Abstract
OBJECTIVE
Suboptimal nutrition in pregnancy is associated with worse offspring cardiometabolic health. DNA methylation may be an underlying mechanism. We meta-analyzed epigenome-wide association studies (EWAS) of maternal dietary glycemic index and load with cord blood DNA methylation.
RESEARCH DESIGN AND METHODS
We calculated maternal glycemic index and load from food frequency questionnaires and ran EWAS on cord blood DNA methylation in 2,003 mother-offspring pairs from three cohorts. Analyses were additionally stratified by maternal BMI categories. We looked-up the findings in EWAS of maternal glycemic traits and BMI as well as in EWAS of birth weight and child BMI. We examined associations with gene expression in child blood in the online Human Early Life Exposome eQTM catalog and in 223 adipose tissue samples.
RESULTS
Maternal glycemic index and load were associated with cord blood DNA methylation at 41 cytosine-phosphate-guanine sites (CpGs, P < 1.17 × 10−7), mostly in mothers with overweight/obesity. We did not observe overlap with CpGs associated with maternal glycemic traits, BMI, or child birth weight or BMI. Only DNA methylation at cg24458009 and cg23347399 was associated with expression of PCED1B and PCDHG, respectively, in child blood, and DNA methylation at cg27193519 was associated with expression of TFAP4, ZNF500, PPL, and ANKS3 in child subcutaneous adipose tissue.
CONCLUSIONS
We observed multiple associations of maternal glycemic index and load during pregnancy with cord blood DNA methylation, mostly in mothers with overweight/obesity; some of these CpGs were associated with gene expression. Additional studies are required to further explore functionality, uncover causality, and study pathways to offspring health.
Introduction
Glycemic index and glycemic load are measures to classify carbohydrate-containing foods according to the blood glucose response they evoke after consumption (1). Glycemic index represents the postprandial rise in blood glucose in response to a specific carbohydrate- containing food, fed as 50 g of net carbohydrate and compared with 50 g of pure glucose, and expressed as a percentage of the postprandial response to glucose. A higher glycemic index corresponds to a higher blood glucose–raising property of the food. Glycemic load incorporates both the glucose-raising properties of a food and the consumed amount of that food (2). Thus, a certain food always has the same glycemic index, but the glycemic load differs based on the amount consumed.
Individuals with a low glycemic–index diet or a low glycemic–load diet have a decreased risk of type 2 diabetes (3) and improved metabolic health (2,4,5). Additionally, compared with a conventional or high glycemic–index diet, a low glycemic–index diet reduces glycated hemoglobin (HbA1c) in individuals with diabetes (6).
Evidence on the association of prenatal dietary glycemic index or load with neonatal outcomes is mixed (7–9). Of three previous observational studies, one reported a positive association of maternal dietary glycemic index with offspring adiposity measures at birth (8), one found negative associations (7), and one found no associations (9). One study in obese pregnant women did not find associations of a diet and physical activity intervention with offspring large-for-gestational-age status, despite, among other factors, a reduced dietary glycemic load (10).
A potential biological mechanism underlying associations of maternal diet with offspring health is differential DNA methylation. Some large-scale meta-analyses of epigenome-wide association studies (EWAS) have previously linked adverse maternal nutrition-related exposures during pregnancy (e.g., BMI [11] and plasma folate concentrations [12]) with cord blood DNA methylation. However, not much is known about the association of maternal dietary glycemic index or load during pregnancy with offspring DNA methylation. Two previous studies provided some first evidence that maternal dietary glycemic index and load may be related to DNA methylation in placental tissue (13) and in cord blood (14), although sample sizes were rather limited. DNA methylation may have functional consequences via changes in gene expression that potentially lead to changes in health outcomes. We therefore conducted a meta-analysis to investigate the association of maternal dietary glycemic index and load during pregnancy with offspring cord blood DNA methylation. In this EWAS, we took a hypothesis-free approach, analyzing DNA methylation at all cytosine-phosphate-guanine sites (CpGs) measured on the array in relation to glycemic index and load. After running the EWAS, we compared the results with previous EWAS of cardiometabolic phenotypes to examine whether the CpGs identified in our study were also related to cardiometabolic phenotypes. Lastly, because women with overweight are more likely to be insulin resistant (15), we therefore additionally stratified analyses by maternal weight status.
Research Design and Methods
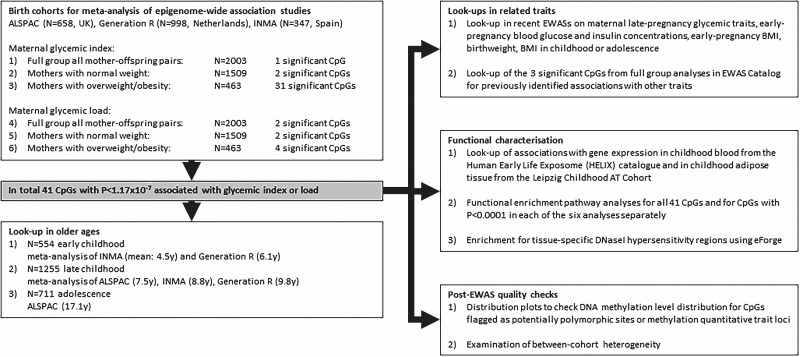
An explanation of the cohorts used for meta-analysis and all following statistical analyses is presented in the flowchart in Fig. 1.
Figure 1.
Flowchart of the study design.
Participants
Three population-based birth cohorts participated in this individual participant meta-analysis: 658 mother-offspring pairs from the Avon Longitudinal Study of Parents and Children (ALSPAC) from the U.K. (16,17), 998 mother-offspring pairs from the Generation R Study (Generation R) from the Netherlands (18), and 347 mother-offspring pairs from the INfancia y Medio Ambiente (INMA) Project from Spain (19). We excluded mothers with gestational diabetes or prepregnancy type 1 or 2 diabetes, because glycemic response changes with insulin resistance and with antidiabetes medication. After cohort-specific quality control on the food frequency questionnaires (FFQs), described in the cohort-specific Supplementary Methods, we additionally excluded mothers with maternal glycemic index and load values outside ±5 SD from the cohort mean. We further excluded multiple pregnancies and if mothers had multiple children (i.e., nontwin siblings) in the cohort. We included only one child per mother, based on completeness of data and, if equal, randomly. Subjects with missing data on any of the covariates were excluded from analyses.
Maternal Dietary Glycemic Index and Glycemic Load During Pregnancy
Cohort-specific methods, including references, are described in detail in the Supplementary Methods. The FFQs were all validated and contained 47, 293, and 101 items in ALSPAC, Generation R, and INMA, respectively. We used FFQ data with information on consumption frequency, and portion size was extracted from the FFQ or from cohort-specific set portion sizes. Cohorts’ calculated total energy intake and carbohydrate content for each food was based on their national food composition tables. We used the total amount of consumed carbohydrates per food (g/100 g) and only calculated glycemic index and load for foods with >1 g total carbohydrate per 100 g. We then linked the foods to the glycemic index values of the country-specific reference database: Diogenes-UK for ALSPAC, Diogenes-NL for Generation R (20), and Atkinson for INMA (21). Foods that could not be found in the country-specific glycemic index reference database were linked to the glycemic index values of reference databases of other countries. Per person, the dietary glycemic index was then calculated as the weighted average of the glycemic index values of all foods consumed (2):
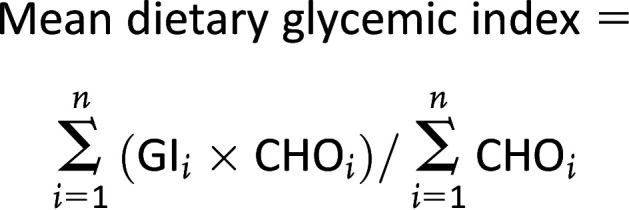 |
GIi (%) = the glycemic index of food i, relative to glucose; CHOi = the amount of food i consumed (g/day, from the raw FFQ data set) × carbohydrate content from food i (g/g, from the reference database); and n = number of foods eaten per day.
