Abstract
2,4,5-Trihydroxytoluene (THT) oxygenase from Burkholderia sp. strain DNT catalyzes the conversion of THT to an unstable ring fission product. Biochemical and genetic studies of THT oxygenase were undertaken to elucidate the mechanism of the ring fission reaction. The THT oxygenase gene (dntD) was previously localized to the 1.2-kb DNA insert subcloned in the recombinant plasmid designated pJS76 (W. C. Suen and J. C. Spain, J. Bacteriol. 175:1831–1837, 1993). Analysis of the deduced amino acid sequence of DntD revealed the presence of the highly conserved residues characteristic of the catechol 2,3-dioxygenase gene family I. The deduced amino acid sequence of DntD corresponded to a molecular mass of 35 kDa. The native molecular masses for the THT oxygenase estimated by using gel filtration chromatography and nondenaturing gel electrophoresis were 67.4 and 77.8 kDa, respectively. The results suggested that the native protein consists of two identical subunits. The colorless protein contained 2 mol of iron per mol of protein. Stimulation of activity in the presence of ferrous iron and ascorbate suggested a requirement for ferrous iron in the active site. The properties of the enzyme are similar to those of the catechol 2,3-dioxygenases (meta-cleavage dioxygenases). In addition to THT, the enzyme exhibited activity towards 1,2,4-benzenetriol, catechol, 3- and 4-methylcatechol, and 3- and 4-chlorocatechol. The chemical analysis of the THT ring cleavage product showed that the product was 2,4-dihydroxy-5-methyl-6-oxo-2,4-hexadienoic acid, consistent with extradiol ring fission of THT.
Bacterial degradation of aromatic compounds under aerobic conditions involves the conversion of the initial substrates into intermediates with two or more hydroxyl groups on the aromatic ring. The intermediates are subject to oxidative attack by the ring cleavage dioxygenases. The ring cleavage enzymes are classified into two groups based on the site of ring fission (23). Enzymes catalyzing ring fission between two hydroxyl groups are designated intradiol (ortho-) cleavage enzymes. Enzymes that cleave at a bond proximal to one of the two adjacent hydroxyl groups are designated extradiol (meta-) cleavage enzymes. The primary structures of many ring cleavage dioxygenases have been determined, and comparison of their amino acid sequences has revealed that they can be divided further into distinct gene families. The ortho-cleavage enzymes are divided into three families designated catechol 1,2-dioxygenase I, catechol 1,2-dioxygenase II, and protocatechuate 3,4-dioxygenase (22). The meta-cleavage enzymes constitute at least three different groups, including the catechol 2,3-dioxygenase I, the protocatechuate 4,5-dioxygenase, and the homoprotocatechuate dioxygenase families (22).
The interest in bacterial metabolism of aromatic compounds has motivated a great amount of research on the biochemistry and molecular genetics of the meta-cleavage enzymes. Harayama et al. provided an early review on the molecular biology of the ring cleavage dioxygenases (22). Since the publication of that review, numerous studies characterizing other meta-ring cleavage enzymes show the broad range of catabolic pathways that use the extradiol dioxygenases. More recent reports have provided additional information and discussion on the phylogeny of the meta-ring cleavage enzymes (14, 36). The molecular genetic analysis can be coupled with the information gained from determinations of the three-dimensional structures for the 2,3-dihydroxybiphenyl dioxygenases from Burkholderia cepacia LB400 (21) and Pseudomonas sp. strain KKS102 (50) to provide a comprehensive overview of the relationships between the primary protein structure and the biochemistry of the meta-ring cleavage enzymes.
Burkholderia sp. strain DNT mineralizes 2,4-dinitrotoluene (2,4-DNT) via an oxidative pathway (Fig. 1) (52). The initial step in the pathway is the dioxygenation of the aromatic ring to form 4-methyl-5-nitrocatechol (MNC), with concomitant release of the first nitrite from the substrate. Subsequently, MNC monooxygenase catalyzes the oxidation of MNC to yield 2-hydroxy-5-methylquinone (HMQ) and nitrite. A quinone reductase then catalyzes the conversion of HMQ to 2,4,5-trihydroxytoluene (THT). THT is oxidized by a ring cleavage enzyme, THT oxygenase. The mode of ring cleavage has not been determined rigorously. Previous results suggested that THT oxygenase catalyzes an extradiol cleavage reaction; however, the preliminary evidence was not sufficient to distinguish among three possible mechanisms of ring fission (Fig. 1) (19).
FIG. 1.
Biodegradation of 2,4-DNT by Burkholderia sp. strain DNT (19). The shaded area indicates the ring cleavage step catalyzed by the THT oxygenase and the potential products of the reaction. The gene designations for the enzymes in the pathway are as follows: dntA, DNT dioxygenase; dntB, MNC monooxygenase; dntC, HMQ reductase; dntD, THT oxygenase.
The cloning and preliminary characterization of the genes encoding DNT dioxygenase (dntA), MNC monooxygenase (dntB), and THT oxygenase (dntD) from strain DNT have been described previously (55). Additional reports outlined analysis of the nucleotide sequences for dntA and dntB (20, 54). The present paper describes biochemical and genetic characterization of the THT oxygenase along with experiments to determine the mechanism of ring fission and identify the product of the reaction catalyzed by THT oxygenase.
(A preliminary report of this work has been presented previously [53].)
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
Burkholderia sp. strain DNT was grown and maintained as previously described (19). Escherichia coli JM109(DE3) (Promega Corp., Madison, Wis.) was used as the host for recombinant plasmids in this work. Plasmid pT7-5 was used as the expression vector (57). E. coli strains were grown at 37°C unless otherwise indicated. Recombinant E. coli strains were cultivated in complex medium containing 20 g of tryptone, 10 g of yeast extract, 5 g of NaCl, and 0.2% glycerol per liter of 50 mM KH2PO4 (pH 7.2). Ampicillin was added at a concentration of 50 μg/ml when required to select for the presence of plasmids.
Enzyme assays.
