubMed] [Google Scholar]
- Komatsu M, Chiba T, Tatsumi K, Iemura S, Tanida I, Okazaki N, Ueno T, Kominami E, Natsume T, Tanaka K (2004) A novel protein‐conjugating system for Ufm1, a ubiquitin‐fold modifier. EMBO J 23: 1977–1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurth I, Pamminger T, Hennings JC, Soehendra D, Huebner AK, Rotthier A, Baets J, Senderek J, Topaloglu H, Farrell SA et al (2009) Mutations in FAM134B, encoding a newly identified Golgi protein, cause severe sensory and autonomic neuropathy. Nat Genet 41: 1179–1181 [DOI] [PubMed] [Google Scholar]
- Lamark T, Svenning S, Johansen T (2017) Regulation of selective autophagy: The p62/SQSTM1 paradigm. Essays Biochem 61: 609–624 [DOI] [PubMed] [Google Scholar]
- Lemaire K, Moura RF, Granvik M, Igoillo‐Esteve M, Hohmeier HE, Hendrickx N, Newgard CB, Waelkens E, Cnop M, Schuit F (2011) Ubiquitin fold modifier 1 (UFM1) and its target UFBP1 protect pancreatic Beta cells from ER stress‐induced apoptosis. PLoS One 6: e18517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennemann NJ, Coyne CB (2017) Dengue and zika viruses subvert reticulophagy by NS2B3‐mediated cleavage of FAM134B. Autophagy 13: 322–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Nakatogawa H (2022) Degradation of nuclear components via different autophagy pathways. Trends Cell Biol 32: 574–584 [DOI] [PubMed] [Google Scholar]
- Liang JR, Lingeman E, Ahmed S, Corn JE (2018) Atlastins remodel the endoplasmic reticulum for selective autophagy. J Cell Biol 217: 3354–3367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang JR, Lingeman E, Luong T, Ahmed S, Muhar M, Nguyen T, Olzmann JA, Corn JE (2020) A genome‐wide ER‐phagy screen highlights key roles of mitochondrial metabolism and ER‐resident UFMylation. Cell 180: 1160–1177.e20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Duan B, Zhang Y, Zhang X, Xia B (2019) Excessive ER‐phagy mediated by the autophagy receptor FAM134B results in ER stress, the unfolded protein response, and cell death in HeLa cells. J Biol Chem 294: 20009–20023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipatova Z, Gyurkovska V, Zhao SF, Segev N (2020) Characterization of constitutive ER‐phagy of excess membrane proteins. PLoS Genet 16: e1009255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipatova Z, Segev N (2015) A role for macro‐ER‐Phagy in ER quality control. PLoS Genet 11: e1005390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipatova Z, Shah AH, Kim JJ, Mulholland JW, Segev N (2013) Regulation of ER‐phagy by a Ypt/Rab GTPase module. Mol Biol Cell 24: 3133–3144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Burgos JS, Deng Y, Srivastava R, Howell SH, Bassham DC (2012) Degradation of the endoplasmic reticulum by autophagy during endoplasmic reticulum stress in Arabidopsis. Plant Cell 24: 4635–4651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Wu H, Wang C, Li Y, Tian H, Siraj S, Sehgal SA, Wang X, Wang J, Shang Y et al (2019) STING directly activates autophagy to tune the innate immune response. Cell Death Differ 26: 1735–1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loi M, Raimondi A (2019) Morone D & Molinari M (2019) ESCRT‐III‐driven piecemeal micro‐ER‐phagy remodels the ER during recovery from ER stress. Nat Commun 101: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Y, Heybrock S, Neculai D, Saftig P (2020) Cholesterol handling in lysosomes and beyond. Trends Cell Biol 30: 452–466 [DOI] [PubMed] [Google Scholar]