The mean dietary glycemic load is then calculated as (2):
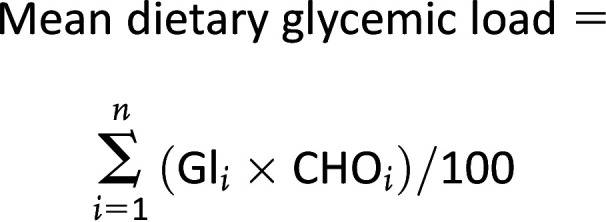 |
Offspring Cord Blood DNA Methylation
Offspring cord blood DNA was isolated from cord blood drawn directly after birth, and DNA methylation was measured using the Illumina 450K array. Each cohort performed their own laboratory analyses, quality control, and normalization (Supplementary Methods). We used untransformed β values, with values between 0 and 1, as the outcome measure and excluded extreme DNA methylation outliers according to the Tukey method: outliers outside the range of (25th percentile − 3∗interquartile range [IQR]) to (75th percentile + 3∗IQR) were excluded. We removed control probes, X and Y chromosome probes, and cross-reactive probes (22,23), resulting in a maximum of 428,328 remaining CpG sites. After meta-analysis, we flagged potentially polymorphic sites (22,23) or methylation quantitative trait loci (mQTLs) (24).
Statistical Analyses
Each cohort ran EWAS using robust linear regression models for glycemic index and load with each CpG site individually, using lmFit() in the limma R package (25), according to a prespecified analysis plan and R script. The fully adjusted models had maternal glycemic index or load as the exposure and cord blood DNA methylation as the outcome and were adjusted for sex, maternal education, age, smoking during pregnancy, total energy intake, batch, and cell types. We additionally ran these models stratified on maternal pre-pregnancy or early pregnancy (≤16 weeks of gestation) normal body weight (BMI 18 to <25 kg/m2) and overweight or obesity (BMI ≥25 kg/m2). Mothers with underweight were removed from these stratified analyses because the sample size was too low.
Sex of the child was obtained from birth records. Maternal educational level was based on self-reported questionnaires during pregnancy and assigned to two (ALSPAC and Generation R) or three (INMA) levels, depending on cohort preference (Supplementary Methods). Maternal age was self-reported in the questionnaire during pregnancy. Maternal smoking during pregnancy was self-reported in the questionnaire during pregnancy and defined as no smoking during pregnancy, as stopped smoking before the second trimester, as sustained smoking in Generation R and INMA, and as sustained smoking versus no smoking or quit before second trimester in ALSPAC. Maternal total energy intake during pregnancy (kcal/day) was calculated from the self-reported FFQ during pregnancy that was also used to calculate the glycemic index and load. Each cohort included their preferred variable to adjust for batch: Generation R used plate number as reported by the laboratory, ALSPAC used surrogate variable analysis (26), and INMA used ComBat (27). We used a cord blood methylation-based reference set to estimate cell type composition (28) (CD8 T cells, CD4 T cells, natural killer cells, B cells, monocytes, granulocytes, nucleated red blood cells).
All cohort EWAS results passed quality control using the QCEWAS R package (29). We then meta-analyzed the cohort-specific EWAS results using the fixed-effects inverse variance weighted method in METAL (30) at Erasmus MC and consecutively shadowed this meta-analysis at ISGlobal using GWAMA (31). Results were confirmed. We used a Bonferroni-corrected P-value threshold of <1.17 × 10−7 to define CpGs to take forward for further analyses, and we also present the false discovery rate (FDR).
Lookup in Older Ages
We tested whether the CpGs that reached the Bonferroni threshold in the full group meta-analyses or in the analyses stratified on maternal BMI showed persistent associations with maternal glycemic index or load during pregnancy when DNA methylation was measured at later ages. We performed lookups in 1) early childhood: meta-analysis of INMA (mean: 4.5 years) and Generation R (6.1 years), 2) late childhood: meta-analysis of ALSPAC (7.5 years), INMA (8.8 years), Generation R (9.8 years), and 3) adolescence: ALSPAC (17.1 years). These analyses were adjusted for the same covariates as the cord blood analyses plus child age at DNA methylation measurement. For cell types, we used the adult blood methylation-based reference (32,33) (CD8 T ells, CD4 T cells, natural killer, B cells, monocytes, and granulocytes). We corrected for multiple testing in these lookups using a Bonferroni P-value cutoff of <0.0012 (0.05/41 tests).
Functional Analyses
We examined potential functionality of the identified CpGs using several analyses. First, we looked up the top hits from the full group and stratified meta-analyses in recently published EWAS on related exposures: maternal late-pregnancy glycemic traits (34), maternal early-pregnancy blood glucose and insulin concentrations (35), and maternal early-pregnancy BMI (11); and in EWAS of cord blood DNA methylation and child health outcomes: birth weight (36) and BMI in childhood or adolescence (37). We corrected for multiple testing in this lookup using a Bonferroni P-value cutoff <0.0012 (0.05/41 tests).
Second, we used the cell type-adjusted Human Early Life Exposome (HELIX) catalog of 13.6 million autosomal expression quantitative trait methylation (cis-eQTMs) in children to check whether the top CpGs from our EWAS were associated with gene expression in childhood blood (38). We additionally tested associations of the top CpGs from our EWAS with RNA transcript levels in subcutaneous adipose tissue in 223 participants (0–21 years old) from the Leipzig Childhood Adipose Tissue Cohort (39). Participating children underwent elective surgery, mostly orthopedic, and were free of severe diseases (diabetes, generalized inflammation, malignant disease, and genetic syndromes, or were not permanently immobilized) and medication potentially influencing adipose tissue biology. We focused on CpGs ≤500 kilobase of the transcription start site of the transcript. We applied Bonferroni correction using the total number of CpGs tested (33, since 8 CpGs were discarded prior to the statistical analysis) and the number of tests per each unique CpG-transcript pair. Detailed methods are described in the Supplementary Methods.
Third, we used missMethyl (40) to run functional enrichment pathway analyses using gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG). For these pathway analyses we used several selections of CpGs: 1) all CpGs that reached Bonferroni threshold in either the full group or stratified analyses for both glycemic index and load combined, to explore common pathways; and 2) CpGs with P < 0.0001 in each of the six analyses of maternal glycemic index or load in the full group or the stratified analyses separately.
Fourth, we explored in the EWAS Catalog (41) if the top hits from the full group meta-analyses were previously identified to be associated with other traits.
Fifth, for those CpGs that reached the Bonferroni threshold in the full group and stratified meta-analyses and that were flagged as potentially polymorphic sites (22,23) or methylation quantitative trait loci (mQTLs) (24), we created distribution plots using the Generation R data to check bimodality of the distribution.
Sixth, using all CpGs associated with glycemic index and load in the full group and stratified meta-analyses, we examined enrichment for tissue-specific DNaseI hypersensitivity regions using eForge version 2.0 (42).
Lastly, we examined between-cohort heterogeneity of the top CpGs with a combination of the I2 statistic, forest plots, and leave-one-out plots.
