Abstract
Fine-tuning cellular physiology in response to intracellular and environmental cues requires precise temporal and spatial control of gene expression. High-resolution imaging technologies to detect mRNAs and their translation state have revealed that all living organisms localize mRNAs in subcellular compartments and create translation hotspots, enabling cells to tune gene expression locally. Therefore, mRNA localization is a conserved and integral part of gene expression regulation from prokaryotic to eukaryotic cells. In this Review, we discuss the mechanisms of mRNA transport and local mRNA translation across the kingdoms of life and at organellar, subcellular and multicellular resolution. We also discuss the properties of messenger ribonucleoprotein and higher order RNA granules and how they may influence mRNA transport and local protein synthesis. Finally, we summarize the technological developments that allow us to study mRNA localization and local translation through the simultaneous detection of mRNAs and proteins in single cells, mRNA and nascent protein single-molecule imaging, and bulk RNA and protein detection methods.
Asymmetric mRNA distribution was first reported in 1983 by Jeffery et al.1, who observed that, during the early stages of ascidian embryonic development, the mRNA encoding actin localized in the cytoplasm, where muscle-forming cells reside1. This observation, along with studies in Xenopus laevis eggs2 and Drosophila melanogaster3,4, led to the hypothesis that, during embryogenesis, specific mRNA pools could be partitioned and anchored into particular cell lineages to determine tissue differentiation. Subcellular mRNA localization was first reported in 1986 by Lawrence and Singer, who observed this phenomenon in chicken fibroblasts using in situ hybridization5, and was subsequently shown to occur in other organisms such as Saccharomyces cerevisiae6, rice plant cells7 (BOX 1), mammalian neurons8 and oligodendrocytes9. Later, mRNA localization was found to encompass a notable percentage of the transcriptome during D. melanogaster10 and X. laevis11 development as well as in Escherichia coli12 and in mammalian tissue13, including at the subcellular level14, in cellular protrusions15 and in organelles such as the endoplasmic reticulum (ER)16. These studies suggested that mRNA localization contributed to the post-transcriptional fine-tuning of gene expression and the control of fundamental processes such as cell migration, polarization and differentiation.
Box 1 |. mRNA localization in plants.
Although subcellular mRNA localization is well characterized in microorganisms and metazoans, less is known for plant cells. Here, we briefly summarize key examples of mRNA localization in plants, and we refer readers to specialized reviews for further details139,251. Studies suggest that plant cells localize mRNAs to the endoplasmic reticulum (ER), mitochondria and the chloroplast. Oryza sativa rice endosperm cells form a tissue inside seeds that surrounds and provides key nutrients to the germinating embryo; these cells have a peculiar pattern of mRNA localization in the ER. Two mRNAs encoding the storage proteins prolamin and glutelin localize to two separate ER compartments, the protein-body ER and the cisternal interconnecting ER, respectively7. Localized mRNAs encoding prolamin are translated on the ER, and the proteins are retained in the ER lumen forming an ER-derived protein body I (PBI). By contrast, mRNAs encoding glutelin localize to the adjacent cis-ER, and the translated proteins are further exported to the Golgi7. The localization of these mRNAs is independent of translation and requires specific mRNA zipcodes and RNA binding proteins (RBPs)251–254. Recent work showed that the mRNA encoding glutelin is transported to the cisternal ER via endosome trafficking mediated by RBP-P, RBP-L and the endosome membrane-bound protein Rab5a255. Besides mRNA localization to the ER, nuclear-encoded mRNAs are also transported to mitochondria. mRNA encoding the voltage-dependent ion channel (VDAC) localizes to mitochondria in Arabidopsis thaliana256. Owing to a cis-element present in the 3′ untranslated region (3′UTR), VDAC and other mitochondria-localized mRNAs are proposed to be transported (via an unknown mechanism) to modulate mitochondria function and number256,257. Finally, the mechanism for localizing RNA to chloroplasts is exploited by plant viruses for replication251,258,259. For instance, the negative strand of Bamboo mosaic virus RNA is imported into the chloroplast via a specific RNA sequence, which is a key step for viral replication260. Further work is required to elucidate the mechanisms controlling the transport of other mRNAs to specific subcellular locations in plants. Furthermore, the field lacks fundamental tools to investigate the mechanisms controlling localized mRNA translation. The recent description of a single-molecule fluorescence in situ hybridization (smFISH) protocol for plant cells261 may trigger the development of novel tools aimed at elucidating localized mRNA translation in plants at high resolution.
In cells, localizing mRNAs, which can be translated tens to hundreds of times in response to local stimuli, is more cost-effective than transporting individual proteins. Furthermore, localized protein synthesis may avoid the ectopic expression of a protein in an undesired compartment and regulate protein function by controlling its local concentration, its chemical environment (including its pH), and its ability to form multi-protein complexes with defined stoichiometry17 and specific post-translational modifications18. Observing mRNAs at high resolution revealed that, across all species, mRNAs localize to specific compartments (for example, dendrites, axons and RNA–protein granules) and organelles (for example, ER, mitochondria and chloroplasts) to control in situ protein synthesis and local cell physiology. In addition, localization events are coordinated with the physiological status of the cell, which can influence whether mRNAs are stored in a translationally repressed state in stress granules or targeted to processing bodies (P-bodies) for degradation. mRNAs are primarily sorted via cis-localization elements called ‘zipcodes’ that, together with RNA binding proteins (RBPs), control active and passive mRNA trafficking19. Furthermore, non-canonical modes of mRNA transport via non-specific interactions with organelles or RNA–RNA interactions are also thought to exist20–23.
The localization and translation of mRNAs influence cell physiology at the subcellular, cellular, tissue and organism level. At the single-cell level, mRNA trafficking dictates cell polarity, motility and differentiation by enabling rapid and localized responses to intracellular and extracellular signals, yet impairment of mRNA localization in single cells rarely causes severe growth phenotypes or lethality. However, in tissues or multicellular organisms, mRNA localization is crucial in homeostasis, differentiation and development. Indeed, blocking mRNA localization in the developing D. melanogaster embryo results in severe developmental defects24, and mRNA localization defects in the central nervous system result in cognitive disorders25. Thus, to determine how mRNA localization influences high order cellular organization and functions, local gene expression should be studied from the subcellular to the multicellular level.
Although cytoplasmic mRNA localization is wide-spread, fundamental questions remain about its functional role and it is unclear if, and how, mRNA localization always controls local protein synthesis. In addition, improving the sensitivity of mRNA imaging technologies to study this process in intact single-cell and multicellular organisms remains challenging.
In this Review, we provide an overview of the rapidly evolving technological advances that drive research into mRNA localization and summarize evidence of mRNA localization and protein synthesis in single-cell organisms (bacteria and fungi) and in the multicellular context, for example, in tissues and whole organisms (D. melanogaster and Caenorhabditis elegans). We then describe key examples of subcellular mRNA localization in different cell types (neurons and fibroblasts) and organelles (mitochondria, ER and centrosomes), before discussing the mechanisms governing mRNA localization and localized translation. Finally, we highlight the importance of studying the composition of mRNA granules to understand how it determines the specificity and fate of an mRNA, and we discuss the future challenges and perspectives in the field.
Localized RNAs across kingdoms of life
Evidence of localized mRNAs and protein synthesis in single-cell and multicellular organisms has revealed that mRNA localization has a range of functions across organisms and has evolved from non-conserved and conserved mechanisms.
Single-cell organisms
Single-cell organisms, such as bacteria and fungi, take advantage of the asymmetric distribution of mRNA and proteins to modulate gene expression and organize cellular functions. Understanding this process in single-cell organisms helps to elucidate the mechanisms that are conserved in more complex models and, from a microbiological perspective, is of both biomedical and biotechnological interest.
RNA compartmentalization in bacteria.
Prokaryotic cells were thought to lack intracellular mRNA localization due to their small size, the lack of membrane-bound organelles and the existence of co-transcriptional translation. However, high-resolution imaging revealed distinct compartmentalization of DNA (nucleoids), RNAs (mRNAs and small regulatory RNAs), RNA polymerases and ribosomes, providing evidence for uncoupling between transcription and translation26–28 (FIG. 1a). Furthermore, specific mRNAs accumulate in subcellular domains where the encoded proteins reside. For instance, cat mRNA (encoding cytoplasmic chloramphenicol acetyltransferase) gathers in helical cytoplasmic structures12, suggesting that the mRNA is localized where the protein is needed. Furthermore, mRNAs encoding the transmembrane transporter lactose permease (ptsC mRNAs)29 or the membrane-bound lactose permease (lacY mRNAs)12 are excluded from the nucleoid region and localize next to the cell membrane. Finally, mRNA granules associated with nucleoids (that is, where the protein is located) were observed for the lacZ mRNA (encoding β-galactosidase) both in E. coli30 and C. crescentus31, further suggesting that, in bacteria, mRNA localization contributes to the timing and localization of protein synthesis. Studies performed over the past 10 years show that RNA localization patterns in bacteria can be dependent on32 or independent of12 translation, suggesting that multiple and yet uncharacterized schemes for spatially regulating gene expression exist in prokaryotic cells.
Fig. 1 |. mRNA localization and local translation in single-cell and multi-cellular organisms.
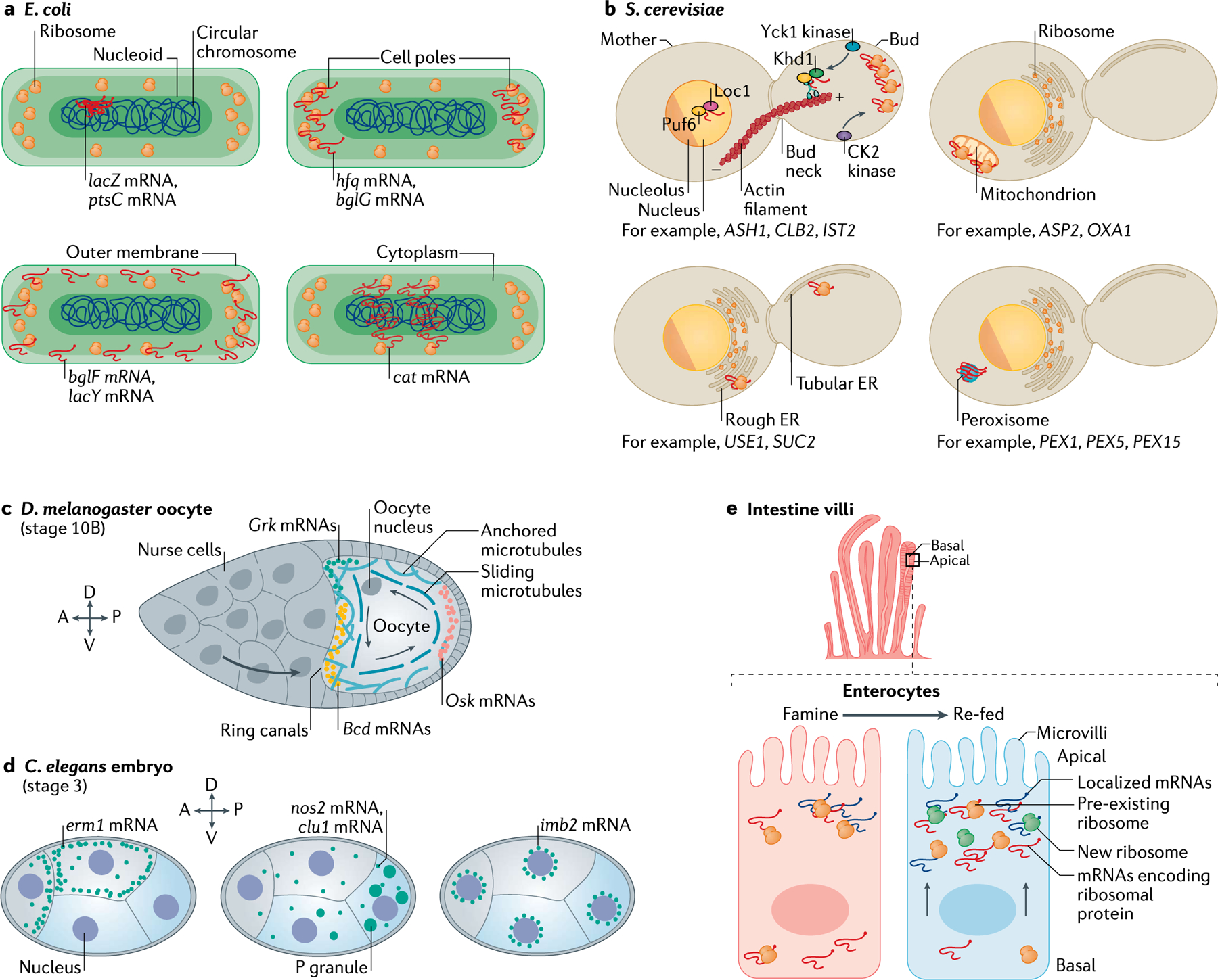
a | In prokaryotes, such as Escherichia coli, the cell is divided into specific sub-compartments, namely the nucleoid (where the circular DNA molecule resides), the cell poles (where ribosomes accumulate) and the outer membrane (where both ribosomes and transporters reside), to which several mRNAs have been shown to localize. ptsC and lacZ mRNAs localize to the nucleoid, hfq and bglG mRNAs localize at the cell poles, bglF and lacY mRNAs localize to the outer membrane, and cat mRNA has a characteristic helical distribution in the cytoplasm. b | In unicellular eukaryotes like Saccharomyces cerevisiae, mRNA is asymmetrically distributed in multiple subcellular compartments. In the growing bud, mRNAs such as ASH1, CLB2 and IST2 are actively transported on actin filaments by the She2–She3–Myo4 complex. Sequences in the 3′ untranslated region (3′UTR) of ASTP2 and OXA1 mRNAs localize these mRNAs to the outer mitochondrial membrane. USE1 and SUC2 mRNAs are localized to the endoplasmic reticulum (ER). PEX1, PEX5 and PEX15 mRNAs are found in peroxisomes. c | During mid-oogenesis in Drosophila melanogaster, the microtubule cytoskeleton of the oocyte is reorganized by cytoplasmic streaming (sliding microtubules) to localize the mRNAs that determine body plan. While Bcd and Grk mRNAs are positioned on the anterior side, Osk mRNAs primarily occupy the posterior side; all three mRNAs are locally translated at their respective positions. d | In Caenorhabditis elegans, maternally inherited transcripts display distinct localization patterns. Transcripts in anterior-biased cells (grey) tend to localize to the cell periphery, where the encoded protein localizes (for example, erm1). mRNAs enriched in posterior cells (blue), such as nos2 and clu1, form clustered granules that overlap with P granules. The imb2 mRNA localizes at the perinuclear region. e | In the intestine, enterocytes lining the villi are polarized cells with distinct apical and basal sides. Components of the translation machinery change their apical–basal distribution in response to nutrient availability. As mRNAs encoding ribosomal proteins move from the basal to the apical side via microtubules, the translation of mRNAs localized at the apical side is boosted to favour nutrient absorption. A, anterior; D, dorsal; P, posterior; V, ventral.
mRNA localization in asymmetrically dividing yeasts.
mRNA localization has been extensively characterized in many fungal species (FIG. 1b), including in the Ascomycota S. cerevisiae (a model organism with biotechnological relevance) and Candida albicans (an opportunistic human pathogen). mRNA transport has also been characterized in the Basidiomycota plant pathogen Ustilago maydis33,34. Tens of mRNAs are localized to the bud of S. cerevisiae by the motor protein SWI5-dependent HO expression protein 2 (She2)–She3 complex35. The best characterized of these mRNAs is ASH1, which accumulates at the bud tip during anaphase6,36–38. This mRNA is transported on actin filaments in a translation-repressed state controlled by RBPs such as Pumilio homology domain family member 6 (also known as Puf6), 60S ribosomal subunit assembly/export protein LOC1 (also known as Loc1) and casein kinase I homologue 1 (also known as Khd1)39,40. Local activation of translation in the bud requires the phosphorylation of Khd1 and Puf6 by the bud-localized casein kinase I homologue 1 (also known as Yck1)40 and CK2 kinase41, respectively. The Ash1 protein is then imported to the daughter nucleus, where it represses the mating-type switching programme by blocking the expression of the homothallic endonuclease42–44. Thus, the asymmetric localization of Ash1 protein ensures that mother and daughter cells acquire opposite mating types to be able to transition to a diploid state. Interestingly, ASH1 mRNA is also localized in C. albicans hyphae in a She3-dependent manner45. About 40 mRNAs localize in these highly polarized and elongated cells, and the inhibition of mRNA transport impairs hyphal development with possible implications on the development and structural stability of biofilms (multicellular structures critical for fungal virulence)34,45.
In addition to ASH1 mRNA, our recent work demonstrated that CLB2 mRNA, encoding the mitotic regulator B-type cyclin, is also localized to the S. cerevisiae bud by the She2–She3 complex and translated in situ to control mitotic entry (E.T. and R.S., unpublished work). Unlike Ash1, Clb2 (also known as G2/mitotic-specific cyclin 2) is not segregated to the daughter cell but is translocated back to the mother nucleus. This observation suggests that mRNA localization can control the temporal expression of proteins as well as their asymmetric distribution. Interestingly, orthologues of the CLB2 mRNA are also symmetrically distributed during mouse46, zebrafish46 and X. laevis47 oocyte development, suggesting that the localization of mRNA encoding cyclin B1 may be functionally conserved during differentiation.
mRNAs are also localized in the ER48–50 and mitochondria51,52 in S. cerevisiae. mRNAs are transported to the ER via signal recognition particle (SRP) and translation-independent pathways that require specific RBPs (for example, She2 and Puf2), and they encode both secreted and non-secreted proteins48–50. mRNAs can also localize to peroxisomes or cytoplasmic membraneless granules such as P-bodies and stress granules (see below)33. Interestingly, mRNAs encoding translation factors (for example, TEF1 and YEF3)53 or glycolytic enzymes (for example, PGK1, ENO1 and ENO2)54 are found in cytoplasmic granules, suggesting that the expression of highly abundant mRNAs can be buffered by controlling their availability.
Finally, work has been conducted to elucidate mRNA transport mechanisms in the filamentous hypha U. maydis33,55,56. This research demonstrated how mRNA trafficking is intertwined with the transport of endosomes (vesicular structures also involved in the asymmetric distribution of proteins and lipids)57. Endosome-mediated mRNA transport requires the RBP Rrm4 (REFs57,58) and the U. maydis PAM2 protein (also called Upa1 protein)59, which couple the mRNA and the associated ribosomes to the endosome and allow their transport on microtubules (see below). This process is important for polarized growth57 and may promote the ability of U. maydis to differentiate from its single-celled form to its multicellular filamentous form when infecting plants.
Multicellular organisms
The importance of timing gene expression by localizing mRNAs is apparent during developmental processes, such as asymmetric cell division and embryonic patterning, and studying mRNA distribution in multicellular organisms has allowed local translation to be linked to physiological changes.
Regulating developmental stages.
Studies in D. melanogaster oocytes and C. elegans early syncytial embryos using single-molecule fluorescence in situ hybridization60,61 (smFISH) (Supplementary Box 1) and the MS2–MS2 coat protein system (MS2–MCP)10,20,62 (BOX 2) have extended previous observations24 showing that localizing mRNAs determine body axes by asymmetrically distributing specification factors of cell fate. During D. melanogaster oogenesis, surrounding nurse cells provide the transcriptionally quiescent oocyte with mRNAs and proteins for its development24. At late oogenesis, nurse cells contract, which squeezes their cytoplasm into the oocyte, depositing hundreds of mRNAs encoding patterning factors through cytoplasmic bridges known as ‘ring-canals’10,24 (FIG. 1c). Upon entering the oocyte, Bcd (encoding homeotic protein bicoid; also known as bicoid) and Grk (encoding Gurken) mRNAs are actively transported along microtubules to the anterior pole, while Osk mRNA (encoding Oskar), along with the RBPs Staufen and Vasa, accumulate at the posterior axis (FIG. 1c). Tagging Twi mRNA (the protein product of which, Twist, is the transcriptional activator of the mesodermal gene network) with the sunTag system (BOX 3) revealed that ‘translation factories’ localize to the basal perinuclear space of living D. melanogaster embryos at nuclear cleavage cycle 14 (REF.63). These factories, which are composed of 2–6 mRNAs, have slow diffusion dynamics, and mRNAs here are preferentially translated (see below).
