Abstract
Objectives
To develop a Deep Learning Artificial Intelligence (AI) model that automatically localizes the position of radiographic stent gutta percha (GP) markers in cone beam computed tomography (CBCT) images to identify proposed implant sites within the images, and to test the performance of the newly developed AI model.
Materials and Methods
Thirty-four CBCT datasets were used for initial model training, validation and testing. The CBCT datasets were those of patients who had a CBCT examination performed wearing a radiographic stent for implant treatment planning. The datasets were exported in Digital Imaging and Communications in Medicine (DICOM), then imported into the software Horos ®. Each GP marker was manually labelled for object detection and recognition by the deep learning model by drawing rectangles around the GP markers in all axial images, then the labelled images were split into training, validation, and test sets. The axial sections of 30 CBCT datasets were randomly divided into training and validation sets. four CBCT datasets were used for testing the performance of the deep learning model. Descriptive statistics were calculated for the number of GP markers present, number of correct and incorrect identifications of GP markers.
Result
The AI model had an 83% true positive rate for identification of the GP markers. Of the areas labelled by the AI model as GP markers, 28 % were not truly GP markers, but the overall false positive rate was 2.8 %.
Conclusion
An AI model for localization of GP markers in CBCT images was able to identify most of the GP markers, but 2.8% of the results were false positive and 17% were missed GP markers. Using only axial images for training an AI program is not enough to give an accurate AI model performance.
Keywords: Artificial intelligence, Deep learning, Dental Implant, Stents, Algorithms, Cone Beam, Computed Tomography
1. Introduction
Nowadays dental implants have become the standard of care in restoring missing teeth. When multiple implants are needed, considerable time may be required to prepare an ideal implant treatment plan, which may delay the surgical implant placement for a patient. The use of machine learning (ML) methods, a branch of artificial intelligence (AI), especially artificial neural networks (ANN), may help in formulating the treatment plan in a shorter period of time, and thus expedite implant placement (Amato, López et al. 2013).
Artificial intelligence has been used to aid in the performance of numerous dental tasks. In a systematic review of 50 studies which reported the use of AI programs in dentomaxillofacial radiology, the studies mainly involved automated localization of cephalometric landmarks, diagnosis of osteoporosis, classification/segmentation of maxillofacial cysts and/ or tumors, and identification of periodontitis/periapical disease (Hung, Montalvao et al. 2020). Other published studies reported the development and use of AI based systems for use in dental implantology. Polášková et al. (2013) presented a web-based tool which utilized patient history and clinical data input into a program and preset threshold levels for various parameters to formulate a decision on whether or not implants may be placed, and if bone grafting is needed, and how long after grafting should implants be placed? (Polášková, Feberová et al. 2013). Sadighpour et al. (2014) developed an ANN model which utilized a number of input factors to formulate a decision regarding the type of prosthesis (fixed or removable) and the specific design of the prosthesis for rehabilitation of the edentulous maxilla (Sadighpour, Rezaei et al. 2014). Lee et al. (2012) applied a decision making system (fuzzy recognition map) for implant abutment selection (Lee, Yang et al. 2012). Additionally, Szejka et al., 2011, Szejka et al., 2013 developed an interactive reasoning system which requires the dentist to select the region of interest within a 3D model of the bone based on computed tomography (CT) images, then aids in selection of the optimum implant length and design (Szejka et al., 2011, Szejka et al., 2013).
Furthermore, AI has been used for implant placement in other areas of the body. In a study performed on 27 subjects, a fully convolutional deep learning model was used to determine the position and orientation of the articular marginal plane of the proximal humerus based on CT scans (Kulyk, Vlachopoulos et al. 2018). Carrillo et al. (2017) generated, in a fully automatic manner, a surgical plan for corrective osteotomies of malunited radius bones (Carrillo, Vlachopoulos et al. 2017).