Results
Participants
We included 2,003 mother-child pairs from the three cohorts. Descriptives per cohort are presented in Table 1, Supplementary Fig. 1A–C, and the Supplementary Methods. The mean maternal glycemic index ranged between 50.8 and 58.3, and the mean maternal glycemic load ranged between 107.6 and 154.6.
Table 1.
Cohort-specific descriptive statistics
| ALSPAC | Generation R | INMA | |
|---|---|---|---|
| All mothers | (n = 658) | (n = 998) | (n = 347) |
| Glycemic index | 58.3 ± 2.8 | 57.62 ± 3.2 | 50.8 ± 2.7 |
| Glycemic load | 126.2 ± 34.4 | 154.6 ± 45.4 | 107.6 ± 31.5 |
| Sex of the child – female | 342 (52.0) | 504 (50.5) | 171 (49.3) |
| Educational level* | |||
| Low | 324 (49.2) | 599 (60.0) | 88 (25.4) |
| Middle | — | — | 149 (42.9) |
| High | 334 (50.8) | 399 (40.0) | 110 (31.7) |
| Maternal age (years) | 29.8 ± 4.3 | 31.7 ± 4.1 | 31.5 ± 4.0 |
| Maternal smoking* | |||
| No smoking during pregnancy | 593 (90.1)* | 769 (77.1) | 246 (70.9) |
| Smoked, stopped before 2nd trimester | — | 97 (9.7) | 54 (15.6) |
| Smoked throughout pregnancy | 65 (9.9) | 132 (13.2) | 47 (13.5) |
| Maternal pre-/early-pregnancy weight status | |||
| BMI < 18 kg/m2 | — | 15 (1.5) | 16 (4.6) |
| BMI ≥ 18 and < 25 kg/m2 | 538 (81.8) | 735 (73.6) | 236 (68.0) |
| BMI ≥ 25 kg/m2 | 120 (18.2) | 248 (24.8) | 95 (27.4) |
| Maternal total energy intake (kcal) | 1,753 ± 453 | 2,152 ± 493 | 2,063 ± 476 |
| Maternal total carbohydrate intake (g) | 216.2 ± 57.3 | 265.5 ± 72.8 | 213.4 ± 59.2 |
| Maternal total carbohydrate intake (energy %) | 49.5 ± 4.7 | 49.2 ± 6.0 | 41.4 ± 6.1 |
| Mothers with pre-/early-pregnancy normal weight | (n = 538) | (n = 735) | (n = 236) |
| Glycemic index | 58.2 ± 2.8 | 57.5 ± 3.2 | 50.6 ± 2.8 |
| Glycemic load | 127.5 ± 35.8 | 156.1 ± 46.0 | 108.1 ± 30.7 |
| Sex of the child – female | 281 (52.2) | 377 (51.3) | 125 (53.0) |
| Educational level* | |||
| Low | 253 (47.0) | 410 (55.8) | 53 (22.5) |
| Middle | — | — | 102 (43.2) |
| High | 285 (53.0) | 325 (44.2) | 81 (34.3) |
| Maternal age (years) | 29.85 ± 4.28 | 31.79 ± 4.10 | 31.43 ± 4.03 |
| Maternal smoking* | |||
| No smoking during pregnancy | 484 (90.0)* | 568 (77.3) | 166 (70.3) |
| Smoked, stopped before 2nd trimester | 75 (10.2) | ||
| Smoked throughout pregnancy | 54 (10.0) | 92 (12.5) | 32 (13.6) |
| Maternal total energy intake (kcal) | 1,772 ± 461 | 2,177 ± 496 | 2,067 ± 474 |
| Maternal total carbohydrate intake (g) | 218.9 ± 59.2 | 268.6 ± 73.5 | 212.9 ± 58.0 |
| Maternal total carbohydrate intake (energy %) | 49.6 ± 4.8 | 49.2 ± 5.9 | 41.2 ± 5.9 |
| Mothers with pre-/early-pregnancy overweight or obesity | (n = 120) | (n = 248) | (n = 95) |
| Glycemic index | 59.0 ± 2.8 | 57.9 ± 3.3 | 50.8 ± 2.6 |
| Glycemic load | 120.1 ± 26.6 | 149.2 ± 43.8 | 105.8 ± 29.8 |
| Sex of the child – female | 61 (50.8) | 120 (48.4) | 38 (40.0) |
| Educational level* | |||
| Low | 71 (59.2) | 180 (72.6) | 30 (31.6) |
| Middle | — | — | 42 (44.2) |
| High | 49 (40.8) | 68 (27.4) | 23 (24.2) |
| Maternal age (years) | 29.66 ± 4.60 | 31.50 ± 4.15 | 31.98 ± 3.97 |
| Maternal smoking* | |||
| No smoking during pregnancy | 109 (90.8)* | 190 (76.6) | 69 (72.6) |
| Smoked, stopped before 2nd trimester | — | 21 (8.5) | 13 (13.7) |
| Smoked throughout pregnancy | 11 (9.2) | 37 (14.9) | 13 (13.7) |
| Maternal total energy intake (kcal) | 1,672 ± 407 | 2,066 ± 476 | 2,013 ± 456 |
| Maternal total carbohydrate intake (g) | 203.9 ± 46.3 | 254.8 ± 70.5 | 208.0 ± 56.5 |
| Maternal total carbohydrate intake (energy %) | 49.1 ± 4.3 | 49.2 ± 6.3 | 41.4 ± 6.5 |
Data are presented as mean ± SD or n (%).
Cohorts used their preferred categories for maternal educational level and maternal smoking during pregnancy. We used two categories for ALSPAC: no smoking or quit before 2nd trimester vs. sustained smoking. Please see the cohort-specific methods for these descriptions.
Meta-analysis of Glycemic Index and Glycemic Load in the Full Group
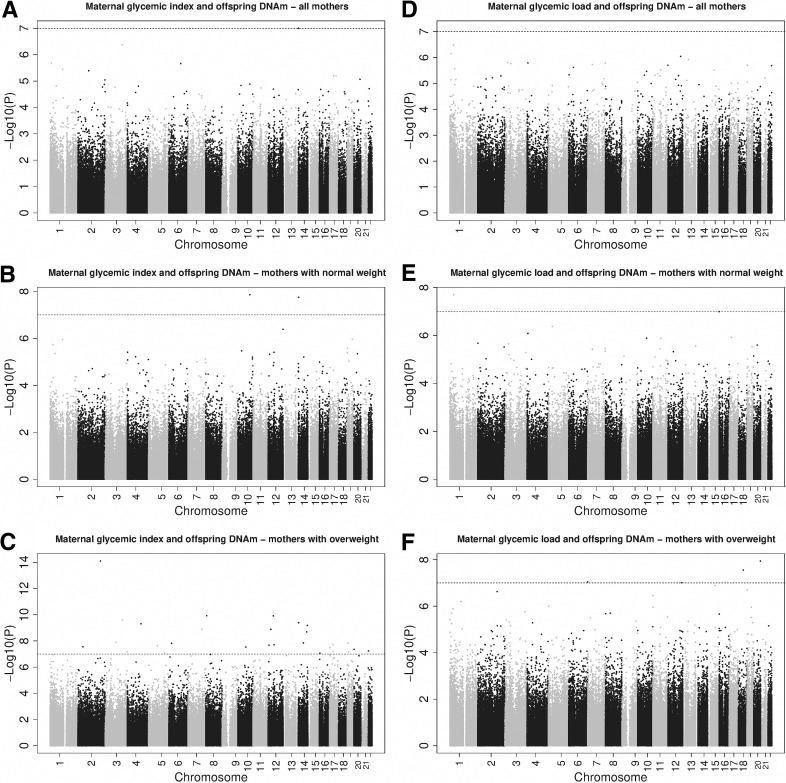
In the full-group analyses, we observed an association between maternal glycemic index and cord blood DNA methylation level of cg21301148 (0.099% increase in DNA methylation per 1-unit increase in glycemic index, SE = 0.019, P = 1.00 × 10−7 (Table 2 and Supplementary Table 1). Maternal glycemic load was associated with cord blood DNA methylation levels of cg09874107, cg21301148, and cg27528695 (Table 2 and Supplementary Table 2). Manhattan plots are presented in Fig. 2.