Box 2 |. Imaging mRNA in living cells.
Here, we summarize the latest methods for imaging mRNAs in live cells; methods for imaging mRNA in fixed cells are described in Supplementary Box 1. We refer the reader to specialized reviews for further details26,248,262–265.
Several fluorescence-based methods allow mRNAs to be visualized and tracked in living cells248,262,266, either via aptamer-based modification of the target mRNA or by detecting endogenous unmodified mRNAs. Aptamer-based mRNA labelling approaches employ RNA stem-loops derived from either bacteriophages (for example, MS2 (REFs36,38,266,267), PP7 (REF.268) or P22 N-peptide for the λ BoxB loop269,270) or from RNA–protein recognition motifs (for example, U1A169,271 or Bgl bacterial anti-terminator272). Arrays consisting of tens of loops are commonly inserted into the 3′ untranslated region (3′UTR) of the target mRNA. Recognition by the cognate RNA binding protein (RBP) fused to a fluorescent protein or a fluorogenic tagging system (for example, HALO273) allows individual mRNAs to be detected in living cells (see the figure, part a). The MS2 and PP7 aptamers are the best-characterized tagging systems and they are well tolerated in transgenic Drosophila melanogaster20,274 and in mouse lines172,275. They have been used for the dual labelling of the same mRNA molecule or of different mRNAs152,276,277 to study the regulation of gene expression. Recently, fluorogenic reporters that rely on RNA aptamers mimicking the structure of GFP have been developed and they become fluorescent upon binding to conditionally fluorescent dyes. Among the best characterized of these reporters are Spinach2 (REF.278), Broccoli279, Corn280 and Mango281,282 (see the figure, part b).
Live-cell imaging reporters that do not require mRNA to be modified include molecular beacons, which are oligonucleotides tagged at their 3′ and 5′ ends with a fluorophore and a quencher283. When the molecular beacon is not bound to the target mRNA, the fluorophore is inactive due to its close proximity to the quencher but the situation is reversed upon molecular beacon–mRNA hybridization (see the figure, part c). Finally, CRISPR–Cas systems such as Cas9-GFP284, Cas9-MS2 or PP7-fusion285 or dCas13-EGFP286 can be exploited to target an mRNA via specific single-guide RNAs (sgRNAs) (see the figure, part d).
To date, stem loop aptamers remain the most widely used technique for labelling mRNAs owing to their higher single-molecule sensitivity and specificity. However, this approach has limitations for tagging highly unstable or small RNAs as it inserts a bulky sequence that can alter the lifecycle of RNA38,287–289. To overcome these problems, MS2 variants that are more easily degraded have been optimized for use in eukaryotic cells38,266,290–292. For both aptamer-based and aptamer-free methods, improvements that increase their brightness and sensitivity are required to minimize the impact of the tagging system on mRNA physiology. For the end-user, the optimal method of choice often depends on the kind of RNA and model organisms they are working with as well as the specific biological question. Further description of these single-molecule methods and their capabilities has been reviewed in REFs248,263,266,293.
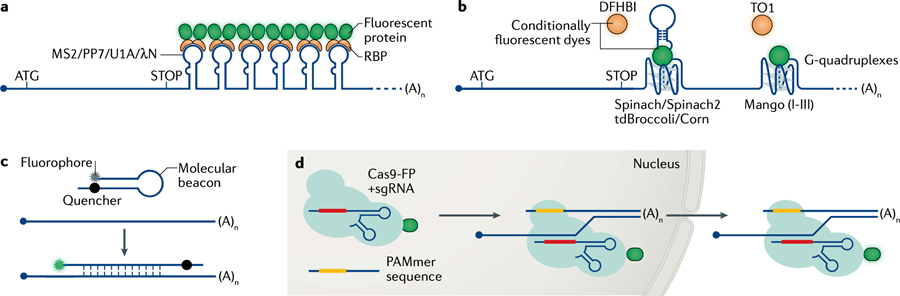
DFHBI, 3,5-difluoro-4-hydroxybenzylidene imidazolinone; FP, fluorescent protein; TO1, thiazole orange-derived fluorophore 1.
Box 3 |. Quantifying translation.
Approaches based on imaging, sequencing and proteomics have been recently combined to determine the site of local translation and quantify its efficiency; the imaging-based approaches are summarized here. Imaging approaches rely on two component systems that tag either different regions of the mRNA and/or the mRNA and the newly synthesized peptide. The translating RNA imaging by coat protein knock-off (TRICK) system detects the first round of translation in real time (see the figure, part a). PP7 and MS2 stem loops are used to label the coding sequence and the 3′ untranslated region (3′UTR), respectively; therefore, the mRNA fluoresces in two colours152. Ribosomes translating the coding sequence knock-off the PP7 coat protein (PCP) from PP7 stem loops, and the mRNA switches from fluorescing in two colours to fluorescing in one colour.
Both nascent chain tracking (NCT) and single-molecule imaging of nascent peptides (SINAP) systems detect nascent peptides as they exit the ribosome tunnel. An array of epitopes, cloned in frame with the open reading frame, recruit multiple copies of antibodies fused to a fluorescent protein93,202,225,294–296. The simultaneous detection of the peptide and the mRNA allows translation heterogeneity to be analysed, translation to be observed in subcellular compartments, and translation elongation and initiation to be quantified in real time (see the figure, part b).
Puromycylation-proximity ligation assay relies on the proximity of two antibodies, one that recognizes the puromycilation modification of nascent chains after puromycin treatment and another that binds to the protein of interest297. The secondary antibodies provide an oligonucleotide platform that is first amplified and then recognized by fluorescent oligonucleotides to generate a specific signal corresponding to the site of translation. However, recent work suggested that this system may provide inaccurate translation measurements at the subcellular scale, prompting caution (see the figure, part c)298,299. Alternatively, the translation state of an mRNA can be extrapolated by measuring the diffusion properties of fluorescently labelled single mRNAs co-moving with ribosomes (see the figure, part d)128. mRNAs loaded with polysomes have a characteristic slow diffusion and corralled movement that is used to deduce the translation state of an mRNA in living cells.
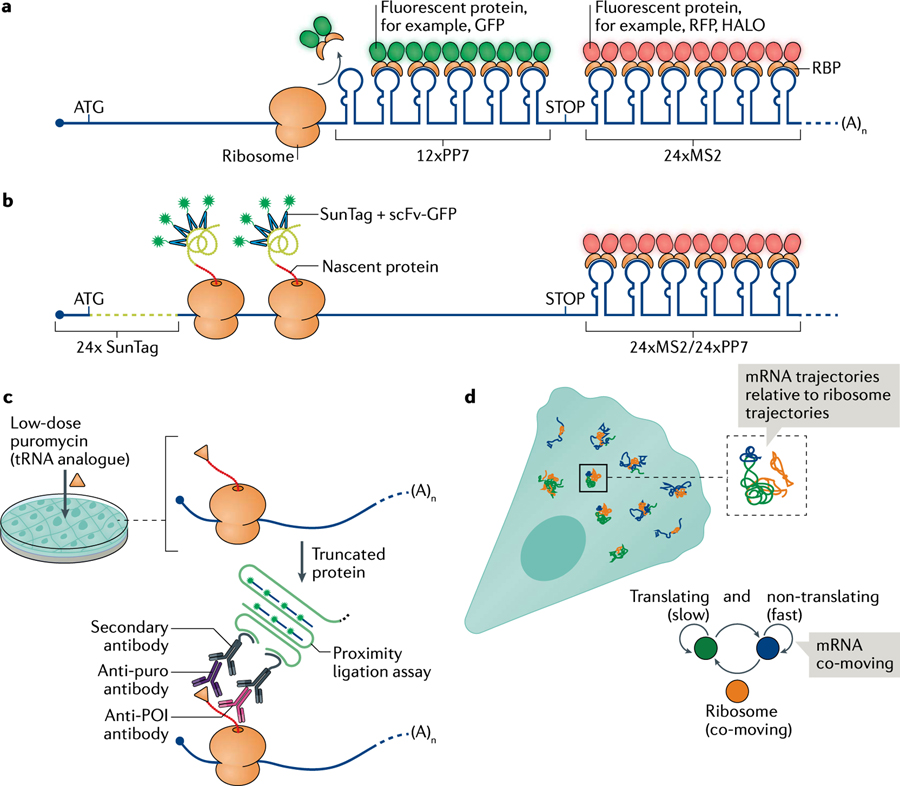
POI, protein of interest.
In C. elegans, essential maternal transcripts are delivered to the germ cells to support cellular differentiation during embryogenesis (FIG. 1d). Specific transcripts asymmetrically localize in subcellular compartments, such as P-granules (for example, nos2 and clu1), to the membrane (for example, erm1) or to the peri-nuclear region (for example, imb2)64–66. P-granules are heterogeneous RNA assemblies that segregate asymmetrically with the P-lineage during embryogenesis. smFISH demonstrated that the repression of mRNA translation is a pre-requisite for, and occurs prior to, the localization of mRNA in P-granules66. Thus, besides mRNA localization, the timing of local translation regulates post-transcriptional gene expression during the development of multicellular organisms.
Tissue functionality and homeostasis in the gut and the brain.
The development of in situ transcriptomics (Supplementary Box 1) has enabled the characterization of mRNA localization patterns within native tissue, advancing our understanding of tissue architecture and functionality. Mammalian gut physiology relies on the apical–basal polarization of epithelial enterocyte cells (FIG. 1e). The apical side absorbs nutrients from the intestinal lumen while the basal side excretes them to the blood. By combining smFISH, in situ transcriptomics and proteomics, researchers demonstrated the asymmetric localization of mRNAs and proteins in the intestinal epithelium and showed that the translation machinery has a preferential apical distribution that can be enhanced upon the refeeding of fasted mice67 (FIG. 1e). Interestingly, the apical localization of mRNAs encoding ribosomal proteins and the consequent synthesis of ribosomes on the apical side of the intestinal epithelium correlates with an increase in the translation efficiency of apically localized mRNAs, boosting nutrient absorption67. Thus, on top of mRNA localization, the dynamic localization of the translation machinery can fine-tune protein expression in response to environmental stimuli.
Cellular organization also determines functionality in the mammalian brain. The local transcriptome in neurons is vast and diverse (~2,500 mRNAs), as revealed by sequencing mRNAs in both the presynaptic (axonal) and postsynaptic (dendritic) sides of neurons13, and by profiling their translation status68,69. The localized pool of translating mRNAs maintains local protein homeostasis to allow for physiological processes such as proper brain wiring, response to injury and activity-driven changes in synaptic strength (synaptic plasticity). Although dendritic protein synthesis has well-documented functions in learning and memory, most studies on axonal translation have been restricted to axonal pathfinding, growth cone steering during development70–72 and regeneration upon axonal injury73,74. Recent technological advancements in visualizing mRNAs and ribosomes in tissue at high resolution are now enabling us to understand the role and mechanisms of translation in the presynaptic terminals of the adult mammalian brain.
In the past few years, combinations of multi-omics approaches and imaging have been developed to identify the localized transcriptome and map the precise location of individual transcripts in the intact brain. The tissue microdissection and biochemical purification of synaptosomes has identified hundreds of localized transcripts13,68. A technique called expansion-FISH (ExFISH) (Supplementary Box 1), which couples physical expansion of the tissue and FISH, reduced RNA crowding and enabled high-resolution mapping of mRNAs to both presynaptic and postsynaptic compartments in the adult mammalian brain68,75. An omics-approach using translating ribosome affinity purification (TRAP) characterized the local axonal ‘translatome’ for the first time, revealing that it comprised 1,000–2,000 mRNAs in developing and mature axons of retinal ganglion cells (RGCs)69. Later, coupling ExFISH with stable isotope labelling by amino acids in cell culture (SILAC) revealed activity-dependent regulation of the brain translatome in a compartment-specific manner68. Polyribosomes have been detected in dendritic shafts and spines76 by electron microscopy and seen to redistribute from shafts to spines in an activity-dependent manner77. Although challenges in visualizing polysomes in adult axons and terminals had raised concerns over the presence of translating mRNAs in these structures, immuno-electron microscopy using genetically tagged ribosomes69 and super-resolution methods have conclusively shown the presence of ribosomes in presynaptic terminals68,78, supporting the occurrence of local translation in axons of the adult brain. Future studies investigating how activity alters ribosome abundance and distribution within these compartments will provide insights into the dynamic regulation of local translation. We are now beginning to elucidate the molecular mechanisms underlying translation regulation in mature axons and its importance in axonal survival79,80, regeneration and synaptic plasticity during learning and memory78,81. As the transcriptome and translatome are dynamically regulated over time and upon activity69, it is possible that the repertoire of localized mRNAs and their translation status changes across different subcellular scales in the intact circuitry of the brain.
Given the increased sensitivity of RNA detection in tissue, it is now possible to investigate whether principles of mRNA localization and local translation in cultured cells and simpler organisms apply to mammalian tissues. The further development of in situ transcriptomics and imaging technologies should enable the discovery of nanoscale structures and RNA localization patterns in healthy and diseased tissues across different organs.
Localized cell biology
Key examples of mRNA localization and protein synthesis have been observed in different cell types (such as neurons and fibroblasts) and in organelles (such as mitochondria, the ER and centrosomes).
Localization in different cell types
The subcellular control of gene expression is often restricted to discrete sites within polarized cells, including in neurons during synaptic connectivity and in fibroblasts during cell migration, owing to the asymmetrical distribution of mRNAs.
Localization in neurons.
In neurons, the long-distance transport of mRNAs to dendrites and axons allows local translation in response to external signals, which is required for proper neuronal wiring during development and in processes such as learning and memory (reviewed in REFs25,82,83). The mRNAs encoding β-actin, ARC, CaMKIIα and BDNF, along with several others, are localized to dendrites and are responsible for the structural and functional remodelling of spines. The delivery of mRNAs to distal dendrites and to axon terminals, which are hundreds of microns away from the cell body, poses a notable challenge. Therefore, the packaging mRNAs into messenger ribonucleoprotein (mRNP) or granules (complexes of RNAs, RBPs, and the motor proteins dynein and kinesin) makes them ‘transport ready’ for movement along microtubules84–86. Although most studies indicate that mRNAs are translationally repressed during transport87, emerging work using SunTag-based reporters indicate that translation may occur during transit88 and/or in association with moving organelles89 (FIG. 2). The accessibility of mRNAs to ribosomes during transport is not well understood, and it is possible that the translational state of different mRNPs during transport may be regulated differently. How neurons achieve the precise and timed localization of each dendritic mRNA to specific synapses for local translation is still unclear (reviewed in REF.25). One idea is the ‘RNA signature’ hypothesis, indicating that each mRNA has a unique signature of regulatory elements that determines its transport, localization and translational control90. This RNA-centric view of how differential sorting is achieved and how the stability of these mRNAs determines transport and translation dynamics needs further investigation.
Fig. 2 |. Subcellular mRNA localization and local translation in neurons.
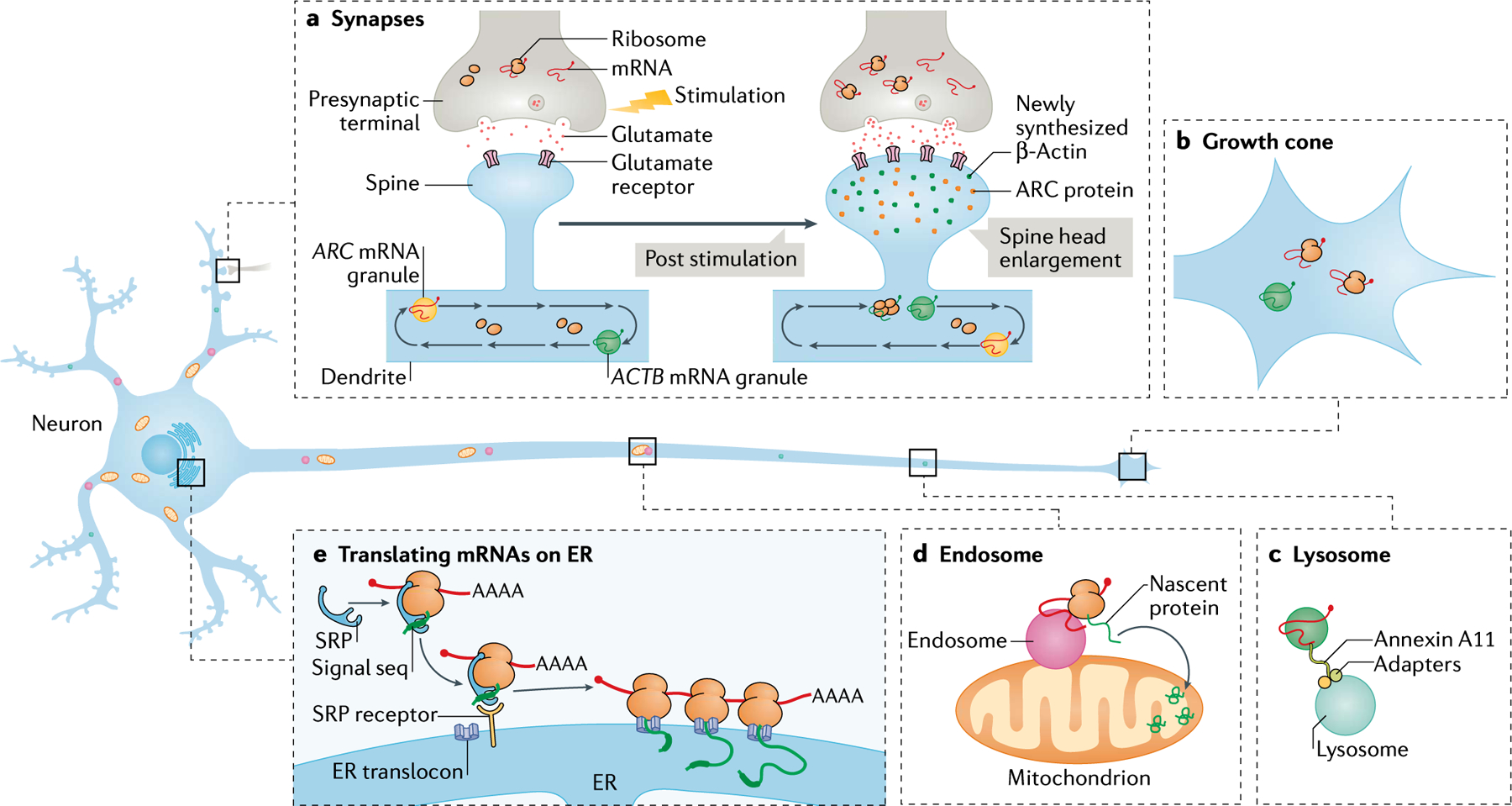
In neurons, mRNA localization and translation occur in processes (dendrites and axons). a | Neuronal transport granules, such as those containing ACTB and ARC mRNA, are trafficked along microtubules in dendrites like a conveyor belt patrolling multiple spines. The activation of specific synapses by stimulating the presynaptic terminal or by direct stimulation of postsynaptic spines using glutamate uncaging increases the binding of glutamate to the glutamate receptors. Synaptic stimulation leads to the capture of the moving mRNAs to the base of the stimulated spine, resulting in the localization and translation of mRNAs (for example ACTB mRNA). The newly synthesized proteins (green and orange dots) participate in enlarging the spine head and strengthening the synapse. Many dendritic mRNAs are localized following activity but it is unknown whether they all move and localize with similar kinetics. mRNA localization and local translation is also observed in the presynaptic compartment in response to stimulation. b | mRNAs such as ACTB mRNA are trafficked along axons to localize and translate in growth cones; this localization has critical roles in development and synaptogenesis. c | Long-distance mRNA transport in axons may also occur via the hitchhiking of mRNAs on lysosomes. The tethering of mRNAs to the lysosomal membrane occurs via proteins such as Annexin A11. d | Endosomes are often closely associated with the mitochondria and may behave as translation platforms for axonal mRNAs such as those encoding Lamin-B2 and VDAC2. The newly synthesized proteins are imported into and contribute to the function of mitochondria. e | mRNAs encoding secretory and membrane proteins are proposed to localize and translate using ribosomes on the endoplasmic reticulum (ER). Translation begins in the cytosol and the ER signal sequence on the nascent peptide gets bound by the signal recognition particle (SRP), which in turn binds to the SRP receptor on the ER membrane. Translation, often engaging polysomes, is resumed on the ER membrane and the nascent protein remains within the ER lumen, where it undergoes further processing.
mRNA encoding β-actin (ACTB mRNA) is the most well-characterized, localized mRNA in neurons. In dendrites, at a given time, only 10–20% of ACTB granules move bi-directionally, and the majority of the remaining granules are stationary91,92. Similar patterns of movement have been observed for other dendritically localized mRNAs, indicative of a ‘sushi-belt’ model, whereby mRNAs patrol multiple spines in a conveyor belt waiting for cues to be captured at a specific spine86 (FIG. 2a). Such cues include local synaptic activity, which can be mimicked in cell culture by uncaging the neurotransmitter glutamate on a subset of spines. This activation leads to the localization of ACTB mRNA to the base of these spines (~40% efficiency), possibly unmasking these granules for translation87,91. Although this model explains how ACTB mRNAs are locally enriched near stimulated spines, it is unclear whether this mechanism is conserved for other mRNAs. In particular, the validity of the model for low copy number and short-lived mRNAs, such as ARC, needs investigation. Once localized, mRNAs are usually translated in bursts of ~17 min, as observed using reporters containing the 3′ untranslated region (3′UTR) of β-actin93, followed by translational shutdown. Newly synthesized actin proteins are proposed to participate in structural changes in the spines91 (FIG. 2a). Again, the translation pattern for other dendritically localized mRNAs may vary, depending on the functions of their respective proteins in the spines.