Bacterial cell extracts of strain DNT and E. coli(pJS77) were prepared from frozen cells suspended in an equal volume of 0.05 M Tris-HCl (pH 7.5) containing 0.5 mg of DNase I. Cell lysis and ultracentrifugation to remove insoluble material were done as previously described (51).
THT oxygenase activity was determined by measuring the rate of oxygen consumption in the presence of THT as previously described (19). For routine purification and analysis, THT oxygenase activity was determined spectrophotometrically by measuring the conversion of 4-methylcatechol (4MC), a structural analog of THT, to the 2-hydroxy-(4- and/or 5-)methylmuconic semialdehyde (19). The assay mixture contained 4MC (10−4 M) and appropriate amounts of protein in 1 ml of KH2PO4 buffer (0.02 M, pH 7.0). The rate of hydroxymethylmuconic semialdehyde accumulation was determined spectrophotometrically at 382 nm. The extinction coefficient of the hydroxymethylmuconic semialdehyde (ɛ382 = 20,000 at pH 7.0) was determined spectrophotometrically after complete conversion of 4MC by THT oxygenase under standard assay conditions. The activity of THT oxygenase towards alternate substrates was initially demonstrated spectrophotometrically. Transformation rates for alternate substrates were also estimated by measuring oxygen consumption in the presence of the test substrate.
Pyruvate formation from THT by cell extracts of Burkholderia sp. strain DNT was estimated by a modification of the method of Friedemann and Haugen (16) and by measuring NADH oxidation in the presence of lactate dehydrogenase. Assays (3-ml mixtures) were performed in 0.02 M KH2PO4 buffer (pH 6.65) with 0.66 mg of cell extract obtained as previously described (20). Reactions were initiated by the addition of 5 × 10−5 to 10 × 10−5 M THT. After a 15- to 20-min incubation at room temperature, 1 ml of the reaction mixture was transferred to a cuvette and pyruvate was quantified by measuring NADH oxidation in the presence of lactate dehydrogenase. Another 1 ml of the assay mixture was transferred to a tube containing 10 μl of phosphoric acid and incubated for an additional 5 min, and then 0.333 ml of 2,4-dinitrophenylhydrazine reagent (16) was added. The precipitated protein was removed by centrifugation at 15,800 × g rpm, and the assay mixture was allowed to incubate for an additional 5 min. After the addition of 2.5 N NaOH (1.667 ml), the assay mixtures were incubated for an additional 10 min and the absorbance at 520 nm was determined.
Protein purification.
All procedures were carried out at 4°C unless stated otherwise. A rapid detection method was used to assay THT oxygenase activity during chromatographic procedures. Samples (10 to 20 μl) of the column fractions were transferred to tubes containing 4-MC (0.1 mM) in 1 ml of KH2PO4 (20 mM, pH 7.0). The presence of the THT oxygenase was indicated by the appearance of the yellow hydroxymethylmuconic semialdehyde resulting from oxidation of 4MC.
High-level expression of dntD was accomplished by subcloning the gene into expression vector pT7-5 and then transferring the plasmid construct to E. coli JM109. Expression of dntD was induced by the addition of IPTG (isopropyl-β-d-thiogalactopyranoside) to the culture medium at 100 μM by standard methods (2).
For protein purification, cell extracts were prepared from 10 g (wet weight) of E. coli(pJS77). The volume of the extract was adjusted to three times the volume of the frozen cell suspension, and ammonium sulfate was added to a final concentration of 25% at 0°C. The solution was subjected to ultracentrifugation at 100,000 × g for 30 min to remove precipitated protein. The supernatant was loaded onto a phenyl-Sepharose CL-4B fast-flow column (5 by 5 cm) (Pharmacia Biotech Inc., Piscataway, N.J.) previously equilibrated with 25% ammonium sulfate in Tris-HCl buffer. Sample loading and elution were at a flow rate of 4 ml/min. The column was washed with 200 ml of Tris-HCl buffer containing 25% ammonium sulfate, and proteins were eluted with a decreasing linear ammonium sulfate gradient (25 to 0%) in Tris-HCl (400 ml). The fractions (6 ml each) that contained maximum THT oxygenase activity were combined and then loaded directly onto a Q-Sepharose fast-flow column (1.6 by 19.5 cm) (Pharmacia). Proteins were eluted with 120 ml of Tris-HCl buffer followed by a 240-ml linear KCl gradient (0 to 0.4 M). The fractions (4 ml each) that contained THT oxygenase activity were pooled and concentrated by using a YM5 filter (Amicon Inc., Beverly, Mass.) under nitrogen at 4°C to yield a final volume of 1 to 2 ml. The solution was loaded in two equal portions onto a Sephacryl S300 HR column (1.6 by 91 cm) (Pharmacia), and proteins were eluted with Tris-HCl buffer at 1 ml/min. Fractions (2 ml each) containing THT oxygenase activity were pooled and loaded onto a DEAE-Sepharose fast-flow column (1.6 by 10 cm) (Pharmacia). Proteins were eluted with a 40-ml buffer wash followed by a continuous gradient of KCl (0 to 0.4 M) in Tris buffer at 2 ml/min. The fractions (1 ml each) that contained maximum activity were combined, and the KCl was removed by dialysis prior to storage at −70°C.
Isolation and identification of THT reaction products.
Purified THT oxygenase (280 μg) was incubated with THT (added in four equal portions at 2-min intervals to give a final concentration of 0.2 mM) in 1 ml of deionized water. Two minutes after the final addition of THT, 100 μl of the assay mixture was transferred to a tube containing 100 μl of acetonitrile. The solution was then analyzed by liquid chromatography (LC)-mass spectrometry (MS) (chemical ionization). Products formed from 1,2,4-benzenetriol and 4MC were analyzed similarly, except that higher concentrations of THT oxygenase (560 and 1,050 μg/ml, respectively) were used in the reactions.
Analytical methods.