Table 2.
CpGs associated with maternal glycemic index or glycemic load in the full group and stratified by maternal weight status
| CpG | Chr | Position# | Nearest gene | Polymorphic | mQTL | Effect* | SE* | P value | Dir† | I 2 |
|---|---|---|---|---|---|---|---|---|---|---|
| Glycemic index—all mothers (n = 2,003) | ||||||||||
| cg21301148 | 14 | 23878195 | MYH6 | Yes | No | 0.099 | 0.019 | 1.00E−07 | +++ | 0 |
| Glycemic load—all mothers (n = 2,003) | ||||||||||
| cg09874107 | 3 | 184870896 | C3orf70 | No | No | 0.002 | 0.000 | 7.50E−08 | +++ | 67.6 |
| cg27528695 | 1 | 109643479 | SCARNA2 | Yes | Yes | 0.010 | 0.002 | 9.74E−08 | +++ | 0 |
| Glycemic index—mothers with normal weight (n = 1,509) | ||||||||||
| cg02079551 | 10 | 107023377 | SORCS3 | Yes | No | −0.085 | 0.015 | 1.38E−08 | −−− | 0 |
| cg21301148 | 14 | 23878195 | MYH6 | Yes | no | 0.120 | 0.021 | 1.78E−08 | −−− | 0 |
| Glycemic index—mothers with overweight or obesity (n = 463) | ||||||||||
| cg18202627 | 2 | 200776533 | C2orf69 | No | No | −0.093 | 0.012 | 7.96E−15 | −−− | 0 |
| cg16169361 | 8 | 8751409 | MFHAS1 | Yes | No | −0.101 | 0.016 | 1.19E−10 | −−− | 24.4 |
| cg24458009 | 12 | 47472775 | AMIGO2 | Yes | Yes | −0.211 | 0.033 | 1.22E−10 | −−− | 72 |
| cg18290075 | 3 | 153840477 | ARHGEF26 | No | Yes | −0.099 | 0.016 | 2.52E−10 | −−− | 54.2 |
| cg25832796 | 14 | 24038103 | JPH4 | Yes | Yes | −0.254 | 0.041 | 4.09E−10 | −−+ | 94.8 |
| cg12972275 | 4 | 122722461 | EXOSC9 | Yes | No | −0.047 | 0.008 | 4.83E−10 | −−− | 48.6 |
| cg18211447 | 14 | 102771717 | MOK | No | No | −0.117 | 0.019 | 6.57E−10 | −−− | 89 |
| cg04351062 | 12 | 24717058 | SOX5 | No | Yes | 0.089 | 0.015 | 1.30E−09 | ++− | 81.8 |
| cg01078248 | 7 | 123197661 | NDUFA5 | No | No | −0.077 | 0.013 | 1.31E−09 | −−+ | 74.8 |
| cg06591466 | 14 | 96505869 | C14orf132 | No | No | −0.107 | 0.018 | 2.05E−09 | −−+ | 74.8 |
| cg26474288 | 3 | 98451287 | ST3GAL6-AS1 | No | No | −0.108 | 0.019 | 1.28E−08 | −−− | 0 |
| cg24688926 | 19 | 6737877 | GPR108 | No | No | −0.074 | 0.013 | 1.41E−08 | −−− | 83 |
| cg00006032 | 14 | 66974439 | GPHN | No | No | −0.126 | 0.022 | 1.41E−08 | −−+ | 69.3 |
| cg06171242 | 6 | 24667490 | ACOT13 | No | No | −0.061 | 0.011 | 1.52E−08 | −−− | 49.5 |
| cg13445358 | 12 | 53661749 | ESPL1 | Yes | No | −0.062 | 0.011 | 1.96E−08 | −−+ | 0 |
| cg17582259 | 17 | 33288330 | CCT6B | No | No | −0.094 | 0.017 | 1.97E−08 | −−+ | 55.3 |
| cg20927656 | 12 | 7863229 | DPPA3 | Yes | Yes | 1.355 | 0.242 | 2.12E−08 | +?+ | 61.3 |
| cg02725014 | 5 | 78809520 | HOMER1 | No | No | −0.113 | 0.02 | 2.30E−08 | −−− | 0 |
| cg13721560 | 2 | 44222568 | LRPPRC | No | No | −0.068 | 0.012 | 2.83E−08 | −−+ | 56.5 |
| cg13358349 | 10 | 71211210 | TSPAN15 | Yes | No | −0.11 | 0.02 | 2.96E−08 | −−− | 28.9 |
| cg16604801 | 17 | 2718310 | RAP1GAP2 | No | No | −0.071 | 0.013 | 3.34E−08 | −−− | 71.1 |
| cg13740771 | 20 | 3026904 | MRPS26 | No | No | −0.113 | 0.021 | 4.36E−08 | −−− | 0.5 |
| cg16031283 | 22 | 20861701 | MED15 | No | No | −0.092 | 0.017 | 5.79E−08 | −−− | 46.3 |
| cg00367659 | 17 | 33288544 | ZNF830 | No | No | −0.132 | 0.024 | 6.40E−08 | −−+ | 40.5 |
| cg26985201 | 3 | 197517876 | LRCH3 | No | No | −0.085 | 0.016 | 6.75E−08 | −−− | 55.4 |
| cg15225042 | 19 | 13044582 | FARSA | No | No | −0.077 | 0.014 | 6.83E−08 | −−+ | 70.5 |
| cg23347399 | 5 | 140766911 | PCDHGA1 | No | Yes | 0.4 | 0.075 | 8.30E−08 | +++ | 17.8 |
| cg27193519 | 16 | 4714443 | MGRN1 | Yes | Yes | 0.292 | 0.055 | 8.58E−08 | +++ | 93.9 |
| cg24340911 | 17 | 5342028 | C1QBP | No | No | −0.077 | 0.014 | 1.06E−07 | −−− | 71.5 |
| cg07254608 | 8 | 41997973 | AP3M2 | No | No | −0.129 | 0.024 | 1.07E−07 | −−− | 11.5 |
| cg09668030 | 1 | 27852173 | AHDC1 | No | No | 0.260 | 0.049 | 1.18E−07 | ++? | 8.7 |
| Glycemic load—mothers with normal weight (n = 1,509) | ||||||||||
| cg02920421 | 1 | 32572534 | KPNA6 | Yes | No | −0.014 | 0.002 | 1.99E−08 | +−− | 74.3 |
| cg26729101 | 16 | 229500 | HBQ1 | Yes | No | −0.007 | 0.001 | 1.01E−07 | +−− | 56.6 |
| Glycemic load—mothers with overweight or obesity (n = 463) | ||||||||||
| cg03223949 | 20 | 60397915 | CDH4 | Yes | Yes | −0.013 | 0.002 | 1.15E−08 | −−+ | 42.7 |
| cg23345004 | 18 | 44526430 | KATNAL2 | No | Yes | −0.035 | 0.006 | 2.81E−08 | −−− | 0 |
| cg25044186 | 6 | 170494475 | LOC154449 | Yes | Yes | −0.013 | 0.002 | 8.99E−08 | −−− | 0 |
| cg09480470 | 12 | 124556975 | ZNF664-RFLNA | No | Yes | −0.084 | 0.016 | 9.74E−08 | −?− | 0 |
Chr, chromosome; Dir, direction of association for each cohort.