Axons can grow up to a metre long and navigate across multiple lengths via growth cones during development to reach their postsynaptic targets. Growth cones require rapid protein synthesis to sense and undergo structural remodelling in response to extracellular cues, which is achieved by the presence of a readily translatable pool of mRNAs as seen in mammalian and X. laevis RGC neurons69,72. Indeed, localized Actb mRNA results in newly synthesized actin that enables growth-cone turning and synaptogenesis70,94 (FIG. 2b). Similar to dendrites, 14% of actb mRNA molecules in X. laevis axons were actively moving, and a bias in the anterograde direction resulted in growth cone localization94. Besides actin, specific ribosomal proteins and mTOR may be locally synthesized in axons, playing roles in axon branching of the developing brain95 and axonal regeneration after injury73. 3′UTR cis-regulatory elements that allow dendrite trafficking are also critical for the localization of mRNA in axons and growth cones, although consensus sequences that target mRNAs specifically to axons have not been identified. mRNAs may also hitchhike on organelles such as lysosomes and endosomes to achieve long-distance axonal transport89,96 (FIG. 2c). However, although this presents an efficient and economical choice for the neuron, the specificity of mRNA localization is difficult to explain with this model.
Neurons must also localize the translation machinery. Although the presence of polyribosomes in dendritic shafts and at the base of spines is well documented76,77, the paucity of these structures in presynaptic terminals97,98 led to uncertainties about the translation state of localized mRNAs in mature axons. A recent study using polysome profiling followed by sequencing demonstrated that notable amounts of protein synthesis in the synaptic neuropil (a region enriched in both axons and dendrites) of the adult rodent brain are likely to occur on monosomes rather than on polysomes99; neuronal cell bodies exhibited more polysome-driven translation than distal processes. Similar monosome-driven translation has been observed in the cue-specific translation of receptor-specific mRNAs, such as dcc and nrp1, in growth cones in RGC axons from X. laevis100. Monosomes potentially allow a diverse set of proteins to be produced from a limited pool of available ribosomes at synapses far away from the cell body83,99.
The translation efficiency in different neuronal compartments can be increased by increasing local mRNA–ribosome interactions via organelles (FIG. 2d,e). For instance, endosomes may bring together the translation machinery and mitochondria, providing ATP for translation (see below)89 (FIG. 2d). A notable number of synaptic proteins belong to the class of membrane proteins and secreted proteins and their translation on the ER is critical for proper folding101 (FIG. 2e). Although mRNAs encoding these proteins have been detected in the processes, their capacity for local translation remains questionable. Finally, the translation machinery may also be delivered to distal extremities via exosomes, as shown for the delivery of ribosomes from Schwann cells to axons of the sciatic nerve102 (reviewed in REF.103). Although neuronal activity leads to exosome release at synaptic terminals104,105, their contribution in effectively transferring genetic material and promoting translation requires further investigation.
The characterization of local translation in neurons has been extensively performed in primary cultures and often using overexpressed exogenous reporters. Although unravelling the molecular regulation of this process is useful, real-time imaging of mRNAs and their translation status in tissue are needed to identify how physiological activity affects the kinetics of their localization and translation. The tagging of endogenous genes with fluorescent proteins has been made possible in primary neurons with ORANGE, an optimized CRISPR–Cas9-based system106. So far, mRNAs encoding CaMKIIα, β-actin and PSD-95 have been tagged with the fluorescent protein Venus to study how chemically induced plasticity influences translation dynamics in real time107. It will be important to characterize the growing repertoire of localized mRNAs in neurons to elucidate which proteins need to be locally synthesized (for example, synaptic, cytoskeletal proteins and trophic peptides) for brain function.
Localization at the leading edge of fibroblasts.
Cell migration requires actin polymerization to promote the formation of lamellipodia and filopodia at the cell leading edge. This was shown in cell types such as fibroblasts108 and mesenchymal cells109, and the discovery that mRNAs coding for β-actin localize in fibroblast protrusions suggested that local translation upholds specific protein networks in subcellular compartments110. In lamellipodia and filopodia, actin filaments are physically coupled to integrins, which are heterodimeric receptors that bind to the extracellular matrix111. Integrins cluster at focal adhesion complexes (FACs), where they provide the mechanical tension required for cell movement and act as a scaffold for signalling molecules and ribosomal proteins. Ribosomes accumulate at the leading edge112, most likely through the interaction of the ribosomal kinase RACK1 with integrins113. Additionally, integrin–extracellular matrix binding is sufficient to recruit mRNAs114 and translation initiation factors115 to FACs to stimulate protein synthesis116.
The most widely studied mRNA localized to protrusions is that encoding β-actin, which has bipartite zipcodes in its 3′UTR110 (reviewed in REF.117) that are bound by zipcode-binding protein 1 (ZBP1; also known as IGF2BP1) with very high affinity118–122. The protein-interactome for ActB mRNA was mapped using a proximity-dependent biotin identification (BioID) assay in murine fibroblasts, which identified the RBP FUBP3 to be essential for the localization of ActB mRNA to the leading edge122. Besides the established roles of ZBP1 in translation regulation, it may also facilitate mRNA– motor binding123 and active transport along actin and microtubules124,125 (see below). In response to external cues or growth-factor stimulation, disassembly of the ZBP1–ActB mRNP occurs by Src-dependent phosphorylation of ZBP1 at the leading edge to allow the local translation of ActB mRNA126. The simultaneous imaging of fluorescently labelled 60S ribosomal subunits and single endogenous ActB mRNAs revealed that, in protrusions, efficiently translated mRNAs are associated with a high number of ribosomes127, namely polysomes. Interestingly, the accumulation of ribosomes next to FACs decreases mRNA diffusion speeds from 0.4 μm2/s to 0.1 μm2/s, indicative of local translation128. Of note, inferring the translation status based on subtle changes in diffusion coefficients is often difficult, and further studies are required to establish a functional link between the local translation of ACTB mRNA at the FACs and cell migration.
A plethora of mRNAs besides ACTB mRNA are differentially distributed in polarized cells, but the extent to which their local translation maintains the front–back asymmetry necessary to promote cell migration is unclear. RNA-sequencing and pulse-SILAC have been combined to determine the relative translation rates (BOX 3) between protrusions and the cell body. The mRNAs encoding for actin cytoskeleton-associated proteins were not enriched at the protrusions despite being heavily translated there. In fact, the translation of mRNAs encoding for mitochondrial and ribosomal proteins was repressed at the protrusions, despite these mRNAs being locally enriched129. Given that the number of mRNAs does not always correspond to translation efficiency, translation rates will have to be determined at single-molecule resolution in living cells in order to fully understand the role of the localization of each mRNA.
Localization at different organelles
Organelles mediate compartmentalization and the regulation of many intracellular processes. Protein targeting to organelles occurs via peptide sequence-based targeting to a specific organelle or via mRNA localization and in situ translation. With increased imaging resolution and proximity labelling assays (BOX 3), how mRNAs are regulated at the subcellular scale on different organelles as well as the potential implications on organelle biogenesis and maintenance have been studied.
Outer mitochondrial membrane.
Mitochondria are essential for ATP synthesis through oxidative phosphorylation, and their activity impacts all aspects of cell physiology. Mitochondria distribution within cells is dynamic, and continuous reshaping of the mitochondrial network supports the local energy demands. For instance, in neurons, mitochondria in dendrites supply the energy required for local translation and synaptic plasticity130. Nuclear transcripts encoding mitochondrial proteins, including those that encode mitochondrial inner membrane proteins, are enriched near the mitochondria52,131,132. Using the MS2–MCP system in S. cerevisiae, up to 24 mRNAs were dynamically visualized in the proximity of mitochondria and in response to changing environments51,133.
The 5′UTR and 3′UTR sequences of mRNA control its localization near mitochondria. In yeast, the 3′UTR contains a cis-regulatory element bound by RBP Puf3, which directs the mRNA to mitochondria51,134. In D. melanogaster and mammalian cells, the 5′UTR interacts with the outer mitochondrial membrane (OMM) kinase PINK1. Interestingly, in mammalian cells, the translation of nuclear-encoded mRNAs coding for mitochondrial proteins is regulated by mTOR135. mTOR phosphorylates a family of inhibitory proteins called 4E-binding proteins (4E-BPs). Phosphorylated 4E-BPs are released from the cap-binding protein eIF4E to promote the assembly of the eIF4F complex, the composition of which is dictated by specific features of the 5′UTRs of nuclear-encoded mitochondrial mRNAs136. Thus, the 5′UTR of nuclear-encoded mitochondrial mRNAs may coordinate their localization and translation.
Although the majority of mitochondrial proteins are synthesized by cytoplasmic polysomes and imported via peptide-dependent targeting137–139, proximity-specific ribosome profiling (BOX 3) revealed the translation of certain nuclear-encoded mitochondrial mRNAs near the OMM in yeast140 and higher eukaryotes141. However, these studies lacked the resolution to elucidate whether ribosomes are tethered to OMM for co-translational import or if they are present in close proximity to the membrane. High-resolution electron cryo-tomography resolved this conundrum by revealing ribosomes on the surface of OMM with the peptidyl exit tunnel oriented to favour the import of elongating polypeptides into the mitochondria. Furthermore, ribosomes appeared to be tethered to the import pore translocase of the outer membrane complex by nascent polypeptides142. Translating ribosomes and mitochondria appear to be associated through an interaction between the ribosome-associated nascent chain-associated complex and the OMM protein (Om14 in yeast) to support co-translational mitochondria import or, as described in D. melanogaster and mammalian cells, through an interaction between the mitochondria-targeting sequence in the nascent peptide and the receptor Tom20 (REF.143).
Single-molecule imaging using the SunTag system (BOX 3) revealed that mitochondrial protein synthesis is also facilitated by the physical association of mitochondria with late endosomes89. This interaction occurs in X. laevis RGC axons, in which late endosomes contact mitochondria for over 2 min and serve as translation sites for mRNAs encoding proteins such as Lamin-B2 and VDAC2. These newly synthesized proteins are likely imported into mitochondria to perform structural functions. Furthermore, the global perturbation of late endosomes by the disruption of Rab7a function impairs local mRNA translation but not mRNA localization. Interestingly, the disruption of the axonal translation of mitochondria mRNAs, as occurs in Charcot–Marie–Tooth type 2B neuropathy, impairs mitochondrial integrity and axonal activity89.
Further work is needed to understand whether the spatiotemporal regulation of local translation at or near the OMM affects mitochondria biogenesis and physiology.
Endoplasmic reticulum.
Electron microscopy studies have identified two major populations of ribosomes in cells: ER-bound ribosomes and those freely diffusing in the cytoplasm. Although cytoplasmic ribosomes are abundant in all cell types144, ER-bound ribosomes are enriched in secretory cells, where they preferentially translate mRNAs encoding secreted and integral membrane proteins. The translation of ER-bound mRNAs begins in the cytoplasm. Once a nascent polypeptide containing a hydrophobic domain (that is, a signal sequence) emerges from the ribosome, it is recognized by the SRP complex. SRP binding halts translation until the mRNA is relocated to the ER145 (FIG. 2e). This dynamic process has been visualized by imaging the nascent polypeptide on translating mRNA at single-molecule resolution93. Briefly, the N-terminal domain of the membrane protein cytochrome p450, which anchors the transmembrane domain into the ER membrane146, was fused to a SunTag peptide array, and the encoding mRNA was simultaneously imaged with the MS2 system. Real-time imaging of this reporter mRNA confirmed that translation starts in the cytoplasm and elongation occurs on the ER93. Thus, mRNA localization to the ER is translation-dependent and SRP peptide sequence-dependent and differs from ‘zipcode’-dependent mRNA localization.
Several studies have challenged the canonical models predicting that the translation of cytoplasmic mRNAs is excluded from ER membranes. Cell fractionation and genome-wide approaches have identified cytoplasmic mRNAs and translation initiation factors in proximity of the ER, suggesting that de novo translation of cytosolic mRNAs can occur on the ER membrane147; however, the physiological relevance is unclear. GAPDH mRNA is a classic example of ER-associated cytosolic mRNA148–150. However, given the abundance of this housekeeping transcript and the resolution of the methodologies used, these findings need further validation. For improved spatial resolution, smFISH (Supplementary Box 1) was employed to detect GAPDH and other cytosolic mRNAs associated with the ER membrane upon a mild digitonin extraction, which removes soluble cytoplasmic molecules while preserving ER integrity151. Imaging with the translation reporter ‘translating RNA imaging by coat protein knock-off ‘ (TRICK)152 (BOX 3) in living cells revealed that a small fraction of cytosolic mRNAs was indeed translated on the ER. Interestingly, the number of ribosomes in the ER extract was higher compared to the cytosolic counterpart, suggesting that the translation of cytoplasmic mRNAs may be more efficient at the ER than in the cytosol151.
Compartmentalizing mRNAs to the ER may play a critical role in maintaining the translation of specific mRNAs under stressful conditions. Viral infection and other cytotoxic stresses suppress cytoplasmic cap-dependent translation while ER membrane-bound mRNAs are able to partially escape this silencing153,154.
Centrosomes.
Centrosomes — membraneless organelles composed of two centrioles surrounded by the pericentriolar material155 — are the microtubule-organizing centre of the cell and participate in chromosome segregation and cell division. The asymmetric localization to and local translation of mRNAs at the centrosome may contribute to asymmetric cell division, leading to embryonic pattering and the selective inheritance of specific transcripts21–23,156,157.
Cell cycle-dependent mRNA localization to centrosomes can be translation dependent or translation independent. The translation-dependent transport of mRNAs is observed in zebrafish and mammalian cells, where mRNAs are co-translationally delivered to centrosomes through a microtubule-dependent and dynein-dependent process22,156–158. Polysomes translating PCNT mRNA, encoding a core component of the pericentriolar material, are attached to the dynein motor complex through the LIC1 domain located in the N-terminal of the PCNT nascent peptide both in zebrafish and mammalian cells22. In mammalian cells, the translation-dependent localization of PCNT mRNA and of the mRNA encoding the microtubule minus-end regulator ASPM to the centrioles enables protein localization within 30 min. ASPM and the microtubule-binding protein NUMA1 were tagged by combining the SunTag and MS2 systems to label the nascent peptides and the mRNA, respectively (BOXES 2,3), further demonstrating that mRNA localization depends on the nascent peptide connecting polysomes to motors. Both mRNAs, when translationally active, were directly and actively transported to centrosomes156,158. Finally, in quiescent, immortalized human retinal pigment epithelial cells, NIN mRNAs, encoding a core component of centrosomes, localize at the centriole basal body found at the base of cilia157. NIN mRNA localization is both translation dependent and exon junction complex dependent157, suggesting a tight connection between the different steps of mRNA processing.
The translation-independent, cell cycle-dependent localization of mRNA to centrosomes occurs via the interdependent localization of two mRNAs to the centrosome. In D. melanogaster, a pair of antisense mRNAs, Ik2 (encoding IκB kinase like 2) and Cen (encoding centrocortin), share complementary sequences in their 3′UTRs, leading to base-pairing and co-transport23. Both mRNAs localize to the centrosome but the localization of Ik2 depends on that of the Cen transcript. The simultaneous localization of Cen mRNA and nascent peptides suggests that Cen mRNA is locally translated at the centrosome. Interestingly, the orthogonal analysis of APEx-seq data indicates that antisense mRNA pairs tend to co-localize in specific subcellular compartments23, suggesting a novel mechanism of coordinated mRNA localization.
Mechanisms of mRNA localization
Three major mechanisms control the localization of mRNAs to subcellular compartments: directed transport, protection from mRNA degradation, and passive diffusion and local entrapment.
Localization by active mRNA transport
Active transport is the most common mode of mRNA localization reported in all eukaryotic cells. Motor-driven transport in cells and organisms occurs on actin filaments or microtubules (FIG. 3a–d). Here, mRNAs are targeted through the binding of RBPs to cis-regulatory elements, which are characterized by unique secondary structures and dubbed ‘zipcodes’. These cis-elements are found in 3′UTRs and 5′UTRs as well as in coding regions (reviewed in REFs19,33,159) and can vary in length and in the diversity of RBPs recognizing them. The recognition is based on the primary sequence (for example, a 54-nulceotide sequence with bipartite motifs in chicken and mouse Actb mRNA is specifically bound by ZBP1 (REF.117)) and also on the basis of complex secondary structures (for example, the zipcode structure of S. cerevisiae ASH1 mRNA160, the helical structure of D. melanogaster bcd mRNAs, and G-quadruplexes in neuronal mRNAs such as those encoding CaMKIIα and PSD95)161. Across different cell types and organisms, mRNAs are moved by myosin, kinesin and dynein motors. How different RBPs recruit motor proteins to form a localization-competent transport granule is unclear and complicated by the fact that a single mRNA may be bound by multiple RBPs with unknown roles in transport regulation. Evidence of an RBP directly binding to motor proteins and bridging the interaction with the mRNA comes from yeast, where She3 binds Myo4 to move mRNAs along actin, although RBPs can also indirectly engage motors via adaptor proteins. Indeed, the APP tail 1 (PAT1) protein is a direct adapter between ZBP1 and the kinesin 1 motor complex, facilitating the neuronal activity-induced transport of ActB mRNA to dendrites162.