Protein determinations, UV-visible spectral analyses, high-pressure LC (HPLC), sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and gel filtration molecular weight determinations were done by standard methods (2) and as described previously (20, 54). Gel filtration was done by both standard LC and HPLC. An S-200 HR column (1.6 by 88 cm) (Pharmacia) with a mobile phase consisting of 0.1 M NaCl and 0.05 M Tris-HCl (pH 7.5 at 4°C) was used for LC. HPLC was done by using a GF-250 gel filtration column (9.4 mm by 25 cm) (Dupont Instruments, Wilmington, Del.) with a mobile phase of 0.3 M Na2H2PO4 (pH 7.0) at 0.5 ml/min. Nondenaturing gel electrophoresis was done in slab gels by the procedure described by the reagent manufacturer (Sigma, St. Louis, Mo.). The iron content of THT oxygenase was determined with o-phenanthroline and Ferene S as described previously (18, 41).
DNA sequencing was done at the genetic engineering core facility at the University of Illinois-Champaign, Urbana. Sequence similarity searches were done with the BLAST programs (1) via the National Center for Biotechnology Information web site. Additional sequence analysis was carried out with the CLUSTAL W software package (61). Multiple-alignment comparisons of amino acid sequences were done by the method of Feng and Doolittle (15), and phylogenetic trees were inferred from the alignments by using the neighbor-joining method of Saitou and Nei (47). Graphics for phylogenetic trees were produced by using the TreeView program designed by Page (45).
Analysis of ring cleavage products was done with a Hewlett-Packard (HP) model 1050 liquid chromatograph coupled with an HP model 5987 mass spectrometer. Reaction products were separated with a Spherisorb C8 column (Altech Bioscience, Deerfield, Ill.) with an acetonitrile-water (70:30) mobile phase at a flow rate of 0.7 ml/min. The LC eluent was passed through an HP model 59980A particle beam interface prior to introduction into the mass spectrometer. Negative chemical ionization was done by using methane as a carrier gas. Nuclear magnetic resonance (NMR) spectra were obtained with a Bruker AC-300 Fourier transform NMR spectrometer by Stephen A. Aubert (Eglin Air Force Base, Fla.) and with a Bruker WM-360 spectrometer by Sol M. Resnick (University of Iowa, Iowa City).
Chemicals.
DNase I, o-phenanthroline, ferrene S, 4-MC, catechol, and 1,2,4-benzenetriol were purchased from Sigma Chemical Co. THT was kindly provided by R. Spanggord (SRI International, Menlo Park, Calif.). 3-Methylmaleylacetate was derived from 3-methyl-cis-dienelactone as previously described (46). 3-Methyl-cis-dienelactone and 3-methyl-trans-dienelactone were generous gifts from D. Pieper (Gesellschaft fur Biotechnologische Forschung, Braunschweig, Germany).
Nucleotide sequence accession number.
The GenBank accession number of dntD from strain DNT is AF076848.
RESULTS
Nucleotide sequence of dntD.
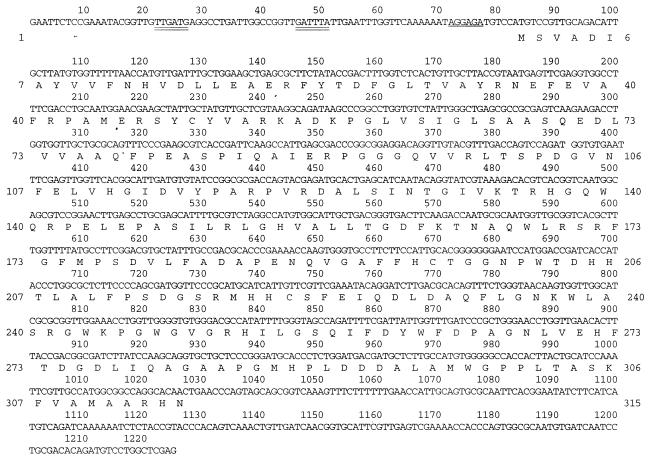
Analysis of the nucleotide sequence of the 1.2-kb DNA insert from pJS75 (55) revealed a 945-bp open reading frame preceded by a ribosome binding site (AGGAG, nucleotides 72 to 76) (Fig. 2). The open reading frame (dntD) encoded a 35-kDa polypeptide composed of 315 amino acids, with a calculated isoelectric point of 5.53. Analysis of the upstream sequence and comparisons with upstream regions from related genes revealed a potential promoter region at positions 22 to 51. The region resembled the canonical sequence motif (TTGACA-17±1-TATAAT) required for recognition by ς70 RNA polymerase holoenzyme (12).
FIG. 2.
Nucleotide sequence of the 1.2-kb DNA insert of pJS76 and deduced amino acid sequence of DntD. The putative promoter regions are underlined twice. The Shine-Dalgarno sequence is underlined.
Comparisons of the deduced amino acid sequence of DntD to sequences stored in the available databases revealed that DntD was similar to previously described extradiol ring cleavage dioxygenases (Fig. 3) used in pathways for the degradation of many aromatic compounds. The most closely related gene products had only 40% identity with DntD. The multiple alignment of THT oxygenase and other ring cleavage enzymes summarizes the sequence similarity among the class I dioxygenases (Fig. 4). Like the other meta-ring cleavage enzymes, the THT oxygenase peptide retained the residues which form the active site (His-154, His-220, and Glu-271) and coordinate the iron cofactors (His-206, His-252, and Tyr-261) (14). In their previous study of extradiol dioxygenases, Eltis and Bolin (14) showed the strict conservation of glycine, leucine, and proline residues which were located along the interface of the N- and C-terminal domains of the protein. For DntD, as well as the most recently described BphC proteins (59), only the Gly-27 and Pro-265 residues are conserved; at position 173, phenylalanine was substituted for the leucine residue.
FIG. 3.