However, none of the previous studies demonstrated use of AI to automatically place simulated implants in the optimum position and angulation within the CT images of the jaws during implant treatment planning. Therefore, the overall goal of the present research group’s project is to develop a deep learning AI model that automatically places simulated implants within cone beam CT (CBCT) images using the optimum size implant and placed within the optimum prosthetically driven position and orientation within the bone. Such a model would expedite and streamline implant treatment planning, especially in cases which require numerous implants. The first step to achieve the above goal is to use an AI model to accurately localize the proposed implant sites in CBCT images. Therefore, the aim of the present study is to use axial CBCT sections to develop an AI model that automatically localizes markers in radiographic stents in order to identify proposed implant sites within CBCT images. This is the first phase in a multi-phase development and validation process in which an AI model will be developed using an increasing number of planes of image sections in all three dimensions to identify GP marker positions in CBCT images.
2. Materials and methods
This study was an experimental study implemented in King Saud University, College of Dentistry (KSUCD) and the College of Computer and Information Sciences (CCIS). Because retrospective patient CBCT images were used to train the AI model, ethical approval was obtained from the King Saud University College of Medicine Institutional Review Board (Project No. E-20–4914). Thirty-four CBCT datasets were used for initial model training, validation and testing. The CBCT datasets were those of patients who had a CBCT examination performed wearing a radiographic stent for implant treatment planning. The list of patients was obtained from:
-
1.
The list of patients from the records of the dental labs and prosthodontic and implant clinics who had radiographic stents requested.
-
2.
A survey of Oral and Maxillofacial Radiologists (OMFR) requesting the list of their patients who had a CBCT interpretation report which indicated patient was wearing a radiographic stent.
The inclusion criteria were any retrievable CBCT datasets for patients who had taken CBCTs with a radiographic stent for implant placement purpose. The exclusion criteria were CBCT datasets with artifacts that degrade the image of the edentulous area, cases in which the radiographic stent was not well-fitted in the patient's mouth, and cases in which the implant site required a bone graft. All the 34 cases were organized in an excel sheet and coded as (A01, A02, A03, ………, A34)
All the CBCT datasets were accessed in Romexis® 3D software program (Planmeca Romexis® 5.2.0.R, Helsinki, Finland) within the server of KSUCD. The datasets were exported in Digital Imaging and Communications in Medicine (DICOM) format in an anonymized manner using the original voxel size and stored in both a Hard Disk and Google Drive for backup. The CBCT datasets were imported into the software Horos ®, and each GP marker was labelled manually by drawing rectangles around the GP markers in all the axial images which demonstrated the GP marker. These labelled images were then used to train the model on localization of the GP marker. Many AI models for object detection are available in the literature. In the present study, the model used to detect the GP markers was Mask R-CNN, (He, Gkioxari et al. 2017) a state-of-the-art object detection deep learning neural network. Transfer learning, a method enabling reuse of a model trained on some dataset to a new dataset, was used to train the model on our dataset.
The CBCT datasets were then converted into a comma-separated values (CSV) file to be further processed in the model. Data preprocessing is an essential step in any prediction model; the data was preprocessed to normalize the grey density value to be in the range [-1,1] which is suitable for the machine learning model. Afterwards, the images were used to train the model to automatically detect the GP markers. In order to train the model, the KERAS open-source software library was used on the Google Colab platform.
Training of the AI model was done by backpropagation, which consists of optimizing the weights using the chain rule to propagate the gradient of the loss function backwards into the model weights. The AI model was trained on 30 cases with 16,272 total number of images, these images were randomly divided into training and validation sets, were 90.2% of images for training and 9.8% for validation. The remaining 4 cases were used to test the model performance.
