Abstract
NK cells utilize a large array of receptors to screen their surroundings for aberrant or virus‐infected cells. Given the vast diversity of receptors expressed on NK cells we seek to identify receptors involved in the recognition of HIV‐1‐infected cells. By combining an unbiased large‐scale screening approach with a functional assay, we identify TRAIL to be associated with NK cell degranulation against HIV‐1‐infected target cells. Further investigating the underlying mechanisms, we demonstrate that TRAIL is able to elicit multiple effector functions in human NK cells independent of receptor‐mediated induction of apoptosis. Direct engagement of TRAIL not only results in degranulation but also IFNγ production. Moreover, TRAIL‐mediated NK cell activation is not limited to its cognate death receptors but also decoy receptor I, adding a new perspective to the perceived regulatory role of decoy receptors in TRAIL‐mediated cytotoxicity. Based on these findings, we propose that TRAIL not only contributes to the anti‐HIV‐1 activity of NK cells but also possesses a multifunctional role beyond receptor‐mediated induction of apoptosis, acting as a regulator for the induction of different effector functions.
Keywords: death receptor, degranulation, HIV‐1, NK cell, TRAIL
Subject Categories: Autophagy & Cell Death; Immunology; Microbiology, Virology & Host Pathogen Interaction
This study shows that TRAIL contributes to the anti‐HIV‐1 activity of NK cells and also possesses a multifunctional role beyond receptor‐mediated induction of apoptosis, acting as a regulator for the induction of various effector functions.
Introduction
Natural killer (NK) cells are innate immune cells which play a pivotal role in antiviral immunity and tumor surveillance (Jost & Altfeld, 2013; Malmberg et al, 2017). NK cells exhibit various effector functions, including direct cellular cytotoxicity and production of pro‐inflammatory cytokines, to eliminate potential target cells and to shape adaptive immune responses (Vivier et al, 2008; Prager & Watzl, 2019). NK cells utilize a large array of germline‐encoded surface receptors to interact with their environment and to recognize virus‐infected or transformed cells (Biassoni & Malnati, 2018). Multiple factors, including host genetics and differentiation stage, impact the receptor repertoire of NK cells, ultimately leading to a remarkable diversity of NK cells within and across individuals (Horowitz et al, 2013; Schwane et al, 2020). The integration of activating and inhibitory signaling through these receptors tightly regulates NK cell activity (Long et al, 2013). The specific receptor profile of an individual NK cell therefore determines the activation threshold of a given cell and impacts the ability to recognize potential target cells.
Given the importance of NK cells in the early control of viral infections, our group has been investigating the contribution of receptors to an effective antiviral response of NK cells, in particular in HIV‐1 infection (Fadda et al, 2012; Körner et al, 2014, 2017). NK cells contribute to the intrinsic control of HIV‐1 infection through NK‐cell‐mediated immune pressure (Alter et al, 2011; Hölzemer et al, 2015). Several NK cell receptors have been attributed to promote NK‐cell‐mediated control of HIV‐1 infection and delay progression to AIDS, most prominent receptors that bind to specific HLA class I molecules (Martin et al, 2002, 2007). Yet, our understanding about receptor profiles that enable NK cells to effectively recognize HIV‐1‐infected cells independent of the host’s HLA class I background is incomplete. We developed an unbiased and comprehensive screening approach that combined a functional readout (degranulation; Alter et al, 2004) and the individual assessment of 327 surface antigens on NK cells in co‐culture with autologous HIV‐1‐infected cells. As a result, we identified the Tumor Necrosis Factor‐Related Apoptosis‐Inducing Ligand (TRAIL) to be associated with increased degranulation of NK cells after exposure to infected cells.
TRAIL is well‐known for its role in receptor‐mediated cytotoxicity, directly inducing apoptosis on target cells upon receptor engagement (Wiley et al, 1995; Pitti et al, 1996). TRAIL has multiple soluble and cell surface interaction partners that differ in their subsequent signaling. The death receptors 4 (DR4, TRAIL‐R1) and 5 (DR5, TRAIL‐R2) contain cytoplasmic death domains that are capable of initiating cell death through the extrinsic apoptotic pathway (Pan et al, 1997b; Sheridan et al, 1997). In contrast, two so‐called decoy receptors, DcR1 and DcR2, lack functional death domains and are therefore not able to trigger the apoptotic cascade (Degli‐Esposti et al, 1997a; Pan et al, 1997a; Sheridan et al, 1997). Finally, TRAIL is able to bind osteoprotegerin (OPG), a soluble protein that has been attributed to inhibit osteoclastogenesis and to increase bone density in vivo (Emery et al, 1998). TRAIL is expressed on a fraction of peripheral blood NK cells and other immune cells and can be induced through various cytokines (Fanger et al, 1999; Griffith et al, 1999; Kayagaki et al, 1999a; Sato et al, 2001). TRAIL‐induced apoptosis has been implicated to play a role in HIV‐1 pathogenesis (Gougeon & Herbeuval, 2012). In this study, we identified TRAIL to be involved in the induction of NK cell degranulation, therefore independent of receptor‐mediated cytotoxicity, and further investigated the underlying mechanisms.
Results
NK cells recognizing HIV‐1‐infected CD4 T cells display elevated TRAIL surface expression
We initially sought to identify NK cell receptors that are involved in the recognition of HIV‐1‐infected CD4 T cells. For this, we used a large‐scale, flow cytometry‐based screening approach to simultaneously assess NK cell degranulation [expression of CD107a (Alter et al, 2004)] and 327 additional individual surface antigens on primary human NK cells after incubating them with autologous in vitro HIV‐1‐infected CD4 T cells. For subsequent analyses, two subsets were defined, responsive CD107a+ NK cells and non‐responsive CD107a‐ NK cells (Fig 1A). As shown in Fig 1B, median percentage of CD107a+ NK cells was higher after exposure to HIV‐1‐infected CD4 T cells (13.8%) than in the presence of mock‐infected CD4 T cells (6.9%) or in the absence of target cells (3.4%). Normalization of the individual values to their corresponding mock control showed an HIV‐1‐specific response for all tested donors (n = 19, P < 0.0001), ranging from 0.9 percentage points (p.p.) up to 20.7 p.p. (Fig 1C). Analyzing 327 surface antigens, TRAIL was one of 48 molecules that were differentially expressed between the CD107a+ and CD107a− NK cell subsets (TRAIL: P < 0.0001; Fig 1D). The relative frequency of TRAIL+ NK cells was consistently higher in CD107a+ NK cells than in their CD107a− counterparts, with a median intra‐donor difference of 15.1 p.p. (P = 0.001, Fig 1E and F). Similar results were observed when median fluorescence intensity (MdFI) was used as an additional metric for TRAIL expression (Fig 1G), with a 2.2‐fold higher MdFI in the responsive NK cell subset (P = 0.001). These data demonstrated that degranulation of NK cells in response to autologous HIV‐1‐infected CD4 T cells was associated with increased TRAIL expression. Based on this observation, we further investigated possible underlying causes and postulated the following three hypotheses: (i) TRAIL acts as an activation marker, being upregulated during or after degranulation; (ii) TRAIL is simply co‐expressed on NK cell subsets with inherently higher antiviral activity but not involved in the induction of degranulation; and (iii) TRAIL is either directly or indirectly involved in degranulation.
Figure 1. NK cell degranulation is associated with increased TRAIL surface expression after co‐incubation with autologous HIV‐1‐infected CD4 T cells.
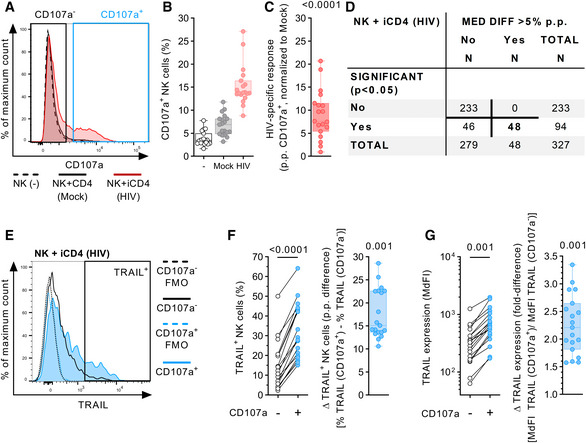
- Representative histograms (overlay) displaying CD107a expression on NK cells after culture with the described culture conditions (NK(−), Mock, HIV). For the HIV‐1 co‐culture condition, subsequent analyses of antigen expression were conducted by gating non‐responsive (CD107a−) and responsive (CD107a+) NK cell subsets.
- Relative frequency of CD107a+ NK cells (y‐axis) after culture in the described conditions (x‐axis) (n = 19 different donors).
- HIV‐specific response of bulk NK cells displayed as percentage points (p.p.) CD107a+ NK cells after normalization to mock CD4 T cells (y‐axis) (n = 19 different donors).
- Summary table showing numeric results of the 327 surface antigens analyzed. A total of 48 surface molecules showed statistically significant intra‐donor differences > 5 p.p. in expression between CD107a+ and CD107a− NK cells after exposure to HIV‐1‐infected CD4 T cells (HIV).
- Representative histograms (overlay) showing TRAIL surface expression as fluorescence intensity on NK cells after co‐incubation with HIV‐1‐infected CD4 T cells. Histograms show TRAIL expression for CD107a+ and CD107a− NK cell subsets (solid lines) as well as their corresponding FMO controls (dotted lines). Gate defining TRAIL+ cells is indicated as well.
- Left panel: Relative frequency of TRAIL+ cells (y‐axis) of CD107a+ and CD107a− NK cell subsets (x‐axis) after co‐incubation with HIV‐1‐infected CD4 T cells. Right panel: Difference in relative frequency of TRAIL+ cells (y‐axis) between CD107a+ and CD107a− cells displayed as p.p. (n = 19 different donors).
- Left panel: TRAIL surface expression (y‐axis) of CD107a+ and CD107a− NK cell subsets (x‐axis) after co‐incubation with HIV‐1‐infected CD4 T cells displayed as median fluorescence intensity. Right panel: Differences in TRAIL surface expression (y‐axis) between CD107a+ and CD107a− cells displayed as fold difference (n = 19 different donors).
Source data are available online for this figure.
TRAIL is neither an activation marker nor simply co‐expressed on antiviral NK cells
In order to investigate whether TRAIL acted as an activation marker or was independently co‐expressed in degranulating NK cells, we repeated the previous experimental setup with 13 different healthy donors. In this workflow, we additionally measured TRAIL surface expression on NK cells in response to the presence of mock‐infected CD4 T cells or in the absence of target cells. Furthermore, we acquired an increased number of cells for each condition to obtain sufficient cell numbers for high resolution of NK cell subsets. As shown in the left panel of Fig 2A, exposure to HIV‐1‐infected CD4 T cells yielded in the highest response rate (median 16.4% CD107a+ cells), followed by the condition with mock‐infected CD4 T cells (8.1%). In the absence of target cells, only 2.2% of bulk NK cells were CD107a+. Stratification of bulk NK cells into CD56Dim and CD56Bright cells (middle panel) or into un‐, KIR‐, or NKG2A‐educated NK cells (right panel) showed a similar hierarchy of NK cell degranulation for each subset. Comparison of these predefined groups in terms of their antiviral capacity revealed a differential ability to degranulate in response to HIV‐1‐infected target cells (Fig 2B). CD56Bright NK cells exhibited a higher HIV‐1‐specific response (15.1 p.p., median) than CD56Dim NK cells (9.4 p.p., P = 0.002). Stratification based on education status, showed that HIV‐1‐specific responses were highest in NKG2A‐educated NK cells (14.5 p.p., median). In contrast, KIR‐educated NK cells (4.8 p.p.) performed rather poorly compared to NKG2A‐educated (P < 0.001) or uneducated NK cells (9.7 p.p., P = 0.005). Next, we investigated whether TRAIL was upregulated after exposure to target cells (Fig 2C). For all tested subsets, surface density of TRAIL on NK cells exposed to HIV‐1‐infected CD4 T cells remained unchanged compared to NK cells cultured in the absence of target cells (Bulk: P = 0.5; Dim: P > 0.99; Bright: P = 0.1; Uneducated: P > 0.99; KIR‐edu.: P > 0.99; NKG2A‐edu.: P > 0.99). Finally, we compared TRAIL surface expression between CD107a+ and CD107a− NK cells across the previously defined subsets. As shown in Fig 2D, in all the investigated subsets, expression of TRAIL was significantly higher on CD107a+ NK cells (Bulk: P < 0.001; Dim: P < 0.001; Bright: P < 0.001; Uneducated: P < 0.001; KIR‐edu.: P < 0.01; NKG2A‐edu.: P < 0.001). The fold difference in TRAIL expression between responsive and non‐responsive NK cells was not different between CD56Bright (1.46) and CD56Dim NK cells (1.68, P = 0.17; Fig 2E). For the KIR‐educated subset, however, median fold difference was significantly lower (1.25) compared to the uneducated (1.49, P = 0.004) or NKG2A‐educated subsets (2.05, P = 0.002). Taken together, our data indicated that TRAIL is neither an activation marker in this experimental setting nor simply co‐expressed in subsets with inherently higher anti‐HIV activity. Instead, in all investigated subsets, NK cell degranulation was associated with increased TRAIL expression, indicating a potential role for the induction of degranulation.
Figure 2. NK cell degranulation is associated with increased TRAIL expression across multiple NK cell subsets.
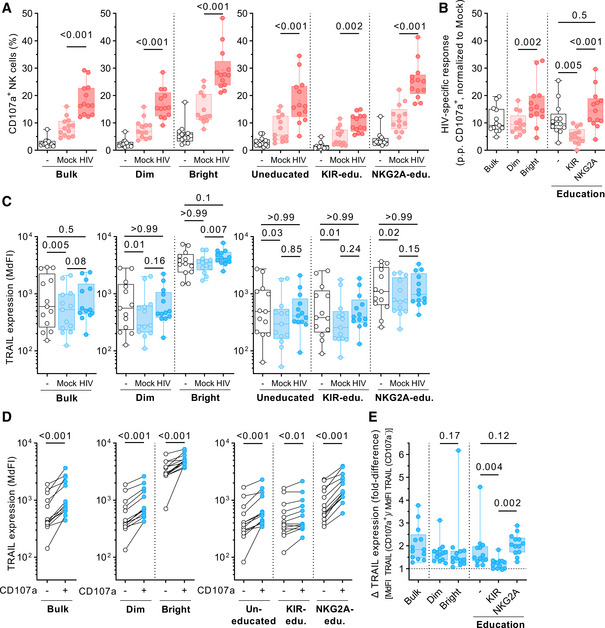
- Relative frequency of CD107a+ cells (y‐axis) of predefined NK cell subsets (x‐axis). Left panel displays data for bulk NK cells, middle panel for CD56Dim and CD56Bright NK cells, right panel for uneducated, KIR‐educated, and NKG2A‐educated cells (n = 13 different donors).
- HIV‐specific responses of predefined NK cell sub‐populations (x‐axis) displayed as percentage points (p.p.) CD107a+ NK cells after normalization to mock CD4 T cells (y‐axis) (n = 13 different donors).
