Abstract
Pathogenicity islands are chromosomal clusters of pathogen-specific virulence genes often found at tRNA loci. We have determined the molecular genetic structure of SPI-3, a 17-kb pathogenicity island located at the selC tRNA locus of Salmonella enterica serovar Typhimurium. The G+C content of SPI-3 (47.5%) differs from that of the Salmonella genome (52%), consistent with the notion that these sequences have been horizontally acquired. SPI-3 harbors 10 open reading frames organized in six transcriptional units, which include the previously described mgtCB operon encoding the macrophage survival protein MgtC and the Mg2+ transporter MgtB. Among the newly identified open reading frames, one exhibits sequence similarity to the ToxR regulatory protein of Vibrio cholerae and one is similar to the AIDA-I adhesin of enteropathogenic Escherichia coli. The distribution of SPI-3 sequences varies among the salmonellae: the right end of the island, which harbors the virulence gene mgtC, is present in all eight subspecies of Salmonella; however, a four-gene cluster at the center of SPI-3 is found in only some of the subspecies and is bracketed by remnants of insertion sequences, suggesting a multistep process in the evolution of SPI-3 sequences.
The gram-negative bacterium Salmonella enterica is responsible for a variety of diseases, which include gastroenteritis and typhoid fever, depending on the nature of the infected host and on the serovar of the infecting bacteria. Salmonella has a complex life cycle in infected animals, and a large number of genes have been implicated in Salmonella virulence. Several of these virulence determinants are clustered within pathogenicity islands, i.e., large segments of horizontally acquired sequences present in pathogenic species but absent from closely related nonpathogenic species (17, 24). Pathogenicity islands constitute major elements in the evolution of bacterial pathogens, because their incorporation can, in a single step, transform a normally benign organism into a pathogen.
In addition to several small pathogenicity islets, five large pathogenicity islands have been identified in Salmonella (21, 51, 52). SPI-1, at 63 min on the S. enterica serovar Typhimurium chromosome, is a 40-kb island that governs the ability to invade epithelial cells (10, 38) and is required for Salmonella-induced macrophage apoptosis (9). The SPI-2 island is also 40 kb in length, maps downstream of a tRNAVal locus at 31 min (25), and harbors genes required for intramacrophage survival and systemic infection (40, 44). The SPI-3 island is located at 82 min, immediately behind selC, a tRNA locus that is the insertion site for distinct pathogenicity islands in enteropathogenic and uropathogenic strains of E. coli (3, 5, 33). Recently, a 27-kb Salmonella-specific DNA fragment at 92 min was designated the fourth Salmonella pathogenicity island because it includes a macrophage survival locus (34). A fifth pathogenicity island, containing genes mediating Salmonella enteropathogenesis, is located downstream of a tRNASer locus at 20 min in the chromosome (52).
The SPI-3 island harbors mgtC, a Salmonella-specific gene that is required for intramacrophage survival, virulence in mice, and growth in low-Mg2+ media (3). The mgtC gene is transcriptionally controlled by the PhoP-PhoQ regulatory system, which governs the adaptation to low-Mg2+ environments (15, 46) and is the major regulator of virulence functions in Salmonella (14, 18). The mgtC gene is cotranscribed with mgtB (45), a Mg2+ transporter gene dispensable for virulence in BALB/c mice (3). SPI-3 is 17 kb long and may contain additional genes that contribute to virulence or to other Salmonella-specific attributes.
In this study, we determined the molecular genetic structure of the SPI-3 island, examined the functions of the genes it carries, and investigated the distribution of SPI-3 sequences among salmonellae. We establish that at least 10 genes are encoded within SPI-3, some of which show similarity to known virulence factors from other bacterial species, and that the evolution of SPI-3 sequences occurred through a multistep process.
MATERIALS AND METHODS
Bacterial strains, plasmids, bacterial genetic techniques, and growth conditions.
Strains used in this study are listed in Table 1. The strains used in this study are derived from 14028s, except for TT10288 and AA3007, which are derived from LT2. Bacteria were grown at 37°C in Luria-Bertani broth (LB) (35). Ampicillin and kanamycin were used at 50 μg/ml, and chloramphenicol was used at 10 μg/ml. Phage P22-mediated transduction was carried out as described previously (12).
TABLE 1.
Salmonella strains used in this study
| Strain | Description | Reference or source |
|---|---|---|
| 14028s | Wild type | American Type Culture Collection |
| TT10288 | hisD9953::MudJ hisA9944::MudI | 20 |
| AA3007 | polA2 ara-9 | 21 |
| MS7953s | phoP7953::Tn10 | 22 |
| EG9527 | mgtCB9232::MudJ | 15 |
| EG9529 | mgtCB9232::MudJ phoP7953::Tn10 | 15 |
| EG10207 | marT1::cat | This work |
| EG10209 | mgtCB9232::MudJ marT1::cat | This work |
| EG10277 | sugR1::MudJ | This work |
| EG10278 | sugR1::MudJ marT1::cat | This work |
| EG10349 | sugR2::MudJ | This work |
| EG10389 | sugR1::MudJ phoP7953::Tn10 | This work |
| EG10755 | misL1::MudJ | This work |
| EG10756 | misL1::MudJ marT1::cat | This work |
| EG10757 | misL1::MudJ phoP7953::Tn10 | This work |
| EG10759 | rhuM1::MudJ | This work |
| EG10760 | rhuM1::MudJ marT1::cat | This work |
| EG10913 | rhuM1::MudJ phoP7953::Tn10 | This work |
| EG11086 | marT2::MudJ | This work |
| EG11087 | marT2::MudJ phoP7953::Tn10 | This work |
Construction of the marT1::cat strain EG10207 was performed as follows. A 2.3-kb HindIII-BspMII fragment from plasmid pEG9106 (3) carrying marT was subcloned into pUC19 between the HindIII and XmaI sites to form plasmid pEG9109. A cat-containing 0.8-kb BamHI fragment from plasmid pKRP10 (43) was introduced into the unique BglII site in marT in plasmid pEG9109. The resulting plasmid (pEG9110) was used to transfer the marT1::cat mutation into the Salmonella chromosome as described previously (23). The structure of the marT gene in the mutant strain was verified by Southern hybridization with both marT- and cat-specific probes (data not shown).