- Michaeli S, Clavel M, Lechner E, Viotti C, Wu J, Dubois M, Hacquard T, Derrien B, Izquierdo E, Lecorbeiller M et al (2019) The viral F‐box protein P0 induces an ER‐derived autophagy degradation pathway for the clearance of membrane‐bound AGO1. Proc Natl Acad Sci U S A 116: 22872–22883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaeli S, Honig A, Levanony H, Peled‐Zehavi H, Galili G (2014) Arabidopsis ATG8‐INTERACTING PROTEIN1 is involved in autophagy‐dependent vesicular trafficking of plastid proteins to the vacuole. Plant Cell 26: 4084–4101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno T, Irie K (2021) Msn2/4 transcription factors positively regulate expression of Atg39 ER‐phagy receptor. Sci Rep 11: 11919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno T, Muroi K, Irie K (2020) Snf1 AMPK positively regulates ER‐phagy via expression control of Atg39 autophagy receptor in yeast ER stress response. PLoS Genet 16: e1009053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochida K, Ohsumi Y, Nakatogawa H (2014) Hrr25 phosphorylates the autophagic receptor Atg34 to promote vacuolar transport of α‐mannosidase under nitrogen starvation conditions. FEBS Lett 588: 3862–3869 [DOI] [PubMed] [Google Scholar]
- Mochida K, Oikawa Y, Kimura Y, Kirisako H, Hirano H, Ohsumi Y, Nakatogawa H (2015) Receptor‐mediated selective autophagy degrades the endoplasmic reticulum and the nucleus. Nature 522: 359–362 [DOI] [PubMed] [Google Scholar]
- Mochida K, Otani T, Katsumata Y, Kirisako H, Kakuta C, Kotani T, Nakatogawa H (2022) Atg39 links and deforms the outer and inner nuclear membranes in selective autophagy of the nucleus. J Cell Biol 221: e202103178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochida K, Yamasaki A, Matoba K, Kirisako H, Noda NN, Nakatogawa H (2020) Super‐assembly of ER‐phagy receptor Atg40 induces local ER remodeling at contacts with forming autophagosomal membranes. Nat Commun 11: 3306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti J, Roy S, Bozec D, Martinez J, Chapman JR, Ueberheide B, Lamming DW, Chen ZJ, Horng T, Yeretssian G et al (2017) STING senses microbial viability to orchestrate stress‐mediated autophagy of the endoplasmic reticulum. Cell 171: 809–823.e13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishita H, Mizushima N (2019) Diverse cellular roles of autophagy. Annu Rev Cell Dev Biol 35: 453–475 [DOI] [PubMed] [Google Scholar]
- Murphy SE, Levine TP (2016) VAP, a versatile access point for the endoplasmic reticulum: review and analysis of FFAT‐like motifs in the VAPome. Biochim Biophys Acta 1861: 952–961 [DOI] [PubMed] [Google Scholar]
- Nakatogawa H (2020) Mechanisms governing autophagosome biogenesis. Nat Rev Mol Cell Biol 21: 439–458 [DOI] [PubMed] [Google Scholar]
- Narendra D, Tanaka A, Suen DF, Youle RJ (2008) Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol 183: 795–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nthiga TM, Shrestha BK, Sjøttem E, Bruun J‐A, Larsen KB, Bhujabal Z, Lamark T, Johansen T (2020) CALCOCO 1 acts with VAMP‐associated proteins to mediate ER ‐phagy. EMBO J 39: e103649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nthiga TM, Shrestha BK, Bruun J‐A, Larsen KB, Lamark T, Johansen T (2021) Regulation of Golgi turnover by CALCOCO1‐mediated selective autophagy. J Cell Biol 220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsumi Y (2014) Historical landmarks of autophagy research. Cell Res 24: 9–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omari S, Makareeva E, Roberts‐Pilgrim A, Mirigian L, Jarnik M, Ott C, Lippincott‐Schwartz J, Leikin S (2018) Noncanonical autophagy at ER exit sites regulates procollagen turnover. Proc Natl Acad Sci U S A 115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto GM, Cheunkarndee T, Leslie JM, Brar GA (2021) Programmed cortical ER collapse drives selective ER degradation and inheritance in yeast meiosis. J Cell Biol 220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallottini V, Pfrieger FW (2020) Understanding and treating Niemann‐pick type C disease: models matter. Int J Mol Sci 21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parashar S, Chidambaram R, Chen S, Liem CR, Griffis E, Lambert GG, Shaner NC, Wortham M, Hay