- TRAIL expression levels (y‐axis) of predefined NK cell subsets (x‐axis). Left panel displays data for bulk NK cells, middle panel for CD56Dim and CD56Bright NK cells, and right panel for uneducated, KIR‐educated, and NKG2A‐educated cells. Expression is displayed as median fluorescence intensity (MdFI) (n = 13 different donors).
- Comparison of TRAIL expression levels (y‐axis) between CD107a− and CD107a+ cells of predefined NK cell subsets. Left panel displays data for bulk NK cells, middle panel for CD56Dim and CD56Bright NK cells, right panel for uneducated, KIR‐educated, and NKG2A‐educated cells. Expression is displayed as MdFI (n = 13 different donors).
- Fold differences in TRAIL surface expression (y‐axis) between CD107a+ and CD107a− cells of predefined NK cell sub‐populations (x‐axis) (n = 13 different donors).
Source data are available online for this figure.
In vitro HIV‐1‐infected CD4 T cells display increased surface expression of the TRAIL receptors DR4/DR5
Next, we investigated whether TRAIL was directly and/or indirectly involved in the induction of NK cell degranulation. A prerequisite for this hypothesis is the presence of TRAIL receptors on HIV‐1‐infected target cells that would engage TRAIL on NK cells and subsequently lead to degranulation. Therefore, we assessed the surface expression levels of two interaction partners of TRAIL, the death receptors 4 and 5 (DR4/5) on CD4 T cells. Exemplary histograms in Fig 3A show the combined expression of DR4/5 on mock CD4 T cells or after infection with five different HIV‐1 strains (NL4‐3, JR‐CSF, CH198, CH236, and WITO). Death receptors 4 and 5 are expressed on uninfected CD4 T cells (Mock) as well as on CD4 T cells that are positive for CD4 and negative for the intracellular HIV protein p24 (CD4+/p24−). DR4/5 expression was increased on HIV‐I‐infected CD4 T cells (CD4−/p24+) compared to their CD4+/p24‐ counterparts. As summarized in Fig 3B, exposure to all tested HIV‐1 strains resulted in an increase in DR4/5 expression on infected cells, measured as relative fluorescence intensity (RFI). Upregulation was independent of the tropism (X4 vs. R5), subtype (clade B vs. C of group M) or origin (cell culture‐adapted vs. primary strains). Of note, uninfected as well as infected cells also showed expression of TRAIL‐R3, which is thought to serve as a decoy receptor (DcR1) for TRAIL (Fig EV1). These results showed that interaction partners for TRAIL were present on CD4 T cells and that DR4 and DR5 were even further upregulated on HIV‐1‐infected target cells.
Figure 3. In vitro HIV‐1‐infected CD4 T cells display increased surface expression of the TRAIL receptors DR4/5.
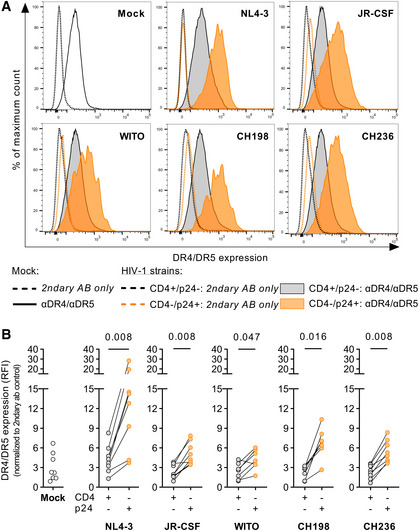
- Histograms (overlay) of one representative donor displaying combined DR4/5 surface expression on CD4 T cells previously incubated with NL4‐3, JR‐CSF, WITO, CH198, or CH236. Expression is displayed as fluorescence intensity (x‐axis).
- Comparison of the combined surface expression of DR4/5 between mock (white circles), uninfected (grey circles), and infected CD4 T cells (orange circles), from left to right: Mock: n = 8; NL4‐3: n = 8; JR‐CSF: n = 8; WITO: n = 7; CH198: n = 7; CH236: n = 8 (7–8 different donors per condition). Expression is displayed as relative fluorescence intensity (RFI) after normalization to the respective secondary AB‐only control (y‐axis).
Source data are available online for this figure.
Figure EV1. DcR1 is expressed on CD4 T cells.
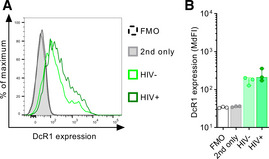
- Representative flow cytometry histogram of DcR1 expression on uninfected (HIV−: CD4+/p24−) and infected (HIV+: CD4−/p24+) cells in comparison to the anti‐goat‐PE control (grey) or FMO control (dashed line).
- Cumulative data of DcR1 expression displayed as MdFI. Data points represent the mean of two technical replicates per condition of three different donors (n = 3). Bar graphs show the median. Error bars show the IQR.
Source data are available online for this figure.
Antibody‐mediated blocking of TRAIL inhibits NK cell degranulation
Then, we investigated whether blocking the interaction between TRAIL and its receptors would affect NK cell degranulation. For this, we co‐cultured NK cells with the MHC‐class I devoid target cell line 721.221 or autologous HIV‐I‐infected CD4 T cells, either in the absence or presence of a mouse anti‐human TRAIL antibody or the respective antibody isotype. As displayed in Fig 4A, co‐culture with 721.221 induced degranulation in up to 41% of NK cells (median, 25.7%). Presence of the isotype antibody did not affect levels of degranulation (26.2%, P = 0.84 vs. .221 alone; median inhibition, 1.4%), while pre‐incubation with αTRAIL led to a reduced number of CD107a+ NK cells (15.5%, P = 0.004 vs. .221 alone) with a median inhibition of 35.4% (P = 0.002 vs. isotype, Fig 4B). Similar results, but less pronounced, were observed when we used autologous HIV‐1‐infected CD4 T cells as target cells (Fig 4C). Here again, presence of the isotype antibody did not affect levels of degranulation (P > 0.99 vs. HIV alone; median inhibition 1.7%), while pre‐incubation with αTRAIL led to a median inhibition of 25.4% (P = 0.008 vs. HIV alone; P = 0.004 vs. isotype, Fig 4D). Next, we tested whether blocking of the TRAIL receptors DR4/5 on target cells would impact NK cell degranulation. 721.221 cells express both DR4 and DR5 to a measurable degree on the cell surface (Fig 4E). Pre‐incubation of 721.221 with DR4/5 antibodies and subsequent co‐culture with NK cells led to reduced degranulation in NK cells from all nine tested donors (median, 22.7% vs. 19.2%, P = 0.008 vs. .221 alone; Fig 4F), while pre‐incubation with the respective isotype control (median, 23.5%, P > 0.99 vs. .221 alone) had no effect on the frequency of CD107a+ NK cells (median inhibition, −0.4% vs. 12.6%, P = 0.012; Fig 4G). These results showed that abrogation of TRAIL binding to two of its receptors negatively impacted NK cell degranulation, indicating that TRAIL is involved in the degranulation process.
Figure 4. Antibody‐mediated blocking of TRAIL inhibits NK cell degranulation.
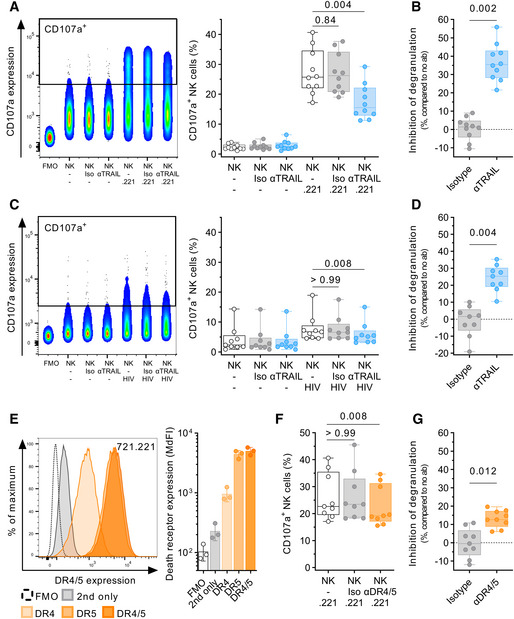
- Comparison of CD107a expression after co‐culture with 721.221 target cells with either 10 µg/ml αTRAIL or isotype control (Iso) using flow cytometry (n = 10 different donors per condition). Each data point represents the mean of two technical replicates. Effector:target ratio was 1:1. Left panel: Concatenated density plot depicting CD107a expression as fluorescence intensity for one representative donor. Right panel: Box plots showing relative frequency of CD107a+ NK cells (y‐axis).
- Box plots display inhibition of degranulation (y‐axis) after co‐culture with 721.221 target cells in presence of either isotype or αTRAIL as relative reduction compared to no antibody (n = 10 different donors per condition).
- Comparison of CD107a expression after co‐culture with autologous HIV‐I‐infected CD4 T cells with either 10 µg/ml αTRAIL or isotype control (Iso) using flow cytometry (n = 9 different donors per condition). Each data point represents the mean of two technical replicates. Left panel: Concatenated density plot depicting CD107a expression as fluorescence intensity for one representative donor. Right panel: Box plots showing relative frequency of CD107a+ NK cells (y‐axis).
- Box plots display inhibition of degranulation (y‐axis) after co‐culture with autologous HIV‐I‐infected CD4 T cells in the presence of either isotype or αTRAIL as relative reduction compared to no antibody (n = 9 different donors per condition).
- Representative histograms (overlay, left panel) and bar graphs (n = 3 independent experiments, right panel) showing the individual and combined surface expression of DR4 and DR5 on 721.221 cells. Each data point represents the mean of three technical replicates.
- Comparison of CD107a expression after co‐culture with 721.221 target cells in the presence of either αDR4/5 (10 µg/ml each) or 20 µg/ml isotype control (Iso) using flow cytometry (n = 9 different donors per condition). Effector:target ratio was 1:1. Box plots showing relative frequency of CD107a+ NK cells (y‐axis).
- Box plots display inhibition of degranulation after co‐culture with 721.221 target cells in the presence of either isotype or αDR4/5 as relative reduction compared to no antibody (n = 9 different donors per condition).
Source data are available online for this figure.
TRAIL engagement directly induces NK cell degranulation
Next, we sought to induce NK cell degranulation through direct engagement of TRAIL. For this, we immobilized (coated) TRAIL antibodies as well as proteins of the different TRAIL receptors on non‐tissue culture‐treated plates, and then cultured NK cells in the coated wells. Plate‐coated αNKG2D and αNKp46 served as positive controls and readily induced degranulation of NK cells in a dose‐dependent manner. Similarly, incubation in αTRAIL‐coated wells also resulted in a significant increase in CD107a+ NK cells compared to the isotype control (P < 0.001 for all concentrations; Fig 5A). To mimic more physiological conditions, we used proteins of the TRAIL receptors DR4 and DR5 to trigger NK cell degranulation. Human IgG served as positive control through crosslinking of CD16 and strongly induced degranulation (10 µg/ml: median 77.9% CD107a+ cells). Both, DR4 and DR5 likewise induced degranulation of NK cells compared to the uncoated negative control (PBS), although DR5 to a lesser extent (PBS: 1.7%, DR4: 1 µg/ml: 4.2%, 5 µg/ml: 14.3%, 10 µg/ml: 13.9%; DR5: 1 µg/ml: 4.6%, 5 µg/ml: 6.7%, and 10 µg/ml: 5.7%, P = 0.002 for all conditions; Fig 5B). In addition to measuring the percentage of CD107a+ NK cells, we also quantified the release of granzyme B into the supernatant (Fig 5C). NK cell cultures in αTRAIL and αNKp46‐coated wells (10 µg/ml) showed increased concentrations of granzyme B in the supernatant compared to the controls (αTRAIL vs. Isotype: P = 0.008). Increased granzyme B levels were also observed for DR4‐coated wells (DR4 vs. PBS: P = 0.027). Spearman rank analyses showed a significant correlation between the frequency of CD107a+ NK cells and granzyme B levels in the respective culture supernatants (rs = 0.69, P < 0.0001, Fig 5D). Lastly, we were interested whether other known interaction partners of TRAIL are able to trigger NK cell degranulation. NK cells cultured in wells coated with either decoy receptor 1 (DcR1) or OPG induced degranulation in a significant number of NK cells (DcR1 vs. PBS: P = 0.008 for 1 µg/ml; OPG vs. PBS: P = 0.008 for all tested concentrations; Fig 5E). Of note, DcR1 was the only TRAIL receptor that showed a dose‐dependent decrease in degranulation, in contrast to OPG, DR4, or DR5. Altogether, our data demonstrated that all tested interaction partners of TRAIL, including decoy receptor I and OPG, triggered degranulation in NK cells; providing further evidence that engagement of TRAIL is able to elicit effector functions in NK cells.
Figure 5. TRAIL engagement directly induces NK cell degranulation.
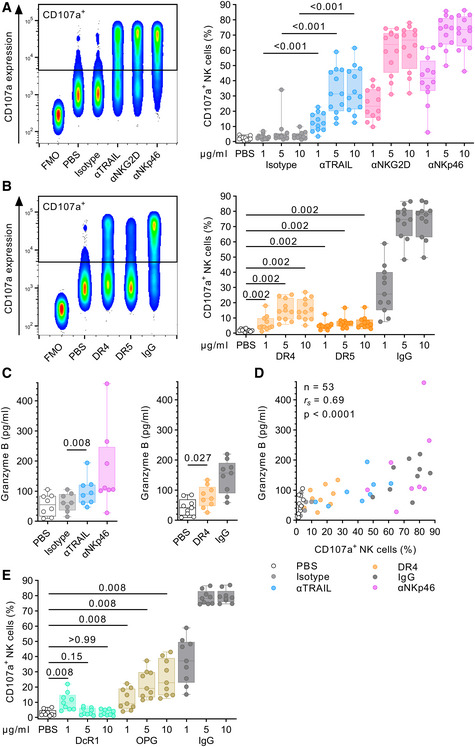
- Comparison of CD107a expression after incubation in either uncoated wells (PBS) or wells coated with αTRAIL, αNKG2D, αNKp46, or isotype using flow cytometry (n = 12 different donors per condition). Left panel: Concatenated density plot depicting CD107a expression as fluorescence intensity (y‐axis) for one representative donor and 10 µg/ml antibody concentration. Right panel: Box plots showing relative frequency of CD107a+ NK cells (y‐axis) after incubation with plate‐coated antibodies of different concentrations (x‐axis).
- Comparison of CD107a expression after incubation with plate‐coated DR4 protein, DR5 protein, or human IgG using flow cytometry (n = 11 different donors per condition). Left panel: Concatenated density plot depicting CD107a expression as fluorescence intensity (y‐axis) for one representative donor and 10 µg/ml protein concentration. Right panel: Box plots showing relative frequency of CD107a+ NK cells (y‐axis) after incubation with plate‐coated proteins of different concentrations (x‐axis).
- Comparison of granzyme B release after incubation with various stimuli (10 µg/ml each). Box plots showing granzyme B concentration in the supernatant as determined by ELISA (left panel: n = 8 different donors per condition, right panel: n = 9 different donors per condition).
- Correlation analysis between relative frequency of CD107a+ NK cells and granzyme B concentration (n = 53, data points obtained from A, B, and C, 11 different donors).