MudJ is a derivative of bacteriophage Mu that harbors a gene conferring resistance to kanamycin and a segment of the lac operon devoid of its promoter sequences (7). To isolate MudJ insertions in SPI-3, a P22 lysate grown in TT10288 was used to infect strain EG10207. A lysate grown on a pool of 25,000 kanamycin-resistant transductants was used to infect 14028s, with selection for both kanamycin and chloramphenicol resistance. To establish the orientation and approximate position of each MudJ insertion, PCR was performed with primers complementary to the ends of MudJ (i.e., attL or attR) and to known SPI-3 sequences.
Molecular biological techniques.
The nucleotide sequence of the 12-kb segment between the selC and mgtB genes was determined on both strands by using plasmid pEG9106 DNA as the template, starting with primers complementary to selC (selC-F) (11) and to the 3′ end of mgtB (3′mgtB-F, 5′-ATCGTCGTGGTTTAACCGCCGTCC-3′) and walking with newly synthesized primers. DNA sequence analysis and protein sequence alignments were performed by using the GeneWorks (IntelliGenetics) and Genetics Computer Group (GCG) (University of Wisconsin) software packages.
PCRs were carried out on purified chromosomal DNA with Taq polymerase (Gibco BRL) according to the manufacturer’s protocol. For amplification of long DNA fragments, we used the TaqPlus Long PCR system (Stratagene). To examine whether SPI-3 sequences are linked to selC in different Salmonella subspecies, we used primers selC-F (11), selC-1-25 (5′-GGAAGATCGTCGTCTCCGGTGAGGC-3′), and slsA-R (5′-TTGTACAAAATCGGCATTATCCCAGGC-3′). To determine whether mgtC and orf307 are linked in different Salmonella subspecies, we used primers mgtC-R (5′-GCCCGCCCCCAGAAAGCCAATCCC-3′) and E07-R (11).
Southern hybridization analysis was carried out with chromosomal DNA as described previously (11). To investigate the distribution of the orf269 gene, a PCR-generated probe corresponding to the Escherichia coli K-12 orf269 open reading frame (ORF) was used for hybridization to DNAs from E. coli K-12, E. coli D, Shigella flexneri, S. enterica serovar Typhimurium, Citrobacter freundii, Enterobacter aerogenes, Enterobacter cloacae, Klebsiella pneumoniae, Serratia odifera, Yersinia enterocolitica, Yersinia pestis, Haemophilus influenzae, Mycobacterium avium, and Pseudomonas aeruginosa. To investigate the distribution of SPI-3 sequences among salmonellae, probes were hybridized to DNAs from strains of the Salmonella Reference Collection C (6) and from E. coli K-12 strain MC1061. Probe 1 (410 bp) was generated from a PCR DNA fragment by using primers selC-415 (5′-AGATGATGTGGCTGGCG-3′) and selC-R (11), probe 2 (780 bp) was generated by using primers described previously (3), and probes 3, 4, 6, 7, 8, and 9 were generated by using primers complementary to the 5′ and 3′ ends of the sugR, rhuM, marT, slsA, mgtB, and mgtC genes, respectively. Probe 5 was generated from a 4-kb EcoRI-HindIII restriction fragment (3).
Virulence and β-galactosidase assays.
Macrophage survival assays with the macrophage-like cell line J774 and invasion assays with canine kidney epithelial (MDCK) cells were conducted as described previously (28). Virulence assays were performed with 7- to 8-week-old female BALB/c mice (10 mice per mutant) inoculated orally with 100 μl of bacteria diluted in phosphate-buffered saline. β-Galactosidase assays were carried out in triplicate with bacteria grown exponentially in LB as described previously (35).
Nucleotide sequence accession number.
The sequence reported in this paper has been deposited in the GenBank database (accession no. AF106566).
RESULTS AND DISCUSSION
Molecular analysis of SPI-3 genes and encoded proteins.
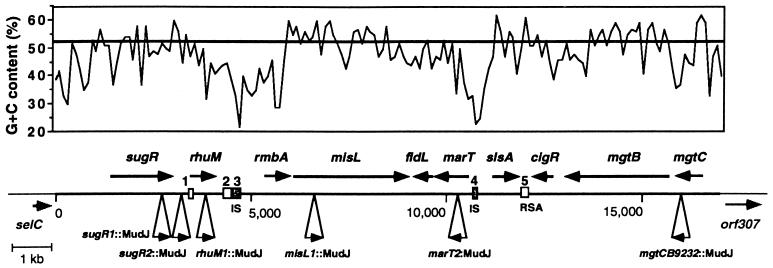
We have previously identified a pathogenicity island downstream of the selC gene in the S. enterica serovar Typhimurium chromosome (3). This island includes the mgtCB operon, which codes for the virulence protein MgtC and the Mg2+ transporter MgtB. To further examine the role of SPI-3 in Salmonella, we determined the molecular genetic structure of the DNA region between the selC and mgtB genes (Fig. 1).
FIG. 1.