Position is annotated using hg19.
Effect size and SE are presented as percentage change in DNA methylation per 1-point increase in the glycemic index or load.
Cohorts are ordered as ALSPAC, Generation R, and INMA.
Figure 2.
Manhattan plots for the associations of maternal glycemic index and load with offspring DNA methylation (DNAm) in the full group and stratified meta-analyses. Maternal glycemic index and offspring with DNAm—all mothers (A), mothers with normal weight (B), and mothers with overweight (C). Maternal glycemic load and offspring with DNAm—all mothers (D), mothers with normal weight (E), and mothers with overweight (F).
Meta-analyses Stratified by Maternal Weight Status
In 1,509 mothers with normal weight, maternal glycemic index was associated with two CpGs, cg02079551 and cg21301148, and maternal glycemic load was associated with two CpGs, cg02920421 and cg26729101 (Table 2 and Supplementary Tables 3 and 4). In 463 mothers with overweight or obesity, maternal glycemic index was associated with DNA methylation of 31 CpGs, and maternal glycemic load was associated with DNA methylation of 4 CpGs (Table 2). Full epigenome-wide results for these stratified analyses are presented in Supplementary Tables 5 and 6. We found no overlap between the hits from these stratified analyses, except for cg21301148, which was associated with the glycemic index in the full group as well as in mothers with normal weight. Of the total of 39 CpGs that were identified in at least one of the stratified analyses, 19 (48.7%) showed opposite directions of association between the mothers with normal weight and those with overweight or obesity. Manhattan plots are presented in Fig. 2. In total, glycemic index or load were associated with 41 CpGs in either the full group or the stratified analyses. Results for all (full group and stratified) meta-analyses for these 41 CpGs are shown in Supplementary Table 7.
Lookup in Older Ages
We tested persistence of associations of maternal glycemic index or load during pregnancy when DNA methylation was measured in early childhood (n = 554), late childhood (n = 1,255), and adolescence (n = 711). None of the CpGs persisted at later ages in relation to the same exposure. Although 7 of the 41 CpGs reached Bonferroni-corrected P < 0.0012 in one or more of the models in early childhood, late childhood, or adolescence (bold and underlined in Supplementary Tables 8–10), these associations were observed in other models than the initial meta-analysis in cord blood (e.g., these were associated with glycemic load in the lookup rather than glycemic index in the cord blood analysis). Additionally, nine CpGs reached nominal significance in the same model as in the main cord blood analyses.
Functional Analyses
None of the 41 CpGs from the full group and stratified analyses was associated with maternal early and late pregnancy glycemic traits or maternal BMI in previous EWAS (all P values >0.0012 [0.05/41]) (Supplementary Table 11). Similarly, none of the 41 CpGs was previously associated with birth weight or childhood or adolescence BMI (Supplementary Table 11).
In the eQTM analyses, we found three CpG-transcript cluster associations in childhood blood for cg24458009, with the PC-esterase domain containing 1B (PCED1B) gene annotated to this transcript cluster. We further found one CpG-transcript cluster association for cg23347399, with the protocadherin γ gene family (PCDHGA, PCDHGB, PCDHGC) annotated to this transcript cluster (Table 3). These two CpGs were both found in the EWAS for associations with glycemic index in mothers with overweight or obesity. In the analyses of associations between DNA methylation and RNA transcript levels in adipose tissue, 4 of the 561 CpG-transcript pairs tested reached statistical significance. These all involved cg27193519, showing a direct association between DNA methylation and expression levels of transcription factor AP-4 (TFAP4), zinc finger protein 500 (ZNF500), periplakin (PPL), and ankyrin repeat and sterile α motif domain containing 3 (ANKS3). Correlation plots of these four CpG-transcript pairs are shown in Supplementary Fig. 2.
Table 3.
Lookup of n = 41 Bonferroni significant CpGs from all EWAS for maternal glycemic index and load in two data sources with DNA methylation and RNA transcript levels
| Child blood (Human Early Life Exposome (HELIX) project) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CpG | Transcript cluster | log2FC | SE | P | CpG chr | CpG position | CpG gene | TC gene start | TC gene end | TC gene TSS | TC gene | ||
| cg24458009 | TC12000334.hg.1 | −0.139 | 0.329 | 3E-05 | 12 | 47472775 | AMIGO2 | 47473386 | 47630443 | 47473386 | PCED1B | ||
| cg24458009 | TC12001432.hg.1 | −0.124 | 0.298 | 4E-05 | 12 | 47472775 | AMIGO2 | 47599681 | 47610239 | 47610239 | PCED1B-AS1 | ||
| cg24458009 | TC12002854.hg.1 | −0.147 | 0.368 | 7E-05 | 12 | 47472775 | AMIGO2 | 47599681 | 47610239 | 47610239 | PCED1B-AS1 | ||
| cg23347399 | TC05000761.hg.1 | 0.011 | 0.023 | 2E-06 | 5 | 140766911 | PCDHGA4/ PCDHGA6/ PCDHGA1/ PCDHGA5/ PCDHGB1/ PCDHGA3/ PCDHGA2/ PCDHGB4/ PCDHGA7/ PCDHGB2/ PCDHGB3 | 140710204 | 140892548 | 140710204 | PCDHGC3; PCDHGA12; PCDHGB4; PCDHGA8; PCDHGA1; PCDHGA10; PCDHGA11; PCDHGA2; PCDHGA3; PCDHGA4; PCDHGA5; PCDHGA6; PCDHGA7; PCDHGA9; PCDHGB1; PCDHGB2; PCDHGB3; PCDHGB5; PCDHGB6; PCDHGB7; PCDHGC4; PCDHGC5 | ||
| Subcutaneous adipose tissue (Leipzig Childhood Adipose Tissue Cohort) | |||||||||||||
| CpG | Transcript cluster | Estimate | SE | P | CpG chr | CpG position | CpG gene | TC gene start | TC gene end | TC gene TSS | TC gene | N tests CpG | N tests total |
| cg27193519 | ILMN_1814657 | 0.091 | 0.014 | 1.6E-09 | 16 | 4664442 | MGRN1 | 4257186 | 4273023 | 4273023 | TFAP4; AP-4 | 24 | 57 |
| cg27193519 | ILMN_1700238 | 0.068 | 0.013 | 7.6E-07 | 16 | 4664442 | MGRN1 | 4748239 | 4767162 | 4767162 | ZNF500; ZKSCAN18 | 24 | 57 |
| cg27193519 | ILMN_1806030 | 0.283 | 0.064 | 1.5E-05 | 16 | 4664442 | MGRN1 | 4882507 | 4937148 | 4937148 | PPL; MGC134872; KIAA0568 | 24 | 57 |
| cg27193519 | ILMN_1751636 | 0.027 | 0.007 | 2.1E-04 | 16 | 4664442 | MGRN1 | 4696511 | 4734271 | 4734271 | ANKS3; FLJ32345; FLJ32767; KIAA1977 | 24 | 57 |
Presented here are the CpG-TC pairs that are significant after multiple testing correction. log2FC = change in expression as log-twofold change per 10% DNA methylation. Estimate is the change in log2 expression per unit change in methylation M-value. P = P value of the CpG-TC association. CpG chr = CpG chromosome annotation. CpG position = CpG position. GpG gene = CpG gene annotation. TC gene start = start position of the transcript cluster (gene) based on the Hg38 build. TC gene end = end position of the Transcript Cluster (gene) based on the Hg38 build. TC gene TSS = position used as Transcription Start Site for the Transcript Cluster (gene) based on the Hg38 build. N tests CpG = the number of tests per each unique CpG-transcript pair. N tests total = total number of tests for this CpG-transcript pair, used for multiple testing correction.TC gene = genes annotated to Transcript Cluster (gene) according to Affymetrix na36 annotation (Illumina HumanHT-12 v4.0 annotations for subcutaneous adipose tissue).