- Comparison of CD107a expression after incubation with plate‐coated DcR1 protein, osteoprotegerin (OPG), or human IgG using flow cytometry (n = 9 different donors). Box plots showing relative frequency of CD107a+ NK cells (y‐axis) after incubation with plate‐coated proteins of different concentrations (x‐axis).
Source data are available online for this figure.
TRAIL engagement induces IFNγ production
Effector functions of NK cells not only comprise granule‐mediated cytotoxicity through degranulation but also the production of pro‐inflammatory cytokines. Using supernatants collected from plate‐coating and TRAIL blocking experiments, we analyzed whether TRAIL interactions may also impact the production of IFNγ. Indeed, NK cells cultured in wells coated with either αTRAIL or DR4 both induced production and release of IFNγ (αTRAIL vs. isotype: P = 0.008; DR4 vs. PBS: P = 0.004; Fig 6A). In turn, blocking of TRAIL in NK cell: 721.221 co‐cultures resulted in reduced concentrations of IFNγ in the supernatant (αTRAIL: P = 0.016), while pre‐treatment with an isotype antibody had no significant effect on IFNγ production (isotype: P > 0.99, median inhibition, −8.2% vs. 32.3%, P = 0.008 vs. isotype; Fig 6B). Next, we tested whether the induction of degranulation and IFNγ production was due to an increased ability to adhere to target cells. Quantifying the ability of NK cells to form conjugates with 721.221 cells in the presence of αTRAIL, we did not observe a significant inhibition of conjugate formation (−0.6% vs. 1.9%, P = 0.148 vs. Isotype; Fig 6C and D). Finally, we sought to investigate intracellular signaling pathways that may be activated after direct TRAIL engagement and thus may provide evidence for reverse signaling of TRAIL. We examined changes in phosphorylation of the kinases p38 MAPK, Akt, Syk, and the phospholipase PLC‐γ2 after direct engagement of TRAIL through immobilized αTRAIL (Fig 6E). All selected molecules are known to be involved in signaling of NKp46, NKG2D, or CD16, which served as positive controls. Phosphorylation occurs rapidly and transiently after receptor engagement with dephosphorylation following shortly thereafter. After an extended incubation time of 30 min, we assessed the changes in phosphorylation of p38 MAPK, Akt, Syk, and PLC‐γ2. First, we observed a high baseline phosphorylation of the examined molecules in the majority of NK cells in the unstimulated conditions, PBS (p‐Syk: 71.3%, p‐p38 MAPK: 71.7%, p‐Akt: 69.2%, p‐PLC‐γ2: 71.1%), and isotype control (p‐Syk: 72.6%, p‐p38 MAPK: 77.8%, p‐Akt: 69.2%, p‐PLC‐γ2: 65.7%). Second, either engagement of the activating receptors NKp46, NKG2D, or CD16 by immobilized antibodies led to a marked reduction in NK cells expressing phosphorylated signaling proteins after the prolonged incubation time indicating the return of the signaling molecules into their dephosphorylated state (αNKp46: p‐Syk: 34.8%, p‐p38 MAPK: 56.1%, p‐Akt: 33.0%, p‐PLC‐γ2: 26.6%; αNKG2D: p‐Syk: 25.5%, p‐p38 MAPK: 31.3%, p‐Akt: 21.5%, p‐PLC‐γ2: 18.3%; αCD16: p‐Syk: 28.5%, p‐p38 MAPK: 36.8%, p‐Akt: 23.1%, and p‐PLC‐γ2: 22.8%). Observed effects of dephosphorylation after the engagement of TRAIL through immobilized αTRAIL were less pronounced. NK cells co‐cultured with αTRAIL did exhibit a noticeable but smaller reduction in phosphorylated Syk+, Akt+, and PLC‐y2+ but not p38 MAPK+ NK cells (p‐Syk: 60.5%, p‐p38 MAPK: 75.4%, p‐Akt: 62.0%, and p‐PLC‐γ2: 56.3%). These results seem to reflect the hierarchy of NK degranulation levels we observed after NK cell receptor cross‐linking in Fig 5.
Figure 6. TRAIL engagement induces Interferon γ production but does not promote cell adhesion.
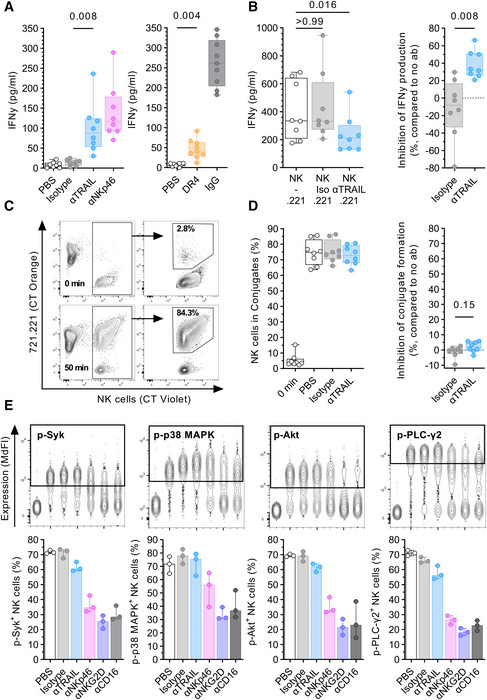
- Comparison of IFNγ production (y‐axis) after incubation with plate‐coated antibodies or proteins (10 µg/ml each) (x‐axis). Box plots showing IFNγ concentration in the supernatant as determined by ELISA (left panel: n = 8 different donors per condition, right panel: n = 9 different donors per condition).
- Comparison of IFNγ levels in the supernatant after co‐culture of NK cells with 721.221 target cells in the presence or absence of either 10 µg/ml αTRAIL or isotype control (Iso) using ELISA (n = 8 different donors per condition). Effector:target ratio was 1:1. Left panel: Box plots showing IFNγ levels (y‐axis) for the described culture conditions (x‐axis). Right panel: Box plots display inhibition of IFNγ production by NK cells (y‐axis) in presence of either isotype or αTRAIL as relative reduction compared to no antibody.
- Representative contour plots showing conjugate formation between NK cells and 721.221 target cells as determined by flow cytometry. Plots showing three distinct populations at the beginning of the co‐culture (0 min) and after 50 min: single 721.221 cells (CT Orange positive), single NK cells (CT Violet positive), and conjugates (double positive). Relative frequency of NK cells in conjugates is calculated based on the total number of gated NK cells.
- Comparison of conjugate formation after co‐culture between NK cells and 722.221 cells after 0 min and after 50 min in the presence or absence of 10 µg/ml αTRAIL or isotype control (n = 8 different donors). Left panel: Box plots showing relative frequency of NK cells in conjugates (y‐axis) for different time points and conditions (x‐axis). Right panel: Box plots display inhibition of conjugate formation (y‐axis) in the presence of either isotype or αTRAIL as relative reduction compared to no antibody.
- Expression levels of phosphorylated signaling proteins. Upper panel: Concatenated contour plots of one donor depicting the expression of phosphorylated signaling proteins Syk, p38 MAPK, Akt, and PLC‐γ2 for the following culture conditions and controls (x‐axis: left to right): FMO, PBS, isotype, αTRAIL, αNKp46, αNKG2D, and αCD16. Lower panel: Bar graphs showing the percentage of p‐Syk+, p‐p38 MAPK+, p‐Akt+, and p‐PLC‐γ2+ NK cells after 30 min of stimulation (n = 3 different donors). Bar graphs represent the median, whiskers display minimum and maximum data points.
Data information: Wilcoxon signed‐rank test. (A, B, D) Samples were acquired in duplicate and the mean was calculated for each donor and condition. Box plots represent the median and 25%/75% percentile. Whiskers indicate minimum and maximum data points.
Source data are available online for this figure.
Altogether, our results demonstrated that the direct engagement of TRAIL elicits multiple effector functions in NK cells, including IFNγ production. Investigation of the underlying mechanisms was less conclusive but did not rule out intracellular signaling at least for some of the signaling molecules we analyzed. In addition, in our experimental setup, TRAIL engagement had limited effects on cell adhesion.
TRAIL contributes to NK‐cell‐mediated killing of target cells
Lastly, we investigated the contribution of TRAIL to NK cell cytotoxicity. For this, we conducted multiple killing assays with various target cells (Fig 7). First, we co‐cultured 722.221 target cells with NK cells in the presence of αTRAIL or an isotype control and quantified the remaining cells relative to the number of target cells alone (Fig 7A). NK cells pre‐treated with soluble αTRAIL showed a significant reduction in their ability to lyse .221 cells compared to isotype‐treated NK cells (median, 18.3% vs. 24.6% remaining cells, P = 0.012 vs. isotype). Next, we quantified the preferential killing of target cells by NK cells compared to control cells in competitive killing assays. .221‐Cas9 (DR4/5 positive) and Raji cells overexpressing DR5 (Raji‐DR5++) served as target cells. The respective control cells were transduced .221 with a DR4 and DR5 gene knockout (.221‐DR4/5KO) or Raji‐pSIP expressing a lower amount of DR5. Calculation of target:control ratios and the specific lysis of target cells showed that .221‐Cas9 cells were preferentially depleted by NK cells (Fig 7B, P = 0.002). A similar outcome was observed for Raji cells with elevated DR5 expression (Fig 7C P = 0.0005). The results of these experiments showed that the target cells expressing death receptors or higher amounts of death receptors are increasingly sensitive to NK‐cell‐mediated cytotoxicity.
Figure 7. TRAIL contributes to NK‐cell‐mediated killing of target cells.
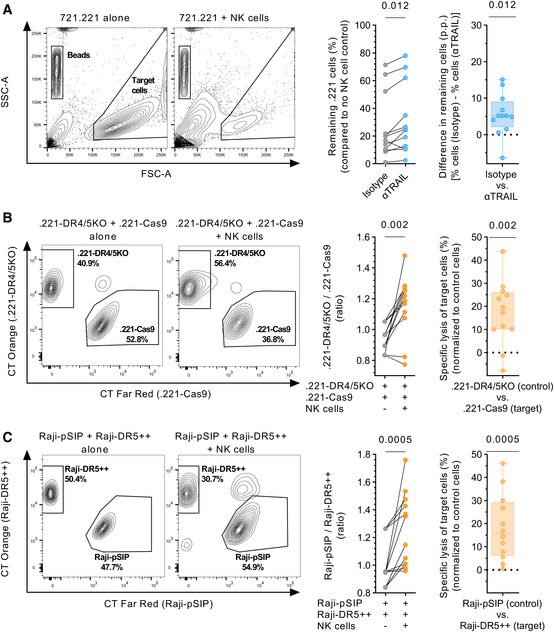
- Left panel: Representative contour plots showing depletion of 721.221 target cells in the presence of NK cells. Middle panel: Percentage of target cells remaining (y‐axis) after co‐culture with NK cells in the presence of either αTRAIL or isotype control, in reference to target cells kept alone. Right panel: Box plots displaying difference in target cells remaining (y‐axis) between αTRAIL and isotype conditions displayed as p.p. (n = 12 different donors). Each data point represents the mean of at least two technical replicates.
- Left panel: Representative contour plots showing the percentage of .221‐DR4/5KO (control) and .221‐Cas9 cells (target) in the presence or absence of NK cells. Middle panel: Ratio between .221‐DR4/5KO and .221‐Cas9 cells (y‐axis) in the presence or absence of NK cells. Right panel: Specific lysis of .221‐Cas9 cells displayed as percent (n = 12 different donors). Each data point represents the mean of at least three technical replicates.
- Left panel: Representative contour plots showing the percentage of Raji‐pSIP (control) and Raji‐DR5++ (target) in the presence or absence of NK cells. Middle panel: Ratio between Raji‐pSIP and Raji‐DR5++ (y‐axis) in the presence or absence of NK cells. Right panel: Specific lysis of Raji‐DR5++ cells displayed as % (n = 12 different donors). Each data point represents the mean of at least three technical replicates.
Source data are available online for this figure.
Discussion
In this study, we provide evidence that TRAIL contributes to the anti‐HIV‐1 activity of NK cells beyond receptor‐mediated cytotoxicity through induction of degranulation. Direct engagement of TRAIL elicited multiple effector functions in human NK cells. Both degranulation and IFNγ production in NK cells were triggered by cross‐linking TRAIL with either its cognate receptors or αTRAIL, as well as inhibited through antibody‐mediated blockade of TRAIL interactions.
The ability of TRAIL to transduce signals after receptor engagement has been debated for a long time with scattered reports on that matter in different species and cell types. In 2001, Chou and colleagues described enhanced proliferation and increased IFNγ production in murine T cells after cross‐linking TRAIL through DR4‐Fc fusion proteins (Chou et al, 2001). However, TRAIL engagement alone was not sufficient to induce proliferation or cytokine production, rather acting as a co‐stimulatory molecule in combination with T cell receptor engagement. In a subsequent study, the group conveyed similar findings in human CD4 T cells but not in cytotoxic CD8 T cells (Tsai et al, 2004). In TRAIL‐deficient mice, murine NK cells showed reduced cytotoxicity but unchanged expression of CD107a after exposure to YAC‐1 target cells (Cardoso Alves et al, 2020). The same study also reported increased numbers and relative frequencies of IFNy+ NK cells upon LCMV infection. This and observations by Diehl et al. indicated that TRAIL may have an inhibitory role in murine viral infections (Diehl et al, 2004). However, these effects may not be linked to reverse signaling but rather caused by other indirect mechanisms during the antiviral immune response, such as altered NK cell–DC cross‐talk (Iyori et al, 2011). These studies hinted at the ability of TRAIL to induce additional effector functions on the expressing immune cells but remain altogether puzzling; TRAIL only acted as a co‐stimulatory molecule in CD4 T cells and not in cytotoxic T cells which are more closely related to NK cells in their role as cytotoxic effectors. Unaltered levels of degranulation in NK cells from TRAIL‐deficient mice may indicate differences in TRAIL signaling between mice and humans.
In this study, we showed that blockade of TRAIL interactions resulted in significantly reduced levels of degranulation after exposure to the MHC class I devoid cell line 721.221 and autologous HIV‐I‐infected CD4 T cells. Similar results were recently described for IL‐18/poly I:C‐treated NK cells after co‐culture with the human hepatic stellate cell (HSC) line LX2 and primary HSCs (Li et al, 2019). The authors observed that blockade of TRAIL inhibited the interaction between NK cells and LX2 cells. Our results, however, did not show a significant contribution of TRAIL in conjugate formation between NK cells and 721.221 cells. Nevertheless, our data also demonstrated that blockade of either side of the interaction between TRAIL and its receptors impacts NK cell degranulation. These observations do not rule out the contribution of TRAIL in target cell adhesion and in the formation of an immunological synapse but indicate that its specific contribution may be target cell specific. The presence of TRAIL receptors on the surface of the respective target cell and their ratio to the expression of ligands for other NK cell receptors may be a determining factor.