Physical and genetic maps of the SPI-3 pathogenicity island. (Top) G+C content of the SPI-3 island. The graph was created by using the program Cricket Graph with data generated by the program Windows (GCG) (window, 100 bp; sliding increment, 100 bp). The line at 52% indicates the overall G+C content estimated for the S. enterica serovar Typhimurium chromosome. (Bottom) Positions and orientations of ORFs encoding products larger than 120 amino acids and containing potential Shine-Dalgarno sequences. DNA sequences reported in Table 2 are indicated by numbers (IS-like sequences are represented by gray squares). The map positions of MudJ insertions in the SPI-3 region are indicated by triangles. RSA refers to a family of protected sequences present in the genomes of members of the family Enterobacteriaceae.
The SPI-3 island is 17 kb long, and in addition to the mgtCB operon, it harbors eight ORFs, all of which contain potential Shine-Dalgarno sequences (Fig. 1). (Additional small ORFs [encoding <120 amino acids] lacking a clear translation start site and similarity to proteins in the sequence databases are not reported here). The 10 genes carried within SPI-3 appear to be organized in six transcriptional units. Characteristics of the SPI-3 ORFs, as well as features of DNA stretches that have identity with sequences in the databases, are described in Table 2. Except for the rmbA gene, which is part of region with a very low G+C content, the codon usages of SPI-3 ORFs do not appear to be significantly different from those of highly expressed E. coli and Salmonella genes.
TABLE 2.
Properties of SPI-3 DNA sequences and SPI-3-encoded proteins
| Sequence | Length | Characteristicsa |
|---|---|---|
| DNAb | ||
| 1 | 150 bp | 83% identical over 125 nucleotides to sequences downstream of E. coli selC (AE000443) |
| 2 | 190 bp | 91% identical to sequences downstream of fimU in S. typhimurium (L19338) and 87% identical to sequences upstream of insA in E. coli (L20943) |
| 3 | 200 bp | 82% identical to IS1351 of S. enteritidis (Z83734) |
| 4 | 100 bp | 95% identical to repetitive sequence 1-3 in E. cloacae (D00952); 76% identical to IS911 of S. dysenteriae (X17613) |
| 5 | 190 bp | RSA repetitive sequence (interrupted by an additional 60 bp) |
| Proteins | ||
| SugR | 519 aac | Predicted cytoplasmic protein; 44% identical over 153 aa to PgaA antigen or P. gingivalis (X95938) and 34% identical over 249 aa to putative ATP binding protein from E. coli clinical isolate (S28007) |
| RhuM | 215 aa | Predicted cytoplasmic protein |
| RmbA | 205 aa | Predicted cytoplasmic protein; 39% identical over 190 aa to 230-aa E. coli ORF product (U29581) |
| MisL | 955 aa | Probable autotransported protein; putative N-terminal signal sequence; C-terminal domain is 43% identical to C-terminal domain of E. coli AIDA-I (Q03155) and 38% identical to C-terminal domain of S. flexneri VirG (A32247) |
| FidL | 154 aa | Predicted inner membrane protein; putative N-terminal signal sequence; 44% identical over 124 aa to 164-aa E. coli ORF product (U29581) |
| MarT | 285 aa | Predicted inner membrane protein (putative transmembrane domain comprises residues 172–188); N-terminal domain is 68% identical to 99 aa of 269-aa E. coli ORF product (U29581) and 32% identical to 98 aa of V. cholerae ToxR protein (P15795); C-terminal domain is 31% identical to 116 aa of 269-aa E. coli ORF product (U29581) |
| SlsA | 226 aa | Predicted inner membrane protein (putative transmembrane domain comprises residues 114–130); 48% identical over 197 aa to E. coli ORF product (P21367) |
| CigR | 159 aa | Predicted membrane protein (putative transmembrane domain comprises residues 134–158); putative cleavable N-terminal signal sequence |
| MgtB | 908 aa | Mg2+ transporter |
| MgtC | 231 aa | Predicted membrane protein; virulence determinant |
Similarities with other sequences as revealed by BLAST searches of sequence databases (http://www.ncbi.nlm.nih.gov/BLAST). Accession numbers in the EMBL database are indicated in parentheses. Prediction of protein localization sites was performed by using the PSORT WWW server (http://psort.nibb.ac.jp/).
For DNA sequences 1 through 5, see Fig. 1.
aa, amino acids.
The first gene of the island, sugR, encodes a protein that exhibits closest similarity to the PgaA antigen of the periodontopathogen Porphyromonas gingivalis (43a) and to a putative ATP binding protein encoded in the genome of a clinical isolate of E. coli (29). The SugR protein contains an imperfect nucleotide-binding Walker A motif (APNGAGKT) that is missing the first conserved G of the consensus Walker sequence (GXXGXGKS/T) (48).
The MisL (for membrane insertion and secretion) protein exhibits similarity to the immunoglobulin A1 protease family of autotransported proteins, which have been found only in pathogenic bacteria (26, 32). These proteins consist of an N-terminal effector domain and a C-terminal conserved domain that forms a pore in the outer membrane through which the N-terminal domain is translocated. The similarity between MisL and the AIDA-I protein from enteropathogenic E. coli and the VirG protein from S. flexneri is limited to the C-terminal region (Fig. 2A), suggesting a similar autotransporter function rather than specific functional similarities with these two proteins, which have been implicated in diffuse adherence to HeLa cells (2) and cell-to-cell spreading (16), respectively. Neither the 955-amino-acid MisL protein nor the 1,286-amino-acid AIDA-I protein contains cysteine residues, a feature that precludes the formation of disulfide bonds and is believed to be crucial for membrane translocation (26). MisL also contains a predicted N-terminal signal sequence required for the translocation of the protein across the inner membrane.
FIG. 2.