JC, Ferro‐Novick S (2021) Endoplasmic reticulum tubules limit the size of misfolded protein condensates. Elife 10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffenwimmer T, Reiter W, Brach T, Nogellova V, Papinski D, Schuschnig M, Abert C, Ammerer G, Martens S, Kraft C (2014) Hrr25 kinase promotes selective autophagy by phosphorylating the cargo receptor Atg19. EMBO Rep 15: 862–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn PMJ, Moreira PI, Ambrósio AF, Alves CH (2020) PINK1/PARKIN signalling in neurodegeneration and neuroinflammation. Acta Neuropathol Commun 8: 189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggio A, Buonomo V, Berkane R, Bhaskara RM, Tellechea M, Peluso I, Polishchuk E, Di Lorenzo G, Cirillo C, Esposito M et al (2021) Role of FAM134 paralogues in endoplasmic reticulum remodeling, ER‐phagy, and collagen quality control. EMBO Rep 22: e52289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez‐Martín P, Komatsu M (2018) p62/SQSTM1 – steering the cell through health and disease. J Cell Sci 131: jcs222836 [DOI] [PubMed] [Google Scholar]
- Schäfer JA, Schessner JP, Bircham PW, Tsuji T, Funaya C, Pajonk O, Schaeff K, Ruffini G, Papagiannidis D, Knop M et al (2020) ESCRT machinery mediates selective microautophagy of endoplasmic reticulum in yeast. EMBO J 39: 1–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck S (2020) Microautophagy – distinct molecular mechanisms handle cargoes of many sizes. J Cell Sci 133: jcs246322 [DOI] [PubMed] [Google Scholar]
- Schuck S, Gallagher CM, Walter P (2014) ER‐phagy mediates selective degradation of endoplasmic reticulum independently of the core autophagy machinery. J Cell Sci 127: 4078–4088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck S, Prinz WA, Thorn KS, Voss C, Walter P (2009) Membrane expansion alleviates endoplasmic reticulum stress independently of the unfolded protein response. J Cell Biol 187: 525–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz ML, Krus KL, Kaushik S, Dang D, Chopra R, Qi L, Shakkottai VG, Cuervo AM, Lieberman AP (2018) Coordinate regulation of mutant NPC1 degradation by selective ER autophagy and MARCH6‐dependent ERAD. Nat Commun 9: 3671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata Y, Shemesh T, Prinz WA, Palazzo AF, Kozlov MM, Rapoport TA (2010) Mechanisms determining the morphology of the peripheral ER. Cell 143: 774–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata Y, Voss C, Rist JM, Hu J, Rapoport TA, Prinz WA, Voeltz GK (2008) The reticulon and Dp1/Yop1p proteins form immobile oligomers in the tubular endoplasmic reticulum. J Biol Chem 283: 18892–18904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima T, Kirisako H, Nakatogawa H (2019) COPII vesicles contribute to autophagosomal membranes. J Cell Biol 218: 1503–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siggel M, Bhaskara RM, Moesser MK, Ikić ID, Hummer G (2021) FAM134B‐RHD protein clustering drives spontaneous budding of asymmetric membranes. J Phys Chem Lett 12: 1926–1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MD, Harley ME, Kemp AJ, Wills J, Lee M, Arends M, von Kriegsheim A, Behrends C, Wilkinson S (2018) CCPG1 is a non‐canonical autophagy cargo receptor essential for ER‐Phagy and pancreatic ER Proteostasis. Dev Cell 44: 217–232.e11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephani M, Picchianti L, Gajic A, Beveridge R, Skarwan E, de Medina S, Hernandez V, Mohseni A, Clavel M, Zeng Y et al (2020) A cross‐kingdom conserved ER‐phagy receptor maintains endoplasmic reticulum homeostasis during stress. eLife 9: 1–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka C, Tan LJ, Mochida K, Kirisako H, Koizumi M, Asai E, Sakoh‐Nakatogawa M, Ohsumi Y, Nakatogawa H (2014) Hrr25 triggers selective autophagy‐related pathways by phosphorylating receptor proteins. J Cell Biol 207: 