Fig. 3 |. Modes of mRNA transport and localization in cells and organisms.
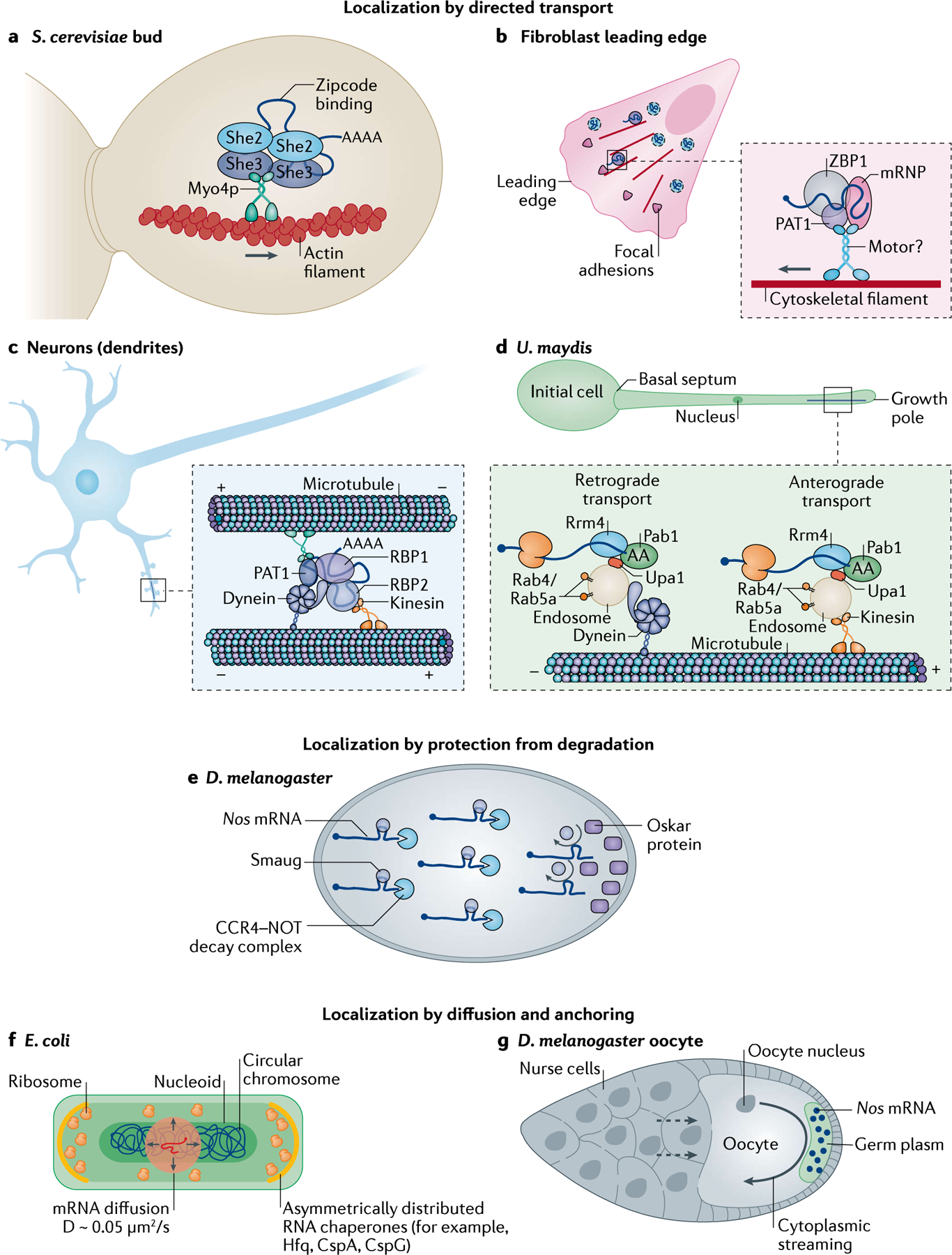
a | Several mRNAs are localized to the bud of Saccharomyces cerevisiae. She2 dimerizes and binds these mRNAs via their zipcodes, before binding She3, which bridges the interaction of the complex with the type V myosin motor Myo4. The ribonucleoparticles are actively transported along actin filaments. b | In mammalian fibroblasts, mRNAs such as those encoding β-actin are localized to the leading edge by RNA binding proteins (RBPs) such as ZBP1, which binds to the zipcode on the 3′ untranslated region (3′UTR) of the mRNAs to form messenger ribonucleoproteins (mRNPs) that associate with unidentified motors. PAT1 acts as a direct adapter between ZBP1 and the motor. This represents a small percentage of mRNA movement as the majority of mRNAs undergo corralled cytoplasmic diffusion (indicated by the dashed boundaries). c | Localization to distal spines is achieved by packaging mRNAs involved in synaptic remodelling into transport granules composed of RBPs, the minus-end-directed motor dynein and the plus-end-directed motor kinesin. Due to the mixed polarity of microtubules in dendrites and the presence of both motors, these granules move bi-directionally (that is, in anterograde and retrograde motion). The net movement is proposed to occur by a ‘tug-of-war’ between the motors determined by their stoichiometry. d | In Ustilago maydis, cells switch from yeast to filamentous growth to promote plant invasion. To sustain asymmetric growth, protein, ribosomes and mRNA are transported to the growth pole. mRNAs are bound to endosomes via the endosome membrane-binding protein Upa1, which mediates the interaction with the RBPs Rrm4 and Pab1. Bi-directional mRNA transport on microtubules occurs via kinesins (anterograde motion) or dyneins (retrograde motion). e | In Drosophila melanogaster embryos, Nos mRNAs are bound by the RBP Smaug, which recruits the CCR4–NOT complex to initiate mRNA decay. At the posterior pole, however, Nos mRNAs are protected from degradation by Oskar proteins, which displace Smaug to increase local concentrations of Nos mRNAs. f | In Escherichia coli, mRNAs localize to ribosome-rich poles or to the membrane by random diffusion at speeds of 0.05 μm2/s, aided by the chaperone proteins that anchor the mRNAs. g | During D. melanogaster oogenesis, several hundreds of mRNAs are deposited to the oocyte by nurse cells (dashed arrows). mRNAs such as nos are localized to the posterior pole of the oocyte by cytoplasmic streaming and entrapped in the germ plasm in an actin-dependent manner.
Transport on the actin cytoskeleton.
In yeast, bud-localized ASH1 mRNA has four zipcodes, distributed across the coding region and the 3′UTR, that are recognized co-transcriptionally by the RBP She2 and its partner Loc1 (REFs163–167). The synergistic binding of She2 dimers to the ASH1 mRNA induces a conformational switch that promotes high-affinity mRNA–She2 binding160. Loc1 is dissociated from the mRNP before export into the cytoplasm168, and the She2–mRNA complex is intercepted by She3, which is constitutively bound to the type V myosin motor Myo4 (REFs160,165–167,169). This interaction promotes the formation of a translocation-competent mRNP that is actively transported to the bud on actin filaments (FIG. 3a). Besides ASH1 mRNA, tens of mRNAs, including CLB2, TCB2, TCB3 and IST2 (REFs35,170), interact with the She2–She3 complex and are actively transported along actin filaments. Further work is required to characterize how their transport and local translation is coordinated. In mammalian cells, the transport of Actb mRNAs to the leading edge of migrating fibroblasts occurs on both actin and microtubules, with ZBP1 playing roles in the transport via its interaction with motor proteins123,124,171 (FIG. 3b). Interestingly, interactions between actin and myosin may contribute to transport efficiency124 but whether ZBP1 results in increased myosin motor engagement needs further study.
Transport on microtubules.
In mammalian cells, motor protein-based active transport is best illustrated by the localization of mRNAs to distal dendrites and axon terminals in neurons via the movement of transport granules (comprising RNAs with RBPs and dynein and/or kinesin) along microtubule tracks84–86 (FIG. 3c). In other cell types, such as in fibroblasts and epithelial cells, active transport localizes a small percentage of mRNAs, and corralled diffusion is often sufficient to move mRNAs128 (FIG. 3b). ACTB mRNA is the best-characterized neuronal transport granule and moves processively at speeds of 0.5–2 μm/sec in both dendrites and axons91,92. Interestingly, these speeds are similar for other microtubule-dependent, dendritically localized mRNAs, such as ARC172, and for exogenous reporters containing 3′UTRs with localization elements173,174. Mostly, dendritic mRNAs exhibit oscillatory movement, switching between anterograde and retrograde directions. This bi-directionality has been attributed to the mixed orientation of the microtubules and a possible ‘tug-of-war’ between the dynein and the kinesin motors, which together determine the net movement direction25,159. One limitation of motor-driven transport is the energy cost for the cell, especially for long axons. To circumvent this problem, mRNAs can hitchhike on organelles such as lysosomes for long-distance transport in axons96 (FIG. 2c). The tethering of mRNAs on actively moving lysosomes is facilitated by Annexin A11 via its intrinsic membrane binding and phase-separating domains. Similar hitchhiking on early and late endosomes has been observed for axonal mRNA transport89.
This hitchhiking strategy also occurs in filamentous fungi, including in the corn plant pathogen U. maydis, in which a close link between mRNA transport and the endocytic pathway was demonstrated56 (FIG. 3d). Endosomes, which are membrane-bound vesicles formed during endocytosis, are important for membrane and lipid trafficking and for transporting cargos such as membrane proteins, cell debris, bacteria and viruses as well as mRNAs175. Work in U. maydis showed that the CDC3 mRNA and the encoded septin protein, a key regulator of unipolar growth, are co-transported on the same endosomes57,176. Ribosomal proteins are also co-transported, suggesting that translation occurs during trafficking176. The RBP Rrm4 is essential for shuttling both the mRNAs and ribosomal proteins, further corroborating the hypothesis of ‘in-motion’ mRNA translation176,177. The mRNA and the RPB Rrm4 as well as the poly-A binding protein Pab1 interact with the endosome via the membrane-bound protein Upa1 (REF.59). This complex is transported bi-directionally on microtubules by the plus-end-directed kinesin Kin3 or by the minus-end-directed dyneins Dyn1 and Dyn2 (REFs178,179). As mRNA localization coupled to vesicle transport (endosomes or lysosomes) also occurs in neuron cells89,96, this active-transport pathway might be highly conserved.
Therefore, future identification of the RBPs and the motors associated with transport granules from different cellular compartments will provide insights into the reorganization of granules (see below). Although motors are bound to mRNAs until they reach their final destination, it remains unclear whether motor proteins are exchanged during granule movement, whether motors are disengaged from mRNAs upon localization and what molecules halt mRNA movement.
Protection from mRNA degradation
In the D. melanogaster embryo, cell-fate specification is achieved by the precise distribution and translation control of mRNAs encoding patterning factors. Cytoplasmic Nos mRNA is targeted by the RBP Smaug (via the Smaug recognition element located in the 3′UTR), which inhibits Nos mRNA translation and recruits the CCR4–NOT complex to trigger Nos mRNA decay in the cytoplasm180,181. Later in development, in the germ plasm at the posterior pole, Oskar displaces Smaug from the Nos mRNAs, thus protecting the mRNA from degradation and relieving translation inhibition181 (FIG. 3e). The Hsp83 mRNA was protected from degradation by a similar mechanism although the Smaug recognition element is located in the coding sequence of Hsp83 mRNA182–184. Around 300 mRNAs are associated with Smaug in D. melanogaster182; however, further investigations are required to elucidate if they are protected from degradation by Smaug and if this regulates their localization.
Passive mRNA diffusion and anchoring
In tiny organisms like bacteria, biomolecules diffuse rapidly within the 1–2 μm long cell26,27. mRNAs typically move with a diffusion coefficient of 0.05 μm2/s. These parameters suggest that the localization to ribosome-rich poles or to the membrane could occur within seconds of mRNA exiting the nucleoid, a timeframe notably shorter than the half-life of bacterial mRNA (~5 min)185. Consistent with these findings, no active mRNA transport mechanism has been observed in bacteria to date, even though the directed transport of proteins has been previously described186. Furthermore, the asymmetric distribution of RNA chaperones (for example, Hfq, CspA and CspG) could partake in the anchoring and localization of mRNA187 (FIG. 3f); however, additional studies are required to elucidate the mechanisms governing these events.
During D. melanogaster oogenesis, nurse cells contract, squeezing their cytoplasm and depositing hundreds of mRNAs into the oocyte, including Nos mRNA10. Nos mRNA diffuses towards the posterior pole owing to the cytoplasmic streaming that microtubules generate for transport20,24. Once at the posterior pole, Nos mRNAs are entrapped in the germ plasm in an actin-dependent manner (FIG. 3g). Nos mRNA localization and localized translation are essential for the anterior–posterior body axis patterning in D. melanogaster62,188.
Regulation of local translation
Although it is clear that the subcellular localization of mRNA regulates gene expression, the mechanisms regulating localized mRNA translation are only just emerging. The development of methods that can simultaneously measure mRNA localization and localized translation in fixed and living cells (BOX 3) are starting to unravel how translation factors and ribosomes are locally regulated to modulate protein synthesis, revealing that a major control step occurs during translation initiation.
Inhibition of translation by RBPs
A number of RBPs that are involved in mRNA transport also play roles in translation repression, thereby coupling the regulation of both processes189. For example, ZBP1-bound ACTB mRNA is packaged into transport granules, which are translationally silent during trafficking in dendrites and fibroblasts. Only upon localization to activated spines in neurons and to the leading edge in fibroblasts ZBP1 is phosphorylated, which unmasks mRNAs for translation117,126. Similarly FMRP, which is widely expressed in human and mouse, plays a role in transporting mRNAs in translationally repressed granules190. In X. laevis, RBPs, such as heterogeneous nuclear RNPs, bind to both ribosomes and mRNAs coding for guidance receptors in the growth cones, thereby keeping the mRNAs in a translationally repressed state under basal conditions. Upon cue stimulation, the binding of the cognate ligand to the specific receptor (DCC or Nrp1) causes the RBP to dissociate and triggers the translation of the specific receptor mRNA100.
All eukaryotic mRNAs possess a 5′-end cap structure, and cap-dependent translation is a multistep and highly regulated process orchestrated by a network of translation initiation factors. At initiation, the cap-binding protein eIF4E recruits eIF4G and the helicase eIF4A to the mRNA and assembles the eIF4F complex191. The eIF4F complex in turn contacts elements of the 48S translational initiation complex192 (FIG. 4a), and polypeptide synthesis starts when small and large ribosomal subunits join the mRNA at the start codon.
Fig. 4 |. Regulation of translation by RNA binding proteins.
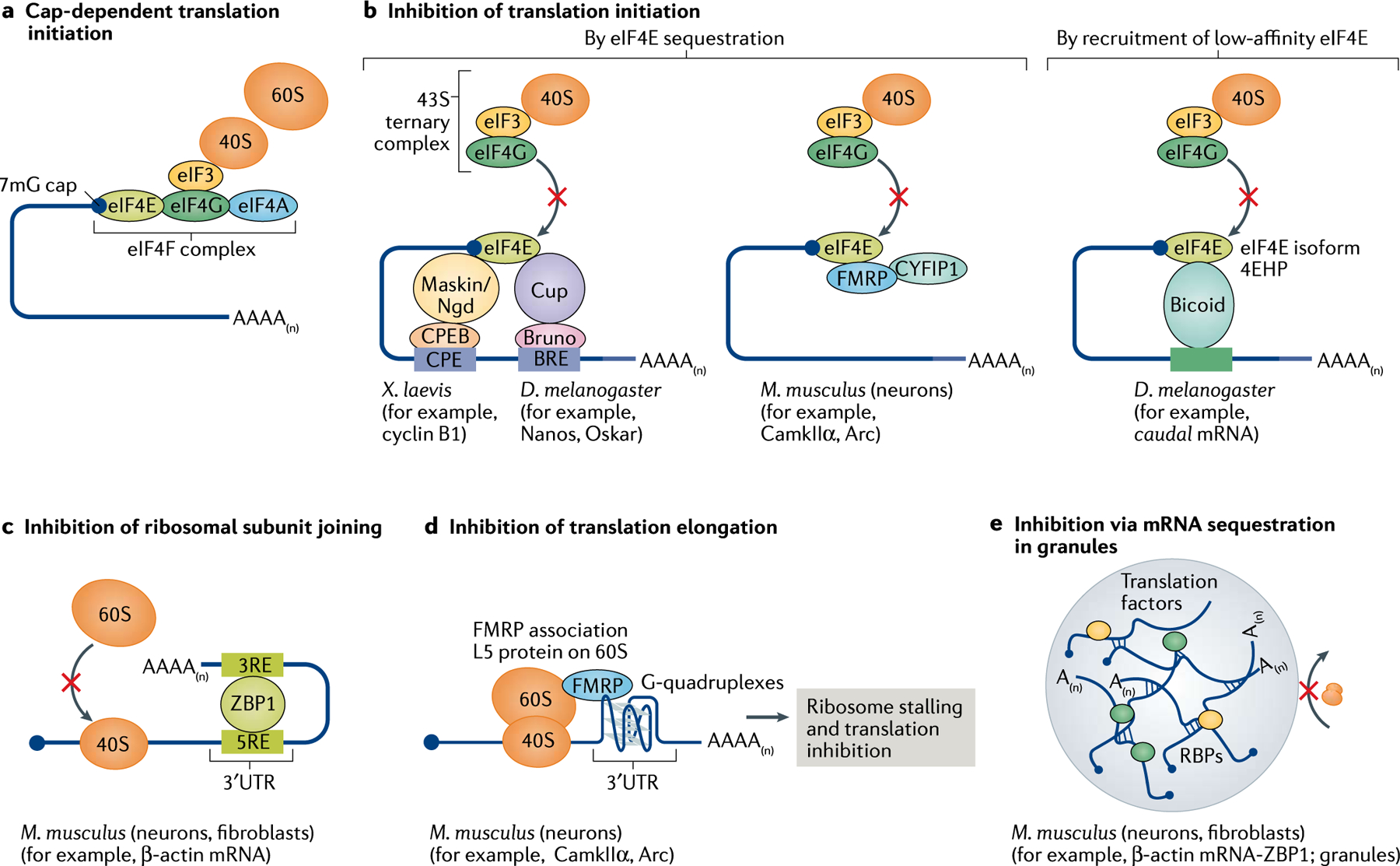
a | Eukaryotic cap-dependent translation initiation occurs when the 40S ribosomal subunit binds to the 7mG-containing cap at the 5’ end of the mRNAs via an interaction involving eIF3 and the eIF4F complex of initiation factors eIF4A–eIF4G–eIF4E. b | Translation initiation is prevented when eIF4E (bound to the cap) is sequestered by 4E-binding proteins (4E-BPs) such as Maskin (in Xenopus laevis) and Cup (Drosophila melanogaster). These 4E-BPs are tethered to the mRNA by RNA binding proteins (RBPs) such as cytoplasmic polyadenylation element binding protein (CPEB) and Bruno, which bind to the cis-regulatory elements CPE and BPE, respectively, in the 3′ untranslated region (3′UTR) of the mRNA. In mammalian cells, RBPs such as FMRP and cytoplasmic FMRP-interacting protein 1 (CYFIP1) may directly interact with eIF4E and prevent it from binding to the preinitiation complex. Also, in D. melanogaster, RBPs such as Bicoid bound to the mRNA recruit an isoform of eIF4E known as 4EHP, which has a low affinity for eIF4G and is therefore unable to initiate translation. c | Some RBPs, such as ZBP1, do not impact the initiation of translation but prevent the 60S ribosomal subunit from joining the 40S subunit to assemble the 80S complex. d | RBPs may also stall elongating ribosomes as seen when FMRP binding to the L5 protein on the 60S subunit halts translation. e | Several RBPs, via protein–protein interactions, may sequester multiple mRNAs into transport granules or stress granules, which are believed to be mostly translationally silent.