(A) Alignment of the C-terminal domains of the S. enterica MisL protein, the plasmid-encoded AIDA-1 protein from enteropathogenic E. coli, and the VirG protein of S. flexneri. An 18-amino-acid duplicated region rich in Pro, Asp, and Val within MisL is indicated by a horizontal line. Amino acids that are identical between the MisL and AIDA-1 proteins or between the AIDA-1 and VirG proteins are linked by vertical lines. Amino acids that are identical between the MisL and VirG proteins are indicated by a short underline in the VirG residue. (B) Alignment of the N-terminal domains of the MarT protein, ORF269 (O269) of E. coli, and the ToxR regulator from V. cholerae. Highly conserved residues among OmpR homologs are indicated by dots (30). Amino acids that have been shown to be important for ToxR function (41) are marked by asterisks. Amino acids that are identical between the MarT and ORF269 proteins or between the ORF269 and ToxR proteins are linked by vertical lines. Amino acids that are identical between the MarT and ToxR proteins are indicated by a short underline in the ToxR residue. Alignments were performed by using the PILEUP program (GCG).
The MarT (for membrane-associated regulator) protein has homology with a protein from E. coli K-12 (ORF269) and exhibits similarity in its N-terminal domain to the ToxR protein from Vibrio cholerae (Fig. 2B). ToxR is a transmembrane regulatory protein that is required for the synthesis of cholera toxin in V. cholerae (36). It consists of an N-terminal cytoplasmic domain, which is homologous to the OmpR family of transcription factors and probably involved in DNA binding, and a C-terminal domain which is thought to be involved in sensing environmental signals (30, 37). Like ToxR, MarT contains a potential transmembrane domain in its central region and exhibits similarity with the putative DNA binding domain of the CadC transcriptional activator of E. coli K-12 (49), another member of the OmpR family, suggesting that the marT gene encodes a regulatory protein.
Finally, the rhuM, rmbA, fidL, slsA, and cigR gene products do not exhibit sequence similarity to proteins with known functions in the sequence databases. FidL and the glycine- and asparagine-rich CigR contain putative signal sequences and might be exported proteins.
Expression of SPI-3-encoded genes.
To examine the expression of the genes encoded within SPI-3, MudJ transposon insertions were isolated in this region of the Salmonella genome, and the β-galactosidase activities produced by the resulting strains were determined (see Materials and Methods) (Fig. 1). When the strains were grown in LB broth, β-galactosidase activity was produced by lac gene fusions to the sugR, rhuM, and marT genes but not by the misL-lac fusion (Table 3), suggesting that the misL gene may respond to signals not present in laboratory media. A lac fusion to the intergenic region between sugR and rhuM, located approximately 200 to 250 bp upstream of rhuM, produced a level of β-galactosidase activity comparable to that of the rhuM-lac fusion, suggesting that sugR and rhuM may constitute an operon even though the distance between sugR and rhuM is 580 bp.
TABLE 3.
Transcriptional activities of SPI-3-encoded genes in wild-type, marT, and phoP strains
| MudJ insertion | β-Galactosidase activitya
|
||
|---|---|---|---|
| Wild type | marT1::cat | phoP7953::Tn10 | |
| sugR1::MudJ | 116 | 114 | 118 |
| sugR2::MudJb | 175 | NDc | ND |
| rhuM1::MudJ | 200 | 205 | 192 |
| misL1::MudJ | Nod | No | No |
| marT2::MudJ | 71 | ND | 74 |
| mgtCB9232::MudJ | 18 | 18 | No |
In Miller units.
Insertion in the intergenic region between sugR and rhuM.
ND, not determined.
No, nondetectable activity.
Expression of horizontally acquired genes is often controlled by regulatory proteins encoded by linked genes within the acquired sequences. For example, several genes in the Salmonella SPI-1 island are regulated by the HilA and InvF proteins, which are also encoded within SPI-1 (1, 27). Likewise, a two-component system encoded in the Salmonella SPI-2 island governs expression of several SPI-2 genes (46–47). Despite its similarity to the regulatory protein ToxR, MarT does not appear to control expression of the sugR, rhuM, misL, and mgtC genes, because similar levels of β-galactosidase were displayed by isogenic marT+ and marT mutant strains when bacteria were grown in LB broth (Table 3). MarT does not appear to regulate its own expression either, because the wild-type marT gene on a multicopy plasmid did not modify the β-galactosidase activity of a marT2::MudJ strain (data not shown). The MarT protein has different amino acids than the ToxR protein at three of four positions shown to be important for ToxR regulatory function (Fig. 2B) (41), which raises the possibility of MarT being involved in a function other than transcriptional regulation. However, it is also possible that the MarT protein governs transcription of other genes within SPI-3 or under different growth conditions.
To coordinate their expression with that of the rest of the genome, foreign sequences often recruit host regulators in addition to those encoded within the acquired sequences. One striking example is provided by the regulatory protein PhoP, which is present in both pathogenic and nonpathogenic bacterial species (19) and controls expression of several horizontally acquired sequences involved in Salmonella virulence, including the SPI-3-carried mgtC gene (18). However, expression of sugR, rhuM, misL, and marT is not dependent on the PhoP regulatory protein, since similar levels of β-galactosidase were displayed by isogenic phoP+ and phoP mutant strains (Table 3).
Virulence properties of SPI-3 mutants.
Because the misL and marT genes encode proteins with similarity to known virulence factors, we investigated whether these genes were required for Salmonella virulence. Strains with a misL1::MudJ mutation (EG10755) or with a marT1::cat mutation (EG10207) exhibited wild-type levels of survival within macrophages and invasion of epithelial cells (data not shown). Moreover, their ability to cause a lethal infection in mice was as efficient as that of the wild-type parent when tested orally on BALB/c mice at doses of 2 × 106 and 1.5 × 107 CFU (the 50% lethal dose of the wild-type strain is 6 × 105 CFU) (22). These results indicate that the misL and marT genes are not essential for virulence under the conditions investigated. However, these genes could be involved in other aspects of pathogenesis, such as chronic infection and host specificity, or they could play a role in processes specific to Salmonella that are unrelated to virulence.