Phylogenetic tree showing the relationships among THT oxygenase and other extradiol ring cleavage enzymes. The scale at the lower right corresponds to a genetic distance of 0.1 substitution/position (10% difference). References for the peptide sequences are as follows: CatA from Rhodococcus rhodochrous, 34; BphC from strain BN6, 27; CmtC from P. putida, 13; BphC from P. stutzeri, 59; BphC from P. mendocina, 59; BphCII from strain BN6, 26; MpcII from Ralstonia eutrophus, 29; CdoA from R. rhodochrous, 5; PheB from Bacillus stearothermophilus, 25; TdnC from P. putida, 39; C23OII from TOL plasmid, 30; XylE from Sphingomonas yanoikuae, 31; CmpE 63; NahH from PpG7, 17; BphE, 6; XylEJI104-1 from P. aeruginosa JI104, 32; XylEJI104-2 from P. aeruginosa JI104, 32; XylETOL from P. aeruginosa JI104, 33; C23O from P. putida KF715, 38; XylE from TOL plasmid, 42; XylE from Bacillus subtilis, 43; XylEII from TOL plasmid, 4; PhlH from P. putida H, 28; C23O from P. putida, 44; C23O from Alcaligenes sp. strain KF711, 40; DmpB from P. putida CF600, 3; and AtdA, 60.
FIG. 4.
Multiple alignment of the amino acid sequences of DntD and selected extradiol ring cleavage enzymes. Alignment of all of the amino acid sequences for the gene products shown in Fig. 3 was done as described in Materials and Methods. From that multiple alignment, individual sequences were selected to show relationships between DntD, its most similar counterparts (BphC from P. stuzeri and P. mendocina), and individual sequences from distant branches of the phylogenetic tree. Amino acid residues conserved in all proteins are shown by the black boxes. The amino acid sequence positions are indicated by the numbers at right; dashes indicate gaps introduced in the original alignment of the 28 peptide sequences. Gene product designations are as in Fig. 3. XylE corresponds to the product of the catechol 2,3-dioxygenase gene from the P. putida mt-2 TOL plasmid (42).
Construction of an E. coli strain for overexpression of THT oxygenase.
To facilitate purification of the THT oxygenase, the 1.2-kb EcoRI-XhoI restriction fragment from pJS76 (55) containing dntD was subcloned into the expression vector pT7-5 to yield pJS77. Subsequently, the plasmid was transferred to E. coli JM109(DE3). The initial induction trials carried out at 37°C led to formation of inclusion bodies that could not be solubilized to obtain THT oxygenase activity. Induction done at 25°C (48) allowed recovery of adequate quantities of soluble THT oxygenase for subsequent purification of the enzyme.
Purification and properties of THT oxygenase.
A homogenous preparation of the THT oxygenase was obtained by using the series of chromatographic methods summarized in Table 1. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis of the enzyme preparation revealed the presence of a single protein band with an estimated molecular weight of 38,000. The native molecular weight was estimated to be 67,400 (by LC) and 79,100 (by HPLC) with the two gel filtration columns and to be 77,800 by using nondenaturing gel electrophoresis. The results suggested that the native enzyme exists as a dimer. The protein was colorless, and the results from iron content assays indicated 2.2 g-atom of iron per mol of protein. The addition of exogenous iron alone did not stimulate activity; however, when ascorbate was also present, the activity increased by 1.8-fold. Incubation of the enzyme with H2O2 or potassium ferricyanide (1 mM) for 1 h resulted in loss of 4MC transformation activity (>95%). Heat treatment of the enzyme (5 min at 50°C) also led to loss of enzyme activity (63%).
TABLE 1.
Purification of THT oxygenase
| Purification step | Protein (mg) | Activity (U)a | Sp act (U/mg) | Recovery (%) | Purification (fold) |
|---|---|---|---|---|---|
| Crude cell extract | 572 | 28 | 0.05 | 100 | 1 |
| Ammonium sulfate | 502 | 26 | 0.05 | 93 | 1 |
| Phenyl-Sepharose | 128 | 20 | 0.16 | 70 | 3 |
| Q-Sepharose | 41 | 15 | 0.37 | 54 | 7 |
| Sephacryl S-300 | 27 | 15 | 0.56 | 54 | 11 |
| DEAE-Sepharose | 24 | 15 | 0.63 | 54 | 13 |
Activity was determined spectrophotometrically by monitoring the appearance of the ring cleavage product of 4-MC at 382 nm. One unit of activity is defined as 1 μmol of product formed per min in the presence of THT oxygenase.
The substrate range of the THT oxygenase was determined by measuring the oxygen consumption stimulated by various aromatic compounds (Table 2). THT oxygenase exhibited low activity with 1,2,4-benzenetriol and 4-MC. Traces of activity toward catechol, 3-MC, and 3- and 4-chlorocatechol were also detected. The spectra and calculated extinction coefficients of the yellow products formed from 1,2,4-benzenetriol, catechol, and 4-MC were consistent with those of the respective hydroxymuconic semialdehydes. No metabolites that corresponded to intradiol cleavage products were detected.
TABLE 2.
Substrate specificity of THT oxygenase
| Substrate | Relative activitya (%) | λmax of product |
|---|---|---|
| THT | 100 | 280, 320–380 |
| 1,2,4-Benzenetriol | 17 | 280, 320–380 |
| Catechol | 1 | 375 |
| 3-MC | 0.5 | 318, 340–420 |
| 4-MC | 3 | 325, 382 |
| 3-Chlorocatechol | 0.7 | 367 |
| 4-Chlorocatechol | 0.8 | 377 |
| 4,5-Dichlorocatechol | NDb | |
| 4-Nitrocatechol | ND | |
| MNC | ND | |
| Protocatechuate | ND |
Oxygen consumption in the presence of 5 × 10−5 M substrate was measured, with the activity for THT (8.633 μmol of oxygen consumed/min/mg of protein) taken as 100% activity.
ND, none detected.
Identification of the THT reaction product.
Preliminary attempts to identify the product of the THT reaction provided misleading results (19). The compound obtained by extraction of the reaction medium with ethyl acetate provided an apparent molecular ion of 154. The proton NMR spectrum of the extracted product gave three sets of peaks at 1.81, 6.81, and 8.23 ppm in a ratio of 3:1:1. The 13C NMR yielded seven resonances at 10.35, 116.6, 126.0, 152.8, 153.8, 161, and 178.5 ppm and indicated that the chemical shifts of the carbonyl resonance are in the general region for acids, esters, and lactones (45a). The chemical shifts are consistent with those of a 3-methyldienelactone; however, the spectra were different from those reported by Prucha et al. for 3-methyl-trans-dienelactone and 3-methyl-cis-dienelactone (46). The extracted compound was not transformed by cell extracts or intact cells of strain DNT. The lack of further transformation suggested that the compound was not an intermediate of the 2,4-DNT pathway but an artifact of the extraction procedure.