Cross‐linking of TRAIL with proteins of the death receptors 4 and 5 induced degranulation and IFNγ production in NK cells. Beyond its potential role in facilitating target cell adhesion, these observations indicate that TRAIL elicits effector functions in NK cells. Interestingly, this was also the case with other interaction partners of TRAIL, the decoy receptor 1 (DcR1), and osteoprotegerin (OPG). DcR1 (TRAIL‐R3) does not contain a death domain and is thus not able to induce apoptosis (Degli‐Esposti et al, 1997b). Hence, DcRI as well as the related DcR2 (TRAIL‐R4) have been attributed to regulate TRAIL sensitivity by sequestering TRAIL in lipid rafts or forming heteromeric complexes with DR5, thus inhibiting caspase‐8 activation (Clancy et al, 2005; Mérino et al, 2006). OPG has been shown to inhibit TRAIL‐mediated apoptosis (Emery et al, 1998) and has been subsequently implicated to play a role in various malignancies (Zauli et al, 2009). In light of our findings, OPG may serve as a double‐edged sword, shielding tumor cells from TRAIL‐induced apoptosis on the one hand (Holen et al, 2002, 2005; Shipman & Croucher, 2003; De Toni et al, 2008) and stimulating TRAIL‐mediated anti‐tumor activity of NK cells (Berg et al, 2009) by triggering degranulation on the other. Overall, our results may add a new perspective on the perceived roles of DcR1 and OPG in the context of NK‐cell‐mediated tumor surveillance.
Although the accumulating evidence of our and previous studies indicates bidirectional signaling of TRAIL, little is known about the underlying signaling pathways. TRAIL is a member of the TNFα superfamily and reverse signaling has been described for some other members (Sun & Fink, 2007; Lee et al, 2019). However, TRAIL contains neither an immunoreceptor tyrosine‐based activation motif (ITAM) in the short cytoplasmic tail or a positively charged amino acid (lysine or arginine) in the transmembrane domain that could confer association with adapter molecules such as DNAX activation protein 12 (DAP12) or CD3ζ (Humphrey et al, 2005; Lanier, 2009). Therefore, the potential signaling pathway may differ from activating receptors like NKG2D, NKp46, or certain KIRs (Campbell et al, 1998; Meza Guzman et al, 2020). Syk, Akt, and PLC‐γ2 play important roles in the signaling pathways of activating NK cell receptors (Spaggiari et al, 2001; Jiang et al, 2002; Sutherland et al, 2002; Upshaw et al, 2005). Our assessment of the phosphorylation of these signaling molecules indicated an increased baseline phosphorylation likely associated with prolonged IL‐2 and IL‐15 treatment, similar to observations in murine NK cells (Luu et al, 2021). Further investigations into the participation of these signaling molecules in TRAIL signaling did not yield conclusive results in our experimental setup, which aimed to measure the dephosphorylation after an elapsed signaling cascade. Since Syk and PLC‐γ2 enable the activation of signaling pathways leading to the activation of the MAP kinases ERK and JNK (Jiang et al, 2002; Chen et al, 2007), TRAIL may also utilize one of these MAPKs for signal transduction. In an earlier study, activation of p38 MAPK after TRAIL‐induced co‐stimulation of CD4 T cells was reported (Tsai et al, 2004). Subsequent investigations in CD4 T cells observed phosphorylation of the upstream TCR‐proximal tyrosine kinases, Lck, and ZAP70 and activation of the NF‐kB pathway after TRAIL co‐stimulation (Huang et al, 2011). ZAP70 binds to phosphorylated ITAMs in the cytoplasmic domain of activating adapter proteins (Vivier et al, 1993; Ting et al, 1995; Lanier et al, 1998; Augugliaro et al, 2003), therefore TRAIL may utilize ITAM‐mediated signaling in an indirect manner. Co‐localization of TRAIL in nanoscale clusters with other immune receptors and DAP12 is therefore one possibility to hijack adapter molecules for downstream signaling (Oszmiana et al, 2016). In this context, it has been shown that CD59 acts as a co‐receptor for the activating receptor NKp46 through physical association, promoting signaling and subsequent cytotoxicity, despite lacking a signaling domain on its own (Marcenaro et al, 2003). So far, the exact mechanism of how TRAIL engagement induces effector functions eventually remains obscure and will require more focused investigations.
We initially observed increased TRAIL expression in NK cells degranulating against HIV‐infected cells indicating the involvement of TRAIL in the anti‐HIV‐1 activity of NK cells. Multiple studies have previously investigated the role of TRAIL and its receptors in immune control and pathogenesis of HIV‐1, reviewed in Gougeon and Herbeuval (2012). TRAIL was upregulated in multiple types of immune cells, in HIV‐1 infection in vivo, in vitro, and in animal models (Herbeuval et al, 2005a, 2005c, 2009; Hardy et al, 2007; Stary et al, 2009; Cheng et al, 2020). Serum levels of soluble TRAIL were elevated in HIV‐1+ individuals and correlated with viral load (Herbeuval et al, 2005a). In turn, increased expression of TRAIL receptors on CD4 T cells was observed in HIV‐1+ donors (Herbeuval et al, 2005b, 2009). In our in vitro infection models, we demonstrated that upregulation of DR4/5 was independent of the tested HIV‐1 strains. This indicates that upregulation of death receptors is a cellular response of the infected cell which HIV‐1 may not be able to prevent through immune evasion mechanisms. Therefore, DR4/5 on infected cells may serve as target molecules for immunotherapeutic HIV‐1 cure approaches (Deeks, 2012), enabling elimination of infected cells through TRAIL+ effector cells after reversal of HIV‐1 latency. Nevertheless, little is known about TRAIL and NK‐cell‐mediated induction of apoptosis in HIV‐1 infection. One study showed that IL‐15‐stimulated NK cells from HIV‐1‐infected donors displayed improved killing of K562 target cells in a TRAIL‐dependent manner (Lum et al, 2004). In addition, in other viral infections, such as HCV infection, in vitro studies indicated that TRAIL+ NK cells may play a crucial part in viral immunity by eliminating virus‐replicating cells by TRAIL‐mediated cytotoxicity (Stegmann et al, 2010). Enhanced TRAIL‐mediated cytotoxic activity by NK cells against murine cells infected with EMCV was observed as well (Sato et al, 2001). Our data confirmed that TRAIL contributes to NK‐cell‐mediated cytotoxicity against target cells expressing death receptors recognized by TRAIL, but did not allow the distinction between granule‐ or death receptor‐mediated target cell lysis. Additional experimental models are therefore needed to determine the overall contribution of TRAIL‐induced degranulation to NK cell cytotoxicity and in comparison to TRAIL‐induced apoptosis via death receptors.
Altogether, our study indicates that TRAIL is involved in NK‐cell‐mediated anti‐HIV‐1 activity by directly triggering degranulation. These observations now raise additional questions related to the underlying mechanisms, including the signaling pathway that links initial TRAIL engagement to downstream effector functions. Based on our findings we propose a multifunctional role for TRAIL beyond receptor‐mediated cytotoxicity, acting as a regulator for the induction of different effector functions in human NK cells. Hence, TRAIL may remain a target of interest for NK‐cell‐based immunotherapeutic approaches in cancer or antiviral therapies that would harness the ability of TRAIL to induce NK‐cell‐mediated target cell killing through multiple avenues.
Materials and methods
Reagents and Tools table
| Reagent/Resource | Reference or Source | Identifier or Catalog Number |
|---|---|---|
| Experimental Models | ||
| Citrate‐treated peripheral blood (H. sapiens) | Institute of Transfusion Medicine, University Medical Center Hamburg‐Eppendorf, Hamburg, Germany | N/A |
| EDTA‐treated peripheral blood (H. sapiens) | Hamburger Gesundkohorte University Medical Center Hamburg‐Eppendorf, Hamburg, Germany | N/A |
| HEK293T/17 (H. sapiens) | ATCC | Cat#CRL‐11268; RRID:CVCL_1926 |
| LCL 721.221 (H. sapiens) | ATCC | Cat#CRL‐1855; RRID:CVCL_6263 |
| .221‐Cas9 | Generated in this study | N/A |
| .221‐DR4/5KO | Generated in this study | N/A |
| Raji (H. sapiens) | Ragon Institute of MGH, MIT and Harvard, Cambridge, MA, USA | RRID:CVCL_0511 |
| Raji‐pSIP | Generated in this study | N/A |
| Raji‐DR5++ | Generated in this study | N/A |
| pNL4‐3 | NIH HIV Reagent Program | Cat#ARP‐114 |
| pYK‐JRCSF | NIH HIV Reagent Program | Cat#ARP‐2708 |
| HIV‐1 WITO | Mary Carrington (National Cancer Institute); Ochsenbauer et al, 2012 | N/A |
| HIV‐1 CH198 | Beatrice Hahn (University of Pennsylvania); Parrish et al, 2013 | N/A |
| HIV‐1 CH236 | Beatrice Hahn (University of Pennsylvania); Fenton‐May et al, 2013 | N/A |
| Recombinant DNA | ||
| pLVX‐SIP | Ragon Institute of MGH, MIT and Harvard, Cambridge, MA, USA; Dr. Thomas Pertel | N/A |
| pCMV‐VSV‐G | Addgene | Cat#8454; RRID:Addgene_8454 |
| psPAX2 | NIH HIV Reagent Program | Cat#ARP‐11348; RRID:Addgene_12260 |
| plentiCas9‐Blast | Addgene | Cat#52962; RRID:Addgene_52962 |
| plentiGuide‐Puro encoding gRNA sequences | GenScript | Cat#SC1678 |
| Antibodies | ||
| Mouse α‐human CD3 PerCP‐Cy5.5 (clone UCHT1) | BioLegend | Cat#300430; RRID:AB_893299 |
| Mouse α‐human CD3 PE/Dazzle594 (clone UCHT1) | BioLegend | Cat#300450; RRID:AB_2563618 |
| Mouse α‐human CD3 BV421 (clone UCHT1) | BioLegend | Cat#300434; RRID:AB_10962690 |
| Mouse α‐human CD3 Pacific blue (clone UCHT1) | BD Biosciences | Cat#558117; RRID:AB_397038 |
| Mouse α‐human CD4 APC (clone RPA‐T4) | BD Biosciences | Cat#555349, RRID:AB_398593 |
| Mouse α‐human CD4 APC (clone OKT4) | BioLegend | Cat#317416; RRID:AB_571945 |
| Mouse α‐human CD4 BV711 (clone RPA‐T4) | BioLegend | Cat#300558, RRID:AB_2564393 |
| Mouse α‐human CD14 PerCP‐Cy5.5 (clone HCD14) | BioLegend | Cat#325622; RRID:AB_893250 |
| Mouse α‐human CD16 BV785 (clone 3G8) | BioLegend | Cat#302046; RRID:AB_2563803 |
| Mouse α‐human CD16 (clone 3G8) | BioLegend | Cat#302014; RRID:AB_314214 |
| Mouse α‐human CD19 PerCP‐Cy5.5 (clone HIB19) | BioLegend | Cat#302230; RRID:AB_2073119 |
| Mouse α‐human CD45 AF700 (clone 2D1) | BioLegend | Cat#368514; RRID:AB_2566374 |
| Mouse α‐human CD45 BV605 (clone 2D1) | BioLegend | Cat#368524; RRID:AB_2715826 |
| Mouse α‐human CD45 PerCP‐Cy5.5 (clone 2D1) | BioLegend | Cat#368503; RRID:AB_2566351 |
| Mouse α‐human CD56 BUV395 (clone NCAM16.2) | BD Biosciences | Cat#563554; RRID:AB_2687886 |
| Mouse α‐human CD57 PE/Dazzle594 (clone HNK‐1) | BioLegend | Cat#359620; RRID:AB_2564063 |
| Mouse α‐human CD107a BV510 (clone H4A3) | BioLegend | Cat#328632; RRID:AB_2562648 |
| Mouse α‐human KIR2DL1/S5 FITC (clone 143211) | R&D Systems | Cat#FAB1844F‐100; RRID:AB_2130402 |
| Mouse α‐human KIR2DL2/L3/S2 BV711 (clone DX27) | BD Biosciences | Cat#745442; RRID:AB_2742987 |
| Human α‐human NKG2C PE (clone REA205) | Miltenyi Biotec | Cat#130‐103‐635; RRID:AB_2655394 |
| Mouse α‐human KIR3DL1 BV421 (clone DX9) | BioLegend | Cat#312714; RRID:AB_2561652 |
| Mouse α‐human NKG2A PE‐Cy7 (clone Z199) | Beckman Coulter | Cat#B10246; RRID:AB_2687887 |
| Mouse α‐human CD253 (TRAIL) APC (clone RIK‐2.1) | Miltenyi Biotec | Cat#130‐097‐314; RRID:AB_2656681 |
| Mouse α‐human CD253 (TRAIL) (clone RIK‐2) | BioLegend | Cat#308214; RRID:AB_2814155 |
| Mouse α‐human TRAIL‐R1 (DR4) (clone HS101) | AdipoGen | Cat#AG‐20B‐0022PF; RRID:AB_2490215 |
| Mouse α‐human CD261 (DR4) Biotin (clone DJR1) | Miltenyi Biotec | Cat#130‐109‐084; RRID:AB_2656741 |
| Mouse α‐human TRAIL‐R2 (DR5) (clone HS201) | AdipoGen | Cat#AG‐20B‐0023; RRID:AB_2490218 |
| Mouse α‐human TRAIL‐R2 (DR5) (clone HS201) | AdipoGen | Cat#AG‐20B‐0023PF; RRID:AB_2490221 |
| Mouse α‐human CD262 (DR5) Biotin (clone DJR2‐4) | Miltenyi Biotec | Cat#130‐097‐303; RRID:AB_2656745 |
| α‐human TRAIL‐R3 (DcR1, CD263) (polyclonal Goat IgG) | R&D Systems | Cat#AF630; RRID:AB_355488 |
| Mouse α‐human CD314 (NKG2D) (clone 1D11) | BioLegend | Cat#320814; RRID:AB_2561488 |
| Mouse α‐human CD314 (NKG2D) PE (clone 1D11) | BioLegend | Cat#320806; RRID:AB_492960 |
| Mouse α‐human CD335 (NKp46) (clone 9E2) | Miltenyi Biotec | Cat#130‐094‐271; RRID:AB_10828456 |
| Mouse α‐human CD335 (NKp46) FITC (clone 9E2) | BioLegend | Cat#331922; RRID:AB_2561965 |
| Mouse α‐human HIV‐1 core antigen FITC (clone KC57) | Beckman Coulter | Cat#6604665 |
| Mouse α‐human p38 MAPK PE (clone 36/p38) | BD Biosciences | Cat#612565; RRID:AB_399856 |
| Mouse α‐human PLC‐γ2 PE (clone K86‐689.37) | BD Biosciences | Cat#558490; RRID:AB_647226 |
| Mouse α‐human Syk PE (clone I120‐722) | BD Biosciences | Cat#558529, RRID:AB_647247 |
| Mouse α‐human Akt PE (clone M89‐61) | BD Biosciences | Cat#560378; RRID:AB_1645328 |
| Mouse IgG1 Isotype Control (clone 15H6) | AdipoGen | Cat#AG‐35B‐0003PF |
| Mouse IgG1, κ Isotype Ctrl (clone MOPC‐21) | BioLegend | Cat#400153 |
| α‐goat IgG PE (polyclonal donkey IgG) | R&D Systems | Cat#F0107; RRID:AB_573123 |
| Human Gamma Globulin | ThermoFisher Scientific | Cat#31879; RRID:AB_2532171 |
| Oligonucleotides and other sequence‐based reagents | ||
| gRNA 5’ATAGTCCTGTCCATATTTGC3‘ | GenScript | Cat# SC1678 |
| gRNA 5’GCGGGGAGGATTGAACCACG3‘ | GenScript | Cat# SC1678 |
| Chemicals, Enzymes and other reagents | ||
| BD CellFIX (10x concentrate) | BD Biosciences | Cat#340181; RRID: AB_2868724 |
| BD Phosflow Fix Buffer I | BD Biosciences | Cat#557870; RRID: AB_2869102 |
| BD Phosflow Perm Buffer III | BD Biosciences | Cat#558050; RRID: AB_2869118 |
| Benzonase Nuclease, Purity >90% | Merck‐Millipore | Cat#71205‐3 |
| Benzonase Nuclease, Purity > 99% | Merck‐Millipore | Cat#70664‐3 |
| Blasticidin | InvivoGen | Cat#ant‐bl‐1 |
| CellTracker Violet BMQC Dye | Invitrogen | Cat#C10094 |
| CellTracker Orange CMTMR Dye | Invitrogen | Cat#C2927 |
| CellTrace Far Red | Invitrogen | Cat#C34564 |
| Immunocult Human CD3/CD28 T Cell Activator | Stemcell | Cat#10971; RRID:AB_2827806 |
| Lenti‐X Concentrator | Clontech Laboratories Inc. | Cat#631232 |
| Lipofectamine 3000 | Invitrogen | Cat#L3000‐150 |
| LIVE/DEAD Fixable Blue Dead Cell Stain Kit | Invitrogen | Cat#L34962 |
| LIVE/DEAD Fixable Near‐IR Dead Cell Stain Kit | Invitrogen | Cat#L34976 |
| Precision Count Beads | BioLegend | Cat#424902 |
| Puromycin dihydrochloride | Sigma‐Aldrich | Cat#P7255 |
| Recombinant human DcR1 protein | Abcam | Cat#ab276266 |
| Recombinant human DR4 protein | Abcam | Cat#ab641 |
| Recombinant human DR5 protein | Abcam | Cat#ab243777 |
| Recombinant human IL‐2 | PeproTech GmbH | Cat#200‐02 |
| Recombinant human IL‐15 | PeproTech GmbH | Cat#200‐15 |
| Recombinant human Osteoprotegerin protein | Abcam | Cat#ab182688 |
| Streptavidin BV421 | BioLegend | Cat#405226 |
| Streptavidin PE | BioLegend | Cat#405204 |
| Software | ||
| Flowjo v10 | BD Life Sciences | RRID:SCR_008520; https://www.flowjo.com/ |
| GraphPad Prism 9 | GraphPad Software | RRID:SCR_002798; https://www.graphpad.com/ |
| Other | ||
| BD Cytofix/Cytoperm Fixation/Permeabilization Kit | BD Biosciences | Cat#554714; RRID:AB_2869008 |
| DNeasy Blood and Tissue Kit | QIAGEN | Cat#69506 |
| EasySep Human CD4 Positive Selection Kit II | Stemcell | Cat#17852 |
| EasySep Human CD4+ T Cell Enrichment Kit | Stemcell | Cat#19052 |
| EasySep Human NK Cell Enrichment Kit | Stemcell | Cat#19055 |
| Granzyme B Human ELISA Kit | ThermoFisher Scientific | Cat#BMS2027 |
| IFN gamma Human ELISA Kit | ThermoFisher Scientific | Cat#EHIFNG |
| MACS Marker Screen, human | Miltenyi Biotec | Cat#130‐110‐055 |
Methods and Protocols
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Human subjects
Peripheral blood was obtained from anonymized healthy human donors (n = 71) at the Institute for Transfusion Medicine, University Medical Center Hamburg‐Eppendorf, Hamburg, Germany. Blood donors gave their general written consent to the usage of their blood samples for scientific studies in an anonymized form. The anonymized use of human material complies with a vote by the ethics committee of the German Medical Association. In addition, peripheral blood samples were obtained from healthy blood donors (n = 27) recruited at the University Medical Center Hamburg‐Eppendorf, Hamburg, Germany. These donors provided written informed consent and studies were approved by the ethical committee of the Ärztekammer Hamburg (PV4780). Information on age and sex was not available for all subjects; however, was not relevant for the purpose of the study.