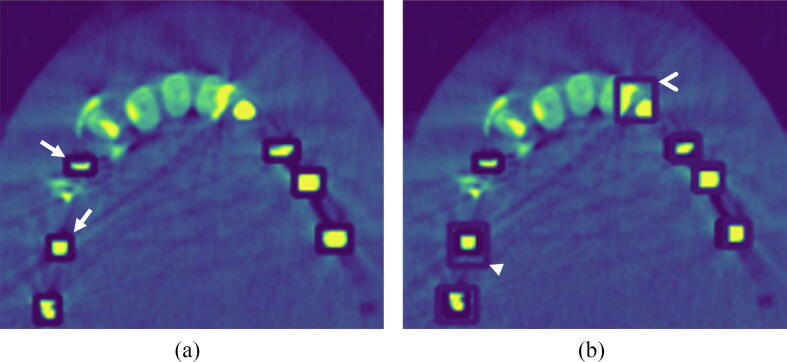
The performance of the AI model was then tested using all the axial sections in the four CBCT datasets. Fig. 1a demonstrates the manually identified GP markers used as the reference, and Fig. 1b demonstrates the AI identification of the GP markers. Descriptive statistics were calculated for the number of GP markers present, and the number of correct and incorrect identifications of GP markers.
Fig. 1.
(a) Sample of CBCT axial section of the maxilla demonstrating boxes placed manually for identification of the GP markers; the manual labelling appears as dark blue boxes (marked by white arrows). (b) The same section is seen with the AI localization of the GP markers; the areas identified by the AI algorithm appear as lighter blue boxes (marked by the arrowheads). A correct identification of a GP marker is seen marked by the closed arrowhead. The restorations in the upper left incisors were incorrectly identified as GP markers by the AI model (marked by open arrowhead). The GP marker in the area of upper right premolar was not identified by the AI model.
The objective of this study was to build a predictive AI model with sensitivity greater than 80%. Based on the literature and pilot testing we expect sensitivity of 88% in this project. Using G*Power tool (version 3.1.9.2) with effect size of 8%, level of significance 0.05 and desired statistical power of 80% we determined a minimum sample size of 135. These number of GP markers can be observed in a sample of 3–4 cases (on average there are 48 GP markers per case). We decided to use 3–4 cases as the testing set and 10-times larger sample to train the AI model. The total sample size of 34 cases was considered sufficient to build and validate an adequately accurate predictive model.
3. Results
Table 1 demonstrates the number of sections and GP markers in each dataset used for testing the AI model, and the number of correct and incorrect identifications. A total of 50 image sections with 193 images of GP markers, and 2284 sections which did not have a GP marker, were included in the testing data. Of the 193 existing images of GP markers, 83% were correctly identified by the algorithm. Furthermore, Of the 223 areas labelled by the AI model as GP markers, 28 % were not truly GP markers. However, if each section without a GP marker (n = 2284) was considered as one potential site for identification of presence or absence of a GP marker, then the false positive performance of the AI model was 2.8 %.
Table 1.
CBCT examinations (testing set instances) used as the testing dataset, and the number of image sections, and GP markers used for testing the AI model, along with correct and incorrect number of identifications achieved by the AI model.
| Code number of the CBCT Examination | Number of sections had no markers | Identification number of the axial section in the dataset | Number of GP markers within image section (identified manually) | Number of GP markers correctly identified by the AI model | Number of GP markers missed by the AI model | Number of areas mistakenly identified as GP by the AI model |
|---|---|---|---|---|---|---|
| A31 | 643 | 368 | 2 | 2 | 0 | 0 |
| 366 | 2 | 2 | 0 | 0 | ||
| 382 | 2 | 2 | 0 | 0 | ||
| 373 | 2 | 2 | 0 | 0 | ||
| 360 | 2 | 2 | 0 | 0 | ||