Similarity of SPI-3 proteins to E. coli proteins encoded by horizontally acquired sequences.
Consistent with the notion the SPI-3 island was acquired by horizontal gene transfer, its overall G+C content is 47.5%, which is much lower than that of the Salmonella chromosome (52%) (39). Moreover, SPI-3 is located next to the selC tRNA gene, and tRNA genes are preferential sites of insertion of foreign sequences, including phages, plasmids, and pathogenicity islands (8, 20, 24). Furthermore, several SPI-3 gene products exhibit similarity with proteins encoded by horizontally acquired DNA sequences in other bacterial species. For example, the sugR gene product exhibits similarity with a protein from a clinical isolate of E. coli encoded by a gene that was probably acquired by lateral gene transfer, because it is part of a region with a low G+C content, located downstream of the thrW tRNA locus (29), and is absent from the E. coli K-12 genome (4).
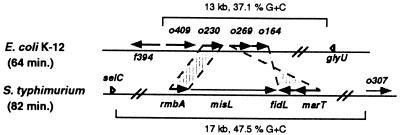
The central region of SPI-3 includes four genes, i.e., rmbA, misL, fidL, and marT, three of which code for proteins with sequence similarity to E. coli K-12 ORF products encoded at 64 min in the chromosome (rmbA, fidL, and marT are similar to orf230, orf164, and orf269, respectively) (4). However, the genetic organizations of these genes are different in E. coli and Salmonella (Fig. 3). The orf230, orf269, and orf164 genes appear to have been acquired horizontally into E. coli K-12, because they have an atypical codon usage and are part of a 13-kb region with a very low G+C content (37.1%) that is located downstream of the glyU tRNA gene (4). Consistent with the notion that this region is not ancestral to enteric bacteria but rather that it was incorporated into E. coli K-12 by horizontal gene transfer, Southern hybridization experiments revealed that orf269-hybridizing sequences are absent from 12 bacterial species, including S. flexneri, which is considered to be part of the E. coli species (see Materials and Methods) (data not shown).
FIG. 3.
Organization of the rmbA, misL, fidL, and marT genes in S. enterica serovar Typhimurium SPI-3 in comparison to the orf230 (o230), orf269 (o269), and orf164 (o164) genes of E. coli K-12. The deduced amino acid sequences of rmbA, fidL, and marT are about 40% identical to the deduced proteins encoded by o230, o164, and o269, respectively (Table 2), suggesting that these proteins are orthologues rather than homologues.
Evolution of SPI-3 sequences.
The G+C content is not uniform along SPI-3 (Fig. 1): genes with the lowest G+C content (rmbA, fidL, and marT, with 37.7, 45.2, and 47.3%, respectively) are located in the central region of SPI-3. However, this region, which is homologous to an E. coli K-12 gene cluster (see above), is interrupted by the misL gene, which has a G+C content of 53%, suggesting that the incorporation of misL was a genetic event separate from that mediating the acquisition of rmbA, fidL, and marT. The central region of SPI-3 is surrounded by DNA segments with a very low G+C content that contain remnants of insertion sequences (Fig. 1): between the rhuM and rmbA genes, a 200-bp segment is homologous to the left inverted repeat and the 40 first residues of the transposase gene of the IS1351 element of Salmonella enteritidis (6a), and the region that separates the marT and slsA genes harbors a 100-bp sequence similar to a repetitive element from E. cloacae that is related to IS10 (31) and to the left inverted repeat of the IS911 element of Shigella dysenteriae (42). Taken together, these data suggest that SPI-3 has a composite structure and that the central region might have an independent origin.
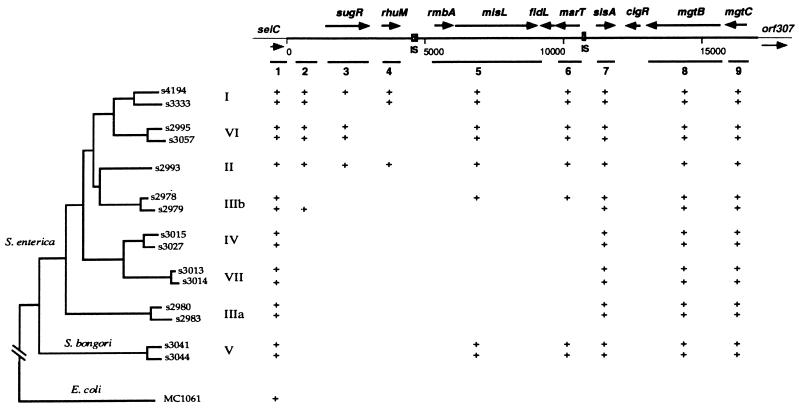
To further examine the evolution of the SPI-3 island, we investigated the Salmonella Reference Collection C, which includes strains that encompass the eight subspecies of the genus Salmonella (6), for the presence of SPI-3 sequences. Southern hybridization experiments established that sequences hybridizing to the 5.3 kb at the right end of the island (including the slsA gene and the mgtCB operon) are present in all eight subspecies of S. enterica (Fig. 4). In contrast, the 5.5-kb central region did not hybridize to DNAs from strains of groups IIIa, IV, and VII and from one representative of group IIIb. Surprisingly, sequences hybridizing to this region were detected in Salmonella bongori (group V), the more divergent form of Salmonella. These results are consistent with the hypothesis that the central region of SPI-3 was incorporated as a separate genetic event, as suggested by the G+C composition and presence of surrounding insertion sequences (ISs). Thus, this portion of SPI-3 might have been acquired independently in S. bongori, or, alternatively, it may have been introduced into the Salmonella lineage or in the donor chromosome from which SPI-3 originated and then have been deleted in a subset of Salmonella subspecies. The left end of SPI-3 (including sugR and rhuM) appears to be less conserved than the central region and might have been the subject of deletions. As expected, a probe complementary to a region outside SPI-3, which included most of the selC gene and 350 bp of upstream sequences, hybridized to DNAs from all Salmonella subspecies and from E. coli K-12.