Cell lines
The HEK293T/17 cell line (ATCC, RRID:CVCL_1926) was used to generate infectious replication‐competent HIV‐1 virions. Cells were maintained in Dulbecco’s modified Eagle medium, high‐glucose, GlutaMAX Supplement, pyruvate (DMEM, Life Technologies) supplemented with 10% (v/v) heat‐inactivated FBS (Biochrom) and 100 U/ml penicillin and 100 µg/ml streptomycin (Sigma‐Aldrich). The sex of the cell line is female.
Raji cells (RRID: CVCL_0511) were used to overexpress DR5. Therefore, the sequence of the receptor was obtained from GeneArt GeneSynthesis (Thermo Fisher) and cloned into a lentiviral transfer vector (pLVX‐SIP) with puromycin resistance. Lipofectamine 3000 (Invitrogen) was used to transfect HEK239T/17 cells with the lentiviral transfer vector encoding the gene of interest, a VSV‐G envelope vector (pCMV‐VSV‐G; Addgene) and a HIV‐1 Gag‐Pol packaging vector (psPAX2; NIH HIV Reagent Program). The supernatant containing the lentivirus was harvested 48 h after transfection and used for transduction of Raji cells. DR5++ Raji cells were selected with 1 μg/ml puromycin (Sigma‐Aldrich) 3 days post‐transduction and later sorted for high DR5 expression with fluorescence‐activated cell sorting. The generated cell line was maintained in complete medium (RPMI‐1640 medium, Life Technologies) supplemented with 10% (v/v) heat‐inactivated FBS (Biochrom), 100 U/ml penicillin, 100 µg/ml streptomycin (Sigma–Aldrich), and 1 μg/ml puromycin (Sigma‐Aldrich) and cultured at 37°C and 5% CO2. The sex of the cell lines is male.
The cell line B‐LCL 721.221 (.221) (RRID: CVCL_6263) (Shimizu & DeMars, 1989) was used to assess the function of NK cells. The cell line was maintained in complete medium [RPMI‐1640 medium (Life Technologies) supplemented with 10% (v/v) heat‐inactivated FBS (Biochrom), 100 U/ml penicillin, and 100 µg/ml streptomycin (Sigma‐Aldrich)]. The sex of the cell line is female. 721.221 DR4/DR5 knockout cells were generated by CRISPR/Cas9‐based gene targeting. First, 721.221 cells were transduced with lentiviral particles containing plentiCas9‐Blast, a lentiviral plasmid construct that delivers hSpCas9 and blasticidin resistance. The lentiviral particles required for this purpose were produced in HEK293/17 cells as described for the transduction of Raji cells. Three days post‐transduction, Cas9+ cells were selected with 5 µg/ml blasticidin (InvivoGen). In the next step, Cas9+ 721.221 cells were transduced with lentivirus containing lentiGuide‐Puro plasmid constructs encoding a puromycin resistance and either a specific gRNA sequence to knockout DR4 or DR5. These plasmid constructs were generated by GenScript Biotech. The gRNA sequences used were as follows: 5’GCGGGGAGGATTGAACCACG3’ (knockout of DR4) and 5’ATAGTCCTGTCCATATTTGC3’ (knockout of DR5). Lentiviral particles were produced by transfection of HEK293/17 cells as described above. Transduced cells were selected with 0.5 µg/ml puromycin (Sigma‐Aldrich) 3 days post‐transduction and were later sorted for the absence of DR4 and DR5 expression. The generated 721.221 DR4/DR5 knockout cell line was maintained in complete medium (RPMI‐1640 medium, Life Technologies) supplemented with 10% (v/v) heat‐inactivated FBS (Biochrom), 100 U/ml penicillin, 100 µg/ml streptomycin (Sigma‐Aldrich), 0.5 μg/ml puromycin (Sigma‐Aldrich), and 5 µg/ml blasticidin (InvivoGen) and cultured at 37°C and 5% CO2. Expression levels of DR4 and DR5 on untransduced and transduced cell lines are displayed in Fig EV2. All cell lines are regularly tested for mycoplasma contamination.
Figure EV2. Expression of TRAIL receptors on transduced 721.221 and Raji cells.
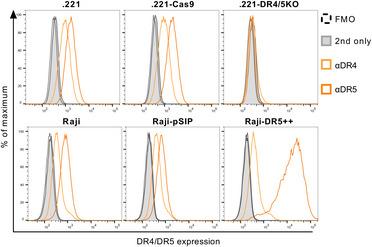
The expression of DR4 and DR5 was assessed by flow cytometry. 721.221 and Raji cells were labeled with LIVE/DEAD Fixable Near‐IR Stain, followed by incubation with biotin‐conjugated mouse anti‐human DR4 or DR5, and then labeled with Streptavidin‐BV421. Expression was quantified as fluorescence intensity. Representative histogram of DR4 (light orange) and DR5 (dark orange) expression in comparison to the Streptavidin‐only control (grey) or the FMO control (dashed line). Upper panel (from left to right): untransduced 721.221 cells, Cas9‐transduced .221s, and DR4/5 double knockout .221s. Lower panel (from left to right): untransduced Raji cells, Raji cells transduced with an empty vector (pSIP), and Raji cells overexpressing DR5.
Method details
Sample processing
Peripheral blood mononuclear cells (PBMCs) were isolated by density gradient centrifugation within a few hours after phlebotomy from peripheral blood of healthy human donors and washed before resuspension in complete medium (RPMI‐1640 medium (Life Technologies) supplemented with 10% (v/v) heat‐inactivated FBS (Biochrom), 100 U/ml penicillin, and 100 µg/ml streptomycin (Sigma‐Aldrich)). PBMCs were directly used for experiments or cryopreserved in FBS supplemented with 10% (v/v) dimethyl sulfoxide (Sigma‐Aldrich) and stored in liquid nitrogen for future experimental applications. Thawing of previously cryopreserved PBMCs was performed by drop‐wise addition of complete medium supplemented with 25 U/ml Benzonase Nuclease (Merck Millipore Novagen), washing and resting of cells at 37°C, 5% (v/v) CO2 for app. 4 h before further use.
Enrichment of CD4 T Cells and NK Cells
Primary human CD4 T cells and NK cells were enriched from PBMCs through negative‐selection strategy using EasySep Human CD4+ T Cell Enrichment Kit (Stemcell) and EasySep Human NK Cell Enrichment Kit (Stemcell), respectively. Isolated CD4 T cells and NK cells were washed and then resuspended in appropriate complete medium for downstream applications. Purity of the enriched cell populations was verified by flow cytometry. Purity of CD4 T cells was 96.8% ± 2.1 (Mean ± SD); purity of NK cells was 86.0% ± 11.8% (Mean ± SD).
Primary infectious molecular clones of HIV‐1
The following reagents were obtained through the NIH HIV Reagent Program, Division of AIDS, NIAID, NIH: Human Immunodeficiency Virus 1 (HIV‐1), Strain NL4‐3 Infectious Molecular Clone (pNL4‐3), ARP‐2852, contributed by Dr. M. Martin (Adachi et al, 1986); Human Immunodeficiency Virus 1 (HIV‐1), Strain JR‐CSF Infectious Molecular Clone (pYK‐JRCSF), ARP‐2708, contributed by Dr. Irvin SY Chen and Dr. Yoshio Koyanagi. Plasmids harboring the proviral genome of infectious molecular clones representing the HIV‐1 primary strains CH198 (Parrish et al, 2013) and CH236 (Fenton‐May et al, 2013) were kindly provided by the Beatrice Hahn Laboratory (University of Pennsylvania). The primary transmitted founder HIV‐1 clone WITO was inferred from plasma viral sequences as reported previously (Ochsenbauer et al, 2012) and kindly provided by Mary Carrington (National Cancer Institute). Infectious viral particles were produced by transfecting HEK293T/17 cells with the respective plasmids using Lipofectamine 3000 (Invitrogen) according to the manufacturer’s protocol. Cell culture supernatants were harvested through centrifugation 72–96 h after transfection and then sterile filtered through either PES‐based 0.45 µl or 0.22 µl Filtropur (Sarstedt) or 0.45 µl Steriflip‐PVDF filter (Merck‐Millipore). Subsequently, Lenti‐X Concentrator (Clontech Laboratories Inc.) was used to increase viral titers according to the manufacturer’s protocol. Lentiviral particles were aliquoted in DMEM (Life Technologies) and stored at −80°C until further use.
Infection of autologous CD4 T cells
After the enrichment of primary CD4 T cells, cells were washed and resuspended in complete medium supplemented with 100 U/ml IL‐2 (PeproTech). Cells were then stimulated for 3 days with αCD3/αCD28 antibodies (Stemcell) at a concentration of 25 μl/ml at 37°C, 5% (v/v) CO2. In vitro infection with various HIV‐1 strains was performed through spinoculation of cells (1,200 rcf, 2 h, 37°C) with infectious viral particles at an MOI between 0.005 and 0.01 (O’Doherty et al, 2000). After centrifugation, cells were incubated at 37°C, 5% (v/v) CO2 for 72–96 h, at 3.5–4 × 106 cells/ml. Cell culture supernatant of HIV‐1‐infected CD4 T cells and Mock CD4 T cells was replaced with fresh complete medium supplemented with 100 U/ml IL‐2 (PeproTech) after 48 h.
Enrichment of HIV‐1‐infected CD4 T cells
Enrichment of HIV‐infected cells (HIV‐1 p24 positive, CD4 negative) was conducted based on previously described protocols (Davis et al, 2011). In brief, HIV‐1‐exposed CD4 T cells were incubated with 25 U/ml Benzonase Nuclease (Merck Millipore Novagen) for 30 min at 37°C, 5% (v/v) CO2, and then washed. Infected cells were subsequently enriched through depletion of CD4‐expressing T cells using EasySep Human CD4 Positive Selection Kit II (Stemcell). CD4(−) cells in the supernatant were collected and used for downstream applications.