Functional enrichment analyses on all 41 CpGs from the full group or stratified meta-analyses did not result in FDR significant GO or KEGG pathways (Supplementary Table 12). We further found no functional enrichment for the 76 and 231 CpGs with P < 0.0001 from the full group meta-analyses on glycemic index and load, respectively. Similarly, there was no functional enrichment for the 116 or 1,176 CpGs with P < 0.0001 from the stratified analyses for glycemic index in mothers with normal weight or overweight or obesity, respectively, nor for the 222 or 265 CpGs with P < 0.0001 from the stratified analyses for glycemic load, respectively.
We looked up the three CpGs from the full-group meta-analyses in the EWAS Catalog and found that cg21301148 was previously associated with (gestational) age, clear cell renal carcinoma, fetal versus adult liver gene expression, and alcohol consumption (Supplementary Table 13). Further, cg27528695 was associated with age and HIV infection, but cg09874107 was not found in the EWAS Catalog.
Distribution plots for all of the 41 CpGs that were flagged as either potentially polymorphic or mQTL mostly showed unimodal distributions. On the basis of visual inspection, only four CpGs had a marginal indication of bimodality: cg00006032, cg26474288, cg02725014, and cg06591466 (Supplementary Figs. 3–5). Further interpretation of these four CpGs needs consideration of potentially polymorphic or mQTL effects.
Finally, using the set of 41 CpGs, we found no evidence of enrichment for tissue-specific DNaseI hypersensitivity regions.
The I2 value was >50 for 20 of the 41 CpGs, which might indicate between-cohort heterogeneity. To examine this in further detail, we created forest plots and leave-one-out plots for all 41 CpGs (Supplementary Figs. 6–17). Based on the forest plots, we found no indication that one cohort consistently caused heterogeneity in the meta-analysis. Leaving out one cohort at a time, the direction of effect never changed, as can be seen in the leave-one-out plots. However, for a limited number of CpGs, a combined exploration of the plots and I2 statistic indicated one cohort had relatively strongly influenced the meta-analysis result. A sensitivity analysis rerunning the functional enrichment analysis only based on the 21 CpGs that reached the Bonferroni threshold and had an I2 <50 also did not result in FDR significant GO or KEGG pathways. Of the three CpGs that were associated with gene expression, two had an I2 >50.
Conclusions
This is the first large-scale EWAS to study associations of maternal dietary glycemic index and load during pregnancy with offspring DNA methylation. Overall, 41 unique CpGs were associated with glycemic index or load. We identified 3 of these in the full group analyses, 4 were identified when the sample was restricted to mothers with a normal weight, and 35 were identified when the sample was restricted to mothers with overweight or obesity. We found no persistence of the associations into childhood or adolescence. There was no overlap between our top CpGs and CpGs associated with maternal glycemic traits or BMI, or with birth weight or childhood BMI. We found no common pathways in functional enrichment analyses. However, two CpGs, associated with glycemic index in mothers with overweight or obesity were associated with gene expression in child blood, and one CpG was associated with gene expression in adipose tissue. On the basis of exploration of I2, forest plots, and leave-one-out plots, some CpGs need careful interpretation. For example, especially interesting for interpretation of our expression findings are cg24458009 and cg27193519; these might have been driven by one cohort.
In the full-group meta-analyses, maternal glycemic index or load were associated with three CpGs: cg21301148, cg09874107, and cg27528695. First, cg21301148 was associated with both glycemic index and glycemic load, and it is annotated to the transcription start site of the myosin heavy chain 6 (MYH6) gene. MYH6 has previously been associated with cardiomyopathy (43). Second, cg09874107, associated with glycemic load, is annotated to the transcription start site of the chromosome 3 open reading frame 70 (C3orf70) gene, which may play a role in neural and neurobehavioral development (44). DNA methylation of another CpG in C3orf70 has been associated with fat-free mass in European children (45). Third, cg27528695, associated with glycemic load, is located close to the small Cajal body-specific RNA 2 (SCARNA2) gene. To the best of our knowledge, SCARNA2 has not been associated with cardiometabolic disease.
Because women with overweight are more likely to be insulin resistant (15) and opposite directions of association with blood glucose and insulin were previously observed between mothers with normal weight versus mothers with overweight or obesity (35), we tested a potential modifying effect of maternal weight status on the association between glycemic index or load and cord blood DNA methylation by running analyses stratified on maternal BMI categories. Only one CpG overlapped between two of our EWAS models: cg21301148 was associated with glycemic index in mothers with normal weight as well as in the full group. We found most hits in mothers with overweight or obesity. In these mothers, maternal glycemic index and load were associated with DNA methylation levels at 31 and 4 CpGs, respectively. This may indicate a stronger effect of glycemic index or load on biological pathways in offspring of mothers with overweight or obesity, for example, through altered insulin sensitivity in these mothers. Indeed, for 20 of the 39 CpGs from the stratified analyses, we observed stronger effect sizes in mothers with overweight or obesity. For 19 of the 39 CpGs, the effect sizes were in opposite directions between strata, explaining the fact that there were no findings for these CpGs in the full group.
Interestingly, the three CpGs that were associated with gene expression were all found in the EWAS for associations with glycemic index in mothers with overweight or obesity. In child blood, cg24458009 was associated with expression of PCED1B, and cg23347399 was associated with expression of PCDHG. These genes do not have a known function in processes associated with cardiometabolic phenotypes (46,47). In subcutaneous adipose tissue, cg27193519 was associated with expression of TFAP4, ZNF500, PPL, and ANKS3. TFAP4 has been associated with lipid metabolism (48). ANKS3 is associated with congenital kidney disease (49). ZNF500 and PPL are not known to be associated with cardiometabolic diseases. However, as mentioned, cg24458009 and cg27193519 showed relatively high heterogeneity and thus need to be carefully interpreted.
We found no overlap between our findings and previously reported associations of maternal glucose, insulin, and the glucose area under the curve for a 2-h oral glucose tolerance test or BMI during pregnancy (11,34,35) with cord blood DNA methylation. This could be because we measured glycemic traits based on dietary intake, while these previous studies measured BMI or different glycemic traits as blood concentrations; moreover, glycemic index values for a food are average for everyone, whereas individual biological responses to specific foods are likely to vary (50). It could also be because we excluded mothers with gestational diabetes, while the two previous glycemic trait EWASs did not (34,41). We could not establish a potentially mediating role of cord blood DNA methylation in the association of maternal glycemic index or load with later child health, since none of the CpGs from our EWAS were previously found to be associated with birth weight (36) or child or adolescent BMI (37). However, power might have been limited, and future studies with larger sample sizes need to investigate this further.