FIG. 4.
Phylogenetic distribution of SPI-3 sequences among salmonellae based on Southern blot experiments carried out as described in Materials and Methods. Positions of the remnants of ISs are indicated. +, presence of a positive hybridization signal with the designated strain. Evolutionary relationships of Salmonella Reference Collection C strains are based on variation in the nucleotide sequences of five housekeeping genes (6). The roman numerals indicate the eight Salmonella subspecific groups.
The selC tRNA locus is the site of insertion of SPI-3 in Salmonella and of the PAI-1 and LEE islands in pathogenic strains of E. coli (3, 5, 33). While this suggests a common mechanism for the acquisition of these foreign sequences, the SPI-3 island does not encode an integrase-like protein or harbor long repeated sequences. This is in contrast to PAI-1, which contains a cryptic integrase gene in its left end and is flanked by short direct repeats (5, 24), and to LEE, which harbors remnants of a transposase gene in its right end (13). The sequences of the Salmonella SPI-1, SPI-2, and SPI-4 islands and their boundaries have, thus far, not revealed long sequence repeats, phage attachment sites, or remnants of integrase genes, which could be responsible for the stability of these regions in the Salmonella genome.
The LEE pathogenicity island was originally identified at the selC locus of enteropathogenic strains of E. coli (33), but recent work indicates that LEE can be found at locations other than selC (50). We investigated whether SPI-3 is located at the selC locus in the different S. enterica subspecies by carrying out PCRs with primers complementary to the selC and slsA genes (these genes are 12 kb apart in S. enterica serovar Typhimurium, and selC- and slsA-hybridizing sequences have been detected in all Salmonella subspecies). The slsA gene is linked to the selC gene in most subspecies, because a 12-kb fragment was obtained when DNAs from Salmonella subspecies I, VI, and II were used as templates, and fragments smaller than 12 kb were amplified from at least one representative of each of the other subspecies (two different primers within selC were used to exclude nonspecific amplifications). At the right end of the island, the mgtC gene appears to be located next to orf307 in all eight subspecies of S. enterica, because PCR experiments with primers complementary to mgtC and orf307 resulted in the amplification of the same 0.76-kb fragment.
Conclusion.
The molecular analysis of SPI-3 sequences and their phylogenetic distribution among the different subspecies that comprise S. enterica indicate that SPI-3 has a mosaic structure, most likely the result of a multistep evolutionary process, and encodes proteins that are not obviously functionally related. This is in contrast to the Salmonella SPI-1 and SPI-2 pathogenicity islands, which were likely acquired through single horizontal gene transfer events and encode functionally related proteins, which include type III export systems and secreted effector proteins (38, 40, 44). The different distribution of SPI-3 sequences may reflect the functional role of encoded genes: sequences present in only a subset of Salmonella subspecies could be involved in host specificity, tissue tropism, and disease manifestation, as recently proposed for a group of Salmonella-specific sequences of atypical base composition recovered by the in vivo expression technology procedure (11).
ACKNOWLEDGMENTS
We thank Matthew Chung-Ying Lo for help in strain construction and Howard Ochman for comments on an earlier version of the manuscript.
This work was supported by NIH grant GM54900 to E.A.G., who is an Associate Investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Bajaj V, Hwang C, Lee C A. hilA is a novel ompR/toxR family member that activates the expression of Salmonella typhimurium invasion genes. Mol Microbiol. 1995;18:715–727. doi: 10.1111/j.1365-2958.1995.mmi_18040715.x. [DOI] [PubMed] [Google Scholar]
- 2.Benz I, Schmidt M A. AIDA-I, the adhesin involved in diffuse adherence of the diarrhoeagenic Escherichia coli strain 2787 (O126:H27), is synthesized via a precursor molecule. Mol Microbiol. 1992;6:1539–1546. doi: 10.1111/j.1365-2958.1992.tb00875.x. [DOI] [PubMed] [Google Scholar]
- 3.Blanc-Potard A-B, Groisman E A. The Salmonella selC locus contains a pathogenicity island mediating intramacrophage survival. EMBO J. 1997;16:5376–5385. doi: 10.1093/emboj/16.17.5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 5.Blum G, Ott M, Lischewski A, Ritter A, Imrich H, Tschäpe H, Hacker J. Excision of large DNA regions termed pathogenicity islands from tRNA-specific loci in the chromosome of an Escherichia coli wild-type pathogen. Infect Immun. 1994;62:606–614. doi: 10.1128/iai.62.2.606-614.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyd E F, Wang F-S, Wittam T S, Selander R K. Molecular genetic relationships of the salmonellae. Appl Environ Microbiol. 1996;62:804–808. doi: 10.1128/aem.62.3.804-808.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6a.Burnens, A. P. GenBank accession no. 283734.