| 386 | 2 | 2 | 0 | 0 | ||
| 352 | 2 | 2 | 0 | 0 | ||
| 374 | 2 | 2 | 0 | 0 | ||
| A32 | 372 | 039 | 4 | 4 | 0 | 1 |
| 067 | 4 | 4 | 0 | 0 | ||
| 056 | 4 | 4 | 0 | 0 | ||
| 082 | 4 | 4 | 0 | 0 | ||
| 058 | 4 | 4 | 0 | 0 | ||
| 089 | 4 | 4 | 0 | 0 | ||
| 048 | 4 | 4 | 0 | 0 | ||
| 083 | 4 | 4 | 0 | 0 | ||
| 090 | 4 | 4 | 0 | 0 | ||
| A33 | 632 | 292 | 6 | 5 | 1 | 1 |
| 338 | 7 | 4 | 3 | 3 | ||
| 319 | 1 | 1 | 0 | 1 | ||
| 336 | 7 | 4 | 3 | 3 | ||
| 327 | 6 | 3 | 3 | 2 | ||
| 330 | 7 | 4 | 3 | 3 | ||
| 341 | 7 | 4 | 3 | 3 | ||
| 307 | 4 | 4 | 0 | 2 | ||
| 343 | 7 | 5 | 2 | 2 | ||
| 302 | 5 | 5 | 0 | 2 | ||
| 287 | 5 | 2 | 3 | 2 | ||
| 360 | 6 | 3 | 3 | 2 | ||
| 274 | 4 | 2 | 2 | 1 | ||
| 354 | 7 | 4 | 3 | 1 | ||
| 285 | 5 | 4 | 1 | 2 | ||
| 309 | 4 | 4 | 0 | 2 | ||
| 282 | 4 | 4 | 0 | 2 | ||
| 288 | 6 | 5 | 1 | 2 | ||
| 311 | 4 | 4 | 0 | 2 | ||
| A34 | 637 | 299 | 3 | 3 | 0 | 1 |
| 281 | 3 | 3 | 0 | 2 | ||
| 313 | 3 | 3 | 0 | 0 | ||
| 264 | 2 | 1 | 1 | 4 | ||
| 308 | 3 | 3 | 0 | 2 | ||
| 276 | 3 | 3 | 0 | 2 | ||
| 284 | 3 | 3 | 0 | 3 | ||
| 303 | 3 | 3 | 0 | 2 | ||
| 269 | 3 | 2 | 1 | 2 | ||
| 297 | 3 | 3 | 0 | 2 | ||
| 281 | 3 | 3 | 0 | 2 | ||
| 316 | 1 | 1 | 0 | 0 | ||
| 295 | 3 | 3 | 0 | 1 | ||
| 304 | 3 | 3 | 0 | 1 | ||
| Total | 2284 | 50 | 193 | 160 | 33 | 63 |
4. Discussion
This study presents the first AI model developed for identification of GP markers used for localizing prospective dental implant sites within CBCT images. The present algorithm correctly identified most of the GP markers, we consider a false positive rate of 2.8% and missed GP marker rate of 17% reasonable for a newly developed AI model. However, we are aiming for a higher model accuracy by using another deep learning algorithm.
A possible reason for the above result may be because the axial images used for training the algorithm did not include a clear and distinct shape of the GP marker in the superior-inferior, buccal-lingual, and mesial-distal perspectives. Additionally, the axial images did not demonstrate the relationship of the GP marker to the bone, a relationship which may aid an AI model in correctly identifying the markers.
As far as the authors are aware, the radiographic stents containing the GP markers were produced by the same laboratory and using the same type of acrylic, but it was evident from the CBCT images that the GP markers had variable diameter and length. However, it is not likely that the variable sizes of the markers had an adverse effect on the resultant model’s accuracy, because such variable GP markers were seen in both the training and the test images.
At the time of writing, and to the authors’ knowledge, there are no other reported AI models for identification of fiducial markers in CT or CBCT images of the maxillofacial region. A systematic review of AI applications in OMF Radiology reported the use of AI with CT and/or CBCT for the detection of the odontoid process, segmentation and measurement of maxillofacial lesions, classification of jaw lesions and tooth types, identification of the root canal, and localization of 3D cephalometric landmarks (Hung, Montalvao et al. 2020). However, the nature of the landmarks being detected, and the techniques used by the previous researchers to localize the anatomic landmarks, were different than the ones used in the present study.