A second approach was used to identify the products formed by the ring fission reaction. THT oxygenase was incubated with THT, as well as the structural analogs 1,2,4-benzenetriol and 4-MC. Following incubation, the reaction mixtures were analyzed directly by using LC-MS. The analyses revealed molecular ion peaks at m/z 172 (THT), 158 (1,2,4-benzenetriol), and 156 (4MC). The mass-to-charge ratios of the molecular ion peaks corresponded to the respective hydroxymuconic semialdehydes identified in the spectrophotometric analyses of reaction products. Authentic 3-methylmaleylacetate, which would be produced by intradiol ring fission of THT, gave a molecular ion peak at m/z 128 (172 − 44 due to decarboxylation during chemical ionization) (46). Cell extracts from strain DNT transformed the ring cleavage product to pyruvate with 1:1 stoichiometry. The subsequent transformation of the 2,4-dihydroxy-5-methyl-6-oxo-2,4-hexadienoic acid indicated that the compound was an intermediate of the DNT pathway and not an experimental artifact like 3-methyldienelactone.
DISCUSSION
The results consistently show that the THT oxygenase is structurally and functionally related to the extradiol dioxygenase gene family I. However, comparisons of the THT oxygenase amino acid sequence to those of previously studied enzymes indicate a distant phylogenetic relationship between THT oxygenase and the other members of the family (Fig. 3). The deep branching seen in the phylogenetic tree indicates a distant origin for the gene family and specialization for the THT oxygenase to accommodate the trihydroxylated substrate.
The bphC genes isolated from Pseudomonas stutzeri and Pseudomonas mendocina have the highest degree of overall identity with dntD. Although the bphC gene products were identified by their ability to catalyze the oxidation of 2,3-dihydroxybiphenyl (59), the bacteria were isolated for the ability to grow on benzoate and could not grow on biphenyl. The enzymes have a high Km toward 2,3-dihydroxybiphenyl, and their physiological substrates are unknown. The next most closely related sequence is that of another homodimeric extradiol dioxygenase involved in the degradation of naphthalenesulfonate (BphCII) by strain BN6 (26). The THT oxygenase from strain DNT and a second ring cleavage enzyme (BphC) from strain BN6 share the uncommon ability to catalyze the extradiol cleavage of 3-chlorocatechol (27). Further functional comparisons between THT oxygenase and its phylogenetic neighbors could reveal characteristics that point to a common ancestor.
The putative promoter found upstream from dntD is consistent with the previous suggestion that the expression of THT oxygenase is controlled by a promoter within the 1.2-kb EcoRI-XhoI fragment of pJS76 (55). However, no expression studies have been completed to confirm the transcription start site or determine whether the region is relevant to expression of dntD.
The properties of the purified THT oxygenase protein were typical of those for previously studied extradiol dioxygenases. The lack of color indicates an absence of flavin or porphyrin cofactors. Most of the enzymes of the catechol 2,3-dioxygenase family consist of four or eight identical subunits (22, 35, 41, 58). THT oxygenase is among the small number of homodimeric extradiol dioxygenases (26, 49). However, the size of the THT oxygenase subunit is similar to those of the other catechol 2,3-dioxygenases. THT oxygenase contains 2 g-atom of Fe(II) per mol of enzyme, consistent with the proposed dimeric configuration and the sequence analysis, which showed one iron binding site per subunit. Metal cofactors play an integral role in ring cleavage mechanisms. The intradiol dioxygenases contain nonheme Fe(III) within the enzyme active site. Most extradiol dioxygenases described to date use Fe(II) to coordinate binding of the activated oxygen molecule used in catalysis (24). An exception is the dihydroxyphenylacetate 2,3-dioxygenase from Arthrobacter globiformis CM-2, which uses manganese as its metal cofactor (62).
Although THT oxygenase catalyzes a meta-cleavage reaction, its ability to transform 1,2,4-benzenetriol and the structural similarities of THT to 1,2,4-benzenetriol suggested that the THT oxygenase might also have sequence similarity with the benzenetriol intradiol dioxygenases. A number of benzenetriol oxygenases have been purified and characterized (11, 37, 56, 64). The gene sequence of the benzenetriol oxygenase from B. cepacia AC1100 (10, 11) and the amino-terminal sequences of the benzenetriol oxygenases from Streptomyces rochei and Azotobacter strain GP-1 have been determined (37, 64). The enzymes catalyze intradiol cleavage and form maleylacetate from 1,2,4-benzenetriol. The amino-terminal sequences of the benzenetriol oxygenases from these strains have 33 to 57% identity (11, 64). The deduced amino-terminal sequence of THT oxygenase had no significant identity to any of the benzenetriol oxygenases (<15%). In addition, THT oxygenase did not catalyze the conversion of 1,2,4-benzenetriol to maleylacetate. Thus, the overlap of substrates for the benzenetriol and THT oxygenases was not reflected in the amino acid sequences of the enzymes.
Our results indicate that THT oxygenase cleaves THT by a mechanism analogous to the extradiol cleavage of 1,2,4-benzenetriol and 2,3,5-trihydroxytoluene by the 2,3,5-trihydroxytoluene 1,2-dioxygenase present in Pseudomonas putida ORC and P. putida O1OC (8, 9). Because of the similarities in the reactions catalyzed by THT oxygenase and 2,3,5-trihydroxytoluene 1,2-oxygenase, it would be interesting to compare the amino acid sequences of these two enzymes. However, the sequence for 2,3,5-trihydroxytoluene 1,2-oxygenase is not available.