- 7.Castilho B A, Olfson P, Casadaban M. Plasmid insertion mutagenesis and lac gene fusion with mini-Mu bacteriophage transposons. J Bacteriol. 1984;158:488–495. doi: 10.1128/jb.158.2.488-495.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheetham B F, Katz M E. A role for bacteriophages in the evolution and transfer of bacterial virulence determinants. Mol Microbiol. 1995;18:201–208. doi: 10.1111/j.1365-2958.1995.mmi_18020201.x. [DOI] [PubMed] [Google Scholar]
- 9.Chen L M, Kaniga K, Galán J E. Salmonella spp. are cytotoxic for cultured macrophages. Mol Microbiol. 1996;21:1101–1115. doi: 10.1046/j.1365-2958.1996.471410.x. [DOI] [PubMed] [Google Scholar]
- 10.Collazo C M, Galán J E. The invasion-associated type-III protein secretion system in Salmonella—a review. Gene. 1997;192:51–59. doi: 10.1016/s0378-1119(96)00825-6. [DOI] [PubMed] [Google Scholar]
- 11.Conner C P, Heithoff D M, Julio S M, Sinsheimer R L, Mahan M J. Differential patterns of acquired virulence genes distinguish Salmonella strains. Proc Natl Acad Sci USA. 1998;95:4641–4645. doi: 10.1073/pnas.95.8.4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davis R W, Botstein D, Roth J R. Advanced bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1980. [Google Scholar]
- 13.Donnenberg M S, Lai L-C, Taylor K A. The locus of enterocyte effacement pathogenicity island of enteropathogenic Escherichia coli encodes secretion functions and remnants of transposons at its extreme right end. Gene. 1997;184:107–114. doi: 10.1016/s0378-1119(96)00581-1. [DOI] [PubMed] [Google Scholar]
- 14.García Véscovi E, Soncini F, Groisman E A. The role of the PhoP/PhoQ regulon in Salmonella virulence. Res Microbiol. 1994;145:473–480. doi: 10.1016/0923-2508(94)90096-5. [DOI] [PubMed] [Google Scholar]
- 15.García Véscovi E, Soncini F C, Groisman E A. Mg2+ as an extracellular signal: environmental regulation of Salmonella virulence. Cell. 1996;84:165–174. doi: 10.1016/s0092-8674(00)81003-x. [DOI] [PubMed] [Google Scholar]
- 16.Goldberg M B, Barzu O, Parsot C, Sansonetti P J. Unipolar localization and ATPase activity of IcsA, a Shigella flexneri protein involved in intracellular movement. J Bacteriol. 1993;175:2189–2196. doi: 10.1128/jb.175.8.2189-2196.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Groisman E A. Bacterial responses to host defense peptides. Trends Microbiol. 1996;4:127–128. doi: 10.1016/0966-842x(96)30013-9. [DOI] [PubMed] [Google Scholar]
- 18.Groisman E A. The ins and outs of virulence gene expression: Mg2+ as a regulatory signal. Bioessays. 1998;20:96–101. doi: 10.1002/(SICI)1521-1878(199801)20:1<96::AID-BIES13>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 19.Groisman E A, Chiao E, Lipps C J, Heffron F. Salmonella typhimurium phoP virulence gene is a transcriptional regulator. Proc Natl Acad Sci USA. 1989;86:7077–7081. doi: 10.1073/pnas.86.18.7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Groisman E A, Ochman H. Pathogenicity islands: bacterial evolution in quantum leaps. Cell. 1996;87:791–794. doi: 10.1016/s0092-8674(00)81985-6. [DOI] [PubMed] [Google Scholar]
- 21.Groisman E A, Ochman H. How Salmonella became a pathogen. Trends Microbiol. 1997;5:343–349. doi: 10.1016/S0966-842X(97)01099-8. [DOI] [PubMed] [Google Scholar]
- 22.Groisman E A, Parra C A, Salcedo M, Lipps C J, Heffron F. Resistance to host antimicrobial peptides is necessary for Salmonella virulence. Proc Natl Acad Sci USA. 1992;89:11939–11943. doi: 10.1073/pnas.89.24.11939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Groisman E A, Sturmoski M A, Solomon F, Lin R, Ochman H. Molecular, functional, and evolutionary analysis of sequences specific to Salmonella. Proc Natl Acad Sci USA. 1993;90:1033–1037. doi: 10.1073/pnas.90.3.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hacker J, Blum-Oehler G, Mühldorfer I, Tschäpe H. Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol Microbiol. 1997;23:1089–1097. doi: 10.1046/j.1365-2958.1997.3101672.x. [DOI] [PubMed] [Google Scholar]
- 25.Hensel M, Shea J E, Bäumler A J, Gleeson C, Blattner F, Holden D W. Analysis of the boundaries of Salmonella pathogenicity island 2 and the corresponding chromosomal region of Escherichia coli K-12. J Bacteriol. 1997;179:1105–1111. doi: 10.1128/jb.179.4.1105-1111.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jose J, Jähnig F, Meyer T F. Common structural features of IgA1 protease-like outer membrane protein autotransporters. Mol Microbiol. 1995;18:377–382. doi: 10.1111/j.1365-2958.1995.mmi_18020378.x. [DOI] [PubMed] [Google Scholar]
- 27.Kaniga K, Bossio J C, Galán J E. The Salmonella typhimurium invasion genes invF and invG encode homologues of the AraC and PulD family of proteins. Mol Microbiol. 1994;13:555–568. doi: 10.1111/j.1365-2958.1994.tb00450.x. [DOI] [PubMed] [Google Scholar]
- 28.Lee C A, Jones B D, Falkow S. Identification of a Salmonella typhimurium invasion locus by selection for hyperinvasive mutants. Proc Natl Acad Sci USA. 1992;89:1847–1851. doi: 10.1073/pnas.89.5.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lim D. Structure and biosynthesis of unbranched multicopy single-stranded DNA by reverse transcriptase in a clinical Escherichia coli isolate. Mol Microbiol. 1992;6:3531–3542. doi: 10.1111/j.1365-2958.1992.tb01788.x. [DOI] [PubMed] [Google Scholar]
- 30.Martínez-Hackert E, Stock A M. Structural relationships in the OmpR family of winged-helix transcription factors. J Mol Biol. 1997;269:301–312. doi: 10.1006/jmbi.1997.1065. [DOI] [PubMed] [Google Scholar]