High‐throughput functional analysis and TRAIL expression in NK cell subsets
NK cells were enriched from previously cryopreserved PBMCs and cultured overnight in complete medium supplemented with 50 U/ml IL‐2 and 5 ng/ml IL‐15 (PeproTech) at 37°C, 5% (v/v) CO2. The next day, NK cells were co‐incubated with autologous enriched HIV‐1‐infected (NL4‐3, MOI = 0.01) CD4(−) T cells in complete‐medium supplemented with 50 U/ml IL‐2 and 5 ng/ml IL‐15 (PeproTech) at an effector:target ratio of 1:2 for 5 h at 37°C, 5% (v/v) CO2, in the presence of αCD107a‐BV510 (BioLegend, clone H4A3). Cells were washed and then incubated with LIVE/DEAD Fixable Blue Dead Cell Stain (Invitrogen) in staining buffer (Dulbecco’s Phosphate Buffered Saline (DPBS, Sigma Aldrich) + 2% (v/v) FBS + 1 mM EDTA (Sigma Aldrich)). Cells from up to four donors were then barcoded using αCD45 (BioLegend, clone 2D1) conjugated to AF700 or BV605: donor A: none; donor B: AF700; donor C: BV605; and donor D: AF700 + BV605 (Akkaya et al, 2016). Multiplexed cells were additionally labeled with αCD3‐PerCP‐Cy5.5 (BioLegend, clone UCHT1), αCD14‐PerCP‐Cy5.5 (BioLegend, clone HCD14), αCD16‐BV785 (BioLegend, clone 3G8), αCD19‐PerCP‐Cy5.5 (BioLegend, clone HIB19), αCD56‐BUV395 (BD Biosciences, clone NCAM16.2), αCD57‐PE‐Dazzle594 (BioLegend, clone HNK‐1), αKIR2DL2/L3/S2‐BV711 (BD Biosciences, clone DX27), αKIR3DL1‐BV421 (BioLegend, clone DX9), αKIR2DL1/KIR2DS5‐FITC (R&D Systems, clone 143211), αNKG2A‐PC7 (Beckman Coulter, clone Z199), and αNKG2C‐PE (Miltenyi Biotec, clone REA205). Cells of each donor were washed, merged together, and then individually stained with pre‐titrated APC‐conjugated antibodies for up to 327 selected surface antigens (Table EV1) and additional controls present in the human MACS marker screen (Miltenyi Biotec, Germany) following the manufacturer’s instructions. Cells were fixed using 1× CellFIX (BD Biosciences) and stored in staining buffer until flow cytometric data acquisition. FMO controls were used to define gates and assess relative frequency of cells positive for each marker.
For independent validation and further in‐depth analyses of the results obtained at the MACS marker screen, the experimental setup was repeated with the following modifications: HIV‐1 in vitro infection of CD4 T cells was performed at an MOI of 0.005. Culture conditions with no target cells and Mock CD4 T cells were added. Antibody labeling for multiplexing, identification of NK cell subsets, and assessment of CD107a expression were performed as described above. LIVE/DEAD Fixable Near‐IR Dead Cell Stain (Invitrogen) was used for discrimination of viable and dead cells. Cells were not distributed for antibody labeling of the MACS marker screen but only labeled with αTRAIL‐APC (Miltenyi Biotec, clone RIK‐2.1). NK cell subsets subject to further analyses comprised CD56Dim, CD56Bright, KIR‐educated, NKG2A‐educated, and uneducated NK cells. Definition of the education status of NK cells was based on the expression of the inhibitory receptors KIR2DL1/L2/L3, KIR3DL1, and NKG2A and the underlying HLA class I genotype. KIR‐educated cells were defined as expressing at least one self‐inhibitory KIR (2DL1/L2/L3, 3DL1) and being negative for NKG2A and NKG2A‐educated cells as expressing NKG2A but lacking self‐inhibitory KIR, and uneducated cells as lacking self‐inhibitory KIR and NKG2A altogether.
Assessment of expression levels of death and decoy receptors
Ninety‐six hours post‐infection with NL4‐3, WITO, CH198, CH236, JR‐CSF, or no virus (Mock) CD4 T cells were treated with 25 U/ml Benzonase Nuclease (Merck Millipore Novagen) for 30 min at 37°C, 5% (v/v) CO2. Cells were washed and then incubated separately in the presence of αCD45 (BioLegend, clone 2D1) conjugated to different fluorochromes for multiplexing (Akkaya et al, 2016). Cells were washed again, the different virus conditions of each donor were merged and stained with LIVE/DEAD Fixable Near‐IR Dead Cell Stain (Invitrogen) as well as αCD3 (BioLegend, clone UCHT1) and αCD4 (BD Biosciences, clone RPA‐T4) at 4°C. Cells were washed and then stained for 30 min with αDR4‐Biotin (Miltenyi Biotech, clone DJR1) and αDR5‐Biotin (Miltenyi Biotech, clone DJR2‐4). Cells were washed before incubation with 0.2 μg/ml Streptavidin‐PE (BioLegend) for 20 min at 4°C. Intracellular staining of HIV gag was performed using BD Cytofix/Cytoperm Fixation/Permeabilization kit (BD Biosciences) and αp24‐FITC (Beckman Coulter, clone KC57). Cells were fixed with 1× CellFIX (BD Biosciences) and stored at 4°C until flow cytometric data acquisition. For the assessment of decoy receptor expression, CD4 T cells were infected with NL4‐3 or no virus (mock). Four days post‐infection, cells were treated with 25 U/ml Benzonase Nuclease (Merck Millipore Novagen) for 30 min at 37°C, 5% (v/v) CO2. Cells were washed and then incubated with LIVE/DEAD Fixable Near‐IR Dead Cell Stain (Invitrogen), αCD3 (BioLegend, clone UCHT1), αCD4 (BD Biosciences, clone RPA‐T4), and goat αDcR1(R&D Systems, polyclonal) at 4°C for 20 min. Cells were washed again and incubated with αgoat‐IgG‐PE (R&D Systems, polyclonal) for additional 20 min at 4°C. Cells were permeabilized and stained for intracellular HIV gag using BD Cytofix/Cytoperm Fixation/Permeabilization kit (BD Biosciences) and αp24‐FITC (Beckman Coulter, clone KC57). Cells were fixed with 1× CellFIX (BD Biosciences) and stored at 4°C until flow cytometric data acquisition.
Antibody‐mediated blocking of TRAIL
Blocking of TRAIL interactions was conducted using a soluble TRAIL antibody (BioLegend, clone RIK‐2). For co‐incubation and blocking experiments with 721.221, isolated NK cells were resuspended in complete medium supplemented with 100 U/ml IL‐2 and 10 ng/ml IL‐15 (PeproTech) and incubated for 3 days to induce TRAIL expression (Kayagaki et al, 1999b; Balzarolo et al, 2013). TRAIL expression was monitored by flow cytometry using αTRAIL‐APC (Miltenyi Biotec, clone RIK‐2.1). TRAIL expression after 3 days was 81.3% ± 8.6% (Mean ± SD). For TRAIL‐blocking experiments, NK cells were resuspended at a concentration of 5 × 105 cells/ml and were pre‐incubated with 20 µg/ml purified αTRAIL or 20 µg/ml purified IgG1, κ isotype control antibody (BioLegend, clone MOPC‐21) in complete medium supplemented with 100 U/ml IL‐2 and 10 ng/ml IL‐15 for 30 min at 37°C, 5% (v/v) CO2. For DR4/DR5‐blocking experiments, 721.221 were pre‐incubated with 20 µg/ml purified αDR4 (AdipoGen, clone HS101) and 20 µg/ml purified αDR5 (AdipoGen, clone HS201) or the same amount of purified IgG1 isotype control antibody (AdipoGen, clone 15H6) for 30 min at 37°C, 5% (v/v) CO2. After pre‐incubation NK cells and 721.221 were co‐incubated at an effector:target ratio of 1:1 in the presence of αCD107a‐BV510 (BioLegend, Clone H4A3) for 5 h at 37°C, 5% (v/v) CO2. During co‐incubation, the respective blocking antibodies also remained present (final concentration per blocking antibody: 10 µg/ml). At the end of the co‐incubation, cells were spun down and supernatants were stored at −20°C for further analysis by ELISA. Cells were stained with the viability dye LIVE/DEAD Fixable Near‐IR (Invitrogen), αCD3‐PerCPCy5.5 (BioLegend, clone UCHT1), αCD14‐PerCPCy5.5 (BioLegend, clone HCD14), αCD16‐BV785 (BioLegend, clone 3G8), αCD19‐PerCPCy5.5 (BioLegend, clone HIB19), and αCD56‐BUV395 (BD Biosciences, NCAM16.2). Thereafter, cells were fixed with 1× CellFIX (BD Biosciences) and analyzed using a BD LSRFortessa (BD Biosciences).
For co‐incubation with autologous HIV‐1‐infected (NL4‐3) CD4 T cells, NK cells, isolated from cryopreserved PBMCs, were stimulated overnight in complete medium supplemented with 100 U/ml IL‐2 and 10 ng/ml IL‐15 (PeproTech) to induce TRAIL expression. This modification was necessary due to the different workflow to generate autologous HIV‐1‐infected CD4 T cells. TRAIL expression was monitored by flow cytometry using αTRAIL‐APC (Miltenyi Biotec, clone RIK‐2.1). TRAIL expression after overnight incubation was 69.8% ± 8.3% (Mean ± SD). According to the previous description, NK cells were resuspended at a concentration of 5 × 105 cells/ml and were pre‐incubated with 20 µg/ml purified αTRAIL or 20 µg/ml purified IgG1, κ isotype control antibody for 30 min at 37°C, 5% (v/v) CO2. Then, NK cells and HIV‐1‐infected CD4 T cells were co‐incubated at an effector:target ratio of 1:2 in the presence of αCD107a‐BV510 (BioLegend, Clone H4A3) for 5 h at 37°C, 5% (v/v) CO2. During co‐incubation, the respective blocking antibodies also remained present (final concentration per blocking antibody: 10 µg/ml). Subsequent antibody labeling and analysis were carried out as described above.
TRAIL cross‐linking
Immobilized (plate‐coated) antibodies and proteins were used to induce clustering of various NK cell receptors and subsequent degranulation. Isolated NK cells were washed, resuspended in complete medium supplemented with 100 U/ml IL‐2 and 10 ng/ml IL‐15 (PeproTech), and incubated for 3 days to induce TRAIL expression (Kayagaki et al, 1999b; Balzarolo et al, 2013). TRAIL expression was monitored by flow cytometry using αTRAIL‐APC (Miltenyi Biotec, clone RIK‐2.1). Mean TRAIL expression after 3 days was 87% ± 6.9% (Mean ± SD). Sterile non‐tissue culture‐treated flat‐bottom 96‐well plates (Corning) were coated with purified αTRAIL (BioLegend, clone RIK‐2), purified αNKG2D (BioLegend, clone 1D11), purified αNKp46 (Miltenyi Biotec, clone 9E2), purified IgG1, κ Isotype control antibody (BioLegend, clone MOPC‐21), human DR4 protein (Abcam), human DR5 protein (Abcam), or human IgG (ThermoFisher Scientific) diluted in PBS at three different concentrations (1, 5, and 10 µg/ml) or were left uncoated (PBS). Coated plates were incubated at 4°C for at least 24 h. Before use, plates were washed six times with PBS. Directly thereafter, isolated NK cells were distributed (2 × 104 cells/well) on coated plates and incubated for 5 h at 37°C, 5% (v/v) CO2 in the presence of αCD107a‐BV510 (BioLegend, Clone H4A3). After incubation, supernatants were collected and stored at −20°C for further analysis. Cells were stained with the viability dye LIVE/DEAD Fixable Near‐IR (Invitrogen), αCD3‐PerCP‐Cy5.5 (BioLegend, clone UCHT1), αCD14‐PerCP‐Cy5.5 (BioLegend, clone HCD14), αCD16‐BV785 (BioLegend, clone 3G8), αCD19‐PerCP‐Cy5.5 (BioLegend, clone HIB19), and αCD56‐BUV395 (BD Biosciences, NCAM16.2). For analysis by flow cytometry, cells were subsequently fixed with 1× CellFIX (BD Biosciences). Acquisition was carried out using a BD LSRFortessa (BD Biosciences).
Conjugate assay
To assess the impact of TRAIL interactions on the ability of NK cells to attach to target cells, we performed a conjugate assay as previously described (Burshtyn et al, 2000). In brief, enriched primary NK cells were incubated for 3 days in complete medium supplemented with 100 U/ml IL‐2 and 10 ng/ml IL‐15 (PeproTech) to induce TRAIL expression. TRAIL expression was assessed by flow cytometry using αTRAIL‐APC (Miltenyi Biotec, clone RIK‐2.1). Effector and target cells were individually labeled with a viability dye (LIVE/DEAD Fixable Near‐IR, Invitrogen) and fluorescent dyes (NK cells: 2 mM CellTracker Violet BMQC, Invitrogen; 721.221: 1 mM CellTracker Orange CMTMR, Invitrogen). After exclusion of dead cells, NK cells were identified as events positive for CellTracker Violet. Conjugates of NK cells and 721.221s were determined as events positive for both, CellTracker Violet and Orange. The relative frequency (%) of NK cells in conjugates was determined as events in conjugates in relation to the total number of NK cells.
Enzyme‐linked immunosorbent assay (ELISA)
Release of granzyme B and the production of IFNγ in in vitro NK cell assays were determined through commercially available ELISA kits. The limit of detection was 0.2 pg/ml for the Granzyme B ELISA kit (Invitrogen) and 2 pg/ml for the Human IFNγ ELISA kit (Invitrogen), respectively.
Phospho‐Epitope staining
Immobilized (plate‐coated) antibodies were used to induce clustering of various NK cell receptors and subsequent signaling cascades. Enriched NK cells were cultured in complete medium supplemented with 100 U/ml IL‐2 and 10 ng/ml IL‐15 (PeproTech) for 4 days to induce TRAIL expression. Sterile non‐tissue culture‐treated flat‐bottom 96‐well plates (Corning) were coated with either purified αTRAIL (BioLegend, clone RIK‐2), purified αNKG2D (BioLegend, clone 1D11), purified αNKp46 (Miltenyi Biotec, clone 9E2), and purified IgG1 κ Isotype control antibody (BioLegend, clone MOPC‐21), diluted in PBS at 10 µg/ml or were left uncoated (PBS). Coated plates were incubated at 4°C for at least 24 h. Before use, plates were washed six times with PBS. Directly thereafter, isolated NK cells were distributed (1 × 105 cells/well) on coated plates, and incubated for 30 min at 37°C, 5% (v/v) CO2. Cells were immediately fixed in BD Phosflow Fix Buffer I (BD Biosciences) for 10 min at 37°C, washed, and then permeabilized by incubating cells in BD Phosflow Perm Buffer III (BD Biosciences) for 30 min on ice. Cells were then washed and individually labeled with the following phospho‐epitope‐specific antibodies: PE mouse αPLC‐γ2 (pY759), PE mouse αSyk (pY348), PE mouse αp38 MAPK (pT180/pY182), and PE mouse αAkt (pS473) (all BD Biosciences). Afterwards, cells were washed and fixed in 1× CellFIX (BD Biosciences). Acquisition of cells was carried out using a BD LSRFortessa (BD Biosciences).