In previous work, gas chromatography-MS analysis of the THT ring cleavage provided a compound that corresponded to the lactonized 2,4-dihydroxy-5-methyl-6-oxo-2,4-hexadienoic acid (19). However, at that time, the possibility that the isolated product was a 3-methyldienelactone could not be ruled out. The 3-methyldienelactone could result from acid-catalyzed dehydration of 3-hydroxy-4-methylmuconate, the product resulting from intradiol cleavage of THT. Recent evidence indicates that the 3-hydroxy-4-methylmuconate does not undergo dehydration but instead undergoes tautomerization to form 4-hydroxy-3-methylmuconolactone and/or 3-methylmaleylacetate (46). The mass spectra of the 3-methyl-cis- and the 3-methyl-trans-dienelactones (46) are different from that of the product formed from the THT ring fission product after acid extraction (19). In addition, the compound formed from THT prior to acid extraction is clearly not 3-methylmaleylacetate (46). The mass spectral results, coupled with the stoichiometric production of pyruvate from THT in reactions with cell extracts from strain DNT, provide strong evidence that ring fission occurs between positions 5 and 6 of the aromatic ring to yield 2,4-dihydroxy-5-methyl-6-oxo-2,4-hexadienoic acid (Fig. 1). We are currently working to determine the subsequent metabolism of the ring cleavage product.
ACKNOWLEDGMENTS
We thank R. J. Spanggord for providing THT, D. Pieper for kind gifts of 3-methyl-cis- and 3-methyl-trans-dienelactones, Mike Henley for gas chromatography and LC-MS spectral determinations, Stephen A. Aubert and Sol Resnick for NMR spectral determinations, and G. J. Zylstra for helpful discussions.
This research was supported in part by an appointment to the Research Participation Program at the Air Force Engineering and Service Laboratory administered by Oak Ridge Associated Universities through an interagency agreement between the U.S. Department of Energy and Tyndall Air Force Base. Funding was provided by the Air Force Office of Scientific Research and the Strategic Environmental Defense Program.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons, Inc.; 1998. [Google Scholar]
- 3.Bartilson M, Shingler V. Nucleotide sequence and expression of the catechol 2,3-dioxygenase-encoding gene of phenol-catabolizing Pseudomonas CF600. Gene. 1989;85:233–238. doi: 10.1016/0378-1119(89)90487-3. [DOI] [PubMed] [Google Scholar]
- 4.Benjamin R C, Voss J A, Kunz D A. Nucleotide sequence of xylE from the TOL pDK1 plasmid and structural comparison with isofunctional catechol-2,3-dioxygenase genes from TOL, pWW0, and NAH7. J Bacteriol. 1991;173:2724–2728. doi: 10.1128/jb.173.8.2724-2728.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Candidus S, van Pee K H, Lingens F. The catechol 2,3-dioxygenase gene of Rhodococcus rhodochrous CTM: nucleotide sequence, comparison with isofunctional dioxygenases and evidence for an active-site histidine. Microbiology. 1994;140:321–330. doi: 10.1099/13500872-140-2-321. [DOI] [PubMed] [Google Scholar]
- 6.Carrington B, Lowe A, Shaw L E, Williams P A. The lower pathway operon for benzoate catabolism in biphenyl-utilizing Pseudomonas sp. strain IC and the nucleotide sequence of the bphE gene for catechol 2,3-dioxygenase. Microbiology. 1994;140:499–508. doi: 10.1099/00221287-140-3-499. [DOI] [PubMed] [Google Scholar]
- 7.Chapman P J. Microbial degradation of halogenated compounds. Biochem Soc Trans. 1976;4:463–476. [PubMed] [Google Scholar]
- 8.Chapman P J, Ribbons D W. Metabolism of resorcinylic compounds by bacteria: alternative pathways for resorcinol catabolism in Pseudomonas putida. J Bacteriol. 1976;125:985–998. doi: 10.1128/jb.125.3.985-998.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chapman P J, Ribbons D W. Metabolism of resorcinylic compounds by bacteria: orcinol pathway in Pseudomonas putida. J Bacteriol. 1976;125:975–984. doi: 10.1128/jb.125.3.975-984.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daubaras D L, Danganan C E, Anette H, Ye R W, Hendrickson W, Chakrabarty A M. Biodegradation of 2,4,5-trichlorophenoxyacetic acid by Burkholderia cepacia strain AC1100: evolutionary insight. Gene. 1996;179:1–8. doi: 10.1016/s0378-1119(96)00326-5. [DOI] [PubMed] [Google Scholar]
- 11.Daubaras D L, Saido K, Chakrabarty A M. Purification of hydroxyquinol 1,2-dioxygenase and maleylacetate reductase: the lower pathway of 2,4,5-trichlorophenoxyacetic acid metabolism by Burkholderia cepacia AC1100. Appl Environ Microbiol. 1996;62:4276–4279. doi: 10.1128/aem.62.11.4276-4279.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deretic V, Konyecsni W M, Mohr C D, Martin D W, Hibler N S. Common denominators of promoter control in Pseudomonas and other bacteria. Bio/Technology. 1989;7:1249–1254. [Google Scholar]
- 13.Eaton R W. p-Cumate catabolic pathway in Pseudomonas putida F1: cloning and characterization of DNA carrying the cmt operon. J Bacteriol. 1996;178:1351–1362. doi: 10.1128/jb.178.5.1351-1362.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eltis L D, Bolin J T. Evolutionary relationships among extradiol dioxygenases. J Bacteriol. 1996;178:5930–5937. doi: 10.1128/jb.178.20.5930-5937.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feng D-F, Doolittle R F. Progressive sequence alignment as a prerequisite to correct phylogenetic trees. J Mol Evol. 1987;25:351–361. doi: 10.1007/BF02603120. [DOI] [PubMed] [Google Scholar]