- 31.Matsutani S. Multiple copies of IS10 in the Enterobacter cloacae MD36 chromosome. J Bacteriol. 1991;173:7802–7809. doi: 10.1128/jb.173.24.7802-7809.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maurer J, Jose J, Meyer T F. Autodisplay: one-component system for efficient surface display and release of soluble recombinant proteins from Escherichia coli. J Bacteriol. 1997;179:794–804. doi: 10.1128/jb.179.3.794-804.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McDaniel T K, Jarvis K G, Donnenberg M S, Kaper J B. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc Natl Acad Sci USA. 1995;92:1664–1668. doi: 10.1073/pnas.92.5.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mecsas J J, Strauss E J. Molecular mechanisms of bacterial virulence: type III secretion and pathogenicity islands. Emerg Infect Dis. 1996;2:270–288. doi: 10.3201/eid0204.960403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 36.Miller V L, Mekalanos J J. Synthesis of cholera toxin is positively regulated at the transcriptional level by toxR. Proc Natl Acad Sci USA. 1984;81:3471–3475. doi: 10.1073/pnas.81.11.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller V L, Taylor R K, Mekalanos J J. Cholera toxin transcriptional activator ToxR is a transmembrane DNA binding protein. Cell. 1987;48:271–279. doi: 10.1016/0092-8674(87)90430-2. [DOI] [PubMed] [Google Scholar]
- 38.Mills D M, Bajaj V, Lee C A. A 40 kilobase chromosomal fragment encoding Salmonella typhimurium invasion genes is absent from the corresponding region of the Escherichia coli K-12 chromosome. Mol Microbiol. 1995;15:749–759. doi: 10.1111/j.1365-2958.1995.tb02382.x. [DOI] [PubMed] [Google Scholar]
- 39.Ochman H, Lawrence J G. Phylogenetics and the amelioration of bacterial genomes. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 2. Washington, D.C: ASM Press; 1996. pp. 2627–2637. [Google Scholar]
- 40.Ochman H, Soncini F C, Solomon F, Groisman E A. Identification of a pathogenicity island required for Salmonella survival in host cells. Proc Natl Acad Sci USA. 1996;93:7800–7804. doi: 10.1073/pnas.93.15.7800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ottemann K M, DiRita V J, Mekalanos J J. ToxR proteins with substitutions in residues conserved with OmpR fail to activate transcription from the cholera toxin promoter. J Bacteriol. 1992;174:6807–6814. doi: 10.1128/jb.174.21.6807-6814.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prère M-F, Chandler M, Fayet O. Transposition in Shigella dysenteriae: isolation and analysis of IS911, a new member of the IS3 group of insertion sequences. J Bacteriol. 1990;172:4090–4099. doi: 10.1128/jb.172.7.4090-4099.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reece K S, Phillips G J. New plasmids carrying antibiotic-resistance cassettes. Gene. 1995;165:141–142. doi: 10.1016/0378-1119(95)00529-f. [DOI] [PubMed] [Google Scholar]
- 43a.Rigg, G. P., and I. S. Roberts. GenBank accession no. 95938.
- 44.Shea J E, Hensel M, Gleeson C, Holden D W. Identification of a virulence locus encoding a second type III secretion system in Salmonella typhimurium. Proc Natl Acad Sci USA. 1996;93:2593–2597. doi: 10.1073/pnas.93.6.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Snavely M D, Miller C G, Maguire M E. The mgtB Mg2+ transport locus of Salmonella typhimurium encodes a P-type ATPase. J Biol Chem. 1991;266:815–823. [PubMed] [Google Scholar]
- 46.Soncini F C, García Véscovi E, Solomon F, Groisman E A. Molecular basis of the magnesium deprivation response in Salmonella typhimurium: identification of PhoP-regulated genes. J Bacteriol. 1996;178:5092–5099. doi: 10.1128/jb.178.17.5092-5099.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46a.Uchiya, K., and E. A. Groisman. Unpublished results.
- 47.Valdivia R H, Falkow S. Fluorescence-based isolation of bacterial genes expressed within host cells. Science. 1997;277:2007–2010. doi: 10.1126/science.277.5334.2007. [DOI] [PubMed] [Google Scholar]
- 48.Walker J E, Saraste M, Runswick M J, Gay N J. Distantly related sequences in the α- and β-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Watson N, Dunyak D S, Rosey E L, Slonczewski J L, Olson E R. Identification of elements involved in transcriptional regulation of the Escherichia coli cad operon by external pH. J Bacteriol. 1992;174:530–540. doi: 10.1128/jb.174.2.530-540.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wieler L H, McDaniel T K, Whittam T S, Kaper J B. Insertion site of the locus of enterocyte effacement in enteropathogenic and enterohemorrhagic Escherichia coli differs in relation to the clonal phylogeny of the strains. FEMS Microbiol Lett. 1997;156:49–53. doi: 10.1111/j.1574-6968.1997.tb12704.x. [DOI] [PubMed] [Google Scholar]
- 51.Wong K-K, McClelland M, Stillwell L C, Sisk E C, Thurston S J, Saffer J D. Identification and sequence analysis of a 27-kilobase chromosomal fragment containing a Salmonella pathogenicity island located at 92 minutes on the chromosome map of Salmonella enterica serovar Typhimurium LT2. Infect Immun. 1998;66:3365–3371. doi: 10.1128/iai.66.7.3365-3371.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wood M W, Jones M A, Watson P R, Hedges S, Wallis T S, Galyov E E. Identification of a pathogenicity island required for Salmonella enteropathogenicity. Mol Microbiol. 1998;29:883–891. doi: 10.1046/j.1365-2958.1998.00984.x. [DOI] [PubMed] [Google Scholar]