91–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsumi K, Sou Y, Tada N, Nakamura E, Iemura S, Natsume T, Kang SH, Chung CH, Kasahara M, Kominami E et al (2010) A novel type of E3 ligase for the Ufm1 conjugation system. J Biol Chem 285: 5417–5427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky A (2019) N‐degron and C‐degron pathways of protein degradation. Proc Natl Acad Sci U S A 116: 358–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vevea JD, Garcia EJ, Chan RB, Di PG, Mccaffery JM, Pon LA, Vevea JD, Garcia EJ, Chan RB, Zhou B et al (2015) Role for lipid droplet biogenesis and Microlipophagy in adaptation to lipid imbalance in article role for lipid droplet biogenesis and Microlipophagy in adaptation to lipid imbalance in yeast. Dev Cell 35: 584–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voeltz GK, Prinz WA, Shibata Y, Rist JM, Rapoport TA (2006) A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell 124: 573–586 [DOI] [PubMed] [Google Scholar]
- Walczak CP, Leto DE, Zhang L, Riepe C, Muller RY, DaRosa PA, Ingolia NT, Elias JE, Kopito RR (2019) Ribosomal protein RPL26 is the principal target of UFMylation. Proc Natl Acad Sci U S A 116: 1299–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Davis S, Menon S, Zhang J, Ding J, Cervantes S, Miller E, Jiang Y, Ferro‐Novick S (2015) Ypt1/Rab1 regulates Hrr25/CK1δ kinase activity in ER–Golgi traffic and macroautophagy. J Cell Biol 210: 273–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Menon S, Yamasaki A, Chou H‐T, Walz T, Jiang Y, Ferro‐Novick S (2013) Ypt1 recruits the Atg1 kinase to the preautophagosomal structure. Proc Natl Acad Sci U S A 110: 9800–9805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Xu Y, Rogers H, Saidi L, Noguchi CT, Li H, Yewdell JW, Guydosh NR, Ye Y (2020) UFMylation of RPL26 links translocation‐associated quality control to endoplasmic reticulum protein homeostasis. Cell Res 30: 5–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M‐J, Ke P‐Y, Hsu JT‐A, Yeh C‐T, Horng J‐T (2014) Reticulon 3 interacts with NS4B of the hepatitis C virus and negatively regulates viral replication by disrupting NS4B self‐interaction. Cell Microbiol 16: 1603–1618 [DOI] [PubMed] [Google Scholar]
- Wu J, Lei G, Mei M, Tang Y, Li H (2010) A novel C53/LZAP‐interacting protein regulates stability of C53/LZAP and DDRGK domain‐containing protein 1 (DDRGK1) and modulates NF‐κB signaling. J Biol Chem 285: 15126–15136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Michaeli S, Picchianti L, Dagdas Y, Galili G, Peled‐Zehavi H (2021) ATI1 (ATG8‐interacting protein 1) and ATI2 define a plant starvation‐induced reticulophagy pathway and serve as MSBP1/MAPR5 cargo receptors. Autophagy 17: 3375–3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H, Kakuta S, Watanabe TM, Kitamura A, Sekito T, Kondo‐Kakuta C, Ichikawa R, Kinjo M, Ohsumi Y (2012) Atg9 vesicles are an important membrane source during early steps of autophagosome formation. J Cell Biol 198: 219–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Klionsky DJ (2010) Eaten alive: A history of macroautophagy. Nat Cell Biol 12: 814–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Ding X, Marshall RS, Paez‐Valencia J, Lacey P, Vierstra RD, Otegui MS (2020) Reticulon proteins modulate autophagy of the endoplasmic reticulum in maize endosperm. eLife 9: e51918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao YG, Liu N, Miao G, Chen Y, Zhao H, Zhang H (2018) The ER contact proteins VAPA/B interact with multiple autophagy proteins to modulate autophagosome biogenesis. Curr Biol 28: 1234–1245.e4 [DOI] [PubMed] [Google Scholar]
- Zhao D, Zou C‐X, Liu X‐M, Jiang Z‐D, Yu Z‐Q, Suo F, Du T‐Y, Dong M‐Q, He W, Du L‐L (2020) A UPR‐induced soluble ER‐Phagy receptor acts with VAPs to confer ER stress resistance. Mol Cell 79: 963–977.e3 [DOI] [PubMed] [Google Scholar]