Cytoplasmic polyadenylation element binding proteins (CPEBs) are a family of well-characterized RBPs that inhibit pre-initiation complex formation. CPEB1 binds near the 3′UTR of mRNA and recruits a set of 4E-BPs, such as Maskin in oocytes193, Cup in D. melanogaster194 and Neuroguidin in neurons195. By binding to eIF4E, these proteins block the interaction between eIF4G and eIF4E, thereby interfering with translation initiation (FIG. 4b). Similarly, the binding of eIF4E to FMRP and the cytoplasmic FMRP-interacting protein 1 (CYFIP1) inhibits eIEF4F complex formation in neurons196, although the role of this interaction in inhibiting translation is debatable as most studies point to a more direct role of FMRP in inhibiting translation elongation197,198. RBPs such as ZBP1 negatively impact the assembly of the 80S complex by impairing the joining of the large and small ribosomal subunits126, a mechanism that is predominant in the perinuclear region of fibroblasts and in neuronal transport granules120 (FIG. 4c). Some RBPs, for example FMRP, may operate at several steps; besides negatively affecting initiation, FMRP blocks elongation by binding to the L5 protein on the 80S ribosome, precluding the binding of tRNAs and translation elongation factors197 (FIG. 4d). Finally, several RBPs can cluster mRNAs into higher order granules via protein–protein interactions (see below) such as in stress granules and transport granules, in which ribosomes and the other components of the translation machinery have limited accessibility (FIG. 4e).
Promoting translation in ‘translation factories’
How translation initiation is regulated upon mRNA localization is not fully understood, and this question is further complicated by the existence of several translation factor homologues as well as of ribosomes with heterogeneous composition199. One concept is that ‘translation factories’, that is, molecular assemblies promoting de novo translation, exist in the cytoplasm156,158. It has been proposed that eIF4F variants cluster together in response to extracellular stimuli200, although it is not clear which fraction of the variants are employed in local factories. A recent translation biosensor designed to compare cap-dependent and IRES translation efficiency at single-molecule resolution demonstrated that translating mRNAs are in close proximity of each other while excluding non-translating mRNAs201. Additionally, mRNAs undergoing cap-independent translation localize more closely to the nucleus than mRNAs translated in a cap-dependent manner201. In X. laevis growth cones, translation factories have been observed close to the membrane, and cue-specific stimulation promotes the synthesis of certain receptor-associated mRNAs, such as ctnnb1 mRNA (encoding β-catenin), that act locally at those specific receptors100. This observation suggests that translation preferentially occurs in some subcellular regions, creating translation hotspots or foci for specific mRNAs202. The concept of translation in specific foci was initially proposed in neuronal dendrites203 and subsequently identified in both shafts and in spines204–206, axonal branch points192 and, recently, in live axon terminals in the intact brain72. In non-neuronal cells, the accumulation of mRNAs in distinct cytoplasmic foci along with their encoded proteins (for example, BUB1, DYNC1H1, β-catenin (encoded by CTNNB1) and ASPM) has been observed to occur in a translation-dependent manner156,158. These foci, in which the translation machinery is localized either separately or along with the mRNAs, have been termed translation factories. Based on these studies, whether ‘hotspots’ and ‘factories’ can be used interchangeably to indicate the clustering of translating mRNAs needs to be elucidated.
Furthermore, in budding yeast, mRNAs coding for translation initiation, elongation and termination factors are packaged in ‘translation factor mRNA granules’. These granules are localized to the bud in a She2–She3-dependent manner, that is, via the same complex that transports bud-localized mRNAs53. Similarly, in neurons, ribosomes may be co-trafficked with mRNAs in dendrites, and assembly of the ribosome subunits occurs once translation repression is relieved by synaptic activity87. It is well known that several translation initiation factors localize to and are stored in stress granules in response to harmful situations in eukaryotes207,208. Finally, translation initiation can be promoted indirectly through the control of ribosome availability. As discussed above, in the intestinal epithelium, the apical localization of mRNAs encoding ribosomal proteins leads to a local increase in ribosome concentration, boosting the translation of apically localized mRNAs67 (FIG. 1e).
Creating translation hotspots and factories is an effective way for a cell to translate rapidly in response to cues and to allow these newly synthesized proteins to act locally without perturbing the protein homeostasis of the entire cell. Further work is required to elucidate the coordinated localization of mRNAs and the translation machinery. In eukaryotes, recent ribosomal profiling has provided a systematic analysis of the mechanisms controlling the co-translational assembly of multi-subunit protein complexes209,210. The development of imaging techniques that allow the simultaneous visualization of multiple translated mRNAs and regulatory factors will be critical to characterize the coordinated translation of multiple components in situ and to determine the composition and function of these translation factories or hotspots.
RNA composition and fate in granules
Across different cell types and organisms, mRNPs are packaged into larger RNA granules. They come in different shapes and sizes and play roles in mRNA trafficking in neurons (transport granules) and in D. melanogaster (germ granules) as well as in storage and translational regulation in stress granules and P-bodies. Most granules are highly dynamic in nature; therefore, a unified model of granule assembly and disassembly is lacking owing to the technological challenges of accurately identifying the stoichiometry of all granule components at a given time. Multiplexed FISH and methods to map RNA–protein interactions using candidate-based approaches such as smFISH-IF211 and global approaches such as APEX-seq132,212 opened avenues for determining the complexity of granule composition (FIG. 5a,b, Supplementary Box 1). The term ‘granule’ has been loosely used to indicate all large assemblies of RNAs and proteins that promote selected mRNPs to come together and distinguish themselves as a granule.
Fig. 5 |. Granule composition and organization.
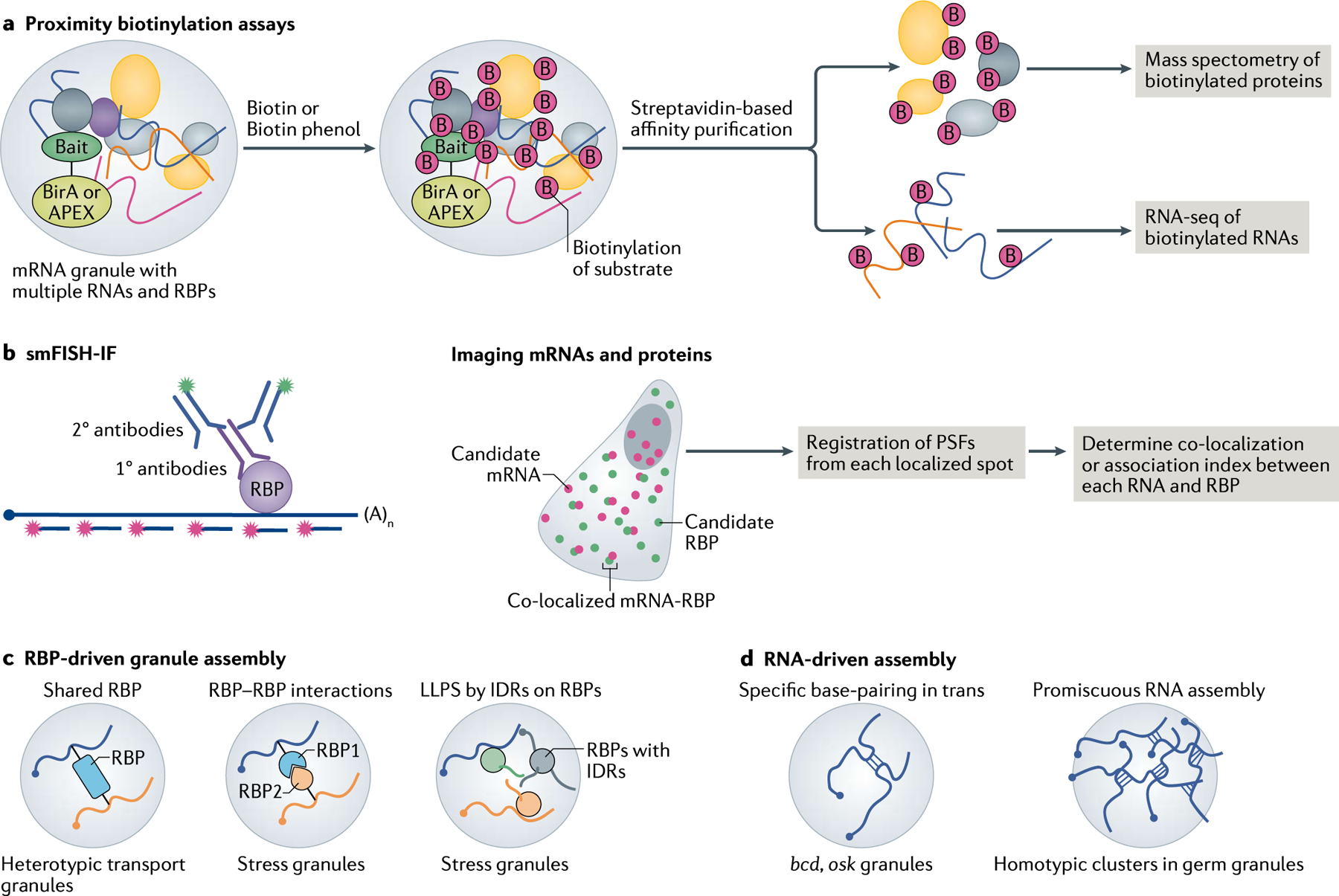
a | Global analysis of mRNA–protein interactions can be performed with proximity-based labelling techniques using ascorbate peroxidase (APEX) or the biotin ligase BioID assay (also known as BirA). A bait protein is fused to the APEX or BirA enzyme, the latter of which biotinylates proteins within a 10 nm radius. In the presence of biotin or biotin phenol, APEX generates short-lived radicals that covalently react with tyrosine and other electron-rich amino acids as well as with amino groups on guanosine. The biotinylated proteins and RNAs are enriched by streptavidin pull-down and identified by mass spectrometry and RNA sequencing (RNA-seq), respectively. b | Imaging-based approaches such as single-molecule fluorescence in situ hybridization (smFISH) combined with immunofluorescence (IF) allow users to simultaneously detect individual mRNAs (using labelled oligonucleotides) and their interacting RNA binding protein (RBP; using antibodies) in situ. By precisely localizing these molecules inside cells and registering the point spread functions (PSFs) of the spots, quantitative measurements of mRNA–protein association are obtained. c | During RBP-driven granule formation, RBPs are shared between multiple mRNAs at the same time, leading to the packaging of these transcripts into heterotypic granules. RBPs engage in two key forms of protein–protein interactions, namely stereospecific interactions between well-folded proteins (such as other RBPs or G3BPs in stress granules) and/or interactions via intrinsically disordered regions (IDRs) of RBPs. The IDRs in turn specifically interact with well-folded proteins and/or undergo non-specific interactions with proteins in their vicinity to form liquid–liquid phase separation (LLPS) condensates or granules. d | RNA–RNA interactions can also promote granule assembly. Specifically, in trans interactions between RNAs, such as bcd and osk mRNAs in Drosophila melanogaster, transport granules. Furthermore, the nature of RNAs to self-assemble may promote promiscuous RNA–RNA interactions that assemble mRNAs, in a dose-dependent manner, into stress granules or germ granules.
The classical view of granule formation relies on the binding of RBPs to the cis-regulatory elements in RNA. Mutations or deletions of the cis-elements or alterations in the secondary structure of the mRNA notably impact RBP binding and granule formation, leading to mRNA mislocalization in yeast160, D. melanogaster213 and neurons214. From a perspective of RBP-driven assembly, the same RBP may be shared between multiple mRNAs or interactions between multiple RBPs may coalesce many individual mRNPs into larger complexes and granules (see REF.215 and FIG. 5c). Furthermore, by virtue of their intrinsically disordered regions, RBPs drive liquid–liquid phase separation (LLPS) and assemble multiple mRNPs indiscriminately such as into stress granules (discussed below) (FIG. 5c). An alternative model is RNA-driven granule assembly, which favours RNA as the primary scaffold for binding several multivalent RBPs, therefore promoting the nucleation of phase-separated assemblies216–218. For example, stress granule assembly is initiated by the stable aggregation of untranslated mRNAs into the core, potentially increasing the concentration of RBPs and therefore driving LLPS216 (FIG. 5d). Additionally, mRNAs engage in both promiscuous219 and specific RNA–RNA interactions220, which is the main driving force for the assembly of stress granules (reviewed in REF.221) (FIG. 5d). This non-specific mechanism is determined by the length of the mRNA (the longer the mRNA, the more interaction sites are available for random base-pairing). In D. melanogaster germ granules, mRNAs can self-assemble and position themselves in granules in a dose-dependent manner222.
Homotypic versus heterotypic granules
The self-assembly of RNAs suggests that RNA granules could be homotypic, constituting the same RNA species, or heterotypic granules formed by mRNAs undergoing base-pairing in trans. D. melanogaster germ plasm contains granules of up to 500 nm in size, the functions of which are determined by the localization and organization of mRNAs within the granule. During oocyte development, specific base-pairing between osk mRNAs or bcd mRNAs promotes the recruitment of both mRNAs to granules, rendering them heterotypic220. Although germ granules are heterotypic, multiple copies of mRNAs from the same gene are organized into discrete homotypic clusters that occupy specific positions within the granule222,223. Interestingly, this self-sorting of clusters within the granules is independent of sequence specificity222,223.
Stress granules and P-bodies are more heterogeneous in their structure and composition. Transcriptome analyses from yeast to mammalian cells have found that >99% of mRNAs enter stress granules; however, only 10% of these mRNAs are stably located within the granules, and there is a preference for longer transcripts224. Although single-molecule imaging has provided tremendous insights into how mRNAs enter and localize in stress granules225, it remains unclear whether each RNA species forms distinct clusters. Similarly, RNA stoichiometry and organization inside transport granules in mammalian neurons are heavily debated. Results from smFISH against several dendritically localized mRNAs indicated that each mRNA species existed as a single, independent mRNP along dendrites226 and may not belong to larger granules. Real-time imaging of endogenous actb mRNAs in X. laevis showed that, in axons, these mRNAs commonly travel as single mRNAs in granules94 whereas, in dendrites, the dynamic merging and splitting of mRNPs have been observed, suggesting the presence of a mixed population of granules containing one or multiple ACTB mRNAs92. Interestingly, real-time imaging of two different mRNAs, Actb and ARC, did not exhibit co-trafficking or coalescing even in crowded dendritic segments (S.D. and R.H.S., unpublished work), furthering the idea that transport granules may be homotypic.
Therefore, several studies suggest that mRNAs might be inherently biased to form homotypic clusters, courtesy of their self-assembling nature. One major advantage of each mRNA species forming its own granule would be the ability to fine-tune the local translation of specific mRNAs without affecting the status of other localized mRNAs. However, heterotypic granules may facilitate the clustering of different mRNAs of multi-subunit protein complexes and promote the co-regulation of their translation according to cellular needs.
mRNAs in stress granules and P-bodies
Cells adapt to adverse conditions by reprogramming translation, primarily by repressing translation initiation in response to stressors such as increased temperature (heat-shock) and nutrient deprivation207. Stressors preclude the formation of the pre-initiation complex by inducing the phosphorylating eIF2α and/or by inhibiting the assembly of the cap-binding complex eIF4F. Untranslated mRNAs are quickly and reversibly rear-ranged with proteins into stress granules and P-bodies227, although stress granule formation can be independent of eIF2α phosphorylation228. Stress granules and P-bodies are heterotypic mRNP granules enriched in translation and decay factors, respectively229. Although stress granules are formed under stress conditions, P-bodies exist in non-stressed mammalian cells and increase in size and number upon stress230. These granules are formed by LLPS, which requires both proteins and RNAs217,231–235, and they can associate with each other during stress. Characterization of the protein composition of stress granules and P-bodies has defined their common features such as the presence of intrinsically disordered regions and RNA-binding activity. These granules lack substrate specificity, with 20% of cellular RNAs localizing to both of them indiscriminately224,236. The release of mRNAs from stalled ribosomes by components of the quality control machinery results in the fast influx of these mRNAs into stress granules, and the inter-molecular interactions that occur facilitate the partitioning of mRNA into condensates237. Interestingly, these granules are enriched in long mRNAs with highly structured 5′UTRs and AU-rich 3′UTRs, which accounts for their low stability and translatability218,238–240.
Recent single-molecule approaches have attempted to visualize the conformation of mRNA within stress granules and P-bodies of mammalian cells as well as their dynamic molecular interactions in vivo225,241,242. The molecular compaction of mRNAs was calculated by measuring the distance of the fluorescence signal using smFISH probes against the 5′ coding sequence and 3′ ends of the same mRNA241,242. Actively translated mRNAs were in an extended conformation as a result of ribosome loading and compacted up to 200-fold upon the stress-induced inhibition of translation (namely, in response to oxidative stress or heat-shock). This compaction after ribosome run-off occurred in the cytoplasm and usually preceded the localization of mRNA to stress granules, indicating that mRNAs are translationally repressed before entering the stress granules241. One caveat to using smFISH methods to assess conformation is that the analysis is restricted to long transcripts (to allow the hybridization of multiple probes); furthermore, compactness may be a product of fixation artefacts. A recent live imaging study visualizing translating mRNAs reported similar stretching of mRNAs upon their increased association with ribosomes in the cytoplasm, validating the observations made in fixed cells201. Real-time imaging also revealed that mRNAs engage in both transient and stable interactions with stress granules and P-bodies and move bi-directionally between them225,243. The nature of the interaction depends on the size of the P-bodies and stress granules as well as on the length and translational status of mRNA (untranslated mRNAs showed stable interactions, whereas translated mRNAs had sporadic interactions). Larger granules retained mRNAs for extended periods of time, as shown for long transcripts, such as that encoding p300 (~8.3 kb), or mTOR-sensitive transcripts, specifically those bearing a terminal oligopyrimidine tract in their 5′UTR225,243. Upon stress withdrawal, stress granules rapidly disappear, and translation is resumed for all kinds of mRNAs trapped inside granules243. Although stress granules are widely believed to contain non-translating mRNAs, a recent single-molecule imaging study challenged this notion, demonstrating that translation continues even after the mRNAs are recruited to stress granules from cytosol during acute cellular stress244. This phenomenon is not limited to mRNAs whose translation is upregulated during stress, but is also true for transcripts that are inhibited during stress. Therefore, deciphering the composition of these granules would provide insight into their transcriptome and could reveal whether all stress granule-associated transcripts are similarly regulated. Recently developed proximity labelling approaches such as APEX-seq are powerful approaches towards understanding the constituents and organization of stress granules212,245. Although a fluid interchange of components with the cytoplasm occurs in both stress granules and P-bodies, their assembly dynamics, composition and intermolecular interactions are cell-type and stress-type dependent228,236,245,246. Likewise, the fate of mRNAs after interacting with P-bodies and stress granules and whether this fate leads to homeostatic reset or pathological outcome depend on the cell type and type of stress. Overall, mRNAs play an essential role in the biogenesis of granules235; nonetheless, the functional relevance of mRNA interactions with stress granules or P-bodies in cell survival during stress or in disease progression is still uncertain. Future work should determine the stress-dependent and context-dependent structural and functional diversity of stress granules and P-bodies.
Conclusion and future perspectives
The localization of mRNAs in subcellular compartments has emerged as a highly conserved mechanism for the spatial and temporal control of protein synthesis. The development of imaging and sequencing-based methods to detect mRNA and translation has made it possible to measure this post-transcriptional regulation across species and cellular scales. Single-gene studies have characterized the molecular events controlling the cytoplasmic localization of mRNA and have started to elucidate the mechanisms of local translation, for instance, at the ER, mitochondria or synaptic sites in dendrites. Large-scale studies have characterized up to thousands of localizing mRNAs and offered a global view of the shared features of trafficking (for example, zipcode elements, functional relationships and the binding sites of RBPs). The impact of mRNA trafficking on the physiology of multicellular tissues and of entire organisms (for example, C. elegans and D. melanogaster) has unravelled how mRNA localization influences high-order cell-to-cell interactions that are important for tissue organization and homeostasis.