- 16.Friedemann T E, Haugen G E. Pyruvic acid. II. The determination of keto acids in blood and urine. J Biol Chem. 1942;147:415–442. [Google Scholar]
- 17.Ghosal D, You I S, Gunsalus I C. Nucleotide sequence and expression of gene nahH of plasmid NAH7 and homology with gene xylE of TOL pWWO. Gene. 1987;55:19–28. doi: 10.1016/0378-1119(87)90244-7. [DOI] [PubMed] [Google Scholar]
- 18.Haigler B E, Gibson D T. Purification and properties of NADH-ferredoxinNAP reductase, a component of naphthalene dioxygenase from Pseudomonas sp. strain NCIB 9816. J Bacteriol. 1990;172:457–464. doi: 10.1128/jb.172.1.457-464.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haigler B E, Nishino S F, Spain J C. Biodegradation of 4-methyl-5-nitrocatechol by Pseudomonas sp. strain DNT. J Bacteriol. 1994;176:3433–3437. doi: 10.1128/jb.176.11.3433-3437.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haigler B H, Suen W-C, Spain J C. Purification and sequence analysis of 4-methyl-5-nitrocatechol oxygenase from Burkholderia sp. strain DNT. J Bacteriol. 1996;178:6019–6024. doi: 10.1128/jb.178.20.6019-6024.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han S, Eltis L D, Timmis K N, Muchmore S W, Bolin J T. Crystal structure of the biphenyl-cleaving extradiol dioxygenase from a PCB-degrading pseudomonad. Science. 1995;270:976–980. doi: 10.1126/science.270.5238.976. [DOI] [PubMed] [Google Scholar]
- 22.Harayama S, Kok M, Neidle E L. Functional and evolutionary relationships among diverse oxygenases. Annu Rev Microbiol. 1992;46:565–601. doi: 10.1146/annurev.mi.46.100192.003025. [DOI] [PubMed] [Google Scholar]
- 23.Harayama S, Rekik M. Bacterial aromatic ring-cleavage enzymes are classified into two different gene families. J Biol Chem. 1989;264:15328–15333. [PubMed] [Google Scholar]
- 24.Harayama S, Rekik M, Wasserfallen A, Bairoch A. Evolutionary relationships between catabolic pathways for aromatics: conservation of gene order and nucleotide sequences of catechol oxidation genes of pWW0 and NAH7 plasmids. Mol Gen Genet. 1987;210:241–247. doi: 10.1007/BF00325689. [DOI] [PubMed] [Google Scholar]
- 25.He, Z. Q., Y. M. Mao, Z. J. Sheng, and R. Q. Shen. Swiss protein database, accession no. P31003.
- 26.Heiss G, Muller C, Altenbuchner J, Stolz A. Analysis of a new dimeric extradiol dioxygenase from a naphthalenesulfonate-degrading sphingomonad. Microbiology. 1997;143:1691–1699. doi: 10.1099/00221287-143-5-1691. [DOI] [PubMed] [Google Scholar]
- 27.Heiss G, Stolz A, Kuhm A E, Muller C, Klein J, Altenbuchner J, Knackmuss H J. Characterization of a 2,3-dihydroxybiphenyl dioxygenase from the naphthalenesulfonate-degrading bacterium strain BN6. J Bacteriol. 1995;177:5865–5871. doi: 10.1128/jb.177.20.5865-5871.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herrmann H, Muller C, Schmidt I, Mahnke J, Petruschka L, Hahnke K. Localization and organization of phenol degradation genes of Pseudomonas putida strain H. Mol Gen Genet. 1995;247:240–246. doi: 10.1007/BF00705655. [DOI] [PubMed] [Google Scholar]
- 29.Kabisch M, Fortnagel P. Nucleotide sequence of the metapyrocatechase II (catechol 2,3-oxygenase II) gene mpcII from Alcaligenes eutrophus JMP 222. Nucleic Acids Res. 1990;18:5543. doi: 10.1093/nar/18.18.5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keil H, Lebens M R, Williams P A. TOL plasmid pWW15 contains two nonhomologous, independently regulated catechol 2,3-dioxygenase genes. J Bacteriol. 1985;163:248–255. doi: 10.1128/jb.163.1.248-255.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim E, Zylstra G J. Molecular and biochemical characterization of two meta-cleavage dioxygenases involved in biphenyl and m-xylene degradation by Beijerinckia sp. strain B1. J Bacteriol. 1995;177:3095–3103. doi: 10.1128/jb.177.11.3095-3103.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kitayama A, Achioku T, Yanagawa T, Kanou K, Kikuchi M, Ueda H, Suzuki E, Nishimura H, Nagamune T, Kawakami Y. Cloning and characterization of extradiol aromatic ring-cleavage dioxygenases of Pseudomonas aeruginosa JI104. J Ferment Bioeng. 1996;82:217–223. [Google Scholar]
- 33.Kitayama, A. Swiss protein database, accession no. .
- 34.Ksenzenko, V. N., A. N. Kulakova, M. J. Larkin, and L. A. Kulakov. GenBank, accession no. L77225.
- 35.Kuhm A E, Stolz A, Ngai K-L, Knackmuss H-J. Purification and characterization of a 1,2-dihydroxynaphthalene dioxygenase from a bacterium that degrades naphthalenesulfonic acids. J Bacteriol. 1991;173:3795–3802. doi: 10.1128/jb.173.12.3795-3802.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kukor J J, Olsen R H. Catechol 2,3-dioxygenases functional in oxygen-limited (hypoxic) environments. Appl Environ Microbiol. 1996;62:1728–1740. doi: 10.1128/aem.62.5.1728-1740.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Latus M, Seitz H-J, Eberspacher J, Lingens F. Purification and characterization of hydroxyquinol 1,2-dioxygenase from Azotobacter sp. strain GP-1. Appl Environ Microbiol. 1995;61:2453–2460. doi: 10.1128/aem.61.7.2453-2460.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee J, Oh J, Min K R, Kim C K, Min K H, Lee K S, Kim Y C, Lim J Y, Kim Y. Structure of catechol 2,3-dioxygenase gene encoded in chromosomal DNA of Pseudomonas putida KF715. Biochem Biophys Res Commun. 1996;224:831–836. doi: 10.1006/bbrc.1996.1108. [DOI] [PubMed] [Google Scholar]
- 39.McClure, N. C., C. P. Saint, and A. S. Weightman. EMBL database, accession no. S31402.