We found no persistence of differential DNA methylation into childhood or adolescence. However, seven CpGs reached the Bonferroni threshold for associations with the other glycemic phenotype (i.e., glycemic index instead of load or in mothers with normal weight instead of mothers with overweight or obesity). This may indicate that DNA methylation at those CpGs is associated with general glycemic processes, potentially with varying strengths at different time points. Additionally, nine CpGs reached nominal significance in the same model as in the main cord blood analyses, which might indicate a lack of power to detect persistence due to lower sample sizes. Future studies might focus on increasing sample size for such a lookup in older ages. We previously observed a similar lack of persistence of differential DNA methylation in relation to birth weight (36). Exposure to maternal factors rapidly diminishes after delivery, and many postnatal exposures may affect DNA methylation, which might explain the lack of persistence. However, even without this persistence, differential DNA methylation in the critical period of organ development may have already initiated structural functional alterations that might have long-term consequences independent of persistent differential DNA methylation or via differential DNA methylation in other tissues.
Limitations of this study include the self-reported dietary data, the difference in number of food items in the FFQs between cohorts, ranging from 47 to 293, and the difference in glycemic index reference databases used in the cohorts. However, the calculation of glycemic index and load and the EWAS analyses were performed in all cohorts according to a prespecified harmonized analysis plan and R script, and with extensive quality control of dietary data. Further, cohorts had comparable distributions of glycemic index and load, and the ranking of mothers based on glycemic index and load will not have differed substantially. Additionally, maternal dietary intake was registered at ∼12 weeks of gestation in Generation R and INMA, with the reference time window being the first trimester, while the FFQ was registered at ∼32 weeks in ALSPAC, with the reference time window “nowadays.” We do not expect this to be a problem, since it has been shown that micronutrient intake and diet quality generally do not change during pregnancy (51). Dietary intake might be influenced by many genetic, environmental, and behavioral factors. We adjusted our analyses for many covariates to exclude potential confounding, but as in all observational studies, residual confounding could still be present. The Illumina 450K array covers <2% of all CpG sites in the human genome. We therefore cannot exclude that maternal glycemic index or load is associated with DNA methylation at unmeasured CpG sites. Further, DNA methylation is tissue specific. Therefore, DNA methylation in pancreatic, liver, or adipose tissue might be more relevant than cord blood, which is also why we did a lookup of associations with gene expression in adipose tissue. However, in the large-scale birth cohorts that we used, cord blood is easily accessible, as opposed to these other tissues. Unfortunately, gene expression data were not available in cord blood. We were also limited in sample size for the stratified analyses, and would therefore recommend future studies to focus on increasing sample size specifically for analyses stratified on maternal weight status.
To conclude, in this meta-analysis we identified associations of maternal glycemic index and load during pregnancy with offspring cord blood DNA methylation levels of 41 CpGs, mostly in mothers with overweight or obesity. These CpGs did not overlap with previously published CpGs associated with maternal glycemic traits or with child birth weight or BMI. Although we found no enrichment for specific biological pathways, three CpGs were associated with expression of PCED1B, PCDHG, TFAP4, ZNF500, PPL, and ANKS3 and might therefore have functional effects. Further study is required to explore functionality in more detail, uncover causality of these findings, and study pathways to offspring health.
Article Information
Acknowledgments. Cohort-specific acknowledgments are stated in the Supplementary Material.
Funding. This work was funded by the Joint Programming Initiative – A Healthy Diet for a Healthy Life to the NutriPROGRAM consortium: ZonMW, the Netherlands (529051022), MRC, U.K. (MR/S036520/1), and Instituto de Salud Carlos III, Spain (AC18/00006), and the PREcisE consortium: ZonMW, the Netherlands (529051023), German Federal Ministry of Education and Research, Germany (FKZ 01EA1905). Information regarding funding for the contributing cohorts and individual authors can be found in the Supplementary Material.
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. L.K.K. and S.F.-B. performed the meta-analysis. L.K.K., S.F.-B., G.M., L.J., R.O., J.V., M.C., K.L., E.W.T., A.K., R.G., J.H.M.d.V., V.W.V.J., M.V., G.C.S., and J.F.F. interpreted the data. L.K.K., S.F.B., G.M., L.J., R.O., J.V., M.C., K.L., E.W.T., A.K., R.G., J.H.M.d.V., V.W.V.J., M.V., G.C.S., and J.F.F. contributed to critical revision for important intellectual content. L.K.K., S.F.B., G.M., L.J., R.O., J.V., M.C., K.L., E.W.T., A.K., R.G., J.H.M.d.V., V.W.V.J., M.V., G.C.S., and J.F.F. gave final approval of the version to be published. L.K.K., S.F.B., G.M., L.J., J.V., K.L., and M.C. conducted cohort-specific analyses. L.K.K. and J.F.F. designed the research. L.K.K. and J.F.F. drafted the manuscript. A.K., R.G., V.W.V.J., M.V., G.C.S., and J.F.F. acquired data. L.K.K. and J.F.F. are the guarantors of this work and, as such, had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
This article contains supplementary material online at https://doi.org/10.2337/figshare.19898830.
L.K.K. and S.F.B. contributed equally as first authors. V.W.V.J., M.V., G.C.S., and J.F.F. contributed equally as senior authors.
References
- 1. Jenkins DJA, Wolever TMS, Taylor RH, et al. Glycemic index of foods: a physiological basis for carbohydrate exchange. Am J Clin Nutr 1981;34:362–366 [DOI] [PubMed] [Google Scholar]
- 2. Du H, van der A DL, van Bakel MME, et al. Glycemic index and glycemic load in relation to food and nutrient intake and metabolic risk factors in a Dutch population. Am J Clin Nutr 2008;87:655–661 [DOI] [PubMed] [Google Scholar]
- 3. Livesey G, Taylor R, Livesey HF, et al. Dietary glycemic index and load and the risk of type 2 diabetes: assessment of causal relations. Nutrients 2019;11:1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chiavaroli L, Lee D, Ahmed A, et al. Effect of low glycaemic index or load dietary patterns on glycaemic control and cardiometabolic risk factors in diabetes: systematic review and meta-analysis of randomised controlled trials. BMJ 2021;374:n1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jenkins DJA, Dehghan M, Mente A, et al.; PURE Study Investigators . Glycemic index, glycemic load, and cardiovascular disease and mortality. N Engl J Med 2021;384:1312–1322 [DOI] [PubMed] [Google Scholar]
- 6. Brand-Miller J, Hayne S, Petocz P, Colagiuri S. Low-glycemic index diets in the management of diabetes: a meta-analysis of randomized controlled trials. Diabetes Care 2003;26:2261–2267 [DOI] [PubMed] [Google Scholar]
- 7. Kizirian NV, Markovic TP, Muirhead R, et al. Macronutrient balance and dietary glycemic index in pregnancy predict neonatal body composition. Nutrients 2016;8:270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Moses RG, Luebcke M, Davis WS, et al. Effect of a low-glycemic-index diet during pregnancy on obstetric outcomes. Am J Clin Nutr 2006;84:807–812 [DOI] [PubMed] [Google Scholar]
- 9. Moses RG, Casey SA, Quinn EG, et al. Pregnancy and Glycemic Index Outcomes study: effects of low glycemic index compared with conventional dietary advice on selected pregnancy outcomes. Am J Clin Nutr 2014;99:517–523 [DOI] [PubMed] [Google Scholar]
- 10. Poston L, Bell R, Croker H, et al.; UPBEAT Trial Consortium . Effect of a behavioural intervention in obese pregnant women (the UPBEAT study): a multicentre, randomised controlled trial. Lancet Diabetes Endocrinol 2015;3:767–777 [DOI] [PubMed] [Google Scholar]