Three major challenges must be overcome to achieve a clear understanding of the molecular events that regulate localized translation. First, few methods are currently available to modulate mRNA localization in vivo for functional studies. Most of these approaches target RBPs or zipcodes and require irreversible loss of function. Recently, the dynamic control of cytoplasmic mRNA localization and possibly translation has been feasible using either the MS2 (REFs92,133,167,171,247) or the Cas9 systems247. However, these multicomponent systems require complex experimental manipulations that may be far from physiological conditions and difficult to reproduce. Second, the development of in situ transcriptomic technologies generated an overwhelming amount of mRNA localization data to be analysed and interpreted. Several groups are at the forefront in the challenge of developing algorithms that define ‘ground-truth’ mRNA localization patterns that can be used as a reference for automatic identification in large datasets156,248–250. Further automation of these machine learning-based tools will allow us to extract meaningful localization patterns from the vast information generated by multiplex in situ transcriptomic data. Finally, the mRNA localization field constantly benefits from developments in imaging such as brighter and more photostable fluorescent proteins and fluorophores and detectors with an increased sensitivity. These improvements will be key to developing RNA imaging reporters for the detection of endogenous unmodified mRNAs at single-molecule resolution. Biophysicists, chemists and biologists will bring this field one-step closer to understanding how the spatial organization of biomolecules in single cells participates in translating a genotype into a phenotype.
Supplementary Material
Acknowledgements
The authors would like to thank the anonymous reviewers for the constructive comments and apologize to collaborators and colleagues whose work could not be cited due to space limitations. This work was supported by NIH grant AG05583 and NSERC grant RGPIN-2019-04767 to M.V., NIH grant NS083085 and NIH Grant GM57071 to R.H.S., and a Rose F. Kennedy IDDRC Pilot Grant to S.D.
Glossary
- Signal recognition particle (sRP)
A cytoplasmic RNA–protein complex required for protein targeting to the eukaryotic endoplasmic reticulum or the plasma membrane of bacteria.
- Single-molecule fluorescence in situ hybridization (smFIsH)
Fluorescence microscopy-based technique that allows the visualization and quantification of single mRNAs in fixed prokaryotic or eukaryotic cells.
- Patterning factors
signalling molecules, the distribution pattern of which within an organism contributes to cell differentiation, the determination of anterior–posterior body axis and development.
- SunTag system
signal amplification system for the visualization of proteins in living or fixed cells. The protein of interest is tagged with a repeated peptide array recognized by a single-chained antibody fused to a fluorescent protein or by immunofluorescence.
- P-lineage
The first cleavage of the fertilized C. elegans zygote produces an anterior and a posterior (P1) blastomere. Asymmetric division of P1, until P4 leads to the production of the germline tissue.
- In situ transcriptomics
Imaging-based gene expression and spatial profiling of fixed single cells based on multiple RNA imaging approaches.
- Aptamer
short RNA oligonucleotide, the sequence and structure of which is specifically recognized by an RNA binding protein (for example, the Ms2 loop).
- Expansion-FISH (ExFIsH)
RNA in-situ imaging approach for fixed cells that combines sample clearing and expansion with smFIsH, multiplexed smFIsH or hybridization chain reaction FIsH.
- Lamellipodia
Thin membrane protrusions filled with a dense actin meshwork. They are required for cell forward projection and chemotaxis.
- Cell leading edge
Protrusion of the cell membrane front formed during the migration of epithelial, endothelial, neuronal and immune cells.
- Proximity-dependent biotin identification (BioID) assay
Tool for the identification of protein–protein interactions based on the in vivo labelling with biotin of interactors located in close proximity to the candidate protein.
- APEX-seq
Proximity labelling of RNA with biotin using the peroxidase enzyme APEX (or APEX2) followed by sequencing.
- Germ granules
RNA-rich membraneless cytoplasmic granules found in the germline of organisms such as X. laevis, D. melanogaster and D. rerio.
Footnotes
Competing interests
The authors declare no competing interests.
Peer review information
Nature Reviews Molecular Cell Biology thanks the anonymous reviewers for their contribution to the peer review of this work.
Supplementary information
The online version contains supplementary material available at https://doi.org/10.1038/s41580-021-00356-8.
References
- 1.Jeffery WR, Tomlinson CR & Brodeur RD Localization of actin messenger RNA during early ascidian development. Dev. Biol 99, 408–417 (1983). [DOI] [PubMed] [Google Scholar]
- 2.Rebagliati MR, Weeks DL, Harvey RP & Melton DA Identification and cloning of localized maternal RNAs from Xenopus eggs. Cell 42, 769–777 (1985). [DOI] [PubMed] [Google Scholar]
- 3.Berleth T et al. The role of localization of bicoid RNA in organizing the anterior pattern of the Drosophila embryo. EMBO J 7, 1749–1756 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bopp D, Burri M, Baumgartner S, Frigerio G & Noll M Conservation of a large protein domain in the segmentation gene paired and in functionally related genes of Drosophila. Cell 47, 1033–1040 (1986). [DOI] [PubMed] [Google Scholar]
- 5.Lawrence JB & Singer RH Intracellular localization of messenger RNAs for cytoskeletal proteins. Cell 45, 407–415 (1986). [DOI] [PubMed] [Google Scholar]
- 6.Long RM et al. Mating type switching in yeast controlled by asymmetric localization of ASH1 mRNA. Science 277, 383–387 (1997). [DOI] [PubMed] [Google Scholar]
- 7.Li X, Franceschi VR & Okita TW Segregation of storage protein mRNAs on the rough endoplasmic reticulum membranes of rice endosperm cells. Cell 72, 869–879 (1993). [DOI] [PubMed] [Google Scholar]
- 8.Knowles RB et al. Translocation of RNA granules in living neurons. J. Neurosci 16, 7812–7820 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carson JH et al. In Cell Polarity and Subcellular RNA Localization (ed. Richter D) 69–81 (Springer, 2001). [Google Scholar]
- 10.Lecuyer E et al. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell 131, 174–187 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Claußen M et al. Global analysis of asymmetric RNA enrichment in oocytes reveals low conservation between closely related Xenopus species. Mol. Biol. Cell 26, 3777–3787 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nevo-Dinur K, Nussbaum-Shochat A, Ben-Yehuda S & Amster-Choder O Translation-independent localization of mRNA in E. coli. Science 331, 1081–1084 (2011). First example of widespread mRNA localization in bacteria. [DOI] [PubMed] [Google Scholar]
- 13.Cajigas IJ et al. The local transcriptome in the synaptic neuropil revealed by deep sequencing and high-resolution imaging. Neuron 74, 453–466 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zivraj KH et al. Subcellular profiling reveals distinct and developmentally regulated repertoire of growth cone mRNAs. J. Neurosci 30, 15464–15478 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mili S, Moissoglu K & Macara IG Genome-wide screen reveals APC-associated RNAs enriched in cell protrusions. Nature 453, 115–119 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xia C, Fan J, Emanuel G, Hao J & Zhuang X Spatial transcriptome profiling by MERFISH reveals subcellular RNA compartmentalization and cell cycle-dependent gene expression. Proc. Natl Acad. Sci. USA 116, 19490–19499 (2019). Example of multiplexed error-robust FISH (MERFISH) demonstrating near-genome-wide, spatially resolved RNA profiling of individual cells.
- 17.Mingle LA et al. Localization of all seven messenger RNAs for the actin-polymerization nucleator Arp2/3 complex in the protrusions of fibroblasts. J. Cell Sci 118, 2425–2433 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lundberg E & Borner GHH Spatial proteomics: a powerful discovery tool for cell biology. Nat. Rev. Mol. Cell Biol 20, 285–302 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Engel KL, Arora A, Goering R, Lo H-YG & Taliaferro JM Mechanisms and consequences of subcellular RNA localization across diverse cell types. Traffic 21, 404–418 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Forrest KM & Gavis ER Live imaging of endogenous RNA reveals a diffusion and entrapment mechanism for nanos mRNA localization in Drosophila. Curr. Biol 13, 1159–1168 (2003). [DOI] [PubMed] [Google Scholar]
- 21.Safieddine A et al. A conserved choreography of mRNAs at centrosomes reveals a localization mechanism involving active polysome transport. Preprint at bioRxiv 10.1101/2020.09.04.282038 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sepulveda G et al. Co-translational protein targeting facilitates centrosomal recruitment of PCNT during centrosome maturation in vertebrates. eLife 7, e34959 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bergalet J et al. Inter-dependent centrosomal co-localization of the cen and ik2 cis-Natural Antisense mRNAs in Drosophila. Cell Rep 30, 3339–3352.e6 (2020). [DOI] [PubMed] [Google Scholar]
- 24.Trcek T & Lehmann R Germ granules in Drosophila. Traffic 20, 650–660 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Das S, Singer RH & Yoon YJ The travels of mRNAs in neurons: do they know where they are going? Curr. Opin. Neurobiol 57, 110–116 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Gijtenbeek LA & Kok J Illuminating messengers: an update and outlook on RNA visualization in bacteria. Front. Microbiol 8, 1161 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bakshi S, Choi H & Weisshaar JC The spatial biology of transcription and translation in rapidly growing Escherichia coli. Front. Microbiol 6, 636 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fei J & Sharma CM RNA localization in bacteria. Microbiol. Spectr 10.1128/microbiolspec.RWR-0024-2018 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Toran P et al. Labeling native bacterial RNA in live cells. Cell Res 24, 894–897 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.So L et al. General properties of transcriptional time series in Escherichia coli. Nat. Genet 43, 554–560 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Montero Llopis P et al. Spatial organization of the flow of genetic information in bacteria. Nature 466, 77–81 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moffitt JR, Pandey S, Boettiger AN, Wang S & Zhuang X Spatial organization shapes the turnover of a bacterial transcriptome. eLife 5, e13065 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Niessing D, Jansen R-P, Pohlmann T & Feldbrügge M mRNA transport in fungal top models. Wiley Interdiscip. Rev. RNA 10.1002/wrna.1453 (2018). [DOI] [PubMed] [Google Scholar]
- 34.McBride AE Messenger RNA transport in the opportunistic fungal pathogen Candida albicans. Curr. Genet 63, 989–995 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shepard KA et al. Widespread cytoplasmic mRNA transport in yeast: Identification of 22 bud-localized transcripts using DNA microarray analysis. Proc. Natl Acad. Sci. USA 100, 11429–11434 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bertrand E et al. Localization of ASH1 mRNA particles in living yeast. Mol. Cell 2, 437–445 (1998). First use of the MS2 system for real-time imaging of single mRNAs in live cells.
- 37.Takizawa PA, Sil A, Swedlow JR, Herskowitz I & Vale RD Actin-dependent localization of an RNA encoding a cell-fate determinant in yeast. Nature 389, 90–93 (1997). [DOI] [PubMed] [Google Scholar]
- 38.Tutucci E et al. An improved MS2 system for accurate reporting of the mRNA life cycle. Nat. Methods 15, 81–89 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gu W, Deng Y, Zenklusen D & Singer RH A new yeast PUF family protein, Puf6p, represses ASH1 mRNA translation and is required for its localization. Genes Dev 18, 1452–1465 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paquin N et al. Local activation of yeast ASH1 mRNA translation through phosphorylation of Khd1p by the casein kinase Yck1p. Mol. Cell 26, 795–809 (2007). [DOI] [PubMed] [Google Scholar]
- 41.Deng Y, Singer RH & Gu W Translation of ASH1 mRNA is repressed by Puf6p-Fun12p/eIF5B interaction and released by CK2 phosphorylation. Genes Dev 22, 1037–1050 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cosma MP Daughter-specific repression of Saccharomyces cerevisiae HO: Ash1 is the commander. EMBO Rep 5, 953–957 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sil A & Herskowitz I Identification of asymmetrically localized determinant, Ash1p, required for lineage-specific transcription of the yeast HO gene. Cell 84, 711–722 (1996). [DOI] [PubMed] [Google Scholar]
- 44.Bobola N, Jansen RP, Shin TH & Nasmyth K Asymmetric accumulation of Ash1p in postanaphase nuclei depends on a myosin and restricts yeast mating-type switching to mother cells. Cell 84, 699–709 (1996). [DOI] [PubMed] [Google Scholar]
- 45.Elson SL, Noble SM, Solis NV, Filler SG & Johnson AD An RNA transport system in Candida albicans regulates hyphal morphology and invasive growth. PLoS Genet 5, e1000664 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kotani T, Yasuda K, Ota R & Yamashita M Cyclin B1 mRNA translation is temporally controlled through formation and disassembly of RNA granules. J. Cell Biol 202, 1041–1055 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Groisman I et al. CPEB, maskin, and cyclin B1 mRNA at the mitotic apparatus: implications for local translational control of cell division. Cell 103, 435–447 (2000). [DOI] [PubMed] [Google Scholar]
- 48.Schmid M, Jaedicke A, Du T-G & Jansen R-P Coordination of endoplasmic reticulum and mRNA localization to the yeast bud. Curr. Biol 16, 1538–1543 (2006). [DOI] [PubMed] [Google Scholar]
- 49.Fundakowski J, Hermesh O & Jansen R-P Localization of a subset of yeast mRNAs depends on inheritance of endoplasmic reticulum. Traffic 13, 1642–1652 (2012). [DOI] [PubMed] [Google Scholar]
- 50.Kraut-Cohen J et al. Translation- and SRP-independent mRNA targeting to the endoplasmic reticulum in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell 24, 3069–3084 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gadir N, Haim-Vilmovsky L, Kraut-Cohen J & Gerst JE Localization of mRNAs coding for mitochondrial proteins in the yeast Saccharomyces cerevisiae. RNA 17, 1551–1565 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Garcia M et al. Mitochondria-associated yeast mRNAs and the biogenesis of molecular complexes. Mol. Biol. Cell 18, 362–368 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pizzinga M et al. Translation factor mRNA granules direct protein synthetic capacity to regions of polarized growth. J. Cell Biol 218, 1564–1581 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Morales-Polanco F et al. Glycolytic mRNAs localize and are translated in core fermentation (CoFe) granules to fuel glucose fermentation. Preprint at bioRxiv 10.1101/741231 (2020). [DOI] [Google Scholar]
- 55.Jansen R-P, Niessing D, Baumann S & Feldbrügge M mRNA transport meets membrane traffic. Trends Genet 30, 408–417 (2014). [DOI] [PubMed] [Google Scholar]
- 56.Kwon S, Tisserant C, Tulinski M, Weiberg A & Feldbrügge M Inside-out: from endosomes to extracellular vesicles in fungal RNA transport. Fungal Biol. Rev 34, 89–99 (2020). [Google Scholar]
- 57.Baumann S, Pohlmann T, Jungbluth M, Brachmann A & Feldbrügge M Kinesin-3 and dynein mediate microtubule-dependent co-transport of mRNPs and endosomes. J. Cell Sci 125, 2740–2752 (2012). [DOI] [PubMed] [Google Scholar]
- 58.König J et al. The fungal RNA-binding protein Rrm4 mediates long-distance transport of ubi1 and rho3 mRNAs. EMBO J 28, 1855–1866 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pohlmann T, Baumann S, Haag C, Albrecht M & Feldbrügge M A FYVE zinc finger domain protein specifically links mRNA transport to endosome trafficking. eLife 4, e06041 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Femino AM, Fay FS, Fogarty K & Singer RH Visualization of single RNA transcripts in situ. Science 280, 585–590 (1998). First demonstration of single mRNA detection in situ by FISH.
- 61.Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A & Tyagi S Imaging individual mRNA molecules using multiple singly labeled probes. Nat. Methods 5, 877–879 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gavis ER & Lehmann R Localization of nanos RNA controls embryonic polarity. Cell 71, 301–313 (1992). [DOI] [PubMed] [Google Scholar]
- 63.Dufourt J et al. Imaging translation dynamics in live embryos reveals spatial heterogeneities. Preprint at bioRxiv 10.1101/2020.04.29.058974 (2020). [DOI] [PubMed] [Google Scholar]
- 64.Seydoux G The P granules of C. elegans: a genetic model for the study of RNA–protein condensates. J. Mol. Biol 430, 4702–4710 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee C-YS et al. Recruitment of mRNAs to P granules by condensation with intrinsically-disordered proteins. eLife 9, e52896 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Parker DM et al. mRNA localization is linked to translation regulation in the Caenorhabditis elegans germ lineage. Development 147, dev186817 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Moor AE et al. Global mRNA polarization regulates translation efficiency in the intestinal epithelium. Science 357, 1299–1303 (2017). Shows that, in the intestinal epithelium, mRNA localization contributes to the polarization of the translation machinery, in turn modulating gene expression changes.
- 68. Hafner A-S, Donlin-Asp PG, Leitch B, Herzog E & Schuman EM Local protein synthesis is a ubiquitous feature of neuronal pre- and postsynaptic compartments. Science 364, eaau3644 (2019). Identified hundreds of mRNA molecules and translation machinery in axonal terminals of adult brain, indicating the existence of local protein synthesis in both presynaptic and postsynaptic compartments of neurons.
- 69. Shigeoka T et al. Dynamic axonal translation in developing and mature visual circuits. Cell 166, 181–192 (2016). The first study to identify the local translatome in the developing and mature nervous system.
- 70.Bassell GJ et al. Sorting of beta-actin mRNA and protein to neurites and growth cones in culture. J. Neurosci 18, 251–265 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Leung K-M et al. Asymmetrical β-actin mRNA translation in growth cones mediates attractive turning to netrin-1. Nat. Neurosci 9, 1247–1256 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wong HH-W et al. RNA docking and local translation regulate site-specific axon remodeling in vivo. Neuron 95, 852–868.e8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Terenzio M et al. Locally translated mTOR controls axonal local translation in nerve injury. Science 359, 1416–1421 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sahoo PK, Smith DS, Perrone-Bizzozero N & Twiss JL Axonal mRNA transport and translation at a glance. J. Cell Sci 131, jcs196808 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Chen F et al. Nanoscale imaging of RNA with expansion microscopy. Nat. Methods 13, 679–684 (2016). This technique allowed the detection of RNAs with nanoscale precision in mammalian tissue by de-crowding of mRNAs and amplification of single-molecule signals.
- 76. Steward O & Levy WB Preferential localization of polyribosomes under the base of dendritic spines in granule cells of the dentate gyrus. J. Neurosci 2, 284–291 (1982). The first study demonstrating the presence of polyribosomes in dendritic spines by electron microscopy and proposing local translation away from the neuronal soma.