Cytotoxicity assays
Three cytotoxicity assays were performed to confirm the impact of TRAIL on NK‐cell‐mediated cytotoxicity. Enriched NK cells were cultured in complete medium supplemented with 100 U/ml IL‐2 and 10 ng/ml IL‐15 (PeproTech) for 3 days to induce TRAIL expression. 721.221 cells (DR4/5 positive), .221‐Cas9 (DR4/5 positive), and Raji‐DR5++ (overexpression of DR5) were used as target cells; and .221‐DR4/5KO (DR4/5 knockout) and Raji‐pSIP (baseline expression of DR5) cells served as control cells for their respective counterparts. Effector, control, and target cells were individually labeled with the following fluorescent dyes: CellTracker Violet BMQC, CellTracker Orange CMTMR, and Celltrace Far Red (all Invitrogen). TRAIL blocking condition: 1 × 105 NK cells were pretreated with either αTRAIL or isotype control (20 µg/ml) for 30 min and then co‐cultured with 721.221 cells at an E:T ratio of 1:1. DR4/5KO condition: 1 × 105 NK cells were co‐cultured with .221‐DR4/5‐KO and .221‐Cas9 cells at an E:C:T ratio of 1:0.5:0.5. DR5 overexpression condition: 1 × 105 NK cells were co‐cultured with Raji‐pSIP and Raji‐DR5++ at an E:C:T ratio of 1:0.5:0.5. Conditions without the addition of NK cells served as references (“No NK” control). Cell suspensions were cultured for 5 h in 5‐ml Polystyrene round‐bottom tubes (Corning) at a final volume of 200 µl at 37°C, 5% (v/v) CO2. After incubation, 25 µl of precision counting beads (BioLegend) were added and cells were immediately counted at a BD LSRFortessa (BD Biosciences). Data analysis: Cell counts were normalized to 10,000 counting beads. For the TRAIL blocking condition, relative frequency of remaining cells compared to the “No NK” control was used as a readout. For the DR5 overexpression and DR4/5KO conditions, specific lysis of designated target cells was calculated as previously described (Stary et al, 2020) with the following equation: [1 − (#control cells/#target cells)no NK cells/(#control cells/#target cells)with NK cells] × 100.
HLA class I genotyping
DNA was extracted from previously cryopreserved PBMCs using DNeasy Blood and Tissue kit (QIAGEN). Samples were sent to DKMS Life Science Lab (Dresden, Germany) where genotyping for HLA class I alleles was performed. Genotypic data of donors displayed in Fig 2 are stated in Table EV2.
Data acquisition and statistical analysis
Acquisition of flow cytometry data was performed on a BD LSRFortessa (BD Biosciences) and further analyzed using FlowJo software v10.7 (FlowJo, LLC, BD Life Sciences). Quantification of granzyme B and IFNγ through ELISAs was performed on a Safire2 microplate reader (Tecan Austria, GmbH). Statistical analyses and graphical display of the data were conducted using GraphPad Prism, version 9 (GraphPad Software, La Jolla, CA, USA). Data of samples, for which processing and technical errors could not be excluded, as well as data of cell populations (% of positive cells), for which the parent population (denominator of the %) was < 50 cells, were set as missing, and analyses performed on available (non‐missing) data. Hence, the total number of individuals analyzed in the high‐throughput screen (n = 19) might not represent the number of individuals analyzed for each single of the 327 surface markers investigated (ranging from n = 13 to n = 19). Non‐parametric statistical tests were applied to test for differences between groups. Wilcoxon signed‐rank test was used for two groups with paired values. For the MACS marker screen, paired comparisons of CD107a+ and CD107a− NK cell subsets were adjusted for multiplicity using the Benjamini and Hochberg false discovery rate (FDR) (Benjamini & Hochberg, 1995). Differential expression was assumed if a median intra‐donor difference of > 5% p.p. between CD107a+ and CD107a− NK cells and simultaneously FDR‐adjusted P‐values < 0.05 were present. For all other subsequent specific analyses, Bonferroni correction was used and applied to comparisons of interest. Spearman’s ρ was used to test for association between two parameters. Calculation of HIV‐I‐specific responses was carried out as follows: HIV‐I‐specific response = [%CD107a+ NK cells (HIV) − %CD107a+ NK cells (Mock)]/[100 − %CD107a+ NK cells (Mock)] * 100. Calculation of relative fluorescence intensity (RFI) was conducted as follows: RFI = [MdFI DR4/5/MdFI secondary ab control] − 1. Statistical parameters are stated in the results section as well as in the figure legends.
Author contributions
Johannes Höfle: Formal analysis; Investigation; Visualization; Methodology; Writing—original draft; Writing—review and editing. Timo Trenkner: Formal analysis; Validation; Investigation; Visualization; Writing—review and editing. Nadja Kleist: Formal analysis; Validation; Investigation; Writing—review and editing. Vera Schwane: Methodology; Writing—review and editing. Sarah Vollmers: Resources; Investigation; Writing—review and editing. Bryan Barcelona: Investigation; Writing—review and editing. Annika Niehrs: Resources; Methodology; Writing—review and editing. Pia Fittje: Resources; Writing—review and editing. Van Hung Huynh‐Tran: Formal analysis; Writing—review and editing. Jürgen Sauter: Resources; Writing—review and editing. Alexander H Schmidt: Resources; Writing—review and editing. Sven Peine: Resources; Writing—review and editing. Angelique Hoelzemer: Supervision; Funding acquisition; Writing—review and editing. Laura Richert: Formal analysis; Supervision; Writing—review and editing. Marcus Altfeld: Resources; Supervision; Funding acquisition; Writing—review and editing. Christian Körner: Conceptualization; Formal analysis; Supervision; Funding acquisition; Investigation; Visualization; Methodology; Writing—original draft; Writing—review and editing.
Disclosure and competing interests statement
The authors declare that they have no conflict of interest.
Supporting information
Expanded View Figures PDF
Table EV1
Table EV2
Source Data for Expanded View
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Source Data for Figure 6
Source Data for Figure 7
Acknowledgements
We would like to thank all blood donors for their participation in this study. We are also grateful to Beatrice Hahn for providing plasmids of the molecular clones CH198 and CH236 and to Mary Carrington for providing the plasmid of the molecular clone WITO. The authors would like to thank the Flow Cytometry technology platform of the Leibniz Institute of Virology for their support and technical assistance. S.V. was supported by the DFG (German Research foundation, KO 5139/3‐1). T.T. received funding by the state of Hamburg, Germany (LFF‐FV74). A.H. was supported by the DZIF (German Center for Infection Research, TTU 01.709; 8009701709). The funders had no influence on the study design, data collection and analysis, the decision to publish, or contents of the manuscript. Open Access funding enabled and organized by Projekt DEAL.
EMBO reports (2022) 23: e54133
Data availability
This study includes no data deposited in external repositories. Raw data storage is performed by the Leibniz Institute of Virology on an internal server. Raw data will be made available upon request and can be shared after confirming that data will be used within the scope of the originally provided informed consent.
References
- Adachi A, Gendelman HE, Koenig S, Folks T, Willey R, Rabson A, Martin MA (1986) Production of acquired immunodeficiency syndrome‐associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol 59: 284–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akkaya B, Miozzo P, Holstein AH, Shevach EM, Pierce SK, Akkaya M (2016) A simple, versatile antibody‐based barcoding method for flow cytometry. J Immunol 197: 2027–2038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alter G, Heckerman D, Schneidewind A, Fadda L, Kadie CM, Carlson JM, Oniangue‐Ndza C, Martin M, Li B, Khakoo SI et al (2011) HIV‐1 adaptation to NK‐cell‐mediated immune pressure. Nature 476: 96–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alter G, Malenfant JM, Altfeld M (2004) CD107a as a functional marker for the identification of natural killer cell activity. J Immunol Methods 294: 15–22 [DOI] [PubMed] [Google Scholar]
- Augugliaro R, Parolini S, Castriconi R, Marcenaro E, Cantoni C, Nanni M, Moretta L, Moretta A, Bottino C (2003) Selective cross‐talk among natural cytotoxicity receptors in human natural killer cells. Eur J Immunol 33: 1235–1241 [DOI] [PubMed] [Google Scholar]
- Balzarolo M, Watzl C, Medema JP, Wolkers MC (2013) NAB2 and EGR‐1 exert opposite roles in regulating TRAIL expression in human Natural Killer cells. Immunol Lett 151: 61–67 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol 57: 289–300 [Google Scholar]
- Berg M, Lundqvist A, McCoy P, Samsel L, Fan Y, Tawab A, Childs R (2009) Clinical grade ex vivo‐expanded human natural killer cells upregulate activating receptors and death receptor ligands and have enhanced cytolytic activity against tumor cells. Cytotherapy 11: 341–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biassoni R, Malnati MS (2018) Human natural killer receptors, co‐receptors, and their ligands. Curr Protoc Immunol 121: e47 [DOI] [PubMed] [Google Scholar]
- Burshtyn DN, Shin J, Stebbins C, Long EO (2000) Adhesion to target cells is disrupted by the killer cell inhibitory receptor. Curr Biol 10: 777–780 [DOI] [PubMed] [Google Scholar]
- Campbell KS, Cella M, Carretero M, López‐Botet M, Colonna M (1998) Signaling through human killer cell activating receptors triggers tyrosine phosphorylation of an associated protein complex. Eur J Immunol 28: 599–609 [DOI] [PubMed] [Google Scholar]
- Cardoso Alves L, Berger MD, Koutsandreas T, Kirschke N, Lauer C, Spörri R, Chatziioannou A, Corazza N, Krebs P (2020) Non‐apoptotic TRAIL function modulates NK cell activity during viral infection. EMBO Rep 21: e48789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Trivedi PP, Ge B, Krzewski K, Strominger JL (2007) Many NK cell receptors activate ERK2 and JNK1 to trigger microtubule organizing center and granule polarization and cytotoxicity. Proc Natl Acad Sci USA 104: 6329–6334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Yu H, Wrobel JA, Li G, Liu P, Hu Z, Xu X‐N, Su L (2020) Identification of pathogenic TRAIL‐expressing innate immune cells during HIV‐1 infection in humanized mice by scRNA‐Seq. JCI Insight 5: 135344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou A‐H, Tsai H‐F, Lin L‐L, Hsieh S‐L, Hsu P‐I, Hsu P‐N (2001) Enhanced proliferation and increased IFN‐γ production in T cells by signal transduced through TNF‐related apoptosis‐inducing ligand. J Immunol 167: 1347–1352 [DOI] [PubMed] [Google Scholar]
- Clancy L, Mruk K, Archer K, Woelfel M, Mongkolsapaya J, Screaton G, Lenardo MJ, Chan FK‐M (2005) Preligand assembly domain‐mediated ligand‐independent association between TRAIL receptor 4 (TR4) and TR2 regulates TRAIL‐induced apoptosis. Proc Natl Acad Sci USA 102: 18099–18104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis ZB, Ward JP, Barker E (2011) Preparation and use of HIV‐1 infected primary CD4+ T‐cells as target cells in natural killer cell cytotoxic assays. J Vis Exp 49: e2668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Toni EN, Thieme SE, Herbst A, Behrens A, Stieber P, Jung A, Blum H, Göke B, Kolligs FT (2008) OPG is regulated by beta‐catenin and mediates resistance to TRAIL‐induced apoptosis in colon cancer. Clin Cancer Res 14: 4713–4718 [DOI] [PubMed] [Google Scholar]
- Deeks SG (2012) HIV: shock and kill. Nature 487: 439–440 [DOI] [PubMed] [Google Scholar]
- Degli‐Esposti MA, Dougall WC, Smolak PJ, Waugh JY, Smith CA, Goodwin RG (1997a) The novel receptor TRAIL‐R4 induces NF‐kappaB and protects against TRAIL‐mediated apoptosis, yet retains an incomplete death domain. Immunity 7: 813–820 [DOI] [PubMed] [Google Scholar]
- Degli‐Esposti MA, Smolak PJ, Walczak H, Waugh J, Huang C‐P, DuBose RF, Goodwin RG, Smith CA (1997b) Cloning and characterization of TRAIL‐R3, a novel member of the emerging TRAIL receptor family. J Exp Med 186: 1165–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl GE, Yue HH, Hsieh K, Kuang AA, Ho M, Morici LA, Lenz LL, Cado D, Riley LW, Winoto A (2004) TRAIL‐R as a negative regulator of innate immune cell responses. Immunity 21: 877–889 [DOI] [PubMed] [Google Scholar]
- Emery JG, McDonnell P, Burke MB, Deen KC, Lyn S, Silverman C, Dul E, Appelbaum ER, Eichman C, DiPrinzio R et al (1998) Osteoprotegerin is a receptor for the cytotoxic ligand TRAIL*. J Biol Chem 273: 14363–14367 [DOI] [PubMed] [Google Scholar]
- Fadda L, Körner C, Kumar S, van Teijlingen NH, Piechocka‐Trocha A, Carrington M, Altfeld M (2012) HLA‐Cw*0102‐restricted HIV‐1 p24 epitope variants can modulate the binding of the inhibitory KIR2DL2 receptor and primary NK cell function. PLoS Pathog 8: e1002805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanger NA, Maliszewski CR, Schooley K, Griffith TS (1999) Human dendritic cells mediate cellular apoptosis via tumor necrosis factor‐related apoptosis‐inducing ligand (TRAIL). J Exp Med 190: 1155–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenton‐May AE, Dibben O, Emmerich T, Ding H, Pfafferott K, Aasa‐Chapman MM, Pellegrino P, Williams I, Cohen MS, Gao F et al (2013) Relative resistance of HIV‐1 founder viruses to control by interferon‐alpha. Retrovirology 10: 146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gougeon M‐L, Herbeuval J‐P (2012) IFN‐α and TRAIL: a double edge sword in HIV‐1 disease? Exp Cell Res 318: 1260–1268 [DOI] [PubMed] [Google Scholar]
- Griffith TS, Wiley SR, Kubin MZ, Sedger LM, Maliszewski CR, Fanger NA (1999) Monocyte‐mediated tumoricidal activity via the tumor necrosis factor‐related cytokine, TRAIL. J Exp Med 189: 1343–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy AW, Graham DR, Shearer GM, Herbeuval J‐P (2007) HIV turns plasmacytoid dendritic cells (pDC) into TRAIL‐expressing killer pDC and down‐regulates HIV coreceptors by Toll‐like receptor 7‐induced IFN‐alpha. Proc Natl Acad Sci USA 104: 17453–17458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbeuval J‐P, Boasso A, Grivel J‐C, Hardy AW, Anderson SA, Dolan MJ, Chougnet C, Lifson JD, Shearer GM (2005a) TNF‐related apoptosis‐inducing ligand (TRAIL) in HIV‐1‐infected patients and its in vitro production by antigen‐presenting cells. Blood 105: 2458–2464 [DOI] [PubMed] [Google Scholar]