- 11. Sharp GC, Salas LA, Monnereau C, et al. Maternal BMI at the start of pregnancy and offspring epigenome-wide DNA methylation: findings from the Pregnancy and Childhood Epigenetics (PACE) consortium. Hum Mol Genet 2017;26:4067–4085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Joubert BR, den Dekker HT, Felix JF, et al. Maternal plasma folate impacts differential DNA methylation in an epigenome-wide meta-analysis of newborns. Nat Commun 2016;7:10577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yan W, Zhang Y, Wang L, et al. Maternal dietary glycaemic change during gestation influences insulin-related gene methylation in the placental tissue: a genome-wide methylation analysis. Genes Nutr 2019;14:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Geraghty AA, Sexton-Oates A, O’Brien EC, et al. A low glycaemic index diet in pregnancy induces DNA methylation variation in blood of newborns: results from the ROLO randomised controlled trial. Nutrients 2018;10:455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Catalano PM, Shankar K. Obesity and pregnancy: mechanisms of short term and long term adverse consequences for mother and child. BMJ 2017;356:j1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Boyd A, Golding J, Macleod J, et al. Cohort profile: the ‘children of the 90s’--the index offspring of the Avon Longitudinal Study of Parents and Children. Int J Epidemiol 2013;42:111–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fraser A, Macdonald-Wallis C, Tilling K, et al. Cohort profile: the Avon Longitudinal Study of Parents and Children: ALSPAC mothers cohort. Int J Epidemiol 2013;42:97–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kooijman MN, Kruithof CJ, van Duijn CM, et al. The Generation R study: design and cohort update 2017. Eur J Epidemiol 2016;31:1243–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Guxens M, Ballester F, Espada M, et al.; INMA Project . Cohort profile: the INMA—INfancia y Medio Ambiente—(Environment and Childhood) Project. Int J Epidemiol 2012;41:930–940 [DOI] [PubMed] [Google Scholar]
- 20. Aston LM, Jackson D, Monsheimer S, et al. Developing a methodology for assigning glycaemic index values to foods consumed across Europe. Obes Rev 2010;11:92–100 [DOI] [PubMed] [Google Scholar]
- 21. Atkinson FS, Foster-Powell K, Brand-Miller JC. International tables of glycemic index and glycemic load values: 2008. Diabetes Care 2008;31:2281–2283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Naeem H, Wong NC, Chatterton Z, et al. Reducing the risk of false discovery enabling identification of biologically significant genome-wide methylation status using the Human Methylation 450 array. BMC Genomics 2014;15:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen YA, Lemire M, Choufani S, et al. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium Human Methylation 450 microarray. Epigenetics 2013;8:203–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Min JL, Hemani G, Hannon E, et al.; BIOS Consortium . Genomic and phenotypic insights from an atlas of genetic effects on DNA methylation. Nat Genet 2021;53:1311–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 2012;28:882–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 2007;8:118–127 [DOI] [PubMed] [Google Scholar]
- 28. Gervin K, Salas LA, Bakulski KM, et al. Systematic evaluation and validation of reference and library selection methods for deconvolution of cord blood DNA methylation data. Clin Epigenetics 2019;11:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Van der Most PJ, Küpers LK, Snieder H, Nolte I. QCEWAS: automated quality control of results of epigenome-wide association studies. Bioinformatics 2017;33:1243–1245 [DOI] [PubMed] [Google Scholar]
- 30. Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 2010;26:2190–2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mägi R, Morris AP. GWAMA: software for genome-wide association meta-analysis. BMC Bioinformatics 2010;11:288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Houseman EA, Accomando WP, Koestler DC, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics 2012;13:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Reinius LE, Acevedo N, Joerink M, et al. Differential DNA methylation in purified human blood cells: implications for cell lineage and studies on disease susceptibility. PLoS One 2012;7:e41361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tobi EW, Juvinao-Quintero DL, Ronkainen J, et al. Maternal glycemic dysregulation during pregnancy and neonatal blood DNA methylation: meta-analyses of epigenome-wide association studies. Diabetes Care 2022;45:614–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Geurtsen ML, Jaddoe VWV, Gaillard R, Felix JF. Associations of maternal early-pregnancy blood glucose and insulin concentrations with DNA methylation in newborns. Clin Epigenetics 2020;12:134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Küpers LK, Monnereau C, Sharp GC, et al. Meta-analysis of epigenome-wide association studies in neonates reveals widespread differential DNA methylation associated with birthweight. Nat Commun 2019;10:1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Vehmeijer FOL, Küpers LK, Sharp GC, et al. DNA methylation and body mass index from birth to adolescence: meta-analyses of epigenome-wide association studies. Genome Med 2020;12:105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ruiz-Arenas C, Hernandez-Ferrer C, Vives-Usano M, et al. Identification of blood autosomal cis-expression quantitative trait methylation (cis-eQTMs) in children. eLife 2022;11:e6531041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Landgraf K, Rockstroh D, Wagner IV, et al. Evidence of early alterations in adipose tissue biology and function and its association with obesity-related inflammation and insulin resistance in children. Diabetes 2015;64:1249–1261 [DOI] [PubMed] [Google Scholar]
- 40. Phipson B, Maksimovic J, Oshlack A. missMethyl: an R package for analyzing data from Illumina’s HumanMethylation450 platform. Bioinformatics 2016;32:286–288 [DOI] [PubMed] [Google Scholar]
- 41. Battram T, Yousefi P, Crawford G, et al. The EWAS Catalog: a database of epigenome-wide association studies. Wellcome Open Res 2022;7:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Breeze CE, Reynolds AP, van Dongen J, et al. eFORGE v2.0: updated analysis of cell type-specific signal in epigenomic data. Bioinformatics 2019;35:4767–4769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Carniel E, Taylor MRG, Sinagra G, et al. α-Myosin heavy chain: a sarcomeric gene associated with dilated and hypertrophic phenotypes of cardiomyopathy. Circulation 2005;112:54–59 [DOI] [PubMed] [Google Scholar]
- 44. Ashikawa Y, Shiromizu T, Miura K, et al. C3orf70 is involved in neural and neurobehavioral development. Pharmaceuticals (Basel) 2019;12:156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rzehak P, Covic M, Saffery R, et al. DNA-methylation and body composition in preschool children: epigenome-wide-analysis in the European Childhood Obesity Project (CHOP)-Study. Sci Rep 2017;7:14349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. National Center for Biotechnology Information . PCED1B PC-esterase domain containing 1B [Homo sapiens (human)] Accessed 16 August 2021. Available from https://www.ncbi.nlm.nih.gov/gene/91523
- 47. National Center for Biotechnology Information . NCBI Gene: PCDHG@ protocadherin gamma cluster [Homo sapiens (human)] Accessed 16 August 2021. Available from https://www.ncbi.nlm.nih.gov/gene/56115
- 48. Li Y, Ding W, Li C-Y, Liu Y. HLH-11 modulates lipid metabolism in response to nutrient availability. Nat Commun 2020;11:5959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Schlimpert M, Lagies S, Budnyk V, Müller B, Walz G, Kammerer B. Metabolic phenotyping of Anks3 depletion in mIMCD-3 cells–a putative nephronophthisis candidate. Sci Rep 2018;8:9022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Berry SE, Valdes AM, Drew DA, et al. Human postprandial responses to food and potential for precision nutrition. Nat Med 2020;26:964–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Looman M, Geelen A, Samlal RAK, et al. Changes in micronutrient intake and status, diet quality and glucose tolerance from preconception to the second trimester of pregnancy. Nutrients 2019;11:E460. [DOI] [PMC free article] [PubMed] [Google Scholar]