- 77.Ostroff LE, Fiala JC, Allwardt B & Harris KM Polyribosomes redistribute from dendritic shafts into spines with enlarged synapses during LTP in developing rat hippocampal slices. Neuron 35, 535–545 (2002). [DOI] [PubMed] [Google Scholar]
- 78.Younts TJ et al. Presynaptic protein synthesis is required for long-term plasticity of GABA release. Neuron 92, 479–492 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cioni J-M, Koppers M & Holt CE Molecular control of local translation in axon development and maintenance. Curr. Opin. Neurobiol 51, 86–94 (2018). [DOI] [PubMed] [Google Scholar]
- 80.Yoon BC et al. Local translation of extranuclear lamin B promotes axon maintenance. Cell 148, 752–764 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ostroff LE et al. Axon TRAP reveals learning-associated alterations in cortical axonal mRNAs in the lateral amgydala. eLife 8, e51607 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Holt CE, Martin KC & Schuman EM Local translation in neurons: visualization and function. Nat. Struct. Mol. Biol 26, 557–566 (2019). [DOI] [PubMed] [Google Scholar]
- 83.Biever A, Donlin-Asp PG & Schuman EM Local translation in neuronal processes. Curr. Opin. Neurobiol 57, 141–148 (2019). [DOI] [PubMed] [Google Scholar]
- 84.Kanai Y, Dohmae N & Hirokawa N Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron 43, 513–525 (2004). [DOI] [PubMed] [Google Scholar]
- 85.Mitsumori K, Takei Y & Hirokawa N Components of RNA granules affect their localization and dynamics in neuronal dendrites. Mol. Biol. Cell 28, 1412–1417 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kiebler MA & Bassell GJ Neuronal RNA granules: movers and makers. Neuron 51, 685–690 (2006). [DOI] [PubMed] [Google Scholar]
- 87.Buxbaum AR, Wu B & Singer RH Single beta-actin mRNA detection in neurons reveals a mechanism for regulating its translatability. Science 343, 419–422 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang C, Han B, Zhou R & Zhuang X Real-time imaging of translation on single mRNA transcripts in live cells. Cell 165, 990–1001 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Cioni J-M et al. Late endosomes act as mRNA translation platforms and sustain mitochondria in axons. Cell 176, 56–72.e15 (2019). This study highlighted that the association of RNA granules with endosomes is important for the local translation of mitochondrial proteins critical for axonal survival.
- 90.Doyle M & Kiebler MA Mechanisms of dendritic mRNA transport and its role in synaptic tagging. EMBO J 30, 3540–3552 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yoon YJ et al. Glutamate-induced RNA localization and translation in neurons. Proc. Natl Acad. Sci. USA 113, E6877–E6886 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Park HY et al. Visualization of dynamics of single endogenous mRNA labeled in live mouse. Science 343, 422–424 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Wu B, Eliscovich C, Yoon YJ & Singer RH Translation dynamics of single mRNAs in live cells and neurons. Science 352, 1430–1435 (2016). Together with refs 202,294–296, a first example of an imaging-based single-molecule mRNA translation reporter.
- 94.Turner-Bridger B et al. Single-molecule analysis of endogenous β-actin mRNA trafficking reveals a mechanism for compartmentalized mRNA localization in axons. Proc. Natl Acad. Sci. USA 115, E9697–E9706 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Shigeoka T et al. On-site ribosome remodeling by locally synthesized ribosomal proteins in axons. Cell Rep 29, 3605–3619.e10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Liao Y-C et al. RNA granules hitchhike on lysosomes for long-distance transport, using annexin A11 as a molecular tether. Cell 179, 147–164.e20 (2019). This study demonstrates co-transport of mRNAs with lysosomes in axons, with implications in amyotrophic lateral sclerosis.
- 97.Bartlett WP & Banker GA An electron microscopic study of the development of axons and dendrites by hippocampal neurons in culture. II. Synaptic relationships. J. Neurosci 4, 1954–1965 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Giuditta A, Cupellot A & Lazzarini G Ribosomal RNA in the axoplasm of the squid giant axon. J. Neurochem 34, 1757–1760 (1980). [DOI] [PubMed] [Google Scholar]
- 99. Biever A et al. Monosomes actively translate synaptic mRNAs in neuronal processes. Science 367, eaay4991 (2020). A first-time demonstration of differential monosome-mediated versus polysome-mediated translation in neurons and the preference for monosome-mediated translation in distal processes.
- 100.Koppers M et al. Receptor-specific interactome as a hub for rapid cue-induced selective translation in axons. eLife 8, e48718 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rangaraju V, tom Dieck S & Schuman EM Local translation in neuronal compartments: how local is local? EMBO Rep 18, 693–711 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Court FA, Hendriks WTJ, MacGillavry HD, Alvarez J & van Minnen J Schwann cell to axon transfer of ribosomes: toward a novel understanding of the role of glia in the nervous system. J. Neurosci 28, 11024–11029 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Twiss JL & Fainzilber M Ribosomes in axons– scrounging from the neighbors? Trends Cell Biol 19, 236–243 (2009). [DOI] [PubMed] [Google Scholar]
- 104.Budnik V, Ruiz-Cañada C & Wendler F Extracellular vesicles round off communication in the nervous system. Nat. Rev. Neurosci 17, 160–172 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Krämer-Albers E-M & Hill AF Extracellular vesicles: interneural shuttles of complex messages. Curr. Opin. Neurobiol 39, 101–107 (2016). [DOI] [PubMed] [Google Scholar]
- 106.Willems J et al. ORANGE: a CRISPR/Cas9-based genome editing toolbox for epitope tagging of endogenous proteins in neurons. PLOS Biol 18, e3000665 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Donlin-Asp PG, Polisseni C, Klimek R, Heckel A & Schuman EM Differential regulation of local mRNA dynamics and translation following long-term potentiation and depression. Preprint at bioRxiv 10.1101/2020.07.08.192369 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wang T, Hamilla S, Cam M, Aranda-Espinoza H & Mili S Extracellular matrix stiffness and cell contractility control RNA localization to promote cell migration. Nat. Commun 8, 896 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Moissoglu K, Yasuda K, Wang T, Chrisafis G & Mili S Translational regulation of protrusion-localized RNAs involves silencing and clustering after transport. Elife 8, e44752 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kislauskis EH, Zhu X & Singer RH Sequences responsible for intracellular localization of beta-actin messenger RNA also affect cell phenotype. J. Cell Biol 127, 441–451 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Michael M & Parsons M New perspectives on integrin-dependent adhesions. Curr. Opin. Cell Biol 63, 31–37 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Boyde A & Bailey A Observations on the marginal ruffles of an established fibroblast-like cell line. Cell Tissue Res 179, 225–234 (1977). [DOI] [PubMed] [Google Scholar]
- 113.Gandin V, Senft D, Topisirovic I & Ronai ZA RACK1 function in cell motility and protein synthesis. Genes Cancer 4, 369–377 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chicurel ME, Singer RH, Meyer CJ & Ingber DE Integrin binding and mechanical tension induce movement of mRNA and ribosomes to focal adhesions. Nature 392, 730–733 (1998). [DOI] [PubMed] [Google Scholar]
- 115.Willett M, Pollard HJ, Vlasak M & Morley SJ Localization of ribosomes and translation initiation factors to talin/beta3-integrin-enriched adhesion complexes in spreading and migrating mammalian cells. Biol. Cell 102, 265–276 (2010). [DOI] [PubMed] [Google Scholar]
- 116.Gorrini C et al. Fibronectin controls cap-dependent translation through β1 integrin and eukaryotic initiation factors 4 and 2 coordinated pathways. Proc. Natl Acad. Sci. USA 102, 9200–9205 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Biswas J et al. Zipcode binding protein 1 (ZBP1; IGF2BP1): a model for sequence-specific RNA regulation. Cold Spring Harb. Symp. Quant. Biol 84, 1–10 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ross AF, Oleynikov Y, Kislauskis EH, Taneja KL & Singer RH Characterization of a beta-actin mRNA zipcode-binding protein. Mol. Cell. Biol 17, 2158–2165 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Patel VL et al. Spatial arrangement of an RNA zipcode identifies mRNAs under post-transcriptional control. Genes Dev 26, 43–53 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wu B, Buxbaum AR, Katz ZB, Yoon YJ & Singer RH Quantifying protein-mRNA interactions in single live cells. Cell 162, 211–220 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Biswas J et al. The structural basis for RNA selectivity by the IMP family of RNA-binding proteins. Nat. Commun 10, 4440 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mukherjee J et al. β-Actin mRNA interactome mapping by proximity biotinylation. Proc. Natl Acad. Sci. USA 116, 12863–12872 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Song T et al. Specific interaction of KIF11 with ZBP1 regulates the transport of beta-actin mRNA and cell motility. J. Cell Sci 128, 1001–1010 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Oleynikov Y & Singer RH Real-time visualization of ZBP1 association with β-actin mRNA during transcription and localization. Curr. Biol 13, 199–207 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Condeelis J & Singer RH How and why does β-actin mRNA target? Biol. Cell 97, 97–110 (2005). [DOI] [PubMed] [Google Scholar]
- 126.Huttelmaier S et al. Spatial regulation of beta-actin translation by Src-dependent phosphorylation of ZBP1. Nature 438, 512–515 (2005). [DOI] [PubMed] [Google Scholar]
- 127.Warner JR, Knopf PM & Rich A A multiple ribosomal structure in protein synthesis. Proc. Natl Acad. Sci. USA 49, 122–129 (1963). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Katz ZB et al. Mapping translation ‘hot-spots’ in live cells by tracking single molecules of mRNA and ribosomes. eLife 5, e10415 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Mardakheh FK et al. Global analysis of mRNA, translation, and protein localization: local translation is a key regulator of cell protrusions. Dev. Cell 35, 344–357 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Rangaraju V, Lauterbach M & Schuman EM Spatially stable mitochondrial compartments fuel local translation during plasticity. Cell 176, 73–84. e15 (2019). [DOI] [PubMed] [Google Scholar]
- 131.Marc P et al. Genome-wide analysis of mRNAs targeted to yeast mitochondria. EMBO Rep 3, 159–164 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Fazal FM et al. Atlas of subcellular RNA localization revealed by APEX-Seq. Cell 178, 473–490.e26 (2019). A spatial transcriptomic approach based on direct proximity labelling to identify localized RNAs in subcellular compartments.
- 133.Tsuboi T et al. Mitochondrial volume fraction and translation duration impact mitochondrial mRNA localization and protein synthesis. eLife 9, e57814 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Saint-Georges Y et al. Yeast mitochondrial biogenesis: a role for the PUF RNA-binding protein Puf3p in mRNA localization. PLoS ONE 3, e2293 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Morita M et al. mTORC1 controls mitochondrial activity and biogenesis through 4E-BP-dependent translational regulation. Cell Metab 18, 698–711 (2013). [DOI] [PubMed] [Google Scholar]
- 136.Gandin V et al. nanoCAGE reveals 5’ UTR features that define specific modes of translation of functionally related MTOR-sensitive mRNAs. Genome Res 26, 636–648 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Becker T, Song J & Pfanner N Versatility of preprotein transfer from the cytosol to mitochondria. Trends Cell Biol 29, 534–548 (2019). [DOI] [PubMed] [Google Scholar]
- 138.Pagliarini DJ et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell 134, 112–123 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Weis BL, Schleiff E & Zerges W Protein targeting to subcellular organelles via mRNA localization. Biochim. Biophys. Acta 1833, 260–273 (2013). [DOI] [PubMed] [Google Scholar]
- 140.Williams CC, Jan CH & Weissman JS Targeting and plasticity of mitochondrial proteins revealed by proximity-specific ribosome profiling. Science 346, 748–751 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Vardi-Oknin D & Arava Y Characterization of factors involved in localized translation near mitochondria by ribosome-proximity labeling. Front. Cell Dev. Biol 7, 305 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Gold VA, Chroscicki P, Bragoszewski P & Chacinska A Visualization of cytosolic ribosomes on the surface of mitochondria by electron cryo-tomography. EMBO Rep 18, 1786–1800 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lesnik C, Golani-Armon A & Arava Y Localized translation near the mitochondrial outer membrane: an update. RNA Biol 12, 801–809 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Palade GE A small particulate component of the cytoplasm. J. Biophys. Biochem. Cytol 1, 59–68 (1955). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Walter P & Johnson AE Signal sequence recognition and protein targeting to the endoplasmic reticulum membrane. Annu. Rev. Cell Biol 10, 87–119 (1994). [DOI] [PubMed] [Google Scholar]
- 146.Costantini LM, Fossati M, Francolini M & Snapp EL Assessing the tendency of fluorescent proteins to oligomerize under physiologic conditions. Traffic 13, 643–649 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Jagannathan S, Reid DW, Cox AH & Nicchitta CV De novo translation initiation on membrane-bound ribosomes as a mechanism for localization of cytosolic protein mRNAs to the endoplasmic reticulum. RNA 20, 1489–1498 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Stephens SB et al. Stable ribosome binding to the endoplasmic reticulum enables compartment-specific regulation of mRNA translation. Mol. Biol. Cell 16, 5819–5831 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Liao G, Ma X & Liu G An RNA-zipcode-independent mechanism that localizes Dia1 mRNA to the perinuclear ER through interactions between Dia1 nascent peptide and Rho-GTP. J. Cell Sci 124, 589–599 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Masibay AS, Qasba PK, Sengupta DN, Damewood GP & Sreevalsan T Cell-cycle-specific and serum-dependent expression of gamma-actin mRNA in Swiss mouse 3T3 cells. Mol. Cell. Biol 8, 2288–2294 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Voigt F et al. Single-molecule quantification of translation-dependent association of mRNAs with the endoplasmic reticulum. Cell Rep 21, 3740–3753 (2017). [DOI] [PubMed] [Google Scholar]
- 152.Halstead JM et al. Translation. An RNA biosensor for imaging the first round of translation from single cells to living animals. Science 347, 1367–1671 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Unsworth H, Raguz S, Edwards HJ, Higgins CF & Yagüe E mRNA escape from stress granule sequestration is dictated by localization to the endoplasmic reticulum. FASEB J 24, 3370–3380 (2010). [DOI] [PubMed] [Google Scholar]
- 154.Lerner RS & Nicchitta CV mRNA translation is compartmentalized to the endoplasmic reticulum following physiological inhibition of cap-dependent translation. RNA 12, 775–789 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Woodruff JB et al. The centrosome is a selective condensate that nucleates microtubules by concentrating tubulin. Cell 169, 1066–1077.e10 (2017). [DOI] [PubMed] [Google Scholar]
- 156.Chouaib R et al. A dual protein-mRNA localization screen reveals compartmentalized translation and widespread co-translational RNA targeting. Dev. Cell 54, 773–791.e5 (2020). [DOI] [PubMed] [Google Scholar]
- 157.Kwon OS et al. Exon junction complex dependent mRNA localization is linked to centrosome organization during ciliogenesis. Nat. Commun 12, 1351 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Safieddine A et al. A conserved choreography of mRNAs at centrosomes reveals a localization mechanism involving active polysome transport. Nat. Commun 12, 1352 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Buxbaum AR, Haimovich G & Singer RH In the right place at the right time: visualizing and understanding mRNA localization. Nat. Rev. Mol. Cell Biol 16, 95–109 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Edelmann FT et al. Molecular architecture and dynamics of ASH1 mRNA recognition by its mRNA-transport complex. Nat. Struct. Mol. Biol 24, 152–161 (2017). [DOI] [PubMed] [Google Scholar]
- 161.Subramanian M et al. G–quadruplex RNA structure as a signal for neurite mRNA targeting. EMBO Rep 12, 697–704 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Wu H, Zhou J, Zhu T, Cohen I & Dictenberg J A kinesin adapter directly mediates dendritic mRNA localization during neural development in mice. J. Biol. Chem 295, 6605–6628 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Chartrand P, Meng X-H, Singer RH & Long RM Structural elements required for the localization of ASH1 mRNA and of a green fluorescent protein reporter particle in vivo. Curr. Biol 9, 333–338 (1999). [DOI] [PubMed] [Google Scholar]
- 164.Gonzalez I, Buonomo SBC, Nasmyth K & von Ahsen U ASH1 mRNA localization in yeast involves multiple secondary structural elementsand Ash1 protein translation. Curr. Biol 9, 337–340 (1999). [DOI] [PubMed] [Google Scholar]
- 165.Jansen RP, Dowzer C, Michaelis C, Galova M & Nasmyth K Mother cell-specific HO expression in budding yeast depends on the unconventional myosin myo4p and other cytoplasmic proteins. Cell 84, 687–697 (1996). [DOI] [PubMed] [Google Scholar]
- 166.Böhl F, Kruse C, Frank A, Ferring D & Jansen R-P She2p, a novel RNA-binding protein tethers ASH1 mRNA to the Myo4p myosin motor via She3p. EMBO J 19, 5514–5524 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Long RM, Gu W, Lorimer E, Singer RH & Chartrand P She2p is a novel RNA-binding protein that recruits the Myo4p-She3p complex to ASH1 mRNA. EMBO J 19, 6592–6601 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Long RM et al. An exclusively nuclear RNA-binding protein affects asymmetric localization of ASH1 mRNA and Ash1p in yeast. J. Cell Biol 153, 307–318 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Takizawa PA & Vale RD The myosin motor, Myo4p, binds Ash1 mRNA via the adapter protein, She3p. Proc. Natl Acad. Sci. USA 97, 5273–5278 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Takizawa PA, DeRisi JL, Wilhelm JE & Vale RD Plasma membrane compartmentalization in yeast by messenger RNA transport and a septin diffusion barrier. Science 290, 341–344 (2000). [DOI] [PubMed] [Google Scholar]
- 171.Katz ZB et al. β-Actin mRNA compartmentalization enhances focal adhesion stability and directs cell migration. Genes Dev 26, 1885–1890 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Das S, Moon HC, Singer RH & Park HY A transgenic mouse for imaging activity-dependent dynamics of endogenous Arc mRNA in live neurons. Sci. Adv 4, eaar3448 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Dynes JL & Steward O Dynamics of bidirectional transport of Arc mRNA in neuronal dendrites. J. Comp. Neurol 500, 433–447 (2007). [DOI] [PubMed] [Google Scholar]
- 174.Bauer KE et al. Live cell imaging reveals 3′-UTR dependent mRNA sorting to synapses. Nat. Commun 10, 3178 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Huotari J & Helenius A Endosome maturation. EMBO J 30, 3481–3500 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Baumann S, König J, Koepke J & Feldbrügge M Endosomal transport of septin mRNA and protein indicates local translation on endosomes and is required for correct septin filamentation. EMBO Rep 15, 94–102 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Higuchi Y, Ashwin P, Roger Y & Steinberg G Early endosome motility spatially organizes polysome distribution. J. Cell Biol 204, 343–357 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Schuster M et al. Kinesin-3 and dynein cooperate in long-range retrograde endosome motility along a nonuniform microtubule array. MBoC 22, 3645–3657 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Steinberg G Endocytosis and early endosome motility in filamentous fungi. Curr. Opin. Microbiol 20, 10–18 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Semotok JL et al. Smaug recruits the CCR4/POP2/NOT deadenylase complex to trigger maternal transcript localization in the early Drosophila embryo. Curr. Biol 15, 284–294 (2005). [DOI] [PubMed] [Google Scholar]
- 181.Zaessinger S, Busseau I & Simonelig M Oskar allows nanos mRNA translation in Drosophila embryos by preventing its deadenylation by Smaug/CCR4. Development 133, 4573–4583 (2006). [DOI] [PubMed] [Google Scholar]
- 182.Chen L et al. Global regulation of mRNA translation and stability in the early Drosophila embryo by the Smaug RNA-binding protein. Genome Biol 15, R4 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Bashirullah A, Cooperstock RL & Lipshitz HD Spatial and temporal control of RNA stability. Proc. Natl Acad. Sci. USA 98, 7025–7028 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Semotok JL et al. Drosophila maternal Hsp83 mRNA destabilization is directed by multiple SMAUG recognition elements in the open reading frame. Mol. Cell. Biol 28, 6757–6772 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Castellana M, Hsin-Jung Li S & Wingreen NS Spatial organization of bacterial transcription and translation. Proc. Natl Acad. Sci. USA 113, 9286–9291 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Sun M, Wartel M, Cascales E, Shaevitz JW & Mignot T Motor-driven intracellular transport powers bacterial gliding motility. Proc. Natl Acad. Sci. USA 108, 7559–7564 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Buskila AA, Kannaiah S & Amster-Choder O RNA localization in bacteria. RNA Biol 11, 1051–1060 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Wang C, Dickinson LK & Lehmann R Genetics of nanos localization in Drosophila. Dev. Dyn 199, 103–115 (1994). [DOI] [PubMed] [Google Scholar]
- 189.Kindler S, Wang H, Richter D & Tiedge H RNA transport and local control of translation. Annu. Rev. Cell Dev. Biol 21, 223–245 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Bassell GJ & Warren ST Fragile X syndrome: loss of local mRNA regulation alters synaptic development and function. Neuron 60, 201–214 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Topisirovic I, Svitkin YV, Sonenberg N & Shatkin AJ Cap and cap-binding proteins in the control of gene expression. Wiley Interdiscip. Rev. RNA 2, 277–298 (2011). [DOI] [PubMed] [Google Scholar]
- 192.Hinnebusch AG eIF3: a versatile scaffold for translation initiation complexes. Trends Biochem. Sci 31, 553–562 (2006). [DOI] [PubMed] [Google Scholar]
- 193.Europe PMC. Maskin is a CPEB-associated factor that transiently interacts with elF-4E. - Abstract - Europe PMC https://europepmc.org/article/med/10635326 (2001). [DOI] [PubMed]
- 194.Richter JD & Sonenberg N Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature 433, 477–480 (2005). [DOI] [PubMed] [Google Scholar]
- 195.Jung M-Y, Lorenz L & Richter JD Translational control by neuroguidin, a eukaryotic initiation factor 4E and CPEB binding protein. Mol. Cell. Biol 26, 4277–4287 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Santini E et al. Reducing eIF4E-eIF4G interactions restores the balance between protein synthesis and actin dynamics in fragile X syndrome model mice. Sci. Signal 10, eaan0665 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Chen E, Sharma MR, Shi X, Agrawal RK & Joseph S Fragile X mental retardation protein regulates translation by binding directly to the ribosome. Mol. Cell 54, 407–417 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Darnell JC & Klann E The translation of translational control by FMRP: therapeutic targets for Fragile X syndrome. Nat. Neurosci 16, 1530–1536 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Genuth NR & Barna M The discovery of ribosome heterogeneity and its implications for gene regulation and organismal life. Mol. Cell 71, 364–374 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Ho JJD & Lee S A cap for every occasion: alternative eIF4F complexes. Trends Biochem. Sci 41, 821–823 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Koch A, Aguilera L, Morisaki T, Munsky B & Stasevich TJ Quantifying the dynamics of IRES and cap translation with single-molecule resolution in live cells. Nat. Struct. Mol. Biol 27, 1095–1104 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Pichon X et al. Visualization of single endogenous polysomes reveals the dynamics of translation in live human cells. J. Cell Biol 214, 769–781 (2016). Together with refs 93,294–296, first example of imaging-based single-molecule mRNA translation reporters.