- Herbeuval J‐P, Grivel J‐C, Boasso A, Hardy AW, Chougnet C, Dolan MJ, Yagita H, Lifson JD, Shearer GM (2005b) CD4+ T‐cell death induced by infectious and noninfectious HIV‐1: role of type 1 interferon–dependent, TRAIL/DR5‐mediated apoptosis. Blood 106: 3524–3531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbeuval J‐P, Hardy AW, Boasso A, Anderson SA, Dolan MJ, Dy M, Shearer GM (2005c) Regulation of TNF‐related apoptosis‐inducing ligand on primary CD4+ T cells by HIV‐1: role of type I IFN‐producing plasmacytoid dendritic cells. Proc Natl Acad Sci USA 102: 13974–13979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbeuval J‐P, Nilsson J, Boasso A, Hardy AW, Vaccari M, Cecchinato V, Valeri V, Franchini G, Andersson J, Shearer GM (2009) HAART reduces death ligand but not death receptors in lymphoid tissue of HIV‐infected patients and simian immunodeficiency virus‐infected macaques. AIDS Lond Engl 23: 35–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holen I, Cross SS, Neville‐Webbe HL, Cross NA, Balasubramanian SP, Croucher PI, Evans CA, Lippitt JM, Coleman RE, Eaton CL (2005) Osteoprotegerin (OPG) expression by breast cancer cells in vitro and breast tumours in vivo–a role in tumour cell survival? Breast Cancer Res Treat 92: 207–215 [DOI] [PubMed] [Google Scholar]
- Holen I, Croucher PI, Hamdy FC, Eaton CL (2002) Osteoprotegerin (OPG) is a survival factor for human prostate cancer cells. Cancer Res 62: 1619–1623 [PubMed] [Google Scholar]
- Hölzemer A, Thobakgale CF, Jimenez Cruz CA, Garcia‐Beltran WF, Carlson JM, van Teijlingen NH, Mann JK, Jaggernath M, Kang S‐G, Körner C et al (2015) Selection of an HLA‐C*03:04‐restricted HIV‐1 p24 gag sequence variant is associated with viral escape from KIR2DL3+ natural killer cells: data from an observational cohort in South Africa. PLoS Med 12: e1001900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz A, Strauss‐Albee DM, Leipold M, Kubo J, Nemat‐Gorgani N, Dogan OC, Dekker CL, Mackey S, Maecker H, Swan GE et al (2013) Genetic and environmental determinants of human NK cell diversity revealed by mass cytometry. Sci Transl Med 5: 208ra145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S‐C, Tsai H‐F, Tzeng H‐T, Liao H‐J, Hsu P‐N (2011) Lipid raft assembly and Lck recruitment in TRAIL costimulation mediates NF‐κB activation and T cell proliferation. J Immunol 186: 931–939 [DOI] [PubMed] [Google Scholar]
- Humphrey MB, Lanier LL, Nakamura MC (2005) Role of ITAM‐containing adapter proteins and their receptors in the immune system and bone. Immunol Rev 208: 50–65 [DOI] [PubMed] [Google Scholar]
- Iyori M, Zhang T, Pantel H, Gagne BA, Sentman CL (2011) TRAIL/DR5 plays a critical role in NK cell‐mediated negative regulation of dendritic cell cross‐priming of T cells. J Immunol 187: 3087–3095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang K, Zhong B, Gilvary DL, Corliss BC, Vivier E, Hong‐Geller E, Wei S, Djeu JY (2002) Syk regulation of phosphoinositide 3‐kinase‐dependent NK cell function. J Immunol 168: 3155–3164 [DOI] [PubMed] [Google Scholar]
- Jost S, Altfeld M (2013) Control of human viral infections by natural killer cells. Annu Rev Immunol 31: 163–194 [DOI] [PubMed] [Google Scholar]
- Kayagaki N, Yamaguchi N, Nakayama M, Eto H, Okumura K, Yagita H (1999a) Type I interferons (IFNs) regulate tumor necrosis factor‐related apoptosis‐inducing ligand (TRAIL) expression on human T cells: A novel mechanism for the antitumor effects of type I IFNs. J Exp Med 189: 1451–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayagaki N, Yamaguchi N, Nakayama M, Takeda K, Akiba H, Tsutsui H, Okamura H, Nakanishi K, Okumura K, Yagita H (1999b) Expression and function of TNF‐related apoptosis‐inducing ligand on murine activated NK cells. J Immunol 163: 1906–1913 [PubMed] [Google Scholar]
- Körner C, Granoff ME, Amero MA, Sirignano MN, Vaidya SA, Jost S, Allen TM, Rosenberg ES, Altfeld M (2014) Increased frequency and function of KIR2DL1‐3+ NK cells in primary HIV‐1 infection are determined by HLA‐C group haplotypes. Eur J Immunol 44: 2938–2948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Körner C, Simoneau CR, Schommers P, Granoff M, Ziegler M, Hölzemer A, Lunemann S, Chukwukelu J, Corleis B, Naranbhai V et al (2017) HIV‐1‐mediated downmodulation of HLA‐C impacts target cell recognition and antiviral activity of NK cells. Cell Host Microbe 22: 111–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanier LL (2009) DAP10‐ and DAP12‐associated receptors in innate immunity. Immunol Rev 227: 150–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanier LL, Corliss BC, Wu J, Leong C, Phillips JH (1998) Immunoreceptor DAP12 bearing a tyrosine‐based activation motif is involved in activating NK cells. Nature 391: 703–707 [DOI] [PubMed] [Google Scholar]
- Lee W‐H, Seo D, Lim S‐G, Suk K (2019) Reverse Signaling of tumor necrosis factor superfamily proteins in macrophages and microglia: superfamily portrait in the neuroimmune interface. Front Immunol 10: 262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Yang Y, Song H, Li H, Cui A, Liu Y, Su L, Crispe IN, Tu Z (2019) Activated NK cells kill hepatic stellate cells via p38/PI3K signaling in a TRAIL‐involved degranulation manner. J Leukoc Biol 105: 695–704 [DOI] [PubMed] [Google Scholar]
- Long EO, Sik Kim H, Liu D, Peterson ME, Rajagopalan S (2013) Controlling natural killer cell responses: integration of signals for activation and inhibition. Annu Rev Immunol 31: 227–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum JJ, Schnepple DJ, Nie Z, Sanchez‐Dardon J, Mbisa GL, Mihowich J, Hawley N, Narayan S, Kim JE, Lynch DH et al (2004) Differential effects of interleukin‐7 and interleukin‐15 on NK cell anti‐human immunodeficiency virus activity. J Virol 78: 6033–6042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luu TT, Schmied L, Nguyen N‐A, Wiel C, Meinke S, Mohammad DK, Bergö M, Alici E, Kadri N, Ganesan S et al (2021) Short‐term IL‐15 priming leaves a long‐lasting signalling imprint in mouse NK cells independently of a metabolic switch. Life Sci Alliance 4: e202000723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmberg K‐J, Carlsten M, Björklund A, Sohlberg E, Bryceson YT, Ljunggren H‐G (2017) Natural killer cell‐mediated immunosurveillance of human cancer. Semin Immunol 31: 20–29 [DOI] [PubMed] [Google Scholar]
- Marcenaro E, Augugliaro R, Falco M, Castriconi R, Parolini S, Sivori S, Romeo E, Millo R, Moretta L, Bottino C et al (2003) CD59 is physically and functionally associated with natural cytotoxicity receptors and activates human NK cell‐mediated cytotoxicity. Eur J Immunol 33: 3367–3376 [DOI] [PubMed] [Google Scholar]
- Martin MP, Gao X, Lee J‐H, Nelson GW, Detels R, Goedert JJ, Buchbinder S, Hoots K, Vlahov D, Trowsdale J et al (2002) Epistatic interaction between KIR3DS1 and HLA‐B delays the progression to AIDS. Nat Genet 31: 429–434 [DOI] [PubMed] [Google Scholar]
- Martin MP, Qi Y, Gao X, Yamada E, Martin JN, Pereyra F, Colombo S, Brown EE, Shupert WL, Phair J et al (2007) Innate partnership of HLA‐B and KIR3DL1 subtypes against HIV‐1. Nat Genet 39: 733–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mérino D, Lalaoui N, Morizot A, Schneider P, Solary E, Micheau O (2006) Differential inhibition of TRAIL‐mediated DR5‐DISC formation by decoy receptors 1 and 2. Mol Cell Biol 26: 7046–7055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meza Guzman LG, Keating N, Nicholson SE (2020) Natural killer cells: tumor surveillance and signaling. Cancers 12: 952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Doherty U, Swiggard WJ, Malim MH (2000) Human immunodeficiency virus type 1 spinoculation enhances infection through virus binding. J Virol 74: 10074–10080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochsenbauer C, Edmonds TG, Ding H, Keele BF, Decker J, Salazar MG, Salazar‐Gonzalez JF, Shattock R, Haynes BF, Shaw GM et al (2012) Generation of transmitted/founder HIV‐1 infectious molecular clones and characterization of their replication capacity in CD4 T lymphocytes and monocyte‐derived macrophages. J Virol 86: 2715–2728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oszmiana A, Williamson DJ, Cordoba S‐P, Morgan DJ, Kennedy PR, Stacey K, Davis DM (2016) The size of activating and inhibitory killer Ig‐like receptor nanoclusters is controlled by the transmembrane sequence and affects signaling. Cell Rep 15: 1957–1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan G, Ni J, Wei Y‐F, Yu G, Gentz R, Dixit VM (1997a) An antagonist decoy receptor and a death domain‐containing receptor for TRAIL. Science 277: 815–818 [DOI] [PubMed] [Google Scholar]
- Pan G, O’Rourke K, Chinnaiyan AM, Gentz R, Ebner R, Ni J, Dixit VM (1997b) The receptor for the cytotoxic ligand TRAIL. Science 276: 111–113 [DOI] [PubMed] [Google Scholar]
- Parrish NF, Gao F, Li H, Giorgi EE, Barbian HJ, Parrish EH, Zajic L, Iyer SS, Decker JM, Kumar A et al (2013) Phenotypic properties of transmitted founder HIV‐1. Proc Natl Acad Sci 110: 6626–6633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitti RM, Marsters SA, Ruppert S, Donahue CJ, Moore A, Ashkenazi A (1996) Induction of apoptosis by Apo‐2 ligand, a new member of the tumor necrosis factor cytokine family. J Biol Chem 271: 12687–12690 [DOI] [PubMed] [Google Scholar]
- Prager I, Watzl C (2019) Mechanisms of natural killer cell‐mediated cellular cytotoxicity. J Leukoc Biol 105: 1319–1329 [DOI] [PubMed] [Google Scholar]
- Sato K, Hida S, Takayanagi H, Yokochi T, Kayagaki N, Takeda K, Yagita H, Okumura K, Tanaka N, Taniguchi T et al (2001) Antiviral response by natural killer cells through TRAIL gene induction by IFN‐alpha/beta. Eur J Immunol 31: 3138–3146 [DOI] [PubMed] [Google Scholar]
- Schwane V, Huynh‐Tran VH, Vollmers S, Yakup VM, Sauter J, Schmidt AH, Peine S, Altfeld M, Richert L, Körner C (2020) Distinct signatures in the receptor repertoire discriminate CD56bright and CD56dim natural killer cells. Front Immunol 11: 2982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheridan JP, Marsters SA, Pitti RM, Gurney A, Skubatch M, Baldwin D, Ramakrishnan L, Gray CL, Baker K, Wood WI et al (1997) Control of TRAIL‐induced apoptosis by a family of signaling and decoy receptors. Science 277: 818–821 [DOI] [PubMed] [Google Scholar]
- Shimizu Y, DeMars R (1989) Production of human cells expressing individual transferred HLA‐A,‐B,‐C genes using an HLA‐A,‐B,‐C null human cell line. J Immunol 142: 3320–3328 [PubMed] [Google Scholar]
- Shipman CM, Croucher PI (2003) Osteoprotegerin is a soluble decoy receptor for tumor necrosis factor‐related apoptosis‐inducing ligand/Apo2 ligand and can function as a paracrine survival factor for human myeloma cells. Cancer Res 63: 912–916 [PubMed] [Google Scholar]
- Spaggiari GM, Carosio R, Pende D, Marcenaro S, Rivera P, Zocchi MR, Moretta L, Poggi A (2001) NK cell‐mediated lysis of autologous antigen‐presenting cells is triggered by the engagement of the phosphatidylinositol 3‐kinase upon ligation of the natural cytotoxicity receptors NKp30 and NKp46. Eur J Immunol 31: 1656–1665 [DOI] [PubMed] [Google Scholar]
- Stary G, Klein I, Kohlhofer S, Koszik F, Scherzer T, Müllauer L, Quendler H, Kohrgruber N, Stingl G (2009) Plasmacytoid dendritic cells express TRAIL and induce CD4+ T‐cell apoptosis in HIV‐1 viremic patients. Blood 114: 3854–3863 [DOI] [PubMed] [Google Scholar]
- Stary V, Pandey RV, Strobl J, Kleissl L, Starlinger P, Pereyra D, Weninger W, Fischer GF, Bock C, Farlik M et al (2020) A discrete subset of epigenetically primed human NK cells mediates antigen‐specific immune responses. Sci Immunol 5: eaba6232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmann KA, Björkström NK, Veber H, Ciesek S, Riese P, Wiegand J, Hadem J, Suneetha PV, Jaroszewicz J, Wang C et al (2010) Interferon‐α–induced TRAIL on natural killer cells is associated with control of hepatitis C virus infection. Gastroenterology 138: 1885–1897 [DOI] [PubMed] [Google Scholar]
- Sun M, Fink PJ (2007) A new class of reverse signaling costimulators belongs to the TNF family. J Immunol 179: 4307–4312 [DOI] [PubMed] [Google Scholar]
- Sutherland CL, Chalupny NJ, Schooley K, VandenBos T, Kubin M, Cosman D (2002) UL16‐binding proteins, novel MHC class I‐related proteins, bind to NKG2D and activate multiple signaling pathways in primary NK cells. J Immunol 168: 671–679 [DOI] [PubMed] [Google Scholar]
- Ting AT, Dick CJ, Schoon RA, Karnitz LM, Abraham RT, Leibson PJ (1995) Interaction between lck and syk family tyrosine kinases in Fc gamma receptor‐initiated activation of natural killer cells. J Biol Chem 270: 16415–16421 [DOI] [PubMed] [Google Scholar]
- Tsai H‐F, Lai J‐J, Chou A‐H, Wang T‐F, Wu C‐S, Hsu P‐N (2004) Induction of costimulation of human CD4 T cells by tumor necrosis factor‐related apoptosis‐inducing ligand: Possible role in T cell activation in systemic lupus erythematosus. Arthritis Rheum 50: 629–639 [DOI] [PubMed] [Google Scholar]
- Upshaw JL, Schoon RA, Dick CJ, Billadeau DD, Leibson PJ (2005) The isoforms of phospholipase C‐gamma are differentially used by distinct human NK activating receptors. J Immunol 175: 213–218 [DOI] [PubMed] [Google Scholar]
- Vivier E, da Silva AJ, Ackerly M, Levine H, Rudd CE, Anderson P (1993) Association of a 70‐kDa tyrosine phosphoprotein with the CD16: zeta: gamma complex expressed in human natural killer cells. Eur J Immunol 23: 1872–1876 [DOI] [PubMed] [Google Scholar]
- Vivier E, Tomasello E, Baratin M, Walzer T, Ugolini S (2008) Functions of natural killer cells. Nat Immunol 9: 503–510 [DOI] [PubMed] [Google Scholar]
- Wiley SR, Schooley K, Smolak PJ, Din WS, Huang C‐P, Nicholl JK, Sutherland GR, Smith TD, Rauch C, Smith CA et al (1995) Identification and characterization of a new member of the TNF family that induces apoptosis. Immunity 3: 673–682 [DOI] [PubMed] [Google Scholar]
- Zauli G, Melloni E, Capitani S, Secchiero P (2009) Role of full‐length osteoprotegerin in tumor cell biology. Cell Mol Life Sci 66: 841–851 [DOI] [PMC free article] [PubMed] [Google Scholar]