- 40.Moon J, Chang H, Min K R, Kim Y. Cloning and sequencing of the catechol 2,3-dioxygenase gene of Alcaligenes sp. KF711. Biochem Biophys Res Commun. 1995;208:943–949. doi: 10.1006/bbrc.1995.1425. [DOI] [PubMed] [Google Scholar]
- 41.Nakai C, Hori K, Kagamiyama H, Nakazawa T, Nozaki M. Purification, subunit structure, and partial amino acid sequence of metapyrocatechase. J Biol Chem. 1983;258:2916–2922. [PubMed] [Google Scholar]
- 42.Nakai C, Kagamiyama H, Nozaki M, Nakazawa T, Inouye S, Ebina Y, Nakazawa A. Complete nucleotide sequence of the metapyrocatechase gene on the TOL plasmid of Pseudomonas putida mt-2. J Biol Chem. 1983;258:2923–2928. [PubMed] [Google Scholar]
- 43.Ng, L. C., C. C. Sze, and C. L. Poh. GenBank, accession no. A14852.
- 44.Ng, L. C., C. C. Sze, and C. L. Poh. GenBank, accession no. G540972.
- 45.Page R D M. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 45a.Pretsch E, Clerc T, Seibel J, Simon W. Tables of spectral data for structure determinations of organic compounds. Berlin, Germany: Springer-Verlag; 1989. [Google Scholar]
- 46.Prucha M, Wray V, Pieper D H. Metabolism of 5-chlorosubstituted muconolactones. Eur J Biochem. 1996;237:357–366. doi: 10.1111/j.1432-1033.1996.00357.x. [DOI] [PubMed] [Google Scholar]
- 47.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 48.Schein C, Noteborn M. Formation of soluble recombinant proteins in Escherichia coli is favored by lower growth temperature. Bio/Technology. 1988;4:291–294. [Google Scholar]
- 49.Schmid A, Rothe B, Altenbuchner J, Ludwid W, Engesser K-H. Characterization of three distinct extradiol dioxygenases involved in mineralization of dibenzofuran by Terrabacter sp. strain DPO360. J Bacteriol. 1997;179:53–62. doi: 10.1128/jb.179.1.53-62.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Senda T, Sugiyama H, Narita H, Yamamoto T, Kimbara K, Fukuda M, Sato M, Yano K, Mitsui Y. Three dimensional structure of free form and two substrate complexes of an extradiol ring-cleavage type dioxygenase, the BphC enzyme from Pseudomonas sp. strain KKS102. J Mol Biol. 1996;255:735–752. doi: 10.1006/jmbi.1996.0060. [DOI] [PubMed] [Google Scholar]
- 51.Spain J C, Nishino S F. Degradation of 1,4-dichlorobenzene by a Pseudomonas sp. Appl Environ Microbiol. 1987;53:1010–1019. doi: 10.1128/aem.53.5.1010-1019.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Spanggord R J, Spain J C, Nishino S F, Mortelmans K E. Biodegradation of 2,4-dinitrotoluene by a Pseudomonas sp. Appl Environ Microbiol. 1991;57:3200–3205. doi: 10.1128/aem.57.11.3200-3205.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Suen W-C, Haigler B E, Spain J C. Abstracts of the IV International Congress on Pseudomonas. 1993. Evidence that 2,4,5-trihydroxytoluene oxygenase is a meta-ring cleavage enzyme; p. 179. [Google Scholar]
- 54.Suen W C, Haigler B E, Spain J C. 2,4-Dinitrotoluene dioxygenase from Burkholderia sp. strain DNT: similarity to naphthalene dioxygenase. J Bacteriol. 1996;178:4926–4934. doi: 10.1128/jb.178.16.4926-4934.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Suen W C, Spain J C. Cloning and characterization of Pseudomonas sp. strain DNT genes for 2,4-dinitrotoluene degradation. J Bacteriol. 1993;175:1831–1837. doi: 10.1128/jb.175.6.1831-1837.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sze I S-Y, Dagley S. Properties of salicylate hydroxylase and hydroxyquinol 1,2-dioxygenase purified from Trichosporon cutaneum. J Bacteriol. 1984;159:353–359. doi: 10.1128/jb.159.1.353-359.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tabor S, Richardson C C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci USA. 1985;82:1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Taira K, Hayase N, Arimura N, Yamashita S, Miyazaki T, Furukawa K. Cloning and nucleotide sequence of the 2,3-dihydroxybiphenyl dioxygenase from the PCB-degrading strain of Pseudomonas paucimobilis Q1. Biochemistry. 1988;27:3990–3996. doi: 10.1021/bi00411a015. [DOI] [PubMed] [Google Scholar]
- 59.Takami H, Kudo T, Horikoshi K. Isolation of extradiol dioxygenase genes that are phylogenetically distant from other meta-cleavage dioxygenase genes. Biosci Biotechnol Biochem. 1997;61:530–532. doi: 10.1271/bbb.61.530. [DOI] [PubMed] [Google Scholar]
- 60.Takeo, M. GenBank, accession no. D86080.
- 61.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Whiting A K, Boldt Y R, Hendrich M P, Wackett L P, Que L., Jr Manganese (II)-dependent extradiol-cleaving catechol dioxygenase from Arthrobacter globiformis CM-2. Biochemistry. 1996;35:160–170. doi: 10.1021/bi951979h. [DOI] [PubMed] [Google Scholar]
- 63.Yrjala K, Paulin L, Kilpi S, Romantschuk M. Cloning of cmpE, a plasmid-borne catechol 2,3-dioxygenase-encoding gene from the aromatic- and chloroaromatic-degrading Pseudomonas sp. HV3. Gene. 1994;138:119–121. doi: 10.1016/0378-1119(94)90792-7. [DOI] [PubMed] [Google Scholar]
- 64.Zaborina O, Latus M, Eberspächer J, Golovleva L A, Lingens F. Purification and characterization of 6-chlorohydroxyquinol 1,2-dioxygenase from Streptomyces rochei 303: comparison with an analogous enzyme from Azotobacter sp. strain GP1. J Bacteriol. 1995;177:229–234. doi: 10.1128/jb.177.1.229-234.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]