- 203.Eberwine J, Miyashiro K, Kacharmina JE & Job C Local translation of classes of mRNAs that are targeted to neuronal dendrites. Proc. Natl Acad. Sci. USA 98, 7080–7085 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Ifrim MF, Williams KR & Bassell GJ Single-molecule imaging of PSD-95 mRNA translation in dendrites and its dysregulation in a mouse model of fragile X syndrome. J. Neurosci 35, 7116–7130 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Kim TK et al. Dendritic glutamate receptor mRNAs show contingent local hotspot-dependent translational dynamics. Cell Rep 5, 114–125 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Spillane M, Ketschek A, Merianda TT, Twiss JL & Gallo G Mitochondria coordinate sites of axon branching through localized intra-axonal protein synthesis. Cell Rep 5, 1564–1575 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Liu B & Qian S-B Translational reprogramming in cellular stress response. Wiley Interdiscip. Rev. RNA 5, 301–315 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Guzikowski AR, Chen YS & Zid BM Stress-induced mRNP granules: form and function of processing bodies and stress granules. Wiley Interdiscip. Rev. RNA 10, e1524 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Aronov S et al. mRNAs encoding polarity and exocytosis factors are cotransported with the cortical endoplasmic reticulum to the incipient bud in Saccharomyces cerevisiae. Mol. Cell. Biol 27, 3441–3455 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Shiber A et al. Cotranslational assembly of protein complexes in eukaryotes revealed by ribosome profiling. Nature 561, 268–272 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Eliscovich C, Shenoy SM & Singer RH Imaging mRNA and protein interactions within neurons. Proc. Natl Acad. Sci. USA 114, E1875–E1884 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Padrón A, Iwasaki S & Ingolia NT Proximity RNA labeling by APEX-Seq reveals the organization of translation initiation complexes and repressive RNA granules. Mol. Cell 75, 875–887.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Jambor H, Mueller S, Bullock SL & Ephrussi A A stem–loop structure directs oskar mRNA to microtubule minus ends. RNA 20, 429–439 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Meer EJ et al. Identification of a cis-acting element that localizes mRNA to synapses. Proc. Natl Acad. Sci. USA 109, 4639–4644 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Treeck BV & Parker R Emerging roles for intermolecular RNA-RNA interactions in RNP assemblies. Cell 174, 791–802 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Wheeler JR, Matheny T, Jain S, Abrisch R & Parker R Distinct stages in stress granule assembly and disassembly. eLife 5, e18413 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Garcia-Jove Navarro M et al. RNA is a critical element for the sizing and the composition of phase-separated RNA–protein condensates. Nat. Commun 10, 3230 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Treeck BV et al. RNA self-assembly contributes to stress granule formation and defining the stress granule transcriptome. Proc. Natl Acad. Sci. USA 115, 2734–2739 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Vourekas A, Alexiou P, Vrettos N, Maragkakis M & Mourelatos Z Sequence-dependent but not sequence-specific piRNA adhesion traps mRNAs to the germ plasm. Nature 531, 390–394 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Jambor H, Brunel C & Ephrussi A Dimerization of oskar 3′ UTRs promotes hitchhiking for RNA localization in the Drosophila oocyte. RNA 17, 2049–2057 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Roden C & Gladfelter AS RNA contributions to the form and function of biomolecular condensates. Nat. Rev. Mol. Cell Biol 22, 183–195 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222. Trcek T et al. Sequence-independent self-assembly of germ granule mRNAs into homotypic clusters. Mol. Cell 78, 941–950.e12 (2020). This paper demonstrates the intrinsic ability of mRNAs to self-organize as homotypic assemblies within Drosophila germ granules.
- 223.Trcek T et al. Drosophila germ granules are structured and contain homotypic mRNA clusters. Nat. Commun 6, 7962 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Khong A et al. The stress granule transcriptom e reveals principles of mRNA accumulation in stress granules. Mol. Cell 68, 808–820.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225. Moon SL et al. Multicolour single-molecule tracking of mRNA interactions with RNP granules. Nat. Cell Biol 21, 162–168 (2019). Multicolour single-molecule mRNA tracking is used to quantify the timing and kinetics of single mRNA translation and transit to ribonucleoprotein granules during stress.
- 226.Batish M, van den Bogaard P, Kramer FR & Tyagi S Neuronal mRNAs travel singly into dendrites. Proc. Natl Acad. Sci. USA 109, 4645–4650 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Matheny T, Rao BS & Parker R Transcriptome-wide comparison of stress granules and P-bodies reveals that translation plays a major role in RNA partitioning. Mol. Cell. Biol 39, e00313–19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Aulas A et al. Stress-specific differences in assembly and composition of stress granules and related foci. J. Cell Sci 130, 927–937 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Youn J-Y et al. High-density proximity mapping reveals the subcellular organization of mRNA-associated granules and bodies. Mol. Cell 69, 517–532.e11 (2018). [DOI] [PubMed] [Google Scholar]
- 230.Kedersha N et al. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol 169, 871–884 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Sanders DW et al. Competing protein-RNA interaction networks control multiphase intracellular organization. Cell 181, 306–324.e28 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Guillén-Boixet J et al. RNA-induced conformational switching and clustering of G3BP drive stress granule assembly by condensation. Cell 181, 346–361.e17 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Yang P et al. G3BP1 is a tunable switch that triggers phase separation to assemble stress granules. Cell 181, 325–345.e28 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Iserman C et al. Condensation of Ded1p promotes a translational switch from housekeeping to stress protein production. Cell 181, 818–831.e19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Tauber D et al. Modulation of RNA condensation by the DEAD-Box protein eIF4A. Cell 180, 411–426.e16 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Hubstenberger A et al. P-body purification reveals the condensation of repressed mRNA regulons. Mol. Cell 68, 144–157.e5 (2017). [DOI] [PubMed] [Google Scholar]
- 237.Moon SL, Morisaki T, Stasevich TJ & Parker R Coupling of translation quality control and mRNA targeting to stress granules. J. Cell Biol 219, e202004120 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Pitchiaya S et al. Dynamic recruitment of single RNAs to processing bodies depends on RNA functionality. Mol. Cell 74, 521–533.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Courel M et al. GC content shapes mRNA storage and decay in human cells. eLife 8, e49708 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Namkoong S, Ho A, Woo YM, Kwak H & Lee JH Systematic characterization of stress-induced RNA granulation. Mol. Cell 70, 175–187.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241. Adivarahan S et al. Spatial organization of single mRNPs at different stages of the gene expression pathway. Mol. Cell 72, 727–738.e5 (2018). smFISH coupled with super-resolved imaging is used to study the mRNA conformation within the mRNP as a function of the translation state.
- 242.Khong A & Parker R mRNP architecture in translating and stress conditions reveals an ordered pathway of mRNP compaction. J. Cell Biol 217, 4124–4140 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Wilbertz JH et al. Single-molecule imaging of mRNA localization and regulation during the integrated stress response. Mol. Cell 73, 946–958.e7 (2019). [DOI] [PubMed] [Google Scholar]
- 244.Mateju D et al. Single-molecule imaging reveals translation of mRNAs localized to stress granules. Cell 183, 1801–1812.e13 (2020). [DOI] [PubMed] [Google Scholar]
- 245.Markmiller S et al. Context-dependent and disease-specific diversity in protein interactions within stress granules. Cell 172, 590–604.e13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Wang C et al. Context-dependent deposition and regulation of mRNAs in P-bodies. eLife 7, e29815 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Kim NY et al. Optogenetic control of mRNA localization and translation in live cells. Nat. Cell Biol 22, 341–352 (2020). [DOI] [PubMed] [Google Scholar]
- 248.Pichon X, Lagha M, Mueller F & Bertrand E A growing toolbox to image gene expression in single cells: sensitive approaches for demanding challenges. Mol. Cell 71, 468–480 (2018). [DOI] [PubMed] [Google Scholar]
- 249.Samacoits A et al. A computational framework to study sub-cellular RNA localization. Nat. Commun 9, 4584 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Park HY, Trcek T, Wells AL, Chao JA & Singer RH An unbiased analysis method to quantify mRNA localization reveals its correlation with cell motility. Cell Rep 1, 179–184 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Tian L, Chou H-L, Fukuda M, Kumamaru T & Okita TW mRNA localization in plant cells. Plant Physiol 182, 97–109 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Washida H et al. RNA targeting to a specific ER sub-domain is required for efficient transport and packaging of α-globulins to the protein storage vacuole in developing rice endosperm. Plant J 70, 471–479 (2012). [DOI] [PubMed] [Google Scholar]
- 253.Washida H et al. Identification of cis-localization elements that target glutelin RNAs to a specific subdomain of the cortical endoplasmic reticulum in rice endosperm cells. Plant Cell Physiol 50, 1710–1714 (2009). [DOI] [PubMed] [Google Scholar]
- 254.Hamada S et al. Dual regulated RNA transport pathways to the cortical region in developing rice endosperm. Plant Cell 15, 2265–2272 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Tian L et al. Zipcode RNA-binding proteins and membrane trafficking proteins cooperate to transport glutelin mRNAs in rice endosperm. Plant Cell 32, 2566–2581 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Michaud M et al. Differential targeting of VDAC3 mRNA isoforms influences mitochondria morphology. Proc. Natl Acad. Sci. USA 111, 8991–8996 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Vincent T et al. A genome-scale analysis of mRNAs targeting to plant mitochondria: upstream AUGs in 5′ untranslated regions reduce mitochondrial association. Plant J 92, 1132–1142 (2017). [DOI] [PubMed] [Google Scholar]
- 258.Gómez G & Pallás V Noncoding RNA mediated traffic of foreign mRNA into chloroplasts reveals a novel signaling mechanism in plants. PLoS ONE 5, e12269 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Tian L & Okita TW mRNA-based protein targeting to the endoplasmic reticulum and chloroplasts in plant cells. Curr. Opin. Plant Biol 22, 77–85 (2014). [DOI] [PubMed] [Google Scholar]
- 260.Cheng S-F, Huang Y-P, Chen L-H, Hsu Y-H & Tsai C-H Chloroplast phosphoglycerate kinase is involved in the targeting of bamboo mosaic virus to chloroplasts in Nicotiana benthamiana plants. Plant Physiol 163, 1598–1608 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Duncan S, Olsson TSG, Hartley M, Dean C & Rosa S A method for detecting single mRNA molecules in Arabidopsis thaliana. Plant Methods 12, 13 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Tutucci E, Livingston NM, Singer RH & Wu B Imaging mRNA in vivo, from birth to death. Annu. Rev. Biophys 47, 85–106 (2018). [DOI] [PubMed] [Google Scholar]
- 263.Sato H, Das S, Singer RH & Vera M Imaging of DNA and RNA in living eukaryotic cells to reveal spatiotemporal dynamics of gene expression. Annu. Rev. Biochem 89, 159–187 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Vera M, Biswas J, Senecal A, Singer RH & Park HY Single-cell and single-molecule analysis of gene expression regulation. Annu. Rev. Genet 50, 267–291 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Schmidt A, Gao G, Little SR, Jalihal AP & Walter NG Following the messenger: recent innovations in live cell single molecule fluorescence imaging. Wiley Interdiscip. Rev. RNA 11, e1587 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Tutucci E, Vera M & Singer RH Single-mRNA detection in living S. cerevisiae using a re-engineered MS2 system. Nat. Protoc 13, 2268–2296 (2018). [DOI] [PubMed] [Google Scholar]
- 267.Wu B et al. Synonymous modification results in high-fidelity gene expression of repetitive protein and nucleotide sequences. Genes Dev 29, 876–886 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Chao JA, Patskovsky Y, Almo SC & Singer RH Structural basis for the coevolution of a viral RNA-protein complex. Nat. Struct. Mol. Biol 15, 103–105 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Lange S et al. Simultaneous transport of different localized mRNA species revealed by live-cell imaging. Traffic 9, 1256–1267 (2008). [DOI] [PubMed] [Google Scholar]
- 270.Daigle N & Ellenberg J λ N -GFP: an RNA reporter system for live-cell imaging. Nat. Methods 4, 633–636 (2007). [DOI] [PubMed] [Google Scholar]
- 271.Brodsky AS & Silver PA Identifying proteins that affect mRNA localization in living cells. Methods 26, 151–155 (2002). [DOI] [PubMed] [Google Scholar]
- 272.Chen J et al. High efficiency of HIV-1 genomic RNA packaging and heterozygote formation revealed by single virion analysis. Proc. Natl Acad. Sci. USA 106, 13535–13540 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Los GV et al. HaloTag: a novel protein labeling technology for cell imaging and protein analysis. ACS Chem. Biol 3, 373–382 (2008). [DOI] [PubMed] [Google Scholar]
- 274.Garcia H & Gregor T Live imaging of mRNA synthesis in Drosophila. Methods Mol. Biol 1649, 349–357 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Lionnet T et al. A transgenic mouse for in vivo detection of endogenous labeled mRNA. Nat. Methods 8, 165–170 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Hocine S, Raymond P, Zenklusen D, Chao JA & Singer RH Single-molecule analysis of gene expression using two-color RNA labeling in live yeast. Nat. Methods 10, 119–121 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Martin RM, Rino J, Carvalho C, Kirchhausen T & Carmo-Fonseca M Live-cell visualization of pre-mRNA splicing with single-molecule sensitivity. Cell Rep 4, 1144–1155 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Strack RL, Disney MD & Jaffrey SR A superfolding Spinach2 reveals the dynamic nature of trinucleotide repeat-containing RNA. Nat. Methods 10, 1219–1224 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Filonov GS, Moon JD, Svensen N & Jaffrey SR Broccoli: rapid selection of an RNA mimic of green fluorescent protein by fluorescence-based selection and directed evolution. J. Am. Chem. Soc 136, 16299–16308 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Song W et al. Imaging RNA polymerase III transcription using a photostable RNA-fluorophore complex. Nat. Chem. Biol 13, 1187–1194 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Dolgosheina EV et al. RNA mango aptamer-fluorophore: a bright, high-affinity complex for RNA labeling and tracking. ACS Chem. Biol 9, 2412–2420 (2014). [DOI] [PubMed] [Google Scholar]
- 282.Mitra J & Ha T Nanomechanics and co-transcriptional folding of Spinach and Mango. Nat. Commun 10, 4318 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Tyagi S & Kramer FR Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol 14, 303–308 (1996). [DOI] [PubMed] [Google Scholar]
- 284.Nelles DA et al. Programmable RNA tracking in live cells with CRISPR/Cas9. Cell 165, 488–496 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Wang S, Su J-H, Zhang F & Zhuang X An RNA-aptamer-based two-color CRISPR labeling system. Sci. Rep 6, 26857 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Yang L-Z et al. Dynamic imaging of RNA in living cells by CRISPR-Cas13 systems. Mol. Cell 76, 981–997.e7 (2019). [DOI] [PubMed] [Google Scholar]
- 287.Garcia JF & Parker R MS2 coat proteins bound to yeast mRNAs block 5′ to 3′ degradation and trap mRNA decay products: implications for the localization of mRNAs by MS2-MCP system. RNA 21, 1393–1395 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Haimovich G et al. Use of the MS2 aptamer and coat protein for RNA localization in yeast: a response to ‘MS2 coat proteins bound to yeast mRNAs block 5′ to 3′ degradation and trap mRNA decay products: implications for the localization of mRNAs by MS2-MCP system’. RNA 22, 660–666 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Heinrich S, Sidler CL, Azzalin CM & Weis K Stem-loop RNA labeling can affect nuclear and cytoplasmic mRNA processing. RNA 23, 134–141 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Vera M, Tutucci E & Singer RH In Imaging Gene Expression: Methods and Protocols (ed. Shav-Tal Y) 3–20 (Springer, 2019). [Google Scholar]
- 291.Tantale K et al. A single-molecule view of transcription reveals convoys of RNA polymerases and multi-scale bursting. Nat. Commun 7, 12248 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Pichon X, Robert M-C, Bertrand E, Singer RH & Tutucci E New generations of MS2 variants and MCP fusions to detect single mRNAs in living eukaryotic cells. Methods Mol. Biol 2166, 121–144 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Braselmann E, Rathbun C, Richards EM & Palmer AE Illuminating RNA biology: tools for imaging RNA in live mammalian cells. Cell Chem. Biol 27, 891–903 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294. Morisaki T et al. Real-time quantification of single RNA translation dynamics in living cells. Science 352, 1425–1429 (2016). Together with refs 93,202,295,296, first example of imaging-based single-molecule mRNA translation reporter.
- 295. Yan X, Hoek TA, Vale RD & Tanenbaum ME Dynamics of translation of single mRNA molecules in vivo. Cell 165, 976–989 (2016). Together with refs 93,202,294,296, first example of imaging-based single-molecule mRNA translation reporter.
- 296. Boersma S et al. Multi-color single-molecule imaging uncovers extensive heterogeneity in mRNA decoding. Cell 178, 458–472.e19 (2019). Together with refs 93,202,294,295, first example of imaging-based single-molecule mRNA translation reporter.
- 297.tom Dieck S et al. Direct visualization of newly synthesized target proteins in situ. Nat. Methods 12, 411–414 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Hobson BD, Kong L, Hartwick EW, Gonzalez RL & Sims PA Elongation inhibitors do not prevent the release of puromycylated nascent polypeptide chains from ribosomes. eLife 9, e60048 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Enam SU et al. Puromycin reactivity does not accurately localize translation at the subcellular level. eLife 9, e60303 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