Neelapu, et al. (2018) applied bone segmentation, standardized the position of the image volumes, extracted contours, and detected landmarks based on the definition on the contours and a template matching algorithm (Neelapu, Kharbanda et al. 2018). Codari, et al. (2017) applied thresholding to segment the regions of interest, registered the images by choosing the most inferior point in the mandibular bone to systematize all the CBCTs, and used an adaptive cluster-based and intensity-based algorithm (Codari, Caffini et al. 2017). Gupta, et al. (2015) used an anatomical reference as a “Seed Point” then applied a knowledge-based algorithm (Gupta, Kharbanda et al. 2015). Montúfar, et al. (2018a) computed digitally reconstructed projections then selected an anatomical structure manually to initialize an active shape model (Montúfar et al., 2018a). Montúfar, et al. (2018b) used a knowledge-based local landmark search after initializing an active shape model (Montúfar et al., 2018b). Shahidi, et al. (2014) used adaptive thresholding and volume matching then applied feature-based and voxel similarity-based algorithms (Shahidi, Bahrampour et al. 2014).
As such, it may be seen that existing AI models described in previous studies were used to identify anatomical landmarks which were known to be present within the dataset, and have characteristic relationships to the surrounding anatomy. The AI model in the present study, on the other hand, was used to identify the presence or absence of fiducial markers (GP), and search for the markers anywhere within the CBCT volume. Furthermore, the target of localization for the present AI model (GP markers) was highly variable in number and relationship to surrounding anatomy. Therefore, due to the above-mentioned differences between the function of the AI models, it is not possible to compare or contrast the performance of the present AI model, with that of previous models reported in the literature.
The present research team is currently working on further refinement of the algorithm through the second phase of research, which is using additional sectional images from the coronal and sagittal planes which include the shape of the marker more clearly, and which include the apical bone with the GP markers within the labelled areas. Also, to reduce the time required for labelling the required GP makers and bone, the number of sections which include the full length of the GP marker may be reduced by exporting the CBCT images using a voxel size of 0.4 mm, which has been reported to provide similar accuracy as 0.2 mm when used for dental implant site analysis (Torres, Campos et al. 2012).
5. Conclusion
An AI model for localization of GP markers in CBCT images was able to identify most of GP markers, but 2.8% of the results were false positive and 17% were missed GP markers. Use of only axial images for training an AI program for localization of GP markers is not enough to give an accurate AI model performance.
CRediT authorship contribution statement
Mona Alsomali: Investigation, Formal analysis, Writing – original draft, Visualization. Shatha Alghamdi: Investigation, Formal analysis, Writing – original draft, Visualization. Shahad Alotaibi: Methodology, Software, Validation. Sara Alfadda: Methodology, Investigation, Validation, Resources, Writing – review & editing, Supervision. Najwa Altwaijry: Methodology, Software, Validation, Resources, Writing – review & editing, Supervision. Isra Alturaiki: Methodology, Software, Validation, Resources, Writing – review & editing. Asma'a Al-Ekrish: Conceptualization, Methodology, Investigation, Validation, Formal analysis, Resources, Writing – review & editing, Supervision, Project administration.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Peer review under responsibility of King Saud University.
Contributor Information
Mona Alsomali, Email: m2alsomali@gmail.com.
Shatha Alghamdi, Email: ffss90075s@gmail.com.
Shahad Alotaibi, Email: 438203473@student.ksu.edu.sa.
Sara Alfadda, Email: Salfadda@gmail.com.
Najwa Altwaijry, Email: ntwaijry@ksu.edu.sa.
Isra Alturaiki, Email: ialturaiki@ksu.edu.sa.
Asma'a Al-Ekrish, Email: aalekrish@ksu.edu.sa, asma.alekrish@gmail.com.
References
- Amato F., López A., Peña-Méndez E.M., Vaňhara P., Hampl A., Havel J. Elsevier; 2013. Artificial neural networks in medical diagnosis. [Google Scholar]
- Carrillo F., Vlachopoulos L., Schweizer A., Nagy L., Snedeker J., Fürnstahl P. International Conference on Medical Image Computing and Computer-Assisted Intervention. 2017. A time saver: Optimization approach for the fully automatic 3D planning of forearm osteotomies. [Google Scholar]
- Codari M., Caffini M., Tartaglia G.M., Sforza C., Baselli G. Computer-aided cephalometric landmark annotation for CBCT data. Int. J. Comput. Assist. Radiol. Surg. 2017;12(1):113–121. doi: 10.1007/s11548-016-1453-9. [DOI] [PubMed] [Google Scholar]
- Gupta A., Kharbanda O.P., Sardana V., Balachandran R., Sardana H.K. A knowledge-based algorithm for automatic detection of cephalometric landmarks on CBCT images. Int. J. Comput. Assist. Radiol. Surg. 2015;10(11):1737–1752. doi: 10.1007/s11548-015-1173-6. [DOI] [PubMed] [Google Scholar]
- He K., Gkioxari G., Dollár P., Girshick R. Proceedings of the IEEE international conference on computer vision. 2017. Mask r-cnn. [Google Scholar]
- Hung K., Montalvao C., Tanaka R., Kawai T., Bornstein M.M. The use and performance of artificial intelligence applications in dental and maxillofacial radiology: A systematic review. Dentomaxillofac Radiol. 2020;49(1):20190107. doi: 10.1259/dmfr.20190107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulyk P., Vlachopoulos L., Fürnstahl P., Zheng G. International Workshop on Computational Methods and Clinical Applications in Musculoskeletal Imaging. 2018. Fully automatic planning of total shoulder arthroplasty without segmentation: A deep learning based approach. [Google Scholar]
- Lee S., Yang J., Han J. Development of a decision making system for selection of dental implant abutments based on the fuzzy cognitive map. Expert Syst. Appl. 2012;39(14):11564–11575. [Google Scholar]
- Montúfar J., Romero M., Scougall-Vilchis R.J. Automatic 3-dimensional cephalometric landmarking based on active shape models in related projections. Am. J. Orthodont. Dentofac. Orthoped. 2018;153(3):449–458. doi: 10.1016/j.ajodo.2017.06.028. [DOI] [PubMed] [Google Scholar]
- Montúfar J., Romero M., Scougall-Vilchis R.J. Hybrid approach for automatic cephalometric landmark annotation on cone-beam computed tomography volumes. Am. J. Orthodont. Dentofac. Orthoped. 2018;154(1):140–150. doi: 10.1016/j.ajodo.2017.08.028. [DOI] [PubMed] [Google Scholar]
- Neelapu B.C., Kharbanda O.P., Sardana V., Gupta A., Vasamsetti S., Balachandran R., Sardana H.K. Automatic localization of three-dimensional cephalometric landmarks on CBCT images by extracting symmetry features of the skull. Dentomaxillofacial Radiol. 2018;47(2):20170054. doi: 10.1259/dmfr.20170054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polášková A., Feberová J., Dostálová T., Kříž P., Seydlová M. Clinical decision support system in dental implantology. MEFANET Journal. 2013;1(1):11–14. [Google Scholar]
- Sadighpour L., Rezaei S.M.M., Paknejad M., Jafary F., Aslani P. The application of an artificial neural network to support decision making in edentulous maxillary implant prostheses. J. Res. Pract. Dentistry. 2014;2014:i1–i10. [Google Scholar]
- Shahidi S., Bahrampour E., Soltanimehr E., Zamani A., Oshagh M., Moattari M., Mehdizadeh A. The accuracy of a designed software for automated localization of craniofacial landmarks on CBCT images. BMC Med. Imag. 2014;14(1):1–8. doi: 10.1186/1471-2342-14-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szejka A.L., Rudek M., Jnr O.C. 41st International Conference on Computers & Industrial Engineering, Los Angeles. Proceedings of the 41st International Conference on Computers & Industrial Engineering. 2011. A Reasoning Method for Determining the Suitable Dental Implant. [Google Scholar]
- Szejka A.L., Rudek M., Junior O.C. 909–919; Springer: 2013. A reasoning system to support the dental implant planning process. Concurrent Engineering Approaches for Sustainable Product Development in a Multi-Disciplinary Environment. [Google Scholar]
- Torres M.G.G., Campos P.S.F., Segundo N.P.N., Navarro M., Crusoé-Rebello I. Accuracy of linear measurements in cone beam computed tomography with different voxel sizes. Implant dentistry. 2012;21(2):150–155. doi: 10.1097/ID.0b013e31824bf93c. [DOI] [PubMed] [Google Scholar]